
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 01 March 2023
Sec. Plant Abiotic Stress
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1089380
This article is part of the Research TopicAbiotic Stress Alleviation in Plants: Morpho-Physiological and Molecular AspectsView all 31 articles
Introduction: A lack of soil phosphorus (P) is a principal factor restricting the normal growth of Lotus corniculatus in the karst area of Guizhou Province, China, but the response mechanism of L. corniculatus under low-phosphorus stress remains unclear.
Methods: Therefore, we treated two selected L. corniculatus lines (low-P-intolerant line 08518 and low-P-tolerant line 01549) from 13 L. corniculatus lines with normal phosphorus (0.5 mmol/L KH2PO4, NP) and low phosphorus (0.005 mmol/L KH2PO4, LP) concentrations to study changes in morphological, physiological and transcriptome data under low-phosphorus stress.
Results: The low-P-tolerant line 01549 exhibited better performance under low-phosphorus stress. Compared with the NP treatment, all root morphological indicators of the low-P-tolerant line 01549 increased, and those of the low-P-intolerant line 08518 decreased under low-P stress. Compared with the NP treatment, acid phosphatase (ACP), catalase (CAT), superoxide dismutase (SOD), and peroxidase (POD) activities, and the malondialdehyde (MDA), soluble sugar (SS), soluble protein (SP) and proline (Pro) contents of the two L. corniculatus lines increased under low-P stress. A transcriptome analysis of L. corniculatus showed that a total of 656 and 2243 differentially expressed genes (DEGs) were identified in line 01549 and line 08518, respectively. Meanwhile, the main pathways, such as carbohydrate metabolism, acid phosphatases, phosphate transporters and biosynthesis of secondary metabolites, as well as related genes were also screened by performing a KEGG enrichment analysis.
Discussion: The findings provide an essential point of reference for studying the physiological and molecular mechanism of the response to low-P stress in L. corniculatus.
Phosphorus (P), one of the essential elements for plants, plays a key role not only in promoting plant growth and development, but also in regulating physiological metabolism and resistance to stress (Ding et al., 2021; Zipfel and Oldroyd, 2017). The soil P concentration is high, but about 80% of the phosphorus is organic phosphorus which is difficult to be used by plants, while inorganic phosphate in soil is easily fixed by iron, aluminum and calcium ions to form insoluble phosphate which cannot be used by plants directly (Chiou et al., 2006). Usually, the concentration of available phosphate in soil does not exceed 2.0 uM, and even in fertile soils it rarely exceeds 10 uM, far from meeting the needs of normal plant growth(Yuan and Liu, 2008). In order to improve the efficiency of P acquisition and utilization by plants, many scholars have studied the mechanisms of plant adaptation and tolerance to low-P stress(Nilsson et al., 2010; Ried et al., 2021).
Plants have been shown to have evolved a range of complex and tightly controlled mechanisms for maintaining cellular Pi concentrations (Tesfaye et al., 2007). One of the most important mechanisms is the ability of the plant root system to improve phosphorus uptake from the soil and autologous phosphorus utilization through a series of collaborative processes involving changes in plant morphology, physiology and molecular biology (Verma et al., 2021). Some studies have reported that plant biomass decreases significantly under low-P conditions and that the root surface area, total root number, lateral root length, number of lateral roots and root volume increase, expanding the area of P uptake by the root system (Hernández et al., 2007; Raya-González et al., 2021). O'Rourke et al. (2013) found that when P is deficient in the soil, white lupin forms cluster roots, while the row roots secrete organic acids to activate the fixed P in the soil, thereby increasing the effective P concentration in the medium. Simultaneously, the plant activates the fixed phosphorus in the soil through its own physiological and biochemical changes, such as the secretion of organic acids, and increases the effective phosphorus content through chemical action (Plaxton and Tran, 2011; Ciereszko et al., 2011). Li et al. (2016) found that Leymus chinensis protected PSII against Pi-induced photooxidative stress in the organism by enhancing the activity of anti-reactive oxygenase. Under P-deprived conditions, chlorophyll contents, soluble protein contents and root vigor are reduced, while malondialdehyde (MDA), soluble sugar and free proline contents are increased, as are peroxidase (POD), superoxide dismutase (SOD) and catalase (CAT) activities (Rohman et al., 2019).
The morphological, physiological, and biochemical changes exhibited by plants under low-P stress are achieved by the synergistic expression of phosphate starvation-response (PSR) genes in plant tissues (Zhang H. et al., 2014). Sun and Zheng (2022) found ZmPHT1;1, ZmPHT1;9 and Zmphytase 2 genes play a key role in phosphorus recovery in maize. Hu et al. (2021) found P deficit increases the expression of genes related to glycerolipid synthesis, while the expression of genes related to phospholipid breakdown is inhibited. An increase in the expression of the genes encoding the PSII protein complex, the cytochrome b/f protein complex, the PSI protein complex, photosynthetic electron transport proteins, and several members of the F-type ATPase family was observed. Müller et al. (2007) found that chronic low-P stress affected primary and secondary plant metabolism, disrupted the balance of carbon metabolism, inhibited the binding of proteins and chlorophyll a or b, and reduced the photosynthetic capacity of plants, while the expression of genes related to sucrose and glucose metabolism were upregulated. Jeong et al. (2017) studied the rice transcriptome under low-P stress and found that kinase and phosphotransferase-related genes were upregulated, and the expression of genes involved in ribosome composition, transcription and translation was downregulated. In addition, in-depth transcriptomics analyses of oil palm (Kong et al., 2021), barley (Ren et al., 2018), wheat (Wang et al., 2019) and sorghum (Zhang J. et al., 2019) under low-P stress have also been performed, and the corresponding drought stress-responsive genes have been identified.
Lotus corniculatus is a perennial herb of the genus Lotus with strong tolerance of cold, barren and drought, developed root system, creeping stem and branch growth, strong grip, wide coverage, and is an excellent grass species for karst ecological environment management (Knežević et al., 2021). Second, the stems and leaves of L. corniculatus are soft and juicy, rich in carbohydrates, with high yield, good palatability, a high saponin content and enrichment of condensed tannins, which may reduce the occurrence of swelling disease in livestock (Antonelli et al., 2021). Its root system has many rhizomes and has good nitrogen fixation, which can increase soil organic matter and improve soil structure, and is a high-quality forage plant resource and green manure (Jiang et al., 2021). Genetic diversity analysis of accession resources is the basis for new variety selection and breeding, and since the 1980s, some cultivars of L. corniculatus have been introduced from abroad in China, and production trials have shown that these varieties have strong adaptability and high production performance and are widely planted in northwest China (Xie et al., 2023). In the karst region of southwest China, the research on the selection and breeding of varieties of L. corniculatus is not systematic, and there are few new varieties of L. corniculatus. It is of great practical significance for the large-scale cultivation of L. corniculatus and the development of local ecological animal husbandry in karst mountainous areas of southwest China by introducing new excellent L. corniculatus line resources and breeding accession varieties suitable for different ecological zones and uses. Moreover, the karst region of southwest China with severe regional rock desertification and effective phosphorus deficiency, and low phosphorus concentrations can impair plant growth and development (Baghbani-Arani et al., 2021; Zhao et al., 2022a), it is of great significance and application value to cultivate phosphorus-efficient plants and improve the utilization of phosphorus in soil by plants. Therefore, in the present study, we used the line resources of L. corniculatus as the object of the study and established two treatments, normal and low P concentrations, to screen two line species of L. corniculatus that are tolerant or sensitive to low P concentrations. The aim was to elucidate the adaptive mechanisms and identify the genes regulating the low-P tolerance in the two genotypes of L. corniculatus. And it is intended to provide a preliminary reference for the innovative utilization of excellent L. corniculatus line resources and the selection and breeding of new varieties.
We conducted sand culture experiments to evaluate the P tolerance of 13 L. corniculatus lines (Table S1) provided by the National Germplasm Forage Medium Term Repository, Institute of Grassland Research, Chinese Academy of Agricultural Sciences. The experiment was performed at Guizhou University College of Animal Science from May to August 2021. For the tested L. corniculatus lines, 4-8 cm cuttings were planted in different pots, the plants were grown and divided, the roots were washed with distilled water, and the uniformly growing L. corniculatus lines was selected and planted in seedling trays filled with quartz sand (width: high: long = 28 cm: 7.5 cm: 54 cm) and precultured with 15 seedlings per tray in a nutrient solution on 14 h/10 h light/dark cycle at 26°C/22°C. Three trays were included for one treatment. The nutrient solution was prepared with deionised water, which was formulated as described in our previous research article (Zhao et al., 2022b). Plants were incubated with the complete nutrient solution for one week and then divided into two treatment groups consisting of normal (0.5 mmol/L KH2PO4, NP) and low phosphorus (0.005 mmol/L KH2PO4, LP) concentrations, with the nutrient solution replenished every 2 d. KH2PO4 was used as the phosphorus source, the K+ concentration was balanced by K2SO4, and the pH of the nutrient solution was adjusted to 6.00 using NaOH and H2SO4. After 21 d, root tissues from the control and low-P stress groups were collected to measure their growth characteristics and whole plant P concentration.
A straightedge was used to measure the plant height. The aboveground and belowground sections of the plant were separated and the fresh weights were measured before these parts were placed in an oven at 105°C for 30 min and dried to a constant weight at 65°C to determine the dry weights of the aboveground and belowground portions. The molybdenum blue technique was used to measure the total P concentration of L. corniculatus (Chapman and Pratt, 1962). Fresh leaves and roots were scanned with an Epson Perfection V800 Photo scanner, and the resulting images were analyzed with WinFOLIA Pro 2015 and WinRHIZO Pro 2015 software (Regent Instructions, Canada Inc.) to obtain leaf indicators (surface area, length, width, length/width and perimeter) and root indicators (total length, total surface area, diameter, volume, tip number and hair number), respectively.
An evaluation of the low-P tolerance of different L. corniculatus lines was conducted. Low-P tolerance coefficients (based on root morphology, leaf morphology, biomass, plant height and whole plant P concentration of the different L. corniculatus lines under low-P stress) were first calculated, followed by a comprehensive evaluation using principal component analysis and affiliation function for low-P tolerance to eliminate differences related to the accessions (Zhu, 2018). The equations were as follows.
Note that Xi denotes the mean value of an indicator, μ(Xj) denotes the affiliation function value of the jth composite indicator, Xj denotes the value of the jth composite indicator, Xmin denotes the minimum value of the jth composite indicator, Xmax denotes the maximum value of the jth composite indicator, Wj denotes the weight of the jth composite indicator to all composite indicators, and Pj denotes the contribution of the jth composite indicator. D denotes the composite evaluation value of the low-P tolerance of each L. corniculatus line.
The experimental design of the two screened L. corniculatus lines (01549 and 08518) was carried out according to the line Selection part, with 15 seedlings per pot and 3 replicates per treatment. After 7 d of low phosphorus stress, the seedlings were removed from the seedling tray, the roots were washed with distilled water and fresh tissue parts were obtained for the determination of physiological indexes and transcriptome sequencing.
Physiological indices, including MDA, soluble protein (SP), proline (Pro) and soluble sugar (SS) contents, as well as SOD, POD, CAT and acid phosphatase (ACP) activities, were determined using an assay kit from Beijing Solarbio Technology Co., Ltd., China.
The methods for transcriptome sequencing and gene expression profiling were the same as those described in our previous study (Zhao et al., 2022c). Briefly, total RNA was extracted from the root samples collected from lines 01549 and 08518, and cDNA libraries were constructed, followed by sequencing using the Illumina NovaSeq6000 platform with a PE150 sequencing strategy. Raw reads obtained from sequencing were quality controlled, and low-quality reads were removed. The quality-controlled clean reads were mapped to the L. corniculatus reference genome (Ensembl, http://www.kazusa.or.jp/lotus/summary3.0.html) using Bowtie2, and then the RSEM was used to count the number of reads mapped to each reference genome for each sample and to calculate the FPKM (fragments per kilobase of transcript per million mapped reads) for each gene. Paired-end reads from the same fragment were counted as one fragment to obtain the expression levels of genes and transcripts. A differential expression analysis was performed using the R package EdgeR. Genes with FDR (false discovery rate)< 0.05 and |log2FC (fold change)| >1 were defined as differentially expressed genes (DEGs). The GO enrichment analysis is an international standard classification system for gene function. DEGs were functionally annotated using the Gene Ontology database (http://www.geneontology.org/), and GO terms with FDR ≤ 0.05 were selected as significantly enriched GO entries for analysis. Functional annotation of DEGs was performed using the same approach with the KEGG database (http://www.genome.jp/kegg/pathway.html) to analyze pathways significantly correlated with DEGs. The number of genes in each pathway was then counted, and the significantly enriched pathways were analyzed using hypergeometric tests.
Eight DEGs (4 upregulated and 4 downregulated) were randomly selected for the experiment using the RNA obtained as described in Table S3 as the template to confirm the accuracy of the RNA-seq results. cDNA was synthesized from the same RNA samples used for transcriptome sequencing. We performed qRT-PCR on a CFX Connect™ Real-Time System (Applied Biosystems) using UltraSYBR mixture (CWBiotech). The thermocycle parameters were 10 min at 95°C, 40 cycles of 10 s at 95°C, 30 s at 60°C, and 32 s at 72°C, followed by 15 s at 95°C, 1 min at 60°C, 15 s at 95°C, and 15 s at 60°C, in a 20 μl reaction mixture (Zhao et al., 2022c). As an internal reference for normalization, the Lotus corniculatus-actin gene was used. Each sample was analyzed in triplicate.
Data were analyzed using Student’s t test in SPSS 25.0 (IBM, Chicago, IL, USA), with a significance level of p<0.05. Relative root length=root length of plants under low-P stress/root length of the control plants, and all morphological and physiological parameters were obtained using similar methods. All data are presented as the means ± standard errors (SE) of three replicates and were plotted graphically using Origin 2021 software.
Previous studies have shown that differences in plant line tolerance to low-P are due to their increased ability to acquire P. (Zhang J. et al., 2019). We speculate that low-P-tolerant plants will have higher P concentration under low-P treatment than low-P-intolerant plants. Thirteen L. corniculatus lines were cultured with the LP and NP treatments, which showed the whole-plant P concentration of all 13 L. corniculatus lines were significantly deceased (P< 0.05) in the range of 9.09%-140.74%, with 08518 (140.74%), 00116 (128.57%), and 01885 (85.71%) showing larger reductions and 01549 showing the smallest reduction (9.09%) under low phosphorus stress (Table S2). The value of the affiliation function of each composite indicator was computed using the formula, and the weight of the indicator was computed according to the contribution rate of each indicator. Finally, the comprehensive value of the low-P tolerance of each accession was obtained. The low-P tolerance of different L. corniculatus lines varied, where the low-P-tolerance line 01549 was strongest, and the low-P-intolerant line 08518 was weakest (Table S3). Therefore, we selected 01549 and 08518 as the low-P-tolerant and low-P-sensitive lines, respectively, for subsequent analyses. At the same time, the root morphology of each accession was examined. The relative P concentration in whole plants was significantly positively correlated with the relative length, relative volume and relative surface area of roots (Table 1), and most values of root morphology differed significantly between lines 01549 and 08518 under low phosphorus stress (Table 2).
The MDA, Pro, SS, and SP contents and SOD, POD, CAT and ACP activities of both lines 01549 and 08518 increased following LP treatment compared with NP treatment. Compared with those of line 01549, the MDA and SS contents and CAT activity of line 08518 increased under low-P stress, but only the MDA content significantly increased. The Pro and SP contents and SOD, POD and ACP activities of line 08518 were decreased under low-P stress, and the Pro content and SOD, POD and ACP activities were significantly decreased (Table 3).
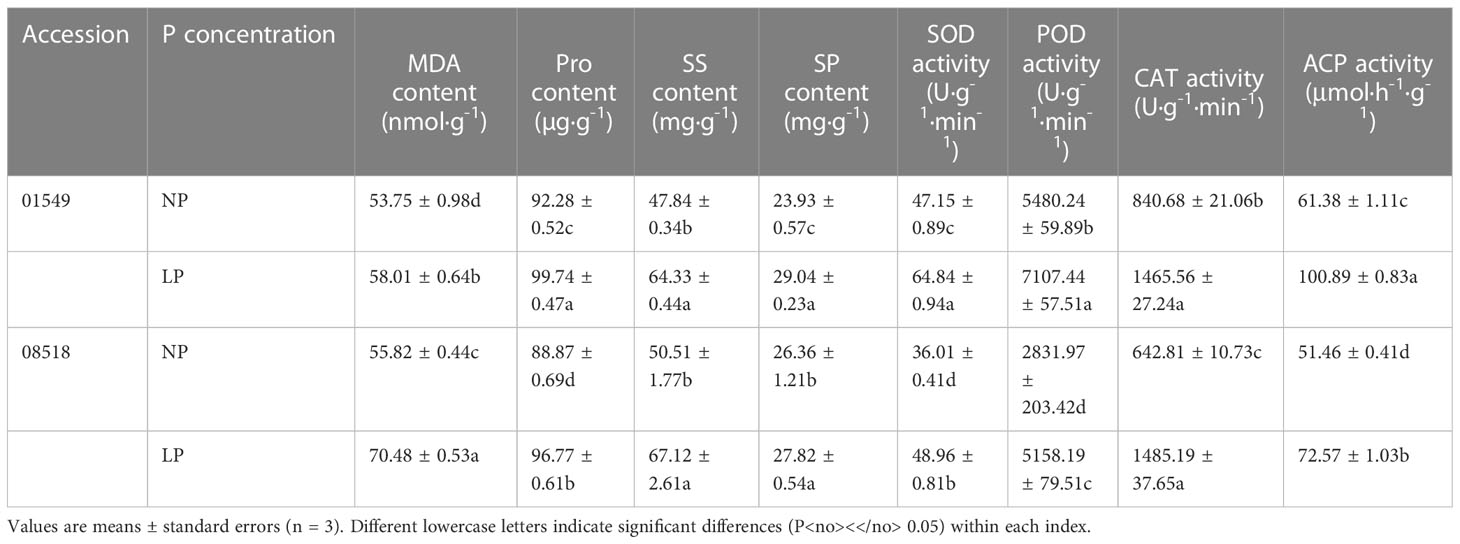
Table 3 Effect of low-P stress on the root physiological traits of L. corniculatus lines 01549 and 08518.
Twelve cDNA libraries were sequenced from the roots of lines 01549 and 08518 grown for 7 d under LP and NP treatments using Illumina high-throughput sequencing technology, generating a total of 554 million high-quality clean reads from three replicates of 12 libraries. The sequencing results were submitted to NCBI (SRA accession: PRJNA871253), and the Q30 values of the samples reached 93%. Approximately 80% of the total clean reads (Ensembl; http://www.kazusa.or.jp/lotus/summary3.0.html) were mapped to the L. corniculatus reference sequence lotus/summary3.0 (Table S4). Eight DEGs were randomly chosen for qRT-PCR validation (Figure 1). Their expression profiles were significantly associated with the RNA-seq data FPKM, validating the trustworthiness of the transcriptomic data (Table S5).
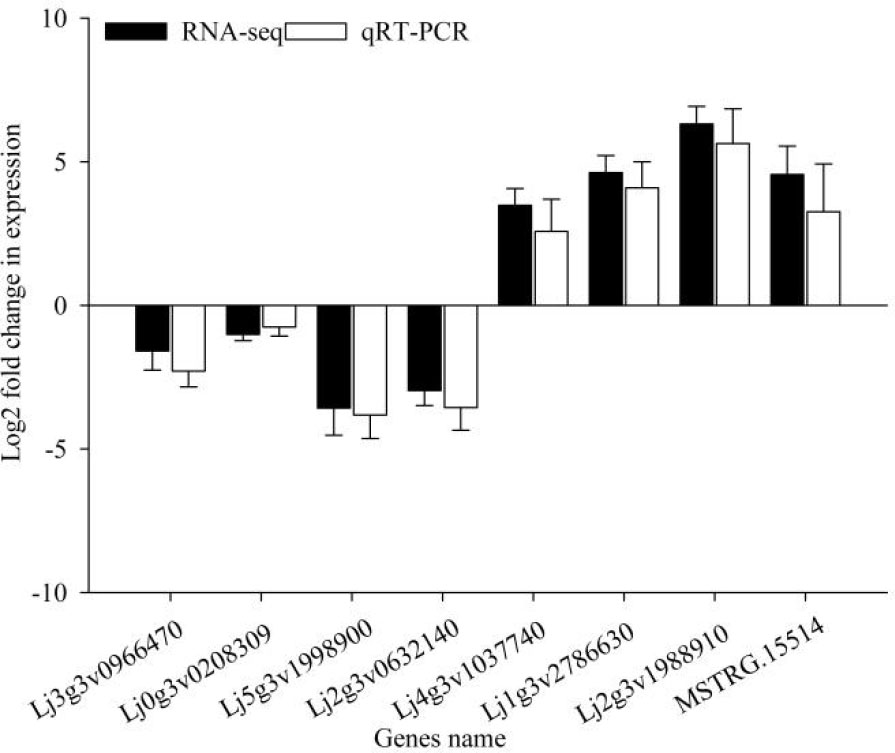
Figure 1 Validation of the transcript abundance of eight selected genes obtained from RNA-seq using qRT-PCR.
When the transcripts of L. corniculatus were cultivated with the LP treatment 656 DEGs (243 upregulated and 413 downregulated) were identified in line 01549, and 2243 DEGs (1,455 upregulated and 788 downregulated) were identified in line 08518 compared with the NP treatment (Figure 2). Surprisingly, the number of DEGs in line 01549 was fewer than that in line 08518, regardless of whether they were upregulated or downregulated. A total of 5,596 DEGs (3,277 upregulated and 2,319 downregulated) were discovered in the low-P-tolerant line 01549 compared with the low-P-intolerant line 08518 under low-P stress. Only 58 common genes were discovered among these DEGs, indicating that these genes were considered valuable candidate genes for improving low-P tolerance (Figure 2B).
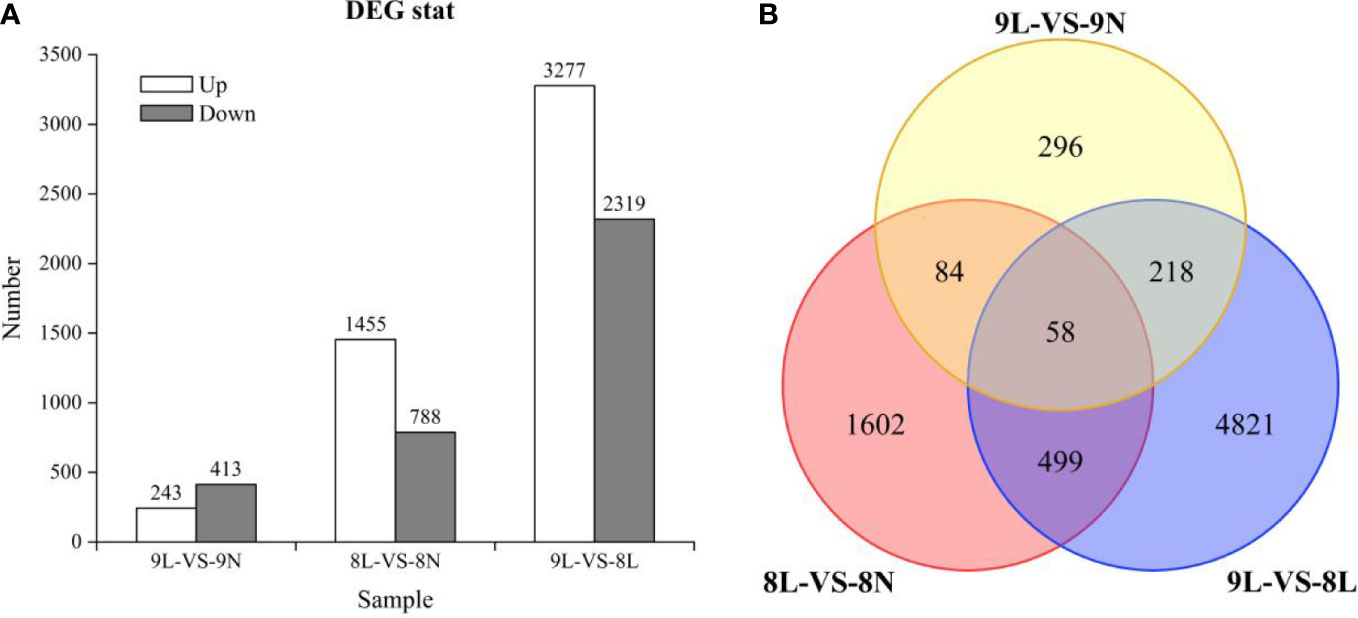
Figure 2 (A) The number of diferentially expressed genes in each comparison. (B) Venn diagram illustrating the genes of L. corniculatus in response to low-P stress.DEGs are shown in Venn diagrams. 9: Low-P-tolerant line 01549; 8: low-P-intolerant line 08518; L: low P; N: normal P.
We performed GO and KEGG pathway enrichment analyses of the DEGs identified in L. corniculatus. In total, 278 of the 656 DEGs in line 01549, 1,055 of the 2,243 DEGs in line 08518 and 1,445 of the 5,595 DEGs in the comparison of lines 01549 and 08518 under low-P stress were identified in the GO database. All DEGs identified in the GO database from the three comparison groups were assigned to cellular components, biological processes and molecular functions. In addition, oxidoreductase activity and oxidation-reduction process were the two GO terms with the largest number of DEGs in the three comparison groups. Second, chlorophyllide oxygenase activity, tetrapyrrole binding, enzyme inhibitor activity, endopeptidase inhibitor activity, tetrapyrrole binding and heme binding were also significantly enriched in the three comparison groups (Table S6).
Ten, 24, and 17 significantly enriched pathways were identified among the enriched KEGG pathways for line 01549, 08518, and 01549 vs. 08518 under low-P stress, respectively (Table S7). As shown in Figures 3A-C, we screened the top 20 enriched KEGG pathways for the analysis of line 01549, line 08518 and lines 01549 vs. 08518 under low-P stress. For line 01549, phenylpropanoid biosynthesis (ko00940, 11 genes), stilbenoid, diarylheptanoid and gingerol biosynthesis (ko00945, 4 genes), and nitrogen metabolism (ko00910, 3 genes) were the three enriched pathways with the highest number of DEGs (Figure 3A). For line 08518, phenylpropanoid biosynthesis (ko00940, 43 genes), linoleic acid metabolism (ko00591, 12 genes), and ubiquinone and other terpenoid-quinone biosynthesis (ko00130, 11 genes) were the three main pathways with a large number of DEGs (Figure 3B). For the comparison of line 01549 with line 08518, phenylpropanoid biosynthesis (ko00940, 51 genes), stilbenoid, diarylheptanoid and gingerol biosynthesis (ko00945, 16 genes), and glycolysis/gluconeogenesis (ko00010, 30 genes) were the three main pathways (Figure 3C). The enriched KEGG pathways therefore revealed that genes in L. corniculatus were considerably differentially expressed, suggesting that the various lines activated various molecular mechanisms in response to low-P stress.
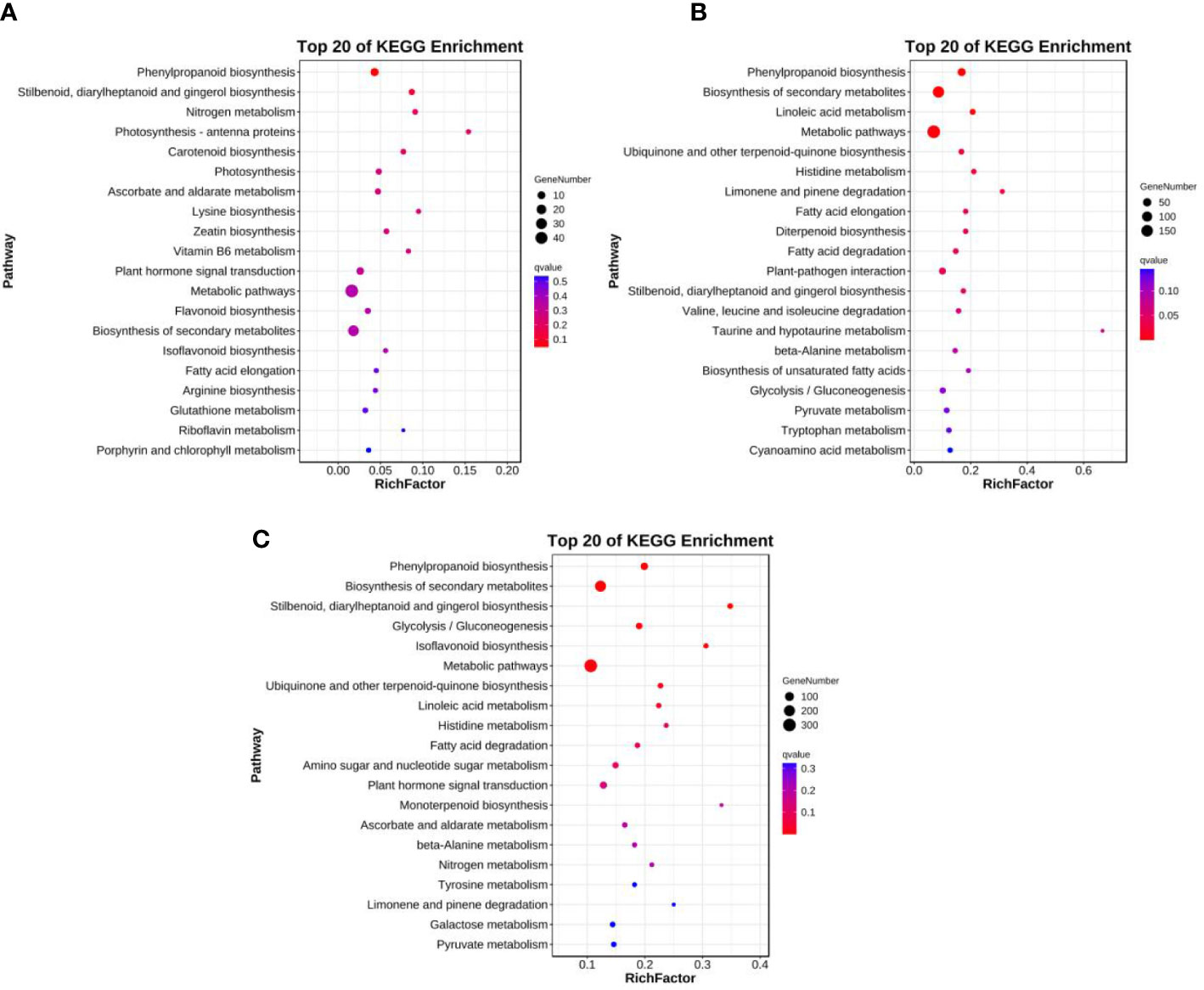
Figure 3 Top 20 enriched KEGG pathways in (A) line 01549, (B) line 08518, and (C) the comparison of lines 01549 and 08518 under low-P stress.
From the transcriptome data, 6 differentially expressed acid phosphatase genes were identified; among those, 4 PAP genes (1 PAP1, 1 PAP17, 1 PAP22 and 1 PAP27) were significantly upregulated in different lines, but 2 APS1 genes were downregulated in the comparison of lines 01549 and 08518 under low-P stress. Eight differentially expressed phosphate transporter genes from 6 families were identified; among those, 2 PHT1-3 genes, 2 PHT1-4 genes, 1 PHT1-7 gene, 1 PHO1-H1 gene, and 1 PHT1-11 gene were significantly upregulated, but 1 PHO1-H10 gene was significantly downregulated in line 01549 compared with line 08518 under low-P stress (Figure 4 and Table S8).
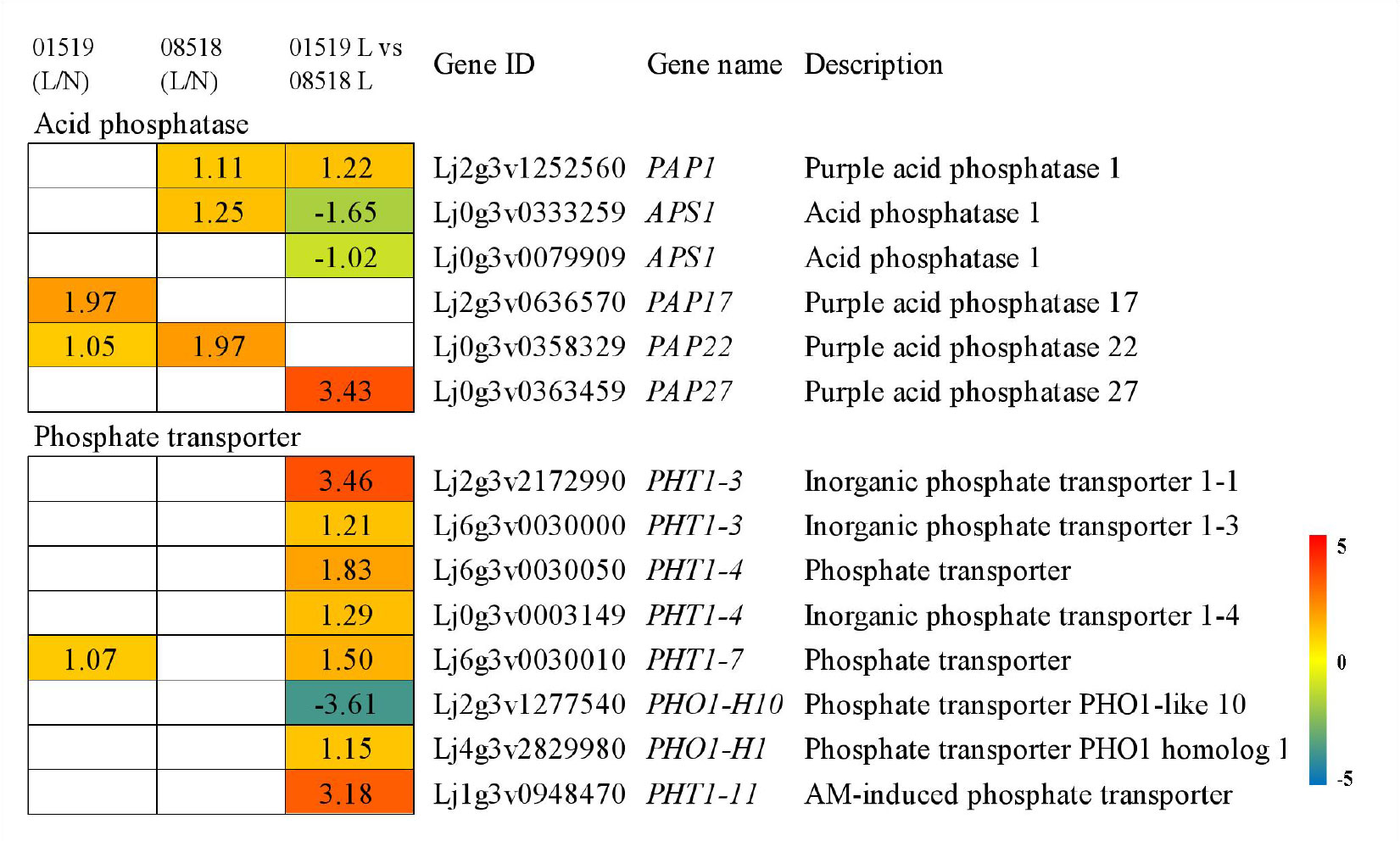
Figure 4 Acid phosphatase and phosphate transporter genes identified in the RNA-seq analysis that showed changes in expression during low-P stress. Log2(fold-changes) of the comparisons are shown by the numbers in the gray rectangles. The comparisons show that each of the genes in the picture is significantly differentially expressed.
Sixty-one DEGs, including genes involved in auxin, abscisic acid, brassinosteroid, cytokinin, ethylene, gibberellin, jasmonic acid, and salicylic acid signal transduction, were discovered to be associated with plant hormones (Figure 5 and Table S9). In the auxin signaling pathway, 2 ARF, 3 GH3 and 4 SAUR genes were significantly upregulated, but 1 GH3 and 6 SAUR genes were significantly downregulated in line 01549 compared with line 08518 under low-P stress. Two GH3, 3 SAUR and 3 IAA genes were significantly upregulated, but 1 GH3 and 2 SAUR genes were significantly downregulated in line 08518 under low-P stress. One GH3 gene was significantly upregulated in line 01549 under low-P stress. In the abscisic acid signaling pathway, 1 PYL and 3 SNRK genes were significantly upregulated, while 2 PP2C and 1 SNRK genes were significantly downregulated in line 01549 compared with line 08518 under low-P stress. One ABF gene and 1 PYL gene were significantly upregulated in lines 01549 and 08518, respectively. In the brassinosteroid signaling pathway, 2 BZR1_2 and 4 TCH4 genes were significantly downregulated in line 01549 compared with line 08518 under low-P stress, and 1 BZR1_2 and 3 TCH4 genes were significantly downregulated in line 08518 under low-P stress. In the cytokinin signaling pathway, 1 AHP and 1 ARR-A gene were significantly upregulated in line 01549 compared with line 08518 under low-P stress, and 1 AHP gene was significantly upregulated and 1 AHK2_3_4 gene was significantly downregulated in line 08518 under low-P stress. In the ethylene signaling pathway, 1 ETR and 2 ERF1 genes were significantly upregulated, while 1 EIN3 and 1 ERF1 gene were significantly downregulated in lines 01549 and 08518 under low-P stress. One ERF1 gene was significantly upregulated, while 1 ERF1 gene was significantly downregulated in line 08518 under low-P stress. One ERF1 gene was significantly downregulated in line 01549 under low-P stress. In the gibberellin signaling pathway, 1 GID2 and 1 PIF3 gene were significantly upregulated, while 1 PIF4 gene was significantly downregulated in lines 01549 and 08518 under low-P stress, 1 PIF4 gene was significantly downregulated in line 08518 under low-P stress, and 1 DELLA gene was significantly upregulated in line 01549 under low-P stress. In the jasmonic acid signaling pathway, 2 JAZ genes were significantly downregulated, while 1 MYC2 and 1 JAR1_4_6 gene were significantly upregulated in line 01549 compared with line 08518 under low-P stress, 1 JAZ gene was significantly downregulated in line 08518 under low-P stress, and 1 JAZ gene was significantly downregulated in line 01549 under low-P stress. In the salicylic acid signaling pathway, 2 TGA genes were significantly upregulated in line 01549 compared with line 08518 under low-P stress.
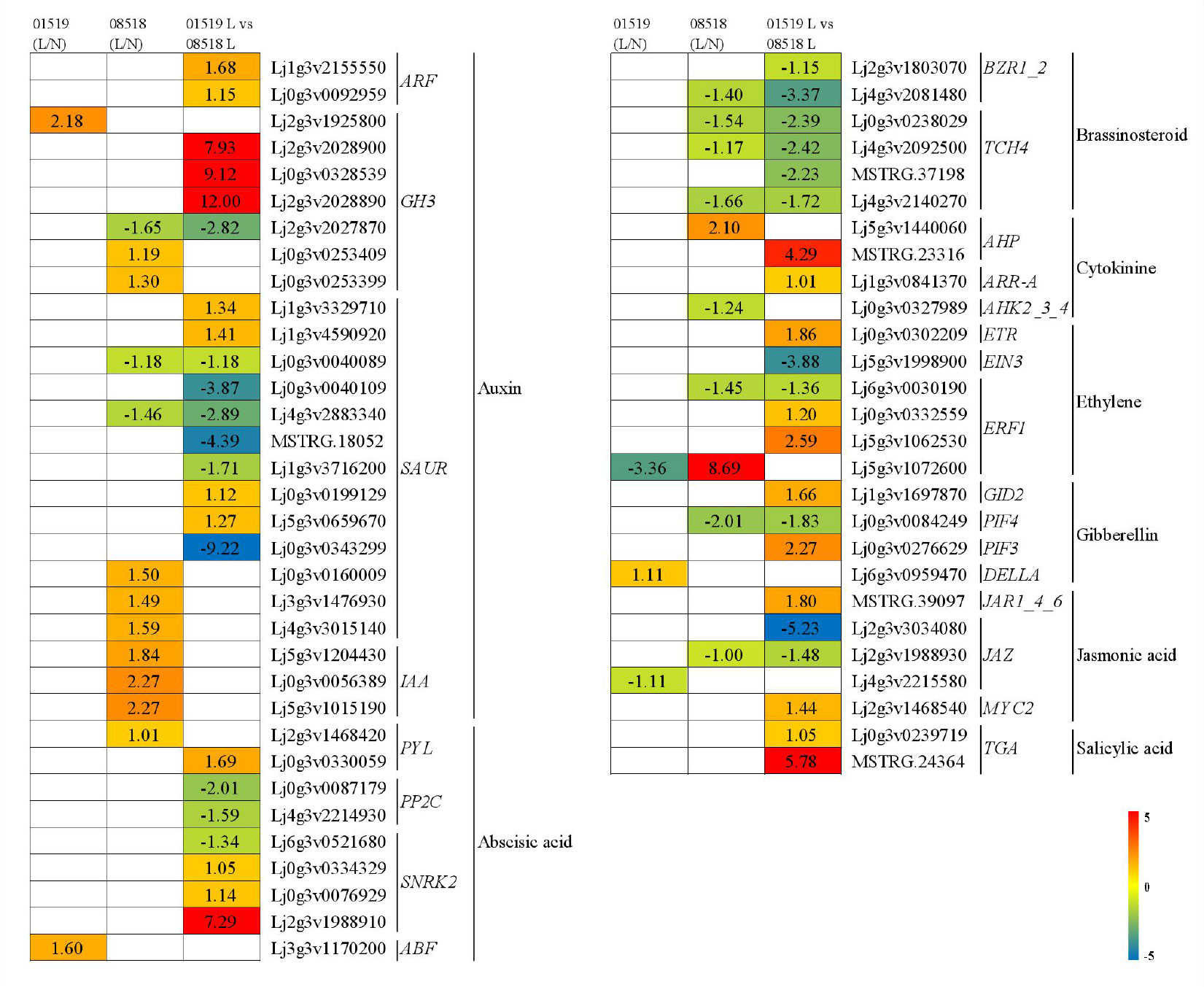
Figure 5 Plant hormone signal transduction-related genes identified in the RNA-seq analysis that showed changes in expression during low-P stress. Log2(fold-changes) of the comparisons are shown by the numbers in the gray rectangles. The comparisons show that each of the genes in the picture is significantly differentially expressed.
In the phenylpropanoid, flavonoid and anthocyanin biosynthesis pathways, 29 genes, including PAL, CYP73A, 4CL, CHS, E5.5.1.6, DFR, E2.3.1.133, CYP98A, E2.1.1.104 and BZ1, were differentially accumulated in the L. corniculatus roots under low-P stress (Figure 6 and Table S10). Overall, the majority of the structural genes implicated in the pathways were significantly downregulated under low-P stress, repressing secondary metabolism in L. corniculatus roots. Among them, 3 PAL, 3 CYP73A, 4 4CL, 1 CHS, 1 DFR, 4 E2.3.1.133, 1 CYP98A and 1 E2.1.1.104 genes were significantly downregulated, while 2 E5.5.1.6, 1 E2.3.1.133 and 2 E2.1.1.104 genes were significantly upregulated in line 01549 compared with 08518 under low-P stress. 2 4CL, 3 CHS and 2 E2.3.1.133 were significantly downregulated, while 2 PAL, 1 4CL, 1 CHS, 1 E2.3.1.133, 1 E2.1.1.104 and 1 BZ1 genes were significantly upregulated in line 08518 under low-P stress. Two CYP73A and 2 E2.3.1.133 genes were significantly downregulated in line 01549 under low-P stress. These results imply that the defense mechanisms of L. corniculatus against low-P stress involved the induction of phenylpropanoid and secondary metabolism.
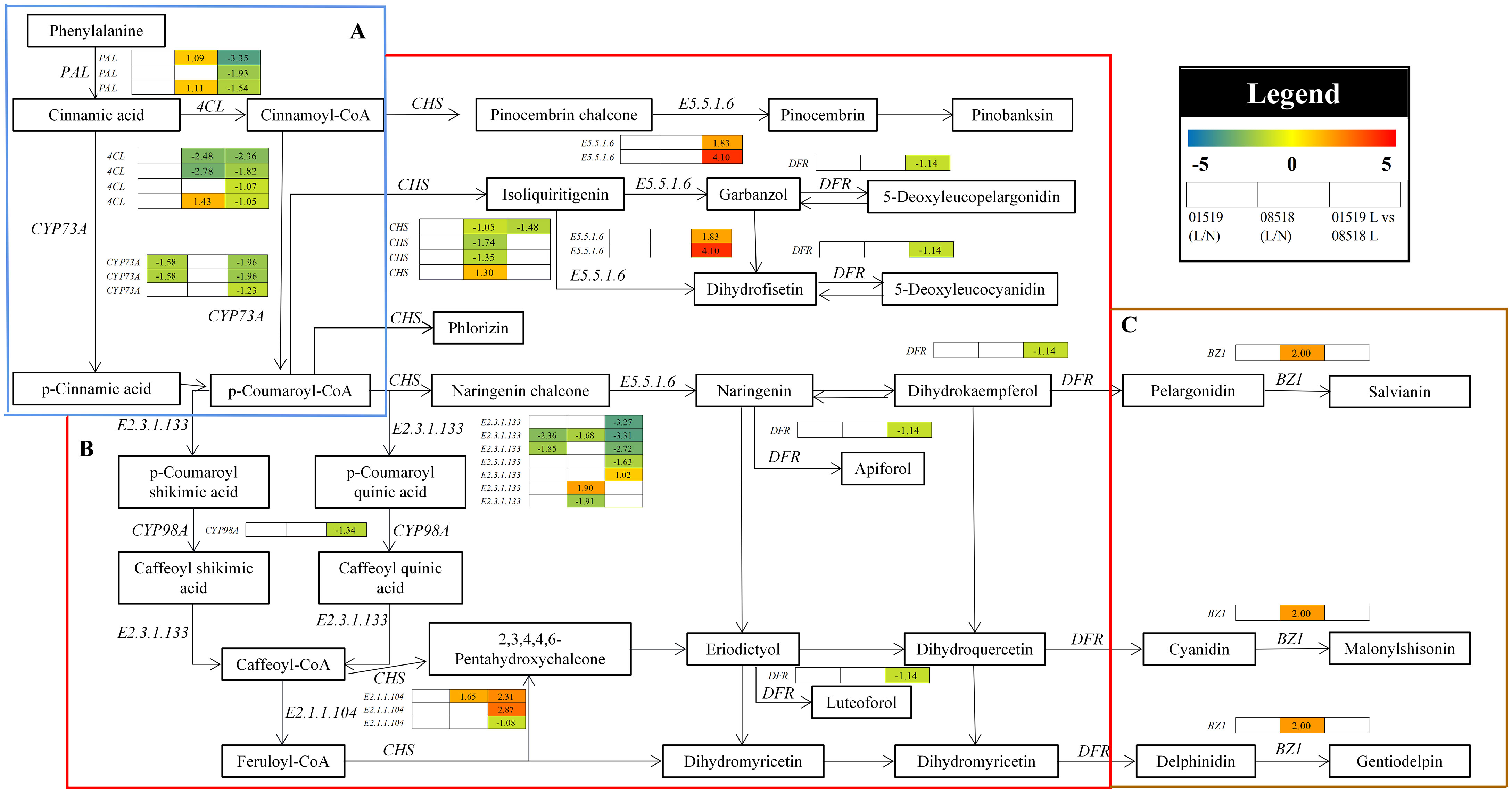
Figure 6 The pathways of phenylpropanoid biosynthesis (A), flavonoid biosynthesis (B) and anthocyanin biosynthesis (C) enriched in the KEGG analysis. Log2(fold-changes) of the comparisons are shown by the numbers in the gray rectangles. The comparisons show that each of the genes in the picture is significantly differentially expressed.
In the glycolysis/gluconeogenesis and citrate cycle pathways, 41 enzyme genes, including HK, GALM, E5.1.3.15, PFK, TPI, GAPDH, ENO, PK, aceE, PDC, ADH1_7, PGAM, DLAT, ACSS1_2, ALDH, AKR1A1, PFP and ACO, were differentially accumulated in L. corniculatus roots under low-P stress (Figure 7 and Table S11). Among them, 2 HK, 1 PFK, 3 GAPDH, 2 ENO, 2 PK, 4 ADH1_7 and 2 AKR1A1 genes were significantly upregulated, while 1 GALM, 1 E5.1.3.15, 1 PFK, 1 TPI, 1 ENO, 2 PK, 2 PDC, 3 ADH1_7 and 1 ACO genes were significantly downregulated in line 01549 compared with line 08518 under low-P stress. 1 GALM, 1 GAPDH, 2 PK, 1 ADH1_7, 1 PGAM, 1 DLAT and 2 ALDH were significantly upregulated, while 1 ENO, 1 PDC, 3 ADH1_7, 1 ACSS1_2 and 1 ALDH genes were significantly downregulated in line 08518 under low-P stress. The 1 PFP gene was significantly downregulated in line 01549 under low-P stress. These results imply that the reaction of L. corniculatus to low-P stress was mediated by activating the citrate cycle pathway and glycolysis/gluconeogenesis as defense mechanisms.
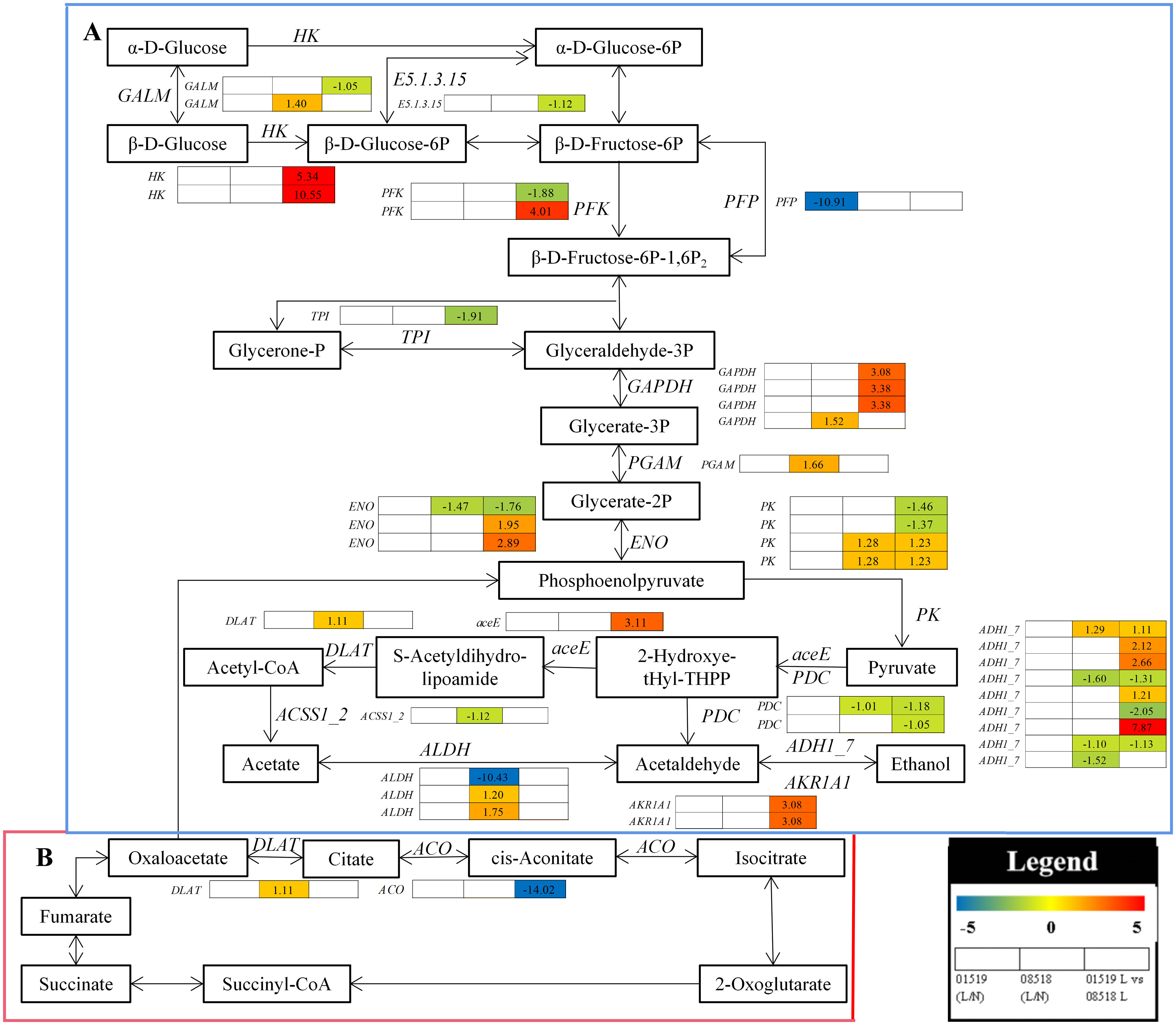
Figure 7 Analysis of the carbohydrate metabolism pathway in plants under low-P stress. (A) Glycolysis/gluconeogenesis. (B) Citrate cycle. Log2(fold-changes) of the comparisons are shown by the numbers in the gray rectangles. The comparisons show that each of the genes in the picture is significantly differentially expressed.
Plant response to low-P stress is a complex process regulated by multiple genes interacting with each other, and therefore, manipulation of a set of genes or individual genes is not sufficient to study plant tolerance to low-P (Sun et al., 2021; Zhao et al., 2022b). In this study, two selected genotypes (01549 and 08518) of L. corniculatus seedlings to better highlight the differences in resistance between the two L. corniculatus genotypes by systematically studying the morphological, physiological, and transcriptional responses of L. corniculatus to low-P stress. In this study, low phosphorus stress caused extensive changes in morphological traits, physiological indicators, and gene expression in the L. corniculatus roots.
In the study, low-P stress increased the MDA content in the roots of both lines 01549 and 08518 compared to the NP treatment, and the MDA content of line 01549 was significantly lower than that of line 08518 during low-P treatment. Based on these results, low-P stress caused a large number of superoxide radicals to be produced in the L. corniculatus roots, which caused an imbalance in plant metabolic oxygen production and subsequent oxidative damage to the plant (Tarumoto et al., 2022). However, the membrane system of line 08518 was damaged to a greater extent by low-P stress. Plants scavenge the accumulated O2- through the antioxidant enzyme system consisting of SOD, CAT and POD to avoid cell membrane damage and keep the free radicals in plants at a low level under adverse conditions (Niu et al., 2013; Xu et al., 2018). In this study, we found that the antioxidant enzyme activities in both lines 01549 and 08518 were increased to enhance the adaptation to the low-P environment. Osmoregulatory substances in plants play an important role in reducing plant damage caused by stress(Anawar et al., 2018). In this study, we found that the proline, soluble sugar and soluble protein contents in lines 01549 and 08518 were increased under low phosphorus stress, indicating that the root cells of L. corniculatus were able to reduce the intracellular osmotic potential by regulating the content of their organic osmoregulatory substances, thus achieving self-protection and enhancing the tolerance to low phosphorus (Sun and Zheng, 2022). However, the antioxidant enzyme activities and osmoregulatory substance contents of line 01549 was higher than that of line 08518, indicating that line 01549 was more adapted to the low-P environment. Meanwhile, these data show very small differences in some cases between the two treatments in P treatment concentrations, this could be the different phosphorus uptake capacity and phosphorus utilization efficiency of different lines of L. corniculatus. However, the exact reasons need further analysis.
By studying the transcriptional responses of L. corniculatus to low-P stress, we have revealed different patterns of L. corniculatus to response low-P stress, including carbohydrate metabolism, acid phosphatases and phosphate transporters, biosynthesis of secondary metabolites, and plant hormone signal transduction.
Anthocyanin biosynthesis is catalyzed by phenylpropanoid and flavonoid biosynthetic pathways through related genes such as PAL, CHS, CHI, F3H, DFR, and ANS (Liu et al., 2016; Farooq et al., 2016). The expression of these genes involved in secondary metabolite biosynthesis were differentially expressed in L. corniculatus lines under low-P stress in the present study, with PAL, 4CL, CHS and DFR expression downregulated in line 01549 compared with line 08518 under low-P stress, while line 08518 showed upregulated PAL expression and decreased total gene expression of the 4CL and CHS genes in the anthocyanin biosynthetic pathway under low-P stress, and the BZ1 gene was upregulated in line 08518 under low-P stress. Although the expression of these genes does not fully reflect the antioxidant characteristics of L. corniculatus and requires further verification in cells and in vivo tissues, it nevertheless provides an indication of the tolerance of L. corniculatus. Regardless of the comparative group, line 08518 was more sensitive to low-P stress, suggesting that a low P concentration may have suppressed gene transcription or translation in L. corniculatus.
Acid phosphatase and high-affinity phosphate transporter family genes play key roles in the activation of phosphorus uptake and efficient phosphorus transport under low-P stress (Kuerban et al., 2020). Phosphatase is an adaptation-inducible enzyme that promotes the mineralization and decomposition of soil organic P (Zhu et al., 2020). In our study, two acid phosphatase 1 (APS1) genes were downregulated in lines 01549 and 08518 under low-P stress, and four purple acid phosphatase (PAP) genes were significantly upregulated by low-P stress. Fita et al. (2012) found that Cm-PAP10.1 and Cm-PAP10.2, genes encoding purple acid phosphatases, were upregulated in melon under low-P stress, indicating that the activity of APS secreted by plant roots under low-P stress was positively correlated with the degree of P deficiency. Meanwhile, genes encoding phosphorus transporter proteins were screened in transcriptome data, among which eight DEGs of high-affinity phosphorus transporter systems (Pht1 and Pho1 families) were screened, all of which were differentially expressed to varying degrees in the two L. corniculatus lines treated with low P. Hu et al. (2021) found that the PHO1 and PHT1 genes were upregulated in the roots of Zygophyllum xanthoxylum, suggesting that these phosphorus transporter protein genes may play a vital role in regulating the distribution, transport and maintenance of dynamic homeostasis of P in the plant body during P deficiency (Hu et al., 2021), but their functions in response to low-P stress remain to be further investigated.
Several respiratory metabolic pathways have been identified in plants, including the glycolytic pathway (EMP), the tricarboxylic acid cycle (TCA), and the mitochondrial electron transport chain (Van Dongen et al., 2011). Plants produce ATP and CO2 through the breakdown of photosynthetic products (e.g., glucose) by respiration to promote root growth and development or maintain root activity for nutrient and water uptake and translocation (Chen et al., 2021). The differential expression of 41 key glycolysis and TCA cycle-related genes was discovered in this study. Among these genes, the glycolytic pathway-related genes HK, PFK, GAPDH, ENO, PK, ADH1 7, and AKR1A1 were elevated, whereas the TCA cycle-related aconitate hydratase gene ACO3 was downregulated in line 01549 compared with line 08518 under low-P conditions. The expression of some genes related to the glycolytic pathway, namely, GALM, PK, ADH1_7, and ALDH, was downregulated, and the expression of DLAT, a pyruvate dehydrogenase E2 component of the TCA cycle, was upregulated in line 01549 compared with line 08518 under low-P stress, suggesting that low-P stress suppresses both pathways while increasing electron transport, and the L. corniculatus root maintains an ATP supply by constantly changing the transcript levels of genes encoding critical enzymes in the glycolytic/gluconeogenic pathway. (Du et al., 2020).
Low-P stress affects the synthesis and distribution of hormones in plants, which further regulates root conformation, such as root morphology and structure (Zhang Z. et al., 2014; Kumar et al., 2021; Martínez-Andújar et al., 2017). In this study, the expression genes involved in plant hormone signal transduction was significantly changed under low-P conditions, which may indicate that hormones play a crucial role in root development under low-P conditions (Zhang X. et al., 2019; Bhosale et al., 2018). In our study, the expression of the ARF, GH3, PYL, SNRK2, ABF, AHP, ARR-A, ERF1, GID, PIF3, JAR1_4_6, MYC2 and TGA genes involved in in auxin, abscisic acid, cytokinin, ethylene, gibberellin, jasmonic acid and salicylic acid signaling, respectively, was upregulated in line 01549 compared with line 08518 under low-P stress, suggesting that these genes associated with phytohormone signaling are involved in regulating the root morphology of L. corniculatus in response to low-P stress. Gene expression was increased in line 01549, and the values of root morphological indicators were significantly higher in line 01549 than in line 08518, indicating that line 01549 regulates root development more quickly in response to stress. Brassinosteroids (BRs) are plant hormones that promote cell elongation and division and play an important role in plant growth and development (Planas-Riverola et al., 2019). In our study, the expression of all genes related to brassinosteroid signaling was downregulated, potentially due to the negative regulation of brassinosteroids in response to low-P stress in the root, but further analyses are needed to determine the exact cause. Similarly, transcription factors related to hormone metabolism and signal transduction play a crucial role. Studies have shown that the growth hormone-responsive transcription factor SAUR also regulates root morphology and promotes lateral root development (Xie et al., 2000). Arabidopsis RLK (receptor-like protein kinase) regulates root hair development (Wei and Li, 2018). In this study, SAUR genes were both upregulated and downregulated in line 01549 compared with line 08518 under low-P stress, indicating that both lines suffered from low-P stress, regardless of the accession. Two SAUR genes were downregulated in line 08518, and no difference in SAUR expression was identified in line 01549 under low-P stress, indicating that line 08518 has a reduced root morphology and requires the regulation of SAUR genes to respond to low-P stress.
By determining morphological and physiological features, we verified that the phosphorus-resistant genotype 01549 performed better under low-P stress, which was also highly supported by transcriptome data. However, previous studies have shown that these genes do not always coincide with changes in physiological indicators, and it is possible that other unknown genes are involved in the response of L. corniculatus to low-P stress.
Key KEGG pathways and their candidate genes were evaluated to explore changes in the morphological and physiological characteristics of the two L. corniculatus lines under low-P stress. We found that the root morphology of phosphorus-tolerant line 01549 increased and that of phosphorus-sensitive line 08518 decreased under low-P stress. L. corniculatus adapted to the low-P environment by increasing enzyme activity and the contents of osmoregulatory substances to rapidly regulate its physiological and metabolic functions as a mechanism to mitigate the injury. In addition, a multilevel analysis of changes in gene expression in L. corniculatus was conducted. Low-P stress activated important pathways, such as plant hormone signal transduction, and we identified key candidate genes, such as PAP, PHT1 and PHO1. The combination of morphological, physiological, and transcriptome data reported in this work provides a theoretical foundation for future studies of the complex processes driving low-P responses in L. corniculatus and other plant species.
The data presented in the study are deposited in the BCBI repository, accession number PRJNA870253.
XZ and K-KC: experimental design, experimental performance, experimental data collection, data analysis, manuscript writing and revision. L-LZ: seed provision, manuscript writing, resource provision and funding acquisition. L-TW: experimental performance, experimental data collection and data analysis. P-CW: manuscript writing and revision. All authors contributed to the article and approved the submitted version.
This work was financially supported by Guizhou Provincial Department of Education Natural Science Research Project (QJJ[2022]076), Guizhou Provincial Science and Technology Projects (QKHPTRC[2021]5636), and the National Natural Science Foundation of China (32060391, 31560664).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1089380/full#supplementary-material
Anawar, H. M., Rengel, Z., Damon, P., Tibbett, M. (2018). Arsenic-phosphorus interactions in the soil-plant-microbe system: Dynamics of uptake, suppression and toxicity to plants. Environ. pollut. (Barking. Essex. 1987). 233, 1003–1012. doi: 10.1016/j.envpol.2017.09.098
Antonelli, C. J., Calzadilla, P. I., Campestre, M. P., Escaray, F. J., Ruiz, O. A. (2021). Contrasting response of two lotus corniculatus l. lines to combined waterlogging-saline stress. Plant Biol. (Stuttgart. Germany). 23 (2), 363–374. doi: 10.1111/plb.13216
Baghbani-Arani, A., Modarres-Sanavy, S. A. M., Poureisa, M. (2021). Improvement the soil physicochemical properties and fenugreek growth using zeolite and vermicompost under water deficit conditions. J. Soil Sci. Plant Nutr. 21, 1213–1228. doi: 10.1007/s42729-021-00434-y
Bhosale, R., Giri, J., Pandey, B. K., Giehl, R., Hartmann, A., Traini, R., et al. (2018). A mechanistic framework for auxin dependent arabidopsis root hair elongation to low external phosphate. Nat. Commun. 9 (1), 1409. doi: 10.1038/s41467-018-03851-3
Chapman, H. D., Pratt, P. F. (1962). Methods of analysis for soils, plants and waters. Soil Sci. 93, 68. doi: 10.1097/00010694-196201000-00015
Chen, W., Zhou, M., Zhao, M., Chen, R., Tigabu, M., Wu, P., et al. (2021). Transcriptome analysis provides insights into the root response of Chinese fir to phosphorus deficiency. BMC Plant Biol. 21 (1), 525. doi: 10.1186/s12870-021-03245-6
Chiou, T. J., Aung, K., Lin, S. I., Wu, C. C., Chiang, S. F., Su, C. L. (2006). Regulation of phosphate homeostasis by MicroRNA in arabidopsis. Plant Cell 18 (2), 412–421. doi: 10.1105/tpc.105.038943
Ciereszko, I., Szczygla, A., Zebrowska, E. (2011). Phosphate deficiency affects acid phosphatase activity and growth of two wheat varieties. J. Plant Nutr. 34 (6), 815–829. doi: 10.1080/01904167.2011.544351
Ding, W., Cong, W. F., Lambers, H. (2021). Plant phosphorus-acquisition and -use strategies affect soil carbon cycling. Trends Ecol. Evol. 36 (10), 899–906. doi: 10.1016/j.tree.2021.06.005
Du, Y., Zhao, Q., Chen, L., Yao, X., Zhang, W., Zhang, B., et al. (2020). Effect of drought stress on sugar metabolism in leaves and roots of soybean seedlings. Plant Physiol. Biochem.: PPB 146, 1–12. doi: 10.1016/j.plaphy.2019.11.003
Farooq, M. A., Gill, R. A., Islam, F., Ali, B., Liu, H., Xu, J., et al. (2016). Methyl jasmonate regulates antioxidant defense and suppresses arsenic uptake in brassica napus l. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.00468
Fita, A., Bowen, H. C., Hayden, R. M., Nuez, F., Picó, B., Hammond, J. P. (2012). Diversity in expression of phosphorus (P) responsive genes in cucumis melo l. PloS One 7 (4), e35387. doi: 10.1371/journal.pone.0035387
Hernández, G., Ramírez, M., Valdés-López, O., Tesfaye, M., Graham, M. A., Czechowski, T., et al. (2007). Phosphorus stress in common bean: root transcript and metabolic responses. Plant Physiol. 144 (2), 752–767. doi: 10.1104/pp.107.096958
Hu, X., Zhang, L., Niu, D., Nan, S., Wu, S., Gao, H., et al. (2021). Transcriptome analysis of zygophyllum xanthoxylum adaptation strategies to phosphate stress. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.723595
Jeong, K., Baten, A., Waters, D. L., Pantoja, O., Julia, C. C., Wissuwa, M., et al. (2017). Phosphorus remobilization from rice flag leaves during grain filling: an RNA-seq study. Plant Biotechnol. J. 15 (1), 15–26. doi: 10.1111/pbi.12586
Jiang, W., Ma, L., Yi, D., Pang, Y. (2021). ). the complete chloroplast genome sequence of lotus corniculatus l. mitochondrial DNA. Part B. Resour. 6 (1), 189–190. doi: 10.1080/23802359.2020.1860708
Knežević, M., Berić, T., Buntić, A., Delić, D., Nikolić, I., Stanković, S., et al. (2021). Potential of root nodule nonrhizobial endophytic bacteria for growth promotion of lotus corniculatus l. and dactylis glomerata l. J. Appl. Microbiol. 131 (6), 2929–2940. doi: 10.1111/jam.15152
Kong, S. L., Abdullah, S., Ho, C. L., Musa, M., Yeap, W. C. (2021). Comparative transcriptome analysis reveals novel insights into transcriptional responses to phosphorus starvation in oil palm (Elaeis guineensis) root. BMC Genomic. Data 22 (1), 6. doi: 10.1186/s12863-021-00962-7
Kuerban, M., Jiao, W., Pang, J., Jing, J., Qiu, L. J., Ding, W., et al. (2020). Targeting low-phytate soybean genotypes without compromising desirable phosphorus-acquisition traits. Front. Genet. 11. doi: 10.3389/fgene.2020.574547
Kumar, S., Chugh, C., Seem, K., Kumar, S., Vinod, K. K., Mohapatra, T. (2021). Characterization of contrasting rice (Oryza sativa l.) genotypes reveals the pi-efficient schema for phosphate starvation tolerance. BMC Plant Biol. 21 (1), 282. doi: 10.1186/s12870-021-03015-4
Li, L. Y., Yang, H. M., Ren, W. B., Liu, B., Cheng, D. M., Wu, X. H., et al. (2016). Physiological and biochemical characterization of sheepgrass (Leymus chinensis) reveals insights into photosynthetic apparatus coping with low-phosphate stress conditions[J]. J. Plant Biol. 59, 336–346. doi: 10.1007/s12374-016-0117-1
Liu, W., Yin, D., Li, N., Hou, X., Wang, D., Li, D., et al. (2016). Influence of environmental factors on the active substance production and antioxidant activity in potentilla fruticosa l. and its quality assessment. Sci. Rep. 6, 28591. doi: 10.1038/srep28591
Liu, X., Zhao, X., Zhang, L., Lu, W., Li, X., Xiao, K. (2013). TaPht1;4, a high-affinity phosphate transporter gene in wheat (Triticum aestivum), plays an important role in plant phosphate acquisition under phosphorus deprivation. Funct. Plant Biol. FPB. 40 (4), 329–341. doi: 10.1071/FP12242
Martínez-Andújar, C., Ruiz-Lozano, J. M., Dodd, I. C., Albacete, A., Pérez-Alfocea, F. (2017). Hormonal and nutritional features in contrasting rootstock-mediated tomato growth under low-phosphorus nutrition. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.00533
Müller, R., Morant, M., Jarmer, H., Nilsson, L., Nielsen, T. H. (2007). Genome-wide analysis of the arabidopsis leaf transcriptome reveals interaction of phosphate and sugar metabolism. Plant Physiol. 143 (1), 156–171. doi: 10.1104/pp.106.090167
Nilsson, L., Müller, R., Nielsen, T. H. (2010). Dissecting the plant transcriptome and the regulatory responses to phosphate deprivation. Physiol. Plant. 139 (2), 129–143. doi: 10.1111/j.1399-3054.2010.01356.x
Niu, Y. F., Chai, R. S., Jin, G. L., Wang, H., Tang, C. X., Zhang, Y. S. (2013). Responses of root architecture development to low phosphorus availability: a review. Ann. Bot. 112 (2), 391–408. doi: 10.1093/aob/mcs285
O'Rourke, J. A., Yang, S. S., Miller, S. S., Bucciarelli, B., Liu, J., Rydeen, A., et al. (2013). An RNA-seq transcriptome analysis of orthophosphate-deficient white lupin reveals novel insights into phosphorus acclimation in plants. Plant Physiol. 161 (2), 705–724. doi: 10.1104/pp.112.209254
Planas-Riverola, A., Gupta, A., Betegón-Putze, I., Bosch, N., Ibañes, M., Caño-Delgado, A. I. (2019). “Brassinosteroid signaling in plant development and adaptation to stress,” in Development, vol. 146. (England: Cambridge), dev151894. doi: 10.1242/dev.151894
Plaxton, W. C., Tran, H. T. (2011). Metabolic adaptations of phosphate-starved plants. Plant Physiol. 156 (3), 1006–1015. doi: 10.1104/pp.111.175281
Raya-González, J., Ojeda-Rivera, J. O., Mora-Macias, J., Oropeza-Aburto, A., Ruiz-Herrera, L. F., López-Bucio, J., et al. (2021). MEDIATOR16 orchestrates local and systemic responses to phosphate scarcity in arabidopsis roots. New Phytol. 229 (3), 1278–1288. doi: 10.1111/nph.16989
Ren, P., Meng, Y., Li, B., Ma, X., Si, E., Lai, Y., et al. (2018). Molecular mechanisms of acclimatization to phosphorus starvation and recovery underlying full-length transcriptome profiling in barley (Hordeum vulgare l.). Front. Plant Sci. 9. doi: 10.3389/fpls.2018.00500
Ried, M. K., Wild, R., Zhu, J., Pipercevic, J., Sturm, K., Broger, L., et al. (2021). Inositol pyrophosphates promote the interaction of SPX domains with the coiled-coil motif of PHR transcription factors to regulate plant phosphate homeostasis. Nat. Commun. 12 (1), 384. doi: 10.1038/s41467-020-20681-4
Rohman, M. M., Islam, M. R., Monsur, M. B., Amiruzzaman, M., Fujita, M., Hasanuzzaman, M. (2019). Trehalose protects maize plants from salt stress and phosphorus deficiency. Plants (Basel. Switzerland). 8 (12), 568. doi: 10.3390/plants8120568
Sun, T., Zhang, J., Zhang, Q., Li, X., Li, M., Yang, Y., et al. (2021). Transcriptome and metabolome analyses revealed the response mechanism of apple to different phosphorus stresses. Plant Physiol. Biochem. PPB 167, 639–650. doi: 10.1016/j.plaphy.2021.08.040
Sun, Y., Zheng, H. (2022). Divergent molecular and physiological response of two maize breeding lines under phosphate deficiency. Plant Mol. Biol. Rep. 40, 197–207. doi: 10.1007/s11105-021-01310-w
Tarumoto, M. B., de Campos, M., Momesso, L., do Nascimento, C., Garcia, A., Coscolin, R., et al. (2022). Carbohydrate partitioning and antioxidant substances synthesis clarify the differences between sugarcane varieties on facing low phosphorus availability. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.888432
Tesfaye, M., Liu, J., Allan, D. L., Vance, C. P. (2007). Genomic and genetic control of phosphate stress in legumes. Plant Physiol. 144 (2), 594–603. doi: 10.1104/pp.107.097386
Van Dongen, J. T., Gupta, K. J., Ramírez-Aguilar, S. J., Araújo, W. L., Nunes-Nesi, A., Fernie, A. R. (2011). Regulation of respiration in plants: a role for alternative metabolic pathways. J. Plant Physiol. 168 (12), 1434–1443. doi: 10.1016/j.jplph.2010.11.004
Verma, S. K., Sahu, P. K., Kumar, K., Pal, G., Gond, S. K., Kharwar, R. N., et al. (2021). Endophyte roles in nutrient acquisition, root system architecture development and oxidative stress tolerance. J. Appl. Microbiol. 131 (5), 2161–2177. doi: 10.1111/jam.15111
Wang, J., Qin, Q., Pan, J., Sun, L., Sun, Y., Xue, Y., et al. (2019). Transcriptome analysis in roots and leaves of wheat seedlings in response to low-phosphorus stress. Sci. Rep. 9 (1), 19802. doi: 10.1038/s41598-019-56451-6
Wei, Z., Li, J. (2018). Receptor-like protein kinases: Key regulators controlling root hair development in arabidopsis thaliana. J. Integr. Plant Biol. 60 (9), 841–850. doi: 10.1111/jipb.12663
Xie, Q., Frugis, G., Colgan, D., Chua, N. H. (2000). Arabidopsis NAC1 transduces auxin signal downstream of TIR1 to promote lateral root development. Genes Dev. 14 (23), 3024–3036. doi: 10.1101/gad.852200
Xie, W., Zhao, W., Wang, L., Zhao, L., Wamg, P. (2023). Genetic diversity analysis of 22 lotus corniculatus germplasm resources based on phenotypic quantitative traits. Acta agrestia sinica 31(1), 173–179. doi: 10.11733/j.issn.1007-0435.2023.01.020
Xu, C., Zhang, H., Sun, J., Guo, Z., Zou, C., Li, W. X., et al. (2018). Genome-wide association study dissects yield components associated with low-phosphorus stress tolerance in maize. TAG. theoretical and applied genetics. Theoretische. Und. Angewandte. Genetik. 131 (8), 1699–1714. doi: 10.1007/s00122-018-3108-4
Yuan, H., Liu, D. (2008). Signaling components involved in plant responses to phosphate starvation. J. Integr. Plant Biol. 50 (7), 849–859. doi: 10.1111/j.1744-7909.2008.00709.x
Zhang, J., Jiang, F., Shen, Y., Zhan, Q., Bai, B., Chen, W., et al. (2019). Transcriptome analysis reveals candidate genes related to phosphorus starvation tolerance in sorghum. BMC Plant Biol. 19 (1), 306. doi: 10.1186/s12870-019-1914-8
Zhang, Z., Liao, H., Lucas, W. J. (2014). Molecular mechanisms underlying phosphate sensing, signaling, and adaptation in plants. J. Integr. Plant Biol. 56 (3), 192–220. doi: 10.1111/jipb.12163
Zhang, H., Uddin, M. S., Zou, C., Xie, C., Xu, Y., Li, W. X. (2014). Meta-analysis and candidate gene mining of low-phosphorus tolerance in maize. J. Integr. Plant Biol. 56 (3), 262–270. doi: 10.1111/jipb.12168
Zhang, X., Wang, B., Zhao, Y., Zhang, J., Li, Z. (2019). Auxin and GA signaling play important roles in the maize response to phosphate deficiency. Plant Sci. 283, 177–188. doi: 10.1016/j.plantsci.2019.02.011
Zhao, X., Huang, L. J., Sun, X. F., Zhao, L. L., Wang, P. C. (2022c). Differential physiological, transcriptomic, and metabolomic responses of paspalum wettsteinii under high-temperature stress. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.865608
Zhao, X., Huang, L. J., Zhao, L. L., Wang, P. C., Sun, X. F. (2022b). Transcriptome analysis of sophora davidii leaves in response to low-phosphorus stress. J. Plant Growth Regul. 41, 1241–1253. doi: 10.1007/s00344-021-10382-7
Zhao, H., Wu, Y., Shen, L., Hou, Q., Wu, R., Li, Z., et al. (2022a). Cross-talk between transcriptome analysis and physiological characterization identifies the genes in response to the low phosphorus stress in malus mandshurica. Int. J. Mol. Sci. 23 (9), 4896. doi: 10.3390/ijms23094896
Zhu, T. Q. (2018). Study on establishment of alfalfa phosphorus efficiency evaluation system and difference metabolism in nitrogen and phosphorus (Gansu: Gansu Agricultural University).
Zhu, S., Chen, M., Liang, C., Xue, Y., Lin, S., Tian, J. (2020). Characterization of purple acid phosphatase family and functional analysis of GmPAP7a/7b involved in extracellular ATP utilization in soybean. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.00661
Keywords: Lotus corniculatus, low-phosphorus stress, physiological, morphological, transcriptomic
Citation: Zhao X, Chen K-k, Wang L-t, Zhao L-L and Wang P-C (2023) Transcriptome analysis provides insights into the response of Lotus corniculatus roots to low-phosphorus stress. Front. Plant Sci. 14:1089380. doi: 10.3389/fpls.2023.1089380
Received: 04 November 2022; Accepted: 17 February 2023;
Published: 01 March 2023.
Edited by:
Mehdi Rahimi, Graduate University of Advanced Technology, IranReviewed by:
John Hammond, University of Reading, United KingdomCopyright © 2023 Zhao, Chen, Wang, Zhao and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Li-Li Zhao, emhhb2xpbGlfMDUwOEAxNjMuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.