- 1National Engineering Research Center for Sugarcane & Guangxi Key Laboratory of Sugarcane Biology, Fujian Agriculture and Forestry University, Fuzhou, China
- 2State Key Laboratory for Conservation and Utilization of Subtropical Agro-Bioresources & Guangxi Key Laboratory of Sugarcane Biology, Guangxi University, Nanning, China
Introduction: Plant-specific Class III peroxidases (PRXs) play a crucial role in lignification, cell elongation, seed germination, and biotic and abiotic stresses.
Methods: The class III peroxidase gene family in sugarcane were identified by bioinformatics methods and realtime fluorescence quantitative PCR.
Results: Eighty-two PRX proteins were characterized with a conserved PRX domain as members of the class III PRX gene family in R570 STP. The ShPRX family genes were divided into six groups by the phylogenetic analysis of sugarcane, Saccharum spontaneum, sorghum, rice, and Arabidopsis thaliana. The analysis of promoter cis-acting elements revealed that most ShPRX family genes contained cis-acting regulatory elements involved in ABA, MeJA, light responsiveness, anaerobic induction, and drought inducibility. An evolutionary analysis indicated that ShPRXs was formed after Poaceae and Bromeliaceae diverged, and tandem duplication events played a critical role in the expansion of ShPRX genes of sugarcane. Purifying selection maintained the function of ShPRX proteins. SsPRX genes were differentially expressed in stems and leaves at different growth stages in S. spontaneum. However, ShPRX genes were differentially expressed in the SCMV-inoculated sugarcane plants. A qRT-PCR analysis showed that SCMV, Cd, and salt could specifically induce the expression of PRX genes of sugarcane.
Discussion: These results help elucidate the structure, evolution, and functions of the class III PRX gene family in sugarcane and provide ideas for the phytoremediation of Cd-contaminated soil and breeding new sugarcane varieties resistant to sugarcane mosaic disease, salt, and Cd stresses.
1 Introduction
Sugarcane, one of the critical sugar and energy crops, is often subjected to various biotic stresses, including Sugarcane mosaic virus (SCMV), Sorghum mosaic virus (SrMV), Sugarcane streak mosaic virus (SCSMV), and abiotic stress, including salt, heavy metal, and drought stress. Sugarcane mosaic disease, caused by SCMV, SrMV, and SCSMV, is currently one of the most severe sugarcane diseases worldwide that adversely affect the healthy and sustainable development of the sugarcane industry (Yao et al., 2017; Moradi et al., 2018; Rice et al., 2019; Rice and Hoy, 2020). Under salt stress, the growth of sugarcane is hindered, seriously affecting the quality of sugarcane and even causing a large area of yield reduction or crop failure. Soil heavy metal pollution, one of the leading environmental stresses affecting plant growth and development, is becoming the prime concern of various terrestrial ecosystems worldwide. Among heavy metals, cadmium (Cd), one of the most dangerous toxic elements for plants, inhibits various physiological processes of plants, including seed germination, seedling growth, photosynthesis, and antioxidant system (Zhu et al., 2021). The contents of chlorophyll and soluble protein decreased significantly, whereas the content of carotenoids increased significantly in the Cd-treated sugarcane. The activity of ascorbate peroxidase, peroxidase (PRX), and catalase (CAT) increased significantly (Yousefi et al., 2018). Sugarcane, one of the cultivated crops with the highest biomass and solid tolerance to Cd, can be used as a candidate crop for the phytoremediation of Cd pollution in soil (Sereno et al., 2007; Yousefi et al., 2018). Therefore, it is one of the main challenges to improve the yield and quality of sugarcane and the ability to repair Cd pollution in soil by improving the resistance of sugarcane to biotic and abiotic stresses.
The PRXs, the critical enzymes of peroxisomes, widely exist in animals, plants, and microorganisms. According to the protein structural and functional characteristics, PRXs are divided into heme PRXs and non-heme PRXs. Heme PRXs are further subdivided into animal PRXs and non-animal PRXs. According to the sequence and catalytic characteristics of proteins, non-animal heme PRXs comprise Class I, II, and III PRXs, all containing a heme group consisting of protoporphyrin IX and iron (III). Class I PRXs widely exist in most organisms, such as plants, fungi, bacteria, and protozoa. However, Class II PRXs exist in fungi, and class III PRXs only exist in plants (Piontek et al., 2001; Zamocky, 2004; Zamocky et al., 2010; Shigeto and Tsutsumi, 2016). Class III PRXs are classical plant secretory PRXs that play a crucial role in lignification, cell elongation, and seed germination (Lee, 1977; Piontek et al., 2001; Shigeto and Tsutsumi, 2016; Kidwai et al., 2020). Numerous class III PRXs were present in the cell walls, and the balance of cell wall loosening and stiffening could be precisely controlled by the antagonistic activities of class III PRXs during plant growth (Francoz et al., 2015). Class III PRXs was identified as an essential enzyme for lignin biosynthesis in plants. Coniferyl alcohol and sinapyl alcohol, precursors for the synthesis of lignin monomers, could be catalyzed by the class III PRX gene PbPRX2 in Chinese pear fruit (Vanholme et al., 2019; Zhu et al., 2021). In addition, class III PRXs were involved in the internal browning of pineapple (Hou et al., 2022) and closely related to pollen fertility in Gossypium hirsutum (Chen et al., 2022).
The class III PRX genes play a vital role in responses to various biotic and abiotic stresses throughout the plant life cycle. The wheat class III PRX gene (TaPRX-2A) increased the activities of superoxide dismutase (SOD), PRX, and CAT to scavenge reactive oxygen species (ROS) (Su et al., 2020). In sweet potato, the B-box family transcription factor IbBBX24 activated the expression of the class III PRX gene IbPRX17 by binding to the promoter of IbPRX17, and the overexpression of IbPRX17 significantly improved the tolerance to salt and drought stresses by scavenging ROS (Zhang et al., 2022). Overexpressed the class III PRX gene (OsPRX38) in Arabidopsis thaliana exposed to arsenic stress increased SOD, PRX, and GST activities to reduce the content of hydrogen peroxide (H2O2), electrolyte leakage, and malondialdehyde (Kidwai et al., 2019). In rice, the overexpression of the class III PRX OsPRX30, maintaining a high level of PRX activity and reducing the content of H2O2, reduced bacterial blight resistance (Liu et al., 2021). In Citrus sinensis, the class III CsPRX family genes, induced by salicylic acid and methyl jasmonate, were involved in citrus bacterial canker disease (Li et al., 2020).
The publication of the genome of Saccharum hybrid cultivar R570 (R570) BAC clone (BAC) and single tiling path (STP) has necessitated the study of the function of the sugarcane class III PRX gene. At present, the class III PRX gene family has been studied in various plant species, including A. thaliana (Tognolli et al., 2002), rice (Passardi et al., 2004), maize (Wang et al., 2015), potato (Yang et al., 2020), soybean (Aleem et al., 2022), Brachypodium distachyon (Zhu et al., 2019), and allotetraploid cotton (Duan et al., 2019). However, it has yet to be reported on the identification and characterization of the class III PRX gene family in sugarcane.
A genome-wide search was carried out on class III PRXs in R570 STP, R570 BAC, Saccharum spontaneum AP85-441 (S. spontaneum AP85-441), sorghum, A. thaliana, and rice. The functions of the class III PRX genes in sugarcane were analyzed concerning the gene structure, conserved motif, cis-acting elements, codon usage bias, and evolutionary analysis. The expression level of ShPRXs was studied under SCMV, Cd, and salt stress response. The results from this study help elucidate the structure, evolution, and functions of the class III PRX gene family.
2 Materials and methods
2.1 Identification of PRX family members
Genome data of R570 BAC and R570 STP were obtained from the sugarcane genome hub (https://sugarcane-genome.cirad.fr/; Garsmeur et al., 2018). Genome data of A. thaliana TAIR10, Oryza sativa (IRGSP-1.0), S. bicolor (NCBIv3), and S. spontaneum AP85-441 were obtained from the Ensembl Plants database (http://plants.ensembl.org/index.html; Yates et al., 2022).
The hidden Markov model (HMM) file of the PRX domain (PF00141) was downloaded from the Pfam database (https://pfam-legacy.xfam.org/; Sonnhammer et al., 1998; Blom et al., 1999; Mistry et al., 2021). The HMMER software (version 3.1b1; http://www.hmmer.org/) was used to search against the whole genome protein file of R570, A. thaliana, O. sativa, S. bicolor, and S. spontaneum under the condition of e-value < 1×10−20. Multiple sequence alignment was performed using ClustalW (http://www.clustal.org/), and a new HMM matrix file was constructed. The PRX domain-containing proteins were searched on the new HMM matrix file and screened by e-value < 0.001. The PRX protein Ref-seq of all plants was downloaded to build a library from the NCBI database (https://www.ncbi.nlm.nih.gov/). The putative PRX genes were searched using BLASTP and screened by e-value < 1×10−10 and identity > 75%. The protein-conserved domains were verified by Pfam and NCBI CDD (https://www.ncbi.nlm.nih.gov/Structure/bwrpsb/bwrpsb.cgi), and the genes without the PRX domain were deleted (Lu et al., 2020; Mistry et al., 2021). In addition, the PRX gene family was identified based on the sugarcane transcriptome data.
The physicochemical properties of the ShPRX family proteins were calculated using ExPASy-ProtParam (https://web.expasy.org/protparam/; Wilkins et al., 1999). The subcellular localization of the ShPRX family proteins was predicted using Plant-mPLoc (http://www.csbio.sjtu.edu.cn/bioinf/plant-multi/; Chou and Shen, 2010). The transmembrane domain of the ShPRX family proteins was predicted using TMHMM 2.0 (https://services.healthtech.dtu.dk/service.php?TMHMM-2.0; Sonnhammer et al., 1998). The signal peptide of the ShPRX family proteins was predicted using SignalP 6.0 (https://services.healthtech.dtu.dk/service.php?SignalP; Teufel et al., 2022). The phosphorylation sites of the ShPRX family proteins were predicted using NetPhos 3.1 (https://services.healthtech.dtu.dk/service.php?NetPhos-3.1; Blom et al., 1999). The secondary structure of the ShPRX family proteins was analyzed using SOPMA (https://npsa-prabi.ibcp.fr/cgi-bin/npsa_automat.pl?page=npsa_sopma.html; Geourjon and Deleage, 1995).
2.2 Phylogenetic analysis
The multiple protein sequences of the PRX gene family in R570 STP, A. thaliana, O. sativa, S. bicolor, and S. spontaneum were compared using the MEGA-X MUSCLE software (https://www.megasoftware.net/; Kumar et al., 2018). The phylogenetic tree was constructed using the neighbor-joining method with the Jones–Taylor–Thornton model, 1,000 bootstrap replications, gamma distributed (G), and partial deletion gaps by MEGA-X. The phylogenetic tree was visualized using the online tool iTOL (https://itol.embl.de/; Letunic and Bork, 2021).
2.3 Chromosomal distribution, gene structure, and conserved motif analysis of the ShPRXs
The positional and structural information of ShPRX genes on the R570 STP chromosomes was extracted from the Generic Feature Format Version 3 (GFF3) (Garsmeur et al., 2018). The chromosomal locations of the ShPRX family genes were drawn using the online tool MG2C v2.1 (http://mg2c.iask.in/mg2c_v2.1/index.html). The conserved motifs of the ShPRX family proteins were identified using the online MEME with optimized parameters: minimum width, 6; maximum width, 50; and the number of motifs, 26 (Bailey and Elkan, 1994). The phylogenetic tree, gene structure, and conserved motifs of the ShPRX family genes were visualized using TBtools (Chen et al., 2020).
2.4 Analysis of cis-acting elements and codon usage bias of the ShPRXs
The sequence 2000 bp upstream from the start codon of the ShPRX family gene was obtained using the hybrid cultivar R570 reference genome (Garsmeur et al., 2018). The cis-acting elements of ShPRX family genes in promotor regions were predicted using the PlantCAR (Lescot et al., 2002). The phylogenetic tree and cis-acting elements of the ShPRX family genes were visualized using GSDS 2.0 (http://gsds.cbi.pku.edu.cn; Hu et al., 2015). The codon usage bias of the ShPRX family genes was analyzed using CodonW (https://codonw.sourceforge.net/) and EMBOSS: chips (https://www.bioinformatics.nl/cgi-bin/emboss/chips; Rice et al., 2000). The effective number of codon (ENc) plots and Parity rule 2 (PR2) plot analysis was performed as described by Wright (1990) and Chakraborty et al. (2020).
2.5 Syntenic and selection pressure analysis of the PRX family genes
The syntenic relationships of the PRX family genes were analyzed and visualized using MCScanX and Circos, respectively (Krzywinski et al., 2009; Wang et al., 2012). The frequencies of synonymous (Ks) and nonsynonymous (Ka) mutations, along with their ratios, were calculated to analyze the selection pressure of PRX duplicated gene pairs using TBtools (Chen et al., 2020). The divergence time (T) of class III PRX family genes was calculated as T = Ks/(2 × λ) × 10−6 Mya (λ = 6.5 × 10−9 for grasses; (Gaut et al., 1996). The divergence time values were estimated using TimeTree (http://www.timetree.org/; Kumar et al., 2022).
2.6 Expression profiles of the PRX family genes
The RNA-seq expression data of the growth and development of S. spontaneum were downloaded from the Saccharum Genome Database (SGD; http://sugarcane.zhangjisenlab.cn/sgd/html/index.html; Zhang et al., 2018). The transcriptome data of sugarcane were obtained by sequencing (Akbar et al., 2021). The gene expression data of rice under Cd (GSE35502) and salt (GSE60287) stresses were downloaded from the GEO database (https://www.ncbi.nlm.nih.gov/geo/; Ishimaru et al., 2012; Garg et al., 2015; Clough and Barrett, 2016; Shankar et al., 2016). The expression pattern of the PRX family genes was drawn based on FPKM values or log2 fold change (log2FC) using the R software (v4.0.5).
2.7 SCMV infection, Cd treatment, salt stress treatment, and qRT-PCR analysis
Badila and B48, grown to leaf stages 6–8 in the field, was inoculated using SCMV crude extract following Yao’s method (Yao et al., 2017), and the −3 and +1 leaves were collected at day 21 post-inoculation. The control plants were rubbed using 0.1 M phosphate buffer (pH 7.0). Sugarcane plants of Zhongzhe 1 were watered using 4.3-mM Cd solution or 0.5-L 1.0% sodium chloride solution when Zhongzhe 1 grew to leaf stage 3 in barrels containing 16 kilograms of soil. The control plants were watered using 0.5-L double-distilled water. The +1 leaves of sugarcane were collected at different time after treatment (0, 4, 8, 12, and 24 h).
Total RNA was extracted using Eastep® Super Total RNA Extraction Kit (Promega (Beijing) Biotech Co. Ltd., Beijing, China) following the manufacturer’s instructions. The first strand of cDNA was synthesized using PrimeScript™ II 1st Strand cDNA Synthesis Kit (Takara Biomedical Technology Co. Ltd., Beijing, China). The qRT-PCR experiment was conducted in three independent replicates using ChamQ Universal SYBR qPCR Master Mix (Nanjing Vazyme Biotech Co., Ltd., Nanjing, China) with the Bio-Rad CFX96 fluorescence quantitative PCR Instrument. The primers for qRT-PCR of 10 PRX genes were designed using Primer Premier 6. The relative expression levels of the PRX family genes were calculated using the 2−ΔΔCt method, and statistical significance was analyzed using ordinary one-way ANOVA in GraphPad Prism 7.
3 Results
3.1 Identification and chromosomal distribution of the PRX gene family in sugarcane
Eighty-two PRX proteins with a conserved PRX domain were characterized as members of the class III PRX gene family in R570 STP and named ShPRX1–ShPRX82 based on their respective locations on the chromosomes (Tables S1 , S2). The total number of PRX family genes in the sugarcane R570 cultivar was lower than that in the S. spontaneum (113), sorghum (150), and rice (126), but slightly higher in A. thaliana (80) (Table S2).
The physical and chemical properties analysis revealed that the ShPRX family genes were predicted to encode polypeptides from 235 to 472 amino acids, with predicted molecular weights ranging from 25.11 to 49.76 kD. The theoretical pI ranged from 4.59 to 10.08, and the grand average of the hydropathicity values of 52 ShPRX proteins was negative, ranging from −0.51 to −0.002, indicating a hydrophilic characteristic. The grand average of the hydropathicity values of 30 ShPRX proteins was positive, ranging from 0.004 to 0.297, indicating a hydrophobic characteristic. The predicted number of negatively charged residues (Asp + Glu) in the ShPRXs was 14–59, and the number of positively charged residues (Arg + Lys) was 20–54. The instability index of 46 ShPRX proteins was less than 40, ranging from 25.38 to 39.76, indicating a stable characteristic. The instability index of 36 ShPRX proteins was greater than 40, ranging from 40.36 to 56.05, indicating an unstable characteristic (Table S1).
The subcellular localization prediction analysis showed that most ShPRX genes were located in the cytoplasm, and some were located in the vacuole, chloroplast, peroxisome, mitochondrion, cell membrane, and nucleus. Most ShPRX genes had signal peptides, 42 ShPRX genes had no transmembrane domain, and other ShPRX genes had only one transmembrane domain. The number of phosphorylation sites ranged from 16 to 47, while four ShPRX genes had no tyrosine phosphorylation site. The secondary structure prediction analysis showed that ShPRX proteins were mainly composed of alpha helix and random coil (Table S1).
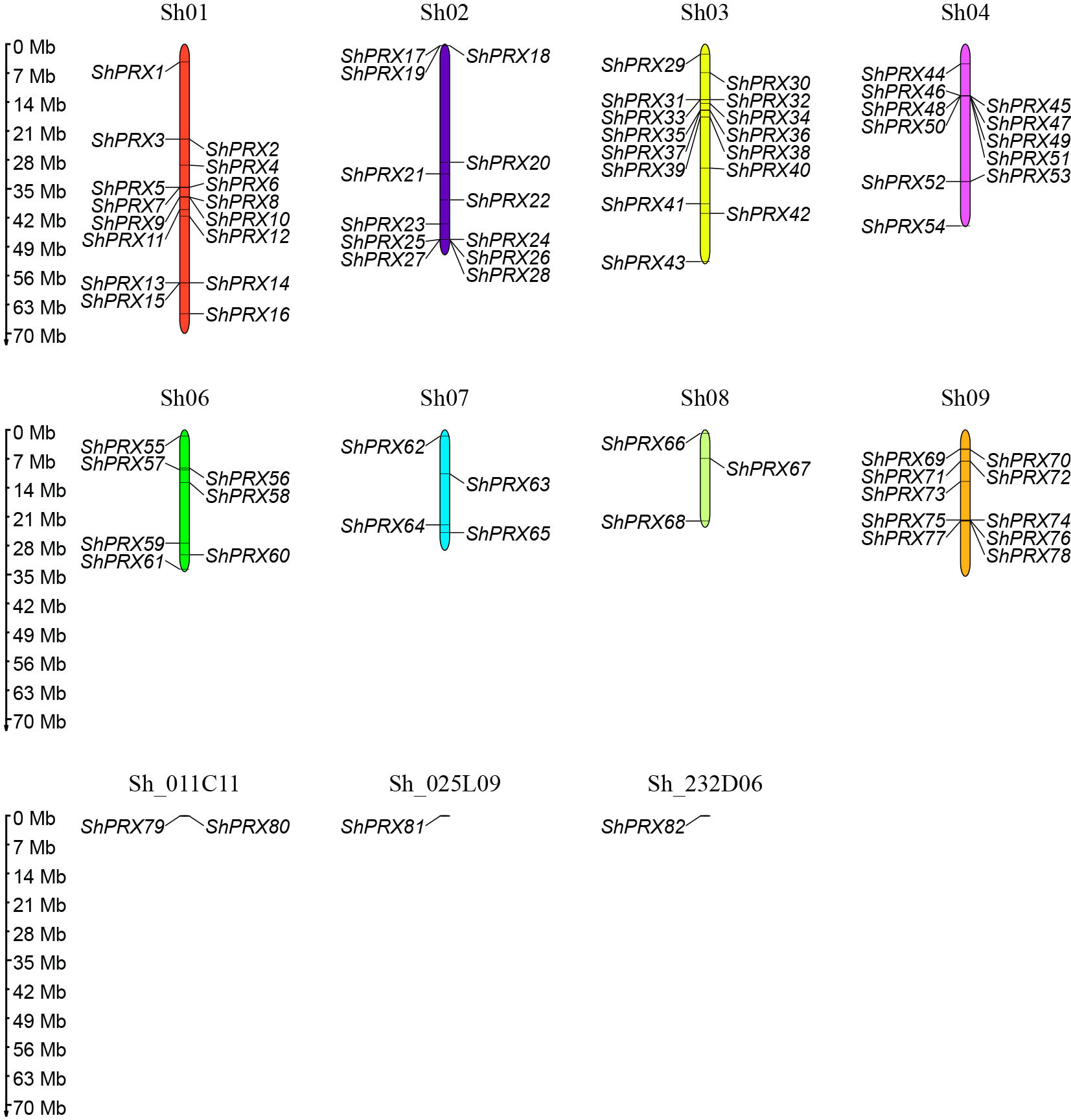
The 82 ShPRXs were unevenly mapped onto the eight chromosomes of R570 STP (see Figure 1) based on the annotation information of the R570 STP genome. The ShPRX genes distributed in Sh01 (16), Sh02 (12), Sh03 (15), Sh04 (11), Sh06 (7), and Sh09 (10), while chromosomes Sh07 and Sh08 had only 4 and 3 ShPRX genes, respectively. Four ShPRX genes were distributed on scaffolds (Sh_011C11, Sh_025L09, and Sh_232D06).
3.2 Phylogenetic of the PRX gene family
The phylogenetic tree from five plants was constructed on PRX amino acid to clarify the evolutionary relationship of the PRX gene family of sugarcane, as shown in Figure 2. The ShPRX family genes were divided into six groups. The phylogenetic tree based on the amino acid sequences of ShPRX indicated that the ShPRX family genes in groups 1, 2, 3, 4, 5, and 6 from five plants were clustered into groups I, II, VI, V, III and IV from sugarcane, besides ShPRX1 (group 3), ShPRX34 (group 3), ShPRX22 (group 4), and ShPRX62 (group 2). ShPRX1 and ShPRX34 were divided into group V, and ShPRX22 and ShPRX62 were divided into group IV (Figure 2; Figure S1). Group 1 comprised the lowest number of PRX genes. The PRX family genes of sugarcane shared high homology with the PRX family genes in sorghum and rice (Figure 2). The ShPRX family genes in other groups shared high homology to S. spontaneum or sorghum (Figure 2).
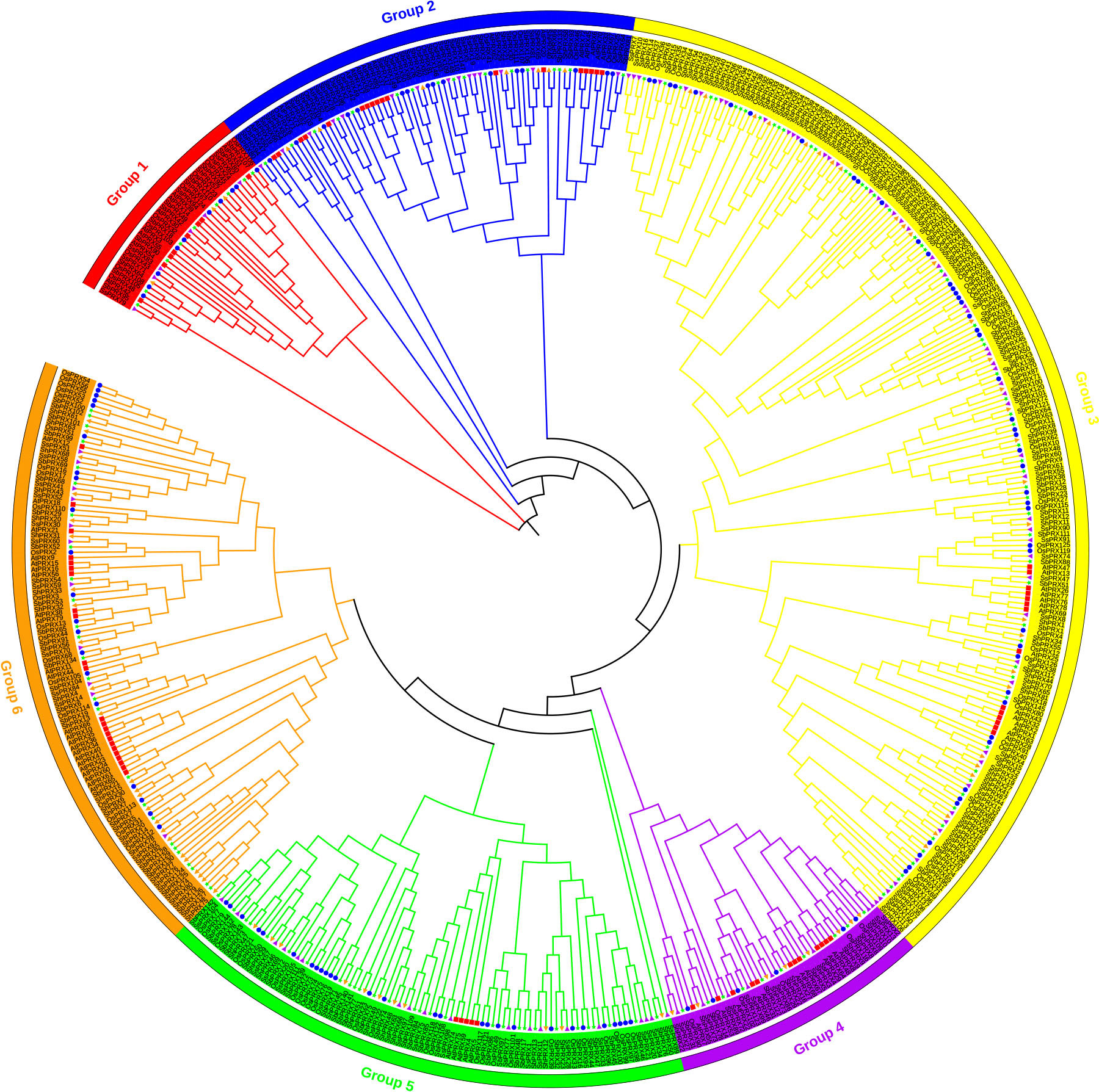
Figure 2 Phylogenetic analysis of PRX proteins from A. thaliana, O. sativa, S. bicolor, S. spontaneum, and sugarcane. AtPRX, red rectangle in the figure; OsPRX, blue circle in the figure; SbPRX, green star in the figure; SsPRX, purple right-pointing triangle in the figure; ShPRX, orange left-pointing triangle in the figure.
3.3 Gene structure and conserved motif analyses of the ShPRX gene family
The conservative structure of the PRX gene family was deciphered through the evolutionary relationship, motif, and structure of the PRX family genes in sugarcane (Figure 3). MEME was used to analyze the motif distribution within the ShPRX gene family, and 26 motifs were identified (p < 0.05) (Table S3). The ShPRX genes in the same group had the same motifs: the genes in Group I had motifs 3, 8, 18, and 19; Group II had motifs 3, 4, 5, 8, 10, and 12; Group III had motifs 3, 8, and 11; Group IV had motifs 2, 4, 5, 6, and 7; group V had motifs 1, 4, 7, and 8; and Group VI had motif 3, and most ShPRX genes have top 14 motifs (Table S4; Figures 3A, B). The ShPRX family genes had motif 3 except for ShPRX10 and ShPRX43, and most genes had motif 8 except for ShPRX43 and ShPRX44 (Figure 3B).

Figure 3 Analysis of gene structure and conserved motif of the PRX gene family in each group of sugarcane. (A) Phylogenetic tree of the ShPRX proteins. (B) Motif composition in the ShPRX proteins. (C) Gene structure of the ShPRXs.
According to the structural analysis of the PRX family genes, the length of these genes ranged from 723 to 7812 bp, where ShPRX67 had the most considerable length and 11 coding regions, ShPRX47 had the most petite length (Table S1; Figure 3C, S2). The respective structures of these genes were found to be similar in the same group, and 10 PRX genes had no introns. Most PRX genes contained 1–3 introns, but ShPRX57, ShPRX64, and ShPRX67 in Group I contained 13, 8, and 10 introns, respectively, and ShPRX20 (Group IV), ShPRX39 (Group VI), and ShPRX69 (Group VI) had long introns (Figures 3A, C).
3.4 Cis-acting elements and codon usage bias of PRX gene family in sugarcane
Cis-acting elements can participate in gene expression and regulation, and members of the ShPRX gene family play a key role in biotic and abiotic stress. The function of the ShPRX family genes can be predicted by analyzing the cis-acting elements in the 2-kb upstream region of the ShPRX family genes. Thirty-one cis-acting elements were identified and involved in hormones, abiotic stress, tissue-specific cell cycle, and circadian control by analyzing and selecting cis-acting elements of the ShPRX family genes (Table S5). The differences were detected in the variety and number of cis-acting elements across the ShPRX family genes (Table S5). ShPRX15, ShPRX41, ShPRX48, and ShPRX62 had the maximum variety of cis-acting elements (15), but the number of cis-acting elements in ShPRX52 was the greatest (37). ShPRX22, ShPRX38, ShPRX49, and ShPRX62 contained plant hormone-responsive elements, including IAA-, GA-, ABA-, SA-, and MeJA-responsive elements. We inferred that plant hormones might regulate these ShPRX family genes. ShPRX31 and ShPRX66 did not contain ABA and light-responsive elements but contained seed-specific regulation elements and MYB binding sites involved in drought inducibility (Table S5). The cis-acting regulatory elements of most ShPRX family genes were involved in ABA, MeJA, light responsiveness, anaerobic induction, and drought inducibility (Figure 4A).
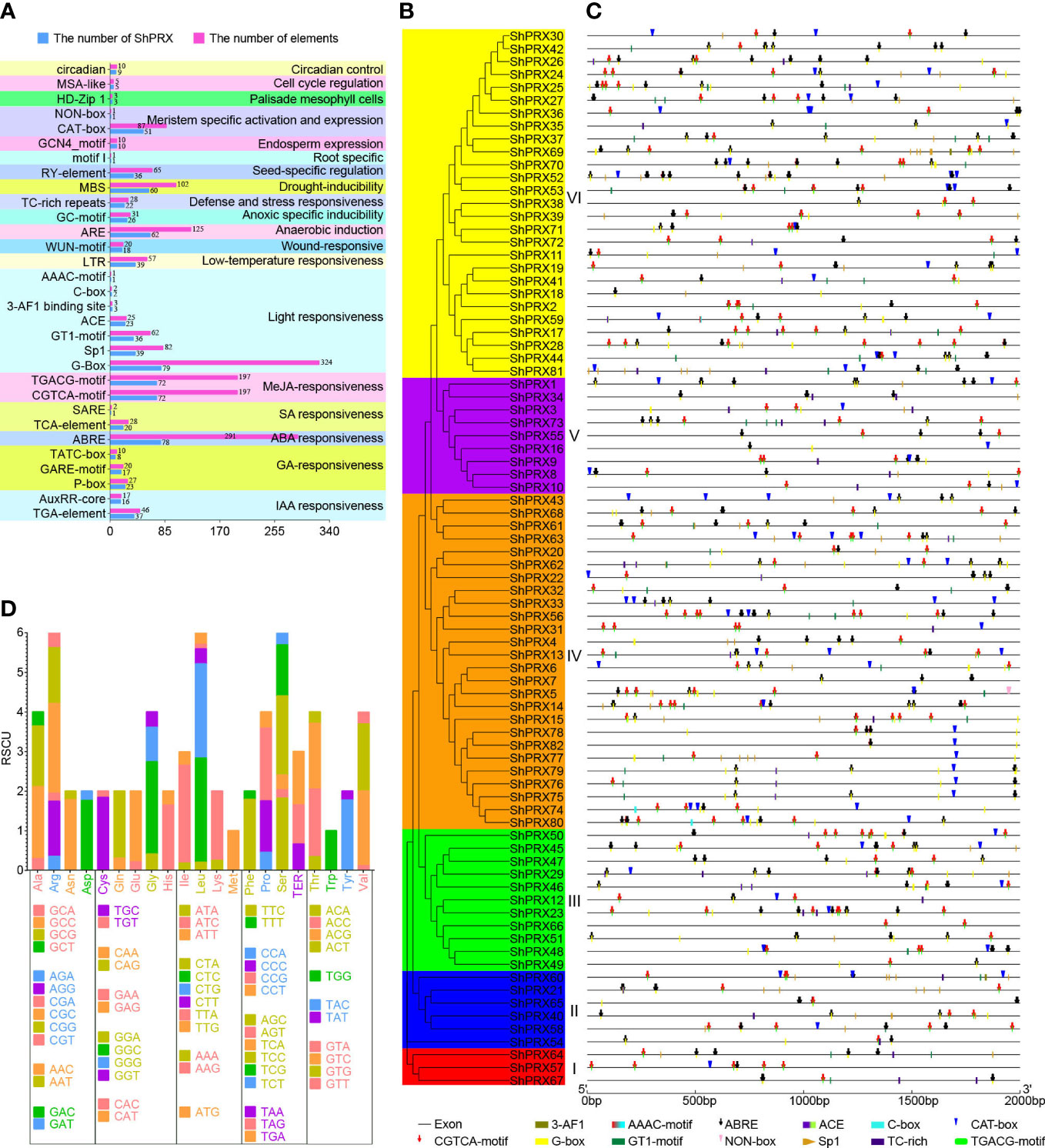
Figure 4 Analysis of cis-acting elements and codon usage bias of the PRX gene family in sugarcane. (A) The analysis of cis-acting elements of the PRX gene family in sugarcane. (B) The phylogenetic tree of the ShPRX proteins. (C) The analysis of cis-acting elements involved in the ABA, MeJA, light, defense, and stress responsiveness, meristem-specific activation, and expression in the PRX gene family of each group. (D) The codon usage bias analysis of PRX gene family in sugarcane.
The cis-acting elements in Groups III, IV, V, and VI, were involved in the meristem-specific activation and expression. Over half of those in the Group I, II, and V were involved in the defense and stress responsiveness (Figures 4B, C). The cis-acting regulatory elements in Group 1 involved in the IAA, ABA, SA, MeJA, light, low-temperature, and anaerobic induction, and group IV in the seed-specific regulation (Table S6). These results suggested that the ShPRX family genes might participate in the response of sugarcane to biotic and abiotic stresses and tissue-specific responses.
The codon usage bias and base composition analysis of coding sequences of the PRX family genes were calculated (Table S7, S8). The mean values of the codon composition at the third position from high to low were C3s (0.589), G3s (0.440), T3s (0.093), and A3s (0.080), and the mean content of the GC (65.1%) was also higher than AT (34.9%), suggesting a GC-rich composition of coding sequences of the PRX family genes (Table S7). The ENC values of coding sequences ranged from 28.25 to 58.62, with a mean of 38.059 (ENC < 40), and most ENC values of coding sequences were below 40, indicating a solid codon usage bias (Table S7). The RSCU revealed that 22 codons of 28 high-frequency codons (mean RSCU value > 1.0) were over-represented (mean RSCU value > 1.6). In comparison, 31 codons of 34 low-frequency codons (mean RSCU value < 1.0) were under-represented (mean RSCU value < 0.6) (Figure 4D; Table S8). The ENc plot (Figure S3A) and PR2 plot (Figure S3B) analyses showed that the codon use of the class III PRX family genes in sugarcane was affected by mutation and selection pressure.
3.5 Syntenic and selection pressure analysis of the ShPRX gene family
The syntenic relationships of the ShPRXs were analyzed to explore the genomic expansion of the PRX gene family in sugarcane. In total, 32 of the 82 ShPRXs had syntenic relationships, and 10 ones in five syntenic pairs underwent segmental duplication, while 22 genes in 13 syntenic pairs underwent tandem duplication (Figure 5A; Table S9). The Ka/Ks ratios of 17 of the 18 syntenic pairs were < 1, which might have undergone purifying selection, indicating that the evolution of these pairs was slow (Figure 5A; Table S9).
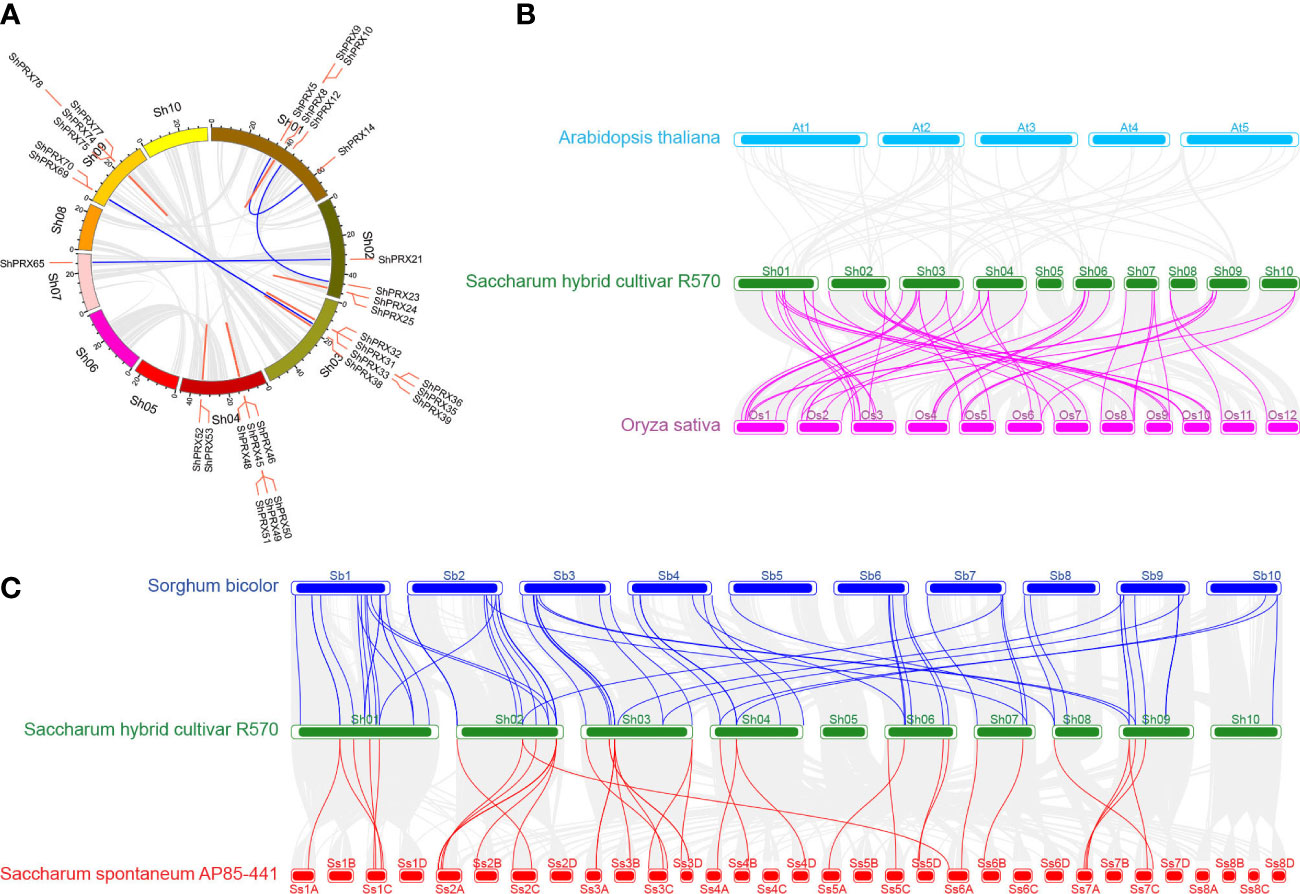
Figure 5 Syntenic analysis of PRX gene family in Sugarcane, A. thaliana, O. sativa, S. bicolor, and S. spontaneum. (A) Syntenic analysis of PRX gene family in sugarcane. Red lines represent tandem duplication PRX gene pairs, and blue lines represent segmental duplication PRX gene pairs. (B) Syntenic analysis of PRX gene family in Sugarcane, A. thaliana, and O. sativa. (C) Syntenic analysis of the PRX gene family in Sugarcane, S. bicolor, and S. spontaneum.
The syntenic relationships of the PRX family genes from S officinarum, A. thaliana, O. sativa, S. bicolor, and S. spontaneum were analyzed to better understand duplication and evolution of PRX gene. The results revealed that 52, 68, and 37 syntenic gene pairs of PRX genes were detected in S officinarum versus O. sativa, S officinarum versus S. bicolor, and S officinarum versus S. spontaneum, respectively (Figures 5B, C; Table S9). The Ka/Ks ratio of 1 of the 157 syntenic gene pairs was> 1, indicating that the syntenic gene pair had been strongly positively selected during evolutionary history (Table S9). The Ka/Ks ratios of 153 from the 157 syntenic gene pairs were < 1, which might have undergone purifying selection, indicating that the evolution of these pairs was slow. The duplication events of homologous collinearity gene pairs of the class III PRX occurred 4.094–85.36 Mya for sugarcane and rice, 21.992–83.6 Mya for sugarcane and sorghum, and 3.568–50.095 Mya for sugarcane and its wild relative of S. spontaneum. The class III PRX gene family has been identified in at least 29 plants in the whole genome. The duplication events occurred 159.9 Mya for 12 plants in monocotyledon from 21 plants in dicotyledon. Moreover, 104.7 Mya in Poaceae diverged from Bromeliaceae (Figure S3C; Table S10).
3.6 Expression profiles of the PRX family genes in different tissue
Tissue-specific expression patterns of the SsPRX genes were analyzed based on the transcriptome data of S. spontaneum to explore the functions of the PRX gene family in sugarcane further. The 113 SsPRX genes were expressed as FPKM values in leaf roll, leaf, the third, sixth, and ninth stem nodes of sugarcane at the seedling, elongation and maturity stage (Figure 6A; Table S11). Four SsPRX genes (SsPRX42, SsPRX62, SsPRX79, and SsPRX83) were highly expressed in all tissues, and 69 SsPRX genes had a low expression or no expression, and the rest of the highly expressed SsPRX genes had tissue specificity.
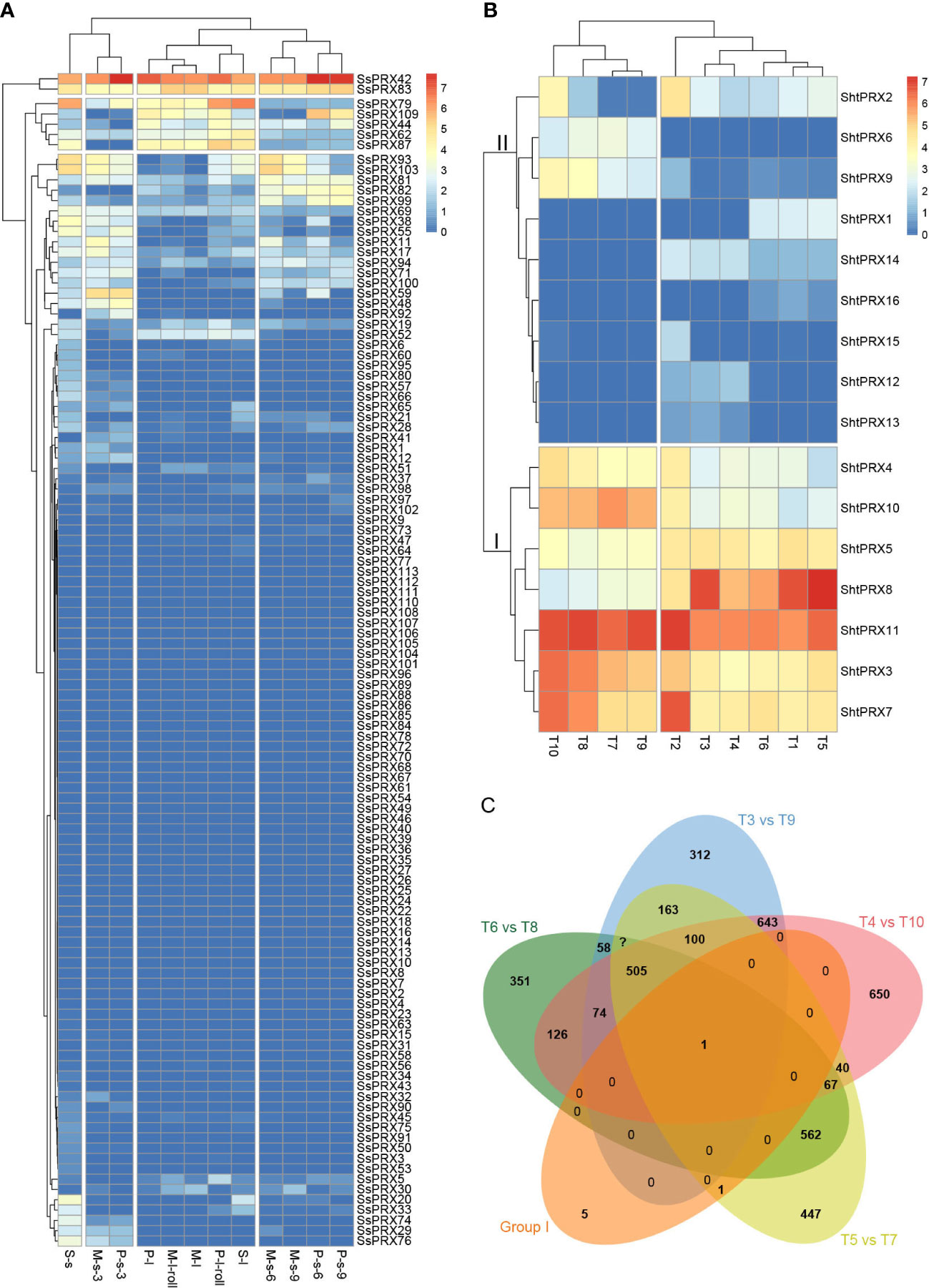
Figure 6 Expression pattern of the PRX genes in sugarcane. (A) Tissue-specific expression patterns of PRX gene family in S. spontaneum. S represents seedling; s represents stem; l represents leaf; P represents early maturity stage; M represents mature stage; 3, 6, and 9 represent sugarcane’s third, sixth, and ninth stem nodes. (B) Expression patterns of PRX gene family of sugarcane in response to SCMV infection. T1, the +1 leaf of Badila; T2, the −3 leaf of Badila; T3, the +1 leaf of virus-free Badila; T4, the −3 leaf of virus-free Badila; T5, the +1 leaf of B48; T6, the −3 leaf of B48; T7, the +1 leaf of B48 post-SCMV infection; T8, the −3 leaf of B48 post-SCMV infection, T9, the +1 leaf of virus-free Badila post-SCMV infection; T10, the −3 leaf of virus-free Badila post-SCMV infection. (C) The number of co-differentially expressed genes in groups I, T6_vs_T8, T3_vs_T9, T4_vs_T10, and T5_vs_T7.
3.7 Expression profiles of PRX family genes under SCMV, Cd, and salt stresses
Sixteen PRX proteins with a conserved PRX domain were characterized as members of the sugarcane class III PRX gene family based on the transcriptome data of Badila and B48 in response to SCMV infection. We named them ShtPRX1–ShtPRX16 based on their Unigene IDs (Table S2). The expression patterns of the ShtPRX genes were analyzed on transcriptome data (Table S12). The ShtPRX genes were divided into two groups according to the clustering of expression patterns. Genes in Group I was primarily highly expressed in all samples. Genes in Group II, except for ShtPRX13 and ShtPRX16, were primarily highly expressed in some samples (Figure 6B). ShtPRX8 was only one co-differentially expressed gene (|log2FC| ≥ 3, FDR < 0.05) in five groups (Figure 6C), indicating that ShtPRX8 might participate in SCMV stress in sugarcane.
Homologous class III PRX genes of R570 were identified using class III PRX family protein sequences of rice as a library, and screened bu BLASTP at the e-value less than 1e-5. (Table S13). The expression profiles of rice under Cd and salt stresses were analyzed to infer the function of PRX homologous genes in sugarcane (Tables S14, S15; Figure S4). In groups #I, III, and VII, except for OsPRX10, the |log2FC| values for (Cd/CK)-WR or (Cd/CK)-WS was more than 3, and homologous genes, including ShPRX38, ShBAC.PRX23, ShBAC.PRX41, ShBAC.PRX42, ShBAC.PRX43, ShBAC.PRX51, ShBAC.PRX52, and ShBAC.PRX75 might participate in Cd stress in sugarcane (Figure S4A; Table S13). In group 2, the |log2FC| of OsPRX56 for PK-SS/PK-CK was more than 2, and homologous genes of ShBAC.PRX20 might participate in salt stress in Sugarcane (Figure S4B; Tables S13, S14).
qRT-PCR analysis was performed to analyze the responses of the PRX family genes in sugarcane exposed to SCMV, Cd, and salt stresses. The primer sequences were listed in Table S16. The expression levels of ShtPRX8 showed a significant decrease in Badila after SCMV infection. After applying 4.3 mM Cd2+ stress to sugarcane, the expression levels of ShBAC.PRX23, ShBAC.PRX41, ShBAC.PRX42, ShBAC.PRX43, ShBAC.PRX51, ShBAC.PRX52, ShBAC.PRX75 and ShPRX38 showed a significant increase from 0 to 24 h (Figure 7). After applying 1% sodium chloride solution stress to sugarcane, the expression levels of ShBAC PRX20 significantly increased from 0 to 24 h (Figure 7).
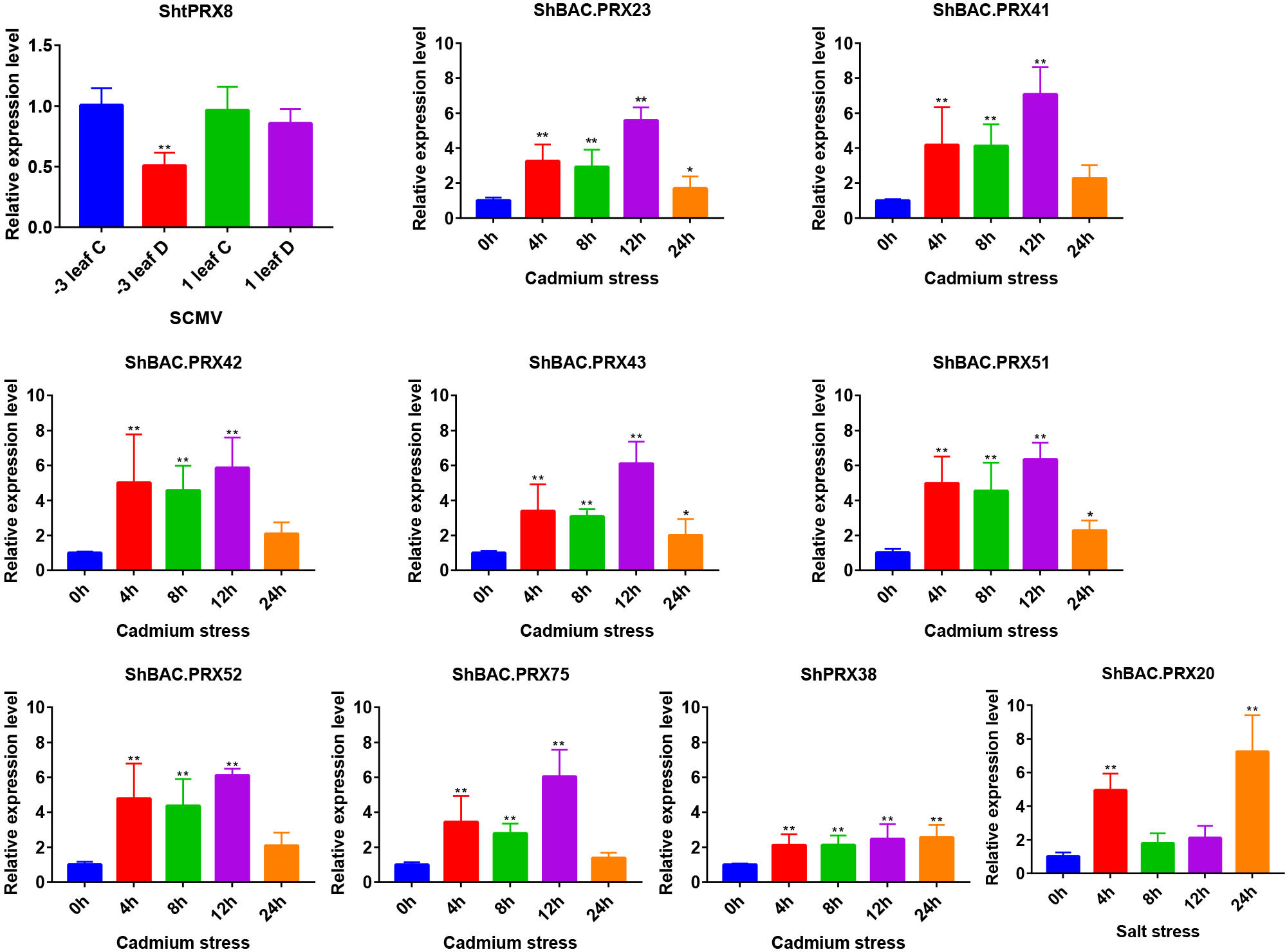
Figure 7 Analysis of SCMV, Cd, and salt stress differentially expressed PRX gene family members in sugarcane. The black bar graphs represent the relative expression levels of PRX family genes in the leaves under SCMV, Cd, and salt stresses. The −3 leaf C, the −3 leaf of virus-free Badila; the 1 leaf C, the +1 leaf of virus-free Badila; −3 leaf D, the -3 leaf of post-SCMV infection Badila (symptomatic leaf); 1 leaf D, the +1 leaf of post-SCMV infection Badila (asymptomatic leaf); *: 0.01 < p < 0.05, **: p < 0.01; The mean and SD were calculated from three biological and three technical replicate samples.
4 Discussion
Class III PRXs are essential in response to various biotic and abiotic stresses during plant growth and development. The class III PRX gene family has been identified in at least 29 plants in the whole genome, but the class III PRX gene family has yet to be studied in sugarcane. The number of PRX family genes identified in R570 BAC and STP was fewer than in other monocotyledons and dicotyledons. The class III PRX family genes were divided into six groups based on the phylogenetic analysis of sugarcane, sorghum, S. spontaneum, rice, and A. thaliana and the structure and conservative motif of class III PRX genes were similar in each group. The class III PRX genes in the same group were highly conserved. The identified class III PRXs in sugarcane had no collinear gene pair in A. thaliana. This is the same as Cesarino’s results, showing three sugarcane PRXs, being monocot-specific sequences, had no clear ortholog in A. thaliana (Cesarino et al., 2012).
Poaceae diverged from Bromeliaceae at 104.7 Mya. The duplication events of homologous collinearity gene pairs of the class III PRX for sugarcane and rice, sugarcane and sorghum, and sugarcane and its wild relative of S. spontaneum occurred at 0–85.36 Mya. These results revealed that class III PRXs in sugarcane was formed after Poaceae and Bromeliaceae diverged. Tandem duplication events play a leading role in the expansion of the class III PRX gene family in sugarcane, which was the same as B. distachyon (Zhu et al., 2019), foxtail millet (Ma et al., 2022), and grapevine (Xiao et al., 2020). Purifying selection essentially maintained the function of class III PRX proteins in sugarcane. However, tandem and segmental duplications are the primary reasons for expanding the class III PRX gene family in maize (Wang et al., 2015) and pineapple (Hou et al., 2022). Segmental duplication is the main reason for the expansion of the class III PRX gene family in tobacco (Cheng et al., 2022), soybean (Aleem et al., 2022), and potato (Yang et al., 2020). These results indicated that tandem and segmental duplications were pivotal in expanding plants’ class III PRX gene family.
Phylogenetic analysis revealed that ShPRX family genes shared high homology with the PRX family genes in S. spontaneum and sorghum, which was consistent with the genetic relationship between species. We inferred that ShPRX family genes might have the same function in the same group. GO enrichment analysis of motifs using InterProScan revealed that motifs 1, 2, 3, 4, 5, 18, 19, and 21 are essential in response to oxidative stress (GO:0006979) and have peroxidase activity (GO:0004601). However, there are some motifs with unknown functions in each group; these motifs might play a key role in the ShPRX family genes.
Gene expression patterns and cis-acting elements can provide important information regarding gene function. More than 87% of class III ShPRX genes were involved in light (G-box), ABA (ABRE), and MeJA (CGTCA and TGACG motifs) response, and more than 73% of class III PRX genes participated in anaerobic (ARE) and drought (MBS) responses. Also, over 62% of class III PRX genes participated in meristem-specific (CAT box) responses (Table S5). We inferred that class III PRX genes of sugarcane played an essential role in plant growth and development. The highest enzymatic activity was presented in the pith and rind of mature internodes at three different developmental stages of sugarcane (young, developing, and mature) (Cesarino et al., 2012). The expression of class III PRX proteins in susceptible sugarcane was inhibited for 72 h after Sporisorium scitamineum inoculation (Peters et al., 2017). Of 113 SsPRX genes, 69 exhibited little or no expression in tissues, indicating that SsPRX genes might be expressed under specific conditions or at specific developmental stages. Of 44 highly expressed genes, 26 and 43 were expressed in leaves and stems, respectively, and most of them might play an essential role in the leaf and stem of sugarcane.
In rice, the overexpression of OsPrx30 contributed to maintaining a high level of PRX activity and reducing H2O2 content, thereby enhancing the rice plant’s susceptibility to Xoo (Liu et al., 2021), suggesting that class III PRX genes might have similar functions in sugarcane. The concentrations of H2O2 increased significantly in B48. The genes related to ROS-producing and scavenging pathways were differentially expressed on the ScMV-inoculated plants at days 3, 6, 9, and 12 (Akbar et al., 2020). The expression of ShtPRX8 was significantly reduced post-SCMV inoculation based on the transcriptional data from our previous study. PRX, primarily existing in peroxisomes, could reduce the ROS level. SCMV could target intracellular peroxisomes for replication (Xie et al., 2021). The class III PRX family genes might play an essential role in sugarcane mosaic disease by activating the antioxidant system and regulating ROS and H2O2 content. Plants produced ROS under salt and Cd stresses, and the class III PRX family genes play an essential role in plant Cd and salt stress by scavenging ROS (Chiang et al., 2006; Kidwai et al., 2020; Su et al., 2020). The overexpression of the class III PRX family gene of TaPRX-2A in wheat activated the ABA pathway and antioxidant enzymes, leading to reduced ROS accumulation and increased osmotic metabolites, thereby enhancing salt tolerance (Su et al., 2020). The expression levels of class III PRXs significantly increased after applying 4.3 mM Cd and 1% NaCl solution stress to sugarcane. We inferred that the class III PRX family genes could enhance the tolerance of Cd and salt stresses in sugarcane by activating the antioxidant system and scavenging ROS.
5 Conclusion
Tandem duplication events play a leading role in the expansion of ShPRX genes, and purifying selection essentially maintains the function of ShPRX proteins. In this study, 82 ShPRX genes were identified in the R570 STP genome and divided them into six groups. Expression profile and qRT-PCR analyses showed that SCMV, Cd, and salt could specifically induce the expression of PRX genes of sugarcane. These results help understand the structure, evolution, and functions of the class III PRX gene family in sugarcane with a view of providing ideas for the phytoremediation of Cd-contaminated soil and breeding of new sugarcane varieties resistant to sugarcane mosaic disease, salt, and Cd in the future.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
MZ, WY, and JX designed the research. HS wrote the draft manuscript. LF, LQ, HJ, ZD, HZ, and ZY performed the experiments and data analyses. GC and YB conducted the genomic analysis. JX and WY edited the first draft. MZ conceived the idea, provided supervision, revised the manuscript, and provided funds for this investigation. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the National Natural Science Foundation of China (32001603), the Science and Technology Major Project of Guangxi (Gui Ke AA22117007, Gui Ke AB21238008), and the China Agriculture Research System of MOF and MARA (CARS-170109, CARS-170726). The authors would like to thank the reviewers for their helpful comments on this manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1101665/full#supplementary-material
Supplementary Figure 1 | Phylogenetic analysis of ShPRX proteins.
Supplementary Figure 2 | Analysis of gene structure and conserved motif of the PRX gene family.
References
Akbar, S., Wei, Y., Yuan, Y., Khan, M. T., Qin, L., et al. (2020). Gene expression profiling of reactive oxygen species (ROS) and antioxidant defense system following sugarcane mosaic virus (SCMV) infection. BMC Plant Biol. 20, 532. doi: 10.1186/s12870-020-02737-1
Akbar, S., Yao, W., Qin, L., Yuan, Y., Powell, C. A., et al. (2021). Comparative analysis of sugar metabolites and their transporters in sugarcane following sugarcane mosaic virus (SCMV) infection. Int. J. Mol. Sci. 22. doi: 10.3390/ijms222413574
Aleem, M., Riaz, A., Raza, Q., Aleem, M., Aslam, M., et al. (2022). Genomewide characterization and functional analysis of class iii peroxidase gene family in soybean reveal regulatory roles of gspod40 in drought tolerance. Genomics 114, 45–60. doi: 10.1016/j.ygeno.2021.11.016
Bailey, T. L., Elkan, C. (1994). Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf Intell. Syst. Mol. Biol. 2, 28–36.
Blom, N., Gammeltoft, S., Brunak, S. (1999). Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J. Mol. Biol. 294, 1351–1362. doi: 10.1006/jmbi.1999.3310
Cesarino, I., Araujo, P., Sampaio, M. J., Paes, L. A., Mazzafera, P. (2012). Enzymatic activity and proteomic profile of class iii peroxidases during sugarcane stem development. Plant Physiol. Biochem. 55, 66–76. doi: 10.1016/j.plaphy.2012.03.014
Chakraborty, S., Yengkhom, S., Uddin, A. (2020). Analysis of codon usage bias of chloroplast genes in oryza species: codon usage of chloroplast genes in oryza species. Planta 252, 67. doi: 10.1007/s00425-020-03470-7
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., et al. (2020). Tbtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, Y., Feng, J., Qu, Y., Zhang, J., Zhang, L., et al. (2022). Genomewide identification and functional analysis of class iii peroxidases in gossypium hirsutum. Peer J. 10, e13635. doi: 10.7717/peerj.13635
Cheng, L., Ma, L., Meng, L., Shang, H., Cao, P., et al. (2022). Genomewide identification and analysis of the class iii peroxidase gene family in tobacco (Nicotiana tabacum). Front. Genet. 13. doi: 10.3389/fgene.2022.916867
Chiang, H. C., Lo, J. C., Yeh, K. C. (2006). Genes associated with heavy metal tolerance and accumulation in Zn/Cd hyperaccumulator arabidopsis halleri: a genomic survey with cDNA microarray. Environ. Sci. Technol. 40, 6792–6798. doi: 10.1021/es061432y
Chou, K. C., Shen, H. B. (2010). Plant-mploc: a top-down strategy to augment the power for predicting plant protein subcellular localization. PloS One 5, e11335. doi: 10.1371/journal.pone.0011335
Clough, E., Barrett, T. (2016). The gene expression omnibus database. Methods Mol. Biol. 1418, 93–110. doi: 10.1007/978-1-4939-3578-9_5
Duan, P., Wang, G., Chao, M., Zhang, Z., Zhang, B. (2019). Genomewide identification and analysis of class iii peroxidases in allotetraploid cotton (Gossypium hirsutum l.) and their responses to pk deficiency. Genes (Basel) 10. doi: 10.3390/genes10060473
Francoz, E., Ranocha, P., Nguyen-Kim, H., Jamet, E., Burlat, V., et al. (2015). Roles of cell wall peroxidases in plant development. Phytochemistry 112, 15–21. doi: 10.1016/j.phytochem.2014.07.020
Garg, R., Narayana, C. V., Shankar, R., Jain, M. (2015). Divergent DNA methylation patterns associated with gene expression in rice cultivars with contrasting drought and salinity stress response. Sci. Rep. 5, 14922. doi: 10.1038/srep14922
Garsmeur, O., Droc, G., Antonise, R., Grimwood, J., Potier, B., et al. (2018). A mosaic monoploid reference sequence for the highly complex genome of sugarcane. Nat. Commun. 9, 2638. doi: 10.1038/s41467-018-05051-5
Gaut, B. S., Morton, B. R., McCaig, B. C., Clegg, M. T. (1996). Substitution rate comparisons between grasses and palms: synonymous rate differences at the nuclear gene adh parallel rate differences at the plastid gene rbcl. Proc. Natl. Acad. Sci. U S 93, 10274–10279. doi: 10.1073/pnas.93.19.10274
Geourjon, C., Deleage, G. (1995). Sopma: significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments. Comput. Appl. Biosci. 11, 681–684. doi: 10.1093/bioinformatics/11.6.681
Hou, X., Lu, Z., Hong, K., Song, K., Gu, H., et al. (2022). The class iii peroxidase gene family is involved in ascorbic acid induced delay of internal browning in pineapple. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.953623
Hu, B., Jin, J., Guo, A. Y., Zhang, H., Luo, J., et al. (2015). Gsds 2.0: an upgraded gene feature visualization server. Bioinformatics 31, 1296–1297. doi: 10.1093/bioinformatics/btu817
Ishimaru, Y., Takahashi, R., Bashir, K., Shimo, H., Senoura, T., et al. (2012). Characterizing the role of rice nramp5 in manganese, iron, and cadmium transport. Sci. Rep. 2, 286. doi: 10.1038/srep00286
Kidwai, M., Ahmad, I. Z., Chakrabarty, D. (2020). Class iii peroxidase: an indispensable enzyme for biotic/abiotic stress tolerance and a potent candidate for crop improvement. Plant Cell Rep. 39, 1381–1393. doi: 10.1007/s00299-020-02588-y
Kidwai, M., Dhar, Y. V., Gautam, N., Tiwari, M., Ahmad, I. Z., et al. (2019). Oryza sativa class iii peroxidase (osprx38) overexpression in arabidopsis thaliana reduces arsenic accumulation due to apoplastic lignification. J. Hazard Mater 362, 383–393. doi: 10.1016/j.jhazmat.2018.09.029
Krzywinski, M., Schein, J., Birol, I., Connors, J., Gascoyne, R., et al. (2009). Circos: an information aesthetic for comparative genomics. Genome Res. 19, 1639–1645. doi: 10.1101/gr.092759.109
Kumar, S., Stecher, G., Li, M., Knyaz, C., Tamura, K. (2018). Mega x: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Kumar, S., Suleski, M., Craig, J. M., Kasprowicz, A. E., Sanderford, M., et al. (2022). Timetree 5: an expanded resource for species divergence times. Mol. Biol. Evol. doi: 10.1093/molbev/msac174
Lee, T. T. (1977). Role of phenolic inhibitors in peroxidase-mediated degradation of indole-3-acetic acid. Plant Physiol. 59, 372–375. doi: 10.1104/pp.59.3.372
Lescot, M., Dehais, P., Thijs, G., Marchal, K., Moreau, Y., et al. (2002). Plantcare, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Letunic, I., Bork, P. (2021). Interactive tree of life (itol) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 49, W293–W296. doi: 10.1093/nar/gkab301
Li, Q., Dou, W., Qi, J., Qin, X., Chen, S., et al. (2020). Genomewide analysis of the ciii peroxidase family in sweet orange (Citrus sinensis) and expression profiles induced by xanthomonas citri subsp. citri and hormones. J. Genet. 99. doi: 10.1007/s12041-019-1163-5
Liu, H., Dong, S., Li, M., Gu, F., Yang, G., et al. (2021). The class iii peroxidase gene osprx30, transcriptionally modulated by the at-hook protein osath1, mediates rice bacterial blight-induced ROS accumulation. J. Integr. Plant Biol. 63, 393–408. doi: 10.1111/jipb.13040
Lu, S., Wang, J., Chitsaz, F., Derbyshire, M. K., Geer, R. C., et al. (2020). Cdd/sparcle: the conserved domain database in 2020. Nucleic Acids Res. 48, D265–D268. doi: 10.1093/nar/gkz991
Ma, X., Xu, R., Suo, X., Li, J., Gu, P., et al. (2022). Genomewide identification of the class iii prx gene family in foxtail millet (Setaria italica l.) and expression analysis under drought stress. Acta Agronomica Sin. 10, 2517–2532.
Mistry, J., Chuguransky, S., Williams, L., Qureshi, M., Salazar, G. A., et al. (2021). Pfam: the protein families database in 2021. Nucleic Acids Res. 49, D412–D419. doi: 10.1093/nar/gkaa913
Moradi, Z., Mehrvar, M., Nazifi, E. (2018). Genetic diversity and biological characterization of sugarcane streak mosaic virus isolates from Iran. Virus Dis. 29, 316–323. doi: 10.1007/s13337-018-0461-5
Passardi, F., Longet, D., Penel, C., Dunand, C. (2004). The class iii peroxidase multigenic family in rice and its evolution in land plants. Phytochemistry 65, 1879–1893. doi: 10.1016/j.phytochem.2004.06.023
Peters, L. P., Carvalho, G., Vilhena, M. B., Creste, S., Azevedo, R. A., et al. (2017). Functional analysis of oxidative burst in sugarcane smut-resistant and -susceptible genotypes. Planta 245, 749–764. doi: 10.1007/s00425-016-2642-z
Piontek, K., Smith, A. T., Blodig, W. (2001). Lignin peroxidase structure and function. Biochem. Soc. Trans. 29, 111–116. doi: 10.1042/0300-5127:0290111
Rice, J. L., Hoy, J. W. (2020). Recovery from mosaic caused by sorghum mosaic virus in sugarcane and impact on yield. Plant Dis. 104, 3166–3172. doi: 10.1094/PDIS-02-20-0376-RE
Rice, J. L., Hoy, J. W., Grisham, M. P. (2019). Sugarcane mosaic distribution, incidence, increase, and spatial pattern in Louisiana. Plant Dis. 103, 2051–2056. doi: 10.1094/PDIS-01-19-0099-RE
Rice, P., Longden, I., Bleasby, A. (2000). Emboss: the European molecular biology open software suite. Trends Genet. 16, 276–277. doi: 10.1016/s0168-9525(00)02024-2
Sereno, M. L., Almeida, R. S., Nishimura, D. S., Figueira, A. (2007). Response of sugarcane to increasing concentrations of copper and cadmium and expression of metallothionein genes. J. Plant Physiol. 164, 1499–1515. doi: 10.1016/j.jplph.2006.09.007
Shankar, R., Bhattacharjee, A., Jain, M. (2016). Transcriptome analysis in different rice cultivars provides novel insights into desiccation and salinity stress responses. Sci. Rep. 6, 23719. doi: 10.1038/srep23719
Shigeto, J., Tsutsumi, Y. (2016). Diverse functions and reactions of class iii peroxidases. New Phytol. 209, 1395–1402. doi: 10.1111/nph.13738
Sonnhammer, E. L., von Heijne, G., Krogh, A. (1998). A hidden Markov model for predicting transmembrane helices in protein sequences. Proc. Int. Conf Intell. Syst. Mol. Biol. 6, 175–182.
Su, P., Yan, J., Li, W., Wang, L., Zhao, J., et al. (2020). A member of wheat class iii peroxidase gene family, taprx-2a, enhanced the tolerance of salt stress. BMC Plant Biol. 20, 392. doi: 10.1186/s12870-020-02602-1
Teufel, F., Almagro, A. J., Johansen, A. R., Gislason, M. H., Pihl, S. I., et al. (2022). Signalp 6.0 predicts all five types of signal peptides using protein language models. Nat. Biotechnol. 40, 1023–1025. doi: 10.1038/s41587-021-01156-3
Tognolli, M., Penel, C., Greppin, H., Simon, P. (2002). Analysis and expression of the class iii peroxidase large gene family in arabidopsis thaliana. Gene 288, 129–138. doi: 10.1016/s0378-1119(02)00465-1
Vanholme, R., De Meester, B., Ralph, J., Boerjan, W. (2019). Lignin biosynthesis and its integration into metabolism. Curr. Opin. Biotechnol. 56, 230–239. doi: 10.1016/j.copbio.2019.02.018
Wang, Y., Tang, H., Debarry, J. D., Tan, X., Li, J., et al. (2012). Mcscanx: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40, e49. doi: 10.1093/nar/gkr1293
Wang, Y., Wang, Q., Zhao, Y., Han, G., Zhu, S. (2015). Systematic analysis of maize class iii peroxidase gene family reveals a conserved subfamily involved in abiotic stress response. Gene 566, 95–108. doi: 10.1016/j.gene.2015.04.041
Wilkins, M. R., Gasteiger, E., Bairoch, A., Sanchez, J. C., Williams, K. L., et al. (1999). Protein identification and analysis tools in the expasy server. Methods Mol. Biol. 112, 531–552. doi: 10.1385/1-59259-584-7:531
Wright, F. (1990). The 'effective number of codons' used in a gene. Gene 87, 23–29. doi: 10.1016/0378-1119(90)90491-9
Xiao, H., Wang, C., Khan, N., Chen, M., Fu, W., et al. (2020). Genomewide identification of the class iii pod gene family and their expression profiling in grapevine (vitis vinifera l). BMC Genomics 21, 444. doi: 10.1186/s12864-020-06828-z
Xie, J., Jiang, T., Li, Z., Li, X., Fan, Z., et al. (2021). Sugarcane mosaic virus remodels multiple intracellular organelles to form genomic RNA replication sites. Arch. Virol. 166, 1921–1930. doi: 10.1007/s00705-021-05077-z
Yang, X., Yuan, J., Luo, W., Qin, M., Yang, J., et al. (2020). Genomewide identification and expression analysis of the class iii peroxidase gene family in potato (Solanum tuberosum l.). Front. Genet. 11. doi: 10.3389/fgene.2020.593577
Yao, W., Ruan, M., Qin, L., Yang, C., Chen, R., et al. (2017). Field performance of transgenic sugarcane lines resistant to sugarcane mosaic virus. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.00104
Yates, A. D., Allen, J., Amode, R. M., Azov, A. G., Barba, M., et al. (2022). Ensembl genomes 2022: an expanding genome resource for non-vertebrates. Nucleic Acids Res. 50, D996–D1003. doi: 10.1093/nar/gkab1007
Yousefi, Z., Kolahi, M., Majd, A., Jonoubi, P. (2018). Effect of cadmium on morphometric traits, antioxidant enzyme activity, and phytochelatin synthase gene expression (sopcs) of saccharum officinarum var. Cp48-103 in vitro. Ecotoxicol Environ. Saf. 157, 472–481. doi: 10.1016/j.ecoenv.2018.03.076
Zamocky, M. (2004). Phylogenetic relationships in class I of the superfamily of bacterial, fungal, and plant peroxidases. Eur. J. Biochem. 271, 3297–3309. doi: 10.1111/j.1432-1033.2004.04262.x
Zamocky, M., Furtmuller, P. G., Obinger, C. (2010). Evolution of structure and function of class I peroxidases. Arch. Biochem. Biophys. 500, 45–57. doi: 10.1016/j.abb.2010.03.024
Zhang, H., Wang, Z., Li, X., Gao, X., Dai, Z., et al. (2022). The ibbbx24-ibtoe3-ibprx17 module enhances abiotic stress tolerance by scavenging reactive oxygen species in sweet potato. New Phytol. 233, 1133–1152. doi: 10.1111/nph.17860
Zhang, J., Zhang, X., Tang, H., Zhang, Q., Hua, X., et al. (2018). Allele-defined genome of the autopolyploid sugarcane saccharum spontaneum l. Nat. Genet. 50, 1565–1573. doi: 10.1038/s41588-018-0237-2
Zhu, X., Jiang, L., Cai, Y., Cao, Y. (2021). Functional analysis of four class iii peroxidases from Chinese pear fruit: a critical role in lignin polymerization. Physiol. Mol. Biol. Plants 27, 515–522. doi: 10.1007/s12298-021-00949-9
Zhu, T., Li, L., Duan, Q., Liu, X., Chen, M. (2021). Progress in our understanding of plant responses to the stress of heavy metal cadmium. Plant Signal Behav. 16, 1836884. doi: 10.1080/15592324.2020.1836884
Keywords: sugarcane, class III peroxidase, sugarcane mosaic virus, cadmium, salt stress
Citation: Shang H, Fang L, Qin L, Jiang H, Duan Z, Zhang H, Yang Z, Cheng G, Bao Y, Xu J, Yao W and Zhang M (2023) Genome-wide identification of the class III peroxidase gene family of sugarcane and its expression profiles under stresses. Front. Plant Sci. 14:1101665. doi: 10.3389/fpls.2023.1101665
Received: 18 November 2022; Accepted: 09 January 2023;
Published: 30 January 2023.
Edited by:
Galal Bakr Anis, Field Crops Research Institute, EgyptReviewed by:
Sandhya Verma, Shri Vaishnav Vidyapeeth Vishwavidyalaya, Indore, IndiaLongbiao Guo, China National Rice Research Institute (CAAS), China
Copyright © 2023 Shang, Fang, Qin, Jiang, Duan, Zhang, Yang, Cheng, Bao, Xu, Yao and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Muqing Zhang, em11cWluZ0AxNjMuY29t; Wei Yao, eWFvd2VpbWFpbEAxNjMuY29t; Jingsheng Xu, eHVqaW5nc2hlbmdAMTI2LmNvbQ==
 Heyang Shang
Heyang Shang Linqi Fang2
Linqi Fang2 Jingsheng Xu
Jingsheng Xu Wei Yao
Wei Yao Muqing Zhang
Muqing Zhang