
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 27 March 2023
Sec. Plant Abiotic Stress
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1123856
This article is part of the Research Topic Dealing With Salinity Stress: Understanding the Mechanism of Plant Adaptation and Resistance View all 7 articles
 Mengchao Zhang1,2
Mengchao Zhang1,2 Xueli Lu2
Xueli Lu2 Tingting Ren2
Tingting Ren2 Prince Marowa3
Prince Marowa3 Chen Meng2
Chen Meng2 Juying Wang4
Juying Wang4 Hui Yang4
Hui Yang4 Chunhua Li5
Chunhua Li5 Li Zhang1*
Li Zhang1* Zongchang Xu2*
Zongchang Xu2*Salt stress is a serious abiotic stress that primarily inhibits plant growth, resulting in severe yield losses. Our previous research found that flavonoids play important roles in A. venetum salt stress tolerance. In response to salt stress, we noted that the flavonoid content was depleted in A. venetum. However, the detailed mechanism is still not clear. In this study, the expression patterns of three flavonoids synthetase genes, AvF3H, AvF3’H, and AvFLS were systemically analyzed under salt stress in A. venetum seedlings. The salt tolerance of transgenic Arabidopsis plants was improved by heterologous overexpression of these synthetase genes. The NBT and DAB staining results as well as H2O2 and O2•- content analysis revealed that under salt stress, ROS molecules were reduced in transgenic plants compared to WT plants, which corresponded to the activation of the antioxidant enzyme system and an increase in total flavonoid content, particularly rutin, eriodictyol, and naringerin in transgenic plants. External application of flavonoids reduced ROS damage in WT plants just like what we observed in the transgenic plants (without the external application). Additionally, our transcriptome analysis demonstrated that auxin and jasmonic acid biosynthesis genes, as well as signaling transduction genes, were primarily activated in transgenic plants under salt stress, leading to activation of the cell wall biosynthesis or modification genes that promote plant growth. As a result, we investigated the mechanism through flavonoids enhance the salt tolerance, offering a theoretical foundation for enhancing salt tolerance in plants.
Various environmental stresses, such as strong winds, extreme temperatures, soil salinity, drought, and floods, have impacted agricultural crop production and cultivation. Soil salinity is one of the most devastating environmental stresses, causing significant reduction in cultivated land area, crop productivity, and crop quality (Shahbaz and Ashraf, 2013). High salinity is estimated to affect 20% of total cultivated and 33% of irrigated agricultural lands worldwide. Furthermore, the salinized areas are increasing at a rate of 10% annually due to various reasons, including low precipitation, high surface evaporation, native rocks weathering, poor irrigation systems and irrigation with saline water as well as due to poor agronomic practices (Shrivastava and Kumar, 2015). It has been estimated that more than 50% of the arable land would be salinized by the year 2050. Average yields for all important crops are only a fraction of record yields, ranging between 20% and 50% (Shrivastava and Kumar, 2015) and these losses are primarily due to drought and high soil salinity, environmental conditions that will worsen in many regions due to global climate change.
Salt stress has complex negative effects on many aspects of plant growth, including morphological, physiological, and biochemical processes (Akbarimoghaddam et al., 2011). In general, the two catastrophic primary effects of salt stress are physiological drought (osmotic stress) and ion toxic effects caused by ionic imbalance (Keisham et al., 2018), which could rapidly lead to cell death (Munns, 2002). Similarly, salt-tolerant plants have evolved various mechanisms to resist salt stress, such as the accumulation of small molecular osmoregulators to increase osmotic potential, a systematic ion transport system, and the separation of ion cellular compartments (Keisham et al., 2018). However, salt stress is always detrimental to plant growth, including ROS production (Das and Roychoudhury, 2014). Though a small amount of ROS can act as signaling molecules, coordinating an incredible range of diverse plant processes (Saed-Moucheshi et al., 2014), excessive ROS levels can cause cell death (Gill and Tuteja, 2010). Plants have evolved an elaborate enzymatic and non-enzymatic antioxidant system that, in conjunction with the ROS-producing enzymes, maintains ROS homeostasis in all cellular compartments to reduce the fatal damage caused by excessive ROS and to ensure the accurate execution of their signaling functions. SOD, catalase (CAT), ascorbate peroxidase (APX), and glutathione peroxidase (GPX) are the most common enzymatic systems (Apel and Hirt, 2004). Among the enzymatic systems, SOD can quickly convert O2•- to H2O2, and the generated H2O2 is then converted to water and dioxygen by peroxidase and CAT (Gechev et al., 2006; Mittler, 2017). The non-enzymatic systems are mainly mediated by low molecular mass antioxidants, such as glutathione, ascorbic acid (AsA) and flavonoids, which are known to remove hydroxyl radicals and singlet oxygen (Gechev et al., 2006).
Flavonoids are a diverse group of plant secondary metabolites that are widely distributed throughout the plant kingdom. They are produced as a protective response to environmental stresses as a member of non-enzymatic systems. Flavonoid biosynthesis is upregulated under a wide range of abiotic stresses, such as ultraviolet (UV) radiation (Hectors et al., 2014) and slat stress (Colla et al., 2013). A previous study reported that NaCl and UV-B treatments increased the expression of genes involved in flavonol biosynthesis as well as flavonol content in Reaumuria trigyna (Zhang et al., 2017). Furthermore, flavonoids were significantly accumulated in leaves of several varieties of artichoke and cardoon treated exposed to salt stress (Colla et al., 2013). In addition, Chen et al. (2019) pointed that exogenous rutin supplementation effectively scavenged O2•- and H2O2 and improved the salt tolerance of the rutin-reduced transgenic tobacco plants (Chen et al., 2019). All these previous studies have established and confirmed the role of flavonoids in salt tolerance.
Because of the abundance of flavonoids in Apocynum venetum leaves, the plant has been used in China to treat angiocardiopathies by regulating blood pressure, lowering blood fat, healing depression, and calming nerves (Xie et al., 2012). As a result, A. venetum has gradually evolved into an important plant used in traditional Chinese medicine with significant economic value. However, previous research found that A. venetum is not a typical halophyte because there is no suitable growth promoting salt concentration, and salt stress inhibited the normal growth and development of A. venetum seedlings and seed germination (Xu et al., 2020). Further investigation revealed that the leaf had the highest Na+ content in A. venetum, and the content of total flavonoids was negatively correlated with the content of sodium ions (Xu et al., 2021), confirming the importance of flavonoids in A. venetum salt tolerance. It was also confirmed in tobacco and Arabidopsis by overexpressing AvF3H, AvF3′H, AvFLS, and AvF3GT. Transgenic plants were more resistant to salt stress and contained more flavonoids than control plants (Wang et al., 2021; Xu et al., 2021). Though total flavonoid levels in A. venetum decreased under salt stress, some compounds, such as quercetin and kaempferol, increased significantly (Xu et al., 2020), and the enzyme synthase genes AvF3’H, AvF3H, and AvFLS showed multiple expression profiles (Xu et al., 2020; Xu et al., 2021). While we have preliminary data showing that these genes may improve tobacco and Arabidopsis thaliana’s ability to withstand salt stress under various salt stress concentrations and times, the precise mechanism by which they do so is still unknown. In this study, we systematically studied the expression profiling of major genes of flavonoid metabolic pathway in response to salt stress with the exposure time, and the mechanism of AvF3’H, AvF3H, and AvFLS enhancing plant salt tolerance was analyzed in Arabidopsis thaliana. This research highlights the roles of AvF3’H, AvF3H, and AvFLS in enhancing salinity stress tolerance, and provides a foundation for subsequent application.
Apocynum venetum seeds collected from Dongying City in China’s Shandong Province and stored in the laboratory were used in this study. In transgenic experiments, Arabidopsis thaliana Columbia-0 (Col-0) was used as the wild type. Our previous study reported on these transgenic plants (AvF3’H-OE, AvF3H-OE, and AvFLS-OE) (Xu et al., 2021).
A. venetum seeds were grown in vermiculite pots in a growth chamber at 24 °C with a 14-hour photoperiod and watered with 10% liquid MS. To investigate the expression patterns of AvF3’H, AvF3H, and AvFLS, six-week-old seedlings were watered with a 100 mM NaCl solution and the second completely spread (full grown) leaf from the top was collected at 0 min, 5 min, 15 min, 30 min, 1 h, 2 h, 4 h, 8 h, 12 h, 24 h, 48 h, and 72 h after watering with NaCl and immediately frozen in liquid nitrogen before storing them at -80 °C.
The sterile transgenic (AvF3’H-OE, AvF3H-OE, and AvFLS-OE) and wild type Arabidopsis thaliana seeds were grown in the growth chamber described above on 1/2 MS growth medium. After 10 days, some of these seedlings were transplanted into plastic trays. Three-week-old seedlings on plastic trays were evenly divided into two groups; one group was watered with water, while the other was watered with a 100 mM NaCl solution. After a week of treatment, the aerial phenotype was photographed with a digital camera (Canon 5D Mark III, Japan) and data was collected with Image J software. The seedlings were immediately frozen in liquid nitrogen and stored at -80 °C for further analysis, which included total flavonoid and compound composition content analysis, Na+ and K+ content detection, antioxidant enzymes activity test, ROS staining, and transcriptome analysis. The remainder of the original lot’s seedlings were transferred to 1/2 MS solid medium with or without 100 mM NaCl. Photographs of the seedlings were taken two weeks later, and data was collected using Image J software. Transgenic and WT seedlings were also transferred to 1/2 MS medium containing 100 mM NaCl with or without 100 µM rutin and eriodictyol for the flavone component complementation experiment. ROS staining was performed on both treated and untreated plants after a week. The flow chart of the experimental design is shown in Supplementary File 1.
The A. venetum genome database (https://www.ncbi.nlm.nih.gov/genome/?term=txid377125[orgn]) was used to extract the 2000-bp sequences of flavonoid synthetase enzyme gene promoters from the NCBI database. The promoter Cis-acting elements were detected and identified using the PLACE database (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/).
The total flavonoids extraction method was based on a previous method (Zhang et al., 2015) with minor modifications. In brief, 50 mg dry sample powder was added to 3 mL 80% (w/v) methyl alcohol extracting in a water bath at 95 °C overnight. In a total of 110 µL supernatant, 440 µL NaNO2 (0.066 M, room temperature for 5 minutes), 60 µL AlCl3 (0.75 M, 6 minutes), and 400 µL NaOH (0.5 M) were added in turns. The absorbance of the mixture was measured at 510 nm. The total flavonoids were presented as mg rutin equivalents, i.e. mg/g DW. HPLC analysis method was used to detect the main flavonoid compounds content in transgenic Arabidopsis plants (Xu et al., 2020).
The Na+ and K+ concentration measurement method was based on a previous method (Xu et al., 2020) with minor modifications. In brief, 0.25 g dried sample powder was digested in 5 mL of HNO3 at 110°C for about 6 hours until a colorless liquid was obtained. The cooled liquid mixture was diluted to 10 mL with deionized water. The Na+ and K+ content was then determined using an optical emission spectrometer (ICP, Optima 8000, PerkinElmer, USA)
The SOD activity detection method was based on a previous method (Stewart and Bewley, 1980). SOD activity was defined as the ability of inhibiting the photochemical reduction of nitro blue tetrazolium. CAT activity was determined according to the method of (Patra et al., 1978), in which the ability of CAT was related to the extinction of H2O2. POD activity was measured by referring to the methodology of (Nickel and Cunningham, 1969).
The histochemical staining methods using 3′3′-diaminobenzidine (DAB) and nitro blue tetrazolium (NBT) to detect H2O2 and O2•-, respectively, was referred (Zhao et al., 2016). Likewise, the H2O2 content detection method was based on a previous method reported by (Libik et al., 2005). The O2•- content was detected with a specific kit (SA-2-G; Cominbio, Suzhou, China) following the manufacturer’s instructions.
IAA and JA detection was done according to (Du et al., 2013) with miner modification. In brief, twelve seedlings of transgenic plants or WT samples under salt stress were ground into powder. 100 mg of powder were extracted twice with 900 µL of extraction buffer [methanol:H2O:acetonitrile = 90:9:1 (v/v)]. Quantification was performed in an ABI Qtrap6500 LC-MS system (Applied Biosystems, USA) with stable-isotope-labeled ABA and auxin as standards (Sigma).
Total RNA was extracted from samples with the EasyPure Plant RNA Kit (TransGen, ER301-01, Beijing, China) and genomic DNA contamination was removed using RNase-free DNase I (TransGen, K21109). First-strand cDNA synthesis and qRT-PCR amplification were performed as previously described by (Wang et al., 2018). Supplementary File 2 lists the gene-specific primers that were used for qRT-PCR. The relative expression levels of target genes were calculated using the 2-ΔΔCt method (Livak and Schmittgen, 2001) and normalized to those of the AtTUBULIN reference gene (Xu et al., 2017). For transcriptome analysis, the RNA purification, reverse transcription, library construction and sequencing were performed at Shanghai Majorbio Bio-pharm Biotechnology Co., Ltd. (Shanghai, China). Transcriptome sequencing data were analyzed on their online platform (www.majorbio.com) (Ren et al., 2022). The transcripts per million reads (TPM) was used to calculate the expression levels of each gene using RSEM (http://deweylab.biostat.wisc.edu/rsem/). Differential expression genes (DEGs) analysis was performed using DESeq2 (Love et al., 2014). The Q value ≤ 0.05 and |log2(Fold Change)| > 1 were used to identify DEGs. The GO and KEGG functional enrichment pathway analyses were performed using GO (https://github.com/tanghaibao/Goatools) and KOBAS (http://kobas.cbi.pku.edu.cn/home.do) tools.
All charts (bar charts, boxplots, bubble charts, Venn diagrams and heatmap, so on) were drawn using Adobe Illustrator cs5 software and Majorbio Cloud Platform (www.majorbio.com). Significant differences among all the experimental treatments were determined using a one-way analysis of variance (ANOVA) followed by Tukey’s test at the probabilities of p < 0.05 level with SPSS version 17.0 software (SPSS, Chicago, IL, USA).
The main biosynthesis pathways of flavonoid metabolism are integrated in Figure 1A. The red marked genes (AvF3’H, AvF3H, and AvFLS) were the main focus in this study. The qRT-PCR results revealed that the expression patterns of these three genes in response to salt stress were similar. Under salt stress, they were induced to express at a high level for four hours. The expression level peaked at 5 minutes and 2 hours, with AvF3’H showing the highest level of expression compared to the other two. Salt stress inhibited the expression of three genes after four hours of exposure (Figure 1B). The expression patterns of other important genes in the flavonoid metabolic pathway were also studied, and the expression profiles were divided into two distinct groups. The expression patterns of AvCHI, Av4CL3, AvF3’5’H, AvRT, AvCHS, AvPAL1, and AvC4H2 were similar to that of AvFLS, AvF3H, and AvF3’H, which showed a higher expression level in the first 4 hours, and reached the peak within 2 to 4 hours (Supplementary File 3). The expression patterns of others genes such as Av4CL1, Av4CL2, AvC4H1, AvC4H3, AvPAL2, Av4CL4, and Av4CL were clustered together. They showed rapid response to salt stress. The expression of these genes was rapidly induced in the first 30 minutes and then started to decline. It reached a low point at 4 hours, after which it gradually began to rise again (Supplementary File 3). The Cis-acting elements in the gene promoter, which usually respond to the types of binding transcription factors, were identified in the PLACE database to help elucidate the potential regulatory mechanism of these genes’ expression under salt stress. From the 2 kb promoters of AvF3’H, AvF3H, and AvFLS, a total of 133 different non-repetitive Cis-acting elements were identified (Supplementary File 4). These Cis-acting elements were found to be primarily involved in abiotic stress (32%), miscellaneous function (17%), hormones (16%), biotic stress (7%), and seed development (6%). (Figure 1C). The top ten Cis-acting elements in each gene were listed. “DOFCOREZM,” “CACTFTPPCA1,” and “ROOTMOTIFTAPOX1” were found in the highest frequency in all three genes, indicating that they may be involved in basic functions. The frequency of a salt-stress response element “GT1GMSCAM4” in AvF3’H, on the other hand, was significantly higher than in AvF3H and AvFLS (Figure 1D), suggesting that AvF3’H may play an important role in response to salt stress.
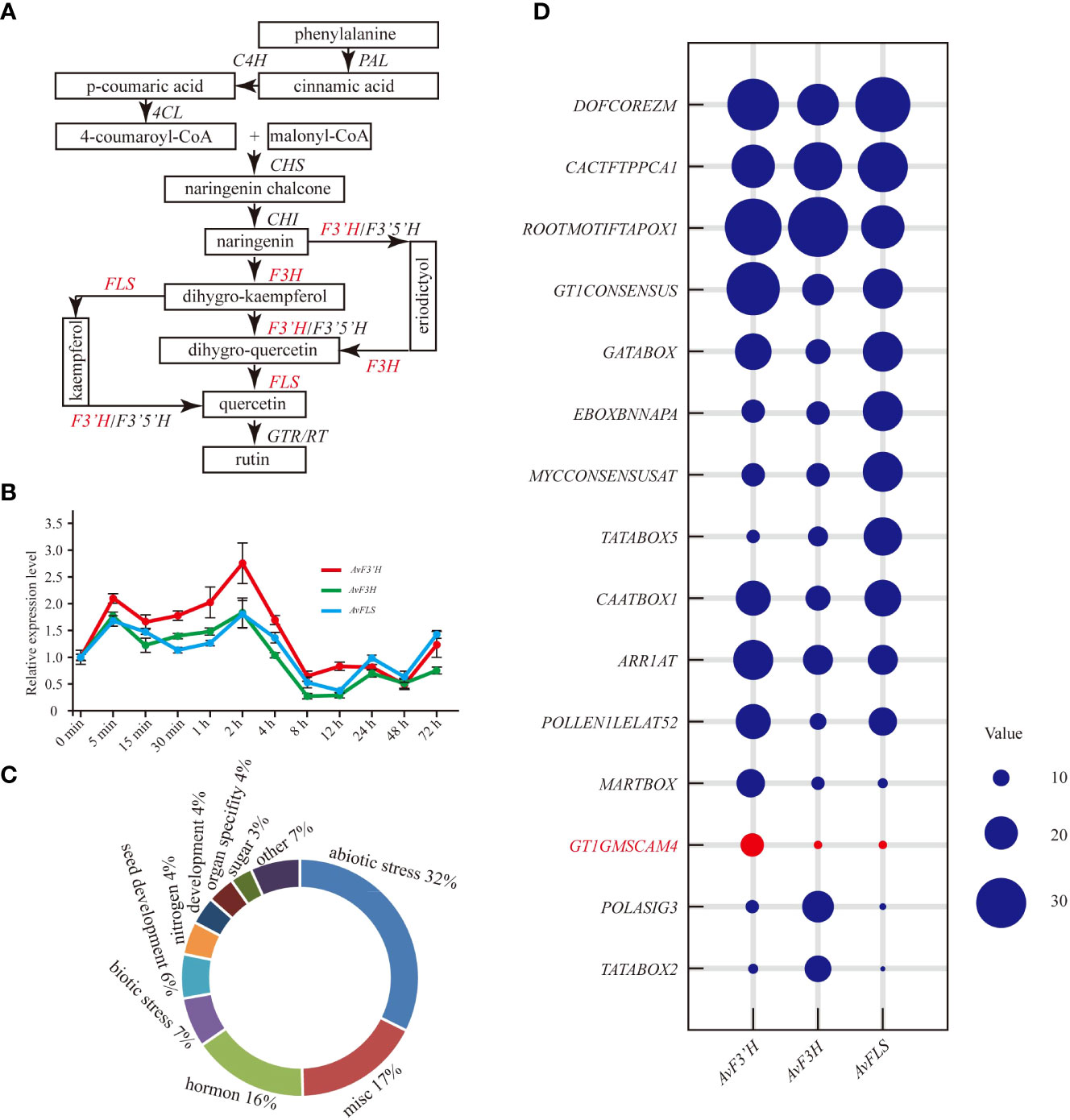
Figure 1 Expression patterns of AvF3’H, AvF3H, and AvFLS under salt stress. (A) Schematic of total flavonoid biosynthesis pathway (Koes et al., 2005; Yin et al., 2012) showed the catalytic action of AvF3’H, AvF3H, and AvFLS. (B) Expression levels of AvF3’H, AvF3H, and AvFLS in A. venetum leaves treated with 100 mM NaCl at different time. (C) Functional classification of cis-acting elements of AvF3’H, AvF3H, and AvFLS promoters. (D) Schematic illustration of the cis-acting elements distribution frequency of AvF3’H, AvF3H, and AvFLS promoters.
OE-transgenic Arabidopsis plants were developed to investigate the potential roles of AvF3’H, AvF3H, and AvFLS in the response to salt stress. Semi-quantitative PCR was used to confirm the expression of AvF3’H, AvF3H, and AvFLS in transgenic plants (Supplementary File 5). Under normal growth conditions, the aerial parts of all three transgenic lines had longer petioles and larger leaves than WT, however, the statistical analysis results were not significant. Under salt stress, these parameters were significantly larger in transgenic plants than WT (Figures 2A, D–F), indicating that these three genes have a salt tolerance function. Phenotypic and statistical analysis revealed that there was no significant difference in the main root length of transgenic plants and WT under normal conditions (Figures 2B, G). Under 100 mM NaCl stress, however, transgenic plant roots were significantly longer than WT roots (Figures 2C, G). Furtherly, the endogenous flavonoid content results of these plants revealed that transgenic plants owed the higher flavonoid than WT both under normal and salt stress conditions (Figure 2H), suggesting a potential link between flavonoid content and salt tolerance. HPLC analysis was used to detect the flavonoids ingredients content. The content of rutin, eriodictyol, and naringerin in transgenic plants and WT were somewhat complex under normal conditions. However, all the transgenic plants showed significantly higher content compared to WT under salt stress (Figures 2I–K), suggesting the potential positive roles of them in response to salt stress.
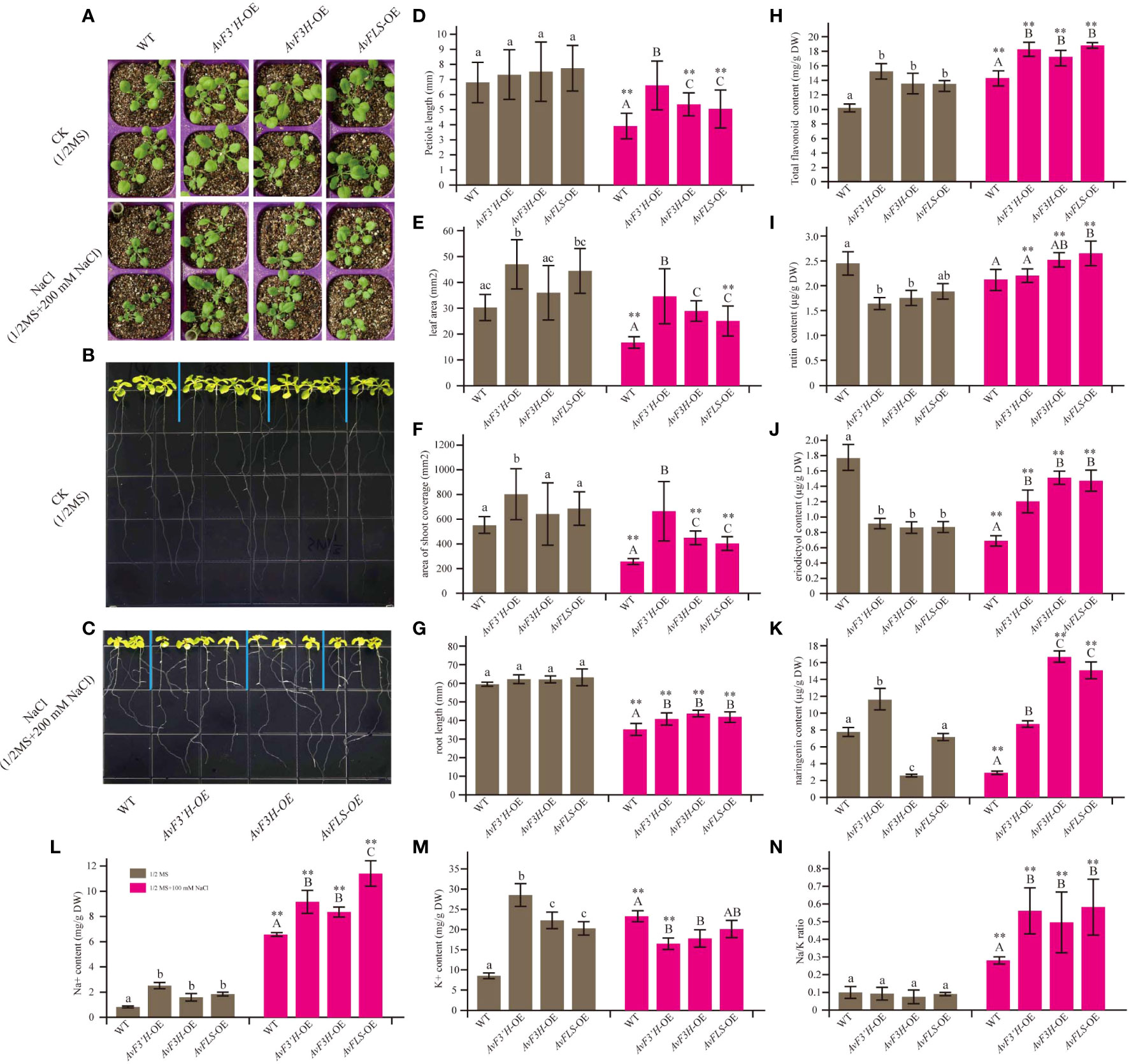
Figure 2 Overexpression of AvF3’H, AvF3H, and AvFLS increased the salt tolerance ability of Arabidopsis. (A) Seedlings aerial phenotype of transgenic plants and WT with and without salt stress. The root phenotype growing on normal 1/2 MS medium of transgenic plants and WT (B) and with 100 mM NaCl (C). (D) Petiole length. (E) Leaf area. (F) The shoot coverage area. (G) Root length. (H) Total flavonoid contents. (I) Rutin contents. (J) Eriodictyol content. (K) Naringin content. (L) Na+ content. (M) K+ content. (N) Na/K ratio. Different lowercase letters indicated significant differences between transgenic plants and WT under normal condition (P < 0.05); different uppercase letters indicated significant differences between transgenic plants and WT under salt stress (P < 0.05), and ** indicated that there was a significant difference between the plants under normal condition and salt stress (P < 0.05).
Transgenic plants had significantly higher Na+ content under normal and salt stress than WT plants. Furthermore, the Na+ content of WT and transgenic plants was 3.6 to 7.9 times higher in saline than in normal conditions (Figure 2L). Interestingly, the K+ content in WT was 2.7 times higher under salt stress than under normal conditions, whereas the K+ content in transgenic plants decreased by 39.86%, 18.54%, and 33.46% in the three transgenic lines under salinity compared to normal conditions (Figure 2M), resulting in a higher Na/K ratio in transgenic plants under salt stress (Figure 2N). These findings suggested that overexpression of the flavonoid synthetase genes could increase the Na+ content and reduce the K+ content in transgenic plants, resulting in the ion homeostasis changes.
Salt stress is one of the detrimental environmental stresses inducing factors that lead to ROS accumulation. Thus, using 3,3’-diaminobenzidine (DAB) and nitrotetrazolium blue chloride (NBT) staining, histochemical analysis was used to investigate the effect of two ROS species, hydrogen peroxide (H2O2) and superoxide anion (O2•-). Under salt stress, the accumulation of brown and blue precipitates (showing DAB and NBT staining, respectively) in transgenic lines was much lower than in WT plants (Figures 3A, B). These findings suggest that transgenic plants’ improved salt stress tolerance may be due to reduced ROS production, which correlates with higher endogenous flavonoid levels in the three transgenic plants (Figure 2H). As expected, control plants with exogenous rutin and naringerin were significantly less damaged by ROS than those without under salt stress (Figures 3A, B). According to the spectrophotometry analysis results, the content of H2O2 and O2•- that accumulated in the leaves of these transgenic plants showed no significant difference with that of WT under normal condition, however, significantly reduced compared with the WT plants under salt stress (Figures 3C, D). To evaluate the plants abiotic stress damage caused by salt stress, the MDA content was measured. As shown in Figure 3E, there was no significant difference in MDA content between the transgenic and WT plants under normal conditions. However, the MDA content in transgenic plants was significantly reduced (28.39% to 32.32%) compared to WT under salt stress. The antioxidant scavenging system, on the other hand, was activated in the transgenic plants. Under salt stress, the activity of CAT in both WT and transgenic plants was significantly higher than that under normal conditions, with the transgenic plants showing significantly higher activities than the control. Although the activities of SOD and POD were inhibited in WT and transgenic plants under salt stress (except SOD in AvF3’H-OE) compared to normal conditions, the activities of SOD and POD in transgenic plants were significantly higher than those in WT plants under salt stress (Figures 3F, H).
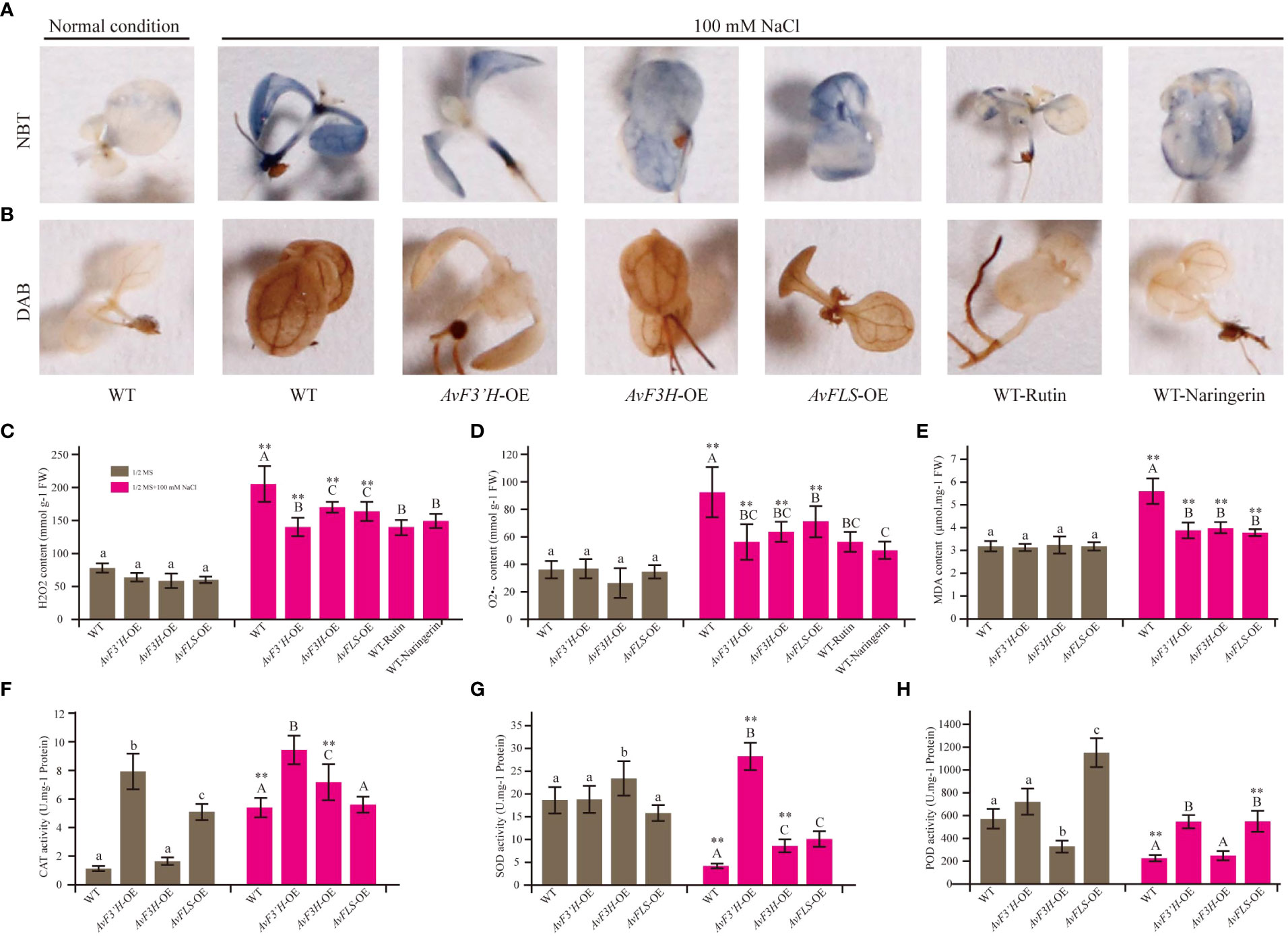
Figure 3 Transgenic plants and exogenous addition of rutin and naringerin alleviated ROS damage under salt stress. (A) NBT staining of transgenic plants and exogenous flavonoids application to plants under salt stress. (B) DAB staining. (C) H2O2 content. (D) O2•- content. (E) MDA content. The CAT (F), SOD (G) and POD (H) activity of transgenic plants and WT under normal and salt stress conditions. Different lowercase letters indicated significant differences between transgenic plants and WT under normal condition (P < 0.05); different uppercase letters indicated significant differences between transgenic plants and WT under salt stress (P < 0.05), and ** indicated that there was a significant difference between the plants under normal condition and salt stress (P < 0.05).
In three independent experiments, we performed transcriptome deep sequencing (RNA-seq) analysis on transgenic and WT plants treated with 100 mM NaCl to further investigate the molecular mechanism of endogenous flavonoid accumulation in reducing the effect of salt stress. Three biological replicates of each treatment were compactly gathered together in the PCA diagram, indicating that the RNA-seq results were stable and reliable. Furthermore, while the four treatment samples were clearly separated, AvF3H-OE was separated from the other three samples, indicating that AvF3H overexpressing plants had a relatively distinct transcriptome profile from the other three samples (Supplementary File 6). We then identified differentially expressed genes (DEGs). A total of 1299 non-repeat genes were identified in transgenic plants when compared with WT (Supplementary File 7), of which 289 were identified in AvF3’H vs WT (148 up-regulated and 141 down-regulated) (Figure 4A), 1028 were identified in AvF3H vs WT (844 up-regulated and 184 down-regulated) (Figure 4B), 235 were identified in AvFLS vs WT (122 up-regulated and 113 down-regulated) (Figure 4C), suggesting that AvF3H overexpressing had a significant effect on gene regulatory networks. A venn map was used to identify shared DEGs among different comparison groups in order to investigate the general rule of flavonoid metabolism pathway related genes in response to salt stress. A total of 43 shared DEGs were identified, with 30 up-regulated and 13 down-regulated (Figure 4D, Supplementary File 7). GO enrichment analysis revealed that some JA related terms such as “Response to jasmonic acid”, “Jasmonic acid hydrolase”, and “Regulation of jasmonic acid mediated signaling pathway” were significantly enriched in these shared DEGs (Figure 4E). The most enriched KEGG pathway was “Nitrogen metabolism” pathway (Figure 4F). These findings suggested that JA and nitrogen metabolism play important roles in the response to salt stress. Furthermore, the top 10 enrichment GO and KEGG terms from each comparison group were integrated. Finally, a total of 23 non-repeated GO terms (Supplementary File 8A) and 18 KEGG pathways (Supplementary File 8B) were listed. We noticed that GO and KEGG terms of the AvF3H vs WT comparison group possessed more DEGs compared to the other two, which is consistent with the highest DEGs identified in this comparison group. GO terms “cell wall” and KEGG pathways “MAPK signaling pathway-plant”, “plant-pathogen interaction”, and “cyanoamino acid metabolism” were shared in all three comparison groups indicating the importance of these genes’ involvement in these terms. Furthermore, hormone related GO terms and KEGG pathways, such as “regulation of jasmonic acid mediated signaling pathway”, “jasmonic acid hydrolase”, “response to jasmonic acid”, “Plant hormone signal transduction”, and “alpha-Linolenic acid metabolism”, cell wall related terms such as “cell wall” and “plant-type cell wall”, flavonoid metabolism related terms such as “flavonoid biosynthesis”, “isoflavonoid biosynthesis”, and “phenylpropanoid biosynthesis” were significantly enriched in the transgenic vs WT groups, which suggests their involvement in salt tolerance. A total of 465 DEGs were involved in this enrichment GO or KEGG terms, and most of them were up-regulated in transgenic plants under salt stress compared to WT plants (Supplementary File 8C).
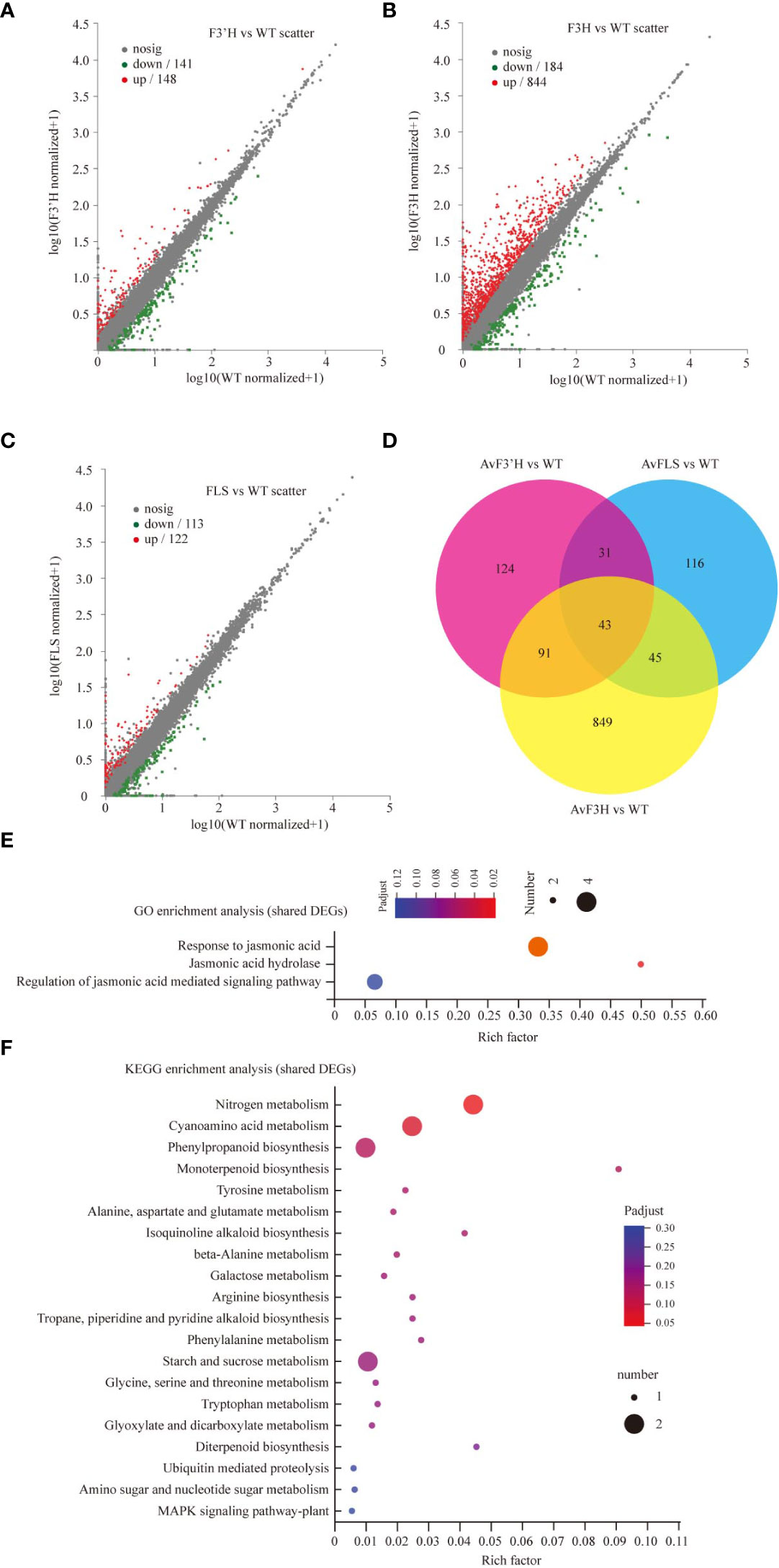
Figure 4 DEGs data annotation and enrichment analysis. The volcano plots showing the up- and down-regulated DEGs in comparisons AvF3’H vs WT (A), AvF3H vs WT (B), and AvFLS vs WT (C). (D) Venn diagrams showing specific and shared DEGs in the 3 comparisons. GO (E) and KEGG (F) enrichment analysis of shared DEGs in the 3 comparisons (p < 0.05). The X-axis (Rich Factor) represents the percentage of DEGs belonging to the corresponding pathway.
The cell membrane acts as a barrier to material movement into and out of the cell. To investigate the cause of Na+ accumulation in transgenic plants, Na+ influx-related genes were investigated using transcriptome data. Despite the fact that no shared Na+ influx DEGs were identified, we discovered that a nonselective cation channel (NSCC) gene GLR1.2 and an aquaporin gene PIP2-3 were up-regulated in transgenic plants (Table 1), which corresponds to transgenic plants’ higher Na+ accumulation compared to WT plants (Figure 2L).
Among the shared DEGs, we discovered that two cell wall related DEGs PRX52 and EXLA2 were significantly up-regulated in transgenic plants under salt stress. In addition, eight cell wall expansin genes including three expansin-like genes (EXL1, EXLA1 and EXT4), and five xyloglucan endotransglucosylase/hydrolase gene (XTH18, XTH22, XTH23, XTH24, and XTH30) were found to be significantly up-regulated in one or two comparison groups (Table 1).
Jasmonic acid and IAA hormones are important plant growth regulators which also play a key role in plant response to abiotic stress. The synthesis and signaling transduction related genes of IAA and JA were screened from the transcriptome data in light of some IAA and JA related GO and KEGG terms enriched in transgenic plants compared to WT.As expected, YUC, TRA, and TAA genes in IAA synthesis pathway, and LOX, AOS, AOC, OPR and DAD genes in JA synthesis pathway were all identified. Although their expression levels tended to increase in transgenic plants (Supplementary Files 9B, D), only four (LOX3, LOX4, AOC3, and OPR3, Figure 5A) and two (TAR2 and NIT2, Figure 5B) key synthesis genes among the nine and fourteen genes of JA and IAA synthesis related genes were identified as DEGs, respectively, and showed up-regulation in transgenic plants compared to WT (Supplementary File 9A, Supplementary File 7). A total of 13 IAA polarity distribution and transporter related genes such as PIN, LAZY, and LAX were identified, however, none of them were DEGs (Supplementary File 9C). Among 69 IAA signaling transduction related genes, only SAUR22, SAUR36 and IAA27 which were the early auxin-responsive genes showed significantly up-regulated in transgenic plants (Supplementary File 9A, Supplementary File 7). Three JA signaling transduction genes JAZ10, JAZ13, and MYC2 were found to be significantly up-regulated in transgenic plants (Supplementary File 9E, Supplementary File 7). To validate the reliability of RNA-Seq data, the expression level of these twelve DEGs was examined by qRT-PCR. The similar expression trends of these DEGs were consistent with the Illumina sequencing (Figures 5C–E), indicating the dependability of the RNA-Seq data. The up-regulated of IAA and JA biosynthesis and signaling transduction pathway related genes indicated the accumulation of IAA and JA content in transgenic plants under salt stress. To confirm this hypothesis, IAA and JA levels were measured. Under normal growth condition, there was no significant difference in the amount of IAA and JA in transgenic plants and WT (Figures 5F, G). However, under salt stress, transgenic plants’ levels of IAA and JA significantly increased in comparison to WT, with an increase in IAA content of between 42.91% and 53.56% (Figures 5F, G). This was consistent with the up-regulated expression of IAA and JA synthetic pathway genes. On the other hand, under both normal and salt stress conditions, there was no significant different in the IAA and JA content of WT (Figures 5F, G). Accordingly, this indicates that the IAA and JA biosynthesis pathways were probably co-activated by the increase in flavonoid content in transgenic plants and salt stress.
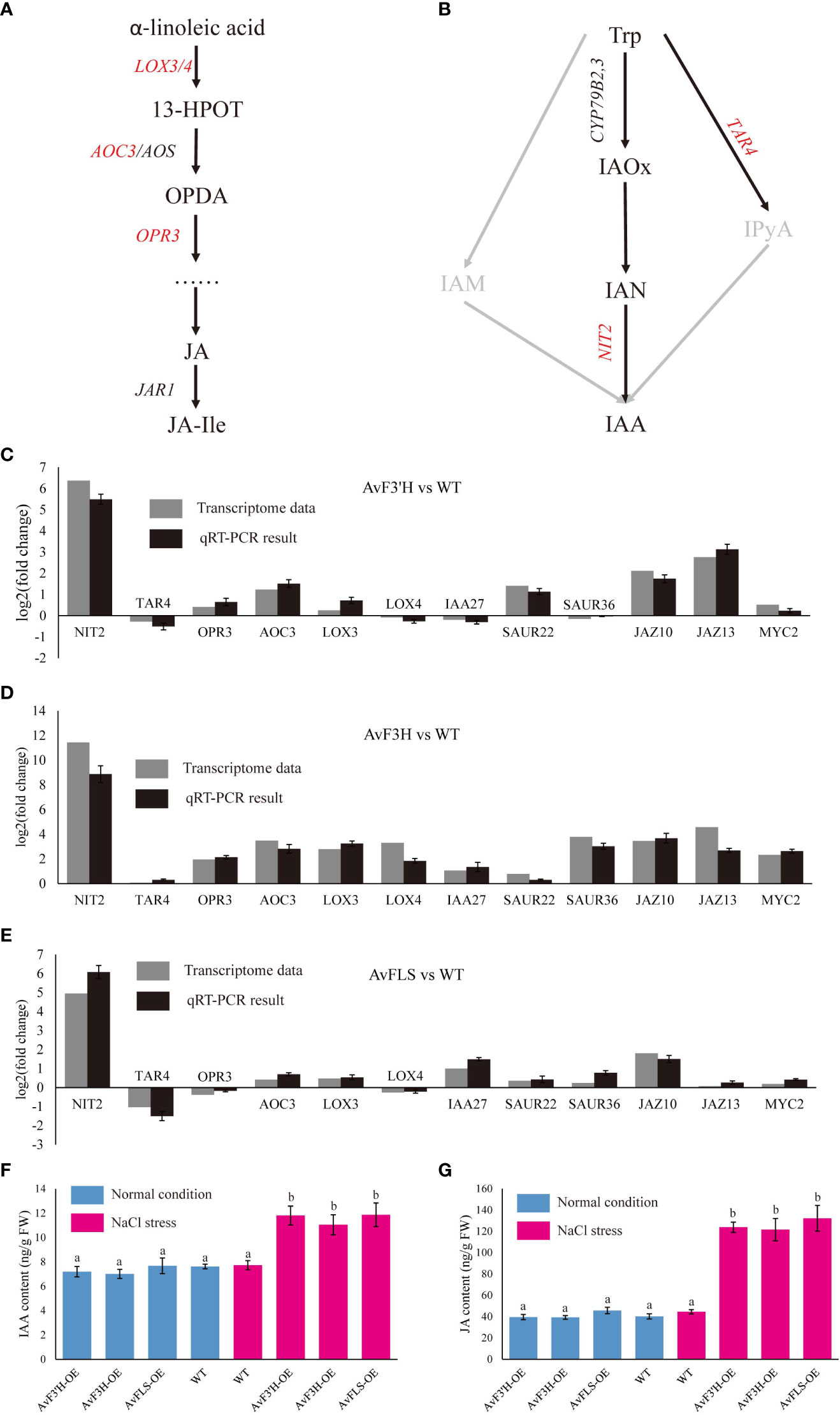
Figure 5 DEGs related to the plant hormone biosynthesis and signal transduction pathway. Diagram of jasmonic acid (A) and auxin (B) biosynthesis pathways. Validation of DEGs expression by qRT-PCR analysis of twelve DEG genes (C–E). The IAA (F) and JA (G) content in transgenic plants and WT under normal growth and salt stress conditions. Different lowercase letters indicated significant differences between transgenic plants and WT (P < 0.05).
Flavonoids, as an important member of the non-enzymatic antioxidant system, could directly eliminate the excess ROS produced by salt stress, working synergistically with the enzymatic antioxidant system. Importantly, flavonoids can also induce the expression of jasmonic acid and auxin synthesis genes via unknown pathways, resulting in the IAA and JA content increase, and activating the JA and IAA signal transduction pathways to promote the expression of cell wall biosynthesis or modification genes that promote cell wall expansion (Figure 6).
Flavonoids are found throughout the plant kingdom and are present in high concentrations in the epidermis of leaves and the skin of fruits. They are mainly classified into nine major subgroups: the chalcones, aurones, isoflavonoids, flavones, flavonols, flavandiols, anthocyanins, condensed tannins, and phlobaphene pigments (Winkelshirley, 2001). The biosynthesis of flavonoids is a very complex process that begins with the phenylpropanoid pathway. Chalcone synthase is the first committed enzyme in the biosynthesis of all flavonoids, producing a yellow-colored compound called chalcone. In most plants, chalcones are not the end-products, but the pathway proceeds with several enzymatic steps to other classes of flavonoids. The catalytic enzyme genes F3H, FLS, and F3’H are crucial for flavonoid branching. F3H catalyzes the formation of dihydroflavonol from flavanone. F3’H takes dihydrokaempferol or kaempferol as a substrate and converts it to dihydroquercetin and quercetin. FLS is the first dedicated enzyme for the biosynthesis of flavonols, branching from the main trunk route to the branch for anthocyanin formation (Pelletier et al., 1997). It could catalyze dihydroflavonols to form flavonols (Saito et al., 2013). Although all of the substrates are important secondary metabolites, the expression patterns of catalytic enzyme genes under salt stress were unclear. A 72-hour long expression experiment revealed multi expression level peaks at collecting time points for many flavonoid synthesis genes (Supplementary File 3), indicating the complex dynamic response of these genes to salt stress. Consistent with expression levels of these genes under salt stress, AvF3’H had a higher frequency of the salt-stress response element “GT1GMSCAM4” in the promoter region (Figure 1D). The cis-element “GT1GMSCAM4” was NaCl-induced and regulated by the GT-1-like transcription factor (Park et al., 2004). In general, salt stress would inhibit plant growth, resulting in shorter roots and stunted aerial parts. Our previous research found that 50 to 400 mM NaCl stress inhibited seed germination, plant height, root length, and leaf length of seedlings (Xu et al., 2020). The ion toxicity caused by the influx and accumulation of Na+ is a major cause of this phenomenon. Na+ influx across the plasma membrane in plants occurs primarily through nonselective cation channels (NSCCs) such as cyclic nucleotide-gated channels (CNGCs) and glutamate receptors (GLRs) (Keisham et al., 2018), as well as transporters such as HKTs and HAKs (Tester and Davenport, 2003; Deinlein et al., 2014). In addition, aquaporins have also been recently reported to be involved in Na+ uptake in plants (Byrt et al., 2017). Some of these Na+ influx genes were reported to be up-regulated in A. venetum under salt stress resulting in the Na+ excessive accumulation in leaves and dwarf plants caused by many cell wall biosynthesis or modification genes that were down-regulated (Xu et al., 2021). In this study, we also observed two potential Na+ influx genes GLR1.2 and PIP2-3 which were induced by salt stress in all the three transgenic lines (Table 1). Consistent with that, the Na+ contents in the transgenic plants were significantly higher than WT plants under salt stress (Figure 2L). Unlike our previous study, two types of cell wall modification genes (xyloglucan endotransglucosylase/hydrolases and cell wall expansin-related genes) were up-regulated in transgenic plants (Table 1). Plant cell walls provides mechanical strength to withstand the turgor pressure and determine the shape, size and function of cells. It is a complex dynamic structure and mainly composed by carbohydrate polymers such as cellulose, hemicellulose (mainly xyloglucan), and pectin, and structural proteins in variable amounts (Bellincampi et al., 2014). The xyloglucan endotransglucosylase/hydrolases (XTH) proteins have two distinct catalytic activities; xyloglucan endo-transglucosylase (XET activity) and xyloglucan endo-hydrolase (XEH activity). The XET domain cleaves a xyloglucan chain and rejoins the reducing end to another xyloglucan molecule, resulting in the elongation of xyloglucan. The XEH domain rejoins the xyloglucan reducing end to a water molecule resulting in irreversible xyloglucan chain shortening (Rose et al., 2002). Thus XTHs are considered key enzymes in the regulation of cell wall extensibility during cell growth (Cosgrove, 2005). Expansin related genes play a similar role (Marowa et al., 2020). The up-regulation of these cell wall modification related genes offered a reasonable explanation for the increased growth of transgenic plants under salt stress (Figures 2A–G).
The most significant difference between transgenic plants and other plants under salt stress is the high content of flavonoids. Flavonoids have been the subject of intense research interest because they were shown to have diverse functions in plants including oxidative damage protection, pathogen defense, interspecies communication, and even in auxin transport (Kuhn et al., 2011). Previous research suggested that flavonoids could influence auxin polar transport by competing for binding to auxin transport proteins with the auxin transport inhibitor 1-N-naphthylphthalamic acid (Murphy et al., 2002). Auxin transport is enhanced in the absence of flavonoids and decreased in the presence of excess flavonols (Peer et al., 2004). Furthermore, a previous study discovered that the accumulation of auxin in rol1-2 seedlings, a flavonols aberrant accumulation mutant, was caused by a flavonol-induced modification of auxin transport (Kuhn et al., 2011). These studies confirmed the role of flavonoids in auxin transport. In this study, though no auxin transport related DEGs were identified, we found that the IAA synthase gene NIT2 which was involved in an important IAA synthase branch hydrolysing indole-3-acetonitile (IAN) into IAA was up-regulated in transgenic plants (Figures 5B–E), which indicates that flavonoids are also involved in the regulation of auxin synthesis pathway. In fact, transgenic plants had higher levels of IAA than WT plants under salt stress (Figure 5F). Previous study have reported that when exogenous IAA was used in plants, the cell wall related genes that function on cellulose synthesis or modification, XyGs, pectins, structural proteins (EXPs), and peroxidases were up-regulated (Nemhauser et al., 2006). These results could explain why the expression of cell wall related genes was up-regulated in transgenic plant. However, the mechanism through which flavonoids promote the synthesis of IAA remains to be explored.
Jasmonic acid (JA) is an essential hormone involved in plant defense against herbivory and in responses to abiotic stress. Large-scale transcriptomic studies have shown that some JA-biosynthesis genes (e.g. AOC1, AOC2, AOS, LOX3 and OPR3) are up-regulated in roots under salt stress (Kilian et al., 2007; Geng et al., 2013). In our study, LOX3, AOC3, and OPR3 were also up-regulated in transgenic plants under salt stress (Figure 5B, Supplementary File 7). Some studies reported that exogenous JA could improve salt tolerance in many plants, which is consistent with the upregulation of JA synthetic genes. Maize seedlings treated with JA were found to significantly reduce the toxic effects of excess Na2CO3 on photosynthesis and plant growth parameters (Mir et al., 2018). In rice, post-application with exogenous JA can improve the leaf water potential, leaf photosynthetic rate, and maximum quantum yield of photosystem II (PSII) resulting in salt-stress alleviation, particularly in salt-sensitive cultivars (Kang et al., 2005). In Arabidopsis, overexpression of the wheat JA-biosynthesis gene OPR1 reduces salt-mediated root growth inhibition (Dong et al., 2013). Furthermore, JA could effectively protect wheat seedlings from salt stress damage by increasing the activities of antioxidant enzymes and the concentration of antioxidative compounds to quench the excessive reactive oxygen species caused by salt stress, and this has practical implications for wheat cultivation in salt-affected soils (Qiu et al., 2014). These findings suggest that salt stress activates the JA biosynthesis and signaling pathway, causing physiological and growth changes in plants, which is consistent with this study.
Plant cells employ multiple mechanisms to regulate the levels of ROS to modulate signaling and prevent oxidative stress. Besides the antioxidant enzymes, synthesis of flavonoid metabolites that function as antioxidants in vitro is another important mechanism (Gayomba et al., 2017).Flavonoids are found in a variety of plant organs as well as different cells and cellular compartments, such as the cell wall, the vacuole of epidermal cells, and the external surface organs (Agati et al., 2012). Flavonoids occur as glycosides in healthy leaf cells, so the ROS scavenger capacity of flavonoids is dependent on the presence of the catechol group in the B-ring of the flavonoid skeleton(Rice-Evans et al., 1997). This also explained why the exogenous addition of rutin and naringerin reduced the ROS content of the control under salt stress (Figure 3).
Flavonoids are important secondary metabolites of non-antioxidant enzyme system members that help to eliminate ROS generated by various abiotic stresses. Overexpression of flavonoids synthetase enzyme genes AvF3’H, AvF3H, and AvFLS increased not only total flavonoids content in transgenic plants, but also synergistically improved the antioxidant enzyme system, which improved salt tolerance. Similar to transgenic plants under salt stress, exogenous addition of flavonoids compounds like rutin and naringerin reduced ROS damage in plants. Furthermore, the biosynthesis and signaling transduction genes of IAA and JA was mainly up-regulated in transgenic plants under salt stress compared with WT, and the IAA and JA levels were significantly increased. Thus, the activated IAA and JA signaling pathway may activate cell wall biosynthesis or modification genes. As a result, the aerial part and root length of transgenic plants were greater than those of the control under salt stress.
The original contributions presented in the study are included in the article/Supplementary Material. The raw data of RNA-seq was uploaded into National Center for Biotechnology Information (NCBI) database with the BioProject accession number PRJNA945093 (https://www.ncbi.nlm.nih.gov/sra/PRJNA945093). Further inquiries can bedirected to the corresponding authors.
ZX and LZ designed the study, TR, XL, JW, HY, CL, and CM carried out the study. MZ, ZX and PM wrote the manuscript. ZX and CM applied for funding. All authors contributed to the article and approved the submitted version.
We gratefully acknowledge financial support from the Fundamental Research Funds for Central Non-profit Scientific Institution (1610232022006), National Natural Science Foundation of China (31900276, 32171948), Agricultural Science and Technology Innovation Program (ASTIP No. CAAS-ZDRW202201), Demonstration and Guidance Program for Technology People-Benefit in Qingdao (20-3-4-7-nsh), Science Foundation for Young Scholars of the Tobacco Research Institute of the Chinese Academy of Agricultural Sciences (2020A02). The funders had no role in designing the study, data collection and analysis, publication decision, or manuscript preparation.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1123856/full#supplementary-material
Supplementary File 1 | The graphical representation of experimental design.
Supplementary File 2 | Primers used in this study.
Supplementary File 3 | Heatmap of key genes expression in flavonoid synthesis pathway.
Supplementary File 4 | Cis-acting elements of AvF3’H, AvF3H and AvFLS identified from the 2 kb length promoters.
Supplementary File 5 | The expression level identification of AvF3’H, AvF3H and AvFLS in transgenic plants by semi-quantitative PCR.
Supplementary File 6 | Principal component analysis (PCA) of the genes.
Supplementary File 7 | Summary of transcriptome sequencing.
Supplementary File 8 | The top 10 enrichment GO (A) and KEGG (B) terms of 3 comparison groups, respectively, and heatmap analysis of enrichment DEGs (C).
Supplementary File 9 | Expression level heatmap of genes related the biosynthesis, transport, and signaling transduction of IAA and JA.
Agati, G., Azzarello, E., Pollastri, S., Tattini, M. (2012). Flavonoids as antioxidants in plants: location and functional significance. Plant Science 196, 67–76. doi: 10.1016/j.plantsci.2012.07.014
Akbarimoghaddam, H., Galavi, M., Ghanbari, A., Panjehkeh, N. (2011). Salinity effects on seed germination and seedling growth of bread wheat cultivars. Trakia J. Sci. 9 (1), 43–50.
Apel, K., Hirt, H. (2004). Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 55 (1), 373–399. doi: 10.1146/annurev.arplant.55.031903.141701
Bellincampi, D., Cervone, F., Lionetti, V. (2014). Plant cell wall dynamics and wall-related susceptibility in plant-pathogen interactions. Front. Plant Sci. 5, 228. doi: 10.3389/fpls.2014.00228
Byrt, C. S., Zhao, M., Kourghi, M., Bose, J., Henderson, S. W., Qiu, J., et al. (2017). Non-selective cation channel activity of aquaporin AtPIP2;1 regulated by Ca2+ and pH. Plant Cell Environ. 40 (6), 802–815. doi: 10.1111/pce.12832
Chen, S., Wu, F., Wang, Y., Li, Y., Wu, Z., Qian, Y., et al. (2019). NtMYB4 and NtCHS1 are critical factors in the regulation of flavonoid biosynthesis and are involved in salinity responsiveness. Front. Plant Sci. 10, 178. doi: 10.3389/fpls.2019.00178
Colla, G., Rouphael, Y., Cardarelli, M., Svecova, E., Rea, E., Lucini, L. (2013). Effects of saline stress on mineral composition, phenolic acids and flavonoids in leaves of artichoke and cardoon genotypes grown in floating system. J. Sci. Food Agric. 93 (5), 1119–1127. doi: 10.1002/jsfa.5861
Cosgrove, D. J. (2005). Growth of the plant cell wall. Nat. Rev. Mol. Cell Biol. 6 (11), 850–861. doi: 10.1038/nrm1746
Das, K., Roychoudhury, A. (2014). Reactive oxygen species (ROS) and response of antioxidants as ROS-scavengers during environmental stress in plants. Front. Environ. Sci. 2. Article 53. doi: 10.3389/fenvs.2014.00053
Deinlein, U., Stephan, A. B., Horie, T., Luo, W., Xu, G., Schroeder, J. I. (2014). Plant salt-tolerance mechanisms. Trends Plant Sci. 19 (6), 371–379. doi: 10.1016/j.tplants.2014.02.001
Dong, W., Wang, M., Xu, F., Quan, T., Peng, K., Xiao, L., et al. (2013). Wheat oxophytodienoate reductase genetaopr1confers salinity tolerance via enhancement of abscisic acid signaling and reactive oxygen species scavenging. Plant Physiol. 161 (3), 1217–1228. doi: 10.1104/pp.112.211854
Du, H., Liu, H., Xiong, L. (2013). Endogenous auxin and jasmonic acid levels are differentially modulated by abiotic stresses in rice. Front. Plant Sci. 4, 1–10. doi: 10.3389/fpls.2013.00397
Gayomba, S. R., Watkins, J. M., Muday, G. K. (2017). Flavonols regulate plantgrowth and development throughregulation of auxin transportand cellular redox status Recent Advances in Polyphenol Research (y John Wiley & Sons, Ltd), 143–170.
Gechev, T. S., Van Breusegem, F., Stone, J. M., Denev, I., Laloi, C. (2006). Reactive oxygen species as signals that modulate plant stress responses and programmed cell death. BioEssays. 28 (11), 1091–1101. doi: 10.1002/bies.20493
Geng, Y., Wu, R., Wee, C. W., Xie, F., Wei, X., Chan, P. Y., et al. (2013). A spatio-temporal understanding of growth regulation during the salt stress response in arabidopsis. Plant Cell. 25 (6), 2132–2154. doi: 10.1105/tpc.113.112896
Gill, S. S., Tuteja, N. (2010). Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 48 (12), 909–930. doi: 10.1016/j.plaphy.2010.08.016
Hectors, K., Van Oevelen, S., Geuns, J., Guisez, Y., Jansen, M., Prinsen, E. (2014). Dynamic changes in plant secondary metabolites during UV acclimation in Arabidopsis thaliana. Physiologia Plantarum 152 (2), 219–230. doi: 10.1111/ppl.12168
Kang, D. J., Seo, Y. J., Lee, J. D., Ishii, R., Kim, K. U., Shin, D. H., et al. (2005). Jasmonic acid differentially affects growth, ion uptake and abscisic acid concentration in salt-tolerant and salt-sensitive rice cultivars. J. Agron. Crop Science. 191, 273—82. doi: 10.1111/j.1439-037X.2005.00153.x
Keisham, M., Mukherjee, S., Bhatla, S. C. (2018). Mechanisms of sodium transport in plants-progresses and challenges. Int. J. Mol. Sci. 19 (3), 647. doi: 10.3390/ijms19030647
Kilian, J., Whitehead, D., Horak, J., Wanke, D., Weinl, S., Batistic, O., et al. (2007). The AtGenExpress global stress expression data set: Protocols, evaluation and model data analysis of UV-b light, drought and cold stress responses. Plant J. 50 (2), 347–363. doi: 10.1111/j.1365-313X.2007.03052.x
Koes, R., Verweij, W., Quattrocchio, F. (2005). Flavonoids: a colorful model for the regulation and evolution of biochemical pathways. Trends Plant Science. 10 (5), 236–242. doi: 10.1016/j.tplants.2005.03.002
Kuhn, B. M., Geisler, M., Bigler, L., Ringli, C. (2011). Flavonols accumulate asymmetrically and affect auxin transport in Arabidopsis. Plant Physiol. 156 (2), 585–595. doi: 10.1104/pp.111.175976
Libik, M., Konieczny, R., Pater, B., Slesak, I., Miszalski, Z. (2005). Differences in the activities of some antioxidant enzymes and in H2O2 content during rhizogenesis and somatic embryogenesis in callus cultures of the ice plant. Plant Cell Rep. 23 (12), 834–841. doi: 10.1007/s00299-004-0886-8
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods. 25 (4), 402–408. doi: 10.1006/meth.2001.1262
Love, M. I., Huber, W., Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15 (12), 550. doi: 10.1186/s13059-014-0550-8
Marowa, P., Ding, A., Xu, Z., Kong, Y. (2020). Overexpression of NtEXPA11 modulates plant growth and development and enhances stress tolerance in tobacco. Plant Physiol. Biochem. 151, 477–485. doi: 10.1016/j.plaphy.2020.03.033
Mir, M. A., John, R., Alyemeni, M. N., Alam, P., Ahmad, P. (2018). Jasmonic acid ameliorates alkaline stress by improving growth performance, ascorbate glutathione cycle and glyoxylase system in maize seedlings. Sci. Rep. 8 (1), 2831. doi: 10.1038/s41598-018-21097-3
Mittler, R. (2017). ROS are good. Trends Plant Science. 22 (1), 11–19. doi: 10.1016/j.tplants.2016.08.002
Munns, R. (2002). Comparative physiology of salt and water stress. Plant Cell Environment. 25, 239–250. doi: 10.1046/j.0016-8025.2001.00808.x
Murphy, A. S., Hoogner, K. R., Peer, W. A., Taiz, L. (2002). Identification, purification, and molecular cloning of n-1-naphthylphthalmic acid-binding plasma membrane-associated aminopeptidases from Arabidopsis. Plant Physiol. 128 (3), 935–950. doi: 10.1104/pp.010519
Nemhauser, J. L., Hong, F., Chory, J. (2006). Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. cell. 126 (3), 467–475. doi: 10.1016/j.cell.2006.05.050
Nickel, K. S., Cunningham, B. A. (1969). Improved peroxidase assay method using leuco 2,3′,6-trichloroindophenol and application to comparative measurements of peroxidatic catalysis. Anal. Biochem. 27, 292–299. doi: 10.1016/0003-2697(69)90035-9
Park, H. C., Kim, M. L., Kang, Y. H., Jeon, J. M., Yoo, J. H., Kim, M. C., et al. (2004). Pathogen- and NaCl-induced expression of the SCaM-4 promoter is mediated in part by a GT-1 box that interacts with a GT-1-Like transcription factor. Plant Physiol. 135 (4), 2150–2161. doi: 10.1104/pp.104.041442
Patra, H. K., Kar, M., Mishra, D. (1978). Catalase activity in leaves and cotyledons during plant development and senescence. Biochem. Physiol. 172, 385–390. doi: 10.1016/S0015-3796(17)30412-2
Peer, W. A., Bandyopadhyay, A., Blakeslee, J. J., Makam, S. N., Chen, R. J., Masson, P. H., et al. (2004). Variation in expression and protein localization of the pin family of auxin efflux facilitator proteins in flavonoid mutants with altered auxin transport in Arabidopsis thaliana. Plant Cell 16 (7), 1898–1911. doi: 10.1105/tpc.021501
Pelletier, M. K., Murrell, J. R., Shirley, B. W. (1997). Characterization of flavonol synthase and leucoanthocyanidin dioxygenase genes in Arabidopsis. Plant Physiol. 11 (3), 1437–1 445. doi: 10.1104/pp.113.4.1437
Qiu, Z., Guo, J., Zhu, A., Zhang, L., Zhang, M. (2014). Exogenous jasmonic acid can enhance tolerance of wheat seedlings to salt stress. Ecotoxicol Environ. Saf. 104, 202–208. doi: 10.1016/j.ecoenv.2014.03.014
Ren, Y., Yu, G., Shi, C., Liu, L., Guo, Q., Han, C., et al. (2022). Majorbio cloud: A one-stop, comprehensive bioinformatic platform for multiomics analyses. iMeta. 1, (2). doi: 10.1002/imt2.12
Rice-Evans, C. A., Miller, N. J., Paganga, G. (1997). Antioxidant properties of phenolic compounds. Trends Plant Science. 2 (4), 152–159. doi: 10.1016/S1360-1385(97)01018-2
Rose, J. K., Braam, J., Fry, S. C., Nishitani, K. (2002). The XTH family of enzymes involved in xyloglucan endotransglucosylation and endohydrolysis: Current perspectives and a new unifying nomenclature. Plant Cell Physiol. 43 (12), 1421–1435. doi: 10.1093/pcp/pcf171
Saed-Moucheshi, A., Pakniyat, H., Pirasteh-Anosheh, H., Azooz, M. M. (2014). Role of ROS as signaling molecules in plants. n Oxidative damage to plants,Academic Press. 585–620. doi: 10.1016/B978-0-12-799963-0.00020-4
Saito, K., Yonekura-Sakakibara, K., Nakabayashi, R., Higashi, Y., Yamazaki, M., Tohge, T., et al. (2013). The flavonoid biosynthetic pathway in Arabidopsis: Structural and genetic diversity. Plant Physiol. Biochem. 72, 21–34. doi: 10.1016/j.plaphy.2013.02.001
Shahbaz, M., Ashraf, M. (2013). Improving salinity tolerance in cereals. Crit. Rev. Plant Sci. 32 (4), 237–249. doi: 10.1080/07352689.2013.758544
Shrivastava, P., Kumar, R. (2015). Soil salinity: A serious environmental issue and plant growth promoting bacteria as one of the tools for its alleviation. Saudi J. Biol. Sci. 22 (2), 123–131. doi: 10.1016/j.sjbs.2014.12.001
Stewart, R. R. C., Bewley, J. D. (1980). Lipid peroxidation associated with accelerated aging of soybean axes. Plant Physiol. 65, 245–248. doi: 10.1104/pp.65.2.245
Tester, M., Davenport, R. (2003). Na+ tolerance and na+ transport in higher plants. Ann. Bot. 91, 503–527. doi: 10.1093/aob/mcg058
Wang, M., Ren, T., Huang, R., Li, Y., Zhang, C., Xu, Z. (2021). Overexpression of an Apocynum venetum flavonols synthetase gene confers salinity stress tolerance to transgenic tobacco plants. Plant Physiol. Biochem. 162, 667–676. doi: 10.1016/j.plaphy.2021.03.034
Wang, M., Xu, Z., Ding, A., Kong, Y. (2018). Genome-wide identification and expression profiling analysis of the xyloglucan endotransglucosylase/hydrolase gene family in tobacco (Nicotiana tabacum l.). Genes. 9 (6), 273. doi: 10.3390/genes9060273
Winkelshirley, B. (2001). Flavonoid biosynthesis. a colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol. 126 (2), 485–493. doi: 10.1104/pp.126.2.485
Xie, W., Zhang, X., Wang, T., Hu, J. (2012). Botany, traditional uses, phytochemistry and pharmacology of Apocynum venetum l. (Luobuma): A review. J. Ethnopharmacol. 141 (1), 1–8. doi: 10.1016/j.jep.2012.02.003
Xu, Z., Wang, M., Ren, T., Li, K., Li, Y., Marowa, P., et al. (2021). Comparative transcriptome analysis reveals the molecular mechanism of salt tolerance in Apocynum venetum. Plant Physiol. Biochem. 167, 816–830. doi: 10.1016/j.plaphy.2021.08.043
Xu, Z., Wang, M., Shi, D., Zhou, G., Niu, T., Hahn, M. G., et al. (2017). DGE-seq analysis of MUR3-related Arabidopsis mutants provides insight into how dysfunctional xyloglucan affects cell elongation. Plant Sci. 258, 156–169. doi: 10.1016/j.plantsci.2017.01.005
Xu, Z., Zhou, J., Ren, T., Du, H., Liu, H., Li, Y., et al. (2020). Salt stress decreases seedling growth and development but increases quercetin and kaempferol content in Apocynum venetum. Plant Biol. 22 (5), 813–821. doi: 10.1111/plb.13128
Yin, R., Messner, B., Faus-Kessler, T., Hoffmann, T., Schwab, W., Hajirezaei, M.-R., et al. (2012). Feedback inhibition of the general phenylpropanoid and flavonol biosynthetic pathways upon a compromised flavonol-3-O-glycosylation. J. Exp. Bot. 63 (7), 2465–2478. doi: 10.1093/jxb/err416
Zhang, B., Deng, Z., Ramdath, D. D., Tang, Y., Chen, P. X., Liu, R., et al. (2015). Phenolic profiles of 20 Canadian lentil cultivars and their contribution to antioxidant activity and inhibitory effects on alpha-glucosidase and pancreatic lipase. Food Chem. 172, 862–872. doi: 10.1016/j.foodchem.2014.09.144
Zhang, H., Wu, Z., Suo, Y., Wang, J., Zheng, L., Wang, Y. (2017). Gene expression and flavonol biosynthesis are induced by ultraviolet-b and salt stresses in Reaumuria trigyna. Biol. Plantarum 61 (2), 246–254. doi: 10.1007/s10535-017-0725-8
Keywords: apocynum venetum, salt stress, ROS, flavonoids, JA, IAA
Citation: Zhang M, Lu X, Ren T, Marowa P, Meng C, Wang J, Yang H, Li C, Zhang L and Xu Z (2023) Heterologous overexpression of Apocynum venetum flavonoids synthetase genes improves Arabidopsis thaliana salt tolerance by activating the IAA and JA biosynthesis pathways. Front. Plant Sci. 14:1123856. doi: 10.3389/fpls.2023.1123856
Received: 14 December 2022; Accepted: 22 February 2023;
Published: 27 March 2023.
Edited by:
Jianfeng Zhang, Chinese Academy of Forestry, ChinaReviewed by:
Yingchun Wang, Inner Mongolia University, ChinaCopyright © 2023 Zhang, Lu, Ren, Marowa, Meng, Wang, Yang, Li, Zhang and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zongchang Xu, eHV6YzExMTBAMTYzLmNvbQ==; Li Zhang, emhhbmdsaTc5MTJAMTYzLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.