- 1State Key Laboratory of Desert and Oasis Ecology, Key Laboratory of Ecological Safety and Sustainable Development in Arid Lands, Xinjiang Institute of Ecology and Geography, Chinese Academy of Sciences, Urumqi, China
- 2College of Resources and Environment, University of Chinese Academy of Sciences, Beijing, China
- 3Xinjiang Key Lab of Conservation and Utilization of Plant Gene Resources, Xinjiang Institute of Ecology and Geography, Chinese Academy of Sciences, Urumqi, China
- 4Turpan Eremophytes Botanical Garden, Chinese Academy of Sciences, Turpan, China
Syntrichia caninervis is a desiccation tolerant moss and is the dominant bryophyte found in biological soil crusts in the Gurbantunggut desert. In this study, we assessed the transcriptome profiles of S. caninervis gametophytes during the dehydration-rehydration (D-R) process (across 9 time points) using Illumina sequencing. In total, 22489 transcripts were identified, including 5337 novel transcripts, that mapped to the reference genome. A total of 12548 transcripts exhibited significant alterations in the D-R samples compared with the control samples. The differentially expressed transcripts (DETs) possessed several enriched Gene Ontology terms, such as “water stress response”, “oxidation-reduction process”, “membrane metabolism”, “photosynthesis”, and “transcription factor activity”. Moreover, during early dehydration stress, the DETs were significantly enriched in stress-related pathways from the Kyoto Encyclopedia of Genes and Genomes, such as “phenylpropanoid biosynthesis”, “alpha-linolenic acid metabolism”, and “fructose and mannose metabolism”. Photosynthesis-related transcripts (e.g., ScPsa H, ScRubisco, and ScLhcb1) were inhibited during the dehydration treatment and significantly accumulated during the late rehydration period. Most transcripts from the late embryogenesis abundant proteins (LEA) and early light-inducible protein (ELIP) families strongly accumulated at the late dehydration stage. These pathways were positively correlated with the content changes of absolute water content and Fv/Fm values, alongside peroxidase and superoxide dismutase activities. Seven transcription factor families, including AP2-ERF, bHLH, G2-like, MYB, NAC, WRKY, and bZIP, were enriched in DETs during D-R treatment. This study is the first transcriptome analysis using the S. caninervis genome for gene annotation and multigroup D-R treatment points. Our results demonstrated the detailed dynamic changes in the transcriptome of S. caninervis during the D-R process. These results also improve understanding of desiccation tolerant plants’ adaptations to desiccation stress at the transcription level and provide promising gene resources for transgenic crop breeding.
Introduction
Syntrichia caninervis is a moss with extremely vegetative desiccation tolerance ability. This moss is a dominant component of biological soil crusts in the Gurbantunggut Desert of China (Zhang, 2005). It is frequently exposed to desiccation and the extreme high/low temperatures of the desert (Silva et al., 2021). S. caninervis has been classified as category “A” for desiccation tolerance (DT) (Wood, 2007), thereby exhibiting strong cell protection and repair adaptability with DT regulation at a morphological (Pan et al., 2016), physiological, biochemical (Wu et al., 2009; Yin et al., 2017), and molecular level (Li et al., 2016; Liang et al., 2017; Liu et al., 2020; Li et al., 2023). Many genes isolated from S. caninervis have been observed to enhance abiotic and biotic stress resistance in model plants. Additionally, some of these genes have been successfully used to create stress-tolerant cotton, tobacco, and Medicago sativa plants (Yang et al., 2015; Yang et al., 2016; Yang et al., 2021; Zhang et al., 2022). For example, ScALDH21 overexpression enhanced drought and salt tolerance in transgenic Arabidopsis, tobacco, and cotton plants, and others (Yang et al., 2012; Yang et al., 2015; Yang et al., 2016; Wang et al., 2020; Yang et al., 2021). Furthermore, ScDREBs and ScABI3 confer osmotic and salt tolerance to Arabidopsis (Li et al., 2016; Liang et al., 2017; Li et al., 2017; Zhang et al., 2018; Li et al., 2019a; Liu et al., 2022). Additionally, ScAPD1-like enhanced resistance to Verticillium wilt in transgenic S. caninervis and Arabidopsis (Li et al., 2023). Finally, ScELIPs have been observed to improve the photosynthetic apparatus protection of transgenic Arabidopsis under high light stress (Liu et al., 2020).
Transcriptomes of plant responses to dehydration-rehydration (D-R) have been established for several DT plants, including angiosperms, ferns, and bryophyte species. Specifically, the transcriptomes related to DT bryophytes include Marchantia inflexa (Marks et al., 2021), Marchantia polymorpha (Godinez-Vidal et al., 2020), Physcomitrella patens (Hiss et al., 2014), Tortula ruralis (Oliver et al., 2004; Oliver et al., 2009), Bryum argenteum (Gao et al., 2017), and Grimmia pilifera (Liu et al., 2022). However, most of these bryophyte transcriptomes were annotated using information from other species, e.g., P. patens, rather than their own genomes. Additionally, very little transcriptome data are available for DT moss. Previous transcriptomic analyses have been conducted on S. caninervis (Gao et al., 2014). However, this analysis of S. caninervis has only been conducted with a mixed transcriptome of diverse moss samples collected under D-R treatment and the dynamic transcriptional changes occurring during the D-R processes were not evaluated. Moreover, gene annotation of this transcriptome has been performed using the genome of P. patens. Recently, the S. caninervis genome has been sequenced at the chromosomal level (Silva et al., 2021), thereby providing a robust model for studies concerning plant DT mechanisms, including transcriptional regulation, and an excellent source of novel stress related genes.
Although many gene functions of S. caninervis have been studied, there is little information regarding the DT mechanism of S. caninervis at the integral transcriptional level in S. caninervis during the D-R process. In this study, a series of temporal transcriptome profiles of S. caninervis, involving nine D-R treatment time points, were investigated using the S. caninervis genome for gene annotation. Our results found several key pathways and critical transcription factors (TFs) were modified in response to desiccation stress in S. caninervis. We also provided a detailed picture of the dynamic changes of the S. caninervis transcript during the D-R process, improved understanding of plant adaptations to DT, as well as providing promising candidate gene resources for crop stress tolerance breeding.
Materials and methods
Plant material and dehydration-rehydration treatment
Dry S. caninervis samples were collected from the Gurbantunggut Desert in Xinjiang, Northwest China (Fukang County, 44°32′30″N, 88°6′42″E). The collected wild moss samples were stored at 25°C under dark conditions. The gametophytes of these samples were completely desiccated and kept in a dormant state. For experiments, the dried wild gametophytes were, first, fully hydrated with ultrapure water for 24 h; then, the slow-dried method was used to keep S. caninervis samples at a relative humidity (RH) of 66.67% (-57 MPa) at 25°C, as described previously (Liang et al., 2021). Samples were collected after 0, 2, 6, and 24 h of dehydration (0 h as the control). Early dehydration occurred at 2 and 6 h (D2h and D6h), and late dehydration occurred at 24 h (D24h). The dehydrated samples were subsequently rehydrated by transferring the dehydrated gametophytes to new Petri dishes at 25°C with filter paper that was saturated with ultrapure water. Rehydrated samples were then harvested at 0.5, 2, 6, 24, and 48 h. Early rehydration was considered to be 0.5, and 2 h (R0.5h and R2h), whereas late rehydration times were 6, 24, and 48 h (R6h, R24h, and R48h, respectively). Fully rehydrated samples without dehydration (0 h) served as the reference control. All samples were frozen in liquid nitrogen immediately after harvest and stored at -80°C. The prepared samples were subjected to transcriptome sequencing and physiological analysis. Photographic records of S. caninveris phenotypes, in addition to the measurement of the absolute water content (AWC) and Fv/Fm (optimal/maximal photochemical efficiency of PSII) were performed at each treatment time point.
Physiological indicator measurements
AWC was measured in gametophytes of S. caninervis at various time points during the D-R process. AWC curves, plotting the water content on a dry weight basis, were produced using the following formula (Rathnayake et al., 2019): AWC (g g-1 DW) = (FW-DW)/DW; FW indicates the fresh weight measured at every time point during the D-R process, and DW indicates the weight measured after drying for 48 h at 80°C in an oven.
Fv/Fm was measured using a portable modulated fluorometer (PAM-2500; Heinz, Walz, Germany). The saturation pulse method was used to calculate Fv/Fm, which was measured in the dark after the box was covered for > 30 min. Parameter settings were based on the recommendations of Zhang et al. (2011).
The activities of the physiological indicators H2O2, malondialdehyde (MDA), peroxidase (POD), and superoxide dismutase (SOD) were measured using detection assay kits (Nanjing Jiancheng Bioengineering Institute, Nanjing, China), according to the manufacturer’s instructions.
Library construction and transcriptome sequencing
A total of 27 samples, consisting of 3 biological replicates from 9 time points of D-R treatment (0 h, D2h, D6h, D24h, R0.5h, R2h, R6h, R24h, R48h) were used for the transcriptome analysis. Illumina sequencing was performed separately for these 27 samples, which was combined with UMI-RNA sequencing technology by the Novogene Company (Beijing, China). First, 1 µg of RNA was used as input material per sample for RNA sample preparation. Briefly, mRNA was purified from the total RNA using poly T oligo-attached magnetic beads. First-strand cDNA was synthesised using random hexamer primers and RNase H (M0297, NEB, Beijing) at 37°C in first-strand synthesis reaction buffer (E7530, NEB, Beijing). Later, the cDNA library fragments were purified with the AMPure XP system (Beckman Coulter, Beverly, USA) to select cDNA fragments of 100-200 bp in length. Adapter ligation was conducted at 25°C for 10 min and performed prior to PCR. Then, PCR was performed using the Phusion High Fidelity DNA polymerase(M0530S, NEB, Beijing). Finally, the transcriptome was sequenced using an Agilent Bioanalyzer 2100 system platform (Agilent Bioanalyzer, Germany).
Quantification of transcript abundance and identification of differentially expressed transcripts
Qualified clean reads were mapped to the S. caninervis genome sequence (Silva et al., 2021) using HISAT v2.0.4 software (Kim et al., 2015). The genome positioning results of all sequenced reads were assembled into transcripts using the Cufflinks software. HTSeq v0.9.1 (Putri et al., 2022) was used to count the read numbers mapped to each gene. Next, fragments per kilobase of exon model per million mapped fragments (FPKM) of each transcript were calculated based on the length of the transcript; then, the read count was mapped to this transcript (Trapnell et al., 2010). Differentially abundant transcripts between the two experimental groups were analysed using the DESeq R package (1.18.0) (Wang et al., 2010). DETs were screened from transcriptome data; p ≤ 0.05, and Log2 fold change ≥ 1 were set as the thresholds for significantly differential abundance. Heatmap analysis of the transcript abundance patterns and circos of the transcriptome was performed using TBtools software (Chen et al., 2020).
Abundance patterns of differential transcripts during D-R using GO, KEGG, and WGCNA-trait analysis
Cluster analysis of the transcript abundance patterns was performed using the TCseq R package using the K-means method. In K-means clustering, the FPKM value of the transcript must first be Z-score standardised. Gene Ontology (GO) enrichment analysis of DETs was conducted using the GOseq R package with significant enrichment being defined by a p-value ≤ 0.05.
Pathway enrichment analysis was performed using the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (Kanehisa et al., 2007) (http://www.genome.jp/kegg). We used KOBAS software (Mao et al., 2005) to test the statistical enrichment of DETs in KEGG pathways. Pathways with p ≤ 0.05 were considered to be significant. Additionally, weighted correlation network analysis-trait (WGCNA-trait) analysis (Langfelder and Horvath, 2008) was conducted using the BMKcloud platform (www.biocloud.net).
RT-qPCR analysis of the expression of desiccation tolerance-related transcripts
RT-qPCR was performed to verify the transcriptome data of the selected DETs. Total RNA was extracted using a Plant RNA Kit (Omega Bio-Tek, Guangzhou, China). First-strand cDNA was synthesised with 1 μg of total RNA using the PrimeScript RT reagent Kit with gDNA Eraser (RR047A, Takara, Japan). For RT-qPCR, the TB Green TM Premix Ex Taq TM II kit (TaKaRa, Dalian, China) was used to produce 20 μL reactions consisting of 2 μL cDNA (50 ng/L), 0.5 μL forward primer (10 μmol/L), 0.5 μL reverse primer (10 μmol/L), 10 μL TB Green Premix Ex Taq II (2×) (RR820Q, Takara, Japan) and 7 μL ddH2O. For PCR, denaturation was conducted at 95°C for 30 s, followed by 40 cycles of the following conditions: denaturation at 95°C for 5 s, annealing at 60°C for 30 s, and final extension step at 95°C for 15 s. RT-qPCR was performed using a CFX96 Real-Time PCR System (Bio-Rad, Hercules, CA, USA). ScTubulin was used as the reference gene for normalisation (Li et al., 2015). Relative abundance levels were calculated using the 2-ΔΔCt method (Livak and Schmittgen, 2001), with three biological replicates and three technical replicates. Primers used for RT-qPCR were designed using the NCBI primer BLAST program and are listed in Supplementary Table S1.
Statistical analysis
Statistical analyses were performed using the GraphPad Prism 9 software for Windows (version 9.0.0, 2020). All data were analysed using an analysis of variance (ANOVA) test at a 95% confidence level. Significant differences were determined using Fisher’s least significant difference multiple comparison test. The data shown are as the mean ± standard deviation (SD) of three replicates; the significance level relative to the controls was assessed at *p < 0.05, **p < 0.01, and ***p < 0.001. Column graphs were generated using GraphPad Prism 9 software. Adobe Photoshop CC 2019 was used for image processing.
Results
Changes in phenotype and physiological parameters during the D-R process
In a closed atmosphere at 67% RH for dehydration treatment, the gametophytes of S. caninervis curled and shrunk. These gametophytes completely dried and equilibrated osmotic potential with the surrounding air after 24 h. Once rehydrated, the gametophytes quickly recovered to their initial states (Figure 1A). Prior to desiccation (0 h), the mean AWC of S. caninervis gametophytes was 2.33 g g-1 DW (Figure 1B and Table S2). This steadily declined to a stable AWC of approximately 0.045 g g-1 DW at D24h. Overall, these gametophytes exhibited a loss of approximately 98.06% of bulk water in 24 h of dehydration, at which point they were considered to be desiccated. Upon rehydration, desiccated gametophytes recovered fresh weight rapidly, to 77% of the control AWC within 5 min. Full rehydration occurred 6 h after the addition of water to desiccated gametophytes (Figure 1B and Table S2). Fv/Fm values decreased as dehydration progressed; after 2 h and 6 h of dehydration, Fv/Fm values decreased by 7.89% and 23.59%, respectively, compared to the control levels (Figure 1C and Table S2). As desiccation reached 24 h, the Fv/Fm declined to values close to zero. Nonetheless, Fv/Fm recovered rapidly to 77.10% of the control value within 5 min of rehydration, and continued to recover, approaching control levels by 24 h of rehydration (Figure 1C and Table S2).
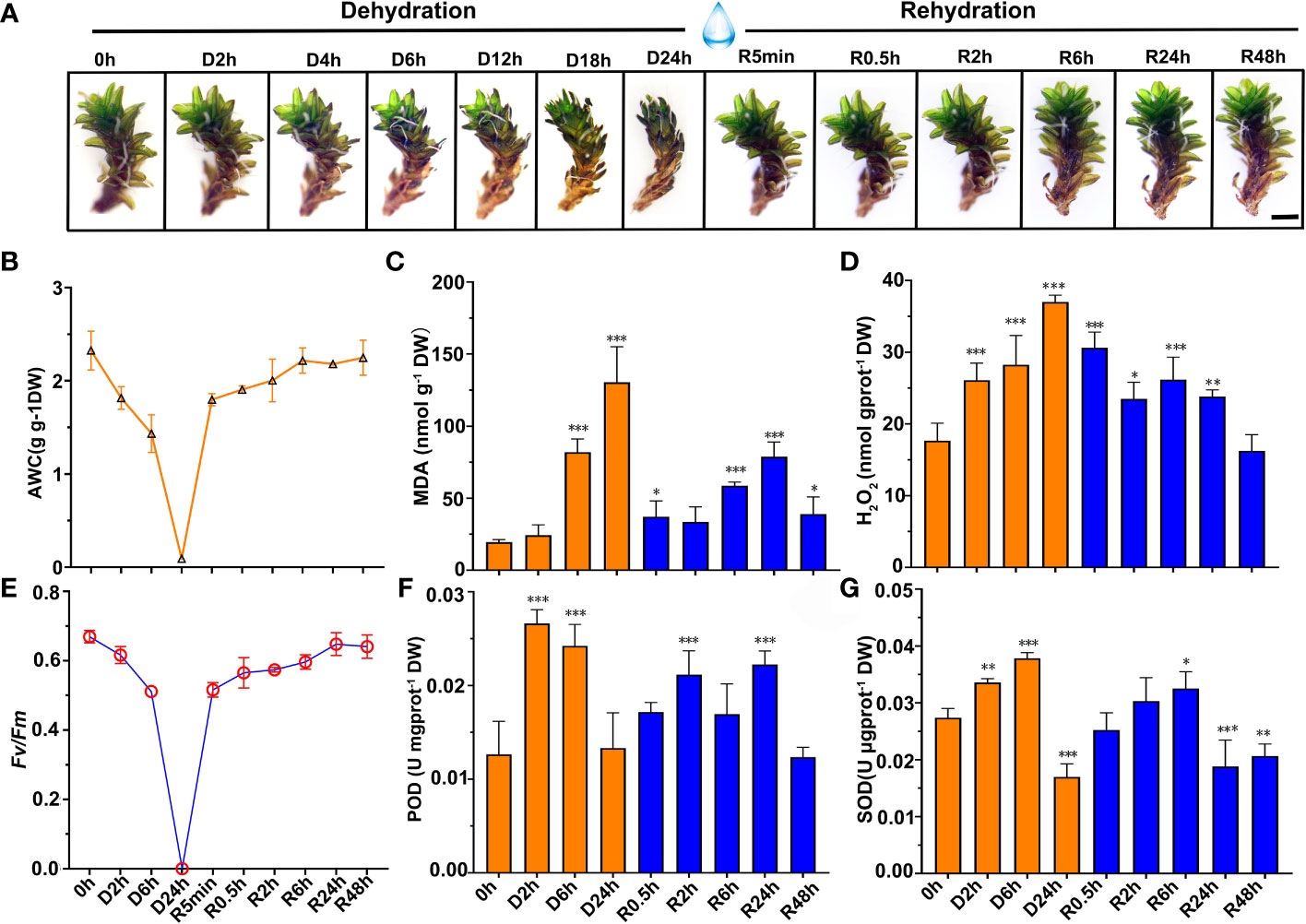
Figure 1 Phenotype, physiological parameters change of S. caninervis under D-R treatments. (A) Phenotype of S. caninervis under D-R conditions, scale bars: 2 mm; (B) AWC; (C) Fv/Fm; (D) MDA content; (E) H2O2 content; (F) POD activity and (G) SOD activity; treatments during 24 h of desiccation (D h) and 48 h of rehydration (R h); data represents the mean ± SD from six biological replicates.*p < 0.05, **p < 0.01, ***p < 0.001, Fisher’s least significant difference test.
To better quantify the oxidative status of the gametophytes, the contents of MDA, a marker of lipid peroxidation and membrane damage, and H2O2 were measured in gametophytes during the D-R process (Figures 1D, E). Our results demonstrated that MDA content exhibited a biphasic response to the D-R process, peaking at 24 h of dehydration, increasing to 133.79 ± 22.09 nmol g-1 DW, followed by a decline to a minimum at 0.5 h of rehydration; a second peak was, then, observed at 24 h of rehydration and recovered to the control (0 h) at 48 h (Figure 1D). The MDA content, like H2O2, reached a peak value (36.9 9 ± 2.00 nmol gprot-1 DW) 24 h after dehydration, and gradually decreased after rehydration (Figure 1E). Additionally, the activities of the antioxidant enzymes POD and SOD in D-R-treated gametophytes were also measured (Figures 1F, G). POD activity increased 2.1-fold compared with that in the control after 2 h of dehydration. SOD activity significantly increased to a peak after 6 h of dehydration (0.037 ± 0.0007 U gprot-1 DW). Upon rehydration, POD and SOD activity levels declined compared to the desiccation treatment. The POD activity was significantly higher than that of the controls at 0.5, 2, 6, and 24 h after rehydration; specifically, POD activity was 35.6%, 67.2%, 33.8%, and 75.8% higher in these rehydrated gametophytes than the control levels, respectively. Therefore, this increased expression allowed the plants to mount a robust antioxidant response, and recover to levels approaching the fully hydrated state (0 h) when rehydrated over 48 h (Figures 1F, G).
Transcriptome assembly, assessment, and differentially expressed transcript analysis
Transcriptome analysis of S. caninervis was performed using its corresponding genome as a reference during the D-R process. In the present study, nucleotide data in the transcriptome ranged from 5.08-9.92 Gb per the sequencing run in each treatment course (Additional File 1). After quality filtering, the number of clean reads was modified from 33-48×106 in 27 samples (Additional File 2), which had an average read mapping rate of 80.86% for each sample on the S. caninervis reference genome. In total, 22489 transcripts were identified in the transcriptome of S. caninervis, of which 17152 transcripts could be mapped to the genome of S. caninervis. However, 5337 novel transcripts could not be mapped to the genome of S. caninervis (Additional File 3). Using principal component analysis (PCA) of S. caninervis transcriptomes, the treatment time points of D-R could be separated on the PCA axis, where PC1 explained 50.9% of the variation (Figure 2A).
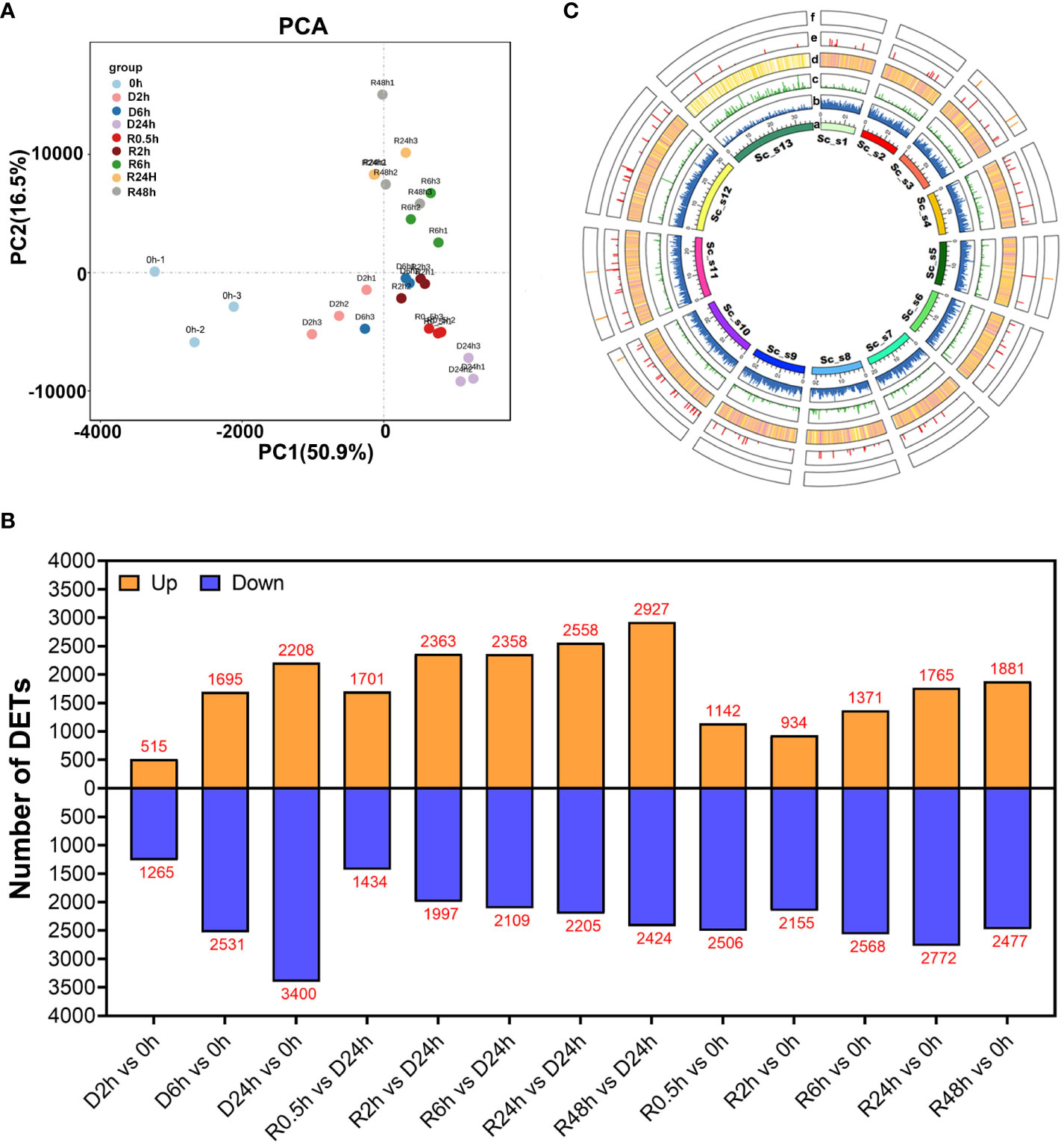
Figure 2 PCA, Circos, DETs of S. caninervis transcriptome during D-R treatment (A) PCA of RNA-Seq data obtained from S. caninervis of full hydrated (0 h), dehydrated (D h) and rehydrated (R h) gametophyte. (C) Circos visualization of genomic and transcriptomic features of S. caninervis under D-R process. a: Chromosomes; b: Gene distribution density, and the height of the column represents the number of genes at the within unit region; c: Novel transcripts distributions from transcriptome, the green column and length show its density mapping on chromosome; d: Transcript density distribution, the heatmap represent transcript density with white (min), yellow (middle) and red (max); e and f: Alternative splice position, the column of red and yellow represent SE and ME, the column of length represent their number of occurrences; (B) Number of DETs among different comparison pairs. |Log2 (Fold Change)| ≥1 and p-value ≤0.05 were used as the threshold to select DETs.
In this study, the number of DETs was evaluated from 13 different comparison pairs (Figure 2B), including different D-R treatment time points (D/R h) compared to 0 h and rehydration treatment time points (R h) compared to D24h. Additionally, R h and 0 h comparison pairs were used to explore the changes in the transcripts of the moss after a round of desiccation compared to 0 h. Furthermore, R h compared to D24h was used to analyse the change in transcript differences between rehydration and absolute desiccation of D24h moss. During the dehydration process, 1810 DETs were in the D2h and 0 h comparison pair; this was the lowest number of DETs among all comparison pairs, of which 507 were up-regulated and 1203 were down-regulated transcripts. As dehydration progressed, the number of DETs gradually increased, with 4226 and 5608 DETs in comparison pairs D6h with 0 h and D24h with 0 h, respectively. Upon rehydration, the number of DETs exhibited a sharp decreasing trend in two comparison pairs (R0.5 h with 0 h and R2h with 0 h). Of the 3089 DETs detected in the R0.5h and 0 h comparison pair, 1142 transcripts were upregulated and 2506 transcripts were downregulated. Furthermore, the number of DETs increased gradually within the comparison pairs from R6h to R48h (Figure 2B). After a round of desiccation stress (R48h), the S. caninervis gametophytes were rehydrated; this process caused more differential changes in the number of transcripts than at 0 h (Figure 2B).
The common and specific DETs were analysed for each comparison pair, and 310 transcripts were found to be differentially abundant at all time points compared to that at 0 h (Figure S1A). Specifically, 47 transcripts were differentially abundant in all dehydration treatments compared with 0 h, while 46 were common in all rehydration treatments compared with 0 h (Figure S1A). There were 165 common DETs in the R h and D24h comparison pair (Figure S1B), and, overall, 19 DETs were present throughout all 13 different comparison pairs (Figure S1C).
We attached the transcripts obtained from the transcriptome sequencing to the chromosomes of the reference genome (Figure 2C). A total of 255 alternative splicing events were identified, of which only exon skips and mutually exclusive exons were detected 247 and 8 times, respectively.
Transcriptome profiles of the gametophyte upon the D-R process
A total of 12548 transcripts (representing 55.7% of protein-coding genes) exhibited significantly altered abundance levels (p ≤ 0.05) in the D-R samples in comparison to the abundance levels in the hydrated control samples (Additional File 4). To investigate the abundance profile of the transcriptome during the D-R process, K-means clustering analysis was performed on DETs to identify clusters with similar transcript abundance patterns. Five distinct clusters, named Cluster 1–5, were revealed; then, GO analysis was performed across the DETs from these 5 clusters (Figure 3; Additional File 5).
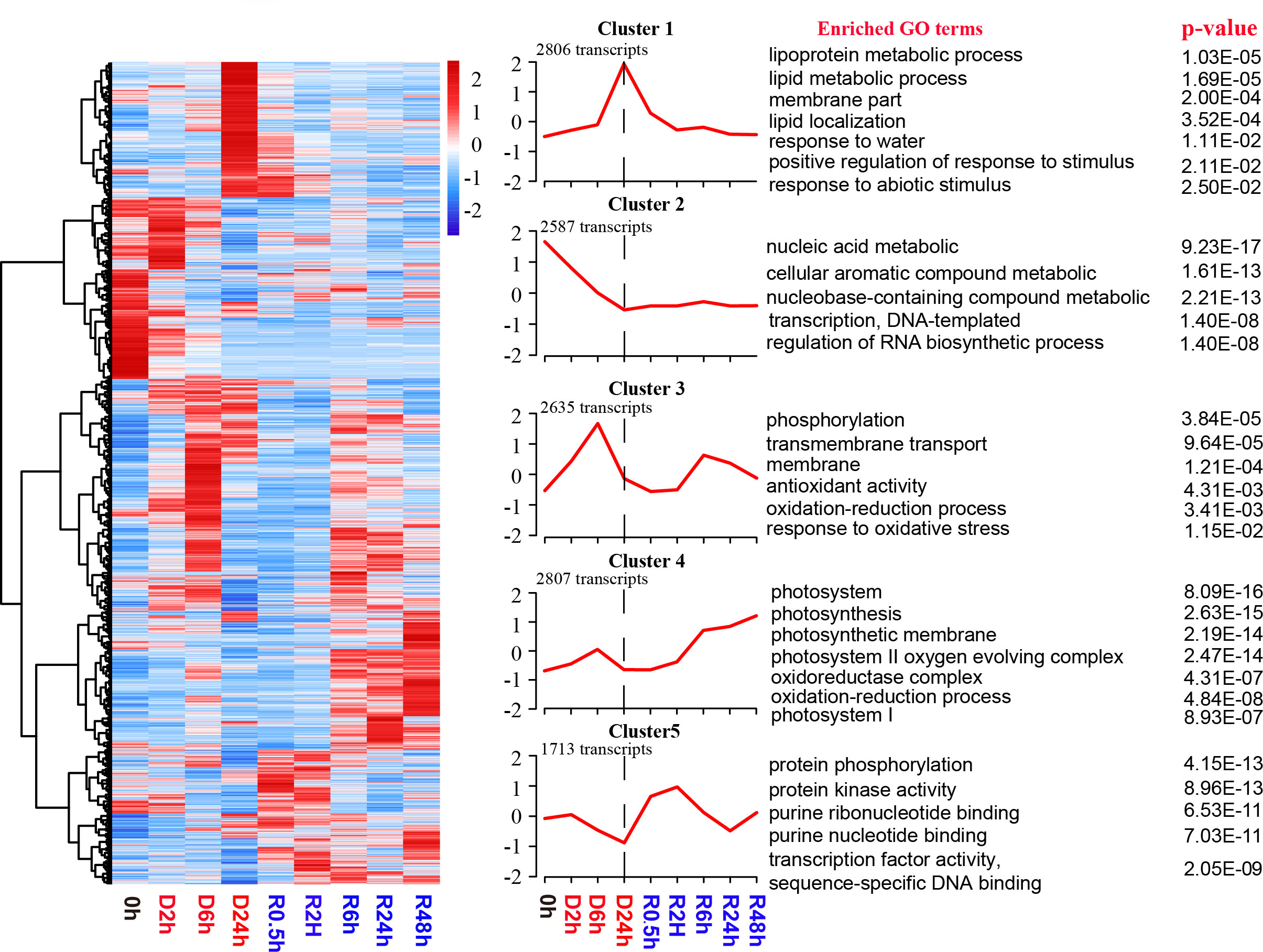
Figure 3 Global transcript abundance profiling of S. caninervis gametophyte during D-R process. |Log2(Fold Change)| ≥1 and p-value ≤ 0.05 were used as the threshold to select DETs; FPKM of DETs values are used for the heatmap, which up- and down-regulated are indicated in red and blue respectively; K-means clustering graph of DETs based on Z-score standardization of differential transcripts; DETs were clustered into five clusters by the K-means clustering (Cluster1-Cluster5). The number of transcripts in each cluster was showed at the top right-hand corner of each cluster. GO enrichment analysis of the five clusters and displaying significant GO terms with p ≤0.05.
During desiccation stress, the transcripts were significantly accumulated into two clusters: Cluster 1 and Cluster 3. Cluster 1 contained 2806 transcripts. These Cluster 1 transcripts exhibited maximal abundance during the late stage of dehydration (D24h) and decreased immediately after re-watering. Additionally, this cluster was predominantly enriched in GO terms such as “lipoprotein metabolic”, “lipid metabolic process”, “membrane part”, “response to water”, “positive regulation of response to stimulus”, and “response to abiotic stimulus”. Cluster 3 showed two peaks in abundance at 6 h after early dehydration and 6 h after late rehydration. This cluster involved 2635 transcripts and was enriched in the GO terms “phosphorylation”, “membrane”, “membrane transport”, “antioxidant activity”, “oxidation-reduction process”, and “response to oxidative stress”. A total of 2587 transcripts were established within the Cluster 2 profile. The transcript abundances persistently declined during the whole desiccation process and maintained consistent transcript abundance levels in the late dehydration (D24h) and rehydration processes. Functional enrichment results demonstrated that the transcript abundance of Cluster 2 is associated with “nucleic acid metabolic”, “transcription, DNA-templated”, and “regulation of RNA biosynthetic process”. Alternatively, the transcript abundances of Cluster 4 continued to increase from 6 h to 48 h after rehydration and were primarily enriched in “photosystem I”, “photosystem II oxygen-evolving complex”, and “oxidation-reduction process”. This indicated that a large number of transcripts related to photosynthesis and antioxidation were transcribed after 6 hours of rehydration. In total, 1713 transcripts were observed within Cluster 5. These transcripts were significantly induced during the early rehydration period (R0.5h and R2h). Furthermore, the abundance of these transcripts decreased significantly in the late rehydration period (R6h-R48h). Transcripts in Cluster 5 were enriched in GO terms such as “protein phosphorylation”, “protein kinase activity”, and “transcription factor activity, sequence-specific DNA binding”.
The KEGG pathways of DETs and WGCNA-trait analysis of desiccation tolerance
All upregulated and downregulated DETs in S. caninervis under D-R conditions were examined using the KEGG database. As shown in Figure 4A, the up-regulated DETs were enriched in various significant KEGG pathways, including “phenylpropanoid biosynthesis”, “alpha-linolenic acid metabolism”, “fructose and mannose metabolism”, “photosynthesis-antenna proteins”, and “carbon fixation in photosynthetic organism”. Among them, “phenylpropanoid biosynthesis”, “alpha-linolenic acid metabolism”, and “fructose and mannose metabolism” were enriched in the D2h with 0 h and D6h with 0 h comparison pairs, whereas the pathways related to photosynthesis were enriched after 6h of rehydration. Contrastingly, the downregulated DETs were enriched in “spiceosome”, “RNA polymerase, ribosome biogenesis in eukaryotes”, “protein processing in endoplasmic reticulum”, and “glutathione metabolism” (Figure 4B).
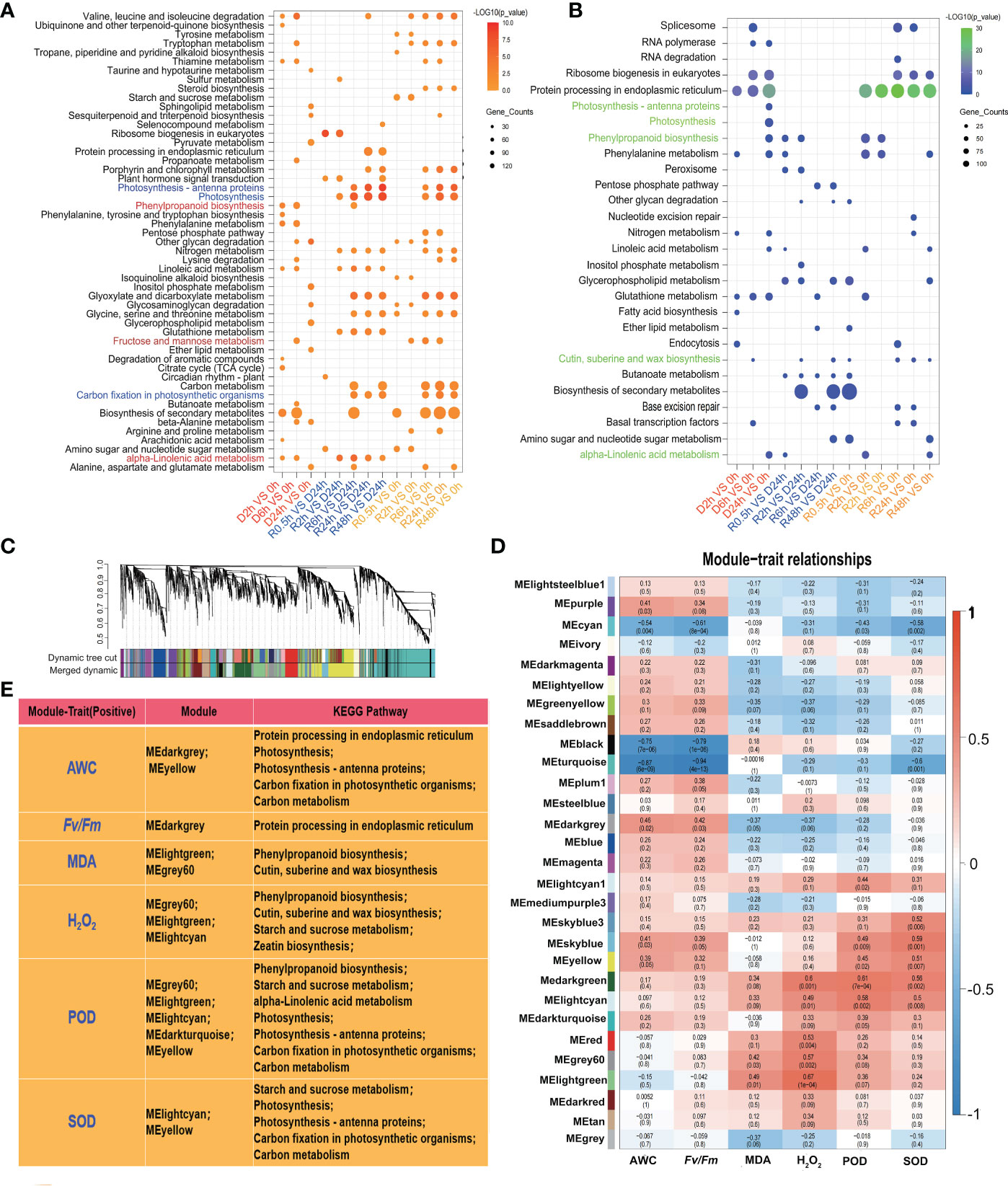
Figure 4 KEGG analysis and identification of modules related with the traits during D-R treatment in S. caninervis. (A) KEGG analysis of up-regulated DETs; Dots indicate the KEGG pathway with significant difference, and color indicates the degree of significance (From orange to red, the higher the significance) and the size of the circle indicates the number of transcripts. (B) KEGG analysis of down-regulated DETs; Dots indicate the KEGG pathway with significant difference, and color indicates the degree of significance (From blue to green, the higher the significance) and the size of the circle indicates the number of transcripts. (C) Transcripts dendrogram with cluster. Each module was assigned different colors. (D) Module-trait relationships. Each row corresponds to a colour module and column corresponds to a trait (AWC, Fv/Fm, MDA, H2O2, POD, SOD). Each block contains the correlation value and p-value; (E) KEGG analysis for screening positive correlation module with traits (AWC,Fv/Fm, MDA, H2O2, POD, SOD) and p ≤ 0.05.
To better investigate the key transcripts which regulate desiccation stress responses in S. caninervis, FPKM of transcripts was used to construct WGCNA. The transcripts were divided into different gene sets (modules), and each module was assigned a different color (Figure 4C). The correlation between each module and six physiological indexes (AWC, Fv/Fm, MDA, H2O2, POD, and SOD) was assessed by plotting a heatmap of module-trait relationships (Figure 4D). As shown in Figure 4E and Figure S2, the AWC had a positive correlation with MEyellow (r = 0.39, p = 0.05) and MEdarkgrey modules (r = 0.46, p = 0.02); the transcripts of these two modules were enriched in KEGG pathways such as photosynthesis related pathways, “protein processing in endoplasmic reticulum”, and “phenylpropanoid biosynthesis”. The Fv/Fm values were positively associated with the MEdarkgrey module (r = 0.42, p = 0.03); these transcripts were enriched in the “protein processing in endoplasmic reticulum” KEGG pathways. MDA was positively correlated with two modules: MEgrey60 (r = 0.42, p = 0.03) and MElightgreen (r = 0.49, p = 0.01). H2O2 was positively correlated with 5 modules, of which MElightcyan (r = 0.49, p = 0.01), MElightgreen (r = 0.67, p = 1e-04), and MEgrey60 (r = 0.57, p = 0.002) were significant; the transcripts found in these modules were enriched in “phenylpropanoid biosynthesis”, “cutin, suberin, and wax biosynthesis”, “zeatin biosynthesis” and “starch and sucrose metabolism”. Moreover, POD and SOD activities had a strong correlation with MElightcyan, MEdarkturquose, MEyellow, MElightgreen, and MEgrey60; the transcripts in these modules were related to “starch and sucrose metabolism”, “alpha-linolenic acid metabolism”, “phenylpropanoid biosynthesis”, and photosynthesis related pathways.
Molecular dynamics of pathways related to DT
KEGG and WGCNA analyses revealed that the differentially expressed transcripts were enriched in phenylpropanoid biosynthesis, alpha-linolenic acid, fructose and mannose metabolism, photosynthesis, and carbon fixation-related pathways. Lignin biosynthesis, a major branch of phenylpropanoid metabolism, has been reported to play an important role in water deficit conditions (Sharma et al., 2019; Dong and Lin, 2021). The transcripts in lignin biosynthesis, including ScPAL (phenylalanine ammonia-lyase), ScHCT (p-hydroxycinnamoyl-CoA shikimate/quinate hydroxycinnamoyl transferase), and ScPOD (peroxidase) transcripts, were significantly expressed at 2 h and 6 h in the early stage of dehydration. Although not significant, ScC4H (cinnamate 4-hydroxylase), Sc4CL (4-coumaric acid: CoA ligase), and ScCAD (cinnamyl alcohol dehydrogenase) transcripts also accumulated during dehydration (Figure 5A).
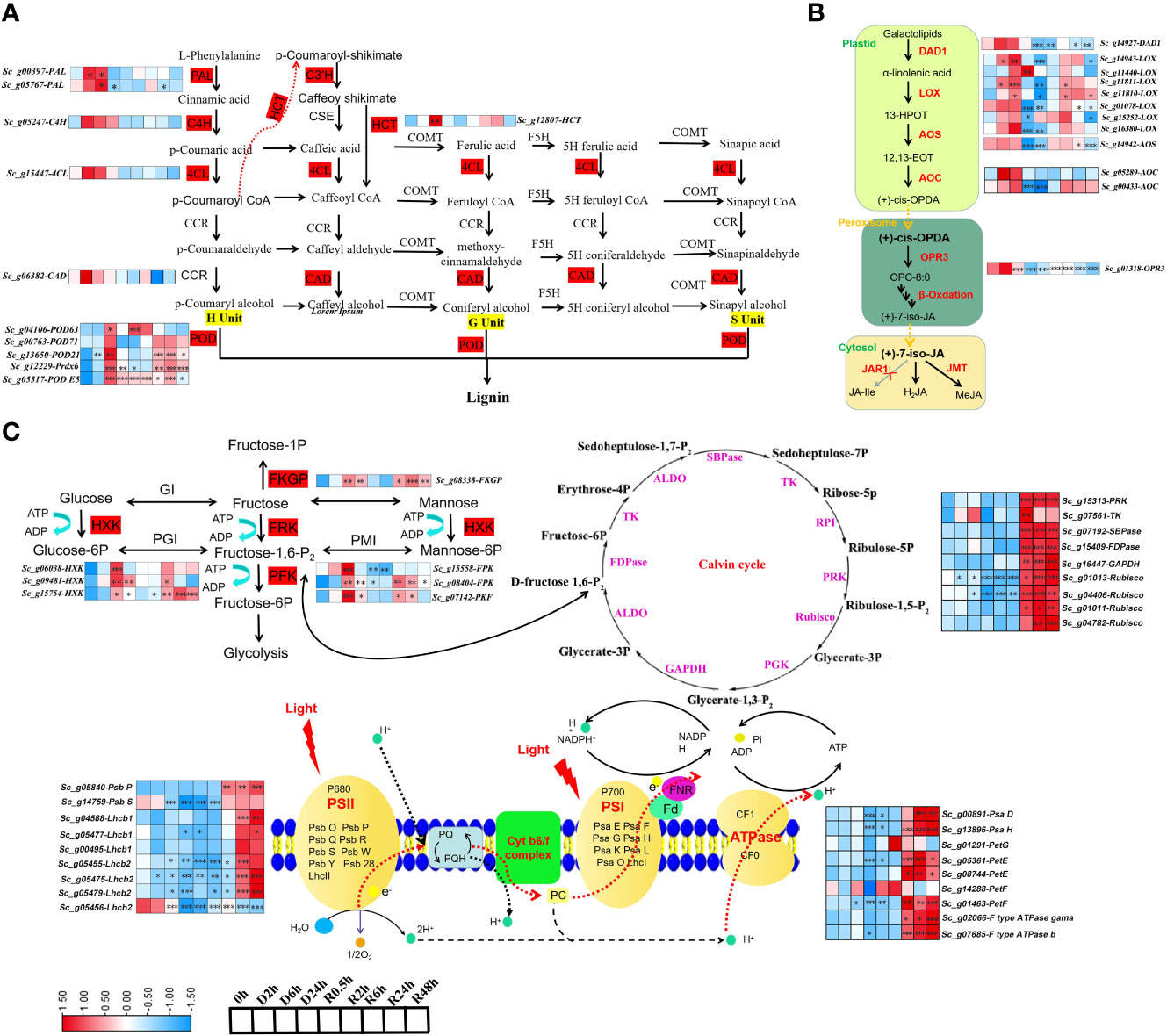
Figure 5 The dynamic change of transcript abundance in Key pathway during D-R process (A) alpha-linolenic acid; (B) phenylpropanoid biosynthesis; (C) fructose and mannose metabolism, photosynthesis and calvin cycle in S. caninervis during D-R process. DETs that up-and down-regulated are indicated in red and blue, respectively;FPKM values were used to heatmap; |Log2(fold change)|≥1 (*p<0.05, **p<0.01, ***p<0.001). Treatments during 24 h of desiccation (D h) and 48 h of rehydration (R h).
Jasmonic acid (JA) is synthesised from alpha-linolenic acid, a major fatty acid in plant cell membranes. The DETs in this pathway, including ScDAD1 (defective in anther dehiscence 1), ScAOC (allene oxide cyclase), and ScLOX (lipoxygenase), significantly increased in expression at 2 h and 6 h of dehydration compared with the control (0 h), and sharply decreased following 24 h of dehydration. The transcript abundance of ScAOS (allene oxide synthase) was stable in the early stage of dehydration from 0-6 h. Alternatively, the ScOPR3 (12-oxophytodienoate reductase 3) transcript accumulated at 2 h and 6 h of dehydration, but was not significant compared with the control (0 h). Moreover, ScLOX, ScAOC, and ScAOS were induced during the rehydration stages (R6h, R24h, and R48h) (Figure 5B).
ScHXK (hexokinase), ScFRK (fructokinase), ScPFK (phosphofructokinase), and ScFKGP (L-fucokinase/GDP-L-fucose pyrophosphorylase), are key components in fructose and mannose metabolism. These transcripts significantly accumulated in the early stage of drying (6 h) and decreased after 24 h of dehydration (Figure 5C). Additionally, S. caninervis DETs involved in photosynthesis sharply declined in expression at 24 h of dehydration and significantly increased from 6-48 h of rehydration; these DETs included subunits of photosystems I and II (PSI and PSII) complexes (ScPsa D, ScPsaH, ScPsbP, and ScPsbS), photosynthetic electron transport (ScPetG, ScPetE, and ScPetF), F-type ATP synthase gamma/b, and the Calvin cycle (ScRuBisCO [rubisco ribulose-1, 5-bishosphate carboxylase], ScTK [transketolase], ScRPI [ribose 5-phosphate isomerase], ScGAPDH [glyceraldehyde-3-phosphate dehydrogenase], ScFDPase [fructose 1, 6-diphosphatase], and ScRPK [phosphoribulokinase]) (Figure 5C). Interestingly, 22 transcripts in the ELIP family and 45 ScLEA transcripts significantly accumulated in the late stage of dehydration (D24h) (Figure S3), indicating that they are important for the tolerance of late dehydration of S. caninervis. These results suggest that alteration of transcript abundance in these key pathways related to desiccation stress, such as ELIP and LEA, could contribute to improving the DT of S. caninervis during the different stages of D-R treatment.
Differentially accumulated TFs under the D-R process
In the present study, the GO term enrichment of DETs in “transcription factor activity, sequence-specific DNA binding” indicated that TFs play important roles in the response of S. caninervis to DT (Figure 3). AP2-ERF, bHLH, MYB, and C3H were the largest groups of TFs during D-R treatments, accounting for 10%, 6%, 5%, and 5% of the total TFs, respectively. Differential analysis showed that 306 TFs were differentially altered during D-R treatment. These differentially abundant TFs belonged to 52 TF families. K-means clustering was performed on differential TFs to identify clusters with similar accumulated transcript patterns. Four distinct clusters, named Cluster 1-4, were identified (Figure 6A; Additional File 6). Clusters 1 and 2 both contained 74 differential TFs, of which Cluster 1 was downregulated and Cluster 2 was upregulated after 24 h of dehydration. The two clusters exhibited opposite responses during dehydration treatments. The top five TF families with the largest gene numbers in Cluster 2 were Orphans, SNF2, MYB, AP2-ERF, and bZIP, which constituted 19%, 7%, 7%, 5%, and 5% of Cluster 2 TF families, respectively. There were 63 differential TFs in Cluster 3, which showed little change within 6 h of dehydration and decreased to the lowest expression level at 24 h of dehydration. Additionally, this expression of Cluster 3 TFs increased rapidly after rehydration and gradually decreased to the control level (0 h) after 0.5 h of dehydration. Cluster 4 contained 95 differential TFs; the expression of these TFs reached a moderate peak after 6 h of dehydration and reached the lowest level after 24 h of dehydration. After re-watering, Cluster 4 transcript abundance continued to rise and reached an accumulation balance at 6 h of rehydration. The largest number of TFs in Clusters 3 and 4 were AP2-ERF, accounting for 8% and 22% of the total differential TFs in these clusters, respectively. Additionally, through TF enrichment analysis, we determined that AP2-ERF, bHLH, G2-like, NAC, WRKY, and bZIP were significantly enriched in the D-R process (Figure 6B). Of these, WRKY, NAC, and G2-like were enriched in the rehydration treatment, whereas bZIP was enriched only in the dehydration treatment. Furthermore, AP2-ERF and bHLH were strongly enriched in both the dehydration and rehydration treatments.
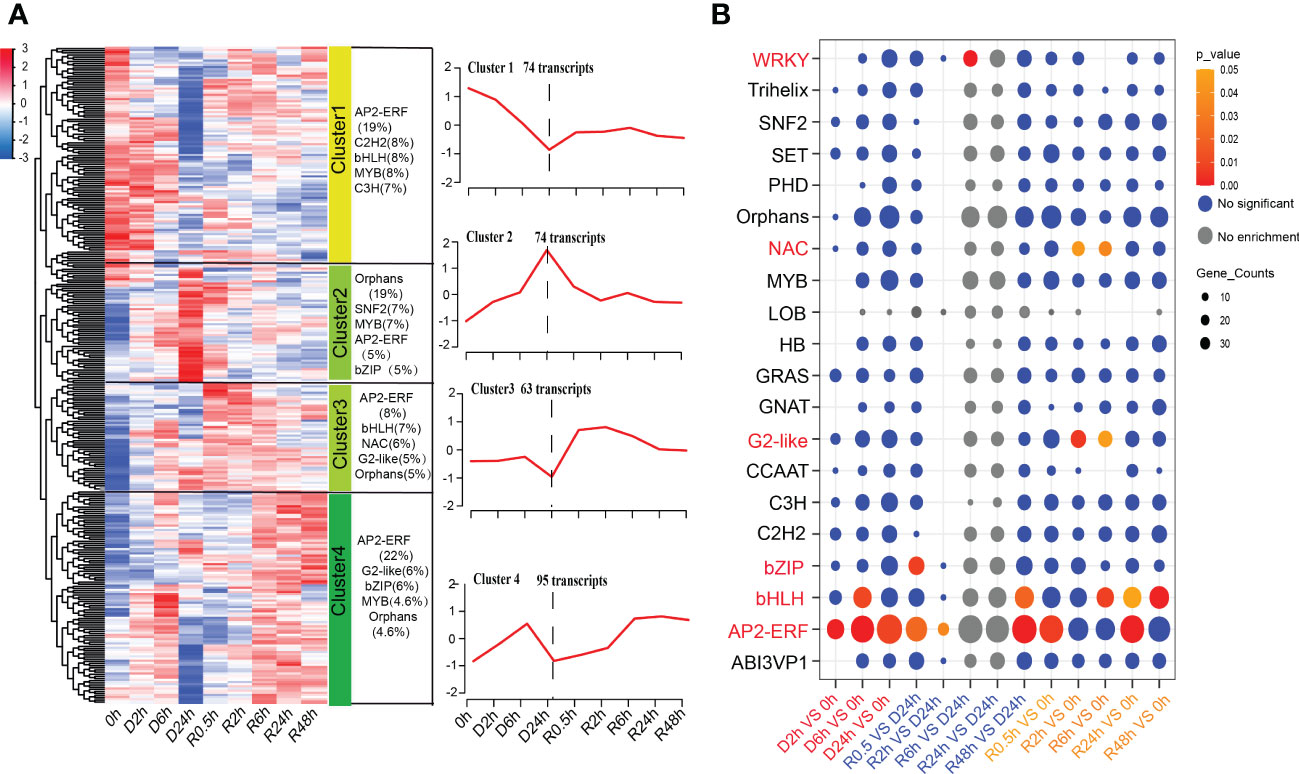
Figure 6 Global transcript abundance profiling and enrichment analysis of TFs during D-R process. (A) Heatmap and the dynamic accumulation patterns of the TFs; FPKM values are used for the heatmap and Clusters, (Cluster 1-Cluster 4) were generated by the K-means. The category and number of transcripts are marked in the upper part of each line chart. (B) Enrichment analysis of TFs in different comparison pairs. Circle size is correlated with number of TFs. TF family names are listed on the left, and different comparison pairs are listed on the bottom; red and orange circle represent p ≤ 0.05, bule circle represent no significant, grey circle indicated no enrichment; Treatments during 24 h of desiccation (D h) and 48 h of rehydration (R h).
Confirmation of DT-related transcriptional changes of represented DETs by RT-qPCR
To validate the RNA-seq results, we used RT-qPCR to analyse the transcript accumulation of 15 selected genes from four desiccation stress response KEGG pathways and two desiccation stress response marker genes, LEA2 and ELIP10 (Figure 7). The changes in the abundance of the selected DETs at different desiccation treatment points were in strong agreement with those of the RNA-seq results, indicating that transcriptome data accurately reflect in vivo transcript expression in the present study. The transcripts involved in phenylpropanoid biosynthesis, alpha-linolenic acid, and fructose and mannose metabolism pathways, including ScPAL (Sc_g00397), ScHCT (Sc_g12807), ScPOD21 (Sc_g13650), ScLOX (Sc_g11811), ScAOC1 (Sc_g05289), ScAOS (Sc_g14942), and ScHXK (Sc_g12069), were markedly upregulated in the early stage of D-R treatment. Furthermore, photosynthesis-related transcripts, such as ScPsa H (Sc_g13896), ScRubisco (Sc_g01013), and ScLhcb1 (Sc_g04588), were significantly upregulated after 6 h of rehydration. Additionally, the abundance of three TFs, ScAP2-ERF (Sc_g12069), ScbHLH (Sc_g07539), and ScbZIP (Sc_g13394), significantly increased under both dehydration and rehydration conditions.
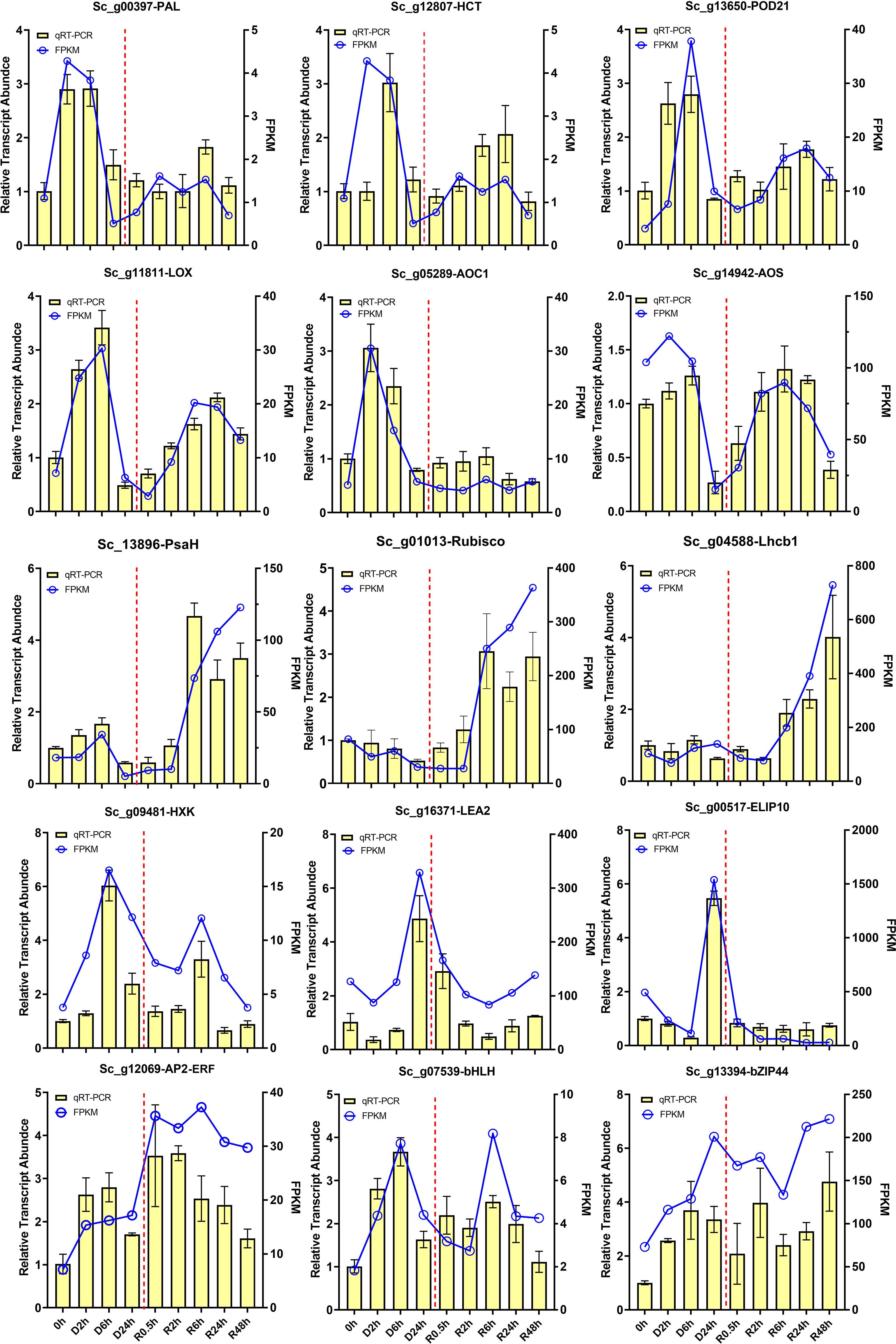
Figure 7 RT-qPCR validation of interested transcripts in S. caninervis during D-R process. Selected transcript abundance profiles were validated using RT-qPCR, including phenylpropanoid biosynthesis: ScPAL (Sc_g00397), ScHCT (Sc_g12807) and ScPOD21 (Sc_g13650); α-linolenic acid pathway: ScLOX (Sc_g11811), ScAOC1 (Sc_g05289) and ScAOS (Sc_g14942); photosynthesis and fructose and mannose metabolism: ScPsaH (Sc_g13896), ScRubisco (Sc_g01013), ScLHcb1 (Sc_g04588), and ScHXK (Sc_g09481); DT-related trancripts: ScLEA2 (Sc_g16371) and ScELIP10 (Sc_g00517). TFs: ScAP2-ERF (Sc_g12069), ScbHLH (Sc_g07539), and bZIP (Sc_g13394). RT-qPCR data were showed as the mean ± SD. Line chart represents FPKM value, and bar chart represents RT-qPCR value.
Discussion
Bryophytes belong to basal plant lineages that first colonised land and have developed adaptive mechanisms to cope with desiccation stress. In the wild, the DT moss S. caninervis from the desert is often subjected to unpredictable D-R events; consequently, this moss species has evolved remarkable constitutive and inducible mechanisms of DT to survive in these adverse desiccation environments (Li et al., 2010). In contrast to vascular plants, particularly the model plant Arabidopsis thaliana, little is known about the water deficit responses in DT moss. Transcriptional profiles during the D-R process have been performed in DT moss, such as T. ruralis and B. argenteum (Wood et al., 1999; Gao et al., 2017). However, previous transcriptomes for desert mosses have limitations; these include gene annotations using genomes of other species and a low sequencing depth. Although our research group has previously generated a mixed transcriptome of S. caninervis, it was impossible to investigate detailed transcriptional dynamic changes. In this study, the D-R transcriptome of S. caninervis was optimised and improved; overall, this made our results more accurate and comprehensive. Our study obtained transcriptome data at nine time points, including one full hydration, three dehydration, and five rehydration time points. Further, we used the newly acquired genomic data from S. caninervis for gene annotation. These improvements in transcriptional analysis helped improve our understanding of the S. caninervis transcriptome, which allowed further exploration of the molecular mechanism of DT moss and resulted in the production of an extensive genetic resource.
The positive effects of phenylpropanoid biosynthesis and alpha-linolenic acid pathways on the DT response of S. caninervis in the early stage of dehydration
The phenylpropanoid biosynthesis pathway is activated under drought stress, resulting in an accumulation of various phenolic compounds which have the potential to scavenge harmful ROS. This process results in the reduction of cell membrane peroxidation, thereby protecting plant cells from the destructive effects of oxidative stress (Sharma et al., 2019). Earlier transcriptomic studies carried out on DT plants, such as B. argenteum, Myrothamnus flabellifolia, and Boea hygrometrica, confirmed that the enhanced abundance of transcripts in the phenylpropanoid pathway help provide desiccation resistance (Ma et al., 2015; Xiao et al., 2015; Gao et al., 2017). In the current study, the peak abundance of phenylpropanoid pathway transcripts in S. caninervis occurred in the early stage of dehydration, aligning with the trend observed in Myrothamnus flabellifolia (Ma et al., 2015). Nonetheless, the main change in expression in the phenylpropanoid biosynthesis pathway of S. caninervis focused on lignin biosynthesis pathway. However, in other DT plants, such as G. pilifera, the flavonoid biosynthesis of phenylpropanoid is activated (Liu et al., 2022), suggesting that different DT plants have specific response pathways in phenylpropanoid biosynthesis.
Alpha-linolenic acid contributes to lipid metabolism as a strong antioxidant and as a precursor to the synthesis of JA, which acts as a phytohormone signalling molecule that stimulates the downstream anti-stress response (Creelman and Mullet, 1997; Wasternack and Feussner, 2018; Zi et al., 2022). JA is significantly induced in the early stages of dehydration in H. rhodopensis (Djilianov et al., 2013). Furthermore, H. rhodopensis desiccation stress transcripts were enriched in the early stages of dehydration (Liu et al., 2018). In addition, for B. hygrometrica and Marchantia inflexa, the GO terms of JA biosynthetic and signalling pathways have been observed to be enriched among the differentially abundant transcripts in response to dehydration (Zhu et al., 2015; Marks et al., 2021). In the present study, we found that these JA synthesis pathway transcripts in the S. caninervis transcriptome were significantly altered during the D-R process and elevated at the early stage of dehydration (2 and 6 h); this suggested that the JA synthesis pathway may have a positive effect on water-deficient stress in S. caninervis. Overall, these results indicate that the JA biosynthesis pathway may be an evolutionarily conserved response to DT, particularly in the early stages of desiccation response in DT plants.
Photosynthesis pathway changes during the DT response of S. caninervis
Photosynthesis, which can be significantly affected by dehydration, is the most important photochemical reaction in plants (Challabathula et al., 2018). DT plants can reduce linear electron transport flux via the reorganisation of the photosynthetic apparatus, thus preventing oxidative stress (Liu et al., 2018). It has been observed that upon dehydration photosynthetic genes are downregulated, photosynthesis stops, and chloroplasts undergo reversible ultrastructural changes (Giarola and Bartels, 2015; Liu et al., 2018). In the DT moss S. caninervis, the abundance patterns of photosynthesis-related transcripts, such as PSI and PSII, light-harvesting complexes I and II, cytochrome b6/f complex, and electron transport proteins, were generally inhibited during dehydration treatments (Figure 5C); these findings are in agreement with other DT species, such as C. plantagineum (Giarola and Bartels, 2015) and H. rhodopensis (Liu et al., 2018). Interestingly, at the transcription level, the transcripts related to S. caninervis photosynthesis were not induced immediately after rehydration but were upregulated after 6 h of rehydration. This phenomenon may be caused by the fact that it is not necessary to synthesise all transcripts immediately during rehydration of S. caninervis; instead, this moss may be able to use mRNA stored in messenger ribonucleoproteins for initial protein translation (Wood and Oliver et al., 1999). Overall, at the transcriptional level, S. caninervis exhibited strong photoprotection capabilities during the dehydration process and rapid photosynthesis recovery during the rehydration process.
ELIP proteins are also important for photoprotection (Silva et al., 2021) and accumulate during the desiccation of many DT species (Zeng et al., 2002; Gechev et al., 2013; Yobi et al., 2017; VanBuren et al., 2019). LEA proteins are associated with drying plant tissues and play important roles in maintaining cellular integrity when water is reintroduced (Oliver et al., 2004). In the angiosperms, the H. rhodopensis and C. plantagineum expression levels of LEA transcripts increase upon desiccation stress (Piatkowski et al., 1990; Michel et al., 1994; Rodriguez et al., 2010; Gechev et al., 2013). Similarly, in the moss P. patens (Hiss et al., 2014) and B. argenteum (Gao et al., 2017), LEA transcripts are more abundant during the dehydration process. Additionally, in S. caninervis, ELIP, and LEA were determined to strongly accumulate following dehydration, especially during late-stage dehydration. Therefore, ELIP and LEA protein functions are conserved as key components in response to DT in both bryophytes and angiosperms.
Desiccation tolerance related TFs in S. caninervis
TFs are important regulators involved in signal transduction and gene expression regulation under environmental stresses, such as water deficiency, temperature, salinity, and wounding stress (Singh and Laxmi, 2015; Yoon et al., 2020). In this study, desiccation stress caused differential changes in the abundance of 306 TF family members in S. caninervis. Among these TF families, 62 and 33 were associated with early dehydration and rehydration, respectively. In the DT moss B. argenteum, 404 TF-coding transcripts differentially accumulated during the time course of D-R. In total, 27 and 23 families were associated with dehydration and early rehydration, respectively (Gao et al., 2014). This indicates that S. caninervis possesses more TF families involved in DT than B. argenteum. Approximately 66% of differential TFs in S. caninervis were altered by desiccation stress. Specifically, seven key TF families (AP2-ERF, WRKY, G2-like, MYB, bZIP, bHLH, and NAC) were the most enriched families in D-R treatments of S. caninervis, indicating that these TFs are involved in the DT response. Similarly, in B. argenteum, AP2-ERF, MYB, bZIP, and bHLH were the most enriched TF families in response to D-R. AP2-ERF is a large family of plant TFs that play important roles in the control of plant metabolism and development, alongside various biotic and abiotic stress responses (Licausi et al., 2013). AP2-ERF is the largest TF family with the most differential abundant TFs in P. patens (Reboledo et al., 2022) and B. argenteum (Gao et al., 2017). Based on our transcriptome analysis, AP2-ERF was also the most abundant TF family in S. caninervis (Gao et al., 2014; Li et al., 2017). Many AP2-ERF family TFs are involved in the response of S. caninervis to DT and other biotic or abiotic stresses. For example, ScDREB5, ScDREB8, and ScDREB10 have been observed to significantly improve drought and salt tolerance in Arabidopsis (Li et al., 2016; Li et al., 2019b). Additionally, ScAPD1-like enhances resistance to Verticillium wilt in transgenic S. caninervis and Arabidopsis (Li et al., 2023). Further, during the rehydration process in the current study, NAC transcript abundance rapidly and significantly increased in S. caninervis. NAC also contributes to water conduction in P. patens (Xu et al., 2014). Similarly, NAC in Arabidopsis regulates xylem vessel differentiation, thereby affecting water conduction. Previous reports have shown that bHLH genes isolated from angiosperms enhance the drought tolerance of plants (Gu et al., 2021; Qian et al., 2021; Gao et al., 2022). Specifically, bHLH is an important component in the stomatal development of P. patens, and is, therefore, involved in the regulation of drought tolerance (Caine et al., 2020). In our study, bHLH genes were significantly enriched in both the dehydration and rehydration processes. Our results, and those of previous studies, have shown that TFs play critical and evolutionarily conserved roles in the DT response of moss and angiosperms.
Data availability statement
The dataset presented in the study are deposited in the China National GeneBank DataBase (CNGBdb) repository (https://db.cngb.org/), accession number CNP0003370.
Author contributions
DZ, XL, and RY conceived the study. RY conducted the experiments and collected the data with the help of WB, YL, XJL and BG. RY performed all the analyses with the help of QY and MZ and wrote the manuscript. DZ and XL revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Key Research Program of Frontier Sciences, Chinese Academy of Sciences (ZDBS-LY-SM009), Natural Science Foundation of Xinjiang Uygur Autonomous Region (Grant No.2022D01E96), and the Third Xinjiang Scientific Expedition Program (Grant No.2021xjkk0500).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1127541/full#supplementary-material
References
Caine, R. S., Chater, C. C. C., Fleming, A. J., Gray, J. E. (2020). Stomata and sporophytes of the model moss Physcomitrium patens. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.00643
Challabathula, D., Zhang, Q. W., Bartels, D. (2018). Protection of photosynthesis in desiccation-tolerant resurrection plants. J. Plant Physiol. 227, 84–92. doi: 10.1016/j.jplph.2018.05.002
Chen, C. J., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y. H., et al. (2020). TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13 (8), 1194–1202. doi: 10.1016/j.molp.2020.06.009
Creelman, R. A., Mullet, J. E. (1997). Biosynthesis and action of jasmonates in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48 (1), 355–381. doi: 10.1146/annurev.arplant.48.1.355
Djilianov, D. L., Dobrev, P. I., Moyankova, D. P., Vankova, R., Georgieva, D. T., Gajdosova, S., et al. (2013). Dynamics of endogenous phytohormones during desiccation and recovery of the resurrection plant species Haberlea rhodopensis. J. Plant Growth Regul. 32 (3), 564–574. doi: 10.1007/s00344-013-9323-y
Dong, N. Q., Lin, H. X. (2021). Contribution of phenylpropanoid metabolism to plant development and plant-environment interactions. J. Integr. Plant Biol. 63 (1), 180–209. doi: 10.1111/jipb.13054
Gao, S. Z., Li, C. H., Chen, X. H., Li, S. D., Liang, N. S., Wang, H. T., et al. (2022). Basic helix-loop-helix transcription factor PxbHLH02 enhances drought tolerance in populus (Populus simonii × P. nigra). Tree Physiol. 43 (1), 185–202. doi: 10.1093/treephys/tpac107
Gao, B., Li, X. S., Zhang, D. Y., Liang, Y. Q., Yang, H. L., Chen, M. X., et al. (2017). Desiccation tolerance in bryophytes: The dehydration and rehydration transcriptomes in the desiccation-tolerant bryophyte Bryum argenteum. Sci. Rep. 7 (1), 7571. doi: 10.1038/s41598-017-07297-3
Gao, B., Zhang, D. Y., Li, X. S., Yang, H. L., Wood, A. J. (2014). De novo assembly and characterization of the transcriptome in the desiccation-tolerant moss Syntrichia caninervis. BMC Res. Notes. 7 (1), 490. doi: 10.1186/1756-0500-7-490
Gechev, T. S., Benina, M., Obata, T., Tohge, T., Sujeeth, N., Minkov, I., et al. (2013). Molecular mechanisms of desiccation tolerance in the resurrection glacial relic Haberlea rhodopensis. Cell. Mol. Life Sci. 70 (4), 689–709. doi: 10.1007/s00018-012-1155-6
Giarola, V., Bartels, D. (2015). What can we learn from the transcriptome of the resurrection plant Craterostigma plantagineum? Planta 242 (2), 427–434. doi: 10.1007/s00425-015-2327-z
Godinez-Vidal, D., López-Leal, G., Covarrubias, A. A., Reyes, J. L. (2020). Early events leading to water deficit responses in the liverwort. Marchantia polymorpha. Environ. Exp. Bot. 178, 104172. doi: 10.1016/j.envexpbot.2020.104172
Gu, X. Y., Gao, S. X., Li, J., Song, P. Y., Zhang, Q., Guo, J. F., et al. (2021). The bHLH transcription factor regulated gene OsWIH2 is a positive regulator of drought tolerance in rice. Plant Physiol. Biochem. 169, 269–279. doi: 10.1016/j.plaphy.2021.11.031
Hiss, M., Laule, O., Meskauskiene, R. M., Arif, M. A., Decker, E. L., Erxleben, A., et al. (2014). Large-Scale gene expression profiling data for the model moss Physcomitrella patens aid understanding of developmental progression, culture and stress conditions. Plant J. 79 (3), 530–539. doi: 10.1111/tpj.12572
Kanehisa, M., Araki, M., Goto, S., Hattori, M., Hirakawa, M., Itoh, M., et al. (2007). KEGG for linking genomes to life and the environment. Nucleic Acids Res. 36, D480–D484. doi: 10.1093/nar/gkm882
Kim, D., Langmead, B., Salzberg, S. L. (2015). HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 12 (4), 357–360. doi: 10.1038/nmeth.3317
Langfelder, P., Horvath, S. (2008). WGCNA: an r package for weighted correlation network analysis. BMC Bioinform. 9, 559. doi: 10.1186/1471-2105-9-559
Li, X. S., Liang, Y. Q., Gao, B., Mijiti, M., Bozorov, T. A., Yang, H. L., et al. (2019b). ScDREB10, an a-5c type of DREB gene of the desert moss Syntrichia caninervis, confers osmotic and salt tolerances to Arabidopsis. Genes 10 (2), 146. doi: 10.3390/genes10020146
Li, C. C., Liu, S. H., Zhang, W., Chen, K. S., Zhang, P. Y. (2019a). Transcriptional profiling and physiological analysis reveal the critical roles of ROS-scavenging system in the Antarctic moss pohlia nutans under ultraviolet-b radiation. Plant Physiol. Biochem. 134, 113–122. doi: 10.1016/j.plaphy.2018.10.034
Li, Y., Wang, Z. B., Xu, T. H., Xu, T. H., Tu, W. F., Liu, C., et al. (2010). Reorganization of photosystem II is involved in the rapid photosynthetic recovery of desert moss Syntrichia caninervis upon rehydration. J. Plant Physiol. 167 (16), 1390–1397. doi: 10.1016/j.jplph.2010.05.028
Li, X. S., Yang, R. R., Liang, Y. Q., Gao, B., Li, S. M., Bai, W. W., et al. (2023). The ScAPD1-like gene from the desert moss Syntrichia caninervis enhances resistance to Verticillium dahliae via phenylpropanoid gene regulation. Plant J. 113 (1), 75–91. doi: 10.1111/tpj.16035
Li, X. S., Zhang, D. Y., Gao, B., Liang, Y. Q., Yang, H. L., Wang, Y. C., et al. (2017). Transcriptome-wide identification, classification, and characterization of AP2/ERF family genes in the desert moss syntrichia caninervis. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.00262
Li, X. S., Zhang, D. Y., Li, H. Y., Gao, B., Yang, H. L., Zhang, Y. M., et al. (2015). Characterization of reference genes for RT-qPCR in the desert moss Syntrichia caninervis in response to abiotic stress and desiccation/rehydration. Front. Plant Sci. 6. doi: 10.3389/fpls.2015.00038
Li, H. Y., Zhang, D. Y., Li, X. S., Guan, K. Y., Yang, H. L. (2016). Novel DREB a-5 subgroup transcription factors from desert moss (Syntrichia caninervis) confers multiple abiotic stress tolerance to yeast. J. Plant Physiol. 194, 45–53. doi: 10.1016/j.jplph.2016.02.015
Liang, Y. Q., Li, X. S., Zhang, D. Y., Gao, B., Yang, H. L., Wang, Y. C., et al. (2017). ScDREB8, a novel a-5 type of DREB gene in the desert moss Syntrichia caninervis, confers salt tolerance to Arabidopsis. Plant Physiol. Biochem. 120, 242–251. doi: 10.1016/j.plaphy.2017.09.014
Liang, Y. Q., Li, X. S., Zhang, J., Zhuo, L., Liu, X. J., Yang, R. R., et al. (2021). Dehydration rates impact physiological, biochemical and molecular responses in desert moss Bryum argenteum. Environ. Exp. Bot. 183, 104346. doi: 10.1016/j.envexpbot.2020.104346
Licausi, F., Ohme-Takagi, M., Perata, P. (2013). APETALA/Ethylene responsive factor (AP2/ERF) transcription factors: mediators of stress responses and developmental programs. New Phytol. 199 (3), 639–649. doi: 10.1111/nph.12291
Liu, Y., Ge, B. K., Zhang, D. Q., Yi, Y. J. (2022). De novo transcriptome assembly and analysis of genes involved in desiccation tolerance in Grimmia pilifera. Gene 847, 146841. doi: 10.1016/j.gene.2022.146841
Liu, J., Moyankova, D., Lin, C. T., Mladenov, P., Sun, R. Z., Djilianov, D., et al. (2018). Transcriptome reprogramming during severe dehydration contributes to physiological and metabolic changes in the resurrection plant Haberlea rhodopensis. BMC Plant Biol. 18, 351. doi: 10.1186/s12870-018-1566-0
Liu, J. Y., Yang, R. R., Liang, Y. Q., Wang, Y., Li, X. S. (2022). The DREB a-5 transcription factor ScDREB5 from syntrichia caninervis enhanced salt tolerance by regulating jasmonic acid biosynthesis in transgenic Arabidopsis. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.857396
Liu, X. J., Zhang, Y. G., Yang, H. L., Liang, Y. Q., Li, X. S., Oliver, M. J., et al. (2020). Functional aspects of early light-induced protein (ELIP) genes from the desiccation-tolerant moss Syntrichia caninervis. Int. J. Mol. Sci. 21 (4), 1411. doi: 10.3390/ijms21041411
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods 25 (4), 402–408. doi: 10.1006/meth.2001.1262
Ma, C., Wang, H., Macnish, A. J., Estrada-Melo, A. C., Lin, J., Chang, Y. H., et al. (2015). Transcriptomic analysis reveals numerous diverse protein kinases and transcription factors involved in desiccation tolerance in the resurrection plant Myrothamnus flabellifolia. Hortic. Res. 2, 15034. doi: 10.1038/hortres.2015.34
Mao, X. Z., Cai, T., Olyarchuk, J. G., Wei, L. P. (2005). Automated genome annotation and pathway identification using the KEGG orthology (KO) as a controlled vocabulary. Bioinformatics 21 (19), 3787–3793. doi: 10.1093/bioinformatics/bti430
Marks, R. A., Smith, J. J., VanBuren, R., McLetchie, D. N. (2021). Expression dynamics of dehydration tolerance in the tropical plant Marchantia inflexa. Plant J. 105 (1), 209–222. doi: 10.1111/tpj.15052
Michel, D., Furini, A., Salamini, F., Bartels, D. (1994). Structure and regulation of an ABA-and desiccation-responsive gene from the resurrection plant Craterostigma plantagineum. Plant Mol. Biol. 24 (4), 549–560. doi: 10.1007/BF00023553
Oliver, M. J., Dowd, S. E., Zaragoza, J., Mauget, S. A., Payton, P. R. (2004). The rehydration transcriptome of the desiccation-tolerant bryophyte Tortula ruralis: Transcript classification and analysis. BMC Genom. 5, 89. doi: 10.1186/1471-2164-5-89
Oliver, M. J., Hudgeons, J., Dowd, S. E., Payton, P. R. (2009). A combined subtractive suppression hybridization and expression profiling strategy to identify novel desiccation response transcripts from Tortula ruralis gametophytes. Physiol. Plant 136 (4), 437–460. doi: 10.1111/j.1399-3054.2009
Pan, Z., Pitt, W. G., Zhang, Y. M., Wu, N., Tao, Y., Truscott, T. T. (2016). The upside-down water collection system of Syntrichia caninervis. Nat. Plants. 2 (7), 16076. doi: 10.1038/nplants.2016.76
Piatkowski, D., Schneider, K., Salamini, F., Bartels, D. (1990). Characterization of five abscisic acid-responsive cDNA clones isolated from the desiccation-tolerant plant craterostigma plantagineum and their relationship to other water-stress genes. Plant Physiol. 94 (4), 1682–1688. doi: 10.1104/pp.94.4.1682
Putri, G. H., Anders, S. P., Pyl, P. T., Pimanda, J. E., Zanini, F. (2022). Analysing high-throughput sequencing data in Python with HTSeq 2.0. Bioinformatics 10 (38), 2943–2945. doi: 10.1093/bioinformatics/btac166
Qian, Y. C., Zhang, T. Y., Yu, Y., Gou, L. P., Yang, J. T., Xu, J., et al. (2021). Regulatory mechanisms of bHLH transcription factors in plant adaptive responses to various abiotic stresses. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.677611
Rathnayake, K. N., Nelson, S., Seeve, C., Oliver, M. J., Koster, K. L. (2019). Acclimation and endogenous abscisic acid in the moss Physcomitrella patens during acquisition of desiccation tolerance. Physiol. Plant 167 (3), 317–329. doi: 10.1111/ppl.12892
Reboledo, G., Agorio, A., De Leon, I. P. (2022). Moss transcription factors regulating development and defense responses to stress. J. Exp. Bot. 73 (13), 4546–4561. doi: 10.1093/jxb/erac055
Rodriguez, M. C. S., Edsgard, D., Hussain, S. S., Alquezar, D., Rasmussen, M., Gilbert, T., et al. (2010). Transcriptomes of the desiccation-tolerant resurrection plant Craterostigma plantagineum. Plant J. 63 (2), 212–228. doi: 10.1111/j.1365-313X.2010
Sharma, A., Shahzad, B., Rehman, A., Bhardwaj, R., Landi, M., Zheng, B. S. (2019). Response of phenylpropanoid pathway and the role of polyphenols in plants under abiotic stress. Molecules 24 (13), 2452. doi: 10.3390/molecules24132452
Silva, A. T., Gao, B., Fisher, K. M., Mishler, B. D., Ekwealor, J. T. B., Stark, L. R., et al. (2021). To dry perchance to live: Insights from the genome of the desiccation-tolerant biocrust moss Syntrichia caninervis. Plant J. 105 (5), 1339–1356. doi: 10.1111/tpj.15116
Singh, D., Laxmi, A. (2015). Transcriptional regulation of drought response: A tortuous network of transcriptional factors. Front. Plant Sci. 6. doi: 10.3389/fpls.2015.00895
Trapnell, C., Williams, B. A., Pertea, G., Mortazavi, A., Kwan, G., van Baren, M. J., et al. (2010). Transcript assembly and quantification by RNA-seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28 (5), 511–515. doi: 10.1038/nbt.1621
VanBuren, R., Pardo, J., Wai, C. M., Evans, S., Bartels, D. (2019). Massive tandem proliferation of ELIPs supports convergent evolution of desiccation tolerance across land plants. Plant Physiol. 179 (3), 1040–1049. doi: 10.1104/pp.18.01420
Wang, L. K., Feng, Z. X., Wang, X., Wang, X. W., Zhang, X. G. (2010). DEGseq: An r package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 26 (1), 136–138. doi: 10.1093/bioinformatics/btp612
Wang, J. C., Yang, H. L., Gao, B., Bozorov, T. A., Li, X. S., Zhang, D. Y., et al. (2020). Analysis and characterization of the aldehyde dehydrogenase (ALDH) gene superfamily in the desert moss Syntrichia caninervis in response to abiotic stress. Environ. Exp. Bot. 178, 104176. doi: 10.1016/j.envexpbot.2020.104176
Wasternack, C., Feussner, I. (2018). The oxylipin pathways: Biochemistry and function. Annu. Rev. Plant Biol. 69 (1), 363–386. doi: 10.1146/annurev-arplant-042817-040440
Wood, A. J. (2007). Invited essay: New frontiers in bryology and lichenology: The nature and distribution of vegetative desiccation-tolerance in hornworts, liverworts and mosses. Bryologist 110 (2), 163–177. doi: 10.1639/0007-2745(2007)110[163:IENFIB]2.0.CO;2
Wood, A. J., Duff, R. J., Oliver, M. J. (1999). Expressed sequence tags (ESTs) from desiccated Tortula ruralis identify a large number of novel plant genes. Plant Cell Physiol. 40 (4), 361–368. doi: 10.1093/oxfordjournals.pcp.a029551
Wood, A. J., Oliver, M. J. (1999). Translational control in plant stress: The formation of messenger ribonucleoprotein particles (mRNPs) in response to desiccation of Tortula ruralis gametophytes. Plant J. 18 (4), 359–370. doi: 10.1046/j.1365-313X.1999.00458.x
Wu, X. L., Shiroto, Y., Kishitani, S., Ito, Y., Toriyama, K. (2009). Enhanced heat and drought tolerance in transgenic rice seedlings overexpressing OsWRKY11 under the control of HSP101 promoter. Plant Cell Rep. 28 (1), 21–30. doi: 10.1007/s00299-008-0614-x
Xiao, L. H., Yang, G., Zhang, L. C., Yang, X. H., Zhao, S., Ji, Z. Z., et al. (2015). The resurrection genome of Boea hygrometrica: A blueprint for survival of dehydration. PNAS 112 (18), 5833–5837. doi: 10.1073/pnas.1505811112
Xu, B., Ohtani, M., Yamaguchi, M., Toyooka, K., Wakazaki, M., Sato, M., et al. (2014). Contribution of NAC transcription factors to plant adaptation to land. Science 343 (6178), 1505–1508. doi: 10.1126/science.1248417
Yang, H. L., Bozorov, T. A., Chen, X. P., Zhang, D. W., Wang, J. C., Li, X. S., et al. (2021). Yield comparisons between cotton variety xin nong mian 1 and its transgenic ScALDH21 lines under different water deficiencies in a desert-oasis ecotone. Agronomy 11 (5), 1019. doi: 10.3390/agronomy11051019
Yang, H. L., Zhang, D. Y., Li, H. Y., Dong, L. F., Lan, H. Y. (2015). Ectopic overexpression of the aldehyde dehydrogenase ALDH21 from Syntrichia caninervis in tobacco confers salt and drought stress tolerance. Plant Physiol. Biochem. 95, 83–91. doi: 10.1016/j.plaphy.2015.07.001
Yang, H. L., Zhang, D. Y., Li, X. S., Li, H. Y., Zhang, D. W., Lan, H. Y., et al. (2016). Overexpression of ScALDH21 gene in cotton improves drought tolerance and growth in greenhouse and field conditions. Mol. Breed. 36 (3), 34. doi: 10.1007/s11032-015-0422-2
Yang, H. L., Zhang, D. Y., Wang, J. C., Wood, A. J., Zhang, Y. M. (2012). Molecular cloning of a stress-responsive aldehyde dehydrogenase gene ScALDH21 from the desiccation-tolerant moss Syntrichia caninervis and its responses to different stresses. Mol. Biol. Rep. 39 (3), 2645–2652. doi: 10.1007/s11033-011-1017-6
Yin, B. F., Zhang, Y. M., Lou, A. R. (2017). Impacts of the removal of shrubs on the physiological and biochemical characteristics of Syntrichia caninervis mitt: In a temperate desert. Sci. Rep. 7, 45268. doi: 10.1038/srep45268
Yobi, A., Schlauch, K. A., Tillett, R. L., Yim, W. C., Espinoza, C., Wone, B. W., et al. (2017). Sporobolus stapfianus: Insights into desiccation tolerance in the resurrection grasses from linking transcriptomics to metabolomics. BMC Plant Biol. 17 (1), 67. doi: 10.1186/s12870-017-1013-7
Yoon, Y., Seo, D. H., Shin, H., Kim, H. J., Kim, C. M., Jang, G. (2020). The role of stress-responsive transcription factors in modulating abiotic stress tolerance in plants. Agronomy 10 (6), 788. doi: 10.3390/agronomy10060788
Zeng, Q., Chen, X. B., Wood, A. J. (2002). Two early light-inducible protein (ELIP) cDNAs from the resurrection plant Tortula ruralis are differentially expressed in response to desiccation, rehydration, salinity, and high light. J. Exp. Bot. 53 (371), 1197–1205. doi: 10.1093/jexbot/53.371.1197
Zhang, Y. M. (2005). The microstructure and formation of biological soil crusts in their early developmental stage. Chin. Sci. Bulletin. 50 (2), 117–121. doi: 10.1360/982004-559
Zhang, Y. G., Liu, X. J., Zhang, K. C., Zhang, D. Y., Guan, K. Y. (2018). An ABSCISIC ACID INSENSITIVE3-like gene from the desert moss Syntrichia caninervis confers abiotic stress tolerance and reduces ABA sensitivity. Plant Cell Tissue Organ Cult. 133 (3), 417–435. doi: 10.1007/s11240-018-1394-9
Zhang, Y., Wang, C., Huang, M. Q., Zhang, Y. G. (2022). Functional analysis of ScABI3 from Syntrichia caninervis mitt. in Medicago sativa l. Agronomy 12 (9), 2238. doi: 10.3390/agronomy12092238
Zhang, J., Zhang, Y. M., Downing, A., Wu, N., Zhang, B. C. (2011). Photosynthetic and cytological recovery on remoistening Syntrichia caninervis mitt., a desiccation-tolerant moss from northwestern China. Photosynthetica 49 (1), 13–20. doi: 10.1007/s11099-011-0002-6
Zhu, Y., Wang, B., Phillips, J., Zhang, Z. N., Du, H., Xu, T., et al. (2015). Global transcriptome analysis reveals acclimation-primed processes involved in the acquisition of desiccation tolerance in Boea hygrometrica. Plant Cell Physiol. 56 (7), 1429–1441. doi: 10.1093/pcp/pcv059
Keywords: Syntrichia caninervis, desiccation tolerance, phenylpropanoid biosynthesis, alpha-linolenic acid, photosynthesis, transcription factor, transcriptome
Citation: Yang R, Li X, Yang Q, Zhao M, Bai W, Liang Y, Liu X, Gao B and Zhang D (2023) Transcriptional profiling analysis providing insights into desiccation tolerance mechanisms of the desert moss Syntrichia caninervis. Front. Plant Sci. 14:1127541. doi: 10.3389/fpls.2023.1127541
Received: 19 December 2022; Accepted: 02 February 2023;
Published: 23 February 2023.
Edited by:
John Paul Délano-Frier, Instituto Politécnico Nacional de México (CINVESTAV), MexicoReviewed by:
Shenghao Liu, Ministry of Natural Resources, ChinaLihong Xiao, Zhejiang Agriculture and Forestry University, China
Copyright © 2023 Yang, Li, Yang, Zhao, Bai, Liang, Liu, Gao and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaoshuang Li, bGl4c0Btcy54amIuYWMuY24=; Daoyuan Zhang, emhhbmdkeUBtcy54amIuYWMuY24=
 Ruirui Yang
Ruirui Yang Xiaoshuang Li
Xiaoshuang Li Qilin Yang
Qilin Yang Mingqi Zhao
Mingqi Zhao Wenwan Bai1,2
Wenwan Bai1,2 Yuqing Liang
Yuqing Liang Bei Gao
Bei Gao Daoyuan Zhang
Daoyuan Zhang