- 1ICAR-All India Coordinated Research Project (ICAR-AICRP) on Small Millets, PC Unit, University of Agricultural Sciences, Gandhi Krishi Vigyana Kendra (GKVK), Bengaluru, Karnataka, India
- 2Department of Plant Biotechnology, University of Agricultural Sciences, Gandhi Krishi Vigyana Kendra (GKVK), Bengaluru, Karnataka, India
- 3Department of Plant Pathology, Vivekananda Parvatiya Krishi Anusandhan Sansthan, Almora, Uttarakhand, India
- 4ICAR-National Rice Research Institute, Cuttack, Odisha, India
- 5ICAR-All India Coordinated Research Project (ICAR-AICRP) on Small Millets Zonal Agril. Research Station, Vishweshwaraiah Canal (V.C.) Farm, Mandya, Karnataka, India
- 6Regional Agricultural Research Station, Assam Agriculture University, Gossaigaon, Assam, India
- 7Department of Plant Pathology, Agricultural Research Station, Gajularega, Vizianagaram, Andra Pradesh, India
- 8Department of Plant Pathology, Uttarakhand University of Hort. and Forestry, Ranichauri, Uttarakhand, India
- 9Department of Plant Pathology, Center for Excellence in Millets, Athiyandal, Tiruvannamalai, Tamil Nadu, India
- 10Department of Plant Pathology, Zonal Agricultural Research Station, Kumharwand Farm, Jagdalpur, Chhattisgarh, India
- 11Indian Council of Agricultural Research ICAR-Indian Institute of Millets Research, Rajendranagar, Hyderabad, Telangana, India
- 12Hill Millet Research Station, Navasari Agricultural University, Waghai, Dangs, Gujarat, India
- 13Department of Plant Pathology, College of Agriculture, Rewa, Madhya Pradesh, India
- 14ICAR-All India Coordinated Research Project (ICAR-AICRP) on Small Millets, Regional Agricultural Research Station, Nandyal, Andhra Pradesh, India
- 15Institute of Excellence, Vijnana Bhavan, University of Mysuru, Manasagangotri, Karnataka, India
- 16ICAR-Indian Agricultural Research Institute, New Delhi, India
Blast pathogen, Magnaporthe spp., that infects ancient millet crops such pearl millet, finger millet, foxtail millet, barnyard millet, and rice was isolated from different locations of blast hotspots in India using single spore isolation technique and 136 pure isolates were established. Numerous growth characteristics were captured via morphogenesis analysis. Among the 10 investigated virulent genes, we could amplify MPS1 (TTK Protein Kinase) and Mlc (Myosin Regulatory Light Chain edc4) in majority of tested isolates, regardless of the crop and region where they were collected, indicating that these may be crucial for their virulence. Additionally, among the four avirulence (Avr) genes studied, Avr-Pizt had the highest frequency of occurrence, followed by Avr-Pia. It is noteworthy to mention that Avr-Pik was present in the least number of isolates (9) and was completely absent from the blast isolates from finger millet, foxtail millet, and barnyard millet. A comparison at the molecular level between virulent and avirulent isolates indicated observably large variation both across (44%) and within (56%) them. The 136 Magnaporthe spp isolates were divided into four groups using molecular markers. Regardless of their geographic distribution, host plants, or tissues affected, the data indicate that the prevalence of numerous pathotypes and virulence factors at the field level, which may lead to a high degree of pathogenic variation. This research could be used for the strategic deployment of resistant genes to develop blast disease-resistant cultivars in rice, pearl millet, finger millet, foxtail millet, and barnyard millet.
Introduction
Millets are one of the earliest food crops that humans have grown, but due to urbanization and industrialization, they were neglected in favor of wheat and rice, which are most widely consumed as food and forage crops in the semi-arid regions and is highly crucial for the global food security and farmers livelihood. Globally, 30.73 million tons of these ancient millets are produced, and of which 11.42 million tons (or 37% of global production) is contributed by India (http://www.fao.org). These millets are considered as the most promising grains for preventing hunger and malnutrition besides the guaranteeing their widespread access across the globe (Gupta, 2006). Considering the importance of millets to the farmers’, consumers and the environment, Government of India has laid an initiative for popularization of these crops by celebrating the international year of Millets, through the United Nations, and this has garnered support from many countries. The support from Government of India in promoting millets production and increasing the area under millets reflects that these are the future crops for nutritional and food security.
Production and productivity of millets has been majorly succumbing to various abiotic and biotic factors that are set to be increasing day by day due to present global climate change scenario. The blast disease, which is incited by Magnoporthe spp., is a most destructive biotic constraint affecting the millets, including rice. It affects in all the growth stages of plants especially different aerial parts of the plants, from the seedling stage to seed setting in finger millet, causing neck and/or finger blast, while it is restricted to the leaf only in the case of pearl millet, foxtail millet, and barnyard millet (Palanna et al., 2021; Rajashekara et al., 2021). Blast is found in most of the regions where millets are grown. According to Nagaraja et al. (2007), yield loss in finger millet due to this disease ranges from 28 to 36%, and can reach as high as 80 to 90% in some locations. Panicle blast is the most severe form of this disease, and damages the crop at all growth phases, from seedling to grain formation (Takan et al., 2004 and Babu et al., 2013). More than 50 graminaceous hosts, including rice, wheat, finger millet, pearl millet, and foxtail millet are susceptible to M. grisea (Ou, 1985 and Rossman et al., 1990). Considerable yield loss of pearl millet grain (Timper et al., 2002) and forage (Wilson and Gates, 1993) has been reported due to blast disease and it has occurred as a serious constraint-causing huge yield loss in both grain and fodder of pearl millet hybrids in India (Lukose et al., 2007; Anonymous, 2009). The yield loss as high as 70 to 80 per cent has been reported in blast disease, when the predisposition factors like temperature (25-27 °C), high humidity (>85%), excessive use of nitrogen fertilizer favored the blast diseases epidemics crop (Piotti et al., 2005). Further, the disease is also on rise in majority of the foxtail millet growing regions of India.
The disease severity depends both on the favorable environment factors and also on the pathogen virulence. However, there is lack of information on the diversity of virulence and avirulent gene in Magnoporthe spp., pathogen populations infecting millets in India. However, efforts are put forth to know the aggressiveness and the genetic diversity of the blast pathogen populations (Takan et al., 2012; Babu et al., 2013). Lack of knowledge on finger millets and other millets -blast pathosystem, genetic diversity of the infected millet population, and host plant resistance (HPR) has hampered the efforts to create and deploy blast-resistant cultivars that are adaptable to various agro-ecological systems. However, the morpho-molecular characterization, and genetic diversity analysis of avirulence genes in the Magnaporthe oryzae in rice was studied using different molecular markers (Yadav et al., 2019; Amoghavarsha et al., 2022) Hence, an effort has been made to use specific genetic markers linked to specific trait to know the genetic diversity of virulent and avirulent genes in the Magnaporthe population that infects the millets and rice crops throughout India.
In this study, diversity and distribution of 10 virulent (vir) genes (ERG2, Cut1, PWL2, MPG1, MPS1, Slp1, Exo70, ABC1, Mlc and Tps1) and four avirulent (Avr) genes (Z41/Z42, Z23/Z24, YL169/YL149, Z21/Z22 and Z27/28) has been analyzed in 136 blast isolates infecting millets, such as finger millet, barnyard millet, foxtail millet and pearl millet, and rice across the different millet occupied regions in India.
Material and methods
Study area and sampling collection
The blast infected samples (leaf, neck and finger blast) from millet crops such as finger millet, barnyard millet, foxtail millet and pearl millet, and rice were collected from major millet growing areas in India including Bengaluru, Mandya, Hassan, Chitrdurga, Bangarpet, Ballari, Tumakuru and Kolar in Karnataka state; Perumallapalli and vizianagaram in Andhra Pradesh state; Waghai in Gujarat state; Athiyandal and Gudalur in Tamil Nadu state; Almora, Ranichauri, Udhamsingh Nagar and Gaja in Uttarakhand state; Jagdalpur in Chhattisgarh state; Ranchi in Jharkhand state; Rewa in Madhya Pradesh state; Dhule, Aurangabad and Kolhapur in Maharashtra state; Mandure and Durgapur in Rajasthan state; Hissar and Rohtak in Haryana state; Ghaziabad in Uttar Pradesh state; Berhampur in Odisha state, Palashquri, Jakappur, Gossaigaon and Rargiaghota in Assam state, Goba in Arunachal Pradesh; Kulai in Tripura and Hyderabad in Telangana; Khundwani in Kashmir; Madhubani in Bihar; Mohali in Punjab (Table 1; Figure 1). The samples were stored at -20 °C and used for fungal isolation in the later steps.
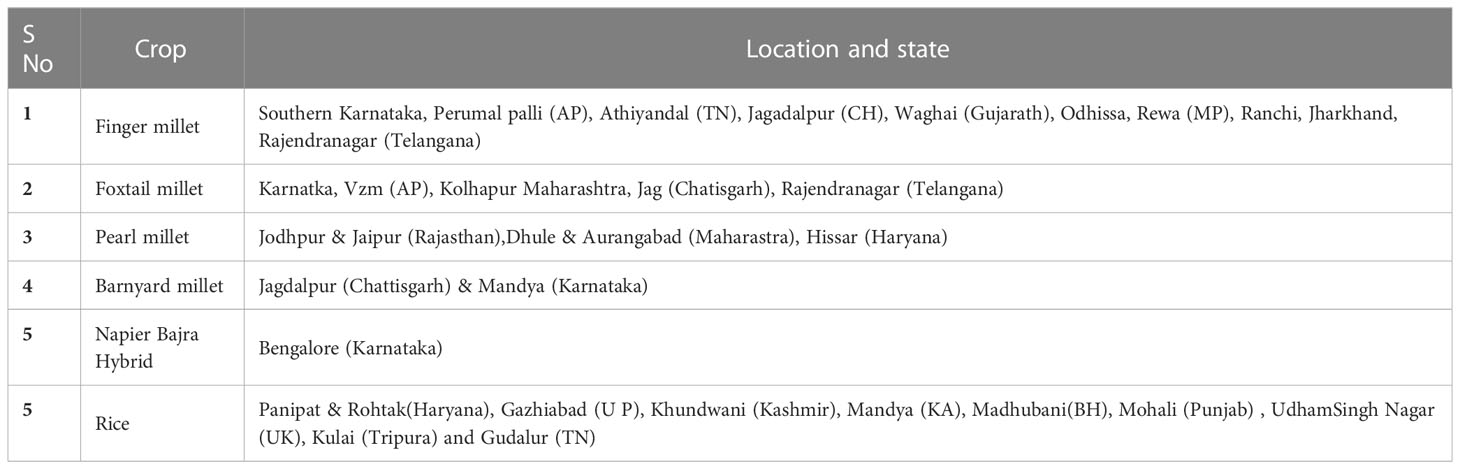
Table 1 Details of sample collection area from different crops across different growing regions of India.
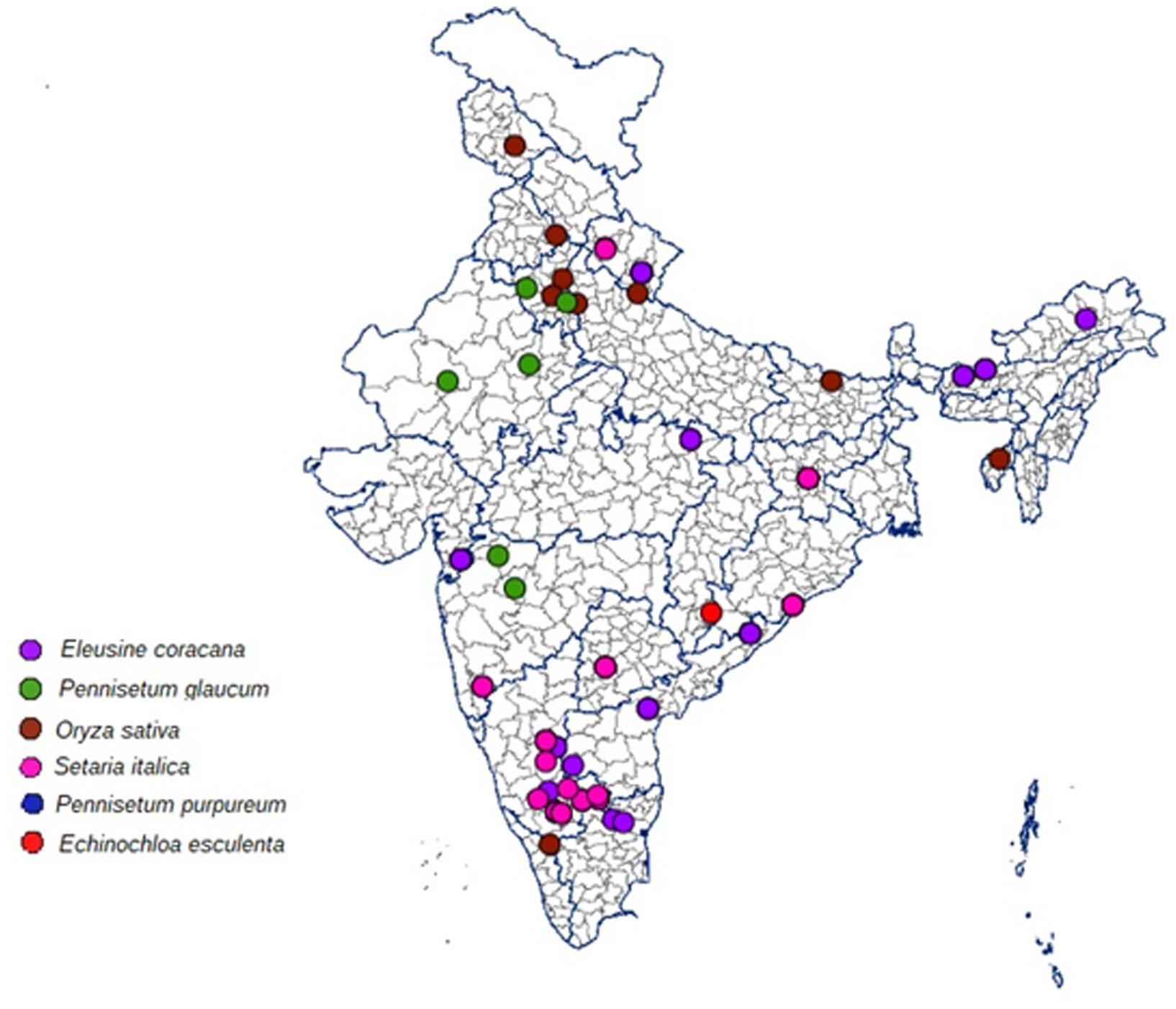
Figure 1 Details of geographical locations Magnaporthe isolates collected and isolated from major millets growing states of India.
Isolation and establishment of pure cultures
Two hundred blast-infected samples of millets (finger millet, barnyard millet, foxtail millet, and pearl millet), rice, and napier bajra hybrid grass were collected from various millet-growing districts in India. One hundred and thirty-six mono-sporidial isolates were established from 200 blast-infected samples using an efficient spore drop technique (Rajashekara et al., 2016), and single spore pure cultures were used for studying their morphology, molecular assays, and cultural aspects. These pure cultures were also preserved for future work in filter paper disc at -20 °C.
Identification of Magnaporthe isolates
The established pure isolates of pathogen were identified based on its mycelial and conidial characteristics by comparing with standard descriptions of Magnaporthe species given by Saccardo (1880) and Hebert (1971).
Studies on morphological and cultural parameters of pathogen
Morphological and cultural characteristics including growth characters, colony texture, surface topography and pigmentation etc. were studied on rice straw extract agar medium (rice straw-100 g, agar-20 g, agar, sucrose-20 g and water-1000 ml). For conidial characteristics for micro morphological traits, such as shape, length, and width, a minimum of twenty conidia per isolate were taken into consideration.
Fungal genomic DNA isolation and PCR confirmation
The genomic DNA from blast isolates (136) was extracted by adopting CTAB (cetyl trimethyl ammonium bromide) method (Murray and Thompson, 1980). The quality and quantity of isolated genomic DNA of blast isolates were determined by spectrophotometer (Nanodrop ND 1000). After quantification, the isolated DNA samples were diluted to a final concentration of 25 ng/µl using nuclease-free water and used for PCR amplification. For confirmation of blast isolates, Internal Transcribed Spacer (ITS) region amplification [ITS-5: 5’-GGAAGTAAAAGTCGTAACAAGG-3’ and ITS-4: 5’-TCCTCCGCTTATTGATATGC-3’] was used in the study (White et al., 1990). PCR reactions were performed with a total volume of 20 µl reaction mix containing 5 ng of template DNA (2 µl), 1 pmol/µl each of forward (1 µl) and reverse primer (1µl), Taq® enzyme and Takara Master Mix containing 25 mM MgCl2, 2 mM of each dNTPs, 1X Taq buffer(10µl), and nuclease free water (6µl). The thermal cycling program was: initial denaturation at 94 °C for 5 minutes followed by 35 cycles of denaturation for 45 seconds at 94°C, primer annealing for 45 seconds at appropriate temperature (Table 2), extension for 1 minute at 72 °C, and final extension for 5 minutes at 72 °C. The PCR amplicons were resolved using gel electrophoresis using 1.4% agarose in 1X TBE (Tris Borate EDTA) buffer. For electrophoresis we used 65 V for 1.5 h. The gel was documented using gel documentation unit by exposing to UV light.
Sequencing and phylogenetic analysis
The PCR amplified DNA fragments were sequenced through outsourcing (Chromous Biotech Pvt. Ltd., Bengaluru), and used for sequence alignment against Magnaporthe genome sequences using blast search tool at NCBI (National Centre for Biotechnology Information: https://blast.ncbi.nlm.nih.gov/Blast.cgi). The sequence reads were then used for phylogenetic analysis. Also, the ITS DNA sequences of all 136 isolates were deposited in NCBI GenBank (http://www.ncbi.nlm.nih.gov), for accession numbers.
PCR assay of Magnaporthe isolates for virulence and avirulence genes
The Magnaporthe isolates genomic DNA was extracted from the single spore pure cultures of fungal mycelia. DNA fragment was amplified using the primers sets specific to virulent and avirulent genes. We studied ten virulence (vir) (ERG2, Cut1, PWL2, MPG1, MPS1, Slp1, Exo70, ABC1, Mlc and Tps1) and four avirulen ce(Avr) (Avr-Pii, Avr-Pik, Avr-Pizt and Avr-Pia) genes (Table 2). PCR reactions were performed with a total volume of 20 µl reaction mix containing 5 ng of template DNA (2 µl), 1 pmol/µl each of forward (1 µl) and reverse primer (1µl), Taq® enzyme and Takara Master Mix containing 25 mM MgCl2, 2 mM of each dNTPs, 1X Taq buffer (10µl), and nuclease free water (6µl). The thermal cycling program was: initial denaturation at 94 °C for 5 minutes followed by 35 cycles of denaturation for 45 seconds at 94°C, primer annealing for 45 seconds at appropriate temperature (Table 2), extension for 1 minute at 72 °C, and final extension for 5 minutes at 72 °C. PCR amplicons were resolved in 1.5% (w/v) agarose gel through electrophoresis. The PCR amplicons were recorded using binary code of 1 and 0 for presence and absence, respectively.
Statistical analysis
The binary data of PCR-based amplicons was used for genetic diversity analysis of 136 blast isolates for 10 virulent and 4 avirulent genes. Using Power Marker version 3.25, the polymorphic information content (PIC), gene diversity, and the major allele frequency were calculated (Liu and Muse, 2005). In the GenAlEx v. 6.502 software, pairwise “F” statistics (FST) and Nei’s genetic distance (Nei, 1987) were computed (Peakall and Smouse, 2012).
Analysis of population structure and AMOVA
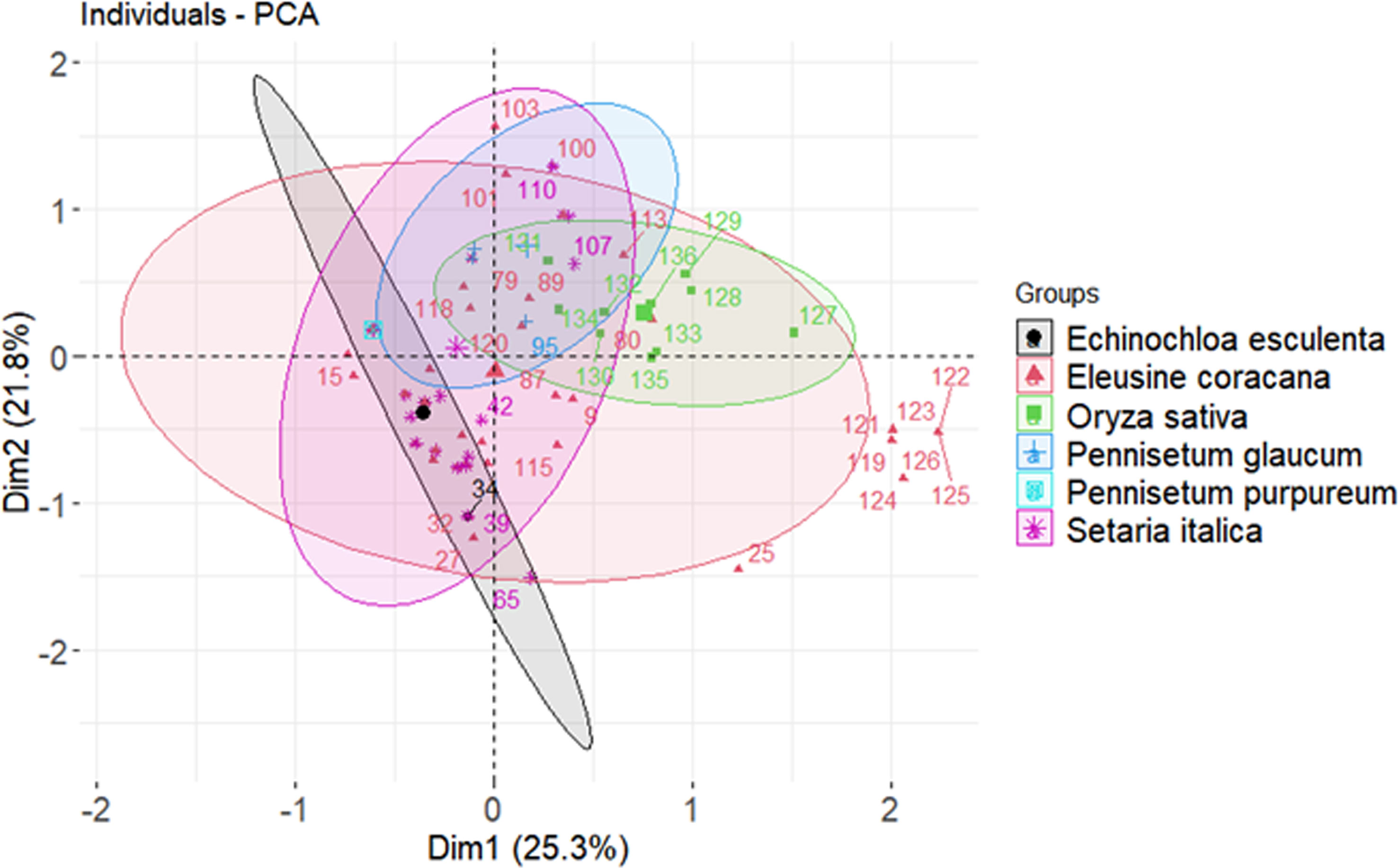
The STRUCTURE (v. 2.3.4) software was used to analyze the population structure and familial ties of 136 blast isolates in relation to the target virulent and avirulent genes (Pritchard et al., 2000). The actual number of subpopulations, designated as K value was determined by admixture ancestry model with ad hoc statistic ΔK. The run length given was 50,000 burning period length followed by 50,0000 Markov Chain Monte Carlo (MCMC) replications. Further each K value was run 10 times with K investigated from 1 to 10. The true number of K was further identified using the STRUCTURE harvester (Earl, 2012) and the best K value was determined on the basis of LnP(D) and Evanno’s ΔK proposed by Evanno et al. (2005). Further, the number of subpopulations with the isolates was confirmed by drawing a cladogram using interactive tree of life (iTOL v6.6) online tool (https://itol.embl.de/). The molecular variation between subpopulations was examined and reported as Analysis of molecular variance (AMOVA) by using the GenAlEx version 6.502 software (Peakal and Smouse, 2006).
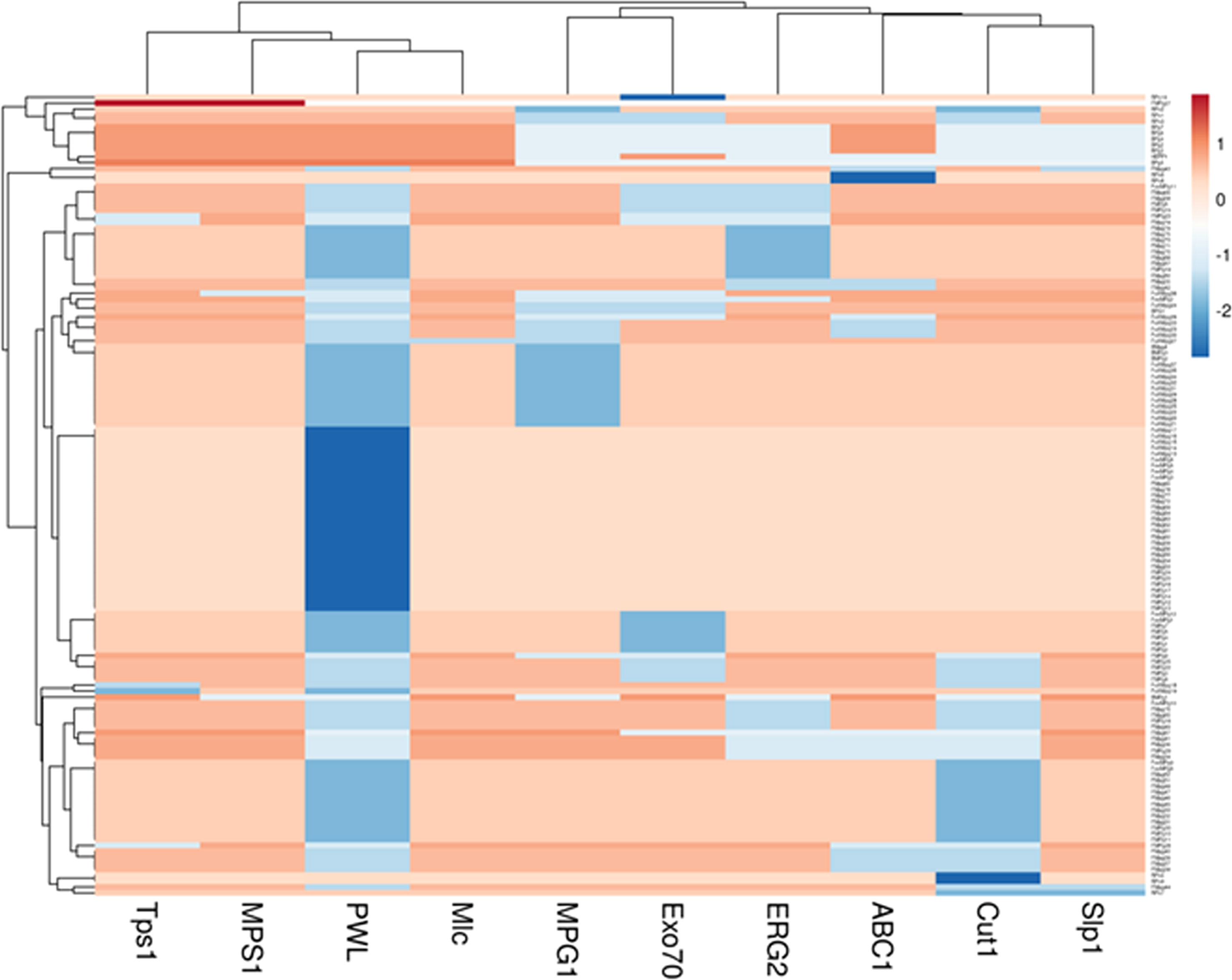
Furthermore, by performing principal component analysis (PCA) with the help of the R software’s “factoextra” package, the virulent genes of isolates were categorized according to host plants, tissues affected, and the site of collection (Kassambara and Mundt, 2017). The contribution of each pathogenic gene to the overall diversity of Magnaporthe was also evaluated by PCA analysis.
Results
Collection samples and establishment of blast isolates
One hundred and thirty-six pure blast pathogen isolates from finger millet, foxtail millet, barnyard millet, pearl millet, rice and napier bajra hybrid were established from the blast infected samples collected from major millet growing regions of India. Among these 136 blast isolates, 78 were from finger millet, 36 from foxtail millet, four from barnyard millet, seven from pearl millet, ten from rice, and one isolate from napier bajra hybrid.
Cultural and morphological characterization
Regardless of the crop or the geographical location of samples collected, variations were seen in the different isolates with respect to colony characters, pigmentation, and micromorphological characteristics. Cultural and morphological factors were also evaluated. White, whitish grey, creamish grey, and grey are some of the colors recorded (Figure 2). Regardless of the type of crop and location, the melanin pigment, indicated by the colony pigmentation, varied across the different isolates. Microscopic observation of conidia based on their length and shape divided them into three classes: short pyriform shape (25 μm), pyriform shape (25–30 μm), and long pyriform shape (>30μm). Conidia sizes in finger millet isolates 17.50×7.94 μm (FMPg 71) to 27.83×9.50 μm (FMPg 72) in representative isolates whereas foxtail millet isolates accounted 21.65×8.87 μm (FoxM Ps15) to 22.13×9.32 μm (FoxM Ps16) and in barnyard millet it ranged from 22.12×9.25 μm (BMPg1) to 23.41×9.15 μm (BMPg4). Similar variation was seen in isolates of pearl millet and rice (Table 3). The pathogen identification based on culture morphologyy and micromorphological characteristics was done following the standard descriptions of the Magnaporthe species provided by Saccardo (1880) and Hebert (1971).
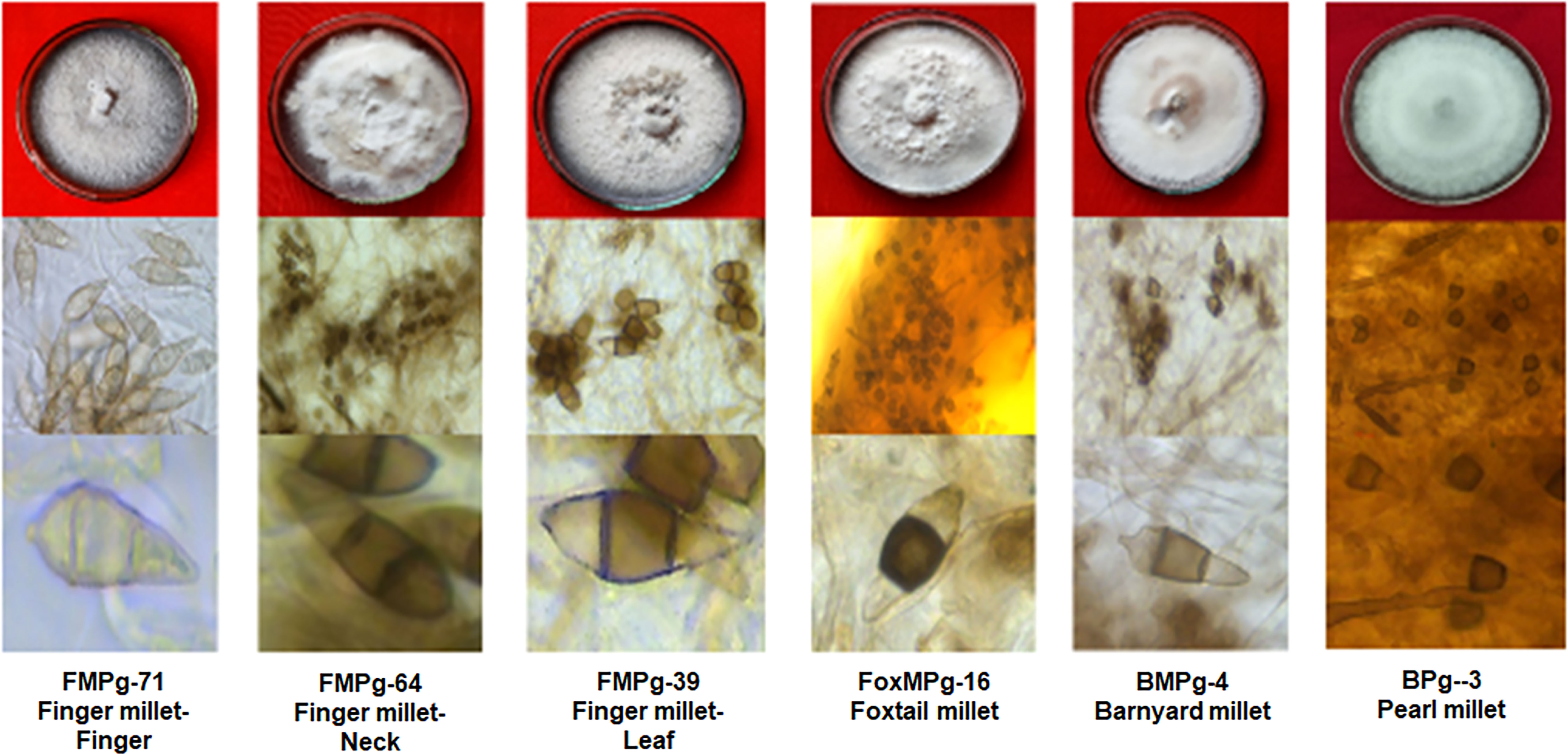
Figure 2 Representative pictures of blast isolates from different millet crops and their conidial morphology.
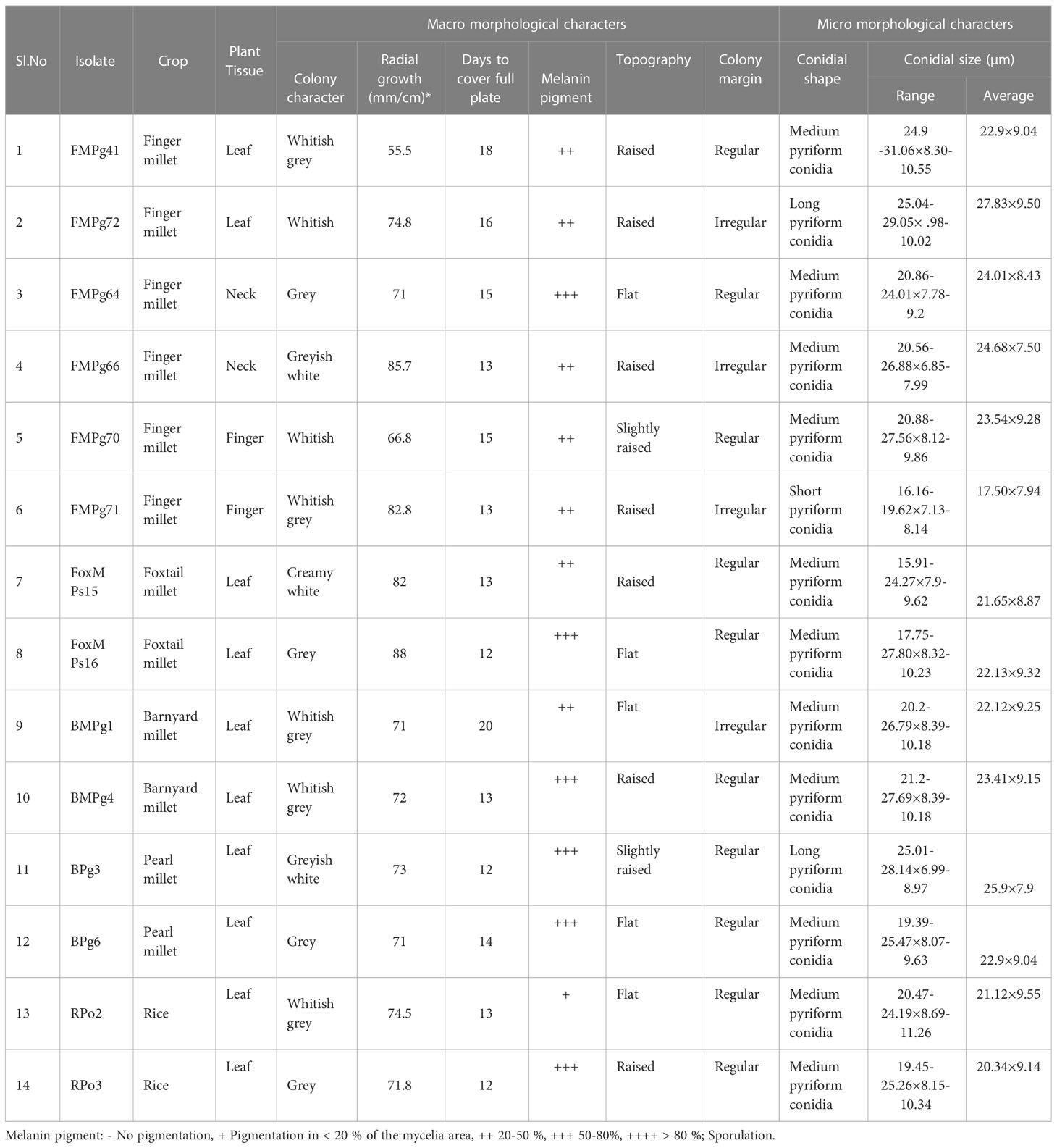
Table 3 Cultural and morphological characteristics of representing Pyricularia isolates across the geographical locations of India.
PCR based molecular characterization of Magnaporthe isolates
The universal primers ITS5 and ITS4 amplified a 600 bp DNA fragment. The PCR amplification was then purified and sequenced. The high-quality gene sequences were then submitted to NCBI and were given GenBank accession numbers (Supplementary Table 1). All the isolates showed maximum matching similarity to Magnaporthe spp. in NCBI database and confirming purely a blast pathogen infecting different cereal hosts.
Analysis of virulent genes in blast isolates
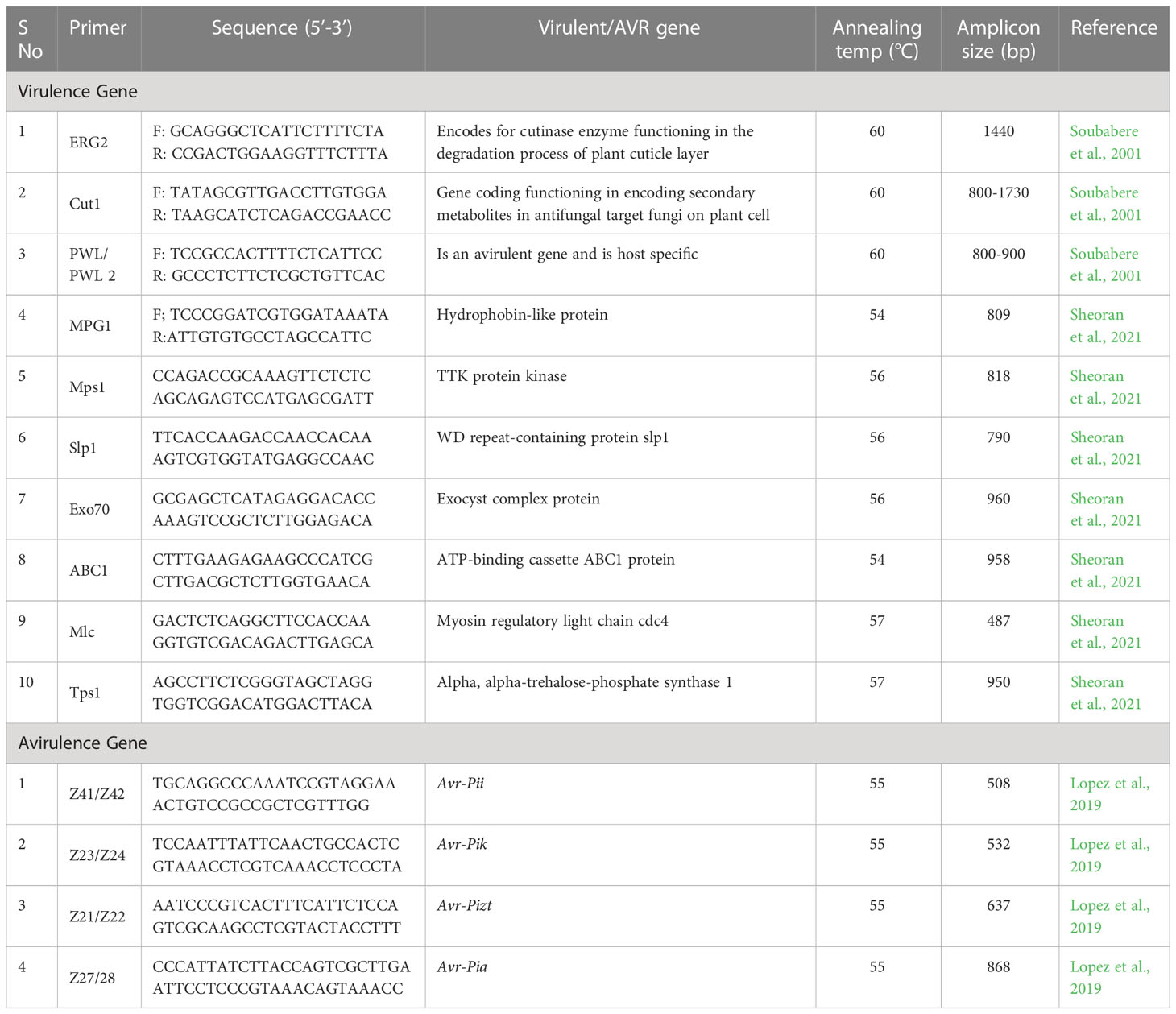
Virulent genes such as ERG2, Cut1, PWL2, MPG1, MPS1, Slp1, Exo70, ABC1, Mlc, and Tps1 were PCR amplified. These genes encode for enzymes, proteins, and secondary metabolites having role in pathogenicity/virulence. Among them, MPS1 and Mlc were amplified in 134 of the 136 isolates (98.53%) with corresponding band sizes of 809 bp and 487 bp, respectively. Tps1 (950 bp) and Slp1 (750 bp), were amplified in 131 (96.33%) and 125 (91.92%) isolates, respectively (Supplementary Figure 1). Only 17 isolates or 12.5% of them were found positive for PWL with DNA amplicons in the range of 800-900 bp. The Cut1 (800-1730bp) marker could amplify in 85 isolates (62.5%) and ERG 2 (1440 bp) marker was amplified in 98 isolates (72.05%). The presence of MPG1 (809 bp), Exo70 (960 bp), and ABC1 (958 bp) in 100, 101, and 114 isolates, respectively, accounted for 73.53, 74.26, and 83.83 percent (Table 4; Figure 3).
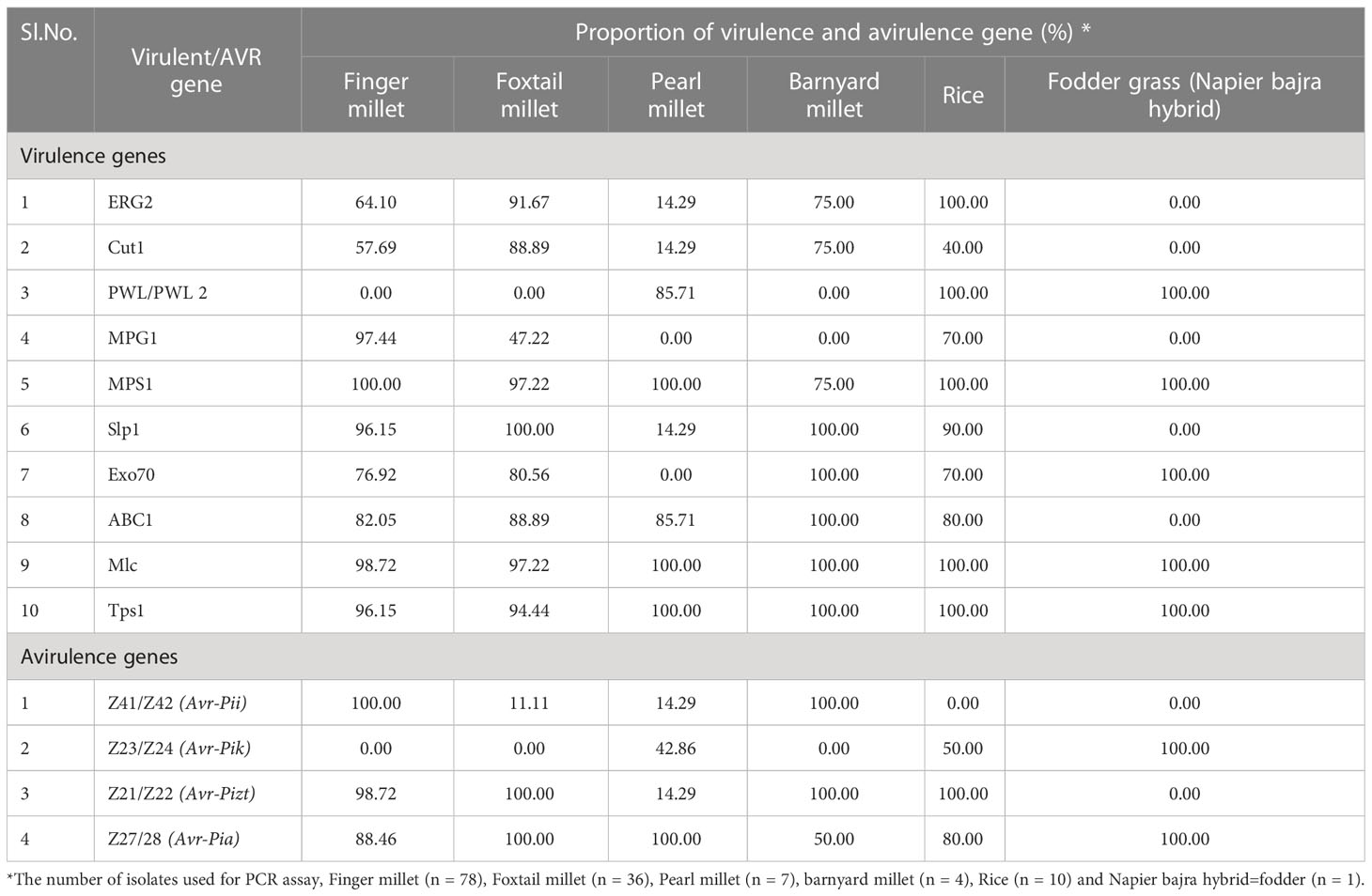
Table 4 Distribution of virulence and avirulence genes in blast fungus isolated from millets and rice.
Avirulent genes in blast isolates
Avr-Pizt, one of the four AVR genes examined, was found in the majority (94.12%) of the isolates of finger millet, foxtail millet, barnyard millet, and rice but was absent from all but one isolate of pearl millet (BPg1). The other Avr genes have frequencies of 91.92% (Avr-Pia), 63.97% (Avr-Pii), and 6.62%. (Avr-Pik). Avr-Pik was only detected in the rice and pearl millet isolates, While Avr-Pik was solely found in rice and pearl millet isolates, Avr-Pii was found in the majority of isolates of finger millet and pearl millet isolates (Table 4; Supplementary Figures 2, 3).
Gene distribution and diversity
Nearly 50% of the genetic diversity of the Magnaporthe population was described by the first two components of the principal component analysis (Supplementary Figure 4). It was evaluated for how pathogenicity genes are distributed among the Magnaporthe isolates from different host species. Since many of the virulent genes were found in isolates from several host plants, there was no distinct pattern of distribution of these genes in different host plants (Figure 4). There was no distinct categorization of isolates based on gene distribution with respect to the tissue of infection as well (Figure 5). A similar pattern was seen when categorizing these isolates by their geographic location (Supplementary Figure 5).
Population structure analysis
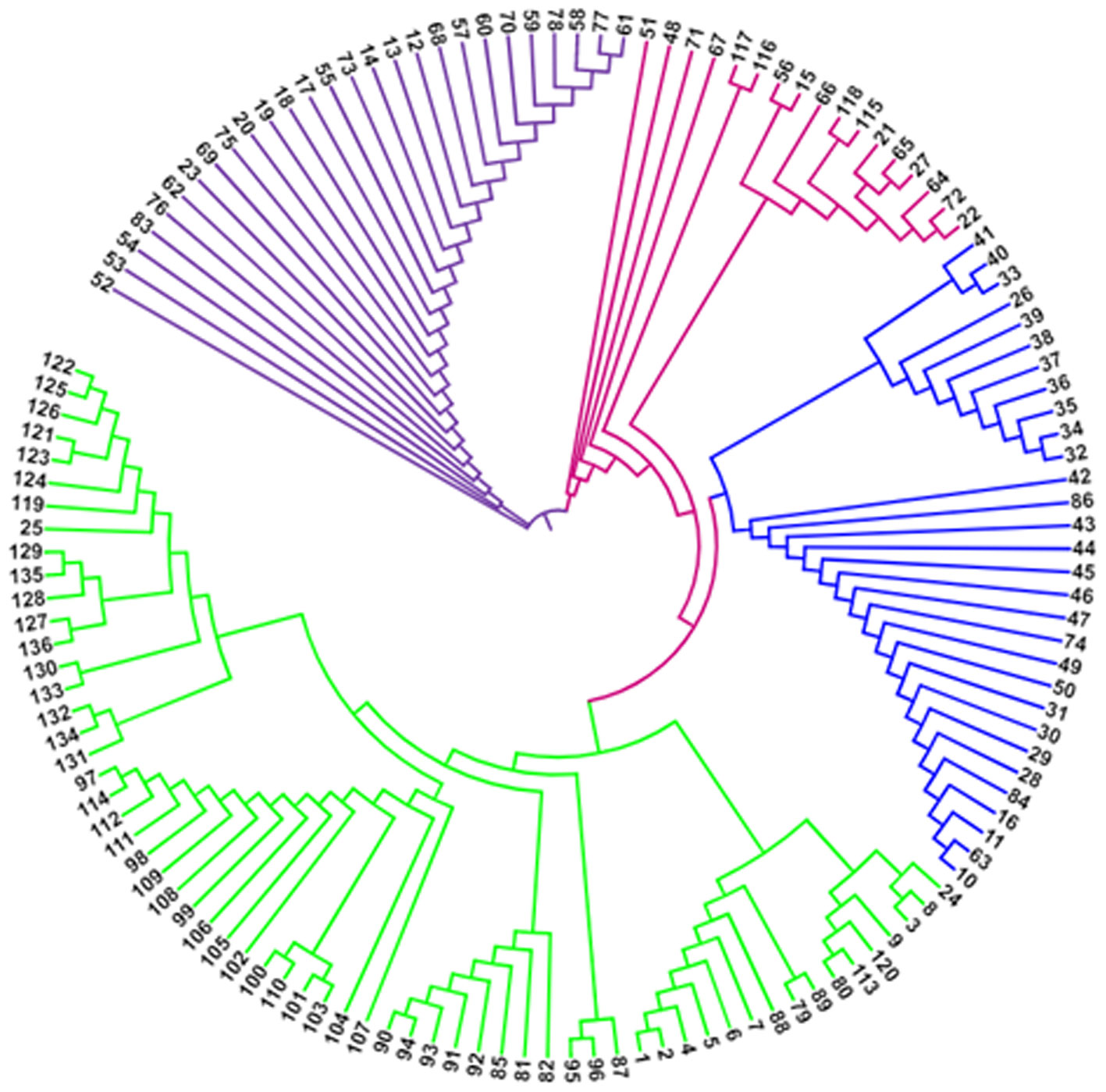
The classification of Magnaporthe populations into four subpopulations based on their family links was done using population structure analysis by utilizing gene-based functional markers data of 136 Magnaporthe isolates and was also validated using K analysis (Figure 6). These 136 blast isolates from various locations of India were shown to contain four subpopulations. Additionally, the cladogram generated using these molecular data also indicated the existence of four subpopulations within the original population of 136 isolates (Figure 7).
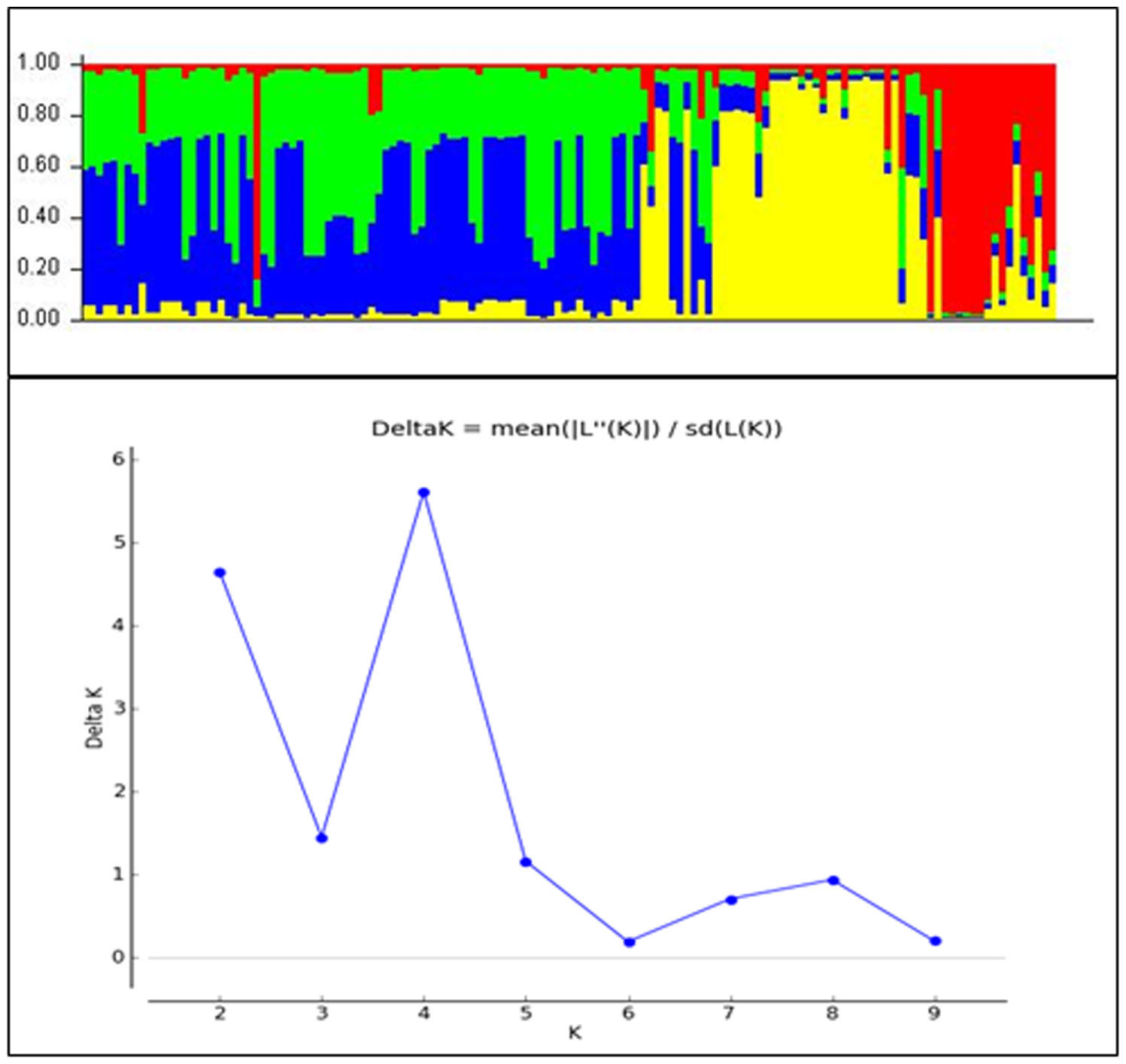
Figure 6 Population clustering of Magnaporthe isolates infecting millets at estimated membership fraction for K = 4.
AMOVA analysis
AMOVA analysis was used to evaluate the existing genetic diversity both within and between the 136 Magnaporthe isolates. The results indicated presence of more molecular variation within-population (56%) of isolates infecting various millet crops, rice, and napier hybrid bajra than the among the population (44%) (Figure 8).
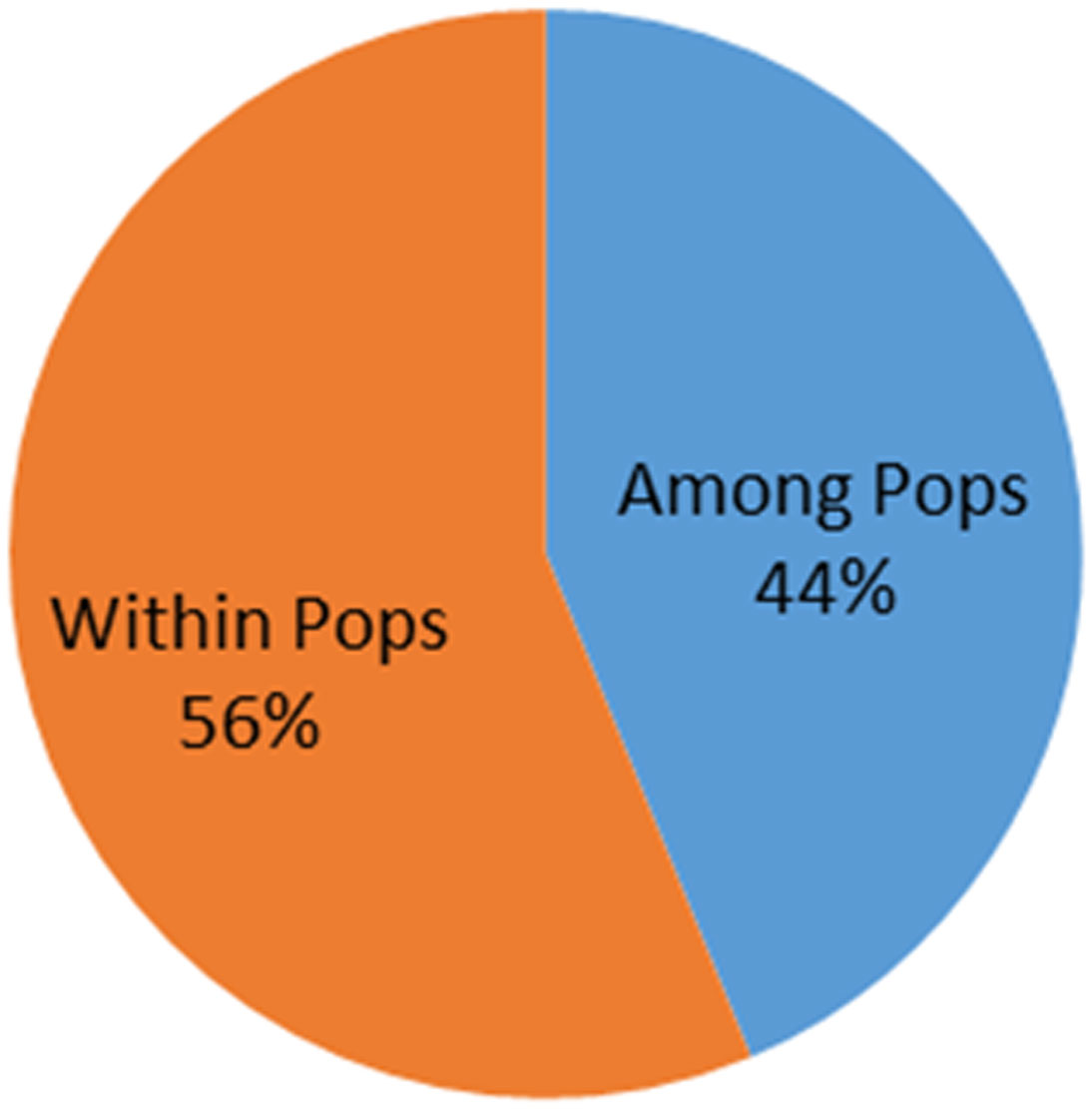
Figure 8 Percentage of molecular variance within and among the Magnaporthe population infecting millets.
Discussion
For a long time, plant pathologists and crop improvement scientist especially plant breeders have tried to understood how important is the pathogen variance in terms of efficacy and endurance of host plant resistance (HPR). The virulent pathogen may interact with a particular host genotype, which could result in the “breakdown” of resistance extremely quickly (Brown, 1995). The identification of diversity with respect to virulence variation (races) within the pathogen population has traditionally been used to detect pathogen variation. To do this, a set of pathogen isolates were inoculated onto a seedlings of different host plants with specific resistance genes (host differentials), and the disease reactivity (resistant or susceptible) for each isolate was recorded visually. The development and application of host resistance has greatly benefited from this method of pathogen population monitoring (Roelfs, 1985; Wolfe and Limpert., 1987; Andrivon and De Vallavieille-Pope, 1993). It has also provides an insight into the evolution of prevailing Magnaporthe populations in response to host resistance (R) gene selection (Kolmer, 1989; Andrivon and De Vallavieille-Pope, 1993; Wolfe and McDermott, 1994). In many pathosystems today, pathotype monitoring is still widely utilized to convey on-time information about the makeup of blast pathogen populations which is very much essential for breeding initiatives and the deployment of resistance crop plant genotypes.
An understanding of the composition, host jump and distribution of Magnaporthe populations in small millet hosts and their geographical locations across India is important in highlighting our knowledge of the biology of the pathogen and potentially adaptive ability and genotypic diversity in these pathogen population. In this study, the analysis of morphogenesis could capture various growth characteristics of Magnaporthe sp. infecting pearl millet, finger millet, foxtail millet, barnyard millet, and rice plants.
The current study evaluated the cultural- morphological and molecular characterization, and diversity of virulent and avirulent genes in the Magnaporthe population from small millets, rice, and napier hybrid bajra hosts representing geographical locations across India. This is a prerequisite to understand the distribution and variation of pathogenicity determinant genes across the geographical locations in various millets, and this information is currently lacking. The knowledge gained through this study from these populations will be very beneficial for understanding pathotypes and developing and deployment of disease-resistant millets cultivars.
Due to the overlapping cultural and morphological characteristics, we employed universal DNA marker (ITS) sequencing to precisely identify blast pathogens. Previous studies relied on the importance of ITS markers in characterization of Pyricularia isolates (Hirata et al., 2007; Abed-Ashtiani et al., 2016). ITS gene sequence could identify blast pathogen Pyricularia at the species level (Hirata et al., 2007). In this study, using the ITS specific ITS4 and ITS5 primers we could confirm that all the test pathogen isolates employed in the present research belonged to the Magnaporthe spp.
During the process of plant infection, pathogens secrete a set of molecules termed as effectors. These effectors are the key determinants of pathogen virulence. The products of plant resistance (R) gene play an important role in the detection and interaction with pathogen effects called avirulence (Avr) proteins and induce R-Avr mediated incompatible disease reaction (Sharma et al., 2012). In this study, out of the 10 investigated virulent genes, majority of 136 Magnaporthe isolates could amplified for MPS1 (TTK Protein Kinase) and Mlc (Myosin Regulatory Light Chain edc4) genes. This amplification pattern was independent of the crop and location of sampling, suggesting these genes may have significant role virulence determination. Additionally, among Avr-Pii, Avr-Pik, Avr-Pizt, and Avr-Pia genes, the Avr-Pizt had the highest frequency of amplification followed by Avr-Pia. Interestingly, Pizt and Pita are among the most broad-spectrum blast resistance genes (Devanna et al., 2022). Also, it’s intriguing to note that the Avr-Pik gene was completely lacking in the blast isolates obtained from finger millet, foxtail millet, and barnyard millet while being present in the fewest isolates from the other three, indicating noticeably significant variation both across and within them.
Population structure of Magnaporthe isolates both within the same species and across geographic locations are probably related to variations in evolutionary history and ecology (Varshney et al., 2009). Similar findings were made by Tosa et al. (2007), who discovered that Setaria and Oryza isolates were genetically more similar and shared the avirulence genes PWT1 and PWT2. The prediction of new races and their interactions in a regional agro-ecology may be made possible by using the information of the local pathotype diversity in Pyricularia oryzae and the virulence genes. The discovery of virulence genes may serve as a crucial foundation for the identification and generation of resistance genetic sources to rice blast (Dubina et al., 2020). In the current study, Mlc and MPs1 were found in 98.53% of isolates, followed by TPs1 (96.33%). Whereas, this frequency was 91.92% for Slp1, 83.83% for ABC1, 74.26% for Xxo70, 73.53% for MPG1, 72.05% for ERG2, 62.50% for Cut1, and 12.5% for PWL. While PWL was amplified only in isolates from pearl millet and rice, and was completely absent in finger millet, foxtail millet, and barnyard millet isolates, the Mlc and MPs1 genes are dispersed regardless of the geographical location and host crop.
The lack of amplification of Cut1, ERG 2, MPG1 Exo 70, and PWL in most of the isolates can be attributed to evolving pattern of these genes. Both pathogenic virulence factors and a diversity of pathogen populations are caused by the mutation. The prevalence of transposon elements may contribute to the P. oryzae pathogen’s mutation (Kito et al., 2002; Singh et al., 2018).
The Cut1 which codes for cutinase enzyme involved in the plant cuticle layers degradation process (Sweigard et al., 1992), whereas Pwl2, avirulent gene which is a host specific in nature (Valent and Chumley, 1994). Erg2 is an antifungal target for blast pathogen and also secretes secondary metabolite in plant cells (Keon et al., 1994). Reflinur et al. (2005) characterized 5 races of P. oryzae distributed among 230 isolates with virulence gene-coding specific markers and found eight haplotypes among the population studied. Li et al. (2010), P. oryzae isolates are genetically stable but have high pathogenic variations. According to Leung et al. (1993), host plant selection, is an evolutionary process that significantly influences the development of pathogen genetic variants. Leung et al. (2003) reported DNA fingerprint studies of P. oryzae strains and showed that deployment monogenic(one) variety has low genetic variation when compared to isolates of P. oryzae collected from areas planted with polygenic varieties. Among Avr genes, Avr-Pizt was more frequently (94.12%) amplified in Magnaporthe isolates of millets followed by Avr-Pia (91.92%). However, Lopez et al. (2019) reported that Avr-Pik, Avr-Pita and Avr-Pii were more frequently occurring genes with 81.50%, 64.16% and 47.98% frequency, respectively. Whereas, the frequency of Avr-Pizt and Avr-Pia was 19.08% and 5.20%, respectively. It was also laid out that the presence of Avr genes in M. oryzae is strongly associated with agro-ecosystems where the complementary resistant (R) genes exist.
Jia et al. (2012) studied the Avr genes from the blast isolates collected from the southern US during (1970 to 2009), and found that majority of the isolates had Avr-Pita (65.7%). The relative frequencies of other Avr genes ranged from 0.3% to 57.1%. Similarly, in Yunnan province of China, AvR-Pii was present in 82 out of 454 field isolates of M. oryzae studied (Lu et al., 2019). In Eastern India, Imam et al. (2015) reported that the occurrence Avr-Pizt and Avr-Pik were in highest frequency (100%), whereas Avr1-CO39 was in the lowest (2%) frequency. In a similar study, the frequency of PWL-2, Avr-Pii and Avr-Pizt genes was 100%, 60% and 54%, respectively in Thailand (Sirisathaworn et al., 2017). These studies infer that there is a lot of diversity in the frequency of blast pathogen avirulence genes in millet isolates of India, as evidenced by Avr- Pizt and Avr-Pia having the highest frequency of 94.12% and 91.92%, respectively. Though Avr-PWL3 is rarely present in M. oryzae isolates from rice (Kang et al., 1995), but in our study, its frequency is more than Avr-Pia.
The distribution of molecular variance in the population also indicates the diverse evolutionary history, both temporal and spatial. In the present study, we observed comparatively higher molecular variance within the population in comparison to among the population. This indicates that isolates within a given geographical regions have evolved more. This implies that there is more chance of evolution of new virulent strains from these populations, and this might affect the present R-Avr balance in these crop growing areas.
Conclusions
Present study provides some novel insight into the genetics of adaptation of Magnaporthe populations to millet crops. The genetic diversity with respect to virulent and avirulent gene observed in the populations adapted to millet might show the pathogenic variations among the Magnaporthe populations. Thus, understanding the genetic structure and pathogenic nature of the Magnaporthe populations belonging to different clonal-lineages will help in designing the strategies for millet blast management programs especially by deployment of resistant genes for eco-friendly management of blast through host plant resistance.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
KP, HV, and SP conceived the study, designed the experiments, and participated in drafting the manuscript. BJ, HRR, FK, CB, VU, TP, LR, MR, PS, PN, GR, ID, HP, AJ, SS, SN, GP, and TN collected the data and performed the analysis. HR, BD, SP, and CA performed the diversity analyses. KP, HR, BD, CA, and SP wrote the manuscript. KP, ID, TN, BD, and SN provided resources and improved the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This research was funded by DST-Science and Engineering Research Board (SERB-EEQ/2019/000678) Government of India, New Delhi - 110 070.
Acknowledgments
The authors express their sincere gratitude to ICAR-AICRP (small millets), University of Agricultural Sciences, GKVK, Bangalore and ICAR-AICRP coordinating centers of small millets and ICAR-IIMR, Hyderabad for their cooperation and support during the collection of samples and the data.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1131315/full#supplementary-material
References
Abed-Ashtiani, F., Kadir, J., Nasehi, A., Hashemian-Rahaghi, S., Vadamalai, G., Rambe, S. (2016). Characterisation of Magnaporthe oryzae isolates from rice in peninsular Malaysia. Czech J. Genet. Plant Breed. 52, 145–156. doi: 10.17221/102/2016-CJGPB
Amoghavarsha, C., Pramesh, D., Naik, G. R., Naik, M. K., Yadav, M. K., Ngangkham, U., et al. (2022). Morpho-molecular diversity and avirulence genes distribution among the diverse isolates of Magnaporthe oryzae from southern India. J. Appl. Microbiol. 132 (2), 1275–1290. doi: 10.1111/jam.15243
Andrivon, D., De Vallavieille-Pope, C. (1993). Racial diversity and complexity in regional populations of Erysiphe graminis f.sp. hordei in france over a 5-year period. Plant Pathol. 42, 443–464. doi: 10.1111/j.1365-3059.1993.tb01523.x
Anonymous (2009). Annual report, all India coordinated pearl millet improvement program (Mandore, Jodhpur: Indian Council of Agricultural Research).
Babu, T. K., Sharma, R., Upadhyaya, H. D., Reddy, P. N., Deshpande, S., Senthilvel, S., et al. (2013). Evaluation of genetic diversity in magnaporthe grisea populations adapted to finger millet using simple sequence repeats (SSRs) markers. Physiol. Mol. Plant Pathol. 84, 10–18. doi: 10.1016/j.pmpp.2013.06.001
Brown, J. K. M. (1995). “Pathogens’ responses to the management of disease resistance genes,” in Advances in plant pathology, vol. vol. 11 . Eds. Andrews, J. H., Tommerup, I. C. (New York: Academic Press), 75–102.
Devanna, B. N., Jain, P., Solanke, A. U., Das, A., Thakur, S., Singh, P. K., et al. (2022). Understanding the dynamics of blast resistance in rice-magnaporthe oryzae interactions. J. Fungi 8, 584. doi: 10.3390/jof8060584
Dubina, E. V., Alabushev, A. V., Kostylev, P. I., Kharchenko, E. S., Ruban, M. G., Aniskina, Y. V., et al. (2020). Biodiversity of pyricularia oryzae cav. in rice-growing regions of the south of russia using PCR method. Physiol. Mol. Biol. Plants 26, 289–303.
Earl, D. A. (2012). Structure harvester: a website and program for visualizing structure output and implementing the evanno method. Conser Gene Resour. 4 (2), 359–361. doi: 10.1007/s12686-011-9548-7
Evanno, G., Regnaut, S., Goudet, J. (2005). Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol. Ecol. 14 (8), 2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x
Gupta, A. (2006). Improvement of millets and pseudo cereals in hill region. sustainable production from watershed in NWH (Uttarakhand: Vivekananda Parvatiya Krishi Anusandhan Sansthan), p.163–p.174.
Hebert, T. T. (1971). The perfect stage of Pyricularia grisea. Phytopathology 61, 83–87. doi: 10.1094/Phyto-61-83
Hirata, K., Kusaba, M., Chuma, I., Osue, J., Nakayashiki, H., Mayama, S. H., et al. (2007). Speciation in pyricularia inferred from multilocus phylogenetic analysis. Mycol. Res. 111, 799–808. doi: 10.1016/j.mycres.2007.05.014
Imam, J., Alam, S., Mandal, N. P., Shukla, P., Sharma, T. R., Variar, M. (2015). Molecular identifification and virulence analysis of AVR genes in rice blast pathogen, Magnaporthe oryzae from Eastern India. Euphytica 206, 21–31. doi: 10.1007/s10681-015-1465-5
Jia, Y., Zhang, Z., Xing, J., Wang, J., Correll, J. C., Cartwright, R. (2012) Natural variation and evolution of the avirulence genes in magnaporthe oryzae. Available at: https://www.ars.usda.gov/research/publications/publication/?seqNo115=279490.
Kang, S., Sweigard, J. A., Valent, B. (1995). The PWL host specificity gene family in the blast fungus Magnaporthe grisea. Mol. Plant Microbe Interact. 8, 939–948. doi: 10.1094/MPMI-8-0939
Kassambara, A., Mundt, F. (2017) Factoextra: extract and visualize the results of multivariate data analyses. Available at: https://mirrors.sjtug.sjtu.edu.cn/cran/web/packages/factoextra/index.html.
Keon, J. P., James, C. S., Court, S., Baden-Daintree, C., Bailey, A. M., Burden, R. S., et al. (1994). Isolation of Erg2 gene, encoding sterol delta 8 to delta isomerase, from the rice blast fungus Magnaporthe grisea and its expression in the maize smut pathogen ustilago maydis. Curr. Genet. 25 (6), 531–537. doi: 10.1007/BF00351674
Kito, H., Sato, J., Takahashi, Y., Fukiya, S., Sone, T., Tomita, F. (2002). Occan, a new transposon-like sequence in magnaporthe grisea abstracts 3rd international rice blast conference (Ibaraki) (Japan: Tsukuba International Congress Center-Epochal Tsukuba- Tsukuba Science City), p 15.
Kolmer, J. A. (1989). Virulence and race dynamics of Puccinia recondita f.sp. tritici in Canada during 1956-1987. Phytopathology 79, 349–356. doi: 10.1094/Phyto-79-349
Leung, H., Fan, J. X., Zhu, Y., Chen, H., Revilla-Monila, I., Pangga, I., et al. (2003). Using genetic diversity to achieve sustainable rice diseases management. Plant Dis. 87 (10), 1156–1169. doi: 10.1094/PDIS.2003.87.10.1156
Leung, H., Nelson, R. J., Leach, J. E. (1993). Population structure of plant pathogenic fungi and bacteria. Plant Pathol. 10, 157–205.
Li, Y., Liang, S., Yan, X., Wang, H., Li, D., Soanes, D. M., et al. (2010). Characterization of MoLDB1 required for vegetative growth, infection-related morphogenesis and pathogenicity in the rice blast fungus magnaporthe oryzae. Mol. Plant-Microbe Interactions 23 (10), 1260–1274. doi: 10.1094/MPMI-03-10-0052
Liu, K., Muse, S. V. (2005). PowerMarker: an integrated analysis environment for genetic marker analysis. Bioinformatics 21 (9), 2128–2129. doi: 10.1093/bioinformatics/bti282
Lopez, A. L. C., Yli-Matilla, T., Cumagun, C. J. R. (2019). Geographic distribution of avirulence genes of the rice blast fungus magnaporthe oryzae in the philippines. Microorganisms 7 (1), 23.
Lu, L., Wang, Q., Jia, Y., Bi, Y. Q., Li, C. Y., Fan, H. C., et al. (2019). Selection and mutation of the avirulence gene AVR-pii of the rice blast fungus magnaporthe oryzae. Plant Pathol. 68 (1), 127–134.
Lukose, C. M., Kadvani, D. L., Dangaria, C. J. (2007). Effificacy of fungicides in controlling blast disease of pearl millet. Indian Phytopathol. 60, 68–71.
Murray, H. G., Thompson, W. F. (1980). Rapid isolation of high molecular weight DNA. Nucleic Acids Res. 8, 4321–4325. doi: 10.1093/nar/8.19.4321
Nagaraja, A., Jagadish, P. S., Ashok, E. G., Krishne Gowda, K. T. (2007). Avoidance of finger millet blast by ideal sowing time and assessment of varietal performance under rainfed production situations in karnataka. J. Mycopathol. Res. 45, 237–240.
Palanna, K. B., Rajashekara, H., Ramesh, G. V., Malikarjuna, B., Praveen, B., Raveendra, H. R., et al. (2021). “Current scenario and integrated approaches for management of finger millet blast (Magnaporthe grisea),” in Blast disease of cereal crops. Eds. Nayaka, S. C., Hosahatti, R., Prakash, G., Satyavathi, C. T., Sharma, R. (Cham: Springer), p 27–p 49.
Peakal, R., Smouse, P. E. (2006). GENALEX 6: genetic analysis in excel. population genetic software for teaching and research. Mol. Ecol. Notes 6 (1), 288–295. doi: 10.1111/j.1471-8286.2005.01155.x
Peakall, R., Smouse, P. E. (2012). GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics 28, 2537–2539.
Piotti, E., Rigano, M. M., Rodino, D., Rodolfifi, M., Castiglione, S., Picco, A. M., et al. (2005). Genetic structure of pyricularia grisea (Cooke) sacc. isolates from Italian paddy fields. J. Phytopathol. 153, 80–86. doi: 10.1111/j.1439-0434.2005.00932.x
Pritchard, J. K., Stephens, M., Donnelly, P. (2000). Inference of population structure using multilocus genotype data. Genetics 155 (2), 945–959. doi: 10.1093/genetics/155.2.945
Rajashekara, H., Jeevan, B., Mishra, K. K., Subbanna, A. R. N. S., Kant., L. (2021). “Blast disease: historical importance, distribution, and host infectivity across cereal crops,” in Blast disease of cereal crops. Eds. Nayaka, S. C., Hosahatti, R., Prakash, G., Satyavathi, C. T. (Cham: Springer), p 1–p13.
Rajashekara, H., Prakash, G., Pandian, R. T. P., Sarkel, S., Dubey, A., Sharma, P., et al. (2016). An efficient technique for isolation and mass multiplication of Magnaporthe oryzae from blast infected samples. Indian Phytopathol. 69, 260–265.
Reflinur, B. M., Widyastuti, U., Aswidinnoor, H. (2005). Keragaman genetik cendawan Pyricularia oryzae berdasarkan primer spesifik gen virulensi. J. Biotek. Pertanian 10 (2), 55–60.
Roelfs, A. P. (1985). “Race specificity and methods of study,” in The cereal rusts, vol. vol. 1 . Eds. Bushnell, W. R., Roelfs, A. P. (Orlando: Academic Press), 132–164.
Rossman, A. Y., Howard, R. J., Valent, B. (1990). Pyricularia grisea The correct name for the rice blast disease fungus. Mycologia 2, 509e12. doi: 10.1080/00275514.1990.12025916
Sharma, T. R., Rai, A. K., Gupta, S. K., Vijayan, J., Devanna, B. N., Ray, S. (2012). Rice blast management through host-plant resistance: retrospect and prospects. Agric. Res. 1, 37–52.
Sheoran, N., Ganesan, P., Mughal, N. M., Yadav, I. S., Kumar, A. (2021). Genome assisted molecular typing and pathotyping of rice blast pathogen, magnaporthe oryzae, reveals a genetically homogenous population with high virulence diversity. Fungal Biol. 125 (9), 733–747. doi: 10.1016/j.funbio.2021.04.007
Singh, P. K., Ray, S., Thakur, S., Rathour, R., Sharma, V., Sharma, T. R. (2018). Co-Evolutionary interactions between host resistance and pathogen avirulence genes in rice-magnaporthe oryzae pathosystem. Fungal Genet. Biol. 115, 9–19. doi: 10.1016/j.fgb.2018.04.005
Sirisathaworn, T., Srirat, T., Longya, A., Jantasuriyarat, C. (2017). Evaluation of mating type distribution and genetic diversity of three Magnaporthe oryzae avirulence genes, PWL-2, AVR-Pii and Avr-Piz-t, in Thailand rice blast isolates. Agric. Nat. Resour. 51, 7–14. doi: 10.1016/j.anres.2016.08.005
Soubabere, O., Jorge, V., Notteghem, J. L., Lebrun, M. H., Tharreau., D. (2001). Sequence characterized amplified region markers for the rice blast fungus. Magnaporthe Grisea Mol. Ecol. Notes 1, 19–21. doi: 10.1046/j.1471-8278.2000.00008.x
Sweigard, J. A., Chumley, F. G., Valent, B. (1992). Disruption of magnaporthe grisea cutinase gene mol. Gene Genet. 232 (2), 183–190.
Takan, J. P., Akello, B., Esele, P., Manyasa, E. O., Obilana, A. B., Audi, P. O., et al. (2004). Finger millet blast pathogen diversity and management in East Africa:A summary of project activities and outputs, ISMN 45. (CORE, ICRISAT).
Takan, J. P., Chipili, J., Muthumeenakshi, S., Talbot, N. J., Manyasa, E. O., Bandyopadhyay, R., et al. (2012). Magnaporthe oryzae populations adapted to finger millet and rice exhibit distinctive patterns of genetic diversity, sexuality and host interaction. Mol. Biotechnol. 50 (2), 145–158. doi: 10.1007/s12033-011-9429-z
Timper, P., Wilson, J. P., Johnson, A. W., Hanna, W. W. (2002). Evaluation of pearl millet grain hybrids f or resistance to Meloidogyne spp. and leaf blight caused by Pyricularia grisea. Plant Dis. 86, 909–914. doi: 10.1094/PDIS.2002.86.8.909
Tosa, Y., Uddin, W., Viji, G., Kang, S., Mayama, S. (2007). Comparative genetic analysis of magnaporthe oryzae isolates causing gray leaf spot of perennial ryegrass turf in the united states and Japan. Plant Dis. 91, 517e2. doi: 10.1094/PDIS-91-5-0517
Valent, B., Chumley, F. G. (1994). Avirulence genes and mechanisms of genetic instability in rice blast fungus rice blast disease. Eds. Zeigler, R. S., Leong, S. A., Teng, P. S. (Wallingford: CAB International-IRRI, Manila, Philippines), p 111–p 134.
Varshney, R., Pande, S., Kannan, S., Mahendar, T., Sharma, M., Gaur, P., et al. (2009). Assessment and comparison of AFLP and SSR based molecular genetic diversity in Indian isolates of ascochyta rabiei, a causal agent of ascochyta blight in chickpea (Cicer arietinum l.). Mycol. Prog. 8 (2), 87–97. doi: 10.1007/s11557-008-0581-1
White, T. J., Bruns, T., Lee, S., Taylor, J. (1990). “Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics,” in PCR protocols, a guide to methods and applications. Eds. Innis, M. A., Gelfand, D. H., Sninsky, J. J., White, T. J. (San Diego, California: Academic Press), p.315–p.322.
Wilson, J. P., Gates, R. N. (1993). Forage yield losses in hybrid pearl millet due to leaf blight caused primarily by Pyricularia grisea. Phytopathology 83, 739–743. doi: 10.1094/Phyto-83-739
Wolfe, M. S., Limpert., E. (1987). Integrated control of cereal powdery mildews: monitoring the pathogen. Ed. Nijhoff, M. (Dordrecht: Springer Science & Business Media).
Wolfe, M. S., McDermott, J. M. (1994). Population genetics of plant pathogen interactions: the example of the erysiphe graminis-hordeum vulgare pathosystem. Annu. Rev. Phytopathol. 32 (1), 89–113.
Yadav, M. K., Aravindan, S., Raghu, S., Prabhukarthikeyan, S. R., Keerthana, U., Ngangkham, U., et al. (2019). Assessment of genetic diversity and population structure of Magnaporthe oryzae causing rice blast disease using SSR markers. Physiol. Mol. Plant Pathol. 106, 157–165. doi: 10.1016/j.pmpp.2019.02.004
Keywords: millet crops, Magnaporthe, virulent and avirulent genes, blast pathogen, PCR
Citation: Palanna KB, Vinaykumar HD, Prasanna SK, Rajashekara H, Devanna BN, Anilkumar C, Jeevan B, Raveendra HR, Khan F, Bhavana CHS, Upadhyay V, Patro TSSK, Rawat L, Rajesh M, Saravanan PT, Netam P, Rajesha G, Das IK, Patil HE, Jain AK, Saralamma S, Nayaka SC, Prakash G and Nagaraja TE (2023) Exploring the diversity of virulence genes in the Magnaporthe population infecting millets and rice in India. Front. Plant Sci. 14:1131315. doi: 10.3389/fpls.2023.1131315
Received: 24 December 2022; Accepted: 03 April 2023;
Published: 09 May 2023.
Edited by:
Salej Sood, ICAR- Central Potato Research Institute, IndiaReviewed by:
Pramesh Devanna, University of Agricultural Sciences Raichur, IndiaPankaj Kumar Singh, National Agri-Food Biotechnology Institute, India
Copyright © 2023 Palanna, Vinaykumar, Prasanna, Rajashekara, Devanna, Anilkumar, Jeevan, Raveendra, Khan, Bhavana, Upadhyay, Patro, Rawat, Rajesh, Saravanan, Netam, Rajesha, Das, Patil, Jain, Saralamma, Nayaka, Prakash and Nagaraja. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: K. B. Palanna, a2JwYWxhbm5hQGdtYWlsLmNvbQ==; H. Rajashekara, cmFqYWlhcmlwYXRoQGdtYWlsLmNvbQ==; B. N. Devanna, ZGV2bm92YTI0NjBAZ21haWwuY29t
†These authors have contributed equally to this work
 K. B. Palanna
K. B. Palanna H. D. Vinaykumar1†
H. D. Vinaykumar1† H. Rajashekara
H. Rajashekara B. N. Devanna
B. N. Devanna C. Anilkumar
C. Anilkumar B. Jeevan
B. Jeevan C. H. Sai Bhavana
C. H. Sai Bhavana M. Rajesh
M. Rajesh I. K. Das
I. K. Das A. K. Jain
A. K. Jain S. Chandra Nayaka
S. Chandra Nayaka G. Prakash
G. Prakash T. E. Nagaraja
T. E. Nagaraja