- 1Heilongjiang Bayi Agricultural University, Key Laboratory of Soybean Mechanized Production, Ministry of Agriculture and Rural Affairs, Daqing, China
- 2Agricultural College, Northeast Agricultural University, Harbin, China
- 3National Coarse Cereals Engineering Research Center, Heilongjiang Bayi Agricultural University, Daqing, China
- 4Institute of Industrial Crops, Heilongjiang Academy of Agricultural Sciences, Harbin, China
- 5Institute of Crop Cultivation and Tillage, Heilongjiang Academy of Agricultural Sciences, Harbin, China
Groundnut or peanut (Arachis hypogaea) is a legume crop. Its seeds are rich in protein and oil. Aldehyde dehydrogenase (ALDH, EC: 1.2.1.3) is an important enzyme involved in detoxification of aldehyde and cellular reactive oxygen species, as well as in attenuation of lipid peroxidation-meditated cellular toxicity under stress conditions. However, few studies have been identified and analyzed about ALDH members in Arachis hypogaea. In the present study, 71 members of the ALDH superfamily (AhALDH) were identified using the reference genome obtained from the Phytozome database. A systematic analysis of the evolutionary relationship, motif, gene structure, cis-acting elements, collinearity, Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment, and expression patterns was conducted to understand the structure and function of AhALDHs. AhALDHs exhibited tissue-specific expression, and quantitative real-time PCR identified significant differences in the expression levels of AhALDH members under saline-alkali stress. The results revealed that some AhALDHs members could be involved in response to abiotic stress. Our findings on AhALDHs provide insights for further study.
1 Introduction
Aldehyde molecules are essential intermediate compounds generated in catabolic and biosynthetic pathways during biological development and growth (Vasiliou et al., 2000). In response to stress, aldehyde accumulates in cells, causing an imbalance and interfering with cellular homeostatic metabolic responses (Bartels, 2001), and becomes toxic if present in excess (Carmona-Molero et al., 2021) Aldehyde dehydrogenase (ALDH, EC: 1.2.1.3) as a scavenger of aldehyde molecules contributes to their homeostasis (Yoshida et al., 1998). The ALDH family is composed of a variety of NAD(P)+-dependent enzymes that irreversibly oxidize endogenously and exogenously derived aldehyde molecules to carboxylic acids (Yoshiba et al., 1997). ALDH enzymes also function in intermediary metabolism by providing protection from osmotic stress and generating NAD(P)H (Kelly and Gibbs, 1973; Ishitani et al., 1995; Brocker et al., 2010). ALDHs have been reported to improve stress resistance in crops (Bartels and Sunkar, 2005). ALDH family members are found in prokaryotic and eukaryotic organisms and are highly conserved and well represented in virtually all plant species (Brocker et al., 2013). Research on the ALDH gene family in prokaryotes and mammals is abundant (Kirch et al., 2004). The ALDH members associated with ALDH enzyme activity are linked to diseases such as cataracts, hyperprolinaemia, and cancers (Jackson et al., 2011). However, the functional and structural characterization of plant ALDHs and gene duplication events underlying their diversification have lagged behind that of their mammalian and bacterial counterparts (Zhang et al., 2012).
ALDH family members, which are found in almost all plant species, are variable, widespread in plant tissues, and regulated developmentally (Chen C, et al., 2014; Guo et al., 2017). ALDH members participate in plant growth and development and play a vital role in catabolic and bio-synthetic pathways, such as carnitine biosynthesis (Marchitti et al., 2008), glycolysis/gluconeogenesis (Tylichová et al., 2010), and amino-acid metabolism (Yang et al., 2011). The ALDH2B2 (rf2) gene has ALDH domain functions and is a male fertility restorer in maize (Skibbe et al., 2002). ALDH7s in Arabidopsis and soybean are involved in aldehyde detoxification (Shin et al., 2009), whereas OsALDH7 is essential for seed maturation, and its mutation leads to seed browning during seed drying and storage of rice (Shin et al., 2009). An increasing number of studies have shown that some ALDH members indirectly function in plant cell protection under various abiotic stresses through detoxification of cellular reactive oxygen species (ROS) and/or reduction of lipid peroxidation (Shin et al., 2009; Singh et al., 2013). Betaine aldehyde dehydrogenases (BADH) are a type of ALDH10 enzyme that catalyze the oxidation of betaine aldehyde into glycine betaine (a major cellular osmolyte) and thereby improve plant resistance to environmental stress (Le Rudulier et al., 1984); the BADH gene has been shown to improve salt tolerance in plants (Zhou et al., 2008). Furthermore, the ectopic expression of ALDH3I1 and ALDH7B4 significantly reduced malondialdehyde (MDA) levels and lipid peroxidation in transgenic Arabidopsis, revealing the role of these two genes in increasing plant tolerance to drought and salt stress (Kotchoni et al., 2006). VvALDH2B4, an ALDH member from the Chinese wild grapevine (Vitis pseudoreticulata), lowered MDA levels and enhanced plant resistance to salt stress and pathogenic bacteria in over-expressed transgenic Arabidopsis (Wen et al., 2012). ScALDH21, an ALDH member isolated from Syntrichia caninervis, enhanced the activity of antioxidant enzymes, increased the proline content, and lowered the MDA content in transgenic tobacco under salt and drought stress, which is likely the reason for increased germination ratios and root lengths in tobacco plants (Yang et al., 2015).
ALDH members have been identified in Arabidopsis (Kirch et al., 2001), rice (Oryza sativa) (Gao and Han, 2009), maize (Zea mays) (Jimenez-Lopez et al., 2010), soybean (Glycine max) (Kotchoni et al., 2012), and cotton (Gossypium hirsutum) (Guo et al., 2017), but few studies have examined their presence in groundnut (Arachis hypogaea). Groundnut (peanut), a member of the legume family, originated in southern Bolivia (Krapovickas and Gregory, 1994). It is cultivated in more than 100 countries on 26 million hectares (ha) of land for its seeds that are a rich source of dietary fiber, minerals, vitamins, and bioactive compounds, especially proteins and oil (Desmae et al., 2019). The growth of groundnut is affected by abiotic stresses, and research has been conducted to increase its stress resistance through molecular breeding (Desmae et al., 2019). In the present study, ALDH members in groundnut were identified, and a comprehensive analysis (location, evolution, motif, gene structure, cis-acting elements, and expression patterns) was conducted. The results can be utilized to breed groundnut with improved stress resistance.
2 Materials and methods
2.1 ALDH members of Arachis hypogaea
The reference genome (Arachis hypogaea v1.0) and adjoining information (protein sequence, mRNA, coding sequence, and DNA) were obtained from the Phytozome database (Goodstein et al., 2012). The ALDH domain (accession number PF00171) was downloaded from the PFAM database. The ALDH members were identified through hmmsearch and hmmbuild using the perl script in the Linux system, in which 1e−15 was set as a filter threshold (Zhang et al., 2020). The SMART software (Letunic and Bork, 2018) was used to confirm the ALDH domain and remove duplicates.
2.2 Analysis of ALDH members
The evolutionary relationship of ALDH members was analyzed using neighbor-joining methods and a Poisson model implemented in MEGA X, with 1000 bootstrap repetitions (Kumar et al., 2018). The motif of ALDH members was analyzed using the MEME platform, in which the length of motifs was 10–15 amino acids, while the e-value of motifs was less than e−5 (Bailey et al., 2009). The gene structure of the ALDH members was analyzed using the GSDS software (Hu et al., 2015). The cis-acting elements were identified and predicted using PlantCare, in which the function of each cis-acting element was predicted (Lescot et al., 2002). The expression of ALDH members and their location on chromosomes was extracted from the Phytozome database (Goodstein et al., 2012). The results were plotted using TBtool (Chen et al., 2020). The Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) databases were used for the gene annotation of ALDH members (Chen et al., 2017); Majorbio Cloud provided the platform for enrichment analysis (Shanghai, China).
2.3 Plant materials and conditions
The locally grown groundnut cultivar Silihong was used in this study. The seeds were provided by the Institute of Economic Botany, Heilongjiang Academy of Agricultural Sciences (Harbin, Hei Longjiang, China). Seeds of the same size were sterilized with 5% NaClO solution for 15 min, rinsed with distilled water, and placed in a Petri dish lined with double-layer filter paper. The seeds were covered with a single filter paper. Three replicates of 10 seeds per Petri dish were prepared for saline-alkali and control treatments. Petri dishes were placed in a GXZ intelligent light incubator (Ningbo Jiangnan Instrument Factory, Zehjiang, China) at 25°C in the dark. When the length of the peanut buds reached half the length of the peanut seeds, the seeds were sampled and used for expression pattern analysis. Control seeds were treated with distilled water. The rest of the seeds were subjected to saline-alkali treatment by adding saline-alkali solution (pH, 8.9) to Petri dishes. The sprouts in each Petri dish were set as the experimental unit, while each treatment had three experimental units as biological replicates.
2.4 Physiological and expression level analysis
The radicle was used as samples in this study, with biological and technological replicates. ALDH enzyme activity was determined under water and saline-alkali treatment at 0, 12, 24, 48, 72, and 96 h using a Multiskan FC microplate reader (Thermo Scientific Company, Wilmington, DE, USA) and the Elisa Kit (M0608, Michy Biology, Suzhou, China) according to the manufacturer’s instructions. Radicles were used as samples. The samples at 0 and 48 h from both treatments were used for quantitative real-time PCR (qRT-PCR) analysis. Total RNA was extracted using a MolPure® Plant Plus RNA Kit (19292ES, Yeason, Shanghai, China). The qualified RNA was used for reverse transcription using a kit (11139ES, Yeason) after detection using a Nanodrop OneC (Thermo Fisher Scientific, Waltham, MA, USA) and 1% agarose for RNA quality. The primers for ALDH amplification were designed using DNAMAN (Lynnon Biosoft, San Ramon, CA, USA) (Nong et al., 2019), and the UKN1 gene was used as the reference gene (Table S2). qRT-PCR was performed on the Roche platform (480 II, Roche, Basel, Switzerland) using Hieff UNICON® Universal Blue qPCR SYBR Green Master Mix (11184ES, Yeason) according to the manufacturer’s instructions. The expression levels of ALDH members were calculated using the 2−ΔCt method (Livak and Schmittgen, 2001). Data were analyzed in the SPSS software (Bezzaouha et al., 2020). Duncan’s multiple method was used to test differences between groups.
3 Results
3.1 Identification and evolution of ALDH members
One hundred and five ALDH members were identified from the reference genome (Arachis hypogaea v1.0) from the Phytozome database. The sequences were confirmed using the Smart software (Letunic and Bork, 2018), and after removing the duplicates, 71 members of the ALDH family were identified. The evolutionary relationships of the AhALDH members were analyzed using the MEGA X software (Kumar et al., 2018), while ALDH members in Arachis duranensis and Arachis ipaensis were used to correct evolutionary relationships. All these ALDH members were classified into 10 subfamilies, labelled I–X according to the order of AhALDH evolution. Subfamilies III and VI had the fewest AhALDH members, only two, while subfamily X had the highest number of AhALDH members at 17. These AhALDH members were named according to the evolutionary results and Brockers’ method (Brocker et al., 2013). ALDH members in A. duranensis and A. ipaensis, along with AhALDH members, were also divided into 10 subfamilies, which proved the accuracy of the evolutionary analysis (Figure 1). Moreover, detailed information about the proteins (including protein length, molecular weight, and isoelectric point) is shown in Table S1. The protein length varied from 303 to 791 amino acids, the molecular weight was 32.48–88.83 kDa, and theisoelectric point was 4.91–9.61. This information revealed that AhALDH members in different subgroups had diversified features, indicating that different members might perform different functions.
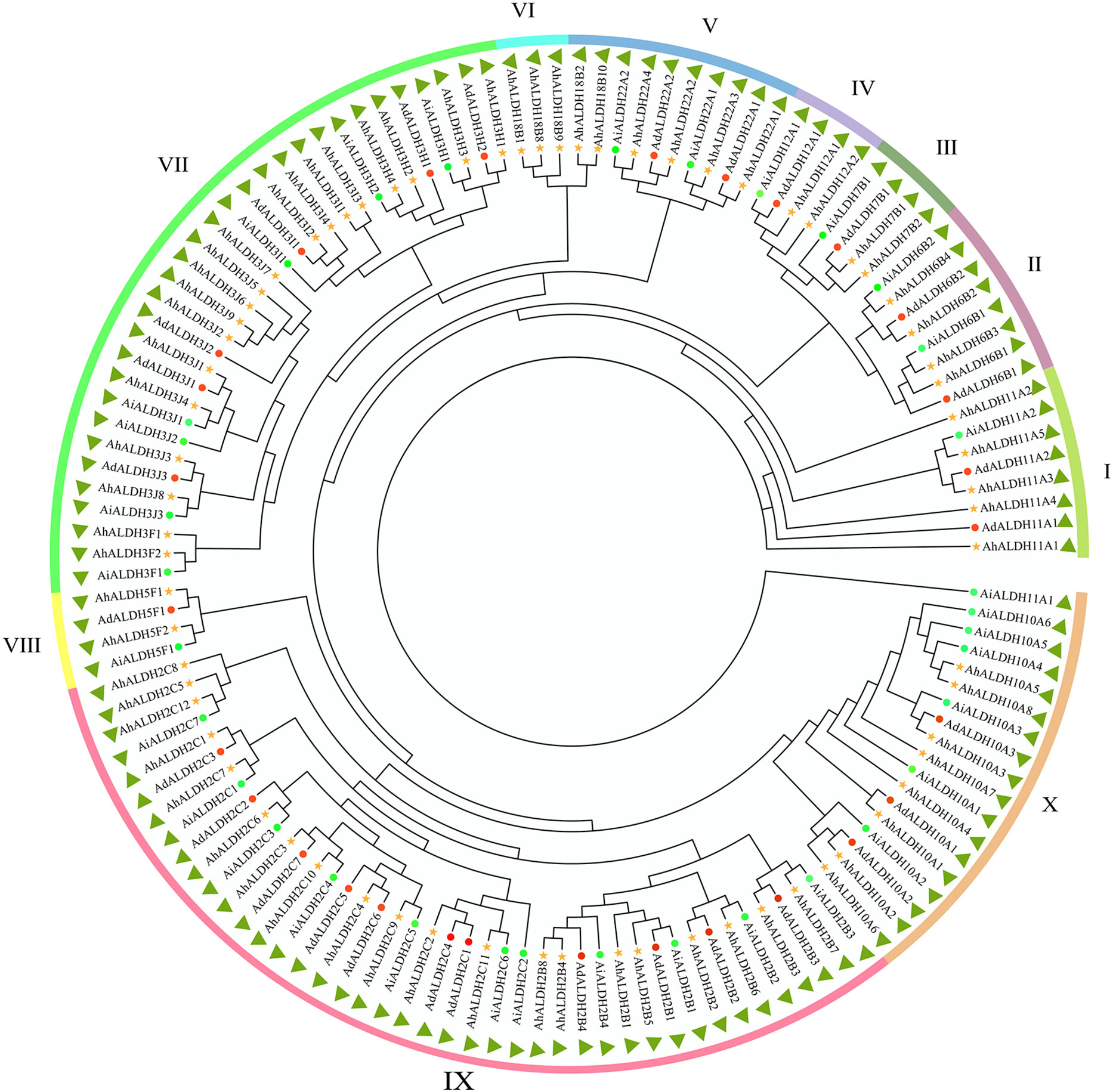
Figure 1 Evolutionary relationship of AhALDH members. Green circles represent ALDH members in Arachis hypogaea, while red circles and yellow stars represent ALDH members in Arachis duranensis and Arachis ipaensis. Outer ring bands in different colors represent the subfamilies I–X.
3.2 Location analysis of AhALDHs
Location information was obtained from databases and drawn using Tbtools (Figure 2). All chromosomes had AhALDH members, except for Chr 10 and Chr 20. The number of AhALDH members on each Chr differed: Chr13 and Chr15 had the most AhALDH members (9), while Chr03 and Chr11 had seven members; Chr1, Chr7, and Chr12, with only one member each, had the fewest, while only two AhALDH members were found on Chr16 and Chr19. These results indicated that the distribution of AhALDH members on groundnut chromosomes is not uniform, indicating that Chr13 and Chr15 might be central to AhALDH evolution.
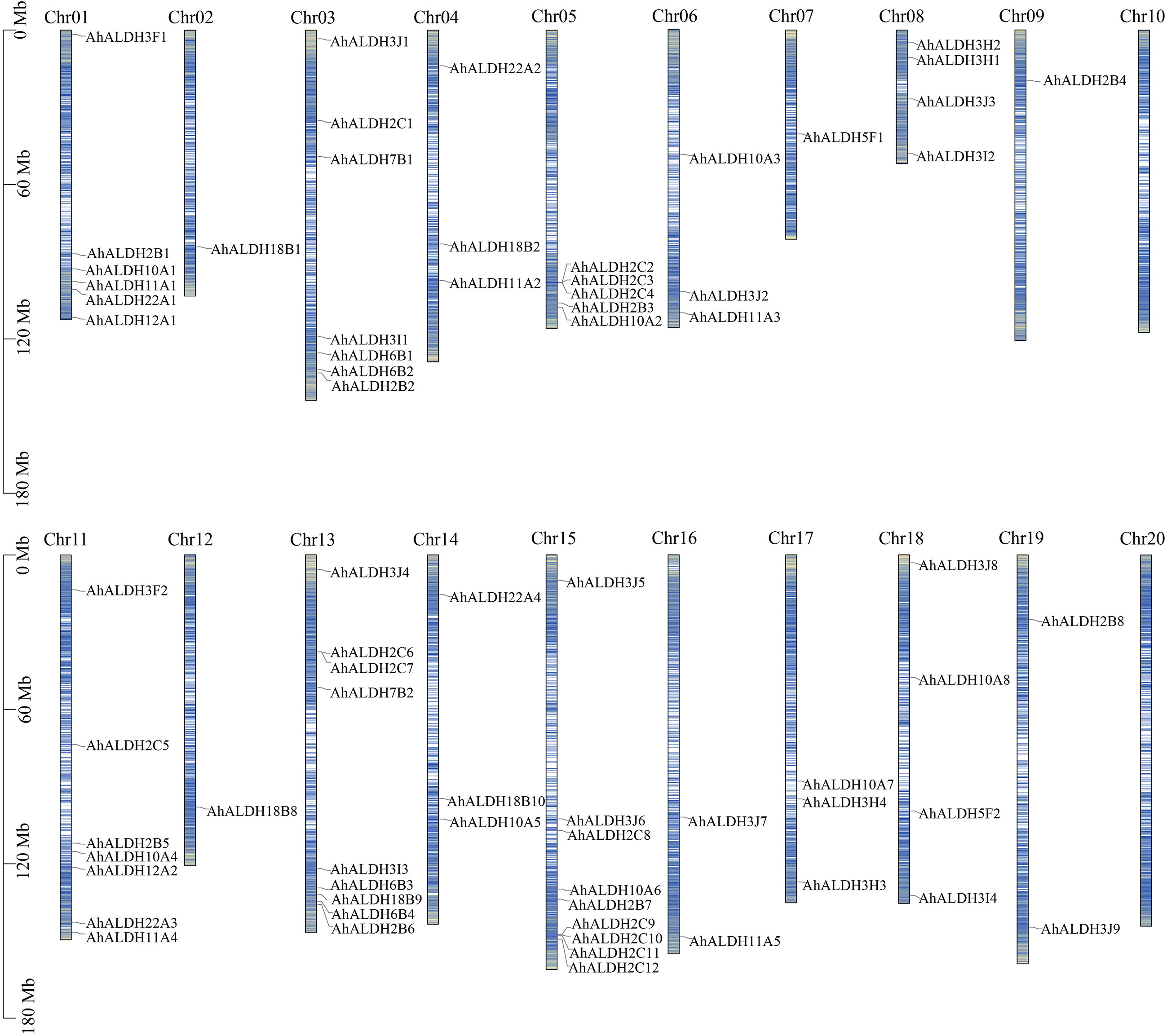
Figure 2 Location of AhALDH members on chromosomes (Chr1–Chr20). The length of the columns represents the size of the chromosomes. The black scale on the left represents the position, and blue lines inside the columns represent the gene density on chromosomes.
3.3 Motif and gene structure analysis of AhALDHs
The motif and gene structure of AhALDHs were analyzed using MEME and GSDS, respectively, to explore their structure and properties. The results are shown in Figure 3. The length of the motifs varied from 10 to 50, and the e-value was lower than 1e−20. AhALDHs in the same subfamily shared similar kinds of motifs, although some motifs also varied within the same subfamily, such as motif 1 (Figure 3). Members of the same subfamily had a similar gene structure, with the longest AhALDH member being in subfamily X. Moreover, the motif and gene structure of ALDH members in A. duranensis and A. ipaensis also revealed that members in each subfamily had similar characteristics, indicating that they might perform similar functions.
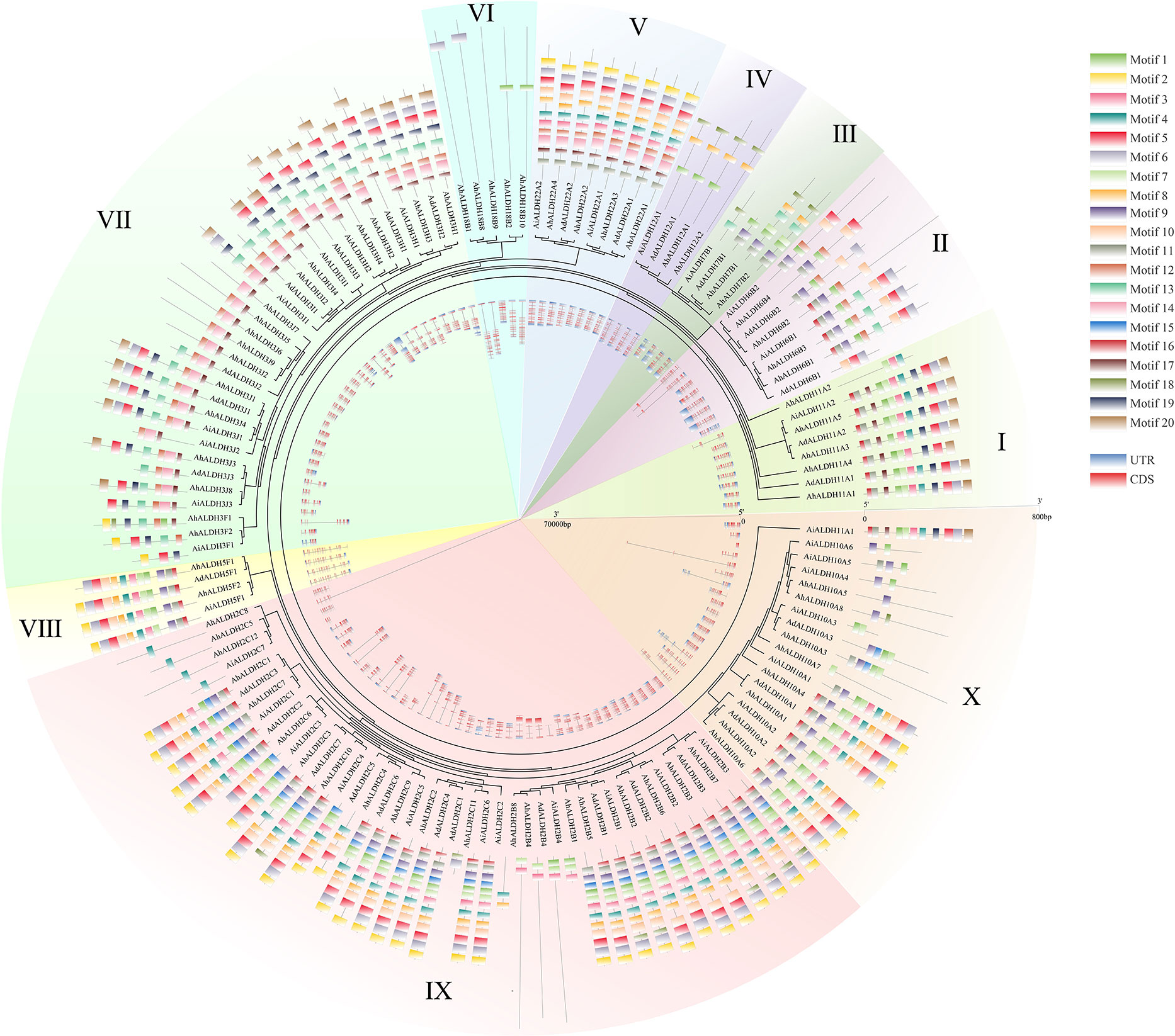
Figure 3 Motif and gene structure analysis of AhALDH members. The middle ring represents the evolutionary relationship of AhALDHs. The 10 families are highlighted with differently colored backgrounds. The outer ring shows the motif analysis of AhALDHs. Motifs from 1 to 20 are marked with differently colored boxes. The inner ring displays the gene structure analysis of AhALDHs. Blue boxes are untranslated regions (UTRs) and pink boxes are coding sequences (CDSs), black lines are the intron region.
3.4 Comparison of ALDHs in different species
In this study, 260 ALDH members in seven species, namely groundnut, Arabidopsis, rice, human, cotton, soybean, and grape (Vitis vinifera), were used for the evolutionary and motif analysis. These 260 members were divided into 10 subfamilies, in which the members within each subfamily shared similar motifs. The evolutionary results were similar to those shown in Figure 2. Under the same analysis threshold, the AhALDHs had similar kinds of motifs to ALDH members in other species (Figure 4).
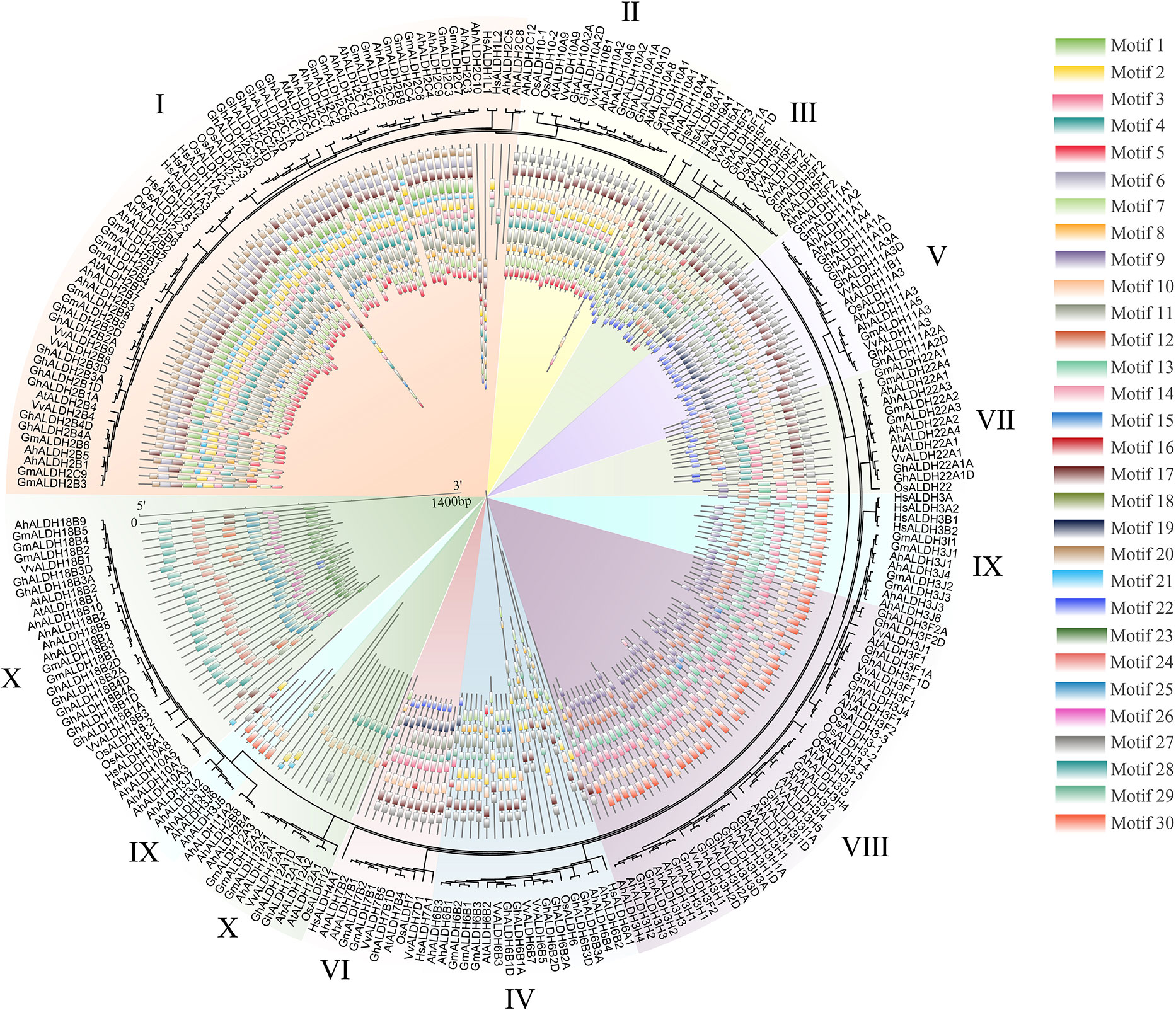
Figure 4 Analysis of ALDH members in seven species. The seven species were groundnut (Ah), Arabidopsis (At), rice (Os), human (Hs), cotton (Gh), soybean (Gm), and grape (Vv). Differently colored backgrounds represent different subfamilies. The inner circle represents the motifs of ALDH members; different squares show different motifs.
3.5 Cis-acting element analysis of AhALDHs
The cis-acting elements of AhALDHs were analyzed and their function predicted using the Plantcare software (Figure 5; Table S3). Three kinds of elements were recognized based on their function. The cis-acting elements TGA-element, ABRE, P-box, GARE-motif, TCA-element, AT-rich sequence, SARE, TATC-box, and AuxRR-core (marked in red in Figure 4) were predicted to be related to hormones; LTR, MBS, ARE, and GC-motif (marked in blue) were related to abiotic stress. The members within each subfamily had similar cis-acting elements, and the analysis revealed that AhALDHs might be associated with hormones and abiotic stress; in particular, some members had stress-related cis-acting elements.
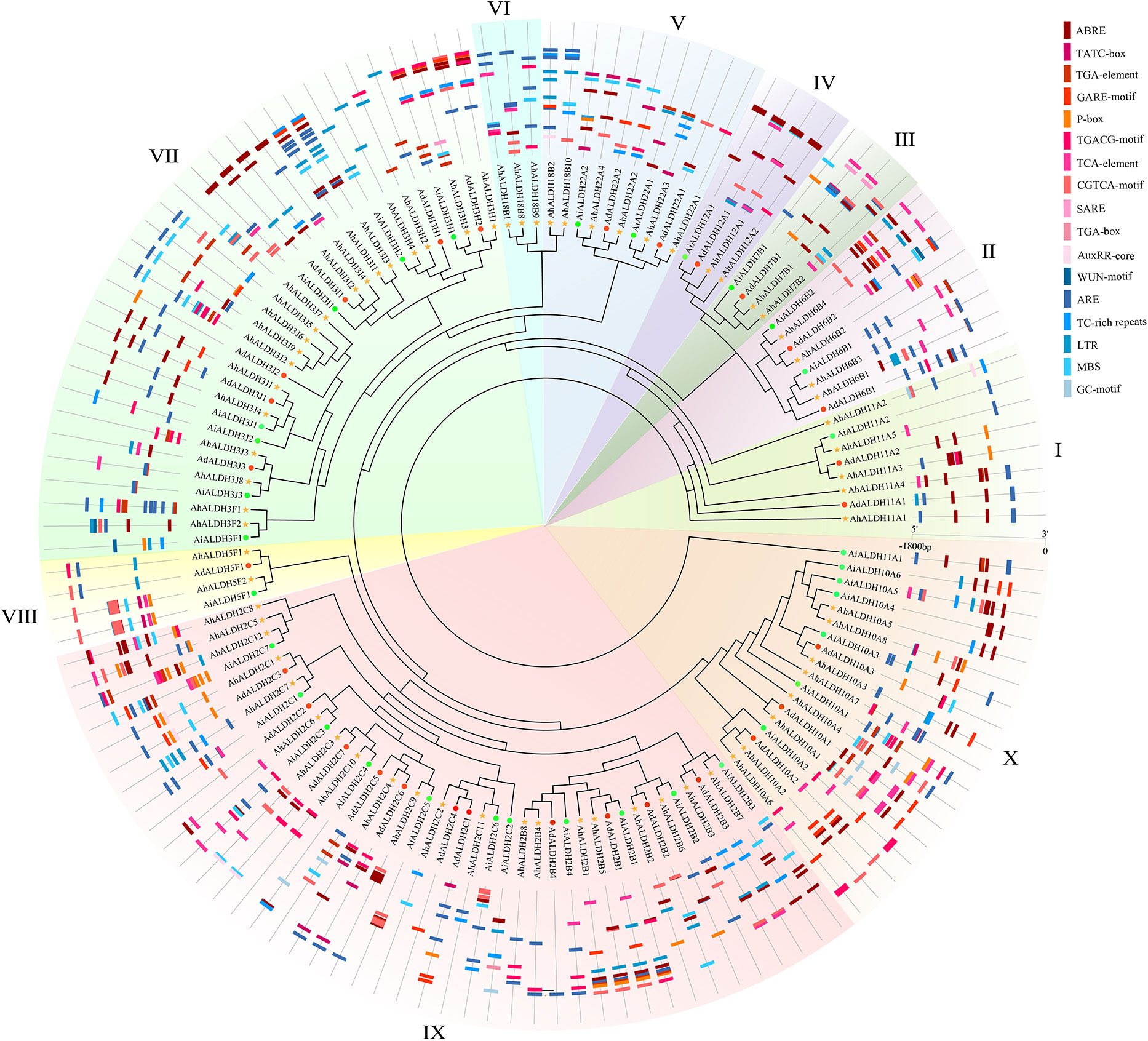
Figure 5 Cis-acting element analysis of AhALDH members. The inner ring shows the evolutionary relationship of AhALDHs; differently colored backgrounds represent different subfamilies. The outer ring shows the Cis-acting elements analysis. Red, orange, and pink boxes in different shapes represent cis-acting elements associated with hormones, while the blue boxes are cis-acting elements responding to abiotic stress.
3.6 Collinearity analysis of AhALDHs
Fifty-two pairs of collinear AhALDHs were identified. Most of the AhALDH members had more than two collinear pairs; AhALDH2B3 had five collinear pairs, which revealed that AhALDH2B3 might have a longer and more pivotal evolutionary relationship (Figure 6A). Compared with the Arabidopsis members, AhALDHs had 24 collinear pairs and likely similar functions to those of Arabidopsis collinear genes. Interestingly, both collinear Arabidopsis members and AhALDH members were not distributed on the last chromosome, which potentially reveals some characteristics of the ALDH members (Figures 2, 6B).
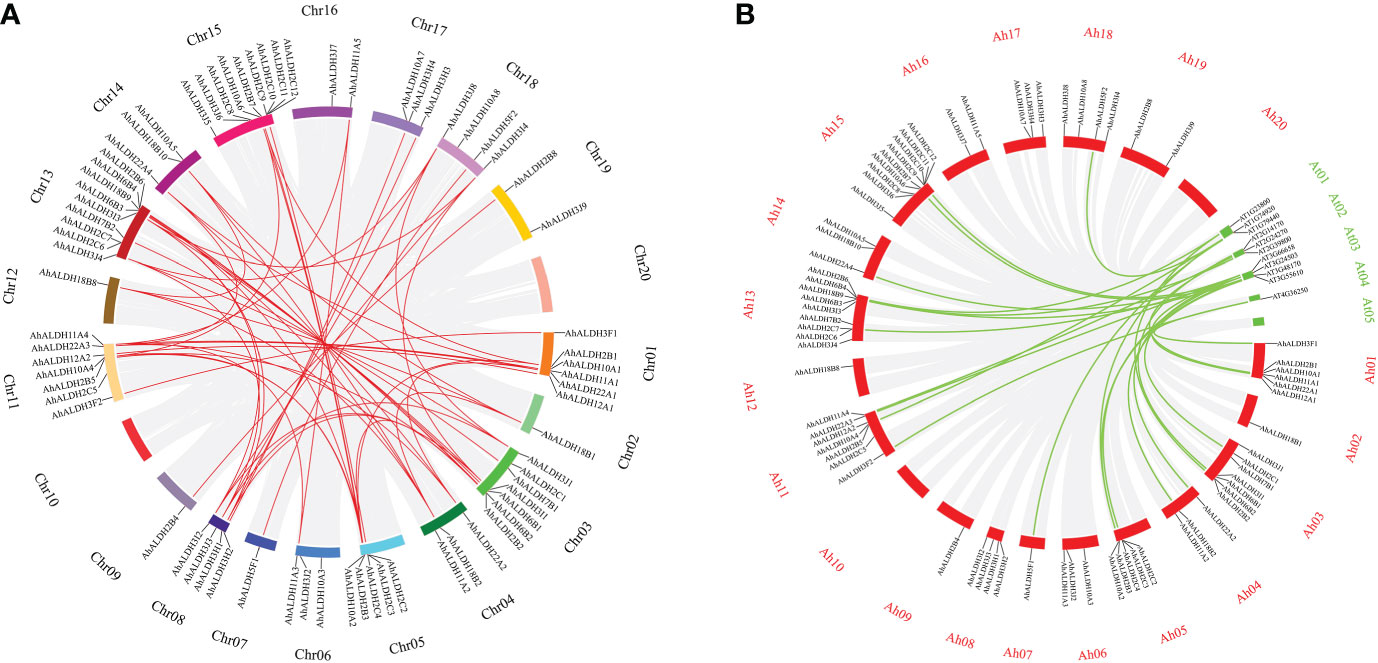
Figure 6 Collinear analysis of AhALDH members in groundnut and Arabidopsis. (A) Collinear analysis of AhALDH members in Arachis hypogaea. The circles of different colors represent different chromosomes, while red lines represent collinear pairs in AhALDHs, and a gray background represents all collinear blocks. (B) Collinear analysis of AhALDHs with Arabidopsis members. The red circles represent groundnut chromosomes, while green circles represent Arabidopsis chromosomes, and the green lines represent collinear gene pairs.
3.7 GO and KEGG enrichment analysis of AhALDHs
The GO and KEGG enrichment analyses (Figure 7) were conducted to understand the function of AhALDHs. The AhALDHs were significantly (P < 0.05) enriched in 81 GO terms (Table S4); the top-10 GO terms were enriched in semialdehyde dehydrogenase activity (6) and amino acid-related terms (4), suggesting that AhALDHs might participate in the growth of plants (Figure 7A). The analysis further revealed that the AhALDHs were significantly (P < 0.05) enriched in 20 pathways (Table S5); eight pathways were related to amino acid metabolism, indicating the role of AhALDHs in the biosynthesis of amino acids (Figure 7B). These results indicated that AhALDHs might exercise function through amino acid pathways.
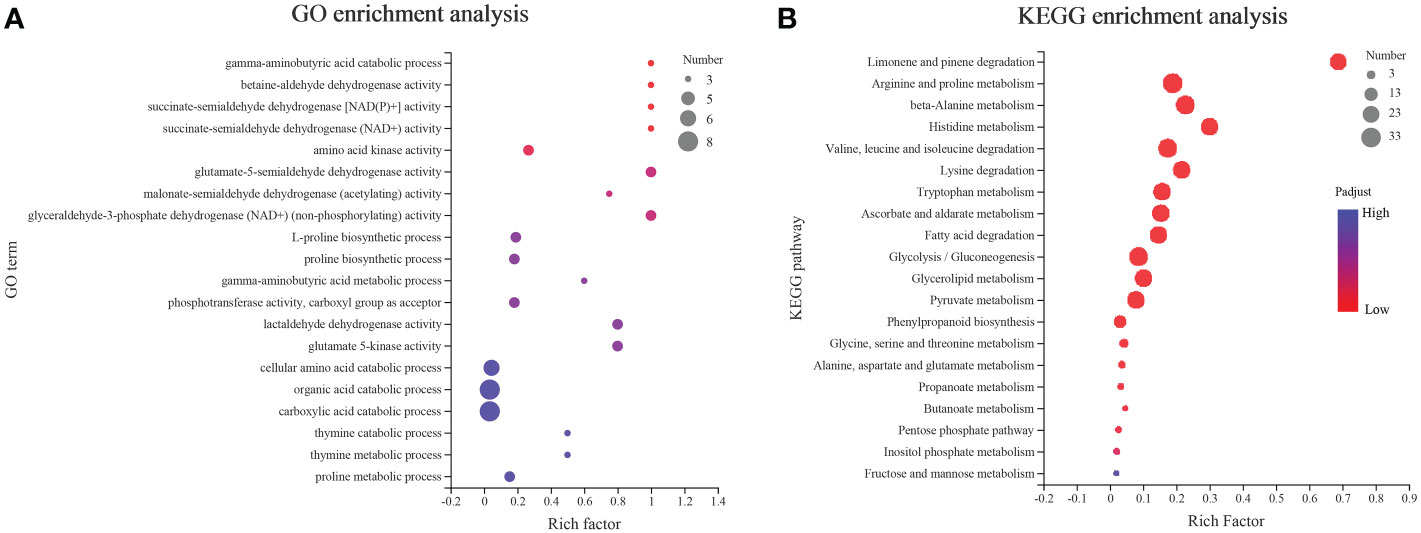
Figure 7 GO and KEGG enrichment analysis of AhALDH members. The size of each circle represents the number of AhALDHs. The transition in color from blue to red represents the P-value from high to low. (A) GO enrichment analysis of AhALDHs. (B) KEGG enrichment analysis of AhALDHs.
3.8 Tissue-specific expression pattern analysis of AhALDHs
The expression pattern of 28 AhALDH members from different tissues encompassing all subfamilies was extracted from the Phytozome database, and a heatmap was drawn using TBtools. The results showed that the expression of AhALDHs was tissue-specific (Figure 8). Some AhALDHs had a higher expression level (such as AhALDH12A1, AhALDH18B1, AhALDH6B2, AhALDH2C7, AhALDH6B4, AhALDH2C11, and AhALDH11A5) in roots than in other tissues; therefore, roots should be used as a target tissue of AhALDHs for further study.
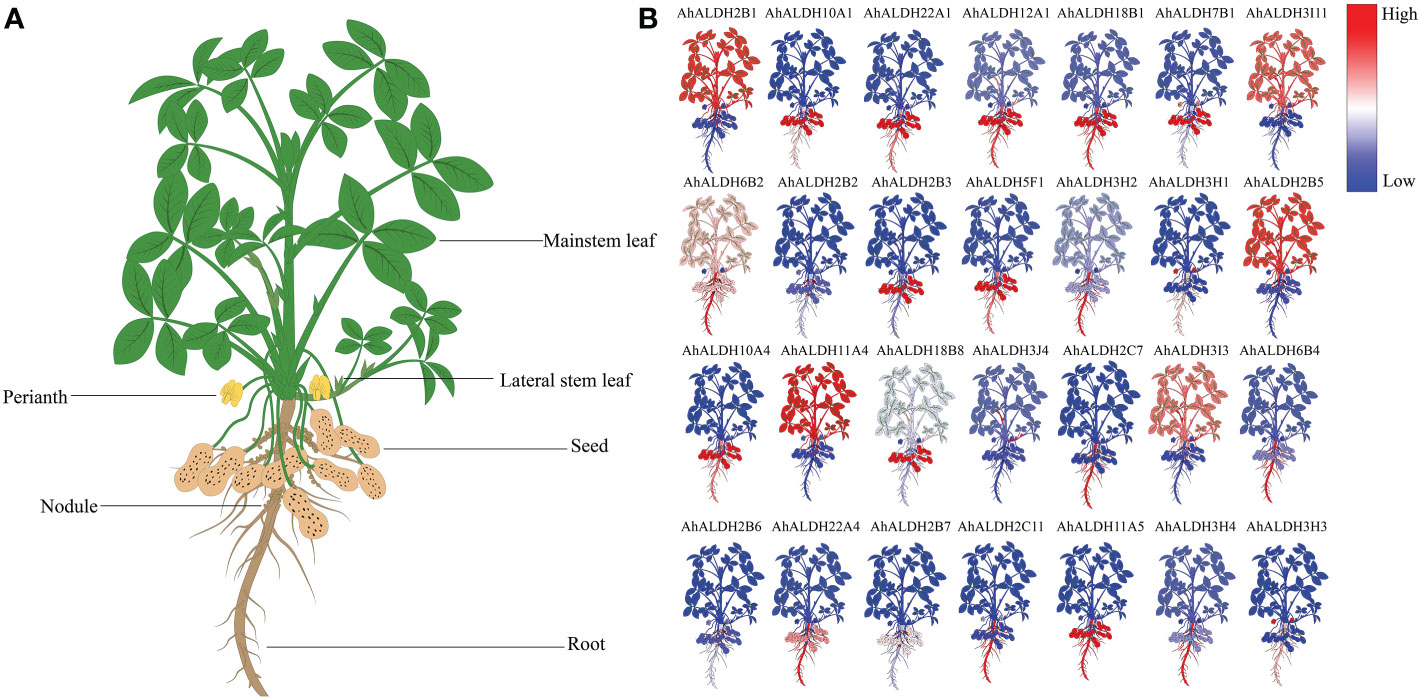
Figure 8 Expression pattern analysis of AhALDHs in different tissues. (A) Schematic diagram of different groundnut tissues; plants were divided into root, nodule, seed, perianth, and leaf. (B) Expression levels of AhALDHs. A change in color from blue to red represents ALDH levels from low to high.
3.9 ALDH activity under saline-alkali stress
The ALDH activity was determined under control and stress conditions at 0, 12, 24, 48, 72, and 96 h (Figure 9). The ALDH activity showed no obvious trend under the control conditions, but the data of activity under saline-alkali stress had increased. The results showed that the ALDH activity began to significantly change at 48 h (P < 0.05). All these results illustrated that 48 h could be used as an simulated stress time for testing the expression of AhALDH members according to ALDH Activity.
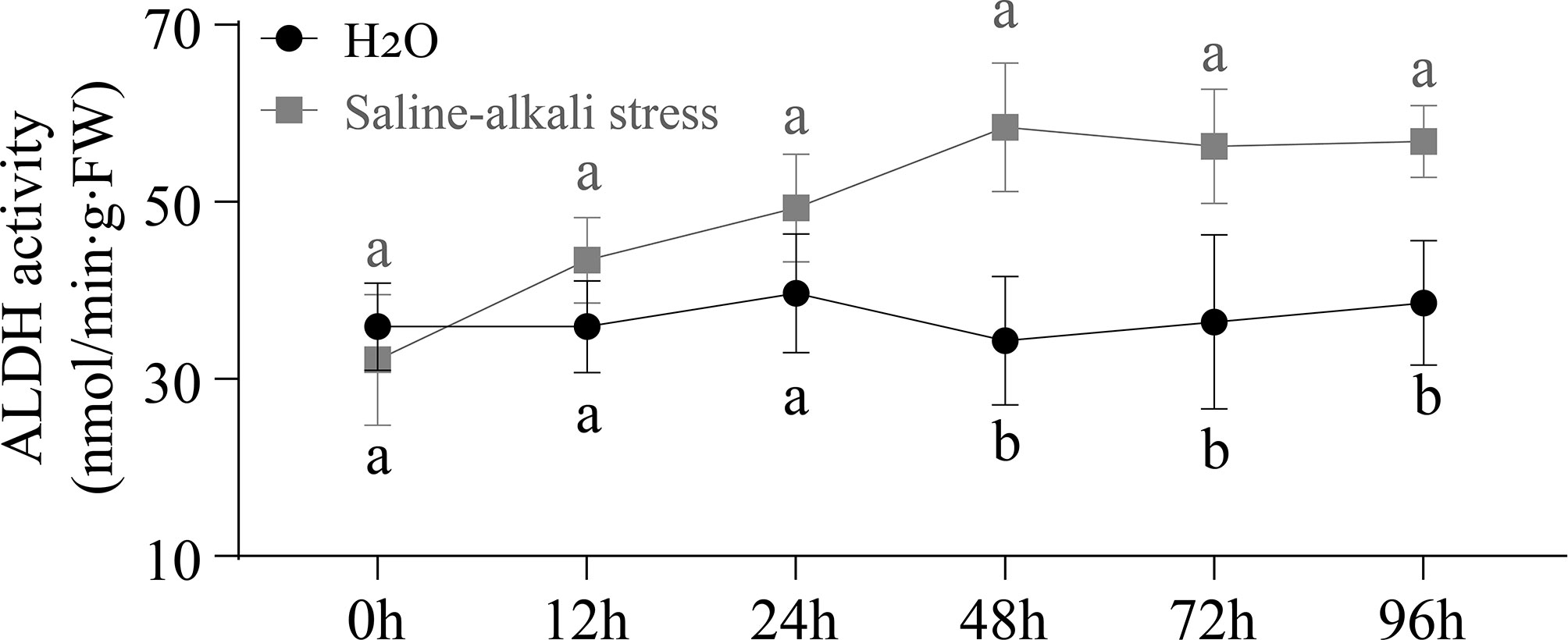
Figure 9 Variation curve of ALDH enzyme activity under control and saline-alkali stress treatment. Black line represents ALDH enzyme activity in H2O treatment at the sprout stage, while gray line represents ALDH enzyme activity in saline-alkali stress treatment.
3.10 Expression analysis under saline-alkali stress
Twelve AhALDHs with higher expression in the roots at the sprout stage, based on the tissue-specific expression pattern analysis, were selected for qRT-PCR analysis under stress, and 48 h was used as the sampling time. The expression of these members significantly differed between the control and stress treatment (P < 0.05). Some AhALDH members, AhALDH10A1, AhALDH22A1, AhALDH12A1, AhALDH6B2, AhALDH3H2, AhALDH3H1, AhALDH10A4, AhALDH2C11, AhALDH11A5, and AhALDH3H3, were upregulated under saline-alkali stress, while others, AhALDH22A4 and AhALDH3H4, were downregulated. These results suggested the role of these members in plant response to abiotic stress; some might be involved in positive regulation, while some might be involved in negative regulation (Figures 10A-L).
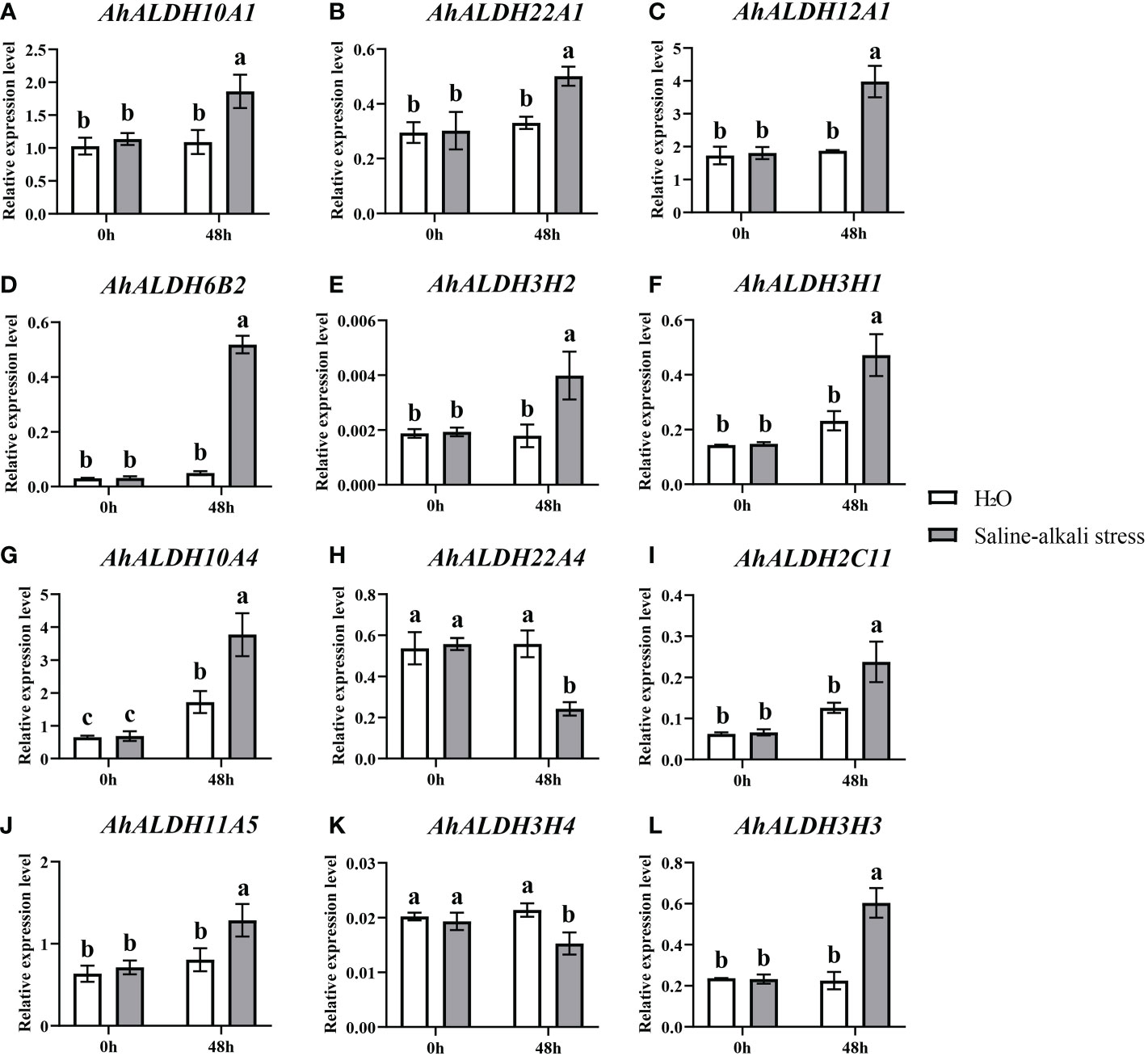
Figure 10 Expression pattern analysis of 12 AhALDHs under saline-alkali stress at 0h and 48h. White columns show the expression in the H2O at the sprout stage, while gray columns show the expression in the saline-alkali stress treatment. (A–L) The expression analysis of 12 AhALDH members.
4 Discussion
Members of the ALDH superfamily have previously been identified in several species (Brocker et al., 2013): 9 ALDH members were identified in Arabidopsis (Kirch et al., 2004), 20 were reported for rice (Gao and Han, 2009), 23 for grape (Zhou et al., 2012), and 19 for tomato (Solanum lycopersicum) (Zhang et al., 2012); 28 for maize (Zhou et al., 2012), 19 for sorghum (Sorghum bicolor) (Islam et al., 2022), and 53 for soybean (Wang et al., 2017). In the present study, 71 ALDH members were identified through the reference genome (Arachis hypogaea v1.0). Such a large number of ALDH members might be related to the large size of the genome and the evolution of the genes (Wang et al., 2017). The evolutionary relationship analysis resolved these members into 10 subfamilies (Man et al., 2020); similarly, VvALDHs, ZmALDHs, and GhALDHs were divided into 10 subfamilies (Zhang et al., 2012; Zhou et al., 2012; Dong et al., 2017), which reveals that 10 subfamilies express the evolutionary relationship characteristics of ALDH members. The evolutionary relationship of seven species confirmed the accuracy of the member classification into 10 subfamilies.
Motif analysis can be used to reveal the function of members according to protein sequences, in which the similar kinds of motifs may have a similar function (Bailey et al., 2015; Liu et al., 2021). The motif analysis of AhALDHs showed that the members in each subfamily shared similar kinds of motifs; these results are in accordance with those reported for ALDH members in soybean (Wang et al., 2017). In cis-acting element analysis, AhALDHs with TGA-element, ABRE, P-box, GARE-motif, TCA-element, AT-rich sequence, SARE, TATC-box, and AuxRR-core were related to hormones, whereas LTR, MBS, ARE, and GC-motif elements were associated with abiotic stress response, suggesting that AhALDHs are related to abiotic stress and hormones. Similarly, the elements LTR, MBS, ABRE, P-box, TCA-element, and AuxRR-core were found in SbALDHs (Islam et al., 2022), whereas LTR, MBS, ARE, ARBE, and TCA-element were predicted in GhALDHs (Guo et al., 2017; Yang et al., 2019). Thus, cis-acting elements such as LTR, MBS, ARE, ARBE, and TCA-element are common cis-acting elements among ALDH members, contributing to the role of AhALDHs in plant response to abiotic stress and hormone interactions (Chen Z, et al., 2014). Some AhALDHs were colinear with Arabidopsis genes; for example, AhALDH10A2 was colinear with AT1G74920, which is involved in plant response to salt and drought (Missihoun et al., 2011); AhALDH18B9 was colinear with AT2G39800 (P5CS1), which acts in abiotic stress response (salt, oxidative, ABA, desiccation, and water deprivation) and participates in proline biosynthesis (Yoshiba et al., 1995; Yoshiba et al., 1999; Huang et al., 2008; Székely et al., 2008); and AhALDH2B2 and AhALDH2B6 were colinear with AT1G23800 (ALDH2B7), a gene that responds to drought and abscisic acid signal (Depuydt and Vandepoele, 2021). These relationships indicated the role AhALDHs play in plant response to abiotic stresses. AhALDHs were enriched in semialdehyde dehydrogenase activity (GO results) and amino acid metabolism (GO and KEGG analysis). Semialdehyde dehydrogenase activity is closely related with abiotic stress and increases stress conditions (Wei et al., 2021; Zhang et al., 2021). Amino acid metabolism usually intensifies during the growth and development of plants especially under stress, such as GST family members (Batista-Silva et al., 2019; Yang et al., 2019). Some amino acids, such as proline, have been used as a parameter for assessing damage to plants under abiotic stress (Ghosh et al., 2022). The enrichment analysis further corroborated the correlation between AhALDHs and abiotic stress.
The expression of several ALDH members, such as GhALDHs, GmALDHs, and SiALDHs, shows a tissue-specific pattern and is higher in roots (Dong et al., 2017; Wang et al., 2017). A similar pattern was observed in the present study, suggesting that the root system is a suitable target for studying ALDH members. Some ALDH members, such as BrALDH7B2, were predicted to function in stress response (Gautam et al., 2019). Similar to that of VvALDHs, SiALDHs, and CaALDHs (Zhang et al., 2012; Chen C, et al., 2014; Carmona-Molero et al., 2021), the expression of some AhALDHs (such as AhALDH10A1, AhALDH22A1, AhALDH12A1, or AhALDH6B2) was significantly upregulated, while it was downregulated in others (such as AhALDH22A4 and AhALDH3H4) under alkali-stress conditions, which these upregulated six members could be considered as cluster genes involved in resistance to abiotic stress.
5 Conclusion
In this study, 71 ALDH members were identified from the groundnut reference genome and classified into 10 subfamilies, with similar motifs and gene structure. AhALDHs were associated with abiotic stress response and hormones via cis-acting elements. The results of collinearity and enrichment analysis (including GO and KEGG pathway analysis) revealed that AhALDHs are involved in plant response to stress and their expression is tissue-specific. The root system is a target tissue suitable for studying AhALDHs, and upregulated members can be used as candidate AhALDHs members (such AhALDH10A1, AhALDH22A1, AhALDH12A1, or AhALDH6B2) involved in resistance to abiotic stress in future research. This study provides insights into AhALDHs and the basis for further research of groundnut.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
Conceptualization: XZ and JZ; methodology: LCa and GY; software: CR; validation: YG, JR, HR, and SZ; formal analysis and investigation: LCh and YZ; writing—review and editing: LW, XZ and YZ; funding acquisition: YZ. All authors contributed to the article and approved the submitted version.
Funding
This research was funded by Heilongjiang Province’s “Revealing the List and Commanding the Leaders” scientific and technological research project (2021ZXJ05B02); China Agriculture Research System of MOF and MARA (CARS-04-PS18); Heilongjiang Bayi Agricultural University Support Program for San Heng San Zong (TDJH202001); Natural Science Foundation of Heilongjiang Province (LH2022115).
Acknowledgments
We thank Heilongjiang Academy of Agricultural Sciences for providing plant materials and Northeast Agricultural University for providing instrument platforms.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1097001/full#supplementary-material
Supplementary Table 1 | The primers sequence of ALDH members in groundnut.
Supplementary Table 2 | The detailed information of ALDH members in groundnut.
Supplementary Table 3 | The information of cis-acting elements.
Supplementary Table 4 | The Gene Ontology (GO) terms enriched in AhALDHs.
Supplementary Table 5 | The Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways enriched in AhALDHs.
References
Bailey, T. L., Boden, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., et al. (2009). MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208. doi: 10.1093/nar/gkp335
Bailey, T. L., Johnson, J., Grant, C. E., Noble, W. S. (2015). The MEME suite. Nucleic Acids Res. 43, W39–W49. doi: 10.1093/nar/gkv416
Bartels, D. (2001). Targeting detoxification pathways: an efficient approach to obtain plants with multiple stress tolerance? Trends Plant Sci. 6, 284–286. doi: 10.1016/s1360-1385(01)01983-5
Bartels, D., Sunkar, R. (2005). Drought and salt tolerance in plants. Crit. Rev. Plant Sci. 24, 23–58. doi: 10.1080/07352680590910410
Batista-Silva, W., Heinemann, B., Rugen, N., Nunes-Nesi, A., Araújo, W. L., Braun, H. P., et al. (2019). The role of amino acid metabolism during abiotic stress release. Plant Cell Environ. 42, 1630–1644. doi: 10.1111/pce.13518
Bezzaouha, A., Bouamra, A., Ammimer, A., Ben Abdelaziz, A. (2020). Non-parametric tests on SPSS to compare two or more means on matched samples. Tunis. Med. 98 (12), 932–941.
Brocker, C., Lassen, N., Estey, T., Pappa, A., Cantore, M., Orlova, V. V., et al. (2010). Aldehyde dehydrogenase 7A1 (ALDH7A1) is a novel enzyme involved in cellular defense against hyperosmotic stress. J. Biol. Chem. 285, 18452–18463. doi: 10.1074/jbc.M109.077925
Brocker, C., Vasiliou, M., Carpenter, S., Carpenter, C., Zhang, Y., Wang, X., et al. (2013). Aldehyde dehydrogenase (ALDH) superfamily in plants: gene nomenclature and comparative genomics. Planta 237, 189–210. doi: 10.1007/s00425-012-1749-0
Carmona-Molero, R., Jimenez-Lopez, J. C., Caballo, C., Gil, J., Millán, T., Die, J. V. (2021). Aldehyde dehydrogenase 3 is an expanded gene family with potential adaptive roles in chickpea. Plants (Basel) 10 (11), 2429. doi: 10.3390/plants10112429
Chen, Z., Chen, M., Xu, Z. S., Li, L. C., Chen, X. P., Ma, Y. Z. (2014). Characteristics and expression patterns of the aldehyde dehydrogenase (ALDH) gene superfamily of foxtail millet (Setaria italica l.). PloS One 9, e101136. doi: 10.1371/journal.pone.0101136
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, C. H., Ferreira, J. C., Gross, E. R., Mochly-Rosen, D. (2014). Targeting aldehyde dehydrogenase 2: new therapeutic opportunities. Physiol. Rev. 94, 1–34. doi: 10.1152/physrev.00017.2013
Chen, L., Zhang, Y. H., Wang, S., Zhang, Y., Huang, T., Cai, Y. D. (2017). Prediction and analysis of essential genes using the enrichments of gene ontology and KEGG pathways. PloS One 12, e0184129. doi: 10.1371/journal.pone.0184129
Depuydt, T., Vandepoele, K. (2021). Multi-omics network-based functional annotation of unknown arabidopsis genes. Plant J. 108, 1193–1212. doi: 10.1111/tpj.15507
Desmae, H., Janila, P., Okori, P., Pandey, M. K., Motagi, B. N., Monyo, E., et al. (2019). Genetics, genomics and breeding of groundnut (Arachis hypogaea l.). Plant Breed 138, 425–444. doi: 10.1111/pbr.12645
Dong, Y., Liu, H., Zhang, Y., Hu, J., Feng, J., Li, C., et al. (2017). Comparative genomic study of ALDH gene superfamily in gossypium: A focus on gossypium hirsutum under salt stress. PloS One 12, e0176733. doi: 10.1371/journal.pone.0176733
Gao, C., Han, B. (2009). Evolutionary and expression study of the aldehyde dehydrogenase (ALDH) gene superfamily in rice (Oryza sativa). Gene 431, 86–94. doi: 10.1016/j.gene.2008.11.010
Gautam, R., Ahmed, I., Shukla, P., Meena, R. K., Kirti, P. B. (2019). Genome-wide characterization of ALDH superfamily in brassica rapa and enhancement of stress tolerance in heterologous hosts by BrALDH7B2 expression. Sci. Rep. 9, 7012. doi: 10.1038/s41598-019-43332-1
Ghosh, U. K., Islam, M. N., Siddiqui, M. N., Cao, X., Khan, M. A. R. (2022). Proline, a multifaceted signalling molecule in plant responses to abiotic stress: understanding the physiological mechanisms. Plant Biol. (Stuttg) 24, 227–239. doi: 10.1111/plb.13363
Goodstein, D. M., Shu, S., Howson, R., Neupane, R., Hayes, R. D., Fazo, J., et al. (2012). Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 40, D1178–D1186. doi: 10.1093/nar/gkr944
Guo, X., Wang, Y., Lu, H., Cai, X., Wang, X., Zhou, Z., et al. (2017). Genome-wide characterization and expression analysis of the aldehyde dehydrogenase (ALDH) gene superfamily under abiotic stresses in cotton. Gene 628, 230–245. doi: 10.1016/j.gene.2017.07.034
Hu, B., Jin, J., Guo, A. Y., Zhang, H., Luo, J., Gao, G. (2015). GSDS 2. 0: an upgraded gene feature visualization server. Bioinformatics 31, 1296–1297. doi: 10.1093/bioinformatics/btu817
Huang, D., Wu, W., Abrams, S. R., Cutler, A. J. (2008). The relationship of drought-related gene expression in arabidopsis thaliana to hormonal and environmental factors. J. Exp. Bot. 59, 2991–3007. doi: 10.1093/jxb/ern155
Ishitani, M., Nakamura, T., Han, S. Y., Takabe, T. (1995). Expression of the betaine aldehyde dehydrogenase gene in barley in response to osmotic stress and abscisic acid. Plant Mol. Biol. 27, 307–315. doi: 10.1007/bf00020185
Islam, M. S., Mohtasim, M., Islam, T., Ghosh, A. (2022). Aldehyde dehydrogenase superfamily in sorghum: genome-wide identification, evolution, and transcript profiling during development stages and stress conditions. BMC Plant Biol. 22, 316. doi: 10.1186/s12870-022-03708-4
Jackson, B., Brocker, C., Thompson, D. C., Black, W., Vasiliou, K., Nebert, D. W., et al. (2011). Update on the aldehyde dehydrogenase gene (ALDH) superfamily. Hum. Genomics 5, 283–303. doi: 10.1186/1479-7364-5-4-283
Jimenez-Lopez, J. C., Gachomo, E. W., Seufferheld, M. J., Kotchoni, S. O. (2010). The maize ALDH protein superfamily: linking structural features to functional specificities. BMC Struct. Biol. 10, 43. doi: 10.1186/1472-6807-10-43
Kelly, G. J., Gibbs, M. (1973). A mechanism for the indirect transfer of photosynthetically reduced nicotinamide adenine dinucleotide phosphate from chloroplasts to the cytoplasm. Plant Physiol. 52, 674–676. doi: 10.1104/pp.52.6.674
Kirch, H. H., Bartels, D., Wei, Y., Schnable, P. S., Wood, A. J. (2004). The ALDH gene superfamily of arabidopsis. Trends Plant Sci. 9, 371–377. doi: 10.1016/j.tplants.2004.06.004
Kirch, H. H., Nair, A., Bartels, D. (2001). Novel ABA- and dehydration-inducible aldehyde dehydrogenase genes isolated from the resurrection plant craterostigma plantagineum and arabidopsis thaliana. Plant J. 28, 555–567. doi: 10.1046/j.1365-313x.2001.01176.x
Kotchoni, S. O., Jimenez-Lopez, J. C., Kayodé, A. P., Gachomo, E. W., Baba-Moussa, L. (2012). The soybean aldehyde dehydrogenase (ALDH) protein superfamily. Gene 495, 128–133. doi: 10.1016/j.gene.2011.12.035
Kotchoni, S. O., Kuhns, C., Ditzer, A., Kirch, H. H., Bartels, D. (2006). Over-expression of different aldehyde dehydrogenase genes in arabidopsis thaliana confers tolerance to abiotic stress and protects plants against lipid peroxidation and oxidative stress. Plant Cell Environ. 29, 1033–1048. doi: 10.1111/j.1365-3040.2005.01458.x
Krapovickas, A., Gregory, W. C. (1994). Taxonomıa del genero arachis (Leguminosae). Bonplandia 8, 1–186. doi: 10.30972/bon.160158
Kumar, S., Stecher, G., Li, M., Knyaz, C., K. Tamura (2018). MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Le Rudulier, D., Strom, A. R., Dandekar, A. M., Smith, L. T., Valentine, R. C. (1984). Molecular biology of osmoregulation. Science 224, 1064–1068. doi: 10.1126/science.224.4653.1064
Lescot, M., Déhais, P., Thijs, G., Marchal, K., Moreau, Y., Van de Peer, Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Letunic, I., Bork, P. (2018). 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 46, D493–d496. doi: 10.1093/nar/gkx922
Liu, Y., Zeng, Z., Zhang, Y. M., Li, Q., Jiang, X. M., Jiang, Z., et al. (2021). An angiosperm NLR atlas reveals that NLR gene reduction is associated with ecological specialization and signal transduction component deletion. Mol. Plant 14, 2015–2031. doi: 10.1016/j.molp.2021.08.001
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Man, J., Gallagher, J. P., Bartlett, M. (2020). Structural evolution drives diversification of the large LRR-RLK gene family. New Phytol. 226, 1492–1505. doi: 10.1111/nph.16455
Marchitti, S. A., Brocker, C., Stagos, D., Vasiliou, V. (2008). Non-P450 aldehyde oxidizing enzymes: the aldehyde dehydrogenase superfamily. Expert Opin. Drug Metab. Toxicol. 4, 697–720. doi: 10.1517/17425255.4.6.697
Missihoun, T. D., Schmitz, J., Klug, R., Kirch, H. H., Bartels, D. (2011). Betaine aldehyde dehydrogenase genes from arabidopsis with different sub-cellular localization affect stress responses. Planta 233, 369–382. doi: 10.1007/s00425-010-1297-4
Nong, X., Zhong, S. N., Li, S. M., Yang, Y. J., Liang, Z., Xie, Y. (2019). Genetic differentiation of pseudoregma bambucicola population based on mtDNA COII gene. Saudi J. Biol. Sci. 26, 1032–1036. doi: 10.1016/j.sjbs.2019.04.016
Shin, J. H., Kim, S. R., An, G. (2009). Rice aldehyde dehydrogenase7 is needed for seed maturation and viability. Plant Physiol. 149, 905–915. doi: 10.1104/pp.108.130716
Singh, S., Brocker, C., Koppaka, V., Chen, Y., Jackson, B. C., Matsumoto, A., et al. (2013). Aldehyde dehydrogenases in cellular responses to oxidative/electrophilic stress. Free Radic. Biol. Med. 56, 89–101. doi: 10.1016/j.freeradbiomed.2012.11.010
Skibbe, D. S., Liu, F., Wen, T. J., Yandeau, M. D., Cui, X., Cao, J., et al. (2002). Characterization of the aldehyde dehydrogenase gene families of zea mays and arabidopsis. Plant Mol. Biol. 48, 751–764. doi: 10.1023/a:1014870429630
Székely, G., Abrahám, E., Cséplo, A., Rigó, G., Zsigmond, L., Csiszár, J., et al. (2008). Duplicated P5CS genes of arabidopsis play distinct roles in stress regulation and developmental control of proline biosynthesis. Plant J. 53, 11–28. doi: 10.1111/j.1365-313X.2007.03318.x
Tylichová, M., Kopecný, D., Moréra, S., Briozzo, P., Lenobel, R., Snégaroff, J., et al. (2010). Structural and functional characterization of plant aminoaldehyde dehydrogenase from pisum sativum with a broad specificity for natural and synthetic aminoaldehydes. J. Mol. Biol. 396, 870–882. doi: 10.1016/j.jmb.2009.12.015
Vasiliou, V., Pappa, A., Petersen, D. R. (2000). Role of aldehyde dehydrogenases in endogenous and xenobiotic metabolism. Chem. Biol. Interact. 129, 1–19. doi: 10.1016/s0009-2797(00)00211-8
Wang, W., Jiang, W., Liu, J., Li, Y., Gai, J., Li, Y. (2017). Genome-wide characterization of the aldehyde dehydrogenase gene superfamily in soybean and its potential role in drought stress response. BMC Genomics 18, 518. doi: 10.1186/s12864-017-3908-y
Wei, M. Y., Liu, J. Y., Li, H., Hu, W. J., Shen, Z. J., Qiao, F., et al. (2021). Proteomic analysis reveals the protective role of exogenous hydrogen sulfide against salt stress in rice seedlings. Nitric. Oxide 111-112, 14–30. doi: 10.1016/j.niox.2021.04.002
Wen, Y., Wang, X., Xiao, S., Wang, Y. (2012). Ectopic expression of VpALDH2B4, a novel aldehyde dehydrogenase gene from Chinese wild grapevine (Vitis pseudoreticulata), enhances resistance to mildew pathogens and salt stress in arabidopsis. Planta 236, 525–539. doi: 10.1007/s00425-012-1624-z
Yang, J., An, D., Zhang, P. (2011). Expression profiling of cassava storage roots reveals an active process of glycolysis/gluconeogenesis. J. Integr. Plant Biol. 53, 193–211. doi: 10.1111/j.1744-7909.2010.01018.x
Yang, Q., Han, X. M., Gu, J. K., Liu, Y. J., Yang, M. J., Zeng. Functional, Q. Y. (2019). And structural profiles of GST gene family from three populus species reveal the sequence-function decoupling of orthologous genes. New Phytol. 221, 1060–1073. doi: 10.1111/nph.15430
Yang, H., Zhang, D., Li, H., Dong, L., Lan, H. (2015). Ectopic overexpression of the aldehyde dehydrogenase ALDH21 from syntrichia caninervis in tobacco confers salt and drought stress tolerance. Plant Physiol. Biochem. 95, 83–91. doi: 10.1016/j.plaphy.2015.07.001
Yoshiba, Y., Kiyosue, T., Katagiri, T., Ueda, H., Mizoguchi, T., Yamaguchi-Shinozaki, K., et al. (1995). Correlation between the induction of a gene for delta 1-pyrroline-5-carboxylate synthetase and the accumulation of proline in arabidopsis thaliana under osmotic stress. Plant J. 7, 751–760. doi: 10.1046/j.1365-313x.1995.07050751.x
Yoshiba, Y., Kiyosue, T., Nakashima, K., Yamaguchi-Shinozaki, K., Shinozaki, K. (1997). Regulation of levels of proline as an osmolyte in plants under water stress. Plant Cell Physiol. 38, 1095–1102. doi: 10.1093/oxfordjournals.pcp.a029093
Yoshiba, Y., Nanjo, T., Miura, S., Yamaguchi-Shinozaki, K., Shinozaki, K. (1999). Stress-responsive and developmental regulation of Delta(1)-pyrroline-5-carboxylate synthetase 1 (P5CS1) gene expression in arabidopsis thaliana. Biochem. Biophys. Res. Commun. 261, 766–772. doi: 10.1006/bbrc.1999.1112
Yoshida, A., Rzhetsky, A., Hsu, L. C., Chang, C. (1998). Human aldehyde dehydrogenase gene family. Eur. J. Biochem. 251, 549–557. doi: 10.1046/j.1432-1327.1998.2510549.x
Zhang, Y., Jiang, D., Yang, C., Deng, S., Lv, X., Chen, R., et al. (2021). The oxidative stress caused by atrazine in root exudation of pennisetum americanum (L. ) k. schum. Ecotoxicol Environ. Saf. 211, 111943. doi: 10.1016/j.ecoenv.2021.111943
Zhang, Y., Mao, L., Wang, H., Brocker, C., Yin, X., Vasiliou, V., et al. (2012). Genome-wide identification and analysis of grape aldehyde dehydrogenase (ALDH) gene superfamily. PloS One 7, e32153. doi: 10.1371/journal.pone.0032153
Zhang, Q., Zhang, W. J., Yin, Z. G., Li, W. J., Zhao, H. H., Zhang, S., et al. (2020). Genome- and transcriptome-wide identification of C3Hs in common bean (Phaseolus vulgaris l. ) and structural and expression-based analyses of their functions during the sprout stage under salt-stress conditions. Front. Genet. 11. doi: 10.3389/fgene.2020.564607
Zhou, S., Chen, X., Zhang, X., Li, Y. (2008). Improved salt tolerance in tobacco plants by co-transformation of a betaine synthesis gene BADH and a vacuolar Na+/H+ antiporter gene SeNHX1. Biotechnol. Lett. 30, 369–376. doi: 10.1007/s10529-007-9548-6
Keywords: aldehyde dehydrogenase, evolutionary, cis-acting elements, expression pattern, saline-alkali stress
Citation: Zhang X, Zhong J, Cao L, Ren C, Yu G, Gu Y, Ruan J, Zhao S, Wang L, Ru H, Cheng L, Wang Q and Zhang Y (2023) Genome-wide characterization of aldehyde dehydrogenase gene family members in groundnut (Arachis hypogaea) and the analysis under saline-alkali stress. Front. Plant Sci. 14:1097001. doi: 10.3389/fpls.2023.1097001
Received: 13 November 2022; Accepted: 02 February 2023;
Published: 16 February 2023.
Edited by:
Zhengjun Xia, Chinese Academy of Sciences (CAS), ChinaReviewed by:
Rongxia Guan, Chinese Academy of Agricultural Sciences (CAAS), ChinaDayong Zhang, Nanjing Agricultural University, China
Copyright © 2023 Zhang, Zhong, Cao, Ren, Yu, Gu, Ruan, Zhao, Wang, Ru, Cheng, Wang and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yuxian Zhang, MTM4MzY5NjIyMTFAMTI2LmNvbQ==
 Xiaoming Zhang
Xiaoming Zhang Jingwen Zhong1
Jingwen Zhong1 Chunyuan Ren
Chunyuan Ren Gaobo Yu
Gaobo Yu