
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 17 January 2023
Sec. Plant Symbiotic Interactions
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.995589
This article is part of the Research TopicMolecular and Cellular Mechanisms of the Legume-Rhizobia Symbiosis, Volume IIView all 5 articles
 Meng Liu1,2*
Meng Liu1,2* Hiromu Kameoka1†
Hiromu Kameoka1† Akiko Oda1
Akiko Oda1 Taro Maeda1†
Taro Maeda1† Takashi Goto1,2
Takashi Goto1,2 Koji Yano1
Koji Yano1 Takashi Soyano1,2
Takashi Soyano1,2 Masayoshi Kawaguchi1,2*
Masayoshi Kawaguchi1,2*Legumes develop root nodules in association with compatible rhizobia to overcome nitrogen deficiency. Rhizobia enter the host legume, mainly through infection threads, and induce nodule primordium formation in the root cortex. Multiple transcription factors have been identified to be involved in the regulation of the establishment of root nodule symbiosis, including ERF Required for Nodulation1 (ERN1). ERN1 is involved in a transcription network with CYCLOPS and NODULE INCEPTION (NIN). Mutation of ERN1 often results in misshapen root hair tips, deficient infection thread formation, and immature root nodules. ERN1 directly activates the expression of ENOD11 in Medicago truncatula to assist cell wall remodeling and Epr3 in Lotus japonicus to distinguish rhizobial exopolysaccharide signals. However, aside from these two genes, it remains unclear which genes are regulated by LjERN1 or what role LjERN1 plays during root nodule symbiosis. Thus, we conducted RNA sequencing to compare the gene expression profiles of wild-type L. japonicus and Ljern1-6 mutants. In total, 234 differentially expressed genes were identified as candidate LjERN1 target genes. These genes were found to be associated with cell wall remodeling, signal transduction, phytohormone metabolism, and transcription regulation, suggesting that LjERN1 is involved in multiple processes during the early stages of the establishment of root nodule symbiosis. Many of these candidate genes including RINRK1 showed decreased expression levels in Ljnin-2 mutants based on a search of a public database, suggesting that LjERN1 and LjNIN coordinately regulate gene expression. Our data extend the current understanding of the pleiotropic role of LjERN1 in root nodule symbiosis.
Legumes are able to overcome nitrogen deficiency by establishing root nodule symbiosis with nitrogen-fixing bacteria known as rhizobia. Inside the unique organs of symbiosis, root nodules, rhizobia convert atmospheric nitrogen into ammonium for plants’ benefits, in exchange for carbon source. Rhizobia enter legumes through cracks on the epidermis or, more commonly, through root hairs (Guinel and Geil, 2002). In Lotus japonicus and Medicago truncatula—two model legumes primarily used for studying root nodule symbiosis—the attachment of compatible rhizobia to the surface of a host root hair is observed to induce polar growth of the root hair tip, resulting in the formation of a “shepherd’s crook” structure. Rhizobia entrapped in the crook of the root hair multiply and form microcolonies. Subsequently, the rhizobia enter the root hair cell through the inward growth of tubular structures, which are called infection threads. The formation of infection threads is accompanied by the modification of the root hair cell wall, plasma membrane, and cytoskeletal structure. Concomitant with infection thread progression, cortical cells underneath the infection sites re-enter the cell cycle, divide, and form nodule primordia. Eventually, the infection threads reach the nodule primordia and release rhizobia into the nodule cells (reviewed by Oldroyd, 2013; Roy et al., 2019). One of the fundamental questions on the topic of root nodule symbiosis is how the legume–rhizobium association is established through root nodule symbiosis signal transduction.
The root nodule symbiosis signaling pathway has been observed to be initiated through the detection of rhizobia by the host legume. The host legume distinguishes Nod factors from compatible rhizobia via receptor kinases, including Nod Factor Receptor1 (LjNFR1)/LysM Receptor Kinase3 (MtLYK3), LjNFR5/Nod Factor Perception (MtNFP), and LjNFRe (Amor et al., 2003; Limpens et al., 2003; Madsen et al., 2003; Radutoiu et al., 2003; Murakami et al., 2018). These receptors transduce the signal to the nucleus and activate calcium signaling, which is then decoded by calcium and calmodulin-dependent protein kinase (LjCCaMK)/Doesn’t Make Infections 3 (MtDMI3) (Ehrhardt et al., 1996; Lévy et al., 2004; Miwa et al., 2006; Tirichine et al., 2006). Downstream of CCaMK activation, several transcription factors, including LjCYCLOPS/Interacting Protein of DMI3 (MtIPD3), Nodulation Signaling Pathway1 (NSP1), NSP2, NODULE INCEPTION (NIN), and ERF Required for Nodulation 1 (ERN1), form a network to reprogram gene transcription (Catoira et al., 2000; Kaló et al., 2005; Heckmann et al., 2006; Marsh et al., 2007; Messinese et al., 2007; Middleton et al., 2007; Cerri et al., 2016; Fonouni-Farde et al., 2016; Jin et al., 2016; Cerri et al., 2017; Murakami et al., 2006; Schauser et al., 1999; Smit et al., 2005; Yano et al., 2008; Singh et al., 2014; Yano et al., 2017). Among these transcription factors, NIN plays a central role in regulating the expression of genes associated with cell wall remodeling, cytoskeleton rearrangement, and cell division. Target genes of NIN encode proteins such as NODULATION PECTATE LYASE (LjNPL), an enzyme that mediates cell wall degradation during infection thread initiation (Xie et al., 2012; Liu et al., 2019a); SCAR-Nodulation (LjSCARN), a component of the actin regulatory complex that promotes the formation of new actin filaments in root hairs during infection thread development (Qiu et al., 2015); and Nuclear Factor-YA1 (LjNF-YA1/MtNF-YA1), a transcription factor that promotes cortical cell division for nodule organogenesis (Combier et al., 2006; Soyano et al., 2013). MtNF-YA1 also regulates infection thread formation via direct activation of MtERN1 expression in M. truncatula (Laloum et al., 2014). Additionally, phytohormones are involved in root nodule symbiosis signaling (Buhian and Bensmihen, 2018; Lin et al., 2020). For example, auxin has been determined to positively affect infection thread formation (Nadzieja et al., 2018). Auxin can be detected in infected root hairs and dividing cortical cells (Suzaki et al., 2014). Mutation of Auxin Response Factor 16a (MtARF16a) leads to a reduced number of infection threads in M. truncatula (Breakspear et al., 2014). More recently it has been shown that IAA carboxyl methyltransferase 1 (IAMT1), which converts auxin (IAA) to its methyl ester (MeIAA), is required for nodule development and its metabolite MeIAA can induce NIN expression (Goto et al., 2022). Cytokinin is found to promote cortical cell division but represses infection in the epidermis. Exogenous application of cytokinin or gain-of-function mutations of the cytokinin receptor gene LjLHK1 (snf2 and snf5) results in spontaneous root nodules, while Ljlhk1 (hit1) mutants exhibit a reduced number of nodules and an increased number of infection threads (Murray et al., 2007; Tirichine et al., 2007; Heckmann et al., 2011; Miri et al., 2016; Liu et al., 2018). Another phytohormone, gibberellin (GA), has been identified to suppress root hair deformation and infection thread formation by degrading DELLA, a protein that interacts with CYCLOPS and NSP1-NSP2 to enhance symbiotic gene expression (Maekawa et al., 2009; Fonouni-Farde et al., 2016; Jin et al., 2016). In summary, root nodule symbiosis signaling involves various genes related to signal transduction, gene transcription regulation, and phytohormone metabolism.
Previously, we reported the function of LjERN1 in root nodule symbiosis signaling through the characterization of two allelic symbiotic mutant lines, Ljern1-5 and Ljern1-6, which show deficiencies in their response to rhizobial infection. Like Mtern1 mutants, Ljern1 mutants display abnormal balloon-shaped root hair tips, a decreased number of infection threads, and immature root nodules (Cerri et al., 2017; Kawaharada et al., 2017a; Yano et al., 2017). Gain-of-function CCaMK or application of cytokinin does not induce spontaneous nodule production in Ljern1 mutants (Kawaharada et al., 2017a). The corresponding gene ERN1 encodes an AP2/ERF transcription factor, which is expressed in infected root hairs and developing nodules (Middleton et al., 2007; Cerri et al., 2016; Cerri et al., 2017; Kawaharada et al., 2017a; Yano et al., 2017). The phenotype of ern1 mutants and expression pattern of ERN1 suggest that ERN1 is needed for infection thread formation and promote nodule organogenesis. Two genes have been identified as targets of ERN1: M. truncatula Early Nodulin11 (MtENOD11), which is involved in cell wall modification, and L. japonicus Exopolysaccharide Receptor3 (LjEpr3), which is responsible for compatible rhizobial recognition (Andriankaja et al., 2007; Kawaharada et al., 2017b). To determine the role of ERN1 in L. japonicus, we conducted RNA sequencing (RNA-seq) to compare the gene expression profiles of wild-type (WT) plants and Ljern1-6 mutant roots. Ljern1-6 is a null allele mutant isolated from L. japonicus accession Miyakojima MG-20 and lacks approximately 10 kb including the entire length of ERN1 (Yano et al., 2017). Although a transcriptome study has already been conducted in Mtern1 mutants, the relationship between ERN1 and NIN differs between M. truncatula and L. japonicus and L. japonicus lacks an ERN2 ortholog gene, suggesting that ERN1 may function differently between these two species (Andriankaja et al., 2007; Yano et al., 2017; Liu et al., 2019a; Liu et al., 2019b). In this present study, we found that of 3,763 genes induced by rhizobial infection in WT plants, 234 were significantly decreased in Ljern1-6 mutants. These genes were found to be involved in processes including cell wall modification, signal transduction, phytohormone metabolism, and regulation of gene transcription. The differentially expressed genes (DEGs) with high fold change in Ljern1-6 encoded expansins, pectin methylesterases (PMEs), and PME inhibitors (PMEIs), which are related to cell wall loosening and extensity. The Ljern1 mutation was also found to reduce the expression of several LjNIN-targeting genes, which is consistent with our previous finding that LjERN1 and LjNIN coordinately affect downstream gene expression. This study extends our understanding of the regulatory network governed by LjERN1.
L. japonicus accession Miyakojima MG-20 (Kawaguchi, 2000) was used as the WT. Ljern1-6 mutant was generated from a MG-20 background by ion beam mutagenesis and carry approximately 10 kb deletion including the entire length of ERN1 (Yano et al., 2017). Ljnin-9 was isolated from MG-20 by EMS mutagenesis (Suzaki et al., 2012). The mutant lines Ljern1-1 and Ljnin-2 were generated from a Gifu Background (Schauser et al., 1999; Kawaharada et al., 2017a). L. japonicus seeds were surface-sterilized in 10% NaClO and germinated in 1/2 B5 medium in a growth chamber at 24°C (16 hr light/8 hr dark). Four-day-old seedlings were transferred to vermiculite with B&D medium (Broughton and Dilworth, 1971). Two days after adaptation, seedlings were inoculated with M. loti MAFF303099. For the inoculation, 15 mL of M. loti liquid culture (OD600 = 1.8-2.0) was diluted in 1 L of B&D medium. Each cultivation pot containing 10 plants was poured twice with 50 mL of the medium.
For each set of sampling conditions, three biological replicates were harvested (20 plants/sample) for total RNA isolation. Total RNA was isolated using the RNeasy Plant Mini Kit (QIAGEN). Genomic DNA was removed by treatment with DNase I (QIAGEN). The integrity of the RNA samples was determined using a bioanalyzer (Agilent). A 350 ng sample of RNA from each replicate was used for library preparation. Library construction was performed using the NEBNext® Ultra™ II RNA Library Prep Kit for Illumina (NEB) and the NEBNext® Poly(A) mRNA Magnetic Isolation Module (NEB). The concentration of the library was measured using a bioanalyzer (Agilent). RNA-seq was conducted on an Illumina HiSeq 2000 platform by single-end sequencing (read length = 50 bp).
For qRT-PCR validation, 50 ng of the total RNA from the extractions described above was used for each sample. The primer sequences were as follows: Ubiquitin_fwd, ACGGCTCTTATCAAGGGACCA; Ubiquitin_rev, CACTTGAGGTGGTTGTAGAGG; EXPB2_fwd, GGAGCTACGAAATGCTGGAA; EXPB2_rev, CACCATCCCCATCCTCATAC; Epr3_fwd, GTCTTCAGCGGGGTATTTGA; Epr3_rev, TGGCAGCAGTTTTGAACAAG; LOG4_fwd, CCTTGAAGAACTGTTGGAAATCATC; LOG4_rev, TCAAGCTTGCACATGAGGTCTTG; RINRK1(ALB1)_fwd, TATGCCTTTGGTGTGATGCT; RINRK1(ALB1)_rev, TCCACAGTCCATTCCTCTCT; NIN_fwd, AGCAAAGAGCATTGGTGTATGT; NIN_rev, AGCACCCTGCACTGAATCAA; Lj0g3v0070749_fwd, GGTTTGGAATTGGATGGTGTTG; Lj0g3v0070749_rev, AGGGACAAAATCAGAAGCACC; Lj0g3v0320499_fwd, GGTGCTGTTGATTTTATCTTTGGTG; Lj0g3v0320499_rev, GGTGCTGTTGATTTTATCTTTGGTG; Lj2g3v3339140_fwd, GGGAACGAACCCAAATGAAGAG; Lj2g3v3339140_rev, TCTCCTGTTACAAACTTGACCTTTG; Lj3g3v3751920_fwd, CAAGTGGTGGAGGATTGCTTTG; Lj3g3v3751920_rev, AGGTCAGCAACATCAAGACGT; Lj5g3v0642670_fwd, GGAGCTACGAAATGCTGGAA; Lj5g3v0642670_rev, CACCATCCCCATCCTCATAC.
After sequencing, 66 bp of each sample was trimmed by trimmomatic (v0.33) and aligned to the L. japonicus genome assembly (v3.0) using Tophat2 (v2.1.0; Kim et al., 2013). Raw counts were calculated using HTSeq (v0.6.0; Anders et al., 2015) and analyzed using the edgeR package (v3.26.8; Robinson et al., 2010; McCarthy et al., 2012) in R (version 3.6.1; R Core Team, 2020; v1.1.453; R Studio Team, 2020). After filtering, genes that met our criteria (FC > 1.5 and FDR < 0.05) were defined as DEGs. For K-means clustering, the package factoextra was used to estimate the optimal number of clusters (Kassambara and Mundt 2020). A heat map was generated using Z-scores with the pheatmap package (Kolde, 2019). A Venn diagram was produced by BioVenn (Hulsen et al., 2008) and the gplots package (Warnes et al., 2022). BLAST and GO enrichment were conducted using BLAST® command line applications (NCBI) and Blast2GO (Götz et al., 2008). FASTA files for BLAST were generated using the packages Biostrings (Pagès et al., 2020) and seqRFLP (Ding and Zhang, 2012). Protein kinase domains were predicted using SMART (Letunic et al., 2015; Letunic and Bork, 2018). The classification of transcription factors was based on PlantTFDB (Jin et al., 2014; Jin et al., 2015; Jin et al., 2017; Tian et al., 2020).The R packages openxlsx (Schauberger and Walker, 2020) and tidyverse (Wickham et al., 2019) were used to import and sort data.
For promoter-GUS analysis, the modified binary vector, pCAMBIA1300 whose HPTII was replaced with GFP and AscI site was introduced into the SmaI site was used (Kumagai and Kouchi, 2003). RINRK1 promoter region (2,976 bp) was amplified using a primer set (5’-ATGGTACCCGCAATATGAGCCACTGCTA-3’, 5’-ATGGCGCGCCTTTTTGCTCTGTATTTTTTTGTTGAATTGTGAAGTTAG-3’). The promoter fragment was digested with KpnI and AscI, and ligated with the vector. ALB1 terminator region (1,309 bp) was amplified using a primer set (5’-ATGGCGCGCCCCCAGAGTTTAGTTACCATGGAC-3’, 5’-ATGTCGACTGAACTTGCAGGAGGAGATG-3’). The terminator fragment was digested with AscI and SalI, and ligated with the vector. The reading frame cassette C.1 of the Gateway vector conversion system (Invitrogen) was inserted into AscI site of the vector. GUSPlus gene in pCAMBIA1305.1 was amplified by 2 rounds PCR using 1st primer set (5’-AAAAAGCAGGCTACCATGGTAGATCTGAGGGTAA-3’, 5’-AGAAAGCTGGGTTCACACGTGATGGTGATGGT-3’) and 2nd primer set (5’-GGGGACAAGTTTGTACAAAAAAGCAGGCT-3’, 5’-GGGGACCACTTTGTACAAGAAAGCTGGGT-3’). The GUSPlus fragment was inserted into pDONR/ZEO (Invitrogen) via Gateway BP reaction (Invitrogen). The GUSPlus gene was transferred between the promoter region and the terminator region via Gateway LR reaction (Invitrogen).
For complementation analysis, the RINRK1 (ALB1) cDNA amplified using a specific primer set (5’-AAGTCGACATGAGCCTAAAACCATTCTGGGC-3’, 5’-AAGCGGCCGCCATGTGTCAAATGATATGGATTTTTCATCT-3’) was inserted between SalI and NotI sites of pENTR-1A (Thermo Fisher Scientific), and subsequently transferred to pUb-GW-GFP (Maekawa et al., 2008) by the LR clonase reaction. L. japonicus hairy root transformation was basically performed with Rhizobium rhizogenes AR1193 as described previously (Díaz et al., 2005). Seedlings removed roots by cutting hypocotyls were co-cultured with R. rhizogenes harboring either pUb-RINRK1-GFP or its empty vector for four days, and then cultured on B5 ager plates for 13 days to generate hairy roots. Seedlings that formed hairy roots were inoculated with M. loti expressing DsRed (Maekawa et al., 2009) three days after transferring to sterile vermiculite. The number of nodules and infection threads formed in hairy roots displaying fluorescence from the GFP selection marker were counted at 22 dpi and 7 dpi, respectively, under an SZX16 stereomicroscope (Olympus).
To gain a deeper understanding of the function of LjERN1 in the transcription network, we conducted an RNA-seq to compare gene expression in WT plants and Ljern1-6 mutants, a strong allele of Ljern1, during the early stages of nodulation. L. japonicus wild type (WT; MG-20) and Ljern1-6 were inoculated with Mesorhizobium loti MAFF 303099. Four time points were selected; samples from 0 day post rhizobial infection (dpi) were used as controls, while samples from 1, 2, and 3 dpi covered the period from root hair deformation initiation to infection threads and nodule primordia becoming visible. Three biological replicates, each consisting of 20 plants, were utilized for the sequencing. Reads were mapped to version 3.0 of the L. japonicus MG-20 genome. According to the L. japonicus Gifu genome recently released (Kamal et al., 2020), many gene IDs in MG-20 may correspond to the same gene in Gifu. We thus annotated corresponding Gifu gene IDs in Table S1.
A multidimensional scaling plot demonstrated the separation of the WT and Ljern1-6 samples collected at 0 and 1−3 dpi (Figure 1A). After quality control and filtering, 47,232 genes were detected by RNA-seq. The expression of 3763 of these genes was up-regulated in response to rhizobial infection [fold change (FC) > 1.5, false discovery rate (FDR) < 0.05; Table S1]. We then subsequently grouped these 3,763 infection-induced DEGs into 7 clusters based on a K-means method (Figure S1). Genes in Cluster 2 and 7 showed lower transcript levels in Ljern1-6 than in the WT. To achieve better separation of genes affected by the LjERN1 mutation, we selected genes with higher fold change in Ljern1-6 compared with the WT (FC > 1.5) from Cluster 2 and 7 for further analyses. In total, 234 genes were identified as DEGs, with decreased expression in Ljern1-6 (Figure 1B; Table S2). To verify the RNA-seq result, we selected a few genes that were observed to change in expression by a large amount and checked their expression levels using qRT-PCR; NIN and LjEpr3 were used as positive controls. The fold changes indicated by qRT-PCR were comparable to those indicated by RNA-seq (Figure S2A). We also selected several known symbiosis genes from a recent review by Roy et al. (2019) and further examined their expression levels in the WT and Ljern1-6 plants (Figure S2B). The expression of genes that are essential for root nodule symbiosis, such as LjCCaMK, LjNSP1, and LjCYCLOPS, were increased in the WT, further confirming the result of RNA-seq. Notably, a few symbiosis genes have exhibited reduced expression levels in the Ljern1-6 mutants, including LjNPL, LONELY GUY4 (LjLOG4), Cytokinin oxidase/dehydrogenase3 (LjCKX3), LjCHIT5, LjNIN, LjNF-YA1, LjASL18, LjNOOT, LjCBS1, and LjRPG.
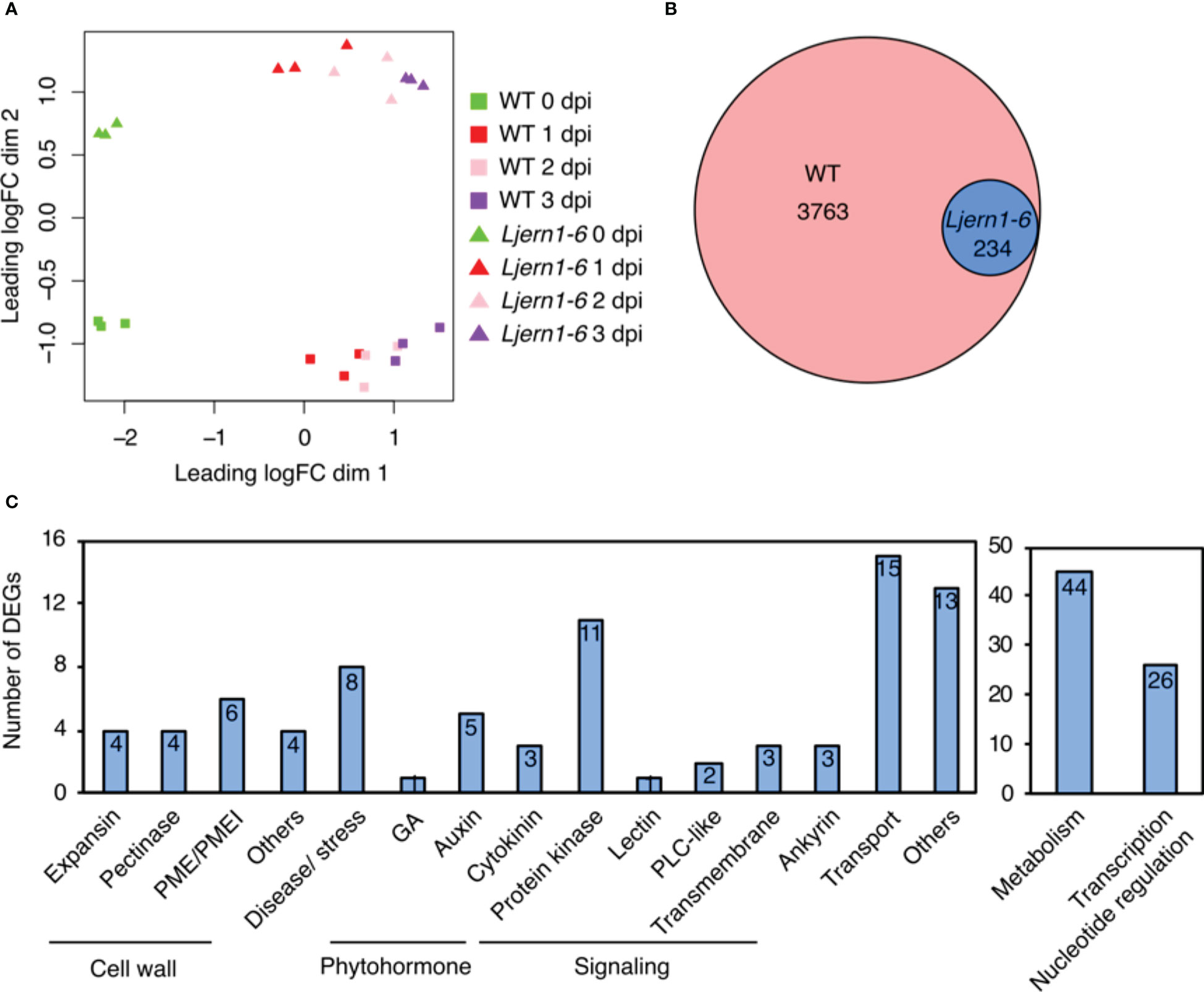
Figure 1 DEGs detected by RNA-seq in wild-type L. japonicus (MG-20) and Ljern1-6 mutants. (A) Multidimensional scaling plot of the effect of rhizobial infection on WT plants and Ljern1-6 mutants. (B) Proportional Venn diagram (Hulsen et al., 2008) shows the number of rhizobial infection-induced genes in WT and DEGs with decreased expression in Ljern1-6 mutants. (C) Classification of DEGs with decreased expression in Ljern1-6 mutants.
Gene Ontology (GO) analysis revealed that DEGs with decreased expression in Ljern1-6 were enriched in a variety of functions, including cell wall modification, signal transduction, transcription, and response to phytohormones (Table S3). Based on this result and BLASTP hits in M. truncatula and Arabidopsis thaliana, we manually classified the functions of 234 DEGs with decreased expression in Ljern1-6 into 17 categories (Figure 1C; Table S4; genes without any annotation were removed).
Root hair curling and subsequent infection thread formation require the synthesis and degradation of the cell wall. Nodule primordium development also necessitates the synthesis of new cell walls (Brewin, 2004; Gage, 2004; Lohar et al., 2006). Transcriptome analyses identified cell wall-associated genes encoding expansin, peroxidases, and proteases that were induced by rhizobial infection in both M. truncatula and Glycine max (Breakspear et al., 2014). We found that LjERN1 mutation affected the expression of 13 genes encoding expansins, pectinase (pectate and pectin lyase), PMEs, and PMEIs post rhizobial infection (Figure 2A), suggesting that LjERN1 may be involved in regulating cell wall-associated processes.
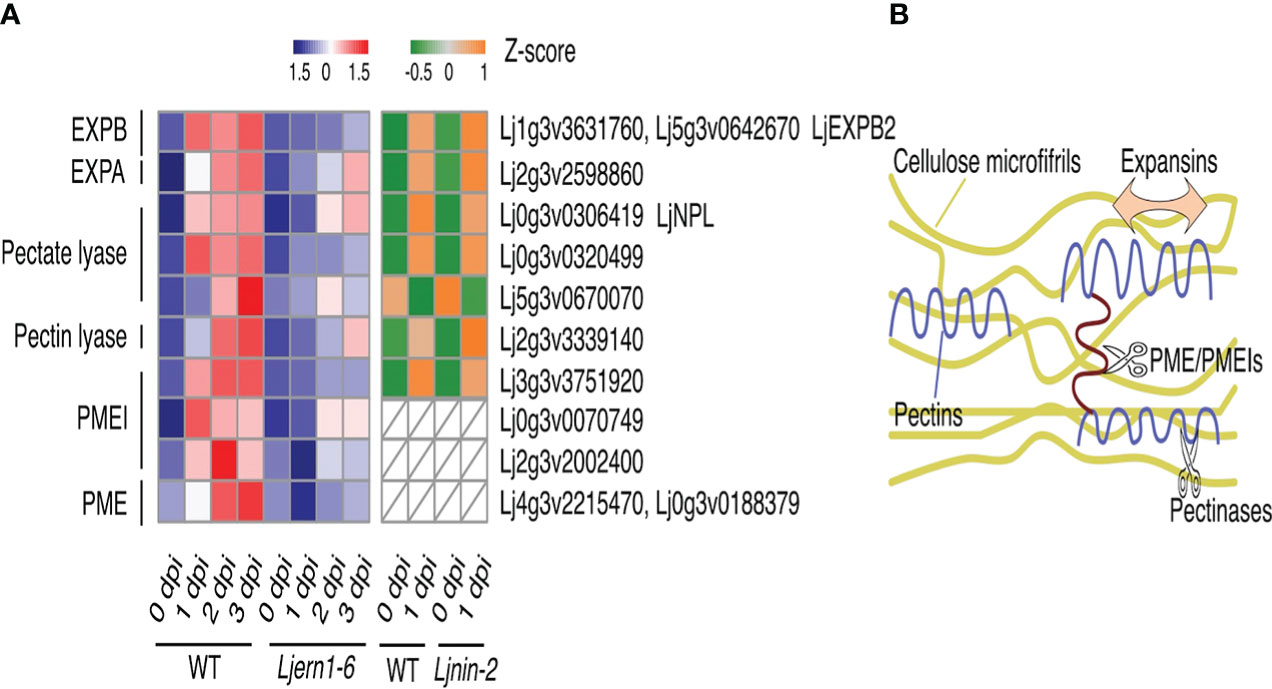
Figure 2 Cell wall-associated DEGs in WT plants and mutants. (A) Heatmap showing the expression levels of cell wall-associated DEGs in the WT and mutants at different time points. Left grid, Z-scores of genes in the WT (MG-20) and Ljern1-6 mutants at 0, 1, 2, and 3 dpi; right grid, Z-scores of genes in the WT (Gifu) and Ljnin-2 mutants at 0 and 1 dpi. (B) Illustration of the role of expansins, PEMs, PMEIs, and pectinases in the cell wall modification process.
Among these cell wall-related genes, the expression level of β-expansin2 (LjEXPB2) was highly decreased in Ljern1-6 mutants (Figure 2A; Figure S3). Expansins loosen the cellulose microfibrils by disintegrating the polysaccharide network, causing cell wall creep during cell growth (Figure 2B; Majda and Robert, 2018; Mohanty et al., 2018). Three expansin-encoding genes were induced by rhizobial infection in the WT, and LjEXPB2 expression was the most affected in Ljern1-6 mutants, especially at 1 dpi. LjEXPB2 may be involved in promoting root hair growth and infection thread formation through loosening of the cell wall at an early infection stage. It has been speculated that expansin plays a role in root nodule symbiosis. Previously, increased expression of EXP1 was detected in infected roots and nodules in Melilotus albus (Giordano and Hirsch, 2004). Immunoblotting showed that in Pisum sativum EXP1 is localized to infection thread walls (Sujkowska et al., 2006). Additionally, overexpression of EXPB2 in G. max increased the number of root hairs, infection threads, and root nodules (Li et al., 2015).
Pectins are embedded in the cellulose microfibrils of cell walls, which enhance cell wall strength (Majda and Robert, 2018; Figure 2B). The degradation of cell walls is necessary for the continuous growth of infection threads therefore involves the removal of pectins (Allan Downie and Xie, 2015). LjNPL encodes a pectate lyase and is required for rhizobia to penetrate root hair cell walls. In Ljnpl mutants, rhizobia are entrapped in the root hair tip and cannot develop into infection threads (Xie et al., 2012). The expression of one pectin lyase gene, two pectate lyase genes, and LjNPL was decreased in Ljern1-6 mutants (Figure 2A); Suppression of these genes may interrupt cell wall degradation in Ljern1-6 mutants.
The Ljern1 mutation also strongly affected genes encoding two cell wall-associated enzymes, PMEs, and their inhibitors, PMEIs (Figure 2A). PMEs have been identified to modify the crosslinks among different pectin domains through demethylesterification, thus softening the cell wall (Majda and Robert, 2018). This activity is negatively regulated by PMEIs (Figure 2B). Among the six rhizobial infection-induced PME- and PMEI-encoding genes, Lj3g3v3751920 and Lj0g3v0070749 showed the highest fold change in Ljern1-6 mutants at 1 dpi, suggesting that PMEI may function during an early stage of infection. In A. thaliana, AtPMEI2 interacts with AtPME1 to regulate cell wall stability at the apex of the pollen tube. Transient expression of AtPMEI2 was observed to increase the pollen tube length in tobacco, suggesting a role of PMEI in promoting polar cell growth (Röckel et al., 2008). In M. truncatula, a PME gene, MtPER, was proposed to have been recruited from the pollen tube elongation process to root nodule symbiosis (Rodríguez-Llorente et al., 2004), which suggests that PME and PMEIs may share similar functions in pollen tube growth and infection thread formation. The reduced expression of LjPMEIs may disrupt the cell wall extensity of the Ljern1-6 root hair and lead to abnormal tip growth.
Because the expression of LjNIN was decreased in Ljern1-6 mutants (Figure S2; Liu et al., 2019b), we compared the transcription profile of Ljern1-6 obtained from the present RNA-seq analysis with DNA array data of the WT (Gifu) and Ljnin-2 from the Lotus japonicus Gene Expression Atlas (Lotus Base; Mun et al., 2016) to determine whether the reduced expression of these cell wall-related genes was a secondary effect of LjNIN. Of all DEGs with decreased expression in Ljern1-6 mutants, corresponding probes of 128 were detected in the DNA array. The expression of 32 genes was LjNIN-dependent (FC of wild type/nin > 1.5 at 1 dpi) (Table S5), while the remaining 32 genes were LjNIN-independent (Table S6). The expression of pectinase-, PME- and PMEI-encoding genes was reduced in Ljnin-2 mutants, whereas the expansin genes showed a comparable expression level in the wild type (Gifu) (Figure 2A). The regulation of LjEXPB2 may be mainly dependent on LjERN1, making LjEXPB2 a candidate LjERN1 target. We examined LjEXPB2 expression in MG-20-derived Ljern1-6 and Ljnin-9 roots using qRT-PCR and found that the induction of LjEXPB2 expression in response to rhizobial infection appeared to be LjERN1-dependent and LjNIN-independent (Figure S3). Cell wall-associated genes such as LjEXPB2 may contribute to cell wall loosening, degradation, and reconstruction during infection thread formation and nodule development. The reduced expression of LjEXPB2 could interrupt cell wall dynamics in Ljern1-6 mutants.
Nine protein kinase genes showed decreased expression in Ljern1-6 mutants, including two well-studied receptor-like kinase (RLK) genes, LjEpr3 and LjRINRK1 (Figure 3A; Figure S3). LjEPR3 recognizes rhizobial exopolysaccharides and regulates rhizobial passage through the host epidermal cell layer (Kawaharada et al., 2015; Kawaharada et al., 2017b). LjRINRK1 is likely involved in positive feedback with LjNIN and amplifies the infection signal (Li et al., 2019). In order to confirm whether ERN1 up-regulates the expression of RINRK1, the transcript accumulation of RINRK1 was compared by qRT-PCR in the background of MG-20 and Ljern1-6. In MG-20, RINRK1 was significantly induced 1 dpi, and its expression was further increased 3 dpi. On the other hand, the induction decreased at about 1/5 to 1/7 of MG-20 in the Ljern1-6 background (Figure 4A). In addition, constitutive expression of ERN1 tended to increase the expression of RINRK1 in Ljnin-2 mutants, under both uninfected and infected roots (Figure 4B). On the other hand, UBp : NIN also induced RINRK1 expression in uninfected and infected roots of Ljern1-1 (Figure S4). These results suggest that transcriptional activation of RINRK1 is NIN- and ERN1-dependent. Indeed, UBp : ERN1 failed to fully restore RINRK1 expression in the nin-2 mutant, suggesting that both NIN and ERN1 are required for full RINRK1 expression.
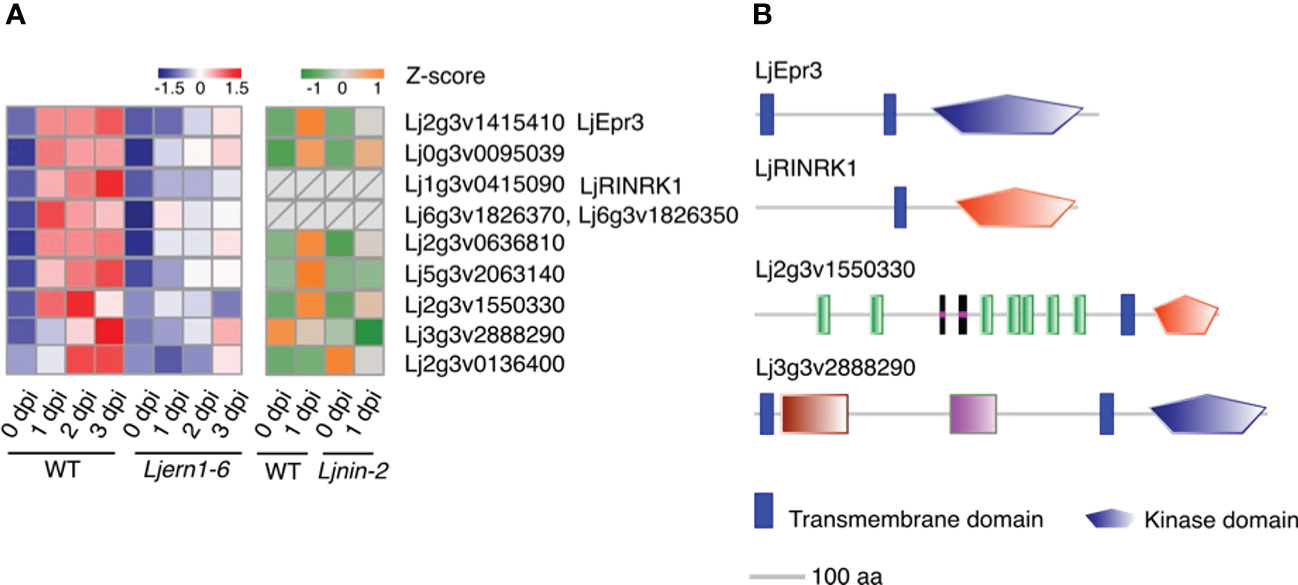
Figure 3 Expression of differentially expressed kinase genes in WT and mutants. (A) Heatmap of changes in the expression of differentially expressed kinase genes in the WT (MG-20) and Ljern1-6 mutants (left grid) and WT (Gifu) and Ljnin-2 mutants (right grid). (B) Receptor-like kinase prediction. Extracellular, transmembrane, and kinase domains were predicted using SMART (Letunic et al., 2015; Letunic and Bork, 2018); LjEpr3 and LjRINRK1 were used as positive controls. Blue stands for S_TKc, Serine/Threonine protein kinases, catalytic domain while red stands for STYKc, protein kinase; unclassified specificity.
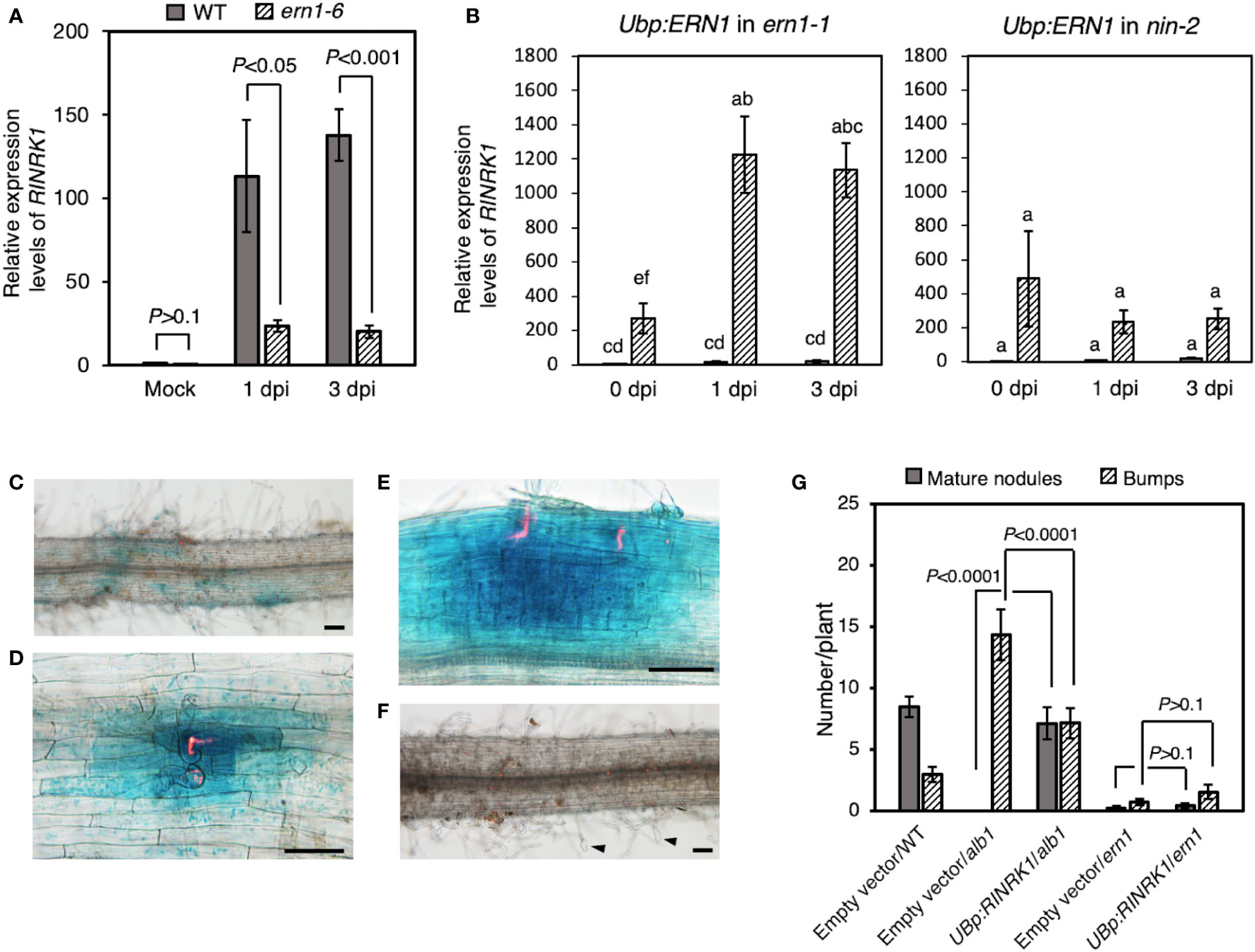
Figure 4 Positive regulation of RINRK1 by ERN1. (A) Relative expression levels of RINRK1 in WT and Ljern1-6. (B) Relative expression levels of RINRK1 in Ljern1-1 and Ljnin-2. Roots transformed with an empty vector (gray bar) or UBp : ERN1 (stripe bar). Data are means of fold changes normalized to Ubiquitin and displayed relative to the empty vector control of Ljern1-1 at 0 dpi (non-inoculation). Error bars indicate SE (n = 3, sample size = 12 plants). Statistical analysis was performed by ANOVA followed by Tukey's HSD test (P<0.05). GUS expression from RINRK promoter after inoculation with DsRed-labeled M. loti. Roots of WT (C–E) or the Ljern1-6 mutants (F). Scale bars, 200μm. Arrow heads: balloon-shaped root hairs. (G) Effects of overexpression of RINRK1 on nodulation in the hairy roots of the alb1 and Ljern1-1 mutants.
Then, to analyze the spatial expression pattern of RINRK1, we constructed a GUS reporter carrying its promoter (2,976 bp), and transformed to the hairy roots via R. rhizogenes. Infection threads are visualized when infected with DsRED-labeled rhizobia. RINRK1 was expressed in the region where the infection threads were formed (Figure 4C), and then strongly expressed in the divided cortical cells and nodule primordia (Figures 4D, E). On the other hand, in the Ljern1-6 background, no expression of GUS was observed even in the region where the characteristic root hair deformation was observed after the infection (Figure 4F). Subsequently, we investigated whether the failure of the infectious process in Ljern1-6 could be suppressed by constitutive expression of RINRK1. Prior to the experiment, we confirmed that the efficiency of formation of mature nodules was increased by introducing RINRK1 into the alb1 mutant carrying a mutation in RINRK1 (Figure 4G). When RINRK1 was overexpressed in the hairy roots of the Ljern1-6 mutant, nodule primordia and mature nodules tended to increase slightly compared to the lines in which the empty vector was introduced (Figure 4G). On the other hand, the average numbers of ITs (± SD) was 1.40 ± 1.62 (n = 15) in the Ljern1-1 hairy roots transfected with the empty vector, and 1.47 ± 2.06 (n = 15) in the hairy roots transfected with the UBp : RINRK1. Phenotypic suppression was not sufficient, suggesting that host factors other than RINRK1 regulate downstream of ERN1 also contributed significantly to the infection process.
In addition to these two kinases, the SMART analysis has revealed that the predicted proteins of Lj2g3v1550330 and Lj3g3v2888290 feature RLK structures (extracellular domain + transmembrane domain + kinase domain; Figure 3B; Shiu and Bleecker, 2001; Letunic et al., 2015; Letunic and Bork, 2018). Lj0g3v0095039 did not contain typical RLK domains, but its best BLASTP hit in A. thaliana was predicted to be RLKs (Shiu and Bleecker, 2001). The prediction that these three DEGs encode RLKs implies that they play a role in signal transduction. The reduced expression of these kinase genes may inhibit the ability of Ljern1-6 mutants to identify compatible rhizobia and transduce signals to the nucleus. Similarly, in Mtern1 mutants, the expression of 30 kinase genes was reduced (Liu et al., 2019a), suggesting that ERN1 may have similar functions in signal transduction in two species.
Previous studies have shown that Ljern1 alleles are deficient in response to cytokinin and auxin signaling during root nodule symbiosis (Kawaharada et al., 2017a; Nadzieja et al., 2018), which implies a role of LjERN1 in phytohormone signaling. In the present Ljern1-6 dataset, we found that four genes associated with phytohormones showed decreased expression levels (Figure 5). GIBBERELLIN 3 BETA-HYDROXYLASE1 (LjGA3ox1) encodes an enzyme that converts GA to a bioactive form. GA negatively regulates root nodule symbiosis through the degradation of DELLA proteins (Fonouni-Farde et al., 2016; Jin et al., 2016). The decreased expression of LjGA3ox1 in Ljern1-6 mutants may be a secondary effect of decreased numbers of infection threads in these mutants.
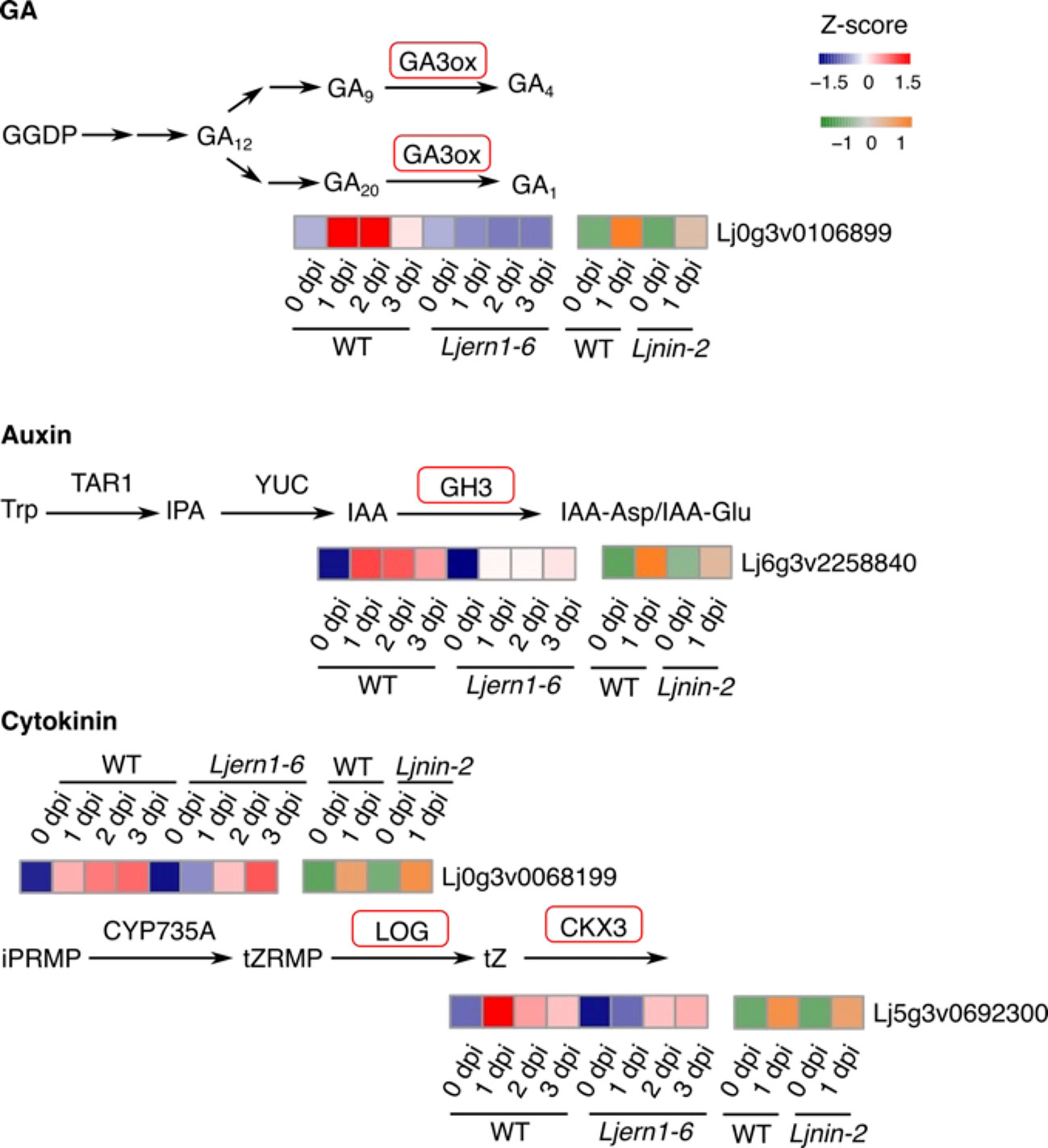
Figure 5 Phytohormone-associated DEGs in WT and mutants. Simplified GA, auxin, and cytokinin biosynthesis pathways were shown. Genes encoding enzymes with red frames were significantly reduced in Ljern1-6 mutants. Heatmaps show the expression patterns of corresponding genes in WT and mutants.
Nadzieja et al. (2018) showed that auxin is necessary for infection thread formation in L. japonicus. The expression of an auxin-related genes, Gretchen Hagen3 (LjGH3), was reduced in Ljern1-6 mutants in the present study. GH3 balances auxin levels by catalyzing the conjugation of auxin. It is possible that LjERN1 is related to auxin homeostasis, which, in turn, affects root nodule symbiosis.
Consistent with a previous report (Reid et al., 2017), we found that LjLOG4, which encodes an enzyme catalyzing the conversion of cytokinin precursors to a bioactive form, showed decreased expression levels in Ljern1-6 mutants (Figure S3). Subsequently, the expression level of LjCKX3, which was involved in the breakdown of cytokinin, was also decreased (Figure 5). Cytokinin has been identified to suppress infection thread formation but promotes cortical cell division (Murray et al., 2007; Tirichine et al., 2007; Miri et al., 2016). Although LjERN1 is not essential for nodule organogenesis as all Ljern1 allele lines are able to produce root nodules, LjERN1 mediates the formation of cytokinin-induced spontaneous root nodules (Kawaharada et al., 2017a). The decreased expression of LjLOG4 and LjCKX3 may explain the lack of spontaneous root nodules in Ljern1 lines.
The expression levels of all four phytohormone-related DEGs were increased in Ljern1-6 mutants to levels comparable with those in the wild type after 2 dpi, suggesting that LjERN1 may be involved in their regulation at an early stage. LjGA3ox1 and LjGH3 have displayed decreased expression levels in Ljnin-2 lines, suggesting that their regulation may be dependent on both LjERN1 and LjNIN. LjERN1 may be more involved in the regulation of the cytokinin pathway, since LjLOG4 and LjCKX3 expression was decreased in Ljern1-6 mutants but not in Ljnin-2 mutants.
To gain a better understanding of the transcription network downstream of LjERN1, we also examined the expression of transcription factor-encoding genes in WT plants and Ljern1-6 mutants. Based on the classification of transcription factor families and gene IDs from PlantTFDB (Jin et al., 2014; Jin et al., 2015; Jin et al., 2017; Tian et al., 2020), we analyzed the expression of 14 transcription factor genes that were decreased in Ljern1-6 mutants (Figure 6). The reduced expression levels of LjNIN and its target genes, such as LjNF-YA1, were consistent with our previous findings (Liu et al., 2019b). The expression of LjNF-YA1 and other LjNIN target genes may be solely dependent on LjNIN or may require regulation by both LjERN1 and LjNIN. Based on a combination of our RNA-seq results and the DNA array data, it seemed that the expression of the Myb transcription factor gene Lj5g3v2013880 was dependent on LjERN1 but not NIN. It is possible that together with a few other transcription factor genes in the Wox, WRKY, and bHLH families, these transcription factors are involved in mediating the regulation of LjNIN expression downstream of LjERN1.
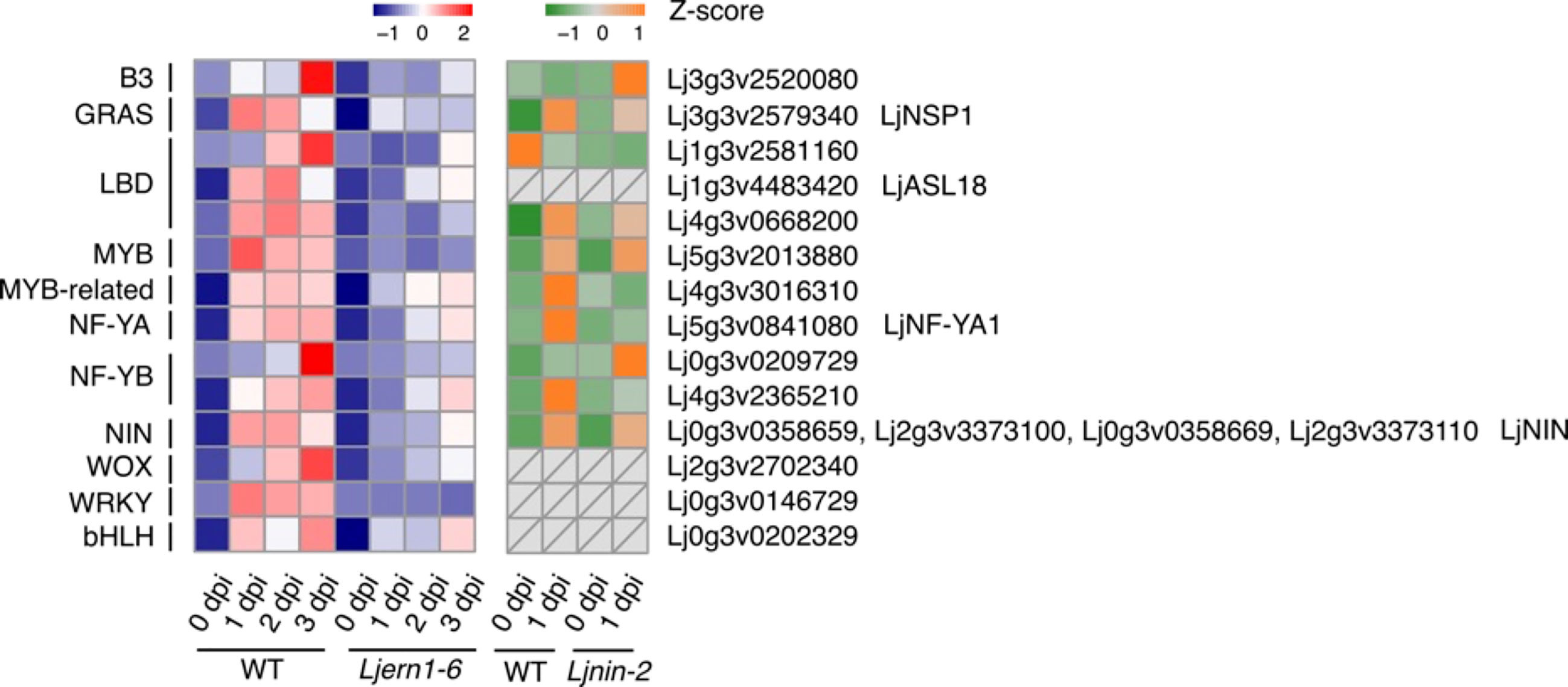
Figure 6 Transcript response of transcription factor genes in WT and mutants Heatmap showing expression patterns of differentially expressed transcription factor genes in WT (MG-20) and Ljern1-6 mutants (left grid) and WT (Gifu B-129) and Ljnin-2 mutants (right grid).
In this present study, we transcriptionally compared gene expression profiles between WT L. japonicus and Ljern1-6 mutants in response to rhizobial infection. LjERN1 affected cell wall remodeling via expansin, pectinases, PMEs, and PMEIs to affect infection thread formation and cortical cell division. LjERN1 may also be involved in mediating signal transduction through protein kinases, including LjEpr3. During root nodule symbiosis, phytohormone signaling is finely tuned, which may also require the involvement of LjERN1. Many of the DEGs with decreased expression in Ljern1-6 mutants have also showed decreased expression levels in Ljnin-2 mutants, suggesting that they were also regulated by LjNIN. This supports the theory that LjERN1 and LjNIN may have a close relationship in the regulation of gene expression. However, the present study did not determine whether the multifunctional LjERN1 is involved in rhizobial infection, nodule organogenesis, or both processes. Future work is needed to resolve this concern.
RNA sequencing data are available in the SRA database in the BioProject PRJDB13938; https://ddbj.nig.ac.jp/resource/sra-submission/DRA014481.
ML, TS and MK conceived and designed the analysis; ML, AO, KY and TG collected data and performed analyses; HK and TM contributed analysis tools and performed analysis; TG deposited RNA sequence data to INSCD. ML, TS and MK wrote the paper. All authors contributed to the article and approved the submitted version.
This work was supported by Ministry of Education, Culture, Sports, Science and Technology Grants-in-Aid for Scientific Research [grant number 17H03702, 20H03283 to MK].
We appreciate the technical support from Dr. Shuji Shigenobu, and Dr. Katsushi Yamaguchi at Functional Genomics Facility and Data Integration and Analysis Facility, NIBB Core Research Facilities. We thank Dr. Shusei Sato at Tohoku University for sharing the genome information of L. japonicus Gifu, Dr. Jens Stougaard at Aarhus University for nin-2 seeds and LORE1 mutant resource for ern1-1 seeds. We also thank Dr. Bihe Kong at Meiji University for her suggestions on coding.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.995589/full#supplementary-material
Allan Downie, J., Xie, F. (2015). A pectate lyase required for plant cell-wall remodeling during infection of legumes by rhizobia. Biol. Nitrogen Fixat. 2–2, 575–582. doi: org/10.1002/9781119053095.ch58
Amor, B., Shaw, S. L., Oldroyd, G. E. D., Maillet, F., Penmetsa, R. V., Cook, D., et al. (2003). The NFP locus of Medicago truncatula controls an early step of nod factor signal transduction upstream of a rapid calcium flux and root hair deformation. Plant J. 34, 495–506. doi: 10.1046/j.1365-313x.2003.01743.x
Anders, S., Pyl, P. T., Huber, W. (2015). HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169. doi: 10.1093/bioinformatics/btu638
Andriankaja, A., Boisson-Dernier, A., Frances, L., Sauviac, L., Jauneau, A., Barker, D. G., et al. (2007). AP2-ERF transcription factors mediate nod factor-dependent MtENOD11 activation in root hairs via a novel cis-regulatory motif. Plant Cell 19, 2866–2885. doi: 10.1105/tpc.107.052944
Breakspear, A., Liu, C., Roy, S., Stacey, N., Rogers, C., Trick, M., Murray, J. D., et al. (2014). The root hair “infectome” of Medicago truncatula uncovers changes in cell cycle genes and reveals a requirement for auxin signaling in rhizobial infection. Plant Cell 26, 4680–4701. doi: 10.1105/tpc.114.133496
Brewin, N. J. (2004). Plant cell wall remodelling in the rhizobium-legume symbiosis. CRC. Crit. Rev. Plant Sci. 23, 293–316. doi: org/10.1080/07352680490480734
Broughton, W. J., Dilworth, M. J. (1971). Control of leghaemoglobin synthesis in snake beans. Biochem. J. 125, 1075–1080. doi: 10.1042/bj1251075
Buhian, W. P., Bensmihen, S. (2018). Mini-review: Nod factor regulation of phytohormone signaling and homeostasis during rhizobia-legume symbiosis. Front. Plant Sci. 9 1247. doi: 10.3389/fpls.2018.01247
Catoira, R., Galera, C., De Billy, F., Penmetsa, R. V., Journet, E. P., Maillet, F., et al. (2000). Four genes of Medicago truncatula controlling components of a nod factor transduction pathway. Plant Cell 12, 1647–1665. doi: 10.1105/tpc.12.9.1647
Cerri, M. R., Frances, L., Kelner, A., Fournier, J., Middleton, P. H., Auriac, M. C., et al. (2016). The symbiosis-related ERN transcription factors act in concert to coordinate rhizobial host root infection. Plant Physiol. 171, 1037–1054. doi: 10.1104/pp.16.00230
Cerri, M. R., Wang, Q., Stolz, P., Folgmann, J., Frances, L., Katzer, K., et al. (2017). The ERN1 transcription factor gene is a target of the CCaMK/CYCLOPS complex and controls rhizobial infection in Lotus japonicus. New Phytol. 215, 323–337. doi: 10.1111/nph.14547
Combier, J. P., Frugier, F., De Billy, F., Boualem, A., El-Yahyaoui, F., Moreau, S., et al. (2006). MtHAP2-1 is a key transcriptional regulator of symbiotic nodule development regulated by microRNA169 in Medicago truncatula. Genes Dev. 20, 3084–3088. doi: 10.1101/gad.402806
Díaz, C. L., Grønlund, M., Schlaman, H. R. M., Spaink, H. P. (2005). “Induction of hairy roots for symbiotic gene expression studies,” in Lotus japonicus handbook. Ed. Márquez, A. J. (Springer), pp261–pp277.
Ding, Q., Zhang, J. (2012)seqRFLP: Simulation and visualization of restriction enzyme cutting pattern from DNA sequences. Available at: https://CRAN.R-project.org/package=seqRFLP.
Ehrhardt, D. W., Wais, R., Long, S. R. (1996). Calcium spiking in plant root hairs responding to rhizobium nodulation signals. Cell 85, 673–681. doi: 10.1016/s0092-8674(00)81234-9
Fonouni-Farde, C., Tan, S., Baudin, M., Brault, M., Wen, J., Mysore, K. S., et al. (2016). DELLA-mediated gibberellin signalling regulates nod factor signalling and rhizobial infection. Nat. Commun. 7, 12636. doi: 10.1038/ncomms12636
Gage, D. J. (2004). Infection and invasion of roots by symbiotic, nitrogen-fixing rhizobia during nodulation of temperate legumes. Microbiol. Mol. Biol. Rev. 68, 280–300. doi: 10.1128/MMBR.68.2.280-300.2004
Giordano, W., Hirsch, A. M. (2004). The expression of MaEXP1, a Melilotus alba expansin gene, is upregulated during the sweetclover-Sinorhizobium meliloti interaction. Mol. Plant-Microbe Interact. 17, 613–622. doi: 10.1094/MPMI.2004.17.6.613
Goto, T., Soyano, T., Liu, M., Mori, T., Kawaguchi, M. (2022). Auxin methylation by IAMT1, duplicated in the legume lineage, promotes root nodule development in Lotus japonicus. Proc. Natl. Acad. Sci. U. S. A. 119, e2116549119. doi: 10.1073/pnas.2116549119
Götz, S., García-Gómez, J. M., Terol, J., Williams, T. D., Nagaraj, S. H., Nueda, M. J., et al. (2008). High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 36, 3420–3435. doi: 10.1093/nar/gkn176
Guinel, F. C., Geil, R. D. (2002). A model for the development of the rhizobial and arbuscular mycorrhizal symbioses in legumes and its use to understand the roles of ethylene in the establishment of these two symbioses. Can. J. Bot. 80, 695–720. doi: org/10.1139/b02-066
Heckmann, A. B., Lombardo, F., Miwa, H., Perry, J. A., Bunnewell, S., Parniske, M., et al. (2006). Lotus japonicus nodulation requires two GRAS domain regulators, one of which is functionally conserved in a non-legume. Plant Physiol. 142, 1739–1750. doi: 10.1104/pp.106.089508
Heckmann, A. B., Sandal, N., Bek, A. S., Madsen, L. H., Jurkiewicz, A., Nielsen, M. W., et al. (2011). Cytokinin induction of root nodule primordia in Lotus japonicus is regulated by a mechanism operating in the root cortex. Mol. Plant-Microbe Interact. 24, 1385–1395. doi: 10.1094/MPMI-05-11-0142
Hulsen, T., de Vlieg, J., Alkema, W. (2008). BioVenn–a web application for the comparison and visualization of biological lists using area-proportional Venn diagrams. BMC Genomics 9, 488. doi: 10.1186/1471-2164-9-488
Jin, J., He, K., Tang, X., Li, Z., Lv, L., Zhao, Y., et al. (2015). An Arabidopsis transcriptional regulatory map reveals distinct functional and evolutionary features of novel transcription factors. Mol. Biol. Evol. 32, 1767–1773. doi: 10.1093/molbev/msv058
Jin, Y., Liu, H., Luo, D., Yu, N., Dong, W., Wang, C., et al. (2016). DELLA proteins are common components of symbiotic rhizobial and mycorrhizal signalling pathways. Nat. Commun. 7, 12433. doi: 10.1038/ncomms12433
Jin, J., Tian, F., Yang, D. C., Meng, Y. Q., Kong, L., Luo, J., et al. (2017). PlantTFDB 4.0: Toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 45, D1040–D1045. doi: 10.1093/nar/gkw982
Jin, J., Zhang, H., Kong, L., Gao, G., Luo, J. (2014). PlantTFDB 3.0: A portal for the functional and evolutionary study of plant transcription factors. Nucleic Acids Res. 42, 1182–1187. doi: 10.1093/nar/gkt1016
Kaló, P., Gleason, C., Edwards, A., Marsh, J., Mitra, R. M., Hirsch, S., Oldroyd, G. E. D., et al. (2005). Nodulation signaling in legumes requires NSP2, a member of the GRAS family of transcriptional regulators. Science 308, 1786–1789. doi: 10.1126/science.1110951
Kamal, N., Mun, T., Reid, D., Lin, J., Akyol, T. Y., Sandal, N., et al. (2020). Insights into the evolution of symbiosis gene copy number and distribution from a chromosome-scale Lotus japonicus gifu genome sequence. DNA Res. 27, dsaa015. doi: 10.1093/dnares/dsaa015
Kassambara, A., Mundt, F. (2020) Factoextra: Extract and visualize the results of multivariate data analyses. Available at: https://CRAN.R-project.org/package=factoextra.
Kawaguchi, M. (2000). Lotus japonicus “Miyakojima“ MG-20: An early-flowering accession suitable for indoor handling. J. Plant Res. 113, 507–509. doi: org/10.1007/PL00013961
Kawaharada, Y., James, E. K., Kelly, S., Sandal, N., Stougaard, J. (2017a). The ERF required for Nodulation1 (ERN1) transcription factor is required for infection thread formation in Lotus japonicus. Mol. Plant-Microbe Interact. 30, 194–204. doi: 10.1094/MPMI-11-16-0237-R
Kawaharada, Y., Kelly, S., Nielsen, M. W., Hjuler, C. T., Gysel, K., Muszyński, A., et al. (2015). Receptor-mediated exopolysaccharide perception controls bacterial infection. Nature 523, 308–312. doi: 10.1038/nature14611
Kawaharada, Y., Nielsen, M. W., Kelly, S., James, E. K., Andersen, K. R., Rasmussen, S. R., et al. (2017b). Differential regulation of the Epr3 receptor coordinates membrane-restricted rhizobial colonization of root nodule primordial. Nat. Commun. 8, 14534. doi: 10.1038/ncomms14534
Kim, D., Pertea, G., Trapnell, C., Pimentel, H., Kelley, R., Salzberg, S. L. (2013). TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene functions. Genome Biol. 14, R36. doi: 10.1186/gb-2013-14-4-r36
Kolde, R. (2019)Pheatmap: Pretty heatmaps. Available at: https://CRAN.R-project.org/package=pheatmap.
Kumagai, H., Kouchi, H. (2003). Gene silencing by expression of hairpin RNA in lotus japonicus roots and root nodules. Mol. Plant-Microbe Interact. 16, 663–668. doi: 10.1094/MPMI.2003.16.8.663
Laloum, T., Baudin, M., Frances, L., Lepage, A., Billault-Penneteau, B., Cerri, M. R., et al. (2014). Two CCAAT-box-binding transcription factors redundantly regulate early steps of the legume-rhizobia endosymbiosis. Plant J. 79, 757–768. doi: 10.1111/tpj.12587
Letunic, I., Bork, P. (2018). 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 46, D493–D496. doi: 10.1093/nar/gkx922
Letunic, I., Doerks, T., Bork, P. (2015). SMART: Recent updates, new developments and status in 2015. Nucleic Acids Res. 43, D257–D260. doi: 10.1093/nar/gku949
Lévy, J., Bres, C., Geurts, R., Chalhoub, B., Kulikova, O., Duc, G., et al. (2004). A putative Ca2+ and calmodulin-dependent protein kinase required for bacterial and fungal symbioses. Science 303, 1361–1364. doi: 10.1126/science.1093038
Limpens, E., Franken, C., Smit, P., Willemse, J., Bisseling, T., Geurts, R. (2003). LysM domain receptor kinases regulating rhizobial nod factor. Science 302, 630–633. doi: 10.1126/science.1090074
Lin, J., Frank, M., Reid, D. (2020). No home without hormones: How plant hormones control legume nodule organogenesis. Plant Commun. 1, 1–20. doi: 10.1016/j.xplc.2020.100104
Liu, C. W., Breakspear, A., Guan, D., Cerri, M. R., Jackson, K., Jiang, S., et al. (2019a). NIN acts as a network hub controlling a growth module required for rhizobial infection. Plant Physiol. 179, 1704–1722. doi: 10.1104/pp.18.01572
Liu, H., Sandal, N., Andersen, K. R., James, E. K., Stougaard, J., Kawaharada, Y. (2018). A genetic screen for plant mutants with altered nodulation phenotypes in response to rhizobial glycan mutants. New Phytol. 220, 526–538. doi: 10.1111/nph.15293
Liu, M., Soyano, T., Yano, K., Hayashi, M., Kawaguchi, M. (2019b). ERN1 and CYCLOPS coordinately activate NIN signaling to promote infection thread formation in Lotus japonicus. J. Plant Res. 132, 641–653. doi: 10.1007/s10265-019-01122-w
Li, X., Zhao, J., Tan, Z., Zeng, R., Liao, H. (2015). GmEXPB2, a cell wall β-expansin gene, affects soybean nodulation through modifying root architecture and promoting nodule formation and development. Plant Physiol. 169, 2640–2653. doi: 10.1104/pp.15.01029
Li, X., Zheng, Z., Kong, X., Xu, J., Qiu, L., Sun, J., et al. (2019). Atypical receptor kinase RINRK1 required for rhizobial infection but not nodule development in Lotus japonicus. Plant Physiol. 181, 804–816. doi: 10.1104/pp.19.00509
Lohar, D. P., Sharopova, N., Endre, G., Peñuela, S., Samac, D., Town, C., et al. (2006). Transcript analysis of early nodulation events in Medicago truncatula. Plant Physiol. 140, 221–234. doi: 10.1104/pp.105.070326
Madsen, E. B., Madsen, L. H., Radutoiu, S., Olbryt, M., Rakwalska, M., Szczyglowski, K., et al. (2003). A receptor kinase gene of the LysM type is involved in legumeperception of rhizobial signals. Nature 425, 637–640. doi: 10.1038/nature02045
Maekawa, T., Kusakabe, M., Shimoda, Y., Sato, S., Tabata, S., Murooka, Y., et al. (2008). Polyubiquitin promoter-based binary vectors for overexpression and gene silencing in Lotus japonicus. Mol. Plant Microbe Interact. 21, 375–382. doi: 10.1094/MPMI-21-4-0375
Maekawa, T., Maekawa-Yoshikawa, M., Takeda, N., Imaizumi-Anraku, H., Murooka, Y., Hayashi, M. (2009). Gibberellin controls the nodulation signaling pathway in Lotus japonicus. Plant J. 58, 183–194. doi: 10.1111/j.1365-313X.2008.03774.x
Majda, M., Robert, S. (2018). The role of auxin in cell wall expansion. Int. J. Mol. Sci. 19, 951. doi: 10.3390/ijms19040951
Marsh, J. F., Rakocevic, A., Mitra, R. M., Brocard, L., Sun, J., Eschstruth, A., et al. (2007). Medicago truncatula NIN is essential for rhizobial-independent nodule organogenesis induced by autoactive Calcium/Calmodulin-dependent protein kinase. Plant Physiol. 144, 324–335. doi: 10.1104/pp.106.093021
McCarthy, D. J., Chen, Y., Smyth, G. K. (2012). Differential expression analysis of multifactor RNA-seq experiments with respect to biological variation. Nucleic Acids Res. 40, 4288–4297. doi: 10.1093/nar/gks042
Messinese, E., Mun, J. H., Li, H. Y., Jayaraman, D., Rougé, P., Barre, A., et al. (2007). A novel nuclear protein interacts with the symbiotic DMI3 calcium- and calmodulin-dependent protein kinase of Medicago truncatula. Mol. Plant-Microbe Interact. 20, 912–921. doi: 10.1094/MPMI-20-8-0912
Middleton, P. H., Jakab, J., Penmetsa, R. V., Starker, C. G., Doll, J., Kaló, P., Oldroyd, G. E. D., et al. (2007). An ERF transcription factor in Medicago truncatula that is essential for nod factor signal transduction. Plant Cell 19, 1221–1234. doi: 10.1105/tpc.106.048264
Miri, M., Janakirama, P., Held, M., Ross, L., Szczyglowski, K. (2016). Into the root: how cytokinin controls rhizobial infection. Trends Plant Sci. 21, 178–186. doi: 10.1016/j.tplants.2015.09.003
Miwa, H., Sun, J., Oldroyd, G. E. D., Allan Downie, J. (2006). Analysis of calcium spiking using a cameleon calcium sensor reveals that nodulation gene expression is regulated by calcium spike number and the developmental status of the cell. Plant J. 48, 883–894. doi: 10.1111/j.1365-313X.2006.02926.x
Mohanty, S. K., Arthikala, M. K., Nanjareddy, K., Lara, M. (2018). Plant-symbiont interactions: the functional role of expansins. Symbiosis 74, 1–10.
Mun, T., Bachmann, A., Gupta, V., Stougaard, J., Andersen, S. U. (2016). Lotus base: an integrated information portal for the model legume Lotus japonicus. Sci. Rep. 6, 39447. doi: 10.1038/srep39447
Murakami, E., Cheng, J., Gysel, K., Bozsoki, Z., Kawaharada, Y., Hjuler, C. T., et al. (2018). Epidermal LysM receptor ensures robust symbiotic signalling in Lotus japonicus. Elife 7, e33506. doi: 10.7554/eLife.33506
Murakami, Y., Miwa, H., Imaizumi-Anraku, H., Kouchi, H., Downie, J. A., Kawaguchi, M., et al. (2006). Positional cloning identifies Lotus japonicus NSP2, a putative transcription factor of the GRAS family, required for NIN and ENOD40 gene expression in nodule initiation. DNA Res. 13, 255–265. doi: 10.1093/dnares/dsl017
Murray, J. D., Karas, B. J., Sato, S., Tabata, S., Amyot, L., Szczyglowski, K. (2007). A cytokinin perception mutant colonized by rhizobium in the absence of nodule organogenesis. Science 315, 101–104. doi: 10.1126/science.1132514
Nadzieja, M., Kelly, S., Stougaard, J., Reid, D. (2018). Epidermal auxin biosynthesis facilitates rhizobial infection in Lotus japonicus. Plant J. 95, 101–111. doi: 10.1111/tpj.13934
Oldroyd, G. E. D. (2013). Speak, friend, and enter: signalling systems that promote beneficial symbiotic associations in plants. Nat. Rev. Microbiol. 11, 252–263. doi: 10.1038/nrmicro2990
Pagès, H., Aboyoun, P., Gentleman, R., DebRoy, S. (2020). Biostrings: Efficient manipulation of biological strings. Bioconductor version: Release 3.15). doi: 10.18129/B9.bioc.Biostrings
Qiu, L., Lin, J., Xu, J., Sato, S., Parniske, M., Wang, T. L., et al. (2015). SCARN a novel class of SCAR protein that is required for root-hair infection during legume nodulation. PloS Genet. 11, e1005623. doi: 10.1371/journal.pgen.1005623
Radutoiu, S., Madsen, L. H., Madsen, E. B., Felle, H. H., Umehara, Y., Grønlund, M., et al. (2003). Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature 425, 585–592. doi: 10.1038/nature02039
R Core Team (2020). R: A language and environment for statistical computing (Vienna, Austria: R Foundation for Statistical Computing). Available at: https://www.R-project.org/.
Reid, D. E., Nadzieja, M., Novak, O., Heckmann, A. B., Sandal, N., Stougaard, J. (2017). Cytokinin biosynthesis promotes cortical cell responses during nodule development. Plant Physiol. 175, 361–375. doi: 10.1104/pp.17.00832
Robinson, M. D., McCarthy, D. J., Smyth, G. K. (2010). edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. doi: 10.1093/bioinformatics/btp616
Röckel, N., Wolf, S., Kost, B., Rausch, T., Greiner, S. (2008). Elaborate spatial patterning of cell-wall PME and PMEI at the pollen tube tip involves PMEI endocytosis, and reflects the distribution of esterified and de-esterified pectins. Plant J. 53, 133–143. doi: 10.1111/j.1365-313X.2007.03325.x
Rodríguez-Llorente, I. D., Pérez-Hormaeche, J., El Mounadi, K., Dary, M., Caviedes, M. A., Cosson, V., et al. (2004). From pollen tubes to infection threads: recruitment of Medicago floral pectic genes for symbiosis. Plant J. 39, 587–598. doi: 10.1111/j.1365-313X.2004.02155.x
Roy, S., Liu, W., Nandety, R. S., Crook, A. D., Mysore, K. S., Pislariu, C. I., et al. (2019). Celebrating 20 years of genetic discoveries in legume nodulation and symbiotic nitrogen fixation. Plant Cell 32, 15–41. doi: 10.1105/tpc.19.00279
R Studio Team (2020). RStudio: Integrated development for r (Boston, MA: RStudio, PBC). Available at: http://www.rstudio.com/.
Schauberger, P., Walker, A. (2020)Openxlsx: Read, write and edit xlsx files. Available at: https://CRAN.R-project.org/package=openxlsx.
Schauser, L., Roussis, A., Stiller, J., Stougaard, J. (1999). A plant regulator controlling development of symbiotic root nodules. Nature 402, 191–195. doi: 10.1038/46058
Shiu, S.-H., Bleecker, A. B. (2001). Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinase. Proc. Natl. Acad. Sci. U. S. A. 98, 10763–10768. doi: 10.1073/pnas.181141598
Singh, S., Katzer, K., Lambert, J., Cerri, M., Parniske, M. (2014). CYCLOPS, a DNA-binding transcriptional activator, orchestrates symbiotic root nodule development. Cell Host Microbe 15, 139–152. doi: 10.1016/j.chom.2014.01.011
Smit, P., Raedts, J., Portyanko, V., Debellé, F., Gough, C., Bisseling, T., et al. (2005). NSP1 of the GRAS protein family is essential for rhizobial nod factor-induced transcription. Science 308, 1789–1791. doi: 10.1126/science.1111025
Soyano, T., Kouchi, H., Hirota, A., Hayashi, M. (2013). NODULE INCEPTION directly targets NF-y subunit genes to regulate essential processes of root nodule development in Lotus japonicus. PloS Genet. 9, e1003352. doi: 10.1371/journal.pgen.1003352
Sujkowska, M., Borucki, W., Golinowski, W.l. (2006). Localization of expansin-like protein in apoplast of pea (Pisum sativum l.) root nodules during interaction with rhizobium leguminosarum bv. viciae 248. Acta Soc Bot. Pol. 76, 17–26.
Suzaki, T., Ito, M., Yoro, E., Sato, S., Hirakawa, H., Takeda, N., et al. (2014). Endoreduplication-mediated initiation of symbiotic organ development in Lotus japonicus. Development 141, 2441–2445. doi: 10.1242/dev.107946
Suzaki, T., Yano, K., Ito, M., Umehara, Y., Suganuma, N., Kawaguchi, M. (2012). Positive and negative regulation of cortical cell division during root nodule development in lotus japonicus is accompanied by auxin response. Development 39, 3997–4006. doi: 10.1242/dev.084079
Tian, F., Yang, D. C., Meng, Y. Q., Jin, J., Gao, G. (2020). PlantRegMap: charting functional regulatory maps in plants. Nucleic Acids Res. 48, D1104–D1113. doi: 10.1093/nar/gkz1020
Tirichine, L., Imaizumi-Anraku, H., Yoshida, S., Murakami, Y., Madsen, L. H., Miwa, H., et al. (2006). Deregulation of a Ca2+/calmodulin-dependent kinase leads to spontaneous nodule development. Nature 441, 1153–1156. doi: 10.1038/nature04862
Tirichine, L., Sandal, N., Madsen, L. H., Radutoiu, S., Albrektsen, A. S., Sato, S., et al. (2007). A gain-of-function mutation in a cytokinin receptor triggers spontaneous root nodule organogenesis. Science 315, 104–107. doi: 10.1126/science.1132397
Warnes, G. R., Bolker, B., Bonebakker, L., Gentleman, R., Huber, W., Liaw, A., et al. (2022)Gplots: Various r programming tools for plotting data. Available at: https://CRAN.R-project.org/package=gplots.
Wickham, H., Averick, M., Bryan, J., Chang, W., McGowan, L., François, R., et al. (2019). Welcome to the tidyverse. J. Open Source Software 4, 1686. doi: org/10.21105/joss.016861
Xie, F., Murray, J. D., Kim, J., Heckmann, A. B., Edwards, A., Oldroyd, G. E. D., et al. (2012). Legume pectate lyase required for root infection by rhizobia. Proc. Natl. Acad. Sci. U. S. A. 109, 633–638. doi: 10.1073/pnas.1113992109
Yano, K., Aoki, S., Liu, M., Umehara, Y., Suganuma, N., Iwasaki, W., et al. (2017). Function and evolution of a Lotus japonicus AP2/ERF family transcription factor that is required for development of infection threads. DNA Res. 24, 193–203. doi: 10.1093/dnares/dsw052
Keywords: LjERN1, Lotus japonicus, RNA sequencing, root nodule symbiosis, rhizobial infection
Citation: Liu M, Kameoka H, Oda A, Maeda T, Goto T, Yano K, Soyano T and Kawaguchi M (2023) The effects of ERN1 on gene expression during early rhizobial infection in Lotus japonicus. Front. Plant Sci. 13:995589. doi: 10.3389/fpls.2022.995589
Received: 16 July 2022; Accepted: 19 December 2022;
Published: 17 January 2023.
Edited by:
Jianping Wang, University of Florida, United StatesReviewed by:
Senjuti Sinharoy, National Institute of Plant Genome Research (NIPGR), IndiaCopyright © 2023 Liu, Kameoka, Oda, Maeda, Goto, Yano, Soyano and Kawaguchi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Masayoshi Kawaguchi, bWFzYXlvc2lAbmliYi5hYy5qcA==; Meng Liu, bGl1bWVuZzYxMUBob3RtYWlsLmNvbQ==
†Present address: Hiromu Kameoka, CAS-JIC Centre of Excellence for Plant and Microbial Science (CEPAMS), Center for Excellence in Molecular Plant Sciences (CEMPS), Chinese Academy of Sciences, Shanghai, China
Taro Maeda, Institute for Advanced Biosciences, Keio University, Tsuruoka, Yamagata, Japan
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.