- 1School of Life Sciences, Liaocheng University, Liaocheng, China
- 2Institute of Urban Agriculture, Chinese Academy of Agricultural Sciences, Chengdu, China
- 3Hainan Yazhou Bay Seed Laboratory, Sanya, China
- 4National Nanfan Research Institute (Sanya), Chinese Academy of Agricultural Sciences, Sanya, China
- 5Department of Forest Mycology and Plant Pathology, Swedish University of Agricultural Sciences, Uppsala, Sweden
Glycogen synthase kinase 3 (GSK3) family members are evolutionally conserved Ser/Thr protein kinases in mammals and plants. In plants, the GSK3s function as signaling hubs to integrate the perception and transduction of diverse signals required for plant development. Despite their role in the regulation of plant growth and development, emerging research has shed light on their multilayer function in plant stress responses. Here we review recent advances in the regulatory network of GSK3s and the involvement of GSK3s in plant adaptation to various abiotic and biotic stresses. We also discuss the molecular mechanisms underlying how plants cope with environmental stresses through GSK3s-hormones crosstalk, a pivotal biochemical pathway in plant stress responses. We believe that our overview of the versatile physiological functions of GSK3s and underlined molecular mechanism of GSK3s in plant stress response will not only opens further research on this important topic but also provide opportunities for developing stress-resilient crops through the use of genetic engineering technology.
1 Introduction
As a major source of food, fuel, and fiber, plant supports human society and sustains the global ecosystem by photosynthesis (Waadt et al., 2022; Zhang et al., 2022a). Plants are challenged throughout their life cycles by adverse environmental conditions including abiotic stresses such as drought, salinity, extreme temperatures, nutrient deficiency, and toxic metal levels in the soil as well as biotic stresses such as pathogen infection and herbivore attack (Peck and Mittler, 2020). These adverse environmental conditions limit the distribution of plants, threaten their growth, and reduce crop productivity, eventually resulting in devastating impacts on our economy. To reduce the impacts, it is necessary to understand how plants adapt to these adverse environmental conditions. There is growing evidence that plants have evolved sophisticated mechanisms to respond to environmental stresses, with recent results from some model and crop species the involvement of glycogen synthase kinase 3 (GSK3) protein in such adaptation.
2 Identification of plant GSK3s
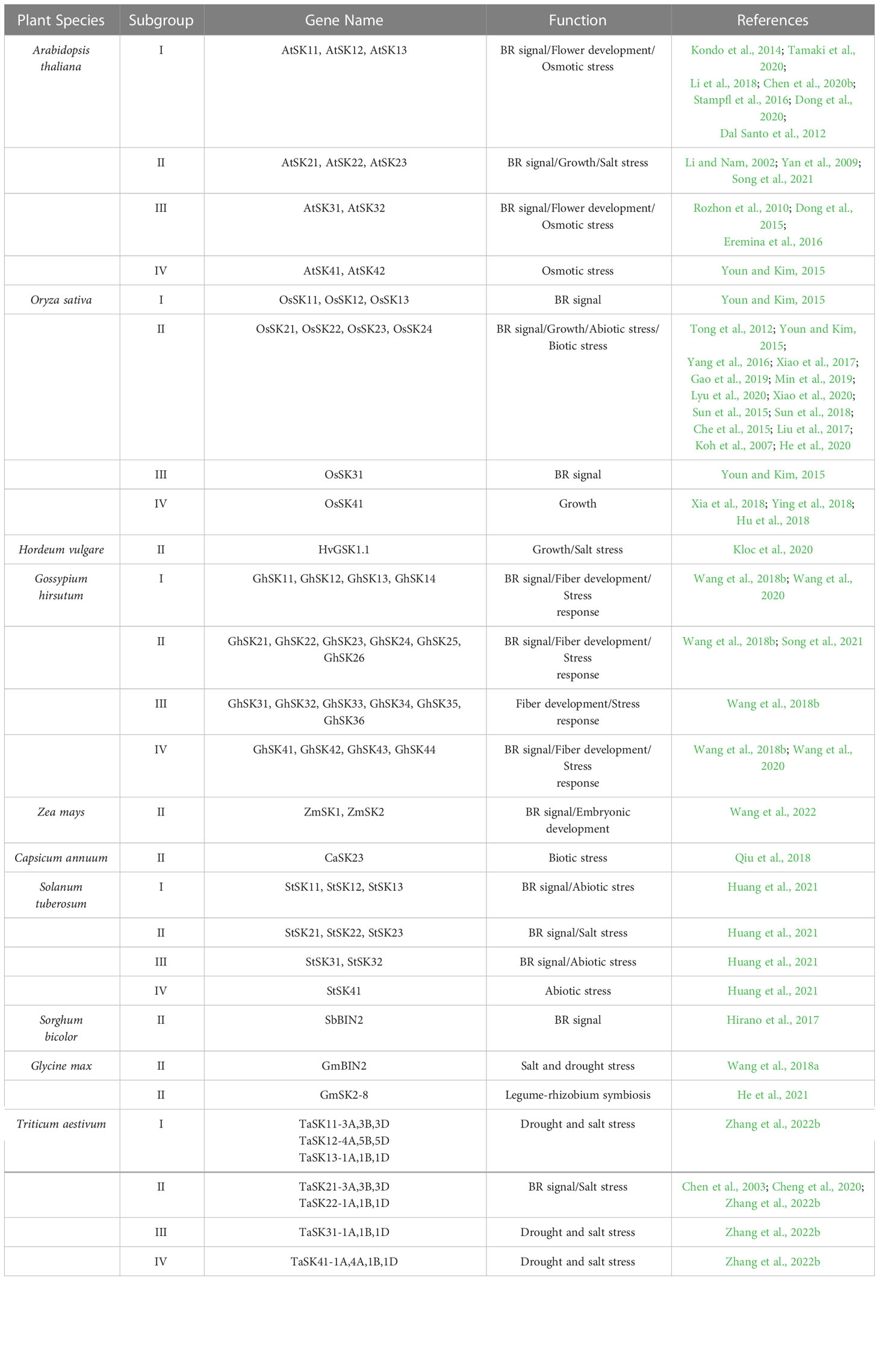
The glycogen synthase kinase GSK3, also known as shaggy-like kinase (SK), was first identified in humans and functions as a regulator of glycogen synthase (Woodgett, 1991; Li et al., 2021). Only two GSK3 isoforms (i.e., GSK3α and GSK3β) are present in human genomes. They regulate diverse biochemical and cellular processes (Beurel et al., 2015; Patel and Woodgett, 2017). In contrast to mammalian GSK3s, GSK3-like kinases in plants are encoded by multiple homologs of the genes and are groiped into four subfamilies (Table 1) (Li et al., 2021). For example, there are ten and nine GSK3-like kinases in the dicot arabidopsis (Arabidopsis thaliana) and the monocot rice (Oryza sativa), respectively (Qi et al., 2013; Youn and Kim, 2015). The GSK3-like genes have also been identified in several important crop species, such as barley (Hordeum vulgare), cotton (Gossypium hirsutum), maize (Zea mays), pepper (Capsicum annuum), potato (Solanum tuberosum), sorghum (Sorghum bicolor), soybean (Glycine max), wheat (Triticum aestivum) (Table 1) (Chen et al., 2003; Hirano et al., 2017; Qiu et al., 2018; Wang et al., 2018b; Cheng et al., 2020; Kloc et al., 2020; Wang et al., 2020; He et al., 2021; Huang et al., 2021; Hou et al., 2022; Wang et al., 2022; Zolkiewicz and Gruszka, 2022; Zhang et al., 2022b). Multiple copies of GSK3 in plant genomes indicate that this gene plays important and diverse roles in the evolutionary adaptation and life strategies of plants (Qi et al., 2013; Li et al., 2021). Indeed, despite evolutionary conservation, the specific function of GSK3 in plants can vary among species.
In plants, the best-known representative GSK3 is brassinosteroid insensitive 2 (BIN2/SK21). It is a key negative regulator of the plant steroid hormones brassinosteroid (BR) response in Arabidopsis (Li and Nam, 2002). The BIN2 gene has two closest homologs i.e., BIN2-like 1 (BIL1) and BIN2-like 2 (BIL2) functioning redundantly with BIN2 to negatively regulate the BR signaling. The bin2 bil1 bil2 triple mutants showed a constitutive BR-activation phenotype (Yan et al., 2009). The BR signaling has been well elucidated and proved to be an important regulator of plant physiological and biological processes including seed germination, cell division, elongation and differentiation, leaf senescence and response to biotic and abiotic stresses (Rozhon et al., 2010; Kondo et al., 2014; Dong et al., 2015; Anwar et al., 2018; Peres et al., 2019; Ackerman-Lavert and Savaldi-Goldstein, 2020; Tamaki et al., 2020; Zolkiewicz and Gruszka, 2022). BRs are recognized by the leucine-rich repeat receptor-like (LRR-RLK) protein brassinosteroid-insensitive 1 (BRI1) and its co-receptor BRI1 associated receptor kinase 1 (BAK1) at the membrane (Wang et al., 2008; Santiago et al., 2013). Moreover, BRI1 phosphorylates its inhibitor, BRI1 kinase inhibitor 1 (BKI1), guides the dissociation of BKI1 from the membrane, and enables the formation of the BRI1-BAK1 receptor complex to fully initiate the signaling cascade (Wang and Chory, 2006). The active receptor complex phosphorylates downstream proteins, including the BR signaling kinases (BSKs), constitutive differential growth 1 (CDG1) protein, and finally bri1 suppressor1 phosphatase (BSU1) (Tang et al., 2008; Kim et al., 2011). BSU1 and its homologs then dephosphorylate and inhibit the glycogen synthase kinase 3 kinase BIN2, which is a major repressor of the BR signaling (Kim et al., 2009). Without BR, BIN2 phosphorylates two key transcription factors bri1-EMS-suppressor 1 (BES1) and brassinazole-resistant 1 (BZR1), which eventually inhibits the BR downstream signaling (Li and Nam, 2002). Upon initiation of the BR signaling, as for the BIN2 inactivation, BZR1 and BES1 proteins bind to the promoters of the BR-responsive genes (Nolan et al., 2020).
In the monocot model rice, several numbers of OsSKs/OsGSK3s proteins are also found to function as negative regulators of BR signaling by phosphorylating and regulating the activity of several transcription factors involved in the BR-dependent gene expression (Tong et al., 2012; Zhang et al., 2012; Sun et al., 2015; Yang et al., 2016; Qiao et al., 2017; Xiao et al., 2017; Sun et al., 2018; Xia et al., 2018; Ying et al., 2018; Gao et al., 2019; Min et al., 2019; Lyu et al., 2020; Xiao et al., 2020). For example, OsSK22 can phosphorylate and stabilize OsPUB24 to promote the degradation of OsBZR1, which is similar to what was discovered in Arabidopsis (Min et al., 2019). Thus, OsSK22 was named as OsGSK2 and is widely considered as the rice ortholog of BIN2. In conclusion, the GSK3 proteins function as negative regulators of BR signaling, and their function is generally conserved among dicots and monocots.
3 Upstream regulators and downstream substrates of GSK3s
Emerging evidence has shown that GSK3s proteins can perceive upstream signals and be regulated at the post-translational level. Moreover, the downstream targets phosphorylated by the GSK3s proteins have also been extensively elucidated. In this section, we review how the GSK3s proteins are modulated by the known upstream regulators and how they regulate the downstream substrates through phosphorylation (Figure 1).
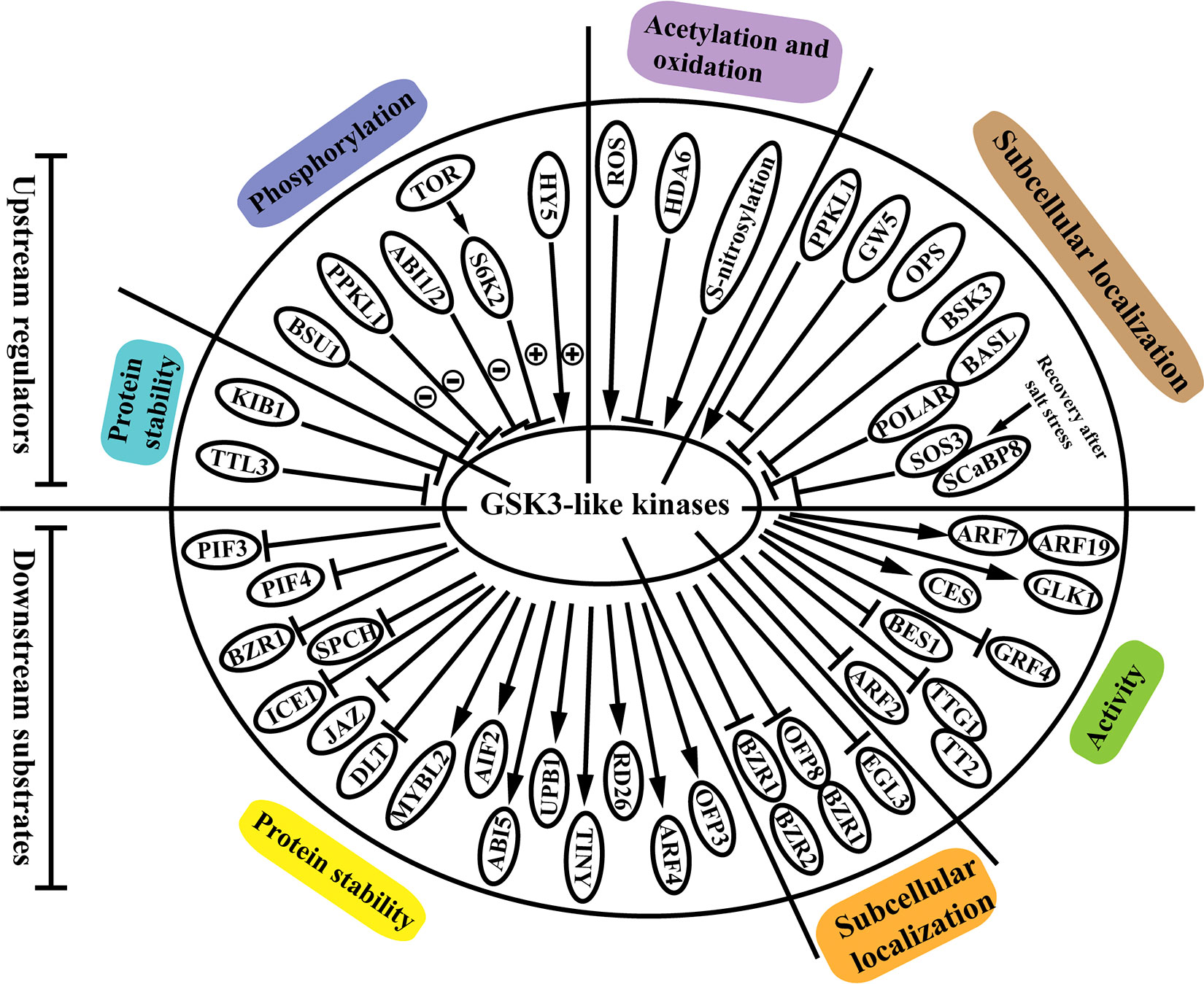
Figure 1 Upstream regulators and downstream substrates of GSK3-like kinases in plants. The GSK3s-like kinases are regulated via modulation of protein stability, protein activity via various modifications including phosphorylation, acetylation and oxidation, and subcellular localization. Meanwhile, GSK3s-like kinases modulate downstream substrates through phosphorylation, and affects the substrates stability, localization, and activity of these substrates. Circled plus or circled minus signs indicate phosphorylation and dephosphorylation, respectively. Arrows and bar ends indicate stimulatory and inhibitory action, respectively. Abbreviations: TTL3: tetratricopeptide thioredoxin-like 3; KIB1: kink suppressed in bzr1-1D 1; BSU1: bri1 suppressor1 phosphatase; PPKL1: phosphatase with Kelch-like repeat domains; ABI1/2: ABA insensitive 1/2; TOR: target of rapamycin; S6K2: ribosomal protein S6 kinase 2; HY5: elongated hypocotyl 5; ROS: reactive oxygen species; HDA6: histone deacetylase 6; GW5: grain width 5; OPS: octopus; BSK3: brassinosteroid signaling kinase 3; BASL: breaking of asymmetry in the stomatal lineage; POLAR: polar localization during asymmetric division and redistribution; SOS3: salt overly sensitive 3; SCaBP8, SOS3-like calcium binding protein 8; PIF3: phytochrome interacting factor 3; PIF4: phytochrome interacting factor 4; BZR1: brassinazole-resistant 1; SPCH: speechless; ICE1: inducer of CBF expression 1; JAZ: jasmonate ZIM domain; DLT: dwarf and low-tillering; MYBL2: myeloblastosis family transcription factor-like 2; AIF2: ATBS1-interacting factor 2; ABI5: abscisic acid insensitive 5; UPB1: upbeat 1; TINY: AP2 family transcription factor; RD26: NAC transcription factor; ARF4: auxin response factor 4; OFP3: OVATE family protein 3; BZR2: brassinazole-resistant 2; OFP8: OVATE family protein 8; EGL3: enhancer of glabra 3; ARF2: auxin response factor 2; TTG1: transparent testa glabra 1; TT2: transparent testa 2; BES1: bri1-EMS-suppressor 1; GRF4: growth regulating factor; CES: cesta; GLK1: golden2-like 1; ARF7: auxin response factor 7; ARF19: auxin response factor 19.
3.1 Upstream regulators of GSK3s
Numerous studies have revealed that the GSK3s proteins are mainly regulated at the post-translational level via modulation of protein stability, protein activity via various modifications (i.e., phosphorylation, acetylation and oxidation), and subcellular localization (Li et al., 2021). A series of GSK3s proteins, such as AtBIN2, AtBIL1, AtBIL2, AtSK11, AtSK12, OsGSK2, and TaSG-D1, is degradated through proteasome-mediated pathway (Li and Nam, 2002; Peng et al., 2008; Yan et al., 2009; Tong et al., 2012; Li et al., 2018; Cheng et al., 2020; Chen et al., 2020b). Moreover, the proteasome-mediated regulation of GSK3s is modulated via a conserved C-terminal TREE motif, and mutations in this motif cause the gain-of-function effects. For example, the mutation in the TREE motif impaired the ubiquitylation-dependent proteasomal degradation of BIN2 mediated by the E3 ubiquitin ligase kink suppressed in bzr1-1D 1 (KIB1). Moreover, homologous proteins of KIB1 are also involved in the BR-induced proteasomal degradation of BIN2, and they function redundantly in the suppression of the GSK3 kinase activity (Zhu et al., 2017). But how KIB1 and its homologous proteins interact with other proteins of the BR signaling remains elusive (Li et al., 2021).
Evidence that the activity of GSK3s is altered by protein modifications including phosphorylation, acetylation, and oxidation is also accumulated. For example, dephosphorylation and phosphorylation of GSK3s, which are the most common ways in the regulation of GSK3s (Figure 1), significantly change the activities of the proteins in various species. The suppressor of bri1 (BSU1) dephosphorylates BIN2 at phospho-tyrosine 200, and deactivates BIN2 kinase activity in the BR signaling transduction (Kim et al., 2009). Similarly, the BIN2 ortholog in rice OsGSK3 (OsSK23) is dephosphorylated by the rice ortholog of BSU1, OsPPKL1 (phosphatase with Kelch-like repeat domains), and affectes BR signaling (Gao et al., 2019). However, the effect of OsPPKL1 on OsGSK3 activity is completely different with that caused by AtBSU1. The dephosphorylation of AtBIN2 kinase resulted in the BIN2 degradation, but in rice, OsPPKL1 stabilized the OsGSK3 protein. The opposite effect on GSK3s protein stabilization may be related with its specific role in different species, which requires further investigation. The bZIP transcription factor, elongated hypocotyl 5 (HY5), binds to BIN2 and enhances the autophosphorylation of BIN2 at the tyrosine 200 residue (Li et al., 2020a). Ribosomal protein S6 kinase 2 (S6K2) phosphorylates BIN2 at serines 187 and 203, and inhibites BIN2 activity in Arabidopsis (Xiong et al., 2017). They also find that the phosphorylation of BIN2 depends on the upstream TOR-S6K2 signaling, which plays a vital role in coordinating plant growth and stress responses. Histone deacetylase 6 (HDA6) can also interact with and deacetylate BIN2 at lysine 189 to repress its kinase activity (Hao et al., 2016). The histone acetylation modification of GSK3s proteins is also identified in rice (Hou et al., 2022). The histone deacetylase1 (OsHDAC1) directly interactes with and deacetylates OsGSK2, and eventually inhibits OsGSK2 activity. The above results suggest the conserved regulation mechanism of GSK3 by acetylation in Arabidopsis and rice. The expression pattern of HDA6 gene when exposed to BR is opposite with that of OsHDAC1 gene, which needs further research. Moreover, it has been found that the interaction of BIN2 and BES1 is oxygen-dependent, and the activity of BIN2 is regulated by the oxidation of several cysteine (Cys) residues including C59, C95, C99, and C162 (Song et al., 2019).
Emerging evidence has indicated that regulation of the subcellular localization of GSK3s significantly influences their biological functions in plants. Plasma membrane recruitment of GSK3s modulates BR signaling in Arabidopsis and rice. In Arabidopsis, several proteins including octopus (OPS), tetratricopeptide thioredoxin-like 3 (TTL3), and brassinosteroid signaling kinase 3 (BSK3), interact with and recruit BIN2 to the plasma membrane, and prevent BIN2 inhibitory role in the nucleus in the BR signaling pathway. Among these proteins, TTL3 affects BIN2 subcellular localization and promotes the degradation of BIN2 (Anne et al., 2015; Amorim-Silva et al., 2019; Ren et al., 2019). As mentioned above, OsPPKL1 dephosphorylated and stabilized OsGSK3 in the cytoplasm to modulate BR signaling (Gao et al., 2019). We find that TTL3 not only influences BIN2 stability, but also affects the BIN2 subcellular localization. OsPPKL1 not only influences BIN2 through protein modifications, but also alters the BIN2 subcellular localization. These results together indicates that the upstream proteins can regulate GSK3s proteins in more ways than one. In rice, a calmodulin binding protein grain width 5 (GW5) can also physically interact with and recruit OsGSK32 to the plasma membrane, thus resulting in accumulation of unphosphorylated OsBZR1 and dwarf and low-tillering (DLT) to promote BR signaling (Liu et al., 2017). Moreover, GSK3s subcellular localization mediates plant transition from the salt stress response to growth recovery. BIN2 is mainly locialized in the nucleus to regulate BR signaling and plant growth under salt stresses. In the recovery phase, BIN2 was recruited to the plasma membrane by the salt-triggered calcium sensors salt overly sensitive 3 (SOS3) and SOS3-like calcium binding protein 8 (SCaBP8) to regulate the salt stress response (Li et al., 2020b). Taking the above aspects into account, we can conclude that the altered subcellular localization of GSK3s proteins contributes to realize GSK3s proteins functions in the balance between plant growth and stress response. GSK3s subcellular localization also influences stomatal patterning. Stomatal cell lineage is an archetypal example of asymmetric cell division (ACD). The scaffold membrane protein polar localization during asymmetric division and redistribution (POLAR) can confine BIN2 and AtSK12 to the cytosol to drive ACD. After ACDs, BIN2 and AtSK12 phosphorylate and dissociate from POLAR (Houbaert et al., 2018).
3.2 Downstream targets of GSK3s
Like upstream regulation, GSK3s also intensively modulate downstream substrates through phosphorylation to accomplish their functions in regulating plant development and stress responses. GSK3s can phosphorylate a series of substrates including transcription factors, cofactors, kinases, scaffold proteins, cytoskeleton proteins, cyclins, metabolic enzymes, and components of the ubiquitin-proteasome system (UPS). The role of GSK3s in modulating proteins including kinases, scaffold proteins, cytoskeleton proteins, cyclins, metabolic enzymes, and components of the ubiquitin-proteasome system (UPS), has been extensively discussed in several excellent recent reviews (Youn and Kim, 2015; Li et al., 2021; Mao et al., 2021; Zolkiewicz and Gruszka, 2022). In this section, we mainly focus on the discoveries about effects of GSK3s on the transcription factors and cofactors, which play essential roles in modulating stress responsive gene expression.
GSK3s-mediated phosphorylation of the transcription factors and cofactors, eventually affects the protein stability, localization, and activity of these substrates. Recent reports have revealed that GSK3-mediated phosphorylation not only promotes the degradation of the substrates but also stabilizes downstream targets. During stomatal patterning, BIN2 and MAPKs phosphorylate and promote the degradation of speechless (SPCH), which is a bHLH transcription factor in the nucleus required for stomatal initiation (Gudesblat et al., 2012). Phosphorylation and degradation of phytochrome interacting factor 3 (PIF3) and 4 (PIF4) are facilitated by BIN2 to control skotomorphogenesis and hypocotyl elongation, respectively (Bernardo-García et al., 2014; Ling et al., 2017). In plants, GSK3s also stabilizes transcription factors through phosphorylation. A series of transcription factors, such as myeloblastosis family transcription factor-like (MYBL2; a MYB transcription factor), ATBS1-interacting factor 2 (AIF2; a bHLH transcription factor), abscisic acid insensitive 5 (ABI5; a bZIP transcription factor), upbeat 1 (UPB1; a bHLH transcription factor), TINY (AP2 family transcription factor) and RD26 (NAC family transcription factor), are phosphorylated and stabilized by BIN2 (Ye et al., 2012; Hu and Yu, 2014; Kim et al., 2017; Jiang et al., 2019; Xie et al., 2019; Li et al., 2020c). However, the underlying molecular mechanisms of substrates degradation or accumulation associated with BIN2-catalyzed phosphorylation have not been investigated.GSK3s-mediated regulation of subcellular localization in substrates is ubiquitous in plants. BIN2-catalyzed phosphorylation of BZR1/BZR2 promotes their binding to the 14-3-3 proteins, resulting in cytoplasmic retention and inhibition of BR-regulated gene expression (Gampala et al., 2007). In rice, phosphorylation of OsBZR1 and transcription factor ovate family protein 8 (OFP8) by OsGSK2 also results in their nuclear export (Yang et al., 2016). Furthermore, GSK3s-mediated phosphorylation can restrict the nuclear localization of their substrates. For example, the bHLH transcription factor enhancer of glabra 3 (EGL3) is phosphorylated by BIN2, thus remaining nuclear localization and participating in the BR-regulated root epidermal cell patterning (Cheng et al., 2014).
In addition, GSK3s have been early documented to affect the activity of transcription factors or cofactors. For example, phosphorylation of auxin response factors ARF7 and ARF19 by BIN2 suppresses their interaction with the AUX/IAAs repressors and enhances auxin signaling export in enhancing lateral root development (Cho et al., 2014). AtSK11 and AtSK12 phosphorylate the WD40 motif-containing transcriptional cofactor transparent testa glabra 1 (TTG1), influencing the interaction between TTG1 and a MYB domain transcription factor transparent testa 2 (TT2), which eventually influence the carbon partitioning between various parts of developing seed (Li et al., 2018). GSK3s also affect the DNA-binding ability or transcriptional activity of transcription factors, such as BES1, BZR1, ARF2, rice growth regulating factor GRF4 (reduced), golden 2-like 1 (GLK1) and cesta CES (enhanced) (Vert and Chory, 2006; Vert et al., 2008; Khan et al., 2014; Che et al., 2015; Duan et al., 2015; Zhang et al., 2021). The transcription factors or cofactors targeted by GSK3s proteins regulate the expression of downstream plant stress responsive genes, and eventually allow GSK3s to participate in plant response to environmental stresses.
4 Crosstalk between the GSK3s and phytohormones
Plant hormones are essential signaling compounds in regulating the interactions between plants and their complex biotic and abiotic environments. GSK3s have been found to mediate the crosstalk between BR and other hormones including auxin, abscisic acid (ABA), jasmonic acid (JA) and salicylic acid (SA). GSK3s coordinate actions of auxin through phosphorylating the auxin transcription factors (ARFs) including ARF2, ARF5, ARF7, ARF9, and OsARF4 (Vert et al., 2008; Cho et al., 2014; Han et al., 2018; Hu et al., 2018). The involvement of GSK3s proteins in the crosstalk with auxin mainly regulates plant developmental processes. ABA is a major stress related hormone that integrates a wide range of stress signals such as cold, salinity, osmolarity, and drought (Zhu, 2016; Chen et al., 2020a). Recent studies have uncovered that there are multifaceted interactions between GSK3s and ABA. Subgroup III snf1-related kinase 2s (SnRK2s) are key positive regulators in the ABA signaling pathway (Umezawa et al., 2009). SnRK2.2 and SnRK2.3 can interact with, be phosphorylated and thus be activated by BIN2, providing significant insight into the modulation of ABA signaling by GSK3s (Cai et al., 2014). BIN2 phosphorylates and stabilizes the bZIP-type transcription factor ABI5 of the ABA signaling pathway to mediate the antagonism of ABA by BR (Hu and Yu, 2014). But its activity on BES1 phosphorylation is affected by ABA insensitive 1 (ABI1) and 2 (ABI2), which are negative regulators of ABA signaling (Wang et al., 2018a). ABI1 and ABI2 interact with and dephosphorylate BIN2, thereby forming a PP2Cs-BIN2-SnRK2s module in the ABA signaling pathway. JA is another essential stress related hormone in plants, and the jasmonate ZIM domain (JAZ) proteins are key repressors of the JA pathway. GSK3s kinase proteins interact with and promote the degradation of JAZ proteins, and the interaction is ubiquitously present in plants including Arabidopsis, rice and cotton (He et al., 2020; Song et al., 2021). SA is a representative plant defense hormone that plays pivotal roles in immunity and systemic acquired resistance. The TGACG motif-binding transcription factors TGAs are known to mediate SA signaling. BIN2 phosphorylates and inhibits the activity of redox-sensitive clade I TGA4 during plant response to Pst DC3000. BR inactivates BIN2 and promotes SA responses by inactivating BIN2 (Kim et al., 2022). Moreover, recent results indicate that SA activates BIN2 which phosphorylates TGA3, enhancing TGA3 DNA binding ability, thereby activating PR gene expression and promoting disease resistance in Arabidopsis (Han et al., 2022). The advances displayed in this section indicates that GSK3s proteins function as hubs of phytohormones signaling pathways mainly through interacting with the transcription factors of these pathways, which eventually influence the expression of downstream responsive genes of these phytohormones. The interaction between GSK3s and other key node genes of these phytohormones needs further investigation.
5 Involvement of the GSK3s in plant response to multiple stresses
Plants are challenged by various types of environmental stresses throughout their life cycles, which have profound effects on plant growth and survival. Accumulated evidence from recent findings has deciphered that GSK3s play a pivotal role in plant stress responses in model and crop species (Figure 2).
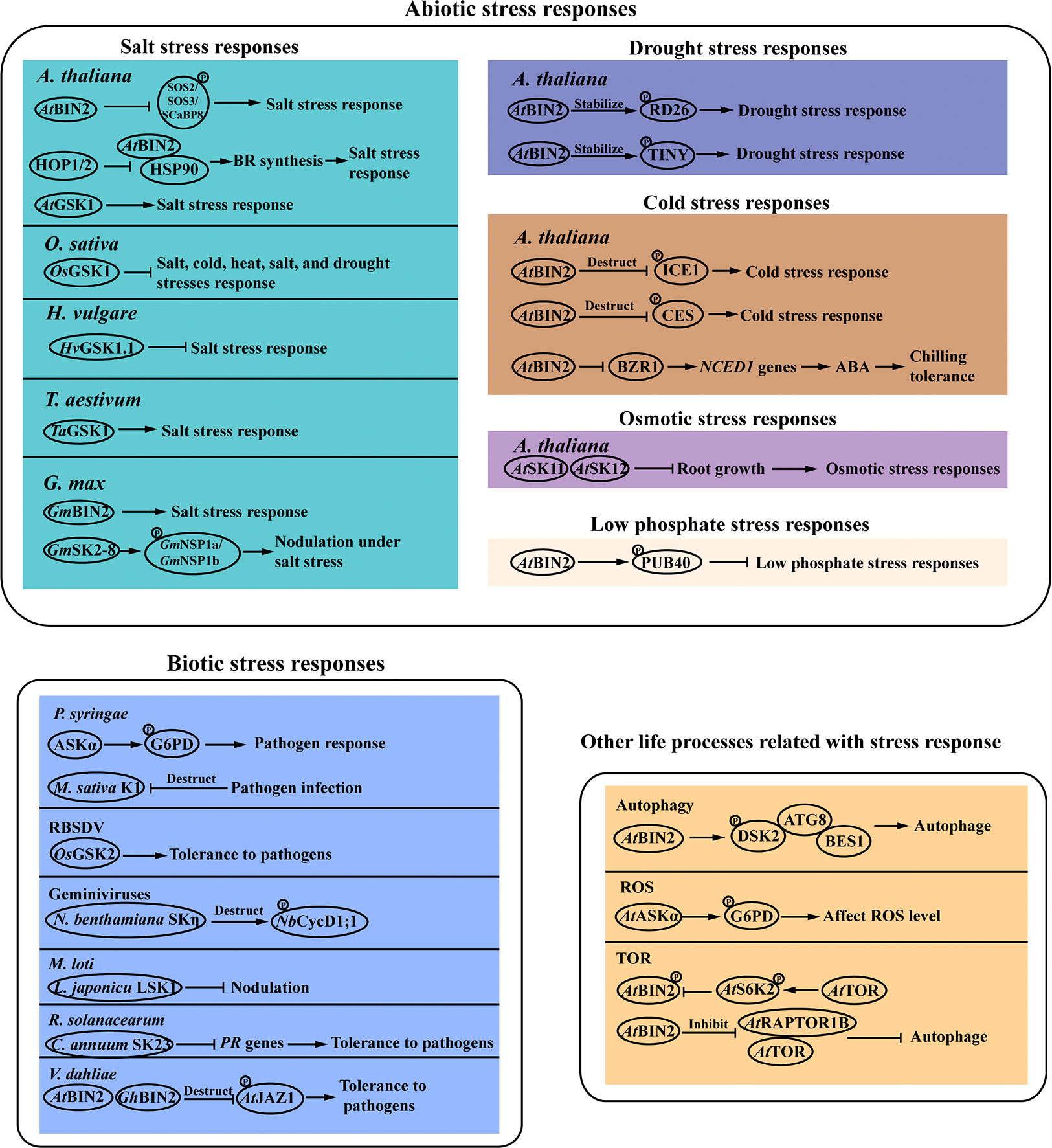
Figure 2 Plant stress responses mediated by GSK3-like kinases. GSK3s participated in various stress response pathways. The detailed description is given in the text. Arrows and bar ends indicate stimulatory and inhibitory action, respectively. Abbreviations: GSK3, glycogen synthase kinase 3; BIN2, brassinosteroid insentisive 2; SOS, salt overly sensitive; SCaBP8, SOS3-like calcium binding protein 8; HOP 1/2: co-chaperone heat shock protein (HSP)70-HSP90 organizing protein 1/2; HSP90, heat shock protein 90; SK2, shaggy-like kinase 2; NSP1, nodulation signaling pathway 1; RD26, NAC family transcription factor; TINY, AP2 family transcription factor; PUB40, plant U-box 40 protein; ICE1, inducer of CBF expression 1; CES, cesta; CycD1;1, Cyclin D 1.1; LSK1, shaggy -like kinase 1; DSK2, dominant suppressor of KAR 2; ROS, reactive oxygen species; G6PD, glc-6-phosphate dehydrogenase; TOR, target of rapamycin; RAPTOR1B, egulatory-associated protein of TOR 1B; PR: pathogenesis-related; JAZ: jasmonate ZIM domain; ATG8: autophagy 8; BES1: bri1-EMS-suppressor 1; S6K2: ribosomal protein S6 kinase 2.
5.1 GSK3s in abiotic stresses
Soil salinization is a growing problem for agriculture worldwide. It is crucial to uncover the key components of the plant salt tolerance network. Recent studies have shown that GSK3s play important roles in regulating plant salinity stress responses. In Arabidopsis, overexpression of AtGSK1 enhances plant resistance to sodium chloride (NaCl) stress, and induces the expression of some NaCl stress-responsive genes including the Ca2+-binding protein (AtCP1), the desiccation 29A (RD29A) and the chalcone synthase gene (CHS1), suggesting that AtGSK1 is involved in the signal transduction pathway of NaCl stress responses (Piao et al., 2001). BIN2 is found to function as a molecular switch between plant salt stress response and growth through phosphorylating and affecting the activity of calcium sensors, such as SOS3, SCaBP8, and SOS2 (Li et al., 2020b). Heat shock proteins (HSPs), a group of highly conserved chaperone protein, are involved in the regulation of plant responses to salinity. HSP90 regulates the activity of the BIN2 kinase by modulating its subcellular localization (Samakovli et al., 2014). The co-chaperone heat shock protein (HSP)70-HSP90 organizing protein 1 (HOP1) and 2 (HOP2) affect HSP90-BIN2 interaction and are involved in plant salt tolerance by affecting BR signaling (Zhang et al., 2022). In rice, knockout mutants of OsGSK1 showed enhanced tolerance to salt stress (Koh et al., 2007). Heterologous overexpression of a GSK3 gene from soybean GmBIN2 enhanced plant tolerance to salt in Arabidopsis. GSK3-like kinase GmSK2-8 is strongly induced in soybean under salt stress. GmSK2-8 interactes with two nodulation signaling pathway 1 (GmNSP1) proteins GmNSP1a and GmNSP1b, which are key transcription factors involved in legume-rhizobium symbiosis. GmSK2-8 phosphorylates the GmNSP1a protein, and thus suppresses nodule formation under salt stress (He et al., 2021). It is reported that the GSK3 family shows altered expression in response to salt stress treatments in Solanum tuberosum L. Overexpression of StSK21 provides enhanced sensitivity to salt stress in Arabidopsis (Huang et al., 2021). In addition, the response of the GSK3 family to salt stress is also identified in cotton (Wang et al., 2018b). In barley (Hordeum vulgare L.), the RNAi-mediated silencing of the target HvGSK1.1 gene enhances the BR-dependent signaling, and generates plants with improved agricultural traits under salt stress conditions (Kloc et al., 2020). Researches show that the wheat (Triticum aestivum L.) TaGSK1 gene is induced by NaCl stress, and expresses more strongly in salt-stress resistant lines than in salt-stress sensitive lines (Chen et al., 2003). From the above results we can see that some of the GSK3s proteins function as positive regulators of plant salt tolerance such as in soybean and wheat, while the GSK3s proteins reported in rice, potato, barley negatively regulate plant salt tolerance. In addition, different members in the Arabidopsis and cotton GSK3s family proteins show different response to the salt treatment. These results indicate that molecular mechanism of the GSK3s proteins in response to salt stress in different species or different members in the protein family is specific.
Drought stress causes a decline in the quantity of crop yields, and has become more accentuated recently due to climatic change. The GSK3 proteins are involved in drought stress tolerance. The stress-responsive NAC transcription factor RD26 participates in the interaction between growth and drought stress signaling by the phosphorylation of BIN2. BIN2 directly interacted with and phosphorylated RD26, which is required for RD26 transcriptional activation on drought-responsive genes (Jiang et al., 2019). Arabidopsis stress-inducible AP2/ERF transcription factor TINY positively regulates drought responses by activating drought-responsive genes. The BR negative regulator BIN2 phosphorylates and stabilizes TINY, which provides a mechanism for BR-mediated down-regulation of TINY to prevent activation of stress responses (Xie et al., 2019).
GSK3s are also involved in osmotic, low temperature and low phosphate stresses. AtSK11 and AtSK12 are involved in the mild osmotic stress (-0.4 MPa) response in Arabidopsis thaliana. They negatively regulate the induction of root growth in response to mild osmotic stress (Dong et al., 2020). Plant U-box 40 protein (PUB40) is a ubiquitin E3 ligase and mediates the proteasomal degradation of BZR1 in Arabidopsis. The interaction between PUB40 and BZR1 influences root tolerance to the low phosphate stress. BIN2 phosphorylates and stabilizes PUB40 to promote BZR1 degradation and enhances the interaction between PUB40 and BZR1, thus reducing plant tolerance to the low phosphate stress (Kim et al., 2019). Moreover, BIN2 is also regulated by the E3 ubiquitin ligase KIB1 as is mentioned above (Zhu et al., 2017). These results indicate that the E3 ubiquitin ligases play pivotal roles in the BIN2-mediated regulation of BR signaling pathway transduction. BR signaling and downstream transcriptional cascades are reported to be involved in regulating plant cold tolerance (Khan et al., 2014; Eremina et al., 2016; Li et al., 2017). As repressors of BR signaling, BIN2 and its homologs mediate the phosphorylation of BZR1 and inducer of CBF expression 1 (ICE1) to facilitate their degradation, and thus affect downstream transcriptional cascades related to cold stress response (Ye et al., 2019). BIN2 negatively regulates chilling tolerance in tomato. BIN2 also regulates the accumulation of BZR1, which controls the expression of ABA biosynthesis gene 9-cis-epoxycarotenoid dioxygenase 1 (NCED1) in tomato. These results demonstrate that BR signaling positively regulates chilling tolerance via ABA biosynthesis in tomato (An et al., 2023). Moreover, cesta (CES) is a bHLH transcription factor of BR signaling and affects plant cold tolerance (Eremina et al., 2016). CES degradation is also promoted by BIN2-mediated phosphorylation, but whether and how this protein turnover is associated in vivo with cold stress response should be further explored.
5.2 GSK3s in biotic stresses
Biotic stresses including bacteria, fungi, oomycetes, viruses and insects, wreak havoc on agricultural products worldwide and increase the risk of starvation in many areas. Advancing researches have shown that GSK3s play multifaceted roles in plant responses to various kinds of biotic stresses and is tightly regulated during plant response to pathogen infection (Wrzaczek et al., 2007). It is reported that the GSK3-like kinase ASKα is identified as a positive regulator of plant immune signaling (Stampfl et al., 2016). Loss of ASKα attenuates, whereas its overexpression enhances, diverse pattern-triggered immunity (PTI) responses, which is the first layer of plant immunity against pathogenic microbes. The bacterial pathogen Pseudomonas syringae glucose-6-phosphate dehydrogenase (G6PD) is the key enzyme of the oxidative pentose phosphate pathway and is phosphorylated by ASKα. Rice black-streaked dwarf virus (RBSDV), a double-stranded RNA virus, causes acute growth abnormalities in plants and results in serious yield losses in cereal crops. Plants over-expressing OsGSK2 display milder symptoms than the control, suggesting a positive role of OsGSK2 in suppressing RBSDV infection in rice (He et al., 2020). Geminiviruses transmitted by whiteflies cause severe developmental abnormalities in plants. The C4 protein encoded by geminiviruses induces abnormal cell division that determines viral symptoms. The tomato leaf curl Yunnan virus (TLCYnV) C4 protein interacts with and affects the subcellular localization of Nicotiana benthamiana GSK3 protein NbSKη, eventually impairing GSK3-mediated degradation of cell division associated protein NbCycD1;1 (Mei et al., 2018; Mei et al., 2020). In Medicago sativa, the GSK3-like kinase MsK1 is important for innate immunity and limits the severity of infection caused by virulent bacterial pathogen P. syringae. MsK1 activity is downregulated by the elicitor cellulase. Cellulase treatment also triggeres the degradation of the MsK1 protein (Wrzaczek et al., 2007). In Lotus japonicu, nodulation is mainly formed through the symbiotic nitrogen-fixing bacterium Mesorhizobium loti infection. Studies show that lotus SHAGGY-like kinase 1 (LSK1) is required to suppress nodulation (Garagounis et al., 2019; Solovou et al., 2021). Ralstonia solanacearum is a devastating soil-borne bacterium that causes wilting disease in over 200 economically-important plant species. The GSK3/SHAGGY-like kinase CaSK23 negatively regulates plant response to R. solanacearum attack in Capsicum annuum (Qiu et al., 2018). We also find that BIN2 negatively regulates plant defence against Verticillium dahliae in Arabidopsis and cotton, consistent with others reports on the regulatory function of GSK3s proteins (Song et al., 2021).
5.3 GSK3s in other life processes related to stress response
Autophagy is a highly conserved quality control mechanism in which harmful or unwanted cellular components are delivered into lytic vacuoles for recycling, and can promote plant resistance to various stresses (Signorelli et al., 2019; Yang et al., 2020). The GSK3-like kinase is one of the key regulators of autophagy (Nolan et al., 2017). The dominant suppressor of KAR 2 (DSK2) is a ubiquitin receptor protein that targets BES1 to the autophagy pathway by interacting with ATG8, a ubiquitin-like protein that directs autophagosome formation and cargo recruitment. BIN2 can phosphorylate DSK2 and promote DSK2-ATG8 interaction, which ultimately targets BES1 for degradation (Nolan et al., 2017). Reactive oxygen species (ROS) play a key role in the acclimation process of plants to abiotic and biotic stresses (Choudhury et al., 2017). Researchers have found that the interaction of BIN2 and BES1 is dependent on oxygen, which can directly modify BIN2 (Song et al., 2019). In Arabidopsis, AtASKα regulates plant stress tolerance by activating glc-6-phosphate dehydrogenase (G6PD) responsible for maintaining cellular redox balance. Plants overexpressing ASKα have low levels of ROS in stress responses and are more tolerant to salt stress (Dal Santo et al., 2012; Stampfl et al., 2016). The target of rapamycin (TOR) is an atypical Ser/Thr protein kinase that is evolutionally conserved among yeasts, plants, and mammals. TOR signaling is involved in plant adaptation to nutrient deficiency and various abiotic stresses (Fu et al., 2020). The TOR downstream effector S6K2 can phosphorylate BIN2 protein, suggesting that BIN2 acts as a downstream effector of TOR signaling (Xiong et al., 2017). The egulatory-associated protein of TOR 1B (RAPTOR1B) is an important component of plant TOR complex. In the absence of BR, the BIN2 kinase directly phosphorylates and inhibits the activity of RAPTOR1B. Furthermore, autophagy is negatively regulated by TOR. Phosphorylation of RAPTOR1B by BIN2 thus activates the autophagy pathway. In the presence of BR, the inhibition effect of BIN2 on RAPTOR1B is attenuated, and results in increased TOR activity and ATG13a phosphorylation, and decreased autophagy activity (Liao et al., 2022). These results suggest that BIN2 functions as a key hub in the crosstalk between TOR and BR signaling pathways. Further studies are required to fully elucidate whether and how BIN2 interacts with other components of TOR signaling pathway.
6 Conclusion
The glycogen synthase kinase 3 protein was first identified in animals, and functions as an ancestral kinase of the stress response in eukaryotes. With the identification of additional proteins and other novel substrates that regulate their functions, our knowledge of plant GSK3s has inceased significantly over the last decades. It is now clear that the GSK3 proteins are involved in numerous stress-response pathways that influence animal health and plant reproduction. In animals, the GSK3s are activated under stress conditions, and determine cell fate based on its subcellular localization and specific partners (Racaud-Sultan and Vergnolle, 2021). GSK3s are found to be activated in pathologies such as inflammation and cancer, where adult stem cells are downregulated (Murata, 2018). Stem cells control tissue regeneration throughout the life of organisms, and should be protected from stressors. The microenvironment of adult stem cells is named “niche”. Numerous studies have shown that the GSK3s proteins are found to function as a sensor of the adult stem cell niche. Besides, the GSK3s are nodes of signaling pathways controlling survival, proliferation, adhesion and differentiation in adult stem cells (Racaud-Sultan and Vergnolle, 2021). Plant stem cells reside within the meristems and are also defined by their ability to self-renew and to generate new tissues (Heidstra and Sabatini, 2014). During plant growth, procambial and cambial cells in the vascular system self-proliferate and differentiate into xylem cells, and are mainly regulated by a peptide ligand and its receptor; tracheary element differentiation inhibitory factor (TDIF) and TDIF receptor (TDR) (Ito et al., 2006). Plant GSK3 proteins are crucial downstream components of TDIF–TDR signaling and regulate xylem cell differentiation (Kondo et al., 2014).
Although our understanding of GSK3s is advanced, many fundamental questions related to the gene family and its associated proteins remain unknown. For example, plants contain divergent GSK3-like kinases, but our current knowledge of the proteins comes mainly from subgroups I and II from a limited number of species. It requires understanding how GSK3s in other subgroups (e.g., III and IV) and species and what other regulatory substrates are involved in plant response to diverse environmental stimuli. The subgroups III and IV GSK3-like kinases and substrates of GSK3-like kinases reviewed here potentially provide new alleles to improve stress resistance in crops through engineering, which is an effective strategy for crop breeding processes. As mentioned above, GSK3s primarily affect hormonal signaling. Whether and how GSK3s directly regulate hormone biosynthesis is unclear and needs to be addressed. Such knowledge gap can be effectively bridged by applying recently emerging technologies such as genome-editing systems, single cell transcriptomics and deep tissue proteomics for research. Deciphering these molecular mechanisms controlling GSK3s function in detail will contribute to a better understanding of how internal and external signals integrate and branch for plant adaptation to the environment, which can eventually be transferred to stress breeding to ensure food production and ecological sustainability.
Author contributions
MR, JsZ, YSo and YW conceived and planned this review paper. YSo and YW prepared and drafted the manuscript. JsZ, QY, YSu and JlZ revised the manuscript. All authors contributed to the article and approved the submitted version. All authors contributed to the article and approved the submitted version.
Funding
This research was funded by Shandong Provincial Natural Science Foundation, grant number ZR2021QC200, ZR2020QC037, ZR2020QB154, ZR2021MC079; the Local Financial Funds of Chengdu National Agricultural Science and Technology Center, grant number NASC2021ST08, NASC2021PC04, NASC2021KR03 and NASC2020AR08; Hainan Yazhou Bay Seed Laboratory (B21Y10210); Nanfan Special Project, CAAS (YBXM12).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Ackerman-Lavert, M., Savaldi-Goldstein, S. (2020). Growth models from a brassinosteroid perspective. Curr. Opin. Plant Biol. 53, 90–97. doi: 10.1016/j.pbi.2019.10.008
Amorim-Silva, V., García-Moreno, Á., Castillo, A. G., Lakhssassi, N., Esteban Del Valle, A., Pérez-Sancho, J., et al. (2019). TTL Proteins scaffold brassinosteroid signaling components at the plasma membrane to optimize signal transduction in Arabidopsis. Plant Cell. 31 (8), 1807–1828. doi: 10.1105/tpc.19.00150
An, S., Liu, Y., Sang, K., Wang, T., Yu, J., Zhou, Y., et al. (2023). Brassinosteroid signaling positively regulates abscisic acid biosynthesis in response to chilling stress in tomato. J. Integr. Plant Biol. 65 (1), 10–24. doi: 10.1111/jipb.13356
Anne, P., Azzopardi, M., Gissot, L., Beaubiat, S., Hématy, K., Palauqui, J. C. (2015). OCTOPUS negatively regulates BIN2 to control phloem differentiation in Arabidopsis thaliana. Curr. Biol. 25 (19), 2584–2590. doi: 10.1016/j.cub.2015.08.033
Anwar, A., Liu, Y., Dong, R., Bai, L., Yu, X., Li, Y. (2018). The physiological and molecular mechanism of brassinosteroid in response to stress: A review. Biol. Res. 51 (1), 46. doi: 10.1186/s40659-018-0195-2
Bernardo-García, S., de Lucas, M., Martínez, C., Espinosa-Ruiz, A., Davière, J. M., Prat, S. (2014). BR-Dependent phosphorylation modulates PIF4 transcriptional activity and shapes diurnal hypocotyl growth. Genes Dev. 28 (15), 1681–1694. doi: 10.1101/gad.243675.114
Beurel, E., Grieco, S. F., Jope, R. S. (2015). Glycogen synthase kinase-3 (GSK3): regulation, actions, and diseases. Pharmacol. Ther. 148, 114–131. doi: 10.1016/j.pharmthera.2014.11.016
Cai, Z., Liu, J., Wang, H., Yang, C., Chen, Y., Li, Y., et al. (2014). GSK3-like kinases positively modulate abscisic acid signaling through phosphorylating subgroup III SnRK2s in Arabidopsis. Proc. Natl. Acad. Sci. U S A. 111 (26), 9651–9656. doi: 10.1073/pnas.1316717111
Che, R., Tong, H., Shi, B., Liu, Y., Fang, S., Liu, D., et al. (2015). Control of grain size and rice yield by GL2-mediated brassinosteroid responses. Nat. Plants. 2, 15195. doi: 10.1038/nplants.2015.195
Chen, K., Li, G. J., Bressan, R. A., Song, C. P., Zhu, J. K., Zhao, Y. (2020a). Abscisic acid dynamics, signaling, and functions in plants. J. Integr. Plant Biol. 62 (1), 25–54. doi: 10.1111/jipb.12899
Chen, G., Ma, W., Huang, Z., Xu, T., Xue, Y., Shen, Y. (2003). Isolation and characterization of TaGSK1 involved in wheat salt tolerance. Plant Science. 165 (6), 1369–1375. doi: 10.1016/S0168-9452(03)00365-0
Chen, Y., Song, S., Gan, Y., Jiang, L., Yu, H., Shen, L. (2020b). SHAGGY-like kinase 12 regulates flowering through mediating CONSTANS stability in Arabidopsis. Sci. Adv. 6 (24), eaaw0413. doi: 10.1126/sciadv.aaw0413
Cheng, X., Xin, M., Xu, R., Chen, Z., Cai, W., Chai, L., et al. (2020). A single amino acid substitution in STKc_GSK3 kinase conferring semispherical grains and its implications for the origin of Triticum sphaerococcum. Plant Cell. 32 (4), 923–934. doi: 10.1105/tpc.19.00580
Cheng, Y., Zhu, W., Chen, Y., Ito, S., Asami, T., Wang, X. (2014). Brassinosteroids control root epidermal cell fate via direct regulation of a MYB-bHLH-WD40 complex by GSK3-like kinases. Elife. 3, e02525. doi: 10.7554/eLife.02525
Cho, H., Ryu, H., Rho, S., Hill, K., Smith, S., Audenaert, D., et al. (2014). A secreted peptide acts on BIN2-mediated phosphorylation of ARFs to potentiate auxin response during lateral root development. Nat. Cell Biol. 16 (1), 66–76. doi: 10.1038/ncb2893
Choudhury, F., Rivero, R., Blumwald, E., Mittler, R. (2017). Reactive oxygen species, abiotic stress and stress combination. Plant J. 90 (5), 856–867. doi: 10.1111/tpj.13299
Dal Santo, S., Stampfl, H., Krasensky, J., Kempa, S., Gibon, Y., Petutschnig, E., et al. (2012). Stress-induced GSK3 regulates the redox stress response by phosphorylating glucose-6-phosphate dehydrogenase in Arabidopsis. Plant Cell. 24 (8), 3380–3392. doi: 10.1105/tpc.112.101279
Dong, X., Nou, I. S., Yi, H., Hur, Y. (2015). Suppression of ASKβ (AtSK32), a clade III Arabidopsis GSK3, leads to the pollen defect during late pollen development. Mol. Cells 38 (6), 506–517. doi: 10.14348/molcells.2015.2323
Dong, L., Wang, Z., Liu, J., Wang, X. (2020). AtSK11 and AtSK12 mediate the mild osmotic stress-induced root growth response in Arabidopsis. Int. J. Mol. Sci. 21 (11), 3991. doi: 10.3390/ijms21113991
Duan, P., Ni, S., Wang, J., Zhang, B., Xu, R., Wang, Y., et al. (2015). Regulation of OsGRF4 by OsmiR396 controls grain size and yield in rice. Nat. Plants. 2, 15203. doi: 10.1038/nplants.2015.203
Eremina, M., Unterholzner, S. J., Rathnayake, A. I., Castellanos, M., Khan, M., Kugler, K. G., et al. (2016). Brassinosteroids participate in the control of basal and acquired freezing tolerance of plants. Proc. Natl. Acad. Sci. U S A. 113 (40), E5982–e5991. doi: 10.1073/pnas.1611477113
Fu, L., Wang, P., Xiong, Y. (2020). Target of rapamycin signaling in plant stress responses. Plant Physiol. 182 (4), 1613–1623. doi: 10.1104/pp.19.01214
Gampala, S., Kim, T., He, J., Tang, W., Deng, Z., Bai, M., et al. (2007). An essential role for 14-3-3 proteins in brassinosteroid signal transduction in Arabidopsis. Dev. Cell. 13 (2), 177–189. doi: 10.1016/j.devcel.2007.06.009
Gao, X., Zhang, J., Zhang, X., Zhou, J., Jiang, Z., Huang, P., et al. (2019). Rice qGL3/OsPPKL1 functions with the GSK3/SHAGGY-like kinase OsGSK3 to modulate brassinosteroid signaling. Plant Cell. 31 (5), 1077–1093. doi: 10.1105/tpc.18.00836
Garagounis, C., Tsikou, D., Plitsi, P. K., Psarrakou, I. S., Avramidou, M., Stedel, C., et al. (2019). Lotus SHAGGY-like kinase 1 is required to suppress nodulation in Lotus japonicus. Plant J. 98 (2), 228–242. doi: 10.1111/tpj.14207
Gudesblat, G., Schneider-Pizoń, J., Betti, C., Mayerhofer, J., Vanhoutte, I., van Dongen, W., et al. (2012). SPEECHLESS integrates brassinosteroid and stomata signalling pathways. Nat. Cell Biol. 14 (5), 548–554. doi: 10.1038/ncb2471
Han, S., Cho, H., Noh, J., Qi, J., Jung, H. J., Nam, H., et al. (2018). BIL1-mediated MP phosphorylation integrates PXY and cytokinin signalling in secondary growth. Nat. Plants. 4 (8), 605–614. doi: 10.1038/s41477-018-0180-3
Han, Q., Tan, W., Zhao, Y., Yang, F., Yao, X., Lin, H., et al. (2022). Salicylic acid-activated BIN2 phosphorylation of TGA3 promotes Arabidopsis PR gene expression and disease resistance. EMBO J. 41 (19), e110682. doi: 10.15252/embj.2022110682
Hao, Y., Wang, H., Qiao, S., Leng, L., Wang, X. (2016). Histone deacetylase HDA6 enhances brassinosteroid signaling by inhibiting the BIN2 kinase. Proc. Natl. Acad. Sci. U S A. 113 (37), 10418–10423. doi: 10.1073/pnas.1521363113
He, C., Gao, H., Wang, H., Guo, Y., He, M., Peng, Y., et al. (2021). GSK3-mediated stress signaling inhibits legume-rhizobium symbiosis by phosphorylating GmNSP1 in soybean. Mol. Plant 14 (3), 488–502. doi: 10.1016/j.molp.2020.12.015
He, Y., Hong, G., Zhang, H., Tan, X., Li, L., Kong, Y., et al. (2020). The OsGSK2 kinase integrates brassinosteroid and jasmonic acid signaling by interacting with OsJAZ4. Plant Cell. 32 (9), 2806–2822. doi: 10.1105/tpc.19.00499
Heidstra, R., Sabatini, S. (2014). Plant and animal stem cells: similar yet different. Nat. Rev. Mol. Cell Biol. 15 (5), 301–312. doi: 10.1038/nrm3790
Hirano, K., Kawamura, M., Araki-Nakamura, S., Fujimoto, H., Ohmae-Shinohara, K., Yamaguchi, M., et al. (2017). Sorghum DW1 positively regulates brassinosteroid signaling by inhibiting the nuclear localization of BRASSINOSTEROID INSENSITIVE 2. Sci. Rep. 7 (1), 126. doi: 10.1038/s41598-017-00096-w
Hou, L., Li, Z., Shaheen, A., Zhang, K., Wang, J., Gao, X., et al. (2022). Zea mays GSK2 gene is involved in brassinosteroid signaling. Plant Growth Regulation. 97 (1), 117–125. doi: 10.1007/s10725-022-00806-z
Hou, J., Zheng, X., Ren, R., Shi, Q., Xiao, H., Chen, Z., et al. (2022). The histone deacetylase 1/GSK3/SHAGGY-like kinase 2/BRASSINAZOLE-RESISTANT 1 module controls lateral root formation in rice. Plant Physiol. 189 (2), 858–873. doi: 10.1093/plphys/kiac015
Houbaert, A., Zhang, C., Tiwari, M., Wang, K., de Marcos Serrano, A., Savatin, D. V., et al. (2018). POLAR-guided signalling complex assembly and localization drive asymmetric cell division. Nature. 563 (7732), 574–578. doi: 10.1038/s41586-018-0714-x
Hu, Z., Lu, S. J., Wang, M. J., He, H., Sun, L., Wang, H., et al. (2018). A novel QTL qTGW3 encodes the GSK3/SHAGGY-like kinase OsGSK5/OsSK41 that interacts with OsARF4 to negatively regulate grain size and weight in rice. Mol. Plant 11 (5), 736–749. doi: 10.1016/j.molp.2018.03.005
Hu, Y., Yu, D. (2014). BRASSINOSTEROID INSENSITIVE2 interacts with ABSCISIC ACID INSENSITIVE5 to mediate the antagonism of brassinosteroids to abscisic acid during seed germination in Arabidopsis. Plant Cell. 26 (11), 4394–4408. doi: 10.1105/tpc.114.130849
Huang, S., Liu, Y., Deng, R., Lei, T., Tian, A., Ren, H., et al. (2021). Genome-wide identification and expression analysis of the GSK gene family in Solanum tuberosum l. under abiotic stress and phytohormone treatments and functional characterization of StSK21 involvement in salt stress. Gene. 766, 145156. doi: 10.1016/j.gene.2020.145156
Ito, Y., Nakanomyo, I., Motose, H., Iwamoto, K., Sawa, S., Dohmae, N., et al. (2006). Dodeca-CLE peptides as suppressors of plant stem cell differentiation. Science. 313 (5788), 842–845. doi: 10.1126/science.1128436
Jiang, H., Tang, B., Xie, Z., Nolan, T., Ye, H., Song, G. Y., et al. (2019). GSK3-like kinase BIN2 phosphorylates RD26 to potentiate drought signaling in Arabidopsis. Plant J. 100 (5), 923–937. doi: 10.1111/tpj.14484
Khan, M., Rozhon, W., Unterholzner, S. J., Chen, T., Eremina, M., Wurzinger, B., et al. (2014). Interplay between phosphorylation and SUMOylation events determines CESTA protein fate in brassinosteroid signalling. Nat. Commun. 5,4687. doi: 10.1038/ncomms5687
Kim, T., Guan, S., Burlingame, A., Wang, Z. (2011). The CDG1 kinase mediates brassinosteroid signal transduction from BRI1 receptor kinase to BSU1 phosphatase and GSK3-like kinase BIN2. Mol. Cell. 43 (4), 561–571. doi: 10.1016/j.molcel.2011.05.037
Kim, T., Guan, S., Sun, Y., Deng, Z., Tang, W., Shang, J., et al. (2009). Brassinosteroid signal transduction from cell-surface receptor kinases to nuclear transcription factors. Nat. Cell Biol. 11 (10), 1254–1260. doi: 10.1038/ncb1970
Kim, E., Lee, S., Park, C., Kim, S., Hsu, C., Xu, S., et al. (2019). Plant U-Box40 mediates degradation of the brassinosteroid-responsive transcription factor BZR1 in Arabidopsis roots. Plant Cell. 31 (4), 791–808. doi: 10.1105/tpc.18.00941
Kim, Y., Song, J., Park, S., Jeong, Y., Kim, S. (2017). Brassinosteroid-induced transcriptional repression and dephosphorylation-dependent protein degradation negatively regulate BIN2-interacting AIF2 (a BR signaling-negative regulator) bHLH transcription factor. Plant Cell Physiol. 58 (2), 227–239. doi: 10.1093/pcp/pcw223
Kim, Y., Youn, J., Roh, J., Kim, J., Kim, S., Kim, T. (2022). Brassinosteroids enhance salicylic acid-mediated immune responses by inhibiting BIN2 phosphorylation of clade I TGA transcription factors in Arabidopsis. Mol. Plant 15 (6), 991–1007. doi: 10.1016/j.molp.2022.05.002
Kloc, Y., Dmochowska-Boguta, M., Zielezinski, A., Nadolska-Orczyk, A., Karlowski, W. M., Orczyk, W. (2020). Silencing of HvGSK1.1-a GSK3/SHAGGY-like kinase enhances barley (Hordeum vulgare l.) growth in normal and in salt stress conditions. Int. J. Mol. Sci. 21 (18), 6616. doi: 10.3390/ijms21186616
Koh, S., Lee, S., Kim, M., Koh, J., Lee, S., An, G., et al. (2007). T-DNA Tagged knockout mutation of rice OsGSK1, an orthologue of Arabidopsis BIN2, with enhanced tolerance to various abiotic stresses. Plant Mol. Biol. 65 (4), 453–466. doi: 10.1007/s11103-007-9213-4
Kondo, Y., Ito, T., Nakagami, H., Hirakawa, Y., Saito, M., Tamaki, T., et al. (2014). Plant GSK3 proteins regulate xylem cell differentiation downstream of TDIF-TDR signalling. Nat. Commun. 5, 3504. doi: 10.1038/ncomms4504
Li, T., Lei, W., He, R., Tang, X., Han, J., Zou, L., et al. (2020c). Brassinosteroids regulate root meristem development by mediating BIN2-UPB1 module in Arabidopsis. PloS Genet. 16 (7), e1008883. doi: 10.1371/journal.pgen.1008883
Li, J., Nam, K. H. (2002). Regulation of brassinosteroid signaling by a GSK3/SHAGGY-like kinase. Science. 295 (5558), 1299–1301. doi: 10.1126/science.1065769
Li, J., Terzaghi, W., Gong, Y., Li, C., Ling, J., Fan, Y., et al. (2020a). Modulation of BIN2 kinase activity by HY5 controls hypocotyl elongation in the light. Nat. Commun. 11 (1), 1592. doi: 10.1038/s41467-020-15394-7
Li, H., Ye, K., Shi, Y., Cheng, J., Zhang, X., Yang, S. (2017). BZR1 positively regulates freezing tolerance via CBF-dependent and CBF-independent pathways in Arabidopsis. Mol. Plant 10 (4), 545–559. doi: 10.1016/j.molp.2017.01.004
Li, C., Zhang, B., Chen, B., Ji, L., Yu, H. (2018). Site-specific phosphorylation of TRANSPARENT TESTA GLABRA1 mediates carbon partitioning in Arabidopsis seeds. Nat. Commun. 9 (1), 571. doi: 10.1038/s41467-018-03013-5
Li, C., Zhang, B., Yu, H. (2021). GSK3s: nodes of multilayer regulation of plant development and stress responses. Trends Plant Sci. 26 (12), 1286–1300. doi: 10.1016/j.tplants.2021.07.017
Li, J., Zhou, H., Zhang, Y., Li, Z., Yang, Y., Guo, Y. (2020b). The GSK3-like kinase BIN2 is a molecular switch between the salt stress response and growth recovery in Arabidopsis thaliana. Dev. Cell. 55 (3), 367–380.e366. doi: 10.1016/j.devcel.2020.08.005
Liao, C., Pu, Y., Nolan, T., Montes, C., Guo, H., Walley, J., et al. (2022). Brassinosteroids modulate autophagy through phosphorylation of RAPTOR1B by the GSK3-like kinase BIN2 in arabidopsis. Autophagy. 24, 1–18. doi: 10.1080/15548627.2022.2124501
Ling, J., Li, J., Zhu, D., Deng, X. (2017). Noncanonical role of Arabidopsis COP1/SPA complex in repressing BIN2-mediated PIF3 phosphorylation and degradation in darkness. Proc. Natl. Acad. Sci. U S A. 114 (13), 3539–3544. doi: 10.1073/pnas.1700850114
Liu, J., Chen, J., Zheng, X., Wu, F., Lin, Q., Heng, Y., et al. (2017). GW5 acts in the brassinosteroid signalling pathway to regulate grain width and weight in rice. Nat. Plants. 3, 17043. doi: 10.1038/nplants.2017.43
Lyu, J., Wang, D., Duan, P., Liu, Y., Huang, K., Zeng, D., et al. (2020). Control of grain size and weight by the GSK2-LARGE1/OML4 pathway in rice. Plant Cell. 32 (6), 1905–1918. doi: 10.1105/tpc.19.00468
Mao, J., Li, W., Liu, J., Li, J. (2021). Versatile physiological functions of plant GSK3-like kinases. Genes (Basel). 12 (5), 697. doi: 10.3390/genes12050697
Mei, Y., Yang, X., Huang, C., Zhang, X., Zhou, X. (2018). Tomato leaf curl yunnan virus-encoded C4 induces cell division through enhancing stability of cyclin d 1.1 via impairing NbSKη -mediated phosphorylation in Nicotiana benthamiana. PloS Pathog. 14 (1), e1006789. doi: 10.1371/journal.ppat.1006789
Mei, Y., Zhang, F., Wang, M., Li, F., Wang, Y., Zhou, X. (2020). Divergent symptoms caused by geminivirus-encoded C4 proteins correlate with their ability to bind NbSKη. J. Virol. 94 (20), e1307-20. doi: 10.1128/jvi.01307-20
Min, H., Cui, L., Oh, T., Kim, J., Kim, T., Kim, W. T. (2019). OsBZR1 turnover mediated by OsSK22-regulated U-box E3 ligase OsPUB24 in rice BR response. Plant J. 99 (3), 426–438. doi: 10.1111/tpj.14332
Murata, M. (2018). Inflammation and cancer. Environ. Health Prev. Med. 23 (1), 50. doi: 10.1186/s12199-018-0740-1
Nolan, T., Brennan, B., Yang, M., Chen, J., Zhang, M., Li, Z., et al. (2017). Selective autophagy of BES1 mediated by DSK2 balances plant growth and survival. Dev. Cell. 41 (1), 33–46. doi: 10.1016/j.devcel.2017.03.013
Nolan, T., Vukašinović, N., Liu, D., Russinova, E., Yin, Y. (2020). Brassinosteroids: multidimensional regulators of plant growth, development, and stress responses. Plant Cell. 32 (2), 295–318. doi: 10.1105/tpc.19.00335
Patel, P., Woodgett, J. R. (2017). Glycogen synthase kinase 3: a kinase for all pathways? Curr. Top. Dev. Biol. 123, 277–302. doi: 10.1016/bs.ctdb.2016.11.011
Peck, S., Mittler, R. (2020). Plant signaling in biotic and abiotic stress. J. Exp. Bot. 71 (5), 1649–1651. doi: 10.1093/jxb/eraa051
Peng, P., Yan, Z., Zhu, Y., Li, J. (2008). Regulation of the Arabidopsis GSK3-like kinase BRASSINOSTEROID-INSENSITIVE 2 through proteasome-mediated protein degradation. Mol. Plant 1 (2), 338–346. doi: 10.1093/mp/ssn001
Peres, A., Soares, J. S., Tavares, R. G., Righetto, G., Zullo, M. A. T., Mandava, N. B., et al. (2019). Brassinosteroids, the sixth class of phytohormones: a molecular view from the discovery to hormonal interactions in plant development and stress adaptation. Int. J. Mol. Sci. 20 (2), 331. doi: 10.3390/ijms20020331
Piao, H., Lim, J., Kim, S., Cheong, G., Hwang, I. (2001). Constitutive over-expression of AtGSK1 induces NaCl stress responses in the absence of NaCl stress and results in enhanced NaCl tolerance in Arabidopsis. Plant J. 27 (4), 305–314. doi: 10.1046/j.1365-313x.2001.01099.x
Qi, X., Chanderbali, A. S., Wong, G. K., Soltis, D. E., Soltis, P. S. (2013). Phylogeny and evolutionary history of glycogen synthase kinase 3/SHAGGY-like kinase genes in land plants. BMC Evol. Biol. 13, 143. doi: 10.1186/1471-2148-13-143
Qiao, S., Sun, S., Wang, L., Wu, Z., Li, C., Li, X., et al. (2017). The RLA1/SMOS1 transcription factor functions with OsBZR1 to regulate brassinosteroid signaling and rice architecture. Plant Cell. 29 (2), 292–309. doi: 10.1105/tpc.16.00611
Qiu, A., Wu, J., Lei, Y., Cai, Y., Wang, S., Liu, Z., et al. (2018). CaSK23, a putative GSK3/SHAGGY-like kinase of Capsicum annuum, acts as a negative regulator of pepper's response to Ralstonia solanacearum attack. Int. J. Mol. Sci. 19 (9), 2698. doi: 10.3390/ijms19092698
Racaud-Sultan, C., Vergnolle, N. (2021). GSK3β, a master kinase in the regulation of adult stem cell behavior. Cells. 10 (2), 225. doi: 10.3390/cells10020225
Ren, H., Willige, B. C., Jaillais, Y., Geng, S., Park, M. Y., Gray, W. M., et al. (2019). BRASSINOSTEROID-SIGNALING KINASE 3, a plasma membrane-associated scaffold protein involved in early brassinosteroid signaling. PloS Genet. 15 (1), e1007904. doi: 10.1371/journal.pgen.1007904
Rozhon, W., Mayerhofer, J., Petutschnig, E., Fujioka, S., Jonak, C. (2010). ASKtheta, a group-III Arabidopsis GSK3, functions in the brassinosteroid signalling pathway. Plant J. 62 (2), 215–223. doi: 10.1111/j.1365-313X.2010.04145.x
Samakovli, D., Margaritopoulou, T., Prassinos, C., Milioni, D., Hatzopoulos, P. (2014). Brassinosteroid nuclear signaling recruits HSP90 activity. New Phytol. 203, 743–757. doi: 10.1111/nph.12843
Santiago, J., Henzler, C., Hothorn, M. (2013). Molecular mechanism for plant steroid receptor activation by somatic embryogenesis co-receptor kinases. Science. 341 (6148), 889–892. doi: 10.1126/science.1242468
Signorelli, S., Tarkowski Ł, P., Van den Ende, W., Bassham, D. C. (2019). Linking autophagy to abiotic and biotic stress responses. Trends Plant Sci. 24 (5), 413–430. doi: 10.1016/j.tplants.2019.02.001
Solovou, T., Garagounis, C., Kyriakis, E., Bobas, C., Papadopoulos, G. E., Skamnaki, V. T., et al. (2021). Mutagenesis of a lotus japonicus GSK3β/Shaggy-like kinase reveals functionally conserved regulatory residues. Phytochemistry. 186, 112707. doi: 10.1016/j.phytochem.2021.112707
Song, S., Wang, H., Sun, M., Tang, J., Zheng, B., Wang, X., et al. (2019). Reactive oxygen species-mediated BIN2 activity revealed by single-molecule analysis. New Phytol. 223 (2), 692–704. doi: 10.1111/nph.15669
Song, Y., Zhai, Y., Li, L., Yang, Z., Ge, X., Yang, Z., et al. (2021). BIN2 negatively regulates plant defence against Verticillium dahliae in Arabidopsis and cotton. Plant Biotechnol. J. 19 (10), 2097–2112. doi: 10.1111/pbi.13640
Stampfl, H., Fritz, M., Dal Santo, S., Jonak, C. (2016). The GSK3/Shaggy-like kinase ASKα contributes to pattern-triggered immunity. Plant Physiol. 171 (2), 1366–1377. doi: 10.1104/pp.15.01741
Sun, S., Chen, D., Li, X., Qiao, S., Shi, C., Li, C., et al. (2015). Brassinosteroid signaling regulates leaf erectness in Oryza sativa via the control of a specific U-type cyclin and cell proliferation. Dev. Cell. 34 (2), 220–228. doi: 10.1016/j.devcel.2015.05.019
Sun, S., Wang, T., Wang, L., Li, X., Jia, Y., Liu, C., et al. (2018). Natural selection of a GSK3 determines rice mesocotyl domestication by coordinating strigolactone and brassinosteroid signaling. Nat. Commun. 9 (1), 2523. doi: 10.1038/s41467-018-04952-9
Tamaki, T., Oya, S., Naito, M., Ozawa, Y., Furuya, T., Saito, M., et al. (2020). VISUAL-CC system uncovers the role of GSK3 as an orchestrator of vascular cell type ratio in plants. Commun. Biol. 3 (1), 184. doi: 10.1038/s42003-020-0907-3
Tang, W., Kim, T., Oses-Prieto, J., Sun, Y., Deng, Z., Zhu, S., et al. (2008). BSKs mediate signal transduction from the receptor kinase BRI1 in Arabidopsis. Science. 321 (5888), 557–560. doi: 10.1126/science.1156973
Tong, H., Liu, L., Jin, Y., Du, L., Yin, Y., Qian, Q., et al. (2012). DWARF AND LOW-TILLERING acts as a direct downstream target of a GSK3/SHAGGY-like kinase to mediate brassinosteroid responses in rice. Plant Cell. 24 (6), 2562–2577. doi: 10.1105/tpc.112.097394
Umezawa, T., Sugiyama, N., Mizoguchi, M., Hayashi, S., Myouga, F., Yamaguchi-Shinozaki, K., et al. (2009). Type 2C protein phosphatases directly regulate abscisic acid-activated protein kinases in Arabidopsis. Proc. Natl. Acad. Sci. U S A. 106 (41), 17588–17593. doi: 10.1073/pnas.0907095106
Vert, G., Chory, J. (2006). Downstream nuclear events in brassinosteroid signalling. Nature. 441 (7089), 96–100. doi: 10.1038/nature04681
Vert, G., Walcher, C. L., Chory, J., Nemhauser, J. L. (2008). Integration of auxin and brassinosteroid pathways by auxin response factor 2. Proc. Natl. Acad. Sci. U S A. 105 (28), 9829–9834. doi: 10.1073/pnas.0803996105
Waadt, R., Seller, C. A., Hsu, P. K., Takahashi, Y., Munemasa, S., Schroeder, J. I. (2022). Plant hormone regulation of abiotic stress responses. Nat. Rev. Mol. Cell Biol. 23 (10), 680–694. doi: 10.1038/s41580-022-00479-6
Wang, L., Cheng, H., Xiong, F., Ma, S., Zheng, L., Song, Y., et al. (2020). Comparative phosphoproteomic analysis of BR-defective mutant reveals a key role of GhSK13 in regulating cotton fiber development. Sci. China Life Sci. 63 (12), 1905–1917. doi: 10.1007/s11427-020-1728-9
Wang, X., Chory, J. (2006). Brassinosteroids regulate dissociation of BKI1, a negative regulator of BRI1 signaling, from the plasma membrane. Science. 313 (5790), 1118–1122. doi: 10.1126/science.1127593
Wang, X., Kota, U., He, K., Blackburn, K., Li, J., Goshe, M. B., et al. (2008). Sequential transphosphorylation of the BRI1/BAK1 receptor kinase complex impacts early events in brassinosteroid signaling. Dev. Cell. 15 (2), 220–235. doi: 10.1016/j.devcel.2008.06.011
Wang, L., Su, A., Wu, X., Chen, Q., Xin, D., Qi, Z., et al. (2018). Overexpression of GmBIN2, a soybean glycogen synthase kinase 3 gene, enhances tolerance to salt and drought in transgenic Arabidopsis and soybean hairy roots. J. Integr. Agric. 17 (9), 13. doi: 10.1016/S2095-3119(17)61863-X
Wang, H., Tang, J., Liu, J., Hu, J., Liu, J., Chen, Y., et al. (2018a). Abscisic acid signaling inhibits brassinosteroid signaling through dampening the dephosphorylation of BIN2 by ABI1 and ABI2. Mol. Plant 11 (2), 315–325. doi: 10.1016/j.molp.2017.12.013
Wang, Y., Xu, J., Yu, J., Zhu, D., Zhao, Q. (2022). Maize GSK3-like kinase ZmSK2 is involved in embryonic development. Plant Sci. 318, 111221. doi: 10.1016/j.plantsci.2022.111221
Wang, L., Yang, Z., Zhang, B., Yu, D., Liu, J., Gong, Q., et al. (2018b). Genome-wide characterization and phylogenetic analysis of GSK gene family in three species of cotton: evidence for a role of some GSKs in fiber development and responses to stress. BMC Plant Biol. 18 (1), 330. doi: 10.1186/s12870-018-1526-8
Woodgett, J. R. (1991). A common denominator linking glycogen metabolism, nuclear oncogenes and development. Trends Biochem. Sci. 16 (5), 177–181. doi: 10.1016/0968-0004(91)90071-3
Wrzaczek, M., Rozhon, W., Jonak, C. (2007). A proteasome-regulated glycogen synthase kinase-3 modulates disease response in plants. J. Biol. Chem. 282 (8), 5249–5255. doi: 10.1074/jbc.M610135200
Xia, D., Zhou, H., Liu, R., Dan, W., Li, P., Wu, B., et al. (2018). GL3.3, a novel QTL encoding a GSK3/SHAGGY-like kinase, epistatically interacts with GS3 to produce extra-long grains in rice. Mol. Plant 11 (5), 754–756. doi: 10.1016/j.molp.2018.03.006
Xiao, Y., Liu, D., Zhang, G., Tong, H., Chu, C. (2017). Brassinosteroids regulate OFP1, a DLT interacting protein, to modulate plant architecture and grain morphology in rice. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.01698
Xiao, Y., Zhang, G., Liu, D., Niu, M., Tong, H., Chu, C. (2020). GSK2 stabilizes OFP3 to suppress brassinosteroid responses in rice. Plant J. 102 (6), 1187–1201. doi: 10.1111/tpj.14692
Xie, Z., Nolan, T., Jiang, H., Tang, B., Zhang, M., Li, Z., et al. (2019). The AP2/ERF transcription factor TINY modulates brassinosteroid-regulated plant growth and drought responses in Arabidopsis. Plant Cell. 31 (8), 1788–1806. doi: 10.1105/tpc.18.00918
Xiong, F., Zhang, R., Meng, Z., Deng, K., Que, Y., Zhuo, F., et al. (2017). Brassinosteriod insensitive 2 (BIN2) acts as a downstream effector of the target of rapamycin (TOR) signaling pathway to regulate photoautotrophic growth in Arabidopsis. New Phytol. 213 (1), 233–249. doi: 10.1111/nph.14118
Yan, Z., Zhao, J., Peng, P., Chihara, R. K., Li, J. (2009). BIN2 functions redundantly with other Arabidopsis GSK3-like kinases to regulate brassinosteroid signaling. Plant Physiol. 150 (2), 710–721. doi: 10.1104/pp.109.138099
Yang, C., Luo, M., Zhuang, X., Li, F., Gao, C. (2020). Transcriptional and epigenetic regulation of autophagy in plants. Trends Genet. 36 (9), 676–688. doi: 10.1016/j.tig.2020.06.013
Yang, C., Shen, W., He, Y., Tian, Z., Li, J. (2016). OVATE family protein 8 positively mediates brassinosteroid signaling through interacting with the GSK3-like kinase in rice. PloS Genet. 12 (6), e1006118. doi: 10.1371/journal.pgen.1006118
Ye, K., Li, H., Ding, Y., Shi, Y., Song, C., Gong, Z., et al. (2019). BRASSINOSTEROID-INSENSITIVE2 negatively regulates the stability of transcription factor ICE1 in response to cold stress in Arabidopsis. Plant Cell. 31 (11), 2682–2696. doi: 10.1105/tpc.19.00058
Ye, H., Li, L., Guo, H., Yin, Y. (2012). MYBL2 is a substrate of GSK3-like kinase BIN2 and acts as a corepressor of BES1 in brassinosteroid signaling pathway in Arabidopsis. Proc. Natl. Acad. Sci. U S A. 109 (49), 20142–20147. doi: 10.1073/pnas.1205232109
Ying, J., Ma, M., Bai, C., Huang, X., Liu, J., Fan, Y., et al. (2018). TGW3, a major QTL that negatively modulates grain length and weight in rice. Mol. Plant 11 (5), 750–753. doi: 10.1016/j.molp.2018.03.007
Youn, J., Kim, T. (2015). Functional insights of plant GSK3-like kinases: multi-taskers in diverse cellular signal transduction pathways. Mol. Plant 8 (4), 552–565. doi: 10.1016/j.molp.2014.12.006
Zhang, K., Duan, M., Zhang, L., Li, J., Shan, L., Zheng, L., et al. (2022). HOP1 and HOP2 are involved in salt tolerance by facilitating the brassinosteroid-related nucleo-cytoplasmic partitioning of the HSP90-BIN2 complex. Plant Cell Environ. 45 (12), 3551–3565. doi: 10.1111/pce.14441
Zhang, D., Tan, W., Yang, F., Han, Q., Deng, X., Guo, H., et al. (2021). A BIN2-GLK1 signaling module integrates brassinosteroid and light signaling to repress chloroplast development in the dark. Dev. Cell. 56 (3), 310–324.e317. doi: 10.1016/j.devcel.2020.12.001
Zhang, C., Xu, Y., Guo, S., Zhu, J., Huan, Q., Liu, H., et al. (2012). Dynamics of brassinosteroid response modulated by negative regulator LIC in rice. PloS Genet. 8 (4), e1002686. doi: 10.1371/journal.pgen.1002686
Zhang, P., Zhang, L., Chen, T., Jing, F., Liu, Y., Ma, J., et al. (2022b). Genome-wide identification and expression analysis of the GSK gene family in wheat (Triticum aestivum l.). Mol. Biol. Rep. 49 (4), 2899–2913. doi: 10.1007/s11033-021-07105-2
Zhang, H., Zhu, J., Gong, Z., Zhu, J. K. (2022a). Abiotic stress responses in plants. Nat. Rev. Genet. 23 (2), 104–119. doi: 10.1038/s41576-021-00413-0
Zhu, J. (2016). Abiotic stress signaling and responses in plants. Cell. 167 (2), 313–324. doi: 10.1016/j.cell.2016.08.029
Zhu, J., Li, Y., Cao, D., Yang, H., Oh, E., Bi, Y., et al. (2017). The f-box protein KIB1 mediates brassinosteroid-induced inactivation and degradation of GSK3-like kinases in Arabidopsis. Mol. Cell. 66 (5), 648–657.e644. doi: 10.1016/j.molcel.2017.05.012
Keywords: glycogen synthase kinase 3 (GSK3), plant, hormone, abiotic stress, biotic stress
Citation: Song Y, Wang Y, Yu Q, Sun Y, Zhang J, Zhan J and Ren M (2023) Regulatory network of GSK3-like kinases and their role in plant stress response. Front. Plant Sci. 14:1123436. doi: 10.3389/fpls.2023.1123436
Received: 14 December 2022; Accepted: 17 February 2023;
Published: 01 March 2023.
Edited by:
Sergio J. Ochatt, INRA UMR1347 Agroécologie, FranceReviewed by:
Wei Wang, Fujian Agriculture and Forestry University, ChinaSaroj Kumar Sah, Brookhaven National Laboratory (DOE), United States
Copyright © 2023 Song, Wang, Yu, Sun, Zhang, Zhan and Ren. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jiasui Zhan, Smlhc3VpLnpoYW5Ac2x1LnNl; Maozhi Ren, cmVubWFvemhpMDFAY2Fhcy5jbg==
†These authors have contributed equally to this work and share first authorship
 Yun Song
Yun Song Ying Wang2,3,4†
Ying Wang2,3,4† Jianling Zhang
Jianling Zhang Maozhi Ren
Maozhi Ren