
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 22 March 2023
Sec. Plant Breeding
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1133024
This article is part of the Research Topic Generating Useful Genetic Variation in Crops by Induced Mutation, Volume III View all 6 articles
 Qingguo Wang1,2†
Qingguo Wang1,2† Hongchun Xiong2†
Hongchun Xiong2† Huijun Guo2
Huijun Guo2 Linshu Zhao2
Linshu Zhao2 Yongdun Xie2
Yongdun Xie2 Jiayu Gu2
Jiayu Gu2 Shirong Zhao2
Shirong Zhao2 Yuping Ding2
Yuping Ding2 Luxiang Liu2*
Luxiang Liu2*Plant height is one of the most important agronomic traits that affects yield in wheat, owing to that the utilization of dwarf or semi-dwarf genes is closely associated with lodging resistance. In this study, we identified a semi-dwarf mutant, jg0030, induced by γ-ray mutagenesis of the wheat variety ‘Jing411’ (wild type). Compared with the ‘Jing411’, plant height of the jg0030 mutant was reduced by 7%-18% in two years’ field experiments, and the plants showed no changes in yield-related traits. Treatment with gibberellic acid (GA) suggested that jg0030 is a GA-sensitive mutant. Analysis of the frequency distribution of plant height in 297 F3 families derived from crossing jg0030 with the ‘Jing411’ indicated that the semi-dwarf phenotype is controlled by a major gene. Using the wheat 660K SNP array-based Bulked Segregant Analysis (BSA) and the exome capture sequencing-BSA assay, the dwarf gene was mapped on the long arm of chromosome 2B. We developed a set of KASP markers and mapped the dwarf gene to a region between marker PH1 and PH7. This region encompassed a genetic distance of 55.21 cM, corresponding to a physical distance of 98.3 Mb. The results of our study provide a new genetic resource and linked markers for wheat improvement in molecular breeding programs.
Bread wheat (Triticum aestivum L.) is one of the most important food crops in the world. It is urgent to increase grain yield to meet the demands of the expanding human population and to mitigate the effects of climate change. Plant height significantly correlates with lodging resistance and therefore plays an important role in yield stability in wheat (Fan et al., 2012). Identification of novel QTLs or genes for plant height is of great significance for improving grain yield in wheat.
In the 1960s, the dwarfing genes Rht1(Rht-B1b) and Rht2(Rht-D1b) were introduced into wheat varieties in breeding, which significantly improved lodging resistance and harvest index, resulting in the well-known ‘Green Revolution’ (Peng et al., 1999; Würschum et al., 2017). Single-base mutations in the nucleotide sequences coding for the DELLA domains in Rht1 and Rht2 produced truncated proteins which resulted in the dwarf phenotypes. These genes do not respond to GA treatment and are thus categorized as GA-insensitive dwarfing genes (Hedden, 2003). Generally, the Rht1/Rht2 genes and their allelic variants (Rht3, Rht10, Rht11, Rht17) are classified as GA-insensitive. The green revolution genes Rht1 and Rht2 derived from the wheat cultivar ‘Norin 10’ are located on the short arms of chromosomes 4B and 4D, respectively. The effect of Rht1 was to reduce plant height by approximately 20%, and that of Rht2 was to reduce height ~24% (Miralles and Slafer, 1995; Sial et al., 2002). Rht1 and Rht2 also affect heading date, spikelet development, tiller number, and the number of kernels per spike (Würschum et al., 2015; Mo et al., 2018). Rht3 has a stronger effect on height reduction than either Rht1 or Rht2 due to a 2-kb retrotransposon insertion in the coding region of the DELLA protein (Flintham and Gale, 1983; Wen et al., 2013). The presence of two copies of the Rht2 tandem segmental duplication in Rht10 causes an extremely dwarf phenotype (Li Y., et al., 2012). Rht11 and Rht17 are allelic variations of Rht1, which also have premature stop codons present in the DELLA domain (Divashuk et al., 2012; Li A., et al., 2012; Bazhenov et al., 2015).
Along with the refinement of wheat genome sequences (Appels et al., 2018), several GA-sensitive dwarfing genes have been cloned in recent years including Rht8, Rht12, Rht13, Rht18, Rht23, and Rht24. Rht8, which is located on the short arm of chromosome 2D, has a frameshift mutation in an RNase H-like protein and regulates plant height through modification of the bioactive GA content (Chai et al., 2022; Xiong et al., 2022). Mutation analysis suggests that the gene encoding GA2oxidaseA13 on chromosome 5A contributes to dwarf phenotype of Rht12 plants (Buss et al., 2020). Rht13 is located on the long arm of chromosome 7B (Ellis et al., 2005), and a recent study found that the dwarf phenotype of Rht13 results from single nucleotide variation in a NB-LRR protein (Borrill et al., 2022). Rht18 is located on chromosome 6A, and the increased expression of GA2oxA9 in Rht18 results in decreased plant height (Ford et al., 2018). Rht23 is located on the long arm of chromosome 5D, and it has been reported that 5Dq is a candidate gene for Rht23 (Chen et al., 2015; Zhao et al., 2018). Rht24 is located on the long arm of chromosome 6A, and encodes a gibberellin (GA) 2-oxidase; it reduces plant height without affecting yield and also increases photosynthetic rate and nitrogen use efficiency (Tian et al., 2022).
It is important to note that a number of dwarf genes have only been located to the chromosomal regions and have not yet been cloned. Rht4 is located on the long arm of chromosome 2B; a 36 Mb deletion derived from the dwarf line DD399 was identified in this region (Wu et al., 2021). Rht5 (Ellis et al., 2005), Rht6 (Mohan et al., 2021), and Rht22 (Peng et al., 2011) are located on chromosomes 3BS, 4D, and 7AS, respectively. Rht7 and Rht21 are located on 2AS (Lu et al., 2015), while Rht9 and Rht12 are located on 5AL (Sutka and Kovács, 1987; Ellis et al., 2005; Sun et al., 2019); Rht14, Rht16 and Rht25 are all located on 6AS (Mo et al., 2018; Vikhe et al., 2019; Gong et al., 2021).
Although more than 20 dwarf genes have been documented in wheat, only a few genes are presently utilized in wheat breeding. More loci or genes that regulate plant height without influencing yield components need to be investigated. In this study, we identified a semi-dwarf mutant named jg0030 that shows no yield penalty. BSA and genetic mapping in an F2 population derived from crossing jg0030 with the ‘Jing411’ showed that a QTL located on the long arm of chromosome 2B was responsible for height reduction in the mutant. The identification of the semi-dwarf mutant and the QTL for height regulation provide important resources for the development of wheat varieties with a high degree of yield stability.
The semi-dwarf mutant jg0030 was induced by γ-ray mutagenesis and the wheat variety ‘Jing411’ was the wildtype. The field experiment was carried out at the Beijing Zhongpuchang and Changping Experimental Stations of the Institute of Crop Sciences, Chinese Academy of Agricultural Sciences. We measured 1,000-grain weight, number of kernels per spike, and effective spike number in the ‘Jing411’ and mutant jg0030 in plants collected from the plots in 2015 and 2017. We also measured plant height in F1 plants produced by crossing jg0030 with the conventional cultivar ‘Nongda5181’ and also with the ‘Jing411’. Plant heights of ‘Jing411’, ‘Nongda5181’, and jg0030 were measured at maturity in 2017 and 2018. Four segregating F2 populations were produced by self-pollinating the Jing411/jg0030, jg0030/Jing411, Nongda5181/jg0030, and jg0030/Nongda5181 F1 hybrids. The F3 families were derived from crossing Jing 411 with jg0030, and the plant heights were measured at maturity.
The seeds of the two parents were selected and soaked in 1% H2O2 solution for 12 hours to break dormancy. Fifteen seeds were placed on the germination rack in a light incubator with a 12 h light/dark photoperiod at a temperature of 21°C. After two days of culture with water, 2 mg L-1 GA3 was added to the experimental group. After 8 days of growth, five seedlings with consistent growth were taken from three replicates to measure seedling length.
Genomic DNA was extracted from ~ 0.1 g of leaf tissue from individual plants. The quality of DNA was assessed via electrophoresis on a 1% agarose gel stained with GelRed to observe the integrity of the DNA. The DNA concentration was measured using a NanoDrop One spectrophotometer (Thermo Scientific). The DNA samples were diluted to ~200 ng/uL and stored at -80°C.
For wheat 660K SNP array-based BSA, 40 extremely tall plants and 40 extremely dwarf plants were selected from the ‘Nongda5181’ × jg0030 F2 segregating population to construct extreme bulks. In addition, 10 individual plants of ‘Nongda5181’ and jg0030 were used to construct the parental pool, and the mixing pools were constructed by combining equal amounts of DNA from each plant. The Axiom® Wheat 660K SNP Array (Thermo) was used to genotype the two pools and the parental DNA samples. Genotyping was performed by China Gold Marker (Beijing) Biotechnology Co., Ltd. The resulting microarray data were screened and analyzed (Dish QC>0.82; call Rate >94). Finally, mapping of the dwarf gene was performed using the high-quality SNP data obtained from the 660K SNP array.
The F3 families derived from the cross between the ‘Jing411’ and the jg0030 mutant were sampled. We selected four plants from one tall or dwarf F3 line, and a total of 20 F3 lines were used for construction of extremely tall or dwarf bulks; two extremely tall or dwarf bulks, and two parental pools containing 10 plants were constructed to exome capture sequencing. Exome capture sequencing mainly includes the following steps; first, the DNA libraries for the extreme bulks and the parental lines were constructed, and the exon probe solution was hybridized with single-stranded DNA. The WheatPanExomeV2 (Tcuni, Chengdu, China) was used as exon probes, which includes 2,574,323 probes covering 137 Mb CDS of Chinese Spring RefSeq V2.1, and also containing specific CDS based on genome sequences from other modern wheat cultivars such as KN9204, AK58 and YZ4110. The uncaptured DNA fragments were then washed away, and the enriched exons were amplified by PCR. After quality control, high-throughput DNA sequencing was performed using the Illumina platform. Filtering the SNPs obtained by mutation detection was performed using the SNP-index algorithm to calculate the genotype frequency of the extreme bulks, and the sites with significant differences between the extreme bulks were statistically screened. The Euclidean Distance (ED) values between the two extreme bulks were calculated by using the depth of each allele, and the candidate region was determined according to the fitting result of the ED.
According to the exome capture sequencing data, the loci with homozygous differences between the two parents and higher sequencing depth (5<DP<300) were screened. The flanking sequences 100 bp in length on both sides of the loci were downloaded from the wheat genome website IWGSC (http://www.wheatgenome.org/) and the relevant data were submitted to the online primer design website Polymarker (http://polymarker.tgac.ac.uk/) to design genome-specific primers. First, genotype analysis was performed in the two parents to verify the specificity of the markers. These specific markers were then used to identify the genotypes of plants in the F2 population. The phenotypic and genotypic data from the F2 plants was combined, the linkage relationships between the marker loci and the target gene were analyzed, and the approximate position of the target gene on the chromosome was preliminarily determined.
QTL mapping of the semi-dwarf gene was performed using 288 F2 individuals obtained from a cross between the ‘Jing411’ and the jg0030 mutant. The genotypes of F2 population were obtained by KASP assay and the genetic map was constructed by QTL IciMapping4.0. The genetic distance between the linked marker loci was estimated using the Kosambi function based on the recombination frequency.
From our γ-ray-induced mutant library, we identified a semi-dwarf wheat mutant, jg0030, that displayed no negative effects with respect to yield components. Based on the results of field experiments conducted in 2017 and 2018, the average plant height in the ‘Jing411’ was 94 cm (2017) and 89 cm (2018), while plant height in jg0030 was 87 cm and 73 cm, respectively. Compared with the ‘Jing411’, plant height in the jg0030 was reduced by 7%-18% (Figures 1A–C). For the yield-related traits, there were no significant differences in 1,000-grain weight (Figure 1D, Figures S1A, D), number of kernels per spike (Figure 1E, Figures S1B, E), and effective spike number (Figure 1F, Figures S1C, F) between the ‘Jing411’ and jg0030, suggesting that jg0030 reduced plant height without imposing a yield penalty.
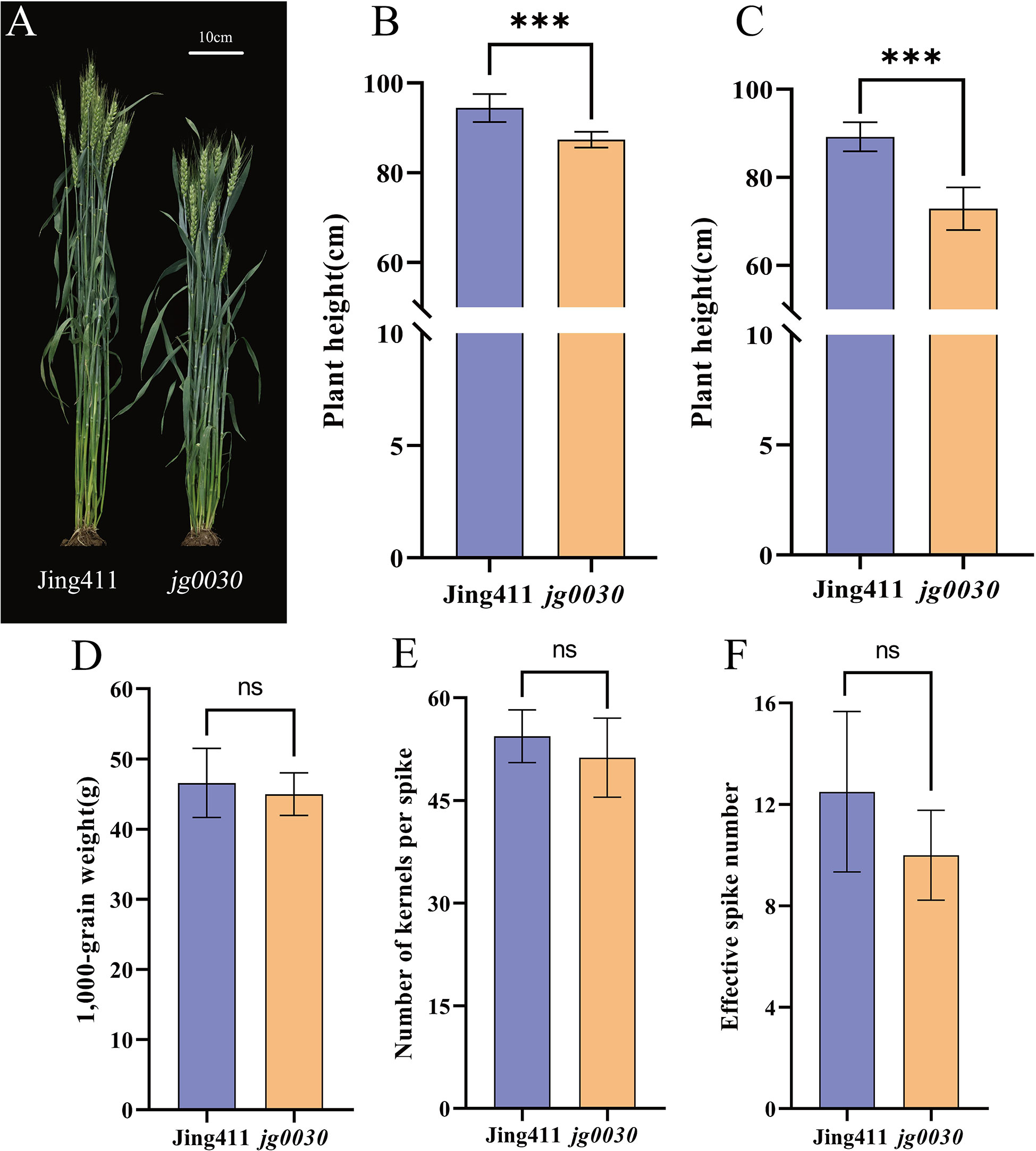
Figure 1 Comparisons of whole-plant phenotypes between the wild type ‘Jing411’ and the dwarf mutant jg0030. (A) Mature plants of ‘Jing411’ and jg0030. Scale bar=10 cm. B-G. statistical analyses of (B) mean plant height of ‘Jing411’ and jg0030 in 2017. (C) mean plant height of ‘Jing411’ and jg0030 in 2018. (D) 1000-grain weight in 2017. (E) number of kernels per spike in 2017. (F) effective spike number in 2017. Bars show the mean SD (n=8). ns p>0.05; *** p<0.001.
In order to clarify the GA sensitivity of the dwarf gene in jg0030, exogenous GA treatment was performed on hydroponically grown ‘Jing411’ and jg0030 seedlings (Figure 2A). The results showed that seedling lengths in the ‘Jing411’ and jg0030 increased by 6.2% and 17.2%, respectively, after exogenous GA treatment (Figure 2B), indicating that jg0030 is a GA-sensitive semi-dwarf mutant.
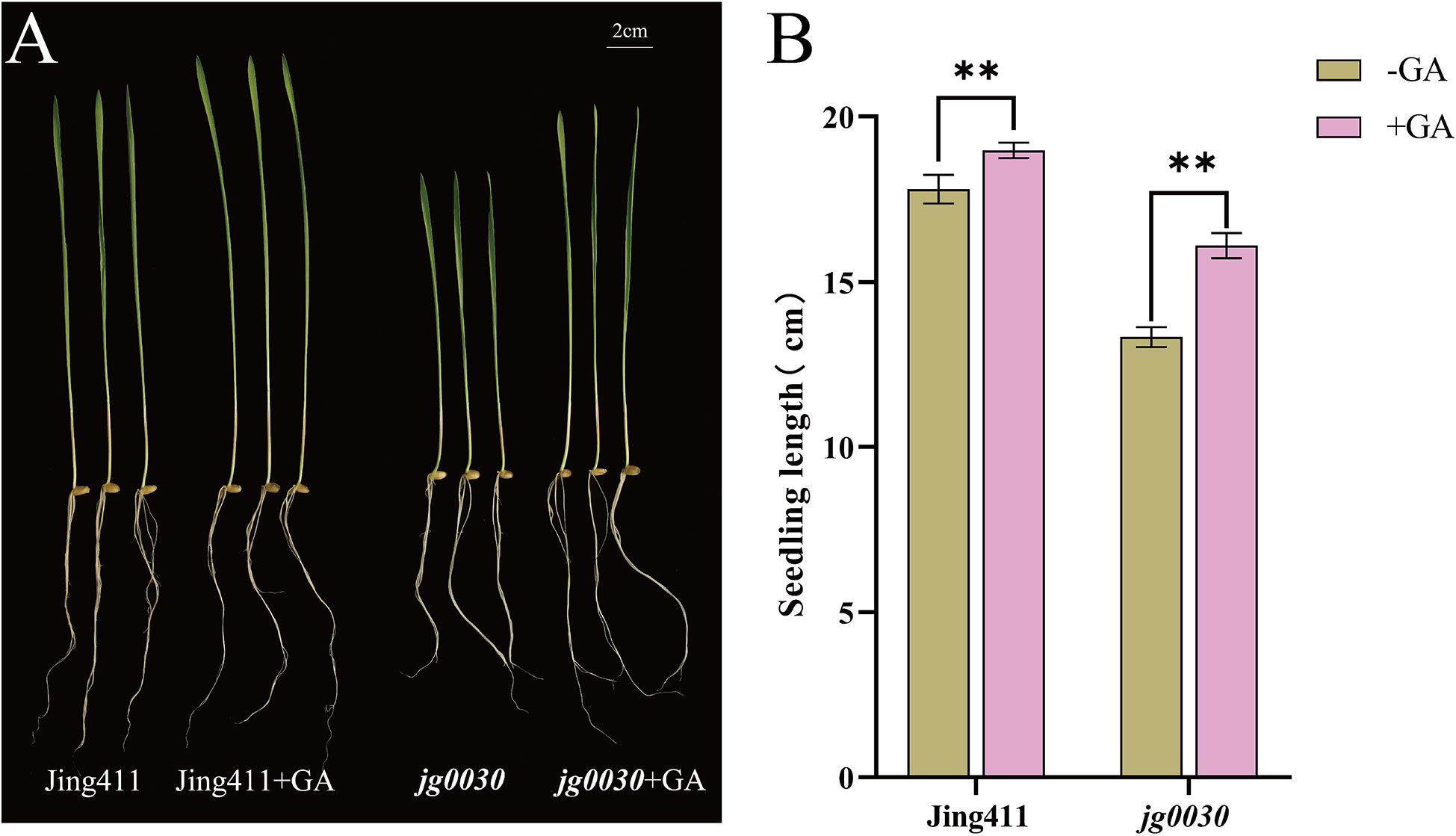
Figure 2 GA sensitivity between the wild type ‘Jing411’ and the dwarf mutant jg0030. (A) Phenotype of wild type ‘Jing411’ and jg0030 before and after GA treatment. (B) seedling length of ‘Jing411’ and jg0030 in response to GA treatment. Bars show the mean SD (n=8). **p< 0.01.
To analyze the genetic characteristics of the semi-dwarf gene in jg0030, we reciprocally crossed the mutant with the ‘Jing411’ and also the relatively tall variety ‘Nongda5181’. The F1 plants from all four reciprocal crosses were significantly taller than the jg0030 plants, and were similar in height to the tall parental lines (Figure 3A). These results suggested that the semi-dwarf gene is genetically recessive. We then analyzed the frequency distribution of plant height in 297 F3 families derived from crossing Jing411 with jg0030, and observed a bimodal distribution (Figure 3B), suggesting that the variation of plant height in this population is controlled by a major gene.
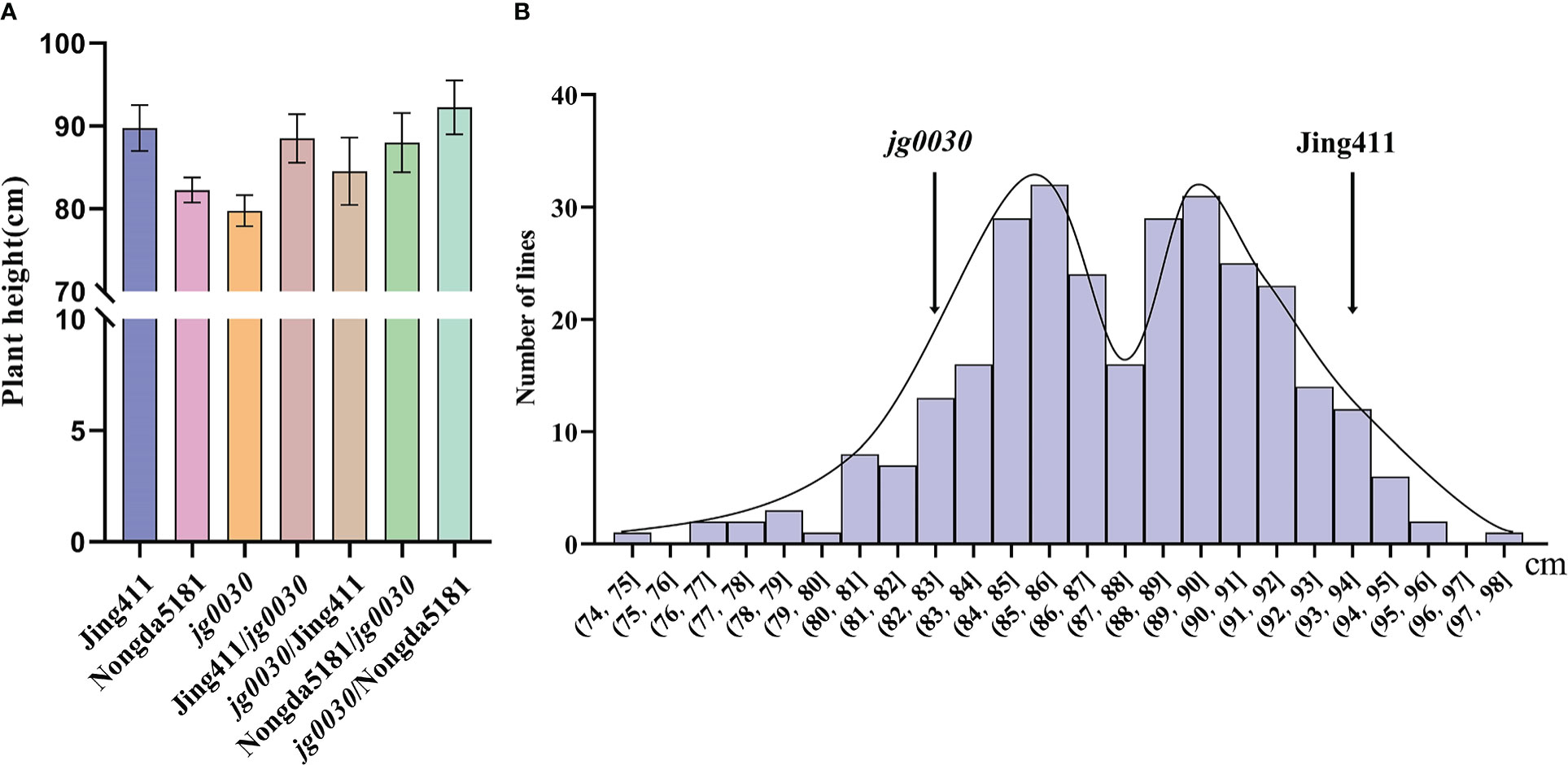
Figure 3 Plant height in F1 plants and four F2 populations. (A) height comparison of the parental lines and F1 plants at maturity. Jing411/jg0030 indicates the F1 hybrid in which ‘Jing411’ was the female parent and jg0030 was the male parent; jg0030/Jing411 indicates the F1 hybrid in which jg0030 was the female parent and ‘Jing411’ was the male parent; ‘Nongda5181’/jg0030 indicates the F1 hybrid with ‘Nongda5181’ as the female parent and jg0030 as the male parent; jg0030/Nongda5181 indicates the F1 hybrid with jg0030 as the female parent and ‘Nongda5181’ as the male parent. (B) Frequency distributions of plant heights in the F3 families derived from crossing Jing411 with jg0030.
To investigate the locus/gene that controls the semi-dwarf phenotype in the mutant, we constructed two F2 populations from reciprocal crosses of jg0030 and Nongda5181 consisting of 289 individuals (Nongda5181/jg0030), and 304 individuals (jg0030/Nongda5181), and selected extremely tall and dwarf F2 plants to construct the tall and dwarf bulks for genotyping using the 660K SNP array for BSA. We identified 79,593 SNPs between the two parental lines ‘Nongda5181’ and jg0030. After filtration of the SNPs based on the genotypes and phenotypes from the two bulks and the two parental lines, we found that the largest number of SNPs associated with plant height were enriched on chromosome 2B in the two populations, indicating that the gene influencing plant height was located on chromosome 2B (Figures 4A, B). We then analyzed the distribution of the associated SNPs on chromosome 2B and identified a large peak between positions 600 and 800 Mb (Figure 4C), suggesting that the candidate gene is located on this region.
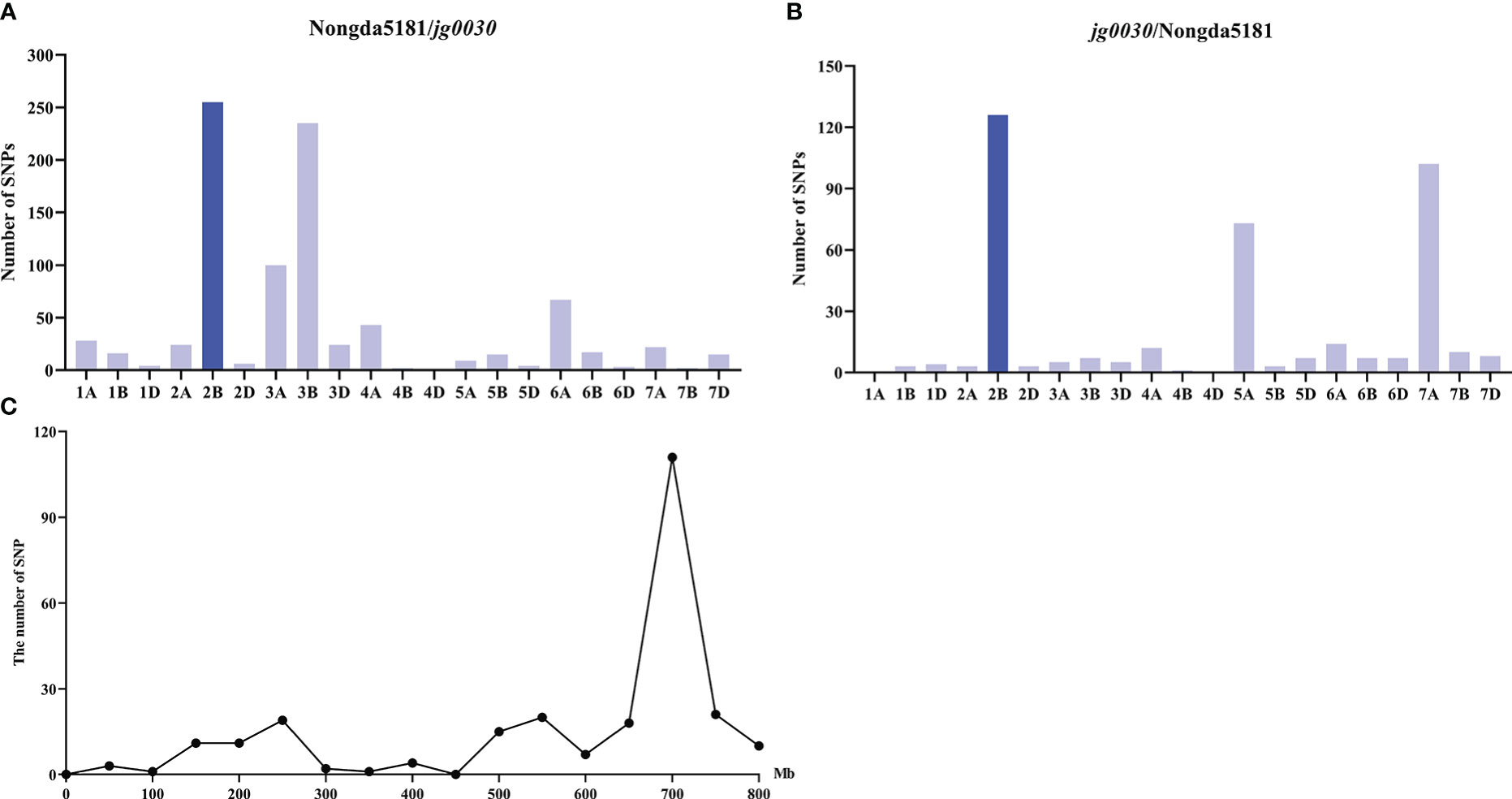
Figure 4 The distribution of SNPs across the 21 T. aestivum chromosomes identified by 660K SNP array-based BSA. (A) the Nongda5181/jg0030 F2 population. (B) the jg0030/Nongda5181 F2 population. (C) The distribution of SNPs on chromosome 2B.
We next selected homozygous dwarf and tall plants in the F3 lines derived from the Jing411/jg0030 populations to construct dwarf and tall bulks and performed exome capture sequencing. According to the genotypes and read depths of the obtained SNPs from the two bulks and the two parental lines, Euclidean Distance (ED) association analysis was performed to determine the linkage strength between each SNP locus and the dwarf gene. Consistent with the results of the 660K SNP array-based BSA, a clear peak was identified at the end of the long arm of chromosome 2B by analysis of the exome capture sequencing data. This result also showed that the gene controlling plant height in the jg0030 mutant is located on the long arm of chromosome 2B (Figure 5).
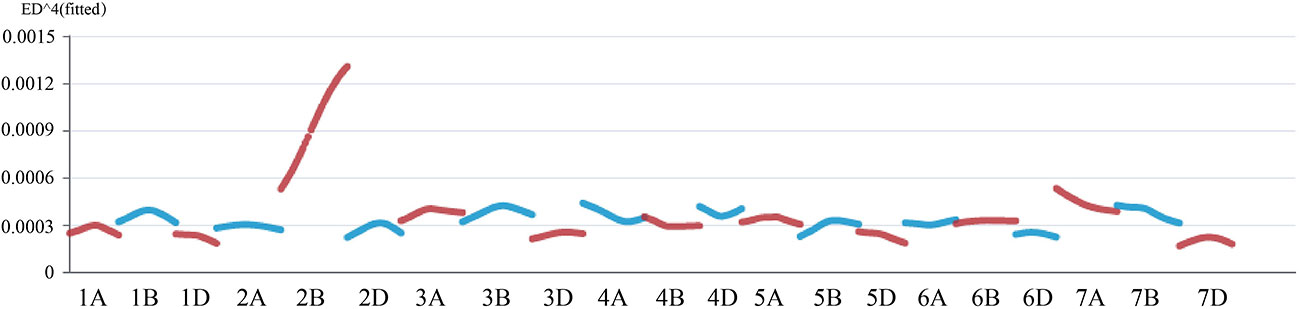
Figure 5 Euclidean Distance correlation analysis by the exome capture sequencing-BSA assay in the Jing411/jg0030 F3 population.
Based on the homozygous SNPs between the ‘Jing411’ and the jg0030 mutant by exome capture sequencing, we designed 35 KASP markers on the long arm of chromosome 2B. After validation in the two parental lines, a total of seven markers were successfully developed and used to genotype the jg0030/Jing411 F2 population (Figures S2A–G). Based on the genotypes and phenotypes of plants from the jg0030/Jing411 F2 population, a QTL with a LOD score of 2.5 was detected in a 55.21 cM chromosomal region between marker PH1 and PH7, corresponding to a physical interval of 98.3 Mb (Figure 6).
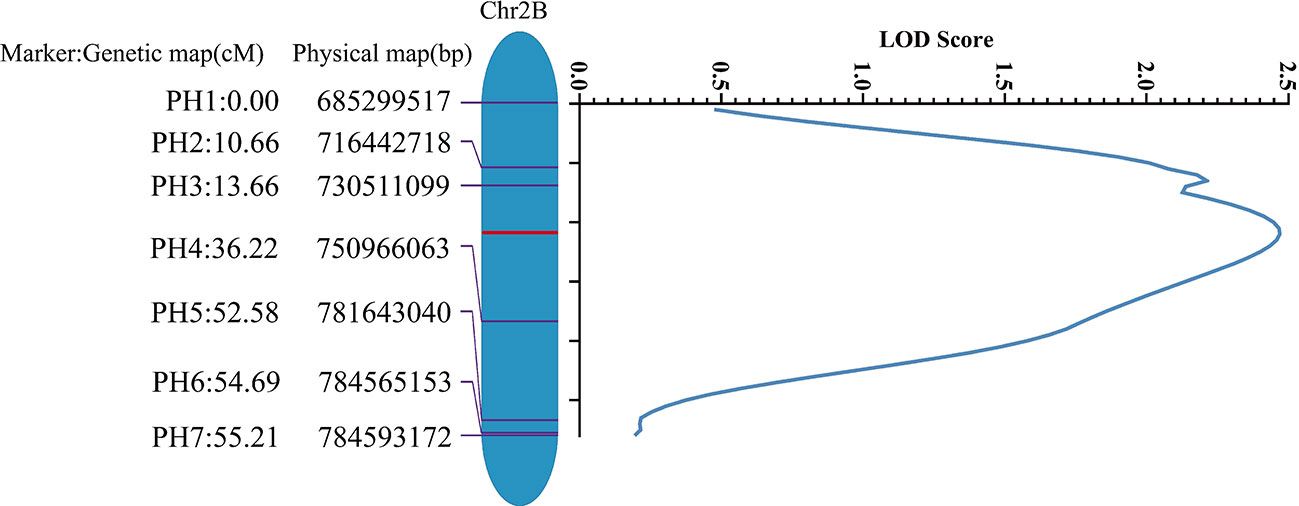
Figure 6 Localization of the detected QTL for dwarf plant stature on the long arm of chromosome 2B in the Jing411/jg0030 F2 population by genetic mapping analysis.
Plant height is one of the important agronomic traits that is closely associated with lodging resistance and is thus important for yield stability in wheat. The utilization of dwarf or semi-dwarf genes is the key component for development of new high-yielding wheat varieties with lodging resistance. At present, the dwarf genes utilized in wheat breeding are mainly Rht1, Rht2, Rht8, Rht9, and Rht24 (Zhong et al., 2018). In addition to reduced plant height, Rht1 and Rht2 can increase grain number per spike to improve grain yield (Peng et al., 1999), while Rht8 (Chai et al., 2022; Xiong et al., 2022) and Rht24 (Tian et al., 2017) have no negative effects on 1,000-grain weight. In this study, we identified a semi-dwarf wheat mutant, jg0030, with high yield potential. Compared with the ‘Jing411’, the height of jg0030 plants was reduced by 7-18% in two years’ field experiments, and there was no significant change in 1,000-grain weight, grain number per spike, and effective spike number in jg0030, demonstrating that the jg0030 mutation reduces plant height without affecting yield components. Therefore, the dwarf gene in jg0030 can be used to reduce plant height and thus improve lodging resistance in wheat breeding.
Results of a previous study indicated that the Rht4 gene is located on the long arm of chromosome 2B and is linked to the SSR marker locus Xwmc317 (Ellis et al., 2005). Using a dwarf mutant, DD399, a locus for reducing plant height was delimited to a deleted region of 36 Mb (763-799Mb) at the end of the long arm of chromosome 2B, and this locus is thought to be the same as Rht4 (Wu et al., 2021). In this study, the dwarf gene in the mutant jg0030 was mapped to the terminus of 2BL using 660K SNP array-based BSA and exome capture sequencing-BSA. By developing seven KASP markers on 2BL, the dwarf gene was mapped between marker PH1 and PH7 that ranged from 685.3 to 784.6 Mb of the physical position on IWGSC RefSeq v2.0, which overlaps with the mapped region of Rht4, suggesting that the gene causing the semi-dwarf phenotype in jg0030 is likely to be the Rht4 gene. Previous studies have suggested that the original Rht4 allele derived from the dwarf mutant Burt ert 937 significantly decreased 1,000-grain weight (Du et al., 2018; Lu et al., 2022). In contrast, the jg0030 mutation showed no significant change in 1,000-grain weight, indicating that this mutation induced by γ-ray irradiation is different from that of Rht4 allele.
It is well documented that Rht1/Rht2 and its homologous genes or allelic variations are insensitive to GA, while the other dwarf genes are sensitive to GA treatment (Peng et al., 1999). GA affects plant growth by promoting cell elongation and cell division (Hedden, 2020). For the GA-sensitive dwarf mutants, degradation of the growth-inhibiting DELLA protein is induced and therefore normal plant height is restored after exogenous application of GA (Hedden and Sponsel, 2015). Several studies have indicated that the GA sensitivity of dwarf genes affects plant growth at the seedling stage, and these genes that differ with respect to GA sensitivity showed distinct effects on coleoptile length in wheat (Tang et al., 2009). Long coleoptiles are beneficial for absorbing enough water from the soil to ensure the early growth of wheat seedlings in the field, allowing them to adapt well to stress (Condon, 2004). A previous study showed that Rht4 gene is a GA-sensitive dwarf gene, and the spike number, yield per plant, and 1,000-grain weight increased significantly after exogenous GA3 treatment of Rht4 dwarf plants (Lu et al., 2022). Consistently, the length of jg0030 seedlings increased significantly after exogenous GA treatment, suggesting that jg0030 is sensitive to GA. Taken together, the results of our study show that the GA-sensitive dwarf gene present in jg0030 represents a new genetic resource for the development of high-yielding semi-dwarf wheat cultivars with good resistance to lodging.
In conclusion, our study identified an elite semi-dwarf wheat mutant jg0030 without yield penalty induced by γ-ray irradiation of the cultivar ‘Jing411’. Exogenous GA treatment suggested that jg0030 was a GA-sensitive semi-dwarf mutant. We mapped the dwarf gene to the terminal region of 2BL using bulked segregant analysis. Further genetic mapping by development of KASP markers in an F2 population derived from crossing the ‘Jing411’ with the jg0030 mutant indicated that the dwarf gene was located on a 55.21 cM interval between marker PH1 and PH7 on 2BL. The results of our study provide important resources and linked molecular markers for breeding wheat varieties with high lodging resistance and good yield stability.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: NCBI BioProject (https://www.ncbi.nlm.nih.gov/bioproject/), PRJNA916817.
LL conceived the project and revised the manuscript. HX designed the experiment, QW performed most of the experiments and data analysis. HG developed the mutant. YX, LZ, JG, SZ, and YD participated in field trials. All authors contributed to the article and approved the submitted version.
This work was financially supported by the National Key Research and Development Program (Grant No. 2022YFD1200700), the Crop Varietal Improvement and Insect Pests Control by Nuclear Radiation, the China Agriculture Research System of MOF and MARA (Grant No. CARS-03), the Agricultural Science and Technology Innovation Program (Grant No. CAAS-ZDRW202109), and the Central Public-interest Scientific Institution Basal Research Fund (No. Y2022GH06).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1133024/full#supplementary-material
Appels, K., E., C., F., Keller, B. (2018). Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 361. doi: 10.1126/science.aar7191
Bazhenov, M. S., Divashuk, M. G., Amagai, Y., Watanabe, N., Karlov, G. I. (2015). Isolation of the dwarfing Rht-B1p (Rht17) gene from wheat and the development of an allele-specific PCR marker. Mol. Breed. 35. doi: 10.1007/s11032-015-0407-1
Borrill, P., Mago, R., Xu, T., Ford, B., Williams, S. J., Derkx, A., et al. (2022). An autoactive NB-LRR gene causes Rht13 dwarfism in wheat. Proc. Natl. Acad. Sci. U. S. A. 119, e2085092177. doi: 10.1073/pnas.2209875119
Buss, W., Ford, B. A., Foo, E., Schnippenkoetter, W., Borrill, P., Brooks, B., et al. (2020). Overgrowth mutants determine the causal role of gibberellin GA2oxidaseA13 in Rht12 dwarfism of wheat. J. Exp. Bot. 71, 7171–7178. doi: 10.1093/jxb/eraa443
Chai, L. L., Xin, M. M., Dong, C. Q., Chen, Z. Y., Zhai, H. J., Zhuang, J. H., et al. (2022). A natural variation in ribonuclease h-like gene underlies Rht8 to confer "green revolution"trait in wheat. Mol. Plant 15, 377–380. doi: 10.1016/j.molp.2022.01.013
Chen, S., Gao, R., Wang, H., Wen, M., Xiao, J., Bian, N., et al. (2015). Characterization of a novel reduced height gene (Rht23) regulating panicle morphology and plant architecture in bread wheat. Euphytica 203, 583–594. doi: 10.1007/s10681-014-1275-1
Condon, A. G. (2004). Breeding for high water-use efficiency. J. Exp. Bot. 55, 2447–2460. doi: 10.1093/jxb/erh277
Divashuk, M. G., Vasil'Ev, A. V., Bespalova, L. A., Karlov, G. I. (2012). Identity of the Rht-11 and Rht-B1e reduced plant height genes. Genetika 48, 897–900. doi: 10.1134/S1022795412050055
Du, Y., Chen, L., Wang, Y., Yang, Z., Saeed, I., Daoura, B. G., et al. (2018). The combination of dwarfing genes Rht4 and Rht8 reduced plant height, improved yield traits of rainfed bread wheat (Triticum aestivum l.). Field Crops Res. 215, 149–155. doi: 10.1016/j.fcr.2017.10.015
Ellis, M. H., Rebetzke, G. J., Azanza, F., Richards, R. A., Spielmeyer, W. (2005). Molecular mapping of gibberellin-responsive dwarfing genes in bread wheat. Theor. Appl. Genet. 111, 423–430. doi: 10.1007/s00122-005-2008-6
Fan, M. S., Shen, J. B., Yuan, L. X., Jiang, R. F., Chen, X. P., Davies, W. J., et al. (2012). Improving crop productivity and resource use efficiency to ensure food security and environmental quality in china. J. Exp. Bot. 63, 13–24. doi: 10.1093/jxb/err248
Flintham, J. E., Gale, M. D. (1983). The tom thumb dwarfing gene Rht3 in wheat : 2. effects on height, yield and grain quality. Theor. Appl. Genet. 66, 249–256. doi: 10.1007/BF00251155
Ford, B. A., Foo, E., Sharwood, R., Karafiatova, M., Vrana, J., MacMillan, C., et al. (2018). Rht18 semidwarfism in wheat is due to increased GA 2-oxidaseA9 expression and reduced GA content. Plant Physiol. 177, 168–180. doi: 10.1104/pp.18.00023
Gong, Y., Shuhong, W., Zhengsong, P., Zaijun, Y., Mingzhi, Z., Jiamin, Z., et al. (2021). Genetic study on plant height and its components,partial yield traits in durum wheat’ANW16F’. Southwest China J. Agric. Sci. 34, 229–235. doi: 10.16213/j.cnki.scjas.2021.2.001
Hedden, P. (2003). The genes of the green revolution. Trends Genet. 19, 5–9. doi: 10.1016/S0168-9525(02)00009-4
Hedden, P. (2020). The current status of research on gibberellin biosynthesis. Plant Cell Physiol. 61, 1832–1849. doi: 10.1093/pcp/pcaa092
Hedden, P., Sponsel, V. (2015). A century of gibberellin research. J. Plant Growth Regul. 34, 740–760. doi: 10.1007/s00344-015-9546-1
Li, Y. Y., Xiao, J. H., Wu, J. J., Duan, J. L., Liu, Y., Ye, X., et al. (2012). A tandem segmental duplication (TSD) in green revolution gene Rht-D1b region underlies plant height variation. New Phytol. 196, 282–291. doi: 10.1111/j.1469-8137.2012.04243.x
Li, A. X., Yang, W. L., Guo, X. L., Liu, D. C., Sun, J. Z., Zhang, A. M., et al. (2012). Isolation of a gibberellin-insensitive dwarfing gene, Rht-B1e, and development of an allele-specific PCR marker. Mol. Breed. 30, 1443–1451. doi: 10.1007/s11032-012-9730-y
Lu, Q., Lu, S., Wang, M., Cui, C., Condon, A. G., Jatayev, S., et al. (2022). The exogenous GA3 greatly affected the grain-filling process of semi-dwarf gene Rht4 in bread wheat. Physiologia Plantarum 174. doi: 10.1111/ppl.13725
Lu, Y., Xing, L., Xing, S., Hu, P., Cui, C., Zhang, R., et al. (2015). Characterization of a putative new semi-dominant reduced height gene, Rht_NM9, in wheat (Triticum aestivum l.). J. Genet. Genomics 42, 685–698. doi: 10.1016/j.jgg.2015.08.007
Miralles, D. J., Slafer, G. A. (1995). Yield, biomass and yield components in dwarf, semi-dwarf and tall isogenic lines of spring wheat under recommended and late sowing dates. Plant Breed. 114, 392–396. doi: 10.1111/j.1439-0523.1995.tb00818.x
Mo, Y., Vanzetti, L. S., Hale, I., Spagnolo, E. J., Guidobaldi, F., Al-Oboudi, J., et al. (2018). Identification and characterization of Rht25, a locus on chromosome arm 6AS affecting wheat plant height, heading time, and spike development. Theor. Appl. Genet. 131, 2021–2035. doi: 10.1007/s00122-018-3130-6
Mohan, A., Grant, N. P., Schillinger, W. F., Gill, K. S. (2021). Characterizing reduced height wheat mutants for traits affecting abiotic stress and photosynthesis during seedling growth. Physiologia Plantarum 172, 233–246. doi: 10.1111/ppl.13321
Peng, Z. S., Li, X., Yang, Z. J., Liao, M. L. (2011). A new reduced height gene found in the tetraploid semi-dwarf wheat landrace aiganfanmai. Genet. Mol. Res. 10, 2349–2357. doi: 10.4238/2011.October.5.5
Peng, J. R., Richards, D. E., Hartley, N. M., Murphy, G. P., Devos, K. M., Flintham, J. E., et al. (1999). 'Green revolution' genes encode mutant gibberellin response modulators. Nature 400, 256–261. doi: 10.1038/22307
Sial, M. A., Arain, M. A., Javed, M. A., Jamali, K. D. (2002). Genetic impact of dwarfing genes (Rht1 and Rht2) for improving grain yield in wheat. Asian J. Plant Sci. 1, 254–256. doi: 10.3923/ajps.2002.254.256
Sun, L., Yang, W., Li, Y., Shan, Q., Ye, X., Wang, D., et al. (2019). A wheat dominant dwarfing line with Rht12, which reduces stem cell length and affects gibberellic acid synthesis, is a 5AL terminal deletion line. Plant J. 97, 887–900. doi: 10.1111/tpj.14168
Sutka, J., Kovács, G. (1987). Chromosomal location of dwarfing gene Rht12 in wheat. Euphytica 36, 521–523. doi: 10.1007/bf00041496
Tang, N. T., Ying, J., Beiru, H., Yingang, H. (2009). Effects of dwarfing genes of Rht-B1b, Rht-D1b and Rht8 with different response to GA3 on coleoptile length and plant height of wheat. Scientia Agricultura Sin. 42, 3774–3784. doi: 10.3864/j.issn.0578-1752.2009.11.003
Tian, X., Wen, W., Xie, L., Fu, L., Xu, D., Fu, C., et al. (2017). Molecular mapping of reduced plant height gene Rht24 in bread wheat. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.01379
Tian, X., Xia, X., Xu, D., Liu, Y., Xie, L., Hassan, M. A., et al. (2022). Rht24b, an ancient variation of TaGA2ox-A9, reduces plant height without yield penalty in wheat. New Phytol. 233, 738–750. doi: 10.1111/nph.17808
Vikhe, P., Venkatesan, S., Chavan, A., Tamhankar, S., Patil, R. (2019). Mapping of dwarfing gene Rht14 in durum wheat and its effect on seedling vigor, internode length and plant height. Crop J. 7, 187–197. doi: 10.1016/j.cj.2018.11.004
Wen, W., Deng, Q. Y., Jia, H. Y., Wei, L. Z., Wei, J. B., Wan, H. S., et al. (2013). Sequence variations of the partially dominant DELLA gene Rht-B1c in wheat and their functional impacts. J. Exp. Bot. 64, 3299–3312. doi: 10.1093/jxb/ert183
Wu, Q., Chen, Y., Xie, J., Dong, L., Wang, Z., Lu, P., et al. (2021). A 36 Mb terminal deletion of chromosome 2BL is responsible for a wheat semi-dwarf mutation. Crop J. 9, 873–881. doi: 10.1016/j.cj.2020.06.015
Würschum, T., Langer, S. M., Longin, C. F. (2015). Genetic control of plant height in European winter wheat cultivars. Theor. Appl. Genet. 128, 865–874. doi: 10.1007/s00122-015-2476-2
Würschum, T., Langer, S. M., Longin, C. F. H., Tucker, M. R., Leiser, W. L. (2017). A modern green revolution gene for reduced height in wheat. Plant J. 92, 892–903. doi: 10.1111/tpj.13726
Xiong, H. C., Zhou, C. Y., Fu, M. Y., Guo, H. J., Xie, Y. D., Zhao, L. S., et al. (2022). Cloning and functional characterization of Rht8, a "green revolution"replacement gene in wheat. Mol. Plant 15, 373–376. doi: 10.1016/j.molp.2022.01.014
Zhao, K., Xiao, J., Liu, Y., Chen, S., Yuan, C., Cao, A., et al. (2018). Rht23 (5Dq′) likely encodes a Q homeologue with pleiotropic effects on plant height and spike compactness. Theor. Appl. Genet. 131, 1825–1834. doi: 10.1007/s00122-018-3115-5
Keywords: wheat, plant height, dwarf mutant, QTL, γ-ray
Citation: Wang Q, Xiong H, Guo H, Zhao L, Xie Y, Gu J, Zhao S, Ding Y and Liu L (2023) Genetic analysis and mapping of dwarf gene without yield penalty in a γ-ray-induced wheat mutant. Front. Plant Sci. 14:1133024. doi: 10.3389/fpls.2023.1133024
Received: 28 December 2022; Accepted: 28 February 2023;
Published: 22 March 2023.
Edited by:
Apichart Vanavichit, Kasetsart University, ThailandReviewed by:
Cheng Liu, Shandong Academy of Agricultural Sciences, ChinaCopyright © 2023 Wang, Xiong, Guo, Zhao, Xie, Gu, Zhao, Ding and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Luxiang Liu, bGl1bHV4aWFuZ0BjYWFzLmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.