- 1Key Laboratory of Genetics and Breeding in Forest Trees and Ornamental Plants of Ministry of Education, College of Biological Sciences and Technology, National Engineering Research Center of Tree Breeding and Ecological Restoration, Engineering Technology Research Center of Black Locust of National Forestry and Grassland Administration, Beijing Forestry University, Beijing, China
- 2Natural Resources and Planning Bureau of Yanshan County, Cangzhou, Hebei, China
- 3Shandong Academy of Forestry, Jinan, Shandong, China
- 4Cangzhou Municipal Forestry Seeding and Cutting Management Center, Cangzhou, China
- 5Forest Biotechnology Group, Department of Forestry and Environmental Resources, North Carolina State University, Raleigh, NC, United States
The AlkB homologs (ALKBH) gene family regulates N6-methyladenosine (m6A) RNA methylation and is involved in plant growth and the abiotic stress response. Poplar is an important model plant for studying perennial woody plants. Poplars typically have a long juvenile period of 7–10 years, requiring long periods of time for studies of flowering or mature wood properties. Consequently, functional studies of the ALKBH genes in Populus species have been limited. Based on AtALKBHs sequence similarity with Arabidopsis thaliana, 23 PagALKBHs were identified in the genome of the poplar 84K hybrid genotype (P. alba × P. tremula var. glandulosa), and gene structures and conserved domains were confirmed between homologs. The PagALKBH proteins were classified into six groups based on conserved sequence compared with human, Arabidopsis, maize, rice, wheat, tomato, barley, and grape. All homologs of PagALKBHs were tissue-specific; most were highly expressed in leaves. ALKBH9B and ALKBH10B are m6A demethylases and overexpression of their homologs PagALKBH9B and PagALKBH10B reduced m6A RNA methylation in transgenic lines. The number of adventitious roots and the biomass accumulation of transgenic lines decreased compared with WT. Therefore, PagALKBH9B and PagALKBH10B mediate m6A RNA demethylation and play a regulatory role in poplar growth and development. Overexpression of PagALKBH9B and PagALKBH10B can reduce the accumulation of H2O2 and oxidative damage by increasing the activities of SOD, POD, and CAT, and enhancing protection for Chl a/b, thereby increasing the salt tolerance of transgenic lines. However, overexpression lines were more sensitive to drought stress due to reduced proline content. This research revealed comprehensive information about the PagALKBH gene family and their roles in growth and development and responsing to salt stress of poplar.
Introduction
AlkB homologs (ALKBH) are a family of specific demethylases which belong to the dioxygenase superfamily and depend upon Fe2+ and α-ketoglutarate to catalyze demethylation of diverse substrates such as proteins, unique nucleic acids (tRNA, snRNA, bubbled DNA, bulged DNA, ds/ss DNA, and mRNA) (Trewick et al., 2002; Xu et al., 2020). About 40 years ago (Kataoka et al., 1983), the Escherichia. coli AlkB (EcAlkB) protein was found to repair damage from alkylating agents by dealkylation. While E. coli has only a single gene encoding ALKB, animals and plants have a family of homologs (ALKBH) (Wei et al., 1996; Kurowski et al., 2003; Chen et al., 2019; Wakisaka et al., 2019; Liang et al., 2020; Marcinkowski et al., 2020; Cai et al., 2021).
As a type of demethylase, the ALKBH gene family was proposed to be an eraser for epigenetic modification. Some N6-methyladenosine (m6A) RNA demethylases have been identified in Arabidopsis such as ALKBH9B (Martínez-Pérez et al., 2017), ALKBH10B (Duan et al., 2018), and SLALKBH2 in tomato (Zhou et al., 2019). In Arabidopsis thaliana, ALKBH10B, an m6A RNA demethylase can demethylate m6A-based mRNA modifications and mediate the m6A demethylation of transcripts of the key flowering time genes flowering locus T (FT), squamosa promoter binding protein-like 3 (SPL3), and SPL9 to specifically regulate the floral transition process (Duan et al., 2018). In addition, karrikins (a group of plant regulators present in smoke after burning) and cesium (Nelson et al., 2009) can induce the up-regulation of ALKBH10B (Hampton et al., 2004; Nelson et al., 2009, 2010). The expression of ALKBH10B decreased in the Arabidopsis dpa1 mutant, in which the γ subunit of the plastid ATP synthase has been inactivated (Bosco et al., 2004). ALKBH10B-mediated m6A demethylation affects the transcriptional level of stress response genes and regulates seed germination, seedling growth, and survival of Arabidopsis thaliana in response to salt stress or ABA (Shoaib et al., 2021). SlALKBH2 bind to the mRNA of the DNA demethylase gene SlDML2 in tomatoes and regulates its m6A modification and stability for fruit maturation (Zhou et al., 2019). ALKBH9-mediated m6A methylation may be a regulatory strategy to control cytoplasmic-replicating RNA viruses. ALKBH9B accumulated in cytoplasmic granules, colocalized with siRNA bodies, and was associated with P bodies and acted in mRNA silencing and/or mRNA decay (Martínez-Pérez et al., 2017). In Arabidopsis, the demethylation activity of AtALKBH9B positively regulates the infectivity of alfalfa mosaic virus (AMV), inhibition of AtALKBH9B increased the relative abundance of m6A in the AMV genome and inhibited systemic invasion of plants (Martínez-Pérez et al., 2017). The AtALKBH6 protein can bind to both m6A-labeled and m5C-labeled RNAs in Arabidopsis thaliana and an Atalkbh6 mutant showed a much lower survival rate than the wild-type under salt, drought, or heat stress, the transcript levels of ABA signaling-related genes may be down-regulated in the Atalkbh6 mutant (Huong et al., 2020). The role of ALKBH1D as a demethylase in chloroplasts may be similar to that of human ALKBH1 in mitochondria, where it is essential for tRNA biogenesis (Kawarada et al., 2017). ALKBH8 may be involved in the mcm5U and 5-methoxycarbonylmethyl-2′-O-methyluridine-2′-O-ribose methylated derivative process in Arabidopsis (Leihne et al., 2011; Endres et al., 2015). In ALKBH8B-overexpressing transgenic plants, global m6A levels were decreased and higher salt-tolerant phenotypes were exhibited compared to wild-type (Huong et al., 2022). Salt stress may induce the expression of ALKBH9 in roots, while high temperature inhibits the expression of ALKBH10A (Ma et al., 2006; Merret et al., 2015).
Soil salinization is a long-term abiotic stressor in extreme environments, causing huge losses to agriculture and forestry production (Janz and Polle, 2012; Zhao et al., 2020). Such extreme environmental conditions can affect the growth and development of plants (Hatfield and Prueger, 2015; Yu et al., 2016), weakening them so that they are more susceptible to disease or insect attacks. Salt stress causes osmotic stress, ion toxicity, and reactive oxygen species damage, resulting in blocked respiration, nutrient deficiency, decreased photosynthesis and transpiration, and inhibited growth and development (Tester and Davenport, 2003; Yu et al., 2016). Plants have evolved complex mechanisms to respond to salt stress, including osmotic regulation, ion balancing, the oxidative stress response, signal transduction via osmotic and ionic homeostasis signaling pathways, organelle stress responses, hormonal changes, and gene expression regulation (Zhao et al., 2020).
Poplar is an important woody model in plant molecular biology and it is also an important economic genus of forest trees (Bradshaw et al., 2000). However, the functions of the ALKBH family in Populus are not yet adequately defined, especially in response to abiotic stresses. Poplar 84K (Populus alba × Populus tremula var. glandulosa) is a well-known hybrid between species of white poplar (Qiu et al., 2019). The completion of the Populus genome sequence has allowed us to comprehensively characterize the ALKBH gene family in poplar (Tuskan et al., 2006; Ma et al., 2014; Qiu et al., 2019; Hofmeister et al., 2020; An et al., 2021). Here, we identified 23 PagALKBHs from poplar 84K, compared the collinearity, amino acid sequence, gene structure, and conserved motifs among homologs, and then classified them into six groups based on the conserved sequence. We described the expression patterns of the PagALKBHs in different tissues. Transgenic experiments showed that PagALKBH9B and PagALKBH10B are m6A RNA demethylases in poplar and that they inhibit adventitious root formation and biomass accumulation. Overexpression of PagALKBH9B and PagALKBH10B can reduce the accumulation of H2O2 and oxidative damage by increasing the activities of SOD, POD, and CAT, and increase the content of proline to reduce osmotic stress, thereby increasing the salt tolerance of transgenic lines. These results provide a molecular basis for further research on the adaptation and evolution of the ALKBH family in Populus.
Materials and methods
Plant material and growth conditions
The poplar 84K (Populus alba × Populus tremula var. glandulosa, a hybrid white poplar, hybrid female parent is P. alba and the hybrid male parent is P. glandulosa) plantlets were propagated by tissue culture, about 3 cm shoot tips were cut and placed in rooting medium (1/2 MS + 0.01 mg/l NAA + 0.1 mg/l IBA + 30 g/l sucrose +6.5 g/l agar) as described in the previous study (Zhao et al., 2022). The subculture plantlets were placed in 25°C culture chamber with a photoperiod of 16 h light (12,000 lux) and relative humidity of approximately 70% for 4 weeks and transferred to Hoagland’s nutrient solution for 1 week. The hydroponic plantlets were treated with 100 mM NaCl for 0, 6, 12, 24, and 48 h, and leaves were collected for detection of PagALKBH9B and PagALKBH10B expression abundance. In order to detect the tissue expression specificity of PagALKBHs, different tissues and organs were collected and immediately frozen in liquid nitrogen, and then stored at −80°C. At least six plantlets were pooled together as a biological replicate and three replicates were used for each experiment.
Poplar (Populus tomentosa) was used for genetic transformation as described earlier (Du et al., 2012). The adventitious roots of different transgenic lines growing in a rooting medium for 10 days were collected to detect the expression abundance of adventitious root (AR) regulatory genes. For phenotypic evaluation and the analysis of physiological indexes, transgenic plants and WT were grown in the rooting medium for 3 weeks for observation. For phenotypic observation and the analysis of physiological indexes, the 3 weeks old WT and transgenic lines were taken out from the medium and transplanted into mixed soil (perlite: vermiculite = 1: 3) for 2 weeks. Different lines were treated with a certain concentration (0, 100, and 200 mM) of salt solution and observed continuously. After 3 days of treatment, the second and third leaves fully stretched from top to bottom were collected for physiological indexes detection. The responses of each line to drought stress were compared under natural drought conditions, the second and third leaves fully stretched from top to bottom were collected for physiological indexes detection after 9 days. At least six plantlets were pooled together as biological replicates and three replicates were used to count the growth indexes and phenotypic observation.
Identification of PagALKBH genes and synteny analysis
Chromosome-level genomes of poplar 84K (Populus alba × P. tremula var. glandulosa) were referenced as published (Qiu et al., 2019). The genomes of Arabidopsis (Arabidopsis thaliana), maize (Zea mays), and rice (Oryza sativa) were downloaded from the Ensemblm1 and Phytozome database.2 We searched the poplar 84K genome with the ALKBH protein sequences of Arabidopsis and rice and selected all sequences with an e-value <= 1e-10 and then used Pfam3 and SMART tools4 to determine whether each candidate 2OG-Fe (II) and Fe2+ domains, identified ALKBH family genes (Letunic et al., 2009). Collinear blocks were identified by PagALKBHs duplication events in the MCScanX (Wang et al., 2012). We integrated and plotted the data using Circos (Krzywinski et al., 2009). DNAMAN 6.0 was used to align amino acid sequences (Nong et al., 2019).
Phylogenetic analysis, gene structure, and protein motifs
The ALKBH protein sequences of different species were imported into MEGA 7.0 software to construct a phylogenetic tree by neighbor-joining (NJ). A bootstrap value was set to 1,000 replicates (Gascuel, 2006; Tamura et al., 2013) and visualized using Evolview (Zhang et al., 2012). The Gene Structure Display Server (GSDS)5 was used to recognize the exons and introns of PagALKBH genes (Zhang et al., 2012). The conserved motifs of PagALKBH proteins were identified using MEME Suite Web server6 (Bailey et al., 2009) and the maximum number of motifs was specified as 10. TBtools was used to visualize gene structure and conserved domains (Chen et al., 2020).
RNA isolation and reverse transcription-quantitative real-time polymerase chain reaction analysis
Total RNA was isolated using TRIzol reagent (TransGen Biotech, Beijing, China). The PrimeScript™ RT kit with gDNA eraser was then used to reverse-transcribe the total RNA into cDNA (TaKaRa, Beijing, China) and detected by 1% agarose gel electrophoresis, qualified cDNA is diluted with different concentrations according to the Ct value of the pre-experiment. Reverse transcription-quantitative real-time polymerase chain reaction (RT-qPCR) was performed with the SYBR Green master mix (TaKaRa). All primers used were detected by PCR and listed in Supplementary Table S1. The 2–ΔΔCt method was used to calculate the relative abundance of PagALKBH transcripts (Livak and Schmittgen, 2001) with the reference gene PagActin as an internal standard (Wang et al., 2021). Three biological replicates with three technical replicates were performed for each sample.
Vector construction, poplar transformation, and characterization
The primers PagALKBH9B-F, PagALKBH9B-R, and PagALKBH10B-F, PagALKBH10B-R were used to amplify the nucleotide sequences PagALKBH9B and PagALKBH10B, respectively. The PagALKBH9B and PagALKBH10B were subcloned into the pBI121 vector under the control of the CaMV35S promoter. Agrobacterium-mediated stable transformation was performed as previously described (Du et al., 2012). In sterile conditions, take 4–6 weeks of P. tomentosa tissue culture plantlets leaves, cut the main vein with a knife, and placed in the OD600 = 0.4 Agrobacterium infection for 10 min. The leaves were taken out, dried with sterile filter paper, and the back of leaves was laid down on the differentiation culture medium (MS + 1.0 mg/l 6-BA +0.2 mg/l NAA + 80 μM acetosyringone), and cultured at 25°C in dark for 2 days. After co-culture, the explants were suctioned with sterile filter paper to remove the suction residual bacteria liquid, transferred to the screening medium (differentiation medium +250 mg/l cephalosporin +30 mg/l kanamycin), light cycle 16 h/8 h, 25°C, every 10 days to replace the selection medium. The length of equal transformed buds was about 2 cm, cut and inserted into rooting medium (1/2 MS + 0.2 mg/l NAA + 0.2 mg/l IBA + 250 mg/l cephalosporin +40 mg/l Kana). Rooted kanamycin-resistant plantlets were screened based on the fusion fragment of the CaMV35S promoter, PagALKBH9B, and PagALKBH10B using genomic PCR amplification with the forward primer 35S-F and reverse primers PagALKBH9B-R and PagALKBH10B-R. Then, the transgenic plants were analyzed using RT-qPCR with the primers PagALKBH9B-qF, PagALKBH9B-qR, and PagALKBH10B-qF, PagALKBH10B-qR. Primers used are listed in Supplementary Table S1.
Determination of the m6A RNA methylation level
Total RNA was isolated from leaves of transgenic lines and WT as previously described using the TRIzol reagent (TransGenBiotech, Beijing, China). RNA quality was assessed using a NanoDrop 2000 spectrophotometer (Thermo Scientific, Waltham, MA, United States) and agarose gel electrophoresis. Oligo (dT) magnetic beads were used to capture the mRNA, and liquid washing was used to remove the non-mRNA, so as to obtain the complete mRNA in the total RNA (TIANGEN, Beijing, China). The m6A level was determined with an Aderr kit (Epigentek, Farmington, NY, United States). In this assay, RNA is bound to strip wells using RNA high binding solution. m6A is detected using capture and detection antibodies. The detected signal is enhanced and then quantified by reading the absorbance in a microplate spectrophotometer. The amount of m6A is proportional to the OD intensity measured. m6A (%) = (Sample OD-NC OD)/S/(PC OD-NC OD)/P*100% (NC: negative control; PC: positive control; S: RNA sample inputs, ng; P: positive control inputs, ng).
Physiological indices analysis
After 3 days of salt stress at different concentrations, the leaves were rounded with a diameter of 1 cm and immersed in 3,3′-diaminobenzidine (DAB) solution (1 mg/m L, 50 mM Tris–HCl, pH 3.8) at 28°C in the dark for 16 h. Then, incubate with 95% ethanol at 70°C for 5 min to remove chlorophyll. After cooling, the leaves were transferred to fresh ethanol at room temperature and photographed. The H2O2 content of each line was detected according to the H2O2 content detection kit (NJJCbio, China) following the manufacturer’s instructions. The MDA and proline contents in the fresh leaves were measured spectrophotometrically according to Zhao et al. (2022). The leaf discs were punched with a 1 cm diameter puncher and placed in a clean test tube. After adding 5 ml deionized water containing Tween 20, initial (S1) and final (S2) conductivity were measured after sealing boiling water bath for 8 min and cooling balance for 3 min. At the same time, the blank conductivity (S0) of distilled water was measured. Electrolyte leakage rate (%) = (S1-S0)*100/(S2-S0). The contents of Chl and Chl b and antioxidant enzyme activity levels of SOD, POD, and CAT were evaluated using the corresponding assay kits (NJJCbio, Nanjing, China).
m6A-IP-qPCR
m6A-IP-qPCR was carried out as previously described with minor modifications (Zhou et al., 2021). Total RNA extracted was preheated at 75°C for 5 min, and then cooled immediately on ice. 200 μg total RNA was fragmented into 300 nt short fragments using RNA fragmentation buffer (10 mM Tris–HCl, pH 7.0, 10 mM ZnCl2) at 95°C for 3 min, and then 50 mM EDTA was added to terminate the fragmentation reaction and then cooled immediately on ice. Add 2.5 times volume of alcohol, 0.1 times volume of 3 M sodium acetate (pH 5.3), 100 μg/ml glycogen, overnight at −80°C. 15,000 g centrifugal 20 min, precipitation using pre-cooling 75% alcohol wash twice, 15,000 g centrifugal 10 min, and ventilation cabinet drying 5 min. Then resuspension with 100 μl DEPC-treated water. Reservation of 2 μg total RNA as input and incubation of 100 μg total RNA with 2–5 μg of anti-m6A polyclonal antibody (Synaptic Systems, 202003, Germany) at 4°C for 2 h. The washed immunomagnetic beads protein A was added to the RNA solution and incubated at 4°C for 2 h. The antibody bound to the protein beads and the RNA complex were collected by the magnetic force frame, and washed twice with 1 × IP buffer, high salt buffer (50 mM Tris–HCl, 1 M NaCl, 1 mM EDTA, 1% NP-40, and 0.1% SDS), respectively. 200 μl elution buffer (6.7 mM m6A in 1 × IP buffer, 300 U mL–1 RNase inhibitor) was added to the mixture for 2 h with continuous shaking at 4°C. After RNA was precipitated by ethanol, centrifuged at 15,000 g for 20 min at 4°C, washed twice with 75% ethanol, centrifuged at 15,000 g for 5 min at 4°C, then dried in the ventilating cabinet for 5 min, and resuspended with 10 μl RNase-free water. Then, both the m6A-containing total RNA and the input total RNA were submitted to RT-qPCR and primers listed in Supplementary Table S1. The 2–ΔΔCt method was used to calculate relative m6A enrichment. Three biological replicates with three technical replicates were performed for each sample.
Statistical analysis
All results were expressed as the mean ± standard error (SE) in triplicate or more. Statistical analyses were performed using SPSS 22.0 software (IBM, Armonk, NY, United States), and significant differences in the measured parameters were inferred according to Duncan’s multiple range test. Significant differences between the two groups of data were evaluated for comparisons (p < 0.05).
Results
Genome-wide identification of PagALKBHs
To identify all the ALKBH members in poplar 84K (Populus alba × Populus tremula var. glandulosa), 14 Arabidopsis and 9 rice ALKBH amino acid sequences were used as queries (Yue et al., 2019) and then screened against Pfam and SMART databases. Twenty-three PagALKBHs were identified in poplar 84K, among them, 11 PagALKBHs were present in the female hybrid parent (P. alba), and 12 PagALKBHs were present in the male hybrid parent (P. glandulosa). There was a difference in the number of ALKBH genes among hybrid parents, hybrid female parent lost one located on chromosome 3 (Figure 1A; Supplementary Table S2). Further sequence alignment revealed high sequence similarity of homologs from hybrid parents of poplar 84K (Supplementary Figure S1). Then we used Circos to draw whole-genome synteny blocks and studied the whole-genome for duplication (WGD) events in the PagALKBH gene family among hybrid parents of poplar 84K (Figure 1B). Collinearity was conserved for 14 pairs of genes among hybrid parents and most homologous genes have a one-to-one correspondence across species.
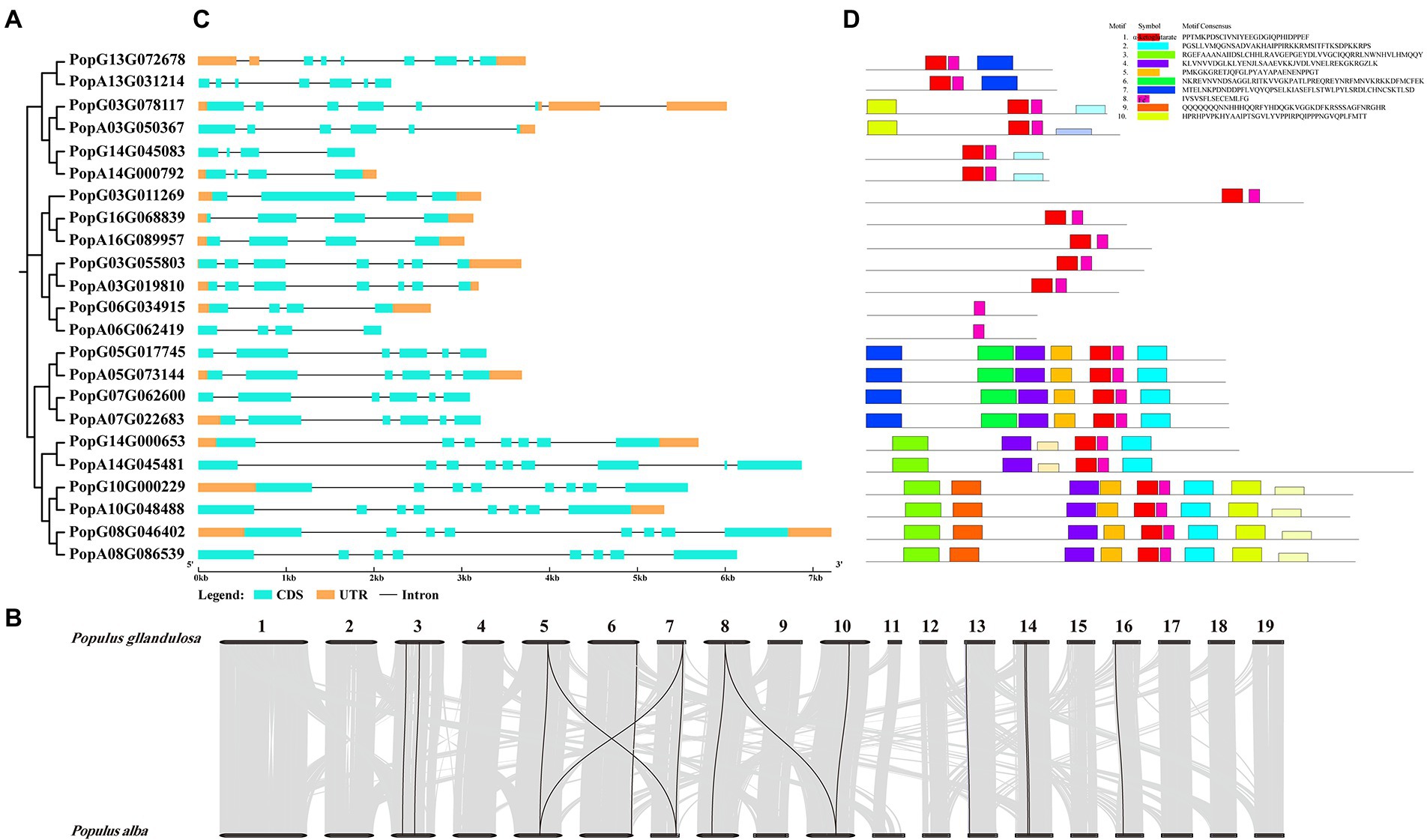
Figure 1. (A) Phylogeny of PagALKBH proteins identified in poplar 84K. (B) Genome distribution and collinearity of the PagALKBH family in poplar 84K. Black lines indicate the collinear relationships of PagALKBHs, gray lines represent other than PagALKBHs collinear relationships. (C) Gene structures of PagALKBHs. The exons, untranslated regions (UTR), and introns were represented by amber and cyan rectangles, and the blank line, respectively. (D) Conserved motifs of PagALKBHs. Different motifs are represented by different colors.
Gene structure and conserved motifs of PagALKBHs
As shown in Figure 1C, similar exon and intron structures exist between genes that are evolutionarily closely related, there were identical intron and exon structures between homologs except PopG14G000653 and PopA14G045481, PopA14G045481 had two extra exons. On the contrary, there were differences in the untranslated region (UTR) between homologs between different hybrid parents. Except for the homologous genes PopG16G068839 and PopA16G089957 which contain the same UTR structure, most of the other homologous genes that exist in pairs only have one of them. Different from gene structure, conserved sequences were highly consistent among homologs. In addition, genes close to each other on the evolutionary tree often contain the same conserved domains and similar motifs suggesting conservation in function of PagALKBH family proteins, and unique motifs may predict specific biological functions. Among the PagALKBHs, motif 8 harboring the Fe2+ binding site was conserved, which was characteristic of the ALKBH gene family. However, another conserved domain of the ALKBH gene family, α-ketoglutarate, was missing in PopG06G034915 and PopA06G062419, which may indicate differences in function.
Phylogenetic analysis of PagALKBHs
To demonstrate the relationships between species, and to illustrate their evolution, a phylogenetic tree was constructed among human (Homo sapiens), Arabidopsis (A. thaliana), maize (Zea mays), rice (Oryza sativa), wheat (Triticum aestivum), tomato (Solanum lycopersicum), barley (Hordeum vulgare), grape (Vitis vinifera), and poplar 84 K (Populus alba × P. tremula var. glandulosa; Figure 2). The proteins clustered into six groups (ALKBH1, ALKBH2, ALKBH6, ALKBH8, ALKBH9, ALKBH10) following earlier nomenclature (Marcinkowski et al., 2020). ALKBHs showed differences in numbers within species. Of all ALKBH proteins, the largest amount of ALKBHs was found in wheat, while the lowest was found in barley which lost the member of the ALKBH2 group. The ALKBH10 group was the most abundant group, with 15 members that displayed differences in number between different species, ALKBH2 and ALKBH6 had the smallest number of members. The ALKBH9 and ALKBH10 groups are relatively closely related to the human m6A RNA demethylation modification enzyme ALKBH5, while another demethylase FTO, although classified in the ALKBH8 group, is farther related to shuffling.
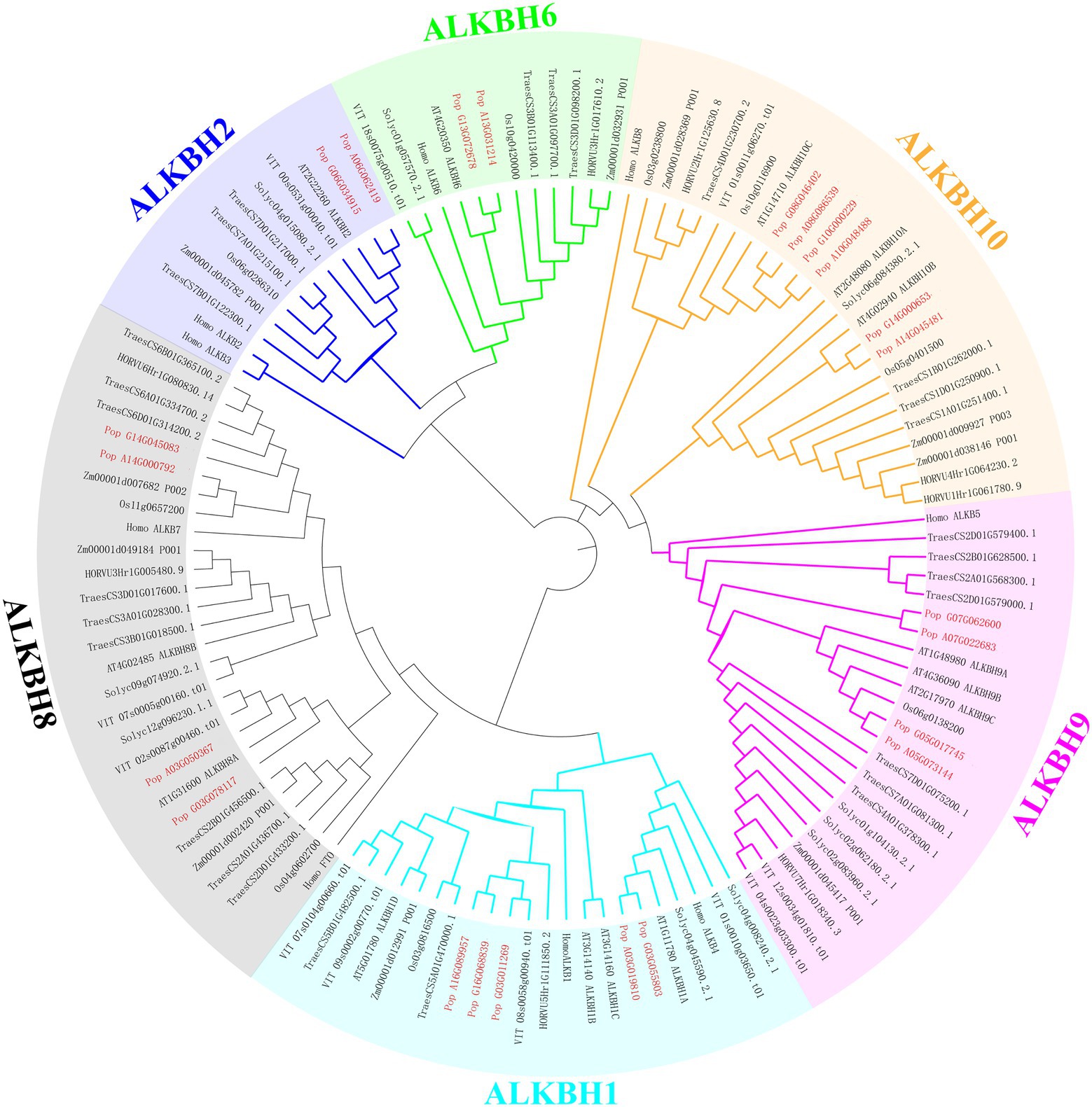
Figure 2. Phylogeny of ALKBH proteins. Phylogenetic relationships of human (Homo sapiens) Arabidopsis (A. thaliana), maize (Zea mays), rice (Oryza sativa), wheat (Triticum aestivum) tomato (Solanum lycopersicum) barley (Hordeum vulgare) grape (Vitis vinifera), and poplar 84K (Populus alba × P. tremula var. glandulosa) were estimated using maximum likelihood. Different colors represent different groups.
Tissue-specific expression of the PagALKBH genes in poplar 84K
To explore the biological functions of PagALKBH genes, RT-qPCR was used to survey the relative abundance of PagALKBH transcripts in different tissues (root, stem, leaf, and stem apex). The relative abundance of PagALKBH transcripts can be determined for various tissues and organs of poplar to identify tissue-specific differences (Figure 3). All the PagALKBHs had similar expression patterns where the transcript level in leaves was relatively high and the abundance in other tissues was low, with the exception of ALKBH10C, which was lowest in leaves.
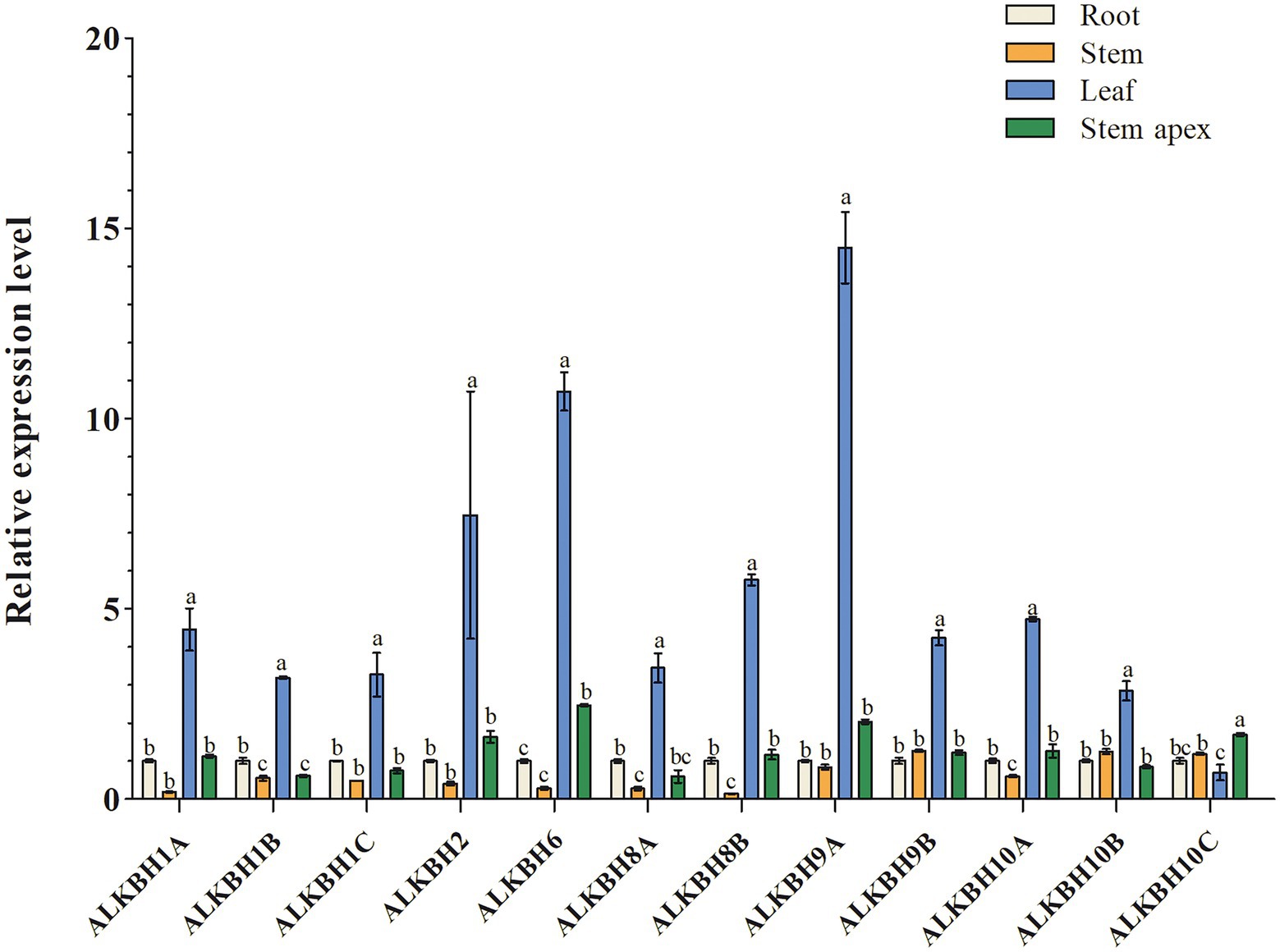
Figure 3. Tissue-specific expression patterns of PagALKBHs poplar 84K. The roots, stems, leaves, and stem apex of 84K poplars under normal growth conditions were collected, different tissues of at least six 84K poplar were collected as a replicate. The data represented averages ± SE of three replicates. The different letter indicated significant differences between different tissue.
Overexpression of PagALKBH9B and PagALKBH10B reduced m6A RNA methylation
ALKBH9B and ALKBH10B were m6A RNA methylation demethylases, and overexpression of ALKBH9B and ALKBH10B reduces the level of m6A RNA methylation in Arabidopsis (Martínez-Pérez et al., 2017; Duan et al., 2018). To determine whether PagALKBH9B and PagALKBH10B regulate m6A methylation in poplar, transgenic poplar harboring the PagALKBH9B and PagALKBH10B genes under the control of CaMV35S promoter (35S::PagALKBH9B and 35S::PagALKBH10B) were generated by Agrobacterium-mediated transformation. After kanamycin screening, genomic PCR identification, and RT-qPCR analysis, then, four transgenic lines (OE9B-4 and OE9B-27 overexpressing of PagALKBH9B and OE10B-7 and OE10B-21 overexpressing of PagALKBH10B), were used for further analyses (Supplementary Figure S2). By detecting the overall level of m6A in the leaves of WT and transgenic lines under normal growth conditions, it was found that overexpression of PagALKBH9B and PagALKBH10B reduced the m6A RNA modification level on total RNA in transgenic poplars (Figure 4A) and overexpression of PagALKBH9B did not significantly affect m6A modification on mRNA (Figure 4B).
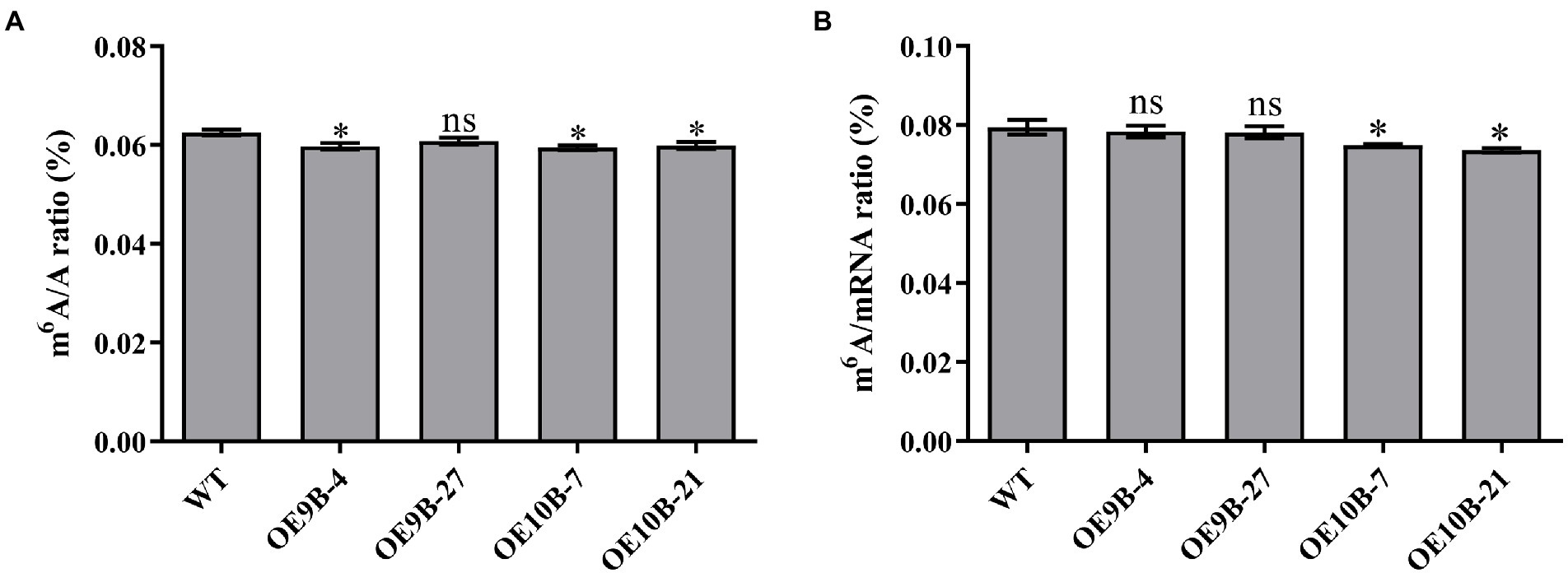
Figure 4. N6-methyladenosine (m6A) RNA methylation level. (A) Global N6-methyladenosine (m6A) RNA methylation level in WT and transgenic lines under normal conditions. (B) N6-methyladenosine (m6A) RNA methylation level on mRNA in WT and transgenic lines under normal conditions. At least six 84K poplars were collected as a replicate. The asterisks indicated significant differences between WT and transgenic plants (*p < 0.05).
Overexpressing of PagALKBH9B and PagALKBH10B inhibited adventitious root formation in poplar 84K
To determine whether PagALKBH9B and PagALKBH10B participated in ARs formation, we compared the AR phenotypic changes of overexpressing transgenic plantlets and WT plantlets under normal conditions (Figure 5A). When the stem segments were inserted into the rooting medium for 7 days, WT had formed adventitious roots, while all transgenic lines did not. After 13 days, although all transgenic lines had formed ARs, the number of ARs was significantly less than that of WT, which continued to 21 days. After culture in rooting medium for 21 days, we took out different lines for phenotype observation and growth index statistical analysis. The observation of the ARs phenotype and the statistics of ARs number showed that the number of ARs of transgenic lines overexpressing of PagALKBH9B and PagALKBH10B was significantly less than that of WT, which was consistent with the observations in tissue culture (Figures 5B,C). There was no significant difference in the average length of ARs among different lines (Figure 5D). Compared with WT, the biomass of ARs (fresh weight and dry weight) in transgenic lines decreased significantly (Figures 5E,F). The expression of ARF6, ARF8, GH3.5, YUCCA6, and ABCB19 that have been provedto regulate the formation of ARs was inhibited in transgenic lines overexpressing PagALKBH9B and PagALKBH10B (Li, 2021), and IAA28 was inhibited in transgenic lines overexpressing PagALKBH10B (Supplementary Figure S3A). However, we found that the m6A levels of these genes were not significantly different in transgenic lines, and they may not be directly regulated by PagALKBH9B or PagALKBH10B (Supplementary Figure S3B). Overexpression of PagALKBH9B did not reduce the plant height of transgenic lines, while the plant height of transgenic lines overexpressing PagALKBH10B was significantly inhibited compared with WT, although the biomass accumulation between transgenic lines was similar and less compared with WT (Supplementary Figure S4). It was therefore reasonable to infer that PagALKBH9B and PagALKBH10B regulate the demethylation of common genes and specific targets involved in ARs formation and plant growth.
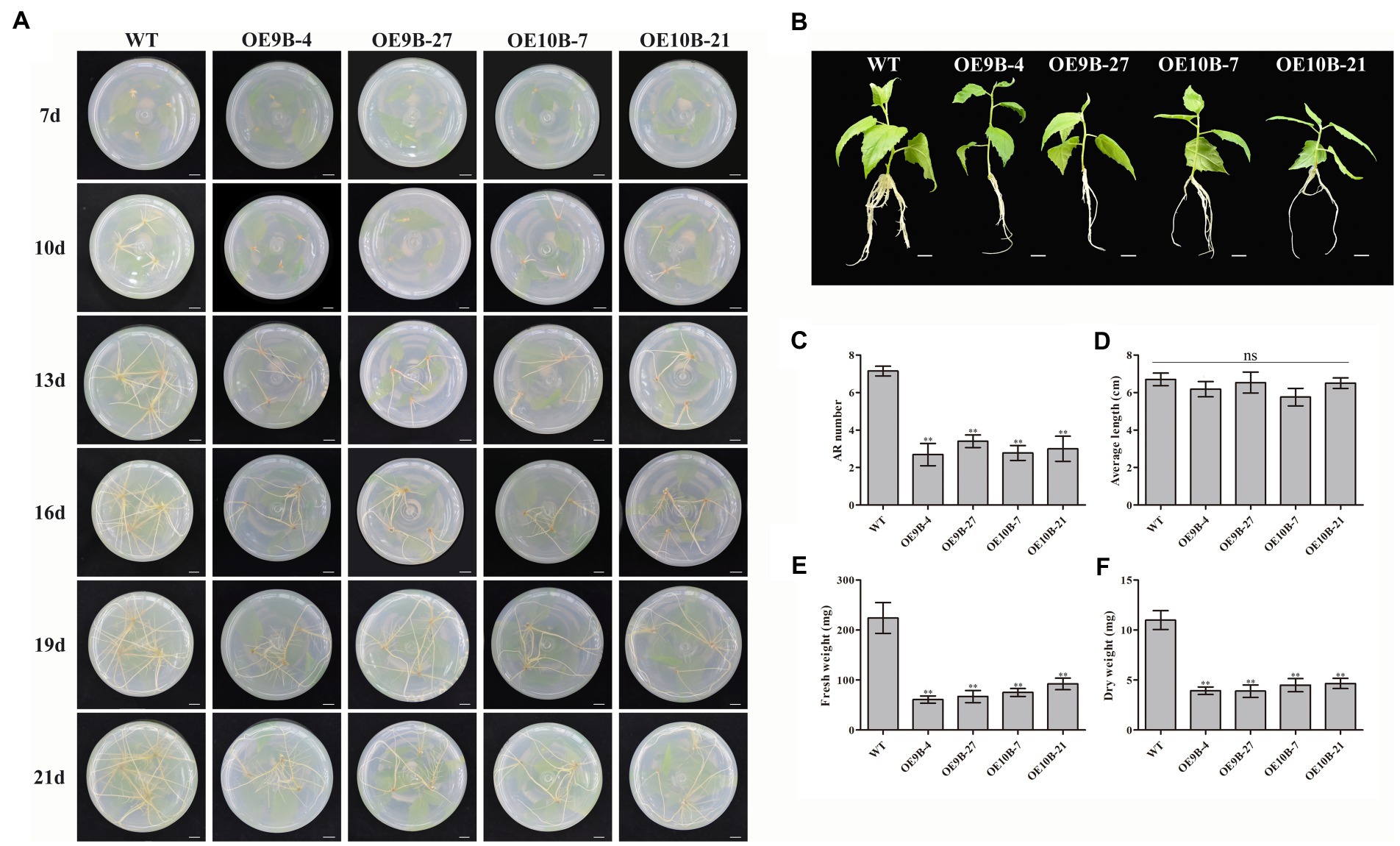
Figure 5. Phenotypes of transgenic lines and WT cuttings 3 weeks after establishment in rooting medium. (A) The adventitious roots of transgenic lines and WT formed and developed over time. Bar = 1 cm. (B) Images of cuttings from different lines 3 weeks after establishment. Bar = 1 cm. (C) AR number between different lines. (D) Average length of ARs between different lines. (E) Fresh weight of ARs between different lines. (F) Dry weight of ARs between different lines. The experiments were performed three times and the similar results were obtained, at least six 84K poplars were collected as a replicate. The bars represent means ± SE (n = 6) and the asterisks indicate significant differences between WT and transgenic plants (*p < 0.05, **p < 0.01).
Overexpressing of PagALKBH9B and PagALKBH10B improved salt tolerance
m6A methyltransferase can improve salt tolerance in Arabidopsis by increasing the overall level of m6A methylation modification (Hu et al., 2021). We speculated whether PagALKBH9B and PagALKBH10B had the opposite function in response to salt stress in poplar. We first analyzed the overall level of m6A between different lines under salt stress, and found that salt stress could significantly induce higher up-regulation of m6A levels in WT (Figure 6). To determine the role of PagALKBH9B and PagALKBH10B in response to salt stress in poplar, we examined the phenotypic changes of overexpressing transgenic lines and WT under different concentrations of salt treatment. As shown in Figure 7A, there was no visible morphological difference between transgenic lines and WT under control (0 mM). When exposed to different concentrations of salt (100 and 200 mM), the WT rapidly showed different degrees of pressure injury with leaf wilting and fresh weight decreased (Figure 7C), while the transgenic lines were less sensitive to salt stress than WT plants. These results indicated that overexpression of PagALKBH9B and PagALKBH10B in poplar alleviated the phenotypic injury induced by salt stress.

Figure 6. Global N6-methyladenosine (m6A) RNA methylation level in WT and transgenic lines under normal conditions and salt treatment. The data represented averages ± SE of three replicates. The different letters indicated significant differences between different tissue (*p < 0.05, **p < 0.01).
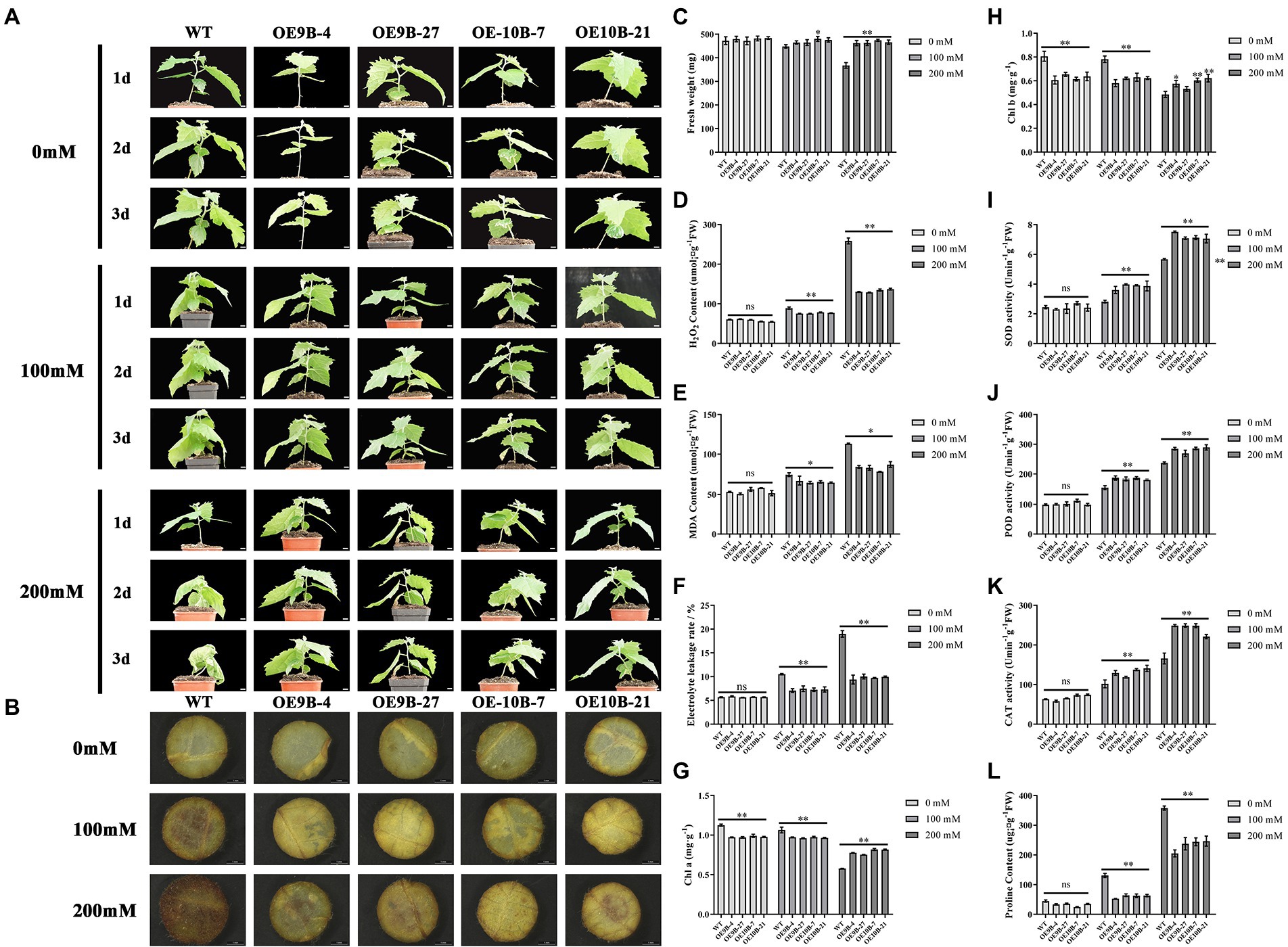
Figure 7. Overexpressing PagALKBH9B and PagALKBH10B enhanced salt tolerance in transgenic poplars. (A) The phenotype of transgenic lines and WT under 0, 100, and 200 mM NaCl treatment. Bar = 1 cm. (B) Histochemical detection of H2O2 accumulation of WT and transgenic lines under a certain concentration salt treatment by DAB staining. (C) The fresh weight of leaves of WT and transgenic lines under a certain concentration salt treatment. (D) Histochemical detection of H2O2 accumulation of WT and transgenic lines under a certain concentration salt treatment. (E) Histochemical detection of MDA contents of WT and transgenic lines under a certain concentration salt treatment. (F) The electrolyte leakage rates of WT and transgenic lines under a certain concentration salt treatment. (G) The Chl content of WT and transgenic lines under a certain concentration salt treatment. (H) The Chl b content of WT and transgenic lines under a certain concentration salt treatment. (I–K) Antioxidant enzyme activity in transgenic lines compared with WT after salt treatment, respectively. (L) Histochemical detection of proline contents of WT and transgenic lines under a certain concentration salt treatment. The experiments were performed three times and the similar results were obtained, at least six 84K poplars were collected as a replicate. The bars represent means ± SE (n = 6) and the asterisks indicate significant differences between WT and transgenic plants (*p < 0.05, **p < 0.01).
Salt stress can lead to the accumulation of reactive oxygen species (ROS) in plant leaves, which is an important factor that cause oxidative damage in plants (Yu et al., 2020). Therefore, we analyzed the H2O2 content of each line treated with different concentrations (Figures 7B,D). Under normal conditions, there was no significant difference in H2O2 content between WT and overexpression lines. However, after different concentrations of salt treatment, discs of PagALKBH9B and PagALKBH10B -overexpressing lines exhibited lighter and less intense brown patterns than those of the WT, the accumulation of H2O2 in WT was significantly higher than that in transgenic lines. This indicated that overexpression of PagALKBH9B and PagALKBH10B both could reduce the accumulation of ROS under salt stress.
Under salt stress, the accumulation of ROS in plants can cause or aggravate membrane lipid peroxidation, resulting in cell membrane system damage and even cell death. MDA is one of the products of membrane lipid peroxidation and an important indicator of the degree of membrane lipid peroxidation, which can indirectly reflect the degree of membrane system damage and plant stress resistance (Yu et al., 2020). As shown in Figure 7E, there was no significant difference in MDA content between overexpression transgenic lines and WT under normal conditions. After stress treatment, the MDA content in both plants increased, but the increase of over-expression transgenic plants was significantly lower than that of WT, and the results were consistent in different concentrations of salt treatment. After salt treatment, the electrolyte leakage rate gradually increased with treatment concentration. Although there was no significant difference in electrolyte leakage rate between WT and transgenic lines under normal conditions, the electrolyte leakage rate of WT was significantly higher than that of transgenic lines after salt stress treatment (Figure 7F). The above results showed that the overexpression of PagALKBH9B and PagALKBH10B reduced the membrane lipid peroxidation caused by salt stress.
Salt stress can affect plant photosynthesis by reducing Chl a/b (Tang C. et al., 2018), therefore, we compared the Chl a/b content of WT and transgenic lines (Figures 7G,H). Although WT contained more Chl a/b than transgenic lines under normal conditions, the content of Chl a/b in WT decreased significantly with the increase of salt concentration. Finally, the content of Chl a/b in WT was significantly lower than that in transgenic lines at 200 mM salt concentration.
In order to further evaluate the oxidative damage of plants after salt stress, we measured the protective enzyme activities of transgenic lines and WT after salt stress, including SOD, POD, and CAT activities. Under normal growth conditions, there was no significant difference in SOD, POD, and CAT activities between WT and overexpression transgenic plants (Figures 7I–K). However, after salt stress, the activities of SOD, POD, and CAT in WT and overexpression transgenic plants were significantly increased. The contents of SOD, POD, and CAT in WT and transgenic lines treated with 100 and 200 mM NaCl were significantly higher than those in WT. The expression abundance detection of SOD, POD, and CAT synthetic regulatory-related genes (Cu/Zn-SOD, POD-20, and CAT) had higher expression in transgenic lines, but there was no significant difference in m6A modification level (Supplementary Figure S6). Overexpression of PagALKBH9B and PagALKBH10B increased the activities of SOD, POD, and CAT in transgenic plants, enhanced the scavenging ability of reactive oxygen species, and reduced oxidative damage.
Salt stress can lead to osmotic stress, and proline is an important osmotic adjustment substance, which is up-regulated after stress, so its content reflects plant stress resistance (Yu et al., 2020). There was no significant difference in proline content between transgenic lines and WT under control. However, with the increase of salt concentration, the proline content increased, but at all salt concentrations, the proline content in overexpression transgenic plants was significantly lower than that in WT (Figure 7L). P5C5 has been shown to regulate proline synthesis (Kubala et al., 2015) and the expression abundance of P5C5 lower in overexpression transgenic plants and m6A enrichment level was no significant difference (Supplementary Figure S6). Proline is a key marker for drought stress response in plants (Chen et al., 2022), so we compared the drought resistance between WT and transgenic lines. The results showed that overexpression of PagALKBH9B and PagALKBH10B increased the sensitivity of transgenic lines to drought stress (Supplementary Figure S7A). Consistent with the proline content under salt stress, the proline content of transgenic lines was lower than WT under drought stress (Supplementary Figure S7B).
Discussion
RNA demethylation mediated by ALKBHs acting as eraser proteins was considered to be an important part of the epigenetic regulatory network for plant growth, development, and abiotic stress responses (Huong et al., 2020; Liang et al., 2020). Yet, the molecular mechanisms of ALKBH regulation need to be further explored, especially in woody plants. Unlike the known AtALKHs and OsALKBHs (Liang et al., 2020), more PagALKBHs were identified from the poplar 84K hybrid (Populus alba × P. tremula var. glandulosa) by comparison of the alignments of homologous ALKBH protein sequences in A. thaliana (Liang et al., 2020). Function is often conserved between paralogs (Shao et al., 2019; Cheng et al., 2021). Sequence alignments between homologs of PagALKBHs (Figure 1A; Supplementary Figure S1) showed high similarity and high collinearity (Figure 1B). Orthologous proteins generally perform analogous biological functions in diverse species within the genus (Schlicker et al., 2006).
Homologs of PagALKBHs contain similar exon/intron structure and conserved motifs (Figures 1C,D), thus, the homologs of PagALKBH between hybrid parents may have functional similarity. The catalytic activity of ALKBH demethylase depends on Fe2+ (Falnes et al., 2002; Trewick et al., 2002; Fedeles et al., 2015) and all PagALKBHs contained an Fe2+ binding domain (Figure 1C) so that PagALKBHs may mediate in the oxidative demethylation of nucleic acids. Although there were homologous catalytic centers in the conserved sequences, substrate selectivity was characteristic of the ALKBH family (Yi et al., 2009; Ougland et al., 2015). Phylogenetic analysis was a useful way to identify functional orthologous proteins (Bauwens et al., 2018). The ALKBHs were divided into six phylogenetic groups, consistent with previous studies (Figure 2). Among the groups, ALKBH9 and ALKBH10 were homologs of human demethylase ALKBH5 (Tang Y. et al., 2018), which can remove m6A methylation in Arabidopsis, and the ALKBH9 group was shown to have functional redundancy (Duan et al., 2018). Similar to the results from other plants, the human demethylase FTO (fat mass and obesity-associated protein) also has no homologs in poplar (Chen et al., 2012).
N6-methyladenosine (m6A) RNA methylation was the most abundant chemical modification in eukaryotic cells, accounting for 80% of RNA base modifications (Kierzek, 2003). RNA methylation is a dynamic process and requires the participation of methyltransferases (writers), demethylation (erasers), and recognition proteins (readers). MTA, MTB, FIP37, VIRILIZER, and HAKAI were considered to be the main components of the m6A methyltransferase complexes in plants (Bodi et al., 2012; Ra et al., 2017; Yue et al., 2019). The m6A readers were members of the YTH family, and the members YTHDF1, YTHDF2, and YTHDF3 recognize m6A modification sites (Dominissini et al., 2012; Wang et al., 2015). In mammals, the first identified m6A demethylase was FTO. Later, ALKBH5 (alkylation repair homolog 5) was shown to be a mRNA m6A demethylase (Jia et al., 2011; Mauer et al., 2017). ALKBH9B and ALKBH10B are homologs of human demethylase ALKBH5 in plants (Figure 2). m6A can revert to adenosine through the action of ALKBH9B and ALKBH10B (Martínez-Pérez et al., 2017; Duan et al., 2018). The human ALKBH5 homologs of PagALKBH9B and PagALKBH10B were also confirmed to regulate the global level of m6A RNA methylation. Overexpression of PagALKBH9B and PagALKBH10B reduced the global m6A RNA methylation in transgenic plants (Figure 4A). However, the m6A methylation level on mRNA of transgenic lines overexpressing PagALKBH9B did not change significantly (Figure 4B). Martínez-Pérez et al. (2017) reported that ALKBH9B can remove m6A from single-stranded RNA (ssRNA) in vitro, the analysis of Duan et al. (2018) showed that the methylation level of m6A of mRNA in alkbh9b mutant did not change significantly in its highly expressed organs. It is possible that ALKBH9B was not involved in the removal of m6A modifications on mRNA. Considering that PagALKBH9B and PagALKBH10B transgenics were driven by CaMV35S, the activity of the overexpressed enzymes may increase demethylated RNAs or demethylate RNAs that were not normally recognized by these proteins, which may lead to novel phenotypes in the overexpressed lines.
m6A RNA methylation plays a crucial and dynamic role in many processes, including development of organs (Vespa et al., 2004; Bodi et al., 2012; Chen et al., 2018a,b; Arribas-Hernández and Brodersen, 2020), seed germination (Zhong et al., 2008) and fruit maturation (Zhou et al., 2019, 2021). The homologs ALKBH9 and ALKBH10 were shown to regulate fruit maturation in tomatoes (Zhou et al., 2019), and the floral transition process in A. thaliana (Duan et al., 2018). Overexpression of PagALKBH9B and PagALKBH10B inhibited the formation of ARs and reduced biomass accumulation of transgenic lines. The transcriptional abundance of the genes involved in the regulation of adventitious roots was inhibited in the overexpression lines (Sorin et al., 2006; Kim et al., 2007; Sukumar et al., 2013), but their m6A enrichment levels were not significantly different between different lines (Supplementary Figure S3). Perhaps there was no m6A modification in their transcription mRNA, and they were regulated by the indirect inhibition of PagALKBH9B and PagALKBH10B, thereby inhibiting the formation of adventitious roots. Plant height was also inhibited by overexpression of PagALKBH10B transgenic lines (Figure 5; Supplementary Figure S4). The result may depend on their regulation of global m6A RNA methylation. Transgenic expression is highly variable and such variation may be the basis for the difference between the expression of PagALKBH9B and PagALKBH10B. Whether ALKBH9B and ALKBH10B have the same target RNA, remains to be determined. Overexpression of m6A RNA methyl transferase (MTA) increased m6A RNA methylation, which could result in a more elaborate root system (Lu et al., 2020). The formation of ARs is a complex process, in which RNA m6A modification sites and their roles should be studied further using high-throughput sequencing.
High salt stress is one of the main abiotic stresses, plants evolved a series of adaptive physiological mechanisms, such as the synthesis of osmotic adjustment substances, the regulation of ion balance in vivo, and the removal of excessive accumulation of reactive oxygen species (Zhao et al., 2020). m6A methylation of RNA has also been shown to be involved in plant response to salt stress. By mapping m6A modification in two sorghum genotypes (salt-tolerant M-81E and salt-sensitive Roma) with different salt tolerance, it is indicated that the quantity and extent of m6A modification in salt-tolerant gene transcripts may be an important factor in salt tolerance (Zheng et al., 2021). Previous studies have confirmed that m6A methyltransferase mutants, including mta, mtb, virilizer, and hakai, display salt-sensitive phenotypes in m6A-dependent manner. VIR-mediated m6A methylation regulates the homeostasis of reactive oxygen species by negatively regulating the mRNA stability of several negative regulatory factors ATAF1, GI, and GSTU17 under salt stress (Hu et al., 2021). In the present study, we found that salt stress induced upregulation of total RNA m6A level, whereas overexpression of PagALKBH9B and PagALKBH10 partially inhibited salt stress-induced m6A level (Figure 6). In addition, salt treatment activated the expression of PagALKBH9B and PagALKBH10B (Supplementary Figure S5). This indicates that m6A methylation in poplar may also be involved in the response to salt stress.
The ALKBH gene family has been shown to be widely involved in salt stress regulation. Under salt stress, the survival rate of alkbh6 mutant was significantly lower than that of wild type (Huong et al., 2020), overexpression of ALKBH8B improved the salt tolerance of transgenic Arabidopsis plants (Huong et al., 2022). In Arabidopsis, ALKBH10B can regulate salt stress resistance and ABA signaling by mediating m6A modification levels of ATAF1, BGLU22, MYB73, ABI3, and ABI4, germination of alkbh10b mutant seeds was significantly delayed and seedling growth and survival rate were enhanced under salt stress (Shoaib et al., 2021). In our present study, we found that overexpression of PagALKBH9B and PagALKBH10B increased salt tolerance (Figure 7A). Further comparison of WT and transgenic lines confirmed that PagALKBH9B and PagALKBH10B could reduce salt stress-induced H2O2 accumulation and oxidative damage by increasing the activities of antioxidant enzyme systems SOD, POD, and CAT, and reduce the damage of salt stress on chloroplasts (Figures 7B–K). The expression abundance of antioxidant enzyme synthesis genes in transgenic lines was higher, however, we did not detect differences in the m6A enrichment level of mRNA between different lines (Supplementary Figure S6), they may not be the direct target genes of PagALKBH9B and PagALKBH10B. Overexpression of PagALKBH9B and PagALKBH10B increased sensitivity to drought stress by inhibiting proline synthesis (Figure 7L; Supplementary Figure S7), although the expression of proline synthesis gene P5C5 decreased, the level of m6A enrichment did not change significantly (Supplementary Figure S6). Discovery of direct target genes for PagALKBH9B and PagALKBH10B may depend on sequencing technologies such as MeRIP-seq.
m6A methyltransferase and demethylase regulatory genes have the same function that has been confirmed in fruit ripening. Mutation of SlALKBH2 (the ALKBH9B and ALKBH10B homologous gene) reduced the mRNA abundance of SlDML2 and delays fruit ripening (Zhou et al., 2019). MTA and MTB can regulate fruit ripening by mediating the mRNA stability and translation efficiency of ABA pathway genes, overexpression of MTA and MTB will promote fruit ripening, while gene silencing of MTA and MTB will delay fruit ripening (Zhou et al., 2021). The reason may be that there is a certain deviation between the known target genes of methyltransferase and demethylase, and no research has yet proved that methyltransferase and demethylase of a target gene were known. In the current study, we found that overexpression of PagALKBH9B and PagALKBH10B could inhibit ARs formation (Figure 5) and improve salt tolerance (Figure 7), which was closely related to their homologous genes of ALKBH5 (Figure 2) and their function of reducing total RNA m6A methylation (Figure 4A). However, the difference in domain determined that their functions have certain specificity, PagALKBH9B was not involved in the regulation of m6A methylation on mRNA (Figure 4B) and had no significant effect on IAA28 expression (Supplementary Figure S3) and plant height (Supplementary Figure S4). In addition, the relationship between them in regulating RNA m6A methylation needs to be further determined. Although we preliminarily verified the similarity between PagALKBH9B and PagALKBH10B in regulating adventitious root formation and salt tolerance, whether the target genes of the two demethylases are consistent needs to be further explored by MeRIP-seq and other technologies.
Conclusion
We identified the PagALKBH family of poplar 84K, analyzed its sequences, and studied its role in growth and development and response to salt stress. Twenty-three PagALKBHs were identified and similar amino acid sequence, gene structure, and conserved domains were determined between homologs. Due to high conservation of PagALKBH, the homologs of poplar 84K can be divided into six groups based on their phylogeny. Expression analysis revealed the tissue specificity for leaves of most of the PagALKBHs, PagALKBH10C was the exception. Overexpression of PagALKBH9B and PagALKBH10B both inhibited adventitious root formation and reduced biomass accumulation of transgenic plants by reducing the m6A RNA methylation, furthermore, PagALKBH10B can also negatively regulate plant height. Overexpression of PagALKBH9B and PagALKBH10B can reduce the accumulation of H2O2 and oxidative damage by increasing the activities of SOD, POD, and CAT, and reducing the damage of salt stress on chloroplasts. But overexpression line reduced the content of proline, thereby increased sensitivity to drought stress. This research helped to better understand the biological functions of ALKBHs and m6A RNA methylation in Populus and suggests new candidate genes and feasible strategies for tree species improvement.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
Author contributions
YZ and YL designed the experiments and wrote the manuscript. YZ, QG, SC, YT, and KH performed the experiments. YZ and YS analyzed the data. JL, QY, and QJ provided guidelines for the research. RS provided advice and amendments to the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Fundamental Research Funds for the Central Universities (2015ZCQ-SW-03), the Major National Science and Technology Projects (2018ZX08020002-003-002), and the National Science Foundation of China (31971675).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.994154/full#supplementary-material
Footnotes
1. ^http://plants.ensembl.org/index.html
2. ^https://phytozome.jgi.doe.gov/pz/portal.html
4. ^http://smart.embl-heidelberg.de/
References
An, X., Gao, K., Chen, Z., Li, J., Yang, X., Yang, X., et al. (2021). High quality haplotype-resolved genome assemblies of Populus tomentosa Carr., a stabilized interspecific hybrid species widespread in Asia. Mol. Ecol. Resour. 22, 786–802. doi: 10.1111/1755-0998.13507
Arribas-Hernández, L., and Brodersen, P. (2020). Occurrence and functions of m6A and other covalent modifications in plant mRNA. Plant Physiol. 182, 79–96. doi: 10.1104/pp.19.01156
Bailey, T. L., Boden, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., et al. (2009). MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208. doi: 10.1093/nar/gkp335
Bauwens, E., Joosten, M., Taganna, J., Rossi, M., Debraekeleer, A., Tay, A., et al. (2018). In silico proteomic and phylogenetic analysis of the outer membrane protein repertoire of gastric helicobacter species. Sci. Rep. 8:15453. doi: 10.1038/s41598-018-32476-1
Bodi, Z., Zhong, S., Mehra, S., Song, J., Graham, N., Li, H., et al. (2012). Adenosine methylation in Arabidopsis mRNA is associated with the 3′ end and reduced levels cause developmental defects. Front. Plant Sci. 3:48. doi: 10.3389/fpls.2012.00048
Bosco, C. D., Lezhneva, L., Biehl, A., Leister, D., Strotmann, H., Wanner, G., et al. (2004). Inactivation of the chloroplast ATP synthase γ subunit results in high non-photochemical fluorescence quenching and altered nuclear gene expression in Arabidopsis thaliana. J. Biol. Chem. 279, 1060–1069. doi: 10.1074/jbc.M308435200
Bradshaw, H. D., Ceulemans, R., Davis, J., and Stettler, R. (2000). Emerging model systems in plant biology: poplar (Populus) as a model forest tree. J. Plant Growth Regul. 19, 306–313. doi: 10.1007/s003440000030
Cai, Y., Wu, G., Peng, B., Li, J., Zeng, S., Yan, Y., et al. (2021). Expression and molecular profiles of the AlkB family in ovarian serous carcinoma. Aging 13, 9679–9692. doi: 10.18632/aging.202716
Chen, B., Ye, F., Yu, L., Jia, G., Huang, X., Zhang, X., et al. (2012). Development of cell-active N6-Methyladenosine RNA demethylase FTO inhibitor. J. Am. Chem. Soc. 134, 17963–17971. doi: 10.1021/ja3064149
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, L., Shen, Y., Yang, W., Pan, Q., Li, C., Sun, Q., et al. (2022). Comparative metabolic study of two contrasting chinese cabbage genotypes under mild and severe drought stress. Int. J. Mol. Sci. 23:5947. doi: 10.3390/ijms23115947
Chen, L., Wang, P., Bahal, R., Manautou, J. E., and Zhong, X. (2019). Ontogenic mRNA expression of RNA modification writers, erasers, and readers in mouse liver. PLoS One 14:e227102. doi: 10.1371/journal.pone.0227102
Chen, M., Urs, M. J., Sánchez-González, I., Olayioye, M. A., Herde, M., and Witte, C. P. (2018a). M6A RNA degradation products are catabolized by an evolutionarily conserved N6-methyl-AMP deaminase in plant and mammalian cells. Plant Cell 30, 1511–1522. doi: 10.1105/tpc.18.00236
Chen, M., Wei, L., Law, C., Tsang, F. H., Shen, J., Cheng, C. L.-H., et al. (2018b). RNA N6-Methyladenosine Methyltransferase-like 3 promotes liver cancer progression through YTHDF2- dependent posttranscriptional silencing of SOCS2. Hepatology 6, 2254–2270. doi: 10.1002/hep.29683/suppinfo
Cheng, S., Jia, K., Liu, H., Zhang, R., Li, Z., Zhou, S., et al. (2021). Haplotype-resolved genome assembly and allele-specific gene expression in cultivated ginger. Hortic. Res. 8:188. doi: 10.1038/s41438-021-00599-8
Dominissini, D., Moshitch-Moshkovitz, S., Schwartz, S., Salmon-Divon, M., Ungar, L., Osenberg, S., et al. (2012). Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485, 201–206. doi: 10.1038/nature11112
Du, N., Liu, X., Li, Y., Chen, S., Zhang, J., Ha, D., et al. (2012). Genetic transformation of Populus tomentosa to improve salt tolerance. Plant Cell Tissue Organ Cult. 108, 181–189. doi: 10.1007/s11240-011-0026-4
Duan, H., Wei, L., Zhang, C., Wang, Y., Chen, L., Lu, Z., et al. (2018). ALKBH10B is an RNA N6-Methyladenosine demethylase affecting Arabidopsis floral transition. Plant Cell 29, 2995–3011. doi: 10.1105/tpc.16.00912
Endres, L., Begley, U., Clark, R., Gu, C., Dziergowska, A., Małkiewicz, A., et al. (2015). Alkbh8 regulates Selenocysteine-protein expression to protect against reactive oxygen species damage. PLoS One 10:e131335. doi: 10.1371/journal.pone.0131335
Falnes, P. O., Johansen, R. F., and Seeberg, E. (2002). AlkB-mediated oxidative demethylation reverses DNA damage in Escherichia coli. Nature 419, 178–182. doi: 10.1038/nature01048
Fedeles, B. I., Singh, V., Delaney, J. C., Li, D., and Essigmann, J. M. (2015). The AlkB family of Fe (II)/α-Ketoglutarate-dependent dioxygenases: repairing nucleic acid alkylation damage and beyond. J. Biol. Chem. 290, 20734–20742. doi: 10.1074/jbc.R115.656462
Gascuel, O. (2006). Neighbor-joining revealed. Mol. Biol. Evol. 23, 1997–2000. doi: 10.1093/molbev/msl072
Hampton, C. R., Bowen, H. C., Broadley, M. R., Hammond, J. P., Mead, A., Payne, K. A., et al. (2004). Cesium toxicity in Arabidopsis. Plant Physiol. 136, 3824–3837. doi: 10.1104/pp.104.046672
Hatfield, J. L., and Prueger, J. H. (2015). Temperature extremes: effect on plant growth and development. Weather Clim. Extremes 10, 4–10. doi: 10.1016/j.wace.2015.08.001i
Hofmeister, B. T., Denkena, J., Colomé-Tatché, M., Shahryary, Y., Hazarika, R., Grimwood, J., et al. (2020). A genome assembly and the somatic genetic and epigenetic mutation rate in a wild long-lived perennial Populus trichocarpa. Genome Biol. 21:259. doi: 10.1186/s13059-020-02162-5
Hu, J., Cai, J., Park, S. J., Lee, K., Li, Y., Chen, Y., et al. (2021). N6-Methyladenosine mRNA methylation is important for salt stress tolerance in Arabidopsis. Plant J. 106, 1759–1775. doi: 10.1111/tpj.15270
Huong, T. T., Ngoc, L. N. T., and Kang, H. (2020). Functional characterization of a putative RNA demethylase ALKBH6 in Arabidopsis growth and abiotic stress responses. Int. J. Mol. Sci. 21:6707. doi: 10.3390/ijms21186707
Huong, T. T., Yang, Z., Ngoc, L. N. T., and Kang, H. (2022). ALKBH8B, a putative RNA demethylase, plays a role in the response of arabidopsis to salt stress and abscisic acid. J. Plant Biol. 65, 319–330. doi: 10.1007/s12374-022-09351-8
Janz, D., and Polle, A. (2012). Harnessing salt for woody biomass production. Tree Physiol. 32, 1–3. doi: 10.1093/treephys/tpr127
Jia, G., Fu, Y., Zhao, X., Dai, Q., Zheng, G., Yang, Y., et al. (2011). N6-Methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 7, 885–887. doi: 10.1038/nchembio.687
Kataoka, H., Yamamoto, Y., and Sekiguchi, M. (1983). A new gene (AlkB) of Escherichia coli that controls sensitivity to methyl methane sulfonate. J. Bacteriol. 153, 1301–1307. doi: 10.1128/jb.153.3.1301-1307.1983
Kawarada, L., Suzuki, T., Ohira, T., Hirata, S., Miyauchi, K., and Suzuki, T. (2017). ALKBH1 is an RNA dioxygenase responsible for cytoplasmic and mitochondrial tRNA modifications. Nucleic Acids Res. 45, 7401–7415. doi: 10.1093/nar/gkx354
Kierzek, E. (2003). The thermodynamic stability of RNA duplexes and hairpins containing N6-alkyladenosines and 2-methylthio-N6-alkyladenosines. Nucleic Acids Res. 31, 4472–4480. doi: 10.1093/nar/gkg633
Kim, J. I., Sharkhuu, A., Jin, J. B., Li, P., Jeong, J. C., Baek, D., et al. (2007). yucca6, a dominant mutation in Arabidopsis, affects auxin accumulation and auxin-related phenotypes. Plant Physiol. 145, 722–735. doi: 10.1104/pp.107.104935
Krzywinski, M., Schein, J., Birol, I., Connors, J., Gascoyne, R., Horsman, D., et al. (2009). Circos: an information aesthetic for comparative genomics. Genome Res. 19, 1639–1645. doi: 10.1101/gr.092759.109
Kubala, S., Wojtyla, Ł., Quinet, M., Lechowska, K., Lutts, S., and Garnczarska, M. (2015). Enhanced expression of the proline synthesis gene P5CSA in relation to seed osmopriming improvement of Brassica napus germination under salinity stress. J. Plant Physiol. 183, 1–12. doi: 10.1016/j.jplph.2015.04.009
Kurowski, M. A., Bhagwat, A. S., Papaj, G., and Bujnicki, J. M. (2003). Phylogenomic identification of five new human homologs of the DNA repair enzyme AlkB. BMC Genomics 4:48. doi: 10.1186/1471-2164-4-48
Leihne, V., Kirpekar, F., Vågbø, C. B., van den Born, E., Krokan, H. E., Grini, P. E., et al. (2011). Roles of Trm9- and ALKBH8-like proteins in the formation of modified wobble uridines in Arabidopsis tRNA. Nucleic Acids Res. 39, 7688–7701. doi: 10.1093/nar/gkr406
Letunic, I., Doerks, T., and Bork, P. (2009). SMART 6: recent updates and new developments. Nucleic Acids Res. 37, D229–D232. doi: 10.1093/nar/gkn808
Li, S. (2021). Molecular bases for the regulation of adventitious root generation in plants. Front. Plant Sci. 12:614072. doi: 10.3389/fpls.2021.614072
Liang, Z., Riaz, A., Chachar, S., Ding, Y., Du, H., and Gu, X. (2020). Epigenetic modifications of mRNA and DNA in plants. Mol. Plant 13, 14–30. doi: 10.1016/j.molp.2019.12.007
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.126
Lu, L., Zhang, Y., He, Q., Qi, Z., Zhang, G., Xu, W., et al. (2020). MTA, an RNA m6A methyltransferase, enhances drought tolerance by regulating the development of trichomes and roots in poplar. Int. J. Mol. Sci. 21:2462. doi: 10.3390/ijms21072462
Ma, S., Gong, Q., and Bohnert, H. J. (2006). Dissecting salt stress pathways. J. Exp. Bot. 57, 1097–1107. doi: 10.1093/jxb/erj098
Ma, T., Wang, J., Zhou, G., Yue, Z., Hu, Q., Chen, Y., et al. (2014). Erratum: genomic insights into salt adaptation in a desert poplar. Nat. Commun. 5:3454. doi: 10.1038/ncomms4454
Marcinkowski, M., Pilžys, T., Garbicz, D., Steciuk, J., Zugaj, D., Mielecki, D., et al. (2020). Human and Arabidopsis alpha-ketoglutarate-dependent dioxygenase homolog proteins-new players in important regulatory processes. IUBMB Life 72, 1126–1144. doi: 10.1002/iub.2276
Martínez-Pérez, M., Aparicio, F., López-Gresa, M. P., Bellés, J. M., Sánchez-Navarro, J. A., and Pallás, V. (2017). Arabidopsis m6A demethylase activity modulates viral infection of a plant virus and the m6A abundance in its genomic RNAs. Proc. Natl. Acad. Sci. U. S. A. 114, 10755–10760. doi: 10.1073/pnas.1703139114
Mauer, J., Luo, X., Blanjoie, A., Jiao, X., Grozhik, A. V., Patil, D. P., et al. (2017). Reversible methylation of m6Am in the 5′ cap controls mRNA stability. Nature 541, 371–375. doi: 10.1038/nature21022
Merret, R., Nagarajan, V. K., Carpentier, M., Park, S., Favory, J., Descombin, J., et al. (2015). Heat-induced ribosome pausing triggers mRNA co-translational decay in Arabidopsis thaliana. Nucleic Acids Res. 43, 4121–4132. doi: 10.1093/nar/gkv234
Nelson, D. C., Flematti, G. R., Riseborough, J. A., Ghisalberti, E. L., Dixon, K. W., and Smith, S. M. (2010). Karrikins enhance light responses during germination and seedling development in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U. S. A. 107, 7095–7100. doi: 10.1073/pnas.0911635107
Nelson, D. C., Riseborough, J., Flematti, G. R., Stevens, J., Ghisalberti, E. L., Dixon, J., et al. (2009). Karrikins discovered in smoke trigger Arabidopsis seed germination by a mechanism requiring gibberellic acid synthesis and light. Plant Physiol. 149, 863–873. doi: 10.1104/pp.108.131516
Nong, X., Zhong, S., Li, S., Yang, Y., Liang, Z., and Xie, Y. (2019). Genetic differentiation of Pseudoregma bambucicola population based on mtDNA COII gene. Saudi J. Biol. Sci. 26, 1032–1036. doi: 10.1016/j.sjbs.2019.04.016
Ougland, R., Rognes, T., Klungland, A., and Larsen, E. (2015). Non-homologous functions of the AlkB homologs. J. Mol. Cell Biol. 7, 494–504. doi: 10.1093/jmcb/mjv029
Qiu, D., Bai, S., Ma, J., Zhang, L., Shao, F., Zhang, K., et al. (2019). The genome of Populus alba x Populus tremula var. Glandulosa clone 84K. DNA Res. 26, 423–431. doi: 10.1093/dnares/dsz020
Ra, M. A., Zhang, M., Campilho, A., Bodi, Z., Kashif, M., Saleh, M., et al. (2017). Identification of factors required for m6A mRNA methylation in Arabidopsis reveals a role for the conserved E3 ubiquitin ligase HAKAI. New Phytol. 215, 157–172. doi: 10.1111/nph.14586
Schlicker, A., Domingues, F. S., Rahnenführer, J., and Lengauer, T. (2006). A new measure for functional similarity of gene products based on gene ontology. BMC Bioinf. 7:302. doi: 10.1186/1471-2105-7-302
Shao, L., Xing, F., Xu, C., Zhang, Q., Che, J., Wang, X., et al. (2019). Patterns of genome-wide allele-specific expression in hybrid rice and the implications on the genetic basis of heterosis. Proc. Natl. Acad. Sci. U. S. A. 116, 5653–5658. doi: 10.1073/pnas.1820513116
Shoaib, Y., Hu, J., Manduzio, S., and Kang, H. (2021). Alpha-ketoglutarate-dependent dioxygenase homolog 10B, an N6-methyladenosine mRNA demethylase, plays a role in salt stress and abscisic acid responses in Arabidopsis thaliana. Physiol. Plant. 173, 1078–1089. doi: 10.1111/ppl.13505
Sorin, C., Negroni, L., Balliau, T., Corti, H., Jacquemot, M., Davanture, M., et al. (2006). Proteomic analysis of different mutant genotypes of Arabidopsis led to the identification of 11 proteins correlating with adventitious root development. Plant Physiol. 140, 349–364. doi: 10.1104/pp.105.067868
Sukumar, P., Maloney, G. S., and Muday, G. K. (2013). Localized induction of the ATP-binding cassette b19 auxin transporter enhances adventitious root formation in Arabidopsis. Plant Physiol. 162, 1392–1405. doi: 10.1104/pp.113.217174
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Tang, C., Klukovich, R., Peng, H., Wang, Z., Yu, T., Zhang, Y., et al. (2018). ALKBH5-dependent m6A demethylation controls splicing and stability of long 3’-UTR mRNAs in male germ cells. Proc. Natl. Acad. Sci. U. S. A. 115, E325–E333. doi: 10.1073/pnas.1717794115
Tang, Y., Yuan, Y., Shu, S., and Guo, S. (2018). Regulatory mechanism of NaCl stress on photosynthesis and antioxidant capacity mediated by transglutaminase in cucumber (Cucumis sativus L.) seedlings. Sci. Hortic. 235, 294–306. doi: 10.1016/j.scienta.2018.02.045
Tester, M., and Davenport, R. (2003). Na+ tolerance and Na+ transport in higher plants. Ann. Bot. 91, 503–527. doi: 10.1093/aob/mcg058
Trewick, S. C., Henshaw, T. F., Hausinger, R. P., Lindahl, T., and Sedgwick, B. (2002). Oxidative demethylation by Escherichia coli AlkB directly reverts DNA base damage. Nature 419, 174–178. doi: 10.1038/nature00908
Tuskan, G. A., DiFazio, S., Jansson, S., Bohlmann, J., Grigoriev, I., Hellsten, U., et al. (2006). The genome of black cottonwood, Populus trichocarpa (Torr & gray). Science 313, 1596–1604. doi: 10.1126/science.1128691
Vespa, L., Vachon, G., Berger, F., Perazza, D., Faure, J., and Herzog, M. (2004). The Immunophilin-interacting protein AtFIP37 from Arabidopsis is essential for plant development and is involved in trichome endoreduplication. Plant Physiol. 134, 1283–1292. doi: 10.1104/pp.103.028050
Wakisaka, K. T., Muraoka, Y., Shimizu, J., Yamaguchi, M., Ueoka, I., Mizuta, I., et al. (2019). Drosophila alpha-ketoglutarate-dependent dioxygenase AlkB is involved in repair from neuronal disorders induced by ultraviolet damage. Neuroreport 30, 1039–1047. doi: 10.1097/WNR.0000000000001323
Wang, Y., Tang, H., DeBarry, J. D., Tan, X., Li, J., Wang, X., et al. (2012). MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40:e49. doi: 10.1093/nar/gkr1293
Wang, L. Q., Wen, S. S., Wang, R., Wang, C., Gao, B., and Lu, M. Z. (2021). PagWOX11/12a activates PagCYP736A12 gene that facilitates salt tolerance in poplar. Plant Biotechnol. J. 19, 2249–2260. doi: 10.1111/pbi.13653
Wang, X., Zhao, B. S., Roundtree, I. A., Lu, Z., Han, D., Ma, H., et al. (2015). N6-methyladenosine modulates messenger RNA translation efficiency. Cells 161, 1388–1399. doi: 10.1016/j.cell.2015.05.014
Wei, Y. F., Carter, K. C., Wang, R. P., and Shell, B. K. (1996). Molecular cloning and functional analysis of a human cDNA encoding an Escherichia coli AlkB homolog, a protein involved in DNA alkylation damage repair. Nucleic Acids Res. 24, 931–937. doi: 10.1093/nar/24.5.931
Xu, B., Liu, D., Wang, Z., Tian, R., and Zuo, Y. (2020). Multi-substrate selectivity based on key loops and non-homologous domains: new insight into ALKBH family. Cell. Mol. Life Sci. 78, 129–141. doi: 10.1007/s00018-020-03594-9
Yi, C., Yang, C., and He, C. (2009). A non-Heme iron-mediated chemical demethylation in DNA and RNA. Acc. Chem. Res. 42, 519–529. doi: 10.1021/ar800178j
Yu, C., Xu, S., and Yin, Y. (2016). Transcriptome analysis of the Taxodium ‘Zhongshanshan 405’ roots in response to salinity stress. Plant Physiol. Biochem. 100, 156–165. doi: 10.1016/j.plaphy.2016.01.009
Yu, Z., Duan, X., Luo, L., Dai, S., Ding, Z., and Xia, G. (2020). How plant hormones mediate salt stress responses. Trends Plant Sci. 25, 1117–1130. doi: 10.1016/j.tplants.2020.06.008
Yue, H., Nie, X., Yan, Z., and Weining, S. (2019). N6-methyladenosine regulatory machinery in plants: composition, function and evolution. Plant Biotechnol. J. 17, 1194–1208. doi: 10.1111/pbi.13149
Zhang, H., Gao, S., Lercher, M. J., Hu, S., and Chen, W. (2012). EvolView, an online tool for visualizing, annotating and managing phylogenetic trees. Nucleic Acids Res. 40, W569–W572. doi: 10.1093/nar/gks576
Zhao, Y., Xin, D., Lu, W., Zong, X., Niu, Y., Guo, X., et al. (2022). PeMPK7 is induced in an ROS-dependent manner and confers poplar Para-hydroxybenzoic acid stress resistance through the removal of ROS. Ind. Crop. Prod. 182:114861. doi: 10.1016/j.indcrop.2022.114861
Zhao, C., Zhang, H., Song, C., Zhu, J. K., and Shabala, S. (2020). Mechanisms of plant responses and adaptation to soil salinity. Innovations 1:100017. doi: 10.1016/j.xinn.2020.100017
Zheng, H., Sun, X., Li, J., Song, Y., Song, J., Wang, F., et al. (2021). Analysis of N6-methyladenosine reveals a new important mechanism regulating the salt tolerance of sweet sorghum. Plant Sci. 304:110801. doi: 10.1016/j.plantsci.2020.110801
Zhong, S., Li, H., Bodi, Z., Button, J., Vespa, L., Herzog, M., et al. (2008). MTA is an Arabidopsis messenger RNA adenosine methylase and interacts with a homolog of a sex-specific splicing factor. Plant Cell 20, 1278–1288. doi: 10.1105/tpc.108.058883
Zhou, L., Tang, R., Li, X., Tian, S., Li, B., and Qin, G. (2021). N6-methyladenosine RNA modification regulates strawberry fruit ripening in an ABA-dependent manner. Genome Biol. 22:168. doi: 10.1186/s13059-021-02385-0
Keywords: AlkB homologs, poplar 84K, m6A RNA methylation, demethylase, salt
Citation: Zhao Y, Guo Q, Cao S, Tian Y, Han K, Sun Y, Li J, Yang Q, Ji Q, Sederoff R and Li Y (2022) Genome-wide identification of the AlkB homologs gene family, PagALKBH9B and PagALKBH10B regulated salt stress response in Populus. Front. Plant Sci. 13:994154. doi: 10.3389/fpls.2022.994154
Edited by:
Binying Fu, Institute of Crop Sciences (CAAS), ChinaReviewed by:
Hunseung Kang, Chonnam National University, South KoreaLei Zhang, Chinese Academy of Forestry, China
Copyright © 2022 Zhao, Guo, Cao, Tian, Han, Sun, Li, Yang, Ji, Sederoff and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yun Li, eXVubGlAYmpmdS5lZHUuY24=
†ORCID: Yun Li http://orcid.org/0000-0002-8426-565X
 Ye Zhao
Ye Zhao Qi Guo1
Qi Guo1 Sen Cao
Sen Cao Yanting Tian
Yanting Tian Ronald Sederoff
Ronald Sederoff Yun Li
Yun Li