
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 21 March 2023
Sec. Plant Development and EvoDevo
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1097158
Low temperatures restrict the growth and geographic distribution of plants, as well as crop yields. Appropriate transcriptional regulation is critical for cold acclimation in plants. In this study, we found that the mutation of Leaf and flower related (LFR), a component of SWI/SNF chromatin remodeling complex (CRC) important for transcriptional regulation in Arabidopsis (Arabidopsis thaliana), resulted in hypersensitivity to freezing stress in plants with or without cold acclimation, and this defect was successfully complemented by LFR. The expression levels of CBFs and COR genes in cold-treated lfr-1 mutant plants were lower than those in wild-type plants. Furthermore, LFR was found to interact directly with ICE1 in yeast and plants. Consistent with this, LFR was able to directly bind to the promoter region of CBF3, a direct target of ICE1. LFR was also able to bind to ICE1 chromatin and was required for ICE1 transcription. Together, these results demonstrate that LFR interacts directly with ICE1 and activates ICE1 and CBF3 gene expression in response to cold stress. Our work enhances our understanding of the epigenetic regulation of cold responses in plants.
Cold stress profoundly limits the growth, development, and geographical distribution of plants, and it can significantly threaten crop production and quality (Shi et al., 2018; Ding et al., 2020; Ding and Yang, 2022). However, plants have evolved precise cellular and molecular mechanisms to improve their chance of survival under extreme low temperature conditions (Shi et al., 2018; Ding and Yang, 2022). Understanding how plants adapt to and tolerate cold stress by sensing, transducing, and responding to cold signals is paramount for the breeding of climate-resilient crops and has long been a topic of interest (Shi et al., 2018; Chen et al., 2021). The transmembrane protein COLD1 was identified as a sensor that perceives cold signals and forms a complex with G-protein α subunit1 (RGA1) to mediate cold-induced influx of intracellular Ca2+, subsequently leading to the expression of cold-regulated (COR) genes in rice (Oryza sativa) (Ma et al., 2015; Shi et al., 2018). COR genes, including COR15A and Responsive to Desiccation 29, encode proteins that can stabilize membranes against freezing-induced injury (Jaglo-Ottosen et al., 1998; Thomashow, 1999). C-repeat binding factors (CBFs), also known as dehydration-responsive element (DRE)-binding factors (DREBs), are key transcription factors (TFs) that bind to CRT/DRE motifs in the promoters of target COR genes, thus inducing their expression (Stockinger et al., 1997; Gilmour et al., 1998; Jaglo-Ottosen et al., 1998; Liu et al., 1998; Song et al., 2021). CBFs are themselves rapidly upregulated by non-freezing low temperatures (Gilmour et al., 1998). Over the past 20 years, multiple TFs have been shown to regulate CBF expression (Shi et al., 2018). For example, inducer of CBF expression 1 (ICE1) is the one of the well-characterized transcriptional activators of CBF genes (Chinnusamy et al., 2003; Kim et al., 2015; Shi et al., 2018; Ding et al., 2019).
ICE1 is a MYC-like basic helix-loop-helix (bHLH) TF that binds directly to a MYC recognition sequence (CANNTG) in the promoters of CBF genes under conditions of cold stress (Chinnusamy et al., 2003; Kim et al., 2015; Tang et al., 2020). In contrast to CBFs and CORs, ICE1 is a constitutively expressed gene with no obvious response to cold stress at the transcriptional level (Chinnusamy et al., 2003; Shi et al., 2018). However, multiple post-translational modifications, including ubiquitination (Dong et al., 2006), sumoylation (Miura et al., 2007), and phosphorylation (Ding et al., 2015; Li et al., 2017; Zhao et al., 2017; Ye et al., 2019), play fundamental roles in the lifespan and turnover of ICE1 at low temperatures (Shi et al., 2018). Interacting partners that are important for the regulation of ICE1 transcriptional activity toward CBFs have also been reported. For example, MYB15 (Agarwal et al., 2006), JASMONATE ZIM-DOMAIN proteins (Hu et al., 2013), and MYC67 and MYC70 (Ohta et al., 2018) were separately isolated as ICE1 interactors; however, they mainly inhibit CBF gene transcription. Interacting factors that are important for ICE1 to activate downstream CBFs are largely unknown.
ATP-dependent chromatin remodeling complexes (CRCs) activate or inhibit transcription by modifying the architecture of chromatin (Clapier and Cairns, 2009; Ho and Crabtree, 2010; Hargreaves and Crabtree, 2011; Clapier et al., 2017; Yang et al., 2022). For example, the SWItch/Sucrose Non-Fermentable (SWI/SNF) CRC uses energy from ATP hydrolysis to change the structure and position of nucleosomes at target genes to activate or repress transcription; this represents a conserved transcriptional regulatory mechanism across species (Hirschhorn et al., 1992; Peterson and Herskowitz, 1992; Khavari et al., 1993; Chiba et al., 1994; Sarnowska et al., 2016; Ojolo et al., 2018; Shu et al., 2021). In the model plant Arabidopsis (Arabidopsis thaliana), the SWI/SNF CRC is a multi-subunit machine containing four core catalytic ATPases [BRAHMA (BRM), SPLAYED (SYD), Chromatin Remodeling Factor 12/MINUSCULE 1 (CHR12/MINU1), and CHR23/MINU2] (Wagner and Meyerowitz, 2002; Farrona et al., 2004; Bezhani et al., 2007; Sang et al., 2012), a single SNF5 subunit called BUSHY (BSH) (Brzeski et al., 1999), four SWI3 proteins (SWI3A, SWI3B, SWI3C, and SWI3D) (Sarnowski et al., 2005), two SWI/SNF ASSOCIATED PROTEINS 73 (SWP73A and SWP73B) (Jégu et al., 2014; Sacharowski et al., 2015; Jégu et al., 2017), two ACTIN-RELATED PROTEINS (ARP4 and ARP7) (Kandasamy et al., 2003), and a single Leaf and Flower Related (LFR) protein (Wang et al., 2009; Lin et al., 2021). In addition, two homologous BRM‐interacting proteins (BRIP1 and BRIP2) (Yu et al., 2020), three bromodomain‐containing proteins (BRD1, BRD2, and BRD13) (Jarończyk et al., 2021; Yu et al., 2021), ANGUSTIFOLIA3 (Vercruyssen et al., 2014), and two TRIPLE PHD FINGERS proteins (TPF1 and TFP2) (Diego-Martin et al., 2022) have been identified as components of the SWI/SNF CRC.
Components of the SWI/SNF CRC are involved in various biological processes, including plant development, phase transitions, DNA methylation, transposon silencing, pre-miRNA processing, and DNA double-strand breaks (Sarnowska et al., 2016; Ojolo et al., 2018; Thouly et al., 2020). The molecular mechanisms of BRM or SWI3B were unraveled in abscisic acid pathway (Saez et al., 2008; Han et al., 2012; Peirats-Llobet et al., 2016) and environmental stress response, such as salt stress (Vicente et al., 2017) and heat-shock (HS) stress (Brzezinka et al., 2016). Recently, SWI3C was reported to bind and alter the nucleosome occupancy and expression of key factors in cold acclimation, including ICE1, MYB15, and CBF1, under low temperature conditions, revealing a role for SWI/SNF components in cold acclimation (Gratkowska-Zmuda et al., 2020). However, which TFs play important roles in recruiting SWI/SNF complexes to target genes during cold acclimation is unknown.
The LFR gene is essential for leaf and flower development in Arabidopsis (Wang et al., 2012; Lin et al., 2018; Lin et al., 2021). LFR encodes an Armadillo (ARM)-repeat domain-containing protein that is predicted to be a subunit of the SWI/SNF CRC (Wang et al., 2009; Lin et al., 2021; Diego-Martin et al., 2022; Shang and He, 2022). In this study, we found that the disruption of LFR led to reduced freezing tolerance in Arabidopsis. We demonstrated that LFR interacts directly with ICE1 in yeast and plant. Meanwhile, chromatin immunoprecipitation (ChIP)-qPCR and Real Time Quantitative PCR (RT-qPCR) assays demonstrated that LFR can directly bind to and activate the transcription of CBF3, an ICE1 target gene, and, in turn, the COR15A gene. Moreover, ICE1 is a direct target of LFR and LFR is required for ICE1 expression. Together, our results show that LFR directly interacts with ICE1 and activates CBFs expression, revealing the molecular mechanism of the SWI/SNF CRC during cold acclimation in plants.
The wild-type (WT) and mutant Arabidopsis plants used in this study were of the Columbia-0 (Col-0) background. The lfr-1 and lfr-2/+ mutants are described elsewhere (Wang et al., 2009). ice1-2 (SALK_003155; Kanaoka et al., 2008) was a gift from Prof. Shuhua Yang’s laboratory. Seeds were surface-sterilized in 75% ethanol and 1% NaClO, washed three times with sterile water, plated on half-strength Murashige and Skoog (1/2 MS) medium with 1% sucrose and 0.36% agar, and stratified at 4°C in the dark for 3 days. All plants were grown under long-day conditions (16 h of light/8 h of dark) at 22°C.
To map the domains of ICE1 (AT3G26744) that are involved in its interaction with LFR, full-length and truncated versions of ICE1 were fused to the GAL4 binding domain (BD) in pGBKT7. Meanwhile, LFR was fused to the GAL4 activation domain (AD) in pGADT7 (Lin et al., 2018). For our Bimolecular Fluorescence Complementation (BiFC) and firefly luciferase (LUC) complementation imaging (LCI) experiments, the full-length coding sequence (CDS) of ICE1 and LFR was amplified by PCR from Col-0 cDNA, cloned into pENTRY/D/SD-TOPO (Takara Bio Inc., Shiga, Japan), and then fused with the C-terminal half of cyan fluorescent protein (CFP) (cCFP), the C-terminal half of LUC (cLUC), the N-terminal half of yellow fluorescent protein (YFP) (nYFP), or the N-terminal half of LUC (nLUC). The full-length CDS of LFR was fused to nYFP (Lin et al., 2018).
To obtain the construct of pLFR : LFR-enhancedYFP (eYFP) and pLFR : LFR-3FLAG, the native promoter of LFR (1600 bp), the genomic sequence of LFR, and the coding sequences of eYFP and 3×FLAG were amplified and digested using appropriate restriction endonucleases and subsequently cloned into pCAMBIA1300. After being verified by DNA sequencing, the plasmid was introduced into lfr-1 and/or lfr-2/+ by Agrobacterium tumefaciens-mediated floral dip transformation (Clough and Bent, 1998). The homologous transgenic plants of pLFR : LFR-eYFP in lfr-2 background (pLFR : LFR-eYFP/lfr-2) and pLFR : LFR-3FLAG in lfr-1 and lfr-2 background (pLFR : LFR-3FLAG/lfr-1 and pLFR : LFR-3FLAG/lfr-2) were identified in the T4 generation and used for further phenotypic observation.
Total RNA was extracted from WT and mutant plants using Trizol reagent (Takara). First-strand cDNA was prepared from 500 ng of total RNA using a PrimeScipt RT Reagent Kit (Takara) and quantified using a CFX96™ Connect Real-Time PCR System (Bio-Rad, Hercules, CA, USA) with a SYBR Green Kit (Takara). The eIF4A1 gene was used as an internal control. The primers used are listed in Supplementary Table S1.
A freezing tolerance assay was performed as described previously (Kim et al., 2015). Approximately 36 plants grown on 1/2 MS medium for 10 days were subjected to freezing conditions; the temperature was set to decrease 1°C/h until it reached -6°C (seedlings did not undergo cold acclimation before freezing) or -9°C (seedlings underwent cold acclimation at 4°C for 4 days before freezing). The plants were then incubated at 4°C overnight in the dark. Next, the plants were allowed to recover for 2 days at 22°C.
The chlorophyll content measurement was performed as described previously (Arnon, 1949; Mostofa et al., 2015). After freezing treatment, the seedlings were allowed to recover for three days at 22°C. Approximately 0.5 g of seedlings from a pool of plants from different plates were dipped in 50 mL aqueous acetone (80%) at 4°C in the dark for 48 h. The light absorption by acetone extracts of chlorophyll at 663 nm and 645 nm are detected in a spectrophotometer. The chlorophyll content was calculated as (20.2 × D645 + 8.02 × D663) × V/(1000 × W); where V = volume of 80% (v/v) acetone (mL), W = weight of sample (g).
Y2H assays were carried out using the yeast GAL4 system following a standard protocol (Clontech Laboratories, Inc., Mountain View, CA, USA). To verify its interaction with LFR, truncated versions of ICE1 [ICE1-2 (the C-terminal region of ICE1, corresponding to amino acids 298–494) and ICE1-3 (the C-terminal region of ICE1, corresponding to amino acids 358–494)] were cloned into pGBKT7 and LFR was fused to the AD in pGADT7. Following verification of the constructs by DNA sequencing, pairs of the plasmids were co-transformed into yeast strain AH109, followed by incubation at 30°C for 3 days. The co-transformed yeast cells were grown on solid medium lacking leucine (Leu) and tryptophan (Trp) (SD/-L-W). Only when the two proteins interacted with each other could the co-transformed yeast cells grow on solid medium lacking Leu, Trp, histidine (His), and adenine (Ade) (SD/-L-W-H-A).
BiFC and LCI assays were performed using transiently transformed tobacco leaves (Chen et al., 2008; Ou et al., 2011). For the BiFC assay, cCFP-ICE1 and nYFP-LFR constructs were co-transformed and co-expressed for about 48 h in tobacco epidermal cells. Fluorescent green fluorescent protein (GFP) (excitation 488 nm; emission 500–550 nm) and 4’,6-diamidino-2-phenylindole (DAPI) (excitation 405 nm; emission 430–480) signals were then detected using a Zeiss 710 Confocal Microscope (Carl Zeiss AG, Jena, Germany). For the LCI assay, tobacco leaves co-expressing the cLUC-ICE1 and LFR-nLUC constructs were incubated at 22°C for about 48 h. Then the luciferase substrate (1 mM D-luciferin) was sprayed onto the surface of the leaves and images were taken using a low-light cooled imaging apparatus (Fusion FX7; Vilber, Marne-la-Valle, France). The LUC activity was analyzed at 48 hpi using the NEWTONZ.0 Bio.Plant Imaging System (Vilber Bio lmaging, France).
ChIP was performed as described previously (Yamaguchi et al., 2014). Briefly, 600 mg of 8-day-old seedlings were cross-linked using 1% formaldehyde; the reaction was stopped by the addition of 0.125 M glycine. The cross-linked chromatin was then sheared to an average size of ~500 bp, and 20 μL of the supernatant was used as the input control. Anti-FLAG antibodies (F3165; Sigma-Aldrich, St. Louis, MO, USA) were added to the remaining chromatin for immunoprecipitation followed by qPCR. Data are presented as the fold change relative to the wild type. The primers used are shown in Supplementary Table S1.
Primary roots from 8-day-old pLFR : LFR-enhancedYFP (eYFP)/lfr-2 transgenic seedlings with or without cold treatment were stained with 10 μg/ml of propidium iodide (PI) for 5 min and then imaged using an Olympus FV 3000 Confocal Microscope (Olympus Corp., Tokyo, Japan) with the following settings: equal laser power, gain, and pinhole. Line scan analyses of the YFP and PI signals in the root tip were measured using Image J. For the relative eYFP fluorescence signals quantification, all the eYFP fluorescence signals in each picture were quantified by image J. Twenty roots were measured for each genotype. The signals for cold-treated roots are analyzed relative to the average fluorescence intensity of at time 0 h. Data were analyzed using Prism 8.0.
To determine the function of the SWI/SNF component LFR in freezing tolerance, we evaluate the freezing tolerance phenotype of 10-day-old wild-type (WT, Col-0) and lfr-1 seedlings grown on 1/2 MS medium under non-acclimated (NA) or cold-acclimated (CA) conditions. The NA seedlings were subjected to freezing stress at -6°C for 1 h. Under this condition, the lfr-1 mutant showed reduced freezing tolerance with a significantly lower survival rate and chlorophyll content compared to WT after freezing treatment (Figures 1A–C). This defect was successfully rescued in pLFR : LFR-3FLAG/lfr-1 transgenic lines, which showed no obvious difference compared to the WT control (Figures 1A–C). The CA seedlings were acclimated at 4°C for 4 days and then subjected to freezing stress at -9°C for 1 h. The lfr-1 mutant also displayed freezing-sensitive defects under CA conditions (Figures 1D–F), and the defects were complemented in pLFR : LFR-3FLAG/lfr-1 transgenic lines (Figures 1D–F). Thus, LFR is essential for freezing tolerance at the seedling stage under both CA and NA conditions.
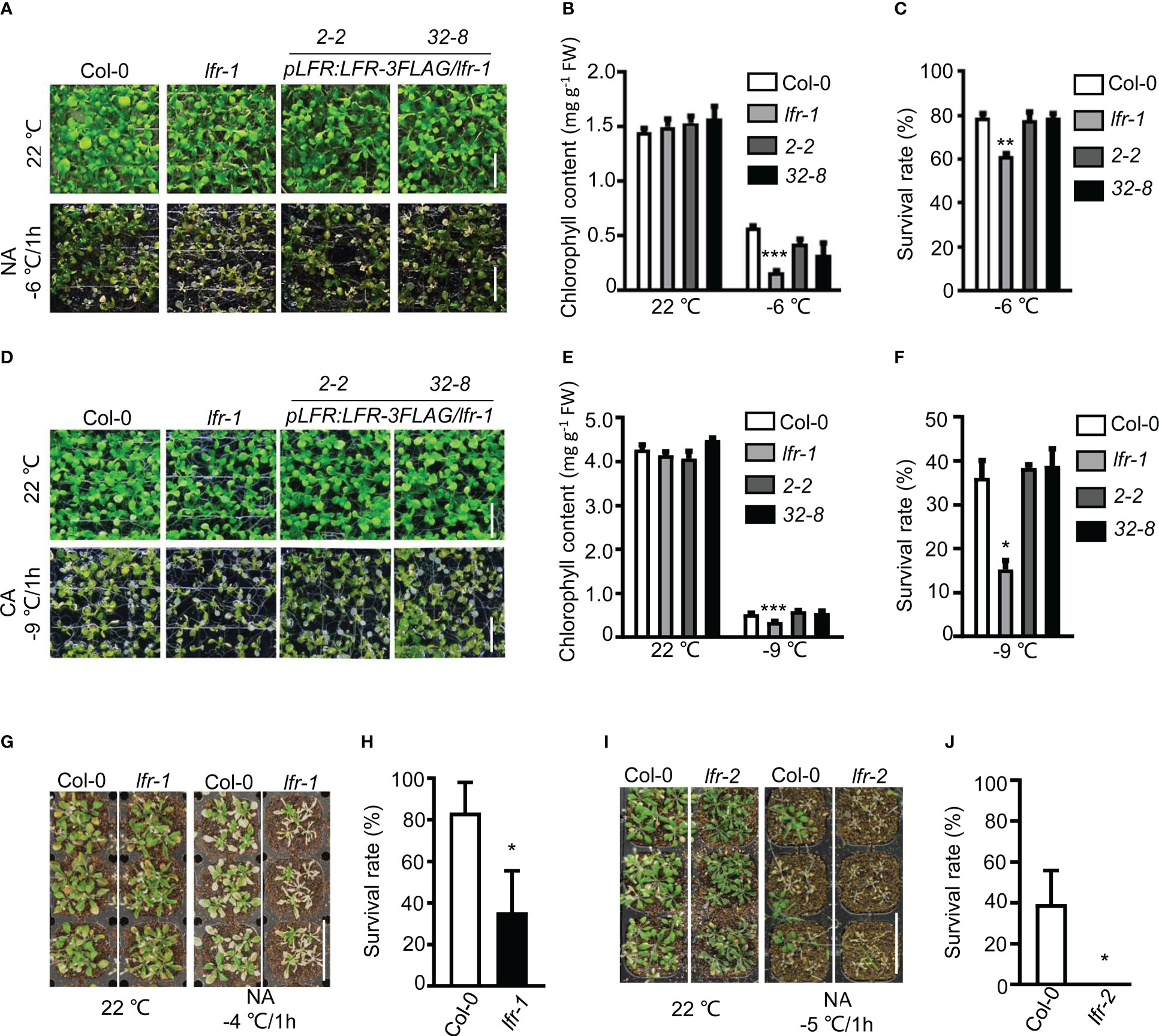
Figure 1 Phenotype analysis of lfr mutant plants and transgenic rescue lines exposed to freezing stress. (A, D) The lfr-1 mutant is sensitive to freezing stress. Representative pictures of 10-day-old seedlings of Col-0, lfr-1, and two complementary lines of pLFR : LFR-3FLAG/lfr-1 (2-2 and 32-8) were subjected to freezing stress under NA (A) or CA (D) conditions. After freezing stress, seedlings were incubated at 4°C overnight in the dark and then were allowed to recover for 2 days at 22°C. Scale bar=1 cm. (B, E) The chlorophyll contents of Col-0, lfr-1, and two complemented transgenic lines (2-2 and 32-8) treated with or without freezing stress under NA (B) and CA (E) conditions are shown. (C, F) The survival rates of Col-0, lfr-1, and two complemented transgenic lines (2-2 and 32-8) after being subjected to freezing stress under NA (C) or CA (F) conditions are shown. (G) and (I) Representative examples of the reduced freezing tolerance observed in soil-grown lfr-1 (G) and lfr-2 (I) plants. Scale bar=2 cm. (H, J) The survival rates of soil-grown Col-0 and lfr-1 (H) and Col-0 and lfr-2 (J) plants that were subjected to freezing stress (-4 or -5°C) and then allowed to recover are shown. Error bars indicate the SD of three biological replicates. Statistically significant differences are indicated by asterisks (*P<0.05, **P < 0.01, and ***P < 0.001; Student’s t-test).
To confirm the function of LFR in freezing tolerance, we subjected 3-week-old lfr-1 and lfr-2 (a strong sterile allele of LFR, segregated from lfr-2+/-) mutant plants to freezing stress in soil. The mutants were subjected to freezing stress at -4 or -5°C for 1 h and then returned to a growth chamber set at 22°C for 7 days. In terms of survival, lfr-1 and lfr-2 both displayed a freezing-sensitive phenotype (Figures 1G–J).
Taken together, these results indicate that LFR is a positive regulator of freezing tolerance in Arabidopsis.
ICE1, CBFs, and downstream COR genes are important players in the response of plants to cold stress; thus, we next assessed the transcript levels of ICE1, CBF1, CBF2, CBF3, and COR15A in plants following low-temperature (4°C) treatment for 3 hours (3 h). Real time RT-qPCR showed that the expression levels of CBF1, CBF2, CBF3, and COR15A were significantly increased by cold treatment (Figures 2A–D). However, the expression levels of CBF1, CBF2, and CBF3 were significantly reduced in lfr-1 compared to Col-0 after cold treatment (Figures 2A–C). Similarly, the transcript level of COR15A was lower in lfr-1 than in Col-0 after low-temperature treatment (Figure 2D). We also tested the expression levels of these four genes in pLFR : LFR-3FLAG/lfr-1 complemented transgenic line (2-2), and we found that their expression levels were fully or partly rescued in the lfr-1 mutant (Figures 2A–D). We also examined the expression level of ICE1, and found that it was not regulated by cold stress as reported previously (Figure 2E; Chinnusamy et al., 2003; Shi et al., 2018) ICE1 was downregulated in lfr-1 following cold treatment, and the downregulation of ICE1 was partially rescued in complemented transgenic lines (Figure 2E). These results suggest that LFR is involved in cold acclimation via the direct or indirect transcriptional regulation of ICE1, CBFs, and COR15A.
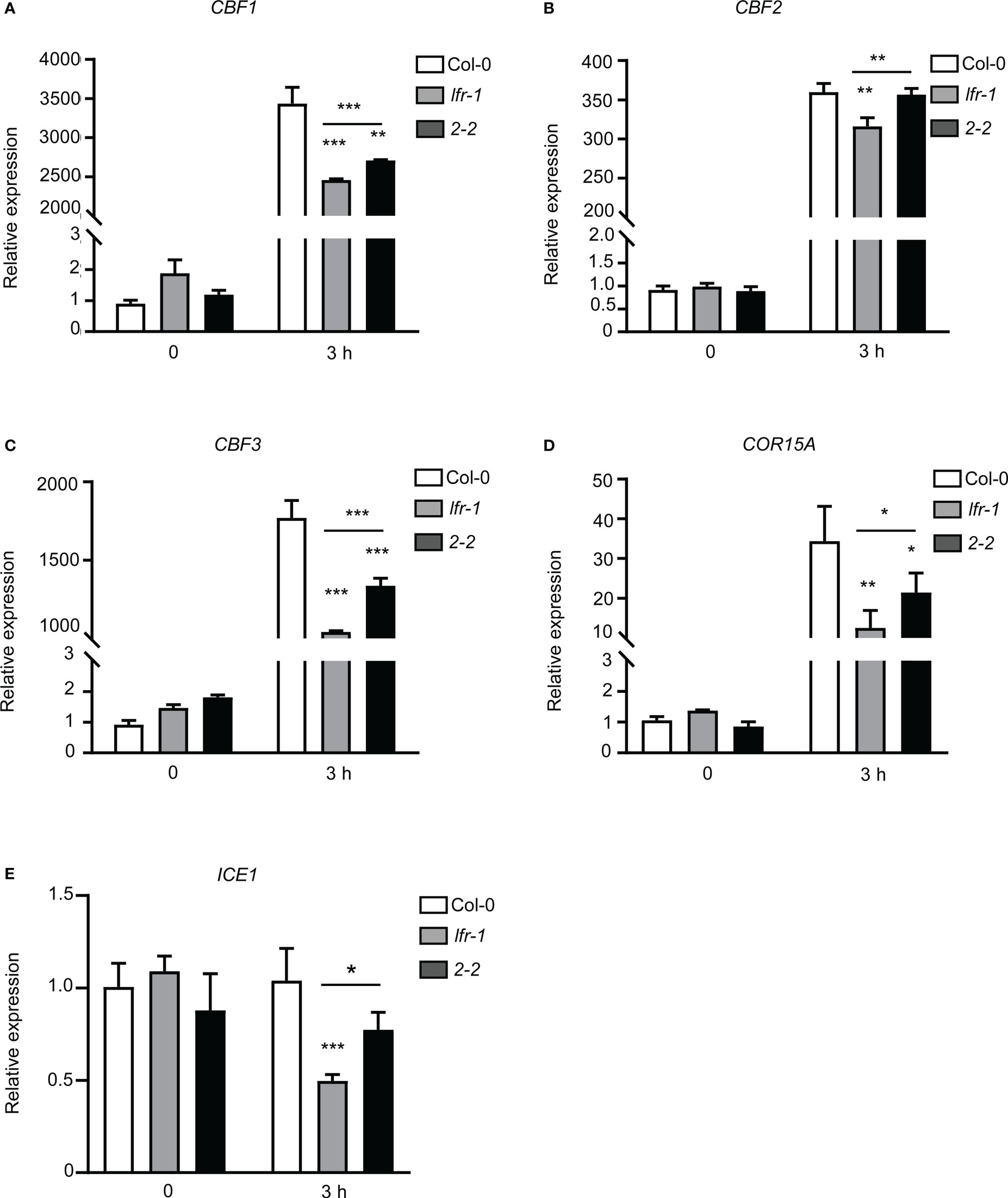
Figure 2 RT-qPCR analysis of CBFs and COR genes expression. (A–E) RT-qPCR data showing the transcript levels of CBF1 (A), CBF2 (B), CBF3 (C), COR15A (D), and ICE1 (E) in Col-0, lfr-1, and complemented transgenic line (2-2). Eight-day-old seedlings were treated at 4°C for the indicated time periods. Error bars indicate the SD of three biological replicates. Statistically significant differences are indicated by asterisks (*P < 0.05, **P < 0.01, and ***P < 0.001; Student’s t-test). eIF4A1 was used as an internal control.
To uncover the molecular mechanism underlying the role of LFR in cold acclimation, we examined Yeast two-hybrid (Y2H) data from a previous study (Lin et al., 2021). Interestingly, we noticed that ICE1 was isolated as an interacting partner of LFR. We confirmed this interaction by Y2H, Bimolecular Fluorescence Complementation (BiFC), and firefly luciferase (LUC) complementation imaging (LCI) assays. Because full-length LFR and ICE1 fused to the GAL4 BD showed obvious autoactivation in yeast (Ohta et al., 2018; Lin et al., 2021), truncated ICE1-2 (including the bHLH and ZIP domains) and ICE1-3 (including the ZIP domain) were fused to the BD to produce BD-ICE1-2 and BD-ICE1-3 (Figure 3A). We then co-transformed yeast strain AH109 with the AD-LFR and BD-ICE1-2 or BD-ICE1-3 plasmids. Yeast colonies co-transformed with AD-LFR/BD-ICE1-2 or AD-LFR/BD-ICE1-3 grew well on selective medium (SD/-L-W-H-A) (Figure 3B). However, negative control cells did not grow on the selective medium under the same conditions (Figure 3B). Thus, ICE1 probably interacts with LFR through its C-terminal ZIP domain.
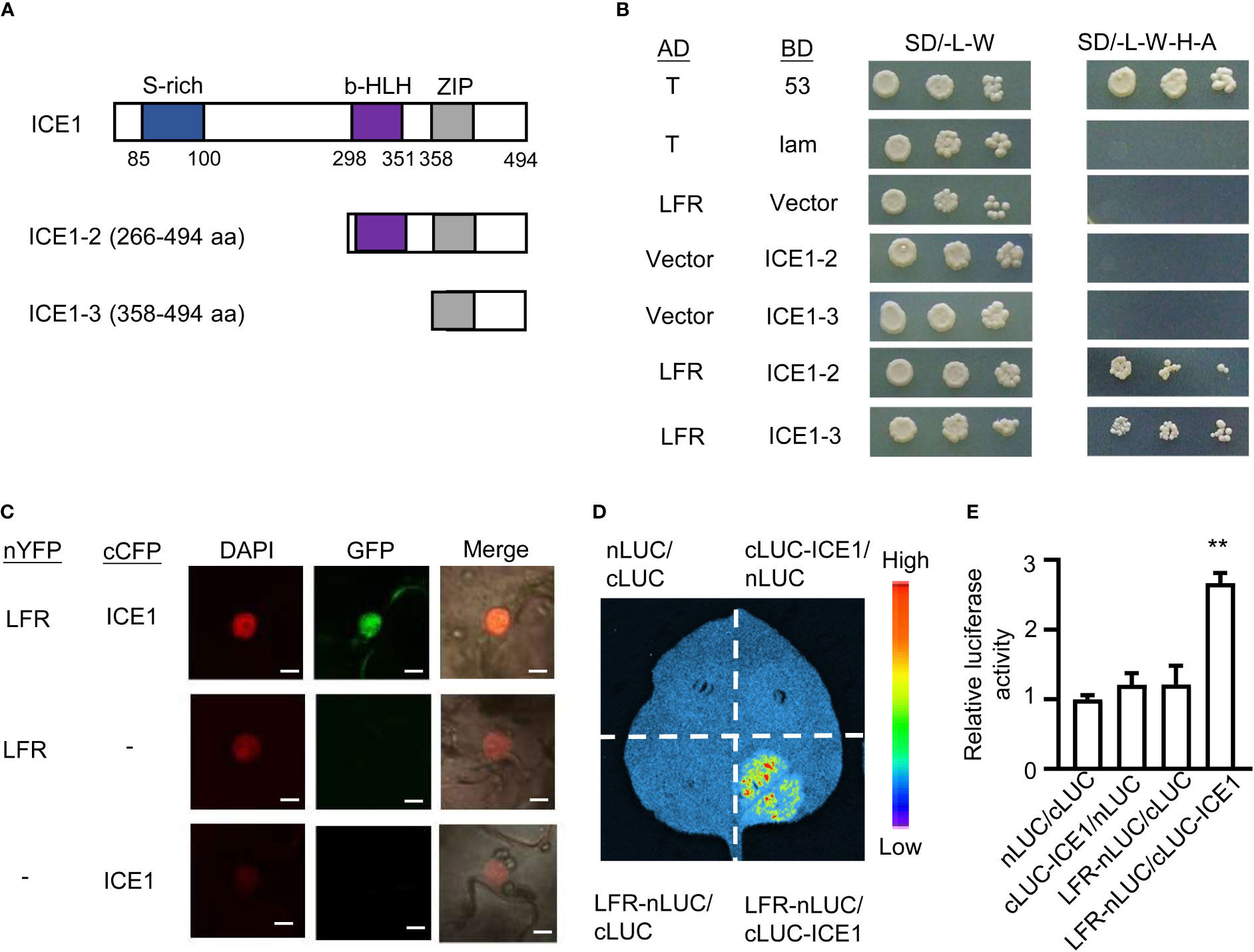
Figure 3 The interaction of LFR with ICE1. (A) Diagrams of the truncated ICE1 proteins used in the Y2H assays shown in (B). ICE1-2 and ICE1-3 include the bHLH and ZIP (amino acids 298–494) and ZIP domains (amino acids 358–494), respectively. (B) LFR interacted with ICE1 in the Y2H assay. The assays were carried out using AH109 yeast cells co-transformed with the indicated plasmid pairs (serially diluted 10-1, 10-2, and 10-3) on synthetic defined medium lacking Ade, His, Leu, and Trp (SD/-L-W-H-A). AD-T/BD-53 and AD-T/BD-Lam were used as positive and negative controls, respectively. (C) nYFP-LFR interacted with cCFP-ICE1 in a BiFC assay carried out in transiently transformed tobacco leaf epidermal cells. Leaves co-transformed with nYFP-LFR/cCFP or nYFP/cCFP-ICE1 served as a negative control. DAPI and GFP fluorescence indicate the nucleus and protein interaction, respectively. Merge corresponds to an overlay of the DAPI and GFP signals. Bar =5 μm. (D) LCI assays were used to test for the interaction of LFR with ICE1 in transiently transformed tobacco leaf epidermal cells. Light emission was observed from tobacco leaves co-expressing LFR-nLUC/cLUC-ICE1, but not from leaves co-expressing the negative controls (LFR-nLUC/cLUC or cLUC-ICE1/nLUC or cLUC/nLUC). (E) The Relative luciferase activity of LCI assay in (D). Error bars indicate the SD of four biological replicates. Statistically significant differences from the nLUC/cLUC control are indicated by asterisks (**P < 0.01, Student’s t-test).
We next conducted a BiFC assay using transiently transformed tobacco (Nicotiana benthamiana) leaves. GFP signals were detected in epidermal cell nuclei co-expressing nYFP-LFR and cCFP-ICE1; however, no GFP signal was detected in negative control cells co-expressing nYFP-LFR/cCFP or nYFP/cCFP-ICE1 (Figure 3C). These results indicate that LFR interacts with ICE1 in plant.
We next verified the LFR–ICE1 interaction using an LCI assay. Tobacco leaf areas co-transformed with LFR-nLUC and cLUC-ICE1 emitted light in the presence of luciferin. However, tobacco leaf areas co-transformed with the negative controls (LFR-nLUC/cLUC or cLUC-ICE1/nLUC or cLUC/nLUC) showed no light emission in the same condition (Figures 3D, E). These results confirm that LFR interacts with ICE1 in plant.
Given that CBF3 is a direct target of ICE1 (Ding et al., 2015; Kim et al., 2015; Tang et al., 2020) and LFR interacts directly with ICE1, we speculated that LFR may also bind to the CBF3 gene. ChIP-qPCR assays of pLFR : LFR-3FLAG/lfr-2 complemented transgenic lines were conducted using anti-FLAG antibodies with Col-0 as a negative control. According to a previous report on the ICE1-binding site of CBF3 (Ding et al., 2015), primers for the amplification of three different fragments (P1–P3) of CBF3 chromatin were designed (Figure 4A). Under normal growth condition, the ChIP-qPCR result showed that LFR binds to CBF3 at the P1 and P2 loci (Figure 4B). After low-temperature treatment, the ChIP-qPCR result showed that LFR binds to CBF3 chromatin at the P2 locus (Figure 4C). The P1 and P2 loci of CBF3 are also binding sites of ICE1 (Ding et al., 2015; Kim et al., 2015; Tang et al., 2020). Thus, these results demonstrated that CBF3 is a direct target of LFR under normal and cold stress conditions.
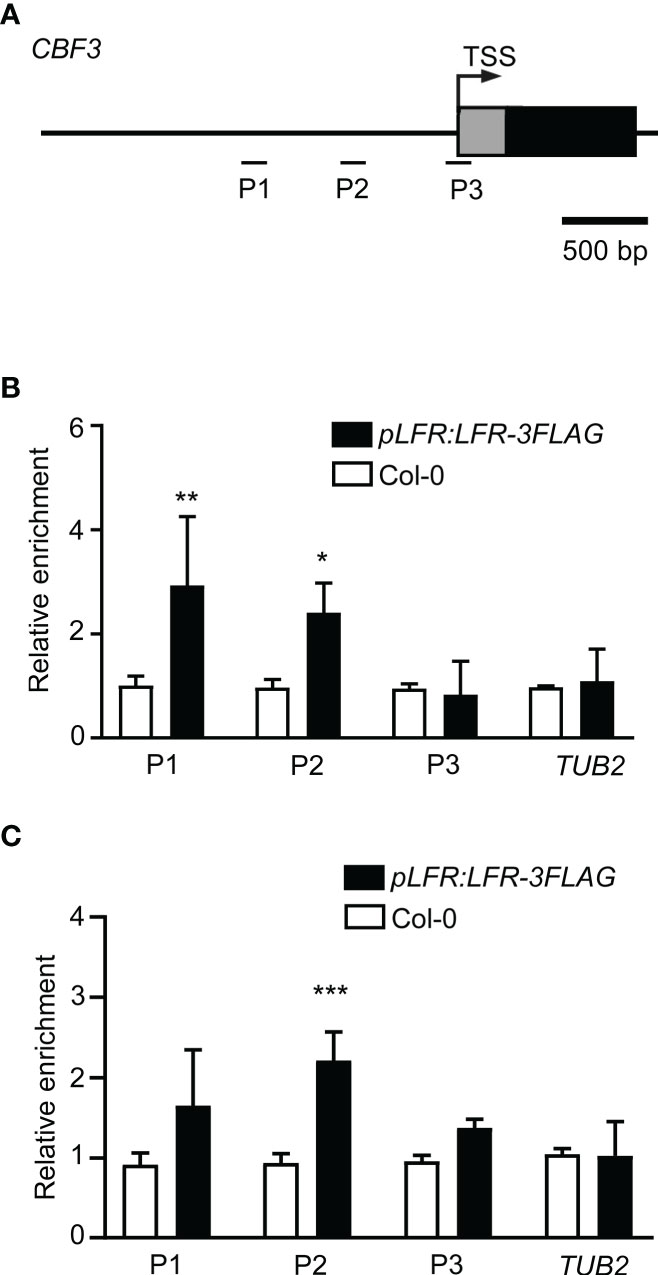
Figure 4 LFR binds to CBF3 chromatin. (A) The gene structure of CBF3. Black boxes indicate exons; lines represent the promoter or intergenic region. Fragments amplified in the ChIP-qPCR assay are marked by short lines below the gene model. TSS, transcription start site. (B, C) ChIP-qPCR assay of LFR binding to CBF3 without (B) and with low-temperature treatment (C). Error bars indicate the SD of three biological replicates. *P < 0.05, **P < 0.01, ***P<0.001, Student’s t-test.
Because we detected reduced expression of ICE1 in lfr-1 plants, we tested whether ICE1 is also a direct target of LFR. A ChIP-qPCR assay was performed using Col-0 and the pLFR : LFR-3FLAG/lfr-2 complemented transgenic line with anti-FLAG antibodies. Different primers for the amplification of different fragments of the ICE1 locus were designed (Figure 5A). The ChIP-qPCR results showed that LFR was able to bind directly to the P2 locus of ICE1 at 22°C (Figure 5B). After cold stress treatment, LFR was associated with the P3 locus of ICE1 (Figure 5C). Thus, these results demonstrated that ICE1 is also a direct target of LFR with or without low-temperature treatment.
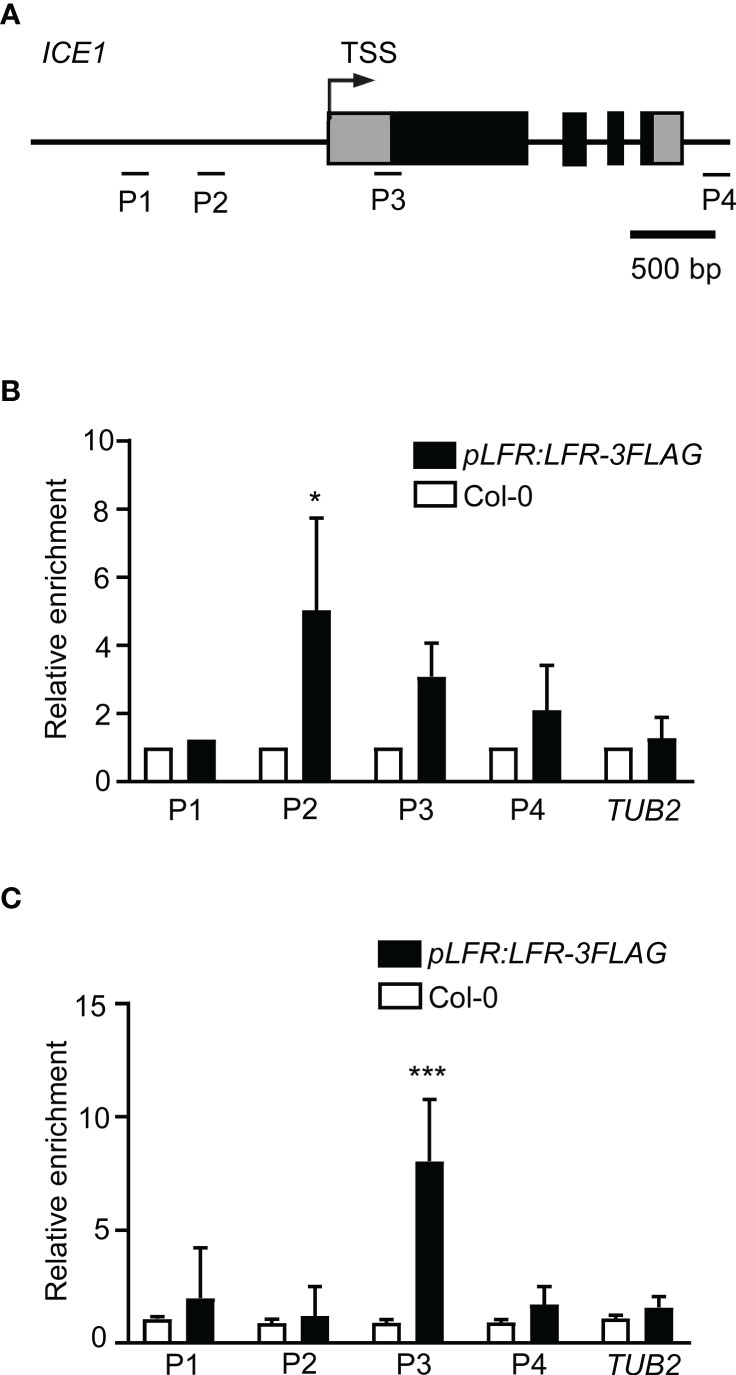
Figure 5 LFR binds to ICE1 chromatin. (A) The gene structure of ICE1. Black boxes indicate exons; lines represent the promoter or introns. Fragments amplified in the ChIP-qPCR assay are marked by short lines below the gene model. TSS, transcription start site. (B, C) ChIP-qPCR assay of LFR binding to ICE1 without (B) and with 4°C treatments (C). Error bars indicate the SD of two (4°C) or three (22°C) biological replicates. *P < 0.05, ***P < 0.001, Student’s t-test.
To elucidate whether LFR is regulated by cold tress at the transcriptional level, we analyzed cis-regulatory elements in the LFR promoter using PLANTCARE. Five low temperature-responsive element (CANNTG) bound by ICE1 was found in the LFR promoter, indicating that LFR may be regulated by ICE1 or a cold-responsive gene (Figure 6A). And we indeed detected significant downregulation of LFR in ice1-2 mutant plant (Supplementary Figure S1A). And we also checked the transcript level of LFR in plants treated with or without cold exposure at different time points. RT-qPCR showed that LFR was downregulated by cold treatment (Figure 6B). We also studied the protein level of LFR using pLFR : LFR-eYFP/lfr-2 complemented lines. Furthermore, when the pLFR : LFR-eYFP/lfr-2 lines were held at 4°C, the eYFP signals were reduced, especially after 6 and 24 h of cold stress (Figures 6C–E). Taken together, these results indicate that LFR may be repressed by cold stress.
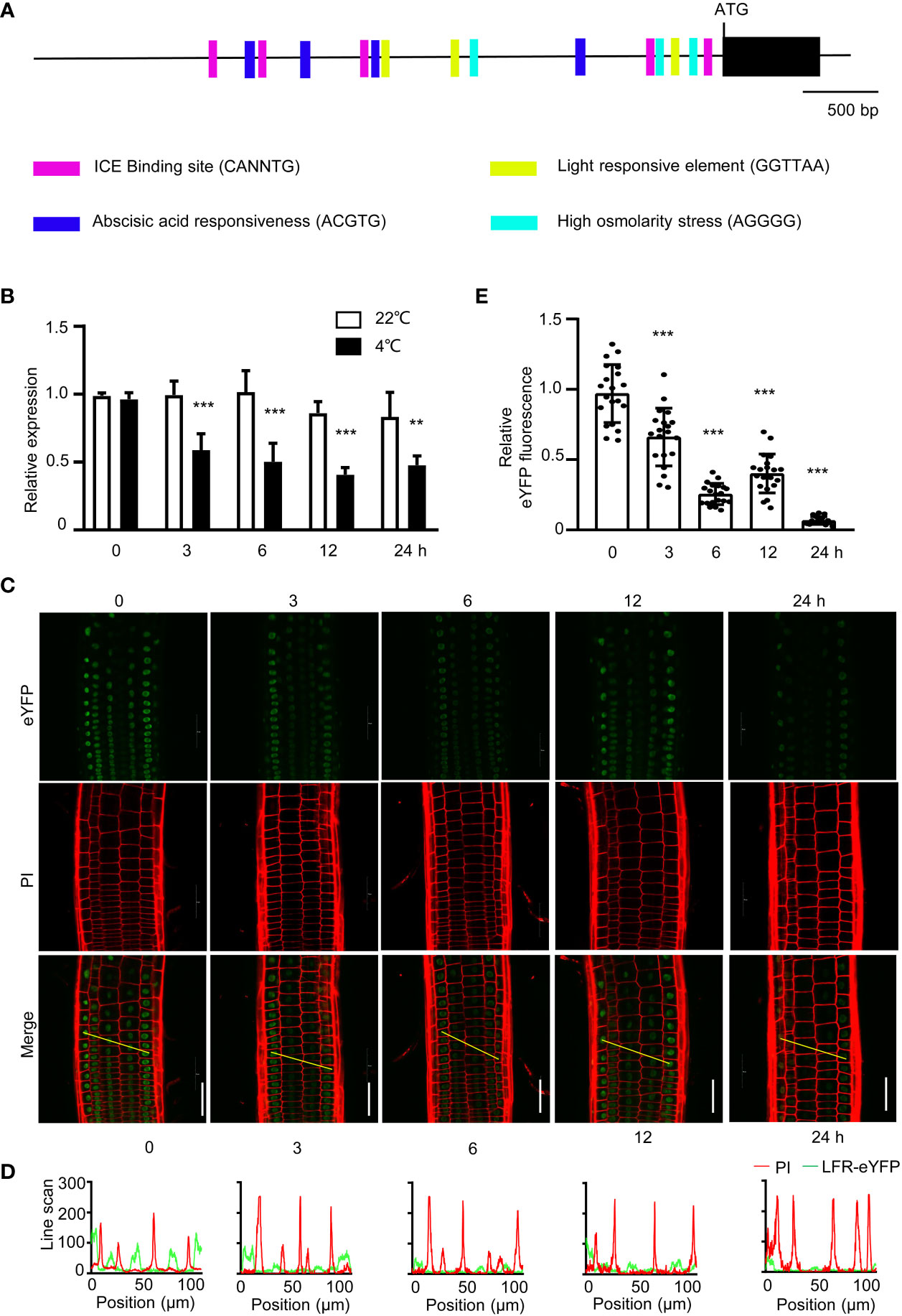
Figure 6 The expression of LFR are regulated by cold stress. (A) Cis-element analysis of the LFR promoter. Different cis-elements are indicated by different symbols. CANNTG is the low temperature-responsive element found in the ICE1 binding site. (B) RT- qPCR data showing the transcript level of LFR with (4°C) or without (22°C) cold treatment. RNA was isolated from 8-day-old Col-0 seedlings with or without cold treatment for 0, 3, 6, 12, or 24 h. eLF4A1 served as an internal control. Error bars indicate the SD of three biological replicates. Statistically significant differences are indicated by asterisks. **P<0.01 and ***P<0.001, Student’s t-test. (C) Representative YFP signals observed in the roots of 8-day-old seedlings of pLFR : LFR-eYFP/lfr-2 complemented transgenic lines as detected by confocal microscopy with (4°C, for 3, 6, 12, or 24 h) or without (0 h) cold treatment are shown. PI staining indicates the cell membrane. Scale bar= 50 μm. (D) Line scan analyses of the YFP (green line) and PI (red line) signals corresponding to the yellow lines indicated in (C) (Merge). YFP signals were significantly reduced after cold treatment. (E) Quantification of the YFP fluorescence in (C). Representative data from one of three biological replicates with similar results are shown. Data are shown as the mean ± SD (n=20). ***P < 0.001; Student’s t-test.
Plants have developed sophisticated mechanisms to survive fluctuating ambient temperatures (Shi et al., 2018; Chen et al., 2021). At chilling or freezing temperatures, plants utilize a cascade of TFs to adapt to low-temperature stress (Shi et al., 2018). Among these TFs, ICE1, a constitutively expressed MYC-type TF, activates the expression of CBFs, which subsequently induce the transcription of downstream cold-responsive genes such as CORs, which promote cold tolerance (i.e., the ICE1–CBF–COR cascade) (Shi et al., 2018). However, the interacting factors that are needed for ICE1 to activate CBFs are largely unknown. In this study, we demonstrated that ICE1 is an LFR-interacting partner using Y2H, BiFC, and LCI assays (Figure 3). Disruption of LFR resulted in reduced cold tolerance under both NA and CA conditions, similar to ice1-2 (Figure 1). In addition, the transcript levels of ICE1–CBF–COR cascade TF genes (e.g., ICE1, CBF1, CBF2, CBF3, and COR15A) were all downregulated after cold treatment in lfr plants (Figure 2). Like ICE1, LFR was able to directly bind to the CBF3 locus (Figure 4). LFR was also found to associate directly with ICE1 chromatin (Figure 5). Together, our results demonstrate that LFR directly interacts with ICE1 and positively regulates the transcription of genes in the ICE1–CBF–COR signaling pathway.
Interestingly, we also detected significant downregulation of LFR in ice1-2 mutant plants, and we found ICE1-binding sites (i.e., CANNTG) in the proximal region of the LFR promoter (Figure 6 and Supplementary Figure S1A). Furthermore, using published ChIP-seq data for GFP-ICE1 (Tang et al., 2020), we found that ICE1 was enriched at the proximal promoter region and gene body of LFR and ICE1 chromatin (Supplementary Figure S1; Tang et al., 2020). Together with our data showing that ICE1 is not only an interacting factor but also a target of LFR, which tends to bind target(s) via interaction with transcription factor (Lin et al., 2018), these results may indicate that LFR and ICE1 interact physically and form transcriptional feedback loops. It will be interesting to test this possibility in the future.
Although LFR may act as a positive regulator of cold signaling, we found that LFR was repressed after prolonged cold treatment (Figures 6). This is similar to ICE1, which is degraded during long-term cold treatment (Dong et al., 2006; Ding et al., 2015; Zhao et al., 2017). The cold-induced downregulation of LFR and ICE1 may help balance plant cold responses and growth by preventing detrimental effects on plant growth resulting from the over-stimulation of critical freezing tolerance factors (e.g., CBFs and CORs) (Zhao et al., 2017).
In addition, we found that the lfr-1 ice1-2 double mutant showed obvious defects in seed germination and greening (Supplementary Figure S2), which may indicate that LFR and ICE1 may also participate in these developmental processes.
Epigenetic factors play critical roles in long-term adaptation to the ambient environment (Miryeganeh, 2021; Chung et al., 2022). However, the exact functions of different CRC components in cold adaptation are unknown. Recently, Gratkowska-Zmuda et al. (2020) provided solid evidence showing that SWI3C of the SWI/SNF complex can directly bind, change the nucleosome occupancy and influence the transcription of ICE1, MYB15, and CBF1 in a temperature-dependent manner. SWI3C was required to activate ICE1 expression at 22°C but not at 14°C, and it repressed the transcription of all three CBF genes. Consistent with the upregulation of CBFs, the swi3c mutant displayed increased freezing tolerance compared to wild type (Gratkowska-Zmuda et al., 2020). However, in this study, we found that ICE1–CBF–COR cascade genes were downregulated in the lfr mutant, which displayed hypersensitivity to freezing. Our previous study showed that LFR interacts with SWI3A/SWI3B, but not SWI3C (Lin et al., 2021). Consistently, LFR together with SWI3A/SWI3B, not SWI3C, were identified in the immunoprecipitation mass spectrometry (IP-MS) data of a recent study of the MINU-associated SWI/SNF (MAS) complex (Diego-Martin et al., 2022; Hernández-García et al., 2022). In addition, a comprehensive characterization of the SWI/SNF complexes by proteomic approaches revealed that SWI3C and LFR are involved in different complexes with specific chromatin accessibility and binding ability for active histone modification (Guo et al., 2022). Thus, the opposite phenotype and differential expression pattern of CBF genes may result from that SWI3C and LFR participate in distinct SWI/SNF complexes with different transcriptional properties. So, it will be interesting to decipher the detailed molecular and genetic interactions between LFR, SWI3C, and the core ATPases of the SWI/SNF complex (BRM, SYD, MINU1, and MINU2), which is of great importance to elucidate the molecular regulatory mechanisms of SWI/SNF complexes in controlling plant response to cold treatment.
Arabidopsis LFR encodes an ARM-repeat domain-containing protein (Wang et al., 2009). Phylogenetic analysis of the ARM domain revealed that plant LFR has a common origin with the signature subunits ARID1/2 of the animal SWI/SNF complex (Hernández-García et al., 2022). Recently, the structural approaches demonstrated that the ARM domain in ARID1 serves as a rigid core in the “Base/Core module” organization and stabilization by directly interacting with the core ATPase and all other Base subunits (He et al., 2020; Mashtalir et al., 2020). Besides, ARID1 is also essential for the efficient nucleosome sliding activity of the SWI/SNF complex (He et al., 2020). However, the molecular weight of Arabidopsis LFR is much lower than its mammalian orthologs, because of the loss of the DNA-interacting AT-rich interaction domain (ARID) (Wang et al., 2009; Hernández-García et al., 2022). Thus, it will be interesting to decipher the biological contribution of plant LFR, to the complex organization, architecture, stability, and nucleosome sliding activity of target genes in plant development and stress in the future.
Taken together, our results demonstrate that the SWI/SNF complex component LFR interacts physically with ICE1 to act as a positive regulator of the ICE1–CBF–COR cascade (Figure 7). This study will aid our understanding of plant-specific epigenetic changes through chromatin remodeling in response to global climate change.
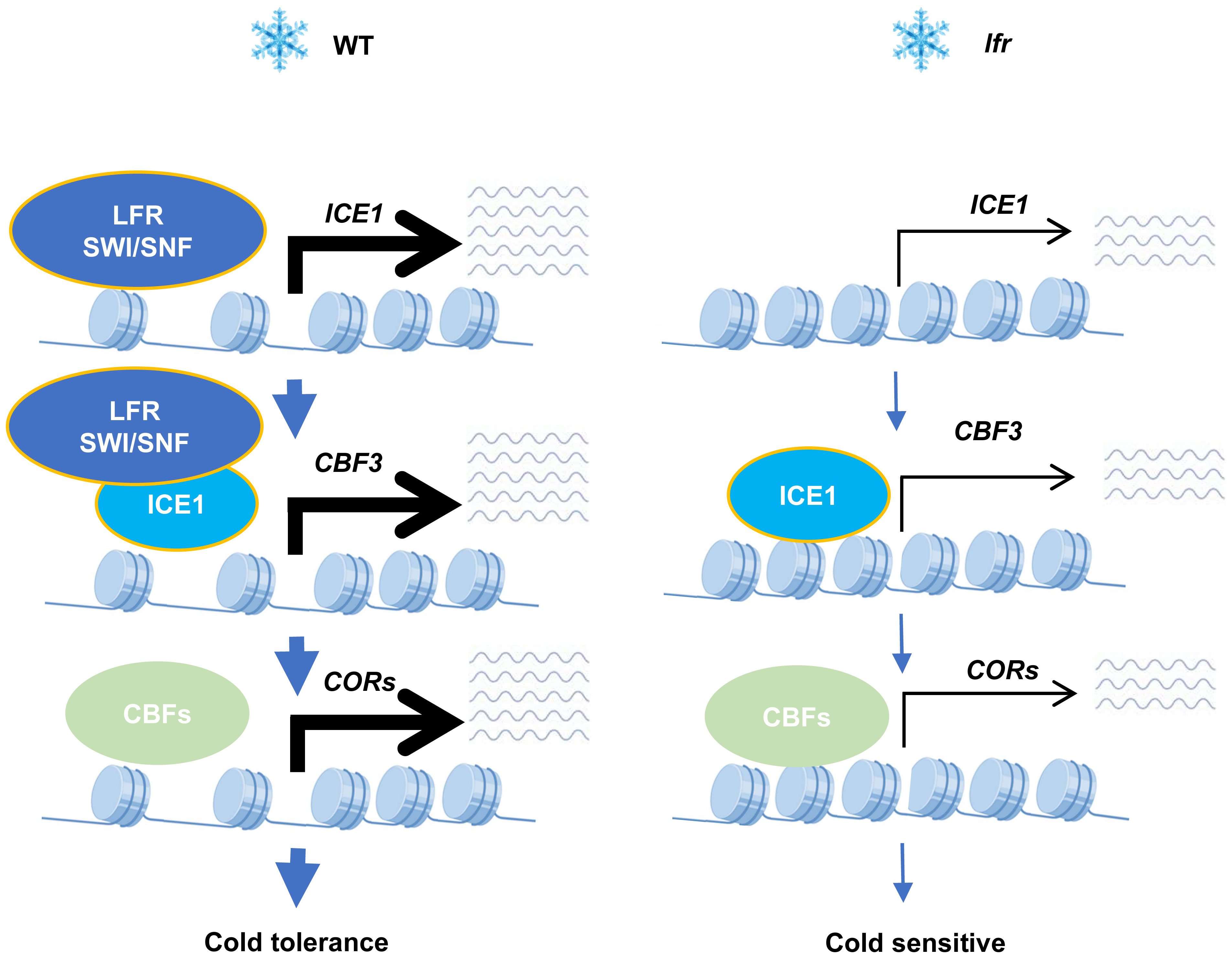
Figure 7 A working model of the function of the SWI/SNF subunit LFR in cold acclimation. Under low-temperature conditions, the SWI/SNF complex component LFR promotes plant survival by acting as a positive regulator of the ICE1–CBF–COR cascade through its physical interaction with ICE1. The depletion of LFR results in the downregulation of ICE1–CBF–COR cascade genes and cold-sensitive defects.
Our findings indicate that the SWI/SNF ATP-dependent CRC component LFR interacts directly with ICE1, and that LFR plays a positive regulatory role in the transcription of ICE1–CBF–COR cascade genes. LFR can bind directly to the promoter regions of ICE1 and CBF3 to regulate their expression and thus participate in the response of plants to low-temperature stress. These results suggest that ICE1 may promote plant survival at freezing temperatures by interacting with the SWI/SNF CRC to activate target gene expression. Our findings provide important information for breeding climate-resilient crops.
Data were deposited in TAIR (Arabidopsis Information Resource) under accession numbers: LFR (AT3G22990), ICE1 (AT3G26744), CBF1 (AT4G25490), CBF2 (AT4G25470), CBF3 (AT4G25480), COR15A (AT2G42540).
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
SC and HZ designed the study. TM constructed the plasmids and performed the Y2H, BiFC, LCI, ChIP-qPCR, phenotype analysis, and RT-qPCR assays. SW and JT conducted the ChIP-qPCR, RT-qPCR, and protein level analyses. CS did the phenotype analyses. HG prepared the pLFR : LFR-3FLAG and pLFR : LFR-eYFP transgenic lines. SW, HZ, and SC wrote the manuscript with input from all the co-authors. All authors contributed to the article and approved the submitted version.
This work was supported by the National Natural Science Foundation of China (NSFC 31400240 and 31771351) and the Hebei Provincial Natural Science Foundation of China (C2020205025).
We thank Prof. Shuhua Yang and Yue Peng (China Agricultural University, Beijing, China) for providing the ice1-2 seeds and many helpful suggestions.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1097158/full#supplementary-material.
Agarwal, M., Hao, Y., Kapoor, A., Dong, C.-H., Fujii, H., Zheng, X., et al. (2006). A R2R3 type MYB transcription factor is involved in the cold regulation of CBF genes and in acquired freezing tolerance. J. Biol. Chem. 281, 37636–37645. doi: 10.1074/jbc.M605895200
Arnon, D. I. (1949). Copper enzymes in isolated chloroplasts. polyphenoloxidase in beta vulgaris. Plant Physiol. 24, 1–15. doi: 10.1104/pp.24.1.1
Bezhani, S., Winter, C., Hershman, S., Wagner, J. D., Kennedy, J. F., Kwon, C. S., et al. (2007). Unique, shared, and redundant roles for the arabidopsis SWI/SNF chromatin remodeling ATPases BRAHMA and SPLAYED. Plant Cell 19, 403–416. doi: 10.1105/tpc.106.048272
Brzeski, J., Podstolski, W., Olczak, K., Jerzmanowski, A. (1999). Identification and analysis of the arabidopsis thaliana BSH gene, a member of the SNF5 gene family. Nucleic Acids Res. 27, 2393–2399. doi: 10.1093/nar/27.11.2393
Brzezinka, K., Altmann, S., Czesnick, H., Nicolas, P., Gorka, M., Benke, E., et al. (2016). Arabidopsis FORGETTER1 mediates stress-induced chromatin memory through nucleosome remodeling. Elife 5, e17061. doi: 10.7554/eLife.17061
Chen, X., Ding, Y., Yang, Y., Song, C., Wang, B., Yang, S., et al. (2021). Protein kinases in plant responses to drought, salt, and cold stress. J. Integr. Plant Biol. 63, 53–78. doi: 10.1111/jipb.13061
Chen, H., Zou, Y., Shang, Y., Lin, H., Wang, Y., Cai, R., et al. (2008). Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol. 146, 368–376. doi: 10.1104/pp.107.111740
Chiba, H., Muramatsu, M., Nomoto, A., Kato, H. (1994). Two human homologues of saccharomyces cerevisiae SWI2/SNF2 and drosophila brahma are transcriptional coactivators cooperating with the estrogen receptor and the retinoic acid receptor. Nucleic Acids Res. 22, 1815–1820. doi: 10.1093/nar/22.10.1815
Chinnusamy, V., Ohta, M., Kanrar, S., Lee, B.-H., Hong, X., Agarwal, M., et al. (2003). ICE1: A regulator of cold-induced transcriptome and freezing tolerance in arabidopsis. Genes Dev. 17, 1043–1054. doi: 10.1101/gad.1077503
Chung, S., Kwon, C., Lee, J.-H. (2022). Epigenetic control of abiotic stress signaling in plants. Genes Genomics 44, 267–278. doi: 10.1007/s13258-021-01163-3
Clapier, C. R., Cairns, B. R. (2009). The biology of chromatin remodeling complexes. Annu. Rev. Biochem. 78, 273–304. doi: 10.1146/annurev.biochem.77.062706.153223
Clapier, C. R., Iwasa, J., Cairns, B. R., Peterson, C. L. (2017). Mechanisms of action and regulation of ATP-dependent chromatin-remodelling complexes. Nat. Rev. Mol. Cell Biol. 18, 407–422. doi: 10.1038/nrm.2017.26
Clough, S. J., Bent, A. F. (1998). Floral dip: a simplified method for agrobacterium-mediated transformation of arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Diego-Martin, B., Pérez-Alemany, J., Candela-Ferre, J., Corbalán-Acedo, A., Pereyra, J., Alabadí, D., et al. (2022). The TRIPLE PHD FINGERS proteins are required for SWI/SNF complex-mediated +1 nucleosome positioning and transcription start site determination in arabidopsis. Nucleic Acids Res. 50, 10399–10417. doi: 10.1093/nar/gkac826
Ding, Y., Li, H., Zhang, X., Xie, Q., Gong, Z., Yang, S. (2015). OST1 kinase modulates freezing tolerance by enhancing ICE1 stability in arabidopsis. Dev. Cell 32, 278–289. doi: 10.1016/j.devcel.2014.12.023
Ding, Y., Shi, Y., Yang, S. (2019). Advances and challenges in uncovering cold tolerance regulatory mechanisms in plants. New Phytol. 222, 1690–1704. doi: 10.1111/nph.15696
Ding, Y., Shi, Y., Yang, S. (2020). Molecular regulation of plant responses to environmental temperatures. Mol. Plant 13, 544–564. doi: 10.1016/j.molp.2020.02.004
Ding, Y., Yang, S. (2022). Surviving and thriving: How plants perceive and respond to temperature stress. Dev. Cell 57, 947–958. doi: 10.1016/j.devcel.2022.03.010
Dong, C.-H., Agarwal, M., Zhang, Y., Xie, Q., Zhu, J.-K. (2006). The negative regulator of plant cold responses, HOS1, is a RING E3 ligase that mediates the ubiquitination and degradation of ICE1. Proc. Natl. Acad. Sci. 103, 8281–8286. doi: 10.1073/pnas.0602874103
Farrona, S., Hurtado, L., Bowman, J. L., Reyes, J. C. (2004). The arabidopsis thaliana SNF2 homolog AtBRM controls shoot development and flowering. Development 131, 4965–4975. doi: 10.1242/dev.01363
Gilmour, S. J., Zarka, D. G., Stockinger, E. J., Salazar, M. P., Houghton, J. M., Thomashow, M. F. (1998). Low temperature regulation of the arabidopsis CBF family of AP2 transcriptional activators as an early step in cold-induced COR gene expression. Plant J. 16, 433–442. doi: 10.1046/j.1365-313x.1998.00310.x
Gratkowska-Zmuda, D. M., Kubala, S., Sarnowska, E., Cwiek, P., Oksinska, P., Steciuk, J., et al. (2020). The SWI/SNF ATP-dependent chromatin remodeling complex in arabidopsis responds to environmental changes in temperature-dependent manner. IJMS 21, 762. doi: 10.3390/ijms21030762
Guo, J., Cai, G., Li, Y.-Q., Zhang, Y.-X., Su, Y.-N., Yuan, D.-Y., et al (2022). Comprehensive characterization of three classes of arabidopsis SWI/SNF chromatin remodelling complexes. Nat. Plants. 8, 1423–1439. doi: 10.1038/s41477-022-01282-z
Han, S.-K., Sang, Y., Rodrigues, A., BIOL425 F2010, Wu, M.-F., Rodriguez, P. L., et al. (2012). The SWI2/SNF2 chromatin remodeling ATPase BRAHMA represses abscisic acid responses in the absence of the stress stimulus in arabidopsis. Plant Cell 24, 4892–4906. doi: 10.1105/tpc.112.105114
Hargreaves, D. C., Crabtree, G. R. (2011). ATP-dependent chromatin remodeling: genetics, genomics and mechanisms. Cell Res. 21, 396–420. doi: 10.1038/cr.2011.32
He, S., Wu, Z., Tian, Y., Yu, Z., Yu, J., Wang, X., et al. (2020). Structure of nucleosome-bound human BAF complex. Science 367, 875–881. doi: 10.1126/science.aaz9761
Hernández-García, J., Diego-Martin, B., Kuo, P. H., Jami-Alahmadi, Y., Vashisht, A. A., Wohlschlegel, J., et al. (2022). Comprehensive identification of SWI/SNF complex subunits underpins deep eukaryotic ancestry and reveals new plant components. Commun. Biol. 5, 549. doi: 10.1038/s42003-022-03490-x
Hirschhorn, J. N., Brown, S. A., Clark, C. D., Winston, F. (1992). Evidence that SNF2/SWI2 and SNF5 activate transcription in yeast by altering chromatin structure. Genes Dev. 6, 2288–2298. doi: 10.1101/gad.6.12a.2288
Ho, L., Crabtree, G. R. (2010). Chromatin remodelling during development. Nature 463, 474–484. doi: 10.1038/nature08911
Hu, Y., Jiang, L., Wang, F., Yu, D. (2013). Jasmonate regulates the inducer of cbf expression-c-repeat binding factor/DRE binding factor1 cascade and freezing tolerance in arabidopsis. Plant Cell 25, 2907–2924. doi: 10.1105/tpc.113.112631
Jaglo-Ottosen, K. R., Gilmour, S. J., Zarka, D. G., Schabenberger, O., Thomashow, M. F. (1998). Arabidopsis CBF1 overexpression induces COR genes and enhances freezing tolerance. Science 280, 104–106. doi: 10.1126/science.280.5360.104
Jarończyk, K., Sosnowska, K., Zaborowski, A., Pupel, P., Bucholc, M., Małecka, E., et al. (2021). Bromodomain-containing subunits BRD1, BRD2, and BRD13 are required for proper functioning of SWI/SNF complexes in arabidopsis. Plant Commun. 2, 100174. doi: 10.1016/j.xplc.2021.100174
Jégu, T., Latrasse, D., Delarue, M., Hirt, H., Domenichini, S., Ariel, F., et al. (2014). The BAF60 subunit of the SWI/SNF chromatin-remodeling complex directly controls the formation of a gene loop at FLOWERING LOCUS c in arabidopsis. Plant Cell 26, 538–551. doi: 10.1105/tpc.113.114454
Jégu, T., Veluchamy, A., Ramirez-Prado, J. S., Rizzi-Paillet, C., Perez, M., Lhomme, A., et al. (2017). The arabidopsis SWI/SNF protein BAF60 mediates seedling growth control by modulating DNA accessibility. Genome Biol. 18, 114. doi: 10.1186/s13059-017-1246-7
Kanaoka, M. M., Pillitteri, L. J., Fujii, H., Yoshida, Y., Bogenschutz, N. L., Takabayashi, J., et al. (2008). SCREAM/ICE1 and SCREAM2 specify three cell-state transitional steps leading to arabidopsis stomatal differentiation. Plant Cell 20, 1775–1785. doi: 10.1105/tpc.108.060848
Kandasamy, M. K., McKinney, E. C., Meagher, R. B. (2003). Cell cycle-dependent association of arabidopsis actin-related proteins AtARP4 and AtARP7 with the nucleus. Plant J. 33, 939–948. doi: 10.1046/j.1365-313x.2003.01691.x
Khavari, P. A., Peterson, C. L., Tamkun, J. W., Mendel, D. B., Crabtree, G. R. (1993). BRG1 contains a conserved domain of the SWI2/SNF2 family necessary for normal mitotic growth and transcription. Nature 366, 170–174. doi: 10.1038/366170a0
Kim, Y. S., Lee, M., Lee, J.-H., Lee, H.-J., Park, C.-M. (2015). The unified ICE-CBF pathway provides a transcriptional feedback control of freezing tolerance during cold acclimation in arabidopsis. Plant Mol. Biol. 89, 187–201. doi: 10.1007/s11103-015-0365-3
Li, H., Ding, Y., Shi, Y., Zhang, X., Zhang, S., Gong, Z., et al. (2017). MPK3- and MPK6-mediated ICE1 phosphorylation negatively regulates ICE1 stability and freezing tolerance in arabidopsis. Dev. Cell 43, 630–642.e4. doi: 10.1016/j.devcel.2017.09.025
Lin, X., Gu, D., Zhao, H., Peng, Y., Zhang, G., Yuan, T., et al. (2018). LFR is functionally associated with AS2 to mediate leaf development in arabidopsis. Plant J. 95, 598–612. doi: 10.1111/tpj.13973
Lin, X., Yuan, C., Zhu, B., Yuan, T., Li, X., Yuan, S., et al. (2021). LFR physically and genetically interacts with SWI/SNF component SWI3B to regulate leaf blade development in arabidopsis. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.717649
Liu, Q., Kasuga, M., Sakuma, Y., Abe, H., Miura, S., Yamaguchi-Shinozaki, K., et al. (1998). Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in arabidopsis. Plant Cell 10, 1391–1406. doi: 10.1105/tpc.10.8.1391
Ma, Y., Dai, X., Xu, Y., Luo, W., Zheng, X., Zeng, D., et al. (2015). COLD1 confers chilling tolerance in rice. Cell 160, 1209–1221. doi: 10.1016/j.cell.2015.01.046
Mashtalir, N., Suzuki, H., Farrell, D. P., Sankar, A., Luo, J., Filipovski, M., et al. (2020). A structural model of the endogenous human BAF complex informs disease mechanisms. Cell 183, 802–817.e24. doi: 10.1016/j.cell.2020.09.051
Miryeganeh, M. (2021). Plants’ epigenetic mechanisms and abiotic stress. Genes (Basel) 12, 1106. doi: 10.3390/genes12081106
Miura, K., Jin, J. B., Lee, J., Yoo, C. Y., Stirm, V., Miura, T., et al. (2007). SIZ1-mediated sumoylation of ICE1 controls CBF3/DREB1A expression and freezing tolerance in arabidopsis. Plant Cell 19, 1403–1414. doi: 10.1105/tpc.106.048397
Mostofa, M. G., Hossain, M. A., Fujita, M., Tran, L.-S. P. (2015). Physiological and biochemical mechanisms associated with trehalose-induced copper-stress tolerance in rice. Sci. Rep. 5, 11433. doi: 10.1038/srep11433
Ohta, M., Sato, A., Renhu, N., Yamamoto, T., Oka, N., Zhu, J.-K., et al. (2018). MYC-type transcription factors, MYC67 and MYC70, interact with ICE1 and negatively regulate cold tolerance in arabidopsis. Sci. Rep. 8, 11622. doi: 10.1038/s41598-018-29722-x
Ojolo, S. P., Cao, S., Priyadarshani, S. V. G. N., Li, W., Yan, M., Aslam, M., et al. (2018). Regulation of plant growth and development: A review from a chromatin remodeling perspective. Front. Plant Sci. 9, 1232. doi: 10.3389/fpls.2018.01232
Ou, B., Yin, K.-Q., Liu, S.-N., Yang, Y., Gu, T., Wing Hui, J. M., et al. (2011). A high-throughput screening system for arabidopsis transcription factors and its application to Med25-dependent transcriptional regulation. Mol. Plant 4, 546–555. doi: 10.1093/mp/ssr002
Peirats-Llobet, M., Han, S.-K., Gonzalez-Guzman, M., Jeong, C. W., Rodriguez, L., Belda-Palazon, B., et al. (2016). A direct link between abscisic acid sensing and the chromatin-remodeling ATPase BRAHMA via core ABA signaling pathway components. Mol. Plant 9, 136–147. doi: 10.1016/j.molp.2015.10.003
Peterson, C. L., Herskowitz, I. (1992). Characterization of the yeast SWI1, SWI2, and SWI3 genes, which encode a global activator of transcription. Cell 68, 573–583. doi: 10.1016/0092-8674(92)90192-f
Sacharowski, S. P., Gratkowska, D. M., Sarnowska, E. A., Kondrak, P., Jancewicz, I., Porri, A., et al. (2015). SWP73 subunits of arabidopsis SWI/SNF chromatin remodeling complexes play distinct roles in leaf and flower development. Plant Cell 27, 1889–1906. doi: 10.1105/tpc.15.00233
Saez, A., Rodrigues, A., Santiago, J., Rubio, S., Rodriguez, P. L. (2008). HAB1-SWI3B interaction reveals a link between abscisic acid signaling and putative SWI/SNF chromatin-remodeling complexes in arabidopsis. Plant Cell 20, 2972–2988. doi: 10.1105/tpc.107.056705
Sang, Y., Silva-Ortega, C. O., Wu, S., Yamaguchi, N., Wu, M.-F., Pfluger, J., et al. (2012). Mutations in two non-canonical arabidopsis SWI2/SNF2 chromatin remodeling ATPases cause embryogenesis and stem cell maintenance defects. Plant J. 72, 1000–1014. doi: 10.1111/tpj.12009
Sarnowska, E., Gratkowska, D. M., Sacharowski, S. P., Cwiek, P., Tohge, T., Fernie, A. R., et al. (2016). The role of SWI/SNF chromatin remodeling complexes in hormone crosstalk. Trends Plant Sci. 21, 594–608. doi: 10.1016/j.tplants.2016.01.017
Sarnowski, T. J., Ríos, G., Jásik, J., Swiezewski, S., Kaczanowski, S., Li, Y., et al. (2005). SWI3 subunits of putative SWI/SNF chromatin-remodeling complexes play distinct roles during arabidopsis development. Plant Cell 17, 2454–2472. doi: 10.1105/tpc.105.031203
Shang, J.-Y., He, X.-J. (2022). Chromatin-remodeling complexes: Conserved and plant-specific subunits in arabidopsis. J. Integr. Plant Biol. 64, 499–515. doi: 10.1111/jipb.13208
Shi, Y., Ding, Y., Yang, S. (2018). Molecular regulation of CBF signaling in cold acclimation. Trends Plant Sci. 23, 623–637. doi: 10.1016/j.tplants.2018.04.002
Shu, J., Chen, C., Li, C., Thapa, R. K., Song, J., Xie, X., et al. (2021). Genome-wide occupancy of arabidopsis SWI/SNF chromatin remodeler SPLAYED provides insights into its interplay with its close homolog BRAHMA and polycomb proteins. Plant J. 106, 200–213. doi: 10.1111/tpj.15159
Song, Y., Zhang, X., Li, M., Yang, H., Fu, D., Lv, J., et al. (2021). The direct targets of CBFs: In cold stress response and beyond. J. Integr. Plant Biol. 63, 1874–1887. doi: 10.1111/jipb.13161
Stockinger, E. J., Gilmour, S. J., Thomashow, M. F. (1997). Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the c-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc. Natl. Acad. Sci. U.S.A. 94, 1035–1040. doi: 10.1073/pnas.94.3.1035
Tang, K., Zhao, L., Ren, Y., Yang, S., Zhu, J.-K., Zhao, C. (2020). The transcription factor ICE1 functions in cold stress response by binding to the promoters of CBF and COR genes. J. Integr. Plant Biol. 62, 258–263. doi: 10.1111/jipb.12918
Thomashow, M. F. (1999). PLANT COLD ACCLIMATION: Freezing tolerance genes and regulatory mechanisms. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 571–599. doi: 10.1146/annurev.arplant.50.1.571
Thouly, C., Le Masson, M., Lai, X., Carles, C. C., Vachon, G. (2020). Unwinding BRAHMA functions in plants. Genes (Basel) 11, 90. doi: 10.3390/genes11010090
Vercruyssen, L., Verkest, A., Gonzalez, N., Heyndrickx, K. S., Eeckhout, D., Han, S.-K., et al. (2014). ANGUSTIFOLIA3 binds to SWI/SNF chromatin remodeling complexes to regulate transcription during arabidopsis leaf development. Plant Cell 26, 210–229. doi: 10.1105/tpc.113.115907
Vicente, J., Mendiondo, G. M., Movahedi, M., Peirats-Llobet, M., Juan, Y.-T., Shen, Y.-Y., et al. (2017). The cys-Arg/N-End rule pathway is a general sensor of abiotic stress in flowering plants. Curr. Biol. 27, 3183–3190.e4. doi: 10.1016/j.cub.2017.09.006
Wagner, D., Meyerowitz, E. M. (2002). SPLAYED, a novel SWI/SNF ATPase homolog, controls reproductive development in arabidopsis. Curr. Biol. 12, 85–94. doi: 10.1016/S0960-9822(01)00651-0
Wang, X., Yuan, C., Yuan, T., Cui, S. (2012). The arabidopsis LFR gene is required for the formation of anther cell layers and normal expression of key regulatory genes. Mol. Plant 5, 993–1000. doi: 10.1093/mp/sss024
Wang, Z., Yuan, T., Yuan, C., Niu, Y., Sun, D., Cui, S. (2009). LFR, which encodes a novel nuclear-localized armadillo-repeat protein, affects multiple developmental processes in the aerial organs in arabidopsis. Plant Mol. Biol. 69, 121–131. doi: 10.1007/s11103-008-9411-8
Yamaguchi, N., Winter, C. M., Wu, M.-F., Kwon, C. S., William, D. A., Wagner, D. (2014). PROTOCOLS: Chromatin immunoprecipitation from arabidopsis tissues. Arabidopsis Book 12, e0170. doi: 10.1199/tab.0170
Yang, T., Wang, D., Tian, G., Sun, L., Yang, M., Yin, X., et al. (2022). Chromatin remodeling complexes regulate genome architecture in arabidopsis. Plant Cell 34, 2638–2651. doi: 10.1093/plcell/koac117
Ye, K., Li, H., Ding, Y., Shi, Y., Song, C., Gong, Z., et al. (2019). BRASSINOSTEROID-INSENSITIVE2 negatively regulates the stability of transcription factor ICE1 in response to cold stress in arabidopsis. Plant Cell 31, 2682–2696. doi: 10.1105/tpc.19.00058
Yu, Y., Fu, W., Xu, J., Lei, Y., Song, X., Liang, Z., et al. (2021). Bromodomain-containing proteins BRD1, BRD2, and BRD13 are core subunits of SWI/SNF complexes and vital for their genomic targeting in arabidopsis. Mol. Plant 14, 888–904. doi: 10.1016/j.molp.2021.03.018
Yu, Y., Liang, Z., Song, X., Fu, W., Xu, J., Lei, Y., et al. (2020). BRAHMA-interacting proteins BRIP1 and BRIP2 are core subunits of arabidopsis SWI/SNF complexes. Nat. Plants 6, 996–1007. doi: 10.1038/s41477-020-0734-z
Keywords: Arabidopsis, cold acclimation, chromatin remodeling, SWI/SNF, LFR, ICE1, CBFs
Citation: Ma T, Wang S, Sun C, Tian J, Guo H, Cui S and Zhao H (2023) Arabidopsis LFR, a SWI/SNF complex component, interacts with ICE1 and activates ICE1 and CBF3 expression in cold acclimation. Front. Plant Sci. 14:1097158. doi: 10.3389/fpls.2023.1097158
Received: 13 November 2022; Accepted: 08 March 2023;
Published: 21 March 2023.
Edited by:
Michael Nicolas, National Center for Biotechnology (CSIC), SpainReviewed by:
Keqiang Wu, National Taiwan University, TaiwanCopyright © 2023 Ma, Wang, Sun, Tian, Guo, Cui and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hongtao Zhao, emhhb2hvbmd0YW9AaGVidHUuZWR1LmNu
†These authors contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.