- 1MARA Key Laboratory of Sustainable Crop Production in the Middle Reaches of the Yangtze River (Co-construction by Ministry and Province)/Engineering Research Center of Ecology and Agricultural Use of Wetland, Ministry of Education/College of Agriculture, Yangtze University, Jingzhou, China
- 2State Key Laboratory for Biology of Plant Diseases and Insect Pests, Institute of Plant Protection, Chinese Academy of Agricultural Sciences, Beijing, China
Background: The CorA / MGT / MRS2 family proteins are an important group of magnesium transporter proteins that maintain magnesium ion homeostasis in plant cells. However, little is known about the MGT functions in wheat.
Methods: The known MGT sequences were used as queries to BlastP against wheat genome IWGSC RefSeq v2.1 assembly (E-value <10–5). Chromosome localization information for each TaMGT gene was obtained from the GFF3 file of the wheat genome data (IWGSCv2.1).The sequence of 1500 bp upstream of the TaMGT genes was extracted from the wheat genome data. The cis-elements were analyzed using PlantCARE online tool.
Result: A total of 24 MGT genes were identified on 18 chromosomes of wheat. After functional domain analysis, only TaMGT1A, TaMGT1B, and TaMGT1D had GMN mutations to AMN, while all the other genes had conserved GMN tripeptide motifs. Expression profiling showed that the TaMGT genes were differentially expressed under different stresses and at different growth and development stages. The expression levels of TaMGT4B and TaMGT4A were significantly up-regulated in cold damage. In addition, qRT-PCR results also confirmed that these TaMGT genes are involved in the wheat abiotic stress responses.
Conclusion: In conclusion, The results of our research provide a theoretical basis for further research on the function of TaMGT gene family in wheat.
1. Introduction
Magnesium (Mg2+) is an important macronutrient (Williams and Salt, 2009). As the most abundant divalent cation in living plant cells, Mg2+ is involved in chloroplast synthesis, regulation of osmotic pressure, and intracellular enzyme activity (Shaul, 2002; Williams and Salt, 2009; Guo et al., 2016). In addition, Mg2+ is essential for the synthesis of proteins and nucleic acids and maintains the cation-anion balance in cell (Marschner and Marschner, 2012). Moreover, magnesium deficiency can adversely affect plant cells, like reducing macromolecular synthesis, photosynthetic capacity, and plant growth (Hermans et al., 2005; Tang et al., 2012; Yang et al., 2012; Peng et al., 2015; Farhat et al., 2016; Li et al., 2016). Therefore, maintaining the balance and stability of Mg is crucial in plants (Shaul, 2002; Kobayashi and Tanoi, 2015).
Magnesium transporter proteins (MGTs), also known as Magnesium Transporter MRS2, play a critical role in maintaining Mg homeostasis in plant cells (Li et al., 2017). ZmMGT10 has the ability to transport Mg under conditions of Mg deficiency in Maize (Li et al., 2016). The transgenic Arabidopsis plants with ZmMGT10 overexpression exhibited vigorous growth, such as larger plant size, longer root length, higher fresh weight and increased chlorophyll content compared with the wild-type plants (Li et al., 2017). Moreover, AtMGT7 can maintain normal growth and development in Arabidopsis under low Mg conditions (Mao et al., 2008).
Among the magnesium transporters, the CorA-type magnesium transporter is the most widely studied magnesium transporter protein. This type of magnesium transporter was originally identified in Salmonella typhimurium, and its mutants exhibit resistance to Co2+ growth inhibition (Silver, 1969). CorA is a funnel-shaped homopentamer with two transmembrane structural domains per monomer, the first of which forms the ion conduction pathway. Mg2+ transport firstly involves the binding of a cation to an extracellular binding loop connecting the transmembrane structural domains (Maguire, 2006). These proteins have a variable conserved N-terminal hydrophilic domain of about 260 amino acids and a fairly conserved hydrophobic domain of 55 amino acids (Maguire, 2006). The MPEL sequence and the YGMNF sequence are the most characteristic features of CorA proteins, both of which are located in the loop between the transmembrane helices. Among them, the (Gly-Met-Asn) GMN sequence is essential for the protein function (Palombo et al., 2013).
CorA-type Mg transport proteins mediate the influx and efflux of Mg2+ (Franken et al., 2022; Papp-Wallace and Maguire, 2008). CorA homologous proteins have been identified in yeast, animals and plants (Knoop et al., 2005). In yeast, the ALR1 family confers Al3+ tolerance and encodes the major plasma membrane Mg2+ uptake system (Liu et al., 2002). Overexpression of AtMGT1 in tobacco plants increases Mg concentration and confers tolerance to low Mg environments (Deng et al., 2006).
Mutations in the GMN motif may eliminate Mg2+ transport, but the naturally occurring variants GVN and GIN may be associated with the transport of other divalent cations (Eshaghi et al., 2006). The GMN motif was changed to AMN in ZmMGT6 in maize, and in functional complementation experiments using Salmonella typhimurium mutant MM281, ZmMGT6 was shown to rescue the susceptibility of Mg2+ in Salmonella typhimurium mutant MM281, but its complementation efficacy was lower than that of other ZmMGTs containing GMN motifs (Li et al., 2016). In rice, the GMN motif was changed to AMN in OsMRS2-4 and OsMRS2-5 and to GIN in OsMRS2-8, and in functional complementation experiments using Yeast mutant CM66, OsMRS2-4, -5, and -8 did not observe complementation ability, but other OsMRS2 genes had obvious complementation ability (Saito et al., 2013). These phenomena suggest that GMN motif mutations affected gene functions. Overall, these features are important markers of Mg2+ transporters. (Szegedy and Maguire, 1999; Knoop et al., 2005).
In plants, MGT proteins have been reported in Arabidopsis thaliana (Schock et al., 2000), maize (Zea mays) (Li et al., 2016), and rice (Oryza sativa) (Saito et al., 2013). Recently, it has been studied in tomato (Solanum lycopersicum) (Regon et al., 2019), sugarcane (Saccharum spontaneum) (Wang et al., 2019), pear (Pyrus bretschneideri) (Zhao et al., 2018), citrus (Poncirus trifoliata) (Liu et al., 2019), Vitis vinifera (Ge et al, 2022), Citrullus lanatus and Cucumis sativus (Heidari et al, 2022), Theobroma cacao, Corchorus capsularis, and Gossypium hirsutum (Heidari et al, 2021), and Triticum turgidum and Camelina sativa (Faraji et al, 2021). Studies have shown that Mg transporter proteins have different functions. The pear PbrMGT7 gene mediates Mg transport between mitochondria and the cytoplasmic matrix (Zhao et al., 2018). The sugarcane MTG6 gene is the main MGT that maintains the concentration of Mg in chlorophyll and transporting Mg ions into the chloroplast stroma, and MGT10 transports Mg from roots to leaves over long distances (Wang et al., 2019). Banana MaMRS2-5 and MaMRS2-7 are involved in the uptake and transport of Mg and its partitioning between different tissues (Tong et al., 2020). The Arabidopsis AtMGT5 and AtMGT9 genes play critical roles in Mg supply during pollen mitosis and pollen in formation (Drummond et al., 2006; Chen et al., 2009; Li et al., 2015). In the Malvaceae family, MGTs appear to be involved in various pathways that control plant growth and development and respond to adverse conditions (Heidari et al, 2021). At the same time, MGTs can also affect the biosynthesis of phytohormones related to stresses by regulating Mg2+ concentration, thereby enhancing plant stress resistance (Guo et al., 2014).
Wheat (Triticum aestivum) is an important food crop worldwide, and Mg2+ deficiency may affect chlorophyll synthesis, multiple enzymes activation, photosynthesis, and partitioning and utilization of photoassimilates, which may inhibit the growth and development of wheat and further lead to yield loss (Shaul, 2002; Cakmak and Yazici, 2010). To date, studies on Mg2+ transporters in wheat have not been reported. In this study, the Mg2+ transporter genes in wheat were identified, their physicochemical properties, chromosomal location and gene structure were systematically analyzed, and their responses to Mg deficiency, Al stress, and abscisic acid treatment were investigated. The present study will provide a basis for further revealing the biological functions of MGT genes in wheat.
2. Materials and methods
2.1. Identification of MGT genes in wheat
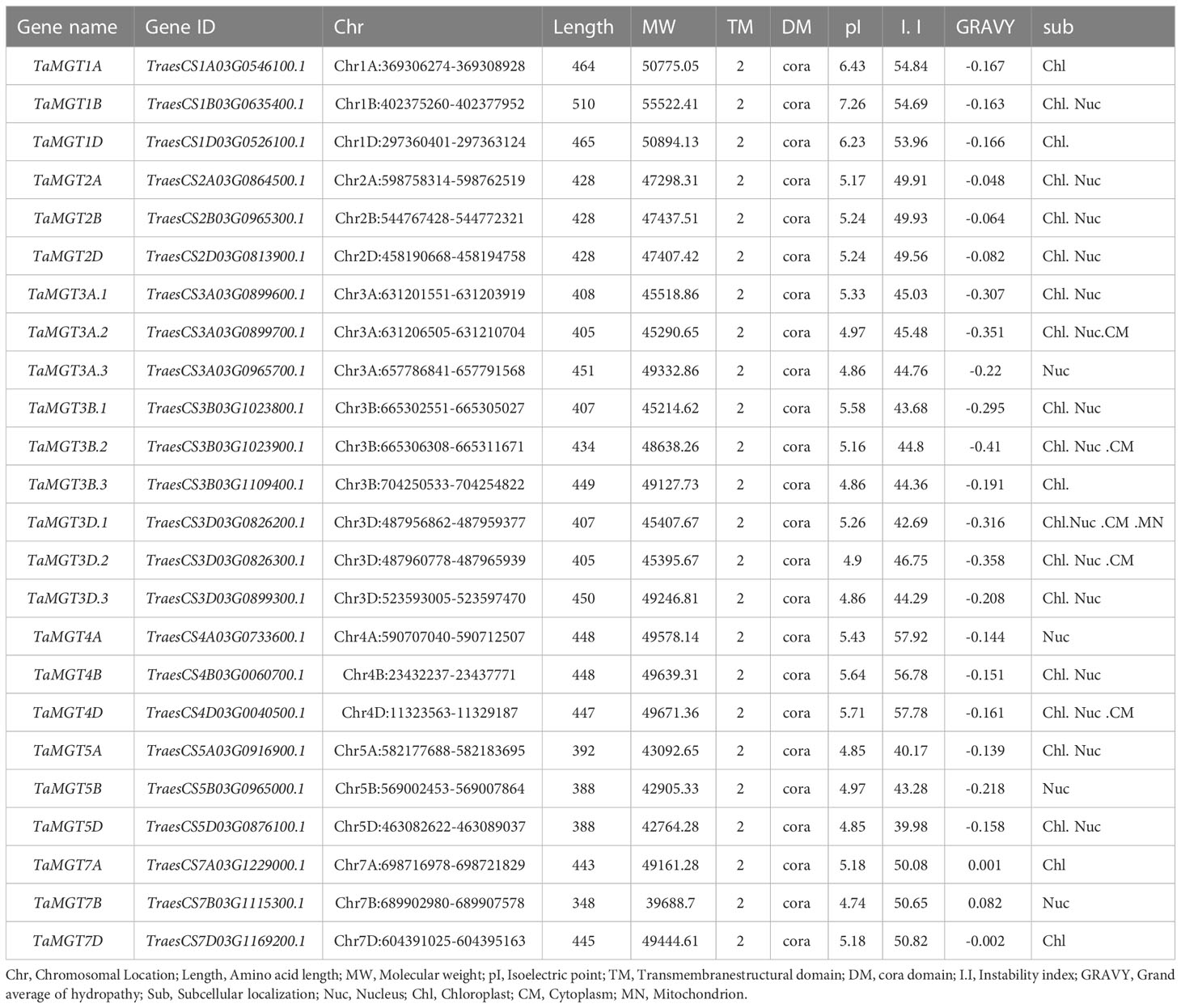
In this experiment, 10 MGT protein sequences from Arabidopsis, 9 from rice, and 12 from maize were obtained from the Arabidopsis genome database (https://www.arabidopsis.org/), the rice genome database (http://Rice.plantbiology.msu.edu), and the maize genome database (https://www.maizegdb.org/), respectively. The known MGT sequences were used as queries to BlastP against wheat genome IWGSC RefSeq v2.1 assembly (E-value <10-5). Duplicates and mismatched candidate proteins were removed. The conserved protein domains were screened through the Pfam database (http://pfam.xfam.org/) (Finn et al., 2006) and the SMART database (http://smart.embl-heidelberg.de/) (Letunic et al., 2004). TaMGT family members were finally determined after excluding the sequences that did not contain the CorA type domain. Subcellular localization prediction of TaMGTs was performed via the Plant-mPLoc online tool (http://www.csbio.sjtu.edu.cn/bioinf/plant-multi/). Potential TM regions in each TaMGT protein were predicted using TMHM Server v2.0 (http://www.cbs.dtu.dk/services/TMHMM/) (Krogh et al., 2001). Protein length, average molecular weight, isoelectric point (pI), instability index, and mean hydrophilicity value (GRAVY) were predicted by ExPASy Server10 (SIB Bioinformatics Resource Portal, https://prosite.expasy.org/PS50011).
2.2. Construction of Phylogenetic tree, gene structure and protein motif analysis
The protein sequences of 10 AtMGTs, 9 OsMGTs, 12 ZmMGTs, and TaMGTs identified in present study were collected. Multiple sequence alignment was performed using ClustalW2 (Thompson et al., 1994). The TM structural domain and conserved GMN motifs were annotated using DNAMAN software (Zhao et al., 2012). The phylogenetic tree was constructed using MEGA7.0 software (version 7.0, Mega Limited, Auckland, New Zealand) using Neighbor-joining (NJ) method (Kumar et al., 2016) with the Bootstrap value setting as 1000. Furthermore, the phylogenetic tree was modified via the online tool iTOL (version 3.2.317, http://itol.embl.de) (Letunic and Bork, 2019).
To study the gene structures of TaMGTs genes, the genome annotation information of TaMGTs was obtained from wheat genome database IWGSC V2.1. And the gene structures were visualized by TBtools (Chen et al., 2020). The conserved protein motifs were analyzed by MEME Suite 5.1.1, with the motif number setting as 20 and other parameters with default values.
2.3. Chromosomal localization and gene duplication events
Chromosome localization information for each TaMGT gene was obtained from the GFF3 file of the wheat genome data (IWGSCv2.1). The chromosome distribution map of the TaMGT gene was subsequently generated using MapInspect software. MCScanX software was used to analyze TaMGT gene duplication events in wheat and the homology of MGT genes between wheat and other selected species(Wang et al., 2012). Duplicate gene pairs among TaMGT members were identified and visualized using the R package “circlize”.
2.4. Cis-elements and transcriptome expression analysis
To identify cis-regulatory elements in the promoter regions, the sequence of 1500 bp upstream of the TaMGT genes was extracted from the wheat genome data. The cis-elements were analyzed using PlantCARE online tool (Lescot et al., 2002). The data was collected and visualized using the R package “pheatmap”.
The RNA-seq data of TaMGTs was downloaded through the NCBI SRA database (The SRA numbers are detailed in Table S3)(Ramírez-González et al., 2018) and the raw data was aligned to the wheat reference genome by Hisat2 (Kim et al., 2015). The genes were then assembled by cufflinks to detect the expression levels of TaMGTs (Fragments per Kilobase Million of exon model per Million mapped fragments, FPKM) (Trapnell et al., 2012). Convert all the transcription data through log2, P<0.05. Heatmaps were created using the R package “pheatmap” to show the expression patterns of TaMGT genes under different conditions.
2.5. Plant materials and stress treatments
Seeds of the hexaploid wheat variety “Yangmai 158” were surface sterilized in 0.5% (w/v) sodium hypochlorite for 15 min. After germinated in a greenhouse at 25°C for 3 d, the seedlings were transferred to Hoagland solution (pH5.8) and were grown under standard greenhouse conditions. The parameters were set as follows: 16h/25°C and 8h/20°C diurnal cycle, 70% relative humidity, and 300 mmolm-2s-1 strong luminosity. The nutrient solution was replaced every two days.
For Mg2+ deficiency treatment, wheat seedlings at two leaves with a bud stage were transferred to Hogeland solution with MgSO4-7H2O deficient. For aluminium chloride stress treatment, wheat seedlings were transferred to a Hoagland solution supplemented with 60uM aluminium chloride (pH4.5). The roots and leaves of treated seedlings were collected at different time points (6h, 12h, 24h, 48h, and 72h after treatment). For hormone treatment, seedlings were treated with 100 mmol/L abscisic acid. Roots and leaves were sampled at 6h, 12h, 24h, and 48h after treatment. Untreated wheat seedlings were used as control. Three biological replicates were set up for each treatment, with each replicate including three technical replicates. The collected samples were immediately frozen in liquid nitrogen and stored at -80°C for subsequent RNA extraction.
2.6. Real-time quantitative RT-PCR analysis
The relative expression levels of TaMGT genes in roots and leaves of wheat under Mg2+ deficiency (-Mg2+), aluminum stress (+Al), and abscisic acid (+ABA) treatments were analyzed by qRT-PCR. Total RNA was extracted from roots and leaves using TRIzol reagent (Life, USA). cDNA was reverse transcribed using the HiScript II Reverse Transcriptase kit (Vazyme, Nanjing, China).Three biological replicates were performed for each sample, with three technical replicates repeated each. The qPCR primers used in present study were listed in Supplementary Table 1. The Ta2291 gene, which was expressed stably under various conditions, was used as the internal reference gene (Paolacci et al., 2009). The expression levels of TaMGT genes were calculated using 2−ΔΔCt method (Livak and Schmittgen, 2001).
3. Results
3.1. Identification of MGT genes in wheat
Twenty-four putative MGT genes were identified in wheat. The candidate genes were named according to their chromosomal location (Table 1). The predicted molecular weights of TaMGTs were ranged from 39688.7D to 55522.41D. The protein isoelectric points were ranged from 4.74 to 7.26, of which TaMGT1A, TaMGT1B, and TaMGT1D had isoelectric points greater than 6, and more interestingly, they were triplet homologous genes. The protein instability index showed that only TaMGT5D was less than 40, which indicated that it was a stable protein. Analysis of the predicted hydrophobicity of the proteins showed that TaMGT7A and TaMGT7B are hydrophobic proteins. The results of subcellular localization analysis of these 24 proteins showed that 20 of them were widely localized in chloroplasts, and might also be localized in mitochondria and cytoplasm. TaMGT3A.3, TaMGT4A, TaMGT5B and TaMGT7B were only localized in the nucleus (Table 1). The TM domain analysis of TaMGT showed that similar to CorA/MRS2/MGT transport proteins in other species (including fungi), all 24 members contain two hypothetical TM domains at the C-terminus (Supplementary Figure 1) and cora-type domain. Multiple sequence alignment of TaMGTs showed that TM1 was more conserved than TM2. Moreover, the GMN tripeptide motif was found to be present in all members except for TaMGT1A, TaMGT1B, and TaMGT1D, which had the mutated motif AMN (alanine-methionine-aspartate) (Figure 1).
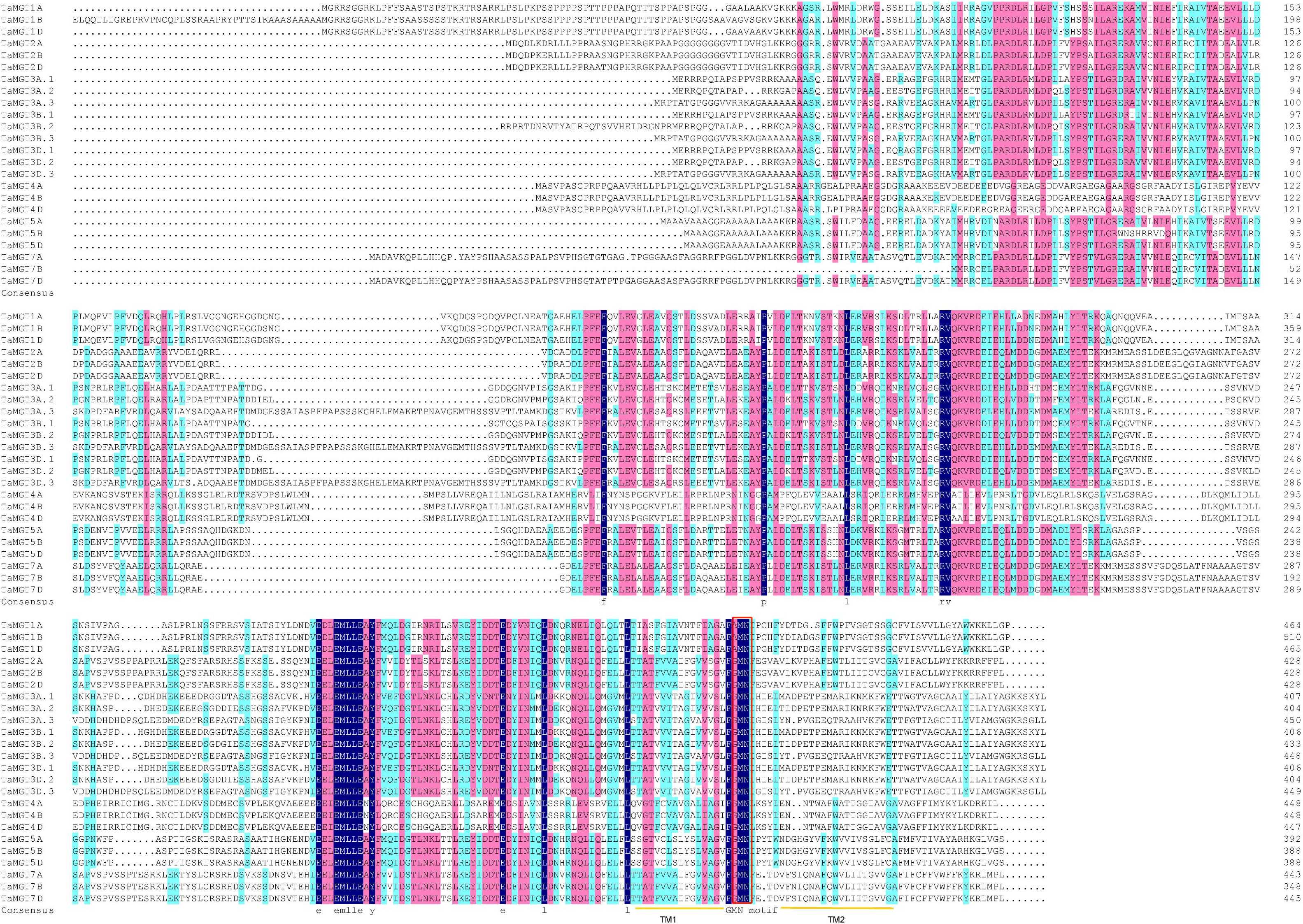
Figure 1 Multiple sequence alignment of TaMGTs. Multiple alignments were performed using DNAMAN software. TM domains are marked with orange lines. Conserved GMN motif is indicated by Red box.
3.2. Construction of phylogenetic tree, protein motif and gene structure analysis
To better understand the evolutionary relationships of TaMGT with other species, phylogenetic trees were constructed for 10 Arabidopsis, 9 rice, 12 maize, and 24 wheat MGTs. As shown in Figure 2, all MGTs were divided into four groups, each group contained members from different plant species. Among them, the second group with the most proteins also has the most members of TaMGTs. The phylogenetic tree (Figure 3) showed that the 24 TaMGTs were also divided into four groups, which is consistent with the phylogenetic tree results of the above four plant species. Moreover, members from the same subgroup have similar motif types and structures. It is worth noting that TaMGT1A, TaMGT1B, TaMGT1D in Group IV lacked Motif 11, TaMGT7B in Group I lacked Motif 12, and TaMGT7A, TaMGT7B, TaMGT7D in Group I contained two Motif 6. The position of the motif varies from subgroup to subgroup, for example, Motif 9 is located at the 5’ end in group 3 and at the 3’ end in other subgroups. Motif 1 and Motif 6 were located in the functional structural domain of TaMGTs.
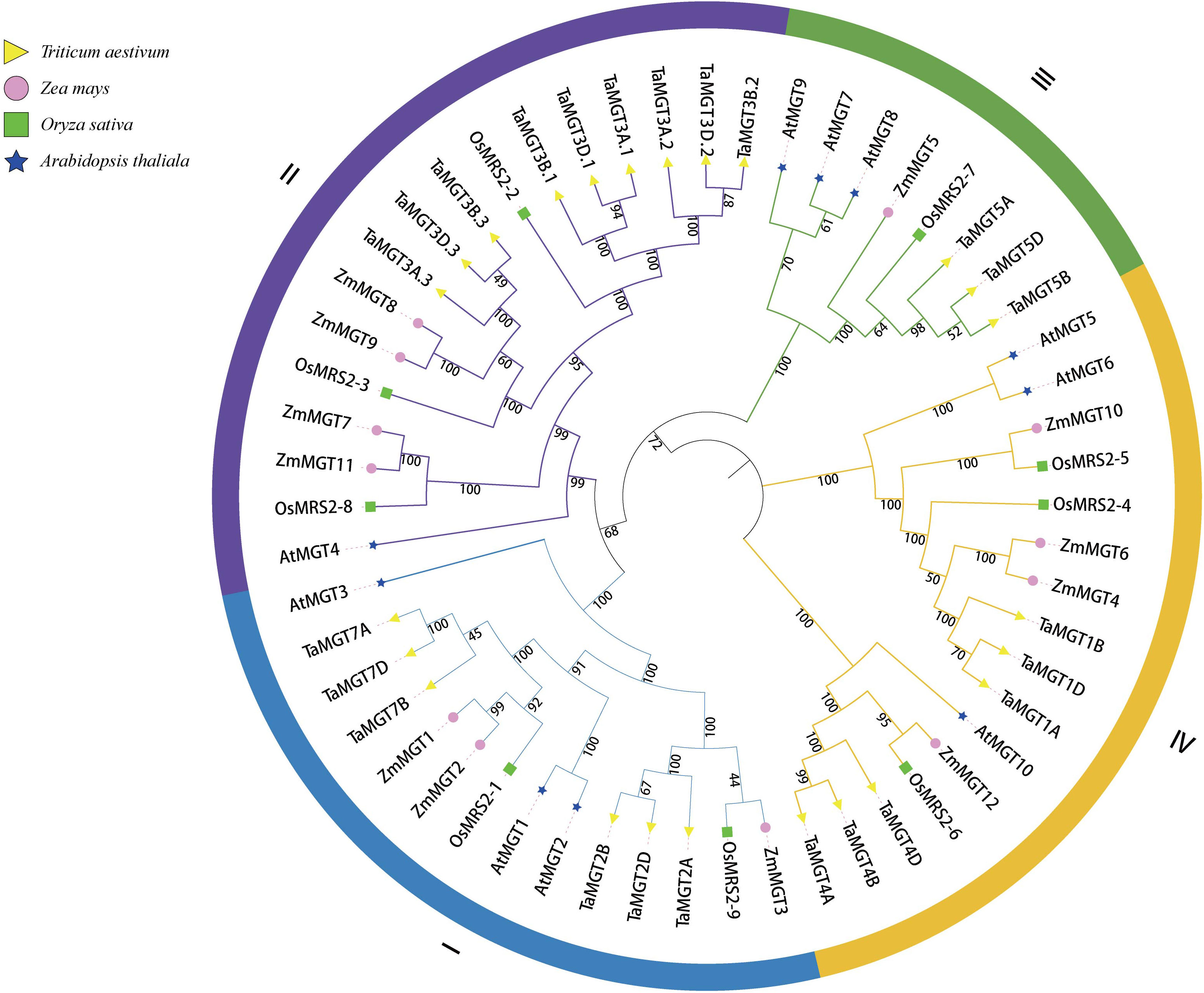
Figure 2 Phylogenetic analysis of Arabidopsis, rice, maize and wheat CorA/MRS2/MGT members. Phylogenetic trees were constructed by the neighbor-Joining method (NJ) using MEGA software for 24 TaMGT proteins, 11 Arabidopsis MRS2/MGT proteins, 9 rice proteins, and 12 maize proteins.
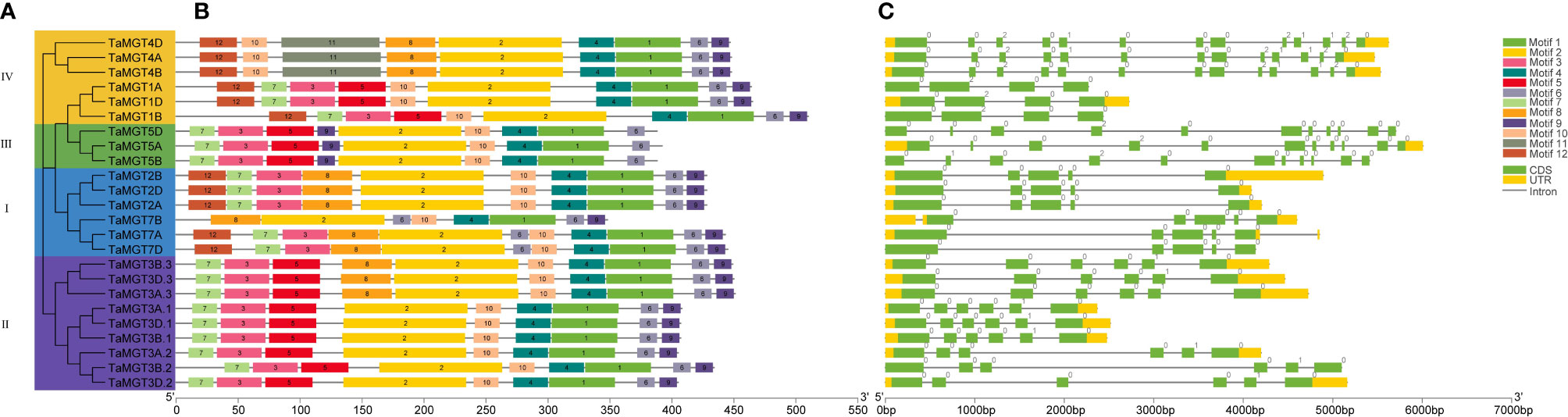
Figure 3 Phylogenetic relationships (A), motif analysis (B) and gene structure (C) of TaMGT family members.
The results of the gene structure analysis (Figure 3C) showed that there are differences in exons/introns among the members in different subgroups. Almost all members of subgroup I and subgroup II contain 4-6 exons/introns and 2 non-coding regions, such as TaMGT2B, TaMGT3B.3, and TaMGT3D.2. Individual TaMGT genes do not have non-coding regions, such as TaMGT7D, TaMG1A, and TaMG1B. While in the fourth subgroup, TaMGT4D, TaMGT4B, and TaMGT4A contain 13 exons, 12 introns and 2 non-coding regions.
3.3. Chromosomal localization and gene duplication events
Based on the GFF3 annotation file, the chromosomal location of TaMGT genes were mapped using MapInspect software (Figure 4A). 24 TaMGT genes were distributed on 18 chromosomes. TaMGT genes were evenly distributed among the three subgenomes of wheat (each subgenome contained 8 genes). However, the distribution of the genes is uneven among the different chromosomes, with three TaMGT genes distributed on chromosome 3 and only one on the other chromosomes.
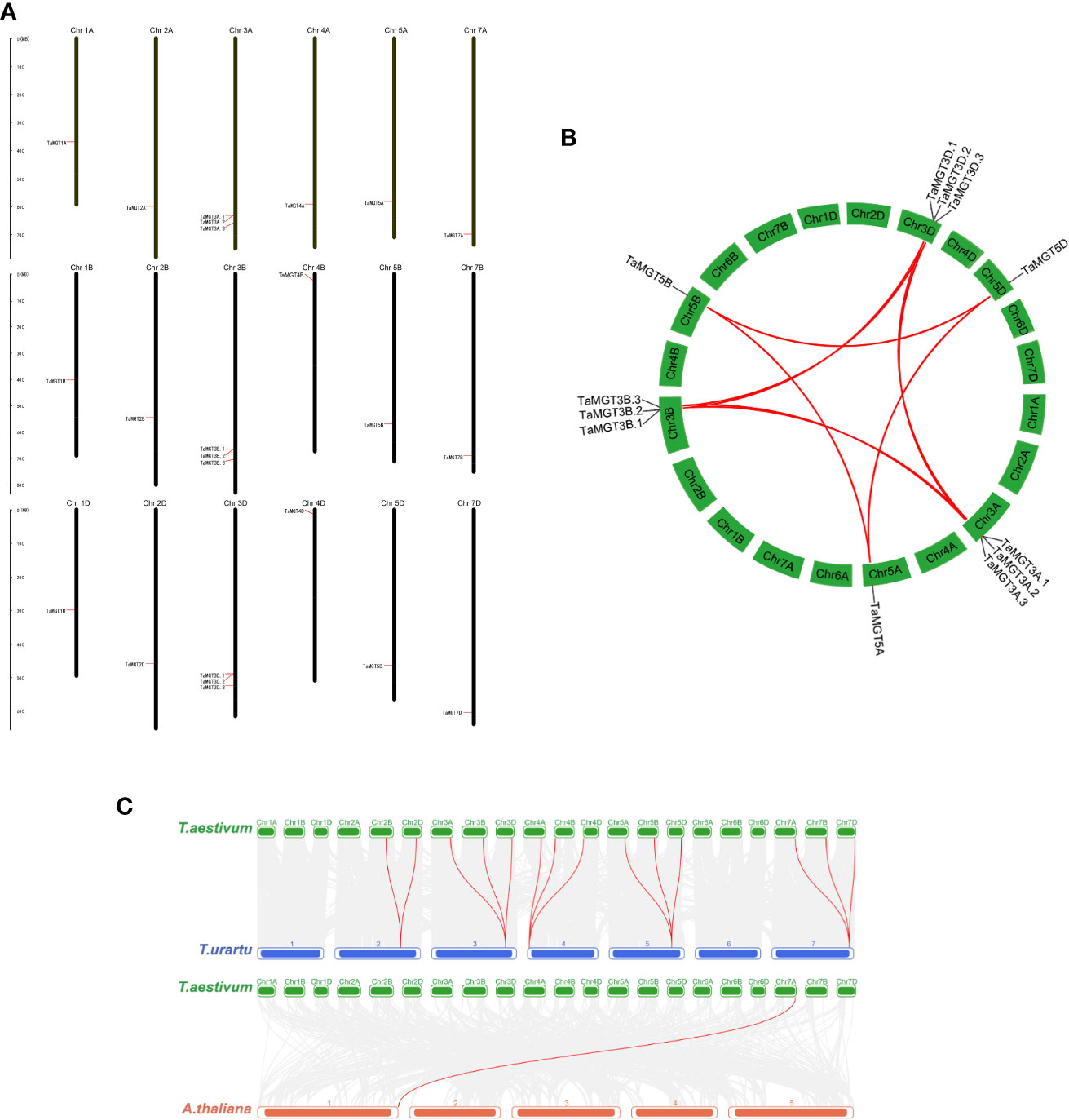
Figure 4 Chromosomal localization and gene duplication of TaMGTs genes. (A) Location of TaMGTs genes on chromosomes, with black representing chromosomes. (B) Gene duplication events in the wheat MGT gene family, where the curves represent gene duplication between chromosomes; the collinearity genes on chromosome 3A, 3B, 3C are highlighted with red lines. (C) Collinearity analysis of wheat with Arabidopsis thaliana and Triticum urartu. The gray line in the background indicates a collinear block in the genome of wheat and Arabidopsis thaliana and Triticum urartu, while the red line highlights the homologous MGT gene pair.
Gene duplication is one of the most important mechanisms by which organisms acquire new genes and create gene novelty (Song et al., 2019). Gene duplication consists of both tandem duplication and segmental duplication (Vision et al., 2000). In order to obtain the amplification mechanism of the TaMGT genes, the collinearity among TaMGT genes was analyzed by MCScanX software (Figure 4B). The results showed that there were twelve pairs of segmental duplication events among TaMGT genes.And most of the duplicated TaMGT genes were located on different chromosomes, and no tandem duplication relationships were found among TaMGT members on the same chromosome. These results suggest that the TaMGT genes may have undergone segmental replications during evolution.
To further analyze the evolutionary and homologous relationships of the MGT family in wheat, collinearity analysis of wheat with Arabidopsis thaliana and Triticum urartu were constructed. The results (Figure 4C) showed that there were 14 MGT homologous gene pairs in wheat and its close relative Triticum urartu, while there were only one MGT homologous gene pair in wheat and dicotyledonous Arabidopsis, which might be due to the short differentiation distance between wheat and its close relatives, with fewer events such as gene loss, insertion and transposition.
3.4. Analysis of Cis-acting elements and transcriptome expression analysis
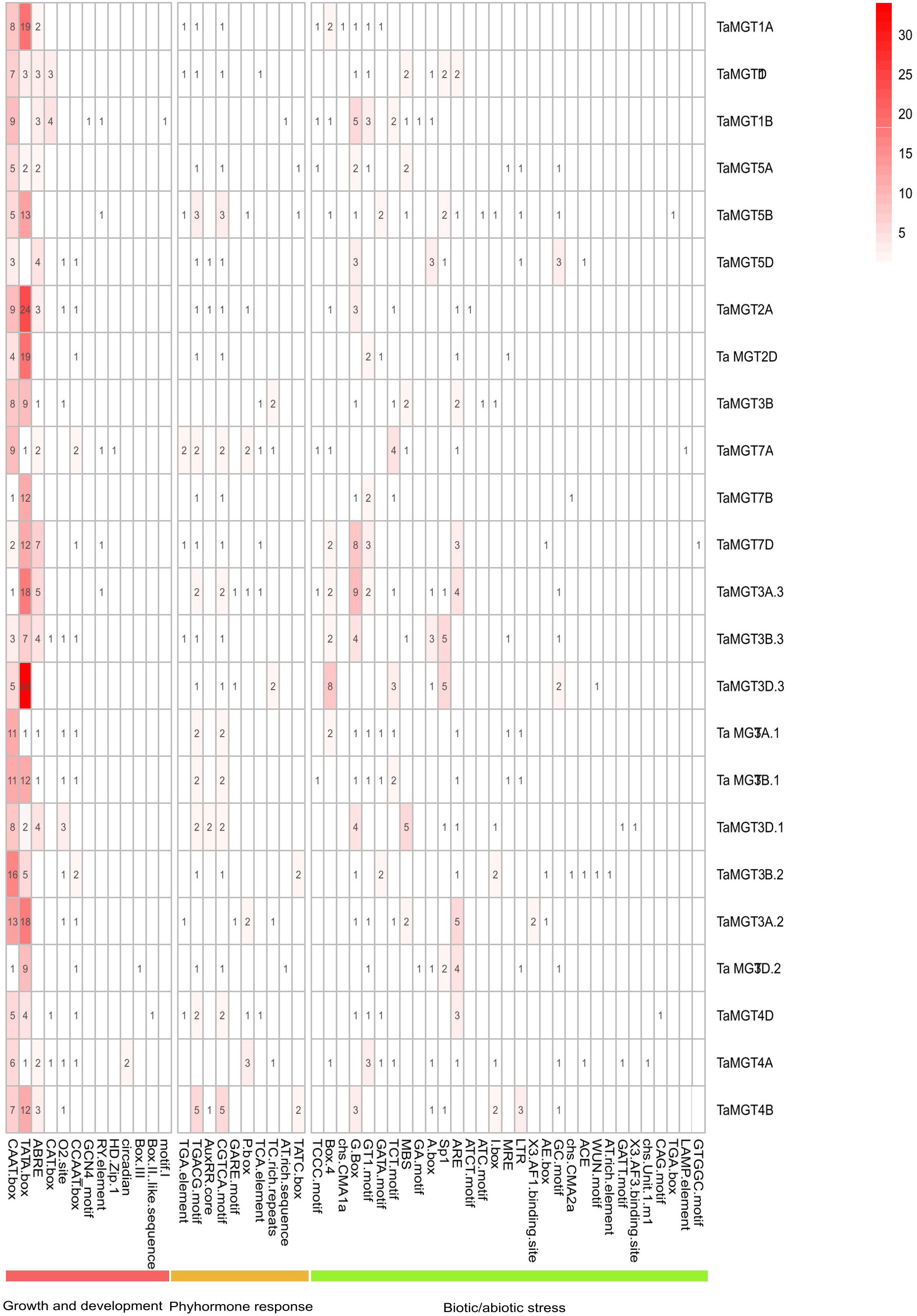
Cis-acting regulatory elements were bound by appropriate transcription factors to control gene transcription (Liu et al., 2013). In this experiment, we identified and summarized the cis-acting elements in the promoter region of the TaMGT gene family related to growth and development, biotic stress and phytohormones (Figure 5 and Supplemental Table 2), and found that there were differences in the number and types of elements contained in the promoters of the TaMGT family. TaMGT3D.3 promoter contained 69 cis-acting elements, while TaMGT7B just contained 20. Among the cis-elements associated with growth and development, the TATA-box was the most. Among the hormone-responsive cis-elements, TGACG-motif and CGTCA-motif were involved in the regulation of the methyl jasmonate response. Among the biotic and abiotic stress-related cis-elements, WUN-motif is a wound responsive element, MYB is a drought-inducibility element, ARE and GC-motif are an anaerobic induction element, and LTR is a low-temperature response element (Bang et al., 2013; Banerjee et al., 2013; Mittal et al., 2009; Kovalchuk et al., 2013; Li et al., 2022). These results suggest that this gene family may also play an important function in the response to adversity stresses.
We used available wheat RNA-seq data (Supplemental information: Tables S3, S4) to analyze the expression levels of the TaMGT gene at different growth and developmental stages of wheat, and the results were shown in Figure 6. These TaMGTs were expressed in grains, headings, stems, leaves, roots, and seeds. According to the clustering results of their expression levels, these TaMGTs can be divided into four groups, in which the genes of Groups 2 and 3 were expressed or lowly expressed in all tissues, while the expression levels of genes in Group 4 were much higher. For example, TaMGT5D, TaMGT5A, and TaMGT5B genes were highly expressed in all five tissues; TaMGT3D.3 and TaMGT3A.3 were abundantly expressed in stems, but not expressed or lowly expressed in other tissues, indicating that TaMGT3D.3 and TaMGT3A.3 may play important roles in stem development. In addition, the expression levels of these genes in the seeds also varied, with ten genes expressed in small amounts and the remaining genes hardly expressed. These results suggest that the differential expression of TaMGTs may play important but distinct roles in different tissues.
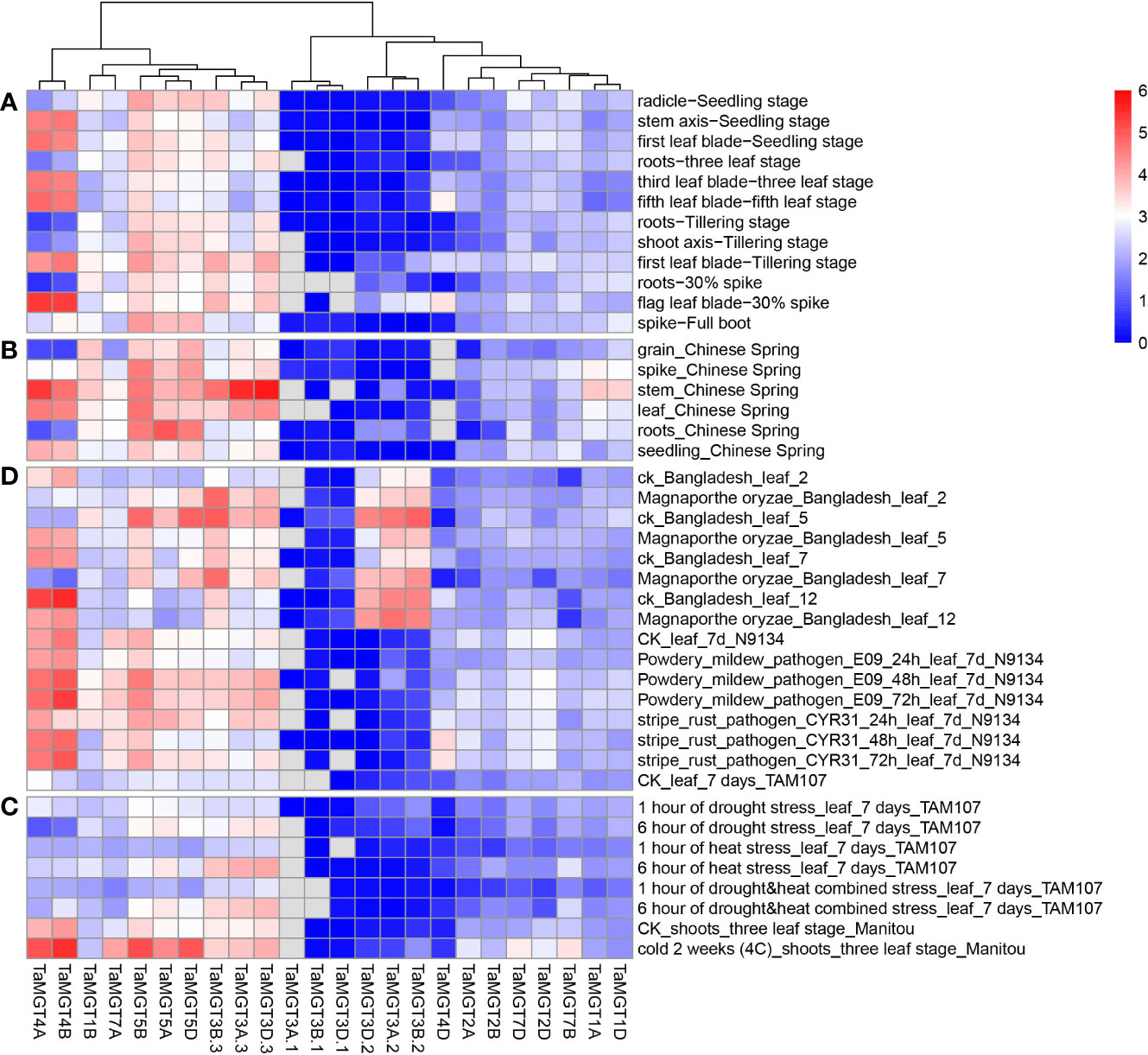
Figure 6 (A) different growth and developmental stages expression analysis (B) different tissues expression analysis (C) abiotic stresses expression analysis (D) biotic stresses expression analysis.
In addition to detecting the expression levels of each TaMGTs in different tissues, we also analyzed their expressions at different developmental stages. Based on the clustering results of their expression levels, TaMGT4A and TaMGT4B were highly expressed at the stem axis-seedling stage, the first leaf-seedling stage, the third leaf-three-leaf stage, the first leaf tillering stage, the root-three-leaf stage, and the flag leaf-30% Ear stage. While almost no expression was found in other periods. These two genes may be mainly involved in the regulation of these six tissue developmental stages in wheat.
Next, we analyzed the expression differences of TaMGTs under biotic and abiotic stresses. As shown in Figure 6C, all TaMGTs genes were not significantly induced under drought stress. In drought stress treatment, the expression level of TaMGT5A, TaMGT3A.3, and TaMGT3D.3 genes gradually increased with the increaseing of treatment time, while the expression of TaMGT4A and TaMGT4B genes gradually decreased. It can be seen that the genes in the 4 groups were affected to a certain extent by the increase of processing time. The expression levels of TaMGT3D.3, TaMGT3A.3, and TaMGT3B.3 increased significantly with the prolongation of drought stress, heat stress, and drought and heat stress, indicating that these genes may be related to the degree of abiotic stress in plants.
After cold treatment, the expression of TaMGT5D, TaMGT5B, TaMGT4A, and TaMGT4B genes was significantly upregulated, which indicated that these genes might play positive roles against cold stress. Among them, TaMGT4A and TaMGT4B were barely expressed in drought stress, thermal stress, and drought and heat stress, and it is speculated that these two genes may be related to the cold resistance of plants.
In addition, under biotic stress, most TaMGTs had different gene expression patterns after Magnaporthe oryzae infection, indicating that these TaMGTs responded differently to rice blast (Figure 6D). The genes in Group 1 and 3 were unexpressed or low-expressed in all tissues, but those in Group 2 and 4 were expressed at much higher levels and responded significantly to different treatments. After the fifth day of infection, the gene expression level of TaMGT3B.2, TaMGT3A.2, TaMGT3D.2, TaMGT3B.3, TaMGT5D, and TaMGT5B were significantly down-regulated. However, with the increase of infection time, the expression levels of TaMGT4A and TaMBT4B were gradually increased. Therefore, we speculate that TaMGT4A and TaMGT4B may be involved in the response to Magnaporthe oryzae infection.
When plants were inoculated with powdery mildew and stripe rust, the expression patterns of TaMGT genes were similar in response to these two diseases. As the time of infection increases, the expression level of some genes were increased significantly. Among them, TaMGT4A and TaMGT4B had significant responses to powdery mildew and stripe rust. Combined with the previous analysis, it is speculated that these two genes are mainly related to the regulation of plant disease resistance.
3.5. Response characteristics of TaMGT family genes to Mg2+ deficiency, AL stress, and ABA treatment
We analyzed the expression of TaMGT gene in roots and shoots under Mg2+ deficiency conditions using qRT-PCR. The results showed that under the condition of Mg2+ deficiency, the expression level of TaMGT1A was strongly upregulated and peaked at 48h in the leaf and peaked at 6h in the root, and TaMGT5B peaked at 24h in leaf. Compared with the untreated group, TaMGT3B.3, TaMGT4B and TaMGT7A showed a downward trend in leaves and roots. These results indicated that the TaMGT genes responded differently to magnesium deficiency.
Compared with the control group, the gene expression levels of TaMGT1B and TaMGT3B reached the maximum at 24h and 48h respectively (Figure 7) after the plant leaves were exposed to aluminum stress, which was about 10 and 2.75 times higher than those of the control group, respectively, indicating that these two genes may be involved in the regulation of aluminium stress. In addition, the gene expression levels of TaMGT4B, TaMGT5B, and TaMGT7A were significantly decreased during each period, and the expression levels of TaMGT4B and TaMGT7A were gradually down-regulated before 48h, which may be caused by the stress response of plants.
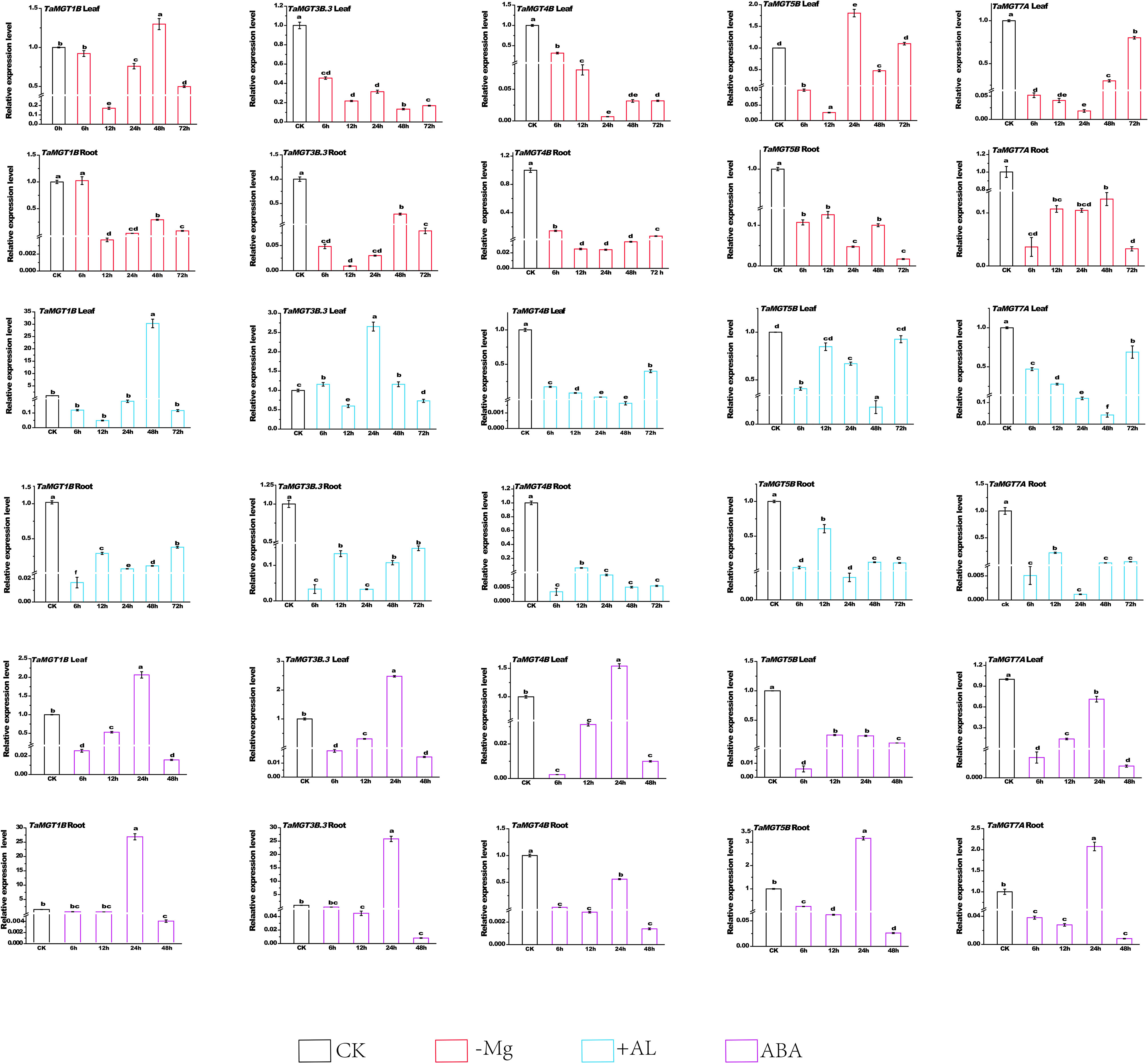
Figure 7 Expression patterns of five TaMGT genes under three different treatments. Three groups of treatments included 60uM aluminum chloride, 100 mmol/L ABA treatment, and magnesium deficiency treatment. The Y-axis represents the relative expression level, and the X-axis represents the time point of treatment. Significant differences at P<0.05 are indicated by letters. The expression levels of TaMGT genes were mapped using Origin software.
Compared with the control group, the expression of all genes was inhibited after the roots of the plant were subjected to aluminum stress. The expression levels of TaMGT5B and TaMGT7A showed a significantly downward trend and began to level-off after 48h. The expression levels of TaMGT3B.3 and TaMGT7A showed the same trend at 6h, 12h, 24h and 48h, with TaMGT7A levelling off after 72h and TaMGT3B.3 having a continued up-regulation trend with increasing treatment time. Gene expression in roots and leaves indicated that gene expression was inhibited under Al stress.
The expression patterns of TaMGTs showed (Figure 7) that after 24 h of abscisic acid treatment, TaMGT5B and TaMGT7A were down-regulated in the leaves and TaMGT4B was down-regulated in the roots. The expression levels of other TaMGT genes peaked in both leaves and roots 24 h after abscisic acid treatment. The expression levels of most TaMGT genes showed a tendency to descend and then rise under abscisic acid treatment. The above results show that the TaMGT family gene has a different role in wheat response to abscisic acid treatment.
4. Discussion
Although the magnesium transporter protein family genes have been extensively studied in a large number of plant species(Schock et al., 2000; Saito et al., 2013; Zhao et al., 2018; Liu et al., 2019; Heidari et al, 2022), no reports in wheat was published. In this study, 24 TaMGT genes were identified for the first time in the wheat genomeusing bioinformatics methods.
The results of gene structure and motif analysis showed that the deletion of Motif 11 in TaMGT1A, TaMGT1B, TaMGT1D and the deletion of Motif 12 in TaMGT7B may be caused by gene recombination (Melamed-Bessudo et al., 2016), while further experiments are needed to prove it. In addition, members of the MGT family had variations in the number of exons. The exon number can increase the diversity of gene-coding proteins by influencing post-transcriptional processes, such as alternative splicing (Koralewski and Krutovsky, 2011). Those with fewer exons can activate rapidly in response to stress, and these genes play a stronger role in adapting to adverse environmental conditions (Jeffares et al., 2008; Jiang et al., 2022).There were segmental duplication relationships between genes involved in gene duplication in TaMGT gene family, with no signs of tandem duplication relationships (Figure 4B), which is consistent with the previous report of interrelationships of genes in the MGT gene family in other species (Yan et al., 2019). Therefore, it is speculated that the expansion of MGT family caused by segmental duplication plays an important role in the adaptation of wheat to external environmental changes.
Cis-regulatory elements in promoter regions play important roles in the regulation of gene expression (Hernandez-Garcia and Finer, 2014). MGT genes have a high potential to respond to stresses, including hormones, growth and development, cis-related elements of biological stress and abiotic stress. It shows that this family plays a major role in the growth and development of wheat and adapts to the external environments (such as changes in light signal, hormones and stress), and also indicates that there are multiple factors regulating the expression of TaMGT gene. Among them, the TaMGT promoter contained the growth and development response elements was the largest, indicating that TaMGT gene may affect the growth and development of wheat.
Differential analysis of the wheat transcriptome revealed significant differences in the TaMGT family of genes under diverse environmental stresses. TaMGT4A and TaMGT4B maintained high expression levels under different stress conditions. However, TaMGT3B.2, TaMGT3A.2, and TaMGT3D.2 were only expressed under the stress of rice blast. There are few studies on magnesium transporters in response to disease stress. Watermelon MSR2 gene were upregulated in response to mosaic virus stress(Heidari et al, 2022). Arabidopsis MGT gene, AT2G21120, which was recognized using a forward genetic screen, was involved in response to viral infection (Guo et al., 2018). Further studies of the functions of these genes are needed to elucidate the specific mechanisms involved in these phenomena.
Magnesium deficiency has been reported to have an effect on MGT gene expression (Li et al., 2016; Tong et al., 2020), so we analyzed the expression levels of the MGT gene in wheat under the condition of magnesium deficiency by qRT-PCR. The results showed that the expression levels of five TaMGT genes were significantly different in wheat leaves and roots. Studies have shown that AtMGT6 and ZmMGT10 are essential for maintaining Mg homeostasis and are highly expressed in roots under Mg deficiency (Mao et al., 2014; Li et al., 2017; Tang and Luan, 2017). Phylogenetic analysis showed that TaMGT1B, a homologue of AtMGT6 and ZmMGT10, was significantly up-regulated in Mg-deficient roots and leaves. AtMGT6 mediates Mg2+ uptake in roots and is required for plant adaptation to a low-Mg2+ environment (Mao et al., 2014). Whether TaMGT1B has the same function needs further experiments. AtMGT7 is involved in Mg uptake and can maintain normal Arabidopsis growth and development under low Mg or Mg-deficient conditions (Mao et al., 2008). TaMGT5B, a homolog of AtMGT7 and OsMRS2-7, was highly upregulated in leaves. It has been shown that branch A of OSMRS2-6 and AtMRS2-11 has been defined as chloroplast transporter group (Saito et al., 2013), TaMGT4A, TaMGT4B, TaMGT4D, OSMRS2-6, and AtMRS2-11 were belonged to the same branch (Figure 2), and they may be related to chloroplast metabolism.
Studies have demonstrated that magnesium transporters can alleviate the toxic of Al in the absence of magnesium. Overexpression of Mg transporters (ALR1 or ALR2) in Saccharomyces cerevisiae confers Al tolerance in yeast (MacDiarmid and Gardner, 1998). The OsMGT1 gene, a plasma membrane-localized transporter of Mg in rice, confers Al tolerance by increasing Mg uptake into cells (Chen et al., 2012). Most of the MGT genes in Arabidopsis were not up-regulated by Al treatment (Hoekenga et al., 2003; Sawaki et al., 2009; Zhao et al., 2009). In this study, it was found that most of the MGT genes in wheat were also not up-regulated by Al. This is consistent with related reports that rice is highly tolerant to Al toxicity (Chen et al., 2012).The transgenic lines for AtMGT1 in Nicotiana benthamiana showed a reduction in Al toxicity (Deng et al., 2006). It seems that increasing MGT activity plays an important role in reducing the negative effects of some elements and ions. In the sugarcane MGT family, different genes were expressed differently under different ABA treatment times. The five TaMGT genes (TaMGT1B, TaMGT3B.3, TaMGT4B, TaMGT5B and TaMGT7A) in wheat have the same expression pattern at 12-48h under abscisic acid treatment, which may indicate that these five genes play some function in response to ABA. However, the exact functions performed need to be further verified.
In summary, this study identified 24 MGT-type magnesium transporter proteins and provided a detailed analysis of their classification, protein structure, evolutionary relationships, gene structure and localization. Changes in the CorA GMN motif in Salmonella typhimurium have been used as zinc transporter proteins, which may mediate the entry and exit of Zn but are unable to transport Mg2+ (Worlock and Smith, 2002). Given this situation, it remains to be demonstrated whether mutation of the GMN motif to AMN in wheat has a function in transporting Mg. Transcriptomic analysis and qRT-PCR analysis showed that TaMGTs play an important role in responding to diverse abiotic stresses. TaMGT genes are differentially expressed under Mg-deficient conditions, aluminium stress and abscisic acid treatment. Among them, TaMGT1B may play a prominent function in maintaining Mg homeostasis, Mg uptake and participation in hormone response in plants. The results of this study provide a theoretical basis for an in-depth study of the function of the TaMGT gene family in wheat.
5. Conclusions
In this study, the classification, protein structure, evolutionary relationship, gene structure and localization of 24 TaMGT genes in wheat were analyzed in detail. The quantitative PCR results suggested that TaMGT plays a role in maintaining magnesium homeostasis, magnesium absorption and hormone response in wheat, as well as in response to biological and non-organic important role in biotic stress. The results provide a theoretical basis for further research on the function of TaMGT gene family in wheat.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
ZL, DM, and WC conceived the study and worked on the approval of the manuscript. YT, XY, and HL performed the experiments and wrote the first draft. WC, DM, and ZL revised the manuscript. YS contributed to data analysis. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by the Open Program of Engineering Research Center of Ecology and Agricultural Use of Wetland Ministry of Education (KFT202103) and the open project program of State Key Laboratory for Biology of Plant Diseases and Insect Pests (NO. SKLOF202008), and the National Natural Science Foundation of China (31672088).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1078299/full#supplementary-material
References
Banerjee, J., Sahoo, D. K., Dey, N., Houtz, R. L., Maiti, I. B. (2013). An intergenic region shared by At4g35985 and At4g35987 in arabidopsis thaliana is a tissue specific and stress inducible bidirectional promoter analyzed in transgenic arabidopsis and tobacco plants. PLos One 8, e79622. doi: 10.1371/journal.pone.0079622
Bang, S. W., Park, S. H., Jeong, J. S., Kim, Y. S., Jung, H., Ha, S. H., et al. (2013). Characterization of the stress-inducible OsNCED3 promoter in different transgenic rice organs and over three homozygous generations. Planta 237, 211–224. doi: 10.1007/s00425-012-1764-1
Cakmak, I., Yazici, A. M. (2010). Magnesium: a forgotten element in crop production. Better. Crops 94, 23–25.
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, J., Li, L. G., Liu, Z. H., Yuan, Y. J., Guo, L. L., Mao, D. D., et al. (2009). Magnesium transporter AtMGT9 is essential for pollen development in arabidopsis. Cell Res. 19, 887–898. doi: 10.1038/cr.2009.58
Chen, Z. C., Yamaji, N., Motoyama, R., Nagamura, Y., Ma, J. F. (2012). Up-regulation of a magnesium transporter gene OsMGT1 is required for conferring aluminum tolerance in rice. Plant Physiol. 159, 1624–1633. doi: 10.1104/pp.112.199778
Deng, W., Luo, K., Li, D., Zheng, X., Wei, X., Smith, W., et al. (2006). Overexpression of an arabidopsis magnesium transport gene, AtMGT1, in nicotiana benthamiana confers Al tolerance. J. Exp. Bot. 57, 4235–4243. doi: 10.1093/jxb/erl201
Drummond, R. S. M., Tutone, A. F., Li, Y.-C., Gardner, R. C. (2006). A putative magnesium transporter AtMRS2-11 is localized to the plant chloroplast envelope membrane system. Plant Sci. 170, 78–89. doi: 10.1016/J.PLANTSCI.2005.08.018
Eshaghi, S., Niegowski, D., Kohl, A., Martinez Molina, D., Lesley, S. A., Nordlund, P. (2006). Crystal structure of a divalent metal ion transporter CorA at 2.9 angstrom resolution. Science 313, 354–357. doi: 10.1126/science.1127121
Faraji, S., Ahmadizadeh, M., Heidari, P. (2021). Genome-wide comparative analysis of mg transporter gene family between Triticum turgidum and Camelina sativa. Biometals 34, 639–660. doi: 10.1007/s10534-021-00301-4
Farhat, N., Elkhouni, A., Zorrig, W., Smaoui, A., Abdelly, C., Rabhi, M. (2016). Effects of magnesium deficiency on photosynthesis and carbohydrate partitioning. Acta Physiol. Plant 38, 145. doi: 10.1007/s11738-016-2165-z
Finn, R. D., Mistry, J., Schuster-Böckler, B., Griffiths-Jones, S., Hollich, V., Lassmann, T., et al. (2006). Pfam: clans, web tools and services. Nucleic Acids Res. 34, D247–D251. doi: 10.1093/nar/gkj149
Franken, G. A. C., Huynen, M. A., Martínez-Cruz, L. A., Bindels, R. J. M., de Baaij, J. H. F. (2022). Structural and functional comparison of magnesium transporters throughout evolution. Cell. Mol. Life Sci. 79, 418. doi: 10.1007/s00018-022-04442-8
Ge, M., Zhong, R., Sadeghnezhad, E., Hakeem, A., Xiao, X., Wang, P., et al. (2022). Genome-wide identification and expression analysis of magnesium transporter gene family in grape (Vitis vinifera). BMC Plant Biol. 22 (1), 217. doi: 10.1186/s12870-022-03599-5
Guo, W., Cong, Y., Hussain, N., Wang, Y., Liu, Z., Jiang, L., et al. (2014). The remodeling of seedling development in response to long-term magnesium toxicity and regulation by ABA-DELLA signaling in arabidopsis. Plant Cell Physiol. 55, 1713–1726. doi: 10.1093/pcp/pcu102
Guo, W., Nazim, H., Liang, Z., Yang, D. (2016). Magnesium deficiency in plants: An urgent problem. Crop J. 4, 83–91. doi: 10.1016/J.CJ.2015.11.003
Guo, Z., Wang, X. B., Wang, Y., Li, W. X., Gal-On, A., Ding, S. W. (2018). Identification of a new host factor required for antiviral RNAi and amplification of viral siRNAs. Plant Physiol. 176, 1587–1597. doi: 10.1104/pp.17.01370
Heidari, P., Abdullah, Faraji, S., Poczai, P. (2021). Magnesium transporter gene family: Genome-wide identification and characterization in Theobroma cacao, Corchorus capsularis, and Gossypium hirsutum of family malvaceae. Agronomy 11, 1651. doi: 10.3390/agronomy11081651
Heidari, P., Puresmaeli, F., Mora-Poblete, F. (2022). Genome-wide identification and molecular evolution of the magnesium transporter (MGT) gene family in Citrullus lanatus and Cucumis sativus. Agronomy 12, 2253. doi: 10.3390/agronomy12102253
Hermans, C., Bourgis, F., Faucher, M., Strasser, R. J., Delrot, S., Verbruggen, N. (2005). Magnesium deficiency in sugar beets alters sugar partitioning and phloem loading in young mature leaves. Planta 220, 541–549. doi: 10.1007/s00425-004-1376-5
Hernandez-Garcia, C. M., Finer, J. J. (2014). Identification and validation of promoters and cis-acting regulatory elements. Plant Sci. 217, 109–119. doi: 10.1016/j.plantsci.2013.12.007
Hoekenga, O. A., Vision, T. J., Shaff, J. E., Monforte, A. J., Lee, G. P., Howell, S. H., et al. (2003). Identification and characterization of aluminum tolerance loci in arabidopsis (Landsberg erecta × Columbia) by quantitative trait locus mapping. a physiologically simple but genetically complex trait. Plant Physiol. 132, 936–948. doi: 10.1104/pp.103.023085
Jeffares, D. C., Penkett, C. J., Bähler, J. (2008). Rapidly regulated genes are intron poor. Trends Genet. 24, 375–378. doi: 10.1016/j.tig.2008.05.006
Jiang, B., Liu, Y., Niu, H., He, Y., Ma, D., Li, Y. (2022). Mining the roles of wheat (Triticum aestivum) SnRK genes in biotic and abiotic responses. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.934226
Jiang, W., Yang, L., He, Y., Zhang, H., Li, W., Chen, H., et al. (2019). Genome-wide identification and transcriptional expression analysis of superoxide dismutase (SOD) family in wheat (Triticum aestivum). PeerJ 7, e8062. doi: 10.7717/peerj.8062
Kim, D., Langmead, B., Salzberg, S. L. (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360. doi: 10.1038/nmeth.3317
Knoop, V., Groth-Malonek, M., Gebert, M., Eifler, K., Weyand, K. (2005). Transport of magnesium and other divalent cations: evolution of the 2-TM-GxN proteins in the MIT superfamily. Mol. Genet. Genom. 274, 205–216. doi: 10.1007/s00438-005-0011-x
Kobayashi, N. I., Tanoi, K. (2015). Critical issues in the study of magnesium transport systems and magnesium deficiency symptoms in plants. Int. J. Mol. Sci. 16, 23076–23093. doi: 10.3390/ijms160923076
Koralewski, T. E., Krutovsky, K. V. (2011). Evolution of exon-intron structure and alternative splicing. PLos One 6, e18055. doi: 10.1371/journal.pone.0018055
Kovalchuk, N., Jia, W., Eini, O., Morran, S., Pyvovarenko, T., Fletcher, S., et al. (2013). Optimization of TaDREB3 gene expression in transgenic barley using cold-inducible promoters. Plant Biotechnol. J. 11, 659–670. doi: 10.1111/pbi.12056
Krogh, A., Larsson, B., von Heijne, G., Sonnhammer, E. L. (2001). Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305, 567–580. doi: 10.1006/jmbi.2000.4315
Kumar, S., Stecher, G., Tamura, K. (2016). MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Lescot, M., Déhais, P., Thijs, G., Marchal, K., Moreau, Y., Van de Peer, Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Letunic, I., Bork, P. (2019). Interactive tree of life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 47, W256–W259. doi: 10.1093/nar/gkz239
Letunic, I., Copley, R. R., Schmidt, S., Ciccarelli, F. D., Doerks, T., Schultz, J., et al. (2004). SMART 4.0: towards genomic data integration. Nucleic Acids Res. 32, D142–D144. doi: 10.1093/nar/gkh088
Li, H., Du, H., Huang, K., Chen, X., Liu, T., Gao, S., et al. (2016). Identification, and functional and expression analyses of the CorA/MRS2/MGT-type magnesium transporter family in maize. Plant Cell Physiol. 57, 1153–1168. doi: 10.1093/pcp/pcw064
Li, J., Huang, Y., Tan, H., Yang, X., Tian, L., Luan, S., et al. (2015). An endoplasmic reticulum magnesium transporter is essential for pollen development in arabidopsis. Plant Sci. 231, 212–220. doi: 10.1016/j.plantsci.2014.12.008
Li, Y., Liu, X., Xiao, Y., Wen, Y., Li, K., Ma, Z., et al. (2022). Genome-wide characterization and function analysis uncovered roles of wheat LIMs in responding to adverse stresses and TaLIM8-4D function as a susceptible gene. Plant Genome 15, e20246. doi: 10.1002/tpg2.20246
Liu, X., Guo, L. X., Luo, L. J., Liu, Y. Z., Peng, S. A. (2019). Identification of the magnesium transport (MGT) family in poncirus trifoliata and functional characterization of PtrMGT5 in magnesium deficiency stress. Plant Mol. Biol. 101, 551–560. doi: 10.1007/s11103-019-00924-9
Liu, G. J., Martin, D. K., Gardner, R. C., Ryan, P. R. (2002). Large Mg(2+)-dependent currents are associated with the increased expression of ALR1 in saccharomyces cerevisiae. FEMS Microbiol. Lett. 213, 231–237. doi: 10.1111/j.1574-6968.2002.tb11311.x
Liu, Y., Wang, L., Xing, X., Sun, L., Pan, J., Kong, X., et al. (2013). ZmLEA3, a multifunctional group 3 LEA protein from maize (Zea mays l.), is involved in biotic and abiotic stresses. Plant Cell Physiol. 54, 944–959. doi: 10.1093/pcp/pct047
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Li, H., Wang, N., Ding, J., Liu, C., Du, H., Huang, K., et al. (2017). The maize CorA/MRS2/MGT-type mg transporter, ZmMGT10, responses to magnesium deficiency and confers low magnesium tolerance in transgenic arabidopsis. Plant Mol. Biol. 95, 269–278. doi: 10.1007/s11103-017-0645-1
MacDiarmid, C. W., Gardner, R. C. (1998). Overexpression of the saccharomyces cerevisiae magnesium transport system confers resistance to aluminum ion. J. Biol. Chem. 273, 1727–1732. doi: 10.1074/jbc.273.3.1727
Maguire, M. E. (2006). Magnesium transporters: properties, regulation and structure. Front. Biosci. 11, 3149–3163. doi: 10.2741/2039
Mao, D., Chen, J., Tian, L., Liu, Z., Yang, L., Tang, R., et al. (2014). Arabidopsis transporter MGT6 mediates magnesium uptake and is required for growth under magnesium limitation. Plant Cell 26, 2234–2248. doi: 10.1105/tpc.114.124628
Mao, D. D., Tian, L. F., Li, L. G., Chen, J., Deng, P. Y., Li, D. P., et al. (2008). AtMGT7: An arabidopsis gene encoding a low-affinity magnesium transporter. J. Integr. Plant Biol. 50, 1530–1538. doi: 10.1111/j.1744-7909.2008.00770.x
Marschner, H., Marschner, P. (2012). Marschner’s mineral nutrition of higher plants (Beijing: Science Press).
Melamed-Bessudo, C., Shilo, S., Levy, A. A. (2016). Meiotic recombination and genome evolution in plants. Curr. Opin. Plant Biol. 30, 82–87. doi: 10.1016/j.pbi.2016.02.003
Mittal, D., Chakrabarti, S., Sarkar, A., Singh, A., Grover, A. (2009). Heat shock factor gene family in rice: genomic organization and transcript expression profiling in response to high temperature, low temperature and oxidative stresses. Plant Physiol. Biochem. 47, 785–795. doi: 10.1016/j.plaphy.2009.05.003
Palombo, I., Daley, D. O., Rapp, M. (2013). Why is the GMN motif conserved in the CorA/Mrs2/Alr1 superfamily of magnesium transport proteins? Biochemistry 52, 4842–4847. doi: 10.1021/bi4007397
Paolacci, A. R., Tanzarella, O. A., Porceddu, E., Ciaffi, M. (2009). Identification and validation of reference genes for quantitative RT-PCR normalization in wheat. BMC Mol. Biol. 10, 11. doi: 10.1186/1471-2199-10-11
Papp-Wallace, K. M., Maguire, M. E. (2008). Magnesium transport and magnesium homeostasis. EcoSal. Plus. 3, 1–18. doi: 10.1128/ecosalplus.5.4.4.2
Peng, H. Y., Qi, Y. P., Lee, J., Yang, L. T., Guo, P., Jiang, H. X., et al. (2015). Proteomic analysis of citrus sinensis roots and leaves in response to long-term magnesium-deficiency. BMC Genom. 16, 253. doi: 10.1186/s12864-015-1462-z
Ramírez-González, R. H., Borrill, P., Lang, D., Harrington, S. A., Brinton, J., Venturini, L., et al. (2018). The transcriptional landscape of polyploid wheat. Science 361, eaar6089. doi: 10.1126/science.aar6089
Regon, P., Chowra, U., Awasthi, J. P., Borgohain, P., Panda, S. K. (2019). Genome-wide analysis of magnesium transporter genes in solanum lycopersicum. Comput. Biol. Chem. 80, 498–511. doi: 10.1016/j.compbiolchem.2019.05.014
Saito, T., Kobayashi, N. I., Tanoi, K., Iwata, N., Suzuki, H., Iwata, R., et al. (2013). Expression and functional analysis of the CorA-MRS2-ALR-type magnesium transporter family in rice. Plant Cell Physiol. 54, 1673–1683. doi: 10.1093/pcp/pct112
Sawaki, Y., Iuchi, S., Kobayashi, Y., Kobayashi, Y., Ikka, T., Sakurai, N., et al. (2009). STOP1 regulates multiple genes that protect arabidopsis from proton and aluminum toxicities. Plant Physiol. 150, 281–294. doi: 10.1104/pp.108.134700
Schock, I., Gregan, J., Steinhauser, S., Schweyen, R., Brennicke, A., Knoop, V. (2000). A member of a novel arabidopsis thaliana gene family of candidate Mg2+ ion transporters complements a yeast mitochondrial group II intron-splicing mutant. Plant J. 24, 489–501. doi: 10.1046/j.1365-313x.2000.00895.x
Shaul, O. (2002). Magnesium transport and function in plants: the tip of the iceberg. Biometals 15, 309–323. doi: 10.1023/a:1016091118585
Silver, S. (1969). Active transport of magnesium in escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 62, 764–771. doi: 10.1073/pnas.62.3.764
Song, S., Hao, L., Zhao, P., Xu, Y., Zhong, N., Zhang, H., et al. (2019). Genome-wide identification, expression profiling and evolutionary analysis of auxin response factor gene family in potato (Solanum tuberosum Group Phureja). Sci. Rep. 9, 1755. doi: 10.1038/s41598-018-37923-7
Szegedy, M. A., Maguire, M. E. (1999). The CorA Mg(2+) transport protein of salmonella typhimurium. mutagenesis of conserved residues in the second membrane domain. J. Biol. Chem. 274, 36973–36979. doi: 10.1074/jbc.274.52.36973
Tang, N., Li, Y., Chen, L. (2012). Magnesium deficiency–induced impairment of photosynthesis in leaves of fruiting citrus reticulata trees accompanied by up-regulation of antioxidant metabolism to avoid photo-oxidative damage. J. Plant Nutr. Soil Sci. 175, 784–793. doi: 10.1002/JPLN.201100329
Tang, R. J., Luan, S. (2017). Regulation of calcium and magnesium homeostasis in plants: from transporters to signaling network. Curr. Opin. Plant Biol. 39, 97–105. doi: 10.1016/j.pbi.2017.06.009
Thompson, J. D., Higgins, D. G., Gibson, T. J. (1994). CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680. doi: 10.1093/nar/22.22.4673
Tong, M., Liu, W., He, H., Hu, H., Ding, Y., Li, X., et al. (2020). Identification and functional analysis of the CorA/MGT/MRS2-type magnesium transporter in banana. PLos One 15, e0239058. doi: 10.1371/journal.pone.0239058
Trapnell, C., Roberts, A., Goff, L., Pertea, G., Kim, D., Kelley, D. R., et al. (2012). Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and cufflinks. Nat. Protoc. 7, 562–578. doi: 10.1038/nprot.2012.016
Vision, T.J., Brown, D.G., Tanksley, S.D. (2000). The origins of genomic duplications in Arabidopsis. Science 290, 2114–2117. doi: 10.1126/science.290.5499.2114
Wang, Y., Hua, X., Xu, J., Chen, Z., Fan, T., Zeng, Z., et al. (2019). Comparative genomics revealed the gene evolution and functional divergence of magnesium transporter families in saccharum. Curr. Opin. Plant Biol. 12, 247–249. doi: 10.1186/s12864-019-5437-3
Wang, Y., Tang, H., Debarry, J. D., Tan, X., Li, J., Wang, X., et al. (2012). MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40, e49. doi: 10.1093/nar/gkr1293
Williams, L., Salt, D. E. (2009). The plant ionome coming into focus. Curr. Opin. Plant Biol. 12 (3), 247. doi: 10.1016/j.pbi.2009.05.009
Worlock, A. J., Smith, R. L. (2002). ZntB is a novel Zn2+ transporter in salmonella enterica serovar typhimurium. J. Bacteriol. 184, 4369–4373. doi: 10.1128/jb.184.16.4369-4373.2002
Yang, G. H., Yang, L. T., Jiang, H. X., Li, Y., Wang, P., Chen, L. S. (2012). Physiological impacts of magnesium-deficiency in citrus seedlings: photosynthesis, antioxidant system and carbohydrates. Trees 26, 1237–1250. doi: 10.1007/s00468-012-0699-2
Yan, H., Wu, F., Jiang, G., Xiao, L., Li, Z., Duan, X., et al. (2019). Genome-wide identification, characterization and expression analysis of NF-y gene family in relation to fruit ripening in banana. Postharvest. Biol. Technol. 151, 98–110. doi: 10.1016/j.postharvbio.2019.02.002
Zhao, L. M. (2016). Bioinformatics analysis of two f-box genes in rice. Hubei Agric. Sci. 9, 2396–2399. doi: 10.14088/j.cnki.issn0439-8114.2016.09.063
Zhao, C. R., Ikka, T., Sawaki, Y., Kobayashi, Y., Suzuki, Y., Hibino, T., et al. (2009). Comparative transcriptomic characterization of aluminum, sodium chloride, cadmium and copper rhizotoxicities in arabidopsis thaliana. BMC Plant Biol. 9, 32. doi: 10.1186/1471-2229-9-32
Zhao, H., Ma, H., Yu, L., Wang, X., Zhao, J. (2012). Genome-wide survey and expression analysis of amino acid transporter gene family in rice (Oryza sativa l.). PLos One 7, e49210. doi: 10.1371/journal.pone.0049210
Keywords: TaMGT, gene structure, abiotic stresses, gene expression, fluorescence quantitative PCR, climate-resilience
Citation: Tang Y, Yang X, Li H, Shuai Y, Chen W, Ma D and Lü Z (2023) Uncovering the role of wheat magnesium transporter family genes in abiotic responses. Front. Plant Sci. 14:1078299. doi: 10.3389/fpls.2023.1078299
Received: 24 October 2022; Accepted: 23 January 2023;
Published: 09 February 2023.
Edited by:
Mehanathan Muthamilarasan, University of Hyderabad, IndiaReviewed by:
Parviz Heidari, Shahrood University of Technology, IranHaifeng Liu, Chonnam National University, Republic of Korea
Huan Luo, Chungnam National University, Republic of Korea
Zeguang Liu, University of Geneva, Switzerland
Copyright © 2023 Tang, Yang, Li, Shuai, Chen, Ma and Lü. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wang Chen, Y2hlbndhbmdjaHdAMTYzLmNvbQ==; Dongfang Ma, bWFkZkB5YW5ndHpldS5lZHUuY24=; Zhichuang Lü, bHZ6aGljaHVhbmdAY2Fhcy5jbg==
 Yanhong Tang
Yanhong Tang Xiaoyue Yang1,2
Xiaoyue Yang1,2 Dongfang Ma
Dongfang Ma Zhichuang Lü
Zhichuang Lü