- School of Life Advanced Agriculture Bioengineering, Yangtze Normal University, Chongqing, China
The major enzyme encoded by the glucosinolate biosynthetic gene AOP2 is involved in catalyzing the conversion of glucoiberin (GIB) into sinigrin (SIN) in Brassicaceae crops. The AOP2 proteins have previously been identified in several Brassicaceae species, but not in Tumorous stem mustard. As per this research, the five identified members of the AOP2 family from the whole genome of Brassica juncea named BjuAOP2.1-BjuAOP2.5 were found to be evenly distributed on five chromosomes. The subcellular localization results implied that BjuAOP2 proteins were mainly concentrated in the cytoplasm. Phylogenetic analysis of the AOP2 proteins from the sequenced Brassicaceae species in BRAD showed that BjuAOP2 genes were more closely linked to Brassica carinata and Brassica rapa than Arabidopsis. In comparison with other Brassicaceae plants, the BjuAOP2 members were conserved in terms of gene structures, protein sequences, and motifs. The light response and hormone response elements were included in the BjuAOP2 genes’ cis-regulatory elements. The expression pattern of BjuAOP2 genes was influenced by the different stages of development and the type of tissue being examined. The BjuAOP2 proteins were used to perform the heterologous expression experiment. The results showed that all the five BjuAOP2 proteins can catalyze the conversion of GIB to SIN with different catalytic activity. These results provide the basis for further investigation of the functional study of BjuAOP2 in Tumorous stem mustard glucosinolate biosynthesis.
1 Introduction
Brassica juncea var. tumida (tumorous stem mustard, TSM) is an important vegetable crop of the Brassica genus of the Cruciferae family that originated in China. The enlarged fleshy stems of TSM are the raw material for Fuling Preserved Szechuan Pickle and can be eaten fresh. Approximately 130 glucosinolates (GSLs), an important class of secondary metabolites that are sulfur- and nitrogen-rich and found primarily in cruciferous plants, have been identified (Blažević et al., 2019; Nguyen et al., 2020). GSLs undergo hydrolysis reactions under their degradation enzyme, myrosinase, to produce products such as isothiocyanates, thiocyanates, and acetonitrile (Chen et al., 2020). The afore-mentioned products are involved in anti-cancer and anti-bacterial (Augustine and Bisht, 2015; Soundararajan and Kim, 2018) processes, resistance to herbivore feeding, pathogenic microbial infestation (Clay et al., 2009; Li et al., 2014; Chen et al., 2020), and the formation of specific flavors in cruciferous vegetables (Engel et al., 2006). The biosynthetic precursors of GSLs include amino acids such as methionine, tryptophan, phenylalanine, and leucine. These amino acids form side chains of different lengths through an extension pathway. GSLs can be categorized as aromatic, aliphatic, and indole types as per the derivation sources of the side chains (Halkier and Gershenzon, 2006). The highest content in TSM is 2-propenyl glucosinolate (sinigrin, SIN), the major aliphatic GSLs present in B. juncea (Li et al., 2011). TSM with high SIN content had a pronounced spicy taste (Mazumder et al., 2016). Therefore, revealing the synthesis process and molecular regulation mechanism of SIN is crucial to improve the flavor of TSM and optimize the GSL fraction in TSM.
Aliphatic GSL biosynthesis in Arabidopsis thaliana comprises three independent steps: elongation of the precursor amino acid-based side chain, core structure formation, and side chain secondary modification (Grubb and Abel, 2006; Sønderby et al., 2010). The current research indicates the involvement of almost 15 transcription factors and 64 structural genes in GSL biosynthesis (Li et al., 2018; Harun et al., 2020; Mitreiter and Gigolashvili, 2021). GSL-Elong is the key site in the side chain elongation step. Three genes, MAM1 (methylthioalkylmalate synthases 1), MAM2, and MAM3, located in a tandem arrangement at this locus (Kroymann et al., 2001; Kroymann et al., 2003; Textor et al., 2007). The enzyme known as MAM1 and MAM2 catalyzes the condensation reaction of the first three elongation cycles whereas MAM3 mediate the condensation reaction of all six elongation cycles (Textor et al., 2007). The functional polymorphism of the MAM gene results in differences in the structure of the GSL among different ecotypes of Arabidopsis thaliana (Wittstock and Halkier, 2002). The cytochrome P450 homolog CYP79 gene family, CYP83 gene family, and UDP-glucosyltransferase 74B1 (UGT74B1) are key genes in the core structure formation step (Grubb et al., 2004; Mikkelsen et al., 2004). The flavin-containing monooxygenases (GSL-OX), 2-oxoglutarate-dependent dioxygenases (GSL-AOP), and 2-oxoacid-dependent dioxygenase (GSL-OH) sites during modification of the GSL side chain determine the diversity of GSL species (Kliebenstein et al., 2001a; Hansen et al., 2008). GSL-AOP includes two closely linked genes, GSL-ALK and GSL-OHP (Kliebenstein et al., 2001a). GSL-ALK (AOP2) and GSL-OHP (AOP3) genes, respectively, catalyze the transformation of methylsulfinyl GSLs into alkenyl GSLs and hydroxyalkyl GSLs (Kliebenstein et al., 2001b). In addition, the enzymes encoded by the AOP2 gene can catalyze the conversion of beneficial GSL glucoraphanin (GRA) with anticancer activity to gluconapin (GNA) and glucoiberin (GIB) to SIN, thus affecting the taste and flavor of cruciferous vegetables.
The AOP gene family in Arabidopsis thaliana includes AOP1, AOP2, and AOP3 (Kliebenstein et al., 2001a). The AOP1 gene may be the ancestor of the other two genes, but its function needs to be further characterized (Neal et al., 2010). AOP genes greatly vary in their expression patterns in various Arabidopsis thaliana ecotypes and under different culture conditions. The AOP2 is expressed in Ler (Landsberg), but AOP3 is absent, while Cvi (Cape Verde Islands), depicts the expression of AOP3, but AOP2 is absent (Kliebenstein et al., 2001a). In addition, an ecotype in which both genes are expressed simultaneously has not been found yet. Studies in Arabidopsis thaliana and Brassica napus suggested that inactivation of the AOP2 gene promotes the accumulation of large amounts of GRA in plants to produce higher amounts of sulforaphane with anticancer activity (Neal et al., 2010). Conversely, increased expression of the AOP2 gene causes the GNA and SIN content to increase in plants (Neal et al., 2010; Liu et al., 2012). In vitro activity analysis of Arabidopsis thaliana AOP2 protein by Kliebenstein et al. suggested that the GRA could be converted to GNA through an induced AOP2 protein solution (Kliebenstein et al., 2001b).
Brassica, a genus of economic and nutritional importance in the Brassicaceae family, consists of six species involving three diploids: B. rapa (2n=20, AA), B. oleracea (2n=18, CC), and B. nigra (2n=16, BB). Mutual hybridization and continuous selective evolution among the three yielded three allotetraploids: B. carinata (2n=34, BBCC), B. juncea (2n=36, AABB), and B. napus (2n=38, AACC). The number and expression pattern of AOP genes in Brassica vary from species to species. For example, the AOP2 gene in both B. rapa and B. oleracea has three homologs (Gao et al., 2004; Liu et al., 2014). Two and four homologs of the AOP2 gene were found in the genomes of cabbage mustard and oil-mustard, respectively (Augustine and Bisht, 2015; Wu et al., 2017). The BoAOP2 gene in B. oleracea can be expressed normally to catalyze the degradation of GRA into GNA (Zheng et al., 2022). However, there is a nonfunctional AOP2 allele in broccoli with a two-base deletion on the exon, which in turn influences the accumulation of glucoraphanin products (Li and Quiros, 2003). Liu et al. reported that B. oleracea contained another non-functional BoAOP2. The translation termination caused by the premature termination codon functioning of this gene also caused the accumulation of glucoraphanin products (Liu et al., 2014). The presence of three alleles of AOP2 in B. rapa, all with catalytic activity, was found to be tissue expression-specific (Wang et al., 2011). Augustine et al. documented a significant reduction in the amount of SIN in transgenic plants than in wild-type plants by constructing an AOP2 silencing vector in oil mustard (Augustine and Bisht, 2015). A recently conducted study in pennycress revealed that the SIN content was significantly reduced in the wild-type F2 population versus the AOP2 mutant (Chopra et al., 2020). Therefore, the AOP2 gene is a bridge between beneficial and deleterious GSLs. Although several relevant studies have been conducted on this gene in B. oleracea and B. rapa, the mechanism that underlies its functions in TSM has not been studied in detail. Therefore, elucidating the mechanism of AOP2 genes in GSL synthesis and degradation remains a great challenge due to the complexity of the Brassica crop genome (Malhotra and Bisht, 2020).
In this study, five genes homologous to Arabidopsis thaliana AOP2 were cloned using the allotetraploid crop TSM (2n=36, AABB) as plant material. In addition, the protein’s physicochemical properties, gene structure, phylogenetic tree, promoter cis-acting elements, subcellular localization, and gene expression were comprehensively analyzed. The gene functions were initially verified using in vitro experiments. This study revealed the role of individual BjuAOP2 genes in the process of SIN synthesis in TSM and the expression differences among different copies. Overall, the data obtained in this research acts as a foundation for revealing the molecular mechanism of SIN synthesis regulation by BjuAOP2 genes in Brassica juncea and offer novel insights into improving the GLS fraction of TSM at the molecular level.
2 Materials and methods
2.1 Sample sources
The TSM high-generation selfing line B186 was sown in the Yangtze Normal University squash trial site in the fall of 2021 after germination. Per the normal growth requirements of TSM, field management was performed during growth. The leaves of B186 were collected at the 4-leaf stage for BjuAOP2 cloning, and the expanded tumorous stems of B186 were collected at 4, 7, 10, 13, 16, 19, and 22 weeks after sowing to analyze the expression pattern of BjuAOP2 at different developmental stages. In addition, roots, stems, leaves, tumorous stems, flowers, and siliques of B186 were collected at the shooting stagegib for analysis of the gene’s tissue expression pattern. Each sample had three biological replicates that were collected and placed in liquid nitrogen and kept at -80°C for subsequent use.
2.2 Identification of the BjuAOP2 gene family
The protein sequence of the Arabidopsis thaliana AOP2 gene was retrieved online from TAIR (http://www.arabidopsis.org/). Subsequently, a BlastP search was performed in the Brassica genome database (http://brassicadb.cn/#/BLAST/) to search for amino acid sequences and nucleic acid sequences of TSM AOP2 gene family members. In addition, HMMsearch (Johnson et al., 2010) was used to identify the 2OG-FeII_Oxy (PF03171) and DIOX-N(PF14226) structural domains for all possible AOP2 genes. Finally, conserved structural domain confirmation was performed through NCBI-CDD (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) in NCBI to exclude sequences without typical structural domains.
2.3 Cloning and sequence analysis of the BjuAOP2 gene
Total RNA of TSM leaves was isolated with Tiangen RNA Extraction Kit TRNzol Universal Reagent (Tiangen Biotech, Beijing, China) and with a subsequent reversal to single-stranded cDNA using TransScript One-Step gDNA Removal and cDNA Synthesis SuperMix kit (TransGen Biotech, Beijing, China). The designing of the primers (Supplementary Table S1) and amplification of the full-length CDS sequences using Primer premier 6.0 software was the next step, using the BjuAOP2 gene sequence obtained from the stem mustard database as a template. The PCR reaction system and reaction procedure were performed according to the instructions of TransStart FastPfu DNA Polymerase from TransGen, and the PCR product recovery and purification were performed according to the instructions of the TransGen Gum Recovery Kit (EG101). In addition, the PCR-recovered products were homologously cloned with the vector pTF101-GFP using the homologous recombination method of the pEASY-Basic Seamless Cloning and Assembly Kit (TransGen Biotech, Beijing, China) from TransGen. Subsequently introducing the recombinant vector into E. coli receptor DH5α, plasmids were extracted from 12 positive clones for validation and sequencing. The protein sequences encoding the BjuAOP2 gene obtained by sequencing were subjected to multiple-sequence alignment with the AOP2 protein sequences of Arabidopsis thaliana using DNAStar 7.1 software. The CDS length and amino acid numbers of the BjuAOP2 gene were obtained from the clone sequencing results. Data such as chromosome position was obtained from the reference genome. Isoelectric points and molecular weights were obtained from the pI/Mw calculation tool on the website ExPASy (http://www.expasy.org/tools/). The prediction of subcellular localization was obtained using online tool BUSCA (http://busca.biocomp.unibo.it/).
2.4 Phylogenetic tree construction of the AOP gene family in Brassicaceae crops
Genomic data of 19 sequenced cruciferous species were obtained from The Brassicaceae Genome Resource (http://www.tbgr.org.cn) (Liu et al., 2022). The protein sequence of the Arabidopsis thaliana AOP gene was accessed at the TAIR website as a reference sequence and the AOP family gene sequence was obtained using the Quick Find Best Homology tool in TBtools software (Chen et al., 2020). Clustal W was employed to carry out the multiple-sequence alignment of AOP family proteins from sequenced species in the Brassicaceae family in MAGE 11 software. Subsequently, utilizing the neighbor-joining (NJ) method phylogenetic trees were drawn, with the Bootstrap method set to 1000 (Tamura et al., 2021). The online tool, GSDS (http://gsds.cbi.pku.edu.cn/index.php) was employed for mapping the intron-exon structure pattern of the AOP gene. The analysis of the conserved motif of the gene was performed using the MEME (http://meme-suite.org/) online tool. The upstream promoter sequences (1.5-kb) of each gene of the identified AOP family were accessed at the Brassica genome website (http://brassicadb.org) with the promotor-bound cis-acting elements were examined by the software PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html). Schematic diagrams of all the above analysis results were illustrated utilizing the software Domain Illustrator software (http://dog.biocuckoo.org/) (Ren et al., 2009).
2.5 Protein subcellular localization of BjuAOP2
Protein subcellular localization of BjuAOP2 was performed by transient expression in tobacco leaf epidermal cells. Agrobacterium tumefaciens GV3101 containing the ProCAMV35S::GFP::BjuAOP2 vector plasmid was activated and enlarge-cultivated. Subsequently, the organisms were collected, resuspended in a resuspension solution (OD600 almost 0.5), left for 2-3 h, and introduced into the lower epidermis of 3-4 weeks old tobacco leaves with a syringe. Observations were made after 3 days of transformation. The leaves transformed with empty pTF101-GFP were used as control. In addition, laser confocal microscopy (Nikon, Tokyo, Japan) was utilized for the observation of GFP fluorescence with excitation light at 488 nm and emission light at 510 nm. Furthermore, chloroplast autofluorescence showed excitation light at 640 nm and emission light at 675 nm.
2.6 Prokaryotic expression and in vitro enzyme activity assay of the BjuAOP2 gene
The design of the primers and amplification of full-length CDS sequences were executed through Primer premier 6.0 software, using the BjuAOP2 gene sequence obtained from the stem mustard database as a template (Supplementary Table S2). The pET-32a expression vector (Novagen. Madison, WI, USA) was constructed using the full-length CDS sequence of the gene through homologous recombination with the pEASY-Basic Seamless Cloning and Assembly Kit (TransGen Biotech, Beijing, China). Subsequently, plasmids were extracted from 12 positive clones for validation and sequencing. The correctly sequenced recombinant plasmids for the prokaryotic expression of the gene were transformed into Escherichia coli strain Transetta (DE3). The recombinant pET vector contains the T7 promoter and capable of expressing a fusion protein containing thioredoxin. The E. coli DE3 strain containing prokaryotic expression of the recombinant plasmid was incubated in LB medium at 37°C while also being shaken until OD600 was approximately = 0.6. Then, a final concentration of 0.5 mM was obtained by adding IPTG, and the expression of recombinant protein was induced by shaking the bacteria overnight at 16°C. The bacteria were isolated after induction by centrifuging the E. coli broth for five minutes at 6000 rpm/min. These bacteria were then suspended in PBS and placed on an ultrasonic cell crusher for 3 min at 100MHz (5-second on-10-second off cycle). The obtained cell-disrupted solution was centrifuged (12000 rpm/min and 20 min) for supernatant collection. Subsequently, the supernatant was purified by fusion protein according to the instruction procedures of Ni IDA Beads 6FF Kit (Changzhou Smart-Lifesciences Biotechnology, Changzhou, China). Finally, the purified fusion protease was detected by SDS-PAGE electrophoresis.
Kliebenstein et al. (2001a) validated the Arabidopsis thaliana AOP2 protein by in vitro experiments. In this procedure, the same method was used to validate the function of BjuAOP2 protein in Arabidopsis thaliana, with a slight modification of the enzyme activity assay. The procedure was as follows: 400 μL of purified BjuAOP2 protein solution, 10 mM ascorbate, 15 mM α-ketoglutarate, 200 mM sucrose, 200 μM FeSO4, and 100 μL GIB were added to five 10 mL centrifuge tubes (total reaction volume: 4 mL). A control 10 mL centrifuge tube was added with 400 μL ddH2O instead of purified BjuAOP2 protein solution, and the other components remained unchanged. Subsequently, the six centrifuge tubes were placed on a shaker at 28°C (110 rpm) for 4 hours. The reaction solution was purified by desulfurization and subjected to high-performance liquid chromatography (HPLC) to analyze the GSL fraction
2.7 GSL extraction and HPLC analysis
GSLs were extracted and assayed per the prior procedure (He et al., 2002). First, 1 mL of the enzyme-activated reaction solution was transferred to a DEAE Sephadex A-25 column for overnight desulfurization at room temperature using lipase sulfate (Sigma, E.C. 3.1.6.) and eluted twice with 750 µL of deionized water. The filtration of the eluent was carried out through a 0.22 μm Hydrophilic polyethersulfone (PES) Syringe Filter (Shanghai Anpel) for HPLC analysis. The elute, the external standard 3-GIB, and SIN were analyzed by HPLC.
2.8 Quantitative real-time fluorescence analysis
Total RNA was obtained from different tissues and developmental stages of tuberous mustard through TRNzol Universal Total RNA Extraction Reagent from Tiangen Biotech (Beijing, China). cDNA was synthesized based on the CDS sequences obtained by homologous recombinant cloning and sequencing using Primer premier 6.0. The afore-mentioned software designed real-time fluorescence quantification primers (Supplementary Table S3). The reaction system was as follows: 2×SuperReal PreMix Plus 10 μL; upstream and downstream primers, 0.5 μL each; cDNA template, 2 μL; ddH2O, 7 μL. In addition, a three-step amplification method was used: 95°C pre-denaturation for 5 min; 95°C for 30 s, 58°C for 30 s, and 72°C for 30 s (40 cycles). Dissociation curve: 65-95°C (0.5°C increase per cycle) for 5 s (1 cycle). The entire fluorescent quantitative PCR reaction was performed on a Roche Light Cycler 480 instrument. The proportionate expression of the five BjAOP2 was calculated using the 2-ΔΔCT calculation method with actin as an internal reference gene (Livak and Schmittgen, 2001). Technical and biological replicates, three each were utilized in each experiment.
2.9 Statistical analysis
One-way analysis of variance (ANOVA), significance analysis, and Duncan’s multiple-comparison were performed on the experimental data using SPSS analysis software, with a significance threshold of p-value < 0.05.
3 Results
3.1 Identification and cloning of BjuAOP2 gene family members
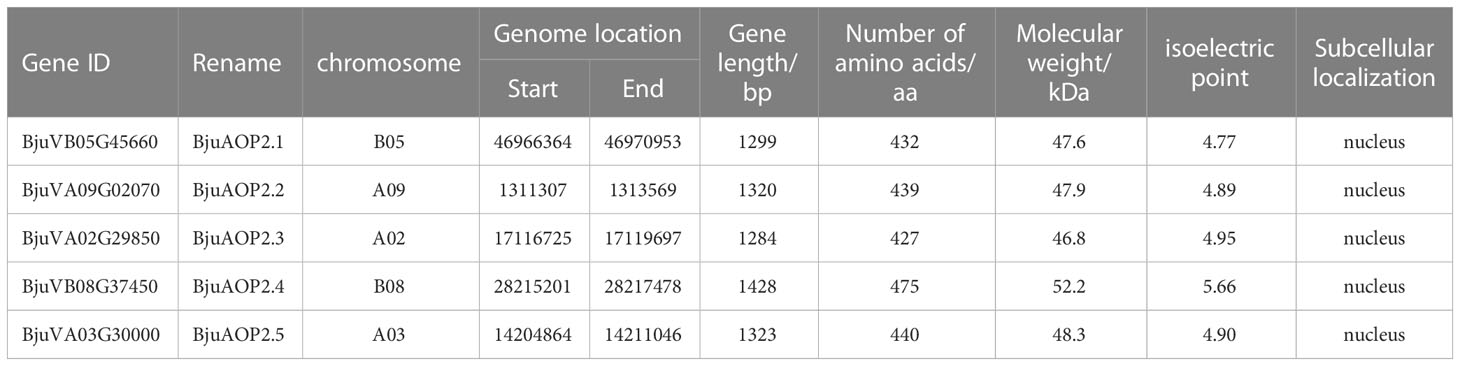
BLAST alignment of the Arabidopsis thaliana AOP2 gene protein sequences with the TSM database was performed. Subsequently, the 2OG-FeII_Oxy (PF14226) structural domain was identified for all possible AOP2 genes. In total, five members of the gene family distributed in different chromosomes, named BjuAOP2.1-BjuAOP2.5, were identified from the TSM genome database (Table 1). The CDS sequences of the genes mentioned above were obtained by PCR using the cDNA obtained by reverse transcription as a template. BjuAOP2 genes have CDS sequences in the length range of 1284-1428 bp, with encoding amino acid numbers between 427-475 aa, molecular weight ranging from 46.8 to 55.2 kDa, and isoelectric point variation ranging from 4.77 to 5.66. The optimal prediction of subcellular localization is nucleus.
Multiple-sequence alignment with the Arabidopsis thaliana AOP2 gene (Figure 1) depicted that the BjuAOP2 gene family’s five members contain the key structural domains, DIOX-N and 2 OG-Fell-Oxy, and the sequences in this range are highly conserved. However, the protein sequence similarity of the middle part of these five members is low, leading to the divergence of gene functions among the members.
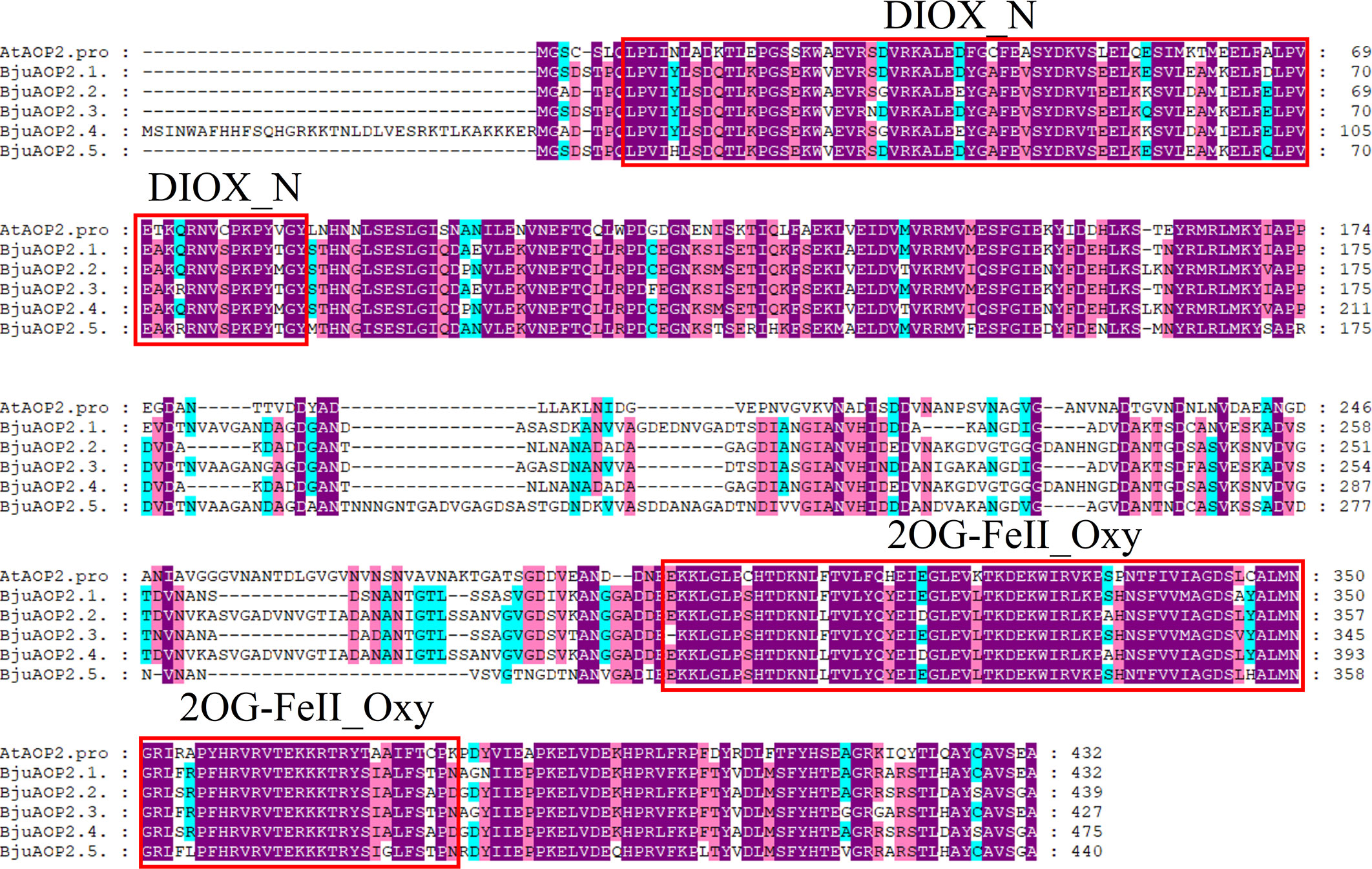
Figure 1 Multiple-sequence alignment of Tumorous stem mustard BjuAOP2 and AtAOP2 protein sequence. Multiple alignments were performed using the MEGA11 software. The red solid boxes represent the structural domains of 2OG-FeII_Oxy and DIOX-N. The purple, blue and pink shading indicates 100%, 80% and 60% conserved percent, respectively.
3.2 Phylogenetic, gene structure, motif and cis-acting regulatory elements analysis of the BjuAOP2 gene family
An AOP gene evolutionary tree was constructed for TSM and 18 other cruciferous crops that have been sequenced to further understand the evolutionary origins of the five members of the BjuAOP2 gene family in TSM (Figure 2; Supplementary Table S4). The AOP gene family is divided into two major categories: (1) AOP1s consisting of 33 AOP1 amino acid sequences; (2) AOP2s and AOP3s consisting of 25 AOP2 amino acids and 3 AOP3 amino acid sequences. In addition, small branches (N ≤ 3) formed by the genes are all derived from different Brassicaceae species, and the small branches further form large branches in the form of clusters containing two and three genes. Members of the TSM AOP gene family are more similar to those in B. carinata and B. rapa than those in Arabidopsis thaliana. The TSM BjuAOP2 gene has the closest affinity to B. rapa, and B. nigra, which is consistent with the evolutionary origin doctrine in U’s Triangle Brassica Species (Cheng et al., 2016).
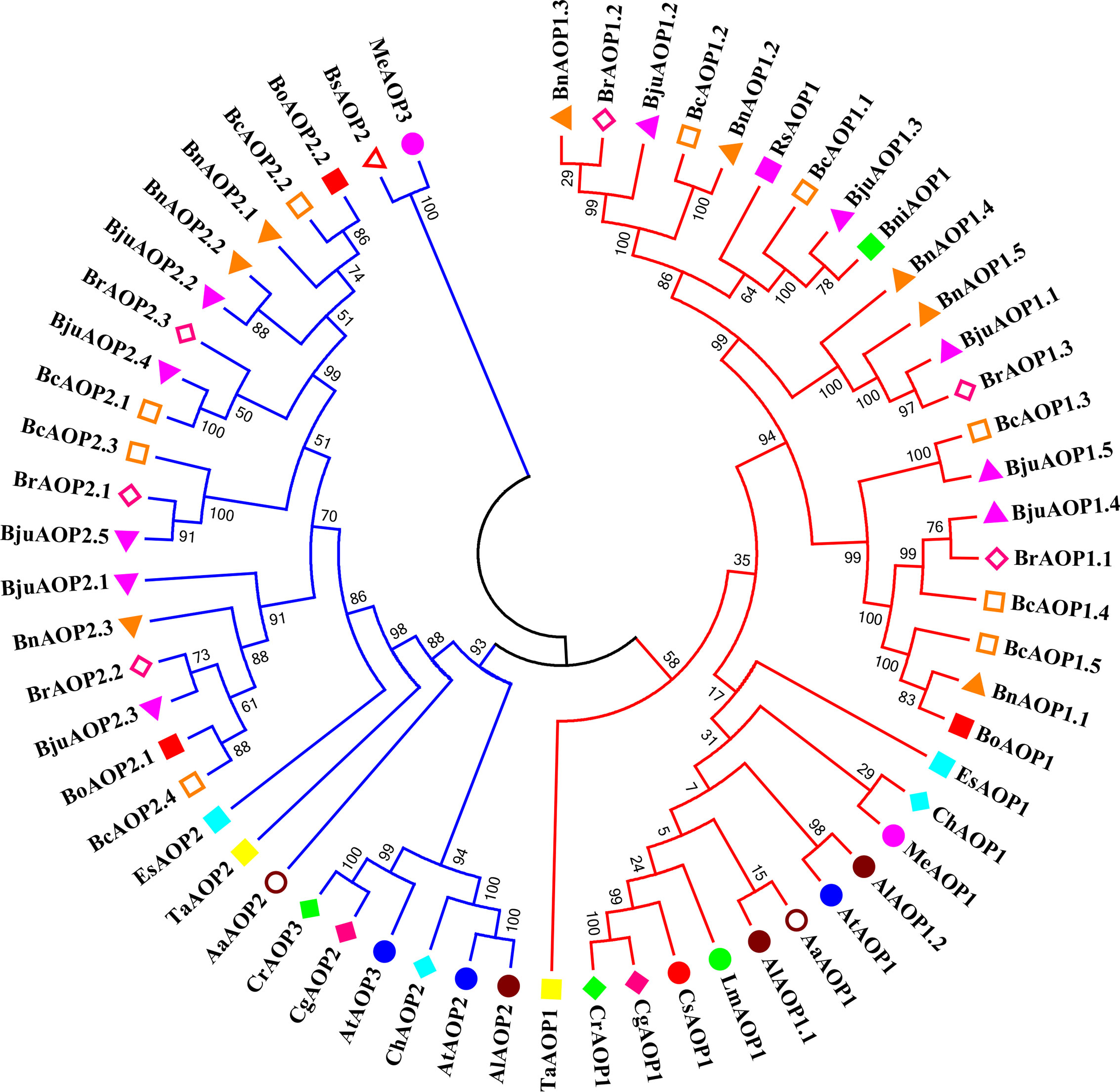
Figure 2 Phylogenetic tree of AOP genes from Brassicaceae species. The phylogenetic tree was built using the neighbor-joining (NJ) method. The unrooted tree was generated using Clustal W in MEGA11 via AOP amino acid sequences from 19 sequenced Brassicaceae species, including: Arabis alpina (Aa.), Arabidopsis lyrate (Al.), Arabidopsis thaliana (At.), Brassica carinata (Bc.), Brassica juncea (Bju.), Brassica napus (Bn.), Brassica nigra (Bni.), Brassica oleracea (Bo.), Brassica rapa (Br.), Boechera stricta (Bs.), Capsella grandiflora (Cg.), Cardamine hirsute (Ch.), Capsella rubella (Cr.), Eutrema salsugineum (Es.), Lepidium meyenii (Lm.), Microthlaspi erraticum (Me.), Raphanus sativus (Rs.), and Thlaspi arvense (Ta.). Genes from each species were labeled with different colors. The numbers at the nodes represent bootstrap percentage values.
The gene structure diagram from the AOP gene family (Figure 3A) revealed that the AOP family is conserved in the number of exons, 53 of the 61 members containing three exons. Despite the similar exon length of most genes, the intron lengths differed significantly. The AOP family was analyzed for conserved motifs using the MEME online tool (Figure 3B). In total, 15 conserved motifs of 8-41 aa in length were contained in the AOP gene (Figure 3C; Supplementary Table S5). In addition, the structural domains of 15 motifs were analyzed using NCBI-CDD online software. The motif 10,4,5 and 7 together form the DIOX_N domain. And the motif 2, 6, 8 and 1 together form the 2OG-FeII_Oxy domain structural of the AOP gene family.To further elucidate the functional element that influences expression of AOP genes, the promoter sequences were analyzed using the PlantCARE database to identify cis-regulatory elements in the promoter region. Nineteen types of stress- and hormone-related cis-acting regulatory elements were detected in the promoters of AOP genes (Figure 4; Supplementary Table S6). All 61 AOP genes contained element related to light. The MYB bingding site were detected in 47. Hormone-related elements (MeJA-responsiveness, salicylic acid responsiveness, zein metabolism regulation, abscisic acid responsiveness, auxin-responsive element, gibberellin-responsive element) were detected in more than 45 members of AOP genes. Other results are shown in the Figure 4.
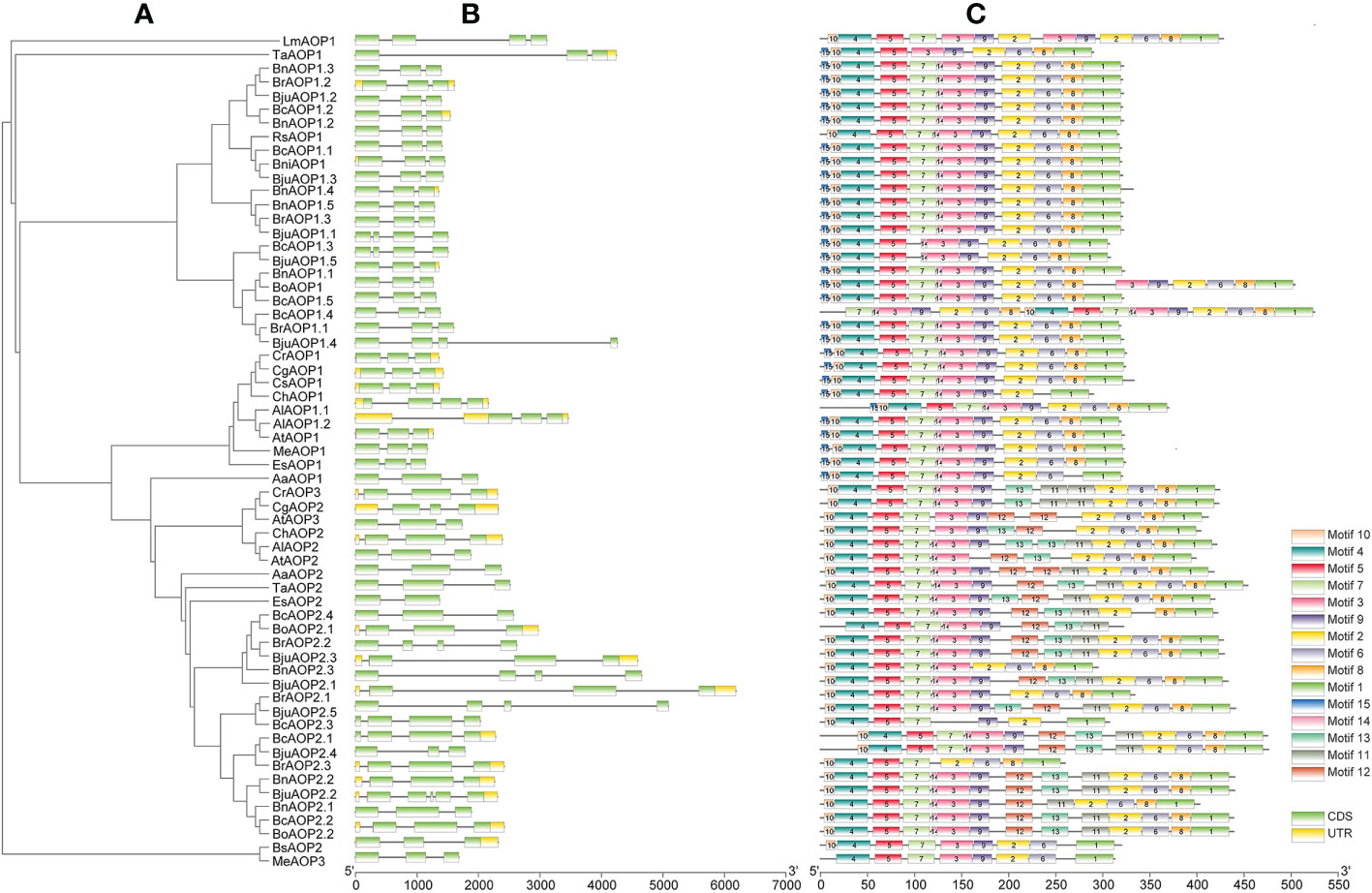
Figure 3 Gene structures and motifs of 61 AOP genes identified in B juncea and other sequenced Brassicaceae crops. (A) unrooted phylogenetic relationships among the AOP protein sequences. (B) exon-intron organization of corresponding AOP gene. Exons are represented by green boxes, while introns are represented by gray lines. (C) conserved motifs of AOP genes. Colored boxes indicated conserved motifs and gray lines represent non-conserved sequences. The length of motifs in each protein was showed proportionally.
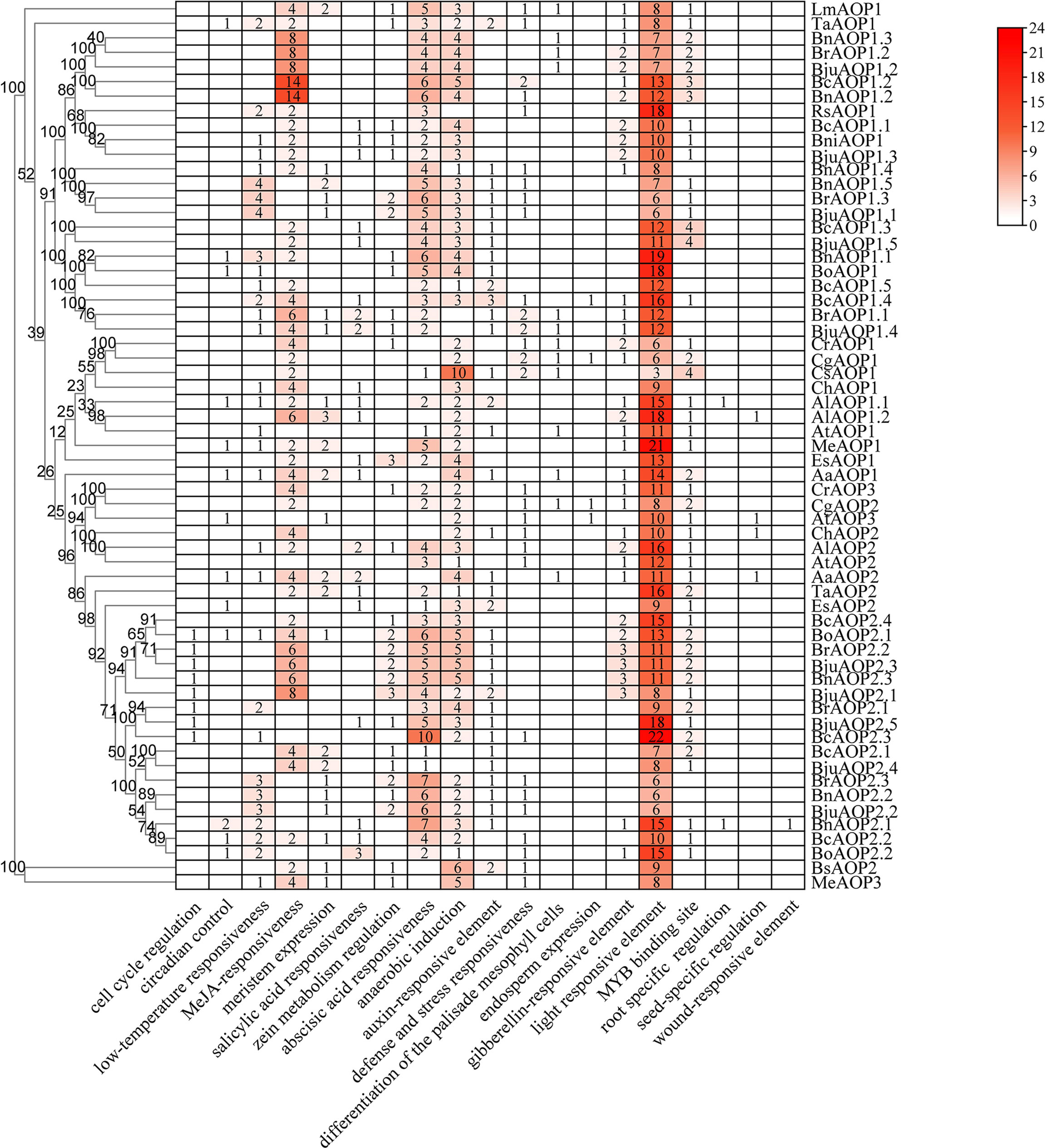
Figure 4 Cis-acting regulatory elements in the promoter region of AOP genes. The numbers and the depth of red represent the frequency of the elements that occur in the promoter region.
3.3 Subcellular localization of the protein encoded by the BjuAOP2 gene
The green fluorescent protein (GFP) fusion transient expression vector containing the BjuAOP2 gene was injected into tobacco leaves and observed by laser confocal microscopy (Figure 5). Except for BjuAOP2.4, where a green fluorescent signal was observed only in the nucleus, the other four family members were observed to fluoresce in both the cytoplasm and nucleus. Thus, BjuAOP2.1, BjuAOP2.2, BjuAOP2.3, and BjuAOP2.5 were expressed in the cytoplasm and nucleus, and BjuAOP2.4 was expressed only in the nucleus, consistent with the prediction results of subcellular localization.
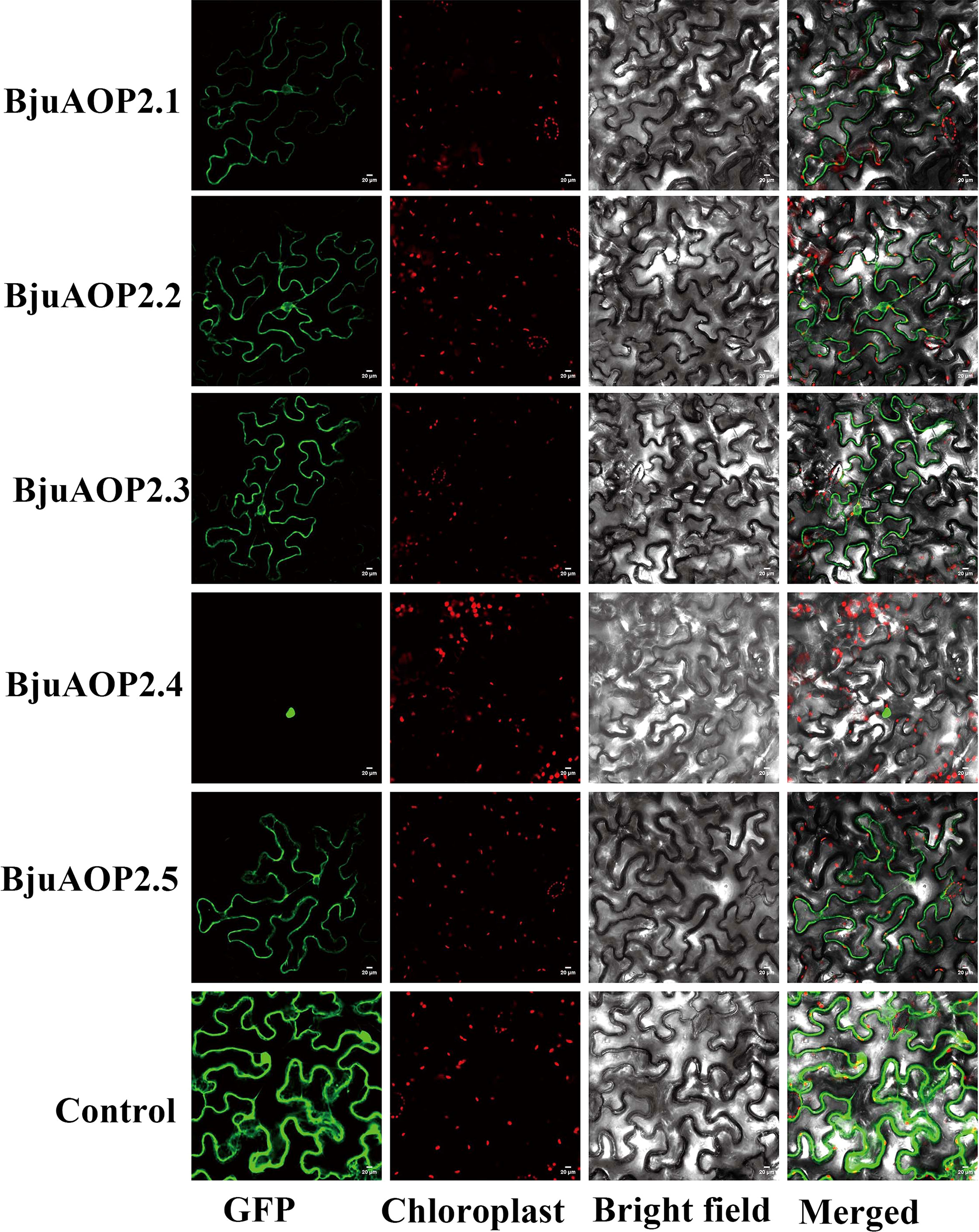
Figure 5 Subcellular localization of BjuAOP2 proteins in Nicotiana benthamiana. These genes were fused with the GFP protein driven by the 35S promoter. Green fluorescence and chloroplast autofluorescence (red) images were captured in a dark field; however, a bright field was used to get images of cell appearance. Bar=20 μm.
3.4 Expression pattern of the BjuAOP2 gene
The expression patterns of five BjAOP2 genes in various tissue parts of B186 and various developmental stages of the tumorous stem were analyzed using real-time PCR (Figure 6). Expression of BjuAOP2 genes was detected in roots, stems, tumorous stems, leaves, flowers and siliques. BjuAOP2 genes had significantly different expression patterns in different tissues (Figure 6A). BjuAOP2.2 were more highly expressed in flowers than in other tissues examined (Figure 6A), whereas BjuAOP2.1, BjuAOP2.3 and BjuAOP2.4 were more highly expressed in stems than in other tissues (Figure 6A). BjuAOP2.5 were more highly expressed in tumorous stems than in other tissues (Figure 6A). Analysis of the expression pattern of the BjuAOP2 gene at different stages of tumorous stem development revealed different expression patterns with the expansion of TSM (Figure 6B). The expression of BjuAOP2.1, BjuAOP2.2 and BjuAOP2.4 peaked at 7 weeks after sowing before falling back to basal levels. BjuAOP2.3 and BjuAOP2.5 showed low expression level throughout the whole growth period.
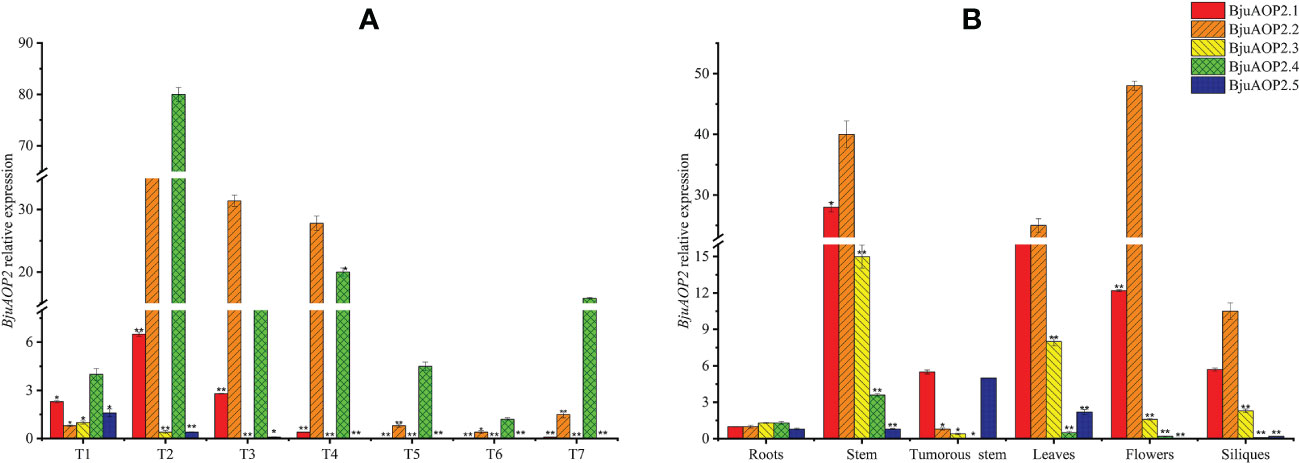
Figure 6 Expression pattern of BjuAOP2 in different tissues and different developmental periods. The vertical axis indicates gene expression levels relative to BjuAOP2.5 expression in roots (A) and BjuAOP2.2 expression in 4 weeks after sowing. T1-T7 represent the seven times 4, 7, 10, 13, 16, 19, and 22 weeks after sowing (B). Error bars represent the standard deviation from three biological repeats. * and ** indicate significant differences at P<0.05 and P<0.01 using ANOVA analysis followed by a Duncan test, respectively.
3.5 In vitro enzyme activity analysis of BjuAOP2
BjuAOP2 prokaryotic expression vector was constructed using PET-32a. The protein expression was induced by IPTG, and the BjuAOP2 fusion protein was obtained by the His-tag purification column. In vitro enzymatic activity analysis of five BjuAOP2 fusion proteins was performed using GIB as substrate. As shown in Figure 7, all five BjuAOP2 proteins successfully catalysed the conversion of GIB to SIN. The standards GIB and SIN were detected at 3.3 min and 4.6 min, where only GIB was detected in the control group. However, both GIB and SIN were detected in the protein elutes of all five members. Therefore, all five BjuAOP2 proteins of TSM have catalytic activity.
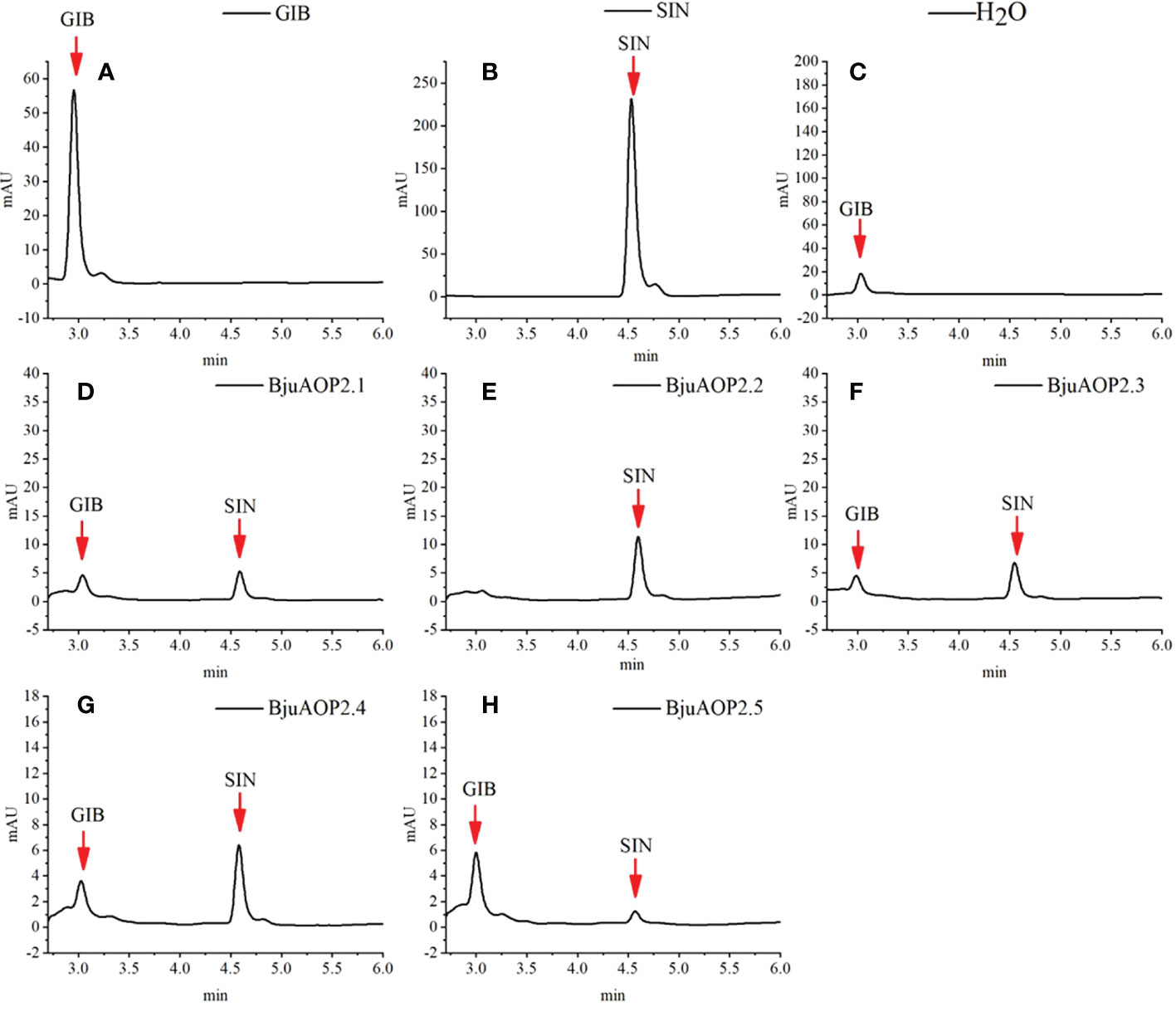
Figure 7 Enzymatic activity of heterologously expressed BjuAOP2. The purified desulfoglucosinolates extracted from E. coli were subjected to HPLC (monitored at 229 nm) and results were documented. (A) indicates desulfated GIB (glucoiberin) standard, (B) indicates desulfated SIN (sinigrin) standard, (C) indicates desulfated GIB standard treated with ddH2O as the negative control; (D–H) indicate desulfated GIB standard treated with BjuAOP2.1, BjuAOP2.2, BjuAOP2.3, BjuAOP2.4 and BjuAOP2.5 enzymes, respectively.
4 Discussion
Increasing the beneficial GSL content has been a major breeding goal for Brassica species in recent years (Ishida et al., 2014; Barba et al., 2016). The hydrolysis products of GRA such as isothiocyanates have anticancer effects in humans (Fahey et al., 2001; Fahey et al., 2002). Furthermore, enzymatic degradation of GRA to PRO inhibits iodine uptake in mammals, leading to thyroid dysfunction. The high content of SIN in TSM adds a strong mustard spiciness, which is derived from the degradation of GIB by the AOP2 gene. Therefore, the focus of breeders’ attention has been the buildup of beneficial GSL components and contents in Brassica vegetables and the reduction of harmful GSL components and contents. In B. juncea, Augustine and Bisht (2015) used RNA interference technology to accumulate a large amount of GRA, a beneficial GSL product, by affecting the expression of AOP2, indicating the significance of AOP genes in the breeding improvement of Brassica. The recent publication of the B. carinata reference genome announced the completion of the reference genome assembly for the U’s Triangle Brassica Species (Song et al., 2021), which facilitates the study of the functional and evolutionary relationships of AOP genes. In this study, five BjuAOP2 genes in TSM were identified and cloned. All five genes possess biological catalytic activity, but the expression and subcellular localization results are tissue-specific and differential, respectively. Therefore, functional divergence may exist in the regulation of SIN synthesis by these five genes. Overall, the findings of this study may provide a reference for breeders to improve the TSM GSL fraction and content.
As a family with large number of members, the Cruciferae contains 338 genera and 3709 species covering many crops that have economic importance (Warwick et al., 2006). The core crops of the Cruciferae family underwent genomic triploidization events during evolution (Lysak et al., 2005). Theoretically, the number of TSM BjuAOP2 should be three times higher (six) than that of Arabidopsis thaliana. However, only five BjuAOP2 family members were retained during the evolution of TSM, indicating the loss of BjuAOP2 genes in this process. The evolutionary analysis of cruciferous AOP genes in this study revealed that AOP1 was in a separate group (Figure 2). Previous studies have shown that the AOP locus in cruciferous species underwent two gene duplication events. The first caused the divergence between AOP1 and AOP2/3 (the AOP1 gene was the ancestor gene of the AOP2/3 gene) while the second event led to the formation of the AOP2 and AOP3 genes (Kliebenstein et al., 2001a). Subsequently, some AOP genes were lost due to the formation of new species or adaptation to variable environments.
Gene duplication enables the tissue-specific expression of genes that undergo replication in response to variable environmental stimuli; in addition, the replicated genes have more diverse expression patterns relative to single genes (Li et al., 2005; Kliebenstein, 2008; Huang et al., 2022). The five BjuAOP2 genes in this study exhibited expression divergence in different tissues and different periods of tumorous stem development, indicating varied regulation of the genes. The BjuAOP2 gene is expressed in trace amounts in roots and in higher amounts in leaves and stems in B186 (Figure 6A), depicting congruency with the expression pattern of the AOP2 gene in Arabidopsis thaliana (Neal et al., 2010). Subcellular localization analysis indicated differences in the expression sites of these five genes. The promoter regions of the BjuAOP2 genes were further analyzed using PLACE software, and each of them had some specific cis-acting elements (Figure 4). These elements were associated with tissue-dependent expression, hormone, biotic and abiotic stress responses, resulting in differential expression of the five BjuAOP2 genes in various tissues and developmental periods. Thereby leading to differences in GSL accumulation in TSM thus improving plant adaptation to the external environment.
Genes may undergo depletion, defunctionalization, functional maintenance, or functional differentiation across copies during replication (Zhang, 2003). In this research, in vitro experiments of prokaryotic expression demonstrated that all five genes in TSM can convert GIB to SIN, leading to abundant SIN in TSM. Therefore, all five genes have biocatalytic activity (Figure 7). The multiple-sequence alignment of the five BjuAOP2 proteins showed that they have two typically conserved structural domains at both their C- and N-terminal ends, whereas the sequence variation outside the conserved structural domain is large (Figure 1). Studies in B. oleracea, B. rapa, and Arabidopsis thaliana have revealed that disruption of the 2OG-FeII_Oxy structural domain leads to loss of AOP2 gene function (Li and Quiros, 2002; Neal et al., 2010; Zhang et al., 2015). The integrity of the 2OG-FeII_Oxy structural domain may be critical for the AOP2 protein to maintain its catalytic activity in a variable environment.
This research dealt with the elucidation of the expression characteristics of the BjuAOP2 gene in TSM and its role in the GSL biosynthesis pathway. In addition, the evolutionary relationships of this gene were analyzed. These findings provide a guide for breeders to improve the aliphatic GSL component of TSM by traditional breeding methods or genetic modification methods.
5 Conclusion
This work identified five BjuAOP2 genes from the TSM genome that were cloned, bioinformatically analyzed, and validated for in vitro activity. First, the protein sequences of the genes were analyzed. All five genes in TSM possessed two conserved structural domains, with increased variation in the variable regions, indicating functional divergence. Subsequently, the evolution of AOP genes in cruciferous species was analyzed. All AOP1 genes clustered into a large independent branch, whereas AOP2 and AOP3 were clustered into a separate large branch. The in vitro activity analysis verified the biocatalytic activity of these five BjuAOP2 genes. In addition, their expression pattern analysis suggested variation in different growth periods or different tissues. Finally, tobacco injection-based subcellular localization analysis using transient expression assay indicated the differences in the cellular localization of these five BjuAOP2 genes.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
Conceived and designed the experiments: JC, BC and YiL. Performed the experiments: BC, YuL, DZ, CX and ZL. Analyzed the data: JC, BC and YiL. Funding acquisition, Project administration: JC and YiL. Writing, reviewing and editing: JC, BC and YiL. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Chongqing University Innovation Research Group Funding Program (CXQT21029), the Chongqing Natural Science Foundation (cstc2019jcyj-msxmX0713), the science and technology research program of Chongqing municipal commission (KJQN202001428), the science and technology research program of Chongqing municipal commission (KJQN202001429).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1111418/full#supplementary-material
References
Augustine, R., Bisht, N. C. (2015). Biofortification of oilseed Brassica juncea with the anti-cancer compound glucoraphanin by suppressing GSL-ALK gene family. Sci. Rep. 5 (1), 1–12. doi: 10.1038/srep18005
Barba, F. J., Nikmaram, N., Roohinejad, S., Khelfa, A., Zhu, Z., Koubaa, M., et al. (2016). Bioavailability of glucosinolates and their breakdown products: Impact of processing. Front. Nutr. 3. doi: 10.3389/fnut.2016.00024.eCollection2016
Blažević, I., Montaut, S., Burcul, F., Olsen, C. E., Burow, M., Rollin, P., et al. (2019). Glucosinolate structural diversity, identification, chemical synthesis and metabolism in plants. Phytochemistry 169, 112100. doi: 10.1016/j.phytochem.2019.112100
Chen, C. J., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y. H., et al. (2020). TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13 (8), 1194–1202. doi: 10.1101/289660
Chen, J. Y., Ullah, C., Reichelt, M., Beran, F., Yang, Z. L., Gershenzon, J., et al. (2020). The phytopathogenic fungus sclerotinia sclerotiorum detoxifies plant glucosinolate hydrolysis products via an isothiocyanate hydrolase. Nat. Commun. 11 (1), 3090. doi: 10.1038/s41467-020-16921-2
Cheng, F., Sun, R. F., Hou, X. L., Zheng, H. K., Zhang, F. L., Zhang, Y. Y., et al. (2016). Subgenome parallel selection is associated with morphotype diversification and convergent crop domestication in Brassica rapa and Brassica oleracea. Nat. Genet. 48 (10), 1218–1224. doi: 10.1038/ng.3634
Chopra, R., Johnson, E. B., Emenecker, R., Cahoon, E. B., Lyons, J., Kliebenstein, D. J., et al. (2020). Identification and stacking of crucial traits required for the domestication of pennycress. Nat. Food 1 (1), 84–91. doi: 10.1038/s43016-019-0007-z
Clay, N. K., Adio, A. M., Denoux, C., Jander, G., Ausubel, F. M. (2009). Glucosinolate metabolites required for an Arabidopsis innate immune response. Science 323 (5910), 95–101. doi: 10.1126/science.11646
Engel, E., Martin, N., Issanchou, S. (2006). Sensitivity to allyl isothiocyanate, dimethyl trisulfide, sinigrin, and cooked cauliflower consumption. Appetite 46 (3), 263–269. doi: 10.1016/j.appet.2006.01.007
Fahey, J. W., Haristoy, X., Dolan, P. M., Kensler, T. W., Scholtus, I., Stephenson, K. K., et al. (2002). Sulforaphane inhibits extracellular, intracellular, and antibiotic-resistant strains of Helicobacter pylori and prevents benzo[a]pyrene-induced stomach tumors. PNAS 99 (11), 7610–7615. doi: 10.1073/pnas.112203099
Fahey, J. W., Zalcmann, A. T., Talalay, P. (2001). The chemical diversity and distribution of glucosinolates and isothiocyanates among plants. Phytochemistry 56 (1), 5–51. doi: 10.1016/S0031-9422(00)00316-2
Gao, M., Li, G., Yang, B., McCombie, W. R., Quiros, C. F. (2004). Comparative analysis of a Brassica BAC clone containing several major aliphatic glucosinolate genes with its corresponding Arabidopsis sequence. Genome 47 (4), 666–679. doi: 10.1139/g04-021
Grubb, C. D., Abel, S. (2006). Glucosinolate metabolism and its control. Trends Plant Sci. 11 (2), 89–100. doi: 10.1016/j.tplants.2005.12.006
Grubb, C. D., Zipp, B. J., Ludwig-Müller, J., Masuno, M. N., Molinski, T. F., Abel, S. (2004). Arabidopsis glucosyltransferase UGT74B1 functions in glucosinolate biosynthesis and auxin homeostasis. Plant J. 40 (6), 893–908. doi: 10.1111/j.1365-313X.2004.02261.x
Halkier, B. A., Gershenzon, J. (2006). Biology and biochemistry of glucosinolates. Annu. Rev. Plant Biol. 57 (1), 303–333. doi: 10.1146/annurev.arplant.57.032905.105228
Hansen, B. G., Kerwin, R. E., Ober, J. A., Lambrix, V. M., Mitchell-Olds, T., Gershenzon, J., et al. (2008). A novel 2-oxoacid-dependent dioxygenase involved in the formation of the goiterogenic 2-hydroxybut-3-enyl glucosinolate and generalist insect resistance in Arabidopsis. Plant Physiol. 148 (4), 2096–2108. doi: 10.1104/pp.108.129981
Harun, S., Abdullah-Zawawi, M. R., Goh, H. H., Mohamed-Hussein, Z. A. (2020). A comprehensive gene inventory for glucosinolate biosynthetic pathway in Arabidopsis thaliana. J. Agric. Food Chem. 68 (28), 7281–7297. doi: 10.1021/acs.jafc.0c01916
He, J. H., Chen, H., Schnitzler, W. H. (2002). Glucosinolate composition and contents in brassica vegetables. Scientia Agric. Sin. 35 (2), 192–197. doi: 10.1006/jfls.2001.0409
Huang, Y., Chen, J., Dong, C., Sosa, D., Xia, S., Ouyang, Y., et al. (2022). Species-specific partial gene duplication in Arabidopsis thaliana evolved novel phenotypic effects on morphological traits under strong positive selection. Plant Cell 34 (2), 802–817. doi: 10.1101/2021.04.05.438504
Ishida, M., Hara, M., Fukino, N., Kakizaki, T., Morimitsu, Y. (2014). Glucosinolate metabolism, functionality and breeding for the improvement of brassicaceae vegetables. Breed Sci. 64 (1), 48–59. doi: 10.1270/jsbbs.64.48
Johnson, L. S., Eddy, S. R., Portugaly, E. (2010). Hidden Markov model speed heuristic and iterative HMM search procedure. BMC Bioinf. 11, 431. doi: 10.1186/1471-2105-11-431
Kliebenstein, D. J. (2008). A role for gene duplication and natural variation of gene expression in the evolution of metabolism. PloS One 3 (3), e1838. doi: 10.1371/journal.pone.0001838
Kliebenstein, D. J., Kroymann, J., Brown, P., Figuth, A., Pedersen, D., Gershenzon, J., et al. (2001a). Genetic control of natural variation in arabidopsis glucosinolate accumulation. Plant Physiol. 126 (2), 811–825. doi: 10.1104/pp.126.2.811
Kliebenstein, D. J., Lambrix, V. M., Reichelt, M., Gershenzon, J., Mitchell-Olds, T. (2001b). Gene duplication in the diversification of secondary metabolism: tandem 2-oxoglutarate-dependent dioxygenases control glucosinolate biosynthesis in Arabidopsis. Plant Cell 13 (3), 681–693. doi: 10.1105/tpc.13.3.681
Kroymann, J., Donnerhacke, S., Schnabelrauch, D., Mitchell-Olds, T. (2003). Evolutionary dynamics of an Arabidopsis insect resistance quantitative trait locus. PNAS 100 (suppl-2), 14587–14592. doi: 10.1073/pnas.1734046100
Kroymann, J., Textor, S., Tokuhisa, J. G., Falk, K. L., Bartram, S., Gershenzon, J., et al. (2001). A gene controlling variation in arabidopsis glucosinolate composition is part of the mcthionine chain elongation pathway. Plant Physiol. 127 (3), 1077–1088. doi: 10.1104/pp.010416
Li, B. H., Gaudinier, A., Tang, M., Taylor-Teeples, M., Nham, N. T., Ghaffari, C., et al. (2014). Promoter-based integration in plant defense regulation. Plant Physiol. 166 (4), 1803–1820. doi: 10.1104/pp.114.248716
Li, W. H., Jing, Y., Xun, G. (2005). Expression divergence between duplicate genes. Trends Genet. 21 (11), 602–607. doi: 10.1016/j.tig.2005.08.006
Li, G., Quiros, C. F. (2002). Genetic analysis, expression and molecular characterizatien of BoGSL-ELONG, a major gene involved in the aliphatic glucosinolate pathway of brassica species. Genetics 162 (4), 1937–1943. doi: 10.1017/S0016672302005906
Li, G., Quiros, C. F. (2003). In planta side-chain glucosinolate modification in arabidopsis by introduction of dioxygenase brassica homolog BoGSL-ALK. Theor. Appl. Genet. 106 (6), 1116–1121. doi: 10.1007/s00122-002-1161-4
Li, B. H., Tang, M., Nelson, A., Caligagan, H., Zhou, X., Clark-Wiest, C., et al. (2018). Network-guided discovery of extensive epistasis between transcription factors involved in aliphatic glucosinolate biosynthesis. Plant Cell 30 (1), 178–195. doi: 10.1105/tpc.17.00805
Li, Y., Wang, X. Y., Wang, Y. H., Meng, Q. F., Sun, J., Wang, B. L. (2011). Studies on composition and contents of glucosinolates in different tuber mustard varieties. Acta Hortic. Sin. 38 (7), 1356–1364. doi: 10.16420/j.issn.0513-353x.2011.07.001
Liu, Z., Hirani, A. H., Mcvetty, P. B., Daayf, F., Quiros, C. F., Li, G. (2012). Reducing progoitrin and enriching glucoraphanin in Braasica napus seeds through silencing of the GSL-ALK gene family. Plant Mol. Biol. 79 (1), 179–189. doi: 10.1007/s11103-012-9905-2
Liu, Z., Li, N., Yu, T., Wang, Z., Wang, J., Ren, J., et al. (2022). The brassicaceae genome resource (TBGR): A comprehensive genome platform for brassicaceae plants. Plant Physiol. 190 (1), 226–237. doi: 10.1093/plphys/kiac266
Liu, S., Liu, Y., Yang, X., Tong, C., Edwards, D., Parkin, I. A., et al. (2014). The Brassica oleracea genome reveals the asymmetrical evolution of polyploid genomes. Nat. Commun. 5 (1), 1–11. doi: 10.1038/ncomms4930
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods 25 (4), 402–408. doi: 10.1006/meth.2001.1262
Lysak, M. A., Koch, M. A., Pecinka, A., Schubert, I. (2005). Chromosome triplication found across the tribe Brassiceae. Genome Res. 15 (4), 516–525. doi: 10.1101/gr.3531105
Malhotra, B., Bisht, N. C. (2020). Glucosinolates: regulation of biosynthesis and hydrolysis. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.620965
Mazumder, A., Dwivedi, A., Du Plessis, J. (2016). Sinigrin and its therapeutic benefits. Molecules 21 (4), 416. doi: 10.3390/molecules21040416
Mikkelsen, M. D., Naur, P., Halkier, B. A. (2004). Arabidopsis mutants in the c-s lyase of glucosinolate biosynthesis establish a critical role for indole-3-acetaldoxime in auxin homeostasis. Plant J. 37 (5), 770–777. doi: 10.1111/j.1365-313x.2004.02002.x
Mitreiter, S., Gigolashvili, T. (2021). Regulation of glucosinolate biosynthesis. J. Exp. Bot. 72 (1), 70–91. doi: 10.1093/jxb/eraa479
Neal, C. S., Fredericks, D. P., Griffiths, C. A., Neale, A. D. (2010). The characterisation of AOP2: a gene associated with the biosynthesis of aliphatic alkenyl glucosinolates in Arabidopsis thaliana. BMC Plant Biol. 10 (1), 1–16. doi: 10.1186/1471-2229-10-170
Nguyen, V. P., Stewart, J., Lopez, M., Ioannou, I., Allais, F. (2020). Glucosinolates: natural occurrence, biosynthesis, accessibility, isolation, structures, and biological activities. Molecules 25 (19), 4537. doi: 10.3390/molecules25194537
Ren, J., Wen, L., Gao, X., Jin, C., Xue, Y., Yao, X. (2009). DOG 1.0: illustrator of protein domain structures. Cell Res. 19 (2), 271–273. doi: 10.1038/cr.2009.6
Sønderby, I. E., Geu-Flores, F., Halkier, B. A. (2010). Biosynthesis of glucosinolates-gene discovery and beyond. Trends Plant Sci. 15 (5), 283–290. doi: 10.1016/j.tplants.2010.02.005
Song, X. M., Wei, Y. P., Xiao, D., Gong, K., Sun, P. C., Ren, Y. M., et al. (2021). Brassica carinata genome characterization clarifies u’s triangle model of evolution and polyploidy in Brassica. Plant Physiol. 186 (1), 388–406. doi: 10.1093/plphys/kiab048
Soundararajan, P., Kim, J. S. (2018). Anti-carcinogenic glucosinolates in cruciferous vegetables and their antagonistic effects on prevention of cancers. Molecules 23 (11), 2983. doi: 10.3390/molecules23112983
Tamura, K., Stecher, G., Kumar, S. (2021). MEGA11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38 (7), 3022–3027. doi: 10.1093/molbev/mst197
Textor, S., Kraker de, J. W., Hause, B., Gershenzon, J., Tokuhisa, J. G. (2007). MAM3 catalyzes the formation of all aliphatic glucosinolate chain lengths in Arabidopsis. Plant Physiol. 144 (1), 60–71. doi: 10.1104/pp.106.091579
Wang, H., Wu, J., Sun, S., Liu, B., Cheng, F., Sun, R., et al. (2011). Glucosinolate biosynthetic genes in Brassica rapa. Gene 487 (2), 135–142. doi: 10.1016/j.gene.2011.07.021
Warwick, S. I., Francis, A., Al-Shehbaz, I. A. (2006). Brassicaceae: Species checklist and database on CD-rom. Plant Systematics Evol. 259 (2), 249–258. doi: 10.1007/s00606-006-0422-0
Wittstock, U., Halkier, B. A. (2002). Glucosinolate research in the Arabidopsis era. Trends Plant Sci. 7 (6), 263–270. doi: 10.1016/S1360-1385(02)02273-2
Wu, S. H., Lei, J. J., Chen, G. J., Chen, H. C., Cao, B. H., Chen, C. M. (2017). De novo transcriptome assembly of Chinese kale and global expression analysis of genes involved in glucosinolate metabolism in multiple tissues. Front. Plant Sci. 8 (92). doi: 10.3389/fpls.2017.00092
Zhang, J. (2003). Evolution by gene duplication: an update. Trends Ecol. Evol. 18 (6), 292–298. doi: 10.1016/S0169-5347(03)00033-8
Zhang, J. F., Liu, Z. Y., Liang, J. L., Wu, J., Cheng, F., Wang, X. W. (2015). Three genes encoding AOP2, a protein involved in aliphatic glucosinolate biosynthesis, are differentially expressed in. Brassica rapa. J. Exp. Bot. 66 (20), 6205–6218. doi: 10.1093/jxb/erv331
Keywords: Brassica juncea, glucosinolate, BjuAOP2, expression pattern, prokaryotic expression, activity analysis
Citation: Chen B, Liu Y, Xiang C, Zhang D, Liu Z, Liu Y and Chen J (2023) Identification and in vitro enzymatic activity analysis of the AOP2 gene family associated with glucosinolate biosynthesis in Tumorous stem mustard (Brassica juncea var. tumida). Front. Plant Sci. 14:1111418. doi: 10.3389/fpls.2023.1111418
Received: 29 November 2022; Accepted: 13 February 2023;
Published: 22 February 2023.
Edited by:
Xiaodong Yang, Yangzhou University, ChinaReviewed by:
Pilar Soengas, Biological Mission of Galicia (CSIC), SpainXiaonan Li, Shenyang Agricultural University, China
Copyright © 2023 Chen, Liu, Xiang, Zhang, Liu, Liu and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yihua Liu, MTgyMjUxMjY4NjZAMTYzLmNvbQ==; Jingjing Chen, Y2hqam1hb0AxNjMuY29t
 Bing Chen
Bing Chen Jingjing Chen
Jingjing Chen