
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 14 April 2023
Sec. Plant Abiotic Stress
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1125324
This article is part of the Research Topic Heat Stress: Response, Mitigation, and Tolerance in Plants View all 14 articles
Global warming causes great thermal stress to macroalgae and those species that can adapt to it are thought to be better able to cope with warmer oceans. Gracilaria bailinae, a macroalgae with high economic and ecological values, can survive through the hot summer in the South China Sea, but the molecular mechanisms underlying its adaptation to high temperatures are unclear. To address this issue, the present study analyzed the growth and transcriptome of G. bailinae after a 7-day exposure to 15°C (LT: low temperature), 25°C (MT: middle temperature), and 35°C (HT: high temperature). Growth analysis showed that the HT group had the highest relative growth rate (RGR = 2.1%) with the maximum photochemical quantum yield of PSII (Fv/Fm = 0.62) remaining within the normal range. Transcriptome analysis showed more differentially expressed genes (DEGs) in the comparison between MT and HT groups than in that between MT and LT, and most of these DEGs tended to be downregulated at higher temperatures. The KEGG pathway enrichment analysis showed that the DEGs were mainly enriched in the carbohydrate, energy, and lipid metabolisms. In addition, the genes involved in NADPH and ATP synthesis, which are associated with photosynthesis, the Calvin cycle, pyruvate metabolism, and the citrate cycle, were downregulated. Downregulation was also observed in genes that encode enzymes involved in fatty acid desaturation and alpha-linolenic acid metabolism. In summary, G. bailinae regulated the synthesis of NADPH and ATP, which are involved in the above-mentioned processes, to reduce unnecessary energy consumption, and limited the synthesis of enzymes in the metabolism of unsaturated fatty acids and alpha-linolenic acid to adapt to high environmental temperatures. The results of this study improve our understanding of the molecular mechanisms underlying the adaptation of G. bailinae to high temperatures.
Since the industrial revolution, increasing anthropogenic CO2 and other greenhouse gas emissions have resulted in an energy imbalance on Earth; specifically, in a positive radiative imbalance at the highest atmospheric layers (Hansen et al., 2011; Von Schuckmann et al., 2016; Association, 2022). As a result, energy began to accumulate as heat in the Earth system, which is what ultimately drives global warming (Hansen et al., 2011; Von Schuckmann et al., 2016; Association, 2022). The global average temperature in 2021 was approximately 1.11 ± 0.13°C warmer than the pre-industrial average recorded between 1850 and 1900, and the 2015–2021 period has been the hottest on record (Association, 2022). Our oceans have absorbed over 90% of the energy accumulated in the Earth system through human-induced greenhouse gas emissions, causing a rise in ocean heat content, which in 2021 was the highest on record (Association, 2022). The Special Report published by the Intergovernmental Panel on Climate Change in 2019 also noted that the oceans have warmed unabated since 2005 (Bindoff et al., 2019). In addition, the frequency of marine heatwaves (MHWs) has increased during the past century, with potential consequences for marine organisms and the communities that depend on them (Smale et al., 2019). Between 1982 and 2020, the duration and frequency of MHWs in the South China Sea increased significantly (Yao and Wang, 2021). The continued warming of the oceans is having an enormous impact on macroalgae. One of the most crucial aspects affecting their dispersion is temperature, since it has an influence on their physiology, growth, and adaptation (Graba-Landry et al., 2018; Ji and Gao, 2021). Ocean warming hinders the growth of some tropical brown algae and subtropical macroalgae, forces tropical species to exist at their maximum temperature limit, and can even cause the entire macroalgal community to retreat (Wernberg et al., 2011; Koch et al., 2013; Bender et al., 2014; Graba-Landry et al., 2018; Ji and Gao, 2021). Increasing sea surface temperatures have already led to the loss of important habitat-forming kelp and macroalgae (Graba-Landry et al., 2018). If global warming persists, the distribution of certain seaweeds will change even more, and some might go extinct (Wernberg et al., 2011; Yao and Somero, 2014). It is thought that macroalgal species that can adapt to heat stress would do better in the increasingly warmer oceans (Ji and Gao, 2021).
The ability of plants to sense temperature stimuli, generate and transmit signals, and activate the proper morpho-anatomical, physiological, and molecular responses plays a vital role in their survival in hot environments (Hasanuzzaman et al., 2013; Zhang et al., 2020). Algae generally employ a variety of coping and repair mechanisms, including the regulation of photosynthesis to balance energy output and consumption, the modification of membrane fluidity by altering the degree of fatty acid saturation, the accumulation of compatible solutes to keep cell osmolality constant, and the activation of the enzymatic antioxidant system to remove excessive reactive oxygen species (Barati et al., 2019). RNA sequencing (RNA-seq) is a comprehensive technique for analyzing gene function and revealing the molecular mechanisms behind certain biological processes (Lim et al., 2016). This technique has steadily been employed in the study of macroalgae’s molecular biology in recent years (Wang et al., 2015; Wenlei et al., 2018; Qin et al., 2021). For example, RNA-seq has been crucial in 1) tracking the expression of essential genes and critical metabolic processes in the growth and development of Gracilariopsis lemaneiformis (Wang et al., 2015; Qin et al., 2021), 2) investigating the molecular processes of G. lemaneiformis’ reaction to low temperature environments (Qin et al., 2021), and 3) understanding Pyropia haitanensis’ thermal adaptation to high temperatures (Wenlei et al., 2018).
Gracilaria bailinae is a red alga generally used for agar extraction, animal farming, and aquaculture water treatments (Elle et al., 2017; Xu et al., 2021). Specifically, this species has the potential to limit the spread of pathogenic Vibrio, reduce unionized ammonia, and promote the steady growth of advantageous phytoplankton (Nannochloropsis and Chlorella) (Elle et al., 2017). Additionally, due to its high agar yield of 29.8% dw (Skriptsova and Nabivailo, 2009), G. bailinae is greatly sought after for uses in the processed food, cosmetic, and pharmaceutical industries, and also in the fields of microbiology and molecular biology (Endoma et al., 2020). Preliminary studies found that G. bailinae can grow at a temperature range between 15 and 35°C and shows several traits associated with high-temperature tolerance (unpublished). Within the context of global warming, this species has a great potential for future applications as it can be farmed on a large scale all year round in tropical areas, continuously providing the raw materials for ecological restoration programs, sewage purification, agar and feed manufacturing, etc. However, the molecular mechanisms behind G. bailinae’s reaction to high temperatures remain unknown. The present study specifically aims to reveal the mechanism of gene network regulation at the transcriptional level in G. bailinae subjected to extreme temperatures using the RNA-seq technique and combining the growth and physiological performances. The results obtained will not only provide a theoretical basis for understanding the molecular mechanisms of adaptation to high temperature in G. bailinae, but also guidance to breed temperature-tolerant seaweed species.
Wild G. bailinae specimens were collected from Wushi (N 20°33′, E 109°51′), Guangdong Province, China, in July 2021.The samples were brought back to the lab in a portable fridge at 4°C. Fresh algae were randomly selected and cleaned thoroughly using sterilized seawater. They were then temporarily cultivated in a tank filled with sterile seawater that had been enhanced with f/2 media at 25°C under a 12 L:12 D photoperiod and light intensity of 100 μmol·m−2·s−1. Three beakers containing 1 L of fresh sterile seawater enriched with f/2 medium were prepared and 3 g of healthy algae was added to each of them. The beakers were then transferred to incubators set at 15°C (LT: low temperature), 25°C (MT: middle temperature), and 35°C (HT: high temperature) for 7 days. Three biological replicates were prepared per treatment and the medium was changed every 2 days. After 7 days of culture, the algae were used for growth and Fv/Fm measurements, as well as transcriptomic analysis.
After the treatment, the fresh weight of G. bailinae was measured and the relative growth rate (RGR) was determined using the formula supplied by Yong (Yong et al., 2013):
where M0 represents the starting weight of the algae and M7 represents their weight after the 7-day treatment.
The maximum photochemical quantum yield of PSII (Fv/Fm) was measured based on Li et al. (Li et al., 2016) after 7 days of treatment at different temperatures using a Hansatech FMS-2 modulated fluorometer (Hansatech Instruments, Norfolk, UK). The formula used was Fv = Fm – F0; the minimum (F0) and maximum (Fm) fluorescence were obtained by exposing algae that had adapted to darkness for 30 minutes to actinic light and saturated light provided by the fluorometer (Li et al., 2016).
Total RNA was isolated from G. bailinae using the Trizol reagent kit (Invitrogen, Carlsbad, CA, USA) in accordance with the manufacturer’s instructions after 7 days of LT, MT, and HT treatments. An Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA) was used to analyze the quality of total RNA and the results were validated via RNase-free agarose gel electrophoresis. Following total RNA extraction, Oligo (dT) beads were used to enrich G. bailinae’s mRNA. Using random primers, the enriched mRNA was reverse transcribed into cDNA after being broken into small fragments by applying fragmentation buffer. Subsequently, DNA polymerase I, RNase H, dNTP, and buffer were used to create second-strand cDNA. Following purification with Qia Quick PCR extraction kit (Qiagen, Venlo, Netherlands), the cDNA fragments were ligated to the Illumina sequencing adapter after end repair and poly(A) addition. The ligation products were size selected via agarose gel electrophoresis, amplified via PCR, and then sequenced using Illumina NovaSeq 6000 (Gene Denovo Biotechnology Co., Guangzhou, China).
Principal component analysis (PCA) was carried out using R (http://www.r-project.org/) on all nine transcriptome datasets to investigate variations in expression patterns between samples. The three duplicates of each temperature treatment were subjected to Pearson correlation analysis in order to evaluate the reproducibility of the transcriptome data.
The raw reads were trimmed by removing those containing adapters, more than 10% of unknown nucleotides, and more than 50% of low quality (Q-value ≤ 20) bases using fastp (version 0.18.0) (Chen et al., 2018). The transcriptome’s de novo assembly was carried out using the short-read assembling program Trinity (Grabherr et al., 2011). BLASTx (http://www.ncbi.nlm.nih.gov/BLAST/) with an E-value threshold of 105 was used to compare transcripts to the NCBI non-redundant protein (Nr) database (http://www.ncbi.nlm.nih.gov), the Swiss-Prot protein database (http://www.expasy.ch/sprot), the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (http://www.genome.jp/kegg), and the COG/KOG database (http://www.ncbi.nlm.nih.gov/COG), and annotate the unigenes. Protein function annotations were then obtained based on the best alignment results.
The differential expression analysis of RNAs between different groups and between samples was performed using DESeq2 (Love et al., 2014) and edgeR (Robinson et al., 2010) software, respectively. The genes with a parameter of false discovery rate (FDR) below 0.05 and absolute fold change ≥ 3 were considered DEGs.
Nine genes associated with temperature changes were randomly selected from the transcriptome results to validate the transcriptome using the 18sRNA (Liu et al., 2021a) gene as a reference gene. The following reaction steps were adopted: 95°C for 300 s, followed by 45 cycles at 95°C for 10 s, 60°C for 15 s, 72°C for 15 s, and reading the fluorescence signal, followed by 1 cycle at 95°C for 10 s, 65°C for 60 s, and 95°C for 1 s. Each reaction was conducted in triplicate. Primer Premier 5.0 (Premier, Toronto, ON, Canada) was used to design each primer. The 2−ΔΔCt (RQ) method was used to determine the relative transcript level (Qin et al., 2021). Primer sequences and qRT-PCR validation results are included in Supplementary Table S1.
The RGR and Fv/Fm data were expressed as means ± SD (n ≥ 3). The statistical significance of different treatments was assessed using one-way ANOVA and Duncan’s test in SPSS 25 (IBM Corp., Armonk, NY, USA) with a significance level of P< 0.05.
G. bailinae maintained a positive growth during the period of culture under the three different temperatures. The HT treatment (35°C) resulted in a maximum growth rate of 2.1%, which was significantly higher than the values obtained at 15°C and 25°C (P< 0.01); in particular, it was 15 times higher than that obtained at 15°C (0.14%) (Figure 1A). The Fv/Fm values at 25°C and 35°C were 0.65 and 0.62, respectively, which were significantly higher than the value at 15°C (0.36) (P< 0.01) (Figure 1B).
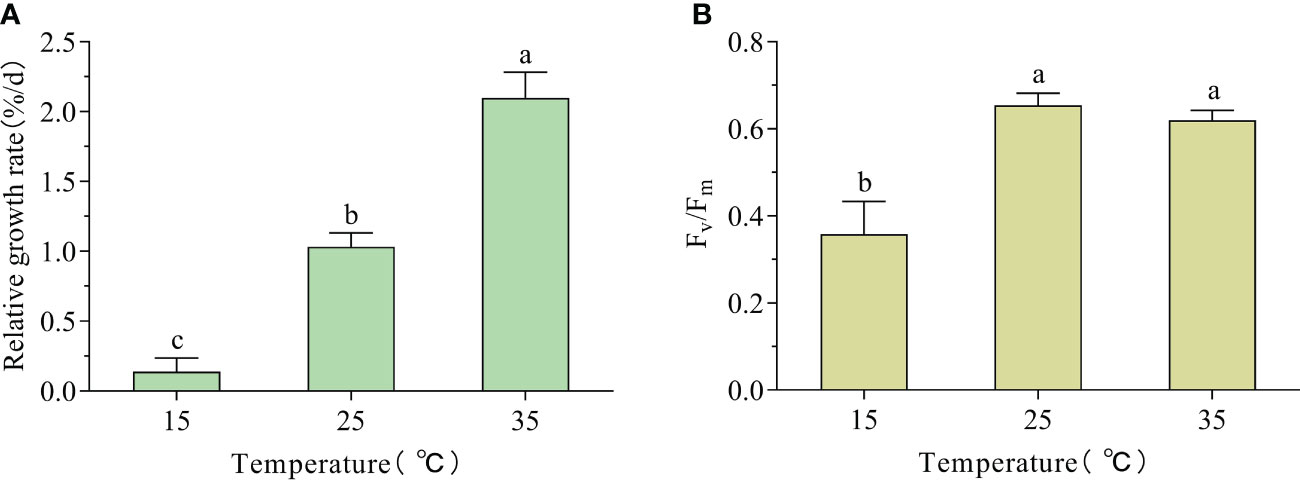
Figure 1 Relative growth rate (RGR) and Fv/Fm changes in G. bailinae exposed to different temperatures. (A) RGR. (B) Maximum photochemical quantum yield of PSII (Fv/Fm). Different small letters above the columns indicate a significant difference between different temperature treatments (P< 0.05).
The raw data obtained from the nine samples sequenced were subjected to quality control to obtain clean data (Supplementary Table S2). Out of a total of 494,707,094 reads 492,443,386 were retained after removing adapters and low-quality sequences. In each sample, the yield of clean reads was above 99%, the GC content was between 50.94% and 51.56%, and the Q20(%) value was above 98%. The clustering of the assembled transcripts of G. bailinae yielded a total of 20,049 single genes (> 200 base pairs) with an N50 count of 4,140 and a length of 1,072 base pairs. The size of single genes ranged from 201 to 8,602 base pairs, with an average length of 769 base pairs and a total length of 15,419,753 base pairs (Supplementary Table S3).
The three duplicate samples in each treatment had Pearson correlation coefficients (r) of less than 0.96 (r > 0.80 indicates a strong correlation) (Supplementary Figure S1A). PCA showed that the expression patterns of G. bailinae differed under the three temperatures (Supplementary Figure S1B). Furthermore, qRT-PCR and mRNA-Seq analyses showed that nine randomly selected genes had a similar expression tendency (Supplementary Table S1).
At present, the whole-genome sequencing of G. bailinae has not been completed, therefore, only 20,049 gene transcript sequences were compared to the NCBI non-redundant protein databases Nr, Swiss-Prot, KEGG, and COG/KOG (evalue< 0.00001) using BLASTx. The proteins with the highest sequence similarity to the given unigene and its functional annotation information were obtained. In total, 20,049 genes were assembled from the transcriptome data in this study and 13,095 (65.31%) of them were compared to the four major databases. Of these 20,049 genes, 12,828 (63.98%), 5,246 (26.17%), 5,027 (25.07%), and 9,714 (48.45%) genes matched the Nr, KEGG, KOG, and GO databases, respectively.
In the Nr database, the main match obtained was Gracilariopsis chorda (84.82%), which belongs to the same genus as G. bailinae, in line with the taxonomic status distinction (Supplementary Figure S2). Cellular processes, environmental information processing, genetic information processing, metabolism, and organic systems were the five primary functional categories of the genes that matched the KEGG database. Each category was subdivided into secondary groups of functions. In particular, the largest number of unigenes were involved in “global and overview maps” (1,445), followed by “translation” (750) and “folding, sorting, and degradation” (409) (Supplementary Figure S3). The genes that matched the KOG database were divided into 25 groups: the largest was “translation, ribosomal structure and biogenesis” (781), followed by “general function prediction only” (701), and the smallest was “cell motility” (5) (Supplementary Figure S4). A total of 5,824 (29.05%) genes were matched to Swiss-Prot database. The number of genes matched in all databases was 4611 (23.00%) (Supplementary Figure S5).
Based on the Nr annotation information, Blast2GO was used to obtain the GO function annotations (Ashburner et al., 2000; Conesa et al., 2005). Three ontologies in GO were used to categorize the genes’ molecular functions (MF), cellular components (CC), and biological processes (BP). The 9,714 unigenes annotated into the GO database by GO annotation were classified into 50 subclasses, accounting for 48.45% of all unigenes, and the same unigenes could be annotated to different subclasses. The top three subclasses of the GO ontologies were: cellular process, metabolic process, and localization with 6,637, 5,845, and 1,497, genes, respectively, for BP; cellular anatomical entity, protein-containing complex, and virion component, with 4,253, 1,958 and 110 genes, respectively, for CC; and catalytic activity, binding, and transporter activity, with 5,066, 4,771, and 802 genes, respectively, for MF (Supplementary Figure S6).
Two comparison groups, MT vs LT and MT vs HT, were established to investigate the variations in gene expression under different temperature treatments using log2(FC) ≥ 1.5 and FDR ≤ 0.05 as screening conditions. A total of 1,868 genes differentially expressed under different temperatures were identified: 1137 DEGs (389 upregulated and 748 downregulated) and 864 DEGs (466 up-regulated and 398 down-regulated) were identified in MT vs HT and MT vs LT, respectively (Figures 2A, B); 133 DEGs were co-regulated under LT and HT treatments (Figure 2D). In addition, to explore the gene expression patterns of G. bailinae under varying temperatures, a trend analysis of DEGs was performed for MT vs LT, and MT vs HT. All DEGs were clustered into eight trend profiles, and 1183 DEGs were significantly enriched in four of them, i.e., 3, 1, 0, and 7 (P< 0.05), which is shown in colored blocks in Figure 2C. Three of the four significantly enriched trends, i.e., about 80% of the significantly enriched DEGs in the trend analysis, showed a decrease with increasing temperature, while only one trend showed an increase with temperature.

Figure 2 Overview of DEGs in G. bailinae exposed to different temperatures. (A) Volcano plots of the DEGs in MT vs LT. (B) Volcano plots of the DEGs in MT vs HT. (C) Trend analysis of DEGs in MT vs LT and MT vs HT. Significant enrichment trends are shown by colored blocks (P < 0.05). The bottom-left number represents the number of genes. (D) Venn diagram depicting the number of DEGs obtained in the MT vs LT and MT vs HT comparisons.
GO and KEGG analyses were conducted to examine the functions of DEGs. The GO classification results showed that DEGs were involved in three groups of functions, i.e., biological processes, molecular functions, and cellular components, and most genes were specifically classified as being involved in cellular processes (LT: 275, HT: 388), metabolic processes (LT: 241, HT: 344), and catalytic activity (LT: 200, HT: 311) both in MT vs LT and MT vs HT (Supplementary Figure S7). KEGG enrichment analysis indicated that DEGs were mainly enriched in four KEGG B classes: carbohydrate metabolism, energy metabolism, global and overview maps, and lipid metabolism (Supplementary Figure S8 and Table 1).
Based on the DEG analysis in KEGG, the following nine pathways related to the energy, carbohydrate, and lipid metabolisms were selected to investigate the specific effects of high and low temperatures on G. bailinae: photosynthesis (ko00195), photosynthesis-antenna proteins (ko00196), carbon fixation in photosynthetic organisms (ko00710), glycolysis/gluconeogenesis (ko00010), pentose phosphate pathway (PPP) (ko00030), pyruvate metabolism (ko00620), citrate cycle (TCA cycle) (ko00020), biosynthesis of unsaturated fatty acids (ko01040), and alpha-linolenic acid metabolism (ko00592).
Comparative transcriptome analysis identified 17 DEGs involved in photosynthesis, which participated in the formation of PS II (psaA, psaB, psaD, psaH), PS I (psbB, psbC, psbD, psbO, psbV), the cytochrome b6/f complex (petB), F-type ATPase (atpA, atpG), and photosynthetic electron transport (petH, petJ) (Figure 3). All these DEGs were significantly downregulated in MT vs HT, while only one significantly downregulated gene (petB) was enriched in MT vs LT. In addition, the analysis identified 10 DEGs associated with photosynthetic antenna proteins and involved in the synthesis of allophycocyanin beta subunit (apcB), phycocyanobilin lyase subunit alpha (cpcE), and light-harvesting complex I chlorophyll a/b binding proteins 1, 2, and 4 (LHCA1, LHCB2, and LHCB4). Most of these DEGs were upregulated in LT and HT treatments, and only three related to LHCB2, apcB, and cpcE were significantly downregulated in MT vs HT.
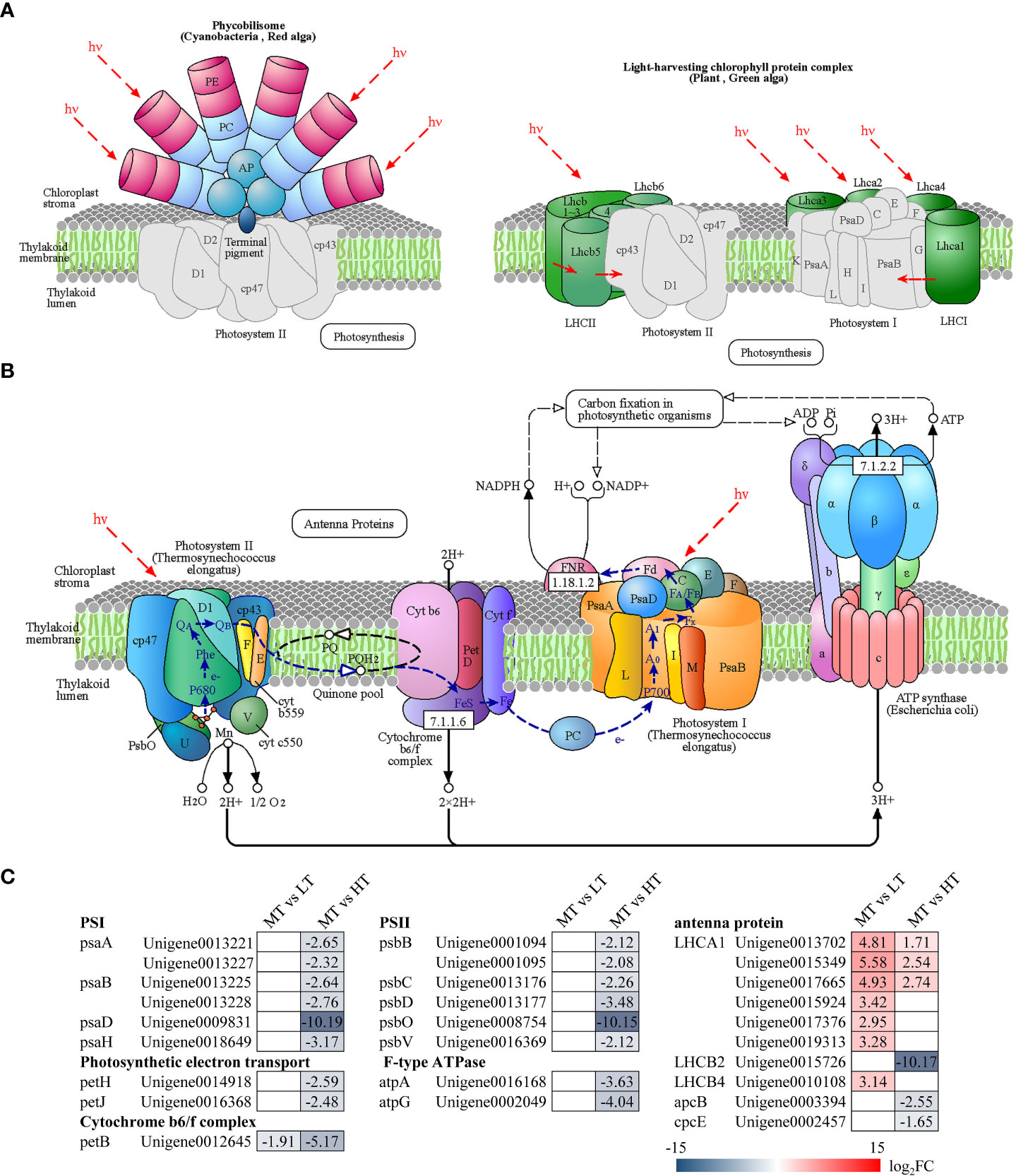
Figure 3 Pathways of photosynthesis and photosynthetic antenna proteins: (A) photosynthetic antenna protein pathway (ko00196), (B) photosynthesis pathway (ko00195), (C) DEGs involved in photosynthesis and photosynthetic antenna proteins. The number in each cell is the log2 fold change (log2FC). Red and blue gradients indicate the upregulation and downregulation of unigenes, respectively, while the white color represents log2FC< 1.5 or FDR > 0.05. The diagrams of photosynthesis and photosynthetic antenna proteins were obtained from the KEGG website.
Transcriptome analysis showed that DEGs were significantly enriched in the pathways related to carbohydrate and energy metabolisms and played a role in the response to temperature changes in G. bailinae. The NADPH and ATP produced by photosynthesis can be used for carbon dioxide fixation (Figure 3B). A total of 11 DEGs were significantly enriched in the pathway of carbon fixation in photosynthetic organisms (Calvin cycle), encoding ALDO, E3.1.3.37, rbcS, FBP, FBA, rpe, TPI, and GAPA. Most of the genes involved in this pathway were significantly downregulated under the HT treatment, while the opposite was observed under the LT treatment (Figure 4). In addition, it is worth noting that the gene encoding rbcS was significantly downregulated under both treatments, which may be associated with different temperature response mechanisms.
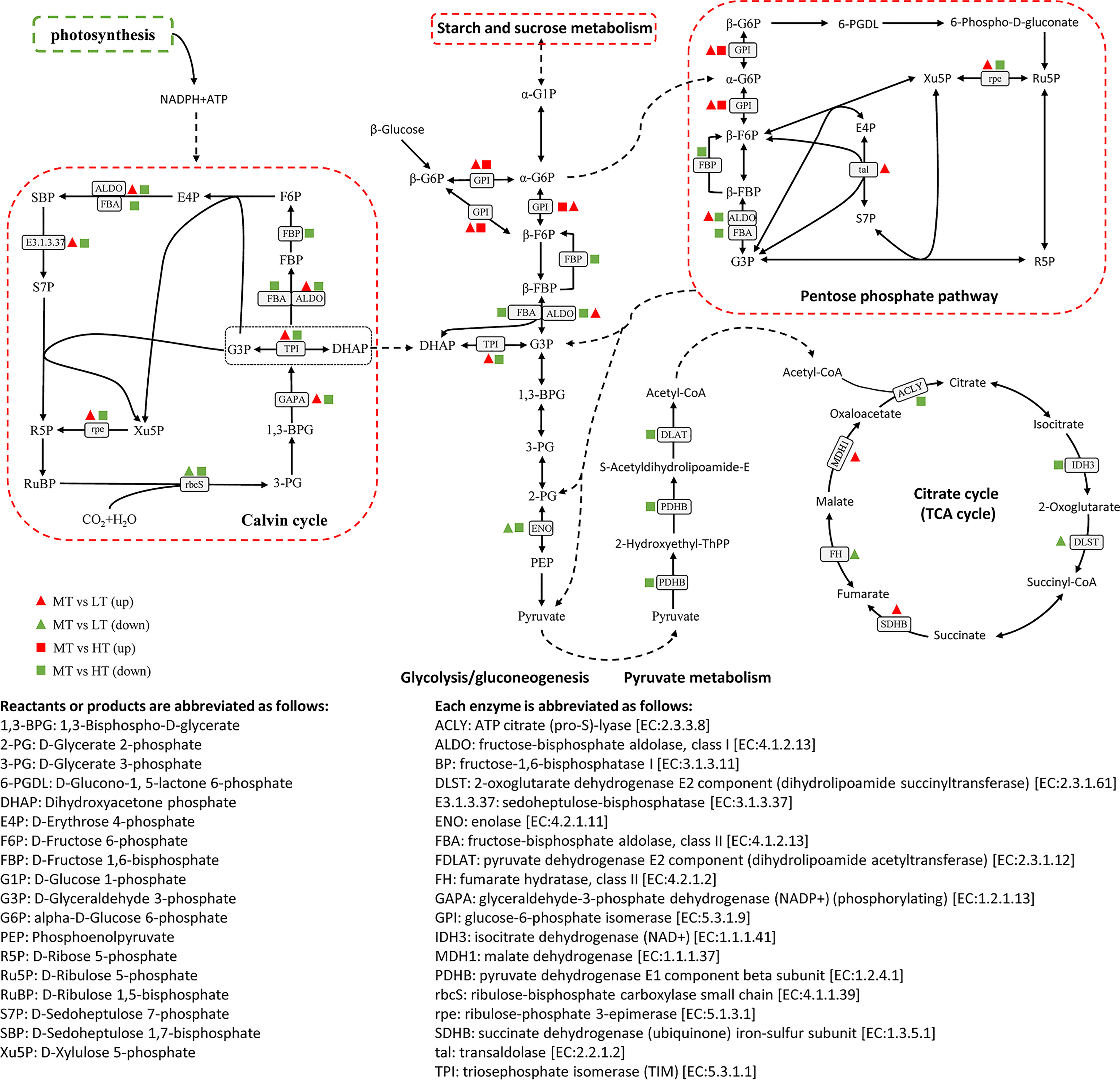
Figure 4 Pathways of carbon fixation in photosynthetic organisms, glycolysis/gluconeogenesis, pyruvate metabolism, pentose phosphate, and citrate cycle (TCA cycle). Detailed descriptions of the DEGs are included in Supplementary Table S4.
The intermediate products synthesized by the Calvin cycle, i.e., propyl phosphates (glyceraldehyde phosphate and dihydroxyacetone phosphate), can be further used in the glycolysis/glycogenesis pathway to synthesize sucrose and starch, enter the TCA cycle to produce energy, or flow to PPP to produce NADPH. In MT vs HT, 15 DEGs were significantly enriched in the above pathways, encoding ALDO, FBA, TPI, FBP, GPI, ENO, IDH3, ACLY, rpe, PDHB, and DLAT. Most of the DEGs were downregulated under the HT treatment, and only one DEG encoding GPI was upregulated (Figure 4). In MT vs LT, a total of 10 DEGs were significantly enriched in the glycolysis/gluconeogenesis, PPP, TCA cycle, and pyruvate metabolism pathways, encoding ALDO, TPI, GPI, ENO, MDH1, SDHB, DLST, FH, rpe, and tal. Specifically, the DEGs encoding ENO, DLST, and FH were significantly downregulated, while the remaining seven DEGs were significantly upregulated.
Transcriptome analysis revealed that temperature induced the differential expression of genes related to lipid metabolism, especially those involved in the biosynthesis of unsaturated fatty acids and alpha-linolenic acid metabolism. Four DEGs encoding desaturation-related enzymes (SCD) and chain extension-related enzymes (KAR) in the biosynthesis of unsaturated fatty acids were significantly differentially expressed in different comparison groups. Both SCD and KAR were upregulated under the LT treatment, while only SCD was downregulated under the HT treatment (Figure 5).
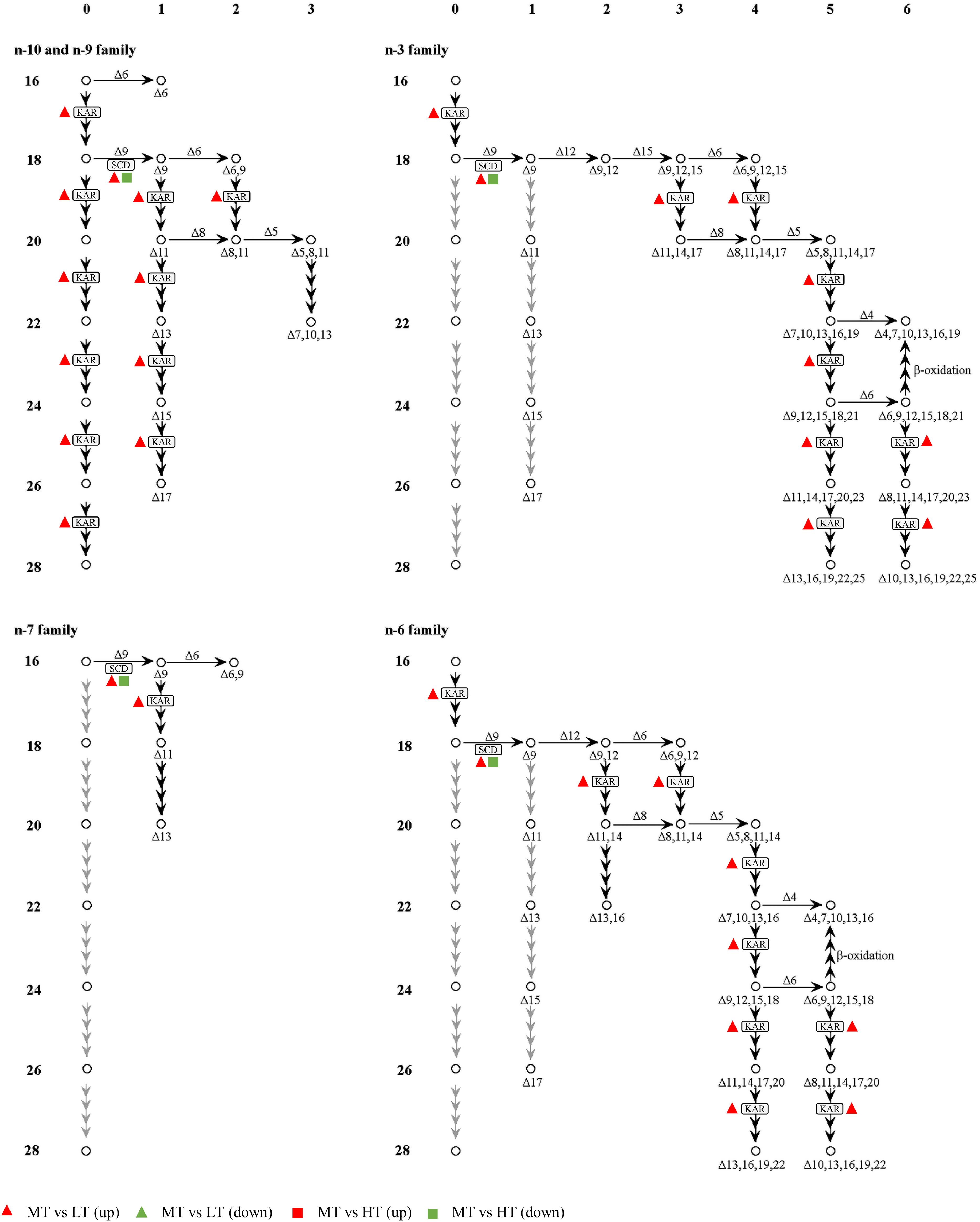
Figure 5 Pathways of biosynthesis of unsaturated fatty acids. The left side and top of the figure show the number of carbon atoms (16~28), and number of double bonds (1~6), respectively. The detailed descriptions of the DEGs are included in Supplementary Table S4.
Figure 6 shows the effect of temperature on the metabolism of alpha-linolenic acid in G. bailinae. Phosphatidylcholine in the plastid membrane is oxidized by lipase to form alpha-linolenic acid, which then undergoes a series of oxidations to form oxo-phytodienoic acid (OPDA) in the plastid. Then, OPDA enters the peroxisomes through the ABC transporter and forms (+)-7-iso-jasmonic acid after reduction and 3-step β-oxidation. In the present study, three DEGs encoding acyl-CoA oxidase (ACX) and 12-oxophytodienoic acid reductase (SCD) were significantly activated after the LT and HT treatments. Under HT, only SCD was significantly downregulated, while under LT both ACX and SCD were significantly upregulated.
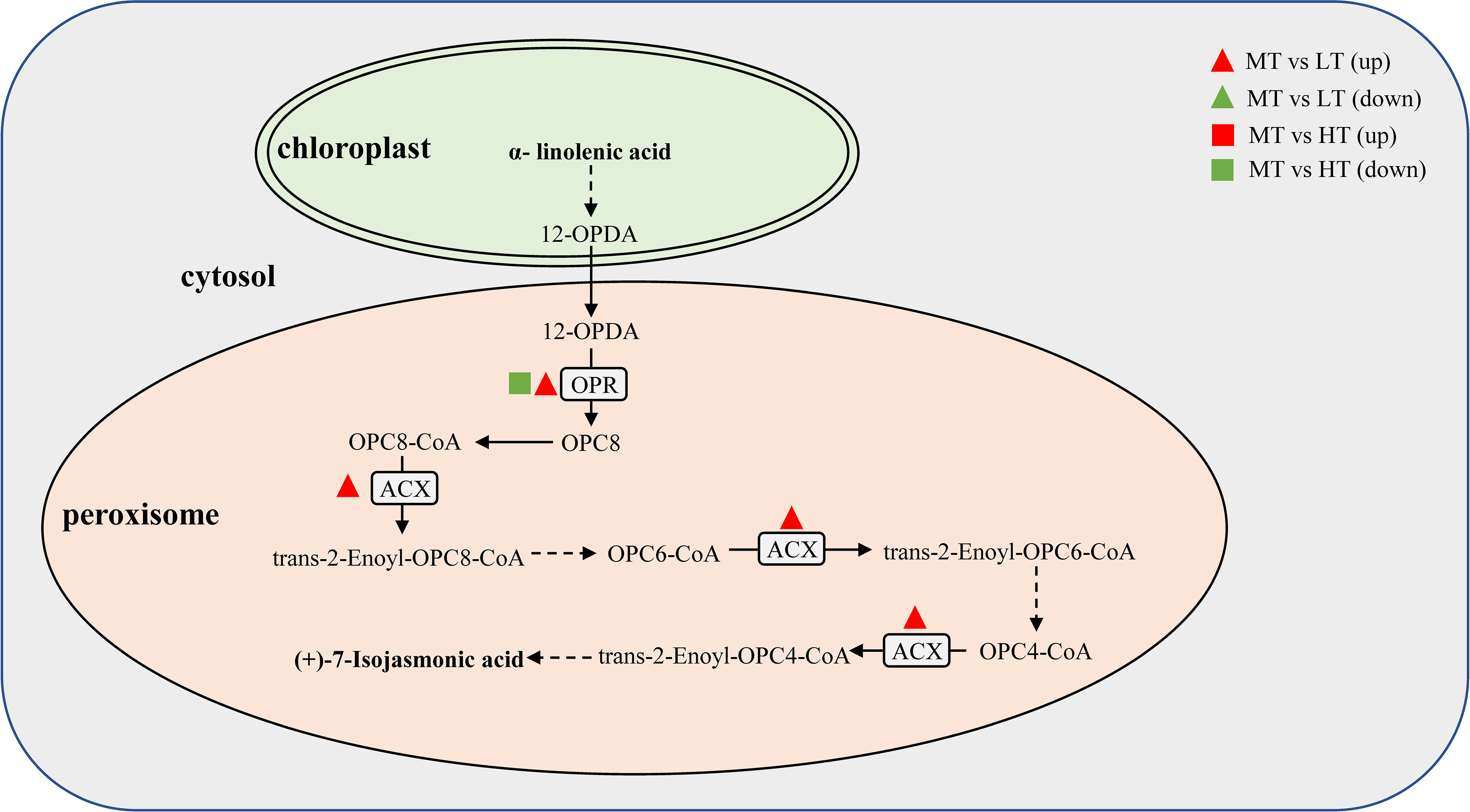
Figure 6 Pathways of the alpha-linolenic acid metabolism. The solid and dashed arrows indicate one-step and multi-step processes, respectively. The detailed descriptions of the DEGs are included in Supplementary Table S4.
As ocean warming continues to increase, there are inevitable consequences for the survival of macroalgae; however, species that can adapt to heat stress are better prepared to cope with the warmer conditions (Ji and Gao, 2021; Association, 2022). The present study found that the optimum growth state of G. bailinae was at 35°C. Based on this observation, transcriptomic analysis was first conducted to reveal the mechanism behind the adaptability of this macroalgae to high temperatures. The results of RNA-seq analysis showed that the process is associated with the regulation of the energy, carbohydrate, and lipid metabolism.
Macroalgae are expected to be especially sensitive to ocean warming since the rates of their biochemical and physiological processes are substantially regulated by ambient temperature (as observed in ectotherms) (Davison, 1991; Brown et al., 2004; Graba-Landry et al., 2018). Typically, within a species’ thermal tolerance range, the rates of physiological processes and other parameters (e.g., growth) increase with temperature until they reach a thermal optimum and rapidly decline thereafter (Brown et al., 2004; Graba-Landry et al., 2018). Different algae show different adaptations to temperature variation, and growth rate is an important indicator that visually reflects their growth status (Yang et al., 2021; Gao et al., 2022): the highest growth rate is recorded at the optimum growth temperature. For example, the highest growth rate of G. lemaneiformis (Liu et al., 2017), Ulva prolifera (Cui et al., 2015), and Caulerpa sertularioides (Zhong et al., 2021) was observed at 23°C, 20°C, and 26°C, respectively. In the present study, the RGR of G. bailinae reached its maximum at 35°C and growth was significantly inhibited under the lower temperatures tested. Fv/Fm is the maximum photochemical quantum yield of PSII, which reflects the potential maximum photosynthetic capacity of macroalgae and can also indirectly reflect their growth state and living environment (Goltsev et al., 2016; Cui et al., 2019; Zhong et al., 2021). Studies have indicated that Fv/Fm is a heat-sensitive indicator, which makes it an important tool for the evaluation of plants’ heat tolerance (Yamada et al., 1996; Wahid et al., 2007; Wu et al., 2019). For example, in G. lemaneiformis, this parameter was shown to decrease under unsuitable temperatures (Liu et al., 2017). A notable drop in Fv/Fm was also observed in Saccharina japonica under high temperature stress, which was caused by the deactivation of PSII (Shindo et al., 2022). In the present study, the analysis of Fv/Fm in G. bailinae indicated that 25°C and 35°C were suitable temperatures, but the parameter decreased significantly at 15°C. Obviously, the changes in RGR and Fv/Fm under different temperatures indicate that G. bailinae has a remarkable capacity to adapt to high temperatures.
It has long been acknowledged that plant photosynthesis is one of the most temperature-sensitive processes (Li et al., 2019). This process depends on electron transport into the photosynthetic system and it gives algae the energy they need to grow and develop (Qin et al., 2021). In the cells of red algae, light energy absorbed by the phycobillisome and the light-harvesting chlorophyll protein complex (LHC) drives the flow of electrons from H2O to NADP+, which promotes the reduction of NADP+ to NADPH (Croce and Van Amerongen, 2014). At the same time, ATP synthase uses the electrochemical gradient generated simultaneously across the thylakoid membrane for ATP synthesis (Croce and Van Amerongen, 2014). NADPH and ATP represent the basis for all subsequent chemical reactions (Croce and Van Amerongen, 2014). Algae can regulate the above processes by controlling gene expression to resist or adapt to environmental changes, that is, regulating the synthesis of NADPH and ATP. Wang et al. (2018) reported that the protein-related genes of the photosynthetic antenna in wild-type P. haitanensis were significantly downregulated under high temperature stress, impeding the absorption of sufficient light to activate photosynthesis, and therefore resulting in insufficient energy to resist heat stress (Wang et al., 2018). Another study on Amphiroa fragilissima revealed that the expression levels of several DEGs involved in photosynthesis were suppressed under high temperature stress compared to those in the control group (Yang et al., 2021). However, in the present study, the photosynthetic activity of G. bailinae remained normal despite the downregulation of photosynthesis-related genes, which was not in line with the results of the above-mentioned studies. The downregulation of genes in most heat-sensitive algae under temperature stress may lead to fatal results, but G. bailinae has a high temperature adaptability, and 35°C is its suitable growth temperature. A study on the high-temperature tolerant P. haitanensis showed that this species can reduce unnecessary energy consumption by decreasing the rate of photosynthesis and energy metabolism (Wenlei et al., 2018). A proteomic analysis similarly indicated that the P. haitanensis strain Z-61 resists high temperature stress by downregulating photosynthesis (Xu et al., 2014). Similarly, G. bailinae regulates NADPH and ATP synthesis by downregulating photosynthesis-related genes to decrease the electron transfer rate and H+ production in the photosynthetic system, thereby potentially reducing unnecessary energy consumption and ultimately adapting to high temperature environments. In addition, as observed for the compensatory reaction of gene expression (Dal Bosco et al., 2004), the high expression of LHCA1-related genes in photosynthetic antenna proteins may compensate for the decreased light capture efficiency caused by the downregulation of genes related to LHCB2, apcB, and cpcE, so as to meet the light energy demand of G. bailinae and achieve a higher growth rate.
In plants, a metabolic regulatory network is frequently engaged in the response to abiotic stimuli (Wenlei et al., 2018). Previous studies have shown that P. haitanensis under stress can reduce unnecessary energy consumption to prevent cell damage (Xu et al., 2014; Xu et al., 2016). However, when stress levels become excessive, the corresponding energy shortage in algae would result in an enhancement of the inherent pathways of carbohydrate metabolism and the induction of alternative pathways, such as glycolysis, to provide energy and the related carbon skeletons for key metabolic processes (Obata and Fernie, 2012; Wenlei et al., 2018). Wang et al. (Wang et al., 2018) corroborated this claim by studying the heat-tolerant P. haitanensis, whose genes showed a tendency to be upregulated under high temperature stress to maintain the energy and associated carbon skeleton for key metabolic processes. In the present study, a joint analysis was performed for the following five pathways related to the carbohydrate and energy metabolisms: Calvin cycle, glycolysis/gluconeogenesis, PPP, pyruvate metabolism, and TCA cycle. In our study, we found that only the DEG encoding GPI was significantly upregulated under HT treatment, while other DEGs enriched in these pathways were downregulated (Figure 4). The accumulation of sugar phosphates associated with glycolysis and the oxidative pentose phosphate pathway (OPPP) indicated that glycolytic carbon flow may be rerouted into the OPPP to produce NADPH for antioxidative action (Obata and Fernie, 2012; Wenlei et al., 2018). GPI catalyzes the interconversion between F6P and G6P. Further oxidation of G6P in the PPP can produce a large amount of NADPH, which may be one of the remedies for the decrease in NADPH synthesis during photosynthesis. The Calvin cycle, which comprises 13 chemical steps and 11 enzymes that catalyze them, is the main mechanism for carbon fixation in C3 plants (Tamoi et al., 2005). The cycle converts atmospheric CO2 into carbon skeletons that are utilized to produce starch and sucrose using the byproducts of the light reactions of photosynthesis (i.e., ATP and NADPH) (Raines, 2003). Gene downregulation during the Calvin cycle is generally thought to lead to a reduced synthesis of related enzyme proteins, especially the key protein Rubisco. However, in this study, the expression of genes related to the Rubisco small subunit rbcS was downregulated under HT treatment, but the biomass of G. bailinae was relatively high. Related research shows that Rubisco activase induced the activation of Rubisco, but the synthesis of this protein (transcription and translation) and enzyme had different sensitivities to long-term heat stress (Almeselmani et al., 2012). Therefore, due to the adaptability of G. bailinae to high temperatures, Rubisco activase may activate Rubisco more obviously under high temperature conditions, thereby improving the carbon sequestration efficiency of the protein and reducing its absolute content during the Calvin cycle. However, further studies are needed to confirm this. The TCA cycle promotes the oxidation of respiratory substrates for ATP generation, and therefore plays a crucial role in plant resistance to abiotic challenges (Sweetlove et al., 2010; Obata and Fernie, 2012; Wenlei et al., 2018). It is worth noting that the genes encoding ACLY and IDH3 were significantly downregulated in the TCA cycle. IDH3 catalyzes the conversion of isocitrate to alpha-ketoglutarate to produce NADH. The downregulation of IDH3 indicated that G. bailinae adapts to high temperatures by reducing unnecessary energy consumption. In addition, the downregulation of other genes in the Calvin cycle, glycolysis/gluconeogenesis, and pyruvate metabolism pathways also indicated that G. bailinae slows the rate of energy or carbohydrate metabolisms to adapt to high temperatures. On the contrary, under low temperature stress, due to energy shortage, this macroalgae needs to produce more energy by overexpressing genes associated with the synthesis of energy molecules, such as MDH1 and SDHB, to maintain the key life activities of cells.
In response to temperature stress, plants typically control the percentage of polyunsaturated fatty acids to alter membrane fluidity (Sun et al., 2015). Two aspects are involved in the production of unsaturated fatty acids: fatty acid peptide chain extension and desaturation (Qin et al., 2021). Fatty acids, which are the major components of membrane phospholipids, induce desaturation to promote cell membrane fluidity and stability, hence minimizing cellular damage (Qin et al., 2021). Transcriptome analysis of G. lemaneiformis revealed that the expression levels of genes associated with fatty acid extension and desaturation under normal temperatures were significantly lower than those under low temperature stress (Qin et al., 2021). Wang et al. (2018) also pointed out that genes related to unsaturated fatty acid synthesis in P. haitanensis were more significantly downregulated under normal temperatures than under high temperature stress. In this study, the genes associated with desaturation (SCD) were significantly downregulated under the HT treatment, while those associated with both fatty acid chain extension (KAR) and desaturation (SCD) were significantly upregulated under the LT treatment. This indicates that G. bailinae may regulate the membrane’s lipid fluidity to adapt to high temperatures by gradually downregulating the genes related to the biosynthesis of unsaturated fatty acids.
Alpha-linolenic acid (a C18 polyunsaturated acid) is a biosynthetic substrate for jasmonic acid (JA) and its diverse derivatives, which are lipid-derived signaling molecules involved in the regulation of numerous plant processes, including growth, reproductive development, photosynthesis, and responses to abiotic and biotic stresses (Bhatla, 2018). In plants, alpha-linolenic acid is perhaps the most prevalent and chemically diverse source of polyunsaturated fatty acids (Gerwick et al., 1991; Leung et al., 2022). JA can further form methyl jasmonate, which is considered to be crucial for algae’s ability to withstand heat (Qin et al., 2021). Exogenous methyl jasmonate has been found to reduce the negative effects of high temperature stress on a number of pathways in G. lemaneiformis (Wang et al., 2017). A transcriptome analysis of this species under low temperature stress also revealed that the activation of crucial genes associated with methyl jasmonate production was dramatically elevated (Qin et al., 2021). Su et al. (Su et al., 2019) reported that heat stress induced the upregulation of genes involved in the metabolism of alpha-linolenic acid in wheat. In the present study, it was revealed that the genes encoding OPR (the enzyme 12-oxophytodienoic acid reductase that catalyzes the conversion of 12-OPDA to OPC8) were significantly downregulated under the HT treatment. In contrast, those encoding OPR and ACX (acyl-CoA oxidase, which plays a crucial role in the three rounds of β-oxidation for the synthesis of JA acid from alpha-linolenic acid (Liu et al., 2021b)) were significantly upregulated in the LT treatment (Figure 6). These results suggest that G. bailinae can adapt to high temperatures by gradually limiting JA synthesis.
In summary, this study proved that the appropriate growth temperature for G. bailinae is 25–35°C, and the optimum is 35°C. It also revealed that this species modifies nine metabolic pathways related to the energy, carbohydrate, and lipid metabolisms to adapt to high temperatures. G. bailinae regulates the synthesis of NADPH and ATP by downregulating genes related to pathways of the energy and carbohydrate metabolisms to reduce unnecessary energy consumption. In addition, it controls the synthesis of unsaturated fatty acids and JA by downregulating the genes related to lipid metabolism to regulate the fluidity of membrane lipids and the content of lipid-derived signaling molecules, respectively. These results provide us with a better understanding of the molecular mechanisms underlying the adaptation of G. bailinae to high temperatures, which may assist in the development of technologies and breeding strategies to improve the heat tolerance of Gracilaria spp.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: BioProject, PRJNA925653.
YH, JC and EX conceived and designed this study. YH conducted the experiments, analyzed the results, and wrote the original manuscript. EX is responsible for obtaining research funding. JC revised and improved the original manuscript. SW and XC conducted a preliminary analysis of the data. JL, YG, RX and BH assisted the completion of the experiment. All authors discussed the results at various stages. All authors contributed to the article and approved the submitted version.
This research was supported by Natural Science Research Project of Guangdong Ocean University (R19049) and Chinese National Key Research and Development Program (2020YFD0901101)
We would like to express our deepest gratitude to all of those who helped us in this paper.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1125324/full#supplementary-material
Almeselmani, M., Deshmukh, P., Chinnusamy, V. (2012). Effects of prolonged high temperature stress on respiration, photosynthesis and gene expression in wheat (Triticum aestivum l.) varieties differing in their thermotolerance. Plant Stress 6, 25–32.
Ashburner, M., Ball, C. A., Blake, J. A., Botstein, D., Butler, H., Cherry, J. M., et al. (2000). Gene ontology: tool for the unification of biology. Nat. Genet. 25, 25–29. doi: 10.1038/75556
Association, W. M. (2022) ). state of the global climate 2021. Available at: https://policycommons.net/artifacts/2434625/1290_statement_2021_en/3456217/.
Barati, B., Gan, S.-Y., Lim, P.-E., Beardall, J., Phang, S.-M. (2019). Green algal molecular responses to temperature stress. Acta Physiol. Plant 41, 1–19. doi: 10.1007/s11738-019-2813-1
Bender, D., Diaz-Pulido, G., Dove, S. (2014). The impact of CO2 emission scenarios and nutrient enrichment on a common coral reef macroalga is modified by temporal effects. J. Phycol. 50, 203–215. doi: 10.1111/jpy.12153
Bhatla, S. C. (2018). “Jasmonic acid,” in Plant physiology, development and metabolism. Eds. Bhatla, S. C., Lal, M. A. (Singapore: Springer), 671–679. doi: 10.1007/978-981-13-2023-1_21
Bindoff, N. L., Cheung, W. W. L., Kairo, J. G., Arístegui, J., Guinder, V. A., Hallberg, R., et al. (2019). “2019: Changing ocean, marine ecosystems, and dependent communities,” in IPCC special report on the ocean and cryosphere in a changing climate. Eds. Pörtner, H.-O., Masson-Delmotte, D. C. R., V., Zhai, P., Tignor, M., Poloczanska, E., Mintenbeck, K., Alegría, A., XXXM. Nicolai, A. O., Petzold, J., Rama, B. (UK and New York: Cambridge), 447–587. doi: 10.1017/9781009157964.007
Brown, J. H., Gillooly, J. F., Allen, A. P., Savage, V. M., West, G. B. (2004). Toward a metabolic theory of ecology. Ecology 85, 1771–1789. doi: 10.1890/03-9000
Chen, S., Zhou, Y., Chen, Y., Gu, J. (2018). Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890. doi: 10.1093/bioinformatics/bty560
Conesa, A., Götz, S., García-Gómez, J. M., Terol, J., Talón, M., Robles, M. (2005). Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21, 3674–3676. doi: 10.1093/bioinformatics/bti610
Croce, R., Van Amerongen, H. (2014). Natural strategies for photosynthetic light harvesting. Nat. Chem. Biol. 10, 492–501. doi: 10.1038/nchembio.1555
Cui, J., Zhang, J., Huo, Y., Zhou, L., Wu, Q., Chen, L., et al. (2015). Adaptability of free-floating green tide algae in the yellow Sea to variable temperature and light intensity. Mar. pollut. Bull. 101, 660–666. doi: 10.1016/j.marpolbul.2015.10.033
Cui, J., Zhang, J., Monotilla, A. P., Huo, Y., Shi, J., Zhao, X., et al. (2019). Assessment of blooming Ulva macroalgae production potential in the yellow Sea, China. Phycologia 58, 535–541. doi: 10.1080/00318884.2018.1551016
Dal Bosco, C., Lezhneva, L., Biehl, A., Leister, D., Strotmann, H., Wanner, G., et al. (2004). Inactivation of the chloroplast ATP synthase γ subunit results in high non-photochemical fluorescence quenching and altered nuclear gene expression in arabidopsis thaliana. J. Biol. Chem. 279, 1060–1069. doi: 10.1074/jbc.M308435200
Davison, I. R. (1991). Environmental effects on algal photosynthesis: temperature. J. Phycol. 27, 2–8. doi: 10.1111/j.0022-3646.1991.00002.x
Elle, B. J., Corre, V., Jr., Felarca, K. G., Pedroso, F. (2017). Potential of Gracilariopsis bailiniae and Oreochromis mossambicus in improving water quality in intensive Litopenaeus vannamei tank culture. AACL Bioflux 10, 1309–1318.
Endoma, L. F., Nuñal, S. N., Traifalgar, R. F. M., Luhan, M. R. J. (2020). Photo-bleached agar extracts from Gracilariopsis heteroclada. Bot. Mar. 63, 559–569. doi: 10.1515/bot-2020-0028
Gao, X., Liu, W., Zhong, Y., Xing, H., Zhang, J., Chang, J., et al. (2022). Effect of different temperatures on the growth and photosynthetic physiology of Zostera marina l. Chin. J. Appl. Environ. Biol. 28, 175–181. doi: 10.19675/j.cnki.1006-687x.2020.10008
Gerwick, W. H., Moghaddam, M., Hamberg, M. (1991). Oxylipin metabolism in the red alga Gracilariopsis lemaneiformis: mechanism of formation of vicinal dihydroxy fatty acids. Arch. Biochem. Biophys. 290, 436–444. doi: 10.1016/0003-9861(91)90563-X
Goltsev, V., Kalaji, H., Paunov, M., Bąba, W., Horaczek, T., Mojski, J., et al. (2016). Variable chlorophyll fluorescence and its use for assessing physiological condition of plant photosynthetic apparatus. Russ. J. Plant Physiol. 63, 869–893. doi: 10.1134/S1021443716050058
Graba-Landry, A., Hoey, A. S., Matley, J. K., Sheppard-Brennand, H., Poore, A. G., Byrne, M., et al. (2018). Ocean warming has greater and more consistent negative effects than ocean acidification on the growth and health of subtropical macroalgae. Mar. Ecol. Prog. Ser. 595, 55–69. doi: 10.3354/meps12552
Grabherr, M. G., Haas, B. J., Yassour, M., Levin, J. Z., Thompson, D. A., Amit, I., et al. (2011). Full-length transcriptome assembly from RNA-seq data without a reference genome. Nat. Biotechnol. 29, 644–652. doi: 10.1038/nbt.1883
Hansen, J., Sato, M., Kharecha, P., Von Schuckmann, K. (2011). Earth's energy imbalance and implications. Atmos. Chem. Phys. 11, 13421–13449. doi: 10.5194/acp-11-13421-2011
Hasanuzzaman, M., Nahar, K., Alam, M. M., Roychowdhury, R., Fujita, M. (2013). Physiological, biochemical, and molecular mechanisms of heat stress tolerance in plants. Int. J. Mol. Sci. 14, 9643–9684. doi: 10.3390/ijms14059643
Ji, Y., Gao, K. (2021). Effects of climate change factors on marine macroalgae: a review. Adv. Mar. Biol. 88, 91–136. doi: 10.1016/bs.amb.2020.11.001
Koch, M., Bowes, G., Ross, C., Zhang, X. H. (2013). Climate change and ocean acidification effects on seagrasses and marine macroalgae. Global Change Biol. 19, 103–132. doi: 10.1111/j.1365-2486.2012.02791.x
Leung, K. S., Oger, C., Guy, A., Bultel-Poncé, V., Vigor, C., Durand, T., et al. (2022). Alpha-linolenic acid, phytoprostanes and phytofurans in plant, algae and food. Adv. Bot. Res. 101, 437–468. doi: 10.1016/bs.abr.2021.09.005
Li, C., Nong, Q., Solanki, M. K., Liang, Q., Xie, J., Liu, X., et al. (2016). Differential expression profiles and pathways of genes in sugarcane leaf at elongation stage in response to drought stress. Sci. Rep. 6, 1–11. doi: 10.1038/srep25698
Li, Q., Zhang, L., Pang, T., Liu, J. (2019). Comparative transcriptome profiling of Kappaphycus alvarezii (Rhodophyta, gigartinales) in response to two extreme temperature treatments: an RNA-seq-based resource for photosynthesis research. Eur. J. Phycol. 54, 162–174. doi: 10.1080/09670262.2018.1536283
Lim, E.-L., Siow, R.-S., Abdul Rahim, R., Ho, C.-L. (2016). Global transcriptome analysis of Gracilaria changii (Rhodophyta) in response to agarolytic enzyme and bacterium. Mar. Biotechnol. 18, 189–200. doi: 10.1007/s10126-015-9680-6
Liu, S., Zhang, J., Hu, C., Chen, X., Sun, X., Xu, N. (2021a). Physiological and transcriptome analysis of exogenous l-arginine in alleviation of high temperature stress in Gracilariopsis lemaneiformis. Front. Mar. Sci. 8, 784586. doi: 10.3389/fmars.2021.784586
Liu, S., Zhang, J., Hu, C., Sun, X., Xu, N. (2021b). Physiological and transcriptome analysis of γ-aminobutyric acid (GABA) in improving Gracilariopsis lemaneiformis stress tolerance at high temperatures. Algal Res. 60, 102532. doi: 10.1016/j.algal.2021.102532
Liu, C., Zou, D., Yang, Y., Chen, B., Jiang, H. (2017). Temperature responses of pigment contents, chlorophyll fluorescence characteristics, and antioxidant defenses in Gracilariopsis lemaneiformis (Gracilariales, rhodophyta) under different CO2 levels. J. Appl. Phycol. 29, 983–991. doi: 10.1007/s10811-016-0971-8
Love, M. I., Huber, W., Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 1–21. doi: 10.1186/s13059-014-0550-8
Obata, T., Fernie, A. R. (2012). The use of metabolomics to dissect plant responses to abiotic stresses. Cell. Mol. Life Sci. 69, 3225–3243. doi: 10.1007/s00018-012-1091-5
Qin, F., Zang, X., Shui, G., Wang, Z. (2021). Transcriptome analysis of Gracilariopsis lemaneiformis at low temperature. J. Appl. Phycol. 33, 4035–4050. doi: 10.1007/s10811-021-02514-z
Raines, C. A. (2003). The Calvin cycle revisited. Photosynth. Res. 75, 1–10. doi: 10.1023/A:1022421515027
Robinson, M. D., McCarthy, D. J., Smyth, G. K. (2010). edgeR: a bioconductor package for differential expression analysis of digital gene expression data. bioinformatics 26, 139–140. doi: 10.1093/bioinformatics/btp616
Shindo, A., Borlongan, I. A., Nishihara, G. N., Terada, R. (2022). Interactive effects of temperature and irradiance including spectral light quality on the photosynthesis of a brown alga Saccharina japonica (Laminariales) from Hokkaido, Japan. Algal Res. 66, 102777. doi: 10.1016/j.algal.2022.102777
Skriptsova, A. V., Nabivailo, Y. V. (2009). Comparison of three gracilarioids: growth rate, agar content and quality. J. Appl. Phycol. 21, 443–450. doi: 10.1007/s10811-008-9389-2
Smale, D. A., Wernberg, T., Oliver, E. C., Thomsen, M., Harvey, B. P., Straub, S. C., et al. (2019). Marine heatwaves threaten global biodiversity and the provision of ecosystem services. Nat. Clim. Change 9, 306–312. doi: 10.1038/s41558-019-0412-1
Su, P., Jiang, C., Qin, H., Hu, R., Feng, J., Chang, J., et al. (2019). Identification of potential genes responsible for thermotolerance in wheat under high temperature stress. Genes 10, 174. doi: 10.3390/genes10020174
Sun, P., Mao, Y., Li, G., Cao, M., Kong, F., Wang, L., et al. (2015). Comparative transcriptome profiling of Pyropia yezoensis (Ueda) MS hwang & HG choi in response to temperature stresses. BMC Genom. 16, 1–16. doi: 10.1186/s12864-015-1586-1
Sweetlove, L. J., Beard, K. F., Nunes-Nesi, A., Fernie, A. R., Ratcliffe, R. G. (2010). Not just a circle: flux modes in the plant TCA cycle. Trends Plant Sci. 15, 462–470. doi: 10.1016/j.tplants.2010.05.006
Tamoi, M., Nagaoka, M., Yabuta, Y., Shigeoka, S. (2005). Carbon metabolism in the Calvin cycle. Plant Biotechnol. 22, 355–360. doi: 10.5511/plantbiotechnology.22.355
Von Schuckmann, K., Palmer, M., Trenberth, K. E., Cazenave, A., Chambers, D., Champollion, N., et al. (2016). An imperative to monitor earth's energy imbalance. Nat. Clim. Change 6, 138–144. doi: 10.1038/nclimate2876
Wahid, A., Gelani, S., Ashraf, M., Foolad, M. R. (2007). Heat tolerance in plants: an overview. Environ. Exp. Bot. 61, 199–223. doi: 10.1016/j.envexpbot.2007.05.011
Wang, W., Li, H., Lin, X., Yang, S., Wang, Z., Fang, B. (2015). Transcriptome analysis identifies genes involved in adventitious branches formation of Gracilaria lichenoides in vitro. Sci. Rep. 5, 1–16. doi: 10.1038/srep17099
Wang, W., Lin, Y., Teng, F., Ji, D., Xu, Y., Chen, C., et al. (2018). Comparative transcriptome analysis between heat-tolerant and sensitive Pyropia haitanensis strains in response to high temperature stress. Algal Res. 29, 104–112. doi: 10.1016/j.algal.2017.11.026
Wang, F., Wang, C., Zou, T., Xu, N., Sun, X. (2017). Comparative transcriptional profiling of Gracilariopsis lemaneiformis in response to salicylic acid-and methyl jasmonate-mediated heat resistance. PloS One 12, e0176531. doi: 10.1371/journal.pone.0176531
Wenlei, W., Fei, T., Yinghui, L., Dehua, J., Yan, X., Changsheng, C., et al. (2018). Transcriptomic study to understand thermal adaptation in a high temperature-tolerant strain of Pyropia haitanensis. PloS One 13, e0195842. doi: 10.1371/journal.pone.0195842
Wernberg, T., Russell, B. D., Thomsen, M. S., Gurgel, C. F. D., Bradshaw, C. J., Poloczanska, E. S., et al. (2011). Seaweed communities in retreat from ocean warming. Curr. Biol. 21, 1828–1832. doi: 10.1016/j.cub.2011.09.028
Wu, J., Lian, W., Liu, Z., Zeng, X., Jiang, J., Wei, Y. (2019). High temperature response of chlorophyll fluorescence parameters and heat tolerance evaluation in different grape cultivars. J. Northwest A F University(Natural Sci. Edition) 47, 80–88. doi: 10.13207/j.cnki.jnwafu.2019.06.011
Xu, Y., Chen, C., Ji, D., Hang, N., Xie, C. (2014). Proteomic profile analysis of Pyropia haitanensis in response to high-temperature stress. J. Appl. Phycol. 26, 607–618. doi: 10.1007/s10811-013-0066-8
Xu, C., Xu, R., Huang, B., Chen, H., Chen, B., Xie, E. (2021). Indoor polyculture of Gracilaria bailinae and Litopenaeus vannamei with zero water exchange. J. Guangdong Ocean Univ. 41, 131–137. doi: 10.3969/j.issn.1673-9159.2021.03.017
Xu, K., Xu, Y., Ji, D., Xie, J., Chen, C., Xie, C. (2016). Proteomic analysis of the economic seaweed Pyropia haitanensis in response to desiccation. Algal Res. 19, 198–206. doi: 10.1016/j.algal.2016.08.010
Yamada, M., Hidaka, T., Fukamachi, H. (1996). Heat tolerance in leaves of tropical fruit crops as measured by chlorophyll fluorescence. Sci. Hortic. 67, 39–48. doi: 10.1016/S0304-4238(96)00931-4
Yang, F., Wei, Z., Long, L. (2021). Transcriptomic and physiological responses of the tropical reef calcified macroalga Amphiroa fragilissima to elevated temperature. J. Phycol. 57, 1254–1265. doi: 10.1111/jpy.13158
Yao, C., Somero, G. N. (2014). The impact of ocean warming on marine organisms. Chin. Sci. Bull. 59, 468–479. doi: 10.1007/s11434-014-0113-0
Yao, Y., Wang, C. (2021). Variations in summer marine heatwaves in the south China Sea. JGR: Oceans 126, e2021JC017792. doi: 10.1029/2021JC017792
Yong, Y. S., Yong, W. T. L., Anton, A. (2013). Analysis of formulae for determination of seaweed growth rate. J. Appl. Phycol. 25, 1831–1834. doi: 10.1007/s10811-013-0022-7
Zhang, X., Wu, C., Hu, C., Li, Y., Sun, X., Xu, N. (2020). Lipid remodeling associated with chitooligosaccharides-induced heat tolerance of marine macroalgae Gracilariopsis lemaneiformis. Algal Res. 52, 102113. doi: 10.1016/j.algal.2020.102113
Keywords: heat-tolerance mechanism, gene regulation, Gracilaria bailinae, macroalgae, temperature, transcriptome
Citation: Huang Y, Cui J, Wang S, Chen X, Liao J, Guo Y, Xin R, Huang B and Xie E (2023) Transcriptome analysis reveals the molecular mechanisms of adaptation to high temperatures in Gracilaria bailinae. Front. Plant Sci. 14:1125324. doi: 10.3389/fpls.2023.1125324
Received: 16 December 2022; Accepted: 28 March 2023;
Published: 14 April 2023.
Edited by:
Nianjun Teng, Nanjing Agricultural University, ChinaReviewed by:
Dilek Ünal, Bilecik Şeyh Edebali University, TürkiyeCopyright © 2023 Huang, Cui, Wang, Chen, Liao, Guo, Xin, Huang and Xie. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jianjun Cui, Y3Vpamlhbmp1bjI5QDE2My5jb20=; Enyi Xie, eGllZW55aUAxNjMuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.