- 1Tobacco Research Institute of Chinese Academy of Agricultural Sciences, Qingdao, China
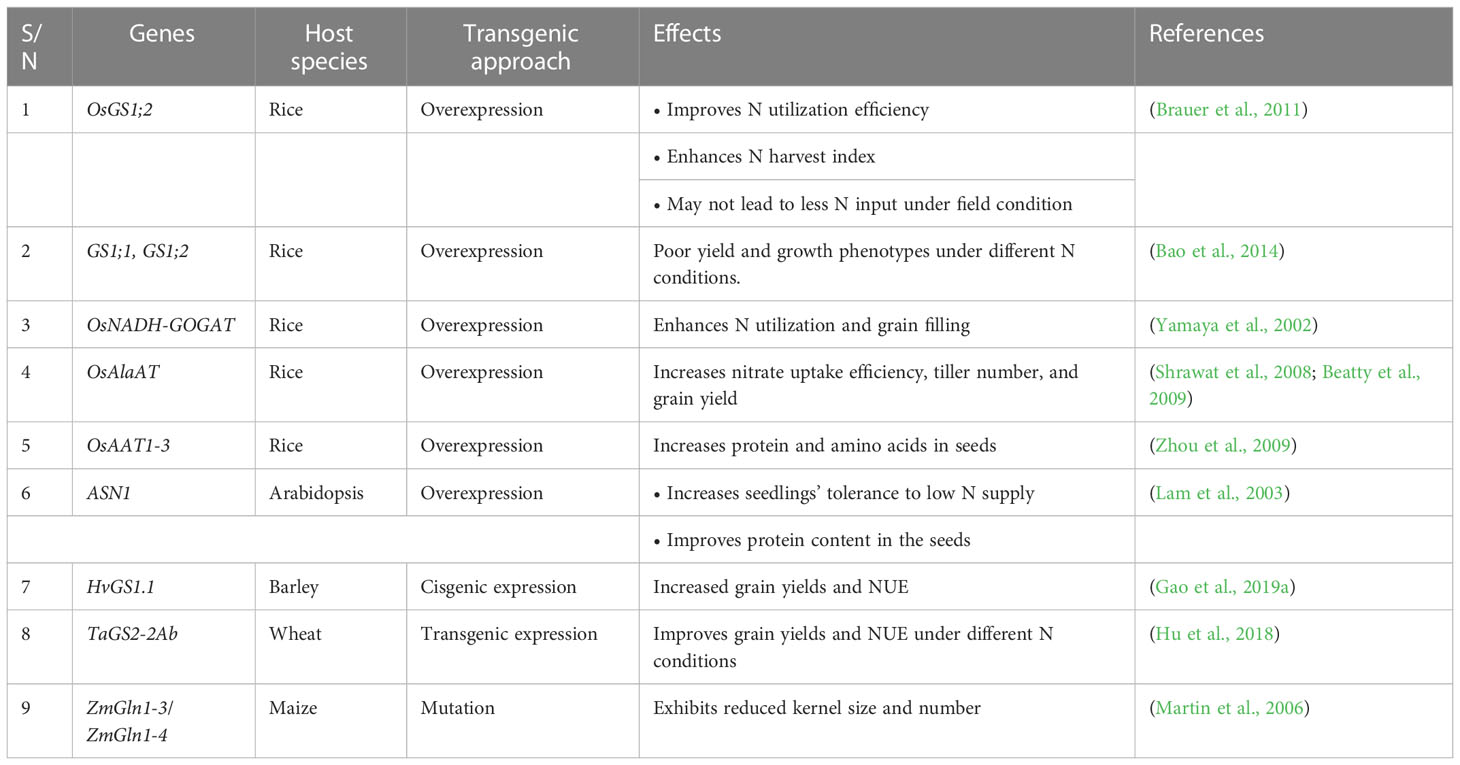
- 2Graduate School of Chinese Academy of Agricultural Sciences, Beijing, China
- 3Agriculture Victoria, Grains Innovation Park, Horsham, VIC, Australia
- 4School of Applied Systems Biology, La Trobe University, Bundoora, VIC, Australia
- 5School of Agriculture, Federal College of Agriculture, Ibadan, Nigeria
Nitrate () transporters have been identified as the primary targets involved in plant nitrogen (N) uptake, transport, assimilation, and remobilization, all of which are key determinants of nitrogen use efficiency (NUE). However, less attention has been directed toward the influence of plant nutrients and environmental cues on the expression and activities of transporters. To better understand how these transporters function in improving plant NUE, this review critically examined the roles of transporters in N uptake, transport, and distribution processes. It also described their influence on crop productivity and NUE, especially when co-expressed with other transcription factors, and discussed these transporters’ functional roles in helping plants cope with adverse environmental conditions. We equally established the possible impacts of transporters on the uptake and utilization efficiency of other plant nutrients while suggesting possible strategic approaches to improving NUE in plants. Understanding the specificity of these determinants is crucial to achieving better N utilization efficiency in crops within a given environment.
1 Introduction
Nitrogen (N) is an essential element required for plant growth and overall yield; hence, the demand and use of N-based chemical fertilizers have consistently increased over the years. Approximately 60-70% of the applied N fertilizers are lost to the environment (Mohanty et al., 2020), causing severe environmental havoc such as pollution, global warming, biodiversity loss, and major plant physiological disorders. Since the increasing rate of N application is becoming increasingly alarming, minimizing fertilizer use while maintaining a high crop yield would be imperative. Thus, improving plants’ nitrogen use efficiency (NUE) is one of the inherent ways of overcoming these crises associated with crop production. Efficient N utilization is a critical factor in crop yield improvement, and research has shown that over 1.0 billion US dollars might be saved with a one percent NUE increment (Kant et al., 2011a).
Crop NUE is the measure of seed yield, grain, or fruit corresponding to a unit of soil N supplied, depending on the individual species of plant. NUE can also be expressed in terms of N uptake efficiency (NUpE), N transport efficiency (NTE), N remobilization efficiency (NRE), and N utilization (assimilation) efficiency (NUtE) (Bharati and Mandal, 2019), all of which are key determinant factors of NUE in plants. N is made available to plants in organic and inorganic forms; nitrate () and ammonium. Due to the mobility nature of , it gets easily leached; thus, its availability to plants becomes limiting (Jin et al., 2015). functions as a signaling molecule, inducing the expression of NO3--related genes involved in its uptake, transport, assimilation, vegetative and reproductive development. Plants take up from the root, assimilate , and subsequently transport it to the shoot, where it can be remobilized to sink organs (Iqbal et al., 2020). transporters are the main drivers involved in the uptake of to the remobilization stage.
Indeed, several studies have discussed the relationship between uptake transport activities in plants while addressing the mechanisms involved in transport, sensing, and signaling processes (Fan et al., 2017; Zuluaga and Sonnante, 2019; Vidal et al., 2020). Therefore, optimizing the activities of transporters is a prerequisite for plants to utilize N supplies. Some studies have elucidated the functional roles of these transporters in plant NUE improvement. However, less is known about the influence of essential nutrients and environmental cues on the expression and activities of transporters. To better understand the extent to which these transporters can function in improving plant NUE, an illustration of their response to changes in plant environmental cues, including salinity, pathogenic and drought stress, and contamination from heavy metals, becomes expedient. Even if these conditions are being optimized, it is crucial to explore the possible aftermath effect of these transporters on the efficiency of other plant nutrient elements and related factors. These necessities ignite a few questions: 1) Does stress affect transporter activities directly or indirectly? and 2) Do the activities of these transporters exert a positive or negative effect on the uptake of other nutrients? To resolve these issues, this review critically summarized the roles of transporters in N uptake, transport, and distribution processes and their functions in crop productivity and NUE, especially when coexpressed with other transcription factors. This review focuses on the functional roles of these nitrate transporters in assisting plants in adverse environmental conditions. We also discussed the impact of these transporters on the uptake and utilization efficiency of other plant nutrients while describing possible strategic approaches to improving NUE in plants. The contribution of nitrate transporters in nitrate and auxin crosstalk for root growth and NUE is also reviewed. Understanding the specificity of all these factors is crucial for better N utilization efficiency of crops.
2 Nitrate uptake and transport systems
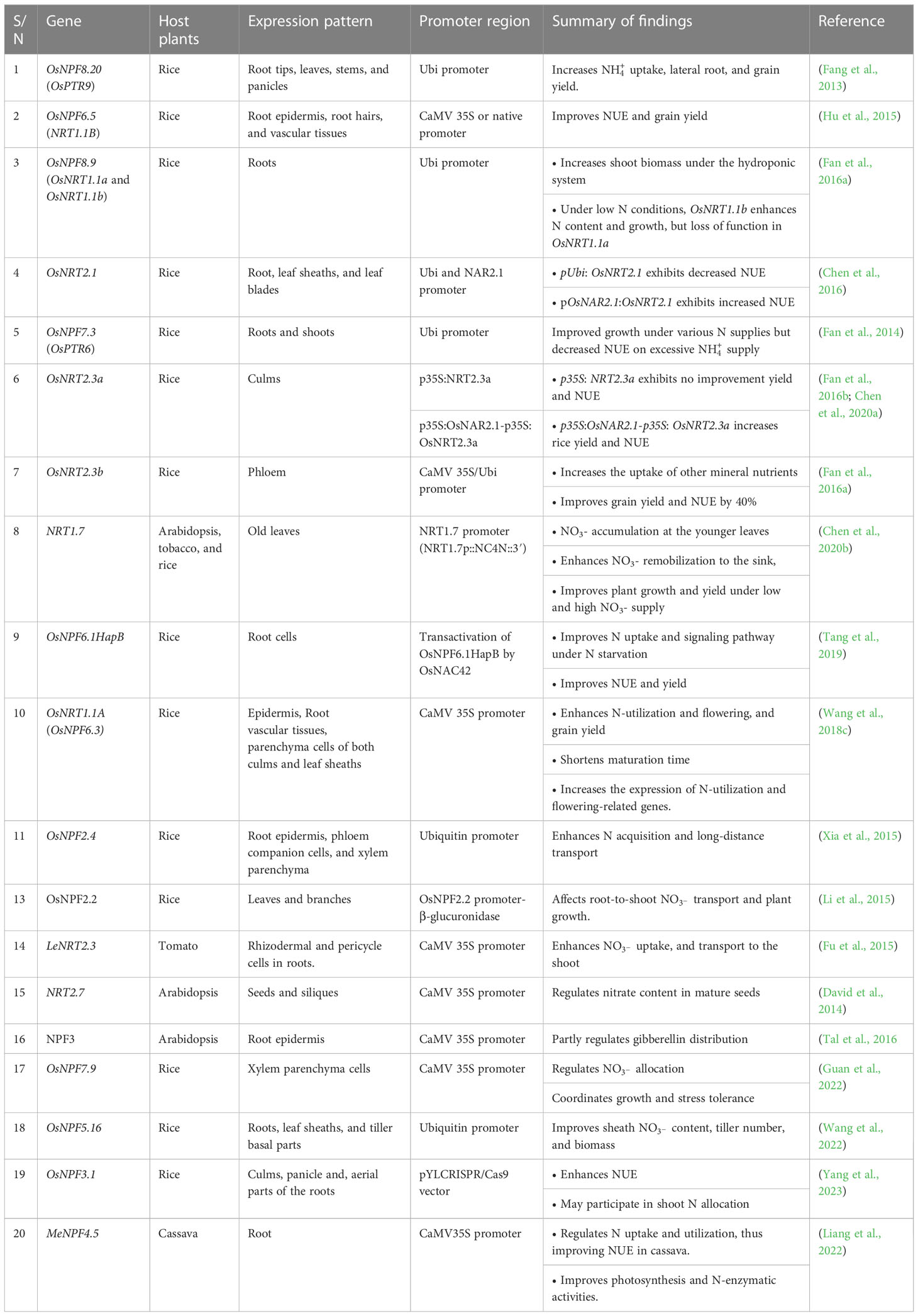
Most agricultural fields, especially, those used for commercial crop production, are deficient with significant spatiotemporal fluctuations, inhibiting N utilization (Kant, 2018). Plants have evolved two major uptake mechanisms to survive. The first is the low-affinity transport system (LATS), which facilitates nitrate uptake under high soil-N (millimolar concentration; > 0.5 mM), while the other is the high-affinity transport system (HATS), which drives nitrate under insufficient soil-N (micromolar range) (Léran et al., 2014; Iqbal et al., 2020; Raddatz et al., 2020). Four families of transporters have been widely known to participate in plant nitrate uptake and transport: nitrate transporter 1/or peptide transporter NPF (NRT1), nitrate transporter 2/nitrate-nitrite-porter NRT2/NNP, slow anion channel-associated homologs (SLAC/SLAH), and chloride channel (CLC) (Tsay et al., 1993; Bergsdorf et al., 2009; Maierhofer et al., 2014; Von Wittgenstein et al., 2014). Among them, NPF (NRT1) and NRT2 and homologs have been identified as the major channels actively involved in root nitrate uptake and long-distance transport between and within plant organs (Hsu and Tsay, 2013; Wang et al., 2021b). In this review, proteins or genes void of prefixes connote Arabidopsis plant species.
Phylogenetic studies revealed that the NPF family comprises 53 identified Arabidopsis genes, and over 130 genes exist in higher plants (Zhang et al., 2020). Generally, NPF transporter genes have low affinity for , except for Chlorate resistant 1/nitrate transporter 1 (CHL1/NRT1.1), also called NPF6.3, a dual-affinity nitrate transporter that operates as both a low- and high-affinity transporter (Liu and Tsay, 2003). The regulatory mechanism involved in the dual-affinity system enables the rapid switch between these two affinity modes. Under a low external supply of , NPF6.3 (CHL1/NRT1.1) functions as a high-affinity transporter and is phosphorylated, whereas it becomes dephosphorylated under a high supply to perform a low-affinity transporter role (Liu and Tsay, 2003; Noguero et al., 2018). Thus, the affinity of the NPF6.3 transporter for uptake depends on the phosphorylation state at the T101 residue, which is subject to the status of N in the medium NPF6.3 (CHL1/NRT1.1) is expressed in various plant tissues, including younger leaves, flower buds, and roots, where it participates in root uptake and translocation (Noguero et al., 2018). In addition to NPF6.3 (CHL1/NRT1.1), NPF4.6 (NRT1.2) and NPF2.7 (NAXT1) are the two putative NPF genes that coordinate influx and efflux in plant roots, respectively (Figure 1). NPF4.6 (NRT1.2) is primarily expressed at the root tip where it takes up (Huang et al., 1999), whereas NPF2.7 (NAXT1), is expressed in the root zone but in the cortex, performs -efflux functions (Segonzac et al., 2007). A considerable amount of NRT1 family members have been identified in other crops, including wheat (Triticum aestivum) (Kumar et al., 2022), rice (Oryza sativa) (Yang et al., 2020), cucumber (Cucumis sativus) (Migocka et al., 2013), potato (Solanum tuberosum) (Zhang et al., 2021a), and apple (Malus × domestica Borkh.) (Wang et al., 2018b), with their unique expression at either the root or shoot of plants. The expression pattern of these transporters is a clear indication of their active involvement in uptake and long-distance transport.
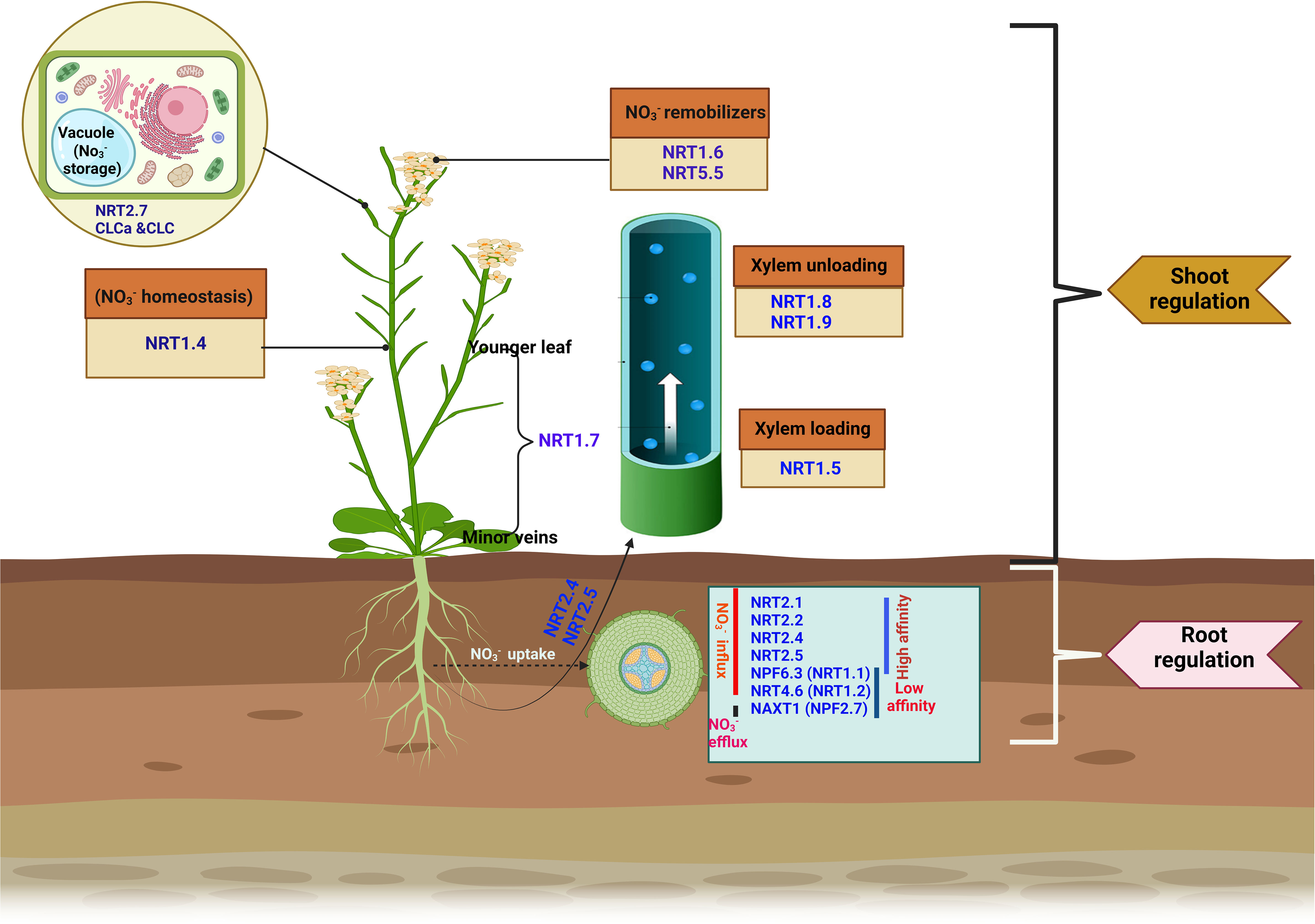
Figure 1 Key nitrate transporters involved in nitrate uptake, transport, and remobilization in plants. Nitrate transporters involved in acquisition from the root include NRT2.1, NRT2.2, NPF4.6 (NRT1.2), NRT2.4, NRT2.5, and NPF6.3 (NRT1.1). NPF2.7 performs the efflux function. In addition to the uptake function, NRT2.4 and NRT2.5 facilitates root-to-shoot transport. NRT1.5 is responsible for xylem loading, while NRT1.8 and NRT1.9 functions to unload from the xylem. NRT1.4 regulates homeostasis, and the expression of NRT1.7 in the phloem of the minor vein promotes nitrate remobilization from mature to younger leaves. At shoot, NRT1.6 and NPF5.5 act as a remobilizer, remobilizing in the embryo. NRT2.7 enhances storage in the seed vacuole.
Unlike the NRT1 family, NRT2 family members are high-affinity transporters (HATs). There are eight identified NRT2 family members, of which seven have been characterized (Von Wittgenstein et al., 2014). Four (NRT2.1, NRT2.2, NRT2.4, and NRT2.5) out of the seven characterized NRT2 transporters have been actively involved in the influx of into Arabidopsis root cells (O’Brien et al., 2016). Detailed functions of these transporters in uptake of are presented in (Figure 1).
Nitrate transporters are the major channels mediating root-to-shoot transport. Transport is predominantly mediated by NRT1 and NRT2 transporters, such as NPF7.3 (NRT1.5), NPF7.2 (NRT1.8), NPF2.3, and NPF2.9 (NRT1.9). NPF7.3 (NRT1.5) is expressed in pericycle cells, where it facilitates xylem loading of (Figure 1). Knockout nrt1.5 mutant plants had reduced amounts of translocated from the roots to the shoots. However, when NRT1.5 was reduced in nrt1.5, no translocation defect was observed, suggesting the existence of another mechanism facilitating nitrate xylem loading (Lin et al., 2008). The low-affinity nitrate transporters NRT1.8 and NRT1.9 perform similar roles of unloading from the xylem (Figure 1), consequently reducing concentration within the xylem. Knockout mutants of such transporters (NRT1.8 and NRT 1.9) exhibited increased amounts of in the xylem and, by implication, accelerated root-shoot transport of nitrate (Li et al., 2010; Wang and Tsay, 2011). In addition, the uptake and transport function of the NRT1 and NRT2 homologs have also been revealed in rice (OsNRT1.1B and OsNRT2.3, respectively) (Tang et al., 2012; Hu et al., 2015; Fan et al., 2017), and tomato, LeNRT2.3 (Fu et al., 2015).
While is relocated to the shoot, a larger proportion of N is delivered to the sink organs (e.g., seeds, fruits, roots, and younger leaves), especially for the anabolic development of new tissues, prioritized by the growth stage or physiological condition of individual plants, a process called N remobilization (Snyder and Tegeder, 2021). NRT1.4, localized in the leaf petiole, regulates accumulation within the petiole while maintaining the homeostasis of available between the leaf lamina and petiole (Figure 1). The nrt1.4 mutant had a low content in its petiole, a major storage organ, indicating the involvement of NRT1.4 in nitrate homeostasis and leaf development (Chiu et al., 2004). Another transporter, NRT1.7, predominantly expressed in the phloem of minor veins, enhances nitrate relocation from older to younger leaves (Figure 1) (Fan et al., 2009). However, the extent of transfer and the proportion of remobilized to the sink organ remain unclear. storage in seeds is mediated by specific transporters that remobilize into embryos during seed formation. The expression of NRT1.6 within the host embryo and seed coat demonstrates a potential role of this transporter in mediating embryonic relocation at the reproductive phase of the parent plant (Figure 1) (Almagro et al., 2008). Similar to NRT1.6, NPF5.5 also mediates transport into the embryo (Figure 1) (Léran et al., 2015; Iqbal et al., 2020). NRT2.7, a high-affinity transporter in the tonoplast, plays specific storage roles in the seed vacuole (Chopin et al., 2007). In the tonoplast, CLCa and CLCb were observed to perform a similar localization pattern, where they also participate in storage (Von Der Fecht-Bartenbach et al., 2010). While accumulation in seed vacuoles has been well documented, relatively less is understood about the characterization of transporter genes involved in efflux out of the vacuole. An in-depth understanding of the specificity of these N transporters, from chronological studies, is the first step toward exploiting and optimizing NUE in plants.
3 Nitrogen assimilation in relation to NUE
For efficient assimilation, a larger proportion of assimilated after root uptake is diverted back to the cytosol, where it is converted to nitrite by nitrate reductase (NR). The nitrite obtained is relocated to plastids for subsequent reduction. At this stage, nitrite is converted to ammonium () by the nitrite-reducing enzyme nitrite reductase (NiR) and then finally incorporated as an amino acid through the glutamine synthetase (GS) and glutamate synthase (GOGAT) cycle (Wilkinson and Crawford, 1993; Li et al., 2017a). Nitrogenous compounds incorporated via glutamine (free amino acid) and glutamate serve as a major checkpoint for regulating N utilization efficiency and are further enhanced by the synergetic expression of NR and transporters (Li et al., 2020; Snyder and Tegeder, 2021). However, a recent study opined an improved grain yield and NUE on concurrent coexpression of OsNRT1.1B and indica OsNR2, indicating the positive regulatory roles of OsNR2 and OsNRT1.1B in uptake of N in rice (Gao et al., 2019b).
The two functionally similar forms of GS, cytosolic GS1, and plastidic GS2, encoded by single or multiple gene families, have been reported to significantly influence N assimilation (Miflin and Habash, 2002). While cytosolic GS1 facilitates root N reassimilation and remobilization during protein turnover, GS2 isoforms primarily assimilate produced during chloroplast photorespiration (Ferreira et al., 2019). Although GS1 is responsible for reassimilation, some GS family members drive N assimilation when is abundant. A good example is GLN1;2 in Arabidopsis, which drives N assimilation when is abundant, compared to the gln1;2 mutant, which exhibits reduced GS activity, rosette biomass, and higher concentration under such conditions. Due to the principal roles of GS in N assimilation, specific focus has been directed toward overexpressing GS family members to improve N assimilation in different plant species, such as Triticum aestivum (Hu et al., 2018), and Oryza sativa (Bao et al., 2014).
Despite the fundamental roles of GS in improving assimilation, seed yield, and NUE (Hu et al., 2018; Gao et al., 2019a), attempts to improve NUE by overexpressing GS1 have yielded inconsistent results (Check Table 1 for details). For instance, TaGS2-2Ab-overexpressing lines in wheat had increased spike number, seed yield, and NUE under poor and rich N supply compared to their wild type, due to an increased root N uptake and remobilization capacity (Hu et al., 2018). Following a similar trend, overexpressing HvGS1-1 using its promoter confers improved grain yield and NUE on barley subjected to low and high N conditions (Gao et al., 2019a). In contrast, Bao et al. (2014) opined a drastic reduction in fresh and dry weight of OsGS1;1- and OsGS1;2-overexpressing lines in rice seedlings, with a further poor growth phenotype at the tillering and heading stages under limited and sufficient N conditions. The results suggest that the GS-overexpressing lines and plant biomass are negatively correlated. Further research is required to understand the underlying mechanisms of GS activity to improve NUE in plants.
Unlike GS, relatively few studies have addressed alterations in the expression of genes encoding NADH-dependent GOGAT (a key enzyme in N assimilation) and plastid-localized ferredoxin-dependent (Fd-GOGAT) (Good et al., 2004; Xu et al., 2012). The two kinds of GOGAT differ in their electron donor specificity. Fd-GOGAT is predominantly involved in the reassimilation of photorespiratory . In contrast, NADH-GOGAT participates in the assimilation of non-photorespiratory and the synthesis of glutamate needed for plant development (Lee et al., 2020). Many attempts have been devoted to studies on the fundamental roles of both NADH-GOGAT and Fd-GOGAT in the growth and seed development of Arabidopsis (Somerville and Ogren, 1980), Hordeum vulgare L. (Kendall et al., 1986), and Oryza sativa (Zeng et al., 2017). However, few research studies have altered the genetic expression of GOGAT to promote seed yield and NUE, while those that focused on NADH-GOGAT had rather limiting outcomes. For example, overexpression of ZmNADH-GOGAT in maize confers drastic reduction on shoot biomass with no considerable alterations in kernel yield when N is abundant (Cañas et al., 2020). Meanwhile, the overexpression lines of OsNADH-GOGAT resulted in an increase in rice grain weight under limited N (Yamaya et al., 2002). Interestingly, Lee et al. (2020) recently revealed that the synergetic expression of OsNADH-GOGAT1 and OsAMT1;2 confers an increase in NUE under both high and low N supply. While transgenic lines had improved seed protein levels without any yield alteration under N-sufficient conditions, seed quality and overall yield increased under N starvation. These observations imply that the combined expression of N-transporters and GOGAT improves N uptake, N assimilation, and NUE rather than the negative effect of the expression of AMT or GOGAT alone. Consequently, understanding the factors involved in the synergetic expression of transporters and GOGAT under rich and poor N conditions in plants is imperative to augment NUE.
4 Nitrate sensing and signaling
In addition to its nutritional roles, functions as a major signaling element regulating several plant physiological processes, such as leaf expansion (Walch‐Liu et al., 2000), induction of root architectural changes (Walch‐Liu and Forde, 2008), regulation of root development, and regulation of floral induction (Marín et al., 2011). The first step in signaling is through external nitrate perception by the dual affinity transporter NPF6.3 (NRT1.1), induced immediately after treatment. NRT1.1 switches between two states of nitrate conditions (low and high conditions) (Wang et al., 1998; Bouguyon et al., 2015; Hu et al., 2015).
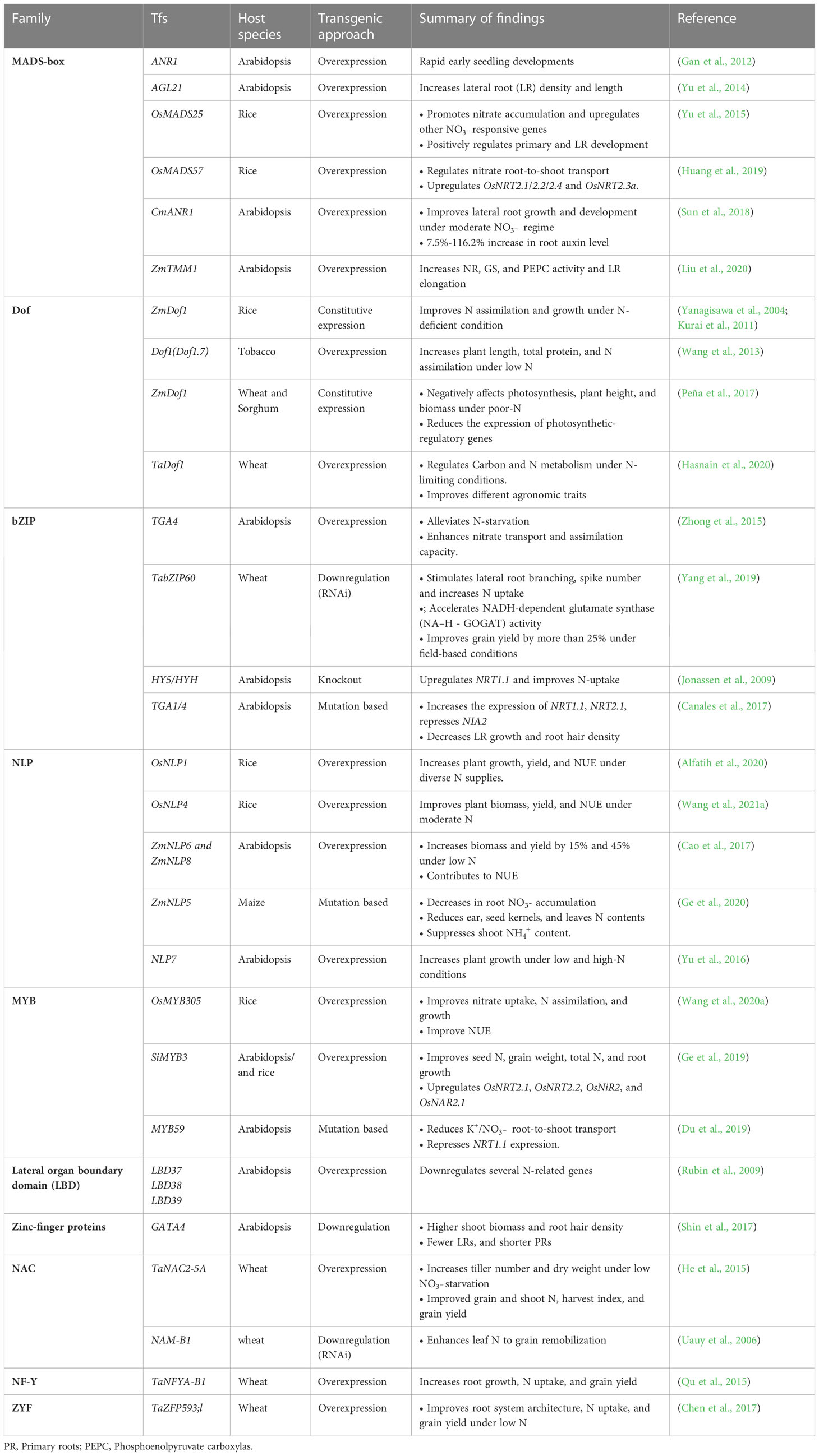
4.1 Roles of transcription factors in N use regulation
Several transcription factors (TFs) have been reported to play critical roles in NUE regulation by modulating the expression of responsive genes. Detailed functions of TFs involved in NUE improvements are outlined in Table 2. DNA binding with one finger (Dof1) TFs increases N use in plants. The transgenic expression of ZmDof1 in A. thaliana (Yanagisawa et al., 2004), TaDof1 in wheat (Hasnain et al., 2020), ZmDof1 in rice (Kurai et al., 2011), wheat and sorghum (Peña et al., 2017) improve N assimilation and plant growth under N starvation.
The key regulators of nitrate assimilatory genes, teosinte branched1-cycloidea-proliferating cell factor1-20 (TCP20) and NIN-like protein (NLP), NLP6 and NLP7 interact with each other under N sufficient and N–starved condition to control response to root growth (Guan et al., 2017), a strong indication of NLP’s involvement in signaling-related responses. Moreover, overexpression of NLP7 results in positive regulation of key nitrate metabolites, total N contents, uptake, and signaling-related genes while improving plant biomass under poor and rich N conditions in Arabidopsis. This peculiar function suggests NLP7 as a master regulator of the primary nitrate response and its importance in plant N use (Yu et al., 2016). Further research on NLP family members reveals that overexpressing ZmNLP6 and ZmNLP8 in Arabidopsis replaces the roles of NLP7 in signaling, and metabolism (Cao et al., 2017). In a recent study by Wu et al. (2021), overexpression of OsNLP4 in rice increased grain yield and NUE by 30% and 47%, respectively, under moderate N conditions. Contrary to NLP, three lateral organ boundary domain TFs (LBD37, LBD38, and LBD39) negatively regulate nitrate uptake and assimilatory genes, and thus could be candidates for improving NUE in plants (Rubin et al., 2009).
A putative MADS-box TF, ANR1, associated with lateral root growth and elongation (Zhang and Forde, 1998), functions as a downstream regulator of NRT1 in response to nitrate (Remans et al., 2006). In addition, AGL21 (AGAMOUS-Like 21) functions in lateral root initiation and growth by regulating auxin biosynthetic genes under N-deficient conditions (Yu et al., 2014). Although, other TFs efficiently utilizing N in Arabidopsis and cereal crops (especially rice) have been identified, the focus on identifying these genes in other crops has been minimal.
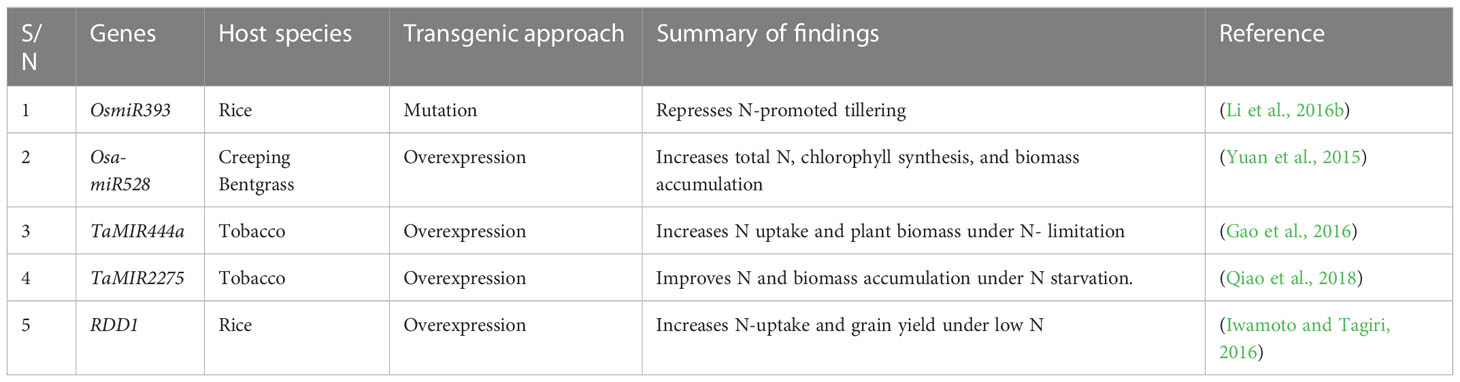
4.2 Nitrate-induced MicroRNA regulation
MicroRNAs (miRNAs) are small noncoding RNAs containing approximately 20-24 nucleotides with diverse regulatory potentials (Zhou et al., 2020). Studies have shown that miRNAs regulate gene expression pathways related to plant growth and developmental processes in response to nitrate (check Table 3 for further details) (Zuluaga and Sonnante, 2019). The upregulation or downregulation of miRNAs primarily anchors on their capacity to regulate key target N-related genes (Zhao et al., 2011). Research has also examined the crucial roles of miR169 family members in cereal crops. A drastic reduction in the expression level of miR169 was observed in N-starved maize (Zhao et al., 2012) and wheat (Qu et al., 2015), upregulating TaNFYA-Bi under such conditions. Despite the numerous miRNA-related NUE phenotypes identified, little is known about the regulatory mechanisms involved. Thus, further research is required to fully understand how N use can be optimized in plants.
5 Nitrate transporters involved in NUE and yield improvement
Nitrate transporters have been shown to play diverse NUE and yield improvement roles in plants (Check Table 4 for details). In Arabidopsis, NRT1.1 transgenic lines habouring Cauliflower Mosaic Virus (CaMV) 35S promoter were observed to increase the uptake of , however, this did not necessarily improve seed yield (Liu et al., 1999). In contrast, the expression of the NRT1.1 homolog OsNRT1.1B driven by the CaMV-35S promoter or its native promoter increased NUE and grain yield in rice. The key regulatory roles in nitrate signaling, absorption, and assimilation enable OsNRT1.1B to be a major contributor of rice NUE (Hu et al., 2015). Although, the crucial roles of OsNRT1.1B in NUE and yield improvement have been well studied, the underlying regulatory mechanism has not been elucidated. Similar to OsNRT1.1B, overexpression of the spliced form OsNRT1.1A also exhibits an approximately 50% grain yield and NUE increase, coupled with shortened maturation times (Wang et al., 2018c). The observations of this latter experiment could be successfully used to develop early maturing and high-yielding varieties in some other crops. The elevated expression of OsNPF8.20 (OsPTR9) leads to increased uptake, better root formation, and ultimately, an increased tiller and panicle number, indicating that OsNPF8.20 improves grain yield and NUE in rice breeding (Fang et al., 2013). Similarly, OsNPF7.20-overexpressing lines exhibited a drastic increase in rice tiller number, fresh weight, dry weight, and grain yield. In contrast, an opposite effect was conferred on the RNA interference (Ri) lines and osnpf7.2 mutant line under mixed nitrate supply (0.5-8 mM ) (Wang et al., 2018a). In their experiment on the modification of transporters in Arabidopsis and rice, Liu et al. (1999) and Hu et al. (2015) reported some discrepancies in the response of these plants to the modified transporters. This may be due to the tolerance and sensitivity of both crops to and . Arabiodopsis thrives under aerobic conditions where the transport system is well optimized, whereas rice thrives best in anaerobic environments where the transport system is optimized. Hence, manipulating and transporters for improved efficiency in Arabidopsis and rice, respectively, would generate little or no effect on their NUE. Several transporter genes in plants whose expression and subcellular localization pattern greatly determine the gene’s function are essential in genetic manipulations of plant traits. As such, deep insight into the function of a gene and the environment to which plants are better adapted can encourage precise manipulation of NUE in crops. The influence of nitrate transporters on crop yield was also reported in tomatoes, where overexpression of LeNRT2.3 improved uptake, root-to-shoot transport, plant biomass, and fruit weight (Fu et al., 2015).
The expression of several NRT2 transporters has also been found to influence yield and NUE under N-starved conditions. NRT2.2 was upregulated to improve N uptake, assimilation, and plant growth under low conditions (Li et al., 2007). Under the same stressed conditions, TaNRT2.5, highly expressed in wheat, increases uptake and root growth (Guo et al., 2014). Chen et al. (2016) conducted a study on transgenic rice and observed that OsNRT2.1, which has the OsNAR2.1 promoter (pOsNAR2.1: OsNRT2.1), was upregulated in the roots and culms. This upregulation significantly increases the overall yield, biomass, and NUE in transgenic lines harboring OsNAR2.1 (pOsNAR2.1: OsNRT2.1). However, the reverse (decrease in NUE) was obtained with the constitutive promoter of OsNRT2.1 (pUbi: OsNRT2.1). These variations could be accrued to alterations in the localization and abundance of OsNRT2.1 in the plant tissue (Chen et al., 2016). Further investigations regarding the importance of the NRT2 gene in NUE showed that two variants, OsNRT2.3a and OsNRT2.3b, were identified in rice. The elevated expression of OsNRT2.3b enhances intracellular pH balance under the synergetic supply of and , thereby increasing the uptake capacity of other nutrients (P, N, and Fe) and ultimately increasing grain yield and NUE by 40% (Fan et al., 2016b). This result demonstrates the importance of pH sensing by OsNRT2.3b in improving plant NUE and adaptation of rice to changes due to different - supplies. However, this N uptake and transport function observed in OsNRT2.3b was lost in OsNRT2.3a (Fan et al., 2016b; Chen et al., 2020a). OsNRT2.3a cannot independently improve crop yield and NUE due to its inability to increase the expression of OsNAR2.1 (Chen et al., 2020a). Thus, the coexpression of OsNRT2.3a with the OsNAR2.1 promoter becomes imperative to enhance rice N use. The literature reviewed thus far has demonstrated a need for most NRT family members to be coexpressed with specific promoters to effectively enhance plant growth, biomass, and NUE, especially in Arabidopsis and rice; however less in known in other crop species.
6 Nitrate transporters and environmental cues: Influence of environmental stress factors and inducers on nitrate allocation to roots
Numerous studies have investigated the crucial roles of transporters in mediating the uptake and long-distance transport of ; however, less is known towards understanding transport systems involved in reallocation under biotic and abiotic stresses. transporters play crucial roles in the plants’ response to adverse environmental conditions. Indeed, plants acclimatize better to environmental stress when less is allocated to the shoot. Thus, this section examines the contribution of transporters in assisting plants to strive in adverse environmental conditions.
The quantity of translocated from roots to shoots varies under diverse environmental conditions, as this could positively or negatively affect plant NUE. Hence, redistribution in plants is a prerequisite to improved plant growth under N shortages and adverse conditions (Fan et al., 2017). Stressed plants tend to uptake and transport less to the shoot while retaining more nitrate in its root than required (Figure 2). Such allocation to the root as induced by environmental fluctuations (including biotic and abiotic stress) is referred to as “stress-initiated nitrate allocation to roots” (SINAR) (Zhang et al., 2018). Over two decades ago, Hernandez et al. (1997) investigated the inherent effects of cadmium (Cd2+) on uptake, and distribution in pea plants. They found that was increasingly retained at the plant root, and fewer were reallocated to the shoot of Cd-treated pea compared with the control, thereby disrupting the NUE of plants (Figure 2). However, the study could not elucidate the mechanism underlying the fluctuation in the root-to-shoot transport of . Many years later, several research investigations have shown the active involvement of transporters in regulating Cd2+ uptake and other SINAR-related stress conditions (Lin et al., 2008; Zhang et al., 2014). Mao et al. (2014) reported NRT1.1 as a potential regulator of Cd2+ uptake in plants. They observed that plants exposed to Cd2+ stress exhibit repression of NRT1.1 and, as such, exert a negative influence on plant N nutrition (Figure 2). Thus, the loss of NRT1.1 function reduced Cd2+ in the roots and shoots, improving plant biomass production under Cd2+ stress (Figure 2). Although the disruption of NRT1.1 activity induced by Cd2+ stress negates uptake, it enhances plant tolerance to Cd2+ stress by reducing Cd2+ influx into the root. A recent study by Jian et al. (2019) opined that overexpression of NRG2 (which functions downstream of NRT1.1) in wild-type and nrt1.1 increased root over shoot nitrate, thus alleviating Cd2+ toxicity. These findings demonstrate the involvement of NRT1.1 in regulating cadmium uptake while coordinating nitrate allocation to the root. NRT1.1 also regulates Zn accumulation in Arabidopsis by improving uptake in the wild type through a dependent pathway under Zn stress (Figure 2) (Pan et al., 2020).
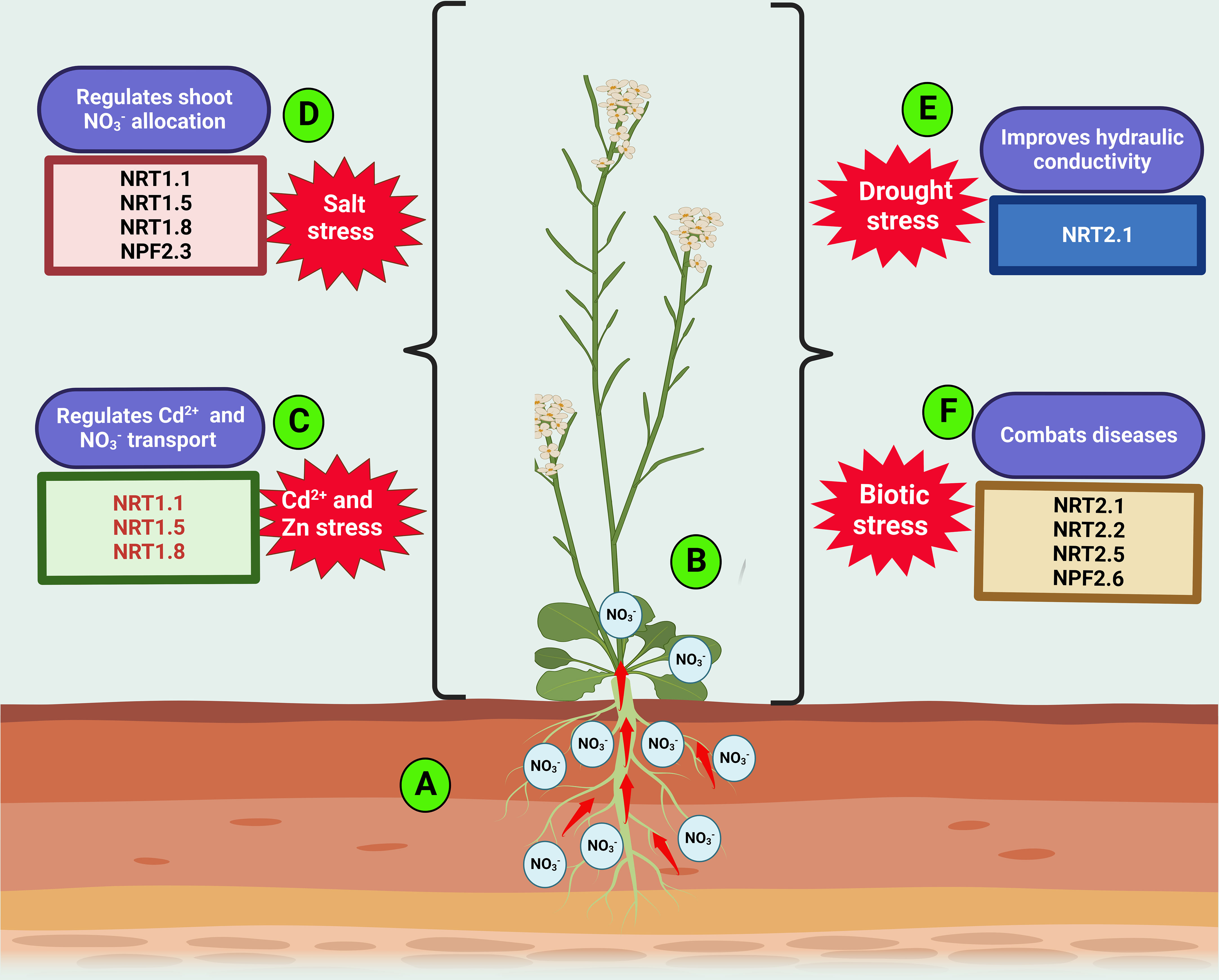
Figure 2 Roles of nitrate transporters in plant response to adverse environmental conditions. Environmental cues including heavy metals (Cd2+ and Zn), salinity, drought, and pathogenic stress engender reduction in plant growth and NUE. The resulting stressed plants accumulate more at the root (A) while retaining less in the shoot (B). Under Cd2+ or Zn stress, nitrate transporters, NRT1.1, NRT1.5 and NRT1.8 concurrently regulates Cd2+ or Zn uptake and allocation to the root (C). The transporters involved in root-to-shoot allocation of under salinity include NPF2.3, NRT1.1, NRT1.5, and NRT1.8 (D). NRT2.1 promotes plants’ tolerance to drought stress (E). In addition to NRT2.1, NRT2.2, NRT2.5 and NRT2.6 are involved in biotic stress regulation (F).
In addition to NRT1.1, NRT1.5 and NRT1.8 regulate the acropetal reallocation of to shoots under cadmium and salinity stress (Fan et al., 2017a). Such stresses activate antagonistic expression of the two latter genes (NRT1.5 and NRT1.8), with reduced expression of NRT1.5/NPF7.3 (Chen et al., 2012) and increased expression of NRT1.8/NPF7.2 (Figure 2) (Li et al., 2010). From the study conducted by Li et al. (2010), loss of NRT1.8 function displays greater sensitivity to Cd2+ stress than wild-type plants under high conditions. However, an opposite effect was observed, with nrt1.5 mutants having greater Cd2+ tolerance in relation to the control. The Cd2+ sensitivity observed with the ntr1.8 mutants could be due to Cd2+ translocation to its shoots, thus counteracting the plant adaptive strategy that supports Cd2+ accumulation in plant roots. The upregulation of NRT1.8 expression triggers nitrate removal from the xylem under Cd2+-stressed conditions. This result suggests a strong link between Cd2+ tolerance and allocation.
In addition to NRT1.5 and NRT1.8, NPF2.3 also contributes to the SINAR response under salt stress. Nitrate allocation to the shoot was drastically reduced under salt-stressed conditions due to the unaltered expression of NPF2.3 and partial expression of the NPF7.3 gene in the root stele. However, the loss of NPF2.3 function led to the reduced root-to-shoot allocation of (Figure 2) (Taochy et al., 2015). These data demonstrate the quantitative and physiological contribution of the efflux transporter NPF2.3 to allocation to the shoot under salinity (Taochy et al., 2015; Chao et al., 2021). Alvarez-Aragon and Rodriguez-Navarro (2017) also found Na+ accumulation to be partially defective in the nrt1.1 mutant, demonstrating the partial contribution of NRT1.1 to dependent Na+ transport (Figure 2). Plants expressing these related genes in response to heavy metal or salt stress exhibit enhanced uptake, plant growth, and tolerance to heavy metal- or salt-stressed environments.
Previous physiological research investigations have shown varying impacts of and availability on water uptake and transport in plants subjected to water stress (Guo et al., 2007). They found that the assimilation rate and stomatal conductance of -fed plants surpassed those of NO3--fed plants; thus, nutrition improves rice seedling tolerance to drought (Guo et al., 2007). Li et al. (2016a) revealed that the high-affinity NO3- transporter NRT2.1 alters accumulation to regulate root hydraulic conductivity (Figure 2). They found NRT2.1 to be a positive regulator of plasma membrane intrinsic protein PIPs. This latter study unraveled the link between use, water stress, and NRT2.1 expression, indicating the potential roles of NRT2.1 in drought tolerance (Li et al., 2016a). However, a more recent investigation has shown how the high-affinity transporter partner protein OsNAR2.1 positively regulates drought-related responses to stress and enhances drought tolerance in rice (Figure 2) (Chen et al., 2019).
Ample agronomic evidence exists regarding the impact of excessive N fertilizer use on the incidence rate of plant diseases (Fagard et al., 2014; Fan et al., 2017). For example, excessive N fertilizer application triggers the severity of powdery mildew caused by a biotrophic pathogen that saps plant nutrients. Interestingly, a reduction in N fertilizer application has been found to reduce Arabidopsis tolerance to Erwinia amylovora. These findings indicate a complex relationship between N uptake, metabolism, and disease infection processes. Thus, it is evident that N status affects plant tolerance or susceptibility to diseases under specific environmental conditions (Fagard et al., 2014). Unfortunately, the molecular mechanism underlying the impact of transporters on fungal infection or pathogenic attack is not fully understood. To investigate the possible mechanisms involved in N uptake by the biotrophic pathogen, Pike et al. (2014) characterized the low-affinity transporter VvNPF3.2 (in grapevine) and cloned Arabidopsis ortholog NPF3.1. In this study, powdery mildew pathogen infection was shown to upregulate the expression of VvNPF3.2 and NPF3.1 in vascular tissues, major and minor veins of leaves. The loss of NRT2.1 and NRT2.2 under N-deficient conditions resulted in increased resistance to Pseudomonas syringae pv tomato DC3000 infection (Figure 2) (Li et al., 2007; Camanes et al., 2012). Additionally, in the NRT2 family, the roles of two putative high-affinity transporters, NRT2.5 and NRT2.6, were investigated in response to rhizospheric bacterium STM196 using single and double Arabidopsis mutants (Kechid et al., 2013). The study revealed that mutations in NRT2.5 and NRT2.6 inhibited plant growth and abolished root system architecture in response to STM196. Hence, Arabidopsis leaves expressing NRT2.5 and NRT2.6 appear to play crucial roles in the plant response to STM196 in a uptake-independent manner (Figure 2). The expression of both genes (NRT2.5 and NRT2.6) is also crucial for promoting plant growth mediated by STM196 (Kechid et al., 2013). Recently, T-DNA mutants of NRT2.5 showed stronger resistance to Pseudomonas syringae pv. tomato DC3000 inoculation compared to its wild-type counterpart, an indication of NRT2.5 role in plant biotic defense (Du Toit et al., 2020; Devanna et al., 2021). These research findings have demonstrated the functional roles of transporters in the plant response to biotic stress, while suggesting safe, innovative, and sustainable means of controlling crop pathogens.Mycorrhizal colonization of rice root also appears to promote the expression of a putative nitrate transporter, OsNPF4.5. This result improved growth and yield properties in host plant (Wang et al., 2020c). However, inactivation of OsNPF4.5 resulted in the reduction of arbuscule incidence, as well as a depletion in symbiotic nitrogen uptake in rice (Wang et al., 2020c).
Another member of the nitrate and peptide transporters family (NPF), OsNPF8.1 (OsPTR7), a putative peptide transporter in rice (localized in the cell plasma membrane), has been reported as permeable to methylated arsenic species, especially, dimethylarsenate (DMA). OsNPF8.1 is involved in long-distance transport of arsenic in rice (Tang et al., 2017). However, the peptide-mediated transport of arsenic species has been linked with imbalance nutrient (especially, phosphate) supply in plants (Finnegan and Chen, 2012). Consequently, it is imperative to investigate the activity of OsNPF8.1 on N uptake, as well as the collateral accumulation of DMA, its clinical significance and nutrient imbalance in economically significant crops.
7 Could nitrate uptake and utilization affect the efficiency of other plant nutrients?
Balanced nutrition is paramount to maintaining good human health, and this is achievable by eating a balanced diet. In plants, maintaining an appropriate nutrient balance is also required because excessive accumulation of a specific nutrient might affect the uptake of the other and vice versa (Aluko et al., 2021). This nutritional balance ultimately affects crop growth and plant nutrient use efficiency (Bouain et al., 2019). Such nutritional crosstalk coexists between phosphorus (P) and N, the most limiting nutrient element required for crop growth and development. Phosphorus starvation reduces nitrate uptake capacity in tobacco (Rufty et al., 1990), maize (De Magalhães et al., 1998), and barley (Lee, 1982). These phenomena demonstrate the mechanisms involved in optimizing nutrient uptake and utilization to maintain plant homeostatic balance. Molecular evidence indicates that nitrogen limitation adaptation (NLA) ubiquitin offsets deficiency induced by excessive P via degradation of PHT1, the phosphate transporter (Kant et al., 2011b). The phenotypic analysis illustrated the functional role of nitrate-inducible garp-type transcriptional repressor 1.2 (NIGT1.2) in integrating N and P signals. Under sufficient P supply, NIGT1.2 was not activated due to the coexpression of PHR1 and SPXs, which are P-sensor proteins and repressors of PHR1, respectively (Medici et al., 2015). However, PHR1 was detached from the inhibitors SPX1/2/3/4 to promote the expression of NIGT1 clade genes under P-starved conditions. Thus, nitrate uptake is suppressed due to P deficiency through the PHR1-NIGT1-NRT2.1 pathway (Maeda et al., 2018). With such development, N uptake regulation via the PHR1-NIGT1 path could be a good adaptative mechanism under P starvation (Maeda et al., 2018). Another recent study found that NIGT1.2 increased the expression of phosphate transporters (PHT1;1 and PHT1;4) but repressed the nitrate transporter NRT1.1, an indication that NIGT1.2 could maintain a balance between N and P to improve N uptake and utilization under (phosphorus) P starvation (Wang et al., 2020b).
The highly inducible NRT1.1-controlled GARP transcription factor, HRS1, and its closest homolog, HHO1, function downstream of NRT1.1, NLP6, and NLP7. However, HRS1 and HHO1 act as major primary root growth inhibitors only when the media is P-starved in the presence of , indicating extensive integration of the N and P signaling networks (Medici et al., 2015). Following the previous discussion on how HRS1 mediates N and P crosstalk, Medici et al. (2019) found that PSR marker gene responses depend on the N supplied. Indeed, transcript levels of PHO2 were coordinated by nitrate availability accumulated during both high and low supplies of nitrate. Notably, this nitrate-induced strategy of PSR regulation is conserved in plants. However, several PSR genes were not regulated by in a pho2 mutant, indicating that PHO2 incorporates nitrate signals into PSR (Medici et al., 2019). Upon P starvation, NRT1.1 is downregulated, while PHO2 functions to positively regulate NRT1.1. In rice, the genes induced by P starvation OsIPS1, OsSPX1, and the P transporter OsPT1 only respond to P starvation when nitrate is present (Medici et al., 2019). On the overall assessment, these findings elucidate the complexity of nitrate and phosphorus responses while emphasizing the principal roles of NRT1.1 in regulating the interaction.
Another macronutrient required for plant health is potassium (K+), as it strongly coordinates nitrate (). Previous reports indicated that NRT1.5 facilitates the long-distance transport of and K+ in a nitrate-dependent manner (Meng et al., 2016; Zheng et al., 2016). NRT1.5, expressed in the pericycle of root cells, participates in the xylem loading of nitrate. When there is a K deficit, NRT1.5 directly triggers the movement of K+ to the root xylem for root-to-shoot transport. This investigation demonstrates the crucial role of NRT1.5 in root-to-shoot K+ transport and its involvement in the synergetic regulation of /K+ distribution in plants (Li et al., 2017b). Another study reported that MYB59 activates the expression of NRT1.5 and binds directly to its promoter to ensure a controlled nutrient distribution from root to shoot. When plants become deficient in /K+, the expression of MYB59 and NRT1.5 is repressed to maintain a balanced /K+ distribution between the roots and shoots (Du et al., 2019).
8 Nitrate transporter regulates nitrate and auxin crosstalk for root growth and nitrogen uptake
Evidence has shown the impact of changes in N status on auxin distribution in plants (Hou et al., 2021). Compared with moderate N supply, limited supply engenders auxin deposition in the roots of Arabidopsis, wheat, soybean, maize, and rapeseed (Caba et al., 2000; Tian et al., 2008; Asim et al., 2020), indicating the importance of in situ auxin synthesis in the root (Yang et al., 2022). Thus, the in situ auxin synthesis and the shoot-to-root polar transport jointly contributes to auxin deposition in the root under N limitation (Yang et al., 2022). In contrast, a 30% reduction in root indole-3-acetic acid (the putative among natural auxins) content was observed when the amount of supplied to rice dropped from 2.5mM to 0.01mM (Sun et al., 2014b). Perhaps, the discrepancies in N induced auxin response stems from varying plant growth conditions and the species involved. Nevertheless, all these findings demonstrate the importance of nitrate and auxin crosstalk in root development, and the mechanism of such responses are triggered by the activities of transporters.
In addition to the transport and signaling function, NRT1.1, among other transporters, facilitates basipetal transport of auxin and negatively regulates auxin biosynthetic genes, TAR2 and LAX3, under deficiency (Maghiaoui et al., 2020). As a consequence, NRT1.1 removes auxin (required for lateral root growth) deposited at the lateral root primordia, inhibiting lateral root growth under such condition. All these inhibitory effects of NRT1.1, including root growth reduction and patchy auxins are alleviated in response to high supply (Maghiaoui et al., 2020). Thus, NRT1.1-mediated auxin transport was disrupted and its (NRT 1.1) expression repressed, to facilitate lateral root growth and auxin accumulation at the root tip under increasing supply (Remans et al., 2006). These findings indicated that NRT1.1 functions in reprogramming root system architecture in response to availability. However, the integrated function of this molecular circuit is yet unraveled.
Although, it is understood that external N status regulates auxin biosynthetic genes and signaling pathways. However, less is known about the identities of auxin-related genes that are N-responsive, and whether these genes reprogram plant N metabolism to improve crop NUE is yet unexplored. To this end, Zhang et al. (2021b) identified DULL NITROGEN RESPONSE1 (DNR1) as an intriguing QTL regulating auxin and N crosstalk for NUE improvement in rice. DNR1 mediates plant N metabolism by counteracting the auxin deposited in response to N availability. This process enhances auxin biosynthesis and induces AUXIN RESPONSE FACTOR, a major regulator of N-responsive genes to improve NUE and grain yield.
Out of the identified transporters, the functions of the transceptor’s (NRT1.1) in auxin regulation has been the most investigated. However, less is known about the versatile functions of other related proteins in regulating other plant developmental traits.
9 Integrated approaches to improve plant NUE
Genetic modification of crops has been a promising strategy for improving plant N use through diverse breeding techniques during the past few decades. Indeed, several transporter genes, their regulators, and other responsive genes regulating NUE have been well studied. However, mechanisms involved in this regulation, which specifically describes the strategies involved in NUE improvement, have been overlooked due to difficulties in identifying N-specific phenotypes. (Hu et al., 2015) revealed that genetic variation of the major quantitative trait locus (QTL) NRT1.1B (OsNPF6.8) promotes NUE divergence between Indica and Japonica rice subspecies. They found that NRT1.1B from indica improved the tiller number, NUE, and grain yield of Japonica rice. Several other QTL-based approaches have generated signaling proteins, transcriptional regulators, and components of hormonal pathways that regulate plant NUE. One of these is a QTL study that used positional cloning and genetic complementation to map out DEP1 (Dense and erect panicles 1), a heterotrimeric G protein that confers a significant yield increase (Sun et al., 2014a). Under moderate N fertilization, plants harboring the dominant allele DEP1-1 display N-insensitive vegetative growth, as well as improved N uptake and assimilation, thereby increasing yield (Sun et al., 2014a). This result implies that modulating the activity of DEP1 could provide a lasting strategy for grain yield increases in rice. Another QTL study showed that the accumulation of the growth inhibitor DELLA confers semi-dwarfism and reduces NUE in rice (Li et al., 2018). However, the NUE and grain yield of green revolution varieties are restored by tilting the GRF4–DELLA stability toward an increased abundance of GRF4. This study indicated that regulating physiological activities and plant growth induced by efficient N use could open up innovative breeding ideas for sustainable food security (Li et al., 2018). Although QTL analysis has also informed the recent NUE gene identification strategy in crop species such as maize (Zhang et al., 2019), the importance of QTL analysis is yet unknown in some other higher plants.
In addition to QTL analysis, other analytical studies involving genome-wide association studies (GWAS) could be used to identify an array of NUE candidate genes in Arabidopsis (Atwell et al., 2010), maize (Li et al., 2013), rice (Si et al., 2016), and other crop species (Korte and Farlow, 2013; Ogura and Busch, 2015). An elite haplotype of the nitrate transporter OsNPF6.1HapB was recently identified using GWAS (Tang et al., 2019). This allele improved nitrate uptake, NUE, and grain yield under N-deficient conditions. In the same study, the NUE-related transcription factor OsNAC4 was used to transactivate OsNPF6.1HapB, thereby increasing plant NUE and grain yield. This result suggests that the NAC42-NPF6.1 signaling cascade is a promising strategy for improving NUE and rice yield (Tang et al., 2019).
To further identify the genes enhancing NUE, Clustered Regularly Interspaced Palindromic Repeats (CRISPR)/Cas9 along with the Cas9 nuclease (CRISPR/CAS9) system was developed. CRISPR/CAS9 has been deployed to facilitate easy and robust technology to edit genes for improved plant N use. Multiple applications of CRISPR/CAS9 technology have been demonstrated in major crops, including sorghum, rice, and tomatoes (Ito et al., 2015; Ma et al., 2015). Notably, CRISPR/CAS9 mostly mutates negative growth regulators instead of overexpressing positive regulators, thereby providing prospects for crop breeding (Tiwari et al., 2020). A related strategy described one of the Bric-a-Brac/Tramtrack/Broad gene family members, BT2, that downregulates the NRT2.1 and NRT2.4 genes (Araus et al., 2016), thus reducing uptake and NUE under low conditions. When this BT2 gene was mutated in Arabidopsis, a 65% increment in nitrate uptake was observed, while mutation of OsBT2 yielded a 20% increase in NUE compared to wild-type under poor supply (Araus et al., 2016). To date, the functions and features of a significant number of negative regulators or inhibitors of nitrate transporters have yet to be functionally characterized in plants. Hence, it is plausible that gene editing or mutating their expression by CRISPR/Cas9 appears to be a promising strategy for achieving future breeding goals (Tiwari et al., 2020).
It is essential to note that incorporating transcriptomics, proteomics, and metabolomics, which characterize the expression profile, could facilitate the identification of agronomically induced genes or pathways. Moreover, computational and system biology could aid in identifying candidate genes during domestication.
10 Conclusion and future perspectives
Nitrate transporters have not only been shown to function in plant uptake and transport capacity; their vital roles and potential in improving plant N use have also guaranteed the possibility of meeting future global food demands. Indeed, improved uptake and utilization ( transport, remobilization, and assimilation) through transporter activity is a prerequisite to attaining increased NUE and overall plant growth. With the understanding that the activities of these transporters are enhanced when co-expressed with their specific promoters or Tfs, it becomes imperative to select and integrate NO3–specific promoters with their transporters for efficient plant N utilization. An excellent way to improve utilization could be to carefully select senescence-specific promoters (primarily expressed in source organs or leaves) to facilitate phloem-expressed nitrate transporters. Most research works have successfully established the impact of nitrate transporters on adverse environmental conditions (biotic and abiotic stress). They have also addressed their relationships with other plant nutrients only under controlled conditions; however, field-based studies affirming these functions are scarce.
Moreover, relatively few transporters performing complex interplay functions have been identified, while the established ones were found to play multiple physiological roles in environmental and nutritional stresses. The underlying mechanisms behind these multipurpose functions are unknown, and the extent to which these transporters can mitigate abiotic stress is unresolved. Thus, to understand and manipulate the functional roles of nitrate transporters in enhancing plant NUE under diverse conditions, future research should address some critical questions, including the following, but not limited to:
● How do the combined effects of biotic/abiotic stressors influence nitrate transporter activities, and to what extent?
● Does the uptake of other macro- and micronutrients alter the expression or impair the prospective function of nitrate transporters and vice versa?
● Is there a possibility of having nutrient imbalance feedback due to alterations in the expression of either nitrate transporters or the transporters of other nutrients (macro- and micronutrients)?
● If the activities of nitrate transporters are eventually established to significantly affect the uptake of other nutrients and vice versa, what molecular techniques could be factored in to recuperate such imbalance?
● Could the crosstalk between N-responsive and auxin biosynthesis genes affect the uptake of other essential nutrients by plants?
● Could specific transporters or related genes function or be expressed differently in diverse crop species?
● Could models be developed to project or predict the possible influence of biotic and abiotic environmental parameters, as well as their complex interplay on the NUE of individual plant species?
Developing profound resolutions to these questions will afford us a better understanding of how nitrate transporters could be maximized to enhance plant NUE under adverse environmental conditions. Knowledge of these factors will also help settle crises related to plant nutritional imbalance and cross-talk, thereby achieving plant breeding goals for quality and sustainable food production.
Author contributions
Conceptualization, OOA, QW, and HL; writing-original draft, OOA; review and editing; SK and OMA; visualization, OOA, GY, and CL; validation, SK, OMA, QW, and HL; supervision, QW and HL; funding acquisition, QW and HL. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Agricultural Science and Technology Innovation Program (ASTIP-TRIC03), the National Natural Science Foundation of China (32170387); the International Foundation of Tobacco Research Institute of Chinese Academy of Agricultural Sciences (IFT202102), the Key Funding of CNTC (No. 110202101035(JY-12)) and YNTI (No. 2022JY03).
Acknowledgments
The authors would like to appreciate E. Sanganyado, and O. Ohore for carefully proofreading the manuscript. We also appreciate V. Ninkuu for helping out with the graphics. Thanks to A. Adegbite, F. Akinde, H. Kaiyan, and T. Nong for their support and encouragement.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Alfatih, A., Wu, J., Zhang, Z.-S., Xia, J.-Q., Jan, S. U., Yu, L.-H., et al. (2020). Rice NIN-LIKE PROTEIN 1 rapidly responds to nitrogen deficiency and improves yield and nitrogen use efficiency. J. Exp. Bot. 71, 6032–6042. doi: 10.1093/jxb/eraa292
Almagro, A., Lin, S. H., Tsay, Y. F. (2008). Characterization of the arabidopsis nitrate transporter NRT1. 6 reveals a role of nitrate in early embryo development. Plant Cell 20, 3289–3299. doi: 10.1105/tpc.107.056788
Aluko, O. O., Li, C., Wang, Q., Liu, H. (2021). Sucrose utilization for improved crop yields: A review article. Int. J. Mol. Sci. 22 (9), 4704. doi: 10.3390/ijms22094704
Alvarez-Aragon, R., Rodriguez-Navarro, A. (2017). Nitrate-dependent shoot sodium accumulation and osmotic functions of sodium in arabidopsis under saline conditions. Plant J. 91, 208–219. doi: 10.1111/tpj.13556
Araus, V., Vidal, E. A., Puelma, T., Alamos, S., Mieulet, D., Guiderdoni, E., et al. (2016). Members of BTB gene family of scaffold proteins suppress nitrate uptake and nitrogen use efficiency. Plant Physiol. 171, 1523–1532. doi: 10.1104/pp.15.01731
Asim, M., Ullah, Z., Oluwaseun, A., Wang, Q., Liu, H. (2020). Signalling overlaps between nitrate and auxin in regulation of the root system architecture: Insights from the arabidopsis thaliana. Int. J. Mol. Sci. 21(8):2880. doi: 10.3390/ijms21082880
Atwell, S., Huang, Y. S., Vilhjálmsson, B. J., Willems, G., Horton, M., Li, Y., et al. (2010). Genome-wide association study of 107 phenotypes in arabidopsis thaliana inbred lines. Nature 465, 627–631. doi: 10.1038/nature08800
Bao, A., Zhao, Z., Ding, G., Shi, L., Xu, F., Cai, H. (2014). Accumulated expression level of cytosolic glutamine synthetase 1 gene (OsGS1; 1 or OsGS1; 2) alter plant development and the carbon-nitrogen metabolic status in rice. PloS One 9, e95581. doi: 10.1371/journal.pone.0095581
Beatty, P. H., Shrawat, A. K., Carroll, R. T., Zhu, T., Good, A. G. (2009). Transcriptome analysis of nitrogen-efficient rice over-expressing alanine aminotransferase. Plant Biotechnol. J. 7, 562–576. doi: 10.1111/j.1467-7652.2009.00424.x
Bergsdorf, E.-Y., Zdebik, A. A., Jentsch, T. J. (2009). Residues important for nitrate/proton coupling in plant and mammalian CLC transporters. J. Biol. Chem. 284, 11184–11193. doi: 10.1074/jbc.M901170200
Bharati, A., Mandal, P. K. (2019). Strategies for identification of genes toward enhancing nitrogen utilization. Nutrient Dynamics Sustain. Crop Production 157, 157–187.
Bouain, N., Krouk, G., Lacombe, B., Rouached, H. (2019). Getting to the root of plant mineral nutrition: combinatorial nutrient stresses reveal emergent properties. Trends Plant Sci. 24, 542–552. doi: 10.1016/j.tplants.2019.03.008
Bouguyon, E., Brun, F., Meynard, D., Kubeš, M., Pervent, M., Leran, S., et al. (2015). Multiple mechanisms of nitrate sensing by arabidopsis nitrate transceptor NRT1. 1. Nat. Plants 1, 1–8. doi: 10.1038/nplants.2015.15
Brauer, E. K., Rochon, A., Bi, Y. M., Bozzo, G. G., Rothstein, S. J., Shelp, B. J. (2011). Reappraisal of nitrogen use efficiency in rice overexpressing glutamine synthetase1. Physiol Plant 141(4), 361–372. doi: 10.1111/j.1399-3054.2011.01443.x
Caba, J. M., Centeno, M. L., Fernández, B., Gresshoff, P. M., Ligero, F. (2000). Inoculation and nitrate alter phytohormone levels in soybean roots: Differences between a supernodulating mutant and the wild type. Planta 211, 98–104. doi: 10.1007/s004250000265
Camanes, G., Pastor, V., Cerezo, M., Garcia-Agustin, P., Flors Herrero, V. (2012). A deletion in the nitrate high affinity transporter NRT2.1 alters metabolomic and transcriptomic responses to pseudomonas syringae. Plant Signal Behav. 7, 619–622. doi: 10.4161/psb.20430
Canales, J., Contreras-López, O., Álvarez, J. M., Gutiérrez, R. A. (2017). Nitrate induction of root hair density is mediated by TGA 1/TGA 4 and CPC transcription factors in arabidopsis thaliana. Plant J. 92, 305–316. doi: 10.1111/tpj.13656
Cañas, R. A., Yesbergenova-Cuny, Z., Belanger, L., Rouster, J., Brulé, L., Gilard, F., et al. (2020). NADH-GOGAT overexpression does not improve maize (Zea mays l.) performance even when pyramiding with NAD-IDH, GDH and GS. Plants 9, 130. doi: 10.3390/plants9020130
Cao, H., Qi, S., Sun, M., Li, Z., Yang, Y., Crawford, N. M., et al. (2017). Overexpression of the maize ZmNLP6 and ZmNLP8 can complement the arabidopsis nitrate regulatory mutant nlp7 by restoring nitrate signaling and assimilation. Front. Plant Sci. 8, 1703. doi: 10.3389/fpls.2017.01703
Chao, H., He, J., Cai, Q., Zhao, W., Fu, H., Hua, Y., et al. (2021). The expression characteristics of NPF genes and their response to vernalization and nitrogen deficiency in rapeseed. Int. J. Mol. Sci. 22, 4944. doi: 10.3390/ijms22094944
Chen, K.-E., Chen, H.-Y., Tseng, C.-S., Tsay, Y.-F. (2020b). Improving nitrogen use efficiency by manipulating nitrate remobilization in plants. Nat. Plants 6, 1126–1135. doi: 10.1038/s41477-020-00758-0
Chen, J., Liu, X., Liu, S., Fan, X., Zhao, L., Song, M., et al. (2020a). Co-Overexpression of OsNAR2. 1 and OsNRT2. 3a increased agronomic nitrogen use efficiency in transgenic rice plants. Front. Plant Sci. 11, 1245. doi: 10.3389/fpls.2020.01245
Chen, C.-Z., Lv, X.-F., Li, J.-Y., Yi, H.-Y., Gong, J.-M. (2012). Arabidopsis NRT1. 5 is another essential component in the regulation of nitrate reallocation and stress tolerance. Plant Physiol. 159, 1582–1590. doi: 10.1104/pp.112.199257
Chen, J., Qi, T., Hu, Z., Fan, X., Zhu, L., Iqbal, M. F., et al. (2019). OsNAR2. 1 positively regulates drought tolerance and grain yield under drought stress conditions in rice. Front. Plant Sci. 10, 197. doi: 10.3389/fpls.2019.00197
Chen, Y., Yang, M., Ding, W., Zhao, Y., Li, X., Xiao, K. (2017). Wheat ZFP gene TaZFP593; l mediates the n-starvation adaptation of plants through regulating n acquisition and the ROS metabolism. Plant Cell Tissue Organ Culture (PCTOC) 129, 271–288. doi: 10.1007/s11240-017-1176-9
Chen, J., Zhang, Y., Tan, Y., Zhang, M., Zhu, L., Xu, G., et al. (2016). Agronomic nitrogen-use efficiency of rice can be increased by driving os NRT 2.1 expression with the os NAR 2.1 promoter. Plant Biotechnol. J. 14, 1705–1715. doi: 10.1111/pbi.12531
Chiu, C.-C., Lin, C.-S., Hsia, A.-P., Su, R.-C., Lin, H.-L., Tsay, Y.-F. (2004). Mutation of a nitrate transporter, AtNRT1: 4, results in a reduced petiole nitrate content and altered leaf development. Plant Cell Physiol. 45, 1139–1148. doi: 10.1093/pcp/pch143
Chopin, F., Orsel, M., Dorbe, M.-F., Chardon, F., Truong, H.-N., Miller, A. J., et al. (2007). The arabidopsis ATNRT2. 7 nitrate transporter controls nitrate content in seeds. Plant Cell 19, 1590–1602. doi: 10.1105/tpc.107.050542
David, L. C., Dechorgnat, J., Berquin, P., Routaboul, J. M., Debeaujon, I., Daniel-Vedele, F., et al. (2014). Proanthocyanidin oxidation of arabidopsis seeds is altered in mutant of the high-affinity nitrate transporter NRT2. 7. J. Exp. Bot. 65, 885–893. doi: 10.1093/jxb/ert481
De Magalhães, J., Alves, V., De Novais, R., Mosquim, P., Magalhães, J., Filho, A. B., et al. (1998). Nitrate uptake by corn under increasing periods of phosphorus starvation. J. Plant Nutr. 21, 1753–1763. doi: 10.1080/01904169809365520
Devanna, B. N., Jaswal, R., Singh, P. K., Kapoor, R., Jain, P., Kumar, G., et al. (2021). Role of transporters in plant disease resistance. Physiologia Plantarum 171, 849–867. doi: 10.1111/ppl.13377
Du Toit, Y., Coles, D. W., Mewalal, R., Christie, N., Naidoo, S. (2020). eCALIBRATOR: a comparative tool to identify key genes and pathways for eucalyptus defense against biotic stressors. Front. Microbiol. 11, 216. doi: 10.3389/fmicb.2020.00216
Du, X.-Q., Wang, F.-L., Li, H., Jing, S., Yu, M., Li, J., et al. (2019). The transcription factor MYB59 regulates K+/NO3– translocation in the arabidopsis response to low k+ stress. Plant Cell 31, 699–714. doi: 10.1105/tpc.18.00674
Fagard, M., Launay, A., Clément, G., Courtial, J., Dellagi, A., Farjad, M., et al. (2014). Nitrogen metabolism meets phytopathology. J. Exp. Bot. 65, 5643–5656. doi: 10.1093/jxb/eru323
Fan, X., Feng, H., Tan, Y., Xu, Y., Miao, Q., Xu, G. (2016a). A putative 6-transmembrane nitrate transporter OsNRT1. 1b plays a key role in rice under low nitrogen. J. Integr. Plant Biol. 58, 590–599. doi: 10.1111/jipb.12382
Fang, Z., Xia, K., Yang, X., Grotemeyer, M. S., Meier, S., Rentsch, D., et al. (2013). Altered expression of the PTR/NRT 1 homologue os PTR 9 affects nitrogen utilization efficiency, growth and grain yield in rice. Plant Biotechnol. J. 11, 446–458. doi: 10.1111/pbi.12031
Fan, S.-C., Lin, C.-S., Hsu, P.-K., Lin, S.-H., Tsay, Y.-F. (2009). The arabidopsis nitrate transporter NRT1. 7, expressed in phloem, is responsible for source-to-sink remobilization of nitrate. Plant Cell 21, 2750–2761. doi: 10.1105/tpc.109.067603
Fan, X., Naz, M., Fan, X., Xuan, W., Miller, A. J., Xu, G. (2017). Plant nitrate transporters: from gene function to application. J. Exp. Bot. 68, 2463–2475. doi: 10.1093/jxb/erx011
Fan, X., Tang, Z., Tan, Y., Zhang, Y., Luo, B., Yang, M., et al. (2016b). Overexpression of a pH-sensitive nitrate transporter in rice increases crop yields. Proc. Natl. Acad. Sci. 113, 7118–7123. doi: 10.1073/pnas.1525184113
Fan, X., Xie, D., Chen, J., Lu, H., Xu, Y., Ma, C., et al. (2014). Over-expression of OsPTR6 in rice increased plant growth at different nitrogen supplies but decreased nitrogen use efficiency at high ammonium supply. Plant Sci. 227, 1–11. doi: 10.1016/j.plantsci.2014.05.013
Ferreira, S., Moreira, E., Amorim, I., Santos, C., Melo, P. (2019). Arabidopsis thaliana mutants devoid of chloroplast glutamine synthetase (GS2) have non-lethal phenotype under photorespiratory conditions. Plant Physiol. Biochem. 144, 365–374. doi: 10.1016/j.plaphy.2019.10.009
Finnegan, P. M., Chen, W. (2012). Arsenic toxicity: the effects on plant metabolism. Front. Physiol. 3, 182. doi: 10.3389/fphys.2012.00182
Fu, Y., Yi, H., Bao, J., Gong, J. (2015). LeNRT2. 3 functions in nitrate acquisition and long-distance transport in tomato. FEBS Lett. 589, 1072–1079. doi: 10.1016/j.febslet.2015.03.016
Gan, Y., Bernreiter, A., Filleur, S., Abram, S., Forde, B. G. (2012). Overexpressing the ANR1 MADS-Box Gene in Transgenic Plants Provides New Insights into its Role in the Nitrate Regulation of Root Development. Plant Cell Physiol 53(6), 1003–1016. doi: 10.1093/pcp/pcs050
Gao, Y., De Bang, T. C., Schjoerring, J. K. (2019a). Cisgenic overexpression of cytosolic glutamine synthetase improves nitrogen utilization efficiency in barley and prevents grain protein decline under elevated CO2. Plant Biotechnol. J. 17, 1209–1221. doi: 10.1111/pbi.13046
Gao, S., Guo, C., Zhang, Y., Zhang, F., Du, X., Gu, J., et al. (2016). Wheat microRNA member TaMIR444a is nitrogen deprivation-responsive and involves plant adaptation to the nitrogen-starvation stress. Plant Mol. Biol. Rep. 34, 931–946. doi: 10.1007/s11105-016-0973-3
Gao, Z., Wang, Y., Chen, G., Zhang, A., Yang, S., Shang, L., et al. (2019b). The indica nitrate reductase gene OsNR2 allele enhances rice yield potential and nitrogen use efficiency. Nat. Commun. 10, 1–10. doi: 10.1038/s41467-019-13110-8
Ge, L., Dou, Y., Li, M., Qu, P., He, Z., Liu, Y., et al. (2019). SiMYB3 in foxtail millet (Setaria italica) confers tolerance to low-nitrogen stress by regulating root growth in transgenic plants. Int. J. Mol. Sci. 20, 5741. doi: 10.3390/ijms20225741
Ge, M., Wang, Y., Liu, Y., Jiang, L., He, B., Ning, L., et al. (2020). The NIN-like protein 5 (ZmNLP5) transcription factor is involved in modulating the nitrogen response in maize. Plant J. 102, 353–368. doi: 10.1111/tpj.14628
Good, A. G., Shrawat, A. K., Muench, D. G. (2004). Can less yield more? is reducing nutrient input into the environment compatible with maintaining crop production? Trends Plant Sci. 9, 597–605. doi: 10.1016/j.tplants.2004.10.008
Guan, Y., Liu, D.-F., Qiu, J., Liu, Z.-J., He, Y.-N., Fang, Z.-J., et al. (2022). The nitrate transporter OsNPF7. 9 mediates nitrate allocation and the divergent nitrate use efficiency between indica and japonica rice. Plant Physiol 189 (1), 215–229. doi: 10.1093/plphys/kiac044
Guan, P., Ripoll, J.-J., Wang, R., Vuong, L., Bailey-Steinitz, L. J., Ye, D., et al. (2017). Interacting TCP and NLP transcription factors control plant responses to nitrate availability. Proc. Natl. Acad. Sci. 114, 2419–2424. doi: 10.1073/pnas.1615676114
Guo, T., Xuan, H., Yang, Y., Wang, L., Wei, L., Wang, Y., et al. (2014). Transcription analysis of genes encoding the wheat root transporter NRT1 and NRT2 families during nitrogen starvation. J. Plant Growth Regul. 33, 837–848. doi: 10.1007/s00344-014-9435-z
Guo, S., Zhou, Y., Shen, Q., Zhang, F. (2007). Effect of ammonium and nitrate nutrition on some physiological processes in higher plants-growth, photosynthesis, photorespiration, and water relations. Plant Biol. 9, 21–29. doi: 10.1055/s-2006-924541
Hasnain, A., Irfan, M., Bashir, A., Maqbool, A., Malik, K. A. (2020). Transcription factor TaDof1 improves nitrogen and carbon assimilation under low-nitrogen conditions in wheat. Plant Mol. Biol. Rep. 38, 441–451. doi: 10.1007/s11105-020-01208-z
He, X., Qu, B., Li, W., Zhao, X., Teng, W., Ma, W., et al. (2015). The nitrate-inducible NAC transcription factor TaNAC2-5A controls nitrate response and increases wheat yield. Plant Physiol. 169, 1991–2005. doi: 10.1104/pp.15.00568
Hernandez, L., Garate, A., Carpena-Ruiz, R. (1997). Effects of cadmium on the uptake, distribution and assimilation of nitrate in pisum sativum. Plant Soil 189, 97–106. doi: 10.1023/A:1004252816355
Hou, M., Wu, D., Li, Y., Tao, W., Chao, L., Zhang, Y. (2021). The role of auxin in nitrogen-modulated shoot branching. Plant Signal Behav. 16, 1885888. doi: 10.1080/15592324.2021.1885888
Hsu, P.-K., Tsay, Y.-F. (2013). Two phloem nitrate transporters, NRT1. 11 and NRT1. 12, are important for redistributing xylem-borne nitrate to enhance plant growth. Plant Physiol. 163, 844–856. doi: 10.1104/pp.113.226563
Huang, S., Liang, Z., Chen, S., Sun, H., Fan, X., Wang, C., et al. (2019). A transcription factor, OsMADS57, regulates long-distance nitrate transport and root elongation. Plant Physiol. 180, 882–895. doi: 10.1104/pp.19.00142
Huang, N.-C., Liu, K.-H., Lo, H.-J., Tsay, Y.-F. (1999). Cloning and functional characterization of an arabidopsis nitrate transporter gene that encodes a constitutive component of low-affinity uptake. Plant Cell 11, 1381–1392. doi: 10.1105/tpc.11.8.1381
Hu, B., Wang, W., Ou, S., Tang, J., Li, H., Che, R., et al. (2015). Variation in NRT1. 1B contributes to nitrate-use divergence between rice subspecies. Nat. Genet. 47, 834–838. doi: 10.1038/ng.3337
Hu, M., Zhao, X., Liu, Q., Hong, X., Zhang, W., Zhang, Y., et al. (2018). Transgenic expression of plastidic glutamine synthetase increases nitrogen uptake and yield in wheat. Plant Biotechnol. J. 16, 1858–1867. doi: 10.1111/pbi.12921
Iqbal, A., Qiang, D., Alamzeb, M., Xiangru, W., Huiping, G., Hengheng, Z., et al. (2020). Untangling the molecular mechanisms and functions of nitrate to improve nitrogen use efficiency. J. Sci. Food Agric. 100, 904–914. doi: 10.1002/jsfa.10085
Ito, Y., Nishizawa-Yokoi, A., Endo, M., Mikami, M., Toki, S. (2015). CRISPR/Cas9-mediated mutagenesis of the RIN locus that regulates tomato fruit ripening. Biochem. Biophys. Res. Commun. 467, 76–82. doi: 10.1016/j.bbrc.2015.09.117
Iwamoto, M., Tagiri, A. (2016). Micro RNA-targeted transcription factor gene RDD 1 promotes nutrient ion uptake and accumulation in rice. Plant J. 85, 466–477. doi: 10.1111/tpj.13117
Jian, S., Luo, J., Liao, Q., Liu, Q., Guan, C., Zhang, Z. (2019). NRT1. 1 regulates nitrate allocation and cadmium tolerance in arabidopsis. Front. Plant Sci. 10, 384. doi: 10.3389/fpls.2019.00384
Jin, Z., Zhu, Y., Li, X., Dong, Y., An, Z. (2015). Soil n retention and nitrate leaching in three types of dunes in the mu us desert of China. Sci. Rep. 5, 1–8. doi: 10.1038/srep14222
Jonassen, E. M., Sévin, D. C., Lillo, C. (2009). The bZIP transcription factors HY5 and HYH are positive regulators of the main nitrate reductase gene in arabidopsis leaves, NIA2, but negative regulators of the nitrate uptake gene NRT1. 1. J. Plant Physiol. 166, 2071–2076. doi: 10.1016/j.jplph.2009.05.010
Kant, S. (2018). Understanding nitrate uptake, signaling and remobilisation for improving plant nitrogen use efficiency. Semin. Cell Dev. Biol. 74, 89–96. doi: 10.1016/j.semcdb.2017.08.034
Kant, S., Bi, Y.-M., Rothstein, S. J. (2011a). Understanding plant response to nitrogen limitation for the improvement of crop nitrogen use efficiency. J. Exp. Bot. 62, 1499–1509. doi: 10.1093/jxb/erq297
Kant, S., Peng, M., Rothstein, S. J. (2011b). Genetic regulation by NLA and microRNA827 for maintaining nitrate-dependent phosphate homeostasis in arabidopsis. PloS Genet. 7, e1002021. doi: 10.1371/journal.pgen.1002021
Kechid, M., Desbrosses, G., Rokhsi, W., Varoquaux, F., Djekoun, A., Touraine, B. (2013). The NRT2.5 and NRT2.6 genes are involved in growth promotion of arabidopsis by the plant growth-promoting rhizobacterium (PGPR) strain phyllobacterium brassicacearum STM196. New Phytol. 198, 514–524. doi: 10.1111/nph.12158
Kendall, A., Wallsgrove, R., Hall, N., Turner, J., Lea, P. (1986). Carbon and nitrogen metabolism in barley (Hordeum vulgare l.) mutants lacking ferredoxin-dependent glutamate synthase. Planta 168, 316–323. doi: 10.1007/BF00392355
Korte, A., Farlow, A. (2013). The advantages and limitations of trait analysis with GWAS: a review. Plant Methods 9, 1–9. doi: 10.1186/1746-4811-9-29
Kumar, A., Sandhu, N., Kumar, P., Pruthi, G., Singh, J., Kaur, S., et al. (2022). Genome-wide identification and in silico analysis of NPF, NRT2, CLC and SLAC1/SLAH nitrate transporters in hexaploid wheat (Triticum aestivum). Sci. Rep. 12, 1–20. doi: 10.1038/s41598-022-15202-w
Kurai, T., Wakayama, M., Abiko, T., Yanagisawa, S., Aoki, N., Ohsugi, R. (2011). Introduction of the ZmDof1 gene into rice enhances carbon and nitrogen assimilation under low-nitrogen conditions. Plant Biotechnol. J. 9, 826–837. doi: 10.1111/j.1467-7652.2011.00592.x
Lam, H. M., Wong, P., Chan, H. K., Yam, K. M., Chen, L., Chow, C. M., et al. (2003). Overexpression of the ASN1 gene enhances nitrogen status in seeds of Arabidopsis. Plant Physiol 132(2), 926–935. doi: 10.1104/pp.103.020123
Lee, R. (1982). Selectivity and kinetics of ion uptake by barley plants following nutrient deficiency. Ann. Bot. 50, 429–449. doi: 10.1093/oxfordjournals.aob.a086383
Lee, S., Marmagne, A., Park, J., Fabien, C., Yim, Y., Kim, S. J., et al. (2020). Concurrent activation of OsAMT1; 2 and OsGOGAT1 in rice leads to enhanced nitrogen use efficiency under nitrogen limitation. Plant J. 103, 7–20. doi: 10.1111/tpj.14794
Léran, S., Garg, B., Boursiac, Y., Corratgé-Faillie, C., Brachet, C., Tillard, P., et al. (2015). AtNPF5. 5, a nitrate transporter affecting nitrogen accumulation in arabidopsis embryo. Sci. Rep. 5, 1–7. doi: 10.1038/srep07962
Léran, S., Varala, K., Boyer, J.-C., Chiurazzi, M., Crawford, N., Daniel-Vedele, F., et al. (2014). A unified nomenclature of NITRATE TRANSPORTER 1/PEPTIDE TRANSPORTER family members in plants. Trends Plant Sci. 19, 5–9. doi: 10.1016/j.tplants.2013.08.008
Liang, Q., Dong, M., Gu, M., Zhang, P., Ma, Q., He, B. (2022). MeNPF4. 5 improves cassava nitrogen use efficiency and yield by regulating nitrogen uptake and allocation. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.866855
Li, J.-Y., Fu, Y.-L., Pike, S. M., Bao, J., Tian, W., Zhang, Y., et al. (2010). The arabidopsis nitrate transporter NRT1. 8 functions in nitrate removal from the xylem sap and mediates cadmium tolerance. Plant Cell 22, 1633–1646. doi: 10.1105/tpc.110.075242
Li, H., Hu, B., Chu, C. (2017a). Nitrogen use efficiency in crops: Lessons from arabidopsis and rice. J. Exp. Bot. 68, 2477–2488. doi: 10.1093/jxb/erx101
Lin, S.-H., Kuo, H.-F., Canivenc, G., Lin, C.-S., Lepetit, M., Hsu, P.-K., et al. (2008). Mutation of the arabidopsis NRT1. 5 nitrate transporter causes defective root-to-shoot nitrate transport. Plant Cell 20, 2514–2528. doi: 10.1105/tpc.108.060244
Li, Y., Ouyang, J., Wang, Y.-Y., Hu, R., Xia, K., Duan, J., et al. (2015). Disruption of the rice nitrate transporter OsNPF2. 2 hinders root-to-shoot nitrate transport and vascular development. Sci. Rep. 5, 1–10. doi: 10.1038/srep09635
Li, H., Peng, Z., Yang, X., Wang, W., Fu, J., Wang, J., et al. (2013). Genome-wide association study dissects the genetic architecture of oil biosynthesis in maize kernels. Nat. Genet. 45, 43–50. doi: 10.1038/ng.2484
Li, S., Tian, Y., Wu, K., Ye, Y., Yu, J., Zhang, J., et al. (2018). Modulating plant growth–metabolism coordination for sustainable agriculture. Nature 560, 595–600. doi: 10.1038/s41586-018-0415-5
Li, G., Tillard, P., Gojon, A., Maurel, C. (2016a). Dual regulation of root hydraulic conductivity and plasma membrane aquaporins by plant nitrate accumulation and high-affinity nitrate transporter NRT2. 1. Plant Cell Physiol. 57, 733–742. doi: 10.1093/pcp/pcw022
Liu, K.-H., Huang, C.-Y., Tsay, Y.-F. (1999). CHL1 is a dual-affinity nitrate transporter of arabidopsis involved in multiple phases of nitrate uptake. Plant Cell 11, 865–874. doi: 10.1105/tpc.11.5.865
Liu, Y., Jia, Z., Li, X., Wang, Z., Chen, F., Mi, G., et al. (2020). Involvement of a truncated MADS-box transcription factor ZmTMM1 in root nitrate foraging. J. Exp. Bot. 71, 4547–4561. doi: 10.1093/jxb/eraa116
Liu, K.-H., Tsay, Y.-F. (2003). Switching between the two action modes of the dual-affinity nitrate transporter CHL1 by phosphorylation. EMBO J. 22, 1005–1013. doi: 10.1093/emboj/cdg118
Li, W., Wang, Y., Okamoto, M., Crawford, N. M., Siddiqi, M. Y., Glass, A. D. (2007). Dissection of the AtNRT2. 1: AtNRT2. 2 inducible high-affinity nitrate transporter gene cluster. Plant Physiol. 143, 425–433. doi: 10.1104/pp.106.091223
Li, X., Xia, K., Liang, Z., Chen, K., Gao, C., Zhang, M. (2016b). MicroRNA393 is involved in nitrogen-promoted rice tillering through regulation of auxin signal transduction in axillary buds. Sci. Rep. 6, 1–12. doi: 10.1038/srep32158
Li, M., Xu, J., Gao, Z., Tian, H., Gao, Y., Kariman, K. (2020). Genetically modified crops are superior in their nitrogen use efficiency-a meta-analysis of three major cereals. Sci. Rep. 10, 1–9. doi: 10.1038/s41598-020-65684-9
Li, H., Yu, M., Du, X.-Q., Wang, Z.-F., Wu, W.-H., Quintero, F. J., et al. (2017b). NRT1. 5/NPF7. 3 functions as a proton-coupled H+/K+ antiporter for k+ loading into the xylem in arabidopsis. Plant Cell 29, 2016–2026. doi: 10.1105/tpc.16.00972
Maeda, Y., Konishi, M., Kiba, T., Sakuraba, Y., Sawaki, N., Kurai, T., et al. (2018). A NIGT1-centred transcriptional cascade regulates nitrate signalling and incorporates phosphorus starvation signals in arabidopsis. Nat. Commun. 9, 1376. doi: 10.1038/s41467-018-03832-6
Maghiaoui, A., Bouguyon, E., Cuesta, C., Perrine-Walker, F., Alcon, C., Krouk, G., et al. (2020). The arabidopsis NRT1. 1 transceptor coordinately controls auxin biosynthesis and transport to regulate root branching in response to nitrate. J. Exp. Bot. 71, 4480–4494. doi: 10.1093/jxb/eraa242
Maierhofer, T., Lind, C., Hüttl, S., Scherzer, S., Papenfuß, M., Simon, J., et al. (2014). A single-pore residue renders the arabidopsis root anion channel SLAH2 highly nitrate selective. Plant Cell 26, 2554–2567. doi: 10.1105/tpc.114.125849
Mao, Q. Q., Guan, M. Y., Lu, K. X., Du, S. T., Fan, S. K., Ye, Y. Q., et al. (2014). Inhibition of nitrate transporter 1.1-controlled nitrate uptake reduces cadmium uptake in arabidopsis. Plant Physiol. 166, 934–944. doi: 10.1104/pp.114.243766
Marín, I. C., Loef, I., Bartetzko, L., Searle, I., Coupland, G., Stitt, M., et al. (2011). Nitrate regulates floral induction in arabidopsis, acting independently of light, gibberellin and autonomous pathways. Planta 233, 539–552. doi: 10.1007/s00425-010-1316-5
Martin, A., Lee, J., Kichey, T., Gerentes, G., Zivy, M., Tatout, C., et al. (2006). Two Cytosolic Glutamine Synthetase Isoforms of Maize Are Specifically Involved in the Control of Grain Production. Plant Cell. 18(11), 3252–3274. doi: 10.1105/tpc.106.042689
Ma, X., Zhang, Q., Zhu, Q., Liu, W., Chen, Y., Qiu, R., et al. (2015). A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol. Plant 8, 1274–1284. doi: 10.1016/j.molp.2015.04.007
Medici, A., Marshall-Colon, A., Ronzier, E., Szponarski, W., Wang, R., Gojon, A., et al. (2015). AtNIGT1/HRS1 integrates nitrate and phosphate signals at the arabidopsis root tip. Nat. Commun. 6, 1–11. doi: 10.1038/ncomms7274
Medici, A., Szponarski, W., Dangeville, P., Safi, A., Dissanayake, I. M., Saenchai, C., et al. (2019). Identification of molecular integrators shows that nitrogen actively controls the phosphate starvation response in plants. Plant Cell 31, 1171–1184. doi: 10.1105/tpc.18.00656
Meng, S., Peng, J.-S., He, Y.-N., Zhang, G.-B., Yi, H.-Y., Fu, Y.-L., et al. (2016). Arabidopsis NRT1. 5 mediates the suppression of nitrate starvation-induced leaf senescence by modulating foliar potassium level. Mol. Plant 9, 461–470. doi: 10.1016/j.molp.2015.12.015
Miflin, B. J., Habash, D. Z. (2002). The role of glutamine synthetase and glutamate dehydrogenase in nitrogen assimilation and possibilities for improvement in the nitrogen utilization of crops. J. Exp. Bot. 53, 979–987. doi: 10.1093/jexbot/53.370.979
Migocka, M., Warzybok, A., Kłobus, G. (2013). The genomic organization and transcriptional pattern of genes encoding nitrate transporters 1 (NRT1) in cucumber. Plant Soil 364, 245–260. doi: 10.1007/s11104-012-1345-x
Mohanty, S., Swain, C. K., Kumar, A., Nayak, A. (2020). “Nitrogen footprint: a useful indicator of agricultural sustainability,” in Nutrient dynamics for sustainable crop production (Springer Singapore: Springer), 135–156.
Noguero, M., Leran, S., Bouguyon, E., Brachet, C., Tillard, P., Nacry, P., et al. (2018). Revisiting the functional properties of NPF6. 3/NRT1. 1/CHL1 in xenopus oocytes. hal-01777543.
O’brien, J. A., Vega, A., Bouguyon, E., Krouk, G., Gojon, A., Coruzzi, G., et al. (2016). Nitrate transport, sensing, and responses in plants. Mol. Plant 9, 837–856. doi: 10.1016/j.molp.2016.05.004
Ogura, T., Busch, W. (2015). From phenotypes to causal sequences: using genome wide association studies to dissect the sequence basis for variation of plant development. Curr. Opin. Plant Biol. 23, 98–108. doi: 10.1016/j.pbi.2014.11.008
Pan, W., You, Y., Weng, Y. N., Shentu, J. L., Lu, Q., Xu, Q. R., et al. (2020). Zn stress facilitates nitrate transporter 1.1-mediated nitrate uptake aggravating zn accumulation in arabidopsis plants. Ecotoxicol Environ. Saf. 190, 110104. doi: 10.1016/j.ecoenv.2019.110104
Peña, P. A., Quach, T., Sato, S., Ge, Z., Nersesian, N., Changa, T., et al. (2017). Expression of the maize Dof1 transcription factor in wheat and sorghum. Front. Plant Sci. 8, 434. doi: 10.3389/fpls.2017.00434
Pike, S., Gao, F., Kim, M. J., Kim, S. H., Schachtman, D. P., Gassmann, W. (2014). Members of the NPF3 transporter subfamily encode pathogen-inducible nitrate/nitrite transporters in grapevine and arabidopsis. Plant Cell Physiol. 55, 162–170. doi: 10.1093/pcp/pct167
Qiao, Q., Wang, X., Yang, M., Zhao, Y., Gu, J., Xiao, K. (2018). Wheat miRNA member TaMIR2275 involves plant nitrogen starvation adaptation via enhancement of the n acquisition-associated process. Acta Physiologiae Plantarum 40, 1–13. doi: 10.1007/s11738-018-2758-9
Qu, B., He, X., Wang, J., Zhao, Y., Teng, W., Shao, A., et al. (2015). A wheat CCAAT box-binding transcription factor increases the grain yield of wheat with less fertilizer input. Plant Physiol. 167, 411–423. doi: 10.1104/pp.114.246959
Raddatz, N., Morales De Los Ríos, L., Lindahl, A. M., Quintero, F. J., Pardo, J. M. (2020). Coordinated transport of nitrate, potassium and sodium. Front. Plant Sci. 11, 247. doi: 10.3389/fpls.2020.00247
Remans, T., Nacry, P., Pervent, M., Filleur, S., Diatloff, E., Mounier, E., et al. (2006). The arabidopsis NRT1. 1 transporter participates in the signaling pathway triggering root colonization of nitrate-rich patches. Proc. Natl. Acad. Sci. 103, 19206–19211. doi: 10.1073/pnas.0605275103
Rubin, G., Tohge, T., Matsuda, F., Saito, K., Scheible, W.-R. D. (2009). Members of the LBD family of transcription factors repress anthocyanin synthesis and affect additional nitrogen responses in arabidopsis. Plant Cell 21, 3567–3584. doi: 10.1105/tpc.109.067041
Rufty, T. W., Jr., Mackown, C. T., Israel, D. W. (1990). Phosphorus stress effects on assimilation of nitrate. Plant Physiol. 94, 328–333. doi: 10.1104/pp.94.1.328
Segonzac, C., Boyer, J.-C., Ipotesi, E., Szponarski, W., Tillard, P., Touraine, B., et al. (2007). Nitrate efflux at the root plasma membrane: identification of an arabidopsis excretion transporter. Plant Cell 19, 3760–3777. doi: 10.1105/tpc.106.048173
Shin, J. M., Chung, K., Sakamoto, S., Kojima, S., Yeh, C.-M., Ikeda, M., et al. (2017). The chimeric repressor for the GATA4 transcription factor improves tolerance to nitrogen deficiency in arabidopsis. Plant Biotechnol. 34, 151–158. doi: 10.5511/plantbiotechnology.17.0727a
Shrawat, A. K., Carroll, R. T., Depauw, M., Taylor, G. J., Good, A. G. (2008). Genetic engineering of improved nitrogen use efficiency in rice by the tissue-specific expression of alanine aminotransferase. Plant Biotechnol. J. 6, 722–732. doi: 10.1111/j.1467-7652.2008.00351.x
Si, L., Chen, J., Huang, X., Gong, H., Luo, J., Hou, Q., et al. (2016). OsSPL13 controls grain size in cultivated rice. Nat. Genet. 48, 447–456. doi: 10.1038/ng.3518
Snyder, R., Tegeder, M. (2021). Targeting nitrogen metabolism and transport processes to improve plant nitrogen use efficiency. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.628366
Somerville, C. R., Ogren, W. L. (1980). Inhibition of photosynthesis in arabidopsis mutants lacking leaf glutamate synthase activity. Nature 286, 257–259. doi: 10.1038/286257a0
Sun, H., Qian, Q., Wu, K., Luo, J., Wang, S., Zhang, C., et al. (2014a). Heterotrimeric G proteins regulate nitrogen-use efficiency in rice. Nat. Genet. 46, 652–656. doi: 10.1038/ng.2958
Sun, H., Tao, J., Liu, S., Huang, S., Chen, S., Xie, X., et al. (2014b). Strigolactones are involved in phosphate-and nitrate-deficiency-induced root development and auxin transport in rice. J. Exp. Bot. 65, 6735–6746. doi: 10.1093/jxb/eru029
Sun, C.-H., Yu, J.-Q., Wen, L.-Z., Guo, Y.-H., Sun, X., Hao, Y.-J., et al. (2018). Chrysanthemum MADS-box transcription factor CmANR1 modulates lateral root development via homo-/heterodimerization to influence auxin accumulation in arabidopsis. Plant Sci. 266, 27–36. doi: 10.1016/j.plantsci.2017.09.017
Tal, I., Zhang, Y., Jørgensen, M. E., Pisanty, O., Barbosa, I. C., Zourelidou, M., et al. (2016). The arabidopsis NPF3 protein is a GA transporter. Nat. Commun. 7, 11486. doi: 10.1038/ncomms11486
Tang, Z., Chen, Y., Chen, F., Ji, Y., Zhao, F. J. (2017). OsPTR7 (OsNPF8.1), a putative peptide pransporter in rice, is pnvolved in dimethylarsenate accumulation in rice grain. Plant Cell Physiol. 58 (5), 904–913. doi: 10.1093/pcp/pcx029
Tang, Z., Fan, X., Li, Q., Feng, H., Miller, A. J., Shen, Q., et al. (2012). Knockdown of a rice stelar nitrate transporter alters long-distance translocation but not root influx. Plant Physiol. 160, 2052–2063. doi: 10.1104/pp.112.204461
Tang, W., Ye, J., Yao, X., Zhao, P., Xuan, W., Tian, Y., et al. (2019). Genome-wide associated study identifies NAC42-activated nitrate transporter conferring high nitrogen use efficiency in rice. Nat. Commun. 10, 1–11. doi: 10.1038/s41467-019-13187-1
Taochy, C., Gaillard, I., Ipotesi, E., Oomen, R., Leonhardt, N., Zimmermann, S., et al. (2015). The arabidopsis root stele transporter NPF2.3 contributes to nitrate translocation to shoots under salt stress. Plant J. 83, 466–479. doi: 10.1111/tpj.12901
Tian, Q., Chen, F., Liu, J., Zhang, F., Mi, G. (2008). Inhibition of maize root growth by high nitrate supply is correlated with reduced IAA levels in roots. J. Plant Physiol. 165, 942–951. doi: 10.1016/j.jplph.2007.02.011
Tiwari, J. K., Buckseth, T., Singh, R. K., Kumar, M., Kant, S. (2020). Prospects of improving nitrogen use efficiency in potato: lessons from transgenics to genome editing strategies in plants. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.597481
Tsay, Y.-F., Schroeder, J. I., Feldmann, K. A., Crawford, N. M. (1993). The herbicide sensitivity gene CHL1 of arabidopsis encodes a nitrate-inducible nitrate transporter. Cell 72, 705–713. doi: 10.1016/0092-8674(93)90399-B
Uauy, C., Distelfeld, A., Fahima, T., Blechl, A., Dubcovsky, J. (2006). A NAC Gene regulating senescence improves grain protein, zinc, and iron content in wheat. Sci. 314(5803):1298–301. doi: 10.1126/science.1133649
Vidal, E. A., Alvarez, J. M., Araus, V., Riveras, E., Brooks, M. D., Krouk, G., et al. (2020). Nitrate in 2020: thirty years from transport to signaling networks. Plant Cell 32, 2094–2119. doi: 10.1105/tpc.19.00748
Von Der Fecht-Bartenbach, J., Bogner, M., Dynowski, M., Ludewig, U. (2010). CLC-b-mediated NO– 3/H+ exchange across the tonoplast of arabidopsis vacuoles. Plant Cell Physiol. 51, 960–968. doi: 10.1093/pcp/pcq062
Von Wittgenstein, N. J., Le, C. H., Hawkins, B. J., Ehlting, J. (2014). Evolutionary classification of ammonium, nitrate, and peptide transporters in land plants. BMC evolutionary Biol. 14, 1–17. doi: 10.1186/1471-2148-14-11
Walch-Liu, P., Forde, B. G. (2008). Nitrate signalling mediated by the NRT1. 1 nitrate transporter antagonises l-glutamate-induced changes in root architecture. Plant J. 54, 820–828. doi: 10.1111/j.1365-313X.2008.03443.x
Walch-Liu, P., Neumann, G., Bangerth, F., Engels, C. (2000). Rapid effects of nitrogen form on leaf morphogenesis in tobacco. J. Exp. Bot. 51, 227–237. doi: 10.1093/jexbot/51.343.227
Wang, X., Cai, X., Xu, C., Wang, Q. (2021b). Identification and characterization of the NPF, NRT2 and NRT3 in spinach. Plant Physiol. Biochem. 158, 297–307. doi: 10.1016/j.plaphy.2020.11.017
Wang, S., Chen, A., Xie, K., Yang, X., Luo, Z., Chen, J., et al. (2020c). Functional analysis of the OsNPF4.5 nitrate transporter reveals a conserved mycorrhizal pathway of nitrogen acquisition in plants. Proc. Natl. Acad. Sci. United States America 117 (28), 16649–16659. doi: 10.1073/pnas.2000926117
Wang, Y., Fu, B., Pan, L., Chen, L., Fu, X., Li, K. (2013). Overexpression of arabidopsis Dof1, GS1 and GS2 enhanced nitrogen assimilation in transgenic tobacco grown under low-nitrogen conditions. Plant Mol. Biol. Rep. 31, 886–900. doi: 10.1007/s11105-013-0561-8
Wang, M., Hasegawa, T., Beier, M., Hayashi, M., Ohmori, Y., Yano, K., et al. (2021a). Growth and nitrate reductase activity are impaired in rice osnlp4 mutants supplied with nitrate. Plant Cell Physiol. 62, 1156–1167. doi: 10.1093/pcp/pcab035
Wang, W., Hu, B., Yuan, D., Liu, Y., Che, R., Hu, Y., et al. (2018c). Expression of the nitrate transporter gene OsNRT1. 1A/OsNPF6. 3 confers high yield and early maturation in rice. Plant Cell 30, 638–651. doi: 10.1105/tpc.17.00809
Wang, R., Liu, D., Crawford, N. M. (1998). The arabidopsis CHL1 protein plays a major role in high-affinity nitrate uptake. Proc. Natl. Acad. Sci. 95, 15134–15139. doi: 10.1073/pnas.95.25.15134
Wang, Q., Liu, C., Dong, Q., Huang, D., Li, C., Li, P., et al. (2018b). Genome-wide identification and analysis of apple NITRATE TRANSPORTER 1/PEPTIDE TRANSPORTER family (NPF) genes reveals MdNPF6. 5 confers high capacity for nitrogen uptake under low-nitrogen conditions. Int. J. Mol. Sci. 19, 2761. doi: 10.3390/ijms19092761
Wang, J., Lu, K., Nie, H., Zeng, Q., Wu, B., Qian, J., et al. (2018a). Rice nitrate transporter OsNPF7. 2 positively regulates tiller number and grain yield. Rice 11, 1–13. doi: 10.1186/s12284-018-0205-6
Wang, Y.-Y., Tsay, Y.-F. (2011). Arabidopsis nitrate transporter NRT1. 9 is important in phloem nitrate transport. Plant Cell 23, 1945–1957. doi: 10.1105/tpc.111.083618
Wang, X., Wang, H.-F., Chen, Y., Sun, M.-M., Wang, Y., Chen, Y.-F. (2020b). The transcription factor NIGT1. 2 modulates both phosphate uptake and nitrate influx during phosphate starvation in arabidopsis and maize. Plant Cell 32, 3519–3534. doi: 10.1105/tpc.20.00361
Wang, J., Wan, R., Nie, H., Xue, S., Fang, Z. (2022). OsNPF5. 16, a nitrate transporter gene with natural variation, is essential for rice growth and yield. Crop J. 10, 397–406. doi: 10.1016/j.cj.2021.08.005
Wang, D., Xu, T., Yin, Z., Wu, W., Geng, H., Li, L., et al. (2020a). Overexpression of OsMYB305 in rice enhances the nitrogen uptake under low-nitrogen condition. Front. Plant Sci. 11, 369. doi: 10.3389/fpls.2020.00369
Wilkinson, J. Q., Crawford, N. M. (1993). Identification and characterization of a chlorate-resistant mutant of arabidopsis thaliana with mutations in both nitrate reductase structural genes NIA1 and NIA2. Mol. Gen. Genet. MGG 239, 289–297. doi: 10.1007/BF00281630
Wu, J., Zhang, Z. S., Xia, J. Q., Alfatih, A., Song, Y., Huang, Y. J., et al. (2021). Rice NIN-LIKE PROTEIN 4 plays a pivotal role in nitrogen use efficiency. Plant Biotechnol. J. 19, 448–461. doi: 10.1111/pbi.13475
Xia, X., Fan, X., Wei, J., Feng, H., Qu, H., Xie, D., et al. (2015). Rice nitrate transporter OsNPF2. 4 functions in low-affinity acquisition and long-distance transport. J. Exp. Bot. 66, 317–331. doi: 10.1093/jxb/eru425
Xu, G., Fan, X., Miller, A. J. (2012). Plant nitrogen assimilation and use efficiency. Annu. Rev. Plant Biol. 63, 153–182. doi: 10.1146/annurev-arplant-042811-105532
Yamaya, T., Obara, M., Nakajima, H., Sasaki, S., Hayakawa, T., Sato, T. (2002). Genetic manipulation and quantitative-trait loci mapping for nitrogen recycling in rice. J. Exp. Bot. 53, 917–925. doi: 10.1093/jexbot/53.370.917
Yanagisawa, S., Akiyama, A., Kisaka, H., Uchimiya, H., Miwa, T. (2004). Metabolic engineering with Dof1 transcription factor in plants: Improved nitrogen assimilation and growth under low-nitrogen conditions. Proc. Natl. Acad. Sci. 101, 7833–7838. doi: 10.1073/pnas.0402267101
Yang, L., Luo, S., Wu, Z., Rong, X., Han, Y. (2022). Low nitrogen stress stimulated nitrate uptake rate modulated by auxin in brassica napus l. J. Soil Sci. Plant Nutr. 22, 3500–3506. doi: 10.1007/s42729-022-00904-x
Yang, X., Nong, B., Chen, C., Wang, J., Xia, X., Zhang, Z., et al. (2023). OsNPF3. 1, a member of the NRT1/PTR family, increases nitrogen use efficiency and biomass production in rice. Crop J 11, 108–118. doi: 10.1016/j.cj.2022.07.001
Yang, J., Wang, M., Li, W., He, X., Teng, W., Ma, W., et al. (2019). Reducing expression of a nitrate-responsive bZIP transcription factor increases grain yield and n use in wheat. Plant Biotechnol. J. 17, 1823–1833. doi: 10.1111/pbi.13103
Yang, X., Xia, X., Zeng, Y., Nong, B., Zhang, Z., Wu, Y., et al. (2020). Genome-wide identification of the peptide transporter family in rice and analysis of the PTR expression modulation in two near-isogenic lines with different nitrogen use efficiency. BMC Plant Biol. 20, 1–15. doi: 10.1186/s12870-020-02419-y
Yuan, S., Li, Z., Li, D., Yuan, N., Hu, Q., Luo, H. (2015). Constitutive expression of rice microRNA528 alters plant development and enhances tolerance to salinity stress and nitrogen starvation in creeping bentgrass. Plant Physiol. 169, 576–593. doi: 10.1104/pp.15.00899
Yu, C., Liu, Y., Zhang, A., Su, S., Yan, A., Huang, L., et al. (2015). MADS-box transcription factor OsMADS25 regulates root development through affection of nitrate accumulation in rice. PloS One 10, e0135196. doi: 10.1371/journal.pone.0135196
Yu, L.-H., Miao, Z.-Q., Qi, G.-F., Wu, J., Cai, X.-T., Mao, J.-L., et al. (2014). MADS-box transcription factor AGL21 regulates lateral root development and responds to multiple external and physiological signals. Mol. Plant 7, 1653–1669. doi: 10.1093/mp/ssu088
Yu, L.-H., Wu, J., Tang, H., Yuan, Y., Wang, S.-M., Wang, Y.-P., et al. (2016). Overexpression of arabidopsis NLP7 improves plant growth under both nitrogen-limiting and-sufficient conditions by enhancing nitrogen and carbon assimilation. Sci. Rep. 6, 1–13. doi: 10.1038/srep27795
Zeng, D.-D., Qin, R., Li, M., Alamin, M., Jin, X.-L., Liu, Y., et al. (2017). The ferredoxin-dependent glutamate synthase (OsFd-GOGAT) participates in leaf senescence and the nitrogen remobilization in rice. Mol. Genet. Genomics 292, 385–395. doi: 10.1007/s00438-016-1275-z
Zhang, J., Fengler, K. A., Van Hemert, J. L., Gupta, R., Mongar, N., Sun, J., et al. (2019). Identification and characterization of a novel stay-green QTL that increases yield in maize. Plant Biotechnol. J. 17, 2272–2285. doi: 10.1111/pbi.13139
Zhang, H., Forde, B. G. (1998). An arabidopsis MADS box gene that controls nutrient-induced changes in root architecture. Science 279, 407–409. doi: 10.1126/science.279.5349.407
Zhang, J., Han, Z., Lu, Y., Zhao, Y., Wang, Y., Zhang, J., et al. (2021a). Genome-wide identification, structural and gene expression analysis of the nitrate transporters (NRTs) family in potato (Solanum tuberosum l.). PloS One 16, e0257383. doi: 10.1371/journal.pone.0257383
Zhang, H., Li, S., Shi, M., Wang, S., Shi, L., Xu, F., et al. (2020). Genome-wide systematic characterization of the NPF family genes and their transcriptional responses to multiple nutrient stresses in allotetraploid rapeseed. Int. J. Mol. Sci. 21, 5947. doi: 10.3390/ijms21175947
Zhang, G.-B., Meng, S., Gong, J.-M. (2018). The expected and unexpected roles of nitrate transporters in plant abiotic stress resistance and their regulation. Int. J. Mol. Sci. 19 (11), 3535. doi: 10.3390/ijms19113535
Zhang, G.-B., Yi, H.-Y., Gong, J.-M. (2014). The arabidopsis ethylene/jasmonic acid-NRT signaling module coordinates nitrate reallocation and the trade-off between growth and environmental adaptation. Plant Cell 26, 3984–3998. doi: 10.1105/tpc.114.129296
Zhang, S., Zhu, L., Shen, C., Ji, Z., Zhang, H., Zhang, T., et al. (2021b). Natural allelic variation in a modulator of auxin homeostasis improves grain yield and nitrogen use efficiency in rice. Plant Cell 33, 566–580. doi: 10.1093/plcell/koaa037
Zhao, M., Ding, H., Zhu, J. K., Zhang, F., Li, W. X. (2011). Involvement of miR169 in the nitrogen-starvation responses in arabidopsis. New Phytol. 190, 906–915. doi: 10.1111/j.1469-8137.2011.03647.x
Zhao, M., Tai, H., Sun, S., Zhang, F., Xu, Y., Li, W.-X. (2012). Cloning and characterization of maize miRNAs involved in responses to nitrogen deficiency. PloS One 7, e29669. doi: 10.1371/journal.pone.0029669
Zheng, Y., Drechsler, N., Rausch, C., Kunze, R. (2016). The arabidopsis nitrate transporter NPF7. 3/NRT1. 5 is involved in lateral root development under potassium deprivation. Plant Signaling Behav. 11, 2832–2847. doi: 10.1080/15592324.2016.1176819
Zhong, L., Chen, D., Min, D., Li, W., Xu, Z., Zhou, Y., et al. (2015). AtTGA4, a bZIP transcription factor, confers drought resistance by enhancing nitrate transport and assimilation in arabidopsis thaliana. Biochem. Biophys. Res. Commun. 457, 433–439. doi: 10.1016/j.bbrc.2015.01.009
Zhou, Y., Cai, H., Xiao, J., Li, X., Zhang, Q., Lian, X. (2009). Over-expression of aspartate aminotransferase genes in rice resulted in altered nitrogen metabolism and increased amino acid content in seeds. Theor. Appl. Genet. 118, 1381–1390. doi: 10.1007/s00122-009-0988-3
Zhou, X., Khare, T., Kumar, V. (2020). Recent trends and advances in identification and functional characterization of plant miRNAs. Acta physiologiae plantarum 42, 1–21. doi: 10.1007/s11738-020-3013-8
Keywords: nitrate transporters, nitrate uptake, nitrate transport and signaling, nitrate remobilization, nitrogen use efficiency, environmental stress
Citation: Aluko OO, Kant S, Adedire OM, Li C, Yuan G, Liu H and Wang Q (2023) Unlocking the potentials of nitrate transporters at improving plant nitrogen use efficiency. Front. Plant Sci. 14:1074839. doi: 10.3389/fpls.2023.1074839
Received: 20 October 2022; Accepted: 16 January 2023;
Published: 21 February 2023.
Edited by:
Mamoru Okamoto, University of Adelaide, AustraliaReviewed by:
Hongmei Cai, Huazhong Agricultural University, ChinaMingyong Zhang, South China Botanical Garden (CAS), China
Copyright © 2023 Aluko, Kant, Adedire, Li, Yuan, Liu and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Haobao Liu, bGl1aGFvYmFvQGNhYXMuY24=; Qian Wang, d2FuZ3FpYW4wMUBjYWFzLmNu
†Present address: Oluwaseun Olayemi Aluko, State Key Laboratory of Cotton Biology, Key Laboratory of Plant Stress Biology, School of Life Sciences, Henan University, Kaifeng, China
 Oluwaseun Olayemi Aluko
Oluwaseun Olayemi Aluko Surya Kant
Surya Kant Oluwafemi Michael Adedire5
Oluwafemi Michael Adedire5 Qian Wang
Qian Wang