- 1Graduate School of Green-Bio Science, Kyung Hee University, Yongin, Republic of Korea
- 2Department of Plant Bioscience, Pusan National University, Miryang, Republic of Korea
- 3Rice Research Institute, Guangdong Academy of Agricultural Sciences, Guangzhou, China
Sucrose controls various developmental and metabolic processes in plants. It also functions as a signaling molecule in the synthesis of carbohydrates, storage proteins, and anthocyanins, as well as in floral induction and defense response. We found that sucrose preferentially induced OsWRKY7, whereas other sugars (such as mannitol, glucose, fructose, galactose, and maltose) did not have the same effect. A hexokinase inhibitor mannoheptulose did not block the effect of sucrose, which is consequently thought to function directly. MG132 inhibited sucrose induction, suggesting that a repressor upstream of OsWRKY7 is degraded by the 26S proteasome pathway. The 3-kb promoter sequence of OsWRKY7 was preferentially induced by sucrose in the luciferase system. Knockout mutants of OsWRKY7 were more sensitive to the rice blast fungus Magnaporthe oryzae, whereas the overexpression of OsWRKY7 enhanced the resistance, indicating that this gene is a positive regulator in the plant defense against this pathogen. The luciferase activity driven by the OsPR10a promoter was induced by OsWRKY7 and this transcription factor bound to the promoter region of OsPR10a, suggesting that OsWRKY7 directly controls the expression of OsPR10a. We conclude that sucrose promotes the transcript level of OsWRKY7, thereby increasing the expression of OsPR10a for the defense response in rice.
1 Introduction
Sugars derived from the photosynthetic process are the basic raw materials for cell vitality and function as intermediate substances to build macromolecules such as nucleic acids and the cell wall (Plaxton, 1996; Wang et al., 2007). Sugars also act as signal molecules in various cellular processes, such as seed germination, seedling growth, storage organ development, flowering, and senescence (Li and Sheen, 2016; Sakr et al., 2018; Yoon et al., 2021). Among various sugars, glucose is widely accepted as a signaling molecule in plants, yeast, and mammals. However, studies on sucrose-specific signaling are scarce, because this sugar is readily metabolized to fructose and glucose in plants. It has been shown that a significant amount of glucose is converted to sucrose in detached tobacco leaves within 8 hours after glucose feeding (Morcuende et al., 1998). Similarly, nourishing detached spinach leaves with glucose rapidly increases their sucrose level (Krapp et al., 1991).
Nonetheless, several studies have also reported that the transcript levels of various genes are preferentially controlled by sucrose (Yoon et al., 2021). For example, the mRNA level of the sucrose transporter BvSUT1 is reduced by a high concentration of sucrose in sweet beet leaves (Nieberl et al., 2017). Sucrose also preferentially induces various genes that encode starch biosynthesis enzymes, such as UDP-glucose pyrophosphorylase, ADP-glucose pyrophosphorylase (AGPase), and granule-bound starch synthase (Müller-Röber et al., 1992; Ciereszko et al., 2001; Wang et al., 2001; Li et al., 2002).
Sucrose functions as a specific signaling molecule to control plant development stages (Yoon et al., 2021). Arabidopsis plants that overexpress the homeodomain–leucine zipper gene, ATH13, show the narrow leaf phenotype when they are grown on sucrose-containing medium (Hanson et al., 2001). Sucrose concentration in the phloem sap increases dramatically upon floral induction in various plant species, such as Sinapis alba, Xanthium strumarium, Lolium temulentum, and Arabidopsis thaliana (Bodson and Outlaw, 1985; Houssa et al., 1991; Corbesier et al., 1998). Exogenous application of sucrose promotes flowering in Brassica campestris, S. alba, and chrysanthemum (Friend et al., 1984; Sun et al., 2017).
Sucrose also functions as a signaling molecule to protect plants from pathogen invasion by modulating the production of peroxidase and pathogen-related proteins (Thibaud et al., 2004; Morkunas et al., 2005). The expression of the maize fungal-inducible PRms gene in transgenic tobacco and rice plants causes the accumulation of a high level of sucrose that provides protection against various pathogens, which suggests that this sugar may promote plant defense (Murillo et al., 2003; Gómez-Ariza et al., 2007). In line with this hypothesis, the exogenous application of sucrose was shown to induce the expression of several PR genes in tobacco and rice (Murillo et al., 2003; Gómez-Ariza et al., 2007). Similarly, sucrose was reported to induce the expression of genes PR-1, PR-2, and PR-5 in Arabidopsis (Thibaud et al., 2004). Although glucose also increases the PR-2 transcript level, the induction is weak and non-metabolizable glucose analogs (i.e., 3-O-methylglucose and 2-deoxyglucose) do not promote gene expression, suggesting that the induction is not hexokinase dependent.
Several WRKY transcription factors play a role in the defense response in various plant species, including Arabidopsis, rice, wheat, grape, pepper, and cotton (Jiang et al., 2016; Jimmy and Babu, 2019). In rice, several WRKY genes are rapidly induced or repressed upon infection with bacterial and fungal pathogens, such as Xanthomonas oryzae pv. oryzae (Xoo) and Magnaporthe oryzae, respectively (Ryu et al., 2006; Jimmy and Babu, 2015). Among these genes, more than 10 were shown to induce resistance against fungal blast (Chen and Wang, 2014). For example, OsWRKY22 appears to be involved in defense responses because the oswrky22 mutant lines are highly susceptible to M. oryzae and Blumeria graminis (Abbruscato et al., 2012). Another WRKY gene, OsWRKY30, which is inducible by salicylic acid (SA) and jasmonic acid (JA), also enhances resistance to M. oryzae, Rhizoctonia solani, and Xoo when it is overexpressed in transgenic rice plants (Peng et al., 2012). Tao et al. (2009) identified a pair of OsWRKY45 alleles that function as a positive factor during M. oryzae infections but play an opposite role in the defense against bacterial pathogens. Whereas OsWRKY45-1 knockout plants are resistant to Xoo and X. oryzae pv. oryzicola, OsWRKY45-2-suppressing plants show increased susceptibility to these pathogens. OsWRKY45 forms a heterodimer with OsWRKY62 that acts as a strong activator, while the OsWRKY62 homodimer acts as a repressor (Fukushima et al., 2016). A number of WRKY factors function negatively in defending the plants against M. oryzae. The transcriptional repressor OsWRKY13 binds to the promoter regions of both the OsWRKY45-1 and OsWRKY45-2 genes, thereby repressing the target gene expression (Xiao et al., 2013). Similarly, OsWRKY42, which functions downstream of WRKY45-2, negatively affects rice resistance by suppressing JA-dependent signaling (Cheng et al., 2015).
Sucrose induces several WRKY genes. For example, it was shown to increase the mRNA level of AtWRKY20, which activates the AGPase gene in Arabidopsis (Nagata et al., 2012). Similarly, sucrose addition to Arabidopsis leaves induces the ectopic expression of an endosperm-specific WRKY transcription factor, SUSIBA2 (Sun et al., 2003). However, the effects of other sugars have not been examined in either of the two above-mentioned studies. In the present study, we investigated the signal roles of sucrose in plant defense.
2 Materials and methods
2.1 Plant materials and growth conditions
The Oc suspension cell line that was established from seedling roots of Oryza sativa L. indica type accession C5928 (Baba et al., 1986) was cultured in 50 mL of R2S liquid medium (Ohira et al., 1973) in a 120-rpm shaking incubator at 28°C, as previously reported (Cho et al., 2016; Yoon et al., 2021). After 2 weeks of culture, 20 mL of the upper liquid layer was decanted, and 10 mL of the remaining cell suspension was transferred to a fresh flask containing 40 mL of R2S culture medium.
The OsWRKY7 mutant (line number 5A-00022) was identified from T-DNA tagging lines generated in Oryza sativa var. japonica, cultivar ‘Dongjin’ (Jeon et al., 2000; Jeong et al., 2002; An et al., 2005a; An et al., 2005b; Jeong et al., 2006). Homozygous mutant plants from the T2 progeny were identified by PCR using genomic DNA isolated from the blade of the second leaf of each plant. The genotyping primers were GTGTCGGGTGCGTATTTAAAAAC (F), GGGAATTGGCGCTAAATCTGC (R), and atccagactgaatgcccacag (NGUS2) (Supplementary Table 1). Null mutations of OsWRKY7 were obtained using the CRISPR/Cas9 method in ‘Donjin’ background. Homozygous mutants at T1 generation were detected through the subcloning and multiple sequencing of the mutated region using the genomic DNA extracted from the leaf blades of individual plants.
For overexpression of OsWRKY7, a full length of the gene was cloned by PCR using the specific primers (Supplementary Table 1) with HindIII and SacI restriction enzyme sites. The gene was subcloned between the maize ubiquitin (ZmUbi) promoter and HA tag in the binary vector pGA3428 (Kim et al., 2009). The resulting vector was used to generate transgenic rice plants overexpressing OsWRKY7-HA. Produced plants were verified by western blot analysis using protein extracts from the leaf blades after 24 hours of M. oryzae infection. All plants were grown in the paddy field or in a controlled growth room under a long day conditions (i.e., 14 hours of light at 28°C/10 hours of darkness at 23°C, humidity of approximately 50%).
2.2 RNA-sequencing and data analysis
Oc cells were grown for 10 days in R2S solution complemented with 3% glucose as a carbon source. They were divided into two equal parts; one was added with sucrose and the other with glucose, obtaining a final sugar concentration of 3% in two replications. Samples were collected immediately before the transfer to new media (0 h) and 4 hours after sugar addition (4 h). Total RNA was extracted using the QIAGEN RNA purification kit (Hilden, Germany), and the purity and concentration of RNA were estimated using a NanoDrop 2000 spectrophotometer (Thermo scientific). RNA-seq analysis was performed based on protocols previously reported in Peng et al. (2021). The gene ontology (GO) terms of the differentially expressed genes were analyzed using the Rice Oligo Array Database (ricephylogenomics-khu.org). All the mapped genes were downloaded and filtered based on the following criteria: hyper p-value < 0.05 and query numbers > 10. Additional significance of the GO terms was denoted by a fold enrichment value > 1.5, which was obtained by dividing the query number by the query expectation value (Hong et al., 2020; Kim et al., 2021). The biotic stress category was obtained via the MapMan analysis toolkit 3.5.1 R2 (Thimm et al., 2004).
2.3 RNA isolation and the expression analysis
Total RNA was extracted from the leaves, roots, and Oc cells using RNAiso Plus (Takara, Japan). After combining 3 µg of RNA with 10 ng of oligo (dT), the first-strand cDNA was synthesized using 100 units of Moloney murine leukemia virus (M-MLV) reverse transcriptase (Promega, Madison, WI, USA), 100 units of RNasin (Promega), and 2.5 mM deoxyribonucleotide triphosphates. The gene expression levels were analyzed by quantitative RT-PCR using SYBR Green I, Prime Q-Matermix (2x) (GENET Bio, Daejeon, Korea) in a Rotor-Gene Q system (Qiagen, Hilden, Germany). Rice Actin1 (OsActin1) was used as an endogenous control to normalize the expression level of the respective genes. All experiments were repeated at least three times, and the relative transcript levels were calculated using the ΔΔCt method (Schmittgen and Livak, 2008; Yoon et al., 2022). The primers used in this study are listed in Supplementary Table 1.
2.4 Generation of CRISPR/Cas9 mutants
After selecting a potential target sequence with the CRISPR web tool, the RNA scaffold structure was analyzed using the RNAfold web server (univie.ac.at), and off-target sites were checked with the web tools (http://crispr.dbcls.jp; Naito et al., 2015 and CRISPOR (tefor.net); Concordet and Haeussler, 2018). A potential target sequence (5’-ATGTCATCGTACTTCTCCCA-3’) within the first exon of OsWRKY7 was cloned into the CRISPR vector pRGEB32 through the BsaI restriction site. The construct was transformed into the Agrobacterium tumefaciens strain LBA4404 using the freeze–thaw method (An, 1987). Transgenic rice plants were obtained via the Agrobacterium-mediated co-cultivation method (Lee et al., 1999).
2.5 Plasmid construction for the reporter assay
The 3.081-kb promoter region (−3098 to −18 bp from the ATG start codon) of OsWRKY7 from cultivar ‘Nipponbare’ was cloned into the HindIII-digested pGA3452 vector (Kim et al., 2009) upstream of the luciferase (LUC) coding region using the 5X In-Fusion HD enzyme Premix (Takara, Japan), which generated the pOsWRKY7-LUC reporter vector. The 2.856-kb promoter portion (−2856 to −1 bp from the start ATG codon) of OsPR10a was inserted into pGA3452, which resulted in pOsPR10a-LUC. The ZmUbi-GUS (Cho et al., 2009) construct was used as the internal control. For effector constructs, the coding regions of OsWRKY7, OsWRKY28, and OsWRKY10 were amplified by PCR and were inserted between the ZmUbi promoter and 6X Myc of pGA3697 (Kim et al., 2009).
2.6 Transient assay
Protoplasts were prepared from Oc cells based on methods described in Cho et al. (2016) and He et al. (2016). In brief, Oc cells were incubated in an enzyme solution (2% cellulose RS, 1% macerozyme, 0.1% CaCl2, 0.4 M mannitol, 0.1% MES, pH 5.7) for 4 hours at 28°C in the dark. After incubation, they were added with an equal volume of KMC solution (117 mM potassium chloride, 82 mM magnesium chloride, and 85 mM calcium chloride). The number of protoplasts was quantified with a hemocytometer to ensure that a cell density of 2 × 106 cells mL−1 was reached. The isolated protoplasts were transformed with 10 µg of each plasmid DNA using the PEG method (Yoo et al., 2007; Cho et al., 2022). ZmUbi : GUS construct (2 µg) was co-transfected as an internal control (Cho et al., 2009). The GUS activity was used to normalize the LUC activity.
2.7 Chromatin immunoprecipitation assay
The chromatin immunoprecipitation (ChIP) assay was performed using the protocols described in Haring et al. (2007) and Yoon et al. (2022) with some modifications. Firstly, the fresh leaves from the OsWRKY7-HA transgenic plants were collected and the proteins and chromatin were cross-linked using 3% formaldehyde. After the isolation of nuclei, DNA was broken into fragments of approximately 200–800 bp in length by sonication. Agarose beads of G and A proteins (Milipore; http://www.emdmilipore.com) were used to preclear the sheared chromatin for 1 hour at 4°C. Anti-HA monoclonal antibody (C29F4, https://www.cellsignal.com) was used for immunoprecipitation together with the precleared agarose beads for 4 hours at 4°C. The washing and reverse cross-linking steps described in Haring et al. (2007) were followed. The enriched chromatin was analyzed by qRT-PCR using the primers listed in Supplementary Table 1. The obtained data were normalized using the fold enrichment method (Haring et al., 2007; Yoon et al., 2022).
2.8 Electrophoresis mobility shift assay (EMSA)
The oligonucleotide (-808 to -831 from the start ATG of OsPR10a) was end labeled with biotin (Microgen, Korea). To produce MBP-tagged OsWRKY7 protein, the coding sequence of OsWRKY7 was amplified by PCR and cloned into the pMAL-c2X vector (Addgene plasmid # 75286; http://n2t.net/addgene:75286; RRID : Addgene_75286; walker et al., 2010) through the EcoR1 and Sal1 enzyme sites. The MBP-OsWRKY7 protein were extracted using amylose resin (E8021S, New England, Biolabs Inc.) The MBP-OsWRKY7 protein (1 µg) and biotin labeled ds-DNA fragment (750 fmol) were incubated in the reagent (KIT 0020148, Thermo scientific) containing 1× binding buffer, 10% glycerol, 5 mM MgCl2, 0.5 µg poly (dI-dC), and 0.05% NP-40 in a final volume of 20 µL for 30 min at 25°C. After electrophoresis in 6% polyacrylamide gel, the DNA-protein complexes were transferred to membrane. The procedure for detection of immobilized nucleic acids was according to the protocol provided by the manufacture.
2.9 Pathogen infection
The Magnaporthe oryzae PO6-6 strain was cultured on an agar medium containing 8% V8 juice (Cambell’s Soup Company, USA) for 2 weeks under fluorescent light. The resulting spores were collected and diluted in distilled water to obtain the concentration of 5 × 106 mL−1 for spot inoculation. A solution containing 2 × 105 spores per mL was sprayed onto the fully expanded healthy leaves of 6-week-old plants. Leaf samples were harvested and photographed to calculate the size of the lesions developed in the susceptible plants (Vo et al., 2019).
3 Results
3.1 Identification of sucrose-induced genes
To identify genes that preferentially responded to exogenously provided sucrose, RNA-seq analysis was performed using Oc cell lines that had been maintained in suspension culture for a long period (Baba et al., 1986). Because the cells were grown in a culture medium containing sucrose, the carbon source was replaced with glucose to deplete sucrose when the cells were sub-cultured. Ten days after subculture, sucrose was added to a culture flask to obtain a final concentration of 3%. As a control, glucose was added to another culture flask to reach the same final concentration. Four hours after adding the sugars, cells were harvested and mRNAs were isolated to construct cDNA libraries. RNA-seq analyses of the libraries showed that the transcript levels of 1,336 genes were at least doubled by the sucrose addition during the 4-h culture period (Supplementary Figure 1A). Among these genes, 871 were also induced by glucose, leaving 465 that were sucrose preferential. The analyses also revealed that 1,148 genes were preferentially suppressed by sucrose (Supplementary Figure 1B). To validate the RNA-seq data, 12 upregulated and 12 downregulated genes were selected for qRT-PCR analysis, which confirmed that eight and 10 genes belonging to the two groups, respectively, were sucrose responsive (Supplementary Table 2). GO analysis of the 465 genes preferentially induced by sucrose showed that the biological terms dominated by sucrose were protein amino acid phosphorylation, defense response, and transport (Supplementary Figure 1C).
3.2 Sucrose preferentially induces the transcription initiation of OsWRKY7
Mapman analysis of all the sucrose up-regulated genes from the RNA-seq analysis showed that three groups of transcription factors were enriched in biotic stress (Supplementary Figure 2). Because WRKY transcription factors are involved in defense response against pathogens, we selected all three WRKY genes (OsWRKY7, OsWRKY26, and OsWRKY108) that were sucrose inducible. qRT-PCR analysis showed that the transcript level of OsWRKY7 was increased by sucrose, reaching the maximum level at 2 hours after sucrose addition, and then rapidly declining to the basal level at 6 hours (Figure 1A). Glucose also induced the transcript level, but not significantly. Mannitol did not increase gene expression during the 8-h incubation time, which indicated that the expression of OsWRKY7 was preferentially induced by sucrose. The level of OsWRKY26 transcript started to increase at 2 hours after sucrose addition, reached the maximum at 4 hours, and then declined (Figure 1B). The transcript level was also increased by glucose addition. However, the gene was not induced at 2 hours after glucose addition, but its transcript was increased to approximately half of the sucrose-induced level at 4 hours and continuously increased at 6 and 8 h. The transcript level was not changed by mannitol. The above observations suggest that OsWRKY26 responded to both sucrose and glucose, but the gene was more rapidly induced by sucrose. Another WRKY gene, OsWRKY108, was also induced by sucrose. Compared to the other two genes discussed above, OsWRKY108 responded more rapidly to sucrose, reaching the maximum level at 0.5 hours after sucrose addition, and declining to the basal level at 2 hours (Figure 1C). This gene was weakly induced by glucose and did not respond to mannitol. The pathogen marker gene OsPR10a was also preferentially induced by sucrose, but not by glucose or mannitol (Figure 1D).
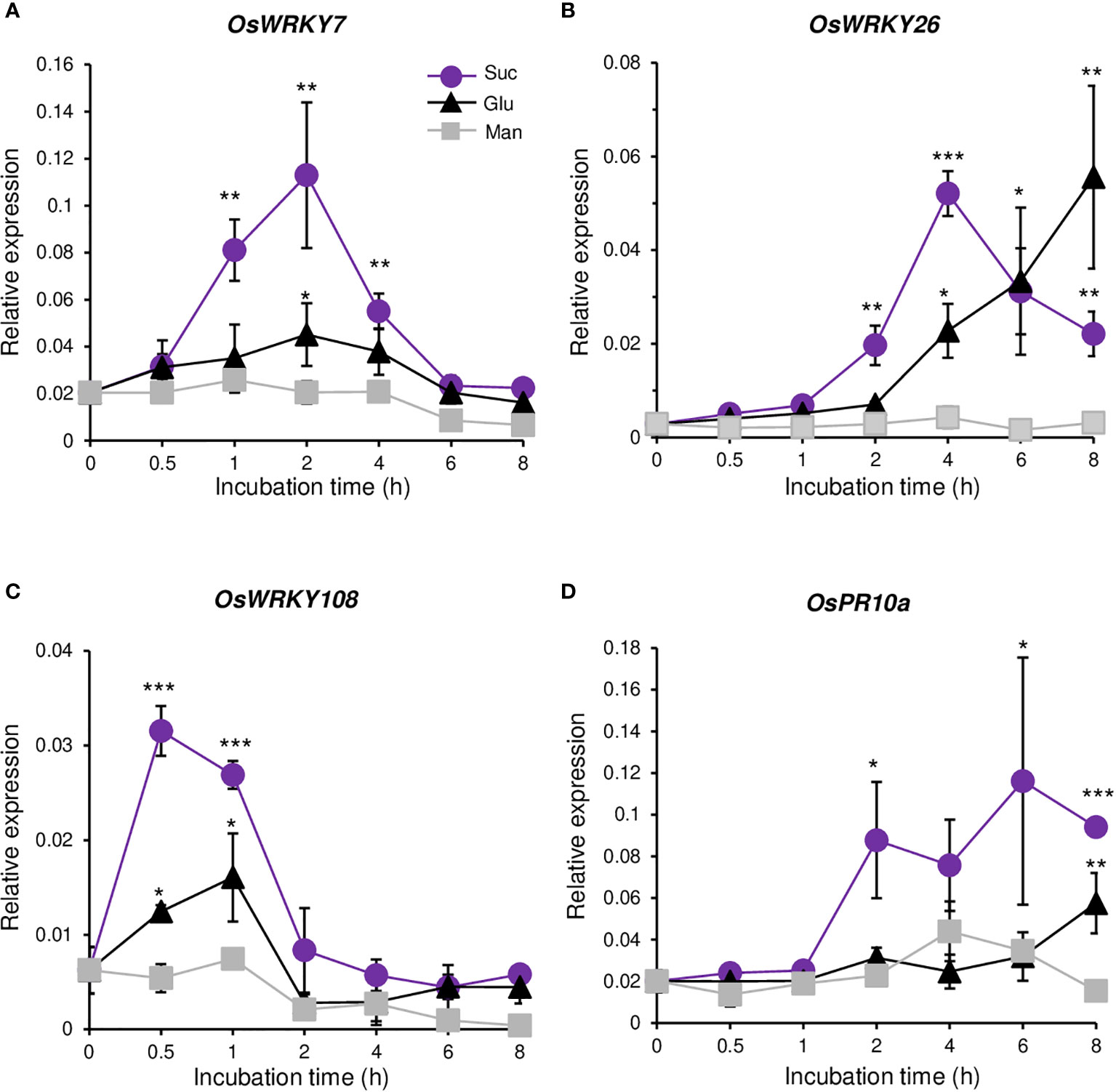
Figure 1 Response to sugars. Quantitative RT-PCR analyses of OsWRKY7 (A), OsWRKY26 (B), OsWRKY108 (C), and OsPR10a (D) in Oc cells after addition of 3% sucrose (Suc), 3% glucose (Glu) or 3% mannitol (Man). The gene expression levels are relative to those of OsActin1. Error bars indicate standard deviations; n = 3 or more. Statistical significance is indicated by *(P < 0.05) **(P < 0.01) and ***(P < 0.001).
Because OsWRKY7 was specifically and transiently induced by sucrose, further tests were conducted to determine whether this gene was induced by other sugars. In addition to glucose, other monosaccharides such as galactose and fructose, as well as glucose and fructose added at the same time, did not induce gene expression above the control level during the 2-h incubation period (Figure 2A). Also, gene expression was not stimulated by maltose, a disaccharide that consists of two glucose molecules. The results suggest that OsWRKY7 is specifically induced by sucrose. Lowering sucrose concentration to 2%, 1%, and 0.5% did not influence the rate and level of OsWRKY7 induction (Figure 2B), however, further reductions to concentrations of 0.25% and 0.1% slightly reduced the sucrose inducibility.
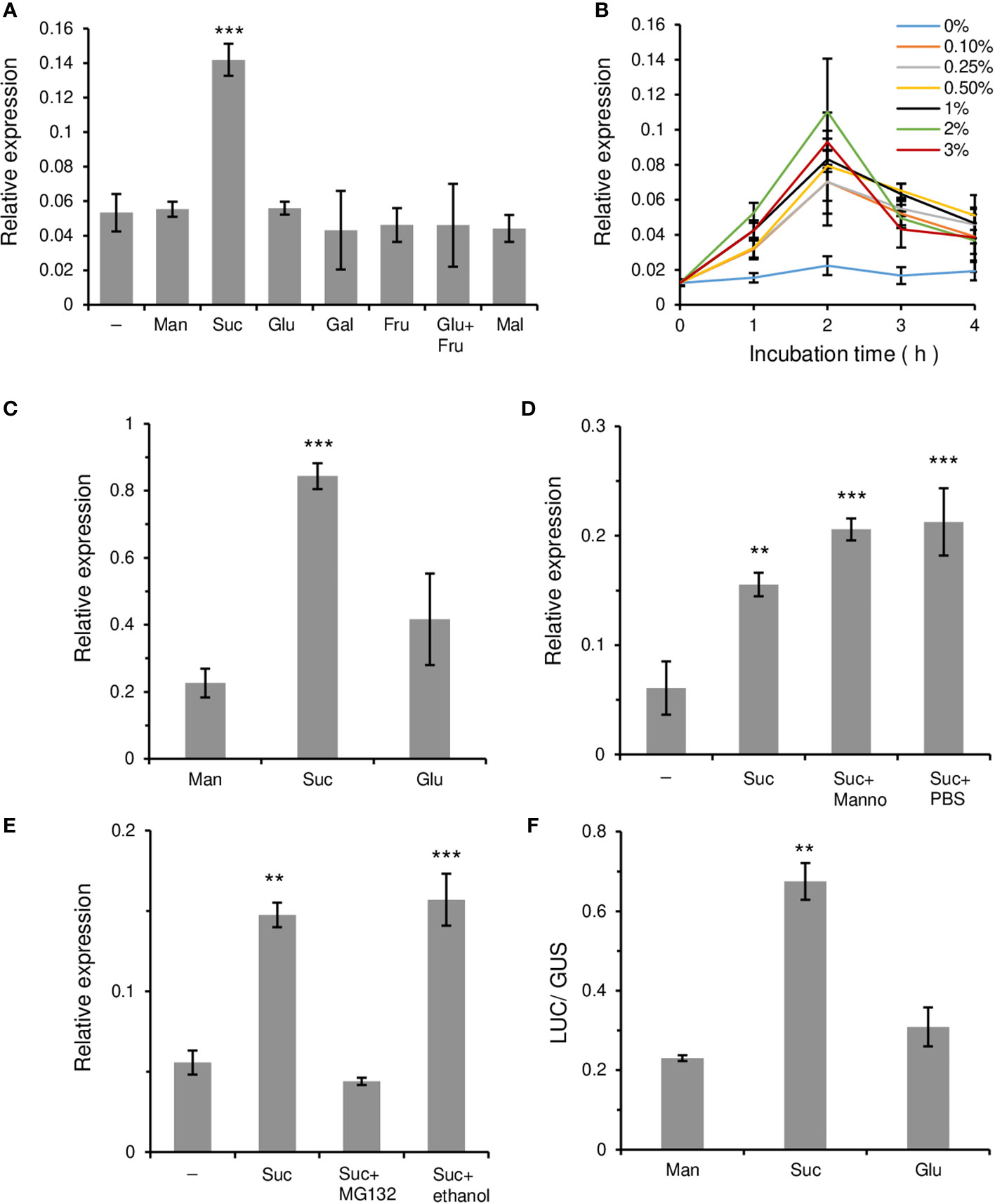
Figure 2 Relative transcripts level of OsWRKY7 affected by various sugars and inhibitors. (A-D) Effects of various sugars and chemicals. (A) Effect of sugars at 3% concentration in Oc cells. Samples were collected at 2 hours after sugar addition. (B) Effect of different sucrose concentrations. (C) Effect of 3% sugars in roots of 14-d-old seedlings. (D) Effects of 5 µM mannoheptulose (Manno). Effect of PBS was tested because mannoheptulose was dissolved in PBS before adding to the sucrose-containing medium. (E) Influence of MG132. Samples were collected at 1 hours after addition of 3% sucrose, 3% sucrose plus 100 µM MG132, or 3% sucrose plus 0.5% ethanol. We tested the effect of ethanol because MG132 was dissolved in ethanol. (F) Luciferase activity driven by the OsWRKY7 promoter and LUC coding region. ZmUbi::GUS construct was used as an internal control. Man, mannitol; Suc, sucrose; Glu, glucose; Gal; galactose; Fru, fructose; Mal, maltose. Minus sign (-) means basal media. Gene expression levels are relative to those of OsActin1. The error bars indicate standard deviations; n = 3 or more. Statistical significance is indicated by **(P < 0.01) and ***(P < 0.001).
To investigate whether gene expression was induced by sucrose in plants, 14-day-old rice seedlings grown hydroponically in Yoshida solution were left in the dark for 48 hours to reduce their endogenous sugar level. After this period, sucrose was added to the culture medium at the final concentration of 3%. Analysis of the OsWRKY7 transcript level in the roots showed that it increased significantly at 8 hours after sucrose addition (Figure 2C). Glucose also induced gene expression in the roots, but the induced levels were only slightly higher than those achieved by mannitol in the control.
The effect of mannoheptulose, which blocks hexose-mediated signaling (Chiou and Bush, 1998), was also tested. The results showed that the addition of 5 µM mannoheptulose to the sucrose solution did not interfere with the sucrose-induced expression of OsWRKY7 (Figure 2D). This observation suggests that hexoses produced from sucrose metabolism are probably not involved in the induction of the gene and that sucrose may function directly to promote OsWRKY7 expression.
To elucidate whether protein stability was involved in the sucrose response, MG132, which is an inhibitor of the 26S proteasomal protease (Genschik et al., 1998), was added to the sucrose solution. Analysis of the OsWRKY7 transcript level showed that the sucrose-induced expression of the gene was suppressed by 100 µM MG132 (Figure 2E). As a control, the sucrose-induced OsWRKY108 gene was also tested and it was shown that the expression of this gene was not affected by MG132 (Supplementary Figure 3). This result suggests that a regulatory protein that represses OsWRKY7 expression is degraded by an E3 ligase during the sucrose treatment.
To examine whether the induction of OsWRKY7 by sucrose occurred at the transcriptional level, the 3.081-kb promoter sequence of OsWRKY7 was placed upstream of the LUC coding region and the construct was introduced into protoplasts prepared from Oc cells. As an internal control, the GUS coding region driven by the ZmUbi promoter was co-transfected. After dividing the treated cells into three portions, sucrose, glucose, and mannitol at the final concentration of 3% were added to each aliquot. After 15 hours of incubation, the samples were analyzed and it was shown that the LUC activity in the sucrose-added cells was considerably higher than that in the mannitol-added cells (Figure 2F). On the contrary, the induction of the reporter gene expression by glucose was only slightly above that achieved by mannitol. This experiment demonstrated that sucrose preferentially induced OsWRKY7 at the transcription initiation level.
3.3 Mutations of OsWRKY7 reduce the defense response against M. oryzae
Expression of OsWRKY7 was induced by M. oryzae 48 hours after infection (Supplementary Figure 4A). To study the functional roles of OsWRKY7, we identified T-DNA tagging line 5A-00022, in which T-DNA was inserted at 375 bp upstream of the start ATG codon of OsWRKY7 (Supplementary Figure 4B). Homozygous progeny of the oswrky7-1 mutant was used for characterization of the gene function. In the mutant line, the expression level of OsWRKY7 was decreased to approximately a half of the wild type (WT) level, which indicated that the inserted T-DNA reduced gene expression (Supplementary Figure 4C). When the plants were infected with M. oryzae, the lesions were longer and broader than those observed in the segregated WT plants (Supplementary Figures 4D, E), suggesting that OsWRKY7 is involved in the defense response against this pathogen.
To confirm this observation, knockout mutations were generated in the first exon of OsWRKY7 via the CRISPR/Cas9 system (Figure 3A). Among 28 independent transgenic plants, five were selected to determine the flanking regions of the target site. Two mutant lines were chosen: oswrky7-2 carrying a single-bp deletion and oswrky7-3 carrying a two-bp deletion (Figure 3A, Supplementary Figures 5A, B). The deletions generated frameshift mutations, causing early termination during translation of the OsWRKY7 transcript. The mutant plants grew normally without any significant phenotypic alteration in the plant height, grain numbers per panicle, panicle length, and 100-grain weight (Figure 3B, Supplementary Figure 6). Interestingly, the OsWRKY7 transcript levels in the frameshift mutants were reduced compared to those in the WT plants, suggesting that the deletion mutations affected either the transcription rate or the stability of the transcript (Figure 3C). When the plants were inoculated with M. oryzae, the mutants displayed a phenotype that was more susceptible to the fungus compared with the WT controls (Figures 3D, E). These experiments confirmed that OsWRKY7 is involved in the defense response against M. oryzae.
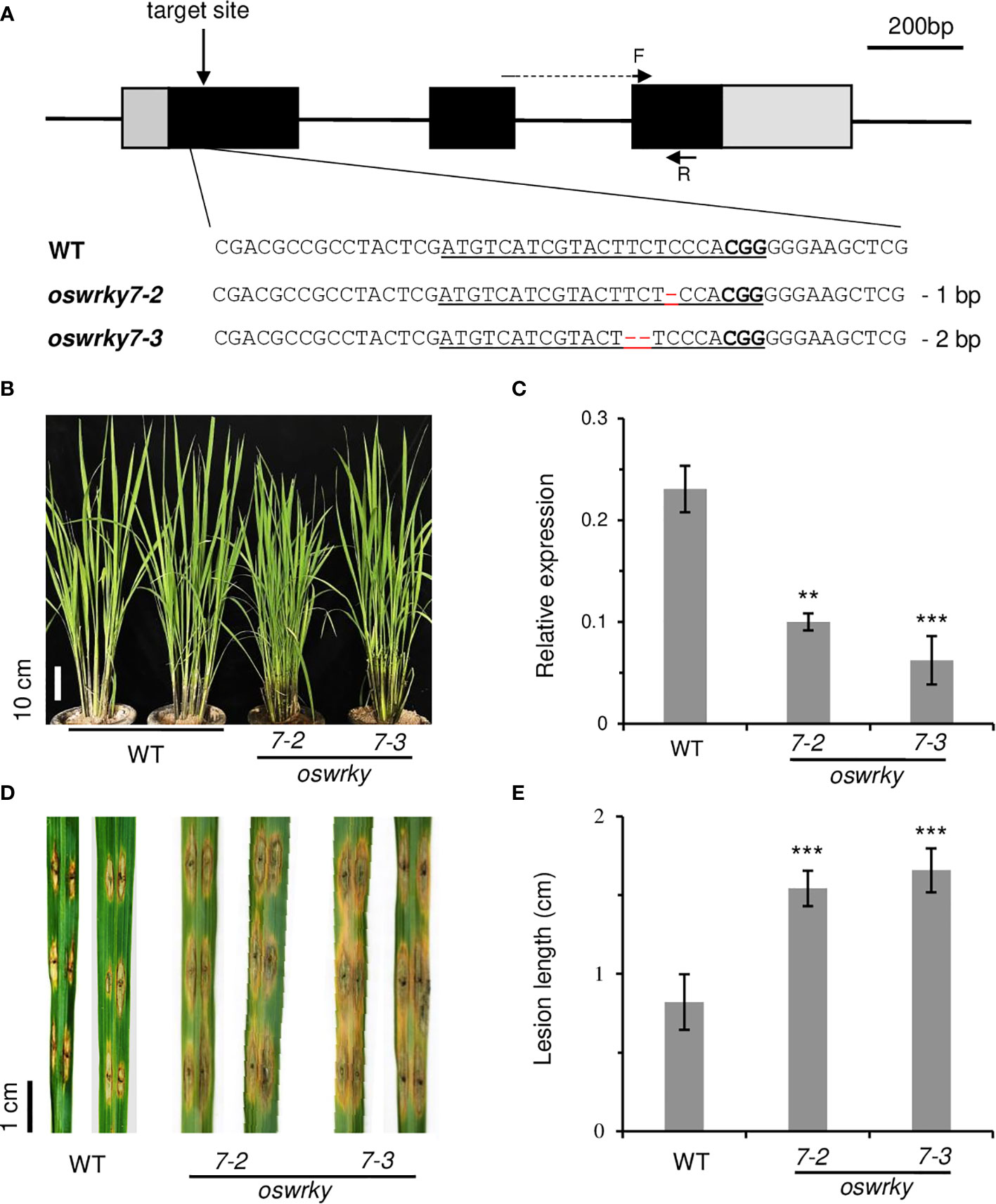
Figure 3 Characterization of oswrky7 mutants generated by the CRISPR/Cas9 method. (A) Schematic diagram of OsWRKY7 gene structure and sequence comparison of sgRNA target regions between WT and mutants. The target sequence and PAM site are underlined. Deleted sequences are indicated with red dashes. In the schematic gene structure, black boxes are exons; gray boxes are 5’- UTR and 3’-UTR regions; lines between the boxes are introns. F and R are primers used for measuring the transcript level of OsWRKY7. Scale bar = 200 bp. (B) Phenotypes of the WT and the knockout mutant plants grown under the paddy field at T1 generation. Scale bar = 10 cm. (C) Transcript level of OsWRKY7 in the WT and knockout mutant plants. Samples were collected at 42 DAG (D) Comparison of the lesions between the WT and mutants. Phenotype was observed at 9 days after M. oryzae infection. Scale bar = 1 cm. (E) The length of the lesion developed by the pathogen. The error bars show standard deviations; n = 10. Statistical significance is indicated by **(P < 0.01) and ***(P < 0.001).
3.4 Overexpression of OsWRKY7 increases the defense response against M. oryzae
To obtain transgenic rice plants that constitutively expressed OsWRKY7, the gene was placed after the ZmUbi promoter and the construct was introduced to rice embryonic calli. However, it was quite difficult to obtain transgenic plants, which suggested that the overexpression of OsWRKY7 was harmful to the plants. To overcome the difficulty, the HA tag was added at the end of the OsWRKY7 coding region, and the fusion molecule was placed after the ZmUbi promoter. In this way, several transgenic plants expressing OsWRKY7-HA were obtained, which indicated that attaching the tag reduced the harmful effect of the gene (Figure 4A). Plants #5 and #7, which expressed the fusion transcript and the HA-tagged protein at high levels, were selected (Figure 4B and Supplementary Figure 7). After inoculating the mature leaves of the transgenic plants with M. oryzae, it was observed that the lesion lengths and widths at the infection sites were significantly smaller in the transgenic plants than in the WT control plants (Figures 4C, D). This observation confirmed that OsWRKY7 is a positive regulatory element in the defense response to M. oryzae.
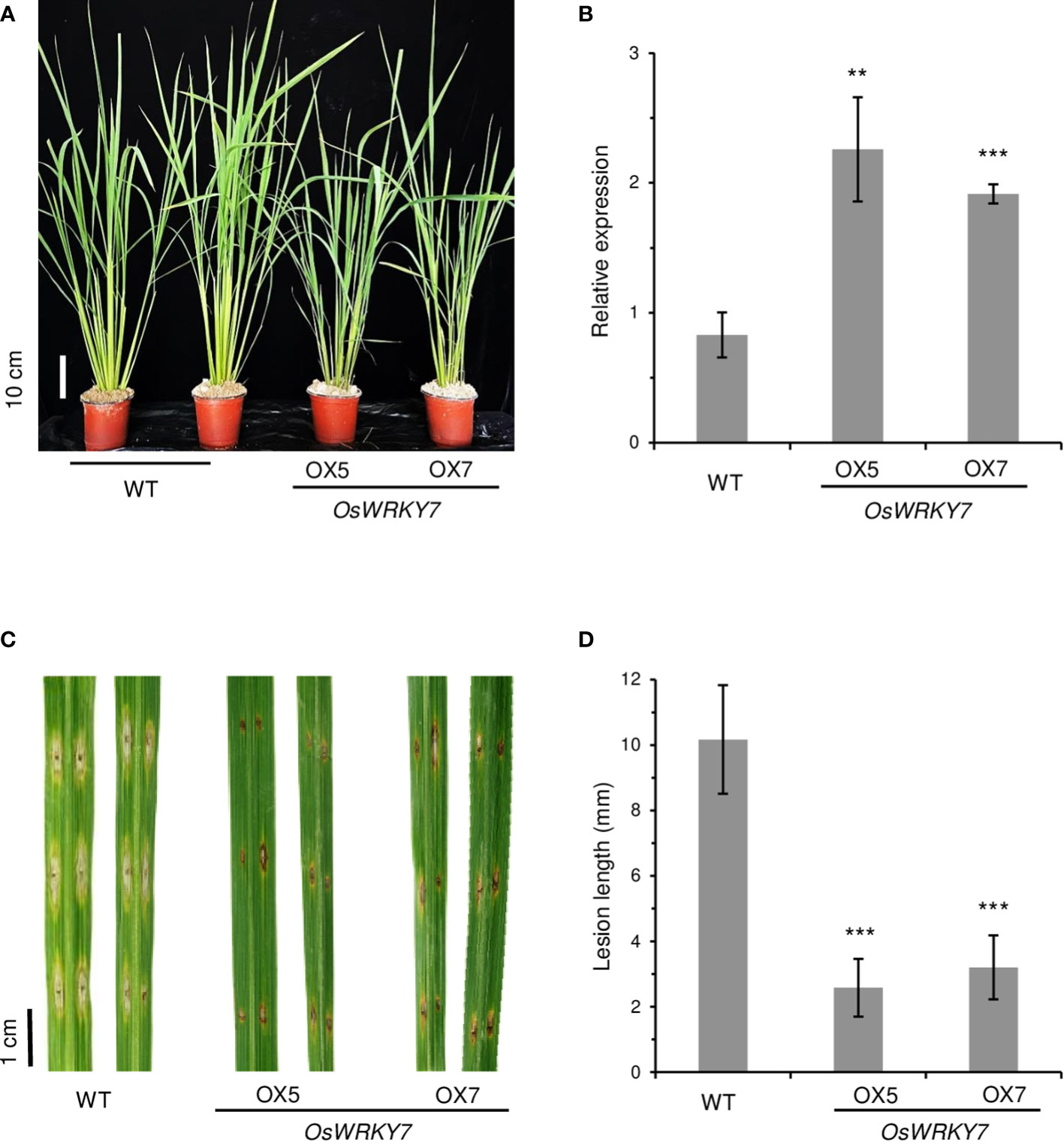
Figure 4 Phenotypes of OsWRKY7 overexpression plants. (A) Phenotype of the WT and transgenic plants overexpressing OsWRKY7. Scale bar = 10 cm. (B) The relative transcript level of OsWRKY7 in the WT and transgenic plants OX5 and OX7 overexpressing OsWRKY7. The gene expression levels are relative to those of OsActin1. Errors bars indicate standard deviation; n = 3. (C) Phenotypes of the lesions at 9 days after infection with M. oryzae. Bar = 1 cm. (D) The length of the lesion developed by the pathogen. Errors bars indicate standard deviation; n = 10. Statistical significance is indicated by **(P < 0.01) and ***(P < 0.001).
The expression level of the genes that were induced in response to pathogen infection were then analyzed. The levels of the pathogen-responsive marker genes OsPR1a, OsPR1b, and OsPR10a, were significantly reduced in oswrky7 mutant plants (Figures 5A-C). On the contrary, expression levels of the pathogen responsive marker genes were markedly increased in the plants of expressing OsWRKY7-HA (Figures 5D-F). This result indicates that the markers function downstream of OsWRKY7. It was then tested whether SA or JA were involved in the induction of the marker genes by OsWRKY7. The mutations of this gene did not affect the expression of phenylalanine ammonia-lyase (OsPAL) or isochorismate synthase 1 (OsICS1), which are involved in SA biosynthesis (Supplementary Figures 8A, B). In addition, a key regulator of systemic acquired resistance, NPR1 homolog 1 (OsNH1), was also not affected in mutant (Supplementary Figure 8C). Likewise, the mutations did not influence the expression of genes encoding enzymes associated with JA biosynthesis, namely allene oxide synthase (OsAOS2), allene oxide cyclase 1 (OsAOC1), and oxophytodienoate reductase 3 (OsOPR3) (Supplementary Figures 8D-F). These results suggest that the signaling pathway of SA or JA did not mediate the induction of the pathogen-responsive marker genes by OsWRKY7.
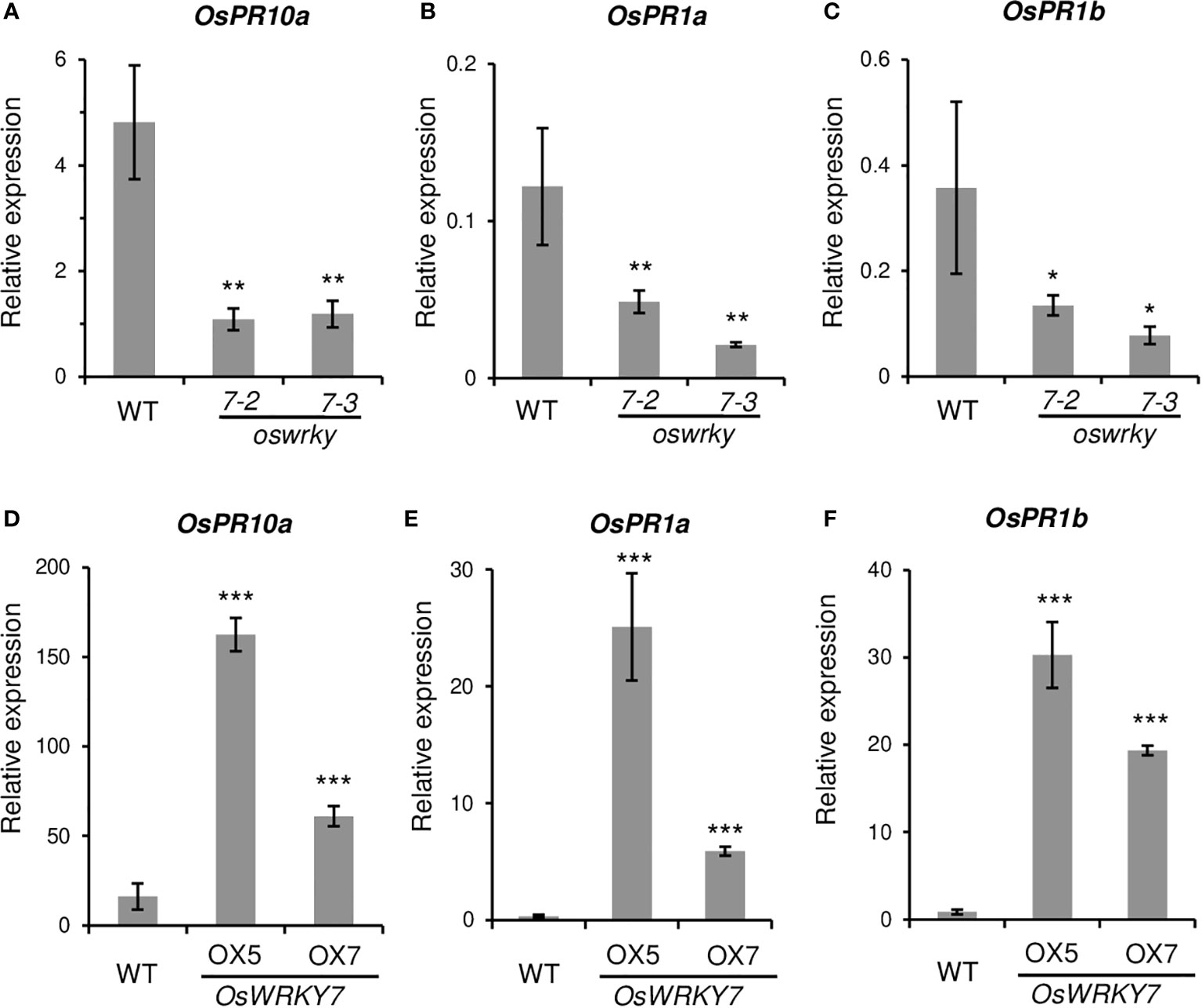
Figure 5 Expression levels of pathogen related genes in the WT and transgenic plants. (A-C) Relative transcript levels of OsPR10a (A), OsPR1a (B) and OsPR1b (C) in the leaves of the WT and oswrky7 mutants. (D-F) Relative transcript levels of OsPR10a (D), OsPR1a (E) and OsPR1b (F) in the leaves of the WT and OsWRKY7 overexpression plants. Samples were collected from the leaf blades at 42 DAG. The gene expression levels are relative to those of OsActin1. Error bars indicate standard deviation; n = 3 or more. Statistical significance is indicated by *(P < 0.05), **(P < 0.01) and ***(P < 0.001).
To evaluate whether the OsWRKY7-induced PR genes were sucrose-inducible, hydroponically grown seedlings were treated with 3% sucrose at 14 days after germination (DAG). After 8 hours of incubation in the solution, the expression levels of the PR genes in the roots were measured. The assay showed that the transcript level of OsPR10a was considerably higher in the sucrose-treated seedlings than in the control seedlings treated with 3% mannitol (Figure 6A). Glucose treatment did not induce gene expression above the level observed in the control. This result is consistent with the data observed in Oc cells (Figure 2D). On the contrary, the expression of OsPR1a was induced by both sucrose and glucose (Figure 6B). The OsPR1b transcript levels in sugar solutions were similar to those observed in mannitol solution (Figure 6C). These experiments indicate that, among the three PR genes tested, only OsPR10a was preferentially induced by sucrose.
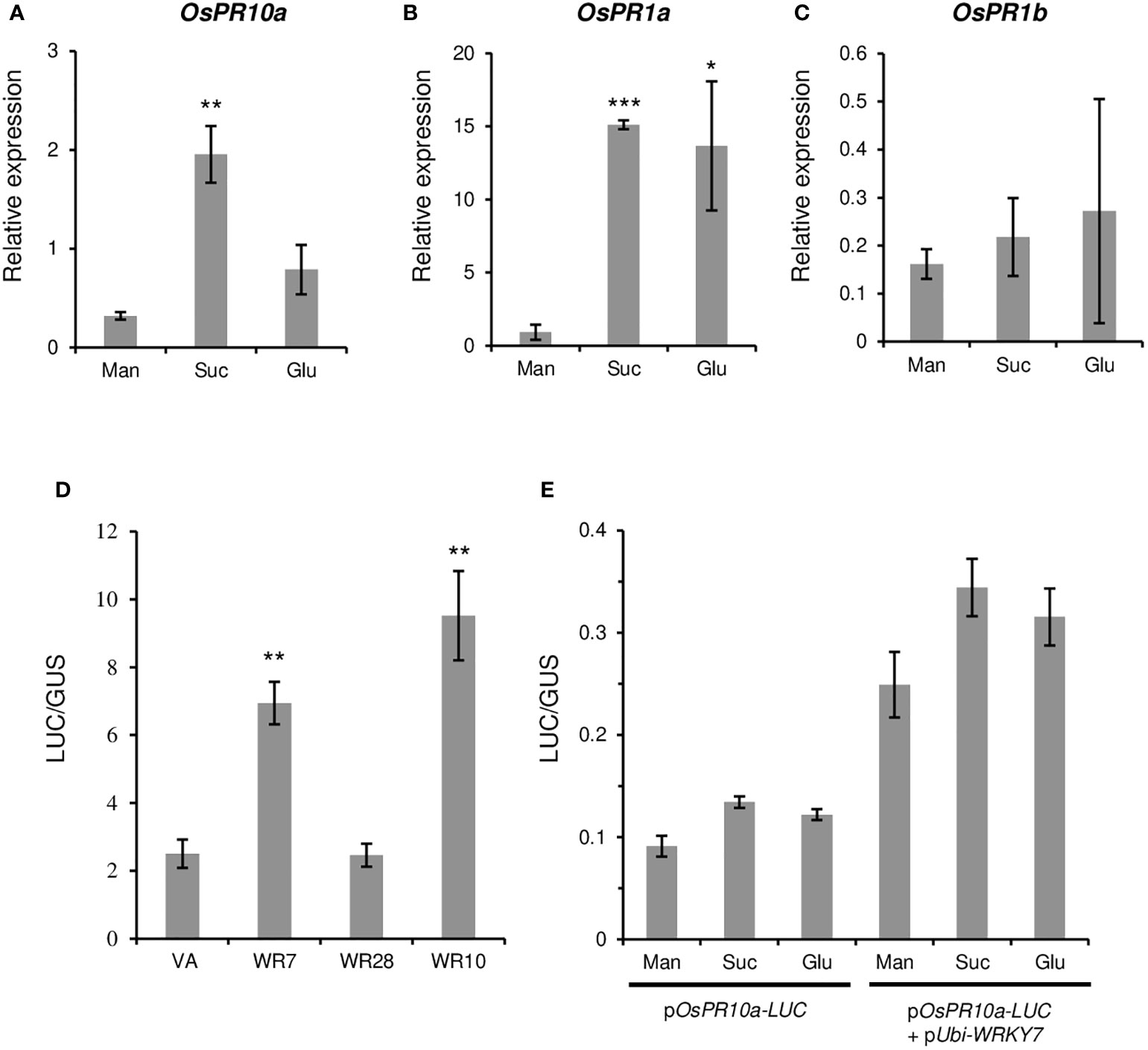
Figure 6 Effect of sugars and WRKY genes on the PR gene expression. (A-C) Effect of sugars on PR gene expression. Fourteen-day-old seedlings were grown in a hydroponic culture medium under constant light and they were transferred to the dark condition for two additional days before sucrose treatment. Mannitol, sucrose, or glucose were added at 3% final concentration. After the treatment for 8 hours, expression levels of OsPR10a (A), OsPR1a (B), and OsPR1b (C) in roots were measured. Gene expression levels are relative to those of OsActin1. Error bars indicate standard deviations; n = 3 or more. (D) Luciferase activity driven by the OsPR10a promoter. The pOsPR10a-LUC construct was transfected to Oc cell protoplasts alone (VA) or with pUbi-OsWRKY7 (WR7), pUbi-OsWRKY10 (WR10), or pUbi-OsWRKY28 (WR28). The LUC activity was assayed 15 hours after transfection of the molecules. ZmUbi-GUS construct was used as an internal control in each combination. (E) Effect of sugars on the OsPR10a promoter activity. pOsPR10a-LUC alone or together with pUbi-WRKY7 were transferred to Oc protoplasts and incubated in a culture medium containing mannitol (Man), sucrose (Suc) or glucose (Glu) at 3% final concentration. Luciferase activity was measured 15 hours after transfection. ZmUbi-GUS construct was used as an internal control in each experiment. Error bars indicate standard deviations; n = 3. Statistical significance is indicated by *(P < 0.05), **(P < 0.01) and ***(P < 0.001).
We tested whether OsPR10a expression in oswrky7 mutant was recovered by the sucrose treatment. The WT and oswrky7 plants that were grown hydroponically in Yoshida solution for 14 days were treated with mannitol or sucrose at the final concentration of 3%. After 9 hours treatment in the solutions, the root were sampled and the expression level of OsPR10a was measured. The experiment showed that sucrose significantly increased OsPR10a expression in the WT as observed in the previous experiments. Similarly, the transcript level of the gene in the mutant was also induced by sucrose, although the induced level was lower compared with WT (Supplementary Figure 9). This observation suggests that there are other sucrose-inducible transcription factors that induce OsPR10a expression.
3.5 Expression of OsPR10a is stimulated by OsWRKY7
Because the expression of PR10a was significantly reduced in OsWRKY7 mutants, PR10a may be directly controlled by the transcription factor. To prove this hypothesis, a construct containing the 2.856-kb promoter region of OsPR10a was placed in front of the LUC gene. We also constructed another molecule in which the coding region of OsWRKY7 was placed under the ZmUbi promoter. Both molecules, pOsPR10a-LUC and pUbi-WRKY7, were co-transfected into the Oc cell protoplasts and incubated for 15 hours to express the introduced genes. The results showed that the LUC expression level in the protoplasts co-transferred with both molecules were significantly higher than that in the protoplasts transferred with pOsPR10a-LUC alone (Figure 6D). As controls, OsWRKY10 and OsWRKY28 were inserted downstream of the ZmUbi promoter. It has been previously reported that OsWRKY10 functions as a positive regulatory element for the expression of PR10a, whereas OsWRKY28 does not influence the PR gene (Ersong et al., 2021). In this study, when pUbi-WRKY28 was co-introduced with pOsPR10a-LUC, the luciferase activity was not increased above the background level that was observed from pOsPR10a-LUC alone (Figure 6D). On the contrary, co-transfection of pOsPR10a-LUC and pUbi-OsWRKY10 significantly induced the reporter gene expression (Figure 6D). The above observations showed that the transient assay resembled the previous results and suggested that OsWRKY7 directly promoted the expression of pOsPR10a-LUC.
To test whether the OsPR10a promoter region responded directly to sucrose, the pOsPR10a-LUC vector was introduced into the protoplasts and was incubated in the culture solution containing three different sugars. Although the LUC expression level in the protoplasts cultured in sucrose-containing medium was slightly higher than that in the mannitol control, a similar expression level was achieved for the protoplasts grown in the glucose-containing medium (Figure 6E). This suggests that sucrose did not induce the gene directly. It was also tested whether the increased expression of the OsPR10a promoter by the OsWRKY7 protein was sucrose dependent. To answer this question, both pOsPR10a-LUC and pUbi-WRKY7 were introduced into the protoplasts and the effects of sucrose were examined. The experiment showed that sucrose did not significantly increase the pOsPR10a promoter activity above the levels obtained from mannitol or glucose (Figure 6E). This suggests that the sucrose induction of PR10a was not due to the stability of the OsWRKY7 mRNA or protein during sucrose treatments.
To confirm that OsWRKY7 directly controlled the OsPR10a promoter, chromatin immunoprecipitation (ChIP) experiments were performed with transgenic plants expressing the OsWRKY7-HA protein. Primers were designed to target W-box elements, W-box like elements 1 (WLE1), and ASF1MOTIF, which all belong to the WRKY protein-binding TGAC core elements (Maleck et al., 2001; Hwang et al., 2008) (Figure 7A). In the ChIP in vivo assay, the P2 and P3 DNA fragments containing the WLE1 element were enriched (Figure 7B).
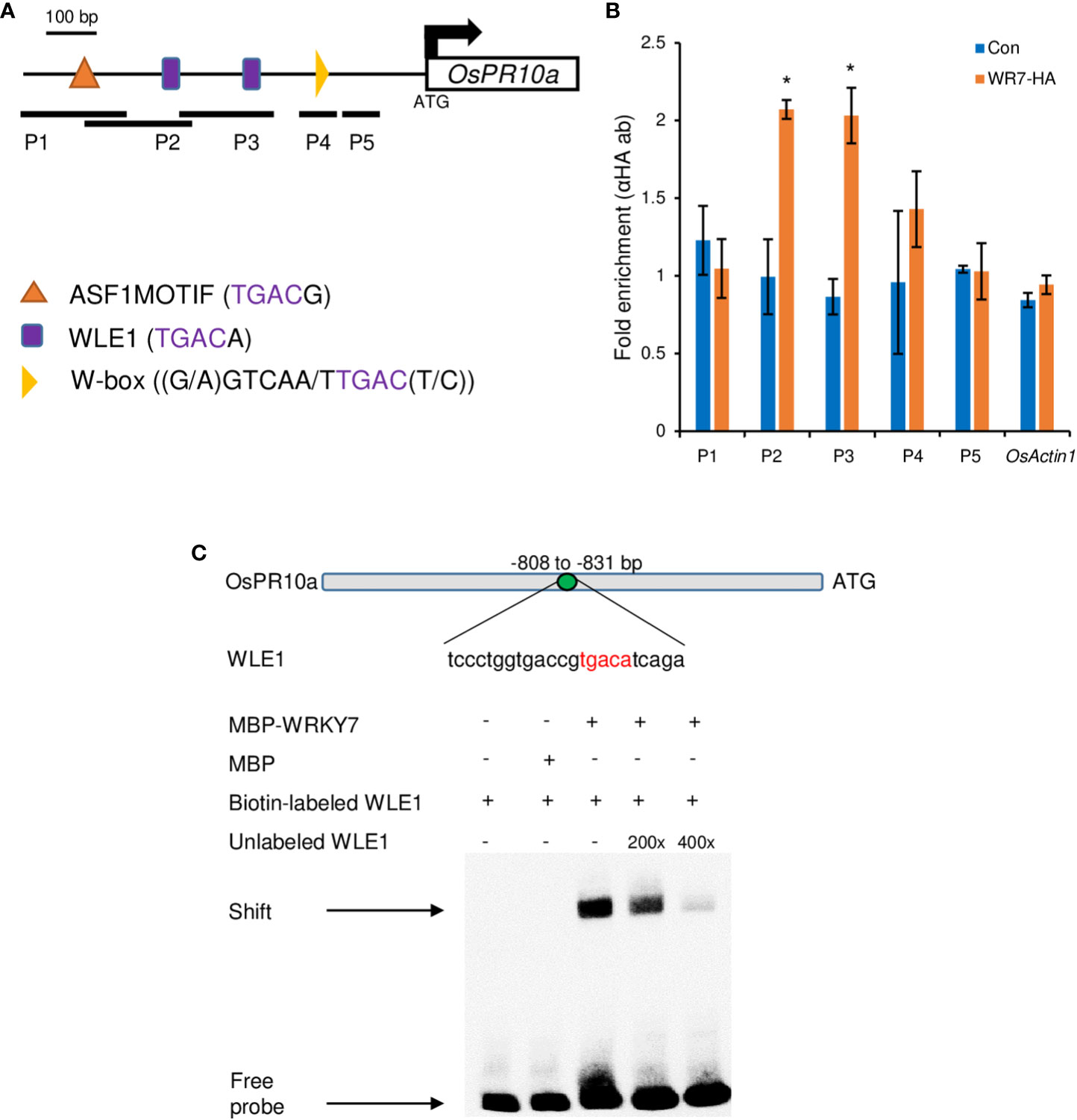
Figure 7 ChIP assay on the OsPR10a chromatin region and EMSA experiment. (A) Promoter region of OsPR10a. The black lines under the promoter region indicate the DNA fragment amplified by RT-PCR. Five pairs of primer set were used in ChIP assay; P1, 849-967 bp from the ATG start codon; P2, 795-946 bp from the ATG start codon; P3, 647-801 bp from the ATG start codon; P4, 207-289 bp from the ATG start codon; P5, 102-205 bp from the ATG start codon. Scale bar = 100 bp. (B) Chromatin enrichment of the OsPR10a promoter region. Transgenic plants expressing OsWRKY7-HA were used for the ChIP assay. Error bars indicate standard deviations; n = 2. Statistical significance is indicated by *(P < 0.05). (C) EMSA experiment using the sequence that contains WLE1 element on the promoter of OsPR10a. Biotin-labeled probe WLE1 without MBP-OsWRKY7 protein and biotin-labeled WLE1 with MBP protein were used as negative control. Excess unlabeled WLE1 was used as competitors.
To further confirm the interaction between OsWRKY7 and the OsPR10a promoter, the oligonucleotide (-808 to - 831 from the start ATG) containing the WLE1 element within the P2 fragment were used for EMSA experiment using MBP tagged OsWRKY7 protein (Figure 7C). The experiment showed that OsWRKY7 protein bind to the sequence and the binding was reduced when the unlabeled probe was used as a competitor. Both ChIP and EMSA assays support the hypothesis that OsWRKY7 directly binds to the promoter region of OsPR10a.
4 Discussion
A hypothesized model for high sugar resistance suggested that a yet unknown sugar receptor senses the elevation of the levels of extracellular sugars and triggers defense through a signaling cascade (Bezrutczyk et al., 2018). Notably, it was shown that rice plants treated with sucrose through the root system exhibited an accumulation of transcripts associated with defense genes in leaves and increased resistance to M. oryzae (Gómez-Ariza et al., 2007). This is consistent with previous results indicating that transgenic tobacco and rice plants expressing PRms accumulated sucrose in leaf tissues and enhanced their defense response (Murillo et al., 2003; Gómez-Ariza et al., 2007). As PRms is localized in plasmodesmata in infected maize radicles (Murillo et al., 1997), the accumulation of sucrose is possibly due to an alteration of sugar distribution. These observations clearly suggest the implication of plants’ sucrose dependent responses in the defense against invading pathogens. However, it remains to be determined how high levels of sucrose can regulate defense genes in plants.
In the present study, to investigate the functions of sucrose in the defense response, we examined OsWRKY7, which is inducible by exogenously applied sucrose. The transcript level of this gene was rapidly increased by the addition of sucrose in both cultured cells and seedlings, suggesting that this sugar functions as a signal molecule to induce gene expression.
Although sucrose controls various processes, such as starch metabolism, storage proteins, and anthocyanin biosynthesis, studies on sucrose responsive transcription factors that may be involved in sucrose signaling are scarce (Yoon et al., 2021). In Arabidopsis, sucrose-induced anthocyanin biosynthesis is mediated by Myb75 (Teng et al., 2005); however, other sugars, such as glucose and fructose, also stimulate anthocyanin accumulation. In addition, mannoheptulose, which is a competitive inhibitor of hexokinase1, inhibits sucrose induction (Neta-Sharir et al., 2000). Therefore, Myb75 may not be a mediator of sucrose-specific signaling. Another transcription factor that is induced by sucrose is AtWRKY20 (Nagata et al., 2012). This regulatory gene controls the expression of ApL3, which encodes AGPase in Arabidopsis. Similarly, the barley starch synthesis gene isoamylase1 is regulated by the sucrose-inducible WRKY gene SUSIBA2 (Sun et al., 2003). However, it has not been tested whether these WRKY genes are specifically induced by sucrose. In this study, we demonstrated that sucrose preferentially controlled the expression of OsWRKY7 because hexoses and mannose were unable to induce this gene. Mannoheptulose did not reduce the sucrose induction of the gene, suggesting that glucose or other hexoses metabolized from sucrose were not responsible for this process.
Sucrose regulates gene expression at the transcriptional or post-transcriptional level. It increases the transcript levels of the WRKY and Myb75 genes, but controls the translation of the bZIP11 transcript post-transcriptionally (Wiese et al., 2004). In the present study, the expression of the LUC reporter gene driven by the 3-kb OsWRKY7 promoter sequence was shown to be increased by sucrose, which indicated that sucrose regulates OsWRKY7 at the transcriptional level.
Because several WRKY genes are involved in plant defense mechanisms, we tested whether OsWRKY7 functions in disease tolerance against M. oryzae using a knockdown mutant generated by T-DNA tagging and knockout mutants created by CRISPR/Cas9. These mutants were more susceptible to the rice blast fungus, however, transgenic plants overexpressing the gene showed reduced disease symptoms. These results suggest that the transcription factor functions positively in the defense against M. oryzae. The transcript levels of the pathogen-related genes OsPR1a, OsPR1b, and OsPR10a were significantly reduced in the mutants and increased in the plants exhibiting overexpression, supporting the hypothesis that OsWRKY7 plays a role in plant defense. However, genes involved in SA synthesis or SA-mediated resistance were not affected by the mutations, and this was the case also for JA biosynthesis genes, suggesting that OsWRKY7 functions independently from the hormones.
Interestingly, the transcript level of OsWRKY7 was significantly reduced when MG132 was added during sucrose induction, which indicates that an upstream repressor protein of OsWRKY7 may be degraded through the proteasome pathway and that sucrose promotes this pathway. AtWRKY50, an ortholog of OsWRKY7, interacts with Botrytis-induced kinase 1 (BIK1), a receptor-like cytoplasmic kinase VII member that is localized in the nucleus and degraded by the proteasome (Lal et al., 2018). This suggests that OsWRKY7 may interact with a kinase. Further study will be needed to investigate how the protein is degraded by a proteasome.
Among the three PR genes that were influenced by OsWRKY7, only OsPR10a was preferentially induced by sucrose. OsPR1a was also induced by glucose, which indicates that other regulatory elements besides OsWRKY7 also control its expression. In contrast, OsPR1b was not induced by either sucrose or glucose. Because OsPR1b also functions downstream of the sucrose-inducible OsWRKY7 gene, the induction of this PR gene by sucrose was expected. It will be interesting to investigate how the sucrose signaling was diminished in the PR gene. To study whether the induction of PR10a expression by sucrose occurred directly or was mediated by OsWRKY7 protein, the promoter of PR10a was tested using the LUC reporter system. It was shown that sucrose did not induce the promoter activity of OsPR10a. Ru et al. (2015) reported that GFP fused OsWRKY7 protein is localized in the nucleus and OsWRKY7 protein has transcriptional activity in yeast hybrid assay, which indicated that the sucrose induction of the PR10a promoter was mediated by OsWRKY7. Direct control of this PR gene by OsWRKY7 was also confirmed by the ChIP and EMSA assays. All the results presented in this study provide a definitive proof that sucrose has a role in defense response in rice by activating the transcription factor OsWRKY7, which triggers the expression of PR10a. Further experiment is needed to investigate what is the upstream element that mediates sucrose signaling and whether OsWRKY7 interacts with a partner to control the downstream gene expression.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
GA, S-WL, K-HJ, J-SJ designed the project. WT, JY, KV, L-HC, TH, XP, E-JK, and KW performed experiments and analyzed data. WT, JY, J-SJ, and GA wrote the paper. All authors contributed to the article and approved the submitted version.
Funding
This work was supported, in part, by grants from the National Research Foundation of Korea (NRF-2020R1A2C2006826 to GA; NRF-2021R1C1C2007906 to JY) and the Rural Development Administration, Korea (PJ01690101 to JSJ).
Acknowledgments
We are thankful to Kyungsook An for maintaining the transgenic T-DNA tagging lines and providing the seed stock.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1117023/full#supplementary-material
References
Abbruscato, P., Nepusz, T., Mizzi, L., Corvo, M., Morandini, P., Fumasoni, I., et al. (2012). OsWRKY22, a monocot WRKY gene, plays a role in the resistance response to blast. Mol. Plant Pathol. 13, 828–841. doi: 10.1111/j.1364-3703.2012.00795.x
An, G. (1987). Binary Ti vectors for plant transformation and promoter analysis. In. Methods Enzymol. 153, 292–305. doi: 10.1016/0076-6879(87)53060-9
An, G., Jeong, D.-H., Jung, K.-H., Lee, S. (2005a). Reverse genetic approaches for functional genomics of rice. Plant Mol. Biol. 59, 111–123. doi: 10.1007/s11103-004-4037-y
An, G., Lee, S., Kim, S.-H., Kim, S.-R. (2005b). Molecular genetics using T-DNA in rice. Plant Cell Physiol. 46, 14–22. doi: 10.1093/pcp/pci502
Baba, A., Hasezawa, S., Syōno, K. (1986). Cultivation of rice protoplasts and their transformation mediated by Agrobacterium spheroplasts. Plant Cell Physiol. 27, 463–471. doi: 10.1093/oxfordjournals.pcp.a077122
Bezrutczyk, M., Yang, J., Eom, J.-S., Prior, M., Sosso, D., Hartwig, T., et al. (2018). Sugar flux and signaling in plant–microbe interactions. Plant J. 93, 675–685. doi: 10.1111/tpj.13775
Bodson, M., Outlaw, W. H. (1985). Elevation in the sucrose content of the shoot apical meristem of Sinapis alba at floral evocation 1. Plant Physiol. 79, 420–424. doi: 10.1104/pp.79.2.420
Cheng, H., Wang, S. (2014). The important player of rice-pathogen interactions: WRKY-type transcription factors. Sci Sin Vitae 44, 784–793.
Cheng, H., Liu, H., Deng, Y., Xiao, J., Li, X., Wang, S. (2015). The WRKY45-2, WRKY13 WRKY42 transcriptional regulatory cascade is required for rice resistance to fungal pathogen. Plant Physiol. 167, 1087–1099. doi: 10.1104/pp.114.256016
Chiou, T. J., Bush, D. R. (1998). Sucrose is a signal molecule in assimilate partitioning. Proc. Natl. Acad. Sci. U.S.A. 95, 4784–4788. doi: 10.1073/pnas.95.8.4784
Cho, J.-I., Ryoo, N., Eom, J.-S., Lee, D.-W., Kim, H.-B., Jeong, S.-W., et al. (2009). Role of rice hexokinases OsHXK5 and OsHXK6 as glucose sensors. Plant Physiol. 149, 745–759. doi: 10.1104/pp.108.131227
Cho, L.-H., Yoon, J., Pasriga, R., An, G. (2016). Homodimerization of Ehd1 is required to induce flowering in rice. Plant Physiol. 170, 2159–2171. doi: 10.1104/pp.15.01723
Cho, L.-H., Yoon, J., Tun, W., Baek, G., Peng, X., Hong, W.-J., et al. (2022). Cytokinin increases vegetative growth period by suppressing florigen expression in rice and maize. Plant J. 10 (6), 1619–1635 doi: 10.1111/tpj.15760
Ciereszko, I., Johansson, H., Kleczkowski, L. A. (2001). Sucrose and light regulation of a cold-inducible UDP-glucose pyrophosphorylase gene via a hexokinase-independent and abscisic acid-insensitive pathway in arabidopsis. Biochem. J. 354, 67–72. doi: 10.1042/bj3540067
Concordet, J.-P., Haeussler, M. (2018). CRISPOR: Intuitive guide selection for CRISPR/Cas9 genome editing experiments and screens. Nucleic Acids Res. 46, W242–W245. doi: 10.1093/nar/gky354
Corbesier, L., Lejeune, P., Bernier, G. (1998). The role of carbohydrates in the induction of flowering in Arabidopsis thaliana: Comparison between the wild type and a starchless mutant. Planta 206, 131–137. doi: 10.1007/s004250050383
Ersong, Z., Xuming, W., Rumeng, X., Feibo, Y., Chao, Z., Yong, Y., et al. (2021). Regulation of OsPR10a promoter activity by phytohormone and pathogen stimulation in rice. Rice Sci. 28, 442–456. doi: 10.1016/j.rsci.2021.07.005
Friend, D. J., Bodson, M., Bernier, G. (1984). Promotion of flowering in Brassica campestris l. cv Ceres by sucrose. Plant Physiol. 75, 1085–1089. doi: 10.1104/pp.75.4.1085
Fukushima, S., Mori, M., Sugano, S., Takatsuji, H. (2016). Transcription factor WRKY62 plays a role in pathogen defense and hypoxia-responsive gene expression in rice. Plant Cell Physiol. 57, 2541–2551. doi: 10.1093/pcp/pcw185
Genschik, P., Criqui, M. C., Parmentier, Y., Derevier, A., Fleck, J. (1998). Cell cycle–dependent proteolysis in plants: Identification of the destruction box pathway and metaphase arrest produced by the proteasome inhibitor MG132. Plant Cell. 10, 2063–2075. doi: 10.1105/tpc.10.12.2063
Gómez-Ariza, J., Campo, S., Rufat, M., Estopà, M., Messeguer, J., Segundo, B. S., et al. (2007). Sucrose-mediated priming of plant defense responses and broad-spectrum disease resistance by overexpression of the maize pathogenesis-related PRms protein in rice plants. Mol. Plant-Microbe Interactions. 20, 832–842. doi: 10.1094/MPMI-20-7-0832
Hanson, J., Johannesson, H., Engström, P. (2001). Sugar-dependent alterations in cotyledon and leaf development in transgenic plants expressing the HDZhdip gene ATHB13. Plant Mol. Biol. 45, 247–262. doi: 10.1023/A:1006464907710
Haring, M., Offermann, S., Danker, T., Horst, I., Peterhansel, C., Stam, M. (2007). Chromatin immunoprecipitation: Optimization, quantitative analysis and data normalization. Plant Methods 3, 11. doi: 10.1186/1746-4811-3-11
He, F., Chen, S., Ning, Y., Wang, G.-L. (2016). Rice (Oryza sativa) protoplast isolation and its application for transient expression analysis. Curr. Protoc. Plant Biol. 1, 373–383. doi: 10.1002/cppb.20026
Hong, W.-J., Jiang, X., Ahn, H. R., Choi, J., Kim, S.-R., Jung, K.-H. (2020). Systematic analysis of cold stress response and diurnal rhythm using transcriptome data in rice reveals the molecular networks related to various biological processes. Int. J. Mol. Sci. 21, 6872. doi: 10.3390/ijms21186872
Houssa, P., Bernier, G., Kinet, J. M. (1991). Qualitative and quantitative analysis of carbohydrates in leaf exudate of the short-day plant, Xanthium strumarium l. during floral transition. J. Plant Physiol. 138, 24–28. doi: 10.1016/S0176-1617(11)80724-8
Hwang, S.-H., Lee, I. A., Yie, S. W., Hwang, D.-J. (2008). Identification of an OsPR10a promoter region responsive to salicylic acid. Planta 227, 1141–1150. doi: 10.1007/s00425-007-0687-8
Jeong, D.-H., An, S., Kang, H.-G., Moon, S., Han, J.-J., Park, S., et al. (2002). T-DNA Insertional mutagenesis for activation tagging in rice. Plant Physiol. 130, 1636–1644. doi: 10.1104/pp.014357
Jeong, D.-H., An, S., Park, S., Kang, H.-G., Park, G.-G., Kim, S.-Y., et al. (2006). Generation of a flanking sequence-tag database for activation-tagging lines in japonica rice. Plant J. 45, 123–132. doi: 10.1111/j.1365-313X.2005.02610.x
Jeon, J.-S., Lee, S., Jung, K.-H., Jun, S.-H., Jeong, D.-H., Lee, J., et al. (2000). T-DNA Insertional mutagenesis for functional genomics in rice. Plant J. 22, 561–570. doi: 10.1046/j.1365-313x.2000.00767.x
Jiang, Y., Guo, L., Liu, R., Jiao, B., Zhao, X., Ling, Z., et al. (2016). Overexpression of poplar PtrWRKY89 in transgenic Arabidopsis leads to a reduction of disease resistance by regulating defense-related genes in salicylate- and jasmonate-dependent signaling. PloS One 11, e0149137. doi: 10.1371/journal.pone.0149137
Jimmy, J. L., Babu, S. (2015). Role of OsWRKY transcription factors in rice disease resistance. Trop. Plant Pathol. 1, 355–361. doi: 10.1007/s40858-015-0058-0
Jimmy, J. L., Babu, S. (2019). Variations in the structure and evolution of rice WRKY genes in indica and japonica genotypes and their co-expression network in mediating disease resistance. Evol. Bioinf. 15, 1–12. doi: 10.1177/1176934319857720
Kim, E.-J., Hong, W.-J., Kim, Y.-J., Jung, K.-H. (2021). Analysis of triple mutant for OsMADS62, OsMADS63, and OsMADS68 reveals the downstream regulatory mechanism for pollen germination in rice (Oryza sativa). Int. J. Mol. Sci. 23, 239. doi: 10.3390/ijms23010239
Kim, S.-R., Lee, D.-Y., Yang, J.-I., Moon, S., An, G. (2009). Cloning vectors for rice. J. Plant Biol. 52, 73. doi: 10.1007/s12374-008-9008-4
Krapp, A., Quick, W. P., Stitt, M. (1991). Ribulose-1,5-bisphosphate carboxylase-oxygenase, other calvin-cycle enzymes, and chlorophyll decrease when glucose is supplied to mature spinach leaves via the transpiration stream. Planta 186, 58–69. doi: 10.1007/BF00201498
Lal, N. K., Nagalakshmi, U., Hurlburt, N. K., Flores, R., Bak, A., Sone, P., et al. (2018). The receptor-like cytoplasmic kinase BIK1 localizes to the nucleus and regulates defense hormone expression during plant innate immunity. Cell Host Microbe 23, 485–497.e5. doi: 10.1016/j.chom.2018.03.010
Lee, S., Jeon, J.-S., Jung, K.-H., An, G. (1999). Binary vectors for efficient transformation of rice. J. Plant Biol. 42, 310–316. doi: 10.1007/BF03030346
Li, L., Sheen, J. (2016). Dynamic and diverse sugar signaling. Curr. Opin. Plant Biol. 33, 116–125. doi: 10.1016/j.pbi.2016.06.018
Li, X., Xing, J., Gianfagna, T. J., Janes, H. W. (2002). Sucrose regulation of ADP-glucose pyrophosphorylase subunit genes transcript levels in leaves and fruits. Plant Sci. 162, 239–244. doi: 10.1016/S0168-9452(01)00565-9
Maleck, K., Levine, A., Eulgem, T., Morgan, A., Schmid, J., Lawton, K., et al. (2001). The transcriptome of Arabidopsis thaliana during systemic acquired resistance. Nat. Genet. 26, 403–410. doi: 10.1038/82521
Morcuende, R., Krapp, A., Hurry, V., Stitt, M. (1998). Sucrose-feeding leads to increased rates of nitrate assimilation, increased rates of α-oxoglutarate synthesis, and increased synthesis of a wide spectrum of amino acids in tobacco leaves. Planta 206, 394–409. doi: 10.1007/s004250050415
Morkunas, I., Marczak, Ł., Stachowiak, J., Stobiecki, M. (2005). Sucrose-induced lupine defense against Fusarium oxysporum: Sucrose-stimulated accumulation of isoflavonoids as a defense response of lupine to Fusarium oxysporum. Plant Physiol. Biochem. 43, 363–373. doi: 10.1016/j.plaphy.2005.02.011
Müller-Röber, B., Sonnewald, U., Willmitzer, L. (1992). Inhibition of the ADP-glucose pyrophosphorylase in transgenic potatoes leads to sugar-storing tubers and influences tuber formation and expression of tuber storage protein genes. EMBO J. 11, 1229–1238. doi: 10.1002/j.1460-2075.1992.tb05167.x
Murillo, I., Cavallarin, L., San Segundo, B. (1997). The maize pathogenesis-related PRms protein localizes to plasmodesmata in maize radicles. Plant Cell. 9, 145–156. doi: 10.1105/tpc.9.2.145
Murillo, I., Roca, R., Bortolotti, C., San Segundo, B. (2003). Engineering photoassimilate partitioning in tobacco plants improves growth and productivity and provides pathogen resistance. Plant J.: For. Cell Mol. Biol. 36, 330–341. doi: 10.1046/j.1365-313X.2003.01880.x
Nagata, T., Hara, H., Saitou, K., Kobashi, A., Kojima, K., Yuasa, T., et al. (2012). Activation of ADP-glucose pyrophosphorylase gene promoters by a WRKY transcription factor, AtWRKY20, in Arabidopsis thaliana l. and sweet potato (Ipomoea batatas lam.). Plant Prod. Sci. 15, 10–18. doi: 10.1626/pps.15.10
Naito, Y., Hino, K., Bono, H., Ui-Tei, K. (2015). CRISPRdirect: Software for designing CRISPR/Cas guide RNA with reduced off-target sites. Bioinformatics 31, 1120–1123. doi: 10.1093/bioinformatics/btu743
Neta-Sharir, I., Shoseyoy, O., Weiss, D. (2000). Sugars enhance the expression of gibberellin-induced genes in developing petunia flowers. Physiol. Plant. 109, 196–202. doi: 10.1034/j.1399-3054.2000.100212.x
Nieberl, P., Ehrl, C., Pommerrenig, B., Graus, D., Marten, I., Jung, B., et al. (2017). Functional characterisation and cell specificity of BvSUT1, the transporter that loads sucrose into the phloem of sugar beet (Beta vulgaris l.) source leaves. Plant Biol. 19, 315–326. doi: 10.1111/plb.12546
Ohira, K., Ojima, K., Fujiwara, A. (1973). Studies on the nutrition of rice cell culture i. a simple, defined medium for rapid growth in suspension culture. Plant Cell Physiol. 14, 1113–1121. doi: 10.1093/oxfordjournals.pcp.a074950
Peng, X., Hu, Y., Tang, X., Zhou, P., Deng, X., Wang, H., et al. (2012). Constitutive expression of rice WRKY30 gene increases the endogenous jasmonic acid accumulation, PR gene expression and resistance to fungal pathogens in rice. Planta 236, 1485–1498. doi: 10.1007/s00425-012-1698-7
Peng, X., Tun, W., Dai, S., Li, J., Zhang, Q., Yin, G., et al. (2021). Genome-wide analysis of CCT transcript factors to identify genes contributing to photoperiodic flowering in Oryza rufipogon. Front. Plant Sci. 12, 736419. doi: 10.3389/fpls.2021.736419
Plaxton, W. C. (1996). The organization and regulation of plant glycolysis. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47, 185–214. doi: 10.1146/annurev.arplant.47.1.185
Ru, L., Jie, Z., Doung-yue, L., Xu-ming, W., Young, Y., Chu-lang, Y., et al. (2015). Expression of OsWRKY7 in rice. Chi. J. Rice Sci., 29 (6), 559–570. doi: 10.3969/j.issn.1001-7216.2015.06.001
Ryu, H.-S., Han, M., Lee, S.-K., Cho, J.-I., Ryoo, N., Heu, S., et al. (2006). A comprehensive expression analysis of the WRKY gene superfamily in rice plants during defense response. Plant Cell Rep. 25, 836–847. doi: 10.1007/s00299-006-0138-1
Sakr, S., Wang, M., Dédaldéchamp, F., Perez-Garcia, M.-D., Ogé, L., Hamama, L., et al. (2018). The sugar-signaling hub: Overview of regulators and interaction with the hormonal and metabolic network. Int. J. Mol. Sci. 19, 2506. doi: 10.3390/ijms19092506
Schmittgen, T. D., Livak, K. J. (2008). Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 3, 1101–1108. doi: 10.1038/nprot.2008.73
Sun, C., Palmqvist, S., Olsson, H., Borén, M., Ahlandsberg, S., Jansson, C. (2003). A novel WRKY transcription factor, SUSIBA2, participates in sugar signaling in barley by binding to the sugar-responsive elements of the iso1 promoter. Plant Cell. 15, 2076–2092. doi: 10.1105/tpc.014597
Sun, J., Wang, H., Ren, L., Chen, S., Chen, F., Jiang, J. (2017). CmFTL2 is involved in the photoperiod- and sucrose-mediated control of flowering time in chrysanthemum. Hortic. Res. 4, 17001. doi: 10.1038/hortres.2017.1
Tao, Z., Liu, H., Qiu, D., Zhou, Y., Li, X., Xu, C., et al. (2009). A pair of allelic WRKY genes play opposite roles in rice- bacteria interactions. Plant Physiol. 151, 936–948. doi: 10.1104/pp.109.145623
Teng, S., Keurentjes, J., Bentsink, L., Koornneef, M., Smeekens, S. (2005). Sucrose-specific induction of anthocyanin biosynthesis in Arabidopsis requires the MYB75/PAP1 gene. Plant Physiol. 139, 1840–1852. doi: 10.1104/pp.105.066688
Thibaud, M.-C., Gineste, S., Nussaume, L., Robaglia, C. (2004). Sucrose increases pathogenesis-related PR-2 gene expression in Arabidopsis thaliana through an SA-dependent but NPR1-independent signaling pathway. Plant Physiol. Biochem. 42, 81–88. doi: 10.1016/j.plaphy.2003.10.012
Thimm, O., Bläsing, O., Gibon, Y., Nagel, A., Meyer, S., Krüger, P., et al. (2004). Mapman: A user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 37, 914–939. doi: 10.1111/j.1365-313X.2004.02016.x
Vo, K. T. X., Lee, S.-K., Halane, M. K., Song, M.-Y., Hoang, T. V., Kim, C.-Y., et al. (2019). Pi5 and Pii paired NLRs are functionally exchangeable and confer similar disease resistance specificity. Mol. Cells 42, 637–645. doi: 10.14348/molcells.2019.0070
Walker, I. H., Hsieh, P. C., Riggs, P. D. (2010). Mutations in maltose-binding protein that alter affinity and solubility properties. Appl. Microbiol. Biotechnol., 88 (1), 187–197. doi: 10.1007/s00253-010-2696-y
Wang, H.-J., Wan, A.-R., Hsu, C.-M., Lee, K.-W., Yu, S.-M., Jauh, G.-Y. (2007). Transcriptomic adaptations in rice suspension cells under sucrose starvation. Plant Mol. Biol. 63, 441–463. doi: 10.1007/s11103-006-9100-4
Wang, S.-J., Yeh, K.-W., Tsai, C.-Y. (2001). Regulation of starch granule-bound starch synthase I gene expression by circadian clock and sucrose in the source tissue of sweet potato. Plant Sci. 161, 635–644. doi: 10.1016/S0168-9452(01)00449-6
Wiese, A., Elzinga, N., Wobbes, B., Smeekens, S. (2004). A conserved upstream open reading frame mediates sucrose-induced repression of translation. Plant Cell. 16, 1717–1729. doi: 10.1105/tpc.019349
Xiao, J., Cheng, H., Li, X., Xiao, J., Xu, C., Wang, S. (2013). Rice WRKY13 regulates cross talk between abiotic and biotic stress signaling pathways by selective binding to different cis-elements. Plant Physiol. 163, 1868–1882. doi: 10.1104/pp.113.226019
Yoo, S.-D., Cho, Y.-H., Sheen, J. (2007). Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2, 1565–1572. doi: 10.1038/nprot.2007.199
Yoon, J., Cho, L.-H., Tun, W., Jeon, J.-S., An, G. (2021). Sucrose signaling in higher plants. Plant Sci. 302, 110703. doi: 10.1016/j.plantsci.2020.110703
Keywords: Magnaporthe oryzae, OsPR10a, OsWRKY7, rice, sucrose
Citation: Tun W, Yoon J, Vo KTX, Cho L-H, Hoang TV, Peng X, Kim E-J, Win KTYS, Lee S-W, Jung K-H, Jeon J-S and An G (2023) Sucrose preferentially promotes expression of OsWRKY7 and OsPR10a to enhance defense response to blast fungus in rice. Front. Plant Sci. 14:1117023. doi: 10.3389/fpls.2023.1117023
Received: 06 December 2022; Accepted: 13 January 2023;
Published: 27 January 2023.
Edited by:
Xiaofeng Zhang, Fujian Agriculture and Forestry University, ChinaReviewed by:
Yifeng Wang, China National Rice Research Institute (CAAS), ChinaMazahar Moin, University of Hyderabad, India
Copyright © 2023 Tun, Yoon, Vo, Cho, Hoang, Peng, Kim, Win, Lee, Jung, Jeon and An. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gynheung An, Z2VuZWFuQGtodS5hYy5rcg==; Jong-Seong Jeon, amplb25Aa2h1LmFjLmty
 Win Tun
Win Tun Jinmi Yoon
Jinmi Yoon Kieu Thi Xuan Vo
Kieu Thi Xuan Vo Lae-Hyeon Cho
Lae-Hyeon Cho Trung Viet Hoang
Trung Viet Hoang Xin Peng
Xin Peng Eui-Jung Kim
Eui-Jung Kim Kay Tha Ye Soe Win1
Kay Tha Ye Soe Win1 Sang-Won Lee
Sang-Won Lee Ki-Hong Jung
Ki-Hong Jung Jong-Seong Jeon
Jong-Seong Jeon Gynheung An
Gynheung An