
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 20 October 2022
Sec. Functional and Applied Plant Genomics
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.998082
This article is part of the Research Topic Research on Brassicaceae Crops Genomics and Breeding View all 10 articles
Crop genomics and breeding CLAVATA3/EMBRYO SURROUNDING REGION-RELATED (CLE) proteins belong to a small peptide family in plants. During plant development, CLE gene family members play a pivotal role in regulating cell-to-cell communication and stem cell maintenance. However, the evolutionary process and functional importance of CLEs are unclear in Brassicaceae. In this study, a total of 70 BnCLEs were identified in Brassica napus (2n = 4x = 38, AnCn): 32 from the An subgenome, 36 from the Cn subgenome, and 2 from the unanchored subgenome. Meanwhile, 29 BrCLE and 32 BoCLE genes were explored in Brassica rapa (2n = 2x = 20, Ar) and Brassica oleracea (2n = 2x = 18, Co). Phylogenetic analysis revealed that 163 CLEs derived from three Brassica species and Arabidopsis thaliana can be divided into seven subfamilies. Homology and synteny analyses indicated whole-genome triplication (WGT) and segmental duplication may be the major contributors to the expansion of CLE family. In addition, RNA-seq and qPCR analysis indicated that 19 and 16 BnCLEs were more highly expressed in immature seeds and roots than in other tissues. Some CLE gene pairs exhibited different expression patterns in the same tissue, which indicated possible functional divergence. Furthermore, genetic variations and regional association mapping analysis indicated that 12 BnCLEs were potential genes for regulating important agronomic traits. This study provided valuable information to understand the molecular evolution and biological function of CLEs in B. napus and its diploid progenitors, which will be helpful for genetic improvement of high-yield breeding in B. napus.
Peptide hormones are active molecules composed of many to several tens of amino acids and serve as signal molecules to exchange information between cells (Grienenberger and Fletcher, 2015). In plants, there are various polypeptides; CLAVATA3/EMBRYO SURROUNDING REGION-RELATED (CLE), one of the largest families of known polypeptides, is generally 12–13 amino acids in length, including a secretion signal peptide in N-terminus and a highly conserved CLE domain in C-terminus. Domain deletion and domain exchange experiments have indicated that the CLE domain of CLV3 in Arabidopsis thaliana plays an independent role in adjacent flanking sequences (Fletcher et al., 1999; Rojo et al., 2002). With the help of matrix-assisted laser desorption ionization–time-of-flight (MALDI-TOF) mass spectrometry, an active 12-amino acid CLV3 polypeptide molecule (corresponding to the CLE domain, with one amino acid removed from each side), is isolated from A. thaliana over-expressing CLV3, and the peptide synthesized in vitro is functional (Tatsuhiko, 2006). Exogenous application of synthetic CLV3, CLE19, and CLE40 polypeptides exhibits a similar phenotype with overexpression of these CLE genes. CLV3 polypeptides also restore the clv3-2 mutant phenotype (Fiers et al., 2006). These results indicate that CLE polypeptides are the active form of CLE family proteins.
It is difficult to clearly identify the function of each CLE peptide owing to its small size and high sequence conservation. However, the application of new technologies such as gene editing makes it easier to study their functions. However, it is easier to study their functions based on the application of new technology, like gene editing. Now, many results show that CLE polypeptides play important roles in plant development and hormone and stress response. In shoot apical meristems (SAMs), the stem cell homeostasis is maintained by a dynamic negative feedback loop involved in the CLV3-WUSCHEL (WUS) pathway. The transcription factor gene WUS, which interacts with SHOOT MERISTEMLESS (STM), can promote the expression of CLV3; meanwhile, overmuch CLV3 will suppress the expression of STM and WUS to maintain the stem cell population in the SAM (Schoof et al., 2000). In addition to participating in the division and differentiation of meristem cells, CLE polypeptides also play a key role in the development of seeds (Fiume, 2010). With the use of the method of promoter fusion GUS, CLE8 and WUSCHEL-related homeobox 8 (WOX8) are expressed in the embryo and endosperm during the early stages of seed development in Arabidopsis (Fiume and Fletcher, 2012). The number of embryo and endosperm cells decreases in the cle8-1 mutant, suggesting that CLE8 promotes the proliferation of embryo and endosperm cells. The length and width of seeds produced by CLE8-overexpressing plants and wox8-1 mutant seeds significantly increase and decrease, respectively, compared with those of wild-type seeds. WOX8 expression significantly increases in CLE8-overexpression lines, indicating that CLE8 promotes its expression (Fiume and Fletcher, 2012; Song et al., 2013). Some CLE peptides likely interact with hormones. For example, CLE26 expressed at the phloem pole, regulated root architecture, and its expression is significantly enhanced by auxin treatment. CLE26 can affect the activity of the polar auxin transporter in the auxin signaling (Czyzewicz et al., 2015). The shoot growth is related to the root-expressed CLE6 under the gibberellin effect (Bidadi et al., 2014). In addition to development and hormone response, CLE also mediates responses to various abiotic stress. The expression of CLE25 is increased when the root is hydropenic, and then the root-derived CLE25 peptide moves into the leaves to modulate the closure of the stoma (Takahashi et al., 2018). In response to low-sulfate conditions, the expression of CLE2 and CLE3 in roots is reduced, which could inhibit lateral root development (Dong et al., 2019).
CLE genes are widely present in plants and even in several plant-parasitic nematodes. Three CLEs expressed in the maize endosperm (Esr1, Esr2, and Esr3) are supposed to be involved in signal transduction between embryo and endosperm during early development (Bonello et al., 2000). ZmCLE7 and ZmFCP1 are CLV3 homologs, and promoter editing performed by CRISPR-Cas9 increases many grain-yield-related traits in maize (Liu et al., 2021). Two CLEs from Lotus japonicus (LjCLE-RS1 and LjCLE-RS2) are repressors of excess root nodulation (Okamoto et al., 2013). An AtCLE19-like gene with high expression in the flower bud, pistil, and embryo of Brassica napus was ectopically expressed in A. thaliana, resulting in large heads (Fiers et al., 2004). The mutations of BnCLV3s induced by CRISPR/Cas9 could result in multilocular siliques and an increase in seed production (Yang et al., 2018). Similarly, BrCLV3 mutations conferred multicellular pods in Brassica rapa (Fan et al., 2014). In Raphanus sativus, overexpression of RsCLE2 and RsCLE19 increased the number of xylem elements (Gancheva et al., 2016). Moreover, CLEs, which may assist the infection process, are identified in cyst nematodes (Wang J. et al., 2011). Current research suggests that the CLEs are highly conserved and involved in biological evolution, especially in plants.
Recent genome-wide analyses have identified CLE genes in many plants (Zhang et al., 2014; Hastwell et al., 2015; Gancheva et al., 2016; Han et al., 2016; Han et al., 2020). The allotetraploid species B. napus (2n = 4x = 38, AnCn), an important oil crop, was formed from the hybridization between B. rapa and Brassica oleracea (2n = 2x = 18, Co) at about 7,500 years ago; therefore, the phylogenetic relationship of Brassica provides a good basis for studying the evolution of gene family (Allender and King, 2010). Based on bioinformatics and comparative genomic approaches, this study performed a multidimensional investigation for CLEs in rapeseed and its diploid progenitors, including genome-wide identification, molecular characterization, phylogenetic analysis, synteny analysis, expression profiling in different tissues, and regional association mapping analysis. The results will provide useful information for biological functions and molecular evolution in Brassica, which also supply candidate genes for genetic improvement in rapeseed breeding.
Multiple TBLASTN and BLASTN searches were performed for CLEs identification in B. napus reference genome (http://brassicadb.org/brad/downloadOverview.php) (Wang et al., 2015) based on A. thaliana CLE [expected threshold (e−10)]. The results were then validated by the Conserved Domain Database (CDD) in the National Center for Biotechnology Information (NCBI) (https://www.ncbi.nlm.nih.gov/cdd) (Lu et al., 2020) and Modular Architecture Research Tool SMART (http://smart.embl-heidelberg.de/) (Letunic et al., 2020) to confirm the authenticity of the CLE domain in the open reading frame. Open reading frames of homologous chromosome regions were confirmed for potential unannotated or truncated duplicates of CLEs. BrCLEs in B. rapa and BoCLEs in B. oleracea (http://brassicadb.org/brad/downloadOverview.php) were obtained as described above to explore the evolution of CLEs in Brassica.
Gene structures and conserved motifs for the CLEs were constructed via TBtools. Logo diagrams used to define consensus sequences were obtained using multiple sequence alignments for each BnCLE peptide group (I–V), including A. thaliana, B. rapa, and B. oleracea by TEXshade (Beitz, 2000). The signal peptides of CLEs were identified via SMART (Letunic et al., 2020). PlantCARE was carried out to predict the promoters in the 2-kb region before the start codon for the cis-element identification (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) (Magali, 2002). Gene Structure Display Server (GSDS 2.0) (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) was used to exhibit the gene structure (Hu et al., 2015), and the heatmap was visualized by R package (https://cran.r-project.org/).
In order to investigate the synteny relationship of CLEs in Brassicaceae, all the CLEs were searched as “syntenic genes” in Brassicaceae Database (BRAD) (http://brassicadb.cn/#/). At the same time, TBtools (Chen et al., 2020) was used to detect gene duplication patterns and verify the synteny relationship, as well as calculate the ratio of non-synonymous to synonymous substitutions (Ka/Ks) of syntenic gene pairs. Orthologous CLEs located on syntenic chromosome blocks were displayed by Circos software (Krzywinski et al., 2009).
Multiple alignments of the CLE peptide sequences from the four Brassicaceae species were performed by the ClustalW (Larkin, 2007). Phylogenetic analysis was generated using the MEGA7 soft with the maximum likelihood (ML) method, 1,000 bootstrap replications, and the JTT+G model (Kumar et al., 2016). The tree was visualized using Evolview (https://www.evolgenius.info/evolview/) (He et al., 2016).
Protein–protein interactions in A. thaliana were obtained in the STRING database (https://www.string-db.org/). The analysis and demonstration of CLEs interactions in B. napus followed the description based on the previous study (Xie et al., 2022).
RNA-seq raw data of siliques, leaves, flowers, and stems for B. napus, B. rapa, and B. oleracea were downloaded from NCBI (ProjectID: PRJNA489323); after being filtered with Trimmomatic (Bolger et al., 2014), the clean data without adapters and low-quality bases were aligned with the reference genome (http://brassicadb.org/brad/downloadOverview.php) using hisat2 (Kim et al., 2015). Based on the mapping results, the gene expressions (fragments per kilobase million (FPKM)) were counted by Stringtie (Pertea et al., 2015), and the heatmaps were drawn by R.
The B. napus ZS11 was used in this study. Tissues like flowers, roots, stems, leaves, immature pods, immature seeds, and apical meristems were collected and extracted using an RNeasy Extraction Kit (Invitrogen, Carlsbad, CA, USA). Quantitative real-time PCR (qRT-PCR) was carried out by referring to a formerly described protocol (Zhao et al., 2021). The expression level was displayed by heatmap in R. Expression patterns of five genes were shown by TBtools-eFP (Chen et al., 2020) (http://yanglab.hzau.edu.cn/BnTIR/eFP).
To investigate the genetic variation of CLE genes in B. napus, a panel of 204 rapeseed accessions was selected for this work (Zhao et al., 2022). The single-nucleotide polymorphism (SNP) information of CLE genes was retained from the previous study. After annotation with SnpEff (Cingolani et al., 2012), the distribution was analyzed to inspect the position of variations. The agronomic traits for three consecutive years (2014–2016) including plant height, branch number, initial branch height, length, silique number, and silique density of main inflorescence, main inflorescence, silique length, seed number per silique, main inflorescence seed density, thousand seed weight, seed weight per silique, and seed weight of the main inflorescence were surveyed and handled with the best linear unbiased prediction (BLUP). To study the potential impact of CLE genes on agronomic traits in B. napus, SNPs within 30 kb upstream and downstream of CLEs were used for the investigation of its potential impact. Regional association analysis considering population structure and relative kinship was conducted by EMMAX (Kang et al., 2010).
A genome-wide analysis of CLE genes in B. napus and its diploid progenitors was performed involving multiple BLAST queries and iterative queries, followed by domain validation and removal of false positives (i.e., no CLE domain). Finally, a total of 29 (BrCLEs), 32 (BoCLEs), and 70 (BnCLEs) genes were identified (Table S2). The total number of BrCLEs and BoCLEs in the two diploid progenitors was lower than that of BnCLEs in the allotetraploid rapeseed, indicating that CLE gene expansion event has occurred in B. napus during polyploidization.
The CLEs in the three species of Brassica species were renamed according to the AtCLEs in Arabidopsis based on the naming conventions for the B. genus (Ostergaard and King, 2008), with the last letter “a” indicating the highest homology with Arabidopsis, and then “b”, and so on. In B. napus, the capital letters A and C following “Bn” represented the An and Cn subgenomes, respectively.
To explore the evolution of the CLEs in Brassica, we constructed a phylogenetic tree using full-length amino acid sequences from Brassicaceae (Figure 1). The results showed CLEs clearly divided into seven subfamilies (I to VII). Subfamily V was the largest (42 members), followed by subfamily I (38 members). There were 17, 11, 23, 17, and 14 members in subfamilies II, III, IV, VI, and VII, respectively. The CLEs of the four species were distributed in all subfamilies. Most AtCLEs in each subfamily matched multiple sets of orthologs from B. napus and its two progenitors. A sister pair demonstrated the closest genetic relationship in a phylogenetic tree. A total of 56 sister pairs were observed. The majority of the sister pairs were orthologous gene pairs between the An (or Cn) subgenomes of B. napus and B. rapa (or B. oleracea), with 18 An–Ar pairs and 19 Cn–Co pairs. These results supported that the gene duplication events happened in the B. napus genome and indicated that CLE orthologous genes of distinct subfamilies were highly conserved in the respective genome.
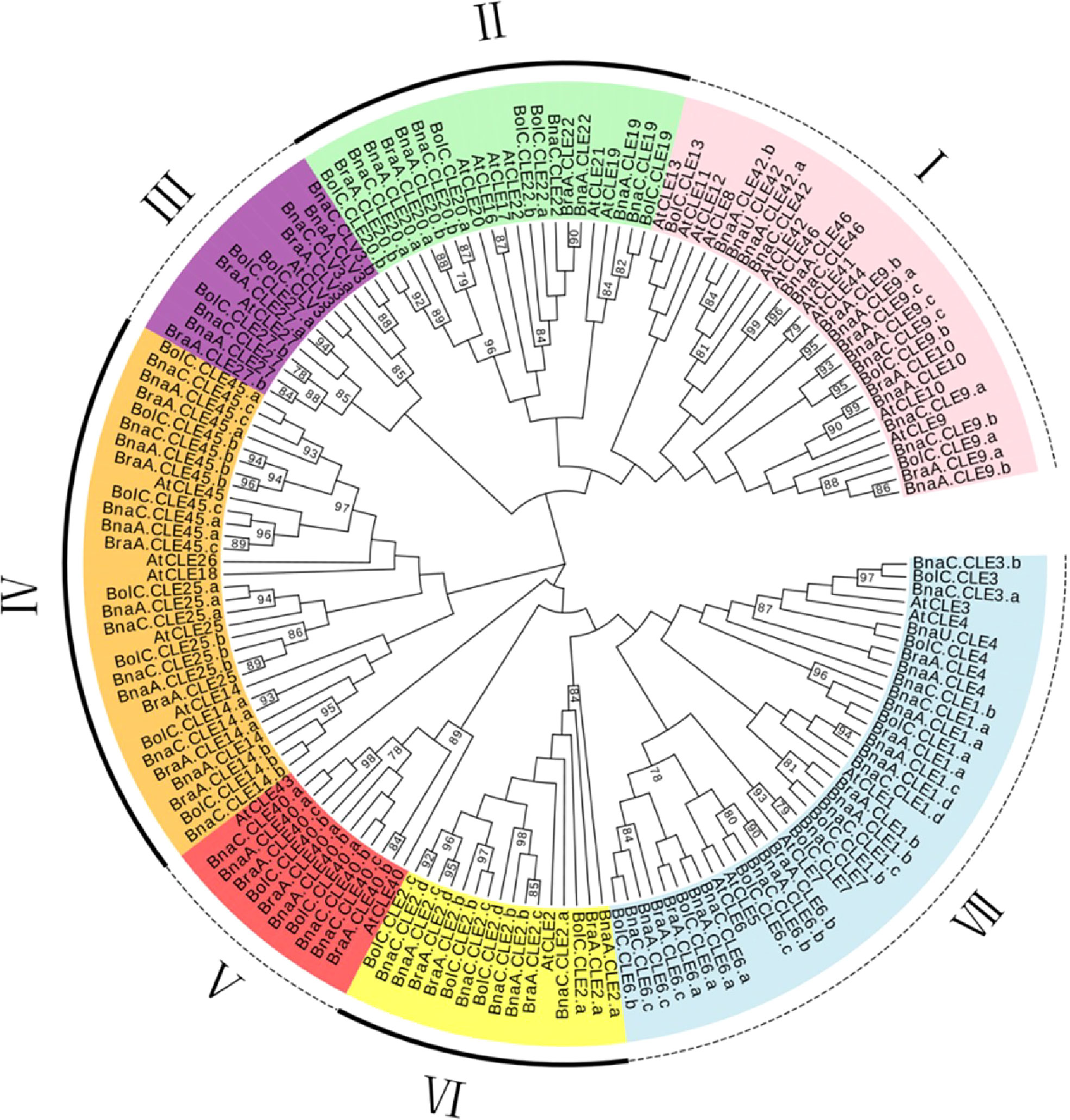
Figure 1 Phylogenetic analysis of Brassica napus, Arabidopsis thaliana, Brassica rapa, and Brassica oleracea CLE genes. This unrooted phylogenetic tree was constructed by MEGA 7.0 using the maximum likelihood method with 1,000 bootstrap replicates. Only values above 50% were displayed.
The chromosomal locations of BnCLEs, BrCLEs, and BoCLEs were investigated according to their physical positions (Figure 2). The BnCLEs were asymmetrically distributed on the 19 chromosomes in B. napus. There were a total of 32 BnCLEs in the An subgenome and 36 in the Cn subgenome, which were similar to those in B. rapa (Ar, 29) and B. oleracea (Co, 32). The remaining two BnCLE genes were located on unanchored scaffolds (Table S2). Each chromosome harbored at least one CLE gene. Chromosome An07 in B. napus carried the most CLEs (seven CLEs). On chromosomes An01, Cn01, and Cn08, only one CLE gene was found. Furthermore, many CLEs retained their relative position in Ar and An, whereas only a portion of CLEs retained their relative position in Co and Cn. For example, the same number of CLEs in Ar01–An01, Ar05–An05, Ar06–An06, and Ar08–An08 was observed, which also showed similar locations in the chromosomes (Figure 2).
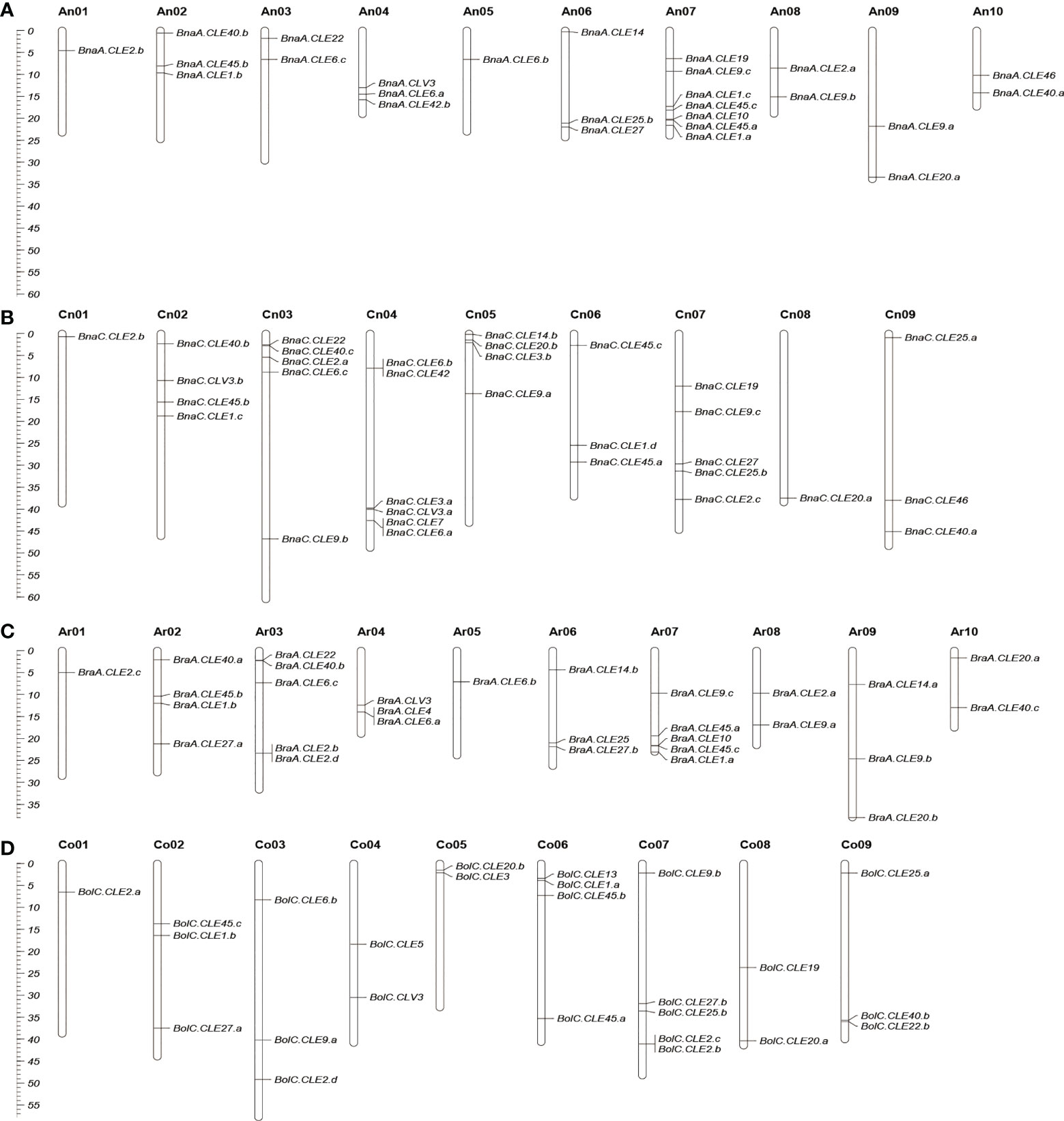
Figure 2 Chromosomal location of CLEs in Brassica napus (A, B), Brassica rapa (C), and Brassica oleracea (D). Partial CLEs in B. oleracea and B. napus located in unassembled scaffolds were not shown. The scale on the left is in megabases (Mbs).
We searched four duplicated types in each Brassicaceae species, including dispersed, proximal, tandem, and whole-genome duplication (WGD) (Table S3). We found that 56 of 70 BnCLEs were derived from segmental duplication/whole-genome triplication (WGT). Therefore, it appeared that segmental duplication/WGT played an important role in the BnCLE expansion. In addition, we examined CLE gene expansion patterns in B. oleracea and B. rapa, finding that most BoCLEs (53.1%; 17/32) and BrCLEs (89.7%; 26/29) were also derived from segmental duplication/WGT (Table S3). One cluster of CLE tandem repeat genes was only present in B. rapa and B. oleracea, but not in B. napus.
A great amount of the identified CLEs lacked predicted introns, except for 20 genes, including one to three introns (Table S2, Figure S1). Like other plants, the multiple protein sequence alignment of CLE pre-propeptides in B. napus contained a signal peptide at the N-terminus, a central variable domain, and a CLE domain at the C-terminus. The signal peptide sequences and the variable domain among distinct genes were not highly conserved, except between orthologous genes. The highly conserved CLE domain that contained approximately 12/13 aa and residues at different positions had different levels of conservation. To study the sequence of the conserved CLE domains and the degree of their conservation in different Brassicaceae species, multiple sequence alignment was used to generate the protein sequence logos in B. rapa, B. oleracea, B. napus, and Arabidopsis (Figure 3). The results showed that conserved residues patterns of the CLE domain were remarkably similar in these four plants (Figure 3). The conservation of residues 1, 4, 6, 8, 9, 11, and 12 suggested that they may be critical to the function of CLE mature peptides. These results indicated that CLE gene family was relatively conserved, while some motif sequences changed slightly during Brassica evolution, which possibly contributed to extended special biological function.
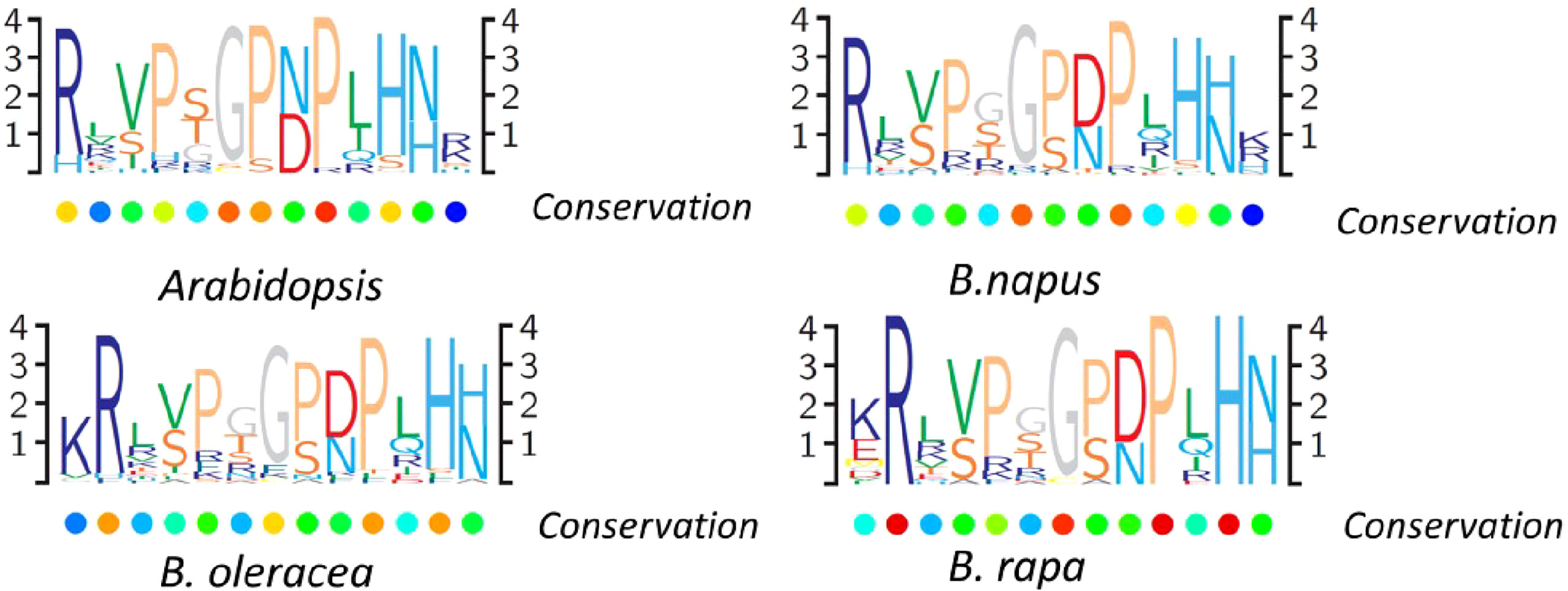
Figure 3 CLAVATA3/EMBRYO SURROUNDING (CLE) domain consensus sequences from Brassica napus, Brassica rapa, Brassica oleracea, and Arabidopsis pre-propeptides. Logo diagrams illustrate the 13 amino acid CLE domain consensus sequences, determined from multiple sequence alignments generated for each species.
Cis-elements in the promoters can affect gene expression (Davuluri et al., 2003; Klaas, 2009). Therefore, CLE gene promoters of these three species were investigated using PlantCARE (Magali, 2002). Four to 12 cis-elements involving development, hormone, and stress were identified in each BnCLE promoter, while 2–12 and 4–13 were found in BrCLEs and BoCLEs, respectively (Figure S2). ARE has the most elements in these three species and existed in 87.1% (61/70) BnCLE promoters, 93.1% (27/29) BrCLEs, and 96.9% (31/32) BoCLEs; this element is essential for anaerobic induction (Figure 4). Hormone-responsive elements involved in abscisic acid responsiveness, methyl jasmonate responsiveness, and ethylene responsiveness were also very common in CLE promoters: 80% (56/70), 86.2% (25/29), and 87.5% (28/32) of BnCLE, BrCLE, and BoCLE promoters included ABRE, respectively. In development elements, GCN4 motif involved in endosperm expression (25.7%, 18/70 in BnCLEs; 37.9%, 11/29 in BrCLEs; and 25%, 8/32 in BoCLEs), O2 site involved in zein metabolism regulation (40%, 28/70 in BnCLEs; 31%, 9/29 in BrCLEs; and 34.4%, 11/32 in BoCLEs), CAT-box related to meristem expression (27.1%, 19/70 in BnCLEs; 31%, 9/29 in BrCLEs; and 37.5%, 12/32 in BoCLEs) and circadian (32.9%, 23/70 in BnCLEs; 24.1%, 7/29 in BrCLEs; and 37.5%, 12/32 in BoCLEs) were common. According to the cluster results, some similar CLE promoters had similar cis-elements, like CLE4, CLE19, and CLE42, while other CLEs were clustered into different groups.
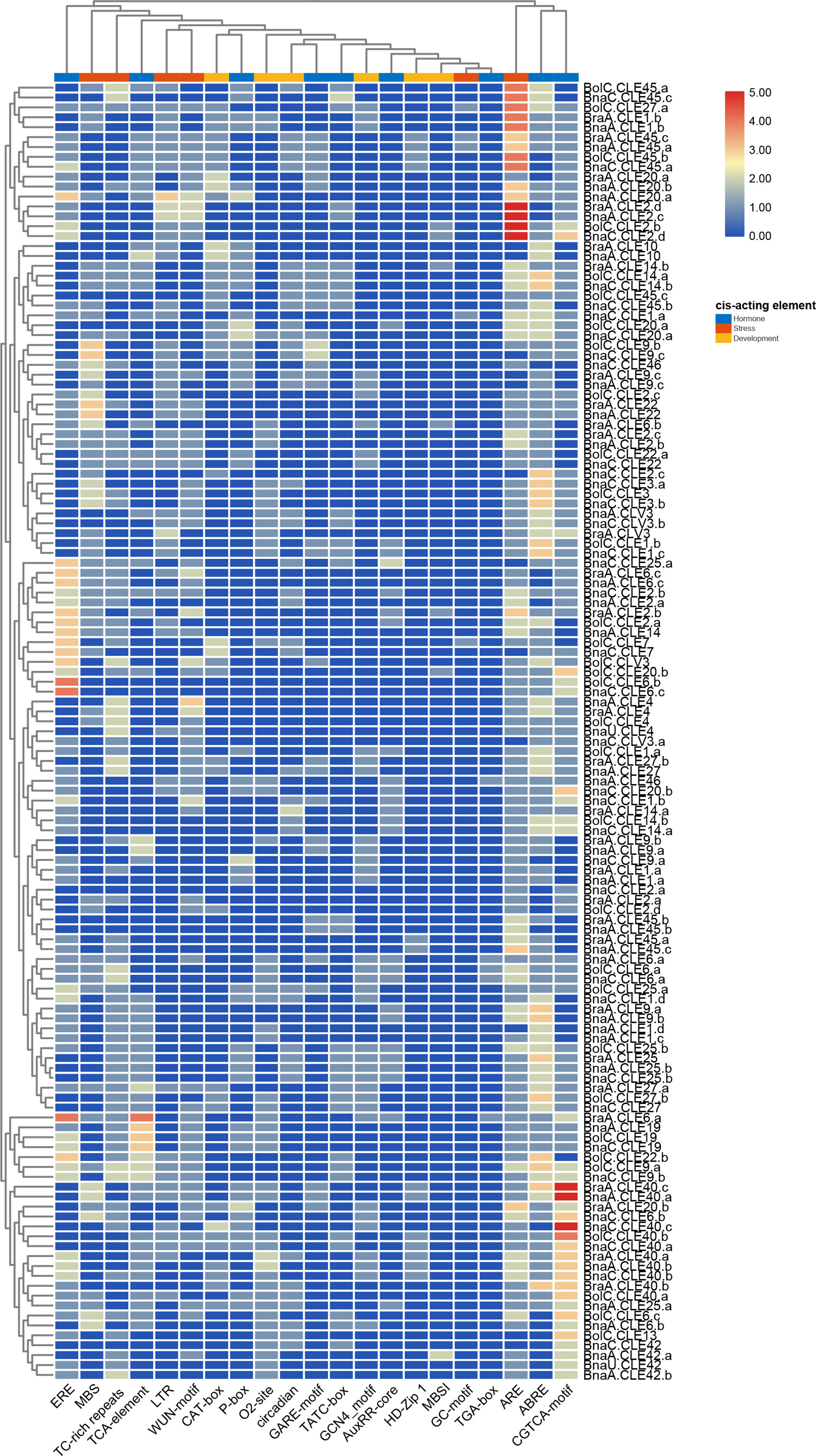
Figure 4 Cis-acting elements analysis of CLEs in Brassica napus, Brassica rapa, and Brassica oleracea. The color scale bar indicates the numbers of cis-acting elements in the promoters.
Syntenic genes are orthologous genes located in syntenic fragments between different species that derive from a shared ancestor. We obtained the syntenic genes of CLE genes of Arabidopsis in three Brassica species by searching “syntenic gene” in BRAD (Wang et al., 2015) and showed these collinearity relationships using Circos (Krzywinski et al., 2009) software between the An and Cn subgenomes of B. napus and its two diploid progenitors (Figure 5, Table S4). A total of 101 CLE genes in three Brassica species showed conserved synteny with those in A. thaliana and were positioned in the same conserved chromosomal blocks, such as A, B, D, E, F, I, J, L, R, U, and Wb (Schranz et al., 2006). In addition, syntenic genes in three Brassica species were divided into three fractionated subgenomes, which were specified as LF (least-fractionated), MF1 (medium-fractionated), and MF2 (most-fractionated) according to the extent of gene retention (Chalhoub et al., 2014). There were 41, 31, and 27 CLE genes caught in the LF, MF1, and MF2 subgenomes, respectively (Table S4). A total of 27 AtCLE genes retained corresponding syntenic genes in the three Brassica species. The existing forms of syntenic genes in the genomes of three Brassica species were different. The first type was that syntenic genes of AtCLE were completely preserved in the same block of synteny in the Ar, Co, An, and Cn subgenomes, such as AtCLV3. The second type was that AtCLE genes were retained in the Ar or/and Co genome but lost in B. napus genomes, such as AtCLE4, AtCLE5, AtCLE6, and AtCLE14. The third type was that AtCLE genes were retained in B. napus genome but lost in B. rapa or B. oleracea genomes, such as AtCLE1, AtCLE9, and AtCLE40. The results showed that the expansion of CLE gene family was also accompanied by gene loss.
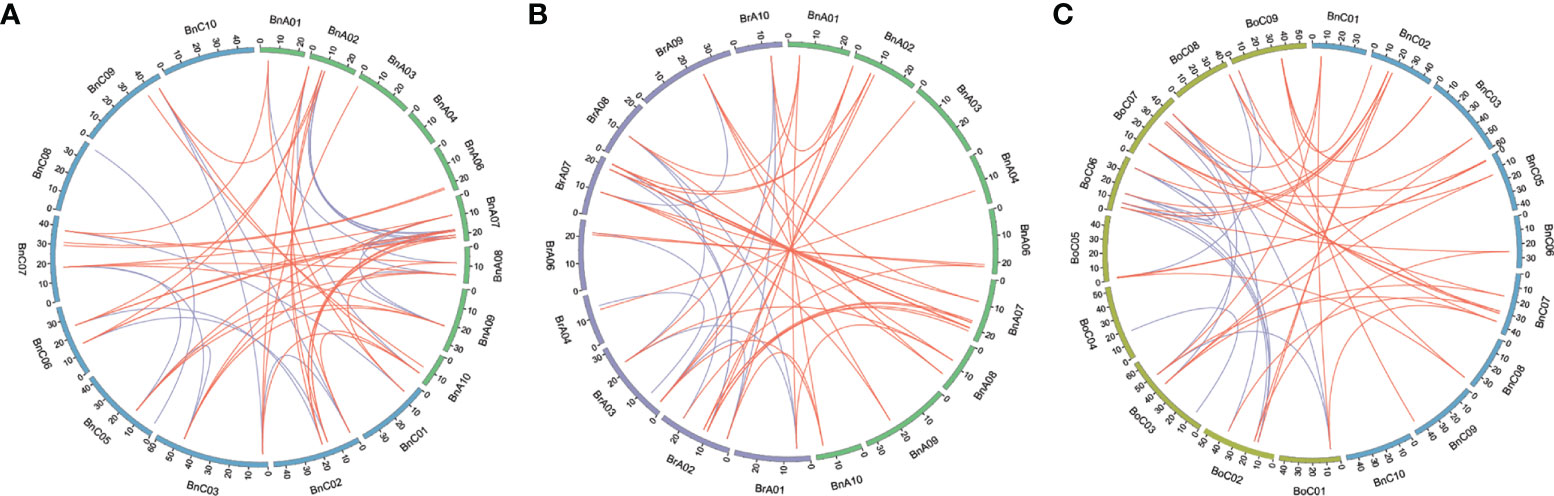
Figure 5 Genome-wide synteny analysis for CLEs among Brassica napus, Brassica rapa, and Brassica oleracea. (A) Synteny analysis of CLEs on An and Cn subgenomes in B. napus. (B) Synteny analysis of CLEs between An subgenome of B. napus and B. rapa. (C) Synteny analysis of CLE genes between Cn subgenome of B. napus and B. oleracea. Inside the circos, brown lines linked the syntenic orthologs, and blue lines linked the syntenic paralogs.
To comprehend whether natural selection acted on the evolution of CLE gene family in B. napus, selection pressure analysis was performed on the syntenic CLE gene pairs between An and Ar, Cn, and Co. The non-synonymous rate (Ka) and synonymous rate (Ks) values were calculated. The Ka/Ks ratio > 1 represents positive selection, the Ka/Ks ratio = 1 represents neutral selection, and the Ka/Ks ratio < 1 represents purifying selection (Nekrutenko, 2002). The Ka/Ks ratios of the syntenic gene pairs are shown in Table S5. The Ka/Ks ratios for some CLE syntenic gene pairs were >1, such as BnaA.CLE.1c and BnaC.CLE.1d, BnaC.CLE.2b and BolC.CLE.2b, BnaA.CLE.10 and BraA.CLE.10, and BnaA.CLE.27 and BnaC.CLE.27, which indicated that these genes were subject to positive selection pressure. Many syntenic gene pairs had no Ka/Ks value in B. napus because these two genes had the same sequence or large differences. The rest were less than one, which indicated that they underwent purifying selection during the evolution process and may preferentially perform conserved functions.
As polypeptide hormones, CLEs need to combine with receptor proteins to transmit signals between cells. To investigate the involved biological process in rapeseed and its diploid progenitors, protein–protein interaction networks were predicted based on known protein interactions in Arabidopsis. A total of 1,061 proteins Arabidopsis proteins interacted with CLEs, resulting in 3,997 proteins in rapeseed (Figure 6A). CLEs interacted with other proteins but also interacted with each other. Taking out the interacted genes for Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis indicated that they played important roles in zeatin biosynthesis, amino acid metabolism (like tryptophan, arginine, and proline), polysaccharide biosynthesis, and ion channels (Figure 6B). Gene Ontology (GO) enrichment analysis showed that their molecular functions mainly were receptor serine/threonine kinase binding and phosphorelay response regulator activity; their biological processes were transmembrane receptor protein tyrosine kinase signaling pathway, polarity specification of adaxial/abaxial axis, stamen development, and lateral root development (Figure 6C). As to CLE genes in B. rapa and B. oleracea, 1,334 and 1,356 proteins were identified as their interacted proteins, respectively. The Gene Ontology enrichment analysis (Figures S3, S4) indicated that these proteins were important in the pattern specification process, shoot system morphogenesis, meristem maintenance, root morphogenesis, and leaf development.
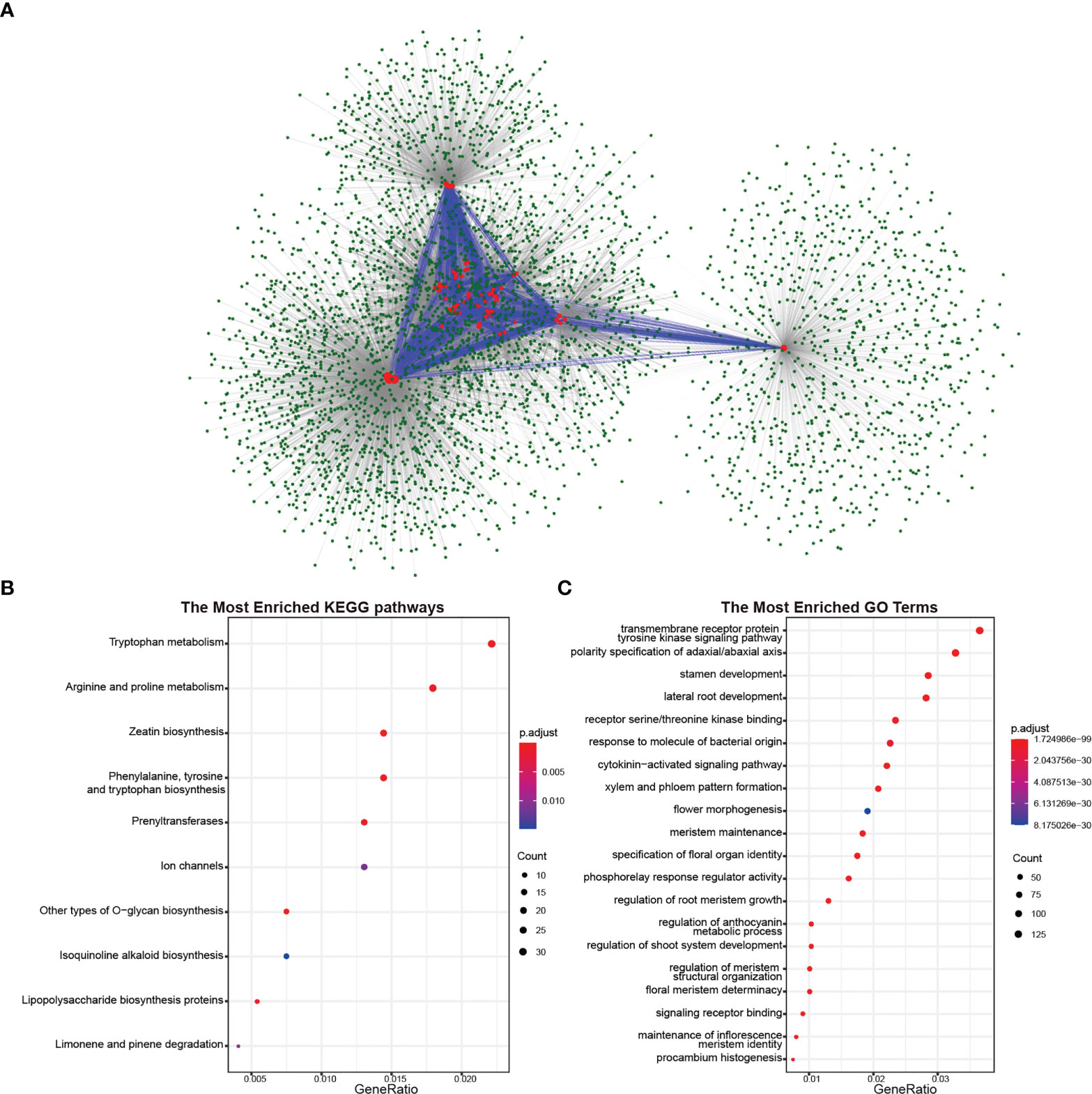
Figure 6 Proteins interacted with CLE proteins in Brassica napus. (A) Protein–protein interaction network of CLE proteins in B. napus. The red circles represent the CLE proteins, and the green circles represent proteins interacting with CLE proteins. The blue lines represent the interaction between CLE proteins, and the gray lines represent the interaction between CLE proteins and other proteins. (B) Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis of proteins interacted with CLE proteins. (C) Gene Ontology enrichment analysis of proteins interacted with CLE proteins.
According to the RNA-seq data in siliques, leaves, flowers, and stems of B. napus, B. rapa, and B. oleracea, the expression patterns of CLEs are illustrated in Figures S5-S7. The majority of CLEs showed low or no expression in all four tissues. Among the expressed CLEs, more CLE genes were highly expressed in stems in three Brassica species, like BnaC.CLE42, BnaA.CLE46, BnaC.CLE46, BolC.CLE20.b, BraA.CLE6.b, and BraA.CLE6.c, whereas BnaA.CLE9.b, BolC.CLE27.a, BolC.CLE14.b, and BraA.CLE14.a were highly expressed in siliques. Only a few paralog CLE genes had a similar expression pattern, like BraA.CLE27.a/BraA.CLE27.b, BnaC.CLE42/BnaA.CLE42.b/BnaA.CLE42.a/BnaU.CLE42, BolC.CLE9.a/BolC.CLE9.b, and BolC.CLE22.a/BolC.CLE22.b. In three Brassica species, orthologs like BnaA.CLE20.b/BraA.CLE20.b/BolC.CLE20.b and BnaC.CLV3.a/BraA.CLV3/BolC.CLV3 displayed a similar expression pattern. Meanwhile, orthologs in B. rapa and B. oleracea showed different expression patterns; for example, BraA.CLE4 was mainly expressed in leaves, whereas BolC.CLE4 was not. These results suggest that some CLE genes have shown functional divergence during their evolution.
Furthermore, we performed a qPCR analysis in B. napus in various tissues—flowers, roots, stems, leaves, immature pods, immature seeds, and apical meristems. Similar to the transcriptome data, BnCLE genes had significantly different expression patterns in different tissues (Figure S8). BnaA.CLE1.b, BnaA.CLE2.b, BnaC.CLE2.a/b/c, BnaA.CLE14, and BnaC.CLE14.b were more highly expressed in roots than in the other tissues examined (Figure 7A), whereas BnaA.CLE6.a/b, BnaC.CLE6.b, BnaC.CLE7, BnaA.CLE4, BnaU.CLE4, BnaA.CLE1.c, BnaC.CLE1.a/c, BnaA.CLE9.a/b/c, and BnaC.CLE9.a/b/c were more highly expressed in immature seeds than in the other tissues (Figure 7B). BnaA.CLE45.c, BnaA.CLE19, BnaC.CLE19, and BnaC.CLV3.a/b were more highly expressed in apical meristems than in the other tissues (Figure 7C). BnaC.CLE2.d and BnaC.CLE46 were expressed at high levels in root and stem (Figure 7D), while BnaA.CLE1.a/b, BnaA.CLE2.a, BnaC.CLE6.a, and BnaC.CLE14.a were expressed at high levels in root and immature seeds (Figure 7E). The materials and sampling time of the qPCR experiments in this study were different from the data downloaded from NCBI; therefore, the results were not completely consistent. However, there was some similarity: BnCLEs showed different expression patterns in different tissues and were modestly expressed in leaves. The expression patterns of most BnCLEs pairs were significantly different, suggesting that their roles were dissimilar. For example, BnaA.CLE1.b was highly expressed in the roots, whereas BnaC.CLE1.c was more highly expressed in immature seeds. BnaA.CLE1.c was highly expressed in immature seeds, whereas BnaA.CLE1.d was similarly expressed in the different tissues. BnaA.CLE22 was highly expressed in the stem, whereas BnaC.CLE22 was more highly expressed in apical meristems. BnaA.CLE46 was highly expressed in the stem, whereas BnaC.CLE46 was the more highly expressed root, followed by stem.
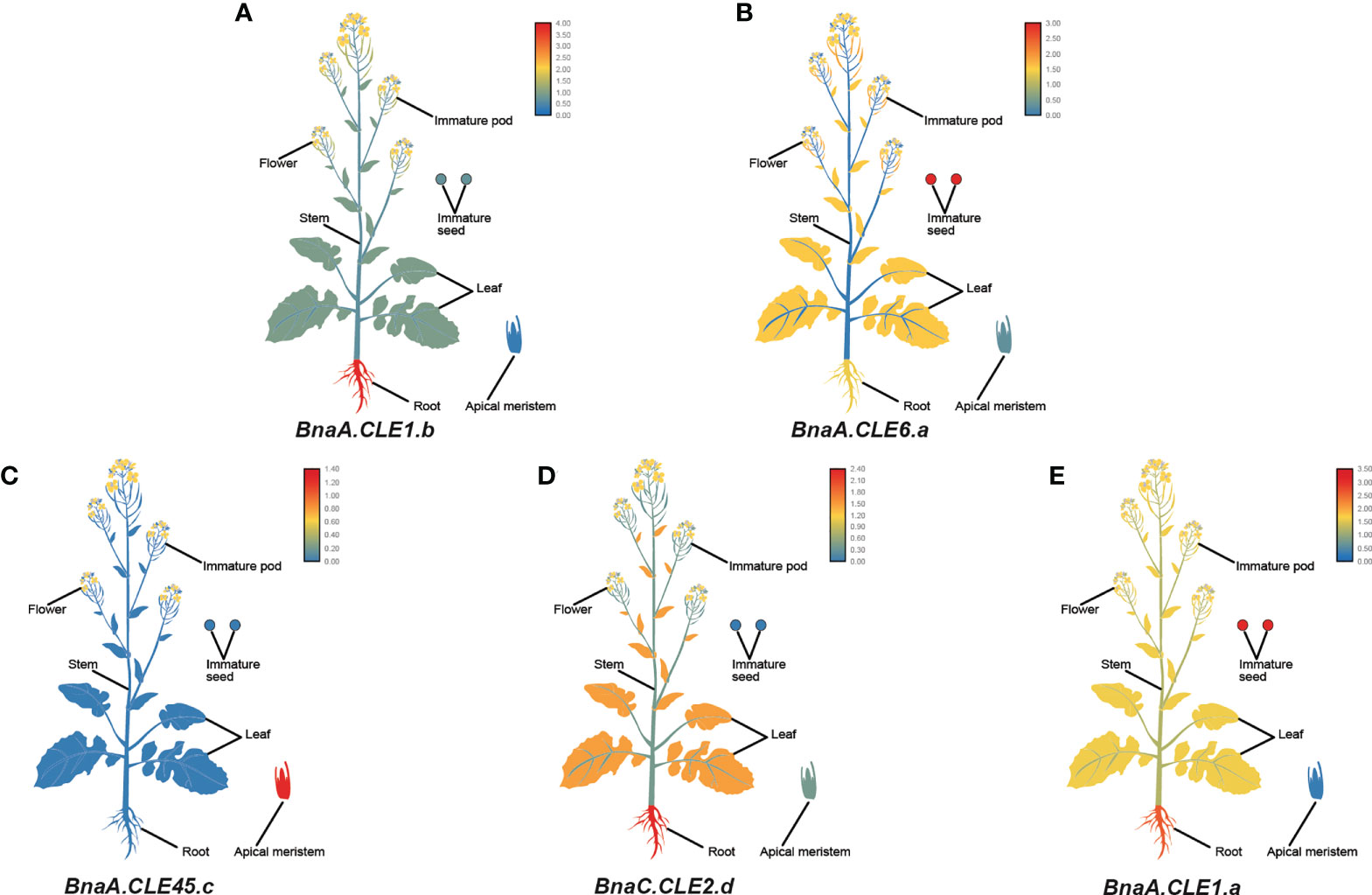
Figure 7 The expression patterns of five selected CLE genes (A–E) in Brassica napus plants. Expression data were processed with log10 normalization. The color scale represents relative expression levels from low (blue color) to high (red color).
A natural population including 204 accessions was selected to analyze the genetic variations of CLEs, and a total of 115 SNPs were identified; on average, 1.6 SNPs were detected for each CLE gene, which was apparently less than the whole genome level (28.9 SNPs per gene). The total and average number of SNPs in the A subgenome (74 SNPs, 2.2 SNPs/gene) were higher than in the C subgenome (41 SNPs, 1.1 SNPs/gene). Considering CLE gene length, the SNP density of the A subgenome was 5.7 SNPs/kb, whereas it was 2.2 SNPs/kb in the C subgenome. Moreover, the SNP distribution among paralogous genes was also unequal; like for BnaA.CLE25.a/BnaA.CLE25.b/BnaC.CLE25.a/BnaC.CLE25.b, the SNP numbers were 9/1/14/0. Finally, SNP annotation implied that 73 SNPs were in exon regions and 37 SNPs could lead to missense mutations.
In this study, 12 agronomic traits (Figure S9) and SNPs within the range of 30 kb upstream and downstream of BnCLEs were used to study the impact of BnCLEs in B. napus. A total of 12 CLEs were significantly associated with at least one agronomic trait (p < 0.0001) (Table S6), including plant height, branch height, main inflorescence length, seed weight per silique, silique number, silique density, seed density, and seed weight of the main inflorescence. For example, there was no variation in the gene sequence of BnaC.CLV3.a, but SNPs in the upstream region (6 kb) were significantly associated with seed weight per silique; the population was clearly divided into two haplotypes based on the SNPs, and the t-test displayed the significant difference in seed weight per silique between these two groups (Figure 8A). According to JASPAR, the SNP region (TCCGTACA) was predicted as the binding site of C2H2 zinc finger factors (SPL3). Moreover, there were another four genes also strongly associated with yield traits, like BnaA.CLE20.b and BnaC.CLE20.b; SNPs in the upstream region (1.4 and 3.4 kb) were associated with main inflorescence seed density (Figures 8B, C). Their interacted proteins were not only enriched in receptor serine/threonine kinase binding (GO:0033612), signaling receptor binding (GO:0005102), and cell–cell signaling involved in cell fate commitment (GO:0045168) but also enriched in GO terms including regulation of meristem structural organization (GO:0009934), maintenance of meristem identity (GO:0010074), and maintenance of root meristem identity (GO:0010078). Therefore, it was speculated that these CLEs could affect the development of rapeseed and result in phenotypic variations.
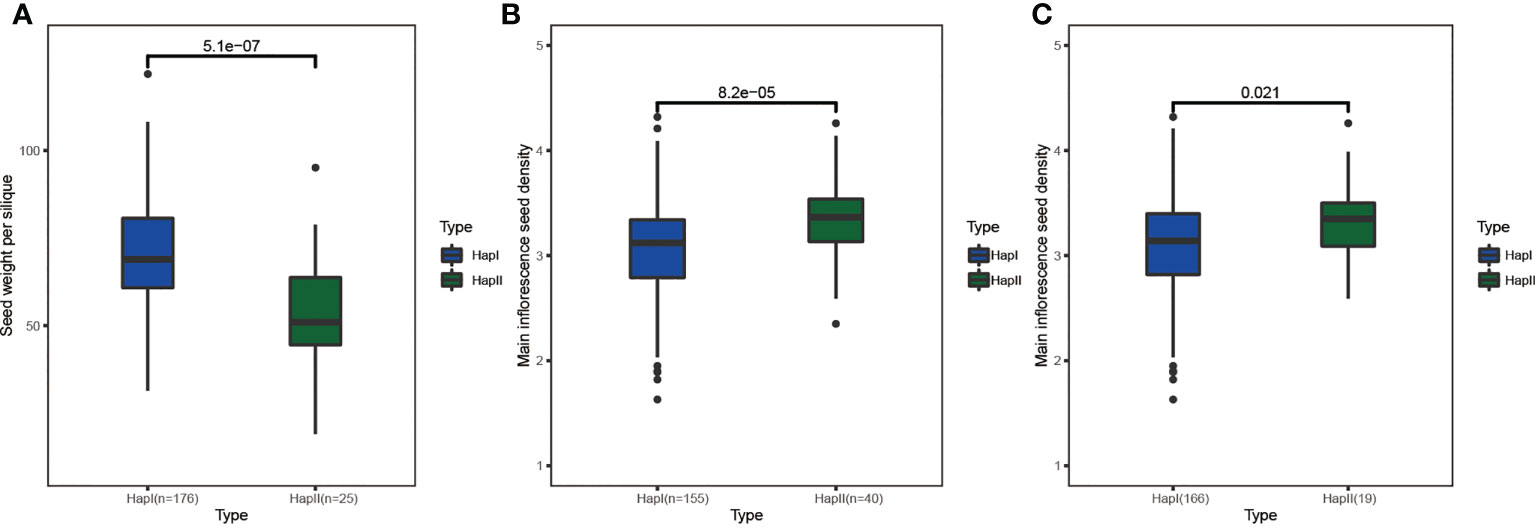
Figure 8 The haplotype analysis of BnCLEs for seed weight per silique and main inflorescence seed density. (A) BnaC.CLV3.a. (B) BnaA.CLE20.b. (C) BnaC.CLE20.b.
CLE peptides are plant-specific peptide hormones that act as mediators of cell-to-cell communication (Fletcher, 2020). Genome-wide studies on CLE gene family have been performed in tomato, soybean, wheat, and populus (Zhang et al., 2014; Hastwell et al., 2015; Han et al., 2016; Li Z. et al., 2019; Han et al., 2020). In this study, systematic identification, classification, evolution, expression, and association mapping analysis were performed in B. napus. In total, we identified 70, 32, and 29 CLEs in B. napus, B. oleracea, and B. rapa, respectively, which were less than those in the previous work (Han et al., 2020), and the possible reason was more strict parameters used in this work. These CLEs were divided into seven subfamilies according to the phylogenetic tree. Furthermore, this work analyzed the relationship of CLEs among three Brassica species and displayed the similarities and differences of cis-acting elements, interacted proteins, and expression patterns. With the aid of a natural population of B. napus, the genetic variations of CLE genes were uncovered, and several CLEs were recognized as candidate genes for important agronomic traits in rapeseed by regional association mapping analysis.
Polyploidization, an important force in the evolution of species (especially magnoliophyte), played an important role in plant adaptation o to new environments (Schemske, 1998; Zhang et al., 2020). Each Brassicaceae genome underwent a WGD event ~35 MYA ago (Bowers, 2003; Yuannian, 2011). Comparative genomic research showed that Brassica species experienced triploidy at the genomic level after they diverged from the Arabidopsis lineage approximately 20 MYA ago (Chalhoub et al., 2014; Liu et al., 2014). Therefore, it is apparent that the Brassica genome underwent paleopolyploidization (Bowers, 2003; Lysak et al., 2005). The differentiation of B. rapa and B. oleracea occurred approximately 4.6 MYA ago (Liu et al., 2014), and their natural hybridization formed the B. napus about 7,500 years ago (Chalhoub et al., 2014). Segmental duplication also leads to increased gene numbers (Flagel and Wendel, 2009). Most plants have undergone polyploidization events and thus retain a large number of duplicated chromosomal blocks at the genome level (Cannon et al., 2004). For example, in B. napus, segmental duplication/WGT is the primary force for WOX expansion (Li M. et al., 2019). In this study, nearly 80% of CLEs were distributed in syntenic blocks, suggesting that segmental duplication/WGT might contribute significantly to CLE expansion in Brassica. Tandem duplicated genes were defined as an array of at least two homologous genes within 50 kb (Cannon et al., 2004). However, we only found one pair of tandem duplicated genes in B. rapa and B. oleracea, suggesting that it is not a major factor for CLE expansion in Brassica.
Not all duplicated genes are retained in plants; gene loss always occurred due to the genomic sequence rearrangement after hybridization or chromosome doubling (Paterson et al., 2004; Ye et al., 2020). Each AtCLE should have three syntenic orthologs in B. rapa and B. oleracea based on the triploidy hypothesis. However, only AtCLE5/6/45 had such a pattern, and the other AtCLEs had only one or two (less than three syntenic orthologs) in both species (Table S3). Hybridization of B. rapa and B. oleracea should theoretically result in six copies for each homologous AtCLE gene in B. napus, and the CLE number in B. napus is equal to the sum of the homologous genes in B. rapa and B. oleracea. In fact, only AtCLE9 has six homologous genes in B. napus. Generally, most CLEs were lost during the formation of B. napus. Orthologs of nine AtCLEs were not found in three Brassica species. B. napus lost 38.02% of genes compared with A. thaliana at the genome-wide level (27,169 × 6 vs. 101,040), while nearly 63.54% of CLEs were lost in B. napus. The significantly higher average gene loss rate suggests that strong selection occurred for CLEs during evolution.
Theoretically, there were three possible means to lose syntenic BnCLEs. First, the loss of the BrCLEs or BoCLEs resulted in the loss of syntenic BnCLEs after Brassica genome triplication. AtCLV3 has one syntenic ortholog (BrCLV3 and BoCLV3) in B. rapa and B. oleracea, so it has two syntenic BnCLV3. Second, the loss of the CLEs occurred during the allopolyploidization process. AtCLE2 has three syntenic orthologs in both B. rapa and B. oleracea, while it has four syntenic orthologs in B. napus. Finally, the two processes together caused the loss of syntenic BnCLEs. AtCLE4 has one syntenic ortholog in both B. rapa and B. oleracea, whereas it has no syntenic ortholog in B. napus. These lost CLEs were redundant, and genetic change through loss could potentially lead to adaptive diversity.
Statistical analysis showed that 22 out of 29 CLE genes (76%) were positioned on the assembled chromosomes in B. rapa, whereas 12 out of 25 (48%) maintained their relative position in B. oleracea during the formation of B. napus. There are two possible reasons for this finding. One possibility is that the Cn subgenome has more abundant transposable elements (TEs) than the An subgenome (Chalhoub et al., 2014). The presence of TEs in the genome can cause the rearrangement of chromosomal sequences, which affects the genomic structure, including deletion, inversion, and translocation. Second, the Cn subgenome underwent more active homologous exchanges than the An subgenome during polyploidization (Chalhoub et al., 2014).
Only a few CLE peptides are studied in Brassica; however, the functions of most AtCLE peptides are characterized. Therefore, Brassica CLE functions can be predicted through sequence similarity and phylogenetic analysis. Seven distinct CLE subfamilies were defined using Arabidopsis and three Brassica species. For example, all CLV3 genes are clustered into subfamily III, while Brclv3 and Bnclv3 cause enlarged meristems and lead to extra organs such as multilocular siliques, similar to the phenotype of Atclv3 (Fletcher et al., 1999; Fan et al., 2014; Yang et al., 2018). This approach could elucidate the function of unknown CLE peptides. However, many genes contain more than one copy in polyploid species (Chalhoub et al., 2014; Li Z. et al., 2019). These duplicated genes are redundant, and variation in promoters could affect their gene expression (Davuluri et al., 2003; Klaas, 2009; Arsovski et al., 2015), For example, BrCLE45 have three highly similar copies, but cis-elements have significant differences, so their expression patterns were different. Distinct expression patterns are a direct sign that duplicated genes may diverge in different directions, indicating the occurrence of non-functionalization, neo-functionalization, or sub-functionalization at the transcriptional level (Gallagher et al., 2016; Cheng et al., 2018). BnCLEs appear to be highly expressed in immature seeds, roots, and stems based on their expression levels in previous studies. Moreover, their interacting proteins are enriched in the development of these organs. Therefore, it is concluded that CLEs perform important functions in the biological process of these organs. However, the majority of CLEs hardly expressed in the tissues analyzed in this study. Therefore, they were either expressed in other developmental stages or conditions, or they were suppressed after duplication. Promoters that regulate the CLEs expression may also play a role. Abundant cis-elements involved in hormone response, development, and stress were identified, and their existence made the regulation more flexible, which helps plants adapt to complex environments.
Genetic variations in CLEs were investigated in the associated B. napus population; the smaller number of SNPs in CLEs indicated that this gene family sequence was very conserved. SNP density was higher in the A subgenome compared with the C subgenome, which is consistent with other published gene families (Zhu et al., 2020; Wahid et al., 2022; Xie et al., 2022). Twelve CLEs were found to be significantly associated with important agronomic traits using regional association mapping analysis. Among them, the variations in the upstream region of BnaC.CLV3.a (the orthologs of the famous gene CLV3) were significantly associated with seed weight per silique. The sequence of variations was predicted as the binding sites of transcription factor SPL3, which can directly bind to the promoters of many genes to affect flower development (Jung et al., 2016). In Brassica, BrCLV3 and BnCLV3 control multilocular silique traits and increase seed production (Fan et al., 2014; Yang et al., 2018). Although no variation was detected in the genetic region of BnaC.CLV3.a in this natural population, the upstream variations that occurred in the binding sites of transcription factor SPL3 may influence the binding ability, which would possibly regulate the expression of BnaC.CLV3.a and affect the yield trait. In summary, the association mapping analysis conducted in this study could provide a better way to explore the significance of CLEs in phenotypic variation and offer candidate genes for further genetic improvement breeding in B. napus.
A total of 29, 32, and 70 CLEs were identified in B. rapa, B. oleracea, and B. napus, respectively, and divided into seven subfamilies in the phylogenetic tree. The conservation of the CLE domain suggested that the CLE family is relatively conserved in its biological function. WGT and segmental duplication were the major contributors to the expansion of CLEs in Brassica species. Transcriptome and qPCR analyses indicated that BnCLEs were highly expressed in immature seeds, roots, and stems. Some CLE pairs exhibited different expression patterns in the same tissue, indicating duplicated CLE differentiation. Moreover, genetic variations and regional association mapping analysis indicated that 12 CLE genes were potential genes for regulating important agronomic traits. In summary, this study was helpful in understanding the molecular evolution and biological function of CLEs in rapeseed and its diploid progenitors, which would aid future genetic improvement for high-yield breeding in B. napus.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
YX, MX, and CZ designed the research, analyzed the data, and wrote the manuscript. MS analyzed the data. YX and CT reviewed the manuscript. All authors contributed to the article and approved the submitted version.
This study was supported by National Natural Science Foundation of China (32070217), Precursor projects of Guizhou province for biological breeding supporting by science and technology in 2022 (Fine identification and evaluation of crop germplasm resources), Subsidy project from NSFC of Guizhou Academy of Agricultural Sciences (No. [2021] 50), Science and technology project of Shandong Education Department to MS (Grant no. J15LE02), China Postdoctoral Science Foundation funded project to MS (Grant no. 2018M632646), the Young Top-notch Talent Cultivation Program of Hubei Province for CT, and the National Natural Science Foundation of China (31770250).
We thank Zhixian Qiao of the Analysis and Testing Center at IHB for the technical support in RNA-seq analysis.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.998082/full#supplementary-material
Allender, C., King, G. (2010). Origins of the amphiploid species brassica napus l. investigated by chloroplast and nuclear molecular markers. BMC Plant Biol. 10, 54. doi: 10.1186/1471-2229-10-54
Arsovski, A. A., Pradinuk, J., Guo, X. Q., Wang, S., Adams, K. L. (2015). Evolution of cis-regulatory elements and regulatory networks in duplicated genes of Arabidopsis. Plant Physiol. 169 (4), 2982–2991. doi: 10.1104/pp.15.00717
Beitz, E. (2000). T(E)Xshade: shading and labeling of multiple sequence alignments using (LTEX)-T-A 2(epsilon). Bioinformatics 16 (2), 135–139. doi: 10.1093/bioinformatics/16.2.135
Bidadi, H., Matsuoka, K., Sage-Ono, K., Fukushima, J., Pitaksaringkarn, W., Asahina, M., et al. (2014). CLE6 expression recovers gibberellin deficiency to promote shoot growth in Arabidopsis. Plant J. 78 (2), 241–252. doi: 10.1111/tpj.12475
Bolger, A. M., Marc, L., Bjoern, U. (2014). Trimmomatic: a flexible trimmer for illumina sequence data. Bioinformatics 15, 2114–2120. doi: 10.1093/bioinformatics/btu170
Bonello, J., Opsahl-Ferstad, H., Perez, P., Dumas, C., Rogowsky, P. (2000). Esr genes show different levels of expression in the same region of maize endosperm. Gene 246, 219–227. doi: 10.1016/s0378-1119(00)00088-3
Bowers, J. E. (2003). Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature 6930, 433-438. doi: 10.1038/nature01521
Cannon, S. B., Mitra, A., Baumgarten, A., Young, N. D., May, G. (2004). The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 4 (1), 10. doi: 10.1186/1471-2229-4-10
Chalhoub, B., Denoeud, F., Liu, S., Parkin, I. A., Tang, H., Wang, X., et al. (2014). Early allopolyploid evolution in the post-neolithic Brassica napus oilseed genome. Science 345 (6199), 950–953. doi: 10.1126/science.1253435
Chen, C., Chen, H., Zhang, Y., Thomas, H., Frank, M., He, Y., et al. (2020). TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13 (8), 1194–1202. doi: 10.1016/j.molp.2020.06.009
Cheng, F., Wu, J., Cai, X., Liang, J., Freeling, M., Wang, X. (2018). Gene retention, fractionation and subgenome differences in polyploid plants. Nat. Plants 4 (5), 258–268. doi: 10.1038/s41477-018-0136-7
Cingolani, P., Platts, A., Wang, L. L., Coon, M., Nguyen, T., Wang, L., et al. (2012). A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of drosophila melanogaster strain w1118; iso-2; iso-3. Fly 6 (2), 80–92. doi: 10.4161/fly.19695
Czyzewicz, N., Shi, C. L., Vu, L. D., Van De Cotte, B., Hodgman, C., Butenko, M. A., et al. (2015). Modulation of Arabidopsis and monocot root architecture by CLAVATA3/EMBRYO SURROUNDING REGION 26 peptide. J. Exp. Bot. 66 (17), 5229–5243. doi: 10.1093/jxb/erv360
Davuluri, R. V., Sun, H., Palaniswamy, S. K., Matthews, N., Molina, C., Kurtz, M., et al. (2003). AGRIS: Arabidopsis gene regulatory information server, an information resource of Arabidopsis cis -regulatory elements and transcription factors. BMC Bioinf. 4, 25. doi: 10.1186/1471-2105-4-25
Dong, W., Wang, Y., Takahashi, H. (2019). CLE-CLAVATA1 signaling pathway modulates lateral root development under sulfur deficiency. Plants (Basel) 8 (4), 103. doi: 10.3390/plants8040103
Fan, C., Wu, Y., Yang, Q., Yang, Y., Meng, Q., Zhang, K., et al. (2014). A novel single-nucleotide mutation in a CLAVATA3 gene homolog controls a multilocular silique trait in Brassica rapa l. Mol. Plant 7 (12), 1788–1792. doi: 10.1093/mp/ssu090
Fiers, M., Golemiec, E., van der Schors, R., van der Geest, L., Li, K., Stiekema, W., et al. (2006). The CLAVATA3/ESR motif of CLAVATA3 is functionally independent from the nonconserved flanking sequences. Plant Physiol. 141 (4), 1284–1292. doi: 10.1104/pp.106.080671
Fiers, M., Hause, G., Boutilier, K., Casamitjana-Martinez, E., Weijers, D., Offringa, R., et al. (2004). Mis-expression of the CLV3/ESR-like gene CLE19 in Arabidopsis leads to a consumption of root meristem. Gene 327 (1), 37–49. doi: 10.1016/j.gene.2003.11.014
Fiume, E. (2010). Expression analysis of the CLE signaling gene family in arabidopsis thaliana and functional characterization of CLE8 in seed development. Electronic Thesis Dissertations.
Fiume, E., Fletcher, J. (2012). Regulation of Arabidopsis embryo and endosperm development by the polypeptide signaling molecule CLE8. Plant Cell 24 (3), 1000–1012. doi: 10.1105/tpc.111.094839
Flagel, L. E., Wendel, J. F. (2009). Gene duplication and evolutionary novelty in plants. New Phytol. 183 (3), 557–564. doi: 10.1111/j.1469-8137.2009.02923.x
Fletcher, J. C. (2020). Recent advances in arabidopsis CLE peptide signaling. Trends Plant Sci. 25, 1005-1016. doi: 10.1016/j.tplants.2020.04.014
Fletcher, J., Brand, U., Running, M., Simon, R., Meyerowitz, E. (1999). Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science 283 (5409), 1911–1914. doi: 10.1126/science.283.5409.1911
Gallagher, J., Grover, C., Hu, G., Wendel, J. (2016). Insights into the ecology and evolution of polyploid plants through network analysis. Mol. Ecol. 25 (11), 2644–2660. doi: 10.1111/mec.13626
Gancheva, M. S., Dodueva, I. E., Lebedeva, M. A., Tvorogova, V. E., Tkachenko, A. A., Lutova, L. A. (2016). Identification, expression, and functional analysis of CLE genes in radish (Raphanus sativus l.) storage root. BMC Plant Biol. 16 Suppl 1, 7. doi: 10.1186/s12870-015-0687-y
Grienenberger, E., Fletcher, J. C. (2015). Polypeptide signaling molecules in plant development. Curr. Opin. Plant Biol. 23, 8–14. doi: 10.1016/j.pbi.2014.09.013
Han, S., Khan, M. H. U., Yang, Y., Zhu, K., Li, H., Zhu, M., et al. (2020). Identification and comprehensive analysis of the CLV3/ESR-related (CLE) gene family in brassica napus l. Plant Biol. (Stuttg) 22 (4), 709–721. doi: 10.1111/plb.13117
Han, H., Zhang, G., Wu, M., Wang, G. (2016). Identification and characterization of the populus trichocarpa CLE family. BMC Genomics 17, 174. doi: 10.1186/s12864-016-2504-x
Hastwell, A., Gresshoff, P., Ferguson, B. (2015). Genome-wide annotation and characterization of CLAVATA/ESR (CLE) peptide hormones of soybean (Glycine max) and common bean (Phaseolus vulgaris), and their orthologues of arabidopsis thaliana. J. Exp. Bot. 66 (17), 5271–5287. doi: 10.1093/jxb/erv351
He, Z., Zhang, H., Gao, S., Lercher, M. J., Chen, W. H., Hu, S. (2016). Evolview v2: an online visualization and management tool for customized and annotated phylogenetic trees. Nucleic Acids Res. 44 (W1), W236–W241. doi: 10.1093/nar/gkw370
Hu, B., Jin, J., Guo, A., Zhang, H., Luo, J., Gao, G. (2015). GSDS 2.0: an upgraded gene feature visualization server. Bioinf. (Oxford England) 31 (8), 1296–1297. doi: 10.1093/bioinformatics/btu817
Jung, J. H., Lee, H. J., Ryu, J. Y., Park, C. M. (2016). SPL3/4/5 integrate developmental aging and photoperiodic signals into the FT-FD module in arabidopsis flowering. Mol. Plant 9 (12), 1647–1659. doi: 10.1016/j.molp.2016.10.014
Kang, H. M., Sul, J. H., Service, S. K., Zaitlen, N. A., Kong, S. Y., Freimer, N. B., et al. (2010). Variance component model to account for sample structure in genome-wide association studies. Nat. Genet. 42 (4), 348–354. doi: 10.1038/ng.548
Kim, D., Langmead, B., Salzberg, S. L. (2015). HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 12 (4), 357–360. doi: 10.1038/nmeth.3317
Klaas, V. (2009). Unraveling transcriptional control in arabidopsis using cis-regulatory elements and coexpression networks. Plant Physiol. 2, 535–536. doi: 10.1104/pp.109.136028
Krzywinski, M., Schein, J., Birol, I., Connors, J., Gascoyne, R., Horsman, D., et al. (2009). Circos: an information aesthetic for comparative genomics. Genome Res. 19 (9), 1639–1645. doi: 10.1101/gr.092759.109
Kumar, S., Stecher, G., Tamura, K. (2016). MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33 (7), 1870–1874. doi: 10.1093/molbev/msw054
Larkin, M. (2007). Clustal W and clustal X v. 2.0. Bioinformatics 23 (21), 2947–2948. doi: 10.1093/bioinformatics/btm404
Letunic, I., Khedkar, S., Bork, P. (2020). SMART: recent updates, new developments and status in 2020. Nucleic Acids Res. 49, D458–D460. doi: 10.1093/nar/gkaa937
Li, Z., Liu, D., Xia, Y., Li, Z., Niu, N., Ma, S., et al. (2019). Identification and functional analysis of the CLAVATA3/EMBRYO SURROUNDING REGION (CLE) gene family in wheat. Int. J. Mol. Sci. 20, 4317. doi: 10.3390/ijms20174319
Liu, L., Gallagher, J., Arevalo, E. D., Chen, R., Skopelitis, T., Wu, Q., et al. (2021). Enhancing grain-yield-related traits by CRISPR-Cas9 promoter editing of maize CLE genes. Nat. Plants 7 (3), 287–294. doi: 10.1038/s41477-021-00858-5
Liu, S., Liu, Y., Yang, X., Tong, C., Edwards, D., Parkin, I., et al. (2014). The brassica oleracea genome reveals the asymmetrical evolution of polyploid genomes. Nat. Commun. 5, 3930. doi: 10.1038/ncomms4930
Li, M., Wang, R., Liu, Z., Wu, X., Wang, J. (2019). Genome-wide identification and analysis of the WUSCHEL-related homeobox (WOX) gene family in allotetraploid brassica napus reveals changes in WOX genes during polyploidization. BMC Genomics 20 (1), 317. doi: 10.1186/s12864-019-5684-3
Lu, S., Wang, J., Chitsaz, F., Derbyshire, M., Geer, R., Gonzales, N., et al. (2020). CDD/SPARCLE: the conserved domain database in 2020. Nucleic Acids Res. 48, D265–D268. doi: 10.1093/nar/gkz991
Lysak, M. A., Koch, M. A., Pecinka, A., Schubert, I. (2005). Chromosome triplication found across the tribe brassiceae. Genome Res. 15 (4), 516–525. doi: 10.1101/gr.3531105
Magali, L. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30 (1), 325–327. doi: 10.1093/nar/30.1.325
Nekrutenko, A. (2002). The K A/K s ratio test for assessing the protein-coding potential of genomic regions: An empirical and simulation study. Genome Res. 12 (1), 198–202. doi: 10.1101/gr.200901
Okamoto, S., Shinohara, H., Mori, T., Matsubayashi, Y., Kawaguchi, M. (2013). Root-derived CLE glycopeptides control nodulation by direct binding to HAR1 receptor kinase. Nat. Commun. 4, 2191. doi: 10.1038/ncomms3191
Ostergaard, L., King, G. J. (2008). Standardized gene nomenclature for the brassica genus. Plant Methods 4 (1), 10–10. doi: 10.1186/1746-4811-4-10
Paterson, A., Bowers, J., Chapman, B. (2004). Ancient polyploidization predating divergence of the cereals, and its consequences for comparative genomics. Proc. Natl. Acad. Sci. U. S. A. 101 (26), 9903–9908. doi: 10.1073/pnas.0307901101
Pertea, M., Pertea, G. M., Antonescu, C. M., Chang, T. C., Mendell, J. T., Salzberg, S. L. (2015). StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 33 (3), 290–295. doi: 10.1038/nbt.3122
Rojo, E., Sharma, V., Kovaleva, V., Raikhel, N., Fletcher, J. (2002). CLV3 is localized to the extracellular space, where it activates the arabidopsis CLAVATA stem cell signaling pathway. Plant Cell 14 (5), 969–977. doi: 10.1105/tpc.002196
Schemske, R. D. W. (1998). Pathways, mechanisms, and rates of polyploid formation in flowering plants. Annu. Rev. Ecol. Systematics 29, 467–501. doi: 10.1146/annurev.ecolsys.29.1.467
Schoof, H., Lenhard, M., Haecker, A., Mayer, K. F. X., Jürgens, G., Laux, T. (2000). The stem cell population of arabidopsis shoot meristems is maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell 100 (6), 635–644. doi: 10.1016/S0092-8674(00)80700-X
Schranz, M. E., Lysak, M. A., Mitchell-Olds, T. (2006). The ABC's of comparative genomics in the brassicaceae: building blocks of crucifer genomes. Trends Plant ence 11 (11), 535–542. doi: 10.1016/j.tplants.2006.09.002
Song, X., Guo, P., Ren, S., Xu, T., Liu, C. (2013). Antagonistic peptide technology for functional dissection of CLV3/ESR genes in arabidopsis. Plant Physiol. 161 (3), 1076–1085. doi: 10.1104/pp.112.211029
Takahashi, F., Suzuki, T., Osakabe, Y., Betsuyaku, S., Kondo, Y., Dohmae, N., et al. (2018). A small peptide modulates stomatal control via abscisic acid in long-distance signalling. Nature 556 (7700), 235–238. doi: 10.1038/s41586-018-0009-2
Tatsuhiko, K. (2006). A plant peptide encoded by CLV3 identified by in situ MALDI-TOF MS analysis. Sci. (New York N.Y.) 5788, 845–848. doi: 10.1126/science.1128439
Wahid, S., Xie, M., Sarfraz, S., Liu, J., Zhao, C., Bai, Z., et al. (2022). Genome-wide identification and analysis of Ariadne gene family reveal its genetic effects on agronomic traits of brassica napus. Int. J. Mol. Sci. 23, 6265. doi: 10.3390/ijms23116265
Wang, J., Replogle, A., Hussey, R., Baum, T. (2011). Identification of potential host plant mimics of CLAVATA3ESR (CLE)-like peptides from the plant-parasitic nematode heterodera schachtii. Mol. Plant Pathol. 12 (2), 177–186. doi: 10.1111/j.1364-3703.2010.00660.x
Wang, X., Wu, J., Liang, J., Cheng, F., Wang, X. (2015). Brassica database (BRAD) version 2.0: Integrating and mining brassicaceae species genomic resources. Database J. Biol. Database Curation 2015, bav093. doi: 10.1093/database/bav093
Xie, M., Zuo, R., Bai, Z., Yang, L., Zhao, C., Gao, F., et al. (2022). Genome-wide characterization of Serine/Arginine-rich gene family and its genetic effects on agronomic traits of brassica napus. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.829668
Yang, Y., Zhu, K., Li, H., Han, S., Meng, Q., Khan, S., et al. (2018). Precise editing of CLAVATA genes in brassica napus l. regulates multilocular silique development. Plant Biotechnol. J. 16 (7), 1322–1335. doi: 10.1111/pbi.12872
Ye, C., Wu, D., Mao, L., Jia, L., Qiu, J., Lao, S., et al. (2020). The genomes of the allohexaploid echinochloa crus-galli and its progenitors provide insights into polyploidization-driven adaptation. Mol. Plant 13 (9), 1298–1310. doi: 10.1016/j.molp.2020.07.001
Yuannian, J. (2011). Ancestral polyploidy in seed plants and angiosperms. Nature 7345, 97-100. doi: 10.1038/nature09916
Zhang, L., Wu, S., Chang, X., Wang, X., Zhao, Y., Xia, Y., et al. (2020). The ancient wave of polyploidization events in flowering plants and their facilitated adaptation to environmental stress. Plant Cell Environ. 43 (12), 2847–2856. doi: 10.1111/pce.13898
Zhang, Y., Yang, S., Song, Y., Wang, J. (2014). Genome-wide characterization, expression and functional analysis of CLV3/ESR gene family in tomato. BMC Genomics 15 (1), 827. doi: 10.1186/1471-2164-15-827
Zhao, C., Safdar, L. B., Xie, M., Shi, M., Dong, Z., Yang, L., et al. (2021). Mutation of the PHYTOENE DESATURASE 3 gene causes yellowish-white petals in Brassica napus. Crop J. 9, 1124–1134. doi: 10.1016/j.cj.2020.10.012
Zhao, C., Xie, M., Liang, L., Yang, L., Han, H., Qin, X., et al. (2022). Genome-wide association analysis combined with quantitative trait loci mapping and dynamic transcriptome unveil the genetic control of seed oil content in brassica napus l. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.929197
Keywords: CLE peptide, Brassica napus, diploid progenitor, orthologous pairs, expression pattern, association mapping analysis, yield traits
Citation: Xie M, Zhao C, Song M, Xiang Y and Tong C (2022) Genome-wide identification and comparative analysis of CLE family in rapeseed and its diploid progenitors. Front. Plant Sci. 13:998082. doi: 10.3389/fpls.2022.998082
Received: 19 July 2022; Accepted: 29 September 2022;
Published: 20 October 2022.
Edited by:
Xiangshu Dong, Yunnan University, ChinaCopyright © 2022 Xie, Zhao, Song, Xiang and Tong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yang Xiang, eGlhbmd5YW5nY2VsbEAxMjYuY29t
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.