- 1Plant Breeding, Wageningen University & Research, Wageningen, Netherlands
- 2Graduate School Experimental Plant Sciences Wageningen University & Research, Wageningen, Netherlands
- 3Cluster of Plant Developmental Biology, Laboratory of Molecular Biology, Wageningen University & Research, Wageningen, Netherlands
- 4Biointeractions & Plant Health, Wageningen University & Research, Wageningen, Netherlands
Tomato bacterial canker caused by Clavibacter michiganensis (Cm) is considered to be one of the most destructive bacterial diseases of tomato. To date, no resistance to the pathogen has been identified. While several molecular studies have identified (Cm) bacterial factors involved in disease development, the plant genes and mechanisms associated with susceptibility of tomato to the bacterium remain largely unknown. Here, we show for the first time that tomato gene SlWAT1 is a susceptibility gene to Cm. We inactivated the gene SlWAT1 through RNAi and CRISPR/Cas9 to study changes in tomato susceptibility to Cm. Furthermore, we analysed the role of the gene in the molecular interaction with the pathogen. Our findings demonstrate that SlWAT1 functions as an S gene to genetically diverse Cm strains. Inactivation of SlWAT1 reduced free auxin contents and ethylene synthesis in tomato stems and suppressed the expression of specific bacterial virulence factors. However, CRISPR/Cas9 slwat1 mutants exhibited severe growth defects. The observed reduced susceptibility is possibly a result of downregulation of bacterial virulence factors and reduced auxin contents in transgenic plants. This shows that inactivation of an S gene may affect the expression of bacterial virulence factors.
Introduction
Plant disease resistance is genetically controlled, mostly by dominantly inherited, race specific resistance (R) genes. In the presence of corresponding pathogen-derived effectors many R genes confer resistance through effector-triggered immunity (ETI) (Jones and Dangl, 2006). In plant-microbe interactions, resistance is a common outcome. In fact, a high degree of adaptation is required for microbes to become pathogenic (Hückelhoven et al., 2013). During their co-evolution pathogens have found ways to target and manipulate plant genes, referred to as susceptibility (S) genes, to promote disease development (Bai et al., 2008; Pavan et al., 2011; Appiano et al., 2015; Pessina et al., 2016; Acevedo-Garcia et al., 2017; Berg et al., 2017). S genes are important for biological functions of plants, which appears to be a significant factor in their retainment across species (Hückelhoven et al., 2013). This is exemplified by the Mildew Locus O (MLO) gene family that has been identified in plant species such as barley, tomato, Arabidopsis, grape, apple and cucumber (Cohn et al., 2014; Cox et al., 2017; Oliva et al., 2019). In contrast to dominant R genes, loss-of-function of S genes can potentially lead to recessively inherited, broad-spectrum, and durable resistance (van Damme et al., 2008; Pavan et al., 2010; Pavan et al., 2011; Huibers et al., 2013; Sun et al., 2016a; Berg et al., 2017; Santillán Martínez et al., 2020). For example, loss-of-function of genes in the glutamate decarboxylases (GADs) family provide enhanced resistance against the vascular bacterium Ralstonia solanacearum in Arabidopsis and tomato (Solanum lycopersicum) (Xian et al., 2020). In addition, mutation of Sugars Will Eventually Be Exported Transporter (SWEET) genes in multiple plant species has been demonstrated to be an effective strategy to obtain resistance to Xanthomonas spp (Cohn et al., 2014; Cox et al., 2017; Oliva et al., 2019).
Bacterial canker of tomato caused by the Gram-positive bacterium Clavibacter michiganensis (Cm), is considered to be one of the most important seed-borne diseases of tomato worldwide (Davis et al., 1984; Gartemann et al., 2008; Nouioui et al., 2018). The pathogen colonizes the vasculature of plants leading to systemic infections that result in wilting of leaves, vascular tissue necrosis and formation of cankers on the stems and petioles of plants, eventually leading to plant death (Eichenlaub and Gartemann, 2011; Sen et al., 2015; Chalupowicz et al., 2017).
On the molecular level of the tomato-Cm interaction, several bacterial factors involved in virulence are known. Full virulence of the Cm reference strain NCPBB382 requires the presence of two native plasmids, pCM1 and pCM2, where the major virulence factors celA and pat-1 are located (Gartemann et al., 2008; Savidor et al., 2014). Loss of either of the plasmids leads to reduced virulence and loss of both results in an endophytic nonvirulent strain (Meletzus et al., 1993; Savidor et al., 2012). Several other proteins are encoded by genes located on the circular chromosome of Cm that are involved in the colonization of plants and induction of disease symptoms (Gartemann et al., 2008; Savidor et al., 2012; Savidor et al., 2014; Thapa et al., 2017). Such genes, include the transcriptional factors vatr1 and vatr2, which act in the regulation of several other virulence genes. vatr1 and vatr2 are involved in the regulation of virulence factors, such as the endo- beta- 1,4- glucanase celA, subtilase proteinase SbtC and the serine proteases pat-1 and PhpA, both on the chromosome and plasmids of Cm (Savidor et al., 2014).
While several molecular studies have identified bacterial factors involved in disease development, the mechanisms associated with susceptibility of tomato to the bacterium remain largely unknown. The only experimentally confirmed plant factor involved in disease development is the phytohormone ethylene (Balaji et al., 2008; Savidor et al., 2012; Savidor et al., 2014). During infection Cm promotes the production of host-derived ethylene by specifically upregulating the ethylene biosynthetic gene ACO1. Mutant Never ripe (Nr) tomato plants with impaired ethylene perception display significant Cm symptom development delay (Balaji et al., 2008). The observation that the nonvirulent Cmm100 strain lacks the ability to induce the production of host derived ethylene further highlights the importance of ethylene in Cm symptom development (Savidor et al., 2012).
Despite extensive screenings of wild germplasm, resistance to the pathogen has not been identified yet (Sen et al., 2015). In our study, we hypothesized that impairment of S genes involved in the tomato-Cm interaction might result in loss-of-susceptibility. Therefore, we set out to identify tomato S genes potentially involved in the interaction.
Recently, the tomato ortholog of Walls Are Thin1 (SlWAT1) was identified and inactivated through RNAi and CRISPR/Cas9 (Hanika et al., 2026). CRISPR/Cas9 mediated knock-out of the gene led to resistance to the vascular fungi Verticillium dahliae, V. albo-altrum and Fusarium oxysporum f. sp. lycopersici (Hanika et al., 2026). The Arabidopsis WAT1 gene encodes for a tonoplast localized plant-specific protein. WAT1 has been shown to be involved in vacuolar auxin transport and secondary cell wall biosynthesis (Ranocha et al., 2010; Ranocha et al., 2013). In Arabidopsis loss-of-function of the gene leads to enhanced resistance to a broad range of vascular pathogens, including the bacterium Ralstonia solanacearum (Denance et al., 2013). In cotton (Gosypium hirsutum) three WAT homologs have been identified. Simultaneous transient silencing of the cotton genes enhanced resistance to the vascular fungus V. dahliae (Tang et al., 2019). In both cotton and Arabidopsis resistance involves the repression of indole metabolism and altered contents of indole-3-acetic acid (IAA) and salicylic acid (SA) (Denance et al., 2013; Tang et al., 2019). In addition, local lignin deposition was associated with V. dahliae resistance in cotton (Tang et al., 2019). In this study, we show that impairment of SlWAT1 through RNAi and CRISPR/Cas9 leads to broad-spectrum reduced susceptibility to genetically different Cm strains. Next to this, we show that downregulation of SlWAT1 reduces auxin content in tomato stems and potentially affects the expression of bacterial virulence factors.
Results
Down-regulation of SlWAT1 leads to broad-spectrum reduced susceptibility to Cm
To study the role of SlWAT1 in susceptibility of tomato to Cm, homozygous T3 progeny of two RNAi lines (RNAi::SlWAT1_1 (TV181036) and RNAi::SlWAT1_2 (TV181034)) derived from two independent transformants in cv. Moneymaker (cv. MM) background were used (Hanika et al., 2026). Expression analysis of the T2 parental lines, revealed that the relative residual expression of lines RNAi::SlWAT1_1 and RNAi::SlWAT1_2 was on average 20% and 54%, respectively (Hanika et al., 2026). To evaluate the spectrum of resistance conferred by silencing of SlWAT1, lines RNAi::SlWAT1_1 and RNAi::SlWAT1_2 were challenged with four genetically diverse Cm strains (Jacques et al., 2012). Wilting symptoms on the infected plants were recorded from 7 to 20 days post inoculation (dpi). Severe wilting symptoms caused by all four genetically distinct Cm stains were observed on MM plants. Aggressiveness of the four strains differed, with NCPBB382 being the most aggressive and CFBP5843 being the least aggressive strain (Figure S1). Both T3 lines used in the disease assays exhibited significant reduction of wilting symptoms to all tested strains (Figure 1). Mild wilting symptoms were observed on RNAi::SlWAT1_1 transgenic plants when inoculated with the most aggressive NCPBB382 strain. For RNAi::SlWAT1_2 mild symptoms were observed for strains NCPBB382 and IPO3356.
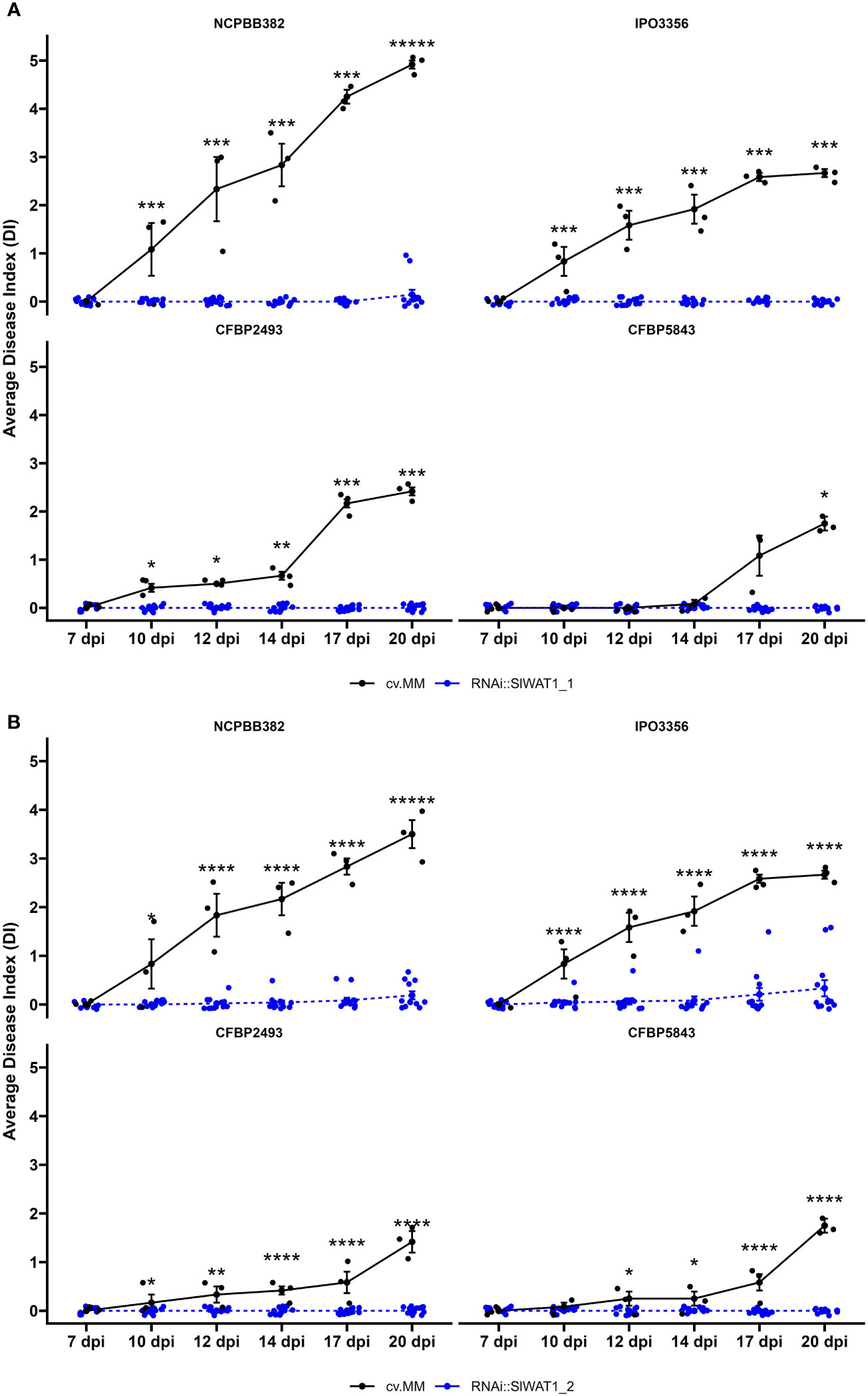
Figure 1 Disease index of SlWAT1 RNAi silenced lines inoculated with genetically diverse Cm strains. Wilting symptom development of (A) RNAi::SlWAT1_1 and (B) RNAi::SlWAT1_2 lines compared to the background donor susceptible control cv. Moneymaker (cv.MM) from 7 days post inoculation (dpi) to 20 dpi. Means of both RNAi::SlWAT1 lines were significantly different from the cv.MM controls, for all strains used in the disease assay (n=12). Bars indicate the standard errors. Asterisks indicate significant differences (Student’s t-test, *p ≤ 0.05; ***p ≤ 0.001,****p ≤ 0.0001).
Bacterial growth is not limited in RNAi::SlWAT1_1 transgenic plants
To determine whether the reduction in observable wilting symptoms in the RNAi lines was correlated with changes in the bacterial growth in planta, the population dynamics of the four different strains were quantified at three time points (4 dpi, 7 dpi and 14 dpi). An estimated 5x105 colony forming units (cfu)/mL was used for the inoculation of the plants. Over the course of infection, all Cm strains reached high population densities (~109 log10(cfu+1/g fresh stem tissue)). No significant statistical differences in population densities were observed between the susceptible cv. MM and transgenic plants for strains NCPBB382, IPO3356, CFBP5843 and CFBPB2493 (Figure 2).
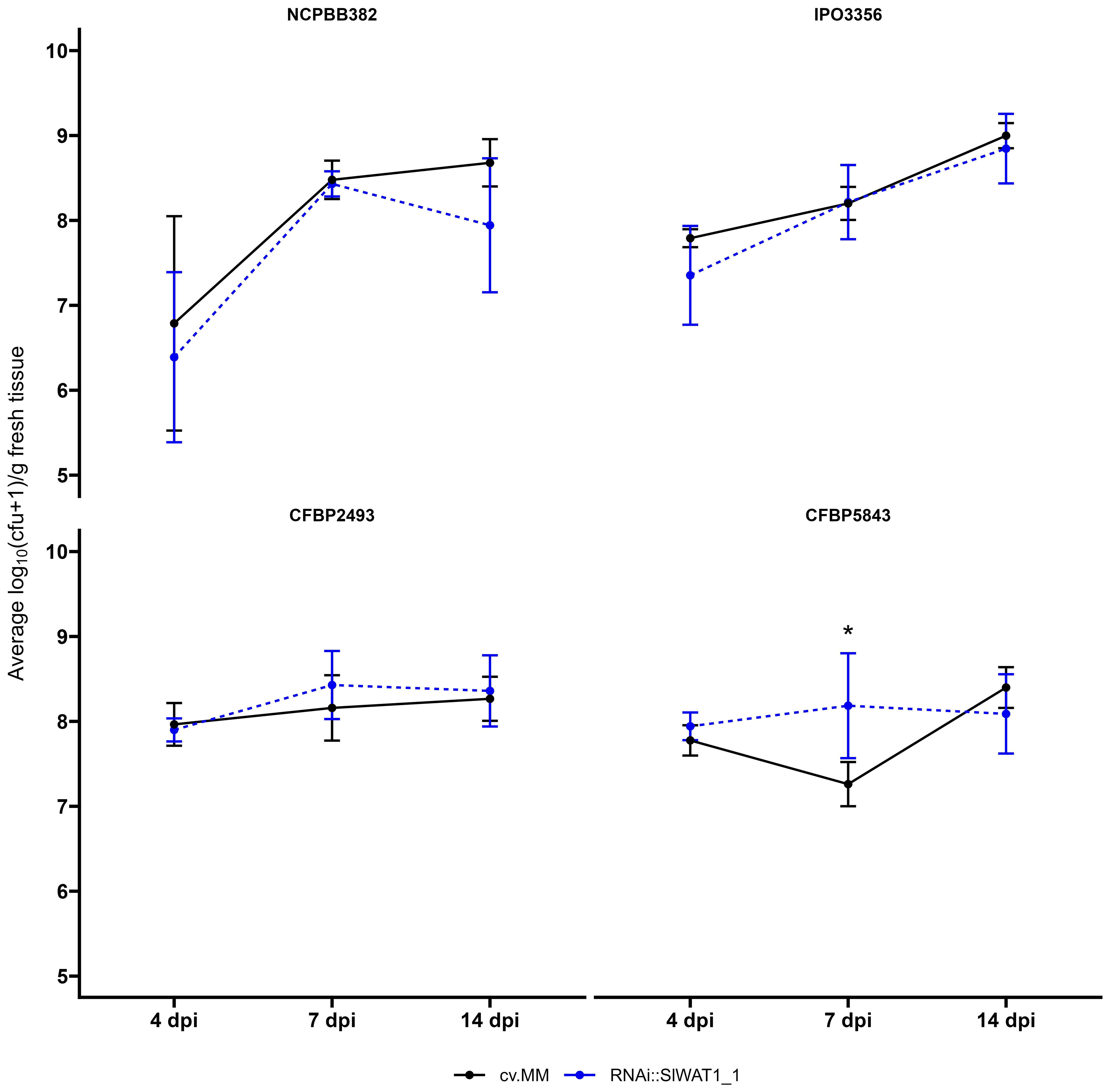
Figure 2 Clavibacter michiganensis population dynamics in cv. Moneymaker (cv.MM) and RNAi::SlWAT1_1 transgenic plants. Bacterial titers of the four bacterial strains used in the experiments were quantified at 4, 7 and 14 days post inoculation (dpi). Five biological replicates (n=5) were used per time point and bacterial strain. Lines represent the average log10(cfu+1/g fresh tissue) ± stdev. The experiments were repeated independently at least twice with similar results. Asterisks indicate statistical differences (Student’s t-test, *p ≤ 0.05).
CRISPR/Cas9- mediated knock-out of SlWAT1 leads to loss-of-susceptibility to Cm without limiting bacterial growth
To exclude the possibility of interference of the residual expression of SlWAT1 in the RNAi lines with the phenotype observed and to confirm our previous results, we decided to include a CRISPR/Cas9 mutant line in the experiments. Silencing of SlWAT1 through RNAi did not lead to any observable adverse pleiotropic effects (Figure 3H). However, for the gene edited mutant line slwat1, severe growth retardation was observed, as previously described (Figure 3) (Hanika et al., 2026). Besides the severe growth retardation, lack of chlorophyll and strong accumulation of anthocyanins at the abaxial side of developing leaves at early developmental stages were also observed. The latter phenotypic abnormalities were alleviated as the plants grew older (Figures 3C, G).
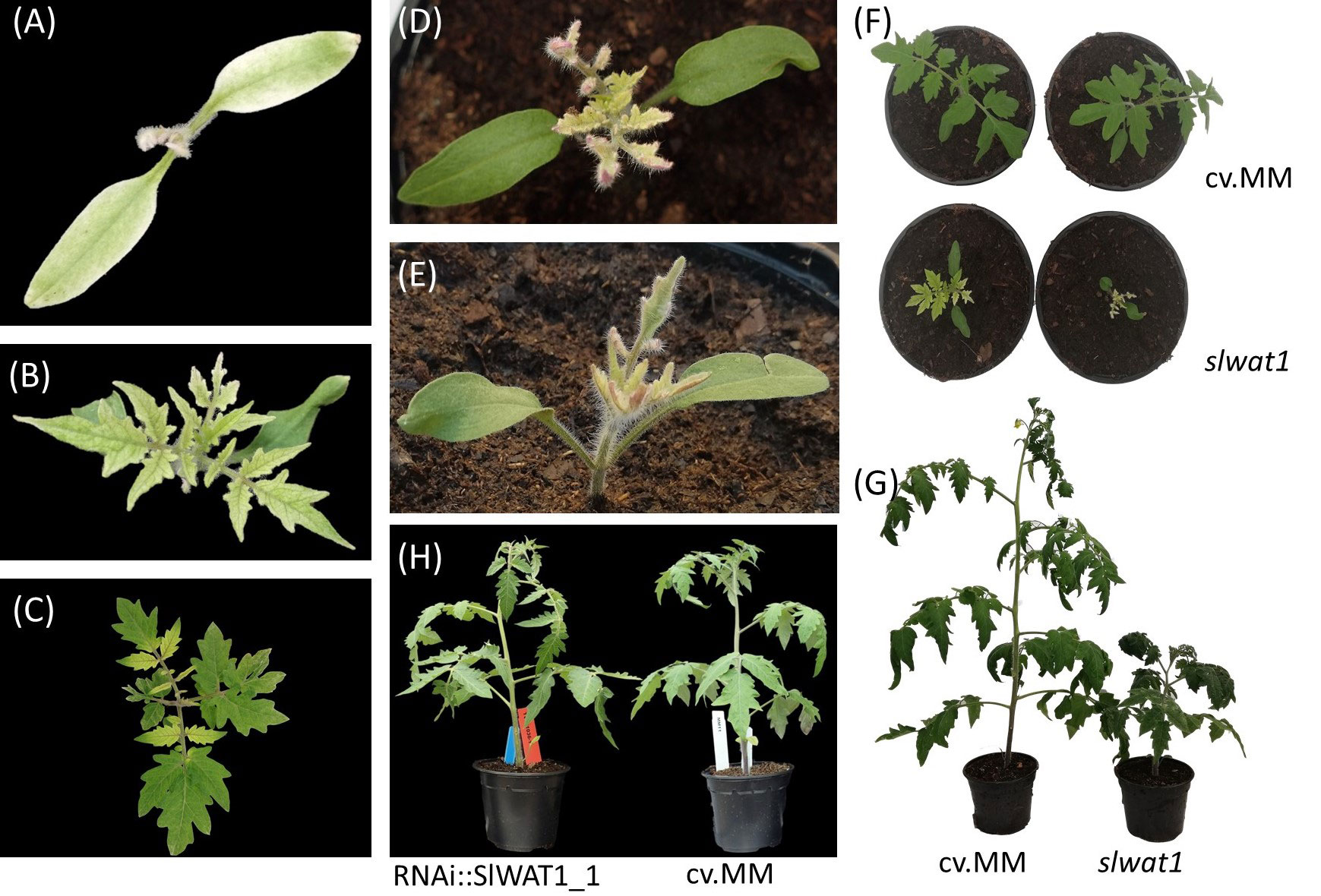
Figure 3 Pleiotropic phenotypes of slwat1 knock-out mutant plants. (A-C) Lack of chlorophyll observed in developing slwat1 mutants. (D, E) Anthocyanin accumulation in the abaxial side of leaves of developing mutants. Severe growth retardation in four weeks old (F) and ten weeks old (G) slwat1 mutants compared to the cv. Moneymaker (cv.MM). (H) phenotype of 6 weeks old RNAi::SlWAT1_1 transgenic plants compared to cv.MM.
Changes of tomato susceptibility in response to Cm due to different developmental stages have previously been reported. Generally, the severity of disease decreases and the incubation period becomes longer with inoculations at later developmental stages (Chang et al., 1992; Sharabani et al., 2013). Therefore, for the inoculation of the plants we decided to use control plants at the same developmental stage as the mutants (4th leaf stage). To achieve synchroneity in the developmental stages of our two genotypes control plants were sown every week. When the control plants and the slwat1 mutants were at the same developmental stage we challenged them with the hypervirulent strain NCPBB382. At 20 dpi, severe wilting symptoms were observed in the susceptible background cv. MM. No symptoms were observed in the slwat1 mutants, confirming our previous results (Figures 4, S2A). When the in planta bacterial titers were quantified, no significant statistical changes were found between the susceptible cv. MM and slwat1 mutants (Figure S2B).

Figure 4 Symptom development of mutants slwat1 in comparison to the susceptible background cv.MM inoculated with strain NCPBB382 at 20 days post inoculation.
Silencing of SlWAT1 reduces auxin content and affects the expression of auxin related genes
Repression of indole metabolism and transcriptional changes of auxin related genes have been reported in Arabidopsis wat1 mutants and cotton WATs silenced plants (Denance et al., 2013; Tang et al., 2019). In our experiments, we monitored the expression of genes involved in auxin transport and auxin responses at different infection time points (Figure 5). To evaluate the distribution of auxin in the stems of transgenic plants the expression of three auxin-related genes was studied at different timepoints. Genes SlPIN1 and LAX4 were selected on the basis of their function as an exporter and an importer of auxin, respectively (Pattison and Catalá, 2012). IAA19 is a useful indicator, as it is primarily expressed in the absence of auxin (Tatematsu et al., 2004).
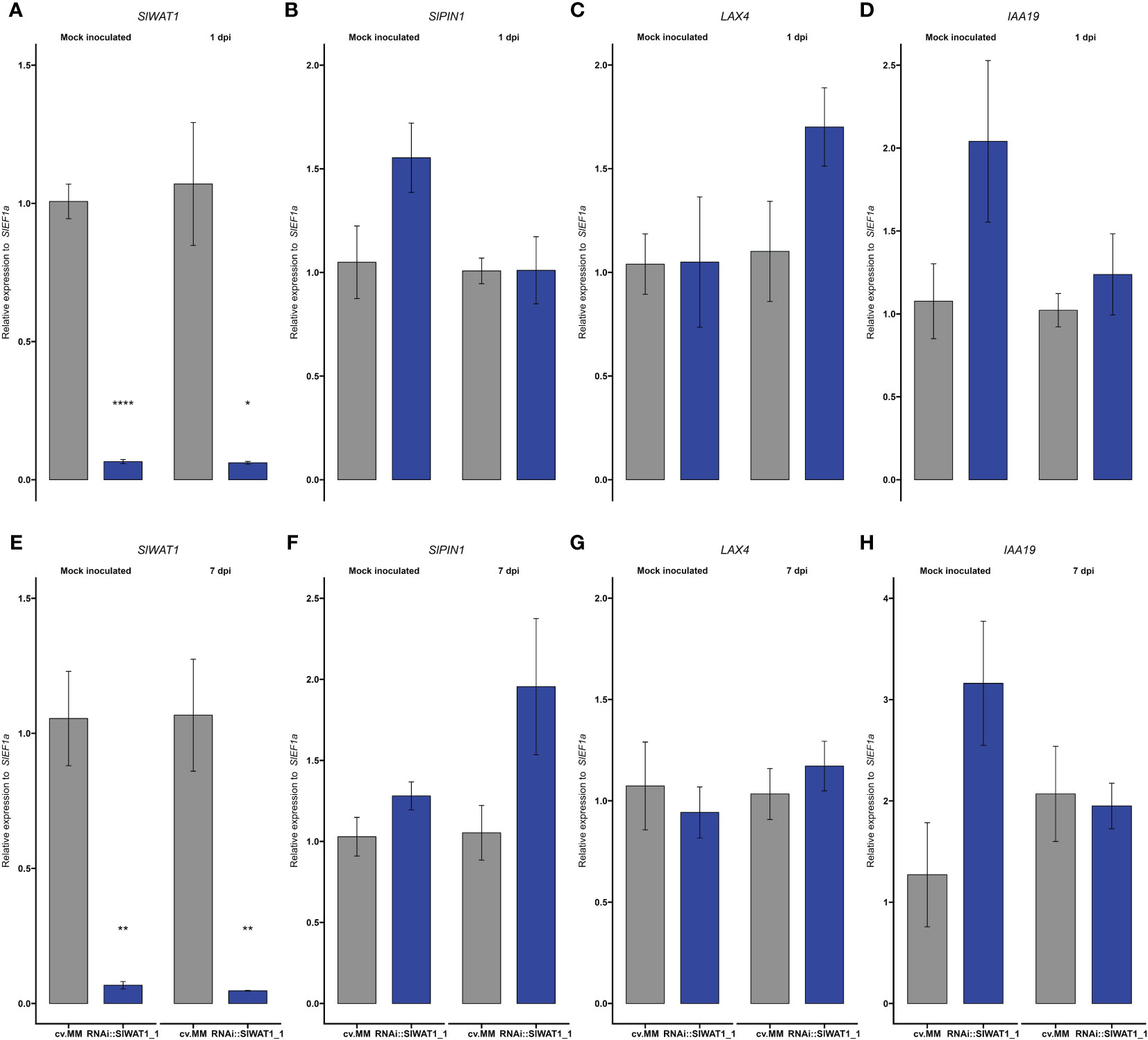
Figure 5 Expression of auxin transporter/signaling genes and auxin content is reprogrammed in RNAi::SlWAT1 plants. Relative expression of genes (A, E) SlWAT1, (B, F) SlPIN1, (C, G) LAX4 and (D, H) IAA19 in mock treated and Cm inoculated plants at 1 day post inoculation (dpi) and 7 dpi. Fold changes were normalized relative to expression of the SlEf1α in cv.MM plants. Bars represent the average fold change over three independent biological replicates (n=5). Error bars indicate standard errors of the mean.
Firstly, we quantified the expression of SlWAT1. As expected, the gene was significantly downregulated in RNAi::SlWAT1_1 plants compared to the cv. MM background (Figures 5A, E). For the rest of the genes we studied no statistically significant differences were found between cv. MM and transgenic plants (Figure 5). At 1 dpi, genes SlPIN1 and IAA19 were upregulated in mock inoculated transgenic plants compared to cv.MM. Upon inoculation, their expression was downregulated in RNAi plants (Figures 5B, D). The same pattern of expression can be observed for gene IAA19 also at 7 dpi (Figure 5H). At 1 dpi, the expression of the auxin importer LAX4 was upregulated in RNAi plants inoculated with Cm (Figure 5C), while at 7 dpi the expression of the gene was comparable to that in cv. MM (Figure 5G). In contrast, the expression of auxin efflux transporter SlPIN1 was upregulated at 7 dpi in infected RNAi plants compared to the cv.MM plants (Figure 5F).
Finally, we quantified the levels of auxin in different parts of tomato stems through LC-MS/MS. Free IAA content was quantified at the apical parts of the stem and hypocotyls of RNAi and cv. MM plants that were mock treated or inoculated (7 dpi). Our results confirm that silencing of SlWAT1 significantly reduces free IAA levels in tomato stems. Consistent with the basipetal auxin transport from source to sink, we also observed a gradient in auxin concentration between the apical meristems and hypocotyls in both genotypes (Figure 6).
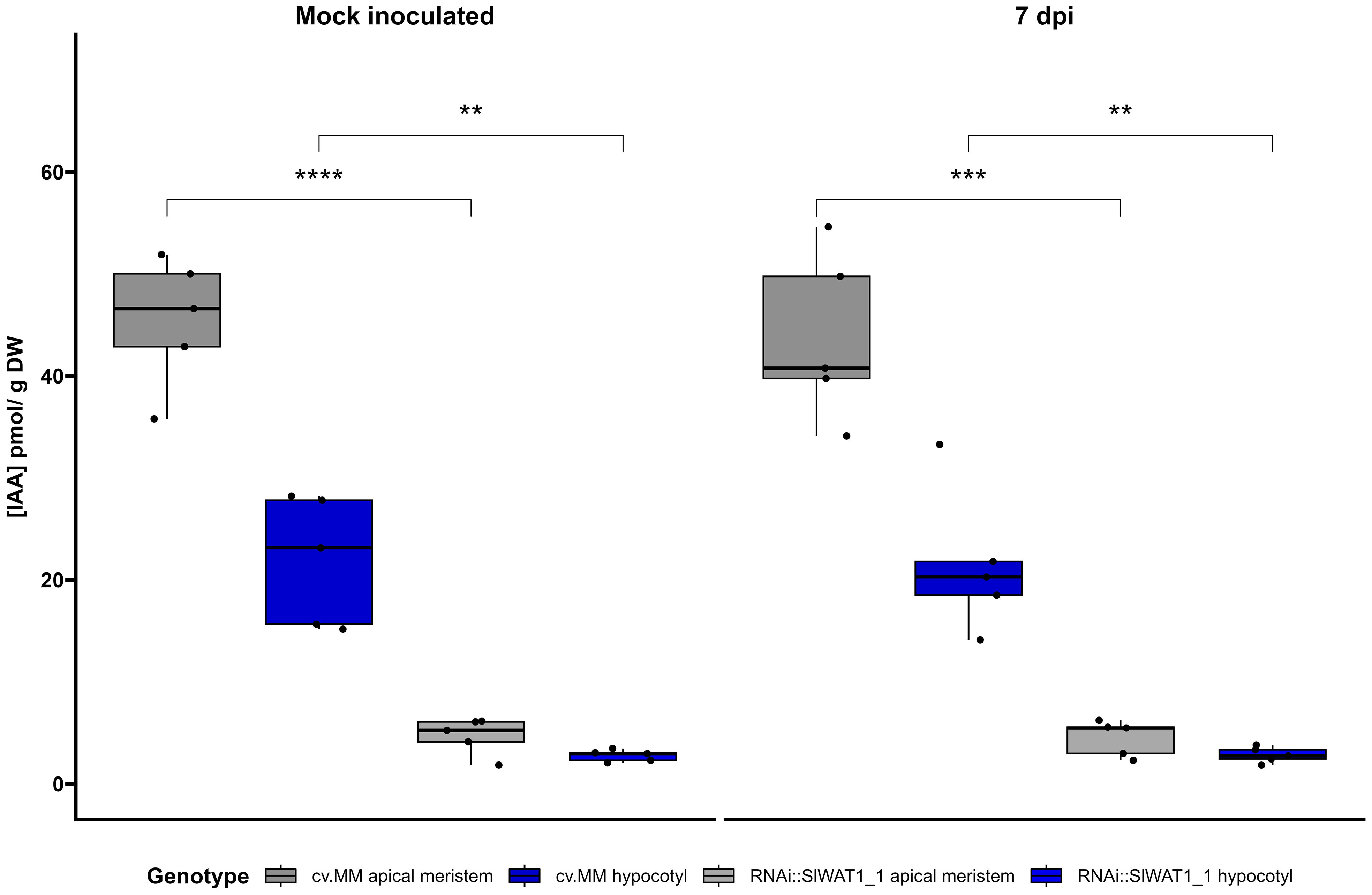
Figure 6 Free IAA content in different stem parts of cv. Moneymaker (cv. MM) and RNAi::SlWAT1_1 plants mock and inoculated at 7 days post inoculation (dpi). Boxplots of IAA concentration (pmol/g DW) in apical meristems and hypocotyls of the two genotypes. Lower and upper box boundaries represent the 25th and 75th percentiles, respectively. Lines in the boxes represent medians of five biological replicates (n=5). (Student’s t-test, **p ≤ 0.01; ***p ≤ 0.001,****p ≤ 0.0001).
Ethylene biosynthesis is downregulated in SlWAT1 silenced plants
Upregulation of ethylene biosynthesis through gene ACO1 has been shown to contribute to the development of wilting symptoms in Cm infected plants (Balaji et al., 2008). Based on our previous observations that silencing of SlWAT1 reduces symptom development on tomato plants, we hypothesized that silencing of the gene will have an effect on ethylene biosynthesis. Therefore, we examined the expression of gene ACO1 in the RNAi::SlWAT1_1 plants. We found that the ACO1 gene is constitutively downregulated in the RNAi plants compared to the cv. MM background, suggesting that ethylene biosynthesis is reduced in the SlWAT1 silenced tomato plants (Figures 7, S3).
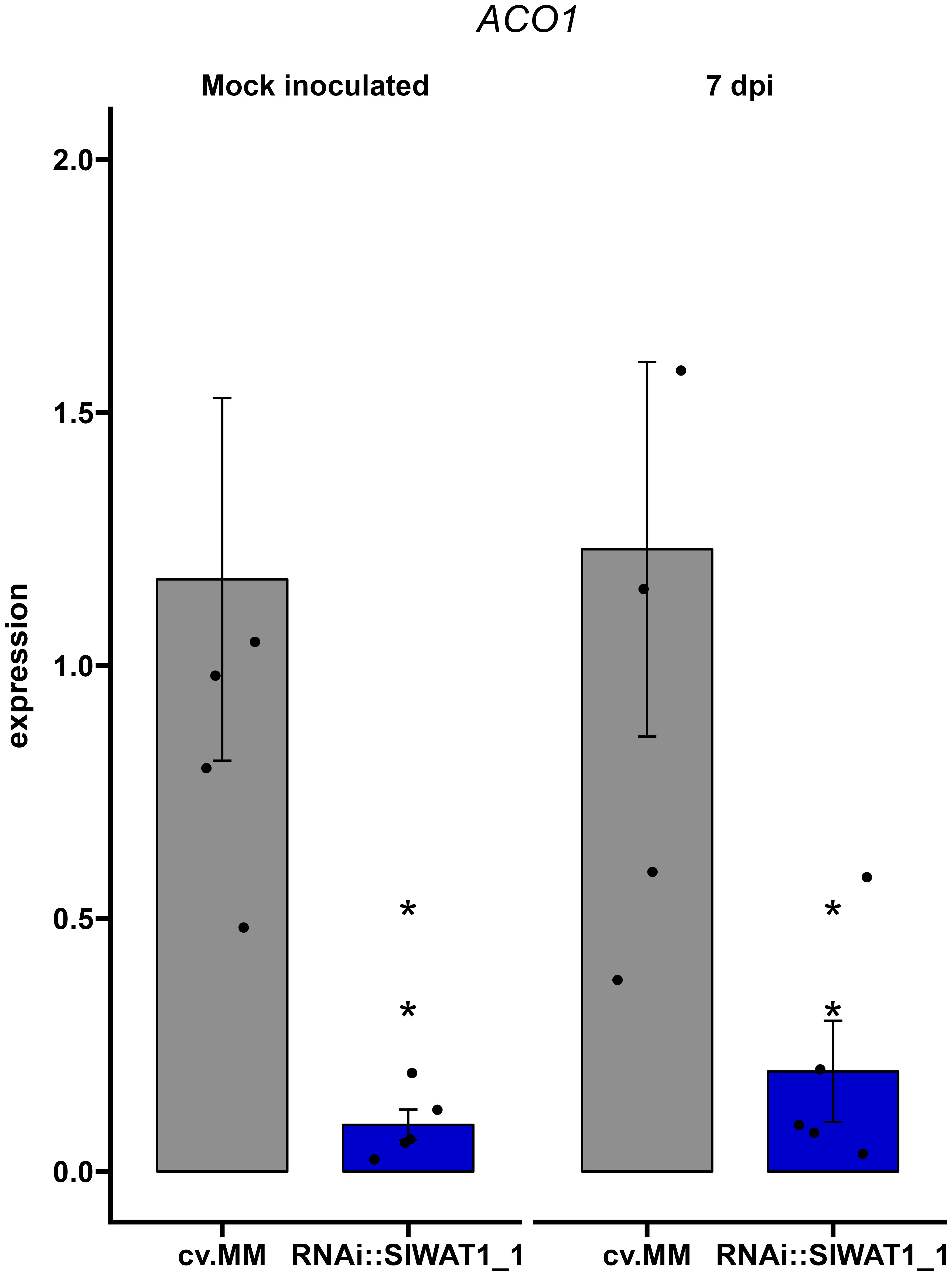
Figure 7 Expression of ethylene biosynthetic gene ACO1 is constitutively downregulated in RNAi::SlWAT1_1 transgenic plants. Relative expression of gene ACO1 in mock treated and Cm inoculated plants at 7 day post inoculation (dpi). Fold changes were normalized relative to expression of the gene in the control plants of cv. Moneymaker (cv.MM). Bars represent the average fold change over five independent biological replicates (n=5). Error bars indicate standard errors of the mean. Asterisks indicate significant differences to the expression prior to inoculation (Student’s T test, *p≤ 0.05).
Inactivation of SlWAT1 affects the expression of pathogen virulence factors
Recent studies in the interaction between Pseudomonas syringae DC3000 and Arabidopsis have revealed a role of IAA in the expression of bacterial virulence factors in planta and in vitro (Kunkel and Harper, 2018). Based on our data that free IAA content in the transgenic plants was significantly lower than in the susceptible cv. MM, we sought to investigate if silencing of SlWAT1 has an effect on the regulation of Cm virulence factors. Plants of cv. MM and RNAi::SlWAT1_1 were inoculated with Cm strain NCPBB382 and stems parts were collected at 1 and 7 dpi. Total RNA from infected plants was isolated and was used to monitor the expression of bacterial virulence genes celA, pat-1, vatr2 (virulence associated transcriptional regulator2) and phpA. At 1 dpi, no amplification of bacterial transcripts was possible due to the low proportion of bacterial mRNA in the total isolated RNA. At 7 dpi, no differences were found in the expression of genes celA and pat-1 on cv. MM and RNAi plants (Figures 8A, B). Expression of transcription factor vatr2 and its target phpA, however, was found to be downregulated in the RNAi plants at 7 dpi, with phpA being significantly downregulated (Figures 8C, D). Overall, our results indicate that silencing of SlWAT1 affects the expression of virulence related genes in Cm during bacterial growth in planta, possibly through reduced IAA content.
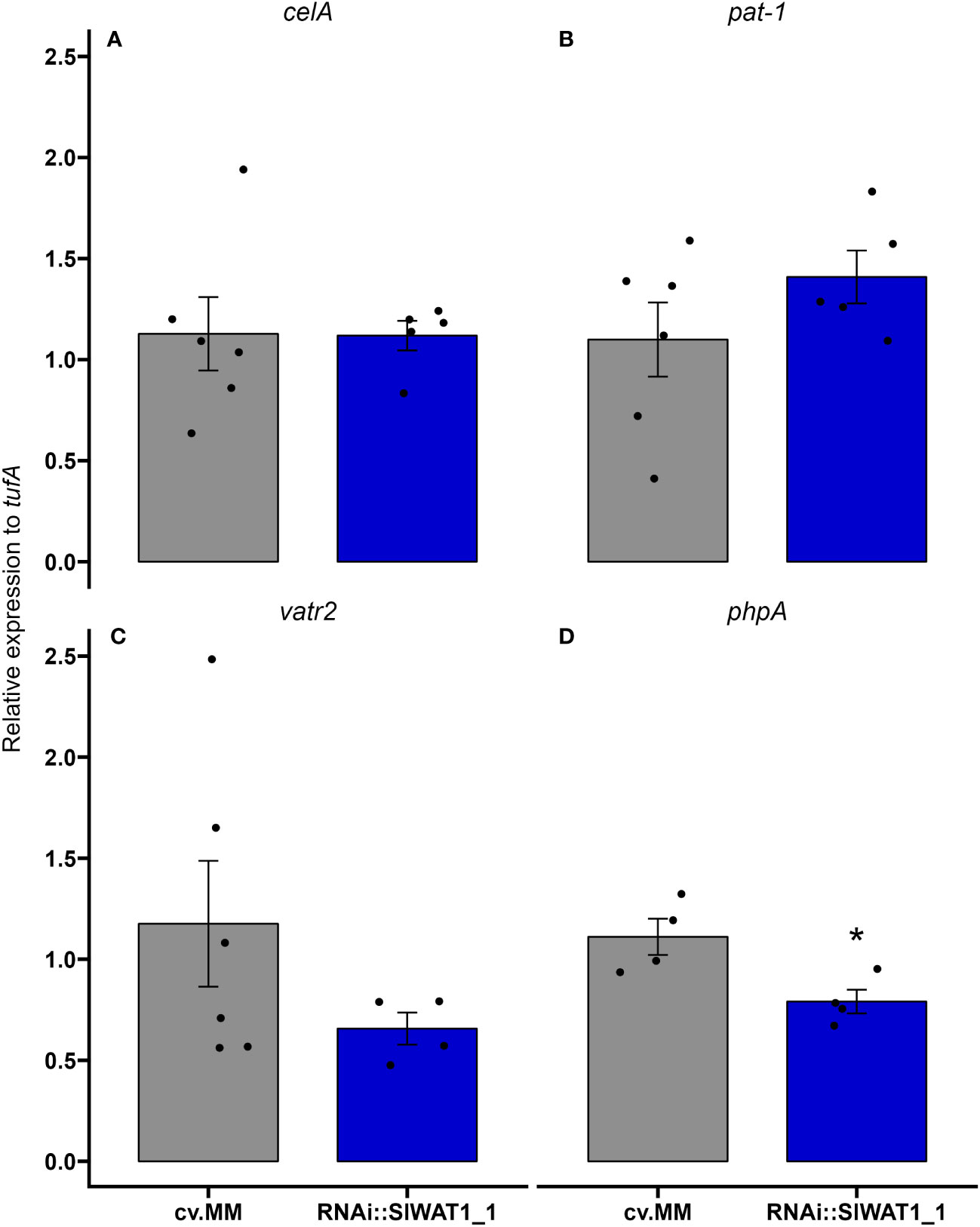
Figure 8 Inactivation of SlWAT1 affects the expression of specific bacterial virulence genes in planta. Relative expression of genes (A) celA, (B) pat-1, (C) vatr2 and (D) phpA on susceptible cv. Moneymaker (cv.MM) and RNAi::SlWAT1_1 plants inoculated with Cm strain NCPBB382 at 7 days post inoculation (dpi). Fold changes were normalized relative to expression of the genes in cv.MM plants. Bars represent the average fold change over five independent biological replicates (n=5). Error bars indicate standard errors of the mean. Asterisks indicate significant differences to the expression of the genes in the different genotypes (Student’s T test, *p≤ 0.05).
Discussion
During their co-evolution with plants many pathogens have evolved the ability to manipulate host S genes to establish a compatible interaction (van Schie and Takken, 2014). Loss-of-function of host S genes can possibly alter a compatible interaction into a non-compatible one, leading to pathogen resistance (Pavan et al., 2010; Gawehns et al., 2013; van Schie and Takken, 2014). Here, we report that the loss-of-function of S gene WAT1 in tomato leads to high tolerance to genetically distinct strains of the bacterial pathogen Clavibacter michiganensis (Figure 1).
WAT1 acts as an S gene that enables the infection process of vascular pathogens (Denance et al., 2013; Tang et al., 2019; Hanika et al., 2026). WAT1 is a tonoplast localized vacuolar auxin transporter, that was first described as a S gene in Arabidopsis thaliana (Ranocha et al., 2010). The arabidopsis wat1-1 mutant was found to be resistant to a broad range of vascular pathogens, including the bacterium Ralstonia solanacearum and the fungi V. dahliae, V. album-altrum and F. oxysporum f. sp. lycopersici. Its function as an S gene to fungal vascular wilts has also been reported in cotton and tomato (Tang et al., 2019; Hanika et al., 2026). In this study, we show that inactivation of tomato homolog SlWAT1 results in strong reduction of symptom development caused by genetically distinct Cm strains (Figure 1). These findings suggest the function of WAT1 as an S gene is possibly conserved across plant species and that its loss-of-function can provide broad-spectrum resistance to vascular pathogens. This is an important trait that has been described for several other S genes (van Damme et al., 2008; Acevedo-Garcia et al., 2017; Oliva et al., 2019; Santillán Martínez et al., 2020; Thomazella et al., 2021).
Despite the strong reduction of wilting symptoms, growth of Cm was not suppressed by inactivation of SlWAT1, in contrast to what it has been reported for other pathogens (Tang et al., 2019; Hanika et al., 2026). According to our initial hypothesis, the residual expression of SlWAT1 in the RNAi lines was possibly responsible for the mild symptoms observed and the sustained growth of the pathogen. To confirm the results we obtained from the disease assays and to study the effect of a full knock-out in the growth of the pathogen, we included a CRISPR/Cas9 mutant line in our experiments. In accordance with our previous results, we observed strong symptom reduction in slwat1 mutant tomato plants. Further, we did not detect any significant differences in the Cm bacterial titers recovered from the slwat1 mutants and the susceptible background. As previously observed virulence of Cm does not always correlate with population size. Strain Cmm100, which lacks the two plasmids involved in pathogenicity, cannot cause symptoms on tomato plants and acts as an endophyte. It can, however, grow to population densities that in some cases are higher than the virulent wild type strain. This has been attributed to plant cell death as symptom development advances (Chalupowicz et al., 2010). The high population densities of Cm in SlWAT1 inactivated plants might be an indication of less cell death, which may be a result of downregulation of bacterial virulence factors. As a result of less cell death the pathogen is able to grow to high population densities. This led us to hypothesize that SlWAT1 is involved in symptom development, rather than sustainment of Cm. A major drawback in the use of mutant S genes to gain resistance to pathogens is the possibility of adverse pleiotropy (Sun et al., 2016b), as also observed in the case of tomato SlWAT1. Although a full knock-out of the gene led to severe growth defects (Figure 3), downregulation of the gene in the RNAi lines resulted in similar tolerance levels to the pathogen without a severe fitness cost on plant growth. In addition to RNAi, alterations in cis-regulatory regions of the gene to change its expression (Alonge et al., 2020), might provide a cost-free strategy to gain tolerance to Cm. Alternatively, the exploration of allelic variation in tomato germplasm may lead to the identification of natural variants that disrupt the compatible host-pathogen interaction without fitness costs, as it was done in the case of gene ROD1 in rice (Gao et al., 2021).
Changes in hormonal homeostasis is a common strategy used by pathogens to promote disease. While upregulation of host derived ethylene has been found to promote wilting development by Cm, the role of other hormones in the infection process remains unclear (Balaji et al., 2008). Resistance conferred by inactivation of WAT1 has been associated with altered crosstalk between auxin and SA in Arabidopsis and cotton (Denance et al., 2013; Tang et al., 2019). According to our findings ethylene biosynthesis and auxin content were reduced in SlWAT1 inactivated plants. We found that the ethylene biosynthetic gene ACO1, that is specifically upregulated by Cm to promote wilting symptoms was constitutively downregulated in RNAi::SlWAT1_1 plants (Figures 6, S3). This is also in accordance with previous studies that found that symptom development, but not Cm bacterial growth was inhibited on Nr ethylene insensitive plants (Balaji et al., 2008).
We also found that the content of free IAA in stem tissues of the RNAi plants was significantly lower than in cv. MM (Figure 6). In addition, expression of auxin related genes was altered by SlWAT1 impaired plants upon Cm inoculation (Figures 5A–H). At 1 dpi, we found that the expression of auxin influx gene LAX4 was upregulated in Cm inoculated RNAi plants, while the expression of auxin efflux gene SlPIN1 was upregulated in mock inoculated RNAI plants. Although rather speculative, these changes in the expression of auxin transporter genes might be induced by Cm in an attempt to increase auxin influx around the inoculation point. The lower expression of IAA19 in infected plants (at 1 and 7 dpi) compared to mock inoculated RNAi plants could also suggest that the auxin contents around the inoculation site are indeed increased during infection, since IAA19 is upregulated in the absence of auxin (Tatematsu et al., 2004). Finally, the upregulation of SlPIN1 in Cm infected SlWAT1 inactivated plants at 7 dpi, might act as a late compensatory mechanism for the absence of SlWAT1, which also facilitates auxin efflux.
Higher contents of SA have been reported for arabidopsis and cotton WAT1 impaired plants (Denance et al., 2013; Tang et al., 2019). This could be a direct consequence of the reduction of free IAA content, as the SA and auxin hormonal pathways are mutually antagonistic (Wang et al., 2007). Although SA is a known regulator of defenses against pathogens, knowledge on its role in resistance against Cm is limited. Recently, it was shown that exogenous application of SA reduces the bacterial populations on tomato cotyledons (Yokotani et al., 2021). The infection of NahG transgenic tomato plants with impaired SA accumulation, however, did not result in higher susceptibility to the pathogen (Barda et al., 2015).
Growing evidence suggests that host derived auxin is an important signaling molecule involved in plant-bacteria interactions (Denance et al., 2013; McClerklin et al., 2018; Djami-Tchatchou et al., 2019; Mashiguchi et al., 2019). Recent studies have reported a direct effect of auxin in the regulation of Pseudomonas syringae DC300 bacterial genes involved in virulence (Aragón et al., 2014; Djami-Tchatchou et al., 2019). Elevated IAA content in Arabidopsis quadruple mutant tir1 afb1 afb4 afb5, as well as the addition of IAA in P. syringae DC300 cultures led to the repression of genes involved in the production of T3SS at early timepoints. The expression of genes involved in late infection stages, however, was significantly upregulated by elevated IAA contents (Djami-Tchatchou et al., 2019). Additionally, auxin produced by bacteria itself can act as a virulence factor (McClerklin et al., 2018). Based on our observations of the significant reduction of symptom development and the significantly lower free IAA content in RNAi::SlWAT1_1 plants, we hypothesized that auxin might play a role in the regulation of Cm virulence genes. Therefore, we monitored the transcript levels of virulence factors celA, pat-1, vatr2 and phpA in planta (Figure 7). Interestingly, we observed that transcription factor vatr2 and its target phpA were downregulated at 7 dpi. This suggests that inactivation of SlWAT1 leads to downregulation of Cm virulence genes, possibly through the reduced contents of free IAA in the stems of transgenic plants. Previous, transcriptomics analysis has shown that the virulence factors celA and pat-1 reach the peak of their expression between 24 and 72 hours post inoculation and gene expression is reduced after that point (Chalupowicz et al., 2010). This might be the reason why we did not detect a difference in the expression of celA and pat-1 isolated from cv.MM and RNAi::SlWAT1_1 plants at 7 dpi. To definitely conclude, however, that auxin directly affects the expression of bacterial genes after supplementation of cultures with IAA could be monitored. Moreover, meta-transcriptomics analysis through RNA-seq on infected SlWAT1 inactivated plants and their susceptible background could be deployed in different experimental timepoints, in order to elucidate the complete pathways involved in the molecular interaction of the organisms (Guo et al., 2021). Future studies on how Cm responds to IAA, as well as the production of IAA non-responsive Cm mutants, could allow us to fully study and understand the role of auxin as a signaling molecule in the pathosystem. Finally, the possibility of changes in the metabolome of the plants due to perturbations in hormonal changes, might influence the expression of bacterial virulence factors, requires further examination.
Materials and methods
Plant materials
The present study included the susceptible Solanum lycopersicum cv. Moneymaker MM as a control, T3 progeny of two independent stable transformants (RNAi::SlWAT1_1, RNAi::SlWAT1_2) in which the SlWAT1 gene was silenced through RNAi in cv. MM background and T2 progeny of a bi-allelic heterozygous CRISPR/Cas9 generated slwat1 mutant line (Hanika et al., 2026). Prior to infection, transgenic plantlets were screened for the presence of the RNAi silencing construct based on the presence of the 35S and NPTII markers. slwat1 mutants were screened for the presence of mutant alleles through PCR based genotyping and sequencing.
Plants were grown in a climate regulated greenhouse compartment at 24°C/18°C under a 12h/12h day/night regime. Relative humidity in the compartment was kept to ~60%.
DNA isolation and genotyping
For the genotyping of the RNAi transgenic and slwat1 mutant plants genomic DNA was isolated using a modified protocol for cetyl trimethylammonium bromide (CTAB) extraction method (Doyle, 1991). PCR was performed with DreamTaq DNA polymerase (Thermo Scientific) and target specific primers (Table S1). The PCR products of the RNAi transgenic plants were visualized on 1% agarose gel for the screening of the presence of NPTII and 35S transgene markers. PCR products of mutant plants were sequenced through Illumina sequencing (Macrogen Europe, Amsterdam).
Bacterial strains and growth conditions
Four genetically diverse Cm strains were used in the experiments, i.e. Cm strains NCPBB382, IPO3356 (rifampicin resistant mutant), CFBP2493 and CFBP5843 were used in the experiments (Jacques et al., 2012). Prior to plant inoculation the strains were grown for two days at 25° C on TBY plates (10 gL-1 tryptone, 5 gL-1 yeast extract, 5 gL-1 sodium chloride, 15 gL-1 bacteriological agar). Plates were supplemented with appropriate antibiotics when needed (25 μl/mL rifampicin).
Disease assay
Tomato plants at the fourth true leaf stage were inoculated by a petiole clipping off method. The petioles of the first two fully expanded leaves were clipped off with razor blades immersed in the bacterial inoculum and 5 μl of the bacterial inoculum were directly pipetted on the lowest wound. Bacterial inocula of the four bacterial strains were prepared by re-suspending cells in Ringer’s buffer to a final concentration of ~108 cfu/ml (OD600 = 0.1). Prior to re-suspension the Cm strains were streaked on TBY plates, supplemented with appropriate antibiotics when needed, and incubated at 25°C for two days. Symptom development (wilting) was monitored up to 20 days post inoculation (dpi). A disease index (DI) scale based on the development of wilting symptoms on the leaves was used (0; no symptoms- 5; all leaves wilting). Per strain, 12 transgenic T3 plants and three susceptible cv. MM control plants were used. The same procedure was used for the disease assays of slwat1 mutants. At least five biological replicates of slwat1 mutants were used in the experiments. Four biological replicates of the susceptible cv. MM were used as controls.
Bacterial quantification
Bacterial quantification was done through serial dilution plating. Whole stems collected ~1 cm above the lowest inoculation point and up to the apical meristem were sampled at three time points; 4 dpi, 7 dpi and 14 dpi. Stems were pulverized and homogenized in Ringer’s solution (Sigma Aldrich). 50 μl of serial dilutions of the homogenate (101 –106) were plated on SCM-F selective plates (Duchefa Biochemie). The medium was supplemented with 1.9 g L-1 yeast extract, 20 μL L-1 nalidixic acid (100 mg mL-1), 8 mL L-1 trimetroprim in MetOH 100% (10 mg/mL), 1 ml L-1 cyclohexamide in MetOH 100% (100 mg mL-1), 1 mL potassium tellurite (1%), 50 ml L-1 nicotinic acid (2 mg/mL). Plates were supplemented with appropriate antibiotics when necessary (25 μl/mL rifampicin). Plates were incubated at 25°C for 7 days. Colonies on the plates were counted 7 days post plating and the log10(cfu+1/g fresh tissue) per plate was calculated. Five biological replicates of RNAi::SlWAT1_1 plants and the susceptible cv. MM were used per time point. Two technical replicates per sample were plated. The same procedure was used for the quantification of in planta bacterial titers of strain NCPBB382 in slwat1 mutants. Five biological replicates per time point were used.
RNA extraction/cDNA synthesis
Stem samples of ~2 cm in length were collected above the inoculation point. The stems were processed using a Precellys Evolution tissue homogenizer (Bertin Technologies) at 7000 RPM for two rounds of 15 sec, with the cryolysis option on. RNA extraction and on column DNase treatment were done using the RNeasy Mini Kit (Qiagen) and RNase-Free DNase Set (Qiagen) following the manufacturer’s instructions. 500 ng of first strand cDNA was synthesized using the iScript cDNA synthesis kit (Bio-rad).
Gene expression analysis
Expression levels of tomato genes SlWAT1, ACO1, IAA19, SlPIN1, LAX4 on mock treated and Cm infected control and RNAi::SlWAT1_1 transgenic plants were monitored through RT-qPCR at 1 and 7 dpi using specific gene primers (Table S1). The expression of bacterial virulence genes celA, pat-1, vatr2 and phpA on infected cv. MM and RNAi::SlWAT1_1 was also assessed at 1 and 7 dpi using target specific primers (Table S1). 5 and 25 ng of cDNA were used as a template for the reactions for the quantification of plant and bacterial transcripts, respectively. Reactions were done in duplicates. At least four biological samples were used per treatment and time point. RT-qPCR was done on a CFX96 Touch Deep Well Real Time PCR Detection system (Bio-Rad).
Prior to cDNA synthesis for the expression analysis of the bacterial virulence genes, the total RNA isolated was run on 1% agarose gel to confirm the absence of gDNA from the samples. The Livak 2-ΔΔCt method was used to normalize and calibrate transcript values relative to the endogenous SlEf1α for tomato and gene tufA for the bacterial genes.
Auxin quantification
Auxin was quantified through Liquid Chromatography- Mass Spectrometry (LC-MS/MS). Plant stem parts were collected and flash frozen in liquid nitrogen. The collected tissue was processed using a Precellys Evolution tissue homogenizer (Bertin Technologies) at 7000 RPM for two rounds of 15 sec, with the cryolysis option on. ~25 mg of tissue were used for the auxin extraction. Ground stem samples were extracted with 1 mL of cold methanol containing [phenyl 13C6]-IAA (0.1 nmol/mL) as an internal standard in a 2-mL eppendorf tube and purified as previously described (Ruyter-Spira et al., 2011; Schiessl et al., 2019). Samples were filtered through a 0.45 μm Minisart SRP4 filter (Sartorius, Goettingen, Germany) and measured on the same day. Auxin was analyzed on a Waters Xevo TQs tandem quadruple mass spectrometer as previously described (Ruyter-Spira et al., 2011; Gühl et al., 2021).
Data availability statement
The data presented in the study are deposited in the National Center for Biotechnology Information (NCBI) repository, accession numbers OP832215 and OP832216.
Author contributions
YB, JW, RV, and EK designed the experiments. YB, JW, and RV supervised the work. EK executed the experiments and performed the data analysis. KH generated the transgenic lines and CRISPR/Cas9 mutants. MMN was involved in the initial characterization of the gene. WK performed the auxin quantification. EK drafted the manuscript. JW, RV, and YB critically reviewed and edited the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This research was (partially) funded by NWO Science domain (NWO-ENW) project 8440590003, which is financed by the Dutch Research Council (NWO).
Acknowledgments
The authors would like to thank Patricia van der Zouwen for the maintenance of the bacterial strains used in this study and her advice about the bacterial quantification.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1082094/full#supplementary-material
References
Acevedo-Garcia, J., Gruner, K., Reinstädler, A., Kemen, A., Kemen, E., Cao, L., et al. (2017). The powdery mildew-resistant arabidopsis mlo2 mlo6 mlo12 triple mutant displays altered infection phenotypes with diverse types of phytopathogens. Sci. Rep. 7 (1). doi: 10.1038/s41598-017-07188-7
Alonge, M., Wang, X., Benoit, M., Soyk, S., Pereira, L., Zhang, L., et al. (2020). Major impacts of widespread structural variation on gene expression and crop improvement in tomato. Cell 182 (1), 145–61.e23. doi: 10.1016/j.cell.2020.05.021
Appiano, M., Pavan, S., Catalano, D., Zheng, Z., Bracuto, V., Lotti, C., et al. (2015). Identification of candidate MLO powdery mildew susceptibility genes in cultivated solanaceae and functional characterization of tobacco NtMLO1. Transgenic Res. 24 (5), 847–858. doi: 10.1007/s11248-015-9878-4
Aragón, I. M., Pérez-Martínez, I., Moreno-Pérez, A., Cerezo, M., Ramos, C. (2014). New insights into the role of indole-3-acetic acid in the virulence ofPseudomonas savastanoipv.savastanoi. FEMS Microbiol. Lett. 356 (2), 184–192. doi: 10.1111/1574-6968.12413
Bai, Y., Pavan, S., Zheng, Z., Zappel, N. F., Reinstadler, A., Lotti, C., et al. (2008). Naturally occurring broad-spectrum powdery mildew resistance in a central American tomato accession is caused by loss of mlo function. Mol. Plant Microbe Interact. 21 (1), 30–39. doi: 10.1094/MPMI-21-1-0030
Balaji, V., Mayrose, M., Sherf, O., Jacob-Hirsch, J., Eichenlaub, R., Iraki, N., et al. (2008). Tomato transcriptional changes in response to clavibacter michiganensis subsp. michiganensis reveal a role for ethylene in disease development. Plant Physiol. 146 (4), 1797–1809. doi: 10.1104/pp.107.115188
Barda, O., Shalev, O., Alster, S., Buxdorf, K., Gafni, A., Levy, M. (2015). Pseudozyma aphidis induces salicylic-Acid-Independent resistance to clavibacter michiganensis in tomato plants. Plant Disease. 99 (5), 621–626. doi: 10.1094/PDIS-04-14-0377-RE
Berg, J. A., Appiano, M., Bijsterbosch, G., Visser, R. G. F., Schouten, H. J., Bai, Y. (2017). Functional characterization of cucumber (Cucumis sativus l.) clade V MLO genes. BMC Plant Biol. 17 (1). doi: 10.1186/s12870-017-1029-z
Chalupowicz, L., Barash, I., Reuven, M., Dror, O., Sharabani, G., Gartemann, K.-H., et al. (2017). Differential contribution of clavibacter michiganensis ssp. michiganensis virulence factors to systemic and local infection in tomato. Mol. Plant Pathol. 18 (3), 336–346. doi: 10.1111/mpp.12400
Chalupowicz, L., Cohen-Kandli, M., Dror, O., Eichenlaub, R., Gartemann, K. H., Sessa, G., et al. (2010). Sequential expression of bacterial virulence and plant defense genes during infection of tomato withClavibacter michiganensissubsp.michiganensis. Phytopathology 100 (3), 252–261. doi: 10.1094/PHYTO-100-3-0252
Chang, R. J., Ries, S. M., Pataky, J. K. (1992). Effects of temperature, plant-age, inoculum concentration, and cultivar on the incubation period and severity of bacterial canker of tomato. Plant Disease. 76 (11), 1150–1155. doi: 10.1094/PD-76-1150
Cohn, M., Bart, R. S., Shybut, M., Dahlbeck, D., Gomez, M., Morbitzer, R., et al. (2014). Xanthomonas axonopodis virulence is promoted by a transcription activator-like effector–mediated induction of a SWEET sugar transporter in cassava. Mol. Plant-Microbe Interactions 27 (11), 1186–1198. doi: 10.1094/MPMI-06-14-0161-R
Cox, K. L., Meng, F., Wilkins, K. E., Li, F., Wang, P., Booher, N. J., et al. (2017). TAL effector driven induction of a SWEET gene confers susceptibility to bacterial blight of cotton. Nat. Commun. 8, 15588. doi: 10.1038/ncomms15588
Davis, M. J., Gillaspie, A. G., Vidaver, A. K., Harris, R. W. (1984). Clavibacter: a new genus containing some phytopathogenic coryneform bacteria, including clavibacter xyli subsp. xyli sp. nov., subsp. nov. and clavibacter xyli subsp. cynodontis subsp. nov., pathogens that cause ratoon stunting disease of sugarcane and be. Int. J. Systematic Bacteriol. 34 (2), 107–117. doi: 10.1099/00207713-34-2-107
Denance, N., Ranocha, P., Oria, N., Barlet, X., Riviere, M. P., Yadeta, K. A., et al. (2013). Arabidopsis wat1 (walls are thin1)-mediated resistance to the bacterial vascular pathogen, ralstonia solanacearum, is accompanied by cross-regulation of salicylic acid and tryptophan metabolism. Plant J. 73 (2), 225–239. doi: 10.1111/tpj.12027
Djami-Tchatchou, A. T., Harrison, G. A., Harper, C. P., Wang, R., Prigge, M. J., Estelle, M., et al. (2019). Dual role of auxin in regulating plant defense and bacterial virulence gene expression during pseudomonas syringae PtoDC3000 pathogenesis. Mol Plant Microbe Interact. 33, 1059–1071. doi: 10.1101/2019.12.29.881581
Eichenlaub, R., Gartemann, K.-H. (2011). The clavibacter michiganensis subspecies: molecular investigation of gram-positive bacterial plant pathogens. Ann. Rev. Phytopathol. 49, 1, 445–464. doi: 10.1146/annurev-phyto-072910-095258
Gao, M., He, Y., Yin, X., Zhong, X., Yan, B., Wu, Y., et al. (2021). Ca2+ sensor-mediated ROS scavenging suppresses rice immunity and is exploited by a fungal effector. Cell 184 (21), 5391–404.e17. doi: 10.1016/j.cell.2021.09.009
Gartemann, K. H., Abt, B., Bekel, T., Burger, A., Engemann, J., Flugel, M., et al. (2008). The genome sequence of the tomato-pathogenic actinomycete clavibacter michiganensis subsp. michiganensis NCPPB382 reveals a Large island involved in pathogenicity. J. Bacteriol. 190 (6), 2138–2149. doi: 10.1128/jb.01595-07
Gawehns, F., Cornelissen, B. J. C., Takken, F. L. W. (2013). The potential of effector-target genes in breeding for plant innate immunity. Microbial. Biotechnol. 6 (3), 223–229. doi: 10.1111/1751-7915.12023
Gühl, K., Holmer, R., Xiao, T. T., Shen, D., Wardhani, T. A. K., Geurts, R., et al. (2021). The effect of exogenous nitrate on LCO signalling, cytokinin accumulation, and nodule initiation in medicago truncatula. Genes 12 (7), 988. doi: 10.3390/genes12070988
Guo, L., Yu, H., Wang, B., Vescio, K., DeIulio, G. A., Yang, H., et al. (2021). Metatranscriptomic comparison of endophytic and pathogenic fusarium-arabidopsis interactions reveals plant transcriptional plasticity. Mol. Plant Microbe Interact. 34 (9), 1071–1083. doi: 10.1094/MPMI-03-21-0063-R
Hanika, K., Schipper, D., Chinnappa, S., Oortwijn, M., Schouten, H. J., Thomma, B. P. H. J., et al. (2021). Impairment of tomato WAT1 enhances resistance to vascular wilt fungi despite severe growth defects. Front. Plant Sci. 2021, 12. doi: 10.3389/fpls.2021.721674
Hückelhoven, R., Eichmann, R., Weis, C., Hoefle, C., Proels, R. K. (2013). Genetic loss of susceptibility: a costly route to disease resistance? Plant Pathol. 62, 56–62. doi: 10.1111/ppa.12103
Huibers, R. P., Loonen, A. E., Gao, D., Van den Ackerveken, G., Visser, R. G., Bai, Y. (2013). Powdery mildew resistance in tomato by impairment of SlPMR4 and SlDMR1. PloS One 8 (6), e67467. doi: 10.1371/journal.pone.0067467
Jacques, M. A., Durand, K., Orgeur, G., Balidas, S., Fricot, C., Bonneau, S., et al. (2012). Phylogenetic analysis and polyphasic characterization of clavibacter michiganensis strains isolated from tomato seeds reveal that nonpathogenic strains are distinct from c. michiganensis subsp michiganensis 78 (23), 8388–8402. doi: 10.1128/AEM.02158-12
Jones, J. D., Dangl, J. L. (2006). The plant immune system. Nature 444 (7117), 323–329. doi: 10.1038/nature05286
Kunkel, B. N., Harper, C. P. (2018). The roles of auxin during interactions between bacterial plant pathogens and their hosts. J. Exp. Botany 69 (2), 245–254. doi: 10.1093/jxb/erx447
Mashiguchi, K., Hisano, H., Takeda-Kamiya, N., Takebayashi, Y., Ariizumi, T., Gao, Y., et al. (2019). Agrobacterium tumefaciensEnhances biosynthesis of two distinct auxins in the formation of crown galls. Plant Cell Physiol. 60 (1), 29–37. doi: 10.1093/pcp/pcy182
McClerklin, S. A., Lee, S. G., Harper, C. P., Nwumeh, R., Jez, J. M., Kunkel, B. N. (2018). Indole-3-acetaldehyde dehydrogenase-dependent auxin synthesis contributes to virulence of pseudomonas syringae strain DC3000. PloS Pathog. 14 (1), e1006811. doi: 10.1371/journal.ppat.1006811
Meletzus, D., Bermphol, A., Dreier, J., Eichenlaub, R. (1993). Evidence for plasmid-encoded virulence factors in the phytopathogenic bacterium clavibacter michiganensis subsp. michiganensis NCPPB382. J. Bacteriol. 175 (7), 2131–2136. doi: 10.1128/jb.175.7.2131-2136.1993
Nouioui, I., Carro, L., García-López, M., Meier-Kolthoff, J. P., Woyke, T., Kyrpides, N. C., et al. (2018). Genome-based taxonomic classification of the phylum actinobacteria. Front. Microbiol. 9. doi: 10.3389/fmicb.2018.02007
Oliva, R., Ji, C., Atienza-Grande, G., Huguet-Tapia, J. C., Perez-Quintero, A., Li, T., et al. (2019). Broad-spectrum resistance to bacterial blight in rice using genome editing. Nat. Biotechnol. 37 (11), 1344–1350. doi: 10.1038/s41587-019-0267-z
Pattison, R. J., Catalá, C. (2012). Evaluating auxin distribution in tomato (Solanum lycopersicum) through an analysis of the PIN and AUX/LAX gene families. Plant J. 70 (4), 585–598. doi: 10.1111/j.1365-313X.2011.04895.x
Pavan, S., Jacobsen, E., Visser, R. G., Bai, Y. (2010). Loss of susceptibility as a novel breeding strategy for durable and broad-spectrum resistance. Mol. Breed. 25 (1), 1–12. doi: 10.1007/s11032-009-9323-6
Pavan, S., Schiavulli, A., Appiano, M., Marcotrigiano, A. R., Cillo, F., Visser, R. G., et al. (2011). Pea powdery mildew er1 resistance is associated to loss-of-function mutations at a MLO homologous locus. Theor. Appl. Genet. 123 (8), 1425–1431. doi: 10.1007/s00122-011-1677-6
Pessina, S., Lenzi, L., Perazzolli, M., Campa, M., Dalla Costa, L., Urso, S., et al. (2016). Knockdown of MLO genes reduces susceptibility to powdery mildew in grapevine. Hortic. Res. 3, 16016. doi: 10.1038/hortres.2016.16
Ranocha, P., Denance, N., Vanholme, R., Freydier, A., Martinez, Y., Hoffmann, L., et al. (2010). Walls are thin 1 (WAT1), an arabidopsis homolog of medicago truncatula NODULIN21, is a tonoplast-localized protein required for secondary wall formation in fibers. Plant J. 63 (3), 469–483. doi: 10.1111/j.1365-313X.2010.04256.x
Ranocha, P., Dima, O., Nagy, R., Felten, J., Corratge-Faillie, C., Novak, O., et al. (2013). Arabidopsis WAT1 is a vacuolar auxin transport facilitator required for auxin homoeostasis. Nat. Commun. 4, 2625. doi: 10.1038/ncomms3625
Ruyter-Spira, C., Kohlen, W., Charnikhova, T., Van Zeijl, A., Van Bezouwen, L., De Ruijter, N., et al. (2011). Physiological effects of the synthetic strigolactone analog GR24 on root system architecture in arabidopsis: another belowground role for strigolactones? Plant Physiol. 155 (2), 721–734. doi: 10.1104/pp.110.166645
Santillán Martínez, M. I., Bracuto, V., Koseoglou, E., Appiano, M., Jacobsen, E., Visser, R. G. F., et al. (2020). CRISPR/Cas9-targeted mutagenesis of the tomato susceptibility gene PMR4 for resistance against powdery mildew. BMC Plant Biol. 20 (1). doi: 10.1186/s12870-020-02497-y
Savidor, A., Chalupowicz, L., Teper, D., Gartemann, K. H., Eichenlaub, R., Manulis-Sasson, S., et al. (2014). Clavibacter michiganensis subsp. michiganensis Vatr1 and Vatr2 transcriptional regulators are required for virulence in tomato. Mol. Plant Microbe Interact. 27 (10), 1035–1047. doi: 10.1094/mpmi-02-14-0061-r
Savidor, A., Teper, D., Gartemann, K.-H., Eichenlaub, R., Chalupowicz, L., Manulis-Sasson, S., et al. (2012). The clavibacter michiganensis subsp. michiganensis –tomato interactome reveals the perception of pathogen by the host and suggests mechanisms of infection. J. Proteome Res. 11, 2, 736–750. doi: 10.1021/pr200646a
Schiessl, K., Lilley, J. L. S., Lee, T., Tamvakis, I., Kohlen, W., Bailey, P. C., et al. (2019). NODULE INCEPTION recruits the lateral root developmental program for symbiotic nodule organogenesis in medicago truncatula. Curr. Biol. 29 (21), 3657–68.e5. doi: 10.1016/j.cub.2019.09.005
Sen, Y., van der Wolf, J., Visser, R. G. F., Van Heusden, S. (2015). Bacterial canker of tomato: current knowledge of detection, management, resistance, and interactions. Plant Disease. 99 (1), 4–13. doi: 10.1094/PDIS-05-14-0499-FE
Sharabani, G., Shtienberg, D., Borenstein, M., Shulhani, R., Lofthouse, M., Sofer, M., et al. (2013). Effects of plant age on disease development and virulence ofClavibacter michiganensissubsp.michiganensison tomato. Plant Pathol. 62 (5), 1114–1122. doi: 10.1111/ppa.12013
Sun, K., Wolters, A.-M. A., Loonen, A. E. H. M., Huibers, R. P., van der Vlugt, R., Goverse, A., et al. (2016a). Down-regulation of arabidopsis DND1 orthologs in potato and tomato leads to broad-spectrum resistance to late blight and powdery mildew. Transgenic Res. 25 (2), 123–138. doi: 10.1007/s11248-015-9921-5
Sun, K., Wolters, A.-M. A., Vossen, J. H., Rouwet, M. E., Loonen, A. E. H. M., Jacobsen, E., et al. (2016b). Silencing of six susceptibility genes results in potato late blight resistance. Transgenic Res. 25 (5), 731–742. doi: 10.1007/s11248-016-9964-2
Tang, Y., Zhang, Z., Lei, Y., Hu, G., Liu, J., Hao, M., et al. (2019). Cotton WATs modulate SA biosynthesis and local lignin deposition participating in plant resistance against verticillium dahliae. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.00526
Tatematsu, K., Kumagai, S., Muto, H., Sato, A., Watahiki, M. K., Harper, R. M., et al. (2004). MASSUGU2 encodes Aux/IAA19, an auxin-regulated protein that functions together with the transcriptional activator NPH4/ARF7 to regulate differential growth responses of hypocotyl and formation of lateral roots in arabidopsis thaliana. Plant Cell. 16 (2), 379–393. doi: 10.1105/tpc.018630
Thapa, S. P., Pattathil, S., Hahn, M. G., Jacques, M.-A., Gilbertson, R. L., Coaker, G. (2017). Genomic analysis of clavibacter michiganensis reveals insight into virulence strategies and genetic diversity of a gram-positive bacterial pathogen. Mol. Plant-Microbe Interactions® 30 (10), 786–802. doi: 10.1094/MPMI-06-17-0146-R
Thomazella, D. P. D. T., Seong, K., Mackelprang, R., Dahlbeck, D., Geng, Y., Gill, U. S., et al. (2021). Loss of function of a DMR6 ortholog in tomato confers broad-spectrum disease resistance. Proc. Natl. Acad. Sci. 118 (27), e2026152118. doi: 10.1073/pnas.2026152118
van Damme, M., Huibers, R. P., Elberse, J., Van den Ackerveken, G. (2008). Arabidopsis DMR6 encodes a putative 2OG-Fe(II) oxygenase that is defense-associated but required for susceptibility to downy mildew. Plant J. 54 (5), 785–793. doi: 10.1111/j.1365-313X.2008.03427.x
van Schie, C. C., Takken, F. L. (2014). Susceptibility genes 101: how to be a good host. Annu. Rev. Phytopathol. 52, 551–581. doi: 10.1146/annurev-phyto-102313-045854
Wang, D., Pajerowska-Mukhtar, K., Culler, A. H., Dong, X. (2007). Salicylic acid inhibits pathogen growth in plants through repression of the auxin signaling pathway. Curr. Biol. 17 (20), 1784–1790. doi: 10.1016/j.cub.2007.09.025
Xian, L., Yu, G., Wei, Y., Rufian, J. S., Li, Y., Zhuang, H., et al. (2020). A bacterial effector protein hijacks plant metabolism to support pathogen nutrition. Cell Host Microbe 28 (4), 548–57.e7. doi: 10.1016/j.chom.2020.07.003
Keywords: susceptibility genes, Clavibacter michiganensis, CRISPR/Cas9, auxin, ethylene, disease, Walls Are Thin1, bacterium
Citation: Koseoglou E, Hanika K, Mohd Nadzir MM, Kohlen W, van der Wolf JM, Visser RGF and Bai Y (2023) Inactivation of tomato WAT1 leads to reduced susceptibility to Clavibacter michiganensis through downregulation of bacterial virulence factors. Front. Plant Sci. 14:1082094. doi: 10.3389/fpls.2023.1082094
Received: 27 October 2022; Accepted: 05 May 2023;
Published: 31 May 2023.
Edited by:
Michelle Teresa Hulin, The Sainsbury Laboratory, United KingdomReviewed by:
Per Hofvander, Swedish University of Agricultural Sciences, SwedenJohn Mansfield, Imperial College London, United Kingdom
Copyright © 2023 Koseoglou, Hanika, Mohd Nadzir, Kohlen, van der Wolf, Visser and Bai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yuling Bai, YmFpLnl1bGluZ0B3dXIubmw=
 Eleni Koseoglou
Eleni Koseoglou Katharina Hanika1
Katharina Hanika1 Jan M. van der Wolf
Jan M. van der Wolf Richard G. F. Visser
Richard G. F. Visser Yuling Bai
Yuling Bai