- 1Research Institute for Dok-do and Ulleung-do Island, Kyungpook National University, Daegu, Republic of Korea
- 2Department of Botany, Graduate School of Science, Kyoto University, Kyoto, Japan
- 3Division of Forest Biodiversity, Korea National Arboretum, Pocheon, Republic of Korea
- 4Department of Biology and Chemistry, Changwon National University, Changwon, Republic of Korea
- 5Department of Biological Sciences, Sungkyunkwan University, Suwon, Republic of Korea
The genus Allium, with over 900 species, is one of the largest monocotyledonous genera and is widely accepted with 15 recognized subgenera and 72 sections. The robust subgeneric and sectional relationships within Allium have long been not resolved. Based on 76 species of Allium (a total of 84 accessions), we developed a highly resolved plastome phylogenetic framework by integrating 18 newly sequenced species (20 accessions) in this study and assessed their subgeneric and sectional relationships, with special emphasis on the two subgenera Anguinum and Rhizirideum. We retrieved the three major evolutionary lines within Allium and found that the two subgenera Anguinum and Rhizirideum are monophyletic whereas others are highly polyphyletic (e.g., Allium, Cepa, Polyprason, and Melanocrommyum). Within the subgenus Anguinum, two strongly supported sublineages in East Asian and Eurasian-American were found. Allium tricoccum in North America belonged to the Eurasian clade. The distinct taxonomic status of A. ulleungense and its sister taxon were further determined. In subg. Rhizirideum, the Ulleung Island endemic A. dumebuchum shared its most recent common ancestor with the species from Mongolia and the narrow Korean endemic A. minus. Two Ulleung Island endemics were estimated to originate independently during the Pleistocene. In addition, a separate monotypic sectional treatment of the east Asian A. macrostemon (subg. Allium) and sister relationship between A. condensatum and A. chinense was suggested.
1 Introduction
The genus Allium L. is one of the most diverse groups in petaloid monocotyledons, with more than 900 species, and some of them are used medicinally or have economic (e.g., onions, garlic, chives, scallions, leeks, shallots) and horticultural values (Fritsch and Friesen, 2002; Fritsch et al., 2010; Li et al., 2010; Herden et al., 2016). The genus has its main center of diversity in southwest and central Asia and a smaller one in North America, and it is characterized by having bulbus enclosed in membranous tunics, free or almost free tepals, and a usual subgynobasic style (Friesen et al., 2006). Although Allium was previously placed in Alliaceae (subfamily Allioideae Herb.) (Fay and Chase, 1996) in the order Amaryllidales (Takhtajan, 1987; Takhtajan, 1997), it is currently recognized as a member of the family Amaryllidaceae in the subfamily Allioideae, which consists of Allium only (including Caloscordum Herb., Milula Prain, and Nectaroscordum Lindl.) (Friesen et al., 2006). The infrageneric classification of Allium has been varied and complicated because of its size and taxonomic complexity, but the widely accepted and recent one includes 15 subgenera and 72 sections (Friesen et al., 2006). Based on molecular phylogenetic analyses of chloroplast and nuclear DNA sequences, the monophyly of Allium has been confirmed, and 15 subgenera and 72 sections have been grouped into three evolutionary lines (Friesen et al., 2006; Li et al., 2010; Wheeler et al., 2013).
Of the 15 subgenera, subg. Anguinum (G. Don. ex W.D.J. Koch) N. Friesen, which consists of ten and several varieties, shows disjunct distribution in high mountains from South West Europe to East Asia and in Northeastern North America (Fritsch and Friesen, 2002) (Figures 1A–D) (also see geographical distribution map of subg. Anguinum in Figures 3 and 4 of Herden et al., 2016). It is one of the strongly supported monophyletic groups and represents a distinct and specialized group, and especially it is adapted to the light regime under deciduous forests within the second evolutionary line (Pistrick, 1992; Li et al., 2010). Subg. Anguinum is characterized by various important characteristics, such as root anatomical traits (Fritsch, 1992a), leaf and bulb structure (Pastor and Valdes, 1985), hypogeal seed germination and seedling type (Druselmann, 1992), and locule and nectary structure (Fritsch, 1992b) and shares the basic chromosome number (x = 8) and karyotype (Jing et al., 1999). For the phylogenetic position of subg. Anguinum in the second evolutionary line, previous studies suggested that Anguinum is closely related to subg. Caloscordum (Herb.) R.M. Fritsch (Nguyen et al., 2008; Li et al., 2010; Herden et al., 2016). Within subg. Anguinum, two major lineages exist: the Eurasian-American Allium victorialis L. alliance, such as A. tricoccum Solander, and the East Asian alliance of A. prattii C.H. Wright, A. ovalifolium Hand.-Mazz., and others (Friesen et al., 2006; Herden et al., 2016). Although the two major lineages within this subgenus were supported based on nuclear rDNA and three non-coding chloroplast DNA sequences, conflicting nuclear and chloroplast phylogenies suggested potential hybrid origin for A. tricoccum in North America (Herden et al., 2016; Choi et al., 2019). The phylogenetic position of A. tricoccum has an implication in correctly understanding the intercontinental disjunct distribution via possible migration routes between the Bering Land Bridge (BLB) and the North Atlantic Land Bridge (NALB) (Tiffney, 1985; Tiffney and Manchester, 2001).

Figure 1 Species of Allium, especially from the two subgenera Anguinum and Rhizirideum, newly sequenced for their plastomes in this study. Subg. Anguimum: A tricoccum (A), A ulleungense (B), A microdictyon (C), and A ochotense (D). Subg. Cepa: A condensatum (E). Subg. Rhizirideum: A tenuissimum (F), A anisopodium (G), A flavescens (H), A angulosum (I), A spirale (J), A prostratum (K), A spurium (L), A dumebuchum (M), A austrosibiricum (N), A senescens (O), and A minus (P). Subg. Allium: A macrostemon (Q-T) show floral variations. Photo credit: Hyeok-Jae Choi.
Another subg. Rhizirideum (G. Don ex Koch) Wendelbo, representing one lineage within the third evolutionary line of Allium, comprised approximately 37 species placed in five sections (Rhizirideum, Caespitosoprason, Tenuissima, Rhizomatosa, and Eduardia) (Friesen et al., 2006) (Figures 1F–P) (also see geographical distribution map of sect. Rhizirideum in Figure 1 of Sinitsyna et al., 2016 and Figure 5 of Jang et al., 2021). These species typically occur in Eurasian steppes, with the greatest diversity in southern Siberia and Mongolia, whereas only a few species are distributed in Europe and East Asia (Korea, Russian Far East, and Japan). Of the five sections, sect. Rhizirideum, represents the most speciose group with 26 species, including the recently published new species A. dumebuchum from Korea (Jang et al., 2021) and A. heterophyllum from China (Xie et al., 2022), which widely occur from Europe to East Asia. Most species are distributed in temperate Asia, whereas only four species occur in Europe, and just two species commonly occur in Europe and reach Western Siberia (Sinitsyna et al., 2016). The species of sect. Rhizirideum share a basic chromosome number of x = 8, with various ploidy levels (2x, 4x, 5x, and 6x), potentially suggesting the important role of polyploidization process (Friesen, 1988; Sinitsyna et al., 2016 and references therein). Section Rhizirideum appears to be monophyletic, but phylogenetic relationships among species within sect. Rhizirideum has been poorly resolved based on traditional markers, such as nrDNA ITS and non-coding chloroplast DNA sequences (Sinitsyna et al., 2016; Jang et al., 2021; Xie et al., 2022).
An infrageneric placement of certain species and section has also been problematic. For example, the phylogenetic position of A. macrostemon (Figures 1Q–T), a wild onion widespread in East Asia (China, Japan, Korea, Mongolia, Tibet, and Russian Far East), has been disputed. In the taxonomic synopsis of the genus Allium in China, A. macrostemon was placed in subg. Allium sect. Allium, along two other species (A. porrum and A. sativum), but it was traditionally included in subg. Allium sect. Scorodon sensu lato (Hanelt, 1992). Friesen et al. (2006) placed it under Allium or suggested placing it to a new section. Based on the combined nuclear ITS and chloroplast rps16 sequence analysis of primarily Chinese Allium species, A. macrostemon was sister to A. caeruleum (subg. Allium sect. Caerulea) (Li et al., 2010). Most recently, the ITS sequence phylogeny showed that the clade containing several accessions of A. macrostemon was sister to A. caeruleum and that A. schoenoprasoides was sister to the A. macrostemon-A. caeruleum clade (Xie et al., 2022). In contrast, three cpDNA region sequences (ndhJ-trnF, psbD-trnT, and psbJ-petA) indicated a sister relationship between A. macrostemon and A. schoenoprasoides (Xie et al., 2022). The complete plastome sequence of A. macrostemon was reported, and the phylogeny suggested that it is sister to the clade of A. cepa–A. fistulosum–A. altaicum (subg. Cepa sect. Cepa) (Xie F.-M. et al., 2019). It was suggested that A. macrostemon shared similar testa sculptures and morphological characters (i.e., bulbil development in inflorescence, pistil morphology, and seed shape) with members of sect. Caerulea, especially with A. caeruleum (Choi et al., 2012). However, the membranous bulb tunics, widely spreading and pinkish perianth, and subulate inner filaments do not fit the description of the sect. Caerulea; hence, its placement in that section could not be justified (Choi and Oh, 2011). In addition, A. macrostemon has been consistently distinguished from its related sections of Allium subg. Allium, warranting the establishment of a new section (Friesen et al., 2006; Li et al., 2010; Xie et al., 2022). Thus, it is necessary to reassess the phylogenetic position of A. macrostemon, with additional sampling, in a much broader plastome phylogenetic framework.
Allium condensatum (Figure 1E), a wild onion with pale yellow perianth commonly occurring in East Asia (China, Mongol, Korea, and Russian Far East), was placed newly in the monotypic sect. Condensatum (subg. Cepa) from its original placement in sect. Oreiprason F. Herm. (subg. Polyprason Radic) (Friesen et al., 2006; Choi et al., 2012). It was shown, based on testa sculpture, that A. condensatum in sect. Condensatum is clearly distinguished from the other members of sect. Oreiprason (e.g., A. hymenorrhizum Ledeb. and A. obliquum L.) and subg. Cepa, supporting its own sectional treatment (Friesen et al., 2006; Choi et al., 2012). However, previous studies suggested that A. condensatum is closely related to species of subg. Allium sect. Codonoprason Rschb., e.g., A. carinatum L., A. flavum L., and A. melanantherum Pancic (Kruse, 1984). Therefore, it has been uncertain whether A. condensatum requires its own sectional treatment or it should belong to subg. Allium, Polyprason, or Cepa. There is a need to evaluate the phylogenetic position of A. condensatum (sect. Condensatum) within the up-to-date Allium plastome phylogenetic framework.
Although concatenated chloroplast non-coding region sequences had limited values, complete plastome sequences have been treasured and used recently to reveal the evolutionary history and adaptive evolution of the genus Allium and related genera (Huo et al., 2019; Xie D.-F. et al., 2019, Xie et al., 2020; Yang et al., 2020; Namgung et al., 2021; Scobeyeva et al., 2021; Chen et al., 2022; Jin et al., 2022). In particular, a total of 39 complete plastomes of Allium, covering 12 subgenera, provided well-supported phylogenetic relationships, identified numerous positively selected genes, and confirmed the monophyly of the genus and the three evolutionary lines (Xie et al., 2020). Furthermore, new species relationships were revealed within the poorly resolved third evolutionary lineage. Recently, nine plastomes of Allium species were assembled and, based on 38 species of Allium and the 11 other Amaryllidaceae species, the functionality loss of rps16, rps2, infA, and ccsA genes was documented within the genus (Scobeyeva et al., 2021).
As an ongoing effort to better understand the fascinating evolutionary history among species of Allium in Korea and neighboring countries in East Asia, we characterized their chloroplast genomes and conducted population genetics and phylogeographic investigations. In this study, we newly sequenced 11 species of Allium from subg. Rhizirideum and seven accessions, representing five species of Allium from subg. Anguinum, and analyzed them within the most comprehensive Allium plastome framework (a total of 84 accessions representing 76 species). We also sequenced two additional species, A. macrostemon and A. condensatum, to assess their phylogenetic positions. The aims of this study were (1) to reevaluate the infrageneric classification of the genus Allium based on the most comprehensive plastome framework, (2) to characterize additional plastome sequences of Allium species primarily from two subgenera, Anguinum and Rhizirideum, (3) to determine species relationships within Anguinum and Rhizirideum, with special emphasis on newly described species in Korea, and (4) to assess the phylogenetic position of A. macrostemon and A. condensatum (sect. Condensatum).
2 Materials and methods
2.1 Plant materials
For subg. Anguinum, we sampled a total of seven accessions, representing five species: one accession of A. tricoccum, two accessions of A. ochotense, two accessions of A. victorialis, one accession of A. ulleungense, and one accession of A. microdictyon (see complete species list in Table 1). Two accessions of previously reported plastome sequences under the names of A. victorialis (NC037240) and A. ochotense (NC057853) from Ulleung Island, Korea, are considered A. ulleungense sequenced in this study (Table 1). Two accessions of A. victorialis were sampled from Austria and Germany, and two accessions of A. ochotense were sampled from China and Japan. We sampled A. microdictyon from Sobaeksan Mountain in Korea. For subg. Rhizirideum, we sampled a total of 11 accessions, representing two sections of Rhizirideum: sect. Tenuissima (A. anisopodium and A. tenuissimum) and sect. Rhizirideum (A. flavescens, A. angulosum, A. senescens, A. spirale, A. dumebuchum, A. spurium, A. prostratum, A. austrosibiricum, and A. minus). Lastly, we sampled one representative of subg. Cepa sect. Condensatum (A. condensatum) from Russian Far East and one representative of subg. Allium sect. Allium (A. macrostemon) from Ulleung Island, Korea.
2.2 DNA isolation, NGS sequencing, and comparative plastome analysis
Fresh leaves were collected and dried using silica gel, and the total genomic DNA was extracted using DNeasy Plant Mini Kit (Qiagen, Carlsbad, CA, USA). The extracted DNA was sequenced using an Illumina HiSeq 4000 (Illumina, Inc., San Diego, CA, USA) at Macrogen Co. (Seoul, Korea), and it yielded a 150-bp paired-end read length. The resulting paired-end reads were assembled de novo using Velvet v1.2.10 with multiple k-mers (Zerbino and Birney, 2008). The complete plastomes were also confirmed by NOVOPlasty v2.6.2. (Dierckxens et al., 2016) using A. cepa (MK335926) and A. sativum (MK335928) as references. tRNAs were confirmed using tRNA scan-SE (Lowe and Eddy, 1997), and the sequences were annotated using Geneious R10 (Kearse et al., 2012). Annotated sequence files in the GenBank format were used to draw a circular map using OGDRAW v1.2 (Greiner et al., 2019). We used DnaSP v6.10 (Rozas et al., 2017) to perform a sliding window analysis with a step size of 200 bp and a window length of 800 bp to determine the nucleotide diversity (Pi) of the plastomes.
2.3 Phylogenetic analysis and molecular dating
The complete plastome sequences were aligned using MAFFT v7 (Katoh and Standley, 2013), and a maximum likelihood (ML) phylogenetic tree was constructed using IQ-TREE, with 1,000 bootstrap replicates (Nguyen et al., 2015). The aligned sequences in FASTA format are available in Supplementary Data Sheet 1. The best-fit evolutionary model for the complete plastome sequences, TVM+F+R6, was selected based on ModelFinder (Kalyaanamoorthy et al., 2017) implemented in IQ-TREE v1.4.2. We used Agapanthus as an outgroup based on a previous study (Xie et al., 2020). We also performed maximum parsimony (MP) and Bayesian inference (BI) analysis to evaluate tree topology based on different phylogenetic methods. For MP analysis, Fitch parsimony was performed with PAUP*4.0b10 (Swofford, 2003) using the HEURISTIC search option with TBR branch swapping and MULPARS on. Gaps were treated as missing, and bootstrap support for groups were determined by 1,000 bootstrap replicates (Felsenstein, 1985) using the HEURISTIC search option from a simple addition sequence with TBR branch swapping. To construct a BI tree, we used the MrBayes v3.2.6 (Ronquist et al., 2012) based on the best-fit model (GTR+G) from MrModeltest v2.2 (Nylander, 2004). The analysis was performed with one million generations initiated with a random starting tree, sampling every 1,000 generations. Tracer v1.7.1 (Rambaut et al., 2018) was used to evaluate the burn-in and to examine log likelihoods, ensuring that the run was in the stationary phase and that adequate effective sample sizes (ESS) were attained. After discarding initial 25% as burn-in, the remaining samples were used to construct a 50% majority-consensus tree with posterior probabilities (PP) for given clades. The consensus tree was finally edited using FigTree v1.4.3. (available online: http://tree.bio.ed.ac.uk/software/giftree/).
Divergence times based on the complete chloroplast genome sequences were estimated using the Bayesian method (Drummond et al., 2006) using the program BEAST v1.10.4 (Suchard et al., 2018). The XML file for the analysis was prepared in the Bayesian Evolutionary Analysis Utility (BEAUTi). As the secondary calibration point, we used the crown Allium clade mean age of 22.18 Myr and standard deviation of 9.93, resulting in a range of 15.23–34.81 Myr (Xie et al., 2020). We used the Yule process speciation prior, a lognormal relaxed clock model, and GTR+G substitution model, and then the ucld. mean parameter was specified to be uniform with 0.333 as the initial value, 0.00 as the lower, and 1 as the upper limit (Drummond et al., 2006). Posterior distributions for each parameter were estimated by means of an MCMC run for 30 million generations with a sampling frequency of every 100,000 generations. The posterior distribution of all statistics was checked using Tracer v1.7.1 (Rambaut et al., 2018) to assess convergence and confirm that the effective sample sizes (ESS) for all parameters were larger than 200 (Drummond et al., 2012). In addition, we used TreeAnnotator version 1.5 (http://beast.bio.ed.ac.uk/TreeAnnotator) to produce a maximum credibility tree of mean divergence time and 95% highest posterior density (HPD) intervals with posterior probability (PP) limit (0.5), after removing the first 25% of trees as burn-in (Drummond et al., 2012).
3 Results
3.1 Plastome characteristics of two subgenera Rhizirideum and Anguinum
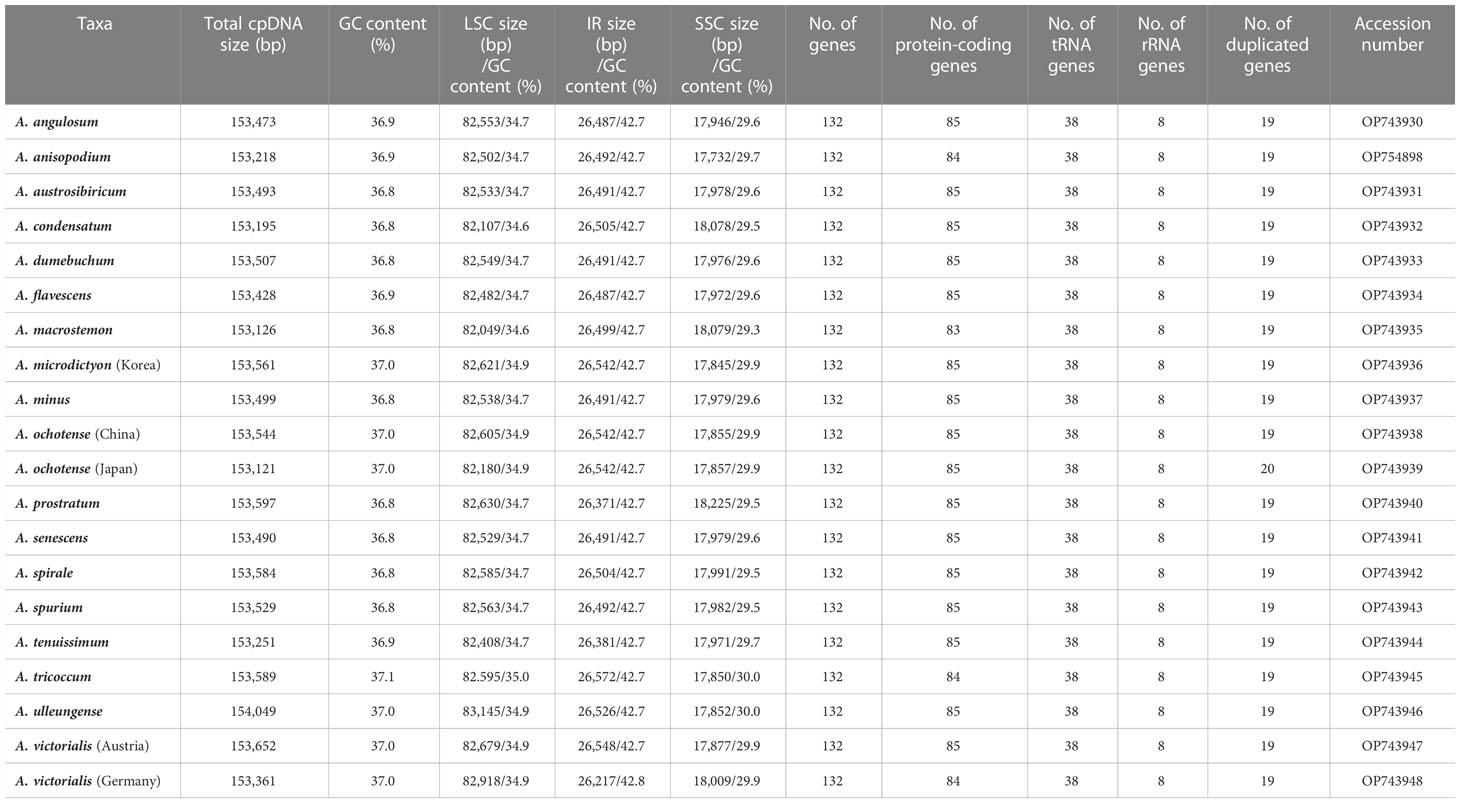
The plastomes of 11 newly sequenced Allium accessions from subg. Rhizirideum were 153,218 (A. anisopodium) to 153,597 bp (A. prostratum) in length and comprised a large single copy (LSC) region of 82,408 (A. tenuissimum) to 82,630 bp (A. prostratum), a small single copy (SSC) region of 17,732 (A. anisopodium) to 18,225 bp (A. prostratum), and two inverted repeat (IR) regions of 26,371 (A. prostratum) to 26,504 bp (A. spirale) (Table 1 and Figure 2). The overall guanine–cytosine (GC) content ranged from 36.8% (A. austrosibiricum, A. dumebuchum, A. minus, A. prostratum, A. senescens, A. spirale, and A. spurium) to 36.9% (A. angulosum, A. anisopodium, and A. tenuissimum). One species, A. anisopodium, had 84 protein-coding genes with two pseudogenized ycf1 genes, and a shortened sequence of 5,248 bp of ycf1 gene in the IR region at the SSC/IRa junction became a pseudogene. Furthermore, a completely functional rps2 gene was observed in all except one species (A. spurium) in subg. Rhizirideum, which contrasts with most Allium species in the third evolutionary line showing pseudogenization.
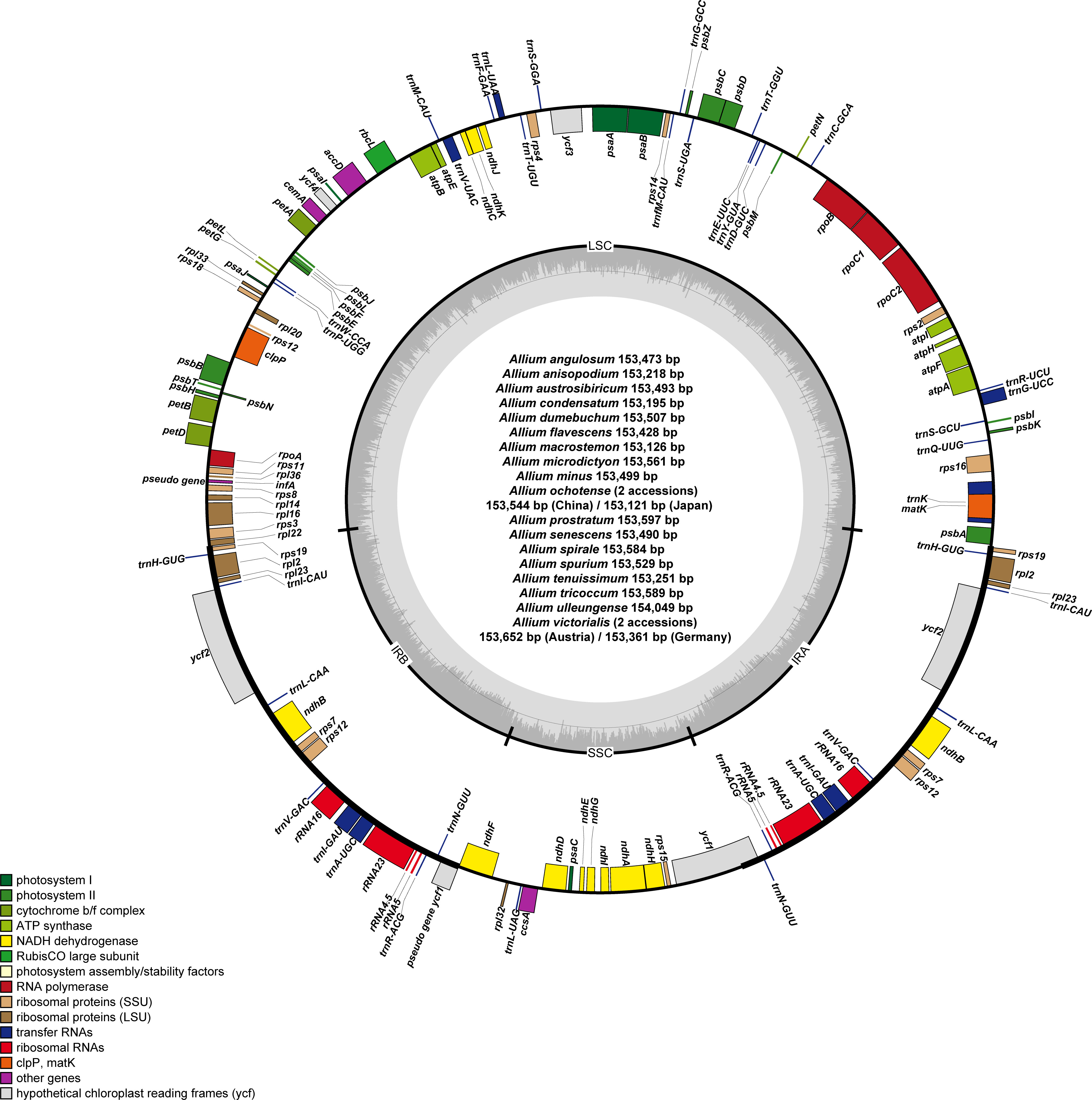
Figure 2 Complete plastome map of newly sequenced Allium species (a total of 20 accessions from 18 species) sequenced in this study. The genes inside and outside of the circle are transcribed in the clockwise and counterclockwise directions, respectively. Genes belonging to different functional groups are shown in different colors. The thick lines indicate the extent of the inverted repeats (IRa and IRb) that separate the genomes into small single copy (SSC) and large single copy (LSC) regions.
The plastomes of seven newly sequenced Allium accessions from subg. Anguinum were 153,121 bp (A. ochotense, Japan), 153,361 bp (A. victorialis, Germany), 153,544 bp (A. ochotense, China), 153,561 bp (A. microdictyon), 153,589 bp (A. tricoccum), 153,652 bp (A. victorialis, Austria), and 154,049 bp (A. ulleungense) in length. The overall guanine–cytosine (GC) content was 37% identical within subg. Anguinum, except for A. tricoccum with 37.1% GC content. Allium victorialis sampled from Germany and A. tricoccum contained 84 protein coding genes with two pseudogenized ycf1 genes: the shortened sequences of 4,254 bp (A. tricoccum) and 4,161 bp (A. victorialis) were in the IR region at the SSC/IRa junction and have become a pseudogene. Like most Allium species in the second evolutionary line, two functional genes, rps2 and rps16, were found in all subg. Anguinum species.
3.2 Plastome characteristics of A. macrostemon and A. condensatum
To further explore the phylogenetic position of A. macrostemon, we sequenced one additional accession sampled from Ulleung Island, Korea. The complete plastome was 153,126 bp in length with an overall GC content of 36.8% and comprised a large single copy (LSC) region of 82,049 bp with 34.6% GC content, a small single copy (SSC) region of 18,079 bp with 29.3% GC content, and two inverted repeat (IR) regions of 26,499 bp with 42.7% GC content (Table 1). We observed that rps2 and rps16 genes had become a pseudogene in A. macrostemon. Thus, a total of 83 protein-coding genes were detected in this species and the remaining Allium plastomes contained a total of 85 protein-coding genes. The complete plastome of A. condensatum sampled from Russia was 153,195 bp in length with an overall GC content of 36.8% and comprised a large single copy (LSC) region of 82,107 bp with a 34.6% GC content, a small single copy (SSC) region of 18,078 bp with a 29.5% GC content, and two inverted repeat (IR) regions of 26,505 bp with a 42.7% GC content. The rps2 gene was shortened in this species, whereas the rps16 gene was a completely functional gene with a completely conserved domain.
3.3 Comparative analysis of plastome structure and mutation hotspots in genus Allium
The position of IR/SC borders were determined in the 84 Allium plastomes, representing the three major evolutionary lines (Supplementary Figure 1). We found gene contents on both sides of the IR/SC borders conserved, and the LSC/IRb border was rps19/rpl22, whereas the IRa/LSC border was rps19/psbA. While the IR/SC borders among the species of Allium in the first evolutionary line (four subgenera Nectaroscordum, Microscordum, Amerallium, and Cyathophora) are conserved, some exceptions included the following: (1) the rpl22 gene of A. montanum (subg. Microscordum) and A. kingdonii (subg. Cyathophora) entirely in the LSC region; (2) the ndhF gene entirely in the SSC region for A. siculum (subg. Nectaroscordum), A. macranthum (subg. Amerallium), A. kingdonii, and A. zebdanense (subg. Amerallium); and (3) the rps19 gene of A. kindgonii in the IRb/LSC border (Supplementary Figure 1). Within the second evolutionary line of Allium (three subgenera, Caloscordum, Melanocrommyum, and Anguinum), some exceptions included the following: (1) the rpl22 gene of subg. Melanocrommyum (A. fetisowii, A. macleanii, and A. karataviense) and A. victorialis, Germany OP743948 (subg. Anguinum), completely in the LSC region; (2) the ndhF gene completely in the SSC region for subg Caloscordum and Melanocrommyum, and subg. Anguinum (A. ovalifolium var. leuconeurum and A. victorialis, Germany, OP743948); and (3) the rps19 gene in the IRb/LSC border for A. fetisowii (subg. Melanocrommyum) and A. victorialis, Germany OP743948 (subg. Anguinum) (Supplementary Figure 1). Lastly, the third evolutionary line included eight subgenera, Butomissa, Cyathophora, Allium, Cepa, Reticulatobulbosa, Rhizirideum, Melanocrommyum, and Polyprason. We found that the rpl22 gene was entirely located in the SSC region for subgenera Cyathophora, Rhizirideum (A. mongolicum, A. tenuissimum, and A. anisopodium), Allium (A. caeruleum), and Polyprason (A. xichuanense, A. rude, and A. chrysanthum). The rps19 gene of subg. Cyathophora (A. mairei and A. spicatum) and subg. Rhizirideum (A. mongolicum and A. tenuissimum) bordered in IRb/LSC. The rps19 gene of A. tenuissimum (subg. Rhizirideum) and A. mairei (subg. Cyathophora) bordered in IRa/LSC.
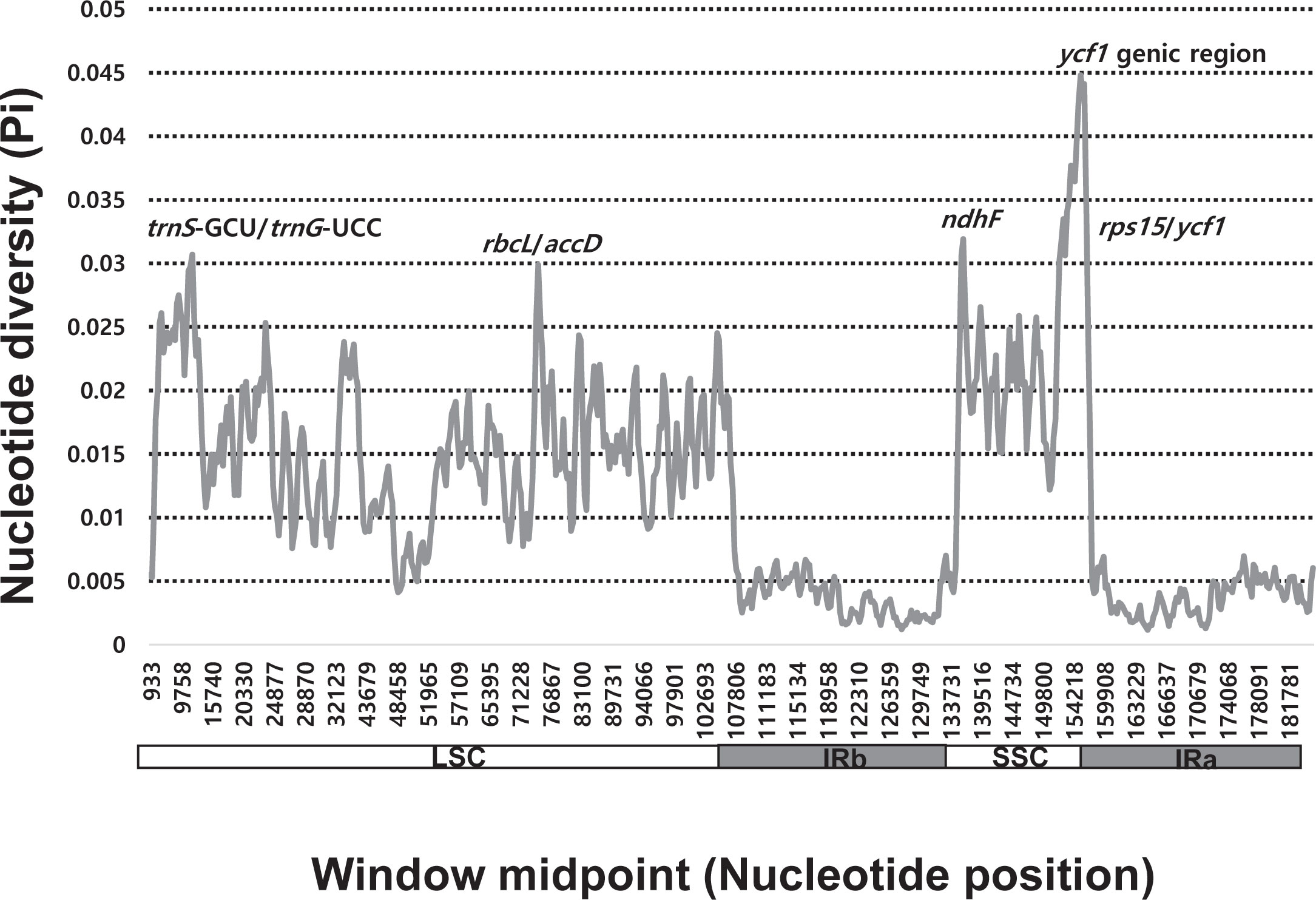
Sliding window analysis revealed highly variable regions in the plastomes of 84 Allium accessions representing the three evolutionary lines (Figure 3). The average nucleotide diversity (Pi) over the entire plastomes was 0.01191, with the most variable region (Pi = 0.0448) being the ycf1 genic region followed by the ndhF genic region (Pi = 0.03192). Three additional intergenic regions, i.e., trnS-GCU/trnG-UCC (Pi = 0.03068), rps15/ycf1 (Pi = 0.03018), and rbcL/accD (Pi = 0.02988), were also highly variable.
3.4 Phylogenetic analysis
The most comprehensive plastome sequences (a total of 84 accessions representing 76 species) provided a well-resolved and highly supported ML phylogenetic framework of the genus Allium (Figures 4A, B). It revealed three major evolutionary lines within the genus and consistently indicated that the first evolutionary line diverged first, followed by the second and third evolutionary lines. Within the first evolutionary line (100% BS), A. siculum, which belongs to subg. Nectaroscordum (sect. Nectaroscordum), diverged first, followed by A. monanthum (subg. Microscordum sect. Microscordum) and the clade of subg. Amerallium (100% BS) (Figure 4A). Allium kingdonii (subg. Cyathophora), which occurs very rarely in southeastern Tibet, was deeply embedded within the subg. Amerallium: A. kingdonii was sister to A. cernuum (100% BS), which belongs to sect. Lophioprason and the most widespread North American species of the genus. In the case of sectional relationships within subg. Amerallium, sect. Briseis (A. paradoxum) is sister to the clade containing two sections, Molium (A. moly and A. zebdanense) and Arctoprasum (A. ursinum) (100% BS).
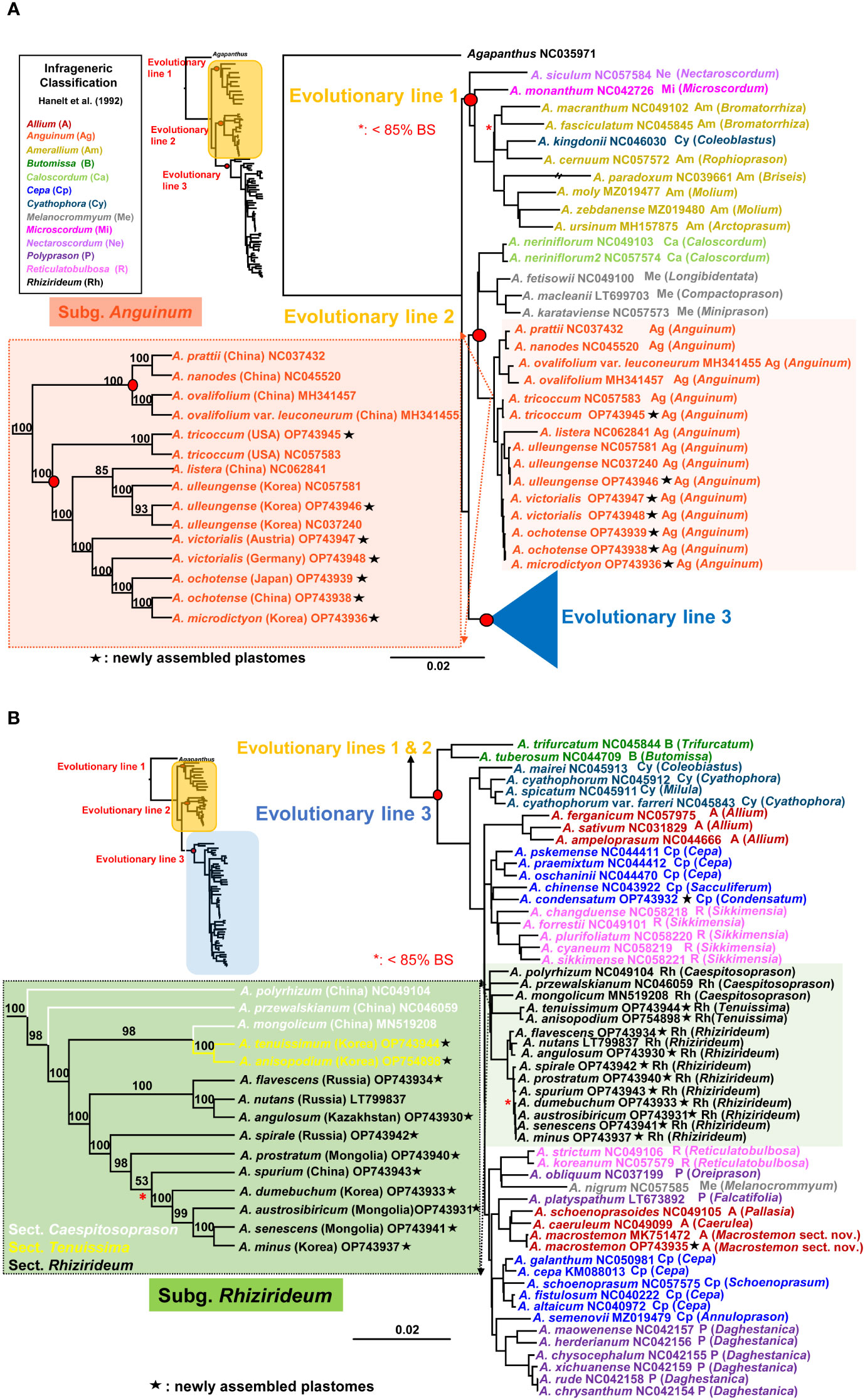
Figure 4 Maximum likelihood (ML) tree of the genus Allium based on 84 accessions representing 76 species. (A) A partial ML tree showing the first and second evolutionary lines of Allium, with enlargement of subg. Anguinum. Node with < 85% bootstrap support (BS) is shown in red asterisk. (B) A partial ML tree showing the third evolutionary line of Allium, with enlargement of subg. Rhizirideum. Node with < 85% bootstrap support (BS) is shown in red asterisk. Newly assembled plastome in this study is shown in black asterisk. Subgenera (Hanelt et al., 1992) are indicated in different colored labels. A species name is followed by a NCBI accession number, a subgeneric abbreviation, and a section in parenthesis.
For the second evolutionary line, the plastome phylogeny showed two major evolutionary lineages: one includes subg. Anguinum (100% BS) and the other (99% BS) includes two reciprocally monophyletic subgenera, Melanocrommyum (100% BS) and Caloscordum (100% BS) (Figure 4A). Within subg. Melanocrommyum, A. karataviense (sect. Miniprason) was sister to A. macleanii (sect. Compactoprason) and A. fetisowii (sect. Longibidentata) was sister to the clade of A. macleanii–A. karataviense (100% BS). For the species relationship within subg. Anguinum, two sublineages were found: one sublineage (100% BS) includes primarily East Asian species (A. nanodes, A. pratii, A. ovalifolium, and A. ovalifolium var. leuconeurum) whereas the other sublineage (100% BS) includes primarily Eurasian-American species (A. tricoccum, A. ochotense, A. victorialis, A. ulleungense, and A. microdictyon) and one exceptional species from East Asian sublineage (A. listera) (Figure 4A). Our result strongly supported that A. tricoccum in North America is sister to the clade containing Eurasian species (A. ochotense, A. ulleungense, A. victorialis, and A. microdictyon) and one East Asian species (A. listera). Both species, A. victorialis and A. ochotense, appeared not to be monophyletic, and A. microdictyon was embedded within A. ochotense. One newly sequenced accession of A. tricoccum in this study was sister to the other accession previously reported (NC057583). In addition, one newly sequenced accession of A. ulleungense (OP743946) in this study formed a clade with other previously reported accessions (NC057583 and NC037240). A distinct species recognition of A. ulleungense on Ulleung Island, Korea, is further supported based on the complete plastome sequences in this study: A. ulleungense was sister to A. listera, which occurs in several provinces in China (Anhui, Hebei, Henan, Jilin, Shaanxi, and Shanxi).
The third most diverse evolutionary line includes several subgenera, such as Allium, Butomissa, Cepa, Cyathophora, Polyprason, Rhizirideum, and Reticulatobulbosa (Figure 4B). Two monophyletic subgenera Butomissa sensu Huang et al. (2014) and Cyathophora diverged first and then followed by the diversification of non-monophyletic subgenera Allium, Cepa, and Polyprason. Two lineages of subg. Allium were found in this study: one group (A. ferganicum, A. ampeloprasum, and A. sativum from sect. Allium; 100% BS) was sister to the remaining clade of the third evolutionary line, whereas the other group of two species (A. macrostemon and A. caeruleum) was closely related to A. schoenoprasoides (subg. Allium sect. Pallasia) and A. platyspathum (subg. Polyprason sect. Falcatifolia). Allium macrostemon sampled from Ulleung Island (Korea; OP743935) in this study and previous accession from China (Jiangsu Province; MK751472) is monophyletic (100% BS) and sister to the clade containing A. schoenoprasoides and A. caeruleum (100% BS). In the case of subg. Cepa, at least four sublineages appeared to exist based on the plastome sequences: (1) A. chinense (sect. Sacculiferum), which is sister to A. condensatum (100% BS), (2) the clade containing sect. Cepa (A. pskemense, A. oschaninii, and A. praemixtum; 100% BS), (3) the clade containing other species of sect. Cepa (A. cepa, A. galanthum, A. altaicum, and A. fistulosum) and sect. Schoenoprasum (A. schoenoprasum) (100% BS), and (4) A. semenovii (sect. Annuloprason), which is sister to one clade of subg. Polyprason (100% BS) (Figure 4B). At least three sublineages of subg. Polyprason seemed to exist: (1) a clade containing species in sect. Daghestanica (A. herderianum, A. maowenense, A. chysocephalum, A. xichuanense, A. rude, and A. chrysanthum; 100% BS), (2) A. obliquum (sect. Oreiprason), which is sister to A. nigrum (subg. Melanocrommyum) (93% BS), and (3) A. platyspathum (sect. Falcatifolia), which is sister to one subg. Allium clade (100% BS). However, subg. Melanocrommyum belongs to the second evolutionary line, and A. nigrum was unexpectedly placed in the third evolutionary line: it is sister to A. obliquum (subg. Polyprason). In the case of subg. Reticulatobulbosa, two plastome lineages exist, one such as sect. Sikkimense (A. changduense, A. forrestii, A. plurifoliatum, A. cyaneum, and A. sikkimense; 100% BS) and the others such as sect. Reticulatobulbosa (A. strictum and A. koreanum) (100% BS).
As one major lineage within the third evolutionary line, subg. Rhizirideum is monophyletic (100% BS) and is sister to the clade containing one small sublineage of Reticulatobulbosa, one clade of Allium, one clade of Cepa, and one major clade of Polyprason (84% BS) (Figure 4B). Within subg. Rhizirideum, sect. Caespitosoprason is paraphyletic whereas sect. Rhizirideum is monophyletic. Section Tenuissima (A. anisopodium and A. tenuissimum) is monophyletic (100% BS), and A. mongolicum (sect. Caespitosoprason) is sister to sect. Tenuissima (98% BS). Within sect. Rhizirideum, two sublineages were identified: one includes A. flavescens, A. angulosum, and A. nutans (100% BS), and the other includes A. senescens, A. spirale, A. dumebuchum, A. spurium, A. prostratum, A. austrosibiricum, and A. minus (100% BS). Two endemic species in Korea, A. minus and A. dumebuchum, are sisters to A. senescens and the clade containing A. austrosibiricum–A. senescens–A. minus, respectively (each 100% BS). Lastly, A. condensatum is sister to A. chinense (subg. Cepa sect. Sacculiferum) (100% BS).
Of a total of 158,090 aligned characters used for MP analysis, we found 123,356 constant characters (78.03%), 19,806 variable sites (12.52%), and 14,928 parsimony informative characters (9.44%). The heuristic search resulted in four equally most parsimonious trees, with a tree length of 59,095, a consistency index (CI) of 0.70, and a retention index (RI) of 0.82 (Supplementary Figure 2). A 50% bootstrap consensus tree was well resolved with high bootstrap support (BS) values and found the same three evolutionary lines identified in ML analysis (Figure 4). The MP tree topology was nearly identical to that of ML in phylogenetic relationships among the three evolutionary lines and subgeneric relationships within each evolutionary line. Within subg. Rhizirideum and Anguinum clade, the species relationships were identical to that of the ML tree, except for the position of A. listera in subg. Anguinum: A. listera is sister to the clade containing A. ulleungense–A. victorialis–A. ochotense–A. microdictyon in the MP tree (Supplementary Figure 2), whereas it is sister to A. ulleungense species only in ML (Figure 4A). As with the ML tree (Figure 4), we found nearly identical tree topology in the BI tree (Supplementary Figure 3), except for the position of A. nigrum. Within two subgenera, Rhizirideum and Anguinum, we found identical tree topology between the ML and BI trees.
3.5 Molecular dating
We estimated the divergence times of major lineages within the genus Allium and several other lineages of our interests. The crown age of the three evolutionary lines within the genus was estimated to be 20.13 Ma (95% HPD, 8.72–30.57 Ma), 13.88 Ma (95% HPD, 6.04–22.43 Ma), and 17.05 Ma (95% HPD, 8.28–25.75 Ma) for the first, second, and third evolutionary line, respectively, suggesting their origins in early Miocene (Figure 5). The crown age of subg. Anguinum was estimated to be 4.85 Ma (95% HPD, 2.08–8.32 Ma), whereas that of subg. Rhizirideum was 8.41 Ma (95% HPD, 3.85–13.49 Ma). The split of subg. Anguinum from its sister lineage (subgenera Caloscordum and Melanocrommyum) was estimated to be 13.88 Ma (95% HPD, 6.04–22.43 Ma). The subg. Rhizirideum was estimated to be a split from two lineages (one lineage of Cepa and another lineage of Reticulatobulbosa) at 10.72 Ma (95% HPD, 5.09–16.57 Ma). Within subg. Anguinum, the crown ages of the two major lineages, representing East Asian and Eurasian-American, were estimated to be 3.05 Ma (95% HPD, 1.22–5.34 Ma) and 2.71 Ma (95% HPD, 1.00–4.86 Ma), respectively. Molecular dating also suggested that the split of Ulleung Island endemic A. ulleungense from its sister taxon (A. listera) was estimated at 1.57 Ma (95% HPD, 0.05–4.43 Ma). In addition, the intercontinental disjunct event between East Asia and Eastern North America (A. tricoccum) was estimated at approximately 2.71 Ma (95% HPD, 1.00–4.86 Ma) in the late Pliocene. Within subg. Rhizirideum, the split of monophyletic sect. Rhizirideum from two early diverged sections, Caespitosoprason and Tenuissima, was estimated at 7.43 Ma (95% HPD, 3.43–12.20 Ma). In addition, the split between European (A. flavescens–A. nutans–A. angulosum) and Asian (A. spirale–A. prostratum–A. spurium–A. dumebuchum–A. austrosibiricum–A. senescens–A. minus) lineages within sect. Rhizirideum was estimated at 2.18 Ma (95% HPD, 0.75–4.13 Ma). Allium dumebuchum, a Ulleung Island endemic, had an estimated divergence time from its continental lineage at 0.29 Ma (95% HPD, 0.08–0.59 Ma). Lastly, the split of A. macrostemon from its sister lineage (A. schoenoprasoides and A. caeruleum in sect. Pallasia and Caerulea, respectively) was estimated at 4.55 Ma (95% HPD, 1.87–7.54 Ma), whereas A. condensatum and A. chinense shared their most recent common ancestor at 7.62 Ma (95% HPD, 3.02–12.36 Ma).
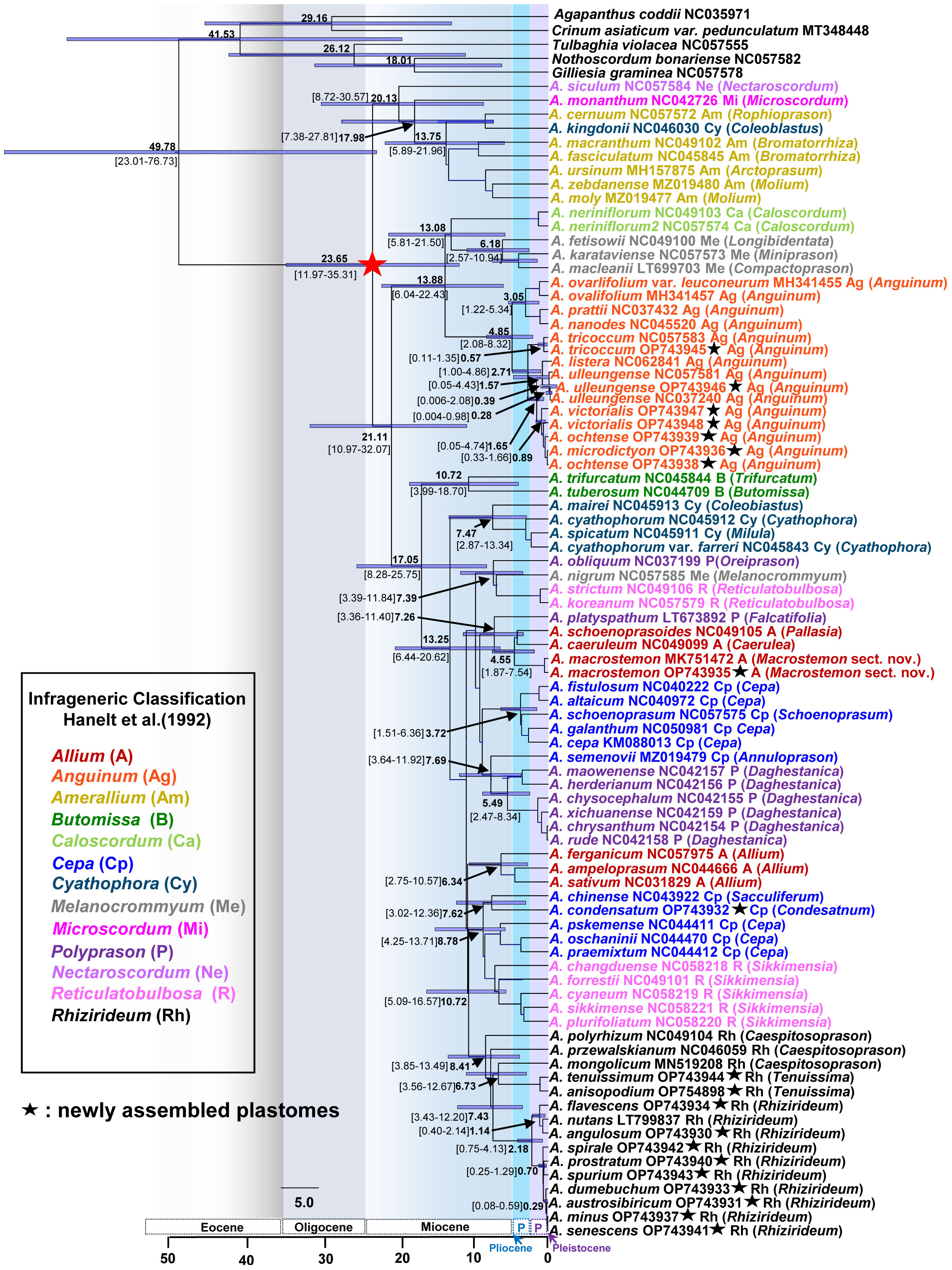
Figure 5 Dated chronogram showing divergence times of major lineages of the genus Allium, including three evolutionary lines identified previously. Estimated mean ages are shown in each node with 95% high posterior density (HPD) in bracket. The secondary calibration point estimated previously are shown as asterisked node (red star); that is, the crown age of the genus Allium was used in this study. Newly assembled plastome in this study is shown in black asterisk. Subgenera (Hanelt et al., 1992) are indicated in different colored labels. A species name is followed by an NCBI accession number, a subgeneric abbreviation, and a section in parenthesis.
4 Discussion
4.1 Allium plastome variation and evolution
In this study, we newly sequenced a total of 18 species (20 accessions) of Allium plastomes, primarily focusing on two subgenera, Rhizirideum and Anguinum, and found them highly conserved in genome structure and organization. The genome size of the newly sequenced 20 plastomes ranged from 153,121 (A. ochotense; Japan) to 154,049 bp (A. ulleungense), which was within the broad range of the genus (145 to 160 kb; Xie et al., 2020 and references therein) (Table 1). The GC contents of the 20 plastomes varied from 36.8% to 37.1%, which was within the range of the genus, that is, from 36.7% to 37.8% (Xie et al., 2020). In addition, the gene number (protein coding, tRNAs, and rRNAs) was very similar to those reported in previous Allium plastomes, suggesting overall high conservation of plastomes as previously shown within infrageneric and infrafamilial levels in angiosperms (e.g., Aroideae, Henriquez et al., 2020; Hosta, Yang et al., 2021; Crassulaceae, Chang et al., 2021; Cotoneaster, Yang et al., 2022; Ocotea, Trofimov et al., 2022).
We examined the position of SC/IR borders across the three evolutionary lines in the 84 Allium plastomes (Supplementary Figure 1). As shown previously in angiosperm plastomes, gene contents on both sides of the SC/IR borders of Allium plastomes were conserved (Downie and Jansen, 2015). Mostly, rpl22 was interrupted by the LSC/IRb border in Allium, but its position entirely within the LSC region has occurred multiple times in different lineages/species of three evolutionary lines (Supplementary Figure 1). In addition, the interrupted position of rps19 in the IRa/LSC border has occurred stochastically in different species within Allium: A. kingdonii (subg. Cyathophora), A. fetisowii (subg. Melanocrommyum), A. victorialis (subg. Anguinum), A. mairei/A. spicatum (subg. Cyathophora), and A. tenuissimum (subg. Rhizirideum). For SSC/IR boundaries, two SSC/IR borders crossed two ycf genes in most of the Allium plastomes and a large part of the ycf1 sequence was mainly positioned in the IRb region. In addition to the ndhF gene’s entire location within the SSC region, its location at the boundary of SSC and IRb was also occurred stochastically. It remains to be determined whether plastome structure reflects phylogenetic or stochastic events, based on broader taxon sampling, ecological analysis, and nuclear-based phylogenomics (Scobeyeva et al., 2021).
We also found five highly variable regions across the three evolutionary lines of Allium (Figure 3). The mutation hotspots included two genic regions, ycf1 (Pi = 0.0448) and ndhF (Pi = 0.03192). We further identified the three intergenic regions (trnS/trnG, rps15/ycf1, and rbcL/accD). Of eight mutation hot spots found among 18 species of Allium (Namgung et al., 2021), the same three regions (i.e., ycf1, ndhF, and rps15/ycf1) were also found in the current study. Two additional intergenic regions, rps15/ycf1 and rbcL/accD, showed high variability. The same three loci (ycf1, trnS/trnQ, and rbcL/accD) were found to be highly variable chloroplast markers for evaluating plant phylogeny at low taxonomic levels and for DNA barcoding in angiosperms (Dong et al., 2012). Three regions, ycf1, trnS/trnG, and rps15/ycf1, have been determined to be highly variable among the 48 Lilium (Liliaceae) plastomes (Sheikh-Assadi et al., 2022). However, different sets of highly variable chloroplast regions (with exception in one or two loci depending on taxa) were identified among the eight species of Fritillaria (Liliaceae; Bi et al., 2018) and the six species of Hosta (Amaryllidaceae; Yang et al., 2021) in monocots, cautiously limiting the wide applicability of universally hypervariable chloroplast regions. Nonetheless, highly variable chloroplast regions found in this study can be used to resolve phylogenies within Allium and for DNA barcoding or phylogeographic study of closely related species or related genera in monocots.
4.2 Inter-subgeneric relationships within Allium
Owing to the insufficient sectional-level sampling in this study, our discussion is based primarily on phylogenetic relationships among subgenera reported in previous studies (Friesen et al., 2006; Li et al., 2010). Our study independently confirmed the three evolutionary lines and their relationships in the genus Allium (Friesen et al., 2006; Li et al., 2010; Xie et al., 2020) (Figure 4A, B). In particular, the previous plastome-based phylogeny (Xie et al., 2020) was robustly recovered in this study. The first evolutionary line includes three subgenera, Nectaroscordum, Microscordum, and Amerallium (Figure 4A). For inter-subgeneric relationships within this lineage, subg. Nectaroscordum diverged first and then was followed by two sister subgenera, Microscordum and Amerallium, corroborating with a previous study (Friesen et al., 2006). This tree topology further supports the view of descending aneuploidy basic chromosome number, Nectaroscordum (x = 9), Microscordum (x = 8), and Amerallium (x = 7): ascending basic chromosome number also evolved independently in several morphologically derived Amerallium groups (Friesen et al., 2006). Although the second evolutionary line was also strongly recovered, inter-subgeneric relationships appeared to be different between the current and previous studies (Figure 4A). For example, despite the lack of representatives from two subgenera (Porphyroprason and Vvedenskya), the complete plastome tree strongly suggests that Caloscordum is closely related to Melanocrommyum and that Anguinum represents another distinct lineage within this evolutionary line. The previous study based on ITS sequences suggests that Caloscordum represents the first diverged subgenus followed by Anguinum, the two sister subgenera Porphyroprason and Vvedenskya, and the massive radiation of Malanocrommyum (Friesen et al., 2006). Subgenus Caloscordum is an oligotypic group with three species in East Asia and shares several characteristics with subg. Melanocrommyuum (that is, multiovulate locules, subterraneous leaf sheaths, and the presence of relatively large inner vascular bundles in the scapes; Friesen et al., 1986; Fritsch, 1993), further supporting our current plastome relationship. In addition, similar characteristics of seed testa cells, slightly verrucose periclinal wall, and straight anticlinal walls shared between Anguinum and Caloscordum might be viewed as either convergent evolution or symplesiomorphies (Friesen et al., 2006). It is required to include the two monotypic subgenera Porphyroprason (A. oreophilum) and Vvedenskya (A. kujukense), which have several autapomorphies, to fully determine the inter-subgeneric relationships within this evolutionary lineage.
The third evolutionary line is rather complex compared with the two former ones (Figure 4B). One consistent relationship found in the current and previous studies is an early divergence of two subgenera, Butomissa and Cyathophora, within this evolutionary line. Subgenus Butomissa, a small group comprising two subgroups, occurs in the Siberian-Mongolian-North Chinese steppes and the mountains from eastern to central Asia up to the borderline of the eastern Mediterranean area (Friesen et al., 2006). The second diverged lineage, subg. Cyathophora, is a small and solely Asian (Tibet and the Himalayas) group, and it shares certain characteristics (i.e., one row of identically oriented vascular bundles in the leaf blades, the presence of palisade parenchyma and subcortical laticifers, and biovulate locules) (Fritsch, 1988; Hanelt, 1992; Friesen et al., 2006). The remaining clade of the five subgenera Cepa, Reticulatobulbosa, Polyprason, Rhizirideum, and Allium (100% BS), is further complicated by non-monophyly of all but one subgenus Rhizirideum (Figure 4B). Nevertheless, after the divergence of one clade of subg. Allium, the following major lineages were identified: (1) one lineage of subg. Cepa and subg. Reticulatobulbosa, (2) subg. Rhizirideum, and (3) one lineage of subg. Reticulatobulbosa, one lineage of subg. Allium, one lineage of subg. Cepa, and two lineages of subg. Polyprason. With much broader sampling, it is necessary to determine phylogenetic relationships among five subgenera to reflect their relationships in the classification system and reevaluate key characteristics in the context of the new phylogenetic framework.
We estimated the crown age of the genus Allium at 23.65 Ma (95% HPD, 11.97–35.31 Ma), which is similar to an earlier estimate at 22.18 Ma (95% HPD, 15.23–34.81 Ma) (Xie et al., 2020) (Figure 5). The crown ages of the three evolutionary lines were estimated to be 20.13 Ma (first line), 13.88 Ma (second line), and 17.05 Ma (third line), which were roughly the same time scale as in previous studies (Xie et al., 2020). These suggest that common ancestors of three evolutionary lines existed during the middle to early Miocene. We further provided some other important divergent times estimated in this study, and they included (1) the split of subg. Microscordum from subg. Amerallium at 17.98 Ma (95% HPD, 7.38–27.81 Ma), (2) crown age of 13. 75 Ma (95% HPD, 5.89–21.96 Ma) for subg. Amerallium, (3) crown age of 13.08 Ma (95% HPD, 5.81–21.50 Ma) for the clade of subgenera Caloscordum and Melanocrommyum, (4) crown age of 10.72 Ma (95% HPD, 3.99–18.70 Ma) for subg. Butomissa, (5) crown age of 7.47 Ma (95% HPD, 2.87–13.34 Ma) for subg. Cyathophora, and (6) crown age of 5.49 Ma (95% HPD, 2.47–8.34 Ma) for the major lineage of subg. Polyprason (sect. Daghestanica). These estimates suggest that major subgenera divergence or speciation events occurred during the Miocene, providing useful information for future studies on subgenera and sections.
4.3 Species relationships within subg. Anguinum
In this study, we confirmed the two major lineages within subg. Anguinum (Herden et al., 2016) (Figure 4A). The novel finding, based on the plastome phylogeny, was the phylogenetic position of A. tricoccum, which exclusively occurs in eastern North America. Unlike previous studies, two accessions of A. tricoccum formed monophyly (100% BS) and clearly belonged to the Eurasian lineage (Group A; 100% BS). These results support the recognition of the Eurasian-American lineage and East Asian lineage but refute the phylogenetic incongruence between nuclear ITS and chloroplast. The previous chloroplast phylogeny based on three non-coding regions (rpl32-trnL spacer, rps16 intron, and atpB-rbcL spacer) showed that A. tricoccum is sister to A. prattii from China, which belongs to the East Asian lineage (group B). The cloning of nrDNA ITS sequences found three ribotypes, that is, A. tricoccum ribotype, the East Asian (Group B) ribotype, and the Eurasian-American (group A) ribotype, and subsequently suggested a hybridization event between the A and B groups (Herden et al., 2016). The frequencies of three ribotypes showed that all but one clone belonged to group A: the first type (two clones) belongs to A. tricoccum (group A), the second type (one clone) belongs to group B, and the third type (13 clones) is related to group A. Therefore, according to the current robust placement of A. tricoccum in group A, incomplete homogenization of different ribotypes based on concerted evolution (i.e., deep coalescence, incomplete lineage sorting) is a more likely explanation than reticulation event between the two groups (Feliner and Rossellö, 2012). Consequently, the congruent tree topology between nuclear and chloroplast phylogeny suggested that the intercontinental disjunction must have occurred soon after the initial divergence between the two major groups of subg. Anguinum and further confirmed the Eurasian-American lineage (Herden et al., 2016). After the initial disjunction event in group A, the lineage of A. ulleungense on Ulleung Island and Chinese A. listera diverged from the remaining lineage, which subsequently diversified to three extant species (A. victorialis, A. ochotense, and A. microdictyon).
There has been considerable confusion on the taxonomy in the subg. Anguinum due to geographical distribution patterns of three species: A. victorialis, A. microdictyon, and A. ochotense (Herden et al., 2016). Nevertheless, those three species are unanimously accepted based on the current species concept and distribution areas given by Prokhanov (1930), with a recent new species recognition of A. ulleungense on Ulleung Island from more widely distributed A. ochotense (Choi et al., 2019). Previous phylogenetic studies provided very few resolutions to fully understand species boundaries and phylogenetic relationships. For example, the ITS tree showed four major clades (A. ulleungense, A. tricoccum, A. victorialis, and A. ochotense–A. microdictyon–A. listera), whereas the combined cpDNA showed A. ulleungense, A. microdictyon, and A. victorialis–A. ochotense (Herden et al., 2016; Choi et al., 2019). The complete plastome sequences in this study suggest the possibility of non-monophyly of two species, A. ochotense and A. microdictyon and that A. microdictyon shares its most common ancestor with A. ochotense, which is geographically much closer than A. victorialis (strictly European) (Figure 4A). The close relationship between A. ochotense and A. microdictyon is further corroborated by the ITS sequences: two species are part of a highly unresolved clade, such as A. listera (88% BS and 0.98 PP), and A. victorialis is sister to this highly unresolved clade (98% BS and 0.99 PP) (Herden et al., 2016).
One more important finding of this study is that A. ulleungense, endemic to Ulleung Island, Korea, is a taxonomically distinct entity (Figure 4A). It was previously recognized as either A. victorialis (Yu et al., 1981) or A. ochotense (Choi and Oh, 2011). Although it also resembles A. microdictyon morphologically, A. ulleungense has much broader leaves, larger whitish perianth, and is diploid (2n = 2x = 16) (Choi et al., 2019). The current study, which is based on the complete plastomes, suggests a sister relationship between A. ulleungense and A. listera, which are both diploid (2n = 2x = 16) (Jing et al., 1999). This clade is in turn sister to the clade containing Eurasian species (A. victorialis, A. microdictyon, and A. ochotense), and because of the phylogenetic incongruence between previous ITS trees (Herden et al., 2016; Choi et al., 2019) and our current plastome tree, further study is required to gain insights into ITS ribotype evolution and determine the precise phylogenetic position of A. ulleungense. Based on the complete plastome sequences in this study, slightly younger crown age of subg. Anguinum at 4.85 Ma (95% HPD, 2.08–8.32 Ma) was estimated compared with that based on ITS sequences (5.44 Ma, 95% HPD, 2.13–9.4 Ma; Herden et al., 2016). The intercontinental disjunct event and the split of Ulleung Island endemic A. ulleungense from its continental sister species were estimated to be 2.71 Ma (95% HPD, 1.00–4.86 Ma) and 1.57 Ma (95% HPD, 0.05–4.43 Ma), which occurred in late Pliocene (Piacenzian) and mid Pleistocene (Calabrian), respectively. Further study is required to (1) test the monophyly of each species and their phylogenetic relationships, (2) assess the polyploidization process in A. ochotense (2n = 4x = 32), and (3) understand geographical disjunct distribution patterns of A. microdictyon in Caucasus, Siberia (from West-to-Central and East Siberia), Southern Ural area, North Mongolia, Kazakhstan, and Korea.
4.4 Evolution within subg. Rhizirideum
The inter-sectional and species relationships within subg. Rhizirideum were not well resolved in previous studies (Nguyen et al., 2008; Li et al., 2010; Sinitsyna et al., 2016; Jang et al., 2021). Although the position of sect. Eduardia in the third evolutionary line of Allium is yet to be determined, it appears that two sections, Tenuissima and Rhizirideum, are monophyletic (Li et al., 2010). The current study, which is based on the complete plastome sequences, showed that sect. Caespitosoprason appears not to be monophyletic, but two major groups within the monophyletic sect. Rhizirideum, “Asiatic” and “European” groups (Sinitsyna et al., 2016), are strongly recognized (100% BS each), except for A. nutans (Figure 4B). It was shown that within the Asiatic group, three subgroups were inferred based on two chloroplast region sequences (trnQ-rps16 and trnL-rpl32; Sinitsyna et al., 2016): (1) unresolved A. spirale (2x)–A. spurium (4x), (2) clade of A. nutans (4x)–A. senescens (4x and 6x)–A. azutavicum (unknown ploidy) (0.88 PP and 68% BS), and (3) A. austrosibiricum (2x)–A. rubens (4x)–A. tytthocephalum (4x)–A. tuvinicum (2x)–A. prostratum (2x)–A. minus (2x)–A. burjaticum (4x)–A. stellerianum (4x) (0.99 PP and 78% BS). ITS phylogeny, which is based on diploid species only, showed that very little was resolved within the Asiatic group, other than three species, A. austrosibiricum, A. prostratum, and A. minus, which shared their most recent common ancestor (0.97 PP and 80% BS). Several incongruences between ITS and chloroplast phylogeny were as follows: (1) A. nutans (4x) is closely related to A. senescens (4x, 6x)/A. azutavicum in the cpDNA tree (0.86 PP and 62% BS), but it is sister to A. austrosibiricum (2x) in the ITS tree (0.89 PP and 89% BS); (2) A. tyttocephalum (4x) is closely related to A. rubens (2x) in the cpDNA tree (0.86 PP and 62% BS), but it is unresolved in the ITS tree; and (3) A. minus is closely related to some accessions of A. prostratum (2x) (0.88 PP and 58% BS), but it is unresolved in the ITS tree (Sinitsyna et al., 2016). The narrow Korean endemic A. minus is sister to A. senescens (Mongolian accession) in this study, suggesting its potential origin from wild onion native to Mongolia. Allium minus, which occurs in the type locality only (Taegisan Mountain, Walhaksan Mountain, Gangwon-do Province, and Yangju, Gyeonggi-do Province) in Korea, was initially recognized as a variety of A. senescens (Yu et al., 1981). Based on the detailed phylogeographic study, it remains to be determined as to how diploid A. minus in Korea is related to tetraploid and hexaploid A. senescens outside of the Korean Peninsula. Recently, populations of A. senescens on Ulleung Island were described as a new tetraploid endemic species (A. dumebuchum) in Korea (Jang et al., 2021). Owing to its distinct diagnostic features (bigger floral parts and late flowering time) from its closely related species (i.e., A. spirale, A. spurium, A. minus, and A. senescens), A. dumebuchum was described as a new taxon; however, its phylogenetic position was unclear (Jang et al., 2021). The current plastome tree suggests strongly that A. dumebucum is sister to the clade containing A. austrosibiricum, A. senescens, and A. minus, suggesting that the former diverged earlier than the three latter ones. The split of A. dumebucum from the continental lineage was estimated to be 0.29 Ma (95% HPD, 0.08–0.59 Ma) in Pleistocene (Ionian), much younger than the other Ulleung Island endemic A. ulleungense (1.54 Ma). The crown age of subg. Rhizirideum was estimated to be 8.41 Ma (95% HPD, 3.85–13.49 Ma) in the late Miocene, much older than that of subg. Anguinum. This estimate is comparable with that based on ITS sequences (7.15 Ma; Sinitsyna et al., 2016). However, the split between “Asian” and “European” groups in this study was estimated to be much younger at 2.18 Ma (95% HPD, 0.75–4.13 Ma) compared with that based on ITS sequences (ca. 3.97 Ma, 95% HPD, 1.13–6.91 Ma; Sinitsyna et al., 2016). Moreover, during the Pleistocene, interspecific hybridization and polyploidization may have led to the diversification of Allium species in sect. Rhizirideum.
4.5 Phylogenetic position of sect. Condensatum
As one of the morphologically “difficult” species in variable sections of Scorodon, Reticulatobulbosa, and Oreiprason (Friesen et al., 2006), the phylogenetic position of A. condensatum (Figure 1E), which occurs from eastern Siberia and Mongolia to North Korea and Russian Far East, has been elusive. Based on one of the four major clades in sect. Oreiprason, the east Asian monotypic section Condensatum was newly recognized along other sections (Cepa, Schoenoprason sensu stricto, Annuloprason, and Sacculiferum) in polyphyletic subg. Cepa (Friesen et al., 2006; Li et al., 2010). However, previous phylogenetic studies have shown the uncertain position of A. condensatum. For example, the nuclear ITS sequence-only tree showed that A. condensatum is sister to the clade containing several sections of subg. Allium (0.52 PP and <50% BS; Li et al., 2010) or highly unresolved compared with other subgenera (Friesen et al., 2006; Nguyen et al., 2008). In addition, the combined nuclear ITS and chloroplast rps16 sequences, which focused on Chinese Allium, placed A. condensatum in the highly unresolved clade of the third evolutionary lineage, such as subgenera Cepa, Polyprason, Allium, and Reticulatobulbosa (Li et al., 2010). Alternatively, the combined and nuclear ITS and chloroplast trnL–trnF, which focused on Korean/northeastern Chinese/Canadian representative species, placed A. condensatum sister to the clade of subg. Allium (<50% BS; Choi et al., 2012). All the previous studies highlighted the uncertain phylogenetic position of A. condensatum, and for the first time, based on the complete plastome sequences, we demonstrated that A. condensatum is closely related to A. chinense, which is widely cultivated as a vegetable in tropical and subtropical China (100% BS). It was also estimated that the two species shared their most recent common ancestor at 7.62 Ma (95% HPD 3.02–12.36 Ma) in the late Miocene. Although the sister relationship is strongly supported based on the current study, the two species have numerous differences, such as perianth color (pale purple to dull purple for A. chinense vs. pale yellow for A. condensatum) and scape (lateral, 20–40 cm in length for A. chinense vs. central, 30–80 cm for A. condensatum), and they are closely related to the species of subg. Reticulatobulbosa (91% BS) (Figure 4B). It was not possible to verify the voucher of A. chinense (NC043922; Yang et al., 2019), and this close relationship could be due to the potential sampling limitation of the current study. Therefore, this preliminary placement of A. condensatum close to subg. Cepa and sect. Sacculiferum rather than Allium species from sect. Cepa. is still tentative and remains to be determined based on extensive sampling of Allium plastomes.
4.6 Phylogenetic position of A. macrostemon
Allium macrostemon (Figures 1Q–T) occurs commonly in sunny lowland meadows, forest margins, and mountain foothills of Russia (Far East), Mongolia, China, Taiwan, Korea, and Japan. The current study showed that A. macrostemon (at least based on accessions from China and Korea) is monophyletic and shared its most recent common ancestor with A. caeruleum (subg. Allium sect. Caerulea) and A. schoenoprasoides (subg. Allium sect. Pallasia). This Allium clade is distinct from the second Allium clade, which includes A. ferganicum, A. ampeloprasum, and A. sativum (Figure 4B). Allium macrostemon was treated as a member of subg. Allium sect. Allium (Li et al., 2010), but potential recognition under a new section was suggested (Friesen et al., 2006). Allium macrostemon shares similar testa sculptures and morphological characteristics (that is, bulbil development in inflorescence, pistil morphology, and seed shape) with members of sect. Caerulea, especially with A. caeruleum (Choi et al., 2012). As two sister lineages were recognized at the sectional level (Caerulea and Pallasia), it is reasonable to propose a new section for A. macrostemon, which is restricted to East Asia. Furthermore, molecular dating suggests that the divergence of A. macrostemon from the central Asian lineage occurred earlier (4.55 Ma, 95% HPD 1.87–7.54 Ma) than the split between the two sections of Caerulea and Pallasia from Central Asia (Figure 5). Thus, in this study, we newly described a monotypic section with a type species of A. macrostemon that occurs in East Asia.
Allium subgen. Allium sect. Macrostemon S.-C.Kim & H.J.Choi, sect. nov.-TYPE: A. macrostemon Bunge
Description. Bulbs solitary with short rhizome, subglobose, sometimes with bulbels; tunics membranous. Leaves fragile, linear, minutely angular, adaxially channeled, hollow in cross section. Umbel globose, sometimes replaced totally or partially by pseudoviviparous bulbils. Perianth widely spreading, whitish pink; inner tepals slightly longer than outer ones; filaments slightly exserted, subulate; anthers elliptical; ovary subcubical, green, with hood-like projections at base; ovules two per locule; style exerted; stigma smooth. Capsules trigonous; seeds oval, flat in cross section.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, OP743930-OP743948 https://www.ncbi.nlm.nih.gov/genbank/, OP754898.
Author contributions
H-JC and S-CK conceived and designed the study; JY and S-HK conducted the experiment; H-JC, H-YG, and S-CK provided experimental support; JY and S-HK prepared the original draft; and H-YG, H-JC, and S-CK revised and finalized the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by research grants from the Korea National Arboretum (Grant No.: KNA1-1-26, 20-1).
Acknowledgments
We thank the reviewers for their constructive and insightful suggestions on the earlier version of the manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1124277/full#supplementary-material
References
Bi, Y., Zhang, M.-F., Xue, J., Dong, R., Du, Y.-P., Zhang, X.-H. (2018). Chloroplast genomic resources for phylogeny and DNA barcoding: a case study on Fritillaria. Sci. Rep. 8, 1184. doi: 10.1038/s41598-018-19591-9
Chang, H., Zhang, L., Xie, H., Liu, J., Xi, Z., Xu, X. (2021). The conservation of chloroplast genome structure and improved resolution of infrafamilial relationships of Crassulaceae. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.631884
Chen, J., Xie, D., He, X., Yang, Y., Li, X. (2022). Comparative analysis of the complete chloroplast genomes in Allium section Bromatorrhiza species (Amaryllidaceae): phylogenetic relationship and adaptive evolution. Genes 13, 1279. doi: 10.3390/genes13071279
Choi, H. J., Giussani, L. M., Jang, C. G., Oh, B. U., Costa-Sánchez, J. H. (2012). Systematics of disjunct northeastern Asian and northern north American Allium (Amaryllidaceae). Botany 90, 491–508. doi: 10.1139/b2012-031
Choi, H. J., Oh, B. U. (2011). A partial revision of Allium (Amaryllidaceae) in Korea and north-eastern China. Bot. J. Lin. Soc 167, 153–211. doi: 10.1111/j.1095-8339.2011.01166.x
Choi, H. J., Yang, S., Yang, J. C., Friesen, N. (2019). Allium ulleungense (Amaryllidaceae), a new species endemic to Ulleungdo Island, Korea. Korean J. Pl. Taxon. 49, 294–299. doi: 10.11110/kjpt.2019.49.4.294
Dierckxens, N., Mardulyn, P., Smits, G. (2016). NOVOPlasty: De novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 4, e18. doi: 10.1093/nar/gkw955
Dong, W., Liu, J., Yu, J., Wang, L., Zhou, S. (2012). Highly variable chloroplast markers for evaluating plant phylogeny at low taxonomic levels and for DNA barcoding. PloS One 7, e35071. doi: 10.1371/journal.pone.0035071
Downie, S. R., Jansen, R. K. (2015). A comparative analysis of whole plastid genomes from the Apiales: expansion and contraction of the inverted repeat, mitochondrial to plastid transfer of DNA, and identification of highly divergent noncoding regions. Syst. Bot. 40, 336–351. doi: 10.1600/036364415X686620
Drummond, A. J., Ho, S. Y., Phillips, M. J., Rambaut, A. (2006). Relaxed phylogenetics and dating with confidence. PloS Biol. 4, 288. doi: 10.1371/journal.pbio.0040088
Drummond, A. J., Suchard, M. A., Xie, D., Rambaut, A. (2012). Bayesian Phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 29, 1969–1973. doi: 10.1093/molbev/mss075
Druselmann, S. (1992). Vergleichende untersuchungen an vertretern der alliaceae agardh. 1. morphologie der keimpflanzen der gattung Allium l. Flora 186, 37–52. doi: 10.1016/S0367-2530(17)30519-4
Fay, M. F., Chase, M. W. (1996). Resurrection of themidaceae for the Brodiaea alliance, and recircumscription of Alliaceae, Amaryllidaceae and Agapanthoideae. Taxon 45, 441–451. doi: 10.2307/1224136
Feliner, G. N., Rossellö, J. A. (2012). “Concerted evolution of multigene families and homeologous recombination,” in Plant genome diversity, vol. 1 . Eds. Wendel, J., Greihuber, J., Dolezel, J., Leitch, I. (Vienna, Austria: Springer). doi: 10.1007/978-3-7091-1130-7_12
Felsenstein, J. (1985). Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39, 783–791. doi: 10.2307/2408678
Friesen, N. (1988). Lukovye sibiri: sistematika, kariologiia, khorologiia (Novosibirsk, Russia: Nauka-Sibirskoe otd).
Friesen, N., Fritsch, R. M., Blattner, F. R. (2006). Phylogeny and intrageneric classification of Allium (Alliaceae) based on nuclear ribosomal DNA ITS sequences. Aliso 22, 372–395. doi: 10.5642/aliso.20062201.31
Friesen, N., Zuev, V. V., Aljanskaja, N. S. (1986). “Krasivoluk nereidocvetnyj – calloscordum neriniflorum Herbert,” in Biologicheskie osobennosti rastenij sibiri, buzhdajushhikhsja v okhrane. Ed. Sobolevskaja, K. A. (Novosibirsk, Russia: Nauka-Sibirskoe Otd), 83–92.
Fritsch, R. M. (1988). Anatomical investigation of the leaf blade of Allium L. (Alliaceae) – I. Species having one row of vascular bundles. Flora 181, 83–100. doi: 10.1016/S0367-2530(17)30354-7
Fritsch, R. M. (1992a). “Infra-subgeneric grouping in subgenus melanocrommyum (Webb et Berth) Rouy,” in The genus allium-taxonomic problems and genetic resources. Eds. Hanelt, P., Hammer, K., Knüpffer, H. (IPK, Gatersleben, Germany), 67–75.
Fritsch, R. M. (1992b). Zur wurzelanatomie in der gattung Allium l. (Alliaceae). Beitr. Biol. Pflanzen 67, 129–160.
Fritsch, R. M. (1993). Anatomische merkmale des blütenschaftes in der gattung Allium l. und ihre systematische bedeutung. Bot. Jahrb. Syst. 115, 97–131.
Fritsch, R. M., Blattner, F. R., Gurushidze, M. (2010). New classification of Allium l. subg. Melanocrommyum (Webb & Berthel) Rouy (Alliaceae) based on molecular and morphological characters. Phyton. 49, 145–220.
Fritsch, R. M., Friesen, N. (2002). “Evolution, domestication and taxonomy,” in Allium crop science: recent advances. Eds. Rabinowitch, H. D., Currah, L. (Wallingford, UK: CABI Publishing), 5–30.
Greiner, S., Lehwark, P., Bock, R. (2019). Organellar genome DRAW (OGDRAW) version 1.3.1: Expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 47, W59–W64. doi: 10.1093/nar/gkz238
Hanelt, P. (1992). “Ovule number and seed weight in the genus Allium I,” in The genus Allium: taxonomic problems and genetic resources. Eds. Hanelt, P., Hammer, K., Knüpffer, H. (IPK, Gatersleben, Germany), 99–105.
Henriquez, C. L., Abdullah, Ahmed, I., Carlsen, M. M., Zuluaga, A., Croat, T. B., et al. (2020). Evolutionary dynamics of chloroplast genomes in subfamily Aroideae (Araceae). Genomics 112, 2349–2360. doi: 10.1016/j.ygeno.2020.01.006
Herden, T., Hanelt, P., Friesen, N. (2016). Phylogeny of Allium l. subgenus Anguinum (G. don. ex W.D.J. Koch) n. friesen (Amaryllidaceae). Mol. Phylogenet. Evol. 95, 79–93s. doi: 10.1016/j.ympev.2015.11.004
Huang, D.-Q., Yang, J.-T., Zhou, C.-J., Zhou, S.-D., He, X.-J. (2014). Phylogenetic reappraisal of Allium subgenus Cyathophora (Amaryllidaceae) and related taxa, with a proposal of two new sections. J. Plant Res. 127, 275–286. doi: 10.1007/s10265-013-0617-8
Huo, Y., Gao, L., Liu, B., Yang, Y., Kong, S., Sun, Y., et al. (2019). Complete chloroplast genome sequences of four Allium species: comparative and phylogenetic analyses. Sci. Rep. 9, 12250. doi: 10.1038/s41598-019-48708-x
Jang, J. E., Park, J. S., Jung, J. Y., Kim, D. K., Yang, S. Y., Choi, H. J. (2021). Notes on Allium section Rhizirideum (Amaryllidaceae) in south Korea and northeastern China: with a new species from Ulleungdo Island. PhytoKeys 176, 1–19. doi: 10.3897/phytokeys.176.63378
Jin, Y., Zhang, T., Liu, B., Zheng, C., Huo, H., Zhang, J. (2022). Comparative and phylogenetic analysis of the complete chloroplast genome sequences of Allium mongolicum. Sci. Rep. 12, 21676. doi: 10.1038/s41598-022-26354-0
Jing, W. C., Xu, J. M., Yang, L. (1999). A study on cytotaxonomy of sect. Anguinum of Allium. Acta Phytotax. Sin. 37, 20–34.
Kalyaanamoorthy, S., Minh, B. Q., Wong, T. K., von Haeseler, A., Jermiin, L. S. (2017). Fast model selection for accurate phylogenetic estimates. Nat. Methods 14, 587–589. doi: 10.1038/nmeth.4285
Katoh, K., Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Kearse, M., Moir, R., Wilson, A., Stones-Havas, S., Cheung, M., Sturrock, S. (2012). Geneious basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. J. Bioinform. 28, 1647–1649. doi: 10.1093/bioinformatics/bts199
Kruse, J. (1984). Rasterelektronenmikroskopische untersuchungen an samen der gattung Allium l. III. Kulturpflanze 36, 355–368. doi: 10.1007/BF02034817
Li, Q.-Q., Zhou, S.-D., He, X.-J., Yu, Y., Zhang, Y.-C., Wei, X.-Q. (2010). Phylogeny and biogeography of Allium (Amaryllidaceae: Allieae) based on nuclear ribosomal internal transcribed spacer and chloroplast rps16 sequences, focusing on the inclusion of species endemic to China. Ann. Bot. 106, 709–733. doi: 10.1093/aob/mcq177
Lowe, T. M., Eddy, S. R. (1997). tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25, 955–964. doi: 10.1093/nar/25.5.955
Namgung, J., Do, H. D. K., Kim, C., Choi, H. J., Kim, J.-H. (2021). Complete chloroplast genomes shed light on phylogenetic relationships, divergence time, and biogeography of Allioideae (Amaryllidaceae). Sci. Rep. 11, 3262. doi: 10.1038/s41598-021-82692-5
Nguyen, N. H., Driscoll, H. E., Specht, C. D. (2008). A molecular phylogeny of the wild onions (Allium; Alliaceae) with a focus on the western north American center of diversity. Mol. Phylogenet. Evol. 47, 1157–1172. doi: 10.1016/j.ympev.2007.12.006
Nguyen, L. T., Schmidt, H. A., von Haeseler, A., Minh, B. Q. (2015). IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenetics. Mol. Biol. Evol. 32, 268–274. doi: 10.1093/molbev/msu300
Nylander, J. A. A. (2004). MrModeltest v2. program distributed by the author (Uppsala, Sweden: Evolutionary Biology Center, Uppsala University).
Pastor, J., Valdes, B. (1985). Bulb structure in some species of Allium (Liliaceae) of the Iberian peninsula. Ann. Mus. Goulandris 7, 249–261. doi: 10.15406/hij.2020.04.00150
Pistrick, K. (1992). “Phenological variability in the genus Allium l,” in The genus Allium – taxonomic problems and genetic resources. Eds. Hanelt, P., Hammer, K., Knüpffer, H. (IPK, Gatersleben, Germany), 243–249.
Rambaut, A., Drummond, A. J., Xie, D., Baele, G., Suchard, M. A. (2018). Posterior summarization in Bayesian phylogenetics using tracer 1.7. Syst. Biol. 67, 901–904. doi: 10.1093/sysbio/syy032
Ronquist, F., Teslenko, M., van der Mark, P., Ayres, D. L., Darling, A., Hönha, S., et al. (2012). MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. doi: 10.1093/sysbio/sys029
Rozas, J., Ferrer-Mata, A., Sánchez-DelBarrio, J., Guirao-Rico, S., Librado, P., Ramos-Onsins, S. E. (2017). DnaSP v6: DNA sequence polymorphism analysis of large datasets. Mol. Biol. Evol. 34, 3299–3302. doi: 10.1093/molbev/msx248
Scobeyeva, V. A., Artyushin, I. V., Krinitsina, A. A., Nikitin, P. A., Antipin, M. I., Kuptsov, S. V., et al. (2021). Gene loss, pseudogenization in plastomes of genus Allium (Amaryllidaceae), and putative selection for adaptation to environmental conditions. Front. Genet. 12, 674783. doi: 10.3389/fgene.2021.674783
Sheikh-Assadi, M., Naderi, R., Kafi, M., Fatahi, R., Salami, S. A., Shariati, V. (2022). Complete chloroplast genome of Lilium ledebourii (Baker) Boiss and its comparative analysis: lights into selective pressure and adaptive evolution. Sci. Rep. 12, 9375. doi: 10.1038/s41598-022-13449-x
Sinitsyna, T. A., Herden, T., Friesen, N. (2016). Dated phylogeny and biogeography of the Eurasian Allium section Rhizirideum (Amaryllidaceae). Plant Syst. Evol. 302, 1–18. doi: 10.1007/s00606-016-1333-3
Suchard, M. A., Lemey, P., Baele, G., Ayres, D. L., Drummond, A. J., Rambaut, A. (2018). Bayesian Phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evol. 4, vey016. doi: 10.1093/ve/vey016
Swofford, D. L. (2003). PAUP*. phylogenetic analysis using parsimony (*and other methods) (Sunderland: Sinauer Associates).
Takhtajan, A. L. (1997). Diversity and classification of flowering plants (New York, USA: Columbia University Press), 643.
Tiffney, B. H. (1985). The Eocene north Atlantic land bridge: its importance in tertiary and modern phytogeography of the northern hemisphere. J. Arnold Arbor. 66, 243–273. doi: 10.5962/bhl.part.13183
Tiffney, B. H., Manchester, S. R. (2001). The use of geological and paleontological evidence in evaluating plant phylogeographic hypotheses in the northern hemisphere tertiary. Int. J. Plant Sci. 162, S3–S17. doi: 10.1086/323880
Trofimov, D., Cadar, D., Schmidt-Chanasit, J., Rodrigues de Moraes, P. L., Rohwer, J. G. (2022). A comparative analysis of complete chloroplast genomes of seven Ocotea species (Lauraceae) confirms low sequence divergence within the Octotea complex. Sci. Rep. 12, 1120. doi: 10.1038/s41598-021-04635-4
Wheeler, E. J., Mashayekhi, S., McNeal, D. W., Columbus, J. T., Pires, J. C. (2013). Molecular systematics of Allium subgenus Amerallium (Amaryllidaceae) in north America. Am. J. Bot. 100, 701–711. doi: 10.3732/ajb.1200641
Xie, D.-F., Cheng, R.-Y., Price, M., Chen, J.-P., Lei, J.-Q., Zhang, Y.-Y., et al. (2022). Allium heterophyllum (Amaryllidaceae), a new species from henan, China. PhytoKeys 190, 53–67. doi: 10.1897/phytokeys.190.77449
Xie, D.-F., Tan, J.-B., Yu, Y., Gui, L.-J., Su, D.-M., Zhou, S.-D., et al. (2020). Insights into phylogeny, age and evolution of Allium (Amaryllidaceae) based on the whole plastome sequences. Ann. Bot. 125, 1039–1055. doi: 10.1093/aob/mcaa024
Xie, D.-F., Yu, H.-X., Price, M., Xie, C., Deng, Y.-A., Chen, J.-P., et al. (2019). Phylogeny of Chinese Allium species in section Daghestanica and adaptive evolution of Allium (Amaryllidaceae, Allioideae) species revealed by the chloroplast complete genome. Front. Plant Sci. 10, 460. doi: 10.3389/fpls.2019.00460
Xie, F.-M., Jiang, Q.-P., Yu, Y., Zhou, S.-D., He, X.-J. (2019). The complete chloroplast genome of Allium macrostemon. Mitochondrial DNA B: Resour. 4, 1938–1939. doi: 10.1080/23802359.2019.1616626
Yang, J. Y., Choi, M.-J., Kim, S.-H., Choi, H.-J., Kim, S.-C. (2021). Plastome characterization and phylogenomic analysis yield new insights into the evolutionary relationships among the species of the subgenus Bryocles (Hosta; Asparagaceae) in East Asia. Plants 10, 1980. doi: 10.3390/plants10101980
Yang, Q.-Q., Jiang, M., Wang, L.-Q., Chen, H.-M., Liu, C., Huang, L.-F. (2019). Complete chloroplast genome of Allium chinense: comparative genomic and phylogenetic analysis. Acta Pharm. Sin. 54, 173–181.
Yang, J. Y., Kim, S.-H., Pak, J.-H., Kim, S.-C. (2022). Infrageneric plastid genomes of Cotoneaster (Rosaceae): Implications for the plastome evolution and origin of C. wilsonii on ulleung island. Genes 13, 728. doi: 10.3390/genes13050728
Yang, X., Xie, D.-F., Chen, J.-P., Zhou, S.-D., Yu, Y., He, X.-J. (2020). Comparative analysis of the complete chloroplast genomes in Allium subgenus Cyathophora (Amaryllidaceae): phylogenetic relationships and adaptive evolution. BioMed. Res. Int. 2020, 1732586. doi: 10.1155/2020/1732586
Yu, S. O., Lee, S., Lee, W. T. (1981). A taxonomic study on the Allium species in Korea. Korean J. Pl. Taxon. 11, 21–41. doi: 10.11110/kjpt.1981.11.1.021
Keywords: Allium ulleungense, Allium dumebuchum, Ulleung Island, plastome evolution, infrageneric classification
Citation: Yang J, Kim S-H, Gil H-Y, Choi H-J and Kim S-C (2023) New insights into the phylogenetic relationships among wild onions (Allium, Amaryllidaceae), with special emphasis on the subgenera Anguinum and Rhizirideum, as revealed by plastomes. Front. Plant Sci. 14:1124277. doi: 10.3389/fpls.2023.1124277
Received: 15 December 2022; Accepted: 01 March 2023;
Published: 21 March 2023.
Edited by:
Francesco Mercati, Institute of Bioscience and Bioresources (CNR), ItalyReviewed by:
Shih-Hui Liu, National Sun Yat-sen University, TaiwanYun-peng Du, Beijing Academy of Agricultural and Forestry Sciences, China
Copyright © 2023 Yang, Kim, Gil, Choi and Kim. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hyeok-Jae Choi, aGpjaG9pMTk3NUBjaGFuZ3dvbi5hYy5rcg==; Seung-Chul Kim, c29uY2h1czk2QHNra3UuZWR1
 JiYoung Yang
JiYoung Yang Seon-Hee Kim
Seon-Hee Kim Hee-Young Gil
Hee-Young Gil Hyeok-Jae Choi
Hyeok-Jae Choi Seung-Chul Kim
Seung-Chul Kim