- 1School of Natural Resource Management, College of Post Graduate Studies in Agricultural Sciences, Central Agricultural University (Imphal), Umiam, Meghalaya, India
- 2Division of System Research and Engineering, Indian Council of Agricultural Research (ICAR) Research Complex for North Eastern Hill Region, Umiam, Meghalaya, India
- 3School of Crop Protection, College of Post Graduate Studies in Agricultural Sciences, Central Agricultural University (Imphal), Umiam, Meghalaya, India
- 4School of Crop Improvement, College of Post Graduate Studies in Agricultural Sciences, Central Agricultural University (Imphal), Umiam, Meghalaya, India
Despite Northeastern India being “Treasure House of Citrus Genetic Wealth,” genetic erosion of citrus diversity poses severe concern with a corresponding loss in seed microbial diversity. The seed microbiome of citrus species unique to the Purvanchal Himalaya is seldom explored for their use in sustainable orchard management. Isolation and characterization of culturable seed microbiomes of eight citrus species, namely, Citrus reticulata Blanco, C. grandis (L.) Osbeck, C. latipes Tanaka, C. megaloxycarpa Lushaigton, C. jambhiri Lush, C. sinensis (L.) Osbeck, C. macroptera Montr, and C. indica Tanaka collected from NE India were carried out. The isolates were then screened for an array of plant growth–promoting (PGP) traits [indole acetic acid (IAA) production, N2 fixation, phosphate and zinc complex dissolution, siderophores, and Hydrogen Cyanide (HCN) production]. The pure culture isolates of seed microbiomes were capable of dissolving insoluble Ca3(PO4)2 (1.31–4.84 µg Pi ml-1 h-1), Zn3(PO4)2 (2.44–3.16 µg Pi ml-1 h-1), AlPO4 (1.74–3.61 µg Pi ml-1 h-1), and FePO4 (1.54–4.61µg Pi ml-1 h-1), mineralized phytate (12.17–18.00 µg Pi ml-1 h-1) and produced IAA-like substances (4.8–187.29 µg ml-1 h-1). A few isolates of the seed microbiome were also able to fix nitrogen, secrete siderophore-like compounds and HCN, and dissolve ZnSO4 and ZnO. The 16S ribosomal Ribonucleic Acid (rRNA)–based taxonomic findings revealed that Bacillus was the most dominant genus among the isolates across citrus species. Isolates CG2-1, CME6-1, CME6-4, CME6-5, CME6-9, CJ7-1, CMA10-1, CI11-3, and CI11-4 were identified as promising bioinoculants for development of microbial consortium having multifaceted PGP traits for nutritional benefits of nitrogen, phosphorus and zinc, and IAA hormonal benefits to citrus crops for better fitness in acid soils.
Introduction
Plants, as metaorganisms, are associated with diverse microbes from various niches (such as the rhizosphere, spermosphere, phyllosphere, and endosphere). The interaction between plant and microbes may be either casual or intimate, but they all furnish an evolving seed microbiome that may carry over to other developmental stages found often in plants growing in natural situations (Hardoim et al., 2015). Bacterial endophytes are all bacteria that may or may not be effectively cultured and can colonize a plant internally without causing apparent harm or creating any obvious exterior structures (Azevedo and Araujo, 2007). Endophytes with plant growth–promoting (PGP) traits are generally the most effective in promoting growth because they live inside plant tissues and can communicate with the host plant and exert their beneficial effect much more effectively (Giauque et al., 2019). Protected from the outside environment, endophytes are much less susceptible to the soil’s frequent chemical–physical biotic and abiotic variations. The beneficial action of these bacteria is primarily expressed by directly promoting nutrient absorption via modulation of plant hormone levels (Marchut-Mikolajczyk et al., 2018). Seed-borne endophytes lay the foundation for the development of plant microbiomes as the pioneer colonizers, and seeds serve as carriers for the advantageous microorganisms for multigenerational preservation of these beneficial relationships (Truyens et al., 2015). Association of seed and endophytes have been documented in a variety of crops including rice (Cottyn et al., 2001), maize (Rijavec et al., 2007), bean (López-López et al., 2010), wheat (Kuźniar et al., 2020), and tomato (Xu et al., 2014). The collection of factors that go into the construction and organization of seed microbiomes is quite complex, with microbes drawn from not only the soil microbiome (Hardoim et al., 2012) but also dispersal agents, pollinator, and floral microbiomes (Gandolfi et al., 2013). Only specialized endophytic strains are able to colonize and flourish in reproductive plant organs (Truyens et al., 2015), and particularly, bacteria with competitive and adaptive colonization traits could dwell in seeds (Okunishi et al., 2005). Through a variety of processes, such as the solubilization of phosphate (Berg, 2009), synthesis of phytohormones such as indole-3-acetic acid (Defez et al., 2017), production of 1-aminocyclopropane deaminase (Glick, 2005), nitrogen fixation (Shabanamol et al., 2018), siderophore generation, and inhibition of phytopathogenic microbes (Hong and Park, 2016), seed bacterial endophytes have been reported to enhance plant development and health. Across a wide range of plant taxa including wheat, Brassica, rice, and maize, seed-associated bacteria have been identified using next-generation sequencing (NGS) techniques and are found largely within the bacterial phyla Acidobacteria, Proteobacteria, Actinobacteria, Firmicutes, Bacteroidetes, and Deinococcus-Thermus (Johnston-Monje and Raizada, 2011; Liu et al., 2012) with Gammaproteobacteria (Hardoim et al., 2015; Herrera et al., 2016) found to be the most prevalent type of bacteria at the class level, followed by Alphaproteobacteria (Hardoim et al., 2015) and Bacilli (Comby et al., 2009). Saving seeds with their indigenous microbes offers an enormous potential to safeguard, explore, and exploit microbial diversity and their untapped metabolic potential for plant, human, and environmental health.
The citrus fruit is a well-known source of biocompounds crucial for human nutrition and among one of the most commercially significant evergreen subtropical fruit crops in the world (Penyalver et al., 2022). North-eastern India is recognized as a significant hotspot of citrus biodiversity renowned as the “Treasure House of Citrus Genetic Wealth” attributable to the region’s special blend of different soil, physiographic, and climatic conditions (Malik et al., 2013). Domestication of wild plant species has been a historical process that has lasted since the dawn of agriculture. In order to meet changing human demands, wild species are domesticated and diversification of the seed microbiome occurs as a consequence of domestication (Abdullaeva et al., 2021). The northeastern region of the country harbors several wild, semiwild, and domesticated species including Citrus indica, C. jambhiri, C. grandis, C. macroptera, C. latipes, and C. megaloxycarpa aside from the commercially cultivated species like Citrus reticulata (Khasi mandarin) and C. sinenis. The rapid destruction of forests in the northeastern region has endangered these naturally occurring citrus species and the microbiota associated with them. These species may have great potential in the improvement of Indian Citrus industry by being a source of genes for combating biotic and abiotic stresses like pests and disease resistance, especially resistance to greening disease in its natural habitat (Yang and Ancona, 2022; Jing et al., 2023), tolerance to psorosis and exocortis virus (Sharma et al., 2004), cold tolerance or resistance (Hazarika, 2012), and drought resistance (Pandey et al., 2008). In citrus, the seed-to-seedling phase is very crucial as the seedling is commonly used as rootstock onto which scion varieties are grafted. Numerous ethnomedic benefits of citrus species, which have been used by the local population for centuries, have been discovered. Traditional medicines frequently employ the peel of the C. grandis fruit to treat epilepsy, edema, and cough (Anmol et al., 2021); fresh fruit, juice, and dried powder from C. indica are used for curing viral diseases, jaundice, and deadly communicable diseases like small pox (Malik et al., 2006). C. macroptera fruits are used medicinally to alleviate digestive disorders and stomach pain (Malik et al., 2013), whereas C. latipes fruits are used to treat viral infections, digestive problems, and jaundice (Malik et al., 2013). In a recent study, microbial profiling was carried out for five different biocompartments, viz. bulk soil, root endosphere, flush, rhizosphere, and flower) of citrus from a single California orchard (Xi et al., 2023). However, except for the work by Newcombe et al. (2018) reporting the presence of one cultivable endophyte per seed in the Rutaceae plant family and the study highlighting Cutibacterium and Acinetobacter as the most prevalent bacterial genera in seeds and shoots of C. limon (Faddetta et al., 2021), seed microbiota of citrus species remain largely unexplored. The present study aimed to explore the culturable members of the seed microbiome of C. reticulata Blanco, C. grandis (L.) Osbeck, C. latipes Tanaka, C. megaloxycarpa Lushaigton, C. jambhiri Lush, C. sinensis (L.) Osbeck, C. macroptera Montr, and C. indica Tanaka collected from the Purvanchal Himalaya region, carried out 16S rRNA–based taxonomic identification, and screened them for multifaceted PGP traits.
Materials and methods
Sampling of fruits
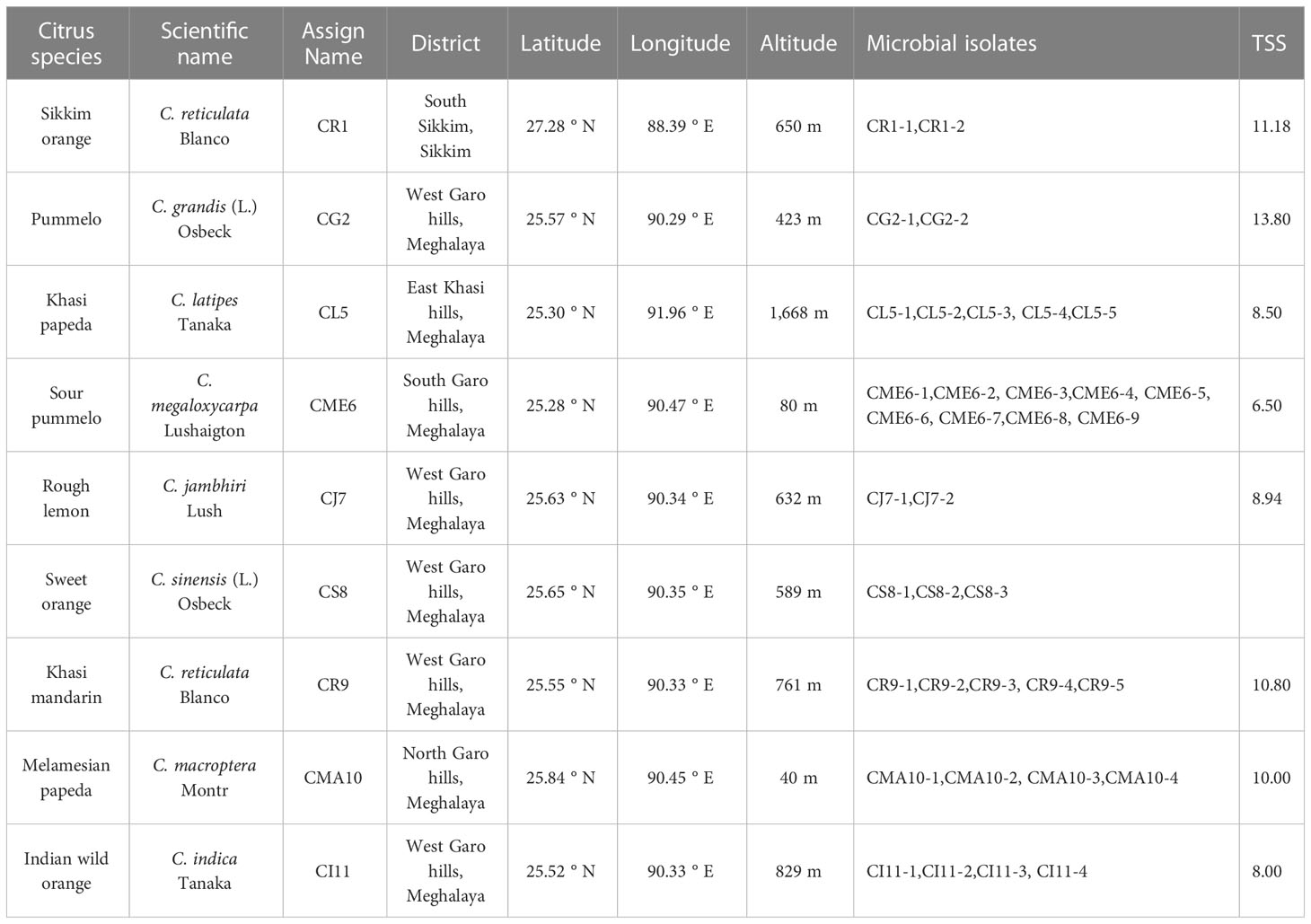
Sampling of citrus fruits from different locations of North East region of India including the Nokrek Biosphere Reserve in the West Garo Hills district of Meghalaya was carried out during October–November, 2021. Approximately 8–10 disease/disorder-free fruits per plant and three plants per species were collected at the physiologically ripen stage (total soluble sugars considered as an indicator; Table 1). Samples were temporarily stored at 4°C for a few hours and then at −20°C until further processing. The citrus species along with their assigned names and details are listed in Table 1.
Extraction of seeds
The fruits were first washed with disinfectant liquid and rinsed with water, followed by sterilization with 70% ethanol under sterile conditions and left to dry. A total of 15 fresh seeds were extracted from each fruit under aseptic conditions using autoclaved tools, which were rinsed with sterilized distilled water for 7–10 times (based on differences in the fibers and flesh coating of the seeds) until the surrounding fibers and mucilage were removed. The extracted seeds were dried and stored in sterile falcon tubes along with the fruit juice at 4°C until use.
Isolation and purification of bacterial microbiota from seeds
Surface sterilization of seeds
Three seeds per citrus species were surface-sterilized, which consisted of the following steps: (1) agitation of the seeds in sodium hypochlorite solution (2.0%) for 2 min, which was repeated twice and (2) immersion in 70% (v/v) ethanol for 1 min, followed by repeatedly rinsing the seeds with autoclaved distilled water. To confirm that the sterilization process was successful, 1 ml aliquot of the last-step washing water was plated on growth agar-media and examined for microbial growth after incubation at 30°C for 3–5 days.
Cultivation of microbiota from the seeds
The surface-sterilized seeds were activated before by incubating them with adequate amount of sterile water overnight at 28°C. Once seed surface sterility was confirmed, three seeds per species were ground gently in an autoclaved mortar using 0.50 ml of sterilized distilled water and the ground seed suspension (100 μl) was plated on nutrient agar media plates followed by incubation for 2–3 days at 28 ± 0.5°C. The colonies obtained were selected on the basis of colony morphology and/or pigmentation and repeatedly streaked on fresh nutrient agar-media to obtain pure cultures. The purified cultures were maintained in nutrient agar slants and stored at 4°C. The pure isolates were finally cultured in Luria-Bertani (LB) broth for glycerol stocks preparation and stored at -20°C.
Morphological characterization
The microorganisms isolated from citrus seed were examined for colony size, shape of colony, color of colony, surface of colony, margin, and elevation. The isolated microbial cultures were streaked on nutrient agar plate, incubated at 28 ± 0.5°C, and observed for colony morphology after 48 h.
In vitro test for multiple plant growth–promoting traits
Production of indole acetic acid–like substances
An aliquot of 0.1 ml suspension of the pure cultures incubated for 24 h at 30 ± 0.5°C was inoculated in nutrient broth containing 1% L-tryptophan and incubated at 30°C for 48 h at 150 rpm followed by determination of IAA in a supernatant. Pink color was developed by the adding van Urk Salkowski reagent, and the intensity is measured in a spectrophotometer (Spectrascan 2600, Thermo Scientific, USA) at 530 nm wavelength. IAA was expressed in μg ml-1 h-1.
N2-fixation test
Nitrogen-free bromothymol blue semi-solid agar was inoculated with the pure isolate broth and incubated along with an uninoculated control at 30°C for 7 days. The color of the media was light green in the start, but due to increase in pH of the medium on fixation of N2 by the bacterial isolate, the media color turns to blue confirming the capacity of the microorganism to fix N2 as described by Okon (1985).
Phosphate-solubilization ability test
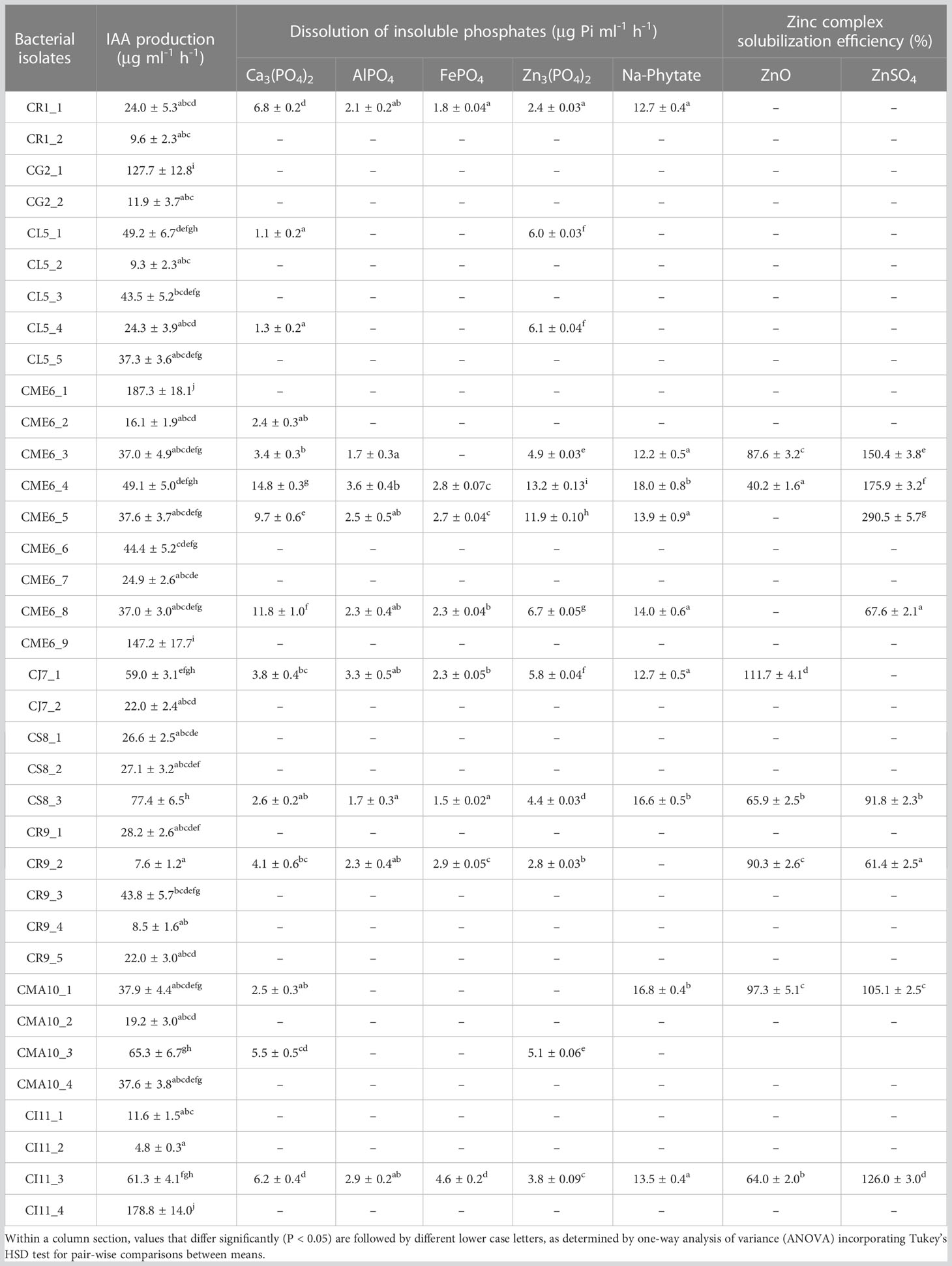
The ability of pure culture isolates to solubilize insoluble forms of phosphates was determined on the Pikovskaya’s agar plates amended with Ca3(PO4)2, AlPO4, FePO4, Zn3(PO4)2, and Na-phytate separately spot-inoculated with culture isolates. Transparent zone formation in the periphery of the individual bacterial colony after 72 h of incubation at 30 ± 0.5°C confirmed the solubilization ability (Pikovskaya, 1948). The quantitative estimation was carried out on Pikovskaya’s broth amended with Ca3(PO4)2, AlPO4, FePO4, Zn3(PO4)2, and Na-phytate separately, inoculated with 10 μl of bacteria pure culture. In brief, 100 μl of clear supernatant was taken and 5 ml 0.03N NH4F in 0.025 N HCl was added. Development of blue color was carried out using Dickman–Bray’s reagent and stannous chloride. Finally, the intensity of blue color was measured at 660 nm (Spectrasca UV- 2600, Thermo Scientific, USA) and concentration of P was read from the P standard curve (0, 0.08, 0.2, 0.4, 0.6, 0.8, and 1.0 ppm). The concentration of P was expressed as μg ml-1 h-1.
Dissolution of insoluble zinc complexes
The zinc-solubilizing efficiency of the pure culture isolates was estimated by the plate assay using modified Bunt and Rovira media–amended 0.1% ZnO and 0.1% ZnSO4 separately as a source of insoluble inorganic zinc with bromophenol blue as an indicator. The isolates were spot-inoculated on the plates, and after 48 h of incubation at 30°C, the plates showing a halo zone around the colonies were considered positive for solubilization of inorganic zinc. The diameter of the clear zone was measured to calculate the solubilizing efficiency (SE) (Ramesh et al., 2014).
Siderophore production
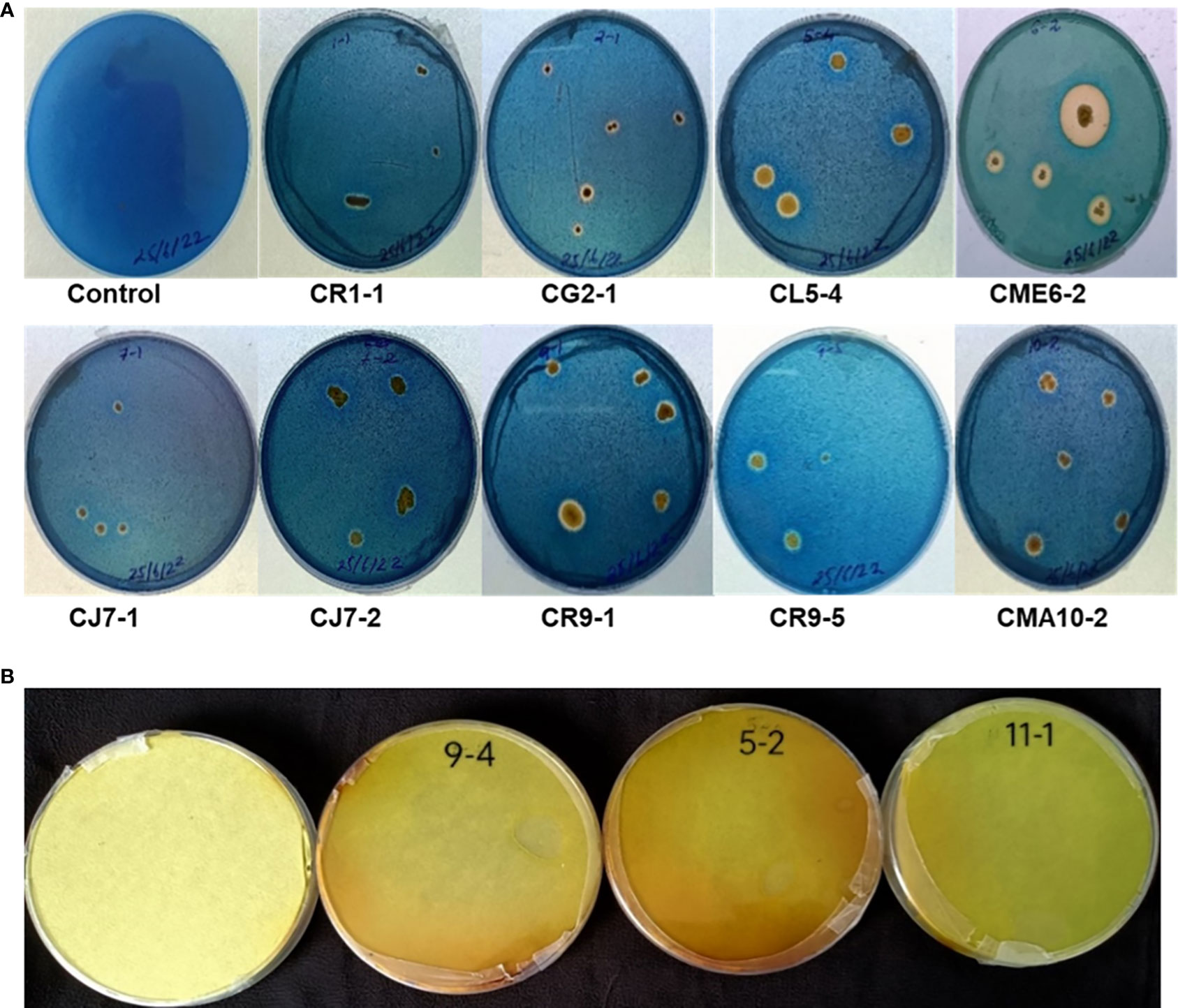
Universal Chrome Azurol S (CAS) agar medium was prepared as described by Schwyn and Neilands (1987) to check siderophore-producing ability of bacterial isolates. The bacterial strains were spot-inoculated on each plate with an uninoculated plate taken as control. After inoculation, plates were incubated at 28°C for 5–7 days and observed for the formation of a clear zone around the bacterial colonies (Louden et al., 2011).
Hydrogen cyanide production
Bacteria were streaked into nutrient agar plates supplemented with glycine (Lorck, 1948). The lid was lined with filter paper (Whatman No. 1) impregnated with 1% picric acid and 10% of sodium carbonate. Petri plates were sealed with parafilm and incubated at 28 ± 1°C for 96 h in inverted condition. Discoloration of the filter paper from yellow to brown after incubation was considered as microbial production of hydrogen cyanide.
Scoring of Plant Growth Promoting Bacteria (PGPB) isolates
After determining the PGP traits for all 36 isolates, a row/column matrix was generated (bacterial isolates in row and PGP traits in column) based on the score assigned to each isolate against each PGP trait. The score to each isolate against each trait was assigned as per the formula given below:
The score against each isolate for a particular trait was given in such a way that the highest trait value among 36 isolates got the maximum score 1 and the least trait got the score of 0. Finally, the scores of seven traits defined as cumulative or total score of multifaceted traits of an isolate was calculated.
Statistical analysis
For each PGP parameter, the significant difference among means of isolates were determined by one-way ANOVA incorporating Levene’s test of homogeneity and test of normality. The multiple pairwise comparison among isolates’ means within a parameter was performed by Tukey’s honestly significant difference test (Tukey’s HSD) test at 95% confidence level (IBM SPSS v.21.0, SPSS Inc., Chicago, IL, USA).
The multivariate statistics was computed using PRIMER v 6.1.9 software (Primer-E Ltd, Plymouth, UK). The data matrix (10 different PGP traits as columns and bacterial isolates as rows) was normalized to eliminate the effects of different units, and square-root-transformed while performing principal component analysis (PCA). The Euclidean distance matrix was used as a measure of dissimilarity between isolates in terms of PGP traits. This data matrix was also subjected to hierarchical cluster analysis using group-average linking incorporating similarity of profile to test the significance difference at 95% confidence limit. Then, the hierarchical cluster profile was superimposed on the PCA plot to form ellipses (Thakuria et al., 2010).
Molecular characterization of isolated microbes
The genomic Deoxyribonucleic acid (gDNA) of the isolated microbes were extracted as per procedure described by Thakuria et al. (2008). Post-extraction gDNA was analyzed on agarose gel electrophoresis, and the quality was checked using a Nano-drop® 2000 spectrophotometer (Thermo Scientific, USA) based on A260/230 and A260/280 ratios. Extracted microbial DNA was amplified in a Gradient Master Cycler 5331 (Eppendorf Make, Germany) with a primer pair (27f and 1492r) specific to bacterial domain yields amplified product size of approximate 1,465 bp. The amplification reactions were carried out in a 25 µl volume containing 50 ng microbial DNA as a template, 100 nM of each oligonucleotide primers [27f (5’-AGAGTTTGATCCTGGCTCAG-3’) and 1492R (5’-GGTTACCTTGTTACGACTT-3’)], 12.5 µl of Go-Taq DNA polymerase (Promega, USA), and the required quantity of PCR grade water (HiMedia, India). The PCR reaction condition included an initial 3 min denaturation at 94°C, and was followed by 35 thermal cycles of 45 s at 94°C, 45 s at 56°C and 45 s at 72°C. Amplification was completed with the final extension step at 72°C for 7 min. All PCR products were examined by electrophoresed in an agarose gel (1% w v-1) using a 100 bp DNA ladder (New England Biolabs, UK). The amplified products were subjected to bi-directional sanger sequencing. Thus, obtained sequences were matched for homolog using the EzTaxon database and BLASTn database, NCBI. The accession numbers for sequences were obtained from the GenBank database. A phylogenetic consensus tree was constructed with reference sequences inferred from the Maximum Parsimony algorithm using MEGA v.11.
Results
Isolation and purification of bacterial microbiota from seeds
A total of 36 pure culture isolates were obtained in nutrient agar plates from surface-sterilized seeds of different citrus species collected from various regions of North East India. The different species with their respective microbial isolates obtained with their assigned name are listed in Table 1. The colonies obtained were selected on the basis of colony morphology and/or pigmentation and repeatedly streaked on fresh nutrient agar-media to obtain pure cultures. The purified cultures were maintained in nutrient agar slants, which were stored at 4°C, and glycerol stocks stored at -20°C. The morphological characteristics such as colony shape, size, color, elevation, margin, and surface were recorded at 48 h of growth on a nutrient agar plate and are presented in Supplementary Table 1 and Supplementary Figure 1.
Screening for multifaceted plant growth promotion properties
Production of indole acetic acid–like substance
The quantitative production of IAA-like substances in tryptophan-amended nutrient broth media by 36 isolates is shown in Figure 1. The range of production of IAA-like substances varied from 4.8 ± 0.3 to 187.3 ± 18.1 µg ml-1 h-1 among the 36 isolates (Table 2). The maximum production of IAA-like substances was possessed by the isolate CME6-1 followed by CI11-4, CME6-9, and CG2-1 in the tune of 187.3, 178.8, 147.1, and 127.6 µg ml-1 h-1, respectively.
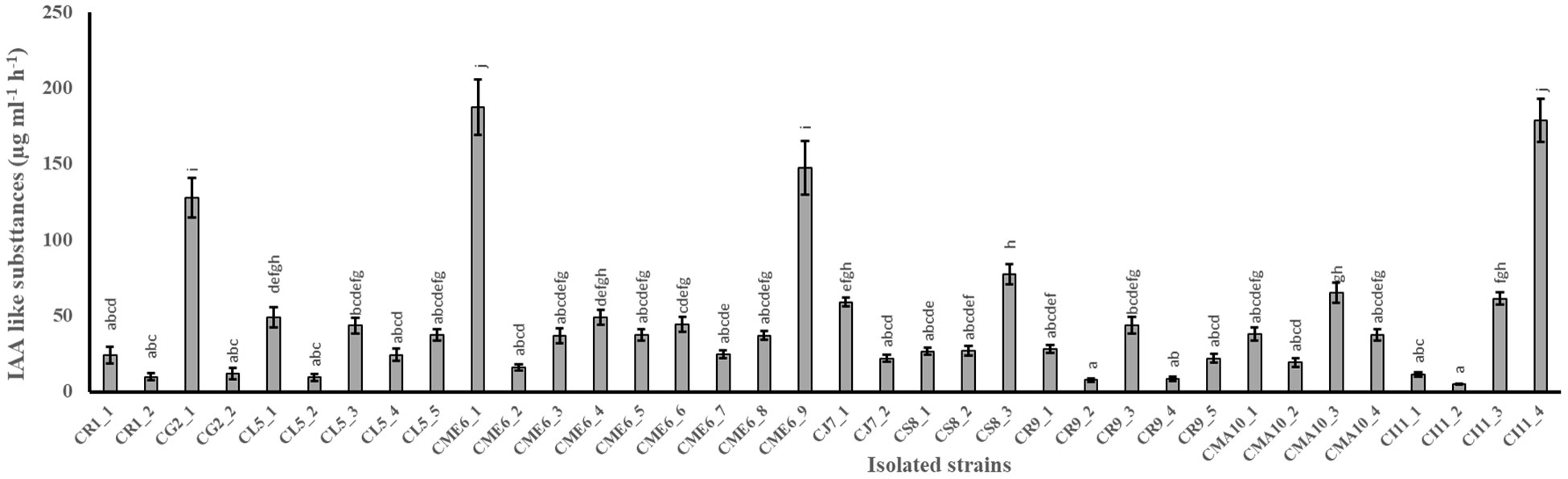
Figure 1 Production of indole acetic acid–like substances in tryptophan-amended nutrient broth media by citrus seed isolates. Within a column section, values that differ significantly (P < 0.05) are followed by different lower case letters, as determined by one-way analysis of variance (ANOVA) incorporating Tukey’s HSD test for pair-wise comparisons between means.
Nitrogen fixation
Out of 36 bacterial isolates, 13 bacterial isolates tested positive for nitrogen fixation by production of blue color from green in the nitrogen-free bromothymol blue medium (Supplementary Figure 2). The bacteria positive for N2 fixation included isolates CG2-1, CL5-1, CL5-5, CME6-1, CME6-5, CME6-8, CR9-1, CR9-3, CMA10-3, CMA10-4, CI11-2, CI11-3, and CI11-4.
Phosphate-solubilization ability test
The ability of all the seed microbial isolates to solubilize different insoluble phosphates and organic phosphate was examined in plate as well as liquid culture. The creation of a solubilization zone surrounding the colony demonstrated the isolates’ phosphate solubilization ability on Pikovskaya’s agar plates modified independently with different phosphate complexes (Table 2; Supplementary Figure 3). A total of 14 bacterial isolates were able to solubilize Pi (inorganic phosphate) on Ca3(PO4)2-amended Pikovskaya’s agar plates, and the range of dissolved inorganic phosphate in liquid culture was from to 1.31 ± 0.2 to 14.84 ± 0.3 µg Pi ml-1 h-1 with isolates CME6-4, CME6-8, and CME6-5 possessing the maximum ability for dissolution of tricalcium phosphate. There were 12 bacterial isolates that were able to solubilize Pi on the Zn3(PO4)2-amended Pikovskaya’s agar, and the range of dissolved inorganic phosphate was from 2.44 ± 0.03 to 13.16 ± 0.13 µg Pi ml-1 h-1 in which isolate CME6-4 possessed the maximum ability for dissolution of zinc phosphate followed by CME6-5 and CME6-8. In case of AlPO4-amended Pikovskaya’s agar plates, nine isolates were able to produce the solubilization zone around the colony to solubilize Pi with the range of dissolved inorganic phosphate was from 1.74 ± 0.3 to 3.61 ± 0.4 µg Pi ml-1 h-1 where isolate CME6-4 possessed the maximum ability for dissolution of aluminum phosphate followed by CJ7-1 and CI11-3. Nine bacterial isolates were able to produce a solubilization zone around the colony on FePO4-amended Pikovskaya’s agar, and the dissolved inorganic phosphate ranged from 1.54 ± 0.02 to 4.61 ± 0.2 µg Pi ml-1h-1 with isolate CI 11-3 possessing the maximum solubilization ability followed by CR 9-2 and CME 6-4. All bacterial isolates were also tested for the ability to release ortho-phosphate on sodium-phytate (Na-phytate)–amended Pikovskaya’s agar plates. Nine bacterial isolates were able to produce a solubilization zone around the colony (Supplementary Figure 3E). The range of ortho-phosphate released from the phytate complex was between 12.17 ± 0.5 and 18.00 ± 0.8 µg Pi ml-1 h-1 among nine isolates with the maximum ability to mineralize the phytate complex possessed by isolate CME6-4 followed by CMA10-1 and CS8-3.
The quantitative data on production of inorganic phosphate by the isolates having a solubilizing ability from Ca3(PO4)2, Zn3(PO4)2, AlPO4, FePO4, and Na-phytate amended separately in Pikovskaya’s broth is shown in Figures 2A–E, respectively.
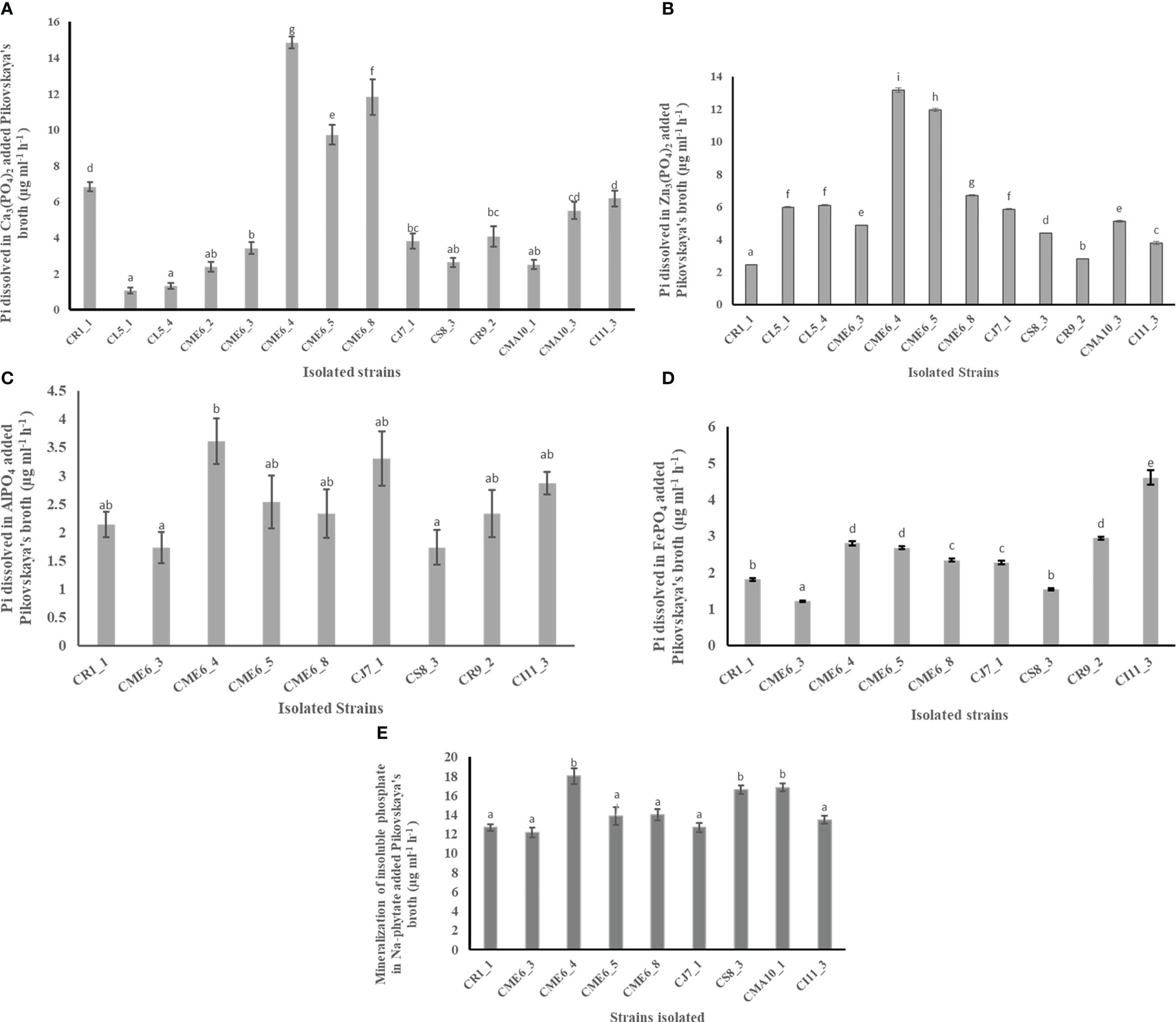
Figure 2 Phosphate solubilization ability of citrus seed isolates in Pikovskaya’s media amended with (A) Ca3(PO4)2), (B) Zn3(PO4)2, (C) AlPO4, (D) FePO4, and (E) Na-phytate. Within a column section, values that differ significantly (P < 0.05) are followed by different lower case letters, as determined by one-way analysis of variance (ANOVA) incorporating Tukey’s HSD test for pair-wise comparisons between means.
Dissolution of insoluble zinc complexes
Solubilization zone development around the colony on zinc-amended Bunt and Rovira media concluded that the isolates have the potential to solubilize zinc. There were 7 of the 36 isolates that showed a positive result by the formation of a solubilization zone around the colony in the zinc oxide–amended Bunt and Rovira media (Supplementary Figure 4A). The zinc oxide– dissolving efficiency ranged from 40.2 ± 1.6 to 111.7 ± 4.1, with isolate CJ7-1 having the highest efficiency, followed by CMA10-1 and CR9-2. There were 8 of the 36 isolates that showed a positive result by the formation of a solubilization zone around the colony in the zinc sulfate–amended Bunt and Rovira media agar plates (Supplementary Figure 4B). The zinc sulfate dissolution efficiency ranged from 61.4 ± 2.5 to 290.5 ± 5.7, with isolate CME 6-5 having the highest efficiency followed by CME 6-4 and CME 6-3. The quantitative data on the zinc oxide solubilization efficiency and zinc sulfate solubilization efficiency is shown in Figures 3A, B, respectively (Table 2).
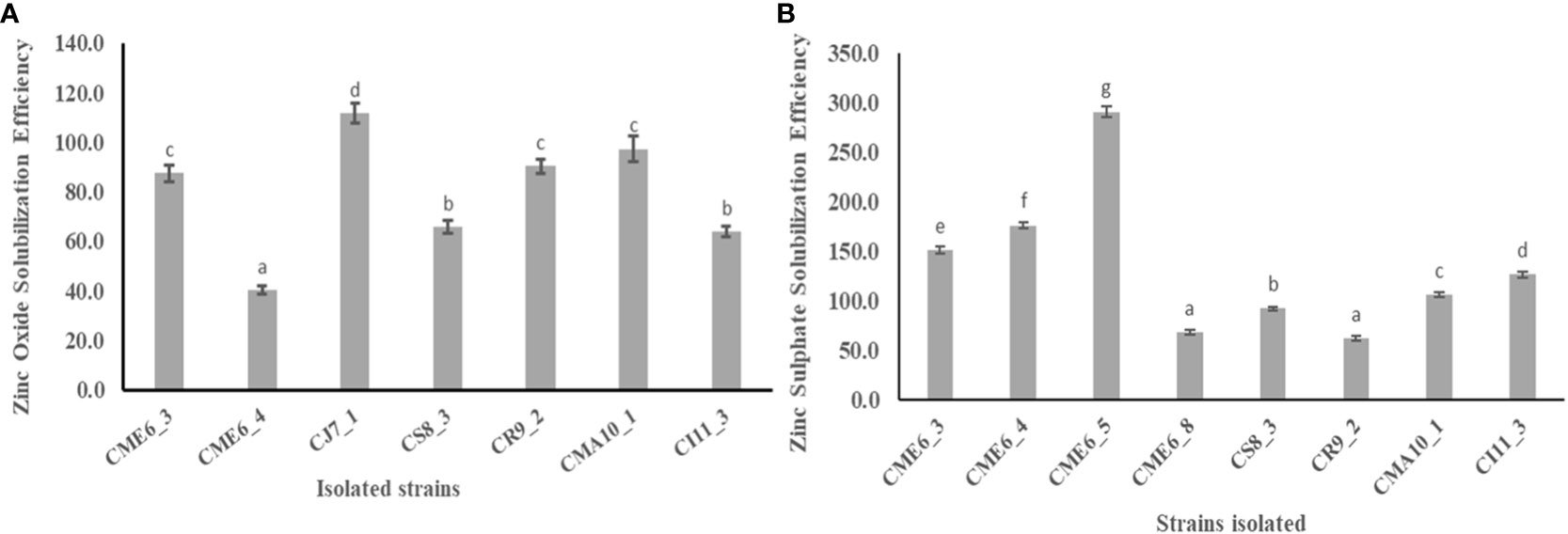
Figure 3 Zinc solubilization efficiency in Bunt and Rovira media amended with (A) 0.1% ZnO and (B) 0.1% ZnSO4. Within a column section, values that differ significantly (P < 0.05) are followed by different lower case letters, as determined by one-way analysis of variance (ANOVA) incorporating Tukey’s HSD test for pair-wise comparisons between means.
Siderophore-like substance
The isolates were tested for siderophore production on CAS blue agar medium. There were 9 bacterial isolates, viz. CR1-1, CG2-1, CL5-4, CME6-2, CJ7-1, CJ7-2, CR9-1, CR9-5, and CMA10-1, out of the 36 bacterial isolates that showed a positive result for siderophore production on CAS agar medium by producing a halo zone around the colony as presented in Figure 4A.
HCN production
Only three isolates, viz. CL5-2, CR9-4, and CI11-1, out of the 36 bacterial isolates that showed positive results for HCN production as the filter paper turned reddish brown from yellow as depicted in Figure 4B.
Scoring of PGPB isolates and statistical analysis
All the 36 microbial isolates were scored for the different PGP properties including production of IAA-like substances, dissolution of insoluble phosphates [Ca3(PO4)2, AlPO4, FePO4, Zn3(PO2)4, and Na-phytate], and dissolution of insoluble zinc complexes (ZnO and ZnSO4). The data on the total (cumulative) score of each of the 36 isolates are presented in Figure 5 (Supplementary Table 2). The isolate CME6-4 (5.16) got the highest score followed by CME6-5 (3.78), CI11-3 (3.25), CJ7-1 (3.00), CME6-8 (2.28), and CMA10-1 (2.08).
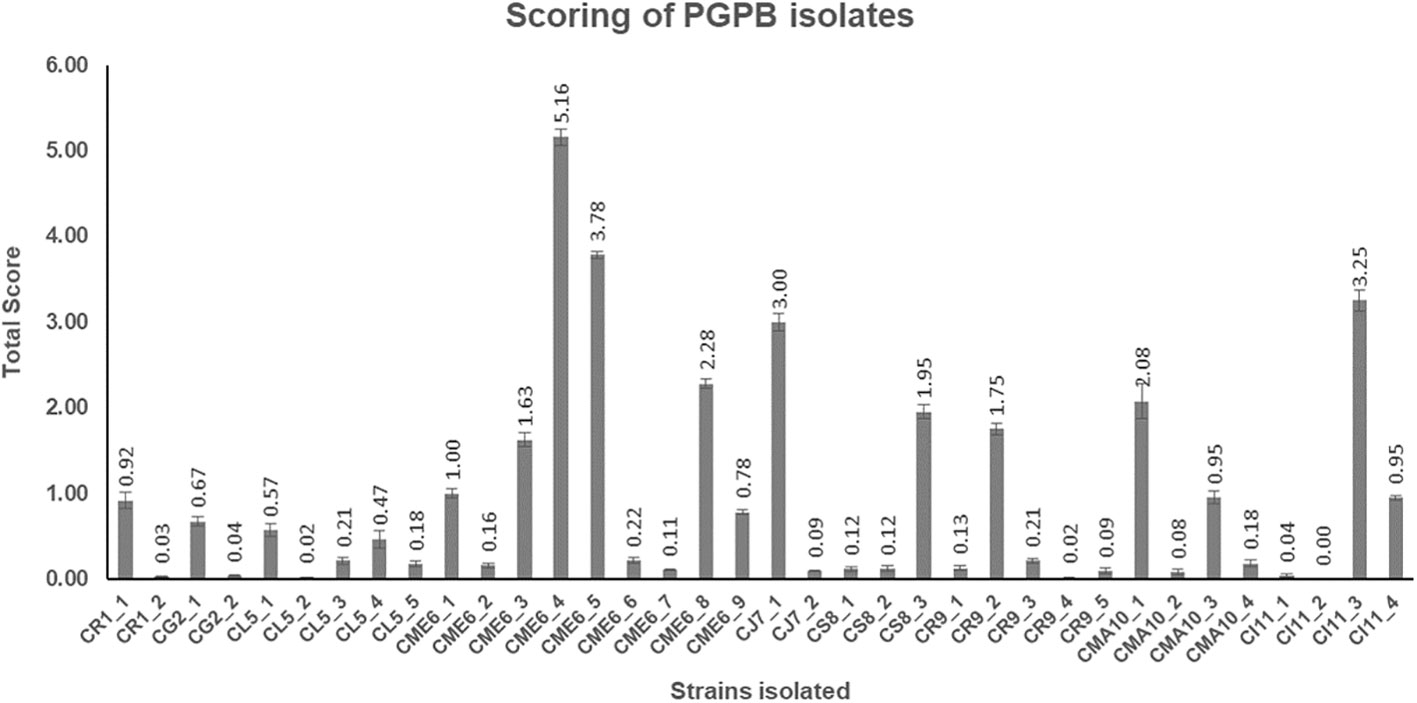
Figure 5 Total score obtained by each microbial isolate on the basis of their multifaceted plant growth–promoting (PGP) properties.
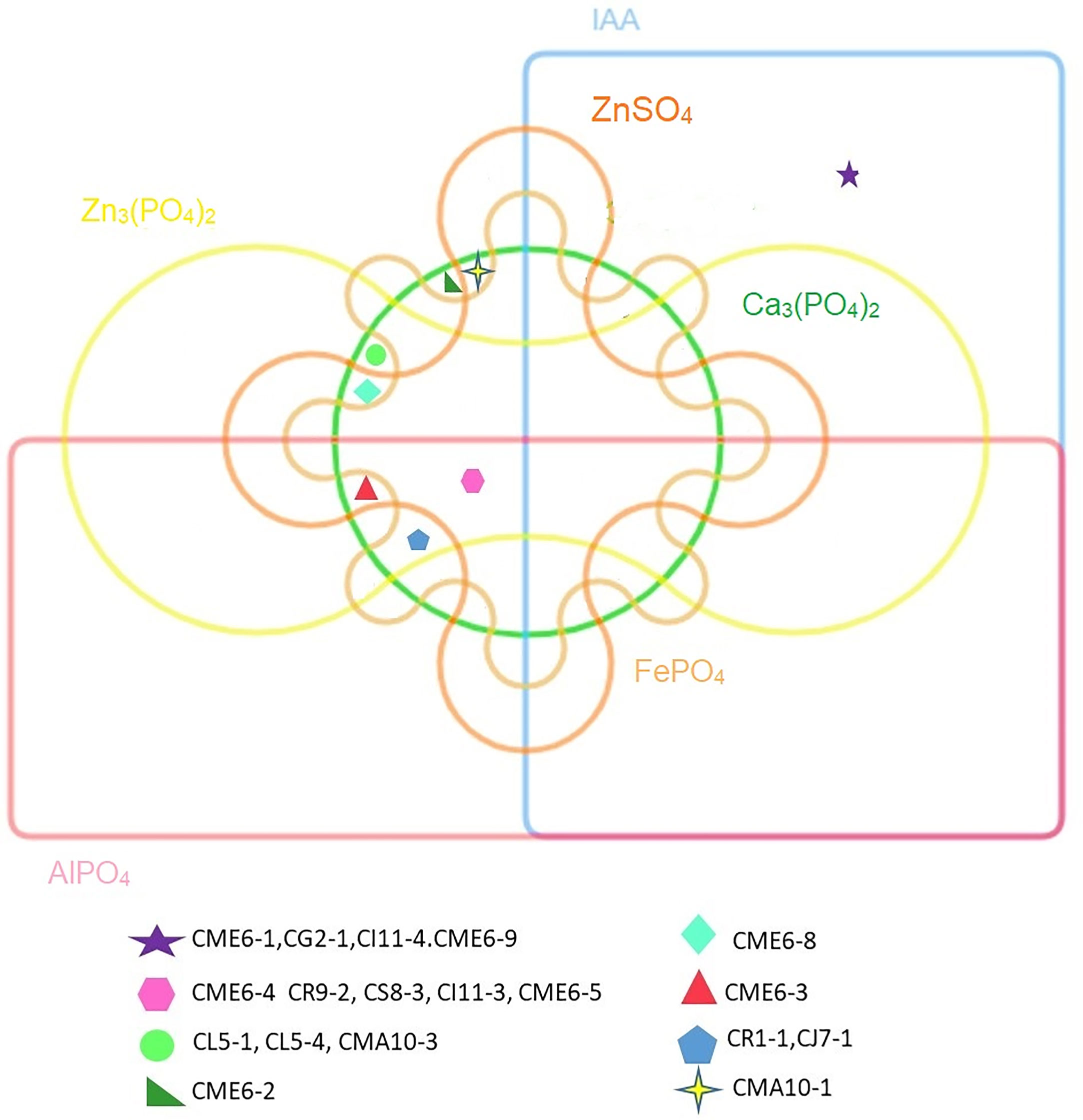
The statistical analysis performed using PCA showed the multivariate interaction between the PGP traits. There were three distinct clusters separated along PC axis 1 that explained 51.1% variability in terms of PGP traits (Figure 6). The variables, namely, dissolution of Ca3(PO4)2, Zn3(PO4)2, ZnSO4, FePO4, and AlPO4 and mineralization of Na-phytate contributed significantly in formation of clusters along PC axis 1. On the other hand, production of IAA-like substances and N2-fixation contributed significant variability (20.6%) in separating bacterial isolates along PC axis 2. The coefficients in the linear combinations of variables making up PC axis 1 were dissolution of Ca3(PO4)2, Zn3(PO4)2, ZnSO4, FePO4, AlPO4, and mineralization of Na-phytate and PC axis 2 were production of IAA-like substances and N2-fixation (Table 3). The Venn diagram in Figure 7 shows the microbial isolates from citrus seeds that exhibit common PGP features such as phosphate solubilization, zinc sulfate solubilization, and IAA production.
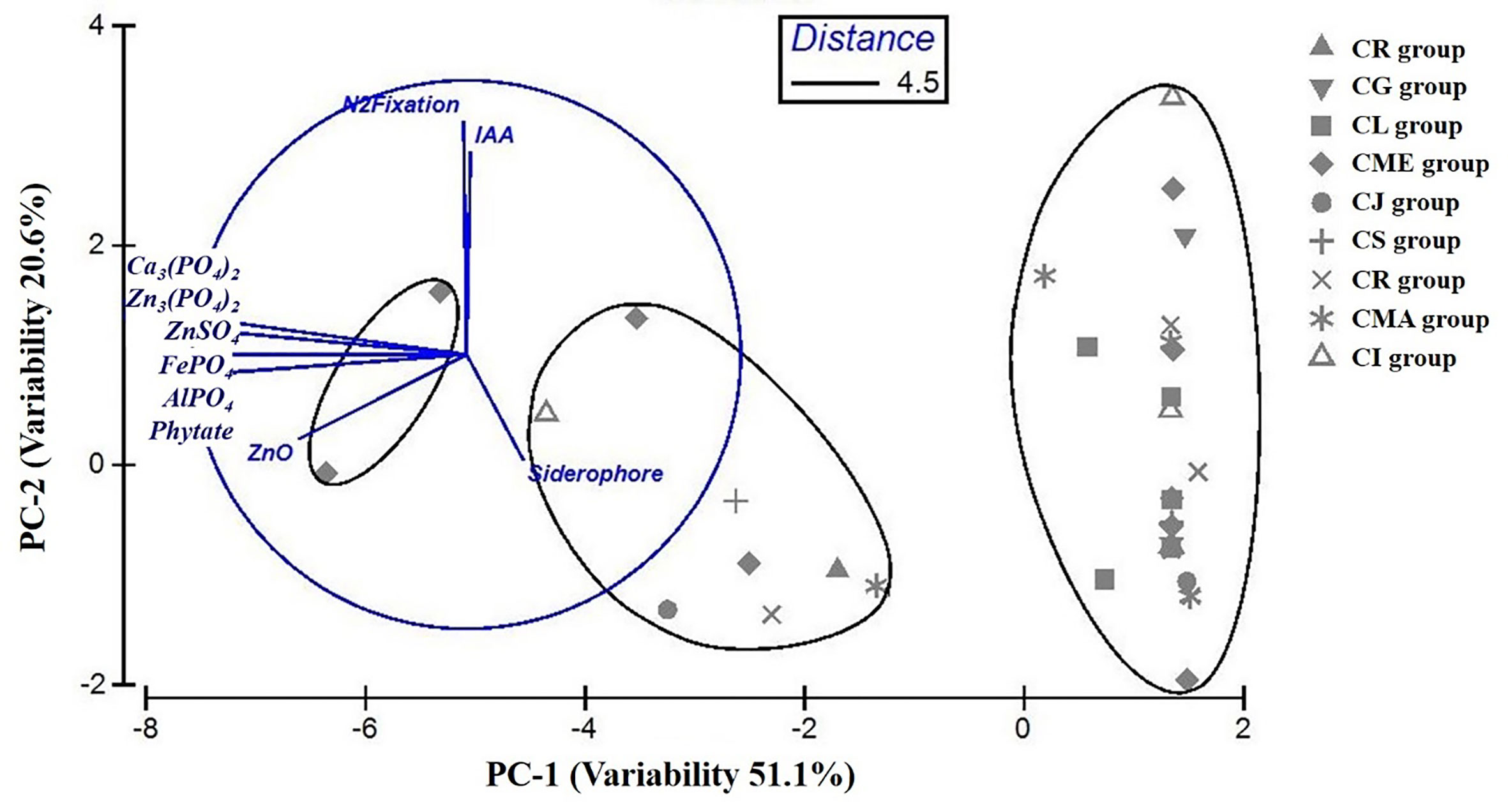
Figure 6 Comparison of variability in PGP traits among bacterial isolates obtained from nine different citrus species performed by principal component analysis (PCA). Ellipses represent superimposed hierarchical clusters (Euclidean distance 4.5) based on the Euclidean distance matrix, deduced using group-average linking incorporating similarity of profile at 95% confidence limit.
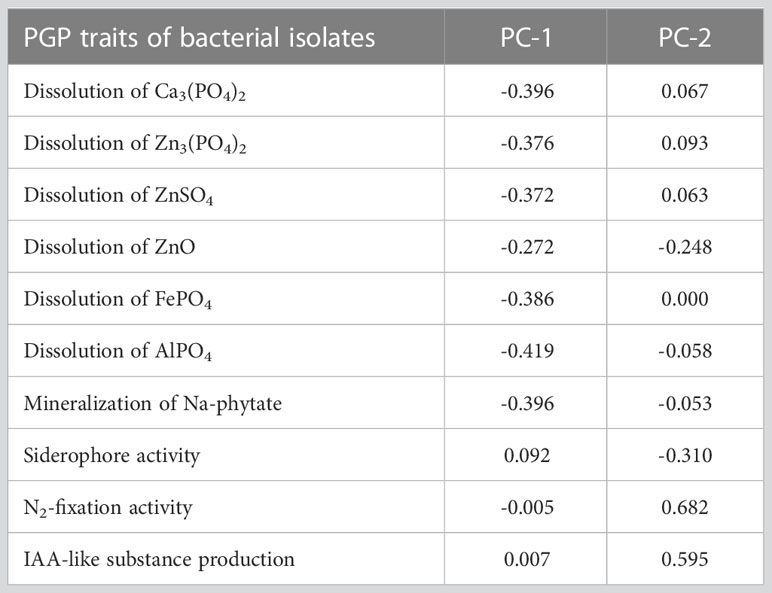
Table 3 Coefficients in the linear combinations of variables (PGP traits) making up principal component axes in the principal component plot.
Molecular characterization of isolated strains
Bacterial isolates characterized for multiple PGP traits were selected for sequencing of 16S rRNA genes for determination of identity. The bacterial isolates were found to have homology with the genus Bacillus and are listed in Table 4. The sequence information was submitted to the GenBank accession. The consensus phylogenetic tree constructed with the nearest member of the genus Bacillus is depicted in Figure 8 along with the bootstrap value.
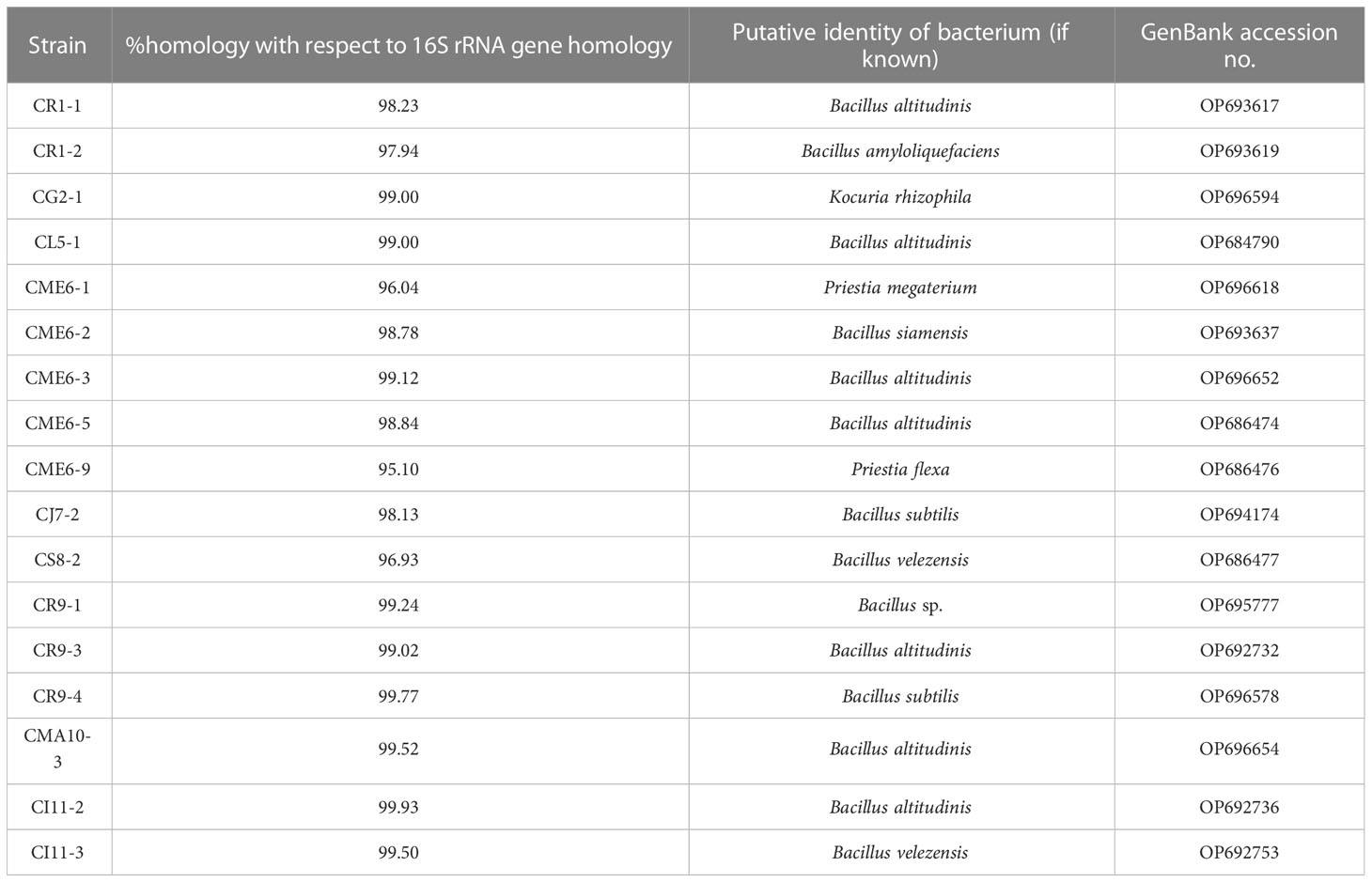
Table 4 Phylogeny of selected culturable bacterial isolates obtained from the seeds of citrus species of the northeastern region.

Figure 8 Phylogenetic consensus tree for the isolated strains with reference sequences inferred from the Maximum Parsimony algorithm using MEGA v.11.
Discussion
The Food and Agriculture Organization of the United Nations estimated that during the past century, agricultural crops have lost approximately 75% of their genetic diversity because of widespread desertion of genetically diverse traditional crops in favor of genetically uniform modern crop types. According to the concept of a “plant holobiont,” a plant is a coevolved species conglomerate made up of an interkingdom community of bacterial, archaeal, fungal, and other eukaryotic species (Vandenkoornhuyse et al., 2015). The domestication and cultivation of agricultural plants may have a detrimental effect on the beneficial microbial community of plants. According to Pérez-Jaramillo et al. (2016), the “back to the roots” outlook calls for investigating the microbiomes of wild relatives of agricultural plant species and their natural environments in order to identify beneficial bacteria that may have been lost during domestication and reintroduce them in seeds of current crop species. The northeastern region is renowned as a treasure house of citrus germplasm and is home to several landraces of the citrus species, many of which remain unexploited due to the lack of commercial cultivation (Singh and Singh, 2006). In the current work, healthy fruits of various citrus species were collected from different parts of northeastern India. Microbial strains were isolated from the surface-sterilized seeds of the fruits, which were further screened for features that promote plant growth. 16S rRNA–based molecular characterization was carried out for taxonomic identification of the seed microbiome.
A total of 36 microbial isolates were recovered from seeds of several citrus species, including 2 from Sikkim orange seeds, 2 from pummelo seeds, 5 from khasi papeda seeds, 9 from sour pummelo seeds, 2 from rough lemon seeds, 3 from sweet orange seeds, 5 from khasi mandarin seeds, 4 from melamesian papeda seeds, and 4 from Indian wild orange seeds. The colony isolates displayed a variety of shapes, of which the majority were circular, while a small number were irregular, most had glistening appearances and dull appearances were rare, flat to elevated surfaces, and white colony colors for the majority of strains, with cream, yellow, and orange pigments for a few. Faddetta et al. (2021) reported 60 endophytes from C. limon seeds that were streaked on a nutrient agar plate on the basis of distinctive colony morphology and pigmentation. Numerous studies have reported the association of microorganisms with the seeds of various crops like maize, rice, wheat, rice cut grass, beans, chickpea, cucurbits, and tomato (Johnston-Monje and Raizada, 2011; Verma et al., 2017;. Herrera et al., 2016; Khalaf and Raizada, 2016; Walitang et al., 2017; Morella et al., 2019). Different strains of epiphytic bacteria such as Bacillus amyloliquefaciens and Pseudomonas parafulva have been found to be associated with the fruits of several citrus species, including C. unshiu, C. reticulata, and C. sinensis and tend to have positive effect on plant health (Jing et al., 2023). PGPR isolated from the rhizospheres of different citrus species was prepared in an aqueous suspension and then used as a foliar spray and soil-drench, which led to increased crop growth (Chakraborty et al., 2016).
The auxin family member that has been investigated the most is indole-3-acetic acid, and tryptophan is the primary IAA precursor in five of the major pathways used by bacteria to produce auxin (Gamalero and Glick, 2011). The primary effects of bacterial IAA on plants include increased lateral and adventitious rooting, which improves nutrient uptake and root exudation, which, in turn, promotes bacterial growth on the roots (Thakuria et al., 2004). The synthesis of IAA-like compounds by the 36 isolates in the current investigation ranged from 4.8 ± 0.3 to 187.3 ± 18.1 µg ml-1 h-1 in the presence of L-tryptophan as a precursor among which CME6-1, CI11-4, CME6-9, and CG2-1 are the highest producers. The results were consistent with those of Verma et al. (2017), who reported that two isolates of rex rice (Oryza sativa L.) seed endophytic bacteria, E. asburiae and P. dispersa, produced 70.81 ± 0.98 and 7.98 ± 0.58 µg ml-1 IAA, respectively. Faddetta et al. (2021) reported IAA production ability by three microbial endophytes isolated from C. limon seeds. Several other researchers have also noted the capacity of seed microbial endophytes to produce IAA in a variety of crops, including cucurbits (Khalaf and Raizada, 2016), rice (Walitang et al., 2017), marama beans (Chimwamurombe et al., 2016), maize (Sandhya et al., 2017), and wheat (Herrera et al., 2016).
Phosphate solubilization is one of the most crucial bacterial physiological features in soil biogeochemical cycles and in plant growth promotion by PGPB. In the current study, the capacity of dissolution of insoluble phosphates was determined on Pikovskaya’s media amended individually with various inorganic phosphate complexes, which include Ca3(PO4)2, AlPO4, FePO4 and Zn3(PO4)2. In the Ca3(PO4)2-amended Pikovskaya’s agar plate, 14 isolates produced solubilization zones with solubilization ranging from 1.31 ± 0.2 to 14.84 ± 0.3 µg Pi ml-1 h-1. Nine bacterial isolates created a solubilization zone with a degree of solubilization between 1.54 ± 0.02 and 4.61 ± 0.2 µg Pi ml-1 h-1 in FePO4-amended Pikovskaya’s media, while nine bacterial isolates produced a solubilization zone with a degree of solubilization ranging from 1.74 ± 0.3 to 3.61 ± 0.4 µg Pi ml-1 h-1 in AlPO4-amended Pikovskaya’s media. The Zn3(PO4)2 dissolution ranged between 2.44 ± 0.03 and 13.16 ± 0.13 µg Pi ml-1 h-1 among 12 isolates that showed a solubilization zone in Zn3(PO4)2-amended Pikovskaya’s agar plate. The findings were supported by Thokchom et al. (2014), who reported that on Ca3(PO4)2-supplemented medium, 54.8% of mandarin orange rhizobacterial strains could solubilize insoluble P with phosphate solubilization ranging from between 6.2 ± 1.1 and 267.9 ± 24.1 µg ml-1. The ability of seed endophytes to solubilize phosphate has been documented in numerous papers Endophytes isolated from Rex Rice seed was found with phosphate solubilization properties (Verma et al., 2017). Johnston-Monje and Raizada (2011) reported 63 isolates obtained from Zea mays that solubilized phosphate. Seed-associated endophytes of cucurbit family reported 21% of strains, primarily Bacillus, lactic acid bacteria and Enterobacteriaceae, showed phosphate solubilization on Ca3(PO4)2-amended plates (Khalaf and Raizada, 2016). The outcomes also agreed with those of Faddetta et al. (2021), who discovered that bacteria cultivated from C. limon seeds could flourish on solid media containing various sources of inorganic phosphate (Ca-, Fe-, and Al-). Gluconacetobacter diazotrophicus was reported to solubilize Zn3(PO4)2 (Saravanan et al., 2007). The primary strategy used by phosphate- solubilizing bacteria to dissolve inorganic P is the production of low-molecular-weight organic acids, which bind phosphate with their hydroxyl and carboxyl groups, chelating cations, and contributing soil acidification, both of which result in the release of soluble phosphate (Rodriguez et al., 2004). Phosphorus is stored in plant seeds and pollen in form of phytate as a primary inositol source. Many seeds, notably those from grains and legumes, store phosphorus mostly in the form of phytate, which promotes the growth of seed endophytes during germination. Bean seed phytate solubilizers include Acinetobacter, Bacillus, Streptomyces, and Rhizobium endophyticum (López-López et al., 2010). Three bacteria isolated from surface-sterilized seeds of rice have been reported to act as phytate solubilizers (Verma et al., 2017), and isolated seed microbial endophytes of C. limon were able to grow on organic phosphate, i.e., Na-phytate-amended agar plates (Faddetta et al., 2021). The results of the current study revealed nine isolates with solubilizing capacities for sodium phytate ranging from 12.17 ± 0.5 to 18.00 ± 0.8 µg Pi ml-1 h-1.
Citrus decline is thought to be a condition with numerous contributing variables. However, a clear causal involvement of zinc nutrition as a contributor to citrus decline/blight have been hypothesized by many. Zinc-solubilizing bacteria are potential substitutes for zinc supplements because they transform applied inorganic zinc into usable forms. In this study, the zinc solubilization efficiency of bacterial isolates was investigated on modified Bunt and Rovira medium amended separately with two insoluble zinc sources, namely, ZnO and ZnSO4. Seven isolates were able to solubilize ZnO (solubilization efficiencies ranged from 64 to 111.68), with isolate CJ7-1 having the largest solubilization diameter of 86 mm. Eight isolates solubilize ZnSO4 (solubilization efficiencies ranged from 60.4 to 290.45, with the isolate CME6-5 (B. altutidinis) having the largest solubilization diameter of 164.4 mm. The findings were consistent with those of Saravanan et al. (2004) who reported 17 bacterial isolates with strain ZSB-O-1 (Bacillus sp.) producing a clearing zone of 2.80 cm on ZnSO4 and 1.80 cm of clearing zone with ZnO-containing medium, and G. diazotrophicus strain produced a solubilization zone diameter of 36 mm (Saravanan et al., 2007).
Since nitrogen makes up approximately 1.5%–2.0% of a plant’s dry matter and 16% of its total protein, plants need more nitrogen than other organisms. Different parts of plant act as microniches for the colonization of endophytic diazotrophs, where without competition with other microorganisms and absence of soil limitations present in the rhizosphere, the endophytes fix nitrogen (Verma et al., 2017). In the present study, 36% of the total microbe isolates were able to fix N2 in N-free bromothymol blue media distinguished by a shift in the medium’s color from green to blue. Other studies have also reported the N2 fixation ability by seed endophytes of Tylosema esculentum (Chimwamurombe et al., 2016), cucurbit family (Khalaf and Raizada, 2016) and Zea species (Johnston-Monje and Raizada, 2011).
Siderophore production is a vital characteristic of endophytic bacteria. These bacteria enhance plant growth and health as they synthesize siderophores that chelate iron in an iron-limited condition making it accessible for the plants and unavailable for phytopathogens (Mercado-Blanco and Bakker, 2007). In the present study, nine isolates of the total isolated strains were able to synthesize siderophores on CAS blue agar medium evident by formation of a halo zone around the colony. The findings were in line with the results of Chimwamurombe et al. (2016) who reported 13 isolates of T. esculentum seed positive for siderophore production and Herrera et al. (2016) who found six bacterial strains isolated from wheat seed positive for siderophore production.
HCN toxicity is influenced by its capacity to inhibit cytochrome c oxidase as well as other significant metalloenzymes (Nandi et al., 2017). While the ability of bacteria to make HCN is often considered as a negative factor, it has been suggested that HCN-producing endophytic bacteria may have important applications for the biocontrol of weeds, soil phytopathogenic fungi, and nematodes. In this study, only three isolates produced HCN in amounts that were positive. Phragmites australis seed endophytes (Verma et al., 2018), maize seed and root endophytes (Sandhya et al., 2017), and endophytes from nodules and roots of chickpea and pea (Maheshwari et al., 2019) have all been documented to be capable of producing HCN.
A fascinating feature in the current study is that the genus Bacillus appears to make up the culturable core seed microbiota across all the examined citrus species. A distinctive characteristic of seed endophytes seen in prior studies is the capacity to produce endospores, which are assumed to protect the inhabitants of the seed from internal changes (Truyens et al., 2015). Munir et al. (2020) reported that B. velezensis, Curtobacterium luteum, Microbacterium testaceum, and B. subtilis as the most prevalent endophytes in the leaves of 24 citrus types, with B. subtilis occurring most frequently in healthy plants. In the leaf tissues of numerous citrus rootstocks, the predominated bacterial species was found to be Pantoea agglomerans and B. pumilus (Araújo et al., 2001). Proteobacteria, Actinobacteria, Bacteroidetes, and Firmicutes were the most prevalent bacterial phyla in the microbial endophytic community of C. limon seeds. (Faddetta et al., 2021). Further, the genus Bacillus has been reported as common member of seed microbiota in different crops such as rice (Cottyn et al., 2001), gurana (e Silva et al., 2016), beans (López-López et al., 2010), and cucurbit family (Khalaf and Raizada, 2016). Members of the genus Bacillus is functionally diverse and one of the most widely utilized bacteria in the field of agrobiotechnology. It is well known that members of the genus Bacillus possess a multitude of beneficial attributes that aid plants either directly or indirectly by acquisition of nutrients, phytohormone production, and shielding plants from biotic and abiotic stress. In this study, various isolates obtained from the citrus seed belonging mainly to the Bacillus genus showed multifaceted plant growth promotion traits like nitrogen fixation, phosphate and zinc solubilization, and IAA production. Nitrogen fixation ability has been reported by B. pumilus and B. altitudinis that were isolated from the rhizosphere of maize and rice, respectively (Habibi et al., 2014). There were 16 of the 19 distinct Bacillus strains screened for the nifH gene that were found to be positive (Yousuf et al., 2017). According to Sharma et al. (2013), many species of Bacillus including B. megaterium M510, which was isolated from the maize rhizosphere in the eastern Himalayan region, were found to be capable of solubilizing iron and aluminum phosphate in addition to moderately solubilizing tricalcium phosphate. B. subtilis, B. thuringiensis, B. aryabhattai, and B. tequilensis are well-known zinc solubilizers (Singh et al., 2017). The isolates isolated from the chickpea rhizosphere that were highly similar to B. altitudinis displayed PGP traits including IAA, ammonia, siderophores, and phosphate solubilization (Kushwaha et al., 2021). K. rhizophila, isolated from citrus seed in this study was found to be a high IAA producer, while K. rhizophila Y1 isolated from maize rhizosphere soil exhibited growth-promoting characteristics such as phosphate solubilization and IAA synthesis (Li et al., 2020).
Microbial strains with PGP properties have the potential to be used in the development of bioinoculants, biofertilizers, and biopreparations that improve crop plant growth and yield. They are committed to being used in organic/natural farming as they increase crop yields enhancing the uptake of nutrition and improving plant vigor, combat pathogens, and increase plant abiotic resistance while conferring habitat-adapted tolerance to the host plant (Pylak et al., 2019). Endophytic isolates were evaluated as bioinoculants for potato tubers and recommended as biofertilizers to reduce dependency on chemical fertilizers (Kuźniar et al., 2019). Works have been focused on the evaluation of compatibility of the different PGP strains and prospecting the optimal combinations of the isolates to develop into bioinoculants for commercialization. Bacillus strains have been most frequently utilized as the common commercially available bioinoculants till date (Kuźniar et al., 2019). For instance, Bacillus species have successfully been used as priming agents in crops such as potato (Gururani et al., 2013), rice, mung bean, and chickpea (Chakraborty et al., 2011). As such, additional research is needed to translate the in vitro PGP activities of the isolates into pot experiments in greenhouse and field trials for improved crop yield.
Conclusion
Our study clearly demonstrated that citrus seeds are an important source of beneficial seed microbiomes and that members belonging to genus Bacilli seem to be the core member as common dominant bacteria across eight citrus species tested. The culturable members of the citrus seed microbiome possessed multifaceted PGP traits like dissolution of insoluble phosphate and zinc complexes, mineralization of the organic phosphate complex, production of IAA-like substances, siderophore-like substances and HCN, and nitrogen fixation. Isolates CG2-1, CME6-1, CME6-4, CME6-5, CME6-8, CME6-9, CJ7-1, CMA10-1, CI11-3, and CI11-4 are promising candidate bioinoculants for development of microbial consortium having multifaceted PGP traits for nutritional benefits of nitrogen, phosphorus, and zinc and IAA hormonal benefits to citrus crops for better fitness in acid soils. Bacterial seed endophytes may be highly conserved in plant species to specific regions, necessitating the exploration of species from different regions. Additionally, research can be conducted using different growth media other than LB to increase the number of cultivable seed microbiomes from different citrus species. The isolated beneficial strains and their consortium will be tested further in greenhouse and field trials under natural environmental conditions.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author contributions
DT conceived and designed the study, validated results, and supervised the final draft of manuscript. SS, CC, and PU executed the experiments and wrote the first draft of manuscript. SH, PD, AS, and BL helped in citrus sample collection and checking the draft manuscript. All authors contributed to the article and approved the submitted version.
Funding
Authors would like to acknowledge the Department of Biotechnology, Govt. of India, New Delhi for granting the project research entitled “Collection conservation and morpho-phenological characterization of citrus germplasm of NE India (BT/PR39933/NER/95/1658/2020)”.
Acknowledgments
The authors would like to acknowledge the Central Agricultural University, Imphal, Manipur for laboratory support.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1104927/full#supplementary-material
References
Abdullaeva, Y., Manirajan, B. A., Honermeier, B., Schnell, S., Cardinale, M. (2021). Domestication affects the composition, diversity, and co-occurrence of the cereal seed microbiota. J. Adv. Res. 31, 75–86. doi: 10.1016/j.jare.2020.12.008
Anmol, R. J., Marium, S., Hiew, F. T., Han, W. C., Kwan, L. K., Wong, A. K. Y., et al. (2021). Phytochemical and therapeutic potential of Citrus grandis (L.) osbeck: a review. J. Evid. Based Integr. Med. 26, 2515690X211043741. doi: 10.1177/2515690X211043741
Araújo, W. L., Maccheroni, W., Jr., Aguilar-Vildoso, C. I., Barroso, P. A., Saridakis, H. O., Azevedo, J. L. (2001). Variability and interactions between endophytic bacteria and fungi isolated from leaf tissues of citrus rootstocks. Can. J. Microbiol. 47 (3), 229–236. doi: 10.1139/w00-146
Azevedo, J. L. D., Araujo, W. L. D. (2007). “Diversity and applications of endophytic fungi isolated from tropical plants,” in Fungi: multifaceted microbes (Boca Raton: CRC Press), 189–207.
Berg, G. (2009). Plant–microbe interactions promoting plant growth and health: perspectives for controlled use of microorganisms in agriculture. Appl. Microbiol. Biotechnol. 84, 11–18. doi: 10.1007/s00253-009-2092-7
Chakraborty, B. N., Allay, S., Chakraborty, A. P., Chakraborty, U. (2016). PGPR in managing root rot disease and enhancing growth in mandarin (Citrus reticulata blanco.) seedlings. J. Hortic. Sci. 11 (2), 104–115.
Chakraborty, U., Roy, S., Chakraborty, A. P., Dey, P., Chakraborty, B. (2011). Plant growth promotion and amelioration of salinity stress in crop plants by a salt-tolerant bacterium. Recent Res. Sci. Technol. 3 (11), 61–70.
Chimwamurombe, P. M., Grönemeyer, J. L., Reinhold-Hurek, B. (2016). Isolation and characterization of culturable seed-associated bacterial endophytes from gnotobiotically grown marama bean seedlings. FEMS Microbiol. Ecol. 92 (6). doi: 10.1093/femsec/fiw083
Comby, M., Lacoste, S., Baillieul, F., Profizi, C., Dupont, J. (2009). Spatial variation of cultivable communities of co-occurring endophytes and pathogens in wheat. Front. Microbiol. 7. doi: 10.3389/fmicb.2016.00403
Cottyn, B., Regalado, E., Lanoot, B., De Cleene, M., Mew, T. W., Swings, J. (2001). Bacterial populations associated with rice seed in the tropical environment. Phytopathology 91 (3), 282–292. doi: 10.1094/PHYTO.2001.91.3.282
Defez, R., Andreozzi, A., Bianco, C. (2017). The overproduction of indole-3-acetic acid (IAA) in endophytes upregulates nitrogen fixation in both bacterial cultures and inoculated rice plants. Microb. Ecol. 74 (2), 441–452. doi: 10.1007/s00248-017-0948-4
e Silva, M. C. S., Polonio, J. C., Quecine, M. C., de Almeida, T. T., Bogas, A. C., Pamphile, J. A., et al. (2016). Endophytic cultivable bacterial community obtained from the Paullinia cupana seed in Amazonas and bahia regions and its antagonistic effects against Colletotrichum gloeosporioides. Microb. Pathog. 98, 16–22. doi: 10.1016/j.micpath.2016.06.023
Faddetta, T., Abbate, L., Alibrandi, P., Arancio, W., Siino, D., Strati, F., et al. (2021). The endophytic microbiota of Citrus limon is transmitted from seed to shoot highlighting differences of bacterial and fungal community structures. Sci. Rep. 11 (1), 1–12. doi: 10.1038/s41598-021-86399-5
Gamalero, E., Glick, B. R. (2011). “Mechanisms used by plant growth-promoting bacteria,” in Bacteria in agrobiology: plant nutrient management. Ed. Maheshwari, D. (Berlin, Heidelberg: Springer). doi: 10.1007/978-3-642-21061-7_2
Gandolfi, I., Bertolini, V., Ambrosini, R., Bestetti, G., Franzetti, A. (2013). Unravelling the bacterial diversity in the atmosphere. Appl. Microbiol. Biotechnol. 97 (11), 4727–4736. doi: 10.1007/s00253-013-4901-2
Giauque, H., Connor, E. W., Hawkes, C. V. (2019). Endophyte traits relevant to stress tolerance, resource use and habitat of origin predict effects on host plants. New Phytol. 221 (4), 2239–2249. doi: 10.1111/nph.15504
Glick, B. R. (2005). Modulation of plant ethylene levels by the bacterial enzyme ACC deaminase. FEMS Microbiol. Lett. 251, 1–7. doi: 10.1016/j.femsle.2005.07.030
Gururani, M. A., Upadhyaya, C. P., Baskar, V., Venkatesh, J., Nookaraju, A., Park, S. W. (2013). Plant growth-promoting rhizobacteria enhance abiotic stress tolerance in solanum tuberosum through inducing changes in the expression of ROS-scavenging enzymes and improved photosynthetic performance. J. Plant Growth Regul. 32, 245–258. doi: 10.1007/s00344-012-9292-6
Habibi, S., Djedidi, S., Prongjunthuek, K., Mortuza, M. F., Ohkama-Ohtsu, N., Sekimoto, H., et al. (2014). Physiological and genetic characterization of rice nitrogen fixer PGPR isolated from rhizosphere soils of different crops. Plant Soil. 379 (1), 51–66. doi: 10.1007/s11104-014-2035-7
Hardoim, P. R., Hardoim, C. C., Van Overbeek, L. S., Van Elsas, J. D. (2012). Dynamics of seed-borne rice endophytes on early plant growth stages. PloS One 7 (2), e30438. doi: 10.1371/journal.pone.0030438
Hardoim, P. R., van Overbeek, L. S., Berg, G., Pirttilla, A. M., Compant, S., Campisano, A., et al. (2015). The hidden world within plants: ecological and evolutionary considerations for defining functioning of microbial endophytes. Microbiol. Mol. Biol. Rev. 79, 293–320. doi: 10.1128/MMBR.00050-14
Hazarika, T. K. (2012). Citrus genetic diversity of north-east India, their distribution, ecogeography and ecobiology. Genet. Resour. Crop Evol. 59 (6), 1267–1280. doi: 10.1007/s10722-012-9846-2
Herrera, S. D., Grossi, C., Zawoznik, M., Groppa, M. D. (2016). Wheat seeds harbour bacterial endophytes with potential as plant growth promoters and biocontrol agents of Fusarium graminearum. Microbiol. Res. 186-187, 37–43. doi: 10.1016/j.micres.2016.03.002
Hong, C. E., Park, J. M. (2016). Endophytic bacteria as biocontrol agents against plant pathogens: current state-of-the-art. Plant Biotechnol. Rep. 10, 353–357. doi: 10.1007/s11816-016-0423-6
Jing, J., Wang, W., Deng, L., Yi, L., Zeng, K. (2023). A core epiphytic bacterial consortia synergistically protect citrus from postharvest disease. Food Chem. 407, 135103. doi: 10.1016/j.foodchem.2022.135103
Johnston-Monje, D., Raizada, M. N. (2011). Conservation and diversity of seed associated endophytes in zea across boundaries of evolution, ethnography and ecology. PloS One 6 (6), e20396. doi: 10.1371/journal.pone.0020396
Khalaf, E. M., Raizada, M. N. (2016). Taxonomic and functional diversity of cultured seed associated microbes of the cucurbit family. BMC Microbiol. 16 (1), 1–6. doi: 10.1186/s12866-016-0743-2
Kushwaha, P., Srivastava, R., Pandiyan, K., Singh, A., Chakdar, H., Kashyap, P. L., et al. (2021). Enhancement in plant growth and zinc biofortification of chickpea (Cicer arietinum l.) by Bacillus altitudinis. J. Soil Sci. Plant Nutr. 21, 922–935. doi: 10.1007/s42729-021-00411-5
Kuźniar, A., Włodarczyk, K., Grządziel, J., Woźniak, M., Furtak, K., Gałązka, A., et al. (2020). New insight into the composition of wheat seed microbiota. Int. J. Mol. Sci. 21 (13), 4634. doi: 10.3390/ijms21134634
Kuźniar, A., Włodarczyk, K., Wolińska, A. (2019). Agricultural and other biotechnological applications resulting from trophic plant-endophyte interactions. Agronomy 9, 779. doi: 10.3390/agronomy9120779
Li, X., Sun, P., Zhang, Y., Jin, C., Guan, C. (2020). A novel PGPR strain Kocuria rhizophila Y1 enhances salt stress tolerance in maize by regulating phytohormone levels, nutrient acquisition, redox potential, ion homeostasis, photosynthetic capacity and stress-responsive genes expression. Environ. Exp. Bot. 174, 104023. doi: 10.1016/j.envexpbot.2020.104023
Liu, Y., Heying, E., Tanumihardjo, S. A. (2012). History, global distribution, and nutritional importance of citrus fruits. Compr. Rev. Food Sci. Food Saf. 11 (6), 530–545. doi: 10.1111/j.1541-4337.2012.00201.x
López-López, A., Rogel, M. A., Ormeno-Orrillo, E., Martínez-Romero, J., Martínez-Romero, E. (2010). Phaseolus vulgaris seed-borne endophytic community with novel bacterial species such as rhizobium endophyticum sp. nov. Syst. Appl. Microbiol. 33 (6), 322–327. doi: 10.1016/j.syapm.2010.07.005
Lorck, H. (1948). Production of hydrocyanic acid by bacteria. Physiol. Plant 1 (2), 142–146. doi: 10.1111/j.1399-3054.1948.tb07118.x
Louden, B. C., Haarmann, D., Lynne, A. M. (2011). Use of blue agar CAS assay for siderophore detection. J. Microbiol. Biol. Educ. 12 (1), 51–53. doi: 10.1128/jmbe.v12i1.249
Maheshwari, R., Bhutani, N., Bhardwaj, A., Suneja, P. (2019). Functional diversity of cultivable endophytes from Cicer arietinum and Pisum sativum: bioprospecting their plant growth potential. Biocatal. Agric. Biotechnol. 20, 101229. doi: 10.1016/j.bcab.2019.101229
Malik, S. K., Chaudhury, R., Dhariwal, O. P., Kalia, R. K. (2006). Collection and characterization of Citrus indica Tanaka and C. macroptera montr.: wild endangered species of northeastern India. Genet. Resour. Crop Evol. 53 (7), 1485–1493. doi: 10.1007/s10722-005-7468-7
Malik, S. K., Kumar, S., Singh, I. P., Dhariwal, O. P., Chaudhury, R. (2013). Socio-economic importance, domestication trends and in situ conservation of wild citrus species of northeast India. Genet. Resour. Crop Evol. 60 (5), 1655–1671. doi: 10.1007/s10722-012-9948-x
Marchut-Mikolajczyk, O., Drożdżyński, P., Pietrzyk, D., Antczak, T. (2018). Biosurfactant production and hydrocarbon degradation activity of endophytic bacteria isolated from chelidonium majus l. Microb. Cell Fact. 17 (1), 1–9. doi: 10.1186/s12934-018-1017-5
Mercado-Blanco, J., Bakker, P. A. (2007). Interactions between plants and beneficial Pseudomonas spp.: exploiting bacterial traits for crop protection. Antonie Van Leeuwenhoek 92 (4), 367–389. doi: 10.1007/s10482-007-9167-1
Morella, N. M., Zhang, X., Koskella, B. (2019). Tomato seed-associated bacteria confer protection of seedlings against foliar disease caused by Pseudomonas syringae. Phytobiomes J. 3 (3), 177–190. doi: 10.1094/PBIOMES-01-19-0007-R
Munir, S., Li, Y., He, P., Huang, M., He, P., He, P., et al. (2020). Core endophyte communities of different citrus varieties from citrus growing regions in China. Sci. Rep. 10 (1), 1–12. doi: 10.1038/s41598-020-60350-6
Nandi, M., Selin, C., Brawerman, G., Fernando, W. G. D., de Kievit, T. (2017). Hydrogen cyanide, which contributes to Pseudomonas chlororaphis strain PA23 biocontrol, is upregulated in the presence of glycine. Biol. Control. 108, 47–54. doi: 10.1016/j.biocontrol.2017.02.008
Newcombe, G., Harding, A., Ridout, M., Busby, P. E. (2018). A hypothetical bottleneck in the plant microbiome. Front. Microbiol. 9. doi: 10.3389/fmicb.2018.01645
Okon, Y. (1985). Azospirillum as a potential inoculant for agriculture. Trends Biotech. 3 (9), 223–228. doi: 10.1016/0167-7799(85)90012-5
Okunishi, S., Sako, K., Mano, H., Imamura, A., Morisaki, H. (2005). Bacterial flora of endophytes in the maturing seed of cultivated rice (Oryza sativa). Microbes Environ. 20 (3), 168–177. doi: 10.1264/jsme2.20.168
Pandey, A., Tomer, A. K., Bhandari, D. C., Pareek, S. K. (2008). Towards collection of wild relatives of crop plants in India. Genet. Resour. Crop Evol. 55, 187–202. doi: 10.1007/s10722-007-9227-4
Penyalver, R., Roesch, L. F., Piquer-Salcedo, J. E., Forner-Giner, M. A., Alguacil, M. D. M. (2022). From the bacterial citrus microbiome to the selection of potentially host-beneficial microbes. New Biotechnol. 70, 116–128. doi: 10.1016/j.nbt.2022.06.002
Pérez-Jaramillo, J. E., Mendes, R., Raaijmakers, J. M. (2016). Impact of plant domestication on rhizosphere microbiome assembly and functions. Plant Mol. Biol. 90 (6), 635–644. doi: 10.1007/s11103-015-0337-7
Pikovskaya, R. I. (1948). Mobilization of phosphorus in soil in connection with vital activity of some microbial species. Mikrobiologiya 17, 362–370.
Pylak, M., Oszust, K., Frąc, M. (2019). Review report on the role of bioproducts, biopreparations, biostimulants and microbial inoculants in organic production of fruit. Rev. Environ. Sci. Biotechnol. 18 (3), 597–616. doi: 10.1007/s11157-019-09500-5
Ramesh, A., Sharma, S. K., Sharma, M. P., Yadav, N., Joshi, O. P. (2014). Inoculation of zinc solubilizing Bacillus aryabhattai strains for improved growth, mobilization and biofortification of zinc in soybean and wheat cultivated in vertisols of central India. App. Soil Ecol. 73, 87–96. doi: 10.1016/j.apsoil.2013.08.009
Rijavec, T., Lapanje, A., Dermastia, M., Rupnik, M. (2007). Isolation of bacterial endophytes from germinated maize kernels. Can. J. Microbial. 53 (6), 802–808. doi: 10.1139/W07-048
Rodriguez, H., Gonzalez, T., Goire, I., Bashan, Y. (2004). Gluconic acid production and phosphate solubilization by the plant growth-promoting bacterium Azospirillum spp. Naturwissenschaften 91, 552–555. doi: 10.1007/s00114-004-0566-0
Sandhya, V., Shrivastava, M., Ali, S. Z., Sai Shiva Krishna Prasad, V. (2017). Endophytes from maize with plant growth promotion and biocontrol activity under drought stress. Russ. Agric. Sci. 43 (1), 22–34. doi: 10.3103/S1068367417010165
Saravanan, V. S., Madhaiyan, M., Thangaraju, M. (2007). Solubilization of zinc compounds by the diazotrophic, plant growth promoting bacterium Gluconacetobacter diazotrophicus. Chemosphere 66 (9), 1794–1798. doi: 10.1016/j.chemosphere.2006.07.067
Saravanan, V. S., Subramoniam, S. R., Raj, S. A. (2004). Assessing in-vitro solubilization potential of different zinc solubilizing bacterial (ZSB) isolates. Braz. J. Microbiol. 35, 121–125. doi: 10.1590/S1517-83822004000100020
Schwyn, B., Neilands, J. B. (1987). Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 160, 47–56. doi: 10.1016/0003-2697(87)90612-9
Shabanamol, S., Dibya, K., George, T., Rishad, K., Sreekumar, T., Jisha, M. (2018). Characterization and in plant nitrogen fixation of plant-growth promoting endophytic diazotrophyc Lysinibacillus sphæricus isolated from rice (Oryza sativa). Physiol. Mol. Plant Pathol. 102, 46–54. doi: 10.1016/j.pmpp.2017.11.003
Sharma, B. D., Hore, D. K., Gupta, S. G. (2004). Genetic resources of citrus of north-eastern India and their potential use. Genet. Resour. Crop Evol. 51 (4), 411–418. doi: 10.1023/B:GRES.0000023456.70577.3d
Sharma, S. B., Sayyed, R. Z., Trivedi, M. H., Gobi, T. A. (2013). Phosphate solubilizing microbes: sustainable approach for managing phosphorus deficiency in agricultural soils. SpringerPlus 2 (1), 1–14. doi: 10.1186/2193-1801-2-587
Singh, D., Rajawat, M. V. S., Kaushik, R., Prasanna, R., Saxena, A. K. (2017). Beneficial role of endophytes in biofortification of zn in wheat genotypes varying in nutrient use efficiency grown in soils sufficient and deficient in zn. Plant Soil 416 (1), 107–116. doi: 10.1007/s11104-017-3189-x
Singh, I. P., Singh, S. (2006). “Citrus genetic resources and cultivars,” in Citrus: production and marketing of khasi mandarin, extension bulletin no. 35. Eds. Singh, S., Bujarbaruah, K. M. (NRCC, ICAR), 6–23.
Thakuria, D., Schmidt, O., Finan, D., Egan, D., Doohan, F. M. (2010). Gut wall bacteria of earthworms: a natural selection process. ISME J. 4 (3), 357–366. doi: 10.1038/ismej.2009.124
Thakuria, D., Schmidt, O., Mac Siúrtáin, M., Egan, D., Doohan, F. M. (2008). Importance of DNA quality in comparative soil microbial community structure analyses. Soil Biol. Biochem. 40 (6), 1390–1403. doi: 10.1016/j.soilbio.2007.12.027
Thakuria, D., Talukdar, N. C., Goswami, C., Hazarika, S., Boro, R. C., Khan, M. R. (2004). Characterization and screening of bacteria from rhizosphere of rice grown in acidic soils of Assam. Curr. Sci. 978-985.
Thokchom, E., Kalita, M. C., Talukdar, N. C. (2014). Isolation, screening, characterization, and selection of superior rhizobacterial strains as bioinoculants for seedling emergence and growth promotion of mandarin orange (Citrus reticulata blanco). Can. J. Microbiol. 60 (2), 85–92. doi: 10.1139/cjm-2013-0573
Truyens, S., Weyens, N., Cuypers, A., Vangronsveld, J. (2015). Bacterial seed endophytes: genera, vertical transmission and interaction with plants. Environ. Microbiol. Rep. 7, 40–50. doi: 10.1111/1758-2229.12181
Vandenkoornhuyse, P., Quaiser, A., Duhamel, M., Le Van, A., Dufresne, A. (2015). The importance of the microbiome of the plant holobiont. New Phytol. 206, 1196–1206. doi: 10.1111/nph.13312
Verma, S. K., Kingsley, K., Bergen, M., English, C., Elmore, M., Kharwar, R. N., et al. (2018). Bacterial endophytes from rice cut grass (Leersia oryzoides l.) increase growth, promote root gravitropic response, stimulate root hair formation, and protect rice seedlings from disease. Plant Soil 422, 223–238. doi: 10.1007/s11104-017-3339-1
Verma, S. K., Kingsley, K., Irizarry, I., Bergen, M., Kharwar, R. N., White, J. F. (2017). Seed vectored endophytic bacteria modulate development of rice seedlings. J. Appl. Microbiol. 122, 1680–1691. doi: 10.1111/jam.13463
Walitang, D. I., Kim, K., Madhaiyan, M., Kim, Y. K., Kang, Y., Sa, T. (2017). Characterizing endophytic competence and plant growth promotion of bacterial endophytes inhabiting the seed endosphere of rice. BMC Microbiol. 17 (1), 1–13. doi: 10.1186/s12866-017-1117-0
Xi, M., Deyett, E., Stajich, J. E., El-Kereamy, A., Roper, M. C., Rolshausen, P. E. (2023). Microbiome diversity, composition and assembly in a California citrus orchard. Front. Microbiol. 14. doi: 10.3389/fmicb.2023.1100590
Xu, M., Sheng, J., Chen, L., Men, Y., Gan, L., Guo, S., et al. (2014). Bacterial community compositions of tomato (Lycopersicum esculentum mill.) seeds and plant growth promoting activity of ACC deaminase producing Bacillus subtilis (HYT-12-1) on tomato seedlings. World J. Microbiol. Biotechnol. 30 (3), 835–845. doi: 10.1007/s11274-013-1486-y
Yang, C., Ancona, V. (2022). An overview of the mechanisms against “Candidatus liberibacter asiaticus”: virulence targets, citrus defenses, and microbiome. Front. Microbiol. 13. doi: 10.3389/fmicb.2022.850588
Keywords: phosphate complexes dissolution, zinc complexes dissolution, IAA production, N2 fixation, siderophore, microbial consortium
Citation: Sinha S, Thakuria D, Chaliha C, Uzir P, Hazarika S, Dutta P, Singh AK and Laloo B (2023) Plant growth–promoting traits of culturable seed microbiome of citrus species from Purvanchal Himalaya. Front. Plant Sci. 14:1104927. doi: 10.3389/fpls.2023.1104927
Received: 22 November 2022; Accepted: 30 May 2023;
Published: 10 July 2023.
Edited by:
Pramod Ramteke, Sam Higginbottom University of Agriculture, Technology and Sciences, IndiaReviewed by:
Dewa Ngurah Suprapta, Udayana University, IndonesiaTanvir Kaur, Eternal University, India
Copyright © 2023 Sinha, Thakuria, Chaliha, Uzir, Hazarika, Dutta, Singh and Laloo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dwipendra Thakuria, dGhha3VyaWEuZHdpcGVuZHJhQHlhaG9vLmNvLmlu
 Sakshi Sinha
Sakshi Sinha Dwipendra Thakuria
Dwipendra Thakuria Chayanika Chaliha
Chayanika Chaliha Panchali Uzir1
Panchali Uzir1 Samarendra Hazarika
Samarendra Hazarika Pranab Dutta
Pranab Dutta Bingiala Laloo
Bingiala Laloo