- 1Institute of Plant Breeding and Biotechnology, Muhammad Nawaz Shareef (MNS) University of Agriculture, Multan, Pakistan
- 2Department of Biosciences, Commission on Science and Technology for Sustainable Development in the South (COMSATS) University Islamabad, Islamabad, Pakistan
- 3Faculty of Agricultural Sciences and Technologies, Sivas University of Science and Technology, Sivas, Turkey
- 4State Key Laboratory of Cotton Biology, Cotton Research Institute of Chinese Academy of Agricultural Sciences, Anyang, China
- 5Department of Biotechnology, Commission on Science and Technology for Sustainable Development in the South (COMSATS), University Islamabad, Abbottabad Campus, Abbottabad, Pakistan
- 6Institute of Food Crops, Yunnan Academy of Agricultural Sciences, Kunming, China
Cysteine-rich poly comb-like protein (CPP) is a member of cysteine-rich transcription factors that regulates plant growth and development. In the present work, we characterized twelve CPP transcription factors encoding genes in soybean (Glycine max). Phylogenetic analyses classified CPP genes into six clades. Sequence logos analyses between G. max and G. soja amino acid residues exhibited high conservation. The presence of growth and stress-related cis-acting elements in the upstream regions of GmCPPs highlight their role in plant development and tolerance against abiotic stress. Ka/Ks levels showed that GmCPPs experienced limited selection pressure with limited functional divergence arising from segmental or whole genome duplication events. By using the PAN-genome of soybean, a single nucleotide polymorphism was identified in GmCPP-6. To perform high throughput genotyping, a kompetitive allele-specific PCR (KASP) marker was developed. Association analyses indicated that GmCPP-6-T allele of GmCPP-6 (in exon region) was associated with higher thousand seed weight under both water regimes (well-water and water-limited). Taken together, these results provide vital information to further decipher the biological functions of CPP genes in soybean molecular breeding.
Introduction
Identification and genome-wide characterization of plant transcription factors (TFs) bear vital significance (Qanmber et al., 2019). In plants, TFs play a central role in various developmental processes as well as a stress response (Green et al., 1987).
Cysteine-rich polycomb-like proteins (CPP-like) belong to a small TFs family characterized by the presence of one or two similar Cys-rich domains known as the CXC domain (also known as the CRC domain), and the TCR motif (Cvitanich et al., 2000; Hauser et al., 2000; Sijacic et al., 2011). Plants and animals contain members of this family, but prokaryotes, yeasts, and fungi lack them. CXC domains of CPP-like proteins are highly conserved in different genera and species (Song et al., 2000; Andersen et al., 2007). CPP-like genes are involved in plant development, and in cell division control. CPP TFs is a small gene family that includes tesmin/TSO1-like CXC (TCX) proteins (Andersen et al., 2007). In many plant species, CPP TFs have been discovered to have a variety of functions. TSO1, the first CPP TFs, were identified and characterized in Arabidopsis thaliana using map-based cloning, and its biological functions were explored through mutant screening (Riechmann et al., 2000). TSO1 gene is mainly expressed in flowers, in developing ovules and microspores. The tso1 mutants show deficiencies in karyokinesis and cytokinesis, as well as a loss of control over directional cellular expansion and coordination of adjacent cell growth (Riechmann et al., 2000; Sijacic et al., 2011).
Many CPP genes have been identified in various plant species, including A. thaliana (Hauser et al., 1998), Oryza sativa (Yang Z. et al., 2008), Zea mays (Song et al., 2016b), and Glycine max (Zhang et al., 2015). Cucumber (Cucumis sativus) plant is susceptible to abiotic stresses due to its high transpiration rate (Zhou et al., 2017). Gene expression of CsCPP genes is upregulated in response to abiotic stresses like salt, cold, drought, and ABA, suggesting that CsCPPs may play a role in abiotic stress responses (Yang et al., 2019).
Soybean (G. max L.) belongs to the leguminous family and is a prominent source of edible oil and is cultivated in different parts of the world (Fehr and Caviness, 1977; Song et al., 2016a). Soybean seed contains 35% protein and 18% oil contents (Wilson, 2004). Abiotic stress factors are major limiting elements affecting its yield and quality. The role of CPP-like protein has been reported in the growth and development of A. thaliana, O. sativa, Z. mays, and C. sativus. Although CPP-like genes have been identified in soybean, more work is required to further decipher their function in G. max. The availability of the soybean pan-genome is expected to pave the way for molecular breeding in soybean (Schmutz et al., 2010). Although molecular markers are available, their deployment in soybean molecular breeding remains limited because of cost ineffectiveness while exploring large populations. Kompetitive Allele Specific PCR (KASP), is a high-throughput and breeder-friendly genotyping platform (Neelam et al., 2013). KASP offers cost-effective genotyping by eliminating the need for post-PCR handling (Majeed et al., 2018).
In this study, we characterized twelve GmCPP genes and performed systematic analyses using genome-wide structure depiction and sequence polymorphism investigations. We analyzed GmCPPs to explore evolutionary relationships, gene structure, conserved motifs, gene duplication, and association of sequence polymorphism with the studied soybean phenotypic traits under well-water (WW) and water limited (WL) conditions. The present work will assist to underpin the evolution of GmCPPs and provide information on GmCPP genes to be used in soybean molecular breeding.
Materials and methods
Sequence identification
The CPP gene and encoded proteins in various species like G. max, O. sativa, Z. mays, A. thaliana, Brassica rapa, G. soja, Cajanus Cajan, Chlamydomonas reinhardtii, and Selaginella moellendorffi were downloaded from plant transcription database.1 To confirm the retrieved CPP proteins, local BLASTp, NCBI Batch CD-search, Interproscan V. 632 and SMART3 were also used. Non-redundant gene members were selected and the rest were excluded for further analyses. Other biophysical characteristics i.e., protein length, molecular weight (MW), isoelectric point (pI), and gravity values for GmCPPs were extracted using ExPASy Protparam Tool.4 Furthermore, sub-cellular localization of GmCPPs was also identified using the Softberry5 and CELLO V2.5 web-tool.6
Sequence alignment and evolutionary analysis
Full-length amino acid sequences of all studied species were aligned and two phylogenetic trees were generated using MEGA X using the maximum likelihood method (ML) following parameters as reported by Kumar et al. (2018). The bootstrap method (1,000 replications) was used to determine the dependability of clades. Graphical representation of multiple sequence alignment of conserved CPP amino acid residues in G. max and G. soja was performed separately by the Clustal W program (Tamura et al., 2011) and WEBLOG webtool.7
Gene structure, protein motif, and cis-element analyses
To explore exon/intron structure, bed-files from databases were obtained and analyzed using GSDS 2.0.8 Protein motif distributions were determined using the online MEME tool.9 For cis-element analyses, ∼2 kb upstream regions were analyzed in the PlantCARE database (Lescot et al., 2002) and the elements were characterized on the basis of their predicted biological functions, and graphical representation was done by using TBtool software.
Gene duplication and synteny analysis
To determine the chromosomal distribution of GmCPPs, extracted gff3-files of soybean genome annotation were downloaded from SoyBase (SoyBase.org). Gene duplication analyses were performed following the methods as reported previously (Yang et al., 2017). CIRCOS was used to create the figure and Ka/Ks values were calculated using PAL2NAL (Suyama et al., 2006; Krzywinski et al., 2009).
Soybean plant material and phenotyping
A set of 46 soybean accessions were planted at MNS University of Agriculture, Multan in the springs of 2021 (2021-UAM). Field experiments were carried out under WL and WW experimental units following Augmented design (Check = UAMSB200). The WL experimental units were subjected to drought especially at the flowering stage, whereas, WW experimental units were irrigated after every fortnight (depending upon water requirement). Each soybean genotype was planted on two beds (length × width = 15 × 2.5 ft) on both sides. Plant-to-plant distance was maintained at a distance of 1 ft. Phenotypic data were recorded for plant height, thousand seed weight, pods–1 plant, seeds–1 pod, seed weight–1 pod, seed length, seed thickness, seed width, and pod length from both water regimes.
Development of single nucleotide polymorphism based kompetitive allele-specific polymerase chain reaction markers for GmCPP
Genomic DNA of the investigated soybean germplasm was extracted from young seedling leaves using the CTAB method (Lecharny et al., 2003). The quality of extracted DNA was initially checked by using NANO-Drop (K5800C Micro-Spectrophotometer) followed by running the extracted DNA on 1.0% agarose gel.
The whole genome sequence of three cultivars of soybean (Williams-82, Lee, and Zhonghuang-13) was downloaded from SoyBase.10 Local BLAST was performed to identify the sequences of GmCPPs in the aforementioned soybean genotypes. For the identification of sequence polymorphism, multiple sequence alignment was performed using the Seqman program in the DNASTAR Lasergene package. Standard kompetitive allele-specific PCR (KASP) guidelines11 were followed for the development of KASP primers on the identified single nucleotide polymorphism (SNP) of GmCPP6. Allele-specific primers were developed having standard HEX and FAM tails with a targeted SNP at three prime ends. Two reverse primers (allele-specific) and one common forward primer were designed so that the total fragment length was less than 100 bp. The standard KASP reaction mixture, KASP assay, and PCR conditions were followed as reported by Rasheed et al. (2016), Majeed et al. (2018), Ur Rehman et al. (2019, 2021), and Irshad et al. (2019, 2021).
Statistical analyses
Phenotypic data were analyzed with XLSTAT Software 2014. Student’s t-test at p less than 0.05 was used to check the effect of each allelic variation on the recorded phenotypic traits.
Results
Identification of cysteine-rich polycomb-like protein gene family members in different species
We identified a total of 81 CPP genes in nine investigated species including chlorophytes (C. reinhardtii), lycophytes (S. moellendorffii), Brassicaceae (A. thaliana and B. rapa), Fabaceae (G. max, G. soja, and C. cajan), and Poaceae (Z. mays and O. sativa). Among these, 12 CPP genes were shortlisted in G. max, 10 in G. soja, 16 in B. rapa, 11 each in Z. mays and O. sativa, eight in A. thaliana, six in C. cajan, four in S. meollendorffii, and three in C. reinhardtii. A higher number of CPPs were identified in G. max as compared to chlorophytes and lycophytes indicating a duplication effect on GmCPPs in G. max. These findings also signify that CPPs experienced extension in higher plants. The transcription factor ID, taxonomic ID, and predict sub-cellular localization are presented in Supplementary Table 1. These results showed that the GmCPP coding sequence ranged from 1,656 to 2,715 bp for GmCPP-6 and GmCPP-11, respectively. Similarly, an amino acid number of GmCPP genes ranged from 483 to 904 for GmCPP-5 and GmCPP-11, respectively. Molecular weight ranged from 54,034.67 to 98,476.32 kDa for GmCPP12 and GmCPP-4, respectively. The isoelectric point of GmCPP4 was the highest (9.2) and that of GmCPP-1 was the lowest (5.41). The grand averages of hydropathicity values of all GmCPPs were less than zero and ranged from -0.734 for GmCPP-11 to -0.511 for GmCPP-4. In addition, all GmCPPs are localized in the nucleus, except GmCPP-1 and 10.
Phylogenetic analyses of cysteine-rich polycomb-like protein gene family
The phylostratum analyses of CPP genes identified the primitive lineage as CPP genes, which were also identified in chlorophyte (C. reinhardtii) (Figure 1). Further, the CPP genes were identified in lychophytes, dicots, and monocots. These outcomes signified that CPPs originated from early plants phylostratum and possible orthologs are present throughout kingdom Plantae. An evolutionary tree was generated to determine the phylogenetic relationship among the studied CPPs. To indicate the CPPs from C. reinhardtii, S. moellendorffii, A. thaliana, B. rapa, G. max, G. soja, C. cajan, Z. mays, and O. sativa the prefixes Cr, Sm, At, Br, Gm, Gs, Cc, Zm, and Os were used correspondingly. The phylogenetic analyses divided 81 genes into six clades based on sequence similarities (Figure 1). Clade-I comprised 16 members, Clade-II possessed 13 members, Clade-III contained 14 members, Clade-IV, and V had 15 members each, and Clade-VI contained eight members. Clade-I lacks genes from chlorophytes which suggested the evolution of CPP genes after the split of chlorophyte. Interestingly, CPP genes from monocot and dicot species were unsystematically distributed to all clades. Further, phylogenetic analyses indicated that G. max and B. rapa experienced gene family expansion since both have more CPP genes compared to other studied organisms.
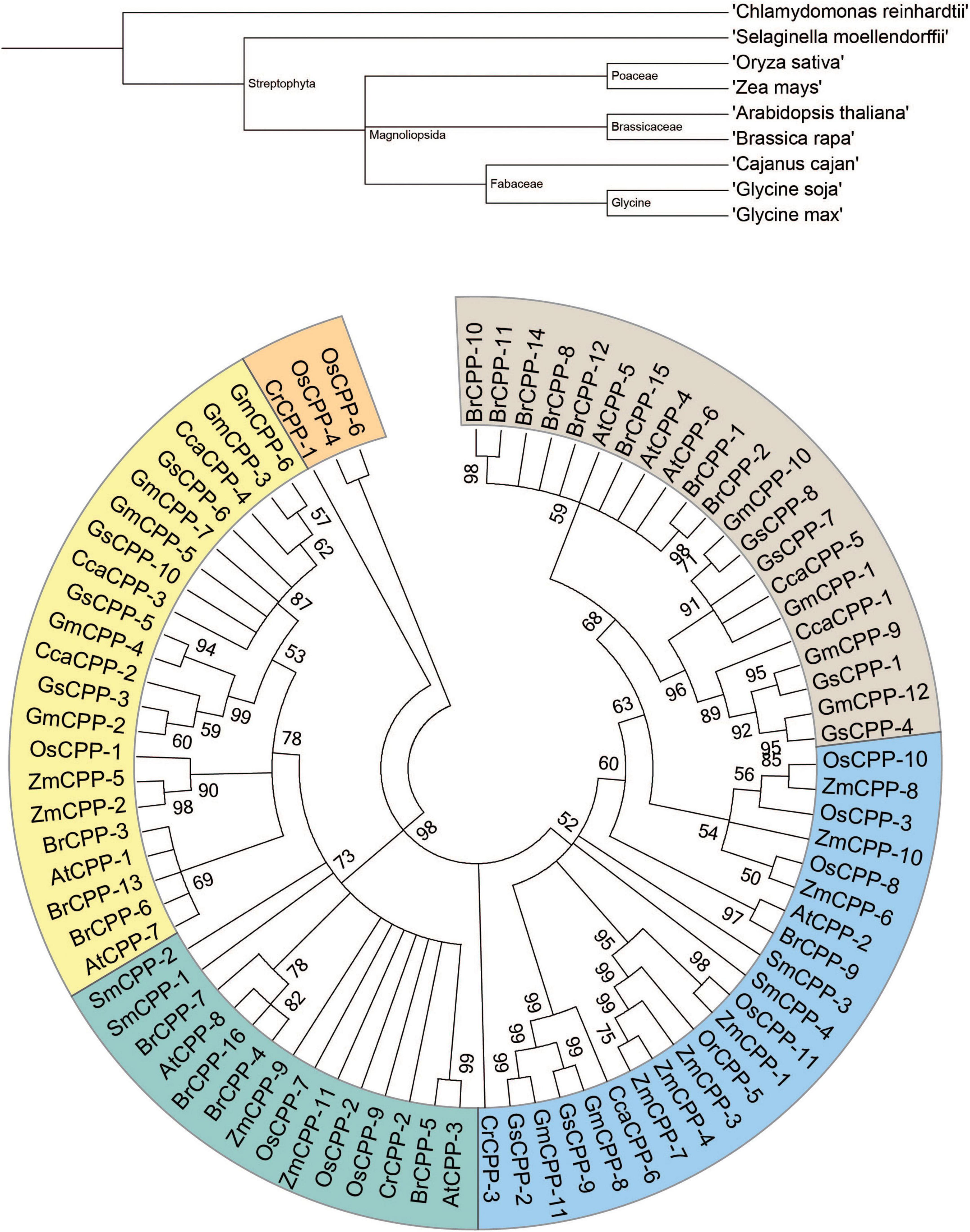
Figure 1. Phylogenetic tree of GmCPPs from nine different species. Phylostartum analyses of CPP gene family (Upper portion). Phylogenetic and evolutionary relationship of CPP gene family in soybean and other plant species (Lower portion).
To explore the conservation of each amino acid residue in GmCPP and GsCPP, multiple sequence alignment was executed to generate sequence logos in G. max and G. soja. The outcomes showed that the amino acid residue distribution was highly similar at most of the loci among the G. max and G. soja. For example, some amino acid residues such as C [6], L [7], Y [8], C [9], C [11], F [12], A [13], N [29], A [34], and so on were found to be highly conserved (Figure 2). Phylogenetic analyses also highlight that GmCPP and GsCPP members lie in close proximity to the evolutionary tree.
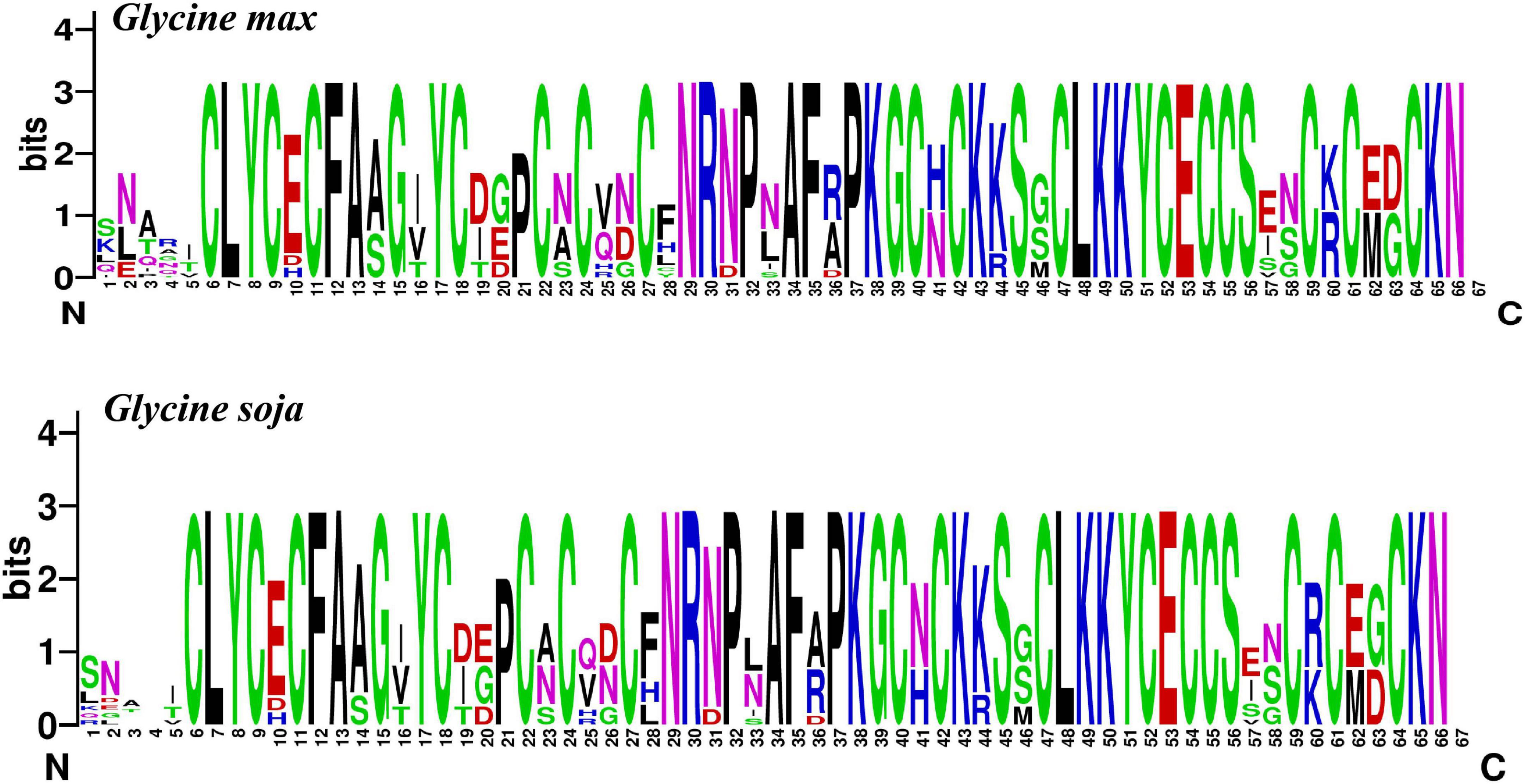
Figure 2. Sequence logo of GmCPP and GsCPP. Amino acid residues shared by two plant species are highly conserved. Each black letter showed the conserved amino acids at a given location.
Gene structure, protein motif, and cis-acting element analysis
It has been well-documented that intron-exon distribution arrangement in a gene is related to its biological function. The intron number in GmCPP ranged from 7 to 9 (Figure 3). Conserved domains in each sequence were identified using the CDD tool of NCBI.12 All members of the GmCPP gene family contain the TCR domain (Supplementary Figure 1). MEME tool was used to explore the conserved motif distributions of GmCPPs. The outcomes indicated that most of the GmCPP proteins exhibited similar motif distribution patterns such as motifs one, two, and eight exist in almost all proteins (Figure 4). We also identified cis-acting elements in the upstream regions of GmCPPs and grouped them on the basis of their functional relevance. All GmCPPs had cis-acting elements related to plant development, stress, and light responses (Supplementary Table 2).
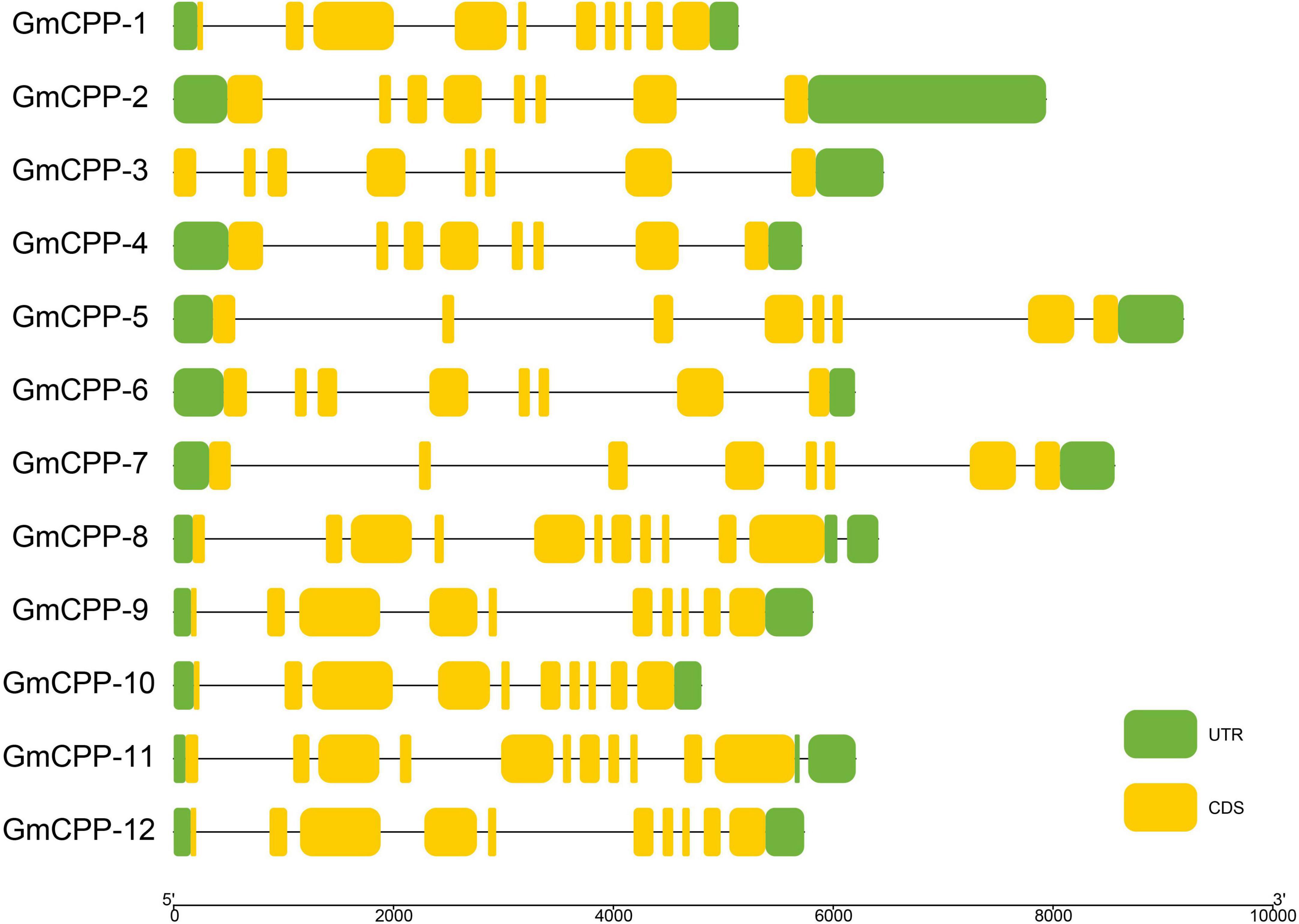
Figure 3. Gene structure display of GmCPP. Exon/intron structure display of GmCPP genes; Green color shows the upstream and downstream region, yellow color shows exons, black lines show introns.
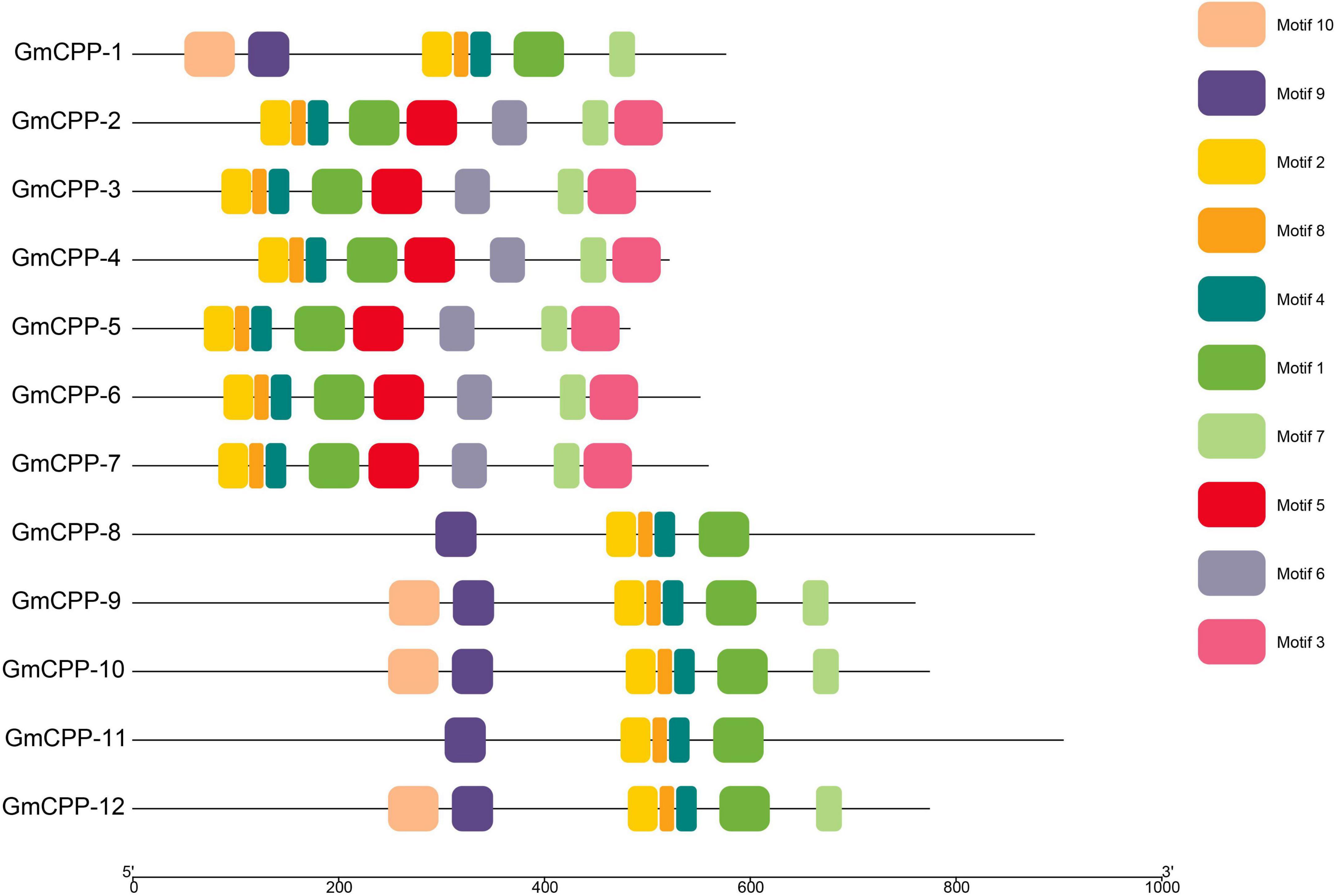
Figure 4. Motifs in all GmCPP genes. Distribution of conserved motifs in GmCPP are presented in different colors.
Chromosomal distribution, gene duplication, and synteny analyses
The 12 GmCPP genes are scattered on five chromosomes, including three of each gene on chromosomes one and four, respectively. Two each gene are on chromosomes five and 10 while one gene is present on chromosome seven (Supplementary Table 3). To investigate the relationship of gene pairs, we explored the gene locus on a chromosome and executed synteny analyses. Their, synteny analyses showed that GmCPP genes were highly conserved among five chromosomes (Figure 5). Whole genome duplication, segmental duplication, and tandem duplication play a vital role in the extension of a gene family (Yang et al., 2017). To investigate the expansion of the GmCPP family in soybean, we executed gene duplication analyses in the soybean genome (Supplementary Table 3). Out of all studied gene pairs, 10 gene pairs were attributed to segmental duplication. We also explored the non-synonymous divergence (Ka) versus synonymous (Ks) values for the GmCPP gene pairs. It was found that nine duplicated gene pairs showed Ka/Ks values <0.5, whereas, two duplicated gene pairs showed Ka/Ks values between 0.5 and 1.0 (Supplementary Table 3). Generally, Ka/Ks of the studied gene pairs were <1, showing that the GmCPP gene family experienced purifying selection pressure with restricted functional differences.
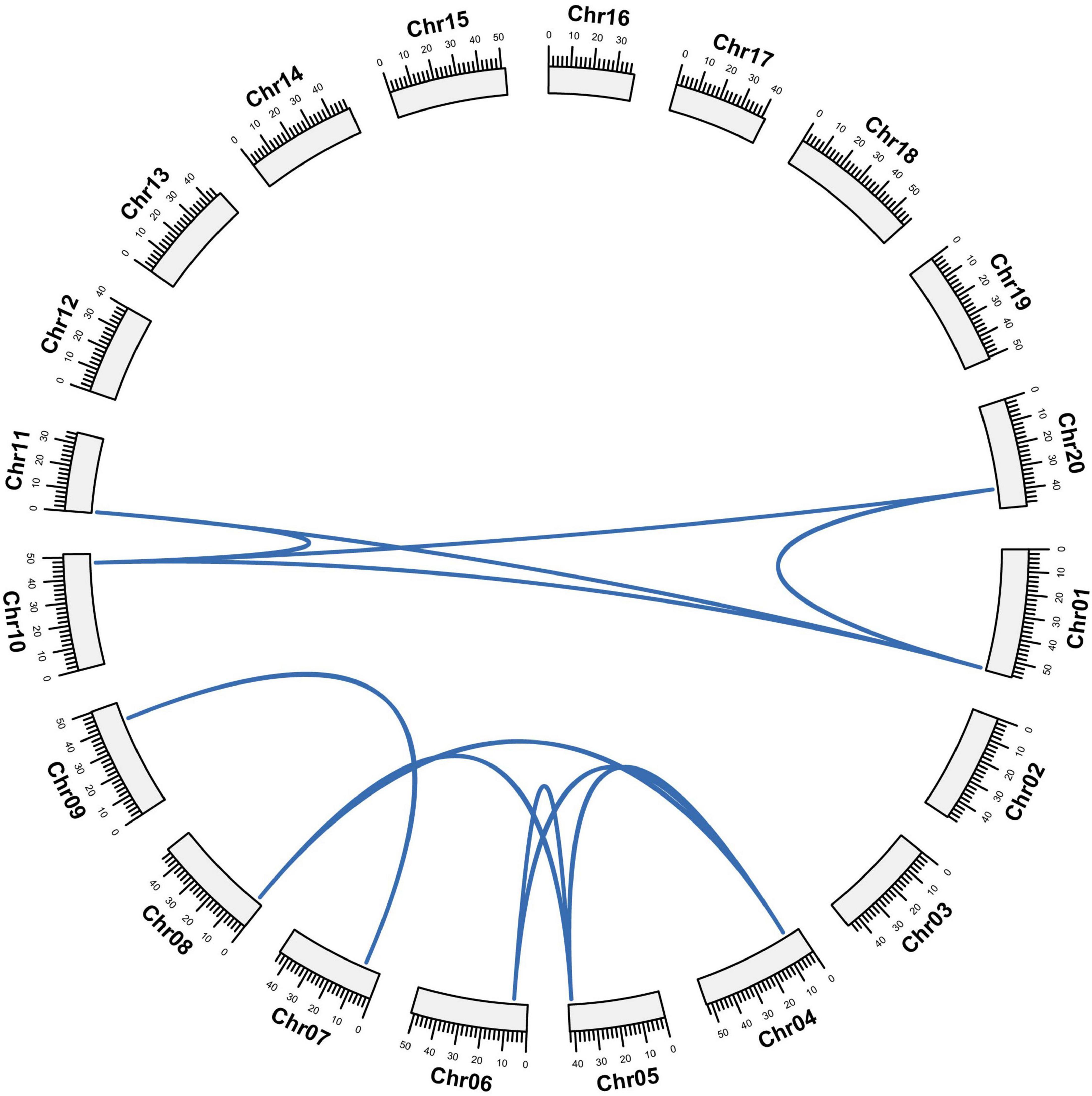
Figure 5. Gene duplication and synteny analysis. Gene duplication among GmCPP genes. Blue line shows orthologous/paralogous pair. Gm01 to Gm20 shows the chromosomes of Glycine max.
Marker trait association analysis
For GmCPP-6, a polymorphic site was identified in the coding region. KASP marker was developed at the SNP site. KASP assay results showed that soybean accession having HEX tail has “C” allele while accession having FAM tail has “T” type allele (Figure 6).
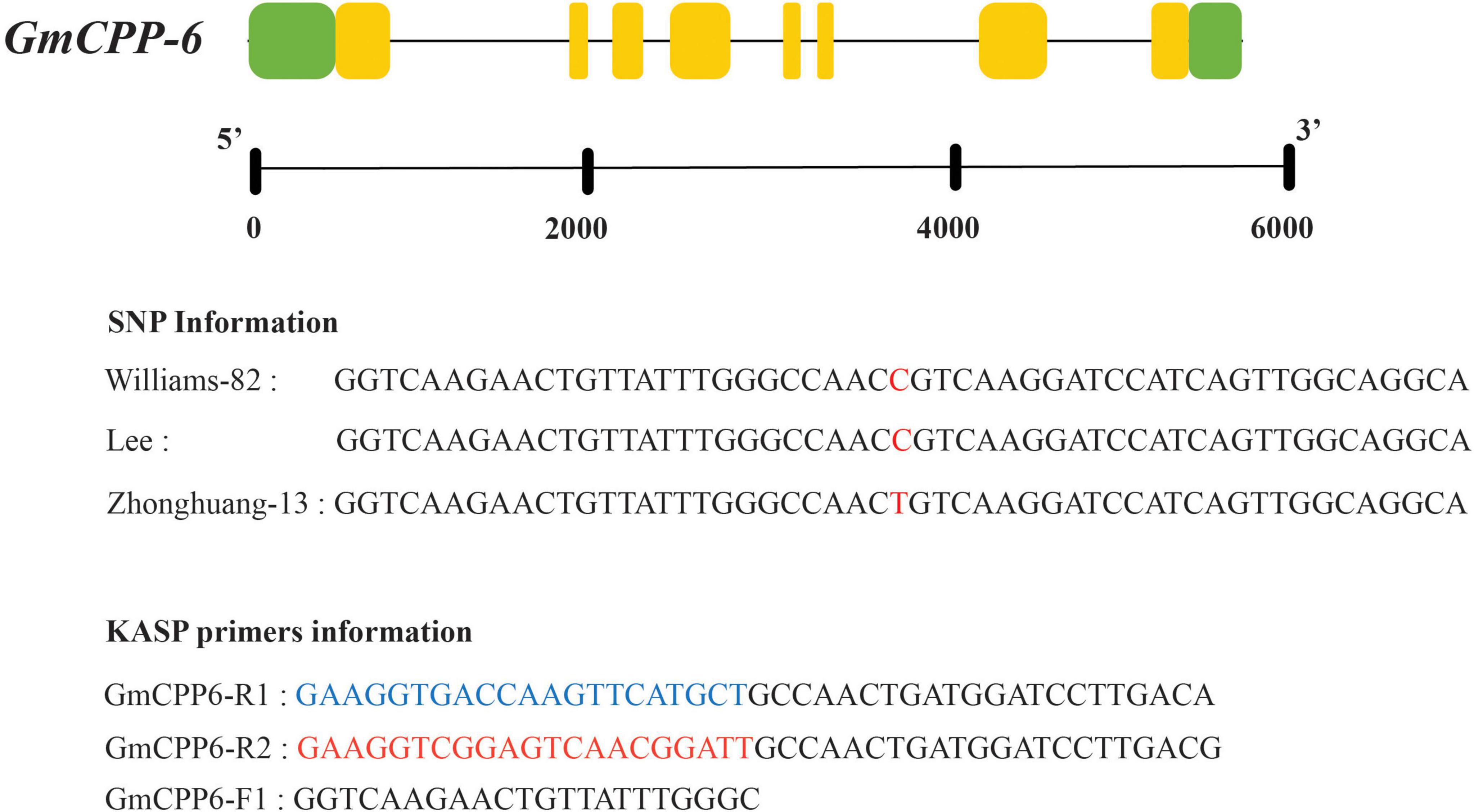
Figure 6. Gene structure and KASP marker development for GmCPP-6. Red colored alphabets indicated sequence polymorphism. Nucleotides color blue indicating FAM where nucleotides colored red indicating HEX.
Two allelic variations of GmCPP-6, i.e., GmCPP-6-T and GmCPP-6-C were identified in the studied soybean germplasm. GmCPP-6-C was the most frequently occurring allelic variation available in 58.6% of studied soybean accessions. Marker trait association analyses exhibited that at unique field sites, GmCPP-6-T was associated with higher thousand seed weight in both environments (Figure 7).
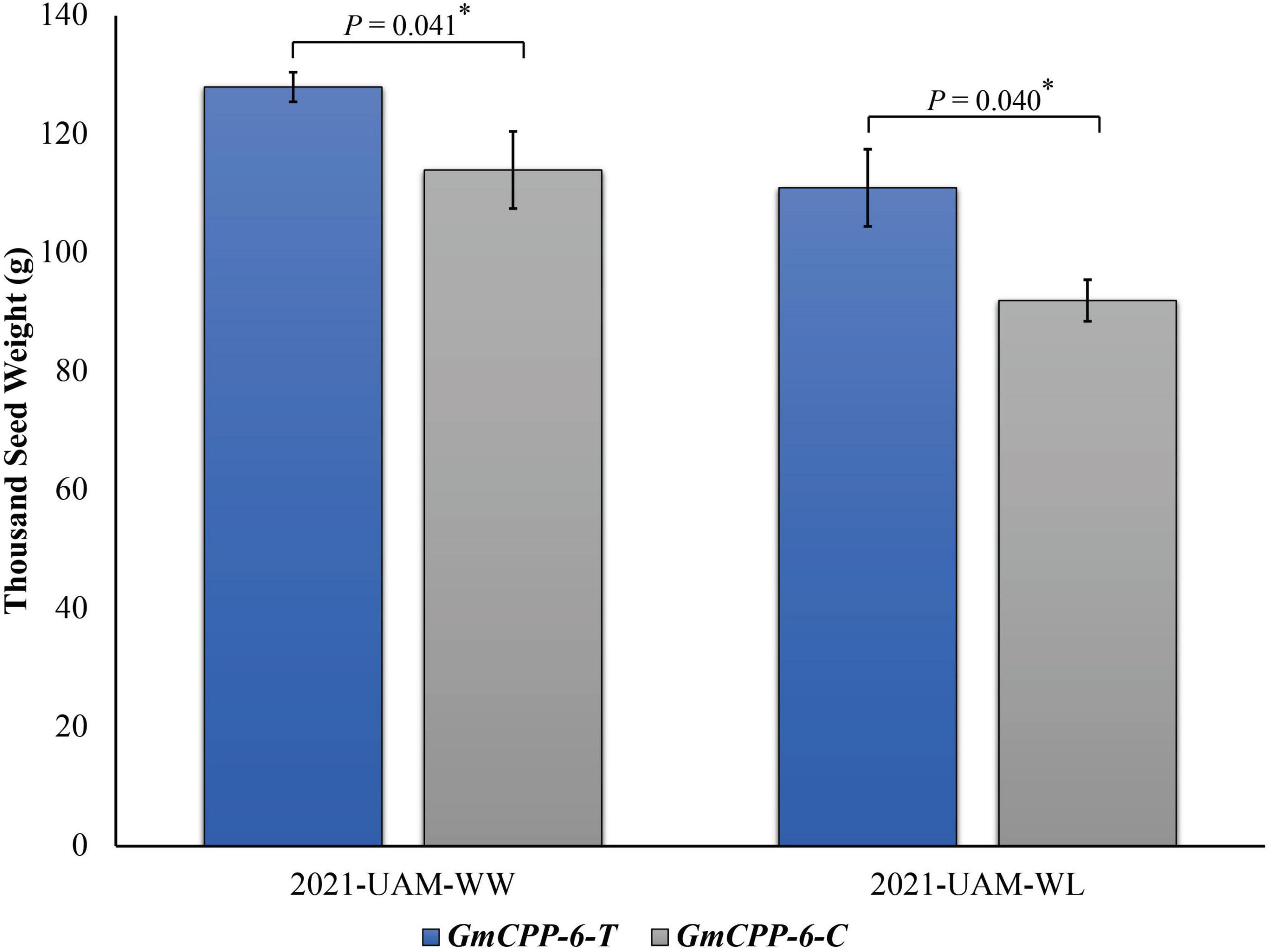
Figure 7. Phenotypic comparison of GmCPP-6 allelic variations under well water (WW) and water limited (WL) conditions. The two environments were at University of Agriculture Multan (UAM) under WW and WL conditions in year 2021. *p less than 0.05. Error bars denote standard error.
Discussion
Cysteine-rich polycomb-like protein TFs are quite a small gene family involved in plant growth and stress responses (Andersen et al., 2007; Lu et al., 2013; Zhang et al., 2015; Zhou et al., 2018; Nan et al., 2021). In earlier studies, identification of the CPP gene family in Camellia sinensis, A. thaliana, C. sativus, and G. max has been performed. But genome-wide characterization, in relation to analyses of sequence polymorphism, has not been performed in soybean. In the present work, a comprehensive identification, characterization, and analysis of sequence polymorphism of GmCPPs was performed.
Soybean cysteine-rich polycomb-like proteins are conserved during evolution
In the present work, we identified 81 CPP genes in nine different organisms i.e., chlorophytes (C. reinhardtii), lycophytes (S. moellendorffii), Brassicaceae (A. thaliana and B. rapa), Fabaceae (G. max, G. soja, and C. cajan), and Poaceae (Z. mays and O. sativa). Phylostratum analyses of CPPs indicated that the primitive plant pedigree as CPPs were existing in chlorophytes, showing that CPP genes originated from early land plants and ortholog genes of CPP are existing across kingdom Plantae. Phylogenetic analyses were performed to establish the evolutionary relationship among the studied species. All CPP genes were divided into six different clades which showed that most of the G. max genes exhibited a close relationship with G. soja genes and indicated that both species share a common ancestry. G. soja genome has shown to have 0.9154 GB consensus sequences, covering ∼98% of G. max genome sequence (Schmutz et al., 2010). Gene structure analyses showed that GmCPPs have a higher number of intron which indicates that GmCPP belong to the primitive gene family group. Sequence logos for conserved amino acid residues were also highly conserved in G. max and G. soja. Both N and C terminals of GmCPPs and GsCPPs are conserved. These results indicate that the GmCPP and GsCPP genes are evolutionarily conserved which might be helpful to underpin the pattern of CPP protein sequence conservation in other members of kingdom Plantae.
Biophysical characteristics
The estimates of biophysical parameters of all GmCPP gene family members delivered helpful information. Biophysical properties predicted that 10 out of 12 GmCPPs were positioned in the nucleus. The values of pI and grand average of hydropathicity of all GmCPPs indicated that all CPP proteins were hydrophilic (<0) and alkaline (∼7) (Supplementary Table 4).
Exon-intron and motif analyses
The structure of the gene is a vital component that might be contributed by deletion and/or insertion incidents (Lecharny et al., 2003). In past, genome-wide studies have demonstrated that the loss or gain of introns during eukaryotic divergence was widespread (Rogozin et al., 2003; Roy and Penny, 2007). Gene structure analyses showed that all GmCPP genes have varied intron lengths that might play crucial roles in the functional divergence of GmCPP genes. It has been well-documented that introns play an important role in the evolution of different species (William Roy and Gilbert, 2006). In the current study, we observed that the number of intron for GmCPPs ranged from seven to nine indicating that G. max has evolved a long time ago (>Million years). Roy and Gilbert also advocated that earlier evolved species have more introns as compared with the newly evolved species (William Roy and Gilbert, 2006). Ten motifs were identified which showed that CPP proteins might function in different biological pathways allied with other co-factors. The motif distribution pattern of CPP proteins indicated that the distribution was relatively conserved and minimal divergence among the proteins from different groups might be linked with the specific biological function associated with soybean development and stress tolerance.
Transcription is governed by the binding of TFs to promoter cis-acting regulatory elements. Various studies have reported the crucial role of cis-acting elements in the processes of plant growth and stress responses (Fankhauser and Chory, 1997). In this study, cis-acting elements related to plant development and stress responses were identified in the upstream region of GmCPPs.
Gene duplication and selection pressure
The uneven distribution of GmCPP genes on chromosomes of G. max shows probable gene loss or addition through whole genome or segmental duplication incidents. It has been reported that gene duplication and divergence generally lead toward evolution (Chothia et al., 2003). Gene duplication creates functional differences, which is crucial for speciation and adaptableness in changing environmental conditions (Conant and Wolfe, 2008). Gene duplication indicates that the aligned sequences share > 70% similarity and coverage length > 80% of the entire length (Yang S. et al., 2008). The two duplicated genes present on the different chromosomes of the same sub-genome might be the consequence of segmental or whole genome duplication, whereas, their presence on the same chromosomes might be the consequence of tandem duplication (He et al., 2012). Tandemly duplicated genes tend to be positioned together on chromosomes whereas, in segmental or whole genome duplication, the duplicated genes are generally distributed throughout the genome (Schauser et al., 2005). Approximately 65 million years ago whole genome and segmental duplications in primitive plant species contributed to the expansion of a number of gene families (Barakat et al., 2009; Wang et al., 2013) and contributed genomic complexity to kingdom Plantae (Cannon et al., 2004).
In the present work, we characterized 12 GmCPP genes, three times the number of CPPs present in chlorophytes, which indicates that CPP experienced expansion during their evolution. As reported previously, expansion in the genome permitted many crop plant species to acclimatize to environmental conditions (Ramsey and Schemske, 1998). We noticed that segmental and whole genome duplication were the major reasons responsible for the expansion CPP gene family in G. max. Segmental type duplication is the main contributor during evolution and it has happened in numerous plant genomes which contain many duplicated chromosomal blocks (Cannon et al., 2004). For instance, many A. thaliana gene families experienced evolutionary dynamics that led toward gene family expansion (Baumberger et al., 2003; Wang et al., 2008). Our results showed that GmCPPs grouped into pairs (Segmental duplication) which shows an ancient expansion in the gene family in G. max. To estimate the selection and environmental pressure, non-synonymous (Ka) and synonymous (Ks) rates of substitution (Ka/Ks) were computed. We noticed that Ka/Ks values of GmCPP genes were <1 illustrating that GmCPP gene family experienced strong purifying selection pressure.
Allelic variations influencing seed weight
Soybean PAN-genome might be helpful for bridging the phenotype to genotype gap in soybean breeding. Recently, PAN-genome has been used to discover genes for flowering time in G. soja (Li et al., 2014). Marker-assisted selection of elite alleles in breeding programs is vital for ongoing soybean breeding. The utilization of elite allelic variations in cultivars can be enriched if effective molecular platforms are available (Rasheed et al., 2017; Majeed et al., 2018; Ur Rehman et al., 2021). In this study, we used the PAN-genome of G. max to explore sequence polymorphism for GmCPP-6. Although, sequence polymorphism was explored in all studied GmCPP genes the allelic variation was only identified in the CDS region of GmCPP-6. Hence, all other genes were excluded for marker-trait association analyses. The absence of polymorphism in all other GmCPP genes is possibly due to allele fixation during evolution or because of the lower number of G. max accessions available for PAN-genomics studies. More work is required for further confirmation or to investigate these two possibilities. Converting sequence polymorphism to gel-free (KASP) markers enable SNPs to be more efficiently applied in selecting desirable alleles in marker-assisted breeding. Moreover, the KASP assay procedure is cost-effective. In the current study, soybean accessions having GmCPP-6-T had higher thousand seed weight under both environmental conditions i.e., WW and WL. Moreover, RNA-Seq Atlas of G. max also reported higher expression of GmCPP-6 in seeds (14–25 days after fertilization) (Severin et al., 2010). Generally, yield-related parameters of crop plants are administered by several genes and are strongly influenced by external stimuli. Pyramiding of favorable alleles might be helpful for continued improvement in soybean. The developed molecular marker will be useful for marker-assisted breeding in soybean which can be used in combination with other molecular markers.
Conclusion
Eighty-one CPP genes were studied in this research, and on the basis of phylogenetic analyses, all genes were divided into six sub-groups. The amino acid residues of G. max and G. soja demonstrated less conservation in web logos. Introns are present in GmCPP genes, and the pattern of protein motif distribution is less consistent across all proteins. Growth regulator cis-acting elements were found in the upstream regions of GmCPP, indicating their role in plant growth and development. Gene duplication and synteny analysis revealed that the GmCPP genes have undergone segmental and whole genome duplication during evolution, resulting in a significant expansion of the GmCPP. The current study also delivers molecular marker associated with higher thousand seed weight in soybean. These findings lay the groundwork for further research into the roles of GmCPP genes in soybean growth, development, and response to external stimuli.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number (s) can be found in the article/Supplementary material.
Author contributions
TN, MT, SI, and SU conceived the idea. TN performed the experiment, analyzed the data, and wrote the original draft of the manuscript. MT, SI, MS, GQ, ZK, ZZ, and ZG guided in the execution of field and laboratory experiments. ZK, ZZ, and ZG assisted in the development of molecular markers. MT, SI, AB, and SU reviewed the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This research was funded by the Key Technology and Science Promotion Cooperation Base on Whole Industrial Chain of Bean, Project number: GHJD-2020025 and Punjab Agricultural Research Board, Project number: PARB-830.
Acknowledgments
The authors are grateful to the Director of University Farms Abdul Ghaffar and Deputy Director University Farms, Mr. Mahmood Alam Khan of MNSUAM for providing the facility to conduct field research. The authors are also grateful to the Graduate Resource Center of MNSUAM for providing training on the construction of high-resolution images for publication.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.996265/full#supplementary-material
Supplementary Figure 1 Showing conserved domain in all GmCPP genes. Conserved domain in GmCPP genes. Yellow color shows TCR family domains.
Supplementary Table 1 Transcription factor ID, taxonomic ID and predict sub-cellular localization.
Supplementary Table 2 cis-acting elements related to plant development, stress, and light responses.
Supplementary Table 3 Gene duplication, collinearity/synteny, and Ka/Ks values.
Supplementary Table 4 Biophysical properties of GmCPP.
Footnotes
- ^ http://planttfdb.gao-lab.org/link.php
- ^ http://www.ebi.ac.uk/InterProScan/
- ^ http://smart.embl-heidelberg.de/
- ^ http://us.expasy.org/tools/protparam.html
- ^ http://www.softberry.com/
- ^ https://mybiosoftware.com/cello-v-2-5-subcellular-localization-predictor.html
- ^ https://weblogo.berkeley.edu/logo.cgi
- ^ http://gsds.gao-lab.org/
- ^ http://memesuite.org
- ^ https://bar.utoronto.ca/eplant/
- ^ http://www.lgcgenomics.com
- ^ https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi
References
Andersen, S. U., Algreen-Petersen, R. G., Hoedl, M., Jurkiewicz, A., Cvitanich, C., Braunschweig, U., et al. (2007). The conserved cysteine-rich domain of a tesmin/TSO1-like protein binds zinc in vitro and TSO1 is required for both male and female fertility in Arabidopsis thaliana. J. Exp. Bot. 58, 3657–70. doi: 10.1093/jxb/erm215
Barakat, A., Bagniewska-Zadworna, A., Choi, A., Plakkat, U., DiLoreto, D. S., Yellanki, P., et al. (2009). The cinnamyl alcohol dehydrogenase gene family in Populus: Phylogeny, organization, and expression. BMC Plant Biol. 9:26. doi: 10.1186/1471-2229-9-26
Baumberger, N., Doesseger, B., Guyot, R., Diet, A., Parsons, R. L., Clark, M. A., et al. (2003). Whole-genome comparison of leucine-rich repeat extensins in Arabidopsis and rice. A conserved family of cell wall proteins form a vegetative and a reproductive clade. Plant Physiol. 131, 1313–1326. doi: 10.1104/pp.102.014928
Cannon, S. B., Mitra, A., Baumgarten, A., Young, N. D., and May, G. (2004). The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 4:10. doi: 10.1186/1471-2229-4-10
Chothia, C., Gough, J., Vogel, C., and Teichmann, S. A. (2003). Evolution of the protein repertoire. Science 300, 1701–1703.
Conant, G. C., and Wolfe, K. H. (2008). Turning a hobby into a job: How duplicated genes find new functions. Nat. Rev. Genet. 9, 938–950. doi: 10.1038/nrg2482
Cvitanich, C., Pallisgaard, N., Nielsen, K. A., Hansen, A. C., Larsen, K., Pihakaski-Maunsbach, K., et al. (2000). CPP1, a DNA-binding protein involved in the expression of a soybean leghemoglobin c3 gene. Proc. Natl. Acad. Sci. U.S.A. 97, 8163–8168. doi: 10.1073/pnas.090468497
Fankhauser, C., and Chory, J. (1997). Light control of plant development. Annu. Rev. Cell Dev. Biol. 13, 203–229.
Green, P. J., Kay, S. A., and Chua, N. H. (1987). Sequence-specific interactions of a pea nuclear factor with light responsive elements upstream of the rbcS-3A gene. EMBO J. 6, 2543–2549.
Hauser, B. A., He, J. Q., Park, S. O., and Gasser, C. S. (2000). TSO1 is a novel protein that modulates cytokinesis and cell expansion in Arabidopsis. Development 127, 2219–2226. doi: 10.1242/dev.127.10.2219
Hauser, B. A., Villanueva, J. M., and Gasser, C. S. (1998). Arabidopsis TSO1 regulates directional processes in cells during floral organogenesis. Genetics 150, 411–423. doi: 10.1093/genetics/150.1.411
He, H., Dong, Q., Shao, Y., Jiang, H., Zhu, S., Cheng, B., et al. (2012). Genome-wide survey and characterization of the WRKY gene family in Populus trichocarpa. Plant Cell Rep. 31, 1199–1217. doi: 10.1007/s00299-012-1241-0
Irshad, A., Guo, H., Rehman, S. U., Wang, X., Gu, Y., Xiong, H., et al. (2021). Identification of single nucleotide polymorphism in TaSBEIII and development of KASP Marker associated with grain weight in wheat. Front. Genet. 12:697294. doi: 10.3389/fgene.2021.697294
Irshad, A., Guo, H., Zhang, S., Gu, J., Zhao, L., Xie, Y., et al. (2019). EcoTILLING reveals natural allelic variations in starch synthesis key gene TaSSIV and its haplotypes associated with higher thousand grain weight. Genes 10:307. doi: 10.3390/genes10040307
Krzywinski, M., Schein, J., Birol, I., Connors, J., Gascoyne, R., Horsman, D., et al. (2009). Circos: An information aesthetic for comparative genomics. Genome Res. 19, 1639–1645. doi: 10.1101/gr.092759.109
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35:1547. doi: 10.1093/molbev/msy096
Lecharny, A., Boudet, N., Gy, I., Aubourg, S., and Kreis, M. (2003). Introns in, introns out in plant gene families: A genomic approach of the dynamics of gene structure. J. Struct. Funct. Genom. 3, 111–116.
Lescot, M., Dehais, P., Thijis, G., Marchal, K., Moreau, Y., Van de Peer, Y., et al. (2002). PlantCare a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acid Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Li, Y. H., Zhou, G., Ma, J., Jiang, W., Jin, L. G., Zhang, Z., et al. (2014). De novo assembly of soybean wild relatives for pan-genome analysis of diversity and agronomic traits. Nat. Biotechnol. 32, 1045–1052. doi: 10.1038/nbt.2979
Lu, T., Dou, Y., and Zhang, C. (2013). Fuzzy clustering of CPP family in plants with evolution and interaction analyses. BMC Bioinform. 14:S10. doi: 10.1186/1471-2105-14-S13-S10
Majeed, U., Darwish, E., Rehman, S. U., and Zhang, X. (2018). Kompetitive allele specific PCR (KASP) a singleplex genotyping platform and its application. J. Agric. Sci. 11, 11–20.
Nan, H., Lin, Y., Wang, X., and Gao, L. (2021). Comprehensive genomic analysis and expression profiling of Cysteine-rich Polycomb-like transcription factor gene family in tea tree. Hortic. Plant J. 7, 469–478.
Neelam, K., Brown-Guedira, G., and Huang, L. (2013). Development and validation of a breeder-friendly KASPar marker for wheat leaf rust resistance locus Lr21. Mol. Breed. 31, 233–237.
Qanmber, G., Liu, J., Yu, D., Liu, Z., Lu, L., Mo, H., et al. (2019). Genome-wide identification and characterization of the PERK gene family in Gossypium hirsutum reveals gene duplication and functional divergence. Int. J. Mol. Sci. 20:1750. doi: 10.3390/ijms20071750
Ramsey, J., and Schemske, D. W. (1998). Pathways, mechanisms, and rates of polyploid formation in flowering plants. Annu. Rev. Ecol. Evol. Syst. 29, 467–501.
Rasheed, A., Hao, Y., Xia, X., Khan, A., Xu, Y., Varshney, R. K., et al. (2017). Crop breeding chips and genotyping platforms: Progress, challenges, and perspectives. Mol. Plant 10, 1047–1064. doi: 10.1016/j.molp.2017.06.008
Rasheed, A., Weie, W., Fengmei, G., Shengnan, Z., Hui, J., Jindong, L., et al. (2016). Development and validation of KASP assays for genes underpinning key economic traits in bread wheat. Theor. Appl. Genet. 10, 1843–1860. doi: 10.1007/s00122-016-2743-x
Riechmann, J. L., Heard, J., Martin, G., Reuber, L., Jiang, C. Z., Keddie, J., et al. (2000). Arabidopsis transcription factors: Genome-wide comparative analysis among eukaryotes. Science 290, 2105–2110. doi: 10.1126/science.290.5499.2105
Rogozin, I. B., Wolf, Y. I., Sorokin, A. V., Mirkin, B. G., and Koonin, E. V. (2003). Remarkable interkingdom conservation of intron positions and massive, lineage-specific intron loss and gain in eukaryotic evolution. Curr. Biol. 13, 1512–1517. doi: 10.1016/s0960-9822(03)00558-x
Roy, S. W., and Penny, D. (2007). Patterns of intron loss and gain in plants: Intron loss–dominated evolution and genome-wide comparison of O. sativa and A. thaliana. Mol. Biol. Evol. 24, 171–181. doi: 10.1093/molbev/msl159
Schauser, L., Wieloch, W., and Stougaard, J. (2005). Evolution of NIN-like proteins in Arabidopsis, rice, and Lotus japonicus. J. Mol. Evol. 60, 229–237.
Schmutz, J., Cannon, S. B., Schlueter, J., Ma, J., Mitros, T., Nelson, W., et al. (2010). Genome sequence of the paleopolyploid soybean. Nature 463, 178–183.
Severin, A. J., Woody, J. L., Bolo, Y. T., Joseph, B., Diers, B. W., Farmer, A. D., et al. (2010). RNA-Seq Atlas of Glycine max: A guide to soybean transcriptome. BMC Plant Biol. 10:160. doi: 10.1186/1471-2229-10-160
Sijacic, P., Wang, W., and Liu, Z. (2011). Recessive antimorphic alleles overcome functionally redundant loci to reveal TSO1 function in Arabidopsis flowers and meristems. PLoS Genet. 7:e1002352. doi: 10.1371/journal.pgen.1002352
Song, J. Y., Leung, T., Ehler, L. K., Wang, C. Z., and Liu, Z. (2000). Regulation of meristem organization and cell division by TSO1, an Arabidopsis gene with cysteine-rich repeats. Development 127, 2207–2217. doi: 10.1242/dev.127.10.2207
Song, L., Nguyen, N., Deshmukh, R. K., Patil, G. B., Prince, S. J., Valliyodan, B., et al. (2016a). Soybean TIP gene family analysis and characterization of GmTIP1; 5 and GmTIP2; 5 water transport activity. Front. Plant Sci. 7:1564. doi: 10.3389/fpls.2016.01564
Song, X., Zhang, Y., Wu, F., and Zhang, L. (2016b). Genome-wide analysis of the maize (Zea may L.) CPP-like gene family and expression profiling under abiotic stress. Genet. Mol. Res. 15:gmr.15038023. doi: 10.4238/gmr.15038023
Suyama, M., Torrents, D., and Bork, P. (2006). PAL2NAL: Robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acid. Res. 2006:W609–W612. doi: 10.1093/nar/gkl315
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. (2011). MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739.
Ur Rehman, S., Sher, M. A., Saddique, M. A. B., Ali, Z., Khan, M. A., Mao, X., et al. (2021). Development and exploitation of KASP assays for genes underpinning drought tolerance among wheat cultivars from Pakistan. Front. Genet. 12:684702. doi: 10.3389/fgene.2021.684702
Ur Rehman, S., Wang, J., Chang, X., Zhang, X., Mao, X., and Jing, R. (2019). A wheat protein kinase gene TaSnRK2.9-5A associated with yield contributing traits. Theor. Appl. Genet. 132, 907–919. doi: 10.1007/s00122-018-3247-7
Wang, D., Guo, Y., Wu, C., Yang, G., Li, Y., and Zheng, C. (2008). Genome-wide analysis of CCCH zinc finger family in Arabidopsis and rice. BMC Genom. 9:44. doi: 10.1186/1471-2164-9-44
Wang, Z., Zhang, H., Yang, J., Chen, Y., Xu, X., Mao, X., et al. (2013). Phylogenetic, expression, and bioinformatic analysis of the ABC1 gene family in Populus trichocarpa. Sci. World J. 2013:785070. doi: 10.1155/2013/785070
William Roy, S., and Gilbert, W. (2006). The evolution of spliceosomal introns: Patterns, puzzles and progress. Nat. Rev. Genet. 7, 211–221. doi: 10.1038/nrg1807
Wilson, R. F. (2004). “Seed composition”, in Soybeans: Improvement, Production, and Uses, eds R. M. Shibles, J. E. Harper, R. F. Wilson, and R. C. Shoemaker (Madison, WI: American Society of Agronomy), 621–677.
Yang, S., Zhang, X., Yue, J. X., Tian, D., and Chen, J. Q. (2008). Recent duplications dominate NBS-encoding gene expansion in two woody species. Mol. Genet. Genom. 280, 187–198. doi: 10.1007/s00438-008-0355-0
Yang, Y., Ahammed, G. J., Wan, C., Liu, H. R., Chen, R., and Zhou, Y. (2019). Comprehensive analysis of TIFY transcription factors and their expression profiles under jasmonic acid and abiotic stresses in watermelon. Int. J. Genom. 2019:6813086. doi: 10.1155/2019/6813086
Yang, Z. E., Gong, Q., Qin, W. Q., Yang, Z. R., Cheng, Y., Lu, L. L., et al. (2017). Genome-wide analysis of WOX genes in upland cotton and their expression pattern under different stresses. BMC Plant Biol. 17:113. doi: 10.1186/s12870-017-1065-8
Yang, Z., Gu, S., Wang, X., Li, W., Tang, Z., and Xu, C. (2008). Molecular evolution of the CPP-like gene family in plants: Insights from comparative genomics of Arabidopsis and rice. J. Mol. Evol. 67, 266–277. doi: 10.1007/s00239-008-9143-z
Zhang, L., Zhao, H. K., Wang, Y. M., Yuan, C. P., Zhang, Y. Y., Li, H. Y., et al. (2015). Genome-wide identification and expression analysis of the CPP-like gene family in soybean. Genet. Mol. Res. 14, 1260–1268. doi: 10.4238/2015.February.13.4
Zhou, Y., Hu, L., Ye, S., Jiang, L., and Liu, S. (2018). Genome-wide identification and characterization of cysteine-rich polycomb-like protein (CPP) family genes in cucumber (Cucumis sativus) and their roles in stress responses. Biologia 73, 425–435.
Keywords: soybean, phylogenetic analyses, kompetitive allele specific PCR, association analyses, drought, GmCPP
Citation: Nisar T, Tahir MHN, Iqbal S, Sajjad M, Nadeem MA, Qanmber G, Baig A, Khan Z, Zhao Z, Geng Z and Ur Rehman S (2022) Genome-wide characterization and sequence polymorphism analyses of cysteine-rich poly comb-like protein in Glycine max. Front. Plant Sci. 13:996265. doi: 10.3389/fpls.2022.996265
Received: 17 July 2022; Accepted: 15 August 2022;
Published: 20 September 2022.
Edited by:
Engin Yol, Akdeniz University, TurkeyReviewed by:
Nastaran Mehri, Ardebil Agriculture and Natural Resources Research Center (AREEO), IranShengwu Hu, Northwest A&F University, China
Sunny Ahmar, University of Silesia in Katowice, Poland
Copyright © 2022 Nisar, Tahir, Iqbal, Sajjad, Nadeem, Qanmber, Baig, Khan, Zhao, Geng and Ur Rehman. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shoaib Ur Rehman, c2hvYWliLnJlaG1hbkBtbnN1YW0uZWR1LnBr; Zhide Geng, Z2VuZ3pkMjAwMkAxNjMuY29t
 Tayyaba Nisar1
Tayyaba Nisar1 Shahid Iqbal
Shahid Iqbal Muhammad Sajjad
Muhammad Sajjad Muhammad Azhar Nadeem
Muhammad Azhar Nadeem Ghulam Qanmber
Ghulam Qanmber Ayesha Baig
Ayesha Baig Zulqurnain Khan
Zulqurnain Khan Shoaib Ur Rehman
Shoaib Ur Rehman