- 1Key Laboratory of Plant Molecular Physiology, Institute of Botany, Chinese Academy of Sciences, Beijing, China
- 2University of Chinese Academy of Sciences, Beijing, China
Fruits are derived from flowers and play an important role in human food, nutrition, and health. In general, flowers determine the crop yield, and ripening affects the fruit quality. Although transcription factors (TFs) only account for a small part of plant transcriptomes, they control the global gene expression and regulation. The plant-specific NAC (NAM, ATAF, and CUC) TFs constitute a large family evolving concurrently with the transition of both aquatic-to-terrestrial plants and vegetative-to-reproductive growth. Thus, NACs play an important role in fruit yield and quality by determining shoot apical meristem (SAM) inflorescence and controlling ripening. The present review focuses on the various properties of NACs together with their function and regulation in flower formation and fruit ripening. Hitherto, we have a better understanding of the molecular mechanisms of NACs in ripening through abscisic acid (ABA) and ethylene (ETH), but how NACs regulate the expression of the inflorescence formation-related genes is largely unknown. In the future, we should focus on the analysis of NAC redundancy and identify the pivotal regulators of flowering and ripening. NACs are potentially vital manipulation targets for improving fruit quantity and quality.
Introduction
Transcription factors (TFs) only account for a small quantity of genes in plant transcriptomes (less than 10%), but they control the global gene expression and regulation in plant development and adaptation. The plant-specific NAC (NAM, ATAF, and CUC) TFs constitute a large family, such as 101, 328, 138, and 280 members in tomato, rice, Arabidopsis and tobacco respectively (Liu G. S. et al., 2022). In general, the role of a TF is dependent on its binding to the cis-elements of the target genes, which are associated with nuclear localization, DNA binding, oligomerization, and gene expression. Because the NAC family exists from aquatic green algae to higher terrestrial plants, its members participate in the formation of the organ boundaries of plants transiting from vegetative growth to reproductive growth and are related to flower development and fruit ripening, which are essential to crop yield and quality (Maugarny-Calès et al., 2016; Ma et al., 2017; Mathew and Agarwal, 2018; Forlani et al., 2021; Singh et al., 2021; Liu G. S. et al., 2022). Thereby, in recent years, much progress has been made toward understanding the molecular mechanisms regulated by NAC TFs in flower development (Guo et al., 2022; Liang et al., 2022; Liu X. et al., 2022; Wang et al., 2022; Ghissing et al., 2023) and fruit ripening (Fu B. L. et al., 2021; Gao et al., 2021; Kou et al., 2021; Gao et al., 2022; Li et al., 2022; Liu B. et al., 2022; Qi et al., 2022). In this review, we mainly summarize up-to-date advances on the basic properties, function, and action mechanisms of NACs in flower formation, flowering, and fruit ripening to improve our understanding of the molecular basis of crop yield and quality mediated by NAC TFs.
Specifically, we highlight that the NAC family includes no apical meristem (NAM), Arabidopsis transcription activator factors (ATAF1/ATAF2), and cup-shaped cotyledon (CUC2), which all contain a conserved amino acid (aa) sequence at the N-terminal with five subdomains, which not only possess versatile regulatory patterns at the DNA, RNA, and protein levels but also serve as activators or repressors through variable transcriptional regulatory regions at the C-terminal. Plant-specific NACs evolved from the streptophyte green algae to higher plants, concomitant with the transition of aquatic-to-terrestrial living and vegetative-to-reproductive growth, with a line of distinct NACs constituting a complex regulatory network linked to the phytohormones abscisic acid (ABA) and ethylene (ETH) that are essential to flowering and ripening. Thus, NACs are potentially important manipulation targets for improving fruit quantity and quality.
Basic traits of NAC transcription factors
In response to the aquatic-to-terrestrial environmental niches, the NAC family has shown gradual amplifications in gene size and diversity for the colony of land concurrently with the evolution of additional gene functions for water conductance, cell support, xylem/phloem differentiation, and vasculature formation, adaptive to land living in a sessile manner. Therefore, the present NACs in higher plants have developed a variety of structural and action traits, including DNA binding, nuclear localization, oligomerization/protein–protein interactions, and hierarchical regulatory patterns, using a conserved N-terminal NAC domain for target binding and a diverse C-terminal flexible region for regulating gene expression as activators or repressors.
NAC domain traits
NAC TFs have the conserved N-terminal NAC region with about 150 aa including both nuclear localization signal (NLS) and DNA binding regions, which are further divided into five subdomains designated from A to E (A, C, and D are more highly conserved compared to B and E) and a C-terminal region as a transcriptional regulatory/activation region (TRR and TAR, respectively) (Figure 1). In particular, members of the NAC family have a conserved domain fold containing one β-sheet flanked by an α-helical element, and the C-subdomain has a DNA recognition conserved motif sequence (WKATGT/NDK) that specifically binds to the developmental core CGT(GA) and the stress core CGTG (Mathew and Agarwal, 2018) (Figures 1B, E). In addition, NACs include activators and repressors for gene expression and repression, respectively, resulting from the NAC repression domain (NARD) in the D subdomain, such as the rice NAC020 and NAC026, which show bifunctionality having both activation and repression properties (Mathew et al., 2016). In addition, NAC proteins have homodimers and heterodimers, and dimerization is required for stable DNA binding to post-transcriptional and translational modifications in target genes (Chen et al., 2016) (Figure 1B). In conclusion, the N-terminal region in NACs contains the repression domain, while the C-terminal region is the transactivation region to a large degree. These properties allow NACs to have multiple regulatory patterns at the transcriptional, post-transcriptional, and translational levels, serving as activators and repressors in response to developmental and environmental cues.
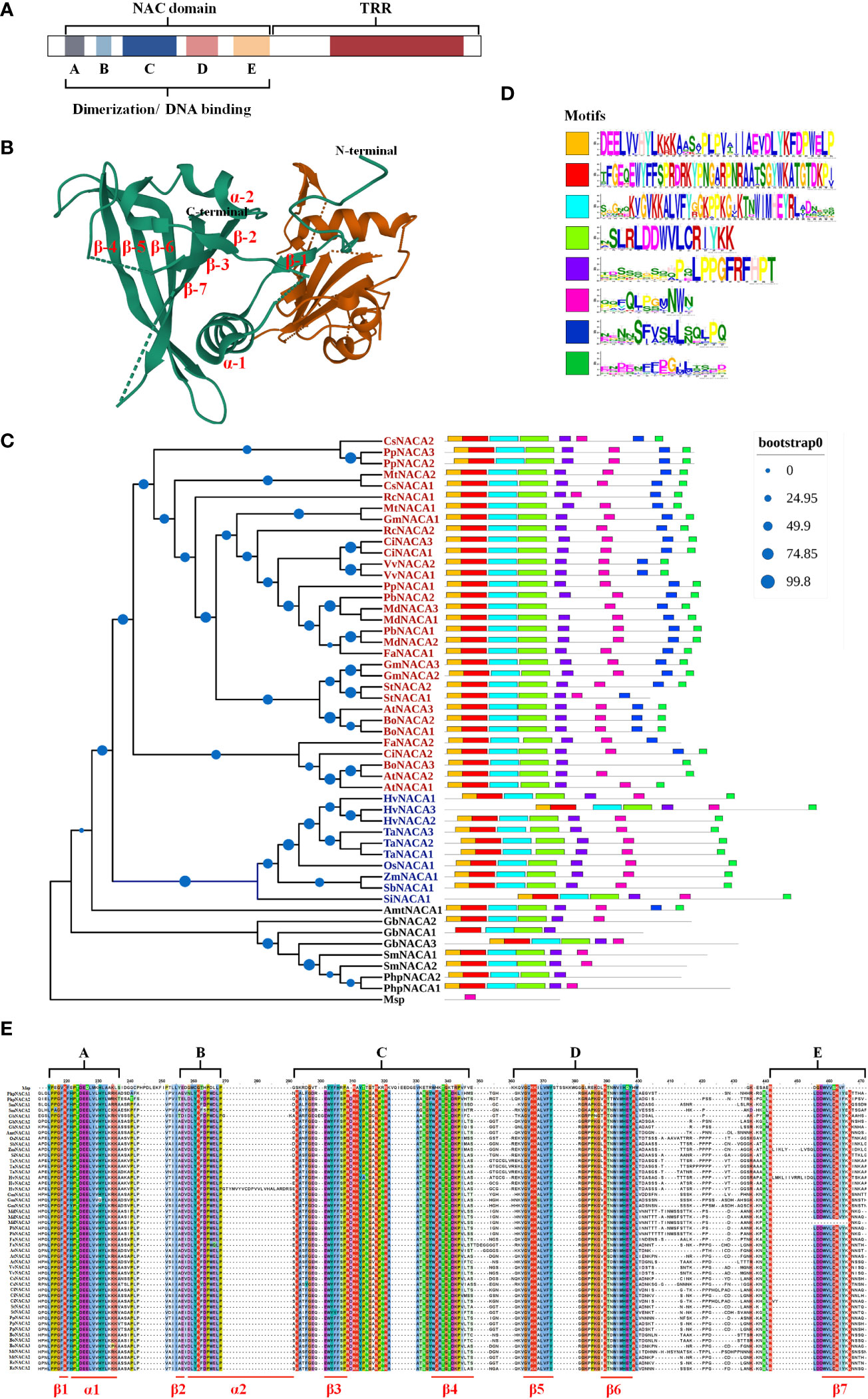
Figure 1 Basic traits of NAC (NAM, ATAF, and CUC) transcription factors (TFs). (A) Structure and function of NACs. (B) Three-dimensional structure of the Arabidopsis AtNAC019 protein homodimer. The AtNAC019 NAC domain crystal form IV is from the PDB database (PDB DOI: 10.2210/pdb4DUL/pdb). (C) Phylogenetic relationship of the NAC TFs. The homologous proteins of the strawberry NAC TF FaNACA2 (FaNAC035) (Martín-Pizarro et al., 2021) were found and verified in 24 species using BLAST and CDD tools, and a phylogenetic tree was constructed with reference to the evolutionary relationship of the NAC TF family in plants. The multiple sequence alignment, sequence pruning, model search, and phylogenetic tree construction were performed in Phylosuite_v1.2.2. The phylogenetic tree was obtained by applying the maximum likelihood method to a matrix of pairwise distances estimated using a JTT (Jones–Taylor–Thornton) model (bootstrap: ultrafast; no. of bootstraps: 10,000). Blue letters represent monocots and red letters represent dicots. Msp, Mougeotia sp.; Php, Physcomitrella patens; Sm, Selaginella moellendorffii; Gb, Ginkgo biloba; Amt, Amborella trichopoda; Si, Setaria italica; Sb, Sorghum bicolor; Zm, Zea mays; Os, Oryza sativa; Ta, Triticum aestivum; Hv, Hordeum vulgare; At, Arabidopsis thaliana; Bo, Brassica oleracea; Ci, Citrus sinensis; Fa, Fragraria ananassa; St, Solanum tuberosum; Gm, Glycine max; MD, Malus domestica; Pb, Pyrus bretschneideri; Pp, Prunus persica; Vv, Vitis vinifera; Rc, Ricinus communis; Mt, Medicago truncatula; Cs, Cucumis sativus. (D) The conserved motifs of all 51 sequences were queried online through the MEME website (https://meme-suite.org/meme/tools/meme). Motif1 corresponds to the A and B subdomains in the NAC domain, Motif2 contains the C subdomain, Motif3 corresponds to the D subdomain, Motif4 corresponds to the E domain, and Motif5–Motif8 are the conserved motifs in the transcription regulatory region (TRR). (E) NAC domain consisting of five subdomains: three highly conserved (A, C, and D) and two diverse (B and E) subdomains.
NAC localization traits
Most of the members of the NAC TF family are localized in the nucleus, although a few of them are situated in other organelles such as the plasma membrane (PM), the cytoplasm, and the endoplasmic reticulum (ER) (Olsen et al., 2005; Kim et al., 2006; Kim et al., 2007; Seo et al., 2008; Wang et al., 2016; Bhattacharjee et al., 2017). Notably, membrane-bound NACs may be anchored in membrane systems, including the PM, ER membrane, and nuclear membrane, in inactive forms. Stimulation by developmental or environmental cues promotes their release and trafficking from the membrane to the nucleus for the control of target gene expression in a common regulatory pattern (Kim et al., 2006; Kim et al., 2007; Seo et al., 2008; Wang et al., 2016; Bhattacharjee et al., 2017). Altogether, the multiple localizations and the translocation traits of NAC TFs implicate various biological functions and regulatory mechanisms in response to developmental and environmental cues.
NAC evolutionary traits
Environmental factors have played a vital role in the diversification of the NAC family, to a great extent, enabling higher plant survival and prosperity on land. Construction of a phylogeny tree based on the NAC family from green algae to higher plants can help to better understand the origin and expansion of NACs from their aquatic to terrestrial evolution (Mathew and Agarwal, 2018). Higher plant-specific NACs are potentially from the ancestral WRKY protein, concomitant with the transition from an aquatic to a terrestrial environment through two landmark expansions—the evolution of bryophytes from streptophytes, followed by the development of flowering angiosperm plants from vascular plants—which highlight the vital role of NACs in plant evolution (Yamasaki et al., 2013; Maugarny-Calès et al., 2016; Mathew and Agarwal, 2018).
The subfamily of FaNAC035 (Moyano et al., 2018; Martín-Pizarro et al., 2021), a key regulator of strawberry fruit development and ripening, is a typical case. Phylogenetic analysis showed that this subfamily also followed the evolutionary relationships from aquatic to terrestrial, and some conserved motifs were discovered to be lost when comparing ferns, gymnosperms, and angiosperms (Figures 1C–E). We supposed that four subdomains (motif1–motif4) appeared after the first evolutionary expansion; subsequently, the angiosperm NACs evolved to the conserved motif7 and motif8, with motif7 being conserved only in dicotyledons rather than in monocotyledons (Figures 1C, D). The number of genes also varies, with monocotyledons having only one member while dicotyledons have more than two members (Figure 1C). In brief, the gene numbers and conserved motifs are species-specific and have developed diverse functions in adapting to developmental and stress processes.
Based on wide phylogenetic analysis, the NAC family of green plants is divided into six major groups (groups I–VI) as follows: 1) the basal group I functions in water conduction and cell support; 2) the developmental group II, which includes CUC1–CUC3 (AtNAC054, AtNAC031, and AtNAC098), function in shoot apical meristem (SAM) formation and cotyledon separation, associated with the ETH–auxin pathways; 3) group III has the transmembrane motif (TMM); 4) group IV functions in the timing control of several developmental processes, including flower evolution, such as AtNAC034 for flowering time, AtNAC009 for cell division, and AtNAC042 for longevity; 5) group V regulates the stress responses; and 6) group VI has neo- and/or sub-functionalization in different plant species. In summary, NACs regulate many developmental and stress processes through phytohormones (Ooka et al., 2003; Ernst et al., 2004).
NAC hierarchical regulatory traits
Higher plants have developed multiple regulatory mechanisms to maintain a balance between the expression and turnover of NACs partly through NAC regulatory loops (Mathew and Agarwal, 2018). In the past years, significant progress has been made toward understanding the regulatory mechanisms of NACs in controlling their homeostasis at the DNA, RNA, and protein levels, including 1) transcriptional regulation by their upstream TFs to form messenger RNA precursors (pre-mRNAs) in the nucleus; 2) post-transcriptional regulation at the nucleotide level, including microRNA (miRNA)-mediated inhibition and/or splicing of pre-mRNAs and their transport from the synthesis site to the action sites; 3) post-translational regulation at the protein level, including the transfer of the cleave-mediated membrane-tethered proteins to the nucleus, which are associated with specific active groups, protein degradation by proteasome machinery, oligomerization, and protein interactions (Mathew and Agarwal, 2018).
Regarding the primary function of NACs in secondary cell wall thickening and xylem vessel element formation, the NAC secondary wall thickening promoting factor2, NST2/AtNAC066, and ORE1 (ORESARA1/AtNAC092) are regulated by WRKY12 and EIN2 (ethylene-insensitive 2) (Wild and Krutzfeldt, 2010; Kim et al., 2014), while the secondary wall-associated NAC domain protein1, SND1/NAC012, appears autoregulatory in nature (Wang et al., 2011). In sweet potato (Ipomoea batatas), the homeostasis of IbNAC1 is regulated by the competitive binding of two upstream TFs (the activator bHLH3 and the repressor bHLH4) to its promoter (Chen et al., 2016). Seven vascular-related NAC domains (AtVNDs) can induce the expression of AtVND7 by binding to its promoter (Endo et al., 2015; Nakano et al., 2015).
NAC TFs have also evolved multiple post-transcriptional regulatory processes, including miRNA-mediated cleavage, alternative splicing, and trans-splicing, in regulating the transcripts of NAC functional genes, such as the microRNA164 (miR164)-mediated and organ boundary formation regulatory genes (e.g., CUC1/AtNAC054, GOBLET/GOB, and SlNAM2) (Samad et al., 2017), the petal size regulatory genes (e.g., RhNAC100 in Rosa hybrid) (Pei et al., 2013), and the cell wall thickening genes (e.g., PtrWND1B-s and PtrWND1B-l in Populus trichocarpa) produced by intron-mediated alternative splicing of the wood-associated NAC TF1B (PtrWND1B) (Zhao et al., 2014).
Nuclear localization is a prerequisite for a protein to function as a TF; thus, NACs contain a bipartite NLS within the D subdomain. However, the Arabidopsis NTL4/AtNAC053 protein accumulates early in the ER, and its C-terminal portion may move to the nucleus after its cleavage from the ER (Kim et al., 2007), as confirmed by several similar reports on NACs targeted to the nucleus by homomerization/heteromerization (Seo et al., 2010; Ng et al., 2013; Lee et al., 2014; Mathew et al., 2016). These findings uncover an important regulatory mechanism of the activity of NACs from early PM/ER localization to nucleus translocation.
In addition, NACs participate in post-translational modifications through reversible acetylation and phosphorylation, protein–protein interactions, and ubiquitin-mediated proteolysis. For instance, the phosphorylation of the membrane-associated NTL6/AtNAC062 by SnRK2.8 (SNF1-related protein kinase) promotes the entry of NTL6 into the nucleus during drought conditions (Robertson, 2004; Kim et al., 2012; Guan et al., 2014). NAC proteins can form both homo- and heterodimers through the conserved β-sheet domain, which is influenced by the TRR/TAR (Zhu et al., 2012). The rice OsNAC29 and OsNAC31 independently interact with OsSLR1 (SLENDER RICE1), a repressor for OsMYB61, which promotes cellulose synthesis by increasing the transcripts of a cellulose synthase gene (Huang et al., 2015). The involvement of NAC1 in auxin signaling is ubiquitinated by SlNAT5, an E3 ubiquitin ligase, which leads to the degradation of NAC1 and the downregulation of the auxin signal in plant cells (Xie et al., 2002; Guo et al., 2015). In summary, NAC TFs have shown varied regulatory mechanisms at the transcriptional, post-transcriptional, translational, and post-translational levels.
NAC transcription factors regulate the transition from vegetative to reproductive growth and flower formation essential to crop yield
Flowering is a critical agricultural trait associated with fruit yield. Flower initiation and fusion congenitally occur within a single whorl of floral organs to form a connation or union, such as the union of sepals to form a calyx tube, the adnation of stamens and petals to form a corolla tube, and the connation of carpels to form the gynoecium (Ma et al., 2017; Phillips et al., 2020). In most angiosperms, the fusion of various floral organs to form diverse flowers and inflorescences with different floral colors, sizes, shapes, scents, inflorescences, and flowering times that are linked to sepals/petals, stamens, and carpels—from free to fused structures, in addition to a spiral arrangement from the whorled corolla and androecium to a single flower or various inflorescences—is closely related to NAM/CUC3-mediated boundary formation through meristematic development and differentiation (Ma et al., 2017; Phillips et al., 2020).
In the primordium, the formation of distinct boundaries is a critical step to floral organ initiation. The meristematic activity between organs allows floral fusion, which is associated with the early identified genes that are key to boundary formation, separate lateral organ development, and floral fusion, such as the Arabidopsis LOB (lateral organ boundaries) and Petunia hybrida NAM/CUC3 (no apical meristem), during the transition from vegetative to reproductive growth (Souer et al., 1996; Husbands et al., 2007). Indeed, variations in the expression of the NAM/CUC3 genes correlate with the variations in fusion within floral organs due to nam mutants failing to initiate a SAM (Zhong et al., 2016). Similarly, the Arabidopsis genes CUC1/CUC2 and ATAF1/ATAF2 possess a mutant phenotype similar to the nam mutant. These mutations are associated with failure to initiate the SAM and defective to boundary formation, leading to the fusion of cotyledons and floral whorls (Aida et al., 1997). A distinct boundary is required for proper leaflet formation, as evidenced by a tomato gob (goblet) mutant similar to the nam (Berger et al., 2009). Taken together, the tomato gob mutant is phenotypically similar to the nam/cuc3 mutants in both Arabidopsis thaliana and P. hybrida, in terms of failing to initiate the SAM and the presence of fused cotyledons and floral organs, suggesting a model to meet the requirements of higher NAM/CUC3 expression to increase the separation of the structures and lower expression to promote organ fusion. To a large extent, the NAM/CUC3 family promotes the evolution of floral fusion phenotypes by controlling organ boundary formation (Phillips et al., 2020).
Interestingly, phylogenetic analyses based on comparative genomics across algae and higher plants showed that the NAC family might date back to streptophyte green algae (Maugarny-Calès et al., 2016). From the view of evolution, after the transition of NACs from algae to land organisms, they subsequently expanded throughout land plants, from 20–30 proteins in mosses and lycophytes to over 100 copies in various angiosperm species (Zhu et al., 2012; Pereira-Santana et al., 2015; Maugarny-Calès et al., 2016), among which NAM/CUC3 became a distinct subfamily while the NAM clade (NAM/CUC1/CUC2) was clustered as a sister group of the CUC3 proteins throughout angiosperms (Maugarny et al., 2016). The function of NACs in the formation of vascular tissues promotes flower diversification and makes plants more adaptable to various land environments through seed propagation, which is a landmark event in the evolution of land plants (Maugarny-Calès et al., 2016). In summary, the NAC family plays a pivotal role in the formation of the SAM, which is fundamental to flowering and post-flowering phylogenetic processes, including organ boundary formation, secondary wall thickening, floral development, flowering, embryo and seed development, and fruit development and ripening.
Indeed, the identification of the nam mutant and the evolution of the later redundant CUC1/AtNAC054, CUC2/AtNAC098, and CUC3/AtNAC031 uncovered the specialization and diversification of NACs in the formation of embryos and flowers (Souer et al., 1996; Hibara et al., 2006; Raman et al., 2008). At the molecular level, CUC1 directly regulates the expression of LSH3/LSH4 (light-dependent short hypocotyls 3/4) in shoot organ boundary cells, which are associated with the transition from shoot vegetative growth to flower reproductive growth (Takeda et al., 2011). It was further found that SlNAM2 participates in establishing the tomato flower whorl and sepal boundaries (Hendelman et al., 2013). In addition, CUC1–CUC3, NAM, and NH16 can suppress vegetable growth and, thus, promote establishment of the boundaries of the floral organs, such as petals, sepals, and stamens, of plants (Vroemen et al., 2003; Zhong et al., 2016). Therefore, NACs are important in the formation and the maintenance of the different meristem tissues for organ boundary formation and are essential to the transition from vegetative to reproductive growth, which is necessary for embryogenesis, seed formation, and fruit development.
Owing to the vital role of NACs in cell wall biosynthesis in higher plants, it is reasonable that some of the NAC TFs, such as NST1/AtNAC043, NST2/AtNAC066, and NST3/AtNAC012 (NAC secondary wall thickening promoting factors 1–3), positively regulate secondary wall thickening and, thus, function in Arabidopsis silique formation and anther dehiscence, which are also negatively regulated by AtNAC053, an anther indehiscence factor (Mitsuda et al., 2005; Shih et al., 2014). In addition, NARS1/NARS2 (NAC-regulated seed morphology1/2), also known as NAC2 and NAM, redundantly mediated Arabidopsis seed embryogenesis by regulating the development and degeneration of ovule integuments (Kunieda et al., 2008). Moreover, the three CUC proteins (CUC1–CUC3) are involved in the initiation and separation of ovules during Arabidopsis reproductive development. CUC1 and CUC2 determine the number of ovules, while CUC2 and CUC3 control organ separation (Gonçalves et al., 2015). Interestingly, NAM-B1 participates in the remobilization of nutrients from the vegetative tissue to grain formation in wheat (Waters et al., 2009). In addition, three rice NAC TFs, namely, OsNAC020, OsNAC026, and OsNAC023, were found to be strongly associated with seed size and grain yield (Mathew et al., 2016), as well as another NAC TF (RhNAC100) in ETH-promoted rose flower opening (Pei et al., 2013). Interestingly, a recent study has found loss of function in tomato OPEN STOMATA 1 (SlOST1), a protein kinase essential for ABA signaling and abiotic stress response. slost1 mutants also exhibited a late flowering phenotype under normal and drought stress conditions through SlOST1 interacting with and phosphorylating the NAC-type TF, VASCULAR PLANT ONE-ZINC FINGER 1 (SlVOZ1), providing insights into a novel strategy to balance drought stress response and flowering (Chong et al., 2022).
It is worth noting that, in the long-day (LD) model plant Arabidopsis, about 180 regulators have been confirmed to be involved in the control of flowering time, among which CONSTANS (CO) is a central activator, FLOWERING LOCUS C (FLC) is a central suppressor, and SQUAMOSA PROMOTER BINDING LIKE (SPL) is a positive TF, together with their downstream floral TFs, including FT (FLOWERING LOCUS T), SOC1 (SUPPRESSOR OF OVEREXPRESSION OF CO1), and AGL24 (AGAMOUS-like24) that activate the meristemic genes LFY (LEAFY), AP1 (APETALA1), SEP3 (SEPALLATA3), and FUL (FRUITFULL) (Blüemel et al., 2015). A more recent study has reported that strawberries (Fragaria sp.), a high-value horticultural crop, initiate flowering during the short photoperiod and low temperatures in the autumn (Liang et al., 2022). With the onset of inflorescence, the SAM may undergo a leaf-to-flower primordium transformation through floral induction, during which SOC1, TFL1, AP1, and CUC2 are vital for floral initiation (Liang et al., 2022). The diversity of NAC proteins is critical for flower formation and fruit development in higher plants. Although much progress has been made toward understanding the molecular mechanisms underlying the formation of a terminal flower through a series of important genes (such as LFY and AP1) to control the transition from SAM to flower formation (Figure 2) (Ma et al., 2017), how NACs regulate the expression of these genes is poorly understood and needs to be urgently explored in the future.
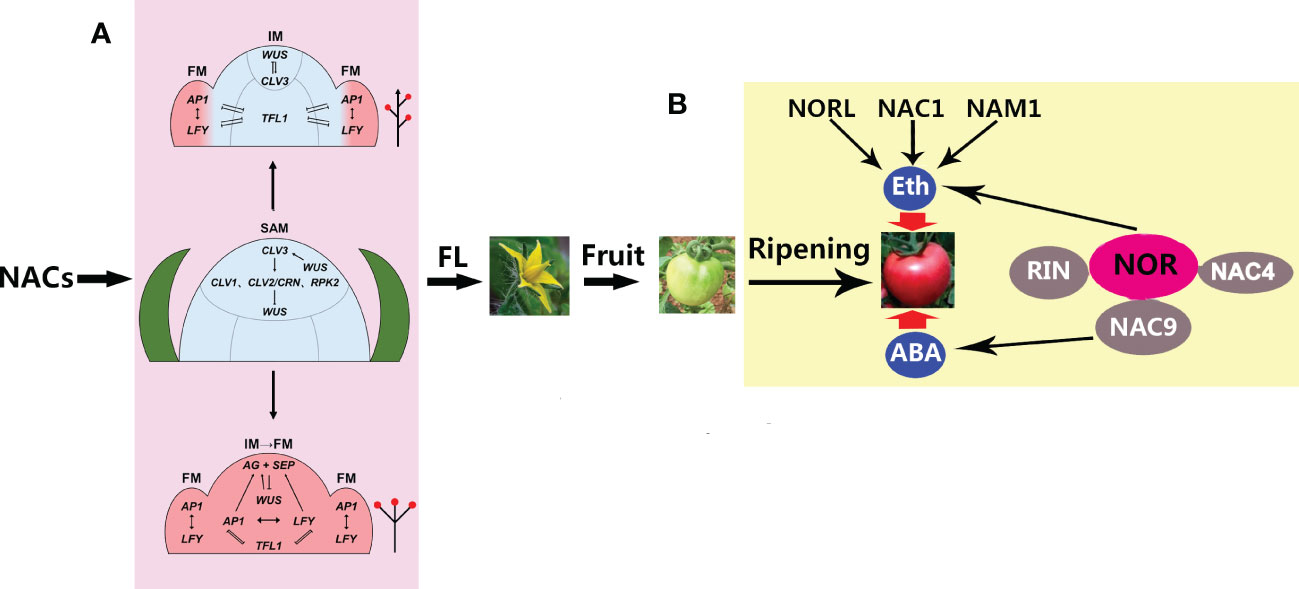
Figure 2 A model for the roles of NACs (NAM, ATAF, and CUC) in flower formation and ripening. (A) NACs control the transition of the shoot apical meristem (SAM) to the floral meristem (FM), including indeterminate inflorescence (up arrow) and determinate inflorescence (down arrow) (Ma et al., 2017). By controlling the relative expression levels of TFL1 (TERMINAL FLOWER 1) and the FM identity genes, AP1 and LFY (APETALA1 and LEAFY, respectively), in the distal inflorescence meristem (IM), the IM can remain meristematic and generate different numbers of FMs before converting to a terminal flower. A higher relative expression ratio prevents the expression of the FM identity genes LFY and AP1 in the distal IM during the onset of indeterminate inflorescence. However, during determinate inflorescence development, the expression of the FM identity genes in the distal IM can evoke the expression of the C class AGAMOUS (AG) and E class SEPALLATA (SEP) genes, thus determining the floral fate of the apical meristem. FL, flowering. (B) NACs control tomato fruit ripening through ethylene (ETH) and abscisic acid (ABA), which are involved in NOR, NORL, RIN, NAM1, and NAC1/4/9. NOR, RIN, and NAC4/9 constitute an interaction complex in which NOR, together with NORL, NAC1, and NAM1, regulates ripening through ETH, while NAC9 regulates ripening through ABA. NOR, non-ripening mutant; NORL, NOR-like; NAM, no apical meristem; RIN ripening inhibitor.
NAC transcription factors regulate fruit ripening fundamental to quality formation
Fruits include seeds with flower accessories, and fruit ripening is essential to human food, nutrition, and health. Ripening is regulated by plant hormones, including ETH in climacteric fruits and ABA in non-climacteric fruits (Bai et al., 2021; Kou et al., 2021; Li S. et al., 2021). Using the model plant for climacteric fruits, tomato, much progress has been made toward understanding the molecular mechanisms of NACs in fruit ripening through the biosynthesis and activity of ETH and ABA (Bai et al., 2021; Kou et al., 2021; Li S. et al., 2021). As a precursor of ETH biosynthesis, methionine (Met) is transformed into S-adenosyl methionine (SAMe) by SAMS (SAMe synthetase), and then SAMe is converted to ETH by 1-aminocyclopropane-1-carboxylic acid synthase (ACS) and 1-aminocylopropane-1-carboxylic acid oxidase (ACO). The ripening signal is relayed through a series of ETH perception and signaling transduction events involving ETH receptors (ETRs), ethylene-insensitive 2 (EIN2), EIN3/EIN3-like, and ETH response factors (ERFs). Similarly, 9-cis-epoxycarotenoid dioxygenase (NCED) and ABA 8′-hydroxylase (CYP707A) are key enzymes in ABA homeostasis. The ABA perception and signaling components include PYR/PYL/RCAR receptors, type 2C protein phosphatase (PP2Cs), SNF1-related kinase subfamily 2 (SnRK2), and downstream TFs, such as the ABA-responsive element binding proteins (AREBs) and ABA-responsive element binding factors (ABFs) (Bai et al., 2021; Kou et al., 2021; Li S. et al., 2021).
Tomato is a model for studying the roles of NACs in fruit ripening
The first landmark finding for NAC function in ripening was from the non-ripening mutant (nor) of tomato (Solanum lycopersicum) fruit, known as NAC-SlNOR, which plays a central role in its ripening (Giovannoni, 2004). However, recent studies have considered that nor is a gain-of-function, rather than a loss-of-function, mutant of SlNOR, and SlNOR positively regulates fruit ripening by binding to the promoter of SlACS2 as an activator (Wang et al., 2019; Gao et al., 2020). In addition, SlNOR is a direct target gene of SlRIN/SlMADS-RIN (RIPENING INHIBITOR) and SlAREB1 (a TF in ABA signaling). SlRIN and SlAREB1 bind to the SlNOR promoter to activate its expression and to promote the expression of SlACS2/SlACS4 and SlACO1 for ETH burst, suggesting that the SlAREB1-mediated transcriptional regulation of SlNOR is involved in the crosstalk between ETH and ABA in ripening (Fujisawa et al., 2013; Mou et al., 2018).
Later, many NAC proteins of SlNOR homologs were identified in ripening, such as SlNOR-like1 (SlNAC3/SlNAC4/SlNAC48) that favors ripening by binding to the promoter of multiple target genes involved in both ETH and ABA biosynthesis and signaling, including SlACS2, SlACS4, SlACS8, SlACO1, SlACO6, SlCYP707A1, SAPK3, and SlPYL9. In particular, SlNAC9 interacts with SlPYL9 and SlAREB1 (Kou et al., 2016; Gao et al., 2018; Yang et al., 2021). In contrast, SlNAC1 (SlNAC033) can directly bind to the promoter regions of SlACS2 and SlACO1 and has multiple effects on tomato fruit ripening by inhibiting the biosynthesis of ETH while enhancing the expression of SlNCED1 and SlNCED2 for the accumulation of ABA (Ma et al., 2014; Meng et al., 2016). It has been reported that SlNAC4 may be upstream of ETH biosynthesis by regulating the activities of SlRIN and SlNOR (Zhu M. et al., 2014). On the other hand, the expression of SlNAC7 was induced by ETH and ABA during color breaking, and, together with SlNAC6, it induced the expression of SlACS2, SlACS4, SlACO1, SlNCED1, SlNCED2, SlABA2, and aldehyde oxidase (SlAAO1/2) (Zhu M. K. et al., 2014; Jian et al., 2021). Similarly, a recent report has shown that the new NAC, SlNAM1, is a positive regulator of ripening initiation by promoting the expression of SlACS2 and SlACS4 (Gao et al., 2021). In addition, an Arabidopsis NAC, JUNGBRUNNEN1 (AtJUB1), was overexpressed in tomatoes and resulted in dwarf plants and delayed ripening fruits by directly binding to a line of gene promoters of GA3ox1 (GA 3-oxidase 1) and DWF4 (DWARF4), which are responsible for the biosynthesis of both gibberellin (GA) and brassinosteroid (BR), and indirectly inhibited the expression of ACS, ACO, SlERF.H15, and SlRIN (Shahnejat-Bushehri et al., 2017).
Notably, a recent study has found that the NAC-NOR target gene encoding lipoxygenase (SlLOXC) is involved in fatty acid-derived volatile synthesis by epigenetics. NAC-NOR activated the expression of SlDML2 (DNA demethylase 2) by directly binding to its promoter both in vitro and in vivo (Gao et al., 2022). On the other hand, the reduced NAC-NOR expression in the sldml2 mutant was accompanied by the hypermethylation of its promoter, demonstrating a relationship between SlDML2-mediated DNA demethylation and NAC-NOR during tomato fruit ripening (Gao et al., 2022).
In summary, a series of NAC TFs constitute a complex regulation network in tomato fruit ripening, mainly through ETH and ABA homeostasis and signaling levels, confirming that the NAC family plays a central role in fruit ripening.
NACs play an important role in the ripening of many climacteric fruits
In addition to tomatoes, the NAC family also plays vital roles in many climacteric fruits. For instance, at least 13 NAC genes have been found during apple (Malus domestica) fruit ripening (Wang et al., 2011; Zhang et al., 2018; Migicovsky et al., 2021). In banana fruits (Musa acuminata), a total of six NACs (MaNAC1–MaNAC6) were associated with fruit ripening (Shan et al., 2012; Li et al., 2020; Shan et al., 2020; Wei et al., 2021). In pear fruits (Pyrus pyrifolia), 185 NACs were found in the genome, among which 1) PpNAC56 may be related to fruit ripening (Ahmad et al., 2018); 2) PpNAP1, PpNAP4, and PpNAP6 may regulate ripening through ETH and ABA; 3) PpNOR controls fruit ripening in a conservative manner, similar to SlNOR; and 4) PpNAC.A59 directly promotes PpERF.A16 by binding to its promoter and facilitating the expression of PpACS1 and PpACO1, confirming that it plays an important role in fruit ripening through ETH biosynthesis and signaling (Li et al., 2016; Ahmad et al., 2018; Guo et al., 2021; Tan et al., 2021).
Moreover, during ripening in papaya fruits (Carica papaya L.), CpEIN3a could interact with CpNAC2 to activate the biosynthesis of carotenoids, while CpNAC3 could interact with CpMADS4 to activate the expression of pERF9 and CpEIL5 (Fu et al., 2017; Fu C. C. et al., 2021). In apricot (Prunus sibirica) fruits, 102 NACs were identified in the genome, among which PsNAC6/13/46/51/41/67/37/59 were highly expressed during the ripening stage (Xu et al., 2021). In kiwifruit (Actinidia deliciosa/Actinidia chinensis), AcNAC2–AcNAC4 are positive regulators of ripening through ETH production, while AdNAC2/AdNAC3 are promoted by methyl jasmonate (MeJA), suggesting a crosstalk between MeJA and ETH via AdNAC2 and AdNAC3 (Wang et al., 2020). In addition, the positive regulators of AdNAC6 and AdNAC7 in ripening are degraded by miR164, while this degradation is inhibited by ETH. Interestingly, the miR164–NAC pathway is conserved in both climacteric and non-climacteric fruits, including apple, pear, banana, peach, strawberry, citrus, and grape (Wang et al., 2020; Wu et al., 2020; Nieuwenhuizen et al., 2021).
Notably, a recent study has found that the NAC TF NOR is a master regulator of climacteric fruit ripening. Melon (Cucumis melo) (Rios et al., 2017) has climacteric and non-climacteric fruit ripening varieties, such as the climacteric and non-climacteric haplotypes CmNAC-NORS,N and CmNAC-NORA,S, respectively. A natural mutation in the transcriptional activation domain of CmNAC-NORS,N contributed to climacteric melon fruit ripening by directly activating its carotenoid, ETH, and ABA biosynthesis genes. In addition, CmNAC-NOR knockout in the climacteric-type melon cultivar “BYJH” completely inhibited fruit ripening, while ripening was delayed by 5–8 days in heterozygous cmnac-nor mutant fruits. Finally, CmNAC-NOR mediated the transcription of the “CmNAC-NOR–CmNAC73–CmCWINV2” module to enhance flesh sweetness. Altogether, the transcriptional activation activity of the climacteric haplotype, CmNAC-NORS,N, on these target genes was significantly higher than that of the non-climacteric haplotype, CmNAC-NORA,S, providing insights into the molecular mechanism of climacteric and non-climacteric fruit ripening in melon (Zhang et al., 2022). In addition, single-cytosine methylome analyses of a DNA demethylase ROS1 (MELO3C024516)-knockout mutant revealed changes in DNA methylation in the promoter regions of the key ripening genes, such as ACS1, ETR1, and ACO1, suggesting the importance of DNA demethylation in melon fruit ripening (Giordano et al., 2022). Thus, the NAC family participates in climacteric fruit ripening at multiple regulation levels.
NACs also play a role in the ripening of many non-climacteric fruits
Strawberry (Fragaria ananassa) is a non-climacteric model plant, and 112 NAC genes have been found in the genome, including six NACs, FaNAC006/021/022/035/042/092, related to fruit development and ripening. FaNAC035/FaRIF regulates ripening via ABA biosynthesis and signaling, demonstrating the presence of not only a FaNAC035-mediated crosstalk among various plant hormones but also a feedback mechanism between the ABA level and its signaling (Moyano et al., 2018; Martín-Pizarro et al., 2021). In addition, FcNAC1 could participate in ripening regulation by responding to upstream hormone signals, such as those of ABA and auxin (Carrasco-Orellana et al., 2018).
In citrus (Citrus reticulata) fruits, an ABA-deficient mutant resulted from the abnormally high expression of CrNAC036, which bound to the CrNCED5 promoter to inhibit its expression. On the other hand, CrMYB68 interacted with CrNAC036 to further inhibit CrNCED5 expression, finally synergistically retarding fruit ripening (Zhu et al., 2020). Although CitNAC62 is located in the cytoplasm, the nucleus-localized CitWRKY1 may interact with CitNAC62 to promote its movement (Li et al., 2017). In jujube (Ziziphus jujuba Mill.) fruits, the NAC LOC107435239 positively regulated the pericarp lignin accumulation during winter jujube fruit coloration by promoting the transcription of F5H (ferulate 5-hydroxylase), and ZjNAC13/14/38/41 showed high expression levels during ripening (Li M. et al., 2021; Zhang Q. et al., 2021). In grape (Vitis vinifera) fruits, VvNAC26 interacted with VvMADS9 to induce the gene expression related to ETH and ABA biosynthesis, resulting in early fruit ripening, suggesting that VvNAC26 promotes ripening by activating the levels of ETH and ABA (Zhang S. et al., 2021). In litchi (Litchi chinensis) fruits, LcNAC13 and LcR1MYB1 may antagonistically regulate the accumulation of anthocyanin during ripening (Jiang et al., 2019). In watermelon (Citrullus lanatus) fruits, ClNAC68 positively regulated sucrose accumulation during ripening by directly binding to the promoters of both the invertase (ClINV) and IAA-amino synthetase (ClGH3.6) to inhibit their expression (Lv et al., 2016; Wang et al., 2021).
In summary, NAC TFs not only directly target genes encoding enzymes related to ripening parameters, softening, sugar, coloring, and aroma but also indirectly affect fruit ripening by regulating the homeostasis of ABA and ETH through conserved and specific mechanisms, which are closely related to fruit quality. ETH-controlled climacteric fruit ripening (such as in tomato) via NAC TFs is directly targeted to the ETH biosynthesis-related genes including ACO and ACS, while also involved in the interaction between ABA and ETH, mostly through ABA to activate NOR transcription and ETH synthesis. With regard to non-climacteric fruit ripening, a homolog of the tomato NOR in strawberry, the NAC TF FaNAC035/FaRIF, can promote ABA biosynthesis via FaNCED3. Thus, NAC TFs play a core role in the two ripening types through the interplay of ETH and ABA.
Conclusion
The plant-specific NAC TFs not only control the aquatic-to-terrestrial evolution through vasculature formation but also govern the transition from vegetative to reproductive growth through directing SAM differentiation. In addition, they constitute a complex regulatory network that regulates fruit ripening via ABA and ETH, improving our understanding of the molecular mechanisms of NACs in fruit ripening (Figure 2). Nevertheless, the mechanisms through which NACs regulate the inflorescence formation-related genes are largely unknown. Given that NAC TFs belong to a large family, more focus should be given to the analysis of their redundancy in order to identify pivotal players in the regulation of flowering and ripening. In addition, more attention should be paid to the regulatory network of NACs regulating the ripening of non-climacteric fruits. Finally, NACs are potentially important manipulation targets for improving fruit quantity and quality in the future, likely through water control.
Author contributions
BH planned the outline of the review. BH and JL collected the available literature and performed re-analyses. BH and JL completed the draft of the paper. YQ and CL compiled and corrected the references. All authors contributed to the article and approved the submitted version.
Funding
The authors gratefully acknowledge the support of the Beijing Natural Science Foundation (6214042) and the National Natural Science Foundation of China (32101769).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Ahmad, M., Yan, X. H., Li, J. Z., Yang, Q. S., Jamil, W., Teng, Y. W., et al. (2018). Genome wide identification and predicted functional analyses of NAC transcription factors in Asian pears. BMC Plant Biol. 18, 214. doi: 10.1186/s12870-018-1427-x
Aida, M., Ishida, T., Fukaki, H., Fujisawa, H., Tasaka, M. (1997). Genes involved in organ separation in arabidopsis: An analysis of the cup-shaped cotyledon mutant. Plant Cell 9, 841–857. doi: 10.1105/tpc.9.6.841
Bai, Q., Huang, Y., Shen, Y. (2021). The physiological and molecular mechanism of abscisic acid in regulation of fleshy fruit ripening. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.619953
Berger, Y., Harpaz-Saad, S., Brand, A., Melnik, H., Sirding, N., Alvarez, J. P., et al. (2009). The NAC-domain transcription factor GOBLET specifes leaflet boundaries in compound tomato leaves. Development 136, 823–832. doi: 10.1242/dev.031625
Bhattacharjee, P., Das, R., Mandal, A., Kundu, P. (2017). Functional characterization of tomato membrane-bound NAC transcription factors. Plant Mol. Biol. 93, 511–532. doi: 10.1007/s11103-016-0579-z
Blüemel, M., Dally, N., Jung, C. (2015). Flowering time regulation in crops-what did we learn from arabidopsis? Curr. Opin. Biotechnol. 32, 121–129. doi: 10.1016/j.copbio.2014.11.023
Carrasco-Orellana, C., Stappung, Y., Mendez-Yanez, A., Allan, A. C., Espley, R. V., Plunkett, B. J., et al. (2018). Characterization of a ripening-related transcription factor FcNAC1 from fragaria chiloensis fruit. Sci. Rep. 8, 10524. doi: 10.1038/s41598-018-28226-y
Chen, S. P., Kuo, C. H., Lu, H. H., Lo, H. S., Yeh, K. W. (2016). The sweet potato NAC-domain transcription factor IbNAC1 is dynamically coordinated by the activator IbbHLH3 and the repressor IbbHLH4 to reprogram the defense mechanism against wounding. PloS Genet. 12, e1006397. doi: 10.1371/journal.pgen.1006397
Chong, L., Xu, R., Huang, P. C., Guo, P. C., Zhu, M. K., Du, H., et al. (2022). The tomato OST1-VOZ1 module regulates drought-mediated flowering. Plant Cell 34, 2001–2018. doi: 10.1093/plcell/koac026
Endo, H., Yamaguchi, M., Tamura, T., Nakano, Y., Nishikubo, N., Yoneda, A., et al. (2015). Multiple classes of transcription factors regulate the expression of VASCULAR-RELATED NAC-DOMAIN7, a master switch of xylem vessel differentiation. Plant Cell Physiol. 56, 242–254. doi: 10.1093/pcp/pcu134
Ernst, H. A., Olsen, A. N., Larsen, S., Lo Leggio, L. (2004). Structure of the conserved domain of ANAC, a member of the NAC family of transcription factors. EMBO Rep. 5, 297–303. doi: 10.1038/sj.embor.7400093
Forlani, S., Mizzotti, C., Masiero, S. (2021). The NAC side of the fruit: Tuning of fruit development and maturation. BMC Plant Biol. 21, 238. doi: 10.1186/s12870-021-03029-y
Fu, C. C., Chen, H. J., Gao, H. Y., Wang, S. L., Wang, N., Jin, J. C., et al. (2021). Papaya CpMADS4 and CpNAC3 co-operatively regulate ethylene signal genes CpERF9 and CpEIL5 during fruit ripening. Postharvest. Biol. Technol. 175, 111485. doi: 10.1016/j.postharvbio.2021.111485
Fu, C. C., Han, Y. C., Kuang, J. F., Chen, J. Y., Lu, W. J. (2017). Papaya CpEIN3a and CpNAC2 co-operatively regulate carotenoid biosynthesis related genes CpPDS2/4, CpLCY-e and CpCHY-b during fruit ripening. Plant Cell Physiol. 58, 2155–2165. doi: 10.1093/pcp/pcx149
Fu, B. L., Wang, W. Q., Liu, X. F., Duan, X. W., Allan, A. C., Grierson, D. (2021). An ethylene-hypersensitive methionine sulfoxide reductase regulated by NAC transcription factors increases methionine pool size and ethylene production during kiwifruit ripening. New Phytol. 2, 237–251. doi: 10.1111/nph.17560
Fujisawa, M., Nakano, T., Shima, Y., Ito, Y. (2013). A large-scale identification of direct targets of the tomato MADS box transcription factor RIPENING INHIBITOR reveals the regulation of fruit ripening. Plant Cell 25, 371–386. doi: 10.1105/tpc.112.108118
Gao, Y., Fan, Z. Q., Zhang, Q., Li, H. L., Liu, G. S., Jing, Y., et al. (2021). A tomato NAC transcription factor, SlNAM1, positively regulates ethylene biosynthesis and the onset of tomato fruit ripening. Plant J. 108, 1317–1331. doi: 10.1111/tpj.15512
Gao, Y., Lin, Y., Xu, M., Bian, H., Zhang, C., Wang, J., et al. (2022). The role and interaction between transcription factor NAC-NOR and DNA demethylase SlDML2 in the biosynthesis of tomato fruit flavor volatiles. New Phytol. 235, 1913–1926. doi: 10.1111/nph.18301
Gao, Y., Wei, W., Fan, Z., Zhao, X., Zhang, Y., Jing, Y., et al. (2020). Re-evaluation of the nor mutation and the role of the NAC-NOR transcription factor in tomato fruit ripening. J. Exp. Bot. 71, 3560–3574. doi: 10.1093/jxb/eraa131
Gao, Y., Wei, W., Zhao, X., Tan, X., Fan, Z., Zhang, Y., et al. (2018). A NAC transcription factor, NOR-like1, is a new positive regulator of tomato fruit ripening. Hortic. Res. 5, 1–18. doi: 10.1038/s41438-018-0111-5
Ghissing, U., Kutty, N. N., Bimolata, W., Samanta, T., Mitra, A. (2023). Comparative transcriptome analysis reveals an insight into the candidate genes involved in anthocyanin and scent volatiles biosynthesis in colour changing flowers of Combretum indicum. Plant Biol. (Stuttg). 25, 85–95. doi: 10.1111/plb.13481
Giordano, A., Santo Domingo, M., Quadrana, L., Pujol, M., Martin-Hernandez, A. M., Garcia-Mas, J. (2022). CRISPR/Cas9 gene editing uncovers the roles of CONSTITUTIVE TRIPLE RESPONSE 1 and REPRESSOR OF SILENCING 1 in melon fruit ripening and epigenetic regulation. J. Exp. Bot. 73, 4022–4033. doi: 10.1093/jxb/erac148
Giovannoni, J. J. (2004). Genetic regulation of fruit development and ripening. Plant Cell 16, S170–S180. doi: 10.1105/tpc.019158
Gonçalves, B., Hasson, A., Belcram, K., Cortizo, M., Morin, H., Nikovics, K., et al. (2015). A conserved role for CUP-SHAPED COTYLEDON genes during ovule development. Plant J. 83, 732–742. doi: 10.1111/tpj.12923
Guan, Q., Yue, X., Zeng, H., Zhu, J. (2014). The protein phosphatase RCF2 and its interacting partner NAC019 are critical for heat stress-responsive gene regulation and thermotolerance in Arabidopsis. Plant Cell. 26, 438–453. doi: 10.1105/tpc.113.118927
Guo, L., Li, Y., Zhang, C., Wang, Z., Carlson, J. E., Yin, W., et al. (2022). Integrated analysis of miRNAome transcriptome and degradome reveals miRNA-target modules governing floral florescence development and senescence across early- and late-flowering genotypes in tree peony. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.1082415
Guo, Z. H., Zhang, Y. J., Yao, J. L., Xie, Z. H., Zhang, Y. Y., Zhang, S. L., et al. (2021). The NAM/ATAF1/2/CUC2 transcription factor PpNAC.A59 enhances PpERF.A16 expression to promote ethylene biosynthesis during peach fruit ripening. Hortic. Res. 8, 209. doi: 10.1038/s41438-021-00644-6
Guo, W., Zhang, J., Zhang, N., Xin, M., Peng, H., Hu, Z., et al. (2015). The wheat NAC transcription factor TaNAC2L is regulated at the transcriptional and post-translational levels and promotes heat stress tolerance in transgenic Arabidopsis. PloS One 10, e0135667. doi: 10.1371/journal.pone.0135667
Hendelman, A., Stav, R., Zemach, H., Arazi, T. (2013). The tomato NAC transcription factor SlNAM2 is involved in flower-boundary morphogenesis. J. Exp. Bot. 64, 5497–5507. doi: 10.1093/jxb/ert324
Hibara, K., Karim, M. R., Takada, S., Taoka, K., Furutani, M., Aida, M., et al. (2006). Arabidopsis CUP-SHAPED COTYLEDON3 regulates postembryonic shoot meristem and organ boundary formation. Plant Cell 18, 2946–2957. doi: 10.1105/tpc.106.045716
Huang, D., Wang, S., Zhang, B., Shang-Guan, K., Shi, Y., Zhang, D., et al. (2015). A gibberellin-mediated DELLA-NAC signaling cascade regulates cellulose synthesis in rice. Plant Cell 27, 1681–1696. doi: 10.1105/tpc.15.00015
Husbands, A., Bell, E. M., Shuai, B., Smith, H. M., Springer, P. S. (2007). LATERAL ORGAN BOUNDARIES defines a new family of DNA-binding transcription factors and can interact with specific bHLH proteins. Nucleic Acids Res. 35, 6663–6671. doi: 10.1093/nar/gkm775
Jian, W., Zheng, Y., Yu, T., Cao, H., Chen, Y., Cui, Q., et al. (2021). SlNAC6, a NAC transcription factor, is involved in drought stress response and reproductive process in tomato. J. Plant Physiol. 264, 153483. doi: 10.1016/j.jplph.2021.153483
Jiang, G., Li, Z., Song, Y., Zhu, H., Lin, S., Huang, R., et al. (2019). LcNAC13 physically interacts with LcR1MYB1 to coregulate anthocyanin biosynthesis-related genes during litchi fruit ripening. Biomolecules 9, 135. doi: 10.3390/biom9040135
Kim, H. J., Hong, S. H., Kim, Y. W., Lee, I. H., Jun, J. H., Phee, B. K., et al. (2014). Gene regulatory cascade of senescence-associated NAC transcription factors activated by ETHYLENE-INSENSITIVE2-mediated leaf senescence signalling in Arabidopsis. J. Exp. Bot. 65, 4023–4036. doi: 10.1093/jxb/eru112
Kim, S. Y., Kim, S. G., Kim, Y. S., Seo, P. J., Bae, M., Yoon, H. K., et al. (2007). Exploring membrane-associated NAC transcription factors in Arabidopsis: implications for membrane biology in genome regulation. Nucleic Acids Res. 35, 203–213. doi: 10.1093/nar/gkl1068
Kim, Y. S., Kim, S. G., Park, J. E., Park, H. Y., Lim, M. H., Chua, N. H., et al. (2006). A membrane-bound NAC transcription factor regulates cell division in Arabidopsis. Plant Cell 18, 3132–3144. doi: 10.1105/tpc.106.043018
Kim, M. J., Park, M. J., Seo, P. J., Song, J. S., Kim, H. J., Park, C. M. (2012). Controlled nuclear import of the transcription factor NTL6 reveals a cytoplasmic role of SnRK2.8 in the drought-stress response. Biochem. J. 448, 353–363. doi: 10.1042/BJ20120244
Kou, X., Liu, C., Han, L., Wang, S., Xue, Z. (2016). NAC transcription factors play an important role in ethylene biosynthesis, reception and signaling of tomato fruit ripening. Mol. Genet. Genomics 291, 1205–1217. doi: 10.1007/s00438-016-1177-0
Kou, X., Zhou, J., Wu, C. E., Yang, S., Liu, Y., Chai, L., et al. (2021). The interplay between ABA/ethylene and NAC TFs in tomato fruit ripening: a review. Plant Mol. Biol. 106, 223–238. doi: 10.1007/s11103-021-01128-w
Kunieda, T., Mitsuda, N., Ohme-Takagi, M., Takeda, S., Aida, M., Tasaka, M., et al. (2008). NAC family proteins NARS1/NAC2 and NARS2/NAM in the outer integument regulate embryogenesis in Arabidopsis. Plant Cell 20, 2631–2642. doi: 10.1105/tpc.108.060160
Lee, S., Lee, H. J., Huh, S. U., Paek, K. H., Ha, J. H., Park, C. M. (2014). The Arabidopsis NAC transcription factor NTL4 participates in a positive feedback loop that induces programmed cell death under heat stress conditions. Plant Sci. 227, 76–83. doi: 10.1016/j.plantsci.2014.07.003
Li, S., Chen, K., Grierson, D. (2021). Molecular and hormonal mechanisms regulating fleshy fruit ripening. Cells 10, 1136. doi: 10.3390/cells10051136
Li, B., Fan, R., Yang, Q., Hu, C., Sheng, O., Deng, G., et al. (2020). Genome-wide identification and characterization of the NAC transcription factor family in Musa acuminata and expression analysis during fruit ripening. Int. J. Mol. Sci. 21, 634. doi: 10.3390/ijms21020634
Li, B. J., Grierson, D., Shi, Y., Chen, K. S. (2022). Roles of abscisic acid in regulating ripening and quality of strawberry, a model non-climacteric fruit. Hortic. Res. 9, uhac089. doi: 10.1093/hr/uhac089
Li, M., Hou, L., Liu, S. S., Zhang, C. X., Yang, W. C., Pang, X. M., et al. (2021). Genome-wide identification and expression analysis of NAC transcription factors in Ziziphus jujuba mill. reveal their putative regulatory effects on tissue senescence and abiotic stress responses. Ind. Crop Prod. 173, 114093. doi: 10.1016/j.indcrop.2021.114093
Li, F., Li, J., Qian, M., Han, M., Cao, L., Liu, H., et al. (2016). Identification of peach NAP transcription factor genes and characterization of their expression in vegetative and reproductive organs during development and senescence. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.00147
Li, S. J., Yin, X. R., Wang, W. L., Liu, X. F., Zhang, B., Chen, K. S. (2017). Citrus CitNAC62 cooperates with CitWRKY1 to participate in citric acid degradation via up-regulation of CitAco3. J. Exp. Bot. 68, 3419–3426. doi: 10.1093/jxb/erx187
Liang, J., Zheng, J., Wu, Z., Wang, H. (2022). Time-course transcriptomic profiling of floral induction in cultivated strawberry. Int. J. Mol. Sci. 23, 6126. doi: 10.3390/ijms23116126
Liu, G. S., Li, H. L., Grierson, D., Fu, D. Q. (2022). NAC transcription factor family regulation of fruit ripening and quality: a review. Cells 11, 525. doi: 10.3390/cells11030525
Liu, B., Santo-Domingo, M., Mayobre, C., Martín-Hernández, A. M., Pujol, M., Garcia-Mas, J. (2022). Knock-out of CmNAC-NOR affects melon climacteric fruit ripening. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.878037
Liu, X., Zhang, L., Yang, S. (2022). Analysis of floral organ development and sex determination in Schisandra chinensis by scanning electron microscopy and rna-sequencing. Life (Basel). 12, 1260. doi: 10.3390/life12081260
Lv, X., Lan, S., Guy, K. M., Yang, J., Zhang, M., Hu, Z. (2016). Global expressions landscape of NAC transcription factor family and their responses to abiotic stresses in Citrullus lanatus. Sci. Rep. 6, 30574. doi: 10.1038/srep30574
Ma, N., Feng, H., Meng, X., Li, D., Yang, D., Wu, C., et al. (2014). Overexpression of tomato SlNAC1 transcription factor alters fruit pigmentation and softening. BMC Plant Biol. 14, 351. doi: 10.1186/s12870-014-0351-y
Ma, Q., Zhang, W., Xiang, Q. Y. J. (2017). Evolution and developmental genetics of floral display–a review of progress. J. Syst. Evol. 55, 487–4515. doi: 10.1111/jse.12259
Martín-Pizarro, C., Vallarino, J. G., Osorio, S., Meco, V., Urrutia, M., Pillet, J., et al. (2021). The NAC transcription factor FaRIF controls fruit ripening in strawberry. Plant Cell. 33, 1574–1593. doi: 10.1093/plcell/koab070
Mathew, I. E., Agarwal, P. (2018). May the fittest protein evolve: Favoring the plant-specific origin and expansion of NAC transcription factors. Bioessays 40, e1800018. doi: 10.1002/bies.201800018
Mathew, I. E., Das, S., Mahto, A., Agarwal, P. (2016). Three rice NAC transcription factors heteromerize and are associated with seed size. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.01638
Maugarny, A., Gonalves, B., Arnaud, N., Laufs, P. (2016). CUC transcription factors: To the meristem and beyond. Plant Transcription. Factors., 229–247.
Maugarny-Calès, A., Gonçalves, B., Jouannic, S., Melkonian, M., Ka-Shu Wong, G., Laufs, P. (2016). Apparition of the NAC transcription factors predates the emergence of land plants. Mol. Plant 9, 1345–1348. doi: 10.1016/j.molp.2016.05.016
Meng, C., Yang, D. Y., Ma, X. C., Zhao, W. Y., Liang, X. Q., Ma, N. N., et al. (2016). Suppression of tomato SlNAC1 transcription factor delays fruit ripening. J. Plant Physiol. 193, 88–96. doi: 10.1016/j.jplph.2016.01.014
Migicovsky, Z., Yeats, T. H., Watts, S., Song, J., Forney, C. F., Burgher-MacLellan, K., et al. (2021). Apple ripening is controlled by a NAC transcription factor. Front. Genet. 12. doi: 10.3389/fgene.2021.671300
Mitsuda, N., Seki, M., Shinozaki, K., Ohme-Takagi, M. (2005). The NAC transcription factors NST1 and NST2 of Arabidopsis regulate secondary wall thickenings and are required for anther dehiscence. Plant Cell. 17, 2993–3006. doi: 10.1105/tpc.105.036004
Mou, W., Li, D., Luo, Z., Li, L., Mao, L., Ying, T. (2018). SlAREB1 transcriptional activation of NOR is involved in abscisic acid-modulated ethylene biosynthesis during tomato fruit ripening. Plant Sci. 276, 239–249. doi: 10.1016/j.plantsci.2018.07.015
Moyano, E., Martinez-Rivas, F. J., Blanco-Portales, R., Molina-Hidalgo, F. J., Ric-Varas, P., Matas-Arroyo, A. J., et al. (2018). Genome-wide analysis of the NAC transcription factor family and their expression during the development and ripening of the Fragaria × ananassa fruits. PloS One 13, e0196953. doi: 10.1371/journal.pone.0196953
Nakano, Y., Yamaguchi, M., Endo, H., Rejab, N. A., Ohtani, M. (2015). NAC-MYB-based transcriptional regulation of secondary cell wall biosynthesis in land plants. Front. Plant Sci. 6. doi: 10.3389/fpls.2015.00288
Ng, S., Ivanova, A., Duncan, O., Law, S. R., Van Aken, O., De Clercq, I., et al. (2013). A membrane-bound NAC transcription factor, ANAC017, mediates mitochondrial retrograde signaling in Arabidopsis. Plant Cell 25, 3450–3471. doi: 10.1105/tpc.113.113985
Nieuwenhuizen, N. J., Chen, X. Y., Pellan, M., Zhang, L., Guo, L., Laing, W. A., et al. (2021). Regulation of wound ethylene biosynthesis by NAC transcription factors in kiwifruit. BMC Plant Biol. 21, 411. doi: 10.1186/s12870-021-03154-8
Olsen, A. N., Ernst, H. A., Lo Leggio, L., Skriver, K. (2005). NAC transcription factors: Structurally distinct, functionally diverse. Trends Plant Sci. 10, 79–87. doi: 10.1016/j.tplants.2004.12.010
Ooka, H., Satoh, K., Doi, K., Nagata, T., Otomo, Y., Murakami, K., et al. (2003). Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. DNA Res. 10, 239–247. doi: 10.1093/dnares/10.6.239
Pei, H., Ma, N., Tian, J., Luo, J., Chen, J., Li, J., et al. (2013). An NAC transcription factor controls ethylene-regulated cell expansion in flower petals. Plant Physiol. 163, 775–791. doi: 10.1104/pp.113.223388
Pereira-Santana, A., Alcaraz, L. D., Castaño, E., Sanchez-Calderon, L., Sanchez-Teyer, F., Rodriguez-Zapata, L. (2015). Comparative genomics of NAC transcriptional factors in angiosperms: implications for the adaptation and diversification of flowering plants. PloS One 10, e0141866. doi: 10.1371/journal.pone.0141866
Phillips, H. R., Landis, J. B., Specht, C. D. (2020). Revisiting floral fusion: the evolution and molecular basis of a developmental innovation. J. Exp. Bot. 71, 3390–3404. doi: 10.1093/jxb/eraa125
Qi, X., Dong, Y., Liu, C., Song, L., Chen, L., Li, M. (2022). The PavNAC56 transcription factor positively regulates fruit ripening and softening in sweet cherry (Prunus avium). Physiol. Plant 174, e13834. doi: 10.1111/ppl.13834
Raman, S., Greb, T., Peaucelle, A., Blein, T., Laufs, P., Theres, K. (2008). Interplay of miR164, CUP-SHAPED COTYLEDON genes and LATERAL SUPPRESSOR controls axillary meristem formation in Arabidopsis thaliana. Plant J. 55, 65–76. doi: 10.1111/j.1365-313X.2008.03483.x
Ríos, P., Argyris, J., Vegas, J., Leida, C., Kenigswald, M., Tzuri, G., et al. (2017). ETHQV6.3 is involved in melon climacteric fruit ripening and is encoded by a NAC domain transcription factor. Plant J. 91, 671–683. doi: 10.1111/tpj.13596
Robertson, M. (2004). Two transcription factors are negative regulators of gibberellin response in the HvSPY-signaling pathway in barley aleurone. Plant Physiol. 136, 2747–2761. doi: 10.1104/pp.104.041665
Samad, A., Sajad, M., Nazaruddin, N., Fauzi, I. A., Murad, A., Zainal, Z., et al. (2017). MicroRNA and transcription factor: Key players in plant regulatory network. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.00565
Seo, P. J., Kim, S. G., Park, C. M. (2008). Membrane-bound transcription factors in plants. Trends Plant Sci. 13, 550–556. doi: 10.1016/j.tplants.2008.06.008
Seo, P. J., Kim, M. J., Park, J. Y., Kim, S. Y., Jeon, J., Lee, Y. H., et al. (2010). Cold activation of a plasma membrane-tethered NAC transcription factor induces a pathogen resistance response in Arabidopsis. Plant J. 61, 661–671. doi: 10.1111/j.1365-313X.2009.04091.x
Shahnejat-Bushehri, S., Allu, A. D., Mehterov, N., Thirumalaikumar, V. P., Alseekh, S., Fernie, A. R., et al. (2017). Arabidopsis NAC transcription factor JUNGBRUNNEN1 exerts conserved control over gibberellin and brassinosteroid metabolism and signaling genes in tomato. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.00214
Shan, W., Kuang, J. F., Chen, L., Xie, H., Peng, H. H., Xiao, Y. Y., et al. (2012). Molecular characterization of banana NAC transcription factors and their interactions with ethylene signalling component EIL during fruit ripening. J. Exp. Bot. 63, 5171–5187. doi: 10.1093/jxb/ers178
Shan, W., Kuang, J. F., Wei, W., Fan, Z. Q., Deng, W., Li, Z. G., et al. (2020). MaXB3 modulates MaNAC2, MaACS1, and MaACO1 stability to repress ethylene biosynthesis during banana fruit ripening. Plant Physiol. 184, 1153–1171. doi: 10.1104/pp.20.00313
Shih, C. F., Hsu, W. H., Peng, Y. J., Yang, C. H. (2014). The NAC-like gene ANTHER INDEHISCENCE FACTOR acts as a repressor that controls anther dehiscence by regulating genes in the jasmonate biosynthesis pathway in Arabidopsis. J. Exp. Bot. 65, 621–639. doi: 10.1093/jxb/ert412
Singh, S., Koyama, H., Bhati, K. K., Alok, A. (2021). The biotechnological importance of the plant-specific NAC transcription factor family in crop improvement. J. Plant Res. 134, 475–495. doi: 10.1007/s10265-021-01270-y
Souer, E., van Houwelingen, A., Kloos, D., Mol, J., Koes, R. (1996). The no apical meristem gene of Petunia is required for pattern formation in embryos and flowers and is expressed at meristem and primordia boundaries. Cell 85, 159–170. doi: 10.1016/s0092-8674(00)81093-4
Takeda, S., Hanano, K., Kariya, A., Shimizu, S., Zhao, L., Matsui, M., et al. (2011). CUP-SHAPED COTYLEDON1 transcription factor activates the expression of LSH4 and LSH3, two members of the ALOG gene family, in shoot organ boundary cells. Plant J. 66, 1066–1077. doi: 10.1111/j.1365-313X.2011.04571.x
Tan, Q., Li, S., Zhang, Y., Chen, M., Wen, B., Jiang, S., et al. (2021). Chromosome-level genome assemblies of five Prunus species and genome-wide association studies for key agronomic traits in peach. Hortic. Res. 8, 213. doi: 10.1038/s41438-021-00648-2
Vroemen, C. W., Mordhorst, A. P., Albrecht, C., Kwaaitaal, M. A., de Vries, S. C. (2003). The CUP-SHAPED COTYLEDON3 gene is required for boundary and shoot meristem formation in Arabidopsis. Plant Cell. 15, 1563–1577. doi: 10.1105/tpc.012203
Wang, R., Tavano, E., Lammers, M., Martinelli, A. P., Angenent, G. C., de Maagd, R. A. (2019). Re-evaluation of transcription factor function in tomato fruit development and ripening with CRISPR/Cas9-mutagenesis. Sci. Rep. 9, 1696. doi: 10.1038/s41598-018-38170-6
Wang, D., Wang, J., Wang, Y., Yao, D., Niu, Y. (2022). Metabolomic and transcriptomic profiling uncover the underlying mechanism of color differentiation in Scutellaria baicalensis georgi. flowers. Front. Plant Sci. 3. doi: 10.3389/fpls.2022.884957
Wang, W. Q., Wang, J., Wu, Y. Y., Li, D. W., Allan, A. C., Yin, X. R. (2020). Genome-wide analysis of coding and non-coding RNA reveals a conserved miR164-NAC regulatory pathway for fruit ripening. New Phytol. 225, 1618–1634. doi: 10.1111/nph.16233
Wang, J., Wang, Y., Zhang, J., Ren, Y., Li, M., Tian, S., et al. (2021). The NAC transcription factor ClNAC68 positively regulates sugar content and seed development in watermelon by repressing ClINV and ClGH3.6. Hortic. Res. 8, 214. doi: 10.1038/s41438-021-00710-z
Wang, D., Yu, Y., Liu, Z., Li, S., Wang, Z., Xiang, F. (2016). Membrane-bound NAC transcription factors in maize and their contribution to the oxidative stress response. Plant Sci. 250, 30–39. doi: 10.1016/j.plantsci.2016.05.019
Wang, H., Zhao, Q., Chen, F., Wang, M., Dixon, R. A. (2011). NAC domain function and transcriptional control of a secondary cell wall master switch. Plant J. 68, 1104–1114. doi: 10.1111/j.1365-313X.2011.04764.x
Waters, B. M., Uauy, C., Dubcovsky, J., Grusak, M. A. (2009). Wheat (Triticum aestivum) NAM proteins regulate the translocation of iron, zinc, and nitrogen compounds from vegetative tissues to grain. J. Exp. Bot. 60, 4263–4274. doi: 10.1093/jxb/erp257
Wei, W., Yang, Y. Y., Su, X. G., Kuang, J. F., Chen, J. Y., Lu, W. J., et al. (2021). MaRTH1 suppression of ethylene response during banana fruit ripening and is controlled by MaXB3-MaNAC2 regulatory module. Postharvest. Biol. Technol. 182, 111707. doi: 10.1016/j.postharvbio.2021.111707
Wild, J. M., Krützfeldt, N. O. (2010). Neocortical-like organization of avian auditory 'cortex'. commentary on Wang y, brzozowska-prechtl a, karten HJ, (2010): Laminar and columnar auditory cortex in avian brain. Proc. natl. acad. sci. U.S.A. 107:12676-12681. Brain Behav. Evol. 76, 89–92. doi: 10.1159/000320215
Wu, Y. Y., Liu, X. F., Fu, B. L., Zhang, Q. Y., Tong, Y., Wang, J., et al. (2020). Methyl jasmonate enhances ethylene synthesis in kiwifruit by inducing NAC genes that activate ACS1. J. Agric. Food Chem. 68, 3267–3276. doi: 10.1021/acs.jafc.9b07379
Xie, Q., Guo, H. S., Dallman, G., Fang, S., Weissman, A. M., Chua, N. H. (2002). SINAT5 promotes ubiquitin-related degradation of NAC1 to attenuate auxin signals. Nature 419, 167–170. doi: 10.1038/nature00998
Xu, W. Y., Chen, C., Gou, N. N., Huang, M. Z., Wuyun, T. N., Zhu, G. P., et al. (2021). Genome-wide identification and expression analysis of NAC transcription factor family genes during fruit and kernel development in Siberian apricot. J. Am. Soc Hortic. Sci. 146, 276–285. doi: 10.21273/JASHS05007-20
Yamasaki, K., Kigawa, T., Seki, M., Shinozaki, K., Yokoyama, S. (2013). DNA-Binding domains of plant-specific transcription factors: structure, function, and evolution. Trends Plant Sci. 18, 267–276. doi: 10.1016/j.tplants.2012.09.001
Yang, S., Zhou, J. Q., Watkins, C. B., Wu, C. E., Feng, Y. C., Zhao, X. Y., et al. (2021). NAC transcription factors SNAC4 and SNAC9 synergistically regulate tomato fruit ripening by affecting expression of genes involved in ethylene and abscisic acid metabolism and signal transduction. Postharvest. Biol. Technol. 178, 111555. doi: 10.1016/j.postharvbio.2021.111555
Zhang, S., Dong, R., Wang, Y., Li, X., Ji, M., Wang, X. (2021). NAC domain gene VvNAC26 interacts with VvMADS9 and influences seed and fruit development. Plant Physiol. Biochem. 164, 63–72. doi: 10.1016/j.plaphy.2021.04.031
Zhang, Q. J., Li, T., Zhang, L. J., Dong, W. X., Wang, A. D. (2018). Expression analysis of NAC genes during the growth and ripening of apples. Hortic. Sci. 45, 1–10. doi: 10.17221/153/2016-HORTSCI
Zhang, J., Li, M., Zhang, H., Gong, G., Wang, M., Xu, Y. (2022). Natural variation in the NAC transcription factor NONRIPENING contributes to melon fruit ripening. J. Integr. Plant Biol. 64, 1448–1461. doi: 10.1111/jipb.13278
Zhang, Q., Wang, L., Wang, Z., Zhang, R., Liu, P., Liu, M., et al. (2021). The regulation of cell wall lignification and lignin biosynthesis during pigmentation of Winter jujube. Hortic. Res. 8, 238. doi: 10.1038/s41438-021-00670-4
Zhao, Y., Sun, J., Xu, P., Zhang, R., Li, L. (2014). Intron-mediated alternative splicing of WOOD-ASSOCIATED NAC TRANSCRIPTION FACTOR1B regulates cell wall thickening during fiber development in Populus species. Plant Physiol. 164, 765–776. doi: 10.1104/pp.113.231134
Zhong, J., Powell, S., Preston, J. C. (2016). Organ boundary NAC-domain transcription factors are implicated in the evolution of petal fusion. Plant Biol. 18, 893–902. doi: 10.1111/plb.12493
Zhu, M., Chen, G., Zhou, S., Tu, Y., Wang, Y., Dong, T., et al. (2014). A new tomato NAC (NAM/ATAF1/2/CUC2) transcription factor, SlNAC4, functions as a positive regulator of fruit ripening and carotenoid accumulation. Plant Cell Physiol. 55, 119–135. doi: 10.1093/pcp/pct162
Zhu, M. K., Hu, Z. L., Zhou, S., Wang, L. L., Dong, T. T., Pan, Y., et al. (2014). Molecular characterization of six tissue-specific or stress-inducible genes of NAC transcription factor family in tomato (Solanum lycopersicum). J. Plant Growth Regul. 33, 730–744. doi: 10.1007/s00344-014-9420-6
Zhu, F., Luo, T., Liu, C., Wang, Y., Zheng, L., Xiao, X., et al. (2020). A NAC transcription factor and its interaction protein hinder abscisic acid biosynthesis by synergistically repressing NCED5 in Citrus reticulata. J. Exp. Bot. 71, 3613–3625. doi: 10.1093/jxb/eraa118
Keywords: flower, fruit ripening, NAC transcription factor, abscisic acid (ABA), ethylene
Citation: Liu J, Qiao Y, Li C and Hou B (2023) The NAC transcription factors play core roles in flowering and ripening fundamental to fruit yield and quality. Front. Plant Sci. 14:1095967. doi: 10.3389/fpls.2023.1095967
Received: 11 November 2022; Accepted: 01 February 2023;
Published: 23 February 2023.
Edited by:
Shifeng Cao, Zhejiang Wanli University, ChinaReviewed by:
Yuanyue Shen, Beijing University of Agriculture, ChinaQiaoli Xie, Chongqing University, China
Copyright © 2023 Liu, Qiao, Li and Hou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bingzhu Hou, aG91YnpAaWJjYXMuYWMuY24=
 Jianfeng Liu
Jianfeng Liu Yuyuan Qiao
Yuyuan Qiao Cui Li1,2
Cui Li1,2 Bingzhu Hou
Bingzhu Hou