
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 16 March 2023
Sec. Plant Metabolism and Chemodiversity
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1112214
This article is part of the Research TopicSecondary Metabolites in Grapevine Stress Response - Women in Plant Science SeriesView all 11 articles
 Robin Nicole Bosman1
Robin Nicole Bosman1 Jessica Anne-Marie Vervalle2
Jessica Anne-Marie Vervalle2 Danielle Lisa November1
Danielle Lisa November1 Phyllis Burger3
Phyllis Burger3 Justin Graham Lashbrooke1*
Justin Graham Lashbrooke1*Volatile organic compounds such as terpenes influence the quality parameters of grapevine through their contribution to the flavour and aroma profile of berries. Biosynthesis of volatile organic compounds in grapevine is relatively complex and controlled by multiple genes, the majority of which are unknown or uncharacterised. To identify the genomic regions that associate with modulation of these compounds in grapevine berries, volatile metabolic data generated via GC-MS from a grapevine mapping population was used to identify quantitative trait loci (QTLs). Several significant QTLs were associated with terpenes, and candidate genes were proposed for sesquiterpene and monoterpene biosynthesis. For monoterpenes, loci on chromosomes 12 and 13 were shown to be associated with geraniol and cyclic monoterpene accumulation, respectively. The locus on chromosome 12 was shown to contain a geraniol synthase gene (VvGer), while the locus on chromosome 13 contained an α-terpineol synthase gene (VvTer). Molecular and genomic investigation of VvGer and VvTer revealed that these genes were found in tandemly duplicated clusters, displaying high levels of hemizygosity. Gene copy number analysis further showed that not only did VvTer and VvGer copy numbers vary within the mapping population, but also across recently sequenced Vitis cultivars. Significantly, VvTer copy number correlated with both VvTer gene expression and cyclic monoterpene accumulation in the mapping population. A hypothesis for a hyper-functional VvTer allele linked to increased gene copy number in the mapping population is presented and can potentially lead to selection of cultivars with modulated terpene profiles. The study highlights the impact of VvTPS gene duplication and copy number variation on terpene accumulation in grapevine.
Terpenes are one of the largest classes of metabolites in plants, where they serve various primary and specialized roles. Volatile terpenes, such as monoterpenes and sesquiterpenes, mainly function as specialized metabolites and are involved in plant-pathogen interactions, protection of plants against herbivores, and are also produced to attract pollinators and seed-dispersing animals (Dudareva et al., 2013; Vranová et al., 2013). Together with additional volatile organic compounds (VOCs) such as short-carbon chain compounds (green leaf volatiles), C13-norisoprenoids and methoxypyrazines, they contribute to the varietal aroma of grape berries (Dunlevy et al., 2009). Indeed, monoterpenes and sesquiterpenes have been extensively studies for their contribution to the distinctive varietal aroma of aromatic cultivars such as Muscat-cultivars (monoterpene alcohols including linalool, geraniol, and α-terpineol), ‘Shiraz’ (the sesquiterpene rotundone), ‘Riesling’ and ‘Gewürztraminer’ (the monoterpene rose-oxide) (Dunlevy et al., 2009).
Biosynthesis of monoterpenes and sesquiterpene occurs via the methyl-erythritol-phosphate (MEP) and mevalonic acid (MVA) pathways, respectively. The first step in the MEP pathway is catalysed by 1-deoxy-D-xylylose-5-phosphate synthase (DXS), an enzyme which is considered to be the vital rate-limiting enzyme in plastidial terpene biosynthesis(Tholl, 2015; Bosman and Lashbrooke, 2023). Several QTL mapping and association studies in grapevine have identified a singular SNP in the active site of VvDXS1 as a causal mutation for increased monoterpene content in Muscat cultivars (Doligez et al., 2006; Battilana et al., 2009; Emanuelli et al., 2010). A SNP at position 1822 of VvDXS1 (G>T) in Muscat cultivars causes a non-synonymous substitution of a lysine (K) with an asparagine (N) at position 284 of the VvDXS1 protein. Functional characterisation of VvDXS1 showed that the non-synonymous amino acid substitution influences enzyme kinetics by increasing the catalytic efficiency of VvDXS1, thereby increasing the total monoterpene content of cultivars carrying this SNP (Battilana et al., 2011).
While VvDXS1 is able to regulate total monoterpene accumulation via biosynthesis of the prenyldiphosphate precursors, terpene synthases (TPSs) are responsible for the formation of specific terpenes (Steele et al., 1998; Chen et al., 2011). Monoterpene synthases catalyse the coupled ionisation, isomerisation and cyclisation of geranyldiphosphate (GPP) leading to the formation of a reactive carbocation intermediate and subsequent reactions e.g. deprotonation or ring closures will form the final monoterpene product (Davis and Croteau, 2000; Degenhardt et al., 2009).
The Vitis vinifera reference genome, PN40024, has a greatly expanded TPS gene family, with an initial prediction of 152 loci and 69 putatively functional TPSs (Jaillon et al., 2007; Martin et al., 2010). However, the recent availability of phased diploid grapevine genomes of various cultivars (Minio et al., 2019; Zhou et al., 2019; Massonnet et al., 2020) reveals that the TPS gene family size varies greatly between cultivars (Smit et al., 2020). Furthermore, VvTPSs are organised in large tandemly duplicated clusters, and a great portion of genes are hemizygous (Martin et al., 2010; Jiang et al., 2019; Smit et al., 2020). Recent research has also shown that grapevine has cultivar-specific TPS genes (Drew et al., 2016; Smit et al., 2019). Cultivar-specific TPSs arise due to small sequence variations, such as single nucleotide polymorphisms (SNPs), which cause a functional change of the enzyme. The extensive level of duplication and functional plasticity of VvTPSs contribute to the neofunctionalisation of these enzymes and results in the large diversity in metabolites formed by VvTPSs (Bosman and Lashbrooke, 2023).
This study utilises a biparental grapevine cross population established by crossing a wine cultivar and a table grape cultivar. A dense linkage map has previously been created for this mapping population (Vervalle et al., 2022) which segregates for various traits, including aromatic profile. Quantification of volatile organic compounds in this population over several seasons was performed and genomic regions associated with these compounds identified. Genomic regions containing multiple TPS genes were further interrogated, and the large variety in cultivar specific TPS copy number associated with accumulation of specific monoterpenes characterised.
82 progenies of the mapping population (‘Deckrot’ x G1-7720), which is held at the Agricultural Research Council (ARC) Nietvoorbij (Stellenbosch, South Africa, 33° 54’ 47.6’’ S, 18° 51’ 54.9’’ E) were used for analysis. G1-7720 is a table grape selection developed by the ARC and is a cross between ‘Black Rose’ and ‘Muscat Seedless’. Grape berries from the progenies and parents were sampled at veraison (EL-stage 35) in January 2019, and at harvest ripeness (EL-stage 38) in February 2021 and 2022. Veraison berries from each bunch were further divided into “pre-veraison” (berries that were still green and firm) and “veraison” (berries which had changed colour and softened). The skin and flesh of all berries (pre-veraison, veraison and harvest ripe) were separated. Additionally, the parent cultivars were sampled at various early developmental stages (EL-stages 19, 23, 26, 29, 31 and 33) between October-December 2020. Three biological replicates were sampled for both cultivars at each developmental stage. Flowers and berries were removed from the rachis, and the rachis was discarded. All plant material was frozen in liquid nitrogen, ground to a fine powder and stored at -80°C.
Approximately 150 mg of ground frozen berry skin tissue was weighed into a 20-mL GC vial and 2 ml tartaric acid buffer (5 g.L-1 tartaric acid, 2 g.L-1, 0.8 g.L-1 and 4.28 M NaCl; pH 3.2), containing 5 g.L-1 internal standard (3-octanol), was added to the vial. Headspace solid phase microextraction (HS-SPME) and gas chromatography mass spectrometry (GCMS) were performed according to the method described in Joubert et al. (2016). Samples were extracted from the vial head space using a 50/30 µm grey divinylbenzene/carboxen/polydimethylsiloxane (DVB/CAR/PDMS) fiber (Supelco, USA). The GCMS analysis was carried out on an Agilent 7890B GC equipped with a 5977B MSD and a PAL RSI 85 autosampler. Chromatographic separation was achieved using a HP-5MSUI capillary column (30m x 0.25 µm x 0.25 mm). The purge flow was 30 mL.min-1 (for 90 seconds). The oven parameters were as follows: initial temperature of 40°C (2 min), a linear increase to a final temperature of 250°C (at a rate of 6°C.min-1), and the temperature was held at 250°C for a final 5 min. The MS detector was operated in scan mode (from 35 to 350 m/z).
Agilent MassHunter Qualitative and Quantitative software packages were used for data analysis. Volatile compounds were identified according to their elution times and masses compared to those of respective authentic standards. Compounds without available authentic standards were identified by matching their mass spectrum with the NIST (Linstrom and Mallard, 2001) mass spectral library, in combination with Kovatz retention indices (RIs). Relative quantification of the compounds was achieved by normalising the peak area of each compound with the peak area of the internal standard. Concentrations are expressed as µg of 3-octanol equivalents per gram fresh weight.
For α-terpineol and geraniol, quantification was achieved through external standard calibration which was done by plotting standard curves using the internal response ratio versus the standard concentration. The resultant concentrations in μg.L-1 were then normalised to the berry fresh weight to obtain the concentration (in µg.g-1 FW).
A total of 137 progeny of the ‘Deckrot’ x G1-7720 mapping population have previously been genotyped with the Vitis18K SNP chip and 92 simple sequence repeat (SSR) markers (Vervalle et al., 2022). The genotyping data were used to construct parental linkage maps for ‘Deckrot’ and G1-7720 with JoinMap®5 (Van Ooijen, 2006). Both maps represented all 19 linkage groups of grapevine and contained 1910 and 2252 markers in the maternal and paternal maps respectively. The maps displayed an average inter-locus gap distance of 0.80 cM. The genetic maps were combined with the metabolomic data to perform QTL analyses in MapQTL®6 (Van Ooijen, 2009). QTL regions were first identified through interval mapping with the maximum likelihood mixture model algorithm. Subsequently, regions were further defined with the multiple-QTL models (MQM) mapping. Genome-wide significant LOD thresholds were determined with permutation tests of 1000 permutations each. All maps were drawn with MapChart v2.32 (Voorrips, 2002).
The position of significant QTLs on the grapevine reference genome (PN40024) version 2 (Jaillon et al., 2007) was determined from the physical positions of the two neighbouring flanking markers. Annotated genes within each QTL were retrieved from URGI (https://urgi.versailles.inra.fr/Species/Vitis/Annotations) and is based on the VCost v.3 structural annotation of the 12X.2 reference genome (Jaillon et al., 2007; Canaguier et al., 2017). Gene function was predicted via BLASTp analysis (https://blast.ncbi.nlm.nih.gov/Blast.cgi) by querying protein sequences against the UniProtKB/SwissProt database and selecting the top hit for each sequence. Candidate genes were selected based on evidence from literature and their function was further investigated through molecular phylogeny. Phylogenetic trees were created on “Geneious Tree Builder” 2022.0.1 (https://www.geneious.com) using the default ‘Geneious Tree Builder” function which used the UPGMA method with the Jukes-Cantor distance measure algorithm and 100 bootstrapping replicates. Multiple sequence alignments were created using the default ‘Geneoius Alignment’ setting. Candidate gene expression was compared to metabolite data using the Transcriptomics & Metabolomics integrated database (TransMetaDb) (Savoi et al., 2016; Savoi et al., 2017) available on the Vitis Visualization platform (VitViz) (http://www.vitviz.tomsbiolab.com/).
A custom TaqMan SNP genotyping assay (ThermoFisher Scientific) was designed for the VvDXS1 SNP and used to genotype the mapping population under investigation. Genomic DNA was extracted from grape berry skins as described in Reid et al. (2006) however the DNase treatment step was substituted with a RNase treatment to eliminate RNA. The custom assay mix consisted of 1X TaqMan® MasterMix (ThermoFisher Scientific), 1X Custom Assay mix which contains the custom primers and probes (Table S1), and 1 µL (10-40 ng) of genomic DNA. Primers and probes were designed using PrimerExpress (Singh and Pandey, 2015). The assay was performed in QuantStudio 3 Real-Time PCR System with the following conditions: a pre-read stage for 30 seconds at 60°C, initial denaturation at 95°C for 5 minutes, 40 cycles of denaturation (95°C for 15 seconds) and annealing/extension (60°C for 1 minute), and then a post-read stage for 30 seconds at 60°C.
Confirmation of the results from the TaqMan assay was performed for a subset (20 progeny and the parents) of the mapping population. VvDXS1 was isolated from gDNA via polymerase chain reaction (PCR). The reaction mixture (25 μL) contained 0.2 μM of the forward (5’-GTCATAGGTGATGGAGCCA-3’) and reverse (5’-ATCTTACCTGTTCTGTCTAGC-3’) primers (Emanuelli et al., 2014), 1 μL DNA (approximately 50 ng) and 1X GoTaq®Green Master Mix (Promega, USA). PCR products were visualised on a 1% agarose gel, excised and purified using Zymoclean Gel DNA recovery kit, and subsequently sequenced via Sanger sequencing.
Total genomic DNA was extracted from pre-veraison samples as described in in Reid et al. (2006), and relative gene copy number was determined via a qPCR-based method described previously (Ma and Chung, 2014; Bhattacharya et al., 2019). qPCR was performed in a QuantStudio 3 Real-Time PCR System (ThermoFisher) in a 15 µL reaction mixture which contained 1X SYBR (Power SYBR Green, Applied Biosystems), 0.2 µM of each primer pair, 2 µL of genomic DNA (approximately 90 ng) and nuclease-free water. Reactions were repeated in technical triplicate for each gene-sample combination, making use of VvActin primers to normalise for the amount of genomic DNA assayed in each sample and either VvGer or VvTer gene specific primers for amplification of the respective gene copies (for primer sequences see Table S2). The PCR conditions were: initial denaturation step at 95°C for 3 minutes, 40 cycles (95°C for 3 seconds and 60°C for 20 seconds), and melt curve analysis from 60 to 95°C. Relative gene copy number was determined with the 2−ΔΔCt method (Ma and Chung, 2014), making use of the QuantStudio Design and Analysis Desktop Software package (v1.5.1) from Applied Biosystems for processing relative copy number and 95% confidence intervals. All gene copy numbers are reported relative to the gene copies present in the ‘Deckrot’ cultivar.
Correlation analysis, Pearson’s correlation (R) and Spearman’s rank correlation (ρ), between expression, copy number and metabolite data was performed with XLStat (version 20213.1) add-on for Excel (Addinsoft, 2023).
Total RNA was extracted from pre-veraison samples using the Spectrum™ Plant Total RNA Kit (Sigma-Aldrich) with the removal of genomic DNA via on-column DNase digestion using the On-Column DNase I Digestion Set (Sigma-Aldrich) according to the manufacturer’s instructions. 1 µg of RNA was converted to cDNA using the GoScript™ Reverse Transcription mix, Oligo(dT) (Promega).
The relative expression of genes was measured via qPCR using primers from literature or newly designed primers (Table S2) with VvActin used as an endogenous control. qPCR was performed as described for gene copy number variation analysis above. Gene expression data for the progeny is reported relative to the individual with the lowest expression, while gene expression data for the parents’ developmental stages are reported relative to gene expression in ‘Deckrot’ green berry stage (EL-stage 33).
Genes were isolated from cDNA samples via polymerase chain reaction (PCR) using primers listed in Table S2. The PCR mixture (25 μL) contained 0.2 μM of the forward primer and reverse primers, 5 μL cDNA (1:10 dilution), 1X ExTaq Buffer and 1U ExTaq polymerase (Takara). The reaction was performed in an Applied Biosystems™ MiniAmp thermal cycler (ThermoFisher Scientific) and conditions were as follows: initial denaturation at 95°C for 3 minutes, then 40 cycles (95°C for 30 seconds, 55°C for 30 seconds, 72°C for 2 minutes) and a final elongation step at 72°C for 7 minutes. PCR products were purified using Zymoclean Gel DNA Recovery Kit (Zymo Research) as per the manufacturer’s protocol.
PCR products were ligated into pGEM® T-Easy Vector System (Promega) overnight at 4°C. Ligated vectors were subsequently transformed into competent DH5α E. coli cells via heat-shock. Transformed cells were plated on LB agar containing ampicillin and after overnight incubation at 37°C, colonies were screened via PCR using M13 primers. Plasmids were isolated from positive colonies using GenElute™ Plasmid Miniprep Kit (Sigma-Aldrich), as per the manufacturer’s protocol. Sanger sequencing was performed by the Central Analytical Facilities at Stellenbosch University (Stellenbosch, South Africa).
68 different volatile compounds were identified from the GCMS analysis: six alcohols, three ketones, ten aldehydes, two esters, four C13-norisoprenoids, 18 monoterpenes and 25 sesquiterpenes (Table S3). The volatile profiles of the parent cultivars were compared (Figure 1) and show clear segregation. Overall, the table grape selection, G1-7720, which is categorised as having a muscat aroma (Vervalle et al., 2022), has a higher volatile content regardless of developmental stage. While, the terpene content of both parents is highest at veraison, G1-7720 has a significant increase in sesquiterpene content from pre-veraison to veraison berries. G1-7720 shows significantly higher terpene levels than that of ‘Deckrot’. Aldehydes and alcohols also make up a major proportion of the volatile composition of each cultivar, specifically ‘Deckrot’, with hexanal, trans-2-hexenal and (E)-3-hexen-1-ol being the most significant contributors.
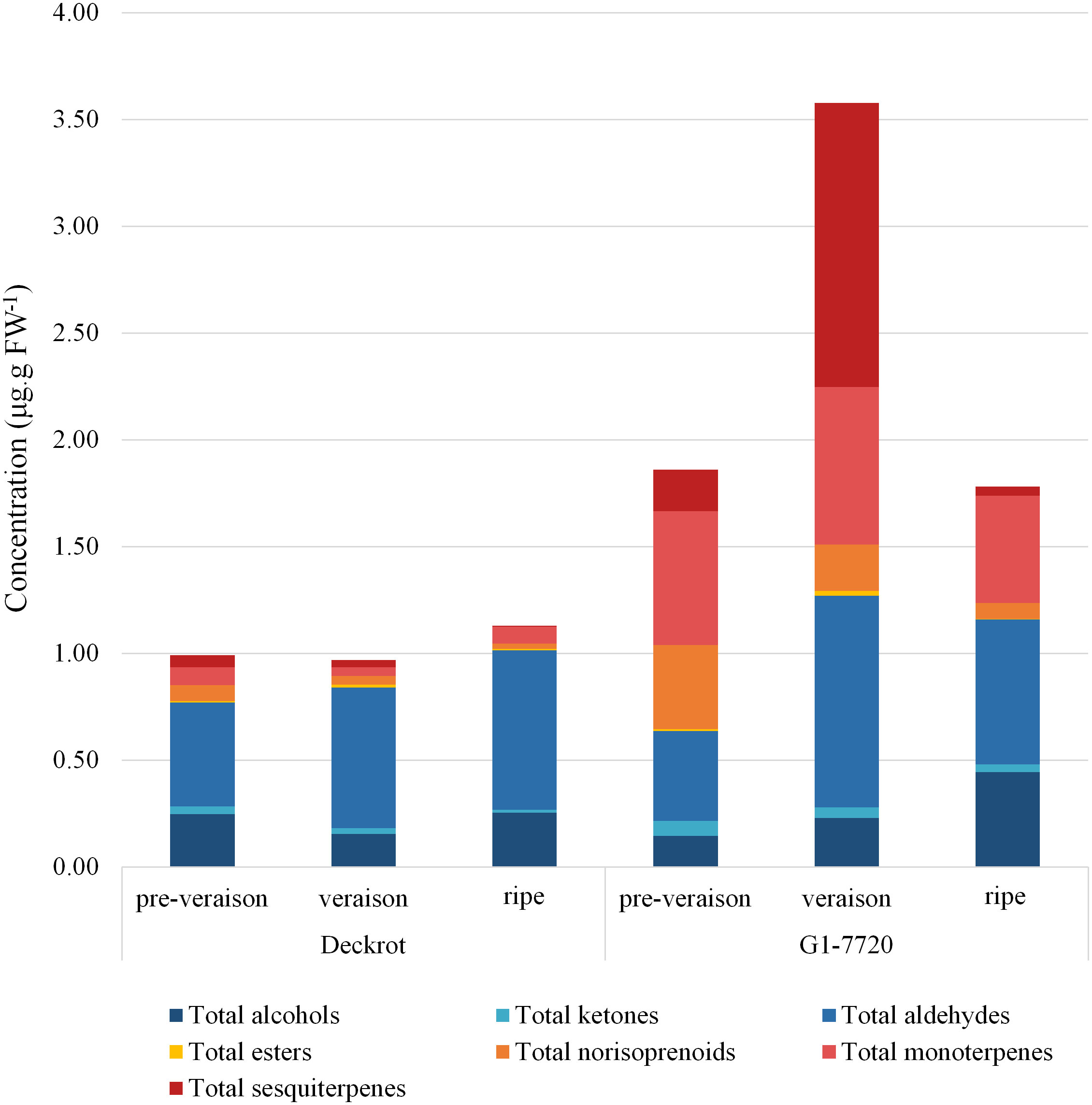
Figure 1 The volatile composition of the parent cultivars ‘Deckrot’ and G1-7720 during pre-veraison, veraison and harvest ripe stage.
The distribution of the various volatile compound classes in the progeny is shown in Figure S1. Aldehydes are the most abundant compound class in the population; the average ratio of total aldehydes to total VOCs ranges between 34-47% across the various developmental stages. Trans-2-hexenal (an aldehyde) is the most abundant compound in the population. In fact, all six-carbon volatiles, also known as green leaf volatiles (GLVs), are among the most abundant VOCs within the population in all development stages. Sesquiterpenes are the most diverse class of compounds identified (26 compounds) in this study however they are present in low concentrations during most years, with the highest concentrations found in unripe veraison berries.
186 Significant QTLs were identified associated with 54 compounds across the various seasons under analysis (Table S4). Consistent QTLs which were identified in at least two stages were mapped to the PN40024 v2 genome and annotated genes in each region were counted and inspected manually for their potential to regulate the associated metabolic phenotypes. QTLs associated with various monoterpenes were identified on chromosomes 5, 12, 13 and 17, while a QTL associated with the sesquiterpenes trans-caryophyllene and α-cubebene was identified on chromosome 19 and a QTL for total aldehydes and trans-2-hexenal was identified on chromosome 2 (Figure 2). Inspection of these genomic regions identified an average of 370 candidate genes in each QTL (Table 1).
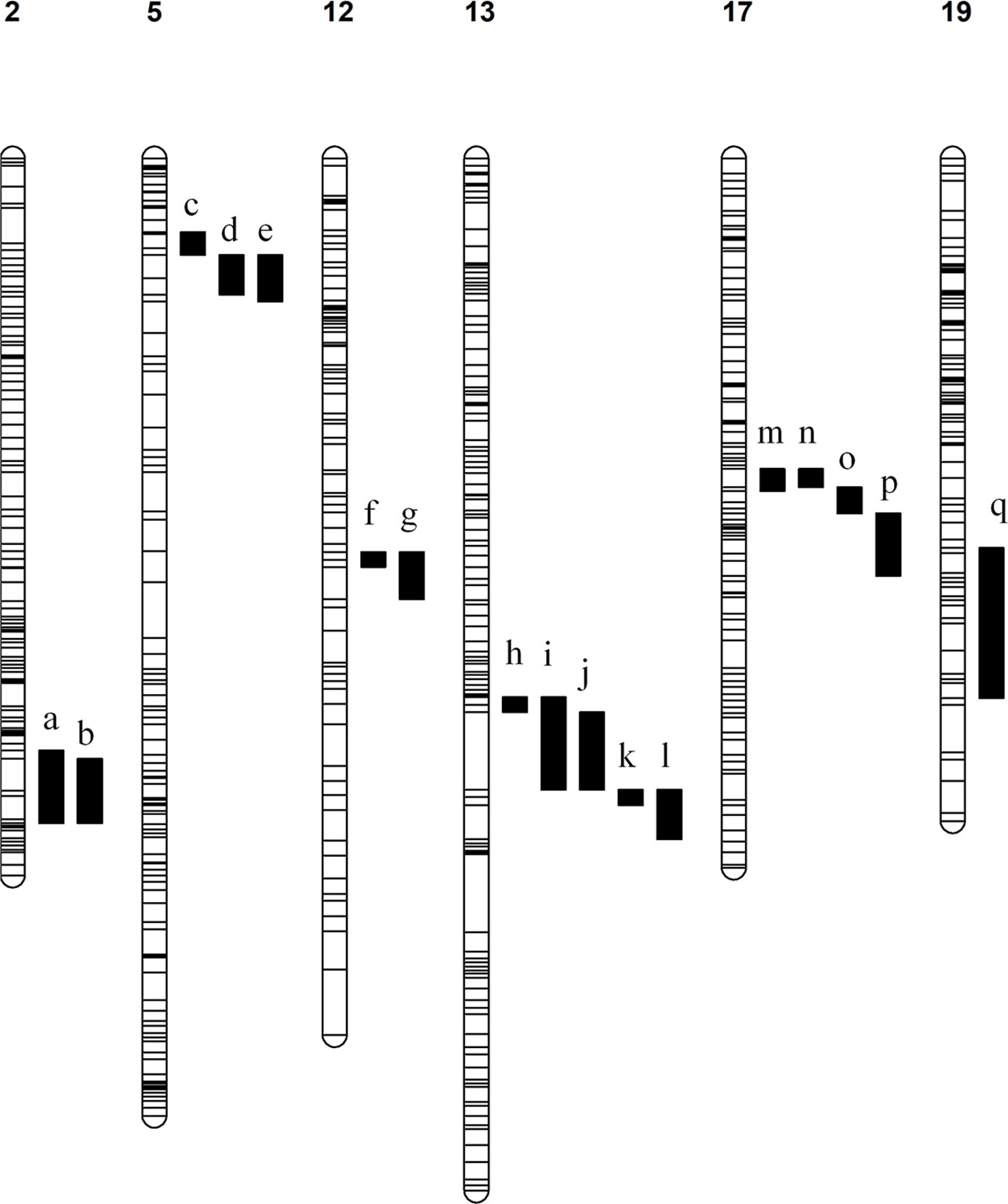
Figure 2 Linkage maps with associated QTLs. The chromosome number is indicated above each map and all QTLs are numbered a – q. The associated compounds for each QTL are listed. a) aldehydes total (veraison 2019) and trans-2-hexenal (veraison 2019). b) aldehydes total (2022) and trans-2-hexenal (2022). c) E-citral (veraison & 2021), β-citronellol (pre-veraison & 2022), cis-rose oxide (veraison, 2021 & 2022), geraniol (2022), limonene (2022), linalool (2022), total monoterpenes (2022), nerol oxide (veraison), p-mentha-1,5-dien-8-ol (veraison & 2021), trans-β-Ocimene (2022), α-terpinene (2022), α-terpineol (2022), α-terpinolene (2022), β-Myrcene (2022), γ-Terpinene (2022), p-Cymene (2022). d) 6-Methyl-5-hepten-2-one (pre-veraison, veraison, 2021), cis-rose oxide (pre-veraison), linalool (pre-veraison), p-Cymene (pre-veraison & veraison), p-mentha-1,5-dien-8-ol (pre-veraison), trans-β-Ocimene (pre-veraison & veraison), α-terpinene (veraison), γ-Terpinene (pre-veraison & veraison). e) β-Citronellol/Nerol (pre-veraison & 2021), C13-norisoprenoids total (veraison & 2021), geraniol (pre-veraison, veraison & 2021), limonene (pre-veraison, veraison & 2021), linalool (2021), linalool oxide (pre-veraison, veraison & 2021), monoterpenes total (pre-veraison, veraison & 2021), nerol oxide (pre-veraison & veraison), p-Cymene (2021), Terpinene-4-ol (pre-veraison, veraison & 2021), trans-β-Ocimene (2021), α-terpinene (veraison & 2021), α-Terpineol (pre-veraison, veraison & 2021), α-terpinolene (pre-veraison, veraison & 2021), β-Myrcene (pre-veraison, veraison & 2021), γ-Terpinene (2021). f) Geraniol (pre-veraison) & β-Myrcene (pre-veraison). g) Geraniol (veraison) & β-Myrcene (veraison). h) α-Terpineol (veraison) & α-terpinolene (veraison). (I) Terpinene-4-ol (2021). j) 1,8-Cineole (pre-veraison & veraison), limonene (veraison), nerol oxide (veraison), p-Cymene (veraison & 2021), p-mentha-1,5-dien-8-ol (pre-veraison & veraison), Terpinene-4-ol (pre-veraison & veraison), α-terpinene (pre-veraison & veraison), γ-Terpinene (pre-veraison). k) limonene (pre-veraison), α-terpinene (2021), γ-Terpinene (veraison). l) p-Cymene (pre-veraison), p-mentha-1,5-dien-8-ol (2021), α-terpinolene (pre-veraison). m) cis-rose oxide (2021) & nerol oxide (veraison). n) nerol oxide (pre-veraison). o) cis-rose oxide (veraison). p) E-citral (2021) & cis-rose oxide (pre-veraison). q) α-Cubebene (pre-veraison & veraison) & trans-caryophyllene (pre-veraison and veraison).
A QTL on chromosome 2 associated with both total aldehydes and trans-2-hexenal and contained 335 genes. A Stearoyl-[acyl-carrier-protein] 9-desaturase 6 (SAD) was identified as candidate gene from this region. The Vitis vinifera SAD (VvSAD) was compared to other functionally characterised SAD enzymes from various species (Figure S2 and Table S5) to infer function.
Three geraniol derived compounds, namely cis-rose oxide, (E)-citral and nerol oxide associated with a QTL on chromosome 17. The enzymes involved in conversion of geraniol to cis-rose oxide, (E)-citral and nerol oxide are not described in grapevine however the reaction likely starts with the reduction of geraniol by an unknown reductase (Lin et al., 2019). To that end five short-chain dehydrogenase/reductases (VvSDR1-5) which co-localises with this QTL were selected as candidate genes. Comparison of VvSDR1-5 expression to metabolite data using TransMetaDb showed that VvSDR4 and VvSDR5 had strong positive correlation with citronellol accumulation (R = 0.71 and 0.70, respectively), while VvSDR1 had a negative correlation (R = -0.54) with citronellol. Additionally, VvSDR3 had a negative correlation (R = -0.62) with nerol. Molecular phylogenetic analysis of the protein sequences of the identified VvSDRs were compared to that of other plant SDRs (Figure S3 and Table S5), and it was found that the VvSDRs cluster with three nepetalactol synthases which catalyse the conversion of 8-oxogeranial to nepetalactol in catmint (Lichman et al., 2019).
While total sesquiterpenes did not produce any significant QTLs, the sesquiterpenes α-cubebene and trans-caryophyllene associated with a QTL on chromosome 19. This QTL co-localises with two predicted sesquiterpene synthases (VvTPS29 and VvTPS69), however VvTPS69 represented a partial gene and was thus not considered a candidate.
The QTLs associated with monoterpenes predominantly localised to chromosomes 5, 12 and 13. The majority of monoterpenes associated with a QTL region on chromosome 5 which co-localises with VvDXS1. Similarly, total monoterpenes and total C13-norisoprenoids, localised to the same region on chromosome 5. While the C13-norisoprenoid 6-Methyl-5-hepten-2-one (MHO) also showed a QTL on the same region on chromosome 5.
The acyclic monoterpenes geraniol and β-myrcene associated with an additional QTL on chromosome 12 while several cyclic monoterpenes associated with an additional QTL on chromosome 13. Genomic inspection of these QTLs found that they co-localise with clusters of terpene synthases (TPSs). When compared to the PN42004 v2 reference genome the QTL on chromosome 12 was found to co-localise with a cluster of eight TPS genes, while the QTL on chromosome 13 was found to co-localise with a TPS cluster that contains 11 terpene synthases (summarised in Table S6).
A phylogenetic tree comparing the VvTPSs in these QTL regions to functionally characterised Vitis vinifera TPSs is shown in Figure 3. VvTPS52 and VvTPS51 fall within a cluster with functionally characterised geraniol synthases (TPS-g), indeed VvTPS52 is the PN40024 gene model for geraniol synthase (VvGer) (Martin et al., 2010) and therefore VvTPS52/VvGer was selected as candidate gene for the QTL associated with geraniol in this mapping population. VvTPS39, VvTPS119 and VvTPS116 cluster with VvTer1 and VvTer2, α-terpineol synthases in the TPS-a clade, however, VvTPS119 and VvTPS16 represent pseudogenes (disrupted by numerous deletions, frameshifts and/or stop codons) in the PN40024 v2 genome. VvTPS39/VvTer was therefore considered the likely candidate gene underpinning the QTL region. Lastly, VvTPS29 clusters with trans-caryophyllene synthases (VviSHTPS27, VviMATPS27 and VvGwECar2) and a germacrene D synthase (VvGerD) in the TPS-a clade.
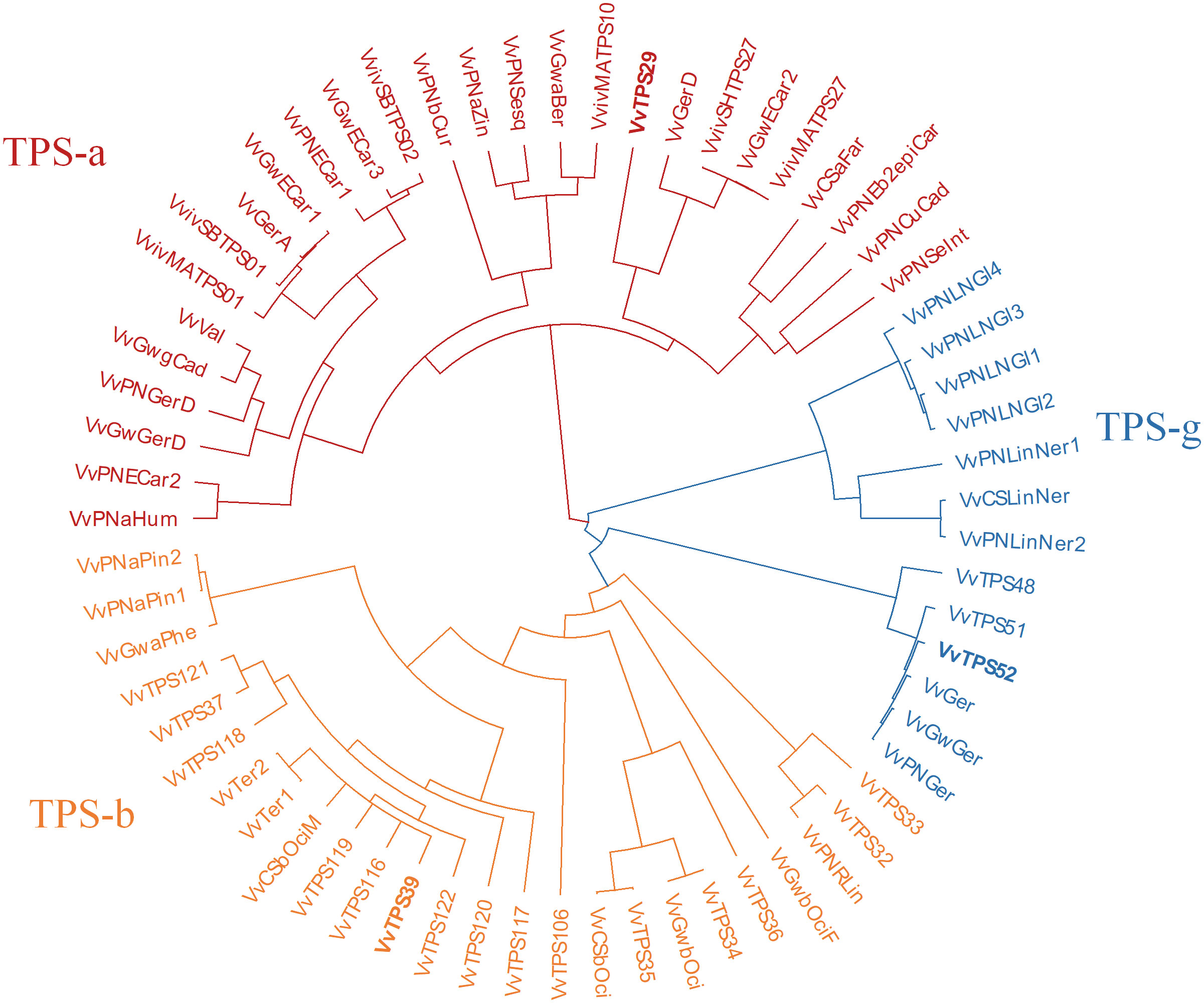
Figure 3 Phylogenetic tree of functionally characterised V. vinifera TPSs and VvTPSs identified in QTLs of interest from this study. The tree is coloured according to TPS-subfamilies: TPS-a subfamily is in red, TPS-b is orange and TPS-g subfamily is blue. Candidate genes are shown in bold typeface.
DNA sequencing of the VvDXS1 alleles revealed that while ‘Deckrot’ was homozygous (GG) for the wild-type allele, G1-7720 was heterozygous (GT) for the muscat aroma linked SNP (Doligez et al., 2006; Battilana et al., 2009; Emanuelli et al., 2010). A TaqMan assay was used to ascertain how the mapping population under investigation segregates for the G>T SNP. Results show that of the 82 progenies genotyped, 50 were heterozygous (GT) and 32 were homozygous (GG) (Table S7). Comparison of the total monoterpene content in the progeny with the occurrence of the G>T SNP found that GG-progeny displayed relatively low monoterpene accumulation, while GT-progeny displayed relatively increased monoterpene accumulation levels, with a continuous variation in distribution (Figure 4). Orthogonal partial least squares (OPLS) analysis of the full volatile dataset across the population and VvDXS1 genotypes show that the population clearly segregates for the SNP (Figure S4). The population segregate for the SNP across principal component 1 (PC1) which explains 27.1% of the variation.
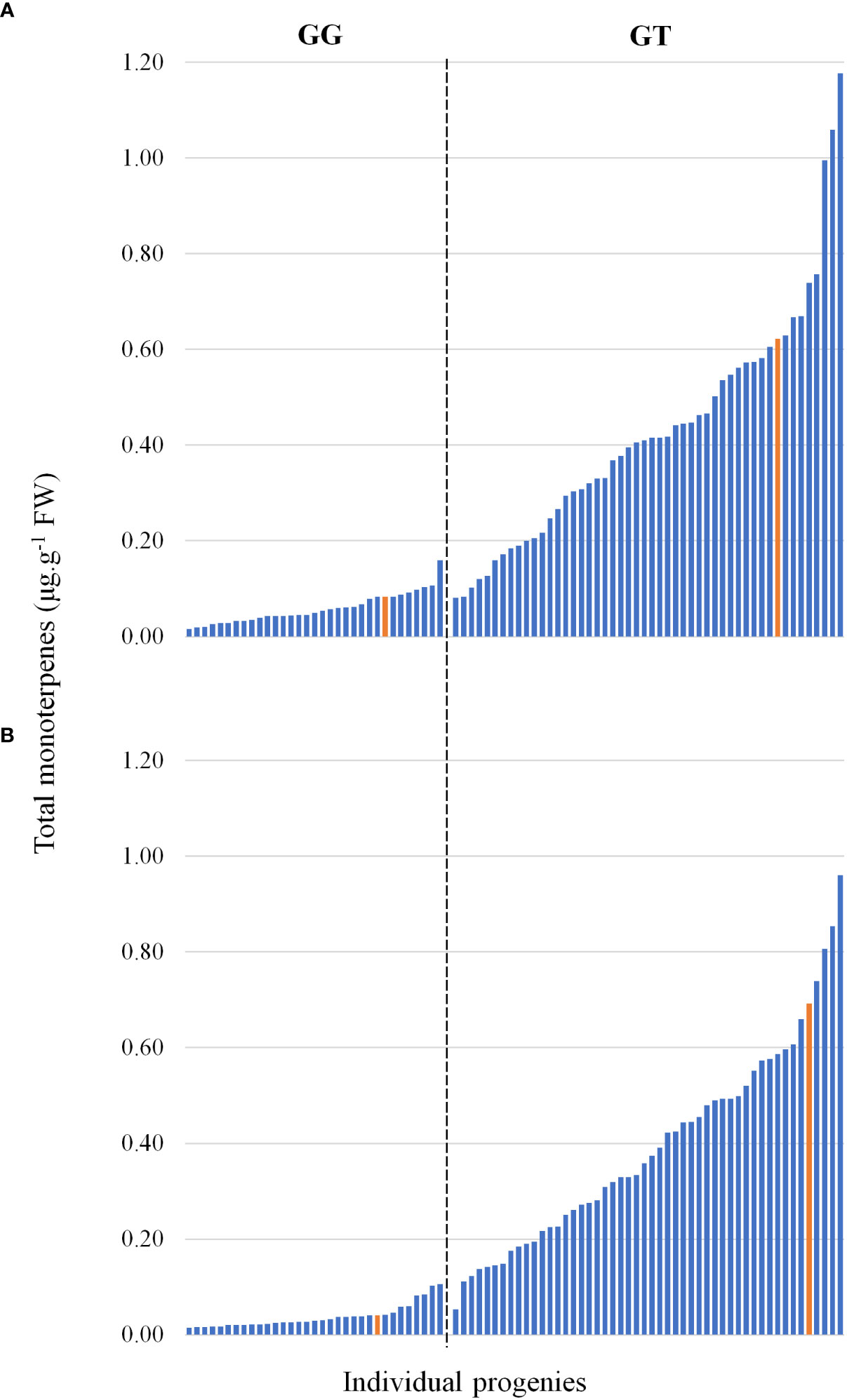
Figure 4 The total monoterpene content in the mapping progeny at (A) pre-veraison and (B) veraison. Each bar represents a member of the progeny, and the parents are shown in yellow. The bars are grouped into absence/presence of the VvDXS1 SNP.
VvTer and VvGer gene expression was quantified during the late flower and early berry development in ‘Deckrot’ and G1-7720 (Figure 5). VvGer showed the highest level of expression in young flowers for both parents, while geraniol content only peaked in the following stage. VvTer expression was highest after fruit-set (EL-29), while α-terpineol concentration peaked earlier in the flowering stages. Both VvTer and VvGer expression, as well as α-terpineol and geraniol concentration, d-ecrease towards later berry development and is lowest in green berries (EL-33). Furthermore, α-terpineol and geraniol concentration is significantly higher in G1-7720 than ‘Deckrot’, irrespective of developmental stage or gene expression (Figure 5).
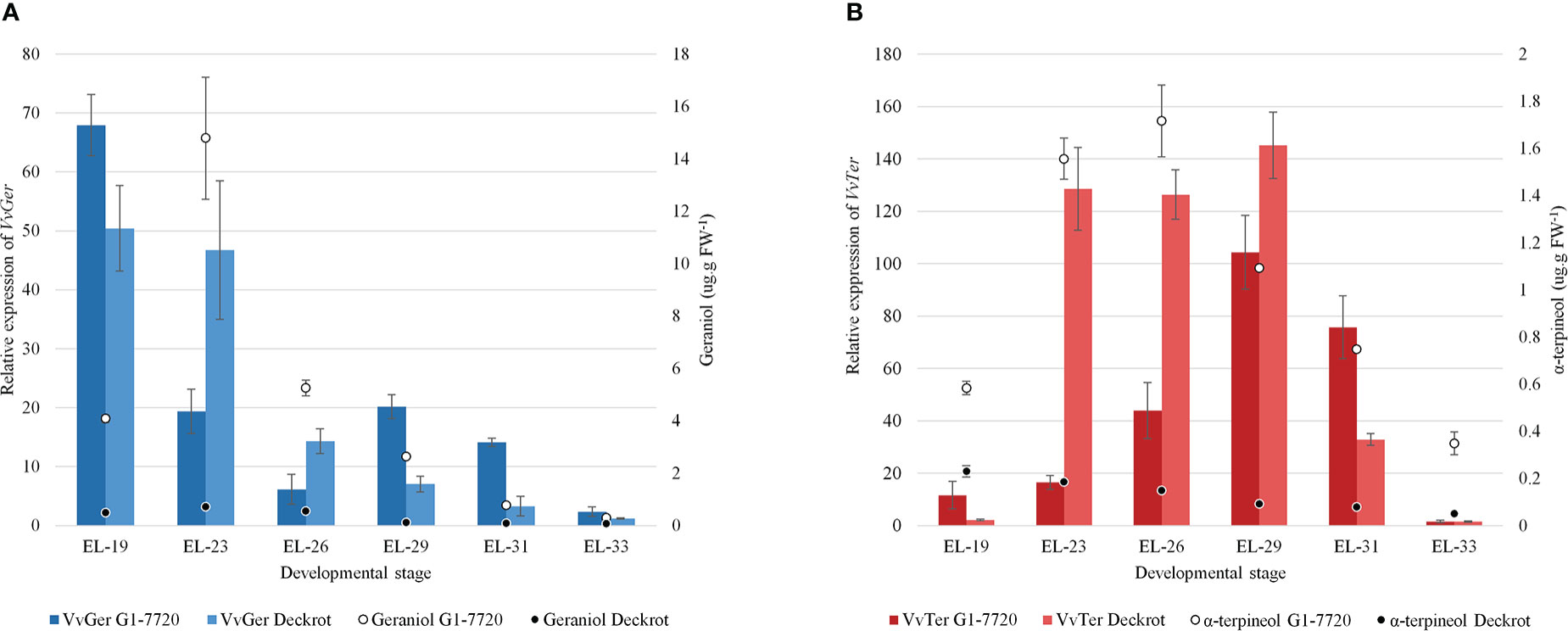
Figure 5 The expression of (A) VvGer and (B) VvTer in the parental cultivars, ‘Deckrot’ and G1-7720, during early berry development. The a-terpineol and geraniol concentrations at each stage is shown (values on right-side y-axis). n = 3; error bars represent standard error.
The expression of VvTer and VvGer was measured in a subset of the progeny to determine whether gene expression correlates with cyclic monoterpene or geraniol accumulation, respectively. The expression was measured in progenies with the VvDXS1 SNP (GT) and without the VvDXS1 SNP (GG). Linear regression analysis showed weak correlations between VvTer expression and cyclic monoterpene accumulation and between VvGer expression and geraniol accumulation (Figure S5). However, in progeny possessing the VvDXS1 SNP, VvTer gene expression and cyclic monoterpene content showed relatively strong correlation (R = 0.78, p < 0.0001), indicating a positive linear relationship between VvTer expression and cyclic monoterpene accumulation (Figure 6).
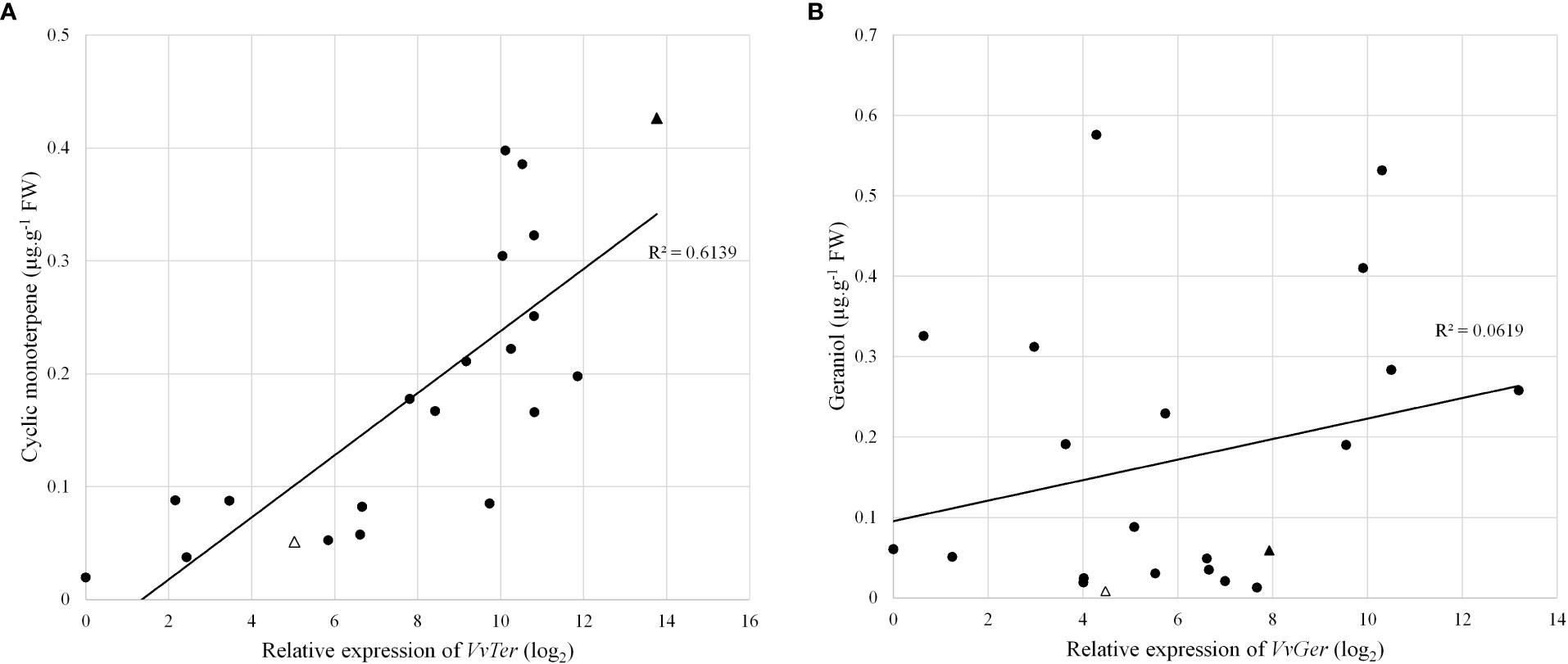
Figure 6 (A) VvTer expression compared to cyclic monoterpene content and (B) VvGer expression compared to geraniol content. Gene expression analysis and volatile quantification was done using pre-veraison berry skins. Circles represent progeny with the VvDXS1 SNP (GT), triangles represent the parents 'Deckrot' (empty) and G1-7720 (filled).
Multiple cDNA clones of VvTer and VvGer from both ‘Deckrot’ and G1-7720 were sequenced in order to identify potential sequence variants. Using single primer pairs for each gene several unique expressed VvTer and VvGer gene copies were isolated from each parent cultivar (Table 2 and Figure S6). The nucleotide sequences of all gene copies were compared to functionally characterised terpene synthases in Vitis vinifera via molecular phylogeny (Table S8 and Figure 7). Regardless of cultivar, all VvTer sequences fall within the TPS-b family cluster while VvGer falls within the TPS-g subfamily. Interestingly, ‘Deckrot’ VvTer genes (DRTer) and G1-7720 VvTer genes (G1Ter) form two distinct clusters. Furthermore, VvGer gene copies from ‘Deckrot’ and G1-7720 share sequence similarity with functionally characterised V. vinifera geraniol synthases (VvGer, VvGwGer and VvPNGer), respectively. DRTer gene copies are similar to functionally characterised α-terpineol synthases (VvTer1 and VvTer2), while G1Ter gene copies share sequence similarity with an (E)-β-ocimene/myrcene synthase (VvCSbOciM). VvTer1, VvTer2 and VvCSbOciM are described as TPS39 genes with VvTer1 and VvTer2 being isolated from ‘Gewürtztraminer’, while VvCSbOciM was isolated from Cabernet Sauvignon (Martin and Bohlmann, 2004; Martin et al., 2010).
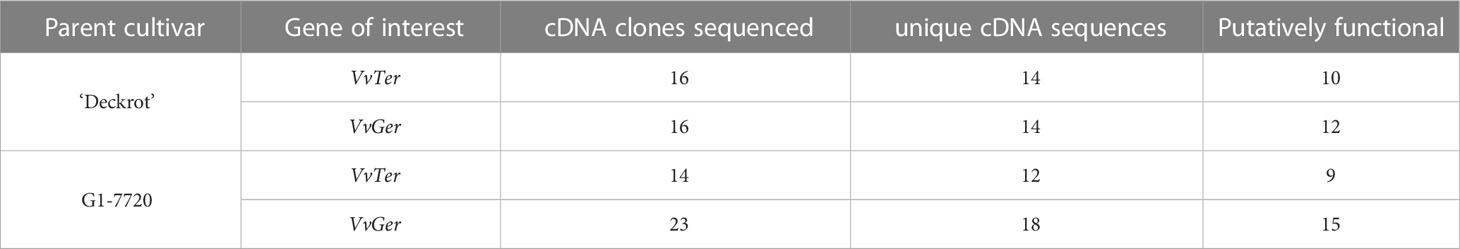
Table 2 The number of unique VvTer and VvGer cDNA clones that were identified for ‘Deckrot’ and G1-7720.
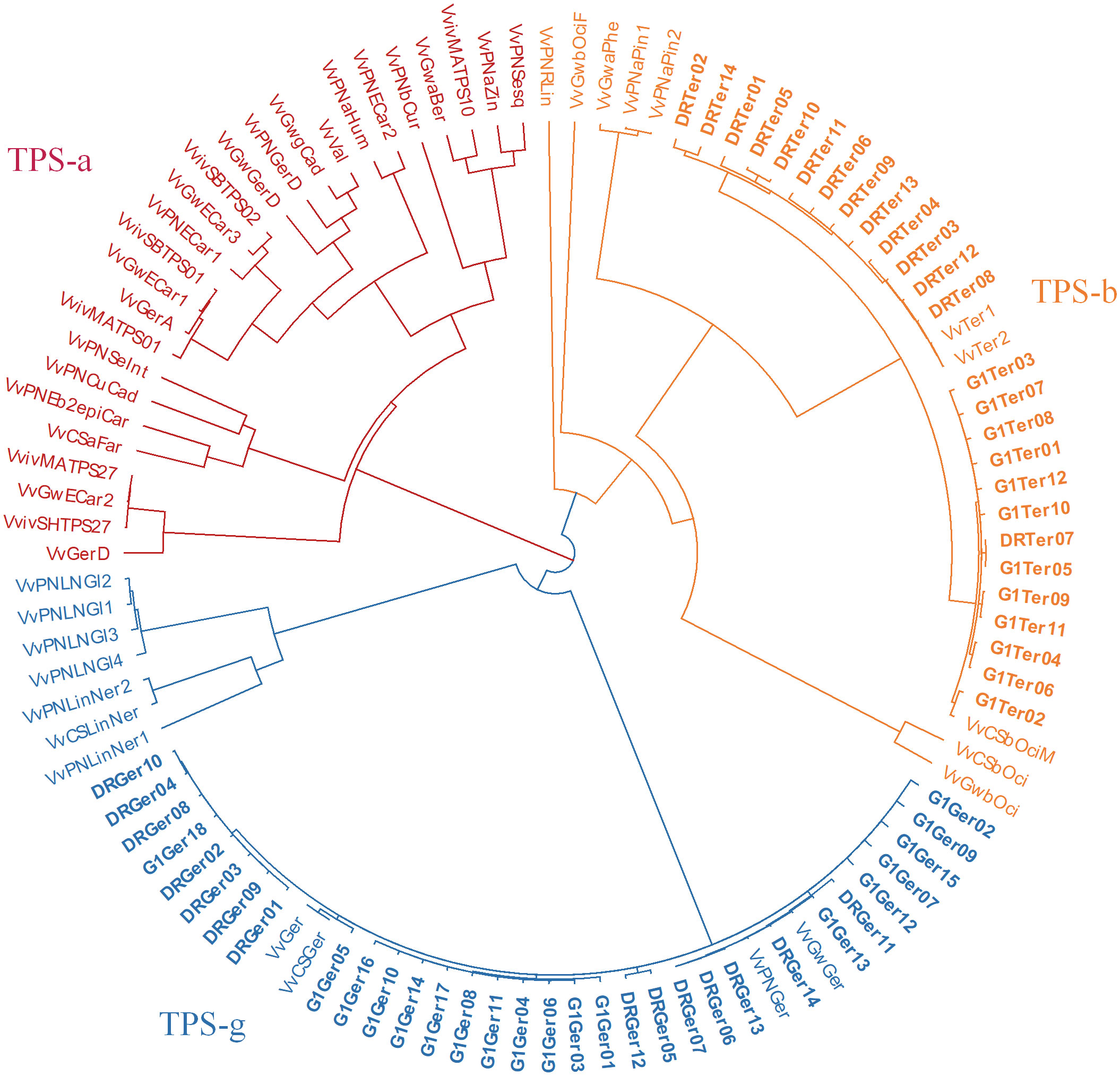
Figure 7 Phylogenetic tree (nucleotide sequences of coding regions) of ‘Deckrot’ and G1-7720’s VvTer and VvGer gene copies compared to functionally characterised V. vinifera terpene synthase genes. Clusters are coloured according to which TPS-subfamily the genes fall under. “G1” and “DR” indicate cultivar origin of isolated gene (G1-7720 and ‘Deckrot’, respectively).
The relative copy numbers of VvTer and VvGer were determined for 82 progenies of the mapping population, as well as the parents (Table S9). Both VvTer and VvGer copy numbers were higher in G1-7720 than in ‘Deckrot’; VvGer had a 10:19 ratio for ‘Deckrot’:G1-7720 while VvTer had a 5:12 ratio. Gene copy number in the population were expressed as relative to ‘Deckrot’ gene copies, such that VvTer copy number ranges between 0.7-2.8 relative copies, while VvGer ranges between 1 – 2.1 relative copies. Additionally, a few individuals from the progeny were outliers. VvGer had the most outliers (6 progenies) with copy numbers up to 12.8 times higher than that observed for ‘Deckrot’.
Spearman’s rank correlation (ρ) analysis found VvTer copy number possessed a strong correlation with VvTer expression (ρ = 0.748), but VvGer copy number and expression had weak correlation (ρ = 0.189). The geraniol and total cyclic monoterpene content was binned into three categories: low, moderate and high (Figure 8A). VvGer copy number shows no discernible pattern between geraniol content and copy number, while total cyclic monoterpene content seems to show a positive correlation with VvTer copy number. The correlation between gene copy number and several monoterpenes was investigated in the progeny (Figure 8B). There was no correlation between VvGer copy number and any monoterpenes in the progeny, regardless of the presence of the VvDXS1 SNP. However, VvTer showed different degrees of correlation with cyclic monoterpenes depending on the presence of the VvDXS1 SNP. Progeny which did not have the G>T VvDXS1 SNP (GG) showed weak correlation (-0.1< ρ < 0.1) between VvTer copy number and cyclic monoterpenes, while in progeny with the VvDXS1 SNP (GT) there was a strong positive correlation (ρ > 0.6).
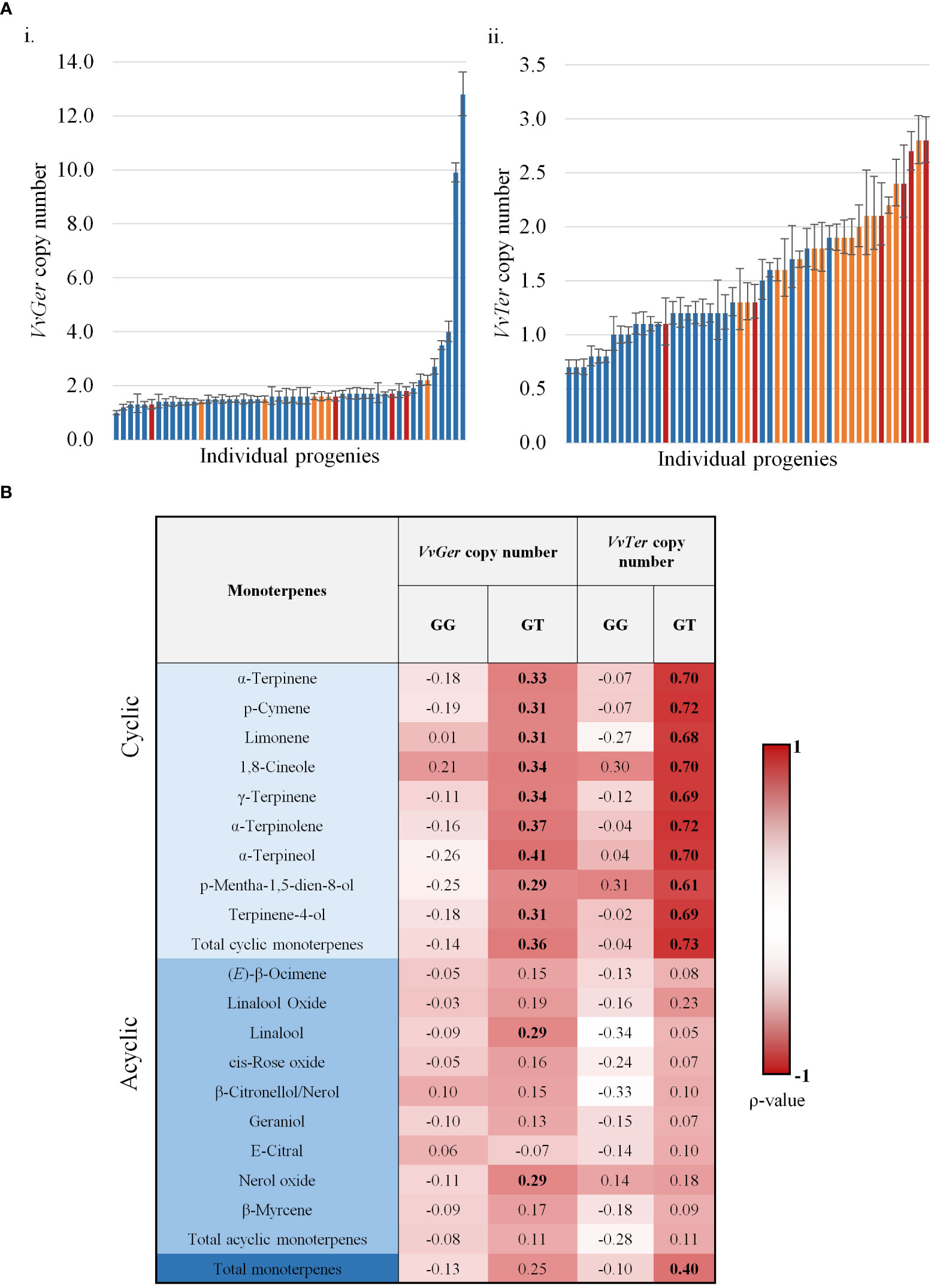
Figure 8 (A) The copy number of VvGer (i) and VvTer (ii) in progenies with the VvDXS1 SNP. Progenies were binned for low (blue), moderate (yellow) and high (red) geraniol (i) or total cyclic monoterpene (ii) content. n = 3; error bars represent standard error. (B) Spearman’s correlation between relative gene copy number (VvGer and VvTer) and monoterpenes. Correlation between gene copy number and monoterpenes were analysed separately for progeny with the VvDXS1 SNP (GT) and without the SNP (GG). Correlation values in boldface were statistically significant; p < 0.001.
Analysis of the draft diploid genome of Cabernet Sauvignon identified 29 putative α-terpineol synthase (VvCSTer) loci, and nine putative geraniol synthase (VvCSGer) loci (Table S10). The majority of α-terpineol synthases were localised to chromosome 13 while geraniol synthases predominantly localised to chromosome 12, and the remaining genes were unplaced (Table S10). The nucleotide coding sequences of VvCSTer genes had a pairwise identity of 90.8% and VvCSGer genes shared 98.2% pairwise identity. Furthermore, the loci appeared in clusters on the chromosomes, with adjacent genes in clusters being less than 100 kb apart (Figure 9). Haplotype 2 of chromosome 13 has three separate VvCSTer clusters which are approximately 1 Mb apart. The high similarity as well as proximity of these genes indicate an extensive level of tandem gene duplication, particularly for VvCSTer genes.
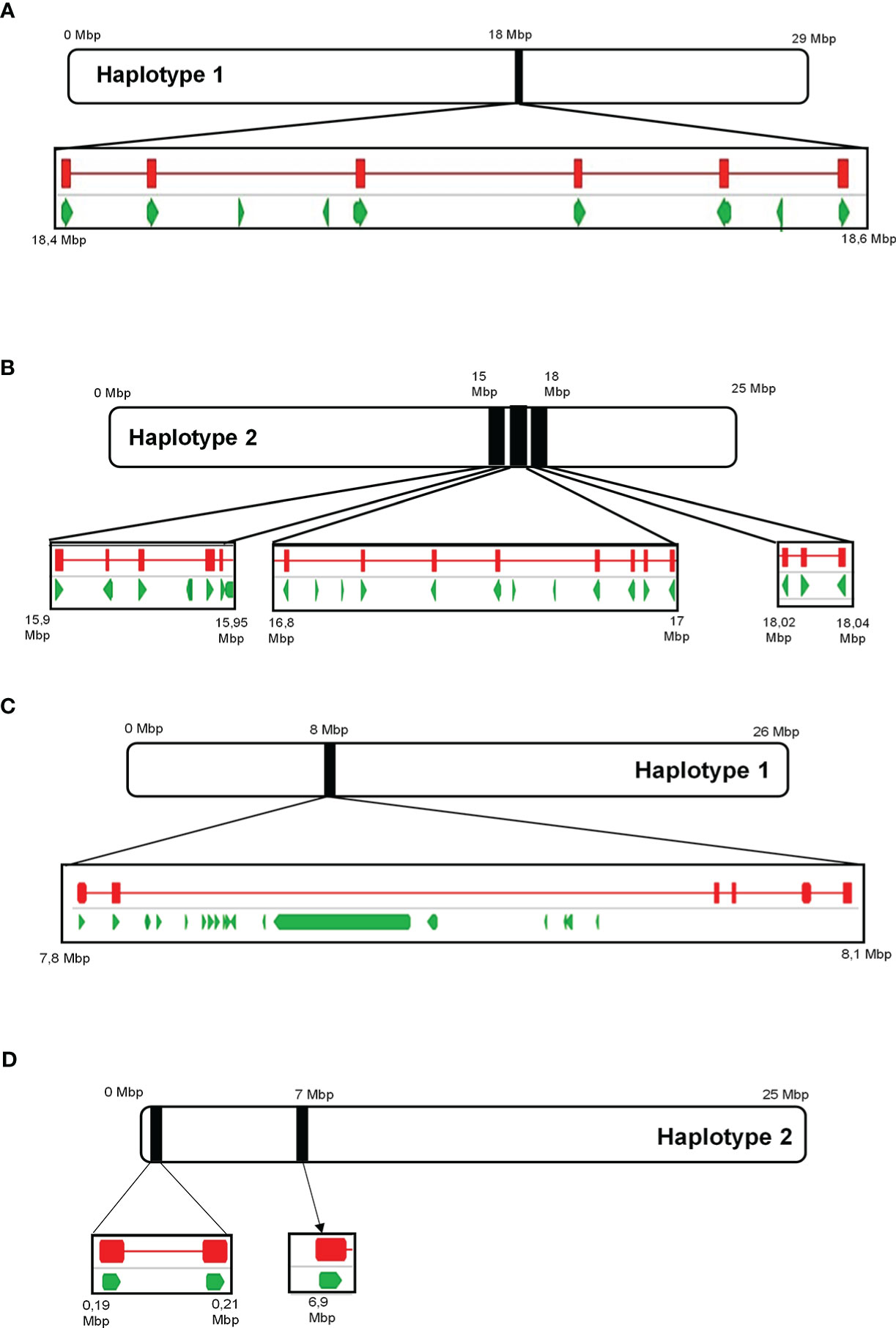
Figure 9 The position of predicted ‘Cabernet Sauvignon’ VvTer and VvGer genes on chromosome 13 (A, B) and chromosome 12 (C, D), respectively. Black blocks indicate the position of the predicted VvTer or VvGer clusters on the chromosomes, and squares underneath provide a magnified view of the region. Green arrows represent predicted genes as annotated in Massonnet et al. (2020), while red lines indicate the position of BLAST hits for VvTer or VvGer.
11 of the 29 predicted Cabernet Sauvignon α-terpineol synthase loci encode for putatively functional enzymes, while 4 of the 9 geraniol synthase loci encode for putatively functional enzymes. A phylogenetic tree (Figure S7) was constructed to compare the putatively functional Cabernet Sauvignon α-terpineol and geraniol synthases with functionally characterised Vitis vinifera terpene synthases. All putatively functional VvCSGer proteins fall within the TPS-g cluster while VvCSTer proteins fall within the TPS-b cluster.
Studies analysing volatile accumulation show mixed results with regards to the accumulation pattern of different VOC classes (Ferrandino et al., 2012; Vilanova et al., 2012; JI et al., 2021; Liu et al., 2022), and it is therefore challenging to compare the metabolite data observed here with previous studies. Importantly, however, the genetic control of VOC accumulation was evident through the observed segregation of compound accumulation in the population under study, and the subsequent associated QTLs that were identified. The significant QTL on chromosome 5 associated with total monoterpene content, houses the well described VvDXS1 gene (Doligez et al., 2006; Battilana et al., 2009; Emanuelli et al., 2010). In the population under investigation here we show that the SNP contributing to high terpene level and a “muscat like aroma” is derived from G1-7720, the aromatic parent. Importantly this SNP was found to be a prerequisite for high monoterpene levels in the progeny, but not an absolute indicator (Figures 4, S4). This is likely due to the multigenic nature of terpene formation, further highlighted by the various other QTLs identified in this study and others (Doligez et al., 2006; Battilana et al., 2009; O’Reilly-Wapstra et al., 2011; Yu et al., 2017; Reichardt et al., 2020; Barbey et al., 2021). Importantly, accumulation of the plastid derived C13-norisoprenoids also associated with this SNP, while the cytosolic derived sesquiterpenes did not. This suggests that, at least in the case of this grapevine population, there is limited isoprenoid crosstalk from plastid to cytosol (Gutensohn et al., 2013).
While the VvDXS1 SNP can be considered a master regulator of monoterpene accumulation, candidate genes which co-localise with QTLs for geraniol, cyclic monoterpenes and sesquiterpenes were also identified in this study. Geraniol shows a QTL on chromosome 12 which co-localised with a cluster of eight terpene synthase genes. VvTPS52, one of the eight VvTPSs in this region, was predicted to function as a geraniol synthase (VvGer) and has indeed been previously functionally characterised as a geraniol synthase in various other cultivars (Martin et al., 2010). Several cyclic monoterpenes showed overlapping associated QTLs on chromosome 13. These QTLs co-localise with a cluster of terpene synthase genes containing putative α-terpineol synthases. VvTPS39 was proposed as candidate gene for this QTL as it was the closest related gene that was not a predicted pseudogene and has been characterised as an α-terpineol synthase (VvTer) via in vitro enzyme assay in ‘Gewurztraminer’ (Martin and Bohlmann, 2004). Interestingly, the authors found that VvTPS39 produced α-terpineol, 1,8-cineole and β-pinene as major products and various minor product including α-thujene, α-pinene, myrcene, sabinene, β-pinene, limonene, and terpinolene. The multi-product nature of terpene synthases may explain why this study found multiple cyclic monoterpenes mapping to the same region on chromosome 13. It is likely that VvTer (and possibly some of the other VvTPSs that clustered in the same region) produce cyclic monoterpenes in varying ratios, with α-terpineol being the major product.
The sesquiterpenes α-cubebene and trans-caryophyllene, associated with a large QTL (approximately 11.65 Mbp) on chrosomome 19. VvTPS29 was proposed as a candidate gene and this was further supported by molecular phylogenetic evidence. VvTPS29 showed to be closely related to functionally characterised trans-caryophyllene synthases and a germacrene D synthase (Figure 3). Additionally, an isocaryophyllene/β-cadinene synthase (VvShirazTPS-Y2) which shares 100% similarity with VvTPS29 has been characterised in ‘Shiraz’ (Dueholm et al., 2019).
Non-terpene VOCs were also found to associate with QTLs in this study. C6-compounds or green leaf volatiles (GLVs) were the most abundant VOCs present in the progeny at all developmental stages, as has been reported in other studies (Kalua and Boss, 2009; Vilanova et al., 2012). A QTL associated with trans-2-hexenal and total aldehydes was identified on chromosome 2 and co-localised with a Stearoyl-[acyl-carrier-protein] 9-desaturase 6 (SAD). SAD catalyses the conversion of stearoyl-ACP to oleoyl-ACP which serves as a precursor for the fatty acids α-linolenic acid and linoleic acid. α-linolenic acid and linoleic acid in turn can be broken down to form green leaf volatiles which are synthesised via the lipoxygenase (LOX) pathway (Matsui and Engelberth, 2022). GLVs are rapidly released upon plant wounding which is thought to play a role in plant signalling and defence (Matsui, 2006; Bouwmeester et al., 2019; Matsui and Engelberth, 2022). C6-aldehydes and -alcohols contribute to the undesirable “green aroma” of wine (Dunlevy et al., 2009), and therefore understanding their metabolism may contribute to improved wine aroma.
A QTL associated with cis-rose oxide, E-citral and nerol oxide was identified in the same region on chromosome 17. A cluster of five short chain dehydrogenase/reductases which co-localised with the QTL were selected as candidate genes. All three of the compounds are derived from geraniol, however importantly geraniol did not associate with this QTL. Geraniol dehydrogenases which catalyse the formation of citral and citronellol have been identified in several plant species but not in grapevine (Iijima et al., 2006; Sato-Masumoto and Ito, 2014; Xu et al., 2017). Lin et al. (2019) summarises a proposed pathway for citronellol, nerol and cis-rose oxide in grapevine but to date none of the proposed enzymes in the pathway have been identified and characterised. Further characterisation of the VvSDRs identified in this QTL may therefore prove promising in elucidating the biosynthesis of these compounds.
In an attempt to isolate the monoterpene synthase genes underlying the QTLs associated with geraniol and cyclic monoterpene accumulation it was discovered that there were more gene copies than represented in the reference genome. Furthermore, as the genes isolated represent cDNA clones, the number of gene copies shown in Table 2 is not exhaustive representation of all the possible VvTer and VvGer copies. Importantly, phylogenetic analysis of the sequenced ‘Deckrot’ and G1-7720 VvTer and VvGer gene copies found strong cultivar specific clustering, indicating this gene duplication has occurred post cultivar diversification.
Due to the somewhat surprising nature of observed VvTPS gene duplication in the cultivars in this study an in silico analysis of VvTer and VvGer in the diploid ‘Cabernet Sauvignon’ genome was performed. This analysis confirmed massive gene duplication and revealed that both genes occur in tandem duplications on chromosome 13 and chromosome 12, respectively. VvTer had the most extensive duplications with 29 copies on the ‘Cabernet Sauvignon’ genome. Analysis of the draft diploid genome further indicated that VvTer genes were unequally distributed between the two haplotypes of chromosome 13. VvGer genes also occurred unequally distributed on the haplotypes of chromosome 12. The distribution VvTer and VvGer loci were uneven on the two haplotypes of chromosomes 12 and 13, revealing a high level of hemizygosity. Furthermore, due to the high sequence similarity between these genes, it is difficult to predict which loci are allelic and which are hemizygous.
Further study is necessary to ascertain the function of the VvTPS candidate genes in this mapping population. High levels of gene duplication contribute to the diverse functions of TPSs through neofunctionalisation and sub-functionalisation (Tholl, 2015). Furthermore, the function of TPSs can be changed by small amino acid substitutions which alter the active site conformation and further contributes to the diverse functions of TPSs (Degenhardt et al., 2009; Karunanithi and Zerbe, 2019) and this has also been shown by the presence of cultivar-specific TPSs in grapevine (Martin et al., 2010; Drew et al., 2016; Smit et al., 2019) therefore it remains a challenge to predict the function of VvTPSs based on sequence similarity to functionally characterised genes alone.
The pattern of VvGer and VvTer expression was similar in both parent cultivars during berry development, but the expression patterns did not correspond with the accumulation of α-terpineol or geraniol. Previous studies have shown that VvTPS developmental expression patterns do not always match the accumulation patterns of their associated metabolites (Martin et al., 2012; Matarese et al., 2013; Matarese et al., 2014). Furthermore, only the volatile fractions of geraniol and α-terpineol were quantified in this study, while monoterpenes are also present in non-volatile glycosylated forms in grapevine (Dunlevy et al., 2009; Hjelmeland and Ebeler, 2015).
However, the expression of VvTer across the mapping population correlated with several cyclic monoterpenes, specifically in progeny containing the VvDXS1 SNP. The high similarity between VvTer paralogs potentially skews qPCR data as the primers have the potential to bind to more than one VvTer gene locus. This fact together with the VvTPS gene duplication data described above led to investigation of the VvTer gene copy number. Importantly, the parent cultivars under investigation showed unequal VvTer gene copy number, with G1-7720 showing 2.4-fold greater number of copies of VvTer than ‘Deckrot’. Relative VvTer gene copy number showed a positive correlation with the accumulation of several individual cyclic monoterpenes as well as total cyclic monoterpenes. The correlation of VvTer gene expression with multiple monoterpenes is in agreement with the fact that this gene cluster co-localises with the QTL for cyclic monoterpenes. Furthermore, VvTer is potentially a multi-product forming enzyme similar to other terpene synthases (Martin et al., 2010; Karunanithi and Zerbe, 2019), and potentially forms several structurally related monoterpene compounds, such as cyclic monoterpenes.
Hypotheses for the correlation of VvTer copy number and cyclic monoterpene accumulation are somewhat more speculative and are detailed in the following sections. The first hypothesis is that VvTer copy number may cause a dosage effect, i.e. the more copies of the gene, the more transcripts produced, and the more enzyme translated. This is supported by the strong correlation between VvTer expression and copy number. However, dosage does not explain why some low copy number individuals have high levels of cyclic monoterpenes and vice versa (Figure 8).
Another possible explanation is that a “hyper-functional” gene/genetic element co-localises with the high copy number allele (Figure 10). “Hyper-functional” is defined as any sequence variation which significantly improves cyclic monoterpene synthase enzyme activity, resulting in increased cyclic monoterpene biosynthesis. In Figure 10 we assume that ‘‘Deckrot’ has 5 copies of VvTer (3 copies on allele A and 2 copies on allele B) and G1-7720 has 12 copies (8 copies on allele A and 4 copies on allele B) as this fulfils the 5:12 (‘Deckrot’:G1-7720) ratio of VvTer copy number in the parents. It is important to note that while the exact gene copy numbers cannot be determined, sequencing of cDNA clones suggests a relatively high VvTer copy number. Furthermore, the fact that the progeny typically presented a higher copy number than ‘Deckrot’, and a lower copy number than G1-7720, indicate that both alleles for G1-7720 have a higher copy number than the ‘Deckrot’ alleles.
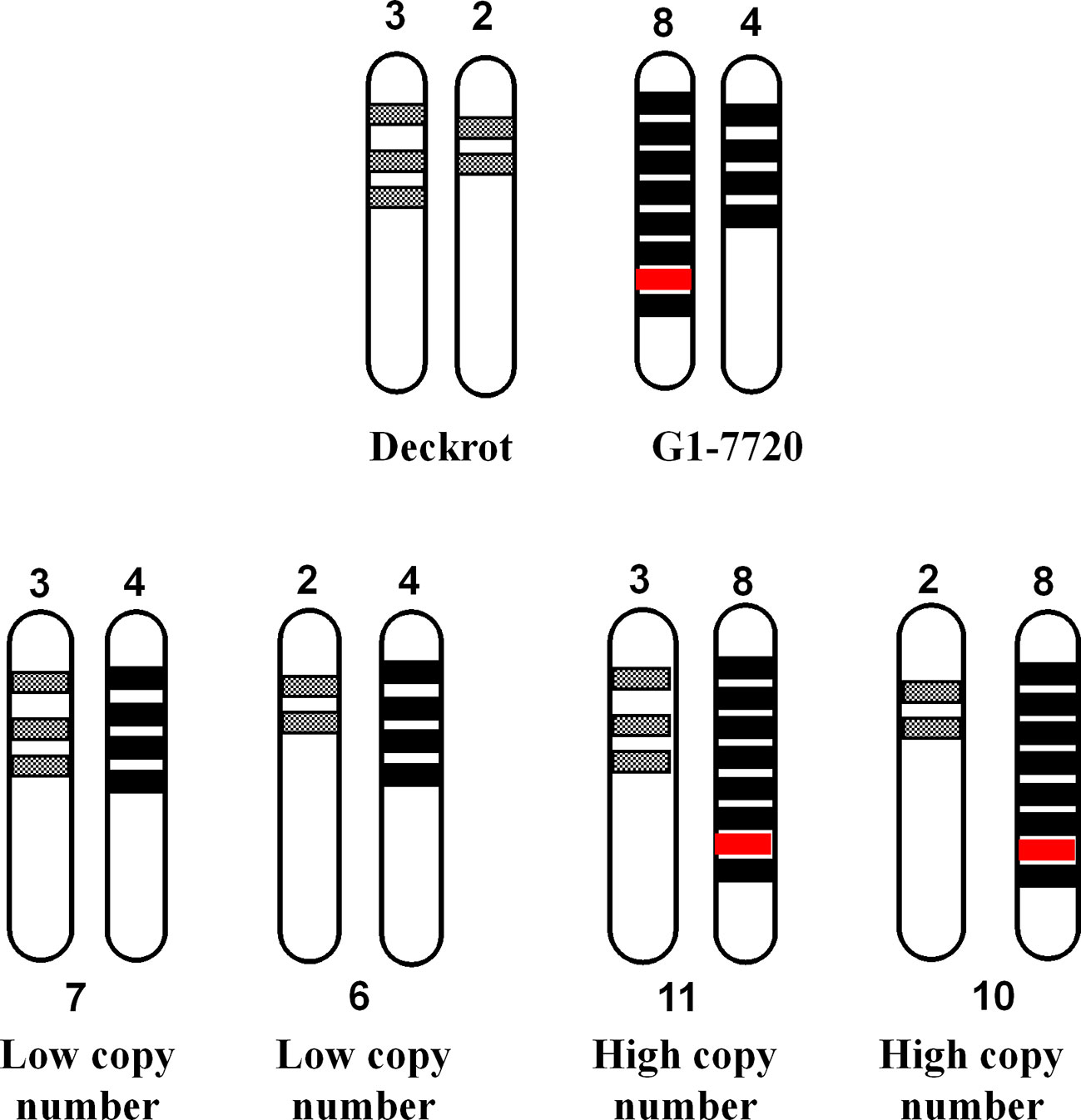
Figure 10 A diagram to explain the correlation between VvTer copy number and cyclic monoterpene accumulation. VvTer genes are represented by shaded blocks and the “hyper-functional” gene is shown in red. The “hyper-functional” gene falls on the parent allele with the highest copy number and is the causal factor for higher cyclic monoterpene content in this parent. The progeny from ‘Deckrot’ x G1-7720 have an equal chance having one of four possible VvTer copies: 6, 7, 10 or 11. Progenies with 6 or 7 copies would be considered “low copy number” individuals and would not inherit the “hyper-functional” gene resulting in lower/normal accumulation of cyclic monoterpenes. Progenies with 10 or 11 copies would be considered “high copy number individuals” and will inherit the “hyper-functional” gene resulting in higher cyclic monoterpene accumulation.
In this hypothesis the progeny has an equal chance of inheriting one allele from each parent in four potential different combinations. If a hyper-functional gene/genetic element co-localizes with the high copy number allele, all individuals that inherited the high copy number allele will have high cyclic monoterpene levels. Therefore, individuals with a high copy number do not have higher levels of cyclic terpenes due to additive effects but due to the co-segregation of high copy number and a “gain-of-function” gene/genetic element. The high level of hemizygosity of VvTer genes in the ‘Cabernet Sauvignon’ genome, as well as the large discrepancy between VvTer copy numbers of ‘Deckrot’ and G1-7720 (G1-7720 has 2.4 times as many VvTer copies as ‘Deckrot’) support this theory. While the exact distribution of VvTer loci in the parent cultivars are unknown, it is safe to assume that both also have high hemizygosity which results in them having either a “high copy number” or “low copy number” allele.
This hypothesis also explains why some individuals have a low copy number but high cyclic monoterpene level. If the hyper-functional element is transferred to a “low copy number” allele during a recombination event, some low copy number individuals will have high cyclic monoterpene levels. Recombination events at a specific locus are rare and would thus only affect a few individuals in the population (Choi, 2017). Furthermore, this hypothesis explains why there is no correlation between VvGer copy number and geraniol levels. The difference in VvGer copy numbers between G1-7720 and ‘Deckrot’ is less prominent, therefore making differentiation between “high copy number” and “low copy number” alleles difficult. The “hyper-functional” element of VvGer thus co-segregates with high copy number similarly to VvTer, but the difference between high and low copy number is too small to make any significant distinction and no correlation is observed.
During berry development VvGer and VvTer gene expression patterns did not align with the accumulation patterns of geraniol and α-terpineol, respectively, indicating that terpene synthase gene expression cannot solely be used to infer gene function. Furthermore, the extensively duplicated nature of the TPS gene family complicates gene expression analysis as well as identification of sequence variants leading to enzyme activity modulation. VvGer and VvTer genes are present in tandemly duplicated gene clusters and these clusters are unequally distributed across chromosome haplotypes indicating extreme levels of hemizygosity for these genes. Furthermore, VvTer is highly duplicated in grapevine genomes and VvTer copy number correlated with cyclic monoterpene accumulation.
Extensive gene duplication is a common feature of TPS-gene families in several plant species and results in the presence of multiple highly similar gene paralogs in these genomes. While the influence of small polymorphisms, such as SNPs and indels, on TPS functions and terpene accumulation are commonly researched, very few studies investigate the influence of this gene copy number variation on terpene accumulation. The knowledge gained from this study further contributes to our understanding of terpene metabolism in grapevine and the potential impact of TPS copy number variation on grapevine terpene accumulation.
The original contributions presented in the study are included in the article/Supplementary materials. Further inquiries can be directed to the corresponding author.
RB designed and performed experiments, analyzed data, and was the primary author of the manuscript. JV performed experiments, analyzed data and co-wrote the manuscript. DN performed experiments. PB designed and performed experiments. JL conceptualized the research, designed experiments, analyzed data, and co-wrote the manuscript. All authors contributed to the article and approved the submitted version.
This work is based on the research supported by grants from the South African Table Grape Industry (SATI), Winetech and The National Research Foundation of South Africa. Grant numbers: S005411, S006681 and 129344, respectively.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1112214/full#supplementary-material
Addinsoft. (2023). XLSTAT statistical and data analysis solution. (New York, USA). https://www.xlstat.com/en.
Barbey, C. R., Hogshead, M. H., Harrison, B., Schwartz, A. E., Verma, S., Oh, Y., et al. (2021). Genetic analysis of methyl anthranilate, mesifurane, linalool, and other flavor compounds in cultivated strawberry (Fragaria × ananassa). Front. Plant Sci. 12, 615749. doi: 10.3389/fpls.2021.615749
Battilana, J., Costantini, L., Emanuelli, F., Sevini, F., Segala, C., Moser, S., et al. (2009). The 1-deoxy-d-xylulose 5-phosphate synthase gene co-localizes with a major QTL affecting monoterpene content in grapevine. Theor. Appl. Genet. 118 (4), 653–669. doi: 10.1007/s00122-008-0927-8
Battilana, J., Emanuelli, F., Gambino, G., Gribaudo, I., Gasperi, F., Boss, P. K., et al. (2011). Functional effect of grapevine 1-deoxy-D-xylulose 5-phosphate synthase substitution K284N on Muscat flavour formation. J. Exp. Bot. 62 (15), 5497–5508. doi: 10.1093/jxb/err231
Bhattacharya, S., Holowka, T., Orner, E. P., Fries, B. C. (2019). Gene duplication associated with increased fluconazole tolerance in candida auris cells of advanced generational age. Sci. Rep. 9 (1), 5052. doi: 10.1038/s41598-019-41513-6
Bosman, R. N., Lashbrooke, J. G. (2023). Grapevine mono- and sesquiterpenes: Genetics, metabolism, and ecophysiology. Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1111392
Bouwmeester, H., Schuurink, R. C., Bleeker, P. M., Schiestl, F. (2019). The role of volatiles in plant communication. Plant J. 100 (5), 892–907. doi: 10.1111/tpj.14496
Canaguier, A., Grimplet, J., Di Gaspero, G., Scalabrin, S., Duchêne, E., Choisne, N., et al. (2017). A new version of the grapevine reference genome assembly (12X.v2) and of its annotation (VCost.v3). Genomics Data 14, 56–62. doi: 10.1016/j.gdata.2017.09.002
Chen, F., Tholl, D., Bohlmann, J., Pichersky, E. (2011). The family of terpene synthases in plants: A mid-size family of genes for specialized metabolism that is highly diversified throughout the kingdom. Plant J. 66 (1), 212–229. doi: 10.1111/j.1365-313X.2011.04520.x
Choi, K. (2017). Advances towards controlling meiotic recombination for plant breeding. Mol. Cells 40 (11), 814. doi: 10.14348/molcells.2017.0171
Davis, E. M., Croteau, R. (2000). Cyclization Enzymes in the Biosynthesis of Monoterpenes, Sesquiterpenes, and Diterpenes. In: Leeper, F. J., Vederas, J. C. (eds) Biosynthesis. Topics in Current Chemistry, vol 209. Berlin, Heidelberg: Springer, 53–95. doi: 10.1007/3-540-48146-X_2
Degenhardt, J., Köllner, T. G., Gershenzon, J. (2009). Monoterpene and sesquiterpene synthases and the origin of terpene skeletal diversity in plants. Phytochemistry 70 (15–16), 1621–1637. doi: 10.1016/j.phytochem.2009.07.030
Doligez, A., Audiot, E., Baumes, R., This, P. (2006). QTLs for muscat flavor and monoterpenic odorant content in grapevine (Vitis vinifera l.). Mol. Breed. 18 (2), 109–125. doi: 10.1007/s11032-006-9016-3
Drew, D. P., Andersen, T. B., Sweetman, C., Møller, B. L., Ford, C., Simonsen, H. T. (2016). Two key polymorphisms in a newly discovered allele of the vitis vinifera TPS24 gene are responsible for the production of the rotundone precursor α-guaiene. J. Exp. Bot. 67 (3), 799–808. doi: 10.1093/jxb/erv491
Dudareva, N., Klempien, A., Muhlemann, J. K., Kaplan, I. (2013). Biosynthesis, function and metabolic engineering of plant volatile organic compounds. New Phytol. 198 (1), 16–32. doi: 10.1111/nph.12145
Dueholm, B., Drew, D. P., Sweetman, C., Simonsen, H. T. (2019). In planta and in silico characterization of five sesquiterpene synthases from vitis vinifera (cv. Shiraz) berries. Planta 249 (1), 59–70. doi: 10.1007/s00425-018-2986-7
Dunlevy, J. D., Kalua, C., Keyzers, R., Boss, P. (2009). The Production of Flavour & Aroma Compounds in Grape Berries. In: Roubelakis-Angelakis, K. A. Ed. Grapevine Molecular Physiology & Biotechnology. (Dordrecht: Springer). doi: 10.1007/978-90-481-2305-6
Emanuelli, F., Battilana, J., Costantini, L., Le Cunff, L., Boursiquot, J. M., This, P., et al. (2010). A candidate gene association study on muscat flavor in grapevine (Vitis vinifera l.). BMC Plant Biol. 10 (1), 1–17. doi: 10.1186/1471-2229-10-241
Emanuelli, FR, Sordo, MA, Lorenzi, SI, Battilana, JU, Grando, M. (2014). Development of user-friendly functional molecular markers for VvDXS gene conferring muscat flavor in grapevine. Mol. Breed. 33 (1), 235–241. doi: 10.1007/s11032-013-9929-6
Ferrandino, A., Carlomagno, A., Baldassarre, S., Schubert, A. (2012). Varietal and pre-fermentative volatiles during ripening of vitis vinifera cv nebbiolo berries from three growing areas. Food Chem. 135 (4), 2340–2349. doi: 10.1016/j.foodchem.2012.06.061
Gutensohn, M., Nagegowda, D. A., Dudareva, N. (2012). Involvement of Compartmentalization in Monoterpene and Sesquiterpene Biosynthesis in Plants. In: Bach, T., Rohmer, M. (eds) Isoprenoid Synthesis in Plants and Microorganisms. New York, NY: Springer. doi: 10.1007/978-1-4614-4063-5
Hjelmeland, A. K., Ebeler, S. E. (2015). Glycosidically bound volatile aroma compounds in grapes and wine: A review. Am. J. Enology Viticulture 66 (1), 1–11. doi: 10.5344/ajev.2014.14104
Iijima, Y., Wang, G., Fridman, E., Pichersky, E. (2006). Analysis of the enzymatic formation of citral in the glands of sweet basil. Arch. Biochem. Biophys. 448 (1–2), 141–149. doi: 10.1016/j.abb.2005.07.026
Jaillon, O., Aury, J. M., Noel, B., Policriti, A., Clepet, C., Casagrande, A., et al. (2007). The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449 (7161), 463–467. doi: 10.1038/nature06148
JI, X. H., Wang, B. L., Wang, X. D., Wang, X. L., Liu, F. Z., Wang, H. B. (2021). Differences of aroma development and metabolic pathway gene expression between kyoho and 87-1 grapes. J. Integr. Agric. 20 (6), 1525–1539. doi: 10.1016/S2095-3119(20)63481-5
Jiang, S.-Y., Jin, J., Sarojam, R., Ramachandran, S. (2019). A comprehensive survey on the terpene synthase gene family provides new insight into its evolutionary patterns. Genome Biol Evol 11 (8), 2078–2098. doi: 10.1093/gbe/evz142
Joubert, C., Young, P. R., Eyéghé-Bickong, H. A., Vivier, M. A. (2016). Field-grown grapevine berries use carotenoids and the associated xanthophyll cycles to acclimate to UV exposure differentially in high and low light (Shade) conditions. Front. Plant Sci. 7 (June2016). doi: 10.3389/fpls.2016.00786
Kalua, C. M., Boss, P. K. (2009). Evolution of volatile compounds during the development of Cabernet Sauvignon grapes (Vitis vinifera L.). J Agric Food Chem 57 (9), 3818–3830. doi: 10.1021/jf803471n
Karunanithi, P. S., Zerbe, P. (2019). Terpene synthases as metabolic gatekeepers in the evolution of plant terpenoid chemical diversity. Front. Plant Sci. 10 (October). doi: 10.3389/fpls.2019.01166
Lichman, B. R., Kamileen, M. O., Titchiner, G. R., Saalbach, G., Stevenson, C. E., Lawson, D. M., et al. (2019). Uncoupled activation and cyclization in catmint reductive terpenoid biosynthesis. Nat. Chem. Biol. 15 (1), 71–79. doi: 10.1038/s41589-018-0185-2
Lin, J., Massonnet, M., Cantu, D. (2019). The genetic basis of grape and wine aroma. Horticul. Res. 6 (1), 81. doi: 10.1038/s41438-019-0163-1
Linstrom, P. J., Mallard, W. G. (2001). The NIST chemistry WebBook: A chemical data resource on the Internet. J. Chem. Eng. Data 46 (5), 1059–1063. doi: 10.1021/je000236i
Liu, X., Fan, P., Jiang, J., Gao, Y., Liu, C., Li, S., et al. (2022). Evolution of volatile compounds composition during grape berry development at the germplasm level. Scientia Hortic. 293 (February 2021), 110669. doi: 10.1016/j.scienta.2021.110669
Ma, L., Chung, W. K. (2014). Quantitative analysis of copy number variants based on real-time LightCycler PCR. Curr. Protoc. Hum. Genet. 80, 7.21.1–7.21.8. doi: 10.1002/0471142905.hg0721s80
Martin, D. M., Bohlmann, J. (2004). Identification of vitis vinifera (–)-α-terpineol synthase by in silico screening of full-length cDNA ESTs and functional characterization of recombinant terpene synthase. Phytochemistry 65 (9), 1223–1229. doi: 10.1016/j.phytochem.2004.03.018
Martin, D. M., Fan, P., Jiang, J., Gao, Y., Liu, C., Li, S., et al. (2010). Functional annotation, genome organization and phylogeny of the grapevine (Vitis vinifera) terpene synthase gene family based on genome assembly, FLcDNA cloning, and enzyme assays. BMC Plant Biol. 10 (1), 226. doi: 10.1186/1471-2229-10-226
Martin, D. M., Chiang, A., Lund, S. T., Bohlmann, J. (2012). Biosynthesis of wine aroma: Transcript profiles of hydroxymethylbutenyl diphosphate reductase, geranyl diphosphate synthase, and linalool/nerolidol synthase parallel monoterpenol glycoside accumulation in gewürztraminer grapes. Planta 236 (3), 919–929. doi: 10.1007/s00425-012-1704-0
Massonnet, M., Cochetel, N., Minio, A., Vondras, A. M., Lin, J., Muyle, A., et al. (2020). The genetic basis of sex determination in grapes. Nat. Commun. 11 (1), 2902. doi: 10.1038/s41467-020-16700-z
Matarese, F., Cuzzola, A., Scalabrelli, G., D’Onofrio, C. (2014). Expression of terpene synthase genes associated with the formation of volatiles in different organs of vitis vinifera. Phytochemistry 105, 12–24. doi: 10.1016/j.phytochem.2014.06.007
Matarese, F., Scalabrelli, G., D’Onofrio, C. (2013). Analysis of the expression of terpene synthase genes in relation to aroma content in two aromatic vitis vinifera varieties. Funct. Plant Biol. 40 (6), 552–565. doi: 10.1071/FP12326
Matsui, K. (2006). Green leaf volatiles: Hydroperoxide lyase pathway of oxylipin metabolism. Curr. Opin. Plant Biol. 9 (3), 274–280. doi: 10.1016/j.pbi.2006.03.002
Matsui, K., Engelberth, J. (2022). Green leaf volatiles–the forefront of plant responses against biotic attack. Plant Cell Physiol. 63 (10), 1378–1390. doi: 10.1093/pcp/pcac117
Minio, A., Massonnet, M., Figueroa-Balderas, R., Castro, A., Cantu, D. (2019). Diploid genome assembly of the wine grape carménère. G3 Genes|Genomes|Genet. 9 (5), 1331–1337. doi: 10.1534/g3.119.400030
O’Reilly-Wapstra, J. M., Freeman, J. S., Davies, N. W., Vaillancourt, R. E., Fitzgerald, H., Potts, B. M. (2011). Quantitative trait loci for foliar terpenes in a global eucalypt species. Tree Genet. Genomes 7 (3), 485–498. doi: 10.1007/s11295-010-0350-6
Reichardt, S., Budahn, H., Lamprecht, D., Riewe, D., Ulrich, D., Dunemann, F., et al. (2020). The carrot monoterpene synthase gene cluster on chromosome 4 harbours genes encoding flavour-associated sabinene synthases. Horticul. Res. 7 (1), 190. doi: 10.1038/s41438-020-00412-y
Reid, K. E., et al. (2006). An optimized grapevine RNA isolation procedure and statistical determination of reference genes for real-time RT-PCR during berry development. BMC Plant Biol. 6 (1), 27. doi: 10.1186/1471-2229-6-27
Sato-Masumoto, N., Ito, M. (2014). Two types of alcohol dehydrogenase from perilla can form citral and perillaldehyde. Phytochemistry 104, 12–20. doi: 10.1016/j.phytochem.2014.04.019
Savoi, S., Wong, D. C., Arapitsas, P., Miculan, M., Bucchetti, B., Peterlunger, E., et al. (2016). Transcriptome and metabolite profiling reveals that prolonged drought modulates the phenylpropanoid and terpenoid pathway in white grapes (Vitis vinifera l.). BMC Plant Biol. 16 (1), 67. doi: 10.1186/s12870-016-0760-1
Savoi, S., Wong, D. C., Degu, A., Herrera, J. C., Bucchetti, B., Peterlunger, E., et al. (2017). Multi-omics and integrated network analyses reveal new insights into the systems relationships between metabolites, structural genes, and transcriptional regulators in developing grape berries (Vitis vinifera l.) exposed to water deficit. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.01124
Singh, A., Pandey, G. K. (2015). Primer design using primer express® for SYBR green-based quantitative PCR. In Basu, C. (eds) PCR Primer Design. Methods in Molecular Biology, vol 1275. (New York, NY: Humana Press), 153–164. doi: 10.1007/978-1-4939-2365-6_11
Smit, S. J., Vivier, M. A., Young, P. R. (2019). Linking terpene synthases to sesquiterpene metabolism in grapevine flowers. Front. Plant Sci. 10 (February). doi: 10.3389/fpls.2019.00177
Smit, S. J., Vivier, M. A., Young, P. R. (2020). Comparative (Within species) genomics of the vitis vinifera l. terpene synthase family to explore the impact of genotypic variation using phased diploid genomes. Front. Genet. 11 (May). doi: 10.3389/fgene.2020.00421
Steele, C. L., Crock, J., Bohlmann, J., Croteau, R. (1998). Sesquiterpene synthases from grand fir (Abies grandis). J. Biol. Chem. 273 (4), 2078–2089. doi: 10.1074/jbc.273.4.2078
Tholl, D. (2015). Biosynthesis and biological functions of terpenoids in plants. Adv. Biochem. engineering/biotechnol. 148 (February), 63–106. doi: 10.1007/10_2014_295
Van Ooijen, J. W. (2006). JoinMap ® 4 software for the calculation of genetic linkage maps in experimental populations. Kyazma BV, Wageningen, 33(10.1371).
Van Ooijen, J. W. (2009). MapQTL 6 - software for the mapping of quantitative trait loci in experimental populations of diploid species. Wageningen: Kyazma BV.
Vervalle, J. A., Costantini, L., Lorenzi, S., Pindo, M., Mora, R., Bolognesi, G., et al. (2022). A high-density integrated map for grapevine based on three mapping populations genotyped by the Vitis18K SNP chip. Theor. Appl. Genet, 1–20. doi: 10.1007/s00122-022-04225-6
Vilanova, M., Genisheva, Z., Bescansa, L., Masa, A., Oliveira, J. M. (2012). Changes in free and bound fractions of aroma compounds of four vitis vinifera cultivars at the last ripening stages. Phytochemistry 74, 196–205. doi: 10.1016/j.phytochem.2011.10.004
Voorrips, R. E. (2002). MapChart: Software for the graphical presentation of linkage maps and QTLs. J. Heredity 93 (1), 77–78. doi: 10.1093/jhered/93.1.77
Vranová, E., Coman, D., Gruissem, W. (2013). Network analysis of the MVA and MEP pathways for isoprenoid synthesis. Annu. Rev. Plant Biol. 64 (1), 665–700. doi: 10.1146/annurev-arplant-050312-120116
Xu, H., Bohman, B., Wong, D. C., Rodriguez-Delgado, C., Scaffidi, A., Flematti, G. R., et al. (2017). Complex sexual deception in an orchid is achieved by Co-opting two independent biosynthetic pathways for pollinator attraction. Curr. Biol. 27 (13), 1867–1877.e5. doi: 10.1016/j.cub.2017.05.065
Yu, Y., Bai, J., Chen, C., Plotto, A., Yu, Q., Baldwin, E. A., et al. (2017). Identification of QTLs controlling aroma volatiles using a “Fortune” x “Murcott” (Citrus reticulata) population. BMC Genomics 18 (1), 646. doi: 10.1186/s12864-017-4043-5
Keywords: terpenes, TPS, grapevine, gene copy number, genomics, QTL, VOC
Citation: Bosman RN, Vervalle JA-M, November DL, Burger P and Lashbrooke JG (2023) Grapevine genome analysis demonstrates the role of gene copy number variation in the formation of monoterpenes. Front. Plant Sci. 14:1112214. doi: 10.3389/fpls.2023.1112214
Received: 30 November 2022; Accepted: 28 February 2023;
Published: 16 March 2023.
Edited by:
Alessandra Ferrandino, University of Turin, ItalyReviewed by:
Dongming Ma, Guangzhou University of Chinese Medicine, ChinaCopyright © 2023 Bosman, Vervalle, November, Burger and Lashbrooke. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Justin Graham Lashbrooke, amdsYXNoQHN1bi5hYy56YQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.