- Key Laboratory of Crop Physiology, Ecology and Genetic Breeding, Ministry of Education, Jiangxi Agricultural University, Nanchang, China
The cultivated Peanut (Arachis hypogaea L.), an important oilseed and edible legume, are widely grown worldwide. The R2R3-MYB transcription factor, one of the largest gene families in plants, is involved in various plant developmental processes and responds to multiple stresses. In this study we identified 196 typical R2R3-MYB genes in the genome of cultivated peanut. Comparative phylogenetic analysis with Arabidopsis divided them into 48 subgroups. The motif composition and gene structure independently supported the subgroup delineation. Collinearity analysis indicated polyploidization, tandem, and segmental duplication were the main driver of the R2R3-MYB gene amplification in peanut. Homologous gene pairs between the two subgroups showed tissue specific biased expression. In addition, a total of 90 R2R3-MYB genes showed significant differential expression levels in response to waterlogging stress. Furthermore, we identified an SNP located in the third exon region of AdMYB03-18 (AhMYB033) by association analysis, and the three haplotypes of the SNP were significantly correlated with total branch number (TBN), pod length (PL) and root-shoot ratio (RS ratio), respectively, revealing the potential function of AdMYB03-18 (AhMYB033) in improving peanut yield. Together, these studies provide evidence for functional diversity in the R2R3-MYB genes and will contribute to understanding the function of R2R3-MYB genes in peanut.
Introduction
MYB transcription factor is one of the most numerous families of transcription factors in plants. They are distinguished by a conserved sequence of four incomplete amino acid repeats (R), with about 52 amino acids serving as the DNA-binding amino acids. Three a-helices are formed by each repeat, and the second and third helices of each repeat create a three-dimensional (3D) HTH structure with three evenly spaced tryptophan (or hydrophobic) residues (Ogata et al., 1996). Each repeat’s third helix serves as a “recognition helix,” coming into touch with the DNA and becoming enmeshed in the main groove (Jia et al., 2004). According to the number of adjacent duplicates, MYB proteins can be classified into several types, with R2R3-MYB proteins accounting for the majority of them in plants (Dubos et al., 2010; Liu et al., 2015).
In many aspects of plant life, including primary and secondary metabolism, cell fate, developmental processes, and response to biotic and abiotic stimuli, R2R3-MYB transcription factors are crucial players (Martin and Paz-Ares, 1997; Jin and Martin, 1999; Dubos et al., 2010; Liu et al., 2015; Cao et al., 2020). The maize C1 gene, related to the mammalian transcription factor C-MYB and is involved in the control of anthocyanin production, is the first MYB gene discovered in plants (Paz-Ares et al., 1987). ErMYB1 and ErMYB2 are regarded as inhibitors and activators, respectively, of the development of secondary cell walls in the eucalyptus (Goicoechea et al. 2005; Legay et al. 2007; Legay et al. 2010). ABA-mediated responses to environmental cues are mediated by the AtMYB13, AtMYB15, AtMYB33, and AtMYB101 (Reyes and Chua, 2007). AdMYB3 has been reported to be involved in anthocyanin biosynthesis and flower development in apples (Vimolmangkang et al., 2013). Members of the transcription factors that resemble MYBMIXTA are involved in starting the formation of cotton seed fiber (Bedon et al. 2014). AhTc1, encoding an R2R3-MYB transcription factor, play important role in regulating anthocyanin biosynthesis in peanut (Zhao et al., 2019). In rice, OsMYB30, an R2R3-MYB transcription factor, regulates the phenylalanine ammonia-lyase pathway to give brown planthopper resistance (He et al. 2020). GmMYB14 controls plant structure via the brassinosteroid pathway, contributing to high-density yield and drought resistance in soybean (Chen et al. 2021). MsMYB741 is involved in alfalfa resistance to aluminum stress by regulating flavonoid biosynthesis (Su et al. 2022). OsMYB60 positively regulates cuticular wax biosynthesis and this helps rice (Oryza sativa) plants tolerate drought stress (Jian et al., 2022).
Cultivated peanut (Arachis hypogaea L.), one of the most widely consumed legumes worldwide, has been used to meet the nutritional needs of developing countries globally (Toomer, 2018). It originated in South America from a heterozygous cross between two diploid ancestors and was domesticated and widely grown in the tropics and subtropics (Bertioli et al., 2016). Most of the Arachis genus is diploid (Sharma and Bhatnagar-Mathur, 2006). However, only tetraploid peanuts have been domesticated and widely grown to meet human nutritional requirements (Bertioli et al., 2016). Polyploid plants often exhibit greater environmental adaptability (Shimizu-Inatsugi et al., 2017). Recently, the contribution of polyploidization to important agronomic traits, including seed quality, fruit shape, and flowering time, has been reported for several crop species lineages such as soybean (Glycine max), wheat (Triticum aestivum), and cotton (Gossypium hirsutum) (Zhang et al., 2015). The large and close subgenomes of cultivated peanut genomes make genome assembly difficult (Bertioli et al., 2016). Due to advanced high-throughput sequencing technology and high-quality assembly and annotation, the sequencing of cultivated peanut was completed in 2019, the cultivated peanut (Arachis hypogaea L.) is of hybrid origin and has a polyploid genome that contains essentially complete sets of chromosomes from two ancestral species. (Bertioli et al., 2019; Chen et al., 2019; Zhuang et al., 2019). Based on high-quality whole-genome sequencing and assembly engineering, genome-wide characterization of the R2R3-MYB gene has been accomplished in various plants, such as Arabidopsis thaliana, rice, maize, soybean, eucalyptus, tomato, Chinese bayberry (Stracke et al., 2001; Jia et al., 2004; Chen et al., 2006; Du et al., 2012a; Soler et al., 2015; Li et al., 2016; Cao et al., 2021).
Most of the previous studies focused on the regulation mechanism of light-inducible anthocyanin (Dubos et al., 2010; Liu et al., 2015; Cao et al., 2020), but peanut is a very special and important crop. Given the fact that this crop possesses the unique characteristics of “aerial flowers and subterranean fruit,” the genes responsible for flavonoid synthesis (in peanut testa) and stress response are likely distinct from those in model plants such as Arabidopsis and rice. In this study, we characterized 196 R2R3-MYB transcription factors genome-wide and analyzed their phylogenetic relationships, motif composition, gene structure, chromosome distribution, gene duplication, tissue and stress response expression pattern and. Furthermore, association analysis identified a candidate gene highly correlated with total branch number (TBN), pod length (PL) and root-shoot ratio (RS ratio).Our study will contribute to the understanding of the function of the R2R3-MYB genes in cultivated peanut and provide candidate genes for development and stress response.
Materials and methods
Identification and conserved DNA-binding domain analysis of R2R3-MYBs in peanut
To identify R2R3-MYBs in peanut, the A. hypogaea cv. Tifrunner protein sequences were retrieved from the PeanutBase (https://peanutbase.org). The MYB DNA-binding domain (PF00249) was exploited for the identification of R2R3-MYB genes in the peanut genome by using the HMMER 3.3.2 program at a standard E value <1×10–5 (http://pfam.xfam.org/search#tabview=tab1). In total, 204 predicted gene models were found with two consecutive repeats of the MYB domain. All but five (199) were also retrieved when performing a BLASTP analysis using the previously identified genes of A. hypogaea against the whole A. thaliana R2R3-MYB gene data set (Dubos et al., 2010) with a cut-off e-value of e-40. Subsequently, protein sequences were evaluated for the presence of the MYB domain against the repository of the NCBI CDD (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) and SMART databases (http://smart.embl-heidelberg.de/). Finally, 196 R2R3-MYB genes were obtained after eliminating incomplete and uncertain sequences. The predicted molecular weights and the theoretical isoelectric point (pI) were obtained by the Expasy proteomics server (https://web.expasy.org/compute_pi/). The prediction of transmembrane helices in AhR2R3-MYB proteins was analyzed by TMHMM - 2.0 (https://services.healthtech.dtu.dk/service.php?TMHMM-2.0).
The 196 R2R3-MYB protein sequences in peanuts were performed by multiple sequence alignment using ClustalX (Larkin et al., 2007). The alignment of 196 peanut R2R3-MYB domains was performed using ClustalX and DNAMAN (Lynnon Biosoft). The amino acid residue distributions of the conserved MYB domains of AhR2R3-MYBs were created using the WebLogo program with default parameters (http://weblogo.berkeley.edu/logo.cgi) (Crooks et al., 2004).
Construction of phylogenetic tree
The protein sequences of the 126 A.thaliana R2R3-MYBs were downloaded from the TAIR (http://www.arabidopsis.org/). The protein sequences of R2R3-MYB proteins from A. hypogaea and A.thaliana (protein sequence information is listed in Supplementary Table 1) were aligned by the MAFFT with the FFT-NS-i algorithm (Katoh et al., 2019), and the multiple sequence alignments were used for phylogenetic analysis. The phylogenetic tree was constructed by the neighbor-joining method of MEGA 7.0 with 1000 bootstrap replicates based on the p-distance model and pairwise deletion for gap treatment (Kumar et al., 2016). The phylogenetic tree was retouched by FigTree (http://tree.bio.ed.ac.uk/software/figtree/). For the construction of the phylogenetic trees of R2R3-MYB proteins from Arachis. hypogaea the same method described above was adopted.
Motif and gene structure analysis
Information on the intron-exon position and splicing sites for each gene model in the corresponding chromosome scaffold was downloaded from PeanutBase. Furthermore, the MEME program (https://meme-suite.org/meme/meme_5.3.3/tools/meme) was used for the identification of motifs of 196 R2R3-MYB protein sequences in peanut. The optimized parameters of MEME were employed as follows: the number of motifs that MEME find, 10; and the optimum width of each motif, 6–60 residues (Bailey et al., 2009). The gene exon-intron pattern and MEME results were also visualized by CFVisual_V2.1.5 (Chen et al., 2022).
Chromosome localization, duplications, and evolutionary analysis of AhR2R3-MYBs
The information on chromosome length and R2R3-MYB gene locations was acquired from the PeanutBase (https://peanutbase.org) (Supplementary Table 2), and the figure was created by TBtools (Chen et al., 2020). The whole-genome sequences and annotation documents of A. hypogaea were downloaded to PeanutBase (https://peanutbase.org). Then, the One Step MCScanx program of TBtools was executed to analyze the synteny relationships of genomes. We identified gene pairs with physical distance within 100 kb, with no more than 10 genes spaced in between, and in the same subgroup as tandem repeat gene pairs according to Hanada et al. (2008). The duplication pattern of the AhR2R3-MYB genes was visualized by the Amazing Super Circos package of TBtools. The Ka/Ks value was completed by the Simple Ka/Ks Calculator program of TBtools. Duplication time was calculated by the following formula as described by Bertioli et al. (2016): T = Ks/2λ (λ = 8.12 × 10−9).
Plant material and stress treatment
The peanut (Arachis hypogaea L.) cultivar ‘Changhua18’, a germplasm resource preserved in our laboratory, was planted in a pot with a 1:1 mixture of nutrient soil and vermiculite, 450 mm long, 335 mm wide, and 170 mm high. When the plants were grown for about 8 weeks, the treatment group was subjected to waterlogging treatment according to Zeng et al. (2021), while the control group was kept under normal growth conditions. The samples (three seedlings per repeat) were collected at 0 h, 6 h, 24 h, 3 days, and 5 days after treatment, respectively. Subsequently, the samples were rapidly frozen using liquid nitrogen and stored at -80°C for RNAseq.
RNA-seq expression analysis
The raw read counts in Arachis hypogaea RNAseq samples were downloaded from PeanutBase (https://peanutbase.org). The data were obtained from 22 tissues at different developmental stages in peanut with three biological repeats (Clevenger et al., 2016) with all raw data deposited as BioSamples SAMN03944933–SAMN03944990. HTSeq was used to generate raw reads that were uniquely mapped on the Arachis hypogaea genome. StringTie and Ballgown were used for the FPKM calculation (Pertea et al., 2016). The transcript profiles for AhR2R3-MYB genes were displayed in TBtools (Chen et al., 2020).
These cDNA libraries generated from the samples were sequenced by Metware Biotechnology Ltd. on the Illumina sequencing platform. (Wuhan, China). Download the reference genome and its annotation files from NCBI (https://ftp.ncbi.nlm.nih.gov/genomes/), use HISAT v2.1.0 to construct the index and compare clean reads to the reference genome. The featureCounts v1.6.2/StringTie v1.3.4d was used to calculate the gene alignment and FPKM. Gene expression patterns were also charted by TBtools. Quantitative RT-PCR (qRT-PCR) was performed to verify the transcriptome data. RNAprep Pure Plant Plus Kit (Tiangen Biotech, Co., Beijing, China) was utilized to extract RNA from control and treated peanut samples. The PrimeScritTM RT Kit with gDNA eraser user manual was used to prepare the cDNA (perfect real-time, Takara Biomedical Technology, Ltd., Beijing, China). The primers were designed by TBtools and were shown in Supplementary Table 3. Subsequently, qRT-PCR was performed using the ABI 7500 qRT-PCR detection system (ABI, United States) with SYBR Green Kit (Tiangen, Beijing, China). The ABI 7500 real-time PCR program was 95°C for 15 min, followed by 40 cycles of 95° C for 10 s, and 60°C for 30 s in a 20 µl volume. Three technical repeats of qRT-PCR were carried out, and the relative expression level was determined using 2-△△Ct technique.The results of differential expression analysis between homeolog pairs of A and B subgenomes across tissues and pod developmental stages in Arachis hypogaea were downloaded from PeanutBase (https://peanutbase.org) (Bertioli et al., 2019). Differential gene expression analysis was performed using the DESeq2 package (v1.14.1) with log2 fold change >= 1 and Benjamini-Hochberg adjusted P-value < 0.05 as the statistical cutoff for differentially expressed genes. Charts generated by GraphPad Prism 9.
Association analysis of the peanut R2R3-MYBs with TBN, PL and RS ratio
Total branch number, length of root and shoot, pod size and RS phenotypes were assessed using a randomized complete block design and replicated in five environments. SNPs of the R2R3-MYBs were obtained from transcriptome data set of a peanut germplasm population with 146 accessions (alleles in each polymorphism with minor allele frequency >0.05). Association analysis was performed with TASSEL 3.0 using an MLM Q + K model. Figures were generated by GraphPad Prism 9.
Results
Identification and conserved DBD analysis of R2R3-MYB genes in peanut
A total of 196 R2R3-MYB genes were obtained after genome-wide screening and exclusion in A. hypogaea cv. Tifrunner. The 196 genes were named AhMYB001-AhMYB196 according to their physical location on chromosome. The mRNA length of the AhR2R3-MYBs ranged from 753 to 3712 bps (Supplementary Table 4); the proteins length ranged from 192 to 916 amino acids, with predicted molecular weights from 21.95 to 103.72 kDa; the theoretical isoelectric point (pI) ranged from 4.60 to 10.32. Furthermore, none of the proteins were predicted to contain transmembrane domains (Supplementary Table 4).
To investigate the MYB conserved domains of R2R3-MYB transcription factors in peanut, we performed WebLogo and multiple alignment (ClustalX) analysis using amino acid sequences of the R2R3 repeats (Supplementary Figure 1). The results showed that the R2 and R3 repeat consisted of two repeats of approximately 51 amino acid residues, and all of them contained highly conserved tryptophan residues (Trp, W) (Supplementary Figure 2). The R2 repeat contains three highly conserved W residues at positions 5, 26, and 47, while only two highly conserved W residues were uncovered at positions 24 and 43 in the R3 repeat, and the W residue at position 5 was generally replaced by phenylalanine or isoleucine (Phe, F or Ile, I). Tryptophan residues formed a hydrophobic core maintaining the stability of the helix-turn-helix (HTH) structure in the DNA binding domain, and the basic and polar amino acids adjacent to that tryptophan were thought to be directly involved in DNA binding (Saikumar et al., 1990). Several highly conserved polar amino acid residues were also found around the third W residue in each repeat, for example, asparagine (Asn, N), arginine (Arg, R), and lysine (Lys, K) around the third tryptophan in the R2 repeat, and K and N near the third W residue in the R3 repeat, which might be directly involved in binding to DNA. These polar amino acids, which are conserved around tryptophan, were also highly conserved in different species, such as Arabidopsis and tomato (Stracke et al., 2001; Li et al., 2016). Overall, most of the conserved amino acid residues were mainly distributed between the second and third conserved tryptophan residues, with the first tryptophan residue being relatively less conserved. Therefore, those conserved residues probably maintain the function of the DNA binding domain together with the conserved tryptophan.
Phylogenetic analysis of AhR2R3-MYBs in cultivated peanut and Arabidopsis
To investigate the evolutionary relationship between the R2R3-MYB genes in peanuts and Arabidopsis, the 196 predicted AhR2R3-MYB proteins were subjected to multiple sequence alignment along with 126 Arabidopsis R2R3-MYB proteins, and their evolutionary relationships were inferred by constructing a neighbor-joining phylogenetic tree. The 322 R2R3-MYB proteins were classified into 48 subgroups based on the topology and bootstrap value of the phylogenetic tree (Figure 1 and Supplementary Figure 3). Since the R2R3-MYB genes have been intensively studied in A. thaliana and most subgroups contained at least one AtR2R3-MYB gene, we named these subgroups according to the nomenclature of (Kranz et al., 1998) revised by Stracke et al. (2001) and Dubos et al. (2010). When a subgroup name was not yet determined in A. thaliana, we named the subgroup after the member of A. thaliana with the most distinct functional characteristics. In general, the phylogenetic characteristics of A. thaliana described in this paper were generally consistent with those described previously (Stracke et al., 2001; Dubos et al., 2010). The only exception was the A. thaliana genes of the subgroups 20 and 25, were split in two respectively (S20a and S20b; S25a and S25b), and the genes from 10 and 24 were merged (S10&24). As shown in Figure 1, 40 subgroups contained at least one gene from peanut, and the other eight subgroups contained genes only from peanut, and they were named as new subgroups 1-8 in this study. The distribution of AhR2R3-MYBs in the 40 subgroups was biased, varying from one (NS-8) to 17 members (S14). Notably, the number of R2R3-MYB genes in almost all subgroups was unbiased in both subgenomes, revealing a close association between two subgenomes of cultivated peanut as described by (Bertioli et al., 2016; Bertioli et al., 2019). The topology of the neighbor-joining tree for AhR2R3-MYB genes was in good agreement with the subgroup described above (Supplementary Figure 4).
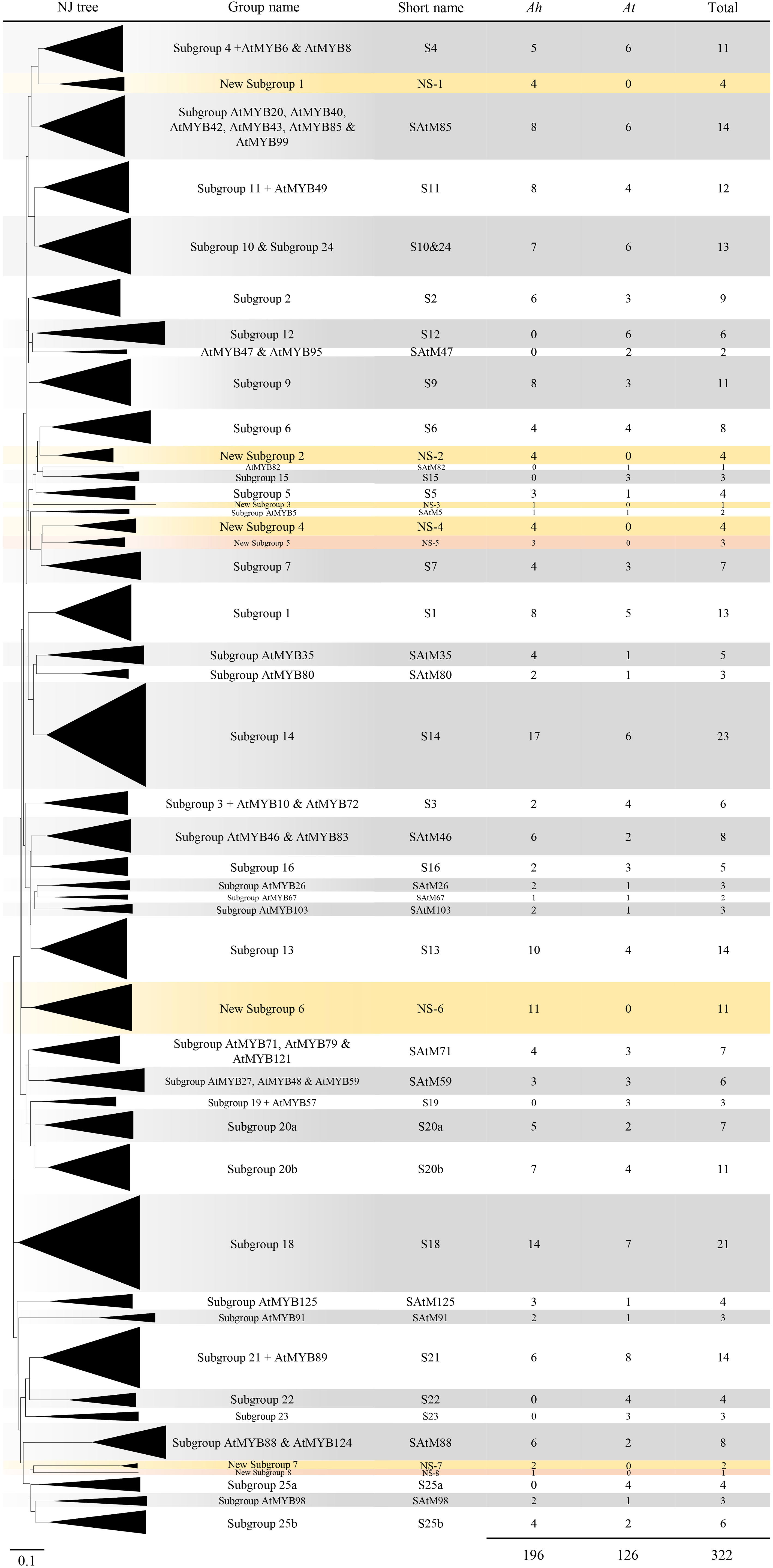
Figure 1 Neighbor-joining phylogenetic tree of R2R3-MYB proteins from peanut (Arachis hypogaea L.) and Arabidopsis. Each triangle represented an R2R3-MYB subgroup, defined based on the topology of the tree and the bootstrap values. Subgroup names were included next to each clade together with a short name to simplify the nomenclature. The number of genes of each species for each subgroup was also included. The eight new subgroups in peanut were marked in yellow or coral, while the other subgroups were in gray and white.
Motif composition and gene structure of the AhR2R3-MYB genes
To investigate the relationship between subgroup classification and function of the peanut R2R3-MYBs, 10 conserved motifs were identified in the AhR2R3-MYBs through MEME program search (Supplementary Figure 5). The DNA binding domain of AhR2R3-MYB was represented by motifs 3, 6, 1, 2. Motifs 1 and 2 contained the amino acid sequence of the third helix forming the MYB domain, which is involved in the recognition and binding of cis-acting elements (Ogata et al., 1996; Jia et al., 2004). Motifs 5 and 8 were only presented in SAtM88, while motif 9 only presented S20a and S20b, suggesting potential specific functions of these subgroups (Figures 2A, B). In general, most of the motif compositions of members in the same subgroup were similar at N-terminal, but differ at C-terminal, and the motif compositions of the members in different subgroups were not identical.
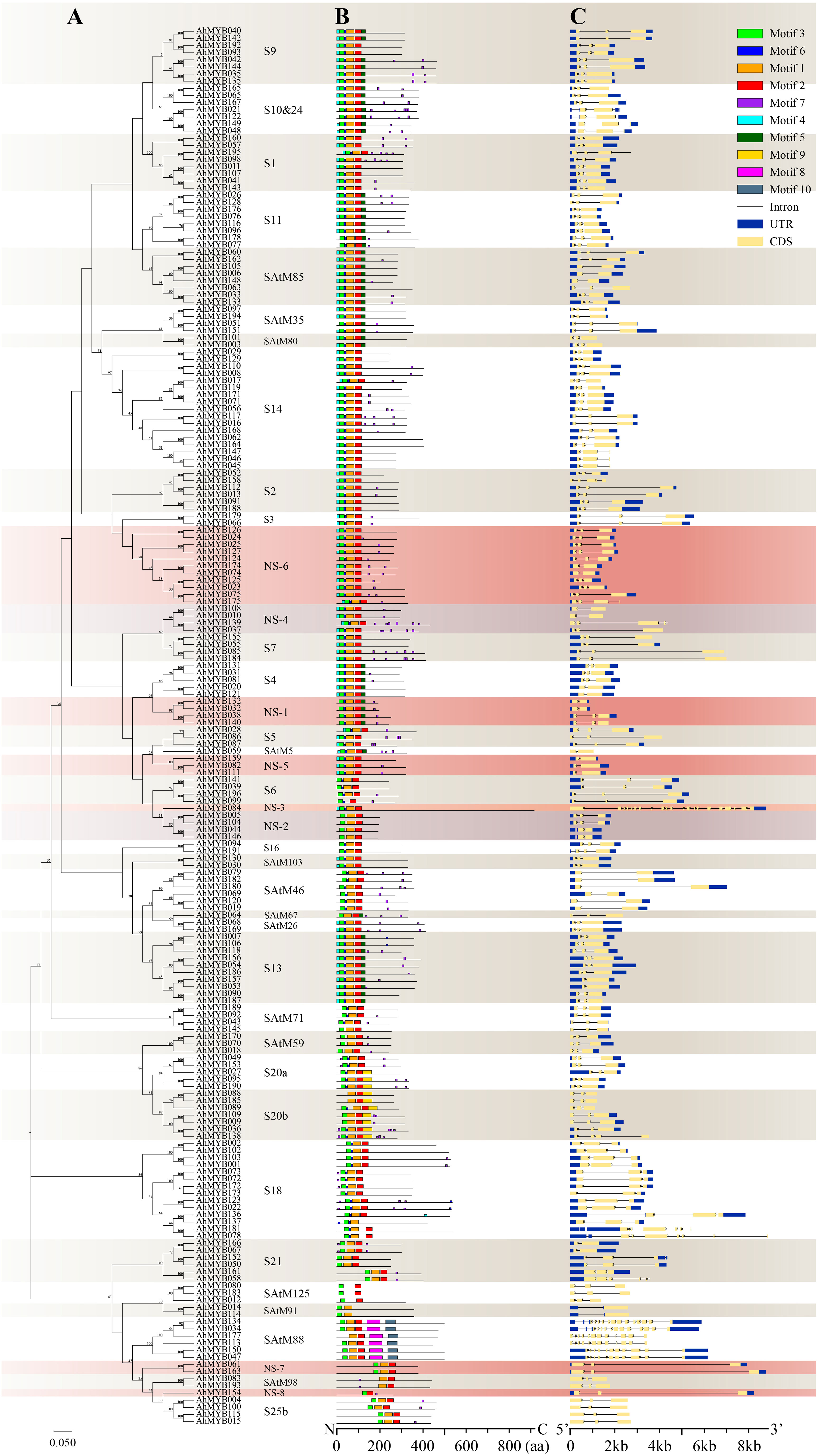
Figure 2 Phylogenetic relationships, conserved motifs, and gene structure analysis of peanut R2R3-MYBs. (A) The neighbor-joining phylogenetic tree was constructed by aligning the full-length amino acid sequences of 196 R2R3-MYBs in peanut. Coral and purple colors mark eight new subgroups, while beige and white indicate other subgroups. (B) The ten conserved motifs were shown in different colors and their specific sequence information was provided in Supplementary Figure S5. (C) The yellow box, blue box, and black line in the gene structure diagram represented CDS, UTR, and introns, respectively.
Exon and intron structure analysis showed that all AhR2R3-MYB genes possessed 1 to 21 exons (Figure 2C). In general, the size of introns was variable, but the locus and phase of introns were relatively conservative among subgroups (Jiang et al., 2004). Most of the R2R3-MYB genes (77%) contained two conserved introns, 14% contained one conserved intron, and the rest showed a different number. Genes in the same subgroup have similar gene structures and highly conserved in intron phasing (Figures 2A, C). The multiple introns of a gene provide the opportunity to selectively splice and provide variant proteins that may play different roles in biological processes (Min et al., 2015). Most members of the subgroups are intron-poor (Contain three or fewer) or intron-less genes. However, we found that genes in subgroups NS-3, S18, and SAtM88 possessed an abundant number of introns, and multiple transcripts were present in all three subgroups except for AhMYB177.
Chromosome localization, duplication, and evolution of the R2R3-MYB genes
Chromosomal localization showed that the A and B subgenomes contained 99 and 97 R2R3-MYB genes, respectively (Figure 3). This suggested that the distributions of R2R3-MYB genes between the two subgenomes were almost not biased. In addition, we found that the distribution of genes on the corresponding chromosomes was similar between A and B subgenomes, except for Chr07 and Chr17, Chr08 and Chr18. This was probably the result of the complex rearrangement event on chromosomes 7, and 8 of two diploid wild ancestors, and subsequent retention of this rearrangement after polyploidization (Bertioli et al., 2016; Bertioli et al., 2019). The R2R3-MYB genes were unevenly distributed among the 20 chromosomes. Chr03 (19), Chr08 (18), Chr13 (18), and Chr18 (15) contained a larger number of R2R3-MYB genes. Chr02 (5), Chr07 (4), Chr10 (4), Chr17 (4), and Chr20 (4) possessed fewer R2R3-MYB genes. The density of R2R3-MYB genes on Chr08 was significantly higher than that on the other 19 chromosomes.

Figure 3 Distribution of 196 R2R3-MYB genes on 20 chromosomes. The name and length of the chromosome are displayed at the top of each chromosome. Yellow is for genes produced by segment duplication, orange is for genes from tandem duplication, and gray indicated genes that have experienced both types of duplication events. Subgroups were annotated to the right of the gene.
The R2R3-MYB genes were far more abundant in A. hypogaea than in lower terrestrial plants, suggesting that a large-scale gene duplication event occurred during the evolution of the plants (Du et al., 2015). To explore the mechanism of R2R3-MYB gene expansion in the cultivated peanut, we further analyzed the syntenic relationships between the peanut R2R3-MYB genes. Based on synteny analyses, 45 genes out of the 33 syntenic pairs in the A subgenome underwent segmental duplication events, while 39 genes out of the 24 syntenic pairs in the B subgenome were undergoing segmental duplication events (Figure 3 and Supplementary Table 5). 12 and eight genes in the A and B subgenomes, respectively, experienced tandem duplication episodes. In addition, significant numbers of orthologous genes were found between the A, and B subgenomes (Figure 4 and Supplementary Table 5). Based on synteny analyses, 173 of the 196 AhR2R3-MYBs had syntenic relationships between the two subgenomes. We found that some subgroups were expanded mainly by segment duplication, such as subgroups S11, NS-2, NS-4, SAtM85, S20a, S20b, S13, S1, S10&24, SAtM88, SAtM46, SAtM59, and NS-5, while tandem duplication occurred mainly in subgroups S5, NS-6, S20b, and S18 (Figure 3).
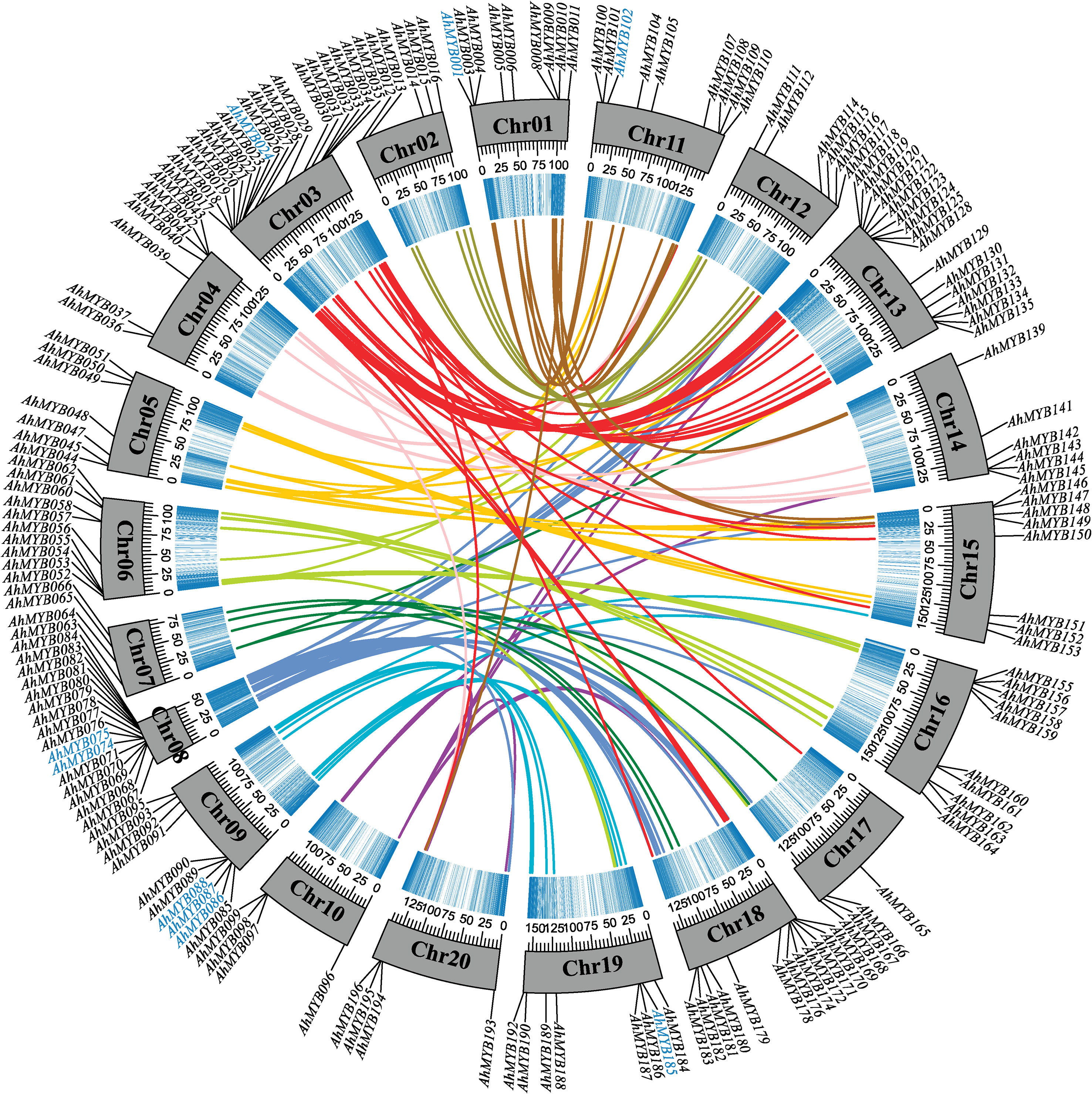
Figure 4 Circos diagram depicting the relationships of the chromosomes of A. hypogaea. Different colored connecting wires were used for different chromosome pairs. The blue color represented the density of genes. The scale for the gray bars was in megabases. Tandem duplication genes were highlighted in blue.
To further understand the evolution of R2R3-MYB duplication in peanut, we analyzed the rate of synonymous (Ks) and nonsynonymous substitutions (Ka) in gene duplication pairs (Supplementary Table 5). The Ks values for direct homologous gene pairs between A and B subgenomes range from 0 to 2.47. The frequency distribution peaks at Ks = 0.03, indicating a massive duplication event of R2R3-MYB genes 1.85 million years ago (Mya). Segment duplication and tandem duplication may occur in 147.75-35.91 and 163.87-2.20 Mya, respectively. Ka/Ks ratios for paralogous and orthologous ranged from 0-2.10 with an average of 0.27, whereas the ratios for tandem duplication ranged from 0.13-1.02 with an average of 0.36. The Ka/Ks analysis showed that the orthologous gene pair AhMYB052-AhMYB158 was neutrally selected and the tandem duplication pair AhMYB173-Arahy.QI53CA and the orthologous gene pairs AhMYB010-AhMYB108, Arahy.M1LASL-AhMYB118, and AhMYB031-AhMYB131 were subjected to positive selection, and all other synteny and tandem duplicated genes were subjected to purifying selection.
Tissue expression profiles of the R2R3-MYBs
To further study the expression pattern of the R2R3-MYB genes in different tissues and explore its function in peanut growth and development, the tissue expression profiles of the R2R3-MYB genes were analyzed by using the transcriptome data of 22 peanut tissues (AhMYB015 and AhMYB100 were not detectable in the dataset) (Figure 5 and Supplementary Table 6). The peanut R2R3-MYBs can be clustered into 10 groups according to their tissue expression pattern (Cluster 1-10). Most genes from same phylogenetic subgroup showed similar tissue expression patterns and were clustered into the same cluster, such as AhMYB023, AhMYB024, AhMYB074, AhMYB125, AhMYB126, AhMYB174, and AhMYB175 in NS-6, which highly expressed in fruit and pericarp, were cluster in cluster 3 (Figure 5). All the members of subgroup SAtM35, for example, were clustered in cluster 9 (Figure 5). However, a certain proportion of the AhMYBs from the same phylogenetic subgroup showed differential tissue expression, such as AhMYB028 and AhMYB087 from S5, AhMYB028 was highly expressed in the reproductive shoot, while AhMYB087 was mostly expressed in seed (Figure 5), this might be because genes belonging to the same phylogenetic subgroup exercise similar functions, but in different tissues responses to different developmental processes or different environmental stimuli ( (Dubos et al., 2010).
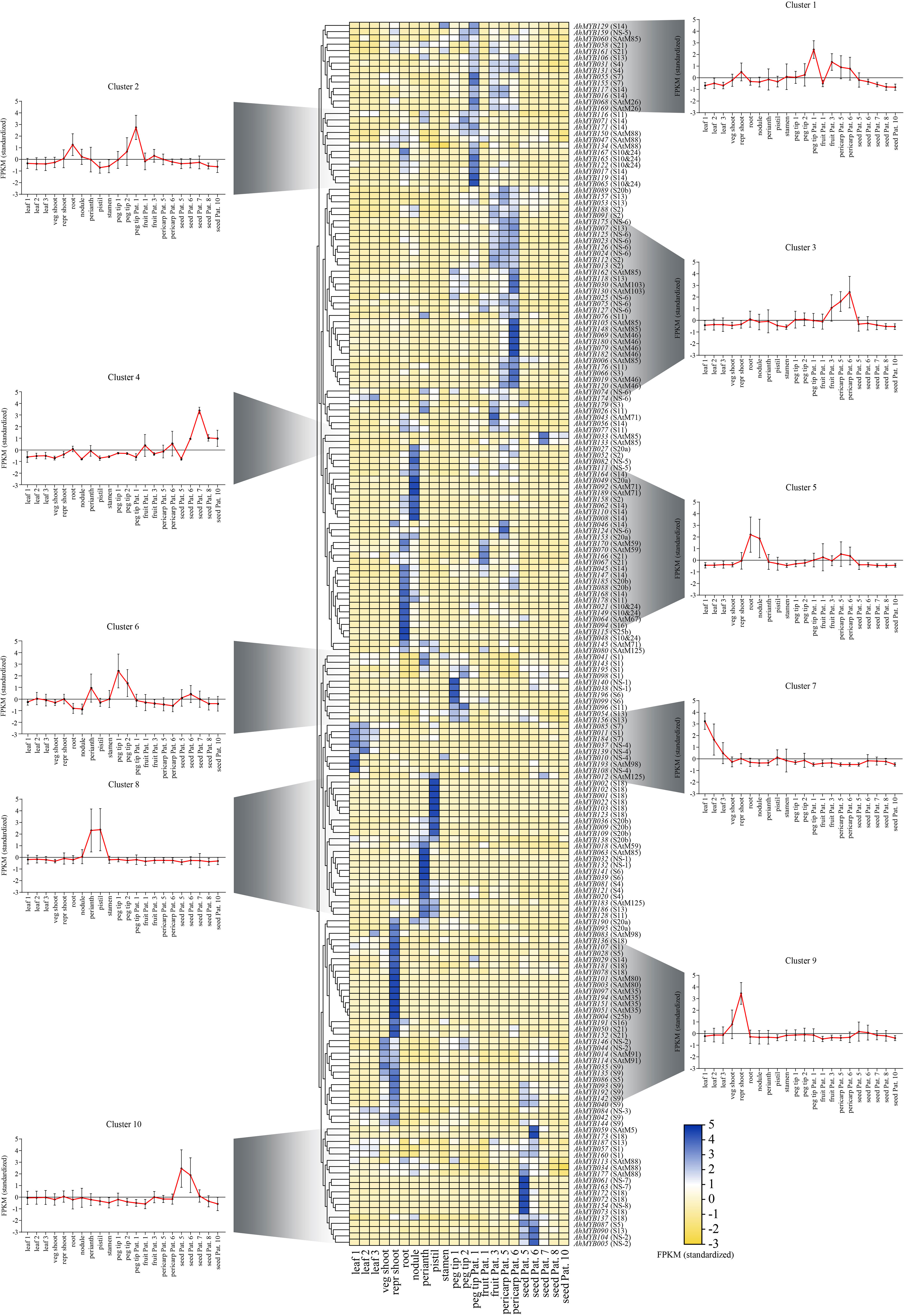
Figure 5 Heat map of the RNAseq transcript abundance pattern of the 194 R2R3-MYB genes in 22 different tissues in 10 expression clusters. For each gene, its name and the subgroup it belongs to were displayed on the right side of the heatmap. The expression pattern was generated based on the fragments per kilobase of exon per million fragments (FPKM) and analyzed by heatmap hierarchical clustering. The color scale (representing −2 to 5) was shown. The meanings of the abbreviations of the 22 tissues were as follows: seedling leaf 10 days post-emergence (leaf 1), main stem leaf (leaf 2), lateral stem leaf (leaf 3), vegetative shoot tip from the main stem (veg shoot), reproductive shoot tip from first lateral (repr shoot), 10-day roots (root), 25-day nodules (nodule), perianth, stamen, pistil, aerial gynophore tip (peg tip 1), subterranean peg tip (peg tip 2), Pattee 1 stalk (peg tip Pat. 1), Pattee 1 pod (fruit Pat. 1), Pattee 3 pod (fruit Pat. 3), Pattee 5 pericarp (pericarp Pat. 5), Pattee 6 pericarp (pericarp Pat. 6), Pat - tee 5 seed (seed Pat. 5), Pattee 6 seed (seed Pat. 6), Pattee 7 seed (seed Pat. 7), Pattee 8 seed (seed Pat. 8), and Pattee 10 seed (seed Pat. 10).
Interestingly, members of the eight peanut specific subgroups (NS-1 to 8) were distributed in all clusters except 2 and 4.The two NS-1 members (AhMYB032 and AhMYB132) were strong expressed in perianth; the two collinear gene pairs (AhMYB005 and AhMYB104, AhMYB044 and AhMYB146) from NS-2 subgroup were highly expressed in seed and shoot, respectively. AhMYB084 (NS-3) had significant expression both in leaf and shoot; the three members in NS-4 (AhMYB010, AhMYB037 and AhMYB139) expressed mainly in leaf; AhMYB082 and AhMYB111 in NS-5 showed higher expression level in nodule, but AhMYB150 was enriched in peg tip; most of the members in NS-6 were clustered in cluster 3 and were highly expressed in pericarp; members in NS-7 (AhMYB061 and AhMYB163) and NS-8 exhibited strong expression in seed. These subgroups contained totally 30 genes, 21 of which were expressed at high levels in reproductive organs, especially the 10 genes in NS-6 that had high transcript abundance in the early pod development stage after peg tip entry, suggesting an important role of these genes in peanut reproductive development.
Comparison of R2R3-MYB gene expression in subgenomes
The expression of homeologous gene pairs from A and B subgenomes was examined in various tissues and developmental phases (Supplementary Table 7 references from (Bertioli et al., 2019)). The total number of homeologous gene pairs with expression biased towards the A subgenome was similar to the previously reported it did not differ significantly from the number with expression biased towards the B subgenome (P = 0.16, binomial test; n = 47 and 42 for A and B, respectively) (Figure 6) (Chalhoub et al., 2014; Zhang et al., 2015). In nine tissues (lateral leaf, seeding leaf, vegetative shoot tip, reproductive shoot tip, perianth, gynoecium, pattee 6 seed, pattee 7 seed, and pattee 8 seed), of the homologous pairs, there were more A subgenome-highly expressed genes rather than B subgenome, whereas the other 10 tissues exhibited the reverse pattern. These differences were more significant in the four reproductive tissues (P < 0.05, binomial test). 14 homologous gene pairs had biased expression in just one tissue whereas 18 homologous gene pairs exhibited the same bias in several tissues (Supplementary Table 8). Interestingly, we identified four homologous gene pairs (AhMYB006 and AhMYB105, AhMYB011 and AhMYB107, AhMYB053 and AhMYB157, AhMYB074 and AhMYB174) showed opposite biased expression in specific tissues, such as homolog pair AhMYB074 and AhMYB174, AhMYB174 owned a higher expression level in lateral leaf {log2FoldChange (B.vs.A homeolog pair comparison):1.906081616}, while AhMYB074 had a stronger expression level in perianth {log2FoldChange (B.vs.A homeolog pair comparison): -2.204691792}.
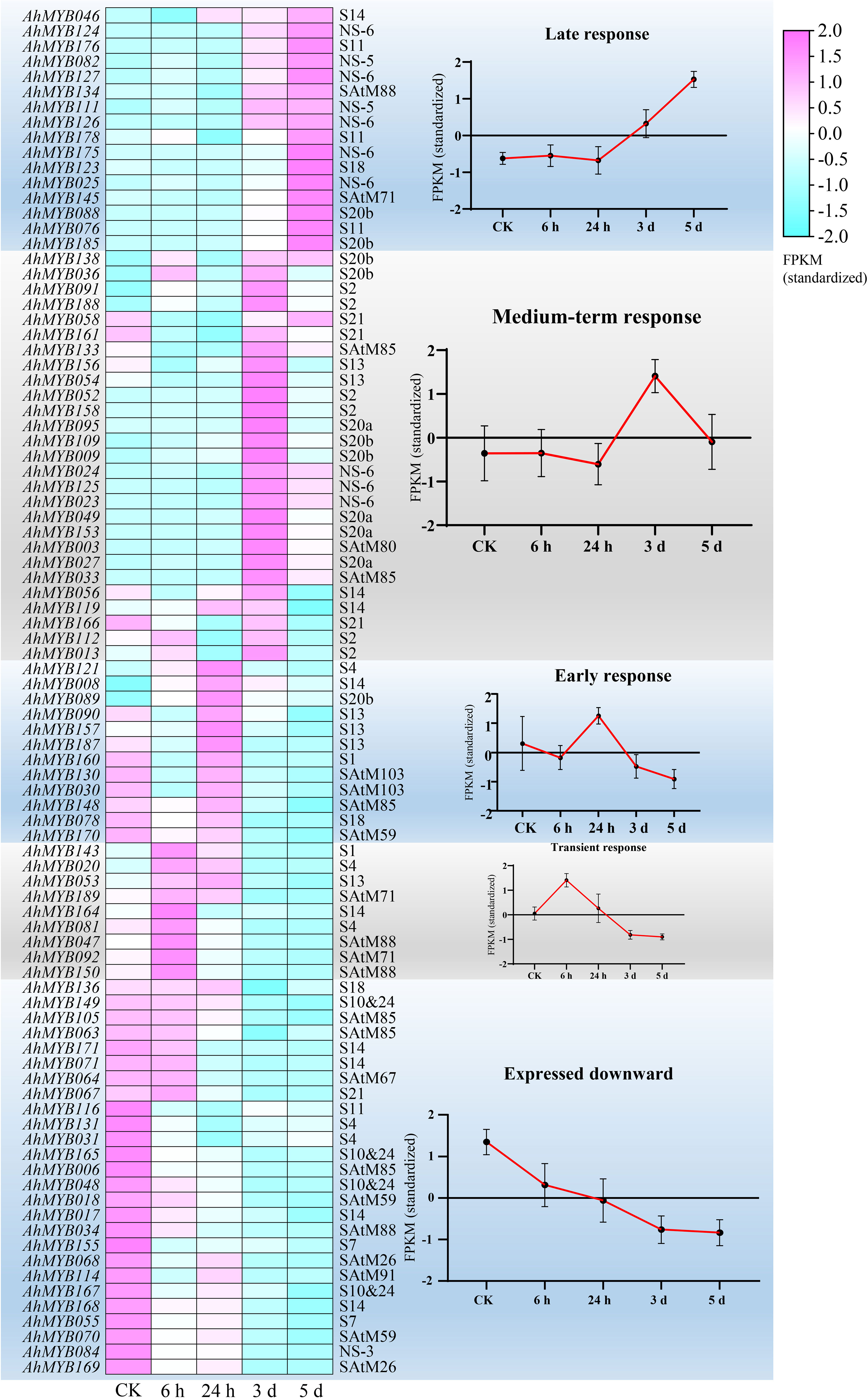
Figure 6 RNAseq transcript abundance patterns of 90 R2R3-MYB genes in root and stem tissues in five expression groups during water logging stress are shown as a heat map. The left and right sides of the heatmap, respectively, showed the names of each gene and the subgroup to which it belonged. Heatmap hierarchical clustering was used to construct the expression pattern based on the fragments per kilobase of exon per million fragments (FPKM). The color scale, which ranged from -2.0 to 2.0, was displayed. The labels at the bottom of the heat map indicate, from left to right, the control group, 6 h, 24 h, 3 days, and 5 days after water logging treatment.
Expression pattern of R2R3-MYB genes in peanut under waterlogging treatment
We examined the transcript abundance of the R2R3-MYB gene in peanut seedlings after water logging treatment to further investigate the function of the genes in water logging response. 90 genes from 24 subgroups (NS-6, S10&24, S11, S13, S14, S2, S20a, S20b, S21, SAtM85, SAtM88, etc.) were found response to waterlogging. According to the chronological order of the respond genes, they were divided into five categories (Figure 7). 26 genes showed a strong tendency to be down-regulated following the water logging treatment. Nine genes in the transitional response group showed strong elevated after 6 hours of treatment. Three genes (AhMYB121, AhMYB008, and AhMYB089) were considerably up-regulated among the 12 early responded genes, whereas the others showed a tendency of down- and subsequently up-regulation. At the time point of 3 and 5 days after treatment, a total of 27 and 16 genes were up-regulated, respectively. In addition, the results of qRT-PCR showed that the expression patterns of the 10 selected genes under water logging treatment were generally consistent with the transcriptome results (Supplementary Figure 6) which verified the results based on transcriptome data analysis.
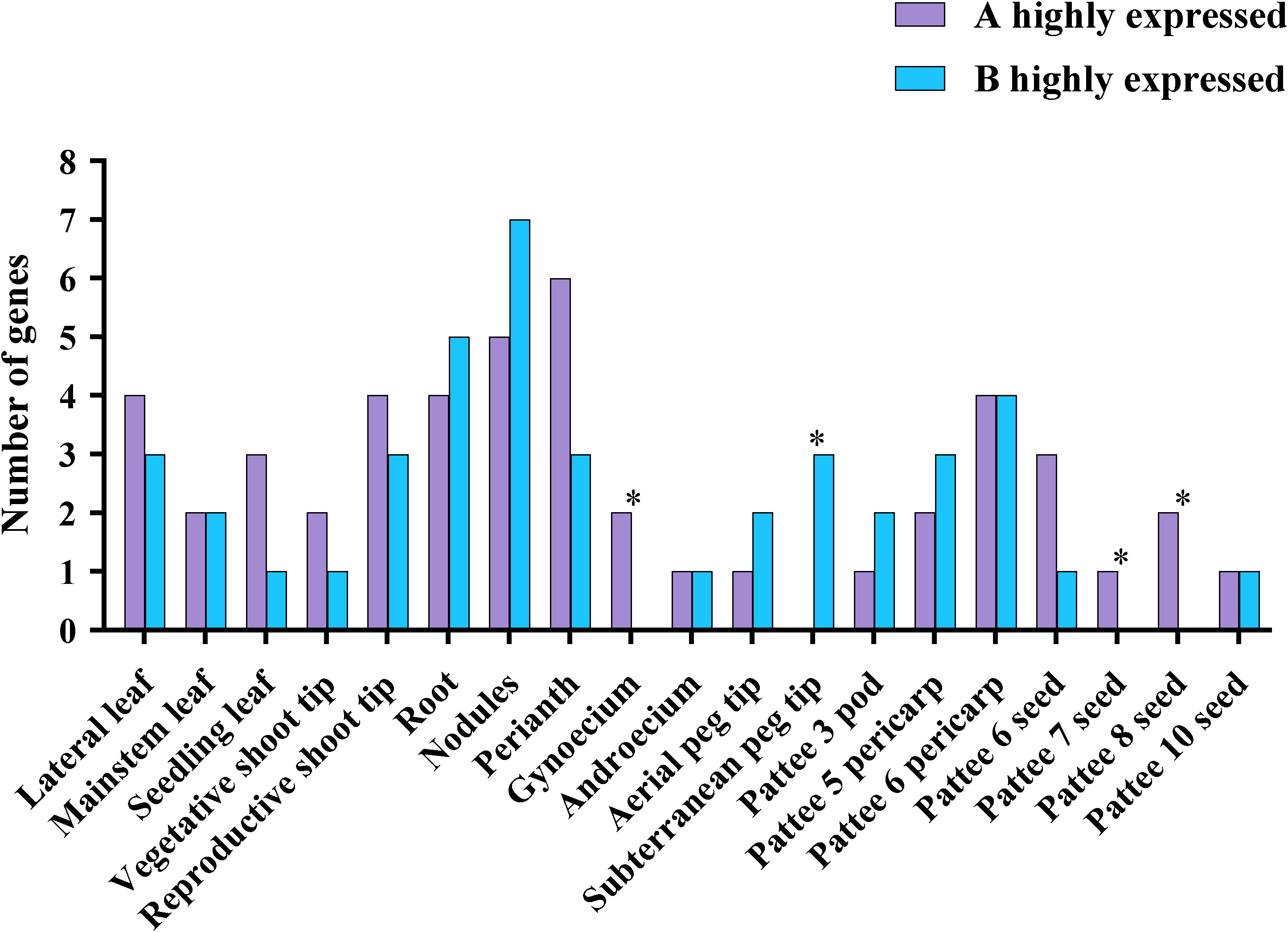
Figure 7 Differential expression of R2R3-MYB homologous genes in A. hypogaea cv. Tifrunner. To look for variations in the levels of gene expression in 19 distinct organs, homeologs were compared. Each subgenome was represented by the number of homeologous genes that were more strongly expressed (log2 fold change ≥ 1, Benjamini-Hochberg adjusted P < 0.05; Wald test) in each subgenome is represented. P-value correspond to binomial test with the odds of A genes being more highly expressed at 0.5 probability. *P < 0.05, others : not significant.
Association analysis of R2R3-MYBs with pod size, total branch number and root-shoot ratio of peanut
To determine the role of R2R3-MYB genes in peanut, we conducted candidate gene association analysis using 59 single-nucleotide polymorphisms in Ad and AiR2R3-MYBs identified from transcriptome data of 146 peanut varieties (Supplementary Table 9) and phenotype of total branch number (TBN), pod length (PL) and root-shoot ratio (RS ratio) variation collected in five environments (Supplementary Table 10). One polymorphic site [A03_122181829 (C/S/G)] was uncovered highly associated with TBN, PL and RS ratio variation in five environments (P < 0.01) (Supplementary Table 11 and Figure 8A), located in the third exon region of AdMYB03-18 (AdMYB03-18) (Figure 8B). A03_122181829 mainly formed three haplotypes (A03_122181829 (C/S/G)) in the associated population (Figure 8C). Analysis results indicated that TBN in haplotype G was notably higher than those in haplotype C, while PL in haplotype C was higher than PL in haplotype G and S, RS ratio in haplotype S was higher than RS ratio in haplotype C and G (Figure 8D).
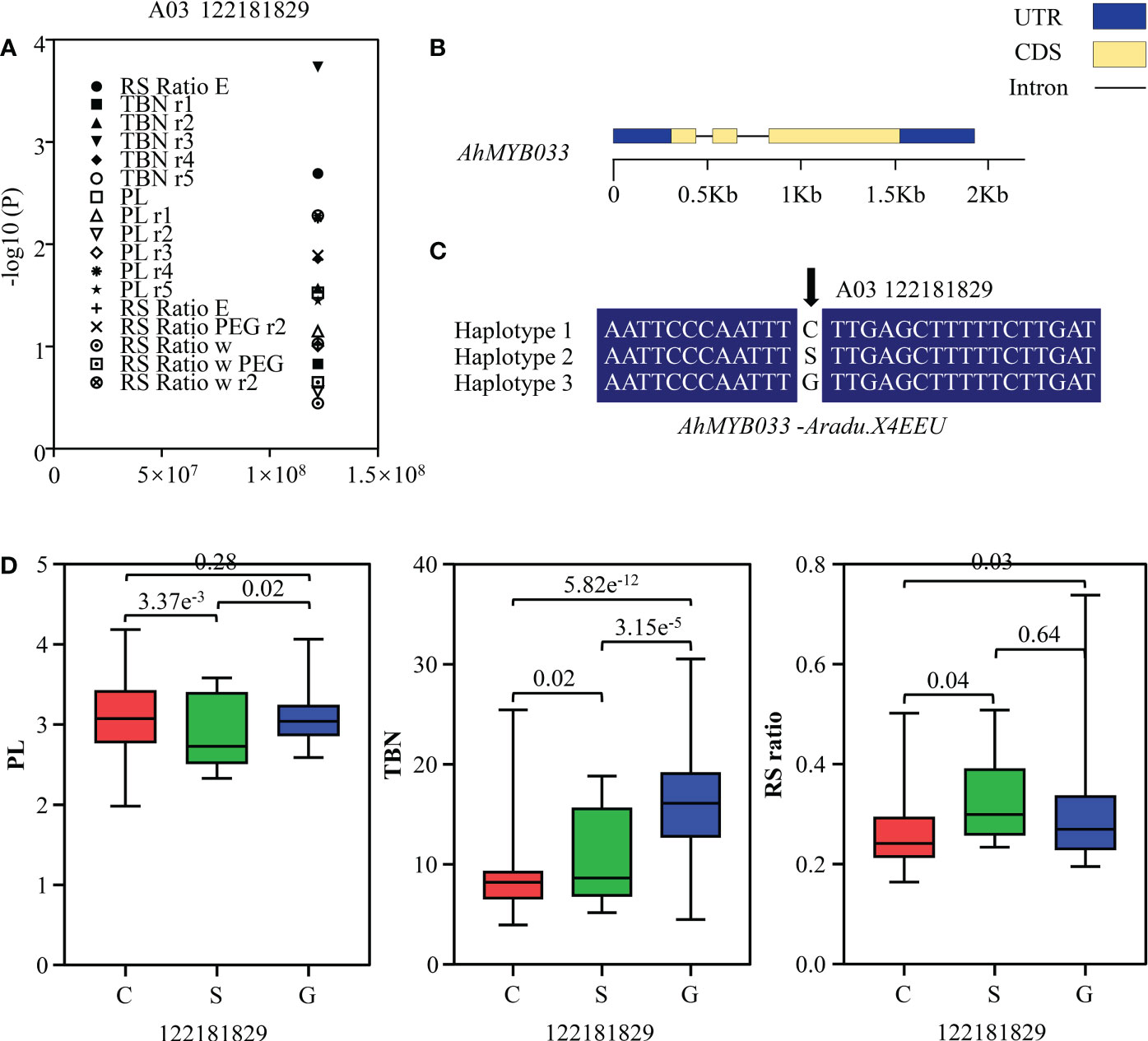
Figure 8 Association mapping results and the phenotypes of the polymorphic sites of peanut R2R3-MYBs associated with TBN, PL and RS ratio variation. (A) Association results between TBN/PL/RS ratio and the polymorphisms in AhMYB033. The vertical and horizontal axes of the scatter plot indicated the P value and the position of the SNP, respectively, and the phenotypes under different environmental treatments were represented by various patterns. (B) Gene structure of AhMYB033. The yellow box, blue box, and black line were CDS, UTR, and intron, respectively. (C) Sequences of three sites were significantly associated with TBN, PL and RS ratio variation. (D) Phenotypic comparison of haplotypes of the three associated sites with TBN, PL and RS ratio in five environments of the population. Three different colored boxes were used to indicate the phenotypes of the three haplotypes.
Discussion
The R2R3-MYB genes have been identified in various species, such as Arabidopsis, rice, maize, soybean, eucalyptus, tomato, and Chinese bayberry (Stracke et al., 2001; Jia et al., 2004; Chen et al., 2006; Du et al., 2012a; Du et al., 2012b; Soler et al., 2015; Li et al., 2016; Cao et al., 2021). In this study, 196 R2R3-MYB genes were identified from cultivated peanut (Arachis hypogaea L.), which were further characterized by co-phylogenetic analysis with the corresponding genes of A. thaliana. According to the topology of the resulting tree and the bootstrap values, a total of 322 R2R3-MYBs were divided into 48 subgroups. Among these subgroups, the Arabidopsis genes of the subgroups S20 and S25 were split into two subgroups (S20a and S20b; S25a and S25b), and the genes from subgroups S10 and S24 were merged (S10&24). The combination of the two subgroups into one group suggested that the genes in these subgroups may have close evolutionary relationships, and there is differentiation in function. In some subgroups (S12, S15, S19, S22, S23, S25a, SAtM47, AtMYB82), only genes in Arabidopsis were present, indicating that after peanut differentiated from Arabidopsis, genes were either lost in peanut or acquired in Arabidopsis. Moreover, in our results, there were eight subgroups (NS-1, 2, 3, 4, 5, 6, 7, 8) specific to peanut, indicating these genes might be newly expanded genes after differentiation from Arabidopsis. Several other genetic characteristics must be considered when performing subfamily classifications, including the presence of highly conserved intron patterns and motif distribution within each subfamily (Du et al., 2015). In this study, exon-intron pattern and motif distribution within each subgroup were also highly conserved, which independently supports our phylogeny analysis and classification results.
As genes in the same subgroup are believed to have relatively similar roles, sequence-based homology classification is crucial for developing hypotheses about the functions of R2R3-MYB genes that have not yet been studied in model species. It is also crucial for defining putative ortholog relationships with known genes in model species to ascertain the functions of genes in nonmodal species (Jiang et al., 2004). The function of novel genes in Arabidopsis and other species can be inferred by protein structure and expression patterns (Dubos et al., 2010). Arabidopsis members in subgroup S6 were involved in the phenylpropanoid pathway, which can activate the late biosynthetic genes (LBGs) leading to both anthocyanin and PA biosynthesis (Gonzalez et al., 2008), suggesting a potential regulatory function of the peanut R2R3-MYB genes in S6 in the phenylpropanoid metabolic pathway. (Zhao et al., 2019) identified the AhTc1 gene encoding an R2R3-MYB transcription factor controlling peanut purple testa color by whole-genome resequencing-based QTL-seq. In this work, AhTc1 was given the name AhMYB099 and belongs to subgroup S6. The identification of AhTc1 supports the above hypothesis, suggesting that these theories help to speculate on unknown gene functions. Similarly, in Arabidopsis, many studies have shown that members of S18 are involved in abscisic acid-mediated responses to environmental signals and in promoting anther and pollen development (Millar and Gubler, 2005; Reyes and Chua, 2007). AtMYB37, AtMYB38, and AtMYB84, members of subgroup S14, partly redundantly control axillary meristem tissue development in Arabidopsis (Keller et al., 2006; Muller et al., 2006). Under adverse circumstances, the root growth-specific regulator AtMYB68 (subgroup 14) has an impact on the development of the entire plant. Members of subgroup S18 were grouped in gene expression clusters 8, 9, and 10. The AhR2R3-MYB genes in subgroup S14 were highly expressed in the subterranean peg tips, roots, and reproductive shoot tips. Therefore, it can be inferred that these genes perform similar functions as the same subgroup of Arabidopsis genes.
Gene expression profiles provided important threads for the study of gene function. In the present study, we also explored the role of the R2R3-MYB gene in water logging stress. A total of 90 genes from 24 subgroups responded to water logging stress in roots and stems. However, some of these 90 genes were not found to be highly expressed in roots and stems in the previous expression analysis of 22 tissues. For example, AhMYB076 and AhMYB176 were only expressed at high levels in the pericarp, and AhMYB071 and AhMYB171 were expressed at high levels in the underground part of the peg tip and the early developing pods. This mi ght be because these genes perform similar functions but have different expression patterns or all respond to certain environmental stimuli (Dubos et al., 2010). In addition, the peg tip of the underground parts becomes more root-like after reaching into the soil, which seems to explain the presence of these genes in the roots or stems in response to water logging stress (Kumar et al., 2019). The important roles of R2R3-MYBs in response to abiotic stress have been reported several times before in Arabidopsis, jatropha, sesame and maize (Dubos et al., 2010; Thirunavukkarasu et al., 2013; Juntawong et al., 2014; Mmadi et al., 2017; Yu et al., 2020). For example, AtMYB60 in subgroup S1 is involved in ABA-mediated control of stomatal opening and closing in response to drought (Cominelli et al., 2005). AtMYB15 in subgroup S2 is involved in cold stress response (Agarwal et al., 2006). AtMYB41 in subgroup S11 probably affects dehydration response after osmotic stress (Cominelli et al., 2008; Lippold et al., 2009). Several MYBs were significantly induced in jatropha, sesame and maize (Thirunavukkarasu et al., 2013; Juntawong et al., 2014; Mmadi et al., 2017; Yu et al., 2020). However, the expression pattern of R2R3-MYB genes under water logging stress exhibited significant temporal specificity. 18 genes showed a continuous down-regulation after treatment, while the remaining genes showed an up-regulation after 6 h, 24 h, 3 days, and 5 days after treatment, respectively. This stress response with a high number of participating genes and a certain temporal pattern disclosed a complex and ordered regulatory network involving R2R3-MYB genes in response to water logging stress.
Whole genome duplication or polyploidization occurs frequently in angiosperms and provides a great deal of material for plant evolution (Jiao, 2018). Most of the replicated genes from WGD are eventually lost (Lynch and Conery, 2000; Conant et al., 2014), and those that are retained are often biased toward certain functional gene taxa (Panchy et al., 2016). In this research, the R2R3-MYB gene also had a high homology ratio (88.3 %) between the two subgenomes of tetraploid peanut (Arachis hypogaea). As previously reported in Arabidopsis and Brassica, the genes that are typically retained (and therefore enriched) are kinases, transcriptional proteins, transcription factors, and genes functioning in transcriptional regulation (Maere et al., 2005; Liu et al., 2014). High retention of the R2R3-MYB homolog after polyploidization in peanut reveals functional conservation of the R2R3-MYB gene. Under natural selection, the ploidized genes experience different fates, such as partial copy loss and loss of function (pseudogenization), partial copy gaining new function, or each exercising part of the function of the ancestral gene (Conant and Wolfe, 2008; Panchy et al., 2016). The results showed that, in general, the homologous gene pairs were not significantly biased to be expressed between the two subgenomes. However, considering only one tissue, homologous pairs showed biased expression among subgenomes in the four reproductive tissues. A total of 32 homologous gene pairs exhibited biased expression between subgenomes in the same or different tissues, suggesting that some genes in these homologous pairs may be lost in function. More homologous genes did not have significantly biased expression and they may have been sub-functionalized, each exercising part of the function of the ancestral gene. Moreover, we identified four homologous pairs exhibiting different biases in different tissues, which reveal a novel functionalization of the R2R3-MYB gene. In conclusion, these findings revealed the fate of the AhR2R3-MYB genes after undergoing polyploidization and collectively maintaining a functional dosage balance.
Conclusion
In this study, 196 R2R3-MYB genes were identified in cultivated peanut genome. A phylogenetic study with Arabidopsis divided the 196 genes into 40 subgroups. Motif composition and gene structure analyses independently confirmed the subgroup delineation. According to the synteny analysis, polyploidization, facilitated in the expansion of the AhR2R3-MYB genes. Tissue expression pattern of R2R3-MYB genes, subgroup functional conservation, and diversification were discovered using gene expression analysis. The varied outcomes of homologous genes following polyploidization were revealed by the biased expression of homologous pairs in the two sub genomes. 90 genes exhibited a clearly time-specific expression pattern when stressed by waterlogging and AhMYB33 was identified by association analysis highly correlated with total branch number (TBN), pod length (PL) and root-shoot ratio (RS ratio). In conclusion, our research advances knowledge of the role of R2R3-MYB transcription factors in cultivated peanut, particularly in response to waterlogging stress.
Data availability statement
The raw data generated in this study have been deposited in the NCBI repository, accession number PRJNA291488.
Author contributions
LW conceived the idea of the paper, SW and ZX carried out all the experiments and data analyses. SW, YY, and WR prepared the figures and tables. SW wrote the manuscript, LW and JF made modifications to the article. All authors contributed to the article and approved the submitted version.
Funding
This research was funded by grants from the National Natural Science Foundation of China (Nos. 31971820, 32160433, and 32160101) and the Jiangxi Agriculture Research System (No. JCARS-18). Funders did not participate in the design of the study, analysis of the results and writing of the manuscript, but provided financial support for the manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1102174/full#supplementary-material
Supplementary Figure 1 | Consistent sequence alignment of the R2R3-MYB domain in Arachis hypogaea. The alignment of 196 AhR2R3-MYB domains was performed using ClustalX and DNAMAN. The shading of the alignment represents different degrees of conservation among sequences, respectively. The sites of five highly conserved tryptophan residues (W) were indicated by pentacles.
Supplementary Figure 2 | Consensus sequence and the level of conservation of R2R3-type MYB domains from peanut. The sequence logos of the R2 and R3 MYB repeats were based on multiple alignment analyses of 196 typical AhR2R3-MYB domains performed with ClustalX 2.1. The vertical axis indicated the degree of amino acid conservation, and the horizontal axis indicated the position of the amino acid on each repeat. The conserved tryptophan residues (Trp, W) in the MYB domain were marked with black asterisks. The replaced residues in the R3 repeat were shown by blue asterisks.
Supplementary Figure 3 | Phylogenetic NJ tree constructed with 126 Arabidopsis and 196 peanut R2R3-MYB proteins. Bootstrap values were on the branch node. The subgroup labels were marked on the right side of the tree.
Supplementary Figure 4 | Phylogenetic NJ tree constructed using 196 AhR2R3-MYB proteins. Bootstrap values were displayed at the branch nodes. The subgroup labels were labeled on the right side of the tree.
Supplementary Figure 5 | 10 MEME motif sequence logos in AhR2R3-MYBs.
Supplementary Figure 6 | qRT-PCR verification of the expression of AhR2R3-MYBs after water logging stress.
References
Agarwal, M., Hao, Y., Kapoor, A., Dong, C. H., Fujii, H., Zheng, X., et al. (2006). A R2R3 type MYB transcription factor is involved in the cold regulation of CBF genes and in acquired freezing tolerance. J. Biol. Chem. 281, 37636–37645. doi: 10.1074/jbc.M605895200
Bailey, T. L., Boden, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., et al. (2009). MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208. doi: 10.1093/nar/gkp335
Bedon, F., Ziolkowski, L., Walford, S. A., Dennis, E. S., Llewellyn, D. J. (2014). Members of the MYBMIXTA-like transcription factors may orchestrate the initiation of fiber development in cotton seeds. Front. Plant Sci. 5, 179. doi: 10.3389/fpls.2014.00179
Bertioli, D. J., Cannon, S. B., Froenicke, L., Huang, G., Farmer, A. D., Cannon, E. K. S., et al. (2016). The genome sequences of Arachis duranensis and Arachis ipaensis, the diploid ancestors of cultivated peanut. Nat. Genet. 48, 438–446. doi: 10.1038/ng.3517
Bertioli, D. J., Jenkins, J., Clevenger, J., Dudchenko, O., Gao, D., Seijo, G., et al. (2019). The genome sequence of segmental allotetraploid peanut Arachis hypogaea. Nat. Genet. 51, 877–884. doi: 10.1038/s41588-019-0405-z
Cao, Y., Jia, H., Xing, M., Jin, R., Grierson, D., Gao, Z., et al. (2021). Genome-wide analysis of MYB gene family in Chinese bayberry (Morella rubra) and identification of members regulating flavonoid biosynthesis. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.691384
Cao, Y., Li, K., Li, Y., Zhao, X., Wang, L. (2020). MYB transcription factors as regulators of secondary metabolism in plants. Biol. (Basel) 9 (3), 61. doi: 10.3390/biology9030061
Chalhoub, B., Denoeud, F., Liu, S., Parkin, I. A. P., Tang, H., Wang, X., et al. (2014). Early allopolyploid evolution in the post-neolithic Brassica napus oilseed genome. Science 345, 950–953. doi: 10.1126/science.1253435
Chen, L., Yang, H., Fang, Y., Guo, W., Chen, H., Zhang, X., et al. (2021). Overexpression of GmMYB14 improves high-density yield and drought tolerance of soybean through regulating plant architecture mediated by the brassinosteroid pathway. Plant Biotechnol. J. 19, 702–716. doi: 10.1111/pbi.13496
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, X., Lu, Q., Liu, H., Zhang, J., Hong, Y., Lan, H., et al. (2019). Sequencing of cultivated peanut, Arachis hypogaea, yields insights into genome evolution and oil improvement. Mol. Plant 12, 920–934. doi: 10.1016/j.molp.2019.03.005
Chen, H., Song, X., Shang, Q., Feng, S., Ge, W. (2022). CFVisual: an interactive desktop platform for drawing gene structure and protein architecture. BMC Bioinf. 23. doi: 10.1186/s12859-022-04707-w
Chen, Y., Yang, X., He, K., Liu, M., Li, J., Gao, Z., et al. (2006). The MYB transcription factor superfamily of Arabidopsis: Expression analysis and phylogenetic comparison with the rice MYB family. Plant Mol. Biol. 60, 107–124. doi: 10.1007/s11103-005-2910-y
Clevenger, J., Chu, Y., Scheffler, B., Ozias-Akins, P. (2016). A developmental transcriptome map for allotetraploid Arachis hypogaea. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.01446
Cominelli, E., Galbiati, M., Vavasseur, A., Conti, L., Sala, T., Vuylsteke, M., et al. (2005). A guard-cell-specific MYB transcription factor regulates stomatal movements and plant drought tolerance. Curr. Biol. 15, 1196–1200. doi: 10.1016/j.cub.2005.05.048
Cominelli, E., Sala, T., Calvi, D., Gusmaroli, G., Tonelli, C. (2008). Over-expression of the arabidopsis AtMYB41 gene alters cell expansion and leaf surface permeability. Plant J. 53, 53–64. doi: 10.1111/j.1365-313X.2007.03310.x
Conant, G. C., Birchler, J. A., Pires, J. C. (2014). Dosage, duplication, and diploidization: clarifying the interplay of multiple models for duplicate gene evolution over time. Curr. Opin. Plant Biol. 19, 91–98. doi: 10.1016/j.pbi.2014.05.008
Conant, G. C., Wolfe, K. H. (2008). Turning a hobby into a job: How duplicated genes find new functions. Nat. Rev. Genet. 9, 938–950. doi: 10.1038/nrg2482
Crooks, G. E., Hon, G., Chandonia, J. M., Brenner, S. E. (2004). WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190. doi: 10.1101/gr.849004
Dubos, C., Stracke, R., Grotewold, E., Weisshaar, B., Martin, C., Lepiniec, L. (2010). MYB transcription factors in Arabidopsis. Trends Plant Sci. 15, 573–581. doi: 10.1016/j.tplants.2010.06.005
Du, H., Feng, B., Yang, S., Huang, Y., Tang, Y., Wu, K. (2012a). The R2R3-MYB transcription factor gene family in maize. PloS One 7, e37463–e37463. doi: 10.1371/journal.pone.0037463
Du, H., Liang, Z., Zhao, S., Nan, M., Tran, L. P., Lu, K., et al. (2015). The evolutionary history of R2R3-MYB proteins across 50 eukaryotes: New insights into subfamily classification and expansion. Sci. Rep. 5. doi: 10.1038/srep11037
Du, H., Yang, S., Liang, Z., Feng, B., Liu, L., Huang, Y., et al. (2012b). Genome-wide analysis of the MYB transcription factor superfamily in soybean. BMC Plant Biol. 12, 106–106. doi: 10.1186/1471-2229-12-106
Goicoechea, M., Lacombe, E., Legay, S., Mihaljevic, S., Rech, P., Jauneau, A., et al. (2005). EgMYB2, a new transcriptional activator from Eucalyptus xylem, regulates secondary cell wall formation and lignin biosynthesis. Plant J. 43, 553–67. doi: 10.1111/j.1365-313X.2005.02480.x
Gonzalez, A., Zhao, M., Leavitt, J. M., Lloyd, A. M. (2008). Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J. 53, 814–827. doi: 10.1111/j.1365-313X.2007.03373.x
Hanada, K., Zou, C., Lehti-Shiu, M. D., Shinozaki, K., Shiu, S. (2008). Importance of lineage-specific expansion of plant tandem duplicates in the adaptive response to environmental stimuli. Plant Physiol. 148, 993–1003. doi: 10.1104/pp.108.122457
He, J., Liu, Y., Yuan, D., Duan, M., Liu, Y., Shen, Z., et al. (2020). An R2R3 MYB transcription factor confers brown planthopper resistance by regulating the phenylalanine ammonia-lyase pathway in rice. Proceedings of the National Academy of Sciences 117, 271–277. doi: 10.1073/pnas.1902771116
Jia, L., Clegg, M. T., Jiang, T. (2004). Evolutionary dynamics of the DNA-binding domains in putative R2R3-MYB genes identified from rice subspecies indica and japonica genomes. Plant Physiol. 134, 575–585. doi: 10.1104/pp.103.027201
Jiang, C., Gu, X., Peterson, T. (2004). Identification of conserved gene structures and carboxy-terminal motifs in the myb gene family of Arabidopsis and Oryza sativa l. ssp. indica. Genome Biol. 5, R46–R46. doi: 10.1186/gb-2004-5-7-r46
Jian, L., Kang, K., Choi, Y., Suh, M. C., Paek, N. C. (2022). Mutation of OsMYB60 reduces rice resilience to drought stress by attenuating cuticular wax biosynthesis. Plant J. 112 (2), 339–351. doi: 10.1111/tpj.15947
Jiao, Y. (2018). Double the genome, double the fun: Genome duplications in angiosperms. Mol. Plant 11, 357–358. doi: 10.1016/j.molp.2018.02.009
Jin, H., Martin, C. (1999). Multifunctionality and diversity within the plant MYB-gene family. Plant Mol. Biol. 41, 577–585. doi: 10.1023/A:1006319732410
Juntawong, P., Sirikhachornkit, A., Pimjan, R., Sonthirod, C., Sangsrakru, D., Yoocha, T., et al. (2014). Elucidation of the molecular responses to waterlogging in jatropha roots by transcriptome profiling. Front. Plant Sci. 5. doi: 10.3389/fpls.2014.00658
Katoh, K., Rozewicki, J., Yamada, K. D. (2019). MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 20, 1160–1166. doi: 10.1093/bib/bbx108
Keller, T., Abbott, J., Moritz, T., Doerner, P. (2006). Arabidopsis REGULATOR OF AXILLARY MERISTEMS1 controls a leaf axil stem cell niche and modulates vegetative development. Plant Cell 18, 598–611. doi: 10.1105/tpc.105.038588
Kranz, H. D., Denekamp, M., Greco, R., Jin, H., Leyva, A., Meissner, R. C., et al. (1998). Towards functional characterisation of the members of the R2R3-MYB gene family from Arabidopsis thaliana. Plant J. 16, 263–276. doi: 10.1046/j.1365-313x.1998.00278.x
Kumar, R., Pandey, M. K., Roychoudhry, S., Nayyar, H., Kepinski, S., Varshney, R. K. (2019). Peg biology: Deciphering the molecular regulations involved during peanut peg development. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.01289
Kumar, S., Stecher, G., Tamura, K. (2016). MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., McGettigan, P. A., McWilliam, H., et al. (2007). Clustal W and clustal X version 2.0. Bioinformatics 23, 2947–2948. doi: 10.1093/bioinformatics/btm404
Legay, S., Lacombe, E., Goicoechea, M., Brière, C., Séguin, A., Mackay, J., et al. (2007). Molecular characterization of EgMYB1, a putative transcriptional repressor of the lignin biosynthetic pathway. Plant Sci. 173, 542–549. doi: 10.1016/j.plantsci.2007.08.007
Legay, S., Sivadon, P., Blervacq, A. S., Pavy, N., Baghdady, A., Tremblay, L., et al. (2010). EgMYB1, an R2R3 MYB transcription factor from eucalyptus negatively regulates secondary cell wall formation in Arabidopsis and poplar. New Phytol. 188, 774–86. doi: 10.1111/j.1469-8137.2010.03432.x
Li, Z., Peng, R., Tian, Y., Han, H., Xu, J., Yao, Q. (2016). Genome-wide identification and analysis of the MYB transcription factor superfamily in solanum lycopersicum. Plant Cell Physiol. 57, 1657–1677. doi: 10.1093/pcp/pcw091
Lippold, F., Sanchez, D. H., Musialak, M., Schlereth, A., Scheible, W. R., Hincha, D. K., et al. (2009). AtMyb41 regulates transcriptional and metabolic responses to osmotic stress in Arabidopsis. Plant Physiol. 149, 1761–1772. doi: 10.1104/pp.108.134874
Liu, S., Liu, Y., Yang, X., Tong, C., Edwards, D., Parkin, I. A., et al. (2014). The brassica oleracea genome reveals the asymmetrical evolution of polyploid genomes. Nat. Commun. 5, 3930. doi: 10.1038/ncomms4930
Liu, J., Osbourn, A., Ma, P. (2015). MYB transcription factors as regulators of phenylpropanoid metabolism in plants. Mol. Plant 8, 689–708. doi: 10.1016/j.molp.2015.03.012
Lynch, M., Conery, J. S. (2000). The evolutionary fate and consequences of duplicate genes. Science 290, 1151–1155. doi: 10.1126/science.290.5494.1151
Maere, S., De Bodt, S., Raes, J., Casneuf, T., Van Montagu, M., Kuiper, M., et al. (2005). Modeling gene and genome duplications in eukaryotes. Proc. Natl. Acad. Sci. U. S. A. 102, 5454–5459. doi: 10.1073/pnas.0501102102
Martin, C., Paz-Ares, J. (1997). MYB transcription factors in plants. Trends Genet. 13, 67–73. doi: 10.1016/s0168-9525(96)10049-4
Millar, A. A., Gubler, F. (2005). The Arabidopsis GAMYB-like genes, MYB33 and MYB65, are microRNA-regulated genes that redundantly facilitate anther development. Plant Cell 17, 705–721. doi: 10.1105/tpc.104.027920
Min, X. J., Powell, B., Braessler, J., Meinken, J., Yu, F., Sablok, G. (2015). Genome-wide cataloging and analysis of alternatively spliced genes in cereal crops. BMC Genomics 16. doi: 10.1186/s12864-015-1914-5
Mmadi, M. A., Dossa, K., Wang, L., Zhou, R., Wang, Y., Cisse, N., et al. (2017). Functional characterization of the versatile MYB gene family uncovered their important roles in plant development and responses to drought and waterlogging in sesame. Genes (Basel). 8 (12), 362. doi: 10.3390/genes8120362
Muller, D., Schmitz, G., Theres, K. (2006). Blind homologous R2R3 myb genes control the pattern of lateral meristem initiation in Arabidopsis. Plant Cell 18, 586–597. doi: 10.1105/tpc.105.038745
Ogata, K., Kanei-Ishii, C., Sasaki, M., Hatanaka, H., Nagadoi, A., Enari, M., et al. (1996). The cavity in the hydrophobic core of myb DNA-binding domain is reserved for DNA recognition and trans-activation. Nat. Struct. Biol. 3, 178–187. doi: 10.1038/nsb0296-178
Paz-Ares, J., Ghosal, D., Wienand, U., Peterson, P. A., Saedler, H. (1987). The regulatory cl locus of Zea mays encodes a protein with homology to myb proto-oncogene products and with structural similarities to transcriptional activators. The EMBO journal 6, 3553–3558
Panchy, N., Lehti-Shiu, M., Shiu, S. H. (2016). Evolution of gene duplication in plants. Plant Physiol. 171, 2294–2316. doi: 10.1104/pp.16.00523
Pertea, M., Kim, D., Pertea, G. M., Leek, J. T., Salzberg, S. L. (2016). Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and ballgown. Nat. Protoc. 11, 1650–1667. doi: 10.1038/nprot.2016.095
Reyes, J. L., Chua, N. H. (2007). ABA induction of miR159 controls transcript levels of two MYB factors during Arabidopsis seed germination. Plant J. 49, 592–606. doi: 10.1111/j.1365-313X.2006.02980.x
Saikumar, P., Murali, R., Reddy, E. P. (1990). Role of tryptophan repeats and flanking amino acids in myb-DNA interactions. Proc. Natl. Acad. Sci. U. S. A. 87, 8452–8456. doi: 10.1073/pnas.87.21.8452
Sharma, K. K., Bhatnagar-Mathur, P. (2006). Peanut (Arachis hypogaea l.). Methods Mol. Biol. 343, 347–358. doi: 10.1385/1-59745-130-4:347
Shimizu-Inatsugi, R., Terada, A., Hirose, K., Kudoh, H., Sese, J., Shimizu, K. K. (2017). Plant adaptive radiation mediated by polyploid plasticity in transcriptomes. Mol. Ecol. 26, 193–207. doi: 10.1111/mec.13738
Soler, M., Camargo, E. L., Carocha, V., Cassan-Wang, H., San, C. H., Savelli, B., et al. (2015). The eucalyptus grandis R2R3-MYB transcription factor family: evidence for woody growth-related evolution and function. New Phytol. 206, 1364–1377. doi: 10.1111/nph.13039
Stracke, R., Werber, M., Weisshaar, B. (2001). The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 4, 447–456. doi: 10.1016/s1369-5266(00)00199-0
Su, L., Lv, A., Wen, W., Fan, N., Li, J., Gao, L., et al. (2022). MsMYB741 is involved in alfalfa resistance to aluminum stress by regulating flavonoid biosynthesis. Plant J. 112, 756–771. doi: 10.1111/tpj.15977
Thirunavukkarasu, N., Hossain, F., Mohan, S., Shiriga, K., Mittal, S., Sharma, R., et al. (2013). Genome-wide expression of transcriptomes and their co-expression pattern in subtropical maize (Zea mays l.) under waterlogging stress. PloS One 8 (8), e70433. doi: 10.1371/journal.pone.0070433
Toomer, O. T. (2018). Nutritional chemistry of the peanut (Arachis hypogaea). Crit. Rev. Food Sci. Nutr. 58, 3042–3053. doi: 10.1080/10408398.2017.1339015
Vimolmangkang, S., Han, Y., Wei, G., Korban, S. S. (2013). An apple MYB transcription factor, MdMYB3, is involved in regulation of anthocyanin biosynthesis and flower development. BMC Plant Biol. 13, 176. doi: 10.1186/1471-2229-13-176
Yu, F., Tan, Z., Fang, T., Tang, K., Liang, K., Qiu, F. (2020). A comprehensive transcriptomics analysis reveals long non-coding RNA to be involved in the key metabolic pathway in response to waterlogging stress in maize. Genes (Basel). 11 (3), 267. doi: 10.3390/genes11030267
Zeng, R., Chen, T., Wang, X., Cao, J., Li, X., Xu, X., et al. (2021). Physiological and expressional regulation on photosynthesis, starch and sucrose metabolism response to waterlogging stress in peanut. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.601771
Zhang, T., Hu, Y., Jiang, W., Fang, L., Guan, X., Chen, J., et al. (2015). Sequencing of allotetraploid cotton (Gossypium hirsutum l. acc. TM-1) provides a resource for fiber improvement. Nat. Biotechnol. 33, 531–537. doi: 10.1038/nbt.3207
Zhao, Y., Ma, J., Li, M., Deng, L., Li, G., Xia, H., et al. (2019). Whole-genome resequencing based QTL seq identified AhTc1 gene encoding a R2R3-MYB transcription factor controlling peanut purple testa colour. Plant Biotechnol. J. 18, 96–105. doi: 10.1111/pbi.13175
Keywords: cultivated peanut, R2R3-MYB transcription factors, allotetraploid, gene duplications, gene expression, functional diversity
Citation: Wang S, Xu Z, Yang Y, Ren W, Fang J and Wan L (2023) Genome-wide analysis of R2R3-MYB genes in cultivated peanut (Arachis hypogaea L.): Gene duplications, functional conservation, and diversification. Front. Plant Sci. 14:1102174. doi: 10.3389/fpls.2023.1102174
Received: 18 November 2022; Accepted: 06 January 2023;
Published: 14 February 2023.
Edited by:
Weijian Zhuang, Fujian Agriculture and Forestry University, ChinaReviewed by:
Xin Wei, Shanghai Normal University, ChinaQing Zhang, Agricultural Genomics Institute at Shenzhen, Chinese Academy of Agricultural Sciences (CAAS), China
Yunpeng Cao, Wuhan Botanical Garden, Chinese Academy of Sciences (CAS), China
Copyright © 2023 Wang, Xu, Yang, Ren, Fang and Wan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jiahai Fang, ZmFuZ2ppYWhhaTIwMTlAMTI2LmNvbQ==; Liyun Wan, d2FubGl5dW4yMDE5QDE2My5jb20=
 Sijian Wang
Sijian Wang Weifang Ren
Weifang Ren Liyun Wan
Liyun Wan