- 1Hainan Academy of Forestry, Hainan Mangrove Research Institute, Haikou, China
- 2Mangrove Institute, Lingnan Normal University, Zhanjiang, China
Sesuvium portulacastrum has a strong salt tolerance and can grow in saline and alkaline coastal and inland habitats. This study investigated the physiological and molecular responses of S. portulacastrum to high salinity by analyzing the changes in plant phytohormones and antioxidant activity, including their differentially expressed genes (DEGs) under similar high-salinity conditions. High salinity significantly affected proline (Pro) and hydrogen peroxide (H2O2) in S. portulacastrum seedlings, increasing Pro and H2O2 contents by 290.56 and 83.36%, respectively, compared to the control. Antioxidant activities, including superoxide dismutase (SOD), peroxidase (POD), and catalase (CAT), significantly increased by 83.05, 205.14, and 751.87%, respectively, under high salinity. Meanwhile, abscisic acid (ABA) and gibberellic acid (GA3) contents showed the reverse trend of high salt treatment. De novo transcriptome analysis showed that 36,676 unigenes were matched, and 3,622 salt stress-induced DEGs were identified as being associated with the metabolic and biological regulation processes of antioxidant activity and plant phytohormones. POD and SOD were upregulated under high-salinity conditions. In addition, the transcription levels of genes involved in auxin (SAURs and GH3), ethylene (ERF1, ERF3, ERF114, and ABR1), ABA (PP2C), and GA3 (PIF3) transport or signaling were altered. This study identified key metabolic and biological processes and putative genes involved in the high salt tolerance of S. portulacastrum and it is of great significance for identifying new salt-tolerant genes to promote ecological restoration of the coastal strand.
Introduction
Salinity often has a significant effect on the survival and growth of plants. Salt stress causes ionic and osmotic stresses, which inhibits photosynthesis, protein synthesis, osmoregulation, and energy and lipid metabolism in plants (Zhu, 2001; Liska et al., 2004). Plants regulate many biological pathways to adapt to salt stress, including antioxidant defense systems and the perception and transduction of plant phytohormone signals (Liska et al., 2004; Hasanuzzaman et al., 2021). The antioxidant defense system (enzymatic and non-enzymatic systems) protects the plant from salt-induced oxidative damage by detoxifying the reactive oxygen species (ROS) and also by maintaining the balance of ROS generation under salt stress (Hasanuzzaman et al., 2021). The enzymatic system including superoxide dismutase (SOD), catalase (CAT), and peroxidase (POD) can scavenge ROS to protect the plant from oxidative stress damage, which is proved to act in reducing ROS under salt stress (Hasanuzzaman et al., 2020). Plant phytohormones including abscisic acid (ABA), auxins, cytokinins, gibberellic acid (GA), and ethylene are chemicals in small amounts but have a great impact to help plants to adapt under salinity (Ribba et al., 2020). Phytohormone signaling pathways also interact with ROS scavenging systems to mount a stress response (Hasanuzzaman et al., 2018). Several physiological processes are regulated by auxins during plant development under salt stress (Ribba et al., 2020). Auxin positively regulates antioxidant enzymes (SOD, CAT, and POD) and confers effective ROS detoxification, thus improving plant abiotic stress resistance (Shi et al., 2014). ABA-dependent pathways may be positively regulated by auxins in response to abiotic stress (Liu et al., 2013; Shi et al., 2014). The ABA-induced hydrogen peroxide (H2O2; non-radical ROS) generation activates mitogen-activated protein kinase (MAPK), which in turn induces the expression level and activity of antioxidant enzymes to scavenge ROS (Zhang A. et al., 2006; Lu et al., 2009; Fahad et al., 2015). A previous study has suggested that GA plays a significant role in regulating the stress response process (Kirungu et al., 2019). Growth inhibition caused by salt stress may be an active adaptation mechanism in plants that regulates GA levels to achieve an optimal growth rate in response to environmental changes. The specific concentration of GA3 can regulate the metabolic process as a function of sugar signaling and antioxidative enzymes under salt stress (Mohammed, 2007; Iqbal et al., 2011). ABA and GA can affect the germination rate of saline plants, and there is a significant change in GA concentration in plants under abiotic stress, indicating an interaction between ABA and GA (Yu et al., 2020).
Sesuvium portulacastrum (Aizoaceae, Sesuvium), a perennial herb with thick, smooth stems (Alsherif et al., 2021), is a well-known halophyte plant with strong drought and salt tolerance that can grow in saline and alkaline coastal and inland habitats (El-Awady et al., 2015). Sesuvium is a “salt accumulator” plant that accumulates high concentrations of salts in its cells and tissues and overcomes salt toxicity by developing succulence (Meng et al., 2018). S. portulacastrum exhibits high photosynthetic activity, succulence, biomass, and antioxidant defense mechanisms at the cellular, organ, and plant levels (Lokhande et al., 2013; Muchate et al., 2016). In China, it is found in mangrove forests. To study their salt response mechanism, seedlings were cultivated in high salt concentrations of 500 mM for 28 days (Nikalje et al., 2018), 500 mM and 600 mM for 28 days (Kannan et al., 2013), and 400 and 600 mM for 14 days (Peng et al., 2019). These studies have focused on propagation, photosynthesis, ion homeostasis, antioxidant defense, accumulation mechanisms, salt-responsive genes, and proteins of S. portulacastrum under salt stress (Rabhi et al., 2010; Lokhande et al., 2013; Yi et al., 2014; Nikalje et al., 2018; Fan et al., 2019; Peng et al., 2019; He et al., 2021), but the signaling pathway involved in salt stress is not known at the transcriptome level, even though one transcriptome profiling study was reported for the same genus plant Sesuvium verrucosum under salinity stress (Thayale Purayil et al., 2022).
The understanding of plant responses to salt stress has expanded with the development of next-generation sequencing technologies. Studies on plant genomics have extensively used transcriptome sequencing techniques (Das et al., 2020; Meera and Augustine, 2020; Elnaggar et al., 2021). Therefore, a high-salinity response mechanism was studied using the RNA-seq technology to screen for salt-tolerance genes. Differentially expressed genes (DEGs) are predominantly enriched in cellular amino acid catabolic processes and responses to ROS biosynthetic processes (Yue et al., 2021). Plant hormone signal transduction genes have been discovered in salt-treated Solenostemma argel (Ahmad et al., 2021).
Transcription factor genes such as MYBs, NAC, B3, and AP2/ERF were found to be differentially expressed and helped the plant adapt to high-salinity conditions in Arabidopsis (Ma et al., 2022) and other non-model plants, Kandelia candel, Bruguiera gymnorhiza (Basyuni et al., 2012), and Sonneratia caseolaris (Zhou et al., 2022). In this study, we analyzed the physiological response of S. portulacastrum to salt stress. Meanwhile, de novo transcriptome patterns of S. portulacastrum in the leaves identified genes associated with high-salinity tolerance. The results of this study elucidate the mechanisms underlying high-salinity tolerance in S. portulacastrum and provide an important role in coastal soil remediation.
Materials and methods
Plant materials and different NaCl treatments
Sesuvium portulacastrum plants were collected from mangrove areas (110.39° N, 21.27° E) in Zhanjiang, Guangdong Province, South China, and propagated by cutting. Notably, 6-cm-long stem segments with two nodes and two opposite leaves on the top node were taken from mother plants and cultivated in tap water under natural conditions for 30 days. Then, the plants were cultivated weekly with a half-strength modified Hoagland nutrient solution (Wang et al., 2012; Yi et al., 2014) and with different NaCl concentrations of 0 (control), 100, 200, 400, 600, and 800 mmol/L. A month later, leaves from each treatment were collected for further experiments. The sample treated with 800 mmol/L NaCl perished. Each sample contained three biological replicates.
Determination of proline, hydrogen peroxide, antioxidant enzyme activity, and plant phytohormone content
Proline (Pro) content was determined following a previous study using the ninhydrin colorimetric method, with some modifications (Rosen, 1957). H2O2 content was determined spectrophotometrically at 410 nm, as described previously (Zhou et al., 2017; Ali et al., 2019). To assay SOD (EC 1.15.1.1), the inhibition of the photochemical reduction of nitro-blue tetrazolium was monitored, as described previously (El-Shabrawi et al., 2010). POD (EC1.11.1.7.) activity was measured at 470 nm, as described by Zhou et al. (2017). CAT (EC 1.11.1.6.) activity was assayed at 240 nm using the method described by Hasanuzzaman et al. (2011). Gas chromatography-mass spectrometry (GC-MS) analysis was used to determine the contents of auxin (IAA) and ABA in a previous study (Zörb et al., 2013). The samples were freeze-dried and ground to a powder to determine the gibberellin (GA3) content by GC-MS following the method of a previous study (Ptošková et al., 2022).
RNA extraction, library preparation, and Illumina NovaSeq 6000 sequencing
Total RNA was extracted from the leaf tissues using TRIzol® Life Reagent (Invitrogen, Carlsbad, CA, USA). RNA was quantified, and a sequencing library was built using the present method (Zhang et al., 2020). RNA purification, reverse transcription, library construction, and sequencing were performed following the manufacturer’s instructions (Illumina, San Diego, CA, USA and sequencing). The Illumina TruSeq™ RNA sample preparation kit (San Diego, CA, USA) was used to prepare the RNA-seq libraries. Briefly, oligo-dT-attached magnetic beads were used to purify poly(A) mRNA and further synthesize double-stranded cDNA. Later, end-repair was performed on the synthesized cDNA, and the addition of “A” bases was followed by PCR amplification. A total of 6 RNA-seq libraries were sequenced using an Illumina NovaSeq 6000 sequencer (Illumina, San Diego, CA, USA). Accession number PRJNA859253 was assigned to the raw reads generated for this study in the NCBI database.
De novo assembly and annotation
De novo assembly was performed on cleaned data using Trinity (Grabherr et al., 2011). BLASTX with an e-value of lower than 1.0 × 10–5 was used to predict the unigenes from the protein non-redundant (NR), Clusters of Orthologous Groups (COG), Kyoto Encyclopedia of Genes and Genomes (KEGG), and NCBI databases. The BLAST2GO program was used to obtain gene ontology (GO) annotations of uniquely assembled transcripts to describe potentially associated biological processes, molecular functions, and cellular components (Conesa et al., 2005). KEGG was used for the metabolic pathway analysis (Ogata et al., 1999).
Differentially expressed genes identification and functional enrichment
Differentially expressed genes of two different samples were identified by calculating the expression levels of each transcript using the transcripts per million reads method. Gene abundance was calculated using RSEM (Li and Dewey, 2011). EdgeR (Robinson et al., 2010) was used to analyze gene expression and DEGs with a | log2FC| > 1 and Q-value ≤ 0.05. GO and KEGG analyses were enriched in DEGs to identify relevant pathways and functions (Xie et al., 2011). A Bonferroni-corrected P-value of ≤ 0.05 was used when compared to the background comprising the whole transcriptome.
Quantitative real-time PCR analysis
Quantitative real-time PCR analysis uses our present method (Zhang et al., 2020). The program Primer Premier version 5.0 was used to create the primers (Supplementary Table 1). The length of the amplified PCR products ranged from 80 to 400 bp. The relative expression levels of the genes in various samples were estimated using the 2–Δ Δ Ct method, with actin as a reference to calculate the relative expression of the DEGs.
Statistical analysis
Data from each group were analyzed individually using the SPSS software (version 19.0). Tukey’s test value of P < 0.05 indicated a statistical significance, and significant differences were indicated by different letters above bars.
Results
Effects of salt stress on Pro, H2O2 content, and antioxidant enzyme activities
Compared with the control, Pro content was significantly increased under different NaCl concentrations (Figure 1A). With increasing salt concentration, the Pro content first decreased, then increased, and finally decreased. Pro content increased by 613.25% under 400 mM NaCl treatment and by 290.56% under the highest concentration (600 mM) NaCl treatment. The H2O2 content was significantly increased by 83.36% under the 600 mM NaCl treatment compared with the control (Figure 1B). Compared with the control, SOD, POD, and CAT activities were significantly increased by 83.05, 205.14, and 751.87%, respectively, under the 600 mM NaCl treatment (Figures 1C–E). The low-concentration NaCl treatment did not significantly affect POD and CAT activities in S. portulacastrum.
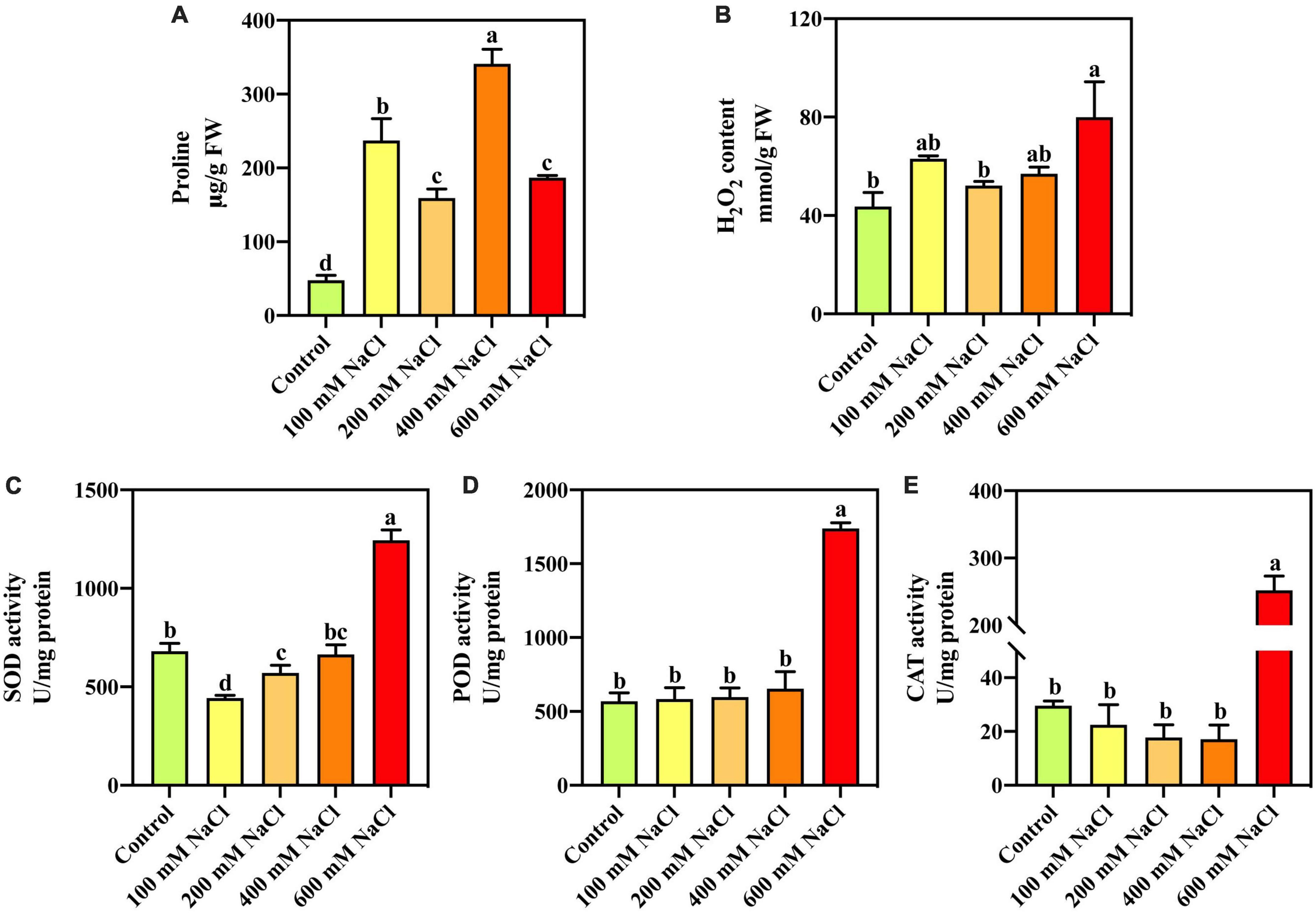
Figure 1. The proline (A), H2O2 content (B), and activities of SOD (C), POD (D), and CAT (E) in the leaves of S. portulacastrum under salt stress.
Effects of salt stress on IAA, ABA, and GA3 content
With an increase in salt concentration, IAA content decreases with high salinity but piques to a normal level as part of homeostasis (Figure 2A). Compared with the control, the ABA content was significantly increased under different NaCl treatments (Figure 2B). The ABA content in S. portulacastrum leaves treated with 600 mM NaCl was 3.36 times that of the control. Compared with the control, GA3 content was significantly decreased under the different NaCl treatments (Figure 2C). The GA3 content in S. portulacastrum leaves treated with 600 mM NaCl was used as a control.
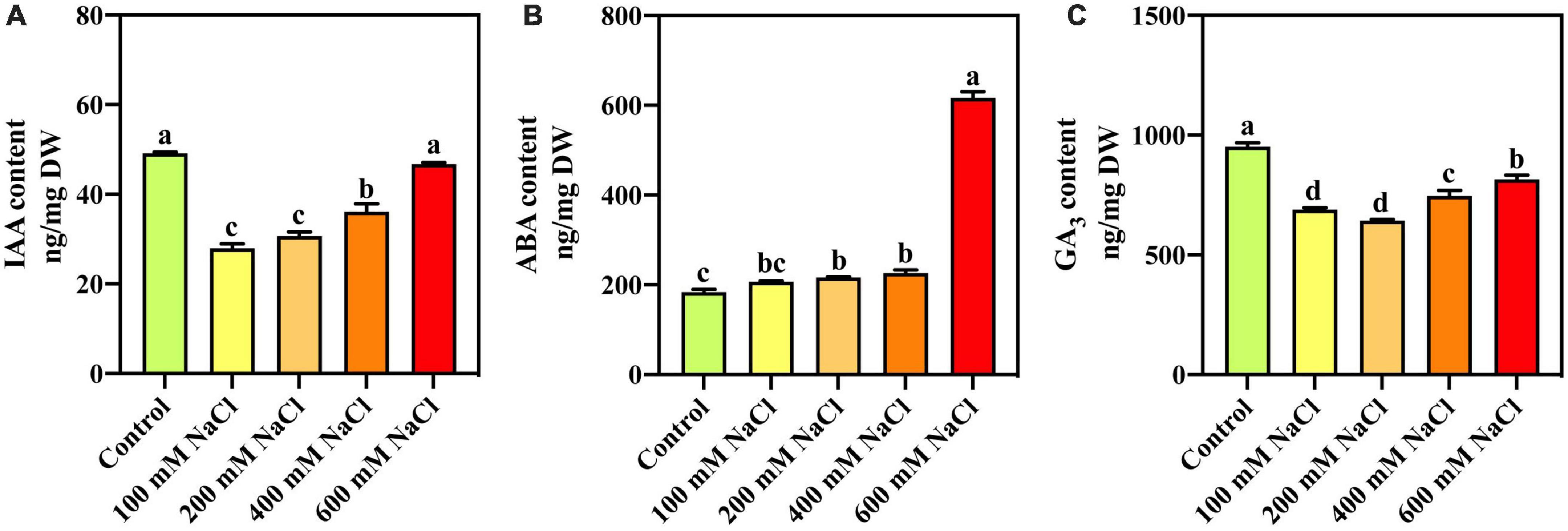
Figure 2. The content of IAA (A), ABA (B), and GA3 (C) in the leaves of S. portulacastrum under salt stress.
RNA sequencing, de novo assembly, and transcriptome annotation
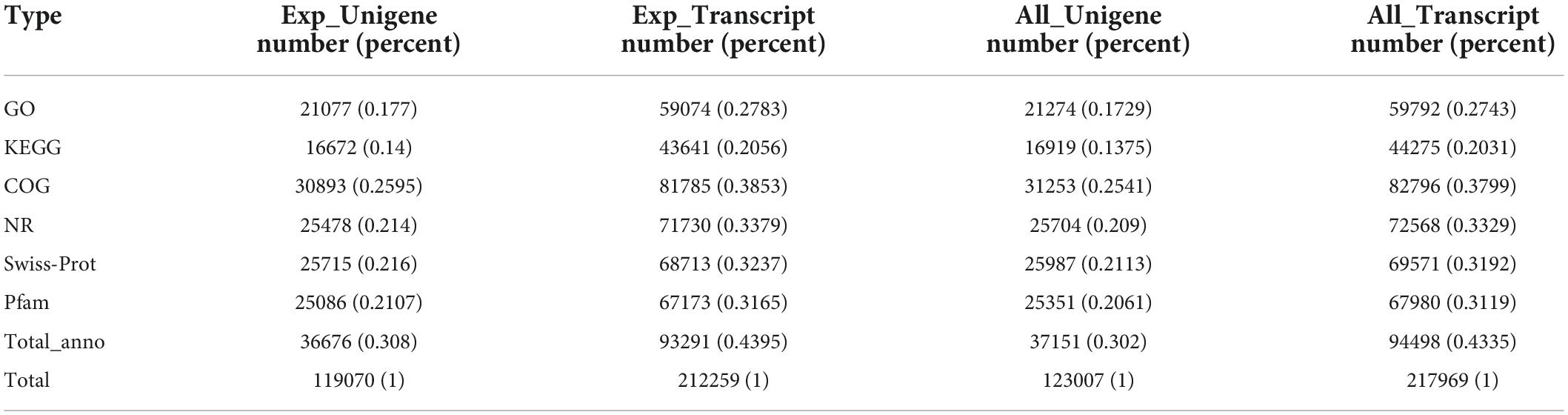
Compared with the control, S. portulacastrum seedling leaves under the 600 mM NaCl treatment had higher SOD, POD, and CAT activities to scavenge H2O2 and higher IAA and ABA contents in response to high salinity. Thus, we performed a transcriptomic analysis of the leaves of S. portulacastrum seedlings following control and 600 mM NaCl treatments. A total of 38.19 Gb of data were obtained from six cDNA libraries. After trimming low-quality reads and adapters, 256,867,510 clean reads with a Q30 higher than 92.50% were obtained (Supplementary Table 2). In total, 217,969 transcripts and 123,007 unigenes were obtained, with an N50 length of 1,184 bp and an average length of 734.24 bp (Table 1). Unigene lengths of 200–500, 501–1,000 bp, 1,001–1,500, and > 1,500 bp accounted for 60, 21, 7, and 12%, respectively (Supplementary Table 3), and 23,431 unigenes had lengths of > 1,000 bp.
A total of 36,676 (30.80%) unigenes were matched to known the genes at least once in the listed databases. Of note, 25,478 (21.40%) and 30,893 (25.95%) unigenes were the best hits in the NR and COG databases, respectively, followed by 21,077 (17.70%) in the GO database, 25,715 (21.60%) in the Swiss-Prot database, 25,086 (21.07%) in the Pfam database, and 16,672 (14.00%) in the KEGG database (Table 2).
A total of 21,077 unigenes were the best hits in the GO database and were enriched for 49 GO terms that classified as biological processes, cellular components, and molecular functions. In biological processes, most of the unigenes were enriched for “cellular process” (9,443), “metabolic process” (8,624), “biological regulation” (2,312), “cellular component organization or biogenesis” (1,497), “response to stimulus” (1,400), and “localization” (1,340) terms, accounting for 36.93, 33.72, 9.04, 5.85, 5.47, and 5.23%, respectively. The other two processes accounted for less than 9% of the gene enrichment. For the cellular component category, the largest subcategories were “cell part” (8,605; 28.63%), “membrane part” (6,900; 22.96%), “organelle” (4,877; 16.23%), “protein-containing complex” (3,200; 10.65%), and “membrane” (1,784; 5.94%). Regarding molecular function, most of the unigenes were enriched for “binding” (12,067; 44.82%) and “catalytic activity” (10,568; 39.25%) (Figure 3A).
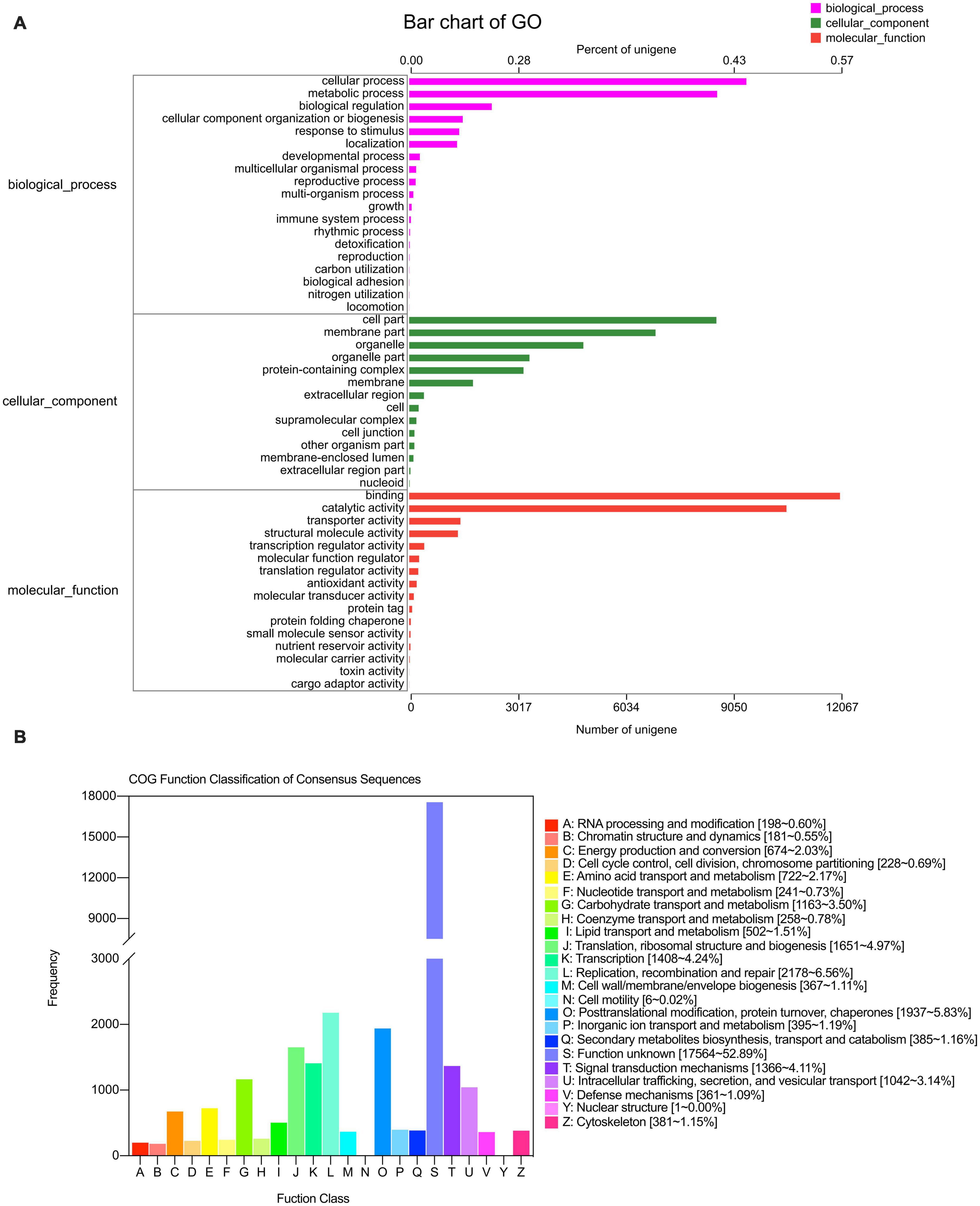
Figure 3. GO (A) and COG (B) annotation of transcriptome in the leaves of S. portulacastrum under high salinity.
Analysis of differentially expressed genes (differentially expressed genes) and gene co-expression clusters
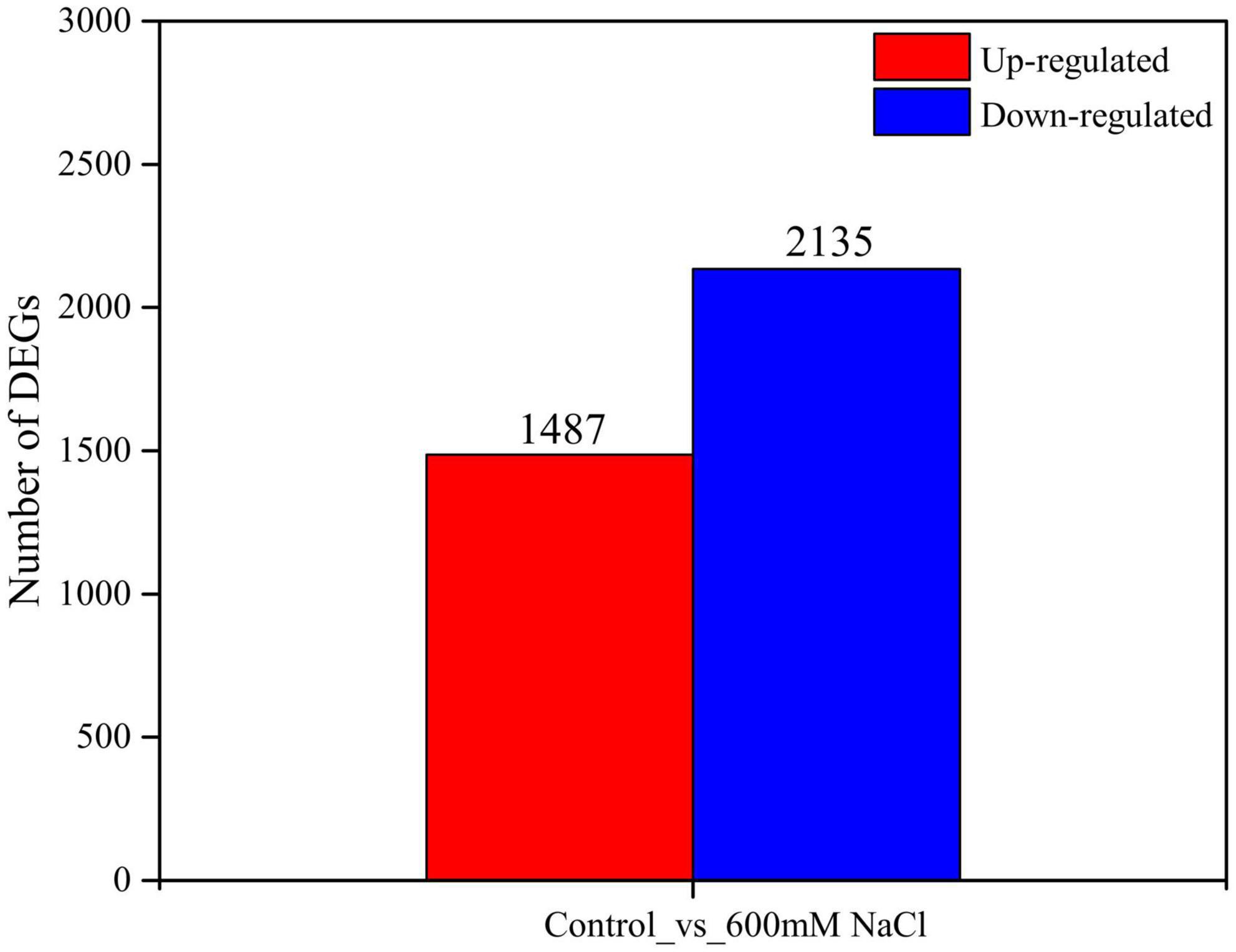
A de novo assembled reference transcriptome of S. portulacastrum was aligned with the RNA-seq reads from the 600 mM NaCl treatment group and the control group; the average mapping rate was 78.44% (Supplementary Table 4). The Pearson’s correlation coefficient between the three biological replicates of the 600 mM NaCl treatment and the control ranged from 0.925 to 1.0 (Supplementary Figure 1). Principal component analysis indicated that the control and 600 mM NaCl treatments displayed distinct transcriptome characteristics (Supplementary Figure 2). A total of 10 clusters of gene co-expression patterns were identified, including a total of 3,622 DEGs in the control vs. 600 mM NaCl comparison (Figure 4 and Supplementary Datasets 1–3). The genes in Clusters 1, 4, 6, and 7 were significantly upregulated in the 600 mM NaCl group compared to the control group, with 841, 120, 278, and 137 genes, respectively (Figure 5A and Supplementary Dataset 1). The GO analysis showed that these upregulated genes were involved in catalytic activity, oxidoreductase activity, Pro metabolic process, gibberellin biosynthetic and metabolic processes, cellular biosynthetic processes, and regulation of stress response. The genes were significantly downregulated in Clusters 2 (858), 3 (801), and 5 (459) under the 600 mM NaCl treatment compared to the control (Supplementary Dataset 2). These genes were associated with DNA-binding transcription factor activity and response to gibberellin and salicylic acid and were involved in nitrogen compound and protein metabolic processes, oxidation-reduction processes, and signal transduction (Figure 5B).
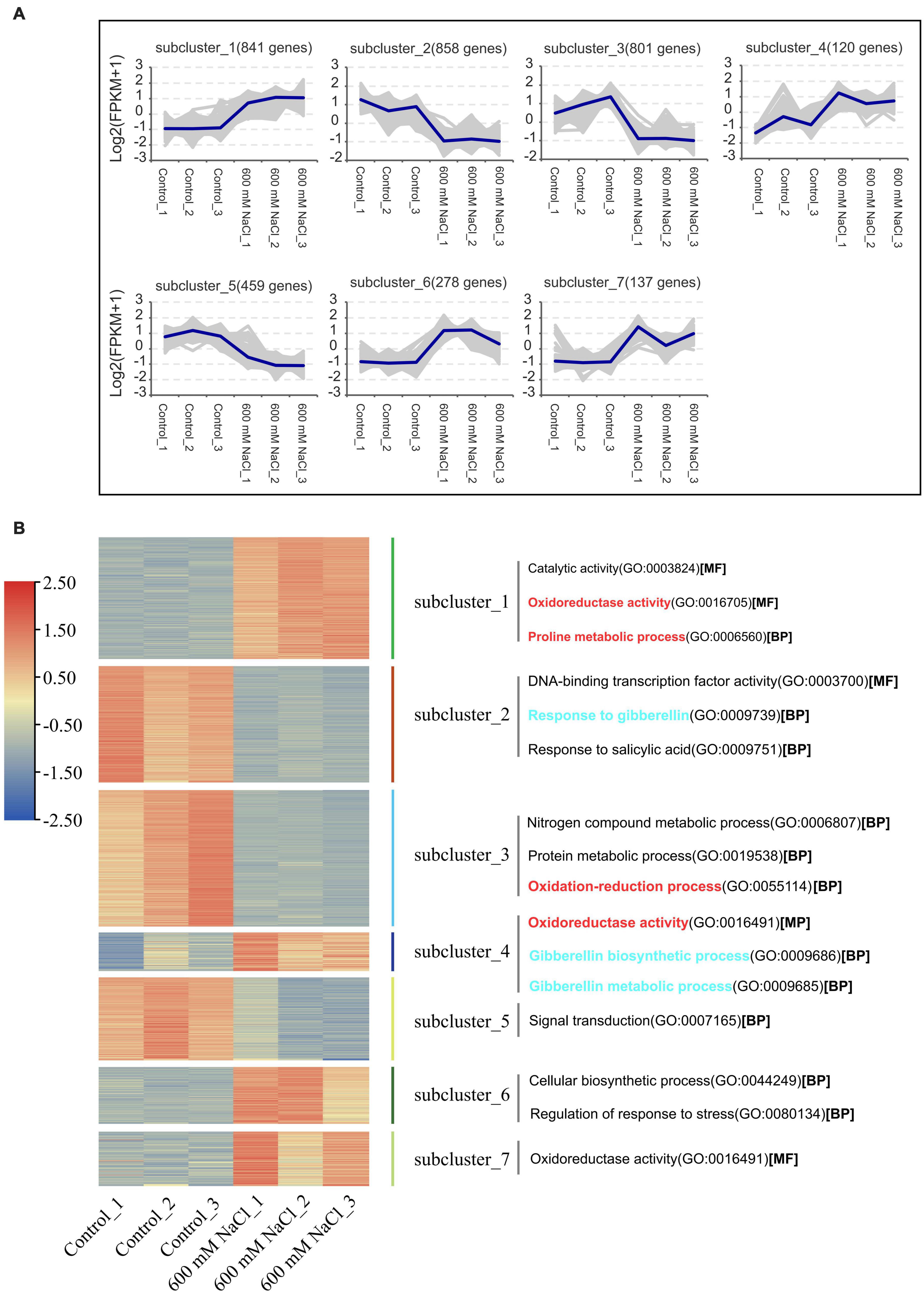
Figure 5. Gene co-expression clusters (A) and heatmap analysis (B) of DEGs in the leaves of S. portulacastrum under high salinity.
The upregulated genes (1,487) in the control vs. 600 mM NaCl were significantly enriched in oxidoreductase activity (GO:0016491), carbohydrate metabolic process (GO:0005975), structural molecule activity (GO:0005198), catabolic process (GO:0009056), and tetrapyrrole binding (GO:0046906). Notably, many pathways were related to response to stress, such as POD activity (GO:0004601), antioxidant activity (GO:0016209), ROS metabolic process (GO:0072593), and Pro metabolic process (GO:0006560). Many pathways were related to plant phytohormones, response to auxin (GO:0009733), and gibberellin biosynthetic and metabolic processes (GO:0009686; GO:0009685) (Figure 6A and Supplementary Dataset 4). The downregulated genes (2135) were significantly enriched in the organic substance metabolic process (GO:0071704), cellular metabolic process (GO:0044237), primary metabolic process (GO:0044238), macromolecule metabolic process (GO:0043170), and organelles (GO:0043226) (Figure 6B and Supplementary Dataset 5).
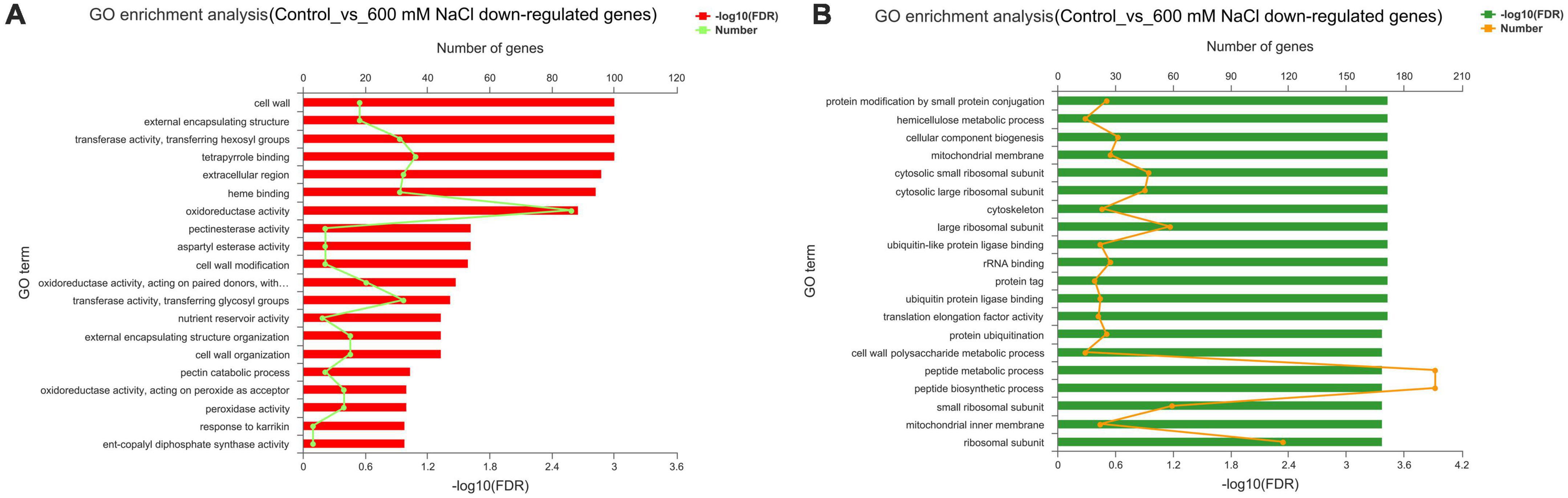
Figure 6. GO enrichment analysis of DEGs in the leaves of S. portulacastrum under high salinity. (A) Upregulated genes, (B) downregulated genes.
Responses of transcription factors, plant phytohormones, and antioxidant activity to salt stress
The candidate 12 DEGs were randomly selected to verify the accuracy of the RNA-seq data of S. portulacastrum under high-salinity conditions. Furthermore, qRT-PCR primers were used to determine expression levels (Supplementary Table 1). These results indicated that the expression patterns of these genes were highly consistent with the results of RNA-seq analysis (Figure 7). The results indicated that the RNA-seq data had a high level of reliability and that these DEGs were suitable for further analysis in S. portulacastrum under high salinity.
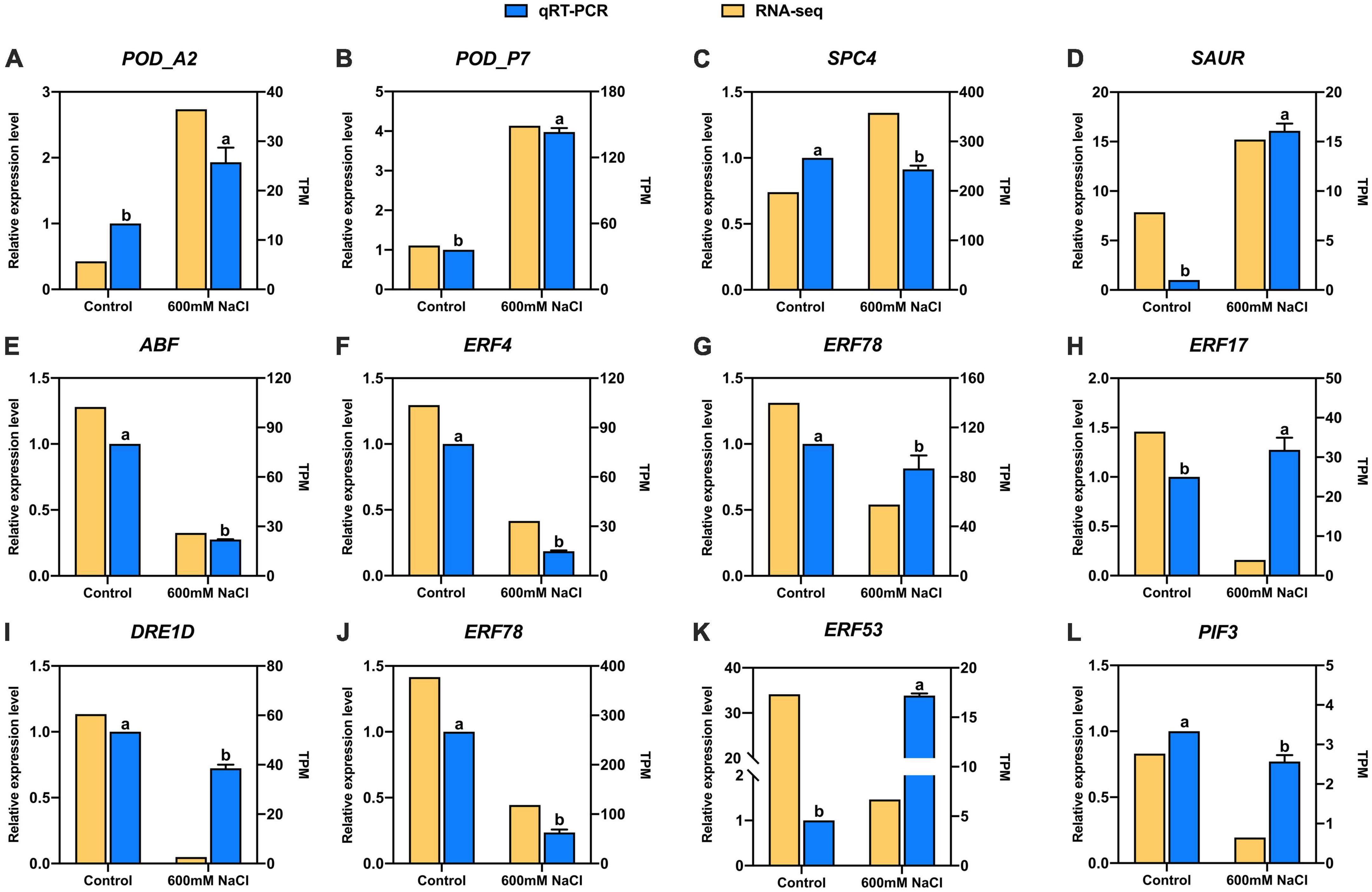
Figure 7. Expression pattern validation of 12 selected DEGs in S. portulacastrum determined by RNA-seq and qRT-PCR. (A) POD_A2, (B) POD_P7, (C) SPC4, (D) SAUR, (E) ABF, (F) ERF4, (G) ERF78, (H) ERF17, (I) DRE1D, (J) ERF78, (K) ERF53, and (L) PIF3.Values are means ± SD (n = 3). Values with a different letter within a sampling date are significantly different (P < 0.05).
In the assembled transcriptome of S. portulacastrum, 802 TFs were predicted. Among them, more than 40 members belonged to the MYB, AP2/ERF, bHLH, C2C2, NAC, and B3 families (Supplementary Figure 3). In addition, we identified a number of phytohormone transport or synthesis-related genes in the DEGs of S. portulacastrum after high salinity, such as four AP2/ERF gene families that were upregulated, and 11 AP2/ERF gene families that were downregulated (Figure 8A and Supplementary Dataset 6). Auxin-responsive protein SAURs and GH3 auxin-responsive promoters were also upregulated (Figure 8B). One gene related to the ethylene response factor (ERF1) was upregulated. Three protein phosphatases 2C (PP2C) and one ABA-insensitive were downregulated, which were involved in ABA signal transduction. One transcription factor, PIF3, was downregulated, which is associated with gibberellin metabolism (Figure 8B). DEGs related to antioxidant activity were identified in S. portulacastrum under 600 mM NaCl treatment, such as PODs, L-ascorbate peroxidase (APX), and peroxiredoxin (Prx) (Figure 8C). A total of 10 PODs, one Prx, and one APX gene were upregulated; and six POD genes, one Prx, and one SOD were downregulated (Figure 8C).
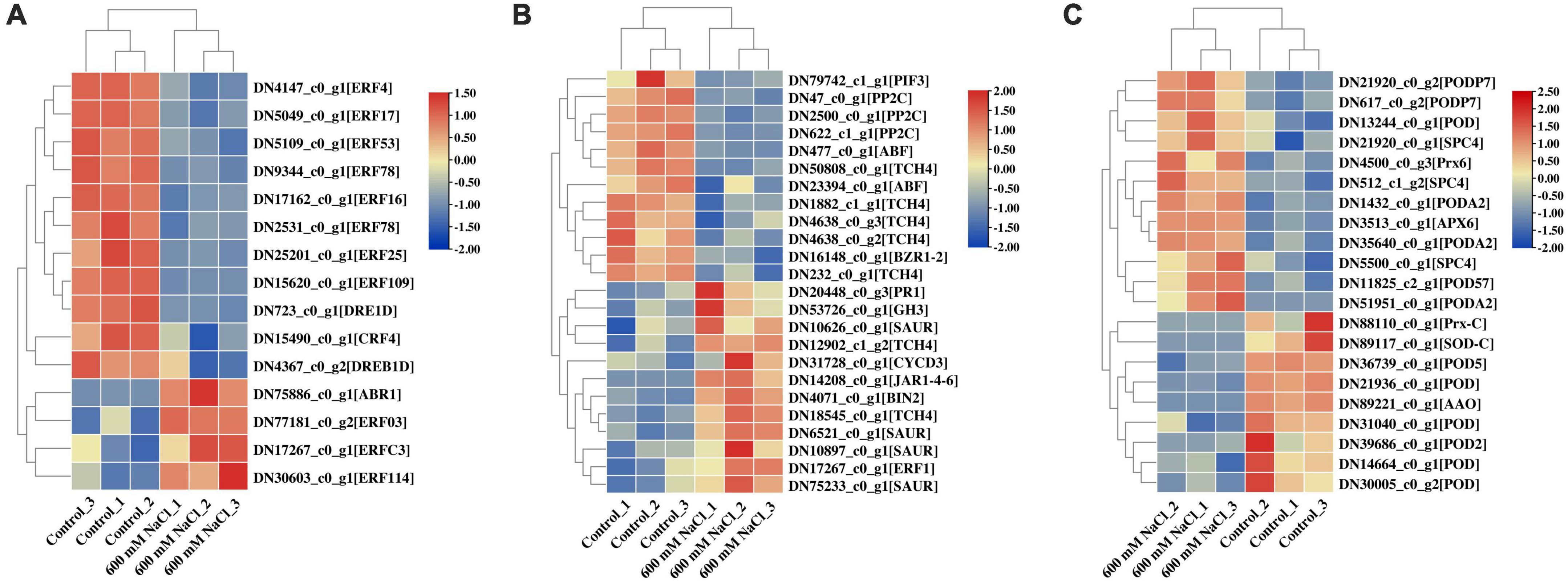
Figure 8. Heatmap analysis of AP2 gene family (A), plant phytohormones (B), and antioxidant activity (C) related to DEGs in the leaves of S. portulacastrum under high salinity.
Discussion
It is becoming increasingly common for soils to become salinized worldwide, negatively influencing plant growth (Parmar et al., 2020). The use of salt-tolerant plants reduces seawater erosion on the coast by creating an environment that facilitates plant growth, aids in the development of a bioreactor system, and assists in the bioremediation process (Duarte et al., 2013; Fariña et al., 2018). The herb S. portulacastrum can tolerate high salt levels without salt glands and bladders in its succulent leaves, indicating the existence of a specialized salt tolerance mechanism (Yi et al., 2014). Although salt-responsive physiological effects have been documented previously in this plant, insight into salinity-induced antioxidant activity and phytohormone alterations is essential to further understand the salt adaptation mechanisms (Lokhande et al., 2013; Muchate et al., 2016; Nikalje et al., 2018). To date, the current understanding of the complex molecular mechanisms underlying salt tolerance in S. portulacastrum remains limited (Fan et al., 2019). High-throughput RNA sequencing is required to identify candidate genes involved in salt stress tolerance and will also facilitate a better understanding of the salt stress tolerance mechanisms in S. portulacastrum (Bhadauria, 2017). In addition, it has been used in the Chinese traditional medicine for centuries. Despite this, less research has been conducted on the molecular basis of physiological functions (Rabhi et al., 2010; He et al., 2021; Zhang C. et al., 2022). Salt stress resistance has not been studied at the transcriptome level. Therefore, investigating the molecular mechanisms of acclimation to salt stress and identifying new salt-tolerant genes that promote ecological restoration of the coastal strand are essential.
Sesuvium portulacastrum could rapidly scavenge ROS to salinity stress at physiological and transcriptome levels
Osmotic stress and ion toxicity caused by salt stress can cause a series of secondary stresses, such as oxidative stress, significantly increasing ROS levels in plants. Various adaptations have been observed in S. portulacastrum in response to salt stress, including changes in morphological and anatomical growth, water use efficiency, and physiological and biochemical changes (Moseki and Buru, 2010; Lokhande et al., 2013). S. portulacastrum has a steady adaptation to unfavorable conditions because of its high antioxidant capacity, which also enhances its stress tolerance (Rabhi et al., 2010). Pro is the main osmotic regulator in many non-salt plants and has an essential function in maintaining the water content and enzyme activity of cells (Ullah et al., 2018; Jannesar et al., 2020). The intracellular Pro content of S. portulacastrum significantly increased under high salt stress, indicating that S. portulacastrum also adopts the same osmotic adjustment mechanism to adapt to external salt stress. The increased H2O2 illustrated that excess ROS was produced in plant cells under salt stress (Sachdev et al., 2021). Plants eliminate excess ROS produced by salt-induced stress by initiating an enzymatic system, which mainly results in an increase in antioxidant enzyme activity (Shafi et al., 2015). SOD, POD, and CAT activities were significantly increased under high-salinity conditions in S. portulacastrum.
Many genes involved in oxidoreductase activity and antioxidant activity were significantly enriched in DEGs in the transcriptome of S. portulacastrum exposed to high salinity. GO enrichment analysis revealed that these genes were related to antioxidant activity. In salt-stressed plants, SOD, POD, APX, and Prx play critical roles in maintaining redox homeostasis. One manifestation of the disruption of cellular redox homeostasis is the accumulation of oxidized Prxs (Poynton and Hampton, 2014). This study found that PODs (PODP7, PODA2, POD, and POD57) and Prx6 were significantly upregulated, and SOD-C was significantly downregulated. POD family members respond differently to salt stress. The expression of seven POD genes increased after salt stress, whereas the transcripts of two POD genes decreased in grapevines (Xiao et al., 2020). The high expression levels of PODs contributed to the conference on salt tolerance in S. portulacastrum, indicating the same trend as the physiological level.
Phytohormone in Sesuvium portulacastrum responses to high salinity at physiological and transcriptome levels
Salt stress affects plants at all stages of their life cycle, from inhibiting seed germination to modifying plant growth and development. Water deficits, ion toxicity, and ion imbalance are the three main reasons that salt negatively affects plants (Ribba et al., 2020). Thus, plants have evolved mechanisms to cope with stress, including stress-sensing and downstream response mechanisms controlled primarily by plant hormones. High-salinity exposure has been associated with the release of ABA, auxins, GA, and ethylene, playing essential roles (Egamberdieva et al., 2018). Several studies have demonstrated that plants under salt stress have lower auxin levels and decreased expression of auxin transporters (Liu et al., 2015). In this study, IAA content was controlled under high-salinity conditions. According to earlier research, genes involved in auxin transport and responsiveness, such as SAURs, were increased in response to severe salt stress (Wu et al., 2012). Gh_A08G1120 (GH3.5) is upregulated in cotton under salt stress (Kirungu et al., 2019). Plant responses to salt stress are strongly influenced by ethylene, the most significant stress-responsive hormone (Tao et al., 2015). The expression levels of genes in S. portulacastrum, which are involved in the ethylene response, as well as ethylene signaling, changed significantly. Genes related to ethylene response and transcription factors, such as ERFs, were downregulated in response to salt stress. ERF1, ERF3, ERF114, and ABR1 were upregulated under high-salinity conditions in S. portulacastrum. Overexpression of ERF1 improves salinity tolerance by amplifying the ROS-activated MAPK cascade signal (Schmidt et al., 2013). In response to salt and drought stress, TaERF3 positively regulates wheat adaptation (Rong et al., 2014). Overexpression of ERF114 enhances resistance to disease by positively modulating lignin and salicylic acid accumulation induced by PevD1 (Li et al., 2022). The ability of ERF114 in S. portulacastrum to help plants under salt stress requires further research. The expression of ABR1 was responsive to ABA and high salt stress. It has been shown that ABA induces hypersensitivity to seed germination and root growth in the absence of ABR1 (Pandey et al., 2005). Promoter analysis of the BjABR1 gene showed that there were many hormones and stress-related cis-acting elements, and expression analysis of this gene showed that it was induced under high salt stress (Xiang et al., 2018).
Abscisic acid is defined as a stress hormone due to its rapid accumulation in response to stress and its role in mediating many stress responses that assist plants in survival (Zhang J. et al., 2006). ABA was high in response to high salinity in S. portulacastrum. ABA has been shown to inhibit ethylene production during stress as part of its function (Zhang J. et al., 2006). PP2C genes have been shown to regulate ABA signaling via modulation of SnRK or MAPK kinase activity during abiotic stress responses (Yang et al., 2018). PP2C genes were downregulated under high-salinity conditions in S. portulacastrum at the transcriptome level. PP2C1 is a member of the PP2C family. BpPP2C1 loss of function resulted in lower salt tolerance, but its overexpression increased SOD and POD activities to improve salt tolerance (Xing et al., 2021). S. portulacastrum, as a halophyte, has different PP2C gene expression levels compared to the control plants. CsPP2C genes in different groups have different responses to different stressors (Zhang G. et al., 2022), and these results provide a reference for the study of PP2C under salt stress. Salt stress has been alleviated using GA3 as a growth regulator, which has been utilized to accelerate the growth of wheat and rice (Forghani et al., 2018). GA3 decreased under different salt treatments but was high in S. portulacastrum treated with 600 mM NaCl. GO enrichment analysis revealed that the genes involved in gibberellin biosynthesis and metabolic processes were upregulated. In summary, S. portulacastrum can maintain its growth under salt stress, with GA playing a vital role (Zhu et al., 2021). In coordination with the circadian clock and GA3, phytochrome-interacting factor 3 (PIF3) modulates plant growth and development by activating the light-responsive transcriptional network genes. Overexpression of ZmPIF3 in rice enhances its tolerance to salt stress without causing growth retardation (Gao et al., 2015). The interaction between plant phytohormones requires further study.
Conclusion
Pro and H2O2 contents increased in S. portulacastrum. SOD, POD, and CAT activities also increased to decompose H2O2 to reduce ROS and minimize the oxidative stress caused by high salinity. Genes encoding antioxidant enzymes, such as PODs and SOD, were upregulated, and Prx6 was downregulated in response to high salinity. ABA content increased and GA3 content decreased under high-salinity conditions in S. portulacastrum. At the transcriptome level, auxin signal receptors SAURs and GH3 were upregulated in S. portulacastrum and played critical roles in its adaptation to high salinity. ERF1, ERF3, ERF114, and ABR1, associated with ethylene signaling, were upregulated in response to high salinity. PP2C and PIF3, which are involved in ABA and GA3 signal transduction, respectively, were downregulated under high-salinity conditions in S. portulacastrum. This study focused on the mechanism of salt tolerance at the physiological and transcriptome levels while laying the foundation for further study of the molecular mechanism of salt tolerance in S. portulacastrum and exploring new salt-tolerant genes.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA859253.
Author contributions
YaZ defined the research theme and wrote the manuscript. YiZ and YiC designed methods and experiments, and carried out interpreted the results. YuC, YF, CZ, and ZF co-designed the experiments, carried out the laboratory experiments, and discussed the analyses and interpretation. All authors have read and approved the final manuscript.
Funding
This research was supported by the Key Science and Technology Program of Hainan Province (Grant No. ZDKJ202008).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.995855/full#supplementary-material
Supplementary Figure 1 | Heatmap and Pearson correlation coefficients for RNA-seq replicates of S. portulacastrum under high salinity. Three biological replicates are shown.
Supplementary Figure 2 | The PCA of transcriptomes of S. portulacastrum under high salinity and control.
Supplementary Figure 3 | The assembling transcriptome of S. portulacastrum revealed a number of different transcription factor families.
References
Ahmad, H., Maher, M., Abdel-Salam, E. M., Li, Y., Yang, C., ElSafty, N., et al. (2021). Integrated de novo analysis of transcriptional and metabolic variations in salt-treated Solenostemma argel desert plants. Front. Plant Sci. 12:744699. doi: 10.3389/fpls.2021.744699
Ali, A., Maggio, A., Bressan, R., and Yun, D.-J. (2019). Role and functional differences of HKT1-type transporters in plants under salt stress. Int. J. Mol. Sci. 20:1059. doi: 10.3390/ijms20051059
Alsherif, E. A., Al-Shaikh, T. M., Almaghrabi, O., and AbdElgawad, H. (2021). High redox status as the basis for heavy metal tolerance of Sesuvium portulacastrum L. inhabiting contaminated soil in Jeddah, Saudi Arabia. Antioxidants 11:19. doi: 10.3390/antiox11010019
Basyuni, M., Baba, S., Kinjo, Y., Putri, L. A. P., Hakim, L., and Oku, H. (2012). Salt-dependent increase in triterpenoids is reversible upon transfer to fresh water in mangrove plants Kandelia candel and Bruguiera gymnorrhiza. J. Plant Physiol. 169, 1903–1908. doi: 10.1016/j.jplph.2012.08.005
Bhadauria, V. (2017). Next-generation Sequencing and Bioinformatics for Plant Science. Norfolk: Caister Academic Press.
Conesa, A., Gotz, S., Garcia-Gomez, J. M., Terol, J., Talon, M., and Robles, M. (2005). Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21, 3674–3676. doi: 10.1093/bioinformatics/bti610
Das, R. R., Pradhan, S., and Parida, A. (2020). De-novo transcriptome analysis unveils differentially expressed genes regulating drought and salt stress response in Panicum sumatrense. Sci. Rep. 10:21251. doi: 10.1038/s41598-020-78118-3
Duarte, C. M., Losada, I. J., Hendriks, I. E., Mazarrasa, I., and Marbà, N. (2013). The role of coastal plant communities for climate change mitigation and adaptation. Nat. Clim. Change 3, 961–968. doi: 10.1038/nclimate1970
Egamberdieva, D., Wirth, S., and Abd_Allah, E. F. (2018). “Plant hormones as key regulators in plant-microbe interactions under salt stress,” in Plant Microbiome: Stress Response Microorganisms for Sustainability, eds D. Egamberdieva and P. Ahmad (Singapore: Springer Singapore), 165–182.
El-Awady, M. A. M., Hassan, M. M., and Al-Sodany, Y. M. (2015). Isolation and characterization of salt tolerant endophytic and rhizospheric plant growth-promoting bacteria (PGPB) associated with the halophyte plant (Sesuvium verrucosum) grown in KSA. Int. J. Appl. Sci. Biotechnol. 3, 552–560. doi: 10.3126/ijasbt.v3i3.13440
Elnaggar, A., Mosa, K. A., Ramamoorthy, K., El-Keblawy, A., Navarro, T., and Soliman, S. S. M. (2021). De novo transcriptome sequencing, assembly, and gene expression profiling of a salt-stressed halophyte (Salsola drummondii) from a saline habitat. Physiol. Plant. 173, 1695–1714. doi: 10.1111/ppl.13591
El-Shabrawi, H., Kumar, B., Kaul, T., Reddy, M. K., Singla-Pareek, S. L., and Sopory, S. K. (2010). Redox homeostasis, antioxidant defense, and methylglyoxal detoxification as markers for salt tolerance in Pokkali rice. Protoplasma 245, 85–96. doi: 10.1007/s00709-010-0144-6
Fahad, S., Hussain, S., Matloob, A., Khan, F. A., Khaliq, A., Saud, S., et al. (2015). Phytohormones and plant responses to salinity stress: a review. Plant Growth Regul. 75, 391–404. doi: 10.1007/s10725-014-0013-y
Fan, Y., Yin, X., Xie, Q., Xia, Y., Wang, Z., Song, J., et al. (2019). Co-expression of SpSOS1 and SpAHA1 in transgenic Arabidopsis plants improves salinity tolerance. BMC Plant Biol. 19:74. doi: 10.1186/s12870-019-1680-7
Fariña, J. M., He, Q., Silliman, B. R., and Bertness, M. D. (2018). Biogeography of salt marsh plant zonation on the Pacific coast of South America. J. Biogeogr. 45, 238–247. doi: 10.1111/jbi.13109
Forghani, A. H., Almodares, A., and Ehsanpour, A. A. (2018). Potential objectives for gibberellic acid and paclobutrazol under salt stress in sweet sorghum (Sorghum bicolor [L.] Moench cv. Sofra). Appl. Biol. Chem. 61, 113–124. doi: 10.1007/s13765-017-0329-1
Gao, Y., Jiang, W., Dai, Y., Xiao, N., Zhang, C., Li, H., et al. (2015). A maize phytochrome-interacting factor 3 improves drought and salt stress tolerance in rice. Plant Mol. Biol. 87, 413–428. doi: 10.1007/s11103-015-0288-z
Grabherr, M. G., Haas, B. J., Yassour, M., Levin, J. Z., Thompson, D. A., Amit, I., et al. (2011). Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29, 644–652. doi: 10.1038/nbt.1883
Hasanuzzaman, M., Bhuyan, M. H. M., Zulfiqar, F., Raza, A., Mohsin, S., Mahmud, J., et al. (2020). Reactive oxygen species and antioxidant defense in plants under abiotic stress: revisiting the crucial role of a universal defense regulator. Antioxidants 9:681. doi: 10.3390/antiox9080681
Hasanuzzaman, M., Hossain, M. A., and Fujita, M. (2011). Nitric oxide modulates antioxidant defense and the methylglyoxal detoxification system and reduces salinity-induced damage of wheat seedlings. Plant Biotechnol. Rep. 5, 353–365.
Hasanuzzaman, M., Nahar, K., and Fujita, M. (eds) (2018). Plants Under Metal and Metalloid Stress: Responses, Tolerance and Remediation. Singapore: Springer Singapore.
Hasanuzzaman, M., Raihan, M. R. H., Masud, A. A. C., Rahman, K., Nowroz, F., Rahman, M., et al. (2021). Regulation of reactive oxygen species and antioxidant defense in plants under salinity. Int. J. Mol. Sci. 22:9326. doi: 10.3390/ijms22179326
He, W., Wang, D., Yang, N., Cao, D., Chen, X., Chen, J., et al. (2021). In vitro shoot culture of Sesuvium portulacastrum: an important plant for phytoremediation. Agriculture 12:47. doi: 10.3390/agriculture12010047
Iqbal, N., Nazar, R., and Mir, K. (2011). Role of gibberellins in regulation of source-sink relations under optimal and limiting environmental conditions. Curr. Sci. 100, 998–1007.
Jannesar, M., Seyedi, S. M., Moazzam Jazi, M., Niknam, V., Ebrahimzadeh, H., and Botanga, C. (2020). A genome-wide identification, characterization and functional analysis of salt-related long non-coding RNAs in non-model plant Pistacia vera L. using transcriptome high throughput sequencing. Sci. Rep. 10:5585. doi: 10.1038/s41598-020-62108-6
Kannan, P. R., Deepa, S., Kanth, S. V., and Rengasamy, R. (2013). Growth, osmolyte concentration and antioxidant enzymes in the leaves of Sesuvium portulacastrum L. under salinity stress. Appl. Biochem. Biotechnol. 171, 1925–1932. doi: 10.1007/s12010-013-0475-9
Kirungu, J. N., Magwanga, R. O., Lu, P., Cai, X., Zhou, Z., Wang, X., et al. (2019). Functional characterization of Gh_A08G1120 (GH3.5) gene reveal their significant role in enhancing drought and salt stress tolerance in cotton. BMC Genet. 20:62. doi: 10.1186/s12863-019-0756-6
Li, B., and Dewey, C. N. (2011). RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12:323. doi: 10.1186/1471-2105-12-323
Li, Z., Zhang, Y., Ren, J., Jia, F., Zeng, H., Li, G., et al. (2022). Ethylene-responsive factor ERF114 mediates fungal pathogen effector PevD1-induced disease resistance in Arabidopsis thaliana. Mol. Plant Pathol. 23, 819–831. doi: 10.1111/mpp.13208
Liska, A. J., Shevchenko, A., Pick, U., and Katz, A. (2004). Enhanced photosynthesis and redox energy production contribute to salinity tolerance in Dunaliella as revealed by homology-based proteomics. Plant Physiol. 136, 2806–2817. doi: 10.1104/pp.104.039438
Liu, W., Li, R.-J., Han, T.-T., Cai, W., Fu, Z.-W., and Lu, Y.-T. (2015). Salt stress reduces root meristem size by nitric oxide-mediated modulation of auxin accumulation and signaling in Arabidopsis. Plant Physiol. 168, 343–356. doi: 10.1104/pp.15.00030
Liu, X., Zhang, H., Zhao, Y., Feng, Z., Li, Q., Yang, H.-Q., et al. (2013). Auxin controls seed dormancy through stimulation of abscisic acid signaling by inducing ARF-mediated ABI3 activation in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 110, 15485–15490. doi: 10.1073/pnas.1304651110
Lokhande, V. H., Gor, B. K., Desai, N. S., Nikam, T. D., and Suprasanna, P. (2013). Sesuvium portulacastrum, a plant for drought, salt stress, sand fixation, food and phytoremediation. A review. Agron. Sustain. Dev. 33, 329–348. doi: 10.1007/s13593-012-0113-x
Lu, S., Su, W., Li, H., and Guo, Z. (2009). Abscisic acid improves drought tolerance of triploid bermudagrass and involves H2O2- and NO-induced antioxidant enzyme activities. Plant Physiol. Biochem. 47, 132–138. doi: 10.1016/j.plaphy.2008.10.006
Ma, L., Liu, X., Lv, W., and Yang, Y. (2022). Molecular mechanisms of plant responses to salt stress. Front. Plant Sci. 13:934877. doi: 10.3389/fpls.2022.934877
Meera, S. P., and Augustine, A. (2020). De novo transcriptome analysis of Rhizophora mucronata Lam. furnishes evidence for the existence of glyoxalase system correlated to glutathione metabolic enzymes and glutathione regulated transporter in salt tolerant mangroves. Plant Physiol. Biochem. 155, 683–696. doi: 10.1016/j.plaphy.2020.08.008
Meng, X., Zhou, J., and Sui, N. (2018). Mechanisms of salt tolerance in halophytes: current understanding and recent advances. Open Life Sci. 13, 149–154. doi: 10.1515/biol-2018-0020
Mohammed, A. H. M. A. (2007). Physiological aspects of mung bean plant (Vigna radiata L . wilczek) in response to salt stress and gibberellic acid treatment. Res. J. Agr. Biol. Sci. 3, 200–213.
Moseki, B., and Buru, J. C. (2010). Ionic and water relations of Sesuvium portulacastrum (L.). Sci. Res. Essays 5, 035–040.
Muchate, N. S., Nikalje, G. C., Rajurkar, N. S., Suprasanna, P., and Nikam, T. D. (2016). Plant salt stress: adaptive responses, tolerance mechanism and bioengineering for salt tolerance. Bot. Rev. 82, 371–406. doi: 10.1007/s12229-016-9173-y
Nikalje, G. C., Variyar, P. S., Joshi, M. V., Nikam, T. D., and Suprasanna, P. (2018). Temporal and spatial changes in ion homeostasis, antioxidant defense and accumulation of flavonoids and glycolipid in a halophyte Sesuvium portulacastrum (L.) L. PLoS One 13:e0193394. doi: 10.1371/journal.pone.0193394
Ogata, H., Goto, S., Sato, K., Fujibuchi, W., Bono, H., and Kanehisa, M. (1999). KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 27, 29–34. doi: 10.1093/nar/27.1.29
Pandey, G. K., Grant, J. J., Cheong, Y. H., Kim, B. G., Li, L., and Luan, S. (2005). ABR1, an APETALA2-domain transcription factor that functions as a repressor of ABA response in Arabidopsis. Plant Physiol. 139, 1185–1193. doi: 10.1104/pp.105.066324
Parmar, S., Gharat, S. A., Tagirasa, R., Chandra, T., Behera, L., Dash, S. K., et al. (2020). Identification and expression analysis of miRNAs and elucidation of their role in salt tolerance in rice varieties susceptible and tolerant to salinity. PLoS One 15:e0230958. doi: 10.1371/journal.pone.0230958
Peng, C., Chang, L., Yang, Q., Tong, Z., Wang, D., Tan, Y., et al. (2019). Comparative physiological and proteomic analyses of the chloroplasts in halophyte Sesuvium portulacastrum under differential salt conditions. J. Plant Physiol. 232, 141–150. doi: 10.1016/j.jplph.2018.10.028
Poynton, R. A., and Hampton, M. B. (2014). Peroxiredoxins as biomarkers of oxidative stress. Biochim. Biophys. Acta BBA Gen. Subj. 1840, 906–912. doi: 10.1016/j.bbagen.2013.08.001
Ptošková, K., Szecówka, M., Jaworek, P., Tarkowská, D., Petøík, I., Pavloviæ, I., et al. (2022). Changes in the concentrations and transcripts for gibberellins and other hormones in a growing leaf and roots of wheat seedlings in response to water restriction. BMC Plant Biol. 22:284. doi: 10.1186/s12870-022-03667-w
Rabhi, M., Giuntini, D., Castagna, A., Remorini, D., Baldan, B., Smaoui, A., et al. (2010). Sesuvium portulacastrum maintains adequate gas exchange, pigment composition, and thylakoid proteins under moderate and high salinity. J. Plant Physiol. 167, 1336–1341. doi: 10.1016/j.jplph.2010.05.009
Ribba, T., Garrido-Vargas, F., and O’Brien, J. A. (2020). Auxin-mediated responses under salt stress: from developmental regulation to biotechnological applications. J. Exp. Bot. 71, 3843–3853. doi: 10.1093/jxb/eraa241
Robinson, M. D., McCarthy, D. J., and Smyth, G. K. (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. doi: 10.1093/bioinformatics/btp616
Rong, W., Qi, L., Wang, A., Ye, X., Du, L., Liang, H., et al. (2014). The ERF transcription factor TaERF3 promotes tolerance to salt and drought stresses in wheat. Plant Biotechnol. J. 12, 468–479. doi: 10.1111/pbi.12153
Rosen, H. (1957). A modified ninhydrin colorimetric analysis for amino acids. Arch. Biochem. Biophys. 67, 10–15. doi: 10.1016/0003-9861(57)90241-2
Sachdev, S., Ansari, S. A., Ansari, M. I., Fujita, M., and Hasanuzzaman, M. (2021). Abiotic stress and reactive oxygen species: generation, signaling, and defense mechanisms. Antioxidants 10:277. doi: 10.3390/antiox10020277
Schmidt, R., Mieulet, D., Hubberten, H.-M., Obata, T., Hoefgen, R., Fernie, A. R., et al. (2013). SALT-RESPONSIVE ERF1 regulates reactive oxygen species-dependent signaling during the initial response to salt stress in rice. Plant Cell 25, 2115–2131. doi: 10.1105/tpc.113.113068
Shafi, A., Chauhan, R., Gill, T., Swarnkar, M. K., Sreenivasulu, Y., Kumar, S., et al. (2015). Expression of SOD and APX genes positively regulates secondary cell wall biosynthesis and promotes plant growth and yield in Arabidopsis under salt stress. Plant Mol. Biol. 87, 615–631. doi: 10.1007/s11103-015-0301-6
Shi, H., Chen, L., Ye, T., Liu, X., Ding, K., and Chan, Z. (2014). Modulation of auxin content in Arabidopsis confers improved drought stress resistance. Plant Physiol. Biochem. 82, 209–217. doi: 10.1016/j.plaphy.2014.06.008
Tao, J.-J., Chen, H.-W., Ma, B., Zhang, W.-K., Chen, S.-Y., and Zhang, J.-S. (2015). The role of ethylene in plants under salinity stress. Front. Plant Sci. 6:1059. doi: 10.3389/fpls.2015.01059
Thayale Purayil, F., Sudalaimuthuasari, N., Li, L., Aljneibi, R., Al Shamsi, A. M. K., David, N., et al. (2022). Transcriptome profiling and functional validation of RING-Type E3 ligases in halophyte Sesuvium verrucosum under salinity stress. Int. J. Mol. Sci. 23:2821. doi: 10.3390/ijms23052821
Ullah, A., Sun, H., Hakim, Yang, X., and Zhang, X. (2018). A novel cotton WRKY gene, GhWRKY6-like, improves salt tolerance by activating the ABA signaling pathway and scavenging of reactive oxygen species. Physiol. Plant. 162, 439–454. doi: 10.1111/ppl.12651
Wang, D., Wang, H., Han, B., Wang, B., Guo, A., Zheng, D., et al. (2012). Sodium instead of potassium and chloride is an important macronutrient to improve leaf succulence and shoot development for halophyte Sesuvium portulacastrum. Plant Physiol. Biochem. 51, 53–62. doi: 10.1016/j.plaphy.2011.10.009
Wu, J., Liu, S., He, Y., Guan, X., Zhu, X., Cheng, L., et al. (2012). Genome-wide analysis of SAUR gene family in Solanaceae species. Gene 509, 38–50. doi: 10.1016/j.gene.2012.08.002
Xiang, L., Liu, C., Luo, J., He, L., Deng, Y., Yuan, J., et al. (2018). A tuber mustard AP2/ERF transcription factor gene, BjABR1, functioning in abscisic acid and abiotic stress responses, and evolutionary trajectory of the ABR1 homologous genes in Brassica species. PeerJ 6:e6071. doi: 10.7717/peerj.6071
Xiao, H., Wang, C., Khan, N., Chen, M., Fu, W., Guan, L., et al. (2020). Genome-wide identification of the class III POD gene family and their expression profiling in grapevine (Vitis vinifera L). BMC Genomics 21:444. doi: 10.1186/s12864-020-06828-z
Xie, C., Mao, X., Huang, J., Ding, Y., Wu, J., Dong, S., et al. (2011). KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 39, W316–W322. doi: 10.1093/nar/gkr483
Xing, B., Gu, C., Zhang, T., Zhang, Q., Yu, Q., Jiang, J., et al. (2021). Functional study of BpPP2C1 revealed its role in salt stress in Betula platyphylla. Front. Plant Sci. 11:617635. doi: 10.3389/fpls.2020.617635
Yang, Q., Liu, K., Niu, X., Wang, Q., Wan, Y., Yang, F., et al. (2018). Genome-wide identification of PP2C genes and their expression profiling in response to drought and cold stresses in Medicago truncatula. Sci. Rep. 8:12841. doi: 10.1038/s41598-018-29627-9
Yi, X., Sun, Y., Yang, Q., Guo, A., Chang, L., Wang, D., et al. (2014). Quantitative proteomics of Sesuvium portulacastrum leaves revealed that ion transportation by V-ATPase and sugar accumulation in chloroplast played crucial roles in halophyte salt tolerance. J. Proteomics 99, 84–100. doi: 10.1016/j.jprot.2014.01.017
Yu, Z., Duan, X., Luo, L., Dai, S., Ding, Z., and Xia, G. (2020). How plant hormones mediate salt stress responses. Trends Plant Sci. 25, 1117–1130. doi: 10.1016/j.tplants.2020.06.008
Yue, J., Wang, Y., Jiao, J., and Wang, H. (2021). Comparative transcriptomic and metabolic profiling provides insight into the mechanism by which the autophagy inhibitor 3-MA enhances salt stress sensitivity in wheat seedlings. BMC Plant Biol. 21:577. doi: 10.1186/s12870-021-03351-5
Zhang, A., Jiang, M., Zhang, J., Tan, M., and Hu, X. (2006). Mitogen-activated protein kinase is involved in abscisic acid-induced antioxidant defense and acts downstream of reactive oxygen species production in leaves of maize plants. Plant Physiol. 141, 475–487. doi: 10.1104/pp.105.075416
Zhang, C., Wang, D., He, W., Liu, H., Chen, J., Wei, X., et al. (2022). Sesuvium portulacastrum-mediated removal of nitrogen and phosphorus affected by sulfadiazine in aquaculture wastewater. Antibiotics 11:68. doi: 10.3390/antibiotics11010068
Zhang, G., Zhang, Z., Luo, S., Li, X., Lyu, J., Liu, Z., et al. (2022). Genome-wide identification and expression analysis of the cucumber PP2C gene family. BMC Genomics 23:563. doi: 10.1186/s12864-022-08734-y
Zhang, J., Jia, W., Yang, J., and Ismail, A. M. (2006). Role of ABA in integrating plant responses to drought and salt stresses. Field Crops Res. 97, 111–119. doi: 10.1016/j.fcr.2005.08.018
Zhang, Y., Chen, Y., Zhou, Y., Zhang, J., Bai, H., and Zheng, C. (2020). Comparative transcriptome reveals the genes’ adaption to herkogamy of Lumnitzera littorea (Jack) Voigt. Front. Genet. 11:584817. doi: 10.3389/fgene.2020.584817
Zhou, Y., Wen, L., Liao, L., Lin, S., Zheng, E., Li, Y., et al. (2022). Comparative transcriptome analysis unveiling reactive oxygen species scavenging system of Sonneratia caseolaris under salinity stress. Front. Plant Sci. 13:953450. doi: 10.3389/fpls.2022.953450
Zhou, Y., Wen, Z., Zhang, J., Chen, X., Cui, J., Xu, W., et al. (2017). Exogenous glutathione alleviates salt-induced oxidative stress in tomato seedlings by regulating glutathione metabolism, redox status, and the antioxidant system. Sci. Hortic. 220, 90–101. doi: 10.1016/j.scienta.2017.02.021
Zhu, J.-K. (2001). Cell signaling under salt, water and cold stresses. Curr. Opin. Plant Biol. 4, 401–406. doi: 10.1016/S1369-5266(00)00192-8
Zhu, Y., Wang, Q., Gao, Z., Wang, Y., Liu, Y., Ma, Z., et al. (2021). Analysis of phytohormone signal transduction in sophora alopecuroides under salt stress. Int. J. Mol. Sci. 22:7313. doi: 10.3390/ijms22147313
Keywords: Sesuvium portulacastrum, plant phytohormones, antioxidant activity, transcription factor, salt stress
Citation: Chen Y, Zhou Y, Cai Y, Feng Y, Zhong C, Fang Z and Zhang Y (2022) De novo transcriptome analysis of high-salinity stress-induced antioxidant activity and plant phytohormone alterations in Sesuvium portulacastrum. Front. Plant Sci. 13:995855. doi: 10.3389/fpls.2022.995855
Received: 16 July 2022; Accepted: 29 August 2022;
Published: 23 September 2022.
Edited by:
Mirza Hasanuzzaman, Sher-e-Bangla Agricultural University, BangladeshReviewed by:
Navdeep Kaur, Guru Nanak Dev University, IndiaMd Ahasanur Rahman, Howard University, United States
Copyright © 2022 Chen, Zhou, Cai, Feng, Zhong, Fang and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ying Zhang, emhhbmd5aW5ncmVkQGxpbmduYW4uZWR1LmNu
†These authors have contributed equally to this work
 YiQing Chen
YiQing Chen Yan Zhou
Yan Zhou Yuyi Cai
Yuyi Cai Yongpei Feng
Yongpei Feng Cairong Zhong
Cairong Zhong ZanShan Fang
ZanShan Fang Ying Zhang
Ying Zhang