- Key Laboratory of Sweet Potato Biology and Biotechnology, Ministry of Agriculture and Rural Affairs/Beijing Key Laboratory of Crop Genetic Improvement/Laboratory of Crop Heterosis and Utilization, Ministry of Education, College of Agronomy and Biotechnology, China Agricultural University, Beijing, China
Leaves comprise one of the most important organs for plant growth and development. Although there have been some reports on leaf development and the establishment of leaf polarity, their regulatory mechanisms are not very clear. In this study, we isolated a NAC (NAM, ATAF, and CUC) transcription factor (TF), i.e., IbNAC43, from Ipomoea trifida, which is a wild ancestor of sweet potato. This TF was highly expressed in the leaves and encoded a nuclear localization protein. The overexpression of IbNAC43 caused leaf curling and inhibited the growth and development of transgenic sweet potato plants. The chlorophyll content and photosynthetic rate in transgenic sweet potato plants were significantly lower than those in wild-type (WT) plants. Scanning electron microscopy (SEM) and paraffin sections showed that the ratio of cells in the upper and lower epidermis of the transgenic plant leaves was unbalanced; moreover, the abaxial epidermal cells were irregular and uneven in transgenic plants. In addition, the xylem of transgenic plants was more developed than that of WT plants, while their lignin and cellulose contents were significantly higher than those of WT. Quantitative real-time PCR (qRT-PCR) analysis showed that the overexpression of IbNAC43 upregulated the genes involved in leaf polarity development and lignin biosynthesis in transgenic plants. Moreover, it was found that IbNAC43 could directly activate the expression of the leaf adaxial polarity-related genes IbREV and IbAS1 by binding to their promoters. These results indicate that IbNAC43 might play a critical role in plant growth by affecting the establishment of leaf adaxial polarity. This study provides new insights regarding leaf development.
Introduction
Plant yield and quality cannot be achieved without normal growth and development (Gong et al., 2020). Leaves comprise one of the most important plant organs where CO2 is fixed to produce carbohydrates through photosynthesis (Guo and Gan, 2014). The morphology of leaves is an important aspect of plant architecture that affects light interception, water loss, photosynthesis, and respiration (Otoole and Cruz, 1980; Hou et al., 2020). Moderate leaf curling helps increase the light-receiving space and improves the utilization rate of light energy, thereby increasing the yield of plants (Xu et al., 2018). Leaf curling also enhances drought tolerance by reducing water loss (Li et al., 2017). However, severe leaf curling often leads to growth retardation and developmental defects (Yuan et al., 2015). The shape of the leaves generally depends on the establishment of leaf polarity, in which adaxial–abaxial polarity plays an important role in leaf curling (Manuela and Xu, 2020) and is regulated by complex molecular mechanisms. Therefore, understanding the established mechanisms of leaf adaxial–abaxial polarity is extremely important for improving plant growth and resistance to abiotic stress.
Recently, genetic studies have reported that different genes affect leaf curling in various species, such as in Arabidopsis thaliana (Husbands et al., 2015), tomato (Pulungan et al., 2018), soybean (Yang et al., 2019), and rice (Sun et al., 2020; Xu et al., 2021). Leaf curling can be differentiated into two types based on the classification of plants, i.e., monocotyledons and dicotyledons. In monocotyledonous plants, such as rice, the phenotype of leaf curling, in general, is caused by the change of bulliform cells (Li et al., 2010; Fang et al., 2012; Li et al., 2016; Sun et al., 2020; Xu et al., 2021). On the other hand, leaf curling in dicotyledonous plants is mainly related to the adaxial–abaxial leaf polarity establishment, cutin and cuticular wax biosynthesis, and hormone responses (Wan et al., 2021). The first gene associated with leaf polarity, phantastica (PHAN), was required for the elaboration of the proximodistal axis (Waites and Hudson, 1995; Waites et al., 1998). Since then, extensive evidence has indicated that the regulation pathway of leaf polarity is extremely complex and that many leaf development-associated genes are involved in regulatory processes. The members of the HD-bZIP III family—REVOLUTA (REV), PHABULOSA (PHB), and PHAVOLUTA (PHV)—are involved in the control of leaf adaxial cells (McConnell et al., 2001; Otsuga et al., 2001). In addition, ASYMMETRIC LEAVES1/AtMYB91 (AS1) physically interacts with ASYMMETRIC LEAVES2 (AS2) to maintain the normal development of leaves together with the trans-acting small interfering RNA (ta-siRNA) pathway (Iwakawa et al., 2007; Husbands et al., 2015; Iwakawa et al., 2020). On the other hand, the members of the YABBY family—AtYAB3/FIL, AtYAB5, BcYAB3, and GmFILa—regulate the fate of leaf abaxial cells (Sawa et al., 1999; Siegfried et al., 1999; Yang et al., 2019; Hou et al., 2020). At present, there are only a few reports on NAC (NAM, ATAF, and CUC) transcription factors (TFs) involved in the regulation of leaf curling.
The NAC family is one of the important TF families in plants, and NACs play important roles in plant growth, development, and abiotic and biotic stress responses (Puranik et al., 2012; Nuruzzaman et al., 2013; Kim et al., 2016). Genetic studies have revealed that most NACs, including ANAC092/ORESARA1 (AtORE1), ANAC029/Arabidopsis NAC-LIKE Activated by AP3/PI (AtNAP), ANAC059/ORESARA1 SISTER1 (AtORS1), AtATAF2, AtANAC016, and NaNAC29, as positive regulators, and ANAC042/JUNGBRUNNEN1 (AtJUB1) and ANAC083/VND-INTERACTING2 (AtVNI2), as negative regulators, are mainly involved in the control of leaf senescence (Balazadeh et al., 2011; Yang et al., 2011; Wu et al., 2012; Kim et al., 2013; Kim et al., 2016; Nagahage et al., 2020; Ma et al., 2021). Furthermore, the NAC TFs CUC1, CUC2, and CUC3 specify the formation of leaf boundary (Vroemen et al., 2003; Laufs et al., 2004; Hibara et al., 2006). At present, there is no report on the function of NAC43 in plants.
Sweet potato is a globally important food crop known for its strong adaptability, abundant nutrient content, high yield, and diverse uses (Ahn et al., 2010). In general, the morphology and photosynthetic activity of leaves can affect plant growth and crop yield (Zhu et al., 2010). Although it is important to understand the mechanism of leaf polarity development, there have been no reports on the leaf development-related genes in sweet potato plants, particularly the leaf curling-related genes. In this study, we isolated the NAC TF IbNAC43 from Ipomoea trifida, which is a wild ancestor of sweet potato. The overexpression of this TF caused leaf curling, reduced the photosynthetic rate, and retarded growth in transgenic sweet potato. These results indicate that IbNAC43 plays an important role in leaf polarity development and plant growth.
Materials and methods
Plant materials and growth conditions
I. trifida was used to isolate the IbNAC43 gene. The sweet potato cultivar Shangshu 19 was employed to analyze the function of IbNAC43. In vitro-grown transgenic sweet potato plants (Shangshu 19) were cultured on Murashige and Skoog (MS) medium at 27 ± 1°C under a photoperiod consisting of 13 h of cool white fluorescent light at 54 μmol m−2 s−1 and 11 h darkness. Subsequently, the sweet potatoplants were cultivated in a field or glasshouse (25 ± 3°C, natural illumination) at the campus of China Agricultural University, Beijing, China.
Cloning and sequence analysis of IbNAC43
Total RNA of sweet potato plants was extracted using the RNAprep Pure Plant Kit (Tiangen Biotech, Beijing, China), and first-strand complementary DNA (cDNA) synthesis was performed using PrimeScript™ II 1st Strand cDNA Synthesis Kit (TaKaRa, Beijing, China). Primer pairs (IbNAC43-ORF-F/R) were used to amplify the full-length cDNA of IbNAC43 (Supplementary Table S1). The protein of IbNAC43 was analyzed using online BLAST (TBLASTX, RRID : SCR_011823; https://blast.ncbi.nlm.nih.gov/Blast.cgi). The DNAMAN software (LynnonBiosoft, San Ramon, CA, USA) was utilized to align the amino acid (aa) sequence of IbNAC43 with those of the NAC43 proteins from different plant species. A phylogenetic tree was constructed using MEGA 5.0 software (downloaded from https://megasoftware.net/) with the neighbor-joining method, which had 1,000 bootstrap replications (Kumar et al., 2018).
Expression analysis of IbNAC43 in sweet potato
The transcript levels of IbNAC43 in the roots, stems, and leaves were measured with Shangshu 19 using quantitative real-time PCR (qRT-PCR). The expression levels were normalized to those of Ibactin (AY905538). The related qRT-PCR primer pairs are listed in Supplementary Table S1.
Subcellular localization of IbNAC43
The IbNAC43 open reading frame (ORF) without a stop codon, amplified with a primer pair (1300-IbNAC43-F/R) (Supplementary Table S1), was ligated into pCAMBIA1300-GFP. The recombinant vectors IbNAC43-GFP, NLR-RFP (a nuclear localization marker), and 35S:GFP were transformed into the Agrobacterium tumefaciens strain EHA105 using the heat shock method. The translational fusion construct and NLS-RFP were transiently expressed in Nicotiana benthamiana leaf epidermal cells via infiltration of A. tumefaciens (Verweij et al., 2008). The construct vectors 35S:IbNAC43-GPF and 35S:GFP were transformed into rice protoplasts isolated from etiolated shoots via treatment with polyethylene glycol (Du et al., 2021; Sun et al., 2022). Subsequently, the green and red fluorescent protein (GFP and RFP, respectively) signals were collected using confocal laser scanning microscopy (LSM880; Zeiss, Oberkochen, Germany).
Transactivation activity assay
Full-length IbNAC43 and 1–167, 167–372, 1–201, 1–235, 1–269, and 1–303 aa of the IbNAC43 coding sequence were amplified by PCR using different primer pairs (Supplementary Table S1) and inserted into the NdeI/EcoRI-digested pGBKT7 vector (Oebiotech; https://www.oebiotech.com/) to produce the fusion constructs. The fusion plasmids, a negative control (pGBKT7-empty), and a positive control (pGAL4) were separately transformed into the yeast strain AH109. The transformed yeast was streaked on SD/-Trp and SD/-Trp/-His/X-α-Gal plates for two-three days at 30°C to observe yeast growth.
Production of transgenic sweet potato plants
The expression vector pCAMBIA1300-IbNAC43, containing the IbNAC43 sequence, was controlled by the CaMV 35S promoter and an NOS terminator. The recombinant plasmids were then transformed into EHA105. Embryogenic suspension cultures of Shangshu 19 were prepared as described by Liu et al. (2001). Transformation and plant regeneration were performed with A. tumefaciens-mediated transformation, as previously described by Zhai et al. (2016) and Sun et al. (2022). Putative transgenic sweet potato plants were identified by PCR and qRT-PCR analyses with the primer pairs 35s-F/JD-IbNAC43-R and qRT-IbNAC43-F/R (Supplementary Table S1). The transgenic lines and wild-type (WT) plants were transferred in soils in a greenhouse and then grown in a field for phenotypic observation.
Phenotype identification of transgenic plants
The leaf rolling index (LRI) of the WT and transgenic plants was measured after two and eight weeks in the greenhouse. These measurements are consistent with the method of Sun et al. (2020), as follows: the leaf blade was stretched out, and its maximum width (Lw) calculated. At the same site, the natural distance of the leaf blade margins (Ln) was measured. The LRI was calculated as follows: LRI (%) = (Lw − Ln)/Lw × 100.
Scanning electron microscopy
Transgenic and WT plants were grown in a greenhouse for four weeks, and then the features of the adaxial and abaxial surfaces in the same regions were observed during the mature leaf stage. Leaves were immediately fixed with 2.5% (v/v) glutaraldehyde for 2.5 h. After rinsing three times with 0.1 mol L−1 phosphoric acid buffer, the leaves were fixed in 1% (v/v) osmic acid for 2 h, followed by dehydration with the LEICA EM CPD 300 (Leica, Wetzlar, Germany) critical point dryer. EIKO IB-3 (Variable Electron Microscope, S3400N; Hitachi, Tokyo, Japan) was used to sputter gold on each sample, and surface images of the leaves were collected using a scanning electron microscope.
Leaf and stem paraffin sections
For the paraffin sections, fully unfolded leaves and stems at the same position were detached from the transgenic and WT plants grown in the greenhouse, fixed with FAA [5% (v/v) glacial acetic acid, 5% (v/v) formalin, and 50% (v/v) ethanol] solution, and evacuated for 30 min with a vacuum pump. The fixed materials were then dehydrated with gradient ethanol (50%, 70%, 80%, 90%, 95%, and 100%, v/v) and gradient xylene (25%, 50%, 75%, and 100% xylene in ethanol, v/v). Subsequently, the samples were soaked in paraffin at 60°C for three days (the paraffin solution was replaced three times during this period), after which they were embedded in a paraffin block. Approximately 8-μm sections were obtained using a microtome (LEICA RM2265, Leica, Germany), dewaxed in xylene, and then stained with safranin and Fast Green FCF. Finally, the images of these sections were observed using an Echo Revolve light microscope (ECHO, San Diego, CA, USA).
Lignin content measurement
The methods for the measurement of the lignin content followed those of Suzhou Comin Biotechnology Co., Ltd. (http://www.cominbio.com/a/shijihe/shenghuashiji/qitaxilie/2016/0502/969.html). The main steps were as follows: 1) for sample preparation, the leaves and stems were dried at 80°C to a constant weight, crushed, and weighed to about 5 mg; 2) in accordance with the instructions in the Suzhou Comin Lignin Kit (MZS-2-G), various reagents were added in sequence, and the supernatant was taken for examination; and 3) for sample determination, a UV–visible spectrophotometer was used to measure the absorbance at 280 nm.
Cellulose content measurement
Similarly, the cellulose content was measured according to Suzhou Comin Biotechnology Co., Ltd. (http://www.cominbio.com/a/shijihe/shenghuashiji/tangdaixiexilie/2016/0324/843.html). The main steps were as follows: 1) for sample preparation, the leaves and stems were dried at 80°C to a constant weight, crushed, and weighed to about 10 mg; 2) according to the instructions in the Suzhou Comin Cellulose Kit (CLL-1-Y), various reagents were added in sequence and the supernatant taken for examination; and 3) for sample determination, a UV–visible spectrophotometer was used to measure the absorbance at 620 nm.
qRT-PCR analysis
Transgenic and WT plants were grown in a greenhouse for four weeks and their leaves were used to measure the expression of the leaf polarity-related genes. qRT-PCR was performed to quantify the messenger RNA (mRNA) expression levels. qRT-PCR was carried out using a QuantStudio 6 Flex (Applied Biosystems by Thermo Fisher Scientific, Waltham, MA, USA) with the SYBR Fast Universal qPCR Kit (Forscience, Beijing, China). The primers used for qRT-PCR are listed in Supplementary Table S1. Relative gene expression was quantified using the ΔΔCT method (Pfaffl, 2001).
Chromatin immunoprecipitation assay
The chromatin immunoprecipitation (ChIP) assay was accomplished as previously described (Bowler et al., 2004; Zheng et al., 2021). Briefly, fresh in vitro-grown materials were collected and subjected to vacuum infiltration in 1% (v/v) formaldehyde for 15 min to crosslink the chromatin proteins to the DNA. Fixation was stopped by adding glycine to a final concentration of 0.125 M. The fixed samples were washed three times with ddH2O and ground to a powder in liquid nitrogen. Chromatin was extracted and sonicated using an Ultrasonic Homogenizer (SCIENTZ, JY 92-IIDN, Ningbo, China). The anti-GFP antibody was purchased from Abmart Shanghai Co., Ltd. (M20004M, Shanghai, China). The immunoprecipitated DNA was then analyzed with quantitative PCR (qPCR), and the amplified DNA from the chromatin fractions prior to antibody incubation was used as the control. PCR reactions were performed in triplicate for each sample, and the enrichments were normalized to the input sample. The β-actin gene was used as an endogenous control. The gene-specific primers used for ChIP–qPCR are listed in Supplementary Table S1.
Electrophoretic mobility shift assay
The full-length IbNAC43 cDNA was amplified with specific primers and fused into the BamHI and EcoRI sites of the expression vector pET-28a (Novagen; https://www.merckmillipore.com/CN/zh). The construct vector was introduced into Escherichia coli BL21 (DE3) cells to produce recombinant 6×His-IbNAC43 proteins induced by 0.03 mM isopropylthio-β-galactoside (IPTG) and then grown at 16°C. The 6×His-IbNAC43 protein was purified with Ni-NTA Agarose (Forscience, Beijing, China) as described previously (Zhang et al., 2022). Labeled probes with biotin at their 5′-ends were used as binding probes, while unlabeled probes were used as competitors. The primer and probe sequences are listed in Supplementary Table S1. The electrophoretic mobility shift assay (EMSA) was performed using a LightShift Chemiluminescent EMSA Kit (Thermo Fisher, Waltham, MA, USA) according to the manufacturer’s instructions.
Dual-luciferase assay
The coding sequence of IbNAC43 was cloned into the PstI and KpnI sites of the pGreenII 62-SK vector, which was used as an effector. The empty pGreenII 62-SK vector was used as a negative control. The IbREV and IbAS1 promoters (KpnI and PstI sites) were inserted upstream of luciferase (LUC) into the pGreenII 0800-LUC vector to generate the IbREVpro : LUC and IbAS1pro:LUC reporter constructs. Rice protoplasts were isolated and used for dual-LUC assays as described previously (Zhang et al., 2020). The activity levels of firefly and Renilla (REN) LUC were measured using the Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA). LUC activity was normalized to REN activity. The specific primers used for the dual-LUC assay are listed in Supplementary Table S1. Three biological replicates were performed for this analysis.
Y1H assay
For the yeast one-hybrid (Y1H) assay, the partial IbCCoAoMT and IbPAL promoter sequences were cloned, recombined with the pAbAi vector, and then integrated with the genome of the Y1H-Gold yeast strain. The ORF of IbNAC43 was amplified, cloned into the pGADT7 vector, and transferred into the bait yeast strain. The detailed process was in accordance with the Yeast One-Hybrid System—Matchmaker Gold Kit (catalog no. 630491; Clontech, Mountain View, CA, USA). The co-transformed yeast cells were cultured on the SD/-Leu plate with or without aureobasidin A (AbA) at 30°C for 48 h.
Statistical analysis
The values are presented as the mean ± SD. A Student’s t-test was used to examine significant differences. Significance levels were determined at *p < 0.05 and **p < 0.01.
Results
Cloning and sequence analysis of IbNAC43 and its promoter
The IbNAC43 cDNA sequence was 1,492 bp in length and contained a 1,119 bp ORF encoding a 372-aa protein with a predicted molecular weight of 41.78 kDa, and the IbNAC43 protein had a conserved NAM (no apical meristem) domain. The genomic sequence of IbNAC43 was 2,501 bp in length and contained three exons and two introns (Figure 1). To elaborate on the difference in NAC43 between the WT and cultivated sweet potato, Shangshu 19 was also used to isolate the IbNAC43 gene. The protein of NAC43 in I. trifida showed a high degree of sequence identity with that in Shangshu 19 (98.92%), and the genomic sequence of IbNAC43 in Shangshu 19 also contained three exons and two introns (Supplementary Figure S1). Phylogenetic analysis showed that IbNAC43 had a close relationship with Durio zibethinus (XP_022773628.1, 55.07%), Nicotiana attenuata (XP_019229017.1, 50.53%), and Ipomoea nil (XP_019168629.1, 47.58%) (Figure 1). A 1,618-bp fragment corresponding to the promoter of IbNAC43 was cloned from I. trifida, and this promoter region contained numerous types of cis-acting elements, such as the abscisic acid-responsive element (ABRE), G-box, gibberellin-responsive element (GARE)-motif, MYB, and MYC, which are associated with different types of stresses (Supplementary Table S2).
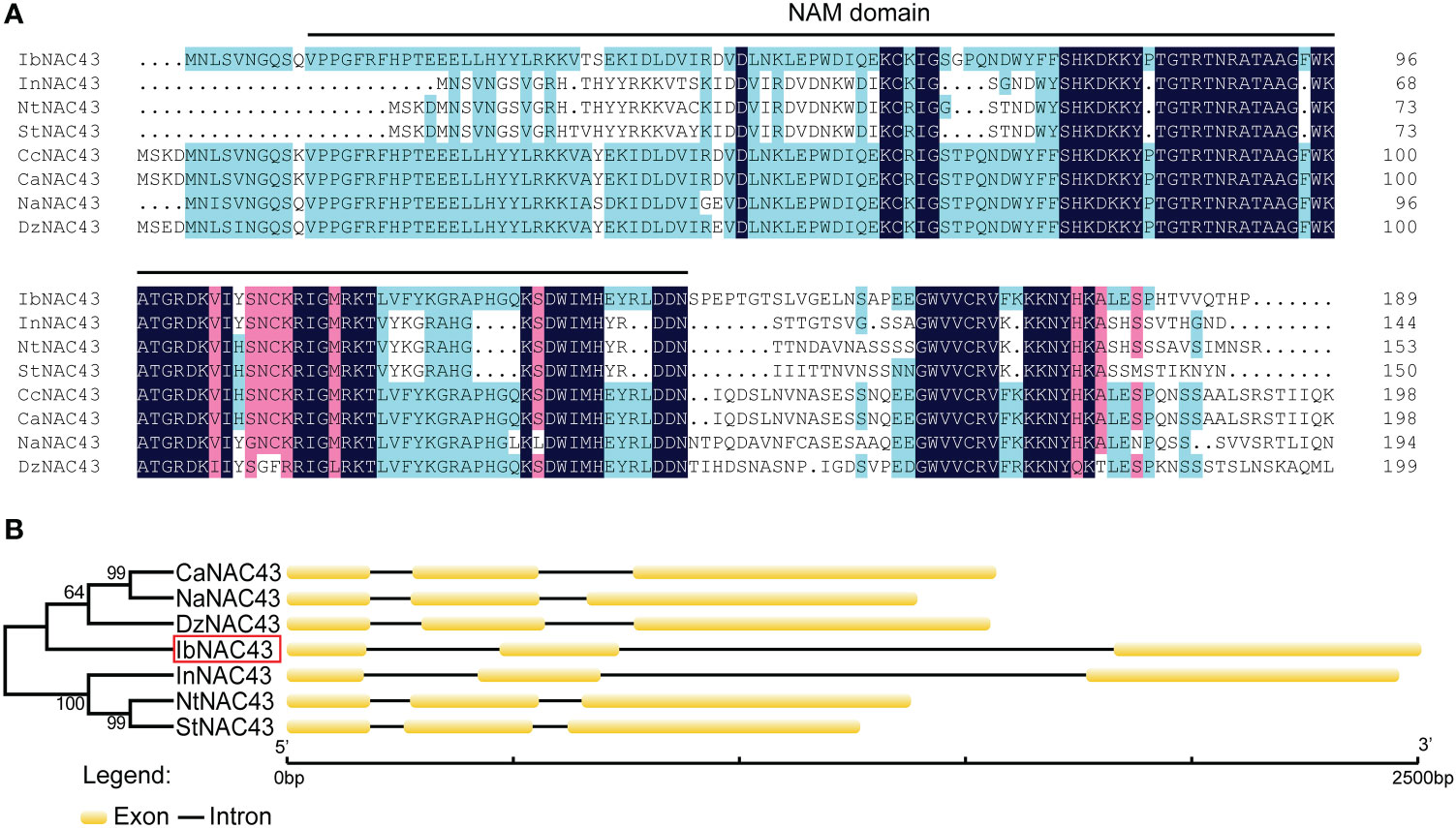
Figure 1 Sequence analysis of IbNAC43. (A) Multiple sequence alignment of the bNAC43 protein and its closest homologs from different plant species. The NAM (no apical meristem) domain is represented by black lines. (B) Phylogenetic relationships and genomic structures of IbNAC43 and its closest homologs from different plant species. The IbNAC43 cloned in this study is indicated by a red box. Exons are represented by boxes, while introns are represented by lines.
Expression of IbNAC43 in different issues
A qRT-PCR analysis was performed to examine the expression level of IbNAC43 in sweet potato. The results showed that IbNAC43 was more highly expressed in the leaves and stems than in the roots (Figure 2). This result indicates that IbNAC43 might be involved in the development and function of leaves.
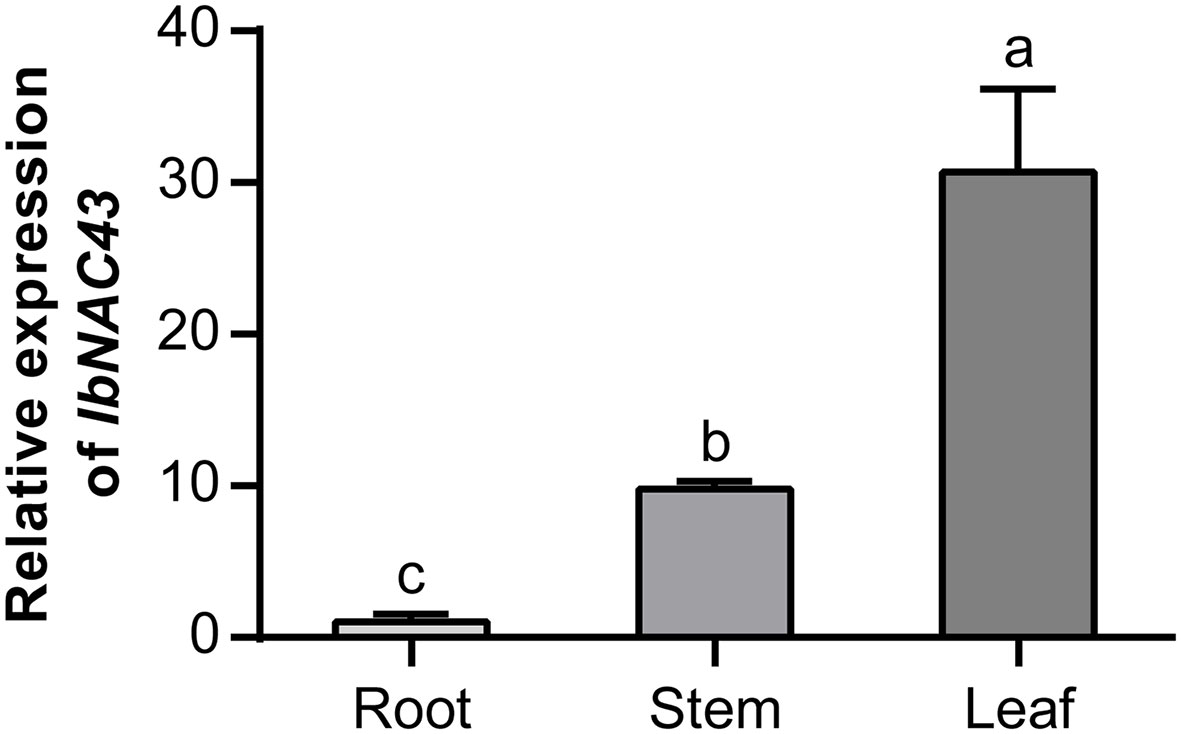
Figure 2 Analysis of the expression of IbNAC43 in the root, stem, and leaf tissues of Shangshu 19. Data are presented as the mean ± SE (n = 3). Different lowercase letters indicate significant differences (p < 0.05, one-way ANOVA).
IbNAC43 is a nuclear localization protein and has self-activation in yeast cells
To study the subcellular localization of IbNAC43, the ORF of IbNAC43 was fused with GFP, generating the fusion construct IbNAC43–GFP. The fusion construct and a nuclear localization protein, NLS-RFP, were then transiently co-expressed in N. benthamiana leaf epidermal cells using A. tumefaciens-mediated transformation. Microscopic visualization demonstrated that the fluorescence of the IbNAC43 fusion protein perfectly overlapped with that of NLS-RFP, indicating that IbNAC43 was localized in the nucleus (Figure 3A). We further analyzed the subcellular localization of IbNAC43 in rice protoplasts, and the result was consistent with the above-mentioned result (Figure 3B).
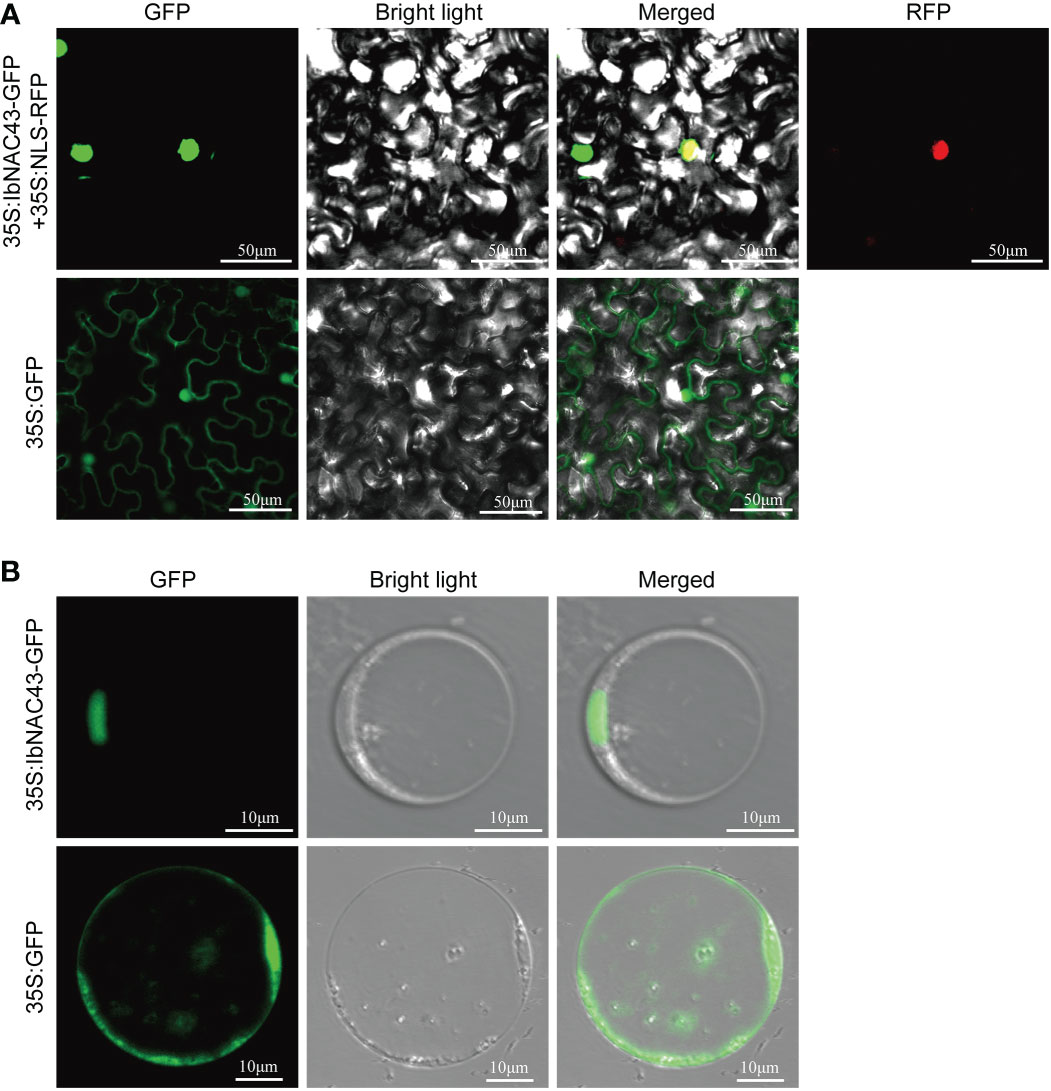
Figure 3 Subcellular localization of IbNAC43 in tobacco leaf epidermal cells (A) and rice protoplasts (B). Confocal scanning microscopy images show that the 35S:IbNAC43-GFP fusion protein is localized in the nucleus, whereas the control 35S:GFP is not.
To investigate the transactivation activity of IbNAC43, the different pGBKT7–IbNAC43 (1–372, 1–166, and 167–372 aa) fusion constructs and the pGBKT7 (negative control) and pGAL4 (positive control) vectors were separately transformed into the yeast strain AH109. Yeast cells containing any of the five vectors grew well on the SD/-Trp medium. On the other hand, yeast cells containing the N0 (1–372 aa), N2 (167–372 aa), and pGAL4 vectors grew well on the SD/-Trp/-His/X-α-Gal medium, while those containing the pGBKT7-IbNAC43 (1–166 aa) and pGBKT7 vectors did not grow (Figure 4A; Supplementary Figure S2). These results indicate that IbNAC43 acted as a transcription activator and that the self-transcriptional activation domain was located in the C-terminal region. To determine whether any of the regions in IbNAC43 are responsible for TF activity, a series of fusion constructs, including N3 (1–201 aa), N4 (1–235 aa), N5 (1–269 aa), and N6 (1–303 aa) were transformed into yeast cells (Figure 4A). Results showed that yeast cells only harboring N6 or pGAL4 grew well on the SD/-Trp/-His/X-α-Gal medium (Figure 4B). The above results indicate that the region of 270–303 aa at the C-terminus was critical for the transactivation activity.
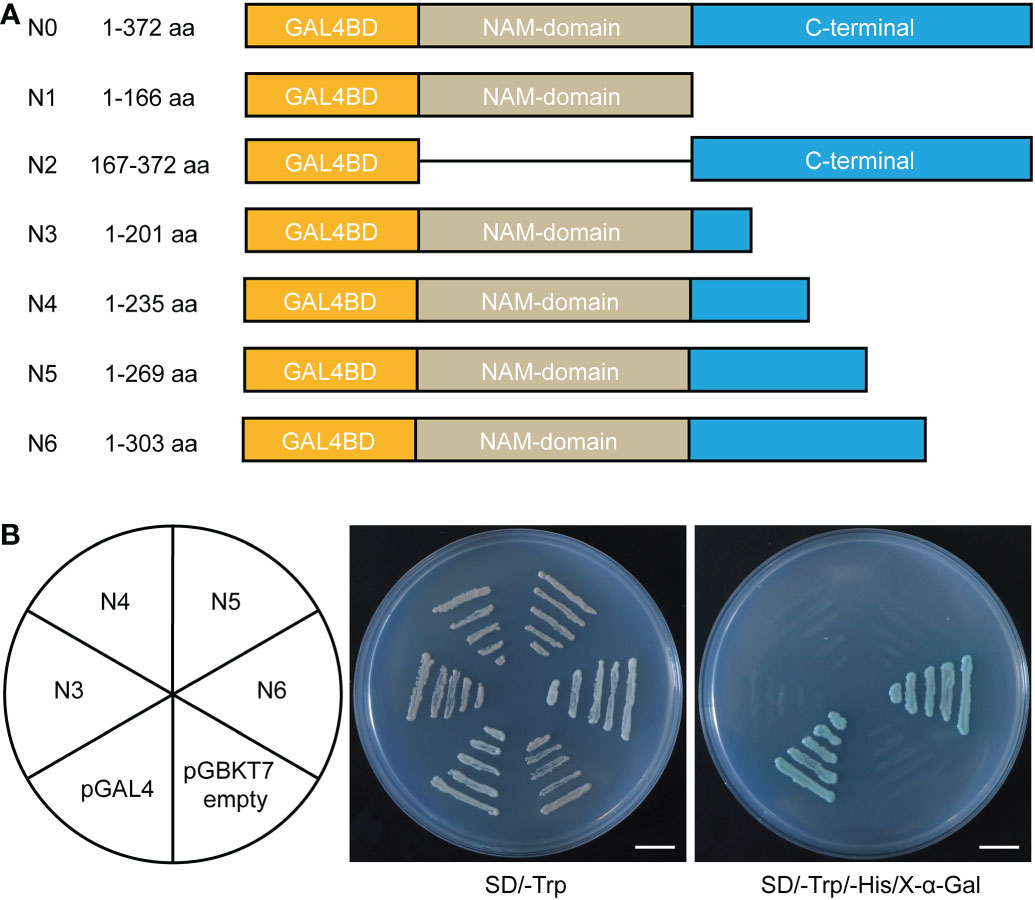
Figure 4 Transactivation activity assay of full-length and truncated IbNAC43 in yeast. (A) Schematic diagrams representing the IbNAC43 fragments encoding different parts of IbNAC43 that were cloned into the pGBKT7 vector. (B) Different fusion constructs were introduced into the yeast strain AH109 and examined on SD/-Trp and SD/-Trp/-His/X-α-Gal selection media. The pGBKT7 empty vector and pGAL4 were used as negative and positive controls, respectively. Scale bar, 1 cm.
Overexpression of IbNAC43 causes leaf curling and plant dwarfism in sweet potato
To evaluate the function of IbNAC43, this TF was transformed into the sweet potato cultivar Shangshu 19 using the A. tumefaciens-mediated transformation method. A total of six positive transgenic lines were obtained and confirmed by PCR. The qRT-PCR analysis showed that transgenic plants exhibited significantly higher expression levels of IbNAC43 compared to WT plants (Supplementary Figure S3). The overexpression lines of IbNAC43 showed a constitutively inward rolling phenotype from in vitro growth in MS medium to growth in the field (Figure 5A). In addition, we found that the degree of leaf curling increases with the growth of the plant. Compared to that in WT plants, the LRI in transgenic plants added up to ~0.25 at two weeks and to ~0.45 at eight weeks (Figure 5B; Supplementary Figure S4).
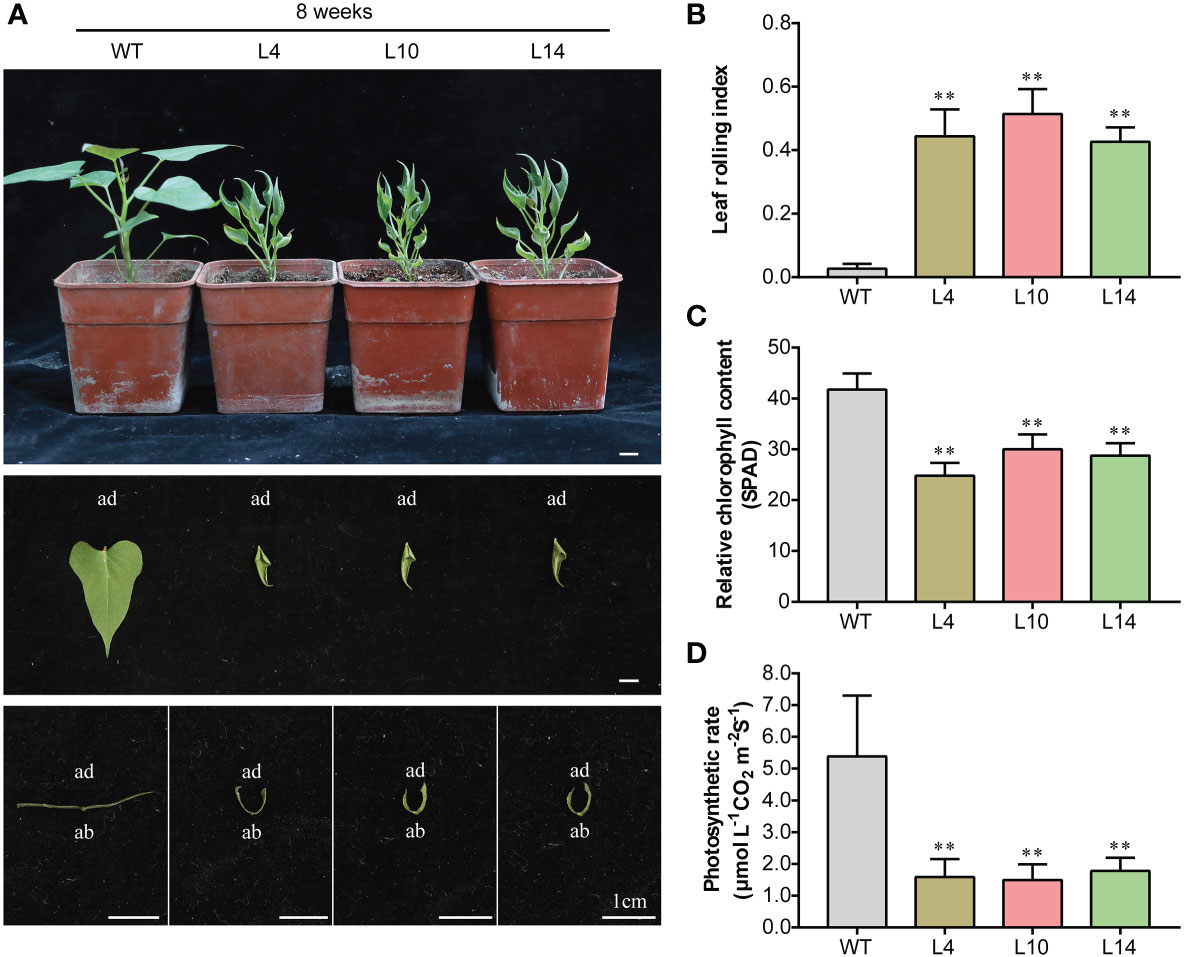
Figure 5 Phenotypes of transgenic and wild-type (WT) sweet potato plants in the greenhouse. (A) Plant stature and leaf phenotypes of both WT and transgenic plants. Scale bar, 1 cm. ad, adaxial side; ab, abaxial side. (B) Leaf rolling index (LRI). Data are presented as the mean ± SE (n = 6). (C) Relative chlorophyll content. (D) Photosynthetic rate. Data are presented as the mean ± SE (n = 3). Asterisks denote significant differences based on a Student’s t-test. **p < 0.01.
On the contrary, transgenic plants appeared dwarfed and showed growth retardation, with the size of their leaves being smaller compared to that of WT plants (Supplementary Figure S3E; Figure 5A); moreover, transgenic plants showed abnormal growth in the field (Supplementary Figure S3I). To elaborate on the reason for the dwarf phenotype, the analysis revealed that transgenic plants had a lower relative chlorophyll content and photosynthetic rate than WT plants (Figures 5C, D). These results indicate that the overexpression of IbNAC43 caused leaf curling and dwarfism in transgenic sweet potato plants.
Subsequently, the expression of some cell cycle genes (e.g., CYCA1;1, CYCA2;2, CYCB2;3, and CYCB3;1) and cell expansion genes (e.g., EXPA1, EXPA5, and EXPA15) (Xu et al., 2016) was analyzed with qRT-PCR. The results showed that the cell cycle-related genes CYCA1;1, CYCA2;2, CYCB2;3, and CYCB3;1 were downregulated in transgenic plants (Supplementary Figure S5). There were no significant differences between the cell expansion-related genes in WT and transgenic plants (Supplementary Figure S5). These results indicate that overexpression of IbNAC43 might inhibit plant growth by negatively regulating a number of cell cycle genes.
IbNAC43 affects the balance of the adaxial and abaxial epidermis
As the morphological development of leaves depends on the morphology of the leaf epidermal cells, we thus compared the epidermal cells of transgenic and WT plants. The adaxial epidermal cells of the leaves were regular and uniform in both transgenic and WT plants. However, the size of the adaxial epidermal cells in WT plants was larger than that in transgenic plants (Figure 6A). In WT plants, the abaxial epidermal cells were regularly shaped and flat, whereas those of transgenic plants were irregular and uneven (Figure 6A). Thereafter, we further observed the cross-sections of the leaf blades in WT and transgenic plants. Compared to WT plants, transgenic plants displayed an interesting phenomenon in which the palisade mesophyll appeared to have a disorderly distribution (Figure 6B). These results indicate that the overexpression of IbNAC43 caused leaf curling by affecting the ratio of adaxial to abaxial epidermal cells in transgenic sweet potato plants (Figure 6C).
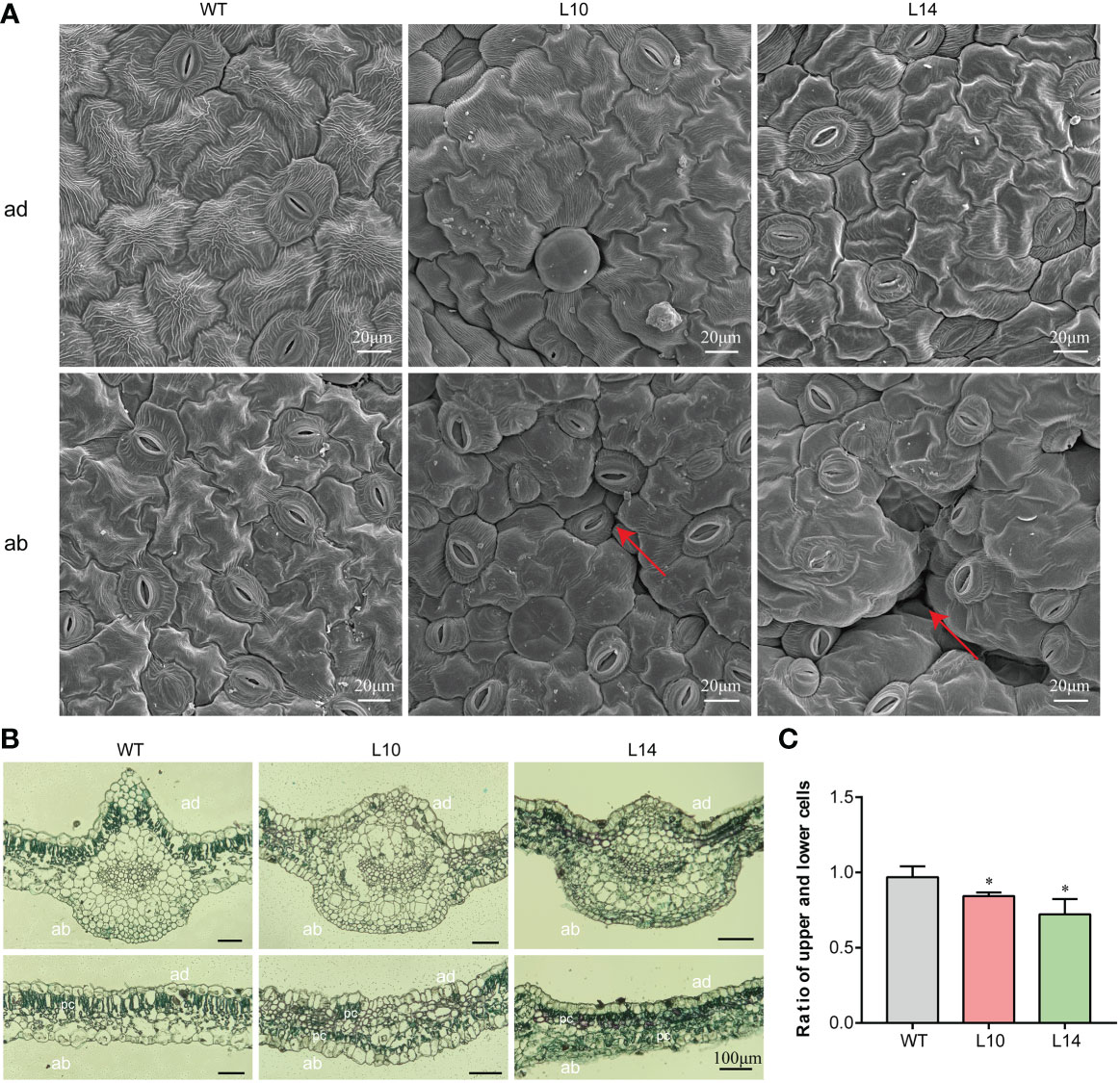
Figure 6 Cytological observations of the IbNAC43-overexpressed and wild-type (WT) plants. (A) The adaxial and abaxial sides of the leaf were subjected to SEM. ad, adaxial side; ab, abaxial side. Scale bar, 20 μm. (B) Paraffin sections of leaves. pc, palisade cells. Scale bar, 100 μm. (C) The ratio of the upper to lower epidermal cells. Data are values from three independent experiments. Asterisk indicates a significant difference compared to the WT based on a Student’s t-test. *p < 0.05.
IbNAC43 upregulates leaf adaxial polarity genes
Previous reports showed that the establishment of leaf adaxial–abaxial polarity is associated with different genes in dicotyledonous plants, including the leaf adaxial (REV, AS1, ATHB8, and HAT1) and leaf abaxial (FILa) polarity genes. In addition, the plant hormone auxin was also found to cause leaf development defects, and AUX1 was reported to influence leaf polarity by facilitating the movement of auxin from outside to inside the cells (Yang and Murphy, 2009). We examined the relative expression of these genes in WT and transgenic sweet potato plants. The adaxial-specific genes IbREV, IbAS1, IbATH8, and IbHAT1 were significantly upregulated in the transgenic plants compared to WT plants (Figure 7). Specifically, the expression level of IbAUX1 was significantly increased in transgenic plants (Figure 7).
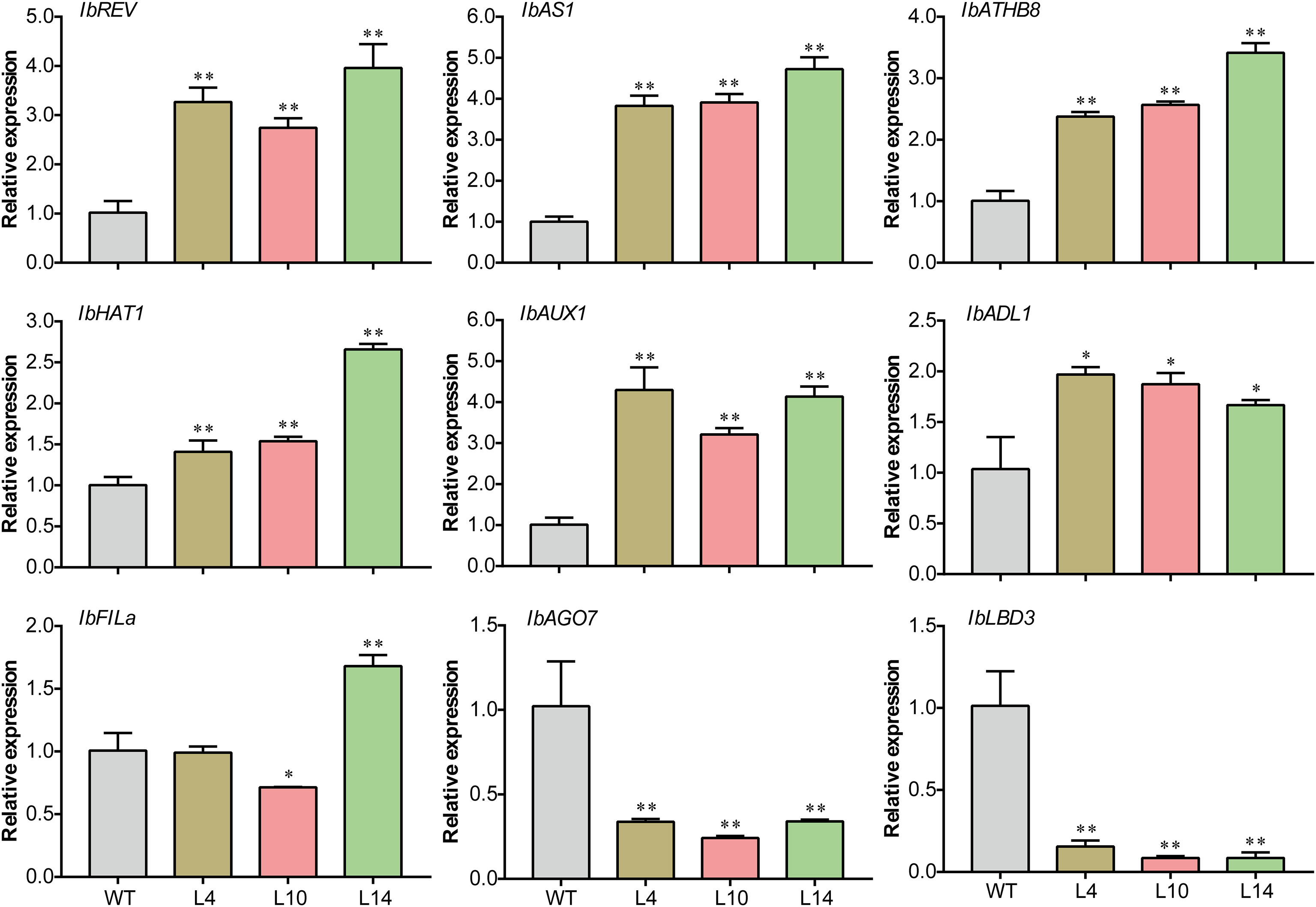
Figure 7 Expression of the genes associated with leaf polarity in both IbNAC43-overexpressed and wild-type (WT) sweet potato plants. Data are presented as the mean ± SE (n = 3). Asterisks indicate significant differences compared to the WT based on a Student’s t-test. *p < 0.05, **p < 0.01.
Moreover, several genes that regulate leaf rolling have been identified in monocotyledonous plants such as rice (Sun et al., 2020; Xu et al., 2021). Genes homologous to the rice leaf rolling genes were detected in transgenic and WT plants, including IbAGO7, IbLBD3, and IbADL1. Compared to that in WT plants, the expression of IbADL1 in transgenic plants was upregulated, whereas that of IbAGO7 and IbLBD3 was downregulated (Figure 7). These results indicate that IbNAC43 might affect the development of leaves by regulating the expression of polarity-related genes.
IbNAC43 participates in the establishment of leaf adaxial polarity by binding to the promoters of IbREV and IbAS1
NAC TFs function mainly by directly binding to the NAC binding sequences (NBS) containing CGTG/A/C and/or CACG (Nakashima et al., 2012; Kim et al., 2016; Diao et al., 2020). To investigate whether IbNAC43 can directly regulate the expression of the leaf adaxial polarity-related genes (i.e., REV, AS1, ATHB8, AUX1, and HAT1), we first established whether these genes contain the NBS in their promoter regions. It was found that the promoters of REV and AS1 had different numbers of NBS elements (Figures 8A, B). This result indicates that IbNAC43 might bind to the promoters of REV and AS1. To confirm this possibility, we then performed the ChIP–qPCR assay using samples from OE-6 (overexpressed line 6). In this experiment, it was revealed that the association of IbNAC43 with the genomic regions of REV and AS1 was significantly increased compared to the empty vector WT (Figures 8A, B).
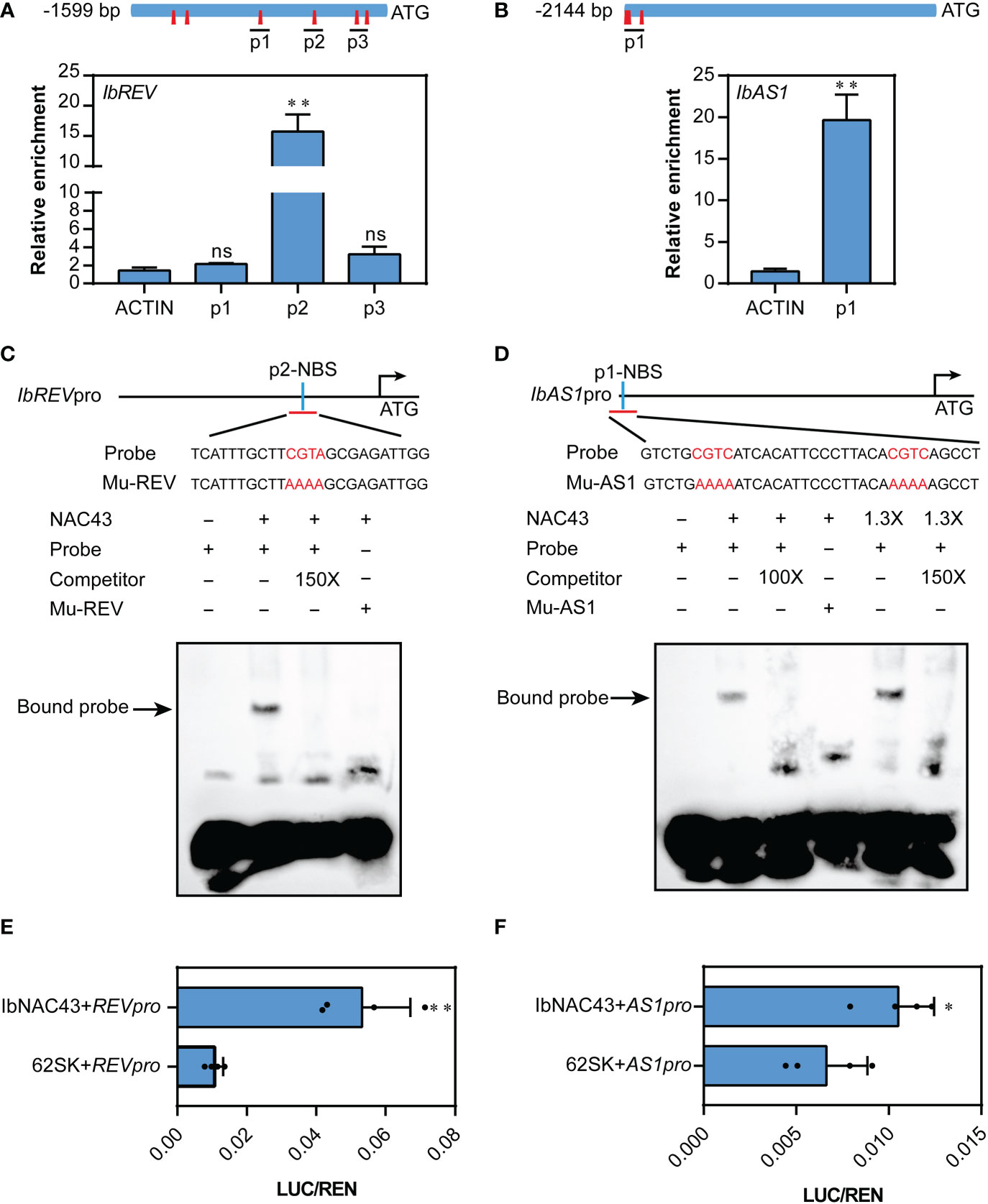
Figure 8 IbNAC43 directly binds to the promoters of IbREV and IbAS1. (A, B) Locations of chromatin immunoprecipitation (ChIP)–quantitative PCR (qPCR) fragments and qPCR analysis of three biological replicates for the binding enrichment of IbNAC43 in IbREV and IbAS1 promoter regions. The PCR primers listed in Supplementary Table S1 were designed to amplify the IbREV and IbAS1 promoter regions. The NAC binding sequences (NBS) are represented by red prisms. (C, D) Electrophoretic mobility shift assays (EMSAs) demonstrate the interactions between IbNAC43 and the DNA motifs. The recombinant protein 6×His-IbNAC43 retarded the shift of the probe. Both 100× and 150× unlabeled probes were used as competitors. The plus sign and minus sign indicate presence and absence, respectively. NBS, NAC binding sequences containing CGTG/A/C; Mu-REV, CGTA was mutated as AAAA; Mu-AS1, CGTG was mutated as AAAA. (E, F) IbNAC43 triggered the activity of IbREVpro-LUC and IbAS1pro-LUC, as determined by the dual-luciferase assays in rice protoplasts. The expression level of Renilla (REN) was used as an internal control. Data are values from four independent experiments and are presented as the mean ± SE (n = 4). Asterisks indicate significant differences according to a Student’s t-test. *p < 0.05, **p < 0.01. ns, not significant.
Subsequently, we performed EMSA to further confirm the binding of IbNAC43 to these promoters in vitro. IbNAC43 bound directly to the IbREV or IbAS1 promoter through the NBS motif, and binding was completed by excess unlabeled probes (Figures 8C, D). These results indicate that IbNAC43 directly binds to the promoters of REV and AS1.
In addition, we also examined whether IbNAC43 could directly regulate the expression levels of REV and AS1 through transient dual-luciferase assays in protoplasts. LUC/REN levels were significantly increased when IbREVpro-LUC was co-transformed with 62SK-IbNAC43 and IbAS1pro-LUC was co-transformed with 62SK-IbNAC43 (Figures 8E, F), indicating that IbNAC43 could activate the expression levels of IbREV and IbAS1.
Overexpression of IbNAC43 promotes vascular bundle development
Previous studies showed that ATHB8 can regulate leaf and vascular development and that its overexpression promotes vascular cell differentiation and lignin biosynthesis (Baima et al., 2001; Gardiner et al., 2011). In this study, the overexpression of IbNAC43 upregulated the expression level of IbATHB8. We speculated that vascular cells might be affected in transgenic plants. Therefore, the paraffin sections of the leaves and stems were used for cytological observations. Stronger red staining was found in transgenic plants compared to WT plants (Figure 9A). The content of lignin in the leaves and stems was then determined. Compared to those of WT plants, the lignin and cellulose contents of transgenic plants were significantly increased (Figures 9B, C). The expression levels of the lignin synthesis-related genes (i.e., IbPAL, IbCoMT, and IbCCoAOMT) were significantly upregulated in the transgenic sweet potato plants compared to the WT (Figures 9D, E). We investigated whether IbNAC43 can directly bind the promoters of IbPAL, IbCoMT, and IbCCoAoMT. Firstly, bioinformatics analysis revealed that the promoters of IbPAL and IbCCoAoMT had NBS motifs. Secondly, the Y1H assay indicated that IbNAC43 was unable to bind to the NBS of their promoters (Supplementary Figure S6). The above results suggest that the overexpression of IbNAC43 promoted vascular bundle development in the leaves and stems of transgenic sweet potato plants.
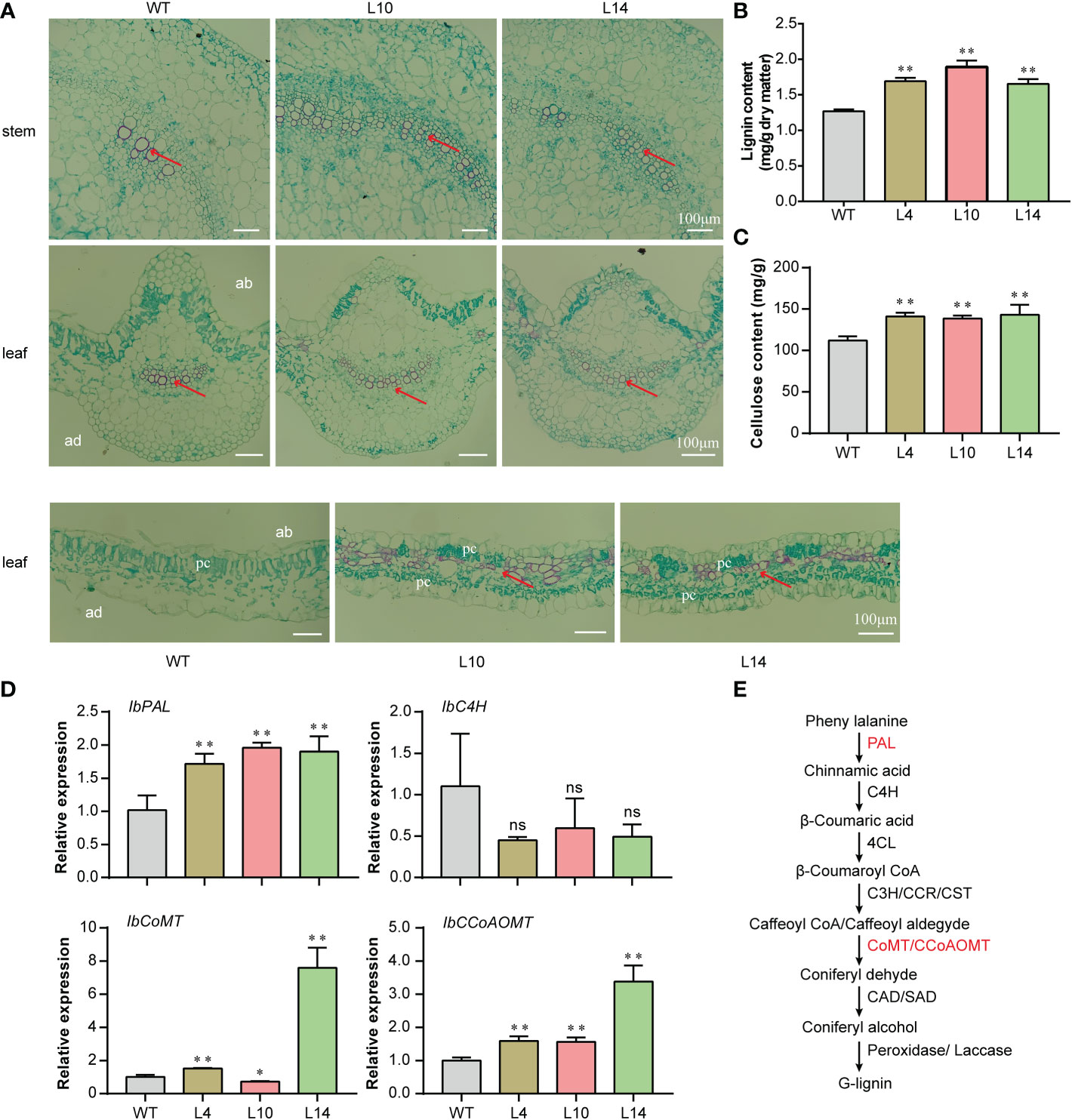
Figure 9 Histological observation of the IbNAC43-overexpressed and wild-type (WT) plants. (A) Paraffin sections. ad, adaxial side; ab, abaxial side; pc, palisade cells. Scale bar, 100 μm. (B) Lignin content. Data are presented as the mean ± SE (n = 3). (C) Mean cellulose content from IbNAC43-overexpressed and non-overexpressed sweet potato plants. Data are presented as the mean ± SE (n = 3). (D) Expression levels of the lignin biosynthesis-related genes. (E) Biosynthesis pathway of lignin. PAL, phenylalanine ammonia-lyase; C4H, cinnamate-4-hydroxylase; 4CL, 4-coumarate-CoA ligase; C3H, p-coumaroyl-CoA3-hydroxylase; CoMT, caffeicacid-5-O-methyltransfenase; CCoAOMT, caffeoyl-CoA 5-O-methyltransfenase; CAD, cinnamyl alcohol dehydrogenase. Asterisks indicate significant differences compared to the WT based on a Student’s t-test. **p < 0.01. ns, not significant.
Discussion
The growth of plants depends on the normal development of their organs. Among the plant organs, the leaves play important roles in photosynthesis, yield, and abiotic stress response. The morphology of the leaves generally affects the photosynthetic efficiency and, consequently, plant growth. Inhibition of the expression of DNL-4 reduced the plant height and leaf blade width and affected the productivity of transgenic rice plants (Bae et al., 2021). Leaf development originates from the shoot apical meristem (SAM), and the leaf primordium initiates growth and proliferates from the SAM. Adaxial–abaxial polarity is formed during this process. Its establishment is a complex process and has been revealed to be restricted via complicated regulatory networks (Du et al., 2018). However, there are no reports on the leaf development of sweet potato. In this study, we isolated the NAC TF IbNAC43 from I. trifida, which is a wild ancestor of a sweet potato cultivar. IbNAC43 was highly expressed in the leaves (Figure 2). Like most of the NAC proteins, IbNAC43 is a nuclear protein with self-transactivation activity in yeast (Figure 3). The overexpression of IbNAC43 caused leaf curling and plant dwarfism, suggesting that it negatively regulated the growth of transgenic sweet potato plants (Figure 5).
The nutrients for plant growth and development come mainly from photosynthesis (Ramamoorthy et al., 2018). Chlorophyll is the most important plant pigment involved in photosynthesis. It absorbs and transmits light energy, initiates primary photochemical reactions, and directly affects photosynthetic efficiency (Tanaka and Tanaka, 2006; Zhao et al., 2019). An increased chlorophyll content enhances the photosynthetic efficiency, while its decrease reduces it (Chen and Xu, 2006). In general, enhanced photosynthesis can promote the growth and yield of plants. The overexpression of SlSDH2.2 (succinate dehydrogenase gene) increased biomass and promoted growth by enhancing the rate of photosynthesis and reducing the rate of the tricarboxylic acid (TCA) cycle in transgenic tomato (Araujo et al., 2011). In contrast, reduced photosynthesis inhibits plant growth. AtHXK1 encodes a hexokinase that senses the endogenous levels of sugars, and its overexpression reduced the chlorophyll content of leaves and the photosynthesis rates, inhibiting the growth of transgenic tomato (Dai et al., 1999). In this study, we found that the overexpression of IbNAC43 decreased the photosynthesis rate by reducing the relative chlorophyll content in transgenic sweet potato plants (Figure 5).
Following the discovery of the first gene (PHAN) reported to influence leaf development, most of the TFs have been identified to serve important roles in partially redundant pathways. AS1 (an ARP MYB domain TF) and AS2 [a LATERAL ORGAN BOUNDARIES (LOB) domain gene] were found to directly bind to the promoters of KNOX1 and KNAT2 to repress their expression, which promoted leaf adaxial polarity formation (Guo et al., 2008). In addition, REV, PHV, PHB, HAT1, ATHB4, and ATHB8, which belong to the homeodomain leucine zipper (HD-ZIP) family, play positive roles in adaxial domain identification in Arabidopsis. However, the control of adaxial–abaxial leaf polarity is still complex, and the molecular regulatory mechanisms remain largely unknown. In this study, we found that the adaxial epidermal cells of WT plants were bigger than those of transgenic plants (Figure 6A), and the palisade mesophyll of transgenic plants appeared to have a disorderly distribution (Figure 6B). These results indicate that the overexpression of IbNAC43 resulted in an imbalance in the number of upper and lower epidermal cells. qRT-PCR analysis showed that the overexpression of IbNAC43 promoted leaf adaxial polarity by upregulating the expression levels of REV, HAT1, ATHB8, and AS1 (Figure 7). In addition, our study revealed that IbNAC43 regulated the expression of REV and AS1 by directly binding to their promoters (Figure 8).
In rice, a number of genes have been reported to regulate leaf rolling. Overexpression of OsLBD3-7 and OsAGO7 induced upward curling of the leaf blade (Shi et al., 2007; Li et al., 2016). The adl1 mutant showed apparent adaxial leaves, indicating that ADL1 has a negative role in adaxial leaf identification in rice (Hibara et al., 2009). Therefore, we further examined the expression levels of these homologous genes in transgenic sweet potato plants. Interestingly, in sweet potato, the overexpression of IbNAC43 upregulated IbADL1, but downregulated IbLBD3 and IbAGO7 (Figure 7). These results indicate that similar/homologous genes in monocotyledons and dicotyledons might play different roles in leaf adaxial–abaxial polarity.
In general, the morphology and development of leaves are closely associated with lignin. Lignin is a complex organic polymer and is important for the structure and repair process of plant cell walls (Halpin, 2019). Proper lignin deposition in specialized leaf cells is important for leaf development (Yoon et al., 2015). The silencing of PtrARF2.1 resulted in asymmetric leaf blades, reduced leaf area, lignin deposition, and a dwarf phenotype in transgenic Paulownia tomentosa plants (Fu et al., 2019). A lot of TFs have been reported to regulate lignin content and vascular bundle development. The NAC TFs NST1 and NST3 are key regulators of the formation of secondary walls and vascular vessels (Mitsuda et al., 2007). Extensive studies have found that the establishment of leaf polarity also affects the deposition of lignin. The crm1-D mutant (a gain-of-function mutant) showed leaf curling and a high lignin content, indicating that OsRoc8 positively mediated lignin biosynthesis (Sun et al., 2020). Compared to the WT, the overexpression of AtATHB8 caused plant dwarfism and showed a higher production of lignified tissue in transgenic plants (Baima et al., 2001). Moreover, AtATHB8 can regulate leaf and vascular development and promote vascular cell differentiation (Gardiner et al., 2011). In this study, the overexpression of IbNAC43 enhanced lignin biosynthesis and vascular bundle development by upregulating the expression of the lignin biosynthesis genes in transgenic sweet potato (Figure 9).
In conclusion, this is the first report on IbNAC43 playing an important role in the establishment of leaf polarity and plant growth. The overexpression of this TF caused leaf curling, reduced the photosynthetic rate, and retarded plant growth by upregulating the leaf adaxial polarity-related genes. Furthermore, we identified a new regulatory mechanism in IbNAC43, which promotes the transcription of IbREV and IbAS1 by directly binding to their promoters. This study provides additional insights into understanding the effects of leaf adaxial–abaxial development on plant growth in dicotyledons.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation, to any qualified researcher.
Author contributions
SS and HoZ: Conceptualization. XL and NN: Data curation. XL and YC: Formal analysis. SS and HoZ: Writing-original draft. SG, HuZ, SH, QL, and HoZ: Writing-review and editing. All authors contributed to the article and approved the submitted version.
Funding
This research was funded by the National Natural Science Foundation of China (31872878), the National Key R&D Program of China (grant nos. 2019YFD1001300 and 2019YFD1001301), and the Earmarked Fund for CARS-10-Sweetpotato.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1095977/full#supplementary-material
References
Ahn, Y. O., Kim, S. H., Kim, C. Y., Lee, J. S., Kwak, S. S., Lee, H. S. (2010). Exogenous sucrose utilization and starch biosynthesis among sweetpotato cultivars. Carbohydr Res. 345 (1), 55–60. doi: 10.1016/j.carres.2009.08.025
Araujo, W. L., Nunes-Nesi, A., Osorio, S., Usadel, B., Fuentes, D., Nagy, R., et al. (2011). Antisense inhibition of the iron-sulphur subunit of succinate dehydrogenase enhances photosynthesis and growth in tomato via an organic acid-mediated effect on stomatal aperture. Plant Cell 23 (2), 600–627. doi: 10.1105/tpc.110.081224
Bae, K. D., Um, T. Y., Yang, W. T., Park, T. H., Hong, S. Y., Kim, K. M., et al. (2021). Characterization of dwarf and narrow leaf (dnl-4) mutant in rice. Plant Signal Behav. 16 (2), 1849490. doi: 10.1080/15592324.2020.1849490
Baima, S., Possenti, M., Matteucci, A., Wisman, E., Altamura, M. M., Ruberti, I., et al. (2001). The Arabidopsis ATHB-8 HD-zip protein acts as a differentiation-promoting transcription factor of the vascular meristems. Plant Physiol. 126 (2), 643–655. doi: 10.1104/pp.126.2.643
Balazadeh, S., Kwasniewski, M., Caldana, C., Mehrnia, M., Zanor, M. I., Xue, G. P., et al. (2011). ORS1, an H2O2-responsive NAC transcription factor, controls senescence in Arabidopsis thaliana. Mol. Plant 4 (2), 346–360. doi: 10.1093/mp/ssq080
Bowler, C., Benvenuto, G., Laflamme, P., Molino, D., Probst, A. V., Tariq, M., et al. (2004). Chromatin techniques for plant cells. Plant J. 39 (5), 776–789. doi: 10.1111/j.1365-313X.2004.02169.x
Chen, Y., Xu, D. Q. (2006). Two patterns of leaf photosynthetic response to irradiance transition from saturating to limiting one in some plant species. New Phytol. 169 (4), 789–797. doi: 10.1111/j.1469-8137.2005.01624.x
Dai, N., Schaffer, A., Petreikov, M., Shahak, Y., Giller, Y., Ratner, K., et al. (1999). Overexpression of Arabidopsis hexokinase in tomato plants inhibits growth, reduces photosynthesis, and induces rapid senescence. Plant Cell 11 (7), 1253–1266. doi: 10.1105/tpc.11.7.1253
Diao, P. F., Chen, C., Zhang, Y. Z., Meng, Q. W., Lv, W., Ma, N. N. (2020). The role of NAC transcription factor in plant cold response. Plant Signaling Behav. 15 (9), 1785668. doi: 10.1080/15592324.2020.1785668
Du, F., Guan, C. M., Jiao, Y. L. (2018). Molecular mechanisms of leaf morphogenesis. Mol. Plant 11 (9), 1117–1134. doi: 10.1016/j.molp.2018.06.006
Du, B., Nie, N., Sun, S. F., Hu, Y. F., Bai, Y. D., He, S. Z., et al. (2021). A novel sweetpotato RING-H2 type E3 ubiquitin ligase gene IbATL38 enhances salt tolerance in transgenic Arabidopsis. Plant Sci. 304, 110802. doi: 10.1016/j.plantsci.2020.110802
Fang, L. K., Zhao, F. M., Cong, Y. F., Sang, X. C., Du, Q., Wang, D. Z., et al. (2012). Rolling-leaf14 is a 2OG-fe (II) oxygenase family protein that modulates rice leaf rolling by affecting secondary cell wall formation in leaves. Plant Biotechnol. J. 10 (5), 524–532. doi: 10.1111/j.1467-7652.2012.00679.x
Fu, Y. Y., Win, P. P., Zhang, H. J., Li, C. F., Shen, Y., He, F., et al. (2019). PtrARF2.1 is involved in regulation of leaf development and lignin biosynthesis in poplar trees. Int. J. Mol. Sci. 20 (17), 4141. doi: 10.3390/ijms20174141
Gardiner, J., Donner, T. J., Scarpella, E. (2011). Simultaneous activation of SHR and ATHB8 expression defines switch to preprocambial cell state in Arabidopsis leaf development. Dev. Dyn. 240 (1), 261–270. doi: 10.1002/dvdy.22516
Gong, Z. Z., Xiong, L. M., Shi, H. Z., Yang, S. H., Herrera-Estrella, L. R., Xu, G. H., et al. (2020). Plant abiotic stress response and nutrient use efficiency. Sci. China-Life Sci. 63 (5), 635–674. doi: 10.1007/s11427-020-1683-x
Guo, Y., Gan, S. S. (2014). Translational researches on leaf senescence for enhancing plant productivity and quality. J. Exp. Bot. 65 (14), 3901–3913. doi: 10.1093/jxb/eru248
Guo, M. J., Thomas, J., Collins, G., Timmermans, M. C. P. (2008). Direct repression of KNOX loci by the ASYMMETRIC LEAVES1 complex of Arabidopsis. Plant Cell 20 (1), 48–58. doi: 10.1105/tpc.107.056127
Halpin, C. (2019). Lignin engineering to improve saccharification and digestibility in grasses. Curr. Opin. Biotechnol. 56, 223–229. doi: 10.1016/j.copbio.2019.02.013
Hibara, K., Karim, M. R., Takada, S., Taoka, K. I., Furutani, M., Aida, M., et al. (2006). Arabidopsis CUP-SHAPED COTYLEDON3 regulates postembryonic shoot meristem and organ boundary formation. Plant Cell 18 (11), 2946–2957. doi: 10.1105/tpc.106.045716
Hibara, K., Obara, M., Hayashida, E., Abe, M., Ishimaru, T., Satoh, H., et al. (2009). The ADAXIALIZED LEAF1 gene functions in leaf and embryonic pattern formation in rice. Dev. Biol. 334 (2), 345–354. doi: 10.1016/j.ydbio.2009.07.042
Hou, H. L., Lin, Y., Hou, X. L. (2020). Ectopic expression of a pak-choi YABBY gene, BcYAB3, causes leaf curvature and flowering stage delay in Arabidopsis thaliana. Genes 11 (4), 370. doi: 10.3390/genes11040370
Husbands, A. Y., Benkovics, A. H., Nogueira, F. T. S., Lodha, M., Timmermans, M. C. P. (2015). The ASYMMETRIC LEAVES complex employs multiple modes of regulation to affect adaxial-abaxial patterning and leaf complexity. Plant Cell 27 (12), 3321–3335. doi: 10.1105/tpc.15.00454
Iwakawa, H., Iwasaki, M., Kojima, S., Ueno, Y., Soma, T., Tanaka, H., et al. (2007). Expression of the ASYMMETRIC LEAVES2 gene in the adaxial domain of Arabidopsis leaves represses cell proliferation in this domain and is critical for the development of properly expanded leaves. Plant J. 51 (2), 173–184. doi: 10.1111/j.1365-313X.2007.03132.x
Iwakawa, H., Takahashi, H., Machida, Y., Machida, C. (2020). Roles of ASYMMETRIC LEAVES2 (AS2) and nucleolar proteins in the adaxial-abaxial polarity specification at the perinucleolar region in Arabidopsis. Int. J. Mol. Sci. 21 (19), 7314. doi: 10.3390/ijms21197314
Kim, H. J., Nam, H. G., Lim, P. O. (2016). Regulatory network of NAC transcription factors in leaf senescence. Curr. Opin. Plant Biol. 33, 48–56. doi: 10.1016/j.pbi.2016.06.002
Kim, Y. S., Sakuraba, Y., Han, S. H., Yoo, S. C., Paek, N. C. (2013). Mutation of the Arabidopsis NAC016 transcription factor delays leaf senescence. Plant Cell Physiol. 54 (10), 1660–1672. doi: 10.1093/pcp/pct113
Kumar, S., Stecher, G., Li, M., Knyaz, C., Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35 (6), 1547–1549. doi: 10.1093/molbev/msy096
Laufs, P., Peaucelle, A., Morin, H., Traas, J. (2004). MicroRNA regulation of the CUC genes is required for boundary size control in Arabidopsis meristems. Development 131 (17), 4311–4322. doi: 10.1242/dev.01320
Li, L., Shi, Z. Y., Li, L., Shen, G. Z., Wang, X. Q., An, L. S., et al. (2010). Overexpression of ACL1 (abaxially curled leaf 1) increased bulliform cells and induced abaxial curling of leaf blades in rice. Mol. Plant 3 (5), 807–817. doi: 10.1093/mp/ssq022
Li, W. Q., Zhang, M. J., Gan, P. F., Qiao, L., Yang, S. Q., Miao, H., et al. (2017). CLD1/SRL1 modulates leaf rolling by affecting cell wall formation, epidermis integrity and water homeostasis in rice. Plant J. 92 (5), 904–923. doi: 10.1111/tpj.13728
Li, C., Zou, X. H., Zhang, C. Y., Shao, Q. H., Liu, J., Liu, B., et al. (2016). OsLBD3-7 overexpression induced adaxially rolled leaves in rice. PloS One 11 (6), e0156413. doi: 10.1371/journal.pone.0156413
Liu, Q. C., Zhai, H., Wang, Y., Zhang, D. P. (2001). Efficient plant regeneration from embryogenic suspension cultures of sweetpotato. In Vitro Cell. Dev. Biol.-Plant 37 (5), 564–567. doi: 10.1007/s11627-001-0098-7
Ma, L., Li, R. P., Ma, L. Y., Song, N., Xu, Z., Wu, J. S. (2021). Involvement of NAC transcription factor NaNAC29 in Alternaria alternata resistance and leaf senescence in Nicotiana attenuata. Plant Divers. 43 (6), 502–509. doi: 10.1016/j.pld.2020.11.003
Manuela, D., Xu, M. L. (2020). Patterning a leaf by establishing polarities. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.568730
McConnell, J. R., Emery, J., Eshed, Y., Bao, N., Bowman, J., Barton, M. K. (2001). Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature 411 (6838), 709–713. doi: 10.1038/35079635
Mitsuda, N., Iwase, A., Yamamoto, H., Yoshida, M., Seki, M., Shinozaki, K., et al. (2007). NAC transcription factors, NST1 and NST3, are key regulators of the formation of secondary walls in woody tissues of Arabidopsis. Plant Cell 19 (1), 270–280. doi: 10.1105/tpc.106.047043
Nagahage, I. S. P., Sakamoto, S., Nagano, M., Ishikawa, T., Mitsuda, N., Kawai-Yamada, M., et al. (2020). An Arabidopsis NAC domain transcription factor, ATAF2, promotes age-dependent and dark-induced leaf senescence. Physiol. Plant 170 (2), 299–308. doi: 10.1111/ppl.13156
Nakashima, K., Takasaki, H., Mizoi, J., Shinozaki, K., Yamaguchi-Shinozaki, K. (2012). NAC transcription factors in plant abiotic stress responses. Biochim. Biophys. Acta 1819 (2), 97–103. doi: 10.1016/j.bbagrm.2011.10.005
Nuruzzaman, M., Sharoni, A. M., Kikuchi, S. (2013). Roles of NAC transcription factors in the regulation of biotic and abiotic stress responses in plants. Front. Microbiol. 4. doi: 10.3389/fmicb.2013.00248
Otoole, J. C., Cruz, R. T. (1980). Response of leaf water potential, stomatal-resistance, and leaf rolling to water-stress. Plant Physiol. 65 (3), 428–432. doi: 10.1104/pp.65.3.428
Otsuga, D., DeGuzman, B., Prigge, M. J., Drews, G. N., Clark, S. E. (2001). REVOLUTA regulates meristem initiation at lateral positions. Plant J. 25 (2), 223–236. doi: 10.1046/j.1365-313x.2001.00959.x
Pfaffl, M. W. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29 (9), e45. doi: 10.1093/nar/29.9.e45
Pulungan, S. I., Yano, R., Okabe, Y., Ichino, T., Kojima, M., Takebayashi, Y., et al. (2018). SlLAX1 is required for normal leaf development mediated by balanced adaxial and abaxial pavement cell growth in tomato. Plant Cell Physiol. 59 (6), 1170–1186. doi: 10.1093/pcp/pcy052
Puranik, S., Sahu, P. P., Srivastava, P. S., Prasad, M. (2012). NAC proteins: regulation and role in stress tolerance. Trends Plant Sci. 17 (6), 369–381. doi: 10.1016/j.tplants.2012.02.004
Ramamoorthy, R., Vishal, B., Ramachandran, S., Kumar, P. P. (2018). The OsPS1-f gene regulates growth and development in rice by modulating photosynthetic electron transport rate. Plant Cell Rep. 37 (2), 377–385. doi: 10.1007/s00299-017-2235-8
Sawa, S., Watanabe, K., Goto, K., Kanaya, E., Morita, E. H., Okada, K. (1999). FILAMENTOUS FLOWER, a meristem and organ identity gene of Arabidopsis, encodes a protein with a zinc finger and HMG-related domains. Genes Dev. 13 (9), 1079–1088. doi: 10.1101/gad.13.9.1079
Shi, Z. Y., Wang, J., Wan, X. S., Shen, G. Z., Wang, X. Q., Zhang, J. L. (2007). Over-expression of rice OsAGO7 gene induces upward curling of the leaf blade that enhanced erect-leaf habit. Planta 226 (1), 99–108. doi: 10.1007/s00425-006-0472-0
Siegfried, K. R., Eshed, Y., Baum, S. F., Otsuga, D., Drews, G. N., Bowman, J. L. (1999). Members of the YABBY gene family specify abaxial cell fate in Arabidopsis. Development 126 (18), 4117–4128. doi: 10.1242/dev.126.18.4117
Sun, J., Cui, X. E., Teng, S. Z., Zhao, K. N., Wang, Y. W., Chen, Z. H., et al. (2020). HD-ZIP IV gene Roc8 regulates the size of bulliform cells and lignin content in rice. Plant Biotechnol. J. 18 (12), 2559–2572. doi: 10.1111/pbi.13435
Sun, S. F., Li, X., Gao, S. P., Nie, N., Zhang, H., Yang, Y. F., et al. (2022). A novel WRKY transcription factor from Ipomoea trifida, ItfWRKY70, confers drought tolerance in sweet potato. Int. J. Mol. Sci. 23 (2), 686. doi: 10.3390/ijms23020686
Tanaka, A., Tanaka, R. (2006). Chlorophyll metabolism. Curr. Opin. Plant Biol. 9 (3), 248–255. doi: 10.1016/j.pbi.2006.03.011
Verweij, W., Spelt, C., Di Sansebastiano, G. P., Vermeer, J., Reale, L., Ferranti, F., et al. (2008). An h+ p-ATPase on the tonoplast determines vacuolar pH and flower colour. Nat. Cell Biol. 10 (12), 1456–1462. doi: 10.1038/ncb1805
Vroemen, C. W., Mordhorst, A. P., Albrecht, C., Kwaaitaal, M. A. C. J., de Vries, S. C. (2003). The CUP-SHAPED COTYLEDON3 gene is required for boundary and shoot meristem formation in Arabidopsis. Plant Cell 15 (7), 1563–1577. doi: 10.1105/tpc.012203
Waites, R., Hudson, A. (1995). Phantastica: a gene required for dorsoventrality of leaves in antirrhinum majus. Development 121 (7), 2143–2154. doi: 10.1242/dev.121.7.2143
Waites, R., Selvadurai, H. R. N., Oliver, I. R., Hudson, A. (1998). The PHANTASTICA gene encodes a MYB transcription factor involved in growth and dorsoventrality of lateral organs in Antirrhinum. Cell 93 (5), 779–789. doi: 10.1016/s0092-8674(00)81439-7
Wan, S. B., Qin, Z. P., Jiang, X. M., Yang, M., Chen, W. J., Wang, Y. M., et al. (2021). Identification and fine mapping of a locus related to leaf up-curling trait (Bnuc3) in Brassica napus. Int. J. Mol. Sci. 22 (21), 11693. doi: 10.3390/ijms222111693
Wu, A. H., Allu, A. D., Garapati, P., Siddiqui, H., Dortay, H., Zanor, M. I., et al. (2012). JUNGBRUNNEN1, a reactive oxygen species-responsive NAC transcription factor, regulates longevity in Arabidopsis. Plant Cell 24 (2), 482–506. doi: 10.1105/tpc.111.090894
Xu, P. Z., Ali, A., Han, B. L., Wu, X. J. (2018). Current advances in molecular basis and mechanisms regulating leaf morphology in rice. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.01528
Xu, P. P., Chen, H. Y., Ying, L., Cai, W. M. (2016). AtDOF5.4/OBP4, a DOF transcription factor gene that negatively regulates cell cycle progression and cell expansion in Arabidopsis thaliana. Sci. Rep. 6, 27705. doi: 10.1038/srep27705
Xu, Y., Kong, W. Y., Wang, F. Q., Wang, J., Tao, Y. J., Li, W. Q., et al. (2021). Heterodimer formed by ROC8 and ROC5 modulates leaf rolling in rice. Plant Biotechnol. J. 19 (12), 2662–2672. doi: 10.1111/pbi.13690
Yang, H. B., Murphy, A. S. (2009). Functional expression and characterization of Arabidopsis ABCB, AUX 1 and PIN auxin transporters in Schizosaccharomyces pombe. Plant J. 59 (1), 179–191. doi: 10.1111/j.1365-313X.2009.03856.x
Yang, S. D., Seo, P. J., Yoon, H. K., Park, C. M. (2011). The Arabidopsis NAC transcription factor VNI2 integrates abscisic acid signals into leaf senescence via the COR/RD genes. Plant Cell 23 (6), 2155–2168. doi: 10.1105/tpc.111.084913
Yang, H., Shi, G. X., Li, X., Hu, D. Z., Cui, Y. M., Hou, J. F., et al. (2019). Overexpression of a soybean YABBY gene, GmFILa, causes leaf curling in Arabidopsis thaliana. BMC Plant Biol. 19, 234. doi: 10.1186/s12870-019-1810-2
Yoon, J., Choi, H., An, G. (2015). Roles of lignin biosynthesis and regulatory genes in plant development. J. Integr. Plant Biol. 57 (11), 902–912. doi: 10.1111/jipb.12422
Yuan, S., Li, Y., Peng, S. B. (2015). Leaf lateral asymmetry in morphological and physiological traits of rice plant. PloS One 10 (6), e0129832. doi: 10.1371/journal.pone.0129832
Zhai, H., Wang, F. B., Si, Z. Z., Huo, J. X., Xing, L., An, Y. Y., et al. (2016). A myo-inositol-1-phosphate synthase gene, IbMIPS1, enhances salt and drought tolerance and stem nematode resistance in transgenic sweet potato. Plant Biotechnol. J. 14 (2), 592–602. doi: 10.1111/pbi.12402
Zhang, H., Wang, Z., Li, X., Gao, X. R., Dai, Z. R., Cui, Y. F., et al. (2022). The IbBBX24-IbTOE3-IbPRX17 module enhances abiotic stress tolerance by scavenging reactive oxygen species in sweet potato. New Phytol. 233 (3), 1133–1152. doi: 10.1111/nph.17860
Zhang, H., Zhang, Q., Zhai, H., Gao, S. P., Yang, L., Wang, Z., et al. (2020). IbBBX24 promotes the jasmonic acid pathway and enhances Fusarium wilt resistance in sweet potato. Plant Cell 32 (4), 1102–1123. doi: 10.1105/tpc.19.00641
Zhao, T., Tan, L. J., Huang, W. Q., Wang, J. T. (2019). The interactions between micro polyvinyl chloride (mPVC) and marine dinoflagellate karenia mikimotoi: The inhibition of growth, chlorophyll and photosynthetic efficiency. Environ. pollut. 247, 883–889. doi: 10.1016/j.envpol.2019.01.114
Zheng, M., Lin, J. C., Liu, X. B., Chu, W., Li, J. P., Gao, Y. J., et al. (2021). Histone acetyltransferase TaHAG1 acts as a crucial regulator to strengthen salt tolerance of hexaploid wheat. Plant Physiol. 186 (4), 1951–1969. doi: 10.1093/plphys/kiab187
Keywords: sweet potato, IbNAC43, plant growth, leaf curling, photosynthetic rate
Citation: Sun S, Li X, Nie N, Chen Y, Gao S, Zhang H, He S, Liu Q and Zhai H (2023) Sweet potato NAC transcription factor NAC43 negatively regulates plant growth by causing leaf curling and reducing photosynthetic efficiency. Front. Plant Sci. 14:1095977. doi: 10.3389/fpls.2023.1095977
Received: 11 November 2022; Accepted: 31 January 2023;
Published: 21 February 2023.
Edited by:
Zhongxiong Lai, Fujian Agriculture and Forestry University, ChinaReviewed by:
Shengcai Liu, Fujian Agriculture and Forestry University, ChinaZhiguo Zhang, Institute of Biotechnology (CAAS), China
Copyright © 2023 Sun, Li, Nie, Chen, Gao, Zhang, He, Liu and Zhai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hong Zhai, emhhaWhvbmdAY2F1LmVkdS5jbg==
 Sifan Sun
Sifan Sun Shaopei Gao
Shaopei Gao Huan Zhang
Huan Zhang Shaozhen He
Shaozhen He Qingchang Liu
Qingchang Liu Hong Zhai
Hong Zhai