
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 27 January 2023
Sec. Plant Pathogen Interactions
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1077301
This article is part of the Research TopicRegulation of Plant Immunity by Immune ReceptorsView all 7 articles
 Anna Mamaeva1†
Anna Mamaeva1† Irina Lyapina1†
Irina Lyapina1† Andrey Knyazev1
Andrey Knyazev1 Nina Golub1
Nina Golub1 Timur Mollaev2
Timur Mollaev2 Elena Chudinova2
Elena Chudinova2 Sergey Elansky2,3
Sergey Elansky2,3 Vladislav V. Babenko4
Vladislav V. Babenko4 Vladimir A. Veselovsky4
Vladimir A. Veselovsky4 Ksenia M. Klimina4
Ksenia M. Klimina4 Tatiana Gribova4
Tatiana Gribova4 Daria Kharlampieva4
Daria Kharlampieva4 Vassili Lazarev4,5
Vassili Lazarev4,5 Igor Fesenko1*
Igor Fesenko1*Background: RAPID ALKALINIZATION FACTOR (RALFs) are cysteine-rich peptides that regulate multiple physiological processes in plants. This peptide family has considerably expanded during land plant evolution, but the role of ancient RALFs in modulating stress responses is unknown.Results: Here, we used the moss Physcomitrium patens as a model to gain insight into the role of RALF peptides in the coordination of plant growth and stress response in non-vascular plants. The quantitative proteomic analysis revealed concerted downregulation of M6 metalloprotease and some membrane proteins, including those involved in stress response, in PpRALF1, 2 and 3 knockout (KO) lines. The subsequent analysis revealed the role of PpRALF3 in growth regulation under abiotic and biotic stress conditions, implying the importance of RALFs in responding to various adverse conditions in bryophytes. We found that knockout of the PpRALF2 and PpRALF3 genes resulted in increased resistance to bacterial and fungal phytopathogens, Pectobacterium carotovorum and Fusarium solani, suggesting the role of these peptides in negative regulation of the immune response in P. patens. Comparing the transcriptomes of PpRALF3 KO and wild-type plants infected by F. solani showed that the regulation of genes in the phenylpropanoid pathway and those involved in cell wall modification and biogenesis was different in these two genotypes.
Conclusion: Thus, our study sheds light on the function of the previously uncharacterized PpRALF3 peptide and gives a clue to the ancestral functions of RALF peptides in plant stress response.
Plants utilize small secreted peptides as important mediators of many processes, from growth and development to response to stress conditions (Olsson et al., 2019). One of such regulators is the conservative 5 kDa RALF (Rapid Alkalinization Factor) peptide family, which is widely present in terrestrial plants (Campbell and Turner, 2017). The mature RALF peptide contains four cysteine amino acid residues, which form two S-S bonds (Frederick et al., 2019). RALF peptides are cleaved from an inactive protein precursor and S1P protease is shown to be involved in the cleavage of a mature RALF peptide from a nonfunctional precursor at the conserved dibasic RR site (Srivastava et al., 2009; Stegmann et al., 2017).
The tandem duplication is considered to play a dominant role in the evolution of RALFs and this peptide family has expanded considerably during land plant evolution (Cao and Shi, 2012; Campbell and Turner, 2017). For example, 37 members of this family were found in Arabidopsis thaliana, 25 in Arabidopsis halleri, 20 in Zea mays, but only 3 in Physcomitrium patens (Campbell and Turner, 2017; Ginanjar et al., 2022). RALF peptides are diverged into four clades based on mature peptide region sequence features (Campbell and Turner, 2017). RALF peptides from I, II and III clades contain a specific protease cleavage site and a conserved YISY motif, which is important for their recognition through receptors (Campbell and Turner, 2017; Xiao et al., 2019). However, RALF peptides from clade IV lack specific cleavage site, in addition, the conserved motif also changes, thus suggesting diverse functions for the representatives from this clade.
Unlike most other plant peptide hormones, RALF peptides bind to membrane-localized receptor-like kinases with a malectin-like extracellular domain instead of a leucine-rich repeat domain (Franck et al., 2018). Receptors of RALF peptides belong to the Catharanthus roseus receptor-like kinase (CrRLK1L) family and include FERONIA (FER), CrRLK1Ls ANXUR1 (ANX1), ANX2, and BUDDHA’S PAPER SEAL (BUPS) 1 and BUPS2 (Ge et al., 2017; Ge et al., 2019). In addition, LRE LIKE GPI-AP1 (LLG1) was proposed to function as a coreceptor for FER in AtRALF1 perception in Arabidopsis (Li et al., 2015). In bryophytes, such as Marchantia polymorpha L., P. patens, and Sphagnum fallax, 1, 5 and 7 CrRLK1Ls were identified, respectively (Solis-Miranda et al., 2020). Also, LEUCINE-RICH REPEAT EXTENSINS (LRX) proteins are shown to bind RALF peptides with high affinity (Mecchia et al., 2017; Zhao et al., 2018; Moussu et al., 2020).
The diversity of receptors and co-receptors that can bind different RALF peptides results in their multiple functions and activation of different signaling pathways (Abarca et al., 2021). RALF peptides are involved in the regulation of root growth, pollen tube elongation, the formation of nitrogen-fixing nodules, and inulin accumulation (Murphy and De Smet, 2014; Tena, 2016; Wieghaus et al., 2019). Also, they are involved in the processes of intercellular communication between sporophyte and gametophyte in land plants (Chevalier et al., 2013; Mecchia et al., 2017; Loubert-Hudon et al., 2020). The possible functions of RALF genes that have been identified in bryophyte genomes are still being studied. PpRALF1 and PpRALF2 peptides identified in P. patens have been shown to promote protonema tip growth and elongation (Ginanjar et al., 2022). Although the function of the PpRALF3 peptide is still unknown. The Marchantia polymorpha genome encodes three RALF peptides (Bowman et al., 2017). The single CrRLK1L gene of Marchantia polymorpha (MpFER) was recently characterized (Mecchia et al., 2022). Defects in vegetative growth and decreased male fertility characterize plants with reduced MpFER levels. Mpfer null mutants and MpFER overexpression lines show dramatic defects in gametophyte cell integrity and morphogenesis (Mecchia et al., 2022).
Besides the regulation of growth and development, RALF peptides are also involved in the regulation of responses to abiotic and biotic stresses (Blackburn et al., 2020). For example, AtRALF8 plays a role in simultaneously controlling responses to drought and nematode attack by allegedly regulating cell wall remodeling (Atkinson et al., 2013). However, as has been shown in Arabidopsis, different RALF peptides may have opposite effects. For example, AtRALF17 increases reactive oxygen species (ROS) production and resistance to Pseudomonas syringae pv. tomato, while AtRALF23 has the opposite effect (Stegmann et al., 2017). AtRALF33, a close relative of AtRALF23, was shown to inhibit pathogen elicitor-induced ROS production (Stegmann et al., 2017). In addition, AtRALF23 and AtRALF22 were also shown to participate in regulation of growth and salt stress tolerance by cell wall remodeling (Zhao et al., 2018). The exogenous treatment with synthetic AtRALF1, AtRALF4, AtRALF19 and AtRALF22 reduces elicitor-induced ROS production (Abarca et al., 2021). The corresponding precursors of these AtRALFs contain an S1P cleavage site, but AtRALF6-13, AtRALF15-17, AtRALF20, AtRALF24, AtRALF29-32, AtRALF35-36, which reported to increase elf18-induced ROS production, are not cleaved by this protease (Abarca et al., 2021). Perhaps, the presence of S1P cleavage sites is linked to the negative regulation of the immunity, with some exceptions (Stegmann et al., 2017; Abarca et al., 2021).
The participation of RALF peptides in modulation of the response to biotic stress has been shown not only in Arabidopsis, but also in other plants, including crops (Stegmann et al., 2017; Merino et al., 2019). Generally, RALF peptides are considered negative regulators of the immune response. For example, the RALF–FER complex modulates complex formation between the immune receptor kinase FLAGELLIN-SENSING 2 (FLS2) and its co-receptor BRASSINOSTEROID INSENSITIVE 1-ASSOCIATED RECEPTOR KINASE 1 (BAK1) to inhibit plant immunity (Stegmann et al., 2017; Xiao et al., 2019). Additionally, RALF-induced pH changes may regulate pathogenicity. For instance, the respiratory burst oxidase homologs (RBOHs), that are involved in apoplastic ROS production, have been found to be regulated by FER and related proteins (Franck et al., 2018; Zhang et al., 2020b). FERONIA-like receptor 1 (FLR1) from rice is involved in the regulation of Ca2+ homeostasis in response to rice blast resistance (Luo et al., 2022). However, it is currently unknown how the RALF–CrRLK1L complex regulates ROS production and interferes with calcium signaling during immune response (Liao et al., 2017).
Haruta et al. (2014) demonstrated that the RALF1–FER pathway phosphorylates the proton pump (e.g., AHA2), resulting in transient alkalinization of the extracellular matrix and inhibition of primary root cell elongation (Haruta et al., 2014). Considering that many pathogens, especially fungi, prefer alkaline conditions, the ability of RALFs to alkalize the environment came in very handy (Tena, 2016; Thynne et al., 2017). Many plant pathogens synthesize RALF-like peptides that enhance the development of infection (Tena, 2016; Thynne et al., 2017). F-RALF from Fusarium oxysporum f. so. lycopersici, which mimics plant RALFs, induces the alkalinization of apoplasts, which activates the orthologous MAPK FMK1 kinase, inhibits root growth and promotes virulence in fungi (Masachis et al., 2016). In tomato and Nicotiana benthamiana, a synthetic RALF-like peptide from F. oxysporum was also able to induce ROS burst, alkalinization, and activation of MAPKs, as well as inhibit the seedlings growth (Thynne et al., 2017). Moreover, RALF-like peptides from plant root-knot nematodes facilitate the process of infection in Arabidopsis and rice (Zhang et al., 2020a). Although these homologs may have an arguable impact on the potency of pathogen infection (Wood et al., 2020). Alternatively, the plant can suppress pathogen-associated molecular pattern (PAMP)-triggered immunity (PTI) by activating its own RALFs expression to shape the root microbiome under phosphate starvation (Tang et al., 2022). However, determining how the RALF–FER pathway modulates immune responses and cell growth by inducing pH changes in a context-dependent manner requires additional research (Zhang et al., 2020b).
The analysis of different plant lineages allows us to gain insight into the possible roles of RALF peptides in the regulation of stress responses (Blackburn et al., 2020). However, the role of ancient RALF peptides in response to stress conditions, particularly phytopathogens, is unknown. Physcomitrium patens is a model host plant that is frequently used to study stress response and the evolution of the plant immune system (Ponce de León, 2011; Ponce de León and Montesano, 2013). A diverse range of phytopathogens, such as Botrytis cinerea, Phytophthora infestans, Colletotrichum gloeosporioides, Altenaria brassicicola, Fusarium spp. (Akita et al., 2011; Ponce de León, 2011; Lehtonen et al., 2012; Ponce De León et al., 2012; Reboledo et al., 2015; Bressendorff et al., 2016; Overdijk et al., 2016; Marttinen et al., 2020), the bacterium Pectobacterium carotovorum (Andersson et al., 2005; Ponce de León et al., 2007), the oomycete Pythium (Oliver et al., 2009; Castro et al., 2016) have been shown to infect P. patens. Using quantitative proteomic and transcriptomic analysis along with infection with Pectobacterium carotovorum and Fusarium solani, we studied the role of three PpRALF peptides from the model bryophyte Physcomitrium patens in the immune stress response. Our study showed the specific role of PpRALF3 in negative regulation of plant stress response.
Physcomitrella patens subsp. patens (Physcomitrium patens “Gransden 2004”, Freiburg) protonemata were grown on BCD medium supplemented with 5 mM ammonium tartrate (BCDAT) and/or 0.5% glucose with 1.5% agar (Helicon, Moscow, Russian Federation) in a Sanyo Plant Growth Incubator MLR-352H (Panasonic, Osaka, Japan) with a photon flux of 61 μM/m2•s during a 16-hour photoperiod at 24°C in 9 cm Petri dishes (Nishiyama et al., 2000). For proteomic and qRT-PCR analyses, the protonemata were grown in liquid BCDAT medium and collected on day 7. The gametophores were grown on free-ammonium tartrate BCD medium under the same conditions, and 8-week-old gametophores were used for analysis. For analysis of abiotic stress resistance, protonemata were grown on BCD medium without ammonium tartrate supplemented with 150 mM NaCl or 2 μM paraquat (PQ). For PpRALFs expression analysis, 7-day-old protonemata were treated with 100 mM hydrogen peroxide for 2 hours.
For morphological analysis, protonema tissue 2 mm in diameter was planted on 9 cm Petri dishes on BCD and BCDAT media.
For growth rate measurements, photographs were taken at day 30 of subcultivation. Protonemal tissues and cells were photographed using a Microscope Digital Eyepiece DCM-510 attached to a Stemi 305 stereomicroscope (Zeiss, Germany).
PpRALF1 (Pp3c3_15280V3), PpRALF2 (Pp3c6_7200V3) and PpRALF3 (Pp3c25_4180V3) knockout lines were created using the CRISPR/Cas9 system (Collonnier et al., 2017). The coding sequences were used to search for guide sequences preceded by a Streptococcus pyogenes Cas9 PAM motif (NGG) using the web tool CRISPOR (http://crispor.tefor.net/). The guide sequence closest to the translation start site (ATG) was selected for cloning (Supplementary Table S1 and Figure S1). These sequences were cloned into plasmid pBB (Fesenko et al., 2019), yielding the final complete sgRNA expression cassette. Protoplasts were obtained from protonemata as described previously (Fesenko et al., 2015) and transformed by the PEG transformation protocol (Schaefer and Zrÿd, 1997) using a mixture of three plasmids 1) one of the pBB plasmid carrying guide RNA expression cassette; 2) pACT-CAS9 carrying the CAS9 gene; 3) pBNRF plasmid carrying resistance gene to G418. The plasmids pACT-CAS9 and pBNRF were kindly provided by Dr. Fabien Nogué. Independent knockout mutant lines have been obtained. Double knockout lines were obtained by simultaneous transformation of two plasmids with the corresponding guide RNAs.
Genomic DNA from gametophores was isolated using a commercial kit (Biolabmix, Russia), according to the manufacturer’s recommendations. Total RNA from gametophores and protonemata was isolated using TRIzol™ Reagent according to the manufacturer’s recommendations. RNA quality and quantity were evaluated using electrophoresis on agarose gel with SYBR Green (Biolabmix, Russia). Total RNA concentration of samples was precisely measured using a Nanodrop™One (Thermo Fisher Scientific, USA).
cDNA was synthesized using the MMLV RT kit (Evrogen, Russian) according to the manufacturer’s recommendations. OligodT primers were used to prepare cDNA from 2 µg total RNA after DNase treatment.
Real-time PCR was performed using the HS-qPCR SYBR Blue (2x) (Biolabmix, Russia) on a LightCycler®96 (Roche, Germany). Three biological and three technical replicates were used for the qPCR. Primers for the target genes could be found in Supplementary Table S1.
For qPCR analysis of infection severity, primers were designed for F. solani or selected from previous research (Kabir et al., 2020) for P. carotovorum to specifically amplify pathogenic DNA from the infected moss plants (Supplementary Table S1).
Phytopathogens Pectobacterium carotovorum subsp. atrosepticum (strain ECPA16 NCBI № OL677456) and Fusarium solani (20 МККК1 NCBI № OQ073458) were used to infect moss plants. Bacterial culture of P. carotovorum was grown for 18 hours, after which the optical density of the bacterial suspension was measured on a spectrophotometer at 600 nm. Based on preliminary results, a concentration of ~107 cfu/mL was chosen for the further experiments. To obtain this concentration the initial bacterial suspension was diluted 1000 times with sterile water. Therefore, sterile water-treated plants were used as a control. For plant treatment, 15 µl of prepared overnight culture with a final concentration 9.5×106 cfu/mL were used. After each experiment, a diluted bacterial suspension was grown on medium for subsequent CFU counts. F. solani spores were obtained from a solid medium culture and suspension with final concentration 8.3×105 spores/mL was applied. Fusarium solani conidia were also diluted with sterile water, and the concentration of conidia was counted under a microscope. Petri dishes with infected plants were cultivated at room temperature under standard conditions (16 hours day/8 hours night). Infected plants were collected after 7 days and frozen in liquid nitrogen.
The three independent biological repeats for each genotype were used for comparative proteomic analysis. Protein extraction and trypsin digestion we conducted as described previously (Faurobert et al., 2007; Fesenko et al., 2021b). iTRAQ labeling (Applied Biosystems, Foster City, CA, USA) was conducted according to the manufacturer’s manual. Proteins were labeled with the iTRAQ tags as follows: wild-type biological replicates – 113, 114, 115 isobaric tags; PpRALF1, PpRALF2 and PpRALF3 KO biological replicates – 116, 117, 118 isobaric tags for each mutant line.
The LC–MS/MS analysis was performed as described earlier (Fesenko et al., 2021a). Tandem mass spectra were searched with PEAKS Studio version 8.0 software (Bioinfor Inc., CA, USA) against a custom database containing 32 926 proteins from annotated genes in the latest version of the moss genome v3.3 (Lang et al., 2018), 85 chloroplast proteins, and 42 mitochondrial proteins. The search parameters were the following: a fragmentation mass tolerance of 0.05 Da; parent ion tolerance of 10 ppm; fixed modification – carbamidomethylation; variable modifications - oxidation (M), deamidation (NQ), and acetylation (protein N-term). The results were filtered by a 1% FDR, but with a significance threshold of not less than 20 (equivalent to a P-value of less than 0.01). The results were filtered by a 1% false discovery rate (FDR). PEAKS Q was used for iTRAQ quantification. Normalization was performed by averaging the abundance of all peptides. The median values were used for averaging. Although iTRAQ quantification usually underestimates the amount of real fold changes between two samples (Ow et al., 2009), we used a very strict filter for differentially expressed proteins. The threshold for calling a protein differentially abundant was calculated based on an s0 parameter (s0 = 0.1) as described previously (Schessner et al., 2022) with a permutation test repeated 100 times and Benjamini and Hochberg FDR correction (FDR_BH < 1%).
DNase treatment was carried out with TURBO DNA-free kit (Thermo Fisher Scientific, Waltham, MA, USA), in volumes of 50 µl. RNA cleanup was performed with the Agencourt RNA Clean XP kit (Beckman Coulter, Brea, USA). The concentration and quality of the total RNA were checked by the Quant-it RiboGreen RNA assay (Thermo Fisher Scientific) and the RNA 6000 Pico chip (Agilent Technologies, Santa Clara, CA, USA), respectively.
RNA libraries were prepared using NEBNext Poly(A) mRNA Magnetic Isolation Module and the NEBNext Ultra II Directional RNA Library Prep Kit (NEB), according to the manufacturer’s protocol. The library underwent a final cleanup using the Agencourt AMPure XP system (Beckman Coulter) after which the libraries’ size distribution and quality were assessed using a high sensitivity DNA chip (Agilent Technologies). Libraries were subsequently quantified by Quant-iT DNA Assay Kit, High Sensitivity (Thermo Fisher Scientific). Finally, equimolar quantities of all libraries (10 pM) were sequenced by a high throughput run on the Illumina HiSeq 2500 using 2 × 100 bp paired-end reads and a 1% Phix spike-in control.
The adaptors and low-quality sequences were removed from raw reads by Trimmomatic v0.39 (Bolger et al., 2014). Clean reads were aligned to the P. patens v3.3 reference genome (download from the website: https://phytozome-next.jgi.doe.gov) using HISAT2 v2.1.0 (Kim et al., 2015) and the alignments were sorted with Samtools (Li et al., 2009). The expression abundances of mapped reads were counted by the FeatureCounts tool (Liao et al., 2014). Differential expression analysis was performed by EdgeR package (Robinson et al., 2010). The genes were defined as differentially expressed genes (DEGs) with adjusted p-value ≤ 0.05 and fold change ≥1.0.
Protoplasts were prepared from protonemata as described previously (Fesenko et al., 2015) and incubated for 48 hours at solid BCD agar medium. Regenerated protoplasts were stained with 10 µg/ml Calcofluor White (Fluorescent Brightener 28) for 5 minutes. After that, protoplasts were analyzed using fluorescent microscope (Axio Imager M2, Zeiss, Germany) at λex = 365 nm, BS FT 395, and λem = 445nm/50 nm (Filter set 49 DAPI, Zeiss, Germany).
The fluorescent dye 2,7-Dichlorofluorescin Diacetate (DCFH-DA, Sigma-Aldrich, USA) was used to identify intracellular ROS. Using a spatula, seven-day-old protonema filaments were removed from the agar surface and transferred to mQ. Protonemata were then treated with 0.0025% driselase (diluted in mQ) for 1 minute or mQ water as a control and incubated with 10 µM DCFH-DA for 15 minutes in total. The No. 44 filter (λex BP 475 nm/40 nm; λem BP 530 nm/50 nm) was used for DCFH-DA fluorescence detection on the fluorescent microscope Axio Imager M2 (Zeiss) with an AxioCam 506 mono digital camera. Data on the fluorescence intensity were obtained from the related Zeiss software Zen.
Protein–protein interaction networks were constructed using STRING v.10 (www.string-db.org) with the default options (Szklarczyk et al., 2014). The visualization of the protein interaction was performed with Cytoscape software (Shannon et al., 2003). The GO enrichment analysis was conducted by g:Profiler (Raudvere et al., 2019). Multiple alignments were created using the MAFFT algorithm (Katoh et al., 2019) and visualized using Jalview software (Waterhouse et al., 2009). IQ-TREE multicore version 2.2.0 (Nguyen et al., 2015) was used to conduct a maximum likelihood (ML) analysis with 1000 ultrafast bootstrap replicates (Minh et al., 2013). The model FLU+F+G4 was chosen as the best-fit model by the in-built ModelFinder program (Kalyaanamoorthy et al., 2017) according to the Bayesian Information Criterion (BIC). Principal Component Analysis (PCA) was performed using the iFeature tool (Chen et al., 2018).
Statistical analysis and visualization were made in Python v. 3.7.5 (Van Rossum and Drake, 1995) using modules scipy 1.5.2 (Virtanen et al., 2020), seaborn 0.11.1 (Waskom, 2021), numpy 1.20.1, pandas 1.2.3 (McKinney, 2012). For two- or more-way analysis of variance (ANOVA), Tukey’s honestly significant difference (HSD) tests based on multiple comparisons of means were applied to determine which pairwise comparisons were statistically significant. Differences were considered to be significant at p < 0.05.
The members of a RALF peptide family possess functional heterogeneity in vascular plants (Blackburn et al., 2020), including the role in modulation of immune response. In Arabidopsis, exogenous application of synthetic AtRALF23, AtRALF33, AtRALF34 are shown to inhibit production of reactive oxygen species (ROS) induced upon treatment with immune elicitors (such as elf18) and inhibit seedling and root growth, whereas AtRALF17, AtRALF24, AtRALF32 and some others were able to induce ROS production (Abarca et al., 2021). The previous phylogenetic analysis divided the RALFs into four clades, and peptide sequences from different clades could be distinguished based on analysis of their physico-chemical properties (Campbell and Turner, 2017). To test how the overall amino acid composition of RALF peptides is related to their possible functions, we used Principal Component Analysis (PCA). The results of PCA analysis showed a distinct pattern in which AtRALF peptides that inhibit the production of pathogen-induced ROS (Abarca et al., 2021) were clustered together. We next found that PpRALFs were clustered with AtRALFs that inhibit elicitor-induced ROS production (Figure 1A; Supplementary Table S2). This result is consistent with previous phylogenetic analysis (Ginanjar et al., 2022), where PpRALFs were grouped with AtRALF22, AtRALF23, AtRALF33, and AtRALF34. These findings suggest that the overall amino acid composition of RALF peptides, to some extent, reflects their functional biases, and PpRALFs are related to AtRALFs that negatively regulate immune response.
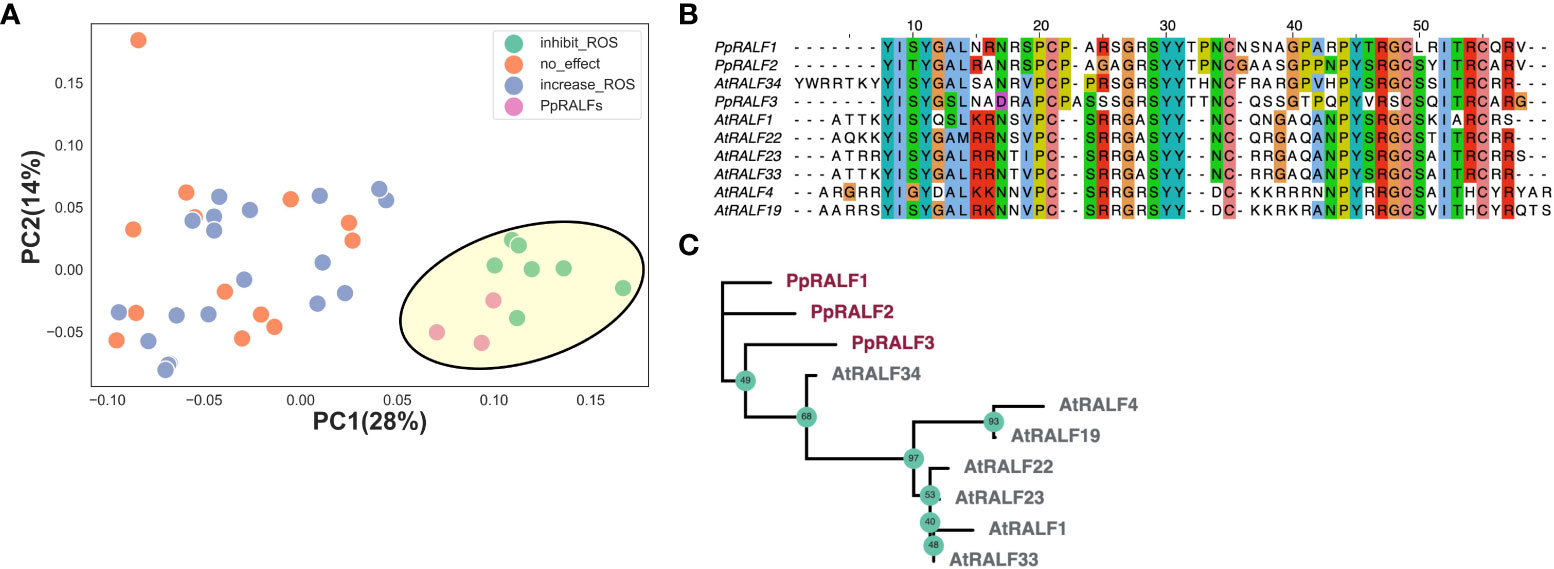
Figure 1 (A) Principal Component Analysis (PCA) of amino acid composition, calculated by iFeature tool (Chen et al., 2018) for AtRALF and PpRALF peptides. Moss PpRALFs were clustered with reported immune-related RALF peptides from Arabidopsis, such as AtRALF1, AtRALF4, AtRALF19, AtRALF22, AtRALF23, AtRALF33, and AtRALF34. (B) The multiple pairwise alignment of PpRALFs and immune-related AtRALFs (AtRALF1, AtRALF4, AtRALF19, AtRALF22, AtRALF23, AtRALF33, AtRALF34). (C) An unrooted phylogenetic tree with RALF amino acid sequences from Physcomitrium patens (PpRALF1, PpRALF2, PpRALF3) and immune-related AtRALFs (AtRALF1, AtRALF4, AtRALF19, AtRALF22, AtRALF23, AtRALF33, AtRALF34); Ultra-fast bootstrap (1000 replicates) support values are shown.
In Physcomitrium patens, PpRALF1 and PpRALF2, which are members of Clade III (Campbell and Turner, 2017), have shown to promote protonema tip growth and elongation (Ginanjar et al., 2022). PpRALF3, the third member of the RALF peptide family in P. patens, contains a substitution in the conserved “RGC” motif (Figure 1B). Our phylogenetic analysis revealed that PpRALF3 diverged from PpRALF1 and PpRALF2 and formed a group with AtRALF peptides (Figure 1C). The AtRALFs that were clustered together with PpRALF peptides (Figure 1A) belong to Clades 1, 2 and 3 (Campbell and Turner, 2017), implying the absence of the correlation between RALF functions in immune response and division into clades in this case. However, the role of PpRALFs in responding to stress conditions is unknown.
PpRALF1 and PpRALF2 peptides are previously shown to promote tip growth and elongation of protonemata filaments in P. patens, but the functions of PpRALF3 peptide are currently unknown (Ginanjar et al., 2022). The chemical synthesis of RALF peptides, as well as the generation of recombinant peptides, is associated with certain difficulties. Proper bonding and folding of peptides require the right conditions, otherwise peptides may not work the way they do, or a higher concentration could be required (Abarca et al., 2021). Since this may not reflect the actual effect of the peptides, we decided to use only knockout lines. At least two independent knockout lines for each PpRALF gene and double knockout lines - PpRALF1 and PpRALF3; PpRALF2 and PpRALF3 were generated by CRISPR/Cas9 technology (Table 1; Supplementary Figure S1). However, we failed to obtain triple and double PpRALF1 and PpRALF2 knockout lines after several attempts. Probably, this is due to their important roles in regulation of moss growth and development.
It has been previously shown that knockout of PpRALF genes results in reducing the formation of caulonema filaments (Ginanjar et al., 2022). Therefore, we tested the growth of our mutants on medium with and without ammonium tartrate (BCDAT/BCD; Figure 2A). The ammonium tartrate reduces the transition into caulonemal cells and promotes chloronemal branching (Vidali and Bezanilla, 2012). Similar to the previous study (Ginanjar et al., 2022), we observed the reduced formation of caulonema filaments in mutant genotypes (Supplementary Figure S2). The morphology of wild-type and single knockout mutants was similar, but the diameter of PpRALF3 KO plants was significantly larger than wild-types on the BCDAT medium (Figures 2A, B; ANOVA with post-hoc Tukey HSD P < 0.001). In contrast, the growth rate of PpRALF1 KO lines did not differ from wild-type on all media examined (Figures 2A, B). In addition, PpRALF2 KO plants were significantly smaller than wild-type plants on the medium with ammonium tartrate (Figure 2B, ANOVA with post-hoc Tukey HSD P < 0.001). Both double knockouts showed significant inhibition of the growth rate on medium with and without ammonium tartrate (Figures 2A, B; ANOVA with post-hoc Tukey HSD P < 0.001). PpRALF1,3 KO plants differed from other knockout plants, and cell death was observed on both mediums.

Figure 2 (A) Phenotype of 30-d-old plants of wild-type and the obtained knockout lines grown on two mediums with and without ammonium tartrate. Scale bar: 10 mm; (B) Wild-type and knockout lines’ plant diameters when grown on media containing (BCDAT) and without ammonium tartrate (BCD). For each experiment, the diameters of the wild-type and knockout plants n = 12 for each genotype) are displayed. Analysis of variance and Tukey’s HSD post hoc tests were performed ***P < 0.001 (compared to wild-type plants); (C) Volcano plots of the entire set of proteins quantified during iTRAQ analysis of PpRALF1, PpRALF2 and PpRALF3 KO lines, respectively. Proteins significantly changed in abundance are depicted in orange; Pp3c15_830 M6 family metalloprotease domain-containing protein, Pp3c14_22870 SHORT-LEAF protein, Pp3c26_120 bryoporin, Pp3c15_13040 12-lipoxygenase, Pp3c16_14730 Stigma-specific protein, Stig1, Pp3c5_6620 DUF1682, Pp3c3_10250 Ricin B, lectin-containing protein.
In addition, the gametophores were significantly longer in both PpRALF3 KO lines in comparison to other genotypes (Supplementary Figure S3; ANOVA with post-hoc Tukey HSD P < 0.001). Thus, our results are in line with previously obtained data on morphology and phenotypes of PpRALF knockouts (Ginanjar et al., 2022). In addition, our results suggest that all PpRALFs are functional and might play different roles, beyond the regulation of only growth processes.
Using isobaric tags for relative and absolute quantification (iTRAQ), we further compared the proteomes of PpRALF1, PpRALF2, and PpRALF3 knockout lines and wild-type plants. In total, we identified 2723 protein groups in PpRALF1 KO, 3074 protein groups in PpRALF2 KO, and 3078 protein groups in PpRALF3 KO (Supplementary Table S3). Although iTRAQ quantification usually underestimates the amount of real fold changes between two samples (Ow et al., 2009), we used a very strict cut-off to reliably identify differentially abundant protein groups (DAPs), such as an s0 parameter equal to 0.1 and multiple hypothesis correction that was performed based on a permutation test (see Methods). In contrast to the transcriptomes of the PpRALF knockout lines that have already been published (Ginanjar et al., 2022), we have not found any major differences between the knockout and wild-type proteomes. This suggests that PpRALF peptides have mostly specific regulatory roles. In PpRALF1 KO plants only 8 protein groups were significantly changed (FDR BH < 1%, s0 = 0.1; Figure 2C; Supplementary Table S3) in comparison to wild-type plants and all of these DAPs were downregulated. The knockout of PpRALF2 gene resulted in downregulation of 15 protein groups (FDR_BH < 1%, s0 = 0.1; Figure 2C; Supplementary Table S3) and upregulation of an uncharacterized 257 aa protein Pp3c16_14730, containing predicted signal peptide. In PpRALF3 KO plants only 5 protein groups were significantly downregulated (FDR_BH < 1%, s0 = 0.1; Figure 2C; Supplementary Table S3).
We further compared these DAPs from all knockout lines and found a core set of four proteins that were changed in at least two single-gene knockout lines. For example, we revealed that a protein Pp3c15_830 (M6 family metalloprotease domain-containing protein) was downregulated in PpRALF1, PpRALF2 and PpRALF3 KO lines. However, the role of such proteins in RALF signaling has not been previously described. Another DA protein that belonged to M6 metalloproteases - Pp3c13_22390 was downregulated only in the PpRALF1 KO line. We also identified two proteins that were downregulated in both PpRALF1 and PpRALF2 KO lines. One of them - Pp3c26_120 (Bryoporin) is known to be important for a response to osmotic stress in mosses (Hoang et al., 2009). The other protein, Pp3c14_22870 (SHORT-LEAF), is encoded by a bryophyte-specific gene that represents a family of near-perfect tandem direct repeat (TDR)-containing proteins and has been shown to regulate gametophore development in moss (Mohanasundaram et al., 2021). We also identified a LOX3 protein (Pp3c15_13040) that was downregulated in PpRALF2 and PpRALF3 KO lines, which might be involved in the arachidonic acid metabolism, suggesting the possible role of these paralogs in biotic stress response.
Thus, the knockout of PpRALF genes resulted in downregulation of some proteases and previously uncharacterized membrane proteins. To determine whether these changes in proteomes of mutant lines affected the cell wall regeneration processes, we explored the process of protoplast regeneration in the knockout and wild-type genotypes. The protoplasts were dyed with Calcofluor White fluorescent dye that is widely used to visualize cellulose, callose, and other β-glucans in the plant cell wall (Maksimov et al., 2016; Herrera-Ubaldo and de Folter, 2018). On average, the cell wall regenerated 80% faster in PpRALF2 KO lines (chi-square P < 0.001) and 46% faster in PpRALF3 KO lines (chi-square P < 0.001) than in wild-type cells after two days of regeneration process (Supplementary Figure S4). We found no significant differences in the cell wall regeneration rate between PpRALF1 KO-1 and wild-type plants.
Some members of the RALF peptide family are shown to modulate the abiotic stress response in angiosperms (Zhao et al., 2018). Therefore, we next sought to expand our understanding of PpRALFs functions under abiotic stress conditions. At first, we used previously obtained RNA-seq data (Khraiwesh et al., 2015) to analyze the expression of PpRALF genes under salt, drought, and cold treatments. According to this study, PpRALF1 was significantly upregulated in all stress conditions after 0.5 hours and under cold treatment after 4h but downregulated under drought and salt treatment after 4 hours. In addition, the PpRALF2 gene was significantly upregulated at drought after 0.5 hours and at cold after 4 hours as well. The PpRALF3 transcripts were detected only under salt and drought treatment, suggesting its possible role in stress response.
We also assessed the transcriptional level of all PpRALFs under hydrogen peroxide treatment for 2 hours. According to qRT-PCR, all PpRALFs were significantly downregulated under this treatment (Figure 3A; ANOVA with post-hoc Tukey HSD P < 0.001). However, the transcriptional level of PpRALF3 decreased less than that of other PpRALFs.
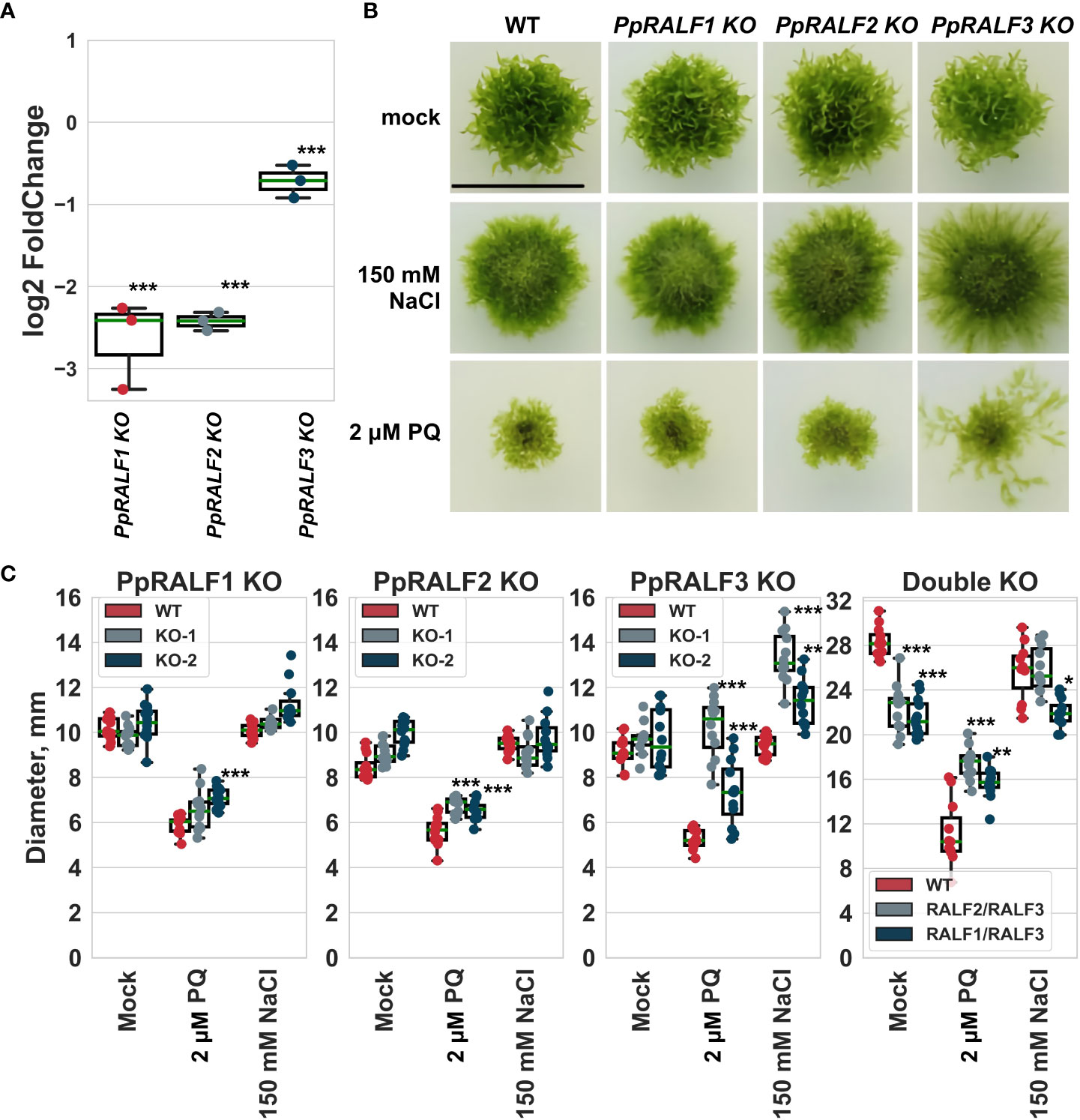
Figure 3 (A) The results of quantitative reverse transcription PCR (RT-qPCR) analysis of the transcriptional level of PpRALFs under hydrogen peroxide treatment for 2 hours. Analysis of variance and Tukey’s HSD post hoc tests were performed ***P < 0.001. Log2FoldChanges represent the logarithmic fold change of the PpRALF genes transcription in treated samples relative to control samples, normalized to a reference gene (2-ΔΔCT values); (B) Phenotype of 30-d-old plants of wild-type and the knockout lines grown on medium supplemented with 150 mM NaCl or 2 μM paraquat. Scale bar: 10 mm; (C) The measured diameter of moss plants grown on medium supplemented with 150 mM NaCl or 2 μM paraquat. Analysis of variance and Tukey’s HSD post hoc tests were performed ***P < 0.001, ** P<0.01, * P<0.05 (comparison to the corresponding wild-type plants).
Taking into account the role of RALF peptides, such as AtRALF22/23, in regulation of salt stress tolerance in Arabidopsis (Zhao et al., 2018), we further assessed the growth rate of PpRALF KO plants upon the oxidative (2 μM paraquat) and salt (150 mM NaCl) treatments. We have chosen the salt concentration of 150 mM NaCl for these experiments based on our assessment of wild-type plant phenotypes at different salt concentrations (Supplementary Figure S5). Our experiments showed that wild-type and knockout plants were affected by 150 mM NaCl, confirming the adverse effect of high salt concentration on moss growth and development (Figure 3B). Herewith, the diameters of wild-type and PpRALF1 and 2 KO plants were similar under salt treatment, except for PpRALF3 KO lines. The diameter of PpRALF3 KO lines was significantly larger (ANOVA with post-hoc Tukey HSD P < 0.001) under salt treatments compared to mock-treated and wild-type plants due to diffusion of protonemal filaments (Figures 3B, C). Under salt treatment, there were some differences in the diameters of the PpRALF3 KO-1 and KO-2 lines, but the protonemal filaments of both knockouts tended to be longer than those of wild-type plants. The plant size of double knockouts was unaffected by 150 mM NaCl, and only the diameter of PpRALF1,3 KO was slightly, albeit significantly smaller than that of the wild-type (Figure 3C).
Paraquat is a commonly used herbicide that significantly increases the production of reactive oxygen species (ROS) and inhibits the regeneration of reducing equivalents and compounds required for the antioxidant system’s activity (Lascano et al., 2012). Under the oxidative stress conditions induced by 2 μM paraquat, both wild-type and mutant plants were severely affected (Figure 3B). Despite the similarly affected phenotypes, the diameter of PpRALF2 KO plants was slightly, albeit significantly, larger (Figures 3B, C; ANOVA with post-hoc Tukey HSD P < 0.001). PpRALF3 KO-1 and KO-2 lines exhibited a phenotype characterized by protonemal segments that were much longer than in wild-type plants and other knockouts (Figure 3B). Because of this, the diameter of PpRALF3 KO plants was significantly larger than that of other genotypes (Figure 3C; ANOVA with post-hoc Tukey HSD P < 0.001). In addition, both double KO mutant lines were less sensitive to paraquat in comparison to wild-type plants (Figure 3C). Thus, our results showed that PpRALF3 is responsive to adverse conditions, and its knockout lines are more tolerant to growth inhibition during an abiotic stress response.
As has been shown for angiosperm RALFs, these results point to the role of PpRALF3 in the response to abiotic stress factors. Previously it has been shown that the overexpression of AtRALF22 or AtRALF23 resulted in increased sensitivity to salt stress (Zhao et al., 2018). However, further research will be required on this topic.
It has been previously shown that some RALF peptides can modulate immune response in vascular plants (Stegmann et al., 2017; Zhang et al., 2020b; Abarca et al., 2021). For example, treatment with AtRALF peptides increased or reduced ROS production under induction of immune response by elf18 (Abarca et al., 2021). To determine whether the knockout of PpRALF genes interfere with immune response in bryophytes, we next analyzed ROS production in P. patens wild-type and knockout lines under driselase treatment. Driselase is a mix of cell wall–degrading enzymes from Basidiomycetes sp. and this enzyme mix is comparable to the enzyme cocktail released by fungal pathogens during infection (Engelsdorf et al., 2018).
We found that ROS production was significantly reduced in PpRALF1 and PpRALF2 KOs in comparison to wild-type plants and PpRALF3 knockout line after driselase treatment (Figure 4A; ANOVA with post-hoc Tukey HSD P < 0.001). Moreover, the background level of ROS production in the PpRALF3 knockout line without treatment was significantly higher than in the other genotypes (Figure 4A; ANOVA with post-hoc Tukey HSD P < 0.001). Background ROS production was also increased in the second PpRALF3 knockout line relative to wild-type plants (Supplementary Figure S6A). Additionally, we analyzed ROS production in double knockout lines following driselase treatment. We found that the background level of ROS production in the PpRALF 1,3 KO line was significantly higher than in the wild-type and PpRALF 2,3 knockout lines (ANOVA with post-hoc Tukey HSD P < 0.001; Supplementary Figure S6B). Double knockouts and wild-type plants did not differ in their ROS elevation in response to driselase treatment. Based on these results, we suggest that PpRLAFs modulate the immune response of P. patens.
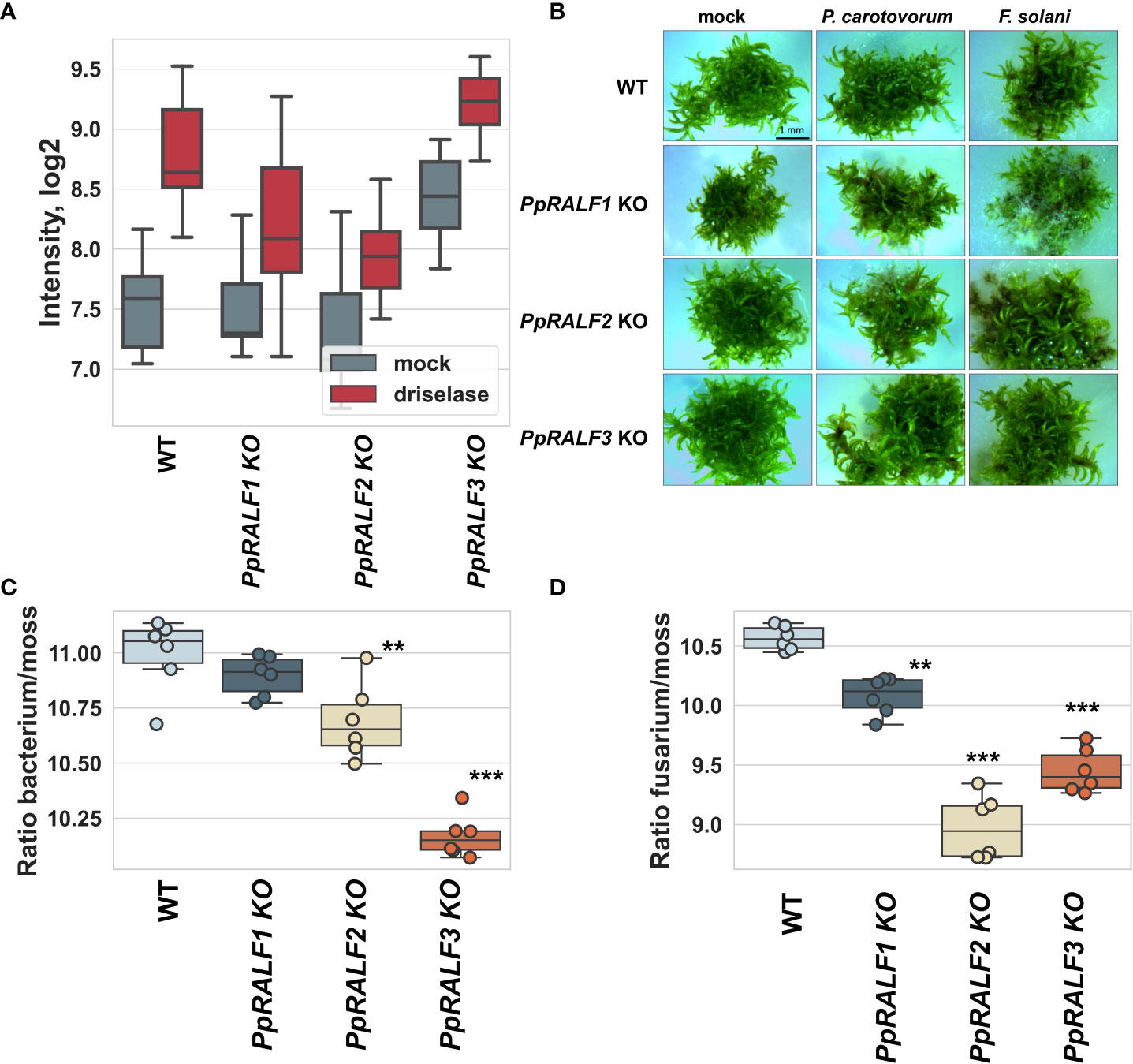
Figure 4 (A) Effect of PpRALF gene knockouts on ROS production in wild-type and knockout lines treated with 0.0025% driselase; (B) Symptom development at 7 dpi in wild-type, PpRALF1, PpRALF2 and PpRALF3 KO moss plants. Scale bar: 1 mm; (C) Ratios of P. carotovorum DNA levels to P. patens genomic DNA that were estimated by qPCR analysis. (D) Ratios of F. solani DNA levels to P. patens genomic DNA that were estimated by qPCR analysis. The results of six independent experiments are shown. Analysis of variance and Tukey’s HSD post hoc tests were performed for qPCR data. **P < 0.01, ***P < 0.001.
To test whether PpRALFs contribute to the P. patens defense response, we used two well-known phytopathogens: Pectobacterium carotovorum subsp. atrosepticum and Fusarium solani. P. carotovorum has been previously shown to infect P. patens (Andersson et al., 2005; Ponce de León et al., 2007). The phytopathogenic fungus with a wide range of hosts, Botrytis cinerea, is commonly used for the study of immune response in bryophytes (Ponce De León et al., 2012; Reboledo et al., 2021). However, B. cinerea is aggressive, and moss plants rapidly deteriorated after inoculation (Ponce De León et al., 2012; Reboledo et al., 2021). Therefore, to compare the immune responses of different genotypes, we selected Fusarium solani, which caused only mild symptoms (Figure 4B). These pathogens also show distinct symptoms of infection, such as brown spots caused by tissue necrosis (Akita et al., 2011; Alvarez et al., 2016). To test whether the colonization and growth of P. carotovorum and F. solani differ between wild-type and knockout lines, moss plants were collected at 7 days post inoculation (dpi), and the ratio of pathogen DNA to plant DNA concentrations was measured using quantitative PCR analysis as was described previously (Castro et al., 2016).
According to our experiments, P. carotovorum growth rate was significantly higher in wild-type plants compared to PpRALF2 KO and PpRALF3 KO mutant lines (Figure 4C; ANOVA with post-hoc Tukey HSD P < 0.001). However, the difference in P. carotovorum growth rate between wild-type and PpRALF1 KO plants was not significant (Figure 4C). The growth of F. solani was significantly reduced in all PpRALF knockouts in comparison to wild-type plants (Figure 4D; ANOVA with post-hoc Tukey HSD P < 0.001). PpRALF2 KO and PpRALF3 KO mutant lines were found to be the most resistant to F. solani infection. These findings suggested that knocking out the PpRALF genes increased P. patens resistance to phytopathogens. Apparently, PpRALF2 and PpRALF3 play a key role in negative regulation of immune response in P. patens, as was shown for some members of the RALF family in Arabidopsis.
We found that both P. carotovorum and F. solani propagated much more slowly in PpRALF2 and 3 KO genotypes than in wild-type plants and PpRALF1 knockout lines. This suggests that these knockout genotypes are more tolerant to phytopathogens. To expand our understanding of the roles of PpRALFs in immune stress responses, we then used RNA sequencing (RNA-seq) analysis to compare the transcriptome responses of PpRALF3 KO and wild-type plants during F. solani infection. Twelve paired-end RNA-Seq libraries were generated from three biological replicates of mock-inoculated and F. solani-infected wild-type and PpRALF3 KO plants (Supplementary Table S4). Overall, we detected more than 1700 differentially regulated genes (DEGs; -1 ≤ log2 fold change ≥ 1, Padj < 0.05) in both genotypes, and 474 DEGs were commonly regulated in wild-type and knockout plants (Figure 5A; Supplementary Table S5). At first, we analyzed these 474 common DEGs and found that their expression changed in a similar way (Figure 5B). Among them we found the well-known pathogenesis-related genes, such as Pp3c11_1420 (the precursor of the antifungal peptide Hevein), Pp3c7_19850, Pp3c17_5160 (pathogenesis-related proteins), and Pp3c6_6560 (DIRIGENT PROTEIN). These DEGs were significantly up-regulated in wild-type and PpRALF3 KO-infected plants (Supplementary Table S5). In addition, the most up-regulated common DEGs in infected plants included Pp3c3_14700 (MLRQ subunit of the NADH-ubiquinone reductase complex), Pp3c7_12870 (expansin 5-related), and Pp3c6_14470 (WRKY TRANSCRIPTION FACTOR 38-RELATED; Supplementary Table S6). These genes are shown to positively regulate plant immune response (Phukan et al., 2016; An et al., 2022). Most common DEGs that were down-regulated in infected plants were Pp3c14_15640 (S-TYPE ANION CHANNEL SLAH2-RELATED), Pp3c20_17620 (Asparagine synthase), Pp3c3_35020 (CCT MOTIF FAMILY PROTEIN-RELATED), and genes with unknown functions (Supplementary Table S5). Using the g:Profiler (Raudvere et al., 2019), we found that commonly regulated DEGs were enriched in GO terms related to response to reactive oxygen species, such as oxidoreductase activity (GO:0016491), response to reactive oxygen species (GO:0000302), and antioxidant activity (GO:0016209; Figure 5C; Supplementary Table S6). Among these genes, we found the superoxide dismutase (Pp3c17_14510), which scavenges ROS and was significantly up-regulated in both infected wild-type and knockout plants.
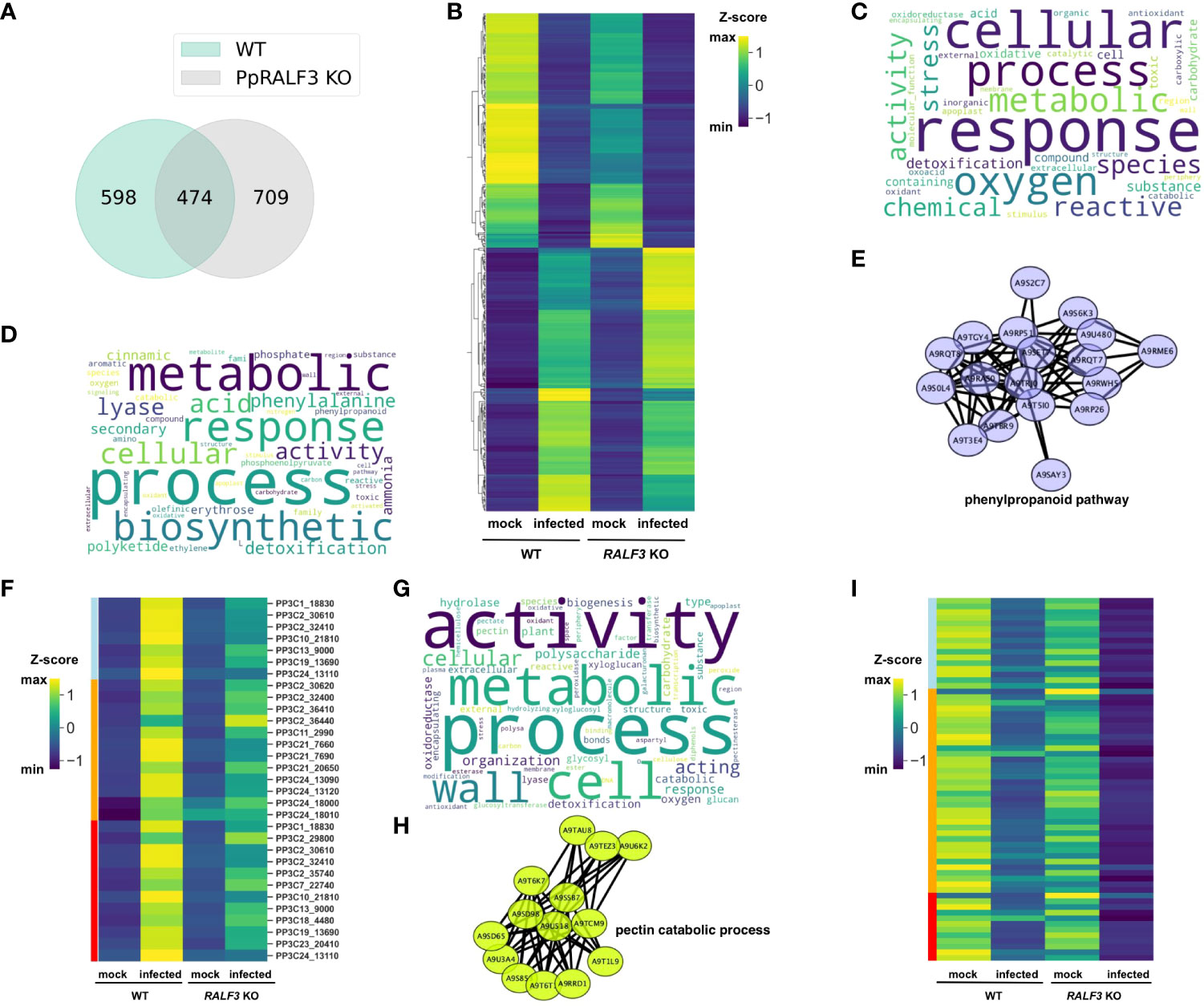
Figure 5 (A) Venn diagram depicting overlapping differentially regulated genes (DEGs) between wild-type and PpRALF3 KO lines; (B) A heat map depicting transcript levels of overlapping between wild-type and PpRALF3 KO differentially expressed genes. Normalized transcripts per million (TPM) values are shown; (C) Gene ontology (GO) terms bias word cloud. Word cloud of enriched GO terms for overlapped DEGs between wild-type and PpRALF3 KO lines is shown (false discovery rate (FDR) adjusted P-values ≤ 0.05); (D) GO terms bias word cloud. Word cloud of enriched GO terms in wild-type plants is shown (false discovery rate (FDR) adjusted P-values ≤ 0.05); (E) Protein–protein interaction networks of a group of upregulated differentially expressed genes in wild-type plants that belong to phenylpropanoid metabolic process. Genes are indicated with nodes, and interactions between proteins are represented by edges; (F) A heat map depicting transcript levels of DEGs that belong to “phenylpropanoid biosynthetic process” (lightblue), “polyketide metabolic process” (orange), “aromatic amino acid family metabolic process” (red). Normalized transcripts per million (TPM) values are shown; (G) GO terms bias word cloud. Word cloud of enriched GO terms in PpRALF3 KO plants is shown (false discovery rate (FDR) adjusted P-values ≤ 0.05); (H) Protein–protein interaction networks of a group of down regulated differentially expressed genes in PpRALF3 KO plants that belong to the pectin catabolic process. Genes are indicated with nodes, and interactions between proteins are represented by edges. (I) A heat map depicting transcript levels of DEGs that belong to “pectin metabolic process” (lightblue), “cell wall organization or biogenesis” (orange), “xyloglucan:xyloglucosyl transferase activity” (red). Normalized transcripts per million (TPM) values are shown.
We next compared the GO terms of all DEGs from wild-type and knockout plants to find differences between genotypes in response to infection. In wild-type plants, the top 10 significantly enriched GO terms belonged to secondary metabolite biosynthetic processes, including polyketide metabolic process (GO:0030638), phenylpropanoid metabolic process (GO:0009698), cinnamic acid metabolic process (GO:0009803; Figure 5D; Supplementary Table S6). The upregulated DEGs belonged to these GO terms included histidine and phenylalanine ammonia-lyases (e.g., Pp3c13_9000) and chalcone synthases (e.g., Pp3c11_9040). We then used the STRING database to create association networks of DEGs from wild-type plants. Based on this analysis, the very distinct cluster of genes involved in the phenylpropanoid pathway was identified (Figure 5E). The transcriptional level of the genes involved in the “phenylpropanoid biosynthetic process”, “polyketide metabolic process” and “aromatic amino acid metabolic process” were significantly increased in wild-type plants in comparison to knockout line (Figure 5F). Among them, a group of histidine-ammonia lyases (e.g., Pp3c2_30610). These findings are in line with the previous study on the interaction between P. patens and B. cinerea (Reboledo et al., 2021). Using qRT-PCR, we additionally examined the transcription of phenylalanine ammonia-lyase (Pp3c13_9000, PAL1), pathogenesis-related protein 10 (Pp3c2_27350, PR10), and dirigent protein (Pp3c6_6545, DIR) in infected PpRALF1 and PpRALF2 KO lines. The induction of the PAL1 gene in the knockout genotypes was significantly lower in comparison to wild-type plants (Supplemental Figure S7). This is consistent with RNA-seq results on the PpRALF3 KO genotype and implies that the phenylpropanoid pathway is suppressed in the PpRALF knockouts. The induction of the PR10 gene was significantly lower only in the PpRALF2 KO line (Supplemental Figure S7). It should be noted that despite a similar pattern of defense gene transcription, the knockout genotypes showed different levels of F. solani propagation (Figure 4D).
Compared to wild-type plants, the GO terms of downregulated genes in PpRALF3 KO-infected plants were mostly enriched for cell wall organization and biogenesis processes, such as pectin catabolic process (GO: 0045490), xyloglucan metabolic process (GO: 0010411), and polysaccharide metabolic process (GO: 0005976; Figure 5G). Based on the STRING analysis, the distinct groups of pectate lyases (e.g., Pp3c21_16990) and pectin esterases were identified in this set of DEGs (Figure 5H). The transcriptional level of DEGs that belonged to “pectin metabolic process”, “cell wall organization or biogenesis”, “xyloglucan:xyloglucosyl transferase activity” was decreased more pronouncedly than in wild-type plants (Figure 5I). This suggests a link between the role of RALF peptides in cell wall regulation and stress response. The upregulated DEGs were enriched by such GO terms as oxidoreductase activity (GO:0016491) and cell wall organization, or biogenesis (GO:0071554). However, the latter GO term included different glycosyl hydrolases and expansins. In conclusion, transcriptomic changes in wild-type plants are in line with previous data on P. patens response to phytopathogens (Reboledo et al., 2021) and alter such processes as ROS production and detoxification, biosynthesis of secondary metabolites with different roles in defense, and some others. The remarkable differences between PpRALF3 KO and wild-type transcriptomes were related to genes involved in cell wall modification and biogenesis processes. Considering the known role of RALF peptides in cell wall remodeling, it can be suggested that these changes during infection result from knockout of the PpRALF3 gene.
RAPID ALKALINIZATION FACTOR (RALF) peptides are ubiquitous for land plants, including Physcomitrium patens and Selaginélla moellendorffii, but were not identified in chlorophyte species (Campbell and Turner, 2017). In angiosperms, RALFs are shown to modulate responses to biotic and abiotic stresses (Blackburn et al., 2020), but it is currently unknown if ancient RALF peptides helped plants to cope with different stress factors or if the immune-related RALFs appeared later as a result of tandem duplication and diversification (Cao and Shi, 2012). Here, we investigated the role of three PpRALF peptides from the model bryophyte P. patens, named as PpRALF1 (Pp3c3_15280), PpRALF2 (Pp3c6_7200) and PpRALF3 (Pp3c25_4180) in the immune response. Previously, PpRALF1 and PpRALF2 have been shown to promote protonema tip growth and elongation (Ginanjar et al., 2022). Although double knockouts were smaller than single knockouts, the phenotype of the mutant lines obtained was similar to those from Ginanjar et al. (2022). Herewith, PpRALF1,3 KO plants were not observed in the previous study (Ginanjar et al., 2022) and we cannot compare these phenotypes. To explain the differences between single- and double-knockout RALF phenotypes, however, additional research is required.
According to our findings, the knockout of all three PpRALF genes led to consistent changes in plant proteomes, suggesting that all three PpRALFs are functional. For example, a predicted metalloprotease Pp3c15_830 was significantly downregulated in all PpRALF knockout lines. Plant metalloproteases are shown to be involved in growth, development, and immunity (Flinn, 2008; Mishra et al., 2021).
The role of RALF peptides in stress response is mainly explored in the context of Arabidopsis biology. In Arabidopsis, several RALF peptides have shown the ability to modulate elf18-induced ROS production (Abarca et al., 2021). Based on the amino acid compositions, we clustered PpRALFs with AtRALFs, such as AtRALF23 and AtRALF33, that negatively regulate immune response and inhibit elf18-induced ROS production in Arabidopsis (Stegmann et al., 2017; Abarca et al., 2021). However, the analysis of amino acid composition was not sufficient to distinguish PpRALF3 from PpRALF1 and 2.
In addition, the AtRALF23 and AtRALF33 were shown to interfere PAMP-triggered immunity (PTI) by binding to receptor FER-LLG1 complex and inhibition of its scaffold function (Shen et al., 2017; Rzemieniewski and Stegmann, 2022). AtRALF23 overexpression increases susceptibility to Pseudomonas syringae pv. tomato DC3000 and to the fungus Plectosphaerella cucumerina (Stegmann et al., 2017) and LRX3/4/5-RALF22/23-FER module negatively regulates the levels of jasmonic acid (JA), salicylic acid (SA) and abscisic acid (ABA) in Arabidopsis (Zhao et al., 2021). The hallmark of AtRALFs, that negatively regulate immune response, is the dibasic site “RR” for subtilase S1P (Srivastava et al., 2009; Stegmann et al., 2017; Abarca et al., 2021). However, there are some exceptions from this rule in Arabidopsis. In P. patens, only PpRALF1 protein precursor contains the corresponding “RRLL” motif (Ginanjar et al., 2022). According to our data, the PpRALF1 peptide has no role in stress response, but the knockouts of two PpRALFs (2 and 3) resulted in increasing resistance to bacterial and fungal pathogens - P. carotovorum and F. solani, suggesting the negative regulation of immune response by these peptides. These PpRALFs are also placed in a distinct group on the phylogenetic tree (Ginanjar et al., 2022). Our phylogenetic analysis clearly indicates that PpRALF3 diverged from PpRALF1 and PpRALF2 (Figure 1C).
In land plants, the immune response resulted in extensive transcriptome reprogramming (Bjornson et al., 2021; Campos et al., 2021; Reboledo et al., 2021). The early transcriptome changes include upregulation of genes involved in response to chitin and wounding at 5 minutes and participated in response to hydrogen peroxide and cell wall modification at 180 minutes after treatment by known elicitors in the model plant Arabidopsis (Bjornson et al., 2021). In our study, we found that transcriptome reprogramming in wild-type P. patens plants at 7 dpi affected genes that participate in response to reactive oxygen species (GO:0000302), polyketide metabolic process (GO:0030638), phenylpropanoid metabolic process (GO:0009698), cinnamic acid metabolic process (GO:0009803). The moss P. patens reacts to the fungal pathogen B. cinerea by reinforcing the cell wall, upregulating genes involved in the defense response, and activating the shikimate and phenylpropanoid pathways (Ponce De León et al., 2012; Castro et al., 2016; Reboledo et al., 2021). The Marchantia response to oomycete infection is also based on the phenylpropanoid-mediated biochemical defenses that suggest this mechanism as a hallmark of an ancestral pathogen deterrence strategy (Carella et al., 2019).
Even though the difference in the pathogenic agent - B. cinerea (necrotrophic fungus) vs F. solani (facultative parasite) and time after inoculation when samples were collected - 24-h vs 7 dpi, about 40% of DEGs in wild-type plants from our data were identical to the Reboledo dataset (Reboledo et al., 2021). However, the overlap between DEGs from PpRALF3 KO plants and the aforementioned dataset was only 27%, suggesting specific immune responses in the knockout line.
Because our ultimate goal was to understand the role of PpRALFs in stress response, we concentrated on the comparison of the transcriptomes between wild-type and PpRALF3 knockout plants during F. solani infection. About 40% of PpRALF3 KO DEGs were identical to wild-type DEGs and changed in similar manner. The corresponding GO terms belonged to oxidoreductase activity (GO:0016491), response to reactive oxygen species (GO:0000302), antioxidant activity (GO:0016209). This finding is in line with previous studies on vascular and non-vascular plants in which increased expression of oxidative stress related genes encoding peroxiredoxins, thioredoxins, ferredoxins during infection was detected (Ponce de León, 2011; Bjornson et al., 2021; Reboledo et al., 2021). According to our results, the PpRALF3 KO DEGs were not enriched in phenylpropanoid and cinnamic acid metabolic processes as we observed in wild-type plants, but some genes involved in the flavonoid biosynthetic process, such as chalcone synthases, were upregulated in mutant lines. Importantly, the PpRALF2 KO and PpRALF3 KO lines were less infected by F. solani than wild-type plants and PpRALF1 KO genotype suggest their increased resistance. Therefore, the transcriptome differences between wild-type and PpRALF3 KO plants might reflect reduced propagation of F. solani in knockout lines. In contrast to wild-type plants, a group of genes involved in the pectin catabolic process, such as pectate lyases and pectin esterases, were significantly downregulated in the PpRALF3 KO plants under infection. Pectins are structural heteropolysaccharides and major components of the plant primary cell wall involved in maintaining plant growth and development, morphogenesis, defense responses, etc. (Hongo et al., 2012; Leng et al., 2017). The pectin-degrading enzymes cause plant tissue maceration, cell lysis and modification of the cell wall structure (Atanasova et al., 2018). Our findings is corroborate with the previous findings that salt stress resulted in downregulation of genes involved in cell wall organization and modification processes, such as pectin lyase-like superfamily proteins, expansins, xyloglucan hydrolases in lrx345 (lrx3, 4, 5 triple mutants) Arabidopsis plants (Zhao et al., 2021). LRX8-LRX11 proteins are shown to interact with RALF4/19 and regulate pollen germination and pollen tube growth in Arabidopsis (Mecchia et al., 2017). The downregulation of similar cell wall related genes in our PpRALF3 knockout line under stress conditions suggest the role of RALF peptides and the cognate receptors, such as FERONIA or LRXs, as modules that integrate plant growth and stress tolerance regulation in land plants.
In addition, it has been shown that pectin methylesterification is a subject of regulation during response to phytopathogens and a high level of pectin methylesterification correlated with an increased resistance to pathogens (Wydra and Beri, 2006; Lionetti et al., 2012). In Arabidopsis, downregulation of some pectin methylesterases during B. cinerea infection probably represent a defense mechanism to limit pectin demethylesterification and its subsequent degradation by fungal enzymes (Lionetti et al., 2017). Together with our results on the increased cell wall regeneration in PpRALF2 and PpRALF3 KO protoplasts relative to wild-type cells, it may point to changes in cell wall composition as an important factor of increased resistance to phytopathogens in the corresponding knockout lines. There is no direct evidence how different RALF peptides influence cell wall composition in plants, but knockout of some LRXs in Arabidopsis resulted in the increase of mannose and lignin if compared with wild-type plants (Draeger et al., 2015). Taken together, our findings suggest the role of PpRALF2 and PpRALF3 peptides in negative regulation of P. patens immune response.
We observed that PpRALF3 and both double KO mutant lines were more tolerant to growth inhibition under salt and oxidative stress conditions. Previously, the role of AtRALF22 or AtRALF23 and their cognate receptors in coordinated regulation of cell wall integrity, growth and salt stress response was demonstrated in Arabidopsis (Zhao et al., 2018; Zhao et al., 2021). The overexpression of AtRALF22 or AtRALF23 are shown to increase sensitivity to salt stress (Zhao et al., 2018). Moreover, LRX3/4/5-RALF22/23-FER module is shown to regulate hormonal homeostasis and ROS accumulation in Arabidopsis. Thus, our findings are in line with previous studies on flowering plants and show that RALF peptides in the non-vascular plants can also participate in abiotic stress response, modulating plant growth in such conditions. The detailed mechanisms through which ancient RALF and their cognate receptors coordinated plant growth, cell wall integrity and response to distinct environmental changes (e.g., pathogen invasion) is unknown. However, the future study on genetically redundant plants, such as P. patens, can help elucidate the evolution and exact mechanisms of RALF peptides signaling.
The nucleotide sequence reported in this paper has been submitted to NCBI Sequence Reads Archive (SRA) with accession number PRJNA879762. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (Perez-Riverol et al., 2022) partner repository with the dataset identifier PXD037111.
AM, IL, IF wrote the manuscript. AM, IL, NG conducted all experiments and contributed to data analysis. IF performed data analysis and supervised the study. AK, VL and DK conducted generation of knockout moss lines, also AK contributed to growth and morphological experiments. TM, EC, SE performed pathogens inoculation and moss treatment. VV, KK, TG and VB performed RNA-seq analysis. All authors contributed to the article and approved the submitted version.
This work was supported by the Russian Foundation for Basic Research (project no. 20-04-00938).
We thank the Center for Precision Genome Editing and Genetic Technologies for Biomedicine, Federal Research and Clinical Center of Physical-Chemical Medicine of the Federal Medical Biological Agency for the expertise and guidance in genetic engineering.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1077301/full#supplementary-material
Abarca, A., Franck, C. M., Zipfel, C. (2021). Family-wide evaluation of RAPID ALKALINIZATION FACTOR peptides. Plant Physiol. 187, 996–1010. doi: 10.1093/plphys/kiab308
Akita, M., Lehtonen, M. T., Koponen, H., Marttinen, E. M., Valkonen, J. P. T. (2011). Infection of the sunagoke moss panels with fungal pathogens hampers sustainable greening in urban environments. Sci. Total Environ. 409, 3166–3173. doi: 10.1016/j.scitotenv.2011.05.009
Alvarez, A., Montesano, M., Schmelz, E., Ponce de León, I. (2016). Activation of shikimate, phenylpropanoid, oxylipins, and auxin pathways in pectobacterium carotovorum elicitors-treated moss. Front. Plant Sci. 7, 328. doi: 10.3389/fpls.2016.00328
Andersson, R. A., Akita, M., Pirhonen, M., Gammelgård, E., Valkonen, J. P. T. (2005). Moss-erwinia pathosystem reveals possible similarities in pathogenesis and pathogen defense in vascular and nonvascular plants. J. Gen. Plant Pathol. 71, 23–28. doi: 10.1007/s10327-004-0154-3
An, L., Zhang, S., Guo, P., Song, L., Xie, C., Guo, H., et al. (2022). RIR1 represses plant immunity by interacting with mitochondrial complex I subunit in rice. Mol. Plant Pathol. 23, 92–103. doi: 10.1111/mpp.13145
Atanasova, L., Dubey, M., Grujić, M., Gudmundsson, M., Lorenz, C., Sandgren, M., et al. (2018). Evolution and functional characterization of pectate lyase PEL12, a member of a highly expanded clonostachys rosea polysaccharide lyase 1 family. BMC Microbiol. 18, 178. doi: 10.1186/s12866-018-1310-9
Atkinson, N. J., Lilley, C. J., Urwin, P. E. (2013). Identification of genes involved in the response of arabidopsis to simultaneous biotic and abiotic stresses. Plant Physiol. 162, 2028–2041. doi: 10.1104/pp.113.222372
Bjornson, M., Pimprikar, P., Nürnberger, T., Zipfel, C. (2021). The transcriptional landscape of arabidopsis thaliana pattern-triggered immunity. Nat. Plants 7, 579–586. doi: 10.1038/s41477-021-00874-5
Blackburn, M. R., Haruta, M., Moura, D. S. (2020). Twenty years of progress in physiological and biochemical investigation of RALF peptides. Plant Physiol. 182, 1657–1666. doi: 10.1104/pp.19.01310
Bolger, A. M., Lohse, M., Usadel, B. (2014). Trimmomatic: a flexible trimmer for illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Bowman, J. L., Kohchi, T., Yamato, K. T., Jenkins, J., Shu, S., Ishizaki, K., et al. (2017). Insights into land plant evolution garnered from the marchantia polymorpha genome. Cell 171, 287–304.e15. doi: 10.1016/j.cell.2017.09.030
Bressendorff, S., Azevedo, R., Kenchappa, C. S., Ponce de León, I., Olsen, J. V., Rasmussen, M. W., et al. (2016). An innate immunity pathway in the moss physcomitrella patens. Plant Cell 28, 1328–1342. doi: 10.1105/tpc.15.00774
Campbell, L., Turner, S. R. (2017). A comprehensive analysis of RALF proteins in green plants suggests there are two distinct functional groups. Front. Plant Sci. 8, 37. doi: 10.3389/fpls.2017.00037
Campos, M. D., Félix, M., do, R., Patanita, M., Materatski, P., Varanda, C. (2021). High throughput sequencing unravels tomato-pathogen interactions towards a sustainable plant breeding. Hortic. Res. 8, 171. doi: 10.1038/s41438-021-00607-x
Cao, J., Shi, F. (2012). Evolution of the RALF gene family in plants: Gene duplication and selection patterns. Evol. Bioinform. Online 8, 271–292. doi: 10.4137/EBO.S9652
Carella, P., Gogleva, A., Hoey, D. J., Bridgen, A. J., Stolze, S. C., Nakagami, H., et al. (2019). Conserved biochemical defenses underpin host responses to oomycete infection in an early-divergent land plant lineage. Curr. Biol. 29, 2282–2294.e5. doi: 10.1016/j.cub.2019.05.078
Castro, A., Vidal, S., Ponce de León, I. (2016). Moss pathogenesis-Related-10 protein enhances resistance to pythium irregulare in physcomitrella patens and arabidopsis thaliana. Front. Plant Sci. 7, 580. doi: 10.3389/fpls.2016.00580
Chen, Z., Zhao, P., Li, F., Leier, A., Marquez-Lago, T. T., Wang, Y., et al. (2018). iFeature: a Python package and web server for features extraction and selection from protein and peptide sequences. Bioinformatics 34, 2499–2502. doi: 10.1093/bioinformatics/bty140
Chevalier, E., Loubert-Hudon, A., Matton, D. P. (2013). ScRALF3, a secreted RALF-like peptide involved in cell-cell communication between the sporophyte and the female gametophyte in a solanaceous species. Plant J. 73, 1019–1033. doi: 10.1111/tpj.12096
Collonnier, C., Epert, A., Mara, K., Maclot, F., Guyon-Debast, A., Charlot, F., et al. (2017). CRISPR-Cas9-mediated efficient directed mutagenesis and RAD51-dependent and RAD51-independent gene targeting in the mossPhyscomitrella patens. Plant Biotechnol. J. 15, 122–131. doi: 10.1111/pbi.12596
Draeger, C., Ndinyanka Fabrice, T., Gineau, E., Mouille, G., Kuhn, B. M., Moller, I., et al. (2015). Arabidopsis leucine-rich repeat extensin (LRX) proteins modify cell wall composition and influence plant growth. BMC Plant Biol. 15, 155. doi: 10.1186/s12870-015-0548-8
Engelsdorf, T., Gigli-Bisceglia, N., Veerabagu, M., McKenna, J. F., Vaahtera, L., Augstein, F., et al. (2018). The plant cell wall integrity maintenance and immune signaling systems cooperate to control stress responses in arabidopsis thaliana. Sci. Signal. 11.eaao3070 doi: 10.1126/scisignal.aao3070
Faurobert, M., Pelpoir, E., Chaïb, J. (2007). Phenol extraction of proteins for proteomic studies of recalcitrant plant tissues. Methods Mol. Biol. 355, 9–14. doi: 10.1385/1-59745-227-0:9
Fesenko, I. A., Arapidi, G. P., Skripnikov, A. Y., Alexeev, D. G., Kostryukova, E. S., Manolov, A. I., et al. (2015). Specific pools of endogenous peptides are present in gametophore, protonema, and protoplast cells of the moss physcomitrella patens. BMC Plant Biol. 15, 87. doi: 10.1186/s12870-015-0468-7
Fesenko, I., Kirov, I., Kniazev, A., Khazigaleeva, R., Lazarev, V., Kharlampieva, D., et al. (2019). Distinct types of short open reading frames are translated in plant cells. Genome Res. 29, 1464–1477. doi: 10.1101/gr.253302.119
Fesenko, I., Shabalina, S. A., Mamaeva, A., Knyazev, A., Glushkevich, A., Lyapina, I., et al. (2021a). A vast pool of lineage-specific microproteins encoded by long non-coding RNAs in plants. Nucleic Acids Res. 49, 10328–10346. doi: 10.1093/nar/gkab816
Fesenko, I., Spechenkova, N., Mamaeva, A., Makhotenko, A. V., Love, A. J., Kalinina, N. O., et al. (2021b). Role of the methionine cycle in the temperature-sensitive responses of potato plants to potato virus y. Mol. Plant Pathol. 22, 77–91. doi: 10.1111/mpp.13009
Flinn, B. S. (2008). Plant extracellular matrix metalloproteinases. Funct. Plant Biol. 35, 1183–1193. doi: 10.1071/FP08182
Franck, C. M., Westermann, J., Boisson-Dernier, A. (2018). Plant malectin-like receptor kinases: From cell wall integrity to immunity and beyond. Annu. Rev. Plant Biol. 69, 301–328. doi: 10.1146/annurev-arplant-042817-040557
Frederick, R. O., Haruta, M., Tonelli, M., Lee, W., Cornilescu, G., Cornilescu, C. C., et al. (2019). Function and solution structure of the arabidopsis thaliana RALF8 peptide. Protein Sci. 28, 1115–1126. doi: 10.1002/pro.3628
Ge, Z., Bergonci, T., Zhao, Y., Zou, Y., Du, S., Liu, M.-C., et al. (2017). Arabidopsis pollen tube integrity and sperm release are regulated by RALF-mediated signaling. Science 358, 1596–1600. doi: 10.1126/science.aao3642
Ge, Z., Dresselhaus, T., Qu, L.-J. (2019). How CrRLK1L receptor complexes perceive RALF signals. Trends Plant Sci. 24, 978–981. doi: 10.1016/j.tplants.2019.09.002
Ginanjar, E. F., Teh, O.-K., Fujita, T. (2022). Characterisation of rapid alkalinisation factors in physcomitrium patens reveals functional conservation in tip growth. New Phytol. 233, 2442–2457. doi: 10.1111/nph.17942
Haruta, M., Sabat, G., Stecker, K., Minkoff, B. B., Sussman, M. R. (2014). A peptide hormone and its receptor protein kinase regulate plant cell expansion. Science 343, 408–411. doi: 10.1126/science.1244454
Herrera-Ubaldo, H., de Folter, S. (2018). Exploring cell wall composition and modifications during the development of the gynoecium medial domain in arabidopsis. Front. Plant Sci. 9, 454. doi: 10.3389/fpls.2018.00454
Hoang, Q. T., Cho, S. H., McDaniel, S. F., Ok, S. H., Quatrano, R. S., shin, J. S. (2009). An actinoporin plays a key role in water stress in the moss physcomitrella patens. New Phytol. 184, 502–510. doi: 10.1111/j.1469-8137.2009.02975.x
Hongo, S., Sato, K., Yokoyama, R., Nishitani, K. (2012). Demethylesterification of the primary wall by PECTIN METHYLESTERASE35 provides mechanical support to the arabidopsis stem. Plant Cell 24, 2624–2634. doi: 10.1105/tpc.112.099325
Kabir, M. N., Taheri, A., Dumenyo, C. K. (2020). Development of PCR-based detection system for soft rot pectobacteriaceae pathogens using molecular signatures. Microorganisms 8.358 doi: 10.3390/microorganisms8030358
Kalyaanamoorthy, S., Minh, B. Q., Wong, T. K. F., von Haeseler, A., Jermiin, L.S. (2017). ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods 14, 587–589. doi: 10.1038/nmeth.4285
Katoh, K., Rozewicki, J., Yamada, K. D. (2019). MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 20, 1160–1166. doi: 10.1093/bib/bbx108
Khraiwesh, B., Qudeimat, E., Thimma, M., Chaiboonchoe, A., Jijakli, K., Alzahmi, A., et al. (2015). Genome-wide expression analysis offers new insights into the origin and evolution of physcomitrella patens stress response. Sci. Rep. 5, 17434. doi: 10.1038/srep17434
Kim, D., Langmead, B., Salzberg, S. L. (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360. doi: 10.1038/nmeth.3317
Kolberg, L., Raudvere, U., Kuzmin, I., Vilo, J., Peterson, H. (2020). gprofiler2 – an r package for gene list functional enrichment analysis and namespace conversion toolset g:Profiler. F1000Res 9.709 doi: 10.12688/f1000research.24956.2
Lang, D., Ullrich, K. K., Murat, F., Fuchs, J., Jenkins, J., Haas, F. B., et al. (2018). The physcomitrella patens chromosome-scale assembly reveals moss genome structure and evolution. Plant J. 93, 515–533. doi: 10.1111/tpj.13801
Lascano, R., Munoz, N., Robert, G., Rodriguez, M., Melchiorre, M., Trippi, V., et al. (2012). Paraquat: An oxidative stress inducer. Herbicides - properties, synthesis and control of weeds. doi: 10.5772/32590
Lehtonen, M. T., Marttinen, E. M., Akita, M., Valkonen, J. P. T. (2012). Fungi infecting cultivated moss can also cause diseases in crop plants. Ann. Appl. Biol. 160, 298–307. doi: 10.1111/j.1744-7348.2012.00543.x
Leng, Y., Yang, Y., Ren, D., Huang, L., Dai, L., Wang, Y., et al. (2017). A rice PECTATE LYASE-LIKE gene is required for plant growth and leaf senescence. Plant Physiol. 174, 1151–1166. doi: 10.1104/pp.16.01625
Liao, Y., Smyth, G. K., Shi, W. (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930. doi: 10.1093/bioinformatics/btt656
Liao, H., Tang, R., Zhang, X., Luan, S., Yu, F. (2017). FERONIA receptor kinase at the crossroads of hormone signaling and stress responses. Plant Cell Physiol. 58, 1143–1150. doi: 10.1093/pcp/pcx048
Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., et al. (2009). The sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079. doi: 10.1093/bioinformatics/btp352
Lionetti, V., Cervone, F., Bellincampi, D. (2012). Methyl esterification of pectin plays a role during plant–pathogen interactions and affects plant resistance to diseases. J. Plant Physiol. 169, 1623–1630. doi: 10.1016/j.jplph.2012.05.006
Lionetti, V., Fabri, E., De Caroli, M., Hansen, A. R., Willats, W. G. T., Piro, G., et al. (2017). Three pectin methylesterase inhibitors protect cell wall integrity for arabidopsis immunity to botrytis. Plant Physiol. 173, 1844–1863. doi: 10.1104/pp.16.01185
Li, C., Yeh, F.-L., Cheung, A. Y., Duan, Q., Kita, D., Liu, M.-C., et al. (2015). Glycosylphosphatidylinositol-anchored proteins as chaperones and co-receptors for FERONIA receptor kinase signaling in arabidopsis. Elife 4. doi: 10.7554/eLife.06587
Loubert-Hudon, A., Mazin, B. D., Chevalier, É., Matton, D. P. (2020). The ScRALF3 secreted peptide is involved in sporophyte to gametophyte signalling and affects pollen mitosis I. Plant Biol. 22, 13–20. doi: 10.1111/plb.13046
Luo, X., Wang, L., Fu, Y., Liu, Q., Chen, G., Liu, Y., et al. (2022). FERONIA-like receptor 1-mediated calcium ion homeostasis is involved in the immune response. Front. Plant Sci. 13, 934195. doi: 10.3389/fpls.2022.934195
Maksimov, N. M., Breigin, M. A., Ermakov, I. P. (2016). Regulation of ion transport across the pollen tube plasmalemma by hydrogen peroxide. Cell Tissue Biol. 10, 69–75. doi: 10.1134/S1990519X16010077
Marttinen, E. M., Niemi-Kapee, J., Laaka-Lindberg, S., Valkonen, J. P. T. (2020). Fungal pathogens infecting moss green roofs in Finland. Urban For. Urban Greening 55, 126812. doi: 10.1016/j.ufug.2020.126812
Masachis, S., Segorbe, D., Turrà, D., Leon-Ruiz, M., Fürst, U., El Ghalid, M., et al. (2016). A fungal pathogen secretes plant alkalinizing peptides to increase infection. Nat. Microbiol. 1, 16043. doi: 10.1038/nmicrobiol.2016.43
McKinney, W. (2012). Python For data analysis: Data wrangling with pandas, NumPy, and IPython (“O’Reilly Media, Inc.” California, USA).
Mecchia, M. A., Rövekamp, M., Giraldo-Fonseca, A., Meier, D., Gadient, P., Vogler, H., et al. (2022). The single marchantia polymorpha FERONIA homolog reveals an ancestral role in regulating cellular expansion and integrity. Development 149.dev200580 doi: 10.1242/dev.200580
Mecchia, M. A., Santos-Fernandez, G., Duss, N. N., Somoza, S. C., Boisson-Dernier, A., Gagliardini, V., et al. (2017). RALF4/19 peptides interact with LRX proteins to control pollen tube growth in arabidopsis. Science 358, 1600–1603. doi: 10.1126/science.aao5467
Merino, M. C., Guidarelli, M., Negrini, F., De Biase, D., Pession, A., Baraldi, E. (2019). Induced expression of the fragaria × ananassa rapid alkalinization factor-33-like gene decreases anthracnose ontogenic resistance of unripe strawberry fruit stages. Mol. Plant Pathol. 20, 1252–1263. doi: 10.1111/mpp.12837
Minh, B. Q., Nguyen, M. A. T., von Haeseler, A. (2013). Ultrafast approximation for phylogenetic bootstrap. Mol. Biol. Evol. 30, 1188–1195. doi: 10.1093/molbev/mst024
Mishra, L. S., Kim, S.-Y., Caddell, D. F., Coleman-Derr, D., Funk, C. (2021). Loss of arabidopsis matrix metalloproteinase-5 affects root development and root bacterial communities during drought stress. Physiol. Plant 172, 1045–1058. doi: 10.1111/ppl.13299
Mohanasundaram, B., Bhide, A. J., Palit, S., Chaturvedi, G., Lingwan, M., Masakapalli, S. K., et al. (2021). The unique bryophyte-specific repeat-containing protein SHORT-LEAF regulates gametophore development in moss. Plant Physiol. 187, 203–217. doi: 10.1093/plphys/kiab261
Moussu, S., Broyart, C., Santos-Fernandez, G., Augustin, S., Wehrle, S., Grossniklaus, U., et al. (2020). Structural basis for recognition of RALF peptides by LRX proteins during pollen tube growth. Proc. Natl. Acad. Sci. U.S.A. 117, 7494–7503. doi: 10.1073/pnas.2000100117
Murphy, E., De Smet, I. (2014). Understanding the RALF family: A tale of many species. Trends Plant Sci. 19, 664–671. doi: 10.1016/j.tplants.2014.06.005
Nguyen, L.-T., Schmidt, H. A., von Haeseler, A., Minh, B. Q. (2015). IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274. doi: 10.1093/molbev/msu300
Oliver, J. P., Castro, A., Gaggero, C., Cascón, T., Schmelz, E. A., Castresana, C., et al. (2009). Pythium infection activates conserved plant defense responses in mosses. Planta 230, 569–579. doi: 10.1007/s00425-009-0969-4
Olsson, V., Joos, L., Zhu, S., Gevaert, K., Butenko, M. A., De Smet, I. (2019). Look closely, the beautiful may be small: Precursor-derived peptides in plants. Annu. Rev. Plant Biol. 70, 153–186. doi: 10.1146/annurev-arplant-042817-040413
Overdijk, E. J. R., de Keijzer, J., de Groot, D., Schoina, C., Bouwmeester, K., Ketelaar, T., et al. (2016). Interaction between the moss physcomitrella patens and phytophthora : a novel pathosystem for live-cell imaging of subcellular defence. J. Microsc. 263, 171–180. doi: 10.1111/jmi.12395
Ow, S. Y., Salim, M., Noirel, J., Evans, C., Rehman, I., Wright, P. C. (2009). iTRAQ underestimation in simple and complex mixtures: “The good, the bad and the ugly”. J. Proteome Res. 8, 5347–5355. doi: 10.1021/pr900634c
Perez-Riverol, Y., Bai, J., Bandla, C., García-Seisdedos, D., Hewapathirana, S., Kamatchinathan, S., et al. (2022). The PRIDE database resources in 2022: a hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res. 50, D543–D552. doi: 10.1093/nar/gkab1038
Phukan, U. J., Jeena, G. S., Shukla, R. K. (2016). WRKY transcription factors: Molecular regulation and stress responses in plants. Front. Plant Sci. 7, 760. doi: 10.3389/fpls.2016.00760
Ponce de León, I. (2011). The moss physcomitrella patens as a model system to study interactions between plants and phytopathogenic fungi and oomycetes. J. Pathog. 2011, 719873. doi: 10.4061/2011/719873
Ponce de León, I., Montesano, M. (2013). Activation of defense mechanisms against pathogens in mosses and flowering plants. Int. J. Mol. Sci. 14, 3178–3200. doi: 10.3390/ijms14023178
Ponce de León, I., Oliver, J. P., Castro, A., Gaggero, C., Bentancor, M., Vidal, S. (2007). Erwinia carotovora elicitors and botrytis cinerea activate defense responses in physcomitrella patens. BMC Plant Biol. 7, 52. doi: 10.1186/1471-2229-7-52
Ponce De León, I., Schmelz, E. A., Gaggero, C., Castro, A., Álvarez, A., Montesano, M. (2012). Physcomitrella patens activates reinforcement of the cell wall, programmed cell death and accumulation of evolutionary conserved defence signals, such as salicylic acid and 12-oxo-phytodienoic acid, but not jasmonic acid, upon botrytis cinerea infection. Mol. Plant Pathol. 13, 960–974. doi: 10.1111/j.1364-3703.2012.00806.x
Raudvere, U., Kolberg, L., Kuzmin, I., Arak, T., Adler, P., Peterson, H., et al. (2019). g:Profiler: a web server for functional enrichment analysis and conversions of gene lists 2019 Update). Nucleic Acids Res. 47, W191–W198. doi: 10.1093/nar/gkz369
Reboledo, G., Agorio, A. D., Vignale, L., Batista-García, R. A., Ponce De León, I. (2021). Transcriptional profiling reveals conserved and species-specific plant defense responses during the interaction of physcomitrium patens with botrytis cinerea. Plant Mol. Biol. 107, 365–385. doi: 10.1007/s11103-021-01116-0
Reboledo, G., Del Campo, R., Alvarez, A., Montesano, M., Mara, H., Ponce de León, I. (2015). Physcomitrella patens activates defense responses against the pathogen colletotrichum gloeosporioides. Int. J. Mol. Sci. 16, 22280–22298. doi: 10.3390/ijms160922280
Robinson, M. D., McCarthy, D. J., Smyth, G. K. (2010). edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. doi: 10.1093/bioinformatics/btp616
Rzemieniewski, J., Stegmann, M. (2022). Regulation of pattern-triggered immunity and growth by phytocytokines. Curr. Opin. Plant Biol. 68, 102230. doi: 10.1016/j.pbi.2022.102230
Schaefer, D. G., Zrÿd, J. P. (1997). Efficient gene targeting in the moss physcomitrella patens. Plant J. 11, 1195–1206. doi: 10.1046/j.1365-313X.1997.11061195.x
Schessner, J. P., Voytik, E., Bludau, I. (2022). A practical guide to interpreting and generating bottom-up proteomics data visualizations. Proteomics 22, e2100103. doi: 10.1002/pmic.202100103
Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., et al. (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504. doi: 10.1101/gr.1239303
Shen, Q., Bourdais, G., Pan, H., Robatzek, S., Tang, D. (2017). Arabidopsis glycosylphosphatidylinositol-anchored protein LLG1 associates with and modulates FLS2 to regulate innate immunity. Proc. Natl. Acad. Sci. U.S.A. 114, 5749–5754. doi: 10.1073/pnas.1614468114
Solis-Miranda, J., Fonseca-García, C., Nava, N., Pacheco, R., Quinto, C. (2020). Genome-wide identification of the CrRLK1L subfamily and comparative analysis of its role in the legume-rhizobia symbiosis. Genes 11.793 doi: 10.3390/genes11070793
Srivastava, R., Liu, J.-X., Guo, H., Yin, Y., Howell, S. H. (2009). Regulation and processing of a plant peptide hormone, AtRALF23, in arabidopsis. Plant J. 59, 930–939. doi: 10.1111/j.1365-313X.2009.03926.x
Stegmann, M., Monaghan, J., Smakowska-Luzan, E., Rovenich, H., Lehner, A., Holton, N., et al. (2017). The receptor kinase FER is a RALF-regulated scaffold controlling plant immune signaling. Science 355, 287–289. doi: 10.1126/science.aal2541
Szklarczyk, D., Franceschini, A., Wyder, S., Forslund, K., Heller, D., Huerta-Cepas, J., et al. (2014). STRING v10: protein–protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 43, D447–D452. doi: 10.1093/nar/gku1003
Tang, J., Wu, D., Li, X., Wang, L., Xu, L., Zhang, Y., et al. (2022). Plant immunity suppression via PHR1-RALF-FERONIA shapes the root microbiome to alleviate phosphate starvation. EMBO J. 41, e109102. doi: 10.15252/embj.2021109102
Thynne, E., Saur, I. M. L., Simbaqueba, J., Ogilvie, H. A., Gonzalez-Cendales, Y., Mead, O., et al. (2017). Fungal phytopathogens encode functional homologues of plant rapid alkalinization factor (RALF) peptides. Mol. Plant Pathol. 18, 811–824. doi: 10.1111/mpp.12444
Van Rossum, G., Drake, F. L. (1995). Python Tutorial: Centrum voor wiskunde en informatica Amsterdam. Netherlands. C.OMMAJ.R.X.X.X.
Vidali, L., Bezanilla, M. (2012). Physcomitrella patens: a model for tip cell growth and differentiation. Curr. Opin. Plant Biol. 15, 625–631. doi: 10.1016/j.pbi.2012.09.008
Virtanen, P., Gommers, R., Oliphant, T. E., Haberland, M., Reddy, T., Cournapeau, D., et al. (2020). SciPy 1.0: fundamental algorithms for scientific computing in Python. Nat. Methods 17, 261–272. doi: 10.1038/s41592-019-0686-2
Waskom, M. (2021). Seaborn: Statistical data visualization. J. Open Source Software 6 3021. doi: 10.21105/joss.03021
Waterhouse, A. M., Procter, J. B., Martin, D. M. A., Clamp, M., Barton, G. J. (2009). Jalview version 2–a multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191. doi: 10.1093/bioinformatics/btp033
Wieghaus, A., Prüfer, D., Schulze Gronover, C. (2019). Loss of function mutation of the rapid alkalinization factor (RALF1)-like peptide in the dandelion taraxacum koksaghyz entails a high-biomass taproot phenotype. PLoS One 14, e0217454. doi: 10.1371/journal.pone.0217454
Wood, A. K. M., Walker, C., Lee, W.-S., Urban, M., Hammond-Kosack, K. E. (2020). Functional evaluation of a homologue of plant rapid alkalinisation factor (RALF) peptides in fusarium graminearum. Fungal Biol. 124, 753–765. doi: 10.1016/j.funbio.2020.05.001
Wydra, K., Beri, H. (2006). Structural changes of homogalacturonan, rhamnogalacturonan I and arabinogalactan protein in xylem cell walls of tomato genotypes in reaction to ralstonia solanacearum. Physiol. Mol. Plant Pathol. 68, 41–50. doi: 10.1016/j.pmpp.2006.06.001
Xiao, Y., Stegmann, M., Han, Z., DeFalco, T. A., Parys, K., Xu, L., et al. (2019). Mechanisms of RALF peptide perception by a heterotypic receptor complex. Nature 572, 270–274. doi: 10.1038/s41586-019-1409-7
Zhang, X., Peng, H., Zhu, S., Xing, J., Li, X., Zhu, Z., et al. (2020a). Nematode-encoded RALF peptide mimics facilitate parasitism of plants through the FERONIA receptor kinase. Mol. Plant 13, 1434–1454. doi: 10.1016/j.molp.2020.08.014
Zhang, X., Yang, Z., Wu, D., Yu, F. (2020b). RALF–FERONIA signaling: Linking plant immune response with cell growth. Plant Commun. 1, 100084. doi: 10.1016/j.xplc.2020.100084
Zhao, C., Jiang, W., Zayed, O., Liu, X., Tang, K., Nie, W., et al. (2021). The LRXs-RALFs-FER module controls plant growth and salt stress responses by modulating multiple plant hormones. Natl. Sci. Rev. 8, nwaa149. doi: 10.1093/nsr/nwaa149
Keywords: RALF, peptide, stress response, immunity, phytopathogens
Citation: Mamaeva A, Lyapina I, Knyazev A, Golub N, Mollaev T, Chudinova E, Elansky S, Babenko VV, Veselovsky VA, Klimina KM, Gribova T, Kharlampieva D, Lazarev V and Fesenko I (2023) RALF peptides modulate immune response in the moss Physcomitrium patens. Front. Plant Sci. 14:1077301. doi: 10.3389/fpls.2023.1077301
Received: 22 October 2022; Accepted: 13 January 2023;
Published: 27 January 2023.
Edited by:
Zhaoyang Zhou, China Agricultural University, ChinaReviewed by:
Deepak D. Bhandari, Michigan State University, United StatesCopyright © 2023 Mamaeva, Lyapina, Knyazev, Golub, Mollaev, Chudinova, Elansky, Babenko, Veselovsky, Klimina, Gribova, Kharlampieva, Lazarev and Fesenko. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Igor Fesenko, ZmVzaWdvckBnbWFpbC5jb20=
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.