
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 20 February 2023
Sec. Plant Pathogen Interactions
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1069971
This article is part of the Research Topic Women in Plant Science - Focus On Plant-Microbe Interaction View all 8 articles
 Virginie Korangi Alleluya1,2*
Virginie Korangi Alleluya1,2* Anthony Argüelles Arias1
Anthony Argüelles Arias1 Bianca Ribeiro3
Bianca Ribeiro3 Barbara De Coninck3
Barbara De Coninck3 Catherine Helmus1
Catherine Helmus1 Pierre Delaplace4
Pierre Delaplace4 Marc Ongena1*
Marc Ongena1*Introduction: Peanut (Arachis hypogaea L.) is a widespread oilseed crop of high agricultural importance in tropical and subtropical areas. It plays a major role in the food supply in the Democratic Republic of Congo (DRC). However, one major constraint in the production of this plant is the stem rot (white mold or southern blight) disease caused by Athelia rolfsii which is so far controlled mainly using chemicals. Considering the harmful effect of chemical pesticides, the implementation of eco-friendly alternatives such as biological control is required for disease management in a more sustainable agriculture in the DRC as in the other developing countries concerned. Bacillus velezensis is among the rhizobacteria best described for its plant protective effect notably due to the production of a wide range of bioactive secondary metabolites. In this work, we wanted to evaluate the potential of B. velezensis strain GA1 at reducing A. rolfsii infection and to unravel the molecular basis of the protective effect.
Results and discussion: Upon growth under the nutritional conditions dictated by peanut root exudation, the bacterium efficiently produces the three types of lipopeptides surfactin, iturin and fengycin known for their antagonistic activities against a wide range of fungal phytopathogens. By testing a range of GA1 mutants specifically repressed in the production of those metabolites, we point out an important role for iturin and another unidentified compound in the antagonistic activity against the pathogen. Biocontrol experiments performed in greenhouse further revealed the efficacy of B. velezensis to reduce peanut disease caused by A. rolfsii both via direct antagonism against the fungus and by stimulating systemic resistance in the host plant. As treatment with pure surfactin yielded a similar level of protection, we postulate that this lipopeptide acts as main elicitor of peanut resistance against A. rolfsii infection.
Peanut (Arachis hypogaea L.) is one of the most widely grown legumes in the warm zones of the world cultivated for its nutritious and oil-rich pods (Arya et al, 2016). In the DRC, this crop plays a major role in the food supply and is considered as the main source of income for the producing households (Nyabyenda, 2005). However, the production of this plant is drastically impacted by various diseases including stem rot (white mold or southern blight) caused by Athelia (Sclerotium, anamorph) rolfsii (Curzi) Tu & Kimbrough. The stem rot disease is characterized by yellowing and wilting of the main stem, lateral branches or the whole plant. It can also attack pods in the soil leading to losses estimated at 10-40% according to the production area (Thiessen and Woodward, 2012; Iquebal et al., 2017; Bosamia et al., 2020). This disease is very difficult to eradicate notably because the fungal pathogen forms sclerotia which can remain in the soil for a considerable time and because this fungus has a wide variety of host plant species (Woodward et al., 2013; Iquebal et al., 2017; Bosamia et al., 2020). Chemical fungicides are mostly used to control this soil-borne pathogen but with limited beneficial effects due to serious problems for humans and environment (Bosamia et al., 2020; Shah, 2020). The development of more environmentally friendly alternatives is needed to manage stem rot in an efficient and sustainable way. In that context, exploiting the so-called plant-beneficial bacteria mostly isolated from the rhizosphere is considered as one of the promising alternative to unwanted chemicals (Backer et al., 2018; Mohanty et al., 2021). These rhizobacteria may promote plant growth and alleviate abiotic stresses (Kashyap et al., 2017) but in many instances, their most desired trait is to protect the host plant from infection by pathogenic microbes referred as biocontrol of plant diseases. This protective effect relies on two main mechanisms, which are direct antagonism of the pathogen and systemically expressed reinforcement of the defensive capacity of the host plant.
Isolates identified as plant-beneficial rhizobacteria with biocontrol potential belong to multiple genera including Pseudomonas, Azospirillum, Azotobacter, Klebsiella, Burkholderia, Serratia and Bacillus (Mohanty et al., 2021). Most of the Bacillus species reported as effective biocontrol agents belong to the so-called Bacillus subtilis complex and typically dwell into the soil/rhizosphere niche (Cawoy et al., 2011; Fan et al., 2017). Among these species, B. velezensis is considered as model for plant-associated bacilli and is the best valorized as microbial inoculant for reducing diseases caused by a wide range of pathogens with different lifecycles and modes of virulence (biotrophic, hemi-biotrophic or necrotrophic) (Fira et al., 2018; Miljaković et al., 2020; Saxena et al., 2020). B. velezensis also distinguishes from other species of the B. subtilis complex considering its richness in biosynthetic gene clusters (representing up to 13% of the whole genome) responsible for the biosynthesis of bioactive secondary metabolites (BSMs). This secondary metabolome largely contributes to the biocontrol activities of the strains and is chemically diverse, including volatiles, terpenes, non-ribosomal peptides (NRP), cyclic lipopeptides (CLPs) and polyketides (PKs), but also ribosomally-synthesized and post translationally-modified bacteriocins and lantibiotics (RiPPs) (Grubbs et al., 2017; Harwood et al., 2018). Cyclic lipopeptides are interesting amphiphilic molecules consisting of a fatty acid chain bound to a cyclic peptide ring (Harwood et al., 2018). B. velezensis produces three families of CLPs: surfactins, fengycins and iturins (Rabbee et al., 2019). They consist of a cyclized peptide of 7 L- or D-amino acids (iturins, surfactins) or 10 residues (fengycins) linked to an acyl chain of variable length (Caulier et al., 2019). These CLPs exhibit a strong antifungal activity but can also trigger systemic resistance in the host plant against a broad spectrum of pathogens (Romero et al., 2007; Ongena and Jacques, 2008; Pršić and Ongena, 2020)
With this work, our objective was to evaluate the understudied potential of B. velezensis for the biocontrol of peanut disease caused by A. rolfsii. To that end, we used the genetically amenable, natural rhizosphere isolate GA1 as representative of the species. Indeed, based on genome mining, this strain produces the whole panoply of bioactive secondary metabolites typically formed by members of this species (Andrić et al., 2021). Therefore, we also wanted to identify the CLPs mainly responsible for the biocontrol activity of the strain by combining different approaches including i) the determination of secondary metabolites secreted upon growth in peanut root exudates, ii) the test of various mutants repressed in CLP synthesis in in vitro antifungal assays and iii) the test of pure CLP in planta for assessing the efficiency to reduce plant infection via the stimulation of systemic resistance.
Bacillus velezensis strains used in this study was B. velezensis GA1 and its mutants (Table 1). Before each experiment, GA1 WT or mutants were pre-cultured as follows. Five µL of bacterial glycerol stock preserved at -80°C was streaked on Luria- Bertani (LB) medium (10 g of Tryptone/l; 5 g of yeast extract/l; 5 g of NaCl/l; 20 g of agar when needed) and incubated overnight at 30°C. One colony was used to inoculate flasks containing five times dilute LB liquid medium and agitated (160 rpm) overnight (16h) at 30°C. Pre-cultures were centrifuged at 10000 rpm for 10 minutes and bacterial cells were washed three times with LB medium. Cell concentration was evaluated (OD600nm) and adjusted to the desired concentration for all subsequent experiments.
Bradyrhizobium arachidis (provided by the Belgian Co-Ordinated Collections of Microorganisms “BCCM”/Bacteria Collection LMG26795) was used in this study. Before each experiments, the bacteria were streaked on Yeast Extract Mannitol Agar (YEMA) (Per liter: 10 g of Mannitol; 0.5 g of K2HPO4; 0;5 g of Sodium glutamate; 50 mg of NaCl; 1 g of yeast extract; 10 ml of Solution A (Per 100 ml:1 g of MgSO4.7H2O); 1 ml of Solution B (Per 100 ml: 5.28 g of CaCl2.2H2O); 1 ml of Solution C (Per 100 ml: 666 mg of FeCl3.6H2O); 20 g of agar). One colony was used to inoculate Yeast Extract Mannitol broth (YEM) for 3 to 7 days at 30°C and the pre-culture was washed with YEM three times. Cell concentration was evaluated (OD600nm) and adjusted to the desired concentration for all subsequent experiments
Athelia rolfsii (Curzi) Tu & Kimbrough (provided by the Belgian Co-Ordinated Collections of Microorganisms “BCCM”/Agro-Food and Environmental fungi MUCL 051031) was reactivated on V8 juice agar (200 ml of V8 juice; 3 g of CaCO3; 15 g of agar; 800 ml of distillated water; pH = 7.2). After this step, fungus was cultivated on Oatmeal agar OMA (Per liter: 60 g of oatmeal; 12.5 g of agar, pH = 7.2 ± 0.2) to produce mycelia and sclerotia at 28°C. Inoculated petri dishes were stored at 4°C or collected sclerotia were retained in peptone water (10 g/l of peptone, 5 g/l of NaCl, 1-2 g of tween 80) at the same temperature for the next experiments.
Fusarium oxysporum, Aspergillus niger and Rhizoctonia solani (lab collection) were cultivated on Potato Dextrose Agar PDA (39 g/l, pH = 5.6 ± 0.2; Merck Germany) to produce mycelia. Cultivated fungi were stored at 4°C for subsequent experiments.
Seeds from healthy and highly quality pods of peanut (Arachis hypogaea L.) cultivar JL24 (provided by National Institute for Agronomic Studies and Research (INERA) - M’vuazi in Kongo Central (DRC)) were used. Peanut seeds were surface disinfected during 2 minutes in 70% ethanol prior to another 2 minutes in 5% Sodium Hypochlorite and finally three times washed in sterile distilled water. The effect of surface sterilization was checked by spotting 50 µl of the final rinse water onto LB plates. After sterilization, seeds were deposited in a petri dish containing sterile filter paper soaked with sterile water and placed in the dark at 22°C during 5 to 7 days until the emergence of an approx. 2 cm-long radicle. Sterile seedlings were then transferred in vials container covered with aluminum foil paper to avoid light filled with 15 ml Murashige and Skoog medium (MS) for hydroponic cultivation (growth chamber conditions: 16h-day and 8h-night cycle at 22°C). After 15 days of cultivation, natural peanut root exudates (NPRE) were filtrated through a membrane (0.22 µm) and stored at 4°C for further experiments.
NPRE were collected, lyophilized, and the dry material (50 mg) was extracted using 1.2 ml of methanol by vortexing 1 minute and then sonicated at room temperature for 20 min. After centrifugation, 500 µL of the supernatant was transferred into two centrifuge tubes for measurement of sugars and acids. For sugars, 20 µL of 3 mg/ml phenyl-β-D-glucopyranoside and 20 µL of 0.1 mg/ml 3-(4-hydroxyphenyl)-propionic acid were added as internal standards. The derivatization of sugars was performed by adding 120 μL methoxyamine-hydrochloride (20 mg/ml in pyridine) for 60 min at 30°C and with 120 μL N,O-bis (trimethylsilyl) trifluoro-acetamide (BSTFA) for 120 min at 45°C, 700 rpm. For amino acids and organic acids, 20 µL of 0.1 mg/ml 3-(4-hydroxyphenyl)-propionic acid was added to the sample as the internal standard and the derivatization was performed by adding 120 µL of methoxylamin-hydrochloride (20 mg/ml in pyridine) for 90 min at 37°C, 700 rpm, followed by addition of 120µl of dimethyl-tert-butylsilyl (TBDMS) and incubated for 30 min at 60°C, 700 rpm. After centrifuging, 100 µL of the supernatant was transferred to vials for detection using TSQ Duo GC-MS/MS (Thermo Fisher Scientific).
Based on NPRE composition, medium mimicking peanut root exudates (MMPRE) was recomposed as follows: Maltose 1.09 g/l; Glucose 0.88 g/l; Arabinose 0.67 g/l; Fructose 0.28 g/l; Trehalose 0.14 g/l; Myo-inositol 0.07 g/l; Lactic acid 2.73 g/l; Succinic acid 0.13 g/l; Malonic acid 0.11 g/l; Fumaric acid 0.07 g/l; Pyruvic acid 0.03 g/l; Pyroglutamic acid 0.02 g/l; Malic acid 0.02 g/l; Citric acid 0.02 g/l; Valine 1.05 g/l; Leucine 0.57 g/l; Threonine 0.57 g/l; Isoleucine 0.42 g/l; Alanine 0.32 g/l; Phenylalanine 0.32 g/l; Tyrosine 0.15 g/l; Serine 0.14 g/l; Glycine 0.11 g/l; Asparagine 0.08 g/l; Glutamic acid 0.03 g/l. Salt and micronutrients were added with these proportions: 0.25 g/l MgSO4·7H2O; 0.34 g/l; K2HPO4; 3-morpholino-1-propanesulfonic acid MOPS 10.5 g/l; 0.25 g/l KCl, 1 g/l (NH4)2SO4; 1.2 mg/l Fe2(SO4)3; 0.4 mg/l MnSO4; 1.6 mg/l CuSO4, 4 mg/l Na2 MoO4, pH adjusted to 7. In case of assay on solid MMPRE, 20 g/l of agar was used for this preparation.
Wild type strain GA1 and GA1-mutants were cultivated in liquid to evaluate bacterial growth and secondary metabolite production on NPRE and liquid MMPRE. Bacterial growth was evaluated by turbidity measurement (OD600nm) after 48 hours culture at 30°C. Relative quantification of CLP production was evaluated by UPLC-MS (Agilent 1290 Infinity II coupled with accurate mass Jet Stream ESI‐Q‐TOF 6530, Agilent) in positive mode. MS parameters were set up as follows: capillary voltage: 3.5 kV; nebulizer pressure: 35 psi; drying gas: 8 L/min; drying gas temperature: 300°C; flow rate of sheath gas: 11 L/min; sheath gas temperature: 350°C; fragmentor voltage: 175 V; skimmer voltage: 65 V; Accurate mass spectra were recorded in the range of m/z = 100-1700. Five µL of samples were injected and separated using a C18 Acquity UPLC BEH column, 2.1 × 50 mm × 1.7 μm (Waters) at a flow rate of 0.6 mL/min and a temperature of 40°C. A gradient of 0.1% formic acid (solvent A) and acetonitrile acidified with 0.1% formic acid (solvent B) was used. First, 10% B was applied during 1 min before increasing to 100% in 20 min and maintained during 3.5 min before going back to initial ratio in 0.5 min. Identification of compounds was based on retention time (in-house database) and accurate mass. Relative quantification was made according to the peak area of each metabolite.
Sterile peanut plantlets (see section 2.3) were immersed during 1 hour in 1 ml GFP tagged GA1 suspension (OD600nm = 0.05) prepared as detailed in section 2.1. After this step, inoculated plantlets were transferred to vials containing 15 ml of sterile liquid Murashige and Skoog for 15 days in a growth chamber (day 16h/night 8h at 22°C). To evaluate the population, plants roots were immersed in 10 ml of peptone water solution supplemented with tween 80 (1 g/L) in which 10-15 sterile glassy beads were added to properly detach the bacterial cells from the root surface. Serial dilutions were prepared and 100 µl of each dilution were plated on solid medium LB. Plated petri dishes were then incubated at 28°C and cell emitting green fluorescence were counted using imaging system (Fusion Fx (Vilber Lourmat), AlexaFluor 488-Blue_AppStd). Bacillus population data were expressed to CFU/g root Dry Matter and calculated from two samples having a similar weight of root dry matter.
Ten µL of GA1 WT or mutants’ strains (OD600nm = 1) prepared as described in section 2.1 were spotted at 4.5 cm of the pathogen (A. rolfsii) on gelosed MMPRE. Fusarium oxysporum, Aspergillus niger and Rhizoctonia solani antagonism were tested in the same conditions on PDA plates. The plates were sealed with parafilm and incubated at 28°C for 6 days. The inhibition of mycelial growth was evaluated by measuring the distance between bacteria and the advancing front of the fungus. This experiment was performed using 6 independent repetitions. Impact of Bacillus volatiles in antagonism was performed using bipartite petri dishes. In this case, one compartment containing 10 ml of gelosed MMPRE was inoculated with 10 µl of GA1 culture (OD600nm = 1) prepared as detailed in section 2.1 and sclerotia were deposited at 4.5 cm from bacteria in the other part of the petri dish. The petri dishes were doubled sealed with parafilm and incubated at 28°C for 6 days. Inhibition rate (IR) was calculated with the formula: IR (%) = (S1-S2/S1) x 100 where S1: represents the fungal area growth without bacteria, and S2 represents the fungal area growth in inoculated plates. This experiment was performed using 6 independent repetitions.
The first experiment was performed in a greenhouse (located at Plant Clinic International/, Kinshasa - DRC). The characteristics of the soil used in this experiment were: pH: 5.35; Organic Carbon (%): 4.60; P available (ppm): 7.38; Nitrogen (%): 0.11; Potassium (ppm): 0.52; conductivity (µS/cm): 23.8. Twenty surface sterilized seeds (see section 2.3) were coated with a Bacillus suspension (50 ml OD600nm = 1) for 10 minutes and then sown directly into plastic pots (12.5 cm diameter, 6.5 cm height) filled with 1 kg of sterilized soil (150°C, 30 hours). Before sowing one seed per pot, 10 ml of a bacterial suspension of B. arachidis (OD600nm = 1) were deposited on the seedbed of each seed of peanut. 14 days after sowing, 3 sclerotia of A. rolfsii were placed on the plant collar and 10 ml of bacterial suspension of B. arachidis were again inoculated at the stem basis. Three days after this step, B. arachidis suspension (10 ml OD600nm = 1/plant) were inoculated again by watering the stem basis and after 5 days, B. velezensis GA1 (50 ml OD600nm = 1/plant) was applied. These two microorganisms were applied once a week for three weeks. Assessment of disease incidence (DI) and disease severity (DS) (severity scale illustrated in Figure 1A) were recorded one week after the last inoculation. The experiment was repeated twice with 50 replicates per treatment.
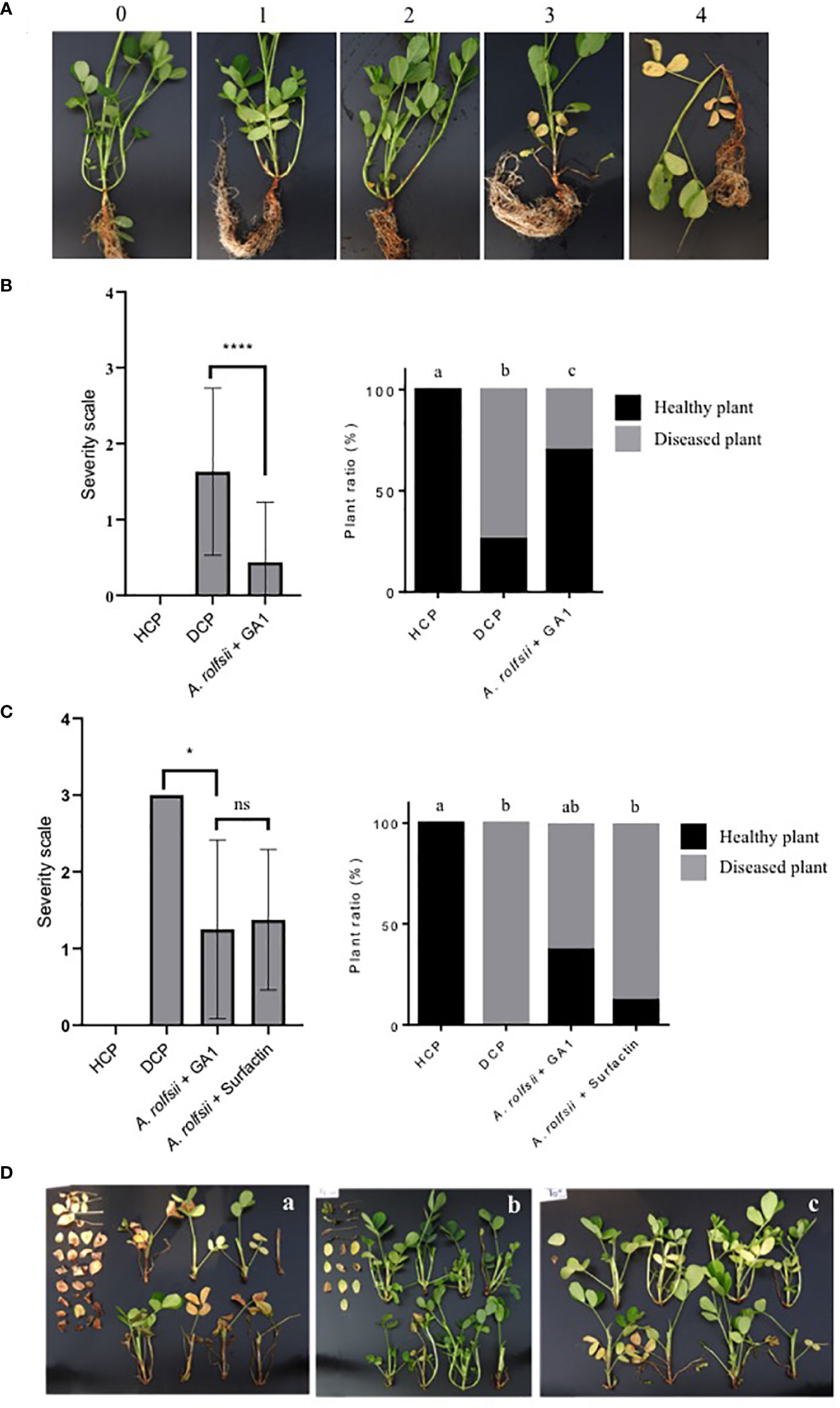
Figure 1 (A) Illustration of the severity scale used for disease rating. 0: no infection on the plant (Healthy plant); 1: mycelium present, and one branch infected; 2: mycelium present, and two branches infected; 3: mycelium present, three branches infected and beginning of leave’s yellowing; 4: three branches infected, wilting and at least 80% of leave’s yellowing. (B) Impact of B. velezensis GA1 direct antagonism against A. rolfsii on disease severity (left panel) and disease incidence (right graph), HCP = not infected plant, DCP = A. rolfsii infected plant; A. rolfsii + GA1 = Infected plants previously treated with B. velezensis GA1. Pearson’s chi-squared test followed by the pairwise tests of independence for nominal data were used to compare DI between modalities. Kruskal-Wallis’s test followed by Dunn’s multiple comparisons were used to compare DS between treatments. Means ± SD from fifty replicates of two experiments are presented. ****, p < 0.0001. Treatments with different letters are significantly different (p < 0.05). (C) Impact of B. velezensis GA1 on the induced resistance of peanuts plants against A. rolfsii. Disease severity (left panel) and disease incidence (right graph) are depicted; A. rolfsii + surfactin = Infected plants previously treated with pure surfactin (10µM/per plant). Pearson’s chi-squared test followed by the pairwise tests of independence for nominal data were used to compare DI between modalities. Kruskal-Wallis’s test followed by Dunn’s multiple comparisons were used to compare DS between treatments. Means ± SD from eight replicates of one experiment are presented. *, p<0.05 and ns (no significant difference), p > 0.05. Treatments with different letters are significantly different (p < 0.05). (D) Pictures showing the extent of disease symptoms as observed on several randomly collected plants for DCP (A), A. rolfsii infected plants treated with GA1 (B) or infected plants treated with surfactin (C). The detached leaves on the left of each image are from branches located near the collar and damaged by A. rolfsii.
The second experiment was conducted in tropical greenhouse at TERRA/Gembloux Agro-Bio Tech in Belgium. To guarantee physical separation between GA1/surfactin and A. rolfsii, a system with two concentric plastic cups was used (adapted from Figueredo et al., 2017 and Rodríguez et al., 2018). Specifically, a large pot (12 x 15 x 11.5 cm) and a small pot (6 x 8.8 x 8 cm) filled with 300 g and 50 g of sterile potting soil respectively were placed on top of each other and a hole was created in the small pot to connect the two containers. Fifty seeds surface sterilized (see section 2.3) were pre-germinated in another container containing twice autoclaved potting soil for one week. Seedlings were then transplanted in the upper pot (the small) in which the roots were introduced into its hole to reach the large pot. One hour before transplanting, a suspension of B. arachidis (10 ml OD600nm = 1/plant) was poured in each large pot. Five days after seedlings transfer, GA1 suspension (50 ml OD600nm = 1/plant) or purified surfactin (10 µM/plant as indicated by the work of Rodríguez et al., 2018, purity of surfactin: 90%) were applied in the large pot. At two days after GA1/surfactin treatment, seedlings were inoculated at the stem basis (in the small pot) by adding 15 sclerotia of A. rolfsii and 2 plugs of agar carrying mycelia and at the same time, B. arachidis (10 ml OD600nm = 1/plant) was applied once again. The application of B. arachidis, GA1 and surfactin was performed once a week for three weeks. Assessment of DI and DS (severity scale illustrated in Figure 1A) were recorded one week after the last inoculation. At the same time, roots nodules were counted on all plants for to evaluate the impact of GA1 inoculation on root nodulation by B. arachidis. The results were expressed by the number of nodules/plant. The experiment was repeated twice with 8 replicates.
One-way ANOVA followed by Tukey’s multiple comparisons test and graphical representatives were performed using GraphPad Prism version 8.1.0 (244) (GraphPad Software, San Diego, California USA, www.graphpad.com). One-way ANOVA (Kruskal-Wallis’s test) followed by Dunn’s multiple comparisons in GraphPad software were used specially to compare DS between treatments. The proportions of diseased plants and healthy plants (DI) after treatment by B. velezensis GA1 were assessed by Pearson’s chi-squared test followed by the pairwise tests of independence for nominal data. These tests were applied with the “pairwiseNominalIndependence” and the “cldList” functions of the R companion package (Mangiafico, 2016) to determine the symmetry between treatments. The significance threshold for all tests was set at 0.05%.
We first wanted to evaluate BSMs production by GA1 growing in root exudates collected from two-week-old peanut plants grown in hydroponics (natural peanut root exudates, NPRE). The amounts of plant chemicals utilizable by bacteria to form biomass are usually quite low in these naturally produced exudates but we observed a substantial development of GA1 with final cell density that reached an OD600nm value of approx. 0.4. We also observed a good colonization potential of the strain on peanut roots as it reaches cell populations of 2.5 x 107 CFU/g root (dry weight) within 4 days post inoculation. We next determined the pattern of BSMs formed by the bacterium in these conditions and UPLC-qTOFMS analysis of the corresponding cell-free crude extract revealed significant amounts of the lipopeptides iturin and surfactin but not fengycin or other non-ribosomal products potentially produced by the strain (Figure 2A). In terms of relative proportions determined by peak area, surfactin production rate is higher than iturin. For both CLPs, different homologues varying in the length of the fatty acid chain are co-produced (Figure 2A). The mass spectral data revealed the presence of iturins with fatty acid chains containing 14 and 15 carbons (C14, C15) and surfactins C13 and C14.
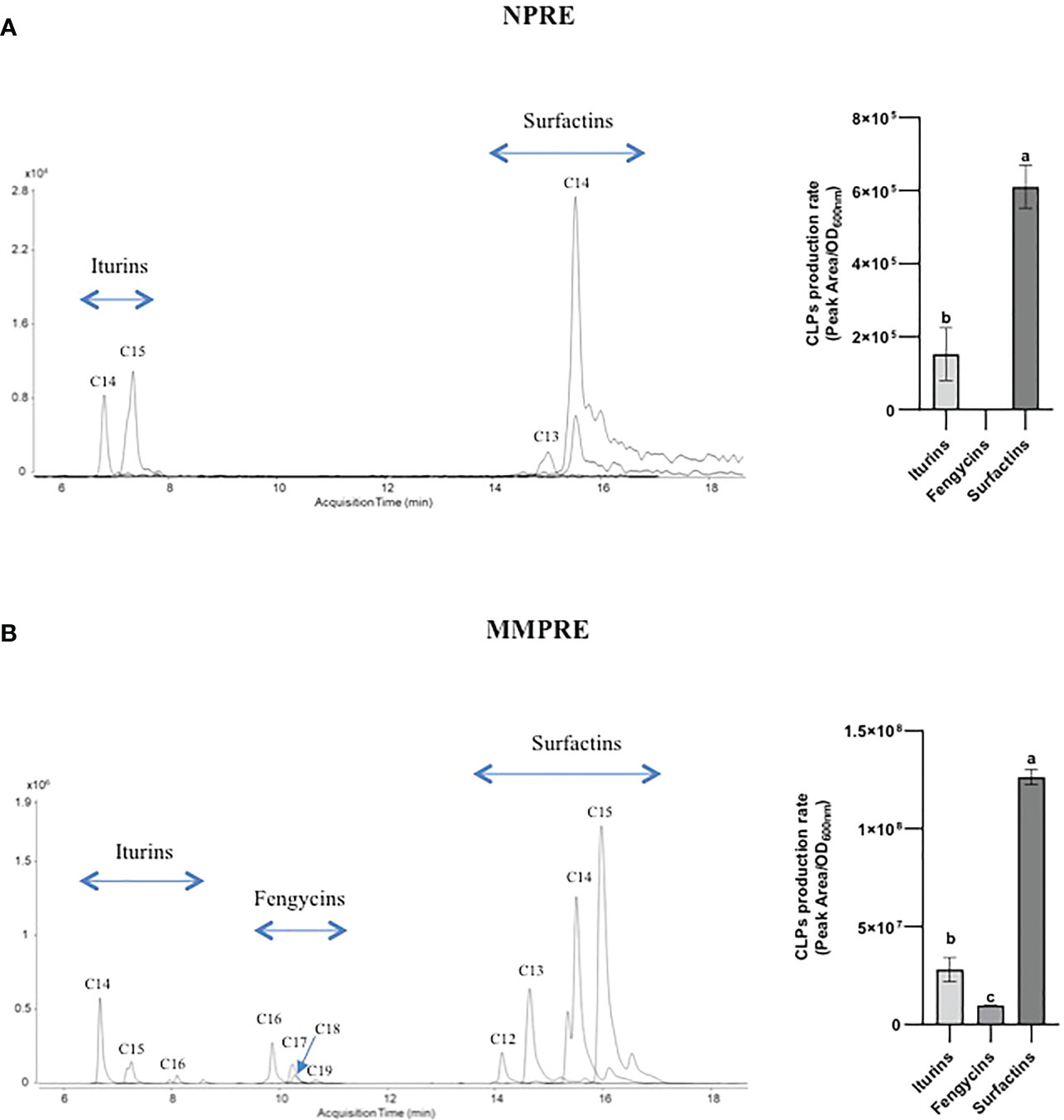
Figure 2 LC-MS extracted ion chromatograms (EICs) of CLPs families produced by B velezensis GA1 in liquid NPRE (A) and MMPRE (B) culture. The different peaks correspond to homologues of the considered family with different fatty acid chain lengths as indicated. For each medium, the relative proportions of the various CLPs have been calculated based on peak area/OD and are also represented in graphs on the right. Means ± SD from two biological replicates (with similar OD) of one experiment are shown. Treatments with different letters are significantly different (p < 0.05).
Using TSQ Duo GC-MS/MS, we identified 6 sugars, 8 organic acids and 11 amino acids as components in the NPRE that can be used as carbon sources by GA1. Maltose, glucose, lactic acid, valine and leucine were identified as the most prevalent compounds (Table 2). We created a synthetic medium based on the relative amounts of all those compounds found in NPRE and which contained 31.25% sugars, 31.25% organic acids and 37.5% amino acids in a total of 10 g/l. Higher biomass concentration was measured (OD600nm = 0.73) upon growth of GA1 in this mimicking NPRE (MMPRE) together with increased amounts of CLPs compared with the production in NPRE. The relative proportions of iturins and surfactins as well as the homologue distribution within each family were similar to NPRE but detectable amounts of fengycins were observed in these conditions (Figure 2B). Since CLPs are quorum-sensing regulated metabolites, a higher biomass most probably led to a higher production of the three CLPs (with the same proportion), therefore allowing to detect fengycin produced above the detection limit in that case (Figure 2B).
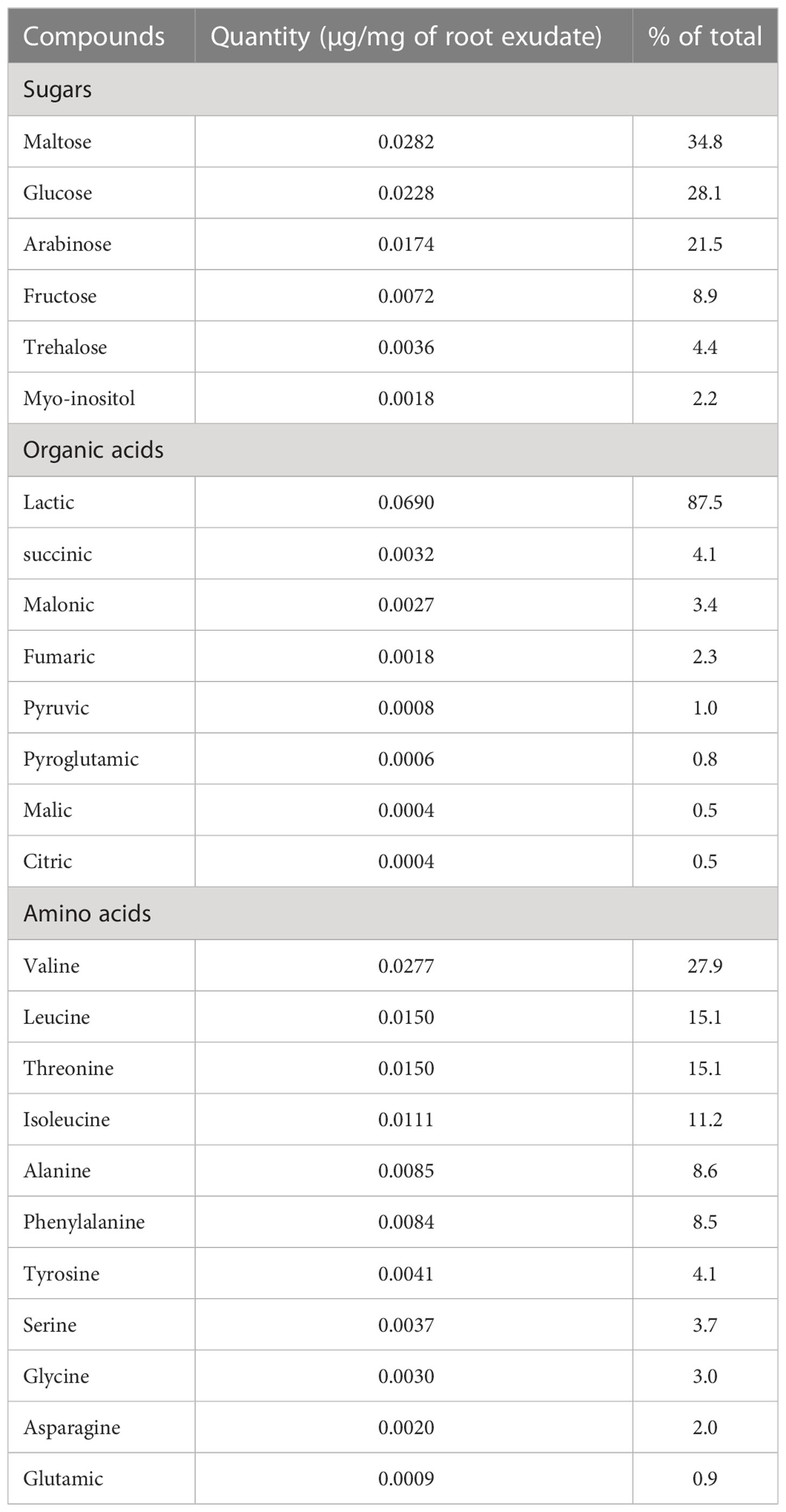
Table 2 Quantity of soluble sugars, organic acids and amino acids detected by GC-MS in root exudates collected from 15 days-old plants of A. hypogea cultivar JL24 grown in hydroponics.
We next used solid MMPRE to evaluate the direct inhibition of A. rolfsii by GA1 in confrontation assays on plates. The wild-type strain displayed a strong inhibitory activity towards the fungus (Figure 3A). To determine the specific involvement of CLPs and other metabolites in bacterial inhibition, we tested a range of single and double knockout mutants of GA1. This included the Δsfp derivative not able to form the 4’-phosphopantetheinyl transferase enzyme, which is essential for proper functioning of non-ribosomal peptide biosynthesis machineries responsible for the synthesis of PKs, CLPs, and bacillibactin. Data revealed that iturin plays a major role in this antifungal activity since the inhibitory effect is significantly reduced in all the mutants repressed in the synthesis of this lipopeptide (Figure 3A). However, another unidentified non-ribosomal compound also contributes to the fungitoxic potential of GA1 as shown by the additional loss of activity of the Δsfp mutant impaired in the synthesis of all NRP- and PK-type BSMs except bacilysin. This dipeptide is non-ribosomally formed but not via a mechanism involving the Sfp protein and is therefore still produced in the Δsfp derivative. Bacilysin was occasionally reported for its antifungal activity but is not active against A. rolfsii as illustrated by the fully conserved antagonistic potential of the ΔbacA mutant and the similar level of activity displayed by the Δsfp and the double ΔsfpΔbacA mutants (Figure 3A). Also, GA1 inhibition is not due to volatiles as shown by the absence of fungal growth reduction in two-compartment plates (Figure 3B). Iturin produced by GA1 is also partially or fully responsible for antagonism towards respectively Fusarium oxysporum and Aspergillus niger as other important peanut pathogens but not Rhizoctonia solani (Figure 3C).
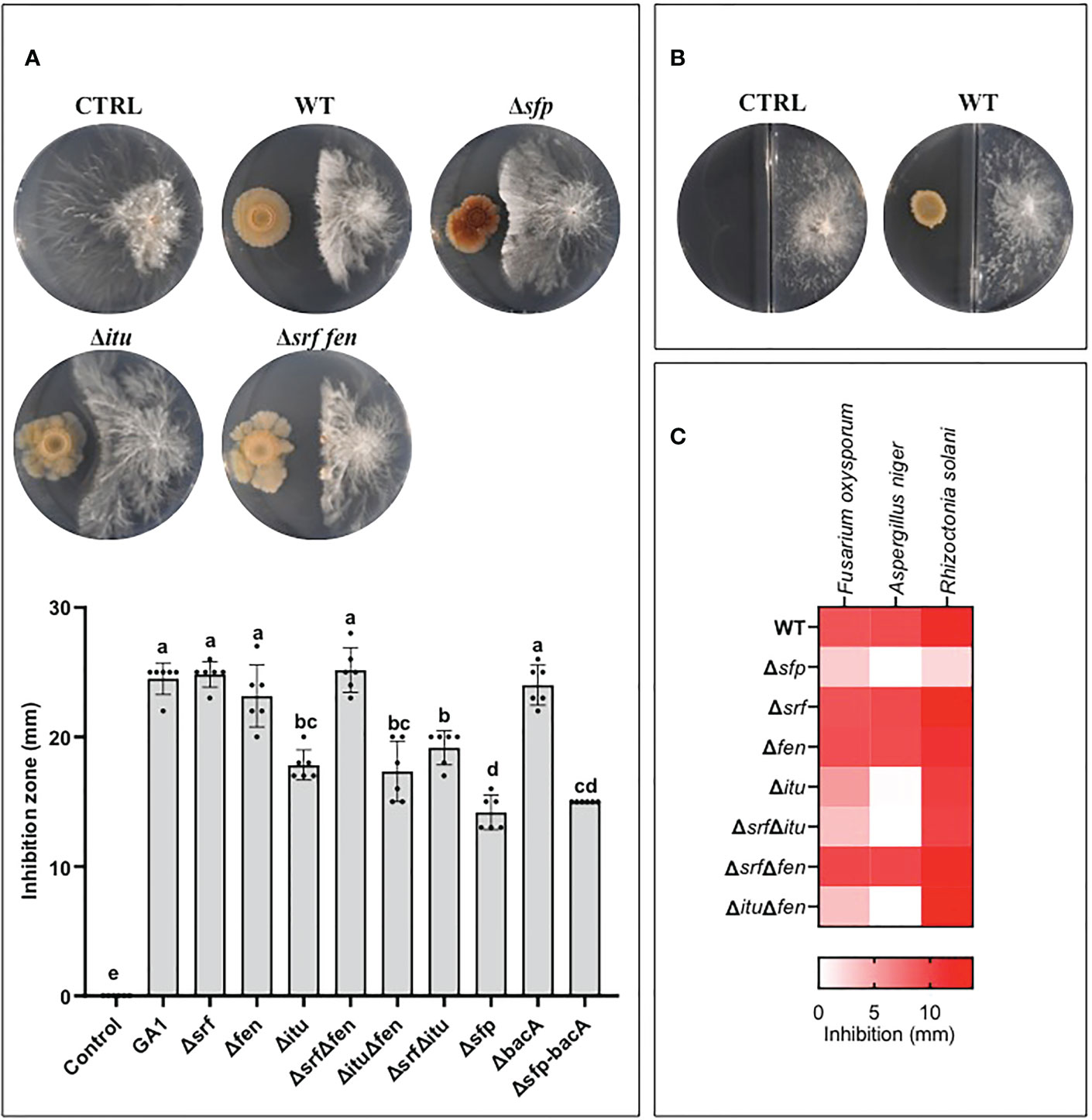
Figure 3 (A) Illustration of the antagonistic activity displayed by B. velezensis GA1 WT and its mutant strains impaired in production of different lipopeptides as observed on MMPRE medium against A. rolfsii. The corresponding quantitative data of inhibition by the different strains are represented in the graph below. Means ± SD from six independent repetitions of one experiment are presented. Treatments with different letters are significantly different (p<0.05). (B) Volatiles secreted by B. velezensis have no impact on the growth of A. rolfsii as visualized on bi-compartmented plates. (C) Heat map representing the level of antagonistic activity of GA1 WT and mutants against some other fungal pathogens of peanuts plants.
The potential of GA1 to reduce the incidence of disease caused by A. rolfsii on peanut plants was next tested in two different types of experiments but in both cases, the pathogen was inoculated as sclerotia on the collar of the plant. In the first type of assays, GA1 was applied both by immersing seeds in a cell suspension just before sowing and by drenching with GA1 cell suspension at the stem basis of two weeks-old plants. Disease rating was done based on macroscopic observation of symptoms as illustrated in Figure 1A. The biocontrol potential of GA1 was evaluated approx. three weeks after infection based on reduction in disease incidence and disease severity compared with control plants infected by A. rolfsii but not treated with the bacterium (Figure 1B). Application of GA1 provided a significant 60% reduction of the disease based on incidence (X2 = 61.166, df = 2, p<0.0001) and severity (p<0.0001) (Figure 1B). As there is no spatial separation between the two microbes in this experimental set-up, disease control is obviously due to direct antagonism of the pathogen. In a second type of assay, we wanted to evaluate the ability of GA1 to protect the host plant via the induction of systemic resistance. We used a system with two concentric pots to guarantee spatial separation between GA1 and the fungal pathogen. Based on disease severity, a significant protective effect (p<0.0001) was observed by comparing A rolfsii-infected plants (disease control) and infected plants previously treated with GA1 (Figures 1C, D). Interestingly, treatment with the purified lipopeptide surfactin (soil drenching with a 10μM solution/per plant) also provided a similar level of disease severity reduction. Considering disease incidence, we observed a reduction in the number of infected plants of 12.5% and 37.5% upon treatment with pure surfactin and with GA1 cells respectively (X2 = 20.267, df = 3, p-value = 0.0001495) (Figures 1C, D).
Peanut belongs to the Fabaceae family which naturally establishes symbiosis with rhizobia helping the plant to acquire nitrogen from atmospheric N2 (Peix et al., 2015). We therefore wanted to test the compatibility of B. velezensis with the development of these important symbionts. Upon in vitro confrontation on Yeast Extract Mannitol Agar, which allows consistent growth of the symbiont, GA1 displays some slight inhibitory activity against B. arachidis selected as model symbiotic rhizobium (Figure 4A). We used the same plants as described above (see section 2.8) to assess whether GA1 may impact the ability of B. arachidis to form nodules on roots of infected plants. Data indicated that the nodulation rate on roots is significantly reduced in presence of GA1 (Figure 4B) but not abolished indicating that B. velezensis does not fully exclude Bradyrhizobium from the rhizosphere niche.
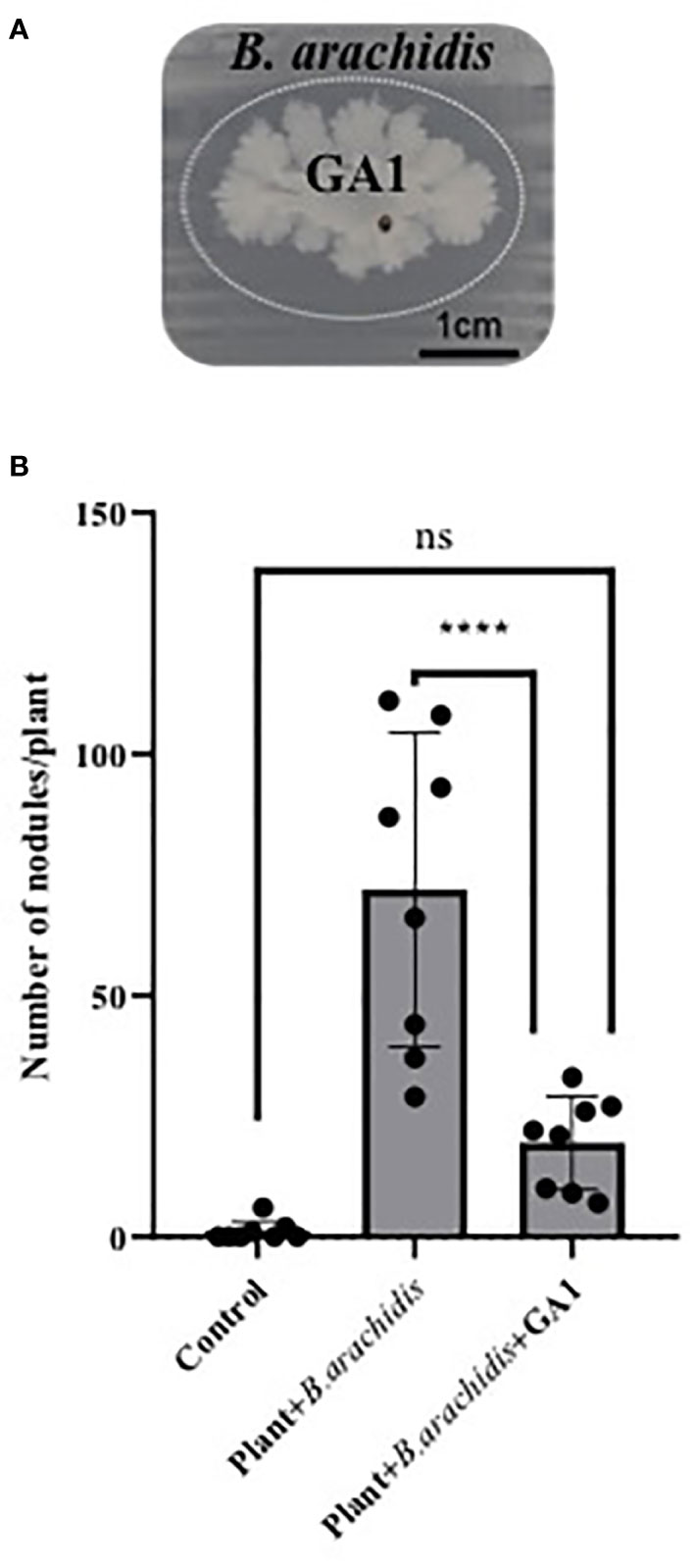
Figure 4 (A) Antagonistic effect of Bacillus velezensis GA1 (growing colony at the center) against Bradyrhizobium arachidis (growing as thin layer of cells over the whole surface) on Yeast Extract Mannitol Agar. It is visualized by the inhibition zone delimited by a dotted elliptic line. (B) Impact of Bacillus inoculation on B. arachidis nodulation on roots evaluated based on the number of nodules in each treatment; Plant: non-inoculated plants; Plant + B. arachidis: A. rolfsii infected peanut plants inoculated with B. arachidis Plant + B. arachidis + GA1: A. rolfsii infected peanut plants inoculated with B. arachidis and B. velezensis GA1.Values are the mean ± SD from eight replicates of one experiment. ****,p < 0.0001 and ns (no significant difference), p > 0.05.
Microbial physiology and production of secondary metabolites is often studied in laboratory conditions upon growth in artificial rich media which do not reflect the nutritional conditions encountered in natural settings. Plant-associated and beneficial rhizobacteria such as B. velezensis primarily use root exuded compounds as carbon and nitrogen sources to multiply and efficiently establish on host tissues and this may strongly impact their behaviour and the synthesis of bioactive small-size metabolites (Fan et al., 2012; Nihorimbere et al., 2012; Qiu et al., 2014; Ogran et al., 2019). It is why we performed an in-depth analysis of the chemicals released by roots of A. hypogaea plants, allowing to provide a first detailed profiling of exuded products for this species. We show that B. velezensis efficiently form cyclic lipopeptides of the iturin and surfactin families by growing on these exudates as sole nutrient source. In addition, the bacterium readily uses the naturally exuded compounds as source of nutrients and display high root colonization potential of peanut roots. Collectively, these data strongly suggest that a consistent production of CLPs also occur in planta.
Iturins are well known for their toxicity against a broad range of phytopathogenic fungi and oomycetes (Gong et al., 2014; Luna-Bulbarela et al., 2018; Jin et al., 2020; Wang et al., 2020). Here, we show that this type of CLP is the main antifungal compound secreted by GA1 responsible for direct inhibition of the peanut pathogen A. rolfsii. This indicates that iturin production also plays a key role for the protection observed in planta when GA1 and the fungal pathogen develop in contact or in close vicinity. Nevertheless, our data show that another non-ribosomal compound also contributes to the antagonistic activity of the GA1 strain. This rules out a possible role for bacteriocins and terpenes which can also be formed by GA1 based on the presence of corresponding biosynthetic gene clusters (BGCs) in the genome (Arguelles-Arias et al., 2009; Caulier et al., 2019). The polyketides bacillaene, difficidin and macrolactin are other non-ribosomal sfp-dependent secondary metabolites known for their antimicrobial activity. However, possible involvement of those compounds in the GA1 antagonistic potential is not obvious since i) they were not detected in cell-free extracts upon growth on MMPRE and because these BSMs are almost exclusively described as anti-bacterial agents not active on fungi (Müller et al., 2014; Wu et al., 2015; Im et al., 2020; Anckaert et al., 2021). Other candidate molecules are linear lipopeptides displaying antifungal properties as recently reported in B. subtilis (Chakraborty et al., 2020) but no BGC encoding such kind of molecule has been identified in the genome of the strain and, except those corresponding to iturins, surfactins and fengycins, no other molecular feature displaying fragmentation pattern typical of (lipo)peptidic compounds were detected in the GA1 cell-free extracts upon UPLC-qTOF-MS analysis (not shown). Identification of this possibly new metabolite certainly deserves further investigation as it may be important in the broad context of natural product discovery but also more specifically as additional weapon driving the biocontrol performance of the B. velezensis species.
Surfactin is also readily produced by B. velezensis developing in the specific nutritional context established by peanut root exudation. In general, this lipopeptide does not display remarkable antimicrobial activity when tested at the biologically relevant low micromolar concentrations (Andric et al, 2020; Anckaert et al., 2021). According to our data, surfactin is also not involved in the antibiosis activity of GA1 toward A. rolfsii. However, as recently reported (Rodríguez et al., 2018), we show that this CLP retains a huge potential to stimulate systemic resistance in peanut when tested as root treatment on plants infected by the pathogen on aerial parts. Such a plant resistance-triggering activity of surfactin has been previously reported in various plants and is associated with the priming of specific defense pathways that remain to be deciphered in the case of peanut (Ongena et al., 2007; García-Gutiérrez et al., 2013; Chowdhury et al., 2015; Debois et al., 2015; Pršić and Ongena, 2020). Our data strongly suggest that disease reduction observed upon treatment with GA1 in a set-up where it remains spatially separated from A. rolfsii is mediated via a surfactin-triggered boost of peanut plant immunity.
Our results show the high potential of B. velezensis strain GA1 to protect peanut from disease caused by A. rolfsii, an economically important pathogen causing stem rot. Other Bacillus isolates belonging to various species of the subtilis subgroup have also been reported as strong antagonists of this phytopathogen in vitro (Ahmad et al., 2019; Chen et al., 2020; Xu et al., 2020; Kumari et al., 2021). However, studies demonstrating biocontrol of this disease under greenhouse or field conditions are rare (Tonelli et al., 2011; Figueredo et al., 2017) and the mechanisms underlying this protective effect are poorly documented even if some involvement of lipopeptides and volatiles were evoked based on the potential of the tested strains to form these compounds in vitro (Munjal et al., 2016; Agisha et al., 2019; Chen et al., 2019). With this work, we thus provide new insights into the role played by CLPs in direct antibiosis and induction of systemic resistance in the Arachis hypogaea/A. rolfsii pathosystem.
Importantly, a successful biocontrol agent must meet a number of criteria, including being ecologically compatible with other plant-beneficial bacteria such as rhizobia in leguminous plants (Mohanty et al., 2021). Upon in vitro confrontation on solid medium, we observed some inhibition of Bradyrhizobium by GA1 but to a quite limited extent compared with what is commonly observed when B. velezensis develops its antagonistic potential against phytopathogenic fungi or bacteria. Still this reflects that some exoproducts formed by B. velezensis are toxic for Bradyrhizobium. The antagonistic effect of GA1 was more clearly observed. In planta, this negative effect was more marked and root-treatment with GA1 led to a significant reduction in the density of rhizobial nodules. It may be due to niche exclusion since B. velezensis, as good colonizer, may outcompete the rhizobium for root invasion but it may also rely on an antagonistic interaction involving secondary metabolites. In vitro confrontation tests were performed on a medium very different from the MMPRE medium mimicking root exudates and we assume that the natural nutritional conditions may be favorable for a higher production of compounds responsible for rhizobium inhibition in the peanut rhizosphere compared to in vitro on plates. That said, inoculation of B. velezensis did not fully impaired Bradyrhizobium development in planta indicating that the two beneficial bacteria may co-exist in accordance with previous studies reporting such compatibility between soil bacilli and various rhizobial isolates on peanut (Abd-Allah and El-Didamony, 2007; Figueredo et al., 2014) and soybean plants (Glycine max L.) also belonging to the Fabaceae family (Sibponkrung et al., 2020).
A better understanding of the molecular processes driving such multitrophic interactions between the host plant and mutualistic, symbiotic and pathogenic microbes is necessary, but our work provide new insights into the mechanisms used by B. velezensis to protect peanut against an important fungal disease. This further support the potential of the species as powerful biocontrol agent keeping in mind that rational management of its mode of application is required to avoid negative impact on natural symbiosis, which is crucial for the ecology and health of such kind of crop.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
VKA performed most of the microbiology and greenhouse experiments with the technical support of CH and advisory help of PD. AAA performed metabolomics using UPLC-MS and data analysis. BR and BC performed the analysis for root exudate composition. VKA, AAA, and MO mainly wrote the manuscript. MO supervised the study. All authors contributed to the article and approved the submitted version.
This work was supported by the EOS project ID 30650620 from the FWO/F.R.S.-FNRS, by the Académie de Recherche et d’Enseignement Supérieur-Commission de Coopération au Développement (ARES-CDD), bourse exceptionnelle program, and by the Schlumberger Foundation Faculty for the Future program. MO is Research Director at the F.R.S.-FNRS.
We acknowledge the Professor Lyna Mukwa Fama Tongo and all the team of Plant Clinic International/Kinshasa which helped us to conduct the experiments in the Democratic Republic of Congo.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abd-Allah, E. F., El-Didamony, G. (2007). Effect of seed treatment of Arachis hypogaea with Bacillus subtilis on nodulation in biocontrol of southern blight (Sclerotium rolfsii) disease. Phytoparasitica 35, 8–12. doi: 10.1007/BF02981055
Agisha, V. N., Kumar, A., Eapen, S. J., Sheoran, N., Suseelabhai, R. (2019). Broad-spectrum antimicrobial activity of volatile organic compounds from endophytic Pseudomonas putida bp25 against diverse plant pathogens. Biocontrol Sci. Technol. 29, 1069–1089. doi: 10.1080/09583157.2019.1657067
Ahmad, A.-G. M., Attia, A.-Z. G., Mohamed, M. S., Elsayed, H. E. (2019). Fermentation , formulation and evaluation of PGPR Bacillus subtilis isolate as a bioagent for reducing occurrence of peanut soil-borne diseases. J. Integr. Agric. 18, 2080–2092. doi: 10.1016/S2095-3119(19)62578-5
Anckaert, A., Arguelles-Arias, A., Hoff, G., Calonne-Salmon, M., Declerck, S., Ongena, M. (2021). “The use of Bacillus spp. as bacterial biocontrol agents to control plant diseases,” in Microbial bioprotectants for plant disease management. Eds. Köhl, J., Ravensberg, W. (Cambridge: Burleigh Dodds Science Publishing), 1–54. doi: 10.19103/as.2021.0093.10
Andrić, S., Meyer, T., Rigolet, A., Prigent-Combaret, C., Höfte, M., Balleux, G., et al. (2021). Lipopeptide interplay mediates molecular interspecies interactions between soil bacilli and pseudomonads. Microbiol. Spectr. 9, 3. doi: 10.1128/spectrum.02038-21
Andric, S., Meyer, T., Ongena, M. (2020) Bacillus responses to plant-associated fungal and bacterial communities.” Front. Microbiol. Front. Media S.A. 11, 1350. doi: 10.3389/fmicb.2020.01350
Arguelles-Arias, A., Ongena, M., Halimi, B., Lara, Y., Brans, A., Joris, B., et al. (2009). Bacillus amyloliquefaciens GA1 as a source of potent antibiotics and other secondary metabolites for biocontrol of plant pathogens. Microbial Cell Factories 8, 63. doi: 10.1186/1475-2859-8-63
Arya, S. S., Akshata, R. S., Chauhan, S. (2016). Peanuts as functional food: A review. J. Food Sci. Technol. 53, 31–41. doi: 10.1007/s13197-015-2007-9
Backer, R., Rokem, J. S., Ilangumaran, G., Lamont, J., Praslickova, D., Ricci, E., et al. (2018). Plant growth-promoting Rhizobacteria : context , mechanisms of action , and roadmap to commercialization of biostimulants for sustainable agriculture. Front. Plant Sci. 9, 1473. doi: 10.3389/fpls.2018.01473
Bosamia, T. C., Dodi, S. M., Mishr, G. P., Ahmad, S., Joshi, B., Thirumalaisam, P. P., et al. (2020). Unraveling the mechanisms of resistance to Sclerotium rolfsii in peanut (Arachis hypogaea l.) using comparative RNA-seq analysis of resistant and susceptible genotypes. PloS One 15, 8. doi: 10.1371/journal.pone.0236823
Caulier, S., Nannan, C., Gillis, A., Licciardi, F., Bragard, C., Mahillon, J. (2019). Overview of the antimicrobial compounds produced by members of the Bacillus subtilis group. Front. Microbiol. 10, 302. doi: 10.3389/fmicb.2019.00302
Cawoy, H., Bettiol, W., Fickers, P., Ongena, M. (2011). “Bacillus-based biological control of plant diseases,” in Pesticides in the modern world–pesticides use and management, ed. Stoytcheva, M. (London: InTechOpen), doi: 10.5772/intechopen.17184
Chakraborty, M., Mahmud, N. U., Gupta, D. R., Tareq, F. S., Shin, H. J., Islam, T. (2020). Inhibitory effects of linear lipopeptides from a marine Bacillus subtilis on the wheat blast fungus Magnaporthe oryzae triticum. Front. Microbiol. 11, 665. doi: 10.3389/fmicb.2020.00665
Chen, L., Shi, H., Heng, J., Wang, D., Bian, K. (2019). Antimicrobial, plant growth-promoting and genomic properties of the peanut endophyte Bacillus velezensis LDO2. Microbiological Res. 218 (October 2018), 41–48. doi: 10.1016/j.micres.2018.10.002
Chen, L., Wu, Y. D., Chong, X. Y., Xin, Q. H., Wang, D. X., Bian, K. (2020). Seed-borne endophytic Bacillus velezensis LHSB1 mediate the biocontrol of peanut stem rot caused by Sclerotium rolfsii. J. Appl. Microbiol. 128, 803–813. doi: 10.1111/jam.14508
Chowdhury, S. P., Uhl, J., Grosch, R., Alquéres, S., Pittroff, S., Dietel, K., et al. (2015). Cyclic lipopeptides of Bacillus amyloliquefaciens subsp. plantarum colonizing the lettuce rhizosphere enhance plant defense responses toward the bottom rot pathogen Rhizoctonia solani. Mol. Plant Microbe Interact. 28, 984–995. doi: 10.1094/MPMI-03-15-0066-R
Debois, D., Fernandez, O., Franzil, L., Jourdan, E., De Brogniez, A., Willems, L., et al. (2015). Plant polysaccharides initiate underground crosstalk with bacilli by inducing synthesis of the immunogenic lipopeptide surfactin. Environ. Microbiol. Rep. 7, 570–582. doi: 10.1111/1758-2229.12286
Fan, B., Blom, J., Klenk, H. P., Borriss, R. (2017). Bacillus amyloliquefaciens, Bacillus velezensis, and Bacillus siamensis form an “operational group B. amyloliquefaciens” within the b. subtilis species complex. Front. Microbiol. 8, 22. doi: 10.3389/fmicb.2017.00022
Fan, B., Carvalhais, L. C., Becker, A., Fedoseyenko, D., Von Wirén, N., Borriss, R. (2012). Transcriptomic profiling of Bacillus amyloliquefaciens FZB42 in response to maize root exudates. BMC Microbiol. 12, 116. doi: 10.1186/1471-2180-12-116
Figueredo, M. S., Tonelli, M. L., Ibáñez, F., Morla, F., Cerioni, G., Tordable, M. C., et al. (2017). Induced systemic resistance and symbiotic performance of peanut plants challenged with fungal pathogens and co-inoculated with the biocontrol agent Bacillus sp. CHEP5 and Bradyrhizobium sp. SEMIA6144. Microbiological Res. 197, 65–73. doi: 10.1016/j.micres.2017.01.002
Figueredo, M. S., Tonelli, M. L., Taurian, T., Angelini, J., Ibañez, F., Valetti, L., et al. (2014). Interrelationships between Bacillus sp. CHEP5 and Bradyrhizobium sp. SEMIA6144 in the induced systemic resistance against Sclerotium rolfsii and symbiosis on peanut plants. J. Biosci. 39, 877–885. doi: 10.1007/s12038-014-9470-8
Fira, D., Dimkić, I., Berić, T., Lozo, J., Stanković, S. (2018). Biological control of plant pathogens by Bacillus species. J. Biotechnol. 285, 44–55. doi: 10.1016/j.jbiotec.2018.07.044
García-Gutiérrez, L., Zeriouh, H., Romero, D., Cubero, J., de Vicente, A., Pérez-García, A. (2013). The antagonistic strain Bacillus subtilis UMAF6639 also confers protection to melon plants against cucurbit powdery mildew by activation of jasmonate- and salicylic acid-dependent defence responses. Microb. Biotechnol. 6, 264–274. doi: 10.1111/1751-7915.12028
Gong, Q., Zhang, C., Lu, F., Zhao, H., Bie, X., Lu, Z. (2014). Identification of bacillomycin d from Bacillus subtilis fmbJ and its inhibition effects against Aspergillus flavus. Food Control 36, 8–14. doi: 10.1016/j.foodcont.2013.07.034
Grubbs, K. J., Bleich, R. M., Santa Maria, K. C., Allen, S. E., Farag, S., Team, A., et al. (2017). Large-Scale bioinformatics analysis of bacillus genomes uncovers conserved roles of natural products in bacterial physiology. mSystems 2, 1–18. doi: 10.1128/msystems.00040-17
Harwood, C. R., Mouillon, J.-M., Pohl, S., Arnau, J. (2018). Secondary metabolite production and the safety of industrially important members of the Bacillus subtilis group. FEMS Microbiol. Rev. 42, 721–738. doi: 10.1093/femsre/fuy028
Im, S. M., Yu, N. H., Joen, H. W., Kim, S. O., Park, H. W., Park, A. R., et al. (2020). Biological control of tomato bacterial wilt by oxydifficidin and difficidin-producing Bacillus methylotrophicus DR-08. Pestic. Biochem. Physiol. 163, 130–137. doi: 10.1016/j.pestbp.2019.11.007
Iquebal, M. A., Tomar, R. S., Parakhia, M. V., Singla, D., Jaiswal, S., Rathod, V. M., et al. (2017). Draft whole genome sequence of groundnut stem rot fungus Athelia rolfsii revealing genetic architect of its pathogenicity and virulence. Sci. Rep. 7, 5299. doi: 10.1038/s41598-017-05478-8
Jin, P., Wang, H., Tan, Z., Xuan, Z., Dahar, G. Y., Li, Q. X., et al. (2020). Antifungal mechanism of bacillomycin d from Bacillus velezensis HN-2 against Colletotrichum gloeosporioides penz. Pestic. Biochem. Physiol. 163, 102–107. doi: 10.1016/j.pestbp.2019.11.004
Kashyap, A. S., Pandey, V. K., Manzar, N., Kannojia, P., Singh, U. B., Sharma, P. K. (2017). “Role of plant growth-promoting rhizobacteria for improving crop productivity in sustainable agriculture,” in Plant-microbe interactions in agro-ecological perspectives, vol. 2 . Eds. Singh, D. P., Singh, H. B., Prabha, R. (Berlin: Springer), 673–693. doi: 10.1007/978-981-10-6593-4
Kumari, P., Bishnoi, S. K., Chandra, S. (2021). Assessment of antibiosis potential of Bacillus sp. against the soil-borne fungal pathogen Sclerotium rolfsii sacc. (Athelia rolfsii (Curzi) tu & kimbrough). Egyptian J. Biol. Pest Control 31, 17. doi: 10.1186/s41938-020-00350-w
Lam, V. B., Meyer, T., Arias, A. A., Ongena, M., Oni, F. E., Höfte, M. (2021). Bacillus cyclic lipopeptides iturin and fengycin control rice blast caused by Pyricularia oryzae in potting and acid sulfate soils by direct antagonism and induced systemic resistance. Microorganisms 9, 1441. doi: 10.3390/microorganisms9071441
Luna-Bulbarela, A., Tinoco-Valencia, R., Corzo, G., Kazuma, K., Konno, K., Galindo, E., et al. (2018). Effects of bacillomycin d homologues produced by Bacillus amyloliquefaciens 83 on growth and viability of Colletotrichum gloeosporioides at different physiological stages. Biol. Control 127, 145–154. doi: 10.1016/j.biocontrol.2018.08.004
Mangiafico, S. S. (2016). Summary and analysis of extension program evaluation in r (New Brunswick, NJ, USA: Rutgers Co).
Miljaković, D., Marinković, J., Balešević-Tubić, S. (2020). The significance of Bacillus spp. in disease suppression and growth promotion of field and vegetable crops. Microorganisms 8, 1037. doi: 10.3390/microorganisms8071037
Mohanty, P., Singh, P. K., Chakraborty, D., Mishra, S., Pattnaik, R. (2021). Insight into the role of PGPR in sustainable agriculture and environment. Front. Sustain. Food Syst. 5, 667150. doi: 10.3389/fsufs.2021.667150
Müller, S., Strack, S. N., Hoefler, B. C., Straight, P. D., Kearns, D. B., Kirby, J. R. (2014). Bacillaene and sporulation protect Bacillus subtilis from predation by Myxococcus xanthus. Appl. Environ. Microbiol. 80, 5603–5610. doi: 10.1128/AEM.01621-14
Munjal, V., Nadakkakath, A. V., Sheoran, N., Kundu, A., Venugopal, V., Subaharan, K., et al. (2016). Genotyping and identification of broad spectrum antimicrobial volatiles in black pepper root endophytic biocontrol agent, Bacillus megaterium BP17. Biol. Control 92, 66–76. doi: 10.1016/j.biocontrol.2015.09.005
Nihorimbere, V., Cawoy, H., Seyer, A., Brunelle, A., Thonart, P., Ongena, M. (2012). Impact of rhizosphere factors on cyclic lipopeptide signature from the plant beneficial strain Bacillus amyloliquefaciens S499. FEMS Microbiol. Ecol. 79, 176–191. doi: 10.1111/j.1574-6941.2011.01208.x
Nyabyenda, P. (2005). Les Plantes cultivées en régions tropicales d ‘altitude d ‘afrique: Généralités, legumineuses alimentaires, Plantes à tubercules et racines, Céréales. Gembloux: Presses Agronomiques de Gembloux.
Ogran, A., Yardeni, E. H., Keren-Paz, A., Bucher, T., Jain, R., Gilhar, O., et al. (2019). The plant host induces antibiotic production to select the most-beneficial colonizers. Environ. Microbiol. 85, :e00519–e519. doi: 10.1128/AEM.00512-19
Ongena, M., Jacques, P. (2008). Bacillus lipopeptides: Versatile weapons for plant disease biocontrol. Trends Microbiol. 16, 115–125. doi: 10.1016/j.tim.2007.12.009
Ongena, M., Jourdan, E., Adam, A., Paquot, M., Brans, A., Joris, B., et al. (2007). Surfactin and fengycin lipopeptides of Bacillus subtilis as elicitors of induced systemic resistance in plants. Environ. Microbiol. 9, 1084–1090. doi: 10.1111/j.1462-2920.2006.01202.x
Peix, A., Ramírez-Bahena, M. H., Velázquez, E., Bedmar, E. J. (2015). Bacterial associations with legumes. Crit. Rev. Plant Sci. 34, 17–42. doi: 10.1080/07352689.2014.897899
Pršić, J., Ongena, M. (2020). Elicitors of plant immunity triggered by beneficial bacteria. Front. Plant Sci. 11, 594530. doi: 10.3389/fpls.2020.594530
Qiu, M., Xu, Z., Li, X., Li, Q., Zhang, N., Shen, Q., et al. (2014). Comparative proteomics analysis of Bacillus amyloliquefaciens SQR9 revealed the key proteins involved in in situ root colonization. J. Proteome Res. 13, 5581–5591. doi: 10.1021/pr500565m
Rabbee, M. F., Md, S., Choi, J., Hwang, B. S., Jeong, S. C., Baek, K.-H. H. (2019). Bacillus velezensis: A valuable member of bioactive molecules within plant microbiomes. Molecules 24, 1046. doi: 10.3390/molecules24061046
Rodríguez, J., Tonelli, M. L., Figueredo, M. S., Ibáñez, F., Fabra, A. (2018). The lipopeptide surfactin triggers induced systemic resistance and priming state responses in Arachis hypogaea l. Eur. J. Plant Pathol. 152, 845–851. doi: 10.1007/s10658-018-1524-6
Romero, D., De Vicente, A., Rakotoaly, R. H., Dufour, S. E., Veening, J.-W., Arrebola, E., et al(2007) The iturin and fengycin families of lipopeptides are key factors in antagonism of Bacillus subtilis toward Podosphaera fusca.” Mol. Plant-Microbe Interact. 20, 430–440. doi: 10.1094/MPMI-20-4-0430
Saxena, A. K., Kumar, M., Chakdar, H., Anuroopa, N., Bagyaraj, D. J. (2020). Bacillus species in soil as a natural resource for plant health and nutrition. J. Appl. Microbiol. 128, 1583–1594. doi: 10.1111/jam.14506
Shah, R. (2020). “Pesticides and human health,” in Emerging contaminants, ed. Nuro, A. (London: IntechOpen), 1–22. doi: 10.5772/intechopen.93806
Sibponkrung, S., Kondo, T., Tanaka, K., Tittabutr, P., Boonkerd, N., Yoshida, K. I., et al. (2020). Co-Inoculation of Bacillus velezensis strain S141 and Bradyrhizobium strains promotes nodule growth and nitrogen fixation. Microorganisms 8, 678. doi: 10.3390/microorganisms8050678
Thiessen, L. D., Woodward, J. E. (2012). Diseases of peanut caused by soilborne pathogens in the southwestern united states. ISRN Agron. 2012, 1–9. doi: 10.5402/2012/517905
Tonelli, M. L., Furlan, A., Taurian, T., Castro, S., Fabra, A. (2011). Peanut priming induced by biocontrol agents. Physiol. Mol. Plant Pathol. 75, 100–105. doi: 10.1016/j.pmpp.2010.11.001
Touré, Y., Ongena, M., Jacques, P., Guiro, A., Thonart, P. (2004). Role of lipopeptides produced by Bacillus subtilis GA1 in the reduction of grey mould disease caused by Botrytis cinerea on apple. Journal of applied microbiology, 96, 1151–1160. doi: 10.1111/j.1365-2672.2004.02252.x
Wang, R., Chang, Y. L., Zheng, W. T., Zhang, D., Zhang, X. X., Sui, X. H., et al. (2013). Bradyrhizobium arachidis sp. nov., isolated from effective nodules of Arachis hypogaea grown in China. Systematic and applied microbiology, 36, 101–105. doi: 10.1016/j.syapm.2012.10.009
Wang, X., Yuan, Z., Shi, Y., Cai, F., Zhao, J., Wang, J., et al. (2020). Bacillus amyloliquefaciens HG01 induces resistance in loquats against anthracnose rot caused by Colletotrichum acutatum. Postharvest Biol. Technol. 160, 1–7. doi: 10.1016/j.postharvbio.2019.111034
Woodward, J. E., Brenneman, T. B., Kemerait, J. R. C. (2013). “Chemical control of peanut diseases: Targeting leaves, stems, roots, and pods with foliar-applied fungicides,” in Fungicides – showcases of integrated plant disease management from around the world, ed. Nita, M. (London: IntechOpen), 56–76. doi: 10.5772/51064
Wu, L., Wu, H., Chen, L., Lin, L., Borriss, R., Gao, X. (2015). Bacilysin overproduction in Bacillus amyloliquefaciens FZB42 markerless derivative strains FZBREP and FZBSPA enhances antibacterial activity. Appl. Microbiol. Biotechnol. 99, 4255–4263. doi: 10.1007/s00253-014-6251-0
Keywords: B. velezensis, secondary metabolites, lipopeptides, Arachis hypogaea L., disease protection, systemic resistance
Citation: Korangi Alleluya V, Argüelles Arias A, Ribeiro B, De Coninck B, Helmus C, Delaplace P and Ongena M (2023) Bacillus lipopeptide-mediated biocontrol of peanut stem rot caused by Athelia rolfsii. Front. Plant Sci. 14:1069971. doi: 10.3389/fpls.2023.1069971
Received: 14 October 2022; Accepted: 17 January 2023;
Published: 20 February 2023.
Edited by:
Paolina Garbeva, Netherlands Institute of Ecology (NIOO-KNAW), NetherlandsReviewed by:
Tofazzal Islam, Bangabandhu Sheikh Mujibur Rahman Agricultural University, BangladeshCopyright © 2023 Korangi Alleluya, Argüelles Arias, Ribeiro, De Coninck, Helmus, Delaplace and Ongena. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Virginie Korangi Alleluya, di5rb3JhbmdpQGRvY3QudWxpZWdlLmJl; dmlyZ2luaWVrb3JhbmdpQGdtYWlsLmNvbQ==; Marc Ongena, bWFyYy5vbmdlbmFAdWxpZWdlLmJl
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.