- 1Key Laboratory of Agroecological Processing and Safety Monitoring of Fujian Province, Fujian Agriculture and Forestry University, Fuzhou, China
- 2College of Tea and Food Science, Wuyi University, Wuyishan, China
- 3College of Life Sciences, Longyan University, Longyan, China
Nitrogen (N) is the first essential nutrient for tea growth. However, the effect of soil acidification on soil N cycle and N forms in tea plantation are unclear. In this study, the nitrogen contents, soil enzyme activity and N mineralization rate in acidified soil of tea plantation were measured. Moreover, the effects of soil acidification on N cycling functional genes and functional microorganisms were explored by soil metagenomics. The results showed that the NH4+-N, available N and net N mineralization rate in the acidified tea soil decreased significantly, while the NO3--N content increased significantly. The activities of sucrase, protease, catalase and polyphenol oxidase in the acidified tea soil decreased significantly. The abundance of genes related to ammonification, dissimilatory N reduction, nitrification and denitrification pathway in the acidified tea soil increased significantly, but the abundance of functional genes related to glutamate synthesis and assimilatory N reduction pathway were opposite. In addition, the abundance of Proteobacteria, Actinobacteria, Chloroflexi, Nitrospirae, Actinomadura, Nitrospira etc. microorganisms related to nitrification, denitrification and pathogenic effect increased significantly in the acidified tea soil. The correlation results showed that soil pH and N forms were correlated with soil enzyme activity, N cycling function genes and microbial changes. In conclusion, soil acidification results in significant changes in enzyme activity, gene abundance and microorganism involved in various N cycle processes in acidified tea soil, which leads to imbalance of soil N form ratio and is not conducive to N transformation and absorption of tea trees.
Introduction
Tea (camellia sinensis) is an important economic plant in tropical and subtropical countries. Tea is a perennial leaf-harvested crop that grows best in acidic soil with pH value of- 4.5-5.5 (Yan et al., 2020). However, soil acidification is major problem in tea plantation. Generally, excessive N fertilization is considered the major driver of soil acidification in tea plantations (Huang et al., 2015a; Yan et al., 2020; Lin et al., 2022). Specifically, the accelerated soil acidification from fertilization is directly caused by the production of protons via the nitrification process after application of ammonium N fertilization (Barak et al., 1997; Zhou et al., 2014). In addition, the nitrate nitrogen (NO3––N) produced by nitrification was washed out of the soil under heavy rainfall and took away a large number of base ions, resulting in the decrease of the acid neutralization ability of the soil (Yang et al., 2018b). Moreover, not all of the large amount of fertilizer applied can be absorbed and utilized by plants, and the loss of N from agroecosystems has led to pollution of groundwater, nitrous oxide (N2O) emissions, and other environmental problems (Sun et al., 2021).
Nitrogen is an essential element for all plants to grow and reproduce. NH4+-N and NO3––N are the main types of available N, but they are limited in the natural environment. Only after N mineralization process, organic N can be converted into effective N (NO3-, NH4+), which can be absorbed and utilized by plants (Robertson et al., 1999). Compared with NO3-, NH4+ could significantly improve the photosynthetic efficiency and quality of tea trees (Ruan et al., 2008). N transformation in soil refers to the process of changing the form or state of N-containing substances in soil by physical chemistry and biochemistry, mainly includes N mineralization, N fixation, nitrification and denitrification etc. (Kuypers et al., 2018). Transformation of soil N forms is carried out by microbiome containing specific functional genes (Zhang et al., 2019). There are many studies focused on soil N transformation, but mainly focuses on the effects of different fertilization management on soil N transformation (Saggar et al., 2013). For example, for acidic soils, long-term application of chemical N fertilizer resulted in increased activity and abundance of Ammonia oxidizing archaea (AOA) (Leininger et al., 2006). Sun et al. (2020) reported that the application of biological fertilizer reduced the abundance of bacterial amoA gene in soil, but enhanced the soil denitrification, reduced the accumulation of NO3––N, thus greatly reduced N leaching loss in runoff. The addition of Oxytetracycline inhibited nitrifying bacteria reduce the conversion of NH4+-N to NO3––N in composting process (Zhang et al., 2021). The use efficiency of soil N can be improved by applying different fertilizers. Although there are a lot of research on soil N genes, most of these studies used qRT-PCR methods, which often lead to biased results and focus on a single gene involved in the N cycle or single N process-related genes. N cycle is a network of interconnected processes. Such an approach lacks information on the complete N-cycle metabolism process, which can lead to the loss of important information.
Soil pH value is an important factor influencing soil N-related transformation and microbial communities (Wang et al., 2017). Previous studies have demonstrated that soil acidification will lead to the loss of soil base ions, the decrease of directly available NH4+-N and NO3––N in soil, the intensification of soil acidification, and then decrease of N conversion efficiency in soil (Yamamoto et al., 2014; Huang et al., 2015b). Soil enzymes play an important role in material cycle and energy flow in soil ecosystem, which regulate the conversion and circulation of soil nutrients. Meanwhile, the study of the composition of soil microbial community is conducive to an in-depth understanding of the soil internal ecological processes. Soil acidification can change the diversity of soil microbial community structure, such as increasing the relative abundance of acidifying bacteria bacillus, and decreasing the number of Proteobacteria, Bacteroides and Actinomycetes (Xu et al., 2016; Goswami et al., 2017). Planctomycetes, Acidobacteria, Nitrospirae etc. Microorganisms are closely related to soil nitrogen cycling (Zhou et al., 2018; Spieck et al., 2020; Hu et al., 2021). Tea plants have a great demand for N as the buds and leaves are the main subjects for tea yield. Then, can soil acidification influence the content of soil N? And the response of the functional genes and microorganisms involved in the N cycle to soil acidification in tea plantation and their relationship with N forms are unclear.
Here, soil N contents, N mineralization rates and soil enzyme activity with different soil pH values were measured. Furthermore, metagenomic methods were used to comprehensively analyze the differences of various functional genes in soil N-cycling process and microbial communities of tea plantation with different pH values. The objective of this study is 1) to analyze the change of N contents and soil enzyme activity in tea soil after acidification; 2) to assess the response of the functional genes involved in the N cycle and functional microorganisms to soil acidification and their relationship with N forms, and 3) to analyze the specific genes involved in different N processes with different acidity of tea soils.
Materials and methods
Sample collection and site description
The experiment site was located in Nanjing County, Fujian Province, China (24°37′04″N, 117°02′80″E), elevation 590-582 m above sea level, The site is a typical monsoonal with a mean annual temperature of 21.40°C. The minimum and maximum mean monthly temperatures are -0.5°C and 38.9°C in January and July, respectively, with a mean annual rainfall of 1821 mm. The site has a haplic arenosol soil (FAO classification) and with a clay loam texture. A 750 kg ha−1 of compound fertilizer (17% N, 17% P, and 17% K) and 600 kg ha−1 of urea were added to the soil in mid-February of every year. In late-May and July, a compound fertilizer and urea were added to the soil surface at the rate of 750 and 300 kg ha-1 each time. The pruning of upper leaves and buds were reserved in tea plantation for surface mulch.
In August 2020, based on the results of previous studies (Lin et al., 2022), tea plantations in the same region with soil pH ranges of 3.5-4.0 (AT) and 5.0-5.5 (ST) were selected as research sites. Each tea plantation plot area is no less than 40 m2 (4 m × 10 m). Then soil samples were randomly collected from three plots in each site. The rhizosphere soil of tea trees was taken from each experimental plot by a five-point sampling method. Briefly, the surface deciduous leaves of the selected trees were removed. The 500 g soils were collected from the tea tree rhizosphere at a radius of 10-15 cm and depth of 10-20 cm, then mixed thoroughly to form one composite sample. Different soil samples were immediately placed into plastic bags, to minimize exposure to O2, and prior to analyses. The fresh soil was sieved in a 2-mm-mesh sieve and divided into three subsamples. One portion dried in air naturally was used to measure chemical properties. One portion was stored at 4°C for enzyme activity determination. And the remainder was immediately frozen at -80°C for DNA analysis.
Chemical properties determining
The pH of soil was determined with a glass electrode (PB-10; Sartorius, Shanghai, China) with a soil to water ratio of 1:2.5. The contents of NO3––N and NH4+-N in soil samples were determined according to the method of Lu (1999). In brief, 5 g soil sample was used to extract and the NO3––N content was determined using dual-wavelength spectrophotometry at 210 nm and 275 nm. The 10 g of soil sample was used to extract and the NH4+-N content was measured using indigophenol blue colorimetric method at 625 nm. The sum of NO3––N and NH4+-N contents was the available N (AN) content. The soil net N mineralization rate (Mitr) was determined by measuring the change of available N (NH4+-N and NO3––N) in the soil samples. In brief, 10 g of tea soils were incubated in a 100 mL culture flask and cultured in the dark at 25°C for 7 d (Li et al., 2020). Soil mineral N (NH4+-N and NO3––N) was determined by colorimetric method at the beginning and the end of the culture, then the N mineralization rate was analyzed. All soil properties were determined in triplicate.
Determination of soil enzyme activity
The soil enzyme activity was analyzed by the reference (Guan et al., 1986). Briefly, sucrase activity (SC) was determined using 3,5-dinitrosalicylic acid colorimetry; urease activity (UR) was determined using sodium phenol colorimetry; protease activity (PR) was determined using Folin colorimetric method; catalase (CAT) activity was determined by potassium permanganate titration; polyphenol oxidase activity (PPO) was determined by purple gallic acid colorimetric method; and phosphomonoesterase activity (PMase) was determined by p-nitrophenylphosphate disodium colorimetric method. All soil properties were determined in triplicate.
DNA extraction and metagenomics sequencing
Soil total DNA extraction were using Bio-Fast Soil Genomic DNA Extraction kit (BioFlux, Hangzhou, China), according to the manufacturer’s instructions with 0.5 g. The integrity of DNA was checked by 1% agarose gels. and the concentration was measured using a NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). The pure DNA in the extraction solution was processed with the Illumina TruSeq Nano DNA LT Library Preparation Kit to construct metagenome shotgun sequencing libraries with insert sizes of 350 bp. Each library was sequenced by using the Illumina HiSeq X-ten platform (Illumina, USA) with the PE150 strategy.
Preprocessing the Raw Data obtained from the Illumina HiSeq sequencing platform were preprocessed by Readfq to obtain the Clean Data for subsequent analysis. After obtaining quality-filtered reads, they were de novo assembled and use MEGAHIT (v1.0.4-beta) to construct the metagenome for each sample. The Scaftigs (> 300 bp) assembled from both single and mixed are all predicted the ORF by MetaGeneMark (Zhu et al., 2010), then were clustered by CD-HIT (V4.5.8) to redundancy and obtain non-redundant gene catalogue. Gene abundance in each sample was estimated by using soap. coverage (http://soap.genomics.org.cn/) based on the number of aligned reads. And then using BLASTP (BLAST Version 2.2.28 + 5) to compare the gene set and NR database (e-value ≤ 1e−5), N cycle microbial species annotation was obtained through the gene taxonomic information database corresponding to the NR database, and then species abundance was calculated by summing the abundances of genes corresponding to the species (Wang et al., 2020).
Statistical analysis
The changes in soil chemical properties, soil enzyme activity and gene abundance were tested for significant differences among tea soils with one-way ANOVA using the SPSS 20.0 (SPSS Inc., Chicago, IL, USA) followed by the least significant difference (LSD) at a 5% level of probability. All experimental data were showed as the mean ± standard error. Graphs were prepared using origin (OriginPro 9.0) unless otherwise indicated. Pearson correlation analysis was used to examine the relationships among the environmental factors, soil enzyme activity and abundance of functional genes for each N transformation, using the Corrplot program package in R software. Redundancy analysis (RDA) was carried out to examine the relationships between environmental factors and microbial abundance at the phylum and genus level, using the vegan package in R software.
Results
Soil physicochemical properties
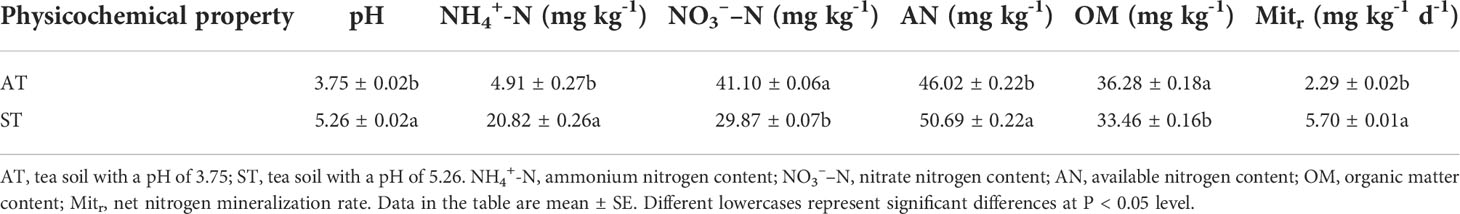
The soil properties varied significantly between the two tea soils (Table 1). The pH of tea soil with AT and ST were 3.75 and 5.26, respectively. With the increase of soil pH value, the contents of NO3––N were 41.10 and 29.87 mg·kg-1, respectively. After acidification, the NO3––N content of tea soil significantly increased (P < 0.05) by 11.23 mg·kg-1. In contrast, relative to ST, the content of NH4+-N in AT was significantly decreased (P < 0.05) by 15.91 mg·kg-1, and corresponding decrease for available N was 4.67 mg·kg-1. With the increase of soil pH value, the rate of net N mineralization were 2.29 and 5.70 mg·kg-1 d-1, respectively. The net N mineralization rate of ST were about 2.5 times as much as that of AT (acidified soil).
Soils enzymatic activity
There were significant differences (P < 0.05) in soil enzyme activities with different pH values (Figure 1). The activity of sucrase was significantly higher than that of other enzymes. Compared with ST, the activities of sucrase, protease, catalase and polyhphenol oxidase decreased by 3.45, 0.44, 0.58 and 0.25 mg g-1 h-1 under AT, respectively. And urease activity of AT was significantly higher than that of ST (P < 0.05). Taken together, the activities of sucrase, protease, catalase and polyphenol oxidase in tea soil decreased significantly, and urease activity increased significantly after soil acidification, but there was no significant difference in phosphomonoesterase activity.
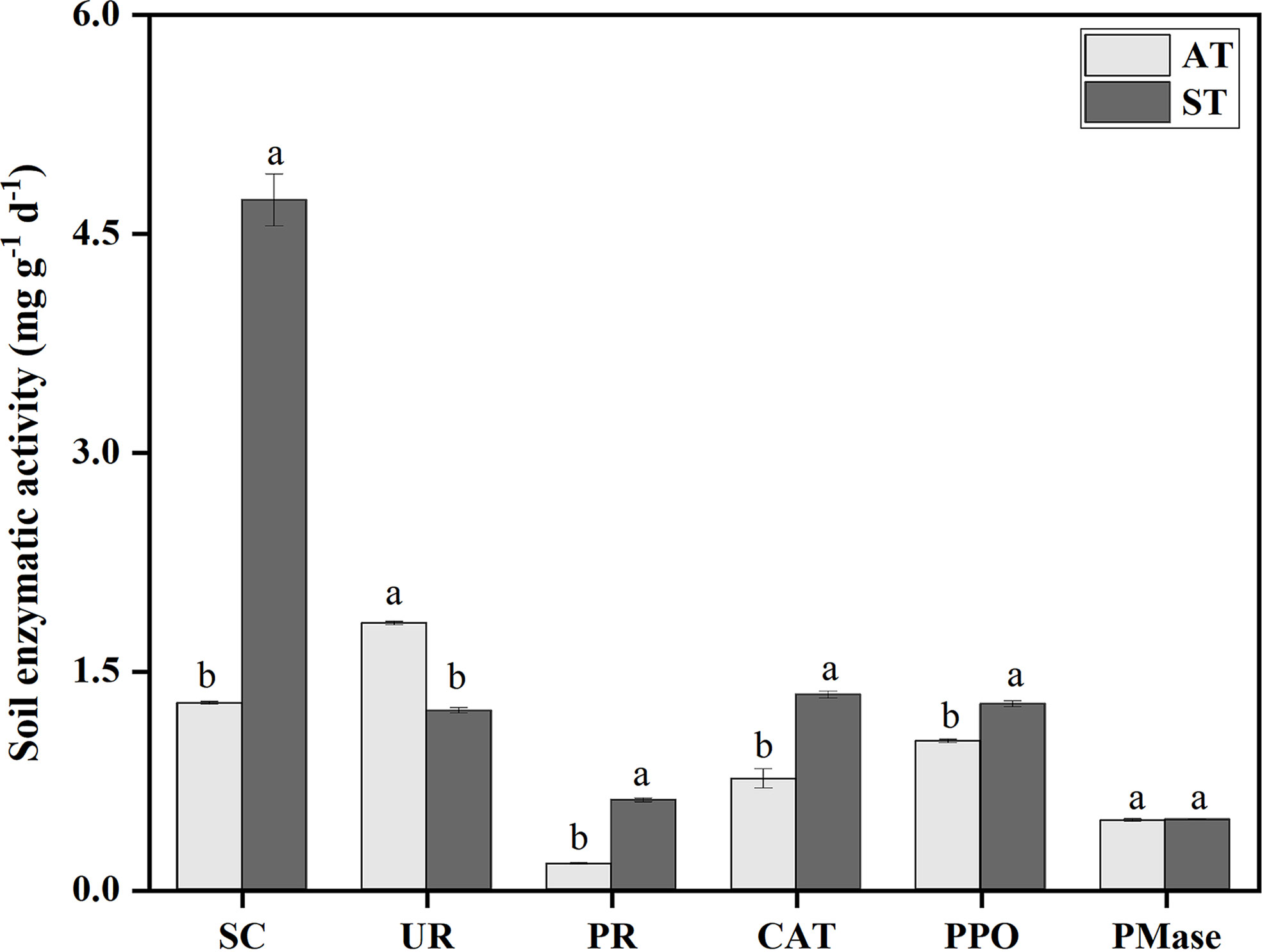
Figure 1 Soil enzyme activities in tea soil with different pH values. SC, sucrase; UR, urease; PR, protease; CAT, catalase; PPO, polyhphenol oxidase; PMase, phosphomonoesterase. AT, tea soil with a pH of 3.75; ST, tea soil with a pH of 5.26. Bars indicate standard error of the mean (n = 3). Different lowercases represent significant differences at P < 0.05 level.
Functional N transformation gene shifts
A total of 76720.18 Mbp of raw reads was obtained from 6 libraries after the Illumina sequencing. After filtering, 76532.51 Mbp of clean reads were identified, and the percentage of clean reads relative to raw reads in each library was above 99.76% (Supplementary Table S1), indicating that the results could comprehensively and truly reflect the composition of soil microbial community. We further summarized and analyzed the frequency and abundance of functional genes involved in the complete N-cycle metabolic process.
A total of 11049 functional gene probes were detected (Supplementary Table S2), which included 28 genes and 6 pathways of N-cycle processes including glutamate synthesis, ammonification, assimilatory N reduction, dissimilatory N reduction, nitrification and denitrification. The processes of N-cycle are shown in Figure 2A. The frequency of genes involved in glutamate synthesis was the highest, with 4114 and 3895 functional genes detected by AT and ST, respectively. In contrast, only 41 and 33 functional genes were detected by nitrification (Figure 2B). However, genes involved in N fixation and anammox were not captured in this study (Figure 2B).
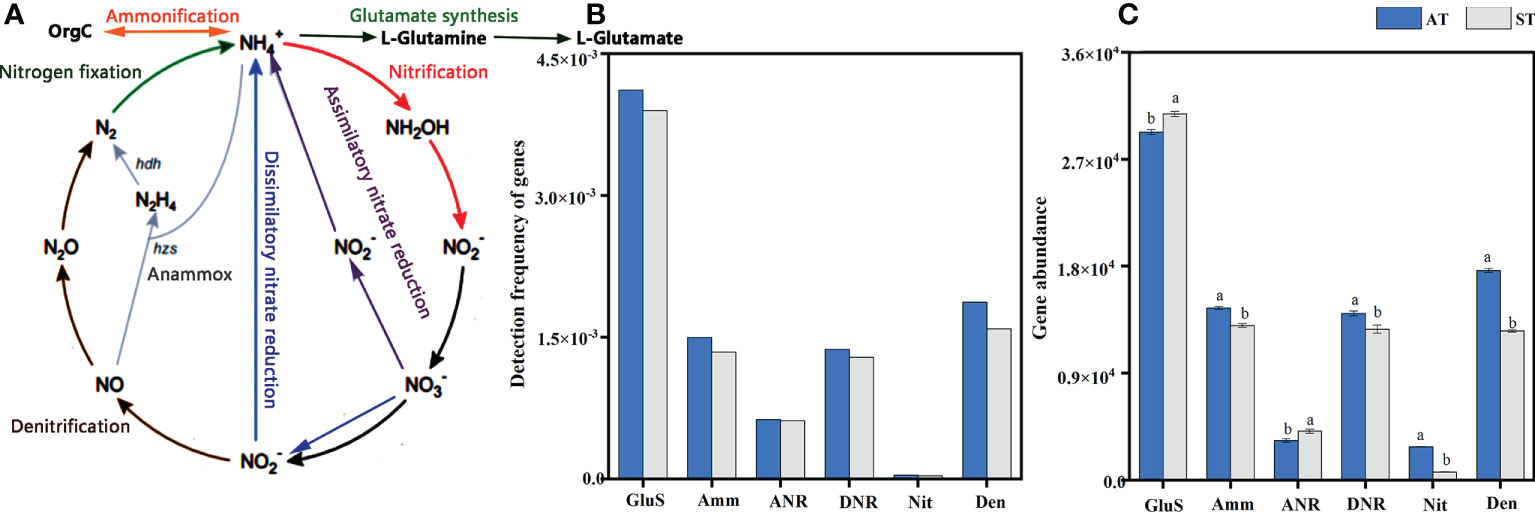
Figure 2 An overall schema illustrating the N-cycle processes (A), detection frequency (B) and abundance (C) of functional genes for each N-cycle with different acidity of tea soils. AT, tea soil with a pH of 3.75; ST, tea soil with a pH of 5.26. GluS, glutamate synthesis; Amm, ammonification; ANR, assimilatory nitrogen reduction; DNR, dissimilatory nitrogen reduction; Nit, nitrification; Den, denitrification. Bars indicate standard error of the mean (n = 3). Different lowercases represent significant differences at P < 0.05 level.
Overall, the total abundance of genes involved in N pathways in AT was significantly higher (P < 0.05) than that in ST (Supplementary Figure S1). With the increase of soil pH, the abundances of genes involved in N pathways were 8.05 × 104 and 7.22 × 104, respectively (Supplementary Figure S1B). For abundance of functional genes for each N-cycle processes, the abundance of functional genes involved in glutamate synthesis pathway was the highest and significantly higher (P < 0.05) than those genes in other N-cycle processes (Figure 2C). After soil acidification, the abundance of genes related to ammonification, dissimilatory N reduction, nitrification and denitrification pathway in tea soils increased significantly (P < 0.05). And the abundance of genes involved in glutamate synthesis and assimilatory N reduction pathways decreased significantly (P < 0.05) under soil acidification (Figure 2C).
From the perspective of individual genes, soil acidification significantly changed the individual gene abundance in tea soils (Figure 3). Overall, soil acidification significantly increased (P < 0.05) the abundances of gdh2 involved in ammonification (Figure 3B), narB and nasB involved in denitrification (Figure 3C), narI, narJ and narH involved in dissimilatory N reduction (Figure 3D), and all individual genes involved in nitrification (amoA_ABC) (Figure 3E) and denitrification (narG, nirK, nirS, norB and nosZ) (Figure 3F). In contrast, the abundances of gltB and gltD involved in glutamate synthesis (Figure 3A), gudB and gdhA involved in ammonification (Figure 3B), nasA and nirA involved in assimilatory N reduction (Figure 3C), and napA, napC, nirB, nirD and nrfA involved in dissimilatory N reduction (Figure 3D) decreased significantly (P < 0.05). The abundances of different individual genes involved in the same pathway were not showed a consistent trend with the change of soil pH value.
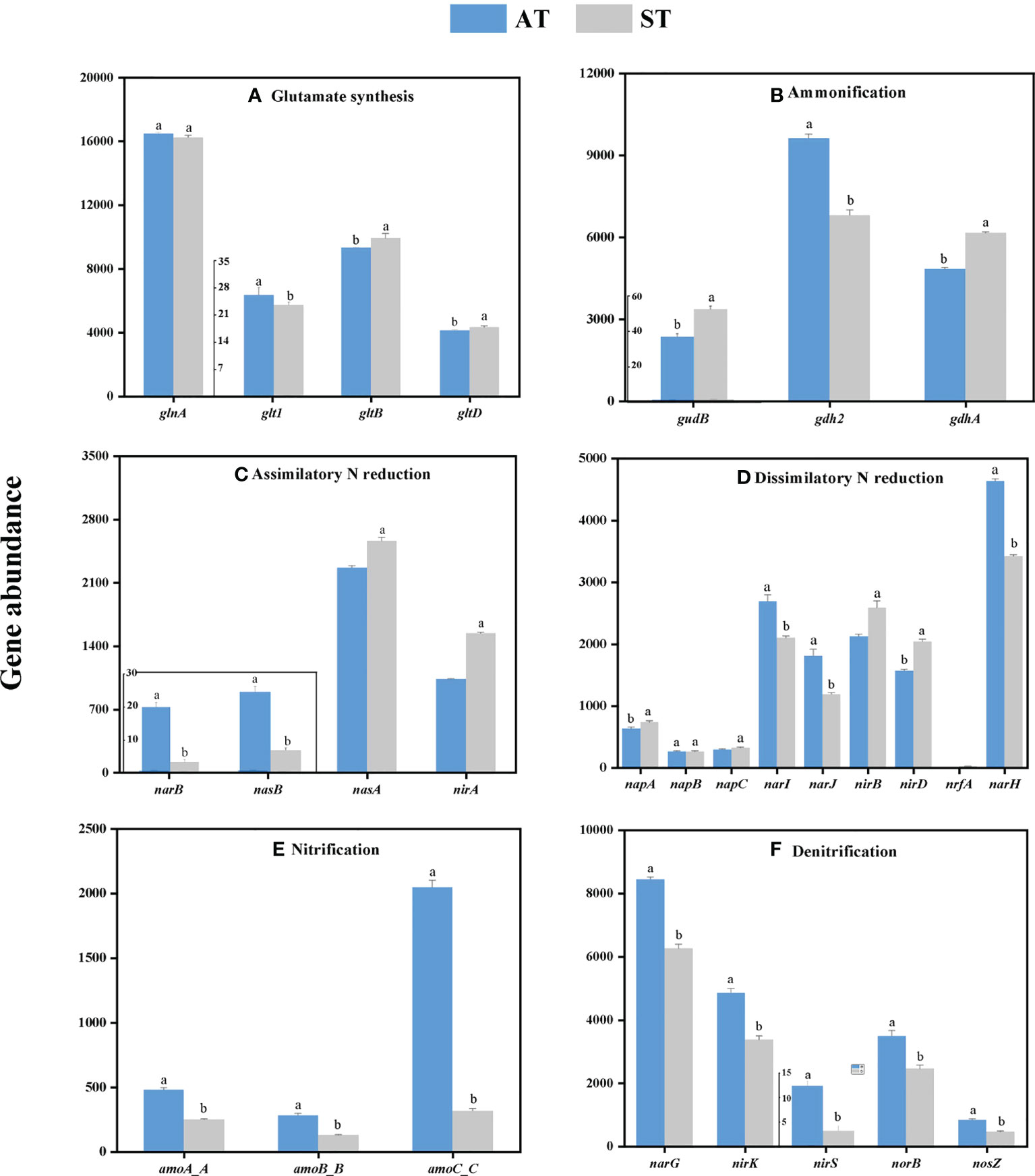
Figure 3 The abundance of individual genes involved in each N-cycle processes (A–F) with different acidity of tea soils. AT, tea soil with a pH of 3.75; ST, tea soil with a pH of 5.26. Bars indicate standard error of the mean (n = 3). Different lowercases represent significant differences at P < 0.05 level.
Microorganisms involved in the nitrogen cycle
The PCA plots indicated that significant differences occurred in soil N-cycling microbial communities with different pH values. The N-cycling microbial were well separated in the first component (PC1) (Supplementary Figure S2), with the three replicates displaying close clustering. At the phylum level, a total of 26 phyla was detected by metagenomic sequencing from two soil samples (Supplementary Figure S3). The top 10 soil microbial communities at the phylum level, accounting for 87.41% of the total soil microorganisms were further selected (Figure 4A). The relative abundances of Proteobacteria, Actinobacteria, Chloroflexi, Nitrospirae and Firmicutes in AT were significantly higher (P < 0.05) than those in ST, while the abundance of Acidobacteria, Armatimonadetes, Verrucomicrobia and Thaumarchaeota were the opposite. Taken together, with the increase of soil pH, the total abundance of top 10 species of soil N-cycling microbial communities at the phylum level accounted for 88.84 and 86.00% of the total, respectively (Supplementary Table S3).
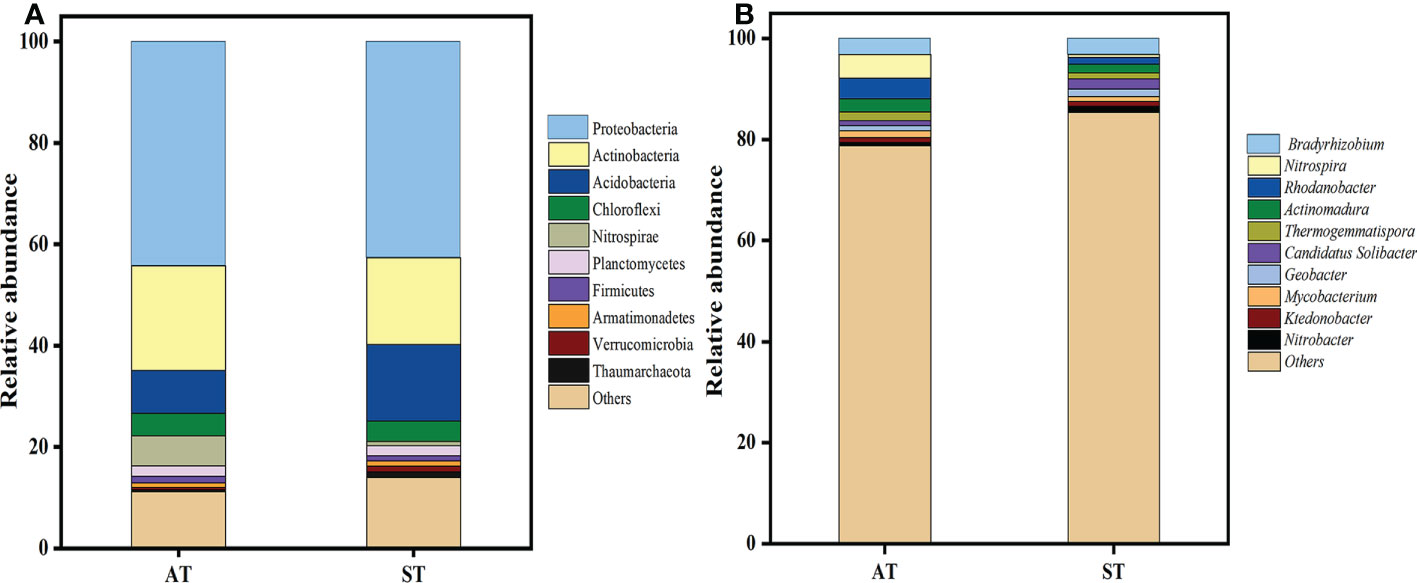
Figure 4 Distribution of nitrogen-related total microbial top 10 phyla (A) and genus (B) with different pH values of tea soils. AT, tea soil with a pH of 3.75; ST, tea soil with a pH of 5.26.
A total of 326 microorganisms genera was obtained at the genus level. As shown in Figure 4B, there were significant differences in soil microbial abundance at genus level in tea soils with different pH values. Compared with ST, the relative abundance of Nitrospira, Rhodanobacter, Actinomadura, Thermogemmatispora and Mycobacterium in AT increased significantly (P < 0.05), while that of Candidatus Solibacter, Geobacter, Ktedonobacter and Nitrobacter decreased significantly (P < 0.05). There was no significant difference in Bradyrhizobium between AT and ST.
Correlation between soil microorganism, soil enzyme activity and environmental parameters
Pearson correlation analysis identified significant correlations (P < 0.05) between physicochemical properties and N-related gene abundances across AT and ST (Figure 5). The pH value, NH4+-N, available N and net N mineralization rate were significantly positively correlated (P < 0.05) with activities of sucrase, protease, catalase, and polyhphenol oxidase, and the gene abundances involved in glutamate synthesis and assimilatory N reduction, whereas they were significantly negatively related (P < 0.05) to urease activity and the gene abundances involved in ammonification, dissimilatory N reduction, nitrification and denitrification. The correlation between NO3––N content and other indexes was contrary to that of NH4+-N and available N (P < 0.05).
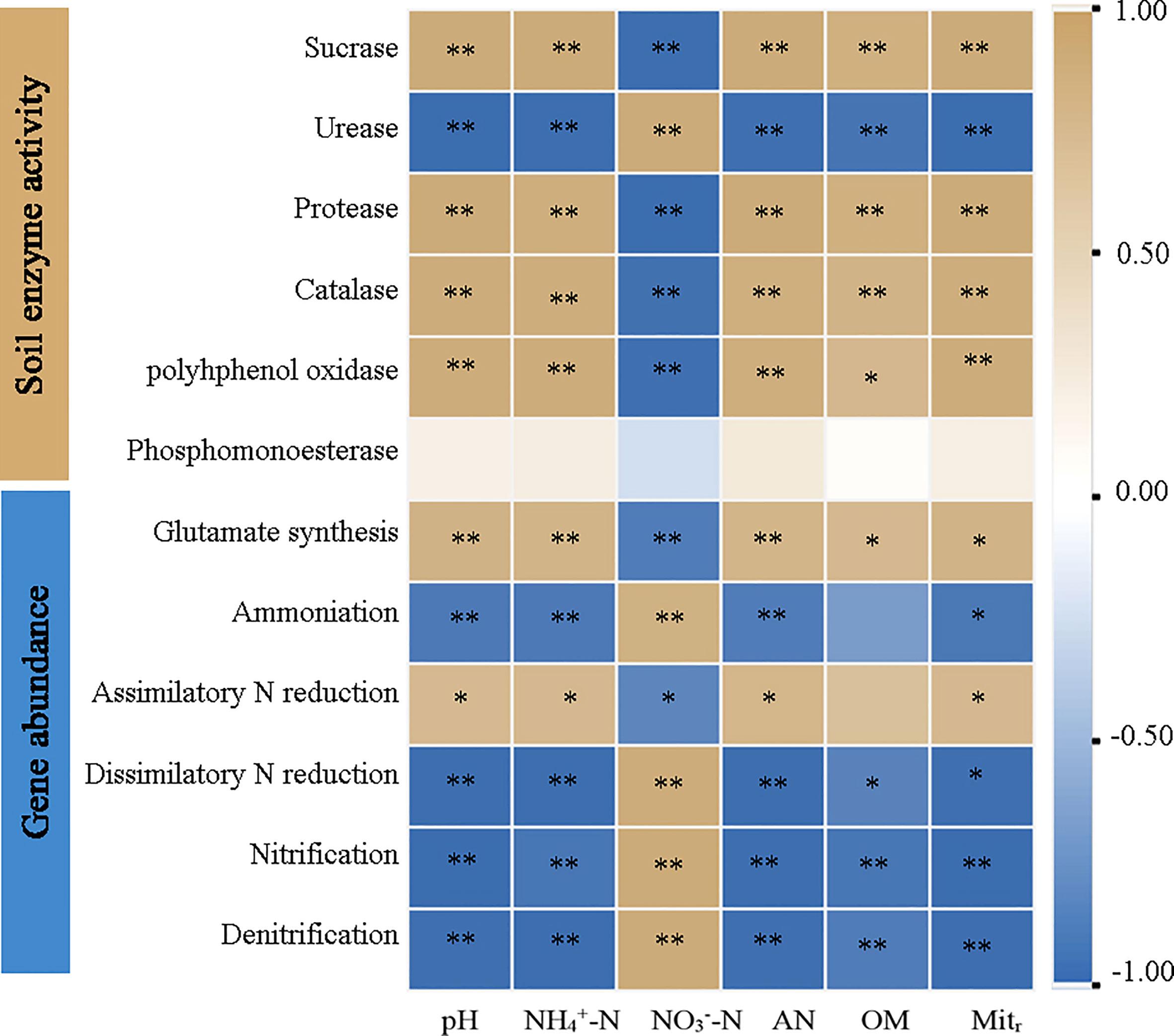
Figure 5 Heatmap of pearson correlation analysis of soil physicochemical properties, enzyme activity and functional gene abundance in tea soil with different pH values. NH4+-N, ammonium nitrogen content; NO3––N, nitrate nitrogen content; AN, available nitrogen content; OM, organic matter content; Mitr, net nitrogen mineralization rate. * and **, significant at P ≤ 0.05 and P ≤ 0.01.
RDA was performed using the top 10 species of microbial taxa at the phylum level and soil physicochemical properties (Figure 6). The results showed that axes 1 and 2 together described 99.94% of the variation in microorganisms at the phylum level and 99.91% of microorganisms at the genus level, indicating a significant correlation between soil physicochemical factors and microbes (P < 0.05) (Figure 6). As for the microorganisms at the phylum level (Figure 6A), soil pH, NH4+-N and available N content and net N mineralization rate were significantly positively correlated (P < 0.05) with the abundances of Acidobacteria, Armatimonadetes, Verrucomicrobia and Thaumarchaeota, and significantly negatively correlated (P < 0.05) with the abundance of Proteobacteria, Actinobacteria, Chloroflexi, Nitrospirae, Planctomycetes and Firmicutes. And the correlation between NO3––N content and other indexes were contrary to that of NH4+-N content (P < 0.05).
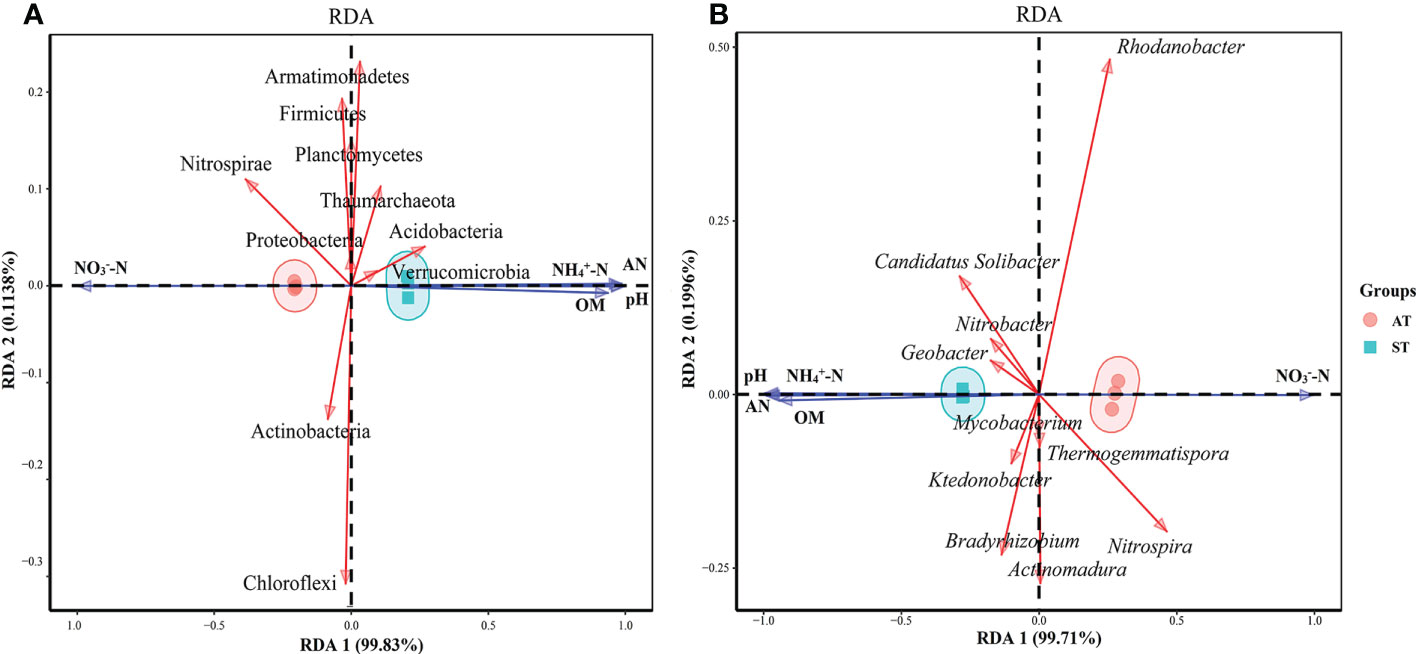
Figure 6 Canonical redundancy analysis (RDA) of soil physicochemical factor and dominant phyla (A) and genera (B) microorganism in three soil sample. AT, tea soil with a pH of 3.75; ST, tea soil with a pH of 5.26.
As for the microorganisms at the genus level (Figure 6B), soil pH, NH4+-N, available N content and net N mineralization rate were positively correlated (P < 0.05) with the abundances of Candidatus Solibacter, Geobacter and Nitrobacter and negatively correlated (P < 0.05) with the abundances of Bradyrhizobium, Nitrospira, Rhodanobacter, Actinomadura, Thermogemmatispora and Mycobacterium. And the correlation between NO3––N content and other indexes were contrary to that of NH4+-N and available N (P < 0.05).
Discussion
Soil nitrogen form and transformation
Nitrogen involved in a lot of biological processes in terms of different level and forms (NH4+ and/or NO3−) (Huang et al., 2018). Tea plants prefer NH4+ rather than NO3−, and NH4+ can promote the growth of tea plants more effectively (Huang et al., 2018; Wang et al., 2021). In our study, the contents of NH4+-N and available N in acidified soil decreased significantly, but the content of NO3−-N was higher than that in soil suitable for tea tree growth (Table 1). These results indicated that soil pH affect the N forms of tea soils, and soil acidification lead to the decrease of available N for tea plant. By further analyzing the soil N transformation capacity, we found that the rate of net N mineralization in acidified soil decreased significantly (Table 1). Consistent with our findings, previous researchers reported that the rate of N mineralization increases as the increase of pH values of 4-8 (Jiang et al., 2015). Mineralization results in increasing the content of available N in plant soil (Robertson and Groffman, 2007), which indicated that acidification would reduce soil mineralization capacity, thus lead to a decline in soil N supply capacity and a decrease of N absorption and utilization capacity of tea trees.
Soil enzyme activity
Soil enzyme activity is an indicator of soil quality, which reflect the direction and intensity of biochemical processes and nutrient cycles in soil (Xu et al., 2021). The activities of N-cycling enzymes such as urease, catalase and protease existed significantly differences under different N fertilizer treatments (He et al., 2021; Li et al., 2021). Our study found that the activities of sucrase, protease, catalase and polyphenol oxidase in acidified tea soil decreased significantly (Figure 2). Soil enzymes such as sucrase and catalase were closely related to soil organic matter accumulation and saprophytic degree, where their debris and subsequent nutrients release into the soil is made available to plants (Dick, 1994; Madejón et al., 2007). Protease can hydrolyze various protein and peptide compounds into amino acids, which is one of the N sources of higher plants (Nie et al., 2020). Polyphenol oxidase has a defense effect against soil pathogens (Mayer, 2006). It could be seen that acidification of tea soil has a negative effect on soil carbon and N conversion, N supply capacity, disease resistance and soil humus content.
Functional N transformation gene shifts
The changes of soil functional gene abundance determine the direction and amplitude of soil N transformation (Chen et al., 2017). Hence, we comprehensively analyzed the genes involved in the grid pathway of N-cycling (Figure 2). Taken together, the frequency and abundance of genes involved in glutamate synthesis were significantly higher than other N-cycle processes, while the abundance of genes involved in nitrification were the lowest (Figure 2C). This finding was similar to the result of Nelson’s metagenomic sequencing (Nelson et al., 2015), indicating that tea soils with different acidity in this study have the same characteristics of N metabolism pathways. However, the abundances of genes involved in N fixation and anammox were below the detection line and not captured (Figure 2B). This may be due to mineral forms of N that were readily available in the environment, requiring no or less metabolic investment according to the principle of resource acquisition (Zheng et al., 2017).
Compared with NO3−, NH4+ can more effectively promote the biosynthesis of free amino acids and catechins in tea leaves and roots, and significantly improve the activity of glutamine synthase, thus improve the tea quality (Huang et al., 2018; Wang et al., 2021). Our results found that the abundance of genes involved in glutamate synthesis in acidified soil decreased significantly, while it was positively correlated with NH4+-N (Figures 2C, 5). This suggested that the content of NH4+-N in acidified soil decreased, which inhibited the synthesis of glutamate. However, it was not conducive to the accumulation and absorption of soil N content, thus reducing the N fertilizer use efficiency (Wang et al., 2020). The presence of NH4+ will improve the absorption rate of NO3- (Ruan et al., 2016). Plants or microorganisms inhale NO3−-N into the body, reduce it to NH4+-N by assimilatory N reduction, and then synthesize N-related organic substances. And then the abundances of genes related to assimilatory N reduction pathway in ST were significantly higher than that in acidified soil (Figure 2C).
Our result was consistent with a previous study that reported that nitrification was negatively correlated with soil pH value (Jin et al., 2012). And high potential nitrification rates lead to low NH4+-N and to high NO3−-N (Jin et al., 2012; Li et al., 2018). Hence, the soil NO3−-N was significantly higher and NH4+-N was less after soil acidification (Table 1). Ammonification is a process in which microorganisms decompose organic matter and release NH4+ or NH3. Soil acidification promotes soil ammonification (Figure 2C), this may result in higher urease activity in acidified soil. But the content of NH4+-N in the acidified tea soil decreased, this suggested that the NH4+-N generated was partly absorbed by tea trees and partly converted to NO3––N due to high nitrification, which also increased the risk of solidification or leaching, thus resulting in less NH4+-N in acidified soil. The results also indicated that the abundances of genes involved in denitrification increased significantly in acidified soil, and were negatively correlated with soil NH4+-N and available N, while were positively correlated with NO3––N (Figures 2C, 5). This was consistent with the results of Sun et al. (Sun et al., 2021). The contribution of denitrification to N2O production was significantly increased with the decrease of soil pH (Saggar et al., 2013; Wu et al., 2017). The transformation of N in soil was a cyclic process. The nitrification and denitrification of tea soil were enhanced after soil acidification, thus resulting in less NH4+-N in soil, which further inhibited glutamate synthesis (Figure 5).
Previous studies on genes related to soil N-cycling mainly focused on the relative abundance of individual genes involved in soil nitrification, denitrification and N-fixation, such as nifH, amoA, nirS and nosZ (Sun et al., 2021). However, other genes involved in N-cycling pathways have received less attention. Hence, we further comprehensively evaluated the abundance of key genes involved in each N-cycling processes. Among the glutamate synthesis-related genes, the abundances of gltB and gltD genes in tea soil with different acidity were consistent with the change trend of glutamate synthesis pathway (Figures 2C, 3A), indicating that gltB and gltD play important roles in glutamate synthesis pathway. The changes in gene abundances in different individuals involved in the same pathway after acidification were not completely consistent (Figure 3), which suggested that the importance of studying the complete gene family involved in N-cycling process. Among the sequential nitrifying genes, we observed that the abundances of amoA/B/C in acidified soil were significantly increased. Soil nitrification begins with the oxidation of ammonia to nitrite, which is dominated by ammonia-oxidizing bacteria (AOB) and ammonia-oxidizing archaea (AOA) (Harter et al., 2014). The key control enzyme of this step is encoded by the amoA gene (Afzal et al., 2019). Yang et al. (2018a) reported that N fertilization stimulated nitrifying bacteria (amoA). Soil denitrification involves multiple N forms and functional genes (Lin et al., 2017). The denitrification process is mainly encoded by functional genes such as narG (NO3- to NO2-), nirS/nirK (NO2- to NO), norB (NO to N2O) and nosZ (N2O to N2) (Grassmann et al., 2022). The decrease of soil pH will reduce the availability of soil mineral N and organic carbon, and then indirectly affect N2O emission (Baggs et al., 2010). Several studies have shown that narG and norB genes significantly promoted N2O emission in acidic tropical forest soil (Tian et al., 2019). After acidification, the abundance of all genes involved in denitrification increased significantly (Figure 3F), which increases the risk of N2O emission.
Soil microbial community composition and correlation analysis
Soil properties could indirectly mediate the N transformation process by affecting soil microbial community structure (Dong et al., 2018). In this research, significant differences were detected in soil N-related microbial community with AT and ST (Figure S2), which implied that soil acidification affected the soil N-related microbial community of tea plantation. Previous studies reported that Proteobacteria, Actinobacteria and Acidobacteria are the predominant microbial community phylum in terrestrial soil ecosystems (Liu et al., 2016; Delgado-Baquerizo et al., 2018). These results were consistent with ours, though the object of our study is the N-related microorganisms (Figure 4). Proteobacteria contains a large number of plant and animal pathogenic bacteria (Wagg et al., 2018). Previous study indicated that the abundance of Proteobacteria in diseased soil was relatively higher (Xue et al., 2015). Zhao et al. (2019) showed that a few parasitic actinomycetes can cause diseases of some plants and animals. Proteobacteria and Actinobacteria were increased after soil acidification (Figure 4A). This suggested that the content of pathogenic bacteria in tea soil increases after acidification. We also found that Chloroflexi, Nitrospirae and Firmicutes were significantly enriched in acidified soil (Figure 4). Several studies have shown that Chloroflexi and Nitrospirae and Firmicutes play nitrification or denitrification function in soil N-cycle (Kartal et al., 2007; Spieck et al., 2020). However, the abundances of Acidobacteria, Armatimonadetes, Verrucomicrobia and Thaumarchaeota in acidified soil decreased significantly (Figure 4A). We suggested that acidification could provide a bad survival environment for some soil beneficial microorganisms. Furthermore, we found that Nitrospira, Rhodoblastus, Actinomadura, Thermogemmatispora and Mycobacterium were significantly enriched in acidified soil (Figure 4B). Our result was consistent with a previous study that the abundance of Nitrospira displayed a significant positive correlation with nitrification potential, and was negatively correlated with soil pH and high ammonia (Hu et al., 2021).
Many documents reported that soil microbial community composition is influenced by physicochemical factors (Zhang et al., 2022). RDA results indicated that some key rhizosphere microorganisms (Proteobacteria, Actinobacteria, Chloroflexi, Nitrospirae, Planctomycetes, Firmicutes, Actinomadura, Nitrobacter) had a positive correlation with soil NO3––N, and had a negative correlation with pH, NH4+-N and available N (Figure 6). These results indicated that pH and soil N level were important factors driving soil microbial diversity and community structure change, which were in agreement with previous reports (Zhou et al., 2018; Li et al., 2020). Results also indicated that the beneficial microorganism in acidified soil decreased and the abundances of microorganisms involved in nitrification and denitrification increased, this increases the risk of soil N2O emission (Huang et al., 2015b).
Conclusion
In conclusion, soil pH value affected the N cycling (Figure 7). After soil acidification, the soil enzyme activities involved in nutrient cycling decreased; the abundances of genes involved in glutamate synthesis and assimilatory N reduction increased significantly. Moreover, the abundances of genes involved in dissimilation N reduction, nitrification and denitrification increased significantly in acidified soil, and then affected the participation in the N-cycle of the microbial community composition, especially increased the amounts of pathogenic microorganism, and microorganisms related to nitrification and denitrification. These factors eventually lead to a decrease in the N supply capacity and the content of NH4+-N in tea soil, thus inhibited the growth of tea trees. In the future, a more comprehensive study is required to explore the response of microorganisms involved in each N transformation process respond to soil acidification in tea plantation.
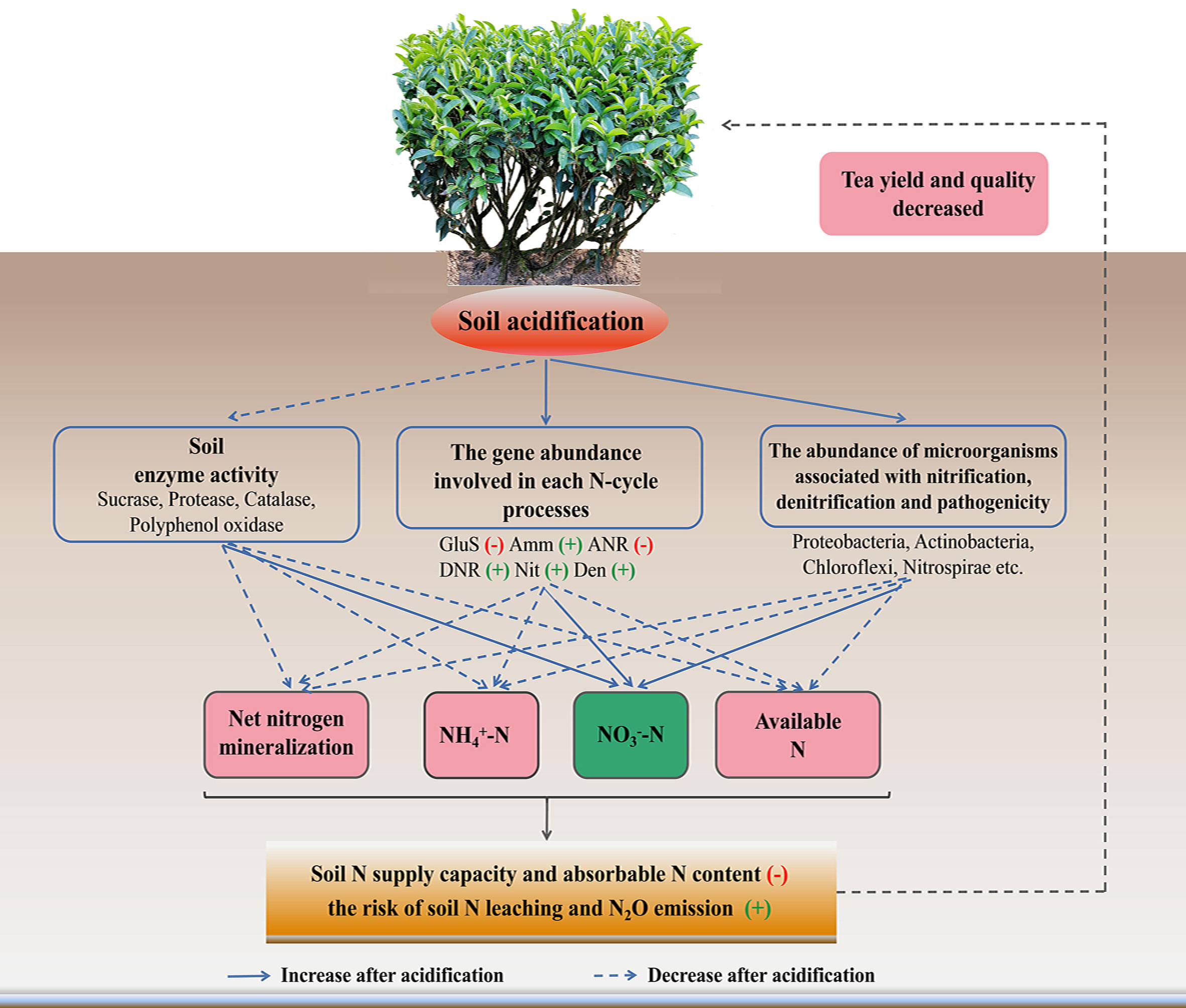
Figure 7 Model showing the mechanism of soil acidification on soil nitrogen form and transformation in tea gardens. The blue dotted and solid lines represent a decrease and increase in an indicator, respectively. Red minus sign indicates upregulated, while green plus sign indicates downregulated. GluS, glutamate synthesis; Amm, ammonification; ANR, assimilatory nitrogen reduction; DNR, dissimilatory nitrogen reduction; Nit, nitrification; Den, denitrification.
Data availability statement
The original contributions presented in the study are publicly available. This data can be found here: NCBI, PRJNA867479.
Author contributions
SL designed and performed the experiments, analyzed the data and writing-original draft. ZL and YW investigation and sampling collection, performed the experiments. JL, GW and JY analyzed the data. HW conceived the experiments, approved the final manuscript. HH supervision, reviewed drafts and approved the final draft. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by National Natural Science Foundation of China (81973412), Natural Science Foundation of Fujian Province (2020J01369, 2020J01408), Project of Scientific Research of Young and Middle-aged Teachers, Fujian Province (JAT170573, JAT190761), Fujian Outstanding Research Talent Cultivation Project and Top Talent Training Program of Longyan University (2019JZ19).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.998178/full#supplementary-material
References
Afzal, M., Yu, M. J., Tang, C. X., Zhang, L. J., Muhammad, N., Zhao, H. C., et al. (2019). The negative impact of cadmium on nitrogen transformation processes in a paddy soil is greater under non-flooding than flooding conditions. Environ. Int. 129, 451–460. doi: 10.1016/j.envint.2019.05.058
Baggs, E. M., Smales, C. L., Bateman, E. J. (2010). Changing pH shifts the microbial source as well as the magnitude of N2O emission from soil. Biol. Fert Soils. 46, 793–809. doi: 10.1007/s00374-010-0484-6
Barak, P., Jobe, B. O., Krueger, A. R., Peterson, L. A., Laird, D. A. (1997). Effects of long-term soil acidification due to nitrogen fertilizer inputs in Wisconsin. Plant Soil. 197, 61–69. doi: 10.1023/A:1004297607070
Chen, J., Xiao, G. L., Kuzyakov, Y., Jenerette, G. D., Ma, Y., Liu, W., et al. (2017). Soil nitrogen transformation responses to seasonal precipitation changes are regulated by changes in functional microbial abundance in a subtropical forest. Biogeosciences 14, 2513–2525. doi: 10.5194/bg-2017-3
Delgado-Baquerizo, M., Oliverio, A. M., Brewer, T. E., Benavent-González, A., Eldridge, D. J., Bardgett, R. D., et al. (2018). A global atlas of the dominant bacteria found in soil. Science 359, 320–325. doi: 10.1126/science.aap9516
Dick, R. P. (1994). Soil enzyme-activities as indicators of soil quality. 35. SSSA Spec Publ. 35, 107–124. doi: 10.2136/sssaspecpub35.c7
Dong, Z. X., Zhu, B., Jiang, Y., Tang, J. L., Liu, W. L., Hu, L. (2018). Seasonal N2O emissions respond differently to environmental and microbial factors after fertilization in wheat–maize agroecosystem. Nutr. Cycl Agroecosys. 112, 215–229. doi: 10.1007/s10705-018-9940-8
Goswami, G., Deka, P., Das, P., Bora, S. S., Samanta, R., Biro, R. C., et al. (2017). Diversity and functional properties of acid-tolerant bacteria isolated from tea plantation soil of Assam. 3 Biotech. 7, 229–244. doi: 10.1007/s13205-017-0864-9
Grassmann, C. S., Mariano, E., Diniz, P. P., Borges, B. M. F., Borges, C. D., Tsai, S. M., et al. (2022). Functional n-cycle genes in soil and N2O emissions in tropical grass-maize intercropping systems. Soil Biol. Biochem. 169, 108655. doi: 10.1016/j.soilbio.2022.108655
Guan, S., Zhang, D., Zhang, Z. (1986). Soil enzyme and its research methods (Beijing: Agricultural Publisher).
Harter, J., Krause, H. M., Schuettler, S., Ruser, R., Fromme, M., Scholten, T., et al. (2014). Linking N2O emissions from biochar-amended soil to the structure and function of the n-cycling microbial community. ISME J. 8, 660–674. doi: 10.1038/ismej.2013.160
He, W. Y., Zhang, M. M., Jin, G. Z., Sui, X., Zhang, T., Song, F. Q. (2021). Effects of nitrogen deposition on nitrogen-mineralizing enzyme activity and soil microbial community structure in a Korean pine plantation. Microb. Ecol. 81, 410–424. doi: 10.1007/s00248-020-01595-6
Huang, Y., Long, X. E., Chapman, S. J., Yao, H. (2015b). Acidophilic denitrifiers dominate the N2O production in a 100-year-old tea orchard soil. Environ. Sci. pollut. R. 22, 4173–4182. doi: 10.1007/s11356-014-3653-6
Huang, H., Yao, Q. Y., Xia, E. H., Gao, L. Z. (2018). Metabolomics and transcriptomics analyses reveal nitrogen influences on the accumulation of flavonoids and amino acids in young shoots of tea plant (Camellia sinensis l.) associated with tea flavor. J. Agr Food Chem. 66, 9828–9838. doi: 10.1021/acs.jafc.8b01995
Huang, P., Zhang, J. B., Xin, X. L., Zhu, A. N., Zhang, C. Z., Ma, D. H., et al. (2015a). Proton accumulation accelerated by heavy chemical nitrogen fertilization and its long-term impact on acidifying rate in a typical arable soil in the Huang-Huai-Hai plain. J. Integr. Agr. 14, 48–157. doi: 10.1016/S2095-3119(14)60750-4
Hu, J. J., Zhao, Y. X., Yao, X. W., Wang, J. Q., Zheng, P., Xi, C. W., et al. (2021). Dominance of comammox Nitrospira in soil nitrification. Sci. Total Environ. 780, 146558. doi: 10.1016/j.scitotenv.2021.146558
Jiang, X. J., Hou, X. Y., Zhou, X., Xin, X. P., Wright, A., Ji, Z. J. (2015). pH regulates key players of nitrification in paddy soils. Soil Biol. Biochem. 81, 9–16. doi: 10.1016/j.soilbio.2014.10.025
Jin, R. C., Yang, G. F., Yu, J. J., Zheng, P. (2012). The inhibition of the anammox process: a review. Chem. Eng. J. 197, 67–79. doi: 10.1016/j.cej.2012.05.014
Kartal, B., Kuypers, M. M. M., Lavik, G. (2007). Anammox bacteria disguised as denitrifiers: nitrate reduction to dinitrogen gas via nitrite and ammonium. Environ. Microbiol. 9, 635–642. doi: 10.1111/j.1462-2920.2006.01183.x
Kuypers, M. M. M., Marchant, H. K., Kartal, B. (2018). The microbial nitrogen-cycling network. Nat. Rev. Microbiol. 16, 263–276. doi: 10.1038/nrmicro.2018.9
Leininger, S., Urich, T., Schloter, M., Schwark, L., Qi, J., Nicol, G. W., et al. (2006). Archaea predominate among ammonia-oxidising prokaryotes in soils. Nature 442, 806–809. doi: 10.1038/nature04983
Li, Y. Y., Chapman, S. J., Nicol, G. W., Yao, H. Y. (2018). Nitrification and nitrifiers in acidic soils. Soil Biol. Biochem. 116, 290–301. doi: 10.1016/j.soilbio.2017.10.023
Lin, Y. X., Ding, W. X., Liu, D. Y., He, T. H., Yoo, G., Yuan, J. J., et al. (2017). Wheat straw-derived biochar amendment stimulated N2O emissions from rice paddy soils by regulating the amoA genes of ammonia-oxidizing bacteria. Soil Biol. Biochem. 113, 89–98. doi: 10.1016/j.soilbio.2017.06.001
Lin, S. X., Liu, Z. J., Wang, Y. C., Li, J. Y., Wang, G. G., Zhang, W., et al. (2022). Soil acidification associated with changes in inorganic forms of n reduces the yield of tea (Camellia sinensis). Arch. Agron. Soil Sci. doi: 10.1080/03650340.2022.2104452
Liu, J. J., Sui, Y. Y., Yu, Z. H., Yao, Q., Shi, Y., Chu, H. Y., et al. (2016). Diversity and distribution patterns of acidobacterial communities in the black soil zone of northeast China. Soil Biol. Biochem. 95, 212–222. doi: 10.1016/j.soilbio.2015.12.021
Li, Y., Wang, C., Gao, S., Wang, P., Qiu, J., Shang, S. (2021). Impacts of simulated nitrogen deposition on soil enzyme activity in a northern temperate forest ecosystem depend on the form and level of added nitrogen. Eur. J. Soil Biol. 103, 103287. doi: 10.1016/j.ejsobi.2021.103287
Li, J. J., Wang, G. L., Yan, B. S., Liu, G. B. (2020). The responses of soil nitrogen transformation to nitrogen addition are mainly related to the changes in functional gene relative abundance in artificial Pinus tabulaeformis forests. Sci. Total Environ. 723, 137679. doi: 10.1016/j.scitotenv.2020.137679
Lu, R. K. (1999). Methods for agricultural chemical analysis of soil (Beijing: China Agricultural Science and Technology Press), 139–162.
Madejón, E., Moreno, F., Murillo, J. M., Pelegrín, F. (2007). Soil biochemical response to long-term conservation tillage under semi-arid Mediterranean conditions. Soil Tillage Res. 94, 346–352. doi: 10.1016/j.still.2006.08.010
Mayer, A. M. (2006). Polyphenol oxidases in plants and fungi: going places? a review. Phytochemistry 67, 2318–2331. doi: 10.1016/j.phytochem.2006.08.006
Nelson, M. B., Berlemont, R., Martiny, A. C., Martiny, J. B. H. (2015). Nitrogen cycling potential of a grassland litter microbial community. Appl. Environ. Microb. 81, 7012–7022. doi: 10.1128/AEM.02222-15
Nie, Z. J., Qin, S. Y., Liu, H. E., Zhao, P., Wu, X. T., Gao, W., et al. (2020). Effects of combined application of nitrogen and zinc on winter wheat yield and soil enzyme activities related to nitrogen transformation. J. Plant Nutr. Fertil. 26, 431–441. doi: 10.11674/zwyf.19207
Robertson, G. P., Groffman, P. M. (2007). “Nitrogen transformation,” in Soil microbiology, ecology and biochemistry. third edition. Ed. Paul, E. (Burlington: Elsevier Academic Press), 341–364.
Robertson, G. P., Wedin, D., Groffman, P. M., Blair, J. M., Sollinset, P. (1999). Soil carbon and nitrogen availability: Nitrogen mineralization, nitrification, and soil respiration potentials. Standard Soil Methods long-term Ecol. Res., 258–271.
Ruan, J. Y., Gerendás, J., Hardter, R., Sattelmacher, B. (2008). Effect of root zone pH and form and concentration of nitrogen on accumulation of quality-related components in green tea. J. Sci. Food Agr. 87, 1507–1516. doi: 10.1002/jsfa.2875
Ruan, L., Wei, K., Wang, L. Y., Cheng, H., Zhang, F., Wu, L. Y., et al. (2016). Characteristics of NH4+ and NO3- fluxes in tea (Camellia sinensis) roots measured by scanning ion-selective electrode technique. Sci. Rep. 6, 38370. doi: 10.1038/srep38370
Saggar, S., Jha, N., Deslippe, J., Bolan, N. S., Tillmanet, R. W. (2013). Denitrification and N2O: N2 production in temperate grasslands: Processes, measurements, modelling and mitigating negative impacts. Sci. Total Environ. 465, 173–195. doi: 10.1016/j.scitotenv.2012.11.050
Spieck, E., Sass, K., Keuter, S., Hirschmann, S., Spohn, M., Indenbirken, D., et al. (20201522). Defining culture conditions for the hidden nitrite-oxidizing bacterium. Nitrolancea. Front. Microbiol. 11, 1522. doi: 10.3389/fmicb.2020.01522
Sun, B., Gu, L., Bao, L., Zhang, S., Wei, Y., Bai, Z., et al. (2020). Application of biofertilizer containing Bacillus subtilis reduced the nitrogen loss in agricultural soil. Soil Biol. Biochem. 148, 107911. doi: 10.1016/j.soilbio.2020.107911
Sun, R. B., Wang, F. H., Hu, C. S., Liu, B. B. (2021). Metagenomics reveals taxon-specific responses of the nitrogen-cycling microbial community to long-term nitrogen fertilization. Soil Biol. Biochem. 156, 0038–0717. doi: 10.1016/j.soilbio.2021.108214
Tian, J., Dungait, J. A. J., Lu, X. K., Yang, Y. F., Hartley, I. P., Zhang, W., et al. (2019). Long-term nitrogen addition modifies microbial composition and functions for slow carbon cycling and increased sequestration in tropical forest soil. Global Change Biol. 25, 3267–3281. doi: 10.1016/j.scitotenv.2020.137679
Wagg, C., Dudenhöffer, J. H., Widmer, F., van der Heijden, M. A. (2018). Linking diversity, synchrony and stability in soil microbial communities. Funct. Ecol. 32, 1280–1292. doi: 10.1111/1365-2435.13056
Wang, Y. S., Li, C. N., Kou, Y. P., Wang, J. J., Tu, B., Li, H., et al. (2017). Soil pH is a major driver of soil diazotrophic community assembly in qinghai-Tibet alpine meadows. Soil Biol. Biochem. 115, 547–555. doi: 10.1016/j.soilbio.2017.09.024
Wang, J., Long, Z. H., Min, W., Hou, Z. N. (2020). Metagenomic analysis reveals the effects of cotton straw-derived biochar on soil nitrogen transformation in drip-irrigated cotton field. Environ. Sci. pollut. R. 27, 43929–43941. doi: 10.1007/s11356-020-10267-4
Wang, Y., Wang, Y. M., Lu, Y. T., Fan, D. M., Wang, X. C., Zheng, X. Q., et al. (2021). Influence of different nitrogen sources on carbon and nitrogen metabolism and gene expression in tea plants (Camellia sinensis l.). Plant Physiol. Bioch. 167, 561–566. doi: 10.1016/j.plaphy.2021.08.034
Wu, J., He, S. Z., Liang, Y., Li, G. X., Li, S., Chen, S. L., et al. (2017). Effect of phosphate additive on the nitrogen transformation during pig manure composting. Environ. Sci. pollut. R. 24, 17760–17768. doi: 10.1007/s11356-017-9285-x
Xue, C., Ryan, R. C., Shen, Z. Z., Zhang, R. F., Huang, Q. W., Li, R., et al. (2015). Manipulating the banana rhizosphere microbiome for biological control of Panama disease. Sci. Rep. 5, 11124. doi: 10.1038/srep14596
Xu, H. W., Qu, Q., Chen, Y. H., Liu, G. B., Xue, S. (2021). Responses of soil enzyme activity and soil organic carbon stability over time after cropland abandonment in different vegetation zones of the loess plateau of China. Catena 196, 104812. doi: 10.1016/j.catena.2020.104812
Xu, N., Tan, G. C., Wang, H. Y., Gai, X. P. (2016). Effect of biochar additions to soil on nitrogen leaching, microbial biomass and bacterial community structure. Eur. J. Soil Biol. 74, 1–8. doi: 10.1016/j.ejsobi.2016.02.004
Yamamoto, A., Akiyama, H., Naokawa, T., Miyazaki, Y., Honda, Y., Sano, Y., et al. (2014). Lime-nitrogen application affects nitrification, denitrification, and N2O emission in an acidic tea soil. Biol. Fert Soils. 50, 53–62. doi: 10.1007/s00374-013-0830-6
Yang, O. Y., Evans, S. E., Friesen, M. L., Tiemann, L. K. (2018a). Effect of nitrogen fertilization on the abundance of nitrogen cycling genes in agricultural soils: a meta-analysis of field studies. Soil Biol. Biochem. 127, 71–78. doi: 10.1016/j.soilbio.2018.08.024
Yang, X. D., Ni, K., Shi, Y. Z., Yi, X. Y., Zhang, Q. F., Fang, L., et al. (2018b). Effects of long-term nitrogen application on soil acidification and solution chemistry of a tea plantation in China. Agr Ecosyst. Environ. 252, 74–82. doi: 10.1016/j.agee.2017.10.004
Yan, P., Wu, L. Q., Wang, D. H., Fu, J. Y. (2020). Soil acidification in Chinese tea plantations. Sci. Total Environ. 715, 136963. doi: 10.1016/j.scitotenv.2020.136963
Zhang, X., Li, S. Z., Cheng, W., Zhao, Y., Cui, H. Y., Xie, X. Y., et al. (2021). Oxytetracycline stress reconstruct the core microbial community related to nitrogen transformation during composting. Bioresource Technol. 319, 124142. doi: 10.1016/j.biortech.2020.124142
Zhang, H. H., Phillip, F. O., Wu, L. N., Zhao, F. Y., Yu, S. L., Yu, K. (2022). Effects of temperature and nitrogen application on carbon and nitrogen accumulation and bacterial community composition in apple rhizosphere soil. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.859395
Zhang, C., Song, Z. L., Zhuang, D. H., Wang, J., Xie, S. S., Liu, G. B. (2019). Urea fertilization decreases soil bacterial diversity, but improves microbial biomass, respiration, and n-cycling potential in a semiarid grassland. Biol. Fert Soils. 55, 229–242. doi: 10.1007/s00374-019-01344-z
Zhao, F., Zhao, M. Z., Wang, Y., Guan, L., Pang, F. H. (2019). Microbial community structures and diversities in strawberry rhizosphere soils based on high-throughput sequencing. Soils 51, 51–60. doi: 10.13758/j.cnki.tr.2019.01.008
Zheng, M. H., Zhang, W., Luo, Y. Q., Mori, T., Mao, Q. G., Wang, S. H., et al. (2017). Different responses of asymbiotic nitrogen fixation to nitrogen addition between disturbed and rehabilitated subtropical forests. Sci. Total Environ. 601, 1505–1512. doi: 10.1016/j.scitotenv.2017.06.036
Zhou, F. W., Cui, J., Zhou, J., Yang, J., Li, Y., Leng, Q. M., et al. (2018). Increasing atmospheric deposition nitrogen and ammonium reduced microbial activity and changed the bacterial community composition of red paddy soil. Sci. Total Environ. 633, 776–784. doi: 10.1016/j.scitotenv.2018.03.217
Zhou, J., Xia, F., Liu, X. M., He, Y., Xu, J. M., Brookes, P. C. (2014). Effects of nitrogen fertilizer on the acidification of two typical acid soils in south China. J. Soil Sediment. 14, 415–422. doi: 10.1007/s11368-013-0695-1
Keywords: tea (Camellia sinensis), soil acidification, nitrogen forms, nitrogen functional genes, microbial composition
Citation: Lin S, Liu Z, Wang Y, Li J, Wang G, Ye J, Wang H and He H (2022) Soil metagenomic analysis on changes of functional genes and microorganisms involved in nitrogen-cycle processes of acidified tea soils. Front. Plant Sci. 13:998178. doi: 10.3389/fpls.2022.998178
Received: 19 July 2022; Accepted: 26 September 2022;
Published: 14 October 2022.
Edited by:
Shusheng Zhu, Yunnan Agricultural University, ChinaReviewed by:
Qiuxia Wang, The Scripps Research Institute, United StatesHongmiao Wu, Anhui Agricultural University, China
Copyright © 2022 Lin, Liu, Wang, Li, Wang, Ye, Wang and He. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Haibin He, YWxleGhoYkAxNjMuY29t; Haibin Wang, dzEzNTk5MDg0ODQ1QHNpbmEuY29t
 Shunxian Lin
Shunxian Lin Zhijun Liu1
Zhijun Liu1 Jiayu Li
Jiayu Li Haibin Wang
Haibin Wang Haibin He
Haibin He