
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci., 10 March 2023
Sec. Plant Symbiotic Interactions
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1132824
This article is part of the Research TopicPlant Probiotics: Recent and Future ProspectsView all 9 articles
Every organism on the earth maintains some kind of interaction with its neighbours. As plants are sessile, they sense the varied above-ground and below-ground environmental stimuli and decipher these dialogues to the below-ground microbes and neighbouring plants via root exudates as chemical signals resulting in the modulation of the rhizospheric microbial community. The composition of root exudates depends upon the host genotype, environmental cues, and interaction of plants with other biotic factors. Crosstalk of plants with biotic agents such as herbivores, microbes, and neighbouring plants can change host plant root exudate composition, which may permit either positive or negative interactions to generate a battlefield in the rhizosphere. Compatible microbes utilize the plant carbon sources as their organic nutrients and show robust co-evolutionary changes in changing circumstances. In this review, we have mainly focused on the different biotic factors responsible for the synthesis of alternative root exudate composition leading to the modulation of rhizosphere microbiota. Understanding the stress-induced root exudate composition and resulting change in microbial community can help us to devise strategies in engineering plant microbiomes to enhance plant adaptive capabilities in a stressful environment.
Being sessile in nature, plants need to withstand changing environmental conditions to ensure their survival. Adapting to fluctuating conditions, plants require the adoption of various sophisticated mechanisms by molecular, physiological, or morphological changes. Emerging research in plant science is very less concerned with the rhizospheric microbial community shaped by the biochemical compounds present in the root exudates delivering adaptive capabilities during stressful environmental conditions. However, it is very interesting that plants themselves invest up to 40% of their photosynthetically assimilated carbon as exudates into the rhizosphere primarily composed of a wide range of compounds, e.g., a blend of amino acids, compound sugars, and organic acids (Canarini et al., 2019; Bhutia et al., 2023). Plants are endowed with the capability to alter plant rhizospheric microbes by natural means such as root exudation, which is crucial for plant growth and development. The root exudation pattern of crop wild relatives is attracting much attention nowadays, as root exudates modulating the rhizosphere microbiome might have the potential to substitute the use of chemicals in agricultural fields (Preece and Peñuelas, 2020). Recently, scientists have developed the concept of entropy-based engineering of the rhizosphere, which targets more production using less input (Zhang et al., 2022). With the advances in understanding the role of root exudates in modulating the rhizospheric microbial community, it can be correlated that under any environmental constraints, root exudation pattern changes, resulting in the alteration of the root-associated microbiome. Understanding the role of root exudates is not possible without discussing various environmental conditions that can play pivotal roles in shaping the rhizospheric microbial community. Changing climatic conditions exert pressure on the plants, which are ultimately executed by the roots in shaping the root system architecture (RSA), depending upon the plant developmental stages and genotypes (Dabhi et al., 2021; Sarita et al., 2022). The RSA helps in the better adaptation of the plants through enhanced nutrient acquisition and by modulating the rhizospheric microbial community. In addition to RSA, various edaphic factors like soil pH, soil temperature, aeration, and physicochemical characteristics have a great influence on determining the microbial community of the rhizosphere (Ling et al., 2022). Root exudation pattern and rhizosphere microbiome structure also change in accordance with the type of soil where the plant grows (İnceoğlu et al., 2012).
In addition to the above-listed edaphic conditions determining the composition and quantity of root exudates, it also firmly depends upon specific plant species or host genotype, prevailing environmental conditions, and the stress status of the plant. The root exudates act as a signalling molecule and help in recruiting beneficial root-associated mutualists, like Pseudomonas, Bacillus, Trichoderma, and mycorrhizal species (Pieterse et al., 2014), and eradicate harmful microbes in the rhizosphere, resulting in shifting the microbial community composition. Plant-associated rhizospheric microbes assist their host in a variety of activities, ranging from nutrient acquisition to defence against herbivores (Malacrinò et al., 2021), by activating induced systemic resistance (ISR) where root-specific transcription factor MYB72, phytohormones like salicylic acid (SA), volatile organic compounds (VOCs), and defence regulatory proteins play a critical role (Pineda et al., 2010; Pieterse et al., 2014). Plants respond robustly against insects and pathogens attacks by priming their leaf tissues with the help of ISR rather than directly activating the defence-related genes (Pineda et al., 2010). Traditionally, the main objectives underlying plant growth-promoting microorganism research include plant growth stimulation and yield enhancement. Understanding the role of root exudation in altering microbial community dynamics has recently emerged and attracted much attention. Further, the rhizospheric microbes and their molecular mechanisms involved in plant–plant interaction for establishing social networking systems have been studied in detail. The mycorrhizal fungal associations and the parasitic plants play significant roles in generating wired social networking systems. In contrast, root exudates and VOCs establish a wireless social networking system among the neighbouring plants (Park and Ryu, 2021; Sharifi and Ryu, 2021).
This review highlights various biotic stress-inducing factors that result in the amendment of rhizospheric microbial community composition. Firstly, we gave an overview of the microbes generally found in the rhizosphere followed by the role of different environmental factors (major biotic factors) influencing the host root exudates composition in the recruitment of either beneficial or pathogenic microbes. Secondly, we discussed the importance of root exudates in influencing the community structure of plant microbiomes. Further, based on various studies, we have tried to decipher the importance of positive and negative below-ground interactions in influencing plant resilience against varied environmental circumstances.
The soil is an overwhelming reservoir of microbial diversity. It has been estimated that 1 g of soil can carry billions of bacterial cells belonging to thousands of different taxa having crucial functions in plants and the environment. These microbial complexes establish an association between plant and rhizosphere and are also considered as the plant’s second genome (Berendsen et al., 2012). Hiltner in 1904 defined rhizosphere as the soil region that is directly associated with plant roots. With a deeper understanding of the rhizosphere as the hotspot of soil biogeochemical cycles, the definition of rhizosphere has been extended. Based on the paradigm of various soluble compounds and gaseous molecules, a new concept of rhizosphere has been developed, which defines it as the portion of soil involving the root system, up to which gaseous exchange by means of diffusion can take place (de la Porte et al., 2020). Contemporary investigations in understanding the rhizosphere microbiome have revealed that the host plant’s genetic makeup is also involved in the alterations of the rhizomicrobiota. The rhizosphere microbiome is very important for the fitness of the host, and the manipulation of the rhizosphere microbiota is presumed to provide a solution for climate-resilient crop production (Pieterse et al., 2016). A competitive battle in the rhizosphere is going on among different types of microorganisms competing for plant-derived nutrients, resulting in their interactions with the roots (Li et al. 2021b). For instance, arbuscular mycorrhizal fungi (AMF) are reported to colonize approximately 71% of terrestrial plants exhibiting complex interaction patterns in various plants except for plants belonging to Brassicaceae, Chenopodiaceae, Polygonaceae, Amaranthaceae, and Caryophyllaceae (Wang and Qiu, 2006; Fernández et al., 2019). Complex chemical signalling between the host plant and AMF results in enhanced plant nutrient acquisition and protection of the plants from pathogen infection (Dowarah et al., 2021). Sensing the varied environmental stimuli, the microbiome structure of the soil changes, which is further interlinked with the nature of the soil and the plant species involved in the recruitment of microbes (Garbeva et al., 2008). İnceoğlu et al. (2012), from their investigation, reported that the type of soil functions as a determining factor in structuring the rhizospheric microbial community. Based on the rhizospheric effect, they concluded that potato cultivars grown on different soil types recruited diverse communities of microbes with diverse functional abilities. Five types of potato cultivars were collected from loamy and sandy soil regions and found that bacterial communities in the loamy soil rhizosphere show more affinity towards carbohydrates and amino acids, while communities reported from the sandy soil rhizosphere have shown a higher affinity towards polymers, alcohols, and nucleotide-based carbon sources, which were secreted out by the roots as exudates. Despite these natural signalling cues, a new concept of “entropy” has been introduced in the area of agricultural research, which tries to emphasize that the lower the entropy, the more ordered the rhizospheric interactions, which can ensure proper utilization of energy and resources by the plant (Zhang et al., 2022).
Plenty of research on soil microbial communities revealed that soil around the root region has a high density of microbes such as Actinomycetes, bacteria, and fungi, with higher activity rather than the bulk soil, a phenomenon known as the “rhizosphere effect” (Philippot et al., 2013; Bakker et al., 2020). However, as compared to bulk soil, the rhizosphere harbours a less diverse group of microbes (Philippot et al., 2013; Bakker et al., 2020). Depending upon environmental stimuli, the rhizosphere acquires selective microbes from the bulk soil. In the rhizosphere, the most prominently reported bacterial groups are the Acidobacteria, Bacteroidetes, Firmicutes, Planctomycetes, and Proteobacteria (Uroz et al., 2010; Bulgarelli et al., 2012; Dastogeer et al., 2020). Moreover, some bacterial communities like Actinobacteria and Xanthomonadaceae are scarcely present in the rhizosphere as compared to the bulk soil (Bulgarelli et al., 2012; Dastogeer et al., 2020). It has also been reported that the rhizosphere is predominantly colonized by mycorrhizal fungi that are estimated to be associated with approximately 80% of all land plants providing N and P for proper growth and development (van der Heijden et al., 2015; Weng et al., 2022). This symbiotic interaction between the mycorrhizae and the roots of higher plants has great significance in shaping the terrestrial ecosystem, as it can regulate the carbon and other nutrient cycles of the earth.
Plant rhizosphere microbiome is a result of interaction among plants, microbes, and associated environmental factors in either biotic or abiotic form or in alliance forming a tritrophic interaction (Badri et al., 2009). The plant species, its genotype, distinct tissue or organ, age and developmental state, ecosystem canopy type, immunity, etc., are some of the factors associated with the host plant, which can determine the rhizospheric microbial community (Dastogeer et al., 2020). Together, several plant-derived compounds like different alkaloids, phenolics, and terpenoids also have an important role in shaping the rhizosphere microbiome. Some of these plant-derived compounds are specific for some plants; e.g., glucosinolates are found predominantly in the Brassicaceae group (Dastogeer et al., 2020), and aglycone benzoxazinoids (BX) are produced by maize, which induces jasmonic acid-dependent resistance against herbivores (Hu et al., 2018).
In addition to the above-ground environmental factors and plant genotype, the rhizospheric microbial community composition relies also upon the soil texture and below-ground abiotic factors such as soil temperature, pH, and CO2 concentration. Fierer et al. (2007) investigated and found that carbon availability in the soil can play a role in the regulation of certain bacterial community structures involving Acidobacteria, Bacteroidetes, and Betaproteobacteria. However, Lauber et al. (2009), in their study, revealed that soil pH has a remarkable effect on the regulation of soil microbiome structure. On the contrary, the temperature was found to modulate plant root interactions with rhizospheric microbes, where the rhizosphere was found to be more responsive to changing temporal conditions than the phyllosphere (Luo et al., 2020); e.g., roots colonized by mycorrhizal fungi shows greater resilience in temperature stressed condition, playing a key role in plant adaptation to high temperature (Usman et al., 2021).
In contrast to various environmental factors, biotic components such as pathogens, parasites, or mutualistic plant microbes, present at the phyllosphere or rhizosphere, perform a crucial role in shaping the rhizospheric microbial community by influencing the survival of plants during or post-stress conditions. Among various biotic factors, AMF actively takes part in shaping the rhizomicrobiota (Dowarah et al., 2021). AMF association with the plant root is a very complex process that requires coordination between plant and fungal signalling molecules. Roots having AMF associations are protected against pathogens as AMF helps in nutrient acquisition and repair of pathogen-damaged cells (Dowarah et al., 2021). Here, the root architecture plays an important role, as it was reported that AMF benefit the tap root system in a better way as compared to the fibrous root system (Yang et al., 2015). Frew and Price (2019) reported that under elevated CO2 levels, both C3 and C4 plants produce a high amount of photosynthetically fixed C compounds. C4 plants allocate their excessive photosynthates in the rhizosphere with the help of root exudation, which leads to better AMF colonization, finally resulting in enhanced nutrient acquisition than that of C3 plants. Data on the underlying mechanism(s) involved in rhizosphere microbiome amendment under the influence of various stress conditions are limited. In this review, we have tried to highlight the role of biotic stresses in altering root exudate secretion patterns and also how the change in root exudate composition can influence the recruitment of the rhizospheric microbes during stress conditions.
Root exudation is the key mechanism by which plants maintain a homeostatic interaction with their below-ground environment. Plants exude various primary and secondary metabolites as root exudates, ultimately shaping the community structure of rhizosphere-dwelling microbes. The exact mechanism of root exudation is not yet clear, but to allocate the photosynthetically fixed C from the aerial plant parts to the soil, a mechanism similar to Munch’s hypothesis (Münch, 1930) is used by the plants, where diffusion plays a critical role (Canarini et al., 2019). According to Munch’s hypothesis, a concentration gradient between the source and sink organs, i.e., phloem and root tip generate turgor differences, eventually resulting in the translocation of metabolites from the phloem to the root tip and then to the soil (De Schepper et al., 2013; Ross-Elliott et al., 2017). Exudation of various metabolites from the root cells to the soil might involve facilitated diffusion, where concentration gradients act as a central force and specific transmembrane proteins help in the efflux of such molecules (Sasse et al., 2018). Specialised efflux carriers and channel proteins facilitate the secretion of primary metabolites like amino acids, organic acids, and sugars into the rhizosphere. For example, for amino acids, UMAMIT (usually multiple acids move in and out transporter), CAT (cationic amino acid transporter), and GDU (glutamine dumper) transporters, for organic acids, ALMT/malate (aluminium-activated malate transporter) and MATE/citrate transporters (multidrug and toxic compound extrusion), and sugar molecules, a family of transporters known as the SWEET transporter family are used by the plants (Canarini et al., 2019). Like primary metabolites, secondary metabolites also constitute a part of root exudates. Various secondary metabolites like phenolics, alkaloids, and terpenoids are transported into the rhizosphere with the help of ATP-binding cassette (ABC) transporters (Badri et al., 2009), whereas most of the primary metabolite transporters use passive transport as a mechanism of exudation, except MATE/citrate transporter having H+ coupled antiport activity (Meyer et al., 2010). A balance in the concentration gradient across the external and internal environment is maintained by the rhizospheric microbes as well as by the plants themselves. The exudates secreted out are consumed by various rhizospheric microbes, and a low concentration level is maintained in the external environment, enhancing root exudation (Canarini et al., 2019). With the help of phloem loading and transport (source activity), phloem unloading (sink activity), regulation of the expression of genes, or post-translational modifications of efflux carriers, plants might control the process of root exudation. Overall, the plant source-sink dynamics fundamentally regulate the allocation of photosynthates into the soil (Farrar and Jones, 2000; Savage et al., 2016). Moreover, plants modify their RSA depending on the concentration of primary metabolites at the root region, thus controlling the exudation process (Canarini et al., 2019; Bhutia et al., 2023). Modification in the efflux or influx of metabolites at the root region critically shapes the RSA where amino acids have a cascading influence in shaping the RSA. The expression of NRT2.1 transporter genes is mainly regulated by the amino acids, indicating the plant-soil N status (Nazoa et al., 2003). Glutamate is an important amino acid that functions similarly to the phytohormone auxin. A rapid increase in cytosolic Ca2+ and depolarization of the membrane is induced by glutamate, where an electrical signal similar to the synapsis of mammals is generated (Baluška and Mancuso, 2013; Ni et al., 2016). Additionally, a receptor family, known as the glutamate receptor-like (GLR) receptors, functions like amino-acid-gated Ca2+ ion channels and acts as a signal to convey the plant N status to the root (Brian and Lea, 2007; Vincill et al., 2012), thus shaping the RSA. The change in RSA, impacts the root exudation pattern by changing the composition of root exudates under various environmental conditions.
Since the exudation of various metabolites involves specialised carriers and takes place through facilitated diffusion, the molecular mechanisms involved in such processes may indirectly govern the mechanisms of root exudation. Further, studies on genes involved in shaping the RSA of plants might help in the detailed understanding of the root exudation mechanism.
The ability of plants to adapt and survive in an environment primarily depends upon their capacity to acquire resources and resist external perturbations, impacting growth and productivity. In non-homogeneous soil conditions, the plant roots perform a crucial function in the absorption of mineral and nutrient resources from the soil, either directly or in association with rhizospheric microbes. These microbes are presumed to be attracted by plant-specific root exudates under normal or stressful circumstances. Thus, nutrient supply effect root exudation mostly from the root apices, ultimately altering the nutrient dynamics and the soil microbiome structure (Paterson et al., 2006).
The quality and quantity of various root exudates with different compositions secreted from different locations of the root are mainly determined by root branching and RSA (Badri and Vivanco, 2009; Canarini et al., 2019). The root meristematic and elongation zone present above the root tip secrete amino acids like asparagine and threonine, while other amino acids like glutamic acid, valine, leucine, and phenylalanine are secreted from the root hairs, along with the whole root exuding aspartic acid (Figure 1B) (Haldar and Sengupta, 2015). Several discrete blends of phytochemicals from various plant species are also released from different parts of the root like the mature root regions and root cap and also from root hairs (Nguyen, 2009).
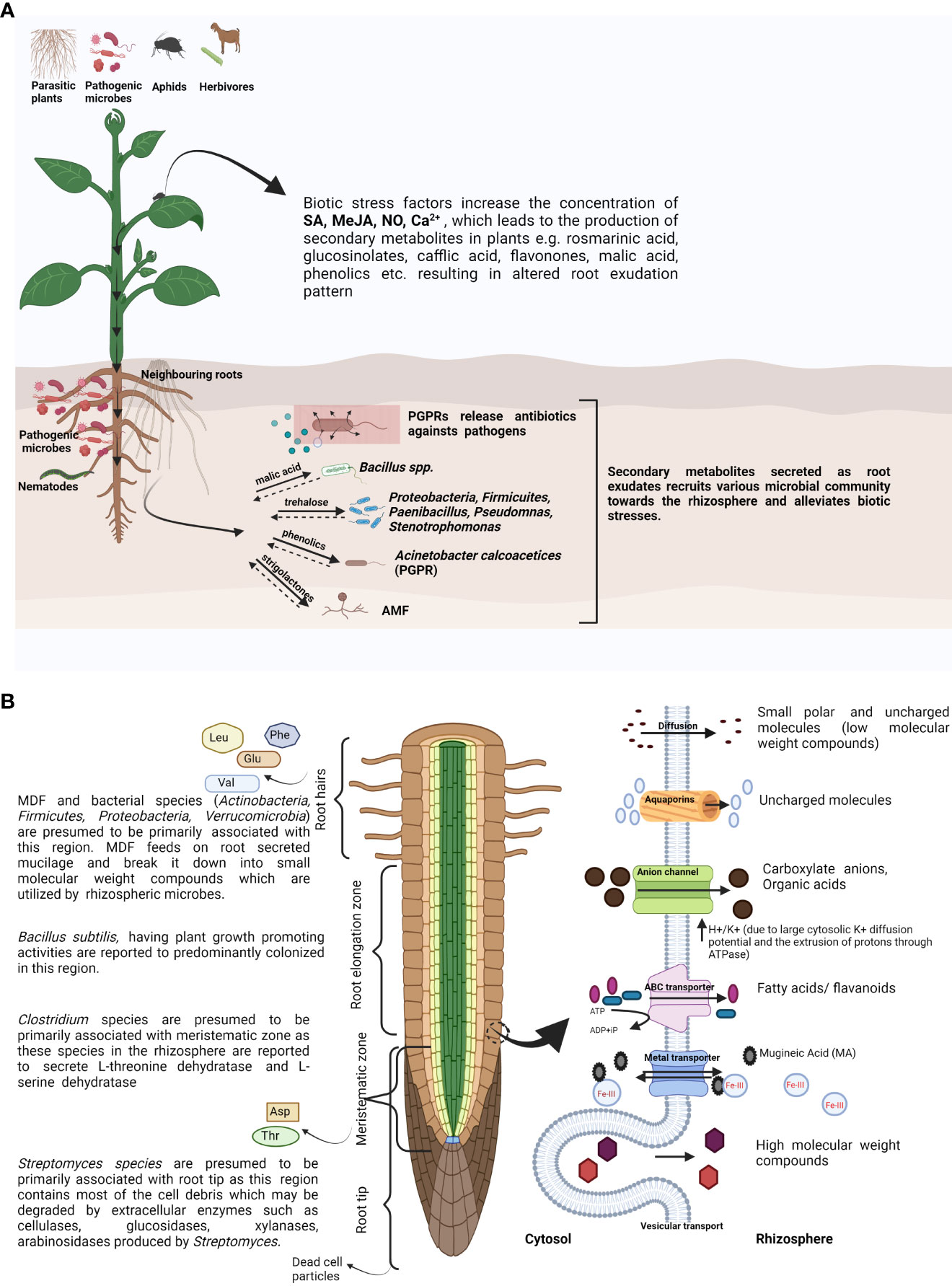
Figure 1 (A) Schematic representation showing the role of root exudates in altering rhizospheric microbial community composition sensing above-ground and below-ground biotic stresses. (B) Schematic representation of distinct root zones secreting varied blends of compounds along with the possible secretion processes across the plasma membrane into the rhizosphere. SA, salicylic acid; MeJA, methyl jasmonate; NO, nitric oxide; AMF, arbuscular mycorrhizal fungi; VOCs, volatile organic compounds; PGPR, plant growth-promoting rhizobacteria; Leu, leucine; Phe, phenylalanine; Glu, glutamic acid; Val, valine; Asp, aspartate; Thr, threonine; ABC transporter, ATP-binding cassette transporter. Created with BioRender.com.
Exudation from the roots contributes to both positive and negative associations in the below-ground environment. Positive root–microbe associations involve mutualistic interactions between root and beneficial microorganisms like plant growth-promoting rhizobacteria (PGPR) and mycorrhizal fungi, while negative associations involve the interaction of roots with harmful organisms like pathogens, invertebrate herbivores, and parasitic plants (Badri et al., 2009). Plants produce diverse arrays of secondary metabolites such as phenolic compounds, alkaloids, and terpenoids, some of which are genus/species-specific (Bednarek and Osbourn, 2009; Dastogeer et al., 2020). For example, barley plants infected with Fusarium graminearum release exudates having a high concentration of phenolic compounds, which alters the rhizospheric microbial community to enhance the plants’ defence against F. graminearum (Phour et al., 2020; Afridi et al., 2022).
Microbial degradation of leaf litter and dead plant substances in the rhizosphere serve as key resources for plant growth and also participate in carbon storage in the ecosystem by releasing nutrients as a decomposition product (Schneider et al., 2012). Studies suggest that mucilage-degrading fungi (MDF) and other microbes actively work upon mucilage or root exudates and break them into small molecular weight compounds, which act as signalling molecules for the recruitment of various PGPR. Artemisia sphaerocephala, a remarkable plant of Asian cold deserts, was used in a study where mucilage-degrading microbes were isolated from the root–soil junction. From the study, it becomes evident that the MDF associated with the roots of A. sphaerocephala recruit bacterial groups like Actinobacteria, Firmicutes, Proteobacteria, and Verrucomicrobia towards the roots and regulate the rhizosphere microbiota (Hu et al., 2021). Further, microfluidics-based investigation for real-time study of plant–microbe interaction revealed that Bacillus subtilis exhibits chemotactic behaviour towards the root elongation zone, which might be due to the secretion of root exudates (Massalha et al., 2017; Sasse et al., 2018). In addition to that, Escherichia coli cells from the rhizosphere were excluded after the colonization of B. subtilis at the rhizospheric region of Arabidopsis thaliana (Massalha et al., 2017). Microbes belonging to the group Clostridium are presumed to be associated with the root meristematic region as they produce l-threonine dehydratase and l-serine dehydratase enzymes, which may catalyze dehydration and isomerization of root-secreted threonine and serine, respectively (Hofmeister et al., 1993), and might alter the root exudation pattern of the host plant. Clostridium spp. are also involved in the degradation of renewable complex compounds like lignocellulose and make it utilizable for the plants as a resource, ultimately maximizing the C substrate exploitation and yield of the plants (Figure 1B) (Du et al., 2020). The root tip actively associates with a number of bacterial groups and withstands numerous obstacles due to which the cells shed off and reform in a continual process. These damaged cell parts contain lignin, cellulose, and various organic compounds and are presumed to be degraded by the Streptomyces species, as it contains a large amount of extracellular enzymes such as cellulase, glucosidase, xylanase, and lignocellulase (Schrempf et al., 2007; Chater et al., 2010).
Since the plants encounter a range of above-ground and below-ground constraints in their lives, to cope with those factors, the plants secrete different blends of chemical compounds in the form of root exudates, which varies in different plants with respect to their age. Nevertheless, it is very difficult to understand the factors responsible for variation in root exudation, as it is an emerging area in plant biology, and more precise research needs to be performed in the future.
Plants, being solitary in nature, have developed sophisticated defence mechanisms to resist the attack of pathogenic microbes as well as herbivorous organisms. It is evident that plants alter their primary and secondary metabolic pathways to withstand biotic stress, but there is very little knowledge that the roots have a key role in the regulation of those pathways for better adaption of the plants. Ongoing research on root exudates revealed that the roots take part in the synthesis and secretion of phytotoxins, helping the plants to sense their surroundings, leading to the activation of defence signals (Erb et al., 2009). Further, secretions from neighbouring plants have effects on one another and can induce changes in root exudate compositions (Ulbrich et al., 2022). Plants encountering biotic challenges in the form of either pathogen attack or herbivory synthesize and release phytohormones and VOCs, which amend the chemical composition of the root exudates and eventually restructure the below-ground microbial community (Erb et al., 2009; Dastogeer et al., 2020). Plants encountering various environmental stimuli either in physical or chemical form release a number of phytochemicals as root exudates, in contrast to the non-stimulated plants (Badri and Vivanco, 2009). Further, variable compounds are released by different plant species encountering the same stimulus at the same time. Several pieces of evidence suggest that the external application of defence-related compounds like SA, methyl jasmonate (MeJA), and nitric oxide (NO) triggers the host plant to accumulate a diverse range of secondary metabolites (Zhao et al., 2005; Badri and Vivanco, 2009), e.g., indole glucosinolates, rosmarinic acid, and caffeic acid (Figure 1A).
In response to recognition of pathogens and pests, plants induce SAR and ISR signalling that helps in defence priming and finally induces the host plant to respond robustly against upcoming pathogen attacks; e.g., SAR and ISR establish a memory mark by activating various defence-related genes or some protein factors like MAPK3 and MAPK6 (Beckers et al., 2009; Doornbos et al., 2012). As a result of biotic stress, root exudates become enriched with phenolic compounds, which can directly suppress the growth of pathogenic microbes (Jousset et al., 2011; Badri et al., 2013; Chaparro et al., 2013) or may indirectly act by changing the expression level of some anti-fungal genes present in the plant beneficial microorganisms (Jousset et al., 2011). For example, barley plants growing in split root systems infected with the pathogenic fungus Pythium ultimum in one side of the root and inoculation of the bacterium Pseudomonas fluorescens on the other side secrete root exudates having a more phenolic compound. This specialized exudation recruits beneficial PGPR towards the roots and inhibits the spore germination of P. ultimum and also results in the activation of antibiotics-related genes present in P. fluorescens against P. ultimum (Jousset et al., 2011). Thus, plants under biotic stress may induce either direct or indirect defence mechanisms, where the exudation of specialised compounds through the roots might have a positive effect.
Capsicum annuum, undergoing herbivory attack due to aphids and also infected with Ralstonia solanacearum SL1931, has shown the attraction of a higher population of beneficial bacteria B. subtilis GB03 in the rhizospheric region of the plant (Lee et al., 2012). Earlier experimental pieces of evidence suggested that the recruitment of B. subtilis FB17 towards the plant rhizospheric region might be due to the secretion of l-malic acid as root exudates (Rudrappa et al., 2008; Badri et al., 2009). Pseudomonas syringae-infected A. thaliana has resulted in the alteration of malic acid concentration by altering the gene expression of root malate transporter, which led to the accumulation of a large amount of the rhizobacterium B. subtilis at the rhizosphere (Lakshmanan et al., 2012; Chen et al., 2013; Dastogeer et al., 2020). Infection of cucumber plants with Fusarium oxysporum f. sp. cucumerinum amended the secretion of fumaric acid and citric acid at the rhizospheric region of the cucumber plants and helped in Bacillus amyloliquefaciens biofilm formation (Dastogeer et al., 2020). In the case of carex plant, Fusarium culmorum infection has resulted in the secretion of monoterpene (Z)-limonene-oxide in the root exudates and attracted a group of microbes including Janthinobacterium, Collimonas, and Paenibacillus towards the rhizospheric region (Schulz-Bohm et al., 2018). This indicates that a distinct blend of signalling molecules are involved in the recruitment of specific rhizobacterial communities. In the Table 1, salient findings from the studies conducted on biotic stress induced change in root exudate composition and modulation of microbial community are enumerated.
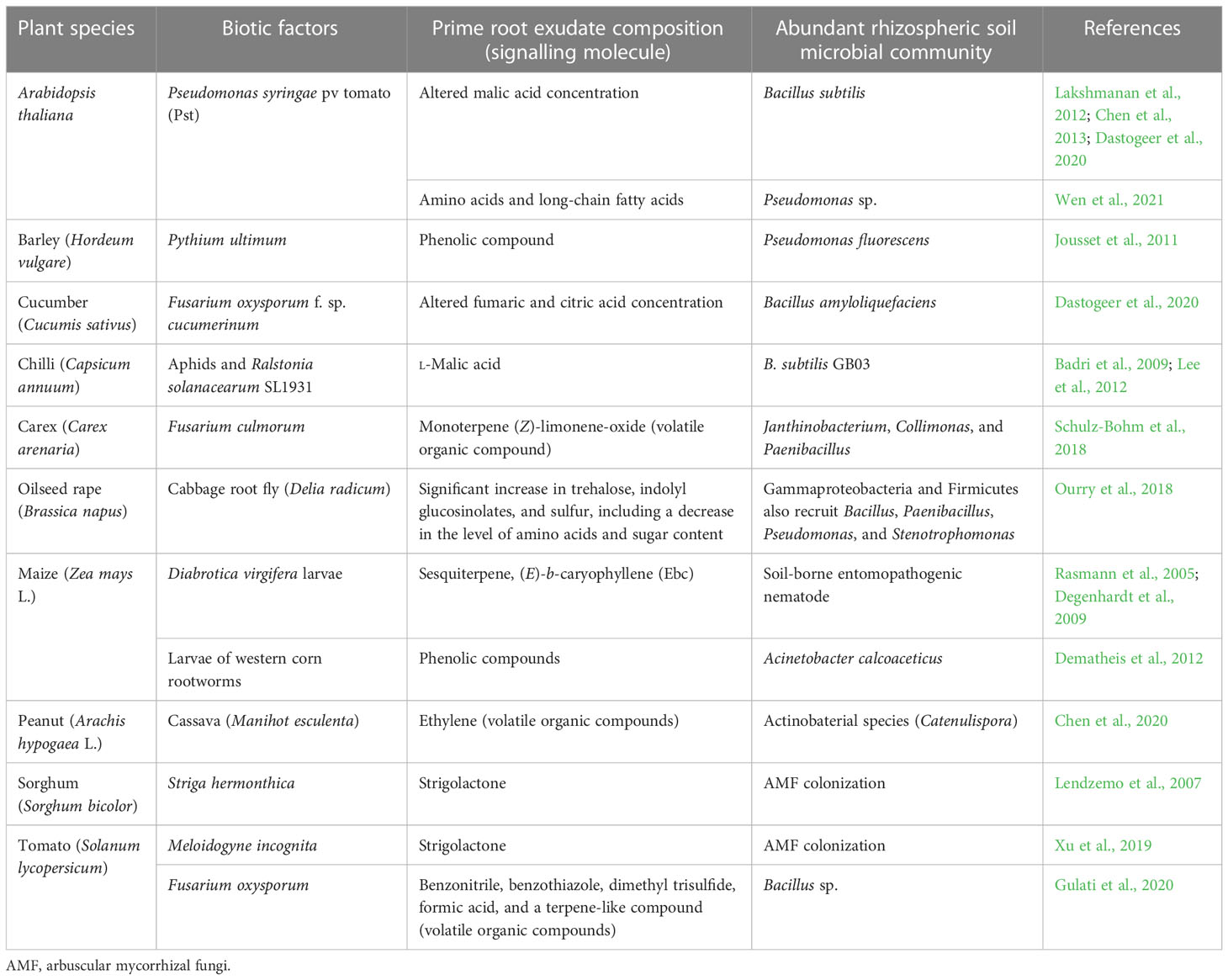
Table 1 Biotic factors modulating rhizospheric microbial community due to altered root exudate compositions.
Plants experience concurrent infestation by various biotrophic or necrotrophic insects in their natural habitat, ultimately inducing the SA or JA pathways for defence. Several studies have revealed that the SA and JA crosstalk may act synergistically or antagonistically (Mur et al., 2006; Wasternack and Hause, 2013). Further, when an herbivore or an insect attacks a plant, they release specific salivary effectors, which directly or indirectly influence the activation of SA or JA pathways (Xu et al., 2018). The molecular mechanisms involved in the regulation of SA and JA crosstalk are not yet very clear. However, according to the studies, NONEXPRESSER OF PATHOGENESIS-RELATED GENES1 (NPR1) acts as a key regulator in SA-induced defence gene expression during the attack of a biotrophic insect, while the SCFCOI1 complex helps in the degradation of jasmonate-ZIM domain (JAZ), allowing the activation of JA-mediated defence-related genes (Xu et al., 2022). For example, potato plants infested with herbivores having varied feeding behaviour induce JA signalling during the infestation by necrotrophic insects, while SA signalling is induced in plants during the attack of biotrophic insects (Bari and Jones, 2009; Malacrinò et al., 2021). A deeper understanding of herbivore infestation with different feeding behaviours revealed that herbivore feeding strategies have a negligible effect on the rhizomicrobiota (Malacrinò et al., 2021). However, altogether herbivore-induced stress affects the root exudation pattern and results in the recruitment of beneficial microbes to alleviate biotic stress (Rasmann and Turlings, 2016; Hoysted et al., 2018; Rolfe et al., 2019). SA in the root exudates helps in the recruitment of siderophore-producing rhizobacteria and takes part in suppressing the disease development in the host plant; further, the threshold concentration of SA in the soil is sufficient to alert the neighbouring plants to withstand upcoming stress (Bakker et al., 2014; Aznar and Dellagi, 2015; Cheol Song et al., 2016; Rolfe et al., 2019). Yuan et al. (2018) inoculated A. thaliana plants with P. syringae pv tomato (Pst) and observed an increase in JA level and improvement in plant immune responses. Plants encountering Pst infection exhibited a significant increase in the amount of amino acids, nucleotides, and long-chain organic acids (LCOAs) and simultaneously decreased the amount of sugars, alcohols, and short-chain organic acids (SCOAs) in the root exudates (Li et al., 2021a; Wen et al., 2021). The modification in the root exudate composition changed the rhizosphere microbiome structure and elicited disease resistance characteristics in A. thaliana. This might be due to the recruitment of beneficial rhizobacterium B. subtilis under the influence of an altered root exudation pattern (Lakshmanan et al., 2012; Chen et al., 2013; Dastogeer et al., 2020). Strigolactone (SL) released as root exudates have a direct impact on structuring the root-associated microbiome, as it enhances AMF colonization in plant roots (Akiyama and Hayashi, 2006). SL synthesis was found to increase in the roots of tomato plants undergoing infestation of parasitic nematode Meloidogyne incognita, augmenting the root–AMF interactions for better resilience of tomato plants (Xu et al., 2019). Further, the use of SL analogue racGR24 in JA-deficient mutants of tomato enhanced plant defence against nematode infection. In addition to that, the secretion of specific molecules like benzonitrile, benzothiazole, dimethyl trisulfide, formic acid, and terpene-like compounds from the roots of tomato plants infected with Fusarium oxysporum was found to recruit Bacillus sp. in the rhizosphere. The recruitment of Bacillus sp. might be due to the change in root exudate composition (Gulati et al., 2020).
Arabidopsis plants treated with MAMP enhance the activation of defence genes CYP71A12 and MYB51 in the root region (Millet et al., 2010; Rolfe et al., 2019), having regulatory effects on the synthesis of tryptophan-derived compounds like camalexin and indolic glucosinolates. These molecules are commonly detected in root exudates and have a direct effect on regulating the rhizosphere microbiome, as they possess both antimicrobial and signalling activities (Stotz et al., 2011; Mönchgesang et al., 2016; Koprivova et al., 2019; Rolfe et al., 2019). Infestation of the Diabrotica virgifera virgifera larvae at the roots of maize plants trigger the secretion of the sesquiterpene, (E)-b-caryophyllene (Ebc), which results in the recruitment of soil-borne entomopathogenic nematode (Rasmann et al., 2005; Degenhardt et al., 2009). The entomopathogenic nematode attracted via the Ebc signalling acts as a natural predator for the beetle D. v. virgifera, showing biocontrol activity. However, Ebc secretion may not always act as a biocontrol strategy, as in a non-yielding maize line, Ebc accumulation in the root–soil junction induced hyphal branching of Colletotrichum graminicola and Fusarium graminearum, which are considered as the key fungal pathogen. Thus, Ebc secretion as root exudates might have the ability to either enhance plant resistance or induce plant susceptibility, depending upon the type of biotic disturbances (Fantaye et al., 2015). In various studies, it has been suggested that the overexpression of the Ebc gene might have a wide-ranging impact on soil microbes (Rolfe et al., 2019). In maize or other cereals, a class of secondary metabolites known as benzoxazinoids has a direct link with the induction of JA-dependent defence signalling against herbivore attack. Again, 6-methoxy-benzoxazolin-2-one (MBOA) and methoxy-2H-1,4-benzoxazin-3(4H)-one (DIMBOA), are the product obtained from the fractionation of benzoxazinoids and helps in the alteration of rhizosphere microbiome structure when released as a root exudate and even enhances the resistance of the next-generation plants against herbivory (Niemeyer, 2009; Hu et al., 2018). Further research is needed on both MBOA and DIMBOA exudation patterns concerning biotic stress and the role of both the compounds and their effects on soil and neighbouring plants. Dematheis et al. (2012) reported the abundance of Acinetobacter calcoaceticus in the rhizosphere of maize under the influence of the larvae of western corn rootworms, which elicits phenolic compounds as exudates and affects mainly the bacterial communities rather than the fungi. The threatening soil herbivorous insect cabbage root fly (Delia radicum) and its major susceptible host oilseed rape (Brassica napus) were used in a study to determine the effect of herbivory in shaping the root–rhizosphere microbiome. Infestation by the insect, attracted more numbers of Gammaproteobacteria and Firmicutes towards the root and rhizospheric region of oilseed rape and also enhanced the abundance of four bacterial groups, namely, Bacillus, Paenibacillus, Pseudomonas, and Stenotrophomonas. Modulation of the bacterial communities in the root and the rhizosphere was due to the increasing trehalose, indolyl glucosinolates, and sulfur including a decrease in the level of amino acids and sugar content in the root exudates (Ourry et al., 2018). Thus, under the influence of various biotic factors, plants release a diverse range of signalling molecules, ultimately shaping the rhizomicrobiota. Biotic factors influencing the rhizospheric microbial community may be due to changes in primary and secondary metabolite concentration gradients along the root and rhizosphere. The altered rhizospheric microbial community can in turn affect the plant root exudation pattern.
As described earlier, the rhizosphere serves as a dwelling place for an enormous number organisms interacting with roots, and it also shows the interaction of root systems with neighbouring plants (Badri et al., 2009). The exudates secreted from the roots establish a chemical crosstalk with the below-ground organisms as well as with their surrounding roots. Root exudates act in a multipurpose way, and this can be easily understood with the example of isoflavone exudation from the roots of soybean plants, as isoflavones secreted out from the soybean roots recruit both mutualistic (Bradyrhizobium japonicum) and pathogenic (Phytophthora sojae) microbes towards them (Bais et al., 2006). The actual mechanisms of how roots interpret a multitude of signals received from surroundings are not yet understood; however, the mechanism of root secretion is assumed (Figure 1B). Small polar and uncharged molecules of low molecular weight are transported through lipid membranes depending on the membrane permeability and pH of the cytosol. In contrast, the protein and ion channels help in the transportation of sugar molecules, amino acids, and carboxylate anions outside the cell membrane depending upon the electrochemical gradient. Compounds having high molecular weight are generally excreted by plants using vesicular transport into the rhizosphere (Badri and Vivanco, 2009). Plants have developed certain mechanisms to alleviate the toxic effect of heavy metals (namely, iron, zinc, manganese, and copper) present in the soil through metal homeostasis (Haydon and Cobbett, 2007). Nutrient accessibilities in the soil modulate the root exudation pattern, which results in changes in the soil pH and the formation of metabolite complexes with the metal ions (Chen et al., 2017). Plants secrete a blend of complex metabolites comprising gaseous molecules, inorganic ions, and carbon-based compounds for the regulation of nutrient availability and to detoxify intolerable metal pollutants in their surroundings. The type of secretion varies among species depending upon species tolerance to nutrient deficiency and heavy metal contamination. Plants use two strategies for the uptake of required nutrients using complex metabolite compositions: reduction-based strategy and chelation-based strategy (Chen et al., 2017). For example, Fe deficiency induces secretion of riboflavin (Rbfl), phenolics, and phytosiderophore compounds via the upregulation of certain enzymes (dimethyl-8-ribityllumazine synthase (DMRL) and GTP cyclohydrolase II (GTPcII) required for flavin biosynthesis) (Rellán-Álvarez et al., 2010, Rodriguez-Clema et al., 2011, Rodríguez-Celma et al., 2013; Hsieh and Waters, 2016), transcription factors (bHLH38 in Arabidopsis and bHLH39 in tobacco and sunflower reported for the Rbfl accumulation and exudation) (Vorwieger et al., 2007), and plasma membrane-localized transporters (pleiotropic drug resistance 9 (PDR9) for phenolics and transporter of mugineic acid1 (TOM1) for phytosiderophores) (Nozoye T et al., 2011, Nozoye T et al., 2013; Fourcroy et al., 2014; Chen et al., 2017).
Recent evidence showed that the secretion of proteins as root exudates is regulated by the microorganisms present in the soil. The presence of a specific microbial community in the rhizosphere can alter the protein constituents of root exudates and vice versa (De-la-Peña et al., 2008). An experiment was conducted with two plants, namely, A. thaliana and Medicago sativa, by using P. syringae (DC3000) and Sinorhizobium meliloti (RM1021) as a pathogen and a symbiont, respectively. Secretions of seven different proteins (categorized as hydrolases, peptidases, and peroxidases) were reported during Medicago–S. meliloti interactions, but no such secretion had been observed when the Medicago plants were exposed to P. syringae. Similarly, Arabidopsis–P. syringae interaction had led to the exudation of several defence-related proteins, but like the alternative result of Medicago–P. syringae, no defence molecules have been secreted during the interaction of Arabidopsis and S. meliloti. This host–microbe-specific result was observed due to the differences in root exudate composition, mainly the protein components secreted out by both the host plants under variable plant–microbe interaction (De-la-Peña et al., 2008; Badri and Vivanco, 2009). Additionally, several studies have also reported the effect of weeds on agricultural fields, where they inhibit the proper growth and development of domesticated plant species. When the weeds migrate from their native region to a new place, they become more invasive and have a consequent effect on the domesticated plant species. As the weeds become exotic, they escape their specialized insects and pathogens including other factors at their native place and change their physiology and biochemistry to become more resilient. Weeds can alter the root exudation profile, which exhibits allelopathy to native plants as well as ensures the recruitment of beneficial microbes (Trognitz et al., 2016; Trivedi et al., 2022). Various rhizospheric microbes contribute to lowering the effect of allelopathy (Mishra et al., 2012a; Mishra et al., 2012b; Trognitz et al., 2016); e.g., invasive plants like Parthenium hysterophorus are widely found in India and some other countries, which secrete parthenin and phenolic acids as allelopathic chemicals. Pseudomonas putida NBRIC19 strain modulates the composition of antioxidant enzymes in root exudates of P. hysterophorus and alleviates the allelopathic effect (Mishra and Nautiyal, 2012; Mishra et al., 2012a; Mishra et al., 2012b). In a population having a large number of Arabidopsis plants, enhanced secretion of glucosinolates was observed, as a result of the neighbouring plant’s effect. However, the secretion of glucosinolates was reduced in an environment with a smaller number of Arabidopsis plants (Wentzell and Kliebenstein, 2008). Similarly, Bressan et al. (2009) in their study, using denaturing gradient gel electrophoresis (DGGE) fingerprint analysis, unearthed the impact of exogenously produced glucosinolate by A. thaliana transgenic line CYP79A1 on rhizospheric soil microbial community where an abundance of Agrobacterium sp., Bosea sp., Rhizobium sp., Mesorhizobium amorphae, and Syncephalis depressa in the rhizospheric region has been found as compared to the wild-type plants. Further, intercropping of peanut (Arachis hypogaea L.) with cassava (Manihot esculenta Crantz) plants triggers the production of ethylene hormone by the peanut plants. The release of ethylene acts as a signalling molecule in the rhizosphere and recruits the actinobacterial species Catenulispora towards the peanut roots, enhancing plant fitness in the intercropped area (Chen et al., 2020). These pieces of evidence suggest that neighbouring plants modulate each other’s exudation patterns and reshape the rhizosphere microbiome, leading to enhanced adaptation of plants under varied environmental conditions.
The rhizosphere harbours infinite numbers of microorganisms and other chemical substances along with plant roots. Complex ecological processes in the rhizosphere involve positive and negative interactions between the microfauna, plant–plant, or plant–microbes, which play a key role in maintaining the soil characteristics (Bais et al., 2006). The exudate secreted out by the roots influence both positive and negative interactions in the rhizosphere (Li et al., 2021b). One of the major constituents of root exudates is the VOCs, which play a critical role in shaping the below-ground microbiota. The VOCs can aid in the detection of neighbours, maintain a homeostatic interaction among neighbouring plants, and enhance plants’ adaptive capabilities under various circumstances (Ninkovic et al., 2016). Further, the rhizobacterial-released VOCs (carbon-based compounds, terpenoids, and sulfur compounds) are well-known organic compounds playing a crucial role in inter and intra specific communications below-ground (Mhlongo et al., 2018). Root exudates can promote plant growth by synergistically acting with the microbes, plants, and soil-dwelling nematodes forming a “tritrophic interaction” (Badri and Vivanco, 2009). Horiuchi et al. (2005), in their study, reported that root exudates of leguminous plants attract the nematode Caenorhabditis elegans, which is a vector as well as the symbiont of S. meliloti. The accumulation of C. elegans along with S. meliloti triggers the host plant to release VOCs, and eventually, the volatiles released recruit rhizobial bacteria, which in turn increases nodule formation in the legumes. Furthermore, insect-infested maize roots released specialized root exudates having Ebc as a major constituent and recruited entomopathogenic nematodes in the rhizosphere (Rasmann et al., 2005). Another example of tritrophic interaction including the host plant (Sorghum), AMF, and parasitic plant Striga hermonthica shows how the root exudate-mediated AMF colonization leads to the inhibition of parasitic plants (Lendzemo et al., 2007). The exudation of strigolactones and their derivatives play an important role in regulating this tritrophic interaction (Bouwmeester et al., 2007; Lendzemo et al., 2007). Therefore, it can be hypothesised that a tritrophic interaction leads to the knockout of harmful microbes by attracting beneficial microorganisms with the help of root exudates.
The rapid evolution of the rhizospheric microbes along a parasitic–mutualistic continuum is evident in some cases, in which root exudates are considered as a determining factor (Li et al., 2021a). Domesticated plants in their native areas interact with diverse co-evolved microorganisms, which may be either beneficial or harmful in nature. However, it has been observed that domesticated plants flourish effectively in an introduced area due to a smaller number of pathogens present in the soil, but if some foreign pathogens enter the ecosystem, it becomes dominant and highly destructive against the entire crop species, as there are no co-evolved competitors against them. The famous potato famine of Ireland (1845-52) is a well-known example of this situation, where the fungus Phytophthora infestans caused havoc in the potato fields of Ireland (Badri and Vivanco, 2009). Exploring the root exudate composition of wild and invasive plants might help in deciphering novel root exudate traits for sustainable agriculture.
Photosynthetic machinery involving C3, C4, and CAM pathways categorizes the plants based on the specific pathway used during photosynthesis. The functioning mechanism of C3 and C4 plants vary based on the site of carbon fixation, and the initial stable compound forms and acts as a substrate during the dark reaction of photosynthesis, where C3 plants convert three carbon molecules (phosphoglycerate) and C4 plants convert four carbon molecules (oxaloacetate) into metabolically active organic compounds. The correlation between the root exudation pattern and the plant’s mode of photosynthesis shows that root exudate composition differs depending upon the mode of photosynthesis. The major constituents of root exudates vary in both the plant groups, and it has been found that C3 plants release a large amount of carbohydrates and organic carbons (Nabais et al., 2011), whereas C4 plants mostly secrete amino acids and organic acids as a major constituent of root exudates. In C3 plants, mainly mannose, maltose, and ribose constitute the exudates (Vranova et al., 2013), but in C4 plants, inositol, erythritol, and ribitol are found (Olanrewaju et al., 2019) (Figure 2). Variability in the exudation pattern of C3 and C4 plants might have contrasting effects in shaping the rhizospheric microbial community. C4 plants in comparison to C3 plants are more tolerant to adverse environmental situations, but the role played by root exudates in C4 plants’ adaptability is not yet clear. It is evident that under elevated CO2 levels, C4 plants allocate their excessive photosynthates in the rhizosphere with the help of root exudation, which leads to better AMF colonization finally resulting in enhanced nutrient acquisition by C4 plants (Frew and Price, 2019), which indicates the reason behind the better resilience of C4 plants in stress condition. Additionally, a large proportion of weeds fall under the category of C4 plants, exhibiting a wide range of adaptive behaviour under various environmental perturbations. Weeds secrete allelochemicals as their root exudates, attracting more beneficial microbes and resulting in better adaptation than the neighbouring crop plants (Trognitz et al., 2016; Trivedi et al., 2022).
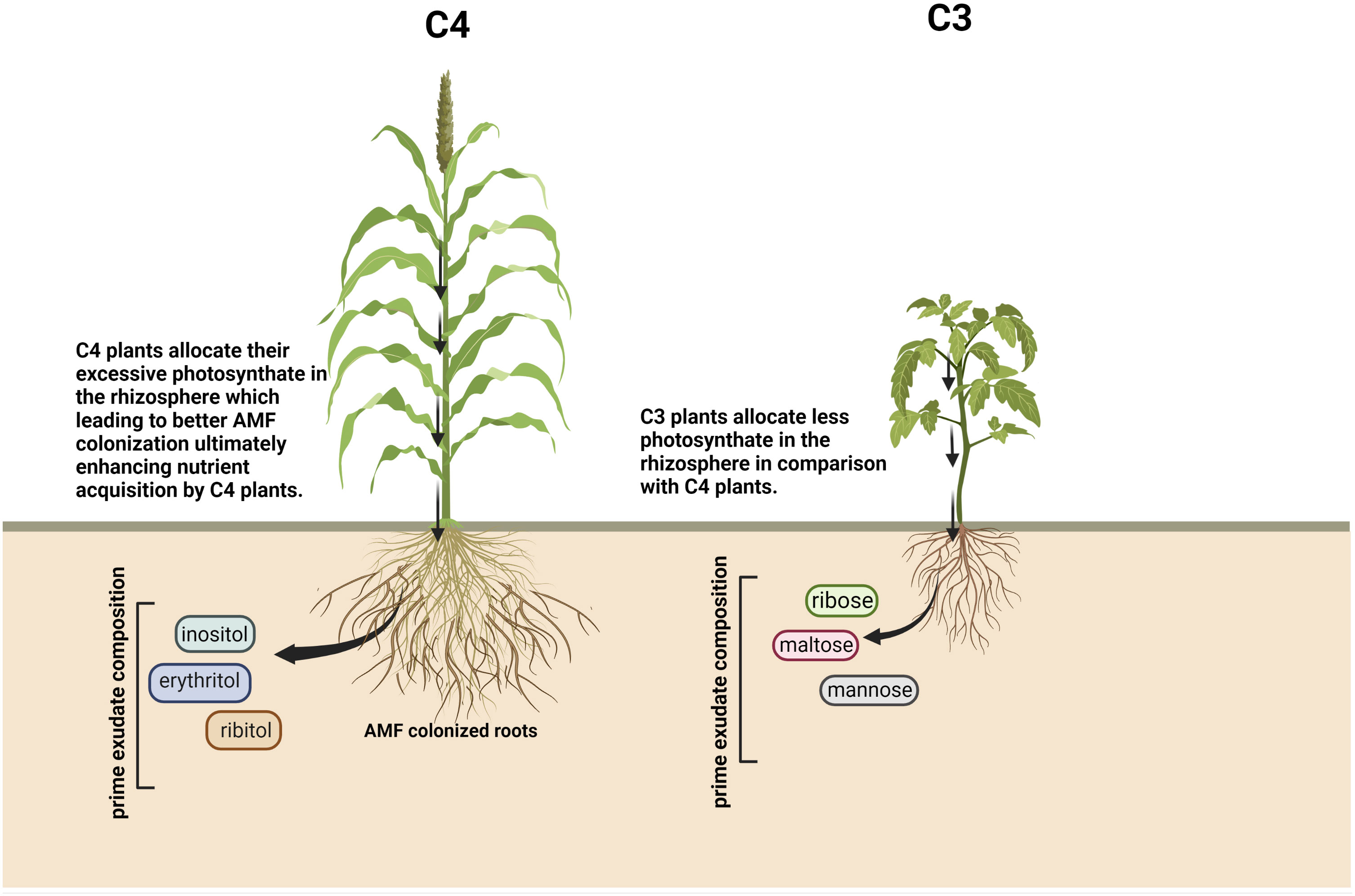
Figure 2 Schematic representation of root exudate compositions according to plants’ mode of photosynthesis. Created with BioRender.com.
C4 plants, like maize, induce JA-dependent resistance against herbivores (Spodoptera littoralis) due to the exudation of BX from the roots, which changes the rhizospheric microbial structure and makes the maize plants more resistant against the caterpillar attack (Hu et al., 2018). In contrast, tobacco plants infected with black shank disease caused by Phytophthora nicotianae can perform a pre-infection prevention strategy by exuding antimicrobial substances and lead to increased disease resistance by eliciting plant defence responses (Zhang et al., 2020). There are very few reports on studies elucidating the composition of the exudates according to the plant’s mode of photosynthesis and their influence on the rhizospheric microbial community. To have a comprehensive understanding, future research work on root exudate secretion mechanisms in C3 and C4 plants is warranted.
The secretion of root exudates as a signalling molecule can help in deciphering the plant’s stress status with the rhizospheric microbes and the neighbouring plants. Root exudates serve as organic carbon sources for rhizospheric microbes and their composition changes during plants’ interaction with pathogens or neighbouring plants either at above-ground or below-ground levels. Specific exudates are released under biotic stress conditions, thereby altering rhizospheric microbial community structure and promoting plant health by inducing SAR or ISR. Maintenance of a low-entropy state in the rhizosphere is important for creating equilibrium and promoting root–soil–microbe interactions by maximizing energy utilization, which optimizes localized nutrient supply. Further, a detailed understanding of entropy-based modulation of root exudates and the rhizomicrobiota in crops and crop wild relatives (CWRs) might pave the way for sustainable crop management. With the advances in molecular biology techniques, the root exudate compositions of various plants might be studied in detail under natural conditions. In addition, traits associated with root exudation of CWRs might have effective agronomic use in the future, as such type of traits might be exploited in the future to generate pest-resistant, fertilizer-independent, or weed-suppressive crops (Preece and Peñuelas, 2020). Furthermore, exploration of the rhizosphere microbiome structure of C3 and C4 plants under varied biotic stresses will help in designing specific agronomic strategies for these groups of plants. The molecular mechanisms governing the root exudation process are not very clear to date, but a detailed understanding of root exudation can provide a solution for effective stress management.
NA was responsible for writing guidance and manuscript preparation. IS and SK were responsible for writing the first draft of the review article. NA revised the article with IS and SK. All authors contributed to the article and approved the submitted version.
Start-Up grant provided by University Grant Commission, Govt. of India (grant number F.30-386/2017) and Scheme for promoting research among young faculty (GU/Acad./YFPGC/50/2018/1738-79/05) provided to NA by Gauhati University.
The authors are thankful to DST, Govt. of India, for providing DST-FIST Support to the Department of Botany, Gauhati University.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
BX, benzoxazinoids; ISR, induced systemic resistance; JA, jasmonic acid; PAMPs, pathogen-associated molecular patterns; PGPR, plant growth-promoting rhizobacteria; RSA, root system architecture; SA, salicylic acid; SAR, systemic acquired resistance; SCOAs, short-chain organic acids; SNS, social networking system; VOCs, volatile organic compounds.
Afridi, M. S., Fakhar, A., Kumar, A., Ali, S., Medeiros, F. H., Muneer, M. A., et al. (2022). Harnessing microbial multitrophic interactions for rhizosphere microbiome engineering. Microbiol. Res. 265, 127199. doi: 10.1016/j.micres.2022.127199
Akiyama, K., Hayashi, H. (2006). Strigolactones: Chemical signals for fungal symbionts and parasitic weeds in plant roots. Ann. Bot. 97 (6), 925–931. doi: 10.1093/aob/mcl063
Aznar, A., Dellagi, A. (2015). New insights into the role of siderophores as triggers of plant immunity: what can we learn from animals? J. Exp. Bot. 66 (11), 3001–3010. doi: 10.1093/jxb/erv155
Badri, D. V., Chaparro, J. M., Zhang, R., Shen, Q., Vivanco, J. M. (2013). Application of natural blends of phytochemicals derived from the root exudates of arabidopsis to the soil reveal that phenolic-related compounds predominantly modulate the soil microbiome. J. Biol. Chem. 288 (7), 4502–4512. doi: 10.1074/jbc.M112.433300
Badri, D. V., Vivanco, J. M. (2009). Regulation and function of root exudates. Plant Cell Environ. 32 (6), 666–681. doi: 10.1111/j.1365-3040.2009.01926.x
Badri, D. V., Weir, T. L., van der Lelie, D., Vivanco, J. M. (2009). Rhizosphere chemical dialogues: Plant–microbe interactions. Curr. Opin. Biotechnol. 20 (6), 642–650. doi: 10.1016/j.copbio.2009.09.014
Bais, H. P., Weir, T. L., Perry, L. G., Gilroy, S., Vivanco, J. M. (2006). The role of root exudates in rhizosphere interactions with plants and other organisms. Annu. Rev. Plant Biol. 57 (1), 233–266. doi: 10.1146/annurev.arplant.57.032905.105159
Bakker, P. A., Berendsen, R. L., Van Pelt, J. A., Vismans, G., Yu, K., Li, E., et al. (2020). The soil-borne identity and microbiome-assisted agriculture: Llooking back to the future. Mol. Plant 13 (10), 1394–1401. doi: 10.1016/j.molp.2020.09.017
Bakker, P. A., Ran, L., Mercado-Blanco, J. (2014). Rhizobacterial salicylate production provokes headaches! Plant Soil 382 (1), 1–16. doi: 10.1007/s11104-014-2102-0
Baluška, F., Mancuso, S. (2013). Root apex transition zone as oscillatory zone. Front. Plant Sci. 4. doi: 10.3389/fpls.2013.00354
Bari, R., Jones, J. D. (2009). Role of plant hormones in plant defence responses. Plant Mol. Biol. 69 (4), 473–488. doi: 10.1007/s11103-008-9435-0
Battey, N. H., Blackbourn, H. D. (1993). The control of exocytosis in plant cells. New Phytol. 125 (2), 307–338. doi: 10.1111/j.1469-8137.1993.tb03883.x
Beckers, G. J., Jaskiewicz, M., Liu, Y., Underwood, W. R., He, S. Y., Zhang, S., et al. (2009). Mitogen-activated protein kinases 3 and 6 are required for full priming of stress responses in arabidopsis thaliana. Plant Cell 21 (3), 944–953. doi: 10.1105/tpc.108.062158
Bednarek, P., Osbourn, A. (2009). Plant-microbe interactions: chemical diversity in plant defense. Science 324 (5928), 746–748.doi: 10.1126/science.1171661
Berendsen, R. L., Pieterse, C. M., Bakker, P. A. (2012). The rhizosphere microbiome and plant health. Trends Plant Sci. 17 (8), 478–486. doi: 10.1016/j.tplants.2012.04.001
Bhutia, D. D., Belbase, S., Paudel, J., Kumar, S. (2023). “Plant exudates and microbial interaction–a change in dynamics,” in Climate change and microbiome dynamics: Carbon cycle feedbacks (Cham: Springer International Publishing), 83–95.
Bouwmeester, H. J., Roux, C., Lopez-Raez, J. A., Becard, G. (2007). Rhizosphere communication of plants, parasitic plants and AM fungi. Trends Plant Sci. 12 (5), 224–230. doi: 10.1016/j.tplants.2007.03.009
Bressan, M., Roncato, M. A., Bellvert, F., Comte, G., Haichar, F. E. Z., Achouak, W., et al. (2009). Exogenous glucosinolate produced by arabidopsis thaliana has an impact on microbes in the rhizosphere and plant roots. ISME J. 3 (11), 1243–1257. doi: 10.1038/ismej.2009.68
Brian, G., Lea, P. J. (2007). Glutamate in plants: Metabolism, regulation, and signaling. J. Exp. Bot. 58 (9), 2339–2358. doi: 10.1093/jxb/erm121
Bulgarelli, D., Rott, M., Schlaeppi, K., Ver Loren van Themaat, E., Ahmadinejad, N., Assenza, F., et al. (2012). Revealing structure and assembly cues for arabidopsis root-inhabiting bacterial microbiota. Nature 488 (7409), 91–95. doi: 10.1038/nature11336
Canarini, A., Kaiser, C., Merchant, A., Richter, A., Wanek, W. (2019). Root exudation of primary metabolites: Mechanisms and their roles in plant responses to environmental stimuli. Front. Plant Sci. 10, 157. doi: 10.3389/fpls.2019.00157
Chaparro, J. M., Badri, D. V., Bakker, M. G., Sugiyama, A., Manter, D. K., Vivanco, J. M. (2013). Root exudation of phytochemicals in arabidopsis follows specific patterns that are developmentally programmed and correlate with soil microbial functions. PloS One 8 (2), e55731. doi: 10.1371/annotation/51142aed-2d94-4195-8a8a-9cb24b3c733b
Chater, K. F., Biró, S., Lee, K. J., Palmer, T., Schrempf, H. (2010). The complex extracellular biology of streptomyces. FEMS Microbiol. Rev. 34 (2), 171–198. doi: 10.1111/j.1574-6976.2009.00206.x
Chen, Y. T., Wang, Y., Yeh, K. C. (2017). Role of root exudates in metal acquisition and tolerance. Curr. Opin. Plant Biol. 39, 66–72. doi: 10.1016/j.pbi.2017.06.004
Chen, Y., Yan, F., Chai, Y., Liu, H., Kolter, R., Losick, R., et al. (2013). Biocontrol of tomato wilt disease by b acillus subtilis isolates from natural environments depends on conserved genes mediating biofilm formation. Environ. Microbiol. 15 (3), 848–864. doi: 10.1111/j.1462-2920.2012.02860.x
Chen, Y., Bonkowski, M., Shen, Y., Griffiths, B. S., Jiang, Y., Wang, X., et al. (2020). Root ethylene mediates rhizosphere microbial community reconstruction when chemically detecting cyanide produced by neighbouring plants. Microbiome 8 (1), 1–17. doi: 10.1186/s40168-019-0775-6
Cheol Song, G., Sim, H. J., Kim, S. G., Ryu, C. M. (2016). Root-mediated signal transmission of systemic acquired resistance against above-ground and below-ground pathogens. Ann. Bot. 118 (4), 821–831. doi: 10.1093/aob/mcw152
Dastogeer, K. M., Tumpa, F. H., Sultana, A., Akter, M. A., Chakraborty, A. (2020). Plant microbiome–an account of the factors that shape community composition and diversity. Curr. Plant Biol. 23, 100161. doi: 10.1016/j.cpb.2020.100161
Degenhardt, J., Hiltpold, I., Köllner, T. G., Frey, M., Gierl, A., Gershenzon, J., et al. (2009). Restoring a maize root signal that attracts insect-killing nematodes to control a major pest. Proc. Natl. Acad. Sci. 106 (32), 13213–13218. doi: 10.1073/pnas.0906365106
De-la-Peña, C., Lei, Z., Watson, B. S., Sumner, L. W., Vivanco, J. M. (2008). Root-microbe communication through protein secretion. J. Biol. Chem. 283 (37), 25247–25255. doi: 10.1074/jbc.M801967200
de la Porte, A., Schmidt, R., Yergeau, É., Constant, P. (2020). A gaseous milieu: extending the boundaries of the rhizosphere. Trends Microbiol. 28 (7), 536–542. doi: 10.1016/j.tim.2020.02.016
Dematheis, F., Zimmerling, U., Flocco, C., Kurtz, B., Vidal, S., Kropf, S., et al. (2012). Multitrophic interaction in the rhizosphere of maize: Root feeding of western corn rootworm larvae alters the microbial community composition. PloS One 7 (5), e37288. doi: 10.1371/journal.pone.0037288
De Schepper, V., De Swaef, T., Bauweraerts, I., Steppe, K. (2013). Phloem transport: A review of mechanisms and controls. J. Exp. Bot. 64 (16), 4839–4850. doi: 10.1093/jxb/ert302
Doornbos, R. F., van Loon, L. C., Bakker, P. A. (2012). Impact of root exudates and plant defense signaling on bacterial communities in the rhizosphere. a review. Agron. Sustain. Dev. 32 (1), 227–243. doi: 10.1007/s13593-011-0028-y
Dowarah, B., Gill, S. S., Agarwala, N. (2021). Arbuscular mycorrhizal fungi in conferring tolerance to biotic stresses in plants. J. Plant Growth Regul., 1–16. doi: 10.1007/s00344-021-10392-5
Du, Y., Zou, W., Zhang, K., Ye, G., Yang, J. (2020). Advances and applications of clostridium co-culture systems in biotechnology. Front. Microbiol. 11, 560223. doi: 10.3389/fmicb.2020.560223
Erb, M., Lenk, C., Degenhardt, J., Turlings, T. C. (2009). The underestimated role of roots in defense against leaf attackers. Trends Plant Sci. 14 (12), 653–659. doi: 10.1016/j.tplants.2009.08.006
Fantaye, C. A., Köpke, D., Gershenzon, J., Degenhardt, J. (2015). Restoring (E)-β-caryophyllene production in a non-producing maize line compromises its resistance against the fungus colletotrichum graminicola. J. Chem. Ecol. 41 (3), 213–223. doi: 10.1007/s10886-015-0556-z
Farrar, J. F., Jones, D. L. (2000). The control of carbon acquisition by roots. New Phytol. 147 (1), 43–53. doi: 10.1046/j.1469-8137.2000.00688.x
Fernández, I., Cosme, M., Stringlis, I. A., Yu, K., de Jonge, R., van Wees, S. M., et al. (2019). Molecular dialogue between arbuscular mycorrhizal fungi and the nonhost plant arabidopsis thaliana switches from initial detection to antagonism. New Phytol. 223 (2), 867–881. doi: 10.1111/nph.15798
Fierer, N., Bradford, M. A., Jackson, R. B. (2007). Toward an ecological classification of soil bacteria. Ecology 88 (6), 1354–1364. doi: 10.1890/05-1839
Fourcroy, P., Sisó-Terraza, P., Sudre, D., Savirón, M., Reyt, G., Gaymard, F., et al. (2014). Involvement of the ABCG 37 transporter in secretion of scopoletin and derivatives by arabidopsis roots in response to iron deficiency. New Phytol. 201 (1), 155–167. doi: 10.1111/nph.12471
Frew, A., Price, J. N. (2019). Mycorrhizal-mediated plant–herbivore interactions in a high CO2 world. Funct. Ecol. 33 (8), 1376–1385. doi: 10.1111/1365-2435.13347
Garbeva, P., Van Elsas, J. D., Van Veen, J. A. (2008). Rhizosphere microbial community and its response to plant species and soil history. Plant Soil 302 (1), 19–32. doi: 10.1007/s11104-007-9432-0
Gulati, S., Ballhausen, M. B., Kulkarni, P., Grosch, R., Garbeva, P. (2020). A non-invasive soil-based setup to study tomato root volatiles released by healthy and infected roots. Sci. Rep. 10 (1), 1–11. doi: 10.1038/s41598-020-69468-z
Haldar, S., Sengupta, S. (2015). Plant-microbe cross-talk in the rhizosphere: insight and biotechnological potential. Open Microbiol. J. 9, 1. doi: 10.2174/1874285801509010001
Haydon, M. J., Cobbett, C. S. (2007). Transporters of ligands for essential metal ions in plants. New Phytol. 174 (3), 499–506. doi: 10.1111/j.1469-8137.2007.02051.x
Hofmeister, A. E., Grabowski, R., Linder, D., Buckler, W. (1993). L-serine and l-threonine dehydratase from clostridium propionicum two enzymes with different prosthetic groups. Eur. J. Biochem. 215 (2), 341–349. doi: 10.1111/j.1432-1033.1993.tb18040.x
Horiuchi, J. I., Prithiviraj, B., Bais, H. P., Kimball, B. A., Vivanco, J. M. (2005). Soil nematodes mediate positive interactions between legume plants and rhizobium bacteria. Planta 222 (5), 848–857. doi: 10.1007/s00425-005-0025-y
Hoysted, G. A., Bell, C. A., Lilley, C. J., Urwin, P. E. (2018). Aphid colonization affects potato root exudate composition and the hatching of a soil borne pathogen. Front. Plant Sci. 9, 1278. doi: 10.3389/fpls.2018.01278
Hsieh, E. J., Waters, B. M. (2016). Alkaline stress and iron deficiency regulate iron uptake and riboflavin synthesis gene expression differently in root and leaf tissue: implications for iron deficiency chlorosis. J. Exp. Bot. 67 (19), 5671–5685. doi: 10.1093/jxb/erw328
Hu, D., Baskin, J. M., Baskin, C. C., Liu, R., Yang, X., Huang, Z. (2021). A seed mucilage-degrading fungus from the rhizosphere strengthens the plant-Soil-Microbe continuum and potentially regulates root nutrients of a cold desert shrub. Mol. Plant-Microbe Interact. 34 (5), 538–546. doi: 10.1094/MPMI-01-21-0014-FI
Hu, L., Robert, C. A., Cadot, S., Zhang, X. I., Ye, M., Li, B., et al. (2018). Root exudate metabolites drive plant-soil feedbacks on growth and defense by shaping the rhizosphere microbiota. Nat. Commun. 9 (1), 1–13. doi: 10.1038/s41467-018-05122-7
İnceoğlu, Ö., Falcão Salles, J., van Elsas, J. D. (2012). Soil and cultivar type shape the bacterial community in the potato rhizosphere. Microb. Ecol. 63 (2), 460–470. doi: 10.1007/s00248-011-9930-8
Jousset, A., Rochat, L., Lanoue, A., Bonkowski, M., Keel, C., Scheu, S. (2011). Plants respond to pathogen infection by enhancing the antifungal gene expression of root-associated bacteria. Mol. Plant-Microbe Interact. 24 (3), 352–358. doi: 10.1094/MPMI-09-10-0208
Koprivova, A., Schuck, S., Jacoby, R. P., Klinkhammer, I., Welter, B., Leson, L., et al. (2019). Root-specific camalexin biosynthesis controls the plant growth-promoting effects of multiple bacterial strains. Proc. Natl. Acad. Sci. 116 (31), 15735–15744. doi: 10.1073/pnas.1818604116
Lakshmanan, V., Kitto, S. L., Caplan, J. L., Hsueh, Y. H., Kearns, D. B., Wu, Y. S., et al. (2012). Microbe-associated molecular patterns-triggered root responses mediate beneficial rhizobacterial recruitment in arabidopsis. Plant Physiol. 160 (3), 1642–1661. doi: 10.1104/pp.112.200386
Lauber, C. L., Hamady, M., Knight, R., Fierer, N. (2009). Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl. Environ. Microbiol. 75 (15), 5111–5120. doi: 10.1128/AEM.00335-09
Lee, B., Lee, S., Ryu, C. M. (2012). Foliar aphid feeding recruits rhizosphere bacteria and primes plant immunity against pathogenic and non-pathogenic bacteria in pepper. Ann. Bot. 110 (2), 281–290. doi: 10.1093/aob/mcs055
Lendzemo, V. W., Kuyper, T. W., Matusova, R., Bouwmeester, H. J., Ast, A. V. (2007). Colonization by arbuscular mycorrhizal fungi of sorghum leads to reduced germination and subsequent attachment and emergence of striga hermonthica. Plant Signaling Behav. 2 (1), 58–62. doi: 10.4161/psb.2.1.3884
Li, E., de Jonge, R., Liu, C., Jiang, H., Friman, V. P., Pieterse, C. M., et al. (2021a). Rapid evolution of bacterial mutualism in the plant rhizosphere. Nat. Commun. 12 (1), 1–13. doi: 10.1038/s41467-021-24005-y
Li, J., Wang, C., Liang, W., Liu, S. (2021b). Rhizosphere microbiome: The emerging barrier in plant-pathogen interactions. Front. Microbiol. 12, 772420. doi: 10.3389/fmicb.2021.772420
Ling, N., Wang, T., Kuzyakov, Y. (2022). Rhizosphere bacteriome structure and functions. Nat. Commun. 13 (1), 1–13. doi: 10.1038/s41467-022-28448-9
Luo, H., Xu, H., Chu, C., He, F., Fang, S. (2020). High temperature can change root system architecture and intensify root interactions of plant seedlings. Front. Plant Sci. 11, 160. doi: 10.3389/fpls.2020.00160
Malacrinò, A., Wang, M., Caul, S., Karley, A. J., Bennett, A. E. (2021). Herbivory shapes the rhizosphere bacterial microbiota in potato plants. Environ. Microbiol. Rep. 13 (6), 805–811. doi: 10.1111/1758-2229.12998
Massalha, H., Korenblum, E., Malitsky, S., Shapiro, O. H., Aharoni, A. (2017). Live imaging of root–bacteria interactions in a microfluidics setup. Proc. Natl. Acad. Sci. 114 (17), 4549–4554. doi: 10.1073/pnas.1618584114
Meyer, S., De Angeli, A., Fernie, A. R., Martinoia, E. (2010). Intra-and extra-cellular excretion of carboxylates. Trends Plant Sci. 15 (1), 40–47. doi: 10.1016/j.tplants.2009.10.002
Mhlongo, M. I., Piater, L. A., Madala, N. E., Labuschagne, N., Dubery, I. A. (2018). The chemistry of plant–microbe interactions in the rhizosphere and the potential for metabolomics to reveal signaling related to defense priming and induced systemic resistance. Front. Plant Sci. 9, 112. doi: 10.3389/fpls.2018.00112
Millet, Y. A., Danna, C. H., Clay, N. K., Songnuan, W., Simon, M. D., Werck-Reichhart, D., et al. (2010). Innate immune responses activated in arabidopsis roots by microbe-associated molecular patterns. Plant Cell 22 (3), 973–990. doi: 10.1105/tpc.109.069658
Mishra, S., Chauhan, P. S., Goel, A. K., Upadhyay, R. S., Nautiyal, C. S. (2012a). Pseudomonas putida NBRIC19 provides protection to neighboring plant diversity from invasive weed parthenium hysterophorus l. by altering soil microbial community. Acta physiol. plantarum 34 (6), 2187–2195. doi: 10.1007/s11738-012-1019-6
Mishra, S., Mishra, A., Chauhan, P. S., Mishra, S. K., Kumari, M., Niranjan, A., et al. (2012b). Pseudomonas putida NBRIC19 dihydrolipoamide succinyltransferase (SucB) gene controls degradation of toxic allelochemicals produced by parthenium hysterophorus. J. Appl. Microbiol. 112 (4), 793–808. doi: 10.1111/j.1365-2672.2012.05256.x
Mishra, S., Nautiyal, C. S. (2012). Reducing the allelopathic effect of parthenium hysterophorus l. @ on wheat (Triticum aestivum l.) by pseudomonas putida. Plant Growth Regul. 66 (2), 155–165. doi: 10.1007/s10725-011-9639-1
Mönchgesang, S., Strehmel, N., Schmidt, S., Westphal, L., Taruttis, F., Müller, et al. (2016). Natural variation of root exudates in arabidopsis thaliana-linking metabolomic and genomic data. Sci. Rep. 6 (1), 1–11. doi: 10.1038/srep29033
Münch, E. (1930). Die stoffbewegungen in der pflanze (Gustav Fischer, jena). Curr. Opin. Plant Biol. 43, 36–42.
Mur, L. A., Kenton, P., Atzorn, R., Miersch, O., Wasternack, C. (2006). The outcomes of concentration-specific interactions between salicylate and jasmonate signaling include synergy, antagonism, and oxidative stress leading to cell death. Plant Physiol. 140 (1), 249–262. doi: 10.1104/pp.105.072348
Nabais, C., Labuto, G., Gonçalves, S., Buscardo, E., Semensatto, D., Nogueira, A. R. A., et al. (2011). Effect of root age on the allocation of metals, amino acids and sugars in different cell fractions of the perennial grass paspalum notatum (bahiagrass). Plant Physiol. Biochem. 49 (12), 1442–1447. doi: 10.1016/j.plaphy.2011.09.010
Nazoa, P., Vidmar, J. J., Tranbarger, T. J., Mouline, K., Damiani, I., Tillard, P., et al. (2003). Regulation of the nitrate transporter gene AtNRT2. 1 in arabidopsis thaliana: responses to nitrate, amino acids and developmental stage. Plant Mol. Biol. 52 (3), 689–703. doi: 10.1023/a:1024899808018
Ninkovic, V., Markovic, D., Dahlin, I. (2016). Decoding neighbour volatiles in preparation for future competition and implications for tritrophic interactions. Perspect. Plant Ecology Evol. Systematics 23, 11–17. doi: 10.1016/j.ppees.2016.09.005
Nguyen, C. (2009). Rhizodeposition of organic c by plant: mechanisms and controls. Sustain. Agric., 97–123. doi: 10.1007/978-90-481-2666-8_9
Ni, J., Yu, Z., Du, G., Zhang, Y., Taylor, J. L., Shen, C., et al. (2016). Heterologous expression and functional analysis of rice GLUTAMATE RECEPTOR-LIKE family indicates its role in glutamate triggered calcium flux in rice roots. Rice 9 (1), 1–14. doi: 10.1186/s12284-016-0081-x
Niemeyer, H. M. (2009). Hydroxamic acids derived from 2-hydroxy-2 h-1, 4-benzoxazin-3 (4 h)-one: Key defense chemicals of cereals. J. Agric. Food Chem. 57 (5), 1677–1696. doi: 10.1021/jf8034034
Nozoye, T., Nagasaka, S., Kobayashi, T., Takahashi, M., Sato, Y., Sato, Y., et al. (2011). Phytosiderophore efflux transporters are crucial for iron acquisition in graminaceous plants. J. Biol. Chem. 286 (7), 5446–5454. doi: 10.1074/jbc.M110.180026
Nozoye, T., Nakanishi, H., Nishizawa, N. K. (2013). Characterizing the crucial components of iron homeostasis in the maize mutants ys1 and ys3. PloS One 8 (5), e62567. doi: 10.1371/journal.pone.0062567
Olanrewaju, O. S., Ayangbenro, A. S., Glick, B. R., Babalola, O. O. (2019). Plant health: feedback effect of root exudates-rhizobiome interactions. Appl. Microbiol. Biotechnol. 103 (3), 1155–1166. doi: 10.1007/s00253-018-9556-6
Ourry, M., Lebreton, L., Chaminade, V., Guillerm-Erckelboudt, A. Y., Hervé, M., Linglin, J., et al. (2018). Influence of belowground herbivory on the dynamics of root and rhizosphere microbial communities. Front. Ecol. Evol. 6, 91. doi: 10.3389/fevo.2018.00091
Park, Y. S., Ryu, C. M. (2021). Understanding plant social networking system: Avoiding deleterious microbiota but calling beneficials. Int. J. Mol. Sci. 22 (7), 3319. doi: 10.3390/ijms22073319
Paterson, E., Sim, A., Standing, D., Dorward, M., McDonald, A. J. S. (2006). Root exudation from hordeum vulgare in response to localized nitrate supply. J. Exp. Bot. 57 (10), 2413–2420. doi: 10.1093/jxb/erj214
Philippot, L., Raaijmakers, J. M., Lemanceau, P., Van Der Putten, W. H. (2013). Going back to the roots: The microbial ecology of the rhizosphere. Nat. Rev. Microbiol. 11 (11), 789–799. doi: 10.1038/nrmicro3109
Phour, M., Sehrawat, A., Sindhu, S. S., Glick, B. R. (2020). Interkingdom signaling in plant-rhizomicrobiome interactions for sustainable agriculture. Microbiol. Res. 241, 126589. doi: 10.1016/j.micres.2020.126589
Pieterse, C. M., de Jonge, R., Berendsen, R. L. (2016). The soil-borne supremacy. Trends Plant Sci. 21 (3), 171–173. doi: 10.1016/j.tplants.2016.01.018
Pieterse, C. M., Zamioudis, C., Berendsen, R. L., Weller, D. M., Van Wees, S. C., Bakker, P. A. (2014). Induced systemic resistance by beneficial microbes. Annu. Rev. Phytopathol. 52, 347–375. doi: 10.1146/annurev-phyto-082712-102340
Pineda, A., Zheng, S. J., van Loon, J. J., Pieterse, C. M., Dicke, M. (2010). Helping plants to deal with insects: the role of beneficial soil-borne microbes. Trends Plant Sci. 15 (9), 507–514. doi: 10.1016/j.tplants.2010.05.007
Preece, C., Peñuelas, J. (2020). A return to the wild: root exudates and food security. Trends Plant Sci. 25 (1), 14–21. doi: 10.1016/j.tplants.2019.09.010
Rasmann, S., Köllner, T. G., Degenhardt, J., Hiltpold, I., Toepfer, S., Kuhlmann, U., et al. (2005). Recruitment of entomopathogenic nematodes by insect-damaged maize roots. Nature 434 (7034), 732–737. doi: 10.1038/nature03451
Rasmann, S., Turlings, T. C. (2016). Root signals that mediate mutualistic interactions in the rhizosphere. Curr. Opin. Plant Biol. 32, 62–68. doi: 10.1016/j.pbi.2016.06.017
Rellán-Álvarez, R., Andaluz, S., Rodríguez-Celma, J., Wohlgemuth, G., Zocchi, G., Álvarez-Fernández, A., et al. (2010). Changes in the proteomic and metabolic profiles of beta vulgaris root tips in response to iron deficiency and resupply. BMC Plant Biol. 10 (1), 1–15. doi: 10.1186/1471-2229-10-120
Rodríguez-Celma, J., Lin, W. D., Fu, G. M., Abadia, J., López-Millán, A. F., Schmidt, W. (2013). Mutually exclusive alterations in secondary metabolism are critical for the uptake of insoluble iron compounds by arabidopsis and medicago truncatula. Plant Physiol. 162 (3), 1473–1485. doi: 10.1104/pp.113.220426
Rodríguez-Celma, J., Vázquez-Reina, S., Orduna, J., Abadía, A., Abadía, J., Álvarez-Fernández, A., et al. (2011). Characterization of flavins in roots of fe-deficient strategy i plants, with a focus on medicago truncatula. Plant Cell Physiol. 52 (12), 2173–2189. doi: 10.1093/pcp/pcr149
Rolfe, S. A., Griffiths, J., Ton, J. (2019). Crying out for help with root exudates: adaptive mechanisms by which stressed plants assemble health-promoting soil microbiomes. Curr. Opin. Microbiol. 49, 73–82. doi: 10.1016/j.mib.2019.10.003
Ross-Elliott, T. J., Jensen, K. H., Haaning, K. S., Wager, B. M., Knoblauch, J., Howell, A. H., et al. (2017). Phloem unloading in arabidopsis roots is convective and regulated by the phloem-pole pericycle. elife 6, e24125. doi: 10.7554/eLife.24125.022
Rudrappa, T., Czymmek, K. J., Paré, P. W., Bais, H. P. (2008). Root-secreted malic acid recruits beneficial soil bacteria. Plant Physiol. 148 (3), 1547–1556. doi: 10.1104/pp.108.127613
Sarita, S., Shaikh, NB, Shaikh, N, Rochlani, A, Dalwani, A, Sharma, S, et al. (2022). Rhizobacteria that promote plant growth and their impact on root system architecture, root development, and function. Acta Sci. Microbiol. 5 (4), 53–62. doi: 10.31080/ASMI.2022.05.1035
Sasse, J., Martinoia, E., Northen, T. (2018). Feed your friends: do plant exudates shape the root microbiome? Trends Plant Sci. 23 (1), 25–41. doi: 10.1016/j.tplants.2017.09.003
Savage, J. A., Clearwater, M. J., Haines, D. F., Klein, T., Mencuccini, M., Sevanto, S., et al. (2016). Allocation, stress tolerance and carbon transport in plants: How does phloem physiology affect plant ecology? Plant Cell Environ. 39 (4), 709–725. doi: 10.1111/pce.12602
Schneider, T., Keiblinger, K. M., Schmid, E., Sterflinger-Gleixner, K., Ellersdorfer, G., Roschitzki, B., et al. (2012). Who is who in litter decomposition? metaproteomics reveals major microbial players and their biogeochemical functions. ISME J. 6 (9), 1749–1762. doi: 10.1038/ismej.2012.11
Schrempf, H., Dworkin, M., Falkow, S., Rosenberg, E., Schleifer, K. H., Stackebrandt, E. (2007). “Biology of streptomycetes,” in The prokaryotes, a handbook on the biology of bacteria, 3rd edn. Eds. Dworkin, M., Falkow, S., Rosenberg, E., Schleifer, K. H., Stackebrandt, E. (New York, NY: Springer Verlag).
Schulz-Bohm, K., Gerards, S., Hundscheid, M., Melenhorst, J., de Boer, W., Garbeva, P. (2018). Calling from distance: attraction of soil bacteria by plant root volatiles. ISME J. 12 (5), 1252–1262. doi: 10.1038/s41396-017-0035-3
Sharifi, R., Ryu, C. M. (2021). Social networking in crop plants: Wired and wireless cross-plant communications. Plant Cell Environ. 44 (4), 1095–1110. doi: 10.1111/pce.13966
Stotz, H. U., Sawada, Y., Shimada, Y., Hirai, M. Y., Sasaki, E., Krischke, M., et al. (2011). Role of camalexin, indole glucosinolates, and side chain modification of glucosinolate-derived isothiocyanates in defense of arabidopsis against sclerotinia sclerotiorum. Plant J. 67 (1), 81–93. doi: 10.1111/j.1365-313X.2011.04578.x
Trivedi, P., Batista, B. D., Bazany, K. E., Singh, B. K. (2022). Plant–microbiome interactions under a changing world: Responses, consequences and perspectives. New Phytol 234 (6), 1951–1959. doi: 10.1111/nph.18016
Trognitz, F., Hackl, E., Widhalm, S., Sessitsch, A. (2016). The role of plant–microbiome interactions in weed establishment and control. FEMS Microbiol. Ecol. 92 (10). doi: 10.1093/femsec/fiw138
Ulbrich, T. C., Rivas-Ubach, A., Tiemann, L. K., Friesen, M. L., Evans, S. E. (2022). Plant root exudates and rhizosphere bacterial communities shift with neighbor context. Soil Biol. Biochem. 172, 108753. doi: 10.1016/j.soilbio.2022.108753
Uroz, S., Buée, M., Murat, C., Frey-Klett, P., Martin, F. (2010). Pyrosequencing reveals a contrasted bacterial diversity between oak rhizosphere and surrounding soil. Environ. Microbiol. Rep. 2 (2), 281–288. doi: 10.1111/j.1758-2229.2009.00117.x
Usman, M., Ho-Plágaro, T., Frank, H. E., Calvo-Polanco, M., Gaillard, I., Garcia, K., et al. (2021). Mycorrhizal symbiosis for better adaptation of trees to abiotic stress caused by climate change in temperate and Boreal forests. Front. Forests Global Change 141. doi: 10.3389/ffgc.2021.742392
van der Heijden, M. G., Martin, F. M., Selosse, M. A., Sanders, I. R. (2015). Mycorrhizal ecology and evolution: the past, the present, and the future. New Phytol. 205 (4), 1406–1423. doi: 10.1111/nph.13288
Vincill, E. D., Bieck, A. M., Spalding, E. P. (2012). Ca2+ conduction by an amino acid-gated ion channel related to glutamate receptors. Plant Physiol. 159 (1), 40–46. doi: 10.1104/pp.112.197509
Vorwieger, A., Gryczka, C., Czihal, A., Douchkov, D., Tiedemann, J., Mock, H. P., et al. (2007). Iron assimilation and transcription factor controlled synthesis of riboflavin in plants. Planta 226 (1), 147–158. doi: 10.1007/s00425-006-0476-9
Vranova, V., Rejsek, K., Skene, K. R., Janous, D., Formanek, P. (2013). Methods of collection of plant root exudates in relation to plant metabolism and purpose: A review. J. Plant Nutr. Soil Sci. 176 (2), 175–199. doi: 10.1002/jpln.201000360
Wang, B., Qiu, Y. L. (2006). Phylogenetic distribution and evolution of mycorrhizas in land plants. Mycorrhiza 16 (5), 299–363. doi: 10.1007/s00572-005-0033-6
Wasternack, C., Hause, B. (2013). Jasmonates: Biosynthesis, perception, signal transduction and action in plant stress response, growth and development. an update to the 2007 review in annals of botany. Ann. Bot. 111 (6), 1021–1058. doi: 10.1093/aob/mct067
Wen, T., Zhao, M., Yuan, J., Kowalchuk, G. A., Shen, Q. (2021). Root exudates mediate plant defense against foliar pathogens by recruiting beneficial microbes. Soil Ecol. Lett. 3 (1), 42–51. doi: 10.1007/s42832-020-0057-z
Weng, W., Yan, J., Zhou, M., Yao, X., Gao, A., Ma, C., et al. (2022). Roles of arbuscular mycorrhizal fungi as a biocontrol agent in the control of plant diseases. Microorganisms 10 (7), 1266. doi: 10.3390/microorganisms10071266
Wentzell, A. M., Kliebenstein, D. J. (2008). Genotype, age, tissue, and environment regulate the structural outcome of glucosinolate activation. Plant Physiol. 147 (1), 415–428. doi: 10.1104/pp.107.115279
Xu, X., Chen, Y., Li, B., Zhang, Z., Qin, G., Chen, T., et al. (2022). Molecular mechanisms underlying multi-level defense responses of horticultural crops to fungal pathogens. Horticulture Res. 9. doi: 10.1093/hr/uhac066
Xu, X., Fang, P., Zhang, H., Chi, C., Song, L., Xia, X., et al. (2019). Strigolactones positively regulate defense against root-knot nematodes in tomato. J. Exp. Bot. 70 (4), 1325–1337. doi: 10.1093/jxb/ery439
Xu, S., Liao, C. J., Jaiswal, N., Lee, S., Yun, D. J., Lee, S. Y., et al. (2018). Tomato PEPR1 ORTHOLOG RECEPTOR-LIKE KINASE1 regulates responses to systemin, necrotrophic fungi, and insect herbivory. Plant Cell 30 (9), 2214–2229. doi: 10.1105/tpc.17.00908
Yang, H., Zhang, Q., Dai, Y., Liu, Q., Tang, J., Bian, X., et al. (2015). Effects of arbuscular mycorrhizal fungi on plant growth depend on root system: a meta-analysis. Plant Soil 389 (1), 361–374. doi: 10.1007/s11104-014-2370-8
Yuan, J., Zhao, J., Wen, T., Zhao, M., Li, R., Goossens, P., et al. (2018). Root exudates drive the soil-borne legacy of aboveground pathogen infection. Microbiome 6 (1), 1–12. doi: 10.1186/s40168-018-0537-x
Zhang, C., Feng, C., Zheng, Y., Wang, J., Wang, F. (2020). Root exudates metabolic profiling suggests distinct defense mechanisms between resistant and susceptible tobacco cultivars against black shank disease. Front. Plant Sci. 11, 559775. doi: 10.3389/fpls.2020.559775
Zhang, K., Rengel, Z., Zhang, F., White, P. J., Shen, J. (2022). Rhizosphere engineering for sustainable crop production: Entropy-based insights. Trends Plant Sci, S1360-1385(22)00307-7. Advance online publication. doi: 10.1016/j.tplants.2022.11.008
Keywords: biotic factors, rhizospheric microbial community, root exudates, stress tolerance, plant microbiome
Citation: Sharma I, Kashyap S and Agarwala N (2023) Biotic stress-induced changes in root exudation confer plant stress tolerance by altering rhizospheric microbial community. Front. Plant Sci. 14:1132824. doi: 10.3389/fpls.2023.1132824
Received: 28 December 2022; Accepted: 31 January 2023;
Published: 10 March 2023.
Edited by:
Pramod Ramteke, Sam Higginbottom University of Agriculture, Technology and Sciences, IndiaReviewed by:
Pranab Dutta, Central Agricultural University, IndiaCopyright © 2023 Sharma, Kashyap and Agarwala. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Niraj Agarwala, bmlyYWpfYm90YW55QGdhdWhhdGkuYWMuaW4=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.