- 1State Key Laboratory of Tree Genetics and Breeding, Beijing Forestry University, Beijing, China
- 2National Engineering Research Center of Tree Breeding and Ecological Restoration, College of Biological Sciences and Technology, Beijing Forestry University, Beijing, China
- 3Key Laboratory of Genetics and Breeding in Forest Trees and Ornamental Plants, Ministry of Education, Beijing Forestry University, Beijing, China
- 4Beijing Laboratory of Urban and Rural Ecological Environment, Beijing Forestry University, Beijing, China
- 5Weixian Eucommia National Forest Tree Germplasm Repository, Weixian Forestry Cultivation Base of Superior Species, Hebei, China
The NAC transcription factor family is a large plant gene family, participating in plant growth and development, secondary metabolite synthesis, biotic and abiotic stresses responses, and hormone signaling. Eucommia ulmoides is a widely planted economic tree species in China that can produce trans-polyisoprene: Eucommia rubber (Eu-rubber). However, genome-wide identification of the NAC gene family has not been reported in E. ulmoides. In this study, 71 NAC proteins were identified based on genomic database of E. ulmoides. Phylogenetic analysis showed that the EuNAC proteins were distributed in 17 subgroups based on homology with NAC proteins in Arabidopsis, including the E. ulmoides-specific subgroup Eu_NAC. Gene structure analysis suggested that the number of exons varied from 1 to 7, and multitudinous EuNAC genes contained two or three exons. Chromosomal location analysis revealed that the EuNAC genes were unevenly distributed on 16 chromosomes. Three pairs of genes of tandem duplicates genes and 12 segmental duplications were detected, which indicated that segmental duplications may provide the primary driving force of expansion of EuNAC. Prediction of cis-regulatory elements indicated that the EuNAC genes were involved in development, light response, stress response and hormone response. For the gene expression analysis, the expression levels of EuNAC genes in various tissues were quite different. To explore the effect of EuNAC genes on Eu-rubber biosynthesis, a co-expression regulatory network between Eu-rubber biosynthesis genes and EuNAC genes was constructed, which indicated that six EuNAC genes may play an important role in the regulation of Eu-rubber biosynthesis. In addition, this six EuNAC genes expression profiles in E. ulmoides different tissues were consistent with the trend in Eu-rubber content. Quantitative real-time PCR analysis showed that EuNAC genes were responsive to different hormone treatment. These results will provide a useful reference for further studies addressing the functional characteristics of the NAC genes and its potential role in Eu-rubber biosynthesis.
Introduction
NAC [for NAM (no apical meristem), ATAF, and CUC (cup-shaped cotyledon)] proteins are one of the largest plant transcription factors (TF) families. NAC TFs are characterized by a conserved N-terminal NAC domain comprising approximately 150 amino acids and a diversified C-terminal (Ng et al., 2018). The DNA binding NAC domain is divided into five sub-domains designated A–E, which are relevant to DNA binding, dimer formation and multiple other functions (Ooka et al., 2003; Ernst et al., 2004). In addition, compared with subdomains B and E, subdomains A, C, and D are highly conserved (Jensen et al., 2010). The C-terminus has transcriptional activation or transcriptional repression activity (Mohanta et al., 2020). With the development of high-throughput sequencing technology, it has become possible to use genome and transcriptome data to identify and screen all NAC family genes in species. The members of the NAC gene family has been widely identified and studied in many species, such as Arabidopsis thaliana (Ooka et al., 2003), Oryza sativa (Nuruzzaman et al., 2010), Vitis vinifera (Wang et al., 2013), Coffea canephora (Dong et al., 2019), and Populus trichocarpa (Hu et al., 2010).
NAC transcription factors play an important role in regulating plant growth and development. For example, secondary wall-associated NAC (SWN) transcription factors play critical roles in regulating secondary cell wall formation and development (Zhou et al., 2014; Endo et al., 2015; Zhang et al., 2020; Zhong et al., 2021a). AtNAC1 and AtNAC2 are involved in lateral root development by downregulating auxin signals in Arabidopsis (Guo et al., 2005). At the same time, NAC genes also play a key role in responses to biotic and abiotic stresses. AtNAC019, AtNAC055, and AtNAC072 can respond to various abiotic stresses and hormonal treatments, such as dehydration, cold, salinity, mechanical damage, and jasmonic acid (JA) and abscisic acid (ABA) (Tran et al., 2004). In addition, the involvement of NAC transcription factors in plant secondary metabolite biosynthesis is attracting increasing attention (Wang et al., 2018). CsNAC7 positively regulates the caffeine synthase gene yhNMT1 and promotes caffeine accumulation in Camellia sinensis (Ma et al., 2022). The overexpression of SmNAC2 reduces the content of tanshinone in Salvia miltiorrhiza, but silencing SmNAC2 promotes the accumulation of tanshinone (Zhang et al., 2021).
Polyisoprene, an isoprene (C5H8) polymer, is the primary chemical constituent in natural rubber. Based on the chemical structures of two isoprene isomers, natural rubber can be classified as cis-polyisoprene (CPI) and trans-polyisoprene (TPI) (Baboo et al., 2011). Over 2000 different plant species produce CPI, including Hevea brasiliensis, Lactuca sativa, Taraxacum kok-saghyz and Parthenium argentatum. (Mooibroek and Cornish, 2000; van Beilen and Poirier, 2007). However, only a few plants can produce trans-rubber, Eucommia ulmoides is a widely known tree species that can produce TPI: Eu-rubber (Roth et al., 1985; Schlesinger and Leeper, 2002). Eu-rubber is especially enriched in the leaves, bark, and peels of E. ulmoides trees (Nakazawa et al., 2009; Wuyun et al., 2018). Compared with CPI, Eu-rubber has unique features, good insulation and resistance to acids and alkalis; moreover, it also has great potential for application in biomedicine, textiles, aerospace and other fields. (Wang et al., 2020). Therefore, E. ulmoides is an ideal material for studying the biosynthesis of TPI. Although the biosynthetic pathway of Eu-rubber has been widely studied, the molecular mechanisms regulating the biosynthesis of Eu-rubber remain unclear. However, the NAC gene family of E. ulmoides has not been systematically analyzed, and its effect on the biosynthesis of TPI is still unclear.
In this study, we identified 71 E. ulmoides NAC genes and divided them into 17 subgroups, including an E. ulmoides-specific subgroup Eu_NAC. A comprehensive analysis of gene structure, motif composition, chromosomal distribution, gene duplication, phylogenetic, and cis-acting elements in promoters and syntenic relationships was completed. In addition, the expression of EuNAC genes in different tissues was also analyzed. The co-expression network of EuNAC genes and Eu-rubber biosynthesis genes and the response to various hormones were also analyzed. The results of this study will provide a platform to identify the biological function of NAC genes in E. ulmoides in the future and will be helpful for further study of the functional characteristics of EuNAC genes in the mechanism of Eu-rubber biosynthesis.
Materials and methods
Identification of EuNAC genes in the E. ulmoides
The protein sequence was obtained from the E. ulmoides genome. The genome accession number is PRJNA599775 in NCBI database (https://www.ncbi.nlm.nih.gov/). First, we downloaded the HMM profile of the NAM domain (PF02365) from the Pfam database (http://pfam.sanger.ac.uk/). Then, the HMMER 3.2.1 program was used to identify the NAC protein in the E. ulmoides genome (Li et al., 2020). The default setting was used, and the cut-off was 0.01. Finally, the NAM domain of all candidate NAC genes was determined through CDD (http://www.ncbi.nlm.nih.gov/cdd) and SMART databases (http://smart.emblheidelberg.de/). Seventy-one putative NAC genes were identified. Meanwhile, the physical and chemical parameters of the EuNAC proteins were predicted by ProtParam (http://web.expasy.org/protparam/), including the CDS (coding sequence) length, protein length, molecular weights (MW), isoelectric points (PI) aliphatic index, and grand average of hydropathicity (GRAVY). Subcellular localization of all EuNAC proteins was performed using Euk-mPLoc 2.0 (http://www.csbio.sjtu.edu.cn/bioinf/euk-multi-2/).
Phylogenetic analysis and multiple alignments
The NAC protein sequences of Arabidopsis were downloaded from the Arabidopsis genome TAIR 11 (https://www.arabidopsis.org/). Multiple sequence alignments of E. ulmoides and Arabidopsis NAC proteins were performed using MUSCLE with default parameters. The phylogenetic and molecular evolutionary analyses were conducted using MEGA (version X) (Sudhir et al., 2018). The neighbor-joining (NJ) method was selected for constructing the phylogenetic tree with 1000 bootstrap replications. The phylogenetic tree was visualized in the EvolView program (https://www.evolgenius.info//evolview/). All the identified EuNAC genes were assigned into different groups based on the classification of the Arabidopsis NACs.
Gene structure and motif analysis
Gene structure was investigated using the TBtools software (version 1.098696). The MEME online program (http://meme.nbcr.net/meme/intro.html) for protein sequence analysis was used to identify conserved motifs in the identified EuNACs proteins, with the maximum number of motifs set to 10 and the width of each motif ranging from 6 to 50. The TBtools software was used to integrate phylogenetic trees, conserved motifs, and gene structure results (Chen et al., 2020).
Chromosomal location, gene duplication, and synteny analysis with several plant species
The chromosomal position of NAC genes was obtained from the E. ulmoides genome annotations using TBtools. The chromosomal map of E. ulmoides NAC genes was visualized by MG2C.10 (http://mg2c.iask.in/mg2c_v2.1/). EuNAC gene duplication events was examined by using MCScanX software with default parameters (Wang et al., 2012). Dual Synteny Plotter software (https://github.com/CJ-Chen/TBtools/) was used to analyze the homology of the NAC gene between E. ulmoides and A.thaliana, C.canephora, V.vinifera, H.brasiliensis, O.sativa, and S.bicolor, respectively. TBtools software was used to visualize the obtained results, the obtained NAC homologous pairs are highlighted (Chen et al., 2020). The parameters non-synonymous mutations (Ka), synonymous mutations (Ks) and estimated evolutionary constraints (Ka/Ks) among the EuNACs genes were calculated using TBtools.
Analysis of cis-acting elements in EuNACs promoters
The promoter refers to the region upstream sequence of the transcription start site (Shafee and Lowe, 2017). At present, we are unable to identify the transcription start site of genes in E.ulmoides, so we used the upstream 2000 bp sequence of the translational start codon (ATG) of EuNAC genes and Eu-rubber biosynthesis pathway genes, and the sequences were submitted to PlantCARE database (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) to predict cis-acting elements. This means that we may have explored 5’ UTR or the sequences analyzed were only part of the promoter regulatory region.
Expression patterns of EuNACs in different tissue based on the public RNA-seq data sets
To survey the expression patterns of EuNAC genes in different tissues, the transcriptome data of E. ulmoides in various tissues (Leaf, Xylem, Seed, and Peel) were obtained from the NCBI sequence read archive (SRX7525252-54, SRX7532003-05, SRX7531725-27, and SRX7533248-50). The transcript abundance of E. ulmoides genes was calculated as fragments per kilobase of exon model per million mapped reads (FPKM). The EuNAC genes expression level was presented based on the transformed data of log2 (FPKM+1) values. A clustered heat map was drawn using Tbtools software, the approach of clustering analysis is hierarchical clustering. (Chen et al., 2020).
Gene Co-Expression Network Construction
To study the relationship between EuNAC genes and Eu-rubber biosynthesis pathway genes in E. ulmoides, we used the FPKM of these genes to construct the network. Pearson’s correlation analysis was performed using the OmicStudio tools at https://www.omicstudio.cn/tool/62 (Lyu et al., 2023). Genes with a Pearson’s correlation coefficient within the appropriate range (r ≥0.6 or ≤−0.6) and p < 0.05 were selected to generate a co-expression network using Cytoscape (version 3.9.0). A positive value represented a positive correlation, and a negative value represented a negative correlation. The connectivity degree of genes was calculated using the Cytoscape software. The node size was positively correlated with the degree of the connectivity of genes. Nodes were colored red for positive regulation and blue for negative regulation.
Plant materials and hormone treatments
The E. ulmoides seeds were collected from 20-year-old diploid E. ulmoides trees at Beijing Forestry University, Beijing, China. The E. ulmoides were cultured in a growth chamber with a 16-h light/8-h dark cycle at 28°C. Studies reported that applications of exogenous hormones, such as 2-(3,4-dichlorophenoxy)-triethylamine (DCPTA), gibberellin (GA3) and brassinolide (BR) increased Eu-rubber concentration in E. ulmoides leaves and spaying BR at 5 mg/L, DCPTA at 500 mg/L and GA3 at 300 mg/L was the optimal treatment concentration (Liu H. et al., 2018). To examine the effect of hormones on the expression of EuNAC genes, 5-month-old seedling leaves of E. ulmoides were sprayed with 300 mg/L GA3, 5 mg/L 1% BR, and 500 mg/L DCPTA until there is liquid dripping. Spray water was used as the control treatment. The leaves were sampled at 0, 3, 6, 12, and 24 h after hormone treatments, frozen in liquid nitrogen, and finally stored at −80°C for RNA extraction. There were three independent replicates for each treatment.
RNA extraction, cDNA synthesis, and qRT-PCR gene expression analysis
The RN38-EASYspin Plus Kit (Aidlab Biotechnologies Co., Ltd) was used to extract total RNA according to the manufacturer’s instructions. A NanoDrop ND-2000 (Thermo Scientific, USA) spectrophotometer and 1% agarose gel electrophoresis were used to detect the RNA quality of all samples. The PC54-TRUEscript RT kit (Aidlab Biotechnologies Co., Ltd) was used to synthesize first-strand cDNA. The real-time polymerase chain reaction (RT-PCR) was accomplished using SYBR® Green Premix Pro Taq HS qPCR Kit (Rox Plus) AG11718 (Accurate Biotechnology (Hunan) Co., Ltd.) using an Applied Biosystems 7500 Fast instrument (AB Ltd., USA). The qRT-PCR master mix included 10 µL 2× SYBR Green Pro Taq HS Premix (ROX Plus), 0.4 µL forward primer, 0.4 µL reverse primer, 2 µL cDNA template, and 7.2 µL RNase-free ddH2O. The RT-PCR was performed using 40 cycles under the following conditions: 95°C for 1 min for pre-degeneration, 95°C for 5 s for degeneration, and 60°C for 30 s for the extension. Afterward, the samples were heated to 95°C for 15 s and then 60°C for 1 min for dissolution curve analysis. Three technical replicates and three biological replicates were used for each sample and randomly selected genes. The primers used in the present study were designed using primer3plus (http://www.primer3plus.com/) and are listed in Supplementary Table S9. UBCE2 was chosen as the reference gene (Ye et al., 2018), and the −2–△△Ct method was used to calculate the relative gene expression levels (Schmittgen and Livak, 2008).
Statistical analysis
The statistical analyses were conducted using SPSS Statistics version 20 software, and the Student t-test was selected for significant difference analysis (* p < 0.05 and ** p < 0.01).
Results
Identification of the EuNAC genes in E. ulmoides
Based on the genomic information of E. ulmoides, 74 putative NAC genes were obtained using HMM. According to the results of CDD and SMART databases, three genes without NAC and NAM domains were eliminated, leaving 71 EuNAC genes, which were named EuNAC1 to EuNAC71 according to their chromosomal position (Supplementary Table S1).
Gene characteristics, including the length of the CDS, the length of the protein sequence, the protein MW, pI, GRAVY, aliphatic index, and the subcellular localization, were examined (Supplementary Table S2). The protein sequence length of all EuNAC proteins ranged from 86 (EuNAC49) to 617 (EuNAC65) amino acids. The MW of the proteins was between 9821.26 (EuNAC49) and 70458.62 (EuNAC65) Da, and the pI ranged from 4.51 (EuNAC20) to 10.01 (EuNAC5).
The aliphatic index varied from 47.7 (EuNAC35) to 100.0 (EuNAC13), which suggested that these predicted EuNAC proteins contained rich aliphatic amino acids. The GRAVY values of the EuNAC proteins were negative, except for EuNAC13, which has a positive value, indicating that most EuNAC proteins were hydrophobic. The predicted subcellular localization results showed that 69 EuNAC proteins were located in the nuclear region, whereas EuNAC13 was located in the nuclear or mitochondrion, and EuNAC60 was located in the nuclear or cytoplasm.
Phylogenetic analysis and classification of EuNAC genes
To explore the evolutionary relationship of the NAC gene family in E. ulmoides, an unrooted neighbor-joining (NJ) tree (with 1000 bootstraps) was constructed using the amino acid sequence alignment of NAC proteins from E. ulmoides and A. thaliana (Figure 1). The 71 E. ulmoides NAC genes could be divided into 17 subgroups: the ONAC022, AtNAC3, ATAF, NAP, SENU5, ONAC003, TIP, OsNAC8, ANAC063, TERN, ANAC006, NAC2, ANAC011, NAC1, NAM, OsNAC7 subgroups and an E. ulmoides-specific subgroup, named Eu_NAC. However, in E. ulmoides, no NAC members were detected from the ANAC001 subgroup. The subgroups AtNAC3, OsNAC8, TERN, and ANAC063 each contained only one EuNAC protein, and only two EuNAC proteins belonged to the subgroups NAP and SENU5 each, whereas the subgroup OsNAC7 contained the greatest number of EuNAC proteins (12).
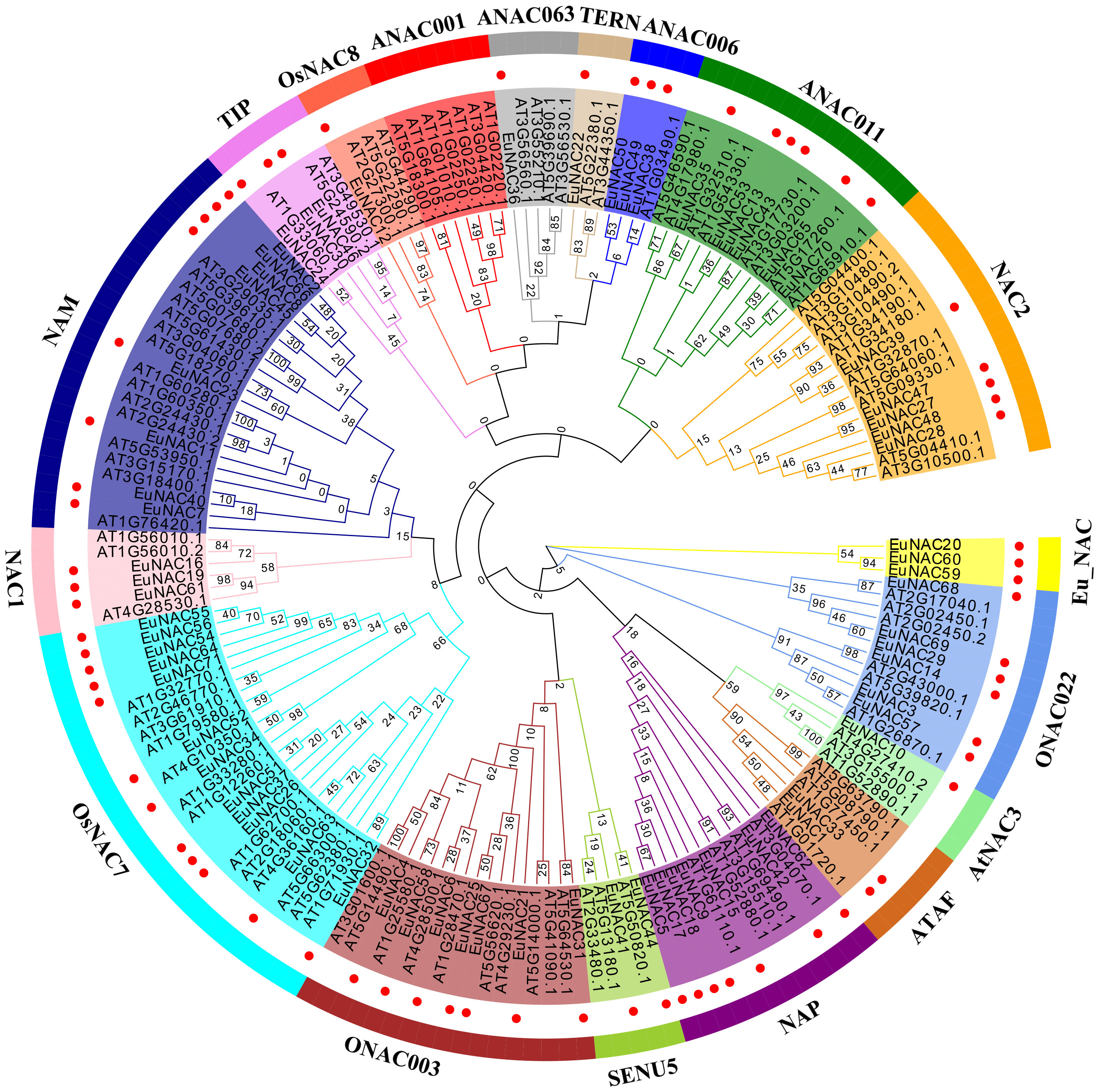
Figure 1 Phylogeny of the NAC TFs of Arabidopsis thaliana and Eucommia ulmoides. The tree branched the NAC proteins into different groups illustrated by different colored clusters within the clade. Markers of genes with red circles were EuNAC. The phylogenetic tree was prepared using the neighbor-joining (NJ) method with 1000 bootstrap replicates using MEGA 6.0.
Gene structures, domain, and putative motifs characterization of EuNAC genes
The NAC proteins comprised an N-terminal NAC domain and a long transcriptional activation region at the C-terminal (Figure 2C; Supplementary Table S3). As shown in Figure 3, All the EuNAC proteins contained five NAC subdomains (A–E), except EuNAC13, EuNAC43, EuNAC5, and EuNAC18, which lacked domains E.
To reveal the protein structural diversification of EuNAC proteins, 10 conserved motifs were identified by MEME (Figure 2B). The amino acid sequences of each motif are listed in Figure S1. The lengths of these conserved motifs ranged from 11 to 57 amino acids and were highly diverse. Motifs 1–6 were the most conserved motifs in EuNAC proteins, Motif 8 and Motif 9 only appeared in ONAC003. Among 71 EuNAC proteins, EuNAC49, EuNAC53, and EuNAC13 only had one type of motif, whereas EuNAC71, EuNAC64, EuNAC54, EuNAC56, EuNAC55, EuNAC65, EuNAC70, and EuNAC42 contain the largest number of motifs (eight types). The motifs of EuNAC members within the same subgroups show similar patterns, indicating that NAC proteins placed in the same group probably have similar functions. However, these motifs’ specific biological functions are unclear and need further study.
To obtain more insights into the evolution of the NAC family in E. ulmoides, the structural features of all the identified EuNAC genes were analyzed. As shown in Figure 2D, among the EuNAC genes, all of these contain exons; only EuNAC5, EuNAC17, EuNAC20, and EuNAC49 had one intron and two exons, and over half (40, 56.34%) had two introns and three exons, and only EuNAC46 genes had seven introns and eight exons (Figure 2D; Supplementary Table S4). Most genes from the same subgroup had a similar exon/intron structure; for example, 87.50% of NAM subgroup genes in E. ulmoides had three exons, and all of the ONAC022 subgroups genes had three exons.
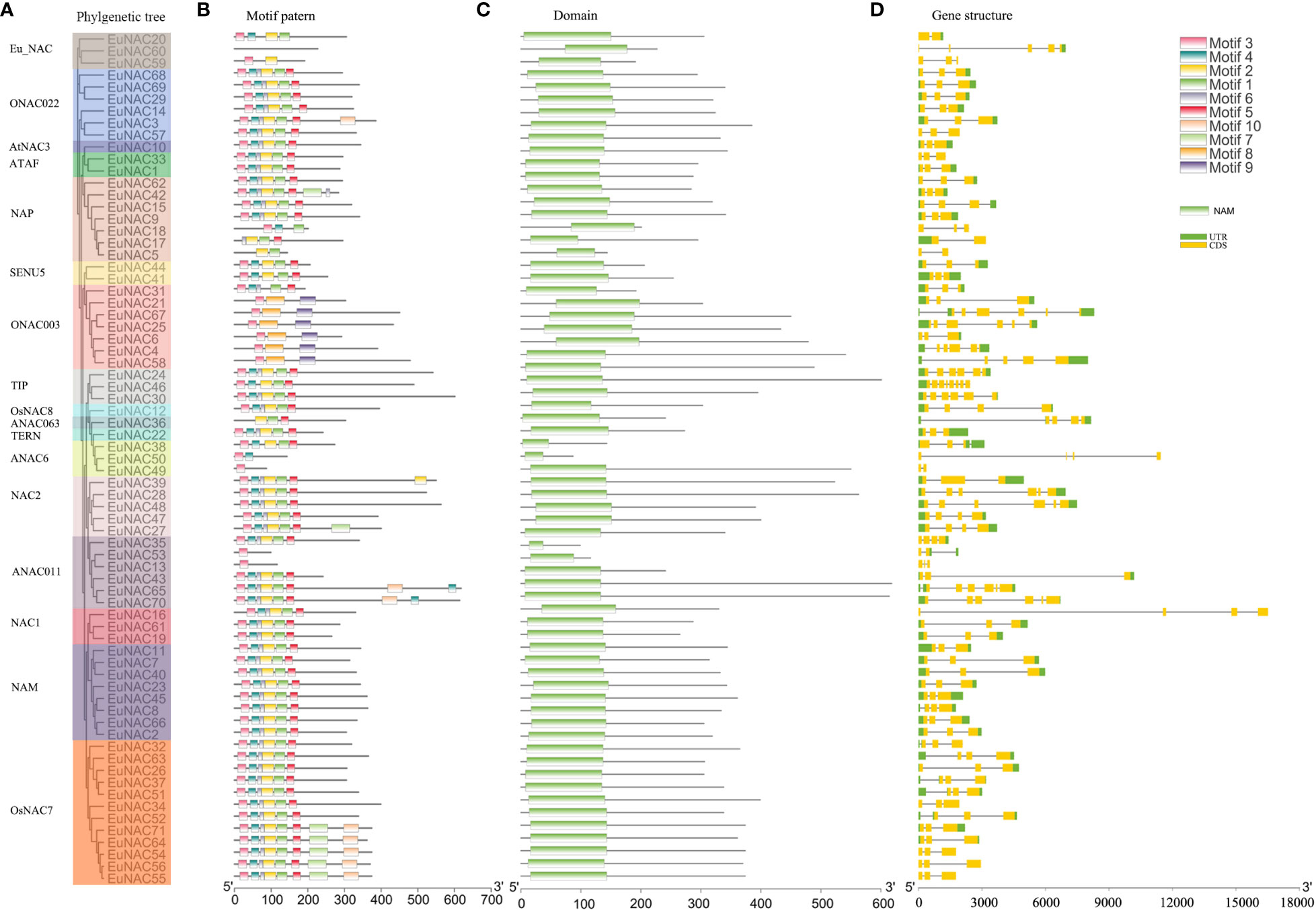
Figure 2 Phylogenetic relationships, domin, gene structure and architecture of conserved protein motifs in NAC genes from E. ulmoides. (A) Estimated phylogeny of EuNAC genes. (B) Different motifs were represented by diferent colors. The black lines represented the non-conserved sequences. Lengths of motifs for each EuNAC protein were displayed proportionally. (C) NAC domains were represented by green. (D) UTR were represented by green, CDS were represented by yellow. Nucleic acid lengths are indicated by the scale at the bottom.
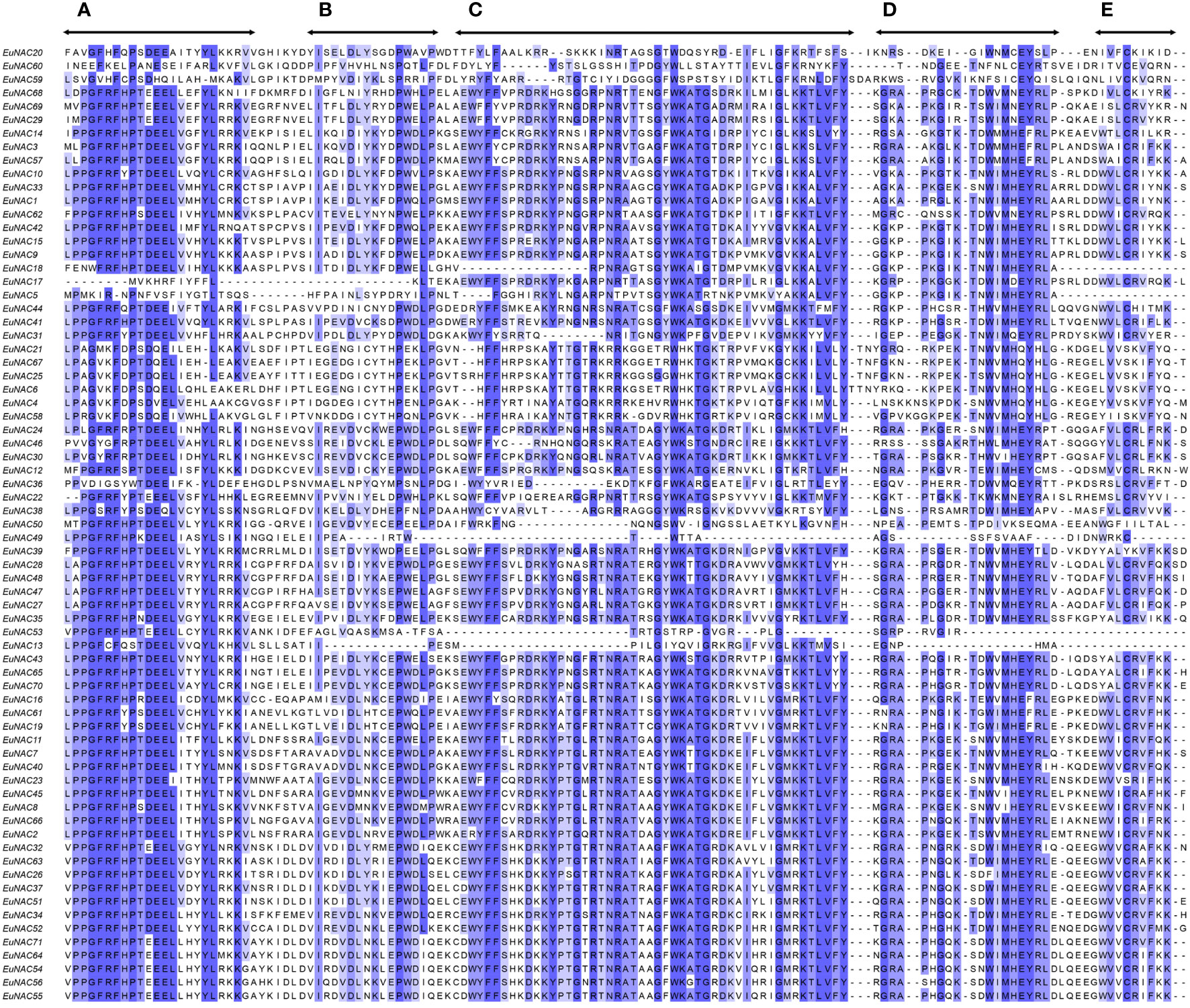
Figure 3 Sequence alignment of two groups of EuNAC domains amino acid sequences. Subdomains (A–E) are shown by arrows above the sequences. Overall conserved amino acids were shaded in blue.
Chromosome distribution and synteny analysis of EuNAC genes
A total of 71 EuNAC genes were unevenly distributed on 16 chromosomes of E. ulmoides, there was no EuNAC located on chromosome 11. Chromosome 8 had the largest number of EuNAC genes (8, 25.35%), and chromosome 3 only harbored EuNAC12 (Figure 4). The E. ulmoides-specific NAC genes are distributed on chromosomes 5 and 14. The EuNAC70 and EuNAC71 genes mapped to the scaffolds of the E. ulmoides genome.
Tandem duplication is an essential source for the origin and evolution of multigene families. In this study, only three pairs of genes of tandem duplicates genes in the EuNAC gene family were identified: EuNAC9/10, EuNAC27/28, and EuNAC 55/56. These are highlighted with a red rectangle (Figure 5). The tandem duplicated genes are presented on chromosomes 2, 7, and 13. There were 12 segmental duplication gene pairs identified in the E. ulmoides NAC gene family. In addition, the Ka/Ks values for the EuNAC genes in tandem and segmental duplications were calculated to determine the selection type that promoted the evolution of the EuNAC family. The Ka/Ks values of segmental duplication gene pairs ranged from 0.11 to 0.33, and those of the three tandem duplication gene pairs varied from 0.15 to 0.55, showing that all the gene pairs have a Ka/Ks ratio <1 (Supplementary Table S5). These results indicated that the evolution of EuNAC genes is mainly affected under purification selection pressure.
To further explore the evolutionary relationship of the NAC gene family in E. ulmoides, we constructed syntenic maps of the E. ulmoides compared with six different species including four dicotyledons (Arabidopsis thaliana, Coffea canephora, Vitis vinifera and Hevea brasiliensis) and two monocotyledons (Oryza sativa and Sorghum bicolor). A total of 58, 46, 26, 87, 5, and 10 similar NAC gene pairs were identified between E. ulmoides and A.thaliana, C.canephora, V.vinifera, H.brasiliensis, O.sativa, and S.bicolor, respectively (Figure 6, Supplementary Table S6).
Cis-acting elements in the promoters of EuNAC genes
To investigate the cis-acting elements of the 71 EuNAC genes, a 2000-bp sequence upstream from the translational start codon was analyzed. The cis-acting elements of the EuNAC genes contained 35 categories, which were related to phytohormone responsive, stress-responsive, light responsiveness, and plant growth and development (Figure 7; Supplementary Table S7).
We found that 757 (28.33%) elements, were involved in light responsiveness, including AE-box, ATC-motif, ATCT-motif, Box 4, CCGTCC-box, circadian, GATA-motif, G-box, GT1-motif, and MRE elements. In addition, 70 (2.62%) elements, including CAT-box, MSA-like, and O2-site, were related to plant growth and development. The promoter regions of many EuNAC genes contain multiple binding sites involved in stress response. A total of 925 (34.62%) elements were involved in stress response, such as ARE, GC-motif, LTR, MBS, MYB, TC-rich repeats, W box, and WUN-motif. The MYB element was related to drought inducibility, 69 EuNAC genes contained the MYB element, which accounted for 16.95% of the total number of cis-acting elements in the EuNAC family. Moreover, more than half of the EuNAC genes contained the W box element. A total of 2672 elements were predicted in the promoter regions of EuNAC genes. Among them, 920 (34.43%) elements were involved in response to plant hormones, such as ABA (ABRE), auxin (AuxRR-core, TGA-element), ethylene (ERE), GA3 (P-box, TATC-box, GARE-motif), salicylic acid (TCA-element), and methyl jasmonate (CGTCA-motif, TGACG-motif, MYC). Except for EuNAC7 and EuNAC8, all EuNAC genes contained MeJA-responsive elements, which accounted for 18.56% of the total number of cis-acting elements in the EuNAC genes. In addition, 58 EuNAC genes contained ABA-responsive elements, accounting for 6.62% of the total number of cis-acting elements in the EuNAC family, followed by 108 (4.04%) ethylene-responsive elements.
Expression profiling of EuNAC genes in various tissues
To determine the expression patterns of individual NAC genes in various tissues, a hierarchical clustering heat map was constructed using the public RNA-seq data obtained from NCBI (Figure 8). A total of 71 EuNAC genes were divided into eight groups based on their expression profiles. The expression levels of EuNAC genes were significantly different in diverse tissues. For instance, the FPKM of 28 EuNAC genes in Group E was less than 0.1 in all examined organizations, wherein EuNAC5, EuNAC9, EuNAC18, EuNAC29, EuNAC53, EuNAC55, EuNAC56, EuNAC58, and EuNAC62 had a FPKM value of 0, reflecting their lack of expression during the sampled stages. In contrast, 11 EuNAC genes in Group A were expressed at high levels in all examined tissues. In addition to low expression in leaves, EuNAC12, EuNAC15, and EuNAC60 were the highest transcript abundances in the peel, seed, and xylem. Some genes exhibited significant trends in different tissues. Ten genes in the xylem in group D, two genes in the seed in group F (EuNAC45 and EuNAC46), and seven genes in the peel in group H presented high transcript abundances and might play a critical role in the development of distinct tissues. The Group C genes were significantly induced in the peel and seed but rarely expressed in the leaf and xylem, meaning that EuNAC10, EuNAC42, and EuNAC8 were associated with fruit development. The transcriptional levels of EuNAC69, EuNAC68, EuNAC1, and EuNAC22 in Group G were highly expressed in leaves and peel but rarely expressed in the xylem and seeds.
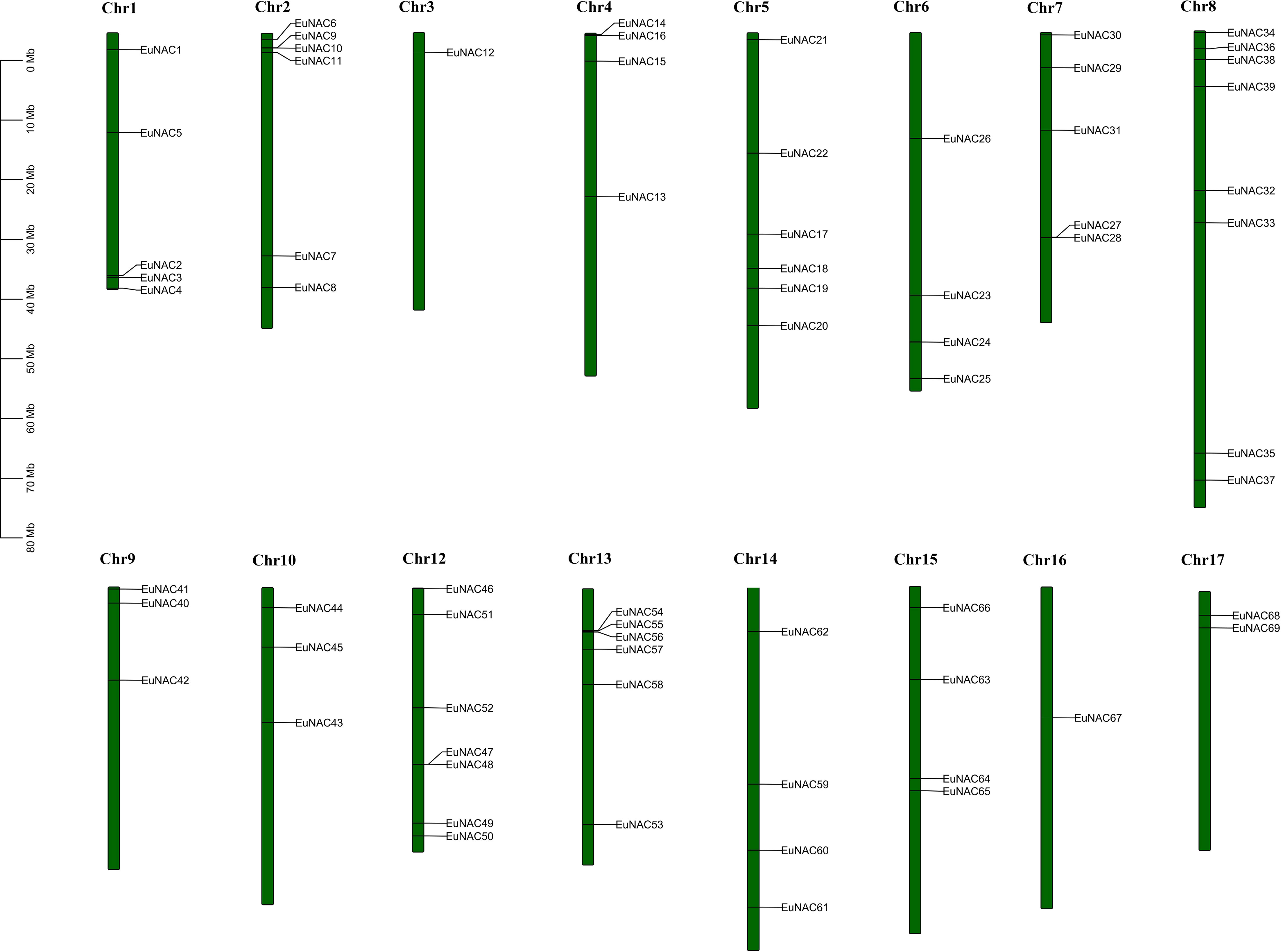
Figure 4 Gene Location on chromosome of E. ulmoides. Vertical bars represent the chromosomes of E. ulmoides. The chromosome number is to the up of each chromosome. The scale on the left represents chromosome length.
Co−expression networks between EuNAC genes and Eu-rubber biosynthesis genes
It has been reported that at least 52 genes in E. ulmoides are involved in the biosynthetic pathway of Eu-rubber (Li et al., 2020). The molecular structure of Eu-rubber is trans-1,4-polyisoprene (TPI), which is synthesized from the precursor ispentenyl diphosphate (IPP) via the MVA and MEP pathways.
To understand the possible relationship between NAC transcription factors in E. ulmoides and Eu-rubber biosynthesis pathway genes, we constructed a co-expression network containing Eu-rubber biosynthesis genes and 71 EuNAC genes (Figure 9). The screening thresholds were |r| ≥0.60 and p < 0.05. The larger nodes have stronger connectivity degrees, indicating that the genes may be more important. In the positive regulatory co-expression network, we identified 345 pairs correlated between 46 Eu-rubber structural genes and 48 EuNAC genes. The degree means the number of Eu-rubber genes associated with EuNAC. Statistical analysis showed that 11 EuNAC genes had a degree of connection greater than 10. Among them, EuNAC22 had the highest degree of connection, followed by EuNAC1, EuNAC68, and EuNAC69, all of which have a degree of connection greater than 20, suggesting that these four genes may play a crucial positive role in regulating Eu-rubber biosynthesis. In the negative regulatory co-expression network, we identified 132 pairs correlated between 37 Eu-rubber structural genes and 35 EuNAC genes, which was significantly less than the number of positive regulatory genes. Only EuNAC12 and EuNAC59 had a connection greater than 10, which might negatively regulate Eu-rubber biosynthesis. The results showed that EuNAC22, EuNAC1, EuNAC68, EuNAC69, EuNAC12, and EuNAC59 have the highest degree of connectivity in the co-expression network, indicating that these six genes are probably important in the regulation of Eu-rubber.
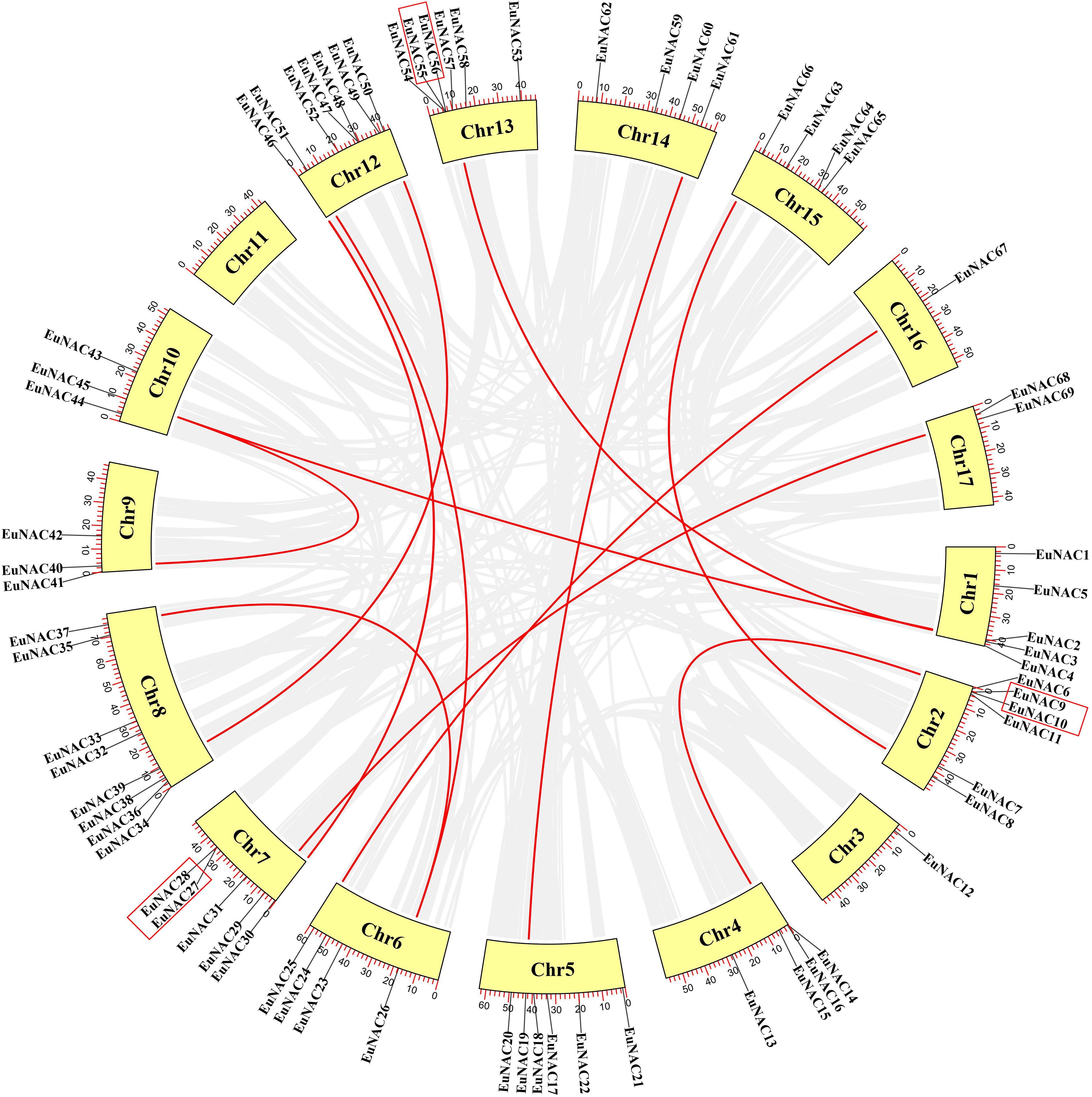
Figure 5 Schematic representations for the chromosomal distribution and interchromosomal relationships of EuNAC genes. Gray lines indicate all synteny blocks in the pineapple genome, and the red lines indicate duplicated NAC gene pairs. The tandem duplicated genes were highlighted with a red rectangle. The red scale bar marked on the chromosome represents the length of the chromosome (Mb). The chromosome number is indicated at the bottom of each chromosome.
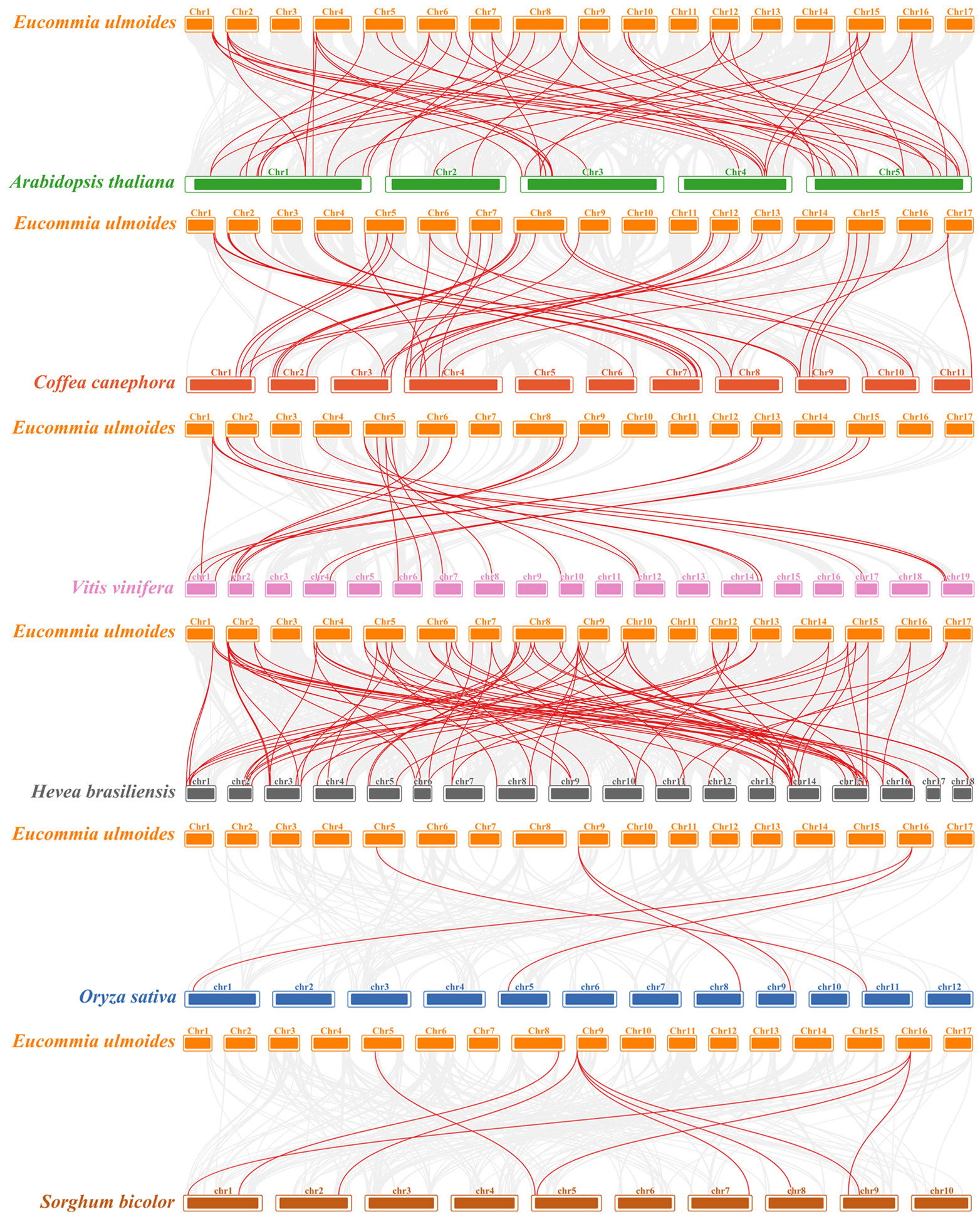
Figure 6 Synteny analysis of NAC genes between E. ulmoides and six representative plant species. Gray lines in the background indicate the collinear blocks within the E. ulmoides and other plant genomes, whereas the red lines highlight the syntenic NAC gene pairs.
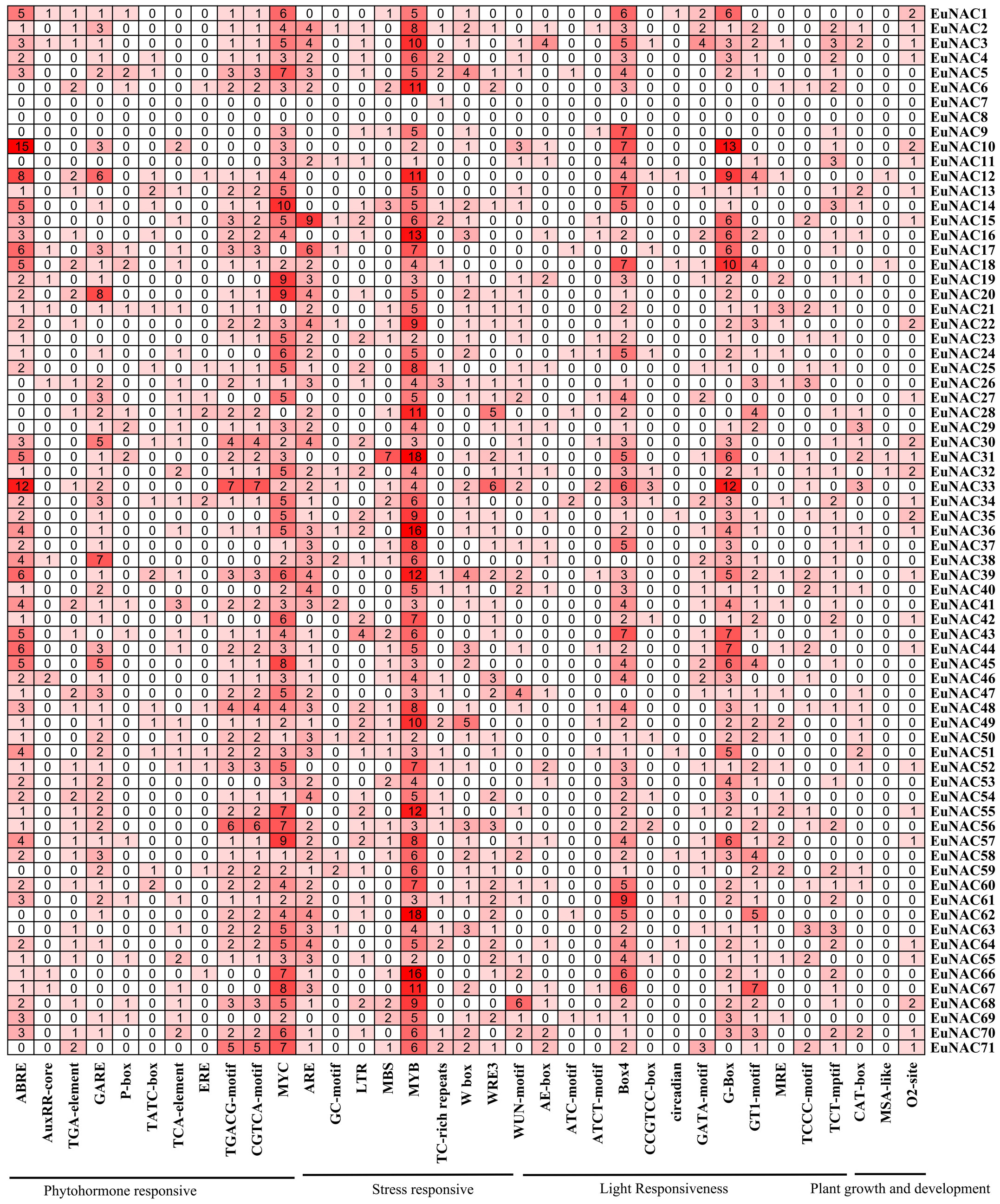
Figure 7 Cis-acting elements in promoter region of EuNAC genes in E. ulmoides. The number and the shade of red indicate the number of cis-acting element.
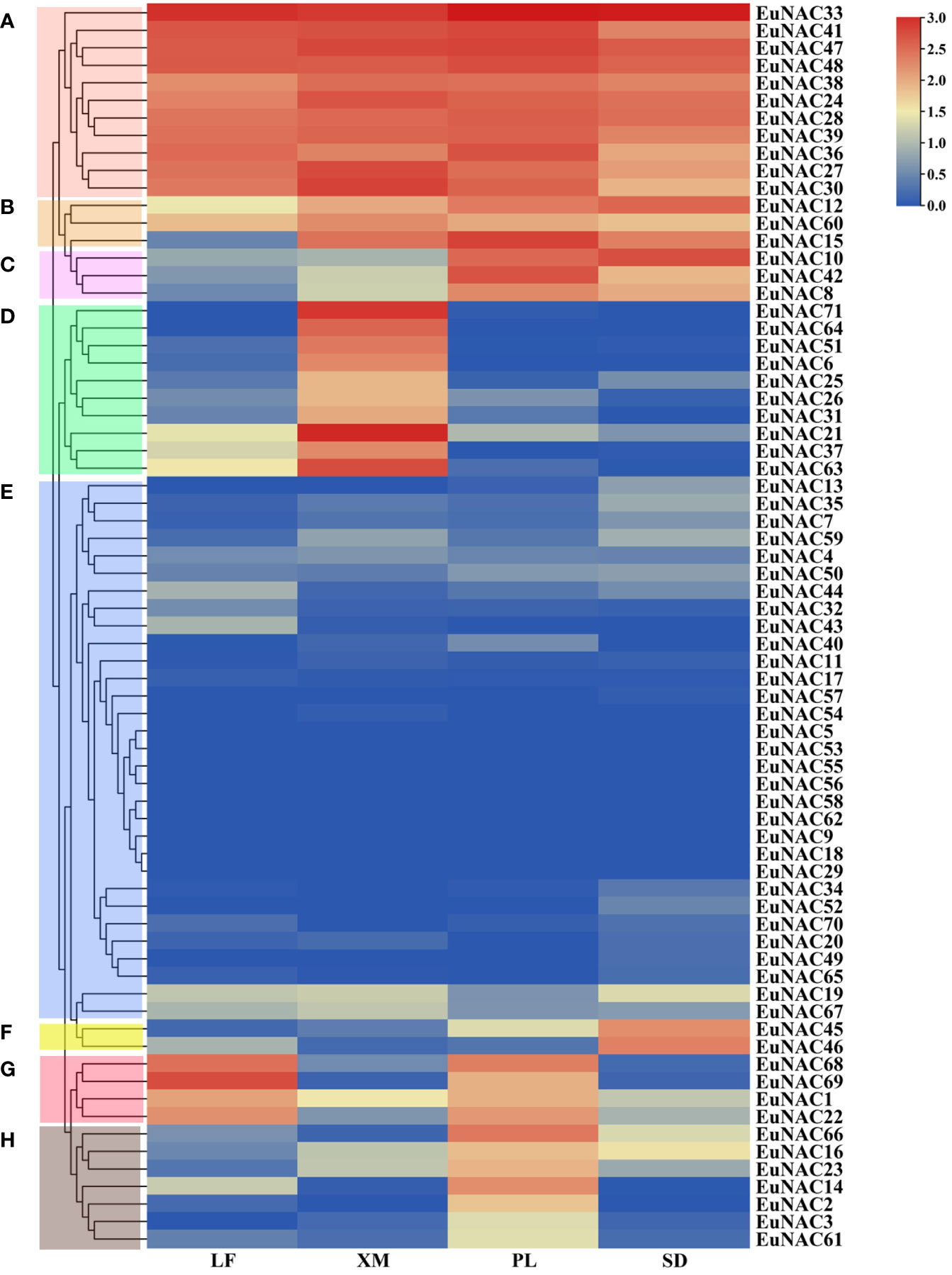
Figure 8 Expression levels of 71 EuNAC genes in different tissues. The expression level was presented based on the transformed data of log2 (FPKM+1) values. A total of 71 EuNAC genes were divided into (A–H) groups based on their expression levels. LF leaf; PL peel; XM xylem; SD seed.
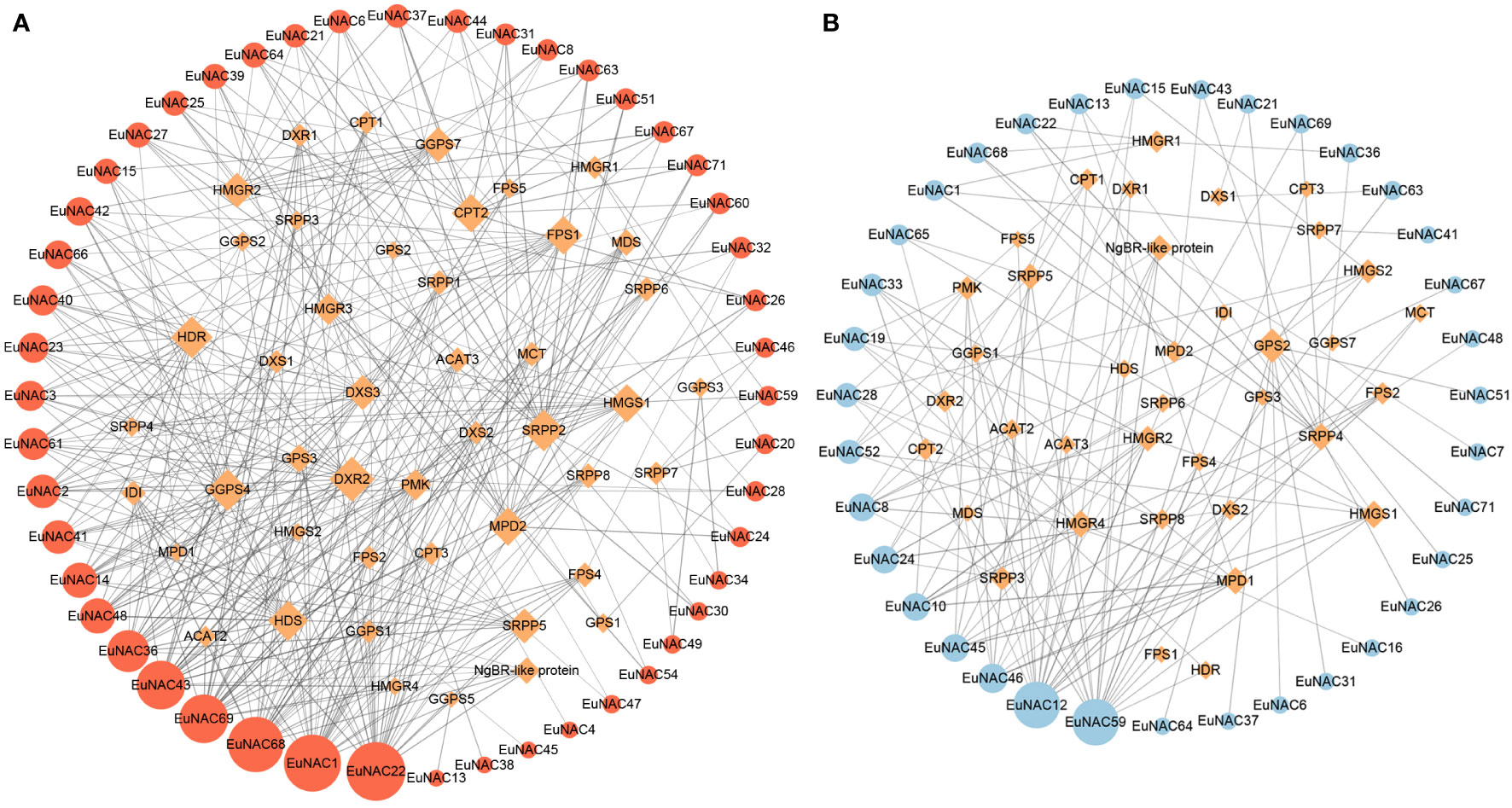
Figure 9 Co-expression networks between EuNAC genes and Eu-rubber biosynthesis genes. (A) Positive regulatory co-expression network between EuNAC genes and Eu-rubber biosynthesis genes. (B) Negative regulatory co-expression network between EuNAC genes and Eu-rubber biosynthesis genes. Orange diamond nodes represent Eu-rubber biosynthetic structural genes. Red circular nodes represent positively regulated EuNAC genes, and blue circular nodes represent negatively regulated EuNAC genes. The node size is positively correlated with the degree of the connectivity of the genes. The width of the connecting line is positively related to the correlation between genes.
EuNAC genes expression in response to hormone treatment
Co-expression analysis was consistent with the results of expression profiles in different tissues, indicating that EuNAC22, EuNAC1, EuNAC68, EuNAC69, EuNAC12, and EuNAC59 may play an important role in the regulation of Eu-rubber biosynthesis. To further explore the potential role of these six genes in Eu-rubber biosynthesis, we selected these six genes as candidate genes and paid attention to the expression levels of these genes after exogenous hormone treatment, the expression levels of the EuNAC gene family were tested in E. ulmoides by qRT-PCR under GA3, BR, and DCPTA treatments.
There were significant expression changes of these genes under different treatments (Figure 10). Among them, the expression levels of EuNAC22, EuNAC68, and EuNAC69 were shown to be significantly up-regulated under GA3 treatment at all detected time points, while the expression levels of EuNAC12 and EuNAC59 were observably decreased, the expression level of EuNAC1 was only significantly increased at 12 h after GA3 treatment. Under BR treatment, the expression level of EuNAC1 significantly increased 6 h and 24 h after treatment, the expression levels of EuNAC22 and EuNAC68 significantly increased at all subsequent time points after treatment (except for 3 h after treatment), and the expression level of EuNAC69 was significantly induced. The expression level of EuNAC59 was not different from the control at 24 h after treatment, and EuNAC12 and EuNAC59 were significantly inhibited at other time points. Under DCPTA treatment, the expression of EuNAC1 and EuNAC22 were highly induced, and EuNAC68 was significantly increased before 24 h of DCPTA treatment. Except for 12 h after treatment, the expression level of EuNAC69 was higher than that of the control after DCPTA treatment.
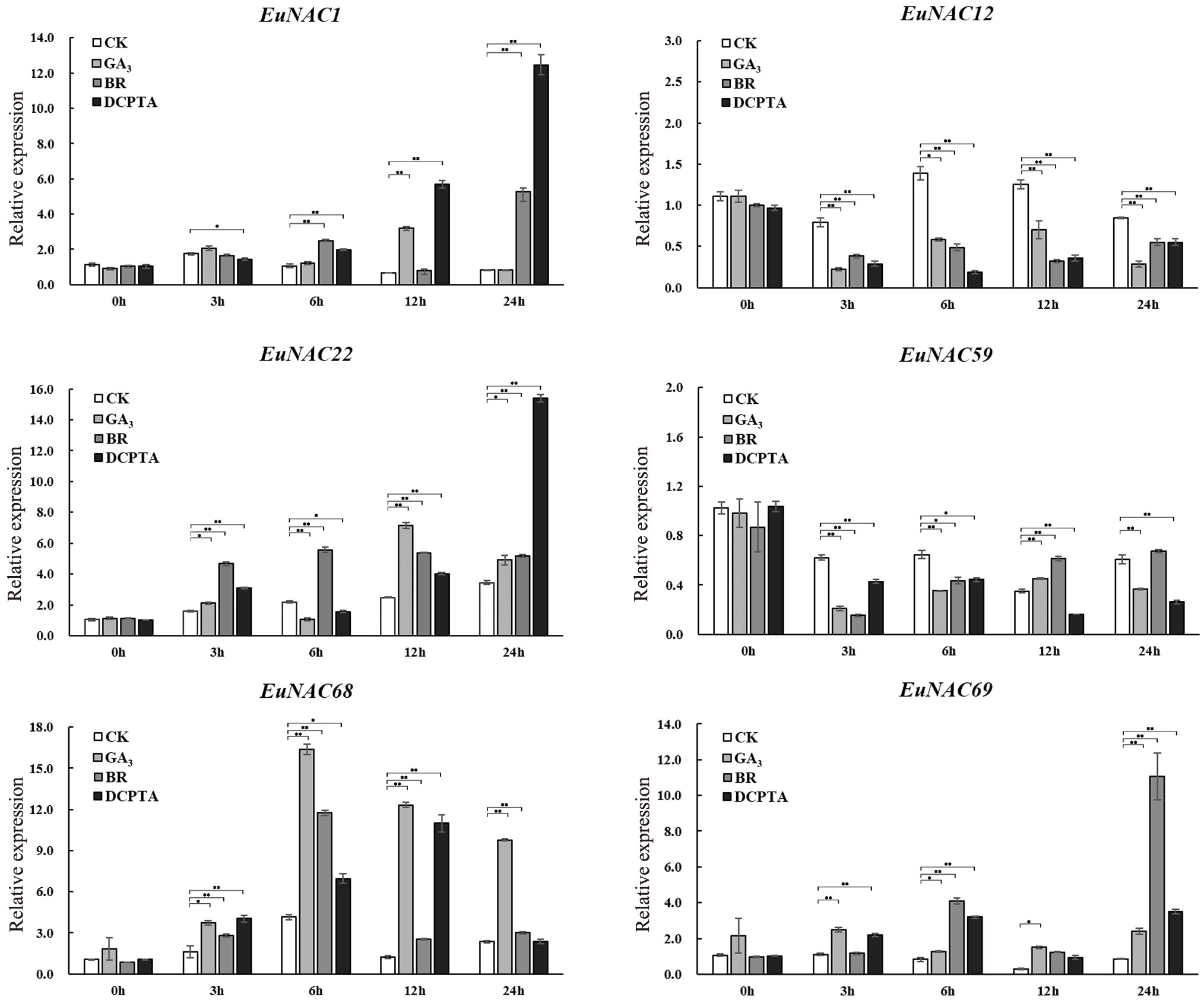
Figure 10 The expression patterns of E. ulmoides NAC genes under control condition and hormone treatments were examined by qRT-PCR. Five-month-old seedling leaves of E. ulmoides were sprayed with 300 mg/L gibberellin (GA3), 5 mg/L 1% brassinolide (BR), and 500 mg/L 2-(3,4-dichlorophenoxy)-triethylamine (DCPTA) until there is liquid dripping. Spray water was used as the control treatment. Error bars were obtained from three measurements. The significance analysis was carried out using Student’s t-test (* p < 0.05, ** p < 0.01).
EuNAC1 and EuNAC22 had the highest expression levels under the DCPTA treatment, EuNAC68 had the highest expression levels under the GA3 treatment, and EuNAC69 had the highest expression levels under the BR treatment. The expression of EuNAC68 quickly reached a maximum at 6 h after GA3 and BR treatments but reached a maximum at 12 h after the DCPTA treatment. The expression of EuNAC1 and EuNAC22 was significantly up-regulated at 24 h after DCPTA treatment. In addition, EuNAC1 and EuNAC69 showed relative expression peaks at 24 h after BR treatment. These results suggest that the EuNAC genes respond to hormonal treatment and that multiple EuNAC genes may play important roles in the different stages of the hormone response.
Discussion
The NAC family is one of the largest plant transcription factor families. The NAC transcription factors play a critical role in plant growth and development, secondary metabolite synthesis, biotic and abiotic stresses, and hormone signaling pathways (Wang et al., 2014a; Wang et al., 2014b; Liu et al., 2016; Chen et al., 2018). Comprehensive investigations of the NAC family genes have been carried out in various plant species, such as Arabidopsis, rice, maize, and soybean. However, a systematic and comprehensive analysis of the E. ulmoides NAC family had not yet been carried out. In this study, we identified and comprehensively analyzed the genes of the NAC family of E. ulmoides.
The number of members of the NAC transcription factor gene family in different species varies significantly. In this study, we used the E. ulmoides genome to identify 71 EuNAC genes; the number of NAC genes was similar to C. canephora (63) (Dong et al., 2019) and V. vinifera (74) (Wang et al., 2013), less than 105 members were identified in A. thaliana (Ooka et al., 2003), 163 members were identified in P. trichocarpa (Hu et al., 2010), 189 members were identified in E. grandis (Hussey et al., 2015). In this study, a total of 71 NAC genes were identified based on the E. ulmoides genome (size of 1.02 G) (Li et al., 2020), while the genome size of Arabidopsis is 125 Mb with 105 NAC members, and the genome size of rice is 466 Mb with 151 NAC members (Ooka et al., 2003). It was indicated that the genome size and the number of NAC family members are not always related. We suspected that some EuNAC genes were lost during evolution, and similar evolutionary loss events occurred in the WRKY and GLK transcription factor family (Liu et al., 2021; Liu et al., 2022). The diversity in the number of NAC gene family members in different species may be influenced genome duplication events, such as whole-genome duplication, segmental duplication, or tandem duplication (Zhang, 2003; Chang and Duda, 2012). In the study, three pairs of genes of tandem duplication and 12 segmental duplication were identified among the EuNAC genes. Therefore, gene duplication, especially segmental duplication, may provide the primary driving force of expansion of EuNAC. This result indicated that, although some EuNAC genes were lost during evolution, the sufficient genetic diversity has been retained in E. ulmoides.
The number of NAC gene family members identified in different plants is varied. We constructed a phylogenetic tree for E. ulmoides and A. thaliana and divided the 71 EuNAC genes into 17 subgroups according to the classification of NAC proteins in A. thaliana, including one E. ulmoides-specific subgroup. But the result is inconsistent with other species, such as tomato (12 subgroups) (Kou et al., 2014), Dactylis glomerata (14 subgroups) (Yang et al., 2021), Dimocarpus longan (12 subgroups) (Munir et al., 2020), and Pyrus bretschneideri (38 subgroups) (Gong et al., 2019). This suggests that although the NAC gene family has similar origins, evolution differs between species. In addition, only five and 10 with the collinear relationship were found in O. sativa and S. bicolor. However, we identified 58, 26, 46, and 87 orthologous pairs in dicotyledonous plants A. thaliana, V. vinifera, C. canephora, and H. brasiliensis, respectively. These results show that the EuNAC genes had higher similarity and a closer evolutionary relationship with dicotyledons.
The various conserved motifs may be related to particular functions (Ooka et al., 2003; Jensen et al., 2010; Chen et al., 2011). The predicted 10 motifs are located at the N-terminal, comprising A–E subdomains (Figures 2B, 3). Motifs 1–6 were the most conserved, among which motifs 2 and 6 were considered as subdomain C, and motif 1 was considered as subdomain D, which may be responsible for DNA binding (Olsen et al., 2005). Moreover, subdomain A was represented by motif 3, which may be involved in dimerization (Ernst et al., 2004; Khedia et al., 2018). Motif 4 is considered subdomain B, and motif 5 is considered subdomain E, which is thought to be responsible for the functional diversity of the NAC proteins (Ooka et al., 2003). Motifs 8 and 9 were only found in ONAC003 (Figure 2B), which was consistent with the results in the NAC family members of kiwifruit (Jia et al., 2021) and orchardgrass (Yang et al., 2021), indicating that it is a typical component of these subgroups and may play an important role in stress responses and secondary cell wall formation (Hussey et al., 2011; Fang et al., 2015; Zhong et al., 2021b). These results confirm the classification of the EuNAC gene family and facilitate further study on the function of EuNAC genes. Most of the EuNAC had two introns and three exons (Figure 2D) and genes within the same phylogenetic clade (Figure 2A) have a similar number of exons. These results are consistent with poplar (Hu et al., 2010) and cucumber (Liu X. et al., 2018) NAC genes suggesting that the genetic makeup of NAC genes are similar with the previously reported species.
The same subgroup may have similar biological activities and functions (Li et al., 2021; Xu et al., 2021). The 71 EuNAC genes were divided into 17 groups, and the gene structure and motif arrangement of the same group of genes were similar (Figures 1, 2). It was possible to predict the functions of E. ulmoides NAC genes based on the functions of their Arabidopsis orthologues, which could also be potentially utilized for further functional studies (Lim et al., 2012). For example, NAP is related to leaf senescence (Guo and Gan, 2006), floral morphogenesis (Sablowski and Meyerowitz, 1998), and salt stress (Seok et al., 2017). Eucommia ulmoides had seven genes (EuNAC5, EuNAC9, EuNAC15, EuNAC17, EuNAC18, EuNAC42 and EuNAC62) in this group, which may also have these features. ANAC019 (AT1G52890), ANAC055 (AT3G15500), and ANAC072 (AT4G27410) belong to the AtNAC3 subgroup, their expression is induced by drought, high salinity, and ABA (Tran et al., 2004). Therefore, we speculate that EuNAC10 in the same subgroup is a drought and high salt responsive gene, which regulates the survival of E. ulmoides under adverse growth conditions.
In addition, transcription factors usually play a key role in controlling the expression of tissue-specific genes (Choi et al., 2006; Lim et al., 2012; Siew et al., 2016). This study provides useful clues for understanding gene function concerning specific processes. For instance, Group D had 10 genes that exhibited a higher expression level in the xylem than in other tissues. It is worth noting that these 10 genes have high homology with secondary wall-associated NAC (SWN) transcription factors (Supplementary Table S2), including SND, NST, and VND, which play critical and dominant roles in secondary cell wall biosynthesis (Hussey et al., 2011; Zhou et al., 2014; Zhang et al., 2020). These results demonstrate that the EuNAC genes in Group D might affect the lignin synthesis of E. ulmoides.
The cis-acting elements which were the binding regions of transcription factors play an important role in regulating gene expression (Liu et al., 2016; Kaur et al., 2017). There are four cis-acting elements in the EuNAC gene promoter: light-responsive elements, stress-responsive elements, hormone-responsive elements, and plant growth and development-related elements. Light-responsive elements are ubiquitous cis-acting elements in the EuNAC promoter, suggesting that light of different colors and intensities may regulate the expression of EuNAC genes through different pathways. Previous studies have shown that MYB26 was the upstream regulator of secondary wall-associated NAC (SWN) genes (Yang et al., 2007). Among the promoters of 71 EuNAC genes, 69 EuNAC genes contained MYB elements and 66 EuNAC genes contained MYC elements, indicating that MYB and MYC may be important upstream regulators. The ubiquitous MYB and MYC elements in EuNAC gene promoters indicate that many other MYBs and MYCs may regulate their expression by combining with EuNAC gene promoters. The regulation of NAC gene expression by plant hormones has been reported for many plants (Ohnishi et al., 2005; Ji et al., 2014; Kou et al., 2014; Nieuwenhuizen et al., 2015). These are inseparable from the fact that the promoter of the NAC gene has corresponding cis-acting elements. For example, a gibberellic acid-responsive element in the PeNAC1 promoter was required for response to gibberellic acid which influenced the salt-stress signaling pathway (Wang et al., 2016). In this study, 920 (34.43%) elements were involved in the response to plant hormones in the promoters of EuNAC genes (Figure 7); EuNAC genes were also associated with different hormone response classes, including 69 genes in MeJA, 58 genes in ABA, 53 genes in ethylene, and 34 genes in GA responsive cis-elements. This implied that the expression of EuNAC genes might be induced by ABA, MeJA, ethylene and GA3, EuNAC genes may play a central role in plant specific hormone signaling responses. Furthermore, the cis-acting elements and qRT-PCR analysis indicated that EuNAC1, EuNAC12, EuNAC22, EuNAC59, EuNAC68 and EuNAC69 responded to GA3 treatment might be dominated by cis-acting elements in the promoter region.
Some studies have reported that NAC transcription factors play important regulatory roles in plant natural rubber biosynthesis. For example, previous studies have found that HbNAC1 regulates natural rubber synthesis by interacting with natural rubber synthesis-related genes in H. brasiliensis (Cao et al., 2017). Eu-rubber is an important natural rubber, similar to H. brasiliensis natural rubber, which is composed of trans-polyisoprene and cis-polyisoprene, respectively. To explore the effect of EuNAC genes on Eu-rubber biosynthesis, a co-expression regulatory network including Eu-rubber biosynthesis genes and EuNAC was constructed. We found that the positive regulation between EuNAC and Eu-rubber biosynthesis genes is dominant. EuNAC22, EuNAC1, EuNAC68, EuNAC69, EuNAC12, and EuNAC59 have the highest degree of connectivity in the co-expression network, and EuNAC22, EuNAC1, EuNAC68, and EuNAC69 had the same expression trend as the Eu-rubber synthetic structural genes of E. ulmoides, but EuNAC12 and EuNAC59 had opposite expression trends. Interestingly, the transcript levels of EuNAC69, EuNAC68, EuNAC1, and EuNAC22 were consistent with the variation in Eu-rubber content (Wuyun et al., 2018; Li et al., 2020), that is, high expression in leaves and peel, but less expression in xylem and seeds. However, the expression level of EuNAC12 and EuNAC59 in the xylem and seeds was significantly higher than in leaves and peels. In summary, co-expression analysis was consistent with the results of expression profiles in different tissues, suggesting that EuNAC22, EuNAC1, EuNAC68, EuNAC69, EuNAC12, and EuNAC59 may play important roles in Eu-rubber biosynthesis.
It has been reported that the application of exogenous hormones, such as DCPTA, GA3 and BR, increases Eu-rubber concentration in E. ulmoides leaves (Liu H. et al., 2018). To explore the role of EuNAC genes in hormone response and Eu-rubber biosynthesis, we focused on the expression levels of EuNAC1, EuNAC12, EuNAC22, EuNAC59, EuNAC68, and EuNAC69 under exogenous GA3, BR, and DCPTA treatment. Six EuNAC genes responded to hormone treatments, but each gene had different expression patterns under different treatments. EuNAC68 had higher expression levels at 6 h with GA3 and 6 h with BR treatment, which then began to decrease, thus indicating that they may play different roles in the early and late stages of the stress response. EuNAC1, EuNAC22 and EuNAC69 reached a maximum at 24 h DCPTA treatment, whereas EuNAC68 had higher expression levels at 12 h, which shows that the EuNAC genes have different sensitivities to hormone treatment. The expression levels of EuNAC1, EuNAC22, EuANC68, and EuNAC69 were observably increased by GA3, BR, and DCPTA treatments. However, EuNAC12 and EuNAC59 expression levels were significantly decreased under all hormone treatments (Figure 9). These results indicate that the expression of EuNAC genes was induced and inhibited to different degrees under different hormone treatment conditions, the phenotypes induced by hormone treatment were consistent with the expression trends of EuNAC1, EuNAC22, EuNAC68, and EuNAC69 but opposite to EuNAC12 and EuNAC59. In conclusion, these findings supported the view that EuNAC may positively or negatively affect Eu-rubber biosynthesis.
Furthermore, many NAC transcription factors have been reported to activate the transcription of target genes by binding to the NACRS core cis-acting element (CACG or CATGT) at the promoter region (Fujita et al., 2004; Xu et al., 2013). ANAC019, ANAC055 and ANAC072 can bind to the core DNA binding element CACG in the promoter region of the drought-induced gene ERD1 (Tran et al., 2004). Moreover, CpNAC1 specifically binds to the NACBS element in the CpPDS2/4 promoter to modulate carotenoid biosynthesis (Fu et al., 2016). HbNAC1 was also found to bind to the cis-acting element CACG in the promoter region of SRPP in H. brasiliensis to regulate natural rubber synthesis (Cao et al., 2017). By analysing the promoters of the genes involved in the biosynthesis of Eu-rubber, it was observed that many genes contain multiple NACRS cis-acting elements, including the CACG element and CATGT element. Exceptions to this include GGPS6 (evm.model.Chr8.585) and FPS5 (Novel08257), which have only two CACG elements, and the NgBR-like protein (evm.model.Chr17.181), which has a single CACG element (Supplementary Table S8). It was deduced that EuNACs might regulate the synthesis of Eu-rubber by combining with the promoter of a gene involved in Eu-rubber biosynthesis. These results provide new insight and can be useful for further verification of the EuNAC gene functions in Eu-rubber biosynthesis.
Conclusion
In this study, 71 EuNAC genes were identifed from the E. ulmoides genome, which were unevenly distributed on 16 chromosomes. Based on the phylogenetic tree, all the EuNAC genes were divided into 17 subfamilies. A comprehensive analysis of gene structure, motif composition, chromosomal distribution, gene duplication, phylogenetic, and cis-acting elements in promoters and homologous relationships were investigated. In addition, the expression of EuNAC genes in different tissues, co-expression network analysis and responds to various phytohormones implied that six EuNAC genes may participate in the biosynthesis of Eu-rubber. In the future, more comprehensive and in-depth studies on the functional properties of the EuNAC genes will be required. The results of this study provides valuable information for further study on the molecular mechanism of EuNAC genes in the biosynthesis of Eu-rubber.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article.
Author contributions
YL and XK conceived and designed the research, SZ performed the experiments. TX, YR and ZL analyzed the data, LS provided plant materials. SZ wrote the manuscript, YL revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by the Fundamental Research Funds for the Central Universities (BLX202118).
Acknowledgments
We thank Charlesworth Author Services (https://www.cwauthors.com.cn/) for editing the English text of a draft of this manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1030298/full#supplementary-material
Abbreviations
Eu-rubber, Eucommia rubber; GA3, Gibberellin; BR, Brassinolide; ABA, Abscisic acid; JA, Jasmonic acid; MeJA, Methyl jasmonate; IAA, Indole acetic acid; CDS, Coding sequence; ORF, Open reading frame; PI, Theoretical isoelectric point; MW, Molecular weight; GRAVY, Grand average of hydropathicity; Ka, Nonsynonymous substitution rates; Ks, Synonymous substitution rates; qRT-PCR, Quantitative real-time polymerase chain reaction; DCPTA, 2-(3, 4-dichlorophenoxy)-triethyl amine; CPI, cis-polyisoprene; TPI, trans-polyisoprene.
References
Baboo, M., Dixit, M., Sharma, K., Saxena, N. S. (2011). Mechanical and thermal characterization of cis-polyisoprene and trans-polyisoprene blends. Polym. Bull. 66, 661–672. doi: 10.1007/s00289-010-0378-7
Cao, Y., Zhai, J., Wang, Q., Yuan, H., Huang, X. (2017). Function of Hevea brasiliensis NAC1 in dehydration-induced laticifer differentiation and latex biosynthesis. Planta 245, 31–44. doi: 10.1007/s00425-016-2589-0
Chang, D., Duda, T. F. J. (2012). Extensive and continuous duplication facilitates rapid evolution and diversification of gene families. Mol. Biol. Evol. 29, 2019–2029. doi: 10.1093/molbev/mss068
Chen, D., Chai, S., McIntyre, C. L., Xue, G. (2018). Overexpression of a predominantly root-expressed NAC transcription factor in wheat roots enhances root length, biomass and drought tolerance. Plant Cell Rep. 37, 225–237. doi: 10.1007/s00299-017-2224-y
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Xia, R. (2020). TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, Q., Wang, Q., Xiong, L., Lou, Z. (2011). A structural view of the conserved domain of rice stress-responsive NAC1. Protein Cell 2, 55–63. doi: 10.1007/s13238-011-1010-9
Choi, S. S., Bush, E. C., Lahn, B. T. (2006). Different classes of tissue-specific genes show different levels of noncoding conservation. Genomics 87, 433–436. doi: 10.1016/j.ygeno.2005.09.013
Dong, X., Jiang, Y., Yang, Y., Xiao, Z., Bai, X., Gao, J., et al. (2019). Identification and expression analysis of the NAC gene family in coffea canephora. Agronomy 9, 670. doi: 10.3390/agronomy9110670
Endo, H., Yamaguchi, M., Tamura, T., Nakano, Y., Nishikubo, N., Yoneda, A., et al. (2015). Multiple classes of transcription factors regulate the expression of VASCULAR-RELATED NAC-DOMAIN7, a master switch of xylem vessel differentiation. Plant Cell Physiol. 56, 242–254. doi: 10.1093/pcp/pcu134
Ernst, H. A., Nina Olsen, A., Skriver, K., Larsen, S., Lo Leggio, L. (2004). Structure of the conserved domain of ANAC, a member of the NAC family of transcription factors. EMBO Rep. 5, 297–303. doi: 10.1038/sj.embor.7400093
Fang, Y., Liao, K., Du, H., Xu, Y., Song, H., Li, X., et al. (2015). A stress-responsive NAC transcription factor SNAC3 confers heat and drought tolerance through modulation of reactive oxygen species in rice. J. Exp. Bot. 66, 6803–6817. doi: 10.1093/jxb/erv386
Fu, C. C., Han, Y. C., Fan, Z. Q., Chen, J. Y., Chen, W. X., Lu, W. J., et al. (2016). The papaya transcription factor CpNAC1 modulates carotenoid biosynthesis through activating phytoene desaturase genes CpPDS2/4 during fruit ripening. J. Agric. Food Chem. 64, 5454–5463. doi: 10.1021/acs.jafc.6b01020
Fujita, M., Fujita, Y., Maruyama, K., Seki, M., Hiratsu, K., Ohme-Takagi, M., et al. (2004). A dehydration-induced NAC protein, RD26, is involved in a novel ABA-dependent stress-signaling pathway. Plant J. 39, 863–876. doi: 10.1111/j.1365-313X.2004.02171.x
Gong, X., Zhao, L., Song, X., Lin, Z., Gu, B., Yan, J., et al. (2019). Genome-wide analyses and expression patterns under abiotic stress of NAC transcription factors in white pear (Pyrus bretschneideri). BMC Plant Biol. 19, 161. doi: 10.1186/s12870-019-1760-8
Guo, Y., Gan, S. (2006). AtNAP, a NAC family transcription factor, has an important role in leaf senescence. Plant J. 46, 601–612. doi: 10.1111/j.1365-313X.2006.02723.x
Guo, H., Xie, Q., Fei, J., Chua, N. (2005). MicroRNA directs mRNA cleavage of the transcription factor NAC1 to downregulate auxin signals for arabidopsis lateral root development. Plant Cell 17, 1376–1386. doi: 10.1105/tpc.105.030841
Hu, R., Qi, G., Kong, Y., Kong, D., Gao, Q., Zhou, G. (2010). Comprehensive analysis of NAC domain transcription factor gene family in Populus trichocarpa. BMC Plant Biol. 10, 145. doi: 10.1186/1471-2229-10-145
Hussey, S. G., Mizrachi, E., Spokevicius, A. V., Bossinger, G., Berger, D. K., Myburg, A. A. (2011). SND2, a NAC transcription factor gene, regulates genes involved in secondary cell wall development in arabidopsis fibres and increases fibre cell area in eucalyptus. BMC Plant Biol. 11, 173. doi: 10.1186/1471-2229-11-173
Hussey, S. G., Saïdi, M. N., Hefer, C. A., Myburg, A. A., Grima-Pettenati, J. (2015). Structural, evolutionary and functional analysis of the NAC domain protein family in eucalyptus. New Phytol. 206, 1337–1350. doi: 10.1111/nph.13139
Jensen, M. K., Kjaersgaard, T., Nielsen, M. M., Galberg, P., Petersen, K., O’Shea, C., et al. (2010). The Arabidopsis thaliana NAC transcription factor family: structure–function relationships and determinants of ANAC019 stress signalling. Biochem. J. 426, 183–196. doi: 10.1042/BJ20091234
Ji, L., Hu, R., Jiang, J., Qi, G., Yang, X., Zhu, M., et al. (2014). Molecular cloning and expression analysis of 13 NAC transcription factors in Miscanthus lutarioriparius. Plant Cell Rep. 33, 2077–2092. doi: 10.1007/s00299-014-1682-8
Jia, D., Jiang, Z., Fu, H., Chen, L., Liao, G., He, Y., et al. (2021). Genome-wide identification and comprehensive analysis of NAC family genes involved in fruit development in kiwifruit (Actinidia). BMC Plant Biol. 21, 44. doi: 10.1186/s12870-020-02798-2
Kaur, A., Pati, P. K., Pati, A. M., Nagpal, A. K. (2017). In-silico analysis of cis-acting regulatory elements of pathogenesis-related proteins of Arabidopsis thaliana and Oryza sativa. PloS One 12, e0184523. doi: 10.1371/journal.pone.0184523
Khedia, J., Agarwal, P., Agarwal, P. K. (2018). AlNAC4 transcription factor from halophyte Aeluropus lagopoides mitigates oxidative stress by maintaining ROS homeostasis in transgenic tobacco. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.01522
Kou, X., Wang, S., Wu, M., Guo, R., Xue, Z., Meng, N., et al. (2014). Molecular characterization and expression analysis of NAC family transcription factors in tomato. Plant Mol. Biol. Rep. 32, 501–516. doi: 10.1007/s11105-013-0655-3
Li, M., Hou, L., Liu, S., Zhang, C., Yang, W., Pang, X., et al. (2021). Genome-wide identification and expression analysis of NAC transcription factors in Ziziphus jujuba mill. reveal their putative regulatory effects on tissue senescence and abiotic stress responses. Ind. Crop Prod. 173, 114093. doi: 10.1016/j.indcrop.2021.114093
Li, Y., Wei, H., Yang, J., Du, K., Li, J., Zhang, Y., et al. (2020). High-quality de novo assembly of the Eucommia ulmoides haploid genome provides new insights into evolution and rubber biosynthesis. Hortic. Res. 7, 183. doi: 10.1038/s41438-020-00406-w
Lim, C. J., Lee, H. Y., Kim, W. B., Lee, B., Kim, J., Ahmad, R., et al. (2012). Screening of tissue-specific genes and promoters in tomato by comparing genome wide expression profiles of arabidopsis orthologues. Mol. Cells 34, 53–59. doi: 10.1007/s10059-012-0068-4
Liu, Y., Sun, J., Wu, Y. (2016). Arabidopsis ATAF1 enhances the tolerance to salt stress and ABA in transgenic rice. J. Plant Res. 129, 955–962. doi: 10.1007/s10265-016-0833-0
Liu, X., Wang, T., Bartholomew, E., Black, K., Dong, M., Zhang, Y., et al. (2018). Comprehensive analysis of NAC transcription factors and their expression during fruit spine development in cucumber (Cucumis sativus l.). Horti Res. 5, 31. doi: 10.1038/s41438-018-0036-z
Liu, J., Wang, X., Chen, Y., Liu, Y., Wu, Y., Ren, S., et al. (2021). Identification, evolution and expression analysis of WRKY gene family in Eucommia ulmoides. Genomics 113, 3294–3309. doi: 10.1016/j.ygeno.2021.05.011
Liu, Z., Xiong, T., Zhao, Y., Qiu, B., Chen, H., Kang, X., et al. (2022). Genome-wide characterization and analysis of golden 2-like transcription factors related to leaf chlorophyll synthesis in diploid and triploid Eucalyptus urophylla. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.952877
Liu, H., Zhu, J., Ding, H., Wang, L., Sun, Z., Liu, P., et al. (2018). Comprehensive analysis of NAC transcription factors and their expression during fruit spine development in cucumber (Cucumis sativus l.). Horti Res. 5. doi: 10.1038/s41438-018-0036-z
Lyu, F., Han, F., Ge, C., Mao, W., Chen, L., Hu, H., et al. (2023). OmicStudio: A composable bioinformatics cloud platform with real-time feedback that can generate high-quality graphs for publication. iMeta, e85. doi: 10.1002/imt2.85
Ma, W., Kang, X., Liu, P., She, K., Zhang, Y., Lin, X., et al. (2022). The NAC-like transcription factor CsNAC7 positively regulates the caffeine biosynthesis-related gene yhNMT1 in Camellia sinensis. Hortic. Res. 9, uhab046. doi: 10.1093/hortre/uhab046
Mohanta, T. K., Yadav, D., Khan, A., Hashem, A., Tabassum, B., Khan, A. L., et al. (2020). Genomics, molecular and evolutionary perspective of NAC transcription factors. PloS One 15, e231425. doi: 10.1371/journal.pone.0231425
Mooibroek, H., Cornish, K. (2000). Alternative sources of natural rubber. Appl. Microbiol. Biot. 53, 355–365. doi: 10.1007/s002530051627
Munir, N., Yukun, C., Xiaohui, C., Nawaz, M. A., Iftikhar, J., Rizwan, H. M., et al. (2020). Genome-wide identification and comprehensive analyses of NAC transcription factor gene family and expression patterns during somatic embryogenesis in Dimocarpus longan lour. Plant Physiol. Biochem. 157, 169–184. doi: 10.1016/j.plaphy.2020.10.009
Nakazawa, Y., Bamba, T., Takeda, T., Uefuji, H., Harada, Y., Li, X., et al. (2009). Production of eucommia-rubber from Eucommia ulmoides oliv. (Hardy rubber tree). Plant Biotechnol. (Tokyo Japan). 26, 71–79. doi: 10.5511/plantbiotechnology.26.71
Ng, D., Abeysinghe, J., Kamali, M. (2018). Regulating the regulators: The control of transcription factors in plant defense signaling. Int. J. Mol. Sci. 19, 3737. doi: 10.3390/ijms19123737
Nieuwenhuizen, N. J., Chen, X., Wang, M. Y., Matich, A. J., Perez, R. L., Allan, A. C., et al. (2015). Natural variation in monoterpene synthesis in kiwifruit: Transcriptional regulation of terpene synthases by NAC and ETHYLENE-INSENSITIVE3-Like transcription factors. Plant Physiol. 167, 1243–1258. doi: 10.1104/pp.114.254367
Nuruzzaman, M., Manimekalai, R., Sharoni, A. M., Satoh, K., Kondoh, H., Ooka, H., et al. (2010). Genome-wide analysis of NAC transcription factor family in rice. Gene 465, 30–44. doi: 10.1016/j.gene.2010.06.008
Ohnishi, T., Sugahara, S., Yamada, T., Kikuchi, K., Yoshiba, Y., Hirano, H., et al. (2005). OsNAC6, a member of the NAC gene family, is induced by various stresses in rice. Genes Genet. Syst. 80, 135–139. doi: 10.1266/ggs.80.135
Olsen, A. N., Ernst, H. A., Leggio, L. L., Skriver, K. (2005). DNA-Binding specificity and molecular functions of NAC transcription factors. Plant Sci. 169, 785–797. doi: 10.1016/j.plantsci.2005.05.035
Ooka, H., Satoh, K., Doi, K., Nagata, T., Otomo, Y., Murakami, K., et al. (2003). Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. DNA Res. 10, 239–247. doi: 10.1093/dnares/10.6.239
Roth, W. B., Carr, M. E., Davis, E. A., Bagby, M. O. (1985). New sources of gutta-percha in garrya flavescens and g. wrightii. Phytochemistry 24, 183–184. doi: 10.1016/S0031-9422(00)80836-5
Sablowski, R. W. M., Meyerowitz, E. M. (1998). A homolog of NO APICAL MERISTEM is an immediate target of the floral homeotic genes APETALA3/PISTILLATA. Cell 92, 93–103. doi: 10.1016/S0092-8674(00)80902-2
Schmittgen, T. D., Livak, K. J. (2008). Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 3, 1101–1108. doi: 10.1038/nprot.2008.73
Schlesinger, W., Leeper, H. M. (2002). Chicle - cis- and trans-polyisoprenes from a single plant species. Ind. Eng. Chem. 43, 398–403. doi: 10.1021/ie50494a034
Seok, H., Woo, D., Nguyen, L. V., Tran, H. T., Tarte, V. N., Mehdi, S. M. M., et al. (2017). Arabidopsis AtNAP functions as a negative regulator via repression of AREB1 in salt stress response. Planta 245, 329–341. doi: 10.1007/s00425-016-2609-0
Shafee, T., Lowe, R. (2017). Eukaryotic and prokaryotic gene structure. WikiJournal Med. 4, 1. doi: 10.15347/wjm/2017.002
Siew, S., Kaneko, M., Mie, M., Kobatake, E. (2016). Construction of a tissue-specific transcription factor-tethered extracellular matrix protein via coiled-coil helix formation. J. Mater. Chem. B 4, 2512–2518. doi: 10.1039/c5tb01579k
Sudhir, K., Glen, S., Li, M., Christina, K., Koichiro, T. (2018). MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Tran, L. P., Nakashima, K., Sakuma, Y., Simpson, S. D., Fujita, Y., Maruyama, K., et al. (2004). Isolation and functional analysis of arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell 16, 2481–2498. doi: 10.1105/tpc.104.022699
van Beilen, J. B., Poirier, Y. (2007). Establishment of new crops for the production of natural rubber. Trends Biotechnol. 25, 522–529. doi: 10.1016/j.tibtech.2007.08.009
Wang, J., Lian, W., Cao, Y., Wang, X., Wang, G., Qi, C., et al. (2018). Overexpression of BoNAC019, a NAC transcription factor from Brassica oleracea, negatively regulates the dehydration response and anthocyanin biosynthesis in arabidopsis. Sci. Rep. 8, 13349. doi: 10.1038/s41598-018-31690-1
Wang, J., Wang, J., Yang, H. (2016). Identification and functional characterization of the NAC gene promoter from populus euphratica. Planta 244, 417–427. doi: 10.1007/s00425-016-2511-9
Wang, Z., Rashotte, A. M., Dane, F. (2014b). Citrullus colocynthis NAC transcription factors CcNAC1 and CcNAC2 are involved in light and auxin signaling. Plant Cell Rep. 33, 1673–1686. doi: 10.1007/s00299-014-1646-z
Wang, Z., Rashotte, A. M., Moss, A. G., Dane, F. (2014a). Two NAC transcription factors from Citrullus colocynthis, CcNAC1, CcNAC2 implicated in multiple stress responses. Acta Physiol. Plant 36, 621–634. doi: 10.1007/s11738-013-1440-5
Wang, Y., Tang, H., Debarry, J. D., Tan, X., Li, J., Wang, X., et al. (2012). MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40, e49. doi: 10.1093/nar/gkr1293
Wang, Q., Xiong, Y., Dong, F. (2020). Eucommia ulmoides gum-based engineering materials: fascinating platforms for advanced applications. J. Mater. Sci. 56, 1855–1878. doi: 10.1007/s10853-020-05345-4
Wang, N., Zheng, Y., Xin, H., Fang, L., Li, S. (2013). Comprehensive analysis of NAC domain transcription factor gene family in Vitis vinifera. Plant Cell Rep. 32, 61–75. doi: 10.1007/s00299-012-1340-y
Wuyun, T. N., Wang, L., Liu, H., Wang, X., Zhang, L., Bennetzen, J. L., et al. (2018). The hardy rubber tree genome provides insights into the evolution of polyisoprene biosynthesis. Mol. Plant 11, 429–442. doi: 10.1016/j.molp.2017.11.014
Xu, Z. Y., Kim, S. Y., Hyeon, D. Y., Kim, D. H., Dong, T., Park, Y., et al. (2013). The arabidopsis NAC transcription factor ANAC096 cooperates with bZIP-type transcription factors in dehydration and osmotic stress responses. Plant Cell 25, 4708–4724. doi: 10.1105/tpc.113.119099
Xu, N., Meng, L., Song, L., Li, X., Du, S., Hu, F., et al. (2021). Identification and characterization of secondary wall-associated NAC genes and their involvement in hormonal responses in tobacco (Nicotiana tabacum). Front. Plant Sci. 12. doi: 10.3389/fpls.2021.712254
Yang, Z., Nie, G., Feng, G., Han, J., Huang, L., Zhang, X. (2021). Genome-wide identification, characterization, and expression analysis of the NAC transcription factor family in orchardgrass (Dactylis glomerata l.). BMC Genomics 22, 178. doi: 10.1186/s12864-021-07485-6
Yang, C., Xu, Z., Song, J., Conner, K., Vizcay Barrena, G., Wilson, Z. A. (2007). Arabidopsis MYB26/MALE STERILE35 regulates secondary thickening in the endothecium and is essential for anther dehiscence. Plant Cell 19, 534–548. doi: 10.1105/tpc.106.046391
Ye, J., Jin, C., Li, N., Liu, M., Fei, Z., Dong, L., et al. (2018). Selection of suitable reference genes for qRT-PCR normalisation under different experimental conditions in Eucommia ulmoides oliv. Sci. Rep. 8, 15043. doi: 10.1038/s41598-018-33342-w
Zhang, J. (2003). Evolution by gene duplication: An update. Trends Ecol. Evol. 18, 292–298. doi: 10.1016/S0169-5347(03)00033-8
Zhang, Q., Luo, F., Zhong, Y., He, J., Li, L. (2020). Modulation of NAC transcription factor NST1 activity by XYLEM NAC DOMAIN1 regulates secondary cell wall formation in arabidopsis. J. Exp. Bot. 71, 1449–1458. doi: 10.1093/jxb/erz513
Zhang, H., Xu, J., Chen, H., Jin, W., Liang, Z. (2021). Characterization of NAC family genes in Salvia miltiorrhiza and NAC2 potentially involved in the biosynthesis of tanshinones. Phytochemistry 191, 112932. doi: 10.1016/j.phytochem.2021.112932
Zhong, R., Kandasamy, M. K., Ye, Z. (2021b). XND1 regulates secondary wall deposition in xylem vessels through the inhibition of VND functions. Plant Cell Physiol. 62, 53–65. doi: 10.1093/pcp/pcaa140
Zhong, R., Lee, C., Haghighat, M., Ye, Z. H. (2021a). Xylem vessel-specific SND5 and its homologs regulate secondary wall biosynthesis through activating secondary wall NAC binding elements. New Phytol. 231, 1496–1509. doi: 10.1111/nph.17425
Keywords: Eucommia ulmoides, NAC transcription factor, gene family, gene expression, Eucommia rubber (Eu-rubber), hormone response
Citation: Zhang S, Xu T, Ren Y, Song L, Liu Z, Kang X and Li Y (2023) The NAC transcription factor family in Eucommia ulmoides: Genome-wide identification, characterization, and network analysis in relation to the rubber biosynthetic genes. Front. Plant Sci. 14:1030298. doi: 10.3389/fpls.2023.1030298
Received: 28 August 2022; Accepted: 13 February 2023;
Published: 03 April 2023.
Edited by:
Gabriella Sonnante, National Research Council (CNR), ItalyReviewed by:
Xiyang Zhao, Jilin Agricultural University, ChinaNunzio D’Agostino, University of Naples Federico II, Italy
Copyright © 2023 Zhang, Xu, Ren, Song, Liu, Kang and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yun Li, bGl5dW5fMzY1QGJqZnUuZWR1LmNu
 Shuwen Zhang
Shuwen Zhang Tingting Xu1,2,3,4
Tingting Xu1,2,3,4 Zhao Liu
Zhao Liu Xiangyang Kang
Xiangyang Kang