
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Plant Sci., 30 January 2023
Sec. Plant Pathogen Interactions
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1127833
This article is part of the Research TopicInduced Plant Resistance Against Pathogens by Application of Bioactive MoleculesView all 7 articles
Accumulating evidence suggests that chloroplasts are an important battleground during various microbe-host interactions. Plants have evolved layered strategies to reprogram chloroplasts to promote de novo biosynthesis of defense-related phytohormones and the accumulation of reactive oxygen species (ROS). In this minireview, we will discuss how the host controls chloroplast ROS accumulation during effector-triggered immunity (ETI) at the level of selective mRNA decay, translational regulation, and autophagy-dependent formation of Rubisco-containing bodies (RCBs). We hypothesize that regulation at the level of cytoplasmic mRNA decay impairs the repair cycle of photosystem II (PSII) and thus facilitates ROS generation at PSII. Meanwhile, removing Rubisco from chloroplasts potentially reduces both O2 and NADPH consumption. As a consequence, an over-reduced stroma would further exacerbate PSII excitation pressure and enhance ROS production at photosystem I.
Chloroplasts are the energy center where solar energy is converted into chemical energy via the photosynthetic electron transport chain (PETC). Oxygenic photosynthesis unavoidably generates a large amount of reactive oxygen species (ROS) in the form of singlet oxygen (1O2), superoxide (O2•-), hydroxyl radicals (OH•) and hydrogen peroxide (H2O2) (Foyer and Hanke, 2022; Li and Kim, 2022). Because of the coordination between photosynthetic control and ROS scavenging systems, chloroplastic ROS (cROS) are maintained at a relatively low level (Foyer and Hanke, 2022; Li and Kim, 2022). cROS accumulation has been observed in pattern-triggered immunity (PTI), effector-triggered immunity (ETI) and various types of host-microbe interactions (Liu et al., 2007; Dong and Chen, 2013; de Torres Zabala et al., 2015; Lu and Yao, 2018; Shang-Guan et al., 2018; Su et al., 2018; Kuźniak and Kopczewski, 2020; Kachroo et al., 2021; Littlejohn et al., 2021). Based on the observation that ETI triggers cROS accumulation whereas effectors of a virulent pathogen suppress this response (de Torres Zabala et al., 2015; Su et al., 2018), it has been widely accepted that cROS is essential for robust ETI (Lu and Yao, 2018; Sowden et al., 2018; Kuźniak and Kopczewski, 2020; Kachroo et al., 2021; Littlejohn et al., 2021). It is worth noting that cROS promotes susceptibility to necrotrophic pathogens (Rossi et al., 2017).
In most cases known to date, ETI is initiated by the activation of intracellular nucleotide-binding leucine-rich repeat receptors (NLRs). According to their biological functions, NLRs are classified into sensor NLRs and helper NLRs (Adachi et al., 2019; Feehan et al., 2020). Upon activation, the majority of sensor NLRs further activate helper NLRs to execute ETI. Activated helper NLRs oligomerize at the plasma membrane to form Ca2+ channels, leading to cytoplasmic Ca2+ influx (Jacob et al., 2021.; Contreras et al., 2022). In the case of the ancient sensor NLR ZAR1, which is functionally conserved across plant species, it is the sensor NLR itself that oligomerizes to form Ca2+ channels (Bi et al., 2021). Cytoplasmic Ca2+ influx triggers multiple downstream immune responses, including activation of calcium-dependent protein kinases (CPKs) and mitogen-activated protein kinases (MPKs), ROS accumulation both at the plasma membrane and in chloroplasts, and reprogramming of the cell at transcriptional, translational, and metabolic levels (Ngou et al., 2022). Rapid chloroplast Ca2+ transients were observed during PTI. Activation of pattern recognition receptors in PTI also leads to cytoplasmic Ca2+ influx, in this case mediated by plasma membrane-resident Ca2+ channels of the cyclic nucleotide-gated channel (CNGC), the glutamate receptor-like (GLR) channel, and the reduced hyperosmolality induced Ca2+ increase (OSCA) channel families (Kim et al., 2022). This cytoplasmic Ca2+ influx in turn is sensed by the chloroplast-localized calcium-sensing receptor (CAS) (Nomura et al., 2012). Because cas mutants showed an impaired ETI-induced hypersensitive response (HR), it is highly possible that cytoplasmic Ca2+ influx could also alter chloroplast Ca2+ dynamics during ETI, which might affect cROS accumulation. This review will discuss how cROS accumulation might be regulated long-term by cytoplasmic processes after the initial events of ETI activation. Because we focus our discussion on events in the cytoplasm, other important contributions to cROS accumulation, such as the chloroplast internal PSII repair cycle and chloroplastic Ca2+ dynamics, are not included in this minireview.
The stability of mRNA varies widely, with deduced half-lives ranging from minutes to more than 24 h (Narsai et al., 2007). The deduced half-lives for many photosynthesis-related transcripts are long-lived, such as LHCB4.2 (t1/2 ≈ 4.8 h), PSAD-2 (t1/2 ≈ 5.5 h), PSBQ-2 (t1/2 ≈ 8.1 h), PSBP-1 (t1/2 ≈ 13.4 h), PSAH-1 (t1/2 ≈ 17.6 h), and PSAK (t1/2 ≈ 20.3 h). An analysis of early microarray data found that mRNA levels of photosynthesis-related genes involved in light reaction, carbon assimilation, and chlorophyll synthesis globally decrease irrespective of the type of host-microbe interaction, suggesting that global downregulation of photosynthesis-related genes is a component of defense responses (Bilgin et al., 2010). Later, drastic downregulation of photosynthesis-related transcripts was observed by activation of MPK3 and MPK6 (Su et al., 2018), two MAP kinases displaying sustained activation during ETI (Tsuda et al., 2013; Su et al., 2018). Surprisingly, many transcripts encoding subunits of PSII and PSI as well as PSII activity regulators drastically decreased after long-term MPK3/MPK6 activation or during ETI (Su et al., 2018; Yoo et al., 2020). Interestingly, genes at each step of the PSII repair cycle were found to be drastically decreased by MPK3/MPK6 activation (Su et al., 2018), for example STN8 kinase required for PSII core protein phosphorylation, metalloproteases FtsH1/2/5 and Deg family of proteases Deg5/8 involved in the degradation of damaged PSII core protein D1, and factors such as LQY1, PAM68, PSB28, LBA1, LBA2, and ALB3 involved in de novo synthesis and assembly of D1 into the PSII core. Thus, the authors proposed that PSII is actively damaged during ETI, which promotes the accumulation of cROS (Su et al., 2018). The drastic decrease of these otherwise highly stable transcripts suggests that photosynthesis-related transcripts are actively degraded during ETI via mRNA decay mechanisms.
mRNA decay plays an important role in fine-tuning mRNA abundance. Eukaryotic mRNAs are characterized by a 5’ m7G cap and a 3’ poly(A) tail. The classical view on mRNA decay was thought to consist of a step-wise process involving mRNA disassociation from ribosomes, progressive removal of the poly(A) tail, 5’ decapping, and exonucleolytic digestion in either the 3’-to-5’ or 5’-to-3’ direction (Abbasi et al., 2013; Łabno et al., 2016; Zhang and Guo, 2017; Li et al., 2018) (Figure 1). Specifically, mRNA catabolism is typically initiated with 3’ poly(A) deadenylation by the 3’-5’ poly(A)-specific ribonuclease (PRAN) complex and the carbon catabolite repression 4-negative on TATA-less (CCR4-NOT) complex. Then, deadenylated mRNA undergoes either 3’-to-5’ decay by the RNA exosome, a multi-subunit exonuclease complex, or 5’-to-3’ decay mediated by the XRN family of exoribonucleases. The 5’-to-3’ decay can only occur after the removal of the 5’ m7G cap, which is catalyzed by the decapping enzyme Decapping 2 (DCP2).
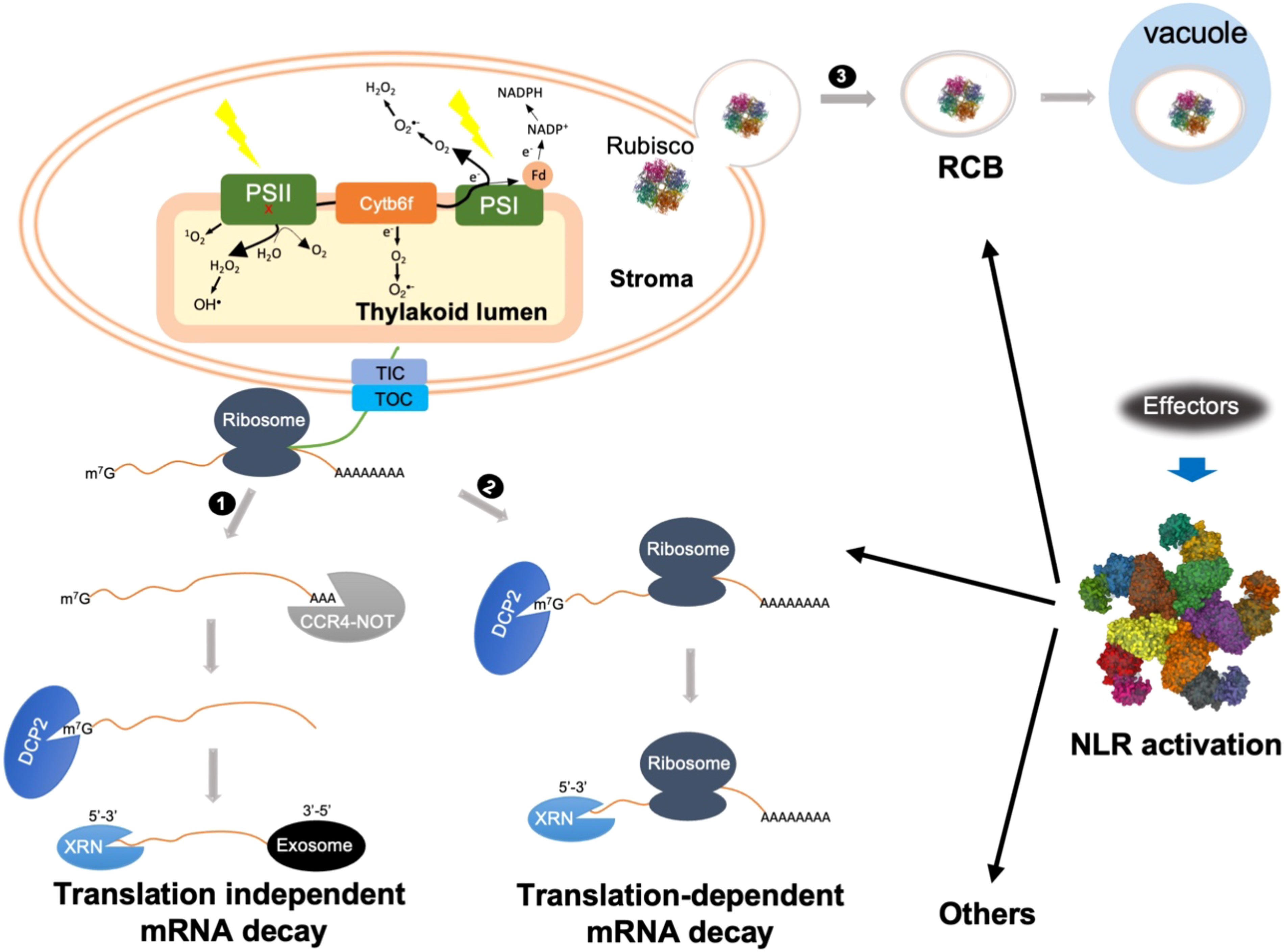
Figure 1 Schematic representations of possible cytoplasmic regulatory mechanisms of cROS accumulation during ETI. ETI activation might trigger both translation-dependent and -independent mRNA decay to actively degrade photosynthetic transcripts. 1) Translation-independent mRNA decay is initiated with CCR4-NOT complex-mediated deadenylation. After 5’ decapping, further degradation is carried out by either 5’-3’ or 3’-5’ decay mediated by XRN or exosome, respectively. 2) Translation-dependent mRNA decay can only occur through XRN-mediated 5’-3’ decay. Bulk mRNA decay of photosynthetic transcripts impairs the repair or turnover cycle of photosynthetic complexes, mainly PSII, resulting in 1O2 and OH• production from damaged PSII, indicated by a red x. 3) NLR activation triggers autophagy-dependent formation of Rubisco-containing body (RCB) through which Rubisco as well as other stroma proteins are delivered into the vacuole for degradation. Removal of Rubisco may cause stroma overreduction and/or reduce O2 consumption, which facilitates electron leakage from PSI as well as cytochrome b6f complex (Cyt b6f) to O2, resulting in O2•- formation at both stroma side and inside thylakoid lumen. Other cytoplasmic mechanisms might also contribute to cROS accumulation during ETI.
In addition to the classical deadenylation-mediated mRNA decay pathway, recent advances highlight the importance of cotranslational mRNA decay, referred to as translation-dependent mRNA decay (Figure 1) (Zhang and Guo, 2017; Tuck et al., 2020; Biasini et al., 2021; Mishima et al., 2022). For a long time, ribosome disassociation was believed to be a prerequisite for mRNA degradation. However, in 2009 Hu and co-workers revealed that mRNA decay could occur on transcripts associated with translating ribosomes (Hu et al., 2009). Subsequent high-throughput experiments revealed that cotranslational mRNA decay is widespread (Pelechano et al., 2015; Yu et al., 2016; Ibrahim et al., 2018). Cotranslational mRNA decay is carried out by components of the translation-dependent cytosolic mRNA surveillance pathways, such as nonsense-mediated decay (NMD), no-go decay (NGD), and no-stop decay (NSD), which are generally thought to remove aberrant transcripts with premature stop codons, transcripts with stalled ribosomes, and transcripts lacking a stop codon, respectively. Nevertheless, a growing body of evidence suggests an important role for these pathways in downregulating functional mRNAs in yeast, mammals, and plants (Łabno et al., 2016; Yu et al., 2016; Simsek et al., 2017; Zhang and Guo, 2017; Heck and Wilusz, 2018; Tuck et al., 2020; Biasini et al., 2021; Morris et al., 2021; Mishima et al., 2022). It is becoming clearer that these translation-dependent mRNA surveillance pathways do not uniquely function in mRNA quality control, but also play an important role in downregulating a large spectrum of functional mRNAs at the transcriptome level. It remains to be determined how translation-independent and translation-dependent mRNA decay are coordinated during development and under stress conditions. In future, it will be interesting to test how classical and cotranslational mRNA decay processes contribute to the degradation of photosynthetic transcripts during ETI.
During PTI, translation is globally reprogrammed, resulting in poor correlation with changes in mRNA levels (Xu et al., 2017). To date, changes of the translatome during ETI have been less studied than those of the transcriptome. With TRAP-seq (translating ribosome affinity purification RNA-seq), one group measured ETI-induced translatome changes at 2 h post AvrRpm1 induction, which triggers RPM1-dependent ETI (Meteignier et al., 2017). No global translational reprogramming was observed during ETI. At the given time point, only three photosynthesis-related differentially expressed genes (DEGs) were found to be downregulated, namely PSAD-2, LHCB2.2 and LHCB4.2. However, differentially translated genes (DTGs) for photosynthesis were highly represented among the 2591 down-regulated DTGs (Meteignier et al., 2017). Hardly any photosynthetic genes were found among the upregulated DEGs and DTGs. Based on this 2 h ETI (early stage) dataset the high number of DTGs and low number of DEGs among photosynthetic transcripts suggest that most of the photosynthetic transcripts are disassociated from polysomes and transferred into RNA-processing bodies for decapping or for short-term storage. Thus, it is highly possible that most photosynthetic transcripts undergoing mRNA decay eventually do so in a classical manner, a promising hypothesis worth testing in the future.
Using Ribo-seq, a protocol entailing polysome enrichment, RNase I digestion, and cDNA library construction to recover ribosome-protected mRNA fragments for further analysis, the work by Yoo et al. measured the ETI-induced traslatome at 8 h post inoculation with Pseudomonas syringae pv. maculicola expressing AvrRpt2, which triggers RPS2-dependent ETI. In this study, 983 upregulated DEGs, 203 downregulated DEGs, 926 upregulated DTGs and 156 downregulated DTGs were discovered (Yoo et al., 2020). At the time point after treatment in this study (8 h post inoculation, approximately middle stage of ETI), ETI-induced changes in transcription and translation are highly correlated (r = 0.92). Consistent with results at 2 h of AvrRpm1-triggered ETI (Meteignier et al., 2017), DEGs and DTGs for both LHCB2.2 and LHCB2.4 were also downregulated (Yoo et al., 2020). Interestingly, photosynthetic transcripts showed varied translation efficiency, with some displaying increased and some decreased efficiency at 8 h after AvrRps2-triggered ETI, though both their DEGs and DTGs were downregulated. For example, the reduction for LHCB2.2 was 2.8-fold by RNA-seq, while it was 2.1-fold by Ribo-seq. This means that polysome-bound LHCB2.2 mRNA decreased less compared to the total decrease of LHCB2.2 mRNA, i.e. polysome-bound LHCB2.2 transcript contributed less to mRNA decay than unbound transcripts or those bound to free 40S or 80S ribosomes. Conversely, the reduction for LHCA6 was 1.5-fold by RNA-seq, while it was 1.9-fold based on Ribo-seq, suggesting that polysome-bound LHCA6 mRNA contributed more to mRNA decay, indicating that translation-dependent decay of LHCA6 mRNA occurs. The varying translation efficiency suggests that translation-dependent and -independent mRNA decay contribute differently to the regulation of different photosynthetic transcripts.
Two major caveats in these considerations are that we are comparing translatome datasets from two different groups, which were generated with different materials, and even with different translatome analysis methods, and that these datasets were derived from single, and different, time points after ETI induction with two different effectors. For a comprehensive understanding of ETI-induced translatome changes, more time points across the whole ETI process are required.
Rubisco (Ribulose-1,5-biphosphate carboxylase-oxygenase), the most abundant protein on earth, accounts for 12%-30% of total leaf protein in C3 plants (Andersson and Backlund, 2008). Rubisco-containing bodies (RCB), small spherical bodies localized both in the cytoplasm and vacuole, were first observed in naturally senescing wheat leaves by immunolocalization of the large subunit of Rubisco (Chiba et al., 2003). Later, RCBs were confirmed to be a type of autophagic body that transfers Rubisco and other stromal proteins to the vacuole for degradation (Chiba et al., 2003; Ishida et al., 2008; Wada et al., 2009). The formation of RCBs is always associated with carbon starvation or senescence, so it is considered to be an efficient nutrient recycling mechanism while maintaining some basal functions of chloroplasts (Izumi et al., 2010; Yoshitake et al., 2021).
RCB induction was also observed during AvrRps4-triggered ETI (Dong and Chen, 2013). The avirulent strain P. syringae pv. tomato DC3000 expressing AvrRps4 [DC3000(AvrRps4)] induced many small RCBs but few large bodies. Conversely, the virulent strain DC3000 induced a small number of RCBs but proportionally more large bodies. The large bodies showed a similar size with chloroplasts, indicating these are possibly whole-chloroplast autophagic bodies. It seems that ETI preferentially induces RCB formation rather than whole-chloroplast autophagy, indicating that RCB formation might play a role during ETI. The authors found that RCB formation and ETI were abolished in atg5-1, a mutant defective in autophagy. Surprisingly, DC3000(AvrRps4)-induced cROS accumulation was also abolished in atg5-1, suggesting that RCB formation facilitates cROS accumulation. If this assumption is true, one would expect less cROS accumulation in response to virulent DC3000.
Rubisco catalyzes both carboxylation and oxygenation, two competing reactions involving CO2 and O2 as substrates, respectively (Andersson and Backlund, 2008). Carboxylation consumes both NAPDH and ATP, which are produced by the photosynthetic light reaction. Because NADP+ is reduced by an electron derived from the photosynthetic electron transport chain (PETC), Rubisco-mediated CO2 reduction relieves PSII excitation pressure under normal conditions, thus minimizing O2•- production at PSI and Cytochrome b6f complex (Cyt b6f). On the other hand, Rubisco-mediated oxygenation consumes O2, and low O2 levels could also minimize cROS production. It is therefore reasonable to hypothesize that RCB-mediated mobilization of Rubisco out of chloroplasts not only reduces the PETC electron sink but also reduces O2 consumption, two processes that synergistically promote cROS accumulation. This could partially explain why ETI induces RCBs, but not whole-chloroplast autophagy. It is worth noting that other stromal proteins, such as glutamine synthetase (Chiba et al., 2003), a key enzyme in nitrogen assimilation, were also detected in RCBs. Blocking chloroplast nitrogen assimilation could also reduce NADPH consumption (Baslam et al., 2021), and thus enhance cROS accumulation.
Under normal conditions, chloroplasts are the center for carbon fixation and nitrogen assimilation. However, during ETI chloroplasts must be reprogrammed to be a center for production of ROS, defense-related hormones, and metabolites (Sowden et al., 2018; Kuźniak and Kopczewski, 2020; Kachroo et al., 2021; Littlejohn et al., 2021). We hypothesize that mRNA decay of photosynthetic transcripts, either translation-dependent or translation-independent, impairs the repair cycle of PSII and thus facilitates the generation of ROS such as 1O2, OH• and H2O2 at PSII. It is possible that decay of nuclear-encoded transcripts alone will lead to an uncoupling of expression of nuclear-encoded and chloroplast-encoded proteins. However, we think this is unlikely since all six sigma factors (SIG1-SIG6) for expression of chloroplast-encoded genes are nuclear-encoded and their transcripts are also destabilized during ETI (Su et al., 2018; Hwang et al., 2022). Meanwhile, removing Rubisco and glutamine synthetase from chloroplasts by RCBs reduces the consumption of both NADPH and O2, causing stromal overreduction. Correspondingly, a highly reduced stroma would further exacerbate PSII excitation pressure and enhance electron leakage at the donor side of PSI or Cyt b6f to O2, resulting in O2•- production. Together, these multi-layered reprogramming mechanisms lead to induction of high cROS accumulation during ETI. In addition to these events discussed above, other cytoplasmic events connected with HR development may also contribute to cROS accumulation. For example, it would be of interest to test whether ETI-induced cytoplasmic Ca2+ influx directly or indirectly affects chloroplast dynamics and thereby promotes cROS accumulation. In the future, it will be interesting to determine the impact of ETI on PSII functioning.
JS wrote the draft. WG revised and edited the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by National Science Foundation grant IOS-1456181 to WG.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbasi, N., Park, Y., Choi, S. B. (2013). RNA Deadenylation and decay in plants. J. Plant Biol. 56, 198–207. doi: 10.1007/s12374-013-0201-8
Adachi, H., Derevnina, L., Kamoun, S. (2019). NLR singletons, pairs, and networks: evolution, assembly, and regulation of the intracellular immunoreceptor circuitry of plants. Curr. Opin. Plant Biol. 50, 121–131. doi: 10.1016/j.pbi.2019.04.007
Andersson, I., Backlund, A. (2008). Structure and function of rubisco. Plant Physiol. Biochem. 46, 275–291. doi: 10.1016/j.plaphy.2008.01.001
Baslam, M., Mitsui, T., Sueyoshi, K., Ohyama, T. (2021). Recent advances in carbon and nitrogen metabolism in C3 plants. Int. J. Mol. Sci. 22, 1–39. doi: 10.3390/ijms22010318
Biasini, A., Abdulkarim, B., Pretis, S., Tan, J. Y., Arora, R., Wischnewski, H., et al. (2021). Translation is required for miRNA-dependent decay of endogenous transcripts. EMBO J. 40, e104569. doi: 10.15252/embj.2020104569
Bilgin, D. D., Zavala, J. A., Zhu, J., Clough, S. J., Ort, D. R., Delucia, E. H. (2010). Biotic stress globally downregulates photosynthesis genes. Plant Cell Environ. 33, 1597–1613. doi: 10.1111/j.1365-3040.2010.02167.x
Bi, G., Su, M., Li, N., Liang, Y., Dang, S., Xu, J., et al. (2021). The ZAR1 resistosome is a calcium-permeable channel triggering plant immune signaling. Cell 184, 3528–3541.e12. doi: 10.1016/j.cell.2021.05.003
Chiba, A., Ishida, H., Nishizawa, N. K., Makino, A., Mae, T. (2003). Exclusion of ribulose-1,5-bisphosphate carboxylase/oxygenase from chloroplasts by specific bodies in naturally senescing leaves of wheat. Plant Cell Physiol. 44, 914–921. doi: 10.1093/pcp/pcg118
Contreras, M. P., Pai, H., Tumtas, Y., Duggan, C., Yuen, E. L. H., Cruces, A. V., et al. (2022). Sensor NLR immune proteins activate oligomerization of their NRC helpers in response to plant pathogens. EMBO J. e111519. doi: 10.15252/embj.2022111519
de Torres Zabala, M., Littlejohn, G., Jayaraman, S., Studholme, D., Bailey, T., Lawson, T., et al. (2015). Chloroplasts play a central role in plant defence and are targeted by pathogen effectors. Nat. Plants 1, 15074. doi: 10.1038/nplants.2015.74
Dong, J., Chen, W. (2013). The role of autophagy in chloroplast degradation and chlorophagy in immune defenses during Pst DC3000 (AvrRps4) infection. PloS One 8, e73091. doi: 10.1371/journal.pone.0073091
Feehan, J. M., Castel, B., Bentham, A. R., Jones, J. D. (2020). Plant NLRs get by with a little help from their friends. Curr. Opin. Plant Biol. 56, 99–108. doi: 10.1016/j.pbi.2020.04.006
Foyer, C. H., Hanke, G. (2022). ROS production and signalling in chloroplasts: cornerstones and evolving concepts. Plant J., 111, 642–661. doi: 10.1111/tpj.15856
Heck, A. M., Wilusz, J. (2018). The interplay between the RNA decay and translation machinery in eukaryotes. Cold Spring Harb. Perspect. Biol. 10, a032839. doi: 10.1101/cshperspect.a032839
Hu, W., Sweet, T. J., Chamnongpol, S., Baker, K. E., Coller, J. (2009). Co-Translational mRNA decay in saccharomyces cerevisiae. Nature 461, 225–229. doi: 10.1038/nature08265
Hwang, Y., Han, S., Yoo, C. Y., Hong, L., You, C., Le, B. H., et al. (2022). Anterograde signaling controls plastid transcription via sigma factors separately from nuclear photosynthesis genes. Nat. Commun. 13, 7440. doi: 10.1038/s41467-022-35080-0
Ibrahim, F., Maragkakis, M., Alexiou, P., Mourelatos, Z. (2018). Ribothrypsis, a novel process of canonical mRNA decay, mediates ribosome-phased mRNA endonucleolysis. Nat. Struct. Mol. Biol. 25, 302–310. doi: 10.1038/s41594-018-0042-8
Ishida, H., Yoshimoto, K., Izumi, M., Reisen, D., Yano, Y., Makino, A., et al. (2008). Mobilization of rubisco and stroma-localized fluorescent proteins of chloroplasts to the vacuole by an ATG gene-dependent autophagic process. Plant Physiol. 148, 142–155. doi: 10.1104/pp.108.122770
Izumi, M., Wada, S., Makino, A., Ishida, H. (2010). The autophagic degradation of chloroplasts via rubisco-containing bodies is specifically linked to leaf carbon status but not nitrogen status in arabidopsis. Plant Physiol. 154, 1196–1209. doi: 10.1104/pp.110.158519
Jacob, P., Kim, H., Wu, F., El-Kasmi, F., Chi, Y., Walton, W. G., et al. (2021). Plant “helper” immune receptors are Ca2+-permeable nonselective cation channels. Science 373, 420–425. doi: 10.1126/science.abg7917
Kachroo, P., Burch-Smith, T. M., Grant, M. (2021). An emerging role for chloroplasts in disease and defense. Annu. Rev. Plant Phyto. 59, 423–445. doi: 10.1146/annurev-phyto-020620
Kim, N. H., Jacob, P., Dangl, J. L. (2022). Con-Ca2+-tenating plant immune responses via calcium-permeable cation channels. New Phytol. 234, 813–818. doi: 10.1111/nph.18044
Kuźniak, E., Kopczewski, T. (2020). The chloroplast reactive oxygen species-redox system in plant immunity and disease. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.572686
Łabno, A., Tomecki, R., Dziembowski, A. (2016). Cytoplasmic RNA decay pathways - enzymes and mechanisms. Biochim. Biophys. Acta 1863, 3125–3147. doi: 10.1016/j.bbamcr.2016.09.023
Li, M., Kim, C. (2022). Chloroplast ROS and stress signaling. Plant Commun. 3. doi: 10.1016/j.xplc.2021.100264
Littlejohn, G. R., Breen, S., Smirnoff, N., Grant, M. (2021). Chloroplast immunity illuminated. New Phytol. 229, 3088–3107. doi: 10.1111/nph.17076
Liu, Y., Ren, D., Pike, S., Pallardy, S., Gassmann, W., Zhang, S. (2007). Chloroplast-generated reactive oxygen species are involved in hypersensitive response-like cell death mediated by a mitogen-activated protein kinase cascade. Plant J. 51, 941–954. doi: 10.1111/j.1365-313X.2007.03191.x
Li, B., Wu, H., Guo, H. (2018). Plant mRNA decay: extended roles and potential determinants. Curr. Opin. Plant Biol. 45, 178–184. doi: 10.1016/j.pbi.2018.08.007
Lu, Y., Yao, J. (2018). Chloroplasts at the crossroad of photosynthesis, pathogen infection and plant defense. Int. J. Mol. Sci., 19. doi: 10.3390/ijms19123900
Meteignier, L. V., El-Oirdi, M., Cohen, M., Barff, T., Matteau, D., Lucier, J. F., et al. (2017). Translatome analysis of an NB-LRR immune response identifies important contributors to plant immunity in arabidopsis. J. Exp. Bot. 68, 2333–2344. doi: 10.1093/jxb/erx078
Mishima, Y., Han, P., Ishibashi, K., Kimura, S., Iwasaki, S. (2022). Ribosome slowdown triggers codon-mediated mRNA decay independently of ribosome quality control. EMBO J. 41. doi: 10.15252/embj.2021109256
Morris, C., Cluet, D., Ricci, E. P. (2021). Ribosome dynamics and mRNA turnover, a complex relationship under constant cellular scrutiny. WIREs RNA 12. doi: 10.1002/wrna.1658
Narsai, R., Howell, K. A., Millar, A. H., O’Toole, N., Small, I., Whelan, J. (2007). Genome-wide analysis of mRNA decay rates and their determinants in arabidopsis thaliana. Plant Cell 19, 3418–3436. doi: 10.1105/tpc.107.055046
Ngou, B. P. M., Ding, P., Jones, J. D. G. (2022). Thirty years of resistance: Zig-zag through the plant immune system. Plant Cell 34, 1447–1478. doi: 10.1093/plcell/koac041
Nomura, H., Komori, T., Uemura, S., Kanda, Y., Shimotani, K., Nakai, K., et al. (2012). Chloroplast-mediated activation of plant immune signalling in arabidopsis. Nat. Commun. 3. doi: 10.1038/ncomms1926
Pelechano, V., Wei, W., Steinmetz, L. M. (2015). Widespread co-translational RNA decay reveals ribosome dynamics. Cell 161, 1400–1412. doi: 10.1016/j.cell.2015.05.008
Rossi, F. R., Krapp, A. R., Bisaro, F., Maiale, S. J., Pieckenstain, F. L., Carrillo, N. (2017). Reactive oxygen species generated in chloroplasts contribute to tobacco leaf infection by the necrotrophic fungus botrytis cinerea. Plant J. 92, 761–773. doi: 10.1111/tpj.13718
Shang-Guan, K., Wang, M., Htwe, N. M. P. S., Li, P., Li, Y., Qi, F., et al. (2018). Lipopolysaccharides trigger two successive bursts of reactive oxygen species at distinct cellular locations. Plant Physiol. 176, 2543–2556. doi: 10.1104/pp.17.01637
Simsek, D., Tiu, G. C., Flynn, R. A., Byeon, G. W., Leppek, K., Xu, A. F., et al. (2017). The mammalian ribo-interactome reveals ribosome functional diversity and heterogeneity. Cell 169, 1051–1065.e18. doi: 10.1016/j.cell.2017.05.022
Sowden, R. G., Watson, S. J., Jarvis, P. (2018). The role of chloroplasts in plant pathology. Ess. Biochem. 62, 21–39. doi: 10.1042/EBC20170020
Su, J., Yang, L., Zhu, Q., Wu, H., He, Y., Liu, Y., et al. (2018). Active photosynthetic inhibition mediated by MPK3/MPK6 is critical to effector-triggered immunity. PloS Biol. 16. doi: 10.1371/journal.pbio.2004122
Tsuda, K., Mine, A., Bethke, G., Igarashi, D., Botanga, C. J., Tsuda, Y., et al. (2013). Dual regulation of gene expression mediated by extended MAPK activation and salicylic acid contributes to robust innate immunity in arabidopsis thaliana. PloS Genet. 9. doi: 10.1371/journal.pgen.1004015
Tuck, A. C., Rankova, A., Arpat, A. B., Liechti, L. A., Hess, D., Iesmantavicius, V., et al. (2020). Mammalian RNA decay pathways are highly specialized and widely linked to translation. Mol. Cell 77, 1222–1236.e13. doi: 10.1016/j.molcel.2020.01.007
Wada, S., Ishida, H., Izumi, M., Yoshimoto, K., Ohsumi, Y., Mae, T., et al. (2009). Autophagy plays a role in chloroplast degradation during senescence in individually darkened leaves. Plant Physiol. 149, 885–893. doi: 10.1104/pp.108.130013
Xu, G., Greene, G. H., Yoo, H., Liu, L., Marqués, J., Motley, J., et al. (2017). Global translational reprogramming is a fundamental layer of immune regulation in plants. Nature 545, 487–490. doi: 10.1038/nature22371
Yoo, H., Greene, G. H., Yuan, M., Xu, G., Burton, D., Liu, L., et al. (2020). Translational regulation of metabolic dynamics during effector-triggered immunity. Mol. Plant 13, 88–98. doi: 10.1016/j.molp.2019.09.009
Yoshitake, Y., Nakamura, S., Shinozaki, D., Izumi, M., Yoshimoto, K., Ohta, H., et al. (2021). RCB-mediated chlorophagy caused by oversupply of nitrogen suppresses phosphate-starvation stress in plants. Plant Physiol. 185, 318–330. doi: 10.1093/plphys/kiaa030
Yu, X., Willmann, M. R., Anderson, S. J., Gregory, B. D. (2016). Genome-wide mapping of uncapped and cleaved transcripts reveals a role for the nuclear mrna cap-binding complex in cotranslational rna decay in arabidopsis. Plant Cell 28, 2385–2397. doi: 10.1105/tpc.16.00456
Keywords: reactive oxygen species (ROS), effector-triggered immunity (ETI), mRNA decay, cotranslational mRNA decay, Rubisco-containing body (RCB)
Citation: Su J and Gassmann W (2023) Cytoplasmic regulation of chloroplast ROS accumulation during effector-triggered immunity. Front. Plant Sci. 14:1127833. doi: 10.3389/fpls.2023.1127833
Received: 20 December 2022; Accepted: 16 January 2023;
Published: 30 January 2023.
Edited by:
Mo Wang, Fujian Agriculture and Forestry University, ChinaReviewed by:
Chanhong Kim, Shanghai Institute for Biological Sciences, Chinese Academy of Sciences (CAS), ChinaCopyright © 2023 Su and Gassmann. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jianbin Su, c3VqQG1pc3NvdXJpLmVkdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.