- 1College of Plant Protection, Henan Agricultural University, Zhengzhou, China
- 2Guangdong Laboratory of Lingnan Modern Agriculture, Genome Analysis Laboratory of the Ministry of Agriculture and Rural Affairs, Agricultural Genomics Institute at Shenzhen, Chinese Academy of Agricultural Sciences, Shenzhen, China
TCP proteins are plant-specific transcription factors, which are involved in a broad range of physiological processes of plant growth and development. However, the origin and evolutionary history of this gene family is not fully resolved. Here, we present a genome-wide survey of TCP genes in 59 species (including 42 genomes and 17 transcriptomes) covering all main lineages of green plants, and reconstruct the evolutionary history of this gene family. Our results suggested that the origin of TCP genes predated the emergence of land plants, possibly in the common ancestor of Phragmoplastophyta. The TCP gene family gradually experienced a continuous expansion and grew from a few members in algae, moss and lycophytes to dozens, and sometimes over 50 members in angiosperms. Phylogenetic analysis indicated that at least four subclades (Class I and three subclades of Class II) have been occurred in the ancestor of spermatophyte (seed plant). Both dispersed duplication and segmental duplication or whole-genome duplication (WGD) contributed significantly to the expansion of the TCP gene family over the course of evolution. Our findings provide a comprehensive evolutionary analysis of the TCP gene family and highlight the importance of gene duplications in the evolution of this plant-specific transcription factors.
Introduction
Transcription factors (TFs) constitute major components of the genetic basis for phenotypic evolution (Wray et al., 2003). Variations of TF, such as the expansion and diversification, play central roles in the evolution of some key innovation of green plants, for example, vascular tissues, megaphylls, roots, reproductive cones, or flowers (Floyd and Bowman, 2007; Lai et al., 2020). With the unprecedented pace of sequencing of genomes and transcriptomes, the repertoire of TFs from a wide variety of green plant species (ranging from algae to angiosperms) will be better characterized. The advent of massive sequence information not only helps to identify the TF family members but also allows a comprehensive investigation of how TF families have expanded and how the expansion has provided members involved in multiple specific innovations.
TCP proteins (TCPs) are plant-specific TFs, which was initially identified and named after TEOSINTE BRANCHED 1 (TB1) in Zea mays (Doebley et al., 1997), CYCLOIDEA (CYC) in Antirrhinum majus (Luo et al., 1996), PROLIFERATING CELL FACTORS 1 and 2 (PCF1 and PCF2) in Oryza sativa (Kosugi and Ohashi, 1997). All known TCPs are characterized by a 59-amino acid basic helix-loop-helix (bHLH) domain known as TCP domain (Cubas et al., 1999). Accumulated evidences indicated that TCP genes were involved in a broad range of growth-related processes, such as flower and leaf shape, axillary meristem development, shoot branching, gametophyte development, hormone signaling, seed germination, regulation of the circadian clock, and defense (Kosugi and Ohashi, 1997, 2002; Martín-Trillo and Cubas, 2009; Danisman et al., 2012; Manassero et al., 2013; Lucero et al., 2017; Shang et al., 2020).
Given the critical roles in diverse biological processes, TCP genes have been identified in various plants, especially in angiosperms, which usually harbors more than 20 members (Liu et al., 2019). The evolutionary history of the TCP gene family (especially CYC clade) have been well characterized in angiosperms. Most previous studies have confirmed that the family can be classified into Class I (also known as PCF class or TCP-P class) and Class II (also known as TCP-C class) in angiosperms (Yao et al., 2007; Mondragón-Palomino and Trontin, 2011; Liu et al., 2019). Class I has a conserved four-amino-acid deletion in the TCP domain. Class II can be divided into the CYC clade and the CINCINNATA (CIN) clade (Martín-Trillo and Cubas, 2009; Liu et al., 2019). The CYC clade underwent two duplication events in the core eudicots, leading to three subgroups: CYC1, CYC2, and CYC3 (Howarth and Donoghue, 2006; Chapman et al., 2008). The CYC1 subclade includes maize TB1, which was related to the control of shoot branching (Doebley et al., 1997). CYC2, which includes Anthirrinum CYC, had a key role in the evolution of floral dorsoventral asymmetry (zygomorphy) (reviewed in Busch and Zachgo, 2009; Hileman, 2014; Fambrini and Pugliesi, 2017). It was reported that the CYC2 subclade has expanded by duplication events in many species-rich taxa with zygomorphic flowers such as Asterales, Dipsacales, Fabales, Lamiales, Proteales, and Ranales (Citerne et al., 2003, 2017; Gübitz et al., 2003; Reeves and Olmstead, 2003; Howarth and Donoghue, 2005; Kölsch and Gleissberg, 2006; Chapman et al., 2008; Howarth et al., 2011; Tähtiharju et al., 2012; Yang et al., 2012; Bello et al., 2017). The CYC3 subclade contains genes such as Arabidopsis BRANCHED2 (BRC2), expressed both in branch and flower primordia, and also appears to be related with the control of shoot branching or the flower development (Martín-Trillo and Cubas, 2009).
Viridiplantae (green plants) contains green algae (Chlorophytes), streptophyte algae (Charophytes), and land plants (Embryophytes). The streptophyte algae is paraphyletic and comprises six lineages, i.e., Klebsormidiophyceae, Chlorokybophyceae, Mesostigmatophyceae, Zygnematophy- ceae, Coleochaetophyceae, and Charophyceae. Of them, Zygnematophyceae has been recognized as the most likely sister group of extant embryophytes (land plants) (Cheng et al., 2019; One Thousand Plant Transcriptomes Initiative, 2019). Land plant harbors five major lineages, i.e., bryophytes, lycophytes, ferns, gymnosperms, and angiosperms (One Thousand Plant Transcriptomes Initiative, 2019; Zhang et al., 2022). In comparison to the vast body of work developed in angiosperms, the evolutionary history of TCP genes in other lineages of green plants has not been widely investigated. Based on the BLAST searches against the EST database or PCR amplification in non-angiosperm plant species, Navaud et al. (2007) firstly reported that TCP genes may have originated before land plant emergence and form small families of no more than 10 members in pluricellular green algae (Cosmarium and Chara), bryophytes (Physcomitrella), ferns (Ceratopteris and Equisetum), lycophytes (Selaginella), and gymnosperms (Pinus, Picea, Cycas, and Gnetum). Liu et al. (2019) investigated the evolutionary history of the TCP gene family of land plant based on the genome-wide analysis and found that TCP genes might emerged at the ancestor of land plants and expanded significantly through whole-genome duplication (WGD). Although 47 species with whole genome sequence (including 25 algae and 22 land plants) were searched in their study, the representative species of streptophyte algae, especially the sister group of land plants (Zygnematophyceae), were overlooked. And also, of 22 land plant species, only 4 non-flowering plant species had been surveyed. Nevertheless, phylogenetic analysis in previous studies indicated that both Class I and CIN TCP genes were identified in all the investigated species, while the CYC clade seemed to be absent in non-flowering species (Navaud et al., 2007; Martín-Trillo and Cubas, 2009; Liu et al., 2019). In addition, the TCP gene family was likely to have undergone a large expansion over the course of evolution (Navaud et al., 2007; Liu et al., 2019). However, due to the sparse sampling or uncompleted genome data in these non-flowering species, when and how the TCP gene family expanded and diversified, especially the divergence of Class II and the origin of CYC, were still controversial.
The recent upward trend in the number of completely sequenced genomes or transcriptomes in different phylogenetic lineages give us the opportunity to revisit the evolutionary history of the TCP gene family and investigate how TCP genes expanded and diverged. In this study, an extensive survey of TCP genes was firstly conducted for some representative species with public genome data across all major lineages of green plants, especially the non-flowering plants. Considering the unevenly distribution of whole genome sequencing across clades, some transcriptomes were also selected for gymnosperms and ferns, which greatly expand coverage across the green plant clade and sampling density within many key clades. Then, a detailed phylogenetic analysis of all available sequence information was performed to infer their origin, diversification and expansion. Lastly, chromosomal locations, gene duplication, and synteny analysis were performed to investigate the expansion mechanism.
Materials and methods
Identification of TCP genes
A total of 59 species, including red algae, green algae, streptophyte algae, bryophytes, lycophytes, ferns, gymnosperms, and angiosperms, were surveyed in our study (Figure 1 and Supplementary Table 1). Firstly, the coding and genomic sequences of TCP genes of 14 species were retrieved according to the gene ID (Supplementary Table 1 in Liu et al., 2019) from phytozome database,1 including 4 non-flowering plants (moss: Physcomitrella patens, Sphagnum fallax, Marchantia polymorpha; lycophyte: Selaginella moellendorffii), and 10 flowering species representing all major lineages of angiosperms according to APG IV (The Angiosperm Phylogeny Group, 2016). Then, the genomic sequences, coding sequences (CDS) and annotations of other 16 genomes of land plants (including 10 gymnosperms, 4 ferns, 1 lycophyte, and 1 bryophyte) were retrieved from phytozome, fernbase,2 CNGB,3 NGDC,4 NCBI,5 TreeGenes,6 Plantgenie,7 or figshare8 database (Supplementary Table 1). To increase the sampling density of gymnosperm and ferns representatives, 17 transcriptomes were also download from NCBI. Thirdly, the Hidden Markov model (HMM) profile of the conserved TCP domain (PF03634) was downloaded from the Pfam database9 and hmmsearch was performed using the hmmer 3.010 with an e-value of 0.01. BLASTP against all protein sequences in each genome with the amino acid sequences of Arabidopsis thaliana, were also conducted with an e-value of 10. In order to infer the origin of the TCP gene family, attempts to identify TCP genes from green algae (Botryococcus braunii, Chlamydomonas reinhardtii, Chromochloris zofingiensis, Coccomyxa subellipsoidea, Dunaliella salina, Micromonas pusilla CCMP1545, Micromonas sp. RCC299, Ostreococcus lucimarinus and Volvox carteri), red algae (Porphyra umbilicalis) and two early diverging streptophyte algae (Spirogloea muscicola and Mesotaenium endlicherianum) were also made. Subsequently, all hits obtained from HMM and BLAST searches were merged together, and the redundant hits were removed. The annotation errors of some TCP genes were manually corrected based on the BLAST results. Finally, all candidate sequences were confirmed to be TCP genes by blast against the Pfam database.
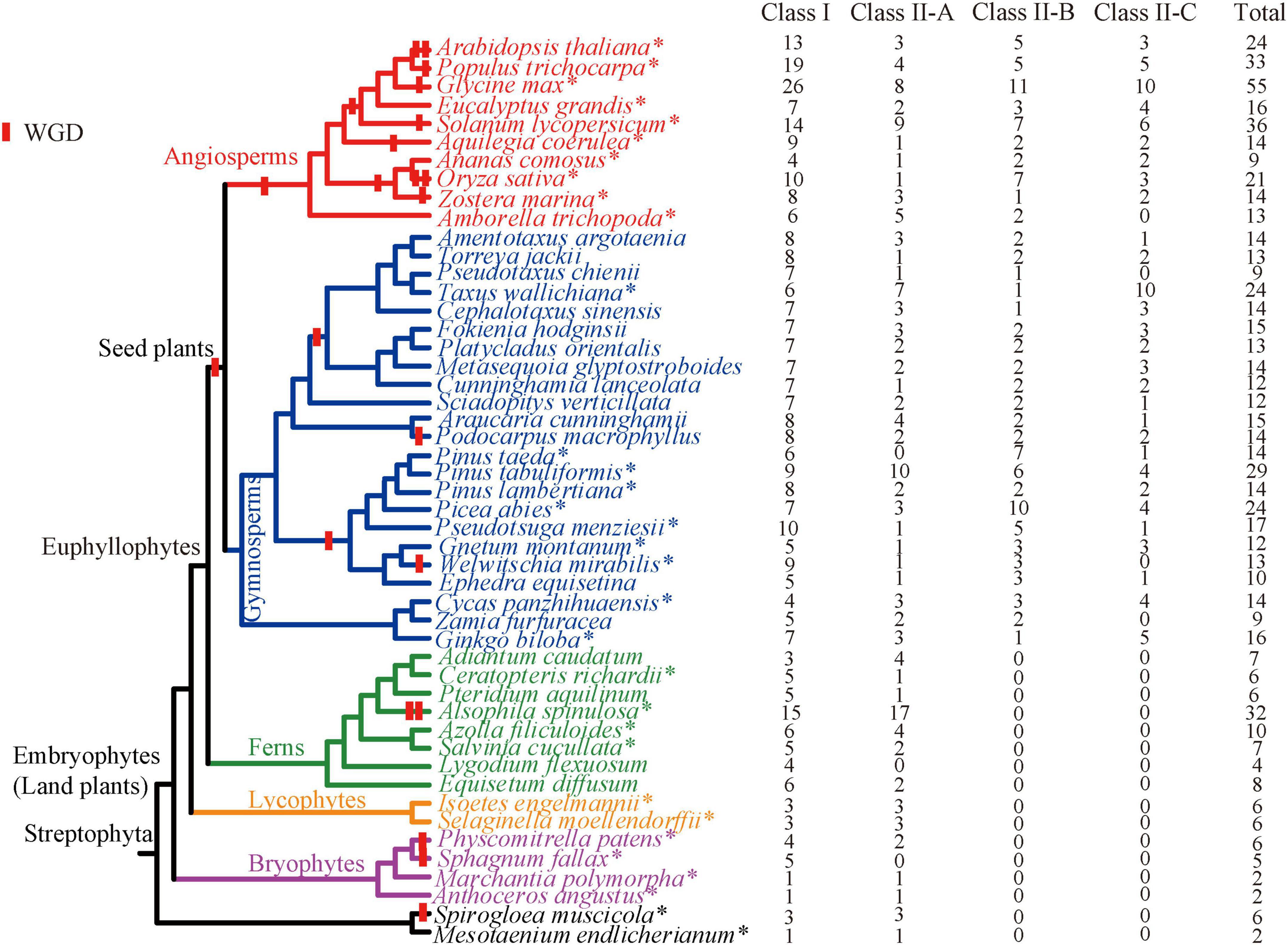
Figure 1. Copy number of the TCP gene family for all investigated species in this study. Species with whole genome sequences are represented by asterisks. The overall topology of these species is based on One Thousand Plant Transcriptomes Initiative (2019), decorated with whole-genome duplication events (Rensing et al., 2007, 2008; Jiao et al., 2011; Li et al., 2015; Ren et al., 2018; Cheng et al., 2019; Stull et al., 2021; Huang et al., 2022).
Sequence alignments and phylogenetic analysis
The coding regions of TCP genes were aligned using Clustal X (Thompson et al., 1997) and manually adjusted in Bioedit v7.0.9 (Hall, 1999). The alignment logos of the TCP domain were generated with SeqLogo in TBtools (Chen et al., 2020). Additionally, the construction of a reliable phylogenetic tree of TCP proteins is problematic due to the small size (59 amino acids maximum) of the conserved TCP domain sequence. Therefore, we aligned the maximum number of amino acids for each protein. To comprehensively explore the evolutionary history of the TCP gene family in green plants, both Maximum-Likelihood (ML) analysis and Bayesian Inference (BI) were performed based on the all conserved nucleotide and amino acid sequences. The ML tree was generated by IQ-TREE v1.6.8 (Nguyen et al., 2014), using the best-fit model selected by ModelFinder (Kalyaanamoorthy et al., 2017) and with 5,000 replications of ultrafast bootstrap support values (UFBoot) (Minh et al., 2013). The BI analysis was conducted with MRBAYES 3.2.6 (Ronquist et al., 2012) by running four simultaneous Markov chain Monte Carlo (MCMC) simulations, sampling every 100 generations, and discarding the first 2,500 trees as “burn-in.” The remaining trees were used to calculate posterior probabilities using a majority-rule consensus. Tracer v1.7 (Rambaut et al., 2018) was used to check for convergence and to ensure that effective sample sizes (ESS) were >200 for all parameters. The final trees were visualized or manually improved by the online program iTOL.11
Chromosomal location, gene duplication, and synteny analysis
To infer the expansion mechanism of the TCP gene family, gene duplication and synteny analysis were performed for six species (ferns: Alsophila spinulosa; gymnosperms: Ginkgo biloba, Cycas panzhihuaensis, and Pinus tabuliformis; and angiosperms: A. thaliana and O. sativa) as follows. Firstly, GFF files, gene files, targeted TCP genes of these six species were downloaded or extracted. Secondly, BLASTP was performed to search homologous sequences in each species with e < 1e−5 and the top five self-BLASTP hits were considered as candidate duplicated genes. Then, the intra-genomic syntenic analysis were conducted using MCScanX (Wang et al., 2012) in TBtools with default parameters through the above BLASTP searches. The “duplicate gene classifier” program implemented in the MCScanX was employed to identify the replication mode. Genes can be classified into singletons, dispersed duplicates, proximal duplicates, tandem duplicates, and segmental duplicates/WGD depending on their copy number and genomic distribution (Wang et al., 2012). Lastly, the location of TCP genes and the duplicated genes caused by tandem and segmental duplications/WGD were displayed in TBtools.
Results
Phylogenetic analyses
The copy number of the TCP gene family is greatly variable in green plants (Figure 1). No homologs were identified in nine green algae and one red algae. Among the other 49 species with TCP genes, a total of 691 TCP genes were obtained (Figure 1 and Supplementary Table 1). Of them, 437 TCPs were newly identified. The two early diverging algae, S. muscicola and M. endlicherianum, possess only two and six members, respectively. The bryophytes, early diverging clade of land plants (including hornworts, liverworts, and mosses) harbors two to six members. The lycophytes S. moellendorffii and Isoetes engelmannii possess six members. In ferns, most species harbor no more than 10 members, with the exception of A. spinulosa, in which 32 members were identified. It should be noted that the copy number of TCP genes in ferns were likely to be underestimated, because only transcriptomes were searched for Adiantum caudatum, Equisetum diffusum, Lygodium flexuosum, and Pteridium aquilinum. There was an obvious expansion for the TCP gene family in seed plants (Figure 1). The gymnosperms possess an average of over 15 members, with Zamia furfuracea, Pseudotaxus chienii having the least (9 genes) and P. tabuliformis having the most (29 genes). The copy number of the TCP gene family of angiosperms vary greatly, ranging from 9 (Ananas comosus) to 55 (Glycine max), with an average of 23.5 members (Figure 1). In general, a gradually expansion history of the TCP gene family was observed during plant evolution, as the gene number continued to increase from algae to angiosperms.
To explore the evolutionary history of the TCP gene family, phylogenetic analysis was firstly performed based on the alignment nucleotide and amino acid sequence of the TCP domain, respectively. Both the unrooted phylogenetic trees inferred from nucleotide and amino acid sequences with ML and BI analysis confirmed the classification of Class I and Class II (Figure 2). The Class I was slightly larger than Class II (348 vs. 343 members). All the investigated Streptophyta species (including land plants and two Zygnematophyceae) had TCP genes from both classes, except for S. fallax and L. flexuosum, which had no Class II TCP genes (Figure 1 and Supplementary Table 1). Class II was mainly divided into three subclades supported by moderate or high bootstrap values (A–C), although their relationship was uncertain (Figure 3). Class II-A contained gene members belonging to all phylogenetic groups from moss to angiosperms, while Class II-B and Class II-C only harbored sequences from seed plants (gymnosperms and angiosperms) (Figures 1, 3). In Class II-A, one subgroup, which harbors the sequence from angiosperms, gymnosperms and ferns, was highly supported (support value = 97/0.94/92/0.72) (Figure 3). In Class II-C, one highly supported subgroup (CYC) was recognized (support value = 100/1/100/1) (Figure 3). It should be noted that all seed plants possess three subclades of Class II, except for Amborella trichopoda, Pinus taeda, P. chienii, Welwitschia mirabilis, and Z. furfuracea, in which Class II-A or Class II-C was absent (Supplementary Table 1). Although the phylogenetic position of TCP genes in lycophytes, and algae were largely unresolved due to the limited informative sites of the conserved domain, the subdivision of TCP genes in euphyllophytes was further supported by the phylogenetic analysis for angiosperms, gymnosperms and ferns, respectively (Supplementary Figures 1–3).
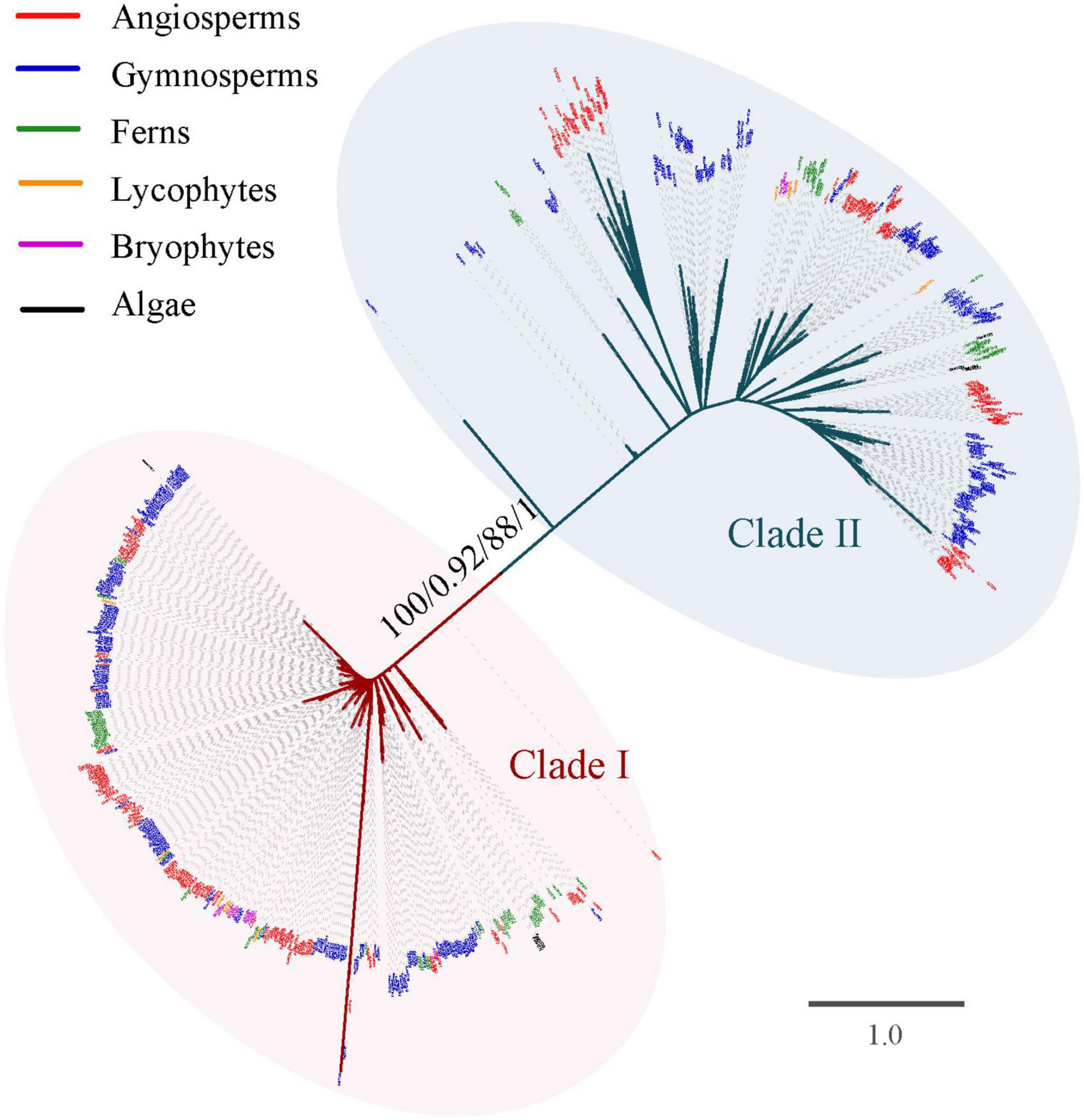
Figure 2. Phylogenetic tree of the TCP gene family constructed based on the amino acid sequences of the TCP domain. The tree was not rooted. Branch lengths indicate the number of amino acid residues substitutions per site and are drawn to scale. Numbers associated with nodes are UFBoot support values and posterior probability in the phylogeny analysis based on amino acid sequences (first and second) and CDS (third and fourth), respectively. All species abbreviations are listed in Supplementary Table 1.
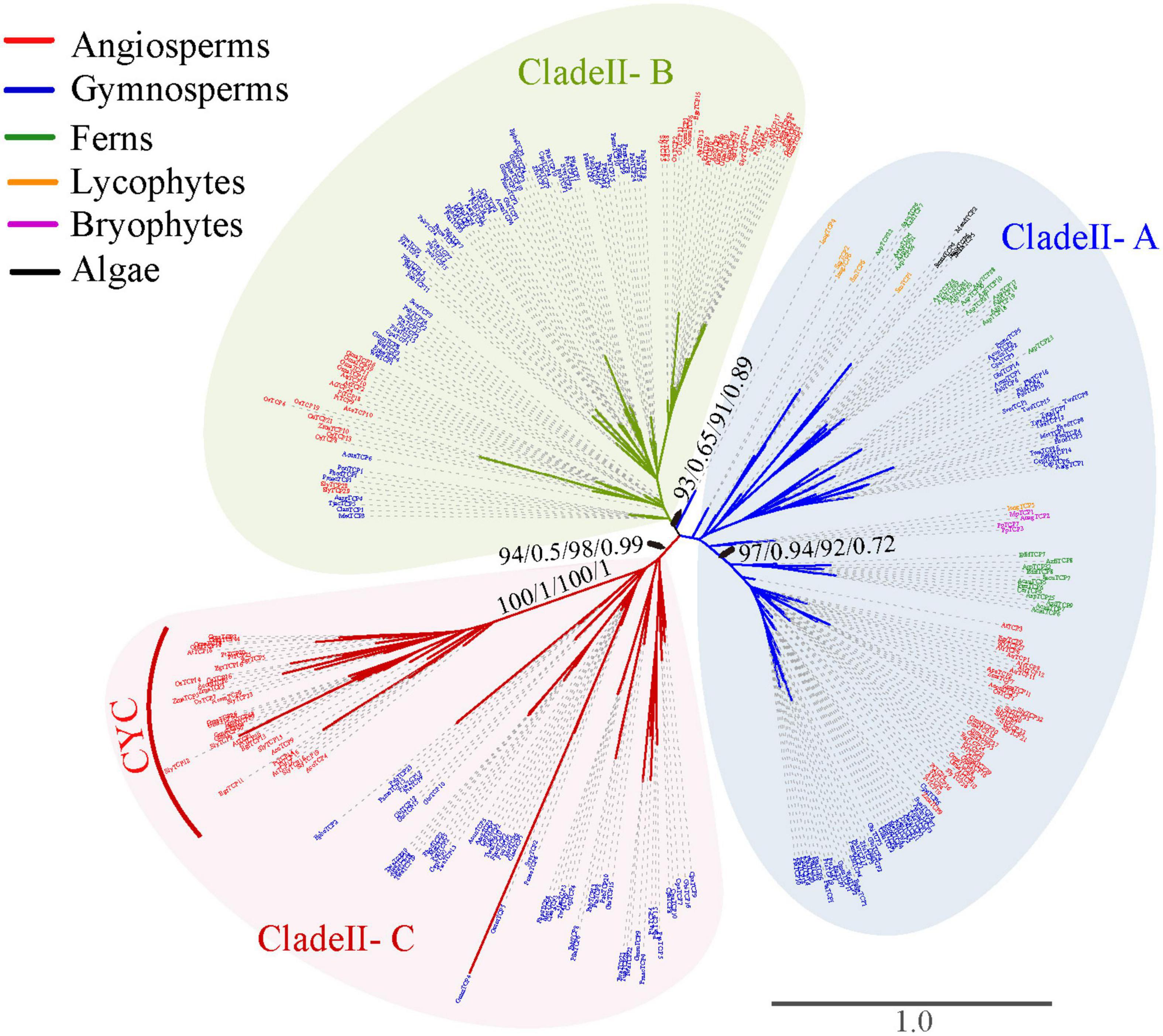
Figure 3. Phylogenetic tree of Class II TCP genes constructed based on the amino acid sequences of the TCP domain and the upstream conserved sequences. The tree was not rooted. Branch lengths indicate the number of amino acid residues substitutions per site and are drawn to scale. Numbers associated with nodes are UFBoot support values and posterior probability in the phylogeny analysis based on amino acid sequences (first and second) and CDS (third and fourth), respectively. All species abbreviations are listed in Supplementary Table 1.
Sequence characteristics of TCP domain
Using SeqLogo, we obtained a graphical representation of sequence variation of TCPs, which provided a more precise description of polymorphism sites and conserved sites (Figure 4). The relative frequency of corresponding amino acids is reflected by the height of the symbol at each position. The TCP domain commonly consisted of a basic region and a helix-loop-helix (HLH) structure. The basic region of the TCP domain, was highly conserved in all family members, with two consensus sequences, DRHxK and RxRRxR (Figure 4). For the HLH region, only five sites were highly conserved, including Ala (A)-25, Leu (L)-35, Gly (G)-36, Trp (W)-46, and L-47 (Figure 4). The most striking difference between these two classes is a four-amino acid deletion in the basic region of Class I relative to Class II. In addition, Class I and Class II contain distinct residues at positions 5, 15, 20, 21, 24, 32, 33, 34, 40, 41, and 58 (Figure 4). In general, the residues in Class I were relatively more conserved than Class II. For Class II, the most striking difference among three subgroups (A, B, and C) was located in the upstream of TCP domain (Supplementary Figure 4). And the CYC subclade and other subclade (Class II-A, Class II-B, and gymnosperms of Class II-C) possessed distinct residues at positions 16, 33, 35, 48, and 49 (Supplementary Figure 4).
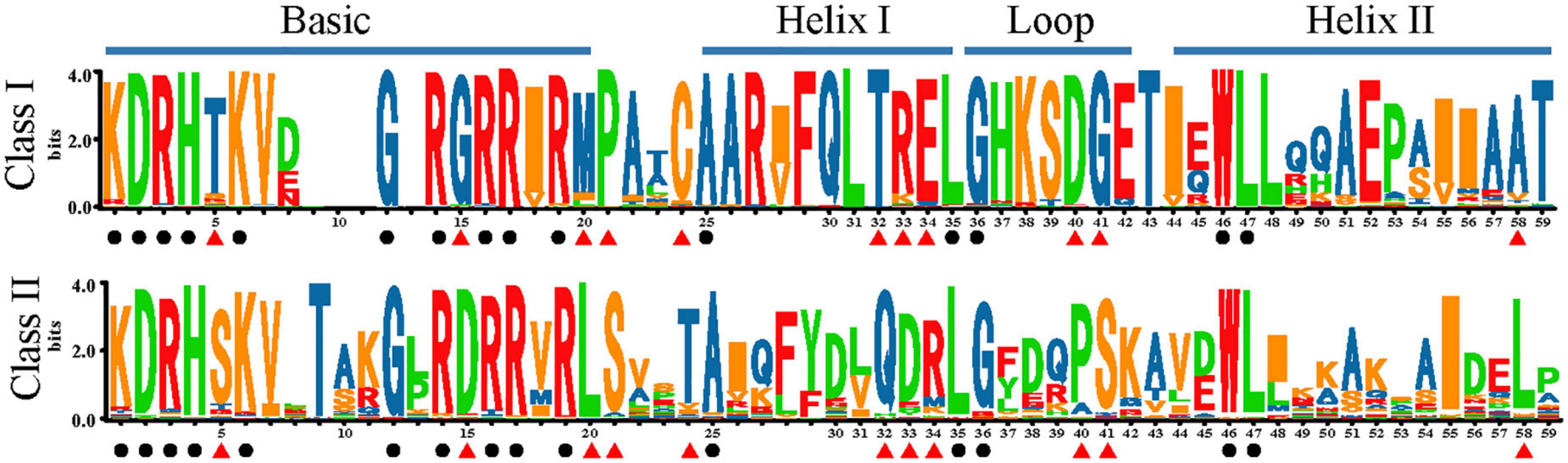
Figure 4. Sequence logos of the plant TCP domains. Bit scores represents the relative frequency of amino acids. The black circle represents conserved loci within the whole family. The red triangle indicates divergent but conserved residues in each subclade.
Chromosomal location, gene duplication, and synteny analysis of TCP genes
All identified TCP genes in A. spinulosa, G. biloba, C. panzhihuaensis, P. tabuliformis, A. thaliana, and O. sativa were mapped to the chromosomes or some scaffolds (Figure 5). TCP genes are widely distributed throughout genomes but are uneven among chromosomes. In A. thaliana, all five chromosomes harbor TCP genes, but eight of the 24 TCP genes are located on Chr (chromosome) 1 and only one gene was detected in Chr 4 (Figure 5E). In other species, TCP genes are not found on all chromosomes. For instance, 32 TCP genes of A. spinulosa were located in 22 chromosomes (a total of 69 chromosomes) (Figure 5A). Chr 2, 3, 5, and 8 of G. biloba (Figure 5B), Chr 4, 6, and 11 of C. panzhihuaensis (Figure 5C), Chr 7 and 8 of P. tabuliformis (Figure 5D), and Chr 10 of O. sativa (Figure 5F) do not carry any TCP genes.
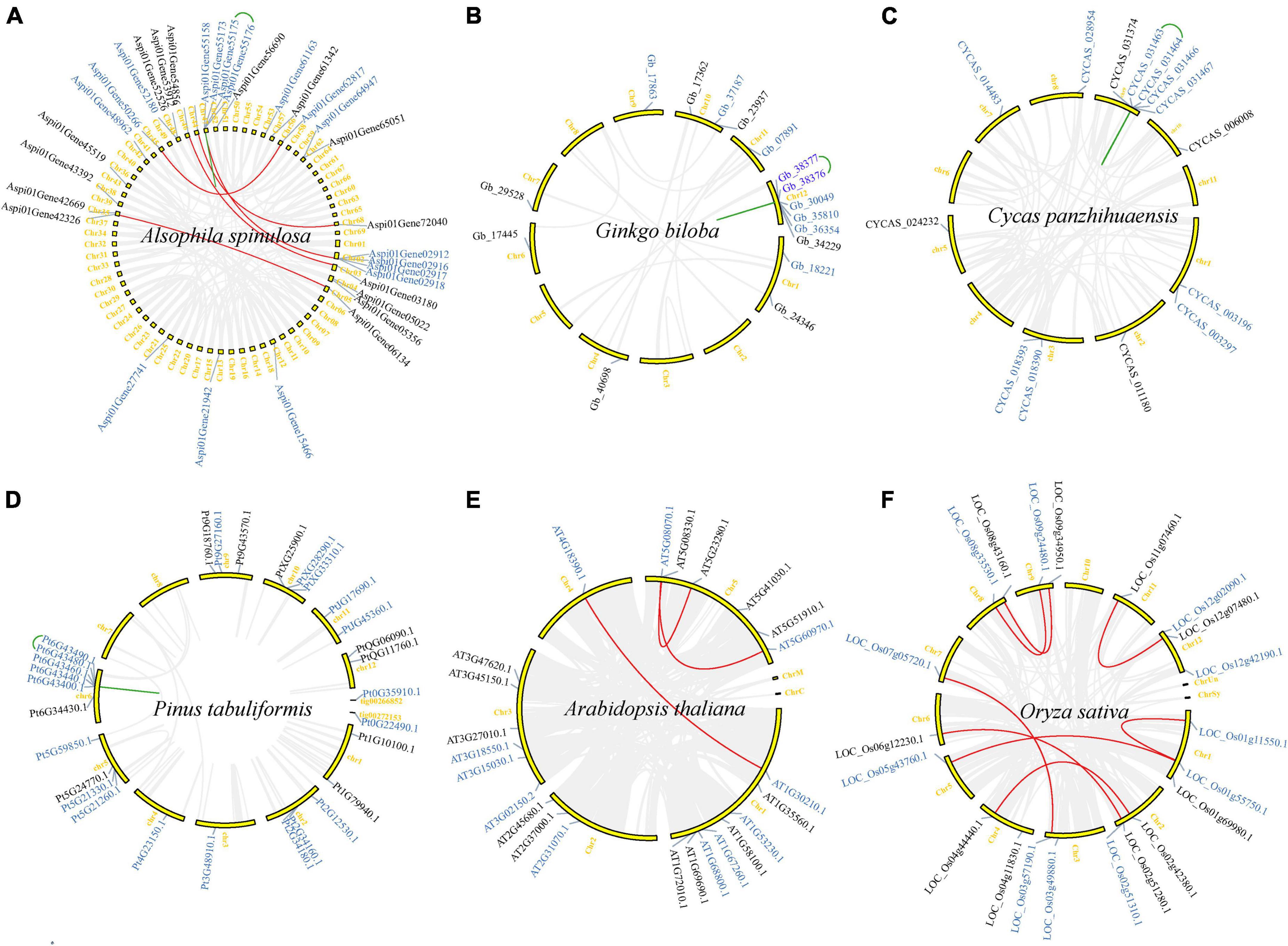
Figure 5. Synteny analysis of TCP genes in six representative species (A–F). Gene names on the side of each chromosome correspond to the approximate locations of each TCP gene. The detailed information is listed in Supplementary Table 1. Gray lines indicate all synteny blocks in the whole genome of each species. Red and green lines suggest TCP gene pairs with segmental/WGD duplication and tandem duplication, respectively. Gene names with black and blue mean Class I and Class II TCP genes, respectively.
Gene duplication and synteny analysis indicated that only a few of TCP genes in A. thaliana (4), G. biloba (2) and C. panzhihuaensis (1) were identified as singleton (Table 1). Most TCP genes (129/136) were generated by gene duplication events. More than half (73/136) of TCP genes were recognized as dispersed duplications and nearly one fourth (32/136) were recognized as segmental duplications/WGD. In A. spinulosa, all four types of gene duplications were detected, and dispersed and segmental duplications/WGD were the two main types, with 16 (50%) and 11 (34%), respectively (Table 1). In G. biloba, 10 (63%), 2 (13%), and 2 (13%) TCP genes are generated by dispersed, proximal, and segmental duplications/WGD, respectively. In C. panzhihuaensis, 9 (64%), 2 (14%), and 2 (14%) TCP genes are generated by dispersed, proximal, and segmental duplications/WGD, respectively. In P. tabuliformis, 18 (62%), 9 (31%), and 2 (7%) TCP genes are generated by dispersed, proximal, and tandem duplications, respectively. In A. thaliana, 14 (58%) and 6 (25%) TCP genes are generated by dispersed and segmental duplications/WGD, respectively. In O. sativa, 6 (29%) and 15 (71%) TCP genes are generated by dispersed and segmental duplications/WGD, respectively. Our results indicated that both dispersed duplications and segmental duplications/WGD play a critical role in the expansion of the TCP gene family.
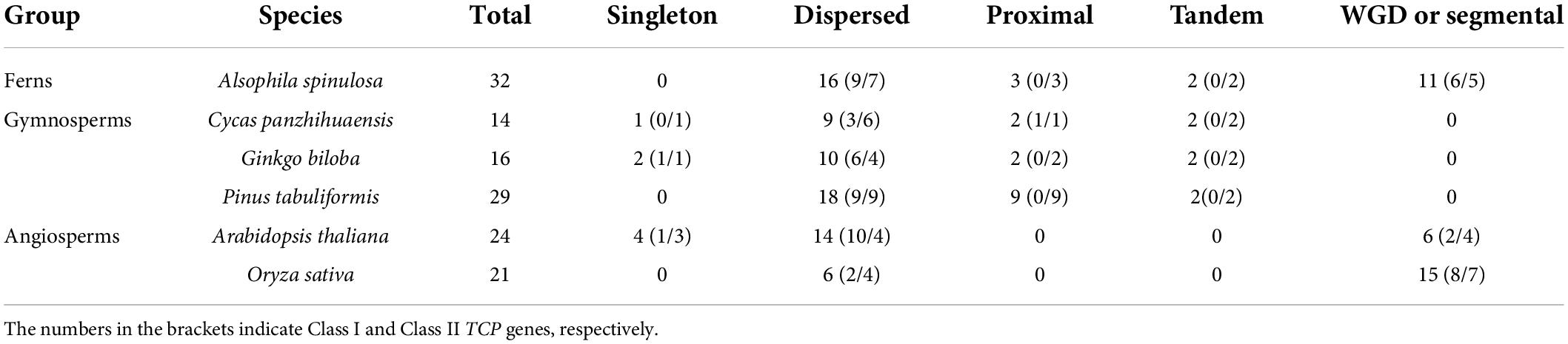
Table 1. Numbers of TCP genes from different origins as classified by duplicate gene classifier in six representative euphyllophytes genomes.
Discussion
The origin and diversification of the TCP gene family
In this study, we present a genome-wide survey of TCP genes in 59 species (42 genomes and 17 transcriptomes), ranging from algae to angiosperms, and reconstruct the evolutionary history of this gene family. It was reported that TCP genes were present in Chara (Charophyceae) and Cosmarium (Zygnematophyceae) (Navaud et al., 2007). In our study, TCP genes were newly identified in other two streptophyte algae (S. muscicola and M. endlicherianum), but no homologs were identified in any species of Chlorophyta and Rhodophyta. Additionally, we also tried to identify TCP genes in the genomes or transcriptomes of other streptophyte algae by TBLASTN in NCBI, and no sequences were identified. All these evidences confirmed that the origin of the TCP gene family predates the emergence of land plants, possibly in the common ancestor of Phragmoplastophyta (Zygnematophyceae, Coleochaetophyceae, Charophyceae, and all extant land plants) (Navaud et al., 2007). However, Classes I and II genes were clearly diverged in all these species, even in the early divergent algal species. Not any possible common ancestor of TCP genes was identified in this study, although extensive searches within prokaryotic and eukaryotic genomes were performed, which renders it difficult to speculate from which TCP genes arose. We tentatively hypothesized that two types of TCP genes were likely generated simultaneously, and then evolved independently. Nevertheless, because of the sparse sampling in the basic group of green plants, an alternative hypothesis could not be completely ruled out. The scenario is that the ancestral TCP gene might have been present in lineages that have since disappeared. For these reasons, we suggest that increasing sampling in early divergent streptophyte algae (such as Zygnematophyceae and Charophyceae) is a requirement for further confirmation of the origin of the TCP genes in the future.
The TCP gene family was generally divided into Classes I and II, with Class II being further divided into CIN and CYC clades in angiosperms (Navaud et al., 2007; Martín-Trillo and Cubas, 2009; Liu et al., 2019). Consistent with most previous studies, the topology of the phylogenetic tree of the 691 TCP genes was also preliminary divided into two Class I and Class II with relatively high support values (Figure 2). Unlike with previous studies, our phylogenetic tree newly recognized three subclades (A, B, and C) in Class II (Figure 3), possibly resulted from the increasing sampling of non-flowering plants. The previously described CYC subclade was embedded in Class II-C, and CIN subclade was assigned into Class II-A and Class II-B subclades (Figure 3). The non-spermatophyte (algae, bryophytes, lycophytes, and ferns) only harbor Class II-A, and seed plants (gymnosperms and angiosperms) possess all three types (Figure 3). Although the relationship of three subclades of Class II was not resolved, the Class II-A subclade was likely more related to the ancestral Class II TCP genes, and two ancient gene duplications predating the divergence of seed plants possibly gave rise to Class II-B and Class II-C. The origin of CYC was most likely coincided with the occurrence and diversification of flowers in angiosperms and the subsequent gene loss likely resulted in the absent of CYC genes in the basal angiosperms A. trichopoda (Figure 2 and Supplementary Table 1). However, we could not totally rule out the possibility that the origin of CYC would take place after the split of Amborella with other angiosperms. The ancient gene duplications of Class II and the origin of CYC were likely related with the ancestral WGD in the common ancestor of extant seed plants (Jiao et al., 2011). Class II TCP genes had crucial roles in the development of reproductive organs (such as gametophyte and flower) (Pagnussat et al., 2005; Martín-Trillo and Cubas, 2009). The retention of three subclades of Class II in the ancestor of seed plants might be related with the origin of seed and flower, ultimately, promoting the origin and rapid diversification of the angiosperms.
Additionally, given the results of sequence comparison (Figure 4) and phylogenetic analysis (Figures 2, 3 and Supplementary Figures 1–3), Class I TCP genes were relatively more conserved than Class II, which suggested that Class I was probably under strong purifying selection and Class II might had gone through prominent functional differentiation over the course of evolution. It has been reported that Class I promote the growth and proliferation in angiosperms. By contrast, the role of Class II was much more complicated and diverged, which was reported to participate in the development of leaf, flower, shoot branching, or even hormone signaling (Doebley et al., 1997; Kosugi and Ohashi, 1997, 2002; Busch and Zachgo, 2009; Martín-Trillo and Cubas, 2009; Danisman et al., 2012; Hileman, 2014; Fambrini and Pugliesi, 2017). In the bryophyte P. patens, PpTCP5 (Class II) was reported to be involved in regulating sporophyte branching (Ortiz-Ramírez et al., 2016), similar to the well-known function of members of the Class II TCP genes of maize in shoot branching (Doebley et al., 1997), although their branching structures (axillary meristems or branch initials) are very different. A wider and more specific expression profile of TCP genes in seed plants indicated their functional divergence. In the bryophytes (M. polymorpha and P. patens), both Class I and Class II had high expression levels in many tissues or organs across various development stages (Supplementary Figures 3A,B of Liu et al., 2019). In seed plants, the vast majority of Class I and Class II-A TCP genes had a broader expression profile (Supplementary Figure 5). However, more members of Class II-B and Class II-C TCP genes tend to had limited expression, such as AthTCP1, AthTCP12, AthTCP18 of Arabidopsis and PabTCP5, PabTCP20, PabTCP23, PabTCP24 of Picea (Supplementary Figure 5), which indicated that they might perform specific roles. As plant complexity increased in the ancestor of seed plants, more intricate regulatory networks were needed. Our study revealed that the expansion and diversification of TCP genes (especially Class II) in green plants might be possibly associated with the development of new organs (e.g., seed, leaf, branching, and flower) or the increasing organ complexity (e.g., floral dorsoventral asymmetry) (Floyd and Bowman, 2007). Additionally, the lineages or species-specific clade was frequently observed in gymnosperms and angiosperms (Figures 2, 3 and Supplementary Figures 1, 2), indicating that TCP genes might have experienced distinct evolution history in each lineage, which was possibly related with the adaptation to the distinct environment. Over the course of evolution, frequent gene and genome duplication events and subsequent functional divergence have significantly contributed to the diversification of TCP genes, which could further facilitate plant diversification, adaptation, and evolution.
The expansion of the TCP gene family
Gene duplication is a major driving force in the expansions and diversification of TF family, which might be related with the evolution of the innovative traits (Taylor and Raes, 2004; Floyd and Bowman, 2007). After gene duplication, plants take up removing most redundant gene copies in a long evolutionary process, but some duplicated copies are retained depending on their environmental adaptation, which is the cause of copy number variation among different species. According to our comprehensive survey, both Classes I and II TCP genes experienced a continuous expansion and grew from a few members in algae, moss and lycophytes to dozens, and sometimes over 50 members in angiosperms (Figure 1). In addition, our comprehensive study from a wide phylogenetic scale and intensive phylogenetic analysis indicated that at least four subclades (Class I and three subclades of Class II) have been occurred in the ancestor of spermatophyte (Figure 3). All these evidences indicated that the TCP gene family gradually expanded and diversified during the diversification of green plants, especially in streptophytes.
Gene duplication and synteny analysis indicated that most TCP genes (129/136) were generated by gene duplication events. In different groups, the expansion of the TCP gene family might be caused by different ways. The relative larger members of the TCP gene family in S. muscicola (algae) and P. patens (moss) than other species in the same lineages (Figure 1) might be result from the recent WGD (Rensing et al., 2007, 2008; Cheng et al., 2019), which confirmed by the monophyly of Classes I and II genes in each species (Figures 2, 3). A. spinulosa, which could have undergone two WGD events (Huang et al., 2022), also harbors the most striking large number of TCP genes in ferns (Figure 3). Our results indicated that both dispersed duplications and segmental duplications/WGD play a critical role in expansion of the TCP gene family in A. spinulosa (Table 1). In gymnosperms, although an ancient WGD has been reported in the common ancestor of extant gymnosperms, few recent WGD events were detected (Li et al., 2015; Guan et al., 2016; Liu et al., 2022; Niu et al., 2022). This was likely associated with the lacking of segmental duplications/WGD in G. biloba, C. panzhihuaensis, and P. tabuliformis, and more than 60% of the gymnosperms TCP genes were generated by the dispersed duplication (Table 1). By contrast, both ancient and recent WGD were occurred in most extant angiosperms (Jiao and Paterson, 2014; Ren et al., 2018), which result in the expansion of the TCP gene family in angiosperms, and this view was also supported by previous study (Liu et al., 2019). It is noteworthy that in some species (e.g., A. trichopoda and A. comosus), the copy number was significantly less than other species of angiosperms, which could be associated with the lacking of recent WGD in these species (Amborella Genome Project, 2013; Ming et al., 2015). Except for segmental duplications/WGD, our study also detected a high dispersed duplication rate in A. thaliana and O. sativa (Table 1). Therefore, it can be concluded that the expansion of the TCP gene family in green plants was mainly driven by dispersed duplication, segmental duplications/WGD.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
Y-YL and S-LK planned and designed this study. J-LW, Y-YL, H-WW, and Y-NC collected and analyzed the data. Y-YL, J-LW, S-LK, and H-WW wrote the manuscript. All authors contributed to the article and agreed to submit this version of the manuscript.
Funding
This study was supported by National Natural Science Foundation of China (Grant No. 32000170) and Science and Technology Planning Project of Henan Province of China (222102110255).
Acknowledgments
We are grateful to Yi-Bo Sun (Henan Agricultural University) and Shu-Qi Song and Meng-Yu Xue (Henan Agricultural University), for their help in the data collection.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.994567/full#supplementary-material
Footnotes
- ^ http://phytozome.jgi.doe.gov/
- ^ https://www.fernbase.org/
- ^ https://db.cngb.org/
- ^ https://ngdc.cncb.ac.cn/
- ^ https://www.ncbi.nlm.nih.gov/
- ^ https://treegenesdb.org/
- ^ https://plantgenie.org/
- ^ https://figshare.com
- ^ http://pfam.xfam.org/
- ^ http://hmmer.org
- ^ https://itol.embl.de/
References
Amborella Genome Project (2013). The Amborella genome and the evolution of flowering plants. Science 342:1241089. doi: 10.1126/science.1241089
Bello, M. A., Cubas, P., Álvarez, I., Sanjuanbenito, G., and Fuertes-Aguilar, J. (2017). Evolution and expression patterns of CYC/TB1 genes in Anacyclus: Phylogenetic insights for floral symmetry genes in Asteraceae. Front. Plant Sci. 8:589. doi: 10.3389/fpls.2017.00589
Busch, A., and Zachgo, S. (2009). Flower symmetry evolution: Towards understanding the abominable mystery of angiosperm radiation. Bioessays 31, 1181–1190. doi: 10.1002/bies.200900081
Chapman, M. A., Leebens-Mack, J. H., and Burke, J. M. (2008). Positive selection and expression divergence following gene duplication in the sunflower CYCLOIDEA gene family. Mol. Biol. Evol. 25, 1260–1273. doi: 10.1093/molbev/msn001
Chen, C. J., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y. H., et al. (2020). TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Cheng, S. F., Xian, W. F., Fu, Y., Marin, B., Keller, J., Wu, T., et al. (2019). Genomes of subaerial Zygnematophyceae provide insights into land plant evolution. Cell 179, 1057–1067.e14. doi: 10.1016/j.cell.2019.10.019
Citerne, H. L., Da, L., Pennington, R. T., and Cronk, C. (2003). A phylogenomic investigation of CYCLOIDEA-like TCP genes in the Leguminosae. Plant Physiol. 131, 1042–1053. doi: 10.2307/4280970
Citerne, H. L., Reyes, E., Guilloux, M. L., Delannoy, E., Simonnet, F., Sauquet, H., et al. (2017). Characterization of CYCLOIDEA-like genes in Proteaceae, a basal eudicot family with multiple shifts in floral symmetry. Ann. Bot. 119, 367–378. doi: 10.1093/aob/mcw219
Cubas, P., Lauter, N., Doebley, J., and Coen, E. (1999). The TCP domain: A motif found in proteins regulating plant growth and development. Plant J. 18, 215–222. doi: 10.1046/j.1365-313x.1999.00444.x
Danisman, S., van der Wal, F., Dhondt, S., Waites, R., de Folter, S., Bimbo, A., et al. (2012). Arabidopsis class I and class II TCP transcription factors regulate jasmonic acid metabolism and leaf development antagonistically. Plant Physiol. 159, 1511–1523. doi: 10.1104/pp.112.200303
Doebley, J., Stec, A., and Hubbard, L. (1997). The evolution of apical dominance in maize. Nature 386, 485–488. doi: 10.1038/386485a0
Fambrini, M., and Pugliesi, C. (2017). CYCLOIDEA 2 clade genes: Key players in the control of floral symmetry, inflorescence architecture, and reproductive organ development. Plant Mol. Biol. Rep. 35, 20–36. doi: 10.1007/s11105-016-1005-z
Floyd, S. K., and Bowman, J. L. (2007). The ancestral developmental tool kit of land plants. Int. J. Plant Sci. 168, 1–35. doi: 10.1086/509079
Guan, R., Zhao, Y., Zhang, H., Fan, G., Liu, X., Zhou, W., et al. (2016). Draft genome of the living fossil Ginkgo biloba. Gigascience 5:49. doi: 10.1186/s13742-016-0154-1
Gübitz, T., Caldwell, A., and Hudson, A. (2003). Rapid molecular evolution of CYCLOIDEA-like genes in Antirrhinum and its relatives. Mol. Biol. Evol. 20, 1537–1544. doi: 10.1093/molbev/msg166
Hall, T. A. (1999). BioEdit: A user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids Symp. Ser. 41, 95–98. doi: 10.1021/bk-1999-0734.ch008
Hileman, L. C. (2014). Trends in flower symmetry evolution revealed through phylogenetic and developmental genetic advances. Philos. Trans. R. Soc. B 369:20130348. doi: 10.1098/rstb.2013.0348
Howarth, D. G., and Donoghue, M. J. (2005). Duplications in CYC-like genes from Dipsacales correlate with floral form. Int. J. Plant Sci. 166, 357–370. doi: 10.1086/428634
Howarth, D. G., and Donoghue, M. J. (2006). Phylogenetic analysis of the “ECE” (CYC/TB1) clade reveals duplications predating the core eudicots. Proc. Natl. Acad. Sci. U. S. A. 103, 9101–9106. doi: 10.1073/pnas.0602827103
Howarth, D. G., Martins, T., Chimney, E., and Donoghue, M. J. (2011). Diversification of CYCLOIDEA expression in the evolution of bilateral flower symmetry in Caprifoliaceae and Lonicera (Dipsacales). Ann. Bot. 107, 1521–1532. doi: 10.1093/aob/mcr049
Huang, X., Wang, W. L., Gong, T., Wickell, D., Kuo, L. Y., Zhang, X. T., et al. (2022). The flying spider-monkey tree fern genome provides insights into fern evolution and arborescence. Nat. Plants 8, 500–512. doi: 10.1038/s41477-022-01146-6
Jiao, Y., Wickett, N. J., Ayyampalayam, S., Chanderbali, A. S., Landherr, L., Ralph, P. E., et al. (2011). Ancestral polyploidy in seed plants and angiosperms. Nature 473, 97–100. doi: 10.1038/nature09916
Jiao, Y. N., and Paterson, A. H. (2014). Polyploidy-associated genome modifications during land plant evolution. Philos. Trans. R. Soc. B 369:20130355. doi: 10.1098/rstb.2013.0355
Kalyaanamoorthy, S., Minh, B. Q., Wong, T. K. F., von Haeseler, A., and Jermiin, L. S. (2017). ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 14, 587–589. doi: 10.1038/nmeth.4285
Kölsch, A., and Gleissberg, S. (2006). Diversification of CYCLOIDEA-like TCP genes in the basal eudicot families Fumariaceae and Papaveraceae s.str. Plant Biol. 8, 680–687. doi: 10.1055/s-2006-924286
Kosugi, S., and Ohashi, Y. (1997). PCF1 and PCF2 specifically bind to cis elements in the rice proliferating cell nuclear antigen gene. Plant Cell 9, 1607–1619. doi: 10.2307/3870447
Kosugi, S., and Ohashi, Y. (2002). DNA binding and dimerization specificity and potential targets for the TCP protein family. Plant J. 30, 337–348. doi: 10.1046/j.1365-313x.2002.01294.x
Lai, X., Chahtane, H., Martin-Arevalillo, R., Zubieta, C., and Parcy, F. (2020). Contrasted evolutionary trajectories of plant transcription factors. Curr. Opin. Plant Biol. 54, 101–107. doi: 10.1016/j.pbi.2020.03.002
Li, Z., Baniaga, A. E., Sessa, E. B., Scascitelli, M., Graham, S. W., Rieseberg, L. H., et al. (2015). Early genome duplications in conifers and other seed plants. Sci. Adv. 1:e1501084. doi: 10.1126/sciadv.1501084
Liu, M. M., Wang, M. M., Yang, J., Wen, J., Guo, P. C., Wu, Y. W., et al. (2019). Evolutionary and comparative expression analyses of TCP transcription factor gene family in land plants. Int. J. Mol. Sci. 20:3591. doi: 10.3390/ijms20143591
Liu, Y., Wang, S., Li, L., Yang, T., Dong, S., Wei, T., et al. (2022). The Cycas genome and the early evolution of seed plants. Nat. Plants 8, 389–401. doi: 10.1038/s41477-022-01129-7
Lucero, L. E., Manavella, P. A., Gras, D. E., Ariel, F. D., and Gonzalez, D. H. (2017). Class I and Class II TCP transcription factors modulate SOC1-dependent flowering at multiple levels. Mol. Plant 10, 1571–1574. doi: 10.1016/j.molp.2017.09.001
Luo, D., Carpenter, R., Vincent, C., Copsey, L., and Coen, E. (1996). Origin of floral asymmetry in Antirrhinum. Nature 383, 794–799. doi: 10.1038/383794a0
Manassero, N. G. U., Viola, I. L., Welchen, E., and Gonzalez, D. H. (2013). TCP transcription factors: Architectures of plant form. Biomol. Concepts 4, 111–127. doi: 10.1515/bmc-2012-0051
Martín-Trillo, M., and Cubas, P. (2009). TCP genes: A family snapshot ten years later. Trends Plant Sci. 15, 31–39. doi: 10.1016/j.tplants.2009.11.003
Ming, R., VanBuren, R., Wai, C. M., Tang, H., Schatz, M. C., Bowers, J. E., et al. (2015). The pineapple genome and the evolution of CAM photosynthesis. Nat. Genet. 47, 1435–1442. doi: 10.1038/ng.3435
Minh, B. Q., Nguyen, M. A., and von Haeseler, A. (2013). Ultrafast approximation for phylogenetic bootstrap. Mol. Biol. Evol. 30, 1188–1195. doi: 10.1093/molbev/mst024
Mondragón-Palomino, M., and Trontin, C. (2011). High time for a roll call: Gene duplication and phylogenetic relationships of TCP-like genes in monocots. Ann. Bot. 107, 1533–1544. doi: 10.1093/aob/mcr059
Navaud, O., Dabos, P., Carnus, E., Tremousaygue, D., and Hervé, C. (2007). TCP transcription factors predate the emergence of land plants. J. Mol. Evol. 65, 23–33. doi: 10.1007/s00239-006-0174-z
Nguyen, L., Schmidt, H. A., von Haeseler, A., and Minh, B. Q. (2014). IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274. doi: 10.1093/molbev/msu300
Niu, S., Li, J., Bo, W., Yang, W., Zuccolo, A., Giacomello, S., et al. (2022). The Chinese pine genome and methylome unveil key features of conifer evolution. Cell 185, 204–217. doi: 10.1016/j.cell.2021.12.006
One Thousand Plant Transcriptomes Initiative (2019). One thousand plant transcriptomes and the phylogenomics of green plants. Nature 574, 679–685. doi: 10.1038/s41586-019-1693-2
Ortiz-Ramírez, C., Hernandez-Coronado, M., Thamm, A., Catarino, B., Wang, M., Dolan, L., et al. (2016). A transcriptome atlas of Physcomitrella patens provides insights into the evolution and development of land plants. Mol. Plant 9, 205–220. doi: 10.1016/j.molp.2015.12.002
Pagnussat, G. C., Yu, H. J., Ngo, Q. A., Rajani, S., Mayalagu, S., Johnson, C. S., et al. (2005). Genetic and molecular identification of genes required for female gametophyte development and function in Arabidopsis. Development 132, 603–614. doi: 10.1242/dev.01595
Rambaut, A., Drummond, A. J., Xie, D., Baele, G., and Suchard, M. A. (2018). Posterior summarisation in Bayesian phylogenetics using Tracer 1.7. Syst. Biol. 67, 901–904. doi: 10.1093/sysbio/syy032
Reeves, P., and Olmstead, R. (2003). Evolution of the TCP gene family in Asteridae: Cladistic and network approaches to understanding regulatory gene family diversification and its impact on morphological evolution. Mol. Biol. Evol. 20, 1997–2009. doi: 10.1093/molbev/msg211
Ren, R., Wang, H., Guo, C., Zhang, N., Zeng, L., Chen, Y., et al. (2018). Widespread whole genome duplications contribute to genome complexity and species diversity in angiosperms. Mol. Plant 11, 414–428. doi: 10.1016/j.molp.2018.01.002
Rensing, S. A., Ick, J., Fawcett, J. A., Lang, D., Zimmer, A., Van de Peer, Y., et al. (2007). An ancient genome duplication contributed to the abundance of metabolic genes in the moss Physcomitrella patens. BMC Evol. Biol. 7:130. doi: 10.1186/1471-2148-7-130
Rensing, S. A., Lang, D., Zimmer, A. D., Terry, A., Salamov, A., Shapiro, H., et al. (2008). The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science 319, 64–69. doi: 10.1126/science.1150646
Ronquist, F. M., Teslenko, P., van der Mark, D. L., Ayres, A., Darling, S., Höhna, B., et al. (2012). MRBAYES 3.2: Efficient Bayesian phylogenetic inference and model selection across a large model space. Syst. Biol. 61, 539–542. doi: 10.1093/sysbio/sys029
Shang, Y., Yuan, L., Di, Z., Jia, Y., Zhang, Z., Li, S., et al. (2020). A CYC/TB1-type TCP transcription factor controls spikelet meristem identity in barley. J. Exp. Bot. 71, 7118–7131. doi: 10.1093/jxb/eraa416
Stull, G. W., Qu, X. J., Parins-Fukuchi, C., Yang, Y. Y., Yang, J. B., Yang, Z. Y., et al. (2021). Gene duplications and phylogenomic conflict underlie major pulses of phenotypic evolution in gymnosperms. Nat. Plants 7, 1015–1025. doi: 10.1038/s41477-021-00964-4
Tähtiharju, S., Rijpkema, A. S., Vetterli, A., Albert, V. A., Teeri, T. H., and Elomaa, P. (2012). Evolution and diversification of the CYC/TB1 gene family in Asteraceae—a comparative study in gerbera (Mutisieae) and sunflower (Heliantheae). Mol. Biol. Evol. 29, 1155–1166. doi: 10.1093/molbev/msr283
Taylor, J. S., and Raes, J. (2004). Duplication and divergence: The evolution of new genes and old ideas. Annu. Rev. Genet. 38, 615–643. doi: 10.1146/annurev.genet.38.072902.092831
The Angiosperm Phylogeny Group (2016). An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV. Bot. J. Linn. Soc. 181, 1–20. doi: 10.1111/j.1095-8339.2009.01000.x
Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F., and Higgins, D. G. (1997). The Clustal_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tool. Nucleic Acids Res. 25, 4876–4882. doi: 10.1093/nar/25.24.4876
Wang, Y. P., Tang, H. B., DeBarry, J. D., Tan, X., Li, J. P., Wang, X. Y., et al. (2012). MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40:e49. doi: 10.1093/nar/gkr1293
Wray, G. A., Hahn, M. W., Abouheif, E., Balhoff, J. P., Pizer, M., Rockman, M. V., et al. (2003). The evolution of transcriptional regulation in Eukaryotes. Mol. Biol. Evol. 20, 1377–1419. doi: 10.1093/molbev/msg140
Yang, X., Pang, H. B., Liu, B. L., Qiu, Z. J., Gao, Q., Wei, L., et al. (2012). Evolution of double positive autoregulatory feedback loops in CYCLOIDEA2 clade genes is associated with the origin of floral zygomorphy. Plant Cell 24, 1834–1847. doi: 10.1105/tpc.112.099457
Yao, X., Ma, H., Wang, J., and Zhang, D. (2007). Genome-wide comparative analysis and expression pattern of TCP gene families in Arabidopsis thaliana and Oryza sativa. J. Integr. Plant Biol. 49, 885–897. doi: 10.1111/j.1744-7909.2007.00509.x
Keywords: TCP, CYC, gene family expansion, gene duplication, green plants
Citation: Wang J-L, Wang H-W, Cao Y-N, Kan S-L and Liu Y-Y (2022) Comprehensive evolutionary analysis of the TCP gene family: Further insights for its origin, expansion, and diversification. Front. Plant Sci. 13:994567. doi: 10.3389/fpls.2022.994567
Received: 15 July 2022; Accepted: 15 August 2022;
Published: 02 September 2022.
Edited by:
Toshi Marie Foster, The New Zealand Institute for Plant and Food Research Ltd., New ZealandReviewed by:
Xianzhao Kan, Anhui Normal University, ChinaYang Dong, Institute of Botany (CAS), China
Wei Zhang, Shandong University, China
Pichang Gong, Institute of Botany (CAS), China
Copyright © 2022 Wang, Wang, Cao, Kan and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yan-Yan Liu, eXlsaXUxMDI1QGhvdG1haWwuY29t; Sheng-Long Kan, c2hlbmdsb25na2FuQDE2My5jb20=
 Jun-Li Wang1
Jun-Li Wang1 Sheng-Long Kan
Sheng-Long Kan Yan-Yan Liu
Yan-Yan Liu