- 1Molecular and Microbiology Department, Sandia National Laboratories, Albuquerque, NM, United States
- 2Bioscience Division, Los Alamos National Laboratory, Los Alamos, NM, United States
- 3Forest Products Laboratory, Forest Biopolymer Science and Engineering, U.S. Forest Service, Madison, WI, United States
- 4Computational Biology and Biophysics Department, Sandia National Laboratories, Albuquerque, NM, United States
The potential benefits of adding raw, non-food, lignocellulosic plant material as a carbon source for mixotrophic growth of microalgae have previously been demonstrated. This approach has advantages over using traditional carbon sources like glucose or acetate due to wide-spread plant biomass availability and substrate recalcitrance to bacterial contamination. Here, we report the overall growth characteristics and explore the metabolic patterns of Scenedesmus obliquus cultured in the presence raw plant substrate. An initial screen of plant substrate candidates showed an increase in specific growth rate and biomass accumulation when S. obliquus was cultured in the presence of switchgrass or yard waste compared to media alone. We observed a near doubling of microalgal dry weight when S. obliquus was grown with 0.2% (w/v) switchgrass under ambient CO2. Scanning electron microscopy (SEM) of corn stem after S. obliquus cultivation exhibited substantial phloem degradation. Transcriptomic analyses of S. obliquus during mid- and late-log phase growth revealed a dynamic metabolic landscape within many KEGG pathways. Notably, differential expression was observed for several potential glycosyl hydrolases. We also investigated the influence of switchgrass on the growth of S. obliquus at 50 L volume in mini raceway ponds to determine the scalability of this approach.
1 Introduction
Select unicellular microalgae can utilize different trophic modes for metabolism and can switch between these modes in response to changing environmental conditions. Growth characteristics of microalgae have been described under photoautotrophic, chemoheterotrophic, and mixotrophic modes, particularly in the context of trying to improve growth rates in industrial applications. Mixotrophy in microalgae has often been described as a synergistic combination of autotrophy and heterotrophy and has been reported to enhance growth rates and biomass accumulation when compared to either trophic mode (Smith et al., 2015; Zhang et al., 2017; Abiusi et al., 2020). The hypothesis for this is that common hinderances experienced in strictly heterotrophic or photoautotrophic cultivations can be overcome by leveraging two energy sources within one cell (Wang et al., 2014; Abiusi et al., 2020). For example, in photoautotrophic cultivation, light availability can be the main growth limiting factor, but with supplementation of an organic carbon source this can be alleviated (Wang et al., 2014; Abiusi et al., 2020).
We have previously investigated mixotrophy of microalgal strains by adding raw, non-food, plant material to microalgal cultures as carbon source (Vogler et al., 2018; Schambach et al., 2020). Not only is this lignocellulosic biomass the most abundant biomaterial in the biosphere (Malhi, 2002), it is inexpensive compared to substrates like glucose, and recalcitrant to bacterial contamination in microalgal cultures; important aspects when considering industrial applicability. Plant substrate utilization has been observed in the freshwater green alga Auxenochlorella protothecoides and in two species of Nannochloropsis (Vogler et al., 2018; Schambach et al., 2020). These strains exhibited increased specific growth rates and biomass accumulation in the presence of switchgrass or corn stover. Glycome profiling of switchgrass after addition to A. protothecoides cultures compared to switchgrass in media alone demonstrated the utilization of the hemicellulose xyloglucan by the alga (Vogler et al., 2018). Moreover, differential expression analysis revealed enzymes likely involved in degradation of lignocellulose, including family 5 and 9 glycosyl hydrolases which catalyze hydrolysis of glycosidic linkages in cellulose (Vogler et al., 2018). SEM analysis and sugar analysis of corn stover revealed signs of phloem degradation and glucan utilization, respectively, by Nannochloropsis gaditana (Schambach et al., 2020).
Here, we examined potential mixotrophic growth of S. obliquus in the presence of plant substrate. Scenedesmus obliquus, a freshwater chlorophyte known for its robust overall growth, has been reported to grow mixotrophically using commonly investigated carbon sources like glucose or acetate (Mandal and Mallick, 2009; Shen et al., 2018; Song et al., 2021). This strain’s high tolerance to a wide range of environmental conditions suggests it is metabolically flexible (Msanne et al., 2020), making it an ideal strain to investigate utilization of lignocellulosic biomass. Through transcriptomic analysis we aimed to describe the molecular mechanisms, particularly regarding glycosyl hydrolase expression, employed by this strain to potentially degrade and utilize switchgrass. We also examine the overarching energy metabolism of S. obliquus based on differential expression of enzymes involved in pathways like oxidative phosphorylation, carbon fixation, and photosynthesis. In addition, we investigated the effects of increased CO2 concentration and scaling up to 50 L volume in mini-raceway ponds on plant substrate utilization.
2 Materials and methods
2.1 Cultivation conditions and initial plant substrate screen
Scenedesmus obliquus DOE0152z (S. obliquus) was obtained from Heliae Development, LLC. Stock cultures were grown in BG-11 media and maintained in exponential phase growth prior to the start of each experiment. The following BG-11 media composition was used: 17.6 mM NaNO3; 0.23 mM K2HPO4; 0.3 mM MgSO4∙7H2O; 0.24 mM CaCl2∙2H2O; 0.031 mM Citric Acid∙H2O; 0.021 mM Ferric Ammonium Citrate; 0.0027 mM Na2EDTA2∙H2O; 0.19 mM Na2CO3; 46 mM H3BO3; 9 mM MnCl2∙4H2O; 0.77 mM ZnSO4; 1.6 mM Na2MoO4; 0.3 mM CuSO4∙5H2O; 0.17 mM Co(NO3)2∙6H2O. Media was filter sterilized using 0.2 µm filters.
For all flask experiments the following conditions were used unless otherwise stated. S. obliquus cultures were grown in a Percival growth chamber (Percival Scientific, Perry, IA) in 100 mL volumes in 250 mL Erlenmeyer flasks and mixed via an orbital shaker (Benchmark Scientific, Sayreville, NJ) at 120 rpm. The light intensity was 100 µmol m-2 s-1 with a 12:12 h light/dark cycle. The temperature was maintained at 25°C. For each experiment three biological replicates were used for each treatment. The cell density of each inoculum was approximately 1-2 x 106 cells mL-1. Growth was monitored by manual cell counts of free-floating cells using a hemocytometer. Note this allowed for differentiation between microalgal cells and microscopic pieces of plant substrate. Cultures were grown until they reached early stationary phase (i.e., when cell counts began to plateau). Plant substrates were separated from microalgal cells using a 40µm Corning nylon cell strainer. Specific growth rates were calculated for log phase of growth using Equation 1:
where N1 and N2 are the cell counts (in cells mL-1) at time 1 (t1) and time 2 (t2), respectively. To determine significant differences between growth rates we employed analysis of variance and least-squares means for multiple comparisons tests with a Tukey p-value adjustment. Data was plotted using R: A Language and Environment for Statistical Computing (https://www.R-project.org).
S. obliquus was inoculated into flasks with corn stover (CS; Zea mays), sugarcane bagasse (SB; Sacharum officinarum), switchgrass (SG; Panicum virgatum), and yard waste (YW) at a concentration of 0.1% weight by volume (w/v) as an initial screen for plant substrate utilization by S. obliquus compared to a control of the microalga alone. Specific growth rates were calculated from day 3 to 12. CS, SB, and SG were provided courtesy of Idaho National Laboratory (INL) Bioenergy Feedstock Library. CS comprises the stalks, leaves, leaf sheaths, tassels, cobs, and cob stems of the corn plant that are left on the field after the corn is harvested. CS was collected by multi-pass harvesting and ground to pass through a 2-in. sieve using a Vermeer BG480 grinder followed by a 1-in. sieve using a Bliss Hammermill (INL Bioenergy Feedstock Library). SB was processed to pass through a 6-in. screen (INL Bioenergy Feedstock Library). SG was ground to pass through a 1-in. sieve using a Vemeer BG480 grinder (INL Bioenergy Feedstock Library). YW was composed mostly of fresh monoculture grass clippings from an untreated lawn in Los Alamos, NM. These substrates were used “raw”, i.e., added to the flask in the format provided by INL (e.g., dried and milled) with no pretreatment other than autoclaving at 15 psi and 121°C for 20 minutes. Based on the results from this experiment reported below, SG was chosen as the primary substrate for further experimentation. We investigated the effect of plant substrate concentration on S. obliquus growth by comparing three concentrations of SG: 0.2%, 0.3%, and 0.4% w/v. Specific growth rates were calculated from day 1 to 12.
To investigate the molecular mechanisms as well as potential effects elevated CO2 may have on substrate utilization, cultures were first grown with and without 0.2% SG in ambient air (0.04% CO2) in the growth chamber. Then, the same stock culture was acclimated to 1% CO2 and the experiment was repeated. The cell density for each inoculum was approximately 5 x 106 cells mL-1. All other cultivation conditions were the same as described above. Specific growth rates were calculated from day 0 to 31 for ambient CO2, and day 0 to 19 for 1% CO2. To determine significant differences between cell density with switchgrass versus without at each timepoint we used the Welch two sample t-test. Samples of microalgal biomass were taken for transcriptomic analysis on day 14 and 35 in air, and day 7 and 21 in 1% CO2 representing mid- and late-log phase growth, respectively. These samples contained 4-9 x 108 cells mL-1, and were immediately pelleted, flash frozen in liquid nitrogen, and stored at -80°C until RNA could be extracted.
2.2 Pond cultivation
To determine how results at laboratory scale translate to a large volume cultivation, S. obliquus was grown with and without 0.2% w/v SG at 50 L volume in 100 L mini-raceway ponds (MicroBio Engineering, San Luis Obispo, CA) in a greenhouse. We had a total of six ponds for three biological replicates each. Each pond had a cylindrical bubble diffuser and was constantly aerated with ambient air (0.04% CO2) at 400 L min-1. CO2 was supplied on demand when pH exceeded 8.1 and turned off when pH fell below 8.0. The paddlewheel speed was 40 rpm. SG and media components for pond cultivation were not autoclaved. The greenhouse had automated light and temperature control. Greenhouse lights would turn on when outdoor light intensity fell below 300 W m-2 between the hours of 6 am and 9 pm. Samples were taken for cell counts, dry weight, and 16S amplicon sequencing. Specific growth rates were calculated from day 1 to 6.
2.3 Light and scanning electron microscopy
To observe physical degradation of the plant substrate when the alga was present, stem sections of SG and corn, at a total concentration of 0.2% w/v, were added to culture with S. obliquus or BG-11 media alone for subsequent SEM and light-microscopy imaging. Treated and untreated stems of a similar developmental stage were compared by microscopy. Because individual SG plants have stems at different degrees of maturation, corn was included due to more easily identifiable morphology compared to switchgrass. The observed surfaces of stem sections were prepared prior to the inoculation experiment according to the following procedure. Fresh stems of similar ontogenic stages, approximately 10 mm (corn) and 5 mm (SG) in diameter were selected. These were cut with a razor blade to 1-µm-thick cross sections and autoclaved before inoculation with algae as described earlier. After harvesting algal cells, plant cross sections were preserved in 1:1:1 glycerol:ethanol:water (Kitin et al., 2020) until they were processed for imaging.
For light microscopy imaging, the sections were stained with acridine orange according to Houtman et al., 2016 and observed by epifluorescence microscopy using a UV (365 nm) excitation and 435 nm long-pass fluorescence, which yielded orange-red fluorescence from unlignified cellulosic walls and green fluorescence from lignified portions of walls (Houtman et al., 2016; Kitin et al., 2020). Some sections were observed by confocal microscopy (LSM, Carl Zeiss 710) using objective lens C-Apochromat 40x/1.20 W Korr, and three excitation/emission channels: 405/460-480; 488/502-562; and 488/580-700.
For SEM imaging, sections were repeatedly rinsed in ethanol series (50%, 25%, 0%), frozen in liquid N2, freeze-dried, sputter-coated with gold, and observed at 10 kV. We observed more than 100 vascular bundles with microalgae and more than 100 vascular bundles without microalgae by both SEM and fluorescence microscopy.
2.4 DNA extraction and 16S amplicon sequencing
DNA was extracted from pond samples during early-log (1 day after inoculation) and late-log (7 days after inoculation) phase growth for 16S amplicon sequencing. DNA was extracted using the Zymo Research Quick-DNA Fungal/Bacterial Miniprep Kit (Zymo Research, Irvine, CA). Samples were homogenized in the ZR BashingBead Lysis Tubes (0.1- and 0.5-mm beads) using the Biospec Mini-Beadbeater-1 set to 4800 oscillations per minute for 30 seconds. Each sample was homogenized three times and rested on ice between runs. The remaining purification procedure followed the manufacturer’s instructions.
Purified DNA was amplified using the Illumina 16S protocol that includes primer pair sequences for the variable V3 and V4 regions with Nextera XT v2 adapters (Klindworth et al., 2013). Sequencing libraries were constructed and sequenced using an Illumina MiSeq (Illumina Inc., San Diego CA) resulting in DNA fragments of 600 pb. Pair-end sequencing was performed using Illumina MiSeq protocol based on instructions specified by the manufacturer resulting in 301 paired end reads. Read analysis was conducted in DADA2 software version 3.9 (Callahan et al., 2016). Pair-end sequencing data corresponding to ribosomal RNA amplicons were merged into contigs and resulting fragments were filtered by quality in non-overlapping regions as having no more than five sites with a Phred-value <= 30. Filtered paired-end sequences were clustered, merged and identified taxonomically using the SILVA 138.1 prokaryotic small subunit taxonomic database formatted specifically for DADA2 (http://arb-silva.de). The amplicon sequence variant (ASV) abundance and number of reads per ASV were integrated into a single table. The V3-V4 sequences with an average length of 200 base pairs were clustered into 569 species-level ASVs from V3-V4 sequences. Species represented by singleton ASVs were removed from the final analysis. A database containing the taxonomic assignment, the ASV table, and sample information was generated using the R package phyloseq (McMurdie and Holmes, 2013). Species represented by singleton ASVs were removed from the final analysis. Normalization and rarefying (less than 1,000 ASV sequences) of the dataset was performed before calculating the proportion of specific species present within each sample.
2.5 RNA extraction and RNA-seq
RNA was extracted using a hybrid method that combined TRIzol and bead beating for homogenization, followed by purification using Omega Bio-tek E.Z.N.A. Plant RNA Kit (Omega Bio-tek Inc., Norcross, GA). Frozen cell pellets were resuspended in 750 mL of TRIzol. The samples were transferred to 2.0 mL Precellys screw cap tubes containing 200 µL of a 1:1 mixture of 0.1 mm silicon carbide and 0.1 mm zirconium beads. The tubes were vortexed to ensure the beads didn’t stick to the bottom of the tube. The Precellys 24 bead beater (Bertin Instruments, Montigny-le-Bretonneux, France) was placed in a cold room and allowed to equilibrate. We used the programmed setting for 2 rounds of bead beating for 30 s at 6000 rpm, followed by a 2 min cooling time. This step was repeated 3-6 times. Homogenization progress was checked by loading a 2 µL aliquot onto a slide and observing lysis under a light microscope. Once it was determined that samples were sufficiently lysed the samples rested for 3 minutes at room temperature and were then transferred to a new 2.0 mL tube carefully avoiding transfer of the beads. 200 µL of chloroform was added and briefly vortexed to mix. The samples were then transferred to 5PRIME Phase Lock Gel Heavy tubes (Quantbio, Beverly, MA). They rested on ice for 15 minutes, inverted by hand every 5 minutes. Phases were separated by centrifugation at 17,000 x g for 20 minutes at 0-4°C. Samples rested at room temperature for 2 minutes after centrifugation. 80% of the top aqueous phase was transferred to a new tube. Equal volumes of 70% ethanol were added and vortexed for 20 seconds to mix. Purification of the RNA continued with step 7 of the Omega Bio-tek E.Z.N.A. Plant RNA kit. Note that during the wash steps we waited 1-2 minutes after adding the wash to the column to improve nucleic acid purity and waited 2 minutes after adding water to the column to improve yield during elution.
RNA was polyA selected using the Illumina Truseq RNA Library Prep kit (V2) according to the manufacturer’s instructions and sequenced on an Illumina NextSeq 500 (Illumina Inc., San Diego, CA). Raw read data (fastq) were aligned to the Scenedesmus obliquus DOE0152Z genome (Joint Genome Institute (JGI), retrieved January 2021). Alignments were performed using STAR v2.7.1a according to encode standard parameters with one exception (Dobin et al., 2013). Specifically, STAR was implemented using the basic two-pass mode to identify novel splice junctions/events given the genomic resources are draft-level and quant mode was used to directly produce gene-level counts data. All gene counts were imported into the R Bioconductor package, EdgeR (Robinson et al., 2010) and trimmed mean of M-values (TMM) normalization size factors were calculated to adjust for sample-specific differences. The TMM size factors and the matrix of counts were then analyzed using the R package LIMMA (Ritchie et al., 2015). Weighted likelihoods based on the observed mean-variance relationships were then calculated for all samples and genes using voomWithQualityWeights (Law et al., 2014) prior to the calculation of differential expression profiles between sample groups using LIMMA. Gene counts and differentially expressed genes were annotated using gene models for Scenedesmus obliquus DOE0152Z from JGI (Starkenburg et al., 2017) and heatmaps were constructed in R.
3 Results
3.1 Growth of S. obliquus with plant substrates
We monitored growth of S. obliquus in the presence of four different plant substrates at 0.1% w/v concentration: corn stover, switchgrass, sugarcane bagasse, and yard waste (Figure 1). On average, we observed a 13% and 33% increase in specific growth rate with switchgrass (µ = 0.169) or yard waste (µ = 0.198) present in the culture, respectively, when compared to that of the microalga alone (µ = 0.149). However, the observed increase in specific growth rate did not reach statistical significance with addition of either substrate compared to the microalga alone (p ≤ 0.05). We continued investigating plant substrate utilization by S. obliquus using switchgrass, as the yard waste continued to introduce contaminating organisms despite autoclaving the substrate.
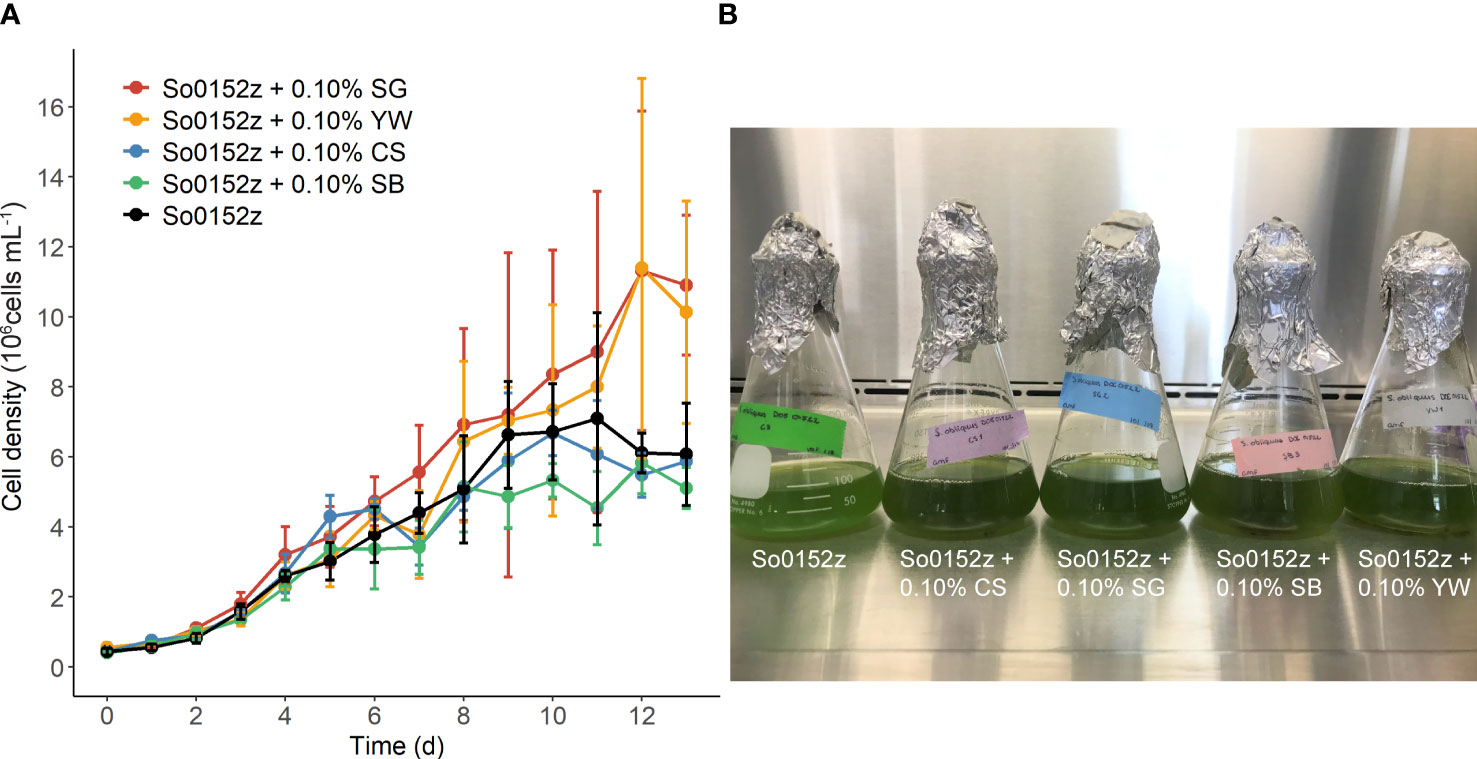
Figure 1 (A) Growth of S. obliquus in the presence of four different plant substrates compared to the microalga alone. Points and error bars indicate mean and standard deviation of three biological replicates. (B) Photo of one replicate from each treatment, from left to right: S. obliquus alone, S. obliquus with corn stover, S. obliquus with switchgrass, S. obliquus with sugarcane bagasse, S. obliquus with yard waste.
To examine the effect of switchgrass concentration on S. obliquus growth, 3 additional concentrations of switchgrass were tested (Supplemental Figure 1). There was no notable difference in overall growth observed between these three concentrations, with specific growth rates of 0.178, 0.120 and 0.162 respectively. In our subsequent experiments we chose 0.2% w/v as the plant substrate concentration, consistent with our previously published work in Nannochloropsis sp. (Schambach et al., 2020).
We investigated growth of S. obliquus with and without 0.2% w/v SG at 50 L volume in mini raceway ponds in a greenhouse (Figure 2). On average, there was no difference in specific growth rate (µ=0.51 algae alone; µ=0.52 algae with SG; p-value=0.79) or dry weight of S. obliquus in the presence of switchgrass when cultured at 50 L scale. Samples were taken during early log phase and late log phase growth for 16S amplicon sequencing to determine if the addition of switchgrass influenced the microbial community composition (Figure 3). The microbial composition between cultures with and without switchgrass were largely similar during early log phase growth. We did observe a shift in composition during late-log phase although there was no obvious pattern regarding the presence or absence of the switchgrass. Paracoccus sp., Allorhizobium-neorhizobium-Pararhizobium-Rhizobium sp., and Rudanellua sp. were present in all cultures regardless of plant substrate addition or timepoint.
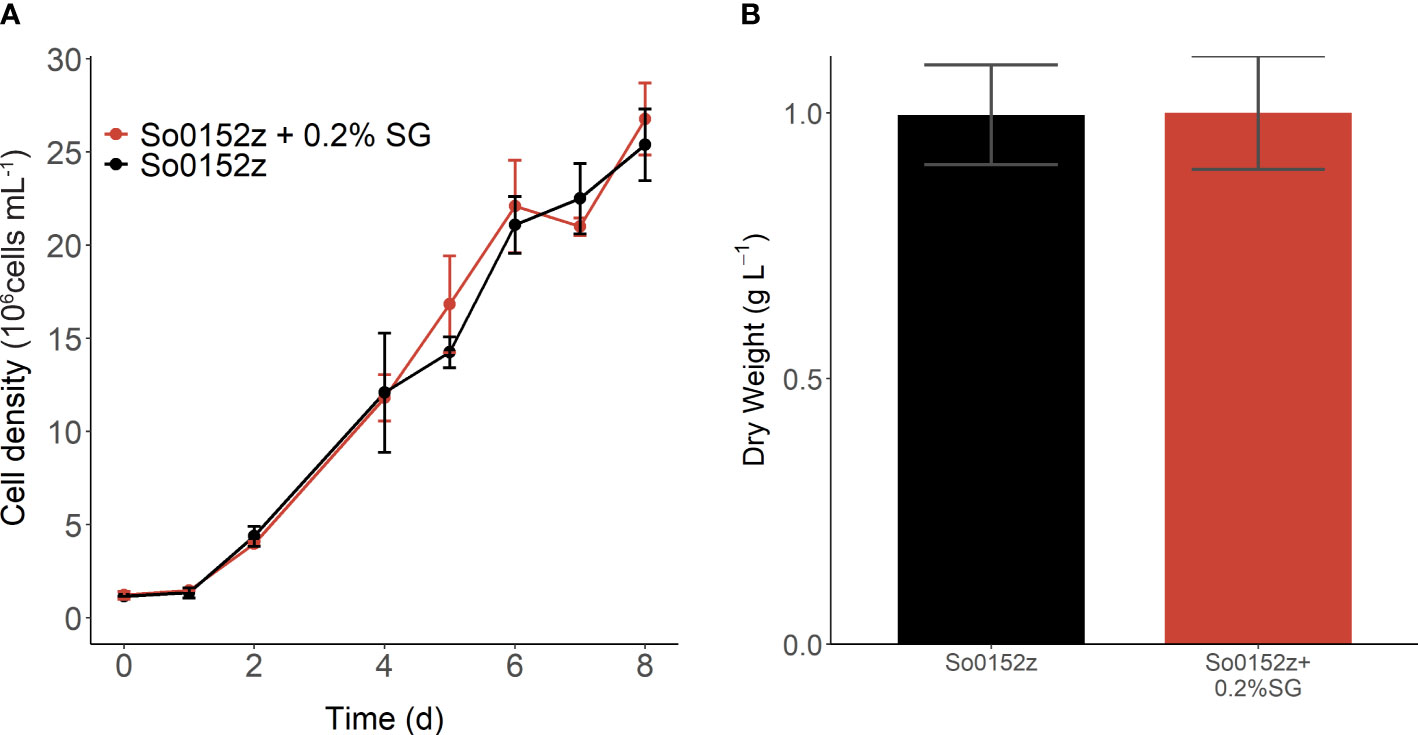
Figure 2 (A) Growth of S. obliquus at 50 L volume in mini-raceway ponds with and without switchgrass addition. (B) Dry weight of microalgal biomass taken on day 8 of cultivation. Points and error bars indicate mean and standard deviation of three biological replicates.
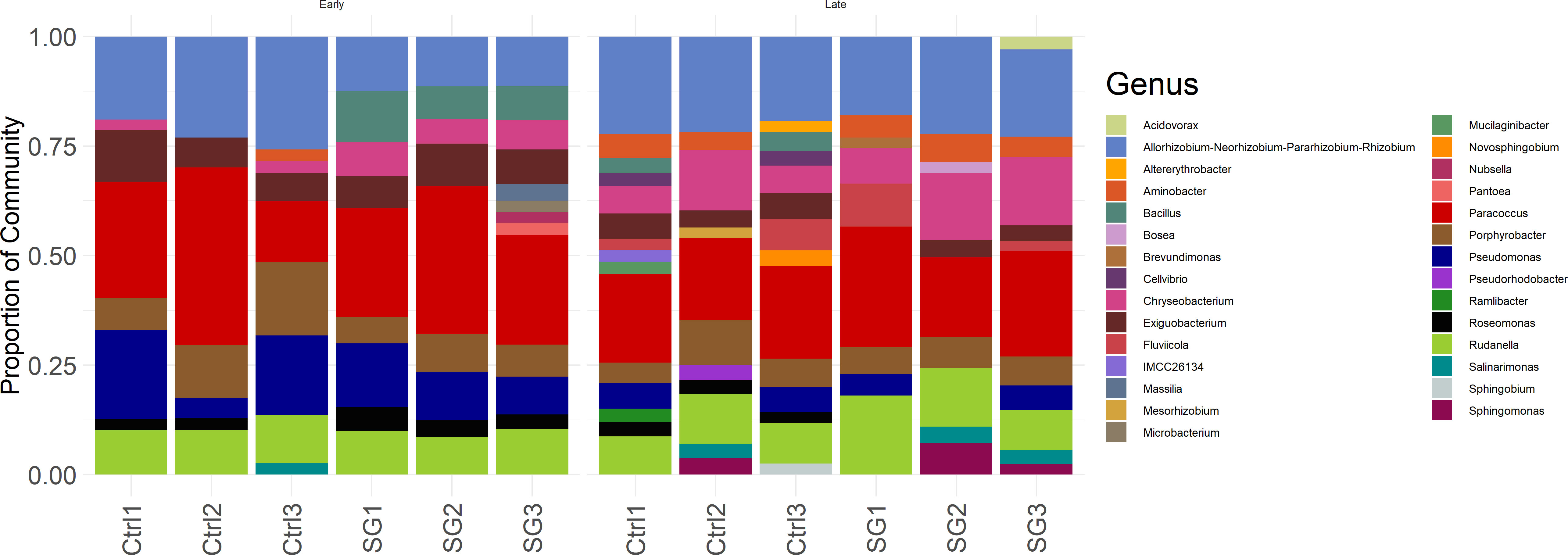
Figure 3 Bacterial community composition during early log phase (“Early”; Left) and late log phase growth (“Late”; Right) of S. obliquus with (“SG”) and without (“Ctrl”) raw switchgrass at 50 L scale in mini raceway ponds. Only genera with an average abundance greater than or equal to 2% were included in this figure.
We investigated the molecular mechanisms for plant substrate utilization by S. obliquus, and how those may be affected by elevated CO2 concentration through RNA-sequencing. S. obliquus was grown with and without 0.2% switchgrass in ambient (i.e., air) and 1% CO2 and samples were taken for RNA sequencing during mid- and late-log phase growth. The growth curve and end-point dry weights for these experiments are shown in Figures 4A–D. There was an increase in specific growth rate in the presence of switchgrass under both ambient (µ=0.06 algae alone; µ=0.09 algae with SG) and 1% CO2 (µ=0.12 algae alone; µ=0.15 algae with SG), although this was only statistically significant under ambient CO2 (p ≤ 0.05). Under ambient CO2, there was a 73% increase in dry weight of S. obliquus when cultured with switchgrass. As expected, under an increased CO2 concentration the dry weight of S. obliquus exceeded that of both conditions tested in air, regardless of switchgrass addition. However, with switchgrass present in the culture we observed an additional 7% increase in dry weight, although this was not statistically significant. Transcript expression of S. obliquus in the presence of switchgrass was compared to that of the alga alone for all four sample types: ambient and 1% CO2; mid-log and late-log timepoints.
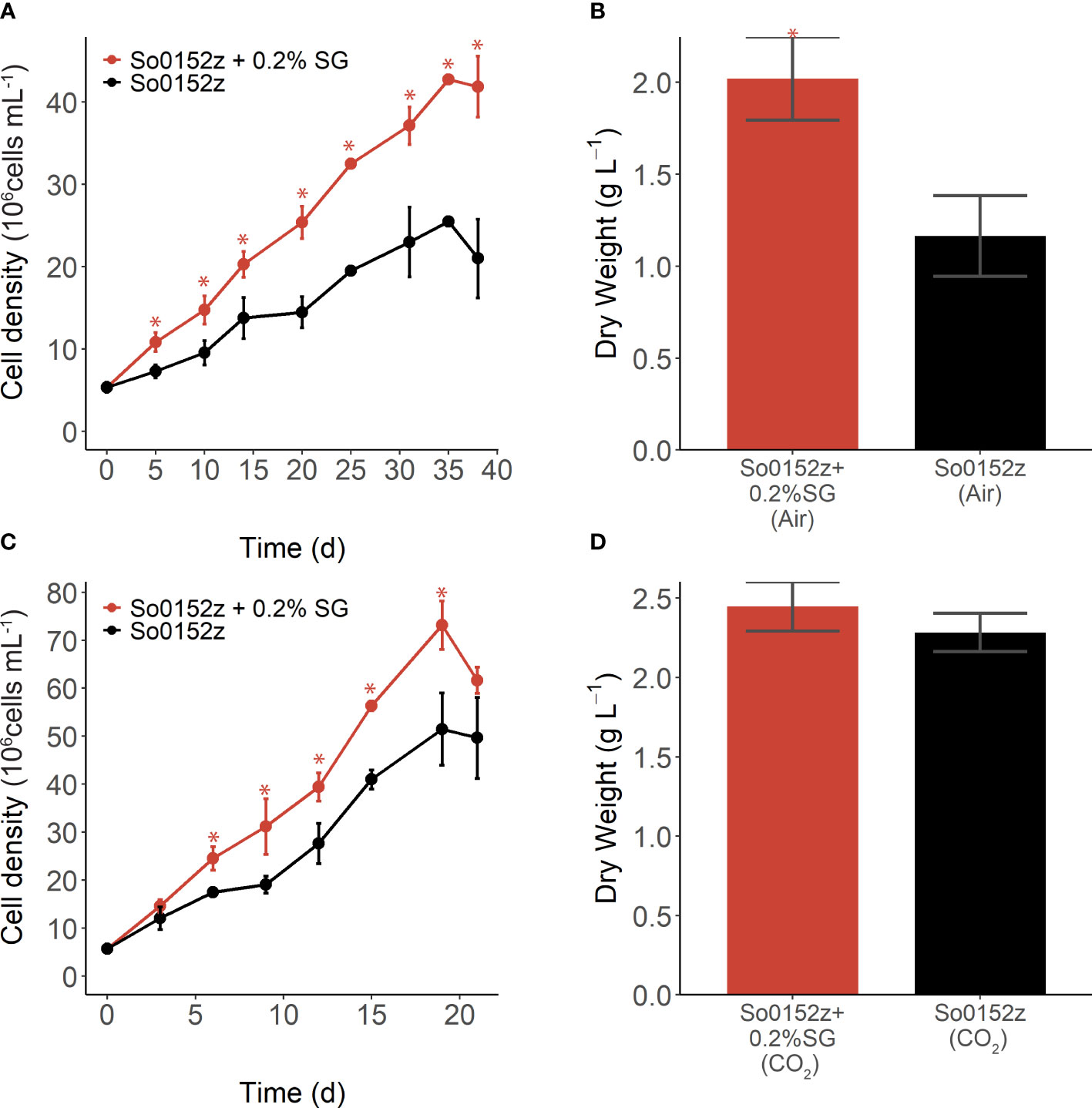
Figure 4 (A) Growth and (B) dry weight of S. obliquus with and without addition of 0.2% w/v switchgrass (SG) grown in air (i.e. ambient CO2). (C) Growth and (D) dry weight of S. obliquus with and without addition of 0.2% w/v switchgrass (SG), grown in l% CO2. Points and error bars indicate mean and standard deviation of three biological replicates. Stars indicate cell density or dry weight of S. obliquus with SG is significantly different from that of the alga alone (p-value < 0.05).
3.2 Plant substrate morphology
Precisely cut cross sections of SG and corn stem were amended to cultures of S. obliquus or to flasks of just BG-11 media alone and incubated for two weeks (Supplemental Figure 2). Following incubation, the preserved cross sections were imaged using confocal and scanning electron microscopy to visualize physical degradation of the corn stem. Note the corn stem provided a better surface for imaging when compared to the SG. Furthermore, the corn stem was acquired from a fresh corn plant, which may be why we observed an increase in growth compared to our previous flask experiment with dried INL corn stover. Vascular bundles were visualized in stem cross sections by confocal microscopy after fluorescence staining with acridine orange (Figures 5A, B, 6A, B and Supplemental Figures 3, 4). The red staining was more intense on cross sections that had been in culture with the microalga which may suggest oxidative degradation (Houtman et al., 2016). Imaging of the vascular bundles revealed phloem cells almost entirely degraded when in culture with the microalga (Figures 7A–D). All vascular bundles displayed some degree of phloem degradation when S. obliquus was present, while all vascular bundles without the microalga present had intact phloem.
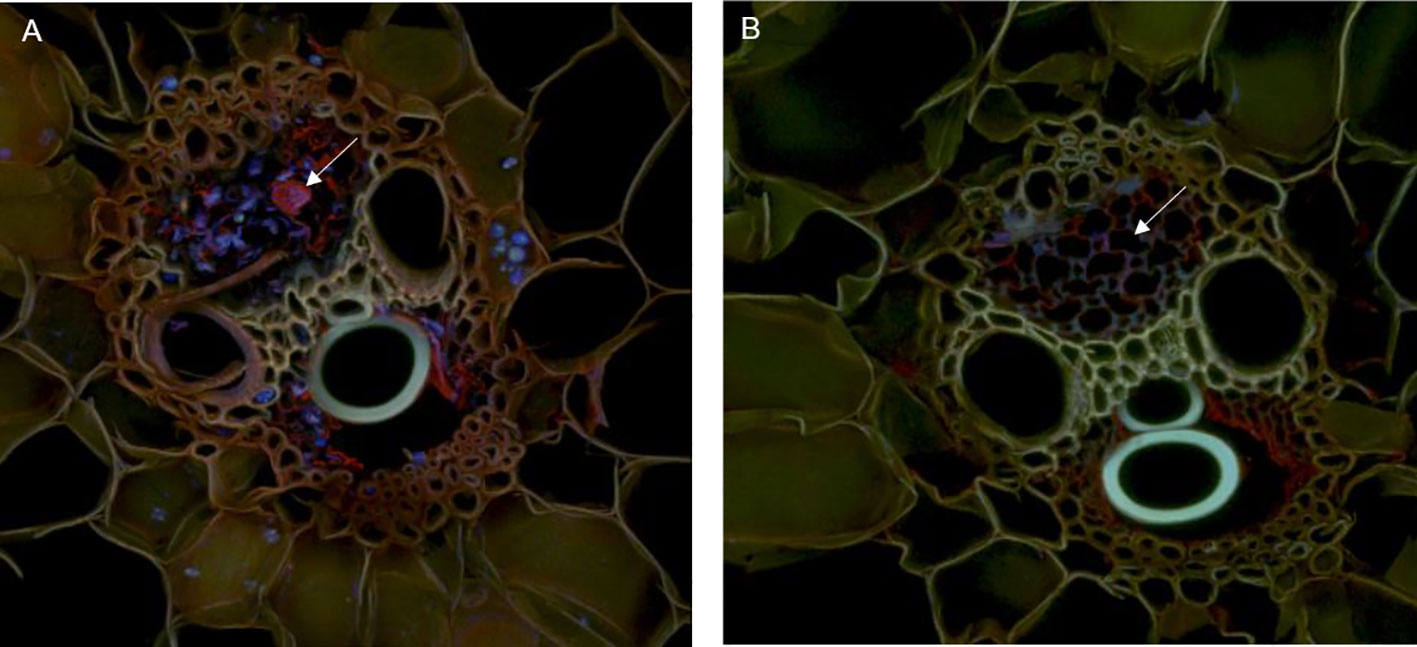
Figure 5 Vascular bundles of corn stem in culture with S. obliquus (A) or incubated with just BG-11 media (B) after fluorescence staining with acridine orange. Arrows point to sieve elements. The blue and green fluorescence in (A) is S. obliquus. In (A), the acridine orange appears shifted to the red spectrum. Maximum-projection images by confocal microscopy.
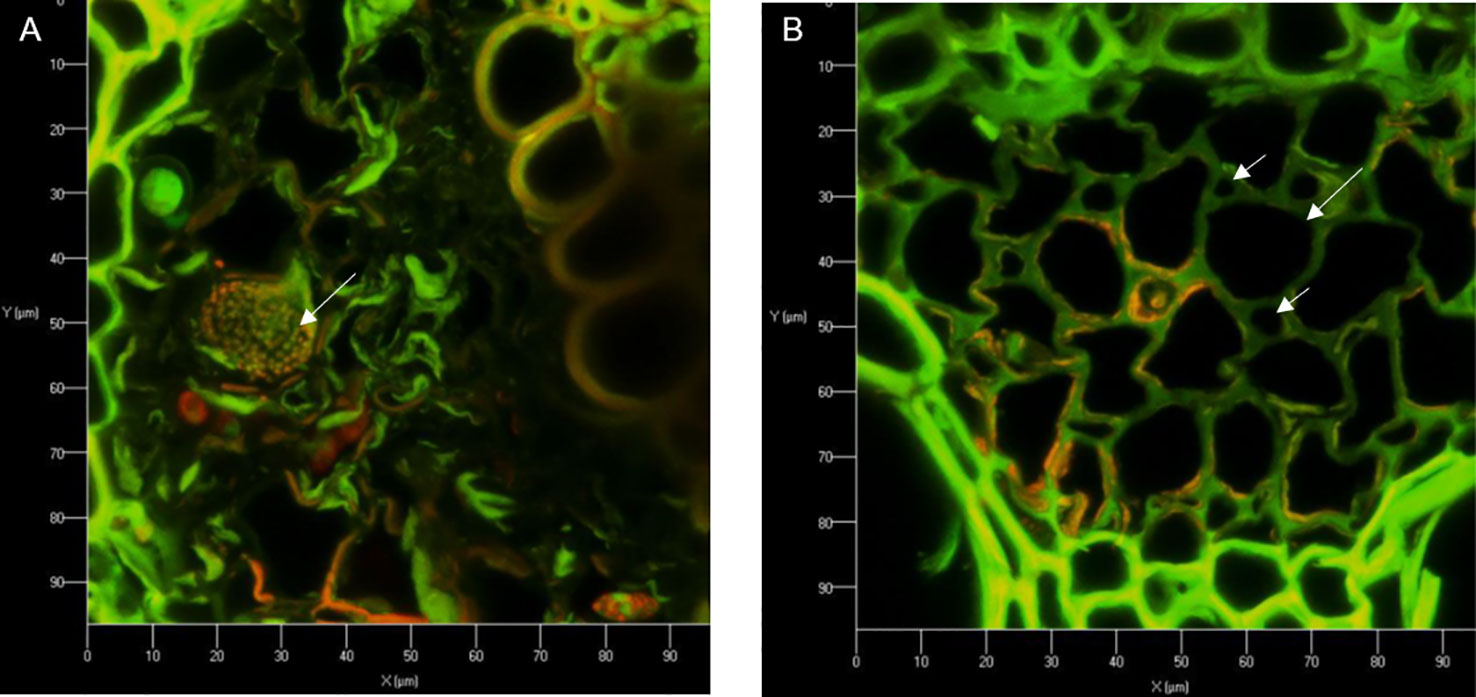
Figure 6 Vascular bundles of corn stem cross sections after being in culture with S. obliquus (A) or incubated with just BG-11 media (B) after fluorescence staining with acridine orange. Long arrows point to sieve elements; short arrows point to companion cells.
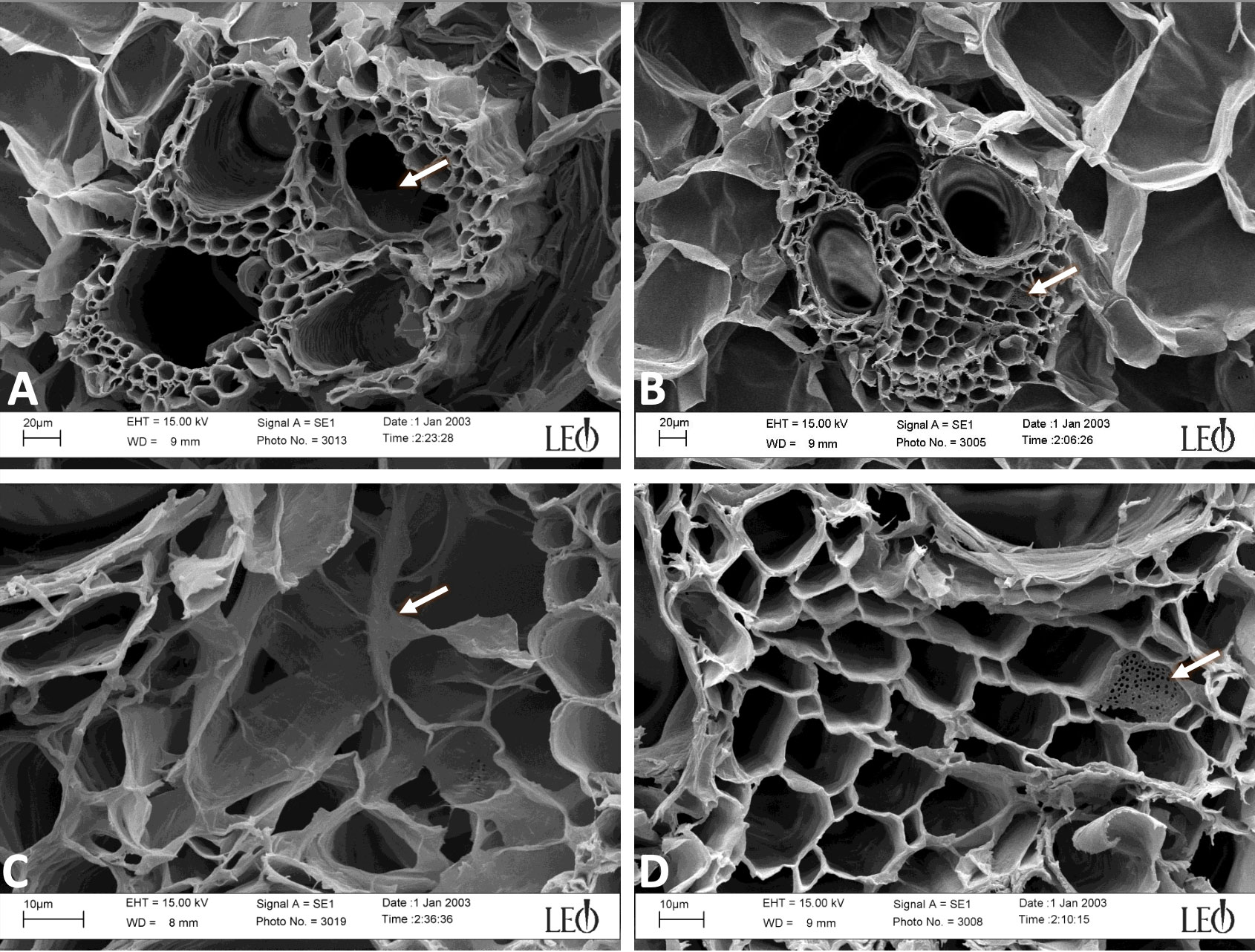
Figure 7 Magnified views of vascular bundles by scanning electron microscopy (SEM) of corn stem cross sections after being in culture with S. obliquus (A, C) or incubated with just BG-11 media (B, D). Arrow in (A) points to the phloem area where the phloem cells are almost entirely degraded. Arrow in (C) shows digested cell walls of phloem. Arrows in (B, C) point to the same sieve plate.
3.3 Transcriptomic analysis of S. obliquus in the presence of switchgrass
3.3.1 Carbohydrate metabolism
We first looked for transcripts encoding potential glycosyl hydrolases, enzymes that hydrolyze glycosidic linkages in cellulose and hemicellulose, to determine if it is indeed S. obliquus that acts on the plant substrate directly. Figure 8 shows a heatmap of the log fold change of transcripts encoding glycosyl hydrolases for all four sample types in the presence of switchgrass compared to the S. obliquus alone. S. obliquus has 14 predicted endo-1,4-beta-D-glucanases (e.g., cellulases; EC 3.2.1.4) which hydrolyze the 1,4-beta-D-glycosidic linkages in cellulose. Of the five expressed transcripts encoding these cellulases, the majority were significantly downregulated in the presence of switchgrass regardless of CO2 condition. However, during mid-log phase in ambient CO2 two beta-glucosidases (EC 3.2.1.21; protein IDs 17466, 32074) not shown on the heatmap, were significantly upregulated 1.56 and 1.21 log fold (adj. p <0.05). Interestingly, four other transcripts encoding beta-glucosidases were significantly downregulated (-2.78, -1.56, -2.52, -2.53) in 1% CO2 (adj. p <0.05).
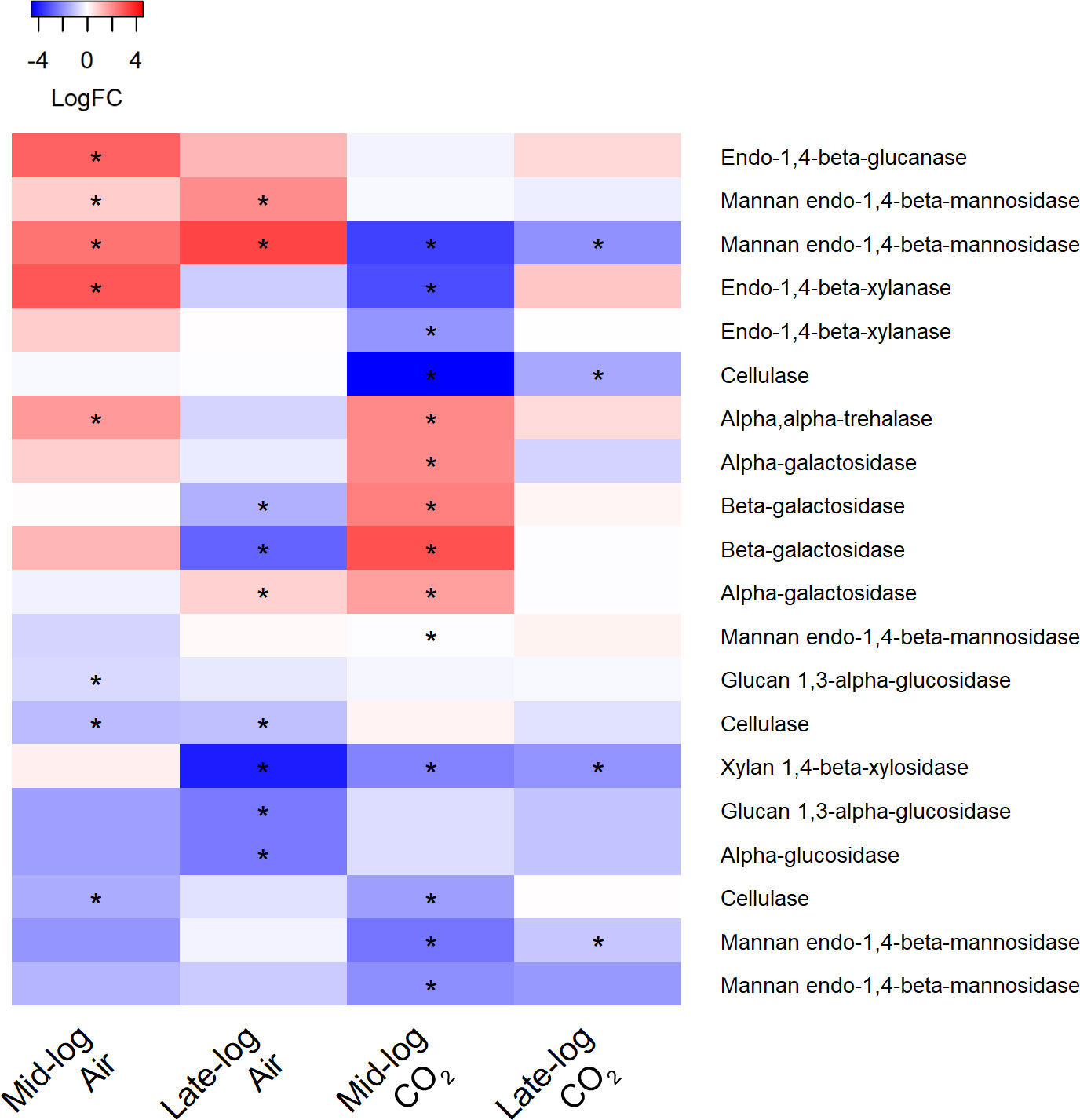
Figure 8 Heat map of differentially expressed transcripts encoding glycosyl hydrolases in S. obliquus grown in the presence of switchgrass in ambient (i.e., air) and 1% CO2 during mid- and late-log phase growth. Differential expression is presented from a mean of triplicate samples as the log fold change of transcripts from cultures containing switchgrass compared to cultures of the algae alone. Stars indicate an adjusted p-value of less than 0.05.
The hemicellulose fraction of switchgrass is made up primarily of xylan. One transcript encoding endo-1,4-beta xylanase (EC 3.2.1.8) was significantly upregulated (2.9 log fold) during mid-log phase growth in ambient CO2. Following a similar trend as the cellulases, the xylanases were significantly downregulated in 1% CO2 during mid-log phase growth with switchgrass (adj. p <0.05).
Mannan is a minor hemicellulose polysaccharide in switchgrass, yet three transcripts encoding extracellular mannan endo-1,4-beta-mannosidases (Protein IDs 31239, 23944, 15946; EC 3.2.1.78) were upregulated in the presence of switchgrass during mid-log phase and late-log phase under ambient CO2. Two of these transcripts (Protein IDs 23944, 16754) were significant at both time points (adj. p <0.05). Of the five mannan endo-1,4-beta-mannosidase transcripts expressed in all sample groups, all but one were downregulated when cultures were grown with switchgrass in 1% CO2 during late log phase. One of the extracellular mannan endo-1,4-beta-mannosidases (Protein ID 23944) was significantly down regulated in 1% CO2 at mid and late log phase times points (adj. p ≤ 0.05).
Galactosidases hydrolyze galactosyl moieties from glycoproteins, glycolipids, and polysaccharides. There was significant upregulation of alpha- and beta-galactosidases (EC 3.2.1.22; 3.2.1.23), most notably in 1% CO2 during mid-log phase with switchgrass present in the flask (adj. p <0.05).
Interestingly, there was a pattern of significant upregulation in transcripts encoding enzymes in both the starch synthesis and degradation pathways in the presence of switchgrass and 1% CO2 during mid-log phase (Figure 9) (adj. p <0.05). Expression of alpha- and beta-amylases (EC 3.2.1.1; 3.2.1.2), which hydrolyze starch producing dextrin and maltose, as well as starch synthase (EC 2.4.1.21) and the 1,4-alpha glucan branching enzyme (EC 2.4.1.18), responsible for elongation of starch, were upregulated. In contrast, many of these transcripts were downregulated in ambient CO2 in the presence of switchgrass at both timepoints.
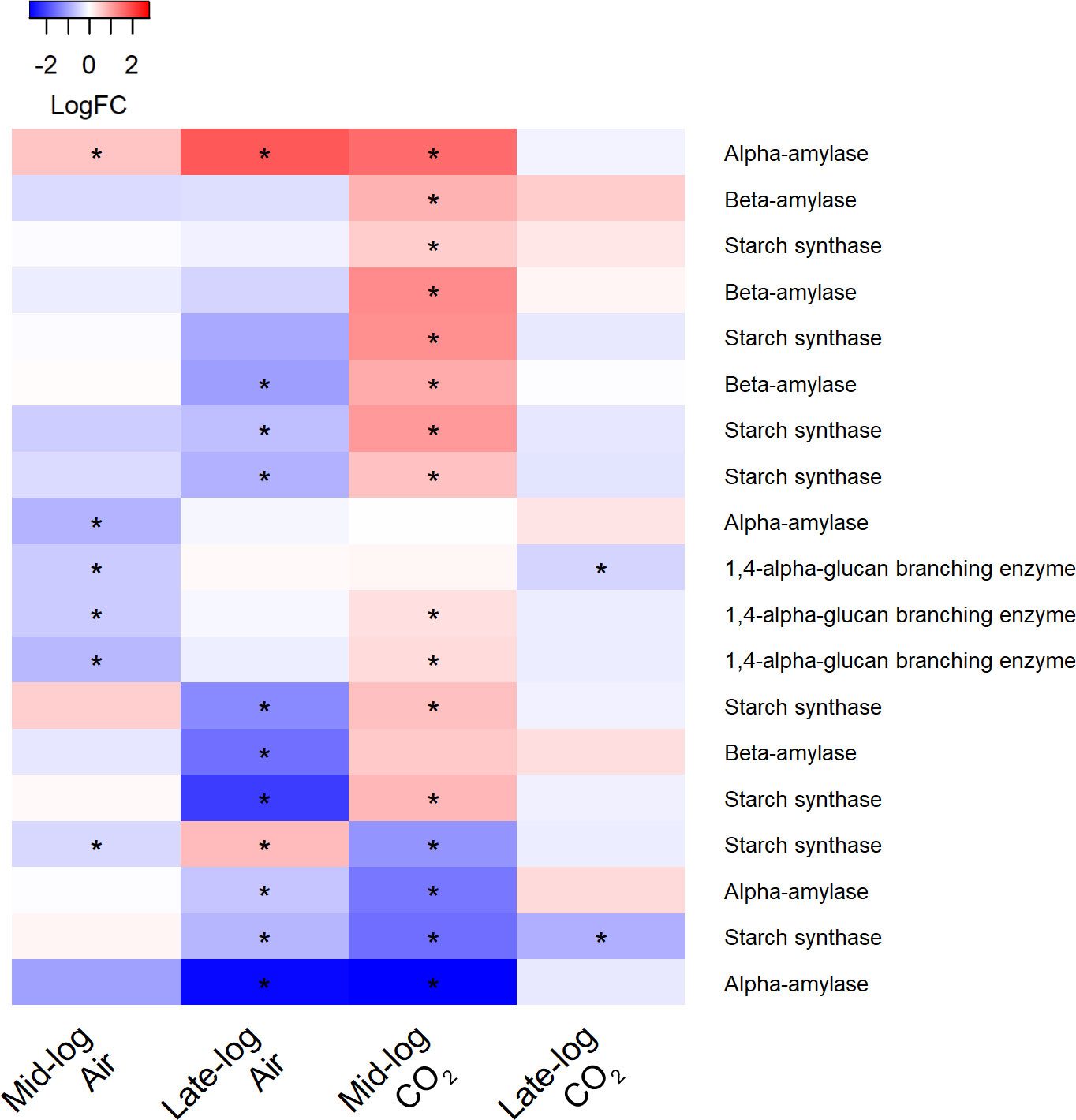
Figure 9 Heat map of differentially expressed transcripts involved in starch metabolism in S. obliquus grown in the presence of switchgrass in ambient (i.e., air) and 1% CO2 during mid- and late-log phase growth. Differential expression is presented from a mean of triplicate samples as the log fold change of transcripts from cultures containing switchgrass compared to cultures of the algae alone. Stars indicate an adjusted p-value of less than 0.05.
3.3.2 Oxidative phosphorylation
Several transcripts encoding inorganic pyrophosphatases associated with ATPases within the oxidative phosphorylation pathway are significantly upregulated in the presence of switchgrass during late log phase in ambient CO2 and mid log phase in 1% CO2 (Figure 10) (adj. p <0.05). Interestingly, during late-log phase in ambient CO2 we found two transcripts encoding proton-exporting ATPases downregulated 2.3 and 2.4 log fold in the presence of switchgrass (adj. p ≤0.01).
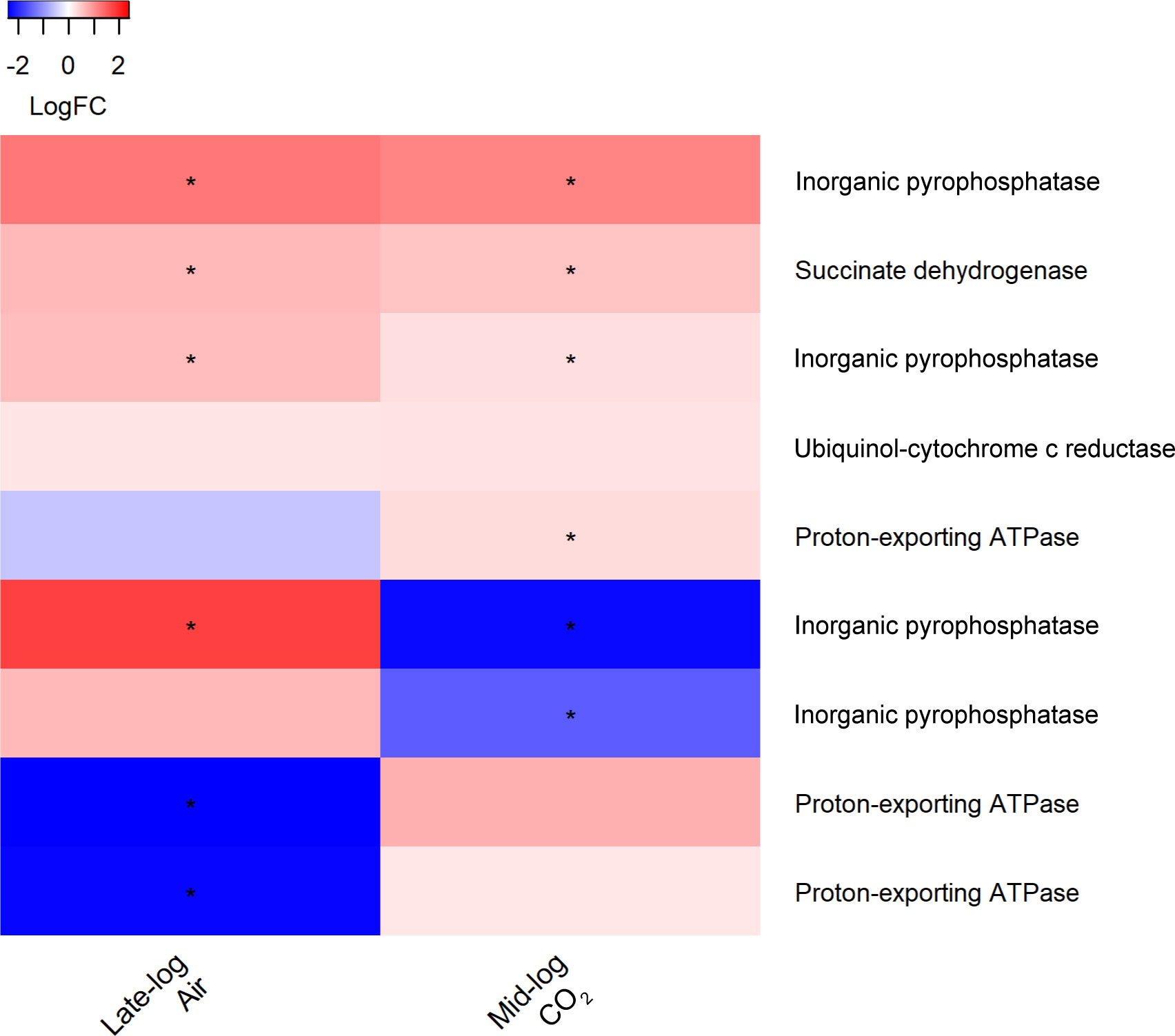
Figure 10 Heat map of differentially expressed transcripts encoding enzymes involved in oxidative phosphorylation in S. obliquus grown in the presence of switchgrass during late-log and mid-log phase growth in ambient (i.e., air) and 1% CO2, respectively. Differential expression is presented from a mean of triplicate samples as the log fold change of transcripts from cultures containing switchgrass compared to cultures of the algae alone. Stars indicate an adjusted p-value of less than 0.05.
3.3.3 Photosynthesis
In cultures grown in ambient CO2 with switchgrass there was significant upregulation of transcripts for subunits in photosystem I (PsaH, PsaI, PsaL, PsaN) and II (Psb27, Psb28, PsbP, PsbR, PsbX), as well as ferredoxin NADP+ reductase (Table 1) (adj. p <0.05). However, in 1% CO2 most transcripts for these or other related photosystem subunits were slightly downregulated in the presence of switchgrass, but not significantly.

Table 1 Log fold changes, adjusted p-values, and protein ID of transcripts encoding photosystem I and II subunits of S. obliquus grown in the presence of switchgrass in ambient and 1% CO2 during mid- and late-log phase growth.
3.3.4 Carbon fixation
One transcript encoding ribulose bisphosphate carboxylase oxygenase (RuBisCO) was significantly upregulated in the presence of switchgrass in all sample types except under 1% CO2 during late-log phase (Table 2). Significant upregulation was observed in other transcripts encoding enzymes involved in Calvin-Benson cycle with switchgrass present in ambient CO2, particularly during late-log phase (Figure 11) (adj. p <0.05). Some of these transcripts were also upregulated in 1% CO2 during mid log phase.

Table 2 Log fold changes and adjusted p-values of transcripts encoding ribulose bisphosphate carboxylase oxygenase (RuBisCO) of S. obliquus grown in the presence of switchgrass in ambient and 1% CO2 during mid- and late-log phase growth.
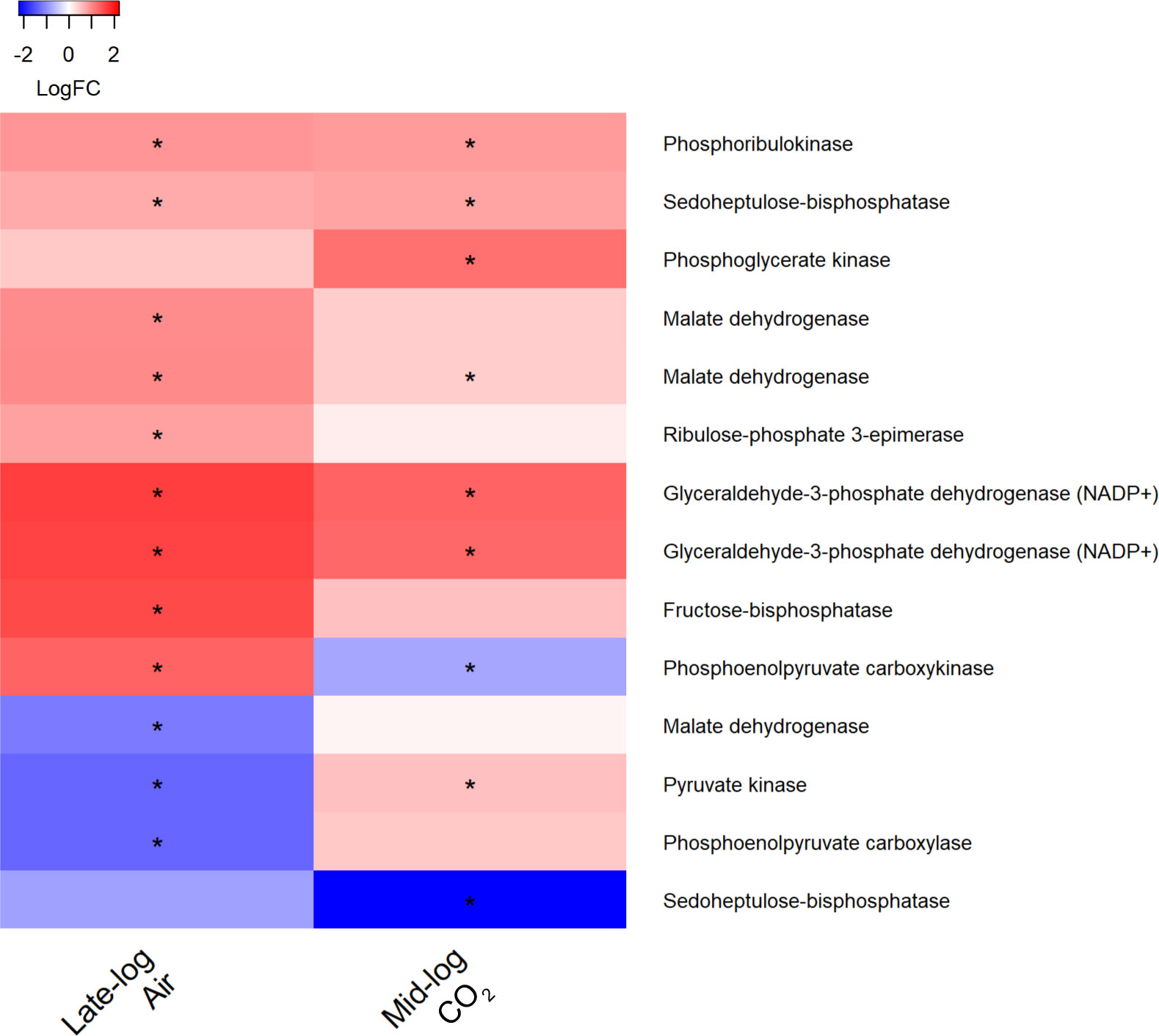
Figure 11 Heat map of differentially expressed transcripts encoding enzymes in the carbon fixation pathway in S. obliquus grown in the presence of switchgrass during late-log and mid-log phase growth in ambient (i.e., air) and 1% CO2, respectively. Differential expression is presented from a mean of triplicate samples as the log fold change of transcripts from cultures containing switchgrass compared to cultures of the algae alone. Stars indicate an adjusted p-value of less than 0.05.
4 Discussion
We grew Scenedesmus obliquus in the presence of raw plant substrates: corn stover, switchgrass, sugarcane bagasse, and yard waste. Compared to photoautotrophic growth of the alga, we found that with switchgrass or yard waste present, the average specific growth rate increased 13% and 33%, respectively, although not statistically significant (p ≤ 0.05). We did observe a statistically significant increase in specific growth rate in the presence of switchgrass under ambient CO2 (p ≤ 0.05), with a 73% increase in dry weight. This difference in results is likely due to the greater concentration of plant substrate and greater inoculation cell density used in the latter experiment. As expected, under an increased CO2 concentration the dry weight of S. obliquus exceeded that of both conditions tested in air, regardless of switchgrass addition. However, with switchgrass present in the culture we observed an additional 7% increase in dry weight, although this was not statistically significant. We have previously reported increased growth rates and biomass accumulation of Auxenochlorella protothecoides and Nannochloropsis sp. when cultured with raw switchgrass or corn stover, respectively, with increases ranging from 13-40% in flask cultures. As with these other species, this overall increase in growth in the presence of plant substrate indicated the potential of S. obliquus to use the raw plant as a carbon source for mixotrophic growth.
When switchgrass utilization was tested at 50 L in mini raceway ponds, we did not see the expected increase in growth of S. obliquus with switchgrass present with no difference observed between treatments. In contrast, when Nannochloropsis gaditana was grown at 50 L in mini raceway ponds, specific growth rate was increased 30% with corn stover present when compared to cultures of the alga alone (Schambach et al., 2020). The discrepancy between laboratory scale results and pond scale results is an ongoing hurdle within applied microalgal research (da Silva and Reis, 2015). Many factors, such as culture depth and mixing, light intensity, temperature range, aeration, and on-demand CO2 for pH balance, can be markedly different from the laboratory experiments and likely play a major role in the outcome. Gaining a better understanding of how these factors play a role in plant substrate utilization and optimizing conditions based on that understanding is necessary to improve scalability of this strategy in the raceway pond setting.
Since we did not see the expected growth increase at 50 L, we hypothesized that we would not see a large difference in microbiome composition, and this was generally the case. We did see more representation from members with low abundance during late log phase. However, Paracoccus sp., Allorhizobium-Neorhizodium-Pararhizobium-Rhizobium sp., and Rudanella sp. were present in all cultures regardless of plant substrate or timepoint. This could indicate that bacteria from these genera could be key members of the microbiome that support growth of S. obliquus. Paracoccus is a genus whose members are ubiquitous to both natural and anthropogenic environments and play important roles in many types of microbiomes (Lasek et al., 2018). This genus was abundant in Nannochloropsis gaditana cultures both with and without plant substrate (Schambach et al., 2020). Allorhizobium-Neorhizodium-Pararhizobium-Rhizobium include diazotrophic bacteria commonly found in association with plant roots as well as some green algae including Scenedesmus (Kim et al., 2014). Rudanella is a genus in the family Spirosomaceae, however there is very little literature about the habitats and ecological functions of members of this genus. Characterizing the microbiome of S. obliquus under favorable conditions for plant substrate utilization is necessary for understanding how the microbiome may be playing a role in utilization and how to leverage it for increased microalgal productivity and culture resilience.
Increases in biomass production of S. obliquus under mixotrophic conditions have been reported with addition of low molecular weight, soluble organic substrates like glucose or acetate to the culture (Mandal and Mallick, 2009; Yang et al., 2014; Shen et al., 2018; Song et al., 2021). In addition, Yang et al., 2014 reported S. obliquus could utilize xylose, and when added at a 4 g L-1 concentration (0.4% w/v), cell density was increased 2.9-fold (Yang et al., 2014). S. obliquus could also successfully grow on filtrate containing soluble product of wheat bran fermented by two fungal species (EL-Sheekh et al., 2012). These studies support the ability of S. obliquus to uptake and assimilate several types of exogenous organic carbon. However, when considering the complexity and heterogeneity of lignocellulosic biomass, it may be necessary for S. obliquus to carry a suite of enzymes (e.g., glycosyl hydrolases and others) that, when working synergistically, hydrolyze the structural carbohydrates into free sugars for uptake and metabolism. Qualitative assessment of corn stem morphology via microscopy after incubation with S. obliquus indicated potential cellulolytic enzymatic production as there was noticeable degradation of the plant when compared to the plant incubated in just algal media. The pattern of degradation in these images was similar to observations in Nannochloropsis gaditana and corn stover, particularly regarding the missing and/or collapsed phloem (Schambach et al., 2020).
Multi-omic analysis of A. protothecoides revealed this alga has a suite of potential glycosyl hydrolases, including those from families 5 and 9, some of which were upregulated when grown in the presence of switchgrass or carboxymethylcellulose (Vogler et al., 2018). Proteins from glycosyl hydrolase families 5 and 9 contain members with known cellulolytic activity, suggesting A. protothecoides could be directly degrading cellulose. In the current study, the consistent downregulation of potential endo-1,4-beta-D-glucanases in the presence of switchgrass, regardless of CO2 concentration or time point, suggests S. obliquus may not be directly acting on the cellulose fraction of the plant. This finding could also indicate a lack of recycling or reorganizing of the cell wall of S. obliquus as it contains cellulose (Voigt et al., 2014). This would allow the alga to maintain cell wall rigidity when in the presence of the switchgrass. Anecdotally, lysis of S. obliquus cells that were in culture with switchgrass required 2 to 3 additional cycles of mechanical homogenization before efficient lysis was seen under the microscope.
Hemicellulose is another major component of plant cell walls, and can consists of xylan, xyloglucan, mannan, glucomannan, and beta-glucan (Scheller and Ulvskov, 2010).Considering that xylans, polysaccharides of beta-1,4 linked xylopyranosyl residues, are the major hemicellulose structure in switchgrass, and that S. obliquus was reported to utilize xylose, we expected to see significant upregulation of xylanases and/or xylosidases. These enzymes are endo- and exoglycosidases that release xylooligosaccharides and xylose, respectively, from the xylan backbone (Terrasan et al., 2016). We only observed one transcript encoding an endo-1,4-beta-xylanase significantly upregulated during mid-log phase in the presence of switchgrass in ambient CO2, whereas one xylan 1,4-beta xylosidase transcript was significantly downregulated in all other sample groups. However, significant upregulation of extracellular mannosidases, and beta-glucosidases under ambient CO2 could suggest S. obliquus is acting upon the mannan fraction of switchgrass hemicellulose. Beta-1,4-mannosidases and beta-1,4-glucosidases are the major enzymes involved in mannan hydrolysis with complete hydrolysis requiring a synergistic effect of these enzymes (Rodríguez-Gacio et al., 2012). While mannose content in most grasses is low, glucomannans are an important seed storage polysaccharide in grass species like switchgrass (Scheller and Ulvskov, 2010; Rodríguez-Gacio et al., 2012). Note that beta-1,4-glucosidases also act on the exposed terminal glycosyl residues of cellulose.
We hypothesized that when S. obliquus was grown in the presence of 1% CO2, there would not be a notable increase in growth rate or dry weight with switchgrass supplementation. The increased CO2 concentration allows for the carboxylation reaction catalyzed by RuBisCO to be more energetically favorable, leading to increased carbon fixation and ultimately contributing to faster overall growth. This is exemplified by the 2-fold increase in dry weight when S. obliquus alone was grown in 1% CO2 versus the ambient condition. Sforza et al., 2012 reported inhibition of mixotrophy in A. protothecoides and Nannochloropsis salina when grown under 5% CO2 likely because CO2 fixation was more energetically favorable under the high CO2 concentration. While not statistically significant, we did see slight increases in specific growth rate and dry weight with switchgrass present under 1% CO2 This could indicate the switchgrass derived carbon could be playing a role.
Despite the lack of a statistically significant growth response to switchgrass under 1% CO2, we saw several transcriptomic responses with switchgrass present, that were also contrary to what was found when compared to ambient CO2, particularly regarding differential expression of potential glycosyl hydrolases. Several mannosidases, and a xylanase were significantly downregulated with switchgrass present under 1% CO2, but several alpha- and beta- galactosidases were significantly upregulated. Furthermore, many alpha- and beta-amylases, starch synthase and a branching enzyme, all of which are involved in starch metabolism, were significantly upregulated in the presence of switchgrass and 1% CO2. Stimulation of starch metabolism in microalgae grown under high CO2 concentrations has been reported (Tanadul et al., 2014), however the presence of switchgrass also appears to influence this response. These transcripts were downregulated in the presence of switchgrass in ambient CO2. These results could indicate that under higher CO2 concentration, different carbon storage metabolic mechanisms are at play in the presence of switchgrass, compared to ambient air.
We explored differential expression within mitochondrial oxidative phosphorylation, photosynthesis, and carbon fixation pathways, particularly during late-log phase growth in ambient CO2 and mid-log in 1% CO2, to determine if there were any discernable patterns in the overarching energy metabolism of S. obliquus when switchgrass was present. It has been reported under mixotrophic conditions, the metabolism of some algae, like N. gaditana and C. reinhardtii, is dominated by respiration with an associated decrease in photosynthetic activity (Chapman et al., 2015; Bo et al., 2021). Differently, photosynthesis in C. sorokiniana is largely unaffected by the addition of acetate (Cecchin et al., 2018). Upregulation of transcripts encoding inorganic pyrophosphatases and proton exporting ATP synthases in S. obliquus in the presence of switchgrass under both CO2 concentrations, could be an indication of increased ATP production via respiration. In addition, significant upregulation of transcripts encoding key enzymes in the carbon fixation pathway, including RuBisCO, under both CO2 concentrations could suggest reutilization of the CO2 produced from respiration (Smith et al., 2015).
Interestingly, upregulation of PSI and PSII subunits in the presence of switchgrass was observed under ambient CO2, whereas, under 1% CO2 these or related subunits were slightly downregulated. It could be that increases in expression of these subunits is a response to increased light attenuation because of the large insoluble switchgrass particles. However, when inorganic carbon is less limited under 1% CO2, this appears to be alleviated and could indicate that under ambient CO2, cells are perhaps more reliant on NADPH and ATP produced from the light reactions of photosynthesis for substrate utilization. Conversely, upregulation of these subunits could be an artifact of increased energy derived from the plant carbon being shuttled into chloroplast production. Cecchin et al., 2018 reported upregulation of Ferredoxin-NADP+ reductase (FNR) when C. sorokiniana was grown with acetate, and we also observed this with S. obliquus in the presence of switchgrass under ambient CO2 (Cecchin et al., 2018). In linear electron flow, electrons are transferred from ferredoxin by FNR to produce NADPH in photosynthesis (Chitnis, 2001). For the case of C. sorokiniana, increased reducing power (i.e., NADH) derived from acetate consumption could be transferred from mitochondria to the chloroplast causing an overreduction of plastoquinones and increasing demand of FNR and ultimately contributing to increased overall growth (Cecchin et al., 2018). Similarly, upregulation of FNR could suggest increased electron transport in photosynthesis in S. obliquus in the presence of switchgrass but only under ambient CO2 and more work would be needed to determine if it is in fact due to increased reducing power derived from switchgrass consumption.
5 Conclusion
We grew S. obliquus in the presence of corn stover, yard waste, sugarcane bagasse, and switchgrass and observed increases in overall growth and dry weight with yard waste and switchgrass. Fluorescence microscopy and SEM of corn stem cross sections in culture with S. obliquus showed degradation of phloem and provides evidence of plant degradation by the microalga. When scaled up to 50 L in mini-raceway ponds we did not see the expected increase in growth nor a substantial difference in the culture microbiome with switchgrass present in the culture. Growth and dry weight of S. obliquus was also increased when we added raw switchgrass to the flask under both ambient and 1% CO2. We observed differential expression of potential glycosyl hydrolases that could suggest S. obliquus is degrading the hemicellulose fraction of switchgrass under ambient CO2. However, under 1% CO2 expression of potential glycosyl hydrolases was largely inversed in the presence of switchgrass when compared to that in ambient CO2 suggesting molecular mechanisms of plant substrate utilization change when inorganic carbon is more available. Upregulation of transcripts encoding PSI and PSII subunits as well as FNR were only observed under ambient CO2 which indicate either that plant substrate utilization is more reliant on photosynthate or that photosynthesis is stimulated because of plant consumption. The transcriptomic analysis employed here only provides a snapshot of the potential metabolic activity and interaction between S. obliquus and switchgrass during mid-photoperiod and how increased CO2 concentration may influence that. Future work, like targeted transcriptomics or proteomics of the potential glycosyl hydrolases found differentially expressed in this study, or tracing stable isotopically labeled plant material into S. obliquus biomass, is necessary to gain a more comprehensive view of the molecular mechanisms of plant substrate utilization and to characterize the energy fluxes and metabolic interactions of photosynthesis and respiration that promote increased overall growth of S. obliquus in the presence of switchgrass.
Data availability statement
The datasets presented in this study can be found in the article/Supplementary Material.
Author contributions
JS designed and executed all experiments, participated in the data analysis for all experiments. CK processed and analyzed the RNA-seq data with oversight by SS. PK prepared plant cross-sections and performed microscopy and imaging with oversight by CH. WM processed and analyzed 16S data. AB conceived and managed the overall project and provided guidance on experimental design. All authors contributed to data analysis and to the writing of the article. All authors declare agreement to authorship and submission of the manuscript for peer review.
Funding
This work was supported under the Leveraging Algae Traits for Fuels (LEAF) project, awarded to AB and the Algal Translational Genomics project awarded to SS by the Department of Energy’s Bioenergy Technologies Office, Office of Energy Efficiency and Renewable Energy, under agreement numbers NL0037289 (WBS 1.3.2.043) and NL0029949 (WBS 1.3.1.600), respectively. This work has been cleared for public release by Los Alamos National Laboratory and Sandia National Laboratories.
Acknowledgments
Thank you to the INL Feedstock Library for milled plant material. Thank you to Steven Pflucker and Heliae Development LLC for providing S. obliquus and scientific discussion. Thank you to Chuck Smallwood for switchgrass substrate and lab space. Thank you to the INL Feedstock Library for milled plant substrates and to Mike Stanek at US Dairy Forage Research Farm, Prairie du Sac, WI, for the corn used to make uniformly cut sections. Thank you to the Applied Genomics Team at Los Alamos National Laboratory for sequencing support.
Conflict of interest
Authors JS, WM, and AB were employed by company Sandia National Laboratories.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher. This article has been authored by an employee of National Technology & Engineering Solutions of Sandia, LLC under Contract No. DE-NA0003525 with the U.S. Department of Energy (DOE). The employee owns all right, title and interest in and to the article and is solely responsible for its contents. The United States Government retains and the publisher, by accepting the article for publication, acknowledges that the United States Government retains a non-exclusive, paid-up, irrevocable, world-wide license to publish or reproduce the published form of this article or allow others to do so, for United States Government purposes. The DOE will provide public access to these results of federally sponsored research in accordance with the DOE Public Access Plan https://www.energy.gov/downloads/doe-public-access-plan
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.992702/full#supplementary-material
Supplementary Figure 1 | Growth of S. obliquus with three different concentrations of switchgrass (SG): 0.20%, 0.30%, 0.40% w/v. Points and error bars indicate mean and standard deviation of three biological replicates.
Supplementary Figure 2 | Growth of S. obliquus with and without corn stem cross sections. Corn stem was harvested at day 14 for confocal microscopy and SEM analysis. Points and error bars indicate mean and standard deviation of three biological replicates.
Supplementary Figure 3 | Magnified views of vascular bundles by scanning electron microscopy (SEM) of corn stem cross sections after being in culture with S. obliquus (A–C) or incubated with just BG-11 media (D). Arrows point to phloem cells.
Supplementary Figure 4 | Magnified views of phloem in vascular bundles by scanning electron microscopy (SEM) of corn stem cross sections after being in culture with S. obliquus (A–C) or incubated with just BG-11 media (D). Long arrows points to a sieve element; small arrow points to a companion cell.
References
Abiusi, F., Wijffels, R. H., Janssen, M. (2020). Doubling of microalgae productivity by oxygen balanced mixotrophy. ACS Sustain. Chem. Eng. 8, 6065–6074. doi: 10.1021/acssuschemeng.0c00990
Bo, D. D., Magneschi, L., Bedhomme, M., Billey, E., Deragon, E., Storti, M., et al. (2021). Consequences of mixotrophy on cell energetic metabolism in microchloropsis gaditana revealed by genetic engineering and metabolic approaches. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.628684
Callahan, B. J., McMurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J. A., Holmes, S. P. (2016). DADA2: High-resolution sample inference from illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869
Cecchin, M., Benfatto, S., Griggio, F., Mori, A., Cazzaniga, S., Vitulo, N., et al. (2018). Molecular basis of autotrophic vs mixotrophic growth in chlorella sorokiniana. Sci. Rep. 8:6465, 1–13. doi: 10.1038/s41598-018-24979-8
Chapman, S. P., Paget, C. M., Johnson, G. N., Schwartz, J.-M. (2015). Flux balance analysis reveals acetate metabolism modulates cyclic electron flow and alternative glycolytic pathways in chlamydomonas reinhardtii. Front. Plant Sci. 6. doi: 10.3389/fpls.2015.00474
Chitnis, P. R. (2001). P HOTOSYSTEM I: Function and physiology. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52, 593–626. doi: 10.1146/annurev.arplant.52.1.593
da Silva, T. L., Reis, A. (2015). Scale-up problems for the Large scale production of algae, in: Algal biorefinery: An integrated approach. Springer Int. Publishing Cham. 1:125–149. doi: 10.1007/978-3-319-22813-6_6
Dobin, A., Davis, C. A., Schlesinger, F., Drenkow, J., Zaleski, C., Jha, S., et al. (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. doi: 10.1093/bioinformatics/bts635
EL-Sheekh, M. M., Bedaiwy, M. Y., Osman, M. E., Ismail, M. M. (2012). Mixotrophic and heterotrophic growth of some microalgae using extract of fungal-treated wheat bran. Int. J. Recycling Organic Waste Agric. 1:12, 1–9. doi: 10.1186/2251-7715-1-12
Houtman, C. J., Kitin, P., Houtman, J. C. D., Hammel, K. E., Hunt, C. G. (2016). Acridine orange indicates early oxidation of wood cell walls by fungi. PloS One 11, e0159715. doi: 10.1371/journal.pone.0159715
Kim, B. H., Ramanan, R., Cho, D. H., Oh, H. M., Kim, H. S. (2014). Role of rhizobium, a plant growth promoting bacterium, in enhancing algal biomass through mutualistic interaction. Biomass Bioenergy 69, 95–105. doi: 10.1016/J.BIOMBIOE.2014.07.015
Kitin, P., Nakaba, S., Hunt, C. G., Lim, S., Funada, R. (2020). Direct fluorescence imaging of lignocellulosic and suberized cell walls in roots and stems. AoB Plants 12, 1–19. doi: 10.1093/aobpla/plaa032
Klindworth, A., Pruesse, E., Schweer, T., Peplies, J., Quast, C., Horn, M., et al. (2013). Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 41, e1–e1. doi: 10.1093/nar/gks808
Lasek, R., Szuplewska, M., Mitura, M., Decewicz, P., Chmielowska, C., Pawłot, A., et al. (2018). Genome structure of the opportunistic pathogen paracoccus yeei (Alphaproteobacteria) and identification of putative virulence factors. Front. Microbiol. 9. doi: 10.3389/fmicb.2018.02553
Law, C. W., Chen, Y., Shi, W., Smyth, G. K. (2014). Voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 15, R29. doi: 10.1186/gb-2014-15-2-r29
Malhi, Y. (2002). Carbon in the atmosphere and terrestrial biosphere in the 21st century. Philos. Transact: Math Phys. Eng. Sci. 360, 2925–2945. doi: 10.1007/s12010-014-0729-1
Mandal, S., Mallick, N. (2009). Microalga scenedesmus obliquus as a potential source for biodiesel production. Appl. Microbiol. Biotechnol. 84, 281–291. doi: 10.1007/s00253-009-1935-6
McMurdie, P. J., Holmes, S. (2013). Phyloseq: An r package for reproducible interactive analysis and graphics of microbiome census data. PloS One 8, e61217. doi: 10.1371/journal.pone.0061217
Msanne, J., Polle, J., Starkenburg, S. (2020). An assessment of heterotrophy and mixotrophy in scenedesmus and its utilization in wastewater treatment. Algal Res. 48:101911, 1–7. doi: 10.1016/j.algal.2020.101911
Ritchie, M. E., Phipson, B., Wu, D., Hu, Y., Law, C. W., Shi, W., et al. (2015). Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47–e47. doi: 10.1093/nar/gkv007
Robinson, M. D., McCarthy, D. J., Smyth, G. K. (2010). edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. doi: 10.1093/bioinformatics/btp616
Rodríguez-Gacio, M., del, C., Iglesias-Fernández, R., Carbonero, P., Matilla, Á.J. (2012). Softening-up mannan-rich cell walls. J. Exp. Bot. 63, 3976–3988. doi: 10.1093/jxb/ers096
Schambach, J. Y., Finck, A. M., Kitin, P., Hunt, C. G., Hanschen, E. R., Vogler, B., et al. (2020). Growth, total lipid, and omega-3 fatty acid production by nannochloropsis spp. cultivated with raw plant substrate. Algal Res. 51:102041, 1–13. doi: 10.1016/j.algal.2020.102041Algal Research 51:102041, 1-13
Scheller, H. V., Ulvskov, P. (2010). Hemicelluloses. Annu. Rev. Plant Biol. 61, 263–289. doi: 10.1146/annurev-arplant-042809-112315
Sforza, E., Cipriani, R., Morosinotto, T., Bertucco, A., Giacometti, G. M.. (2012). Excess CO¬2 supply inhibits mixotrophic growth of Chlorella protothecoides and Nannochloropsis salina. Bioresource Technology, 104, 523–529. doi: 10.1016/j.biortech.2011.10.025
Shen, X. F., Hu, H., Ma, L. L., Lam, P. K. S., Yan, S. K., Zhou, S. B., et al. (2018). FAMEs production from: Scenedesmus obliquus in autotrophic, heterotrophic and mixotrophic cultures under different nitrogen conditions. Environ. Sci: Water Res. Technol. 4, 461–468. doi: 10.1039/c7ew00470b
Smith, R. T., Bangert, K., Wilkinson, S. J., Gilmour, D. J. (2015). Synergistic carbon metabolism in a fast growing mixotrophic freshwater microalgal species micractinium inermum. Biomass Bioenergy 82, 73–86. doi: 10.1016/j.biombioe.2015.04.023
Song, Y., Wang, X., Cui, H., Ji, C., Xue, J., Jia, X., et al. (2021). Enhancing growth and oil accumulation of a palmitoleic acid–rich scenedesmus obliquus in mixotrophic cultivation with acetate and its potential for ammonium-containing wastewater purification and biodiesel production. J. Environ. Manage. 297, 113273. doi: 10.1016/J.JENVMAN.2021.113273
Starkenburg, S. R., Polle, J. E. W., Hovde, B., Daligault, H. E., Davenport, K. W., Huang, A., et al. (2017). Draft nuclear genome, complete chloroplast genome, and complete mitochondrial genome for the Biofuel/Bioproduct feedstock species scenendesmus obliquus strain DOE0152z. Genome Announc 5 (32):1–2. doi: 10.1128/genomea.00617-17
Tanadul, O., VanderGheynst, J. S., Beckles, D. M., Powell, A. L. T., Labavitch, J. M. (2014). The impact of elevated CO 2 concentration on the quality of algal starch as a potential biofuel feedstock. Biotechnol. Bioengin 111, 1323–1331. doi: 10.1002/bit.25203
Terrasan, C. R. F., Guisan, J. M., Carmona, E. C. (2016). Xylanase and β-xylosidase from penicillium janczewskii: Purification, characterization and hydrolysis of substrates. Electronic J. Biotechnol. 23, 54–62. doi: 10.1016/J.EJBT.2016.08.001
Vogler, B. W., Starkenburg, S. R., Sudasinghe, N., Schambach, J. Y., Rollin, J. A., Pattathil, S., et al. (2018). Characterization of plant carbon substrate utilization by auxenochlorella protothecoides. Algal Res. 34, 37–48. doi: 10.1016/j.algal.2018.07.001
Voigt, J., Stolarczyk, A., Zych, M., Malec, P., Burczyk, J. (2014). The cell-wall glycoproteins of the green alga scenedesmus obliquus. the predominant cell-wall polypeptide of scenedesmus obliquus is related to the cell-wall glycoprotein gp3 of chlamydomonas reinhardtii. Plant Sci. 215–216, 39–47. doi: 10.1016/j.plantsci.2013.10.011
Wang, J., Yang, H., Wang, F. (2014). Mixotrophic cultivation of microalgae for biodiesel production: Status and prospects. Appl. Biochem. Biotechnol. 172, 3307–3329. doi: 10.1007/s12010-014-0729-1
Yang, S., Liu, G., Meng, Y., Wang, P., Zhou, S., Shang, H. (2014). Utilization of xylose as a carbon source for mixotrophic growth of scenedesmus obliquus. Bioresource Technol. 172, 180–185. doi: 10.1016/j.biortech.2014.08.122
Keywords: Scenedesmus, mixotrophy, plant substrate, metabolism, transcriptomics
Citation: Schambach JY, Kruse CPS, Kitin P, Mays W, Hunt CG, Starkenburg SR and Barry AN (2022) Metabolism of Scenedesmus obliquus cultivated with raw plant substrates. Front. Plant Sci. 13:992702. doi: 10.3389/fpls.2022.992702
Received: 12 July 2022; Accepted: 12 September 2022;
Published: 02 December 2022.
Edited by:
Ben Lucker, Prosel Biosciences, United StatesReviewed by:
Bo Wang, Vanderbilt University, United StatesJoshua Adam Temple, Michigan State University, United States
Copyright © 2022 Schambach, Kruse, Kitin, Mays, Hunt, Starkenburg and Barry. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Amanda N. Barry, YW5iYXJyeUBzYW5kaWEuZ292
 Jenna Y. Schambach
Jenna Y. Schambach Colin P. S. Kruse
Colin P. S. Kruse Peter Kitin3
Peter Kitin3 Wittney Mays
Wittney Mays Christopher G. Hunt
Christopher G. Hunt Shawn R. Starkenburg
Shawn R. Starkenburg Amanda N. Barry
Amanda N. Barry