
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 01 February 2023
Sec. Crop and Product Physiology
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1119282
This article is part of the Research TopicDevelopment, Stress Response and Metabolism of Sweet PotatoView all 7 articles
 Yiren Su†
Yiren Su† Yang Liu†
Yang Liu† Shizhuo Xiao
Shizhuo Xiao Yuan Wang
Yuan Wang Yitong Deng
Yitong Deng Lukuan Zhao
Lukuan Zhao Yao Wang
Yao Wang Donglan Zhao
Donglan Zhao Xibin Dai
Xibin Dai Zhilin Zhou
Zhilin Zhou Qinghe Cao*
Qinghe Cao*Adverse environmental stress is a major environmental factor threatening food security, which is why improving plant stress resistance is essential for agricultural productivity and environmental sustainability. The NAC (NAM, ATAF, and CUC) transcription factors (TFs) play a dominant role in plant responses to abiotic and biotic stresses, but they have been poorly studied in Ipomoea pes-caprae. In this research, 12 NAC TFs, named IpNAC1–IpNAC12, were selected from transcriptome data. The homologous evolution tree divided IpNACs into four major categories, and six IpNACs were linearly associated with Arabidopsis ANAC genes. From the gene structures, protein domains, and promoter upstream regulatory elements, IpNACs were shown to contain complete NAC-specific subdomains (A–E) and cis-acting elements corresponding to different stress stimuli. We measured the expression levels of the 12 IpNACs under abiotic stress (salt, heat, and drought) and hormone treatment (abscisic acid, methyl jasmonate, and salicylic acid), and their transcription levels differed. IpNAC5/8/10/12 were located in the nucleus through subcellular localization, and the overexpressing transgenic Arabidopsis plants showed high tolerance to salt stress. The cellular Na+ homeostasis content in the mature and elongation zones of the four IpNAC transgenic sweetpotato roots showed an obvious efflux phenomenon. These conclusions demonstrate that IpNAC5/8/10/12 actively respond to abiotic stress, have significant roles in improving plant salt tolerance, and are important salt tolerance candidate genes in I. pes-caprae and sweetpotato. This study laid the foundation for further studies on the function of IpNACs in response to abiotic stress. It provides options for improving the stress resistance of sweetpotato using gene introgression from I. pes-caprae.
Ipomoea pes-caprae (Linn.) R. Br. (Convolvulaceae 2n = 2x = 30) mainly grows on tropical and subtropical beaches (Miryeganeh et al., 2003), and it has anti-inflammatory (Cristiane et al., 2017) and antitumor effects (Manigauha et al., 2015), relieves gout (Akinniyi et al., 2022), and has significant curative effects. Compared with other crops, it has obvious biological characteristics, such as salt tolerance (Liu et al., 2020), as well as drought (Suarez, 2011) and high temperature resistance (Cheng et al., 2021). It is an important medicinal and coastal sand fixation greening plant (Akinniyi et al, 2022). In the plant evolutionary process, variation in the natural environment is one of the most important factors affecting plant growth and development (Hawrot-Paw et al., 2016). To cope with abiotic stress (drought, salt, and high temperature), plants have formed a variety of complex and precise biological resistance mechanisms (Li et al., 2021). In many stress signaling pathways, transcription factors (TFs), as multi-functional proteins, play a crucial role in plant defense mechanisms by controlling gene expression and regulating the basic functions of plants (Falak et al., 2021; Hrmova and Hussain, 2021).
NAC (NAM, ATAF, and CUC) proteins comprise a unique TF family in plants (Wang et al., 2021). It has five highly conserved subdomains A–E (Meng et al., 2018). Domains C and D are responsible for binding to DNA (Du et al., 2020), domain A is involved in the formation of functional dimers (Nakashima et al., 2012), and domains B and E determine the functional diversity of NAC genes (Puranik et al., 2012). With the development of biotechnology, NAC TFs in different species have been identified, including 117 in Arabidopsis thaliana (Ooka et al., 2003), 151 in Oryza sativa (Nuruzzaman et al., 2010), and 152 in Nicotiana tabacum (Rushton et al., 2008). As one of the largest transcription families in plants, NAC TFs have many functions, such as in plant secondary metabolism (Srivastava and Sahoo, 2021), plant development and senescence (Gong et al., 2020), and hormone regulation (Tao et al., 2022). In response to high salt, drought, and high temperatures, NAC TFs play an important regulatory function by activating or inhibiting the expression of target genes and improving abiotic stress tolerance (Trishla and Kirti, 2021; Guo et al., 2022). Ju et al. (2020) overexpressed VvNAC17 in Arabidopsis, enhancing tolerance to salt, osmotic, and freezing stress and increasing ABA sensitivity. Li et al. (2021) suggested that GmNAC06 could cause the accumulation of proline and glycine betaine alleviate the effects of reactive oxygen species (ROS), and maintain the homeostasis of the Na+ ion. Therefore, GmNAC06 overexpression in hairy roots led to significant salt tolerance in the whole composite plant. Duan et al. (2017) verified that MfNACs combine with the MtGlyl promoter to improve plant drought tolerance by regulating glutathione back to the reduction state.
Although the NAC family has been widely studied, there have been few reports about I. pes-caprae and its related species. Many genes in wild relatives provide genetic resources for germplasm improvement (Zhang et al., 2016). Ipomoea pes-caprae, as a wild relative of sweetpotato, can be used as a model plant for studying the salt tolerance mechanism of sweetpotato. China has abundant sweetpotato resources, and its planting area and yield rank first in the world (Kim et al., 2018). However, the increasing occurrence of extreme temperatures, drought, and severe secondary salinization seriously limits sweetpotato growth, development, and yield. Sweetpotato is a salt-sensitive plant (Meng et al., 2020), and there has been little research on its tolerance to salt. It is necessary to utilize related gene resources in I. pes-caprae to improve the cultivated sweetpotato. Therefore, screening wild relatives for salt-tolerance genes to study salt tolerance mechanisms is particularly important to breed new sweetpotato varieties adapted to salt stress and other abiotic stressors. In our study, according to the transcriptome data, 12 I. pes-caprae IpNAC genes encoding NAC proteins were screened and identified. Phylogenetic relationship, gene structure, and protein domain analyses were carried out. Genetic expression profiles under different abiotic stressors and hormone treatments were studied. IpNAC5, IpNAC8, IpNAC10 and IpNAC12 were localized in the nuclei. Through heterologous overexpression in Arabidopsis, the plant phenotype in 180 mM NaCl treatment was observed, and Na+ flow in sweetpotato roots in mature and elongation zones was measured under salt stress. IpNAC5/8/10/12 were preliminarily verified to participate in resistance to abiotic stress, such as salt stress. This study laid a foundation for utilizing I. pes-caprae genes to improve sweetpotato stress resistance.
The experimental materials—I. pes-caprae, ‘ZiShu8’, Nicotiana tabacum, and Arabidopsis thaliana—were cultivated at the Xuzhou SweetPotato Research Center, China. Nicotiana and Arabidopsis were grown at 20–28 °C under a 16 h/8 h dark/light cycle with 50–60% humidity and 500 µmol·m−2·s−1 light intensity. Ipomoea pes-caprae was cultivated in Hoagland nutrient solution. After 3–5 weeks of growth, 7–9 functional leaves and 8–12 cm roots were present, and I. pes-caprae plants were transferred to soil mixed with fine sand and a nutritional substance (3:1). Then, after 4–5 weeks, we selected plants with similar growth characteristics for experiments.
The amount of expressed genes was obtained from transcriptome data (Liu et al., 2020). Combined with the level of I. pes-caprae NAC differentially expressed genes under salt stress, 12 TFs were preliminarily screened as research targets and named IpNAC1–12. The IpNAC nucleotide sequence was converted into an amino acid sequence by NCBI1. The physicochemical properties of the IpNAC1–12 proteins, including the theoretical molecular weight (MW), isoelectric point (pI), and hydrophobicity, were analyzed using the Expasy ProtParam tool2. The full-length amino acid sequences of the IpNAC and ANAC proteins were used to generate a phylogenetic tree by deleting gaps and blanks. The unrooted neighbor-joining tree was constructed after ClustalW alignment using MEGA (Tamura et al., 2011). Bootstrap analysis was performed with 100 replicates to assess the level of statistical support for each tree node (Hu et al., 2015). EvoView was used for subsequent beautification.
The MEME3 online tool was used to identify the protein domains of the IpNAC, with the following parameters: site distribution = zero or one occurrence per sequence, largest number of motifs = 10, and optimum motif width of 6–50. Default settings were used for the other parameters. The results were presented by GSDS 2.04. The gene sequences of IpNACs were compared with the full-length sequences by SnapGene 4.36 to determine the exon–intron structure and were visualized using TBtools software. Collinearity between I. pes-caprae (IpNACs) and A. thaliana (TAIR)5 (ANACs) was determined using MCScanX (Wang et al., 2013), and the figures were generated using Circos (Darzentas, 2010).
Most gene regulatory regions are located 2 kb upstream of the translation start site. The IpNAC promoter sequences were obtained from the I. pes-caprae genome database (unpublished). Then, the PlantCARE6 online database was used to identify the cis-acting elements of these promoters.
We selected plants with similar growth rates for plant hormone and abiotic stress treatments. Untreated plants were used as controls. The leaves or adventitious roots were collected after 0, 3, 6, 12, 24, or 48 h of treatment. Heat treatment was performed by transferring I. pes-caprae to a 45 °C climate box, and the roots were submerged in 20% PEG 6000 for 48 h for dehydration treatment. The leaves and adventitious roots were harvested from the plants. The roots of I. pes-caprae were submerged in a salt solution at three concentrations of 300, 600, and 900 mM NaCl, and adventitious roots were harvested. For hormone treatment, adventitious roots were submerged in 0.1 mM abscisic acid (ABA), 2 mM methyI jasmonate (MeJA), or 2 mM salicylic acid (SA) for 0, 3, 6, 12, 24, or 48 h, and the adventitious roots were collected. The roots were collected from three separate plants at each time point of each treatment and combined to form one sample. All experiments were performed independently and in triplicate. The collected samples were immediately frozen in liquid nitrogen and stored at −80 °C.
Following the manufacturer’s instructions, the FastPure Plant Total RNA Isolation Kit (Vazyme, China) was used to isolate the total RNA in the leaves and roots. The quality of the RNA was determined using a NanoDrop2000c spectrophotometer (Thermo Fisher Scientific, US), and total RNA integrity was identified by electrophoresis on 1.0% agarose gel. The HiScript II 1st Strand cDNA Synthesis kit (Vazyme, China) was used to obtain cDNA from total RNA. Real-time quantitative PCR was carried out using SYBR green (TOYOBO, Japan) on an ABI QuantStudio 6 Flex (Thermo Fisher Scientific, US) with a final volume of 10 µL per reaction. Each reaction mixture contained 5 µL SYBR Green Realtime PCR Master Mix (TOYOBO, Japan), 0.4 µL forward primer (10 μM), 0.4 µL reverse primer (10 μM), 3.2 µL ddH2O, and 1 µL cDNA template (diluted at 1:10). Specific primers for each gene are listed in Supplementary Table S3. The cycling parameters were 95 °C for 1 min and 40 cycles of 95 °C for 15 s, 60 °C for 15 s, and 72 °C for 45 s. The Actin gene of I. pes-caprae was used as an internal control. Each measurement was performed with three biological replicates. Data were analyzed using the 2-ΔΔCt method.
The coding DNA sequences of IpNAC1–12 were obtained from the transcriptome datasets of I. pes-caprae. We designed the specific primers (Supplementary Table S2). The Phanta Max Super-Fidelity DNA Polymerase (Vazyme, China) (18 μL ddH2O; 25 μL 2 × Phanta Max Buffer; dNTP Mix (10 mM each); forward primer (10 μM); reverse primer (10 μM); 1 μL Phanta Max Super-Fidelity DNA Polymerase; 1 μL cDNA) were used to amplify the IpNAC1–12 genes from the cDNA of I. pes-caprae. The PCR parameters were set as follows: initial denaturation at 95 °C for 3 min, 30 cycles of denaturation at 95 °C for 15 s, annealing at 58 °C for 30 s, extension at 72 °C for 1 min 30 s, and final extension at 72 °C for 10 min. The PCR products were subcloned into the pHB-GFP vector with a Hind III restriction site to form pHB-IpNAC1-12-GFP. pHB-IpNAC1-12-GFP and pHB (set as a control) were transformed into Arabidopsis. In addition, using pCAMBIA0390-DsRed as the backbone vector, the coding regions of IpNAC1–12 were inserted into the pCAMBIA0390-DsRed expression vector constructing pUBI.U4:: IpNAC1-12-CaMV35S::DsRed for Agrobacterium rhizogenes-mediated transformation. pCAMBIA0390-DsRed was used as a control. Each experiment was conducted using more than three biological replicates.
The 35S: GFP-IpNAC5/IpNAC8/IpNAC10/IpNAC12 plasmid were transformed into Agrobacterium tumefaciens strain GV3101 for plant transformation. Transient expression in tobacco leaves was determined according to a published method (Zhou et al., 2018). After transformation, tobacco was kept in the light room for 36 h before examination by fluorescence microscopy. Then, the tobacco leaf epidermis was peeled to make a temporary squash. After incubation with phosphate-buffered saline containing 40, 60-diamidino-2-phenylin dole (DAPI), the temporary squashes were observed under a Leica Fluorescence Microscope at 10 × 20 (Leica Microsystems, Wetzlar, GmbH). The image data were processed using Adobe Photoshop (Mountain View, CA, USA). All transient expression assays were repeated at least three times.
The recombinant vectors pHB-IpNAC5/IpNAC8/IpNAC10/IpNAC12-GFP were transferred into Arabidopsis through the Agrobacterium tumefaciens strain GV3101 using the floral dipping method. After transformation, the T3 (third generation) seeds were germinated on 1/2 MS medium with 50 mg L−1 hygromycin for screening. Then, the WT and overexpressing transgenic seeds were cultivated on 1/2 MS medium with 180 mM NaCl at 22 °C under 16 h of daylight. After 10 d, the primary root length was measured and the growth state was observed.
The pUBI.U4:: IpNAC5/IpNAC8/IpNAC10/IpNAC12-CaMV35S::DsRed constructs were introduced into A. rhizogenes strain K599. ZiShu 8 with the same growth state was selected for Agrobacterium infection according to the published method (Yu et al., 2020). Subsequently, the transformed plants were transferred into the soil under a high-humidity (28 °C/65%) environment. After three weeks, the positive plants were selected through the portable fluorescent lamp instrument, immersed in hydroponic solution for 3 d, and then collected.
The WT and IpNAC5/8/10/12 transgenic sweetpotato were cultured in 150 mM NaCl solution for 24 h. First, 3–4 cm of intact root tips were collected and fixed in a buffer solution for 30 min. Then, Na+ flux in the root elongation zone was recorded for 5 min. The root mature zone was measured for another 5 min. Each group had five replicates.
The data are presented as the mean ± standard deviation of the mean (SD), and statistically significant differences were assessed using Dunnett’s test at p < 0.05 as the point of minimal statistical significance in all analyses.
We identified 12 NAC genes in I. pes-caprae, named IpNAC1–12. We found that IpNAC1–12 had high homology with Arabidopsis, sweetpotato, and their wild species. IpNAC2 is one of the reported genes, and NAC1 was screened from the cDNA library of I. pes-caprae, but its function was not clear. We verified that IpNACs contained a complete ORF. Analysis of the physicochemical properties revealed amino acid lengths of IpNACs ranging from 242 to 344. The molecular weight varied from 27.58 to 39.21 KDa, and the pI ranged from 5.24 to 8.52. Except for IpNAC2, IpNAC3, and IpNAC6, IpNAC proteins were weakly acidic and were predicted to be located in the nucleus (Supplementary Table S1). IpNAC proteins contain NAC family characteristic subdomains (A–E) (Figure 1; Supplementary Figure S1).
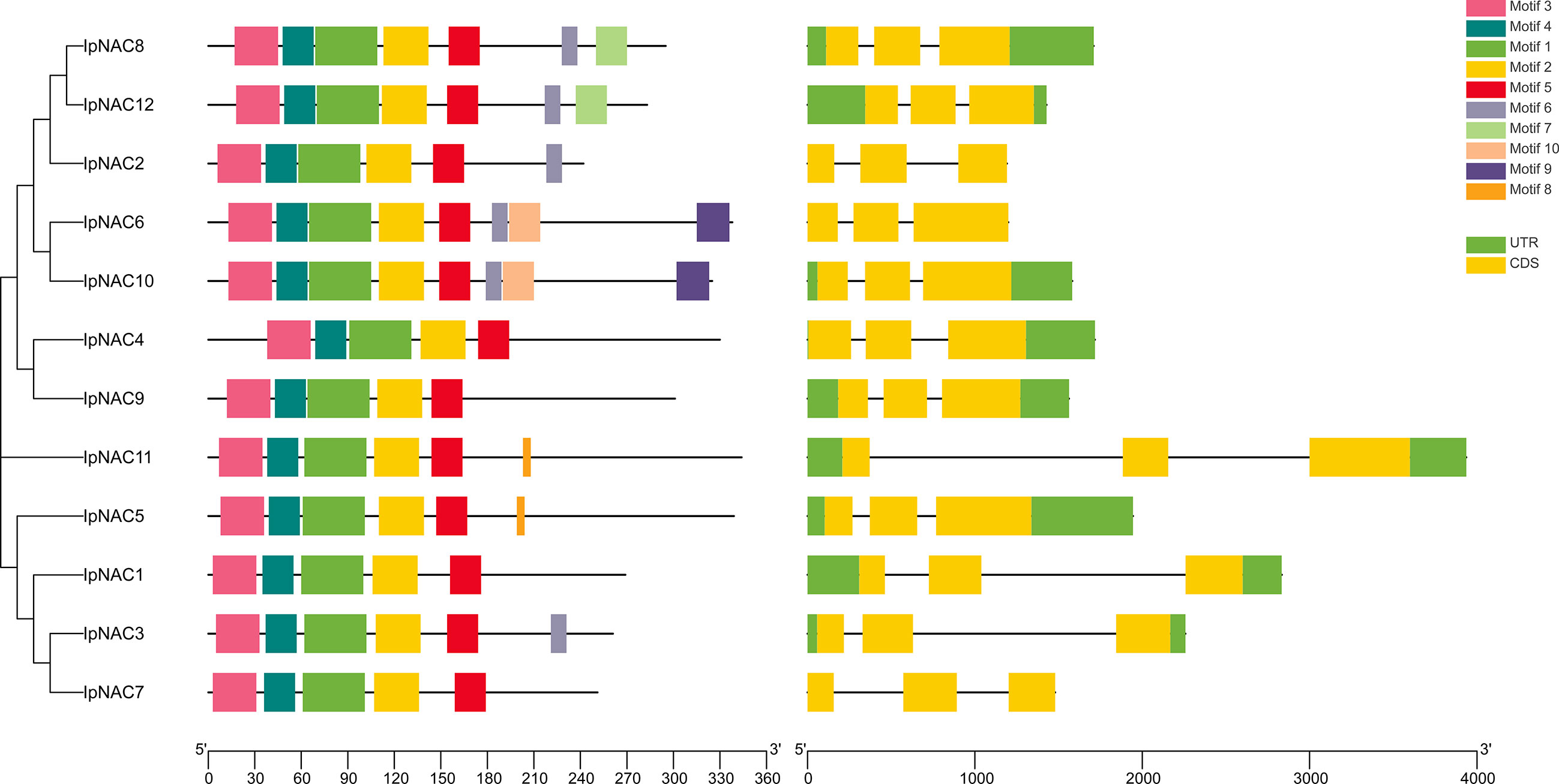
Figure 1 Analysis of the conserved motifs and gene structures of Ipomoea pes-caprae IpNAC genes. The conservative motif distribution and homologous tree of IpNACs are located on the left side, which is expressed by a color box. The black scale lines represent the relative protein length. The gene structure is on the right side, the exon and intron are represented by a yellow rectangle and black line, respectively, and the untranslated region (UTR) is represented by a green rectangle. Gene size can be expressed by the scale.
To investigate the taxonomic and phylogenetic relationships of IpNACs, we used the model plant Arabidopsis to analyze the potential function of the IpNAC protein. Using MEGA-X software, we performed phylogenetic analysis of the IpNAC and ANAC proteins (without duplicated and erroneous sequences) to establish an unrooted phylogenetic tree (Figure 2A). The NAC proteins were classified into nine subfamilies. IpNACs were divided into ONAC22 (including IpNAC1, IpNAC3, IpNAC5, and IpNAC7), ATAF (including IpNAC2, IpNAC6, IpNAC8, IpNAC10, and IpNAC12), OsNAC7 (including IpNAC11), and NAM (including IpNAC4 and IpNAC9). To obtain more information about IpNAC genes, we identified syntenic relationships between IpNACs and ANACs and found six pairs of syntenic NAC genes between I. pes-caprae and Arabidopsis (Figure 2B). Interestingly, one IpNAC gene (IpNAC6) and one ANAC gene (ANAC072) were associated with three or two syntenic blocks, respectively. At the same time, other IpNAC genes have a close relationship with ANAC genes, indicating that their functions may be similar.
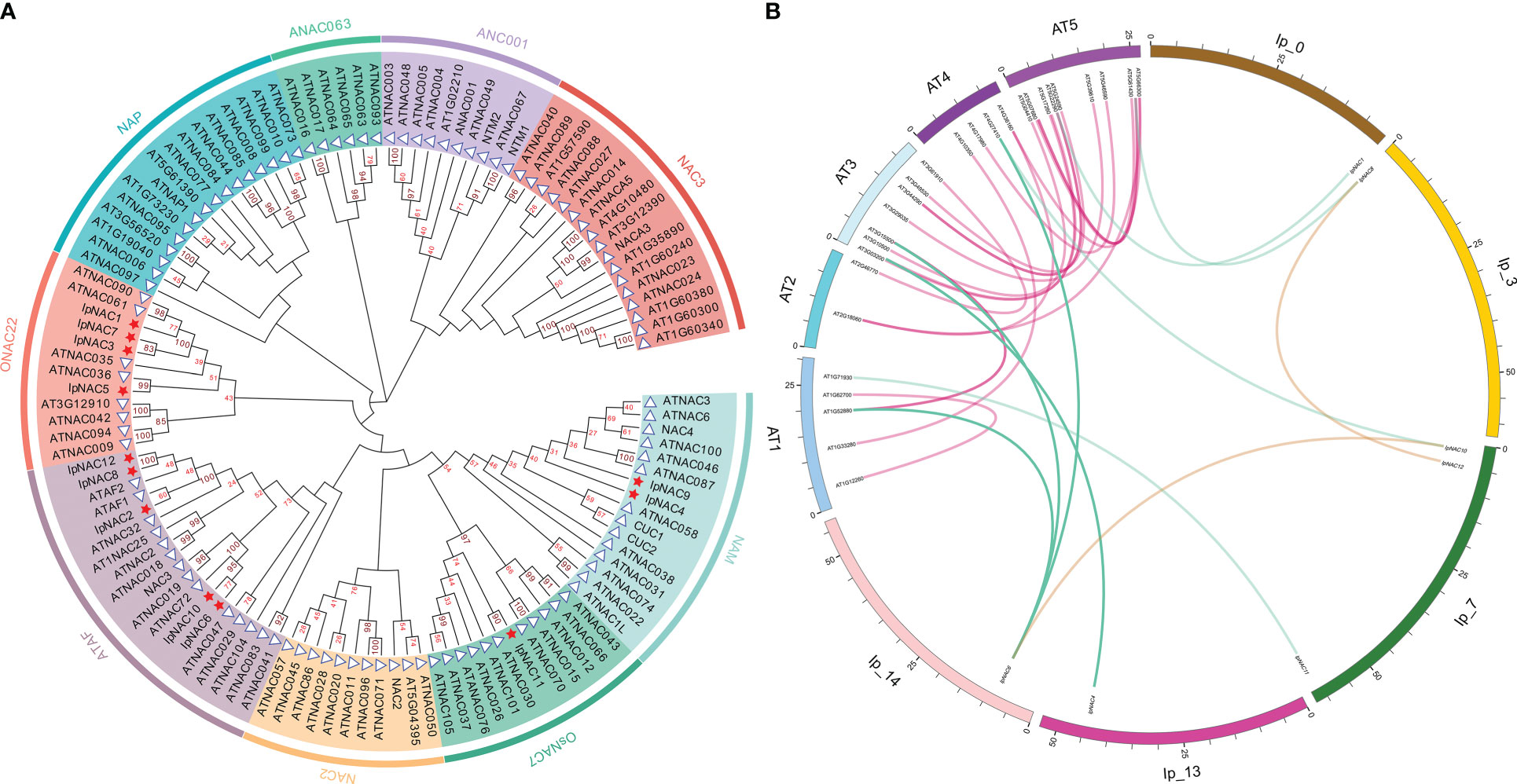
Figure 2 Comparative analysis of Ipomoea pes-caprae IpNACs and Arabidopsis ANAC genes. (A) Phylogenetic tree of IpNAC and ANAC proteins. The full-length amino acid sequences of the IpNAC proteins were used to construct the phylogenetic tree with MEGA X through 100 bootstrap tests and the Poisson model method. The number at the node indicates the bootstrap value. The nine subfamilies, namely ONAC22, NAP, ANAC063, ANAC001, NAC3, NAM, OsNAC7, NAC2, and ATAF, were set in different colors as the background. IpNACs are represented by red pentagons, and ANACs are represented by blue triangles. (AtNAC=ANAC). (B) Collinearity analysis of NAC genes in I pes-caprae and Arabidopsis. The chromosomes of I pes-caprae and Arabidopsis are expressed in different colors, and the positions of IpNAC and ANAC genes in chromosomes are marked. The purple curve represents the collinear region of ANAC genes, the brown curve represents the collinear region of IpNAC genes, and the blue curve represents the collinear region of ANACs and IpNACs.
Motifs are key functional units. The IpNAC motif components were analyzed using MEME online software. All IpNAC proteins contained the NAC family characteristic subdomains (A–E), named motif1/2/3/4/5, located at positions 0–210. Except for IpNAC1/4/7/9, other protein sequences had more than five motifs, with a maximum of eight. The 12 IpNACs had three exons, separated by two introns (Figure 1), forming a highly conserved gene structure. The untranslated region (UTR) lengths differed significantly, indicating a difference in the efficiency of IpNAC gene expression. TFs regulate gene function by binding cis-acting elements to the promoter. The PLACE Online website was used to analyze the 2 kb upstream region of the transcription start site of IpNACs, and a series of stress response elements were found (Figure 3). A cis-acting element involved in drought-inducibility (MBIS) was found in three IpNAC genes, an enhancer-like element involved in anoxic specific inducibility cis-acting element (GC-motif) was detected in four IpNAC genes, and a defense and stress response cis-acting element (TC-rich) was detected in five IpNAC genes. Anaerobic induction cis-acting regulatory elements (AREs) were identified in 11 IpNAC genes. Interestingly, there were five IpNAC genes with low-temperature responsive cis-acting elements (LTRs). Many hormone-responsive elements were found in the IpNAC promoter sequence. GA responsive elements were found in three IpNAC genes. SA responsive elements were found in four IpNAC genes. IAA responsive elements were found in 11 IpNAC genes, and ABA responsive elements were found in 12 IpNAC genes. This indicates that plant hormones play a central role in the regulation of the salt stress response.
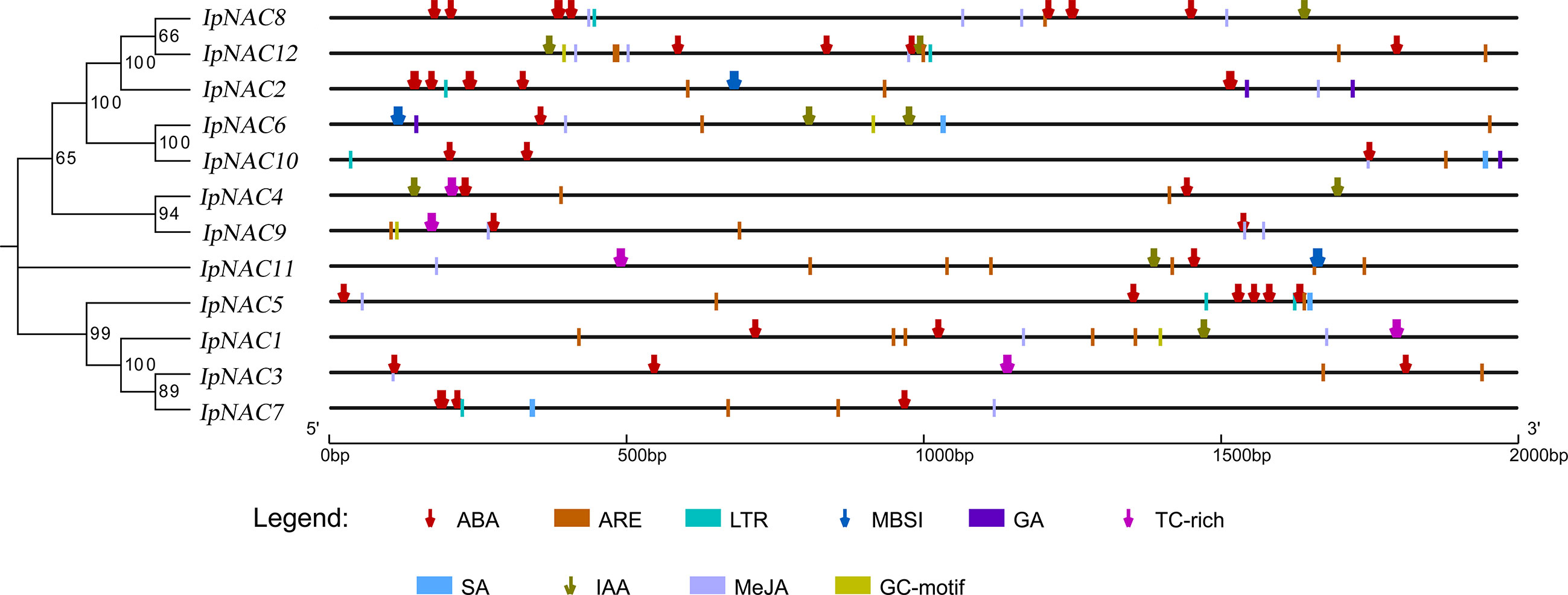
Figure 3 Cis-elements in the 2 kb upstream promoter of Ipomoea pes-caprae IpNACs. The homologous tree of the IpNAC genes is located on the left, and the number at the node indicates the bootstrap value. Symbols of different colors and shapes mark the relative positions of different elements, and the black scale line represents the relative protein length.
Ipomoea pes-caprae is a highly salt-tolerant crop, and the expression pattern of IpNACs was determined after treatment with different concentrations of NaCl solution. Among the 12 genes, there were significant differences (Figure 4). Compared with 600 and 900 mM NaCl solutions, IpNAC genes treated with 300 mM NaCl solution had the highest response to salt induction. Interestingly, IpNAC10 showed the strongest significance after different salt solution treatments and maintained a high mRNA level (>2000-fold) at 6 and 24 h treatment periods. IpNAC11 was upregulated significantly, reaching the maximum change (47-fold) at 3 h, and returned to baseline over time. In contrast, IpNAC2 reached maximum expression at 48 h (6-fold), and the trend was downregulated and only slightly upregulated throughout the treatment period. IpNAC5, IpNAC8, and IpNAC12 expression were induced 19–300-fold at 6 h. The expressions of IpNAC3, IpNAC6, and IpNAC9 were increased 15–300-fold at 12 h, and those of IpNAC4 and IpNAC7 expression reached 25–130-fold at 24 h. Under 600 mM NaCl treatment, the 12 genes were not strongly induced at 3 and 6 h. IpNAC1 and IpNAC2 were upregulated after 12 h (16–19-fold) and remained stable or were downregulated in the early stage. The induction level of IpNAC5, IpNAC8, IpNAC11, and IpNAC12 increased 10- to 60-fold (24 h) and were upregulated steadily throughout the period. The expressions of IpNAC3, IpNAC4, IpNAC6, IpNAC7, and IpNAC9 reached 15- to 280-fold (48 h). Interestingly, IpNAC6, IpNAC7, and IpNAC9 were upregulated, but the trend was high–low–high. The IpNAC genes were generally downregulated after 900 mM NaCl solution treatment, and IpNAC2, IpNAC3, IpNAC4, IpNAC6, IpNAC7, IpNAC8, IpNAC9, IpNAC11, and IpNAC12 reached the maximum significance level at 48 h, among which IpNAC2, IpNAC3, and IpNAC4 were downregulated (3–24 h), and IpNAC5 and IpNAC10 maintained a high induction at 6 h. Interestingly, IpNAC1 did not respond to 900 mM NaCl solution treatment.
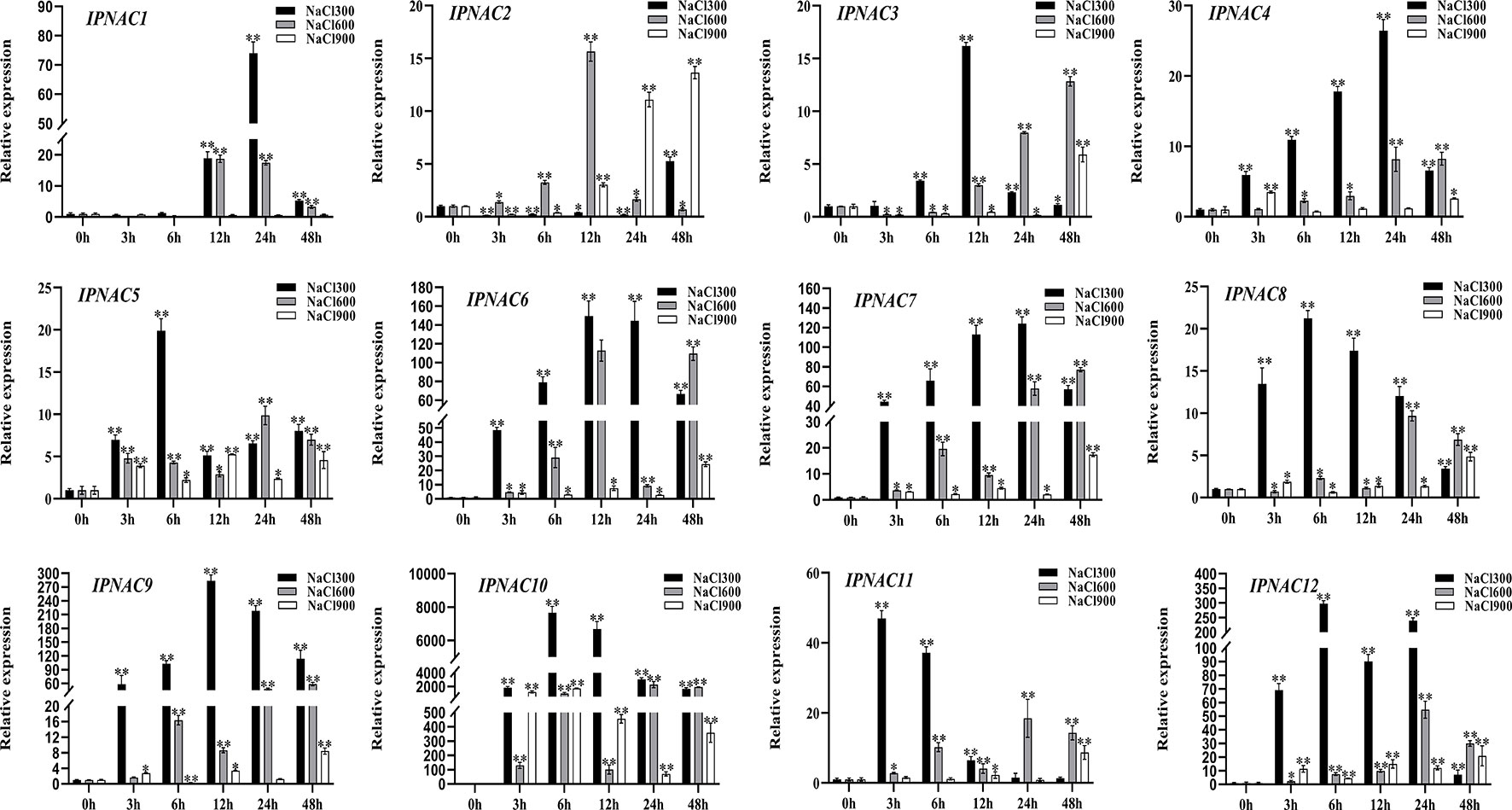
Figure 4 The expression level of IpNAC genes under different salt treatments. The roots were soaked in 300, 600, and 900 mM NaCl solutions and collected at different time points (0, 3, 6, 12, 24, and 48 h). The expression of IpNAC genes under salt treatment was detected by quantitative real-time PCR. Non-stressed plants (0 h) were used as a control, and each experiment was conducted with three biological replicates and three technical replicates. The mean values and SD were obtained from the experiment. The p-values were evaluated using Student’s t-test. Asterisks indicate the level of significance, **p < 0.01 and *0.01 < p < 0.05. Transcript levels at 0 h were set as 1.
Ipomoea pes-caprae is a high temperature- and drought-tolerant crop. The expression level of most IpNAC genes was increased under heat stress (45 °C) and drought stress (20% PEG 6000), but the change in the expression level under drought stress was more obvious than that under high temperature stress (Figure 5). Under a high temperature (45 °C), the IpNAC5 induction level was the highest (451-fold), followed by that of IpNAC1, IpNAC2, IpNAC3, and IpNAC9, which reached the highest significant expression (10–90-fold) at 24 h. However, the maximum induction of IpNAC4, IpNAC6, IpNAC7, IpNAC8, IpNAC9, IpNAC10, IpNAC11, and IpNAC12 was less than 10-fold. Interestingly, all IpNAC genes were strongly induced at 24 h under heat stress. Most IpNAC genes were near or below the baseline at 3–24 h and near the baseline at 48 h. Under drought stress, IpNAC12 reached a maximum induction at 48 h (442-fold), followed by IpNAC1, IpNAC2, IpNAC3, IpNAC5, IpNAC8, IpNAC9, and IpNAC11, reaching a maximum at 12 h (1–80-fold). IpNAC4, IpNAC6, and IpNAC10 increased 10- to 300-fold at 3 and 48 h. However, the expression trends of IpNAC2 and IpNAC3 after high temperature and drought stress were consistent (Figure 5). In addition, compared with high temperature stress, IpNAC2 and IpNAC3 were less responsive to drought stress. Under the same stress, IpNAC6 and IpNAC10 showed similar trends but were less responsive to high temperature stress.
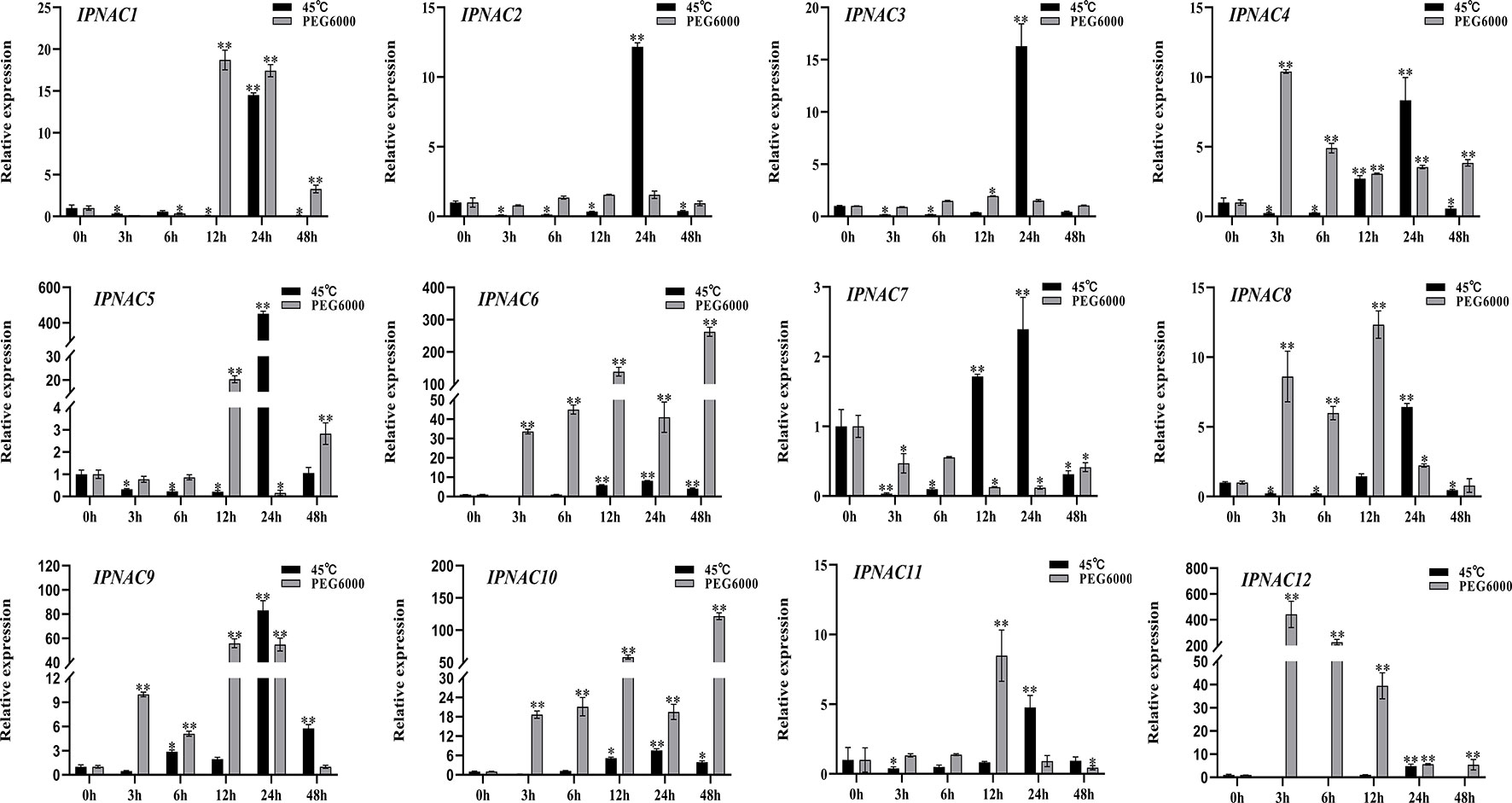
Figure 5 The expression level of IpNAC genes under heat (45 °C) and 20% PEG 6000 solution stress treatments. The roots were soaked in 20% PEG 6000 solutions and collected at different time points (0, 3, 6, 12, 24, and 48 h). Non-stressed plants (0 h) were used as a control, and each experiment was conducted with three biological replicates and three technical replicates. The mean values and SD were obtained from the experiment. The p-values were evaluated using Student’s t-test. Asterisks indicate the level of significance, **p < 0.01 and *0.01 < p < 0.05. Transcript levels at 0 h were set as 1.
In response to internal and external stress, plant hormones (abscisic acid [ABA], salicylic acid [SA], and methyl jasmonate [MeJA]) play an important role in the stress signal network. We studied the relationship between the transcriptional levels of the 12 IpNAC genes under different plant hormones (Figure 6). Gene expression of IpNAC4, IpNAC6, IpNAC7, IpNAC9, IpNAC10, IpNAC11, and IpNAC12 after ABA treatment increased throughout the treatment period, and most genes were rapidly upregulated 5- to 500-fold at 24 and 48 h, while IpNAC10 quickly reached 477-fold at 6 h. Similarly, the induction levels of IpNAC1, IpNAC2, IpNAC3, IpNAC5, and IpNAC8 reached maximum significance (4–60-fold) at 12–24 h, and the early response was weak or downregulated. Under SA stress, IpNAC1, IpNAC8, and IpNAC12 were significantly upregulated throughout the treatment period. The induced expression of IpNAC12 reached 2064-fold at 48 h. IpNAC2, IpNAC3, IpNAC4, IpNAC5, IpNAC6, IpNAC7, IpNAC9, IpNAC10, and IpNAC11 were mainly upregulated (2–500-fold) at 6–24 h, and except for the highest induction time, the induction times were close to baseline or downregulated. Based on the results of the MeJA treatment, the transcript level of IpNAC9 reached a peak of 241-fold at 48 h. IpNAC4, IpNAC6, IpNAC7, IpNAC8, IpNAC11, and IpNAC12 also maintained a high level (5–150-fold) throughout the period. Most trends continuously increased from 3 to 24 h and steadily decreased from 24 to 48 h. In contrast, IpNAC1, IpNAC2, IpNAC3, IpNAC5, and IpNAC10 were downregulated (2–60-fold) in different periods (3–12 h). At the same time, IpNAC5 did not respond to MeJA induction and was not sensitive throughout the period (Figure 6).
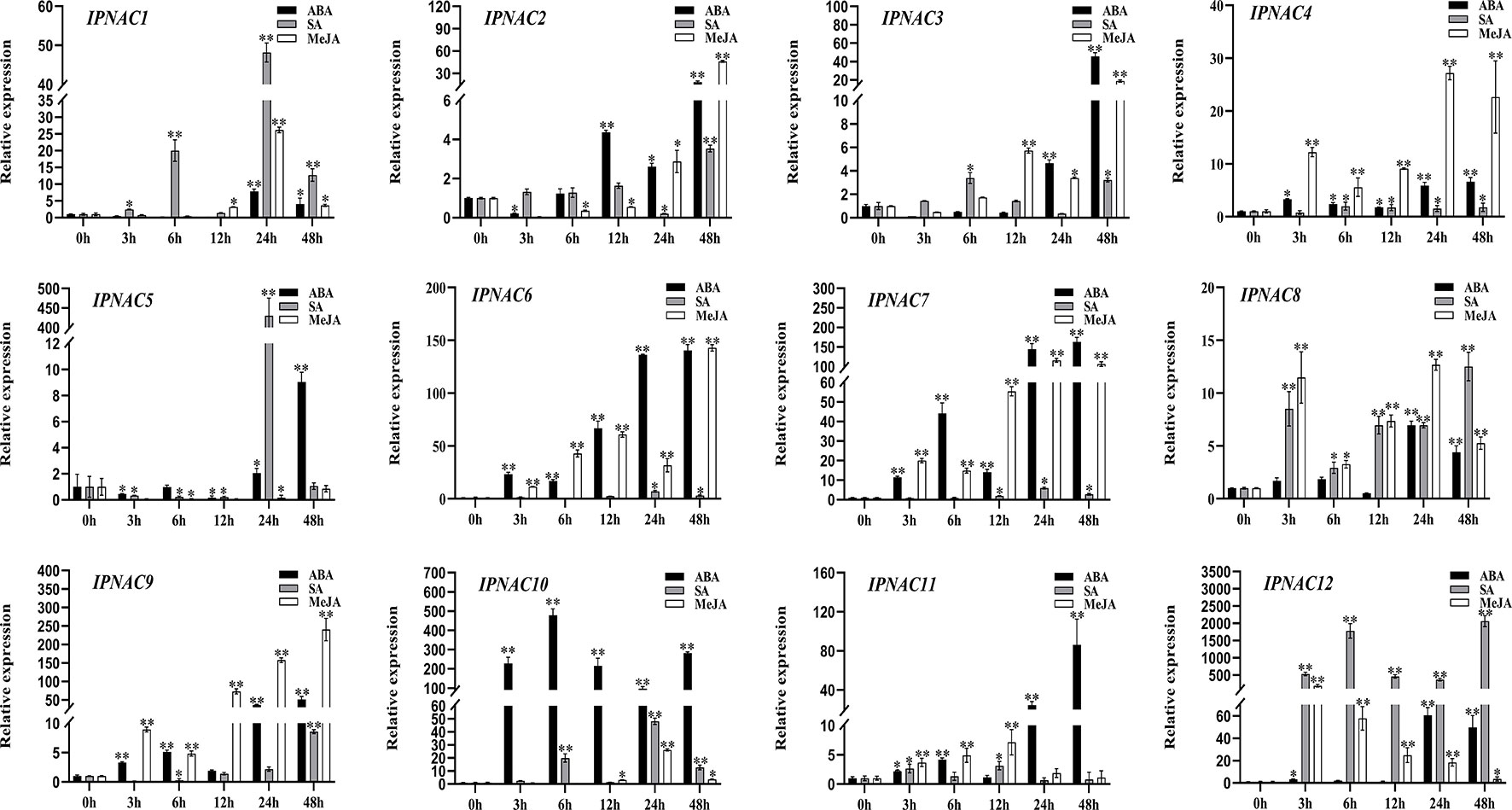
Figure 6 The expression level of IpNAC genes in the plant hormone treatment. The roots were soaked in 0.1 mM abscisic acid (ABA), 2 mM salicylic acid (SA) solution, or 2 mM jasmonic acid (JA) solution, and the roots were collected at different time points. Non-stressed plants (0 h) were used as a control, and each experiment was conducted with three biological replicates and three technical replicates. The mean values and standard deviation (SD) were obtained from the experiment. The p-values were evaluated using Student’s t-test. Asterisks indicate the level of significance, **p < 0.01 and *0.01 < p < 0.05. Transcript levels at 0 h were set as 1.
DNA- and TF-specific binding is involved in transcriptional regulation activities, and most TFs with substantial functions are located in the cell nucleus. We screened four candidate salt-tolerance genes and performed subcellular localization for IpNAC5/8/10/12 in vivo (Figure 7). The positions of IpNAC5/8/10/12 in plant cells were detected by GFP markers under the control of the CaMV 35S promoter. In the transient expression of GFP-IpNAC5/IpNAC8/IpNAC10/IpNAC12 in tobacco mesophyll protoplasts, the GFP signal was located in the nucleus and mixed with nuclear marker dye DAPI (Figure 7). However, GFP was located in the nucleus of tobacco mesophyll cytoplasm when tobacco mesophyll protoplasts carrying GFP alone were used as controls. These results suggest that GFP-IpNAC5, GFP-IpNAC8, GFP-IpNAC10, and GFP-IpNAC12 are located in the nucleus (Figure 7), which is consistent with the results presented in Table 1.
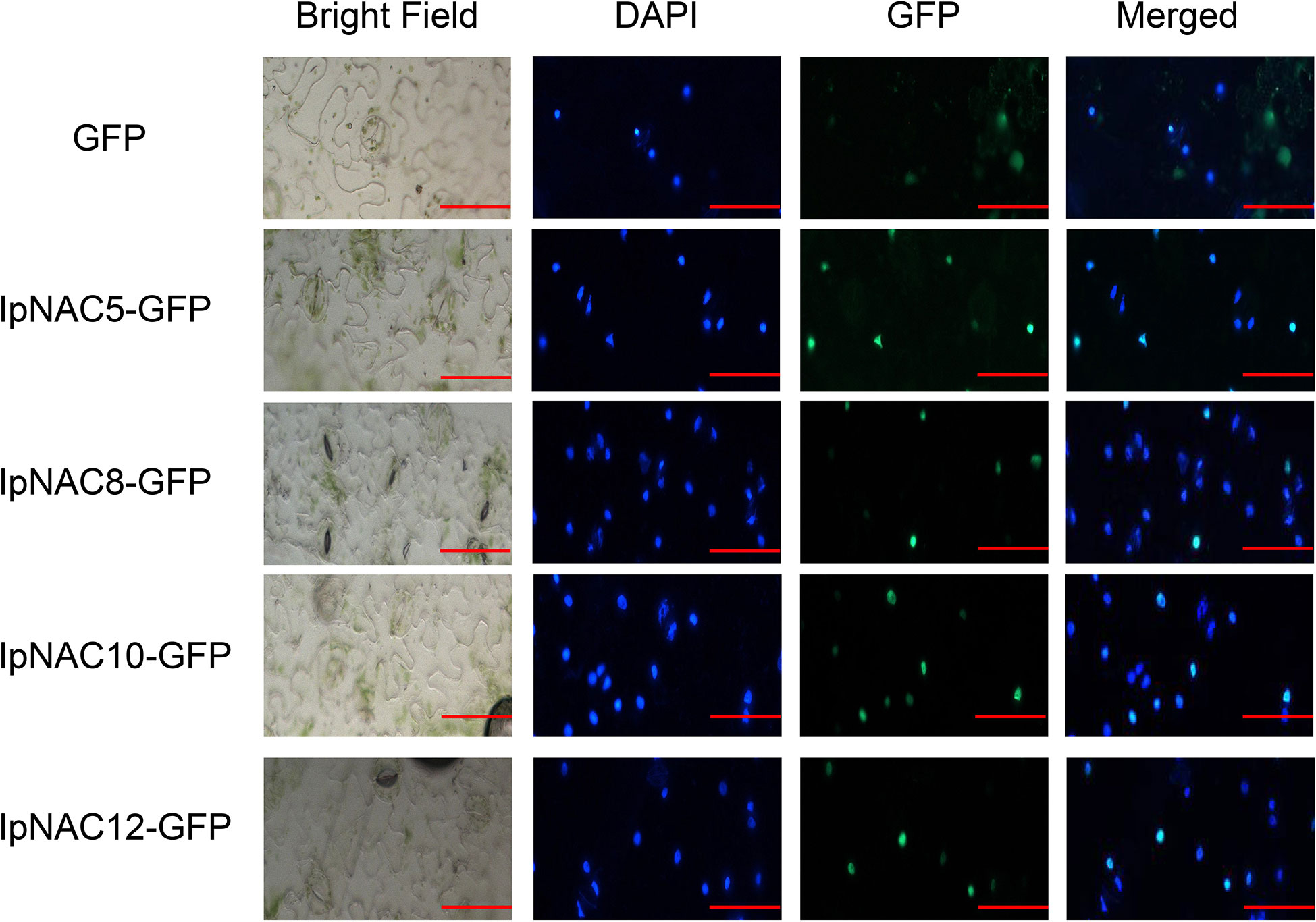
Figure 7 Subcellular localization of IpNAC5/8/10/12 in tobacco cells. IpNAC5/8/10/12-GFP and PHB-GFP were transiently expressed in tobacco. GFP fluorescence was observed 2 d after infection by 10 × 20-fold laser scanning. Photos were taken under bright and dark light for the detection of green fluorescent protein (GFP), diamino-2-phenylindole (DAPI), and binding (merging). Bars = 30 μm.
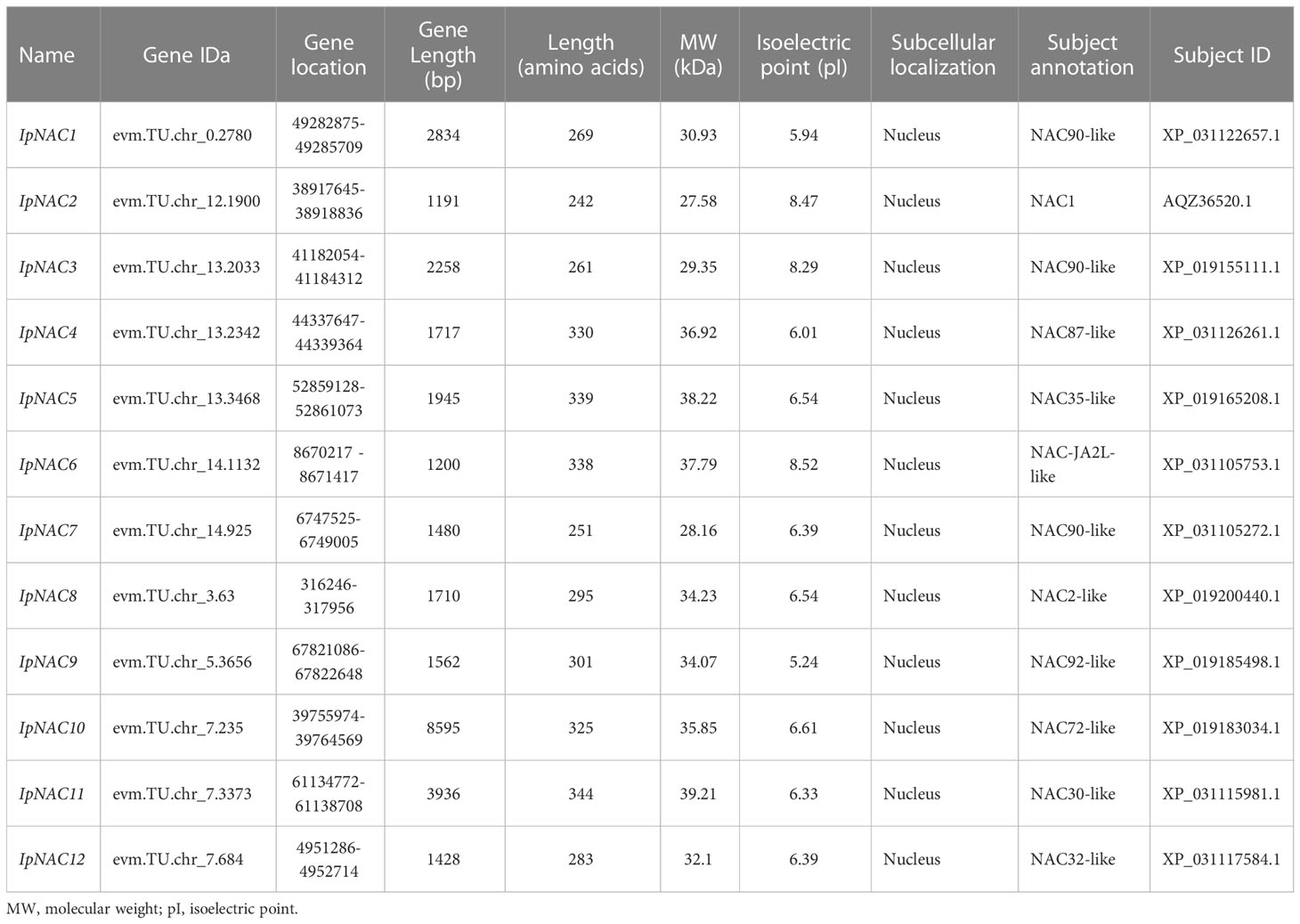
Table 1 The information of screened salt stress-responsive Ipomoea pes-caprae IpNAC genes in this study.
Heterologous overexpression in Arabidopsis is a useful technique for verifying gene function. The physiological phenotypes of the WT and transgenic Arabidopsis on 1/2 MS were similar (Figure 8A). At 180 mM NaCl, the root length of transgenic plants was longer than that of WT plants (Figures 8B, C). Arabidopsis overexpressing IpNAC5/8/10/12 was highly salt tolerant at the post-germination stages. Among these, plants overexpressing IpNAC5 and IpNAC10 showed excellent characteristics.
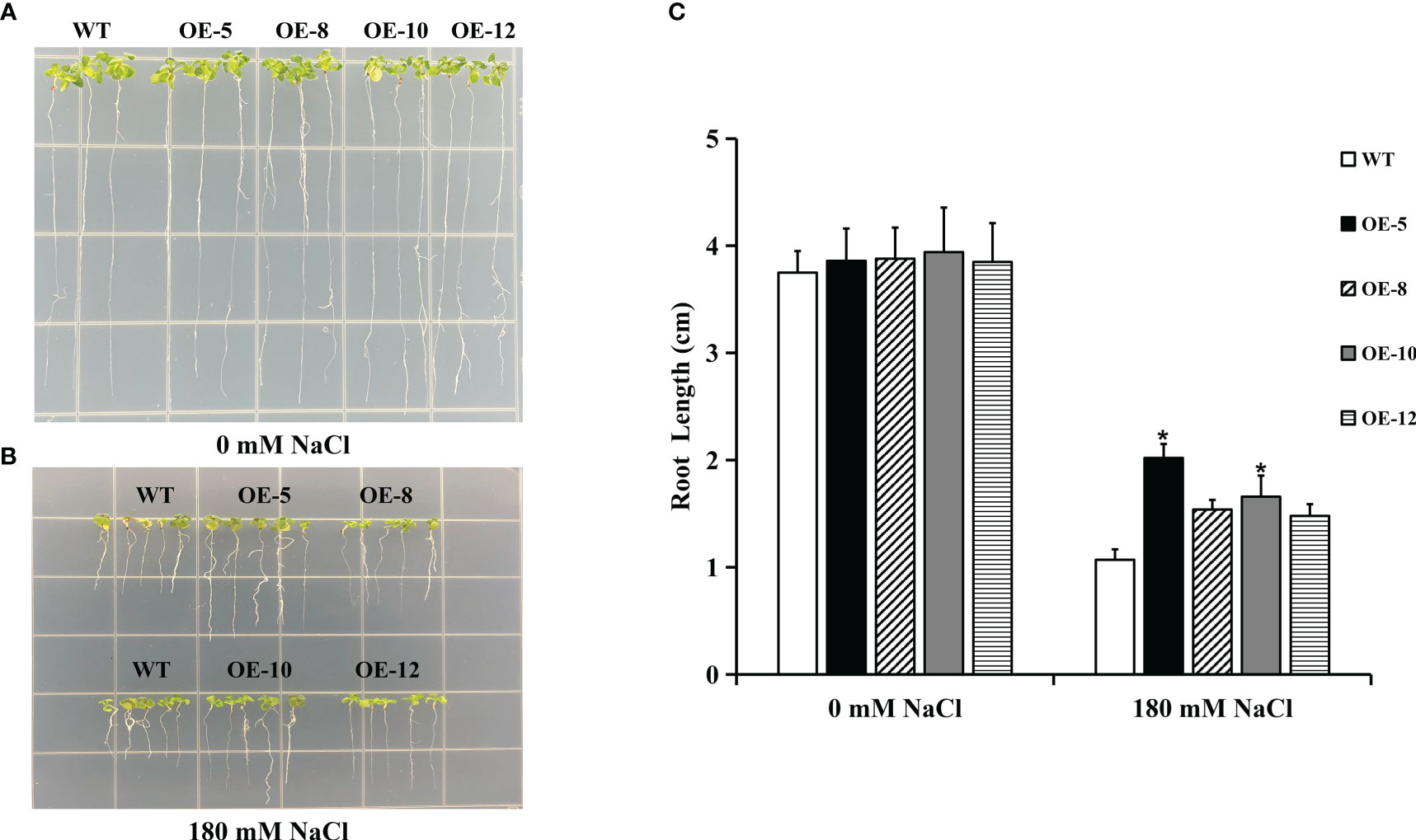
Figure 8 Transgenic plants overexpressing IpNAC5/8/10/12 had improved root length under salt stress. (A-C) Comparisons of the root length between wild type (WT) and transgenic Arabidopsis thaliana plants grown on 1/2 MS + 180 mM NaCl for 10 d. Data are the means ± SE of three independent biological experiments. OE-5 is an IpNAC5-overexpressing plant, OE-8 is an IpNAC8-overexpressing plant, OE-10 is an IpNAC10-overexpressing plant, and OE-12 is an IpNAC12-overexpressing plant. The p-values were evaluated using Student’s t-test. Asterisks indicate the level of significance, The meaning of the symbol “*” is the level of significance, *0.01 < p < 0.05, between the WT and transgenic plants.
To determine whether IpNAC5/8/10/12 affect root Na+ homeostasis, we carried out root transgenics and obtained many complete transgenic roots (Figure 9). The target genes showed higher expression in the positive transgenic roots (TR) than in the non-transgenic roots (AR) (Figures 9B, D, F, H). The salt-induced Na+ fluxes in the elongation and maturation zones of AR and TR were measured using the noninvasive microtest technique (NMT) after 24 h of NaCl stress (150 mM). Na+ efflux in the elongation and mature regions of AR and TR differed (Figure 10, Figure 11). Among the four genes, the Na+ efflux of the IpNAC10-overexpressing plant TR was more obvious than that of AR. The average Na+ efflux rates in the two root zones of TR were 3.2- and 6.7-fold that of AR, respectively (Figures 11A, B). Interestingly, the Na+ efflux of TR in the IpNAC5-overexpressing plant was 3.3-fold that of AR in the elongation zone, but there was no significant difference in the mature zone (Figures 10A, B). The average Na+ efflux rate of TR in IpNAC8 was 1.8-fold that of AR in the mature zone (Figures 10C, D). The average Na+ efflux rate of TR in IpNAC12 was 2.2-fold that of AR in the elongation zone (Figures 11C, D). These results indicate that IpNAC5/8/10/12 overexpression probably activated the ion channel and increased the root Na+ ion efflux efficiency.
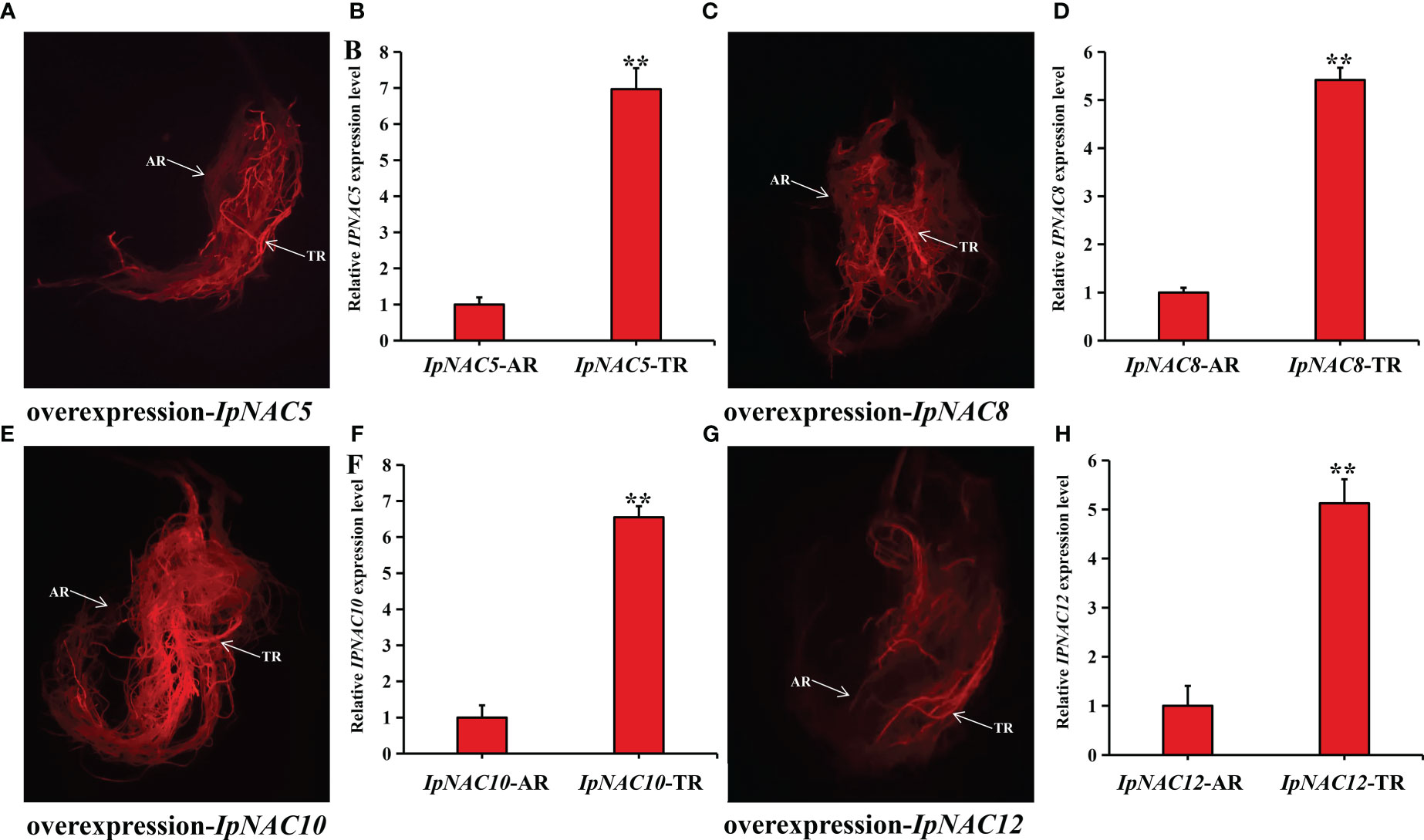
Figure 9 Preparation of IpNAC5/8/10/12 transgenic sweetpotato roots. (A, C, E, G) DsRed fluorescence diagram of transgenic sweetpotato root. TR: positive transgenic roots; AR: non-transgenic roots. (B, D, F, H) Relative expression levels of IpNAC5/8/10/12 in TR and AR. Asterisks indicate the level of significance, **p < 0.01. Gene levels of AR were used as a reference and set as 1.
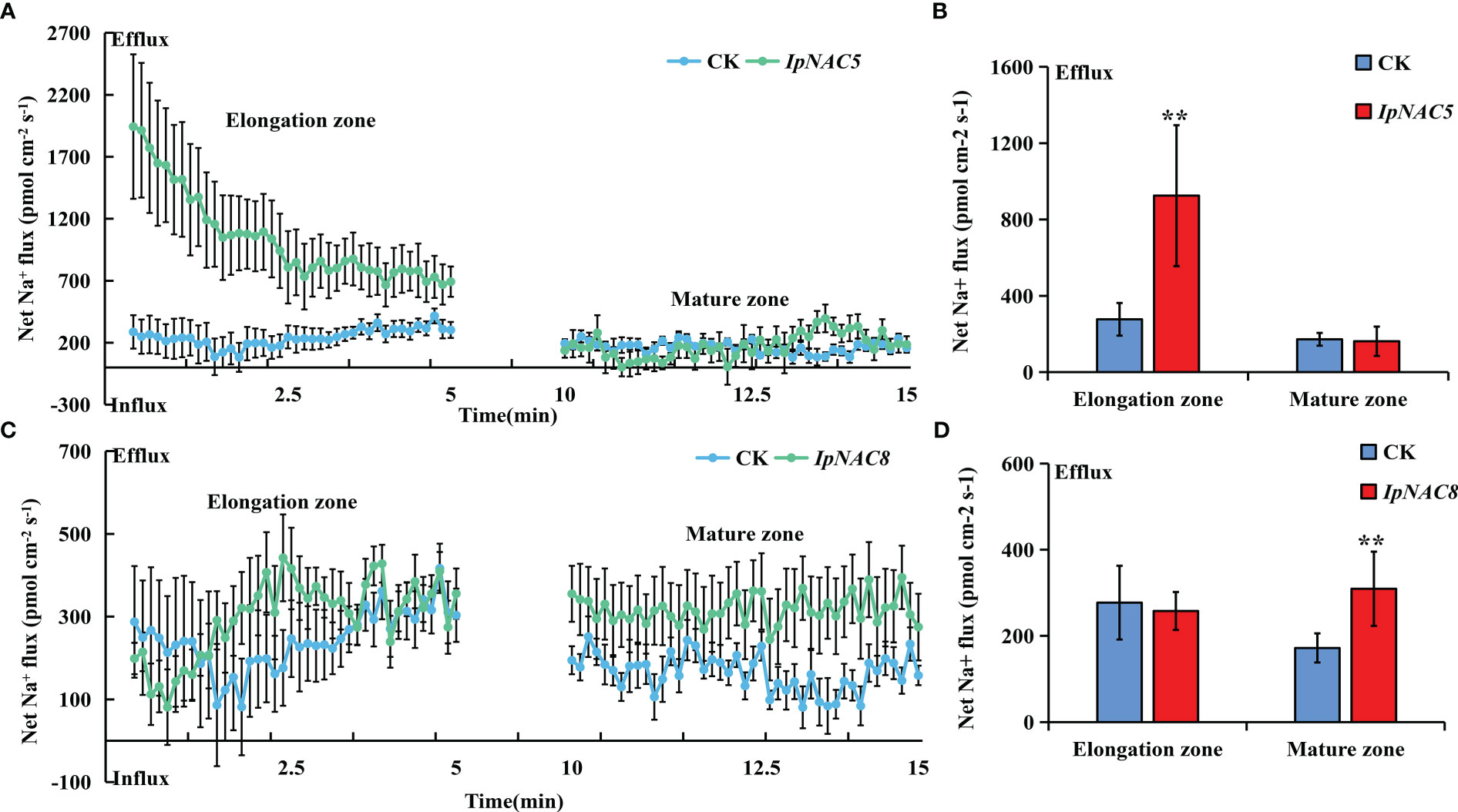
Figure 10 Na+ flux determination in IpNAC5/8-overexpressing sweetpotato roots. (A, C) The steady-state Na+ fluxes in the AR and TR, including the elongation and mature zones, after 24 h of NaCl treatment (150 mM). The standard errors of the means are represented by the bars. (B, D) Mean net Na+ efflux rates in (A, C), respectively. Asterisks indicate the level of significance between the AR and TR groups at **p < 0.01.
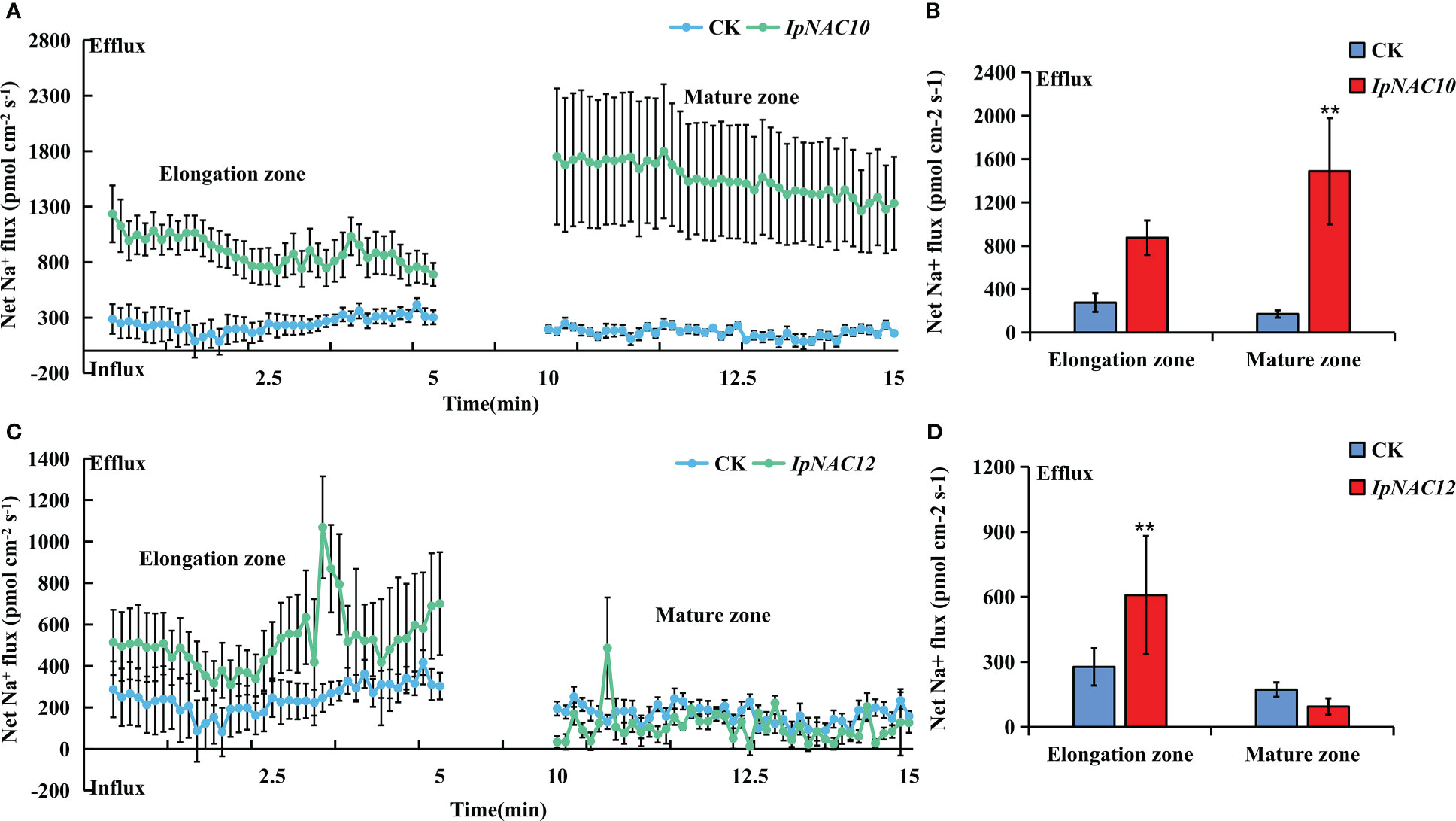
Figure 11 Na+ flux determination in IpNAC10/12-overexpressing sweetpotato roots. (A, C) The steady-state Na+ fluxes in the AR and TR, including the elongation and mature zones, after 24 h of NaCl treatment (150 mM). The standard errors of the means are represented by the bars. (B, D) Mean net Na+ efflux rates in (A, C), respectively. Asterisks indicate the level of significance between the AR and TR groups at **p < 0.01.
The influence of extreme weather is aggravating. Enhancing crop tolerance to respond to various abiotic stressors is essential for improving agricultural production efficiency. Ipomoea pes-caprae, a wild relative of sweetpotato, has significant biological characteristics in saline–alkali tolerance (Liu et al., 2020) and resistance to high temperature, drought (Suarez, 2011), and other abiotic stressors (Zhang et al., 2018). Compared with sweetpotato, I. pes-caprae has a smaller genome and is an excellent model for studying the function of sweetpotato genes. NAC proteins are a vital part of the signal transduction network and play an important regulatory role in plant responses to various stressors (Meng et al., 2022). However, NAC TF-related research reports mainly focus on model plants, and there are few reports on I. pes-caprae. Here, 12 salt-responsive IpNAC genes were isolated. The number of IpNAC genes was less than that of ANAC genes (Table 1). One reason may be that the genome of I. pes-caprae is small, and there were no large-scale repetitive events in the early stages of plant evolution. In Arabidopsis, a model plant, functional verification is clear (Ooka et al., 2003). Based on the classification of the phylogenetic tree of IpNAC and ANAC proteins, IpNACs were divided into four subfamilies (Figure 2A). IpNAC4 and IpNAC9 proteins may belong to the NAM subfamily. NAM are involved in flower formation (Hendelman et al., 2013) and leaf senescence (Wang et al., 2022). IpNAC2, IpNAC6, IpNAC8, IpNAC10, and IpNAC12 were assigned to ATAF (Figure 2A). The ATAF subfamily has many transcriptional activators or inhibitors that are crucial in plant stress resistance and hormone synthesis (Peng and Neff, 2021), indicating that IpNAC2, IpNAC8, and IpNAC12 may have similar roles. Among them, the IpNAC2 protein is NAC1, and its function is unknown. Based on other plants, NAC1 significantly contributes to increasing resistance to stress (You et al., 2014). The OsNAC7 subfamily contains only IpNAC11, which plays a major role in the response to abiotic stress and the regulation of plant growth (Wang et al., 2021). The ONAC22 subfamily significantly reduces the water loss rate and the transpiration rate to reduce drought stress (Peng et al., 2022). In addition, this subfamily improves salt tolerance by adjusting root length (Hong et al., 2016). As shown in Figure 8, the IpNAC5 transgenic roots were relatively long. Collinearity analysis can describe the positional relationships of genes, as well as evolutionary and functional connections on chromosomes. Six IpNAC genes had a collinear relationship with ANACs (Figure 2B). Among them, IpNAC6 had a collinear relationship with three ANAC genes (AT1G52880, AT3G15500, AT4G27410) and had many important regulatory functions. In addition, AT4G27410 had a collinear relationship with IpNAC6 and IpNAC10. At4G27410 encodes a dehydration-induced NAC transcription factor and acts as a transcriptional activator in the ABA-mediated dehydration response (Fujita et al., 2004). IpNAC6 and IpNAC10 may enable stress resistance through ABA hormone regulation. Figure 2 shows that most of the IpNAC genes have a close relationship with ANAC genes, indicating that their functions may be similar. The structure of the gene and the number of amino acid motifs play important roles. The 12 IpNAC proteins contained five NAC-specific motifs (A–E), indicating that IpNACs are highly conserved (Figure 1). Through the multiple sequence alignment of IpNAC proteins, the B and E domains had obvious differences, which may be an important reason for gene function diversification (Supplementary Figure S1). The UTR and intron–exon constitute the gene structure and, ultimately, through biological rules, perform biological functions. IpNAC2, IpNAC6, and IpNAC7 lack an UTR, which indicates that the response expression may be slow (Srivastava et al., 2018) (Figure 1). Cis-acting elements are important molecular switches that work together with stress-inducible regulatory factors (Chanwala et al., 2022). The IpNAC promoter sequence contained some abiotic stress response elements and many hormone-responsive elements (Figure 3). This indicates that plant hormones play a central role in the regulation of the abiotic stress response. Different stress factors directly or indirectly affect gene expression levels. According to the gene expression level, we inferred that most of the IpNAC genes responded to induction under different stress treatments. Compared with different salt solutions, under the 300 mM NaCl solution, the treated IpNACs had the strongest ability to respond to the induction, followed by the 600 mM NaCl solution (Figure 4). With the increase in salinity, the time of the maximum gene expression level was delayed. The IpNAC gene expression level after 900 mM NaCl treatment was generally near the baseline or even downregulated. We speculate that the salt concentration may be too high, and that the plant reduces most of the biological reactions to maintain cell survival. In particular, IpNAC10 showed the highest induction level under different salt solutions. Moreover, IpNAC10 was the strongest inducer of the ABA response (about 500-fold) among IpNACs (Figure 6) and was also highly responsive to drought stress (about 150-fold) (Figure 5). This finding is similar to the results of previous study (Fujita et al., 2004). Plant root cells can absorb water only when their water potential is lower than the surrounding medium water potential (Erlandsson, 1979). We speculated that, because of the high salt solution content, the osmotic potential increases, which makes it difficult for plants to absorb water, leading to the high expression of IpNAC10, and its subsequent functions need further research. The characteristics of IpNAC8 and IpNAC12 were consistent with ATAF (Figure 2). Wu et al. (2009) found that ATAF1 was significantly induced by high salinity and ABA, which enhanced the drought resistance of plants. Whether the function of the IpNAC8 and IpNAC12 genes is similar to that of ATAF1 needs investigation. IpNAC8 and IpNAC12 were significantly higher under salt stress (about 25-fold and 300-fold, respectively) (Figure 4). Among the 12 genes, IpNAC12 was the most strongly induced after drought stress (~500-fold) and SA stress (~2000-fold), but the expression level under high temperature treatment showed low sensitivity (Figure 5). At the same time, IpNAC8 significantly responded to ABA (~200-fold) and MeJA stress (~150-fold). We speculate that IpNAC10 and IpNAC8 accomplish stress signal transduction not only through the ABA pathway but also through the SA and MeJA stress mechanisms. As a subtropical coastal plant, a high temperature is an essential element for I. pes-caprae. Under induction at 45 °C, IpNAC5 had the highest expression (~500-fold). IpNAC5 shared a relatively high homology with ANAC036 (Figure 2A). Kato et al. (2010) suggested that ANAC036 is induced by osmotic stress and salt stress. In terms of natural factors, salt and high temperature are inseparable. As a result, we speculated that high temperatures accelerated the evapotranspiration rate and increased the loss of water, leading the IpNAC5 gene to be highly expressed in response to stress. Therefore, based on this analysis, we speculate that IpNAC5/8/10/12 are important candidate genes for stress response. Further biological proof is necessary. Hence, we performed subcellular localization of IpNAC5, IpNAC8, IpNAC10 and IpNAC12 and found that they were located in the nucleus (Figure 7). Heterologous overexpression in Arabidopsis verified the gene function. We obtained transgenic seeds by overexpressing IpNAC5/8/10/12 in Arabidopsis. T3 homozygous seeds were selected for the salt tolerance test. There was no significant difference between transgenic plants and WT plants in the no-salt medium (Figure 8A). Upon 180 mM NaCl stress, the root length and plant growth status of transgenic plants were greater than those of WT plants (Figure 8B). Compared with other IpNAC5/8/10/12 genes, IpNAC5 and IpNAC10 had the strongest salt tolerance in the post-germination stages of Arabidopsis. Cellular Na+ homeostasis and metabolic processes significantly affect plant salt tolerance (Zhang et al., 2017). By measuring Na+ homeostasis in the transgenic sweetpotato roots, we found that the TR of IpNAC10 had obvious Na+ efflux in the mature and elongation zones compared with AR (Figure 11A). Interestingly, TR of IpNAC5 and IpNAC12 showed 3.3- and 2.2-fold more Na+ efflux in the elongation zone than AR, while there was no significant difference in the maturation zone (Figure 10A, Figure 11C). Similarly, the TR of IpNAC8 showed significant Na+ efflux in the mature region compared to AR (Figure 10C). These results indicate that IpNAC5/8/10/12 are highly responsive and protective in sweetpotato, resisting salt stress through Na+ ion channels, and were activated, leading to Na+ efflux in different regions of the sweetpotato roots.
Using I. pes-caprae as a basis, we utilized biotechnology to explore salt tolerance genes to provide relevant genetic resources and strategies for the genetic improvement of sweetpotato. At the same time, it is important to analyze the molecular regulatory mechanism of salt tolerance in I. pes-caprae to further plant adaptation to extreme climates.
In this study, by combining transcriptome data with bioinformatics analysis, we identified 12 IpNAC genes in I. pes-caprae. Evolutionary collinearity, gene structure, and promoter cis-acting elements of IpNACs were analyzed. Different responses under a variety of abiotic and hormone stressors indicate that IpNAC genes are controlled by a variety of regulatory mechanisms. IpNAC5/8/10/12 were screened as candidate genes. IpNAC5, IpNAC8, IpNAC10 and IpNAC12 localized in the nucleus. In addition, the root length and growth status of transgenic Arabidopsis under salt stress and the Na+ ion content in sweetpotato roots showed that IpNAC5, IpNAC8, IpNAC10 and IpNAC12 significantly enhanced plant salt tolerance. This study lays the foundation for further studies on the function of IpNACs under stress in I. pes-caprae. The candidate genes may contribute to the functional characterization of salt tolerance genes for breeding abiotic stress-resistant varieties of sweetpotato or other species. Taken together, these results provide new insights into the molecular response of I. pes-caprae and sweetpotato to salt stress.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
QC planned and designed the research. YS, YL, and SX wrote the manuscript. YS, YuW, YD, LZ, YaW, DZ, XD, and ZZ conducted the research and analyzed the data. QC supervised the research and the manuscript. All authors read and approved the final manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by the Earmarked Fund for CARS-10-Sweetpotato (CARS-10-GW01) and the “JBGS” Project of Seed Industry Revitalization in Jiangsu Province (JBGS (2021) 010).
We thank Prof. Jian Sun from School of Life Sciences, Jiangsu Normal University for providing technical guidance.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1119282/full#supplementary-material
Supplementary Figure 1 | IpNAC amino acid sequence alignment.
Supplementary Table 1 | The information of screened salt stress-responsive Ipomoea pes-caprae IpNAC genes in this study.
Supplementary Table 2 | IpNAC qRT-PCR primer design.
IpNAC5/8/10/12: IpNAC5, IpNAC8, IpNAC10, and IpNAC1235S:GFP-IpNAC5/IpNAC8/IpNAC10/IpNAC12: 35S: GFP-IpNAC5, 35S:GFP-IpNAC8, 35S:GFP-IpNAC10, 35S:GFP-IpNAC12. GFP-IpNAC5/IpNAC8/IpNAC10/IpNAC12: GFP-IpNAC5, GFP-IpNAC8, GFP-IpNAC10, and GFP-IpNAC12 pHB-IpNAC5/IpNAC8/IpNAC10/IpNAC12-GFP: pHB-IpNAC5-GFP, pHB-IpNAC8-GFP, pHB-IpNAC10-GFP, pHB-IpNAC12-GFP. pUBI.U4::IpNAC5/IpNAC8/IpNAC10/IpNAC12-CaMV35S::DsRed: pUBI.U4::IpNAC5-CaMV35S::DsRed, pUBI.U4::IpNAC8-CaMV35S::DsRed, pUBI.U4::IpNAC8-CaMV35S::DsRed, pUBI.U4::IpNAC12-CaMV35S::DsRed. TR: positive transgenic roots; AR, non-transgenic roots.
Akinniyi, G., Lee, J., Kim, H., Lee, J. G., Yang, I. (2022). A medicinal halophyte Ipomoea pes-caprae (Linn.) r. br.: A review of its botany, traditional uses, phytochemistry, and bioactivity. Mar. Drugs 20 (5), 329. doi: 10.3390/md20050329
Chanwala, J., Jha, D. K., Sandeep, I. S., Dey, N. (2022). The role of transcription factors in response to biotic stresses in pearl millet Vol. 22 (Cham: Springer), 195–211. doi: 10.1007/978-3-031-12990-2-10
Cheng, Y., Zhou, Q., Li, W., Cheng, H., Mohammadi, M. A., Liu, Y., et al. (2021). De novo transcriptome assembly and gene expression profiling of Ipomoea pes-caprae l. under heat and cold stresses. Sci. Hortic. 289 (17), 110379. doi: 10.1016/j.scienta.2021.110379
Cristiane, D. S. B., Guilherme, T. D. S. H., Rocha, L. W., Da Silva, G. F., Dos Anjos, M. F., Pastor, V. D.A., et al. (2017). Ipomoea pes-caprae (L.) r. br (Convolvulaceae) relieved nociception and inflammation in mice - a topical herbal medicine against effects due to cnidarian venom-skin contact. J. Ethnopharmacol. 200, 156–164. doi: 10.1016/j.jep.2017.02.014
Darzentas, N. (2010). Circoletto: visualizing sequence similarity with circos. Bioinformatics 26 (20), 2620–2621. doi: 10.1093/bioinformatics/btq484
Duan, M., Zhang, R., Zhu, F., Zhang, Z., Gou, L., Wen, J., et al. (2017). A lipid-anchored NAC transcription factor is translocated into the nucleus and activates glyoxalase I expression during drought stress. Plant Cell. 29 (7), 1748–1772. doi: 10.1105/tpc.17.00044
Du, X. Y., He, F., Zhu, B., Ren, M., Tang, H. (2020). NAC transcription factors from aegilops markgrafii reduce cadmium concentration in transgenic wheat. Plant Soil. 449, 39–50. doi: 10.1007/s11104-019-04419-w
Erlandsson, G. (1979). Efflux of potassium from wheat roots induced by changes in the water potential of the root medium. Physiol. Plantarum. 47 (1), 1–6. doi: 10.1111/j.1399-3054.1979.tb06501.x
Falak, N., Imran, Q. M., Hussain, A., Yun, B. W. (2021). Transcription factors as the "Blitzkrieg" of plant defense: A pragmatic view of nitric oxide's role in gene regulation. Int. J. Mol. Sci. 22 (2), 522. doi: 10.3390/ijms22020522
Fujita, M., Fujita, Y., Maruyama, K., Seki, M., Hiratsu, K., Ohme-Takagi, M., et al. (2004). A dehydration-induced NAC protein, RD26, is involved in a novel ABA-dependent stress-signaling pathway. Plant J. 39 (6), 863–876. doi: 10.1111/j.1365-313X.2004.02171x
Gong, L., Zhang, H., Liu, X., Gan, X., Nie, F., Yang, W., et al. (2020). Ectopic expression of HaNAC1, an ATAF transcription factor from Haloxylon ammodendron, improves growth and drought tolerance in transgenic Arabidopsis. Plant Physiol. Bioch. 151, 535–544. doi: 10.1016/j.plaphy.2020.04.008
Guo, F., Liu, S., Zhang, C., Dong, T., Meng, X., Zhu, M. (2022). Genome-wide systematic survey and analysis of NAC transcription factor family and their response to abiotic stress in sweetpotato. Sci. Hortic. 299, 27. doi: 10.1016/j.scienta.2022.111048
Hawrot-Paw, M., Wijatkowski, A., Mikiciuk, M. (2016). Influence of diesel and biodiesel fuel-contaminated soil on microorganisms, growth and development of plants. Plant Soil Environ. 61 (5), 189–194. doi: 10.17221/974/2014-pse
Hendelman, A., Stav, R., Zemach, H., Arazi, T. (2013). The tomato NAC transcription factor SlNAM2 is involved in flower-boundary morphogenesis. J. Exp. Bot. 64 (18), 5497–5507. doi: 10.1093/jxb/ert324
Hong, Y., Zhang, H., Huang, L., Li, D., Song, F. (2016). Overexpression of a stress-responsive NAC transcription factor gene ONAC022 improves drought and salt tolerance in rice. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.00004
Hrmova, M., Hussain, S. S. (2021). Plant transcription factors involved in drought and associated stresses. Int. J. Mol. Sci. 22 (11), 1–29. doi: 10.3390/ijms22115662
Hu, Y., Han, Y., Wei, W., Li, Y., Zhang, K., Gao, Y., et al. (2015). Identification, isolation, and expression analysis of heat shock transcription factors in the diploid woodland strawberry Fragaria vesca. Front. Plant Sci. 6. doi: 10.3389/fpls.2015.00736
Ju, Y., Yue, X., Min, Z., Wang, X., Fang, Y., Zhang, J. (2020). VvNAC17, a novel stress-responsive grapevine (Vitis vinifera l.) NAC transcription factor, increases sensitivity to abscisic acid and enhances salinity, freezing, and drought tolerance in transgenic Arabidopsis. Plant Physiol. Bioch. 146, 98–111. doi: 10.1016/j.plaphy.2019.11.002
Kato, H., Motomura, T., Komeda, Y., Saito, T., Kato, A. (2010). Overexpression of the NAC transcription factor family gene ANAC036 results in a dwarf phenotype in Arabidopsis thaliana. J. Plant Physiol. 167 (7), 571–577. doi: 10.1016/j.jplph.2009.11.004
Kim, H. S., Lee, C. J., Kim, S. E., Chang, Y. J., Kwak, S. S. (2018). Current status on global sweetpotato cultivation and its prior tasks of mass production. J. Plant Biotechnol. 45 (3), 190–195. doi: 10.5010/JPB.2018.45.3.190
Li, M., Chen, R., Jiang, Q., Sun, X., Zhang, H., Hu, Z. (2021). GmNAC06, a NAC domain transcription factor enhances salt stress tolerance in soybean. Plant Mol. Biol. 105 (3), 333–345. doi: 10.1007/s11103-020-01091-y
Liu, Y., Dai, X., Zhao, L., Huo, K., Jin, P., Zhao, D., et al. (2020). RNA-Seq reveals the salt tolerance of Ipomoea pes-caprae, a wild relative of sweetpotato. J. Plant Physiol. 255, 153276. doi: 10.1016/j.jplph.2020.153276
Li, S., Zheng, H., Lin, L., Wang, F., Sui, N. (2020). Roles of brassinosteroids in plant growth and abiotic stress response. Plant Growth Regul. 93 (2), 1–10. doi: 10.1007/s10725-020-00672-7
Manigauha, A., Kharya, M. D., Ganesh, N. (2015). In vivo antitumor potential of Ipomoea pes-caprae on melanoma cancer. Pharmacogn Mag. 11 (42), 426–433. doi: 10.4103/0973-1296.153099
Meng, X., Liu, S., Dong, T., Xu, T., Ma, D., Pan, S., et al. (2020). Comparative transcriptome and proteome analysis of salt-tolerant and salt-sensitive sweetpotato and overexpression of IbNAC7 confers salt tolerance in Arabidopsis. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.572540
Meng, X., Liu, S., Zhang, C., He, J., Ma, D., Wang, X., et al. (2022). The unique sweetpotato NAC transcription factor IbNAC3 modulates combined salt and drought stresses. Plant Physiol. 508. doi: 10.1093/plphys/kiac508
Meng, X., Li, G., Yu, J., Cai, J., Dong, T., Sun, J., et al. (2018). Isolation, expression analysis, and function evaluation of 12 novel stress-responsive genes of NAC transcription factors in sweetpotato. Crop Sci. 58 (3), 1328–1342. doi: 10.2135/cropsci2017.12.0738
Miryeganeh, M., Takayama, K., Tateishi, Y., Kajita, T. (2014). Long-distance dispersal by sea-drifted seeds has maintained the global distribution of Ipomoea pes-caprae subsp. brasiliensis (Convolvulaceae). PloS one 9 (4), e91836. doi: 10.1371/journal.pone.0091836
Nakashima, K., Takasaki, H., Mizoi, J., Shinozaki, K., Yamaguchi-Shinozaki, K. (2012). NAC transcription factors in plant abiotic stress responses. Bba-gene Regul. Mech. 1819 (2), 97–103. doi: 10.1016/j.bbagrm.2011.10.005
Nuruzzaman, M., Manimekalai, R., Sharoni, A. M., Satoh, K., Kondoh, H., Ooka, H., et al. (2010). Genome-wide analysis of NAC transcription factor family in rice. Gene 465 (1-2), 30–44. doi: 10.1016/j.gene.2010.06.008
Ooka, H., Satoh, K., Doi, K., Nagata, T., Otomo, Y., Murakami, K., et al. (2003). Comprehensive analysis of NAC family genes in oryza sativa and Arabidopsis thaliana. DNA Res. 10 (6), 239–247. doi. 10.1093/dnares/10.6.239
Peng, X., Feng, C., Wang, Y., Zhang, X., Wang, Y., Sun, Y., et al. (2022). miR164g-MsNAC022 acts as a novel module mediating drought response by transcriptional regulation of reactive oxygen species scavenging systems in apple. Hortic. Res. 9, uhac192. doi: 10.1093/hr/uhac192
Peng, H., Neff, M. M. (2021). Two ATAF transcription factors ANAC102 and ATAF1 contribute to the suppression of cytochrome P450-mediated brassinosteroid catabolism in Arabidopsis. Physiol. Plantarum. 172 (3), 1493–1505. doi: 10.1111/ppl.13339
Puranik, S., Sahu, P. P., Srivastava, P. S., Prasad, M. (2012). NAC proteins: regulation and role in stress tolerance. Trends Plant Sci. 17 (6), 369–381. doi: 10.1016/j.tplants.2012.02.004
Rushton, P. J., Bokowiec, M. T., Han, S., Zhang, H., Brannock, J. F., Chen, X., et al. (2008). Tobacco transcription factors: novel insights into transcriptional regulation in the solanaceae. Plant Physiol. 147 (1), 280–295. doi: 10.1104/pp.107.114041
Srivastava, A. K., Lu, Y., Zinta, G., Lang, Z., Zhu, J. (2018). UTR-dependent control of gene expression in plants. Trends Plant Sci. 23 (3), 248–259. doi: 10.1016/j.tplants.2017.11.003
Srivastava, R., Sahoo, L. (2021). Balancing yield trade-off in legumes during multiple stress tolerance via strategic crosstalk by native NAC transcription factors. J. Plant Biochem. Biot. 30, 708–729. doi: 10.1007/s13562-021-00749-y
Suarez, N. (2011). Comparative leaf anatomy and pressure-volume analysis in plants of Ipomoea pes-caprae experimenting saline and/or drought stress. J. Exp. Bot. 7 (1), 53–62. doi: 10.1016/j.flora.2010.05.006
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., Kumar, S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28 (10), 2731–2739. doi: 10.1093/molbev/msr121
Tao, Y., Wan, J., Liu, Y., Yang, X., Shen, R., Zhu, X. (2022). The NAC transcription factor ANAC017 regulates aluminum tolerance by regulating the cell wall-modifying genes. Plant Physiol. 189 (4), 2517–2534. doi: 10.1093/plphys/kiac197
Trishla, V. S., Kirti, P. B. (2021). Structure-function relationship of gossypium hirsutum NAC transcription factor, GhNAC4 with regard to ABA and abiotic stress responses. Plant Sci. 302, 110718. doi: 10.1016/j.plantsci.2020.110718
Wang, Q., Guo, C., Li, Z., Sun, J., Deng, Z., Wen, L., et al. (2021). Potato NAC transcription factor StNAC053 enhances salt and drought tolerance in transgenic Arabidopsis. Int. J. Mol. Sci. 22 (5) 2568. doi: 10.3390/ims22052568
Wang, H., Li, T., Li, W., Wang, W., Zhao, H. (2021). Identification and analysis of chrysanthemum nankingense NAC transcription factors and an expression analysis of OsNAC7 subfamily members. Peer J. 9, e11505. doi: 10.7717/peerj.11505
Wang, Y., Li, J., Paterson, A. H. (2013). MCScanX-transposed: detecting transposed gene duplications based on multiple colinearity scans. Bioinformatics 29 (11), 1458–1460. doi: 10.1093/bioinformatics/btt150
Wang, X., Rehmani, M. S., Chen, Q., Yan, J., Zhao, P., Li, C., et al. (2022). Rapeseed NAM transcription factor positively regulates leaf senescence via controlling senescence-associated gene expression. Plant Sci. 323, 111373. doi: 10.1016/j.plantsci.2022.111373
Wu, Y., Deng, Z., Lai, J., Zhang, Y., Yang, C., Yin, B., et al. (2009). Dual function of Arabidopsis ATAF1 in abiotic and biotic stress responses. Cell Res. 19 (11), 1279–1290. doi: 10.1038/cr.2009.108
You, J., Zong, W., Hu, H., Li, X., Xiao, J., Xiong, L. (2014). A STRESS-RESPONSIVE NAC1-regulated protein phosphatase gene rice protein phosphatase18 modulates drought and oxidative stress tolerance through abscisic acid-independent reactive oxygen species scavenging in rice. Plant Physiol. 166 (4), 2100–2114. doi: 10.1104/pp.114.251116
Yu, Y., Xuan, Y., Bian, X., Zhang, L., Pan, Z., Kou, M., et al. (2020). Overexpression of phosphatidylserine synthase IbPSS1 affords cellular na+ homeostasis and salt tolerance by activating plasma membrane Na+/H+ antiport activity in sweetpotato roots. Hortic. Res. 7, 131. doi: 10.1038/s41438-020-00358-1
Zhang, H., Mittal, N., Leamy, L. J., Barazani, O., Song, B. H. (2016). Back into the wild-apply untapped genetic diversity of wild relatives for crop improvement. Evol. Appl. 10 (1), 5–24. doi: 10.1111/eva.12434
Zhang, W. D., Wang, P., Bao, Z., Ma, Q., Duan, L. J., Bao, A. K., et al. (2017). SOS1, HKT1;5, and NHX1 synergistically modulate na+ homeostasis in the halophytic grass puccinellia tenuiflora. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.00576
Zhang, M., Zhang, H., Zheng, J. X., Mo, H., Xia, K. F., Jian, S. G. (2018). Functional identification of salt-Stress-Related genes using the FOX hunting system from Ipomoea pes-caprae. Int. J. Mol. Sci. 19 (11), 3446. doi: 10.3390/ijms19113446
Keywords: I. pes-caprae, NAC transcription factor, abiotic stress, expression analysis, salt-tolerance, sweetpotato
Citation: Su Y, Liu Y, Xiao S, Wang Y, Deng Y, Zhao L, Wang Y, Zhao D, Dai X, Zhou Z and Cao Q (2023) Isolation, characterization, and functional verification of salt stress response genes of NAC transcription factors in Ipomoea pes-caprae. Front. Plant Sci. 14:1119282. doi: 10.3389/fpls.2023.1119282
Received: 08 December 2022; Accepted: 19 January 2023;
Published: 01 February 2023.
Edited by:
Mingku Zhu, Jiangsu Normal University, ChinaReviewed by:
Shaozhen He, China Agricultural University, ChinaCopyright © 2023 Su, Liu, Xiao, Wang, Deng, Zhao, Wang, Zhao, Dai, Zhou and Cao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qinghe Cao, Y2FvcWluZ2hlQGphYXMuYWMuY24=
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.