
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 19 January 2023
Sec. Plant Abiotic Stress
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1103340
This article is part of the Research Topic Identification and Functional Analysis of Differentially Expressed Genes in Plant Response to Abiotic Stresses View all 24 articles
Membrane transporters encoded by NITRATE TRANSPORTER 1/PEPTIDE TRANSPORTER (NPF) genes, which play crucial roles in plant growth, development and resistance to various stresses, are involved in the transport of nitrate (NO3-) and peptides. In several plant species, NPF genes are involved in the resistance to abiotic stresses; however, whether the whole NPF gene family in cotton contributes to this resistance has not been systematically investigated. Here, 201 genes encoding NPF proteins with a peptide transporter (PTR) domain were confirmed in three different Gossypium species, namely, Gossypium hirsutum, Gossypium arboreum and Gossypium raimondii. The NPF proteins in these three Gossypium species and Arabidopsis thaliana were classified into three different subfamilies via phylogenetic analysis. Among the genes that encode these proteins, most GhNPF genes in the same subfamily contained similar gene structures and conserved domains. Predictions of the promoters of these genes revealed that the cis-acting elements included phytohormone- and light-responsive elements, indicating that some of these genes might be expressed in response to abiotic stress. Furthermore, 52 common potential candidate genes in 98 GhNPFs were predicted to exhibit specific spatiotemporal expression patterns in different tissues based on two RNA sequencing (RNA-seq) datasets. Finally, the gene expression profiles of abiotic stress indicated that 31 GhNPF genes were upregulated in at least one treatment period. Under abiotic stress for 12 and 24 h, the expression of GhNPF8 was upregulated upon cold treatment but downregulated with heat treatment, salt treatment and drought treatment. Furthermore, the expression of genes GhNPF8, GhNPF54 and GhNPF43 peaked at 6 h after heat and salt treatment. These results indicated that these genes exhibit underlying characteristics related to responses to abiotic stress. The verification of NPFs and analysis of their expression profiles in different tissues and in response to different abiotic stresses of cotton provide a basis for further studying the relationship between abiotic stress resistance and nitrogen (N) transport in cotton, as well as identifying candidate genes to facilitate their functional identification.
Abiotic stressors, such as heat, cold, drought, and salinity, are major threats and can markedly reduce plant quality and productivity (Deinlein et al.,2014; Drechsler et al., 2018; Hossain et al., 2018). In response to these extremely adverse conditions, plants have developed comprehensive signaling systems to counteract and avoid adverse effects of environmental stress (Saeed et al., 2012). Stress sensing and signal transduction, which initiate a transduction cascade likely comprising multiple components, are important parts of plant response mechanisms. Studies have shown that the signaling functions of reactive oxygen species (ROS), reactive nitrogen species (RNS) and reactive carbonyl species (RCS) regulate plant resistance to abiotic stresses by regulating gene expression and protein posttranslational modification (Hossain et al., 2018), such as those of OsAPX2 in Oryza sativa (Chou et al., 2012) and LeNHX3 in Lycopersicon esculentum (Villalta et al., 2008). Additionally, climate change also directly and indirectly affects plant nutrition. Research has shown that when the concentration of CO2 increases, the nitrogen (N) content of plants decreases (Taub and Wang, 2008). Therefore, the pivotal regulatory factors of the nutrient signaling pathway also have a crucial effect on plants (Gong et al., 2020). It has been reported that the phosphate starvation response (PSR) is enhanced by directly enhancing the activity of the phosphate starvation response (PHR) gene in Arabidopsis thaliana (Rubio et al., 2001; Bustos et al., 2010) and that N use efficiency can be improved by NRT1.1B transport in rice (Zhang et al., 2019). These results indicated that NRT1 can improve N use efficiency and thus can improve plant quality and productivity under adverse conditions.
NRT1/PTR, which is also named nitrate transporter 1/peptide transporter (NPF), is a type of low-affinity transport system (LATS) of N or NO3- (Fan et al., 2017). The NPF gene family is the most abundant subfamily that encode NO3- transporters in plants (O’Brien et al., 2016). The earliest cloned plant nitrate (NO3-) transporter gene was NRT1.1 (also known as NPF6.3 or CHL1) in Arabidopsis, which had been involved in both low- and high-affinity NO3- transport (Ho et al., 2009; Wang et al., 2018). The absorption of NO3– and ammonium-N in plants involves a major process mediated by NO3– and ammonium-N transporters, respectively. Assimilation of N includes the reduction of NO3- to ammonium, which eventually is incorporated into amino acids (aa) through an assimilation process (Goel and Singh, 2015). In plants, a number of processes, including N absorption and assimilation, are negatively influenced by extreme temperature, salt and drought (Goel and Singh, 2015). NO3- is redistributed in plants under stress conditions, and this phenomenon occurs partly in response to the decreased expression of NRT1.1 and NRT1.5 (Zhang et al., 2014; Goel and Singh, 2015; Taochy et al., 2015). There is evidence that different stresses cause NO3- assimilation by redistribution, which is transmitted by NO3–transport proteins NRT1.5 and NRT1.8 (Zhang et al., 2014). The expression levels of NRT1.1 and NRT1.5 in Brassica juncea and Arabidopsis are downregulated in response to 24 h of salt and drought stresses (Goel and Singh, 2015; Taochy et al., 2015), and the expression of PtrNPF2.1 and PtrNPF7.4 in Poncirus trifoliata is also induced by salt stress (Zhao et al., 2022). Research has shown that the supply of exogenous N to sorghum and tomato can efficiently moderate Na+ uptake and increase the K+ content in plants (Miranda et al., 2016; Singh et al., 2016). Exogenous N can also alleviate the uptake of Cl- and Na+ in mustard under salinity stress (Jahan et al., 2020). In wheat, drought stress limits N translocation during the grain filling period, resulting in decreased yields (Kirda et al., 2001). In addition, high temperature can also inhibit N absorption and assimilation in wheat, rice and creeping bentgrass (Tahir and Nakata, 2005; Rachmilevitch et al., 2006; Ito et al., 2009; Ercoli et al., 2010), and the expression of BJNRT1.1 is downregulated after 24 h of hot and cold treatment in B. juncea (Goel and Singh, 2015). Taken together, the results of these studies indicated that the NPF genes that are related to N transport may have a potential effect on the growth and development of plants under abiotic stress.
Cotton (Gossypium spp.) is an economically essential crop species in China, and cotton growth and development are intimately tied to water and fertilizer. Moreover, cotton is very sensitive to N (Zheng et al., 2018). Studies have shown that N fertilizer can improve cotton yield and contribute to drought stress tolerance through increased N metabolism (Zhang et al., 2019; Iqbal et al., 2020). Under conditions of salt stress, fertilization can improve the salt resistance of cotton and can substantially increase cotton yields (Dai et al., 2013). These findings suggest that plant growth and productivity under stress conditions can be best achieved by improving N use efficiency. In addition, the GhNPF6.14 gene affects growth and nitrogen uptake and accumulation of cotton (Dong et al., 2022). Nevertheless, the NPF gene family has been poorly characterized in abiotic stress response of cotton. In this study, by performing a whole-genome analysis, we comprehensively identified 201 Gossypium NPF genes (including those in Gossypium arboreum, Gossypium raimondii and Gossypium hirsutum). Then, chromosome distributions, collinearity, motifs, gene structures, cis-acting element compositions and phylogenetic relationships were investigated. Additionally, the expression patterns of 98 GhNPFs in different tissues and under different abiotic stresses were systematically analyzed by RNA sequencing (RNA-seq) performed by staff at Zhejiang University and the Cotton Research Institute of CAAS (CRI) and by quantitative real-time PCR (qRT−PCR) techniques. The results provide a theoretical foundation for further elucidating the role and molecular mechanism of GhNPF genes in the abiotic stress response of cotton.
The amino acid sequences of Arabidopsis NPFs were used as references. The hidden Markov model (HMM) model file (PF00854) for the AtNPF gene was obtained from the Pfam database (El-Gebali et al., 2019). Then, HMME 3.0 software (Finn et al., 2015) was used to search for homologous genes in three Gossypium species (Zhu et al., 2017), with an E-value < 1e-5, and the preliminary candidate genes were identified after we omitted incorrect and redundant members. Finally, SMART (https://smart.embl.de/smart/set_mode.cgi?NORMAL=1), PfamScan (https://www.ebi.ac.uk/Tools/pfa/pfamscan/) and the NCBI Conserved Domain Database (CDD) (https://www.ncbi.nlm.nih.gov/cdd) online websites were used to further confirm whether these candidate NPF proteins contained conserved domains. The physicochemical properties of the GhNPF proteins were predicted using the online website ExPASy (https://web.expasy.org/compute_pi/). The subcellular localizations of GhNPF proteins were predicted using the WoLF PSORT online website (https://wolfpsort.hgc.jp/). Genomic datum for Arabidopsis, G. hirsutum, G. raimondii and G. arboreum were obtained from The Arabidopsis Information Resource (TAIR) (https://www.arabidopsis.org/index.jsp) and Cotton Functional Genomics Database (CottonFGD) (https://cottonfgd.net/about/download.html), respectively. TBtools 1.098745 (Chen et al., 2020) software was used to map the locations of the genes on the chromosomes, and genes were named according to the chromosomal locations of the NPF gene family in G. hirsutum species.
To identify tandem and segmental duplication events of NPF genes, a multiple sequence alignment of full-length NPF proteins was performed by MCScanX; for this, whole-genome sequences of G. hirsutum, G. arboreum and G. raimondii and gene annotations were used. Plots of the data were created using TBtools (Chen et al., 2020) software. To assess the evolutionary constrictions on each gene pair, the non-synonymous (Ka) and synonymous (Ks) substitutions were calculated using the Simple Ka/Ks Calculator (NG) in TBtools (Chen et al., 2020). To further observe the interspecific and intraspecific homology of the NPF genes, phylogenetic trees were constructed based on the NPF protein sequences of Arabidopsis, G. hirsutum, G. arboreum and G. raimondii. The ClustalW tool of MEGA-X software (Kumar et al., 2018) was used to align the protein sequences of the cotton and Arabidopsis NPF gene family members, and then the neighbor-joining (NJ) method was used to construct a phylogenetic tree; the Poisson model was used, and the bootstrap value was 1,000. Finally, the online tool iTOL (https://itol.embl.de/upload.cgi) was used to produce a high-quality phylogenetic tree map.
The structures of the GhNPF genes were investigated on the basis of the G. hirsutum genome annotation data via the Visualize Gene Structure tool in TBtools (Chen et al., 2020). The conserved motifs of the GhNPF genes were explored via the online website MEME (https://meme-suite.org/meme/doc/meme.html), and the maximum base numbers were set to 10, with the default parameters used. The Gene Structure View tool in TBtools (Chen et al., 2020) was used to illustrate the gene structures and construct conserved motifs maps.
To understand the possible regulatory and response mechanisms of GhNPF genes, the promoter region was selected for analysis. For this purpose, the 2,000 bp nucleotide sequence upstream of the start codon of the GhNPF family members were obtained from the CottonFGD (https://cottonfgd.net/about/download.html). The online website PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) was used to screen cis-acting elements in the promoter region. The gene function of the GhNPF family in G. hirsutum was annotated with gene ontology (GO) by using DAVID bioinformatics resources (https://david.ncifcrf.gov/). ChiPlot (https://www.chiplot.online/) online analytical tools was used to plot.
To verify the expression profiles of the GhNPF genes in various tissues of G. hirsutum, the RNA-seq data for 9 tissue-specific samples of upland cotton (TM-1) (root, stem, leaf, sepal, petal, anther, pistil, ovule and fiber) and samples under salt, drought, cold and heat stress were downloaded from Zhejiang University (ZJU) (http://cotton.zju.edu.cn/) (Zhang et al., 2015). The Gossypium Resource and Network Database (GRAND) website (http://grand.cricaas.com.cn/home) was used to obtain the RNA-seq data for 9 different tissues of upland cotton (TM-1) and samples under salt, drought, cold, and heat stress from the CRI. The transcript abundance of GhNPFs in different tissues and in response to different abiotic stresses was calculated according to the fragments per kilobase of transcript per million mapped reads (FPKM) values. Heatmaps of all 98 GhNPF genes were generated using TBtools software, and Venn diagrams of candidate genes were plotted using the hiplot online website (https://hiplot-academic.com/basic/venn2).
The upland cotton cultivar Zhongmian 113 (ZM113) was grown in a greenhouse (25°C; 16 h/8 h light/darkness; humidity of approximately 60%-80%) at Gansu Agricultural University, Lanzhou, Gansu Province, China. The seeds were obtained from the CRI. Nine different organs (roots, stems, leaves, petals, sepals, anther, pistils, ovules and fibers) were collected from ZM113, which was healthy at budding and flowering stage and immediately frozen in liquid nitrogen for subsequent experiments. Healthy ZM113 plants of the same age (4 weeks old) were selected for abiotic stress treatments (heat, cold, salinity and drought). All the plants were grown in a growth chamber at 25°C before stress exposure. Each abiotic stress was applied for 0 h (control), 1 h, 3 h, 6 h, 12 h and 24 h (10 replications per treatment). Some ZM113 seedlings were subjected to cold (12°C) and heat (42°C) stress. For other ZM113 seedlings, their roots were soaked in 200 mmol/L NaCl and 15% polyethylene glycol (PEG-6000) to induce salinity and drought stresses. After the above stresses were applied, the shoot tips and young leaves were collected and immediately frozen in liquid nitrogen for subsequent experiments.
Total RNA was extracted from the shoot tips and young leaf samples collected after the stress treatments and from the tissue of nine different organs via an RNA Prep Pure Plant Kit (Tiangen, China). Two micrograms of total RNA were used to synthesize 20 µl of cDNA using FastKing gDNA Dispelling RT SuperMix (KR118) (Tiangen, China) to analyze the relative expression of the GhNPF genes in the nine organs and under the different abiotic stresses. The GhNPF gene primers used were designed using NCBI Primer-BLAST (a primer design tool) and developed by Sangon Biotech (Shanghai) Co., Ltd.; the primers used are shown in Table S1. Real-time PCR amplification was performed using an LightCycler® 96 Instrument together with SuperReal Premix Plus (SYBR Green) (FP209, Tiangen, China) according to the manufacturers’ instructions. The thermocycle procedure was as follows: 95°C for 3 minutes, followed by 40 cycles of 95°C for 5 seconds and 60°C for 15 seconds. All the data were normalized to those of actin (Wu et al., 2021), which served as an internal reference gene, and the relative expression of all the evaluated GhNPF genes was calculated using the 2-ΔΔCt method (Willems et al., 2008). After normalization of the data from three independent experiments, all the data were expressed as the mean ± standard error. One-way analysis of variance (P<0.05), least significant difference (LSD) was used to evaluate the significance of each sample.
In this study, in total, 98, 52 and 51 NPF genes were verified in G. hirsutum, G. raimondii, and G. arboreum, and the G. hirsutum genes were denoted GhNPF1 to GhNPF98 according to their physical locations on the chromosome (Figure 1). The details of these GhNPF gene family members and their related proteins are listed in Table S2. The interrelated protein length (amino acids [aa]) varied greatly from 537 aa (GhNPF18) to 818 aa (GhNPF81). The predicted molecular weights (MWs) and isoelectric points (pIs) of the proteins ranged from 59,756.73 Da (GhNPF19) to 89,983 Da (GhNPF79) and from 5.38 (GhNPF52) to 9.56 (GhNPF36), respectively. With respect to the secondary structure of the GhNPF proteins, alpha-helices (Hh) and random coils (Cc) accounted for a large proportion, while extended strands (Ee) and beta turns (Tt) constituted a comparatively low proportion. Subcellular localization predictions showed that the great majority of the proteins encoded by the GhNPF genes were located at the plasma membrane, except in the cases of those encoded by GhNPF13 and GhNPF61.
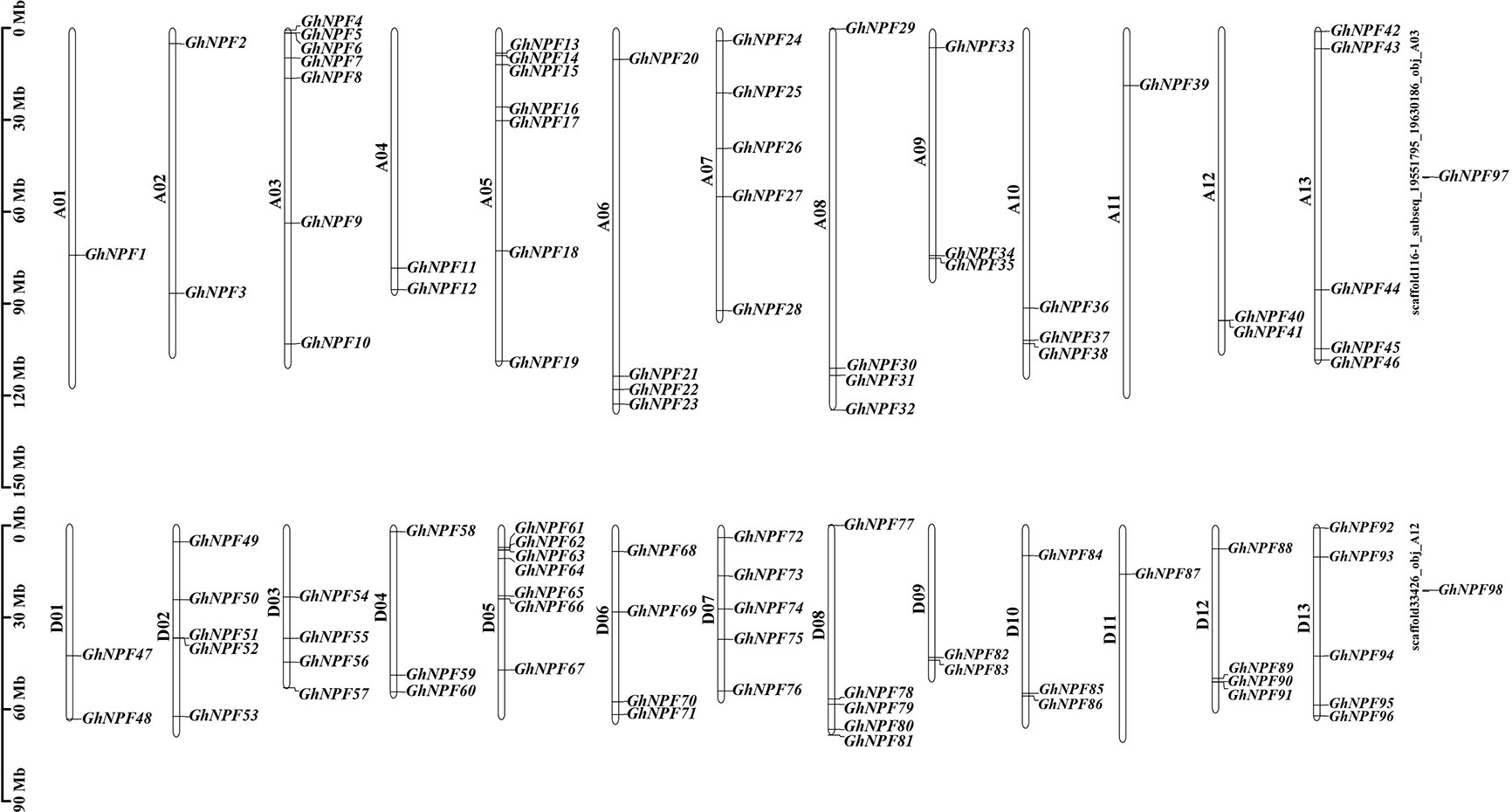
Figure 1 Positions of GhNPFs on the chromosomes of G. hirsutum. Partial GhNPF genes located on scaffolds. The white-colored bars show chromosomes from the G. hirsutum At and Dt subgenomes. A01-A13 and D01-D13 indicate chromosomes of the At and Dt subgenomes, respectively. The chromosomal positions of genes calculated from published genomic data are shown on the left side of each chromosome in the At and Dt subgenomes. The corresponding gene names are written on the right side of each chromosome of the At and Dt subgenomes.
The 96 GhNPF members were disproportionately located across the 26 chromosomes of G. hirsutum, and two genes (GhNPF97 and GhNPF98) were on scaffolds (Figure 1). Chromosomes A03, A05 and D05 contained the greatest numbers of GhNPFs (7 members), while chromosomes A07, A13, D02, D07, D08 and D13 contained 5 GhNPFs, and they accounted for a large portion of the GhNPFs across the 26 chromosomes. In contrast, chromosomes A01, A11 and D11 contained the fewest GhNPF genes (1 member each).
To reveal the homologous locus relationships of the GhNPF gene family members between the At and Dt subgenomes in G. hirsutum, gene duplication events were studied using the MCScan tool to elucidate their amplification patterns. Two pairs of genes with tandem repeats were identified on chromosomes A03 and D05 (GhNPF5/6 and GhNPF63/64), respectively. In addition, 84 segmentally duplicated genes were discovered in the GhNPF gene family of G. hirsutum (Figure 2, Table S3). These results showed that segmental duplication accounted for a large proportion of the evolution of the GhNPF gene family, which reflected the dominant role of segmental repeats relative to tandem repeats in the GhNPF gene family evolution. Moreover, the intergenomic synteny analysis results between G. hirsutum and two other Gossypium species were compared to further the understand homologous gene functions and phylogenetic relationships of NPF genes (Figure S1). The analysis of collinearity among the different species showed that 79 pairs of genes were collinear between G. hirsutum and G. arboreum and between G. hirsutum and G. raimondii. In conclusion, the present results provide evidence that NPF genes might undergo some genomic rearrangements during polyploidy. To better comprehend the evolutionary constraints controlling the functional divergence of the GhNPF gene family, the non-synonymous substitutions (Ka), synonymous substitutions (Ks), and non-synonymous to synonymous substitution (Ka/Ks) ratio were calculated (Table S4). All duplicated GhNPF gene pairs presented a Ka/Ks ratio of <1, suggesting that the GhNPF family genes might have experienced selective pressure throughout their evolution.
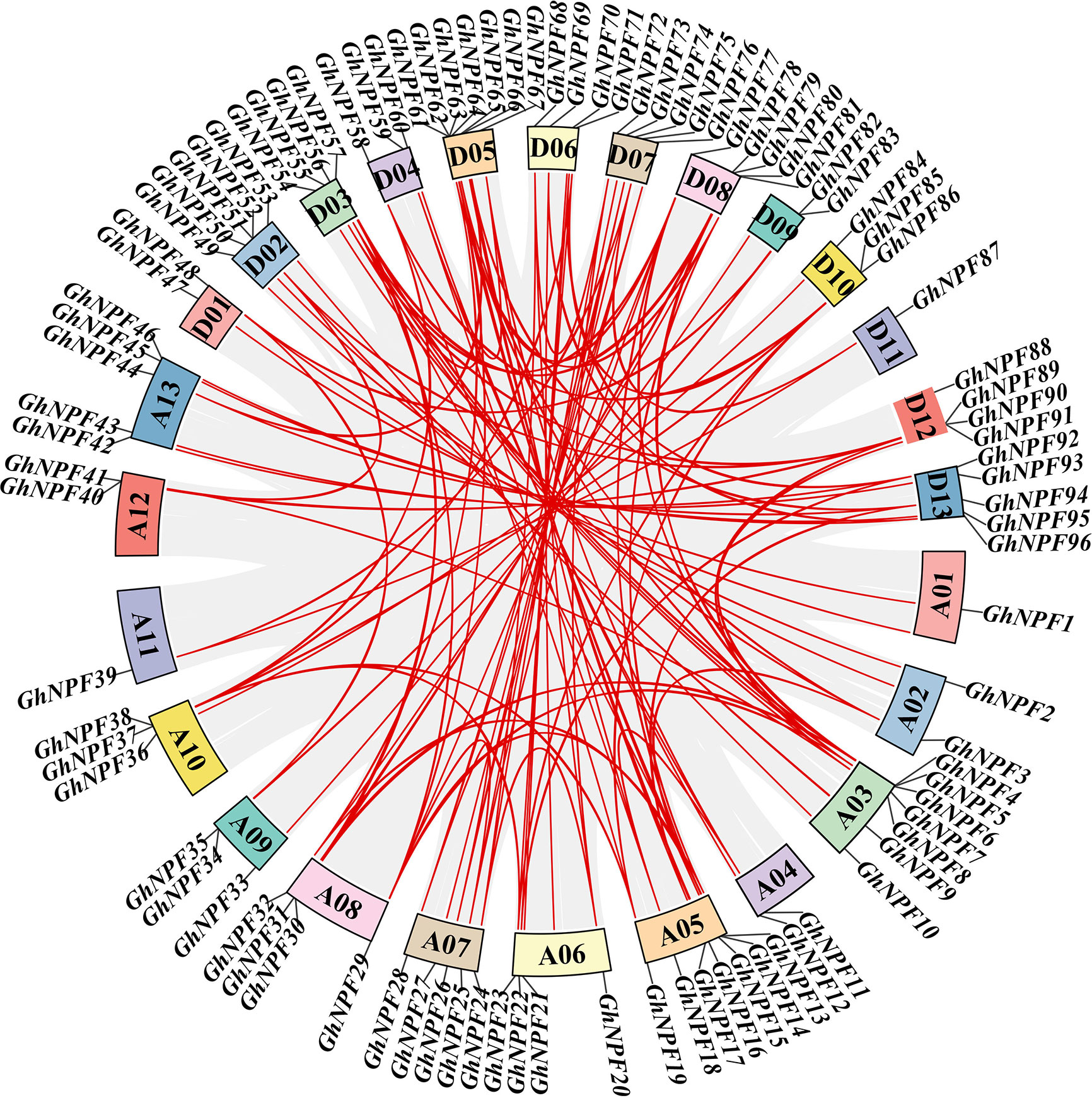
Figure 2 Duplication of GhNPF genes on chromosome 26 of G. hirsutum. The gray lines represent collinear relationships of all genes in the G. hirsutum genome, and the red lines represent gene pairs of GhNPF. The different colored rectangles indicate chromosomes.
To analyze the phylogenetic relationships of the NPFs among G. hirsutum, G. raimondii, G. arboreum and Arabidopsis, a phylogenetic tree comprising the NPF proteins of G. hirsutum (n=98), G. raimondii (n=52), G. arboreum (n=51) and Arabidopsis (n=53) was constructed (Figure 3). The 98 GhNPF proteins clustered into three primary groups (Group I, Group II and Group III) according to bootstrap values (=1,000). There were only two GhNPF genes (GhNPF34 and GhNPF82) in G. hirsutum belonging to Group I. There were eight GhNPF genes in G. hirsutum belonging to Group II. At the same time, Group III was unevenly divided into four subgroups: III-1, III-2, III-3 and III-4. Furthermore, the GhNPF members essentially clustered into subgroups III-2, III-3 and III-4, and the number of GhNPFs in G. hirsutum was two to three times greater than that in Arabidopsis among these subgroups. Among these species, 18 pairs of paralogous genes were found—15 pairs of genes in Arabidopsis, two pairs in G. hirsutum and one pair in G. raimondii. Furthermore, 86 pairs of orthologs from G. hirsutum, G. arboreum and G. raimondii were identified, revealing the paralogous and orthologous connections among these plant species.
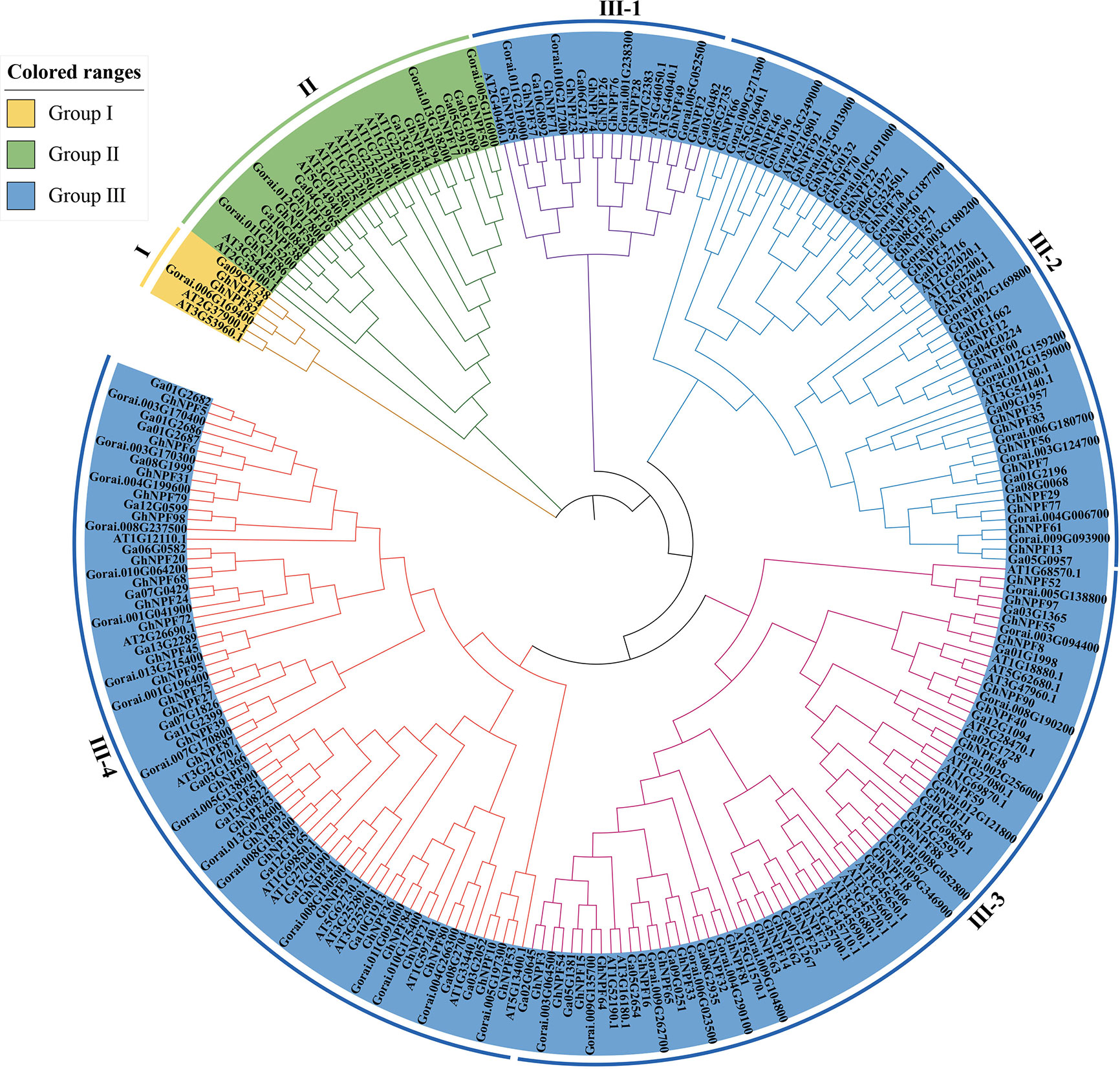
Figure 3 Phylogenetic tree of NPF genes in cotton and Arabidopsis thaliana. The tree was generated using the neighbor connection method of MEGA X software (1,000 bootstrap replicates). The tree was divided into three subfamilies, and the different colors show the following NPF subfamilies: yellow represents group I, green represents group II, and blue represents group III;. Group III was divided into four subgroups, in which different colors of branches represent different subgroups.
To research the gene structure in the evolution of the G. hirsutum gene family, the structures of the GhNPF genes were obtained by analyzing the exon/intron boundaries (Figure 4A). The analysis of exon/intron structure revealed relatively high structural divergence among the GhNPF genes. The number of exons in the 98 GhNPF genes ranged from three to seven, and GhNPF81 contained the most exons (n = 7). Most of the genes in Group I contained three introns, whereas in Group II, the genes contained two and four introns. Most of the genes in Group III contained three and four introns and the genes (GhNPF81) with the most introns were also included in the Group III. A total of 54.08% of all GhNPFs (53 genes) contained three introns each, suggesting that introns were gained and lost as the GhNPF gene family evolved, which might have resulted in functional diversity among the GhNPF genes.
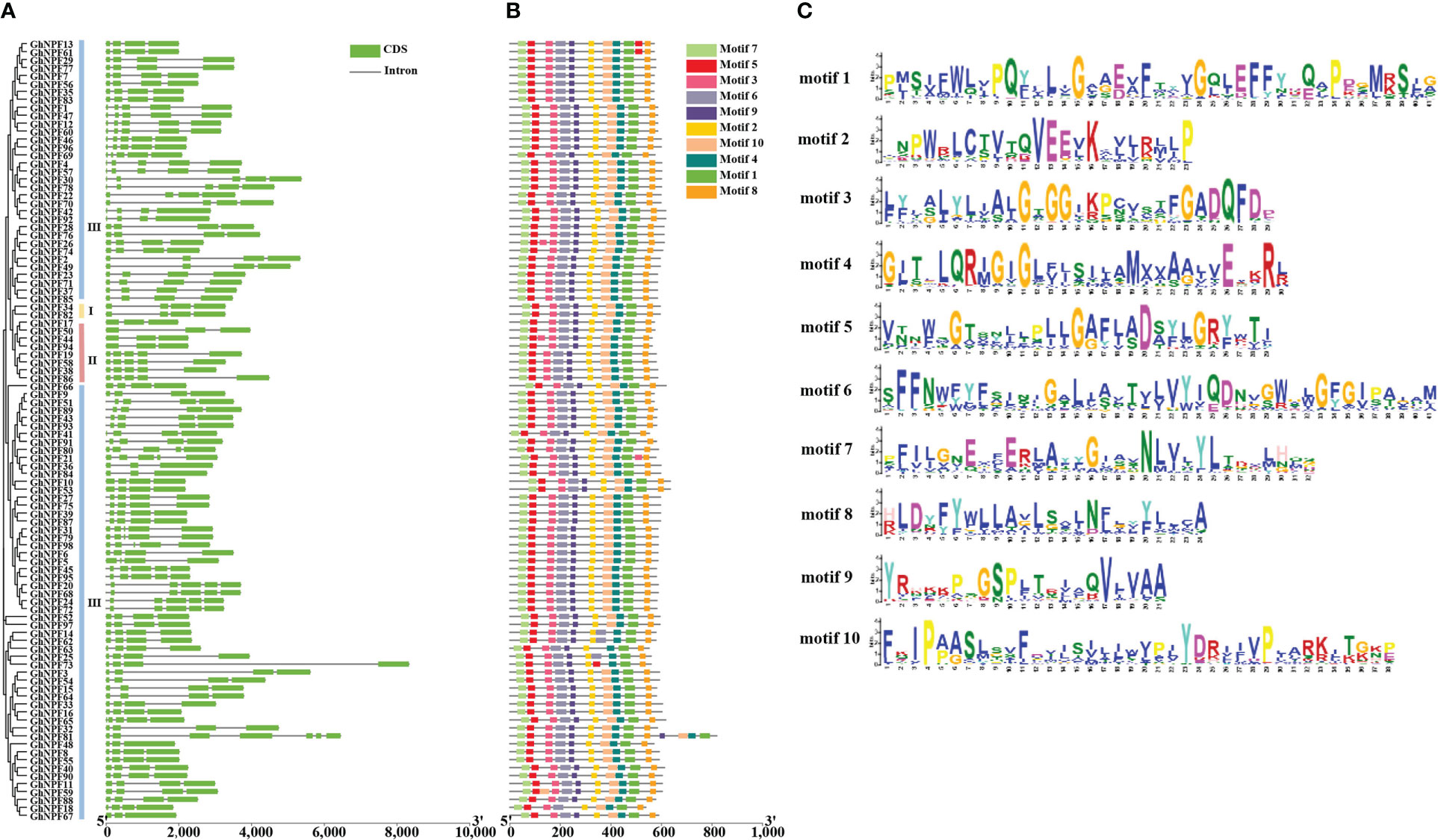
Figure 4 Phylogenetic relationships, structures and motif compositions of GhNPF genes. (A) Unrooted phylogenetic tree and exon/intron structure of GhNPFs. (B) Conserved motifs of 98 GhNPF proteins. (C) Conserved repeat markers of GhNPF genes.
Ten conserved motifs were detected in most GhNPF protein sequences by the use of the online MEME program, which further the similarities and differences in motif composition (Figure 4B). The amino acid numbers of the motifs ranged from 21 to 41 (Figure 4C). The number of motifs for each GhNPF was nine to fourteen (Figures 4B, C). Motifs 1, 2, 3, 4, 5, 6, 7 and 8 were present in all the GhNPF proteins, while motif 9 was not present only in the GhNPF18 protein of Group III. Similarly, motif 10 was not present in the GhNPF14, GhNPF62, GhNPF63, GhNPF25 and GhNPF73 proteins of Group III. In contrast, motifs 1-10 were all present in all the GhNPF members of Groups I and II. In general, almost all the GhNPF proteins within the same subgroup presented very similar motif compositions, suggesting that these GhNPF proteins have similar functions.
The cis-regulatory elements in the 2,000 bp upstream region of the 5’ end of the 98 GhNPF genes were identified and analyzed to reveal their potential response mechanisms (Figure 5). We identified 55 cis-regulatory elements involved in stress responsiveness, tissue-specific expression, phytohormone responsiveness and light responsiveness. Five stress-related elements were identified, namely, DREs, LTRs, MBSs, TC-rich repeats and WUN motif–containing elements. These cis-acting elements were involved in responses to low temperature, salt stress, defense and drought. There were seven cis-acting elements associated with tissue-specific expression, namely, AREs, CAT-boxes, GC-motif, GCN4_motif, HD-Zip 1s, O2-site–and RY elements. Moreover, among these elements AREs were the most common in the GhNPF gene promoters. In addition, eleven hormone-related elements, namely, ABREs, AuxRR-core–containing elements, CGTCA motif–containing elements, GARE motif–containing elements, P-boxes, SAREs, TATC-boxes, TCA elements, TGA-boxes, TGACG motif–containing elements and TGA elements, were also found. This category included abscisic acid-responsive elements (ABREs), auxin-responsive elements (AuxRR-core–containing elements, TGA-boxes and TGA motif–containing elements), methyl jasmonate (MeJA)-responsive elements (CGTCA motif–containing elements and TGACG motif–containing elements), gibberellin-responsive elements (GARE motif–containing elements, P-boxes and TATC motif–containing elements) and salicylic acid-response elements (SAREs and TCA elements). There were also 32 cis-acting elements related to the light response, including Box 4 elements, C-boxes, G-boxes, etc. Box 4 elements and G-boxes were present in relatively high numbers within the light-responsive cis-acting regulatory elements. Interestingly, we found that the GhNPF26 gene does not contain any type of cis-acting element. Taken together, these results showed that GhNPF genes might play an important role in abiotic stress responses, defense-related signal transduction, and phytohormone responses. In addition, the genes might be involved in various light responses during G. hirsutum growth.
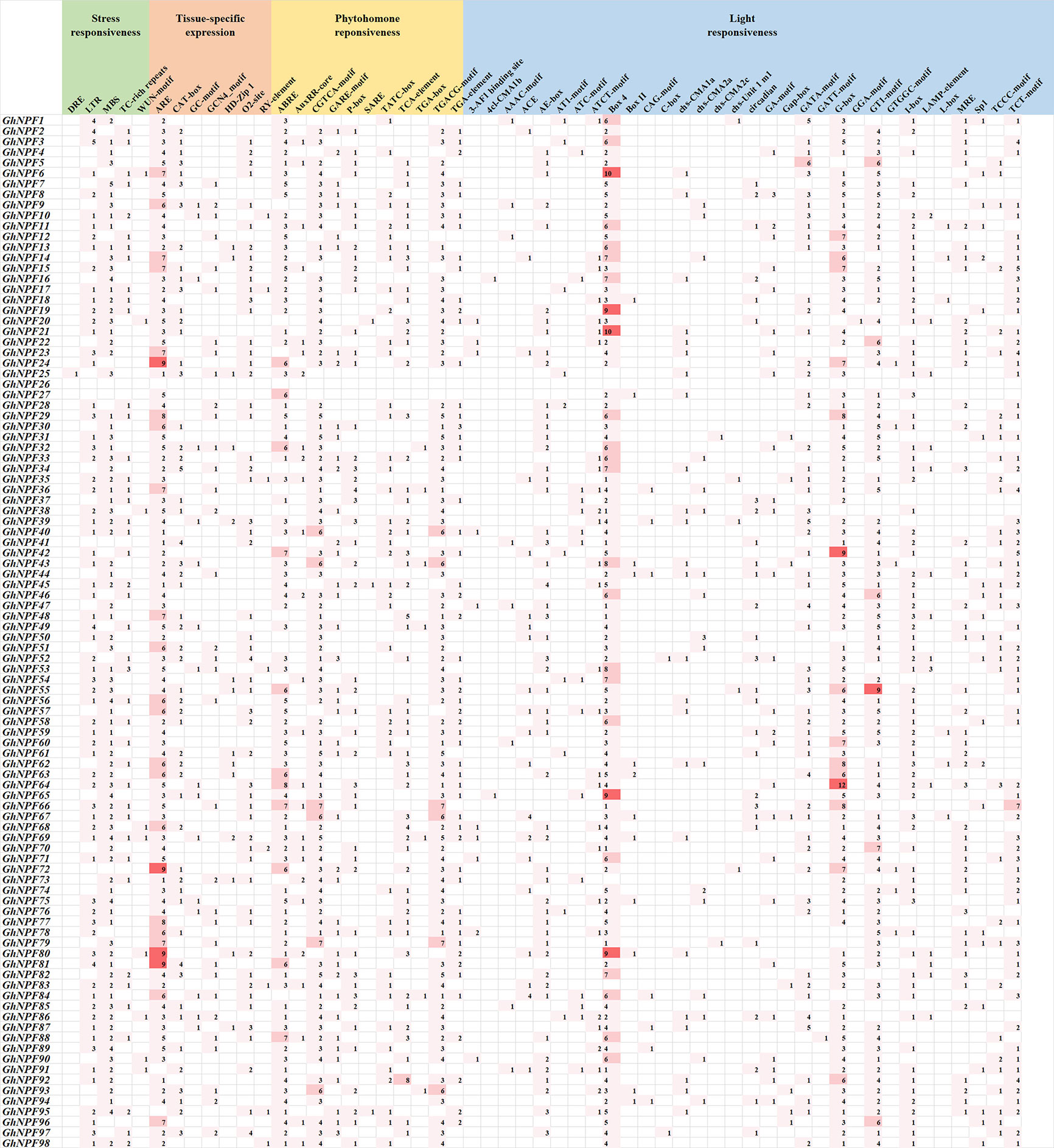
Figure 5 Prediction results of cis-regulatory elements in the promoter regions of GhNPF gene family members. The numbers in the cells represent the numbers of genes.
To analyze the expression patterns of GhNPF genes during G. hirsutum development, RNA-seq data of various G. hirsutum tissues were used in this study. The expression characteristics of all 98 GhNPF genes were determined at varying levels across different tissues and developmental stages (Figure 6). The RNA-seq data of ZJU showed that 55.1% of GhNPF genes were highly expressed in vegetative organs (roots, stems and leaves) and 65.3% of GhNPF genes were highly expressed in reproductive organs (petals, sepals, anther, pistils, ovules and fibers). In addition, according to the FPKM value of CRI’s RNA-seq data, 56 out of 98 GhNPF genes were highly expressed in vegetative organs (roots, stems and leaves), and 71 out of 98 GhNPF genes were highly expressed in reproductive organs (petals, sepals, anther, pistils, ovules and fibers). A total of 52 common genes were identified in vegetative organs from two RNA-seq datasets, which verified the reliability of the data (Figure 6C). The expression of 14 select genes in the tissues of G. hirsutum was examined via qRT−PCR, and the results were essentially consistent with the RNA-seq data (Figure 6D).
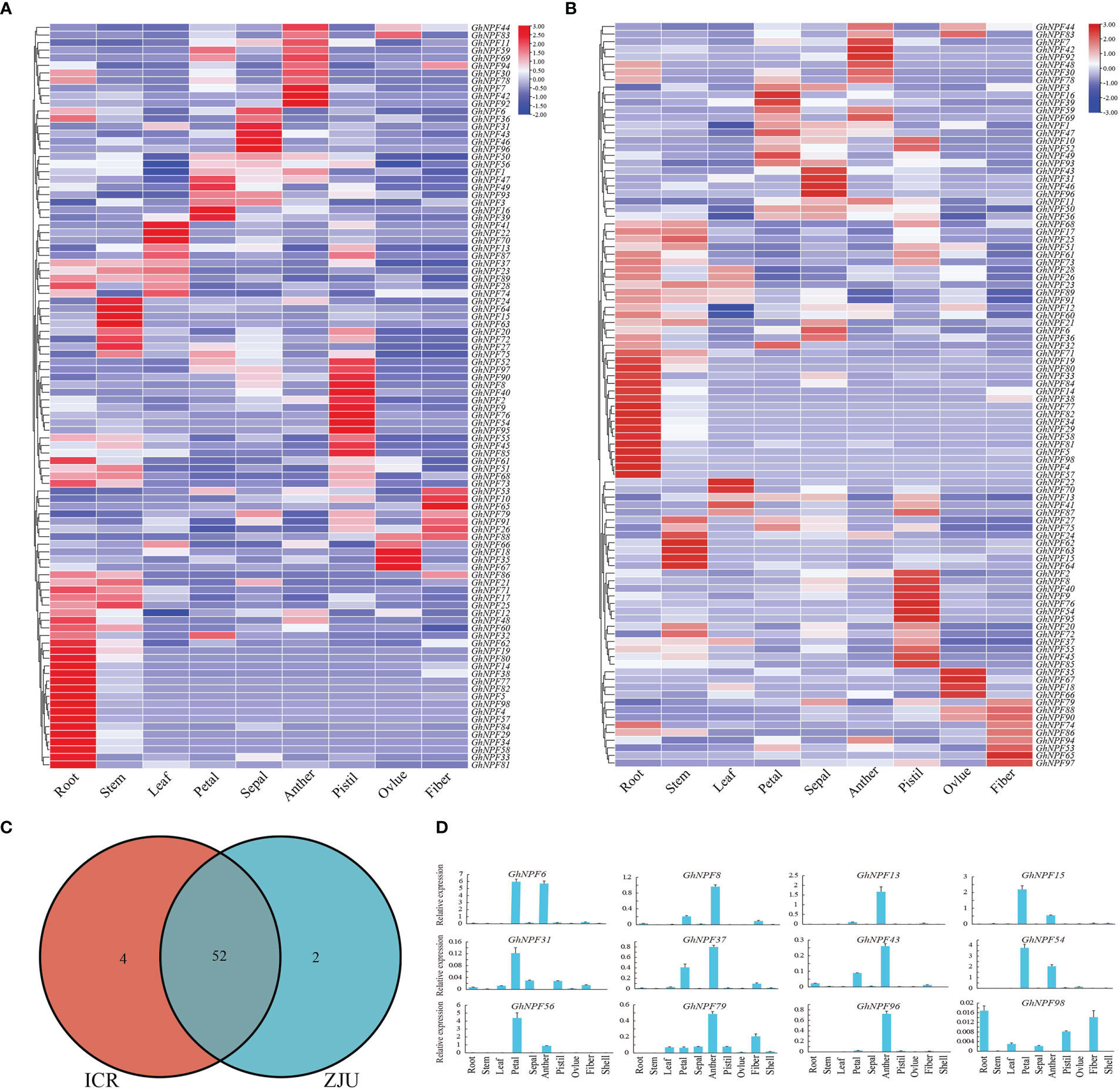
Figure 6 Expression profiles of GhNPF genes in 9 organs of upland cotton. (A, B) The expression patterns of 98 GhNPFs in 9 G. hirsutum (TM-1) tissues were analyzed by RNA-seq at the ZJU and CRI. The red and purple colors indicate high and low expression levels, respectively. (C) Venn diagram of common highly expressed genes in the two RNA-seq datasets (ZJU and CRI). (D) Relative expression levels of 14 GhNPFs in 9 organs of G. hirsutum. The error bars represent the standard deviations of three biological replications.
To further understand the functional segregation of the identified GhNPF genes, GO was performed by DAVID based on three categories: molecular function, biological process and cellular component (Figure 7). A total of 39 GhNPF genes belonged to molecular function, including transmembrane transporter activity (GO:0022857), tripeptide transporter activity (GO:0042937), dipeptide transmembrane transporter activity (GO:0071916), symporter activity (GO:0015293), low-affinity nitrate transmembrane transporter activity (GO:0050054) and nitrate transmembrane transporter activity (GO:0015112). Twenty-six GhNPF genes were involved in nitrate transport (GO:0015706), transmembrane transport (GO:0055085), nitrate assimilation (GO:0042128), oligopeptide transport (GO:0006857), dipeptide transport (GO:0042938), tripeptide transport (GO:0042939), response to nitrate (GO:0010167), response to nematode (GO:0009624), response to wounding (GO:0009611) and response to jasmonic acid (GO:0009753) in biological process. A total of 24 genes can function as an integral component of the membrane (GO:0016021) and plasma membrane (GO:0005886) in cellular component. Interestingly, some GhNPFs exist in different cell components, participate in different biological processes, and have multiple molecular functions.
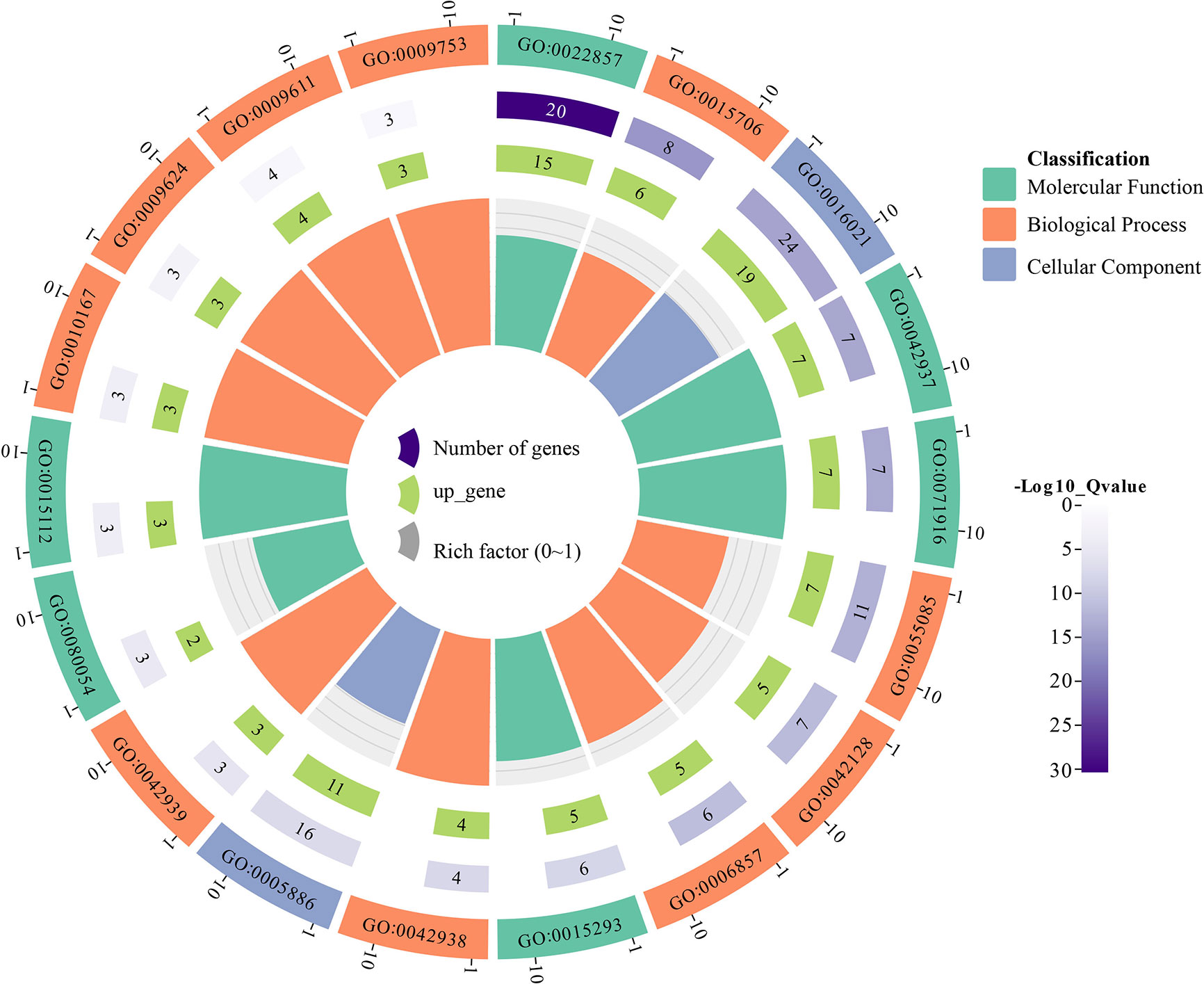
Figure 7 Functional categorization of the GhNPF genes in G. hirsutum. Purple represents the number of genes, light green represents the number of upregulated genes, gray represents the rich factor, green represents molecular function, orange represents biological process, and light purple represents cellular component.
To analyze the potential functions of the GhNPF genes in response to abiotic stresses, the GhNPF expression levels were evaluated via two RNA-seq datasets (those compiled by ZJU and the ICR) corresponding to plants under salt, drought, cold and heat treatments. Analysis of the expression profiles showed that six genes, namely, GhNPF5, GhNPF16, GhNPF18, GhNPF65, GhNPF69 and GhNPF86, were not expressed in any of the four treatments; moreover, GhNPF77 and GhNPF29 were not expressed in response to temperature stress, and GhNPF88 was not expressed in response to drought and salt stress (Figure S2). The expression of several genes was significantly increased or decreased under the cold, heat, NaCl and PEG treatments compared with the control treatment, and the DEGs differed at different treatment time periods. According to the FPKM values of the ZJU RNA-seq dataset, 52, 36, 41 and 48 genes were highly expressed during at least two of the five different time periods (1 h, 3 h, 6 h, 12 h and 24 h) and stress treatment groups (heat, cold, salt and drought) (Figures S3A, C). According to the FPKM values of the CRI RNA-seq dataset, 51, 36, 51 and 31 genes were highly expressed in at least two of the five different time stress treatment groups (heat, cold, salt and drought, respectively) (Figures S3A, C). These results showed that after heat, cold, salt and drought treatment, 59 (heat and cold) and 38 (salt and drought) genes were upregulated, and according to the two datasets (Figures S3B, E), 31 genes common to the temperature (heat and cold) and saline-alkali (salt and drought) treatments could be candidate genes with resistance-related characteristics in cotton (Figure S3F). Among these candidate genes, the GhNPF6, GhNPF37 and GhNPF54 genes were all upregulated after heat and cold treatment, but the expression of the GhNPF64 and GhNPF27 genes was inhibited by the temperature treatments. In addition, the GhNPF28, GhNPF55 and GhNPF78 genes were all upregulated after NaCl and PEG treatment, while the GhNPF74 gene was upregulated only after NaCl treatment; PEG treatment inhibited the expression of the GhNPF39 gene. To further investigate the possible response of GhNPFs to abiotic stress conditions, by performing qRT−PCR, we analyzed the expression of 13 select genes from different tissues of G. hirsutum under different stresses (Figure 8). The heat treatments (at all time points) induced the expression of GhNPF13, GhNPF31, GhNPF43, GhNPF79 and GhNPF96, and cold stress exposure at all time intervals induced the expression of GhNPF15, GhNPF43 and GhNPF56. Similarly, the salinity stress treatments induced the expression of GhNPF6, GhNPF31, GhNPF54, GhNPF57 and GhNPF96 at all time intervals, and the GhNPF31, GhNPG37, GhNPF43, GhNPF79, GhNPF96 and GhNPF98 genes exhibited increased expression in response to drought stress. Taken together, the results of our abiotic stress response gene expression analysis showed that the GhNPF gene family members in upland cotton have potential regulatory roles in the response to abiotic stress.
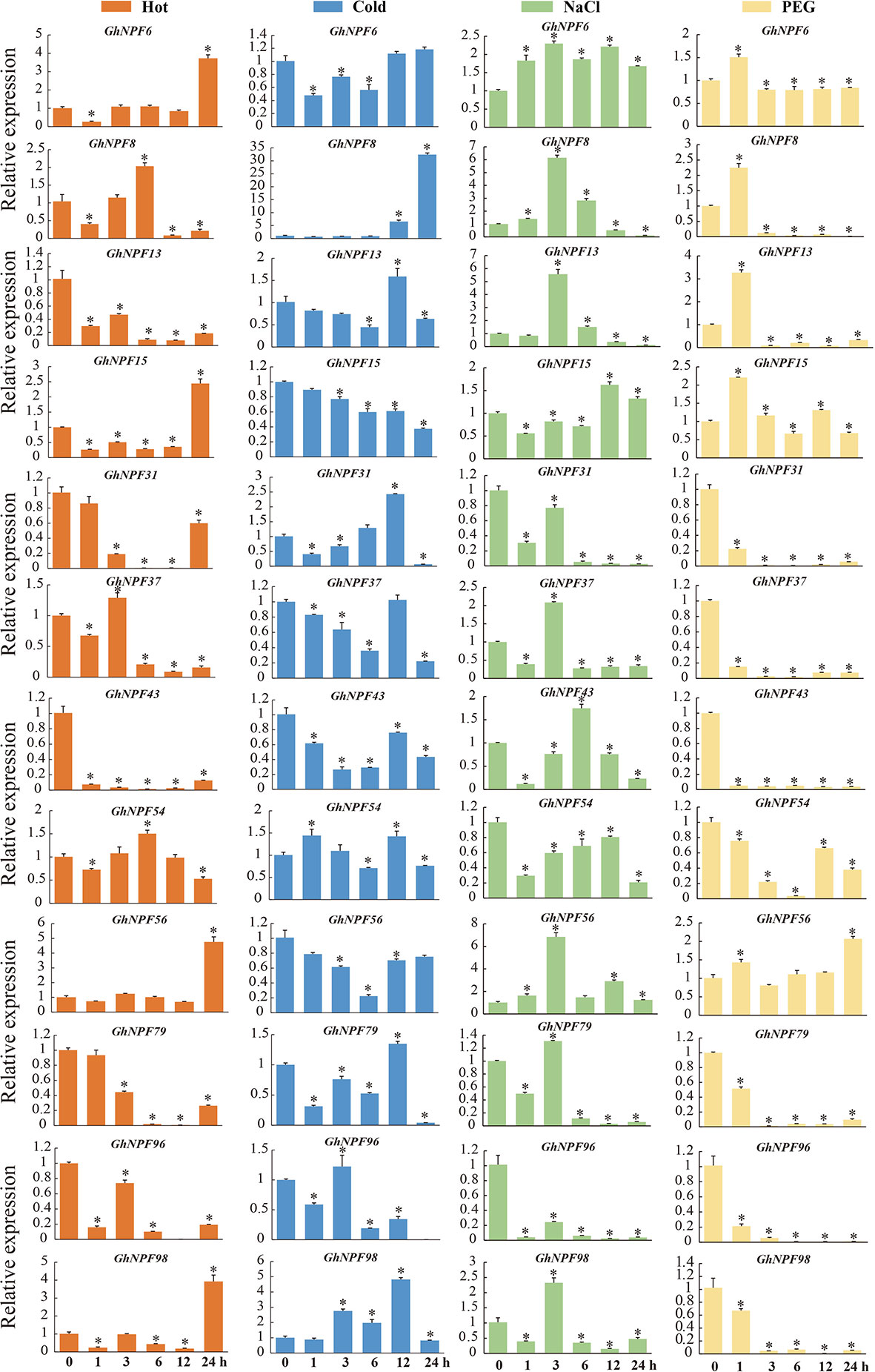
Figure 8 Relative expression levels of 12 GhNPFs in response to cold, salt, and drought treatments. The error bars represent the standard deviations of three biological replications. Orange represents heat stress, blue represents cold stress, green represents salt stress and yellow represents drought stress. Asterisks were used to indicate a significant degree of expression compared to the value of the control (*P < 0.05).
N plays a substantial role in the growth and development of plants under abiotic stresses (Ercoli et al., 2010; Zhang et al., 2014; Goel and Singh, 2015; Taochy et al., 2015). NPFs are LATSs of N or NO3- and compose the largest subfamily of NO3- transporters in plants (O’Brien et al., 2016; Fan et al., 2017). The NPF family members plant species and subspecies such as Arabidopsis, Populus, rice, Brassica napus, soybean, Brassica rapa subsp. pekinensis, Populus tomentosa and P. trifoliata were identified and analyzed to determine their gene structure and transcript accumulation (Tsay et al., 2007; Bai et al., 2013; Drechsler et al., 2018; Zhang et al., 2020; You et al., 2020; Ma et al., 2021; Zhao et al., 2021; Zhao et al., 2022). In this study, 99, 52 and 51 NPF genes were identified in G. hirsutum, G. raimondii, and G. arboreum, respectively, compared to other identified plant species. Fifty-three have been identified in Arabidopsis (Tsay et al., 2007), along with 68 in Populus (Bai et al., 2013), 82 in rice (Drechsler et al., 2018), 193 in B. napus (Zhang et al., 2020), 120 in soybean (You et al., 2020), 72 in B. rapa subsp. pekinensis (Ma et al., 2021), 87 in P. tomentosa (Zhao et al., 2021) and 56 in P. trifoliata (Zhao et al., 2022). The number of genes in G. hirsutum was similar to the number of genes in P. tomentosa and was twice that in Arabidopsis. Genome-wide identification of the NPF genes in cotton was conducted to analyze the phylogenetic relationships of the NPFs between G. hirsutum and two other cotton species as well as A. thaliana. The NPF proteins could be separated into three main groups, namely, I, II, and III, according to the phylogenetic results, of which Group III could be further divided into four subgroups: III-1, III-2, III-3, and III-4. In these species, 18 pairs of paralogous genes were found: there were 15 pairs of genes in Arabidopsis, two pairs in G. hirsutum and one pair in G. raimondii. Furthermore, 86 pairs of orthologs from G. hirsutum, G. arboreum and G. raimondii were identified, suggesting that polyploidy led to the evolution of new cotton-specific ortholog clusters. During long-term natural selection, basic NPF genes were retained in the G. hirsutum genome, while others were lost, which is consistent with the findings of a study involving B. napus (Zhang et al., 2020). Other studies have shown that genes within the same taxa might have similar functions due to sequence similarity (Nan et al., 2021). Analysis of the exon/intron structure revealed a relatively high structural divergence among the GhNPF genes. The results suggested that events in which introns were lost and gained occurred during the evolution of the GhNPF gene family, which might result in functional redundancy among GhNPF genes. Therefore, cotton NPF family members might have differentiated during evolution, which might have resulted in functional differences.
Gene duplication is the main mechanism through which gene families expand. Segmental and tandem duplication are considered to be the two main causes of gene family expansion in plants (Cannon et al., 2004). The number of segmental duplication of GhNPFs in G. hirsutum was lower than that in B. napus (Zhang et al., 2020), and 84 segmentally duplicated genes were discovered in the GhNPF gene family of G. hirsutum, while the number of tandemly duplicated genes of both species was the same. Nevertheless, collinearity analysis of different species is one way to study the gene evolution and relationships (Yu et al., 2020). Therefore, the results of the intergenomic synteny analyses between G. hirsutum and the other two cotton species were compared to further understand the homologous gene functions and phylogenetic relationships of the NPF genes. The results showed that since the number of G. hirsutum genes was slightly greater than the total number of G. arboreum and G. raimondii genes, compared with those in G. hirsutum, the NPF gene duplication events and chromosomal rearrangements in G. arboreum and G. raimondii might be conserved. Likewise, duplication events in the B. napus genome might have facilitated the expansion of the NPF gene family (Zhang et al., 2020). Generally, due to the high diversity and allopolyploid characteristics of the NPF gene family, the members of the NPF gene family might have complex phylogenetic relationships in G. hirsutum. In order to investigate differentiation after gene duplication, non-synonymous substitutions (Ka) and synonymous substitutions (Ks) of replicated GhNPF genes in G. hirsutum were calculated. The present results suggested that GhNPF family genes have experienced selective pressures during evolution.
Cis-acting regulatory elements play paramount roles in regulating gene transcription by coordinating responses to developmental and environmental cues (Schmitz et al., 2022). It has been found that NPF transport is affected by nitrite, auxin, abscisic acid, jasmonoyl-isoleucine, and gibberellins, and NPF transport even participates in flowering time regulation and is negatively affected by abiotic stresses (Sugiura et al., 2007; Krouk et al., 2010; Kanno et al., 2012; Chiba et al., 2015; Goel and Singh, 2015; Saito et al., 2015; David et al., 2016; Teng et al., 2019). In this study, 55 types of cis-acting elements (stress-responsive, tissue-specific, phytohormone-responsive and light-responsive ones) were confirmed in the promoters of GhNPFs. Most GhNPF genes contained stress-responsive elements, hormone-responsive elements and light-responsive elements, which indicated that the expression and regulation of these genes were affected by stress, hormones and light. Like in the P. trifoliata study (Zhao et al., 2022), in the present study, the GhNPF promoters contained MyB-binding sites, indicating that these genes might be regulated by the same transcriptional mechanism. There is direct evidence that NPF genes are affected by salt and drought stress (Zhang et al., 2014). Furthermore, based on two RNA-seq datasets and qRT−PCR analyses, we characterized the spatial and temporal expression profiles of NPF genes and the responses of NPF genes to various stress treatments in G. hirsutum and found that a large number of GhNPF genes were highly expressed in the roots, stems and pistils, suggesting that GhNPF might be important for the functions of those organs. GhNPF37 was expressed in all the tested tissues, while GhNPF5, a member of the NPF6 family, was mainly expressed in the roots, and AtNPF6.3 was also highly expressed in the lateral roots (Guo et al., 2001). Both GhNPF56 and GhNPF13 are members of the NPF8 family and were highly expressed in the petals. However, AtNPF8.2 is mainly expressed in the pollen and ovules (Komarova et al., 2008). GhNPF family members were unevenly expressed across all the evaluated tissues, indicating that they played an important role in controlling the growth and development of G. hirsutum. Interestingly, some GhNPFs exist in different cell components, participate in different biological processes, and have multiple molecular functions. This gene family may play an important role in the growth process and environmental diversity. For example, GhNPF6 is located in the plasma membrane, has membrane boundary functions such as transmembrane transporter activity, and participates in nitrogen compound transport, response to nitrate, response to wounding and response to jasmonic acid. Research has shown that NPF genes respond specifically to abiotic and biotic stressors except N starvation (Fan et al., 2017). For example, GsNRT1.12, GsNRT1.43, GsNRT1.62, and GsNRT1.57 in soybean were shown to be rapidly upregulated after salt treatment (You et al., 2020). Phyllostachys edulis responds to cold and drought treatment through altered expression of PeNPF to a certain extent (Yuan et al., 2021). To further investigate the potential functions of GhNPFs in abiotic stress responses, we analyzed the gene expression profile data. In addition, qRT−PCR was used to analyze the expression of 13 GhNPFs under four abiotic stress conditions: salt, drought, heat and cold. Two genes (GhNPF31 and GhNPF96) were downregulated after three treatments, namely, heat, salinity and drought, and the expression of GhNPF31 was the highest after 12 h of cold treatment, while the expression of GhNPF96 was the highest after 3 h of cold treatment, the results of which implied that GhNPFs might participate in the transduction of different signaling pathways in response to abiotic stress. The expression level of PtrNPF7.3 in P. trifoliata (Zhao et al., 2022), a homologous gene of GhNPF96, was lower in the control group than in the treatment groups, but its transcript level significantly increased after salt treatment. The GhNPF6 gene was upregulated under salt, but cold, heat and drought had little effect on its expression. The expression of the GhNPF79 and GhNPF98 genes was downregulated in response to drought treatment, and their transcript levels increased after 3 h of salt treatment then began to decrease at 6 h, 12 h and 24 h. Similarly, the expression level of NRT1.1, a homolog of GhNPF79 and GhNPF98, was reduced in B. juncea and Arabidopsis after salt and drought stresses (Goel and Singh, 2015; Taochy et al., 2015). High temperature inhibited the expression of 79 genes under heat at 24 h. Interestingly, GhNPF5 was significantly expressed in the roots but not in response to the four biotic stresses, which proved that the GhNPF gene responds to specific stressors, which explains why it was not expressed under any one stress. These results suggested that NO3- uptake might increase the osmotic potential of cells in response to abiotic stress. In general, this study revealed the NPF genes in G. hirsutum and explored their expression profiles in different tissues and under different abiotic stresses, the findings of which provide a theoretical basis for further studies on the function of GhNPFs and plant N use efficiency under abiotic stress.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
JL, CW and JS designed the research. JL, JJ and YL performed the experiments. CL and JP analyzed the data. JL wrote the manuscript. CW and JS revised the manuscript. All authors contributed to the article and approved the submitted version.
This work was funded by the Science and Technology Innovation Funds of Gansu Agricultural University [GAU-KYQD-2018–32], China; Gansu Province Science and Technology Program [20JR10RA531], China; Education Technology Innovation Project of Gansu Province [2022QB-076], China.
We are grateful to Professor Xiongfeng Ma and Shuai Dai (at the Institute of Cotton Research of CAAS), who provided us with a good seed for the experiment.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1103340/full#supplementary-material
Bai, H., Euring, D., Volmer, K., Janz, D., Polle, A. (2013). The nitrate transporter (NRT) gene family in poplar. PloS One 8 (8), e72126. doi: 10.1371/journal.pone.0072126
Bustos, R., Castrillo, G., Linhares, F., Puga, M. I., Rubio, V., Pérez-Pérez, J., et al. (2010). A central regulatory system largely controls transcriptional activation and repression responses to phosphate starvation in Arabidopsis. PloS Genet. 6 (9), e1001102. doi: 10.1371/journal.pgen.1001102
Cannon, S. B., Mitra, A., Baumgarten, A., Young, N. D., May, G. (2004). The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 4, 10. doi: 10.1186/1471-2229-4-10
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13 (8), 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chiba, Y., Shimizu, T., Miyakawa, S., Kanno, Y., Koshiba, T., Kamiya, Y., et al. (2015). Identification of Arabidopsis thaliana NRT1/PTR FAMILY (NPF) proteins capable of transporting plant hormones. J. Plant Res. 128 (4), 679–686. doi: 10.1007/s10265-015-0710-2
Chou, T. S., Chao, Y. Y., Kao, C. H. (2012). Involvement of hydrogen peroxide in heat shock- and cadmium-induced expression of ascorbate peroxidase and glutathione reductase in leaves of rice seedlings. J. Plant Physiol. 169 (5), 478–486. doi: 10.1016/j.jplph.2011.11.012
Dai, J., Lu, H., Li, Z., Duan, L., Dong, H. (2013). Effects of fertilization on cotton growth and nitrogen use efficiency under salinity stress. Chin. J. Appl. Ecol. 24 (12), 3453–3458. doi: 10.13287/j.1001-9332.2013.0579
David, L. C., Berquin, P., Kanno, Y., Seo, M., Daniel-Vedele, F., Ferrario-Méry, S. (2016). N availability modulates the role of NPF3.1, a gibberellin transporter, in GA-mediated phenotypes in Arabidopsis. Planta 244 (6), 1315–1328. doi: 10.1007/s00425-016-2588-1
Deinlein, U., Stephan, A. B., Horie, T., Luo, W., Xu, G., Schroeder, J. I., et al. (2014). Plant salt-tolerance mechanisms. Trends Plant Sci. 19 (6), 371–379. doi: 10.1016/j.tplants.2014.02.001
Dong, Q., Wang, G., Iqbal, A., Muhammad, N., Wang, X., Gui, H., et al. (2022). Identification and expression analysis of the NPF genes in cotton. Int. J. Mol. Sci. 23 (22), 14262. doi: 10.3390/ijms232214262
Drechsler, N., Courty, P. E., Brule, D., Kunze, R. (2018). Identification of arbuscular mycorrhiza-inducible Nitrate transporter 1/Peptide transporter family (NPF) genes in rice. Mycorrhiza 28 (1), 93–100. doi: 10.1007/s00572-017-0802-z
El-Gebali, S., Mistry, J., Bateman, A., Eddy, S. R., Luciani, A., Potter, S. C., et al. (2019). The pfam protein families database in 2019. Nucleic Acids Res. 47 (D1), D427–D432. doi: 10.1093/nar/gky995
Ercoli, L., Arduini, I., Mariotti, M., Masoni, A. (2010). Post-anthesis dry matter and nitrogen dynamics in durum wheat as affected by nitrogen and temperature during grain filling. Cereal Res. Commun. - Cereal Res. Commun. 38, 294–303. doi: 10.1556/CRC.38.2010.2.16
Fan, X., Naz, M., Fan, X., Xuan, W., Miller, A. J., Xu, G. (2017). Plant nitrate transporters: from gene function to application. J. Exp. Bot. 68 (10), 2463–2475. doi: 10.1093/jxb/erx011
Finn, R. D., Clements, J., Arndt, W., Miller, B. L., Wheeler, T. J., Schreiber, F., et al. (2015). HMMER web server: 2015 update. Nucleic Acids Res. 43 (W1), W30–W38. doi: 10.1093/nar/gkv397
Goel, P., Singh, A. K. (2015). Abiotic stresses downregulate key genes involved in nitrogen uptake and assimilation in Brassica juncea l. PloS One 10 (11), e143645. doi: 10.1371/journal.pone.0143645
Gong, Z., Xiong, L., Shi, H., Yang, S., Herrera-Estrella, L. R., Xu, G., et al. (2020). Plant abiotic stress response and nutrient use efficiency. Sci. China Life Sci. 63 (5), 635–674. doi: 10.1007/s11427-020-1683-x
Guo, F. Q., Wang, R., Chen, M., Crawford, N. M. (2001). The Arabidopsis dual-affinity nitrate transporter gene AtNRT1.1 (CHL1) is activated and functions in nascent organ development during vegetative and reproductive growth. Plant Cell 13 (8), 1761–1777. doi: 10.11054/tpc.010126
Ho, C. H., Lin, S. H., Hu, H. C., Tsay, Y. F. (2009). CHL1 functions as a nitrate sensor in plants. Cell 138 (6), 1184–1194. doi: 10.1016/j.cell.2009.07.004
Hossain, M. A., Li, Z. G., Hoque, T. S., Burritt, D. J., Fujita, M., Munné-Bosch, S., et al. (2018). Heat or cold priming-induced cross-tolerance to abiotic stresses in plants: key regulators and possible mechanisms. Protoplasma 255 (1), 399–412. doi: 10.1007/s00709-017-1150-8
Iqbal, A., Dong, Q., Wang, X., Gui, H., Zhang, H., Zhang, X., et al. (2020). High nitrogen enhance drought tolerance in cotton through antioxidant enzymatic activities, nitrogen metabolism and osmotic adjustment. Plants (Basel) 9 (2), 178. doi: 10.3390/plants9020178
Ito, S., Hara, T., Kawanami, Y., Watanabe, T., Khuankaew, T., Norikuni, O., et al. (2009). Carbon and nitrogen transport during grain filling in rice under high-temperature conditions. J. Agron. Crop Sci. 195, 368–376. doi: 10.1111/j.1439-037X.2009.00376.x
Jahan, B., AlAjmi, M. F., Rehman, M. T., Khan, N. A. (2020). Treatment of nitric oxide supplemented with nitrogen and sulfur regulates photosynthetic performance and stomatal behavior in mustard under salt stress. Physiol. Plant 168 (2), 490–510. doi: 10.1111/ppl.13056
Kanno, Y., Hanada, A., Chiba, Y., Ichikawa, T., Nakazawa, M., Matsui, M., et al. (2012). Identification of an abscisic acid transporter by functional screening using the receptor complex as a sensor. Proc. Natl. Acad. Sci. U.S.A. 109 (24), 9653–9658. doi: 10.1073/pnas.1203567109
Kirda, C., Derici, M., Schepers, J. S. (2001). Yield response and n-fertilizer recovery of rainfed wheat growing in the Mediterranean region. Field Crops Res. 71, 113–122. doi: 10.1016/S0378-4290(01)00153-8
Komarova, N. Y., Thor, K., Gubler, A., Meier, S., Dietrich, D., Weichert, A., et al. (2008). AtPTR1 and AtPTR5 transport dipeptides in planta. Plant Physiol. 148 (2), 856–869. doi: 10.1104/pp.108.123844
Krouk, G., Lacombe, B., Bielach, A., Perrine-Walker, F., Malinska, K., Mounier, E., et al. (2010). Nitrate-regulated auxin transport by NRT1.1 defines a mechanism for nutrient sensing in plants. Dev. Cell 18 (6), 927–937. doi: 10.1016/j.devcel.2010.05.008
Kumar, S., Stecher, G., Li, M., Knyaz, C., Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35 (6), 1547–1549. doi: 10.1093/molbev/msy096
Ma, J., Wu, Y., Li, X., Li, M., Hou, L. (2021). Identification and bioinformatics analysis of NPF gene family members in Chinese cabbage (Brassica rapa subsp. pekinensis). J. Henan Agric. Sci. 50 (09), 117–127. doi: 10.15933/j.cnki.10043268.2021.09.014
Miranda, R., Gomes-Filho, E., Prisco, J. T., Alvarez-Pizarro, J. (2016). Ammonium improves tolerance to salinity stress in Sorghum bicolor plants. Plant Growth Regul. 78, 121–131. doi: 10.1007/s10725-015-0079-1
Nan, H., Lin, Y., Wang, X., Gao, L. (2021). Comprehensive genomic analysis and expression profiling of cysteine-rich polycomb-like transcription factor gene family in tea tree. Hortic. Plant J. 7, 469–478. doi: 10.1016/j.hpj.2021.03.001
O’Brien, J. A., Vega, A., Bouguyon, E., Krouk, G., Gojon, A., Coruzzi, G., et al. (2016). Nitrate transport, sensing, and responses in plants. Mol. Plant 9 (6), 837–856. doi: 10.1016/j.molp.2016.05.004
Rachmilevitch, S., Huang, B., Lambers, H. (2006). Assimilation and allocation of carbon and nitrogen of thermal and nonthermal Agrostis species in response to high soil temperature. New Phytol. 170 (3), 479–490. doi: 10.1111/j.1469-8137.2006.01684.x
Rubio, V., Linhares, F., Solano, R., Martin, A. C., Iglesias, J., Leyva, A., et al. (2001). A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev. 15 (16), 2122–2133. doi: 10.1101/gad.204401
Saeed, M., Dahab, A., Wangzhen, G., Tianzhen, Z. (2012). A cascade of recently discovered molecular mechanisms involved in abiotic stress tolerance of plants. OMICS 16 (4), 188–199. doi: 10.1089/omi.2011.0109
Saito, H., Oikawa, T., Hamamoto, S., Ishimaru, Y., Kanamori-Sato, M., Sasaki-Sekimoto, Y., et al. (2015). The jasmonate-responsive GTR1 transporter is required for gibberellin-mediated stamen development in Arabidopsis. Nat. Commun. 6, 6095. doi: 10.1038/ncomms7095
Schmitz, R. J., Grotewold, E., Stam, M. (2022). Cis-regulatory sequences in plants: their importance, discovery, and future challenges. Plant Cell 34 (2), 718–741. doi: 10.1093/plcell/koab281
Singh, M., Singh, V. P., Prasad, S. M. (2016). Responses of photosynthesis, nitrogen and proline metabolism to salinity stress in Solanum lycopersicum under different levels of nitrogen supplementation. Plant Physiol. Biochem. 109, 72–83. doi: 10.1016/j.plaphy.2016.08.021
Sugiura, M., Georgescu, M. N., Takahashi, M. (2007). A nitrite transporter associated with nitrite uptake by higher plant chloroplasts. Plant Cell Physiol. 48 (7), 1022–1035. doi: 10.1093/pcp/pcm073
Tahir, I., Nakata, N. (2005). Remobilization of nitrogen and carbohydrate from stems of bread wheat in response to heat stress during grain filling. J. Agron. Crop Sci. 191, 106–115. doi: 10.1111/j.1439-037X.2004.00127.x
Taochy, C., Gaillard, I., Ipotesi, E., Oomen, R., Leonhardt, N., Zimmermann, S., et al. (2015). The Arabidopsis root stele transporter NPF2.3 contributes to nitrate translocation to shoots under salt stress. Plant J. 83 (3), 466–479. doi: 10.1111/tpj.12901
Taub, D. R., Wang, X. (2008). Why are nitrogen concentrations in plant tissues lower under elevated CO2? a critical examination of the hypotheses. J. Integr. Plant Biol. 50 (11), 1365–1374. doi: 10.1111/j.1744-7909.2008.00754.x
Teng, Y., Liang, Y., Wang, M., Mai, H., Ke, L. (2019). Nitrate transporter 1.1 is involved in regulating flowering time via transcriptional regulation of FLOWERING LOCUS c in Arabidopsis thaliana. Plant Sci. 284, 30–36. doi: 10.1016/j.plantsci.2019.04.002
Tsay, Y. F., Chiu, C. C., Tsai, C. B., Ho, C. H., Hsu, P. K. (2007). Nitrate transporters and peptide transporters. FEBS Lett. 581 (12), 2290–2300. doi: 10.1016/j.febslet.2007.04.047
Villalta, I., Reina-Sánchez, A., Bolarín, M. C., Cuartero, J., Belver, A., Venema, K., et al. (2008). Genetic analysis of na+ and K + concentrations in leaf and stem as physiological components of salt tolerance in tomato. Theor. Appl. Genet. 116 (6), 869–880. doi: 10.1007/s00122-008-0720-8
Wang, Y. Y., Cheng, Y. H., Chen, K. E., Tsay, Y. F. (2018). Nitrate transport, signaling, and use efficiency. Annu. Rev. Plant Biol. 69, 85–122. doi: 10.1146/annurev-arplant-042817-040056
Willems, E., Leyns, L., Vandesompele, J. (2008). Standardization of real-time PCR gene expression data from independent biological replicates. Anal. Biochem. 379 (1), 127–129. doi: 10.1016/j.ab.2008.04.036
Wu, C., Cheng, H., Li, S., Zuo, D., Lin, Z., Zhang, Y., et al. (2021). Molecular cloning and characterization of GhERF105, a gene contributing to the regulation of gland formation in upland cotton (Gossypium hirsutum l.). BMC Plant Biol. 21 (1), 102. doi: 10.1186/s12870-021-02846-5
You, H., Liu, Y., Minh, T. N., Lu, H., Zhang, P., Li, W., et al. (2020). Genome-wide identification and expression analyses of nitrate transporter family genes in wild soybean (Glycine soja). J. Appl. Genet. 61 (4), 489–501. doi: 10.1007/s13353-020-00571-7
Yuan, T., Zhu, C., Yang, K., Song, Z., Gao, Z. (2021). Identification of nitrate transporter gene family PeNPFs and their expression analysis in Phyllostachys edulis. For. Res. 34 (03), 1–12. doi: 10.13275/j.cnki.lykxyj.2021.03.001
Yu, J., Xie, Q., Li, C., Dong, Y., Zhu, S., Chen, J., et al. (2020). Comprehensive characterization and gene expression patterns of LBD gene family in Gossypium. Planta 251 (4), 81. doi: 10.1007/s00425-020-03364-8
Zhang, T., Hu, Y., Jiang, W., Fang, L., Guan, X., Chen, J., et al. (2015). Sequencing of allotetraploid cotton (Gossypium hirsutum l. acc. TM-1) provides a resource for fiber improvement. Nat. Biotechnol. 33 (5), 531–537. doi: 10.1038/nbt.3207
Zhang, H., Li, S., Shi, M., Wang, S., Shi, L., Xu, F., et al. (2020). Genome-wide systematic characterization of the NPF family genes and their transcriptional responses to multiple nutrient stresses in Allotetraploid rapeseed. Int. J. Mol. Sci. 21 (17), 5947. doi: 10.3390/ijms21175947
Zhang, J., Liu, Y. X., Zhang, N., Hu, B., Jin, T., Xu, H., et al. (2019). NRT1.1B is associated with root microbiota composition and nitrogen use in field-grown rice. Nat. Biotechnol. 37 (6), 676–684. doi: 10.1038/s41587-019-0104-4
Zhang, G. B., Yi, H. Y., Gong, J. M. (2014). The Arabidopsis ethylene/jasmonic acid-NRT signaling module coordinates nitrate reallocation and the trade-off between growth and environmental adaptation. Plant Cell 26 (10), 3984–3998. doi: 10.1105/tpc.114.129296
Zhao, L., Chen, P., Liu, P., Song, Y., Zhang, D. (2021). Genetic effects and expression patterns of the nitrate transporter (NRT) gene family in Populus tomentosa. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.661635
Zhao, Z., Li, M., Xu, W., Liu, J. H., Li, C. (2022). Genome-wide identification of NRT gene family and expression analysis of nitrate transporters in response to salt stress in Poncirus trifoliata. Genes (Basel) 13 (7), 1115. doi: 10.3390/genes13071115
Zheng, C., Li, P., Sun, M., Pang, C., Zhao, X., Gui, H., et al. (2018). Effeects of foliar nitrogen applications on the absorption of nitrate nitrogen by cotton roots. Cotton Sci. 30 (04), 338–343. doi: 10.11963/1002-7807.zcsdhl.20180703
Keywords: cotton, NPF genes, genome-wide identification, abiotic stresses, gene expression
Citation: Liu J, Wang C, Peng J, Ju J, Li Y, Li C and Su J (2023) Genome-wide investigation and expression profiles of the NPF gene family provide insight into the abiotic stress resistance of Gossypium hirsutum. Front. Plant Sci. 14:1103340. doi: 10.3389/fpls.2023.1103340
Received: 20 November 2022; Accepted: 06 January 2023;
Published: 19 January 2023.
Edited by:
Libei Li, Zhejiang Agriculture and Forestry University, ChinaReviewed by:
Zhiyong Ni, Xinjiang Agricultural University, ChinaCopyright © 2023 Liu, Wang, Peng, Ju, Li, Li and Su. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Junji Su, c3VqakBnc2F1LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.