
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 27 January 2023
Sec. Plant Metabolism and Chemodiversity
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1114215
This article is part of the Research TopicAdvances in Metabolism and Chemodiversity – Focus – Plant EnzymesView all 7 articles
Pullulanase (EC 3.2.1.41, PUL), a debranching enzyme belonging to glycoside hydrolase family 13 subfamily 13, catalyses the cleavage of α-1,6 linkages of pullulan and β-limit dextrin. The present work studied PUL from cassava Manihot esculenta Crantz (MePUL) tubers, an important economic crop. The Mepul gene was successfully cloned and expressed in E. coli and rMePUL was biochemically characterised. MePUL was present as monomer and homodimer, as judged by apparent mass of ~ 84 - 197 kDa by gel permeation chromatography analysis. Optimal pH and temperature were at pH 6.0 and 50 °C, and enzyme activity was enhanced by the addition of Ca2+ ions. Pullulan is the most favourable substrate for rMePUL, followed by β-limit dextrin. Additionally, maltooligosaccharides were potential allosteric modulators of rMePUL. Interestingly, short-chain maltooligosaccharides (DP 2 - 4) were significantly revealed at a higher level when rMePUL was mixed with cassava isoamylase 3 (rMeISA3), compared to that of each single enzyme reaction. This suggests that MePUL and MeISA3 debranch β-limit dextrin in a synergistic manner, which represents a major starch catabolising process in dicots. Additionally, subcellular localisation suggested the involvement of MePUL in starch catabolism, which normally takes place in plastids.
Starch is a vital carbohydrate reserve in plants and serves as a key component of the food chain for animals. It is synthesised in plastids, including chloroplasts in leaves and amyloplasts in tubers and grain. Starch consists of two α-glucans: amylose and amylopectin. The amylose mainly presents as a linear glucan structure comprised of α-1,4 glycosidic bond with a limited α-1,6-linked branching (less than 1%). This glucan is synthesised by the concerted action of ADP-glucose pyrophosphorylase (AGPase) and granule-bound starch synthase I (GBSSI). In contrast, amylopectin is much more complex since it harbours highly branched structures generated by the action of AGPase, soluble starch synthases (SSs), starch branching enzymes (SBEs), and starch debranching enzymes (DBEs) (Ball and Morell, 2003; Tappiban et al., 2019). In addition, the starch degrading process in chloroplasts also relies on several enzymes, including β-amylases (BAMs), disproportionation enzyme 1 (D-enzyme1 or DPE1) and debranching enzymes (DBEs), which generate residual sugars, glucose and maltose, which are exported to the cytosol to be further metabolised (Zeeman et al., 2010).
Debranching enzymes (DBEs), members of glycoside hydrolase family 13 (GH13), are found in plants and microorganisms. They could be categorised into two major types: isoamylase (EC 3.2.1.68, ISA) and pullulanase (PUL, limit dextrinase, R-enzyme, EC 3.2.1.41) according to their peptide sequences and substrate specificities. Although they both can act on α-1,6 glucosidic linkages (Panpetch et al., 2018b), only PUL shows high specific activity against pullulan, α-1,6-linked maltotriose units, and liberates maltotriose final product. DBEs are required for both starch synthesis and degradation in plants. Generally, plants contain 3 isoforms of ISA (ISA1, ISA2, and ISA3) but only 1 pullulanase (Zeeman et al., 2010). The ISA isoforms 1 and 2 (ISA1 and ISA2) exist as either heterooligo- or homooligo-meric structures, depending on the plant species (Hussain et al., 2003; Panpetch et al., 2018b). They have been proposed to play important roles mainly in starch synthesis by the removal of redundant branches of immature amylopectin. This produces an appropriate structure that is competent to form highly structured granular starch, based on “the glucan trimming model” (Myers et al., 2000; Nakamura, 2002). Loss of DBEs in starch biosynthesis results in the accumulation of phytoglycogen (Delatte et al., 2005), while in starch degradation, a starch-excess or sex phenotype arises (Caspar et al., 1991). Further, PUL together with ISA3 participates in the debranching of so-called β-limit dextrin, starch residues arising from the action of BAM (Zeeman et al., 2010). Deficiency of these debranching enzymes promotes the accumulation of branch-maltooliogosaccharides and sex phenotypes, resulting in a slow growth rate (Delatte et al., 2006). To date, plant starch metabolism has been mainly evaluated in model plants such as Arabidopsis thaliana; additional information on starch metabolism in other plants, however, is also required.
Currently, only plant PUL from barley (Hordeum vulgare) (HvLD), a monocotyledon, has been biochemically and structurally characterised (Møller et al., 2015). The HvLD was classified into GH13 subfamily 13. In addition to the 3 major domains, including N-, C-, and catalytic domains, HvLD structure also haboured an additional carbohydrate-binding module 48 (CBM48) located between N-terminal domain and catalytic domain (Stam et al., 2006; Machovič and Janeček, 2008; Vester-Christensen et al., 2010b; Drula et al., 2022). Here is less data for PUL, especially from dicotyledonous plants. Only partially purified PUL from potato (Solarium tuberosum L.) (StPUL) (Ishizaki et al., 1983) and purified PUL from spinach (Spinacia oleracea L.) (SoPUL) (Renz et al., 1998) were biochemically characterised to date. Moreover, the PUL from cassava, a tropical dicotyledonous plant, has not been studied before (Tappiban et al., 2019).
Cassava (Manihot esculenta Crantz), one of the most important economic crops in Asia, Africa and South America, in particular, possesses a very high starch content in its tubers (Tappiban et al., 2019). Although cassava starch has been widely used for many years, its starch-modifying enzymes have been little studied. Some of these enzymes have been cloned, expressed, and characterised (Tappiban et al., 2019), such as DPE1 (Tantanarat et al., 2014), ISA1/ISA2 (Panpetch et al., 2018b), and ISA3 (Panpetch et al., 2018a). Notably, most studies of plant pullulanases have been conducted in vivo (Dinges et al., 2003; Fujita et al., 2009; Li et al., 2019), which is complicated by the concerted action of several other enzymes and, as a result, specific action information about individual enzymes has rarely been investigated. This study aimed to biochemically characterise pullulanase from Manihot esculenta Crantz (MePUL), with a view to determining its biological role in starch metabolism which remained unclear.
Total RNA was extracted from nine-month-old cassava Manihot esculenta Crantz cultivar ‘KU50’ tubers harvested from the National Research Centre of Millet and Corn, Thailand, using PureLink™ Plant RNA Reagent (Invitrogen™). The RNA was converted into cDNA using a SMARTer® RACE 5′/3′ Kit (Clontech®) followed the manufacturer’s manual. Full length MePUL was amplified by PrimeSTAR® HS DNA Polymerase (Clontech®) using the 5′ RACE cDNA as a template. 5′ RACE forward primer provided in the RACE kit and a gene specific reverse primer (5′ GTAAGCTTAAATTTTCCTAGGCTCAACAAACACAG 3′) were used for PCR. The PCR product was cloned into a pJET1.2/blunt (Thermo Fisher Scientific™) and the putative gene was confirmed by DNA sequencing (1st BASE DNA sequencing). A new forward primer (5′ CTCTA GCTAGCGGTTCCACTCCCACTTCTGAGTTGC 3′) excluding a transit peptide sequence predicted by ChloroP1.1 server and a new reverse primer (5′ AAGGAAAAAATGCGGCCGCAATTTTCCTAGGCTCAACAAACAC 3′) were specifically designed. The gene excluding a stop codon was subcloned into a pET21b vector via NheI and NotI restriction sites in-framed with C-terminal 6x His sequence of the plasmid. The recombinant plasmid (pET21b-Mepul) was transformed into E. coli TOP10 (Invitrogen™). The plasmid was extracted and subjected to DNA sequencing.
The pET21b-Mepul was transformed into E. coli SoluBL21 (DE3) and grown on an LB agar plate containing 100 μg/mL ampicillin. Then, the transformant was cultured in LB broth containing 100 μg/mL ampicillin until OD600 reached ~ 0.6. Cells were induced for protein production by adding 0.4 mM IPTG and continuously cultured at 16°C with shaking at 250 rpm for overnight. Cells were collected by centrifugation at 8,000 x g for 10 min at 4°C. After that, the cell pellet was suspended in buffer A (25 mM phosphate buffer + 500 mM NaCl + 25 mM imidazole, pH 7.2) and lysed by ultrasonication. After that, cell debris was removed by centrifugation at 10,000 x g for 10 min at 4°C. The supernatant was loaded onto a Histrap™ FF column equilibrated with the buffer A. The column was washed by the same buffer and the rMePUL was eluted with a linear gradient of buffer B (25 mM phosphate buffer + 500 mM NaCl + 50 – 300 mM imidazole, pH 7.2). Finally, the purified fractions containing MePUL activity were pooled and subjected to dialysis against 25 mM phosphate buffer pH 7.2 at 4°C for overnight. SDS-PAGE and western blot analysis were used for evaluating protein purity.
The molecular size of purified rMePUL was analysed with an MAbPac™ SEC-1 column (4 x 300 mm, Thermo Fisher Scientific™). The system was run in 25 mM phosphate buffer pH 6.8 containing 300 mM NaCl with a flowrate of 0.2 mL/min at 30°C. The signal was monitored by measuring A280. Gel Filtration Standards (Bio-Rad) were used for molecular weight calibration. To investigate the multimeric state of rMePUL/rMeISA3 (Panpetch et al., 2018a), ~ 0.26 nmol of each enzyme was mixed and incubated in 25 mM phosphate buffer pH 7.2 in 0.1 mL total volume at 4°C overnight prior to analysis.
The purified rMePUL was incubated with 0.75% (w/v) pullulan (Megazyme®) in 25 mM acetate buffer pH 6.0 in the total volume of 0.2 mL at 50°C to determine debranching activity. The reaction was terminated by adding an equal volume of 2,4-dinitrosalicylic (DNS) acid reagent (Miller, 1959) and then boiled for 10 min. The colour development of the reactions was monitored by measuring A540. One unit of MePUL was defined as the amount of enzyme that release 1 μmol of reducing sugar from pullulan substrate in a minute. Standard glucose in range of 0, 0.5, 1, 1.5, 2, 4, 6, and 8 mM was used for a standard curve.
The substrate specificity of rMePUL and the effect of pH, temperature, and metal ions and chemicals, and enzyme kinetics were studied. The characterisation was monitored by relative debranching activity of the purified enzyme determined by modified DNS method as described in the enzymatic assay. All experiments were performed in triplicate.
The purified rMePUL was assayed in the reaction containing 0.75% (w/v) pullulan in 25 mM buffer at various pH values [acetate buffer (pH 4.0 – 6.0) and phosphate buffer (pH 6.0 – 7.5)] at 30°C.
The purified rMePUL was assayed in reactions containing 0.75% (w/v) pullulan in 25 mM acetate buffer pH 6.0 incubated at various temperatures (30 - 60°C).
To evaluate the effect of EDTA, the activity of purified rMePUL was measured in the reaction containing 0.75% (w/v) pullulan in 25 mM acetate buffer pH 6.0 and 1 mM EDTA at 50°C. The activity was compared with the reaction without EDTA.
The effect of various metal ions was observed. The purified rMePUL was dialysed against 1 mM EDTA in 25 mM phosphate buffer pH 7.2 at 4°C overnight. Next, the pretreated enzyme was incubated with 0.75% (w/v) pullulan substrate in 25 mM acetate buffer pH 6.0 at 50°C with the presence of 10 mM metal ion [CaCl2, CoCl2, CuSO4, FeCl2, MnCl2, MgCl2, NiSO4, and ZnCl2].
The purified rMePUL was incubated with 0.75% (w/v) of different types of substrates [pullulan, β-limit dextrin, amylopectin, potato starch, cassava starch, glycogen type II, and maltodextrin] in 25 mM acetate buffer pH 6.0 at 50°C.
To assess enzyme kinetics, the purified rMePUL was incubated with pullulan or β-limit dextrin in 25 mM acetate buffer pH 6.0 at 50°C in 0.1-mL reactions under enzyme initial velocity of 15 min. The reactions were initiated by addition of 0.75 μg of the purified rMePUL and then analysed by DNS method. To determine the effect of some maltooligosaccharides on enzyme activity, 1 - 3 mM maltose or maltotriose were mixed in the reactions. The kinetic parameters were fitted and determined by the OriginPro 2017 software.
The rMePUL was incubated with 0.75% (w/v) pullulan and 0.01% (w/v) β-CD in 25 mM acetate buffer pH 6.0 in the total volume of 0.2 mL at 50°C. The activity was assayed by DNS method. Relative activity was compared with the control reaction without adding β-CD.
To determine the substrate specificity of MePUL, purified rMePUL was incubated with 2.5 mg/mL various oligosaccharides [maltohexaose (G6, Wako®), maltotriosyl maltotriose (G3G3, Megazyme®), glucosyl maltotriose (G1G3), panose (Megazyme®), isopanose (Megazyme®), isomaltotriose (IMO3, TCI®), β-CD (Sigma®), maltosyl β-CD (β-CD-G2, Ensuiko Sugar Reffining Co., Ltd.), glucosyl β-CD (β-CD-G1, Bio Research Coporation of Yokohama), and acarbose (Wako®)] in 25 mM acetate buffer pH 6.0 at 50°C for 2.5 hr.
In addition, to confirm the specificity of pullulan cleavage, products produced by KpPUL (commercial pullulanase, Sigma®) and the purified rMePUL were also compared. 0.2 U/mL of both PULs were incubated with 0.5% (w/v) pullulan in 25 mM acetate buffer pH 6.0 at 25°C and at 50°C for KpPUL and rMePUL, respectively. The products were analysed at 0, 0.5, 1, 3.5, 6, and 24 hr after incubation.
The products from the enzymatic reactions were spotted onto TLC plates (TLC Silica gel 60 F254, Merck). The TLC was run in a TLC tank equilibrated with acetonitrile:ethylacetate:1-propanol:water (85:20:50:60). Finally, the result was visualised by heating at 110°C for 10 min after staining with orcinol solution orcinol solution (sulphuric acid:ethanol:water (5:27:13) containing 0.2%(w/v) orcinol) (Waksmundzka-Hajnos et al., 2008; Wangpaiboon et al., 2021).
To analyse the product pattern according to the debranching activity of PUL on pullulan substrate, The 0.2 U/mL of both PULs were incubated with 0.5% (w/v) pullulan in 25 mM acetate buffer pH 6.0 at 25°C and at 50°C for KpPUL and rMePUL, respectively. The products were analysed at 0.5 hr after incubation.
The enzymatic reactions were analysed by HPAEC-PAD using a CarboPac™ PA-100 column with a flowrate of 1 mL/min at 30°C. The products were eluted with a linear gradient of 0 – 0.5 M sodium acetate in 150 mM NaOH solution for 35 min, followed by 0.5 M sodium acetate in 150 mM NaOH for 5 min.
Each sample obtained from PUL reactions (section 2.6.2) was desalted by using a mixed bed resin (AG 501-X8, Bio-Rad). The samples were diluted 10-fold prior to mixing with 10 mg/mL of 2,5-dihydroxybenzoic acid with the ratio of 1:1 and spotted onto a MALDI target plate. Masses were analysed with a JEOL SpiralTOF MALDI Imaging-TOF/TOF Mass Spectrometer using spiral mode.
To investigate the possible relationship between PUL and ISA3 in starch degradation process, rMeISA3 and rMePUL activities were defined under the same condition; 0.75% (w/v) β-limit dextrin (Megazyme®) in 25 mM acetate buffer pH 6.0 at 37°C. For the individual enzyme reactions, 0.2 U/mL of either rMeISA3 or rMePUL was incubated with 0.5% (w/v) β-limit dextrin in 25 mM acetate buffer pH 6.0 at 37°C. For the mixed-enzyme reaction, 0.1 U/mL of each enzyme was added to the reaction and then incubated under the same conditions. The reactions were collected at the time points of 0, 0.5, 1, 3.5, 6, and 24 hr and analysed by HPAEC-PAD and MALDI-TOF MS.
SWISS-MODEL server was employed to constructed homology model of mature MePUL using crystal structure of barley limit dextrinase (E510A mutant) in complex with a 6II-O-maltotriosylmaltotriose (branched maltohexaose, PDB ID: 4J3W) (Møller et al., 2015) as a template. Structure of branched maltohexaose in complex with MePUL homology model was also extracted from this crystal structure. H++ server was used to protonated all amino acids of this model at pH 6.0 (Gordon et al., 2005; Myers et al., 2006; Aguilar et al., 2012). LEaP module of AMBER20 was employed to solvated the complex in an isomeric truncated octahedral box of TIP3P water model (Jorgensen et al., 1983) with the buffer distance of 13 Å. Sodium ions (Na+) were added to neutralise the systems. PMEMD module of AMBER 20 was then used to minimise and simulate the system under the periodic boundary condition as previously described (Wangpaiboon et al., 2022). In the production run, the systems were simulated at 323 K for 50 ns. The Cpptraj module (Roe and Cheatham, 2013) of AMBER 20 was employed to calculated for Root Mean Square Deviation (RMSD) of the complex, and percentage of Hydrogen bond (H-bond) occupation (%H-bondoc). RMSD value of all atoms of the protein or ligands were calculated with respect to the minimised structure. The structure was found to be stable in the range of 40 to 50 ns. Therefore, this trajectory was used for analysis. Hydrogen bonds will present if the following conditions are met: (i) a donor-acceptor distance of less than 3.5, and (ii) a donor H-acceptor bond angle of less than 120°. H-bond occupations > 50% were taken into account (Klaewkla et al., 2021). The binding affinity of each residue to the ligand was calculated based on molecular mechanics/Generalised Born surface area (MM/GBSA) method (Genheden et al., 2012) using MMPBSA.py modules (Genheden et al., 2012) of AMBER20.
The full-length Mepul was cloned into a pCR™8/GW/TOPO® TA vector (Invitrogen™) based on circular polymerase extension cloning technique (Aslanidis and De Jong, 1990; Quan and Tian, 2011). The gene with LR sequences was then subcloned into a pGWB5 vector fused with the C-terminal green fluorescent protein (GFP) (Nakagawa et al., 2007) using Gateway® LR Clonase® II (Invitrogen™). The pGWB5-Mepul was transformed into an Agrobacterium tumefaciens GV3101 by electroporation.
A. tumefaciens harbouring pGWB5-Mepul and A. tumefaciens containing the silencing suppressor p19 gene were co-infiltrated into 4-week Nicotiana benthamiana leaves (Lindbo, 2007; Panpetch and Sirikantaramas, 2021). A pGWB2 bearing GFP gene was used as a control for infiltration experiments. After 3 days, localisation of MePUL was observed under a FluoView® FV10i-DOC confocal laser scanning microscope (Olympus Corp.). GFP fluorescence signal was monitored at 488/510 nm of excitation/emission, while chloroplast autofluorescence was observed at 633/664 nm.
The full-length Mepul sequence with an open reading frame of 2,904 bp (Supplementary Figure S1) was successfully obtained from 5′ RACE and encoded 967 amino acid residues. According to the full peptide sequence, the first 74 residues belonged to a transit peptide, as predicted by the ChloroP1.1 server (Supplementary Figure S2). For gene expression in E. coli, mature Mepul without the transit peptide was directly cloned into a pET21b expression vector and successfully expressed in soluble form in E. coli SoluBL (DE3) transformant. After purification by a metal ion affinity column, SDS-PAGE and western blot analyses showed a single band of the purified rMePUL. The apparent molecular mass of ~ 100 kDa corresponded to that calculated for monomeric protein the by Expasy Compute pI/Mw tool (https://web.expasy.org/compute.pi/) which was 100776.98 Da (including 6x His). Nevertheless, rMePUL exhibited two protein peaks by gel filtration chromatography with the calculated sizes of 197 and 84 kDa, as shown in Figure 1: the higher MW peak was a major form.
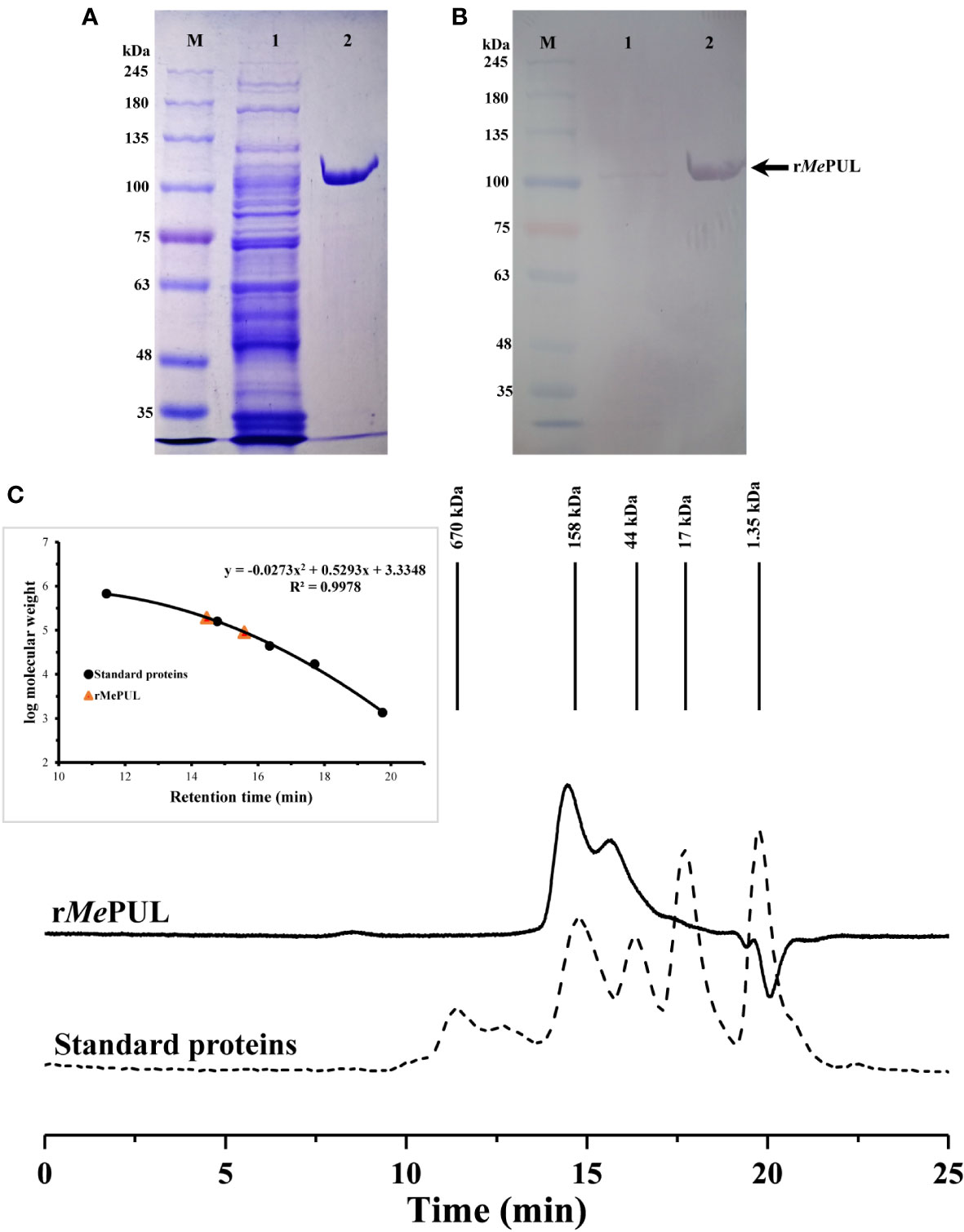
Figure 1 Molecular weight analysis of rMePUL. (A, B) SDS-PAGE and western blot analyses. Crude extract and pooled activity fractions of rMePUL purified by a Histrap™ column were analysed and visualised by SDS-PAGE and Western blot analysis. Lane M: TriColour Broad Protein Ladder (3.5 – 245 kDa) (BiotechRabbit); lane 1: crude protein; lane 2: purified rMePUL (~ 100 kDa). (C) Non-denaturing gel filtration analysis. Concentrated pooled purified fraction was loaded onto a gel filtration column (MAbPac™ SEC-1). Protein standards (shown by dashed line) are as follows; (1) vitamin B12 (1.35 kDa), (2) myoglobin (17 kDa), (3) ovalbumin (44 kDa), (4) γ-globulin (158 kDa), and (5) thyroglobulin (670 kDa).
The rMePUL exhibited optimal temperature and pH at 50 °C and pH 6.0, respectively (Figure 2A, B). In addition, EDTA treatment could increase enzyme activity of around 30%. Only Ca2+ ion could promote rMePUL activity ~ 30% higher than the control reaction. Conversely, Co2+, Cu2+, Zn2+, Mg2+, and Ni2+ ions could apparently inhibit MePUL activity (Figure 2C). Under assay conditions with different branched substrates, rMePUL exhibited the highest activity against pullulan, followed by β-limit dextrin with ~ 80% activity (Figure 2D). rMePUL showed little hydrolysis activity on potato starch and maltodextrin, at ~ 20% compared with that of pullulan. In contrast, amylopectin, cassava starch, and glycogen type II were barely cleaved by rMePUL.
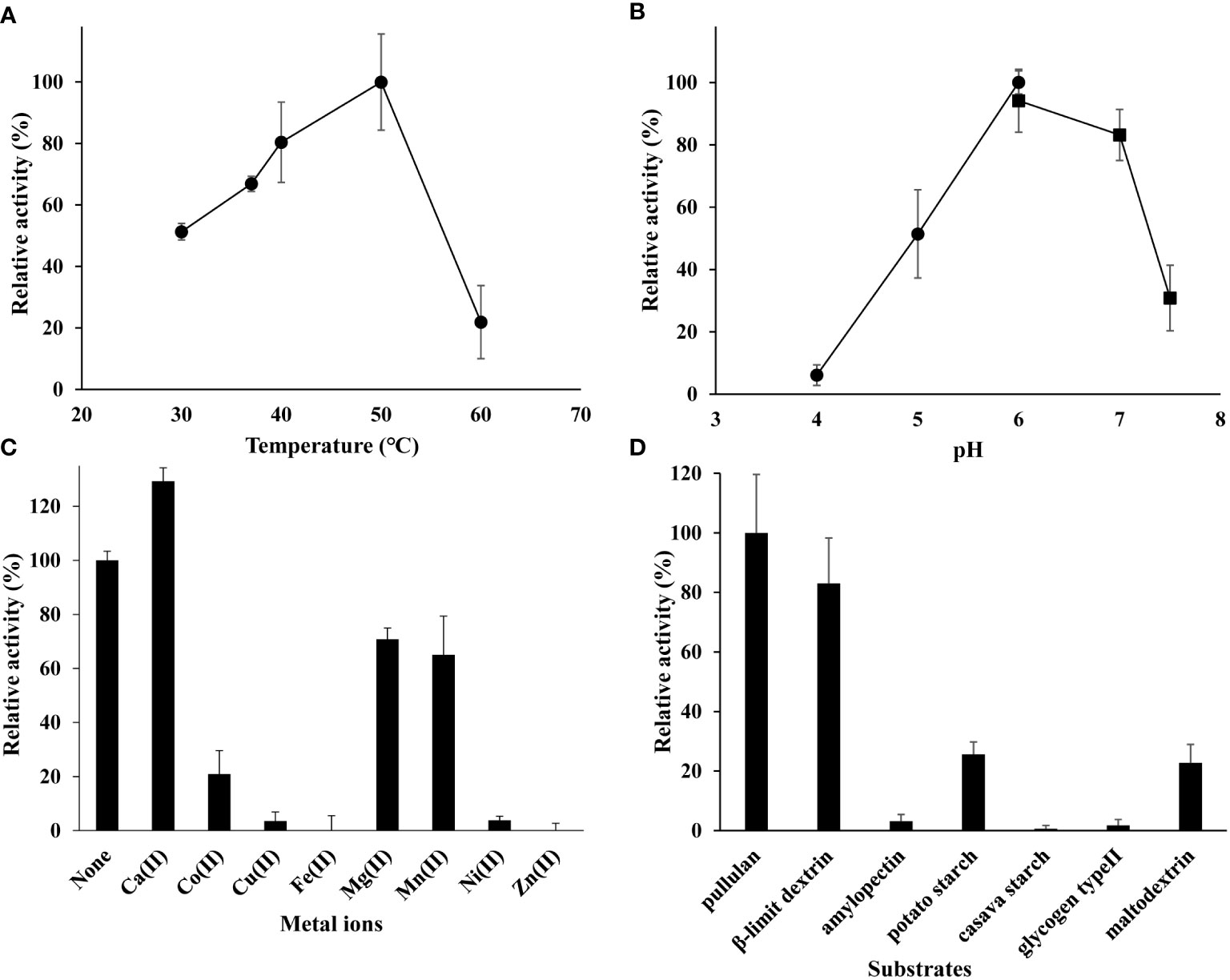
Figure 2 Biochemical characterisations of purified rMePUL. (A) Optimum temperature for debranching activity of rMePUL determined at temperatures ranging from 30 – 60°C. (B) Optimum pH for debranching activity of rMePUL measured at pH 4.0 – 7.5 [acetate buffer (●) (pH 4.0, 5.0, 6.0); potassium phosphate buffer (▪) (pH 6.0, 7.0, 7.5)]. (C) The effect of metal ions [10 mM of Ca2+, Co2+, Cu2+, Fe2+, Mg2+, Mn2+, Ni2+, and Zn2+] on debranching activity of rMePUL. (D) Substrate specificity of the rMePUL on various types of branched substrates (pullulan, β-limit dextrin, potato amylopectin, potato starch, cassava starch, glycogen type II, and maltodextrin). Enzyme activity was analysed by DNS assay.
In addition, the kinetics study showed that rMePUL catalysis could be fitted well with Hill plots, with both pullulan and β-limit dextrin substrates (represent in black lines in Figure 3A, B). In contrast, pullulan and β-limit dextrin kinetics in the presence of maltose or maltotriose could not be fitted with the same kinetic model. Notably, addition of maltotriose could obviously reduce the k value when pullulan was used as substrate (Figure 3A), while both kcat and Km values of β-limit dextrin kinetics in the presence of maltose or maltotriose were highly increased (Figure 3B; Supplementary Figure S3, and Table 1).To further investigate the linkage-specificity of substrates for rMePUL, the enzyme was incubated with different oligosaccharides harbouring various types of linkages. Among those substrates, maltotriosyl-maltotriose was the only substrate that was completely hydrolysed, releasing maltotriose. On the other hand, for other oligosaccharide substrates including maltohexose, β-cyclodextrin, maltosyl-β-cyclodextrin, glucosyl-β-cyclodextrin, panose, isopanose, glucosyl-maltotriose, isomaltotriose, and acarbose, no catalysis was apparently observed even after 2.5-hr incubation with the enzyme (Figure 4A). Additionally, the effect of β-CD on the inhibition of rMePUL in pullulan hydrolysis was also investigated. The result clearly showed that the pullulan cleavage was almost completely inhibited which only the relative activity of 2.4% was shown compared to the control reaction.
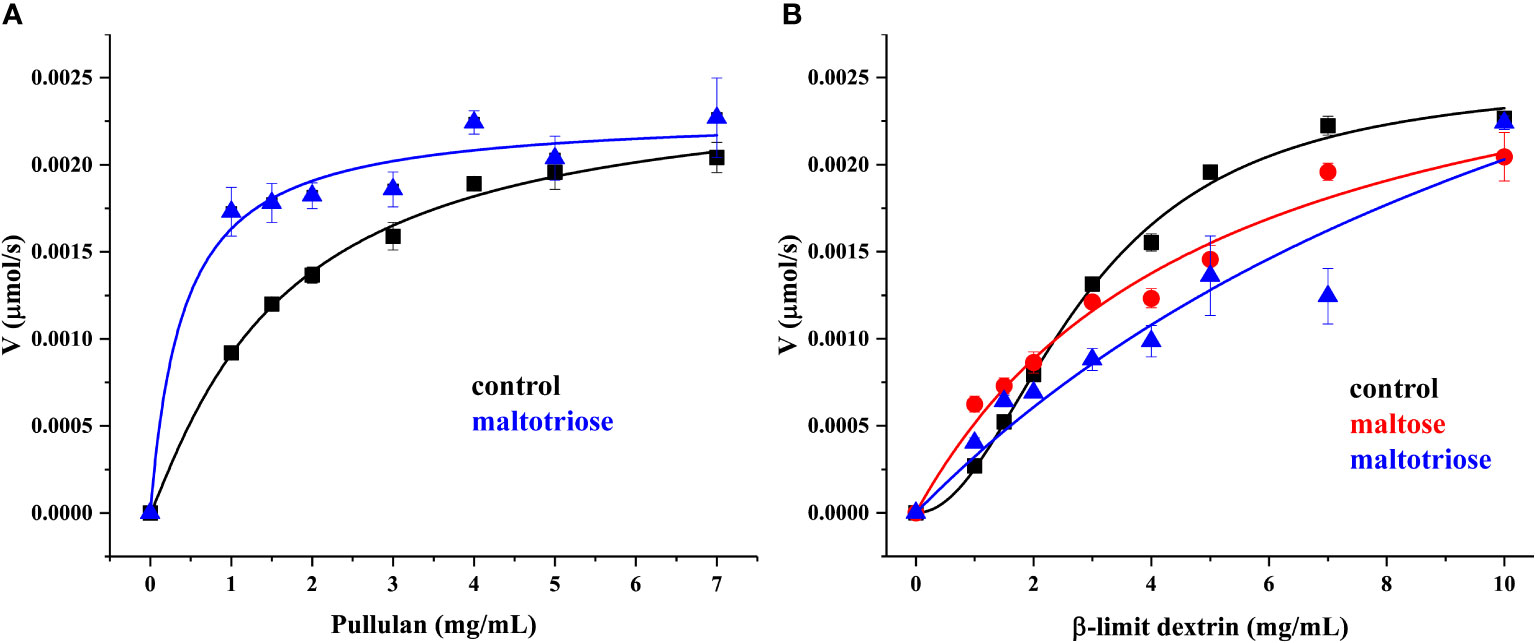
Figure 3 Kinetics study of rMePUL. (A, B) Pullulan and β-limit dextrin were used as substrates, respectively. Black lines represent kinetics without addition of maltooligosaccharide (control) and meanwhile red and blue lines show enzyme kinetics with the presence of 1 mM maltose and maltotriose, respectively. The reactions were carried out in 25 mM acetate buffer pH 6.0 at 50°C. The debranching activity was determined by DNS assay.
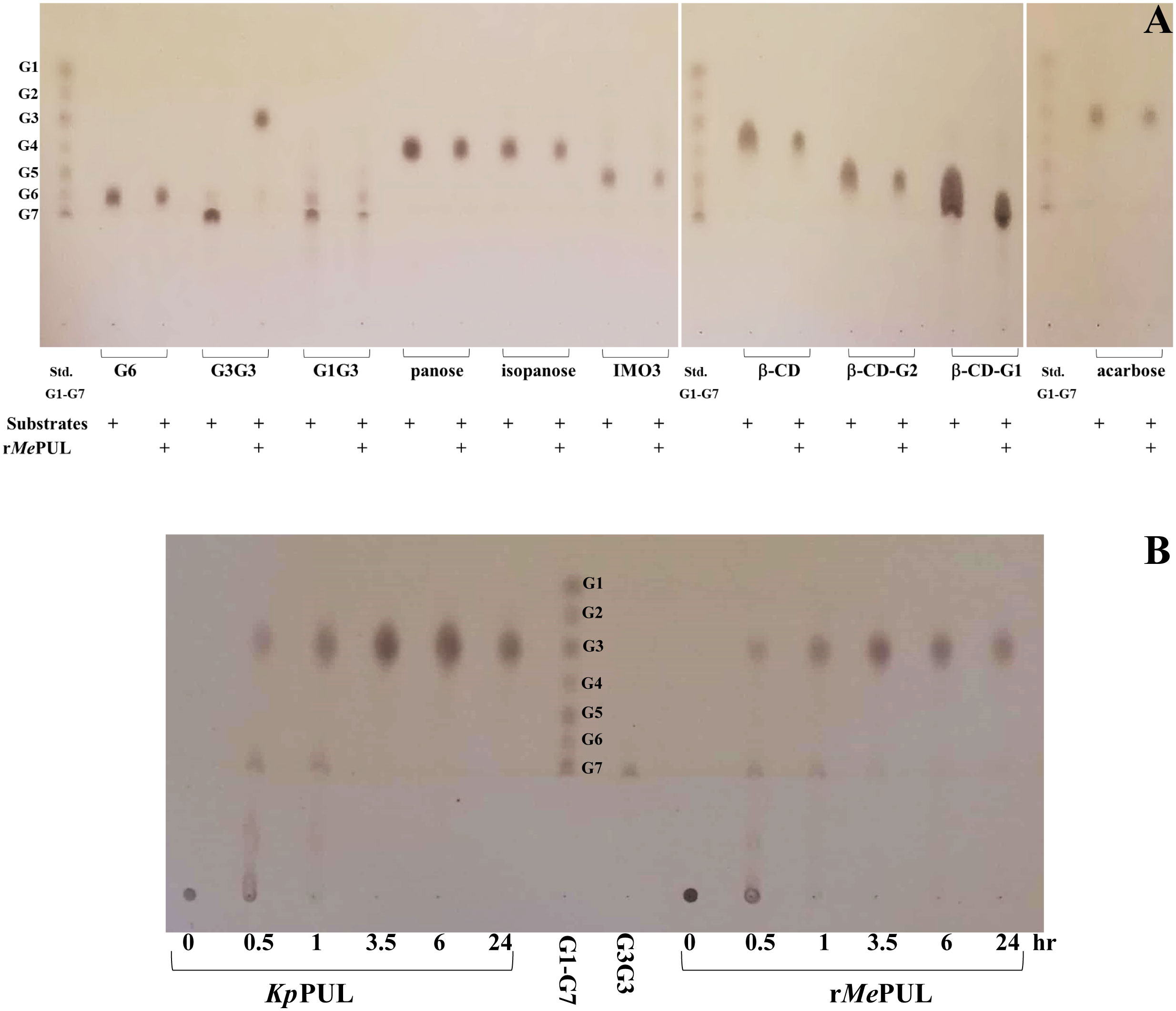
Figure 4 Product analysis by TLC of PUL action. (A) TLC analysis of rMePUL activity on 2.5 mg/mL various oligosaccharides with different linkages after incubating in 25 mM acetate buffer pH 6.0 at 50°C for 2.5 hr. (B) TLC analysis of products produced from KpPUL (commercial pullulanase, Sigma) and rMePUL. The 0.2 U/mL of both PULs were incubated with 5 mg/mL pullulan in 25 mM acetate buffer pH 6.0 at 25°C for KpPUL and at 50°C for rMePUL. The products were analysed at 0, 0.5, 1, 3.5, 6, and 24 hr after incubation.
When pullulan was used as substrate, the patterns of products released from both rMePUL and a commercial pullulanase, Klebsiella pneumoniae pullulanase (KpPUL), were as expected rather similar, as shown in the TLC image (Figure 4B). At 0.5 hr of incubation, series of oligosaccharides harbouring masses of maltotriose building block were apparently released. Notably, at 3.5 h onwards, maltotriose was apparently obtained as a main product (Figure 4B). In addition, the product patterns were also confirmed by HPAEC-PAD and MALDI-TOF MS, where the main peaks corresponded to maltotriose and maltotriosyl-maltotriose (Figures 5A, B).
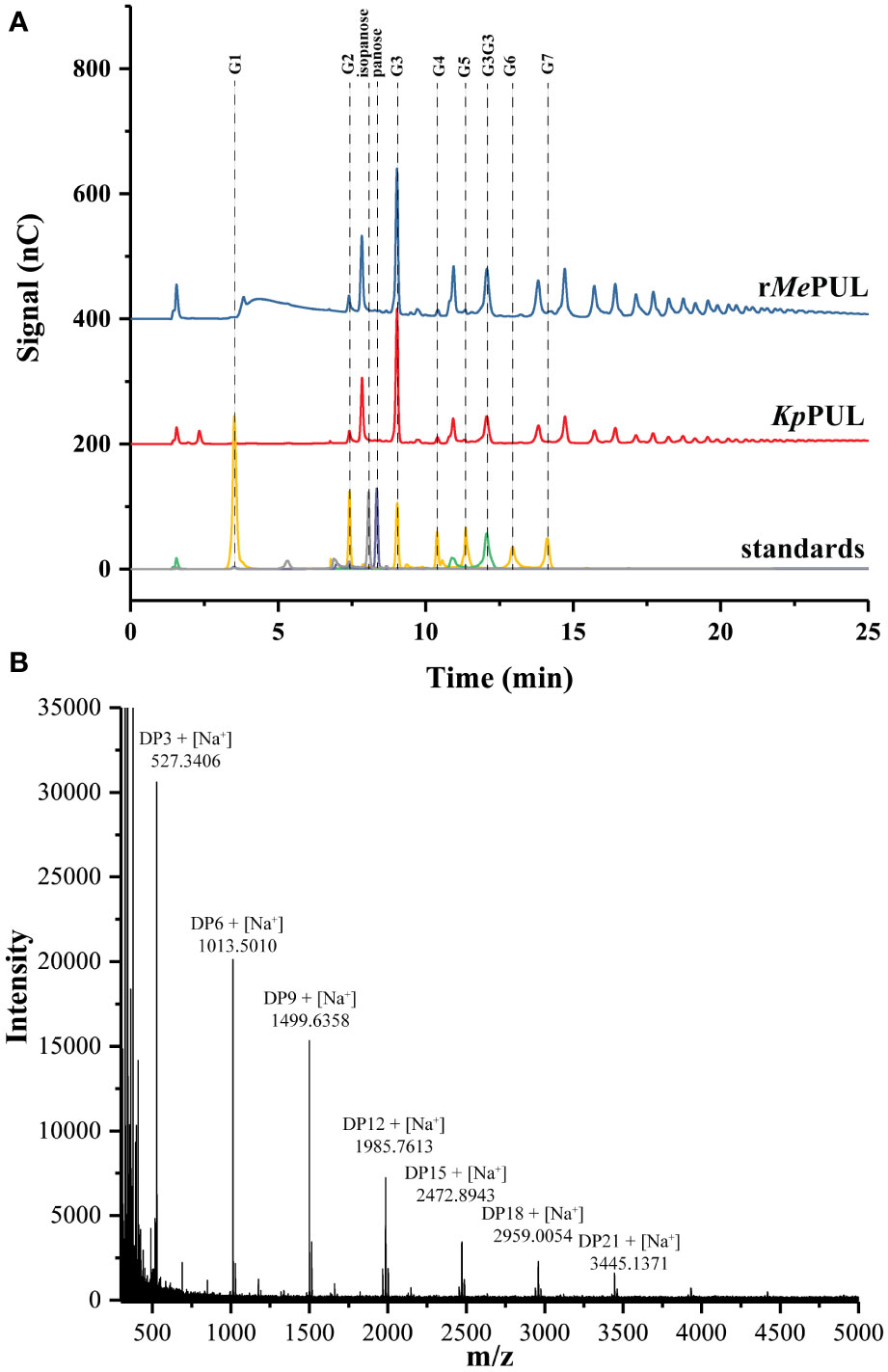
Figure 5 Pullulan hydrolysis. (A) HPAEC-PAD analysis and (B) MALDI-TOF MS analyses of products synthesised from rMePUL. The 0.2 U/mL of rMePUL and KpPUL were incubated with 5 mg/mL pullulan in 25 mM acetate buffer pH 6.0 at 25°C for KpPUL and at 50°C for rMePUL for 0.5 hr.
Determination of a catalytically competent binding conformation of the enzyme is important to describe how the enzyme acts on its specific substrate. Although rMePUL exhibited the highest activity on pullulan (Figure 2D), this substrate is not found in plants but generally obtained from fungi such as Aureobasidium pullulans (Pandey et al., 2021). Additionally, the MePUL active site could not be fully occupied by pullulan substrate since the pullulan structure lacks glucosyl branch point at the position of 0′ subsite in the active site. Thus, 6II-O-maltotriosylmaltotriose or branched maltohexaose which is an interior structure of β-limit dextrin, a more close-to-nature substrate, was chosen instead of pullulan to evaluate the interaction and conformation of mature MePUL when binding to its substrate by using molecular dynamics (MD) simulation analysis. The RMSD plots showed that MePUL was likely stable at 10 ns of simulation, while branched maltohexaose was stable after 20 ns of simulation (Figure 6A). High fluctuation of RMSD value was observed for branched maltohexaose due to its flexibility. Therefore, the last 10 ns trajectories of simulations were selected for further analysis including H-bond network and binding free energy. The binding conformation and H-bond network of branched maltohexaose in MePUL binding site were illustrated in Figure 6B. Branched maltohexaose substrate laid along the MePUL binding track formed H-bonds with N368, D551, R554, D652, N653, and D708. The per-residue decomposition free energy of crucial residues within the substrate-binding track was calculated using the MM/GBSA method (Figure 6C). The result revealed that W365, Y367, N368, H414, C449, V450, L484, W522, F524, D551, R554, F563, H651, D652, N653, D708, Y710, and K737 of MePUL significantly interacted with branched oligosaccharides. Additionally, residue conservation analysis with searching 149 related sequences, such as pullulanases, α-1,6-glucosidases, etc. (Supplementary data II), with the ConSurf server (Ashkenazy et al., 2016) showed that the best predicted binding residues are highly conserved (> 80%), except F563, D708, and K737 (Supplementary Figure S4) which exhibited a lower cutoff (<80%).
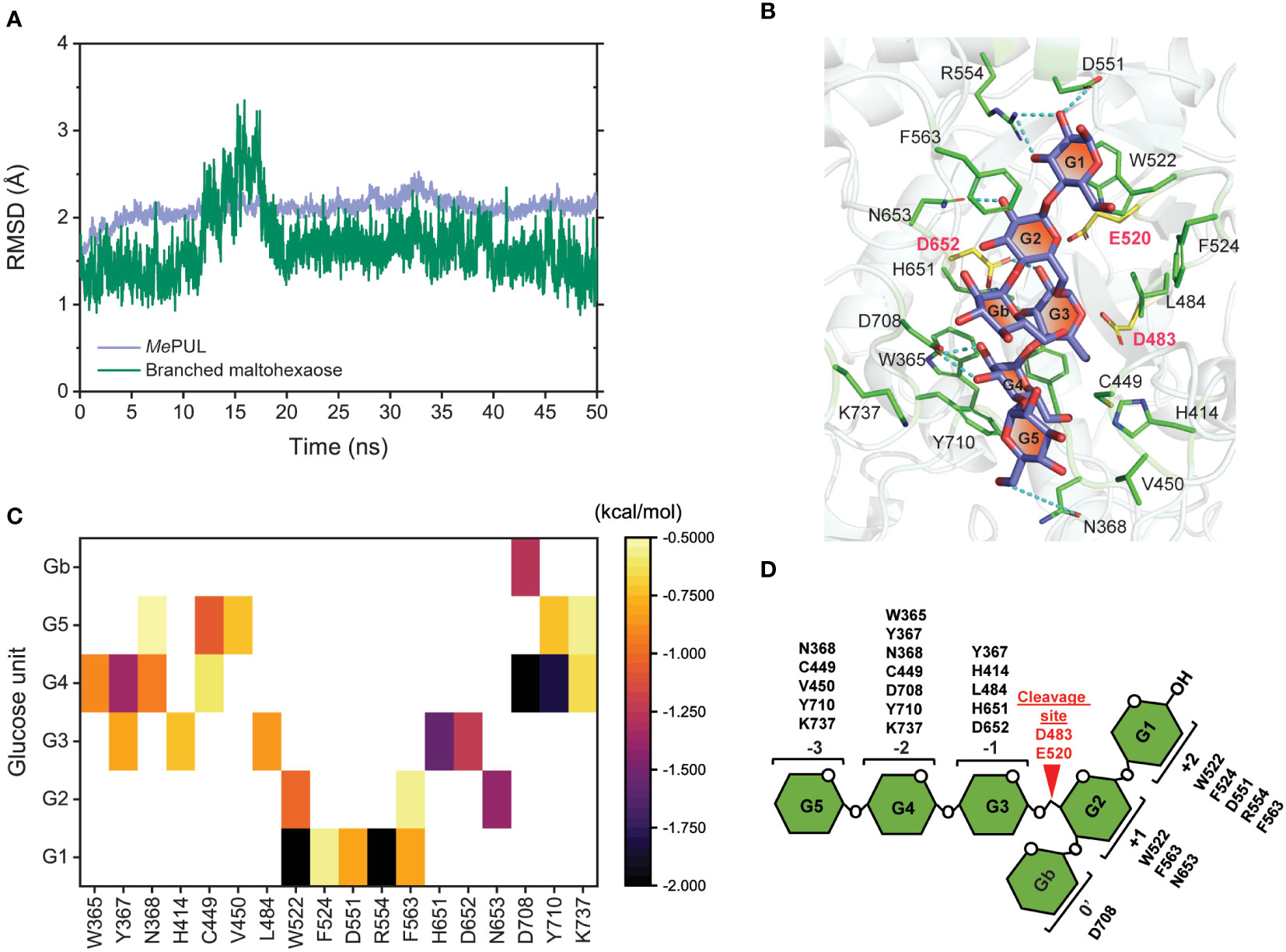
Figure 6 Molecular dynamics simulation analysis of mature MePUL/branched maltohexaose complex. (A) Time evolution of all-atom RMSD. (B) The binding conformation and H-bond network of branched maltohexaose in MePUL binding site. (C) The per-residue decomposition free energy of residues within the substrate-binding track. (D) The proposed oligosaccharide binding subsites of MePUL.
rMePUL could release a broad range of maltooligosaccharides from β-limit dextrin with degree of polymerisation (DP) longer than 7, while products of rMeISA3 were mainly short maltooligosaccharides (DP <5) as shown by HPAEC-PAD (Figure 7) and MALDI-TOF MS (Supplementary Figure S5). When rMePUL and rMeISA3 were co-incubated, the product pattern was similar to that of single rMePUL, however, the peak areas of DP 2 - 4 were obviously greater (>2 times) than a summary of the peak area obtained from each single rMePUL and rMeISA3 reaction (Supplementary Figure S6).
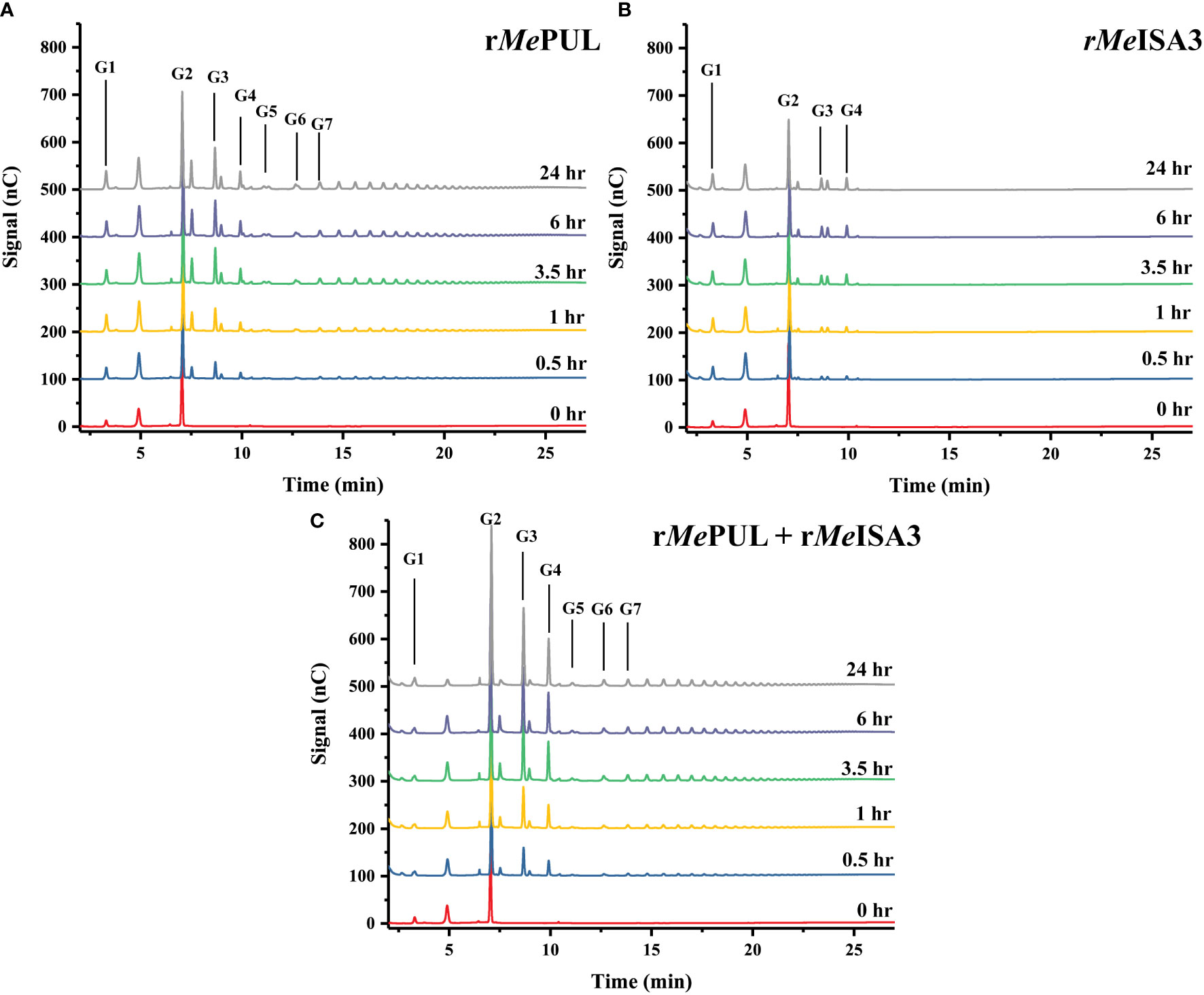
Figure 7 Synergistic activity between rMePUL and rMeISA3 on β-limit dextrin. 0.2 U/mL of (A) rMePUL or (B) rMeISA3 as well as (C) mixture of 0.2 U/mL rMePUL + 0.2 U/mL rMeISA3 were incubated with 0.5% (w/v) β-limit dextrin in 25 mM acetate buffer pH 6.0 at 37°C for 24 hr and products were analysed by HPAEC-PAD.
The pGWB5-Mepul and the silencing suppressor p19 were co-expressed in Agrobacterium tumefaciens GV3101 infiltrated in 4-wk N. benthamiana leaves. The in-planta assay showed that GFP-tagged MePUL fluorescence signals were detected in the chloroplasts (Figure 8, bottom panel, inset). This corresponded to the presence of transit peptide sequence predicted in silico. Conversely, the eGFP protein, as control, was overexpressed and localised throughout the plant cells.
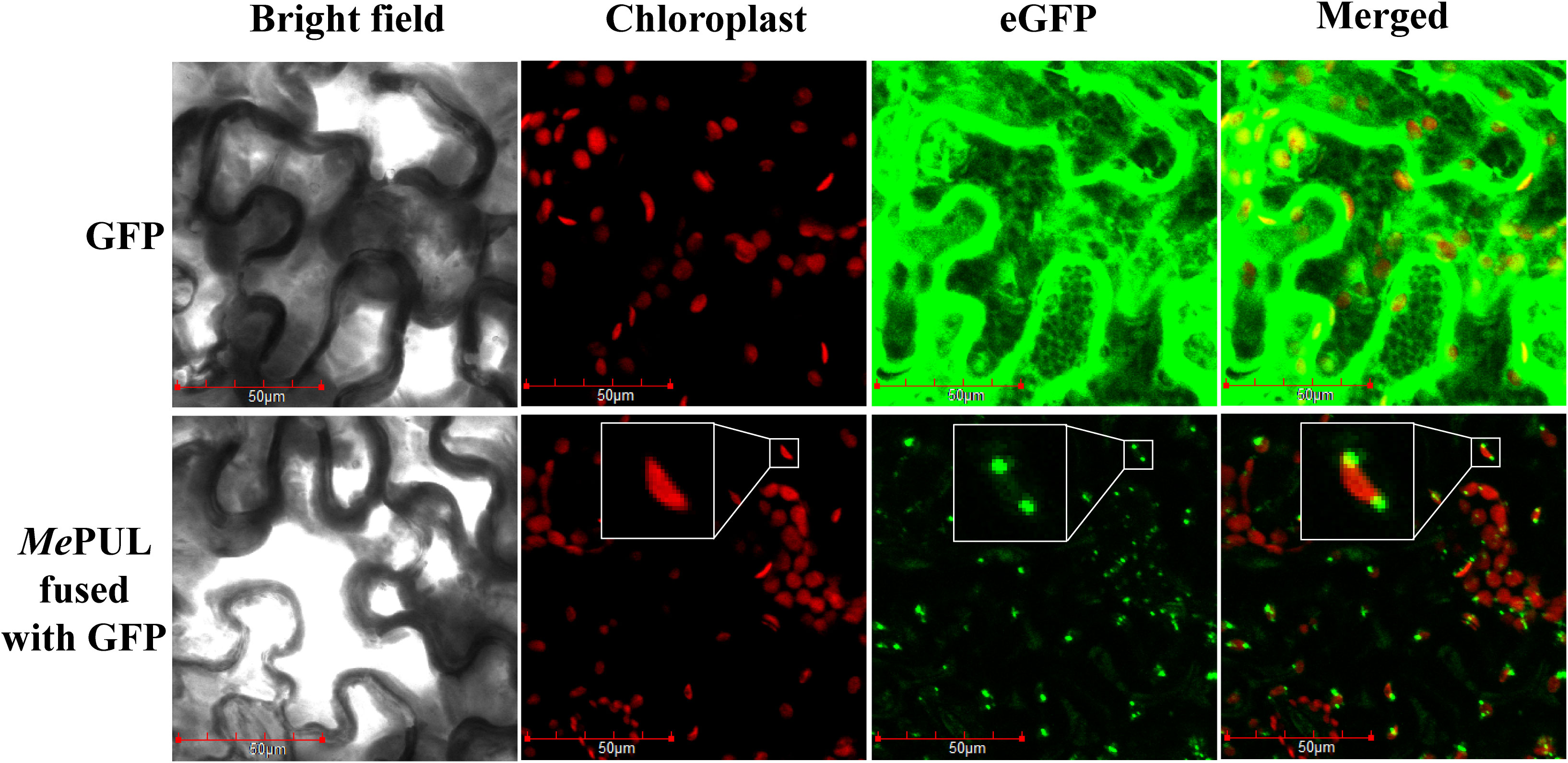
Figure 8 Subcellular GFP-tagged MePUL localisation in Nicotiana benthamiana leaves. Confocal microscopic images of N. benthamiana leaf epidermal cells infiltrated with pGWB2-GFP (control; upper panel) and pGWB5-Mepul (lower panel). Chloroplast autofluorescence (chloroplast), GFP fluorescence (eGFP), and merged images are shown. Plastidial MePUL-GFP localisation is shown as enlargements in the insets. Scale bars: 50 μm.
The protein sequence of rMePUL was identical to the cassava pullulanase in the Phytozome database [Manes.10G051700.1.p]. Comparison of the MePUL to those of other plants and bacteria showed some sequecne similarity - around 37 – 74%, including to Hordeum vulgare pullulanase (HvLD; GenBank AAD04189.1, 65%), Oryza sativa pullulanase (OsPUL; GenBank BAA09167.1, 60%), Solanum tuberosum pullulanase (StPUL; GenBank XP_006361707.1, 71%), Arabidopsis thaliana pullulanase (AtPUL; GenBank NP_196056.2, 72%) Zea mays pullanase (ZmPUL; GenBank NP_001104920.1, 74%), and Spinacia oleracea pullulanase (SoPUL; GenBank CAA58803.1, 71%), and Klebsiella pneumoniae pullulanase, a bacterial pullulanase (KpPUL: PDB 2FGZ, 37%). The sequence alignments are presented in Figures S2, S7. However, MePUL showed very low similarity compared with other types of Manihot esculenta debranching enzymes (MeISA1: GenBank AUZ20772.1, 23%; MeISA2: GenBank AUZ20773.1, 24%; and MeISA3: GenBank AUZ20774.1, 29%) shown in Figure S8. Although they are members of the same Glycoside hydrolase (GH) 13 family, ISAs are subcategorised into subfamily 11. The MePUL belongs to GH 13 subfamily 13 (pullulanase subfamily) based on sequence similarity (Machovič and Janeček, 2008). The MePUL also comprised 4 domains of N-terminal domain, CBM48, catalytic domain, and C-terminal domain similar to that of HvPUL as shown in Figure S2 (Vester-Christensen et al., 2010b).
Fortunately, although the MePUL comprised a total of 14 cysteine residues in the mature protein sequence, it was successfully expressed in soluble form in E. coli SoluBL (DE3) transformant. Noticeably, protein MW obtained from gel filtration chromatography under non-denaturing condition was ~ 2-fold higher than the MW shown by SDS-PAGE analysis (Figure 1), which implied that MePUL could be naturally present as a homodimer. Unlike other debranching enzymes from cassava, the MeISA1 and MeISA2 essentially exist as catalytically active hetermeric enzymes with various molar ratios (Panpetch et al., 2018b), while MeISA3 is a monomeric enzyme (Panpetch et al., 2018a). Additionally, SoPUL was reported as a monomeric protein (Renz et al., 1998) and partially purified StPUL from potato tuber present as a dimer analysed under nondenaturing condition however 3 protein bands were shown on SDS-PAGE gel (Ishizaki et al., 1983).
An in vitro biochemical assay of rMePUL disclosed that rMePUL has pH optimum in the slightly acidic pH range and is a metal ion-dependent enzyme (Figure 2). Surprisingly, the optimum temperature of rMePUL was at 50°C (Figure 2A), while for rMeISA3 it was at only at 37°C (Panpetch et al., 2018a) even though they both originate from the same organism. rMePUL exhibited the highest activity in acetate buffer pH at 6.0 (Figure 2B) like rMeISA3. The activity dropped by up to 50% at pH 5.0 and 7.0, with a sharp drop of activity below 5 or above 7, similar to the reported optimum pH of isoamylases from potato and maize (Ishizaki et al., 1983; Doehlert and Knutson, 1991; Rahman et al., 1998). Nevertheless, both optimal pH and temperature between MePUL and StPUL were similarly (Ishizaki et al., 1983). Moreover, only Ca2+ ion could promote the activity of rMePUL (Figure 2C) corresponded to the presence of embedding of Ca2+ in HvPUL crystal structure (Vester-Christensen et al., 2010b), while the opposite was true for StPUL where Ca2+ ion lowered the enzyme activity by around 40%. However, enzyme activity was completely inhibited by Cu2+ and Zn2+ (Figure 2C), correlating with previous work of Iwaki and Fuwa (1981) (Iwaki and Fuwa, 1981), which reported that 1 mM Cu2+, Zn2+, Ag2+, or Cd2+ inhibited rice endosperm debranching enzyme. Moreover, we also found that rMePUL activity could be enhanced after treating with EDTA. Possibly, there was contamination of some metal ion inhibitors in gene expression and protein purification processes
To evaluate the substrate specificity of rMePUL, the enzyme was incubated with various substrates (Figure 2). rMePUL exhibited the highest activity toward pullulan (Figure 2A). This confirmed that this debranching enzyme was pullulanase-type, while isoamylase-type debranching enzyme clearly showed no activity on pullulan at all (Panpetch et al., 2018b; Panpetch et al., 2018a). Notably, rMePUL was unable to cleave substrates harbouring highly branched structures, especially amylopectin, cassava starch, and glycogen. However, compared to no activity on cassava starch, rMePUL could digest potato starch weakly, with ~20% of its activity on pullulan. This possibly resulted from less complexity of the intrinsic structure of potato starch than that of cassava. Hence, in vitro results could support the trimming model hypothesis of starch granule synthesis, proposing that PUL could partially substitute for ISA1 activity (Dinges et al., 2003; Wattebled et al., 2005; Fujita et al., 2009). Importantly, rMePUL also preferentially digested β-limit dextrin at a high level as shown in Figure 2D. This might be because its structure is rather similar to that of α-limit dextrin or pullulan-like polymers found in plants, which are both produced by amylase action in starch degradation. Moreover, the results from protein localisation indicated that MePUL was specifically localised to the plastid organelles (Figure 8) such as chloroplast and amyloplast, where starch degradation occurs in plant cells (Zeeman et al., 2010). Taken together, these data along with some previous reports on target localisation to plastid organelles support the notion that MePUL principally participates in starch degradation processes (Zeeman et al., 2010). Additionally, unlike other PULs from cereals, such as rice (Yamasaki et al., 2008) and barley (Mcdougall et al., 2004), transglycosylation activity could not be observed in rMePUL.
Enzyme kinetic analysis with rMePUL acting on pullulan and β-limit dextrin was fitted with Hill plots, indicating the cooperative manner of the enzyme. Conversely, it was still unclear why addition of some maltooligosaccharides such as maltose and maltotriose could obviously change the kinetic pattern of rMePUL (from Hill to Michaleis-Menten). Notably, maltotriose, the main product of pullulan hydrolysis, was a potential positive effector of the rMePUL action, since it could clearly promote binding affinity (k or Km) of the enzyme to pullulan substrate. However, maltose and maltotriose could also increase kcat around 1.2 - 1.7 folds for β-limit dextrin hydrolysis (Figure 3B, Supplementary Figure S3, and Table 1). It might be that these maltooligosaccharides produced during starch degradation (Zeeman et al., 2010) act as allosteric modulators to some enzymes participating in the starch degradation process.
Although MePUL exhibited very low sequence similarity to KpPUL (Mikami et al., 2006) (Supplementary Figure S7), their product patterns of on pullulan degradation were rather similar, mainly reflecting the release of maltotriose and maltotriose-containing oligosaccharides (Figure 4B). However, pullulan does not entirely consist of maltotriose building blocks (Catley and Whelan, 1971; Carolan et al., 1983); it also contains maltooligosaccharides with different DPs, such as maltose (DP2) and maltotetraose (DP4), which could also be detected in HPAEC analysis from rMePUL and KpPUL reactions. rMePUL specifically hydrolysed α-1,6 linkage between maltoriose units but no hydrolytic activity on linear α-1,4-linked oligosaccharides was observed (Figure 4A), strongly indicating that MePUL is a pullulanase type I (Møller et al., 2016). Cyclic oligosaccharides such as β-cyclodextrin (β-CD) and α-1,6 branched β-cyclodextrins (β-CD-G1 and β-CD-G2) were not cleaved by rMePUL. Furthermore, the pullulan cleavage was almost completely inhibited by β-CD as judged by DNS assay. This corresponded to the competitive inhibition of debranching activities of KpPUL and HvLD by CDs, where the cyclic structure of cyclic oligosaccharides can productively bind at subsites +1, +2, and 0′, as shown in superimposition structures of MePUL and HvLD (Vester-Christensen et al., 2010a; Møller et al., 2015) (Supplementary Figure S9). Although β-CD-G1 and β-CD-G2 contained an α-1,6 branch, the glucosyl and maltosyl branches were not appropriately positioned at catalytic cleavage site (Supplementary Figure S9), corresponding to complex structure of HvLD with 6-S-(α-D-maltosyl)-6-deoxy-6-thiocyclomaltoheptaose (Møller et al., 2015). Therefore, β-CD-G1 and β-CD-G2 could not be hydrolysed by rMePUL. Interestingly, rMePUL could not digest the α-1,6 linkage of glucosyl-maltotriose (G1G3), panose or isopanose. The experimental data demonstrated that the active site of rMePUL required longer substrates in order to make productive contacts with the longer binding site. This information acorrelated with computational analysis, which showed that the MePUL-branched maltohexaose complex exhibited high binding energy at +2, +1, -1, -2, and -3 sites (Figure 6C). The computational and bioinformatic data revealed that the residues W365, Y367, N368, H414, C449, V450, L484, W522, F524, D551, R554, F563, H651, D652, N653, D708, Y710, and K737 were potentially essential for substrate binding based on the results of the MM/GBSA method and conserved sequence analysis. Most of these residues, which bound with ligands (Figure 6B), were highly conserved at the same positions in the 3D-structures of MePUL, HvLD, (Møller et al., 2015), and KpPUL (Mikami et al., 2006) even though they did not show high similarity of overall protein sequences. Notably, two out of those residues, F563 and D708 located near the branch point of branched maltohexaose, showed 80% lower score of the cutoff conservation. Perhaps these variable residues might play a role in imposing the substrate specificity. Proposed oligosaccharide binding subsites of MePUL are shown in Figure 6D.
To analyse the relationship between MePUL and MeISA3 debranching activities in starch degradation, mixed-enzyme reaction was performed on β-limit dextrin substrate (Figure 7). The results suggested that rMePUL preferentially cleave longer chains in broad ranges of maltooligosaccharides and meanwhile rMeISA3 liberated only short-chain maltooligosaccharides (Figure 7 and Figure S4). This suggested that MePUL might play a major role in starch debranching of cassava starch catabolism. Interestingly, when rMeISA3 and rMePUL were present together in the same reaction, the debranching activity of short-chain oligosaccharides (DP 2 - 4) was apparently increased (Figure 7 and Figure S6). This result implied that MeISA3 and MePUL worked synergistically, possibly by providing favourable substrates for each other by removal of some unfavourable branched points due to their different substrate preferences. Importantly, considering the prospect of an heterocomplex between rMeISA3 and rMePUL, no multimeric rMeISA3/rMePUL complex was observed by gel filtration chromatography (Supplementary Figure S10). This demonstrated that they should work separately for their debranching activities.
Some previous studies in vivo revealed that deficiency of PUL in rice (Fujita et al., 2009) and maize (Wong et al., 2003) slightly effected on an accumulation of water-soluble glucan. Although MeISA3 and MePUL seemed to exhibit overlapping activities in debranching processes, cassava starch catabolism may not be efficient when only either PUL or ISA3 are present, which contrasts with some previous reports from other plant species (Wong et al., 2003; Delatte et al., 2006; Fujita et al., 2009). Importantly, both PUL and ISA3 have been conserved in most plants. Previous in vivo studies showed the relationship between these two enzymes, but they did not clearly unravel the reason for the conservation of both enzymes. Nevertheless, our results show that both PUL and ISA3 are necessary for efficient starch degradation in vitro: their synergistic action may account for why these two types of debranching enzymes are typically present in plant cells.
Pullulanase from Manihot esculenta Crantz ‘KU50’ was successfully cloned, expressed, and biochemically characterised. This debranching enzyme was clearly categorised as pullulanase type I due to its preference for cleaving α-1,6 linkages of pullulan and β-limit dextrin. The debranching action of PUL is conserved in most plants, both mono- and di-cotyledons, and is located in plastids. The present work demonstrates the synergistic action between MePUL and MeISA3, which is important for efficient starch degradation in cassava and which may have implications for biotechnology innovation in relation to harnessing starch for food and materials applications.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
KW, TC, and PP: conceptualization. KW, TC, MK, and PP: methodology. KW, TC, and PP: formal analysis. KW, TC, MK, RF, and PP: investigation. TC and PP: resources. TC and PP: data curation. KW, TC, and MK: writing - original draft. KW, TC, RF, and PP: writing - review & editing. KW, TC, MK, and PP: visualization. KW and PP: supervision. PP: funding acquisition. PP: Project Administration. All authors contributed to the article and approved the submitted version.
This research was supported by Thailand Science research and Innovation Fund Chulalongkorn University [BCG66230003 to PP]. KW received a Second Century Fund (C2F) Postdoctoral Fellowship of Chulalongkorn University. This work [Grant No. RGNS 64-014] is also supported by Office of the Permanent Secretary, Ministry of Higher Education, Science, Research and Innovation (OPS MHESI), Thailand Science Research and Innovation (TSRI) and Chulalongkorn University. This research project is supported by grants for development of new faculty Staff, Ratchadaphiseksomphot Fund, Chulalongkorn University”.
We would like to thank Mr. Prasert Thala from National Research Centre of Millet and Corn (Suwan Farm) in Nakhon Ratchasima province for kindly providing cassava ‘KU50’ tubers. The N. benthamiana plants were kindly provided by Ms. Poorichaya Singcha. We also thanks to Assist. Prof. Dr. Rath Pichyangkura, Assoc. Prof. Dr. Kuakarun Krusong, and Assoc. Prof. Dr. Supaart Sirikantaramas, Department of Biochemistry, Faculty of Science, Chulalongkorn University for some chemicals supports.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1114215/full#supplementary-material
Aguilar, B., Anandakrishnan, R., Onufriev, A. (2012). H++ 3.0: automating pK prediction and the preparation of biomolecular structures for atomistic molecular modeling and simulations. Nucleic Acids Res. 40, W537–W541. doi: 10.1093/nar/gks375
Ashkenazy, H., Abadi, S., Martz, E., Chay, O., Mayrose, I., Pupko, T., et al. (2016). ConSurf 2016: an improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res. 44, W344–W350. doi: 10.1093/nar/gkw408
Aslanidis, C., De Jong, P. J. (1990). Ligation-independent cloning of PCR products (LIC-PCR). Nucleic Acids Res. 18, 6069–6074. doi: 10.1093/nar/18.20.6069
Ball, S. G., Morell, M. K. (2003). From bacterial glycogen to starch: understanding the biogenesis of the plant starch granule. Annu. Rev. Plant Biol. 54, 207–233. doi: 10.1146/annurev.arplant.54.031902.134927
Carolan, G., Catley, B. J., Mcdougal, F. J. (1983). The location of tetrasaccharide units in pullulan. Carbohydr. Res. 114, 237–243. doi: 10.1016/0008-6215(83)88190-7
Caspar, T., Lin, T.-P., Kakefuda, G., Benbow, L., Preiss, J., Somerville, C. (1991). Mutants of arabidopsis with altered regulation of starch degradation. Plant Physiol. 95, 1181–1188. doi: 10.1104/pp.95.4.1181
Catley, B., Whelan, W. (1971). Observations on the structure of pullulan. Arch. Biochem. biophysics 143, 138–142. doi: 10.1016/0003-9861(71)90193-7
Delatte, T., Trevisan, M., Parker, M. L., Zeeman, S. C. (2005). Arabidopsis mutants Atisa1 and Atisa2 have identical phenotypes and lack the same multimeric isoamylase, which influences the branch point distribution of amylopectin during starch synthesis. Plant J. 41, 815–830. doi: 10.1111/j.1365-313X.2005.02348.x
Delatte, T., Umhang, M., Trevisan, M., Eicke, S., Thorneycroft, D., Smith, S. M., et al. (2006). Evidence for distinct mechanisms of starch granule breakdown in plants. J. Biol. Chem. 281, 12050–12059. doi: 10.1074/jbc.M513661200
Dinges, J. R., Colleoni, C., James, M. G., Myers, A. M. (2003). Mutational analysis of the pullulanase-type debranching enzyme of maize indicates multiple functions in starch metabolism. Plant Cell 15, 666–680. doi: 10.1105/tpc.007575
Doehlert, D. C., Knutson, C. A. (1991). Two classes of starch debranching enzymes from developing maize kernels. Journal of Plant Physiology 138(5), 566–572. doi: 10.1016/S0176-1617(11)80242-7
Drula, E., Garron, M.-L., Dogan, S., Lombard, V., Henrissat, B., Terrapon, N. (2022). The carbohydrate-active enzyme database: functions and literature. Nucleic Acids Res. 50, D571–D577. doi: 10.1093/nar/gkab1045
Fujita, N., Toyosawa, Y., Utsumi, Y., Higuchi, T., Hanashiro, I., Ikegami, A., et al. (2009). Characterization of pullulanase (PUL)-deficient mutants of rice (Oryza sativa l.) and the function of PUL on starch biosynthesis in the developing rice endosperm. J. Exp. Bot. 60, 1009–1023. doi: 10.1093/jxb/ern349
Genheden, S., Kuhn, O., Mikulskis, P., Hoffmann, D., Ryde, U. (2012). The normal-mode entropy in the MM/GBSA method: effect of system truncation, buffer region, and dielectric constant. J. Chem. Inf. modeling 52, 2079–2088. doi: 10.1021/ci3001919
Gordon, J., Myers, J., Folta, T., Shoja, V., Heath, L. S., Onufriev, A. (2005). H++: a server for estimating pKas and adding missing hydrogens to macromolecules. Nucleic Acids Res. 33, 368–371. doi: 10.1093/nar/gki464
Hussain, H., Mant, A., Seale, R., Zeeman, S., Hinchliffe, E., Edwards, A., et al. (2003). Three isoforms of isoamylase contribute different catalytic properties for the debranching of potato glucans. Plant Cell 15, 133–149. doi: 10.1105/tpc.006635
Ishizaki, Y., Taniguchi, H., Maruyama, Y., Nakamura, M. (1983). Debranching enzymes of potato tubers (Solanum tuberosum l.) II. purification of a pullulanase (R-enzyme) from potato tubers and comparison of its properties with those of the potato isoamylase. J. Japanese Soc. Starch Sci. 30, 19–29. doi: 10.5458/jag1972.30.19
Iwaki, K., Fuwa, H. (1981). Purification and some properties of debranching enzyme of germinating rice endosperm. Agricultural and Biological Chemistry 45(12), 2683–2688. doi: 10.1271/bbb1961.45.2683
Jorgensen, W., Chandrasekhar, J., Madura, J. (1983). Lmpey, RW; Klein, ML. J. Chem. Phys. 79, 926. doi: 10.1063/1.445869
Klaewkla, M., Charoenwongpaiboon, T., Mahalapbutr, P. (2021). Molecular basis of the new COVID-19 target neuropilin-1 in complex with SARS-CoV-2 S1 c-end rule peptide and small-molecule antagonists. J. Mol. Liquids 335, 116537. doi: 10.1016/j.molliq.2021.116537
Li, E., Hasjim, J., Gilding, E. K., Godwin, I. D., Li, C., Gilbert, R. G. (2019). The role of pullulanase in starch biosynthesis, structure, and thermal properties by studying sorghum with increased pullulanase activity. Starch-Stärke 71, 1900072. doi: 10.1002/star.201900072
Lindbo, J. A. (2007). High-efficiency protein expression in plants from agroinfection-compatible tobacco mosaic virusexpression vectors. BMC Biotechnol. 7, 1–11. doi: 10.1186/1472-6750-7-52
Møller, M. S., Henriksen, A., Svensson, B. (2016). Structure and function of α-glucan debranching enzymes. Cell. Mol. Life Sci. 73, 2619–2641. doi: 10.1007/s00018-016-2241-y
Møller, M. S., Windahl, M. S., Sim, L., Bøjstrup, M., Abou Hachem, M., Hindsgaul, O., et al. (2015). Oligosaccharide and substrate binding in the starch debranching enzyme barley limit dextrinase. J. Mol. Biol. 427, 1263–1277. doi: 10.1016/j.jmb.2014.12.019
Machovič, M., Janeček, Š. (2008). Domain evolution in the GH13 pullulanase subfamily with focus on the carbohydrate-binding module family 48. Biologia 63, 1057–1068. doi: 10.2478/s11756-008-0162-4
Mcdougall, G. J., Ross, H. A., Swanston, J. S., Davies, H. V. (2004). Limit dextrinase from germinating barley has endotransglycosylase activity, which explains its activation by maltodextrins. Planta 218, 542–551. doi: 10.1007/s00425-003-1141-1
Mikami, B., Iwamoto, H., Malle, D., Yoon, H.-J., Demirkan-Sarikaya, E., Mezaki, Y., et al. (2006). Crystal structure of pullulanase: evidence for parallel binding of oligosaccharides in the active site. J. Mol. Biol. 359, 690–707. doi: 10.1016/j.jmb.2006.03.058
Miller, G. L. (1959). Use of dinitrosalicylic acid reagent for determination of reducing sugar. Analytical Chem. 31, 426–428. doi: 10.1021/ac60147a030
Myers, J., Grothaus, G., Narayanan, S., Onufriev, A. (2006). A simple clustering algorithm can be accurate enough for use in calculations of pKs in macromolecules. Proteins: Structure Function Bioinf. 63, 928–938. doi: 10.1002/prot.20922
Myers, A. M., Morell, M. K., James, M. G., Ball, S. G. (2000). Recent progress toward understanding biosynthesis of the amylopectin crystal. Plant Physiol. 122, 989–998. doi: 10.1104/pp.122.4.989
Nakagawa, T., Kurose, T., Hino, T., Tanaka, K., Kawamukai, M., Niwa, Y., et al. (2007). Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J. bioscience bioengineering 104, 34–41. doi: 10.1263/jbb.104.34
Nakamura, Y. (2002). Towards a better understanding of the metabolic system for amylopectin biosynthesis in plants: rice endosperm as a model tissue. Plant Cell Physiol. 43, 718–725. doi: 10.1093/pcp/pcf091
Pandey, S., Shreshtha, I., Sachan, S. G. (2021). “Pullulan: Biosynthesis, production and applications,” in Microbial exopolysaccharides as novel and significant biomaterials (Cham, Switzerland: Springer), 121–141.
Panpetch, P., Field, R. A., Limpaseni, T. (2018a). Cloning of the full-length isoamylase3 gene from cassava manihot esculenta crantz ‘KU50’and its heterologous expression in e. coli. Plant Physiol. Biochem. 132, 281–286. doi: 10.1016/j.plaphy.2018.09.010
Panpetch, P., Field, R. A., Limpaseni, T. (2018b). Heterologous co-expression in e. coli of isoamylase genes from cassava manihot esculenta crantz ‘KU50’achieves enzyme-active heteromeric complex formation. Plant Mol. Biol. 96, 417–427. doi: 10.1007/s11103-018-0707-z
Panpetch, P., Sirikantaramas, S. (2021). Fruit ripening-associated leucylaminopeptidase with cysteinylglycine dipeptidase activity from durian suggests its involvement in glutathione recycling. BMC Plant Biol. 21, 1–14. doi: 10.1186/s12870-021-02845-6
Quan, J., Tian, J. (2011). Circular polymerase extension cloning for high-throughput cloning of complex and combinatorial DNA libraries. Nat. Protoc. 6, 242–251. doi: 10.1038/nprot.2010.181
Rahman, A., Wong, K. S., Jane, J. L., Myers, A. M., James, M. G. (1998). Characterization of SU1 isoamylase, a determinant of storage starch structure in maize. Plant Physiology 117(2), 425–435. doi: 10.1104/pp.117.2.425
Renz, A., Schikora, S., Schmid, R., Kossmann, J., Beck, E. (1998). cDNA sequence and heterologous expression of monomeric spinach pullulanase: multiple isomeric forms arise from the same polypeptide. Biochem. J. 331, 937–945. doi: 10.1042/bj3310937
Roe, D., Cheatham, T. (2013). PTRAJ and CPPTRAJ: Software for processing and analysis of molecular. Dynamics Trajectory Data 9, 3084–3095. doi: 10.1021/ct400341p
Stam, M. R., Danchin, E. G., Rancurel, C., Coutinho, P. M., Henrissat, B. (2006). Dividing the large glycoside hydrolase family 13 into subfamilies: towards improved functional annotations of α-amylase-related proteins. Protein Engineering Design Selection 19, 555–562. doi: 10.1093/protein/gzl044
Tantanarat, K., O’neill, E. C., Rejzek, M., Field, R. A., Limpaseni, T. (2014). Expression and characterization of 4-α-glucanotransferase genes from manihot esculenta crantz and arabidopsis thaliana and their use for the production of cycloamyloses. Process Biochem. 49, 84–89. doi: 10.1016/j.procbio.2013.10.009
Tappiban, P., Smith, D. R., Triwitayakorn, K., Bao, J. (2019). Recent understanding of starch biosynthesis in cassava for quality improvement: A review. Trends Food Sci. Technol. 83, 167–180. doi: 10.1016/j.tifs.2018.11.019
Vester-Christensen, M. B., Abou Hachem, M., Naested, H., Svensson, B. (2010a). Secretory expression of functional barley limit dextrinase by pichia pastoris using high cell-density fermentation. Protein Expression purification 69, 112–119. doi: 10.1016/j.pep.2009.08.016
Vester-Christensen, M. B., Abou Hachem, M., Svensson, B., Henriksen, A. (2010b). Crystal structure of an essential enzyme in seed starch degradation: barley limit dextrinase in complex with cyclodextrins. J. Mol. Biol. 403, 739–750. doi: 10.1016/j.jmb.2010.09.031
Waksmundzka-Hajnos, M., Sherma, J., Kowalska, T. (2008). Thin layer chromatography in phytochemistry (Boca Raton, Florida: CRC Press).
Wangpaiboon, K., Klaewkla, M., Charoenwongpaiboon, T., Vongkusolkit, N., Panpetch, P., Kuttiyawong, K., et al. (2022). Synergistic enzyme cocktail between levansucrase and inulosucrase for superb levan-type fructooligosaccharide synthesis. Enzyme Microbial Technol. 154, 109960. doi: 10.1016/j.enzmictec.2021.109960
Wangpaiboon, K., Laohawuttichai, P., Kim, S.-Y., Mori, T., Nakapong, S., Pichyangkura, R., et al. (2021). A GH13 α-glucosidase from weissella cibaria uncommonly acts on short-chain maltooligosaccharides. Acta Crystallographica Section D: Struct. Biol. 77, 1064–1076. doi: 10.1107/S205979832100677X
Wattebled, F., Dong, Y., Dumez, S., Delvallé, D., Planchot, V., Berbezy, P., et al. (2005). Mutants of arabidopsis lacking a chloroplastic isoamylase accumulate phytoglycogen and an abnormal form of amylopectin. Plant Physiol. 138, 184–195. doi: 10.1104/pp.105.059295
Wong, K.-S., Kubo, A., Jane, J.-L., Harada, K., Satoh, H., Nakamura, Y. (2003). Structures and properties of amylopectin and phytoglycogen in the endosperm of sugary-1 mutants of rice. J. Cereal Sci. 37, 139–149. doi: 10.1006/jcrs.2002.0485
Yamasaki, Y., Nakashima, S., Konno, H. (2008). Pullulanase from rice endosperm. Acta Biochim. Polonica 55, 507–510. doi: 10.18388/abp.2008_3056
Keywords: cassava (Manihot esculenta Crantz), debranching enzyme, pullulan, pullulanase, starch, starch degradation, synergistic debranching
Citation: Wangpaiboon K, Charoenwongpaiboon T, Klaewkla M, Field RA and Panpetch P (2023) Cassava pullulanase and its synergistic debranching action with isoamylase 3 in starch catabolism. Front. Plant Sci. 14:1114215. doi: 10.3389/fpls.2023.1114215
Received: 02 December 2022; Accepted: 11 January 2023;
Published: 27 January 2023.
Edited by:
Mariam Gaid, Independent researcher, Braunschweig, GermanyReviewed by:
Stefan Janecek, Slovak Academy of Sciences (SAS), SlovakiaCopyright © 2023 Wangpaiboon, Charoenwongpaiboon, Klaewkla, Field and Panpetch. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Pawinee Panpetch, UGF3aW5lZS5wQGNodWxhLmFjLnRo
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.