
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 27 January 2023
Sec. Functional and Applied Plant Genomics
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1093913
This article is part of the Research TopicPeanut Genomics and Biotechnology in Breeding ApplicationsView all 11 articles
Plant protein phosphatase 2C (PP2C) play important roles in response to salt stress by influencing metabolic processes, hormone levels, growth factors, etc. Members of the PP2C family have been identified in many plant species. However, they are rarely reported in peanut. In this study, 178 PP2C genes were identified in peanut, which were unevenly distributed across the 20 chromosomes, with segmental duplication in 78 gene pairs. AhPP2Cs could be divided into 10 clades (A-J) by phylogenetic analysis. AhPP2Cs had experienced segmental duplications and strong purifying selection pressure. 22 miRNAs from 14 different families were identified, targeting 57 AhPP2C genes. Gene structures and motifs analysis exhibited PP2Cs in subclades AI and AII had high structural and functional similarities. Phosphorylation sites of AhPP2C45/59/134/150/35/121 were predicted in motifs 2 and 4, which located within the catalytic site at the C-terminus. We discovered multiple MYB binding factors and ABA response elements in the promoter regions of the six genes (AhPP2C45/59/134/150/35/121) by cis-elements analysis. GO and KEGG enrichment analysis confirmed AhPP2C-A genes in protein binding, signal transduction, protein modification process response to abiotic stimulus through environmental information processing. Based on RNA-Seq data of 22 peanut tissues, clade A AhPP2Cs showed a varying degree of tissue specificity, of which, AhPP2C35 and AhPP2C121 specifically expressed in seeds, while AhPP2C45/59/134/150 expressed in leaves and roots. qRT-PCR indicated that AhPP2C45 and AhPP2C134 displayed significantly up-regulated expression in response to salt stress. These results indicated that AhPP2C45 and AhPP2C134 could be candidate PP2Cs conferring salt tolerance. These results provide further insights into the peanut PP2C gene family and indicate PP2Cs potentially involved in the response to salt stress, which can now be further investigated in peanut breeding efforts to obtain cultivars with improved salt tolerance.
Protein phosphorylation/dephosphorylation by protein kinases (PKs) and protein phosphatases (PPs) regulates targeted protein function in signal transduction cascades, leading to alterations in metabolism and/or gene expression in many biological processes, such as the development and stimulation of responses to the external environment (Lessard et al., 1997). PKs primarily phosphorylate serine (Ser), threonine (Thr) and tyrosine (Tyr), whereas PPs reverse this process (Li et al., 2014). PPs can be divided into three categories based on their substrate specificity: serine/threonine phosphatases, protein tyrosine phosphatases and dual specificity phosphatases (Kerk et al., 2008; Cao et al., 2016). Among them, protein tyrosine phosphatases can be further divided into phosphoprotein metallophosphatases and phosphoprotein phosphatases. Mn2+ or Mg2+-dependent protein phosphatases (PP2Cs) are important members of the phosphoprotein metallophosphatase family (Kerk et al., 2002).
PP2Cs are highly conserved in evolution and regulate stress signaling pathways in a targeted manner (Cao et al., 2016). Most PP2Cs in plants have a conserved catalytic region at the C-terminus, while the N-terminus is a poorly conserved region of varying lengths. The N-terminus is important for the definition of the PP2C member’s functionality, since this region can contain transmembrane regions and/or sequence motifs related with intracellular signaling, including those for interaction with kinases (Stone et al., 1994; Carrasco et al., 2003). It has been reported that some plant PP2Cs have significant roles in the plant response to environmental pressures, including salt, through their effects on metabolism, hormone levels and growth factors (Lim et al., 2015; Chu et al., 2021).
Relatively large PP2C gene families have been reported in several plant species, including soybean (103) (Chen et al., 2018), cotton (181) (Shazadee et al., 2019), Arabidopsis (76) (Kerk et al., 2002) and rice (132) (Singh et al., 2010). In Arabidopsis, PP2Cs (AtPP2Cs) are divided into clades A-J (Schweighofer et al., 2004). In clade A, ABA-INSENSITIVE 1 (ABI1) and ABA-INSENSITIVE 2 (ABI2) are important components of ABA signaling (Rodriguez et al., 1998; Kaliff et al., 2007). ABI1 and ABI2 act as negative regulators of ABA signaling, which play a pivotal role in the ABA-dependent salt stress response process. Under normal conditions, the ABA receptor (Pyrabatin Resistance 1-Like proteins; PYLs) is conformationally inactive and certain PP2Cs bind and dephosphorylate the sucrose non-fermenting-1-related protein kinases 2 (SnRK2) to inactivate ABA signaling. However, under osmotic stress ABA levels increase and their binding to the PYL induces conformational changes which enhance PYL-PP2C binding and inactivation of the PP2C. Consequently, the SnRK2 remains active and ABA-mediated stress signaling is initiated (reviewed in Gong et al., 2020; Zhang et al., 2022). Four members of the AtPP2Cs in clade B (APP2C1-4) are mitogen-activated protein kinase (MAPK) phosphatases that are negative regulators of phytohormones and defense responses (Ayatollahi et al., 2022). POLTERGEIST (AtPOL) and POLTERGEIST LIKE 1-5 (AtPLL1-5) of clade C are essential for cell maintenance and differentiation (Song et al., 2020). The functions of some AtPP2Cs in clades A, D, E, F and G have been reported, but the functions of most Arabidopsis PP2Cs remain unknown. The PP2C gene family in rice is divided into 11 clades, and most genes in clade A are triggered by various external abiotic stimuli, indicating that they play an important role in stress tolerance, particularly in salt stress (Xue et al., 2008).
Many PP2C gene family members have been reported to participate in plant salt stress regulation directly. (Leung and Giraudat et al., 1998; Kuhn et al., 2006; Liu et al., 2008). AtPP2CG1 (At2G33700), a PP2C gene from Arabidopsis, is fully expressed under salt stress and positively regulates salt tolerance in an ABA-dependent manner (Liu et al., 2012). At3G63320 and At3G63340 actively respond to external stress stimuli and play a key role in the regulation of stomatal opening and closing, revealing that protein phosphorylation and dephosphorylation of signal regulators are significant for controlling stomatal pore size (Pang et al., 2020). Among OsPP2Cs, OsPP2C53 (Os05g51510) and OsPP2C51 (Os05g49730) are significantly expressed under 150mM NaCl stress (Xue et al., 2008). OsPP2C08 (Os01g46760) is reported to be distinctly expressed in response to saline stress at the seedling stage compared to other PP2Cs (Sun et al., 2019). In rice, ubiquitination and degradation of OsPP2C09 (Os01g62760), a clade A PP2C, by abscisic acid-responsive RING Finger E3 Ligase (OsRF1) induced the salt tolerance of rice. (Kim et al., 2022).
Peanut (Arachis hypogaea L.) is an important oilseed and food crop worldwide. Cultivated peanut evolved from a cross between Arachis duranensis (A) and Arachis ipaensis (B) which subsequently underwent chromosome doubling to form the modern heterotetraploid (AABB genome, 2n=4x=40) with a total genome size of approximately 2.7 Gb (Bertioli et al., 2019). The main obstacles to peanut growth in semi-arid regions, where more than half of the global peanut production takes place, are drought and soil salinity (Banavath et al., 2018). There has been considerable advancement in the study of salt tolerance in peanut. Important examples include the stress-induced expression of Arabidopsis homeodomain-leucine zipper transcription factor (AtHDG11) to enhance salt tolerance, the identification of crucial intrinsic proteins that regulate salt stress in peanuts, the overexpression of the sodium/proton antiporter gene (AtNHX1) to improve salt tolerance, the vacuolar H+-pyrophosphatase gene (AVP1) (Asif et al., 2011; Banavath et al., 2018; Han et al., 2021). Compared to other classes of proteins, the role of AhPP2Cs in the peanut response to saline and other stresses are substantially less well understood. In order to advance the development of peanut salt-tolerant mechanisms, we identified the peanut PP2C gene family and selected genes associated with salt tolerance. In this study, we gained insights into the AhPP2C genes by using some computer analysis, such as characterization, genomic evolution, gene structure, motifs, cis-acting elements, catalytic sites and gene functional annotations, etc., and expression analysis in different tissues and under salt stress for research purposes. 178 AhPP2Cs were identified in the A. hypogea genome. 18 members that clustered together with clade A of Arabidopsis were studied further. Of these, six AhPP2Cs were clustered together with salt tolerant PP2C genes from rice and Arabidopsis. There were total 20 ABA response elements and 21 MYB binding sites in promoter region of the six genes (AhPP2C45/59/134/150/35/121). Moreover, their expression patterns under salt stress were analyzed from publicly available RNA-Seq data and qRT-PCR. AhPP2C45 and AhPP2C134 displayed significantly up-regulated expression under salt stress. They were considered as strong candidates for PP2Cs mediating salt stress signaling in peanut. These results provide theoretical support for biological functions of the peanut PP2C gene family under salt stress. Also, identification of AhPP2C45/134 furnish new research threads for mechanism study of salt response differences between peanut germplasms, which play important roles in selection of salt tolerance germplasms and breeding.
Protein and gene sequences of A. hypogaea cv. Tifrunner and their annotated gene models were downloaded from PeanutBase1 (Version 1). Protein sequences were scanned using the HMMER v3 (Finn et al., 2011) using the hidden Markov model (HMM) for PP2Cs (PF00481) from the Pfam database1 (Finn et al., 2014). Proteins carrying the raw PP2C HMM with an E-value (≤1×10-20) were used to construct a peanut-specific hidden Markov model using hmmbuild from HMMER v3. The resultant peanut-specific HMM was used to search the protein sequences once again, and proteins that conformed to the criteria were submitted to NCBI-CDS2 (Conserved Domain Search) in order to select those containing the tPP2C domain, PF00481.
Phylogenetic trees were constructed with MEGA11 software (Tamura et al., 2021) using the ClustalW algorithm (Thompson et al., 1994) for sequence alignment with the Maximum Likelihood (ML) method with 1000 bootstrap replicates and Pearson correction.
The chromosomal locations of AhPP2Cs were mapped with Map Gene 2 Chrom web v23 (Chao et al., 2021). AhPP2C gene duplications were identified based on a sequence similarity of their CDS ≥75% and a sequence overlap of ≥75% of the longest sequence of the pair. Gene duplications were depicted using a circos diagram (Krzywinski et al., 2009). The Ka/Ks ratios of all AhPP2Cs were predicted via simple Ka/Ks calculator in TBtools (Chen et al., 2020).
AhPP2Cs in clade A were further clustered with PP2C genes associated with salt tolerance in Arabidopsis and rice. The physicochemical properties of AhPP2C-A, salt tolerant PP2C genes from Arabidopsis and rice, including amino acid length, molecular weight (MW), isoelectric point, were calculated using Expasy ProtPrarm tools4 (Wilkins et al., 1999).
The gene structures of clade A AhPP2Cs were predicted and visualized using Gene Structure Display Server 2.05 (Hu et al., 2015). Their conserved protein motifs were analyzed by MEME Suite Version 4.12.0 (Bailey and Elkan, 1994) using a maximum of 10 motifs with an a.a. length of 6-50. The other parameters were set as default. For AhPP2Cs in subclades AI and AII, cis-acting elements were searched for in the 1500 bp region upstream of their start codons using PlantCARE6 (Lescot et al., 2002). Their protein secondary and tertiary structures were predicted using SOPMA7 (Geourjon and Deléage, 1994) and SwissModel8 (Waterhouse et al., 2018), respectively. The protein sequences were submitted to NCBI-Blast9, NetPhos3.110 (Blom et al., 1999; Blom et al., 2004) and Phyre211 (Kelley et al., 2015), for prediction of their catalytic sites.
Through the psRNATarget website, the CDS of all AhPP2Cs was used to predict miRNA target sites (Dai et al., 2018) with default. Figure of the interaction network among miRNAs and AhPP2Cs were mapped by Cytoscape software (Shannon et al., 2003). Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) annotation evaluation was performed by submitting AhPP2C-A protein sequences to eggNOG-mapper12 (Cantalapiedra et al., 2021). GO and KEGG enrichment evaluations were undertaken using TBtools.
In this study, the peanut cultivar Shanhua 11, which was bred and is maintained by our research group, was used as the plant material. Mature seeds were germinated on cotton wool soaked in distilled water within seed cultivation discs placed in darkness for three days at 26°C, after which, intact seedlings were transplanted to a hydroponic box and cultured with 1/5 Hoagland’s nutrient solution under a light/dark cycle of 16/8 h. Salt stress was applied to two-week-old seedlings by the adjustment of the nutrient solution to 200 mM NaCl. Leaves or roots from peanut seedlings were harvested after 72 h of salt or control treatments and were flash-frozen in liquid nitrogen before storage at -80°C until further use. Three biological replicates were used for analysis, each formed from materials obtained from five randomly chosen seedlings.
Total RNA was isolated with the Quick RNA Isolation Kit (Waryong, Beijing, China) and reverse transcribed into cDNA using Advantage RT-for-PCR Kit (TaKaRa, Dalian, China) following the manufacturer’s instructions. The specific primers were designed using Beacon Designer 7.9. SYBR Green real-time PCR was performed according to the guidelines of the PCR machine. The relative expression levels were calculated by the 2–ΔΔCt method (Livak and Schmittgen, 2001). Statistical differences were determined by T-test (**P<0.01, *P<0.05).
The tissue specificity of AhPP2Cs was determined from RNA-Seq data obtained for 22 different tissues at different developmental stages during peanut growth (Clevenger et al., 2016) obtained from the peanut database. The RNA-Seq data from salt-stressed peanut seedlings (Luo et al., 2021) was obtained from the public repository of NCBI13 under the biological project accession number, PRJNA560660. The FPKM values of AhPP2C-A, including subclades AI and AII and other AhPP2Cs were extracted from the data set. The heatmap of was generated by TBtools after log2- transformation of FPKM values.
A total of 183 PP2C-coding candidate genes were identified via HMM searching in peanut (A. hypogaea L.). After screening for the presence of the typical PP2C domain (PF00481), 178 genes were retained. A phylogenetic tree was created after multiple sequence alignment of the 178 AhPP2Cs 76 Arabidopsis PP2C genes. The 178 AhPP2Cs were divided into the 10 clades, A-J, as reported in other plants before, while 6 AhPP2Cs did not belong to any reported PP2Cs clades (Figure 1 and Supplementary Table 1). Of these, clade E was the largest, containing 34 AhPP2Cs, while clade B was the smallest, including 4 AhPP2Cs. The cladistics analysis indicated that the PP2Cs of peanut and Arabidopsis were similarly distributed between the clades, indicating the maintenance of the same types of functional diversity in PP2Cs following their speciation.
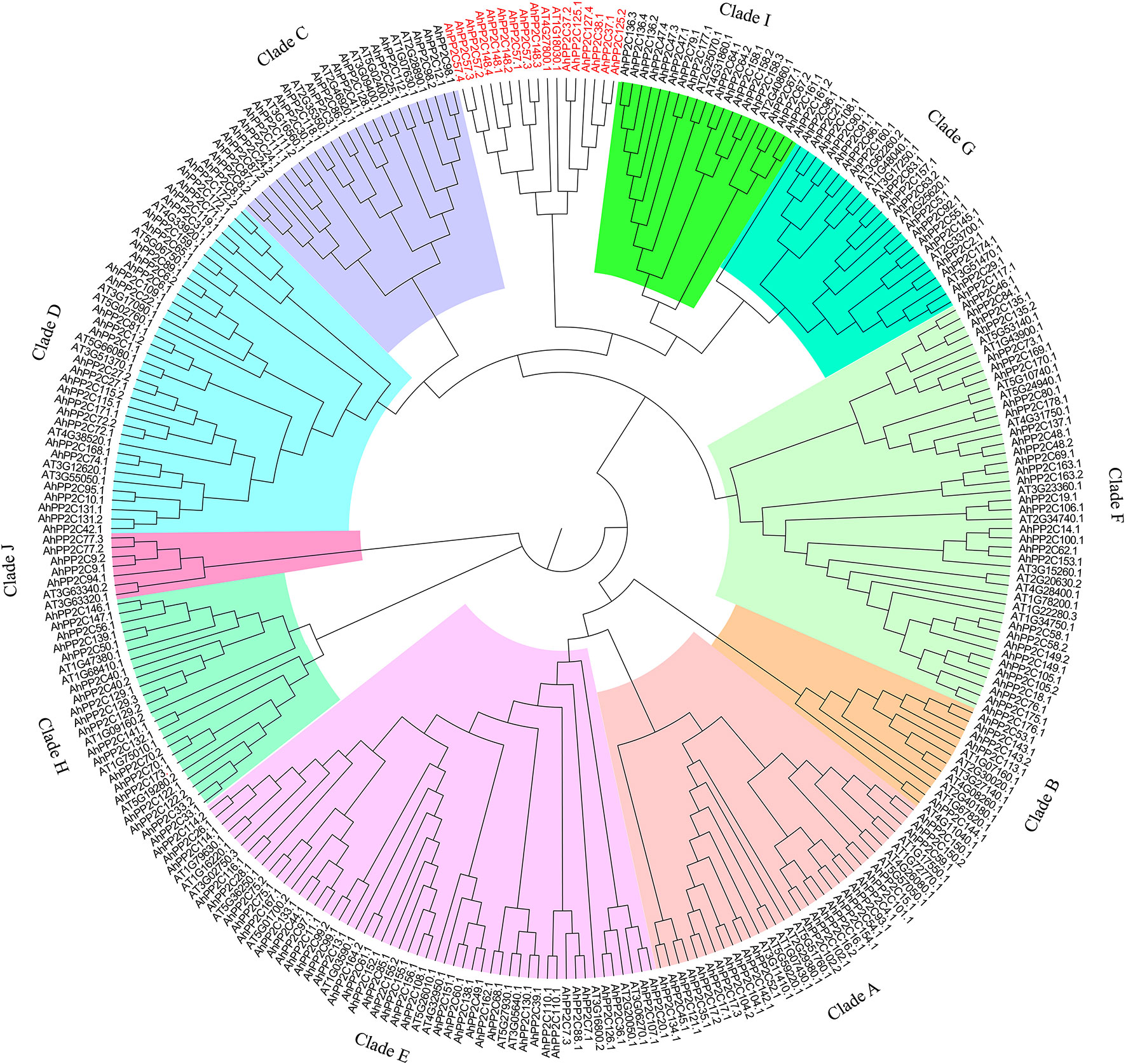
Figure 1 A Phylogenetic tree of PP2Cs from peanut and Arabidopsis. PP2Cs in different clades are labeled with different colors. 6 AhPP2C genes in red were not classified.
As shown in Figure 2, all 178 AhPP2Cs are unevenly distributed on 20 chromosomes, with no more than five genes on chromosomes 2, 7, 10 and 12, while the others are greater than five, with the greatest number of genes on chromosome 20. Most AhPP2Cs were concentrated near the ends of each chromosome, which was similar to the chromosomal distribution of PP2Cs reported for other plant species (Shazadee et al., 2019; Yu et al., 2019). A gene duplication analysis based on sequence similarity of AhPP2Cs indicated 78 genes pairs and found that there was segmental duplication in 78 genes pairs (Figure 3 and Supplementary Table 2). As expected, the paired AhPP2Cs clustered together in the phylogenetic tree. However, their distribution positions on 20 chromosomes were similar. Thus, to understand the mode of selection of the AhPP2Cs, the Ka, Ks, and Ka/Ks ratio was revealed for all gene pairs (Supplementary Table 2). The dataset unveiled that all duplicated AhPP2C gene pairs had a Ka/Ks ratio of <1, except for AhPP2C54/154, suggesting that AhPP2Cs in these groups underwent purifying selective pressure. The Ka/Ks value of two gene pairs (AhPP2C42/131, AhPP2C81/137) was zero, indicating strong purifying selection.
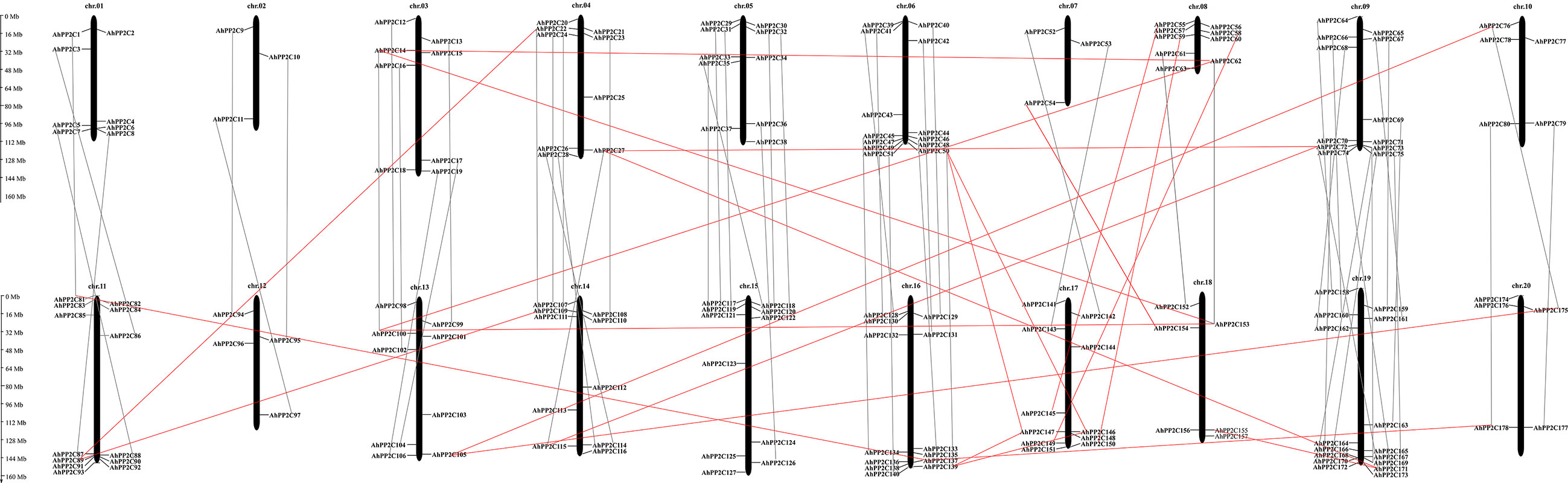
Figure 2 Chromosomal distribution of AhPP2Cs. Orthologous genes were linked by gray lines. Paralogous genes were linked by red lines.
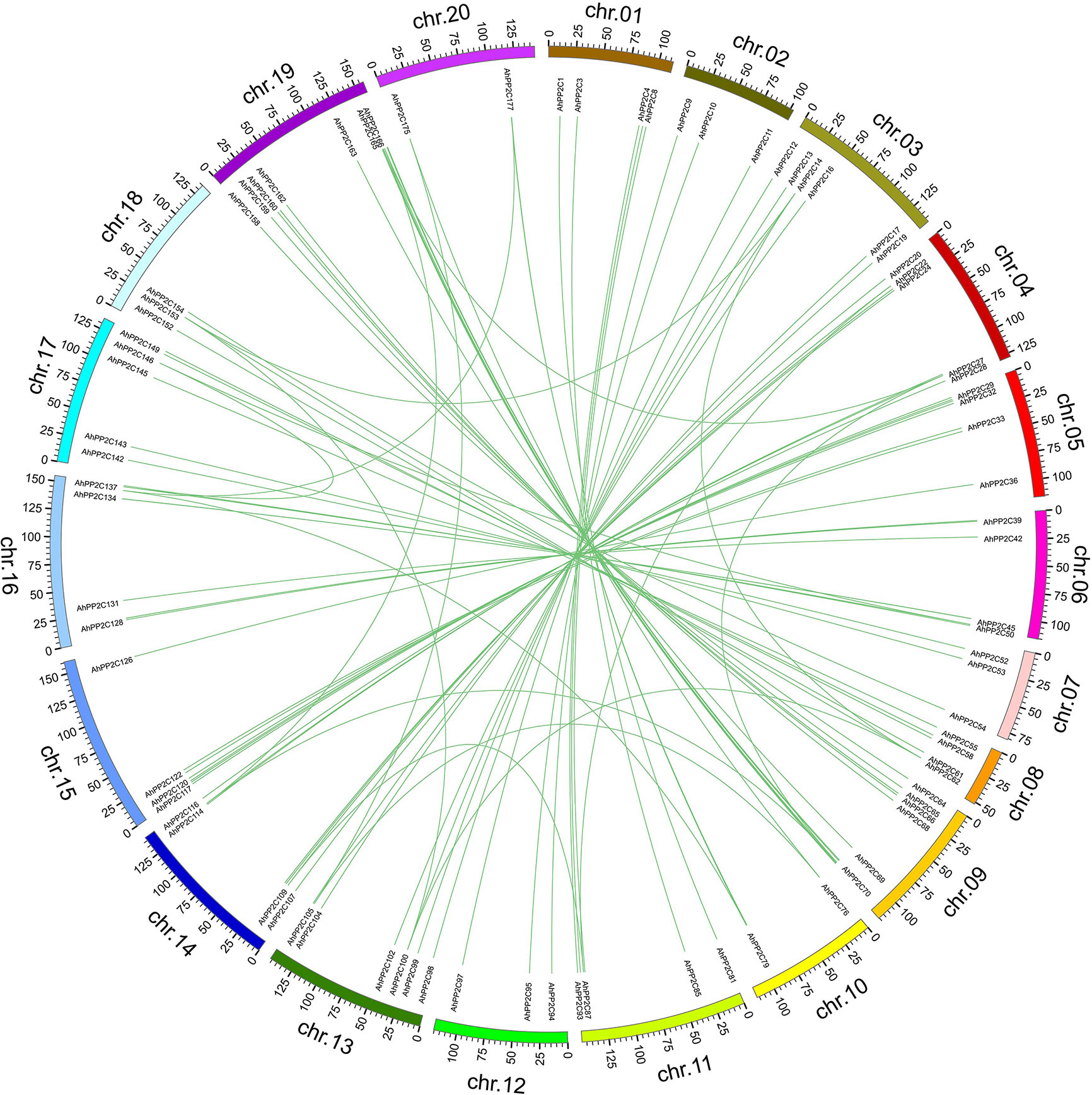
Figure 3 Gene duplication analysis of AhPP2Cs. Chromosomes are drawn in different colors. The scale provided represents the chromosome size (Mbp). Duplicated AhPP2Cs are linked by green lines.
To better investigate how the AhPP2C genes were regulated by miRNAs during translation, we identified 22 miRNAs targeting 57 genes (Figure 4 and Supplementary Table 3). These miRNAs belonged to 14 different families. The result showed that ahy-miR3513-5p targeted the most number (8) of genes, followed by ahy-miR159 that targeted seven genes. Two miRNAs, including ahy-miR156a and ahy-miR3511-5p, targeted four genes, followed by ahy-miR156c, ahy-miR394, ahy-miR3516, ahy-miR3514-3p and ahy-miR3520-5p that targeted three genes, while ahy-miR408-3p, ahy-miR156b-3p, ahy-miR3519, ahy-miR3514-5p, ahy-miR156b-5p and ahy-miR3509-5p targeted two different genes. Only four miRNAs, including ahy-miR3520-3p, ahy-miR167-3p, ahy-miR160-3p and ahy-miR398 targeted one gene, AhPP2C163, AhPP2C34, AhPP2C47 and AhPP2C15 respectively. In AhPP2C-A, only AhPP2C15, AhPP2C35 and AhPP2C121 were targeted by miRNAs. Some genes like AhPP2C30, AhPP2C117 AhPP2C19, AhPP2C41, AhPP2C128 and AhPP2C155 were found to be targeted by more than one miRNA.
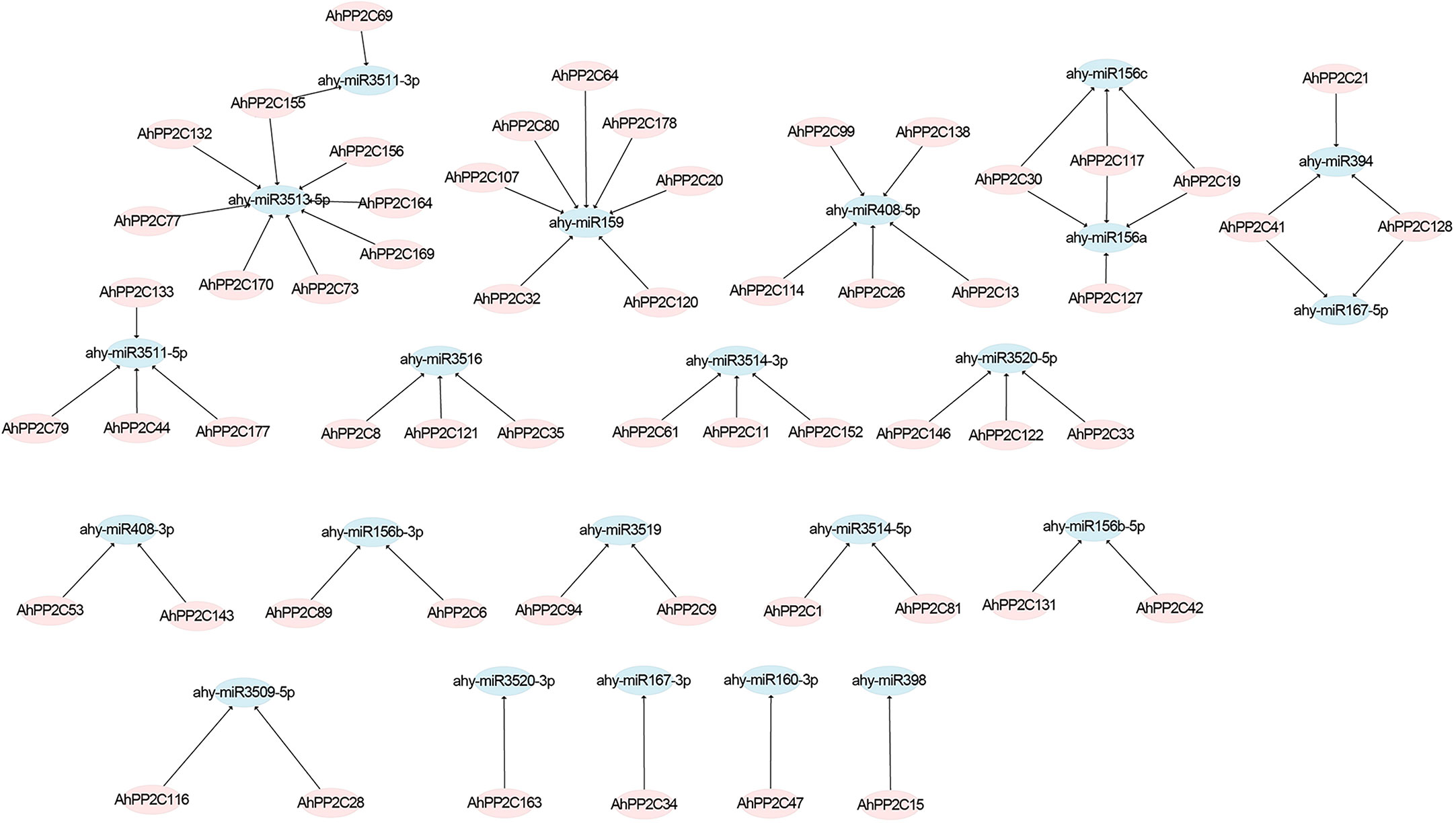
Figure 4 Prediction of putative miRNAs targeting AhPP2Cs. The pink shapes correspond to AhPP2Cs, and blue shapes indicate predicted miRNAs.
Previous studies have demonstrated that clade A PP2C genes has been identified to be associated with salt tolerance in rice and Arabidopsis (Schweighofer et al., 2004; Xue et al., 2008). In peanut, eighteen PP2C genes belong to clade A. To further investigate their biological functions, AhPP2C-A were further compared with the reported salt response PP2C genes in rice and Arabidopsis. As shown in Figure 5, the latter were clustered together with closely related AhPP2Cs in subclades AI and AII. The bootstrap values of these two branches were 74.3% and 74.6%, respectively, indicating a high degree of confidence, suggesting that AhPP2Cs in these subclades may be functionally associated with salt tolerance. Their identification regresses to finding peanut sequences with the high homology with known PP2C genes associated with salt tolerance. The physicochemical properties of subclades AI and AII are shown in Table 1, which indicates that all members of each subclade shared similar physiochemical properties.
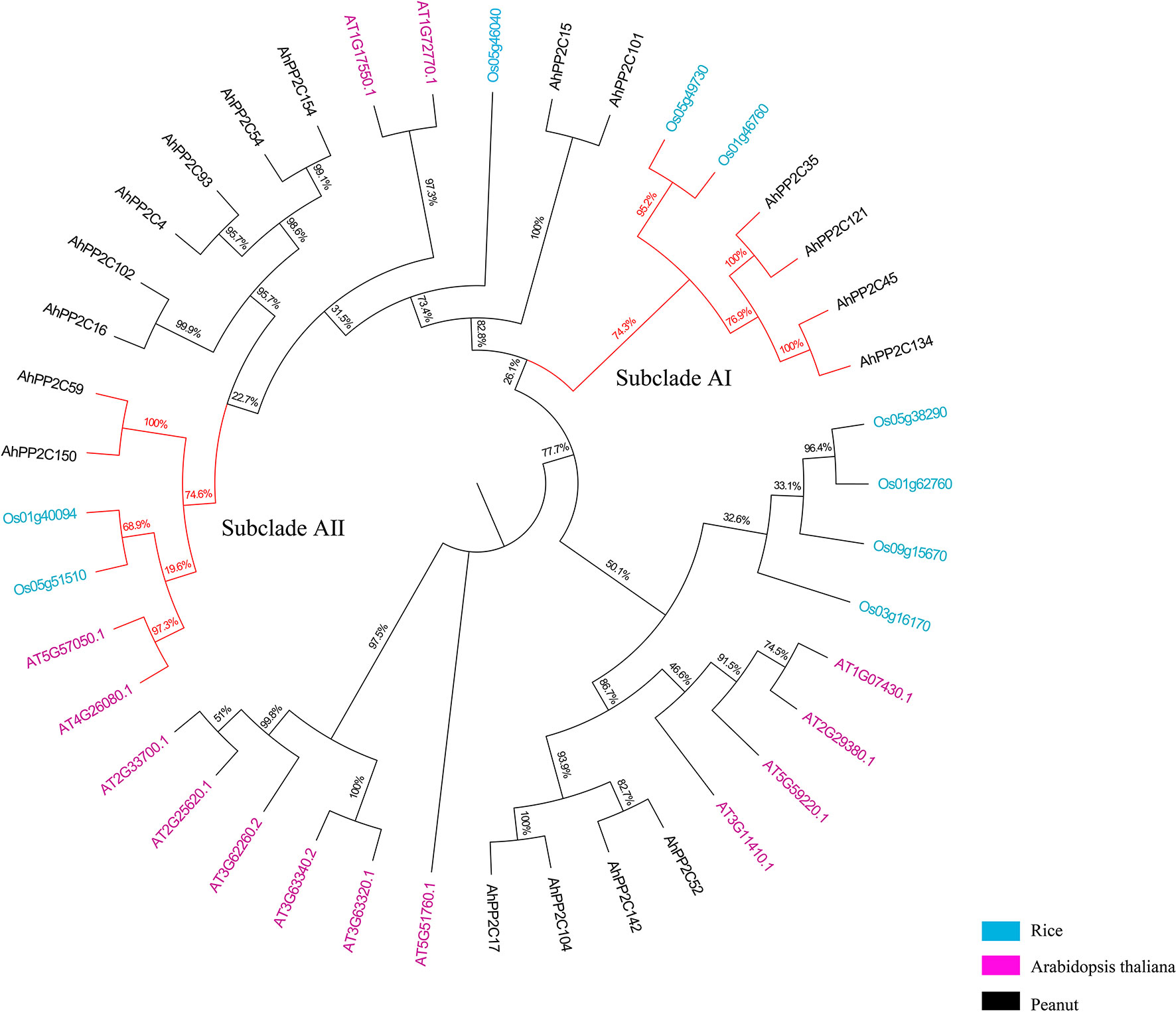
Figure 5 Phylogenetic tree of clade A AhPP2Cs, with salt tolerant genes from Arabidopsis and rice. The branches of subclades AI and AII are indicated in red.
To further analyze the potential roles of AhPP2Cs in subclades AI and AII, their phylogeny was analyzed together with their gene structures and motifs (Figure 6). In subclade AI, all PP2Cs, including the salt tolerant genes from Arabidopsis and rice in the same branch of the phylogenetic tree were similar in structures and contained 4 exons. In subclade AII, AhPP2C134 and 45, and two rice PP2Cs involved in the response to salt stress contained 3 exons, while the remaining two AhPP2Cs contained only 2 exons.
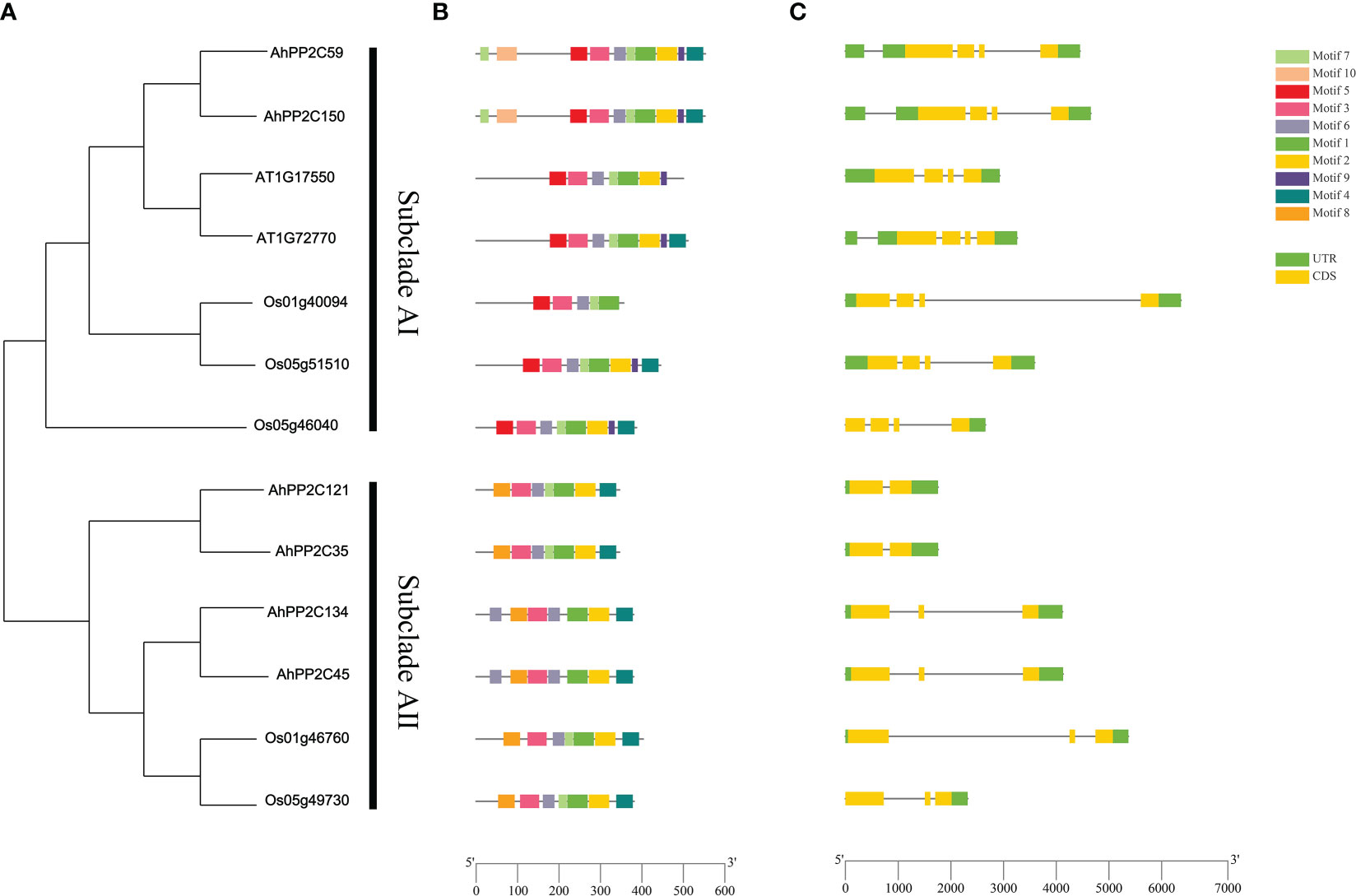
Figure 6 Phylogenetic relationship, gene structures and motifs analysis of PP2Cs in subclades AI and AII. (A) Phylogenetic tree of PP2Cs (B) Motifs analysis of PP2Cs. Colored boxes indicate the different conserved motifs as indicated in the scheme to the right of the figure. (C) Exon-Intron Structure of PP2Cs, where exons are shown as yellow boxes. Where present, 5’ and 3’ UTRs are depicted as green boxes.
A total of 10 motifs was identified using MEME Suite. Motifs 5, 9 and 10 were exclusive to subclade AI, motif 8 was exclusive to subclade AII, while motifs 1, 2, 3, 4, 6 and 7 were present in both subclades. After alignment of protein sequences, conserved domain sequences and motifs, it was found motifs 2 and 4 are located in the catalytic site at the C-terminus (Bork et al., 1996). These sites were highly conserved across the PP2Cs of subclades AI and AII, except Os1g40094 (Figure 6 and Supplementary Figure 1). Motif 10 was located at the N-terminal, but it appeared to be highly conserved, because it was found to exist on fewer number of sequences (Supplementary Table 4). Motif 5 and motif 9 were only present in subclade AI. This may account for the functional differences between the two subclades. AhPP2C45/134 in subclade AII had a greater number of motif 6, where it may confer unique functions to these PP2Cs, which will need be investigated further.
The secondary structures of AhPP2Cs in Subclades AI and AII consisted of 31.16%-43.64% α-helices, 30.26%-46.11% irregular curls, 15.91%-23.68% β-sheets and 5.25%-9.21% β-turns. Analysis of the secondary structures of orthologous gene pairs AhPP2C150/AhPP2C59, AhPP2C134/AhPP2C45 and AhPP2C121/AhPP2C35 showed they were very similar. The tertiary structures were also highly similar (Figure 7). Moreover, their catalytic structural domains all consist of a central β-sandwich that binds Mn2+/Mg2+ and is surrounded by α-helices (Das et al., 1996; Shi, 2009). The phosphorylation sites, which can be found in motifs 4 and 2 within their predicted catalytic sites were also found to be similar: S362 and S309 for AhPP2C134/AhPP2C45, S322 and S276 for AhPP2C35/AhPP2C121, S532 and S473 for AhPP2C59 and S531 and S472 for AhPP2C150.
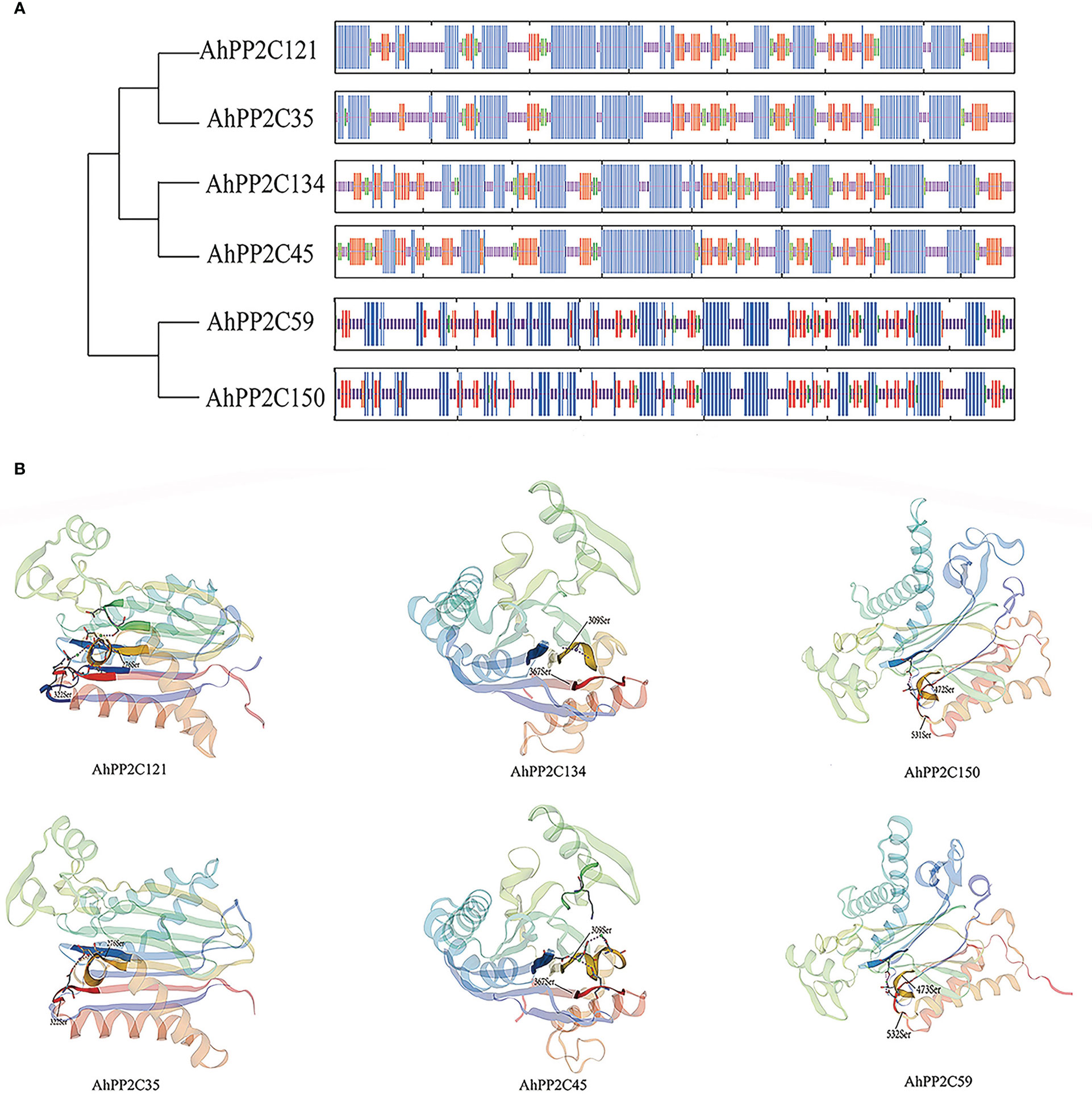
Figure 7 Protein structures and catalytic site prediction of AhPP2Cs in subclades AI and AII. (A) protein secondary structures of AhPP2Cs. α-helices were in blue; irregular curls were in purple; β-sheets were in red; β-turns were in green. (B) protein tertiary structures and catalytic sites of AhPP2Cs in subclades AI and AII.
To further understand the role of AhPP2C-A genes at the molecular level, we performed GO and KEGG enrichment analysis (Supplementary Figure 2). The GO enrichment analysis can be divided into three main categories: biological processes (BP), molecular functions (MF) and cellular components (CC). For instance, in MF class, the most enriched term was protein binding (GO:0005515), followed by binding (GO:0005488) and hydrolase activity (GO:0016787). In CC class, the highly enriched terms were cytosol (GO:0005829) and nucleus (GO:0005634). Whereas in BP class, the highly enriched terms were cell communication (GO:0007154), signal transduction (GO:0007165), regulation of molecular function (GO:0065009), response to endogenous stimulus (GO:0009719), response to abiotic stimulus (GO:0009628), etc. In addition, KEGG pathway enrichment study identified four pathways involved in the different functions of the AhPP2C-A genes. The highly enriched pathways include signal transduction (B09132), plant hormone signal transduction (04075), environmental information processing (A09130), MAPK signaling pathway – plant (04016) (Supplementary Table 5). In brief, it could be concluded that AhPP2C-A genes related to abiotic stresses response, protein modification process and plant hormone signaling.
In higher plants, PP2C is involved in many stress resistance responses and is induced by a variety of abiotic stresses. Cis-acting elements were therefore searched in the 1500 bp sequences upstream of the start codons of peanut PP2Cs of subclades AI and AII (Table 2). There were obvious differences in the types and numbers of cis-acting elements among different AhPP2Cs. The results showed that many phytohormones and stress-related response elements were identified, including ABRE/ABRE4/ABRE3a, Aux RR-core/TGA-element, TCA-element, MYB and CGTCA-motif/TGACG-motifs. AhPP2Cs in subclades AI and AII all contained MYB binding factors, which are essential for regulating how plants respond to abiotic stress, particularly in response to salt stress. AhPP2C150, 35, 134 and 45 contained cis-acting elements involved in the regulation of the ABA response (ABRE/ABRE4/ABRE3a), while AhPP2C121 and 59 did not. AhPP2C45 contained the maximum number of cis-elements (21), while AhPP2C35 contained the minimum number of cis-elements (4). The differences in number and kind of elements may lead to functional difference of AhPP2Cs.
Based on the transcriptome data of 22 peanut tissues (Clevenger et al., 2016), the expression patterns of clade A AhPP2Cs were extracted and shown in Figure 8 and Supplementary Table 6. Clade A AhPP2Cs diverse with differing levels of tissue specificity. AhPP2C17, 59, 150, 134, 45, 15 and 101 expressed at relatively high levels in several of the 22 different tissues. In contrast, the overall expression levels of AhPP2C54, 35 and 121 were lower, and mainly detected in seeds tissues. The remaining genes (7) were moderately expressed in all tissues. Considering AhPP2Cs of subclades AI and AII, AhPP2C121 and 35 were only expressed in seeds, while the other four genes were also expressed in leaves and roots. The above results indicated that clade A AhPP2Cs exhibited different expression patterns.
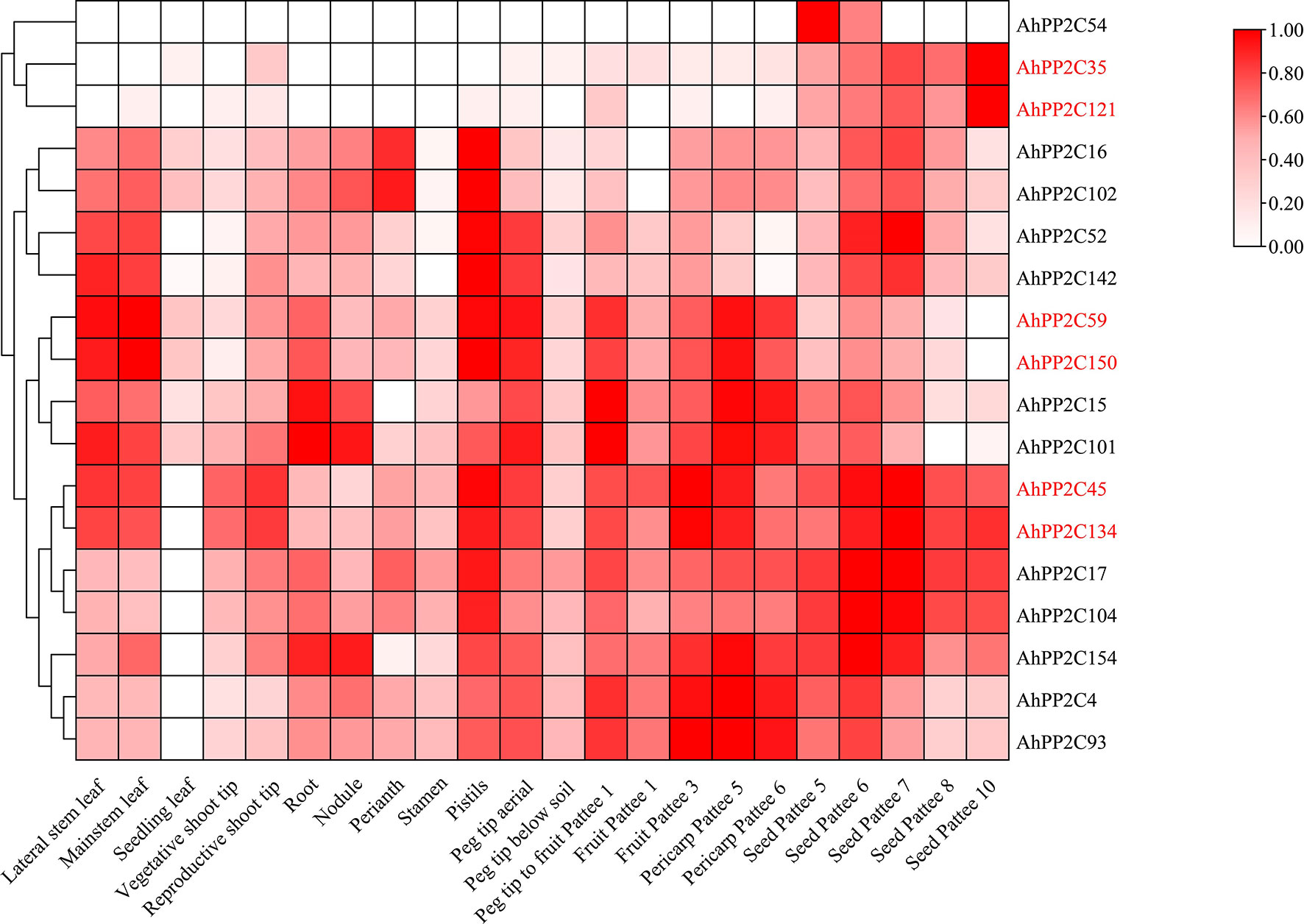
Figure 8 Expression pattern analysis of clade A AhPP2Cs. The heat map of log2-transformed averaged FPKM data was mapped using the intensity scale shown on the right. The visualization and clustering utilized TBtools. The AhPP2Cs of subclades AI and AII are labelled in red.
An initial assay of the expression of subclade AI and AII AhPP2Cs in seedlings under salt stress used publicly available RNA-Seq data from leaves and roots. The heatmap of relative expression is shown in Figure 9A. Under salt stress, in leaves, AhPP2C45, 134, 59 and 150 were up-regulated compared with the control, but this was only significant in AhPP2C45 and 134. In roots, AhPP2C45 and AhPP2C134 were also up-regulated, while AhPP2C59 and AhPP2C150 were down-regulated relative to the control. The expression of AhPP2C121 was not detected in roots or leaves with or without salt stress.
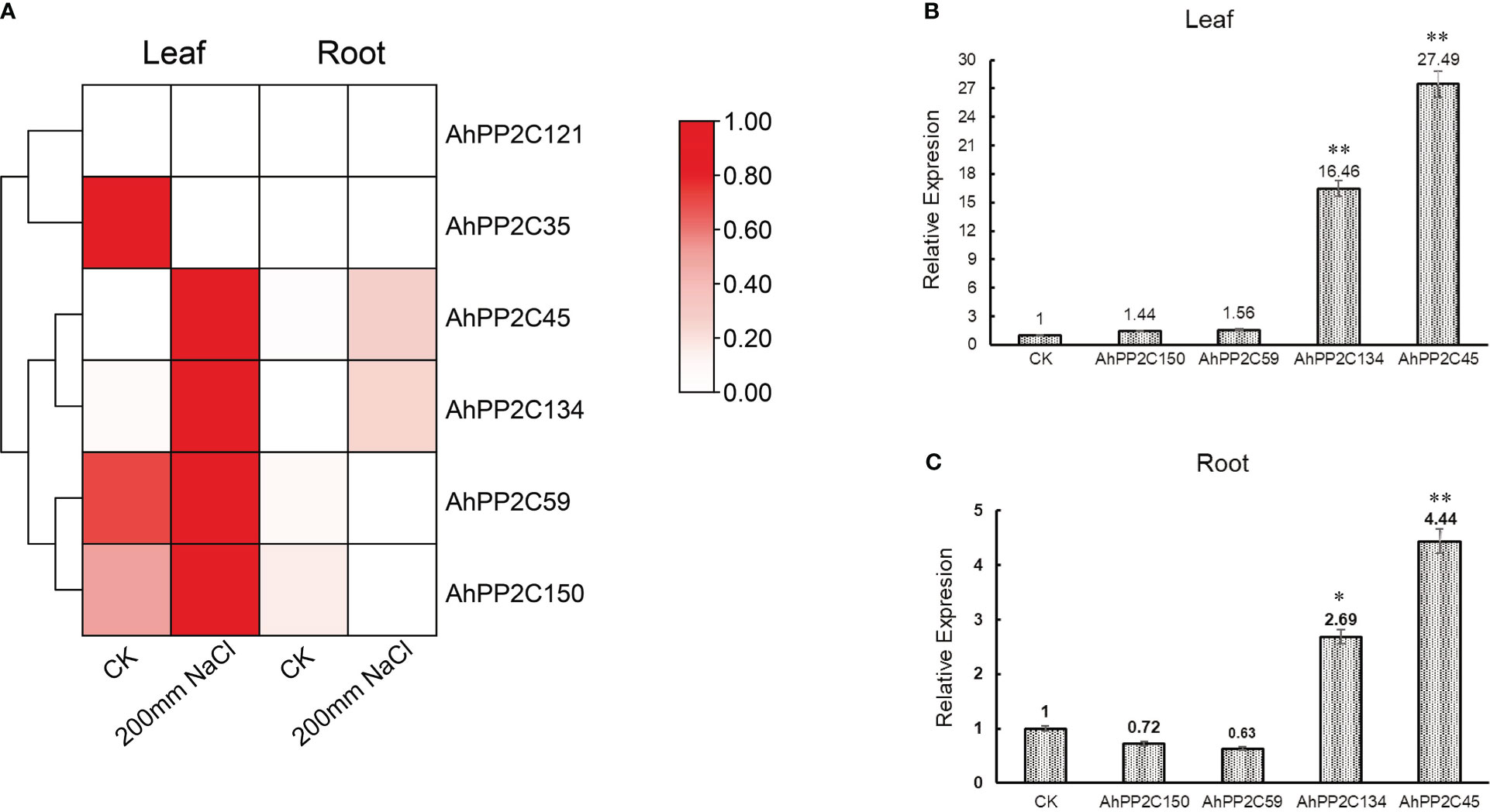
Figure 9 Expression profiles under salt stress. (A) Relative expression pattern of AhPP2Cs in subclades AI and AII in response to salt stress. (B, C) qRT-PCR analysis of AhPP2Cs 45, 59, 134 and 150 under salt stress in leaves (B) and roots (C). “*” indicated that the genes were significant differences compared with the untreated control (**P<0.01, *P<0.05).
To validate the results for the four up-regulated genes under salt stress one step further, the relative expression levels of these AhPP2Cs (45, 134, 59 and 150) in leaves and roots were analyzed by qRT-PCR (Figures 9B, C). After 72 h of salt treatment (200 mM NaCl), the relative expression levels of AhPP2C45 and 134 were up-regulated significantly in both leaves and roots. These results indicated that AhPP2C45 and 134 may play important roles in peanut salt stress response.
Salt stress is the second worst abiotic factor affecting global agricultural productivity, disrupting many physiological, biochemical and molecular processes in plants. Salinity affects plant growth, development and productivity (Raza et al., 2022). Salt stress also has a serious impact on peanut production (Aravind et al., 2022). PP2Cs are widespread and highly conserved in prokaryotes and eukaryotes (Yang et al., 2018). Several studies of plant PP2Cs have indicated that of a subfamily of PP2Cs (clade A) have regulatory roles in stress responses through dephosphorylating the substrate proteins, especially in ABA-dependent responses, such as the drought and salinity response (Xue et al., 2008; Cui et al., 2013; Singh et al., 2015; Krzywińska et al., 2016). However, less information is available for PP2Cs in the economically important crop plant peanut, whose productivity is considerably affected by salinity stresses. This research therefore aimed to provide a systematic analysis of AhPP2Cs and to identify candidate AhPP2Cs participate in salt stress response.
In this study, we identified 178 AhPP2Cs that showed a phylogenetic clustering similar to that of Arabidopsis, including a distinct clade A, which is considered to contain a subfamily of PP2Cs with roles in ABA-mediated salt stress responses (Zhang et al., 2013; Singh et al., 2015; He et al., 2019). Numbers of PP2C genes differ a lot among species (Kerk et al., 2002; Chen et al., 2018; Shazadee et al., 2019). Deviations in the number of PP2C members from different plant species may be attributed to gene duplication events, including tandem duplication and segmental duplication, which play an important role in expanding PP2Cs members. Duplications of PP2C genes have also been found in some plant species (Yang et al., 2018; Zhang et al., 2022). Our results confirmed that AhPP2Cs had underwent segmental duplication strong purifying selective pressure (Figure 3 and Supplementary Table 2). Previous phylogenetic tree of Arabidopsis showed that there were 6 AtPP2Cs could not clustered to any clades (Schweighofer et al., 2004; Bhaskara et al., 2019). In this work, we found 6 AhPP2Cs clustered with 2 of the 6 AtPP2Cs, and we speculated that this may be universal phenomena in plant PP2C gene family. Some AhPP2Cs of clade A clustered together with PP2Cs known to confer salt tolerance in Arabidopsis and rice (Figure 5). The AhPP2C35, 121, 45 and 134 of subclade AI were found to be closely clustered with the OsPP2Cs Os05g49730 (OsPP2C51) and Os01g46760 (OsPP2C08). OsPP2C08 (Os01g46760) was identified as a potential candidate gene conferring improved salt tolerance through the integration of genome-wide polymorphism information with QTL mapping and expression profiling data (Subudhi et al., 2020). The heterogeneous expression of OsPP2C51 (Os05g49730) in Arabidopsis revealed that it could positively regulate ABA-induced rice seed germination under conditions of salt stress (Bhatnagar et al., 2017).
Similarly, the AhPP2C59 and 150 of subclade AII were closely clustered with two rice Os01g40094 (OsPP2C06) and Os05g51510 (OsPP2C53) and two Arabidopsis PP2Cs (AT5G57050.1 and AT4G26080.1). ABI2 (AT5G57050.1) as a SOS2-interacting protein can affect salt stress response in Arabidopsis (Ohta et al., 2003). Mutations ABI1 (AT4G26080.1), affecting ABA-perception in Arabidopsis, can reduce the accumulation of both AtP5CS mRNAs during salt-stress and result in higher proline accumulation. (Strizhov et al., 1997). OsPP2C53 (Os05g51510) negatively regulates OsSLAC1 in stomatal closure and transgenic rice overexpressing OsPP2C53 showed significantly higher water loss than control (Min et al., 2019). Microarray analysis of transgenic rices overexpressing OsNAP in high salinity, drought and low temperature revealed that a stress-related gene OsPP2C06 (Os01g40094) was up-regulated (Chen et al., 2014). Salt tolerant PP2C genes from Arabidopsis and rice are located in both subclades AI and AII, then this functionality occurs in two phylogenetically distinct subclades. This could, for example, be because function of salt tolerant PP2C genes can be achieved in mechanistically different ways and these evolved independently, or these genes differ in other aspects of their sequence (domains/motifs) that have more weight in their phylogenetic placing. In addition, the peanut AhPP2Cs that closely clustered with PP2Cs conferring salt tolerance also shared similar motifs and gene structures with their Arabidopsis and rice counterparts (Figure 6), indicating that they may also share similar functionalities.
Additionally, we found nine cis-elements, two of them, ABRE/ABRE4/ABRE3a and MYB binding factor are related to salt tolerance. ABRE/ABRE4/ABRE3a has been discovered in previous studies of PP2Cs in stress (Xue et al., 2008; Chen et al., 2018), however there is less available information regarding MYB binding factors in this process. Furthermore, AhPP2Cs gene functions were further predicted by GO enrichment analysis, which supported the role of these genes in dephosphorylating the substrate proteins and salt stress response. It is therefore of interest that after our analysis of the expression profiles of AhPP2Cs under salt stress, we observed the PP2C genes with ABRE/ABRE4/ABRE3a and MYB binding factors showed greater relative changes in expression. The genes (AhPP2C45/134) contained both cis-elements and having the maximum number of both cis-elements. The greater number of salt tolerance-related cis-elements AhPP2Cs have, the greater expression levels under salt stress. The number of salt tolerance-related cis-elements may therefore be correlated with either high or low gene expression in response to salt stress.
The potential phosphorylation sites and protein structures of the AhPP2Cs were also examined. The conserved catalytic core domain of PP2Cs contains a central β-sandwich with a pair of α-helices on either side of each β-sheet (Das et al., 1996). Within the conserved catalytic structural domain, the PP2Cs contain three characteristic sequences: DG××G, DG, and G××DN (where x is any amino acid) (Shi, 2009). These characteristic sequences located in motifs 3, 2 and 4, respectively (Supplementary Figure 3). Moreover, phosphorylation sites were predicted to be located in motifs 2 and 4, but not in motif 3. More importantly, some of phosphorylation sites, S362 for AhPP2C134/AhPP2C45, S322 for AhPP2C35/AhPP2C121, S532 for AhPP2C59 and S531 for AhPP2C150, were within the G××DN characteristic sequence. These phosphorylation sites may provide an important direction for future studies of AhPP2Cs in ABA-related signal transduction under salt stress.
Based on phylogenetic, gene structure, motif and cis-acting elements analysis of AhPP2C-A, AhPP2C45 and AhPP2C134 had similar structures to the reported PP2C genes associated with salt tolerance in rice and Arabidopsis and contained more cis-elements that related to salt tolerance. According to the salt stress effects on the expression, AhPP2C45 and AhPP2C134 had significant differences compared with the control. These results indicated that AhPP2C45 and AhPP2C134 could be candidate PP2Cs conferring salt tolerance. The study of AhPP2Cs in response to salt stress may be used in conjunction with phenotypic data to screen for more salt tolerant genotypes and salt tolerant varieties, providing a crucial foundation for the next step breeding.
In this study, we identified 178 AhPP2C genes which distributed across the 20 chromosomes, with segmental duplication in 78 gene pairs. Phylogenetic tree, gene and protein structures, cis-elements and expression analysis indicated that AhPP2C45 and AhPP2C134 could be candidate PP2Cs conferring salt tolerance. These results provide important information for the research of salt-stress mechanism, selection of salt-resistant germplasm and breeding of salt-tolerance cultivar in peanut.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: The public repository of NCBI under the biological project accession number, PRJNA560660.
FL conceived the study, designed the experiments, supervised and complemented the writing. YW provided suggestions and supervision. ZW and LL produced the figures and wrote the original article. All authors discussed the results and commented on the article. All authors contributed to the article and approved the submitted version.
This work was supported by the Peanut Seed Industry Project in Shandong Province, China (No. 2022LZGC007 to FL), and funding from the Agriculture Research System in Shandong Province, China (No. SDAIT-04-03 to FL) and the Chinese Agriculture Research System (CARS-13 to YW).
We gratefully acknowledge students including Hui Yang, Yuanyuan Sang, Yijun Li, Haiyang Yu, and Junjie Zhang for assisting us with this study.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1093913/full#supplementary-material
Supplementary Figure 1 | Motifs analysis of PP2Cs in Subclades AI and AII.
Supplementary Figure 2 | GO and KEGG enrichment analysis of clade A AhPP2Cs. (A) The enriched GO terms from MF, CC, BP classifications in AhPP2Cs. (B) The enriched KEGG pathways in AhPP2Cs.
Supplementary Figure 3 | Sequence alignment of PP2Cs in subclades AI and AII.
Aravind, B., Nayak, S. N., Choudhary, R. S., Gandhadmath, S. S., Prasad, P. V. V., Pandey, M. K., et al. (2022). “Integration of genomics approaches in abiotic stress tolerance in groundnut (L.): An overview,” in Genomic Designing Abiotic Stress Resistant Oilseed Crops, ed. C. Kole (Cham: Springer), 149197. doi: 10.1007/978-3-030-90044-1_4
Asif, M. A., Zafar, Y., Iqbal, J., Iqbal, M. M., Rashid, U., Ali, G. M., et al. (2011). Enhanced expression of AtNHX1, in transgenic groundnut (Arachis hypogaea l.) improves salt and drought tolerence. Mol. Biotechnol. 49 (3), 250–256. doi: 10.1007/s12033-011-9399-1
Ayatollahi, Z., Kazanaviciute, V., Shubchynskyy, V., Kvederaviciute, K., Schwanninger, M., Rozhon, W., et al. (2022). Dual control of MAPK activities by AP2C1 and MKP1 MAPK phosphatases regulates defence responses in Arabidopsis. J. Exp. Bot. 73 (8), 2369–2384. doi: 10.1093/jxb/erac018
Bailey, T. L., Elkan, C. (1994). Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intelligent Syst. Mol. Biol. 2, 28–36.
Banavath, J. N., Chakradhar, T., Pandit, V., Konduru, S., Guduru, K. K., Akila, C. S., et al. (2018). Stress inducible overexpression of AtHDG11 leads to improved drought and salt stress tolerance in peanut (Arachis hypogaea l.). Front. Chem. 6. doi: 10.3389/fchem.2018.00034
Bertioli, D. J., Jenkins, J., Clevenger, J., Dudchenko, O., Gao, D., Seijo, G., et al. (2019). The genome sequence of segmental allotetraploid peanut (Arachis hypogaea l.). Nat. Genet. 51 (5), 877–884. doi: 10.1038/s41588-019-0405-z
Bhaskara, G. B., Wong, M. M., Verslues, P. E. (2019). The flip side of phospho-signalling: regulation of protein dephosphorylation and the protein phosphatase 2Cs. Plant Cell Environ. 42 (10), 2913–2930. doi: 10.1111/pce.13616
Bhatnagar, N., Min, M. K., Choi, E. H., Kim, N., Moon, S. J., Yoon, I., et al. (2017). The protein phosphatase 2C clade a protein OsPP2C51 positively regulates seed germination by directly inactivating OsbZIP10. Plant Mol. Biol. 93 (4-5), 389–401. doi: 10.1007/s11103-016-0568-2
Blom, N., Gammeltoft, S., Brunak, S. (1999). Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J. Mol. Biol. 294 (5), 1351–1362. doi: 10.1006/jmbi.1999.3310
Blom, N., Sicheritz-Pontén, T., Gupta, R., Gammeltoft, S., Brunak, S. (2004). Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics 4 (6), 1633–1649. doi: 10.1002/pmic.200300771
Bork, P., Brown, N. P., Hegyi, H., Schultz, J. (1996). The protein phosphatase 2C (PP2C) superfamily: Detection of bacterial homologues. Protein Sci. A Publ. Protein Soc. 5 (7), 1421–1425. doi: 10.1002/pro.5560050720
Cantalapiedra, C. P., Hernández-Plaza, A., Letunic, I., Bork, P., Huerta-Cepas, J. (2021). eggNOG-mapper v2: Functional annotation, orthology assignments, and domain prediction at the metagenomic scale. Mol. Biol. Evol. 38 (12), 5825–5829. doi: 10.1093/molbev/msab293
Cao, J., Jiang, M., Li, P., Chu, Z. (2016). Genome-wide identification and evolutionary analyses of the PP2C gene family with their expression profiling in response to multiple stresses in brachypodium distachyon. BMC Genomics 17, 175. doi: 10.1186/s12864-016-2526-4
Carrasco, J. L., Ancillo, G., Mayda, E., Vera, P. (2003). A novel transcription factor involved in plant defense endowed with protein phosphatase activity. EMBO J. 22 (13), 3376–3384. doi: 10.1093/emboj/cdg323
Chao, J., Li, Z., Sun, Y., Aluko, O. O., Wu, X., Wang, Q., et al. (2021). MG2C: A user-friendly online tool for drawing genetic maps. Mol. Horticul. 1 (1), 1–4. doi: 10.1186/s43897-021-00020-x
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13 (8), 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, X., Wang, Y., Lv, B., Li, J., Luo, L., Lu, S., et al. (2014). The NAC family transcription factor OsNAP confers abiotic stress response through the ABA pathway. Plant Cell Physiol. 55 (3), 604–619. doi: 10.1093/pcp/pct204
Chen, C., Yu, Y., Ding, X., Liu, B., Duanmu, H., Zhu, D., et al. (2018). Genome-wide analysis and expression profiling of PP2C clade d under saline and alkali stresses in wild soybean and. Arabidopsis. Protoplasma 255 (2), 643–654. doi: 10.1007/s00709-017-1172-2
Chu, M., Chen, P., Meng, S., Xu, P., Lan, W. (2021). The Arabidopsis phosphatase PP2C49 negatively regulates salt tolerance through inhibition of AtHKT1;1. J. Integr. Plant Biol. 63 (3), 528–542. doi: 10.1111/jipb.13008
Clevenger, J., Chu, Y., Scheffler, B., Ozias-Akins, P. (2016). A developmental transcripttome map for allotetraploid Arachis hypogaea. Front. Plant Sci. 7 1446. doi: 10.3389/fpls.2016.01446
Cui, M. H., Yoo, K. S., Hyoung, S., Nguyen, H. T., Kim, Y. Y., Kim, H. J., et al. (2013). An Arabidopsis R2R3-MYB transcription factor, AtMYB20, negatively regulates type 2C serine/threonine protein phosphatases to enhance salt tolerance. FEBS Lett. 587 (12), 1773–1778. doi: 10.1016/j.febslet.2013.04.028
Dai, X., Zhuang, Z., Zhao, P. X. (2018). psRNATarget: a plant small RNA target analysis server, (2017 release). Nucleic Acids Res. 46 (W1), W49–W54. doi: 10.1093/nar/gky316
Das, A. K., Helps, N. R., Cohen, P. T., Barford, D. (1996). Crystal structure of the protein serine/threonine phosphatase 2C at 2.0 a resolution. EMBO J. 15 (24), 6798–6809. doi: 10.1002/j.1460-2075.1996.tb01071.x
Finn, R. D., Bateman, A., Clements, J., Coggill, P., Eberhardt, R. Y., Eddy, S. R., et al. (2014). Pfam: The protein families database. Nucleic Acids Res. 42 (Database issue), D222–D230. doi: 10.1093/nar/gkt1223
Finn, R. D., Clements, J., Eddy, S. R. (2011). HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 39 (web server issue), W29–W37. doi: 10.1093/nar/gkr367
Geourjon, C., Deléage, G. (1994). SOPM: a self-optimized method for protein secondary structure prediction. Protein Eng. 7 (2), 157–164. doi: 10.1093/protein/7.2.157
Gong, Z., Xiong, L., Shi, H., Yang, S., Herrera-Estrella, L. R., Xu, G., et al. (2020). Plant abiotic stress response and nutrient use efficiency. Sci. China Life Sci. 63 (5), 635674. doi: 10.1007/s11427-020-1683-x
Han, Y., Li, R., Liu, Y., Fan, S., Wan, S., Zhang, X., et al. (2021). The major intrinsic protein family and their function under salt-stress in peanut. Front. Genet. 12. doi: 10.3389/fgene.2021.639585
He, Z., Wu, J., Sun, X., Dai, M. (2019). The maize clade a PP2C phosphatases play critical roles in multiple abiotic stress responses. Int. J. Mol. Sci. 20 (14), 3573. doi: 10.3390/ijms20143573
Hu, B., Jin, J., Guo, A. Y., Zhang, H., Luo, J., Gao, G. (2015). GSDS 2.0: an upgraded gene feature visualization server. Bioinf. (Oxford England) 31 (8), 1296–1297. doi: 10.1093/bioinformatics/btu817
Kaliff, M., Staal, J., Myrenås, M., Dixelius, C. (2007). ABA is required for leptosphaeria maculans resistance via ABI1- and ABI4-dependent signaling. Mol. Plant-Microbe interactions: MPMI 20 (4), 335–345. doi: 10.1094/MPMI-20-4-0335
Kelley, L. A., Mezulis, S., Yates, C. M., Wass, M. N., Sternberg, M. J. (2015). The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10 (6), 845–858. doi: 10.1038/nprot.2015.053
Kerk, D., Bulgrien, J., Smith, D. W., Barsam, B., Veretnik, S., Gribskov, M. (2002). The complement of protein phosphatase catalytic subunits encoded in the genome of Arabidopsis. Plant Physiol. 129 (2), 908–925. doi: 10.1104/pp.004002
Kerk, D., Templeton, G., Moorhead, G. B. (2008). Evolutionary radiation pattern of novel protein phosphatases revealed by analysis of protein data from the completely sequenced genomes of humans, green algae, and higher plants. Plant Physiol. 146 (2), 351–367. doi: 10.1104/pp.107.111393
Kim, S., Park, S. I., Kwon, H., Cho, M. H., Kim, B. G., Chung, J. H., et al. (2022). The rice abscisic acid-responsive RING finger E3 ligase OsRF1 targets OsPP2C09 for de-gradation and confers drought and salinity tolerance in rice. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.797940
Krzywińska, E., Kulik, A., Bucholc, M., Fernandez, M. A., Rodriguez, P. L., Dobrowolska, G. (2016). Protein phosphatase type 2C PP2CA together with ABI1 inhibits SnRK2.4 activity and regulates plant responses to salinity. Plant Signaling Behav. 11 (12), e1253647. doi: 10.1080/15592324.2016.1253647
Krzywinski, M., Schein, J., Birol, I., Connors, J., Gascoyne, R., Horsman, D., et al. (2009). Circos: An information aesthetic for comparative genomics. Genome Res. 19 (9), 1639–1645. doi: 10.1101/gr.092759.109
Kuhn, J. M., Boisson-Dernier, A., Dizon, M. B., Maktabi, M. H., Schroeder, J. I. (2006). The protein phosphatase AtPP2CA negatively regulates abscisic acid signal transduction in Arabidopsis, and effects of abh1 on AtPP2CA mRNA. Plant Physiol. 140 (1), 127–139. doi: 10.1104/pp.105.070318
Lescot, M., Déhais, P., Thijs, G., Marchal, K., Moreau, Y., Van de Peer, Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30 (1), 325–327. doi: 10.1093/nar/30.1.325
Lessard, P., Kreis, M., Thomas, M. (1997). Protein phosphatases and protein kinases in higher plants. Comptes rendus l'Academie Des. Sci. Serie III Sci. la vie 320 (9), 675–688. doi: 10.1016/s0764-4469(97)84815-9
Leung, J., Giraudat, J. (1998). Abscisic acid signal transduction. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49, 199–222. doi: 10.1146/annurev.arplant.49.1.199
Li, F., Fan, G., Wang, K., Sun, F., Yuan, Y., Song, G., et al. (2014). Genome sequence of the cultivated cotton Gossypium arboretum. Nat. Genet. 46 (6), 567–572. doi: 10.1038/ng.2987
Lim, C. W., Baek, W., Jung, J., Kim, J. H., Lee, S. C. (2015). Function of ABA in stomatal defense against biotic and drought stresses. Int. J. Mol. Sci. 16 (7), 15251–15270. doi: 10.3390/ijms160715251
Liu, P. F., Chang, W. C., Wang, Y. K., Chang, H. Y., Pan, R. L. (2008). Signaling pathways mediating the suppression of Arabidopsis thaliana Ku gene expression by abscisic acid. Biochimica. Biophy. Acta 1779 (3), 164–174. doi: 10.1016/j.bbagrm.2007.12.005
Liu, X., Zhu, Y., Zhai, H., Cai, H., Ji, W., Luo, X., et al. (2012). AtPP2CG1, a protein phosphatase 2C, positively regulates salt tolerance of Arabidopsis in abscisic acid-dependent manner. Biochem. Biophys. Res. Commun. 422 (4), 710–715. doi: 10.1016/j.bbrc.2012.05.064
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCt method. Methods (San Diego Calif.) 25 (4), 402–408. doi: 10.1006/meth.2001.1262
Luo, L., Wan, Q., Zhang, K., Zhang, X., Guo, R., Wang, C., et al. (2021). AhABI4s negatively regulate salt-stress response in peanut. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.741641
Min, M. K., Choi, E. H., Kim, J. A., Yoon, I. S., Han, S., Lee, Y., et al. (2019). Two clade a phosphatase 2Cs expressed in guard cells physically interact with abscisic acid signaling components to induce stomatal closure in rice. Rice (New York N.Y.) 12 (1), 37. doi: 10.1186/s12284-019-0297-7
Ohta, M., Guo, Y., Halfter, U., Zhu, J. K. (2003). A novel domain in the protein kinase SOS2 mediates interaction with the protein phosphatase 2C ABI2. Proc. Natl. Acad. Sci. United States America 100 (20), 11771–11776. doi: 10.1073/pnas.2034853100
Pang, Q., Zhang, T., Zhang, A., Lin, C., Kong, W., Chen, S. (2020). Proteomics and phosphoproteomics revealed molecular networks of stomatal immune responses. Planta 252 (4), 66. doi: 10.1007/s00425-020-03474-3
Raza, A., Tabassum, J., Fakhar, A. Z., Sharif, R., Chen, H., Zhang, C., et al. (2022). Smart reprograming of plants against salinity stress using modern biotechnological tools. Crit. Rev. Biotechnol., 1–28. doi: 10.1080/07388551.2022.2093695. Advance online publication.
Rodriguez, P. L., Benning, G., Grill, E. (1998). ABI2, a second protein phosphatase 2C involved in abscisic acid signal transduction in Arabidopsis. FEBS Lett. 421 (3), 185–190. doi: 10.1016/s0014-5793(97)01558-5
Schweighofer, A., Hirt, H., Meskiene, I. (2004). Plant PP2C phosphatases: emerging functions in stress signaling. Trends Plant Sci. 9 (5), 236–243. doi: 10.1016/j.tplants.2004.03.007
Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., et al. (2003). Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 13 (11), 2498–2504. doi: 10.1101/gr.1239303
Shazadee, H., Khan, N., Wang, J., Wang, C., Zeng, J., Huang, Z., et al. (2019). Identification and expression profiling of protein phosphatases (PP2C) gene family in Gossypium hirsutum l. Int. J. Mol. Sci. 20 (6), 1395. doi: 10.3390/ijms20061395
Shi, Y. (2009). Serine/threonine phosphatases: mechanism through structure. Cell 139 (3), 468–484. doi: 10.1016/j.cell.2009.10.006
Singh, A., Giri, J., Kapoor, S., Tyagi, A. K., Pandey, G. K. (2010). Protein phosphatase complement in rice: Genome-wide identification and transcriptional analysis under abiotic stress conditions and reproductive development. BMC Genomics 11, 435. doi: 10.1186/1471-2164-11-435
Singh, A., Jha, S. K., Bagri, J., Pandey, G. K. (2015). ABA inducible rice protein phosphatase 2C confers ABA insensitivity and abiotic stress tolerance in Arabidopsis. PloS One 10 (4), e0125168. doi: 10.1371/journal.pone.0125168
Song, S. K., Yun, Y. B., Lee, M. M. (2020). POLTERGEIST and POLTERGEIST-LIKE1 are essential for the maintenance of post-embryonic shoot and root apical meristems as revealed by a partial loss-of-function mutant allele of pll1 in Arabidopsis. Genes Genomics 42 (1), 107–116. doi: 10.1007/s13258-019-00894-8
Stone, J. M., Collinge, M. A., Smith, R. D., Horn, M. A., Walker, J. C. (1994). Interaction of a protein phosphatase with an arabidopsis serine-threonine receptor kinase. Sci. (New York N.Y.) 266 (5186), 793–795. doi: 10.1126/science.7973632
Strizhov, N., Abrahám, E., Okrész, L., Blickling, S., Zilberstein, A., Schell, J., et al. (1997). Differential expression of two P5CS genes controlling proline accumulation during salt-stress requires ABA and is regulated by ABA1, ABI1 and AXR2 in Arabidopsis. Plant J. Cell Mol. Biol. 12 (3), 557–569. doi: 10.1046/j.1365-313x.1997.00557.x
Subudhi, P. K., Shankar, R., Jain, M. (2020). Whole genome sequence analysis of rice genotypes with contrasting response to salinity stress. Sci. Rep. 10 (1), 21259. doi: 10.1038/s41598-020-78256-8
Sun, B. R., Fu, C. Y., Fan, Z. L., Chen, Y., Chen, W. F., Zhang, J., et al. (2019). Genomic and transcriptomic analysis reveal molecular basis of salinity tolerance in a novel strong salt-tolerant rice landrace changmaogu. Rice (New York N.Y.) 12 (1), 99. doi: 10.1186/s12284-019-0360-4
Tamura, K., Stecher, G., Kumar, S. (2021). MEGA11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38 (7), 3022–3027. doi: 10.1093/molbev/msab120
Thompson, J. D., Higgins, D. G., Gibson, T. J. (1994). CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22 (22), 4673–4680. doi: 10.1093/nar/22.22.4673
Waterhouse, A., Bertoni, M., Bienert, S., Studer, G., Tauriello, G., Gumienny, R., et al. (2018). SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 46 (W1), W296–W303. doi: 10.1093/nar/gky427
Wilkins, M. R., Gasteiger, E., Bairoch, A., Sanchez, J. C., Williams, K. L., Appel, R. D., et al. (1999). Protein identification and analysis tools in the ExPASy server. Methods Mol. Biol. (Clifton N.J.) 112, 531–552. doi: 10.1385/1-59259-584-7:531
Xue, T., Wang, D., Zhang, S., Ehlting, J., Ni, F., Jakab, S., et al. (2008). Genome-wide and expression analysis of protein phosphatase 2C in rice and Arabidopsis. BMC Genomics 9, 550. doi: 10.1186/1471-2164-9-550
Yang, Q., Liu, K., Niu, X., Wang, Q., Wan, Y., Yang, F., et al. (2018). Genome-wide identification of PP2C genes and their expression profiling in response to drought and cold stresses in Medicago truncatula. Sci. Rep. 8 (1), 12841. doi: 10.1038/s41598-018-29627-9
Yu, X., Han, J., Wang, E., Xiao, J., Hu, R., Yang, G., et al. (2019). Genome-wide identification and homoeologous expression analysis of PP2C genes in wheat (Triticum aestivum l.). Front. Genet. 10. doi: 10.3389/fgene.2019.00561
Zhang, J., Li, X., He, Z., Zhao, X., Wang, Q., Zhou, B., et al. (2013). Molecular character of a phosphatase 2C (PP2C) gene relation to stress tolerance in Arabidopsis thaliana. Mol. Biol. Rep. 40 (3), 2633–2644. doi: 10.1007/s11033-012-2350-0
Zhang, Z., Luo, S., Liu, Z., Wan, Z., Gao, X., Qiao, Y., et al. (2022). Genome-wide identification and expression analysis of the cucumber PYL gene family. PeerJ 10, e12786. doi: 10.7717/peerj.12786
Keywords: peanut, PP2C gene family, salt stress, genome-wide identification, gene expression analysis
Citation: Wu Z, Luo L, Wan Y and Liu F (2023) Genome-wide characterization of the PP2C gene family in peanut (Arachis hypogaea L.) and the identification of candidate genes involved in salinity-stress response. Front. Plant Sci. 14:1093913. doi: 10.3389/fpls.2023.1093913
Received: 09 November 2022; Accepted: 09 January 2023;
Published: 27 January 2023.
Edited by:
Weijian Zhuang, Fujian Agriculture and Forestry University, ChinaReviewed by:
Benjamin Karikari, University for Development Studies, GhanaCopyright © 2023 Wu, Luo, Wan and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fengzhen Liu, bGl1ZnpAc2RhdS5lZHUuY24=; Yongshan Wan, eXN3YW5Ac2RhdS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.