
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 24 February 2023
Sec. Plant Abiotic Stress
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1124905
This article is part of the Research TopicIdentification and Functional Analysis of Differentially Expressed Genes in Plant Response to Abiotic StressesView all 24 articles
Wheat (Triticum aestivum L., 2n = 6x = 42, AABBDD) is the world’s most widely cultivated crop and an important staple food for humans, accounting for one-fifth of calories consumed. Proteins encoded by the regulator of chromosome condensation 1 (RCC1) are highly conserved among eukaryotes and consist of seven repeated domains that fold into a seven-bladed propeller structure. In this study, a total of 76 RCC1 genes of bread wheat were identified via a genome-wide search, and their phylogenetic relationship, gene structure, protein-conserved domain, chromosome localization, conserved motif, and transcription factor binding sites were systematically analyzed using the bioinformatics approach to indicate the evolutionary and functional features of these genes. The expression patterns of 76 TaRCC1 family genes in wheat under various stresses were further analyzed, and RT-PCR verified that RCC1-3A (TraesCS3A02G362800), RCC1-3B (TraesCS3B02G395200), and RCC1-3D (TraesCS3D02G35650) were significantly induced by salt, cold, and drought stresses. Additionally, the co-expression network analysis and binding site prediction suggested that Myb-7B (TraesCS7B02G188000) and Myb-7D (TraesCS7D02G295400) may bind to the promoter of RCC1-3A/3B and upregulate their expression in response to abiotic stresses in wheat. The results have furthered our understanding of the wheat RCC1 family members and will provide important information for subsequent studies and the use of RCC1 genes in wheat.
The regulator of chromosome condensation 1 (RCC1) genes encode proteins whose sequence is highly conserved among eukaryotes and consists of seven repeated domains that fold into a seven-bladed propeller structure (Renault et al., 1998). RCC1 proteins in mammals act as the guanosine nucleotide exchange factors (GEFs) for a GTPase well known as Ras-related nuclear protein (Ran) and are involved in diverse biological processes, such as spindle assembly, nuclear membrane formation, and nucleocytoplasmic transport during mitosis (Bischoff and Ponstingl, 1991; Hutchins et al., 2004; Li and Zheng, 2004; Chen et al., 2007; Terry et al., 2007). RCC1 proteins are implicated in the initiation and progression of a variety of cancers by promoting nuclear entry and accumulation of β-catenin (Brabletz et al., 2001; Polakis, 2007; Dominguez et al., 2009). Interestingly, several studies have reported that another class of fungal protein, latcripin, which contains the RCC1 domain, can effectively promote the apoptosis of cancer cells (Liu et al., 2012; Ann et al., 2014; Tian et al., 2016; Wang et al., 2016).
RCC1 family genes are also present in plants. However, a few plant RCC1 genes have only been identified successively in the last decade or so. RCC1 family proteins in plants can be divided into two major groups: single-domain proteins (containing only a single RCC1 repeat domain) and multi-domain proteins (containing other domains in addition to the RCC1 repeat domain) (Sun et al., 2022). Single-domain RCC1 family proteins have been identified in both animals and plants, while PRAF (PH, RCC1, and FYVE) proteins, a class of typical multi-domain RCC1 family proteins, are unique to plants and contain four distinctive domains: two lipid‐binding domains, including pleckstrin homology (PH) and FYVE (Fab1, YOTB, Vac 1, and EEA1) zinc‐finger domains, the RCC1 (seven repeats of the regulator of chromosome condensation 1) or alpha‐tubulin suppressor domain1 (ATS1) motif, and a C‐terminal BRX/DZC (brevis radix/disease resistance, zinc finger, chromosome condensation) domain (Sun et al., 2022). In Arabidopsis, there are 24 putative proteins containing the RCC1-like domains, but only five have been functionally studied (Brown et al., 2005; Kuhn et al., 2011; Ji et al., 2015; Su et al., 2017; Ji et al., 2019; Duarte et al., 2021). Arabidopsis UV RESISTANCE LOCUS 8 (UVR8) is the first plant RCC1 family member to be identified as the only UV receptor in plants (Kliebenstein et al., 2002; Rizzini et al., 2011; Christie et al., 2012; Wu et al., 2012; Jenkins, 2017). Upon absorbing UV-B radiation, UVR8 immediately switches from homodimer to monomer and then accumulates in the nucleus through interaction with constitutive photomorphogenic 1 (COP1), triggering a UV-B cascade, thus regulating the expression of downstream genes and plant responses to UV-B (Brown et al., 2005; Kaiserli and Jenkins, 2007; Favory et al., 2009; Rizzini et al., 2011; Yin et al., 2016). RCC1/UVR8/GEF-like 3 (RUG3), another RCC1 family protein, interacts with ataxia–telangiectasia mutant (ATM) protein in mitochondria to synergistically regulate the splicing of nad2 mRNA and its complex function, which is necessary for reactive oxygen species homeostasis and plant development (Kuhn et al., 2011; Su et al., 2017). The third characteristic protein of the RCC1 family protein in Arabidopsis is tolerant to chilling and freezing 1 (TCF1), which is located in the nuclear genome and regulates plant cold adaptation and tolerance through a chromatin-based regulation mechanism (Ji et al., 2015). Under cold stress, TCF1 is upregulated rapidly and affects the expression of the blue copper-binding protein (BCB), which regulates lignin biosynthesis and subsequent cell wall remodeling (Ji et al., 2015). Another RCC1 family protein, sensitive to ABA 1 (SAB1), can bind to the promoter of abscisic acid-insensitive 5 (ABI5) and inhibit its expression by increasing the level of histone H3K27me2 in the ABI5 promoter, thus negatively regulating the seed germination process (Ji et al., 2019). Recent research focused on RCC1 genes in Arabidopsis revealed that another RCC1 family protein, PROTON1, regulates rosette leaf growth in response to nitrogen availability (Duarte et al., 2021). In addition to Arabidopsis, a number of RCC1 genes have successively been identified in other plants. In cotton, two RCC1 family genes showed crucial roles in salt tolerance (Liu et al., 2019). GmTCF1a responds specifically to cold stress and positively regulates cold tolerance in soybean (Dong et al., 2021). In maize, the RCC1 family protein Dek47 can influence the assembly of the mitochondrial complex and maize seed development by regulating the splicing of the nad2 transcript (Cao et al., 2021). SaRCC1, an RCC1 family protein in Spartina alterniflora, was found to negatively regulate salt tolerance in plants by using a heterologous expression assay in Arabidopsis (Li et al., 2022). In Medicago truncatula, PRAF protein MtZR1 (belonging to the multi-domain RCC1 family proteins) is a cytomembrane‐ and nuclear‐located protein that plays a key role in root development and symbiotic root nodules (Hopkins et al., 2014). In rice, another PRAF family protein, OsRLR4, alters OsAUX1 promoter histone H3K4me3 levels by recruiting the histone methyltransferase OsTrx1, which promotes OsAUX1 expression, alters auxin accumulation in root tips, and ultimately affects the root apical meristem (RAM) activity (Sun et al., 2022).
Wheat (Triticum aestivum L., 2n = 6x = 42, AABBDD) is the world’s most widely cultivated crop and an important staple food for humans, accounting for one-fifth of calories consumed (International Wheat Genome Sequencing, C 2018). No study on the RCC1 domain proteins in wheat has been reported, mainly because of the later release of the genome than in other species. Fortunately, with the release of the high-quality reference genome and annotation of the Chinese Spring (CS, a bread wheat cultivar from China) by the Wheat Genome Sequencing Consortium (IWGSC) (International Wheat Genome Sequencing, C 2018), rapid and systematic methods for understanding wheat genomics and genetics have been rapidly developed. In the present study, a total of 76 RCC1 genes of bread wheat were firstly identified with a genome-wide scan on the latest released wheat genome, and then a systematical analysis, including the gene phylogenetic relationship, gene structure, protein-conserved domain, chromosome localization, conserved motif, and transcription factor binding sites, was performed for these genes to indicate their evolutionary and functional features. The tissue-specific and stress-induced expression of these genes was also examined using public RNA-seq data and real-time quantitative PCR (qRT-PCR). The results have furthered our understanding of the wheat RCC1 family members and will provide important information for subsequent studies and use of RCC1 genes in wheat.
The hidden Markov model (HMM) profile of the RCC1 gene family (PF00415) in PFAM (http://pfam.xfam.org/) was downloaded and used to identify the RCC1 genes in the local protein database of bread wheat, emmer wheat (Triticum dicoccoides, 2n = 4x = 28, AABB) and Aegilops tauschii (2n = 2x = 14, DD) (downloaded from Ensembl Plants, http://plants.ensembl.org/index.html) with the hmmsearch tool of HMMER3.1 software (HMMER 3.1; http://hmmer.org/). To avoid missing RCC1 family members, an aligned file of a high-quality protein set (E value < 1 × 10−20) in MEGA X software (Kumar et al., 2018) was used to reconstruct the new HMM profile, which was used as the query to search all the RCC1 members (E value < 0.01) in all bread wheat, emmer wheat, and Aegilops tauschii proteins, respectively. All the detected protein sequences were submitted to the PFAM (http://pfam.xfam.org/), SMART domain search (http://smart.embl.de/smart/batch.pl) (Letunic et al., 2021), and NCBI Batch CD-search database (https://www.ncbi.nlm.nih.gov/Structure/bwrpsb/bwrpsb.cgi) to confirm the structural integrity of the RCC1 domain (Marchler-Bauer et al., 2005). The non-redundant, verified genes encoding proteins with RCC1 domains were assigned as members of the RCC1 gene family.
Multiple alignments of the conserved RCC1 protein sequences of bread wheat, emmer wheat, and Aegilops tauschii were performed using Clustal Omega (Sievers et al., 2011) using default parameters, and a phylogenetic tree was constructed using a maximum-likelihood method with 1,000 bootstrap replications in the RaxML_NG software (Kozlov et al., 2019). Figtree 1.4.4 (http://tree.bio.ed.ac.uk/software/figtree/) was used to visualize and optimize the phylogenetic tree.
The RCC1 gene loci of wheat and its related genome donors were extracted from the corresponding annotated gff3 file (downloaded from Ensembl Plants, http://plants.ensembl.org/index.html) using a perl script. The Multiple Collinearity Scan toolkit (MCScanX) was used to analyze the gene collinearity among wheat, emmer wheat, and Aegilops tauschii with the default parameters (Kozlov et al., 2019).
Homolog analysis of RCC1 genes among the A, B, and D genomes of wheat was performed based on the aligned result. The chromosomal distribution and collinearity of RCC1 genes among the wheat and its donors and of the homoeologous RCC1 genes among A, B, and D genomes were visualized by the circle package in R (Gu et al., 2014).
Clustal Omega (Sievers et al., 2011) was used to analyze RCC1 protein sequences of wheat, and RaxML_NG (Kozlov et al., 2019) was used to construct a phylogenetic tree via a maximum-likelihood method with 1000 bootstrap replications. The domains of the RCC1 gene family in wheat were verified by the SMART domain search (http://smart.embl.de/smart/batch.pl) (Letunic et al., 2021) and the NCBI Batch CD-search database (https://www.ncbi.nlm.nih.gov/Structure/bwrpsb/bwrpsb.cgi) (Marchler-Bauer et al., 2005). The conserved motifs of the RCC1 gene family in wheat were determined by the online Multiple Em for Motif Elicitation (MEME) suite program (http://meme-suite.org) (Bailey et al., 2006). The software TBtools (Chen et al., 2020) was used to visualize the gene structure, protein domains, and motifs of the RCC1 genes according to the annotated GFF files, the genome sequence of wheat, and the protein domain file from the SMART domain search database, as well as the motif result files from the MEME suite.
The promoter sequences (2-kb upstream) of the TaRCC1 genes were extracted from the wheat reference genome (IWGSC RefSeq v1.1, International Wheat Genome Sequencing, C 2018) using the GTF/GFF3 Sequences Extract function of TBtools (Chen et al., 2020), and their potential cis-acting elements were predicted by submitting to PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/).
To analyze the tissue-specific and stress-induced expression of TaRCC1 genes, the RNA-seq expression data of five publicly available studies (International Wheat Genome Sequencing, C 2014; Zhang et al., 2014; Li et al., 2015; Liu et al., 2015; Zhang et al., 2016) were obtained from expVIP Wheat Expression Browser (http://www.wheat-expression.com/) (Ramirez-Gonzalez et al., 2018) and Triticeae Multi-omics Center (http://202.194.139.32/expression/index.html) and then visualized using the pheatmap package of R software.
A KnetMiner web application for wheat (https://knetminer.com/Triticum_aestivum/, HassaniPak and Keywan, 2017) was used to search gene-evidence networks extracted from the knowledge network and predict the transcription factors for the three TaRCC1 genes. The expression pattern of the predicted transcription factors was detected by qRT-PCR.
The full-length coding DNA sequences (CDS) of RCC1-3A (TraesCS3A02G362800), RCC1-3B (TraesCS3B02G395200), RCC1-3D (TraesCS3D02G356500), Myb-7B (TraesCS7B02G188000), and Myb-7D (TraesCS7D02G295400) were inserted into pCambia1300-35S-GFP, creating RCC1-3A::GFP, RCC1-3B::GFP, RCC1-3D::GFP, Myb-7B::GFP, and Myb-7D::GFP fusion vectors. The recombinant plasmids were mixed with the nuclear marker NLS-mCherry and transfected into wheat mesophyll protoplasts as previously described by Yoo et al. (2007). The transfection mixture was induced by PEG-Ca2+, and the protoplasts were cultured for 12 h at 25°C. The protoplasts were observed and photographed with a fluorescence microscope (Zeiss Imager A2, Germany).
The bread wheat cultivar, CS, was grown in a greenhouse with controlled conditions of 26°C/14 h light and 20°C/10 h dark. Three different treatments were applied, namely salt stress, cold, and drought stress induced by polyethylene glycol (PEG). During the two-leaf stage, seedlings were treated with Hoagland liquid medium containing 200 mM NaCl for 1, 3, and 6 h (salt stress), 4°C for 1, 3, and 6 h (cold stress), and 20% PEG4000 for 1, 3, and 6 h (drought stress). Seedlings grown in a normal environment without treatment were set as the control. Three biological replicates were set for all the trials.
The total RNA of plant materials was extracted using the RNAprep Pure Plant Kit (polysaccharide- and polyphenolic-rich) (TIANGEN, Beijing, China), following the manufacturer’s instructions. A NanoDrop One spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA) and agarose gel electrophoresis were used to assess RNA quantity and purity. Complementary DNA (cDNA) was synthesized using Reverse Transcriptase M-MLV (Takara, Beijing, China) according to the manufacturer’s instructions. qRT-PCR was performed using a 7300 Real-Time PCR System (Applied Biosystems, Foster City, CA, USA) according to the supplier’s instructions. A total of 6 L of DNase/RNase-free water, 11 μl of TB Green Real-Time PCR master mix, 2 μl of diluted cDNA product, and 1 μl of gene-specific primer was added to each reaction mixture. Three biological replicates were used for each tissue and three technical repeats for each biological replicate. The thermal cycle was set as follows: denaturing at 95°C for 30 s, then denaturing at 95°C for 15 s, and annealing and elongating at 58°C for 30 s with 45 cycles. The GAPDH gene was used as an internal reference for the normalization of the expression of the TaRCC1 genes. The relative expression levels were calculated using the 2−ΔΔCt method.
To identify RCC1 genes in bread wheat, emmer wheat, and Aegilops tauschii, a genome-wide search was performed by local BLASTP using HMM profiles. In total, 149 RCC1 genes, comprising 76 TaRCC1s, 49 TdRCC1s, and 24 AetRCC1, were identified and verified by detecting the RCC1-conserved domain via the Pfam (http://pfam.xfam.org/), SMART domain search (http://smart.embl.de/smart/batch.pl), and NCBI Batch CD-search database (https://www.ncbi.nlm.nih.gov/Structure/bwrpsb/bwrpsb.cgi). The details of the identified RCC1 genes are listed in Supplementary Table S1. The distribution of the RCC1 genes on chromosomes, different homoeologous groups, and sub-genomes was determined (Figure 1). Emmer wheat (AABB) and Aegilops tauschii (DD) are two genome donors of bread wheat; thus, the RCC1 genes of emmer wheat and Aegilops tauschii were integrated together to compare with bread wheat. The number of RCC1 genes on chromosomes, homoeologous groups, and sub-genomes showed little difference between bread wheat and the combined data for emmer wheat and Aegilops tauschii. Most RCC1 genes were located in homoeologous groups 1, 2, and 3, while no RCC1s were detected in homoeologous group 4. Two RCC1 genes (TraesCSU02G009000LC and TraesCSU02G009100LC) on ChrUn of bread wheat were certificated to belong to Chr1B through phylogenetic and synteny analysis with the RCC1s of emmer wheat and Aegilops tauschii as follows.
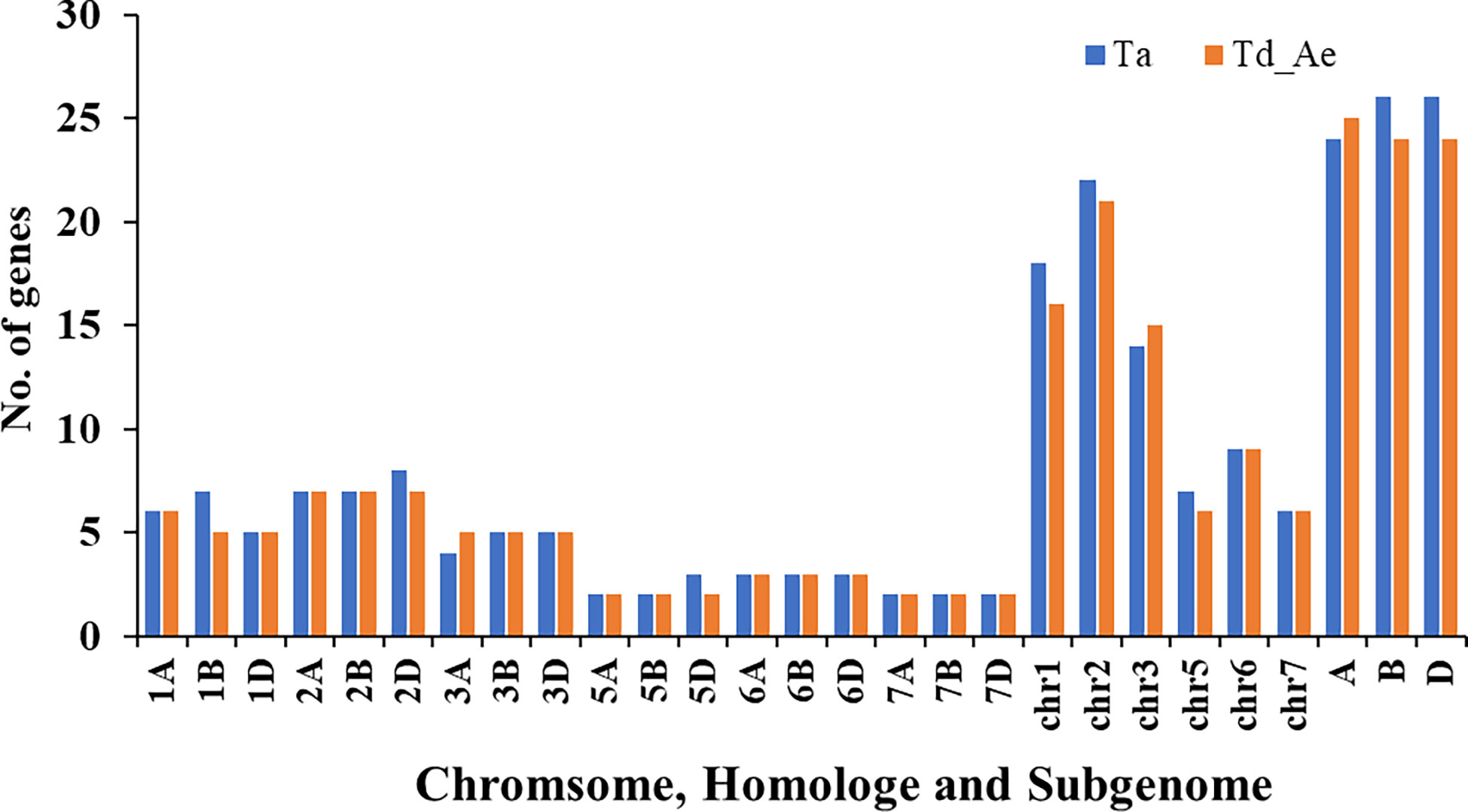
Figure 1 Distribution of RCC1s on chromosomes, different homoeologous groups, and sub-genomes in bread wheat, emmer wheat, and Aegilops tauschii.
To investigate the phylogenetic relationships and compare the evolutionary relationships of RCC1 genes among bread wheat, emmer wheat, and Aegilops tauschii, a maximum-likelihood phylogenetic tree was constructed using the protein sequences of RCC1s (Figure 2). The best-fit model to construct the tree was LG+FC+G8m, and the RCC1s were classified into four subfamilies (sub. I–IV) and named RCC1 I–IV. The RCC1 I, II, III, and IV subfamilies contained 16 (nine for wheat, five for emmer wheat, and two for Aegilops tauschii), 55 (28 for wheat, 18 for emmer wheat and nine for Aegilops tauschii), 24 (12 for wheat, eight for emmer wheat and four for Aegilops tauschii), and 54 RCC1 genes (27, 18, and nine), respectively. Interestingly, in each subgroup, the number of RCC1 genes from bread wheat (AABBDD), emmer wheat (AABB), and Aegilops tauschii (DD) showed approximately a 3:2:1 ratio, which indicated that the RCC1 gene is evolutionarily conserved across the three species.
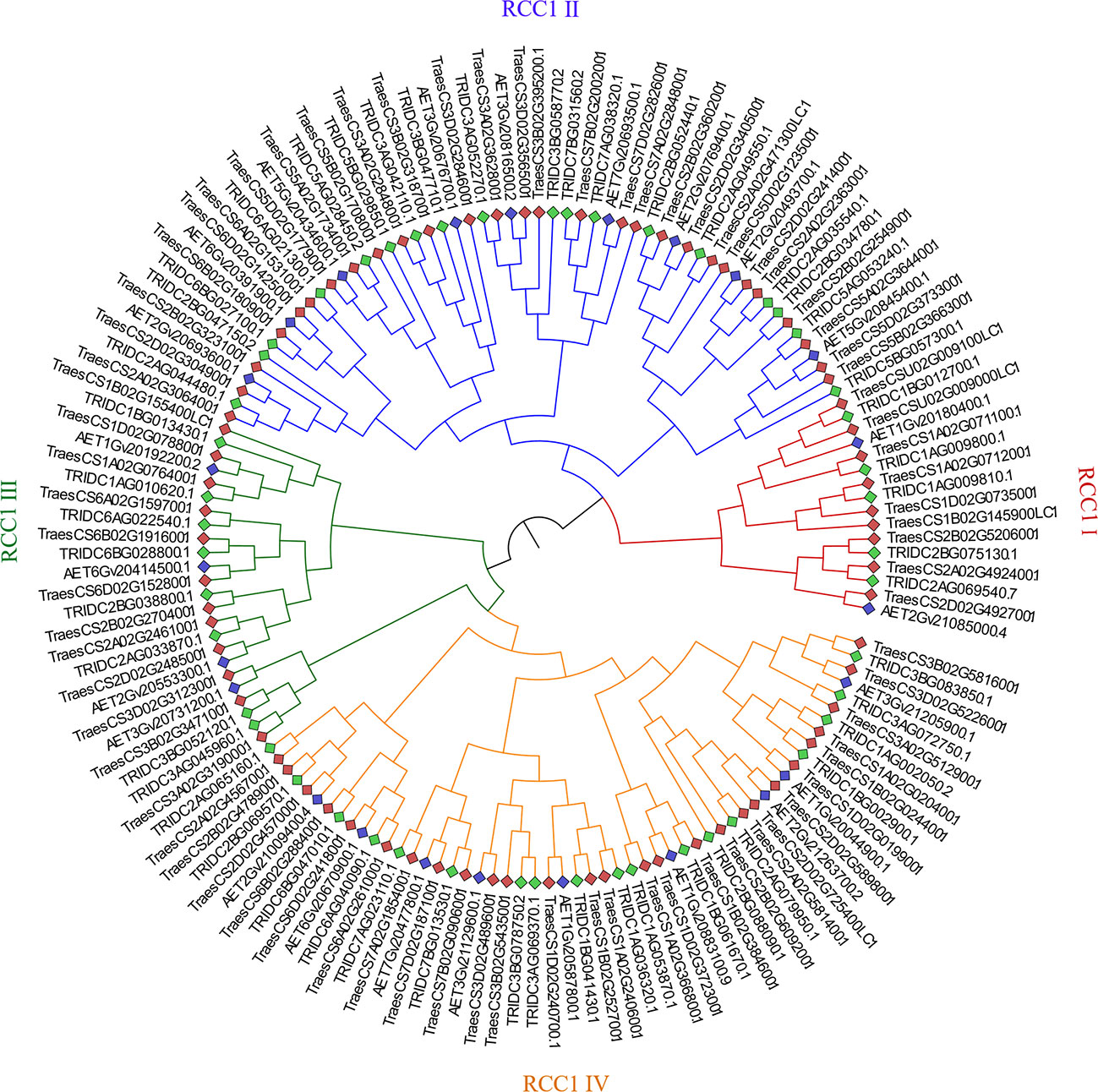
Figure 2 Phylogenetic tree of the RCC1 protein sequences of bread wheat, emmer wheat, and Aegilops tauschii. The phylogenetic tree was built using the maximum-likelihood method in the RaxML_NG web server with 1,000 bootstrap replications.
There are 76 wheat TaRCC1 genes mapped to 18 of the 21 wheat chromosomes, except 4A, 4B, and 4D, according to available annotation information of the wheat genome. Synteny analysis showed that most TdRCC1s (except TRIDC3AG069370.1) and all the AeRCC1s were highly collinear with the TaRCC1s, and phylogenetic analysis indicated the collinear RCC1 genes of the three species were clustered together (Figures 2, 3; Supplementary Table S2). Homologous gene analysis indicated that, except for TraesCS5D02G123500.1, most TaRCC1 genes were homoeologous to each other among the A, B, and D genomes, and clustered together (Figures 2, 3; Supplementary Table S3). It was noteworthy that two unanchored genes, TraesCSU02G009100LC.1 and TraesCSU02G009000LC.1 on ChrUn, should be anchored on chromosome 1B for their high collinearity to TraesCS1D02G073500.1 and TraesCS1A02G071100.1; therefore, we adjusted the positions of TraesCSU02G009100LC.1 and TraesCSU02G009000LC.1 on chromosome 1B for the visualization of collinearity analysis (Figure 3). According to the descriptions, a chromosomal region within 200 kb containing two or more genes is defined as a tandem duplication event. The gene pairs TraesCS1A02G071100.1/TraesCS1A02G071200.1 and TraesCSU02G009000LC.1/TraesCSU02G009100LC.1 were each clustered into one tandem duplication event region on chromosomes 1A and 1B of bread wheat, respectively; moreover, no homoeologous gene on chromosome 1A was found for the two homoeologous genes TraesCS3B02G543500.1 and TraesCS3D02G489600.1. In general, the TaRCC1s, TdRCC1s, and AeRCC1s on corresponding chromosomes show high collinear with each other according to the synteny analysis (Figure 3), and the number of RCC1 genes on corresponding chromosomes, homoeologous groups, and sub-genomes in bread wheat, emmer wheat, and Aegilops tauschii showed approximately a 1:1 ratio, respectively (Figure 1), which indicated that RCC1 genes were highly conserved during the evolution of wheat, and the expansion of RCC1 gene family was mostly due to the genome polyploidization.
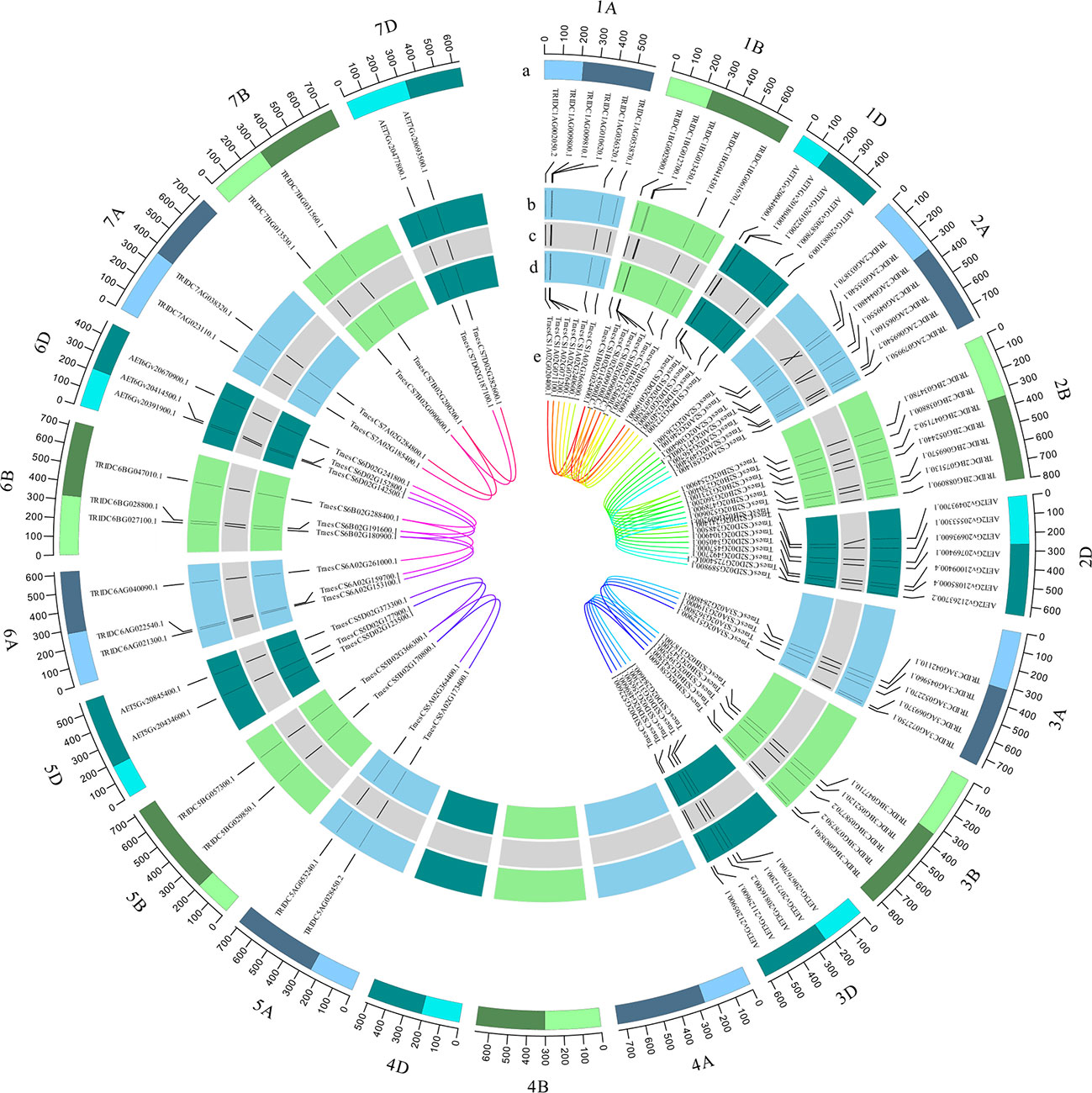
Figure 3 Genomic distribution of TaRCC1 genes and gene homology analysis in wheat. The tracks toward the center of the circle display (a) the chromosome name and size of wheat (100-Mb tick size; three different colors refer to sub-genomes (a, b, d); the light and dark bars indicate the short and long chromosome arms, respectively); (b) the distribution of TdRCC1 and AeRCC1 on chromosomes (the relative positions were adjusted according to the length of the corresponding wheat chromosome; A and B genomes were from emmer wheat; the D genome was from Aegilops tauschii). (c) Collinearity of TaRCC1, TdRCC1, and AeRCC1. (d) Genomic distribution of TaRCC1 genes in the wheat genome. (e) Homoeologous genes among A, B, and D sub-genomes.
To further estimate the gene structure, protein-conserved domains, and motifs of wheat TaRCC1 genes, the full-length protein sequences of 76 TaRCC1s were aligned using Clustal Omega, and the phylogenetic tree was constructed using RaxML_NG (Figure 4A). The TaRCC1s in wheat were classified into four subfamilies named TaRCC1 I–IV. The TaRCC1 I, II, and III subfamilies contained 10, 27, and 12 genes, respectively, and carried only RCC1 domain repeats, while the remaining 27 TaRCC1s of the TaRCC1 IV subfamily contained multiple domains, including RCC1 domain repeats and PH or BRX domains (Figure 4C). The conserved motifs of the TaRCC1 genes were determined by the online MEME suite program (http://meme-suite.org): 20 conserved motifs with lengths from 11 to 41 amino acids were detected among the TaRCC1 genes (Supplementary Table S4; Figure 4B). TaRCC1s in the same cluster shared similar conserved motif compositions, which again indicated that there is high conservation of the RCC1 gene family sequence in wheat. Despite the similarity of motifs among closely related genes, the size of the gene fragments varied widely (390–23,462 bp), such that the TraesCS2D02G725400LC gene fragment was much smaller than TraesCS7B02G200200. The gene structure, including the size and number of intron–exon, varies a lot among different TaRCC1s (such as the number of exons is from 1 to 17) (Figure 4D). It is worth noting that the closely related members, especially homologous genes, showed similar exon–intron structure, and the difference among them was the exon–intron length. The homologous genes TraesCS7A02G284800, TraesCS7B02G200200, and TraesCS7D02G282600 were similar in motif, protein domains, and exon–intron structure, while their exon–intron lengths varied greatly (10,997 bp, 23,461bp and 14,257 bp, respectively) (Figure 4; Supplementary Table S1).
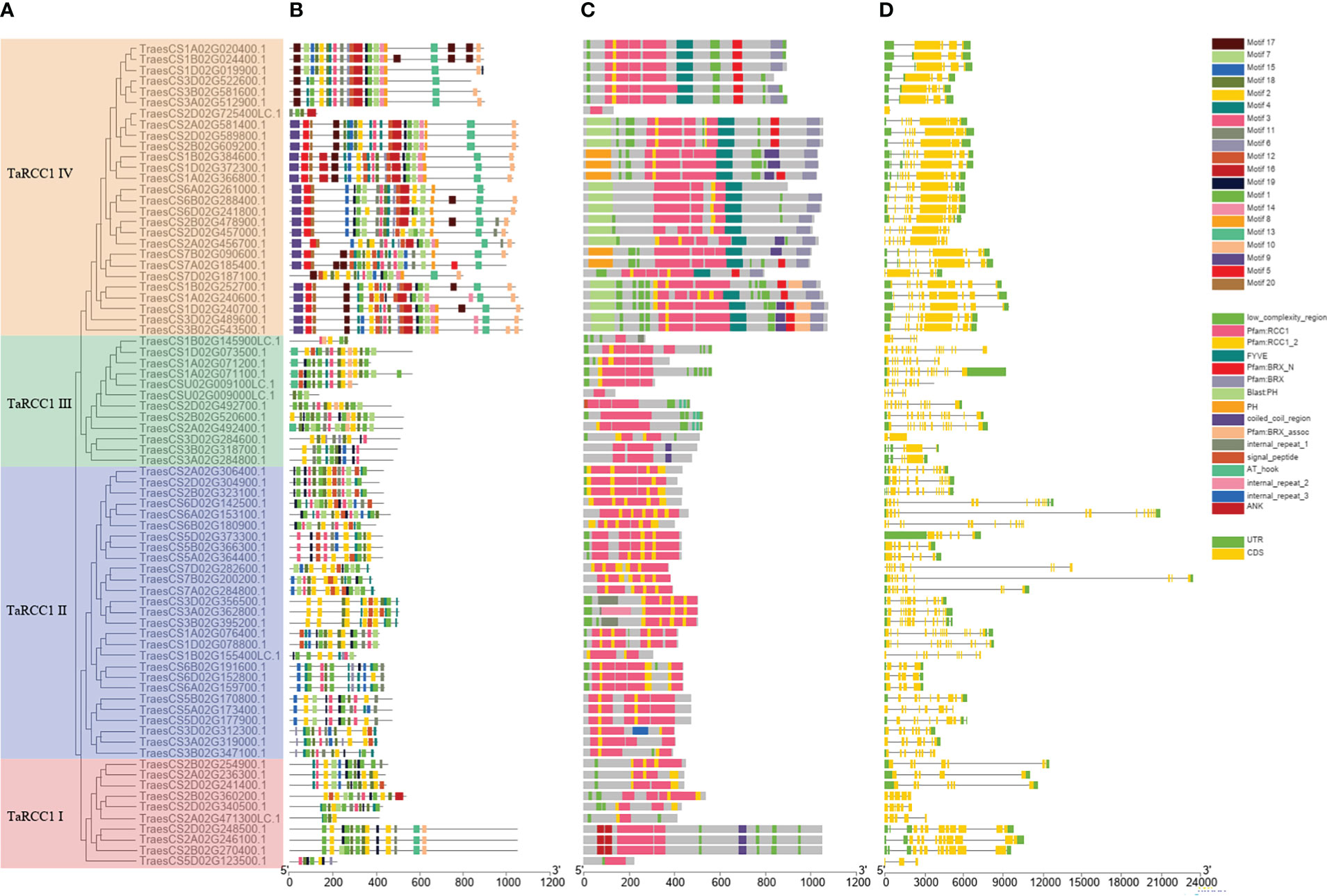
Figure 4 Phylogenetic relationship, conserved motifs, protein-conserved domains, and gene structure analysis of TaRCC1 genes. (A) Phylogenetic tree of 76 TaRCC1 proteins. (B) Conserved motifs of TaRCC1 proteins. (C) Conserved domains of TaRCC1 proteins; different domains are marked with different colors. (D) Exon–intron structures of TaRCC1 genes: exons are represented by orange boxes, introns are represented by black lines, and the upstream/downstream regions are represented by green boxes.
Cis-acting elements in gene promoters are crucial regions for initiating transcription at transcription factor-binding sites, which play an important role in regulating gene expression. The potential cis-acting elements on the promoter regions (2 kb upstream) of TaRCC1s were analyzed by PlantCARE to further explore their possible biological functions (details in Supplementary Table S5). Various potential cis-acting regulatory elements in the promoter regions of TaRCC1 genes were predicted to be related to transcription, cell cycle, development, hormones, and response to stresses (Figure 5; Supplementary Table S5). All of the TaRCC1 genes contained light-responsive elements. A total of 70 and 74TaRCC1s were detected with MeJA-responsive elements and ABA-responsive elements (ABRE), respectively. In addition, many elements were predicted to be involved in various abiotic stresses, such as drought, salt, cold, and light (Figure 5; Supplementary Figure S1; Supplementary Table S5).
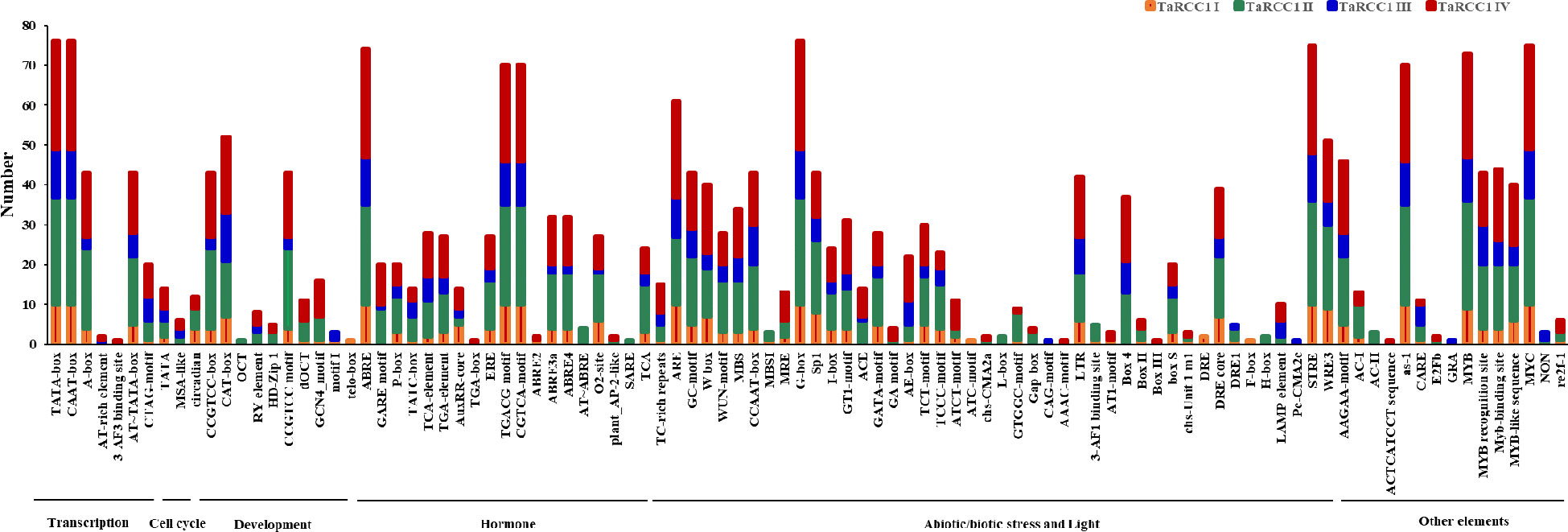
Figure 5 Cis-acting regulatory elements of TaRCC1 genes. The graph was generated using cis-acting element names and functions of TaRCC1 genes; four different subfamilies are represented by different colors.
Using the available RNA-seq database of (International Wheat Genome Sequencing, C 2014) obtained from the expVIP Wheat Expression Browser (http://www.wheat-expression.com/), the temporal and spatial expression patterns of 76 TaRCC1 genes in five different tissues (root, stem, leaf, spike, and grain) (Supplementary Table S6) were visualized using the heatmap package of R software (Supplementary Figure S2). The expression levels of TaRCC1s varied significantly among different tissues. Some TaRCC1s from the same group showed similar expression patterns, while others indicated diverse expression patterns. For example, TraesCS2B02G323100, TraesCS2A02G306400, and TraesCS2D02G304900 from TaRCC1 II were predominantly expressed in the root and stem, while the TraesCS3A02G362800, TraesCS3B02G395200, and TraesCS3D02G356500 from TaRCC1 II were most strongly expressed in the leaf, followed by the spike, root, and early stage of the stem. Similar expression patterns were observed for most homoeologous genes, although others presented diverse patterns. For instance, TraesCS6A02G153100 and TraesCS6D02G142500 were predominantly expressed in the root and stem, while the homoeologous gene TraesCS6B02G180900 presented very low expression in the five tissues. It was worth noting that, in the TaRCC1 I family, a total of four TaRCC1 genes exhibited no expression in the five tissues.
The available RNA-seq data from four studies (Zhang et al., 2014; Li et al., 2015; Liu et al., 2015; Zhang et al., 2016), obtained from the expVIP Wheat Expression Browser (http://www.wheat-expression.com/) and Triticeae Multi-omics Center (http://202.194.139.32/expression/index.html), were used to study the expression of wheat RCC1s in response to salt, drought, heat, cold, and stripe rust stresses. The transcript-per-million-read (TPM) values of TaRCC1 genes are presented in Supplementary Table S7; values were transformed by log2(x+1) and used for visualization with the pheatmap package of R software. The expression patterns of TaRCC1 genes varied a lot under different stresses (Figure 6A). Although some homoeologous genes presented diverse expression patterns, most of them exhibited similar expression patterns. The homoeologous genes TraesCS2A02G306400, TraesCS2B02G323100, and TraesCS2D02G304900 showed similar high expression trends under the four stresses and no significant differential expression under the different stresses, except for salt stress, indicating that these three genes might be induced by salt stress. The gene TraesCS2A02G456700 exhibited lower expression only under salt stress, while the homoeologous genes TraesCS2B02G478900 and TraesCS2D02G457000 presented differential expression, especially under cold stress, suggesting these two genes might participate in the cold tolerance of wheat. The homoeologous genes TraesCS3A02G362800, TraesCS3B02G395200, and TraesCS3D02G356500 showed similar and significant differential expression under the five different stresses. Under salt treatments at 6, 12, 24, and 48 h, the expression level of the three genes were higher than in the control; with drought and heat treatments, the three genes showed significantly higher expression under drought and/or heat treatments for 1 h, then decreasing after treatment for 6 h. Similar trends were detected under cold (4°C) and stripe rust pathogen stresses (Figure 6B). The results indicated that these three genes might be the key genes that participated in the early stress responses of wheat under stress, and might alleviate the stress injury of plants.
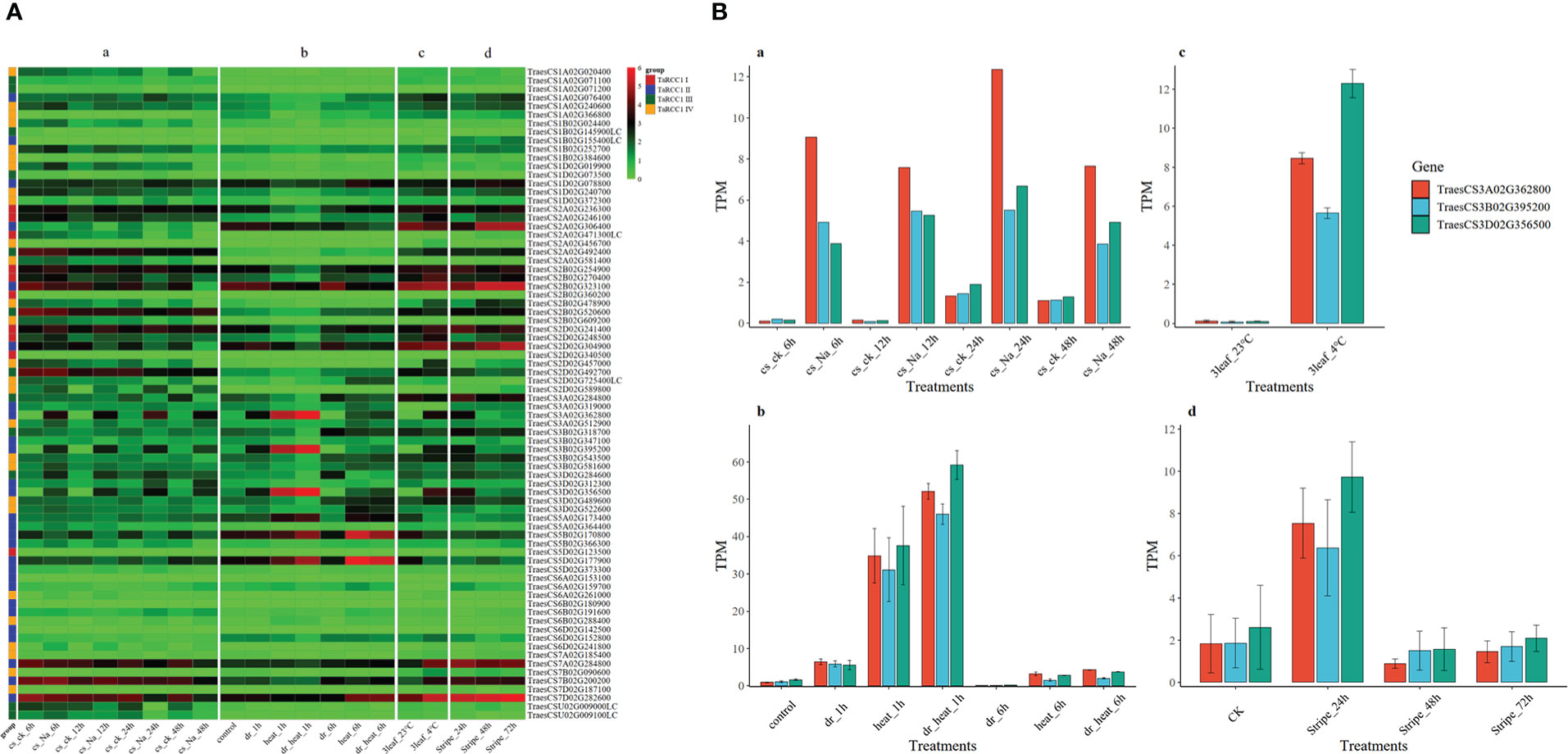
Figure 6 Expression profiles of TaRCC1 genes under salt, drought, heat, cold and stripe rust pathogen stresses. (A) Expression heat map of 76 TaRCC1 genes. Transcript-per-million-read (TPM) values of TaRCC1 genes were transformed by log2(x+1) for visualization using the pheatmap package of R software. TPM values of the three genes were obtained from four published studies: (a) Zhang et al. (2016), (b) Liu et al. (2015), (c) Li et al. (2015), and (d) Zhang et al. (2014). (B) The expression histogram of TaRCC1-3A/B/D genes under salt, drought, heat, cold, and strip rust pathogen stresses. TPM values of the three genes were obtained from four public studies including: (a) the study of Zhang et al. (2016), (b) the study of Liu et al. (2015), (c) the study of Li et al. (2015), and (d) the study of Zhang et al. (2014).
The gene-evidence networks extracted from the knowledge network of wheat through KnetMiner showed several transcription factors for the three TaRCC1 genes (Figure 7A). Five genes were identified as candidates for participating in the regulation of the three TaRCC1s, among which Myb-7B (TraesCS7B02G188000) and Myb-7D (TraesCS7D02G295400) were associated with the regulation of all three TaRCC1s (RCC1-3A, RCC1-3B, and RCC1-3D), suggesting that Myb-7B and Myb-7D might be the regulators of the three TaRCC1s. Myb-7B and Myb-7D encode two Myb-like transcription factors, which were related to the terms stripe rust response and drought tolerance in the KnetMiner knowledge network (Figure 7A). The expression patterns of Myb-7B and Myb-7D under multiple stresses, obtained from the expVIP Wheat Expression Browser (http://www.wheat-expression.com/) and the Triticeae Multi-omics Center (http://202.194.139.32/expression/index.html), showed similar and significant differential expression under different treatments of the five stresses. For instance, the expression level of the two Myb genes was higher than in the control for salt treatments at 6 h and 12 h and lower than in the control at 24 and 48 h. Under cold (4°C) and stripe rust pathogen stresses, the genes showed a similar gene expression pattern. Under drought and heat treatments, the Myb-7B/D showed reduced expression under dr_6h, heat_6h, and dr_heat_6h treatments, compared with the control (Figure 7B). Myb-binding sites on the promoters of TaRCC1-3A, TaRCC1-3B, and TaRCC1-3D were predicted using the PlantRegMap software (http://plantregmap.gao-lab.org/binding_site_prediction.php). A potential Myb-binding site was found in the promoters of TaRCC1-3A (−2119 to −2133) and TaRCC1-3B (−227 to −241), but no predicted Myb-binding sites were detected immediately upstream of the transcription start site of TaRCC1-3D, indicating that TaRCC1-3A/B are most likely directly regulated by the above Myb-like transcription factor, while TaRCC1-3D might be indirectly regulated (Figure 7C).
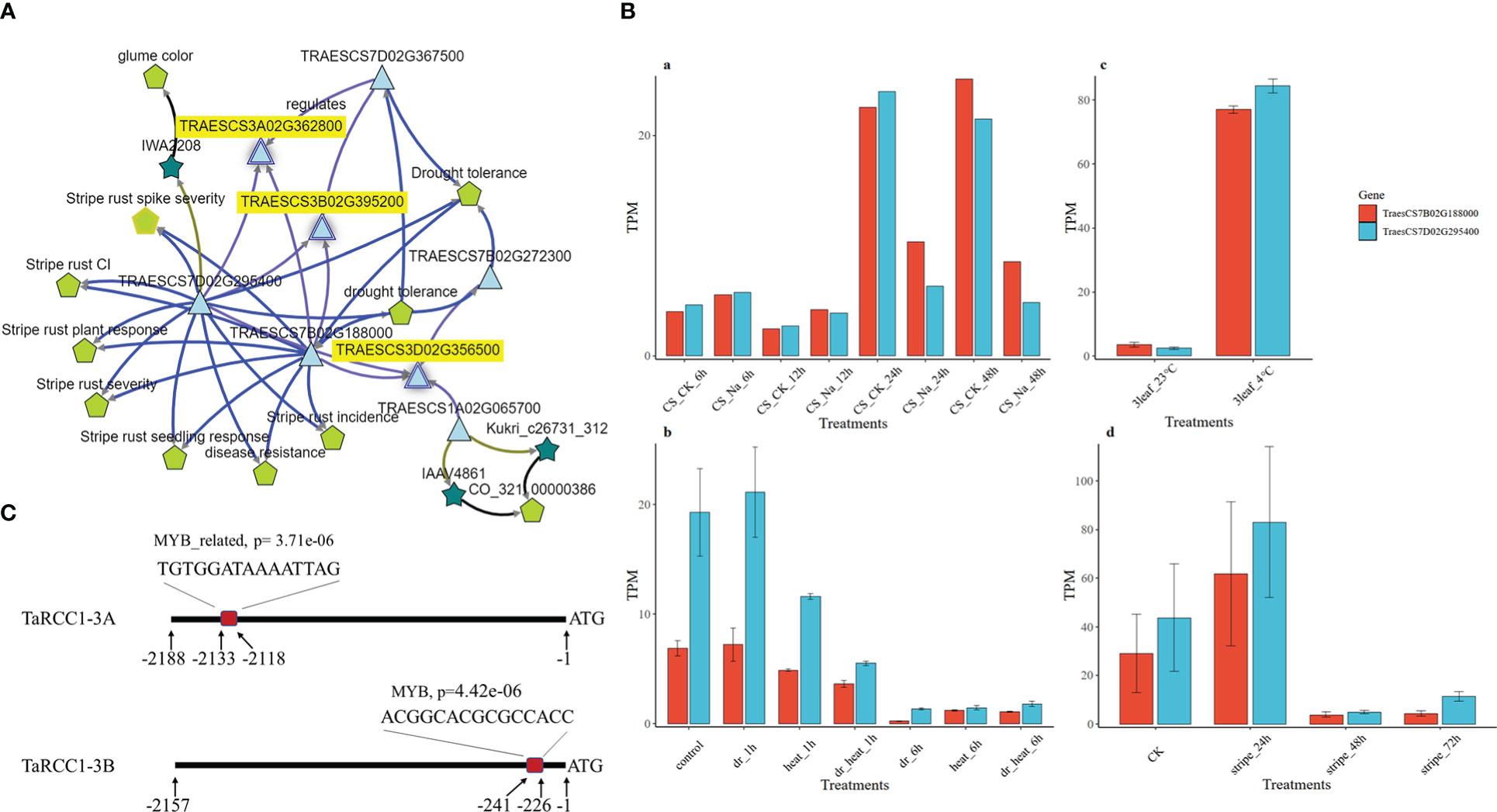
Figure 7 The Myb-7B/7D genes were predicted to regulate RCC1-3A/B/D. (A) Gene-evidence networks extracted from the wheat knowledge network for three TaRCC1 genes through KnetMiner. (B) Expression histogram of Myb-7B/7D genes under salt, drought, heat, cold, and stripe rust pathogen stresses. Transcript-per-million-read (TPM) values of the three genes were obtained from four published studies: (a) Zhang et al. (2016), (b) Liu et al. (2015), (c) Li et al. (2015), and (d) Zhang et al. (2014). (C) The predicted Myb-binding sites upstream of the transcription start site of TaRCC1-3A/B.
In Arabidopsis, RCC1 family proteins, such as UVR8 and TCF1, are located in the nucleus. Similarly, subcellular localization prediction suggested that all of the TaRCC1 family proteins are located in the nucleus. Our experiments to investigate the subcellular localization of the three TaRCC1 proteins (TaRCC1-3A (TraesCS3A02G362800), TaRCC1-3B (TraesCS3B02G395200) and TaRCC1-3D (TraesCS3D02G356500) in wheat protoplasts confirmed the results as predicted and showed that these proteins are located in the nucleus (Figure 8). The subcellular localization of the Myb-7B/D proteins showed that these two Myb proteins are also located in the nucleus (Figure 8). The primers used are listed in Supplementary Table S8.
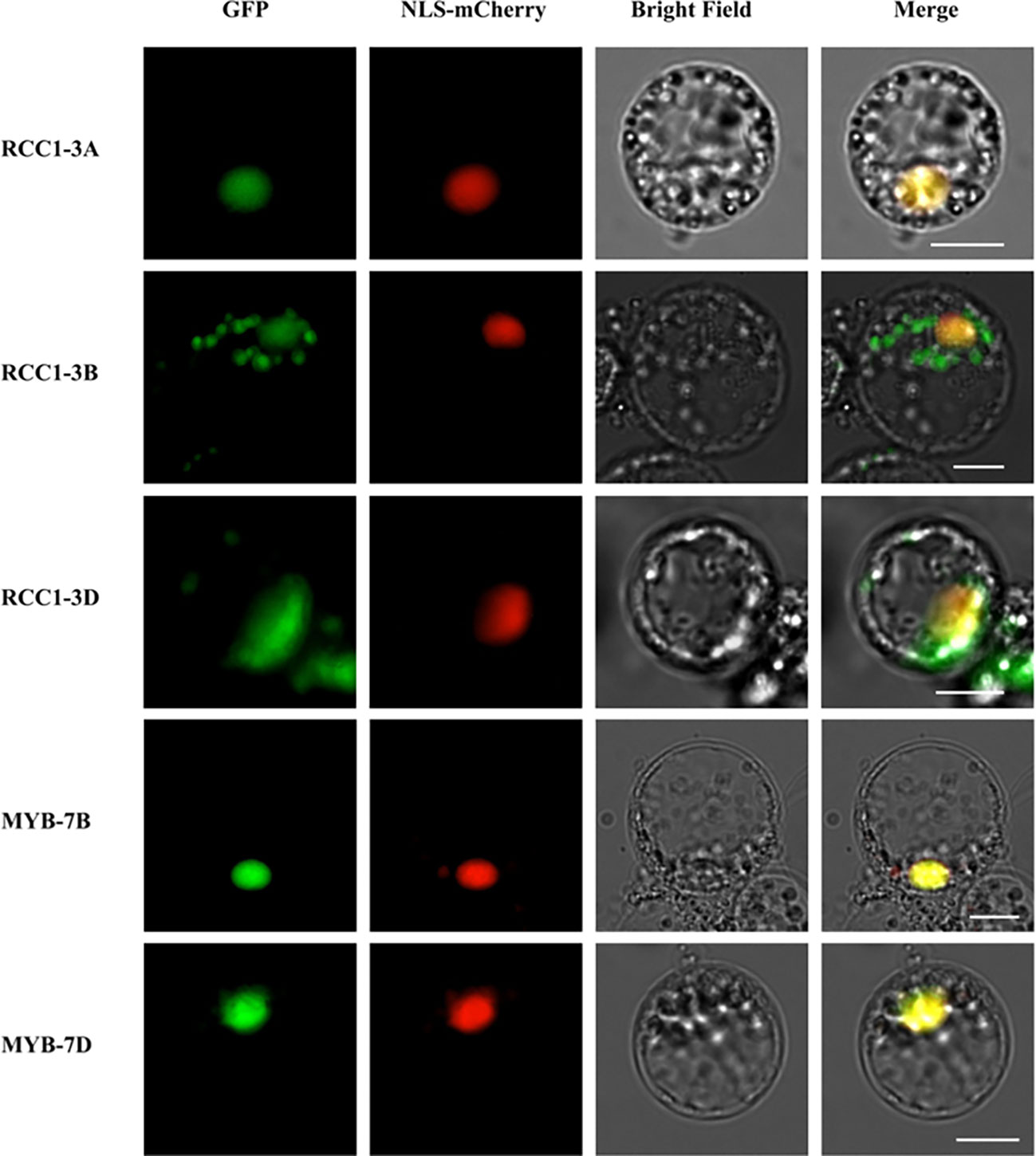
Figure 8 Subcellular localization of TaRCC1-3A (TraesCS3A02G362800), TaRCC1-3B (TraesCS3B02G395200), TaRCC1-3D (TraesCS3D02G356500), Myb-7B (TraesCS7B02G188000), and Myb-7D (TraesCS7D02G295400) proteins. TaRCC1-3A, TaRCC1-3B, TaRCC1-3D, Myb-7B, and Myb-7D were fused with GFP and co-expressed with the nuclear localization signal marker (NLS-mCherry) in wheat protoplasts. Scale bar = 20 μm.
From the available RNA-seq data of several studies, the expression patterns of TaRCC1s in different tissues and multiple stresses had been analyzed, as described above (Supplementary Figure S2; Figure 6). The expression levels of TaRCC1s varied significantly in different tissues and under multiple stresses; several TaRCC1s were induced by different stresses. Homoeologous genes TraesCS3A02G362800, TraesCS3B02G395200, and TraesCS3D02G356500 responded to five stresses (salt, drought, heat, cold, and stripe rust pathogen). We used qRT-PCR to verify the expression patterns of the three TaRCC1s in response to salt, cold, and drought stresses (Figure 9). Overall, these three TaRCC1s were induced by almost all the treatments, showing similar patterns to the RNA-seq results mentioned above. The expression of Myb-7B and Myb-7D was analyzed by qRT-PCR, and they shared a similar gene expression pattern to RCC1-3A, RCC1-3B, and RCC1-3D under salt, drought, and cold treatment (Figure 9), further suggesting that Myb-7B and Myb-7D might be regulators of the three TaRCC1s above. The qRT-PCR primers for RCC1-3A, RCC1-3B, RCC1-3D, Myb-7B, and Myb-7D are listed in Supplementary Table S9.
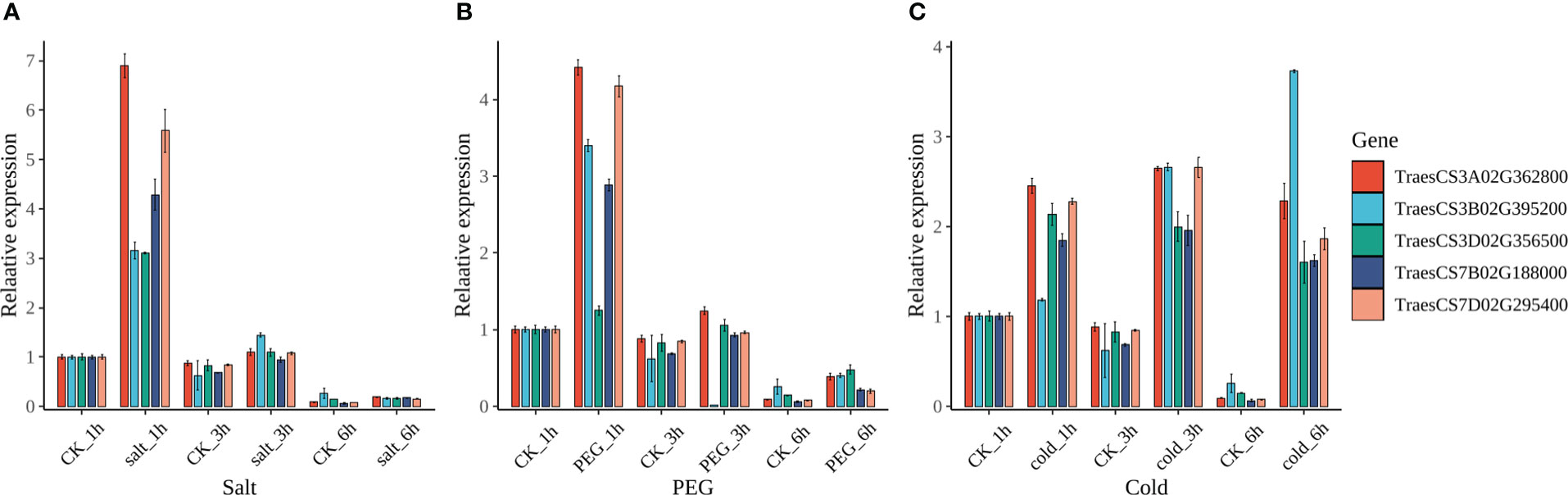
Figure 9 Expression pattern by qRT-PCR of RCC1-3A, RCC1-3B, RCC1-3D, Myb-7B, and Myb-7D under salt, drought, and cold stresses. (A) Salt stress, salt_1h/3h/6h: 1 h/3 h/6 h after watering with 1/2 MS liquid medium containing 200 mM NaCl. (B) Drought stress, PEG_1h/3h/6h: 1 h/3 h/6 h after watering with 1/2 MS liquid medium containing 20% PEG4000. (C) Cold stress, cold_1h/3h/6h: 1 h/3 h/6 h after watering with 1/2 MS liquid medium at 4°C. CK_1h/3h/6h in all three stress: 1 h/3 h/6 h after watering with 1/2 MS liquid medium at room temperature (control). Error bars represent the standard deviation of three biological replicates.
The RCC1 gene family is important in the functioning of the cell cycle. RCC1-like domains have been identified in a variety of proteins that mediate diverse biological processes (Hadjebi et al., 2008). Plant RCC1 proteins can be classified into two major groups, one consisting of six or seven RCC1 repeat units, similar to human RCC1, and the other composed of multi-domains, including the RCC1 repeat domain (Kuhn et al., 2011). In plants, however, the role of the RCC1 family genes is still unknown. UVR8 and TCF1 in Arabidopsis belonging to the single domain RCC1 protein have been found to be involved in the regulation of signal cascades, such as UV-B and cold-induced signaling pathways (Heijde and Ulm, 2012; Li et al., 2013; Tilbrook et al., 2013; Jenkins, 2014; Ji et al.,2015). Wheat is the world’s most cultivated crop and an important staple food for humans, accounting for one-fifth of calories consumed (Abhinandan et al., 2018). The release of a high-quality wheat reference genome has enabled the rapid and systematic study of the function of wheat genes to develop. Sequencing projects provide an opportunity for the isolation of gene families using a genome-wide scan. In wheat, there has been no comprehensive study focusing on the RCC1 genes, therefore, in this study, a comprehensive analysis of the TaRCC1 genes, including studies of phylogenetic relationships, gene structure, conserved motifs, chromosomal location, and expression profiles in different tissues, was performed to characterize the gene family in bread wheat. We first isolated 144 RCC1 genes, including 76 TaRCC1s, 49 TdRCC1s, and 24 AetRCC1s in wheat, emmer wheat, and Aegilops tauschii, respectively, identified from the fully annotated reference genomes. Phylogenetic analysis and synteny analysis showed that the RCC1 genes were clustered into four subfamilies (named RCC1 I–IV). Most TdRCC1s (except TRIDC3AG069370.1) and all the AeRCC1s were high-collinear with the TaRCC1s. The collinear RCC1 genes of the three species were clustered together in one clade (Figures 2, 3), and the number of RCC1s of the three species showed an approximate 3:2:1 ratio. Because of the collinearity among the A, B, and D sub-genomes of wheat, most of the 76 TaRCC1 genes identified were triplet genes (Figure 3). These results indicated that the RCC1 genes are evolutionarily conserved in bread wheat, emmer wheat, and Aegilops tauschii.
Based on phylogenetic and gene structure analyses, the 76 TaRCC1s were clustered into four subfamilies (named TaRCC1 I–IV). The TaRCC1 I, II, and III subfamilies contained 10, 27, and 12 genes, which contained only RCC1 domain repeats. The remaining 27 TaRCC1s of TaRCC1 IV contained multiple domains, including RCC1 domain repeats and PH or BRX domains (Figure 4C). These findings indicated that two different mechanisms might regulate genes in the TaRCC1 family. It appears that most TaRCC1 genes in a subfamily share a similar exon–intron structure, motif, and domain composition (Figure 4), indicating that the evolution might not only affect gene function but also gene structure (Babenko et al., 2004; Roy and Penny, 2007).
Analysis of the cis-acting regulatory elements in the promoter regions of TaRCC1 genes showed that TaRCC1s might be involved in the regulation of various biological processes, and several cis-acting regulatory elements were especially related to responses to hormones and stresses (Figure 5; Supplementary Table S5). Thus, we can speculate that the wheat RCC1 genes participate in specific signaling pathways that regulate growth, development, and defensive responses.
According to the publicly available transcriptome data of several studies, the expression profiles of TaRCC1 genes in wheat varied among different tissues and developmental periods, and the TaRCC1s showed different expression patterns under different stresses, namely salt, drought, heat, cold, and stripe rust. Three homologous TaRCC1s (TraesCS3A02G362800, TraesCS3B02G395200, and TraesCS3D02G356500) were proved to respond to all five different stresses; the genes were induced by almost all the treatments, suggesting that they might participate in regulating the plant responses to numerous stresses. The RCC1 proteins in plants have been implicated in regulating gene expression via epigenetic mechanisms (Ji et al., 2015; Ji et al., 2019; Jiang et al., 2021; Sun et al., 2022). Therefore, we determined whether the three TaRCC1 proteins above were located in the nucleus to investigate the possibility of their involvement in the regulation of downstream gene expression. Our results demonstrated that RCC1-3A, RCC1-3B, and RCC1-3D are all nuclear-localized proteins (Figure 9). At the same time, the green fluorescent protein (GFP) signals in wheat protoplasts were also obtained outside the nucleus. These results revealed the ability of RCC1-3A, RCC1-3B, and RCC1-3D proteins to migrate within cells, as has been reported for the UV-B receptor UVR8 (Kaiserli and Jenkins, 2007; Yin et al., 2016). Moreover, two Myb transcription factor genes (Myb-7B and Myb-7D) that co-expressed with RCC1-3A, RCC1-3B, and RCC1-3D were identified by co-expression and bioinformatics analysis (Figure 7C), which suggested that Myb-7B and Myb-7D might bind to the promoters of RCC1-3A/3B and upregulate their expression in response to abiotic stresses. The roles of RCC1-3A/3B/3D, and their interaction with Myb-7B/D, need to be investigated further by mechanistic studies, for example using transgenic and yeast single hybrid experiments.
From the fully annotated reference genomes, 149 RCC1 genes comprising 76 TaRCC1s, 49 TdRCC1s, and 24 AetRCC1s were identified in wheat, emmer wheat, and Aegilops tauschii, respectively. The 76 TaRCC1s in wheat were comprehensively analyzed in terms of gene structure, chromosome distribution, conserved domains, collinearity, phylogenetic relationship, and expression patterns in different tissues and in response to stresses. The expression patterns of 76 TaRCC1s in wheat under various stresses were further analyzed: qRT-PCR verified that RCC1-3A (TraesCS3A02G362800), RCC1-3B (TraesCS3B02G395200), and RCC1-3D (TraesCS3D02G35650) were significantly induced by salt, cold, and drought stresses. Co-expression network analysis and binding site predictions suggested that transcription factors encoded by Myb-7B (TraesCS7B02G188000) and Myb-7D (TraesCS7D02G295400) bind to the promoter of RCC1-3A/3B and upregulate gene expression in response to abiotic stresses in wheat. Our results provide valuable reference data for further study of RCC1 genes in wheat.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Conceptualization: XA and CS. Methodology: XA and CS. Software: WL. Validation: XA, WL, and TL. Formal analysis: XA. Investigation: XL. Resources: XA. Data curation: XA. Writing–original draft preparation: XA and LZ. Writing—review and editing: XA, CC, SZ, and CS. Visualization: XA. Supervision: XA. Project administration: XA. Funding acquisition: XA and CS. All authors contributed to the article and approved the submitted version.
This research was funded by the International Cooperation Fund of ZAAS (2022) and the National Natural Science Foundation of China (32201710).
We are greatly indebted to the reviewers for their critical, helpful, and constructive comments on this manuscript. We thank Huw Tyson, PhD, from Liwen Bianji (Edanz) (www.liwenbianji.cn) for editing the English text of a draft of this manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1124905/full#supplementary-material
Abhinandan, K., Skori, L., Stanic, M., Hickerson, N. M. N., Jamshed, M., Samuel, M. A. (2018). Abiotic stress signaling in wheat - an inclusive overview of hormonal interactions during abiotic stress responses in wheat. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.00734
Ann, X. H., Lun, Y. Z., Zhang, W., Liu, B., Li, X. Y., Zhong, M. T., et al. (2014). Expression and characterization of protein latcripin-3, an antioxidant and antitumor molecule from lentinula edodes C91-3. Asian Pac J. Cancer Prev. 15, 5055–5061. doi: 10.7314/apjcp.2014.15.12.5055
Babenko, V. N., Rogozin, I. B., Mekhedov, S. L., Koonin, E. V. (2004). Prevalence of intron gain over intron loss in the evolution of paralogous gene families. Nucleic Acids Res. 32, 3724–3733. doi: 10.1093/nar/gkh686
Bailey, T. L., Williams, N., Misleh, C., Li, W. W. (2006). MEME: Discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 34, W369–W373. doi: 10.1093/nar/gkl198
Bischoff, F. R., Ponstingl, H. (1991). Catalysis of guanine nucleotide exchange on ran by the mitotic regulator RCC1. Nature 354, 80–82. doi: 10.1038/354080a0
Brabletz, T., Jung, A., Reu, S., Porzner, M., Hlubek, F., Kunz-Schughart, L. A., et al. (2001). Variable beta-catenin expression in colorectal cancers indicates tumor progression driven by the tumor environment. Proc. Natl. Acad. Sci. U.S.A. 98, 10356–10361. doi: 10.1073/pnas.171610498
Brown, B. A., Cloix, C., Jiang, G. H., Kaiserli, E., Herzyk, P., Kliebenstein, D. J., et al. (2005). A UV-b-specific signaling component orchestrates plant UV protection. Proc. Natl. Acad. Sci. U.S.A. 102, 18225–18230. doi: 10.1073/pnas.0507187102
Cao, S. K., Liu, R., Sayyed, A., Sun, F., Song, R., Wang, X, et al. (2021) Regulator of chromosome condensation 1-domain protein DEK47 functions on the intron splicing of mitochondrial nad2 and seed development in maize. Frontiers in Plant Science 12:695249. doi: 10.3389/fpls.2021.695249
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, T., Muratore, T. L., Schaner-Tooley, C. E., Shabanowitz, J., Hunt, D. F., Macara, I. G. (2007). N-terminal alpha-methylation of RCC1 is necessary for stable chromatin association and normal mitosis. Nat. Cell Biol. 9, 596–603. doi: 10.1038/ncb1572
Christie, J. M., Arvai, A. S., Baxter, K. J., Heilmann, M., Pratt, A. J., O'Hara, A., et al. (2012). Plant UVR8 photoreceptor senses UV-b by tryptophan-mediated disruption of cross-dimer salt bridges. Science 335, 1492–1496. doi: 10.1126/science.1218091
Dominguez, I., Sonenshein, G. E., Seldin, D. C. (2009). Protein kinase CK2 in health and disease: CK2 and its role in wnt and NF-kappaB signaling: Linking development and cancer. Cell Mol. Life Sci. 66, 1850–1857. doi: 10.1007/s00018-009-9153-z
Dong, Z., Wang, H., Li, X., Ji, H. (2021). Enhancement of plant cold tolerance by soybean RCC1 family gene GmTCF1a. BMC Plant Biol. 21, 369. doi: 10.1186/s12870-021-03157-5
Duarte, G. T., Pandey, P. K., Vaid, N., Alseekh, S., Fernie, A. R., Nikoloski, Z., et al. (2021). Plasticity of rosette size in response to nitrogen availability is controlled by an RCC1-family protein. Plant Cell Environ. 44, 3398–3411. doi: 10.1111/pce.14146
Favory, J. J., Stec, A., Gruber, H., Rizzini, L., Oravecz, A., Funk, M., et al. (2009). Interaction of COP1 and UVR8 regulates UV-b-induced photomorphogenesis and stress acclimation in arabidopsis. EMBO J. 28, 591–601. doi: 10.1038/emboj.2009.4
Gu, Z., Gu, L., Eils, R., Schlesner, M., Brors, B. (2014). Circlize implements and enhances circular visualization in r. Bioinformatics 30, 2811–2812. doi: 10.1093/bioinformatics/btu393
Hadjebi, O., Casas-Terradellas, E., Garcia-Gonzalo, F. R., Rosa, J. L. (2008). The RCC1 superfamily: From genes, to function, to disease. Biochim. Biophys. Acta 1783, 1467–1479. doi: 10.1016/j.bbamcr.2008.03.015
Hassani-Pak, Keywan (2017). KnetMiner - an integrated data platform for gene mining and biological knowledge discovery. (Bielefeld: Universität Bielefeld).
Heijde, M., Ulm, R. (2012). UV-B photoreceptor-mediated signalling in plants. Trends Plant Sci. 17, 230–237. doi: 10.1016/j.tplants.2012.01.007
Hopkins, J., Pierre, O., Kazmierczak, T., Gruber, V., Frugier, F., Clement, M., et al. (2014). MtZR1, a PRAF protein, is involved in the development of roots and symbiotic root nodules in medicago truncatula. Plant Cell Environ. 37, 658–669. doi: 10.1111/pce.12185
Hutchins, J. R., Moore, W. J., Hood, F. E., Wilson, J. S., Andrews, P. D., Swedlow, J. R., et al. (2004). Phosphorylation regulates the dynamic interaction of RCC1 with chromosomes during mitosis. Curr. Biol. 14, 1099–1104. doi: 10.1016/j.cub.2004.05.021
International Wheat Genome Sequencing, C (2014). A chromosome-based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Science 345, 1251788. doi: 10.1126/science.1251788
International Wheat Genome Sequencing, C (2018). Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 361, eaar7191. doi: 10.1126/science.aar7191
Jenkins, G. I. (2014). The UV-b photoreceptor UVR8: From structure to physiology. Plant Cell 26, 21–37. doi: 10.1105/tpc.113.119446
Jenkins, G. I. (2017). Photomorphogenic responses to ultraviolet-b light. Plant Cell Environ. 40, 2544–2557. doi: 10.1111/pce.12934
Ji, H., Wang, S., Cheng, C., Li, R., Wang, Z., Jenkins, G. I., et al. (2019). The RCC1 family protein SAB1 negatively regulates ABI5 through multidimensional mechanisms during postgermination in arabidopsis. New Phytol. 222, 907–922. doi: 10.1111/nph.15653
Ji, H., Wang, Y., Cloix, C., Li, K., Jenkins, G. I., Wang, S., et al. (2015). The arabidopsis RCC1 family protein TCF1 regulates freezing tolerance and cold acclimation through modulating lignin biosynthesis. PloS Genet. 11, e1005471. doi: 10.1371/journal.pgen.1005471
Jiang, J., Liu, J., Sanders, D., Qian, S., Ren, W., Song, J., et al. (2021). UVR8 interacts with de novo DNA methyltransferase and suppresses DNA methylation in arabidopsis. Nat. Plants 7, 184–197. doi: 10.1038/s41477-020-00843-4
Kaiserli, E., Jenkins, G. I. (2007). UV-B promotes rapid nuclear translocation of the arabidopsis UV-b specific signaling component UVR8 and activates its function in the nucleus. Plant Cell 19, 2662–2673. doi: 10.1105/tpc.107.053330
Kliebenstein, D. J., Lim, J. E., Landry, L. G., Last, R. L. (2002). Arabidopsis UVR8 regulates ultraviolet-b signal transduction and tolerance and contains sequence similarity to human regulator of chromatin condensation 1. Plant Physiol. 130, 234–243. doi: 10.1104/pp.005041
Kozlov, A. M., Darriba, D., Flouri, T., Morel, B., Stamatakis, A. (2019). RAxML-NG: A fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics 35, 4453–4455. doi: 10.1093/bioinformatics/btz305
Kuhn, K., Carrie, C., Giraud, E., Wang, Y., Meyer, E. H., Narsai, R., et al. (2011). The RCC1 family protein RUG3 is required for splicing of nad2 and complex I biogenesis in mitochondria of arabidopsis thaliana. Plant J. 67, 1067–1080. doi: 10.1111/j.1365-313X.2011.04658.x
Kumar, S., Stecher, G., Li, M., Knyaz, C., Tamura, K. (2018). MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Letunic, I., Khedkar, S., Bork, P. (2021). SMART: Recent updates, new developments and status in 2020. Nucleic Acids Res. 49, D458–D460. doi: 10.1093/nar/gkaa937
Li, W., Wen, J., Song, Y., Yuan, H., Sun, B., Wang, R., et al. (2022). SaRCC1, a regulator of chromosome condensation 1 (RCC1) family protein gene from spartina alterniflora, negatively regulates salinity stress tolerance in transgenic arabidopsis. Int. J. Mol. Sci. 23, 8172. doi: 10.3390/ijms23158172
Li, J., Yang, L., Jin, D., Nezames, C. D., Terzaghi, W., Deng, X. W. (2013). UV-B-induced photomorphogenesis in arabidopsis. Protein Cell 4, 485–492. doi: 10.1007/s13238-013-3036-7
Li, H. Y., Zheng, Y. (2004). Phosphorylation of RCC1 in mitosis is essential for producing a high RanGTP concentration on chromosomes and for spindle assembly in mammalian cells. Genes Dev. 18, 512–527. doi: 10.1101/gad.1177304
Li, Q., Zheng, Q., Shen, W., Cram, D., Fowler, D. B., Wei, Y., et al. (2015). Understanding the biochemical basis of temperature-induced lipid pathway adjustments in plants. Plant Cell 27, 86–103. doi: 10.1105/tpc.114.134338
Liu, X., Wu, X., Sun, C., Rong, J. (2019). Identification and expression profiling of the regulator of chromosome condensation 1 (RCC1) gene family in gossypium hirsutum l. under abiotic stress and hormone treatments. Int. J. Mol. Sci. 20, 1727. doi: 10.3390/ijms20071727
Liu, Z., Xin, M., Qin, J., Peng, H., Ni, Z., Yao, Y., et al. (2015). Temporal transcriptome profiling reveals expression partitioning of homeologous genes contributing to heat and drought acclimation in wheat (Triticum aestivum l.). BMC Plant Biol. 15, 152. doi: 10.1186/s12870-015-0511-8
Liu, B., Zhong, M., Lun, Y., Wang, X., Sun, W., Li, X., et al. (2012). A novel apoptosis correlated molecule: Expression and characterization of protein latcripin-1 from lentinula edodes C(91-3). Int. J. Mol. Sci. 13, 6246–6265. doi: 10.3390/ijms13056246
Marchler-Bauer, A., Anderson, J. B., Cherukuri, P. F., DeWeese-Scott, C., Geer, L. Y., Gwadz, M., et al. (2005). CDD: A conserved domain database for protein classification. Nucleic Acids Res. 33, D192–D196. doi: 10.1093/nar/gki069
Polakis, P. (2007). The many ways of wnt in cancer. Curr. Opin. Genet. Dev. 17, 45–51. doi: 10.1016/j.gde.2006.12.007
Ramirez-Gonzalez, R. H., Borrill, P., Lang, D., Harrington, S. A., Brinton, J., Venturini, L., et al. (2018). The transcriptional landscape of polyploid wheat. Science 361, eaar6089. doi: 10.1126/science.aar6089
Renault, L., Nassar, N., Vetter, I., Becker, J., Klebe, C., Roth, M., et al. (1998). The 1.7 a crystal structure of the regulator of chromosome condensation (RCC1) reveals a seven-bladed propeller. Nature 392, 97–101. doi: 10.1038/32204
Rizzini, L., Favory, J. J., Cloix, C., Faggionato, D., O'Hara, A., Kaiserli, E., et al. (2011). Perception of UV-b by the arabidopsis UVR8 protein. Science 332, 103–106. doi: 10.1126/science.1200660
Roy, S. W., Penny, D. (2007). On the incidence of intron loss and gain in paralogous gene families. Mol. Biol. Evol. 24, 1579–1581. doi: 10.1093/molbev/msm082
Sievers, F., Wilm, A., Dineen, D., Gibson, T. J., Karplus, K., Li, W., et al. (2011). Fast, scalable generation of high-quality protein multiple sequence alignments using clustal omega. Mol. Syst. Biol. 7, 539. doi: 10.1038/msb.2011.75
Su, C., Zhao, H., Zhao, Y., Ji, H., Wang, Y., Zhi, L., et al. (2017). RUG3 and ATM synergistically regulate the alternative splicing of mitochondrial nad2 and the DNA damage response in arabidopsis thaliana. Sci. Rep. 7, 43897. doi: 10.1038/srep43897
Sun, C., Li, D., Gao, Z., Gao, L., Shang, L., Wang, M., et al. (2022). OsRLR4 binds to the OsAUX1 promoter to negatively regulate primary root development in rice. J. Integr. Plant Biol. 64, 118–134. doi: 10.1111/jipb.13183
Terry, L. J., Shows, E. B., Wente, S. R. (2007). Crossing the nuclear envelope: Hierarchical regulation of nucleocytoplasmic transport. Science 318, 1412–1416. doi: 10.1126/science.1142204
Tian, L., Wang, X., Li, X., Liu, B., Zhang, W., Cao, J., et al. (2016). In vitro antitumor activity of latcripin-15 regulator of chromosome condensation 1 domain protein. Oncol. Lett. 12, 3153–3160. doi: 10.3892/ol.2016.5106
Tilbrook, K., Arongaus, A. B., Binkert, M., Heijde, M., Yin, R., Ulm, R. (2013). The UVR8 UV-b photoreceptor: Perception, signaling and response. Arabidopsis Book 11, e0164. doi: 10.1199/tab.0164
Wang, J., Wan, X., Gao, Y., Zhong, M., Sha, L., Liu, B., et al. (2016). Latcripin-13 domain induces apoptosis and cell cycle arrest at the G1 phase in human lung carcinoma A549 cells. Oncol. Rep. 36, 441–447. doi: 10.3892/or.2016.4830
Wu, D., Hu, Q., Yan, Z., Chen, W., Yan, C., Huang, X., et al. (2012). Structural basis of ultraviolet-b perception by UVR8. Nature 484, 214–219. doi: 10.1038/nature10931
Yin, R., Skvortsova, M. Y., Loubery, S., Ulm, R. (2016). COP1 is required for UV-b-induced nuclear accumulation of the UVR8 photoreceptor. Proc. Natl. Acad. Sci. U.S.A. 113, E4415–E4422. doi: 10.1073/pnas.1607074113
Yoo, S. D., Cho, Y. H., Sheen, J. (2007). Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2, 1565–1572. doi: 10.1038/nprot.2007.199
Zhang, Y., Liu, Z., Khan, A. A., Lin, Q., Han, Y., Mu, P., et al. (2016). Expression partitioning of homeologs and tandem duplications contribute to salt tolerance in wheat (Triticum aestivum l.). Sci. Rep. 6, 21476. doi: 10.1038/srep21476
Keywords: wheat, regulator of chromosome condensation 1 (RCC1), gene family, abiotic stress, expression analysis
Citation: An X, Zhao S, Luo X, Chen C, Liu T, Li W, Zou L and Sun C (2023) Genome-wide identification and expression analysis of the regulator of chromosome condensation 1 gene family in wheat (Triticum aestivum L.). Front. Plant Sci. 14:1124905. doi: 10.3389/fpls.2023.1124905
Received: 15 December 2022; Accepted: 13 February 2023;
Published: 24 February 2023.
Edited by:
Libei Li, Zhejiang Agriculture and Forestry University, ChinaReviewed by:
Bin Wang, Donghua University, ChinaCopyright © 2023 An, Zhao, Luo, Chen, Liu, Li, Zou and Sun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xia An, YW54aWFAemFhcy5hYy5jbg==; Chendong Sun, MTAwNTUwOTkxOUBxcS5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.