
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 03 October 2022
Sec. Plant Physiology
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.998664
This article is part of the Research Topic The Plant Cell Wall: Advances and Current Perspectives View all 14 articles
Embryonic and endosperm development are important biological events during Arabidopsis seed development, and are controlled by dynamic changes in a range of gene expression. Nevertheless, the regulatory mechanisms of endosperm cellularization and embryo differentiation remain unclear. Here, we characterized the early embryo and endosperm development of the naa15 mutant that had abnormal embryo differentiation and incomplete endosperm cellularization compared to WT of Arabidopsis, and comparatively investigated the changes of gene expressions in WT seeds at 3, 4, and 5 days after pollination (3W, 4W, and 5W) and the white homozygous aborted naa15 seeds at 5, 6, and 7 DAP (5M, 6M, and 7M) from naa15-1/+ siliques using RNA sequencing and qPCR assays. The transcriptome analyses showed that there were 2040 and 3630 differentially expressed genes (DEGs) in 4W (at endosperm cellularization initiation stage and heart embryo stage) vs 3W (at syncytium stage and globular embryo stage), and 5W (at end of endosperm cellularization stage and torpedo embryo stage) vs 4W, respectively. The KEGG and GO analyses showed that lipid metabolic processes and transmembrane transport related to cell wall biogenesis, cell division and differentiation, the plant hormone signaling pathway, photosynthesis, and transcription regulator activity were evidently enriched in WT and naa15. The heatmap and qPCR analyses showed that auxin response genes (ARFs), auxin transport genes (PINs) cytokinin synthesis genes (LOGs), cytokinin dehydrogenase genes (CKXs), cytokinin receptor, transcription factors (MYB, bHLH, MADS-box, and ERF) were significantly downregulated in naa15 compared to WT. A series of cell wall genes annotated to xyloglucan endotransglycosylase/hydrolase, pectin methyl esterase, and pectin methyl esterase inhibitor were also identified in these DEGs. Moreover, using an immunofluorescent assay, the features of cell walls displayed that cellulose fluorescence signals in the embryo and endosperm of naa15 were significantly decreased, and the signals of low- and high- methyl esterification of pectin were also obviously decreased in the endosperm of naa15. In summary, we identified a large number of DEGs and investigated the features of cell walls during endosperm cellularization and embryonic differentiation, which provided important information on transcription and gene expression to reveal their regulatory mechanisms.
Seed development in angiosperms is triggered by double fertilization, in which sperm cells reach the embryo sac, and then two sperm cells fuse with egg cell and central cell resulting in the formation of diploid embryo and triploid endosperm, respectively (Dresselhaus et al., 2016; Wang et al., 2022). The intact seed contains the embryo, endosperm, and seed coat (Lafon-Placette and Köhler, 2014). In Arabidopsis, the embryo undergoes a series of cell division and organ differentiation, leading to its patterning establishment and maturation. The embryo reaches the pre-globular stage after eight rounds of cell divisions. Meanwhile, the endosperm conducts rapid and successive free nuclear divisions without cell wall formation and enters the syncytial endosperm phase (Sørensen et al., 2002). With the embryo development transition from the globular to heart stage, endosperm cellularization begins from the micropylar zone near the developing embryo with cell wall formation and moves forward the chalazal pole of seed. When the embryo enters torpedo phase, endosperm completes cellularization process (Berger, 2003; Li et al., 2013). When embryo develops from heart to torpedo stage, chloroplast biogenesis initiates and embryo turns green (Mansfield and Briarty, 1991). Endosperm cellularization process is an important milestone in endosperm development and embryogenesis, and it proceeds gradually with embryo differentiation (Wang et al., 2022).
The regulatory mechanisms of embryo differentiation and endosperm cellularization are sophisticated and involve in multiplex pathways of gene regulation. The apetala2 (ap2) mutant endosperm undergoes an early expanded growth period to lead to delayed endosperm cellularization and overgrowth of the endosperm central vacuole (Ohto et al., 2009). MINI3 (WRKY10), a WRKY-type transcription factor, is specifically expressed in endosperm and embryo, and its mutant has a precocious cellularization of endosperm and produces small seeds (Luo et al., 2005). AGAMOUS-LIKE proteins (AGLs) are a large family of MADS-box proteins that form heterodimers or homodimers among the MADS-box family. In the three mutants of agl61, agl62, and agl80, embryogenesis and endosperm cellularization are damaged to varying degrees (Portereiko et al., 2006; Kang et al., 2008; Steffen et al., 2008). In other transcription factors, the genes of the ARF and bHLH families are expressed in seeds, and their deletions also affect development processes of embryo and endosperm, leading to seed abortion (Sun et al., 2010).
Plant hormones, important signaling molecules, play vital roles during the entire plant lifecycle. Auxin is critical in embryo and endosperm development, and its biosynthesis is activated when sperm-egg fuse to form zygotes (Figueiredo et al., 2015). However, the change in auxin homeostasis affects its signal transmission in the seed coat and embryo, eventually leading to seed abortion. Excessive accumulation of auxin delays endosperm cellularization (Batista et al., 2019). In seed formation, auxin and cytokinin are related to cell division and differentiation, as well as organ growth and development (Bishopp et al., 2011; Vanstraelen and Benkova, 2012). The sizes of embryos and seeds are increased owing to disordered cytokinin homeostasis in ahk2 ahk3 ahk4 mutants (Riefler et al., 2006). Brassinosteroids are essential steroid hormones that play critical roles in plant reproductive growth and seed development. In the rice BR-deficient mutant brd2 and Arabidopsis BR deficient mutants dwf5 and det2, there are smaller seeds than WT (Hong et al., 2005; Jiang and Lin, 2013).
In addition to hormones, endosperm and embryo development are also related to cell wall properties. Methyl-esterified pectin is demethylesterified by pectin methylesterases (PMEs), which change the rigidity and stiffening of the cell wall in accordance with the pattern of dimethyl esterification (Levesque-Tremblay et al., 2015).
In 5-6 DAP Arabidopsis brd-1 mutant, the fluorescence signal of low-esterified homogalacturonic acids (HGs) in endosperm cell wall are faint and heterogeneous, but the signal of high-esterified HGs significantly increases (Cruz-Valderrama et al., 2018). In rice, there are seven predicted invertase/PME inhibitor genes, which are exclusively expressed during seed development (French et al., 2014). The overexpression of the pectin methylesterase gene PME5 in Arabidopsis reduces the pectin methylesterification of flower primordia leading to the formation of ectopic primordia, whereas the overexpression of pectin methylesterase inhibitor 3 (PMEI3) significantly increases the pectin methylesterification, which inhibits organ formation (Peaucelle et al., 2011). In the Arabidopsis zou-4 mutant, the seeds retain persistent endosperm growth to lead to abnormal increases in seed size, and the embryo sheath that is located near the surface of embryo at the late heart stage cannot be detected with a JIM12 antibody (Fourquin et al., 2016; Moussu et al., 2017).
N-terminal protein α-acetylation (NTA) is a highly prevalent protein modification that affects multiple cellular functions, such as cell multiplication, regulation of stress stimulation, and immune response (Linster et al., 2015; Xu et al., 2015; Giglione and Meinnel, 2021). Plastids are one of the places where co- and post-translational N-terminal acetylation occurs, and approximately 20-30% of all plastid proteins are affected by NTA (Bischof et al., 2011; Bienvenut et al., 2012). The process of N-terminal acetylation is that N-terminal acetyltransferases (NATs) transfer acetyl groups of acetyl-CoA to α amino acids of nascent polypeptides with 25-50 amino acids (Gautschi et al., 2003; Varland et al., 2015). N-terminal acetyltransferase A (NatA), one of the Nats (NatA to NatF), consists of the catalytic subunit NAA10 and the auxiliary subunit NAA15, and is the predominant acetyltransferase complex in yeast and humans (Arnesen et al., 2009). The auxiliary subunit NAA15 anchored to the ribosome participates in acetyltransferase activity, and/or interacts with nascent polypeptides (Ree et al., 2018).
In Arabidopsis, a point mutation of NAA15 leads to immune receptor suppression of NPR1 protein instability and decreases plant immunity (Xu et al., 2015), and T-DNA insertion mutants of NAA10 and NAA15 in Arabidopsis display embryo abortion and abnormal endosperm (Feng et al., 2016; Chen et al., 2018a). In humans, truncating variants of NAA15 lead to varying degrees of growth defects, including intellectual disability and autism spectrum disorder (Cheng et al., 2018). The artificial microRNAs against NAA10 and NAA15 significantly downregulate their transcripts and show drought stress tolerance by activation of the abscisic acid response in Arabidopsis (Linster et al., 2015). However, the transcriptional regulation of NatA during embryo differentiation and endosperm cellularization in Arabidopsis is unknown to date. Here, Arabidopsis seeds from WT and naa15 at three developmental stages were used for transcriptomic analyses and gene expression assays. Meanwhile, the cell wall features were analyzed by detecting the degrees of pectin methyl esterification and cellulose. In our transcriptome profile, a list of genes associated with plant hormones were also identified and analyzed. These results contributed to understanding the transcriptional regulatory information and network relationship during embryo differentiation and endosperm cellularization.
Arabidopsis thaliana ecotype Columbia-0 (Col-0) was used as wild-type plant (WT), and the T-DNA insertion mutant naa15-1/+ (CS836292) was used as described previously (Chen et al., 2018a). All plants were cultivated at 22 ± 2°C with 16-h light/8-h dark in a growth chamber of Wuhan University. The genotype assay of naa15/+ was shown in Supplementary Figure 1 by using PCR amplification, and the gene specific primers were listed in Supplementary Table 1.
Seeds were quickly and carefully dissected from siliques and then cleared in Hoyer’s solution (chloral hydrate: glycerol: water, 8:1:2) 12 hours at 4°C (Chen et al., 2018a). The transparent seeds were placed onto slides and observed under a Leica SP8 microscope (Leica, Germany) equipped with differential interference contrast (DIC) optics. To observe endosperm cellularization, the seeds were fixed in 4% glutaraldehyde in PBS (pH 7.0) under vacuum conditions for 1 h and then transferred into freshly 4% glutaraldehyde in PBS (pH 7.0) and incubated overnight at 4°C. The samples were progressively dehydrated according to a series of alcohol gradients, 15%, 30%, 50%, 70%, 90%, 100%, and then rehydrated using alcohol gradients of 90%, 70%, 50%, 30%, 15%, each gradient for 20 min. The samples were cleared with Hoyer’s solution overnight at 4°C, and observed under a Leica TCS SP8 confocal microscope. The excitation and emission wavelengths were 488 and 505–535 nm, respectively (Li et al., 2017).
For RNA-seq material, WT seeds at 3, 4, and 5 days after pollination (DAP) (3W, 4W, and 5W) and the homozygous white aborted naa15 seeds at 5, 6, and 7 DAP (5M, 6M, and 7M) were separated under dissecting microscope, respectively. Transcriptome sequencing was performed by Majorbio Biotech Co., Ltd. (Shanghai, China). For RNA-sequencing (RNA-seq), the total RNA of samples was extracted using an OminiPlant RNA Kit (CWBIO, Beijing, China) and digested with DNaseI. RNA quality was analyzed by 1.2% agarose gel electrophoresis. The mRNA was purified using poly-T-oligo-attached magnetic beads. The purified mRNA was broken into 300 bp (± 20 bp) paired-end reads. Raw data were obtained and analyzed on an Illumina HiSeq 2500 platform. More than 6.73 Gb of high-quality reads were generated from each cDNA library.
To obtain high-quality clean reads, we removed some reads with over 10% uncertain bases (N) and the low sequencing quality reads of the raw reads were also cleared. Transcriptome assembly was completed using Cufflinks 2.2.1 (http://cole-trapnell-lab.github.io/cufflinks/) and StringTie software (http://ccb.jhu.edu/software/stringtie/). Principal component analysis (PCA) of RNA-seq samples was implemented to detect the expression variances using the plotPCA function in RSEM software (http://deweylab.github.io/RSEM/). Gene functions were annotated using the NR, Swiss-Prot, Pfam, EggNOG, GO, and KEGG databases.
The sequencing reads were aligned to the Arabidopsis TAIR 10.0 reference genome (http://plants.ensembl.org/Arabidopsisthaliana/Info/Index) using HISAT2 (http://ccb.jhu.edu/software/hisat2/index.shtml). The expression values of each transcript were calculated with the Fragments Per Kilobases per Million reads (FPKM) method. Differentially expressed genes (DEGs) were identified by comparing the gene expression levels between different groups using the DESeq R package. A log2 (Fold Change) ≥ 1 or ≤ −1 and P-value < 0.05 were set as the thresholds to discriminate DEGs. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses were performed to investigate the putative functions of DEGs. The top 30 terms of GO enrichment analysis of enriched DEGs were counted with the enriched P-value < 0.05. Venn diagrams were constructed using Venny 2.1 (http://bioinfogp.cnb.csic.es/tools/venny/) to obtain the intersection targets. The hierarchical clustering of heatmap analyses was generated using the online tool (http://www.heatmapper.ca/expression/) (Babicki et al., 2016). Pairwise distances were measured using Pearson correlation, and average linkage was set as the clustering method in heatmap analyses (Karunakaran et al., 2021; Lemke et al., 2021).
Total RNA was extracted using an OminiPlant RNA Kit (CWBIO, Beijing, China), and cDNA was synthesized using an EasyScript® One-Step gDNA Removal and cDNA Synthesis SuperMix Kit (TransGen, Beijing, China). Quantitative real-time PCR (qPCR) was conducted using Transtart Top Green qPCR SuperMix (TransGen Biotech, Beijing, China) and automatically analyzed by the Bio-Rad CFX Manager 3.1 system (Bio-Rad, USA). AtGAPDH was used as an internal reference to normalize the gene expression levels. The specific gene primers were designed by Primer Premier 5.0 and were listed in Supplementary Table 1. For qPCR analysis, the sample was independently repeated in triplicate, and the relative expression values were calculated by the delta-delta Ct (ΔΔCT) method as described previously (Li et al., 2017).
For semithin sections, seeds of WT and the white aborted seeds of naa15/+ were fixed in 4% (v/v) paraformaldehyde buffered with 100 mM phosphate buffered saline (PBS, pH 7.4), placed under vacuum for 30 min, and then fixed overnight at 4°C. After fixation, the samples were washed four times with PBS. The samples were gradually dehydrated in a series of alcohol gradients (Zou et al., 2021). After dehydration, the samples were embedded in LR-White resin and polymerized at 50°C for two days. The samples were cut into sections with 1 μm thickness under an ultramicrotome (EM UC7, Leica) and then placed on slides.
To observe cell wall features of seeds, the samples were incubated in a mixture of calcofluor white (Sigma−Aldrich, St. Louis, USA) and 10% KOH (1:1, v/v) for 5 min at room temperature, and their fluorescent signals were observed under a Leica SP8 microscope. For immunofluorescent analysis of seeds, the samples were washed with distilled water and treated with the blocking solution (5% BSA buffered with PBS, pH 7.4) for 3 h. The primary rat monoclonal antibodies John Innes Monoclonal 5 (JIM5, antibody of low methyl-esterified HGs) and JIM7 (antibody of high methyl-esterified HGs) (diluted 1:10 in blocking solution) were added to the semithin sections at 4°C for 12 h. After incubation, the sections were washed with PBS solution three times to remove the primary rat antibody solution (Ma et al., 2019). The fluorochrome secondary antibody anti-mouse IgG DyLight 488 (Abbkine, Beijing, China) was added, and the sections were covered for 2 h at room temperature. Finally, the samples were observed under a Leica SP8 microscope with the same microscope magnification and fluorescence intensity. Immunofluorescence assays were conducted under the same conditions. The fluorescence signals were quantitated under a Leica LAS AF Lite v2.6.3. The fluorescence intensity of the cell walls was measured at five to ten points in each seed, and the final value was an average represented by a black dot.
All statistical tests and graphic plotting were performed using GraphPad Prism Software version 8.0 (GraphPad, San Diego, USA). Values were expressed as the mean ± standard deviation (SD). The experiments were conducted with three independent biological replicates. Statistical significance was established according to Student’s t-test analysis, and the significance levels were established at *P<0.05, **P<0.01, and ***P < 0.001.
The embryo and endosperm are the most important components of seeds. In our early research, we obtained an Arabidopsis naa15 mutant that possesses the deficient embryo and endosperm (Chen et al., 2018a). To investigate the characteristics of endosperm cellularization and embryo differentiation in Arabidopsis seeds, we observed the development processes of seeds at different stages. In 3W, the embryo reached the globular stage, and the endosperm presented dispersed free nuclei (Figures 1A, D). Subsequently, the embryos developed to the heart and torpedo stages at 4 DAP and 5 DAP, respectively (Figures 1B, C). In 4W, the cell wall of free endosperm nuclei formed in the middle and micropylar end of seeds, indicating that the endosperm was undergoing a process of cellularization (Figure 1E). In 5W, the cell walls of the free endosperm nuclei around the chalazal end were formed, which implied that the endosperm basically completed the process of cellularization (Figure 1F). We observed the embryo and endosperm phenotypic features of the naa15 mutant using spontaneous fluorescence detection and semithin sections. In 5-7M, embryos stagnated at abnormal globular stages (Figures 1G–I), indicating that embryo differentiation was severely disrupted. In 5M, the endosperm was still in the syncytial stage, showing that its cellularization failed to be normally initiated. In 6 and 7 M, the partial free endosperm nuclei formed cell walls, but seeds produced cavities due to abnormal cellularization. The results indicated that the mutation of NAA15 led to incomplete endosperm cellularization (Figures 1H, I). Therefore, we called this abnormal development occurring in the embryo and endosperm of the naa15 seeds as “Globular embryo abnormal development” and “Endosperm delayed development”. Based on the phenotypic characteristics of endosperm and embryo development described above, the 5M had an abnormal globular embryo and syncytial endosperm (Figure 1G), which was similar to the 3W. The 6M formed the partial endosperm cell wall and was similar to 4W (Figure 1H). The 5W completed cellularization, but the naa15 mutant still had a large cavity and incomplete cellularization. Therefore, the naa15 mutant was a well-experimental material for investigating the transcriptional regulation and gene expression in embryo differentiation and endosperm cellularization of Arabidopsis.
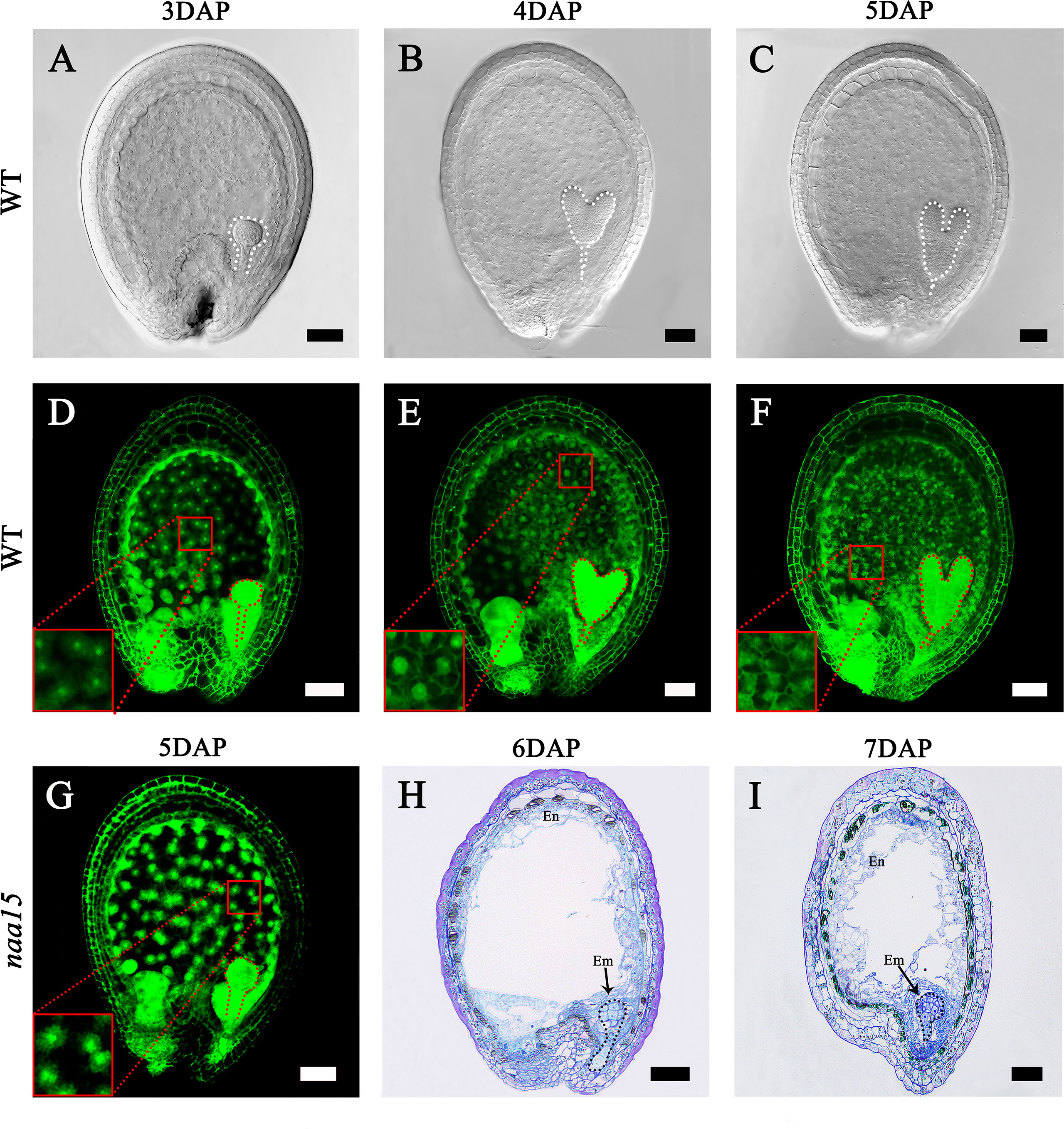
Figure 1 Embryo differentiation and endosperm cellularization in WT and naa15 of Arabidopsis. (A–F) Seeds in WT; (G-I) Seeds in naa15. (A) A seed with a globular embryo at 3 DAP. (B) The seed with a heart embryo at 4 DAP. (C) The seed with a torpedo embryo at 5 DAP. (D) Free nucleus dispersed in syncytium endosperm at 3 DAP. (E) Free endosperm nucleus began to form cell wall at 4 DAP. (F) Endosperm cellularization at 5 DAP basically completed. (G) The seed showed an abnormal globular embryo with free endosperm nucleus without cell walls at 5 DAP. (H, I) Semi-thin sections showed abnormal globular embryo and partially endosperm cells at 6 DAP and 7 DAP, respectively. Embryos were marked with discontinuous lines of different colors. The small red boxes were magnified and shown on the lower left of the corresponding images. Em, embryo; En, endosperm. Scale bars = 50 μm.
The seeds of 5 DAP WT turned green from translucent, while the naa15 seeds at 5DAP were still translucent and white in naa15/+ siliques (Supplementary Figure 1A). To investigate the transcriptional levels during endosperm cellularization and embryo differentiation, we collected normal seeds of 3-5 DAP WT (3W, 4W, and 5W, respectively) and white aborted homozygous seeds of 5-7 DAP naa15 (5M, 6M, and 7M, respectively) as the tested materials and conducted RNA sequencing. The results showed that a total of 137.14 Gb of clean reads was obtained, and the mapping rate to the Arabidopsis genome was over 97%. The statistics of the clean reads in each sample were listed in Supplementary Table 2. The FPKM values of genes were shown in Supplementary Table 3 and the gene expression levels in each library were calculated and represented as the log10 (FPKM) method (Supplementary Figure 2A). The principal components of the PCA score plot (PC1 and PC2) were calculated to be 43.76% and 16.22%, respectively (Supplementary Figures 2B, C). The six sample groups were clearly separated, and three biological replicates of each group were clustered together (Supplementary Figure 2C).
We performed comparisons between 4W vs 3W (heart stage vs globular stage) and 5W vs 4W (torpedo stage vs heart stage) to uncover the changes in the transcriptome profiles during the early stages of embryo and endosperm development. A total of 2040 DEGs (1271 up- and 769 down-regulated) in 4W vs 3W and 3630 DEGs (1922 up- and 1708 down-regulated) in 5W vs 4W groups were obtained, respectively (Figures 2B, C). Venn diagrams showed that 1102 DEGs were shared between the 4W vs 3W and the 5W vs 4W groups (Figure 2A), and these DEGs were distributed in four clusters using the K-means approach, indicating that they had similar expression patterns in 3-5 DAP WT (Figures 2D-G and Supplementary Tables 4-7). Many transcription factors, such as growth-regulating proteins, MYBs, and bZIPs were in cluster I (560 DEGs), showing that their expression levels were distinctly upregulated with seed development (Figure 2D and Supplementary Table 8). In contrast, the transcript levels of auxin response factors (ARF12, ARF13, ARF14, ARF15, ARF20, ARF21, and ARF22) and MADS-box transcription factors (AGL36, AGL48, AGL58, AGL62, AGL90, and AGL 96) were significantly downregulated from 3W to 5W, and were all clustered in cluster II (423 DEGs) (Figure 2E and Supplementary Table 9). The numbers of DEGs in clusters III and IV were 83 and 36, respectively. The genes involved in heat shock proteins and chaperones were enriched in cluster III, while many pectin acetylesterase, methyltransferase, and protein kinase enzymes were concentrated in cluster IV (Supplementary Tables 6, 7). Therefore, we proposed that the differential transcriptional regulation of genes may contribute to early seed development.
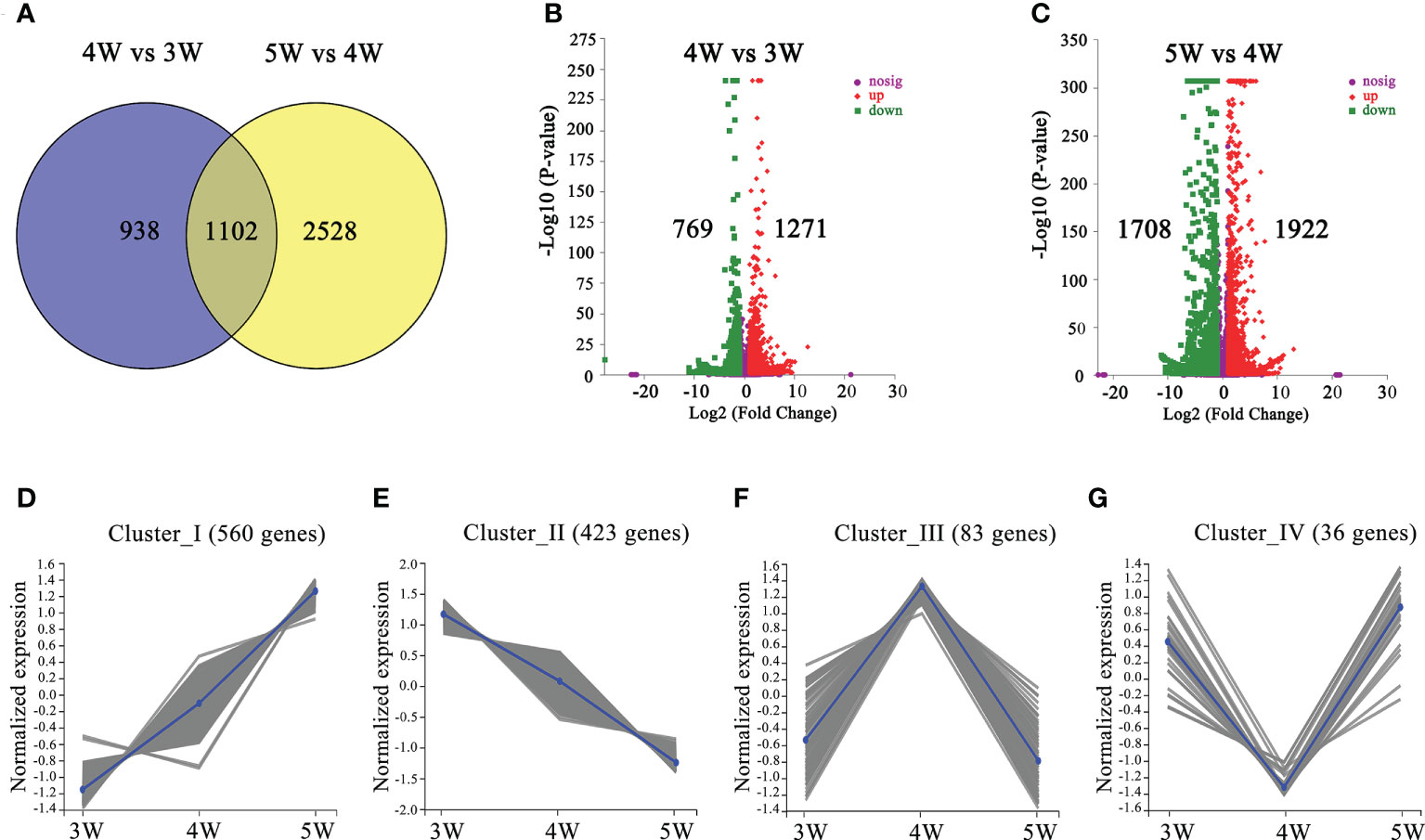
Figure 2 The number and functional annotation of differentially expressed genes (DEGs) during embryo differentiation and endosperm cellularization in WT of Arabidopsis. (A) The number of DEGs from venn diagram analysis in 4W vs 3W and 5W vs 4W groups, respectively. (B, C) Volcano maps of DEGs in 4W vs 3W and 5W vs 4W groups, respectively. Log2 (Fold Change)≥ 1 or ≤ −1, and P-value < 0.05 were used as a standard for screening DEGs. The “-Log10 (P-value)” indicated the statistical significance level, and the larger value showed that the divergence of gene expression was more significant. Significantly up- or down-regulated genes were represented by using red dots or green dots, respectively. (D–G) Clustered gene expression profiles of 1102 DEGs from 4W vs 3W and 5W vs 4W using the statistical method of K-means.
To reveal the regulatory network of seed morphogenesis, DEGs were classified and annotated using GO and KEGG databases. The up-regulated DEGs of KEGG enrichment contained indole alkaloid biosynthesis, plant hormone signal transduction and phenylpropanoid biosynthesis in the 4W vs 3W and 5W vs 4W groups (Figures 3A, B and Supplementary Table 10). A lot of terms related-cell division, the RNA metabolism, the transcription regulator activity, cell cycle terms, and cytoskeleton-related terms, were significantly enriched in up-regulated DEGs of 4W vs 3W (Supplementary Figure 3A), indicating that 3 to 4 DAP seeds might undergo vigorous cell divisions. The photosynthesis, photosynthesis - antenna proteins, starch and sucrose metabolism, and oxidative phosphorylation pathways were significantly enriched in 5W vs 4W (Figure 3B). Within the biological process category of GO enrichment analyses, many up-regulated DEGs were also significantly enriched in photosynthesis terms (GO:0019684, GO:0009768, and GO:0015979) (Supplementary Figure 3B), indicating that they might participate in plastid transformation to the chloroplast process during early seed development and involve in the embryonic transfer from the heterotrophic stage to the autotrophic stage.
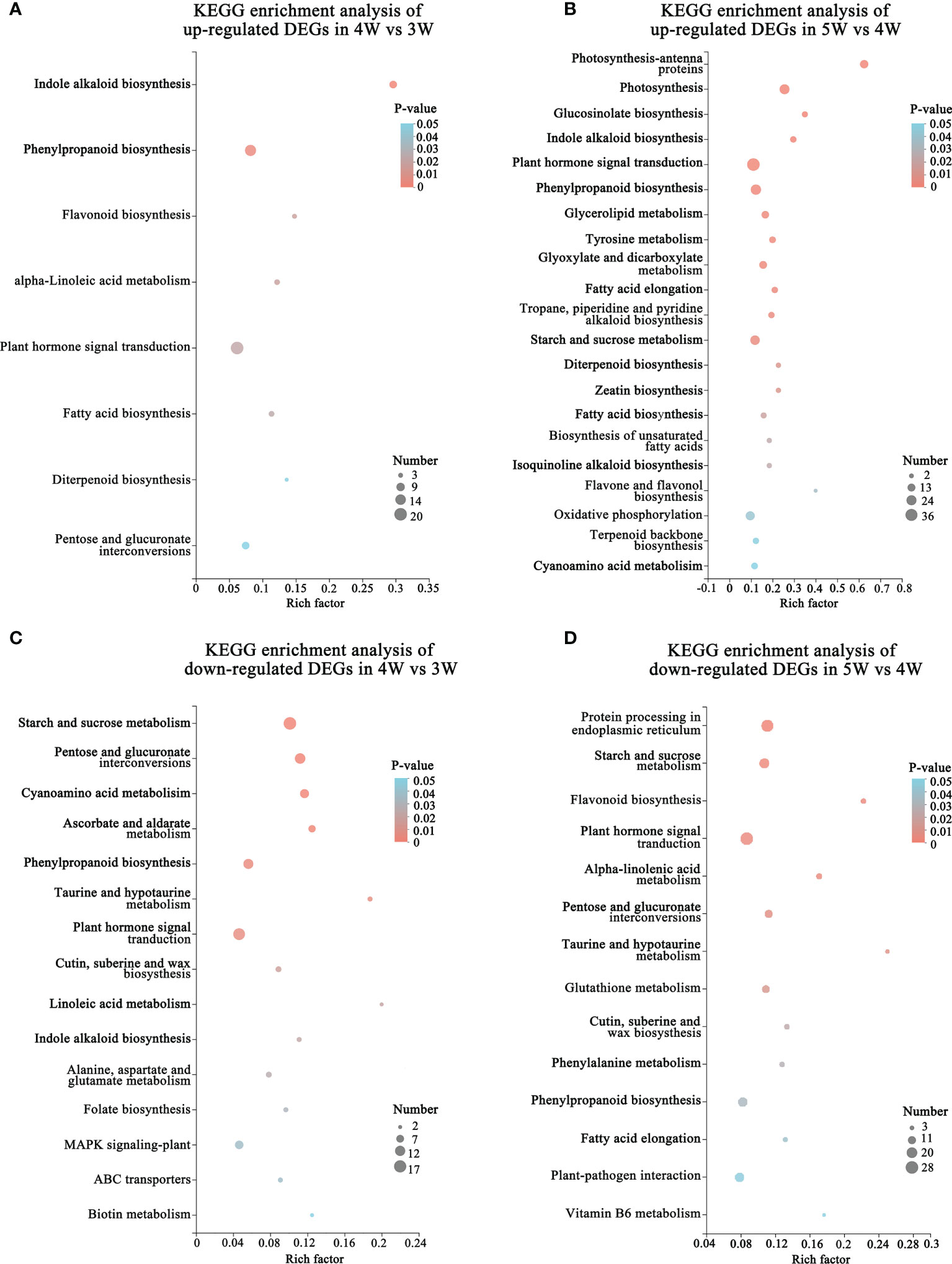
Figure 3 Functional annotation and classification of DEGs during embryo differentiation and endosperm cellularization in WT of Arabidopsis. (A, B) KEGG enrichment analysis of up-regulated DEGs in 4W vs 3W and 5W vs 4W groups, respectively. The “Rich factor” represented the enrichment degree of DEGs in each pathway. The number of the enriched DEGs was represented by the size of each circle. The larger the number, the bigger the circle. (C, D) Diagrams of KEGG enrichment analysis of down-regulated DEGs in 4W vs 3W and 5W vs 4W, respectively.
In addition, the terms related to cell wall (cell wall organization or biogenesis, plant-type cell wall organization or biogenesis, and cell wall) were also significantly enriched in cellular component using GO enrichment analysis of 4W vs 3W and 5W vs 4W groups (Supplementary Figures 3B,C), which mainly included the genes related to cell wall modification (pectinesterase and pectin methylesterase inhibitor), cellulose synthase, and expansins (Supplementary Table 11). The hormone signaling pathways were significantly enriched in up- and down-regulated DEG of GO enriched terms and KEGG enriched pathways during early seed development at 3-5 DAP (Figures 3A–D). These DEGs mainly participated in the auxin, cytokinin, and abscisic acid pathways, indicating that there was wide crosstalk among hormones. The mRNA levels of auxin transport genes (PIN6 and LAX2), auxin response factors (ARF4 and ARF11), and small auxin upregulated RNA 78 (SAUR78) were markedly upregulated from 3 to 5 DAP. Particularly, cytokinin synthesis genes LONELY GUY 4 and 5 (LOG4 and LOG5) were mainly upregulated in 4W vs 3W (Supplementary Table 11), speculating that they might be involved in the formation of embryo organ primordia and cellularization of free endosperm nuclei.
We performed the transcriptome comparative analysis in both WT and naa15, and identified a total of 9633 DEGs in 5M vs 3W, 6M vs 4W and 7M vs 5W. Among these DEGs, the 2195 DEGs were shared in three comparison groups (Figure 4A). The number of DEGs was 5988 DEGs in 5M vs 3W (containing 2743 up- and 3245 down-regulated), 5595 DEGs in 6M vs 4W (containing 2530 up- and 3065 down-regulated), and 5598 DEGs in 7M vs 5W (containing 3063 up- and 2535 down-regulated), respectively (Figures 4B–D). KEGG enriched analysis showed that the up-regulated DEGs of WT and naa15 were significantly enriched in phenylpropanoid biosynthesis, protein processing in endoplasmic reticulum, starch and sucrose metabolism, and amino sugar and nucleotide sugar metabolism (Figures 4E–G and Supplementary Tables 12A–C). The GO enrichment analysis displayed that the up-regulated DEGs of WT and naa15 were evidently aggregated in the terms of cell cycle process, cell division, cytoskeleton part, and related-microtubule terms (including microtubule binding, microtubule-based process, and microtubule-based movement) (Supplementary Figure 6).
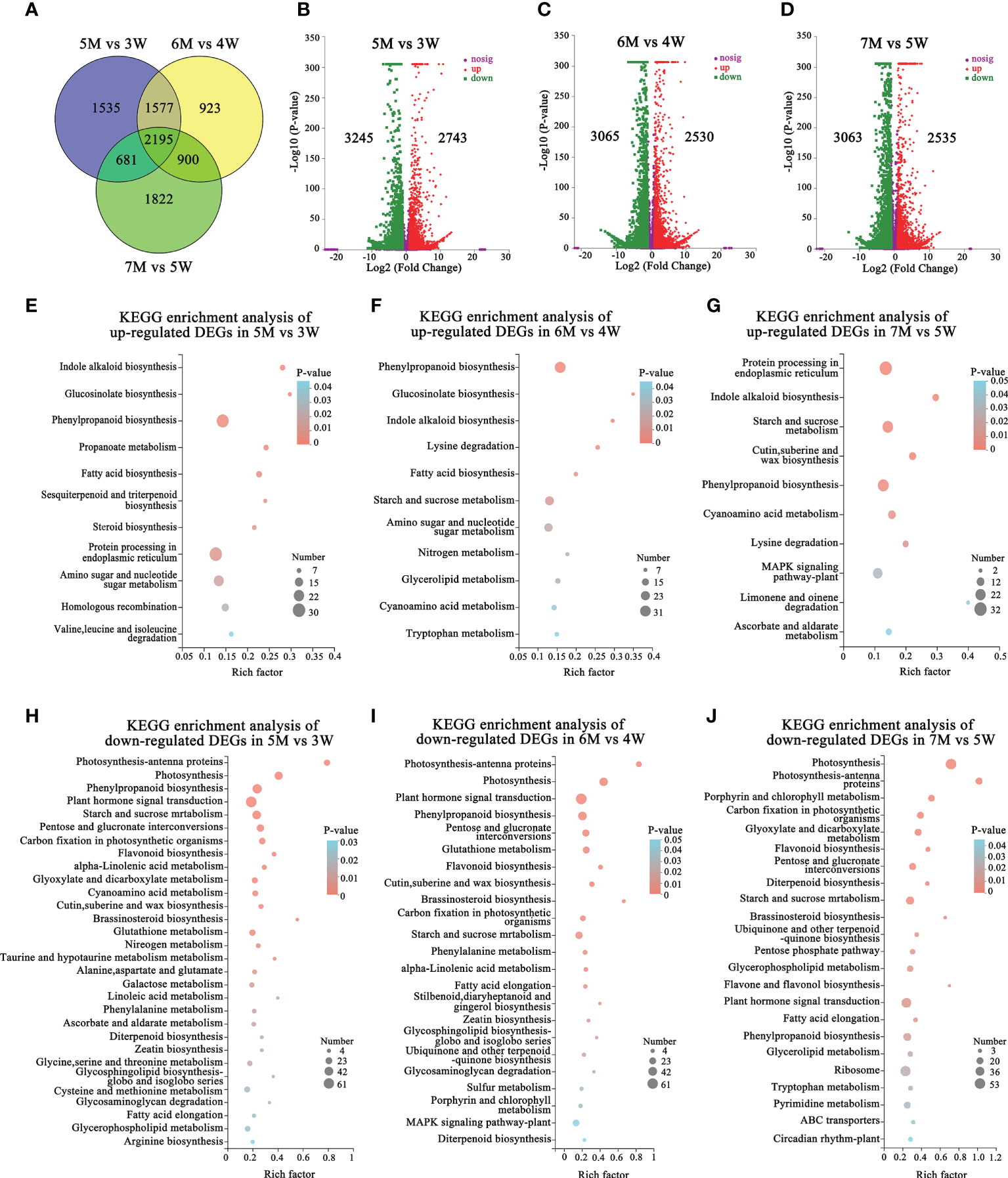
Figure 4 The number and KEGG enrichment pathways of DEGs between Arabidopsis WT and naa15 in 5M vs 3W, 6M vs 4W, and 7M vs 5W, respectively. (A) Venn diagram of DEGs in three comparison groups between WT and naa15. (B–D) The volcano maps of DEGs in three comparison groups between WT and naa15. (E–G) The KEGG enrichment pathways of up-regulated DEGs in 5M vs 3W, 6M vs 4W, and 7M vs 5W, respectively. (H–J) The KEGG enrichment pathways of down-regulated DEGs in 5M vs 3W, 6M vs 4W, and 7M vs 5W, respectively.
The down-regulated DEGs of KEGG enrichment analyses in WT and naa15 were mainly enriched in plant hormone signal transduction photosynthesis, phenylpropanoid biosynthesis, and starch and sucrose metabolism pathways (Figures 4H–J and Supplementary Tables 12D–F). Totally, we identified 137 DEGs involved in auxin, cytokinin, and ethylene signaling pathways in three comparison groups between WT and naa15 (Supplementary Table 13). As shown in Figures 5A–C, the genes annotated to auxin-responsive proteins (IAA), PIN, and auxin response factors (ARF) were identified, and most of them were downregulated in naa15. qPCR analyses showed that the expression of auxin transporter PIN1 was significantly increased in 3W-5W. In contrast, the PIN1 transcript level was markedly decreased in naa15 (Figure 5J). The mRNA levels of ARF5 showed a downregulated pattern in WT and were decreased in 5-7M (Figures 5C, K).
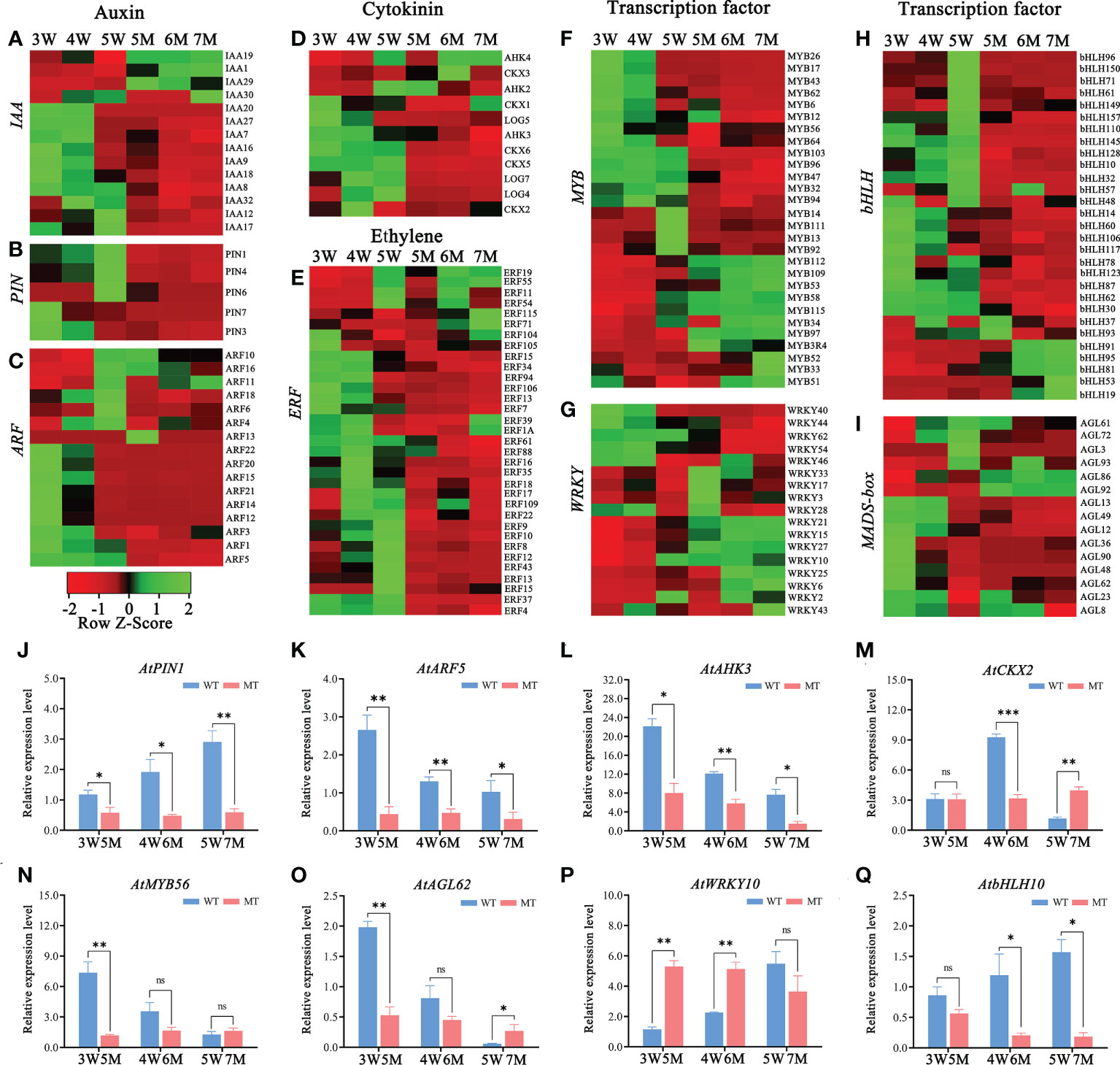
Figure 5 Heatmaps and qPCR analyses of DEGs related to hormones and transcription factors in WT and naa15 seeds of Arabidopsis. (A–D) The relative expression patterns of DEGs involved in auxin and cytokinin signal pathways. (E–I) The relative expression patterns of DEGs involved in transcription factors including ERF, MYB, WRKY, bHLH, and MADS-box. FPKM values were standardized with Z-Score for the heatmap analysis. Red represented down-regulation of gene expression, and green represented up-regulation of gene expression. (J–Q) The qPCR analyses of DEGs related to hormones and transcription factors. Asterisks denoted significant differences basing on Student’s t-test (*P<0.05, **P<0.01, and ***P < 0.001).
In addition, the heatmap showed that the genes related to the cytokinin pathway, cytokinin receptors (ARABIDOPSIS HISTIDINE KINASE, AHK3), cytokinin dehydrogenases (CYTOKININ OXIDASE/DEHYDROGENASE, CKX1, CKX2, CKX5, and CKX6), and cytokinin-activating enzymes (LONELY GUY, LOG4, LOG5, and LOG7) were also downregulated in naa15 (Figure 5D). The cytokinin receptor AHK3 in 3-5 W was gradually downregulated, while its transcript was distinctly decreased in 5-7 M (Figure 5L). CKX2, encoding a limiting enzyme in cytokinin metabolism, is activated by WRKY10 to promote endosperm growth (Li et al., 2013). In our results, the expression of CKX2 was upregulated at 3W and 4W, but down-regulated at 5W; however, it remained at low levels in 5-7M (Figure 5M). These results implied that the gene expression of NAA15 was involved in regulating the hormone signaling pathways during embryo and endosperm development.
Transcription factors (TFs), including MYB, bHLH, WRKY, ERF, and GATA, were identified in GO and KEGG enriched analyses in comparison between WT and naa15. In 5M vs 3W group, there were 89 transcription factors (TFs) (53 down- and 36 up-regulated DEGs), and the most downregulated TFs was the bHLH family (18 DEGs), followed by ERF family (16 DEGs). In 7M vs 5W group, the downregulated TFs were mainly in the MYB, ERF, and bHLH families (Supplementary Table 14). The heatmaps showed that ERF (23 DEGs), MYB (16 DEGs) and bHLH (35 DEGs) were significantly down-regulated in 5-7M (Figures 5E, F, H). The gene expressions of WRKY (17 DEGs) and AGL (15 DEGs) were also significantly changed in 5-7M (Figures 5G, I). qPCR analyses showed that the expressions of AtMYB56 and AtAGL62 gradually decreased in 3-5W, and their expressions distinctly deceased in 5M compared to WT (Figures 5N, O). AtbHLH10 and AtWRKY10 were markedly increased in 3-5W (Figures 5P, Q). Nevertheless, AtWRKY10 in 5 and 6M had higher expression than in 3W and 4W (Figure 5P). Hence, we hypothesized that NAA15 impacted embryo differentiation and endosperm cellularization by directly or indirectly mediating the transcription levels of these genes.
Sucrose controls a wide range of developmental and metabolic processes including cell division, cotyledon development, far-red light signaling, and tuber development (Yoon et al., 2021). In our research, the starch and sucrose metabolism pathway was enriched in the KEGG enriched analysis of 3W-5W (Figures 3B–D). And the expressions of two sucrose synthetases (suc1 and suc2) and eight sucrose transporter genes (sweet3, sweet4, sweet5, sweet7, sweet9, sus1, sus6, and sus4) were significantly decreased in naa15 (Supplementary Figure 4A). Sucrose stimulates the expression of transcription factors such as WRKY and MYB, which function upstream of sucrose-responsive genes. However, the molecular mechanism of sucrose as a signal regulating embryo and endosperm development remains unknown. Our transcriptome data provided some related to sucrose genes that might play vital functions during early seed development.
The photosynthesis pathway was also significantly enriched in the KEGG and GO enriched analyses up-regulated DEGs in 5W vs 4W group (Figure 3B and Supplementary Figure 3B). The GO enrichment analysis of down-regulated DEGs in naa15 was mainly enriched in photosystem and the relevant chloroplast genes (Supplementary Figure 5). The gene expressions from six photosystem I-antenna (LHCI) genes and 12 chlorophyll a/b binding protein complex Ⅱ were decreased in naa15 (Supplementary Figure 4B). Interestingly, Lhca5, Lhcb1.1, PORB, and PORC have been considered potential substrates of Arabidopsis N-terminal acetyltransferase A (NatA), which consists of NAA10 and NAA15 (Linster et al., 2015). Hence, we speculated that plant hormones, photosynthesis, and carbohydrate pathways were critical during early seed development. However, the molecular regulatory function of NAA15 in these pathways was unknown and requires further investigation.
In addition, the abnormal globular embryo in naa15 implied that embryo differentiation was disrupted. Thus, we inspected the expression levels of the genes reported to be associated with embryo differentiation in 3 DAP WT and 5-7 DAP naa15. WOX family had an essential role in body axis of embryo. Among these DEGs from our transcriptome profiles, WOX2, WOX5, and WOX8 in naa15 were significantly downregulated (Supplementary Tables 2, 4), and the qPCR results also showed that the expressions of AtWOX2 and AtWOX5 in 5-7M were significantly decreased compared with that in 3W (Figures 6A, B), implying that the downregulated expression of WOX genes in naa15 might lead to the formation of abnormal globular embryos. CUP-SHAPED COTYLEDON (CUC) and HD-ZIP TFs are involved in bilateral symmetry and SAM establishment. The qPCR results showed that AtCUC2 and ATHB8 were significantly decreased in naa15 (Figures 6C, D). DORNRÖSCHEN (DRN) is an AP2/ERF-type TF that interacts with HD-ZIP III TFs. The expression of AtDRN also decreased significantly in naa15 (Figure 6E). PLETHORA1 (PLT1) and SHORT-ROOT (SHR) are required for hypophysis divided asymmetrically and quiescent center (QC) formation in late globular embryos, respectively (Aida et al., 2004).
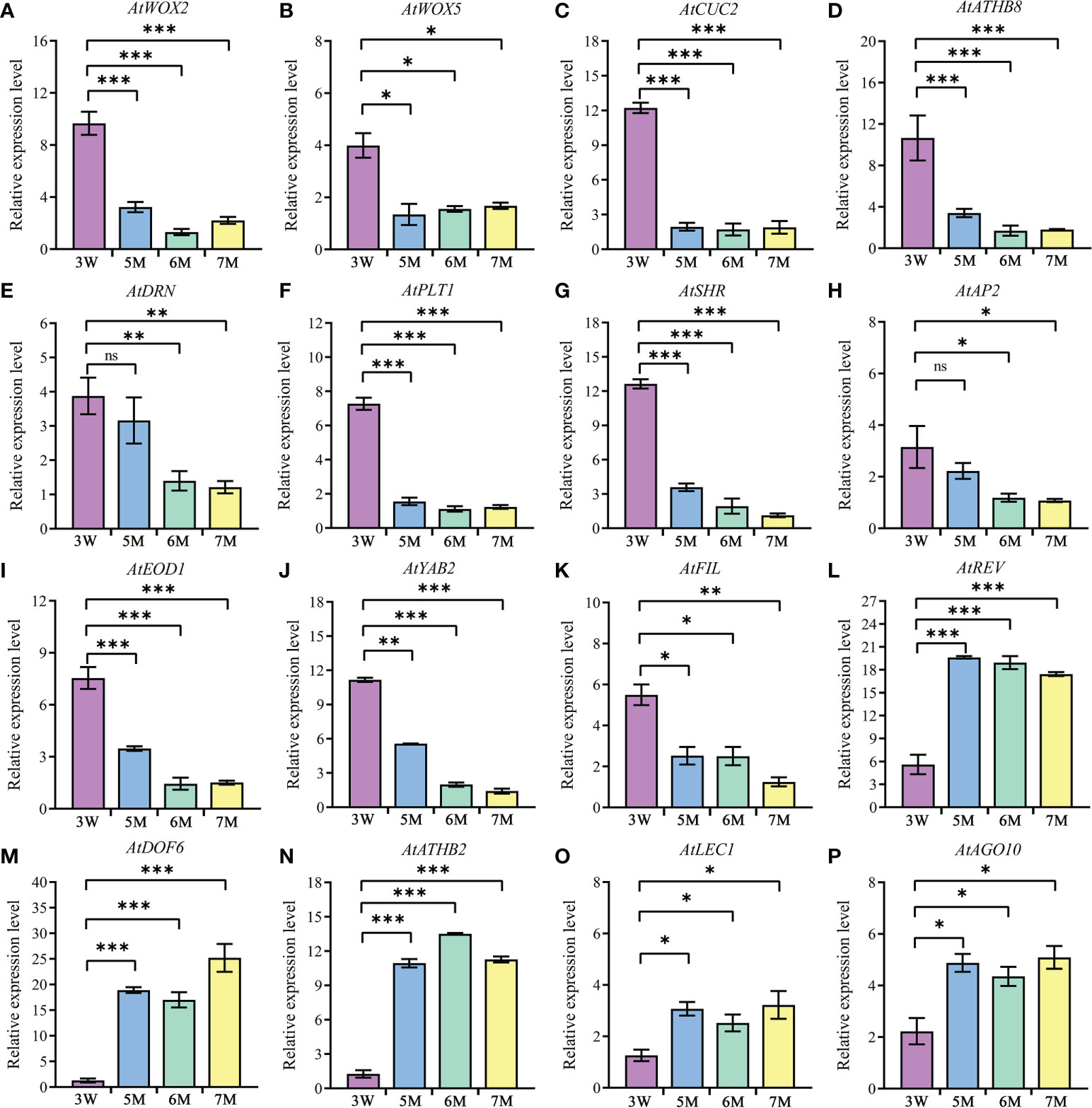
Figure 6 The expression of genes related to embryo differentiation in WT and naa15. (A–P) The qPCR analyses of DEGs in 3W, 5M, 6M, and 7M. 3W, 3 DAP WT; 5M, 5 DAP naa15; 6M, 6 DAP naa15; 7M, 7 DAP naa15. The "ns" represented no significant differences. Asterisks denoted significant differences basing on Student’s t-test *P < 0.05, **P < 0.01, and ***P < 0.001.
In the results, PLT1 and SHR showed high expressions at 3 DAP in WT, but down-regulated in 5-7M (Figures 6F, G), suggesting that it may be one cause of the abnormal globular embryo formation of naa15. Besides, the expressions of APETALA2 (AP2) and EOD1, which influence the sizes of seeds, also significantly decreased in naa15 (Figures 6H, I). YABBY2 (YAB2), FILAMENTOUS FLOWER (FIL), and REVOLUTA (REV) played essential roles in the establishment of abaxial patterning. In our study, the expressions of YAB2 and FIL were significantly downregulated, while the REV was evidently upregulated (Figures 6J-L). In addition, DOF6, ATHB2, LEC1, and AGO10 were observably up-regulated in naa15 (Figures 6M-P), suggesting that the abnormal expression may affect the embryo differentiation of naa15. Therefore, we postulated that the gene expression changes related to embryo differentiation may be one of the causes of abnormal embryonic differentiation of naa15, and the detailed relationship between these DEGs and NAA15 needs to be further investigated.
According to GO and KEGG enrichment analyses from two comparison groups in 3-5W, many DEGs were enriched in cell wall-related pathways (e.g., cell wall organization or biogenesis and plant-typed cell wall) (Figure 3 and Supplementary Figure 3). However, these DEGs were abnormal expressions in naa15. Heatmap analysis showed that 13 genes encoding expansin (EXPA), which functions as the modified proteins of cell wall, were downregulated in naa15 (Figure 7A). The mRNA level of AtEXPA15 was increased in 3W-5W, whereas it was significantly down-regulated in 5M-7M (Figure 7G). qPCR analyses showed that AtEXPB4 was gradually downregulated in 3W-5W, and was also significantly downregulated in 5M and 6M compared to 3W and 4W (Figure 7H), hinting that the decrease in AtEXPA15 and AtEXPB6 mRNA levels in naa15 might be one reason for abnormal endosperm cellularization. In addition, we also found that cell wall modification genes, including 15 xyloglucan endotransglycosylase/hydrolase (XTH) and 27 pectin esterase/pectin methylesterase (PME) genes, were downregulated in naa15 (Figures 7B, C). qPCR analyses demonstrated that the expression of PME12 and PME19 in 5M were downregulated with 7.9-fold and 9.0-fold compared to 3W, respectively (Figures 7K, L). The expressions of XTH6 and XTH14 in 5M and 6M were downregulated compared to 3W and 4W (Figures 7I, J). The genes encoding proteins of cell wall, such as arabinogalactan proteins (AGPs, AGP23/7/6/11/12/40/41/26/22/17/9) and fasciclin-like arabinogalactan proteins (FLAs, FLA10/4/8/12/13/2/1), were also downregulated in naa15 (Figures 7D, E). qPCR analyses confirmed that AGP9, AGP17, and FLA10 were significantly downregulated in 5M and 6M compared to 3W and 4W (Figures 7M–O), which was consistent with the RNA sequencing data (Figures 7D, E). However, the expressions of several cell wall related-genes CESA5, CESA8, CSLD5, CSLB3, CSLD6, and FLA10 were up-regulated in naa15 (Figures 7F, P–R), which might play important role in abnormal cell walls of the endosperm.
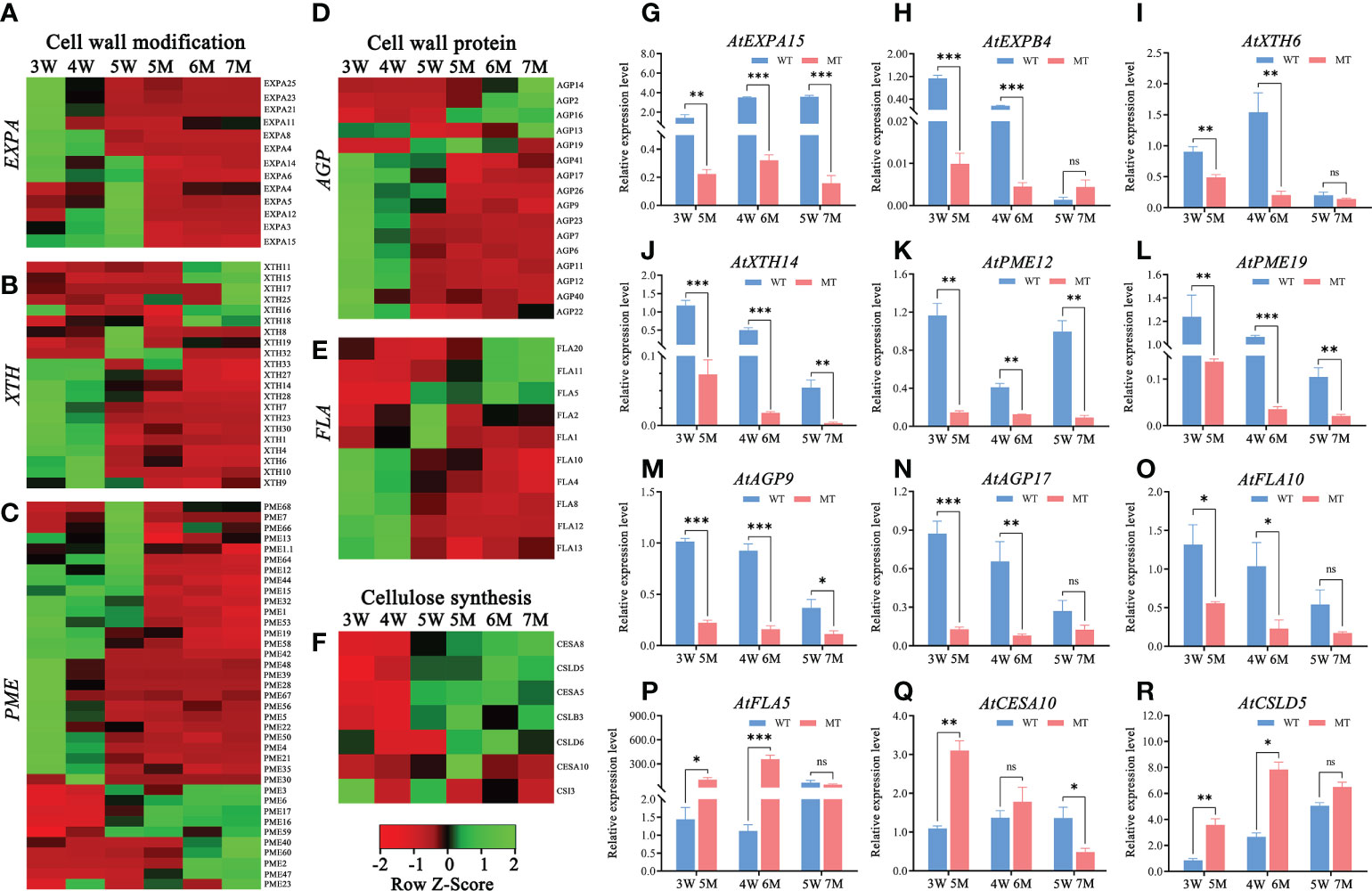
Figure 7 Heatmaps and qPCR analyses of DEGs related to cell wall formation between WT and naa15 in Arabidopsis seeds at different development stages. (A–C) Heatmaps of the cell wall-modification proteins genes including the types of EXP, XTH, and PME/PMEI. (D, E) Heatmaps of the cell wall-protein genes including the types of AGP and FLA. (F) The heatmap of genes related to cellulose synthesis. (G–R) The qPCR analyses of DEGs related to formation of cell wall. Asterisks denoted significant differences basing on Student's t-test *P < 0.05, **P < 0.01, and ***P < 0.001. The "ns" represented no significant difference. AGP, arabinogalactan proteins; EXP, expansin; FLA, fasciclin-like arabinogalactan protein; PME/PMEI, pectinesterase/pectin methylesterase inhibitor; XTH, xyloglucan- endotransglycosylases/hydrolases.
Many DEGs from our transcriptome analysis were involved in cell wall formation and modification; therefore, we analyzed the features of cell wall in the embryo and endosperm of WT and naa15. Histological staining and immunofluorescence assays were performed in the seeds of 4W, 5W, 6M, and 7M using calcofluor white (CW) staining and pectin methyl-esterification monoclonal antibodies of (JIM5 and JIM7). The CW fluorescence gradually increased in cell walls of 4W and 5W embryos, while the signals in 7M embryonic cell walls significantly decreased compared to 5W (Figures 8A–E). The cell wall of naa15 endosperm at 6M and 7M exhibited decreased fluorescence intensity compared to 5W (Figures 8A–D, F), inferring that the decreased cellulose in naa15 might affect the formation of endosperm cell walls.
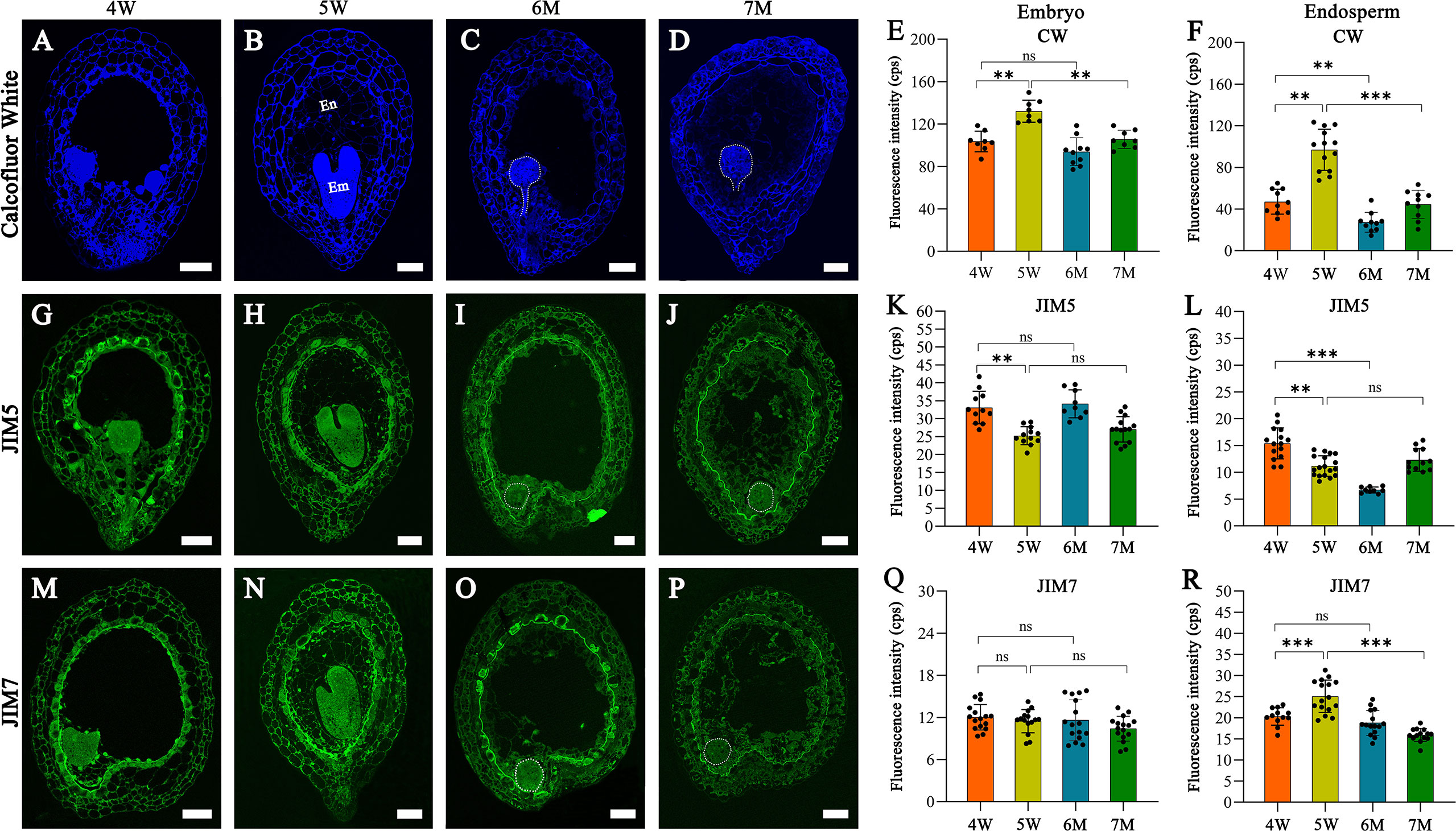
Figure 8 The images of histochemistry and immunofluorescence localization in WT and naa15 seeds at different development stages. (A–D) Semi-thin sections of seeds were stained with calcofluor white in WT and naa15. (E, F) Fluorescence intensity with calcofluor white staining in embryo and endosperm of WT and naa15. (G–J) Semithin sections of seeds were immunolabeled using JIM5 antibody in WT and naa15. (K, L) Fluorescence intensity with JIM5 immunolabelling in embryo and endosperm of WT and naa15. (M–P) Semi-thin sections of seeds were immunolabeled using JIM7 antibody in WT and naa15. (Q, R) Fluorescence intensity with JIM7 immunolabelling of WT and naa15 in embryo and endosperm, respectively. The p-value was calculated by using the Student t-test. Asterisks denoted significant differences, **P < 0.01 and ***P < 0.001. The "ns" represented no significant difference. Each black dot represented a seed. Em, embryo; En, endosperm; cps: counts per second. Scale bars = 50 µm.
Using the JIM5 antibody to detect the low methyl-esterified pectin, the fluorescence signals of embryo cells decreased gradually from 4W to 5W and from 6M to 7M (Figures 8G–K). In cell walls of endosperm at 4W and 5W, JIM5 signals decreased following endosperm cellularization, which might contribute to the rapid development of subsequent embryos (Figures 8G, H, L). JIM5 fluorescence in the cell walls of naa15 endosperm at 6M significantly decreased compared to 4W and 5W (Figures 8G–J, L), while the signal in 7M was similar to 5W. The results indicated the delayed and impaired endosperm growth and development in naa15
Using the JIM7 antibody to detect high methyl-esterified pectin, the results showed that there was no apparent change in the fluorescence signals in 6M and 7M compared to embryos at 4W and 5W (Figures 8M–Q). The fluorescence signals in the cell walls of endosperm at 4W and 5W were obviously increased, while the signal in 6M and 7M evidently decreased (Figures 8M–P, R), indicating that cell wall formation in embryo and endosperm of naa15 was disrupted, resulting in the aberrant embryo differentiation and incomplete endosperm cellularization.
It is well known that seed formation depends on well-coordinated development between endosperm and embryo. The endosperm is considered as a nutrition source to support embryo development, and its cellularization process undergoes nuclear proliferation, formation of endosperm cell wall, and cell differentiation, all of which are essential for embryo growth (Jo et al., 2019; Song et al., 2021). Nevertheless, extensive efforts are required to understand the process of embryo differentiation and endosperm cellularization, including the fundamental regulatory mechanisms of the initial endosperm cellularization process, and the relationship between embryo and endosperm. In this research, we employed RNA-sequencing analyses and observed cell histological features to identify candidate genes and expression pathways involved in embryo differentiation and endosperm cellularization, which provide new insights into seed morphogenesis.
It has been reported that the appearance of endosperm cell wall in the micropyle zone of seeds indicates that endosperm development transforms from the syncytial endosperm to cellularization stages (Otegui and Staehelin, 2000). Cell walls in plants contain various polysaccharides (cellulose, hemicellulose, and pectins), glycoproteins, and aromatic or aliphatic compounds, which form the mechanically strong and extensile network (Caffall and Mohnen, 2009). Cell walls provide mechanical strength for cells and fix the cell shape, which requires the modification and remodeling of the cell wall. The modification gene XTH31, coding a xyloglucan endotransglycosylase/hydrolase, is expressed in endosperm of Arabidopsis and can reinforce their cell walls during seed germination (Endo et al., 2012). ZHOUPI is an endosperm-specific gene in Arabidopsis and affects embryo and endosperm development by regulating the expression of pectin-modifying enzymes. In endosperm cell wall of zou mutant, (1-5)-α-L-arabinan epitopes immunized with the LM6 antibody were abundant, its endosperm cells are more robust than WT, and the cell wall fail to breakdown, resulting in the abnormal size of embryo (Fourquin et al., 2016). The focus of investigation of seed cell walls is mainly the roles of cell wall during seed germination; however, up to date, the transcriptional regulation of cell wall formation during endosperm cellularization is still unclear.
AGPs and FLAs are important proteoglycans that form covalent bonds with both hemicellulose and pectin in cell walls. In rice, AGP protein is widely distribution in tapetum cells, pollen mother cells, and mature pollens (Ma et al., 2019). In FLA3 RNAi transgenic plants of Arabidopsis, pollen grains are shrunken and wrinkled, and cellulose abnormal distribution in pollen walls to lead to reduced male fertility (Li et al., 2010). APAP1 (ARABINOXYLAN PECTIN ARABINOGALACTAN PROTEIN1) encodes an AGP protein, and contributes to covalent linkages between cell wall proteins, pectins, and xylans. In apap1, the covalent linkages of pectin and xylan in cell walls of stem are less tightly (Tan et al., 2013). In our study, cell wall organization/biogenesis and cell wall terms were significantly enriched in embryo differentiation and endosperm cellularization of 3-5 DAP WT (Supplementary Figure 2 and Supplementary Table 10). These DEGs were mainly annotated to genes related to proteins of cell walls (AGP and FLA) and its modification (Supplementary Table 11). Among them, 27 DEGs annotated to pectin esterase/pectin methylesterases (PME) showed lower expression levels in naa15 (Figure 7C). This indicated that these DEGs might be involved in the formation of the cell wall during early seed development.
Meanwhile, through analyzing the distribution of esterified pectin during embryo and endosperm development, the immunolabeled results showed that low-esterification pectin of endosperm cell wall gradually decreased by using JIM5 antibody; in contrast, by using JIM7 antibody, the high-esterification pectin was increased in 4 and 5W (Figures 8K, L, R). Thus, we proposed that decreased low-esterification pectin and increased high-esterification pectin in cell walls might be largely related to endosperm cellularization, which increased the rigidity and stiffness of cell wall. In the endosperm cell walls of naa15, JIM5 and JIM7 signals showed lower fluorescence intensities than in WT (Figures 8L, R), but the JIM7 signals of embryos in WT and naa15 had no significant differences (Figure 8Q). Based on the results, we speculated that the esterification feature change of the endosperm cell wall in naa15 might be one of the causes of the abnormal endosperm. We hypothesized that the defective embryo and endosperm of the naa15 mutant might be due to impairing the cell characteristics, which in turn affected embryo differentiation and endosperm cellularization. It has been reported that the embryo morphology and mucilage extrusion of seeds in Arabidopsis hms-1 mutant were altered owing to methyl esterification degree of cell walls (Levesque-Tremblay et al., 2015). Hence, cell wall formation and modification were vital for embryo and endosperm during early seed development.
In GO and KEGG enriched analysis of these DEGs, starch and sucrose metabolism pathways were enriched in 3-5W (Figures 3B-D). Sucrose is imported into developing embryo by sucrose transporters (suc/sut) and sucrose exporter proteins (SWEET). The sweet11;12;15 triple mutant in Arabidopsis disrupts sugar transport, resulting in delayed embryo development (Chen et al., 2015). In our research, the transcriptome levels of SWEET5, SWEET11, and SWEET15 in 3-5W were upregulated, and sucrose metabolism genes such as AT1G19450, SFP1, and SPS3 were also upregulated. However, their roles in seed development remain unclear. The starch and sucrose pathway was also significantly enriched using KEGG analysis in naa15 compared with WT (Figures 4E–I). The heatmap of sucrose synthesis and sucrose transporter genes showed that SUS1, SUS4, SUC1, SWT3, SWT4, SWT7, and SWT9 were significantly down-regulated in naa15 compared with WT. Taken together, these results implied that sucrose metabolism may provide nutrients and energy for promoting normal cell growth of early seed development. However, the role of NAA15 in sucrose metabolism remained unclear and needed to be further investigated.
From globular-embryo stage, the organ differentiation of embryo begins to occur, and the seed is transformed from the heterotrophic stage to the autotrophic stage. In embryonic cells, proplastids are converted into chloroplasts, accompanied by the biosynthesis of chlorophyll (Tejos et al., 2010). Therefore, lots of gene expressions related to photosynthesis are regulated during embryo differentiation and endosperm cellularization. Transcriptome GO analysis shows that photosynthesis term is significantly enriched in soybean seeds at globular, heart, and cotyledon stages, implying that photosynthesis might play important role in the embryo development (Sun et al., 2020). In many homozygous mutants, the abnormal development of chloroplasts leads to albino seeds and embryonic lethality. EMB1990 and EMB2726 are plastid-localized proteins, and the chloroplasts of EMB1990 and EMB2726 knockout mutants are severely impaired, resulting in defective embryos (Chen et al., 2018b; Li et al., 2021). It remains poorly understood what are intriguing biological functions of embryonic chloroplast in Arabidopsis.
In our transcriptome data, KEGG and GO analyses showed that many DEGs of 4W vs 5W group were enriched in photosynthesis term and chloroplast components (apoplast, thylakoid, and photosynthetic membrane) (Supplementary Figures 4B–D, 6). Interestingly, six photosystem I-antenna (LHCI) genes and 12 chlorophyll a/b binding protein complex Ⅱ genes exhibited up-regulated expression trends at 3-5W, but the photosynthetic genes were dramatically downregulated in naa15 (Supplementary Figure 4B), indicating that embryonic photosystem of the naa15 mutant failed to function, which might be the reason for the white aborted seeds of naa15. However, further studies are needed to investigate the relationship between NAA15 and genes related to photosystems.
Plant hormones are crucial signaling molecules in seed development. It has been reported that auxin is involved in the communication between the endosperm, embryo, and seed coat, and proper embryo development requires the transportation of auxin between endosperm and integument (Batista et al., 2019). In the agl62 mutant of Arabidopsis, the export of auxin is impaired, causing auxin accumulation in the endosperm, and cellularization prematurely occurs (Kang et al., 2008; Figueiredo et al., 2016). In the auxin response factor 2 (arf2) mutant of Arabidopsis, extra cell proliferation occurs in the seed integuments, leading to enlarged seed coats and enhanced seed mass (Schruff et al., 2006). Additionally, it is well documented that auxin has an immediate negative control on cytokinin levels by inhibiting its biosynthesis (Nordström et al., 2004), and the ratio of auxin and cytokinin is critical for tissue proliferation and differentiation (Day et al., 2008). The cytokinin has important roles in root vascular patterning, and the septuple mutant log1 log2 log3 log4 log5 log7 log8 shows severe embryonic vascular tissue development and patterning defects (De Rybel et al., 2014).
In our transcriptome data, the GO and KEGG analyses showed that the signal transduction pathways related to phytohormones were enriched in 3-5W (Figure 3 and Supplementary Table 10), involving in auxin, cytokinin, and abscisic acid (Supplementary Table 11). In naa15, DEGs related to auxin response factor (ARF), auxin-responsive protein (IAA), and auxin efflux proteins were significantly down-regulated in naa15 (Figures 5A-C). Hence, the auxin response and transport in naa15 was disrupted owing to deletion of NAA15. In addition, the genes related to several cytokinin dehydrogenases and cytokinin-activating enzymes were also down-regulated in naa15 (Figure 5D). It has been reported that cytokinin signaling is crucial during syncytial endosperm development (Day et al., 2008), and the homeostasis of auxin and cytokinin is essential for seed formation. Enhancing auxin or reducing cytokinin can disrupt embryo development (Zhang et al., 2017). Based on our transcriptional data, DEG-related hormones might be useful for investigating the roles of hormone regulation in these processes. However, further investigation into the relationship between NAA15 and phytohormones (auxin and cytokinin) were required.
The spatiotemporal expression of different genes is an important foundation during embryo and endosperm development and is regulated by their transcription factors (TFs). In early seed development of maize, transcriptome analysis shows that 110 TFs display high temporal specificity expression, and that MYB131, MYB16, and BZIP109 might be critical for endosperm cellularization (Yi et al., 2019). The WOX, HD-Zip, ARF, and CUC families of the transcription factors have been reported to be crucial in the establishment of embryonic patterning (Verma et al., 2022). WOX2 and WOX8 are co-expressed in the zygote of Arabidopsis, and WOX2 expression in the apical region is activated by WOX8 and WOX9 to regulate proper embryo development (Breuninger et al., 2008). Besides, the rev phb (HD-Zip TF) double mutant displays single or radially symmetric cotyledon embryo patterning defects (Prigge et al., 2005). The WRKY transcription factor MINISEED3 (MINI3) is expressed in both endosperm and globular embryos; hence, the endosperm proliferation in mini3 mutant is retarded, leading to the reduced size of seeds (Luo et al., 2005).
In our research, the transcription regulator activity term was significantly enriched in 3-5W (Supplementary Figure 3A), but genes related to embryonic differentiation, such as WOXs (WOX1, WOX2, WOX5, WOX8), CUCs (CUC2, CUC3), and HD-Zip III (ATHB8, REV), exhibited downregulated expression in naa15 (Figure 6 and Supplementary Table 15). N-terminal acetylation of cellular proteins affects protein interactions, subcellular targeting, protein folding, and protein degradation (Scott et al., 2011; Yang et al., 2013). However, up to date, no correlations between NAA15 and these genes were reported. We proposed that NAA15 mutation in Arabidopsis led to NatA inactivation, which might cause some protein misfolding and excessive accumulation, resulting in abnormal expressions of these DEGs in naa15 seeds. Additionally, 35 MYB, 37 bHLH, 30 WYKY, and 39 ERFs from our transcriptome data showed abnormal expressions in naa15 (Supplementary Table 14). Among these transcription factors, except for MYB56, bHLH95, WRKY10, and WRKY2, which have been reported for their roles in embryo and endosperm (Ueda et al., 2011; Zhang et al., 2013; MacGregor and Zhang., 2019), there were no reports on other transcription factors. The results implied that the transcription factors might have important functions in embryo and endosperm development and could serve as potential regulators for seed morphogenesis.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: NCBI Bioproject accession number is PRJNA856854.
JZ designed the program of studies and experiments, guided the entire process of the study, and revised the manuscript. CL finished the material collection, carried out most of the experiments, analyzed the data, and wrote the paper. FH performed qPCR analyses. HC performed endosperm autofluorescence assay. All authors read and approved the content of the paper. All authors contributed to the article and approved the submitted version.
This work was supported by the National Natural Science Foundation of China (31870303, 32170337).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.998664/full#supplementary-material
Supplementary Figure 1 | Seed characteristics and genotype analysis of wild type and naa15/+ mutants. (A) Seed phenotypes in siliques of the wild type (WT) and naa15/+. The homozygous white aborted naa15 seeds are labeled by white arrows. Bar = 1mm. (B) Genotypic analysis of WT and naa15/+ mutants. FP, forward primer; RP, reverse primer.
Supplementary Figure 2 | Global analysis of the RNA-seq profile. (A) Violin map of gene expression distribution. The vertical axis meant the value change of the gene expression using log10 (FPKM). The horizontal axis represented eighteen biological samples. (B) Principal component analysis (PCA) of eighteen biological samples. (C) Pearson-correlation coefficients showed the repeatability of the samples.
Supplementary Figure 3 | Gene Ontology (GO) enriched terms of DEGs in 4W vs 3W and 5W vs 4W of Arabidopsis. (A,B) GO enrichment analysis of up-regulated DEGs in 4W vs 3W and 5W vs 4W W, respectively. (C,D) GO enrichment analysis of down-regulated DEGs in 4W vs 3W and 5W vs 4W, respectively.
Supplementary Figure 4 | Heatmaps of DEGs related to sucrose and photosynthesis in WT and naa15 of Arabidopsis. (A) Heatmap of DEGs related to sucrose synthesis and transport in WT and naa15. (B) Heatmap of DEGs related to photosynthesis in WT and naa15.
Supplementary Figure 5 | Gene Ontology (GO) enriched terms of down-regulated DEGs from comparisons between WT and naa15 in 5M vs 3W, 6M vs 4W, and 7M vs 5W of Arabidopsis.
Supplementary Figure 6 | Gene Ontology (GO) enriched terms of up-regulated DEGs from comparisons between WT and naa15 in 5M vs 3W, 6M vs 4W, and 7M vs 5W of Arabidopsis.
Aida, M., Beis, D., Heidstra, R., Willemsen, V., Blilou, I., Galinha, C., et al. (2004). The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell 119, 109–120. doi: 10.1016/j.cell.2004.09.018
Arnesen, T., Van Damme, P., Polevoda, B., Helsens, K., Evjenth, R., Colaert, N., et al. (2009). Proteomics analyses reveal the evolutionary conservation and divergence of n-terminal acetyltransferases from yeast and humans. Proc. Natl. Acad. Sci. U.S.A. 106, 8157–8162. doi: 10.1073/pnas.0901931106
Babicki, S., Arndt, D., Marcu, A., Liang, Y., Grant, J. R., Maciejewski, A., et al. (2016). Heatmapper: web-enabled heat mapping for all. Nucleic Acids Res. 44, 147–153. doi: 10.1093/nar/gkw419
Batista, R. A., Figueiredo, D. D., Santos-González, J., Köhler, C. (2019). Auxin regulates endosperm cellularization in Arabidopsis. Genes Dev. 33, 466–476. doi: 10.1101/gad.316554.118
Berger, F. (2003). Endosperm: the crossroad of seed development. Curr. Opin. Plant Biol. 6, 42–50. doi: 10.1016/S1369526602000043
Bienvenut, W. V., Sumpton, D., Martinez, A., Lilla, S., Espagne, C., Meinnel, T., et al. (2012). Comparative large scale characterization of plant versus mammal proteins reveals similar and idiosyncratic n-α-acetylation features. Mol. Cell Proteomics 11, M111.0151311–111.0151314. doi: 10.1074/mcp.M111.015131
Bischof, S., Baerenfaller, K., Wildhaber, T., Troesch, R., Vidi, P. A., Roschitzki, B., et al. (2011). Plastid proteome assembly without Toc159: photosynthetic protein import and accumulation of n-acetylated plastid precursor proteins. Plant Cell 23, 3911–3928. doi: 10.1105/tpc.111.092882
Bishopp, A., Benková, E., Helariutta, Y. (2011). Sending mixed messages: auxin-cytokinin crosstalk in roots. Curr. Opin. Plant Biol. 14, 10–16. doi: 10.1016/j.pbi.2010.08.014
Breuninger, H., Rikirsch, E., Hermann, M., Ueda, M., Laux, T. (2008). Differential expression of WOX genes mediates apical-basal axis formation in the Arabidopsis embryo. Dev. Cell 14, 867–876. doi: 10.1016/j.devcel.2008.03.008
Caffall, K. H., Mohnen, D. (2009). The structure, function, and biosynthesis of plant cell wall pectic polysaccharides. Carbohydr. Res. 344, 1879–1900. doi: 10.1016/j.carres.2009.05.021
Cheng, H., Dharmadhikari, A. V., Varland, S., Ma, N., Domingo, D., Kleyner, R., et al. (2018). Truncating variants in NAA15 are associated with variable levels of intellectual disability, autism spectrum disorder, and congenital anomalies. Am. J. Hum. Genet. 102, 985–994. doi: 10.1016/j.ajhg.2018.03.004
Chen, H., Li, S., Li, L., Hu, H., Zhao, J. (2018b). Arabidopsis EMB1990 encoding a plastid-targeted YlmG protein is required for chloroplast biogenesis and embryo development. Front. Plant Sci. 9, 181. doi: 10.3389/fpls.2018.00181
Chen, H., Li, S., Li, L., Wu, W., Ke, X., Zou, W., et al. (2018a). Nα-acetyltransferases 10 and 15 are required for the correct initiation of endosperm cellularization in Arabidopsis. Plant Cell Physiol. 59, 2113–2128. doi: 10.1093/pcp/pcy135
Chen, L. Q., Lin, I. W., Qu, X. Q., Sosso, D., McFarlane, H. E., Londoño, A., et al. (2015). A cascade of sequentially expressed sucrose transporters in the seed coat and endosperm provides nutrition for the Arabidopsis embryo. Plant Cell 27, 607–619. doi: 10.1105/tpc.114.134585
Cruz-Valderrama, J. E., Jiménez-Durán, K., Zúñiga-Sánchez, E., Salazar-Iribe, A., Márquez-Guzmán, J., Gamboa-deBuen, A. (2018). Degree of pectin methyl esterification in endosperm cell walls is involved in embryo bending in Arabidopsis thaliana. Biochem. Biophys. Res. Commun. 495, 639–645. doi: 10.1016/j.bbrc.2017.11.077
Day, R. C., Herridge, R. P., Ambrose, B. A., Macknight, R. C. (2008). Transcriptome analysis of proliferating Arabidopsis endosperm reveals biological implications for the control of syncytial division, cytokinin signaling, and gene expression regulation. Plant Physiol. 148, 1964–1984. doi: 10.1104/pp.108.128108
De Rybel, B., Adibi, M., Breda, A. S., Wendrich, J. R., Smit, M. E., Novak, O., et al. (2014). Integration of growth and patterning during vascular tissue formation in Arabidopsis. Science 345, 1255215. doi: 10.1126/science.1255215
Dresselhaus, T., Sprunck, S., Wessel, G. M. (2016). Fertilization mechanisms in flowering plants. Curr. Biol. 26, 125–139. doi: 10.1016/j.cub.2015.12.032
Endo, A., Tatematsu, K., Hanada, K., Duermeyer, L., Okamoto, M., Yonekura-Sakakibara, K., et al. (2012). Tissue-specific transcriptome analysis reveals cell wall metabolism, flavonol biosynthesis and defense responses are activated in the endosperm of germinating Arabidopsis thaliana seeds. Plant Cell Physiol. 53, 16–27. doi: 10.1093/pcp/pcr171
Feng, J., Li, R., Yu, J., Ma, S., Wu, C., Li, Y., et al. (2016). Protein n-terminal acetylation is required for embryogenesis in Arabidopsis. J. Exp. Bot. 67, 4779–4789. doi: 10.1093/jxb/erw257
Figueiredo, D. D., Batista, R. A., Roszak, P. J., Hennig, L., Köhler, C. (2016). Auxin production in the endosperm drives seed coat development in Arabidopsis. eLife 5, e20542. doi: 10.7554/eLife.20542.028
Figueiredo, D. D., Batista, R. A., Roszak, P. J., Köhler, C. (2015). Auxin production couples endosperm development to fertilization. Nat. Plants 1, 15184–15190. doi: 10.1038/nplants.2015.184
Fourquin, C., Beauzamy, L., Chamot, S., Creff, A., Goodrich, J., Boudaoud, A., et al. (2016). Mechanical stress mediated by both endosperm softening and embryo growth underlies endosperm elimination in Arabidopsis seeds. Development 143, 3300–3305. doi: 10.1242/dev.137224
French, S., Abu-Zaitoon, Y., Uddin, M., Bennett, K., Nonhebel, H. (2014). Auxin and cell wall invertase related signaling during rice grain development. Plants 3, 95–112. doi: 10.3390/plants3010095
Gautschi, M., Just, S., Mun, A., Ross, S., Rücknagel, P., Dubaquié, Y., et al. (2003). The yeast n(alpha)-acetyltransferase NatA is quantitatively anchored to the ribosome and interacts with nascent polypeptides. Mol. Cell Biol. 23, 7403–7414. doi: 10.1128/MCB.23.20.7403-7414.2003
Giglione, C., Meinnel, T. (2021). Evolution-driven versatility of n terminal acetylation in photoautotrophs. Trends Plant Sci. 26, 375–391. doi: 10.1016/j.tplants.2020.11.012
Hong, Z., Ueguchi-Tanaka, M., Fujioka, S., Takatsuto, S., Yoshida, S., Hasegawa, Y., et al. (2005). The rice brassinosteroid-deficient dwarf2 mutant, defective in the rice homolog of Arabidopsis DIMINUTO/DWARF1, is rescued by the endogenously accumulated alternative bioactive brassinosteroid, dolichosterone. Plant Cell 17, 2243–2254. doi: 10.1105/tpc.105.030973
Jiang, W. B., Lin, W. H. (2013). Brassinosteroid functions in Arabidopsis seed development. Plant Signal. Behav. 8, e259281–e259285. doi: 10.4161/psb.25928
Jo, L., Pelletier, J. M., Harada, J. J. (2019). Central role of the LEAFY COTYLEDON1 transcription factor in seed development. J. Integr. Plant Biol. 61, 564–580. doi: 10.1111/jipb.12806
Kang, I. H., Steffen, J. G., Portereiko, M. F., Lloyd, A., Drews, G. N. (2008). The AGL62 MADS domain protein regulates cellularization during endosperm development in Arabidopsis. Plant Cell 20, 635–647. doi: 10.1105/tpc.107.055137
Karunakaran, K. B., Amemori, S., Balakrishnan, N., Ganapathiraju, M. K., Amemori, K. I. (2021). Generalized and social anxiety disorder interactomes show distinctive overlaps with striosome and matrix interactomes. Sci. Rep. 11, 18392–18417. doi: 10.1038/s41598-021-97418-w
Lafon-Placette, C., Köhler, C. (2014). Embryo and endosperm, partners in seed development. Curr. Opin. Plant Biol. 17, 64–69. doi: 10.1016/j.pbi.2013.11.008
Lemke, M. D., Fisher, K. E., Kozlowska, M. A., Tano, D. W., Woodson, J. D. (2021). The core autophagy machinery is not required for chloroplast singlet oxygen-mediated cell death in the Arabidopsis thaliana plastid ferrochelatase two mutants. BMC Plant Biol. 21, 342–362. doi: 10.1186/s12870-021-03119-x
Levesque-Tremblay, G., Müller, K., Mansfield, S. D., Haughn, G. W. (2015). HIGHLY METHYL ESTERIFIED SEEDS is a pectin methyl esterase involved in embryo development. Plant Physiol. 167, 725–737. doi: 10.1104/pp.114.255604
Li, J., Nie, X., Tan, J. L., Berger, F. (2013). Integration of epigenetic and genetic controls of seed size by cytokinin in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A 110, 15479–15484. doi: 10.1073/pnas.1305175110
Linster, E., Stephan, I., Bienvenut, W. V., Maple-Grødem, J., Myklebust, L. M., Huber, M., et al. (2015). Downregulation of n-terminal acetylation triggers ABA-mediated drought responses in Arabidopsis. Nat. Commun. 6, 7640–7653. doi: 10.1038/ncomms8640
Li, C., Shang, J. X., Qiu, C., Zhang, B., Wang, J., Wang, S., et al. (2021). Plastid-localized EMB2726 is involved in chloroplast biogenesis and early embryo development in Arabidopsis. Front. Plant Sci. 12, 675838. doi: 10.3389/fpls.2021.675838
Li, J., Yu, M., Geng, L. L., Zhao, J. (2010). The fasciclin-like arabinogalactan protein gene, FLA3, is involved in microspore development of Arabidopsis. Plant J. 64, 482–497. doi: 10.1111/j.1365-313X.2010.04344.x
Li, G., Zou, W., Jian, L., Qian, J., Deng, Y., Zhao, J. (2017). Non-SMC elements 1 and 3 are required for early embryo and seedling development in Arabidopsis thaliana. J. Exp. Bot. 68, 1039–1054. doi: 10.1093/jxb/erx016
Luo, M., Dennis, E. S., Berger, F., Peacock, W. J., Chaudhury, A. (2005). MINISEED3 (MINI3), a WRKY family gene, and HAIKU2 (IKU2), a leucine-rich repeat (LRR) KINASE gene, are regulators of seed size in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A 102, 17531–17536. doi: 10.1073/pnas.0508418102
MacGregor, D. R., Zhang, N. (2019). ICE1 and ZOU determine the depth of primary seed dormancy in Arabidopsis independently of their role in endosperm development. Plant J. 98, 277–290. doi: 10.1111/tpj.14211
Ma, T., Dong, F., Luan, D., Hu, H., Zhao, J. (2019). Gene expression and localization of arabinogalactan proteins during the development of anther, ovule, and embryo in rice. Protoplasma 256, 909–922. doi: 10.1007/s00709-019-01349-3
Mansfield, S. G., Briarty, L. G. (1991). Early embryogenesis in Arabidopsis thaliana. II. the developing embryo. Can. J. Bot. 69, 461–476. doi: 10.1139/b91-063
Moussu, S., Doll, N. M., Chamot, S., Brocard, L., Creff, A., Fourquin, C., et al. (2017). ZHOUPI and KERBEROS mediate embryo/endosperm separation by promoting the formation of an extracuticular sheath at the embryo surface. Plant Cell 29, 1642–1656. doi: 10.1105/tpc.17.00016
Nordström, A., Tarkowski, P., Tarkowska, D., Norbaek, R., Astot, C., Dolezal, K., et al. (2004). Auxin regulation of cytokinin biosynthesis in Arabidopsis thaliana: a factor of potential importance for auxin-cytokinin-regulated development. Proc. Natl. Acad. Sci. U.S.A 101, 8039–8044. doi: 10.1073/pnas.0402504101
Ohto, M. A., Floyd, S. K., Fischer, R. L., Goldberg, R. B., Harada, J. J. (2009). Effects of APETALA2 on embryo, endosperm, and seed coat development determine seed size in Arabidopsis. Sex Plant Reprod. 22, 277–289. doi: 10.1007/s00497-009-0116-1
Otegui, M., Staehelin, L. A. (2000). Syncytial-type cell plates: a novel kind of cell plate involved in endosperm cellularization of Arabidopsis. Plant Cell 12, 933–947. doi: 10.1105/tpc.12.6.933
Peaucelle, A., Braybrook, S. A., Le Guillou, L., Bron, E., Kuhlemeier, C., Höfte, H. (2011). Pectin-induced changes in cell wall mechanics underlie organ initiation in Arabidopsis. Curr. Biol. 21, 1720–1726. doi: 10.1016/j.cub.2011.08.057
Portereiko, M. F., Lloyd, A., Steffen, J. G., Punwani, J. A., Otsuga, D., Drews, G. N. (2006). AGL80 is required for central cell and endosperm development in Arabidopsis. Plant Cell 18, 1862–1872. doi: 10.1105/tpc.106.040824
Prigge, M. J., Otsuga, D., Alonso, J. M., Ecker, J. R., Drews, G. N., Clark, S. E. (2005). Class III homeodomain-leucine zipper gene family members have overlapping, antagonistic, and distinct roles in Arabidopsis development. Plant Cell 17, 61–76. doi: 10.1105/tpc.104.026161
Ree, R., Varland, S., Arnesen, T. (2018). Spotlight on protein n-terminal acetylation. Exp. Mol. Med. 50, 1–13. doi: 10.1038/s12276-018-0116-z
Riefler, M., Novak, O., Strnad, M., Schmülling, T. (2006). Arabidopsis cytokinin receptor mutants reveal functions in shoot growth, leaf senescence, seed size, germination, root development, and cytokinin metabolism. Plant Cell 18, 40–54. doi: 10.1105/tpc.105.037796
Sørensen, M. B., Mayer, U., Lukowitz, W., Robert, H., Chambrier, P., Jürgens, G., et al. (2002). Cellularisation in the endosperm of Arabidopsis thaliana is coupled to mitosis and shares multiple components with cytokinesis. Development 129, 5567–5576. doi: 10.1242/dev.00152
Schruff, M. C., Spielman, M., Tiwari, S., Adams, S., Fenby, N., Scott, R. J. (2006). The AUXIN RESPONSE FACTOR 2 gene of Arabidopsis links auxin signalling, cell division, and the size of seeds and other organs. Development 133, 251–261. doi: 10.1242/dev.02194
Scott, D. C., Monda, J. K., Bennett, E. J., Harper, J. W., Schulman, B. A. (2011). N-terminal acetylation acts as an avidity enhancer within an interconnected multiprotein complex. Science 334, 674–678. doi: 10.1126/science.1209307
Song, J. P., Xie, X., Cui, Y., Zou, J. T. (2021). Endosperm-embryo communications: recent advances and perspectives. Plants 10, 2511–2520. doi: 10.3390/plants10112511
Steffen, J. G., Kang, I. H., Portereiko, M. F., Lloyd, A., Drews, G. N. (2008). AGL61 interacts with AGL80 and is required for central cell development in Arabidopsis. Plant Physiol. 148, 259–268. doi: 10.1104/pp.108.119404
Sun, X., Shantharaj, D., Kang, X., Ni, M. (2010). Transcriptional and hormonal signaling control of Arabidopsis seed development. Curr. Opin. Plant Biol. 13, 611–620. doi: 10.1016/j.pbi.2010.08.009
Sun, S., Yi, C., Ma, J., Wang, S., Peirats-Llobet, M., Lewsey, M. G., et al. (2020). Analysis of spatio-temporal transcriptome profiles of soybean (Glycine max) tissues during early seed development. Int. J. Mol. Sci. 21, 7603–7624. doi: 10.3390/ijms21207603
Tan, L., Eberhard, S., Pattathil, S., Warder, C., Glushka, J., Yuan, C., et al. (2013). An Arabidopsis cell wall proteoglycan consists of pectin and arabinoxylan covalently linked to an arabinogalactan protein. Plant Cell 25, 270–287. doi: 10.1105/tpc.112.107334
Tejos, R. I., Mercado, A. V., Meisel, L. A. (2010). Analysis of chlorophyll fluorescence reveals stage specific patterns of chloroplast-containing cells during Arabidopsis embryogenesis. Biol. Res. 43, 99–111. doi: 10.4067/S0716-97602010000100012
Ueda, M., Zhang, Z., Laux., T. (2011). Transcriptional activation of Arabidopsis axis patterning genes WOX8/9 links zygote polarity to embryo development. Dev. Cell. 20, 264–270. doi: 10.1016/j.devcel.2011.01.009
Vanstraelen, M., Benkova, E. (2012). Hormonal interactions in the regulation of plant development. Annu. Rev. Cell Dev. Biol. 28, 463–487. doi: 10.1146/annurev-cellbio-101011-155741
Varland, S., Osberg, C., Arnesen, T. (2015). N-terminal modifications of cellular proteins: The enzymes involved, their substrate specificities and biological effects. Proteomics 15, 2385–2401. doi: 10.1002/pmic.201400619
Verma, S., Attuluri, V., Robert, H. S. (2022). Transcriptional control of Arabidopsis seed development. Planta 255, 90–99. doi: 10.1007/s00425-022-03870-x
Wang, W., Xiong, H., Sun, K., Zhang, B., Sun, M. X. (2022). New insights into cell-cell communications during seed development in flowering plants. J. Integr. Plant Biol. 64, 215–229. doi: 10.1111/jipb.13170
Xu, F., Huang, Y., Li, L., Gannon, P., Linster, E., Huber, M., et al. (2015). Two n-terminal acetyltransferases antagonistically regulate the stability of a nod-like receptor in Arabidopsis. Plant Cell 27, 1547–1562. doi: 10.1105/tpc.15.00173
Yang, D., Fang, Q., Wang, M., Ren, R., Wang, H., He, M., et al. (2013). Na-Acetylated Sir3 stabilizes the conformation of a nucleosome-binding loop in the BAH domain. Nat. Struct. Mol. Biol. 20, 1116–1118. doi: 10.1038/nsmb.2637
Yi, F., Gu, W., Chen, J., Song, N., Gao, X., Zhang, X., et al. (2019). High temporal-resolution transcriptome landscape of early maize seed development. Plant Cell 31, 974–992. doi: 10.1105/tpc.18.00961
Yoon, J., Cho, L. H., Tun, W., Jeon, J. S., An, G. (2021). Sucrose signaling in higher plants. Plant Sci. 302, 110703–110712. doi: 10.1016/j.plantsci.2020.110703
Zhang, Y., Liang, W., Shi, J., Xu, J., Zhang, D. (2013). MYB56 encoding a R2R3 MYB transcription factor regulates seed size in Arabidopsis thaliana. J. Integr. Plant Biol. 55, 1166–1178. doi: 10.1111/jipb.12094
Zhang, Z., Tucker, E., Hermann, M., Laux, T. (2017). A molecular framework for the embryonic initiation of shoot meristem stem cells. Dev. Cell 40, 264–277. doi: 10.1016/j.devcel.2017.01.002
Keywords: Arabidopsis, endosperm cellularization, embryo differentiation, cell wall, transcriptomics, gene expression
Citation: Li C, Hu F, Chen H and Zhao J (2022) Transcriptome characteristics during cell wall formation of endosperm cellularization and embryo differentiation in Arabidopsis. Front. Plant Sci. 13:998664. doi: 10.3389/fpls.2022.998664
Received: 20 July 2022; Accepted: 16 September 2022;
Published: 03 October 2022.
Edited by:
Rowan Andrew Craig Mitchell, Rothamsted Research, United KingdomReviewed by:
Duarte D. Figueiredo, Max Planck Institute of Molecular Plant Physiology, GermanyCopyright © 2022 Li, Hu, Chen and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jie Zhao, anpoYW9Ad2h1LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.