
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 24 January 2023
Sec. Plant Biotechnology
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1044675
This article is part of the Research TopicThe Utilization of Plants in Vaccine ResearchView all 9 articles
Porcine reproductive and respiratory syndrome (PRRS) is a disease leading to spontaneous abortions and stillbirths in sows and lowered life quality and expectancy in growing pigs. PRRS is prevalent worldwide and has significant economic impacts to swine industries around the globe. Co-expression of the two most abundant proteins in the viral envelope, the matrix protein (M) and glycosylated protein 5 (GP5), can produce a neutralizing immune response for the virus providing a potentially effective subunit vaccine against the disease, but these proteins are difficult to express. The goal of this research was to display antigenic portions of the M and GP5 proteins on the surface of tobacco mosaic virus-like particles. A modified tobacco mosaic virus coat protein (TMVc) was transiently expressed in Nicotiana benthamiana leaves and targeted to three subcellular compartments along the secretory pathway to introduce glycosylation patterns important for M-GP5 epitope immunogenicity. We found that accumulation levels in the apoplast were similar to the ER and the vacuole. Because glycans present on plant apoplastic proteins are closest to those present on PRRSV proteins, a TMVc-M-GP5 fusion construct was targeted to the apoplast and accumulated at over 0.5 mg/g of plant fresh weight. TMVc virus-like particles self-assembled in plant cells and surface-displayed the M-GP5 epitope, as visualized by transmission electron microscopy and immunogold localization. These promising findings lay the foundation for immunogenicity and protective-immunity studies in animals to examine the efficacy of this vaccine candidate as a measure to control PRRS.
One of the major diseases affecting the global swine industry is the porcine reproductive and respiratory syndrome (PRRS) (Holtkamp et al., 2013). This disease causes respiratory symptoms in pigs, as well as stunting of growth and reduced life expectancy. In pregnant sows, PRRS leads to reproductive issues such as spontaneous abortions and stillbirths (Charerntantanakul, 2012). PRRS is caused by the porcine reproductive and respiratory syndrome virus (PRRSV), a small enveloped RNA virus from the family Arteriviridae within the order Nidovirales. Because PRRS is a viral disease, antibiotics cannot control its transmission. However, a serious consequence of PRRSV infection is secondary bacterial superinfection. Bacterial pathogens such as Mycoplasma hyopneumoniae cause more severe disease when PRRSV is present and, for this reason, PRRSV outbreaks are often treated with antibiotics (Glass-Kaastra et al., 2013; Chae, 2016), hence contributing to the spread of resistance to antibiotics genes in both pathogenic and commensal bacteria (Modi et al., 2013; Liu et al., 2019). Vaccines are a preferred option for preventing infection with PRRSV. There are a variety of vaccines currently available against PRRS, however, they are either ineffective at preventing transmission or pose safety concerns (Zuckermann et al., 2007; Okuda et al., 2008; Kimman et al., 2009). There is also clear evidence of recombination between PRRS live attenuated vaccines and field strains (Nielsen et al., 2001; Wang et al., 2019; Eclercy et al., 2019). Therefore, there is currently a need for a safer and more effective vaccine to control this disease.
PRRSV is divided into two distinct species: PRRSV-1 (European) and PRRSV-2 (North American). The two species are very heterogeneous, sharing only around 65% genome sequence identity (Rupasinghe et al., 2022). PRRSV-1 is mainly found in Europe, however, PRRSV-2 is the predominant species circulating around the world (Paploski et al., 2019; Jiang et al., 2020). Because PRRSV-2 is the main cause of global PRRS outbreaks, controlling the spread of this species should be a priority. Therefore, this research focuses on vaccine development against PRRSV-2.
The PRRSV envelope contains several structural proteins including the matrix protein (M) and glycosylated proteins 2-5 (Wissink et al., 2005). The glycosylated protein 5 (GP5) and M are the most abundant in the envelope and form heterodimers through a disulfide bond between their N-terminal ectodomains (Mardassi et al., 1996; Zhang et al., 2021). Several PRRSV structural proteins are known to contain neutralizing antibody binding sites, however, anti-GP5 antibodies appear most efficient at neutralizing the virus and preventing infection (Gonin et al., 1999; Popescu et al., 2017; Stoian and Rowland, 2019). Also, while M contains no neutralizing antibody binding sites, its co-expression alongside GP5 was shown to increase the immunogenicity of GP5 (Jiang et al., 2006). Based on this information, the co-expression of GP5 and M, or at least their ectodomains, is an important pairing for investigating vaccine candidates against PRRS.
A newer method of vaccine production involves the display of antigenic epitopes on the surface of unrelated scaffold proteins, such as virus-like particles (VLPs). VLP-based vaccines have been shown to have stronger interaction with the immune system and elicit better protective immunity than antigenic proteins alone (Bachmann and Jennings, 2010; López-Sagaseta et al., 2016). The tobacco mosaic virus (TMV), a member of the Tobamovirus genus, is a virus that targets and infects a wide range of plant species. The TMV virion consists of a long, rigid nanorod structure approximately 300 x 18 nm in size composed of over two thousand identical coat protein subunits (TMVc) surrounding the viral RNA genome (Lomonossoff and Wege, 2018). These nanorods rely on the viral genome for stability, and recombinant production of wild-type TMVc originally resulted in the assembly of only small ring-like structures (Lomonossoff and Wege, 2018). It was later confirmed that repulsive carboxylate groups on TMVc subunits are the cause of this instability, and modifying specific amino acids produced more stable nanorod VLPs (Lu et al., 1996). These modified TMV VLPs can also display foreign peptides on their surface in an organized and repetitive manner (Brown et al., 2013), which can be co-opted for vaccine design.
Plants are an expression system that has emerged in the past thirty years and picked up momentum in the past ten years (MacDonald et al., 2015). Plants are easy and inexpensive to grow, and they act as their own individual bioreactors. While different expression techniques have been developed to allow for recombinant protein expression in all or select plant tissues (Marsian and Lomonossoff, 2016), there has been a shift towards increased use of transient expression in recent years as it can be quickly and easily scaled up to produce large amounts of recombinant proteins (Margolin et al., 2020). Another benefit of plants is that they have more complex protein processing machinery than many other protein production platforms (Strasser et al., 2021). Proteins can be targeted to a wide range of subcellular compartments, allowing for an assortment of possible N-linked glycosylation patterns (Faye et al., 2005; Strasser et al., 2021). Because of this, researchers can mimic the absence, presence, and type of glycan chains on recombinant proteins to their native counterparts.
In this paper, we describe the plant-production and characterization of a modified TMV coat protein displaying a PRRSV epitope. We found that VLPs of various lengths accumulate and surface-display the PRRSV epitope.
To test if modified TMVc nanorods could be produced in plants, the TMVc sequence from Brown et al. (2013) encoding a two amino acid substitution (E50Q and D77N) was used (Figure 1A). This sequence was shown to produce nanorods in Escherichia coli but had not been expressed in a plant system.
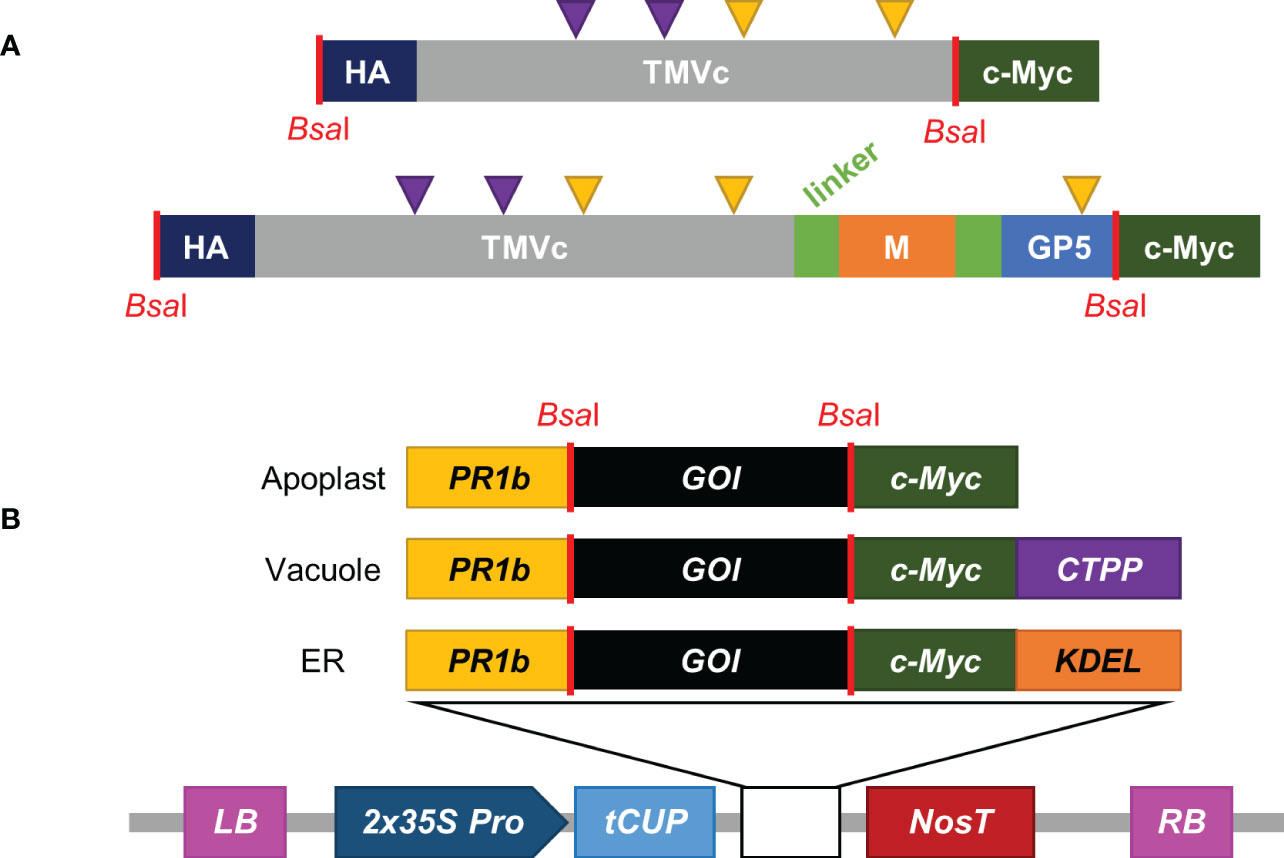
Figure 1 Construct design and pCLGG-X plant protein expression vectors. (A) The modified TMVc monomer is shown in grey, with purple arrows representing the altered amino acids (E50Q and D77N). M (orange), GP5 (light blue) and TMVc are separated by (GGS)4 flexible linkers (light green). The C-terminal c-Myc tag (dark green) and the N-terminal HA tag (dark blue) are used for detection. Yellow triangles represent potential sites for N-linked glycosylation. Sequence lengths not to scale. (B) pCLGG-X plant expression vectors target proteins to the apoplast, vacuole, or ER. All three plasmids contain the double-enhanced 35S promoter (2x35S Pro; dark blue), tCUP translation enhancer (light blue) and Nos terminator (NosT; dark red) between the left and right T-DNA borders (LB and RB; pink). The signal peptides encoded in the gene constructs include the PR1b signal peptide (yellow), the C-terminal propeptide (CTPP) for vacuolar targeting (purple), and KDEL for ER retrieval (orange). GOI indicates the gene of interest. The locations corresponding to BsaI restriction enzyme sites (red lines) used for GoldenGate cloning are noted in both the protein (A) and nucleotide (B) schematics.
To determine if the modified coat protein would express well in plants, transient agroinfiltration was performed into Nicotiana benthamiana plants. The recombinant protein was targeted to organelles along the secretory pathway (apoplast, vacuole, and ER; Figure 1B) because proteins targeted to these subcellular compartments acquire N-linked glycosylation that would later be important when the PRRSV epitope is fused. Because accumulation of some proteins rises steadily with time, while other proteins decrease with time, a time-course was conducted to investigate the accumulation profile of recombinant TMVc for one week after infiltration. This determined the best time to sample the leaf tissue for maximal accumulation of recombinant protein.
A triplet of bands was detected in most TMVc lanes (Figure 2). This can be explained by heterogeneous glycosylation at the two putative glycosylation sites (Figure 1A), as determined by NetNGlyc 1.0 server (Technical University of Denmark). The lower band is likely the unglycosylated protein, with the upper bands containing one or two glycan chains. The apoplast-targeted TMVc has only two bands visible, which may be the result of efficient glycosylation as no band for the unglycosylated monomer is detectable. This experiment was repeated, and in both experiments the bands ran slightly lower in the apoplast-targeted samples than in the ER and vacuole samples (Figure 2). This is likely due to the KDEL ER retrieval tetrapeptide and the C-terminal vacuole targeting peptide that add amino acids to the mature protein. While no quantification or data analysis was performed for these experiments, visual inspection of band intensity indicated that apoplast targeting was suitable for further analysis. Sampling at 7 days post-infiltration (dpi) was selected to provide more time for the proteins to self-assemble into VLPs.
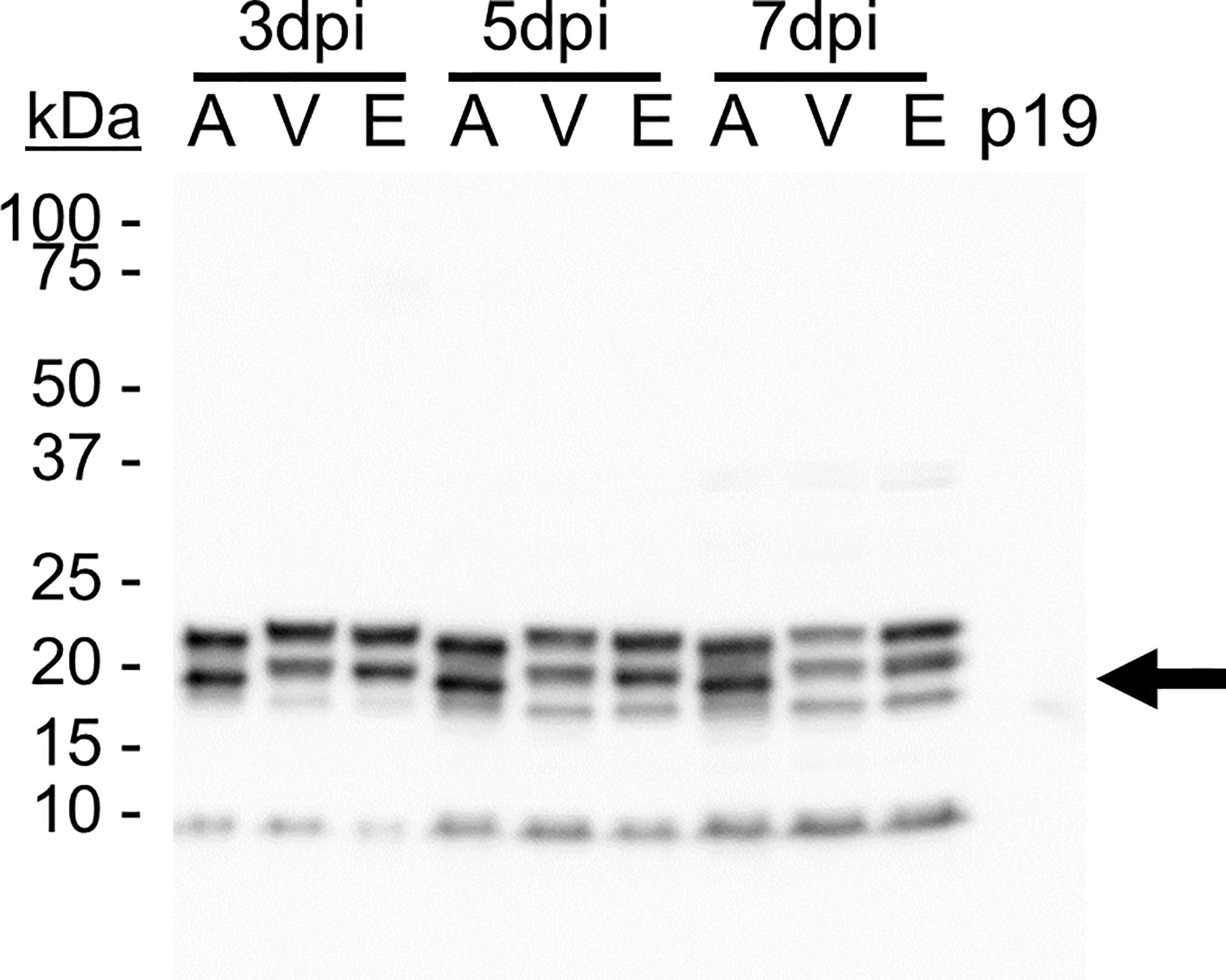
Figure 2 TMVc accumulates in the secretory pathway. Proteins were targeted to the apoplast (A), vacuole (V), and ER (E) and were sampled on 3, 5, and 7 days post-infiltration (dpi). The p19-infiltrated sample acts as a negative control. The black arrow denotes the expected size of unglycosylated TMVc monomer (19.9 kDa). Detection was performed using an anti-HA primary antibody at a 1:5000 dilution. Detection was performed using an anti-HA primary antibody at a 1:5000 dilution, and is representative of two transient expression experiments, and is representative of two transient expression experiments. In each lane 20 µl of total soluble protein were loaded, corresponding to approximately 6.67 mg of leaf fresh weight.
To assess the quaternary assembly and nanorod formation of the modified TMVc, VLPs were extracted and partially purified from infiltrated leaf samples then visualized by TEM. Characteristic nanorod structures with 18 nm diameters and varying lengths were observed (Figure 3), demonstrating that the modified TMVc protein does assemble into VLPs upon plant expression. The variation in nanorod length was expected as the consistent 300 nm of the native TMV is dictated by the encompassed viral genome. Because the VLPs produced here contain no genetic material, uniform nanorod length could not be expected.
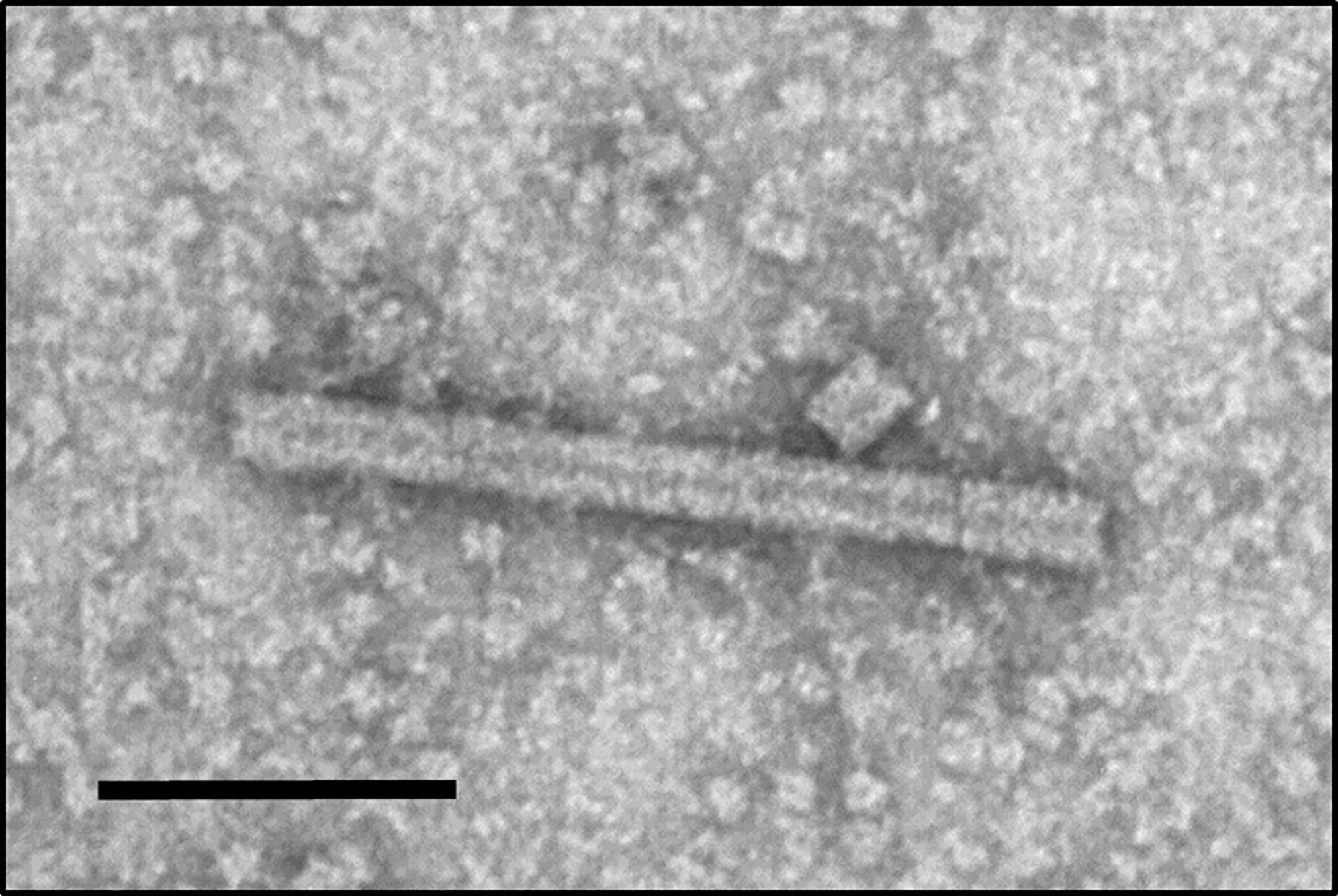
Figure 3 Plant-produced TMVc assemble into nanorod structures. The modified TMVc self-assembles into nanorod structures of varying lengths. Scale bar represents 100 nm. The protein sample was targeted to the apoplast and sampled on 7 dpi. Partially purified VLP extract was negatively stained and examined using a TEM (JEM-1400, JEOL) operated at 80 kV.
The PRRSV GP5 ectodomain was selected for the vaccine epitope because it contains at least one neutralizing antibody binding site within its ectodomain and anti-GP5 antibodies have been found effective at neutralizing this virus, with data suggesting broad-neutralization of heterologous strains (Gonin et al., 1999; Popescu et al., 2017; Stoian and Rowland, 2019). Furthermore, the GP5 ectodomain contains varying numbers of glycosylation sites depending on the viral strain. The PRRSV sequences selected for this study are from the PRRSV-2 reference strain, VR-2332. The GP5 sequence selected for this epitope corresponds to amino acids 30-54 because the first approximately 30 amino acids make up a cleaved signal peptide (Popescu et al., 2017). Also, a neutralizing epitope between residues 37-41 has been previously described (Rupasinghe et al., 2022) so the GP5 ectodomain sequence selected for this research encompasses that epitope, with additional flanking sequences found to be relatively conserved across different PRRSV-2 strains. It was shown that glycan chains at N33 and N51 shield the antibody binding site on GP5 from the immune system (Ansari et al., 2006), while the glycan chain at N44 is important for eliciting antibodies capable of neutralizing wild-type PRRSV (Wei et al., 2012). Therefore, mutations N33A and N51A were introduced in the GP5 ectodomain sequence for improved immunogenicity of this epitope.
Co-expression of M with GP5 was previously shown to amplify the anti-GP5 immune response (Jiang et al., 2006), so pairing the M and GP5 ectodomains together in our designed epitope may have a similar effect. Amino acids 2-18 from M were selected as this corresponds to the entire ectodomain lacking the initial methionine.
The PRRSV M-GP5 epitope was genetically fused to the C-terminus of TMVc as previous work found this terminus is surface-exposed on the nanorods and is amenable to the addition of foreign peptides (Pogue et al., 2007). The M and GP5 ectodomain sequences were separated from each other and the TMVc monomer by flexible (GGS)4 linkers (Figure 1A).
Upon transient expression by agroinfiltration, two TMVc-M-GP5 bands were observed on the immunoblot at around 35 kDa and an additional faint band at about 70 kDa (Figure 4A). To verify that the double banding does not represent C-terminal cleavage, another sample was immunoblotted with anti-c-Myc antibodies. The same pattern was observed (Figure 4B), indicating that both bands represent different glycosylation states of the full length protein. The expected molecular weight of the unglycosylated protein is 25.9 kDa, and that of the fully glycosylated protein is 32.5 kDa. Therefore, it appears as if the dominant protein bands on the immunoblot are from the glycosylated TMVc-M-GP5, and the 70 kDa band may represent a dimer. Deglycosylation analysis on TMVc-M-GP5 resulted in a single band at about 25 kDa confirming that the double-banding pattern is the result of N-linked glycosylation (Figure 4C).
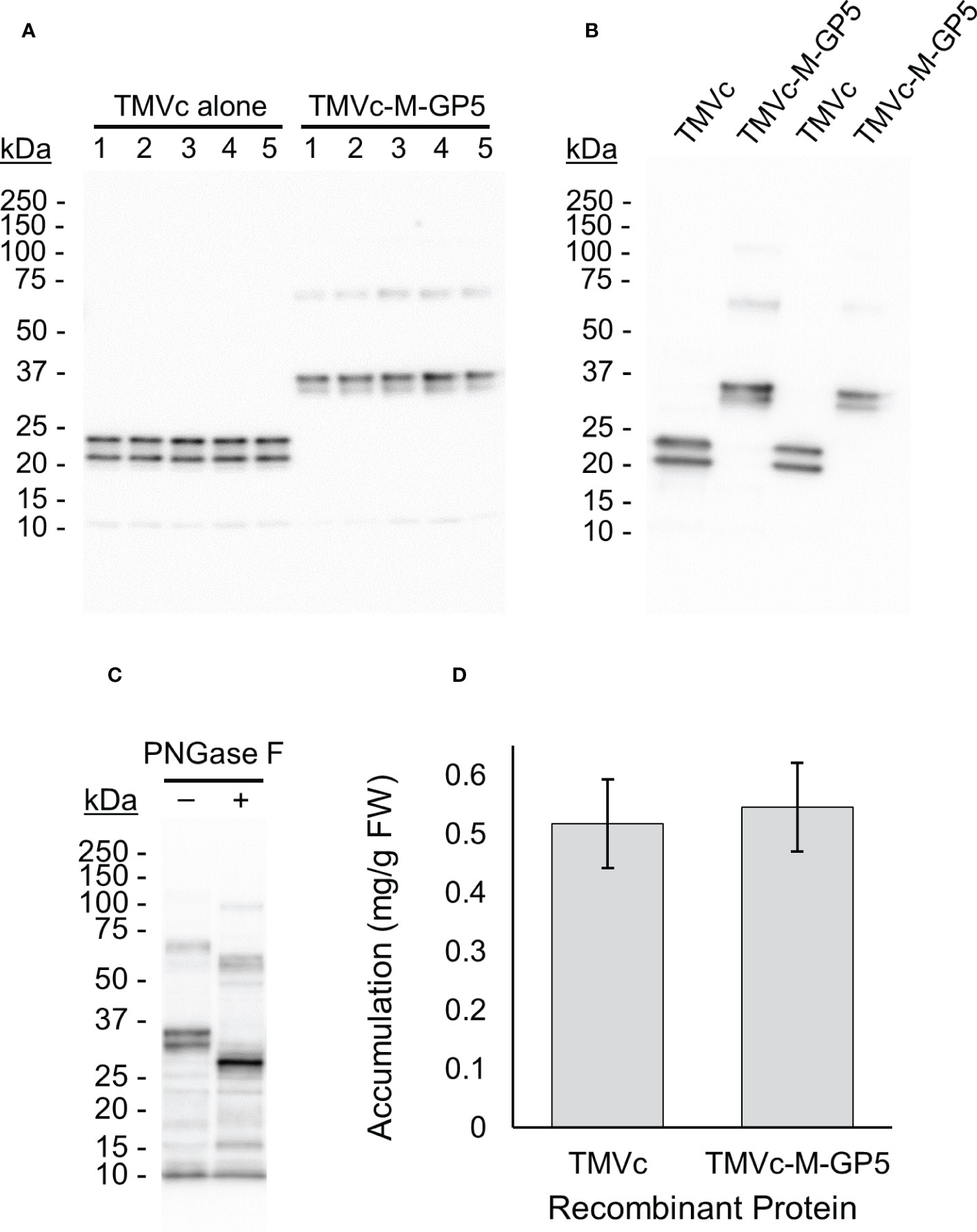
Figure 4 TMVc-M-GP5 accumulates well in plants and is efficiently glycosylated. All proteins were targeted to the apoplast and sampled on 7 dpi. (A) The numbers across the top (1-5) identify different plants considered biological replicates. The expected size of the fully glycosylated TMVc and TMVc-M-GP5 monomers are 24.3 kDa and 32.5 kDa, respectively. The 70 kDa band observed for TMVc-M-GP5 may be due to dimerization. A standard curve was generated with known amounts of a protein standard and used for quantification. In each lane 5 µl of total soluble plant proteins were loaded corresponding to approximately 0.5 mg of plant tissue fresh weight. Exact tissue weights were recorded and considered for accumulation values. This blot was probed with an anti-HA primary antibody, and is representative of three transient expression experiments. (B) TMVc and TMVc-M-GP5 immunoblot probed with an anti-c-Myc primary antibody. Two replicates are shown for each protein. The same double banding pattern as probing with an anti-HA primary antibody is observed. (C) Deglycosylation of TMVc-M-GP5 with PNGase F (+) shows a single band at the expected molecular weight of the unglycosylated protein, compared to the double banding observed for the untreated protein (–). This blot was probed with an anti-HA primary antibody. (D) The mean recombinant protein accumulation levels of 15 biological replicates over three independent experiments (5 biological replicates per experiment) are shown in milligrams of recombinant protein per gram of plant fresh weight (FW), with the bars above and below representing 95% confidence intervals. No significant difference in recombinant protein accumulation was observed, as assessed by ANOVA.
To confirm whether the recombinant proteins accumulate in the apoplast, the apoplastic fluid was isolated from leaves infiltrated with TMVc-M-GP5. The recombinant protein was detected in infiltrated leaves compared to a negative control (Supplementary Figure 1), indicating localization of the recombinant proteins in the apoplast.
For quantifying soluble protein accumulation, a total of three experiments were run with 5 plants each, resulting in 15 biological replicates. The average accumulation levels of the recombinant protein was not significantly different between TMVc and TMVc-M-GP5 (p-value = 0.59) (Figure 4D). Therefore, fusion of the M-GP5 epitope onto TMVc does not appear to alter accumulation upon expression in plants.
To assess whether fusion of the PRRSV M-GP5 epitope affects VLP assembly, TMVc-M-GP5 samples were prepared and examined using a TEM and nanorods with 18 nm diameters and variable lengths were observed similar to TMVc nanorods (Figure 5A). While genetic fusion of the M-GP5 epitope does not inhibit VLP assembly, TMVc-M-GP5 nanorods were generally shorter in length than those of TMVc alone. For example, the average length of 150 TMVc nanorods was 66.42 nm (s.e. 2.79), compared to an average length of 43.48 nm (s.e. 1.32) for 235 TMVc-M-GP5 nanorods. Once assembly of the fusion proteins was confirmed, we assessed whether the PRRSV epitope is displayed on the nanorod surface. Immunogold localization was performed using anti-c-Myc primary antibodies; gold particles were found near the nanorods (Figure 5B), suggesting that M-GP5 epitopes are displayed on the nanorod surface. As a negative control, no gold particles were observed when the primary antibody was omitted (Figure 5C).
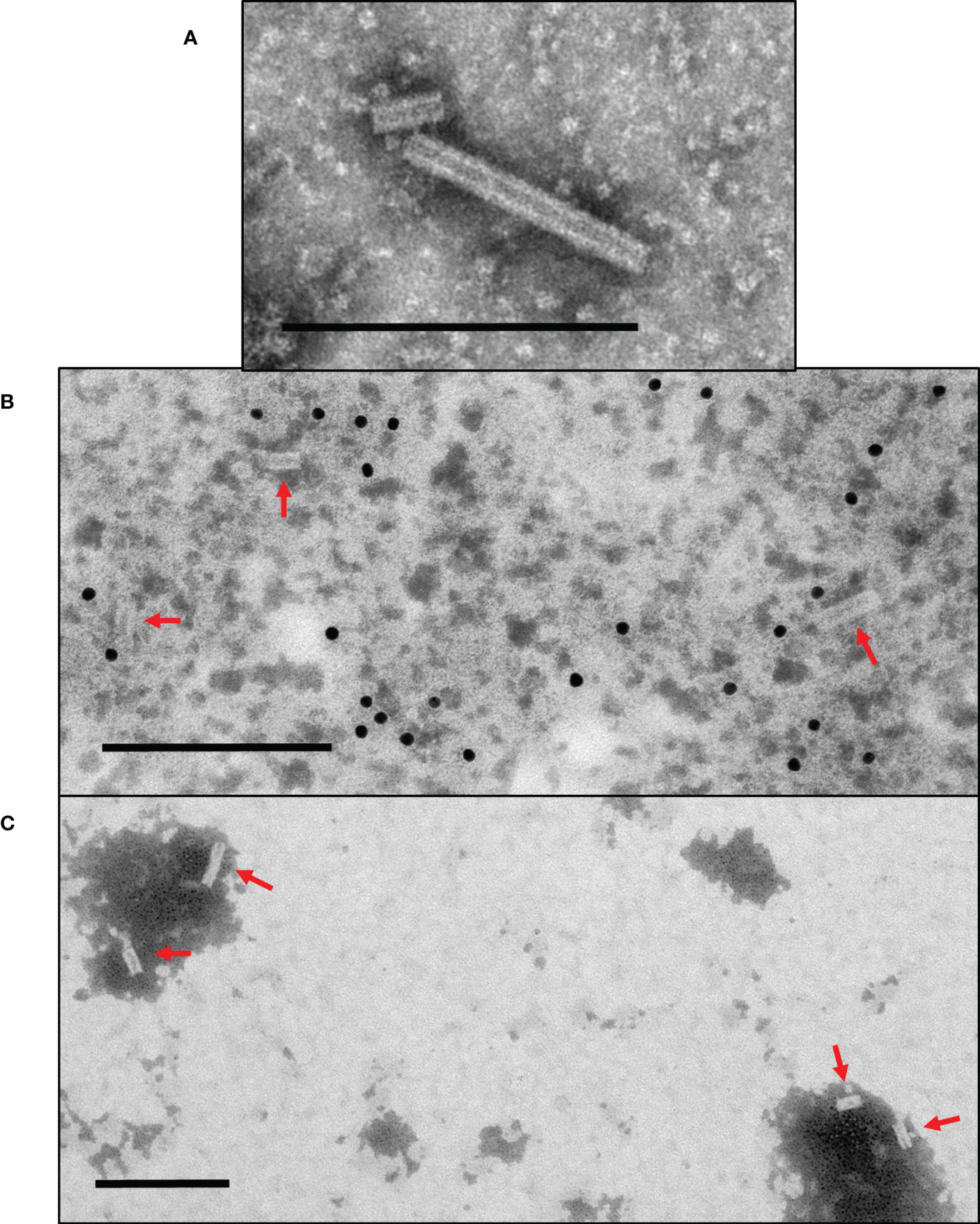
Figure 5 Plant-produced TMVc-M-GP5 assembles into nanorod structures surface-displaying the PRRSV epitope. (A) The modified TMVc-M-GP5 fusion protein self-assembles into nanorod structures of varying lengths. (B) TMVc-M-GP5 with immunogold localization using anti-c-Myc primary antibodies. Gold particles (10 nm) bound close to the nanorod structures indicate that the PRRSV epitope are displayed on the VLP surface. (C) Negative control for immunogold localization of TMVc-M-GP5, using no primary antibody for detection. Nanorods are present but no gold particles are visible surrounding VLPs. Scale bars represent 200 nm, and nanorods are denoted with red arrows. All protein samples were targeted to the apoplast and sampled on 7 dpi. Partially purified VLP extracts were negatively stained and examined using a TEM (JEM-1400, JEOL) operated at 80 kV.
VLP-based vaccines provide safe and effective alternatives to traditional vaccines. Unlike live attenuated vaccines, protein VLPs are unable to replicate or revert to virulence because they lack genetic material (Francis, 2018; Vetter et al., 2018). VLP-based vaccines are also efficient at producing strong and long-lasting immune responses, a benefit they have over many traditional subunit or killed virus vaccine methods (Francis, 2018; Vetter et al., 2018). Because of these reasons, more research has been allocated to studying different VLPs as potential vaccine candidates.
Some recombinantly produced VLPs of pathogenic viruses are now commercially available as vaccines. Two vaccines available for human papillomavirus (HPV) (Gardasil® and Cervarix®) are prime examples of this, where the recombinant expression of a major HPV capsid protein results in self-assembling VLPs able to induce protection against HPV infection in humans (Wang and Roden, 2013). More recently, Medicago Inc. (Quebec, QC) produced the first plant-derived vaccine against SARS-CoV-2 to reach clinical trials (Ward et al., 2021). Enveloped VLPs with a modified viral spike protein were produced using transient expression in N. benthamiana, resulting in VLPs similar in size and shape to the SARS-CoV-2 virus displaying a high density of spike proteins on their surface. After successful phase II and III clinical trials, this vaccine was approved by Canadian Health Authorities in early 2022 (Dubé et al., 2022).
Unlike HPV and SARS-CoV-2, vaccines for many viruses and bacteria cannot easily be produced by recombinant expression of full or slightly modified viral proteins. In these instances, displaying antigenic peptides from the pathogen on the surface of unrelated scaffold proteins can be an effective alternative. The use of scaffold proteins, such as VLPs, may stabilize viral epitopes and allow for higher accumulation of recombinant protein vaccines (López-Sagaseta et al., 2016). Nanoparticle vaccines can also have adjuvant-like properties due to their size and repetitive epitope-display interacting with the immune system to induce robust immune responses (Bachmann and Jennings, 2010; López-Sagaseta et al., 2016). Therefore, displaying PRRSV epitopes on VLPs could be a promising novel approach to vaccine development for this disease.
In this study, TMVc accumulated to high levels and showed no significant change in these levels when fused to the M-GP5 epitope. These high accumulation levels may be due to correct folding of proteins by chaperones and disulfide bond formation in the ER (Strasser, 2018; Robinson and Bulleid, 2020). Also, the introduction of N-linked glycans in the plant secretory pathway may play a role as glycosylation of certain proteins increases their stability (Solá and Griebenow, 2009). For the TMVc-M-GP5 samples, the faint band running at about 70 kDa may represent a dimer of the recombinant protein. Previous work by Castells-Graells and Lomonossoff (2021) found that plant-produced VLPs also contained dimers and further assembly products, which were completely absent when the same proteins were expressed in insect cells. This is due to some component of the plant extract as insect-produced VLPs incubated in wild-type plant extract developed similar banding patterns. It could not be confirmed, however, whether this occurs within the plant cells, or only in extraction conditions (Castells-Graells and Lomonossoff, 2021).
Wild-type TMVc does not produce nanorod structures when the viral RNA is absent due to repulsive carboxylate groups between the rings of the helix, and while these carboxylate groups play an important role in the assembly and disassembly of the native virion (Lu et al., 1996; Brown et al., 2013) they serve no purpose in displaying vaccine epitopes on TMVc VLPs. Therefore, a modified TMVc sequence was used in this project containing a two amino acid substitution (E50Q and D77N) to allow self-assembly into stable nanorod structures even in the absence of RNA, and these modified TMVc nanorods were able to display foreign peptides on their outer surface (Brown et al., 2013). The current work is the first time this modified TMVc sequence was expressed recombinantly in plants. Our results show that nanorod VLPs can form, in accordance with the previous work from E. coli. We have also demonstrated that the PRRSV M-GP5 epitope can be displayed on the outer nanorod surface. While nanorod lengths were shorter on average for the fusion protein, VLP morphology and assembly was not impacted to the extent reported by others (Frolova et al., 2010).
Viral diseases are one of the major hurdles facing the livestock industry, especially because viruses cannot be controlled by antibiotics. Interestingly, PRRSV infection may result in antibiotic treatment due to opportunistic bacterial infections that often accompany this virus (Glass-Kaastra et al., 2013; Chae, 2016). High usage of antibiotics for a wide range of purposes including veterinary medicine has led to the growing issue of antimicrobial resistance, where some disease-causing bacteria are unaffected by most or all of the antibiotics used in their treatment (Liu et al., 2019; Pinto Ferreira et al., 2022). Employing vaccines to control diseases like PRRS would allow for decreased transmission rates, and therefore minimize the use of antibiotics on farms (Pinto Ferreira et al., 2022).
While there are many ways to design and produce vaccines, keeping production costs low is essential for veterinary medicine (Schillberg and Finnern, 2021) as livestock farmers are unlikely to purchase expensive medications for their animals (MacDonald et al., 2015). High accumulation levels of recombinant proteins are a major factor in limiting the production costs. For industrial production of plant-produced pharmaceutical proteins for human use, an unofficial threshold of 1% of total soluble protein, or approximately 0.1 mg/g of recombinant protein per fresh plant weight, is used to evaluate economic viability (Rybicki, 2009). Next to accumulation levels, working with insoluble proteins requires more laborious and costly unfolding and refolding steps. Therefore using soluble proteins, which tend to be correctly folded, is preferred. In the current study, we found high soluble accumulation of TMVc-M-GP5 upon plant-production, with accumulation being over 5x the unofficial industry threshold.
Plant-produced vaccines have the potential to be administered orally to livestock, which provides several benefits over injection. One benefit is avoiding the need to purify the recombinant proteins because the plant tissue can be lyophilized and fed to the animals (Chan and Daniell, 2015; Topp et al., 2016), thereby greatly reducing costs (Schillberg and Finnern, 2021). A second benefit is the technical ease of oral administration, where lyophilized plant tissue could be mixed into livestock feed as opposed to injection of the vaccine by a veterinarian (Chan and Daniell, 2015). This would make it difficult to know the exact dose each pig receives, however, regulations for veterinary medicine are not as strict as for human vaccination and a broader dosage range is acceptable (Topp et al., 2016). A third benefit of oral administration is the direct delivery of the vaccine to the intestinal mucosa, improving the resulting IgA titers (Topp et al., 2016). Unlike parenteral administration which tends to result in higher IgG production and lower IgA levels, oral administration of vaccines can directly stimulate high IgA production in mucosal tissues of the intestinal tract (Topp et al., 2016). Stimulating IgA production in certain mucosal tissues, such as the digestive tract, results in raised specific IgA levels at other mucosal tissues, like the respiratory tract (Hosseini et al., 2015). Because the PRRSV infects at mucosal tissues (Pileri and Mateu, 2016), IgA antibodies are likely better suited for interrupting initial infection of the pigs (Kolotilin et al., 2014; Topp et al., 2016).
Another major benefit of using a plant expression system is the variety of post-translational modifications they can add to proteins. One modification of particular interest, especially for this project, is N-linked glycosylation. Plant and mammalian cells both allow for N-linked glycosylation of proteins. The PRRSV is secreted in pig cells, and acquires the complex N-linked glycan chains characteristic of the mammalian late Golgi (Li et al., 2015). In plants, the complex N-linked glycan chains on apoplast-targeted proteins are the closest to those from the mammalian late Golgi (Margolin et al., 2020; Strasser et al., 2021).
While similar, plant-specific complex glycans decorating apoplast-targeted proteins receive plant-specific β1,2 xylose and α1,3 fucose, compared to the α1,6 fucose and terminal sialic acid sugars found on secreted mammalian proteins (Gomord et al., 2010; Margolin et al., 2020). Because of this, plant-specific sugars may be differentially recognized by the immune system. Early concerns that such plant-produced proteins could cause allergic reactions if injected have been disproved (Margolin et al., 2020) and this would not be a concern for oral administration as animals ingest glycosylated plant proteins regularly with no ill effects. The proteins may, however, be more easily identified as foreign and therefore receive more attention from the immune system (Gomord et al., 2010). For many therapeutic proteins this would be undesirable as their intended purpose does not involve the immune system and being recognized as foreign could increase their turnover rate in the body (Gomord et al., 2010; Margolin et al., 2020). For vaccines, this improved recognition by the host immune system may actually serve to benefit the immunogenicity of the proteins (Gomord et al., 2010). In this study, the plant-produced recombinant proteins are being efficiently decorated with N-linked glycans important for the M-GP5 epitope’s immunogenicity.
Moving forward, animal trials must be conducted to examine the immunogenicity of this particulate vaccine candidate. Adjuvants are routinely used in conjunction with vaccines; however, VLP-based vaccines already show improved interaction with the immune system due to their particulate nature and multivalent display of antigenic epitopes, therefore adjuvants may not be required (Zhao et al., 2014; López-Sagaseta et al., 2016). While there is much work yet to be completed before the vaccine candidate produced here can be used in the field to control PRRS, this work provides a promising foundation for further study. It also adds to the ever-growing evidence that producing protein-based pharmaceuticals in plants is an innovative alternative to more traditional approaches.
The TMVc sequence was based on the entire TMVc monomer from Brown et al. (2013), containing two modifications compared to wild type (E50Q and D77N). For the fusion protein construct, the PRRSV epitope was composed of amino acids 2-18 of the M protein (accession number: AAO13197.1) and amino acids 30-54 of GP5 (accession number: AAO13196.1) from the PRRSV strain VR-2332. The GP5 sequence also encoded a two amino acid substitution (N33A and N51A) described by Ansari et al. (2006). The M and GP5 sequences were separated from one another, as well as from TMVc, by flexible (GGS)4 linkers. Both gene constructs were synthesized by Bio Basic Inc. (Markham, Canada) and included sequences for an N-terminal HA tag and flanking BsaI sites for GoldenGate cloning.
Both genes were cloned into in-house pCLGG-X plant expression vectors targeting proteins to the apoplast, vacuole, or ER using GoldenGate Cloning methodology (Marillonnet and Werner, 2015). All three of these vectors contain a double-enhanced 35S promoter (Kay et al., 1987) and tobacco cryptic upstream promoter translational enhancer (tCUP) (Wu et al., 2001) for high expression, the Nos terminator (Bevan et al., 1983), and a c-Myc tag for detection and purification of the encoded proteins. A PR1b signal peptide from tobacco is also encoded in all three vectors for targeting proteins to the secretory pathway (Cutt et al., 1988). The ER vector encodes a KDEL tetrapeptide for retrieval of the proteins to the ER (Pelham, 1990), and the vacuole targeting vector contains the coding sequence for a C-terminal propeptide (CTPP) vacuolar sorting signal from the tobacco chitinase gene (Vitale and Raikhel, 1999). The resulting plasmids were transformed into Agrobacterium tumefaciens EHA105 cells (Hood et al., 1993). Plasmids were isolated from Agrobacterium cultures and verified by sequencing.
Wild-type N. benthamiana plants were grown at 22°C with 65% humidity on a light:dark cycle of 16:8 h in a walk-in growth chamber. Plants were grown in 4” pots with PRO-MIX BX soil, and were watered as necessary using 20:8:20 fertilizer (N:P:K) at 0.25 g/L of water.
Infiltrations into N. benthamiana leaves were performed as previously described (Miletic et al., 2016). Briefly, Agrobacterium cultures containing the genes of interest as well as p19, a suppressor of posttranscriptional gene silencing from the Cymbidium ringspot virus (Silhavy et al., 2002), were co-infiltrated into the abaxial surface of 6-8 week old N. benthamiana leaves. Agrobacterium containing p19 alone was used as a negative control.
For the time course with TMVc, 3 leaves on 5 plants were infiltrated with the apoplast, vacuole, and ER targeted constructs. One leaf disc was sampled from each infiltrated area on 3, 5, and 7 dpi with samples pooled across all 5 plants each day. This transient expression experiment was repeated twice. For the fusion construct analysis, 3 leaves on 5 plants were infiltrated with the apoplast-targeted TMVc and TMVc-M-GP5 fusion constructs. For sampling, one leaf disc was taken at 7 dpi from each infiltrated area with samples pooled by plant. This experiment was repeated three times to allow for 15 biological replicates per protein. All plant tissue samples were weighed, flash-frozen in liquid nitrogen, and stored at -80°C until processed.
Total soluble proteins were isolated from the plant tissue as described by Miletic et al. (2016). The proteins were transferred to a polyvinylidene difluoride (PVDF) membrane and detected with mouse anti-hemagglutinin (HA) primary antibody (Sigma-Aldrich Cat. No. H3663) or mouse anti-c-Myc primary antibody (Genscript Cat. No. A00864), anti-mouse horseradish peroxidase-conjugated secondary antibodies (Bio-Rad #1706516), and Clarity Western ECL Blotting Substrates (Bio-Rad Cat. No. 1705061). Proteins were visualized on the MicroChemi 4.2 (DNR Bio-Imaging Systems Ltd.) and quantified compared to a standard curve of known concentrations of a protein standard using GelQuant software (DNR Bio-Imaging Systems Ltd.). The protein standard is a synthetic protein designed with multiple detection tags.
TMVc-M-GP5 plant extract was deglycosylated using PNGase F (New England BioLabs Cat. No. P0704L) as per the manufacturer’s instructions, with samples incubated at 37°C for 60-90 minutes. The negative control consisted of the same TMVc-M-GP5 plant extract lacking the PNGase enzyme. The samples then underwent SDS-PAGE and Western blotting for visualization.
Apoplastic fluid isolation was performed as previously described (Hu et al., 2021). Briefly, infiltrated N. benthamiana leaves were sampled between 5-7 dpi. The leaves were submerged and vacuum infiltrated in an apoplastic fluid isolation buffer (20 mM MES, 2 mM CaCl2, 0.1 M NaCl, pH 6). The external surfaces of the leaves were dried, and the leaves were placed into a 30 ml syringe inside a 50 ml tube. The samples were centrifuged at 700 x g for 45-60 minutes at 4°C. The resulting solutions were collected.
To compare the soluble protein accumulation levels of TMVc and TMVc-M-GP5, the values were calculated in mg/g of fresh plant tissue by considering the quantification values from the blots, the volume of plant protein extraction buffer used, the mass of plant tissue per sample, and dilution of samples loaded on gels (i.e. volume of extract loaded per lane). The resulting soluble protein accumulation data (n = 15) was examined using R 4.0.3 (R Core Team, 2020). The analysis began with checking the data for outliers, zero inflation, and balance of the categorical variable. A linear model was used and validated by checking the residual distribution and residuals versus fitted values. Finally, an ANOVA was performed to assess significance in the data.
For visualization of the nanorods using transmission electron microscopy, partial purification of the plant extract was performed as previously described (Dai et al., 2020). Briefly, infiltrated plant tissue was ground in a mortar and pestle with 150 µl of potassium phosphate buffer (pH 7.0) containing 0.1% β-mercaptoethanol per 0.1 g plant leaf tissue. The homogenate was centrifuged at 12,000 x g for 5 minutes at 4°C, and the supernatant was filtered through a 100 µm nylon cell strainer. After adding one-third volume of chloroform, the samples were vortexed at maximum speed for 1 minute. The samples were centrifuged at 12,000 x g for 10 minutes at 4°C, then the aqueous phase was taken and centrifuged again for 30 minutes. The aqueous phase containing the final VLP preparation was then collected.
To examine nanorod assembly, the partially purified VLP preparation was spotted onto formvar carbon-coated copper grids and negatively stained with 2% phosphotungstic acid (PTA). The grids were examined on a TEM (JEM-1400, JEOL Ltd.) operated at 80 kV. For immunogold localization, grids were spotted with the VLP preparation then blocked with 0.3% bovine serum albumin (BSA) for 15 minutes. The grids were incubated with a 1/650 dilution of mouse anti-c-Myc primary antibody (GenScript A00864) for 1 hour. After 5 washes with 0.03% BSA, the grids were incubated for 1 hour with a 1/15 dilution of 10 nm colloidal gold-conjugated goat anti-mouse secondary antibody (Electron Microscopy Sciences Cat. No. 25129). The grids were washed 5 times with 0.03% BSA, then 3 times with distilled water. Finally, the grids were negatively stained with 2% PTA and examined on the JEM-1400 (JEOL Ltd.) TEM.
The original contributions presented in the study are included in the article/Supplementary material. Further inquiries can be directed to the corresponding author.
JV designed the constructs, carried out the experiments, conducted the analysis and wrote the manuscript. OH identified the M and GP5 ectodomain sequences, CPG conceptualized the epitope, SEK co-supervised experimental work, and RM conceptualized the study and edited the manuscript. All authors contributed to the article and approved the submitted version.
This work was funded by AAFC grant # 2351 and 3441 to RM and CPG.
We thank Hong Zhu and Angelo Kaldis at Agriculture and Agri-Food Canada for providing technical support. We also thank Dr. Emily Clayton for technical guidance, and both Carly Charron and Shabnam Shamriz for feedback on protein purification.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1044675/full#supplementary-material
Ansari, I. H., Kwon, B., Osorio, F. A., Pattnaik, A. K. (2006). Influence of n-linked glycosylation of porcine reproductive and respiratory syndrome virus GP5 on virus infectivity, antigenicity, and ability to induce neutralizing antibodies. J. Virol. 80, 3994–4004. doi: 10.1128/JVI.80.8.3994-4004.2006
Bachmann, M. F., Jennings, G. T. (2010). Vaccine delivery: a matter of size, geometry, kinetics and molecular patterns. Nat. Rev. Immunol. 10, 787–796. doi: 10.1038/nri2868
Bevan, M., Barnes, W. M., Chilton, M.-D. (1983). Strcuture and transcription of the nopaline synthase gene region of T-DNA. Nucleic Acids Res. 11, 369–385. doi: 10.1093/nar/11.2.369
Brown, A. D., Naves, L., Wang, X., Ghodssi, R., Culver, J. N. (2013). Carboxylate-directed in vivo assembly of virus-like nanorods and tubes for the display of functional peptides and residues. Biomacromolecules 14, 3123–3129. doi: 10.1021/bm400747k
Castells-Graells, R., Lomonossoff, G. P. (2021). Plant-based production can result in covalent cross-linking of proteins. Plant Biotechnol. J. 19, 1095–1097. doi: 10.1111/pbi.13598
Chae, C. (2016). Porcine respiratory disease complex: interaction of vaccination and porcine circovirus type 2, porcine reproductive and respiratory syndrome virus, and Mycoplasma hyopneumoniae. Vet. J. 212, 1–6. doi: 10.1016/j.tvjl.2015.10.030
Chan, H.-T., Daniell, H. (2015). Plant-made oral vaccines against human infectious diseases - are we there yet? Plant Biotechnol. J. 13, 1056–1070. doi: 10.1111/pbi.12471
Charerntantanakul, W. (2012). Porcine reproductive and respiratory syndrome virus vaccines: immunogenicity, efficacy and safety aspects. World J. Virol. 1, 23–30. doi: 10.5501/wjv.v1.i1.23
Cutt, J. R., Dixon, D. C., Carr, J. P., Klessig, D. F. (1988). Isolation and nucleotide sequence of cDNA clones for the pathogenesis-related proteins PR1a, PR1b and PRIc of Nicotiana tabacum cv. xanthi nc induced by TMV infection. Nucleic Acids Res. 16, 9861–9861. doi: 10.1093/nar/16.20.9861
Dai, Z., He, R., Bernards, M. A., Wang, A. (2020). The cis-expression of the coat protein of turnip mosaic virus is essential for viral intercellular movement in plants. Mol. Plant Pathol. 21, 11941211. doi: 10.1111/mpp.12973
Dubé, C., Paris-Robidas, S., Andreani, G., Gutzeit, C., D'Aoust, M.-A., Ward, B. J., et al. (2022). Broad neutralization against SARS-CoV-2 variants induced by ancestral and B.1.351 AS03-adjuvanted recombinant plant-derived virus-like particle vaccines. Vaccine 40, 4017–4025. doi: 10.1016/j.vaccine.2022.05.046
Eclercy, J., Renson, P., Lebret, A., Hirchaud, E., Normand, V., Andraud, M., et al. (2019). A field recombinant strain derived from two type 1 porcine reproductive and respiratory syndrome virus (PRRSV-1) modified live vaccines shows increased viremia and transmission in SPF pigs. Viruses 11, 296. doi: 10.3390/v11030296
Faye, L., Boulaflous, A., Benchabane, M., Gomord, V., Michaud, D. (2005). Protein modifications in the plant secretory pathway: Current status and practical implications in molecular pharming. Vaccine 23, 1770–1778. doi: 10.1016/j.vaccine.2004.11.003
Francis, M. J. (2018). Recent advances in vaccine technologies. Vet. Clin. North Am. Small Anim. Pract. 48, 231–241. doi: 10.1016/j.cvsm.2017.10.002
Frolova, O. Y., Petrunia, I. V., Komarova, T. V., Kosorukov, V. S., Sheval, E. V., Gleba, Y. Y., et al. (2010). Trastuzumab-binding peptide display by Tobacco mosaic virus. Virology 407, 713. doi: 10.1016/j.virol.2010.08.005
Glass-Kaastra, S. K., Pearl, D. L., Reid-Smith, R. J., McEwen, B., McEwen, S. A., Amezcua, R., et al. (2013). Describing antimicrobial use and reported treatment efficacy in Ontario swine using the Ontario swine veterinary-based surveillance program. BMC Vet. Res. 9, 238. doi: 10.1186/1746-6148-9-238
Gomord, V., Fitchette, A. C., Menu-Bouaouiche, L., Saint-Jore-Dupas, C., Plasson, C., Michaud, D., et al. (2010). Plant-specific glycosylation patterns in the context of therapeutic protein production. Plant Biotechnol. J. 8, 564–587. doi: 10.1111/j.1467-7652.2009.00497.x
Gonin, P., Pirzadeh, B., Gagnon, C. A., Dea, S. (1999). Seroneutralization of porcine reproductive and respiratory syndrome virus correlates with antibody response to the GP5 major envelope glycoprotein. J. Vet. Diagn. Invest. 11, 20–26. doi: 10.1177/104063879901100103
Holtkamp, D. J., Kliebenstein, J. B., Neumann, E. J., Zimmerman, J. J., Rotto, H. F., Yoder, T. K., et al. (2013). Assessment of the economic impact of porcine reproductive and respiratory syndrome virus on united states pork producers. J. Swine Health Prod 21, 72–84.
Hood, E. E., Gelvin, S. B., Melchers, L. S., Hoekema, A. (1993). New Agrobacterium helper plasmids for gene transfer to plants. Transgenic Res. 2, 208–218. doi: 10.1007/BF01977351
Hosseini, M., Dobakhti, F., Pakzad, S. R., Ajdary, S. (2015). Immunization with single oral dose of alginate-encapsulated BCG elicits effective and long-lasting mucosal immune responses. Scand. J. Immunol. 82, 489–497. doi: 10.1111/sji.12351
Hu, S., Yin, Y., Chen, B., Lin, Q., Tian, Y., Song, X., et al. (2021). Identification of viral particles in the apoplast of Nicotiana benthamiana leaves infected by potato virus X. Mol. Plant Pathol. 22, 456–464. doi: 10.1111/mpp.13039
Jiang, W., Jiang, P., Li, Y., Tang, J., Wang, X., Ma, S. (2006). Recombinant adenovirus expressing GP5 and m fusion proteins of porcine reproductive and respiratory syndrome virus induce both humoral and cell-mediated immune responses in mice. Vet. Immunol. Immunopathol. 113, 169–180. doi: 10.1016/j.vetimm.2006.05.001
Jiang, Y., Li, G., Yu, L., Li, L., Zhang, Y., Zhou, Y., et al. (2020). Genetic diversity of porcine reproductive and respiratory syndrome virus (PRRSV) from 1996 to 2017 in China. Front. Microbiol. 11, 618. doi: 10.3389/fmicb.2020.00618
Kay, R., Chan, A., Daly, M., McPherson, J. (1987). Duplication of CaMV 35S promoter sequences creates a strong enhancer for plant genes. Science 236, 1299–1302. doi: 10.1126/science.236.4806.1299
Kimman, T. G., Cornelissen, L. A., Moormann, R. J., Rebel, J. M. J., Stockhofe-Zurwieden, N. (2009). Challenges for porcine reproductive and respiratory syndrome virus (PRRSV) vaccinology. Vaccine 27, 3704–3718. doi: 10.1016/j.vaccine.2009.04.022
Kolotilin, I., Topp, E., Cox, E., Devriendt, B., Conrad, U., Joensuu, J., et al. (2014). Plant-based solutions for veterinary immunotherapeutics and prophylactics. Vet. Res. 45, 117–117. doi: 10.1186/s13567-014-0117-4
Li, J., Tao, S., Orlando, R., Murtaugh, M. P. (2015). N-glycosylation profiling of porcine reproductive and respiratory syndrome virus envelope glycoprotein 5. Virology 478, 8698. doi: 10.1016/j.virol.2015.02.013
Liu, G., Bogaj, K., Bortolaia, V., Olsen, J. E., Thomsen, L. E. (2019). Antibiotic-induced, increased conjugative transfer is common to diverse naturally occurring ESBL plasmids in Escherichia coli. Front. Microbiol. 10, 2119. doi: 10.3389/fmicb.2019.02119
Lomonossoff, G. P., Wege, C. (2018). TMV particles: The journey from fundamental studies to bionanotechnology applications. Adv. Virus Res. 102, 149–176. doi: 10.1016/bs.aivir.2018.06.003
López-Sagaseta, J., Malito, E., Rappuoli, R., Bottomley, M. J. (2016). Self-assembling protein nanoparticles in the design of vaccines. Comput. Struct. Biotechnol. J. 14, 5868. doi: 10.1016/j.csbj.2015.11.001
Lu, B., Stubbs, G., Culver, J. N. (1996). Carboxylate interactions involved in the disassembly of tobacco mosaic tobamovirus. Virology 225, 11–20. doi: 10.1006/viro.1996.0570
MacDonald, J., Doshi, K., Dussault, M., Hall, J. C., Holbrook, L., Jones, G., et al. (2015). Bringing plant-based veterinary vaccines to market: Managing regulatory and commercial hurdles. Biotechnol. Adv. 33, 1572–1581. doi: 10.1016/j.biotechadv.2015.07.007
Mardassi, H., Massie, B., Dea, S. (1996). Intracellular synthesis, processing, and transport of proteins encoded by ORFs 5 to 7 of porcine reproductive and respiratory syndrome virus. Virology 221, 98–112. doi: 10.1006/viro.1996.0356
Margolin, E. A., Strasser, R., Chapman, R., Williamson, A.-L., Rybicki, E. P., Meyers, A. E. (2020). Engineering the plant secretory pathway for the production of next-generation pharmaceuticals. Trends Biotechnol. 38, 1034–1044. doi: 10.1016/j.tibtech.2020.03.004
Marillonnet, S., Werner, S. (2015). Assembly of multigene constructs using golden gate cloning. Methods Mol. Biol. 1321, 269–284. doi: 10.1007/978-1-4939-2760-9_19
Marsian, J., Lomonossoff, G. P. (2016). Molecular pharming - VLPs made in plants. Curr. Opin. Biotechnol. 37, 201–206. doi: 10.1016/j.copbio.2015.12.007
Miletic, S., Simpson, D. J., Szymanski, C. M., Deyholos, M. K., Menassa, R. (2016). A plant- produced bacteriophage tailspike protein for the control of Salmonella. Front. Plant Sci. 6, 1221. doi: 10.3389/fpls.2015.01221
Modi, S. R., Lee, H. H., Spina, C. S., Collins, J. J. (2013). Antibiotic treatment expands the resistance reservoir and ecological network of the phage metagenome. Nature 499, 219–222. doi: 10.1038/nature12212
Nielsen, H. S., Oleksiewicz, M. B., Forsberg, R., Stadejek, T., Bøtner, A., Storgaard, T. (2001). Reversion of a live porcine reproductive and respiratory syndrome virus vaccine investigated by parallel mutations. J. Gen. Virol. 82, 1263–1272. doi: 10.1099/0022-1317-82-6-1263
Okuda, Y., Kuroda, M., Ono, M., Chikata, S., Shibata, I. (2008). Efficacy of vaccination with porcine reproductive and respiratory syndrome virus following challenges with field isolates in Japan. J. Vet. Med. Sci. 70, 1017–1025. doi: 10.1292/jvms.70.1017
Paploski, I. A. D., Corzo, C., Rovira, A., Murtaugh, M. P., Sanhueza, J. M., Vilalta, C., et al. (2019). Temporal dynamics of co-circulating lineages of porcine reproductive and respiratory syndrome virus. Front. Microbiol. 10, 2486. doi: 10.3389/fmicb.2019.02486
Pelham (1990). The retention signal for soluble proteins of the endoplasmic reticulum. Trends Biochem. Sci. 15, 483–486. doi: 10.1016/0968-0004(90)90303-S
Pileri, E., Mateu, E. (2016). Review on the transmission porcine reproductive and respiratory syndrome virus between pigs and farms and impact on vaccination. Vet. Res. 47, 108. doi: 10.1186/s13567-016-0391-4
Pinto Ferreira, J., Battaglia, D., Dorado García, A., Tempelman, K., Bullon, C., Motriuc, N., et al. (2022). Achieving antimicrobial stewardship on the global scale: challenges and opportunities. Microorganisms 10, 1599. doi: 10.3390/microorganisms10081599
Pogue, G. P., Lindbo, J. A., McCulloch, M. J., Lawrence, J. E., Gross, C. S., Garger, S. J. (2007). Inventors; Large scale biology corporation, assignee. production of a parvovirus vaccine in plants as viral coat protein fusions. . United States patent US 7,270, 825 B2.
Popescu, L. N., Trible, B. R., Chen, N., Rowland, R. R. R. (2017). GP5 of porcine reproductive and respiratory syndrome virus (PRRSV) as a target for homologous and broadly neutralizing antibodies. Vet. Microbiol. 209, 90–96. doi: 10.1016/j.vetmic.2017.04.016
R Core Team. (2020). R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. Available at:https://www.R-project.org
Robinson, P. J., Bulleid, N. J. (2020). Mechanisms of disulfide bond formation in nascent polypeptides entering the secretory pathway. Cells 9, 1994. doi: 10.3390/cells9091994
Rupasinghe, R., Lee, K., Liu, X., Gauger, P. C., Zhang, J., Martínez-López, B. (2022). Molecular evolution of porcine reproductive and respiratory syndrome virus field strains from two swine production systems in the midwestern united states from 2001 to 2020. Microbiol. Spectr. 10, e0263421. doi: 10.1128/spectrum.02634-21
Rybicki, E. P. (2009). Plant-produced vaccines: promise and reality. Drug Discovery Today 14, 6–24. doi: 10.1016/j.drudis.2008.10.002
Schillberg, S., Finnern, R. (2021). Plant molecular farming for the production of valuable proteins - critical evaluation of achievements and future challenges. J. Plant Physiol. 258259, 153359. doi: 10.1016/j.jplph.2020.153359
Silhavy, D., Molnár, A., Lucioli, A., Szittya, G., Hornyik, C., Tavazza, M., et al. (2002). A viral protein suppresses RNA silencing and binds silencing-generated, 21- to 25- nucleotide double-stranded RNAs. EMBO J. 21, 3070–3080. doi: 10.1093/emboj/cdf312
Solá, R. J., Griebenow, K. (2009). Effects of glycosylation on the stability of protein pharmaceuticals. J. Pharm. Sci. 98, 1223–1245. doi: 10.1002/jps.21504
Stoian, A. M. M., Rowland, R. R. R. (2019). Challenges for porcine reproductive and respiratory syndrome (PRRS) vaccine design: Reviewing virus glycoprotein interactions with CD163 and targets of virus neutralization. Vet. Sci. 6, 9. doi: 10.3390/vetsci6010009
Strasser, R. (2018). Protein quality control in the endoplasmic reticulum of plants. Annu. Rev. Plant Biol. 69, 147–172. doi: 10.1146/annurev-arplant-042817-040331
Strasser, R., Seifert, G., Doblin, M. S., Johnson, K. L., Ruprecht, C., Pfrengle, F., et al. (2021). Cracking the “sugar code”: a snapshot of n- and O-glycosylation pathways and functions in plants cells. Front. Plant Sci. 12, 640919. doi: 10.3389/fpls.2021.640919
Topp, E., Irwin, R., McAllister, T., Lessard, M., Joensuu, J. J., Kolotilin, I., et al. (2016). The case for plant-made veterinary immunotherapeutics. Biotechnol. Adv. 34, 597–604. doi: 10.1016/j.biotechadv.2016.02.007
Vetter, V., Denizer, G., Friedland, L. R., Krishnan, J., Shapiro, M. (2018). Understanding modern-day vaccines: what you need to know. Ann. Med. 50, 110120. doi: 10.1080/07853890.2017.1407035
Vitale, A., Raikhel, N. V. (1999). What do proteins need to reach different vacuoles? Trends Plant Sci. 4, 149–155. doi: 10.1016/s1360-1385(99)01389-8
Wang, A., Chen, Q., Wang, L., Madson, D., Harmon, K., Gauger, P., et al. (2019). Recombination between vaccine and field strains of porcine reproductive and respiratory syndrome virus. Emerg. Infect. Dis. 25, 2335–2337. doi: 10.3201/eid2512.191111
Wang, J. W., Roden, R. B. S. (2013). Virus-like particles for the prevention of human papillomavirus-associated malignancies. Expert Rev. Vaccines 12, 129–141. doi: 10.1586/erv.12.151
Ward, B. J., Gobeil, P., Séguin, A., Atkins, J., Boulay, I., Charbonneau, P.-Y., et al. (2021). Phase 1 randomized trial of a plant-derived virus-like particle vaccine for COVID-19. Nat. Med. 27, 1071–1078. doi: 10.1038/s41591-021-01370-1
Wei, Z., Lin, T., Sun, L., Li, Y., Wang, X., Gao, F., et al. (2012). N-linked glycosylation of GP5 of porcine reproductive and respiratory syndrome virus is critically important for virus replication in vivo. J. Virol. 86, 99419951. doi: 10.1128/JVI.07067-11
Wissink, E. H., Kroese, M. V., van Wijk, H. A., Rijsewijk, F. A., Meulenberg, J. J., Rottier, P. J. (2005). Envelope protein requirements for the assembly of infectious virions of porcine reproductive and respiratory syndrome virus. J. Virol. 79, 12495–12506. doi: 10.1128/JVI.79.19.12495-12506.2005
Wu, K., Malik, K., Tian, L., Hu, M., Martin, T., Foster, E., et al. (2001). Enhancers and core promoter elements are essential for the activity of a cryptic gene activation sequence from tobacco, tCUP. Mol. Genet. Genomics 265, 763–770. doi: 10.1007/s004380100478
Zhang, M., Han, X., Osterrieder, K., Veit, M. (2021). Palmitoylation of the envelope membrane proteins GP5 and m of porcine reproductive and respiratory syndrome virus is essential for virus growth. PloS Pathog. 17, e1009554. doi: 10.1371/journal.ppat.1009554
Zhao, L., Seth, A., Wibowo, N., Zhao, C. X., Mitter, N., Yu, C., et al. (2014). Nanoparticle vaccines. Vaccine 32, 327–337. doi: 10.1016/j.vaccine.2013.11.069
Zuckermann, F. A., Garcia, E. A., Luque, I. D., Christopher-Hennings, J., Doster, A., Brito, M., et al. (2007). Assessment of the efficacy of commercial porcine reproductive and respiratory syndrome virus (PRRSV) vaccines based on measurement of serologic response, frequency of gamma-IFN-producing cells and virological parameters of protection upon challenge. Vet. Microbiol. 123, 69–85. doi: 10.1016/j.vetmic.2007.02.009
Keywords: PRRS (porcine reproductive and respiratory syndrome virus), PRRSV, subunit vaccine, virus-like particle, tobacco mosaic virus, plant molecular farming
Citation: VanderBurgt JT, Harper O, Garnham CP, Kohalmi SE and Menassa R (2023) Plant production of a virus-like particle-based vaccine candidate against porcine reproductive and respiratory syndrome. Front. Plant Sci. 14:1044675. doi: 10.3389/fpls.2023.1044675
Received: 14 September 2022; Accepted: 10 January 2023;
Published: 24 January 2023.
Edited by:
Linda Avesani, University of Verona, ItalyReviewed by:
Richard Strasser, University of Natural Resources and Life Sciences Vienna, AustriaCopyright © 2023 VanderBurgt, Harper, Garnham, Kohalmi and Menassa. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rima Menassa, cmltYS5tZW5hc3NhQGFnci5nYy5jYQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.