- 1Horticultural Sciences Department, University of Florida, Gainesville, FL, United States
- 2Citrus Research and Education Center, University of Florida, Lake Alfred, FL, United States
- 3Agronomy Department, University of Florida, Gainesville, FL, United States
Plant genomes are comprised of nuclear, plastid and mitochondrial components characterized by different patterns of inheritance and evolution. Genetic markers from the three genomes provide complementary tools for investigations of inheritance, genetic relationships and phenotypic contributions. Plant mitochondrial genomes are challenging for universal marker development because they are highly variable in terms of size, gene order and intergenic sequences and highly conserved with respect to protein-coding sequences. PCR amplification of introns with primers that anneal to conserved, flanking exons is effective for the development of polymorphic nuclear genome markers. The potential for plant mitochondrial intron polymorphisms to distinguish between congeneric species or intraspecific varieties has not been systematically investigated and is possibly constrained by requirements for intron secondary structure and interactions with co-evolved organelle intron splicing factors. To explore the potential for broadly applicable plant mitochondrial intron markers, PCR primer sets based upon conserved sequences flanking 11 introns common to seven angiosperm species were tested across a range of plant orders. PCR-amplified introns were screened for indel polymorphisms among a group of cross-compatible Citrus species and relatives; two Raphanus sativus mitotypes; representatives of the two Phaseolus vulgaris gene pools; and congeneric pairs of Cynodon, Cenchrus, Solanum, and Vaccinium species. All introns were successfully amplified from each plant entry. Length polymorphisms distinguishable by gel electrophoresis were common among genera but infrequent within genera. Sequencing of three introns amplified from 16 entries identified additional short indel polymorphisms and nucleotide substitutions that separated Citrus, Cynodon, Cenchrus and Vaccinium congeners, but failed to distinguish Solanum congeners or representatives of the Phaseolus vulgaris major gene pools. The ability of primer sets to amplify a wider range of plant species’ introns and the presence of intron polymorphisms that distinguish congeners was confirmed by in silico analysis. While mitochondrial intron variation is limited in comparison to nuclear introns, these exon-based primer sets provide robust tools for the amplification of mitochondrial introns across a wide range of plant species wherein useful polymorphisms can be identified.
Introduction
Plant genetic information is distributed among nuclear, plastid and mitochondrial genomes (Mahapatra et al., 2021; Camus et al., 2022), and genetic markers for each genome provide complementary tools for investigations of inheritance and evolution (Qiu et al., 1999; Duminil and Besnard, 2021; Besse, 2021; Camus et al., 2022). Although genome sequencing is the gold standard for such studies, convenient PCR-based markers retain utility and appeal (Egan et al., 2012; Hodel et al., 2016; Besse, 2021). The distinctive features of each genome necessitate different strategies for marker development and create different opportunities for application. Plant plastid genomes are relatively conserved in gene order, moderately conserved in coding sequences and more polymorphic with respect to introns and intergenic spacers (Wicke et al., 2011), facilitating the development of universal primers for the PCR amplification and subsequent characterization of more variable regions. Amplified plastid sequences such as rbcL, matK are the core of the DNA barcoding approach for distinguishing plant species (CBOL Plant Working Group, 2009), with the BOLD database facilitating applications (Ratnasingham and Hebert, 2007), and with intergenic spacer regions proving more variable and useful in distinguishing closer relatives (Shaw et al., 2014). Barcoding is also being combined with plastid genome sequencing for broader applicability and enhanced resolution (Tonti-Filippini et al., 2017). Plant mitochondrial markers provide an important adjunct to plastid DNA markers (Duminil and Besnard, 2021). Mitochondrial genotype can have a significant influence on plant phenotype (Bock et al., 2014; Colombatti et al., 2014; Hu et al., 2014; Dourmap et al., 2020). It is therefore important to be able to track mitochondrial contributions in sexual crosses and somatic cell fusions. Both plastid and plant mitochondrial genomes have uni-parental inheritance patterns, but these are not always concordant with respect to parent of origin (Camus et al., 2022). Moreover, horizontal gene transfer, observed in both organelle genomes, is especially prevalent in plant mitochondria (Keeling, 2009; Archibald and Richards, 2010). Plant mitochondrial gene coding sequences are, with some exceptions (Mower et al., 2007), highly conserved (Wolfe et al., 1987; Drouin et al., 2008), but genome size, gene order and intergenic sequences vary exensively between, and even within, plant species (Sloan, 2013; Gualberto and Newton, 2017). Mitochondrial restriction fragment length polymorphisms (RFLPs) are therefore readily detected within plant species (Levings and Pring, 1977; Palmer and Herbon, 1988), but the development of polymorphic PCR-based mitochondrial markers that work across a wide range of plant species is problematic. The lack of conserved gene order precludes the development of universal primer sets that will anneal to conserved coding sequences and amplify the highly polymorphic intergenic sequences.
Minisatellites and microsatellites (tandem repeats of 10 to 100, or less than 10 base pairs, respectively) identified within the sequenced mitochondrial genomes of some plant species have provided the basis for PCR-based polymorphic markers. Minisatellite repeat number polymorphisms have demonstrated intraspecific variation in Beta vulgaris, B. maritima (Nishizawa et al., 2000; Nishizawa et al., 2007), Picea abies (Sperisen et al., 2001; Bastien et al., 2003), Pinus banksiana (Godbout et al., 2005), and Pinus ponderosa (Mitton et al., 2000), as well as interspecific polymorphisms in Brassica and Oryza species (Honma et al., 2011). Interspecific, but not intraspecific, variation for a Gn microsatellite is present in the genus Pinus (Soranzo et al., 1999), whereas a compound, highly polymorphic microsatellite region reveals both intra- and interspecific variation in Abies (Jaramillo-Correa et al., 2013). Tandemly repeat mitochondrial loci are not generally conserved across diverse plant taxa and are not always polymorphic between related taxa, but recent work has identified extensive mitochondrial microsatellites among plant species (de Freitas et al., 2022; Xiong et al., 2022). These studies and databases of plant mitochondrial microsatellite repeats (Kumar et al., 2014; Sablok et al., 2015) facilitate the experimental search for loci that are polymorphic in specific taxa.
Plant mitochondrial introns present an under-explored approach for the development of more universal, PCR-based mitochondrial genome markers. PCR amplification of polymorphic introns with primers designed to conserved flanking exon sequences (Lessa, 1992) has allowed the development of nuclear genome markers in plant species having limited genomic information (Gupta et al., 2011; Li et al., 2012; Chandra et al., 2013; Kim et al., 2015) or limited genetic variability (Wang et al., 2010; Galeano et al., 2012). Angiosperm mitochondrial genomes encode 20-24 group II introns. Although sporadic intron loss is observed among evolutionary lineages, many of these introns are common to the sequenced angiosperm mitochondrial genomes (Kubo and Mikami, 2007), and flanked by conserved coding sequences that can be exploited for universal primer development. Laroche et al. (1997) surveyed the genomic sequences of six mitochondrial introns that were located within five genes and were common to five different angiosperm species and concluded that plant mitochondrial introns could provide a source of polymorphic markers. Across these species, base substitutions per site were higher within introns than within exons. Insertion-deletion (indel) polymorphisms were observed at 0.2-0.5 times the frequency of base substitutions. These sequence comparisons were made across a small set of diverse angiosperm genera, and so did not determine whether plant mitochondrial introns are commonly polymorphic between cross-compatible species or within species – situations in which polymorphisms could function as useful genetic markers. These points require investigation as correct splicing of plant organelle group II introns depends upon a complex intron secondary structure and upon RNA-protein interactions with multiple, co-evolving, nuclear-encoded splicing factors (Bonen, 2008; de Longevialle et al., 2010; Brown et al., 2014) –requirements that potentially constrain the degree of intron polymorphism that can be found among close relatives.
DNA markers based upon PCR-amplified plant mitochondrial intron sequences have proved useful in some cases. While most plant mitochondrial microsatellite and minisatellite repeats are located in intergenic regions, polymorphic examples are found within introns (Sperisen et al., 2001; Godbout et al., 2005; Jaramillo-Correa et al., 2013; Potter et al., 2013; Xiong et al., 2022). Duminil et al. (2002) designed primer pairs for the amplification of 16 different introns, based upon the mitochondrial genome sequences of Arabidopsis thaliana and Beta vulgaris. These primer sets amplify their corresponding introns in 20-28 of 28 diverse angiosperm species, and some have been investigated for polymorphisms in related species. The PCR amplified NADH dehydrogenase subunit 1 intron 2 (nad1i2), NADH dehydrogenase subunit 4 intron 1 (nad4i1) and intron 2 (nad4i2) are not polymorphic within Quercus robur, but distinguish between Q. robur and Q. rubra (Demesure et al., 1995). Notably, complex mitochondrial SSR loci analyzed across 88 genomes are especially prevalent in the introns of nad2, nad4 and nad7 genes (Xiong et al., 2022).
Mitochondrial intron polymorphisms also have utility in citrus breeding and genetics. Commercial citrus types are complex hybrids with at least three maternal lineages among them - Citrus maxima (pummelo), C. reticulata (mandarin) and C. medica (citron). The genus overall has complex taxonomy (Moore, 2001; Wu et al., 2018). Froelicher et al. (2011) amplified short, internal, regions of Citrus NADH dehydrogenase subunit 2 intron 3 (nad2i3), NADH dehydrogenase subunit 5 intron 2 (nad5i2), and NADH dehydrogenase subunit 7 intron 1 (nad7i1), with primers based upon A. thaliana and B. vulgaris mitochondrial genome sequences. Citrus and citrus relatives are polymorphic for indels in these introns, which collectively identify seven Citrus mitotypes. Intron-flanking primers designed from alignment of conserved DNA sequences flanking introns common to seven sequenced angiosperm mitochondrial genomes (Grosser, 2011) generate intron amplification products that distinguish C. maxima from C. reticulata (Satpute et al., 2015) and C. maxima from C. japonica (kumquat) (Omar et al., 2017). Here, we demonstrate the utility of these primer sets for amplification of their target introns not only in the previously studied C. maxima, C. reticulata and C. Japonica lineages, but also across diverse angiosperm species. We further investigate the amplified introns for indel and single nucleotide polymorphisms (SNPs) that distinguish mitochondrial genomes within a plant species or between congeneric plant relatives, wherein polymorphic mitochondrial markers have potential applications in studies of evolution and inheritance.
Materials and methods
Plant materials and DNA extraction
The plant materials used in this study (Table 1) were selected to explore primer amplification across across six angiosperm orders and to investigate whether intron amplification products could, at least, distinguish congener species of agricultural importance within these orders. These included two commercial Raphanus sativus mitotypes confirmed by PCR markers as described by Kim et al. (2007), representatives of the two major Phaseolus vulgaris gene pools (Bhakta et al., 2017), congener species representatives of Cenchrus, Citrus, Cynodon, Solanum and Vaccinium, along with Poncirus trifoliata, which is cross-compatible with Citrus species (Moreira et al., 2002) and considered by some to fall within the genus Citrus (Ollitrault et al., 2020). Citrus materials were from the University of Florida Citrus Research and Education Center, Lake Alfred, Florida and Harris Citrus Nursery, Lithia, FL. The Cynodon entries were from the USDA National Plant Germplasm System. The Phaseolus, Cenchrus, Solanum and Vaccinium entries were obtained from the University of Florida research programs of Dr. C.E. Vallejos, Dr. L. Sollenberger, Dr. C.E. Vallejos, and Dr. J. Olmstead, respectively. Total cellular DNA was extracted from leaf samples by a modification of the cetyl trimethylammonium bromide (CTAB) method in which 50 mg of tissue was combined with 750 μl of CTAB buffer (Murray et al., 1980) and 10 μg of DNase-free RNase A in a FastPrep™ Lysing Matrix A tube, disrupted for 40 s in a FastPrep®-24 Instrument (MP Biomedicals LLC, Santa Ana, CA) and incubated at 65°C for 5 min. Cellular and lysing matrix debris was removed by centrifugation at 13,000 xg for 10 min at room temperature. Supernatants were extracted with an equal volume of chloroform-isoamyl alcohol mixed in a ratio of 24:1. DNA was precipitated from the aqueous phase by the addition of a 2/3 volume of isopropyl alcohol and recovered by centrifugation at 13,000 xg for 10 min at room temperature. The pellets were washed in 750 μl of 70% ethanol, air dried and rehydrated in 80 ul of 1 mM Trizma base, 0.1 mM di-sodium ethylene diamine tetra acetic acid (Na2EDTA), 1 mM NaCl, pH 8. The concentration of DNA samples was determined from the absorbance at 260 nm.
DNA amplification and fractionation
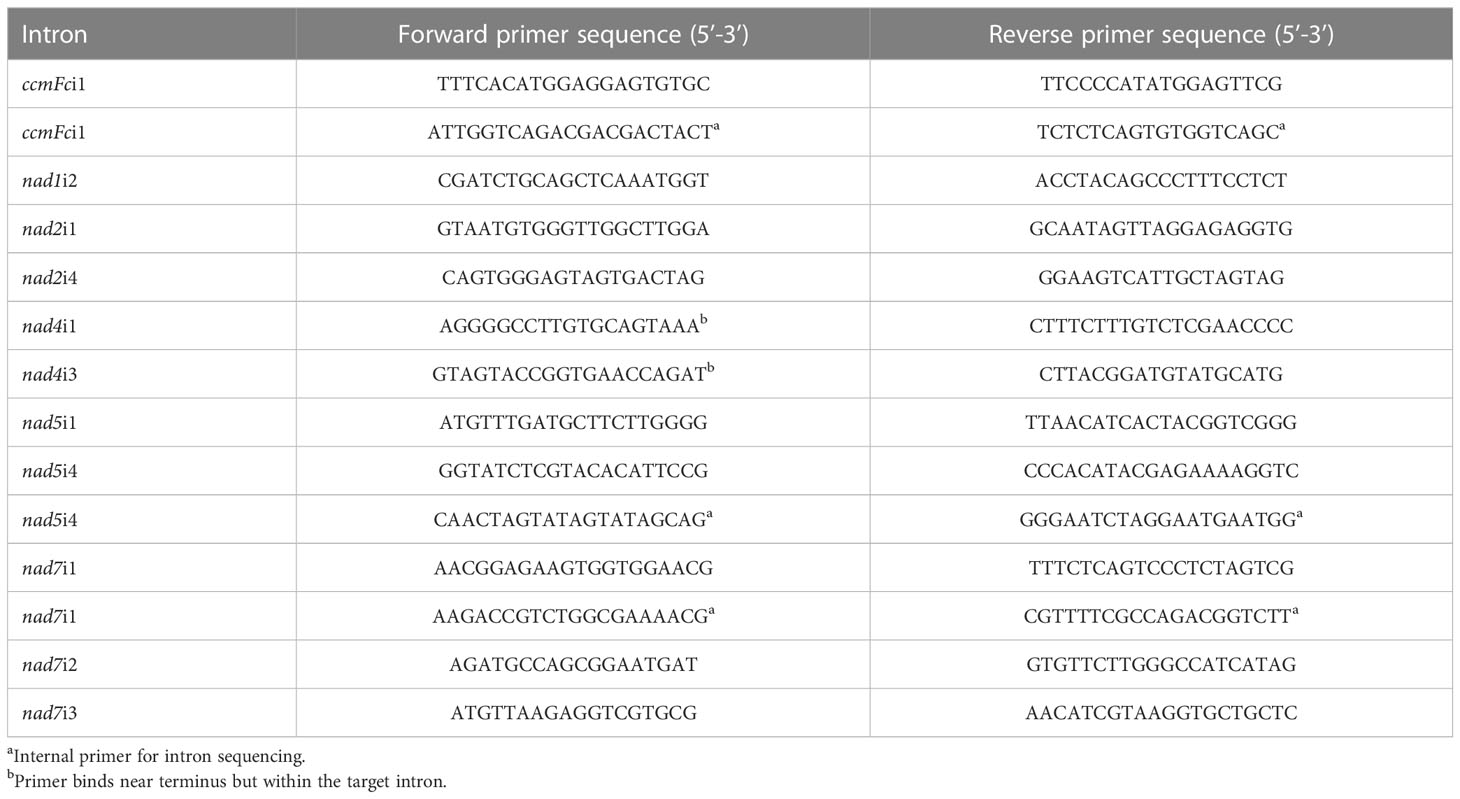
The PCR primers used in this work (Table 2; Grosser, 2011) were designed against introns of the mitochondrial nad1, nad2, nad4, nad5, nad7 and cyctochrome c maturation Fc (ccmFc) genes because these introns were common to seven plant species’ mitochondrial genomes: A. thaliana (Unseld et al., 1997), B. napus (Handa, 2003), B. vulgaris (Kubo et al., 2000), N. tabacum (Sugiyama et al., 2005), O. sativa (Notsu et al., 2002), T. aestivum (Ogihara et al., 2005), and Z. mays (Allen et al., 2007). The National Center for Biotechnology Information (NCBI) accession numbers for these genomes are NC_001284, NC_002511, NC_008285, NC_006581, NC_007886, NC_007579, and NC_007982, respectively (https://www.ncbi.nlm.nih.gov/genome/organelle/, accessed 1/20/2023). Primer pairs were designed manually based upon the highly conserved coding regions flanking intron sequences or, in some cases, from conserved sequences within introns.
PCR amplification reactions were performed on replicate DNA preparations made from different plants of each entry, with the exception of the two Cynodon entries. For these only a single pot culture was available, so replicate DNA extractions were prepared from the single culture of each. PCR reactions of 50 μl contained 25-100 ng of DNA, 0.2 µM of each primer, 0.125 mM dNTPs, 1.25 units of high fidelity, TAKARA EXTAQ Hot Start DNA polymerase (Clontech, Mountain View, CA) in 1X TAKARA Hot Start reaction buffer. This high-fidelity polymerase was selected due to the length of the amplified introns and the intent to sequence PCR products. Amplification was for 30 cycles of 1 min at 94°C, 2 min at 55°C, and 3 min at 72°C. Electrophoresis through 1% agarose gels was performed to survey PCR reactions for successful amplification. The DNA Hyperladder II (Bioline Inc., Cambridge, MA) was used as a size marker. Electrophoresis was at 100V for 100 min in Tris-Borate-EDTA (TBE) buffer (10 mM Trizma base, 10 mM boric acid, 2.5 mM Na2EDTA, pH 8.2). Gels were stained in 0.5 μg/ml ethidium bromide for 20 min and viewed over a UV transilluminator in a Molecular Imager® Gel Doc™ XR System (Bio Rad Laboratories, Inc. Hercules, CA). Gel images were captured with the Quantity One® 1-D Analysis Software (Bio Rad Laboratories, Inc.) and exported as.tif files. The AdvanCE™ FS96 capillary electrophoresis system (Advanced Analytical Technologies Inc., Ames, IA) was used to estimate the length of PCR amplification products in DNA base pairs (bp). Amplification products were diluted 1:15 in TE buffer (10 mM Trizma Base 1 mM Na2EDTA, pH 8) and fractionated by use of the DNF-915 dsDNA 915 Reagent Kit (Advanced Analytical Technologies Inc.) according to the supplier’s instructions. Indel polymorphisms were confirmed by electrophoresis of DNA amplification products, individually and mixed, through Criterion™ precast 5% polyacrylamide gels (Bio-Rad Laboratories Inc., Hercules, CA) run in TBE buffer for 740 Volt-h and imaged as described above.
DNA sequencing and sequence analysis
The amplification products of ccmFci1, nad5i4, nad7i1were purified for DNA sequencing through use of the QIAquick PCR Purification Kit (Qiagen Inc., Valencia, CA) according to supplier’s instructions. Purified amplification products were fully sequenced in both directions by the University of Florida Interdisciplinary Center for Biotechnology Research (ICBR) Sanger Sequencing Core Laboratory in Gainesville, FL or by Eurofins USA. Intron sequences and their corresponding GenBank Accession numbers are listed in Table 1. The sequences were aligned on the MultAlin web server (Corpet, 1988) (http://multalin.toulouse.inra.fr/multalin/, accessed 9/2/2022). Nucleotide substitutions per site (K0) were calculated by the formula of Kimura (1980) based upon pairwise alignments of sequences with all indels removed. Indels per site (I) were calculated as the number of indels in a pairwise alignment divided by the number of nucleotides in the alignment with indels removed (Laroche et al., 1997). Intron sequences found to differ between congener species were also analyzed for potential restriction fragment polymorphisms with the NEB cutter V 2.0 tool (Vincze et al., 2003) (http://nc2.neb.com/NEBcutter2/index.php, accessed 1/26/2023).
In silico prediction of intron amplification products
Prediction of intron amplification products across a wider range of plant taxa was performed through application of the Primer-BLAST tool (Ye et al., 2012) (https://www.ncbi.nlm.nih.gov/tools/primer-blast/) to selected plant mitochondrial genomes in the NCBI organelle genome database (https://www.ncbi.nlm.nih.gov/genome/organelle/) (both accessed 1/20/2023). Genomes queried included early andiosperms Magnolia biondii (NC_049134.1) (Dong et al., 2020) and Magnolia officinalis (NC_064401) (unpublished), which could potentially differ in sequence from later diverged andiosperms. Additional orders of monocots were selected to complement the single order (Poales) investigated experimentally. These included Allium cepa male-sterilizing (KU318712.1) (Kim et al., 2016) and normal (AP018390.1) (Tsujimura and Terachi, 2018) cytoplasms representing monocot order Asparagales; Cocos nucifera (KX028885.1) (Aljohi et al., 2016) representing monocot order Arecacales; and Zostera japonica (NC_068803.1) (Chen et al., 2022) and Zostera marina (KX808392.1)(Petersen et al., 2017) representing monocot order Alismatales. Also included were dicots Silene conica (JF40490.1-JF50629.1), Silene noctiflora (KP053825.1-KP053880.1), Silene latifolia (HM562727.1) and Silene vulgaris (JF750427.1-JF750430.1). Silene is an important model genus that includes species exhibiting unusual patterns of mitochondrial genome expansion and nucleotide substitution, which potentially affect primer performance and utility. Silene conica and Silene noctiflora provide tests of primers on expanded mitochondrial genomes that exhibit accelerated nucleotide substitution rates in comparison to Silene latifolia and Silene vulgaris (Sloan et al., 2012).
Results
Mitochondrial intron amplification across angiosperm taxa
The intron primer sets (Table 2) successfully amplified the target intron in each of the 19 entries investigated (Figure 1, Table 3). PCR reactions generally produced a single major product, although additional products of low abundance were detected for some nad2i1, nad5i4 and nad7i3 amplifications (Figure 1).
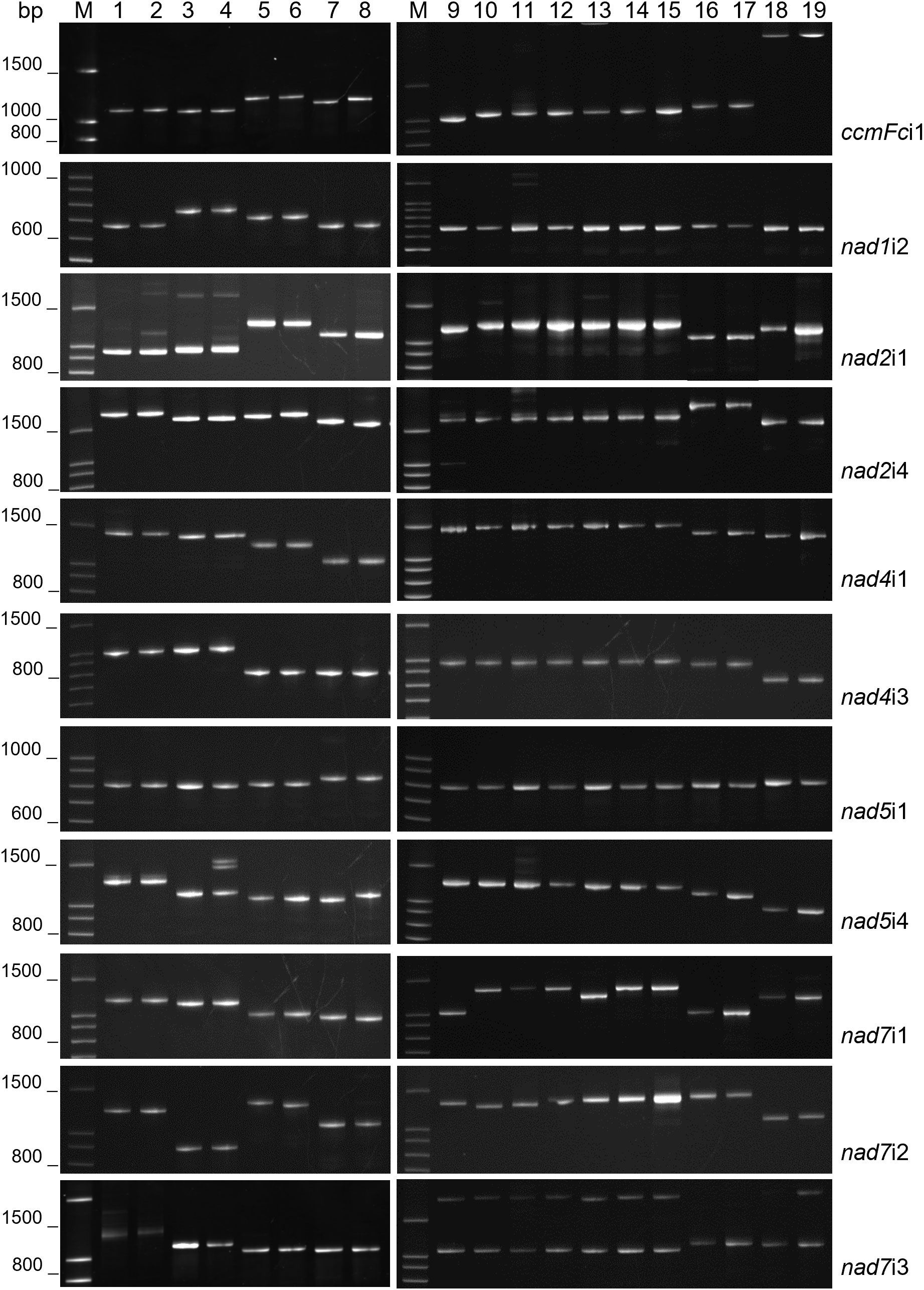
Figure 1 Mitochondrial intron lengths vary between but are conserved within plant genera. PCR amplification products of 11 plant mitochondrial introns were analyzed by polyacrylamide gel electrophoresis. M corresponds to a 100 base pair (bp) DNA ladder. DNA templates for PCR were as follows: 1) Solanum pinnellii LA716, 2) Solanum lycopersicon Bonny Best, 3) Raphanus sativus Red Velvet, 4) Raphanus sativus April Cross, 5) Cynodon dactylon Royal Cape, 6) Cynodon transvaalensis Frankenwald Fine, 7) Cenchrus americanus Tifleaf3, 8) Cenchrus purpureus Merkeron, 9) Poncirus trifoliata English Large Flower, 10) Citrus japonica Meiwa, 11) Citrus medica Etrog, 12) Citrus maxima Hirado Buntan, 13) Citrus reticulata Ponkan, 14) Citrus paradisi Ruby Red, 15) Citrus sinensis Valencia, 16) Vaccinium virgatum Tifblue, 17) Vaccinium corymbosum Blue Crop, 18) Phaseolus vulgaris Calima, 19) Phaseolus vulgaris Jamapa.
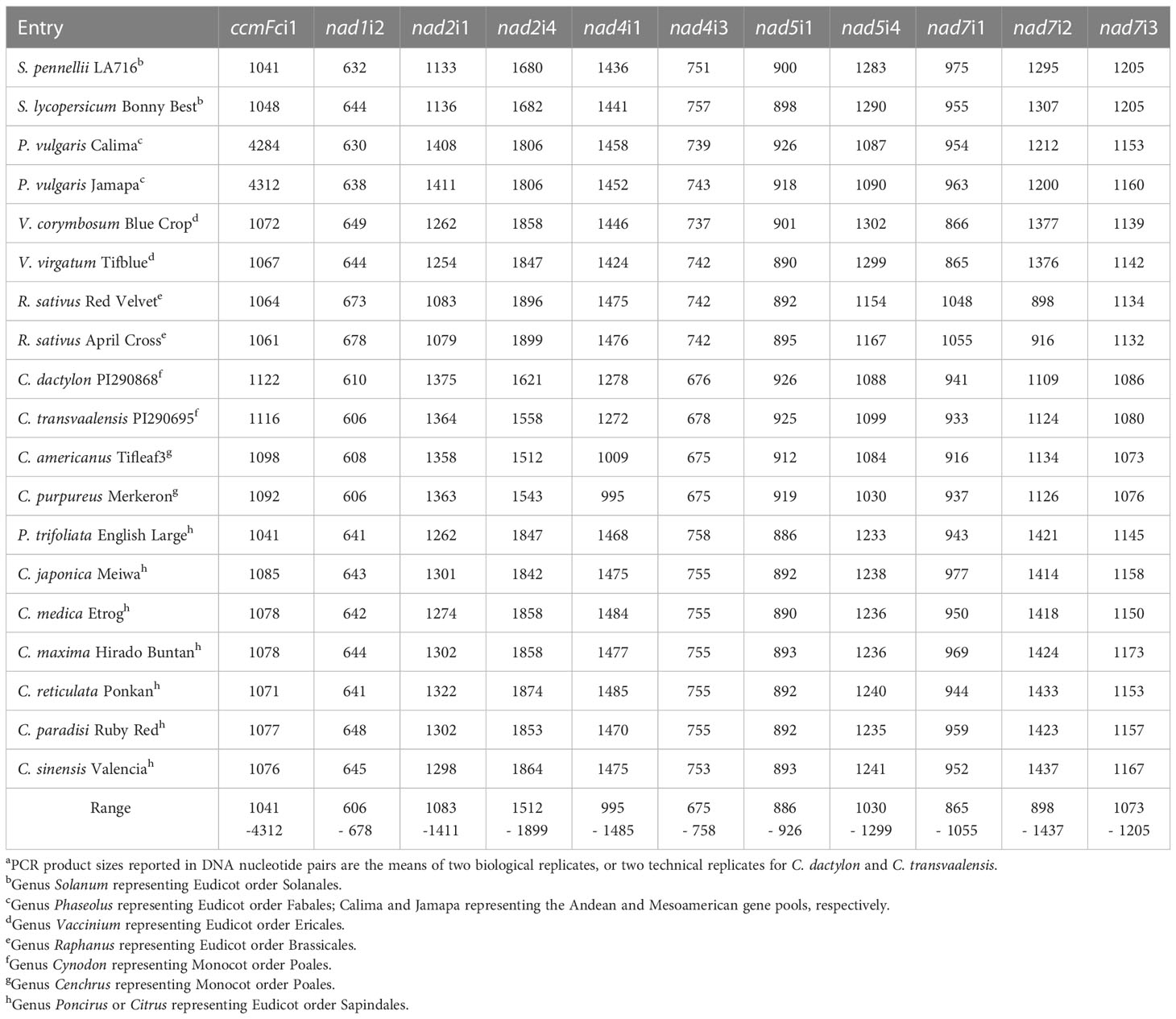
Table 3 Intron PCR product lengtha estimated by Advance™ capillary electrophoresis.
The increasing number of complete plant mitochondrial genome sequences enabled investigation of the potential for these primer sets to amplify target introns in additional taxa. Primer-BLAST analysis of selected fully sequenced mitochondrial genomes predicted successful application of the introns in early angiosperms represented by Magnolia biondii and Magnolia officionalis; additional orders of monocots represented by Allium cepa, Cocos nucifera, Zoster japonica and Zoster marina; and an additional order of dicots represented by Silene conica, Silene latifolia, Silene noctifolora and Silene vulgaris (Table 4). Of the 121 primer-accession combinations tested, 79 predicted a single amplification product produced by perfectly matched primers. An additional 18 combinations predicted a single amplification product produced by primers with only one or two mis-matched nucleotides between the target genome and primer set. The 11 primer sets are therefore predicted to be useful for the amplification of mitochondrial introns across the angiosperms. Primers were predicted to be less effective for plant mitochondrial genomes that exhibit exceptionally high rates of genome expansion and nucleotide substitution. Silene conica and Silene noctiflora represent expanded mitochondrial genomes with accelerated nucleotide substitution rates in comparison to Silene latifolia and Silene vulgaris (Sloan et al., 2012). While all primer sets were predicted to amplify single products in Silene latifolia and Silene vulgaris, most primer sets predicted multiple, weak matches to Silene conica and Silene noctiflora. Nevertheless, 3-4 primer sets were still predicted to work well for these two templates (Table 4).
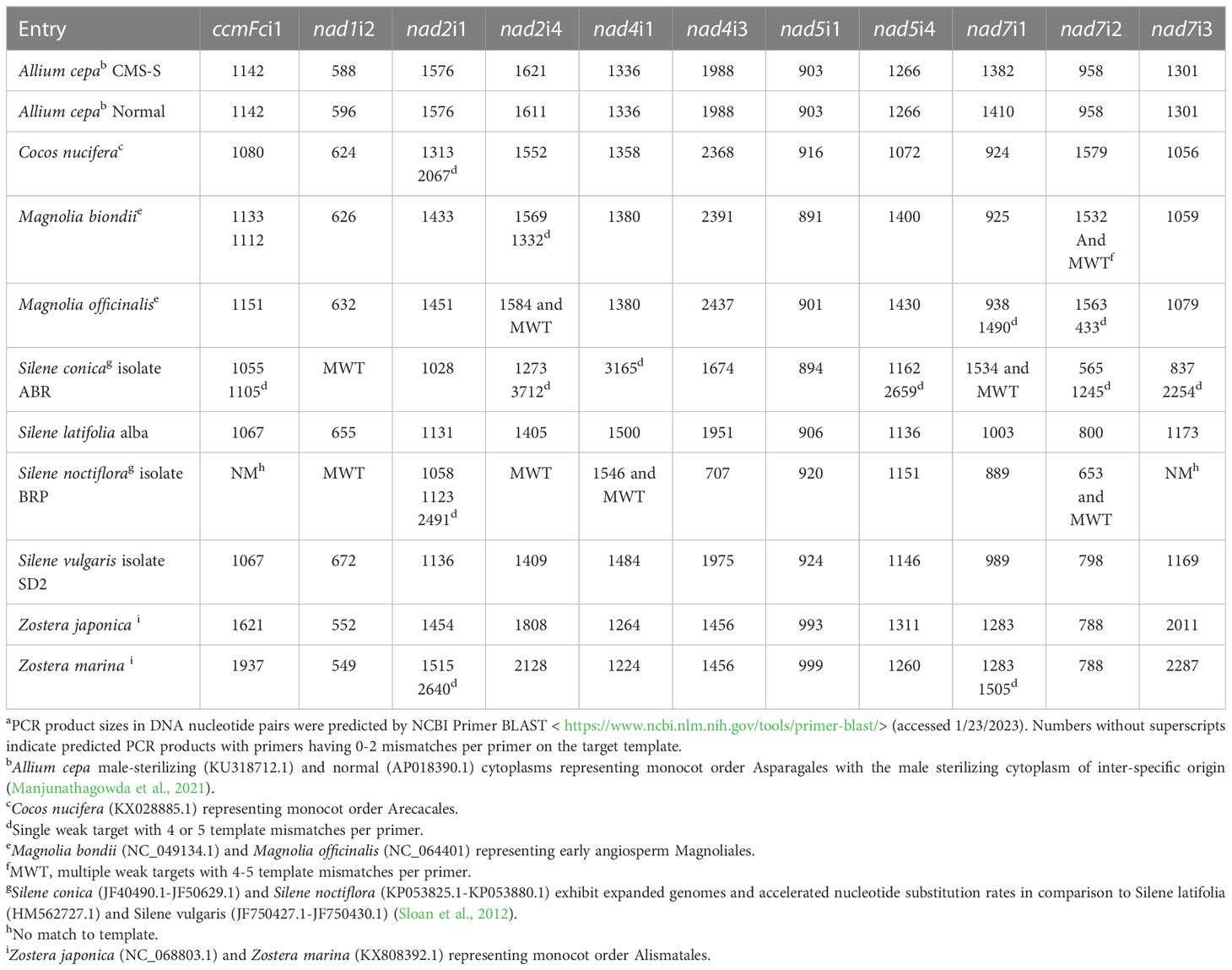
Table 4 Intron PCR product length predicted by Primer-BLASTa.
Intron length polymorphisms
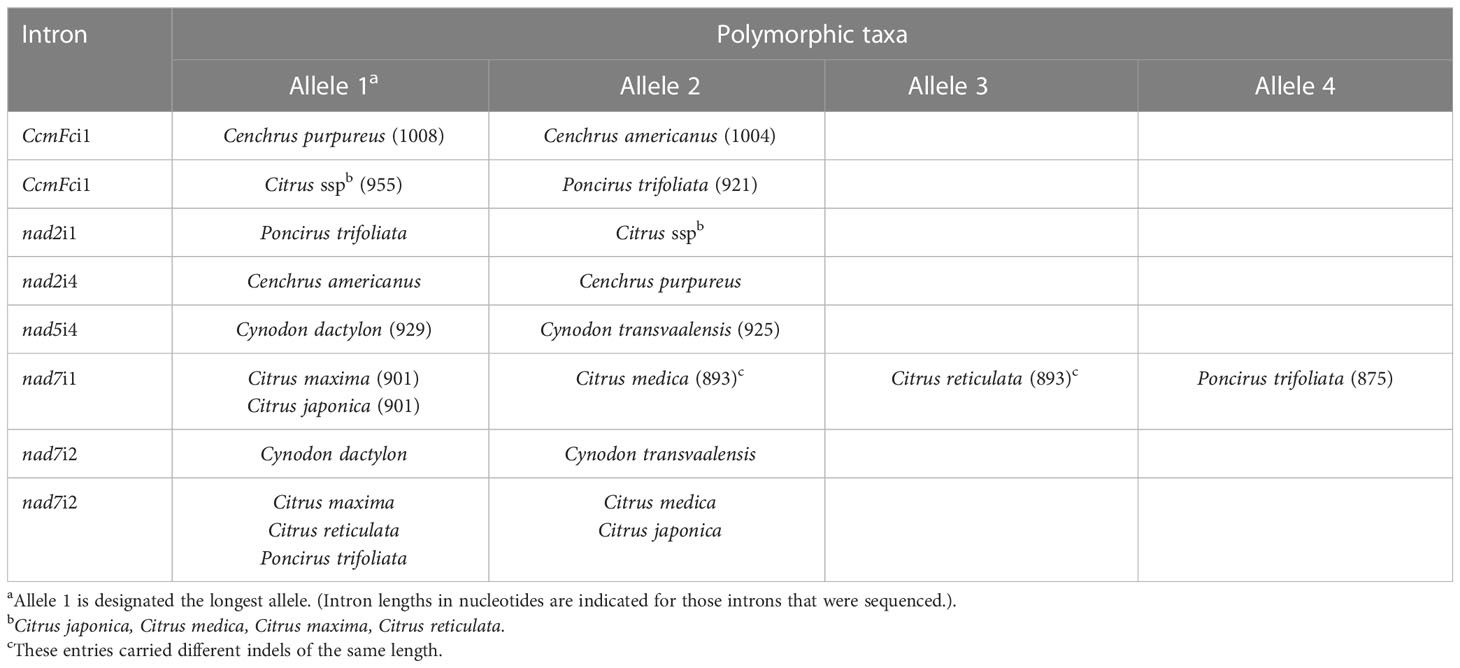
The fractionation of experimentally produced intron amplification products by gel electrophoresis (Figure 1) and AdvanCE™ FS96 capillary electrophoresis (Table 3) demonstrated significant intron length variation among diverse angiosperm genera, in agreement with primer-BLAST observations (Table 4). Intron lengths, as estimated by the AdvanCE™ capillary technique, varied across genera by as few as 40 nucleotides in the case of nad5i1 to as many as 539 nucleotides in the case of nad7i2. This was excluding the extreme size (4284 nucleotides) of Phaseolus ccmFci1, which likely reflects a split intron. Length polymorphisms between congener species were, however, few in number and challenging to detect by electrophoresis. The well-to-well variation of the AdvanCE™ FS96 precluded use of length values to detect small indel polymorphisms in relatively large DNA amplification products. Length polymorphisms were identified by fractionation of amplification products on polyacrylamide gels (Figure 1) and confirmed by acrylamide gel electrophoresis of PCR product mixtures (Figure 2) for congeners of Cenchrus (ccmFci1 and nad2i4), Cynodon (nad7i2) and Citrus (nad7i1 and nad7i2) species. Intron length polymorphisms are summarized in Table 5. The three Citrus maternal lineages and C. japonica were individually distinguished by the combination of nad7i1 and nad7i2 polymorphisms. C. paradisi (grapefruit) and C. sinensis (orange) were not distinguished from their respective C. maxima and C. reticulata maternal lineages. Citrus species were distinguished from P. trifoliata by length polymorphisms in ccmFci1, nad2i1, nad7i1, and nad7i2. Electrophoresis did not, however, distinguish the introns of the two Vaccinium or Solanum species, Phaseolus gene pools, or Raphanus sativus mitotypes.
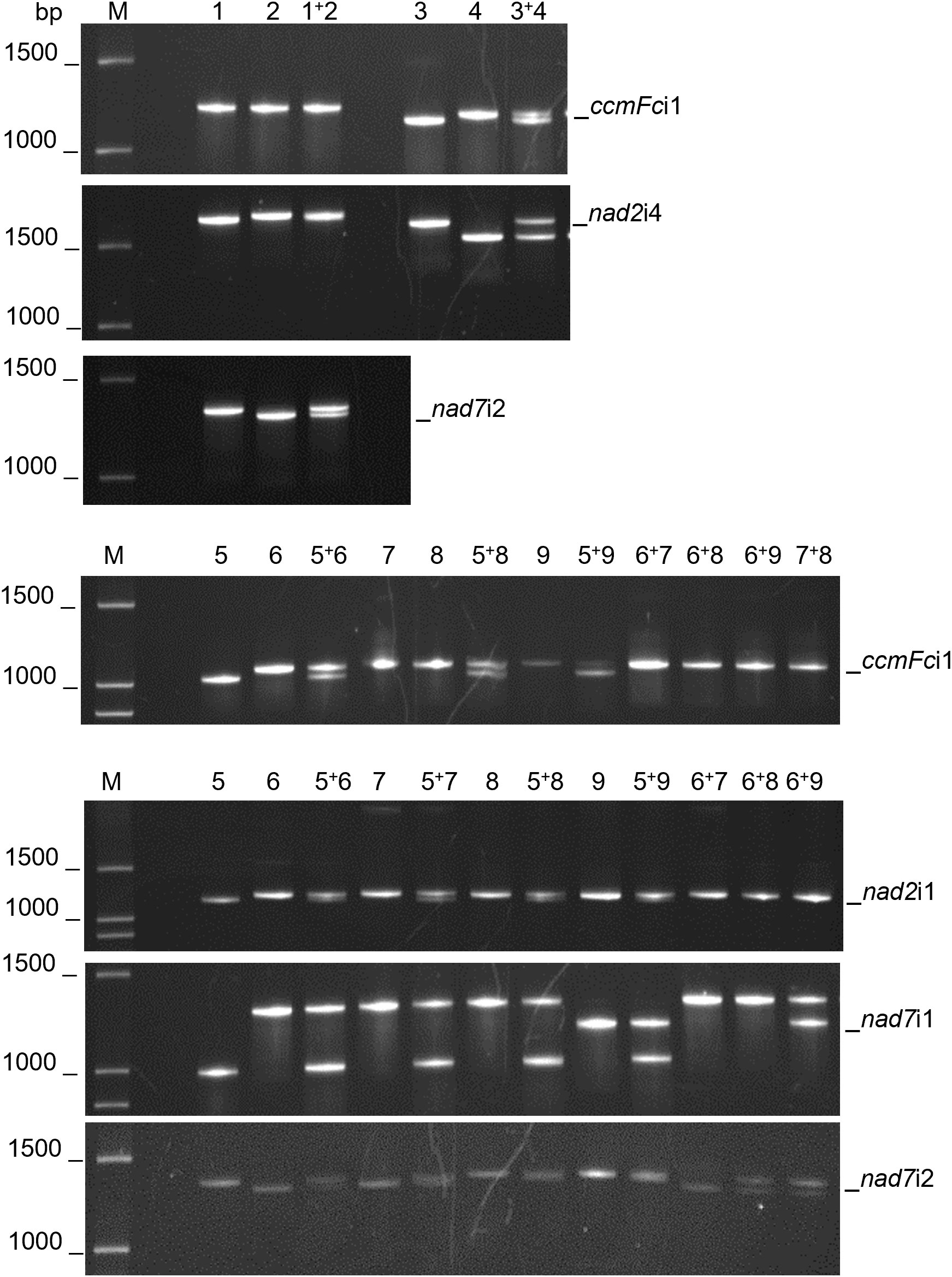
Figure 2 Mitochondrial intron length polymorphisms that distinguish related species. PCR amplification products of mitochondrial introns were separated by polyacrylamide gel electrophoresis. PCR products were analyzed individually and as mixtures to confirm the indel polymorphisms. M corresponds to a 100 base pair (bp) DNA ladder. DNA templates for PCR were as follows: 1) Cynodon dactylon Royal Cape, 2) Cynodon transvaalensis Frankenwald Fine, 3) Cenchrus americanus Tifleaf3, 4) Cenchrus purpureus Merkeron, 5) Poncirus trifoliata English Large Flower, 6) Citrus japonica Meiwa, 7) Citrus medica Etrog, 8) Citrus maxima Hirado Buntan, 9) Citrus reticulata Ponkan. Polymorphisms between Cenchrus spp. were confirmed for ccmFci1 and nad2i4 and between Cynodon spp. for nad7i2. CcmFci1, nad2i1, and nad7i1 polymorphisms differentiated P. trifoliata (5) from Citrus species (7-9). Nad7i1 also distinguished C. reticulata (9) from C. japonica, C. medica and C. maxima (6-8), whereas nad7i2 polymorphisms distinguished P. trifoliata, C. maxima and C. reticulata (5, 8, 9) from C. japonica and C. medica (6, 7).
Primer-BLAST demonstrated that short length polymorphisms often distinguish congener species’ mitochondrial introns (Table 4). In Allium, introns differing by 8, 10 and 28 nucleotides distinguished the male sterilizing cytoplasm, derived by interspecific introgression (Manjunathagowda et al., 2021), from the normal cytoplasm. Magnolia biondii and Magnolia officinalis varied in seven introns with length differences ranging from 6 to 46 nucleotides. Silene vulgaris differed from Silene latifolia in nine introns having length variations ranging from 4-24 nucleotides. Zoster japonica and Zoster marina were polymorphic with respect to length in eight introns. While five of these differences ranged from 3-61 nucleotides, length polymorphisms of 316, 320 and 276 nucleotides were predicted for ccmFci1, nad2i4 and nad7i3, respectively. DNA sequence information clearly allows detection of mitochondrial intron length polymorphisms that distinguish related plant species.
Intron sequence analysis
CcmFci1, nad5i4 and nad7i1 introns amplified from 16 entries were sequenced to further characterize indels detected by electrophoresis and to search for additional indels, along with SNPs (Figures S1, S2, and S3, respectively). Citrus sinensis (sweet orange with the C. reticulata maternal lineage) was not included, and heteroplasmy or seed mixtures in the two commercial Raphanus sativus accessions precluded obtaining quality sequences for comparison of these two mitotypes within this species. With respect to intra-species variation, nad5i4 and nad7i1 sequences did not distinguish the two gene pools of Phaseolus vulgaris. The Phaseolus ccmFci1 shared 632 5’ nucleotides and 133 3’ nucleotides with other species separated by a 3353 nucleotide insertion (Figure S1B). The two Phaseolus accessions were polymorphic for one SNP and a 4 base indel within the 3353 nucleotide insertion, but were not polymorphic with respect to the intron regions. Moreover, the three C. paradisi introns were not polymorphic with respect to those of their C. maxima maternal ancestor. Sequencing further characterized indels detected by gel electrophoresis and revealed additional length polymorphisms (Table 5). The ccmFci1 length polymorphism differentiating P. trifoliata from Citrus entries was due to separate deletions of 8, 9, and 17 nucleotides in P. trifoliata compared to Citrus (Figure S1A). Similarly, the polymorphism in nad7i1 was caused by separate deletions of 9, 8, and 9 nucleotides in P. trifoliata relative to C. maxima, C. medica and C. japonica. C. reticulata shared the 8 nucleotide deletion with P. trifoliata, while C. medica carried a unique 8 nucleotide deletion (Figure S3). The ccmFci1 sequence distinguishing Cenchrus congeners was a 4 nucleotide indel (Figure S1A). Additional length polymorphisms identified by sequencing included a 4 nucleotide nad5i4 indel that distinguished Cynodon congeners (Figure S2) and a 4 nucleotide nad7i1 that distinguished Cenchrus congeners (Figure S3). Sequencing did not reveal indel polymorphisms between Vaccinium or Solanum congeners.
Sequences of three introns identified only nine SNPs that distinguished congener species (Table 6). The nad5i4 sequence alignment (Figure S2) revealed a SNP that distinguished V. corymbosum from V. virgatum. This was the only Vaccinium polymorphism identified in this study. Three nad5i4 SNPs distinguished C. dactylon from C. transvaalensis (Figure S2). In addition to the nad7i1 indels, a nad7i1 SNP was found to distinguish C. reticulata from other Citrus species (Figure S3). Of the nine SNPs, only one was a C/T difference that could possibly be erased at the RNA level by plant mitochondrial C-to-T RNA editing. In these comparisons, the frequency of SNPs per site (K0) within genera was low - zero in the case of ccmFci1. The average K0 for nad5i4 and nad7i1 in congeneric species comparisons was 0.03 and 0.01, respectively, that of comparisons among dicot genera (Table 7). The frequency of indels per site (I) within genera was also low, 0.02-0.10 of I for comparisons among dicot genera (Table 7).
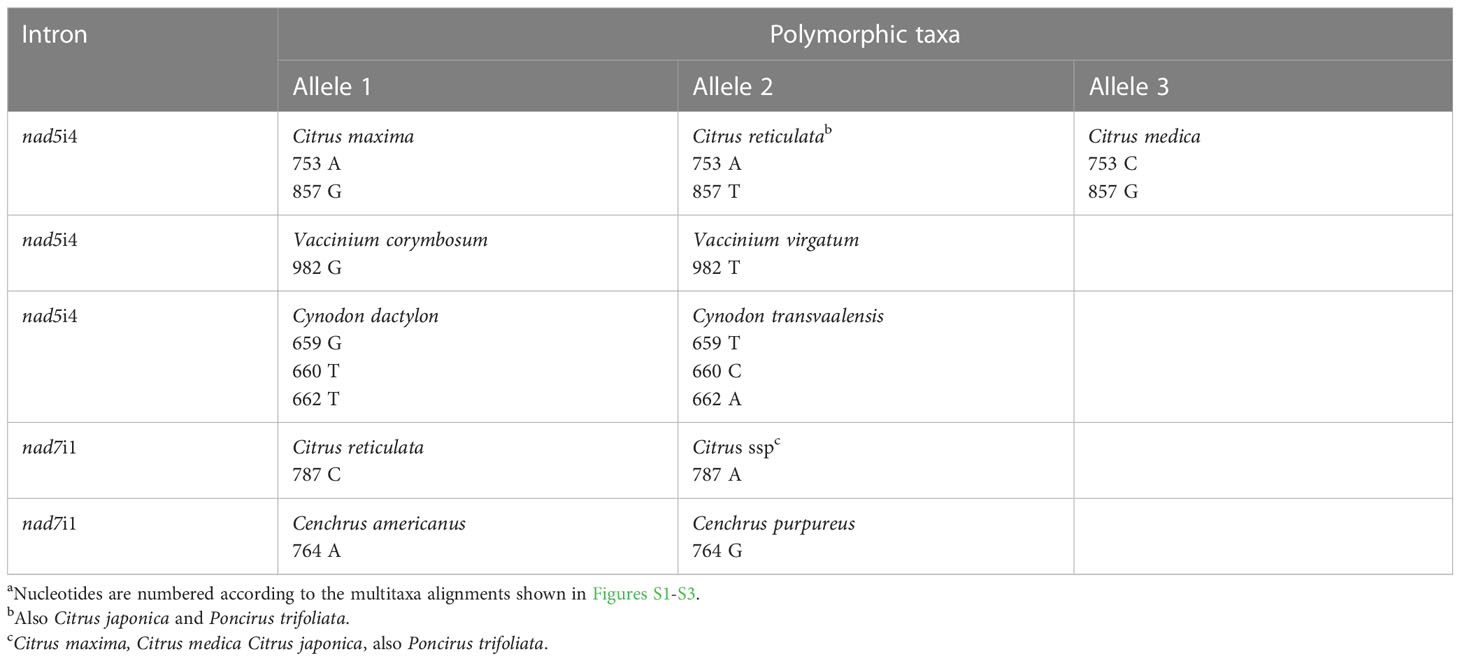
Table 6 Intron nucleotidea polymorphisms between congener species.
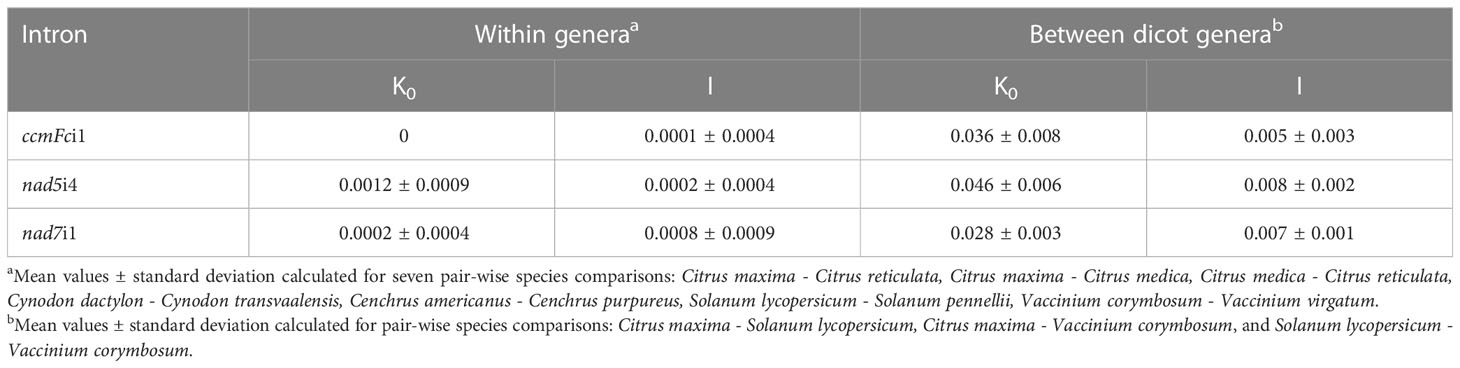
Table 7 Average nucleotide substitutions (K0) and indels (I) per site within genera and between dicot generab.
While sequence analysis is the most direct means of identifying length and SNP variation in amplified introns, these polymorphisms also create restriction pattern differences. Analysis of sequenced introns with NEB Cutter (Table S1) associated unique restriction patterns with the variant alleles reported in Tables 5 and 6. The only exception was the SNP that distinguished Vaccinium corymbosum and Vaccinium virgatum nad5i4 created no RFLPs across the 112 enzymes predicted by the NEB Cutter tool to cut these templates.
Discussion
Universal primers for amplification of plant mitochondrial introns
The 11 PCR primer sets used in this work demonstrated robust amplification of the target mitochondrial introns across 16 species representing eight plant genera and seven plant orders. Primer-BLAST analysis with these same primer sets predicted successful amplification of mitochondrial introns from early angiosperms and additional orders of monocots and dicots. This expands and improves the available universal primers for plant mitochondrial introns (Demesure et al., 1995; Dumolin-Lapegue et al., 1997; Duminil et al., 2002). Aleksić (2016) found limited applicability of previously developed universal mitochondrial primers to legume (Fabaceae) species and suggested family-specific primers as a more practical approach. The primer sets employed here successfully amplified the mitochondrial introns of P. vulgaris as a representative legume. Previous universal primer design strategies (Duminil et al., 2002; Froelicher et al., 2011) utilized mitochondrial sequences conserved between A. thaliana and B. vulgaris only. Primer design based on conserved introns and flanking sequences from seven plant species (Grosser, 2011) likely contributed to the extended applicability of the current primer sets. Although most plant species’ mitochondrial genomes evolve slowly with respect to coding sequences (Wolfe et al., 1987; Palmer and Herbon, 1988), plant genera containing taxa with widely varying rates of mitochondrial nucleotide substitution have been identified (Cho et al., 2004; Parkinson et al., 2005; Mower et al., 2007; Sloan et al., 2009). Primer-BLAST analysis did predict that some, but not all, of the 11 primer sets would work reliably on the Silene species having rapidly evolving mitochondrial coding sequences. A further complication with Silene conica and Silene noctiflora is that their highly expanded genomes apparently contain multiple, degenerate targets for the intron flanking primers (Table 4).
Intron polymorphism between and within genera
Mitochondrial intron length polymorphisms detectable by electrophoretic techniques were frequently observed between genera, whereas comparisons within genera revealed primarily short intron length variations. Large indels are therefore tolerated within introns, but rates of such variation are low within genera. These contrasting observations likely reflect the evolutionary processes that shaped modern plant organellar group II introns from their self-splicing, progenitor introns. On the one hand, altered intron sequences combined with novel nuclear and organelle-encoded splicing factors to maintain competence for splicing while shifting away from the group II ribozymic, self-splicing structures (Bonen, 2008; Brown et al., 2014). At the same time, the requirement for splicing factors to evolve in concert with the intron structure likely constrained variants that can be successfully spliced (de Longevialle et al., 2010; Zimmerly and Semper, 2015). Plant organelle introns retain significant common structural features (Bonen, 2008). Moreover, they reside within genes essential to photosynthesis or respiration, creating selective pressure for the maintenance of efficient splicing (Brown et al., 2014; Zimmerly and Semper, 2015; Best et al., 2020). Arrays of protein factors are required for the splicing of plastid and mitochondrial introns. These include members of the maturase family, descended from the maturases encoded in ribozymic, self-splicing group II introns (Schmitz-Linneweber et al., 2015), along with APO, CRM, PORR, PPR and TERF families of RNA binding proteins. With the exception of one plastid and one mitochondria-encoded maturase, these proteins are encoded by the nuclear genome and imported into the organelles where they act in combinatorial fashion for the splicing of particular introns or groups of introns (de Longevialle et al., 2010; Brown et al., 2014; Zimmerly and Semper, 2015; Wang et al., 2022). The complexity and specificity of this process may explain the lack of large intron indels found within genera.
The plant mitochondrial intron length differences between congener species as predicted by by Primer-BLAST averaged 17 nucleotides, excepting the three Zoster introns with larger differences. The experimentally characterized indels that distinguished congeners or cross-compatible species averaged less than 10 nucleotides in length, necessitating high-resolution acrylamide gels or DNA sequencing for discernment. Gel-resolved intron length polymorphisms differentiated Citrus, Cenchrus, and Cynodon congeners, but intron sequencing provided a more accurate picture of indel polymorphisms. The nad7i1 amplicons of C. medica and C. reticulata, for example, carried different indels of the same length. Even when larger intron size differences were apparent within genera, sequencing revealed them to result from multiple short indels. This is consistent with prior reports that short indels (1-10 bp) comprise greater than 50% of indels in plant mitochondrial introns and probably originate from slipped strand mispairing events during replication (Laroche et al., 1997).
For each of the three introns sequenced in the present study, the average frequency of nucleotide substitutions per site (K0) was also low within genera, but SNPs that distinguished congeners of Citrus, Cynodon, Cenchrus, and Vaccinium were identified. These can serve as useful markers through workflows such as cleaved amplified polymorphic sequence (CAPS), PCR combined with sequencing, or amplification refractory mutation analysis strategies (Lo, 1998; Ciarmiello et al., 2013). With one exception, SNPs and short indels that distinguished congeners’ introns also created CAPS markers (Table S1). More broadly, K0 values for comparisons between Citrus, Vaccinium and Solanum as representative dicot genera (Table 7) were similar to those reported by Laroche et al. (1997) for comparisons of six mitochondrial introns between two to three dicot genera. In the present study, K0 values for intron sequence comparisons between congeneric species were 0.01-0.03 times those for comparisons between dicot genera. The low nucleotide substitution rates likely result from the low frequency of nucleotide substitutions characteristic of most plant mitochondrial genomes, typically three to ten times lower than nuclear nucleotide substitution rates (Wolfe et al., 1987; Palmer and Herbon, 1988; Drouin et al., 2008).
The limited variation of mitochondrial introns within plant genera contrasts with the extensive diversity of nuclear introns, which show a high frequency of length and substitution polymorphisms within species of Oryza (Wang et al., 2005), Solanum (Wang et al., 2010), Allium (Jayaswal et al., 2019) and Medicago (Shilpa and Lohithaswa, 2021) among others. While no mitochondrial intron polymorphisms distinguished Solanum lycopersicum from S. pennellii, two studies document extensive nuclear intron polymorphisms within Solanum lycopersicum (Van Deynze et al., 2007; Wang et al., 2010). Organellar group II introns are considered the ancestors of nuclear introns, which lack folding constraints because they share the use of spliceosomal RNAs that have taken on the functions of the group II intron domains (Sharp, 1991). The spliceosome is a complex that is highly malleable in order to accommodate diverse exon ends and alternative splicing, perhaps permitting more varied intron sequences (Chen and Moore, 2014).
Application of plant mitochondrial intron polymorphisms
When present, organelle intron polymorphisms have valuable applications for determining inheritance in sexual crosses or somatic hybridizations. The markers investigated in this study have proved useful for determining organelle inheritance in Citrus cybrids. Cybrids are produced spontaneously as a by-product of protoplast fusion and are characterized by the diploid nuclear genome of the mesophyll fusion partner, the mitochondrial genome of the embryogenic callus partner, and random inheritance of chloroplast DNA (Grosser et al., 1996; Cabasson et al., 2001; Guo et al., 2004; Guo et al., 2013). This contrasts with typical protoplast fusion products, which possess tetraploid nuclei inherited from both parents. Cybrids provide a means to quickly create novel combinations of nuclear and organellar genotypes and to evaluate their phenotypic consequences. Specific organelle genotypes are associated with beneficial traits in cybrids. For example, grapefruit cybrids with mandarin mitochondrial DNA exhibit an extended season of high-quality fruit (Satpute et al., 2015), whereas grapefruit cybrids with kumquat plastid DNA exhibit increased resistance to citrus canker regardless of mitochondrial origin (Omar et al., 2017). The nad7i1 and nad7i2 primer sets (Table 2) were utilized, respectively, for verification and characterization of mitochondrial DNA inheritance in these two sets of cybrids, illustrating application for mitochondrial intron markers.
The currently reported nad7i1 marker overlaps with and confirms one of the three markers that Froelicher et al. (2011) demonstrated to be polymorphic in Citrus. Because our primer set amplified the entire intron, a new SNP was added to the previously published indels. Moreover, the list of intron markers polymorphic for Citrus species was expanded to include nad7i2, ccmFc, and nad2i1. The additional polymorphic markers did not, however, further distinguish differences within the seven citrus mitotypes identified by Froelicher et al. (2011). For example, C. maxima and its maternal derivative C. paradisi remained indistinguishable for all introns sequenced in this study.
Due to the lack of conserved gene order among plant mitochondrial genomes, often even between closely related taxa, assembling plant mitochondrial genome sequences presents special challenges and can preclude the universal application of intergenic sequences for distinguishing between closely related groups (Duminil and Besnard, 2021). Mitochondrial intron markers have demonstrated applicability in studies of population genetics, genotype characterization, detection of past hybridizations, and biogeographic studies of gene pool distributions (Ciarmiello et al., 2013; Aizawa et al., 2014; Xiang et al., 2014; Kersten et al., 2015). The current study documents a widely applicable set of primers for the mitochondrial marker toolbox and provides insights into the conservation and variation of plant mitochondrial introns.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, OP800658-OP800705.
Author contributions
CC, FG, JGro, and JGra: designed the study and collected the genetic materials. KC, KL, MG, SS, MM and YL: conducted the research. CC and MG: wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This research was funded by the University of Florida Institute of Food and Agricultural Science. MG and SS were supported by the University of Florida University Scholars Program. MMM was supported by the Hunt Brothers Fellowship and the Ciencia sem Fronteiras Award 1245/13-9.
Acknowledgments
We thank Drs. Jim Olmstead, Lynn Sollenberger and Eduardo Vallejos for sharing genetic materials used in this study.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1116851/full#supplementary-material
References
Aizawa, M., Yoshimaru, H., Takahashi, M., Kawahara, T., Sugita, H., Saito, H., et al. (2014). Genetic structure of Sakhalin spruce (Picea glehnii) in northern Japan and adjacent regions revealed by nuclear microsatellites and mitochondrial gene sequences. J. Plant Res. 128, 91–102. doi: 10.1007/s10265-014-0682-7
Aleksić, J. M. (2016). Family-specific vs. universal PCR primers for the study of mitochondrial DNA in plants. Genetika 48, 777–798. doi: 10.2298/GENSR1602777A
Aljohi, H. A., Liu, W., Lin, Q., Zhao, Y., Zeng, J., Alamer, A., et al. (2016). Complete sequence and analysis of coconut palm (Cocos nucifera) mitochondrial genome. PloS One 10. doi: 10.1371/journal.pone.0163990
Allen, J. O., Fauron, C. M., Minx, P., Roark, L., Oddiraju, S., Lin, G. N., et al. (2007). Comparisons among two fertile and three male-sterile mitochondrial genomes of maize. Genetics 177, 1173–1192. doi: 10.1534/genetics.107.073312
Archibald, J. M., Richards, T. A. (2010). Gene transfer: anything goes in plant mitochondria. BMC Biol. 8, 147. doi: 10.1186/1741-7007-8-147
Bastien, D., Favre, J. M., Collignon, A. M., Sperisen, C., Jeandroz, S. (2003). Characterization of a mosaic minisatellite locus in the mitochondrial DNA of Norway spruce [Picea abies (L.) karst.]. Theor. Appl. Genet. 107, 574–580. doi: 10.1007/s00122-003-1284-2
Besse, P. (2021). “Guidelines for the choice of sequences for molecular plant taxonomy,” in Molecular plant taxonomy. methods in molecular biology, vol. vol. 2222 . Ed. Besse, P. (New York: Humana). doi: 10.1007/978-1-0716-0997-2_2
Best, C., Mizrahi, R., Ostersetzer-Biran, O. (2020). Why so complex? the intricacy of genome structure and gene expression, associated with angiosperm mitochondria may relate to the regulation of embryo quiescence or dormancy-intrinsic blocks to early plant life. Plants 9, 598. doi: 10.3390/plants9050598
Bhakta, M. S., Gezan, S. A., Michelangeli, C. J. A., Carvalho, M., Zhang, L., Jones, J. W., et al. (2017). A predictive model for time-to-flowering in the common bean based on QTL and environmental variables. G3: Genes Genomes Genet. 7, 3901–3912. doi: 10.1534/g3.117.300229
Bock, D. G., Andrew, R. L., Rieseberg, L. H. (2014). On the adaptive value of cytoplasmic genomes in plants. Mol. Ecol. 20, 4899–4911. doi: 10.1111/mec.12920
Bonen, L. (2008). Cis-and trans-splicing of group II introns in plant mitochondria. Mitochondrion 8, 26–34. doi: 10.1016/j.mito.2007.09.005
Brown, G. G., des Francs-Small, C. C., Ostersetzer-Biran, O. (2014). Group II intron splicing factors in plant mitochondria. Front. Plant Sci. 5. doi: 10.3389/fpls.2014.00035
Cabasson, C. M., Luro, F., Ollitrault, P., Grosser, J. W. (2001). Non-random inheritance of mitochondrial genomes in Citrus hybrids produced by protoplast fusion. Plant Cell Rep. 20, 604–609. doi: 10.1007/s002990100370
Camus, M. F., Alexander-Lawrie, B., Sharbrough, J., Hurst, G. D. D. (2022). Inheritance through the cytoplasm. Heredity 129, 31–43. doi: 10.1038/s41437-022-00540-2
CBOL Plant Working Group (2009). A DNA barcode for land plants. Proc. Natl. Acad. Sci. U. S. A. 106, 12794–127987. doi: 10.1073/pnas.0905845106
Chandra, A., Jain, R., Solomon, S., Shrivastava, S., Roy, A. K. (2013). Exploiting EST databases for the development and characterisation of 3425 gene-tagged CISP markers in biofuel crop sugarcane and their transferability in cereals and orphan tropical grasses. BMC Res. Notes 6, 47. doi: 10.1186/1756-0500-6-47
Chen, W., Moore, M. J. (2014). The spliceosome: Disorder and dynamics defined. Curr. Opin. Struct. Biol. 24, 141–149. doi: 10.1016/j.sbi.2014.01.009
Chen, J., Zang, Y., Liang, S., Xue, S., Shang, S., Zhu, M., et al. (2022). Comparative analysis of mitochondrial genomes reveals marine adaptation in seagrasses. BMC Genom 23. doi: 10.1186/s12864-022-09046-x
Cho, Y., Mower, J. P., Qiu, Y. L., Palmer, J. D. (2004). Mitochondrial substitution rates are extraordinarily elevated and variable in a genus of flowering plants. Proc. Natl. Acad. Sci. U S A. 101, 17741–17746. doi: 10.1073/pnas.0408302101
Ciarmiello, L. F., Pontecorvo, G., Piccirillo, P., De Luca, A., Carillo, P., Kafantaris, I., et al. (2013). Use of nuclear and mitochondrial single nucleotide polymorphisms to characterize English walnut (Juglans regia l.) genotypes. Plant Mol. Biol. Rep. 31, 1116–1130. doi: 10.1007/s11105-013-0575-2
Colombatti, F., Gonzalez, D. H., Welchen, E. (2014). Plant mitochondria under pathogen attack: a sigh of relief or a last breath? Mitochondrion 19 Pt B, 238–244. doi: 10.1016/j.mito.2014.03.006
Corpet, F. (1988). Multiple sequence alignment with hierarchical clustering. Nucl. Acids Res. 16, 10881–10890. doi: 10.1093/nar/16.22.10881
de Freitas, K. E. J., Busanello, C., Viana, V. E., Pegoraro, C., de Carvalho, V. F., da Maia, L. C., et al. (2022). An empirical analysis of mtSSRs: Could microsatellite distribution patterns explain the evolution of mitogenomes in plants? Funct. Integr. Genomics 22, 35–53. doi: 10.1007/s10142-021-00815-7
de Longevialle, A. F., Small, I. D., Lurin, C. (2010). Nuclearly encoded splicing factors implicated in RNA splicing in higher plant organelles. Mol. Plant 3, 691–705. doi: 10.1093/mp/ssq025
Demesure, B., Sodzi, N., Petit, R. J. (1995). A set of universal primers for amplification of polymorphic non-coding regions of mitochondrial and chloroplast DNA in plants. Mol. Ecol. 4, 129–134. doi: 10.1111/j.1365-294x.1995.tb00201.x
Dong, S., Chen, L., Liu, Y., Wang, Y., Zhang, S., Yang, L., et al. (2020). The draft mitochondrial genome of magnolia biondii and mitochondrial phylogenomics of angiosperms. PloS One 15. doi: 10.1371/journal.pone.0231020
Dourmap, C., Roque, S., Morin, A., Caubrière, D., Kerdiles, M., Béguin, K., et al. (2020). Stress signalling dynamics of the mitochondrial electron transport chain and oxidative phosphorylation system in higher plants. Ann. Bot. 125, 721–736. doi: 10.1093/aob/mcz184
Drouin, G., Daoud, H., Xia, J. (2008). Relative rates of synonymous substitutions in the mitochondrial, chloroplast and nuclear genomes of seed plants. Mol. Phylogenet. Evol. 49, 137–141. doi: 10.1016/j.ympev.2008.09.009
Duminil, J., Besnard, G. (2021). “Utility of the mitochondrial genome in plant taxonomic studies,” in Molecular plant taxonomy. methods mol. biol, vol. 2222 . Ed. Besse, P. (New York: Humana). doi: 10.1007/978-1-0716-0997-2_6
Duminil, J., Pemonge, M. H., Petit, R. J. (2002). A set of 35 consensus primer pairs amplifying genes and introns of plant mitochondrial DNA. Mol. Ecol. Notes 2, 428–430. doi: 10.1046/j.1471-8286.2002.00263.x
Dumolin-Lapegue, S., Pemonge, M. H., Petit, R. J. (1997). An enlarged set of consensus primers for the study of organelle DNA in plants. Mol. Ecol. 6, 393–397. doi: 10.1046/j.1365-294X.1997.00193.x
Egan, A. N., Schlueter, J., Spooner, D. M. (2012). Applications of next-generation sequencing in plant biology. Am. J. Bot. 99, 175–185. doi: 10.3732/ajb.1200020
Froelicher, Y., Mouhaya, W., Bassene, J. B., Costantino, G., Kamiri, M., Luro, F., et al. (2011). New universal mitochondrial PCR markers reveal new information on maternal citrus phylogeny. Tree Genet. Genomes 7, 49–61. doi: 10.1007/s11295-010-0314-x
Galeano, C. H., Cortés, A. J., Fernández, A. C., Soler, Á., Franco-Herrera, N., Makunde, G., et al. (2012). Gene-based single nucleotide polymorphism markers for genetic and association mapping in common bean. BMC Genet. 13, 48. doi: 10.1186/1471-2156-13-48
Godbout, J., Jaramillo-Correa, J. P., Beaulieu, J., Bousquet, J. (2005). A mitochondrial DNA minisatellite reveals the postglacial history of jack pine (Pinus banksiana), a broad-range north American conifer. Mol. Ecol. 14, 3497–3512. doi: 10.1111/j.1365-294X.2005.02674.x
Grosser, M. (2011) Plant mitochondrial introns as genetic markers (Gainesville, Florida: University of Florida). Available at: https://ufdc.ufl.edu/AA00060090/00001/pdf (Accessed 12/1/2022). undergraduate thesis.
Grosser, J. W., Gmitter, F. G., Tusa, N., Recupero, G. R., Cucinotta, P. (1996). Further evidence of a cybridization requirement for plant regeneration from citrus leaf protoplasts following somatic fusion. Plant Cell Rep. 15, 672–676. doi: 10.1007/BF00231922
Gualberto, J. M., Newton, K. J. (2017). Plant mitochondrial genomes: Dynamics and mechanisms of mutation. Annu. Rev. Plant Biol. 68, 225–252. doi: 10.1146/annurev-arplant-043015-112232
Guo, W. W., Prasad, D., Cheng, Y. J., Serrano, P., Deng, X. X., Grosser, J. W. (2004). Targeted cybridization in citrus: Transfer of Satsuma cytoplasm to seedy cultivars for potential seedlessness. Plant Cell Rep. 22, 752–758. doi: 10.1007/s00299-003-0747-x
Guo, W. W., Xiao, S. X., Deng, X. X. (2013). Somatic cybrid production via protoplast fusion for citrus improvement. Sci. Hortic. (Amsterdam). 163, 20–26. doi: 10.1016/j.scienta.2013.07.018
Gupta, S., Kumari, K., Das, J., Lata, C., Puranik, S., Prasad, M. (2011). Development and utilization of novel intron length polymorphic markers in foxtail millet (Setaria italica (L.) p. beauv.). Genome 54, 586–602. doi: 10.1139/g11-020
Handa, H. (2003). The complete nucleotide sequence and RNA editing content of the mitochondrial genome of rapeseed (Brassica napus l.): comparative analysis of the mitochondrial genomes of rapeseed and arabidopsis thaliana. Nucl. Acids Res. 31, 5907–5916. doi: 10.1093/nar/gkg795
Hanna, W. W. (1997). Registration of tift 8593 pearl millet genetic stock. Crop Sci. 37, 1412. doi: 10.2135/cropsci1997.0011183X003700040100x
Hanna, W. W., Hill, G. M., Gates, R. N., Wilson, J. P., Burton, G. W. (1997). Registration of 'Tifleaf 3' pearl millet. Crop Sci. 37, 1388. doi: 10.2135/cropsci1997.0011183X003700040075x
Hodel, R. G. J., Segovia-Salcedo, C., Landis, J. B., Crowl, A. A., Sun, M., Liu, X., et al. (2016). The report of my death was an exaggeration: A review for researchers using microsatellites in the 21st century. Appl. Plant Sci. 41. doi: 10.3732/apps.1600025
Honma, Y., Yoshida, Y., Terachi, T., Toriyama, K., Mikami, T., Kubo, T. (2011). Polymorphic minisatellites in the mitochondrial DNAs of Oryza and Brassica. Curr. Genet. 57, 261–270. doi: 10.1007/s00294-011-0345-3
Hu, J., Huang, W., Huang, Q., Qin, X., Yu, C., Wang, L., et al. (2014). Mitochondria and cytoplasmic male sterility in plants. Mitochondrion 19 Pt B, 282–288. doi: 10.1016/j.mito.2014.02.008
Jaramillo-Correa, J. P., Aguirre-Planter, E., Eguiarte, L. E., Khasa, D. P., Bousquet, J. (2013). Evolution of an ancient microsatellite hotspot in the conifer mitochondrial genome and comparison with other plants. J. Mol. Evol. 76, 146–157. doi: 10.1007/s00239-013-9547-2
Jayaswal, K., Sharma, H., Bhandawat, A., Sagar, R., Yadav, V. K., Sharma, V., et al. (2019). Development of intron length polymorphic (ILP) markers in onion (Allium cepa l.), and their cross-species transferability in garlic (A. sativum l.) and wild relatives. Genet. Resour. Crop Evol. 66, 1379–1388. doi: 10.1007/s10722-019-00808-3
Keeling, P. J. (2009). Role of horizontal gene transfer in the evolution of photosynthetic eukaryotes and their plastids. Methods Mol. Biol. 532, 501–515. doi: 10.1007/978-1-60327-853-9_29
Kersten, B., Voss, M. M., Fladung, M. (2015). Development of mitochondrial SNP markers in different Populus species. Trees 29, 575–582. doi: 10.1007/s00468-014-1136-5
Kim, B., Kim, K., Yang, T. J., Kim, S. (2016). Completion of the mitochondrial genome sequence of onion (Allium cepa l.) containing the CMS-s male-sterile cytoplasm and identification of an independent event of the ccmF n gene split. Curr. Genet. 62, 873–885. doi: 10.1007/s00294-016-0595-1
Kim, J. H., Lee, C., Hyung, D., Jo, Y. J., Park, J. S., Cook, D. R., et al. (2015). CSGM designer: A platform for designing cross-species intron-spanning genic markers linked with genome information of legumes. Plant Methods 11, 1–11. doi: 10.1186/s13007-015-0074-6
Kim, S., Lim, H., Park, S., Cho, K.-H., Sung, S.-K., Oh, D.-G., et al. (2007). Identification of a novel mitochondrial genome type and development of molecular markers for cytoplasm classification in radish (Raphanus sativus l.). Theor. Appl. Genet. 111, 1191–1200. doi: 10.1007/s00122-007-0639-5
Kimura, M. (1980). A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16, 111–120. doi: 10.1007/BF01731581
Kubo, T., Mikami, T. (2007). Organization and variation of angiosperm mitochondrial genome. Physiol. Plant 129, 6–13. doi: 10.1111/j.1399-3054.2006.00768.x
Kubo, T., Nishizawa, S., Sugawara, A., Itchodo, N., Estiati, A., Mikami, T. (2000). The complete nucleotide sequence of the mitochondrial genome of sugar beet (Beta vulgaris l.) reveals a novel gene for tRNACys(GCA). Nucl. Acids Res. 28, 2571–2576. doi: 10.1093/nar/28.13.2571
Kumar, M., Kapil, A., Shanker, A. (2014). MitoSatPlant: Mitochondrial microsatellites database of viridiplantae. Mitochondrion 19 Pt B, 334–337. doi: 10.1016/j.mito.2014.02.002
Laroche, J., Li, P., Maggia, L., Bousquet, J. (1997). Molecular evolution of angiosperm mitochondrial introns and exons. Proc. Natl. Acad. Sci. U. S. A. 94, 5722–5727. doi: 10.1073/pnas.94.11.5722
Lessa, E. P. (1992). Rapid surveying of DNA sequence variation in natural populations. Mol. Biol. Evol. 9, 323–330. doi: 10.1093/oxfordjournals.molbev.a040723
Levings, C. S., Pring, D. R. (1977). Diversity of mitochondrial genomes among normal cytoplasms of maize. J. Hered. 68, 350–354. doi: 10.1093/oxfordjournals.jhered.a108858
Li, C., Riethoven, J.-J. M., Naylor, G. J. P. (2012). EvolMarkers: a database for mining exon and intron markers for evolution, ecology and conservation studies. Mol. Ecol. Resour. 12, 967–971. doi: 10.1111/j.1755-0998.2012.03167.x
Lo, Y. M. D. (1998). The amplification refractory mutation system. Methods Mol. Med. 16, 61–70. doi: 10.1385/0-89603-499-2:61. Clinical Applications of PCR.
Mahapatra, K., Banerjee, S., De, S., Mitra, M., Roy, P., Roy, S. (2021). An insight into the mechanism of plant organelle genome maintenance and implications of organelle genome in crop improvement: An update. Front. Cell Dev. Biol. 10. doi: 10.3389/fcell.2021.671698
Manjunathagowda, D. C., Muthukumar, P., Gopal, J., Prakash, M., Bommesh, J. C., Nagesh, G. C., et al. (2021). Male Sterility in onion (Allium cepa l.): origin: origin, evolutionary status, and their prospectus. Genet. Resour. Crop Evol. 68, 421–439. doi: 10.1007/s10722-020-01077-1
Mitton, J. B., Kreiser, B. R., Latta, R. G. (2000). Glacial refugia of limber pine (Pinus flexilis James) inferred from the population structure of mitochondrial DNA. Mol. Ecol. 9, 91–97. doi: 10.1046/j.1365-294X.2000.00840.x
Moore, G. A. (2001). Oranges and lemons: clues to the taxonomy of Citrus from molecular markers. Trends Genet. 17, 536–540. doi: 10.1016/s0168-9525(01)02442-8
Moreira, C. D., Gmitter, F. G., Jr., Grosser, J. W., Huang, S., Ortega, V. M., Chase, C. D. (2002). Inheritance of organelle DNA sequences in a Citrus-poncirus intergeneric cross. J. Hered. 93, 174–178. doi: 10.1093/jhered/93.3.174
Mower, J. P., Touzet, P., Gummow, J. S., Delph, L. F., Palmer, J. D. (2007). Extensive variation in synonymous substitution rates in mitochondrial genes of seed plants. BMC Evol. Biol. 7, 135. doi: 10.1186/1471-2148-7-135
Murray, M. G., Murray, W. F., Thompson, W. F. (1980). Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 8, 4321–4325. doi: 10.1093/nar/8.19.4321
Nishizawa, S., Kubo, T., Mikami, T. (2000). Variable number of tandem repeat loci in the mitochondrial genomes of beets. Curr. Genet. 37, 34–38. doi: 10.1007/s002940050005
Nishizawa, S., Mikami, T., Kubo, T. (2007). Mitochondrial DNA phylogeny of cultivated and wild beets: Relationships among cytoplasmic male-sterility-inducing and nonsterilizing cytoplasms. Genetics 177, 1703–1712. doi: 10.1534/genetics.107.076380
Notsu, Y., Masood, S., Nishikawa, T., Nubo, K., Akiduki, G., Nakazono, M., et al. (2002). The complete sequence of the rice (Oryza sativa l.) mitochondrial genome: frequent DNA sequence acquisition and loss during the evolution of flowering plants. Mol. Genet. Genom. 268, 434–445. doi: 10.1007/s00438-002-0767-1
Ogihara, Y., Yamazaki, Y., Murai, K., Kanno, A., Terachi, T., Shiina, T., et al. (2005). Structural dynamics of cereal mitochondrial genomes as revealed by complete nucleotide sequencing of the wheat mitochondrial genome. Nucleic Acids Res. 33, 6235–6250. doi: 10.1093/nar/gki925
Ollitrault, P., Curk, F., Krueger, R. (2020). “Citrus taxonomy,” in The citrus genus. Eds. Talon, M., Caruso, M., Gmitter, F. G., Jr. (Amsterdam: Elsevier), 57–81. doi: 10.1016/B978-0-12-812163-4.00004-8
Omar, A. A., Murata, M., Yu, Q., Gmitter, F. G., Chase, C. D., Graham, J. H., et al. (2017). Production of three new grapefruit cybrids with potential for improved citrus canker resistance. Vitr. Cell. Dev. Biol. - Plant 53, 256–269. doi: 10.1007/s11627-017-9816-7
Palmer, J. D., Herbon, L. A. (1988). Plant mitochondrial DNA evolved rapidly in structure, but slowly in sequence. J. Mol. Evol. 28, 87–97. doi: 10.1007/BF02143500
Parkinson, C. L., Mower, J. P., Qiu, Y. L., Shirk, A. J., Song, K., Young, N. D., et al (2005). Multiple major increases and decreases in mitochondrial substitution rates in the plant family geraniaceae. BMC Evol. Biol. 5, 73. doi: 10.1186/1471-2148-5-73
Petersen, G., Cuenca, A., Zervas, A., Ross, G. T., Graham, S. W., Barrett, C. F., et al. (2017). Mitochondrial genome evolution in alismatales: Size reduction and extensive loss of ribosomal protein genes. PloS One 12. doi: 10.1371/journal.pone.0177606
Potter, K. M., Hipkins, V. D., Mahalovich, M. F., Means, R. E. (2013). Mitochondrial DNA haplotype distribution patterns in Pinus ponderosa (Pinaceae): Range-wide evolutionary history and implications for conservation. Am. J. Bot. 100, 1562–1579. doi: 10.3732/ajb.1300039
Qiu, Y.-L., Lee, J., Bernasconi-Quadroni, F., Soltis, D. E., Soltis, P. S., Zanis, M., et al. (1999). The earliest angiosperms: evidence from mitochondrial, plastid and nuclear genomes. Nature. 402, 404–407. doi: 10.1038/46536
Ratnasingham, S., Hebert, P. D. (2007). BOLD: The barcode of life data system. Mol. Ecol. Notes 1, 355–364. doi: 10.1111/j.1471-8286.2007.01678.x
Sablok, G., Raju, G. V. P., Mudunuri, S. B., Prabha, R., Singh, D. P., Baev, V., et al. (2015). ChloroMitoSSRDB 2.00: more genomes, more repeats, unifying SSRs search patterns and on-the-fly repeat detection database update. Database. 84. doi: 10.1093/database/bav084
Satpute, A. D., Chen, C., Gmitter, F. G., Ling, P., Yu, Q., Grosser, M. R., et al. (2015). Cybridization of grapefruit with ‘Dancy’ mandarin leads to improved fruit characteristics. J. Am. Soc Hortic. Sci. 140, 427–435. doi: 10.21273/jashs.140.5.427
Schmitz-Linneweber, C., Lampe, M.-K., Sultan, L. D., Ostersetzer-Biran, O. (2015). Organellar maturases: A window into the evolution of the spliceosome. Biochim. Biophys. Acta 1847, 798–808. doi: 10.1016/j.bbabio.2015.01.009
Shaw, J., Shafer, H. L., Leonard, O. R., Kovach, M. J., Schorr, M., Morris, A. B. (2014). Chloroplast DNA sequence utility for the lowest phylogenetic and phylogeographic inferences in angiosperms: the tortoise and the hare IV. Am. J. .Bot. 101, 1987–2004. doi: 10.3732/ajb.1400398
Shilpa, H. B., Lohithaswa, H. C. (2021). Discovery of SNPs in important legumes through comparative genome analysis and conversion of SNPs into PCR-based markers. J. Genet. 100. doi: 10.1007/s12041-021-01320-3
Sloan, D. B. (2013). One ring to rule them all? genome sequencing provides new insights into the ‘master circle’ model of plant mitochondrial DNA structure. New Phytol. 200, 978–985. doi: 10.1111/nph.12395
Sloan, D. B., Alverson, A. J., Chuckalovcak, J. P., Wu, M., McCauley, D. E., Palmer, J. D., et al. (2012). Rapid evolution of enormous, multichromosomal genomes in flowering plant mitochondria with exceptionally high mutation rates. PloS Biol. 10. doi: 10.1371/journal.pbio.1001241
Sloan, D. B., Oxelman, B., Rautenberg, A., Taylor, D. R. (2009). Phylogenetic analysis of mitochondrial substitution rate variation in the angiosperm tribe sileneae (Caryophyllaceae). BMC Evol. Biol. 9, 260. doi: 10.1186/1471-2148-9-260
Soranzo, N., Provan, J., Powell, W. (1999). An example of microsatellite length variation in the mitochondrial genome of conifers. Genome 42, 158–161. doi: 10.1139/g98-111
Sperisen, C., Buchler, U., Gugerli, F., Matyas, G., Geburek, T., Vendramin, G. G. (2001). Tandem repeats in plant mitochondrial genomes: application to the analysis of population differentiation in the conifer Norway spruce. Mol. Ecol. 10, 257–263. doi: 10.1046/j.1365-294X.2001.01180.x
Sugiyama, Y., Watase, Y., Nagase, M., Makita, N., Yagura, M., Hirai, A., et al. (2005). The complete nucleotide sequence and multipart organization of the tobacco mitochondrial genome: comparative analysis of mitochondrial genomes in higher plants. Mol. Genet. Genom. 272, 603–615. doi: 10.1007/s00438-004-1075-8
Tonti-Filippini, J., Nevill, P. G., Dixon, K., Small, I. (2017). What can we do with 1000 plastid genomes? Plant J. 90, 808–818. doi: 10.1111/tpj.13491
Tsujimura, M., Terachi, T. (2018). “Cytoplasmic genome,” in The allium genomes. Eds. Shigyo, M., Khar, A., Abdelrahman, M. (Switzerland: Springer International Publishing), 89–98. doi: 10.1007/978-3-319-95825-5_6
Unseld, M., Marienfeld, J. R., Brandt, P., Brennicke, A. (1997). The mitochondrial genome of Arabidopsis thaliana contains 57 genes in 366,924 nucleotides. Nat. Genet. 15, 57–61. doi: 10.1038/ng0197-57
Van Deynze, A., Stoffel, K., Robin, C. R., Kozik, A., Liu, J., van der Knaap, E., et al. (2007). Diversity in conserved genes in tomato. BMC Genomics 8, 465. doi: 10.1186/1471-2164-8-465
Vincze, T., Posfai, J., Roberts, R. J. (2003). NEBcutter: A program to cleave DNA with restriction enzymes. Nucleic Acids Res. 31, 368836–368891. doi: 10.1093/nar/gkg526
Wang, Y., Chen, J., Francis, D. M., Shen, H., Wu, T., Yang, W. (2010). Discovery of intron polymorphisms in cultivated tomato using both tomato and arabidopsis genomic information. Theor. Appl. Genet. 121, 1199–1207. doi: 10.1007/s00122-010-1381-y
Wang, X., Wang, J., Li, S., Lu, C., Sui., N. (2022). An overview of RNA splicing and functioning of splicing factors in land plant chloroplasts. RNA Biol. 19, 897–907. doi: 10.1080/15476286.2022.2096801
Wang, X., Zhao, X., Zhu, J., Wu, W. (2005). Genome-wide investigation of intron length polymorphisms and their potential as molecular markers in rice (Oryza sativa l.). DNA Res. 12, 417–427. doi: 10.1093/dnares/dsi019
Wicke, S., Schneeweiss, G. M., dePamphilis, C. W., Müller, K. F., Quandt, D. (2011). The evolution of the plastid chromosome in land plants: gene content, gene order, gene function. Plant Mol. Biol. 76, 273–297. doi: 10.1007/s11103-011-9762-4
Wolfe, K. H., Li, W. H., Sharp, P. M. (1987). Rates of nucleotide substitution vary greatly among plant mitochondrial, chloroplast, and nuclear DNAs. Proc. Natl. Acad. Sci. U. S. A. 84, 9054–9058. doi: 10.1073/pnas.84.24.9054
Wu, G. A., Terol, J., Ibanez, V., López-García, A., Pérez-Román, E., Borredá, C., et al. (2018). Genomics of the origin and evolution of citrus. Nature. 554, 311–316. doi: 10.1038/nature25447
Xiang, Q.-P., Wei, R., Shao, Y.-Z., Yang, Z.-Y., Wang, X.-Q., Zhang, X.-C. (2014). Phylogenetic relationships, possible ancient hybridization, and biogeographic history of abies (Pinaceae) based on data from nuclear, plastid, and mitochondrial genomes. Mol. Phylogenet. Evol. 82 Pt A, 1–14. doi: 10.1016/j.ympev.2014.10.008
Xiong, Y., Yu, Q., Yiong, Y., Zhao, J., Lei, X., Liu, L., et al. (2022). The complete mitogenome of Elymus sibiricus and insights ito its evolutionary pattern based on simple repeat sequences of seed plant mitogenomes. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.802321
Ye, J., Coulouris, G., Zaretskaya, I., Cutcutache, I., Rozen, S., Madden, T. L. (2012). Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinf. 13, 134. doi: 10.1186/1471-2105-13-134
Keywords: group II intron, indel polymorphism, organelle genome, PCR-based markers, plant mitochondria, single nucleotide polymorphism
Citation: Grosser MR, Sites SK, Murata MM, Lopez Y, Chamusco KC, Love Harriage K, Grosser JW, Graham JH, Gmitter FG Jr. and Chase CD (2023) Plant mitochondrial introns as genetic markers - conservation and variation. Front. Plant Sci. 14:1116851. doi: 10.3389/fpls.2023.1116851
Received: 05 December 2022; Accepted: 02 March 2023;
Published: 20 March 2023.
Edited by:
Satoshi Watanabe, Saga University, JapanReviewed by:
Fabio Palumbo, University of Padua, ItalyRuslan Kalendar, University of Helsinki, Finland
Copyright © 2023 Grosser, Sites, Murata, Lopez, Chamusco, Love Harriage, Grosser, Graham, Gmitter and Chase. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Christine D. Chase, Y2RjaGFzZUB1ZmwuZWR1
†Present addresses: Melinda R. Grosser, Department of Biology, University of North Carolina, Asheville, NC, United States Samantha K. Sites, North Carolina Department of Health and Human Services, Raleigh, NC, United States Mayara M. Murata, Stricto Sensu Department, Universidade Norte do Paraná, Londrina, Paraná, Brazil Kyra Love Harriage, Career and Technical Education Department, Polk County Public Schools, Bartow, FL, United States
 Melinda R. Grosser
Melinda R. Grosser Samantha K. Sites1†
Samantha K. Sites1† Mayara M. Murata
Mayara M. Murata Yolanda Lopez
Yolanda Lopez Jude W. Grosser
Jude W. Grosser Fred G. Gmitter Jr.
Fred G. Gmitter Jr. Christine D. Chase
Christine D. Chase