
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 17 March 2023
Sec. Plant Breeding
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1130723
This article is part of the Research TopicInteractive Dynamics of Geminiviruses with Host PlantsView all 8 articles
 Lampros Siskos1
Lampros Siskos1 Maria Antoniou1
Maria Antoniou1 Jose Riado2
Jose Riado2 Montserrat Enciso2
Montserrat Enciso2 Carlos Garcia2
Carlos Garcia2 Daniele Liberti3
Daniele Liberti3 Danny Esselink1
Danny Esselink1 Andrey G. Baranovskiy4
Andrey G. Baranovskiy4 Tahir H. Tahirov4
Tahir H. Tahirov4 Richard G. F. Visser1
Richard G. F. Visser1 Richard Kormelink5
Richard Kormelink5 Yuling Bai1
Yuling Bai1 Henk J. Schouten1*
Henk J. Schouten1*The family of Geminiviridae consists of more than 500 circular single-stranded (ss) DNA viral species that can infect numerous dicot and monocot plants. Geminiviruses replicate their genome in the nucleus of a plant cell, taking advantage of the host’s DNA replication machinery. For converting their DNA into double-stranded DNA, and subsequent replication, these viruses rely on host DNA polymerases. However, the priming of the very first step of this process, i.e. the conversion of incoming circular ssDNA into a dsDNA molecule, has remained elusive for almost 30 years. In this study, sequencing of melon (Cucumis melo) accession K18 carrying the Tomato leaf curl New Delhi virus (ToLCNDV) recessive resistance quantitative trait locus (QTL) in chromosome 11, and analyses of DNA sequence data from 100 melon genomes, showed a conservation of a shared mutation in the DNA Primase Large subunit (PRiL) of all accessions that exhibited resistance upon a challenge with ToLCNDV. Silencing of (native) Nicotiana benthamiana PriL and subsequent challenging with three different geminiviruses showed a severe reduction in titers of all three viruses, altogether emphasizing an important role of PRiL in geminiviral replication. A model is presented explaining the role of PriL during initiation of geminiviral DNA replication, i.e. as a regulatory subunit of primase that generates an RNA primer at the onset of DNA replication in analogy to DNA Primase-mediated initiation of DNA replication in all living organisms.
The family of Geminiviridae consists of more than 500 viral species which infect numerous monocot and dicot plants. Geminiviruses are divided into 14 genera based on genome organization, transmission vectors, and host range. The Begomovirus genus is the most diverse one and contains more than 400 whitefly transmitted species (Fondong, 2013; Fiallo-Olivé et al., 2021), many of them being plant pathogenic, with a major economic importance in the agriculture of tropical and subtropical regions (Fiallo-Olivé and Navas-Castillo, 2020).
The genome of geminiviruses varies in size from ~2.5 to 5 kb, and is organized in one (monopartite) or two (bipartite) circular single stranded (ss)-DNA molecules of approximately 2.5 kb, that are encapsidated into twinned icosahedral protein particles (Krupovic et al., 2009). Only begomoviruses include species with two genomic components, denoted DNA-A and DNA-B, which contain six respectively two open reading frames (ORFs). Proteins encoded by DNA-A are involved in genome replication (REn, replication enhancer protein; Rep, replication-associated protein; TrAP, transcriptional activator protein), particle assembly and insect transmission (CP, coat protein), control of gene expression, and modulation of host defense responses (AC2, AC4), whereas proteins encoded by DNA-B are required for intra-intercellular movement in host plants (MP, movement protein; NSP, nuclear shuttle protein) (Briddon et al., 2010). Except for a sequence of approximately 200 nucleotides, called the common region, A and B components hardly share sequence homology. The common region contains various regulatory elements (e.g. iterons, TATA box) involved in replication and transcription, and a stem-loop structure that is essential for replication. The stem-loop structure contains a highly conserved nonanucleotide sequence (5’-TAATATTCA-3’) at its top, presenting the origin of viral replication, and is found in all geminiviruses (Bhattacharjee and Hallan, 2022). Upon feeding of viruliferous whiteflies onto healthy plants, virions introduced into the host’s cytoplasm uncoat and release their ss-DNA, which traffics to the nucleus for replication (Kumar, 2019).
Geminiviruses replicate their genomes initially with the conversion of ss-DNA into a double stranded (ds) replicative intermediate (RI) that is used as template for rolling circle replication (RCR) which generates progeny single stranded DNA molecules (Bernardi and Timchenko, 2008). Geminiviruses do not encode DNA polymerases for replication of their DNA genome but rely on DNA polymerases encoded by the host. DNA polymerase α is responsible for generation of dsRIs by creating the complementary strand of ss-DNA whereas polymerase δ promotes subsequent accumulation of new geminiviral ss-DNA from the dsRI template (Wu et al., 2021). RCR is initiated by Rep with the introduction of a nick within the nonanucleotide sequence of a dsRI molecule (Ruhel and Chakraborty, 2019). Apart from the essential functional role of Rep in replication initiation, Rep is also responsible for recruiting (Gong et al., 2021; Kushwaha et al., 2017) host factors involved in ubiquitination to promote trimethylation of histones on viral replisomes, leading to enhanced viral genome transcription. Viral genes are clock- and anti-clock wise oriented on the DNA genome and become expressed via bidirectional transcription of dsDNA molecules (Fondong, 2013).
Resistance to the geminivirus ToLCNDV has been investigated in cucurbits (Saez et al., 2021) since the virus is a major threat to these crops (Desbiez et al., 2021; Fortes et al., 2016; Juarez et al., 2014; Panno et al., 2016; Orfanidou et al., 2019; Zaidi et al., 2017). In melon (Cucumis melo) ToLCNDV resistance is attributed to two minor QTLs in chromosomes 2 and 12 and one major recessive QTL in chromosome 11: 30,112,563-30,725,907 nt (melonomics v.4) (Sáez et al., 2017; Roman et al., 2019; Romay et al., 2019; Saez et al., 2022) whereas in Cucurbita moschata the same type of resistance was mapped in a major recessive QTL on chromosome 8 (Saez et al., 2016; Sáez et al., 2020). This region of C. moschata is highly syntenic to chromosome 11 of melon, again at the similar (overlapping) locus ch11:30,045,251-31,108,850 nt. Within this syntenic region, two C. moschata genes (CmoCh08G001790, CmoCh08G001780) showed high impact mutations in the resistant parents, presumably leading to a knock-out of the gene, while another two (CmoCh08G001760, CmoCh08G001720) exhibited moderate changes that could alter the biochemical properties of the protein. The homologues of these genes in melon are MELO3C022313 (putative transmembrane protein), MELO3C022314 (putative F17L21.8), MELO3C022315 (MADS-box transcription factor 8-like) and MELO3C022319 (DNA primase large subunit), respectively. Due to the synteny of the QTLs between C. moschata and melon any changes in the aforementioned melon homologues could also be associated with resistance to ToLCNDV.
In our study, we evaluated these four candidate genes. Our plant genomic and functional data analyses revealed that plant DNA primase is essential for geminivirus pathogenicity. This enzyme is part of the DNA Primase complex, that synthesizes RNA primers for initiation of a DNA strand that is complementary to a single-stranded DNA template. A model is presented to explain the role of melon DNA primase in geminiviral ss-DNA replication.
Leaf material from the ToLCNDV resistant line K18 carrying the recessive QTL in chromosome 11 and the susceptible line of similar genetic background K15 but lacking the ch11 QTL, was provided by Nunhems B.V in order to extract genomic DNA (gDNA) for whole genome sequencing (WGS). 200 mg of tissue per genotype were ground to fine powder using liquid nitrogen. The protocol of Healey et al. (2014) was followed for isolation of highly concentrated, high quality gDNA. 1.2 ug of non-contaminated, non-degraded gDNA, was sent to Novogene Cambridge Genomics Center, United Kingdom, for library preparation and WGS (PE150, Q30≥80%), using the Illumina Novaseq 6000 platform. Raw reads were aligned using Bwa mem with less stringent settings (-A1 -B1 -E1 -O1 -M) to reference genome Melon_v4.0 (Castanera et al., 2020), which includes the chloroplast and mitochondrial genome sequences. Duplicate reads were tagged using Mark Duplicates from Picardtools. Samtools was used for post-processing of the alignment files and Alfred for computing quality statistics of the alignment files (Danecek et al., 2021). Variants of chr 11 of both samples were joined called using FreeBayes (min-mapping-quality 2) and filtered using vcflib (vcffilter -f “QUAL > 1 & QUAL/AO > 10 & SAF > 0 & SAR > 0 & RPR > 1 & RPL >1”). SNPeff was used to predict the effect of protein-coding SNVs on the structural phenotype of proteins (De Baets et al., 2012).
115 N. benthamiana plants were grown at Unifarm, Wageningen University & Research. The greenhouse growth conditions were 24°C - 18°C day/night temperature with a photoperiod of 16 hours and 60% relative humidity (RH) for optimal germination and growth.
A 321bp region of the N. benthamiana DNA primase large subunit (NbPriL) coding sequence targeting both gene transcript variants (Niben101Scf00366g01013.1, Niben101Scf01950g03015.1, Sol Genomics Network) was amplified through PCR with primers (ForwardNbPriL:5’-AGGATGCAACTTGGTCTATTTCTC-3’, ReverseNbPriL:5’-TTGTCTAACACATCCTCAACTGCT-3’). The region was chosen based on the best scoring of Sol Genomics Network VIGS tool for optimal virus induced gene silencing. The PCR product was purified and cloned into a pENTR™/D-TOPO™ vector with the cloning pENTR Directional TOPO® cloning kit. Using the Gateway™ LR Clonase™ Enzyme Mix the cloned insert was incorporated into the pTRV2 vector. Agrobacterium tumefaciens strain C58C1 was transformed using heat shock method with the pTRV2:NbPriL vector for downstream VIGS assays. In the same way the TRV : GUS and TRV : PDS constructs were previously generated.
Agrobacterium tumefaciens cultures transformed with the various TRV2 constructs (TRV : NbPriL, TRV : NbGUS and TRV : NbPDS) were mixed in 1:1 ratio with TRV1 cultures until a final OD600 of 2 was reached, and were eluted into MMA buffer (10 mM MES pH 5.6, 10 mM MgCl2, 200 Mm acetosyringone) (Norkunas et al., 2018). GUS encodes the beta-glucuronidase enzyme and is a known reporter gene that is not found in N. benthamiana genome (Hull and Devic, 1995). Two-week-old N. benthamiana cotyledons were syringe infiltrated with these mixtures. In plants with the TRV: GUS construct, the plant silencing machinery was turned on without any of the native plant genes being silenced. PriL silenced plants were compared to GUS silenced plants. Silencing of the N. benthamiana PDS gene was used as a positive control of silencing as successful silencing of this gene leads to a characteristic clearly visible developmental phenotype (photo bleaching). 40 plants were infiltrated with TRV : NbPriL, 40 plants with TRV : NbGUS and 5 with TRV : NbPDSA. tumefaciens cultures whereas the remaining 30 plants were kept wild type (WT).
As soon as the PDS silenced plants exhibited the photo-bleaching phenotype (2 weeks post infiltration) showing successful silencing, agro-inoculation (Peyret and Lomonossoff, 2015) with various geminiviruses took place. Agrobacteria cultures (OD600 = 2) transformed with viral vectors of BCTV (Stanley et al., 1986), TYLCV (Morilla et al., 2005) or ToLCNDV (Lee et al., 2020) were eluted in MMA buffer and syringe infiltrated into 1-2 fully grown leaves of N. benthamiana. Eight plants were used per virus per VIGS construct, and six plants per virus for the WT controls. Eight TRV : GUS, eight TRV : PriL and 6 WT plants remained non-inoculated as controls. Viruses were maintained in different experimental blocks to prevent cross contamination, and the plants within each block were completely randomized.
Young leaf tissue (non-infiltrated, non-inoculated) from all plants was harvested after the appearance of the first systemic viral infection symptoms. TYLCV inoculated plants were harvested 7 dpi as they started to exhibit symptoms earlier, whereas ToLCNDV and BCTV plants were harvested 10 dpi. The tissue was ground into fine powder using metal beads and Qiagen Tissue Lyser II Cat. No./ID: 85300.
Total DNA was extracted from the collected samples in order to determine viral DNA titers. For DNA extraction 200 mg of ground tissue per sample was mixed with 400 μl working buffer solution. The working buffer solution was prepared for 40 samples and contained: a) 12.5 ml extraction buffer (0.35 M Sorbitol, 0.1M Tris-HCl pH 8.0, 5 mM EDTA pH 8.0), b) 12.5 ml lysis buffer (0.2 M Tris-HCl pH 8.0, 0.5M EDTA pH 8, 2 M NaCl, 2% CTAB), c) 5ml sarcosyl 5% (w/v), d) 0.15g NaHSO3, e) 0.6g PVP-40. RNase A 20μg/ml was also added into the buffer mix working solution, and mixed. Samples were incubated at 65°C for 1h. After incubation, two equal volumes of chloroform: isoamylalcohol (24:1) were added for extraction. After vortexing for 30s and subsequent centrifugation for 5 min at 13.000 rpm, the aqueous phase was collected and transferred to a clean microcentrifuge tube. An equal volume of cold isopropanol was added, and the tube inverted several times for DNA precipitation. DNA was pelleted by centrifugation for 5 min at 13.000 rpm. After isopropanol was discarded, DNA pellets were washed with 70% ethanol, and subsequently dried at room temperature for 15 min. DNA was resuspended in sterilized water and stored at -20°C for further analysis.
RNA was extracted from plants in order to determine transcription levels of NbPriL after different treatments. RNA extraction was carried out using TRIzol according to the manufacturers’ protocol (Invitrogen). In brief, 1 ml of Trizol was added to each ground sample and the samples were vortexed for 30s. Next, 200 μL of chloroform was added, and samples again vortexed for 15s, followed by a centrifugation at 13.000 rpm for 30 min at 4°C. From the aqueous phase 400 μL was transferred to a clean microcentrifuge tube and 400 μL of isopropanol was added and mixed. After incubation for 10 min at room temperature, RNA was pelleted during centrifugation at 13000 rpm for 20 min at 4°C, and the pellet washed with 75% ethanol, air dried and subsequently resuspended in 50-70 μL of sterilized water. Thermofisher DNase I was used to remove (residual) DNA from the purified RNA samples. First strand cDNA synthesis was performed using BioRad iScript cDNA synthesis kit according to the manufacturers’ protocol.
Quantitative real time PCR (qRT-PCR) was performed to determine gene expression levels and viral titers. Quantitative RT-PCRs were performed in technical triplicates, using a Bio-Rad CFX96 C1000 Thermal cycler with BioRad iQ SYBR Green supermix. Two sets of primers for the NbPriL gene were used to quantify the total expression of the gene due to the two transcript variants Niben101Scf00366g01013.1 and Niben101Scf01950g03015.1. Primers for viral titer quantification were designed based on the genomic sequence of each virus (Supplementary Table 1) using total DNA extracted from the plants as input. NbPriL total expression was calculated taking into account the CQ values of both transcript variants. NbPriL expression levels as well as viral titers were normalized to the housekeeping gene NbAPR (Solyc02g093150.2) (Liu et al., 2012). The 2-ΔΔCt method (Livak and Schmittgen, 2001) was applied to assess relative transcript levels and viral titers. 2-ΔΔCt was calculated in Microsoft excel and t-test was used to discern the significant differences between control and test samples CQ values.
Having identified four mutations in PriL, leading to an amino acid substitution in the resistant melon parent K18, their occurrence was analyzed in the PRiL gene of 100 melon accessions, as sequenced by Demirci et al. (2021). Eighteen melon accessions of the Center of Genetic Resources, the Netherlands (CGN), carrying at least one of the SNPs present in K18 were grown in a multispan greenhouse with passive ventilation through zenithal windows and cooling system for temperature control at SAKATA, Almeria, Spain Twelve replicates per accession were used for the assay. The same number of susceptible control “Coliseo” melon plants as well as the resistant control “GRAND RIADO” melon plants were added. Temperature range during the assay was 20-28 °C and relative humidity 60-90%. Each plant genotype was maintained in a different experimental block. Whiteflies collected from zucchini plants infected with ToLCNDV were released onto 1 week old plants (5-10 adults/plant). 20 days post inoculation access of viruliferous whiteflies, plants were scored on disease severity. Taking into account the systemic symptoms of ToLCNDV in melon as well as the overall fitness of the plant, a scale of 1-10 of disease score was used (1: fully asymptomatic – 10: fully symptomatic).
The coordinates of the melon PriS and PriL three-dimensional models were obtained from the AlphaFold database (accession codes A0A5A7T8Y3 and A0A1S3CBL6, respectively) (Jumper et al., 2021). Then melon PriS, PriLNTD and PriLCTD, domains were individually fitted into the model of human primase initiation complex (Baranovskiy et al., 2016b) using the “align to molecule” function in PyMOL (The PyMOL Molecular Graphics System, Version 2.0 Schrödinger, LLC.). The linker between PriLNTD and PriLCTD was rebuild and refined using Coot (Coot3Emsley et al., 2010). The sites for the binding of metal ions and substrates are well conserved between the melon and human primases, providing their straightforward transfer from the human to the melon model.
Plant material of melon accession K18 carrying the ToLCNDV recessive resistance QTL was sequenced in order to examine potential changes in the four top candidate gene coding sequences compared to the susceptible K15 background. No high impact changes that could result into early stop codons and gene knock outs were found in any of the four candidate genes examined in K18. A more detailed look revealed no changes whatsoever in the coding sequence of the first candidate i.e. MELO3C022313 compared to the susceptible reference genome. The coding sequence of the second candidate, MELO3C022314, showed two SNPs, one synonymous T-C 30,276,085 nt and one moderate impact SNP G-C 30,275,808 resulting into a conserved L-V aa change in position 100 of the protein. A high impact, frameshift mutation (TA insertion, ch11: 32734434, v.3.6) that could result to a knock-out protein was previously found in MELO3C022314 of CGN24928 accession. However, subsequent disease assays showed that plants of this accession carrying the frameshift mutation in MELO3C022314 were susceptible to ToLCNDV, indicating that this gene is unlikely the susceptibility gene. As a potentially loss of function MELO3C022314 protein could not confer resistance to the virus, this gene was excluded as candidate susceptibility gene. The coding sequence of the third candidate MELO3C022315 revealed only one synonymous nucleotide substitution (A-G, ch11: 30,297,365 nt) which does not impact the encoded protein sequence. However, and interestingly, five changes were found in the coding sequence of the last candidate gene, MELO3C022319. One of the nucleotide substitutions was synonymous, having no effect on the produced protein, whereas the other four resulted into amino acid changes in the DNA primase large subunit (PriL) protein (Table 1). Taken altogether, changes in the DNA primase large subunit most likely caused for the resistance mapped in chromosome 11.
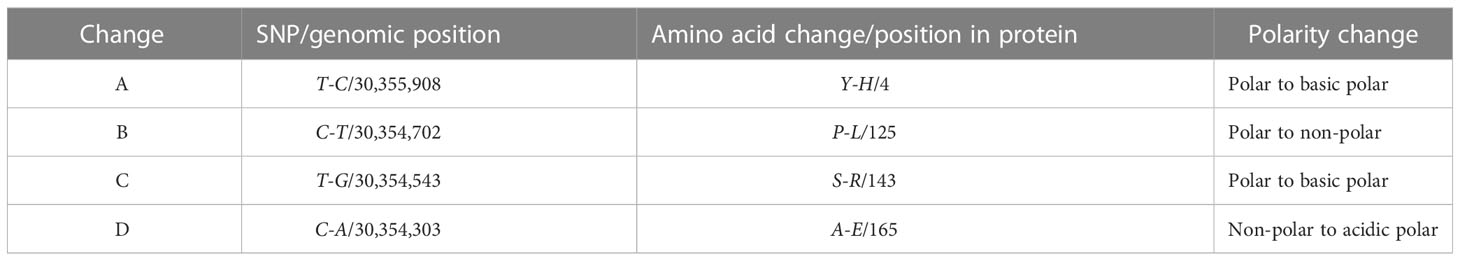
Table 1 Overview on the four types of changes observed in the amino acid sequence of DNA Primase Large subunit (PriL) of melon accession K18, carrying the ToLCNDV recessive resistance QTL on chromosome 11. We used melonomics genome version 4.0.
We knocked down the PriL homologue (NbPriL) by virus induced gene silencing vector in Nicotiana benthamiana, and subsequently challenged the plants with ToLCNDV. In order to investigate the involvement of PriL in the susceptibility to geminiviruses at family and genus level two other distinct geminiviruses were included: Tomato yellow leaf curl virus (TYLCV) (Geminiviridae: begomovirus) and Beet curly top virus (BCTV) (Geminiviridae: curtovirus). N. benthamiana plants verified for a reduction in the level of NbPriL transcripts exhibited a characteristic phenotype that appeared 8-9 days post infiltration with the TRV : NbPriL = construct. This phenotype was consistently observed in all replicates and included shorter internodes and stunting that persisted throughout the experiment. Despite the stunted phenotype of plants infiltrated with TRV : NbPriL, when compared with plants infiltrated with a TRV : NbGUS negative control construct, the former plants were able to survive without any further defects (Figure 1A).
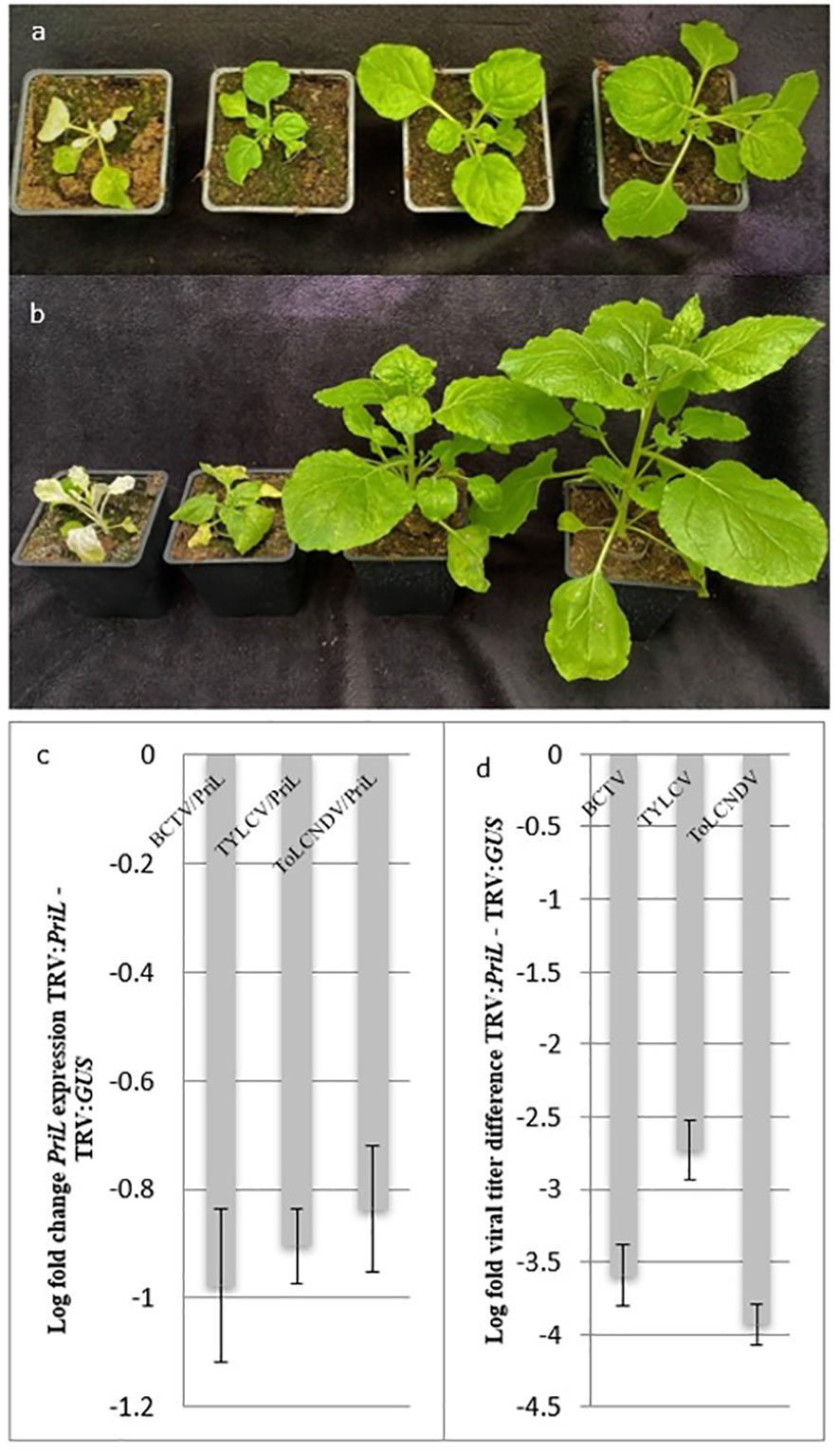
Figure 1 (A). Nicotiana benthamiana plant phenotypes 14 days post infiltration with VIGS constructs. From left to right: TRV: NbPDS, TRV: NbPriL, TRV: GUS, Wild Type (WT); (B) ToLCNDV inoculated N. benthamiana plant phenotypes 10 days post inoculation. From left to right: TRV: NbPDS, TRV: NbPriL, TRV: GUS, Wild Type (WT). Inoculation with the geminiviruses TYLCV and BCTV led to similar phenotypes compared to ToLCNDV. (C) Average log fold PriL expression change of TRV : PriL plants, inoculated with the different viruses, compared to TRV: GUS plants. (D) Average log fold difference in viral titers between TRV: PriL and TRV: GUS plants. Significant difference between TRV : PriL and TRV : GUS (p < 0.05).
Two weeks post infiltration with the VIGS constructs, N. benthamiana plants were agro-inoculated with infectious clones of ToLCNDV, TYLCV and BCTV. In plants that had received the silencing construct TRV-NbGUS disease symptoms started to show up one week post inoculation with TYLCV, consisting of leaf curling and light yellowing of the upper leaves. Symptoms were more prominent in TRV : NbGUS infiltrated plants than in non-silenced wild type N. benthamiana. Three days following the symptoms in TYLCV challenged plants, first symptoms were observed in ToLCNDV (Figure 1B) and BCTV challenged plants, and similarly revealed mild leaf curling and yellowing of the upper leaves, especially in the TRV : GUS plants. However, and in contrast, during this entire period of monitoring no symptoms appeared upon a challenge with any of the geminiviruses in plants silenced on PriL by the TRV-NbPriL construct.
Upon the onset of the first viral symptoms in TRV : GUS plants (i.e. 7 dpi for TYLCV and 10 dpi in ToLCNDV and BCTV), expression levels of NbPriL and viral titers were determined. As expected, PriL expression levels in plants that had received TRV : PriL were significantly reduced compared to the control plants containing TRV : GUS (Figure 1C) and WT plants (p <0.05). When viral titers were determined, significantly lower titers were observed for all viruses in TRV : PriL treated plants compared to the negative control, TRV : GUS treated plants (p<0.05) (Figure 1D). A further look showed that in PriL-silenced plants TYLCV viral titers were higher than the other two viruses (p<0.05). Altogether, silencing of PriL led to significant reduction in viral titers of all three geminiviruses, strengthening its role as a susceptibility factor for geminiviruses.
ToLCNDV disease assays were performed on accessions carrying at least one of the four mutations found in the PriL coding sequence of K18 to investigate whether, and if so which mutations conferred ToLCNDV resistance, similarly to the K18 accession. ToLCNDV disease assay data were collected from greenhouse screenings in Spain using whitefly inoculations. In 18 out of the 100 accessions one or more mutations were present in PriL that were also observed in K18, and leading to an amino acid substitution. Those alleles contained all the mutations as present in K18 (ABCD) or only some of them (ACD, AB, C, D, B, BD, BCD) (Table 1; Supplementary Table 2).
From all four mentioned amino acids changes only mutation A consistently associated with ToLCNDV highly resistant phenotypes (symptomless). All evaluated accessions that harboured the Y to H substitution on position 4 (A) of PriL appeared to exhibit resistant phenotypes to ToLCNDV comparable to the resistant control genotype. Two accessions were carrying the same aa change according to sequencing data, however on a heterozygous state, and appeared to be susceptible to ToLCNDV. One accession carried mutation A in a homozygous state but only about half of the plants that were phenotyped exhibited highly resistant phenotypes to the virus. The rest of the CGN140853 plants were exhibiting fully or intermediately resistant phenotype scores (D.I. with 10 = fully symptomatic and 1 = fully symptomatic: 7.0 in 3/18 plants, 5.0 in 3/18 plants and 3.0 in 2/18 plants, 1.0 which means fully asymptomatic in 10/18 plants). Mutation B exhibited a correlation with phenotypes of intermediate resistance, independently of the presence of mutation A (Supplementary Table 2).
From these observations we conclude that the amino acid substitutions from tyrosine to histidine (Y-H) at position 4, and from proline to leucine (P-L) at position 125 are the most likely changes in PriL in the resistant genotype K18, leading to loss of its function for the virus, but maintenance of the protein’s function for the host plant.
The investigation into the genetic cause of recessive resistance to ToLCNDV on chromosome 11 of melon identified the DNA primase large subunit (PRiL) gene as the most likely candidate. We found non-synonymous mutations in the PRiL gene in the resistant K18 melon line, which were also present in multiple other resistant melon accessions with diverse genotypes. The resistance of these accessions was determined through disease index scoring. In future studies, the quantification of viral titers could be included to confirm that the observed resistance is reflected in reduced viral titers.
To investigate the functional role of PRIL in ToLCNDV susceptibility a VIGS approach was implemented in N. benthamiana (Senthil-Kumar and Mysore, 2014) due to the inefficiency of the method in melon. Efficient silencing of the native PriL homologue in N. benthamianana resulted into characteristic developmental phenotypes but also into significantly reduced viral titers for all evaluated geminiviruses ToLCNDV, TYLCV and BCTV. In future experiments the effect of PriL-silencing will be extended to other viruses (e.g RNA viruses) that do not rely on PriL as a susceptibility factor for their multiplication, to further substantiate the findings from this study.
Our findings indicated a high correlation of melon’s PriL A-type mutation (involving an Y-H substitution, Table 1) with ToLCNDV resistant phenotypes. There was one exception to this correlation, which was observed with accession CGN140853. Although the plants in this accession were homozygous for the A-type mutation, they were still somewhat susceptible to ToLCNDV, although their susceptibility was reduced compared to the susceptible control. Since resistance against ToLCNDV in melon is known to be controlled by more than one QTL (Sáez et al., 2017), it could be that the minor QTLs in chromosome 2 and 12, with an additive effect on resistance, are absent in this particular accession resulting into a more susceptible phenotype. Moreover, the resistance in chromosome 11 could be the result of more than the A-type mutation in PriL. Type B mutation (Table 1) could be another factor of resistant phenotypes. The B mutation gave indications of conferring intermediate resistance (independently of the A presence) in CGN140808 and CGN140870, but the correlation of the mutation with the effect was weaker. Altogether, the results indicate that PriL plays a major role in the pathogenicity of ToLCNDV and other geminiviruses in melon. Eventually, at least two aminoacid changes can alter PriL in a way that still supports the protein’s role for the plant but hamper viral replication.
The plant replication machinery includes several essential genes that are required for DNA replication of the plant. Apart from DNA polymerases, DNA primases are also essential components for DNA replication of the plant. Their role is to synthesize RNA primers that polymerases subsequently elongate on both DNA template strands of the replication fork for the generation of new dsDNA, since polymerases alone are unable for de novo dsDNA synthesis (Frick and Richardson, 2001; Kuchta and Stengel, 2010). Eukaryotic primases are heterodimers consisting of two distinct subunits: a small one, DNA Primase Small subunit (PriS), which carries the active site for RNA primer (~9 nt) synthesis, and a large one, PriL, which determines the activity of the primase and the transfer of the synthesized primer to Polymerase α (Polα) (Baranovskiy et al., 2016b). In organisms with dsDNA, primer synthesis happens only once in the continuous leading strand replication, which becomes extended at its 3’ terminus by Polα and Polϵ for the formation of the complementary strand. In the lagging strand the orientation is opposite. Consequently, RNA primers need to be newly generated constantly, with following extension by Polα and Polδ leading to Okazaki fragments. With this discontinuous strand replication, sequential Okazaki fragments altogether generate the complementary strand of the lagging strand (Zhou et al., 2019).
Considering the fundamental role of PriL in cell replication, the developmental phenotype that was observed after silencing of NbPriL was expected since plant DNA replication was likely inhibited to some extent. Although the development of a normal phenotype was compromised after silencing of NbPriL, the residual expression levels (~50% transcript availability) of NbPriL were still enough to maintain viability and functionality of the plants throughout the experiment. Another indication of the significance of PriL in plant viability was the absence of high impact mutations (e.g coding sequence deletions, insertions) that could lead to a knock-out of the protein in any of the 100 melon genotypes examined. Normally, plants have only one gene copy of PRiL. Loss of function of that gene would be lethal, explaining the absence of high impact mutations in PriL in the 100 melon genomes. This implies that only subtle mutations in PriL would be allowed, such as amino acid substitutions.
Remarkably, despite the thorough studies (Wu et al., 2021) on the role of host polymerases in geminiviral replication there were still no suggestions, comments or experimental data that refer to a possible involvement of host encoded DNA primases in replication of viral DNA. The only study that gets close to this point goes back 30 years ago: Saunders et al. (1992) observed that the conversion of ss- into dsDNA of the geminivirus African cassava mosaic virus (ACMV) initiated in a region very close to the hairpin loop with the conserved nonanucleotide sequence. There, a small RNA nucleotide sequence was preceding the complementary DNA strand, at the start of this complementary strand. This observation pointed to the possibility of priming with a RNA oligonucleotide, of which the origin and generation have remained elusive so far.
Based on findings from this study it is now tempting to hypothesize that PriL together with PriS from the host generates an RNA oligonucleotide as the very first step to prime the complementary strand on the single stranded DNA of the virus, thus enabling the conversion of incoming ssDNA into dsDNA (Figure 2). Whereas the ‘natural’ location of the DNA primase in living organisms is close to the replication fork, the single-stranded DNA geminiviruses do not have such replication fork. However, all geminivirus DNA components contain a hairpin loop in the common region with the conserved nonanucleotide sequence “TAATATTAC” at its top (Figure 2), which resembles a replication fork of DNA replication complexes (Heyraud et al., 1993). The base of this hairpin loop could present the initiation site for DNA Primase to start RNA oligomerization. Docking of the host DNA primase just upstream of the hairpin loop would be in agreement with the findings from Saunders et al. (1992), who mapped an RNA primer at this site. Since all Geminiviruses need the conversion from incoming single-stranded to double-stranded DNA, and all geminiviruses contain the structurally highly conserved region with a hairpin loop, it is likely that priming by DNA primase at the hairpin structure is generic to all geminiviruses, irrespective of the host. This also turns PriL into a susceptibility factor for geminiviruses that do not limit to melon, and opens the possibility for its exploitation in other important crops as well.
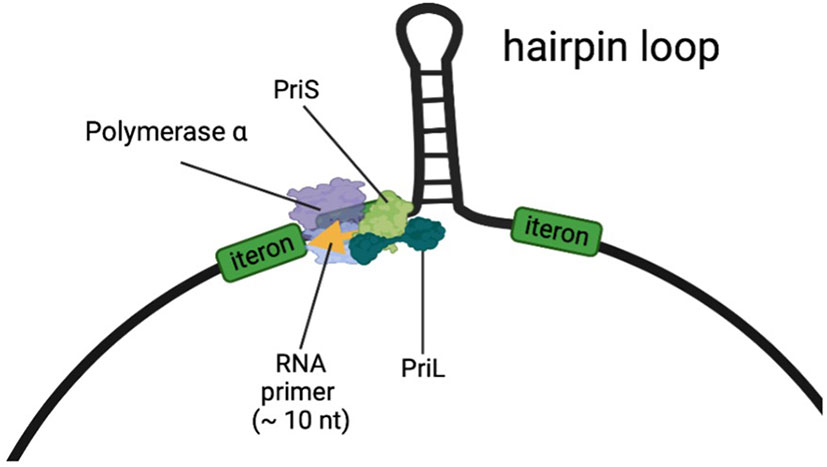
Figure 2 A model for the DNA Primase complex, consisting of PriL, PriS, and DNA polymerase α, producing an RNA primer near the hairpin loop of the ss-DNA of geminiviruses. DNA polymerase α consists of a catalytic subunit (p180) and an accessory subunit (p70). We hypothesize that in the resistant melon one or more mutations in PriL hamper the production or extension of the primer on the viral DNA, but still supporting the primer production in DNA replication forks of the host plant.
Mechanistically, it is yet still unclear how the changes detected in melon PriL specifically would inhibit its functionality towards the virus. The PriL protein comprises of two domains, an N-terminal (PriLNTD) and a C-terminal domain (PriLCTD) with a long, flexible linker of 18 residues in between (Baranovskiy et al., 2016a). PriLCTD is attached to the eukaryotic DNA of the lagging strand, supporting PriS with the initiation of the primer synthesis, while the PriLNTD is attached to PriS which is elongating the primer by adding nucleotides (Baranovskiy et al., 2016a). It is possible that ToLCNDV ss-DNA interacts with the PriL close to the C- terminus of the protein as this is the part that has DNA binding abilities and is unique in higher eukaryotes (Klinge et al., 2007; Weiner et al., 2007; Holzer et al., 2017; Holt et al., 2018). Mutations in this part could inhibit protein-viral DNA affinity and subsequently the efficacy of priming for viral DNA replication. However, the Tyr4His mutation in melon that associates with the resistance to the virus is located in the N-terminal of the protein (position 4). Mutations in the N-terminal could affect the interaction of the protein with the PriS or the Pol α (Baranovskiy et al., 2015), and in case of the latter abrogate extension of the RNA primer by Pol α. Alternatively, binding of other virulence factors (Fondong, 2013) of ToLCNDV to the N-terminal domain of PriL (PriLNTD) could be prevented and affect the functional activity and integrity of the primase/elongation complex. Considering that at this very first step in viral DNA replication, viral genome transcription has not yet occurred, and viral proteins thus are not synthesized, making the latter explanation less likely.
To understand the possible implications of PriL mutations for the discrimination between melon and geminivirus DNA replication, the model of the melon’s primase initiation complex was constructed. The model shows that the 3’-end of template DNA protrudes toward the open space at the junction of PriS and PriLNTD (Figure 3). The major difference between the DNA templates of melon and geminivirus is a conserved 11 bp long stem-loop structure in a geminivirus circular ssDNA. Indeed, the space at the junction of PriS and PriLNTD is a potential site for the docking of the stem-loop upon initiation of primer synthesis on the geminivirus circular DNA. Intriguingly, the Tyr4 and Pro125 are close to this area and their mutations may affect the primase interaction with the stem-loop, causing disruption of a delicate initiation process. The effect could be direct, by disrupting the local docking surface by Pro125Leu mutation or the primase interaction with the stem-loop by the Tyr4His mutation. Alternatively, the other viral or melon protein may facilitate the recruitment and holding of geminivirus stem-loop, and the Tyr4His and Pro125Leu mutations disrupt primase interaction with that accessory protein. Further studies are required to discriminate between these mechanisms.
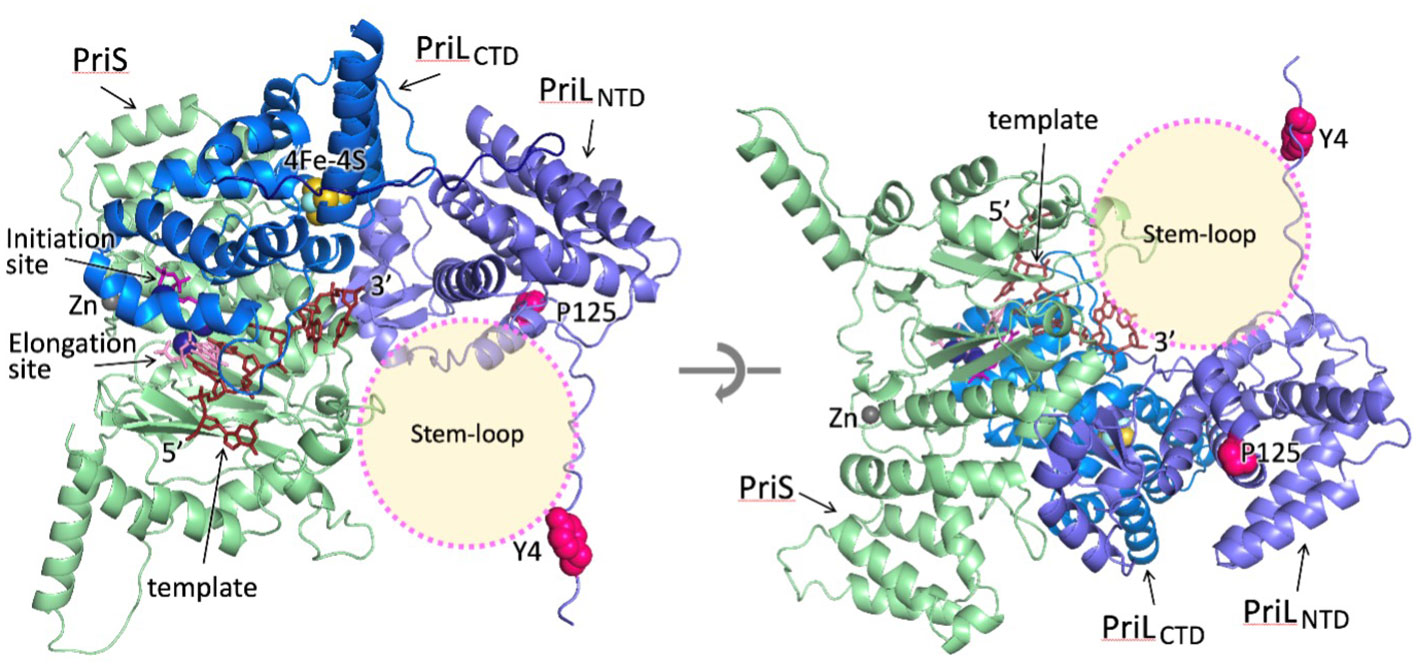
Figure 3 The model of the melon primase initiation complex. For clarity, two different views from opposite directions are shown. The potential stem-loop docking site is indicated by a transparent circle. The two amino acid substitutions in PriL in the resistant melon (Tyr4 and Pro125) are shown as circles in magenta.
The results presented here provide supporting evidence for a role of PriL as an S-gene for geminiviruses. This evidence is based on sequence data analysis of the resistant melon accession K18 with the recessive QTL for resistance on ch11, and on DNA sequences of 100 other melon accessions and concomitant virus challenging assays to analyse their susceptibility or resistance to ToLCNDV. Additional support was obtained by silencing of PriL in N. benthamiana leading to reduced viral titers for three distinct geminivirus species. For almost 30 years the very first step in the conversion of ss-DNA to ds-DNA of geminiviruses has remained unclear. Based on the findings from this study, a role for the host DNA primase in the initiation of geminiviral DNA replication is postulated. How the Tyr4His and Pro125Leu amino acid substitutions within PriL of melon accession K18, carrying the Tomato leaf curl New Delhi virus (ToLCNDV) recessive resistance QTL in chromosome 11, is correlated to the resistance to ToLCNDV remains an experimental challenge for the future.
The original contributions presented in this study are publicly available. The DNA sequence reads of the QTL region of the sequenced melon accessions can be found at European Nucleotide Archive (ENA) (https://www.ebi.ac.uk/ena/) under accession number PRJEB59465.
LS designed, conducted the research and wrote the manuscript, with inputs and supervision from HS, YB, RK and RV. MA contributed to the quantification of viral titers and gene expression for the VIGS experiment. JR, ME and CG contributed to the inoculation and disease scoring of melon accessions. DL provided the ToLCNDV resistant K18 melon for sequencing. TT and AB contributed to protein models for PriL. All authors contributed to the article and approved the submitted version.
The authors declare that this study received funding from Axia Vegetable Seeds, BASF Vegetable Seeds, Bejo Zaden B.V., Hortigenetics Research (S.E. Asia) Limited, Known-You Seed Co. Ltd., Sakata Vegetables Europe, and Syngenta Seeds B.V. Further, BASF provided DNA from a ToLCNDV resistant melon accession together with genetic mapping information of the resistance. The funders were not involved in the study design, analysis, interpretation of data, nor the writing or submission of this article.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1130723/full#supplementary-material
Supplementary Table 1 | Primers used for qRT-PCR.
Supplementary Table 2 | PriL alleles detected in various CGN accessions and the resulted phenotype after a challenge with ToLCNDV. The average ToLCNDV phenotypic disease score is given per accession and refers to a scale of 1 (highly resistant, no symptoms) to 10 (highly susceptible, severe symptoms). The resistant melon control genotype “Coliseo” exhibited a score of 1.5 whereas the susceptible melon control “Grand Riado” exhibited a score of 7.0. DI ≤ 1.5 = Highly resistant, 1.5<DI ≤ 3.0 = Intermediately resistant, 3.0<DI ≤ 5 = Intermediately susceptible, 5<DI ≤ 10 = Fully susceptible.
Baranovskiy, A. G., Babayeva, N. D., Zhang, Y., Gu, J., Suwa, Y., Pavlov, Y. I., et al. (2016a). Mechanism of concerted RNA-DNA primer synthesis by the human primosome. J. Biol. Chem. 291, 10006–10020. doi: 10.1074/jbc.M116.717405
Baranovskiy, A. G., Zhang, Y., Suwa, Y., Babayeva, N. D., Gu, J., Pavlov, Y. I., et al. (2015). Crystal structure of the human primase. J. Biol. Chem. 290 (9), 5635–5646. doi: 10.1074/jbc.M114.624742
Baranovskiy, A. G., Zhang, Y., Suwa, Y., Gu, J., Babayeva, N. D., Pavlov, Y. I., et al. (2016b). Insight into the human DNA primase interaction with template-primer. J. Biol. Chem. 291 (9), 4793–4802. doi: 10.1074/jbc.M115.704064
Bernardi, F., Timchenko, T. (2008). Réplication des géminivirus : le mécanisme du cercle roulant chez les plantes [Geminivirus replication: rolling circle mechanism in plants]. Virologie (Montrouge) 12 (2), 117–135. doi: 10.1684/12-2.2011.10490
Bhattacharjee, B., Hallan, V. (2022). Geminivirus-derived vectors as tools for functional genomics. Front. Microbiol. 13. doi: 10.3389/fmicb.2022.799345
Briddon, R. W., Patil, B. L., Bagewadi, B., Nawaz-Ul-Rehman, M. ,. S., Fauquet, C. ,. M. (2010). Distinct evolutionary histories of the DNA-a and DNA-b components of bipartite begomoviruses. BMC Evol. Biol. 10, 97. doi: 10.1186/1471-2148-10-97
Castanera, R., Ruggieri, V., Pujol, M., Garcia-Mas, J., Casacuberta, J. ,. M. (2020). An improved melon reference genome with single-molecule sequencing uncovers a recent burst of transposable elements with potential impact on genes. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.01815
Danecek, P., Bonfield, J. K., Liddle, J., Marshall, J., Ohan, V., Pollard, M. O., et al. (2021). Twelve years of SAMtools and BCFtools. Gigascience 10 (2), giab008. doi: 10.1093/gigascience/giab008
De Baets, G., Van Durme, J., Reumers, J., Maurer-Stroh, S., Vanhee, P., Dopazo, J., et al. (2012). SNPeffect 4.0: on-line prediction of molecular and structural effects of protein-coding variants. Nucleic Acids Res. 40 (Database issue), D935–D939. doi: 10.1093/nar/gkr996
Demirci, S., Fuentes, R. ,. R., Van Dooijeweert, W., Aflitos, S., Schijlen, E., Hesselink, T., et al. (2021). Chasing breeding footprints through structural variations in cucumis melo and wild relatives. G3 (Bethesda) ;11 (1), jkaa038. doi: 10.1093/g3journal/jkaa038
Desbiez., C., Gentit, P., Cousseau-Suhard., P., Renaudin, I., Verdin, E. (2021). First report of Tomato leaf curl new Delhi virus infecting courgette in France. New Dis. Rep. 43, e12006. doi: 10.1002/ndr2.12006
Emsley, P., Lohkamp, B., Scott, W. G., Cowtan, K. (2010). Features and development of coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501. doi: 10.1107/S0907444910007493
Fiallo-Olivé, E., Lett, J. ,. M., Martin, D. P., Roumagnac, P., Varsani, A., Zerbini, F. ,. M., et al. (2021). ICTV virus taxonomy profile: Geminiviridae 2021. J. Gen. Virol. 102 (12), 1696. doi: 10.1099/jgv.0.001696
Fiallo-Olivé, E., Navas-Castillo, J. (2020). Molecular and biological characterization of a new world mono-/Bipartite Begomovirus/Deltasatellite complex infecting corchorus siliquosus. Front. Microbiol. 11. doi: 10.3389/fmicb.2020.01755
Fondong, V. ,. N. (2013). Geminivirus protein structure and function. Mol. Plant Pathol. 14 (6), 635–649. doi: 10.1111/mpp.12032
Fortes, I. M., Sánchez-Campos, S., Fiallo-Olivé, E., Díaz-Pendón, J. ,. A., Navas-Castillo, J., Moriones, E. (2016). A novel strain of tomato leaf curl new Delhi virus has spread to the Mediterranean basin. Viruses 8 (11), 307. doi: 10.3390/v8110307
Frick, D. N., Richardson, C. C. (2001). DNA Primases. Annu. Rev. Biochem. 70, 39–80. doi: 10.1146/annurev.biochem.70.1.39
Gong, P., Tan, H., Zhao, S., Li, H., Liu, H., Ma, Y., et al. (2021). Geminiviruses encode additional small proteins with specific subcellular localizations and virulence function. Nat. Commun. 12 (1), 4278. doi: 10.1038/s41467-021-24617-4
Healey, A., Furtado, A., Cooper, T., Henry, R. J. (2014). Protocol: a simple method for extracting next-generation sequencing quality genomic DNA from recalcitrant plant species. Plant Methods 10, 21. doi: 10.1186/1746-4811-10-21
Heyraud, F., Matzeit, V., Schaefer, S., Schell, J., Gronenborn, B. (1993). The conserved nonanucleotide motif of the geminivirus stem-loop sequence promotes replicational release of virus molecules from redundant copies. Biochimie 75 (7), 605–615. doi: 10.1016/0300-9084(93)90067-3
Holt, M. ,. E., Salay, L. ,. E., O'brien, E., Barton, J. K., Chazin, W. J. (2018). Functional and structural similarity of human DNA primase [4Fe4S] cluster domain constructs. PloS One 13 (12), e0209345. doi: 10.1371/journal.pone.0209345
Holzer, S., Yan, J., Kilkenny, M. ,. L., Bell, S. ,. D., Pellegrini, L. (2017). Primer synthesis by a eukaryotic-like archaeal primase is independent of its fe-s cluster. Nat. Commun. 8 (1), 1718. doi: 10.1038/s41467-017-01707-w
Hull, G. ,. A., Devic, M. (1995). The beta-glucuronidase (gus) reporter gene system. gene fusions; spectrophotometric, fluorometric, and histochemical detection. Methods Mol. Biol. 49, 125–141. doi: 10.1385/0-89603-321-X:125
Juárez, M., Tovar, R., Fiallo-Olivé, E., Aranda, M. A., Gosálvez, B., Castillo, P., et al. (2014). First detection of tomato leaf curl new Delhi virus infecting zucchini in Spain. Plant Dis. 98 (6), 857. doi: 10.1094/PDIS-10-13-1050-PDN
Jumper, J., Evans, R., Pritzel, A., Green, T., Figurnov, M., Ronneberger, O., et al. (2021). Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589. doi: 10.1038/s41586-021-03819-2
Klinge, S., Hirst, J., Maman, J. ,. D., Krude, T., Pellegrini, L. (2007). An iron-sulfur domain of the eukaryotic primase is essential for RNA primer synthesis. Nat. Struct. Mol. Biol. 14 (9), 875–877. doi: 10.1038/nsmb128
Krupovic, M., Ravantti, J. ,. J., Bamford, D. ,. H. (2009). Geminiviruses: a tale of a plasmid becoming a virus. BMC Evol. Biol. 9, 112. doi: 10.1186/1471-2148-9-112
Kuchta, R. ,. D., Stengel, G. (2010). Mechanism and evolution of DNA primases. BiochimBiophys Acta 1804 (5), 1180–1189. doi: 10.1016/j.bbapap.2009.06.011
Kumar, R. V. (2019). Plant antiviral immunity against geminiviruses and viral counter-defense for survival. Front. Microbiol. 10. doi: 10.3389/fmicb.2019.01460
Kushwaha, N. ,. K., Mansi, Chakraborty, S. (2017). The replication initiator protein of a geminivirus interacts with host monoubiquitination machinery and stimulates transcription of the viral genome. PLoSPathog 13 (8), e1006587. doi: 10.1371/journal.ppat.1006587
Lee, C. ,. H., Zheng, Y. ,. X., Chan, C. ,. H., Ku., H. ,. M., Chang, C. ,. J., Jan, F. ,. J. (2020). A single amino acid substitution in the movement protein enables the mechanical transmission of a geminivirus. Mol. Plant Pathol. 21 (4), 571–588. doi: 10.1111/mpp.12917
Liu, D., Shi, L., Han, C., Yu, J., Li, D., Zhang, Y. (2012). Validation of reference genes for gene expression studies in virus-infected nicotiana benthamiana using quantitative real-time PCR. PloS One 7 (9), e46451. doi: 10.1371/journal.pone.0046451
Livak, K. ,. J., Schmittgen, T. ,. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods 25 (4), 402–408. doi: 10.1006/meth.2001.1262
Morilla, G., Janssen, D., Garcia-Andres, S., Moriones, E., Cuadrado, I. ,. M., Bejarano, E. ,. R. (2005). Pepper (Capsicum annuum) is a dead-end host for tomato yellow leaf curl virus. Phytopathology 95 (9), 1089–1097. doi: 10.1094/PHYTO-95-1089
Norkunas, K., Harding, R., Dale, J., Dugdale, B. (2018). Improving agroinfiltration-based transient gene expression in Nicotiana benthamiana. Plant Methods 14, 71. doi: 10.1186/s13007-018-0343-2
Orfanidou, C. ,. G., Malandraki, I., Beris, D., Kektsidou, O., Vassilakos, N., Varveri, C., et al. (2019). First report of tomato leaf curl new Delhi virus in zucchini crops in Greece. J. Plant Pathol. 101, 799. doi: 10.1007/s42161-019-00265-y
Panno, S., Iacono, G., Davino, M., Marchione, S., Zappardov, B. P., Tomassoli, L., et al. (2016). First report of tomato leaf curl new Delhi virus affecting zucchini squash in an important horticultural area of southern Italy. New Dis. Rep. 33, 6. doi: 10.5197/j.2044-0588.2016.033.006
Peyret, H., Lomonossoff, G. ,. P. (2015). When plant virology met agrobacterium: the rise of the deconstructed clones. Plant Biotechnol. J. 13 (8), 1121–1135. doi: 10.1111/pbi.12412
Román, B., Gómez, P., Picó, B., López, C., Janssen, D. (2019). Candidate gene analysis of tomato leaf curl new Delhi virus resistance in Cucumis melo. Sci. Hortic. 243 (3), 12–20. doi: 10.1016/j.scienta.2018.07.005
Romay, G., Pitrat, M., Lecoq, H., Wipf-Scheibel, C., Millot, P., Girardot, G., et al. (2019). Resistance against melon chlorotic mosaic virus and tomato leaf curl new Delhi virus in melon. Plant Dis. 103 (11), 2913–2919. doi: 10.1094/PDIS-02-19-0298-RE
Ruhel, R., Chakraborty, S. (2019). Multifunctional roles of geminivirus encoded replication initiator protein. Virusdisease 30 (1), 66–73. doi: 10.1007/s13337-018-0458-0
Sáez, C., Ambrosio, L., G., M., Miguel, S. ,. M., Valcárcel, J. ,. V., Díez, M. ,. J., Picó, B., et al. (2021). Resistant sources and genetic control of resistance to ToLCNDV in cucumber. Microorganisms 9 (5), 913. doi: 10.3390/microorganisms9050913
Sáez, C., Esteras, C., Martínez, C., Ferriol, M., Dhillon, N.P.S., López, C., et al. (2017). Resistance to tomato leaf curl new Delhi virus in melon is controlled by a major QTL located in chromosome 11. Plant Cell Rep. 36, 1571–1584. doi: 10.1007/s00299-017-2175-3
Sáez, C., Flores-León, A., Montero-Pau, J., Sifres, A., Dhillon, N.,P.,S., López, C., et al. (2022). RNA-Seq transcriptome analysis provides candidate genes for resistance to tomato leaf curl new Delhi virus in melon. Front. Plant Sci. 18 (12), 798858. doi: 10.3389/fpls.2021.798858
Sáez, C., Martínez, C., Ferriol, M., Manzano, S., Velasco, L., Jamilena, M., et al. (2016). Resistance to tomato leaf curl new Delhi virus in cucurbita spp. Ann. Appl. Biol. 169 (1), 91–105. doi: 10.1111/aab.12283
Sáez, C., Martínez, C., Montero-Pau, J., Esteras, C., Sifres, A., Blanca, J., et al. (2020). A major QTL located in chromosome 8 of Cucurbita moschata is responsible for resistance to tomato leaf curl new Delhi virus. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.00207
Saunders, K., Lucy, A., Stanley, J. (1992). RNA-Primed complementary-sense DNA synthesis of the geminivirus African cassava mosaic virus. Nucleic Acids Res. 20 (23), 6311–6315. doi: 10.1093/nar/20.23.6311
Senthil-Kumar, M., Mysore, K. (2014). Tobacco rattle virus–based virus-induced gene silencing in Nicotiana benthamiana. Nat. Protoc. 9, 1549–1562. doi: 10.1038/nprot.2014.092
Stanley, J., Markham, P. ,. G., Callis, R. ,. J., Pinner, M. ,. S. (1986). The nucleotide sequence of an infectious clone of the geminivirus beet curly top virus. EMBO J. 5 (8), 1761–1767. doi: 10.1002/j.1460-2075.1986.tb04424.x
Weiner, B. ,. E., Huang, H., Dattilo, B. ,. M., Nilges, M. ,. J., Fanning, E., Chazin, W. ,. J. (2007). An iron-sulfur cluster in the c-terminal domain of the p58 subunit of human DNA primase. J. Biol. Chem. 282 (46), 33444–33451. doi: 10.1074/jbc.M705826200
Wu, M, Wei, H, Tan, H, Pan, S, Liu, Q, Bejarano, ER, et al. (2021). Plant DNA polymerases α and δ mediate replication of geminiviruses. Nat Commun. 12 (1), 2780. doi: 10.1038/s41467-021-23013-2
Zaidi, S. ,. S., Martin, D. ,. P., Amin, I., Farooq, M., Mansoor, S. (2017). Tomato leaf curl new Delhi virus: a widespread bipartite begomovirus in the territory of monopartite begomoviruses. Mol. Plant Pathol. 18 (7), 901–911. doi: 10.1111/mpp.12481
Keywords: recessive resistance, ToLCNDV, DNA Primase, DNA replication, melon, geminivirus
Citation: Siskos L, Antoniou M, Riado J, Enciso M, Garcia C, Liberti D, Esselink D, Baranovskiy AG, Tahirov TH, Visser RGF, Kormelink R, Bai Y and Schouten HJ (2023) DNA primase large subunit is an essential plant gene for geminiviruses, putatively priming viral ss-DNA replication. Front. Plant Sci. 14:1130723. doi: 10.3389/fpls.2023.1130723
Received: 23 December 2022; Accepted: 03 March 2023;
Published: 17 March 2023.
Edited by:
Keiichi Okazaki, Niigata University, JapanCopyright © 2023 Siskos, Antoniou, Riado, Enciso, Garcia, Liberti, Esselink, Baranovskiy, Tahirov, Visser, Kormelink, Bai and Schouten. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Henk J. Schouten, aGVuay5zY2hvdXRlbkB3dXIubmw=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.