
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 06 February 2023
Sec. Plant Pathogen Interactions
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1117788
This article is part of the Research TopicLegume Root DiseasesView all 17 articles
Chickpea fields in Saskatchewan, one of the three Canadian prairie provinces, have suffered from major health issues since 2019, but no definitive cause has been determined. Field surveys were conducted in Saskatchewan in 2020 and 2021 in order to develop a better understanding of root rot pathogens associated with chickpea. Root samples were analyzed for the presence of 11 potential chickpea root rot pathogens using end-point PCR. Fusarium redolens, F. solani and F. avenaceum were the most prevalent pathogen species detected in both survey years. The cause of Fusarium wilt in chickpea, F. oxysporum f. sp. ciceris, was not detected in either year, nor were Phytophthora spp. and Verticillium albo-atrum. Berkeleyomyces sp. was detected in one field in each year, and Verticillium dahliae was detected in several fields sampled in 2021. These two pathogens have not been reported previously on chickpea in Saskatchewan. The prevalence of Fusarium species obtained from 2021 root isolations was similar to that determined by molecular tests, with frequent isolation of F. redolens, F. oxysporum, F. avenaceum and F. solani. A series of indoor pathogenicity testing compared root disease severity caused by a selection of 16 isolates of six Fusarium species and single isolates of V. dahliae, Berkeleyomyces sp. and Macrophomina phaseolina. Results showed that select isolates of F. avenaceum were the most aggressive of the Fusarium isolates on chickpea. Despite relatively low inoculum density, a highly aggressive isolate of F. avenaceum caused severe stunting and more root rot symptoms than single isolates of V. dahliae, Berkeleyomyces sp. and M. phaseolina under the test conditions.
Chickpea (Cicer arietinum L.) is an important pulse crop grown in Saskatchewan, one of the three Canadian prairie provinces, accounting for 78% of Canadian chickpea production in 2021 (Government of Saskatchewan, 2021). The majority of chickpeas grown in Saskatchewan are the kabuli type, which have a thin, colorless seed coat, making them susceptible to attack by a variety of soil-borne pathogens. Seed treatment with fungicide, particularly to control damping off caused by Pythium spp., is a routine part of disease management programs in this region. In addition to Pythium and Rhizoctonia spp., several Fusarium spp. can cause economically damaging root rot to chickpea worldwide (Haware, 1998; Infantino et al., 2006; Chen et al., 2011). The intensification of pulse production on the prairies has resulted in increased prominence of root rots, including those caused by Fusarium spp. The wide-spread occurrence of Aphanomyces root rot in this region makes chickpea an attractive alternative to lentil and pea in crop rotations due to their high partial resistance (Moussart et al., 2008). However, it has been suspected that root rots caused by other pathogens have been increasing. To date, the spectrum of root-rot pathogens prevalent in the chickpea cropping system, particularly Fusarium spp. and their potential for causing significant disease, are unknown, while this has been well studied in pea and lentil during the last decade.
Fusarium avenaceum (Fr.) Sacc. and F. solani (Mart.) Sacc. (syn. Neocosmospora solani (Mart.) L. Lombard & Crous) are the predominant Fusarium species in the root rot complex attacking pea and lentil (Esmaeili Taheri et al., 2017, Chatterton et al., 2019). Both of these species are known to impact emergence and cause moderate to severe symptoms on chickpea roots (Kraft, 1969; Westerlund et al., 1974; Safarieskandari et al., 2021). Besides root rot, F. avenaceum also contributes to the development of Fusarium head blight in cereal crops (Tekauz et al., 2000; Xue et al., 2004, Tekauz et al., 2004). In a recent study, isolates of Fusarium redolens Wollenw., F. culmorum (Wm.G. Sm.) Sacc., F. sporotrichioides Sherb. (now Fusarium chlamydosporum Wollenw. & Reinking), F. oxysporum Schltdl. and F. equiseti (Corda) Sacc. obtained from diseased chickpea samples were all confirmed to be pathogenic on chickpea (Zhou et al., 2021). The most aggressive isolates on chickpea were of F. culmorum and F. chlamydosporum, but there were also isolates of these species with low aggressiveness. In addition to root rot, Fusarium wilt of chickpea, caused by F. oxysporum f. sp. ciceris Matuo & K. Sato, can cause devastating losses in many chickpea growing areas, including most of those found in Asia, Africa, southern Europe, and the Americas (Jiménez-Díaz et al., 2015; Jha et al., 2020). This pathogen has not been reported in Canada.
In addition to Fusarium spp., several other chickpea root pathogens have been reported around the globe. Berkeleyomyces basicola (Berk. & Broome) W.J. Nel, Z.W. de Beer, T.A. Duong & M.J. Wingf (formerly Thielaviopsis basicola Berk. & Broome) which causes black streak root rot, was reported from chickpea roots in eastern Washington in 1985 (Bowden et al., 1985). Macrophomina phaseolina (Tassi) Goid. (dry root rot) and Verticillium albo-atrum Reinke & Berthold (Verticillium wilt) have been reported in California (Erwin, 1958; Westerlund et al., 1974). These three pathogens have not been reported from chickpea grown in the North American Prairies. Verticillium wilt of canola caused by V. dahliae Kleb. was recently reported on the prairies (Hwang et al., 2017) but has not been observed in Canadian chickpea (Chen et al., 2011). Similarly, although Phytophthora medicaginis E.M. Hansen & D.P. Maxwell, (Phytophthora root rot) is a pathogen of alfalfa fields in North America, Phytophthora root rot is not common in alfalfa in Saskatchewan (Bill Biligetu, Crop Development Centre/Dept. of Plant Sciences, University of Saskatchewan, personal communication) and it has not been recorded from chickpea crops in the USA or Canada.
Commercial chickpea fields in Saskatchewan were surveyed in June and July of 2020 and 2021. The scope of the survey included 41 rural municipalities with 42 commercial chickpea fields and one research location with chickpea breeder plots in 2020. Rural municipalities where chickpea root rot symptoms were most prevalent in 2020 were chosen for sampling in 2021, which included 19 commercial chickpea fields in 14 rural municipalities. Above-ground disease symptoms were recorded for five plants at each of ten locations in each field in 2020, and for five plants each at five locations in each field in 2021. Disease scoring was performed according to a 1-5 qualitative scale adapted from Infantino et al. (2006), in which 1 = no symptoms, 2 = slight yellowing of lower leaves, 3 = yellowing of the lower leaves up to the 3rd or 4th node and some stunting, 4 = necrosis of at least half or more of the plant with some stunting, 5 = entire plant dead or nearly so. Roots were collected at five locations in each field and submitted to the University of Saskatchewan Pulse Crop Pathology Laboratory for further analysis. Due to laboratory access restrictions during the COVID-19 pandemic, roots submitted in 2020 were immediately frozen and not assessed for visual root symptoms. In 2021, root rot symptoms were rated on dry root samples using the 1-7 scale described by Safarieskandari et al. (2021): 1 = no symptoms, 2 = 0.1–0.2 cm, small reddish brown lesions at seed attachment area, 3 = coalescing of localized tap root lesions approximately 180° around the tap root with lesions from 0.5 to 1 cm, 4 = lesions extending and completely encircling the tap root (1–2 cm), 5 = increasingly discoloured and extended tap root lesions (2–4 cm), 6 = lesions encircling the tap root extending over 4 cm and 7 = tap root completely brown/black.
A total of 208 root samples collected in 2020 and 93 samples collected in 2021 were freeze-dried (FreeZone 6, Labconco Corp., Kansas City MO USA) and ground for DNA extraction. Grinding was performed using custom designed tubes (high strength polycarbonate, Metalshapes Manufacturing, Saskatoon) containing a 1.7 cm diameter stainless steel ball placed in a homogenizer (2010 Geno/GrinderTM, SPEX Sample Prep, Metuchen, NJ USA) at 1400 rpm for 5 min (2020) or at 1000 rpm for 2 min (2021). Ground tissue (approx. 10 mg) was transferred to microcentrifuge tubes along with a 0.6 cm diameter ceramic bead for a second grinding step (1400 rpm for 1 min). Extraction of DNA from ground tissue was conducted using a DNeasy Plant Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions, with elution volume reduced to 60 µL. Concentration and quality of DNA (260/280 nm and 260/230 nm ratios) were assessed using a NanoDrop spectrophotometer (Thermo Scientific, Waltham MA, USA), and DNA concentration was diluted to 20 ng µL-1.
For molecular detection of pathogens, primer sets specific to various root rot pathogens were selected based on their prior use in the scientific literature and successful amplification of DNA of their particular target. Cross-reaction of primers with other closely related pathogens of relevance to the project was evaluated to determine the possibility of false positive results. The primers chosen for pathogen detection and their respective positive controls are listed in Table 1. Of the 12 primer sets, five were originally designed with a central TaqMan probe, but were used as conventional primers without the probe. The IPC primer set, which detects ascomycete fungi (Kulik, 2011), was redesigned using Primer 3 Plus software (Untergasser et al., 2012) and renamed IPC9 (Table 1). This primer set combines part of the probe with the reverse primer and uses an upstream forward primer.
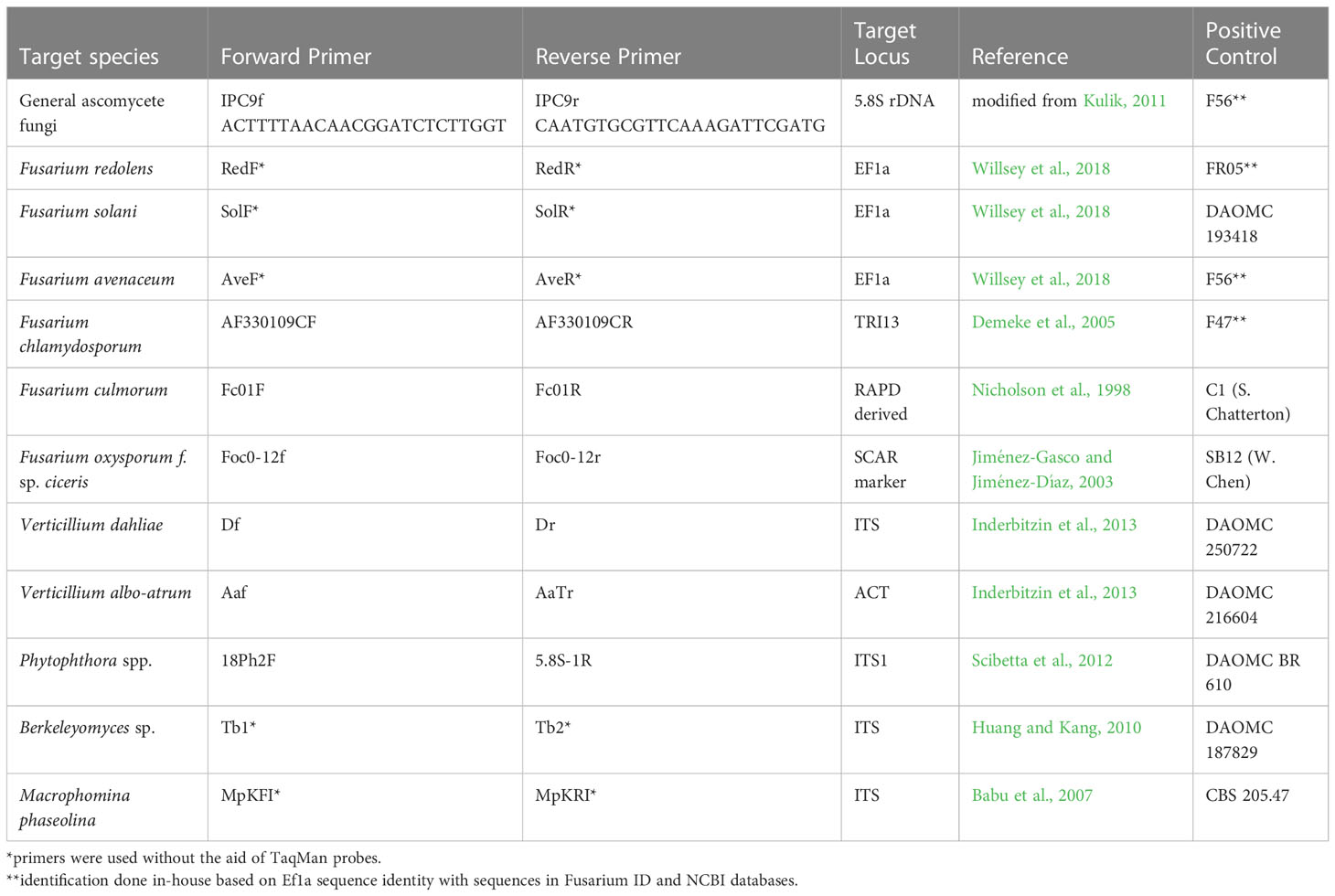
Table 1 Primers used for pathogen detection in DNA samples derived from chickpea roots collected in Saskatchewan in 2020 and 2021.
Detection of pathogens was accomplished through end-point PCR of 20 µL reactions consisting of 1X buffer, 2.5 mM MgCl2, 125 µM dNTP mixture, 0.1 µM of each primer, 1 U of Taq DNA Polymerase (Invitrogen recombinant), and 40 ng of genomic DNA. In order to avoid non-specific bands with primers designed for F. culmorum, the MgCl2 concentration was reduced to 2 mM. Cycler conditions were 95°C for 4 min, followed by 34 cycles of 95°C for 30 s, 60°C for 30 s and 72°C for 30 s, finished with a final extension at 72°C for 7 min. Amplicons were run on 1.5% agarose gel containing GelRed® (Biotium, Freemont CA USA) for 1 h at 120 v and visualized using a ChemiDoc (Bio-Rad, Hercules CA USA).
Detection of previously unreported pathogens by PCR (Berkeleyomyces basicola, Verticillium dahliae and Macrophomina phaseolina) was confirmed by sequencing the band produced by their respective species-specific primers (Table 1). DNA from excised bands was extracted using a monarch gel extraction kit (New England Biolabs, Ipswich MA USA) and samples were sent for sequencing (Eurofins Genomics, Louisville KY USA). Sequence data were used to construct a trimmed consensus contig (DNA Baser, Heracle Biosoft, Arges, Romania) which was compared with sequences in the NCBI Genbank database (Altschul et al., 1990).
Frozen tissues in 2020 and air-dried root tissues in 2021 were used for pathogen isolation. Root segments were surface sterilized for 2 min in 10% bleach solution, rinsed in sterile deionized water and placed on potato dextrose agar (PDA) medium for 7-10 days. Fungal colonies were selected based on colony morphology to exclude common saprophytes. Colonies were purified by transferring single germinated conidia to fresh medium. Culture plugs were stored in milk-glycerol solution at -80°C.
Mycelia were produced for DNA extraction by growing purified isolates in liquid medium (1 g NH4H2PO4 [Millipore Sigma], 0.2 g KCl [Fisher Chemical], 0.2 g MgSO4 x 7 H2O [Millipore Sigma], 10 g D-glucose [Fisher Chemical], 5 g yeast extract [Fisher Chemical], 0.01 g ZnSO4 x 7 H2O [Millipore Sigma], 0.005 g CuSO4 x 5 H2O [Millipore Sigma], 1 L distilled water) on a rotary shaker for 2-4 days, filtered to remove media and freeze dried. Freeze-dried tissues were pulverized inside microcentrifuge tubes containing a 0.6 cm diameter ceramic bead using a custom-made paint can shaker at full speed for 1 min. Extraction, quantification and dilution of DNA were conducted as described above.
Soil was collected from the research field from which Berkeleyomyces sp. had been detected by PCR in 2020. Desi chickpea seedlings were grown in this field soil and the root tissue was used for pathogen isolation as described above. Examination of endoconidia and chlamydospore morphology (Nel et al., 2018) was used to select Berkeleyomyces-like colonies. Identification of a Berkeleyomyces sp. isolate was confirmed with species-specific primers (Huang and Kang, 2010), and sequence data was generated and analyzed as described above.
Since morphological identification of Fusarium species is often unreliable, species-specific primers were also used to identify isolates of common Fusarium species by end-point PCR (Table 1). Reactions were processed as described above, except that 1.5 mM MgCl2 and 1 ng µL-1 of genomic DNA were used. For Fusarium isolates with inconclusive identification using selective primers, the TEF1 locus was sequenced after amplification with primers EF1 and EF2 (Geiser et al., 2004). Extraction of PCR amplicons, sequencing and data analysis were performed as described above. In addition to using the NCBI database, results were submitted to the online Fusarium identification tool (fusarium.mycobank.org, CBS-KNAW Fungal Biodiversity Centre).
A series of three pathogenicity tests were conducted for comparisons among isolates obtained from field surveys and those obtained from culture collections. All experiments were conducted in controlled environment chambers (Conviron model GR-48, Winnipeg, Canada) with 25°C daytime, 10°C night temperature and a 16 h photoperiod. Plants were grown in 10 cm diameter pots of peat-based medium (Sunshine mix #4, Sun Gro Horticulture, Agawam, MA USA or ProMix-BX-general purpose soil mix, Premier Tech Horticulture, Rivière-du-Loup, QC Canada). Cultures were grown on PDA for 5 to 7 days under incandescent lighting at room temperature. To prepare spore suspension of F. culmorum, PDA cultures were rinsed with deionized water and filtered through miracloth (Calbiochem, San Diego, CA USA). For spore production of all other Fusarium species, two plugs cut from the growing edge of the colony on PDA were added to a 250 mL flask containing 100 mL of carboxymethyl cellulose (CMC) medium and incubated under light for 4-5 days at 23°C on a shaker at 150 rpm (Foroud et al., 2012). After filtering through miracloth, conidia in liquid cultures were harvested by centrifugation for 5 min at 3400 rpm, followed by two washes with deionized water. Following re-suspension in deionized water, the resulting suspension was adjusted to 1 x 104 spores mL-1. This suspension was added to moist growth medium at a rate of 3 x 106 conidia per kg (300 mL of 1 x 104 conidia mL-1) prior to planting. Ten days after seeding, fresh conidia suspensions were prepared as described above and adjusted to 1 x 103 conidia mL-1. Aliquots of 5 mL were pipetted to the base of each seedling, henceforth referred to as drenching. Water was pipetted to the base of seedlings in non-inoculated controls. Seedling emergence was recorded 10 days after planting. Root rot severity was assessed 3 weeks after planting by assessing disease development on the hypocotyl. All experiments had four replicates arranged in a randomized complete block design and were conducted twice.
Experiment 1 included three chickpea survey isolates each of F. redolens (FR06, FR08, FR10) and F. solani (FSL01, FSL03, FSL04) as well as one isolate of F. avenaceum (Fav7). Two additional local isolates of F. avenaceum, Fav3 from pea and Fav 5 from lentil, which had previously been used for germplasm screening of various other pulse crops, were included. Seeds of CDC Orkney (kabuli) and CDC Sunset (desi) were surface sterilized in 10% bleach for 2 min and rinsed twice with deionized water prior to seeding in inoculated potting mix and further processed as described above. Disease severity was assessed on a 0-5 scale (modified from Coyne et al., 2019), where 0 indicated no disease symptoms, 1 indicated small hypocotyl lesions, 2 indicated lesions coalescing around epicotyls and hypocotyls, 3 indicated lesions starting to spread into the root system with some root tips infected, 4 indicated epicotyl, hypocotyl and root system almost completely infected and 5 indicated a completely infected root and dead plant.
Experiment 2 compared the most aggressive F. avenaceum (Fav3, Fav5), F. solani (FSL04) and F. redolens (FR06) isolates evaluated in the first experiment to an isolate of F. oxysporum f. sp. ciceris from Washington state (race 1 isolate SB12, W. Chen, USDA ARS, Dept. of Crop and Soil Sciences, and Plant Pathology, Washington State University) and 6 other local Fusarium isolates. These included four F. culmorum from chickpea (FC04, FC05, FC06, FC07), one F. inflexum from chickpea (Fi01) and one F. inflexum from lentil (Fi02). Inoculation of soil at planting and 10 days later was performed as described above. Seeds of kabuli chickpea cultivar CDC Leader were planted as described above, but without surface sterilization. Plant height was measured on 3-week-old plants prior to removing plants from pots for disease assessment. Disease assessment was performed using a 0-10 incremental scale (0 = no symptoms, 1 = 1 to 10% of root tissue affected, 2 = 11 to 20% of root tissues affected, and so on, to 10 = 91 to 100% of root tissues affected) to indicate the degree of damage to the hypocotyl region.
In Experiment 3, disease severity caused by a local, highly aggressive F. avenaceum (Fav5) was compared to disease caused by single isolates of V. dahliae (DAOMC 250722, from soil, Ontario), M. phaseolina (CBS 205.47, from common bean, Italy), and a local isolate of Berkeleyomyces sp. from chickpea (TB02). The kabuli cultivar CDC Leader was planted after seeds were surface sterilized as described above. For F. avenaceum, Berkeleyomyces sp. and M. phaseolina, soil incorporation of inoculum was followed by drenching 10 days after seeding as previously described, with method modifications to suit the biology of each pathogen, including inoculum preparation, concentration, and, for V. dahliae, delivery method. Inoculum preparation and concentrations for F. avenaceum followed the standard protocol described above. Cultures of Berkeleyomyces sp. were grown on PDA and incubated for 10 days at room temperature under continuous incandescent lighting. Chlamydospores and endoconidia (which were the majority of spores) were harvested by flooding the Petri dishes with sterile tap water, scraping with a sterile glass slide, and filtering the suspension through miracloth. Based on prior research, the spore suspension was adjusted to 1 x 104 spores mL-1 for soil incorporation and drenching (Tabachnik et al., 1979). Cultures of M. phaseolina were grown on oatmeal agar medium incubated at room temperature for 10 days under continuous incandescent lighting. Mycelia were harvested by flooding the Petri dishes with sterile distilled water and scraping the culture surface with a sterile glass slide. The mycelia were homogenized in a blender and adjusted to 3 x 104 mycelia fragments per mL for soil incorporation and for drenching (modified from Cota-Barreras et al., 2022). Cultures of V. dahliae were prepared on PDA and in CMC medium for spore production as described above. Based on prior research, spore suspensions were adjusted to 3 x 107 spores mL-1, and a seedling root soaking method was used for inoculation (Jiménez-Fernández et al., 2016). Seedlings were grown in medium horticultural vermiculite (Perlite Canada Inc., Lachine QC Canada) and removed from their pots 7 days after seeding. Seedlings were soaked in spore suspension for 15 min and transplanted into non-inoculated ProMix-BX-general purpose soil mix (Premier Tech Horticulture, Rivière-du-Loup, QC Canada). Spore suspension of 3 x 107 spores mL-1 was used for drenching 10 days after transplanting. Non-inoculated controls were maintained for each isolate (species) treatment in order to capture any effect of seedling dipping and transplanting or the soil incorporation method to help determine relative differences in disease severity. Plant height was measured prior to removing plants from pots for disease assessment. Disease assessment was performed using the 0-10 incremental scale to indicate the degree of damage to the hypocotyl region.
All analyses were conducted using SAS software version 9.4 (SAS Institute, Cary NC, USA). Pooling of experimental runs was performed after ensuring that there was no statistical effect of experimental run. Analysis of ordinal disease rating data from pathogenicity experiment 1 was performed following conversion to rank using the rank procedure. The mid-ranks (r), the default in the rank procedure, were then used in the mixed procedure to calculate the nonparametric test statistics and their significance levels (P-values). Genotype, treatment and genotype by treatment were considered fixed effects. The Wald-type statistic (WTS) was computed using the Chi-squared test. The anovaf option in the mixed procedure was used to generate the calculation of the ANOVA-type statistic (ATS), and the repeated statement was used to specify properties of the variances within experimental units (Shah and Madden, 2004).
For pathogenicity experiments 2 and 3 that had percentage disease data, normality of errors were evaluated with the Shapiro–Wilk test and homogeneity of variance with the Levene’s test before being modelled with the mixed model procedure. Replicate nested in experimental runs and experimental run were considered random effects whereas isolates were considered fixed effects. Heterogeneous variances were modeled with the repeated statement as required. Means were separated based on Fisher’s least significant difference at P = 0.05.
Spring moisture was adequate in the chickpea growing area of Saskatchewan in 2020, but moisture was limited in spring 2021. Summer conditions in both years were characterized by below average rainfall along with hot temperatures and drying winds. Mean disease severity assessments of above-ground symptoms (yellowing, stunting, necrosis) on a 1 to 5 rating scale ranged from 1.1 to 4.1 in 2020 and from 2.2 to 4.4 in 2021 when averaged for each field (Tables S1, S2). The median score for all fields assessed was 1.8 in 2020, indicating slight yellowing on above-ground plant parts in many fields at most assessment locations in each field. It is noteworthy that even in those fields with low average disease scores, 39 of the 43 fields had locations that were rated with scores of 3 and 4, indicating the possibility of serious root rot foci in the majority of fields. Fields in 15 rural municipalities had average ratings of 3 and higher indicating moderate to severe yellowing and stunting, and in four RMs, dead plants were observed (rating score of 5). Rural municipalities where the most severe root rot symptoms were observed in 2020 were selected for sampling in 2021. Of the 19 fields surveyed in 2021, 16 fields had maximum ratings of 4 or 5, indicating that severe symptoms and/or dead plants were observed in most fields. Heat and drought stress likely contributed to these symptoms, as may have other unknown factors.
Assessment of root rot severity in 2021 was performed on dry roots, which made fine features of lesions more difficult to observe. Mean severity of root rot symptoms ranged from 1.6, indicating only very small lesions, to 6.0, indicating extensive lesion development on the taproot. The overall mean root rot severity for the 19 fields was 2.9, which demonstrates that root rot damage was significant despite dry growing conditions. Severe root rot (rating of 4 to 7) was observed in root samples from seven of the 19 fields (Table S2).
Primer testing demonstrated that cross-reactions among the species involved in this study were only observed for F. culmorum primers Fc01F/R (Nicholson et al., 1998). This primer set resulted in cross reaction with several other species at 2.5 mM MgCl2, including V. albo-atrum, F. avenaceum, F. solani, F. redolens, and F. chlamydosporum. Reduction of MgCl2 to 2 mM eliminated most cross-reactions so that only a faint band persisted with F. redolens. Although this primer set has been cited extensively in the literature, it has only been used in the context of cereal pathology, and thus its specificity was not tested against a full spectrum of Fusarium species and other fungi.
In both survey years, Fusarium solani and F. redolens were the most prevalent pathogens detected in root samples, but F. solani was the most frequently detected pathogen in 2020 samples, whereas F. redolens was most frequent in 2021 samples. Fusarium avenaceum was also frequently detected in 2021 samples (73%), whereas it was only present in 33% of samples in 2020. Fusarium chlamydosporum was also detected in both years at relatively low frequency. Fusarium culmorum was not detected in any of the 2020 samples, and at a relatively low frequency (9%) in 2021 samples (Figure 1).
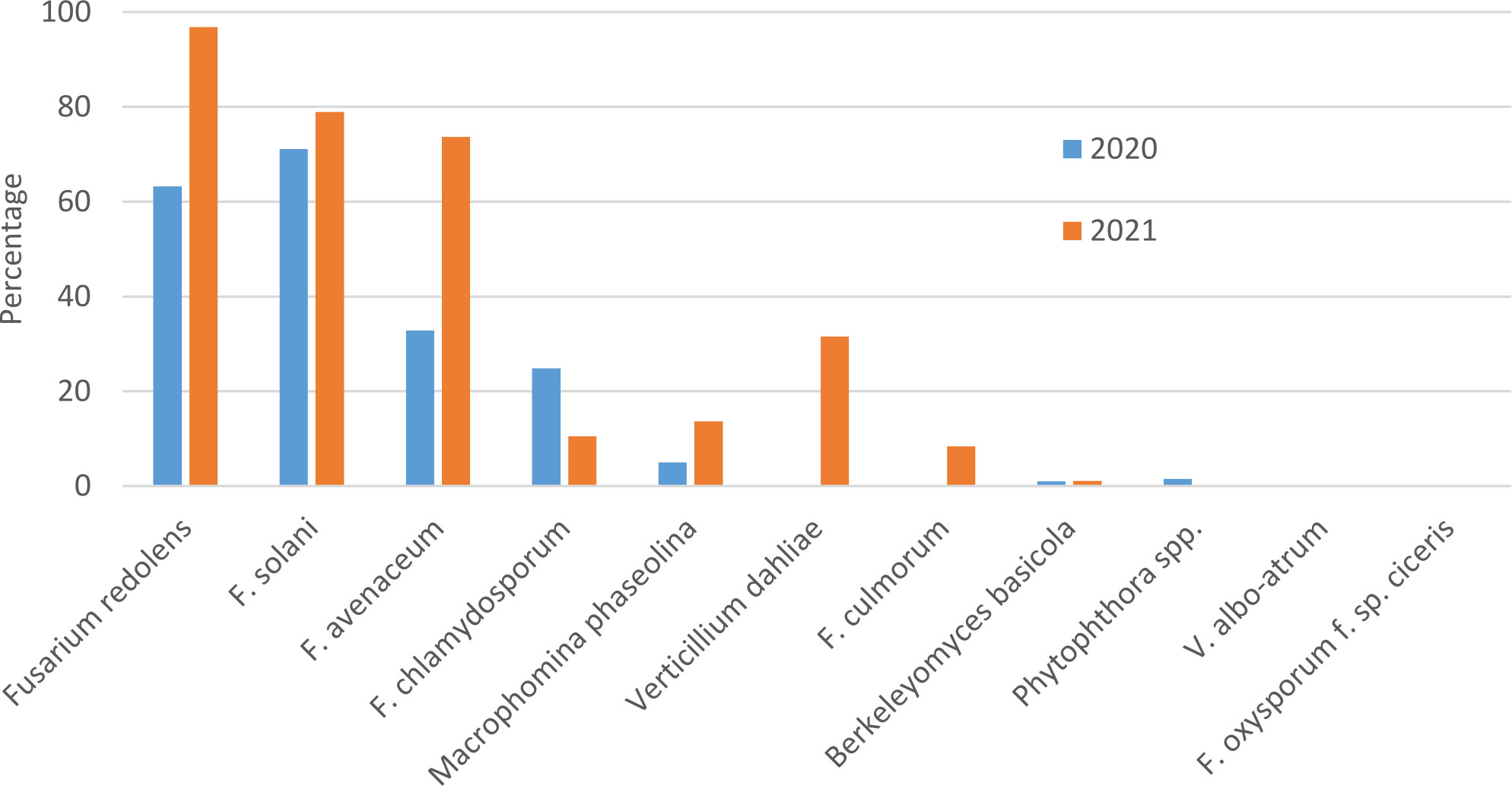
Figure 1 Percentage of chickpea root samples collected in Saskatchewan in 2020 and 2021 in which potential chickpea root pathogens were detected by end-point PCR.
Amplicons of the expected size were obtained with primers designed for detection of Macrophomina phaseolina in 5% of 2020 and 14% of 2021 samples, but attempts to sequence these bands were unsuccessful, suggesting that amplification was non-specific. Berkeleyomyces basicola was detected in two 2020 samples originating from the University of Saskatchewan research farm in Saskatoon, and from 1 sample from a commercial farm in 2021. Although not detected in 2020 samples, Verticillium dahliae was detected in 33% of 2021 samples (Figure 1). Bands amplified with the Berkeleyomyces sp. and V. dahliae-specific primers (Table 1) were sequenced to confirm species identity. A 312 bp consensus sequence generated from the Berkeleyomyces sp. band (OQ183437) had 100% coverage and 100% identity with NCBI sequences for a reference strain of B. basicola (MF952429) and the type strain of B. rouxiae (MF952412.1). Identity with both of these reference strains at an rDNA locus is not surprising, as these organisms were only recently split into two species, B. basicola and B. rouxiae W.J. Nel, Z.W. de Beer, T.A. Duong & M.J. Wingf. (Nel et al., 2018). Bands obtained from two root samples using the V. dahliae primers generated 498 bp (OQ183438) and 508 bp (OQ183439) sequences, which had 99.48% and 97.22% identity with NCBI sequences for the type specimen of V. dahliae (NR_126124.1) with 77% coverage. Higher coverage (99%) was observed for V. dahliae accession HE972025.1, with 99.6% identity for the 498 bp contig and 98.02% identity for the 508 bp contig. Both Berkeleyomyces sp. and V. dahliae have not been reported previously from chickpea in Saskatchewan.
Fusarium oxysporum f. sp. ciceris and V. albo-atrum were not detected in any samples. Detection of members of the Phytophthora genus were rare, with 1% of samples in 2020 but zero in 2021 (Figure 1).
Pathogen isolation from chickpea root tissues resulted in the purification of 7 Fusarium spp. isolates in 2020, and 52 Fusarium spp. isolates in 2021. Of the 59 Fusarium spp. isolates, 58 were identified using a combination of species-specific primers and sequencing at the Ef1a locus. Sequencing was performed for 20 Fusarium spp. isolates, including all putative F. oxysporum (OQ181356 to OQ181375). Seventeen isolates were Fusarium redolens, 13 F. oxysporum, ten F. avenaceum, seven F. solani, five F. culmorum, three F. caucasicum Letov, one F. incarnatum-equiseti complex, one F. acuminatum or F. tricinctum complex and one F. toxicum L. Lombard & J.W. Xia. The identity of one isolate remained undetermined. No F. oxysporum f. sp. ciceris isolates were obtained.
One Berkeleyomyces isolate was obtained from chickpea seedlings grown in field soil collected from a research site in 2020. The amplicon obtained for DNA of this isolate using species-specific primers (Huang and Kang, 2010) was of expected size and matched the amplicon size obtained with a culture collection isolate (DAOMC 187829). A 318 bp consensus sequence generated from the band had 100% coverage and 100% identity with NCBI sequences for a reference strain of B. basicola (MF952429.1) and the type strain of B. rouxiae (MF952412.1). Due changes in fungal taxonomy that occurred after the publication of the primer set and inability to resolve species based on the ITS sequence, we refer to this isolate as Berkeleyomyces sp.
In growth chamber tests comparing three isolates each of F. redolens, F. avenaceum and F. solani (Experiment 1), chickpea cultivars and isolates both had significant effects on root rot severity (P ≤ 0.014), but their interaction was not significant (P = 0.18). Disease severity was highest for two isolates of F. avenaceum (Fav 3, Fav5) on both the desi and kabuli cultivar tested. The remaining isolates caused only limited disease, with mean ratings of less than 1 on both cultivars (Figure 2). The two most aggressive isolates of F. avenaceum (Fav 3, Fav5) originating from pea and lentil caused a 16 to 22% reduction in emergence of the kabuli cultivar (data not shown). These isolates, as well as F. redolens (FR06) and F. solani (FSL04), were chosen to be included in Experiment 2.
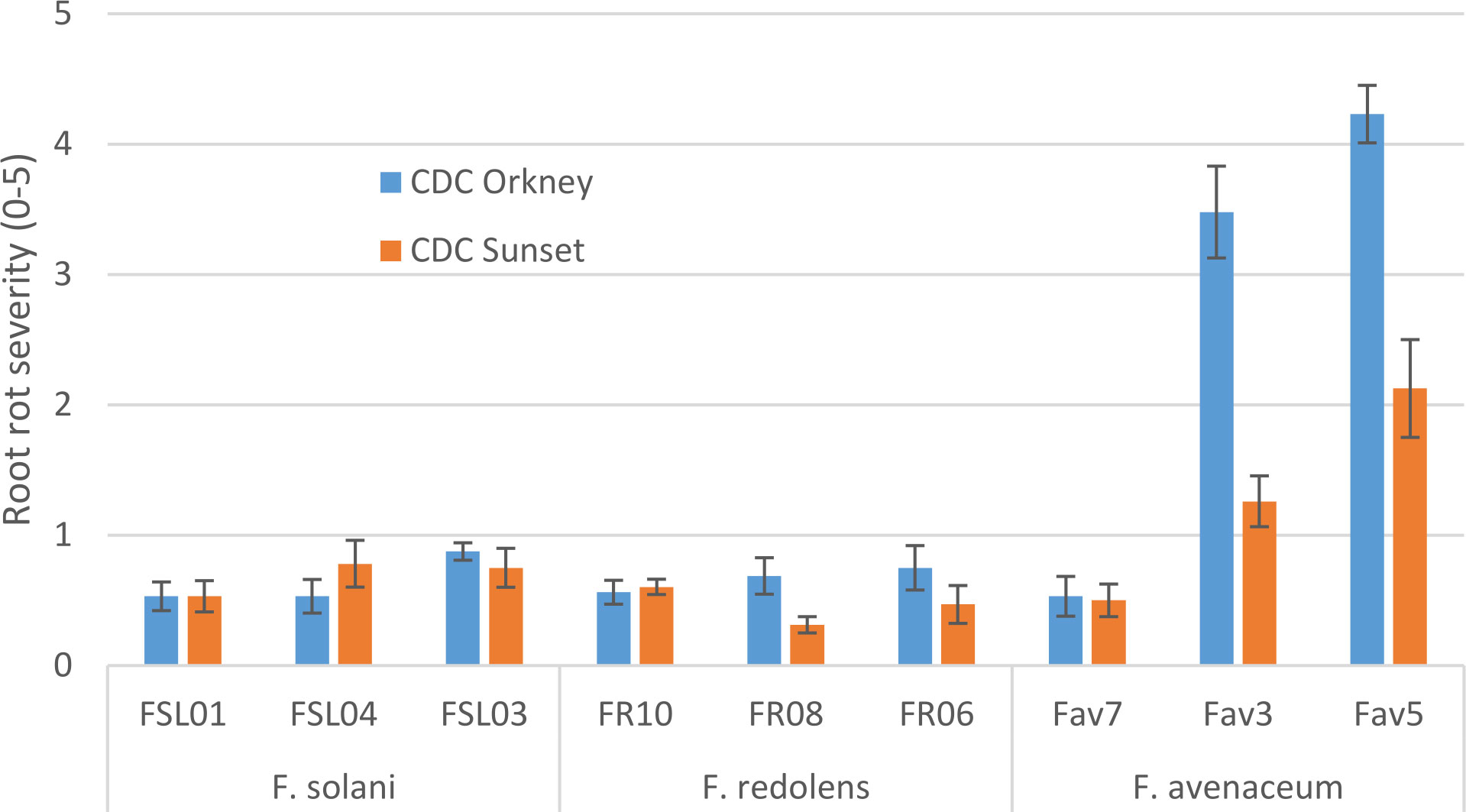
Figure 2 Root rot severity (0-5 scale) caused by three isolates each of three Fusarium species on 3-week-old plants of CDC Orkney kabuli chickpea and CDC Sunset desi chickpea under controlled conditions. Inoculum was incorporated into soil at planting and applied by soil drenching 10 days after planting.
Experiment 2 included nine selected isolates of six different Fusarium species. None of the isolates had a significant effect on emergence of kabuli cultivar CDC Leader (P = 0.075) but isolate significantly impacted plant height and root rot severity (P<0.0001 for both). Single isolates of F. solani, F. culmorum, F. redolens and both F. inflexum isolates caused low disease severity (<24%, Figure 3). Disease severity observed for the Fav3 isolate of F. avenaceum and an isolate of F. oxysporum f. sp. ciceris did not differ from that observed in the non-inoculated controls. This result was not unexpected for F. oxysporum f. sp. ciceris, given that this organism causes vascular wilting rather than root rot symptoms, but this isolate also failed to cause any height reduction of CDC Leader within the 21-day time frame of the experiment (Figure 4). Three of the F. culmorum isolates caused moderate to severe root rot symptoms, ranging from 57% to 82% severity. This was significantly less than the 94% disease severity caused by F. avenaceum isolate Fav5 (Figure 3). These same four isolates (F. culmorum FC04, FC05, FC06, and F. avenaceum Fav5) were the only ones that caused a significant height reduction relative to non-inoculated CDC Leader plants (Figure 4).
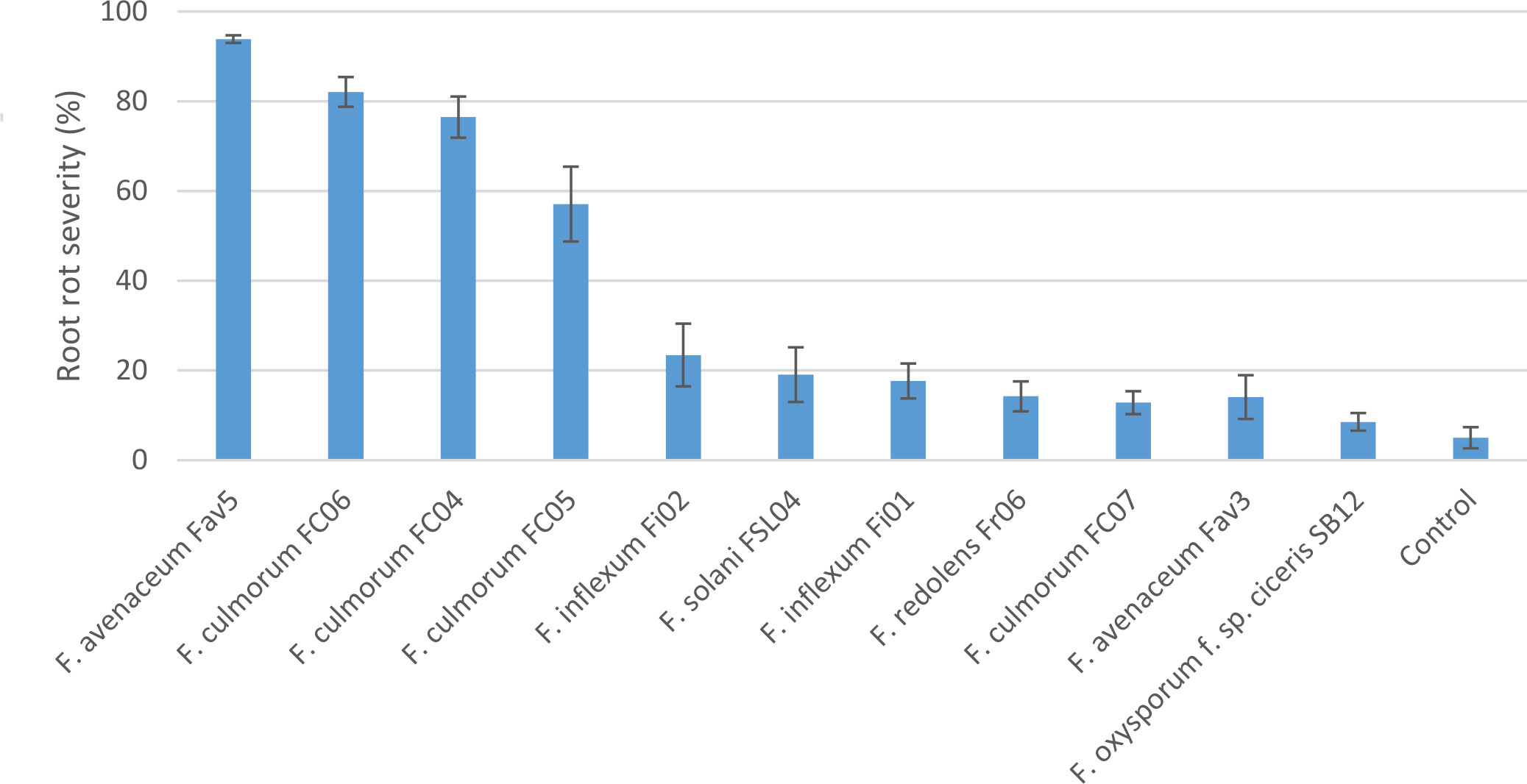
Figure 3 Root rot severity (%) caused by nine isolates of six Fusarium species on 3-week-old plants of CDC Leader kabuli chickpea under controlled conditions. Inoculum was incorporated into soil at planting and applied by soil drenching 10 days after planting.
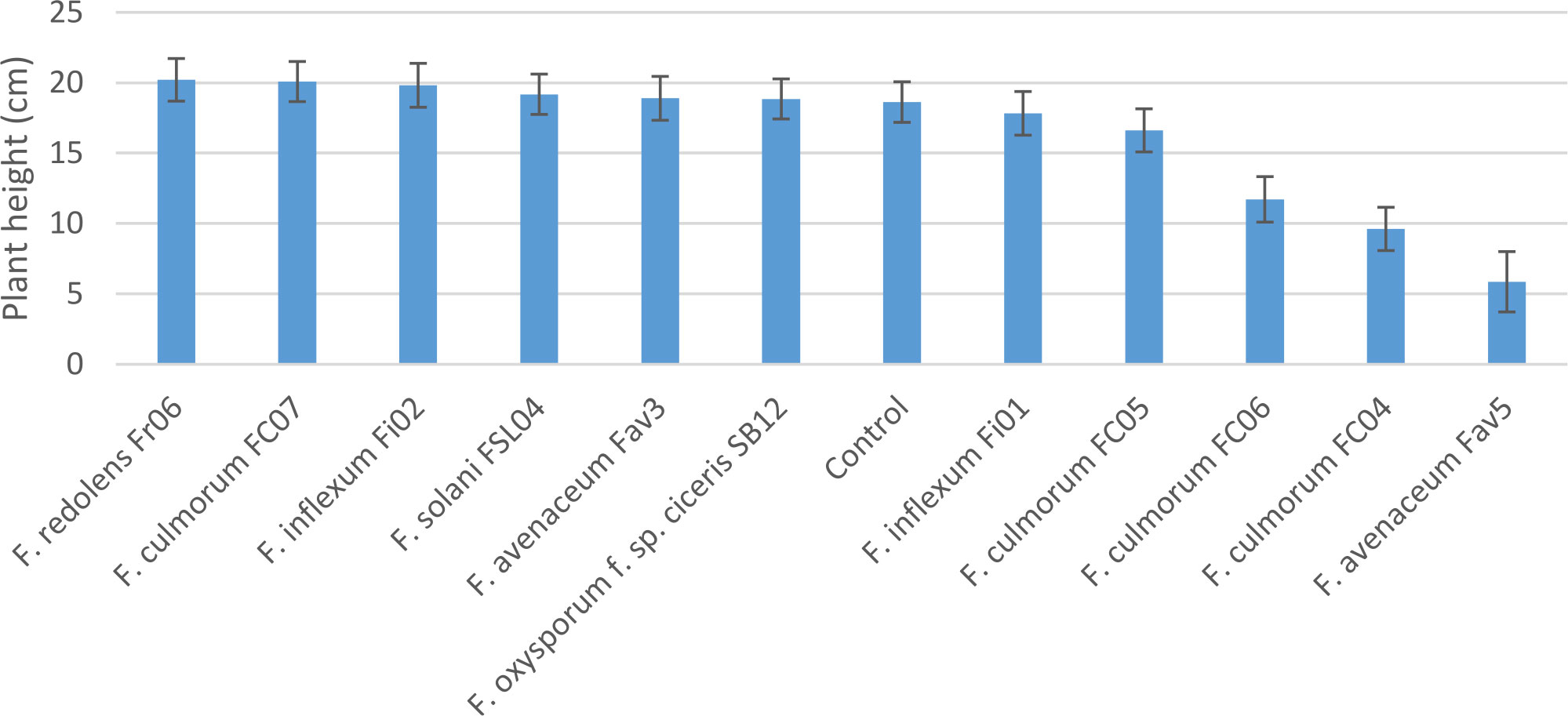
Figure 4 Height (cm) of 3-week-old CDC Leader kabuli chickpea inoculated with nine isolates of six Fusarium species under controlled conditions. Inoculum was incorporated into soil at planting and applied by soil drenching 10 days after planting.
Comparison of pathogenicity of single isolates of M. phaseolina, Berkeleyomyces sp., and V. dahliae to a local, highly aggressive isolate of F. avenaceum (Fav5) used in the two prior experiments in Experiment 3, revealed no significant effect of the isolates on emergence (P = 0.84), but isolate impacted height (P < 0.0001) and root rot severity (P < 0.0001). Plants in the non-inoculated controls accompanying soil-incorporated inoculum treatments showed no root rot, and non-inoculated plants that had been removed from their pots and dipped in deionized water showed only trace root discoloration (<2%, Figure 5). In the combined data analysis, the only isolate to significantly impact plant height was F. avenaceum Fav5, which resulted in 79% height reduction relative to non-inoculated control plants (data not shown). Verticillium dahliae significantly reduced plant height in one of the two experimental runs, but this effect was not statistically supported by means comparisons with combined data. Disease severity differed significantly for the single isolates of the four pathogens, with F. avenaceum causing the most damage to roots, followed by V. dahliae, Berkeleyomyces sp. and M. phaseolina under the test conditions (Figure 5). Infection by F. avenaceum resulted in totally collapsed brown hypocotyls and a small, brown tap root. Plants were severely stunted, dying or dead (Figure 6). Plants infected with V. dahliae developed brown lesions that encircled the hypocotyl. Disease development was more severe in experimental run 2, where V. dahliae infection affected the whole hypocotyl, epicotyl and tap root, and caused significant height reduction (P = 0.009). The impact on height observed in experimental run 2 was not statistically supported in pooled data. Symptoms caused by Berkeleyomyces sp. included black lesions on the hypocotyl that were generally not more than 1 cm in length and did not encircle the hypocotyl. Black lesions were also observed on hypocotyls of M. phaseolina-infected plants, but these were discrete lesions of limited size (Figure 6). Infection by Berkeleyomyces sp. and M. phaseolina did not impact plant height (data not shown).
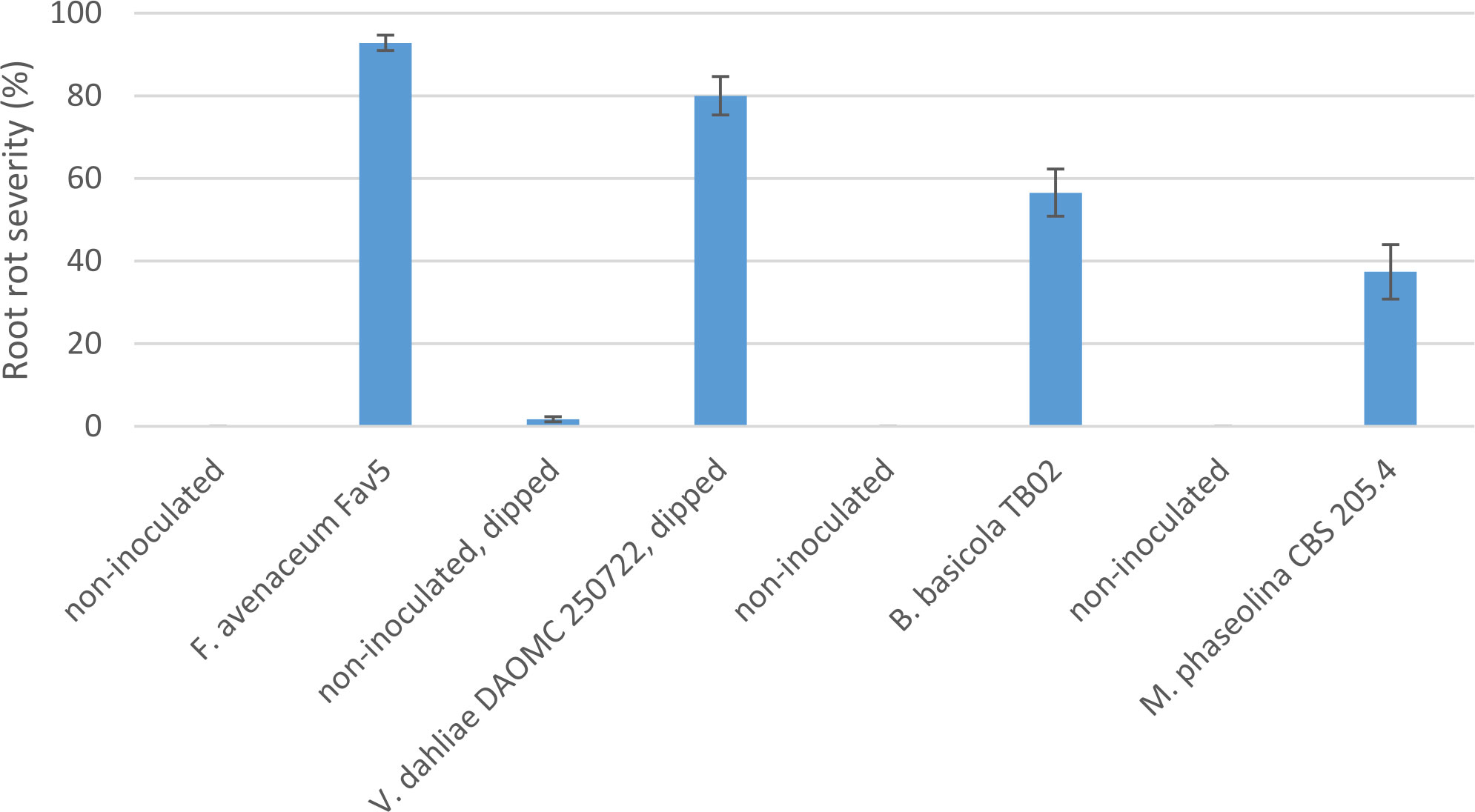
Figure 5 Root rot severity (%) caused by four pathogens on 3-week-old plants of CDC Leader kabuli chickpea under controlled conditions. Inoculum was incorporated into soil at planting or applied as a seedling dip, and applied by soil drenching 10 days after seeding or transplanting.

Figure 6 Root rot symptoms caused by four pathogens on 3-week-old plants CDC Leader kabuli chickpea under controlled conditions. Inoculum was incorporated into soil at planting or applied as a seedling dip, and applied by soil drenching 10 days after seeding or transplanting. From left to right: non-inoculated control, Macrophomina phaseolina, Berkeleyomyces basicola, Verticillium dahliae, Fusarium avenaceum.
The increasing prominence of root rot diseases in pulse crops is an important issue facing agricultural systems on the Canadian prairies. Along with Aphanomyces euteiches, Fusarium avenaceum and F. solani are major pathogens of pea and lentil in this region (Chatterton et al., 2019). Chickpea cultivars grown in this region are susceptible to Fusarium root rot caused by F. avenaceum (S. Banniza, unpublished). During surveys of chickpea fields during 2020 and 2021, the typical pattern of patchy root rot development was observed under the prevailing hot, dry conditions. The 2021 survey focused upon regions where wilting, discoloration and/or stunting of chickpea were observed in 2020, which lead to a higher proportion of symptomatic plants and/or roots observed in 2021 as compared to 2020. No obvious geographical localization of individual root pathogens was detected in the chickpea production area surveyed. In both years, F. redolens, F. avenaceum and F. solani DNA detections were the most prevalent, with a prominent increase in F. avenaceum and F. redolens detection frequency in 2021 over that in 2020. The use of M. phaseolina species-specific primers gave misleading results due to non-specific amplification. Despite using a standard technique implemented successfully by other researchers (Mayék-Pérez et al., 2001; Cota-Barreras et al., 2022), this pathogen was never observed in PDA isolation plates, suggesting that M. phaseolina is not a current threat to chickpea production in Saskatchewan. Previously unreported pathogens V. dahliae and Berkeleyomyces sp. were detected in chickpea root tissue using molecular techniques, but whereas Berkeleyomyces sp. was isolated from chickpea roots, V. dahliae was never observed in isolation plates. While it is possible that isolation of V. dahliae would have benefitted from the use of semi-selective medium, other researchers have successfully used PDA for isolation of V. dahliae (Jabnoun-Khiareddine et al., 2010; Ashraf et al., 2012). Despite molecular detection of V. dahliae in 12 field samples, the pathogen could not be reliably isolated using these standard techniques. It is possible that the use of dry root tissue for isolation also did not favor Verticillium recovery. The impact of V. dahliae infection of chickpea under field conditions warrants further investigation, particularly given the wide host range of this pathogen and its potential for impact on canola, which is widely grown on the Canadian prairies (Hwang et al., 2017).
Pathogenicity studies involving the three most prevalent Fusarium species detected in surveys showed that three F. redolens isolates and three F. solani isolates caused only very mild symptoms on chickpea roots. This is supported by a previous study using two F. redolens isolates from durum which caused low to moderate disease on pea, desi chickpea and durum, with one of these isolates causing more disease on pea than on desi chickpea or durum (Esmaeili Taheri et al., 2011). Of the three F. avenaceum isolates in the current study, two caused severe disease on CDC Orkney kabuli chickpea, whereas only one of these, an isolate obtained from lentil, caused severe disease on CDC Leader kabuli chickpea. The lone chickpea isolate chosen for pathogenicity testing caused only minor symptoms. Some variation in the aggressiveness of 19 F. avenaceum isolates was previously observed on pea and chickpea (Safarieskandari et al., 2021). When compared to isolates of F. solani, F. culmorum, F. redolens, F. oxysporum and F. acuminatum, six of the 19 isolates of F. avenaceum were aggressive enough to kill pea plants, and one isolate caused only moderate symptoms on pea. A mixture of three of these F. avenaceum isolates with high aggressiveness on pea were inoculated onto two kabuli and two desi cultivars. Reduced emergence of one desi and one kabuli cultivar following inoculation using a seed soaking method was observed, and CDC Leader, a kabuli cultivar also used in the current study, did not emerge. Moderate root rot severity was reported for the chickpea cultivars that successfully emerged. The large variation in aggressiveness among F. avenaceum isolates reported by Safarieskandari et al. (2021) and observed in the current study suggests that further work is needed to investigate whether host origin relates to isolate aggressiveness and host preference. Given the potential for F. avenaceum to impact pulse and cereal crops, further research on this topic could be used to improve disease management strategies.
Three of the four F. culmorum isolates tested under controlled conditions caused moderate to severe root rot severity and significant height reduction in inoculated chickpea plants. Research comparing Fusarium sp. isolates from pea showed that isolates of F. culmorum caused root rot symptoms equivalent to that caused by the most aggressive F. avenaceum isolates on pea seedlings (Safarieskandari et al., 2021). During chickpea root rot surveys conducted during two dry, hot seasons, F. culmorum was not detected in root tissues collected in 2020, and only at low frequency in 2021. Isolation of F. culmorum from root tissue also occurred at low frequency in 2021. Given this pathogen’s potential to cause severe root rot on chickpea and pea indoors where conditions are more moist and temperate, it is worthwhile continuing to learn about its role in the root rot complex of pulse and cereal crops.
Conducting disease survey work provided an opportunity to assess the potential threat of several previously unreported pathogens of chickpea in our region. Given that climate change threatens to modify growing conditions and may thereby shift the importance and composition of pathogen species and populations, creating an inventory of potential pathogens and assessing their relative pathogenicity is one small step toward system resiliency. No isolates of F. oxysporum f. sp. ciceris were recovered from diseased chickpea roots collected in disease surveys. An isolate of F. oxysporum f. sp. ciceris race 1 obtained from the US was included in growth chamber testing, where it failed to cause wilting or stunting of CDC Leader. As a pathogen with known impact on chickpea, these results may have been due to unsuitable test conditions, insufficient time for disease development, or resistance of CDC Leader to race 1 Fusarium wilt. Three additional root rot pathogens of chickpea with international significance, Berkeleyomyces sp., V. dahliae and M. phaseolina, were compared to a local, highly aggressive isolate of F. avenaceum. As these pathogens all have distinct biology and infection strategies, a direct, subjective comparison is somewhat difficult to attain. In addition, since these pathogens have not previously been reported in our region, we only had access to a single local isolate of Berkeleyomyces sp. and had to rely on single isolates of V. dahliae and M. phaseolina from culture collections. Despite these limitations, pathogenicity testing conducted using methods tailored to each pathogen provided some interesting insight. Although the inoculum density for the F. avenaceum isolate was the lowest of all four pathogens, this organism caused the most severe damage to chickpea seedlings under the test conditions. Within 20 days, chickpea inoculated with F. avenaceum Fav5 were dead or dying, and those inoculated with V. dahliae in one experimental run exhibited stunting. Chickpea seedlings inoculated with Berkeleyomyces sp. and M. phaseolina still appeared healthy above-ground but had hypocotyl lesions developing. Continued vigilance and assessment of the impact of emerging chickpea pathogens V. dahliae and Berkeleyomyces sp. under field conditions is recommended.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, OQ181356 OQ181357 OQ181358 OQ181359 OQ181360 OQ181361 OQ181362 OQ181363 OQ181364 OQ181365 OQ181366 OQ181367 OQ181368 OQ181369 OQ181370 OQ181371 OQ181372 OQ181373 OQ181374 OQ181375 OQ183436 OQ183437 OQ183438 OQ183439.
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication. Cheryl Armstrong-Cho and Nimllash Thangam Sivachandra Kumar have contributed equally to this work and share first authorship.
Western Grains Research Foundation and the Agricultural Development Fund of the Saskatchewan Ministry of Agriculture, ADF project 20190243.
Technical support for pathogenicity testing was provided by Preetpal Kaur, Madison Muzyka and Dawson George. Our thanks to the many staff at the Saskatchewan Ministry of Agriculture, Saskatchewan Crop Insurance Corporation (SCIC), and Saskatchewan Association of Rural Municipalities (SARM) who coordinated survey activities, conducted field disease assessments, and collected chickpea roots. Isolates of F. oxysporum f. sp. ciceris were obtained from Dr. Weidong Chen at USDA-ARS at Washington State University. An isolate of F. culmorum (C1) was obtained from Dr. Syama Chatterton at AAFC Lethbridge. Identification of F. inflexum from lentil was conducted by Collins Bugingo in Mary Burrows’ lab (Montana State University). The isolate of Verticillium dahliae was obtained from DAOMC, Ottawa and the M. phaseolina isolate was obtained from Westerdijk Fungal Biodiversity Institute, Utrecht, Netherlands. We are grateful for funding of this project from the Western Grains Research Foundation and the Agricultural Development Fund of the Saskatchewan Ministry of Agriculture.
We gratefully acknowledge that this work was conducted on Treaty 4 and Treaty 6 land and commit ourselves to the work of reconciliation in our community.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1117788/full#supplementary-material
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Ashraf, A., Rauf, A., Abbas, M. F., Rehman, R. (2012). Isolation and identification of Verticillium dahliae causing wilt on potato in Pakistan. Pak. J. Phytopathol. 24, 112–116.
Babu, B. K., Saxena, A. K., Srivastava, A. K., Arora, D. K. (2007). Identification and detection of Macrophomina phaseolina by using species-specific oligonucleotide primers and probe. Mycologia 99, 797–803.
Bowden, R. L., Wiese, M. V., Crock, J. E., Auld, D. L. (1985). Root rot of chickpeas and lentils caused by Thielaviopsis basicola. Plant Dis. 69, 1089–1091.
Chatterton, S., Harding, M. W., Bowness, R., Mclaren, D. L., Banniza, S., Gossen, B. D. (2019). Importance and causal agents of root rot on field pea and lentil on the Canadian prairies 2014–2017. Can. J. Plant Pathol. 41, 98–114. doi: 10.1080/07060661.2018.1547792
Chen, W., Sharma, H. C., Muehlbauer, F. J. (2011). Compendium of chickpea and lentil diseases and pests (St Paul, Minnesota, USA: The American Phytopathological Society).
Cota-Barreras, C. I., García-Estrada, R. S., Valdez-Torres, J. B., León-Félix, J., Valenzuela-Herrera, V., Tovar-Pedraza, J. M. (2022). Molecular detection, virulence, and mycelial compatibility of Macrophomina phaseolina isolates associated with chickpea wilt in sinaloa and Sonora, Mexico. Can. J. Plant Pathol 44, 849–857. doi: 10.1080/07060661.2022.2084642
Coyne, C. J., Porter, L. D., Boutet, G., et al. (2019). Confirmation of fusarium root rot resistance QTL Fsp-ps 2.1 of pea under controlled conditions. BMC Plant Biol. 19, 98.
Demeke, T., Clear, R. M., Patrick, S. K., Gaba, D. (2005). Species-specific PCR-based assays for the detection of Fusarium species and a comparison with the whole seed agar plate method and trichothecene analysis. Int. J. Food Micro. 103, 271–284. doi: 10.1016/j.ijfoodmicro.2004.12.026
Erwin, D. C. (1958). Verticillium wilt of Cicer arietinum in southern California. Plant Dis. Rep. 42, 1111.
Esmaeili Taheri, A., Hamel, C., Gan, Y., Vujanovic, V. (2011). First report of Fusarium redolens from Saskatchewan and its comparative pathogenicity. Can. J. Plant Pathol. 33, 559–564. doi: 10.1080/07060661.2011.620631
Esmaeili Taheri, A., Chatterton, S., Foroud, N. A., et al (2017). Identification and community dynamics of fungi associated with root, crown, and foot rot of field pea in western Canada. Eur. J. Plant Pathol. 147, 489–500.
Foroud, N. A., McCormick, S. P., MacMillan, T., Badea, A., Kendra, D. F., Ellis, B. E., et al. (2012). Greenhouse studies reveal increased aggressiveness of emergent Canadian Fusarium graminearum chemotypes in wheat. Plant Dis. 96, 1271–1279. doi: 10.1094/PDIS-10-11-0863-RE
Geiser, D. M., Jiménez-Gasco, M., Kang, S., Makalowska, I., Veeraraghavan, N., Ward, T. J., et al. (2004). FUSARIUM-ID v. 1.0: A DNA sequence database for identifying Fusarium. Eur. J. Plant Pathol. 110, 473–479. doi: 10.1023/B:EJPP.0000032386.75915.a0
Government of Saskatchewan (2021) 2021 specialty crop report. Available at: https://www.saskatchewan.ca/business/agriculture-natural-resources-and-industry/agribusiness-farmers-and-ranchers/market-and-trade-statistics/crops-statistics/specialty-crop-report (Accessed Oct 1 2022).
Haware, M. P. (1998). “Diseases of chickpea,” in The pathology of food and pasture legumes. Eds. Allen, D. J., Lenné, J. M. (UK: CAB International Wallingford), 473–516.
Huang, J., Kang, Z. (2010). Detection of Thielaviopsis basicola in soil with real-time quantitative PCR assays. Microbiol. Res. 165, 411–417. doi: 10.1016/j.micres.2009.09.001
Hwang, S. F., Strelkov, S. E., Ahmed, H. U., Zhou, Q., Fu, H., Fredua-Agyeman, R., et al. (2017). First report of Verticillium dahliae kleb. causing wilt symptoms in canola (Brassica napus l.) in north America. Can. J. Plant Pathol. 39, 514–526. doi: 10.1080/07060661.2017.1375996
Inderbitzin, P., Davis, R. M., Bostock, R. M., Subbarao, K. V. (2013). Identification and differentiation of Verticillium species and v. longisporum lineages by simplex and multiplex PCR assays. PloS One 8, e65990. doi: 10.1371/journal.pone.0065990
Infantino, A., Kharrat, M., Riccioni, L., Coyne, C. J., McPhee, K. E., Grünwald, N. J. (2006). Screening techniques and sources of resistance to root diseases in cool season food legumes. Euphytica 147, 201–221. doi: 10.1007/s10681-006-6963-z
Jabnoun-Khiareddine, H., Daami-Remadi, M., Barbara, D. J., El Mahjoub, M. (2010). Morphological variability within and among Verticillium species collected in Tunisia. Tunis. J. Plant Prot. 5, 19–38.
Jha, U. C., Abhishek, B., Shailesh, P., Kumar, P. S. (2020). Breeding, genetics, and genomics approaches for improving fusarium wilt resistance in major grain legumes. Front. Genet. 11, 1001. doi: 10.3389/fgene.2020.01001
Jiménez-Díaz, R. M., Castillo, P., Jiménez-Gasco, M. M., Landa, B. B., Navas-Cortés, J. A. (2015). Fusarium wilt of chickpeas: Biology, ecology and management. Crop Prot. 73, 16–27. doi: 10.1016/j.cropro.2015.02.023
Jiménez-Fernández, D., Trapero-Casas, J. L., Landa, B. B., Navas-Cortés, J. A., Bubici, G., Cirulli, M., et al. (2016). Characterization of resistance against the olive-defoliating Verticillium dahliae pathotype in selected clones of wild olive. Plant Pathol. 65, 1279–1291. doi: 10.1111/ppa.12516
Jiménez-Gasco, M. M., Jiménez-Díaz, R. M. (2003). Development of a specific polymerase chain reaction-based assay for the identification of Fusarium oxysporum f. sp. ciceris and its pathogenic races 0, 1A, 5, and 6. Phytopathology 93, 200–209. doi: 10.1094/PHYTO.2003.93.2.200
Kraft, J. M. (1969). Chickpea, a new host of Fusarium solani f. sp. pisi. Plant Dis. Rep. 53, 110–111.
Kulik, T. (2011). Development of TaqMan assays for 3ADON, 15ADON and NIV Fusarium genotypes based on Tri12 gene. Cereal Res. Commun. 39, 200–214. doi: 10.1556/CRC.39.2011.2.4
Mayék-Pérez, N., López-Castañeda, C., González-Chavira, M., Garcia-Espinosa, R., Acosta-Gallegos, J., de la Vega, O. M., et al. (2001). Variability of Mexican isolates of Macrophomina phaseolina based on pathogenesis and AFLP genotype. Physiol. Mol. Plant Pathol. 59, 257–264. doi: 10.1006/pmpp.2001.0361
Moussart, A., Even, M. N., Tivoli, B. (2008). Reaction of genotypes from several species of grain and forage legumes to infection with a French pea isolate of the oomycete Aphanomyces euteiches. eur. J. Plant Pathol. 122, 321–333. doi: 10.1007/s10658-008-9297-y
Nel, W. J., Duong, T. A., Wingfield, B. D., Wingfield, M. J., de Beer, Z. W. (2018). A new genus and species for the globally important, multihost root pathogen Thielaviopsis basicola. Plant Pathol. 67, 871–882. doi: 10.1111/ppa.12803
Nicholson, P., Simpson, D. R., Weston, G., Rezanoor, H. N., Lees, A. K., Parry, D. W., et al. (1998). Detection and quantification of Fusarium culmorum and Fusarium graminearum in cereals using PCR assays. Physiol. Mol. Plant Pathol. 53, 17–37. doi: 10.1006/pmpp.1998.0170
Safarieskandari, S., Chatterton, S., Hall, L. M. (2021). Pathogenicity and host range of Fusarium species associated with pea root rot in Alberta, Canada. Can. J. Plant Pathol. 43, 162–171. doi: 10.1080/07060661.2020.1730442
Scibetta, S., Schena, L., Chimento, A., Cacciola, S. O., Cooke, D. E. (2012). A molecular method to assess Phytophthora diversity in environmental samples. J. Microbiol. Methods 88, 356–368. doi: 10.1016/j.mimet.2011.12.012
Shah, D. A., Madden, L. V. (2004). Nonparametric analysis of ordinal data in designed factorial experiments. Phytopathology 94, 33–43. doi: 10.1094/PHYTO.2004.94.1.33
Tabachnik, M., DeVay, J. E., Garber, R. H., Wakeman, R. J. (1979). Influence of soil inoculum concentrations on host range and disease reactions caused by isolates of Thielaviopsis basicola and comparison of soil assay methods. Phytopathology 69, 974–977. doi: 10.1094/Phyto-69-974
Tekauz, A., McCallum, B., Ames, N., Fetch, J. M. (2004). Fusarium head blight of oat - current status in western Canada. Can. J. Plant Pathol. 26, 473–479. doi: 10.1080/07060660409507167
Tekauz, A., McCallum, B., Gilbert, J. (2000). Review: Fusarium head blight of barley in western Canada. Can. J. Plant Pathol. 22, 9–16. doi: 10.1080/07060660009501156
Untergasser, A., Ioana Cutcutache, I., Koressaar, T., Ye, J., Faircloth, B. C., Remm, M., et al. (2012). Primer3–new capabilities and interfaces. Nucleic Acids Res. 40, e115. doi: 10.1093/nar/gks596
Westerlund, F. V., Campbell, R. N., Kimble, K. A. (1974). Fungal root rots and wilts of chickpea in California. Phytopathology 64, 432–436.
Willsey, T. L., Chatterton, S., Heynena, M., Erickson, A. (2018). Detection of interactions between the pea root rot pathogens Aphanomyces euteiches and fusarium spp. using a multiplex qPCR assay. Plant Pathol. 67, 1912–1923. doi: 10.1111/ppa.12895
Xue, A. G., Armstrong, K. C., Voldeng, H. D., Fedak, G., Babcock, C. (2004). Comparative aggressiveness of isolates of fusarium spp. causing head blight on wheat in Canada. Can. J. Plant Pathol. 26, 81–88. doi: 10.1080/07060660409507117
Keywords: disease survey, fusarium avenaceum, fusarium redolens, verticillium dahliae, berkeleyomyces (thielaviopsis) basicola
Citation: Armstrong-Cho C, Sivachandra Kumar NT, Kaur R and Banniza S (2023) The chickpea root rot complex in Saskatchewan, Canada- detection of emerging pathogens and their relative pathogenicity. Front. Plant Sci. 14:1117788. doi: 10.3389/fpls.2023.1117788
Received: 06 December 2022; Accepted: 23 January 2023;
Published: 06 February 2023.
Edited by:
Rachid Lahlali, Ecole Nationale d’Agriculture de Meknès, MoroccoReviewed by:
Malkhan Singh Gurjar, Indian Agricultural Research Institute (ICAR), IndiaCopyright © 2023 Armstrong-Cho, Sivachandra Kumar, Kaur and Banniza. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Cheryl Armstrong-Cho, Y2hlcnlsLmNob0B1c2Fzay5jYQ==
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.