- 1State Forestry and Grassland Administration Key Laboratory of Silviculture in Downstream Areas of The Yellow River, College of Forestry, Shandong Agricultural University, Tai’an, Shandong, China
- 2Mountain Tai Forest Ecosystem Research Station of State Forestry and Grassland Administration, College of Forestry, Shandong Agricultural University, Tai’an, Shandong, China
- 3Department of Life Sciences, The Natural History Museum, London, United Kingdom
Taxa are traditionally identified using morphological proxies for groups of evolutionarily isolated populations. These proxies are common characters deemed by taxonomists as significant. However, there is no general rule on which character or sets of characters are appropriate to circumscribe taxa, leading to discussions and uncertainty. Birch species are notoriously hard to identify due to strong morphological variability and factors such as hybridization and the existence of several ploidy levels. Here, we present evidence for an evolutionarily isolated line of birches from China that are not distinguishable by traditionally assumed taxon recognition proxies, such as fruit or leaf characters. We have discovered that some wild material in China and some cultivated in the Royal Botanic Gardens Edinburgh, formerly recognized as Betula luminifera, differ from other individuals by having a peeling bark and a lack of cambial fragrance. We use restriction site-associated DNA sequencing and flow cytometry to study the evolutionary status of the unidentified Betula samples to assess the extent of hybridization between the unidentified Betula samples and typical B. luminifera in natural populations. Molecular analyses show the unidentified Betula samples as a distinct lineage and reveal very little genetic admixture between the unidentified samples and B. luminifera. This may also be facilitated by the finding that B. luminifera is tetraploid, while the unidentified samples turned out to be diploid. We therefore conclude that the samples represent a yet unrecognized species, which is here described as Betula mcallisteri.
Introduction
Recognizing and naming entities is a fundamental step to manage and use biodiversity. The central unit to handle biodiversity is the species. These units are used to make generalized statements and predictions about its individual members, e.g., about behaviors, reactions to environment, edibility, dangers, or requirements for cultivation.
There are long-standing theoretical debates about what a species is, leading to multiple species concepts (Mayden, 1997; de Queiroz, 1998). Most basically, species are considered as a group of populations with active or at least potential ongoing free gene flow along generations—coined the “biological species concept” (Mayr, 1942). Historically, however, due to the lack of other methods, species have been defined as groups of morphologically similar individuals with significant differences to other groups. Which characters are used to distinguish these different morphological species, or morphospecies, is up to each evaluating researcher (Holstein and Luebert, 2017). However, there is no generalized rule as to which (set of) characters and what degree of similarity fulfill the requirements of a coherent and predictable unit, or “species.” Once one or more characters deemed to distinguish a group from others are found, a representative specimen—the type—would be chosen to define the name of this group in addition to a description or diagnosis (Turland et al., 2018). This methodology, called taxonomy, would serve to create names for these groups.
Many names that represent species were based on the observation of only few individuals or even fragments of individuals. The observation of more individuals often reveals the full morphological scope and may blur and put initial species circumscriptions and the application of names into question. However, since different names have been created using different proxies, species concepts may be incompatible with each other, causing discussions about which (set of) characters might serve best to circumscribe a species. Many species were even described based on morphological characters that turned out to be less reliable, as they vary considerably across environments (Wang et al., 2014a; Beatty et al., 2016). Occasional or localized interspecific gene flow may give rise to hybrids or, further along through introgression, blur morphological species boundaries, which further complicates species delimitation (Tovar-Sánchez and Oyama, 2004; Bardy et al., 2011; Castillo-Mendoza et al., 2019; Beeler et al., 2020; Wehenkel et al., 2020). Some hybrids possess a combination of parental morphological characters and sometimes were regarded as species (Barnes and Dancik, 1985), while hybridization can also lead to such true species by establishing reproductive barriers, e.g., though polyploidization (Ferguson and Sang, 2001; Rieseberg, 2001). In plants, diploid and tetraploid close relatives usually have a strong reproductive isolation as triploids are mostly sterile, impeding interspecific gene flow (Levin, 1975; Fowler and Levin, 1984; Rieseberg, 2001; Husband and Sabara, 2004; Roccaforte et al., 2015). However, polyploids exhibit a mosaic of parental characters or even new ones (Rieseberg et al., 2003; Ahmed et al., 2019). In consequence, the detection of “true species” and analysis of their evolution can be challenging in groups in which the reproductive barriers of once isolated genetic lines are low, e.g., in Salix (Marchenko and Kuzovkina, 2022).
To delineate species more reliably, an integrated approach has been increasingly adopted, combining morphological, molecular, and (or) cytogenetic data (Fujita et al., 2012; Alors et al., 2016; Newton et al., 2020; Giacò et al., 2022; Wang et al., 2022). The advantage of using an integrated approach for species delimitation is that it overcomes the subjective species concepts (Hey, 2006; Hausdorf, 2011; Carstens et al., 2013) . Phylogenomic approaches generate a multitude of characters that are usable for analyzing complex questions. The large number of single-nucleotide polymorphisms (SNPs) obtained from next-generation genotyping allows for robustly inferring the phylogenetic position and ploidy level of a diploid or tetraploid species (Cariou et al., 2013; Zohren et al., 2016; Grewe et al., 2017).
Betula (birch) includes approximately 65 species and subspecies, which are broadly distributed across the Northern Hemisphere (Ashburner and McAllister, 2013). Birches are well known for their frequent hybridization (Anamthawat-Jónsson and Tómasson, 1999; Thórsson et al., 2010; Wang et al., 2014b; Tsuda et al., 2017; Bona et al., 2018; Ding et al., 2021), due to wind pollination, self-incompatibility, and lack of complete reproductive barriers (Ashburner and McAllister, 2013). Betula species exhibit substantial morphological variation. For example, bark color varies considerably among B. pendula and B. pubescens populations (Tarieiev et al., 2019). In addition, Betula species have a series of ploidies, ranging from the diploid to dodecaploid (Ashburner and McAllister, 2013; Wang et al., 2016). Consequently, Betula has a very tough taxonomy, and species misidentification often occurs (Wang et al., 2016).
We came across some individuals during fieldwork in the Qinling-Daba Mountains in central China, which have similar fruits and leaves and an overlapping phenology with the widespread species Betula luminifera H.J.P.Winkl. (Winkler, 1904). However, these individuals have a peeling bark, whereas B. luminifera has a smooth bark according to the Flora of China (Li, 1979; Li and Skvortsov, 1999). We also noticed that these samples lacked the typical wintergreen fragrance when the bark was damaged, a character commonly found in specimens identified as B. luminifera. We termed these individuals as “unidentified samples” hereafter. We also looked at cultivated material of “B. luminifera” in botanical gardens and found an accession (#19933472) at the Royal Botanic Gardens Edinburgh, which shares the characters of the unidentified samples. We integrated multiple lines of evidence to resolve the taxonomic status of the “unidentified sample” and then investigated the amount of gene flow with B. luminifera.
The aims of our study were (1) to resolve the taxonomic status of the “unidentified sample”, (2) to explore if B. luminifera #19933472 and the “unidentified sample” are conspecific, and (3) to investigate the extent of gene flow between the “unidentified sample” and B. luminifera. To this end, we collected 38 individuals of the “unidentified sample” and 48 individuals of the B. luminifera samples from 4 and 13 natural populations, respectively.
Materials and methods
Species identification and sampling
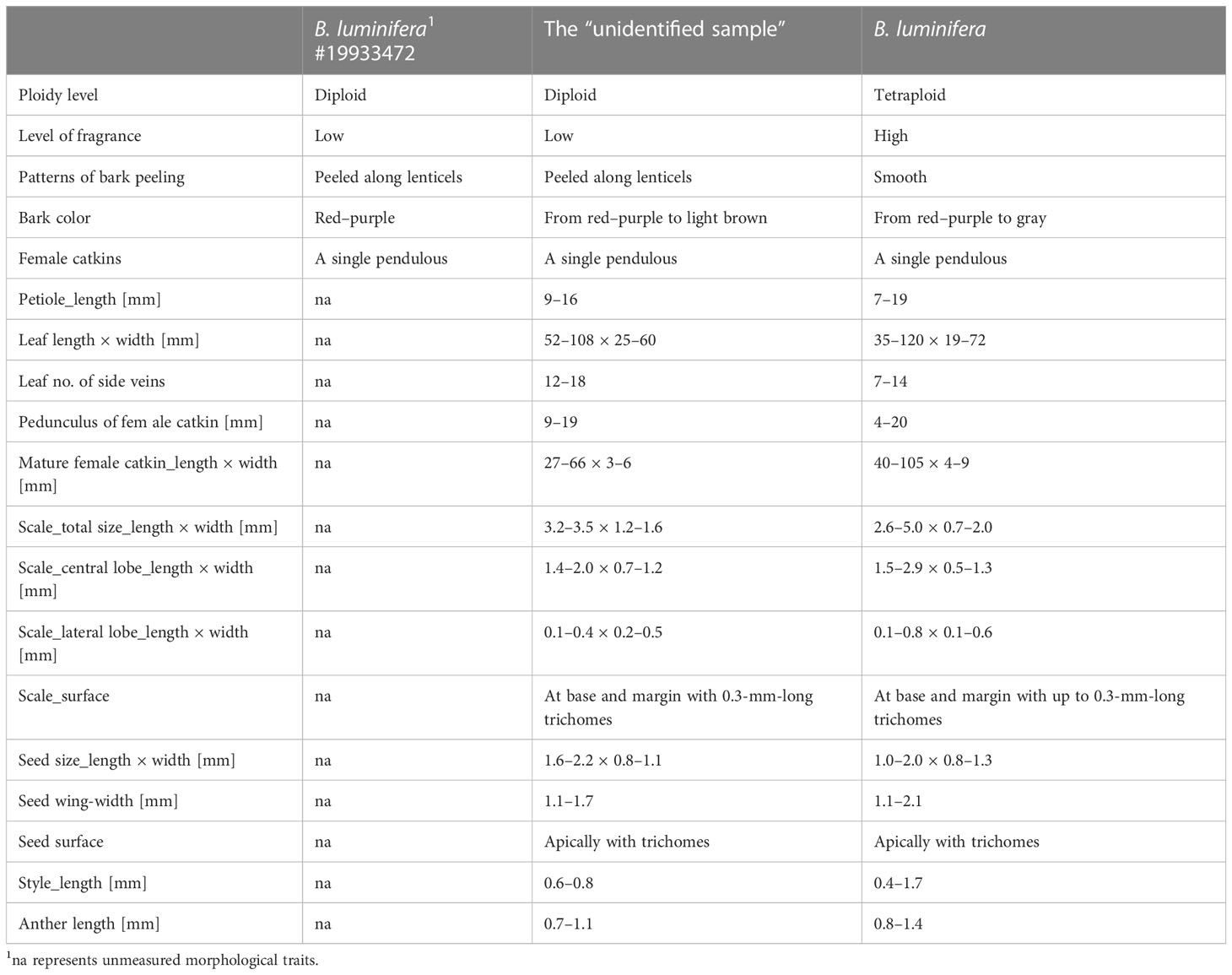
We collected B. luminifera and the “unidentified sample” between May and September of 2019, 2020, and 2021 from 13 and 4 populations, respectively (Figure 1A). Adjacent samples were separated by ~20 m. Betula luminifera was identified based on a morphological description according to Flora of China and the monograph of the Betula species (Li and Skvortsov, 1999; Ashburner and McAllister, 2013). Key features to recognize B. luminifera include a single pendulous female catkin in raceme, a smooth bark, a fruiting period between April and June, and a strong fragrance from fresh cambial tissues (Figure 1B). To further accurately identify B. luminifera, we included samples from Guizhou province where B. luminifera commonly occurs. We grouped individuals as the “unidentified sample”, based on the following characteristics: a single pendulous female catkin in raceme, a peeled bark, a fruiting period between April and June, and no obvious fragrance from fresh cambial tissues (Figure 1C). A GPS system (UniStrong) was used to record the coordinate points of each population. Detailed sampling information is provided in Table S1.
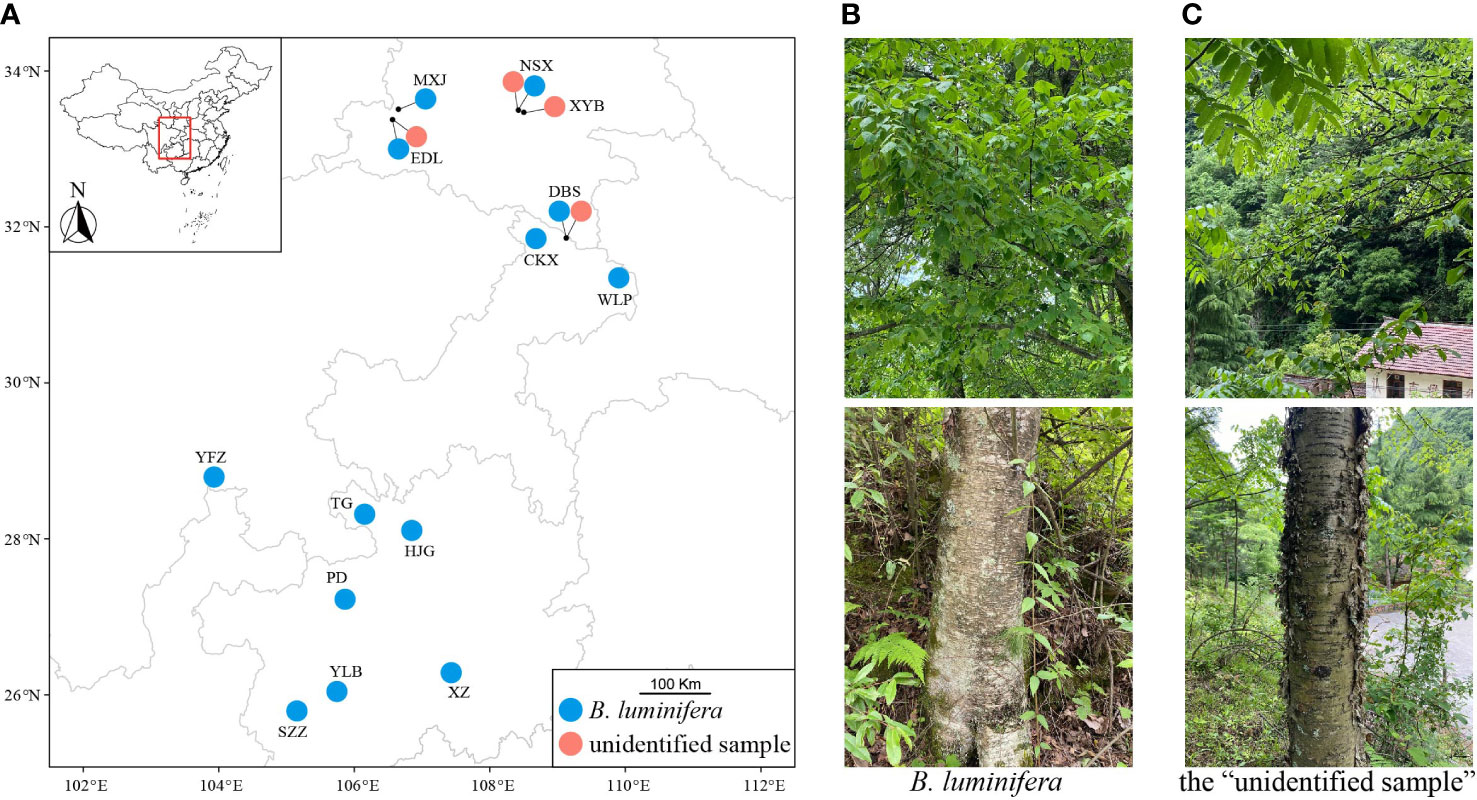
Figure 1 Sampling localities (A), female catkins and barks of B luminifera (B), and the “unidentified sample” (C).
We measured a subset of the material that we collected (deposited in SDAU) and herbarium specimens from BM and K [abbreviations according to Thiers updated continuously (Thiers, 2023)] with bark samples or sufficient annotations of the bark morphology.
DNA extraction, sequencing, and read filtering
We extracted high-quality DNA from cambial tissues following a modified 2× CTAB (cetyltrimethylammonium bromide) protocol (Wang et al., 2013). Extracted DNA was assessed with 1.0% agarose gels.
ITS sequencing
We amplified nuclear ribosomal (nr)ITS for 25 individuals of the “unidentified sample” using primers ITS4 (White et al., 1990) and ITSLeu (Baum et al., 1998). Reactions were performed following Hu et al. (2019). PCR products were sequenced at Tsingke Company (Qingdao, China). ITS sequences were deposited at NCBI with GenBank accession numbers OP263695-OP263719.
RADseq and read filtering
A total of 86 DNA samples were selected for RADseq using an Illumina HiSeq 2500 and 150-bp pair-end sequencing with the restriction enzyme PstI (Personalbio company, Shanghai, China). RADseq data of four samples of closely related Betula species in Wang et al. (2021) were included for population genomic analyses, representing one each of B. alnoides, B. cylindrostachya, B. hainanensis, and B. luminifera #19933472. Raw data were trimmed using Trimmomatic (Bolger et al., 2014) in paired-end mode. Reads with a quality of below 20 within the sliding window of 5 bp and unpaired reads were discarded. Then, a SLIDINGWINDOW step was performed to discard reads shorter than 40 bp.
Read mapping and SNP calling
Filtered reads of each sample were mapped to the whole genome sequence of B. pendula (Salojärvi et al., 2017) using the BWA-MEM v.0.7.17-r1188 algorithm in BWA with default parameters (Li and Durbin, 2009). Alignments were converted to sorted and indexed bam files using SAMtools v1.8 (Li et al., 2009). The MarkDuplicates tool and HaplotypeCaller from GATK v 4.1.4 were used to mark duplicates and call genotypes for each sample, respectively (Mckenna et al., 2010; Depristo et al., 2011). The GenomicsDBImport was used to merge the gVCF files into a combined VCF file, which was used for joint genotyping using the GenotypeGVCFs tool. The SNPs were filtered using a mapping quality (MQ) threshold of 40, a variant confidence (QUAL) of 30, a normalized QUAL score (QD) of 2.0, a maximum symmetric odds ratio (SOR) of 3.0, a minimum depth (DP) of 5 and a maximum depth of 200, a maximum probability of strand bias (FS) of 60.0, an excess heterozygosity (ExcessHet) of 54.69, a minimum Z-score of read mapping qualities (MQRankSum) of −12.5, and a position bias (ReadPosRankSum) of −8.0. The SelectVariants filtering tool was applied to select SNPs present in at least 90 of the samples. BCFtools v1.8 was used to remove SNPs within a 50-kb window with r2 > 0.5 to reduce linkage disequilibrium and then was used to remove SNPs with a minimum allele frequency (MAF) ≤ 0.02 (Li, 2011). All sequences were deposited in the NCBI-Sequence Read Archive (SRA) repository under the BioProjectID PRJNA871086.
Population genomic analyses
A principal component analysis (PCA) was performed on the SNPs of B. luminifera, the “unidentified sample”, and the four samples of closely related Betula species using the “adegenet” R package 2.1.1 (Jombart, 2008). The SNPs were also analyzed in ADMIXTURE v1.3.0 (Alexander and Lange, 2011), setting K from 1 to 10 with 20 replicates for each K value. Cross-validation error estimation was performed in order to assess the most suitable value of K (Alexander and Lange, 2011). Replicate runs were aligned and visualized in pong v1.4.9 with the greedy algorithm (Behr et al., 2016).
Inference of ploidy level based on flow cytometry and SNPs
We performed flow cytometry following Wang et al. (2022) for three accessions of the “unidentified sample” collected from the populations NSX and XYB. Briefly, we co-chopped fresh cambial tissues with internal standards Solanum lycopersicum in 0.5 ml of Extraction Buffer (Cystain PI absolute P, Partec GmbH, Germany) and then filtered them into a tube containing 1 ml of Staining Solution (Cystain PI absolute P, Partec GmbH). We added 50 µl of diluted propidium iodide (PI) to the tube and incubated samples at room temperature in the dark for ~5 min. We analyzed one replicate per sample with >5,000 nuclei measured using flow cytometry (Beckman Coulter CytoFLEX). We also inferred the ploidy of the 86 samples sequenced in this study using the method described in Zohren et al. (2016). Briefly, we plotted the distribution of the allele ratios from read counts at heterozygous sites, and we expected a peak of approximately 0.50 for a diploid, peaks of approximately 0.33 and 0.67 for a triploid, and peaks close to 0.25, 0.50, and 0.75 for a tetraploid.
Phylogenetic analyses based on ITS, ITS secondary structure, and SNPs
Seventy additional ITS sequences from Betulaceae (Wang et al., 2016; Wang et al., 2022) were included to infer the phylogenetic position of the “unidentified sample”. A total of 95 ITS sequences and their secondary structure (ITS1 and ITS2), as previously used by Tarieiev et al. (2021), were aligned separately using BioEdit v.7.0.9.0 (Hall, 1999) with default parameters.
We included the RADseq data of 23 Betula taxa from a previous study (Wang et al., 2021) and of 45 samples generated in the present study for phylogenomic analysis. Alnus inokumae was selected as the outgroup. The SNPs were concatenated into a supermatrix with missing data below 50%. This resulted in 2,309,898 SNPs.
We analyzed the ITS alignment, the ITS1 alignment, the ITS2 alignment, and the supermatrix of SNPs separately, with a rapid bootstrap analysis under a GTR + GAMMA nucleotide substitution model in RAxML v.8.1.16 (Stamatakis, 2006). A total of 100 bootstraps and 10 searches using the ML were performed.
Results
Read mapping and variant calling
The individual read mappings of B. luminifera and the “unidentified sample” from the present study resulted in 8.8% to 95.1% of mapped reads per individual. After filtering, 340,979 variants were present in at least one individual (Table S1).
Population structure
PC1 and PC2 explained 30.7% and 3.2% of the total variation, respectively (Figure 2A). The PCA (Figure 2A) results based on the 40,209 biallelic SNPs revealed a clear separation between B. luminifera and the “unidentified sample” on PC1. Betula luminifera #19933472 formed a cluster with the “unidentified sample”. A putative hybrid between B. luminifera and the “unidentified sample” was positioned in between. A specimen identified as Betula cylindrostachya formed a cluster with B. luminifera. Betula alnoides and B. hainanensis were separated from B. luminifera on PC2.
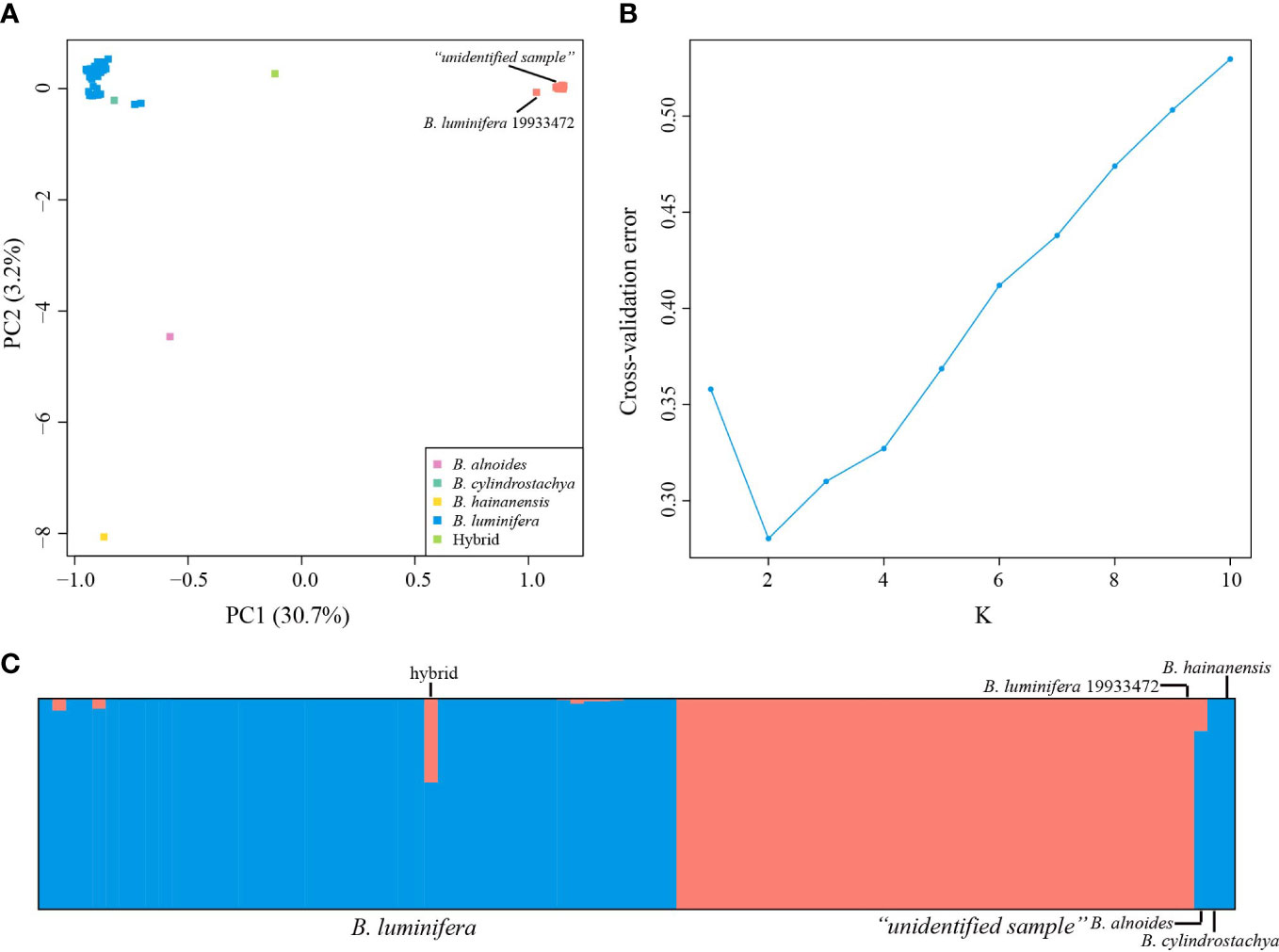
Figure 2 Population structure analyses based on RADseq data. (A) Principal component analysis (PCA) of Betula species of section Acuminate at 40,209 SNPs. (B) Cross-validation error calculated for Ks from 1 to 10. (C) Estimated genetic admixture of 90 Betula samples at 40,209 SNPs. Each individual is represented by a vertical line. Blue and orange represent B luminifera and the “unidentified sample”, respectively. Previously sequenced samples (B. luminifera #19933472, B alnoides, B cylindrostachya, and B hainanensis) are placed on the right side.
Consistent with the PCA results, admixture analyses showed the optimal K value of 2 (Figure 2B), corresponding to B. luminifera and the “unidentified sample”/B. luminifera #19933472. The “unidentified sample”/B. luminifera #19933472 formed a distinct lineage even when the K value was increased to 6 (Figure S1). Very little admixture was detected in the “unidentified sample” into B. luminifera, with the highest admixture value of 5.31% (excluding the putative hybrid). The sample EDL20-022 showed a genetic admixture of 39.76% and 60.24% of the “unidentified sample” and B. luminifera, respectively (Figure 2C).
A high level of genetic differentiation was observed between B. luminifera and the “unidentified sample”, with FST values among populations with at least four individuals ranging from 0.30 to 0.43. Within species, FST values ranged from 0.01 to 0.04 for B. luminifera. The FST value between the two populations of the “unidentified sample” was 0.02.
Ploidy-level analyses
Flow cytometry analyses showed that the genome size of the three “unidentified sample” ranged from 388 to 445 M, indicating that it is diploid (Figure S2). The plot of the allele ratios at heterozygous sites showed that the “unidentified sample” had a peak of approximately 0.50, B. luminifera had peaks near 0.25, 0.50, and 0.75, and the putative hybrid between the “unidentified sample” and B. luminifera had peaks close to 0.33 and 0.67 (Figures 3, S3, S4). One B. luminifera individual (YLB001) had a ploidy level that remains unclear due to the low number of reads (Figure S3).
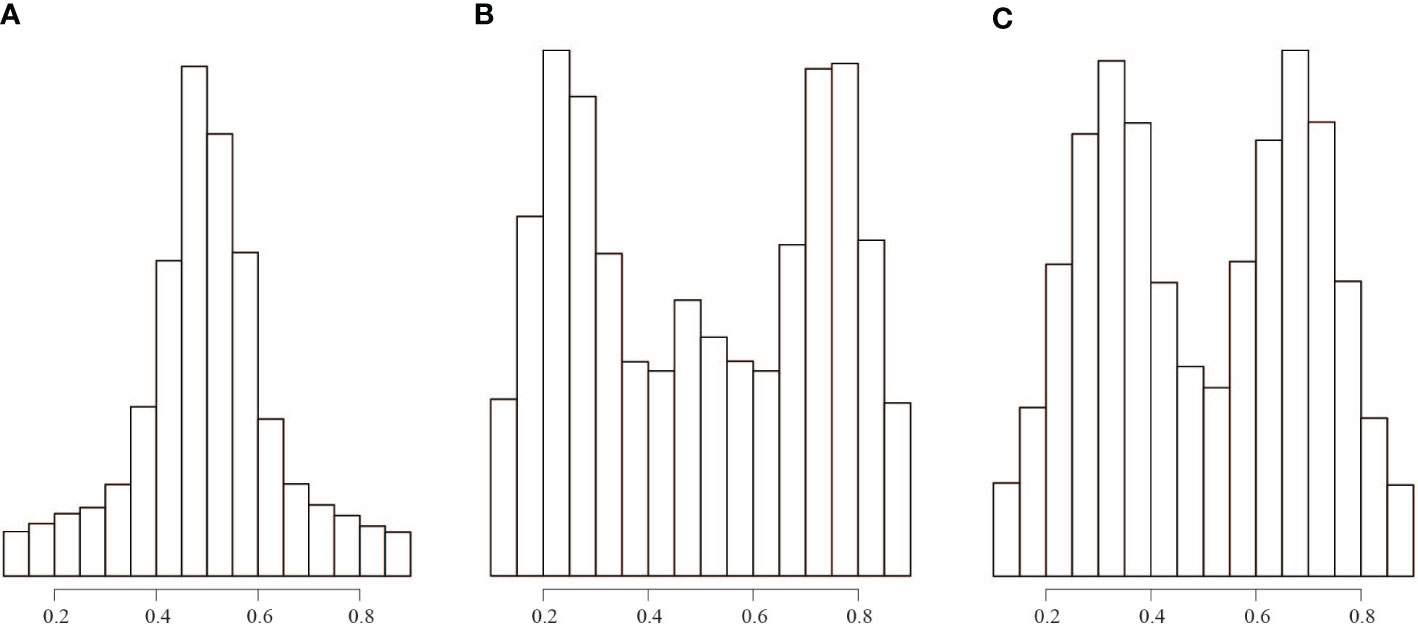
Figure 3 Distribution of read count ratios for heterozygous sites covered by at least 30 reads. (A) All the “unidentified sample” individuals. (B) All B luminifera individuals. (C) Sample number EDL20-022, which is a hybrid between B luminifera and the “unidentified sample”.
Phylogenetic position of the “unidentified sample”
The phylogenetic tree based on ITS1 failed to resolve the phylogenetic position of the “unidentified sample”; however, based on ITS or ITS2, it showed that the “unidentified sample” tended to form a cluster, except for sample EDL20-022, which we interpret as an F1 hybrid between the “unidentified sample” and B. luminifera (Figures S5–S7). The phylogenetic tree based on a supermatrix of 2,309,898 SNPs revealed that the B. luminifera #19933472 and the “unidentified sample” formed a well-supported monophyletic clade, which is a sister to a clade of B. luminifera, B. cylindrostachya, B. alnoides, and B. hainanensis (Figure 4). B. cylindrostachya was nested into a well-supported clade of B. luminifera samples (Figure 4).
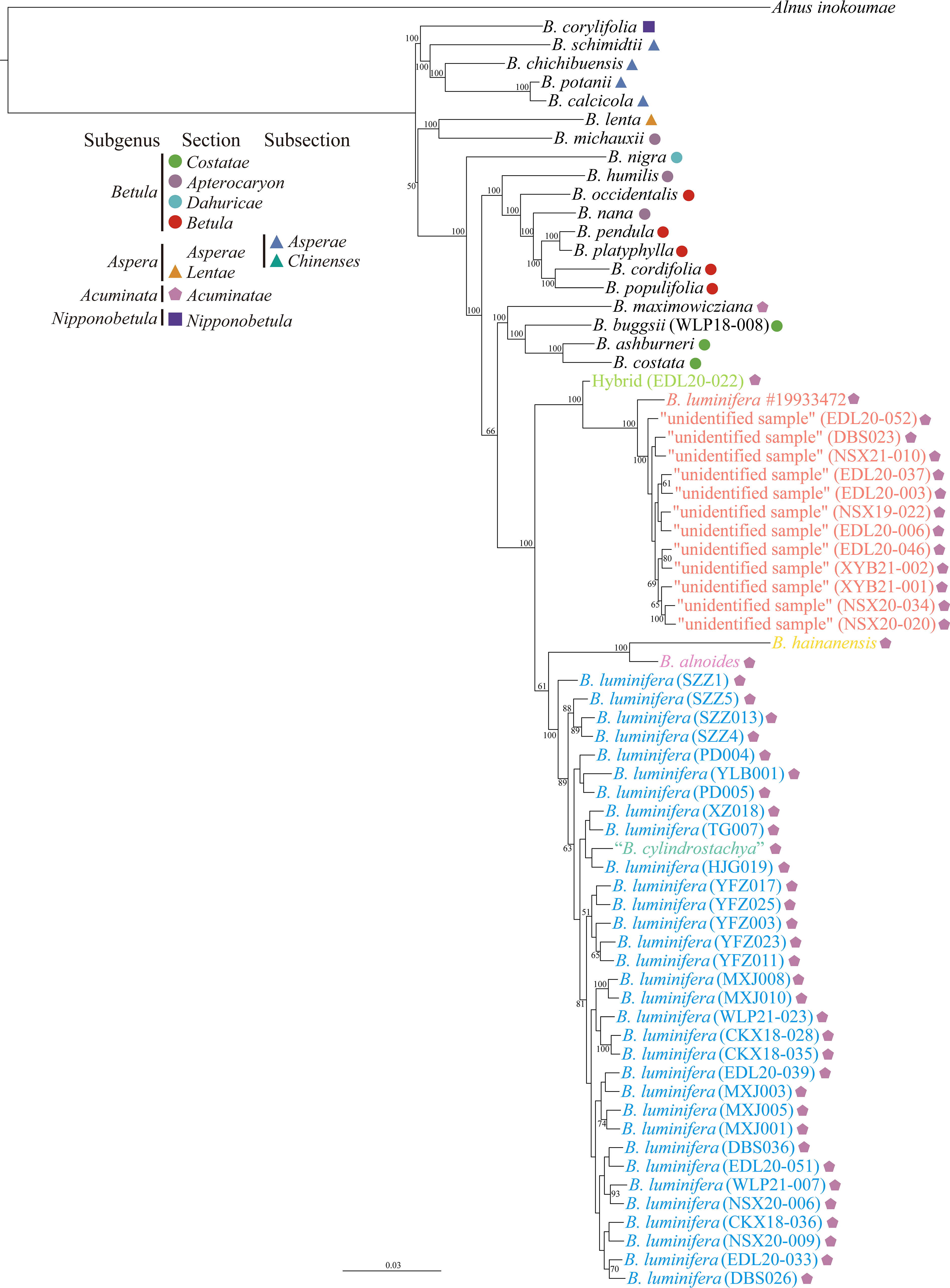
Figure 4 Phylogenetic tree from the maximum likelihood analysis of the “unidentified sample” based on a supermatrix of 2,309,898 SNPs. The scale bar below indicates the mean number of nucleotide substitutions per site. Species were classified according to Ashburner and McAllister (2013). Values close to branches are bootstrap percentages of >50%.
Features of the B. luminifera #19933472, the “unidentified sample”, and B. luminifera
Features of B. luminifera #19933472 and the “unidentified sample” include a peeling bark (Figure S8), no fragrance from fresh cambial tissues, and diploidy, which are distinct from that of B. luminifera. We measured 16 samples of “B. luminifera” with a peeling bark and 6 of the “unidentified samples” and did not find any leaf, flower, or infructescence characters that could be used to distinguish peeling and non-peeling individuals (Table 1). Non-peeling individuals seem to have had a higher number of side veins and larger male flower scales, but the sizes were overlapping.
Discussion
A new diploid species in section Acuminatae
Our results show that B. luminifera #19933472 and the “unidentified samples” with a peeling bark form a distinct genetic cluster based on admixture results at the value of K = 2 and onwards (Figure S1). Phylogenomic analyses strongly support them as a monophyletic clade, which is separated from smooth-barked B. luminifera and other species of section Acuminatae (Figure 4).
The type material of B. luminifera was collected from Chengkou County in the north of the municipality of Chongqing (Winkler, 1904), very much in the vicinity of our “unidentified samples” (northeastern part of Chongqing and just north of Chongqing in the southern part of Shaanxi province). Although we do not know the exact locality where the type specimen of B. luminifera was collected, nor is there any description of the bark in the protologue of B. luminifera, the current understanding of B. luminifera is that it has a smooth bark (Li, 1979; We et al., 1987; Li and Skvortsov, 1999; Fu et al., 2004; Ashburner and McAllister, 2013). Smooth-barked B. luminifera is widespread in the subtropical monsoon climate areas of China (Li and Skvortsov, 1999). While the four wild populations of the “unidentified sample” were from the Qinling-Daba Mountains (Shaanxi, NE Chongqing), B. luminifera #19933472 was collected from Yunnan province in southwest China. This indicates that this group may have a wider distribution, and our sampling shows that it grows even sympatrically with the smooth-barked B. luminifera.
All the samples with a peeling bark are found to be diploid, while the smooth-barked B. luminifera material from our study is tetraploid. Betula luminifera has been reported and considered as diploid in previous studies (Ashburner and McAllister, 2013; Wang et al., 2016; Wang et al., 2021), but the material used for that analysis by Wang et al. (2016) and Wang et al. (2021) is in fact “B. luminifera” #19933472. All the samples from Chengkou County in this study are found to have a smooth bark and to be tetraploid. Hence, the description of B. luminifera as diploid (Ashburner and McAllister, 2013) is likely incorrect. Betula luminifera may also have different cytotypes, like other Betula species, such as B. chinensis (Ashburner and McAllister, 2013). However, we do not find any contradiction from the correlation of ploidy, bark morphology, and clustering in the phylogeny to support different ploidy levels in smooth-barked B. luminifera.
Although we did not conduct pollination experiments, a difference in ploidy indirectly indicates reproductive barriers, especially between the diploid and tetraploid (Borges et al., 2012). We expect that the specimens with a peeling bark also fit the biological species concept that focuses on reproductive isolation (Mayr, 1942). We therefore propose that they are accepted as a new species, which here we name Betula mcallisteri. Aside from our case, a difference in ploidy has also been used to separate other closely related Betula species. For example, B. pendula and B. pubescens were once treated as a single species, B. alba (Linnaeus, 1753), and were later acknowledged to be two species, as B. pendula is a diploid and B. pubescens is a tetraploid (Tuley, 1973; Atkinson, 1992).
Hybridization between B. mcallisteri and B. luminifera
Betula mcallisteri and B. luminifera are closely related. Given the difference in ploidy level, it can be assumed that an ancestor of the diploid B. mcallisteri may have been involved in the evolution of the tetraploid B. luminifera.
Allele sharing among closely related species is usually ascribed to incomplete lineage sorting and/or introgressive hybridization (Twyford and Ennos, 2012). Incomplete lineage sorting is an unlikely explanation for the pattern of allele sharing observed between B. mcallisteri and B. luminifera. In most allopatric populations of B. luminifera, no allele transfer from B. mcallisteri to B. luminifera was detected, whereas in sympatric populations, either hybrid or a low level of genetic admixture was detected (Figure 2C). Such a geographic signal would not be expected from an incomplete lineage sorting (Barton, 2001).
The sample EDL20-022 showed a high level of genetic admixture, which stood out from other samples. We interpret this result as a putative F1 hybrid. The presence of a hybrid in the population EDL where both species occur sympatrically indicates that B. mcallisteri hybridized with B. luminifera, meeting our expectation that interploidy hybridization could occur among Betula species (Zohren et al., 2016; Tsuda et al., 2017; Hu et al., 2019). However, introgression between B. mcallisteri and B. luminifera was not detected in this population. In contrast, in the sympatric population NSX, hybrids between B. mcallisteri and B. luminifera were not detected, but signals can be interpreted as traces of introgression (between 0.3% and 2%) from B. mcallisteri to B. luminifera (Figure 3C).
Asymmetric introgression from the diploid to tetraploid was also observed in other studies (Wang et al., 2014b; Clark et al., 2015; Eidesen et al., 2015; Zohren et al., 2016; Tsuda et al., 2017). However, there is very little introgression from B. mcallisteri to B. luminifera, with the highest admixture value of 5.3%, much lower than the level of introgression from B. nana to B. pubescens, which shows the highest admixture value of 16.9% (Zohren et al., 2016). One possible explanation is that triploids between B. mcallisteri and B. luminifera may produce less viable gametes than triploids between B. nana and B. pubescens, which were reported to produce a small proportion of viable gametes (Anamthawat-Jónsson et al., 2021). Alternatively, we may have failed to sample the introgressed individuals. The fact that we did not detect hybrid swarms indicates a strong reproductive barrier between B. mcallisteri and B. luminifera, likely due to a difference in ploidy.
Taxonomic issues in section Acuminatae
Multiple lines of evidence support that the “unidentified samples” and the cultivated B. luminifera #19933472 represent a yet undescribed species, here described as B. mcallisteri. Betula mcallisteri belongs to Betula sect. Acuminatae (Regel, 1865), with its long pendulous female catkins and widely winged seeds with wings wider than the nutlet (Skvortsov, 1997; Ashburner and McAllister, 2013).
Betula mcallisteri has a single pendulous female catkin, like B. luminifera, and both can be distinguished from most of the other species of section Acuminatae, which have three to five pendulous female catkins in a raceme, such as B. alnoides, B. fujianensis, and B. hainanensis (Zeng et al., 2008; Zeng et al., 2014). However, single female catkins also occur in B. cylindrostachya (Ashburner and McAllister, 2013), including the type material (Wallich, 1830; Kumar et al., 2022), although two or three catkins are more common.
The number of female catkins being one or two may therefore not be a reliable character for species delimitation in members of section Acuminatae. For example, Hugh McAllister identified a sample with two female catkins as B. cylindrostachya, and it was nested in a clade of B. luminifera (Figure 4). This is consistent with Skvortsov’s view that B. cylindrostachya is most closely related to B. luminifera (Skvortsov, 1997). Indeed, in some populations, B. luminifera sporadically has two female catkins in a raceme [e.g., S. Chen 481 (K), S.J. Zhang 4493 (NAS00284530), and S.K. Lai 2577 (KUN0529329, LBG00108762); personal communication with Jie Zeng], and B. cylindrostachya occasionally has a single female catkin (Ashburner and McAllister, 2013). However, B. cylindrostachya is considered to be tetraploid and to have a peeling and fragrant bark (Ashburner and McAllister, 2013; Kumar et al., 2022), which is in contrast to Skvortsov’s hypothesis.
Betula mcallisteri is diploid, and several other samples identified as B. alnoides, B. cylindrostachya, and B. luminifera are tetraploid, based on genome size estimation or inference from reads covering heterozygous SNPs (Figures S2–S4). Betula alnoides was once considered as diploid (Mehra and Sareen, 1973), but it was found to be tetraploid based on the chromosome counting of samples from several populations (personal communication with Hugh McAllister). We did not see the voucher of the material that was used by Mehra and Sareen. Their assumption that the material must have been B. alnoides because B. cylindrostachya (misspelled as “cylindrostachys”) would be only occurring in the Eastern Himalayas is puzzling, because B. cylindrostachya was actually described from the Kumaon district (Wallich, 1830; Kumar et al., 2022), the very same area where their voucher is from. It could mean that (1) there are diploid members of section Acuminatae in India (assuming that the authors studied local material), and (2) the name B. cylindrostachya may have been misapplied currently. However, only B. alnoides and B. cylindrostachya from section Acuminatae are currently accepted. Both species are easily confused and rather weakly differentiated (Ashburner and McAllister, 2013). How the tetraploid species in section Acuminatae evolved, speciation after polyploidization or independent polyploidizations, and how they are morphologically differentiated require further investigation.
We could not find any traditional characters in flower, fruit, or leaf morphology to distinguish B. mcallisteri from B. luminifera. Only bark smooth vs. peeling (Figures 1B, C) and cambial fragrance turned out to be suitable macromorphological and field characters. The number of leaf side veins and male flower scale sizes are different but overlapping (Table 1). Since most herbarium material does not contain any sample or description of the bark, our sampling is rather small. It is likely that these two characters would turn out to be even less helpful in distinguishing the two groups if more material from other regions and habitats were to be examined. This leaves natural history collections with a peculiar problem: herbarium collections commonly do not include or even mention bark characters. Even though the bark is often ornamental in Betula (Ashburner and McAllister, 2013), it is often ignored or imprecisely annotated when collecting birches. This renders the majority of “B. luminifera” collections as indeterminable. Our results exemplify why it is important to document taxa more extensively, including bark samples, or at least to add information using photography. The call to collect more extensive information for a better understanding of the organism is not new (Webster, 2017), especially since new questions about interactions of plants with their environment are given more research focus (Schindel and Cook, 2018). In birches, however, it may make the difference between determinable and indeterminable natural history collections.
Our discovery of B. mcallisteri being in cultivation at RBGE for 30 years is another nice example that botanic gardens may grow undiscovered species. Such cases may exist for other plant species with poorly understood taxonomy. A broad knowledge of morphological variation from wild populations is important in examining species delimitation. Botanic gardens play an important role in cultivating and conserving plant species (O'donnell and Sharrock, 2017) and provide a venue for conducting various research activities (Chen and Sun, 2018). While getting suspected new species into cultivation is not unusual, only few examples of material being cultivated for a long time and turning out to be new to science are known, e.g., Oncidium herrenhusanum (Schlumpberger and Königer, 2011). Also, the recently discovered Victoria boliviana was initially suspected to be Victoria amazonica, and living material helped to resolve its species status (Smith et al., 2022). Living collections in botanic gardens can supplement valuable information for taxonomy and species discovery. However, our finding that the RBGE accession #19933472 was used to determine the ploidy level of B. luminifera (Wang et al., 2016; Wang et al., 2021), despite the morphological difference of the bark from what is commonly understood as B. luminifera, may serve as a reminder that pars pro toto statements for species could lead to incorrect assumptions. It also stresses the importance of the voucher material and detailed descriptions.
Taxonomic treatment
Betula mcallisteri, Nian Wang & Holstein, sp. nov (Figure 5).
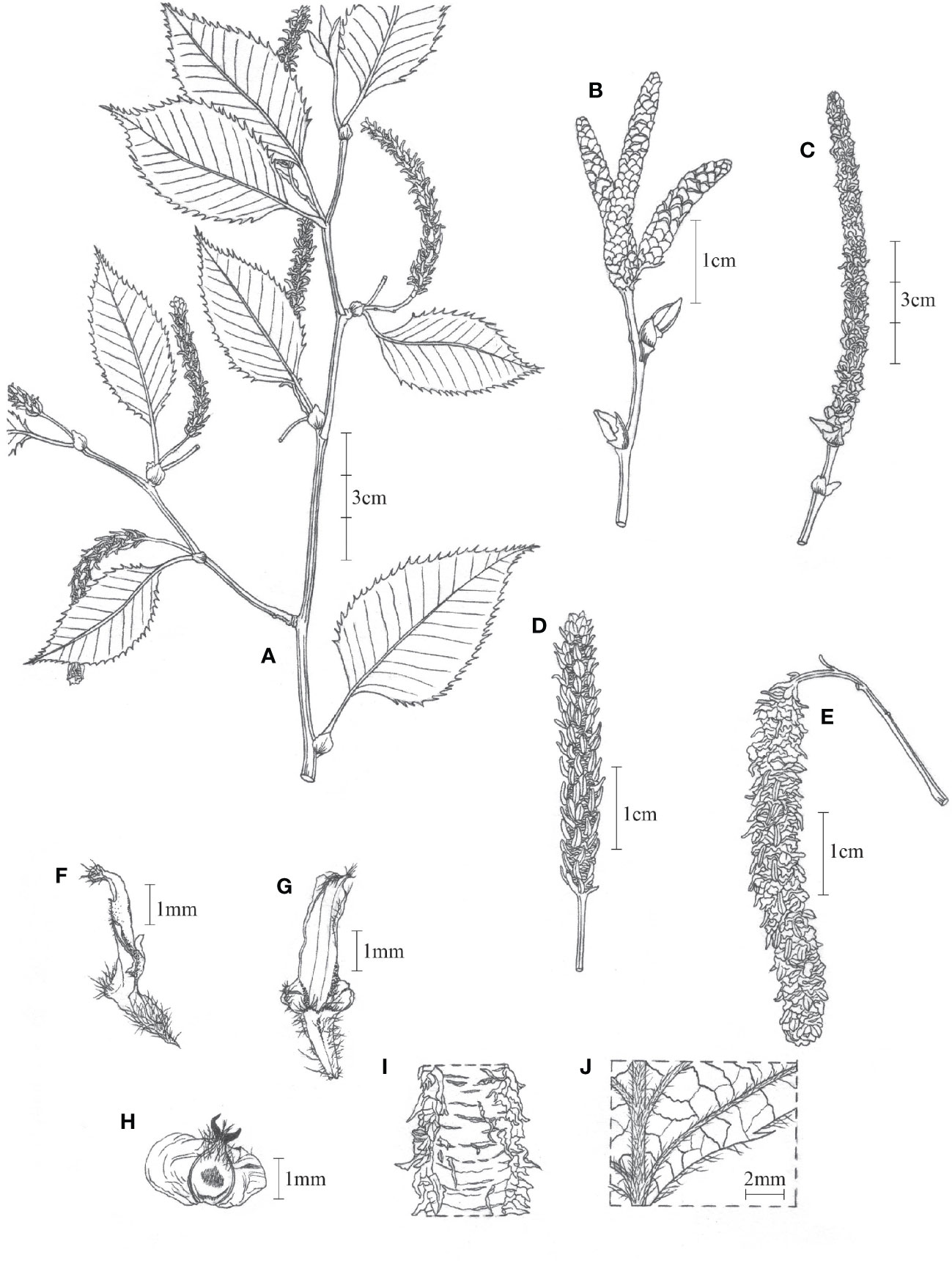
Figure 5 Betula mcallisteri. (A) Specimen with female catkins; (B) male catkins, immature; (C) male catkins, mature; (D) female catkin, immature; (E) female catkin, mature; (F) side view of fruiting catkin scale; (G) fruiting catkin scale, outer face; (H) mature seed; (I) bark; (J) adaxial surface of leaf blade.
Diagnosis
Betula mcallisteri is a tree species with single pendular female catkins, just like B. luminifera (rarely also 2 catkins), but in contrast to other species in section Acuminatae (two or more female catkins, single ones in B. cylindrostachya only exceptionally). It differs from B. luminifera by having a bark that peels off in horizontal strips instead of a smooth, non-peeling bark. Also, its damaged bark does not have a noticeable fragrance of oil of wintergreen (methyl salicylate), while B. luminifera is noticeably fragrant when the bark is damaged.
Type: China. Shaanxi: Mian County, elev. ca. 1,600–2,000 m, 33.4° N, 106.6° E, 6 May 2021, Nian Wang DEDL001 (Figure S9) (holotype IBSC0895006; isotypes HIB0216644, IBSC0895007, HITBC0083551, KUN1571214, K, and PE).
Description
Single trunked tree to 30 m high. Trunk up to 60 cm in diameter; fresh bark red–brown, peeling along transverse lenticels into persistent and tattered sheets. Leaves one to three per node, accessory leaves often smaller. Petiole (3–)7–16 mm, densely beset with short trichomes but glabrescent, sometimes with reddish or whitish glands. Leaves ovate, 52–108 × 25–60 mm, 12–18 side veins, all running into a tooth on the margin. Leaf tip short apiculate, base round to widely cuneate. Leaf margin double-serrate. Upper lamina darker than below, with silky trichomes on lamina between the side veins, glabrescent. Lower lamina with red glands, silky trichomes along the veins, glabrescent. Male catkins in clusters of three to four, pendent when matured, ~10 × 0.3 cm, length/breadth ratio ~30:1. Male flower bracts scale-like, 1.5–1.8 × 1.2–1.5 mm, margin ciliate. Anthers 0.7–1.1 mm long. Female catkins with a 9- to 16-mm-long peduncle. Immature female catkins green, born singly, pendent, up to 5 cm long, and 6.5 mm broad including the scales. Fruiting catkins, borne singly, long cylindrical, up to ~100 × 6–8 mm at maturity; bracts scale-like, 3.2–3.5 × 1.2–1.6 mm, trilobate with central lobe dominating; adaxially at base with a tuft of up to 0.5 mm long trichomes, margin sometimes ciliate, surfaces more or less glabrous; central lobe narrowly oblong or rhombic, 1.4–2.0 × 0.7–1.2 mm; lateral lobes 0.1–0.4 × 0.2–0.5 mm short rounded to triangulate. Seed 1.6–2.2 × 0.8–1.1 mm, wing 1.1–1.7 mm wide on each side, translucent, styles 0.6–0.8 mm; seed surface hairy on apical half.
Distribution and habitat
Betula mcallisteri occurs in the Qinling-Daba Mountains in central China, according to our field survey, and in Yunnan province, SW China, based on the collection information from B. luminifera #19933472. Betula mcallisteri grows sympatrically with B. luminifera at an altitude between 1,400 and 2,100 m. Betula mcallisteri and B. luminifera have no altitudinal separation. We found abundant B. mcallisteri individuals in EDL and NSX populations, with 16 and 30 samples confirmed as B. mcallisteri, respectively.
Conservation status
Habitat destruction and logging are common issues for the survival of woody species. The area where population NSX is located once underwent severe tree logging before China introduced the Natural Forest Protection Program and the Slope Land Conversion Program in 1998 (Delang and Wang, 2013). Since then, the area has been under strict protection, and logging is totally prohibited. Different from the NSX population, the area where the EDL population was located is near a village and was cut through by a road allowing for car traveling. The finding of a cultivated specimen originating from Yunnan suggests that the species is more widely distributed but overlooked. We therefore do not see any indication to assume that B. mcallisteri requires a conservation status and propose to categorize it as least concern (LC).
Etymology
Betula mcallisteri is named after Hugh A. McAllister, a botanist from the Institute of Integrative Biology, University of Liverpool, for his devotion to research on the genus Betula. The Chinese name of B. mcallisteri is “陕南桦” (shǎn nán huà; literally: Shannan birch).
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
NW conceived the project; NW and JD collected the samples; HZ and NH performed the experiments; HZ, JD and NH analyzed the data; HZ, NH and NW wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was funded by the Youth Innovation Team Project for Talent Introduction and Cultivation in University of Shandong Province to NW, National Natural Science Foundation of China (31770230), and the Young Scholars selected by the State Forestry and Grassland Administration, China.
Acknowledgments
We thank Peter Brownless from the Royal Botanic Garden Edinburgh for sharing pictures of B. luminifera #19933472 and Professor Richard Buggs from the Royal Botanic Gardens Kew for the valuable comments on the manuscript. We thank Rongmei Zhang for drawing the botanical illustration of the new species.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1113274/full#supplementary-material
References
Ahmed, D., Comte, A., Curk, F., Costantino, G., Luro, F., Dereeper, A., et al. (2019). Genotyping by sequencing can reveal the complex mosaic genomes in gene pools resulting from reticulate evolution: a case study in diploid and polyploid citrus. Ann. Bot. 123, 1231–1251. doi: 10.1093/aob/mcz029
Alexander, D. H., Lange, K. (2011). Enhancements to the ADMIXTURE algorithm for individual ancestry estimation. BMC Bioinf. 12, 1–6. doi: 10.1186/1471-2105-12-246
Alors, D., Lumbsch, H. T., Divakar, P. K., Leavitt, S. D., Crespo, A. (2016). An integrative approach for understanding diversity in the Punctelia rudecta species complex (Parmeliaceae, ascomycota). PloS One 11, e0146537. doi: 10.1371/journal.pone.0146537
Anamthawat-Jónsson, K., Karlsdóttir, L., Jóhannsson, M. (2021). Naturally occurring triploid birch hybrids from woodlands in Iceland are partially fertile. New Forests 52, 659–678. doi: 10.1007/s11056-020-09816-z
Anamthawat-Jónsson, K., Tómasson, T. (1999). High frequency of triploid birch hybrid by Betula nana seed parent. Hereditas 130, 191–193. doi: 10.1111/j.1601-5223.1999.00191.x
Ashburner, K., McAllister, H. (2013). The genus Betula: a taxonomic revision of birches (London: Kew Publishing).
Atkinson, M. (1992). Betula pendula Roth (B. verrucosa ehrh.) and B. pubescens ehrh. J. Ecol. 80, 837–870. doi: 10.2307/2260870
Bardy, K. E., Schönswetter, P., Schneeweiss, G. M., Fischer, M. A., Albach, D. C. (2011). Extensive gene flow blurs species boundaries among Veronica barrelieri, V. orchidea and V. spicata (Plantaginaceae) in southeastern Europe. Taxon 60, 108–121. doi: 10.1002/tax.601010
Barnes, B. V., Dancik, B. P. (1985). Characteristics and origin of a new birch species, Betula murrayana, from southeastern Michigan. Can. J. Bot. 63, 223–226. doi: 10.1139/b85-025
Barton, N. H. (2001). The role of hybridization in evolution. Mol. Ecol. 10, 551–568. doi: 10.1046/j.1365-294x.2001.01216.x
Baum, D. A., Small, R. L., Wendel, J. F. (1998). Biogeography and floral evolution of baobabs (Adansonia, Bombacaceae) as inferred from multiple data sets. Syst. Biol. 47, 181–207. doi: 10.1080/106351598260879
Beatty, G. E., Montgomery, W. I., Spaans, F., Tosh, D. G., Provan, J. (2016). Pure species in a continuum of genetic and morphological variation: sympatric oaks at the edge of their range. Ann. Bot. 117, 541–549. doi: 10.1093/aob/mcw002
Beeler, R. B., Sharples, M. T., Tripp, E. A. (2020). Introgression among three western north American bilberries (Vaccinium section Myrtillus). Syst. Bot. 45, 576–584. doi: 10.1600/036364420X15935294613383
Behr, A. A., Liu, K. Z., Liu-Fang, G., Nakka, P., Ramachandran, S. (2016). Pong: fast analysis and visualization of latent clusters in population genetic data. Bioinformatics 32, 2817–2823. doi: 10.1093/bioinformatics/btw327
Bolger, A. M., Lohse, M., Usadel, B. (2014). Trimmomatic: a flexible trimmer for illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Bona, A., Petrova, G., Jadwiszczak, K. A. (2018). Unfavourable habitat conditions can facilitate hybridisation between the endangered Betula humilis and its widespread relatives B. pendula and B. pubescens. Plant Ecol. Divers. 11, 295–306. doi: 10.1080/17550874.2018.1518497
Borges, L. A., Souza, L. G. R., Guerra, M., Machado, I. C., Lewis, G. P., Lopes, A. V. (2012). Reproductive isolation between diploid and tetraploid cytotypes of Libidibia ferrea (= Caesalpinia ferrea) (Leguminosae): ecological and taxonomic implications. Plant Syst. Evol. 298, 1371–1381. doi: 10.1007/s00606-012-0643-3
Cariou, M., Duret, L., Charlat, S. (2013). Is RAD-seq suitable for phylogenetic inference? an in silico assessment and optimization. Ecol. Evol. 3, 846–852. doi: 10.1002/ece3.512
Carstens, B. C., Pelletier, T. A., Reid, N. M., Satler, J. D. (2013). How to fail at species delimitation. Mol. Ecol. 22, 4369–4383. doi: 10.1111/mec.12413
Castillo-Mendoza, E., Salinas-Sánchez, D., Valencia-Cuevas, L., Zamilpa, A., Tovar-Sánchez, E. (2019). Natural hybridisation among Quercus glabrescens, Q. rugosa and Q. obtusata (Fagaceae): microsatellites and secondary metabolites markers. Plant Biol. 21, 110–121. doi: 10.1111/plb.12899
Chen, G., Sun, W. B. (2018). The role of botanical gardens in scientific research, conservation, and citizen science. Plant Divers. 40, 181–188. doi: 10.1016/j.pld.2018.07.006
Clark, L. V., Stewart, J. R., Nishiwaki, A., Toma, Y., Kjeldsen, J. B., Jørgensen, U., et al. (2015). Genetic structure of Miscanthus sinensis and Miscanthus sacchariflorus in Japan indicates a gradient of bidirectional but asymmetric introgression. J. Exp. Bot. 66, 4213–4225. doi: 10.1093/jxb/eru511
Delang, C., Wang, W. (2013). Chinese Forest policy reforms after 1998: the case of the natural forest protection program and the slope land conversion program. Int. Forest. Rev. 15, 290–304. doi: 10.1505/146554813807700128
Depristo, M. A., Banks, E., Poplin, R., Garimella, K. V., Maguire, J. R., Hartl, C., et al. (2011). A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 43, 491–498. doi: 10.1038/ng.806
de Queiroz, K. (1998). “The general lineage concept of species, species criteria, and the process of speciation: a conceptual unification and terminological recommendations,” in Endless forms: species and speciation. Eds Howard, D. J., Berlocher, S. H. (New York: Oxford University Press), 57–75.
Ding, J., Hua, D., Borrell, J. S., Buggs, R. J., Wang, L., Wang, F., et al. (2021). Introgression between Betula tianshanica and Betula microphylla and its implications for conservation. Plants People Planet 3, 363–374. doi: 10.1002/ppp3.10182
Eidesen, P. B., Alsos, I. G., Brochmann, C. (2015). Comparative analyses of plastid and AFLP data suggest different colonization history and asymmetric hybridization between Betula pubescens and B. nana. Mol. Ecol. 24, 3993–4009. doi: 10.1111/mec.13289
Ferguson, D., Sang, T. (2001). Speciation through homoploid hybridization between allotetraploids in peonies (Paeonia). Proc. Natl. Acad. Sci. USA 98, 3915–3919. doi: 10.1073/pnas.061288698
Fowler, N. L., Levin, D. A. (1984). Ecological constraints on the establishment of a novel polyploid in competition with its diploid progenitor. Am. Nat. 124, 703–711. doi: 10.1086/284307
Fu, L., Chen, T., Lang, K., Hong, T., Lin, Q. (2004). Higher plants of China (Qingdao: Qingdao Publishing House).
Fujita, M. K., Leaché, A. D., Burbrink, F. T., Mcguire, J. A., Moritz, C. (2012). Coalescent-based species delimitation in an integrative taxonomy. Trends Ecol. Evol. 27, 480–488. doi: 10.1016/j.tree.2012.04.012
Giacò, A., Varaldo, L., Casazza, G., De Luca, D., Caputo, P., Sarigu, M., et al. (2022). An integrative taxonomic study of Santolina (Asteraceae) from southern France and north-eastern Spain reveals new endemic taxa. J. Syst. Evol. doi: 10.1111/jse.12925
Grewe, F., Huang, J. P., Leavitt, S. D., Lumbsch, H. T. (2017). Reference-based RADseq resolves robust relationships among closely related species of lichen-forming fungi using metagenomic DNA. Sci. Rep. 7, 1–11. doi: 10.1038/s41598-017-09906-7
Hall, T. A. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucl. Acids. Symp. Ser. 41, 95–98.
Hausdorf, B. (2011). Progress toward a general species concept. Evol.: Int. J. Organic Evol. 65, 923–931. doi: 10.1111/j.1558-5646.2011.01231.x
Hey, J. (2006). On the failure of modern species concepts. Trends Ecol. Evol. 21, 447–450. doi: 10.1016/j.tree.2006.05.011
Holstein, N., Luebert, F. (2017). Taxonomy: stable taxon boundaries. Nature 548, 158–158. doi: 10.1038/548158d
Hu, Y. N., Zhao, L., Buggs, R. J., Zhang, X. M., Li, J., Wang, N. (2019). Population structure of Betula albosinensis and Betula platyphylla: evidence for hybridization and a cryptic lineage. Ann. Bot. 123, 1179–1189. doi: 10.1093/aob/mcz024
Husband, B. C., Sabara, H. A. (2004). Reproductive isolation between autotetraploids and their diploid progenitors in fireweed, Chamerion angustifolium (Onagraceae). New Phytol. 161, 703–713. doi: 10.1046/j.1469-8137.2004.00998.x
Jombart, T. (2008). Adegenet: a r package for the multivariate analysis of genetic markers. Bioinformatics 24, 1403–1405. doi: 10.1093/bioinformatics/btn129
Kumar, N., Holstein, N., Khuraijam, J. S., Rana, T. S. (2022). Taxonomic synopsis of Betula (Betulaceae) in India and typification of three names. Ann. Bot. Fenn. 59, 75–80. doi: 10.5735/085.059.0113
Levin, D. A. (1975). Minority cytotype exclusion in local plant populations. Taxon 24, 35–43. doi: 10.2307/1218997
Li, P. C. (1979). “Betulaceae,” in Flora reipublicae popularis sinicae. Ed. Wang, W. T. (Beijing: Science Press).
Li, H. (2011). A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27, 2987–2993. doi: 10.1093/bioinformatics/btr509
Li, H., Durbin, R. (2009). Fast and accurate short read alignment with burrows–wheeler transform. Bioinformatics 25, 1754–1760. doi: 10.1093/bioinformatics/btp324
Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., et al. (2009). The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079. doi: 10.1093/bioinformatics/btp352
Li, P. C., Skvortsov, A. K. (1999). “Betulaceae,” in Flora of China. Eds. Wu, Z. Y., Raven, P. H. (St. Louis: Missouri Botanical Garden Press).
Marchenko, А.M., Kuzovkina, Y. A. (2022). Notes on the nomenclature and taxonomy of Salix fragilis (Salicaceae). Taxon 71, 721–732. doi: 10.1002/tax.12685
Mayden, R. L. (1997). “A hierarchy of species concepts: the denouement in the saga of the species problem,” in Species: the units of diversity. Eds. Claridge, M. F., Dawah, H. A., Wilson, M. R. (London: Chapman & Hall), 381–423.
Mayr, E. (1942). Systematic and the origin of species, from the viewpoint of a zoologist (No. 13) (Cambridge, Massachusetts: Harvard University Press).
Mckenna, A., Hanna, M., Banks, E., Sivachenko, A., Cibulskis, K., Kernytsky, A., et al. (2010). The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303. doi: 10.1101/gr.107524.110
Mehra, P., Sareen, T. (1973). Cytology of West Himalayan Betulaceae and Salicaceae. J. Arnold Arboretum 54, 412–418. doi: 10.5962/p.324711
Newton, L. G., Starrett, J., Hendrixson, B. E., Derkarabetian, S., Bond, J. E. (2020). Integrative species delimitation reveals cryptic diversity in the southern Appalachian Antrodiaetus unicolor (Araneae: antrodiaetidae) species complex. Mol. Ecol. 29, 2269–2287. doi: 10.1111/mec.15483
O'donnell, K., Sharrock, S. (2017). The contribution of botanic gardens to ex situ conservation through seed banking. Plant Divers. 39, 373–378. doi: 10.1016/j.pld.2017.11.005
Regel, E. A. (1865). Bemerkungen über die gattungen Betula und Alnus nebst beschreibung einiger neuer arten. Bull. Soc. Naturalists (Moscou) 38, 388–434.
Rieseberg, L. H. (2001). Chromosomal rearrangements and speciation. Trends Ecol. Evol. 16, 351–358. doi: 10.1016/S0169-5347(01)02187-5
Rieseberg, L. H., Raymond, O., Rosenthal, D. M., Lai, Z., Livingstone, K., Nakazato, T., et al. (2003). Major ecological transitions in wild sunflowers facilitated by hybridization. Science 301, 1211–1216. doi: 10.1126/science.1086949
Roccaforte, K., Russo, S. E., Pilson, D. (2015). Hybridization and reproductive isolation between diploid Erythronium mesochoreum and its tetraploid congener E. albidum (Liliaceae). Evol. 69, 1375–1389. doi: 10.1111/evo.12666
Salojärvi, J., Smolander, O. P., Nieminen, K., Rajaraman, S., Safronov, O., Safdari, P., et al. (2017). Genome sequencing and population genomic analyses provide insights into the adaptive landscape of silver birch. Nat. Genet. 49, 904–912. doi: 10.1038/ng.3862
Schindel, D. E., Cook, J. A. (2018). The next generation of natural history collections. PloS Biol. 16, e2006125. doi: 10.1371/journal.pbio.2006125
Skvortsov, A. K. (1997). Taxonomic notes on Betula. 1. section Betulaster. Harvard Papers Bot. 11, 65–70.
Smith, L. T., Magdalena, C., Przelomska, N. A., Pérez-Escobar, O. A., Melgar-Gómez, D. G., Beck, S., et al. (2022). Revised species delimitation in the giant water lily genus Victoria (Nymphaeaceae) confirms a new species and has implications for its conservation. Front. Plant Sci. 2241. doi: 10.3389/fpls.2022.883151
Stamatakis, A. (2006). RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22, 2688–2690. doi: 10.1093/bioinformatics/btl446
Tarieiev, A., Gailing, O., Krutovsky, K. V. (2021). ITS secondary structure reconstruction to resolve taxonomy and phylogeny of the Betula l. genus. PeerJ 9, e10889. doi: 10.7717/peerj.10889
Tarieiev, A., Olshanskyi, I., Gailing, O., Krutovsky, K. V. (2019). Taxonomy of dark-and white-barked birches related to Betula pendula and B. pubescens (Betulaceae) in Ukraine based on both morphological traits and DNA markers. Bot. J. Linn. Soc. 191, 142–154. doi: 10.1093/botlinnean/boz031
Thiers, B. M. (2023)Index herbariorum: a global directory of public herbaria and associated staff. In: New York botanical garden's virtual herbarium. Available at: http://sweetgum.nybg.org/ih/ (Accessed 24 January 2023).
Thórsson, Æ.T., Pálsson, S., Lascoux, M., Anamthawat-Jónsson, K. (2010). Introgression and phylogeography of Betula nana (diploid), B. pubescens (tetraploid) and their triploid hybrids in Iceland inferred from cpDNA haplotype variation. J. Biogeogr. 37, 2098–2110. doi: 10.1111/j.1365-2699.2010.02353.x
Tovar-Sánchez, E., Oyama, K. (2004). Natural hybridization and hybrid zones between Quercus crassifolia and Quercus crassipes (Fagaceae) in Mexico: morphological and molecular evidence. Am. J. Bot. 91, 1352–1363. doi: 10.3732/ajb.91.9.1352
Tsuda, Y., Semerikov, V., Sebastiani, F., Vendramin, G. G., Lascoux, M. (2017). Multispecies genetic structure and hybridization in the Betula genus across Eurasia. Mol. Ecol. 26, 589–605. doi: 10.1111/mec.13885
Tuley, G. (1973). A taxonomic history of the British birch tree. Trans. Bot. Soc. Edinb. 42, 31–41. doi: 10.1080/03746607308685260
Turland, N. J., Wiersema, J. H., Barrie, F. R., Greuter, W., Hawksworth, D. L., Herendeen, P. S., et al. (2018). International Code of Nomenclature for algae, fungi, and plants (Shenzhen Code) adopted by the Nineteenth International Botanical Congress Shenzhen, China, July 2017. Regnum Vegetabile 159. (Glashütten: Koeltz Botanical Books).
Twyford, A. D., Ennos, R. (2012). Next-generation hybridization and introgression. Heredity 108, 179–189. doi: 10.1038/hdy.2011.68
Wallich, N. (1830). Plantae asiaticae rariores; or, descriptions and figures of a select number of unpublished East Indian plants (London: Treuttel and Würtz).
Wang, N., Borrell, J. S., Bodles, W. J., Kuttapitiya, A., Nichols, R. A., Buggs, R. J. (2014b). Molecular footprints of the Holocene retreat of dwarf birch in Britain. Mol. Ecol. 23, 2771–2782. doi: 10.1111/mec.12768
Wang, N., Borrell, J., Buggs, R. (2014a). Is the Atkinson discriminant function a reliable method for distinguishing between Betula pendula and B. pubescens (Betulaceae)? New J. Bot. 4, 90–94. doi: 10.1179/2042349714Y.0000000044
Wang, L. W., Ding, J. Y., Borrell, J. S., Cheek, M., Mcallister, H. A., Wang, F. F., et al. (2022). Molecular and morphological analyses clarify species delimitation in section Costatae and reveal Betula buggsii sp. nov.(sect. Costatae, Betulaceae) in China. Ann. Bot. 129, 415–428. doi: 10.1093/aob/mcac001
Wang, N., Kelly, L. J., Mcallister, H. A., Zohren, J., Buggs, R. J. (2021). Resolving phylogeny and polyploid parentage using genus-wide genome-wide sequence data from birch trees. Mol. Phylogenet. Evol. 160, 107126. doi: 10.1016/j.ympev.2021.107126
Wang, N., Mcallister, H. A., Bartlett, P. R., Buggs, R. J. (2016). Molecular phylogeny and genome size evolution of the genus Betula (Betulaceae). Ann. Bot. 117, 1023–1035. doi: 10.1093/aob/mcw048
Wang, N., Thomson, M., Bodles, W. J., Crawford, R. M., Hunt, H. V., Featherstone, A. W., et al. (2013). Genome sequence of dwarf birch (Betula nana) and cross-species RAD markers. Mol. Ecol. 22, 3098–3111. doi: 10.1111/mec.12131
We, T., Hu, Q., Chen, Z. (1987). Flora of Guangdong (Guangzhou: Guangdong Science and Technology Press).
Webster, M. S. (2017). The extended specimen: emerging frontiers in collections-based ornithological research (Portland: CRC Press).
Wehenkel, C., Del Rocío Mariscal-Lucero, S., González-Elizondo, M. S., Aguirre-Galindo, V. A., Fladung, M., López-Sánchez, C. A. (2020). Tall Pinus luzmariae trees with genes from P. herrerae. PeerJ. 8, e8648. doi: 10.7717/peerj.8648
White, T. J., Bruns, T., Lee, S., Taylor, J. (1990). “Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics,” in PCR protocols: a guide to methods and applications. Eds. Innis, M. A., Gelfland, D. H., Sninsky, J. J., White, T. J. (New York: Academic Press), 315–322.
Winkler, H. (1904). “Betulaceae,” in Das Pflanzenreich Heft. Ed. Engler, A. (Leipzig, Germany), 1–149.
Zeng, J., Li, J. H., Chen, Z. D. (2008). A new species of Betula section Betulaster (Betulaceae) from China. Bot. J. Linn. Soc. 156, 523–528. doi: 10.1111/j.1095-8339.2007.00764.x
Zeng, Z., Ren, B. Q., Zhu, J. Y., Chen, Z. D. (2014). Betula hainanensis (Betulaster, Betulaceae), a new species from Hainan island, China. Ann. Bot. Fenn. 51, 399–402. doi: 10.5735/085.051.0606
Keywords: Betula, botanic garden, introgression, RAD-seq, polyploid
Citation: Zhang H, Ding J, Holstein N and Wang N (2023) Betula mcallisteri sp. nov. (sect. Acuminatae, Betulaceae), a new diploid species overlooked in the wild and in cultivation, and its relation to the widespread B. luminifera. Front. Plant Sci. 14:1113274. doi: 10.3389/fpls.2023.1113274
Received: 01 December 2022; Accepted: 19 April 2023;
Published: 23 May 2023.
Edited by:
Jim Leebens-Mack, University of Georgia, United StatesReviewed by:
Hengchang Wang, Chinese Academy of Sciences (CAS), ChinaLlorenç Saez, Autonomous University of Barcelona, Spain
Copyright © 2023 Zhang, Ding, Holstein and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nian Wang, bmlhbi53YW5nQHNkYXUuZWR1LmNu
 Huayu Zhang
Huayu Zhang Junyi Ding
Junyi Ding Norbert Holstein3
Norbert Holstein3 Nian Wang
Nian Wang