
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci., 10 February 2023
Sec. Plant Pathogen Interactions
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1041868
This article is part of the Research TopicPlant-Virus Interactions: Crop Resistance in FocusView all 5 articles
 Nikolay Zlobin*
Nikolay Zlobin* Vasiliy Taranov
Vasiliy TaranovPotyviruses are the largest group of plant-infecting RNA viruses that affect a wide range of crop plants. Plant resistance genes against potyviruses are often recessive and encode translation initiation factors eIF4E. The inability of potyviruses to use plant eIF4E factors leads to the development of resistance through a loss-of-susceptibility mechanism. Plants have a small family of eIF4E genes that encode several isoforms with distinct but overlapping functions in cell metabolism. Potyviruses use distinct eIF4E isoforms as susceptibility factors in different plants. The role of different members of the plant eIF4E family in the interaction with a given potyvirus could differ drastically. An interplay exists between different members of the eIF4E family in the context of plant–potyvirus interactions, allowing different eIF4E isoforms to modulate each other’s availability as susceptibility factors for the virus. In this review, possible molecular mechanisms underlying this interaction are discussed, and approaches to identify the eIF4E isoform that plays a major role in the plant–potyvirus interaction are suggested. The final section of the review discusses how knowledge about the interaction between different eIF4E isoforms can be used to develop plants with durable resistance to potyviruses.
Plant viruses are microscopic organisms with a small genome that typically encodes a dozen proteins. It is insufficient to complete the different stages of the viral life cycle, i.e., multiplication within cells, cell-to-cell movement, and long-distance systemic movement in the plant. To accomplish this, viruses have to use various host proteins. These proteins function as susceptibility factors that allow a compatible interaction between the virus and the plant. In theory, the unavailability of even one of these factors for the virus in the plant cell will make the interaction incompatible and lead to resistance. This type of resistance, known as loss of susceptibility, has been observed in various plant–pathogen systems, mainly for obligate biotrophic pathogens such as viruses (Pavan et al., 2010). Unlike more well-recognized resistance mechanisms that rely on R-genes to recognize pathogen effectors and mount defense responses, resistance through loss of susceptibility is generally characterized by increased durability and spectrum (Pavan et al., 2010; van Schie and Takken, 2014).
Because loss of susceptibility is based on the nonavailability of a certain cell factor to the pathogen, this resistance is inherited recessively. The proportion of recessively inherited resistance genes to plant viruses is extraordinarily high (Truniger and Aranda, 2009). This is especially true for potyviruses, the largest group of plant-infecting RNA viruses that cause severe crop losses worldwide (Yang et al., 2021). Less than half of the genes that confer resistance to these viruses are inherited recessively (Provvidenti and Hampton, 1992; Truniger and Aranda, 2009). The potential diversity of the host genes that encode susceptibility factors in plant–potyvirus interactions is enormous. The ability to promote potyviral infection was shown for numerous cellular proteins, from enzymes (Beffa et al., 1996; Ouibrahim et al., 2014; Poque et al., 2015) and carrier proteins (Park et al., 2017; Wu et al., 2020; Soler-Garzón et al., 2021a) to transcription factors (Endres et al., 2010; Soler-Garzón et al., 2021b), as well as many other proteins with diverse functions (Hofius et al., 2007; Castelló et al., 2010; Hafrén et al., 2010; Zhu et al., 2014; De et al., 2020; Bruckner et al., 2022; Zuo et al., 2022). However, the study of the genes that confer natural recessive resistance to potyviruses in various crop plants revealed that the molecular basis of this resistance is surprisingly similar in most cases. In different plants, natural recessive resistance to numerous potyviruses was shown to rely on the polymorphism of just a single type of cell factor, i.e., the translation initiation factor eIF4E. In some of these plants, several independently discovered recessive resistance genes were eventually shown to encode the same eIF4E factor (Table 1). Therefore, eIF4E-mediated resistance to potyviruses has evolved numerous times in systematically distinct plant species independently. Moreover, eIF4E-mediated resistance to various groups of viruses other than Potyviruses (Bymoviruses, Potexviruses, Tritimoviruses, Ipomoviruses, Carmoviruses, Carlaviruses, and Cucumoviruses) was discovered in nature (Kanyuka et al., 2005; Stein et al., 2005; Nieto et al., 2006; Nieto et al., 2007) or artificially generated (Yoshii et al., 2004; Keima et al., 2017; Rodríguez-Hernández et al., 2012; Rupp et al., 2019; Chen et al., 2022b).
The recessive nature of eIF4E-mediated resistance implies the inability of viruses to use certain allelic variants of eIF4E as susceptibility factors to complete their life cycles within plants. This type of resistance can manifest on different levels, including the inhibition of virus multiplication within the cell (Deom et al., 1997; Murphy et al., 1998; Lellis et al., 2002; Sato et al., 2005), cell-to-cell movement (Arroyo et al., 1996; Sato et al., 2003; Gao et al., 2004), and long-distance and systemic movement (Acosta-Leal and Xiong, 2008; Contreras-Paredes et al., 2013). This suggests that eIF4Es have a wide range of functions during the viral life cycle, which is presumably one of the causes of their prevalence as susceptibility factors for different viruses.
eIF4E-mediated resistance is often durable and is not overcome by viruses in the field. Some recessive genes encoding eIF4E have been widely used in agriculture for over half a century and have not been broken down (van Schie and Takken, 2014). EIF4E-mediated resistance is also characterized by a broad spectrum, because a single eIF4E allele often provides resistance to different viral strains and isolates or even to different viruses (Rusholme et al., 2007; Andrade et al., 2009; Moury et al., 2014a; Gauffier et al., 2016). In contrast to many other resistance mechanisms mostly discovered in wild relatives of cultivated plants, eIF4E-mediated resistance occurs more frequently in domesticated plant populations than in wild ones (Hofinger et al., 2011; Konečná et al., 2014; Poulicard et al., 2016). This suggests that this type of resistance is particularly suited to cultivated plants and is associated with mild pleiotropic effects on crop growth and productivity. Together, these advantages have led to the widespread use of eIF4E-mediated resistance in agriculture.
eIF4E is one of the factors constituting the translation initiation complex. eIF4E binds to the 5′-cap of mRNA and eIF4G, leading to the formation of the eIF4F complex, which is recruited to the 40S ribosome subunit together with the other translation initiation factors. In addition to translation, eIF4E participates in other mRNA-related processes, in particular the transport of mRNA from the nucleus to the cytoplasm (Wang and Krishnaswamy, 2012).
In plants, there is not a single eIF4E factor; rather, they constitute a small family (Figure 1). In addition to eIF4E, flowering plants contain its isoform, which is known as eIF(iso)4E (Patrick and Browning, 2012). Although the members of this family have a relatively low level of sequence homology, they probably have a similar three-dimensional structure (Estevan et al., 2014) and overlapping functions. eIF4E1 inactivation in Arabidopsis thaliana was associated with mild pleiotropic effects under laboratory conditions; the resulting plants were smaller and had a 7-day delayed bolting and a slightly decreased seed yield (Sato et al., 2005; Bastet et al., 2018). The effects of eIF(iso)4E inactivation on Arabidopsis were even weaker, with only one study reporting some changes in root development (Duprat et al., 2002; Lellis et al., 2002; Sato et al., 2005; Martínez-Silva et al., 2012). Arabidopsis eIF4E1 was also shown to be involved in root growth regulation (Liu et al., 2022). In contrast, double knockout of both eIF4E1 and eIF(iso)4E in Arabidopsis was lethal (Callot and Gallois, 2014). Arabidopsis eIF4E1 or eIF(iso)4E inactivation decreased tolerance to low temperatures (Salazar-Díaz et al., 2021), while had no effect on elf18-induced resistance against the bacteria Pseudomonas syringae pv. maculicola (Wang et al., 2022). Cucumber plants with eIF4E knockout were viable and fertile, and pepper varieties containing inactivated eIF(iso)4E are used in agriculture (Ruffel et al., 2006; Chandrasekaran et al., 2016). In contrast, targeted inactivation of the eIF4E gene in barley leads to a yield penalty and to male sterility in melons (Hoffie et al., 2021; Pechar et al., 2022). eIF4E and eIF(iso)4E participate in the translation of various sets of mRNAs (Dinkova et al., 2011; Martínez-Silva et al., 2012). In Nicotiana benthamiana, eIF(iso)4E was suggested to be primarily associated with rough endoplasmic reticulum, whereas eIF4E was suggested to be primarily associated with free ribosomes (Beauchemin et al., 2007). The difference in eIF4E and eIF(iso)4E functions may be more pronounced in plants grown under stress conditions as opposed to the optimal growth conditions used in most studies (Martínez-Silva et al., 2012).
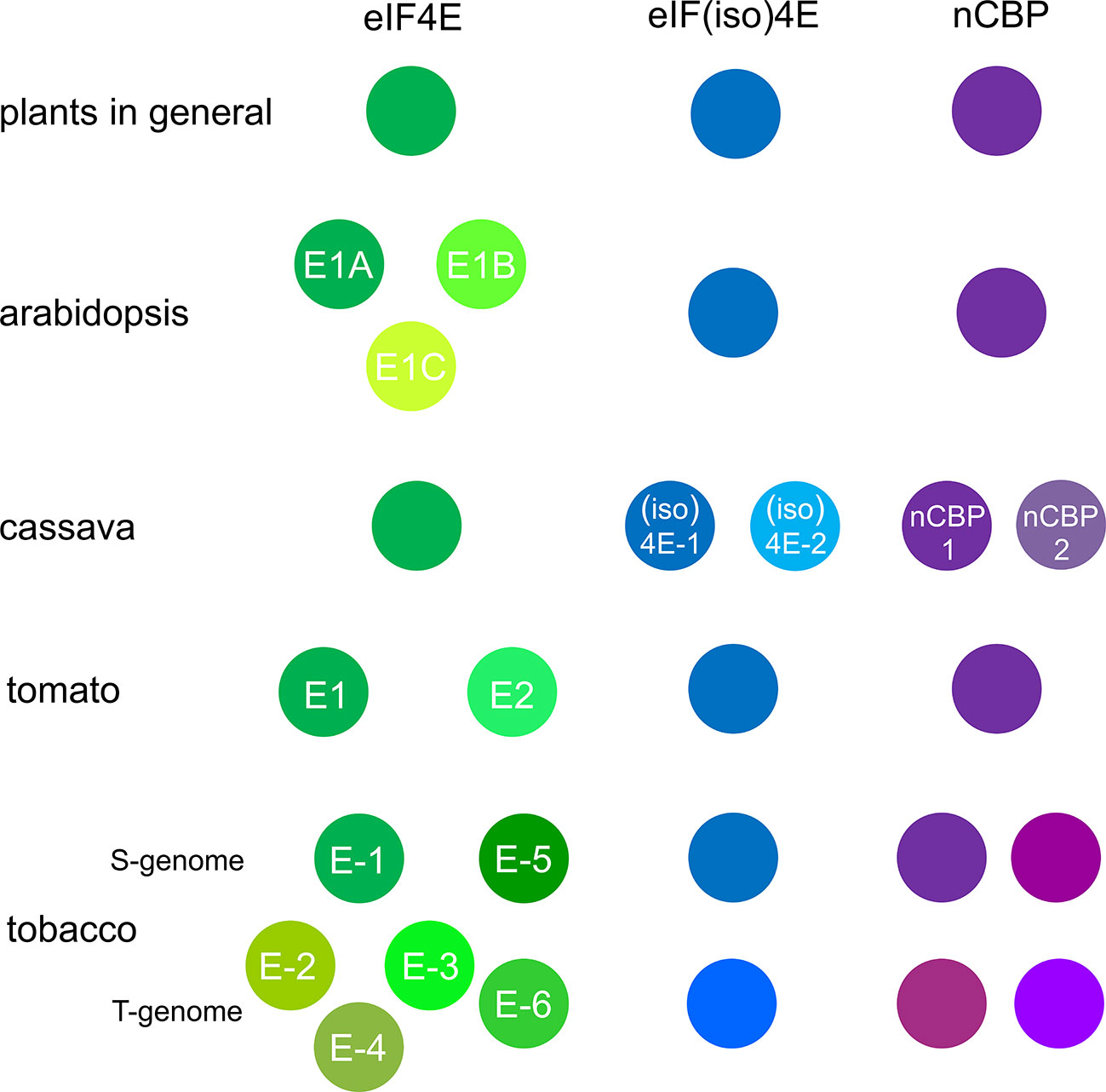
Figure 1 Plants contain several eIF4E isoforms. The names of some additional isoform members were as per those given by Patrick et al. (2014) for Arabidopsis, Gomez et al. (2019) for cassava, Piron et al. (2010) for tomato, and Michel et al. (2019) for tobacco.
In addition to eIF4E and eIF(iso)4E, plants have another eIF4E family member known as nCBP (from novel cap-binding protein) (Ruud et al., 1998; Patrick and Browning, 2012). nCBP binds to the 5′-cap and interacts with eIF(iso)4G in vitro (Ruud et al., 1998; Kropiwnicka et al., 2015). Its functions in the context of plant growth and development have been barely studied (Christie and Igreja, 2021). In potatoes, nCBP knockdown causes deformation of young leaves (Chen et al., 2022a).
Additional eIF4E factors have been discovered in different plants (Figure 1) (Patrick and Browning, 2012; Patrick et al., 2014). In addition to eIF4E1, eIF(iso)4E, and nCBP, Arabidopsis plants have a locus that contains two other eIF4E genes EIF4E1B (also EIF4E3) and EIF4E1C (also EIF4E2) (Patrick et al., 2014). Although these factors have a high sequence homology to eIF4E1 (eIF4E1A), can bind m7GTP and eIF4G, and can initiate translation in yeast cells, they cannot compensate for the depletion of eIF4E1 and eIF(iso)4E in Arabidopsis (Patrick et al., 2014). The Brassica rapa genome comprises three genes encoding eIF4E and three genes encoding eIF(iso)4E, which are denoted by the letters a, b, and c, with one of the eIF4Es (eIF4E.a) being a pseudogene (Jenner et al., 2010; Nellist et al., 2014). Cassava has a single eIF4E gene but two eIF(iso)4E and two nCBP genes, all of which are expressed, albeit at significantly different levels (Shi et al., 2017; Gomez et al., 2019). In the genomes of different plants of the Solanaceae family, i.e., pepper, tomato, and potato, two homologous eIF4E variants, eIF4E1 and eIF4E2, were reported (Piron et al., 2010; Lebaron et al., 2016; Lucioli et al., 2022). In tomato, simultaneous inactivation of eIF4E1 and eIF4E2 genes caused dwarfism, in contrast to the separate inactivation of eIF4E1 or eIF4E2 (Gauffier et al., 2016; Kumar et al., 2022); interestingly, the double knockout of eIF(iso)4E and eIF4E2 in tomato was lethal (Bastet et al., 2017). Cultivated Nicotiana tabacum, which is an allotetraploid derived from the natural interspecific hybridization between Nicotiana sylvestris (S-genome) and Nicotiana tomentosiformis (T-genome), contains as many as 12 genes encoding eIF4Es, i.e., six eIF4E (eIF4E-1 to eIF4E-6), two eIF(iso)4E, and four nCBP (Julio et al., 2015; Michel et al., 2019). In addition, some tobacco varieties have additional copies of some eIF4E genes from N. tomentosiformis and even recombinants between these genes (Michel et al., 2019).
A molecular genetic analysis of recessive genes that determine potyvirus resistance in different crop plants revealed that most of them encode an eIF4E isoform, while some encode eIF(iso)4E (Table 1). Although different potyviruses use different eIF4E isoforms, resistance to certain potyvirus usually relies on the allelic composition of a single gene (Table 1). This suggests that specific potyvirus generally rely on a single eIF4E isoform as a susceptibility factor from several potentially available isoforms in the host plant.
In rare cases, natural eIF4E-mediated resistance is controlled by two recessive genes simultaneously. Pepper resistance to Chilli veinal mottle virus (ChiVMV) or pepper veinal mottle virus (PVMV) requires certain alleles of both eIF4E1 and eIF(iso)4E (Ruffel et al., 2006; Hwang et al., 2009; Rubio et al., 2009). Surprisingly, these viruses use pepper eIF4E1 and eIF(iso)4E but not eIF4E1 and eIF4E2, which share more similar primary structures.
In plants containing several members of some eIF4E isoforms, their roles as susceptibility factors to potyviruses may differ remarkably. For example, tobacco contains as many as six variants of eIF4E inherited from both N. sylvestris and N. tomentosiformis (Figure 1) (Julio et al., 2015; Michel et al., 2019). Although all of these variants are classified as eIF4E, only one is a susceptibility factor for potyviruses potato virus Y (PVY), tobacco vein mottling virus (TVMV), and tobacco etch virus (TEV) (Rufty et al., 1989; Nicolas et al., 1997; Julio et al., 2015). This variant is inherited by tobacco from N. sylvestris and is known as eIF4E-1 (Michel et al., 2019) or eIF4E1-S (Takakura et al., 2018). Similarly, in B. rapa, the roles of the three eIF(iso)4E variants, i.e., eIF(iso)4E.a, b, and c, in turnip mosaic virus (TuMV) infection are also very different (Nellist et al., 2014).
Sometimes a single potyvirus uses different eIF4E isoforms in different plants. For example, TEV and lettuce mosaic virus LMV use eIF4E in pepper and lettuce (Ruffel et al., 2002; Nicaise et al., 2003) but rely on eIF(iso)4E in Arabidopsis (Lellis et al., 2002; Estevan et al., 2014). PVMV uses both eIF4E1 and eIF(iso)4E to infect pepper (Ruffel et al., 2006) but requires eIF4E2 to infect another Solanaceae family member, tomato (Moury et al., 2020).
Currently, a large number of eIF4E allelic variants associated with resistance to potyviruses have been reported. The comparative analysis of these alleles from susceptible and resistant plants revealed that their amino acid sequences are generally highly similar. eIF4E alleles from resistant plants often differ from alleles from susceptible ones by only a few (generally 2–4) amino acid substitutions (Gao et al., 2004; Ruffel et al., 2005; Charron et al., 2008; Hart and Griffiths, 2013). They are mostly nonconserved, leading to considerable changes in the charge or hydrophobicity of the corresponding protein regions (Nicaise et al., 2003; Gao et al., 2004; Charron et al., 2008; Poulicard et al., 2016). These substitutions are located in the region of the eIF4E molecule that interacts directly with the potyviral protein VPg (Gao et al., 2004; Charron et al., 2008; Perez et al., 2012). This protein is covalently bound to the potyviral mRNA and mimics the 5′-cap of cellular mRNAs, thus allowing the translation of the potyviral polyprotein and providing some other eIF4E-mediated functions in the viral life cycle, such as cell-to-cell transport (Wang and Krishnaswamy, 2012; Tavert-Roudet et al., 2017). Nonconserved substitutions in eIF4E disrupt its binding to VPg (Kang et al., 2005; Charron et al., 2008), thus hindering viral multiplication and/or spread in the plant. If the plant only has resistant alleles of the eIF4E isoform, which the virus uses as a susceptibility factor, it is resistant to this virus.
Either under natural infectious conditions or after artificial inoculation, some plants with resistant eIF4E alleles in the homozygous state after viral inoculation eventually display disease symptoms (Ayme et al., 2006; Naderpour et al., 2010; Moury et al., 2014a; Moury et al., 2014b; Congdon et al., 2016; Lebaron et al., 2016). Only a small percentage of plants developed disease, and symptom appearance was significantly delayed. This indicates that potyviruses can overcome the loss of susceptibility mediated by polymorphisms in eIF4E, resulting in resistance breaking (RB). In potyvirus samples isolated from infected plants, point mutations that led to amino acid substitutions in viral proteins were found. Although RB could be achieved through mutations in various potyviral proteins (Hjulsager et al., 2002; Nakahara et al., 2010; Sorel et al., 2014) and possibly in the viral mRNA itself (Krause-Sakate et al., 2005), the most common way to overcome eIF4E-mediated resistance in different plant–potyvirus pathosystems is through single or multiple substitutions in the VPg protein (Borgstrøm and Johansen, 2001; Sato et al., 2003; Moury et al., 2004; Ayme et al., 2007; Bruun-Rasmussen et al., 2007; Perez et al., 2012; Moury et al., 2014a; Moury et al., 2014b; Lebaron et al., 2016). Biochemical studies have shown that these mutations allow the re-interaction of VPg with the resistant eIF4E allele, allowing potyviruses to resume the use of the corresponding eIF4E isoform in planta as a susceptibility factor (Charron et al., 2008; Truniger and Aranda, 2009; Perez et al., 2012).
The appearance of mutations in VPg that allow the overcoming of resistance represents potyvirus evolution, which necessitates viral multiplication in the cells of resistant plants. It was discovered that low level, asymptomatic accumulation of potyviruses can occur in resistant plants (Acosta-Leal and Xiong, 2008; Montarry et al., 2011). This phenomenon is also known as subliminal infection (Acosta-Leal and Xiong, 2008). Although subliminal infections do not cause significant damage to the plant, viral multiplication allows for the emergence of mutations, some of which will lead to changes in VPg and its re-interaction with resistant eIF4E alleles (Montarry et al., 2011). Asymptomatic accumulation of potyviruses in plant tissues, which enables their evolution, has been shown to be the major determinant of the RB (Quenouille et al., 2013; Moury et al., 2016).
Faint multiplication in plants with resistant eIF4E alleles suggests that viruses can use these alleles for multiplication, although with very low efficiency, as indicated by a low infection level. Some natural eIF4E alleles, such as pepper pvr1 or pvr22, appear to be capable of completely blocking viral multiplication at the cellular level (Deom et al., 1997; Murphy et al., 1998) and providing extremely durable resistance (van Schie and Takken, 2014). Seemingly, these alleles are hardly used by potyviruses, even for low-level multiplication. In contrast, other alleles, such as pvr23 or pvr21, which allow for subliminal infection (Arroyo et al., 1996; Montarry et al., 2011), are less durable (Ben Khalifa et al., 2012; Moury et al., 2014a; Moury et al., 2014b).
In all studied cases, potyviruses overcame resistance based on natural polymorphisms in certain eIF4E isoforms via re-interaction with this isoform, which was made possible by acquiring mutations in VPg (Charron et al., 2008; Perez et al., 2012; Lebaron et al., 2016). Thus, the obvious way to develop plants resistant to potyviruses is to inactivate this eIF4E isoform, depriving the virus of this crucial host factor. As mentioned above, plants with an inactivated single eIF4E isoform are often viable and do not exhibit severe abnormalities because of the partial redundancy between eIF4E isoforms.
To date, resistance to diverse potyviruses has been achieved in different plants by inactivating the eIF4E isoform (Duprat et al., 2002; Sato et al., 2005; Piron et al., 2010; Julio et al., 2015; Chandrasekaran et al., 2016; Pechar et al., 2022). However, in many cases, a fraction of plants with inactivated eIF4E eventually develop disease, although at a slower rate than the wild-type plants, indicating the RB process (Duprat et al., 2002; Julio et al., 2015; Chandrasekaran et al., 2016; Pechar et al., 2022). Therefore, rather than providing complete immunity, inactivating the single eIF4E isoform often provides only a nondurable resistance to potyviruses that use this isoform as a susceptibility factor.
Potyviruses require eIF4E to perform various functions related to translation as well as other stages of their life cycle. This means that potyviruses have to use another factor in plants with inactivated eIF4E isoform. Viruses may bypass its absence by using either another eIF4E isoform or non-eIF4E cell factors. Although the latter cannot be excluded (Gallie, 2001; Khan et al., 2008), the main way to bypass the lack of the required eIF4E isoform is to use an alternative variant of the eIF4E isoform. This has been proven by the fact that plants with simultaneous inactivation of two eIF4E isoforms often demonstrate immunity to one or several potyviruses. Arabidopsis plants lacking both eIF4E1 and eIF(iso)4E were resistant to TuMV strains that overcome resistance based on the inactivation of eIF(iso)4E alone (Bastet et al., 2018). In tomato, resistance to multiple viruses was achieved by inactivating both eIF4E1 and eIF4E2 (Gauffier et al., 2016). In tobacco, the knockout of a single eIF4E gene was overcome by PVY (Julio et al., 2015), whereas the inactivation of two or more eIF4E factors from multiple ones in this plant rendered tobacco almost immune to PVY (Udagawa et al., 2021; Le et al., 2022). Therefore, potyviruses overcome resistance based on the absence of a particular eIF4E isoform primarily, if not exclusively, via the use of another eIF4E isoform.
Usually, potyvirus must undergo adaptation to use an alternative variant of the eIF4E isoform. This is accomplished by acquiring mutations in the VPg protein, which allow VPg to interact with the new eIF4E isoform. In the tobacco–PVY pathosystem, the VPg of strains that overcame eIF4E-1 inactivation carried substitutions, most often at position 105 of this protein (Michel et al., 2019), allowing VPg to bind one of the tobacco eIF(iso)4Es (Takakura et al., 2018). In TuMV, substitutions at position 116 or 163 of VPg were required to use eIF4E1 in addition to eIF(iso)4E in Arabidopsis (Gallois et al., 2010; Bastet et al., 2018).
The accumulation of mutations in VPg takes time and manifests as a delay in the development of the symptoms of infection. In plants with inactivated eIF4E isoforms, subliminal infections were detected, allowing potyviral evolution (Acosta-Leal and Xiong, 2008; Acosta-Leal and Xiong, 2013). Subliminal infections indicate that these viruses can use alternative eIF4E isoforms, although inefficiently, because viral accumulation is very low. During these subliminal infections, variants of the virus could appear with mutations in VPg, allowing the virus to use an alternative eIF4E isoform more efficiently. When this happens, the resistance mediated by eIF4E inactivation is broken, the virus accumulates excessively in plant tissues, and disease symptoms will develop.
Surprisingly, at least in some cases, potyviruses can efficiently use an alternative eIF4E isoform in plants directly without any apparent adaptation. This observation has been supported by evidence from the tomato–PVY and tomato–TEV pathosystems. eIF4E1 is the susceptibility factor to these viruses in tomato because its alleles (pot-1 and pot-12, with 4 and 2 amino acid substitutions, respectively) confer resistance to diverse PVY and TEV strains in different tomato species and interspecific hybrids (Ruffel et al., 2005; Gauffier et al., 2016; Lebaron et al., 2016). Strikingly, inactivation of eIF4E1 yields low, if any, resistance to these viruses, with the exception of a single PVY strain (Piron et al., 2010; Gauffier et al., 2016; Kumar et al., 2022). In tomato with inactivated eIF4E1, these viruses use eIF4E2 as a susceptibility factor because double knockout of eIF4E1 and eIF4E2 leads to resistance (Gauffier et al., 2016; Kumar et al., 2022). Interestingly, after infection of eIF4E1 KO tomato, no features of RB, such as infection of only a fraction of plants or delayed development of symptoms, were observed. Accordingly, no VPg mutations were observed (Kumar et al., 2022). This speaks in favor of the ability of different PVY and TEV strains to use eIF4E2 in tomato directly without any adaptation. In fact, the ability of PVY and TEV VPg to physically bind to tomato eIF4E2 as well as eIF4E1 was demonstrated in a yeast-two-hybrid screen (Mazier et al., 2011).
Thus, in different plant–potyvirus systems, in addition to the “main” eIF4E isoform, which is normally used by virus as a susceptibility factor and the allelic composition of which determines resistance in nature, potyviruses can use an alternative eIF4E isoform in plants with inactivation of the “main” susceptibility isoform. According to Moury et al. (2020), this eIF4E isoform can be characterized as a backup susceptibility factor. In the next part of this review, the following terms will be used: main susceptibility isoform, or main eIF4E isoform, and backup susceptibility isoform, or backup eIF4E isoform. Potyviruses normally use the main eIF4E isoform to infect the plant, but in its absence, they use the backup eIF4E isoform. The “main” and “backup” designations do not reflect their importance to normal cell physiology and are only used in the context of the specific plant–potyvirus interaction.
Plants typically have a single primary and a single backup eIF4E because inactivation of two eIF4E genes often provides robust resistance, in some cases to multiple potyviruses (Gauffier et al., 2016; Bastet et al., 2018; Udagawa et al., 2021). This is most likely due to potyviruses limited adaptability as they cannot adapt to use any eIF4E isoform in plant. However, some TEV and PVY strains were shown to partially retain the ability to multiplicate in tomato plants with inactivation of two isoforms (eIF4E1 and eIF4E2), although the level of infection was severely reduced (Gauffier et al., 2016; Kumar et al., 2022). Some potyviruses lack the ability to use the backup eIF4E isoform because the inactivation of the main eIF4E isoform provides a resistance that was not broken down (Piron et al., 2010). It is unclear whether this is due to the virus’s complete inability to adapt to another eIF4E isoform or to experimental conditions such as insufficient infection pressure or sample size. Interestingly, the ability of different strains of the same potyvirus to use the backup eIF4E isoform was shown to differ (Piron et al., 2010; Atarashi et al., 2020).
In some cases, inactivating the main eIF4E isoform did not suppress viral multiplication within cells but restricted its spread in plant tissues (Contreras-Paredes et al., 2013; Julio et al., 2015). Alternatively, potyviral translation rather than replication could be impaired (Kumar et al., 2022). This suggests that potyviruses can use alternative eIF4E factors for some but not all eIF4E-mediated activities in the viral proliferation cycle.
When potyviruses adapt to use the backup eIF4E isoform, they retain the ability to interact with the main eIF4E isoform. However, the efficiency of their application may vary. The accumulation of TuMV RB strains in Arabidopsis was a few-fold lower in plants with an inactivated main eIF(iso)4E isoform than in WT plants (Bastet et al., 2018). This implies that the backup eIF4E isoform is suboptimal for TuMV, even for RB strains with mutations in VPg. In tomato, the main eIF4E1 supports PVY multiplication more efficiently compared to the backup eIF4E2 (Kumar et al., 2022). Conversely, an RB strain of PVY in tobacco uses the backup isoform, i.e., one of the tobacco eIF(iso)4E factors, more efficiently than it uses the main isoform eIF4E-1 (Takakura et al., 2018; Udagawa et al., 2021).
In diverse plant–potyvirus pathosystems, the ability of the potyvirus to infect the plant using a backup eIF4E isoform has been demonstrated. Therefore, it may appear surprising that recessive resistance to potyviruses via an eIF4E-mediated mechanism is so widespread. Apparently, to achieve such resistance, it is necessary to modify not only the main eIF4E isoform but also the backup eIF4E isoform; otherwise, potyvirus will overcome the resistance by using the latter.
However, these findings contradict the observation that, in the vast majority of cases, natural eIF4E-mediated resistance was determined by the allelic composition of a single gene (Table 1), implying that the virus depends on a single eIF4E isoform to infect the plant. The widespread use of such genes in agriculture indicates that the level of resistance provided by them is sufficient for their practical application. Moreover, sometimes single gene eIF4E-mediated resistance can be extremely durable and effective against various potyviruses (van Schie and Takken, 2014; Moury et al., 2014a; Moury et al., 2014b).
One distinguishing feature of the majority of natural resistant eIF4E genes is that they are generally not null alleles but encode apparently functional proteins. A few amino acid substitutions in these proteins prevent their use by viruses but do not usually inhibit their function as translation initiation factors. This was confirmed by the ability of the proteins encoded by resistance eIF4E alleles to perform intrinsic eIF4E functions, such as binding to the cap analog in vitro and initiating translation in yeast cells lacking their own eIF4E (Kang et al., 2005; Charron et al., 2008; Gauffier et al., 2016).
Despite the presence of backup eIF4E isoforms in plants, the resistance conferred by natural eIF4E proteins with point mutations is overcome through re-interaction with them. In tomato, the pot-12 allele of eIF4E1 conferred a low but significant level of resistance to the PVY strain N605, although it could be overcome by single or double mutations in VPg (Lebaron et al., 2016). It is uncertain why PVY must overcome the resistance conferred by pot-12 by accumulating mutations in VPg, despite the fact that tomato contains eIF4E2, which could be directly used as a susceptibility factor by PVY-N605 without any apparent adaptation (Lebaron et al., 2016; Kumar et al., 2022). In pepper, the overcoming of resistant eIF4E1 alleles by PVY and TEV also occurs by the re-acquisition of the ability to use these “resistant” proteins. For some pepper eIF4E1 alleles, it requires multiple VPg mutations, is hardly achieved in laboratory studies, and is not observed in the field (Ayme et al., 2007; Fabre et al., 2012; Ben Khalifa et al., 2012). Concomitantly, the VPgs of PVY and TEV were able to directly bind to pepper eIF4E2 in yeast two-hybrid screening without any mutations (Gauffier et al., 2016). However, for some reason, PVY and TEV are unable to use pepper eIF4E2 as a susceptibility factor in planta.
These observations led to the conclusion that natural resistant alleles of the main eIF4E isoform, which encode a functional protein with point modifications, are not just passive nonparticipants in plant–potyvirus interaction. They repress the use of alternative eIF4E isoforms as susceptibility factors (Gauffier et al., 2016; Bastet et al., 2017). Unlike functional eIF4E alleles, null alleles do not have such a repressive effect, and the resistance they impart can be relatively easily overcome by the viruses by using the backup eIF4E isoform. Hence, functional resistant alleles of the main eIF4E isoform can somehow modulate the functionality of other eIF4E isoforms as susceptibility factors. This conclusion contradicts the established view of eIF4E-mediated resistance being recessive, which requires the lack of susceptibility factors in the plant. Rather, the eIF4E isoform can inhibit the use of the other eIF4E isoforms as susceptibility factors by the virus. Because the different isoforms are encoded by nonallelic genes, it does not represent dominance; instead, it indicates an epistatic interaction between them.
It is obvious that the effectiveness of the binding of two proteins, e.g., eIF4E and VPg, will depend on their concentrations. Accordingly, the efficiency of the use of the eIF4E isoform by potyviruses depends not only on the affinity of its binding to potyviral VPg but also on the intracellular concentration of this isoform. Reducing the concentration of eIF4E in plant cells will lead to a lower efficiency of its use by the virus, whereas its increase will have the opposite effect. In fact, increased eIF4E levels promote potyviral accumulation (Schaad et al., 2000; Nicaise et al., 2003; Hafrén et al., 2013), whereas eIF4E silencing results in resistance or at least significantly suppressed potyviral accumulation in plant tissues (Hwang et al., 2009; Eskelin et al., 2011; Mazier et al., 2011; Takakura et al., 2018; Chen et al., 2022a). Artificial silencing rarely results in complete gene repression; however, the decrease in eIF4E concentration was sufficient to achieve resistance.
Do the eIF4E isoforms regulate each other’s accumulation of in plant cells? As per the abovementioned information, the answer is yes. In Arabidopsis and tobacco, the inactivation of eIF(iso)4E led to an increase in eIF4E accumulation, although the inverse was not true (Duprat et al., 2002; Combe et al., 2005; Zafirov et al., 2021). In tomato, eIF4E1 knockout resulted in an increase in eIF4E2 levels (Gauffier et al., 2016). In the context of potyviral infection, knockout of the main eIF4E isoform will result in the compensatory overaccumulation of the remaining eIF4E isoforms in plant cells. Consequently, higher concentration of the backup eIF4E isoform will facilitate its binding to potyviral VPg and promote its use as a susceptibility factor by virus (Figure 2A). Conversely, in the presence of a functional resistant allele of the main eIF4E isoform, a compensatory increase in the backup eIF4E isoform concentration does not occur, which impedes its use by the virus. Tomato genotypes with a resistant eIF4E1 allele and decreased eIF4E2 concentration exhibited robust resistance to different strains of PVY and TEV, which used eIF4E2 as a backup susceptibility factor (Gauffier et al., 2016). In contrast, tomato plants with inactivated eIF4E1 accompanied by a compensatory increase in eIF4E2 concentration were susceptible to these viruses (Gauffier et al., 2016; Kumar et al., 2022).
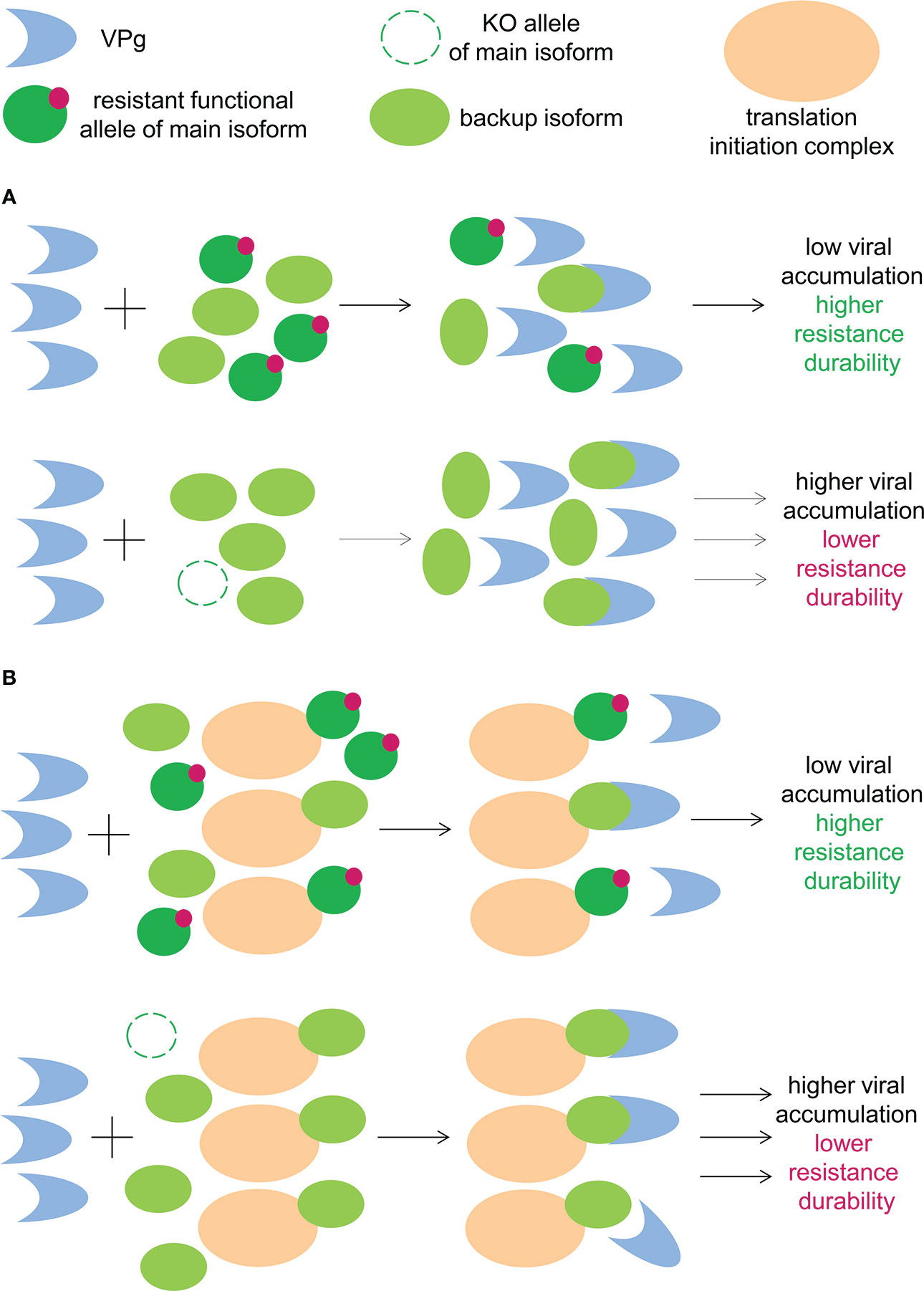
Figure 2 (A) Functional resistant allele of main eIF4E isoforms downregulates the cellular concentration of the backup eIF4E isoform, leading to reduced viral accumulation. (B) The functional resistant allele of the main eIF4E isoform competes with the backup eIF4E isoform for the cellular translation apparatus, leading to reduced viral accumulation.
The initiation of translation requires not only eIF4E but the entire translation initiation complex. Therefore, one possible mechanism underlying the mutual effect of the eIF4E isoforms on the availability to viruses is competition for the remaining elements of the translation initiation complex. In Arabidopsis, the inactivation of eIF(iso)4E increased the eIF4E concentration the polysomal actively translated fraction (Duprat et al., 2002). The functional resistant allele of the main eIF4E isoform competes with other isoforms for the translation apparatus, which reduces the efficiency of mRNA translation associated with other eIF4E isoforms, including potyviral mRNAs associated with the backup eIF4E isoform (Figure 2B). In plants that lack the main eIF4E isoform, this type of competition is absent; this facilitates the translation of viral mRNAs through alternative eIF4E variants. Considering that the potyviral interaction with backup eIF4E isoform is suboptimal, this quantitative increase or decrease in translational efficiency may lead to a qualitative difference in the infection course, i.e., susceptibility or resistance. Potentially, the decreased accumulation and delayed propagation of viruses in plant tissues affords the plants sufficient time to effectively develop diverse antiviral defense mechanisms (Acosta-Leal and Xiong, 2013; Contreras-Paredes et al., 2013; Nicaise, 2014). Although this was not directly observed, different studies have reported in favor of the existence and significance of the competitive effects between eIF4E isoforms in the context of plant–potyvirus interactions (Kang et al., 2007; Michel et al., 2019; Zafirov et al., 2021).
Previously in this review, it was discussed how the functional resistant alleles of the main eIF4E isoform could hamper the use of alternative eIF4E isoforms by the viruses. However, the mutual regulation of the concentration and competition effects should, to a certain extent, exist not only for the main and backup eIF4E isoforms but for all eIF4E factors in plant cells. Accordingly, the presence of each eIF4E factor that potyvirus could not use should promote resistance by decreasing the availability of “susceptible” eIF4E isoforms to this virus to some extent.
To date, evidence has confirmed this assumption in different plant–potyvirus pathosystems. In pepper, the durability of PVY resistance mediated by eIF4E1 alleles substantially decreased with the inactivation of eIF(iso)4E. The gene encoding eIF(iso)4E was the major QTL that defined resistance durability, and the existence of a functional eIF(iso)4E in pepper increased PVY resistance (Quenouille et al., 2014, 2016). Because PVY on pepper overcomes resistant eIF4E1 alleles by re-interacting with them through the accumulating mutations in VPg, the presence of functional eIF(iso)4E in pepper cells obviously suppressed this re-interaction. eIF(iso)4E, which is not used by PVY as a susceptibility factor, apparently downregulated eIF4E1 concentration and/or competed with it for the cellular translational machinery. This hampers the use of resistant eIF4E1 allelic variants by PVY, which is anyway inefficient, thus suppressing subliminal infections and increasing resistance durability.
In tobacco, the durability of PVY resistance conferred by eIF4E-1 deletion depends heavily on an additional genetic locus that contains single or multiple copies of eIF4E-2 (Michel et al., 2019). Although eIF4E-2 is phylogenetically referred to as an eIF4E isoform (Figure 1), similar to eIF4E-1, it was inherited by tobacco from PVY-resistant N. tomentosiformis and is not used by this virus as a susceptibility factor. An excellent link was identified between the eIF4E-2 expression levels in tobacco and the durability of PVY resistance (Michel et al., 2019); therefore, the increased accumulation of eIF4E-2 in tobacco cells qualitatively impeded RB. As RB in plants with an inactivated main eIF4E isoform occurs through the use of the backup eIF4E isoform, the increased concentration of eIF4E-2 appeared to decrease the efficiency of its use by PVY. Michel et al. (2019) suggested that the genome segment containing and expressing multiple eIF4E-2 copies corresponds to the previously identified va2 locus of tobacco PVY resistance. In combination with the inactivation of the main susceptibility factor, eIF4E-1, the va2 locus greatly enhances resistance durability. Moreover, va2 is able to confer partial PVY resistance by itself (Acosta-Leal and Xiong, 2008; Lacroix et al., 2011). The presence of va2 reduces the accumulation of PVY in tobacco cells (Acosta-Leal and Xiong, 2008), possibly because of a decrease in viral translation and replication.
Taken together, these results suggest that eIF4E isoforms that are not directly involved in the plant–potyvirus interaction could affect its outcome. These isoforms could indirectly increase eIF4E-mediated resistance by reducing the efficiency of the use of the main or backup eIF4E isoforms by potyviruses. Therefore, they in some way “dilute” the cell translation complexes that are suitable for the virus, thereby decreasing its multiplication efficiency (Figure 3). Although this effect is unlikely to be adequate to afford plant resistant by itself, the aforementioned examples indicated that it could be significant in the context of resistance durability.
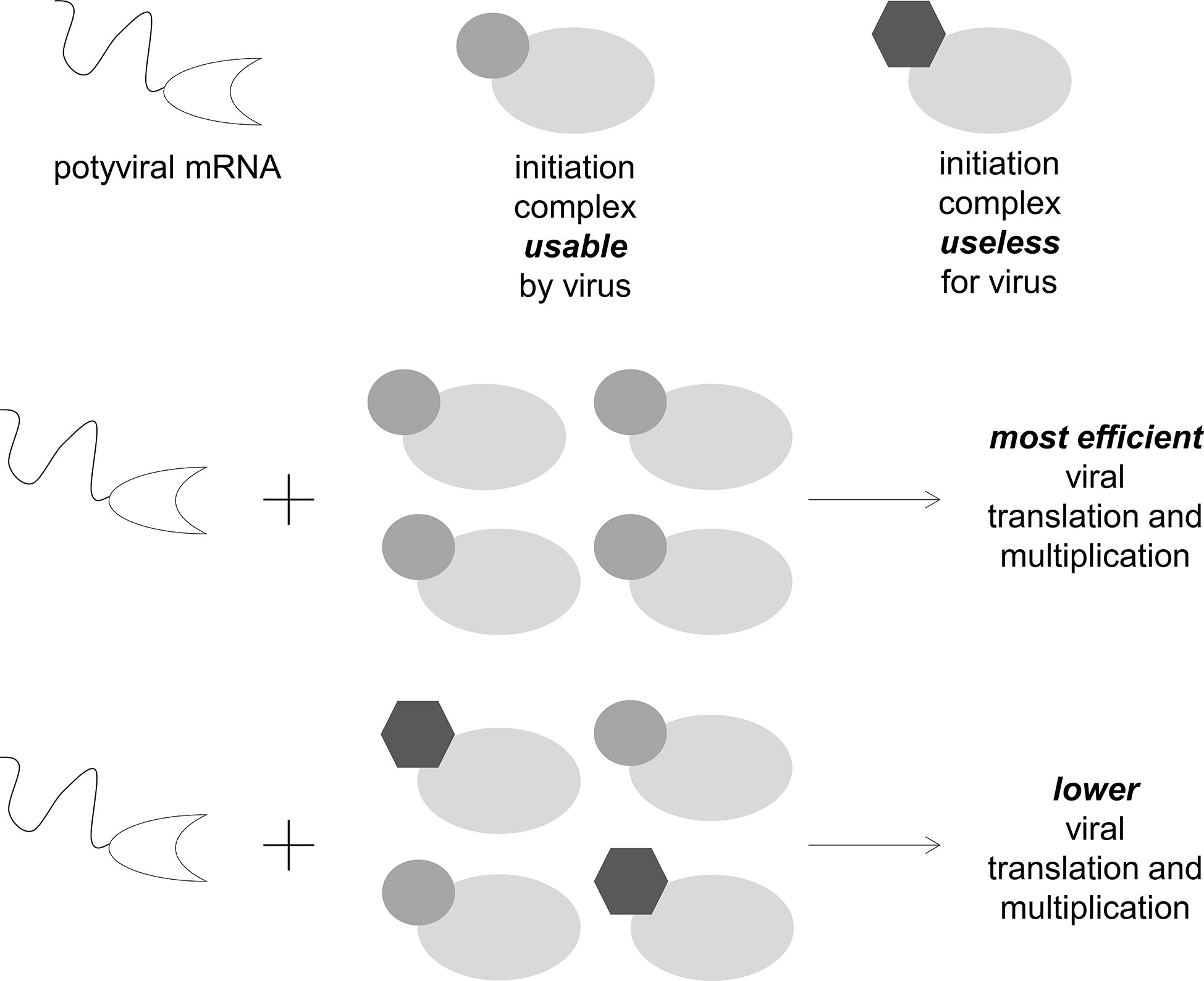
Figure 3 eIF4E isoforms that cannot be used by viruses can decrease their accumulation in plant cells by the mutual regulation of concentration and competition effects.
The main susceptibility eIF4E isoform is also the main resistance isoform because the resistant alleles of this isoform do not support infection and hinder the use of backup eIF4E isoforms by the virus, thus exerting an epistatic effect on them as a susceptibility factors. Considering its importance, what approaches can be used to identify the eIF4E isoform, which is the main isoform for a given potyvirus? This task is most simple in plants, for which there are known eIF4E alleles that confer recessive resistance to the virus of interest (Table 1). However, these alleles for many important crops that are heavily damaged by potyviruses, such as potato, soybean, cassava, or plum, are unknown.
Because the interaction of eIF4E with viral proteins, particularly VPg, is required for potyvirus multiplication, the most obvious way to determine whether the isoform can be used by potyviruses is to analyze the protein–protein interactions. A match between the binding of VPg with eIF4E in model systems (mainly yeast two-hybrid screening) and the ability of this eIF4E to sustain potyviral infection has been proven numerous times (Léonard et al., 2000; Kang et al., 2005; Charron et al., 2008; Mazier et al., 2011; Perez et al., 2012; Takakura et al., 2018). However, in some cases, discrepancies, or at least incomplete consistencies, were observed between the eIF4E isoforms that interacted with VPg on protein–protein interaction screening and isoforms used by the potyviruses in planta as susceptibility factors (Gao et al., 2004; Gallois et al., 2010; Estevan et al., 2014; Moury et al., 2020). Apparently, these discrepancies are explained by the fact that model systems are heterologous and do not consider different factors of plant cells, such as the relative concentrations of each isoform and the competitive effects between them, as a single eIF4E protein is studied in heterologous systems.
Another straightforward approach, i.e., complementation of potyviral infection by the eIF4E isoform expression in resistant plants, could also provide unclear results. B. rapa contains multiple factors belonging to both eIF(iso)4E and eIF4E isoforms. In this plant, TuMV exclusively uses some eIF(iso)4E variants, but not any of the eIF4Es, as susceptibility factors (Rusholme et al., 2007; Qian et al., 2013; Nellist et al., 2014). However, transgenic overexpression of different B. rapa eIF4E factors complement TuMV infection in Arabidopsis plants with inactivated AteIF(iso)4E (Jenner et al., 2010). This is apparently explained by the use of the strong 35S promoter to express eIF4E, leading to its overaccumulation in plant cells and facilitating its use by potyvirus.
The reverse genetics approach can also be used to determine the role of eIF4E isoforms as susceptibility factors. If eIF4E isoform inactivation confers resistance, this proves its role as a main susceptibility factor to a given potyvirus, even if RB occurs eventually. However, in some pathosystems, such as tomato with TEV and different PVY strains, the inactivation of the main isoform (eIF4E1 in these cases) did not result in any significant resistance (Piron et al., 2010; Gauffier et al., 2016). In potato, eIF4E1 inactivation reduced PVY multiplication (Lucioli et al., 2022). Notably, the suppression of nCBP in potato cells has a similar effect to one PVY strain (Chen et al., 2022a). Although these data strongly suggest that at least some PVY strain could unexpectedly use multiple and least-similar eIF4E isoforms in potato, the inactivation of a single eIF4E isoform would likely disturb some regulatory mechanism between different eIF4E isoforms. Consequently, the effect of eIF4E inactivation on susceptibility to potyviruses should be interpreted with some caution.
As studying the role of isoforms in plant–potyvirus interaction using direct approaches could be problematic, indirect approaches are more likely to be useful. Although eIF4E are highly conserved proteins, viral infection in different plants strongly promotes diversifying selection of eIF4E sequences, which leads to the appearance of point mutations (Cavatorta et al., 2008; Hofinger et al., 2011; Konečná et al., 2014; Moury et al., 2014a; Moury et al., 2014b; Poulicard et al., 2016). For example, in pepper, more than two dozen allelic variants of the eIF4E1 isoform, which is the susceptibility factor to different potyviruses (Table 1), were discovered (Charron et al., 2008; Ibiza et al., 2010; Jeong et al., 2012; Moury et al., 2014a; Moury et al., 2014b; Poulicard et al., 2016). Moreover, multiple alleles were identified for pepper eIF(iso)4E (Ibiza et al., 2010), which is also used by some potyviruses as a susceptibility factor (Hwang et al., 2009; Rubio et al., 2009). The selection pressure resulting from viral infection manifests itself in the form of predominance of nonsynonymous substitutions over synonymous ones, mainly in the eIF4E–VPg interaction interface (Charron et al., 2008; Poulicard et al., 2016). The accumulation of such substitutions in the eIF4E isoform strongly suggests that this isoform is a susceptibility factor for the most common potyvirus(es).
Interestingly, the allelic diversity of eIF4E sequences was repeatedly shown to be higher in domesticated populations compared with wild ones (Hofinger et al., 2011; Konečná et al., 2014; Poulicard et al., 2016; Yang et al., 2017). This is presumably caused by cultivation in dense monocultures, which triggers a higher viral pressure and provokes plant evolution toward resistance. According to the law of homologous series in variation (Vavilov, 1922; Nanjundiah et al., 2022), the existence of resistant eIF4E alleles is expected in diverse plants that are widely infected by potyviruses. For autopolyploid species, such as potato, where the spontaneous emergence of eIF4E-mediated resistance is relatively unlikely because of its recessive nature and the presence of several almost identical genes encoding each eIF4E isoform, accessing eIF4E biodiversity in diploid relatives could be more productive. As the occurrence frequency of eIF4E allelic variants could be low in some cases (Nieto et al., 2007), large-scale genotyping studies are warranted.
Although the polymorphisms in the eIF4E isoform suggest that it is used by viruses as a susceptibility factor, the exact virus that caused them cannot be determined using this approach. To determine whether these polymorphisms are involved in resistance to a potyvirus of interest, common methods of studying protein–protein interactions between eIF4E alleles and the VPg of this virus could be used. In such model systems, different alleles of the same isoform instead of different isoforms, are compared. Therefore, the effect of the factors that could lead to misinterpretation, e.g., interplay between isoforms, could be diminished.
Knowledge of the interaction between eIF4E isoforms in plant–virus interactions is required to develop optimal strategies to obtain plants with robust eIF4E-mediated resistance.
This resistance is hardly unimaginable without modifying the main eIF4E isoform. Fortunately, there is only a single main isoform in most plant–virus interactions that facilitates the introgression of this type of resistance. After the identification of the main eIF4E isoform, the best option is to introduce point amino acid changes which will preclude its binding to the viral VPg without having significant negative impacts on eIF4E functionality as a translation initiation factor of cellular mRNAs (Bastet et al., 2017). This will allow us to avoid the possible pleiotropic effects of the loss of eIF4E function as well as to maintain the repressive epistatic effect of the main isoform on other eIF4E isoforms as susceptibility factors in planta.
This task is most simple if the resistant alleles of the main eIF4E isoform in the plant of interest, or at least in its close relatives, are known. Resistance-conferring mutations could be introduced in the main eIF4E isoform of a susceptible cultivar through hybridization or more modern approaches, such as genome editing. However, the use of the latter is significantly hampered by the fact that naturally resistant eIF4E alleles almost always have multiple substitutions relative to susceptibility ones. Moreover, these substitutions are often present at a distance of a few to a few dozen nucleotides (Nicaise et al., 2003; Gao et al., 2004; Charron et al., 2008). Genome editing approaches, which are best suited for the introduction of such mutation combinations in plant genomes (prime editing and editing using homology-directed repair), currently have low efficiency in plants (Čermák, 2021). The use of traditional selection methods to introduce resistant alleles is more developed approach; however, it is hampered by other considerations, for example, low compatibility or cosegregation with undesirable traits.
Natural eIF4E-mediated resistance has not been discovered in many important crops; therefore, resistant eIF4E alleles are unavailable. In such cases, resistant alleles could be artificially created by introducing substitutions that are homologous to resistance-associated mutations in eIF4Es of other plants (Yeam et al., 2007; Bastet et al., 2019) or through using protein engineering approaches (German-Retana et al., 2008; Ashby et al., 2011). However, the effect of such mutations may not be conserved after transferring into eIF4E of the other plant (Cavatorta et al., 2011; Bastet et al., 2019). Moreover, the consequences of such modifications on eIF4E functionality in plants are hardly predictable, especially in the case of artificially designed substitutions.
An alternative variant of modification, which is not accompanied by the aforementioned difficulties, is the knockout of the main eIF4E isoform. It is easier to inactivate the protein than to precisely modify it while retaining its functionality in plant cells but not to the virus. Moreover, novel plant genome editing technologies with limited success in introducing precise substitutions, especially multiple ones, are much more efficient in inactivating genes through the introduction of indel mutations or deletions.
Resistance to potyviruses based on the inactivation of the main eIF4E isoform is currently used in agriculture. Several widely grown tobacco varieties lack the eIF4E-1 factor because of a large genomic deletion or point nonsense mutations and are resistant to PVY and TVMV (Nicolas et al., 1996; Julio et al., 2015; Lin et al., 2021). Also, some varieties of B. rapa display resistance to TuMV because of the miss-splicing of one of the eIF(iso)4E genes (eIF(iso)4E.a) (Nellist et al., 2014). Although the PVY isolates that overcome this resistance in tobacco are common, inactivation eIF4E-1 lowers the probability of a tobacco plant being infected by PVY in the field (Lacroix et al., 2010). In B. rapa, eIF4E-mediated resistance persists and remains effective against multiple TuMV strains (Nellist et al., 2014).
A major potential concern associated with eIF4E inactivation is the negative pleiotropic effects on plant growth and yield, which could be indistinguishable in the laboratory but can have a significant impact under stressful field conditions. Apparently, resistance based on eIF4E inactivation is most suitable for species that carry multiple eIF4E factors belonging to different eIF4E isoforms. Tobacco has six eIF4Es and two eIF(iso)4Es (Julio et al., 2015), whereas B. rapa has three eIF(iso)4Es and three eIF4Es (Jenner et al., 2010) (or even more (Kim et al., 2013; Qian et al., 2013)). Inactivation of the single main eIF4E or even of main and backup eIF4Es in plants with multiple isoforms will likely not have any considerable pleiotropic effects on plant growth and development. This allows the use of cultivars with inactivated eIF4E in agriculture without significant yield penalties.
The second major concern associated with resistance mediated by nonfunctional alleles of the single (main) eIF4E isoform is its low robustness owing to the ability of the virus to switch to another (backup) eIF4E isoform (Bastet et al., 2017). To improve resistance durability, this backup isoform needs to be modified. In tobacco, PVY resistance durability mediated by eIF4E-1 inactivation could be markedly improved by inactivating the backup susceptibility isoform (Udagawa et al., 2021; Le et al., 2022). In B. rapa, durable broad-spectrum resistance to TuMV besides eIF(iso)4E.a inactivation also required a certain resistant allele (named ConTR01) of eIF(iso)4E.c, which is apparently used by TuMV as backup susceptibility factor (Rusholme et al., 2007; Nellist et al., 2014).
Another possible complementary approach to improve resistance durability is the combination of the resistant eIF4E allele with additional resistance-associated QTLs (Naderpour et al., 2010; Quenouille et al., 2013, 2014), which could sometimes be eIF4E. The high significance of such eIF4E-QTLs for resistance to potyviruses has been demonstrated in pepper (Quenouille et al., 2014, 2016) and tobacco (Acosta-Leal and Xiong, 2008; Lacroix et al., 2011; Michel et al., 2019) for PVY and in plum for the plum pox virus (Decroocq et al., 2005; Marandel et al., 2009a, 2009b). Apparently, they represent eIF4E isoforms that could not be used by the virus and decreased its accumulation in plant cells through the abovementioned mechanisms, i.e., downregulating the concentration of the main and backup eIF4E isoforms and competing with them for the cell translational apparatus. It seems appropriate to conduct studies that can thoroughly search for such eIF4Es that quantitatively enhance resistance to potyviruses while considering not only amino acid sequences but also the expression levels. It is expected that plants with increased accumulation of different eIF4Es that cannot be used by potyvirus represent a good genetic background to develop varieties with robust resistance.
Although this review mainly discussed eIF4E-mediated resistance to potyviruses, it is applicable to viruses from other groups as well. Although not studied as thoroughly, this resistance is likely to follow the same rules. Therefore, understanding the interaction between eIF4E isoforms in plant–potyvirus interactions could also be used to develop resistance to other viruses, especially to those systematically close to potyviruses.
A very interesting, albeit almost unstudied, question is why viruses use one or the other eIF4E isoform as a susceptibility factor. Currently, only assumptions can be made regarding this. To function as a susceptibility factor, the eIF4E isoform should be available to potyviruses in sufficient quantity. Most potyviruses use eIF4E as the main susceptibility factor, which is presumably explained by the generally more ubiquitous expression. However, in Arabidopsis, for instance, the majority of the studied potyviruses use eIF(iso)4E as a susceptibility factor (Sato et al., 2005). This could possibly be explained by the increased accumulation of the isoform at certain developmental stages, during which natural infection predominantly occurs. Interestingly, in Arabidopsis, eIF(iso)4E is more abundant in young tissues (Rodriguez et al., 1998).
Even the functions of plant eIF4Es on their own as susceptibility factors to viruses are still far from being well understood. The most investigated topic is the participation of eIF4E in viral translation and replication. However, eIF4E-mediated resistance sometimes acts on different levels, e.g., blocking the cell-to-cell or systemic movement of the virus. Although possible mechanisms of eIF4E involvement in these processes have been proposed a long time ago (Lellis et al., 2002), this subject is currently underexplored.
Considering the variety of eIF4E isoforms that can be used as susceptibility factors by different potyviruses in different plants, it is interesting to know that specific potyvirus generally rely on a single eIF4E isoform in a specific plant. Apparently, different isoforms have different interaction interfaces with VPg, which hampers the ability of the potyvirus to efficiently use multiple eIF4E isoforms. Furthermore, potyviruses have to use eIF4E efficiently to allow its rapid distribution in plants before the plant mounts antiviral defenses, which will lead to its recovery from infection that was observed in some studies (Masuta et al., 1999; Ibiza et al., 2010; Chandrasekaran et al., 2016). This necessitates viral specialization on the single eIF4E isoform, thus allowing single-locus eIF4E-mediated resistance and restricting the number of possible backup eIF4E isoforms.
NZ wrote the manuscript and prepared the figures. VT wrote and edited the manuscript. Both authors have reviewed and approved the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by the Russian Science Foundation Grant №21-76-10050, https://rscf.ru/project/21-76-10050/.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acosta-Leal, R., Xiong, Z. (2008). Complementary functions of two recessive r-genes determine resistance durability of tobacco ‘Virgin a Mutant’(VAM) to Potato virus y. Virology 379, 275–283. doi: 10.1016/j.virol.2008.06.026
Acosta-Leal, R., Xiong, Z. (2013). Intrahost mechanisms governing emergence of resistance-breaking variants of Potato virus y. Virology 437, 39–47. doi: 10.1016/j.virol.2012.12.001
Amano, M., Mochizuki, A., Kawagoe, Y., Iwahori, K., Niwa, K., Svoboda, J., et al. (2013). High-resolution mapping of zym, a recessive gene for Zucchini yellow mosaic virus resistance in cucumber. Theor. Appl. Genet. 126, 2983–2993. doi: 10.1007/s00122-013-2187-5
Andrade, M., Abe, Y., Nakahara, K. S., Uyeda, I. (2009). The cyv-2 resistance to Clover yellow vein virus in pea is controlled by the eukaryotic initiation factor 4E. J. Gen. Plant Pathol. 75, 241–249. doi: 10.1007/s10327-009-0163-3
Arroyo, R., Soto, M. J., Martinez-Zapater, J. M., Ponz, F. (1996). Impaired cell-to-cell movement of potato virus y in pepper plants carrying the y(a) (pr21) resistance gene. Mol. Plant Microbe Interact. 9, 314–318. doi: 10.1094/MPMI-9-0314
Ashby, J. A., Stevenson, C. E., Jarvis, G. E., Lawson, D. M., Maule, A. J. (2011). Structure-based mutational analysis of eIF4E in relation to sbm1 resistance to pea seed-borne mosaic virus in pea. PloS One 6, e15873. doi: 10.1371/journal.pone.0015873
Atarashi, H., Jayasinghe, W. H., Kwon, J., Kim, H., Taninaka, Y., Igarashi, M., et al. (2020). Artificially edited alleles of the eukaryotic translation initiation factor 4E1 gene differentially reduce susceptibility to cucumber mosaic virus and potato virus y in tomato. Front. Microbiol. 11, 564310. doi: 10.1111/j.1365-313X.2006.02792.x
Ayme, V., Petit-Pierre, J., Souche, S., Palloix, A., Moury, B. (2007). Molecular dissection of the potato virus y VPg virulence factor reveals complex adaptations to the pvr2 resistance allelic series in pepper. J. Gen. Virol. 88, 1594–1601. doi: 10.1099/vir.0.82702-0
Ayme, V., Souche, S., Caranta, C., Jacquemond, M., Chadoeuf, J., Palloix, A., et al. (2006). Different mutations in the genome-linked protein VPg of Potato virus y confer virulence on the pvr23 resistance in pepper. Mol. Plant Microbe Interact. 19, 557–563. doi: 10.1094/MPMI-19-0557
Bastet, A., Lederer, B., Giovinazzo, N., Arnoux, X., German-Retana, S., Reinbold, C., et al. (2018). Trans-species synthetic gene design allows resistance pyramiding and broad-spectrum engineering of virus resistance in plants. Plant Biotechnol. J. 16, 1569–1581. doi: 10.1111/pbi.12896
Bastet, A., Robaglia, C., Gallois, J. L. (2017). eIF4E resistance: natural variation should guide gene editing. Trends Plant Sci. 22, 411–419. doi: 10.1016/j.tplants.2017.01.008
Bastet, A., Zafirov, D., Giovinazzo, N., Guyon-Debast, A., Nogué, F., Robaglia, C., et al. (2019). Mimicking natural polymorphism in eIF4E by CRISPR-Cas9 base editing is associated with resistance to potyviruses. Plant Biotechnol. J. 17, 1736–1750. doi: 10.1111/pbi.13096
Beauchemin, C., Boutet, N., Laliberté, J. F. (2007). Visualization of the interaction between the precursors of VPg, the viral protein linked to the genome of Turnip mosaic virus, and the translation eukaryotic initiation factor iso 4E in planta. J. Virol. 81, 775–782. doi: 10.1128/JVI.01277-06
Beffa, R. S., Hofer, R. M., Thomas, M., Meins, F., Jr. (1996). Decreased susceptibility to viral disease of β-1,3-Glucanase-Deficient plants generated by antisense transformation. Plant Cell 8, 1001–1011. doi: 10.1105/tpc.8.6.1001
Ben Khalifa, M., Simon, V., Fakhfakh, H., Moury, B. (2012). Tunisian Potato virus y isolates with unnecessary pathogenicity towards pepper: support for the matching allele model in eIF4E resistance–potyvirus interactions. Plant Pathol. 61, 441–447. doi: 10.1111/j.1365-3059.2011.02540.x
Borgstrøm, B., Johansen, I. E. (2001). Mutations in Pea seedborne mosaic virus genome-linked protein VPg alter pathotype-specific virulence in Pisum sativum. Mol. Plant Microbe Interact. 14, 707–714. doi: 10.1094/MPMI.2001.14.6.707
Bruckner, F. P., Andrade, P. O., de Souza Cascardo, R., Laliberté, J. F., Alfenas-Zerbini, P. (2022). Translationally controlled tumour protein: A protein necessary for potyvirus intracellular multiplication that supports plant infection by unrelated viruses. Ann. Appl. Biol. 180, 90–98. doi: 10.1111/aab.12708
Bruun-Rasmussen, M., Møller, I. S., Tulinius, G., Hansen, J. K. R., Lund, O. S., Johansen, I. E. (2007). The same allele of translation initiation factor 4E mediates resistance against two Potyvirus spp. in Pisum sativum. Mol. Plant Microbe Interact. 20, 1075–1082. doi: 10.1094/MPMI-20-9-1075
Callot, C., Gallois, J. L. (2014). Pyramiding resistances based on translation initiation factors in Arabidopsis is impaired by male gametophyte lethality. Plant Signal. Behav. 9, e27940. doi: 10.4161/psb.27940
Castelló, M. J., Carrasco, J. L., Vera, P. (2010). DNA-Binding protein phosphatase AtDBP1 mediates susceptibility to two potyviruses in arabidopsis. Plant Physiol. 153, 1521–1525. doi: 10.1104/pp.110.158923
Cavatorta, J. R., Perez, K. W., Gray, S. M., Van Eck, J., Yeam, I., Jahn, M. (2011). Engineering virus resistance using a modified potato gene. Plant Biotechnol. J. 9, 1014–1021. doi: 10.1111/j.1467-7652.2011.00622.x
Cavatorta, J. R., Savage, A. E., Yeam, I., Gray, S. M., Jahn, M. M. (2008). Positive Darwinian selection at single amino acid sites conferring plant virus resistance. J. Mol. Evol. 67, 551–559. doi: 10.1007/s00239-008-9172-7
Čermák, T. (2021). Sequence modification on demand: search and replace tools for precise gene editing in plants. Transgenic Res. 30, 353–379. doi: 10.1007/s11248-021-00253-y
Chandrasekaran, J., Brumin, M., Wolf, D., Leibman, D., Klap, C., Pearlsman, M., et al. (2016). Development of broad virus resistance in non-transgenic cucumber using CRISPR/Cas9 technology. Mol. Plant Pathol. 17, 1140–1153. doi: 10.1111/mpp.12375
Charron, C., Nicolaï, M., Gallois, J. L., Robaglia, C., Moury, B., Palloix, A., et al. (2008). Natural variation and functional analyses provide evidence for co-evolution between plant eIF4E and potyviral VPg. Plant J. 54, 56–68. doi: 10.1111/j.1365-313X.2008.03407.x
Chen, R., Tu, Z., He, C., Nie, X., Li, K., Fei, S., et al. (2022a). Susceptibility factor StEXA1 interacts with StnCBP to facilitate potato virus y accumulation through the stress granule-dependent RNA regulatory pathway in potato. Hortic. Res. 9, 1–17. doi: 10.1093/hr/uhac159
Chen, R., Yang, M., Tu, Z., Xie, F., Chen, J., Luo, T., et al. (2022b). Eukaryotic translation initiation factor 4E family member nCBP facilitates the accumulation of TGB-encoding viruses by recognizing the viral coat protein in potato and tobacco. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.946873
Choi, S. H., Nakahara, K. S., Andrade, M., Uyeda, I. (2012). Characterization of the recessive resistance gene cyv1 of Pisum sativum against Clover yellow vein virus. J. Gen. Plant Pathol. 78, 269–276. doi: 10.1007/s10327-012-0383-9
Christie, M., Igreja, C. (2021). eIF4E-homologous protein (4EHP): A multifarious cap-binding protein. FEBS J 290, 266–285. doi: 10.1111/febs.16275
Combe, J. P., Petracek, M. E., van Eldik, G., Meulewaeter, F., Twell, D. (2005). Translation initiation factors eIF4E and eIFiso4E are required for polysome formation and regulate plant growth in tobacco. Plant Mol. Biol. 57, 749–760. doi: 10.1007/s11103-005-3098-x
Congdon, B. S., Coutts, B. A., Renton, M., Banovic, M., Jones, R. A. C. (2016). Pea seed-borne mosaic virus in field pea: Widespread infection, genetic diversity, and resistance gene effectiveness. Plant Dis. 100, 2475–2482. doi: 10.1094/PDIS-05-16-0670-RE
Contreras-Paredes, C. A., Silva-Rosales, L., Daròs, J. A., Alejandri-Ramírez, N. D., Dinkova, T. D. (2013). The absence of eukaryotic initiation factor eIF (iso) 4E affects the systemic spread of a Tobacco etch virus isolate in Arabidopsis thaliana. Mol. Plant Microbe Interact. 26, 461–470. doi: 10.1094/MPMI-09-12-0225-R
De, S., Pollari, M., Varjosalo, M., Mäkinen, K. (2020). Association of host protein VARICOSE with HCPro within a multiprotein complex is crucial for RNA silencing suppression, translation, encapsidation and systemic spread of potato virus a infection. PloS Pathog. 16, e1008956. doi: 10.1371/journal.ppat.1008956
Decroocq, V., Foulongne, M., Lambert, P., Gall, O. L., Mantin, C., Pascal, T., et al. (2005). Analogues of virus resistance genes map to QTLs for resistance to sharka disease in Prunus davidiana. Mol. Genet. Genom. 272, 680–689. doi: 10.1007/s00438-004-1099-0
Deom, C. M., Murphy, J. F., Paguio, O. R. (1997). Resistance to tobacco etch virus in Capsicum annuum: inhibition of virus RNA accumulation. Mol. Plant Microbe Interact. 10, 917–921. doi: 10.1094/MPMI.1997.10.7.917
Dinkova, T. D., Márquez-Velázquez, N. A., Aguilar, R., Lázaro-Mixteco, P. E., de Jiménez, E. S. (2011). Tight translational control by the initiation factors eIF4E and eIF (iso) 4E is required for maize seed germination. Seed Sci. Res. 21, 85–93. doi: 10.1017/S0960258511000043
Duprat, A., Caranta, C., Revers, F., Menand, B., Browning, K. S., Robaglia, C. (2002). The Arabidopsis eukaryotic initiation factor (iso)4E is dispensable for plant growth but required for susceptibility to potyviruses. Plant J. 32, 927–934. doi: 10.1046/j.1365-313X.2002.01481.x
Endres, M. W., Gregory, B. D., Gao, Z., Foreman, A. W., Mlotshwa, S., Ge, X., et al. (2010). Two plant viral suppressors of silencing require the ethylene-inducible host transcription factor RAV2 to block RNA silencing. PloS Pathog. 6, e1000729. doi: 10.1371/journal.ppat.1000729
Eskelin, K., Hafrén, A., Rantalainen, K. I., Mäkinen, K. (2011). Potyviral VPg enhances viral RNA translation and inhibits reporter mRNA translation In planta. J. Virol. 85, 9210–9221. doi: 10.1128/JVI.00052-11
Estevan, J., Maréna, A., Callot, C., Lacombe, S., Moretti, A., Caranta, C., et al. (2014). Specific requirement for translation initiation factor 4E or its isoform drives plant host susceptibility to Tobacco etch virus. BMC Plant Biol. 14, 67. doi: 10.1186/1471-2229-14-67
Fabre, F., Montarry, J., Coville, J., Senoussi, R., Simon, V., Moury, B. (2012). Modelling the evolutionary dynamics of viruses within their hosts: A case study using high-throughput sequencing. PloS Pathog. 8, e1002654. doi: 10.1371/journal.ppat.1002654
Gallie, D. R. (2001). Cap-independent translation conferred by the 5′ leader of tobacco etch virus is eukaryotic initiation factor 4G dependent. J. Virol. 75, 12141–12152. doi: 10.1128/JVI.75.24.12141-12152.2001
Gallois, J. L., Charron, C., Sanchez, F., Pagny, G., Houvenaghel, M. C., Moretti, A., et al. (2010). Single amino acid changes in the turnip mosaic virus viral genome-linked protein (VPg) confer virulence towards Arabidopsis thaliana mutants knocked out for eukaryotic initiation factors eIF (iso) 4E and eIF (iso) 4G. J. Gen. Virol. 91, 288–293. doi: 10.1099/vir.0.015321-0
Gao, Z., Johansen, E., Eyers, S., Thomas, C. L., Noel, E. T. H., Maule, A. J. (2004). The potyvirus recessive resistance gene, sbm1, identifies a novel role for translation initiation factor eIF4E in cell-to-cell trafficking. Plant J. 40, 376–385. doi: 10.1111/j.1365-313X.2004.02215.x
Gauffier, C., Lebaron, C., Moretti, A., Constant, C., Moquet, F., Bonnet, G., et al. (2016). A TILLING approach to generate broad-spectrum resistance to potyviruses in tomato is hampered by eIF4E gene redundancy. Plant J. 85, 717–729. doi: 10.1111/tpj.13136Citations:50
German-Retana, S., Walter, J., Doublet, B., Roudet-Tavert, G., Nicaise, V., Lecampion, C., et al. (2008). Mutational analysis of plant cap-binding protein eIF4E reveals key amino acids involved in biochemical functions and potyvirus infection. J. Virol. 82, 7601–7612. doi: 10.1128/JVI.00209-08
Gomez, M. A., Lin, Z. D., Moll, T., Chauhan, R. D., Hayden, L., Renninger, K., et al. (2019). Simultaneous CRISPR/Cas9-mediated editing of cassava eIF 4E isoforms nCBP-1 and nCBP-2 reduces cassava brown streak disease symptom severity and incidence. Plant Biotechnol. J. 17, 421–434. doi: 10.1111/pbi.12987
Gonzalez-Ibeas, D., Cañizares, J., Aranda, M. A. (2012). Microarray analysis shows that recessive resistance to Watermelon mosaic virus in melon is associated with the induction of defense response genes. Mol. Plant Microbe Interact. 25, 107–118. doi: 10.1094/MPMI-07-11-0193
Hafrén, A., Hofius, D., Rönnholm, G., Sonnewald, U., Mäkinen, K. (2010). HSP70 and its cochaperone CPIP promote potyvirus infection in Nicotiana benthamiana by regulating viral coat protein functions. Plant Cell 22, 523–535. doi: 10.1105/tpc.109.072413
Hafrén, A., Eskelin, K., Mäkinen, K. (2013). Ribosomal protein P0 promotes Potato virus a infection and functions in viral translation together with VPg and eIF (iso) 4E. J. Virol. 87, 4302–4312. doi: 10.1128/JVI.03198-12
Hart, J. P., Griffiths, P. D. (2013). A series of eIF4E alleles at the Bc-3 locus are associated with recessive resistance to Clover yellow vein virus in common bean. Theor. Appl. Genet. 126, 2849–2863. doi: 10.1007/s00122-013-2176-8
Hart, J. P., Griffiths, P. D. (2014). Resistance to Clover yellow vein virus in common bean germplasm. Crop Sci. 54, 2609–2618. doi: 10.2135/cropsci2014.03.0263
Hjulsager, C. K., Lund, O. S., Johansen, I. E. (2002). A new pathotype of Pea seedborne mosaic virus explained by properties of the P3-6k1-and viral genome-linked protein (VPg)-coding regions. Mol. Plant Microbe Interact. 15, 169–171. doi: 10.1094/MPMI.2002.15.2.169
Hoffie, R. E., Otto, I., Perovic, D., Budhagatapalli, N., Habekuß, A., Ordon, F., et al. (2021). Targeted knockout of eukaryotic translation initiation factor 4E confers bymovirus resistance in winter barley. Front. Genome Ed. 3. doi: 10.3389/fgeed.2021.784233
Hofinger, B. J., Russell, J. R., Bass, C. G., Baldwin, T., Dos Reis, M., Hedley, P. E., et al. (2011). An exceptionally high nucleotide and haplotype diversity and a signature of positive selection for the eIF4E resistance gene in barley are revealed by allele mining and phylogenetic analyses of natural populations. Mol. Ecol. 20, 3653–3668. doi: 10.1111/j.1365-294X.2011.05201.x
Hofius, D., Maier, A. T., Dietrich, C., Jungkunz, I., Börnke, F., Maiss, E., et al. (2007). Capsid protein-mediated recruitment of host DnaJ-like proteins is required for Potato virus y infection in tobacco plants. J. Virol. 81, 11870–11880. doi: 10.1128/JVI.01525-07
Hwang, J., Li, J., Liu, W. Y., An, S. J., Cho, H., Her, N. H., et al. (2009). Double mutations in eIF4E and eIFiso4E confer recessive resistance to Chilli veinal mottle virus in pepper. Mol. Cells 27, 329–336. doi: 10.1007/s10059-009-0042-y
Ibiza, V. P., Cañizares, J., Nuez, F. (2010). EcoTILLING in Capsicum species: searching for new virus resistances. BMC Genom. 11, 631. doi: 10.1186/1471-2164-11-631
Jenner, C. E., Nellist, C. F., Barker, G. C., Walsh, J. A. (2010). Turnip mosaic virus (TuMV) is able to use alleles of both eIF4E and eIF(iso)4E from multiple loci of the diploid Brassica rapa. Mol. Plant Microbe Interact. 23, 1498–1505. doi: 10.1094/MPMI-05-10-0104
Jeong, H. J., Kwon, J. K., Pandeya, D., Hwang, J., Hoang, N. H., Bae, J. H., et al. (2012). A survey of natural and ethyl methane sulfonate-induced variations of eIF4E using high-resolution melting analysis in Capsicum. Mol. Breed. 29, 349–360. doi: 10.1007/s11032-011-9550-5
Julio, E., Cotucheau, J., Decorps, C., Volpatti, R., Sentenac, C., Candresse, T., et al. (2015). A eukaryotic translation initiation factor 4E (eIF4E) is responsible for the “va” tobacco recessive resistance to potyviruses. Plant Mol. Biol. Rep. 33, 609–623. doi: 10.1007/s11105-014-0775-4
Kang, B. C., Yeam, I., Frantz, J. D., Murphy, J. F., Jahn, M. M. (2005). The pvr1 locus in Capsicum encodes a translation initiation factor eIF4E that interacts with Tobacco etch virus VPg. Plant J. 42, 392–405. doi: 10.1111/j.1365-313X.2005.02381.x
Kang, B. C., Yeam, I., Li, H., Perez, K. W., Jahn, M. M. (2007). Ectopic expression of a recessive resistance gene generates dominant potyvirus resistance in plants. Plant Biotechnol. J. 5, 526–536. doi: 10.1111/j.1467-7652.2007.00262.x
Kanyuka, K., Druka, A., Caldwell, D. G., Tymon, A., McCallum, N., Waugh, R., et al. (2005). Evidence that the recessive bymovirus resistance locus rym4 in barley corresponds to the eukaryotic translation initiation factor 4E gene. Mol. Plant Pathol. 6, 449–458. doi: 10.1111/j.1364-3703.2005.00294.x
Keima, T., Hagiwara-Komoda, Y., Hashimoto, M., Neriya, Y., Koinuma, H., Iwabuchi, N., et al. (2017). Deficiency of the eIF4E isoform nCBP limits the cell-to-cell movement of a plant virus encoding triple-gene-block proteins in Arabidopsis thaliana. Sci. Rep. 7, 39678. doi: 10.1038/srep39678
Khan, M. A., Miyoshi, H., Gallie, D. R., Goss, D. J. (2008). Potyvirus genome-linked protein, VPg, directly affects wheat germ in vitro translation: interactions with translation initiation factors eIF4F and eIFiso4F. J. Biol. Chem. 283, 1340–1349. doi: 10.1074/jbc.M703356200
Kim, J., Kang, W. H., Yang, H. B., Park, S., Jang, C. S., Yu, H. J., et al. (2013). Identification of a broad-spectrum recessive gene in Brassica rapa and molecular analysis of the eIF4E gene family to develop molecular markers. Mol. Breed. 32, 385–398. doi: 10.1007/s11032-013-9878-0
Konečná, E., Šafářová, D., Navratil, M., Hanáček, P., Coyne, C., Flavell, A., et al. (2014). Geographical gradient of the eIF4E alleles conferring resistance to potyviruses in pea (Pisum) germplasm. PloS One 9, e90394. doi: 10.1371/journal.pone.0090394
Krause-Sakate, R., Redondo, E., Richard-Forget, F., Jadão, A. S., Houvenaghel, M. C., German-Retana, S., et al. (2005). Molecular mapping of the viral determinants of systemic wilting induced by a Lettuce mosaic virus (LMV) isolate in some lettuce cultivars. Virus Res. 109, 175–180. doi: 10.1016/j.virusres.2004.12.001
Kropiwnicka, A., Kuchta, K., Lukaszewicz, M., Kowalska, J., Jemielity, J., Ginalski, K., et al. (2015). Five eIF4E isoforms from Arabidopsis thaliana are characterized by distinct features of cap analogs binding. Biochem. Biophys. Res. Commun. 456, 47–52. doi: 10.1016/j.bbrc.2014.11.032
Kumar, S., Abebie, B., Kumari, R., Kravchik, M., Shnaider, Y., Leibman, D., et al. (2022). Development of PVY resistance in tomato by knockout of host eukaryotic initiation factors by CRISPR-Cas9. Phytoparasitica 50, 743–756. doi: 10.1007/s12600-022-00991-7
Lacroix, C., Glais, L., Kerlan, C., Verrier, J. L., Jacquot, E. (2010). Biological characterization of French Potato virus y (PVY) isolates collected from PVY-susceptible or -resistant tobacco plants possessing the recessive resistance gene va. Plant Pathol. 59, 1133–1143. doi: 10.1111/j.1365-3059.2010.02342.x
Lacroix, C., Glais, L., Verrier, J. L., Jacquot, E. (2011). Effect of passage of a Potato virus y isolate on a line of tobacco containing the recessive resistance gene va 2 on the development of isolates capable of overcoming alleles 0 and 2. Eur. J. Plant Pathol. 130, 259–269. doi: 10.1007/s10658-011-9751-0
Le, N. T., Tran, H. T., Bui, T. P., Nguyen, G. T., Van Nguyen, D., Ta, D. T., et al. (2022). Simultaneously induced mutations in eIF4E genes by CRISPR/Cas9 enhance PVY resistance in tobacco. Sci. Rep. 12, 14627. doi: 10.1038/s41598-022-18923-0
Léonard, S., Plante, D., Wittmann, S., Daigneault, N., Fortin, M. G., Laliberté, J. F. (2000). Complex formation between potyvirus VPg and translation eukaryotic initiation factor 4E correlates with virus infectivity. J. Virol. 74, 7730–7737. doi: 10.1128/JVI.74.17.7730-7737.2000
Lebaron, C., Rosado, A., Sauvage, C., Gauffier, C., German-Retana, S., Moury, B., et al. (2016). A new eIF4E1 allele characterized by RNAseq data mining is associated with resistance to PVY in tomato albeit with a low durability. J. Gen. Virol. 97, 3063–3072. doi: 10.1099/jgv.0.000609
Lellis, A. D., Kasschau, K. D., Whitham, S. A., Carrington, J. C. (2002). Loss-of-susceptibility mutants of Arabidopsis thaliana reveal an essential role for eIF(iso)4E during potyvirus infection. Curr. Biol. 12, 1046–1051. doi: 10.1016/S0960-9822(02)00898-9
Lin, S., Dewey, R. E., Wang, R., Ren, X., Wang, Z., Zhang, J., et al. (2021). Discovery of a novel eIF4E1.S allele conferring PVY resistance in Chinese tobacco (Nicotiana tabacum) landraces. Plant Breed. 140, 693–702. doi: 10.1111/pbr.12919
Ling, K. S., Harris, K. R., Meyer, J. D., Levi, A., Guner, N., Wehner, T. C., et al. (2009). Non-synonymous single nucleotide polymorphisms in the watermelon eIF4E gene are closely associated with resistance to Zucchini yellow mosaic virus. Theor. Appl. Genet. 120, 191–200. doi: 10.1007/s00122-009-1169-0
Liu, T., Liu, Q., Yu, Z., Wang, C., Mai, H., Liu, G., et al. (2022). eIF4E1 regulates arabidopsis embryo development and root growth by interacting with RopGEF7. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.9
Lucioli, A., Tavazza, R., Baima, S., Fatyol, K., Burgyan, J., Tavazza, M. (2022). CRISPR-Cas9 targeting of the eIF4E1 gene extends the potato virus y resistance spectrum of the Solanum tuberosum l. cv. desirée. Front. Microbiol. 13. doi: 10.3389/fmicb.2022.873930
Marandel, G., Pascal, T., Candresse, T., Decroocq, V. (2009a). Quantitative resistance to Plum pox virus in Prunus davidiana P1908 linked to components of the eukaryotic translation initiation complex. Plant Pathol. 58, 425–435. doi: 10.1111/j.1365-3059.2008.02012.x
Marandel, G., Salava, J., Abbott, A., Candresse, T., Decroocq, V. (2009b). Quantitative trait loci meta-analysis of Plum pox virus resistance in apricot (Prunus armeniaca l.): New insights on the organization and the identification of genomic resistance factors. Mol. Plant Pathol. 10, 347–360. doi: 10.1111/j.1364-3703.2009.00535.x
Martínez-Silva, A. V., Aguirre-Martínez, C., Flores-Tinoco, C. E., Alejandri-Ramírez, N. D., Dinkova, T. D. (2012). Translation initiation factor AteIF(iso)4E is involved in selective mRNA translation in arabidopsis thaliana seedlings. PloS One 7, e31606. doi: 10.1371/journal.pone.0031606
Masuta, C., Nishimura, M., Morishita, H., Hataya, T. (1999). A single amino acid change in viral genome-associated protein of potato virus y correlates with resistance breaking in ‘Virgin a mutant’tobacco. Phytopathology 89, 118–123. doi: 10.1094/PHYTO.1999.89.2.118
Mazier, M., Flamain, F., Nicolaï, M., Sarnette, V., Caranta, C. (2011). Knock-down of both eIF4E1 and eIF4E2 genes confers broad-spectrum resistance against potyviruses in tomato. PloS One 6, e29595. doi: 10.1371/journal.pone.0029595
Michel, V., Julio, E., Candresse, T., Cotucheau, J., Decorps, C., Volpatti, R., et al. (2019). A complex eIF4E locus impacts the durability of va resistance to potato virus y in tobacco. Mol. Plant Pathol. 20, 1051–1066. doi: 10.1111/mpp.12810
Montarry, J., Doumayrou, J., Simon, V., Moury, B. (2011). Genetic background matters: a plant–virus gene-for-gene interaction is strongly influenced by genetic contexts. Mol. Plant Pathol. 12, 911–920. doi: 10.1111/j.1364-3703.2011.00724.x
Moury, B., Charron, C., Janzac, B., Simon, V., Gallois, J. L., Palloix, A., et al. (2014a). Evolution of plant eukaryotic initiation factor 4E (eIF4E) and potyvirus genome-linked protein (VPg): A game of mirrors impacting resistance spectrum and durability. Infect. Genet. Evol. 27, 472–480. doi: 10.1016/j.meegid.2013.11.024
Moury, B., Janzac, B., Ruellan, Y., Simon, V., Ben Khalifa, M., Fakhfakh, H., et al. (2014b). Interaction patterns between Potato virus y and eIF4E-mediated recessive resistance in the solanaceae. J. Virol. 88, 9799–9807. doi: 10.1128/JVI.00930-14
Moury, B., Lebaron, C., Szadkowski, M., Khalifa, M. B., Girardot, G., Bi, B. A. B., et al. (2020). Knock-out mutation of eukaryotic initiation factor 4E2 (eIF4E2) confers resistance to pepper veinal mottle virus in tomato. Virology 539, 11–17. doi: 10.1016/j.virol.2019.09.015
Moury, B., Morel, C., Johansen, E., Guilbaud, L., Souche, S., Ayme, V., et al. (2004). Mutations in Potato virus y genome-linked protein determine virulence toward recessive resistances in Capsicum annuum and Lycopersicon hirsutum. Mol. Plant-Microbe Interact. 17, 322–329. doi: 10.1094/MPMI.2004.17.3.322
Murphy, J. F., Blauth, J. R., Livingstone, K. D., Lackney, V. K., Jahn, M. K. (1998). Genetic mapping of the pvr1 locus in Capsicum spp. and evidence that distinct potyvirus resistance loci control responses that differ at the whole plant and cellular levels. Mol. Plant-Microbe Interact. 11, 943–951. doi: 10.1094/MPMI.1998.11.10.943
Naderpour, M., Lund, O. S., Larsen, R., Johansen, E. (2010). Potyviral resistance derived from cultivars of Phaseolus vulgaris carrying bc-3 is associated with the homozygotic presence of a mutated eIF4E allele. Mol. Plant Pathol. 11, 255–263. doi: 10.1111/j.1364-3703.2009.00602.x
Nakahara, K. S., Shimada, R., Choi, S. H., Yamamoto, H., Shao, J., Uyeda, I. (2010). Involvement of the P1 cistron in overcoming eIF4E-mediated recessive resistance against Clover yellow vein virus in pea. Mol. Plant-Microbe Interact. 23, 1460–1469. doi: 10.1094/MPMI-11-09-0277
Nanjundiah, V., Geeta, R., Suslov, V. V. (2022). Revisiting N.I. vavilov’s “The law of homologous series in Variation”(1922). Biol. Theory 2022, 1–10. doi: 10.1007/s13752-022-00403-3
Nellist, C. F., Qian, W., Jenner, C. E., Moore, J. D., Zhang, S., Wang, X., et al. (2014). Multiple copies of eukaryotic translation initiation factors in Brassica rapa facilitate redundancy, enabling diversification through variation in splicing and broad-spectrum virus resistance. Plant J. 77, 261–268. doi: 10.1111/tpj.12389
Nicaise, V. (2014). Crop immunity against viruses: outcomes and future challenges. Front. Plant Sci. 5. doi: 10.3389/fpls.2014.00660
Nicaise, V., German-Retana, S., Sanjuán, R., Dubrana, M. P., Mazier, M., Maisonneuve, B., et al. (2003). The eukaryotic translation initiation factor 4E controls lettuce susceptibility to the potyvirus Lettuce mosaic virus. Plant Physiol. 132, 1272–1282. doi: 10.1104/pp.102.017855
Nicolas, O., Dunnington, S. W., Gotow, L. F., Pirone, T. P., Hellmann, G. M. (1997). Variations in the VPg protein allow a potyvirus to overcome va gene resistance in tobacco. Virology 237, 452–459. doi: 10.1006/viro.1997.8780
Nicolas, O., Pirone, T. P., Hellmann, G. M. (1996). Construction and analysis of infectious transcripts from a resistance-breaking strain of tobacco vein mottling potyvirus. Arch. Virol. 141, 1535–1552. doi: 10.1007/BF01718253
Nieto, C., Morales, M., Orjeda, G., Clepet, C., Monfort, A., Sturbois, B., et al. (2006). An eIF4E allele confers resistance to an uncapped and non-polyadenylated RNA virus in melon. Plant J. 48, 452–462. doi: 10.1111/j.1365-313X.2006.02885.x
Nieto, C., Piron, F., Dalmais, M., Marco, C. F., Moriones, E., Gómez-Guillamón, M. L., et al. (2007). EcoTILLING for the identification of allelic variants of melon eIF4E, a factor that controls virus susceptibility. BMC Plant Biol. 7, 34. doi: 10.1186/1471-2229-7-34
Ouibrahim, L., Mazier, M., Estevan, J., Pagny, G., Decroocq, V., Desbiez, C., et al. (2014). Cloning of the arabidopsis rwm1 gene for resistance to Watermelon mosaic virus points to a new function for natural virus resistance genes. Plant J. 79, 705–716. doi: 10.1111/tpj.12586
Palukaitis, P., Kim, S. (2021). Resistance to turnip mosaic virus in the family Brassicaceae. Plant Pathol. J. 37, 1–23. doi: 10.5423/PPJ.RW.09.2020.0178
Park, S. H., Li, F., Renaud, J., Shen, W., Li, Y., Guo, L., et al. (2017). NbEXPA1, an α-expansin, is plasmodesmata-specific and a novel host factor for potyviral infection. Plant J. 92, 846–861. doi: 10.1111/tpj.13723
Patrick, R. M., Browning, K. S. (2012). The eIF4F and eIFiso4F complexes of plants: an evolutionary perspective. Comp. Funct. Genomics 2012, 1–12. doi: 10.1155/2012/287814
Patrick, R. M., Mayberry, L. K., Choy, G., Woodard, L. E., Liu, J. S., White, A., et al. (2014). Two arabidopsis loci encode novel eukaryotic initiation factor 4E isoforms that are functionally distinct from the conserved plant eukaryotic initiation factor 4E. Plant Physiol. 164, 1820–1830. doi: 10.1104/pp.113.227785
Pavan, S., Jacobsen, E., Visser, R. G., Bai, Y. (2010). Loss of susceptibility as a novel breeding strategy for durable and broad-spectrum resistance. Mol. Breed. 25, 1–12. doi: 10.1007/s11032-009-9323-6
Pechar, G. S., Donaire, L., Gosalvez, B., García-Almodovar, C., Sánchez-Pina, M. A., Truniger, V., et al. (2022). Editing melon eIF4E associates with virus resistance and male sterility. Plant Biotechnol. J 20, 2006–2022. doi: 10.1111/pbi.13885
Perez, K., Yeam, I., Kang, B. C., Ripoll, D. R., Kim, J., Murphy, J. F., et al. (2012). Tobacco etch virus infectivity in capsicum spp. is determined by a maximum of three amino acids in the viral virulence determinant VPg. Mol. Plant Microbe Interact. 25, 1562–1573. doi: 10.1094/MPMI-04-12-0091-R
Piron, F., Nicolaï, M., Minoïa, S., Piednoir, E., Moretti, A., Salgues, A., et al. (2010). An induced mutation in tomato eIF4E leads to immunity to two potyviruses. PloS One 5, e11313. doi: 10.1371/journal.pone.0011313
Poque, S., Pagny, G., Ouibrahim, L., Chague, A., Eyquard, J. P., Caballero, M., et al. (2015). Allelic variation at the rpv1 locus controls partial resistance to Plum pox virus infection in Arabidopsis thaliana. BMC Plant Biol. 15, 159. doi: 10.1186/s12870-015-0559-5
Poulicard, N., Pacios, L. F., Gallois, J. L., Piñero, D., García-Arenal, F. (2016). Human management of a wild plant modulates the evolutionary dynamics of a gene determining recessive resistance to virus infection. PloS Genet. 12, e1006214. doi: 10.1371/journal.pgen.1006214
Provvidenti, R., Alconero, R. (1988a). Inheritance of resistance to a third pathotype of pea seed-borne mosaic virus in Pisum sativum. J. Hered. 79, 76–77. doi: 10.1093/oxfordjournals.jhered.a110457
Provvidenti, R., Alconero, R. (1988b). Inheritance of resistance to a lentil strain of pea seed-borne mosaic virus in Pisum sativum. J. Hered. 79, 45–47. doi: 10.1093/oxfordjournals.jhered.a110444
Provvidenti, R., Hampton, R. O. (1991). “Chromosomal distribution of genes for resistance to seven potyviruses in pisum sativum,” in Pisum genetics, vol. 23 . Ed. Murfet, I. C. (Hobart, Tasmania, Australia: University of Tasmania), 26–28.
Provvidenti, R., Hampton, R. O. (1992). “Sources of resistance to viruses in the potyviridae,” in Potyvirus taxonomy, 5th ed. Ed. Barnett, O. W., Jr. (Wien: Springer-Verlag), 189–211.
Qian, W., Zhang, S., Zhang, S., Li, F., Zhang, H., Wu, J., et al. (2013). Mapping and candidate-gene screening of the novel turnip mosaic virus resistance gene retr02 in Chinese cabbage (Brassica rapa l.). Theor. Appl. Genet. 126, 179–188. doi: 10.1007/s00122-012-1972-x
Quenouille, J., Montarry, J., Palloix, A., Moury, B. (2013). Farther, slower, stronger: how the plant genetic background protects a major resistance gene from breakdown. Mol. Plant Pathol. 14, 109–118. doi: 10.1007/s00122-012-1972-x
Quenouille, J., Paulhiac, E., Moury, B., Palloix, A. (2014). Quantitative trait loci from the host genetic background modulate the durability of a resistance gene: A rational basis for sustainable resistance breeding in plants. Hered. (Edinb.) 112, 579–587. doi: 10.1038/hdy.2013.138
Quenouille, J., Saint-Felix, L., Moury, B., Palloix, A. (2016). Diversity of genetic backgrounds modulating the durability of a major resistance gene. analysis of a core collection of pepper landraces resistant to Potato virus y. Mol. Plant Pathol. 17, 296–302. doi: 10.1111/mpp.12277
Rodriguez, C. M., Freire, M. A., Camilleri, C., Robaglia, C. (1998). The Arabidopsis thaliana cDNAs coding for eIF4E and eIF (iso) 4E are not functionally equivalent for yeast complementation and are differentially expressed during plant development. Plant J. 13, 465–473. doi: 10.1046/j.1365-313X.1998.00047.x
Rodríguez-Hernández, A. M., Gosalvez, B., Sempere, R. N., Burgos, L., Aranda, M. A., Truniger, V. (2012). Melon RNA interference (RNAi) lines silenced for Cm-eIF4E show broad virus resistance. Mol. Plant Pathol. 13, 755–763. doi: 10.1111/j.1364-3703.2012.00785.x
Rubio, M., Nicolaï, M., Caranta, C., Palloix, A. (2009). Allele mining in the pepper gene pool provided new complementation effects between pvr2-eIF4E alleles and pvr6-eIF(iso)4E for resistance to the pepper veinal mottle virus. J. Gen. Virol. 90, 2808–2814. doi: 10.1099/vir.0.013151-0
Ruffel, S., Dussault, M. H., Palloix, A., Moury, B., Bendahmane, A., Robaglia, C., et al. (2002). A natural recessive resistance gene against potato virus y in pepper corresponds to the eukaryotic initiation factor 4E (eIF4E). Plant J. 32, 1067–1075. doi: 10.1046/j.1365-313X.2002.01499.x
Ruffel, S., Gallois, J. L., Lesage, M. L., Caranta, C. (2005). The recessive potyvirus resistance gene pot-1 is the tomato orthologue of the pepper pvr2-eIF4E gene. Mol. Genet. Genom. 274, 346–353. doi: 10.1007/s00438-005-0003-x
Ruffel, S., Gallois, J. L., Moury, B., Robaglia, C., Palloix, A., Caranta, C. (2006). Simultaneous mutations in translation initiation factors eIF4E and eIF (iso) 4E are required to prevent pepper veinal mottle virus infection of pepper. J. Gen. Virol. 87, 2089–2098. doi: 10.1099/vir.0.81817-0
Rufty, R. C., Miller, R. D., Gooding, G. V., Jr. (1989). Effect of tobacco etch and tobacco vein mottling virus on yield of burley tobacco genotypes. Plant Dis. 73, 45–48. doi: 10.1094/PD-73-0045
Rupp, J. S., Cruz, L., Trick, H. N., Fellers, J. P. (2019). RNAi-mediated silencing of endogenous wheat genes EIF(Iso)4E-2 and EIF4G induce resistance to multiple RNA viruses in transgenic wheat. Crop Sci. 59, 2642–2651. doi: 10.2135/cropsci2018.08.0518
Rusholme, R. L., Higgins, E. E., Walsh, J. A., Lydiate, D. J. (2007). Genetic control of broad-spectrum resistance to turnip mosaic virus in Brassica rapa (Chinese cabbage). J. Gen. Virol. 88, 3177–3186. doi: 10.1099/vir.0.83194-0
Ruud, K. A., Kuhlow, C., Goss, D. J., Browning, K. S. (1998). Identification and characterization of a novel cap-binding protein from Arabidopsis thaliana. J. Biol. Chem. 273, 10325–10330. doi: 10.1074/jbc.273.17.10325
Salazar-Díaz, K., Aquino-Luna, M., Hernández-Lucero, E., Nieto-Rivera, B., Pulido-Torres, M. A., Jorge-Pérez, J. H., et al. (2021). Arabidopsis thaliana eIF4E1 and eIF(iso)4E participate in cold response and promote translation of some stress-related mRNAs. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.698585
Sato, M., Masuta, C., Uyeda, I. (2003). Natural resistance to Clover yellow vein virus in beans controlled by a single recessive locus. Mol. Plant Microbe Interact. 16, 994–1002. doi: 10.1094/MPMI.2003.16.11.994
Sato, M., Nakahara, K., Yoshii, M., Ishikawa, M., Uyeda, I. (2005). Selective involvement of members of the eukaryotic initiation factor 4E family in the infection of Arabidopsis thaliana by potyviruses. FEBS Lett. 579, 1167–1171. doi: 10.1016/j.febslet.2004.12.086
Schaad, M. C., Anderberg, R. J., Carrington, J. C. (2000). Strain-specific interaction of the tobacco etch virus NIa protein with the translation initiation factor eIF4E in the yeast two-hybrid system. Virology 273, 300–306. doi: 10.1006/viro.2000.0416
Shi, S., Zhang, X., Mandel, M. A., Zhang, P., Zhang, Y., Ferguson, M., et al. (2017). Variations of five eIF4E genes across cassava accessions exhibiting tolerant and susceptible responses to cassava brown streak disease. PloS One 12, e0181998. doi: 10.1371/journal.pone.0181998
Shopan, J., Mou, H., Zhang, L., Zhang, C., Ma, W., Walsh, J. A., et al. (2017). Eukaryotic translation initiation factor 2B-beta (eIF2Bβ), a new class of plant virus resistance gene. Plant J. 90, 929–940. doi: 10.1111/tpj.13519
Soler-Garzón, A., McClean, P. E., Miklas, P. N. (2021a). Coding mutations in vacuolar protein-sorting 4 AAA+ ATPase endosomal sorting complexes required for transport protein homologs underlie bc-2 and new bc-4 gene conferring resistance to Bean common mosaic virus in common bean. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.769247
Soler-Garzón, A., McClean, P. E., Miklas, P. N. (2021b). Genome-wide association mapping of bc-1 and bc-u reveals candidate genes and new adjustments to the host-pathogen interaction for resistance to bean common mosaic necrosis virus in common bean. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.699569
Sorel, M., Svanella-Dumas, L., Candresse, T., Acelin, G., Pitarch, A., Houvenaghel, M. C., et al. (2014). Key mutations in the cylindrical inclusion involved in Lettuce mosaic virus adaptation to eIF4E-mediated resistance in lettuce. Mol. Plant Microbe Interact. 27, 1014–1024. doi: 10.1094/MPMI-04-14-0111-R
Stein, N., Perovic, D., Kumlehn, J., Pellio, B., Stracke, S., Streng, S., et al. (2005). The eukaryotic translation initiation factor 4E confers multiallelic recessive Bymovirus resistance in Hordeum vulgare (L.). Plant J. 42, 912–922. doi: 10.1111/j.1365-313X.2005.02424.x
Takakura, Y., Udagawa, H., Shinjo, A., Koga, K. (2018). Mutation of a Nicotiana tabacum l. eukaryotic translation-initiation factor gene reduces susceptibility to a resistance-breaking strain of Potato virus y. Mol. Plant Pathol. 19, 2124–2133. doi: 10.1111/mpp.12686
Tavert-Roudet, G., Anne, A., Barra, A., Chovin, A., Demaille, C., Michon, T. (2017). The potyvirus particle recruits the plant translation initiation factor eIF4E by means of the VPg covalently linked to the viral RNA. Mol. Plant Microbe Interact. 30, 754–762. doi: 10.1094/MPMI-04-17-0091-R
Truniger, V., Aranda, M. A. (2009). Recessive resistance to plant viruses. Adv. Virus Res. 75, 119–231. doi: 10.1016/S0065-3527(09)07504-6
Udagawa, H., Koga, K., Shinjo, A., Kitashiba, H., Takakura, Y. (2021). Loss-of-function of Nicotiana tabacum l. eukaryotic translation initiation factors eIF4E1-s and eIF (iso) 4E-T synergistically confers high-level resistance to both Potato virus y (PVY) and resistance-breaking PVY. Breed. Sci. 71, 193–200. doi: 10.1270/jsbbs.20114
van Schie, C. C., Takken, F. L. (2014). Susceptibility genes 101: how to be a good host. Annu. Rev. Phytopathol. 52, 551–581. doi: 10.1146/annurev-phyto-102313-045854
Vavilov, N. I. (1922). The law of homologous series in variation. J. Genet. 12, 47–89. doi: 10.1007/BF02983073
Wang, A., Krishnaswamy, S. (2012). Eukaryotic translation initiation factor 4E-mediated recessive resistance to plant viruses and its utility in crop improvement. Mol. Plant Pathol. 13, 795–803. doi: 10.1111/j.1364-3703.2012.00791.x
Wang, J., Zhang, X., Greene, G. H., Xu, G., Dong, X. (2022). PABP/purine-rich motif as an initiation module for cap-independent translation in pattern-triggered immunity. Cell 185, 3186–3200. doi: 10.1016/j.cell.2022.06.037
Wu, G., Cui, X., Dai, Z., He, R., Li, Y., Yu, K., et al. (2020). A plant RNA virus hijacks endocytic proteins to establish its infection in plants. Plant J. 101, 384–400. doi: 10.1111/tpj.14549
Yang, P., Habekuß, A., Hofinger, B. J., Kanyuka, K., Kilian, B., Graner, A., et al. (2017). Sequence diversification in recessive alleles of two host factor genes suggests adaptive selection for bymovirus resistance in cultivated barley from East Asia. Theor. Appl. Genet. 130, 331–344. doi: 10.1007/s00122-016-2814-z
Yang, X., Li, Y., Wang, A. (2021). Research advances in potyviruses: From the laboratory bench to the field. Annu. Rev. Phytopathol. 59, 1–29. doi: 10.1146/annurev-phyto-020620-114550
Yeam, I., Cavatorta, J. R., Ripoll, D. R., Kang, B. C., Jahn, M. M. (2007). Functional dissection of naturally occurring amino acid substitutions in eIF4E that confers recessive potyvirus resistance in plants. Plant Cell 19, 2913–2928. doi: 10.1105/tpc.107.050997
Yoshii, M., Nishikiori, M., Tomita, K., Yoshioka, N., Kozuka, R., Naito, S., et al. (2004). The arabidopsis Cucumovirus multiplication 1 and 2 loci encode translation initiation factors 4E and 4G. J. Virol. 78, 6102–6111. doi: 10.1128/JVI.78.12.6102-6111.2004
Zafirov, D., Giovinazzo, N., Bastet, A., Gallios, J. L. (2021). When a knockout is an achilles’ heel: Resistance to one potyvirus species triggers hypersusceptibility to another one in Arabidopsis thaliana. Mol. Plant Pathol. 22, 334–347. doi: 10.1111/mpp.13031
Zhu, M., Chen, Y., Ding, X. S., Webb, S. L., Zhou, T., Nelson, R. S., et al. (2014). Maize elongin c interacts with the viral genome-linked protein, VPg, of Sugarcane mosaic virus and facilitates virus infection. New Phytol. 203, 1291–1304. doi: 10.1111/nph.12890
Keywords: plant resistance, recessive resistance, susceptibility gene, potyvirus, eIF4E isoform
Citation: Zlobin N and Taranov V (2023) Plant eIF4E isoforms as factors of susceptibility and resistance to potyviruses. Front. Plant Sci. 14:1041868. doi: 10.3389/fpls.2023.1041868
Received: 11 September 2022; Accepted: 30 January 2023;
Published: 10 February 2023.
Edited by:
Heng-Mu Zhang, Zhejiang Academy of Agricultural Sciences, ChinaReviewed by:
Ming-Bo Wang, Commonwealth Scientific and Industrial Research Organisation (CSIRO), AustraliaCopyright © 2023 Zlobin and Taranov. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nikolay Zlobin, c3RyZXNzbGFiQHlhbmRleC5ydQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.