- 1Laboratory of Microbiology and Molecular Biology, Faculty of Sciences, BioBio Research Center, University Mohammed V in Rabat, Rabat, Morocco
- 2Biodiversity and Crop Improvement Program, International Center for Agricultural Research in the Dry Areas (ICARDA), Rabat, Morocco
- 3Olds College, Field Crop Development Center, Lacombe, AB, Canada
- 4Indian Institute of Wheat and Barley Research (IIWBR), Karnal, Haryana, India
- 5Mount Vernon North-Western Washington Research & Extension Center, Washington State University, Mount Vernon, WA, United States
Barley is an important crop worldwide known for its adaptation to harsh environments and used in multiple forms as feed, food and beverages. Its productivity is affected by major abiotic and biotic stresses. Scald caused by hemibiotrophic fungus Rhynchosporium commune is a major foliar disease in many parts of the world. Host plant resistance is targeted by breeders to efficiently control this disease. An association mapping panel of 316 spring barley genotypes (AM2017) was screened for seedling resistance in greenhouse against three R. commune isolates and for adult plant resistance in three field locations in Morocco. The phenotyping results showed different numbers of entries with resistant and moderately resistant reactions at both seedling and adult plant stages. The reactions differed between the isolates with the highest percentage of resistant genotypes observed for isolate SC-S611 (49.4%) and highest percentage of susceptible genotypes (73.8%) for isolate SC-1122. At adult plant stage, the highest percentage of scald resistant genotypes (64.5%) was observed at Rommani site compared to 56% at Guich site and only 28.8% at Marchouch site. Seven genotypes were resistant at the seedling and adult plant stages. Genome wide association study (GWAS) revealed 102 MTA (15 QTL) at the seedling stage, and 25 MTA (12 QTL) associated with scald resistance at the adult plant stage. In addition, the sequences of 92 out of 102 at SRT, and 24 out of 25 significant SNP markers at APR were located in genomic regions enriched with functional proteins involved in diverse cellular processes including disease resistance. These markers span over all chromosomes with the majority of SNPs located on 3H and 7H. This study has verified 18 QTL reported in previous studies. In addition, it was successful in identifying new sources of resistance and novel genomic regions which could help in enhancing scald resistance in barley breeding programs.
1 Introduction
Barley ranks the second most important temperate cereal crop in the world after Triticum aestivum L. Globally, it was sown over 51 million hectares with a production of 150 million metric tons in 2019 (FAOSTAT, 2020). Barley use is not limited only to feed, food and beverages, but several studies have highlighted it’s nutritional value and health benefits including the potential for reducing colon cancer incidence, and lowering cholesterol (FDA, 2006; Kawka et al., 2019; Li et al., 2021).
The quality and productivity of barley crop are affected by abiotic and biotic stress factors. Among biotic stresses, leaf blotch or scald caused by the hemibiotrophic ascomycete fungus Rhynchosporium commune (formerly known as R. secalis) is one of the most economically important and destructive disease of barley worldwide, which can reduce grain quality and inflict high yield losses (Xi et al., 2000; Perfect and Green, 2001; Zhan et al., 2008). Rhynchosporium commune is known for its high genetic diversity which allow the pathogen population to overcome resistant cultivars, and to develop resistance to fungicides within a short period of time (Stefansson et al., 2012; Stefansson et al., 2013; Mcdonald, 2015; Brunner et al., 2016; Mohd-Assaad et al., 2016). The inherent complexity of the fungus requires an integrated disease management approach (Stefansson et al., 2012; McLean and Hollaway, 2018), and genetic resistance is the most economical, sustainable, and environment friendly strategy for controlling barley scald disease.
Development and deployment of resistant barley cultivars through incorporation and pyramiding of both major and minor genes can provide durable resistance against R. commune (Walters et al., 2012). Scald disease resistance can be classified as horizontal or vertical, both types of resistance are based on the number of “R genes” involved. Vertical resistance is qualitative in nature while horizontal resistance is governed by minor genes.
However, both vertical and horizontal resistance mechanisms are used by breeders. Compared to other barley diseases, relatively a few resistance loci against R. commune have been discovered which can be employed in breeding programs (Dracatos et al., 2019; Clare et al., 2020; Zhang et al., 2020). The qualitative resistance genes, frequently detected using specific strains at the seedling stage, provide a high level of resistance at all growth stages (Zhan et al., 2008). However, quantitative resistance to R. commune identified at the adult plant stage provide a partial level of resistance (Wallwork and Grcic, 2011; Zhan et al., 2008). The first R. commune resistance gene was designated as Rrs1 which conferred resistance under field conditions, and under controlled conditions with specific isolates (Zhan et al., 2008; Looseley et al., 2018). Frequently, barley scald disease resistance loci were detected on chromosome 3H flanking Rrs1 locus, and on chromosome 7H flanking Rrs2 locus (Zhan et al., 2008; Looseley et al., 2018). Across the barley genome, several major and minor R genes, and QTLs for leaf scald resistance have been identified in different barley genotypes scattered across the seven barley chromosomes (Zhang et al., 2020). To date, 11 major loci including Rrs1, Rrs2, Rrs4, Rrs12, Rrs13, Rrs14, Rrs15, Rrs16, Rrs17, (Rrs15 (CI8288), and Rrs18 controlling resistance to R. commune have been identified. Further, the major scald resistance genes have been detected on all chromosomes except for 5H, with Rrs14 on chromosome 1H, Rrs17 on chromosome 2H, Rrs1 and Rrs4 on chromosome 3H, Rrs3 and Rrs16 on chromosome 4H, Rrs13 and Rrs18 on chromosome 6H, and Rrs2, Rrs12 and Rrs15 on chromosome 7H (Bjørnstad et al., 2002; Zhang et al., 2020). Most of the modern barley cultivars share the same allele combinations of resistant loci indicating the limited genetic diversity among the barley germplasm (Williams et al., 2003). Furthermore, owing to the complex virulence spectrum and ability of R. commune populations to overcome resistant cultivars, durable resistance against scald disease requires continuous research for polygenic resistance (Xi et al., 2000).
Breeders and researchers are using landraces and wild relatives, genebank accessions, panels and genetic stocks to identify new sources of resistance. There are various sources of resistance derived from Hordeum vulgare ssp. vulgare (Silvar et al., 2010; Hofmann et al., 2013), from wild barley Hordeum vulgare ssp. spontaneum (von Korff et al., 2005; Yun et al., 2005) and from Hordeum bulbosum (Pickering et al., 2006).
In recent years, with the availability of a high-quality reference genome assembly (Mascher et al., 2017), and of high-density genotyping platforms (Moragues et al., 2010; Comadran et al., 2012), there has been a growing interest in genomic resources of barley. Different statistical methods have been developed and used to identify molecular markers that are linked to traits of interest in bi-parental QTL mapping (linkage mapping) and in genome wide association mapping (Linkage dis-equilibrium mapping). The allelic richness and the genetic mapping resolution in bi-parental mapping populations are mainly affected by the genetic recombination and segregation (Alqudah et al., 2020). Genome-wide association studies (GWAS) have proven to be an efficient tool that captures greater genetic variation and offers high resolution mapping of QTL and associated candidate genes compared to classical linkage analysis (Cockram et al., 2008; Comadran et al., 2012; Bayer et al., 2017). In barley, GWAS has been successfully applied to identify QTLs associated with resistance to net blotch (Amezrou et al., 2018), powdery mildew (Czembor et al., 2022), spot blotch (Visioni et al., 2020), and barley stripe rust (Visioni et al., 2018).
The main objectives of this study were to identify sources of resistance to scald in a diverse panel of barley genotypes at the seedling and at the adult plant stages, and to use GWAS to identify the underlying genomic regions conditioning resistance to R. commune.
2 Materials and methods
2.1 Plant material
A collection of 316 spring barley genotypes designated as association mapping-panel 2017 (AM2017) constructed by barley breeders at the International Center for Agriculture Research in the Dry Areas (ICARDA) was used in this study. It has 143 six-row and 173 two-row type barley genotypes, consisting of 134 advanced breeding lines from ICARDA, 21 landraces from ICARDA genebank, 161 released cultivars from Asia, Africa, Europe, and America (Verma et al., 2021).
2.2 Seedling stage disease resistance test
Scald infected barley leaves were collected from naturally infected barley fields from different agro-ecological zones of Morocco during the disease surveys conducted from 2015 to 2018. Isolation of R. commune was performed by soaking the scald infected barley leaves in sterile distilled water for 15 min followed by surface sterilization using 10% sodium hypochlorite solution for 30 s and 50% ethanol for 15 s, rinsed with sterile distilled water three times, and dried up within two layers of sterile Whatman filter paper. The leaf segments were incubated at 14°C in darkness for two weeks on Lima Bean agar (LMA) supplemented with Kanamycin and Streptomycin (50 mg per liter). After two weeks of incubation, the sporulating white or pinkish colonies were streaked on new LBA plates for the preparation of single conidial isolates.
Four seeds of each genotype were planted in a single cone of 3.8 cm diameter and 14 cm depth containing peat moss supplemented with 14-14-14 NPK fertilizer in 98-Ray Leach containers (Stuewe and Sons, Inc., Oregon, USA) in 3 replications. Seedlings of the AM2017 were grown under controlled conditions with a photoperiod of 16 h light/8 h dark at 20 ± 1 ° C in the growth chamber (Model MC1750; Snijder Scientific, Tilberg, Netherlands) at ICARDA, Rabat, Morocco. Three virulent isolates of R. commune, SC-511, SC-1122 and SC-611 collected from Marchouch (MCH), Sidi Allal Tazi (SAT), and Annoceur (ACR) were used for seedling resistance. The inoculum was prepared from the 10-12 days old LBA plates by rubbing the agar surface with sterile glass slide followed by filtration through double layer of cheese cloth. The inoculation of about 2 weeks old seedlings, with the second leaf fully extended, was performed using spore suspension of 5×105 conidia/ml supplemented with surfactant Tween 20 (0.01%) followed by incubation at 100% relative humidity in dark for 72 h at 15°C in the growth chamber. Then the seedlings were transferred to a greenhouse with day/night temperature regime of 20°C/16°C with a photoperiod of 16h light and 8h of darkness. After 16 days post infection (dpi), the infection types were recorded using disease rating scale of 0 to 5 (Salamati and Tronsmo, 1997) where 0 = I (Immune), 1= R (resistant), 2 = MR (moderately resistant), 3 = MS (moderately susceptible), 4 = S (susceptible), and 5 = HS (highly susceptible).
We also checked the virulence spectrum of three Rhynchosporium commune (Rc) isolates SC-1122, SC-511, and SC-611 on nine barley differential Atlas, Atlas 46, Brier, CI3515, CI4364, Jet, La Masita, Osiris, and Turk. These differentials possess specific resistance genes (R-genes) and have been previously characterized for their response to Rc isolates. In addition, barley differential interactions were examined to determine the compatibility between the Rc isolates and the different host genotypes. The presence or absence of compatible interactions, indicated by disease development, and incompatible interactions, indicated by the absence of disease symptoms, were recorded for each combination.
2.3 Adult plant stage disease assessment
Field trials were conducted in Morocco at the INRA experimental stations of Marchouch (MCH; 33° 56 N, 6° 63 W) and Guich (33° 58 N, 6° 51 W), and at Rommani region during the cropping season of 2018-2019. Each entry was planted as paired row of 1 meter length with row spacing of 0.5 m in an augmented design using two susceptible (Tissa and Tocada) and two resistant checks (Atlas 46 and ICARDA 4). To ensure high disease pressure, each block was surrounded by a border composed of a mixture of scald susceptible genotypes (Tissa, Aglou, Tiddas, Fleet, Adrar, Shepherd, Baudin and Alester).
Starting from Zadoks scale GS30 (Zadoks and Board, 1974) at an interval of 10-12 days, the trials were artificially inoculated four times using a knapsack sprayer with inoculum composed of a mixture of 15 isolates of R. commune which were collected from different agro-ecological zones of Morocco. Furthermore, scald infected barley residues from the previous growing season were uniformly distributed over the trial to ensure disease establishment. In addition, the development and spread of the disease was favoured by applying frequent mist irrigation in late afternoons daily. The disease severity was assessed at GS 73-75 using 0-9 scale (Saari and Prescott, 1975), and the genotypes were categorized into five classes; 0 to 2 as resistant, 3 to 4 as moderately resistant, 5 to 6 as moderately susceptible, 7 to 8 as susceptible and 9 as highly susceptible. The area under the disease progression curve (AUDPC) was calculated using the following equation (Jeger and Viljanen-Rollinson, 2001).
Where RCi = R. commune severity on ith days, ti = time in days at ith observation, and n is the total number of observations.
2.4 Data analysis
The frequencies of different classes of reaction to R. commune at the seedling and adult plant stages were determined using R software (R Core Team, 2019) and the Venn diagrams were developed to show the number of resistant entries common between the three isolates and among the three fields.
To gain a comprehensive understanding of the genetic control of scald resistance and its potential for improvement through breeding efforts, narrow-sense heritability (H2) of scald genetic resistance was estimated for each test at the seedling and adult plant stages using the variance component method. The heritability estimate was calculated as the ratio of additive genetic variance to the total phenotypic variance (Falconer and Mackay, 1996) with the use of R software.
2.5 Genotyping, population structure, and linkage disequilibrium of AM2017
Genomic DNA extraction, genotyping, population structure, and genetic diversity of the 316 genotypes used in this study has been described in an earlier study by Verma et al. (2021). Isolation of genomic DNA was conducted at the growth stage (GS12) using a lyophilized young leaf tissue from a single plant at the Cereal Crop Research Unit, USDA-ARS, Fargo, North Dakota, USA (Slotta et al., 2008). The Illumina iSelect 50K SNP array for barley (Illumina, San Diego, CA, USA) was used to perform SNP genotyping following the manufacturer’s guidelines (Bayer et al., 2017). Further, 36,793 SNP markers were used for genetic analysis after quality control by discarding all monomorphic markers, markers with missing values of more than 20%, and with minor allele frequency (MAF) of less than 5%.
The population structure of AM2017 was assessed using the filtered SNP markers to estimate individual admixture coefficients and was performed using the sparse Non-negative Matrix Factorization (sNMF) algorithm implemented in the R package LEA (Frichot and François, 2015). The cross-entropy criterion was used from sNMF function to evaluate several putative populations (K) ranging from K=2 to K=10 (Alexander and Lange, 2011; Frichot et al., 2014).
For each K we set the number of iterations to 200, the number of runs to 10, alpha to 10, and tolerance to 10-5. The genotypes were declared admixed or were assigned to subgroups using 80% membership criterion. The Principal Component Analysis (PCA) was performed with the 36,793 filtered SNP markers using TASSEL version 5.0. Furthermore, the kinship matrix (K) among 316 genotypes was computed using filtered set of SNP markers in TASSEL 5.0 (Bradbury et al., 2007).
The linkage disequilibrium (LD) was determined for all pairs of loci using SNP markers with known positions. The squared allele-frequency correlations (r2) were computed in PLINK V1.9 using non-linear regression with a threshold set at 0.2 (Amezrou et al., 2018; Visioni et al., 2018; Visioni et al., 2020; Amouzoune et al., 2022). The extent of genome wide LD decay was visualized by plotting intra-chromosomal r2 values against the physical distance in cM (Purcell et al., 2007).
2.6 Genome-wide association study
The association mapping was conducted in Tassel (version 5.2.53) software by combining phenotypic and genotypic data (Bradbury et al., 2007; Lipka et al., 2012). Two general linear models (GLM+Q), and (GLM+ PCA), and two mixed linear models, (MLM+Q+K), and (MLM+PCA+K), were tested by taking into account kinship (K matrix), population structure (Q matrix), and principal coordinate analysis (PCA) as covariates to control false positives.
In addition, several GAPIT version 3.0 models (Lipka et al., 2012) were used to validate significant SNP markers/loci associated to scald resistance. In GAPIT3, GWAS was performed using Settlement of MLM Under Progressively Exclusive Relationship (SUPER) (Wang et al., 2014), Multiple-locus MLM (MLMM) (Segura et al., 2012), Fixed and random model Circulating Probability Unification (FarmCPU) (Zhang et al., 2010), and Bayesian-information and Linkage-disequilibrium Iteratively Nested Keyway (BLINK) (Huang et al., 2019).
A GWAS threshold P-value of < 2.0 x 10-4 [−log10(P value)< 3.7] was used for declaring significant-marker trait associations as reported by Borrego-Benjumea et al. (2021). This hybrid approach was based on the median of two threshold methods: a stringent method proposed by Wang et al. (2014) determines the significance threshold using the equation α = 1/m, where ‘m’ represents the number of markers. In this case, -log10(P-value) < 4.4 was considered significant; and a less stringent approach (Chen et al., 2010) employs the use of bottom 0.1 percentile of P-values distribution [−log10(P value)< 3.0]. It balances the need for stringent control of Type I error (false positives) with the goal of discovering meaningful associations. In addition, we also used LD adjusted Bonferroni correction (Duggal et al., 2008; Visioni et al., 2020) which declared the P-value significance threshold to < 2.0 x 10-4 [−log10(P value) < 3.7]. Furthermore, QTL being validated by our study were also kept with a cut-off value of [−log10(P value)< 3.0].
The additive genetic effect refers to the portion of phenotypic variation that can be attributed to the additive effects of multiple genetic loci. In the output, R2 value explains how much of the phenotypic variation can be accounted for by the significant SNP marker. A higher R2 value indicates a stronger relationship between the marker and the observed trait. While the positive or negative allele effect indicates whether the allele increases susceptibility or provides resistance to the disease, respectively. When the allele effect is positive, it suggests that possessing that allele increases the disease score, indicating susceptibility to the disease. Conversely, a negative allele effect signifies that the allele reduces the disease score, indicating resistance to the disease. The Manhattan plots of GWAS were generated using CMplot package in R 3.3.1.
2.7 QTL alignment and candidate genes
The marker sequences of previously reported QTL were retrieved from Grain Genes database (https://wheat.pw.usda.gov/blast/), and their positions were checked on the barley pseudomolecules Morex v. 2.0 2019 and Morex v3 pseudomolecules (2021) using Barleymap pipeline (Cantalapiedra et al., 2015). For candidate genes (CGs), the sequences of significant SNP markers were subject to BLAST search tool of IPK barley server (https://galaxy-web.ipk-gatersleben.de/) and based on a threshold of BIT score (> 200), sequence identity (90-100%), and an expect value (0 – 1.40). The candidate gene search considered the presence of proteins with functional domains implicated in plant disease resistance.
3 Results
3.1 Seedling resistance of AM2017 panel with three isolates of R. commune
Analyses of the phenotypic data showed a uniform disease establishment and diverse infection responses (IR) depending on the virulence spectrum of each isolate at the seedling stage under controlled conditions. The average IR was 0 and 5, respectively for the resistant (ICARDA 4 and Atlas 46) and susceptible (Tocada and Tissa) checks for all the three isolates.
An average IR of 1.52 for Rc isolate SC-611, 3.6 for isolate SC-1122, and 1.98 for isolate SC-511 was observed in the AM2017 panel. Moreover, the frequency distribution of IR of AM2017 panel followed a normal distribution for the Rc isolates SC-511 and SC-611, whereas the distribution was negatively skewed towards susceptibility for the isolate SC-1122 (Figure 1A). Furthermore, the highest percentages of resistant genotypes were observed for the Rc isolate SC-S611 with 19.1% (60) immune and 30.3% (95) resistant genotypes, followed by the Rc isolate SC-511 with 6.7% (22) immune and 23.4% (74) resistant genotypes, whereas the Rc isolate SC-1122 showed the highest cumulative percentage of 73.8% (233) of susceptible and highly susceptible genotypes (Figure 1A). The Rc isolate SC-511 showed the highest percentage of moderately resistant (23.2%) genotypes followed by SC-S611 and SC-1122 isolate with 23.2% and 4.4%, respectively. Six-row genotypes had higher percentage of resistant barley genotypes with an average IR of 1.25 ± 0.9 for Rc isolate SC-511, followed by IR of 1.4 ± 1.1 for SC-S611, and IR of 2.9 ± 1.5 for SC-1122, whereas, the two-row barley genotypes were prone to susceptibility with an average IR of 1.62 ± 1.3 for SC-S611, followed by 2.6 ± 1.1 for SC-S511, and 4.1 ± 1.2 for SC-1122 isolates. About 11 barley genotypes, AM5, AM19, AM39, AM51, AM53, AM59, AM165, AM221, AM248, AM267, and AM280 were resistant to all the three Rc isolates used (Figure 1B). The heritabilities of scald resistance for all the three isolates at the seedling stage ranged from 0.40 to 0.58 (Table 1).
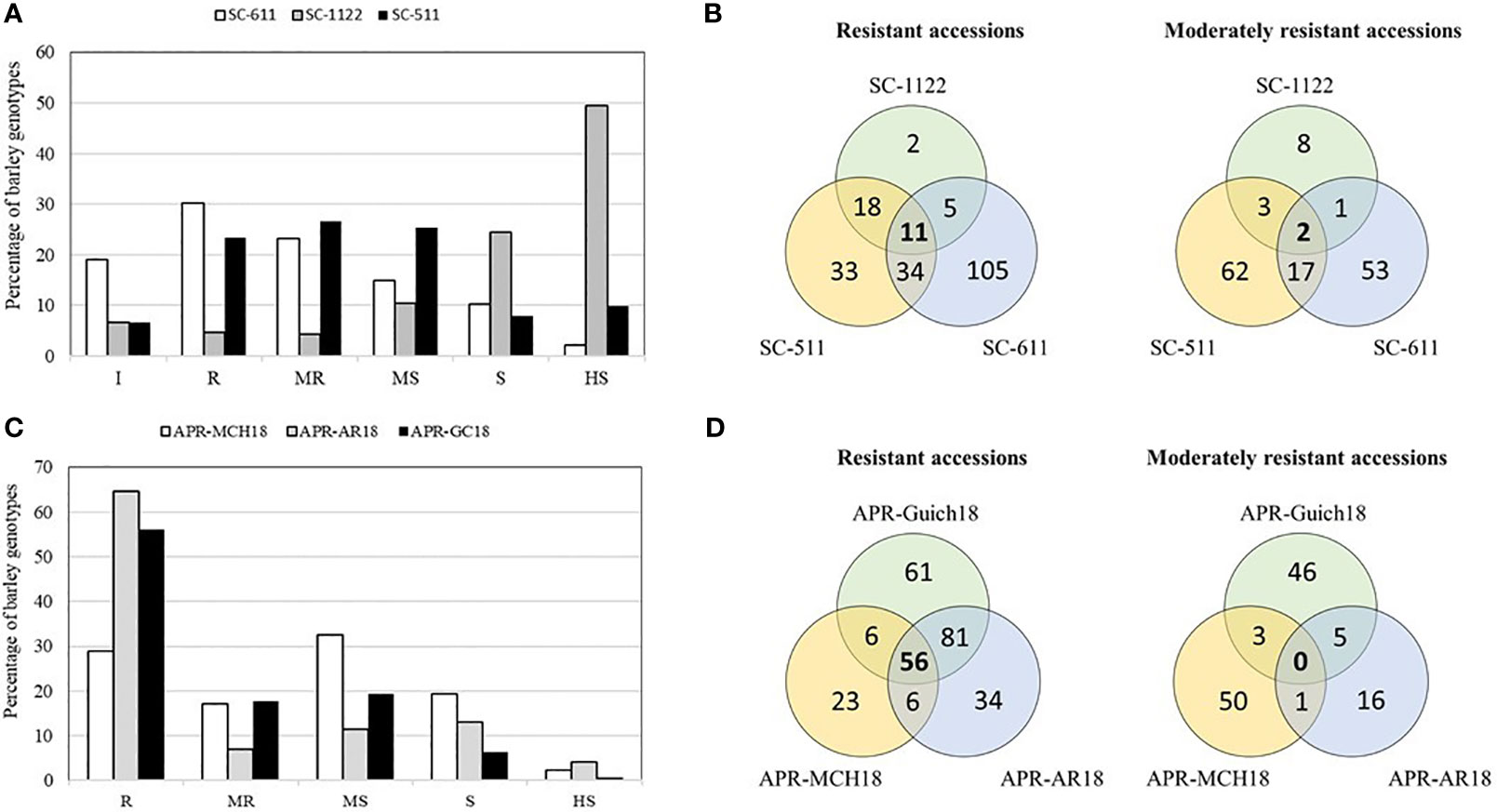
Figure 1 Frequency distribution of leaf scald resistance in 316 barley genotypes of AM2017 mapping panel at the seedling stage against R. commune isolates, SC-511, SC-611, and SC-1122 (A). Venn diagrams showing barley genotypes with combined resistance at the seedling stage to the three R. commune isolates under controlled conditions (B). Frequency distribution of leaf scald resistance in 316 barley genotypes of AM2017 mapping panel at the adult plant stage at Marchouch (MCH18), Rommani (AR18), and Guich (Guich18) (C). Venn diagrams showing barley genotypes with combined resistance at the adult plant stage at three field locations (D).
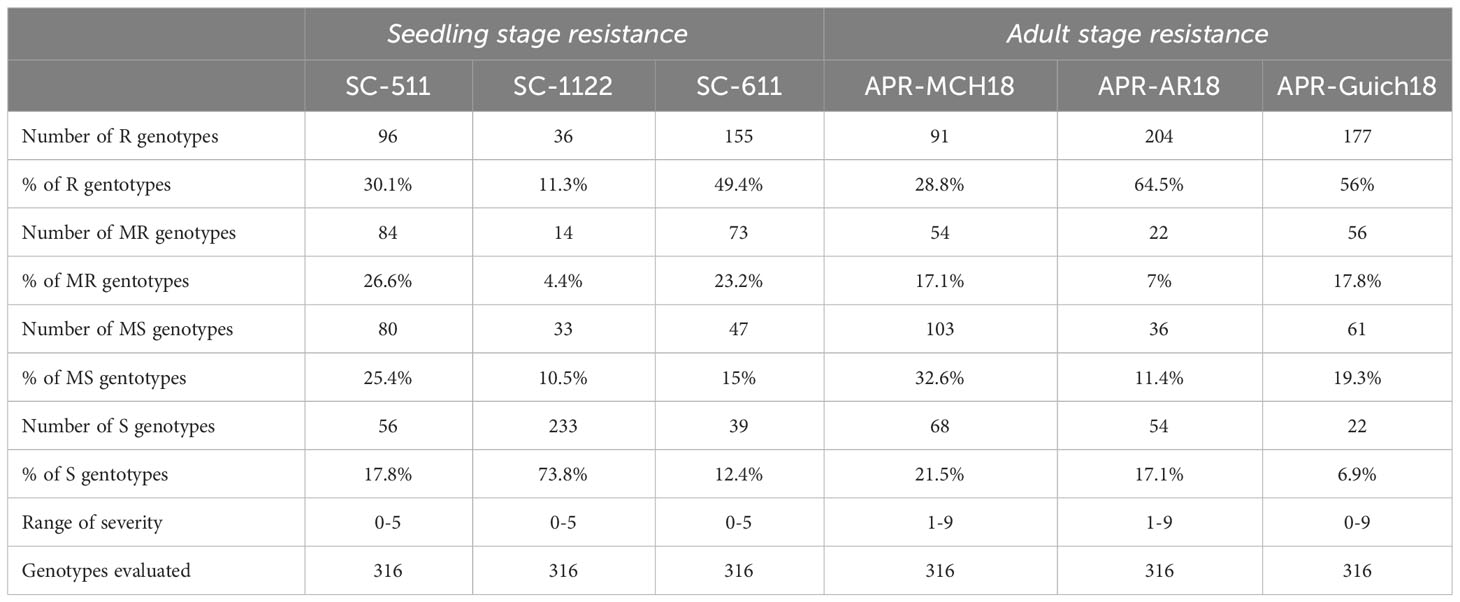
Table 1 The heritability estimates of scald resistance and the response of 316 genotypes of AM2017 panel at the seedling stage against R. commune isolates, SC-511, SC-1122, and SC-611 at the adult plant stage at Marchouch (MCH18), Rommani (AR18), and Guich (Guich18).
The interaction between barley differentials against the three Rc isolates showed that the Rc isolate SC-1122 was avirulent on Atlas (Rrs2), Atlas 46 (Rrs1, Rrs2), Brier (Rrs1), however it was virulent on CI3515 (Rh4, Rh10), and La Masita (Rh10, Rh4). The Rc isolate SC-511 was virulent on Atlas (Rrs2), Brier (Rrs1), CI4364 (rh11), and CI3515 (Rh4, Rh10), but it was avirulent on Atlas 46 (Rrs1, Rrs2), Jet (Rh6, rh7, rh6), Osiris (Rh4, rh6, Rh10, BRR6), and Turk (Rh5, Rrs1Turk, rh6). However, the Rc isolate SC-611 was avirulent on all of the differentials tested except Skiff. A differential interaction was observed for the Rc isolate SC-511 on Atlas, Brier and CI4364. It can be concluded that the Rc isolate SC-511 may be NIP1 deficient as it displayed compatible interaction on Brier (Rrs1) as reported previously (Stefansson et al., 2014; Mohd-Assaad et al., 2019). Interestingly, it also showed compatible interaction on Atlas (Rrs2). Furthermore, four differentials showed incompatible interaction against the three Rc isolates tested; Atlas 46 (Rrs1, Rrs2), Jet (Rh6, rh7, rh, rh6), Osiris (Rh4, rh6, Rh10, BRR6), and Turk (Rh5, Rrs1Turk (Rh3), rh6). A differential interaction was observed for the Rc isolate SC-511 on Atlas, Brier and CI4364. Furthermore, four differentials showed incompatible interaction against the three Rc isolates tested; Atlas 46 (Rrs1, Rrs2), Jet (Rh6, rh7, rh, rh6), Osiris (Rh4, rh6, Rh10, BRR6), and Turk (Rh5, Rrs1Turk (Rh3), rh6).
3.1.1 Adult plant stage phenotyping of AM2017 panel
At the adult plant stage, the disease severity on barley genotypes differed based on the environmental conditions in each location and the local pathogen population (Figure 1C). At Marchouch (MCH18) and Rommani (AR18) in 2018, the average disease severity of 1 on the resistant (ICARDA4 and Atlas 46) and 9 on the susceptible (Tocada and Tissa) checks were observed. While at Guich in 2018 (Guich18), the average disease severity of 0 and 7 was observed on the resistant and susceptible checks respectively.
The highest percentage of scald resistant genotypes was observed at AR18 with 64.5% (204) of genotypes compared to 56% (177) at Guich18 and 28.8% (91) of resistant genotypes at MCH18 (Figure 1C). Likewise, the highest percentage of scald susceptible genotypes were found at MCH18 with 21.5% (68) of genotypes, followed by 17.1% (54) genotypes at AR18, and 6.9% (22) susceptible genotypes at Guich18. About 56 genotypes were resistant across locations, whereas only seven genotypes displayed combined resistance at the seedling and adult plant stages (Figure 1D; Table 2). The heritabilities of scald resistance at the adult plant stage for three environments ranged from 0.89 to 0.99 (Table 1).
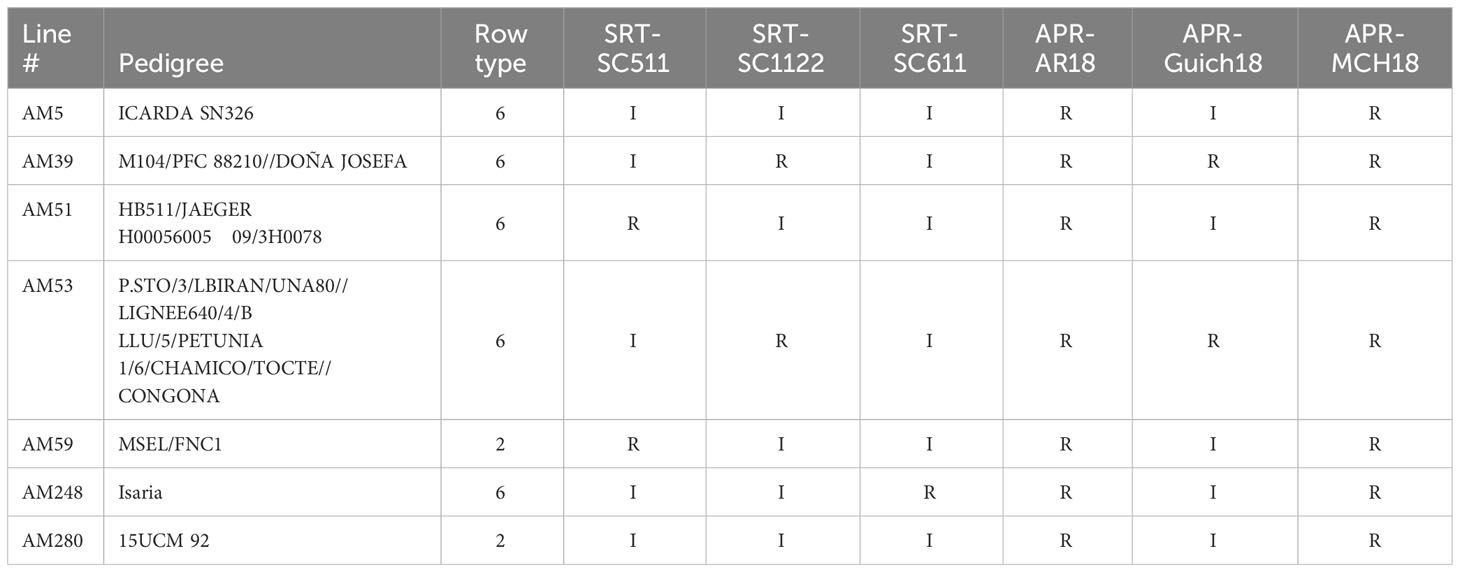
Table 2 Immune (I) and resistant (R) barley genotypes in AM2017 mapping panel to three R. commune isolates at the seedling stage and at the adult plant stage at three field locations in Morocco.
3.1.2 Population structure and linkage disequilibrium
We performed principal component analysis (PCA) using a filtered set of 36,793 SNP markers to evaluate the genetic diversity of the 316 barley genotypes. The PCA of AM2017 categorized the panel into distinct clusters and showed a clear differentiation based on row type and germplasm type. About 10.6% of the genotypic variance was explained in the first axis of the PCA, partitioned the genotypes into two clearly defined groups according to their row type, being the six-rowed genotypes located towards the positive side of the axis and the two-rowed genotypes in the negative side (Figure 2A). The same separation was observed using sNMF, cross-validation and the cross-entropy criterion approaches, indicating that the AM2017 panel at K=2 is partitioned into two major clusters based of row types (Figure 2B). About 40.18% of the two-row genotypes were represented by one cluster and 59.81% of the six-row genotypes were represented by the second cluster. In addition, the PCA of the AM2017 showed a separation according to the germplasm type, but the barley landraces were not grouped into distinct group (Figure 2C). The cross-entropy criterion did not show a clear plateau and steadily decreased at higher K values. However, significant decreases in the criterion were observed from 2 to 10 (Figure 2D). Furthermore, the LD decay of AM2017 was estimated to be ~600 kb (~0.4 cM) (r2 = 0.2; Figure 2E).
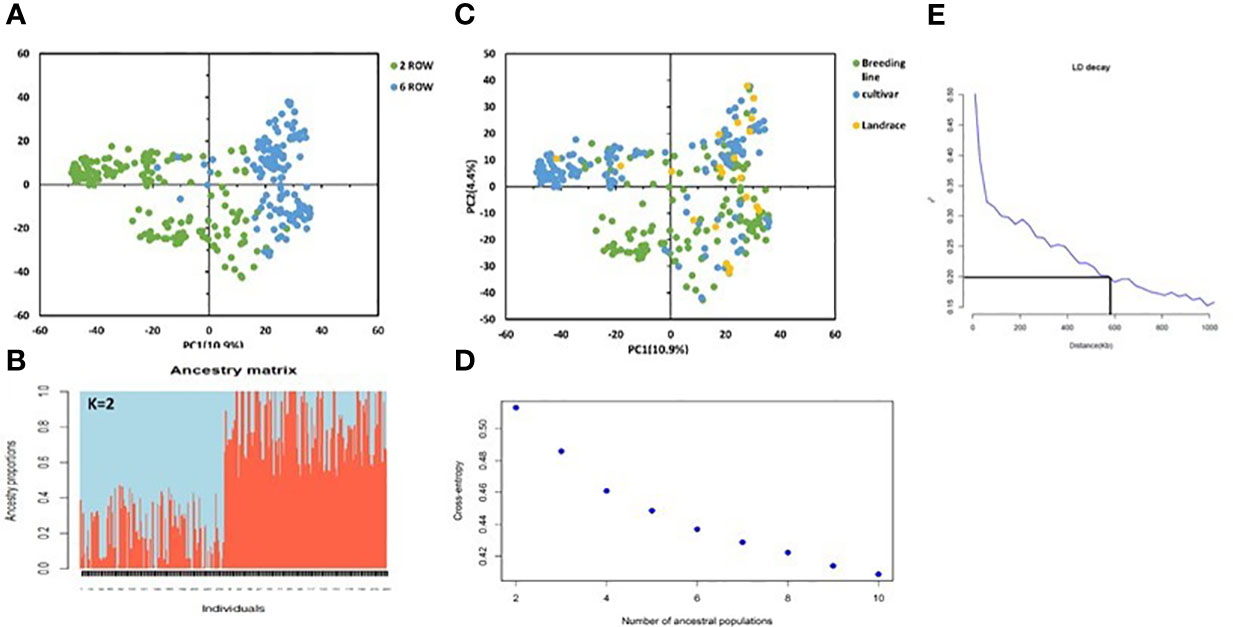
Figure 2 Principal component analysis (PCA) using 36,793 SNP markers displaying the spatial distribution of the 316 genotypes (A, B). Genotypes are shown with colored round symbols based on their germplasm type (A) and row type (B). Population structure using sNMF fonction at K=2 (C). Values of the cross-entropy criterion for SNMF algorithm to evaluate several putative populations (K) ranging from K=2 to K=10 (D). Linkage disequilibrium plot where intra-chromosomal values were plotted against the physical genetic distance (E).
3.2 GWAS of scald resistance at the seedling stage
Performing GWAS for the assessment of resistance to the three R. commune isolates, SC-511, SC-1122 and SC-611 at the seedling stage, the best fitting model was the MLM procedure using PCA, accounting for population structure (Q) and relatedness (K) (Figures 3A, B).
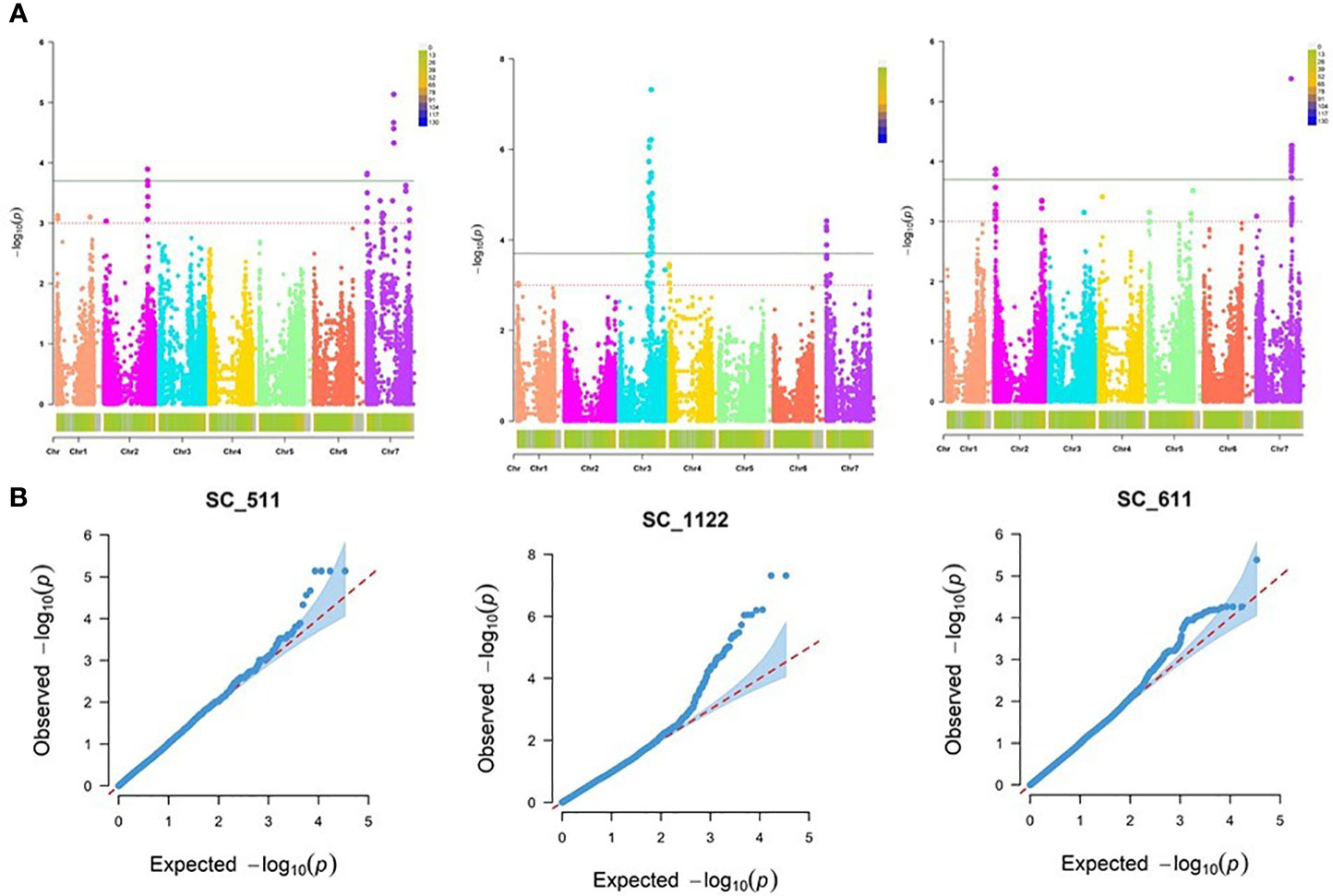
Figure 3 Genome-wide association mapping of barley scald resistance at the seedling stage. (A) The Manhattan plots display significant marker-trait associations on all chromosomes of barley at -log10 of p-values (LOD 3.0 and 3.7 for R. commune isolates SC-511, SC-1122, and SC-611. (B) Quantile-Quantile (Q-Q) plots of marker-trait associations at the seedling stage using MLM + PCA + K model.
Our genome scan detected 102 significant MTAs corresponding to 15 QTL against the three Rc isolates at the seedling stage, with a range of marker R2 from 3.55 – 9.84%, and additive effects ranging from -1.62 to 1.45 (Table 3). In addition, 14 (93%) out of 15, and 6 (50%) out of 12 APR QTL could be validated by more than 3 models. The genome scan for Rc isolate SC-511 detected 15 MTA corresponding to 5 QTL on chromosomes 2H and 7H, with marker R2 and the additive effect ranging from 3.55 to 6.64%, and from -1.02 to 0.905, respectively. The highest phenotypic variation of 6.64% was explained by the SNP marker JHI-Hv50k-2016-483776 associated with qSc.SRT4 on 7H (390.164 Mb; 70.54 cM), reducing the disease severity by -0.886 (17.2%), and by the SNP marker SCRI_RS_226361 associated with qSc.SRT3 on 7H (5.168 Mb; 1.91 cM), reducing the disease severity by -0.637 (21%) and explaining 4.69% of phenotypic variation.
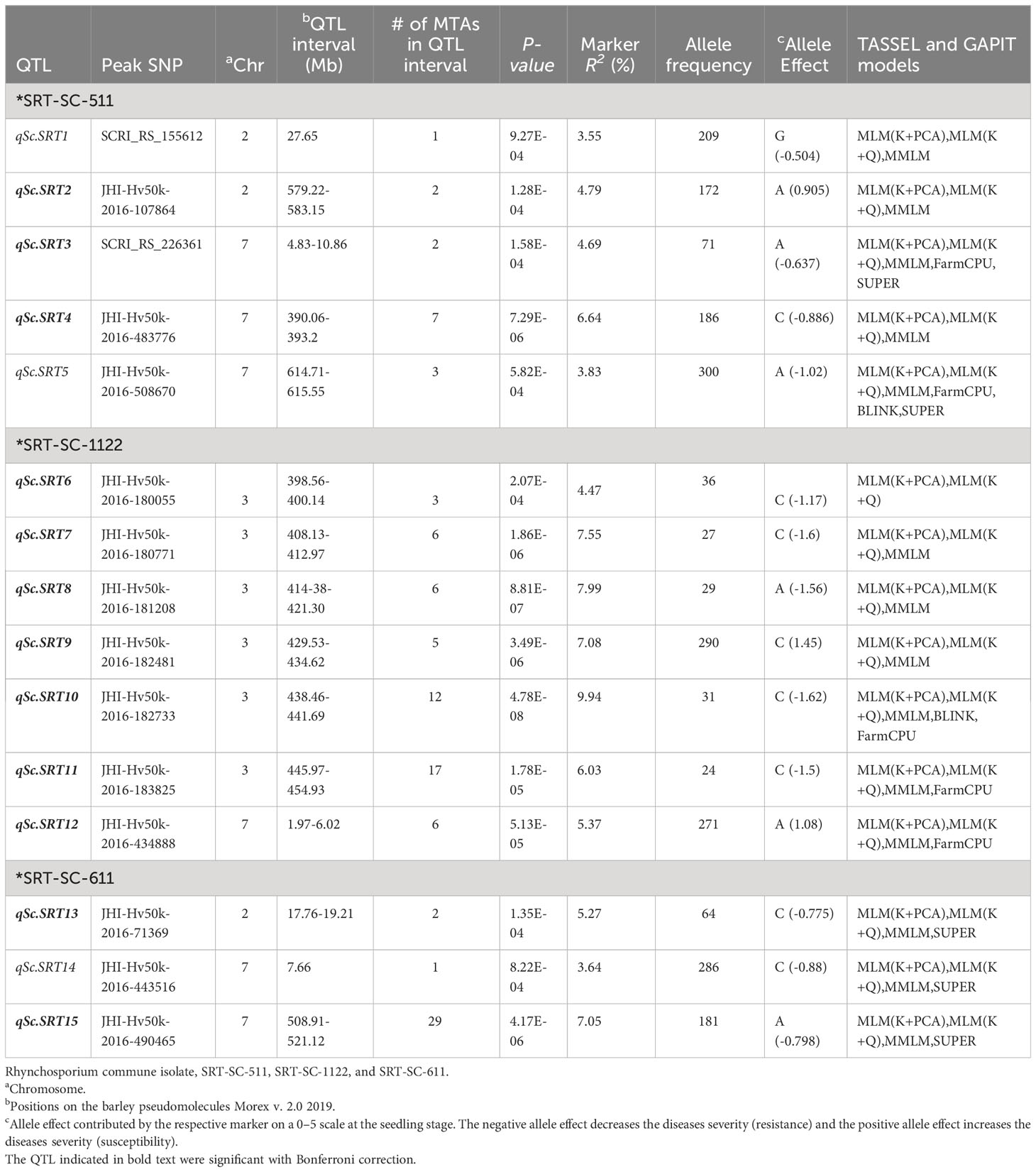
Table 3 Summary of QTL associated with resistance against three isolates of Rhynchosporium commune in barley AM2017 panel at the seedling stage.
For Rc isolate SC-1122, 55 MTAs corresponding to 7 QTL, were detected on chromosomes 3H and 7H, with marker R2 and the additive effect ranging from 4.47 to 9.94%, and from -1.62 to 1.45, respectively. The highest phenotypic variation of 9.94% was explained by the SNP marker JHI-Hv50k-2016-182733 associated with qSC.SRT10 on 3H (438.945 Mb; 51.63 cM), reducing the disease severity by -1.62 (32.4%). In addition, the results demonstrated that several genomic regions were mapped at the vicinity of the previously mapped alleles loci on chromosome 3H (Figure 4).
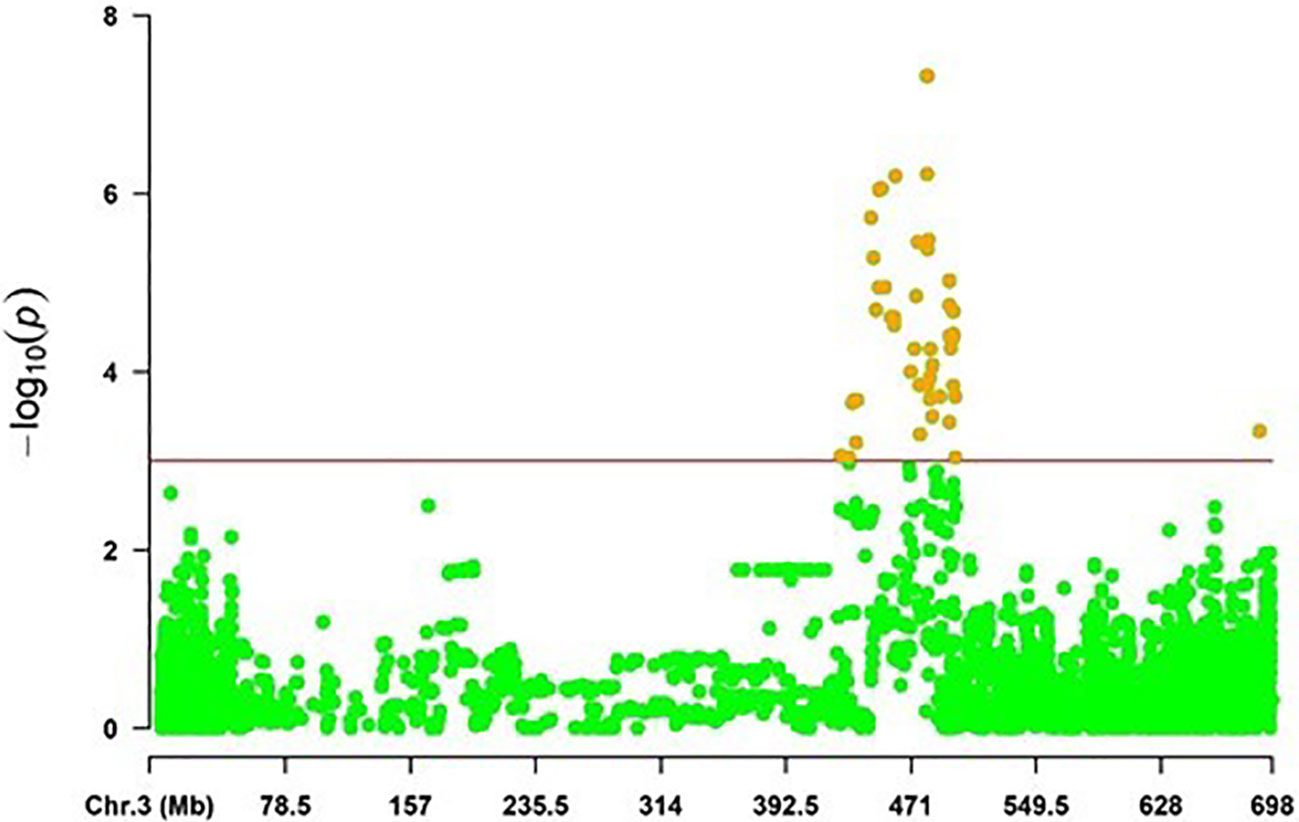
Figure 4 The distribution of significant SNP markers associated with scald resistance at the seedling stage on the chromosome 3H in a region from 400 to 500 Mb.
Likewise, for Rc isolate SC-611, 32 MTAs represented by 3 QTL were detected on chromosomes 2H and 7H, with marker R2 and the additive effect ranging from 3.64 to 7.05%, and from -0.88 to -0.775, respectively (Table 3). The highest phenotypic variation of 7.05% was explained by a SNP marker JHI-Hv50k-2016-490465 associated with qSc.SRT15 on 7H (508.91 Mb; 77.41 cM). However, the highest reduction in disease severity of -0.88 (17.6%) was caused by the SNP marker JHI-Hv50k-2016-443516 associated with qSc.SRT14 on 7H (7.666 Mb; 3.97 cM).
3.3 GWAS of scald resistance at the adult stage
For the resistance at the adult plant stage, MLM procedure accounting for population structure (Q or PCA) and relatedness (K) was identified as the best fitting model (Q-Q plots for all tested models are shown in Figure 5). Our genome scan detected 25 significant MTA corresponding to 12 QTL scattered on all barley chromosomes except the chromosome 2H, with a marker R2 ranging from 4.10 – 6.42%, and additive effects from -2.25 to -1.070 (Table 4). In addition, 19 MTAs, corresponding to 16 QTL with a LOD score of 3.0 – 3.6 have been presented in Supplementary Table S1 as they had been reported in previous studies, thus validating our analysis.
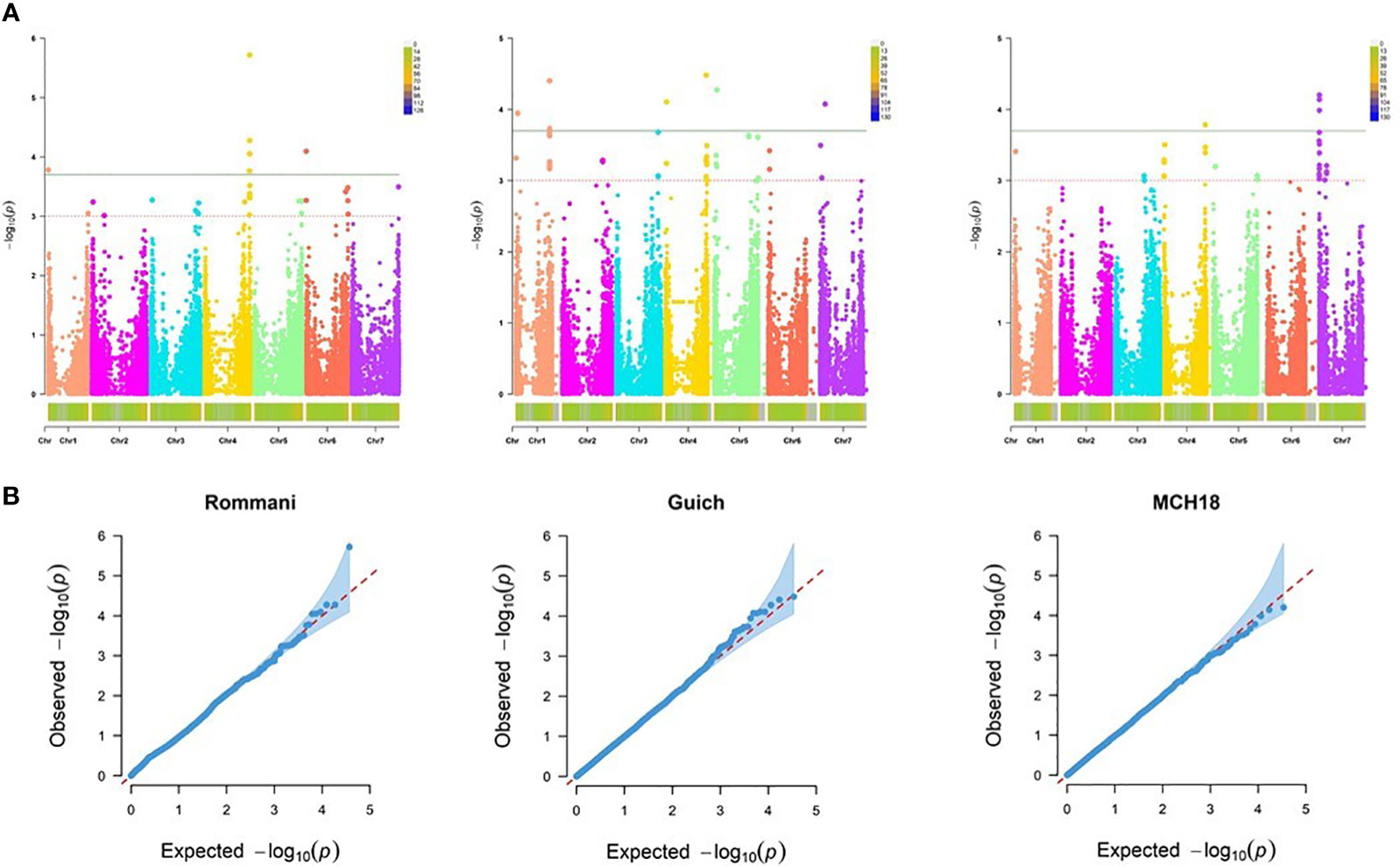
Figure 5 Genome-wide association mapping of barley scald resistance at the adult plant stage. (A) The Manhattan plots display significant marker-trait associations on all chromosomes of barley at -log10 of p-values (LOD 3.0 and 3.7 for three field locations: Marchouch (APR-MCH), Guich (APR-Guich18), and Rommani (APR-Rommani18). (B) Quantile-Quantile (Q- Q) plots of marker-trait associations at the adult plant stage using MLM + PCA + K model.
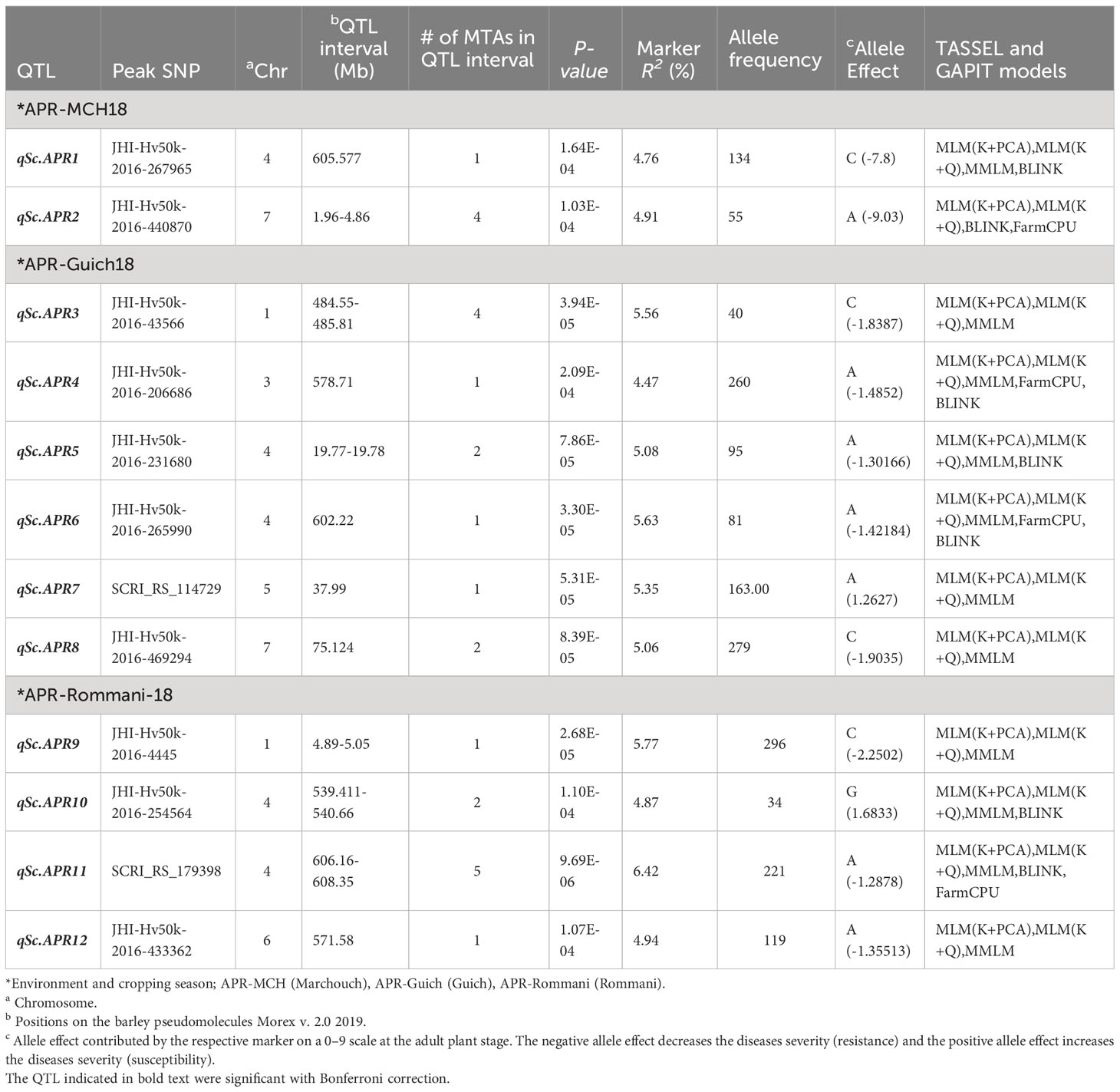
Table 4 Summary of QTL associated with Rhynchosporium commune resistance in barley AM2017 panel at the adult plant stage.
In case of Rommani site (AR18), 9 MTA represented by 4 QTL on the chromosomes 1H, 4H, and 6H were involved in scald disease resistance with a range of marker R2 of 4.55 to 6.42, and additive effect of -2.25 to -1.107. The highest phenotypic variation of 6.42% was explained by a SNP marker SCRI_RS_179398 associated with qSc.APR11 on 4H (606.21 Mb; 103.9 cM), reducing the disease severity by -1.28 (14.2%). However, the highest reduction in disease severity of -2.25 (25%) was caused by a SNP marker JHI-Hv50k-2016-4445 associated with qSc.APR9 on 1H (5.04 Mb; 4.96 cM), explaining a phenotypic variation of 5.7%. For Guich (Guich18), 11 MTA represented by 6 QTL were detected on all barley chromosomes except 2H and 6H with a range of marker R2 of 4.10 to 5.63, and additive effect of -1.903 to -1.262 (Table 4). The highest phenotypic variation of 5.63% was explained by the SNP marker JHI-Hv50k-2016-265990 associated with qSc.APR6 on 4H (602.22 Mb; 99.43 cM), reducing the disease severity by -1.42 (15.7%). However, the highest reduction in disease severity of -1.9 (21.1%) was attributed to the SNP marker JHI-Hv50k-2016-469294 associated with qSc.APR8 on 7H (75.12 Mb; 61.47 cM), explaining a phenotypic variation of 5.06%. For Marchouch (MCH18), 5 MTA represented by 2 QTL were detected on the chromosomes 4H and 7H with a range of marker R2 4.51 to 5.58 and additive effects from -9.03 to -7.22. The highest phenotypic variation of 5.58% was caused by the SNP marker JHI-Hv50k-2016-438638 associated with qSc.APR2 on 7H (3.376 Mb; 1.63 cM). However, the highest reduction in disease severity of -9.03 was caused by the SNP marker JHI-Hv50k-2016-440870 associated with qSc.APR2 on 7H (4.83 Mb; 52.01 cM). Interestingly, 4 SNP markers were associated with Rc resistance at both the seedling and the adult plant stages (Table 5).
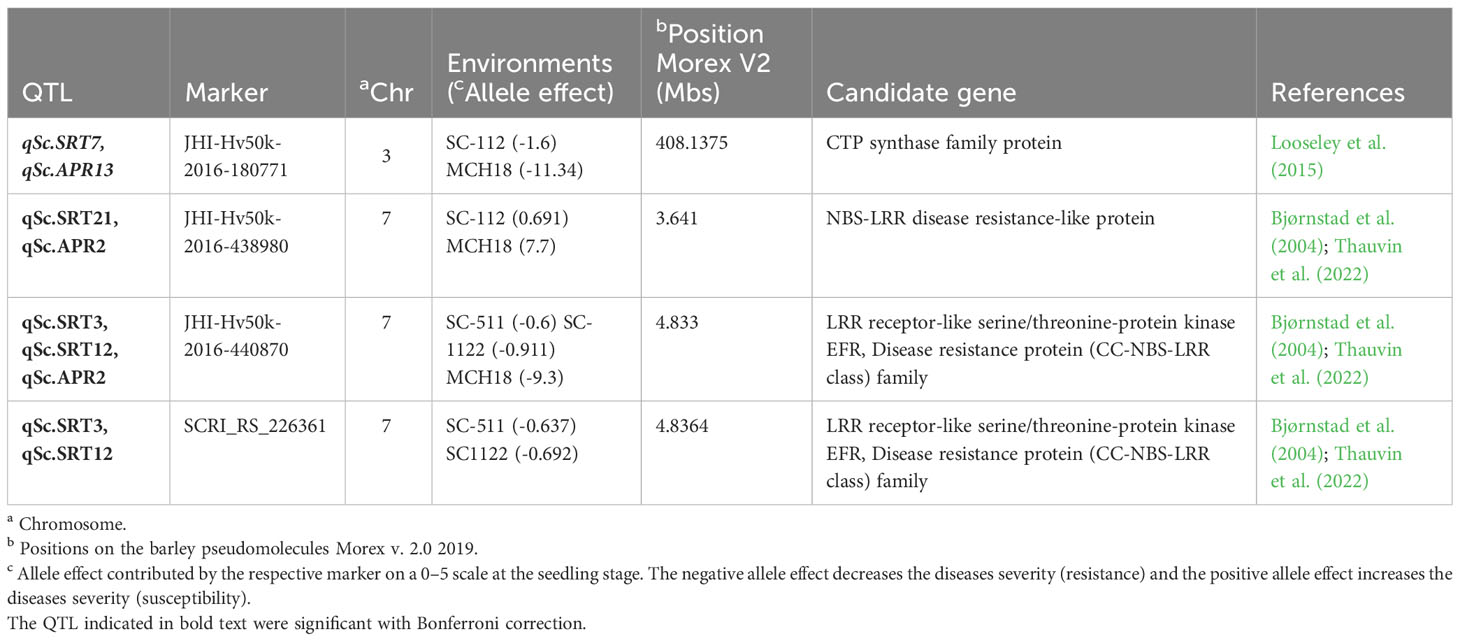
Table 5 A list of common SNP markers and their encoded candidate genes for the seedling and adult plant stage resistance to Rhynchosporium commune in barley AM2017 panel.
3.4 Candidate genes and QTL alignment
Based on the annotation of the sequences in the barley reference genome, the majority of the MTAs were located in genomic regions that are enriched with functional proteins associated with plant disease resistance and defense mechanisms. The BLAST searches revealed their homology with functional proteins/enzymes implicated in host plant disease resistance such as Serine/threonine-protein kinase Receptor-like protein kinase family protein, Disease resistance protein (CC-NBS-LRR class) family, receptor like kinases, Glutathion S-transferase, Nodulin MtN21/EamA-like transporter family protein, Subtilisin-like protease, ABC transporter ATP-binding protein, wall associated kinase, Glycosyltransferase family 92 protein, Hydroxycinnamoyl-CoA shikimate/quinate hydroxycinnamoyl transferase, ankyrin repeat family protein, Pentatricopeptide repeat-containing protein, Sugar transporter protein, and WD repeat domain phosphoinositide-interacting protein. Among 15 SRT QTL, BLAST searches using sequences of peak SNP markers revealed the homology of 4 SNP markers with RLKs (receptor like kinases) and 2 SNP markers with NLR (Nucleotide-binding leucine repeat). Likewise, in case of APR, SNP markers associated with 7 QTL encode RLK (Tables 6, 7; Supplementary Table S2).
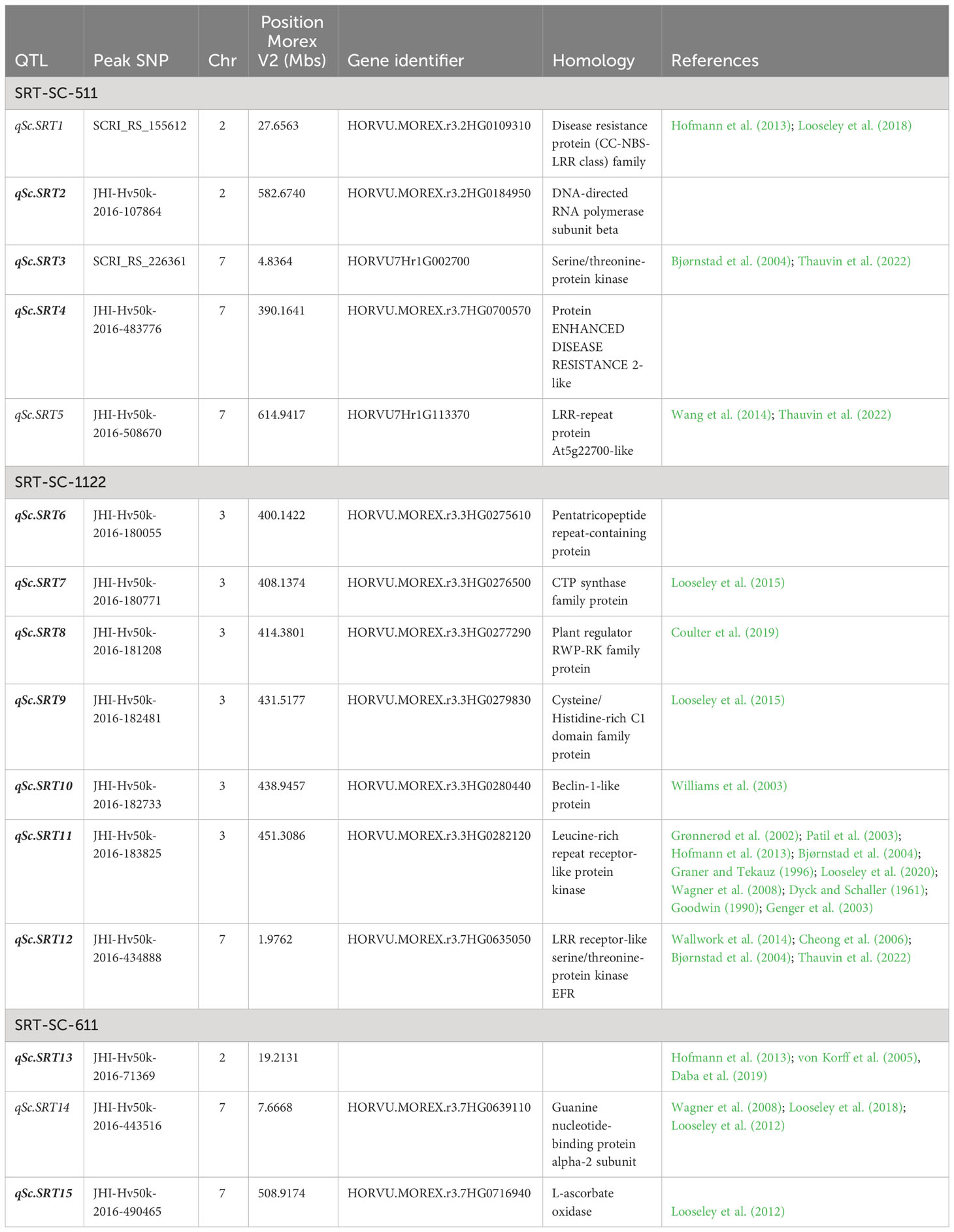
Table 6 QTL alignment and candidate genes identified for the seedling resistance against three isolates of Rhynchosporium commune in barley AM2017 panel.
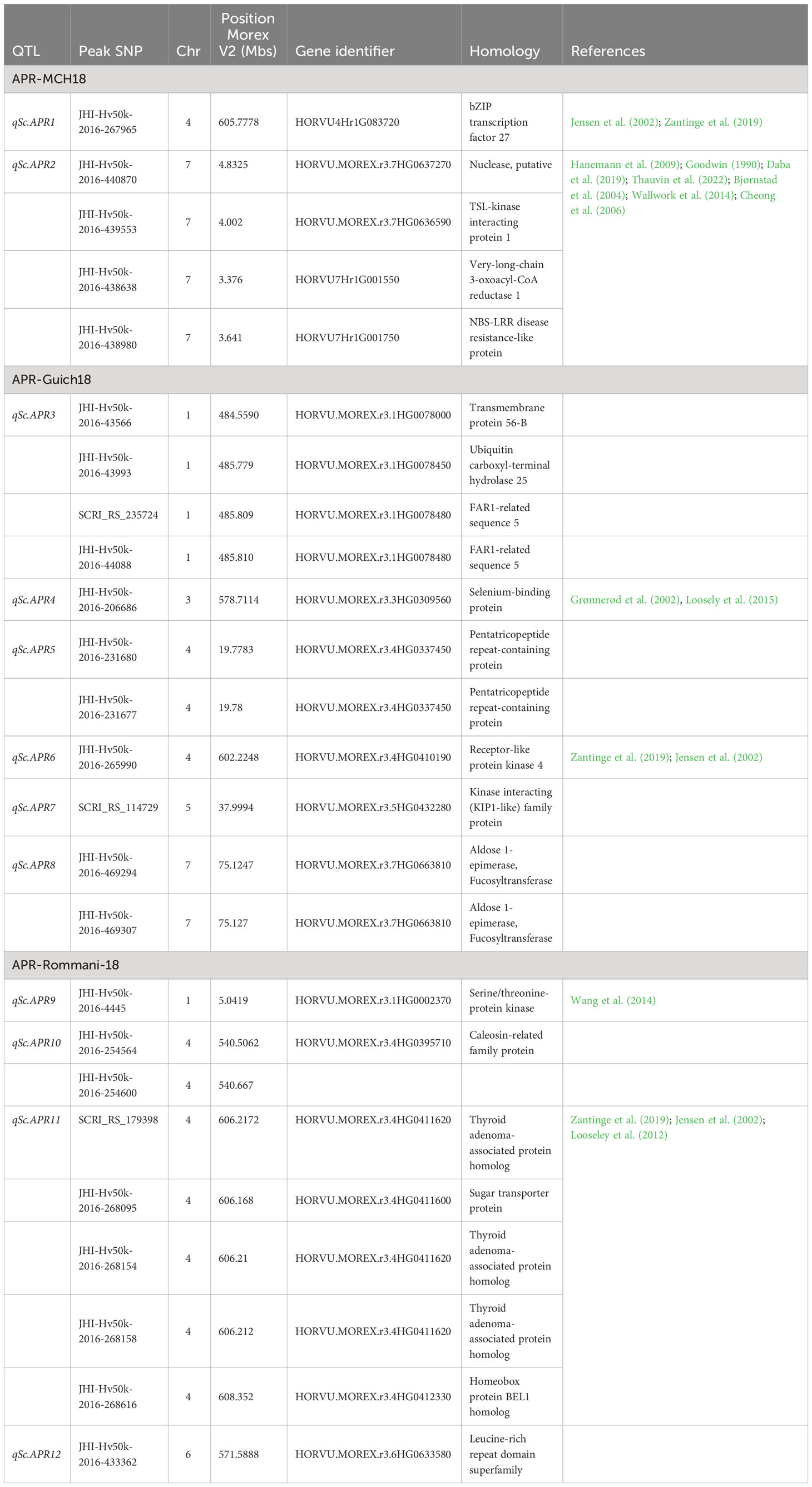
Table 7 QTL alignment and candidate genes identified for the adult plant stage resistance against Rhynchosporium commune in barley AM2017 panel.
Furthermore, 12 out of 15 SRT QTL, and 6 out of 12 APR QTL were validated by our study. However, 9 QTL on chromosomes 1, 2, 3, 4, 5, 6 and 7H reported in this study could be considered as novel (Tables 3, 4).
4 Discussion
Rhynchosporium commune is one of the most destructive diseases of barley worldwide including in parts of North Africa and West Asia, requiring a continuous search for new sources of resistance and the development of associated molecular markers to introgress resistance loci to develop resistant varieties. In this study, we are reporting sources of resistance to Rhyncosporium commue from a collection of diverse barley genotypes at the seedling and at the adult plant stages. To our knowledge, this is the first GWAS study of scald resistance in spring barley from North Africa.
The successful inoculations at greenhouse and at the fields have allowed for efficient screening of AM2017 to scald at seedling and adult plant stages. A large number of sources of resistance were identified with differences in infection responses observed among the three Rhynchosporium commune isolates at the seedling stage and the at the three locations at the adult plant stage. Among the isolates, SC-1122 at the seedling stage, and the adult plant stage testing location APR MCH18 appeared to be the most virulent suggesting their use for further screening of germplasm. The high aggressiveness of SC-1122 could be due to the presence of functional NIP1 gene which affects virulence as reported by several authors (Stefansson et al., 2014; Mohd-Assaad et al., 2019).
The results confirmed the usefulness of AM2017 constructed by breeders in supplying sources of resistance to scald and to leaf rust (Amouzoune et al., 2022). In general, breeders prefer to use as parents, either elite lines or released varieties from different countries included in the constructed panels. However, core collections and other subsets of landraces conserved in genebanks and accessions of wild Hordeum are also bringing sources of resistance (Hiddar et al., 2021).
The monitoring of virulence of the pathogen and the screening of germplasm against the most virulent isolates and field populations could allow for a strategic deployment of effective resistance genes as an important component of integrated management of the disease. Major genes can be either used sequentially or pyramided. However, the screening of the same material at both seedling and adult plant stages could allow the identification of both minor and major genes of resistance which can be deployed for a more durable resistance protecting barley at all stages. However, compilation of these genes require the identification of QTLs associated with resistance.
Our results showed differences in infection response within the genotypes of AM2017 at the seedling stage to the three scald isolates. These differences could be attributed to diverse virulence spectrum of each Rc isolate. This notion was reinforced by conducting a race analysis of the three Rc isolates using barley differentials with known resistance genes. Based on the race analysis, it can be postulated that the Rc isolate SC-511 is NIP1 deficient as it showed compatible reactions both on Brier (Rrs1) and on Atlas (Rrs2).
Successful barley improvement depends on availability of genetic variation for breeders sought traits. Hence, with low genetic diversity in a given gene pool, crops become more vulnerable to biotic and abiotic stress (Sharma, 2017). The availability of the whole genome sequences and genotyping platforms have accelerated the identification, and the use of existing allelic diversity in barley germplasm. Compared with bi-parental mapping which is limited by allelic diversity among parents, GWAS can explore the existing allelic diversity and historical recombination events, resulting in reduced LD and superior mapping resolution. The LD of AM2017 panel was estimated to be 0.6 Mb (~0.4 cM), compared to earlier studies where LD of 3 to 5 cM was reported using different set of barley germplasm (Amezrou et al., 2018; Visioni et al., 2018; Visioni et al., 2020). In addition, AM2017 has an average SNP density of 37.09 SNPs/cM, which is much higher than earlier GWAS studies with 5 SNP/cM (Amezrou et al., 2018), 1 DArT marker per 1.5 cM (Comadran et al., 2009), and 1 SNP marker per 0.72 cM (Cockram et al., 2008; Pasam et al., 2012). Higher genetic diversity found in AM2017 is due to diverse origin of barley genotypes; 134 advance breeding lines from ICARDA’s international spring barley breeding program, 161 registered cultivars from Asia, Africa, Europe, America, and 21 landraces. In addition, very high heritability estimates were observed for scald resistance at the adult plant stage (0.89-0.99) compared with heritability estimates at the seedling stage (0.40 to 0.58). Similarly, Feriani et al. (2012) reported a higher heritability of 64% for all scald isolates. Likewise, Xi et al. (2000) also reported high heritability of scald resistance ranging from 0.57 to 0.91 under field conditions.
Barley QTL conferring resistance to Rhynchosporium commune at the seedling stage have been identified through linkage analysis of biparental mapping populations (Zhang et al., 2020). Though bi-parental mapping has resulted in the identification of eleven major scald disease resistance genes, Rrs14 on chromosome 1H; Rrs1, Rrs3, and Rrs4 on 3H; Rrs6 and Rrs9 on 4H; Rrs13 on 6H; Rrs2, and Rrs12 on 7H, most of the mapping studies converged on loci proximal of Rrs1 (3H) and Rrs2 (7H) (Zhang et al., 2020). Interestingly, the necrosis inducing protein (NIP1) is the product of avirulence gene AvrRrs1 and conditions disease resistance in barley genotypes carrying Rrs1(Rohe et al., 1995; Steiner-Lange et al., 2003; Stukenbrock and McDonald, 2009). Based on the position of flanking markers on Morex genome V2, Rrs1 has been mapped on 445-455 Mbs on the chromosome 3H, and we also detected 15 SNPs in the same genomic region for Rc isolate SC-1122 as reported by numerous studies (Dyck and Schaller, 1961; Goodwin, 1990; Graner and Tekauz, 1996; Grønnerød et al., 2002; Genger et al., 2003; Patil et al., 2003; Wagner et al., 2008; Hofmann et al., 2013; Looseley et al., 2020). Based on these findings, it can be postulated that only Rc isolate SC-1122 seems to have NIP1, whereas Rc isolates SC-611 and SC-511 did not harbor NIP1 as no QTL was detected on 3H. Likewise, Rrs2 has been mapped on 5.2 Mb on 7H, and our genome scan has detected three QTL in the same genomic region; qSc.SRT3 (4.83 Mb) for Rc isolate SC-511, qSc.SRT12 (1.97-6.02 Mb) for SC-1122, and qSc.SRT14 (7.66 Mb) for Rc SC-611. The same genomic region has been reported previously on 5.3 Mb by Daba et al. (2019) on 6.42 Mb, and in many other studies (Grønnerød et al., 2002; Bjørnstad et al., 2004; Cheong et al., 2006; Yan et al., 2009; Wallwork et al., 2014). These results demonstrated that many new genomic regions were mapped at the vicinity of the previously mapped loci on chromosome 3H and 7H.
On chromosome 1H, the major resistance gene Rrs14 introgressed from Hordeum spontaneum was mapped on 2.8 Mb (Garvin et al., 2000). We detected one APR QTL qSc.APR9 (4.89-5.0 Mb) on the short arm of 1H which is in close proximity of Rrs14 locus. Another APR QTL qSc.APR3 on chromosome 1H at 484.55-485.81 Mb seems to be novel as it is located ~482 Mb away from Rrs14 locus. Interestingly, the peak SNP JHI-Hv50k-2016-4445 (5.04 Mb) associated with the APR QTL qSc.APR9 reduced the disease severity by 2.25 units (25%) and validated a QTL reported on 4.28 Mb by Wang et al. (2014).
On chromosome 2H, for Rc isolate SC-511 two SRT QTL qSc.SRT1 (27.65 Mb), qScSRT2 (579.22-583.15 Mb), and for Rc SC-611 one SRT QTL qSc.SRT13 (17.76-19.21 Mb) were detected. Our study has validated a QTL reported by Daba et al. (2019) on 17.12 Mb and the same QTL was reported by Hofmann et al. (2013) and (von Korff et al., 2005). Likewise, one APR QTL qSc.APR21 has been reported by Looseley et al. (2012, 2015) (Supplementary Table S2).
On chromosome 3H, 49 SRT MTAs (48.03%) were grouped into 6 SRT QTL, spanning a physical distance from 394 to 454.93 Mb. Of the 6 SRT QTL from this study, five SRT QTL; qSc.SRT7, qSc.SRT8, qScSRT9, qScSRT10, and qScSRT11 have been reported previously in different studies (Dyck and Schaller, 1961; Goodwin, 1990; Graner and Tekauz, 1996; Grønnerød et al., 2002; Genger et al., 2003; Patil et al., 2003; Cheong et al., 2006; Wagner et al., 2008; Hofmann et al., 2013; Looseley et al., 2015; Looseley et al., 2018; Coulter et al., 2019; Looseley et al., 2020). Whereas the SRT QTL qSc.SRT6 (398.56-400.14 Mb) was novel. Furthermore, 4 APR MTAs were classified into 4 QTL and none of them overlapped with the mapped interval of Rrs1, which may indicate that the other genomic regions on 3H are important in conditioning adult plant stage resistance to scald. Alternatively, the field populations of Rc are either AvrRrs1 deficient or have a mutated version of AvrRrs1. This can further be supported from our seedling screening assay where one of three Rc isolates SC-1122 detected QTL in Rrs1 mapped locus. Three APR QTL; qSc.APR13 (408-414 Mb), qSc.APR4 (578.7-578.8 Mb), and qSc.APR23 (597.95 Mb) have been reported previously (Grønnerød et al., 2002; Cheong et al., 2006; Looseley et al., 2015).
On chromosome 4H, the major resistance gene Rrs16 introgressed from H. bulbosum was mapped on 1.7 Mb (Pickering et al., 2006). Likewise, we detected two MTAs linked to qSc.APR14 spanning a genomic region of 0.39-2.37 Mb with the peak SNP marker JHI-Hv50k-2016-225852 on 1.36 Mb, reducing the disease severity by 7.88 AUDPC units (Supplementary Table S2). Rc QTLs have been reported in the same genomic region (Pickering et al., 2006; Wallwork et al., 2014; Wang et al., 2014). Similarly, qSC.APR1 (605.57-606.16 Mb), qSc.APR6 (602.22 Mb), and qSc.APR11 (606.16-608.35 Mb) have been reported by several authors (Jensen et al., 2002; Looseley et al., 2012; Zantinge et al., 2019).
Five APR QTL were detected on 5H conditioning resistance to scald at two field locations. The two QTL qSc.APR17 (Guich18) and qSc.APR24 (AR18) were located between 550.77 to 553.49 Mb may represent the same QTL, and Looseley et al. (2012) also reported the same genomic region to be associated with scald resistance (Supplementary Table S2). Likewise, Daba et al. (2019) mapped a QTL on 587.44 Mb which coincides with two APR QTL; qSc.APR18 (586.95 Mb) and qSc.APR25 (582-589 Mb) reducing the disease severity by 0.9 (10%) to 1.13 (14%) units. Of the 5 APR QTL, only 1 QTL (qSc.APR7; 37.99 Mb) was novel. Interestingly, the peak SNP JHI-Hv50k-2016-343816 (553.49 Mb) associated with qSc.APR24 reduced the disease severity by 1.78 units (20%) and can be used as a functional marker in Marker Assisted Selection (MAS).
In previous studies, two major scald resistance genes, Rrs13 (11.8 Mb) and Rrs18 (25.6 Mb) were mapped on the chromosome 6H (Abbott et al., 1995; Read and Oram, 1995; Genger et al., 2003; Coulter et al., 2019). Among the three APR QTL identified on 6H, qSc.APR19 (23.712-23.746 Mb) was detected in close proximity of Rrs13 and Shtaya et al. (2006) also mapped a QTL on 21.3 Mb. Further, another APR QTL qSc.APR26 (4.34 Mb) was detected by Jensen et al. (2002) from the cultivar Alexis, and by Cheong et al. (2006) from the cultivar Keel in two different studies. No SRT QTL was detected on 6H for the three Rc isolates tested.
Of the 102 MTAs at SRT, 48 (47%) were mapped on the chromosome 7H and constituted 6 QTL of which 5 QTL were validated by our study and 1 QTL was novel. The SRT QTL qSc.SRT5 (614.71-615.55 Mb) may represent the same QTL reported by Wang et al. (2014) and Thauvin et al. (2022) on 7H.
The markers/QTL stable across environments would be ideal candidates for MAS (marker assisted selection) and genomic selection (GS). Two common SRT QTL were detected. We detected two SNPs JHI-Hv50k-2016-440870 (4.832 Mb) and SCRI_RS_226361 (4.863 Mb) associated with qSc.SRT3 and qSc.SRT12 against Rc SC-511 and SC-1122 on 7H, respectively and may represent the same QTL. About 9 significant SNPs were encompassing a genomic region of 1.97-10.86 Mb on 7H, conditioning resistance to all the three Rc isolates. These may represent different alleles of Rrs2 or different resistance genes. In addition, seven common APR QTL were detected across environments. Seven SNPs encompassed a genomic region of 602.22-608.35 Mb on 4H representing qSc.APR1 (APR-MCH18), qSc.APR16 (Guich18), and qSc.APR11 (AR18) which may represent the same QTL. Likewise, the close proximity of qSc.APR17 (550.77 Mb; Guich18) with qSc.APR24 (553.49 Mb; AR18), and qSc.APR18 (586.95 Mb; Guich18) with qSc.APR25 (582.53-589.44; AR18) on 5H most probably represent the same QTL. A list of 4 common SNPs were shared between SRT and APR. Similarly, qSc.APR20 (10.12 Mb; Guich18) and qSc.APR27 (10.74 Mb; AR18) represent the same QTL. These SNPs can be used for MAS and GS platforms to accelerate gene pyramiding efforts.
The understanding of host-pathogen interaction at genetic level is quite important for identifying and deploying scald resistance. Plants have a multilayered inducible defense system. Their first layer of defense is based on extracellular pattern recognition receptors (PRRs) which recognize conserved microbe/pathogen/damage-associated-molecular patterns (MAMPs/PAMPs/DAMPs). Mostly PRRs are membrane anchored receptors with an ecto-domain for ligand binding for example leucine-rich-repeat (LRR) domain or a chitin binding (LysM) domain (Miya et al., 2007) with or without an endo- kinase domain like in receptor like kinases (RLKs) or receptor-like proteins (RLPs) which require a co-receptor for cellular signaling. The activation of PRRs induces transcriptome reprogramming and biosynthesis of antimicrobial compounds to limit pathogen’s ingress. However, successful pathogens use avirulence gene products (effectors) to subvert host defense responses. The second layer of defense is initiated upon recognition of pathogen secreted effectors by nucleotide-binding leucine-rich repeat (NLR) receptors either directly (gene-for-gene model) or indirectly by guarding and sensing conformational changes in the host virulence targets of the effectors which results in a localized cell death, a phenomenon referred here as effector triggered immunity (ETI) (Jones and Dangl, 2006). R. commune is an hemibiotrophic pathogen of barley and a gene-for-gene interaction has been proposed with its host. Three necrosis inducing proteins (NIP1, NIP2, and NIP3) have been described from Rc, where NIP1 is expressed in spores, but NIP2 and NIP3 are expressed upon host infection (Kirsten et al., 2012). NIP1 serves the function of an effector by inducing leaf necrosis, and as an elicitor to induce Rrs1 dependent defense response (Wevelsiep et al., 1993; Rohe et al., 1995; Steiner-Lange et al., 2003a). That is why NIP1 has been under positive selection as indicated by the presence of two paralogous and copy number variations. From a global collection of 614 Rc isolates, Schürch et al. (2004) reported absence of NIP1 deletion in 45% of the isolates, whereas, NIP2 and NIP3 were present in all isolates. Different Rc isolates either lacking NIP1 or mutated NIP1 can overcome cultivars carrying Rrs1 (Stukenbrock and McDonald, 2009; Mohd-Assaad et al., 2019). Therefore, the identification and deployment of both qualitative and quantitative resistance genes offers durable control of this rapidly evolving pathogen. None of the major resistance genes against scald have been cloned and functionally characterized, hence the molecular mechanism of disease resistance conditioned by Rrs1 and Rrs2 remains elusive. Rrs1 does not encode a receptor for NIP1 but triggers resistance response upon recognition of NIP1 by an unknown co-receptor. With the availability of barley genome and predicted protein repertoire, functional homology-based BLAST searches using marker sequences associated with mapped QTL will help in candidate gene identification. Interestingly, the QTL qSc.SRT11 was co-located within Rrs1 mapped region on the chromosome 3H which was reported in different studies (Dyck and Schaller, 1961; Goodwin, 1990; Graner and Tekauz, 1996; Bjørnstad et al., 2002; Grønnerød et al., 2002; Patil et al., 2003; Wagner et al., 2008; Hofmann et al., 2013; Looseley et al., 2020).
RLKs contain an ectodomain, a transmembrane spanning region, and an intracellular cytoplasmic kinase domain to relay the pathogen recognition signal (Tör et al., 2004). The SNP marker JHI-Hv50k-2016-124224 associated with qSc.SRT15 on chromosome 2H (632.26 Mb) encodes a serine/threonine-protein kinase (HORVU.MOREX.r3.2HG0199700), reducing the disease severity by 1.35 units (27%). Likewise, the SNP marker JHI-Hv50k-2016-4445 associated with qSc.APR9 on chromosome 1H (5.041 Mb) also encodes a serine/threonine-protein kinase (HORVU.MOREX.r3.1HG0002370), reducing the disease severity by 2.25 units (25%). The Serine/threonine protein kinases phosphorylate the OH group of two amino acids, serine or threonine of the target protein, and this protein phosphorylation process plays an important role in the disease resistance pathway. In wheat the Pm21 locus conditions durable resistance to wheat powdery mildew (B.graminis f.sp. tritici (Bgt)) (He et al., 2022). A putative serine/threonine-protein kinase (Stpk-V) was cloned from the Pm21 locus and its transient expression in a susceptible wheat variety Yangmati 158 rendered broad spectrum durable resistance to powdery mildew. Whereas virus induced gene silencing of Stpk-V in resistant wheat cultivar rendered it highly susceptible to Bgt (Chen et al., 1995; Cao et al., 2011). In barley, the leaf rust resistance gene Rph22 encodes a lectin receptor like kinase and some of the RLKs may possibly be involved in basal defense if an analogy is to be drawn with other extracellular PTT receptors (Wang et al., 2019). Likewise, in rice, an RLK (Xa21) confers resistance to bacterial blast caused by Xanthomonas oryzae pv. Oryzae (Song et al., 1995; Tör et al., 2004). We found 6 APR QTL (qSc.APR6, qSc.APR7, qSc.APR18, qSc.APR19, qSc.APR21,and qSc.APR24), and 5 SRT (qSc.SRT2, qSc.SRT3, qSc.SRT11, qSc.SRT12, qSc.SRT15) where several SNP markers encode RLK as candidate genes. The co-location of candidate genes within families with known role in disease resistance such as RLK, NLR and wall associated proteins can help identify new allelic variants of existing genes or completely new resistance genes.
Sugar transporters mediate sugar transport and are one of important players in plant-pathogen interaction (Breia et al., 2021). A SNP marker JHI-Hv50k-2016-268095 is associated with a APR QTL qSc.APR11 on 4H at 606.1682 Mb and encodes a sugar transporter (HORVU.MOREX.r3.4HG0411610). This SNP marker reduced the disease severity by ~20% in MCH18. Recently cloned what leaf rust resistance gene Lr67 encodes a sugar transporter 13 (SAT13), which plays a crucial role in conditioning partial resistance to all wheat rusts and powdery mildew (Milne et al., 2019). Its susceptible variant (Lr67sus) is a functional sugar transporter and its expression is upregulated upon pathogen invasion, whereas its dominant variant (LR67res) is unable to transport sugar which is required by wheat rusts and powdery mildew. Likewise, Xanthomonas oryzae pv. oryzae secreted TAL (Transcription-activator like) effector upregulate the expression of a sugar transporter Os SWEETY11 in rice (Chen et al., 2010). Similar findings have been reported for Arabidopsis-Botrytis cinerea, grape vine interaction with powdery mildew (Erysiphe nectar) and downy mildew (Plamopara viticola) (Hayes et al., 2010; Lemonnier et al., 2014). Sugar transporters are quite conserved in plant kingdom and a similar role in disease resistance can be envisioned for these transporters in barley-Rhynchosporium commune interactions.
The nodulin-like proteins belong to a group of aquaporins and they have been implicated in the transport of various solutes in plants in addition to their interaction with various plant pathogens. In barley, the sequences of two SNPs on 7H, JHI-Hv50k-2016-483974 (qSc.SRT4, 393.195 Mb) encode putative Early Nodulin-like protein. Interestingly, Gyawali et al. (2018) also reported a QTL (Rcs-qtl-7H-32.81) for spot blotch resistance in barley on 7H where the SNP marker 11_20162 encoded an Early Nodulin-93 like protein.
The ABC transporter proteins condition disease resistance by secreting the anti-fungal products into the apoplast like PEN3 in Arabidopsis against B. cinerea (He et al., 2019), and Lr34 against wheat leaf rust (Krattinger et al., 2009). We found that ten adjacent SNP markers (453.535-453.764 Mb), in the interval of qSc.SRT11 on 3H, were flanked by ABC transporter B protein (HORVU.MOREX.r3.3HG0282510). In wheat, Lr34 provided resistance against wheat stem rust, stipe rust, and powdery mildew (Rinaldo et al., 2017) in addition to powdery mildew and rust resistance in barley (Boni et al., 2018), and against rice blast (Krattinger et al., 2016). We speculate an important role of ABC transporter proteins in resistance against R. commune.
Our study detected several SNP markers encoding functional proteins which have been involved in plant defense against diverse fungal pathogens. The newly detected QTLs can be applied in barley breeding program and the information could be employed by breeders for marker assisted selection, fine mapping, and gene cloning for improving resistance to scald disease.
5 Conclusion
The identification of diverse sources of resistance and molecular tagging of new genomic regions associated with resistance to R. commune is crucial for future marker-assisted selection and genomic selection for continuous improvement of barley germplasm. Our GWAS was efficient in genetic dissection of scald resistance. Several SNPs were located within already known QTL associated with scald disease in addition to the identification of novel seedling stage resistance QTL on the chromosomes 2H, 3H, and 7H, and the adult stage resistance QTL on the chromosomes 1H, 4H, 5H, 6H, and 7H. Durable scald resistance requires further understanding of R. commune pathogen population dynamics and efficient introgression of major and minor resistance genes using molecular markers.
Data availability statement
The original contributions presented in the study are publicly available. This data can be found here: https://bms.icarda.org:59395/public/?db=Barley_AM17&pj=AM17.
Author contributions
HH, AA, BB, formulated the research problem and designed the approaches. HH, MJ, SR, BB, raised experiments in fields and recorded phenotyping data. HH, SR, SG, performed statistical analysis of data, AA and SR planned and supervised research activity. HH, SR, wrote the initial draft of the paper All authors contributed to the article and approved the submitted version.
Funding
This study is supported financially through GIZ-attributed funding to ICARDA genebank, the CAIGE-GRDCICA00010 project and the CGIAR-Genebank Platform.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1133404/full#supplementary-material
References
Abbott, D. C., Lagudah, E. S., Brown, A. H. D. (1995). Identification of RFLPs flanking a scald resistance gene on barley chromosome 6. J. Hered. 86, 152–154. doi: 10.1093/oxfordjournals.jhered.a111547
Alexander, D. H., Lange, K. (2011). Enhancements to the ADMIXTURE algorithm for individual ancestry estimation. BMC Bioinf. 12, 246. doi: 10.1186/1471-2105-12-246
Alqudah, A. M., Sallam, A., Stephen Baenziger, P., Börner, A. (2020). GWAS: Fast-forwarding gene identification and characterization in temperate Cereals: lessons from Barley – A review. J. Adv. Res. 22, 119–135. doi: 10.1016/j.jare.2019.10.013
Amezrou, R., Verma, R. P. S., Chao, S., Brueggeman, R. S., Belqadi, L., Arbaoui, M., et al. (2018). Genome-wide association studies of net form of net blotch resistance at seedling and adult plant stages in spring barley collection. Mol. Breed. 38. doi: 10.1007/s11032-018-0813-2
Amouzoune, M., Rehman, S., Benkirane, R., Verma, S., Gyawali, S., Al-Jaboobi, M., et al. (2022). Genome-wide association study of leaf rust resistance at seedling and adult plant stages in a global barley panel. Agric. 12. doi: 10.3390/agriculture12111829
Bayer, M. M., Rapazote-Flores, P., Ganal, M., Hedley, P. E., Macaulay, M., Plieske, J., et al. (2017). Development and evaluation of a barley 50k iSelect SNP array. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.01792
Bjørnstad, Å., Grønnerød, S., Mac Key, J., Tekauz, A., Crossa, J., Martens, H. (2004). Resistance to barley scald (Rhynchosporium secalis) in the Ethiopian donor lines “Steudelli” and “Jet”, analyzed by partial least squares regression and interval mapping. Hereditas 141, 166–179. doi: 10.1111/j.1601-5223.2004.01817.x
Bjørnstad, A., Patil, V., Tekauz, A., Mar\oy, A. G., Skinnes, H., Jensen, A., et al. (2002). Resistance to Scald (Rhynchosporium secalis) in Barley (Hordeum vulgare) Studied by Near-Isogenic Lines: I. Markers and Differential Isolates. Phytopathology® 92, 710–720. doi: 10.1094/PHYTO.2002.92.7.710
Boni, R., Chauhan, H., Hensel, G., Roulin, A., Sucher, J., Kumlehn, J., et al. (2018). Pathogen-inducible Ta-Lr34res expression in heterologous barley confers disease resistance without negative pleiotropic effects. Plant Biotechnol. J. 16, 245–253. doi: 10.1111/pbi.12765
Borrego-Benjumea, A., Carter, A., Min, Z., Tucker, J. R., Zhou, M., Badea, A. (2021). Genome-Wide association study of waterlogging tolerance in barley (Hordeum vulgare L.) under controlled field conditions. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.711654
Bradbury, P. J., Zhang, Z., Kroon, D. E., Casstevens, T. M., Ramdoss, Y., Buckler, E. S. (2007). TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics 23, 2633–2635. doi: 10.1093/BIOINFORMATICS/BTM308
Breia, R., Conde, A., Badim, H., Fortes, A. M., Gerós, H., Granell, A. (2021). Plant SWEETs: from sugar transport to plant–pathogen interaction and more unexpected physiological roles. Plant Physiol. 186, 836–852. doi: 10.1093/plphys/kiab127
Brunner, P. C., Stefansson, T. S., Fountaine, J., RiChina, V., McDonald, B. A. (2016). A global analysis of CYP51 diversity and azole sensitivity in Rhynchosporium commune. Phytopathology 106, 355–361. doi: 10.1094/PHYTO-07-15-0158-R
Cantalapiedra, C. P., Boudiar, R., Casas, A. M., Igartua, E., Contreras-Moreira, B. (2015). BARLEYMAP: physical and genetic mapping of nucleotide sequences and annotation of surrounding loci in barley. Mol. Breed. 35, 13. doi: 10.1007/s11032-015-0253-1
Cao, A., Xing, L., Wang, X., Yang, X., Wang, W., Sun, Y., et al. (2011). Serine/threonine kinase gene Stpk-V, a key member of powdery mildew resistance gene Pm21, confers powdery mildew resistance in wheat. Proc. Natl. Acad. Sci. U. S. A. 108, 7727–7732. doi: 10.1073/pnas.1016981108
Chen, X., Hackett, C. A., Niks, R. E., Hedley, P. E., Booth, C., Druka, A., et al. (2010). An eQTL analysis of partial resistance to Puccinia hordei in barley. PloS One 5. doi: 10.1371/journal.pone.0008598
Chen, P. D., Qi, L. L., Zhou, B., Zhang, S. Z., Liu, D. J. (1995). Development and molecular cytogenetic analysis of wheat-Haynaldia villosa 6VS/6AL translocation lines specifying resistance to powdery mildew. Theor. Appl. Genet. 91, 1125–1128. doi: 10.1007/BF00223930
Cheong, J., Williams, K., Wallwork, H. (2006). The identification of QTLs for adult plant resistance to leaf scald in barley. Aust. J. Agric. Res. 57, 961–965. doi: 10.1071/AR05389
Clare, S. J., Wyatt, N. A., Brueggeman, R. S., Friesen, T. L. (2020). Research advances in the pyrenophora teres-barley interaction. Mol. Plant Pathol. 21 (2), 272–288. doi: 10.1111/mpp.12896
Cockram, J., White, J., Leigh, F. J., Lea, V. J., Chiapparino, E., Laurie, D. A., et al. (2008). Association mapping of partitioning loci in barley. BMC Genet. 9, 1–14. doi: 10.1186/1471-2156-9-16
Comadran, J., Kilian, B., Russell, J., Ramsay, L., Stein, N., Ganal, M., et al. (2012). Natural variation in a homolog of Antirrhinum CENTRORADIALIS contributed to spring growth habit and environmental adaptation in cultivated barley. Nat. Genet. 44, 1388–1391. doi: 10.1038/ng.2447
Comadran, J., Thomas, W. T. B., Van Eeuwijk, F. Á., Ceccarelli, S., Grando, S., Stanca, A. M., et al. (2009). Patterns of genetic diversity and linkage disequilibrium in a highly structured Hordeum vulgare association-mapping population for the Mediterranean basin. Theor. Appl. Genet. 119, 175–187. doi: 10.1007/s00122-009-1027-0
Coulter, M., Büttner, B., Hofmann, K., Bayer, M., Ramsay, L., Schweizer, G., et al. (2019). Characterisation of barley resistance to rhynchosporium on chromosome 6HS. Theor. Appl. Genet. 132, 1089–1107. doi: 10.1007/s00122-018-3262-8
Czembor, J. H., Czembor, E., Suchecki, R., Watson-Haigh, N. S. (2022). Genome-wide association study for powdery mildew and rusts adult plant resistance in european spring barley from polish gene bank. Agronomy 12. doi: 10.3390/agronomy12010007
Daba, S. D., Horsley, R., Brueggeman, R., Chao, S., Mohammadi, M. (2019). Genome-wide association studies and candidate gene identification for leaf scald and net blotch in barley (Hordeum vulgare L.). Plant Dis. 103, 880–889. doi: 10.1094/PDIS-07-18-1190-RE
Dracatos, P. M., Bartos, J., Elmansour, H., Singh, D., Karafiatova, M., Zhang, P., et al. (2019). The coiled-coil NLR Rph1, confers leaf rust resistance in barley cultivar sudan. Plant Physiol. 179, 1362–1372. doi: 10.1104/pp.18.01052
Duggal, P., Gillanders, E. M., Holmes, T. N., Bailey-Wilson, J. E. (2008). skfsffsk Establishing an adjusted p-value threshold to control the family-wide type 1 error in genome wide association studies. BMC Genomics 9, 516. doi: 10.1186/1471-2164-9-516
Dyck, P. L., Schaller, C. W. (1961). Inheritance of resistance in barley to several physiologic races of the scald fungus. Can. J. Genet. Cytol. 3, 153–164. doi: 10.1139/g61-019
Falconer, D. S., Mackay, T. F. C. (1996). Introduction to Quantitative Genetics. 4th Edition (Harlow: Addison Wesley Longman).
FDA (2006) FDA (The Food and Drug Administration, USA) (Health Claim for Barley Products). Available at: https://permanent.access.gpo.gov/lps1609/www.fda.gov/fdac/departs/2006/406_upd.html#barley (Accessed March 14, 2022).
Feriani, W., Cherif, M., Rezgui, S. (2012). Preliminary genetic analysis of resistance to scald in Tunisian barley. Int. J. Plant Breed. 6 (2).
Food and Agriculture Organization (FAO) of the United Nations (2020) Crops. 2020. Available at: http://faostat.fao.org/ (Accessed January 8, 2021).
Frichot, E., François, O. (2015). LEA: An R package for landscape and ecological association studies. Methods Ecol. Evol. 6, 925–929. doi: 10.1111/2041-210X.12382
Frichot, E., Mathieu, F., Trouillon, T., Bouchard, G., François, O. (2014). Fast and efficient estimation of individual ancestry coefficients. Genetics 196, 973–983. doi: 10.1534/genetics.113.160572
Garvin, D. F., Brown, A. H. D., Raman, H., Read, B. J. (2000). Genetic mapping of the barley Rrs14 scald resistance gene with RFLP, isozyme and seed storage protein markers. Plant Breed. 119, 193–196. doi: 10.1046/j.1439-0523.2000.00456.x
Genger, R. K., Brown, A. H. D., Knogge, W., Nesbitt, K., Burdon, J. J. (2003). Development of SCAR markers linked to a scald resistance gene derived from wild barley. Euphytica 134, 149–159. doi: 10.1023/B:EUPH.0000003833.63547.78
Goodwin, S. B. (1990). A nomenclature for Rhynchosporium secalis pathotypes. Phytopathology 80, 1330. doi: 10.1094/phyto-80-1330
Graner, A., Tekauz, A. (1996). RFLP mapping in barely of a dominant gene conferring resistance to scald (Rhynchosporium secalis). TAG Theor. Appl. Genet. 93, 421–425. doi: 10.1007/s001220050297
Grønnerød, S., Marøy, A. G., MacKey, J., Tekauz, A., Penner, G. A., Bjørnstad, A. (2002). Genetic analysis of resistance to barley scald (Rhynchosporium secalis) in the Ethiopian line “Abyssinian” (CI668). Euphytica 126, 235–250. doi: 10.1023/A:1016368503273
Gyawali, S., Chao, S., Viash, S. S., Rehman, S., Vishwakarma, S. R., Verma, R. P. S. (2018). Genome wide association studies (GWAS) of spot blotch resistance at the seedling and the adult plant stages in a collection of spring barley. Mol. Breed. 38, 62. doi: 10.1007/s11032-018-0815-0
Hanemann, A., Schweizer, G. F., Cossu, R., Wicker, T., Röder, M. S. (2009). Fine mapping, physical mapping and development of diagnostic markers for the Rrs2 scald resistance gene in barley. Theor. Appl. Genet. 119 (8), 1507–1522.
Hayes, M. A., Feechan, A., Dry, I. B. (2010). Involvement of abscisic acid in the coordinated regulation of a stress-inducible hexose transporter (VvHT5) and a cell wall invertase in grapevine in response to biotrophic fungal infection. Plant Physiol. 153, 211–221. doi: 10.1104/pp.110.154765
He, H., Guo, R., Gao, A., Chen, Z., Liu, R., Liu, T., et al. (2022). Large-scale mutational analysis of wheat powdery mildew resistance gene Pm21. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.988641
He, Y., Xu, J., Wang, X., He, X., Wang, Y., Zhou, J., et al. (2019). The Arabidopsis pleiotropic drug resistance transporters PEN3 and PDR12 mediate camalexin secretion for resistance to Botrytis cinerea. Plant Cell 31, 2206–2222. doi: 10.1105/tpc.19.00239
Hiddar, H., Rehman, S., Lakew, B., Verma, R. P. S., Al-Jaboobi, M., Moulakat, A., et al. (2021). Assessment and modeling using machine learning of resistance to scald (Rhynchosporium commune) in two specific barley genetic resources subsets. Sci. Rep. 11, 15967. doi: 10.1038/s41598-021-94587-6
Hofmann, K., Silvar, C., Casas, A. M., Herz, M., Büttner, B., Gracia, M. P., et al. (2013). Fine mapping of the Rrs1 resistance locus against scald in two large populations derived from Spanish barley landraces. Theor. Appl. Genet. 126, 3091–3102. doi: 10.1007/s00122-013-2196-4
Huang, M., Liu, X., Zhou, Y., Summers, R. M., Zhang, Z. (2019). BLINK: A package for the next level of genome-wide association studies with both individuals and markers in the millions. Gigascience 8, 1–12. doi: 10.1093/gigascience/giy154
Jeger, M. J., Viljanen-Rollinson, S. L. H. (2001). The use of the area under the disease-progress curve (AUDPC) to assess quantitative disease resistance in crop cultivars. Theor. Appl. Genet. 102, 32–40. doi: 10.1007/s001220051615
Jensen, J., Backes, G., Skinnes, H., Giese, H. (2002). Quantitative trait loci for scald resistance in barley localized by a non-interval mapping procedure. Plant Breed. 121, 124–128. doi: 10.1046/j.1439-0523.2002.00685.x
Jones, J. D. G., Dangl, J. L. (2006). The plant immune system. Nature 444, 323–329. doi: 10.1038/nature05286
Kawka, K., Kinga Lemieszek, M., Rzeski, W., Chemopreventive, R. W. (2019). Chemopreventive properties of young green barley extracts in in vitro model of colon cancer. Ann. Agric. Environ. Med. 26, 174–181. doi: 10.26444/aaem/102624
Kirsten, S., Navarro-Quezada, A., Penselin, D., Wenzel, C., Matern, A., Leitner, A., et al. (2012). Necrosis-inducing proteins of rhynchosporium commune, effectors in quantitative disease resistance. Mol. Plant-Microbe Interact. 25, 1314–1325. doi: 10.1094/MPMI-03-12-0065-R
Krattinger, S. G., Lagudah, E. S., Spielmeyer, W., Singh, R. P., Huerta-Espino, J., McFadden, H., et al. (2009). A putative ABC transporter confers durable resistance to multiple fungal pathogens in wheat. Science 323, 1360–1363. doi: 10.1126/science.1166453
Krattinger, S. G., Sucher, J., Selter, L. L., Chauhan, H., Zhou, B., Tang, M., et al. (2016). The wheat durable, multipathogen resistance gene Lr34 confers partial blast resistance in rice. Plant Biotechnol. J. 14, 1261–1268. doi: 10.1111/pbi.12491
Lemonnier, P., Gaillard, C., Veillet, F., Verbeke, J., Lemoine, R., Coutos-Thévenot, P., et al. (2014). Expression of Arabidopsis sugar transport protein STP13 differentially affects glucose transport activity and basal resistance to Botrytis cinerea. Plant Mol. Biol. 85, 473–484. doi: 10.1007/s11103-014-0198-5
Li, D., Feng, Y., Tian, M., Hu, X., Zheng, R., Chen, F. (2021). Dietary barley leaf mitigates tumorigenesis in experimental colitis-associated colorectal cancer. Nutrients 13, 1–14. doi: 10.3390/nu13103487
Lipka, A. E., Tian, F., Wang, Q., Peiffer, J., Li, M., Bradbury, P. J., et al. (2012). GAPIT: Genome association and prediction integrated tool. Bioinformatics 28, 2397–2399. doi: 10.1093/bioinformatics/bts444
Looseley, M. E., Griffe, L. L., Büttner, B., Wright, K. M., Bayer, M. M., Coulter, M., et al. (2020). Characterisation of barley landraces from Syria and Jordan for resistance to rhynchosporium and identification of diagnostic markers for Rrs1Rh4. Theor. Appl. Genet. 133, 1243–1264. doi: 10.1007/s00122-020-03545-9
Looseley, M. E., Griffe, L. L., Büttner, B., Wright, K. M., Middlefell-Williams, J., Bull, H., et al. (2018). Resistance to Rhynchosporium commune in a collection of European spring barley germplasm. Theor. Appl. Genet. 131, 2513–2528. doi: 10.1007/s00122-018-3168-5
Looseley, M. E., Keith, R., Guy, D., Barral-Baron, G., Thirugnanasambandam, A., Harrap, D., et al. (2015). Genetic mapping of resistance to Rhynchosporium commune and characterisation of early infection in a winter barley mapping population. Euphytica 203, 337–347. doi: 10.1007/s10681-014-1274-2
Looseley, M. E., Newton, A. C., Atkins, S. D., Fitt, B. D. L., Fraaije, B. A., Thomas, W. T. B., et al. (2012). Genetic basis of control of Rhynchosporium secalis infection and symptom expression in barley. Euphytica 184, 47–56. doi: 10.1007/s10681-011-0485-z
Mascher, M., Gundlach, H., Himmelbach, A., Beier, S., Twardziok, S. O., Wicker, T., et al. (2017). A chromosome conformation capture ordered sequence of the barley genome. Nature 544, 427–433. doi: 10.1038/nature22043
Mcdonald, B. A. (2015). How can research on pathogen population biology suggest disease management strategies? The example of barley scald (Rhynchosporium commune). Plant Pathol. 64, 1005–1013. doi: 10.1111/ppa.12415
McLean, M. S., Hollaway, G. J. (2018). Suppression of scald and improvements in grain yield and quality of barley in response to fungicides and host-plant resistance. Australas. Plant Pathol. 47, 13–21. doi: 10.1007/s13313-017-0529-5
Milne, R. J., Dibley, K. E., Schnippenkoetter, W., Mascher, M., Lui, A. C. W., Wang, L., et al. (2019). The wheat LR67 gene from the sugar transport protein 13 family confers multipathogen resistance in barley. Plant Physiol. 179, 1285–1297. doi: 10.1104/pp.18.00945
Miya, A., Albert, P., Shinya, T., Desaki, Y., Ichimura, K., Shirasu, K., et al. (2007). CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 104, 19613–19618. doi: 10.1073/pnas.0705147104
Mohd-Assaad, N., McDonald, B. A., Croll, D. (2016). Multilocus resistance evolution to azole fungicides in fungal plant pathogen populations. Mol. Ecol. 25, 6124–6142. doi: 10.1111/mec.13916
Mohd-Assaad, N., McDonald, B. A., Croll, D. (2019). The emergence of the multi-species NIP1 effector in Rhynchosporium was accompanied by high rates of gene duplications and losses. Environ. Microbiol. 21, 2677–2695. doi: 10.1111/1462-2920.14583
Moragues, M., Comadran, J., Waugh, R., Milne, I., Flavell, A. J., Russell, J. R. (2010). Effects of ascertainment bias and marker number on estimations of barley diversity from high-throughput SNP genotype data. Theor. Appl. Genet. 120, 1525–1534. doi: 10.1007/s00122-010-1273-1
Pasam, R. K., Sharma, R., Malosetti, M., van Eeuwijk, F. A., Haseneyer, G., Kilian, B., et al. (2012). Genome-wide association studies for agronomical traits in a world wide spring barley collection. BMC Plant Biol. 12, 16. doi: 10.1186/1471-2229-12-16
Patil, V., Bjørnstad, Å., Mackey, J. (2003). Molecular mapping of a new gene Rrs4CI11549 for resistance to barley scald (Rhynchosporium secalis). Mol. Breed. 12, 169–183. doi: 10.1023/A:1026076511073
Perfect, S. E., Green, J. R. (2001). Infection structures of biotrophic and hemibiotrophic fungal plant pathogens. Mol. Plant Pathol. 2, 101–108. doi: 10.1046/j.1364-3703.2001.00055.x
Pickering, R., Ruge-Wehling, B., Johnston, P. A., Schweizer, G., Ackermann, P., Wehling, P. (2006). The transfer of a gene conferring resistance to scald (Rhynchosporium secalis) from Hordeum bulbosum into H. vulgare chromosome 4HS. Plant Breed. 125, 576–579. doi: 10.1111/j.1439-0523.2006.01253.x
Purcell, S., Neale, B., Todd-Brown, K., Thomas, L., Ferreira, M. A. R., Bender, D., et al. (2007). PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575. doi: 10.1086/519795
Read, B. J., Oram, R. N. (1995). Register of Australian winter cereal cultivars. Hordeum vulgare (barley) cv. Brindabella. Aust. J. Exp. Agric. 35, 425. doi: 10.1071/EA9950425
Rinaldo, A., Gilbert, B., Boni, R., Krattinger, S. G., Singh, D., Park, R. F., et al. (2017). The Lr34 adult plant rust resistance gene provides seedling resistance in durum wheat without senescence. Plant Biotechnol. J. 15, 894–905. doi: 10.1111/pbi.12684
Rohe, M., Gierlich, A., Hermann, H., Hahn, M., Schmidt, B., Rosahl, S., et al. (1995). The race-specific elicitor, NIP1, from the barley pathogen, Rhynchosporium secalis, determines avirulence on host plants of the Rrs1 resistance genotype. EMBO J. 14, 4168–4177. doi: 10.1002/j.1460-2075.1995.tb00090.x
Saari, E. E., Prescott, J. M. (1975). Scale for appraising the foliar intensity of wheat diseases. Plant Dis. Rep.
Salamati, S., Tronsmo, A. M. (1997). Pathogenicity of Rhynchosporium secalis isolates from Norway on 30 cultivars of barley. Plant Pathol. 46, 416–424. doi: 10.1046/j.1365-3059.1997.d01-20.x
Schürch, S., Linde, C. C., Knogge, W., Jackson, L. F., McDonald, B. A. (2004). Molecular population genetic analysis differentiates two virulence mechanisms of the fungal avirulence gene NIP1. Mol. Plant-Microbe Interact. 17, 1114–1125. doi: 10.1094/MPMI.2004.17.10.1114
Segura, V., Vilhjálmsson, B. J., Platt, A., Korte, A., Seren, Ü., Long, Q., et al. (2012). An efficient multi-locus mixed-model approach for genome-wide association studies in structured populations. Nat. Genet. 44, 825–830. doi: 10.1038/ng.2314
Sharma, S. (2017). Prebreeding using wild species for genetic enhancement of grain legumes at ICRISAT. Crop Sci. 57, 1132–1144. doi: 10.2135/cropsci2017.01.0033
Shtaya, M. J. Y., Marcel, T. C., Sillero, J. C., Niks, R. E., Rubiales, D. (2006). Identification of QTLs for powdery mildew and scald resistance in barley. Euphytica 151, 421–429. doi: 10.1007/s10681-006-9172-x
Silvar, C., Casas, A. M., Kopahnke, D., Habekuß, A., Schweizer, G., Gracia, M. P., et al. (2010). Screening the Spanish Barley Core Collection for disease resistance. Plant Breed. 129, 45–52. doi: 10.1111/j.1439-0523.2009.01700.x
Slotta, T. A. B., Brady, L., Chao, S. (2008). High throughput tissue preparation for large-scale genotyping experiments. Mol. Ecol. Resour. 8, 83–87. doi: 10.1111/j.1471-8286.2007.01907.x
Song, W.-Y., Wang, G.-L., Chen, L.-L., Kim, H.-S., Pi, L.-Y., Holsten, T., et al. (1995). A receptor kinase-like protein encoded by the rice disease resistance gene, xa21. Science 270, 1804–1806. doi: 10.1126/science.270.5243.1804
Stefansson, T. S., Mcdonald, B. A., Willi, Y. (2013). Local adaptation and evolutionary potential along a temperature gradient in the fungal pathogen Rhynchosporium commune. Evol. Appl. 6, 524–534. doi: 10.1111/eva.12039
Stefansson, T. S., Serenius, M., Hallsson, J. H. (2012). The genetic diversity of Icelandic populations of two barley leaf pathogens, Rhynchosporium commune and Pyrenophora teres. Eur. J. Plant Pathol. 134, 167–180. doi: 10.1007/s10658-012-9974-8
Stefansson, T. S., Willi, Y., Croll, D., McDonald, B. A. (2014). An assay for quantitative virulence in Rhynchosporium commune reveals an association between effector genotype and virulence. Plant Pathol. 63, 405–414. doi: 10.1111/ppa.12111
Steiner-Lange, S., Fischer, A., Boettcher, A., Rouhara, I., Liedgens, H., Schmelzer, E., et al. (2003). Differential defense reactions in leaf tissues of barley in response to infection by Rhynchosporium secalis and to treatment with a fungal avirulence gene product. Mol. Plant-Microbe Interact. 16, 893–902. doi: 10.1094/MPMI.2003.16.10.893
Stukenbrock, E. H., McDonald, B. A. (2009). Population genetics of fungal and oomycete effectors involved in gene-for-gene interactions. Mol. Plant-Microbe Interact. 22, 371–380. doi: 10.1094/MPMI-22-4-0371
Thauvin, J. N., Russell, J., Vequaud, D., Looseley, M., Bayer, M., Le Roux, P. M., et al. (2022). Genome-wide association study for resistance to Rhynchosporium in a diverse collection of spring barley germplasm. Agronomy 12, 0–14. doi: 10.3390/agronomy12040782
Tör, M., Brown, D., Cooper, A., Woods-Tör, A., Sjölander, K., Jones, J. D. G., et al. (2004). Arabidopsis downy mildew resistance gene RPP27 encodes a receptor-like protein similar to CLAVATA2 and tomato cf-9. Plant Physiol. 135, 1100–1112. doi: 10.1104/pp.103.037770
Verma, S., Yashveer, S., Rehman, S., Gyawali, S., Kumar, Y., Chao, S., et al. (2021). Genetic and Agro-morphological diversity in global barley (Hordeum vulgare L.) collection at ICARDA. Genet. Resour. Crop Evol. 68, 1315–1330. doi: 10.1007/s10722-020-01063-7
Visioni, A., Gyawali, S., Selvakumar, R., Gangwar, O. P., Shekhawat, P. S., Bhardwaj, S. C., et al. (2018). Genome wide association mapping of seedling and adult plant resistance to barley stripe rust (Puccinia striiformis f. sp. hordei) in India. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.00520
Visioni, A., Rehman, S., Viash, S. S., Singh, S. P., Vishwakarma, R., Gyawali, S., et al. (2020). Genome wide association mapping of spot blotch resistance at seedling and adult plant stages in barley. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.00642
von Korff, M., Wang, H., Léon, J., Pillen, K. (2005). AB-QTL analysis in spring barley. I. Detection of resistance genes against powdery mildew, leaf rust and scald introgressed from wild barley. Theor. Appl. Genet. 111, 583–590. doi: 10.1007/s00122-005-2049-x
Wagner, C., Schweizer, G., Krämer, M., Dehmer-Badani, A. G., Ordon, F., Friedt, W. (2008). The complex quantitative barley-Rhynchosporium secalis interaction: Newly identified QTL may represent already known resistance genes. Theor. Appl. Genet. 118, 113–122. doi: 10.1007/s00122-008-0881-5
Wallwork, H., Grcic, M. (2011). The use of differential isolates of rhynchosporium secalis to identify resistance to leaf scald in barley. Austral. Plant Pathol. 40, 490–496. doi: 10.1007/s13313-011-0065-7
Wallwork, H., Grcic, M., Li, C. D., Hayden, M. J., Chalmers, K., Mather, D. E. (2014). Use of specific differential isolates of Rhynchosporium commune to detect minor gene resistance to leaf scald in barley seedlings. Australas. Plant Pathol. 43, 197–203. doi: 10.1007/s13313-013-0264-5
Walters, D. R., Avrova, A., Bingham, I. J., Burnett, F. J., Fountaine, J., Havis, N. D., et al. (2012). Control of foliar diseases in barley: Towards an integrated approach. Eur. J. Plant Pathol. 133, 33–73. doi: 10.1007/s10658-012-9948-x
Wang, Y., Gupta, S., Wallwork, H., Zhang, X. Q., Zhou, G., Broughton, S., et al. (2014). Combination of seedling and adult plant resistance to leaf scald for stable resistance in barley. Mol. Breed. 34, 2081–2089. doi: 10.1007/s11032-014-0164-6
Wang, Y., Subedi, S., de Vries, H., Doornenbal, P., Vels, A., Hensel, G., et al. (2019). Orthologous receptor kinases quantitatively affect the host status of barley to leaf rust fungi. Nat. Plants 5, 1129–1135. doi: 10.1038/s41477-019-0545-2
Wevelsiep, L., Rüpping, E., Knogge, W. (1993). Stimulation of barley plasmalemma H+-ATPase by phytotoxic peptides from the fungal pathogen Rhynchosporium secalis. Plant Physiol. 101, 297–301. doi: 10.1104/pp.101.1.297
Williams, K., Donnellan, S., Smyl, C., Scott, L., Wallwork, H. (2003). Molecular variation in Rhynchosporium secalis isolates obtained from hotspots. Australas. Plant Pathol. 32, 257–262. doi: 10.1071/AP03008
Xi, K., Xue, A. G., Burnett, P. A., Helm, J. H., Turkington, T. K. (2000). Quantitative resistance of barley cultivars to Rhynchosporium secalis. Can. J. Plant Pathol. 22, 217–223. doi: 10.1080/07060660009500466
Yan, J., Shah, T., Warburton, M. L., Buckler, E. S., McMullen, M. D., Crouch, J. (2009). Genetic characterization and linkage disequilibrium estimation of a global maize collection using SNP markers. PloS One 4. doi: 10.1371/journal.pone.0008451
Yun, S. J., Gyenis, L., Hayes, P. M., Matus, I., Smith, K. P., Steffenson, B. J., et al. (2005). Quantitative trait loci for multiple disease resistance in wild barley. Crop Sci. 45, 2563–2572. doi: 10.2135/cropsci2005.0236
Zadoks, J. C., Board, E. (1974). Data sheet highlights close coupled pumps. World Pumps 1999, 9. doi: 10.1016/s0262-1762(99)80614-2
Zantinge, J., Xue, S., Holtz, M., Xi, K., Juskiw, P. (2019). The identification of multiple SNP markers for scald resistance in spring barley through restriction-site associated sequencing. Euphytica 215, 1–13. doi: 10.1007/s10681-018-2317-x
Zhan, J., Fitt, B. D. L., Pinnschmidt, H. O., Oxley, S. J. P., Newton, A. C. (2008). Resistance, epidemiology and sustainable management of Rhynchosporium secalis populations on barley. Plant Pathol. 57, 1–14. doi: 10.1111/j.1365-3059.2007.01691.x
Zhang, Z., Ersoz, E., Lai, C.-Q., Todhunter, R. J., Tiwari, H. K., Gore, M. A., et al. (2010). Mixed linear model approach adapted for genome-wide association studies. Nat. Genet. 42, 355–360. doi: 10.1038/ng.546
Keywords: barley, Rhynchosporium commune, resistance, SNP, GWAS, quantitative trait loci
Citation: Hiddar H, Rehman S, Belkadi B, Filali-Maltouf A, Al-Jaboobi M, Verma RPS, Gyawali S, Kehel Z and Amri A (2023) Identification of sources of resistance to scald (Rhynchosporium commune) and of related genomic regions using genome-wide association in a mapping panel of spring barley. Front. Plant Sci. 14:1133404. doi: 10.3389/fpls.2023.1133404
Received: 28 December 2022; Accepted: 30 October 2023;
Published: 28 November 2023.
Edited by:
Dilip R. Panthee, North Carolina State University, United StatesReviewed by:
Ahmad H. Sallam, University of Minnesota Twin Cities, United StatesRudolph Fredua-Agyeman, University of Alberta, Canada
Copyright © 2023 Hiddar, Rehman, Belkadi, Filali-Maltouf, Al-Jaboobi, Verma, Gyawali, Kehel and Amri. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ahmed Amri, YS5hbXJpQGNnaWFyLm9yZw==
 Houda Hiddar
Houda Hiddar Sajid Rehman2,3
Sajid Rehman2,3 Bouchra Belkadi
Bouchra Belkadi Abdelkarim Filali-Maltouf
Abdelkarim Filali-Maltouf Muamar Al-Jaboobi
Muamar Al-Jaboobi Ramesh Pal Singh Verma
Ramesh Pal Singh Verma Sanjaya Gyawali
Sanjaya Gyawali Zakaria Kehel
Zakaria Kehel Ahmed Amri
Ahmed Amri