- 1State key Laboratory of Crop Gene Exploration and Utilization in Southwest China, Sichuan Agricultural University, Chengdu, China
- 2College of Agronomy, Sichuan Agricultural University, Chengdu, China
Stripe rust and powdery mildew are devastating diseases that have severe effects on wheat production. Introducing resistant genes/loci from wheat-related species into the wheat genome is an important method to improve wheat resistance. Rye (Secale cereale L.) is a cross-pollinating plant and is the most important related species for wheat genetic improvement. In this study, we developed three 6RS ditelosomic addition lines, three 6RL ditelosomic addition lines, and two 6R disomic addition lines by crossing common wheat cultivar Chuannong 25 and rye inbred line QL2. The chromosome composition of all new lines was confirmed by non-denaturing fluorescence in situ hybridization (ND-FISH) and molecular marker analyses. Disease responses to different Puccinia striiformis f. sp. tritici (Pst) races and Blumeria graminis f. sp. tritici (Bgt) isolates and cytogenetic analysis showed that the resistance of the new lines was derived from the rye chromosome 6R of QL2, and both arms (6RS and 6RL) may harbor resistance genes against Pst and Bgt. These new lines could be used as a promising bridging parent and valuable genetic resource for wheat disease resistance improvement.
Introduction
Wheat (Triticum aestivum L.) is one of the most important food crops in the world and provides ~20% of the protein and calories for human beings (Chaves et al., 2013). However, stripe rust (caused by Puccinia striiformis f. sp. tritici, Pst) and powdery mildew (Blumeria graminis f. sp. tritici, Bgt) are typically devastating diseases and are very detrimental to wheat production in subtropical agricultural zones (Duveiller et al., 2007). Stripe rust and powdery mildew result in the loss of ~5% of production. However, in some of the most prevalent areas, stripe rust and powdery mildew can increase the loss to 20%–30% of production (Ren et al., 2009; Chen et al., 2014). For example, serious outbreaks of stripe rust resulted in a 25% yield loss in the United States from 1958 to 2014 (Chen, 2005, 2014). In China, stripe rust is epidemic almost every year and causes significant yield losses (Wan et al., 2007; Ren et al., 2022). In recent years, with the change in global climate, powdery mildew has affected wheat production more (Huo et al., 2002). For example, in 1990 and 1991, more than 12 million hectares of wheat were attacked by Bgt in China, causing 1.4 million and 0.7 million tons of yield losses, respectively (Hou et al., 2000).
Breeding disease-resistant wheat cultivars represent the most economical, environmentally friendly, and effective control measures at present. Related species of wheat, such as rye (Secale cereale; Schlegel and Korzun, 1997), Haynaldia villosa (Chen et al., 1995), Elytrigia intermedium (Luo et al., 2009), and Agropyron cristatum (Wang et al., 2022), contain a large number of resistance genes and have been used in wheat disease resistance breeding for a long time. Among them, the most valuable related species is rye (Schlegel and Korzun, 1997; Ren et al., 2022). Disease-resistance genes for Pst and Bgt, such as Yr9, Pm8, and Pm17, were introduced into the wheat genome via T1RS·1BL or T1RS·1AL translocation (Mater et al., 2004; Mago et al., 2005). However, due to the changes in the prevalent pathogens, these resistance genes were overcome by new pathogens at the end of the last century (Ren et al., 2009). In the past 10 years, several new 1RS chromosomes have been introduced into the wheat genome and have exhibited resistance to the new Pst and Bgt pathogens (Ren et al., 2017a, 2018, 2022; Han et al., 2020). Additionally, 2R, 4R, 5R, 6R, and 7R chromosomes are also introduced into the wheat genome in the form of chromosome translocation, addition, and substitution, and several of these newly developed lines exhibit resistance to stripe rust or powdery mildew (An et al., 2015, 2019; Li et al., 2016; Schneider et al., 2016; Ren et al., 2017b, 2020; Johansson et al., 2020; Han et al., 2022).
In this study, eight new addition lines, including three 6RS ditelosomic addition lines, three 6RL ditelosomic addition lines, and two 6R disomic addition lines, were developed and selected from a cross between the rye inbred line QL2 and wheat cultivar CN25. All eight addition lines showed high resistance to both Pst and Bgt. The results indicated that both the long arm and short arm of QL2 may contain resistance genes for Pst and Bgt. These eight new lines could represent valuable genetic resources for wheat disease resistance breeding programs in the future.
Materials and methods
Plant materials
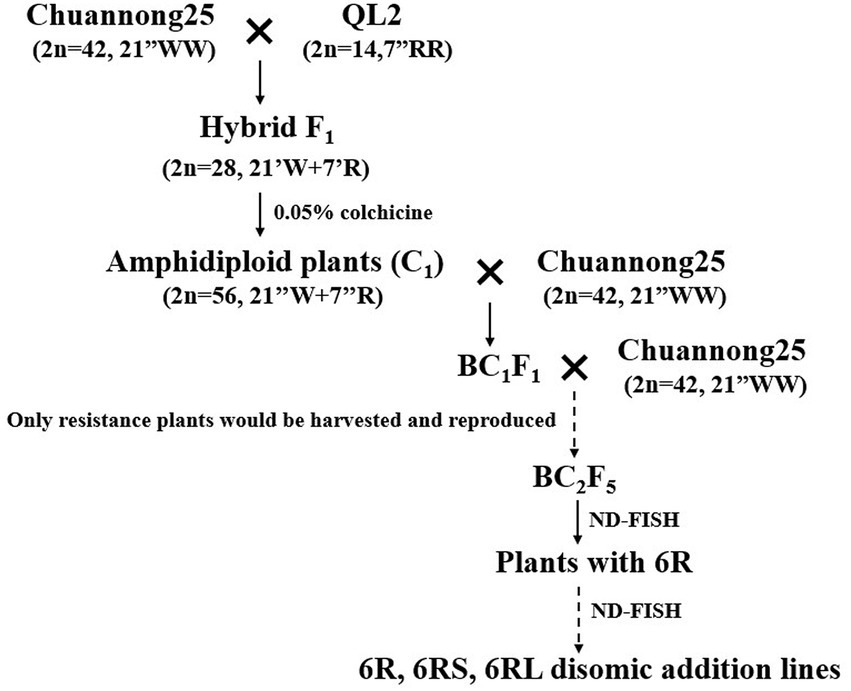
Chuannong 25 (CN25) is a high-yield wheat cultivar that was approved by the Sichuan Provincial Variety Examination and Committee in 2007 and has been widely planted in southwestern China. The pure genetic stocks of CN25 used in this study were bred by single-spike descent over several generations. Rye Qinling was collected by our lab in the 1990s. An inbred line of Qinling rye, QL2, was used in this study. CN25 was crossed directly with QL2 to produce hybrid F1 seeds. Then, the F1 seedlings were treated with 0.05% colchicine plus 3% dimethyl sulfoxide for 8 h to produce the amphidiploid (C1). The C1 plants were backcrossed with wheat parent CN25 twice to produce the BC2F1 plants. From now on, only the plants that showed resistance to diseases in the field were harvested and reproduced in the generations BC2F1 to BC2F4. In the generation BC2F5, the chromosome composition of the plants was examined by non-denaturing fluorescence in situ hybridization (ND-FISH), and then, the plants were transplanted into the field. In this generation, several plants with rye 6R chromosomes were identified. The seeds of these plants were harvested and reproduced by selfing. In BC2F6, 50 seeds were randomly selected from each line and cultured in the lab first, and the chromosome composition of the plants was determined by ND-FISH. Then, all surviving plants were transplanted into the field. From BC2F6 to BC2F8, 6R disomic addition plants, 6RS ditelosomic addition plants, and 6RL ditelosomic addition plants were identified.
Cytogenetic and molecular analyses
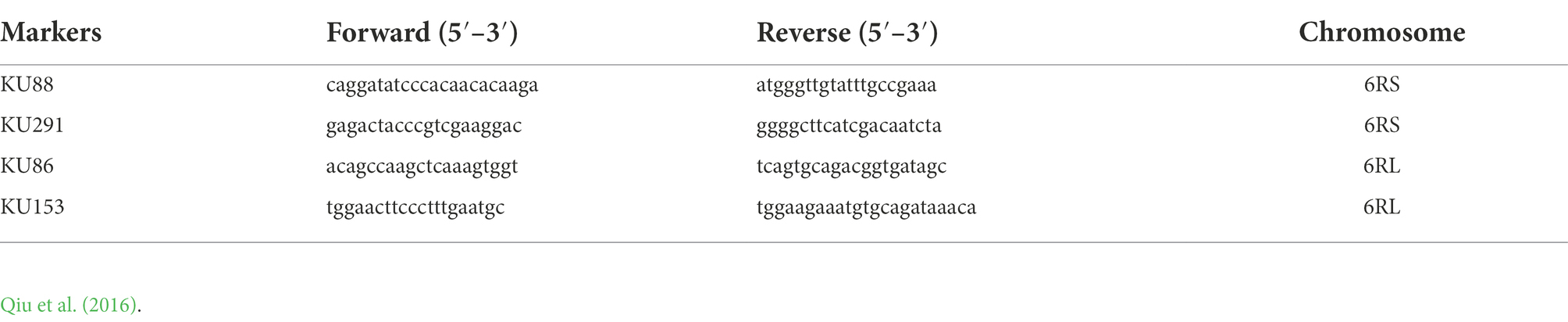
ND-FISH was used to identify the chromosome composition of the plant materials. Five oligonucleotide probes, Oligo-pSc119.2-1, Oligo-pTa535-1, Oligo-Ku, Oligo-pSc200, and Oligo-pSc250, were used in this study (Tang et al., 2014; Ren et al., 2019). The sequences and chromosomes that could be identified by these five probes are listed in Table 1. The wheat and rye chromosomes could be accurately distinguished by the combination of these five probes in one cell. Moreover, the centromeric-specific probe Oligo-CCS1, rye centromeric-specific probe Oligo-PAWRC.1, and telomere-specific probe Oligo-Telo were mixed and used in another ND-FISH experiment (Cuadrado and Jouve, 2010; Tang et al., 2014). Cell images were captured using an epifluorescence microscope (model BX51, Olympus, Center Valley, PA, United States) equipped with a cooled charge-coupled device camera and operated with the software program HCIMAGE Live (version 2.0.1.5, Hamamatsu Corp., Sewickley, PA, United States). The processes of sample preparation (root tips), probe labeling, and ND-FISH were performed according to Tang et al. (2014) and Ren et al. (2019). Molecular markers specific to 6RL and 6RS were also used to confirm the chromosome. The genomic DNA of the plant materials was isolated from young leaves using the surfactant cetyltrimethylammonium bromide (CTAB; Doyle and Doyle, 1987). Two primer pairs, KU88 and KU291, which were specific for the rye 6RS chromosome arm, and two primer pairs, KU86 and KU153, which were specific for the rye 6RL chromosome arm, were used (Qiu et al., 2016; Table 2). These primers can detect rye 6RS or 6RL chromosome arms in the wheat genetic background and can amplify specific ~400-bp fragment bands (Qiu et al., 2016). PCR and electrophoresis were performed according to Qiu et al. (2016).
Stripe rust and powdery mildew tests
The Pst races CYR32 (virulent to Yr1, 2, 3, 4, 6, 7, 8, 9, 17, 25, 27, 28, 31, 32, 43, 44, A, Alba, Cle, Gaby, Res, SD, SO, Exp2, SK, and SP), CYR33 (virulent to Yr1, 2, 3, 4, 6, 7, 8, 9, 17, 25, 28, 31, 32, A, and Su), and CYR34 [virulent to Yr1, 2, 3, 4, 6, 7, 8, 9, 10, 17, 19, 24(=26), 25, 27, 28, 31, 32, 43, 44, Exp2, SP, A, and Sk] were used in this experiment (Ren et al., 2022). These three Pst races were considered the most virulent and frequent Pst races occurring in China and were used in stripe rust resistance tests at the seedling stage (Ren et al., 2022). The inoculation of Pst on wheat seedlings was performed according to Ren et al. (2020). Two Bgt isolates, E20 and No. 15, were used in the powdery mildew resistance tests at the seedling stage. Both Bgt isolates have been virulent to many newly released cultivars in China in recent years (Ren et al., 2017a; Yang et al., 2021). The inoculation of Bgt on wheat seedlings was performed according to Yang et al. (2021). The three Pst races were provided by the Plant Protection Institute, Gansu Academy of Agricultural Sciences, China. The Bgt isolate E20 was provided by the Department of Plant Protection, Sichuan Agricultural University. The Bgt isolate No. 15 was collected by our lab in Ya’an City, Sichuan, China (Ren et al., 2018). The inoculation of Pst and Bgt on wheat seedlings was performed with three replications in the greenhouse. In addition, the disease resistance of all materials identified in this study was tested in the field under severe natural Pst and Bgt infection at the Qionglai Research Station of Sichuan Agricultural University in Cheng Plain, Southwest China (30°25′N, 103°28′E) from 2020 to 2022. The field experiments were performed according to the methods described previously (Ren et al., 2020, 2022). The wheat parent CN25, rye parent QL2, wheat cultivar Mianyang 11, and RT1104-1 were served as controls. Stripe rust reactions were scored at the grain filling stage as infection types (ITs) based on a 0–9 scale as described by Wan et al. (2004) and Ren et al. (2022). Powdery mildew reactions were scored at the heading stage as ITs based on a 0–4 scale as described by Xie et al. (2004) and Ren et al. (2020).
Results
Development of disomic addition lines
The pedigrees of the 6R disomic addition lines, 6RS ditelosomic addition lines, and 6RL ditelosomic addition lines are displayed in Figure 1. The wheat parent CN25 (2n = 42, 21″WW) was directly crossed with QL2 (2n = 14, 7″RR), and several F0 seeds (2n = 28, 21″W + 7″R) were obtained. The chromosome numbers of F1 plants were doubled by colchicine treatment to produce amphidiploid (C0) seeds (2n = 56, 21″WW + 7″RR; Figure 2A). The amphidiploid plants were then backcrossed with the wheat parent CN25 twice to produce the BC2F1 plants. The continued generations were reproduced by selfing, and only the plants that exhibited resistance to diseases were harvested and bred. In the BC2F5 generation, the chromosome composition of the plants was examined by ND-FISH during the seedling stage, and then, the plants were transplanted into the field. In this generation, several plants with rye 6R or partial 6R were identified, and these plants also showed resistance in the field. The seeds of these plants containing alien chromatin were harvested and reproduced by selfing. The seedlings of BC2F6 were also identified by ND-FISH. In this generation, several homozygous 6R disomic addition lines, 6RS ditelosomic addition lines, and 6RL ditelosomic addition lines were identified. On the other hand, several plants were heterozygous or missed the 6R chromosomes. When plants missed the 6R, they also lose disease resistance in the field. The homozygous addition lines were harvested and reproduced the next generation by selfing. The heterozygous plants were also harvested, and we continued to screen the homozygous plants in their progeny in the generation of BC2F7 to BC2F8. Finally, three 6RS ditelosomic addition lines (46-13-3, 46-13-11, and 46-13-16), three 6RL ditelosomic addition lines (48-9-2, 48-9-5, and 48-9-6), and two 6R disomic addition lines (48-9-8 and 48-9-10) were selected.
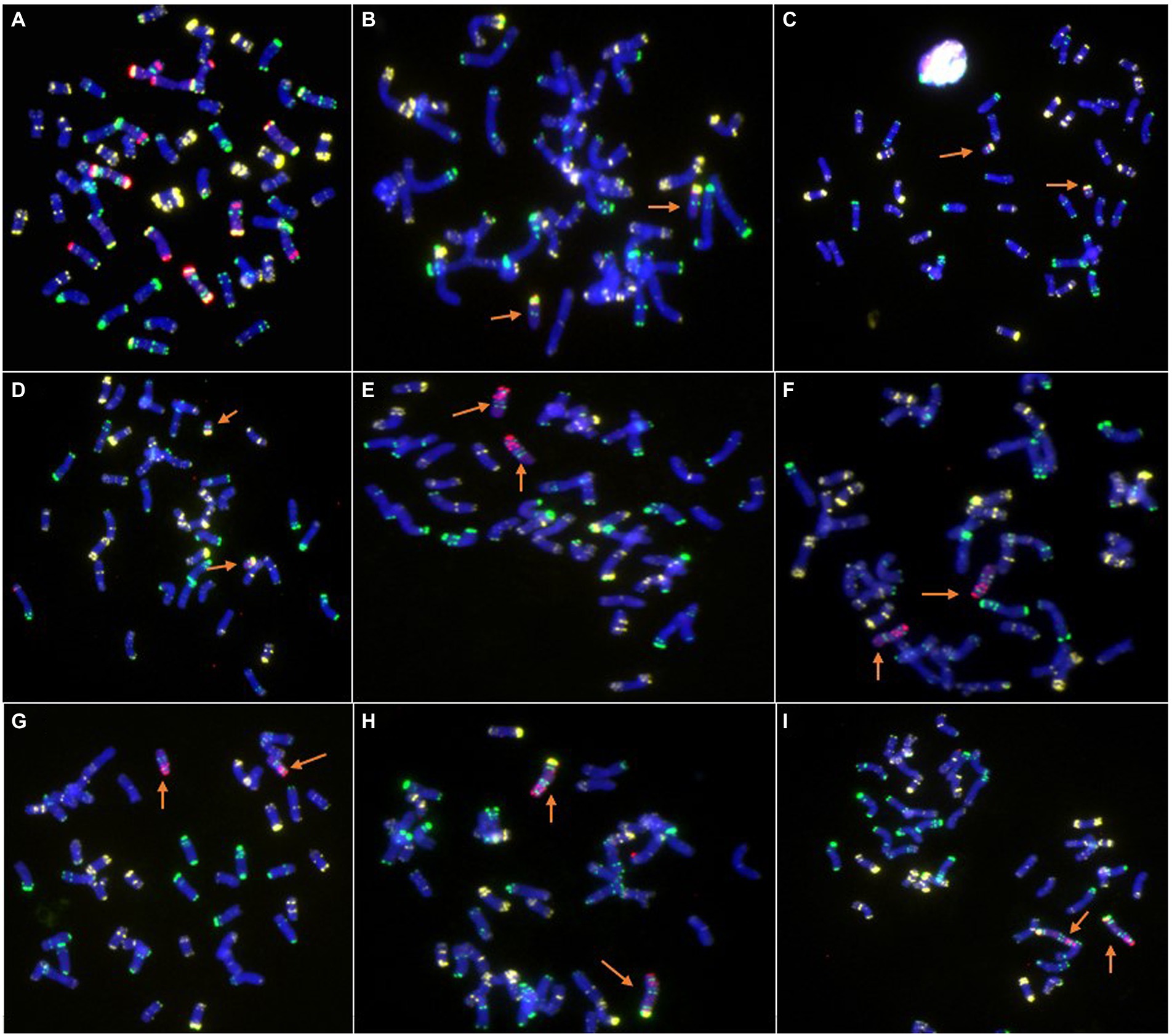
Figure 2. Cytogenetic analysis of new wheat-rye lines by ND-FISH. (A) Amphidiploid plants (2n = 56, 21″WW + 7″RR). (B) 46-13-3, 6RS ditelosomic addition line. (C) 46-13-11, 6RS ditelosomic addition line. (D) 46-13-16, 6RS ditelosomic addition line. (E) 48-9-2, 6RL ditelosomic addition line. (F) 48-9-5, 6RL ditelosomic addition line. (G) 48-9-6, 6RL ditelosomic addition line. (H) 48-9-8, 6R disomic addition line. (I) 48-9-10, 6R disomic addition line. The arrows show the 6R, 6RL, or 6RS chromosomes. Oligo-pSc119.2-1: green; Oligo-pTa535-1: white; Oligo-Ku, Oligo-pSc200, and Oligo-pSc250: red.
Chromosome identification
The combination of the five probes Oligo-pSc119.2-1, Oligo-pTa535-1, Oligo-Ku, Oligo-pSc200, and Oligo-pSc250 can easily and accurately distinguish wheat and rye chromosomes in one cell. The ND-FISH results indicated that three lines, 46-13-3, 46-13-11, and 46-13-16, contained a pair of 6RS chromosome arms (Figures 2B–D). Three lines, 48-9-2, 48-9-5, and 48-9-6, contained a pair of 6RL chromosome arms (Figures 2E–G). Two lines, 48-9-8 and 48-9-10, contained a pair of 6R chromosomes (Figures 2H,I).
The centromere and telomere structure of the new lines was identified using the combinations of the probes Oligo-CCS1, Oligo-PAWRC.1, and Oligo-Telo. The Oligo-CCS1 probe can detect both wheat and rye centromeres, whereas Oligo-PAWRC.1 can detect only rye centromeres. Therefore, the centromere of the 6R chromosome showed both signal patterns of Oligo-PAWRC.1 and Oligo-CCS1 (Figures 3E,F). The probe Oligo-Telo can detect the structure of the telomere and can also detect the telocentric chromosome. Therefore, the centromere of the 6RS and 6RL chromosome arms can show both signal patterns of the probes Oligo-PAWRC.1 and Oligo-Telo. The ND-FISH results showed that both 6RS and 6RL chromosomes showed complex signal patterns of Oligo-PAWRC.1 and Oligo-Telo (Figures 3A–D).
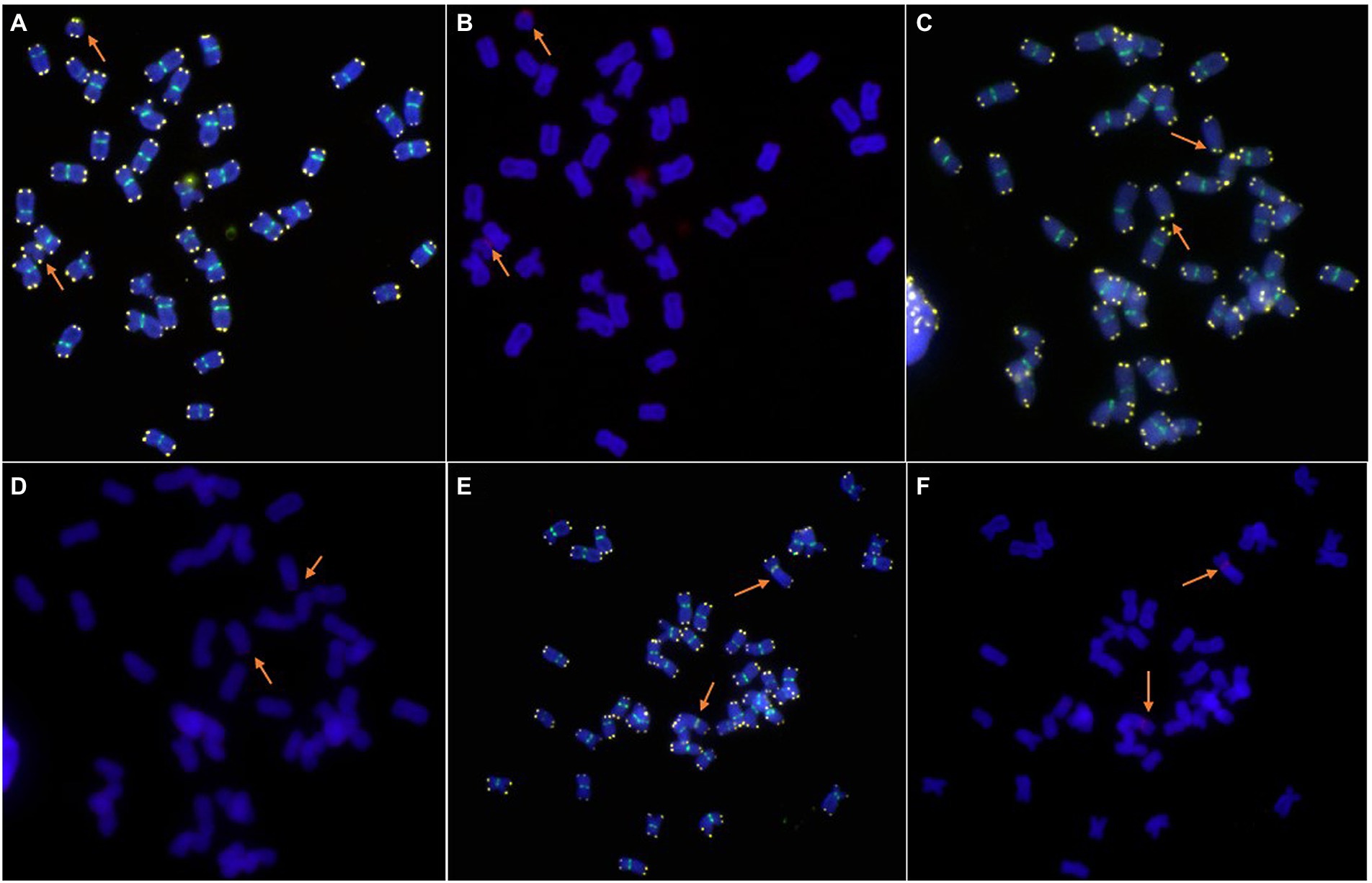
Figure 3. Cytogenetic analysis of the telomeres and centromeres of new lines by ND-FISH. (A,B): 6RS ditelosomic addition line. The signal patterns were derived from one cell. (C,D): 6RL ditelosomic addition line. The signal patterns were derived from one cell. (E,F): 6R disomic addition line. The signal patterns were derived from one cell. Oligo-CCS1 (green), Oligo-PAWRC.1 (red), and Oligo-Telo (white). The arrows show the 6RS, 6RL, or 6R chromosomes.
The chromosome composition of all lines was also confirmed by specific molecular markers. Two primer pairs, KU88 and KU291, were specific for the rye 6RS chromosome arm and amplified a specific band of ~400 bp. The PCR results showed that all DNA of the 6RS ditelosomic and 6R disomic addition lines amplified the expected bands at ~400 bp, whereas the bands were absent from the DNA of the 6RL ditelosomic addition lines and wheat CN25 (Figures 4A,B). On the other hand, the primer pairs KU86 and KU153 were specific for the rye 6RL chromosome arm and could also amplify a specific band at ~400 bp. The PCR results showed that all DNA of 6RL ditelosomic and 6R disomic addition lines can amplify the expected bands of ~400 bp, whereas the bands were absent from the DNA of 6RS ditelosomic addition lines and wheat CN25 (Figures 4C,D; Table 2).
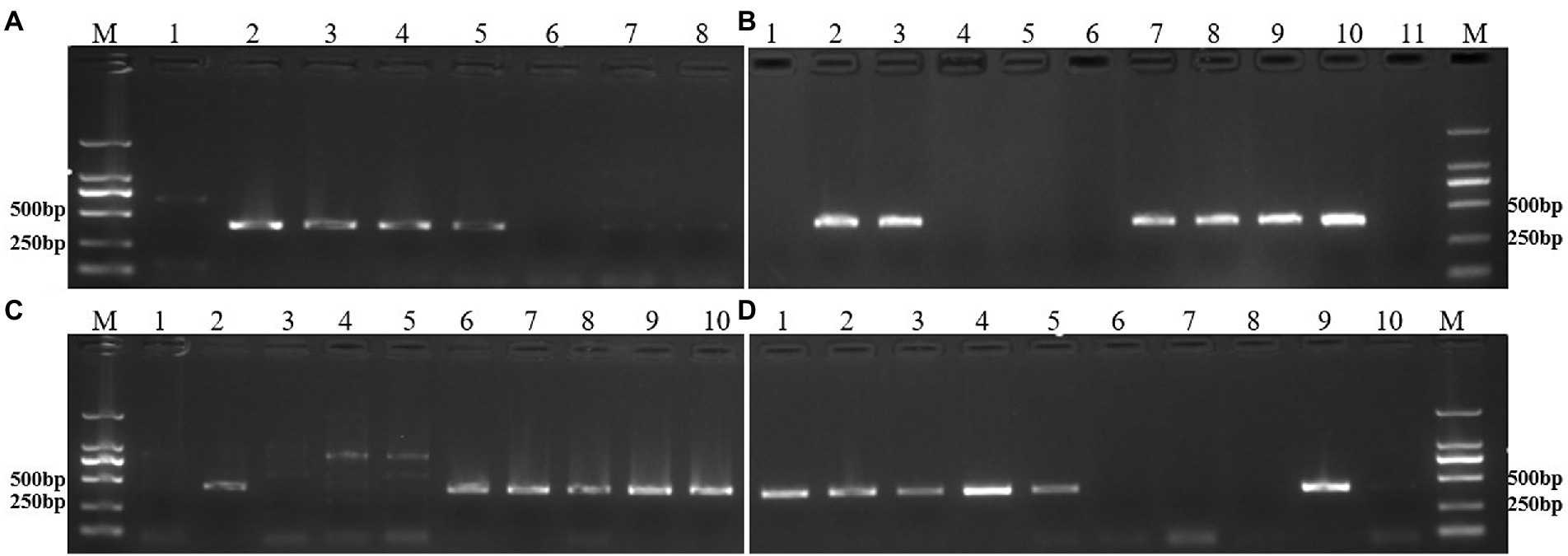
Figure 4. PCR results of specific molecular markers. (A) PCR results of the primer pair KU88. Lane 1: CN25, Lane 2: 46-13-3 (6RS), Lane 3: 46-13-11 (6RS), Lane 4: 46-13-16 (6RS), Lane 5: rye QL2, Lane 6: 48-9-2 (6RL), Lane 7: 48-9-5 (6RL), Lane 8: 48-9-6 (6RL). (B) PCR results of the primer pair KU291. Lane 1: CN25, Lane 2: 48-9-8 (6R), Lane 3: 48-9-10 (6R), Lane 4: 48-9-2 (6RL), Lane 5: 48-9-5 (6RL), Lane 6: 48-9-6 (6RL), Lane 7: 46-13-3 (6RS), Lane 8: 46-13-11 (6RS), Lane 9: 46-13-16 (6RS), Lane 10: rye QL2, Lane 11: CN25. (C) PCR results of the primer pair KU86. Lane 1: CN25, Lane 2: rye QL2, Lane 3: 46-13-3 (6RS), Lane 4: 46-13-11 (6RS), Lane 5: 46-13-16 (6RS), Lane 6: 48-9-2 (6RL), Lane 7: 48-9-5 (6RL), Lane 8: 48-9-6 (6RL), Lane 9: 48-9-8 (6R), Lane 10: 48-9-10 (6R). (D) PCR results of the primer pair KU153. Lane 1: 48-9-2 (6RL), Lane 2: 48-9-5 (6RL), Lane 3: 48-9-6 (6RL), Lane 4: 48-9-8 (6R), Lane 5: 48-9-10 (6R), Lane 6: 46-13-3 (6RS), Lane 7: 46-13-11 (6RS), Lane 8: 46-13-16 (6RS), Lane 9: rye QL2, Lane 10: CN25. Lane M: DNA marker DL2000. 6RS: 6RS ditelosomic addition lines; 6RL: 6RL ditelosomic addition lines; 6R: 6R disomic addition lines.
Resistance to stripe rust and powdery mildew
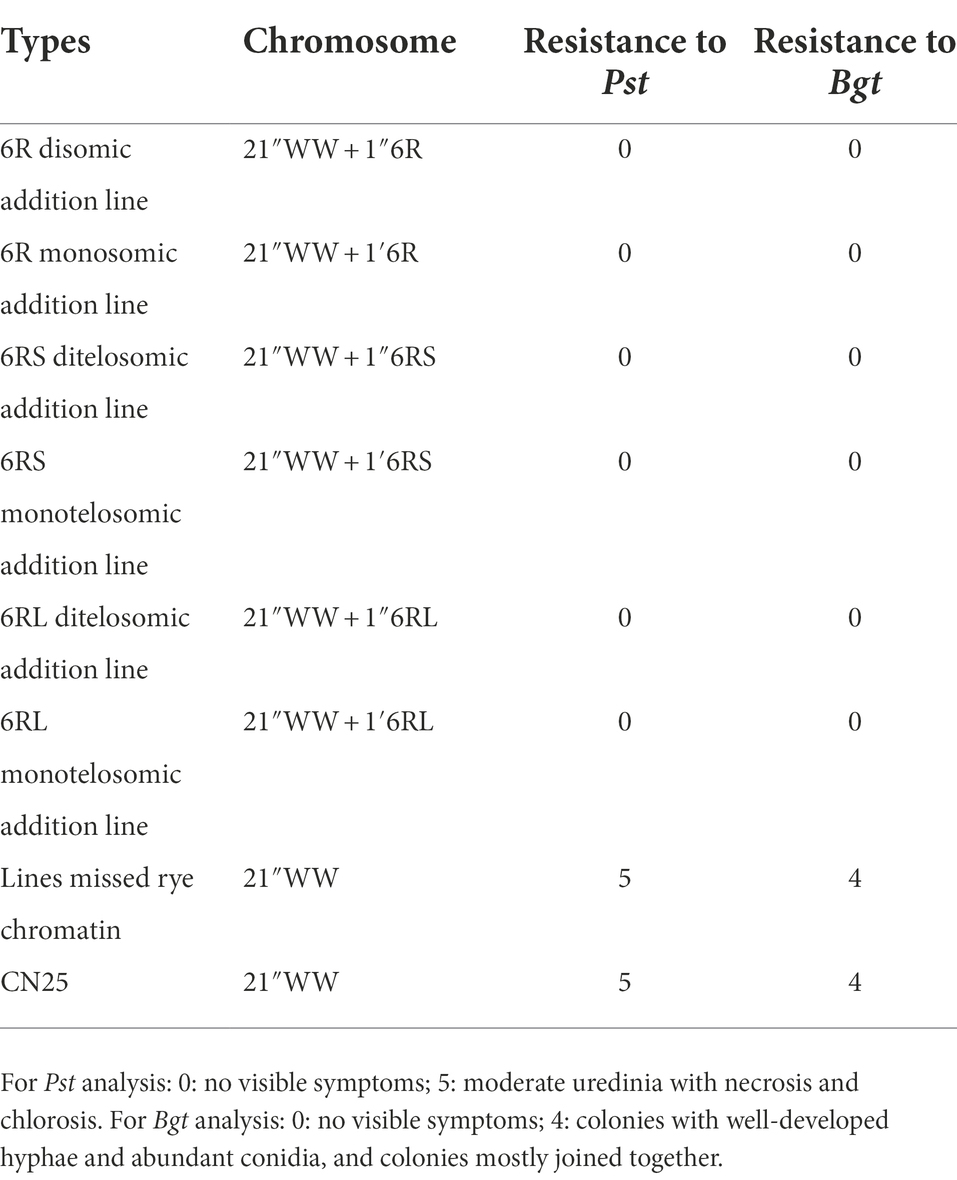
The wheat parent CN25 was intermediately resistant to two Pst races (CYR32 and 34) and highly susceptible to the Bgt isolate E20 and No. 15. CN25 also exhibited intermediate resistance to stripe rust and was highly susceptible to powdery mildew in the field (Table 3). On the other hand, inbred rye line QL2 was highly resistant to three Pst races and two Bgt isolates. QL2 also exhibited high resistance to stripe rust and powdery mildew in the field (Table 3). The control Mianyang 11 was highly susceptible to three Pst races and two Bgt isolates, and also exhibited susceptible to stripe rust and powdery mildew in the field. The control RT1104-1 exhibited resistance to three Pst races and two Bgt isolates, and also showed resistance in the field (Table 3). The 6R disomic, 6RS ditelosomic, and 6RL ditelosomic addition lines showed a high level of resistance to three Pst races and two Bgt isolates at the seedling stages. In addition, these eight lines also showed high resistance to stripe rust and powdery mildew in the field at the adult stage (Table 3).
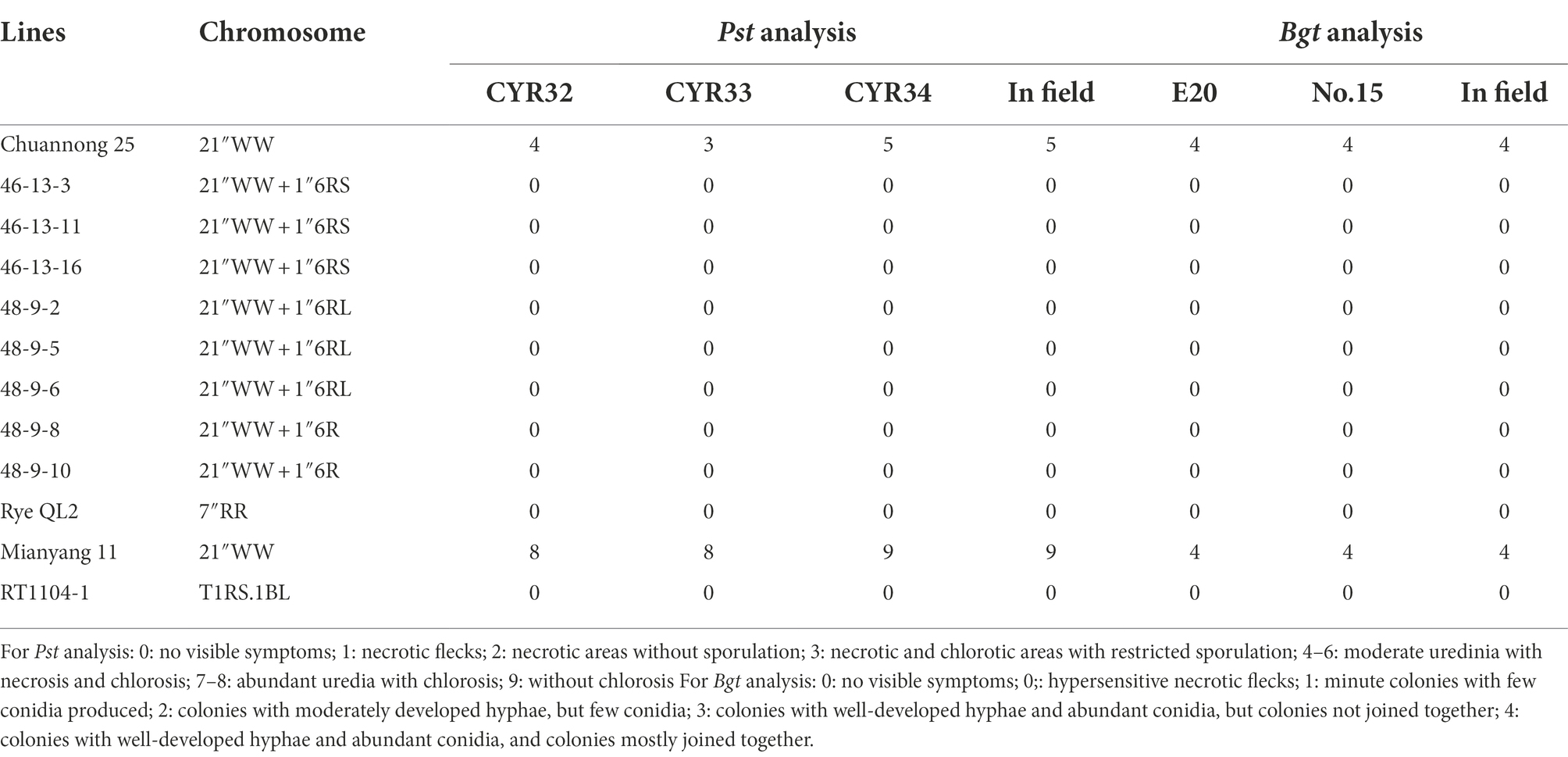
Table 3. Analysis of resistance to stripe rust and powdery mildew when inoculated with prevalent races/isolates of stripe rust and powdery mildew.
Moreover, in addition to these homozygous 6R, 6RS, and 6RL disomic or ditelosomic addition lines, several other types, such as 6R monosomic addition (Figure 5A), 6RL monotelosomic addition (Figure 5B), and 6RS monotelosomic addition (Figure 5C), and several plants that missed 6R, 6RL, or 6RS (Figure 5D), were also identified in the generation of BC2F7–BC2F8. Without exception, plants exhibit disease resistance as long as they have 6R, 6RS, or 6RL in their genome, but disease resistance was lost when 6R, 6RS, or 6RL was missed (Table 4).
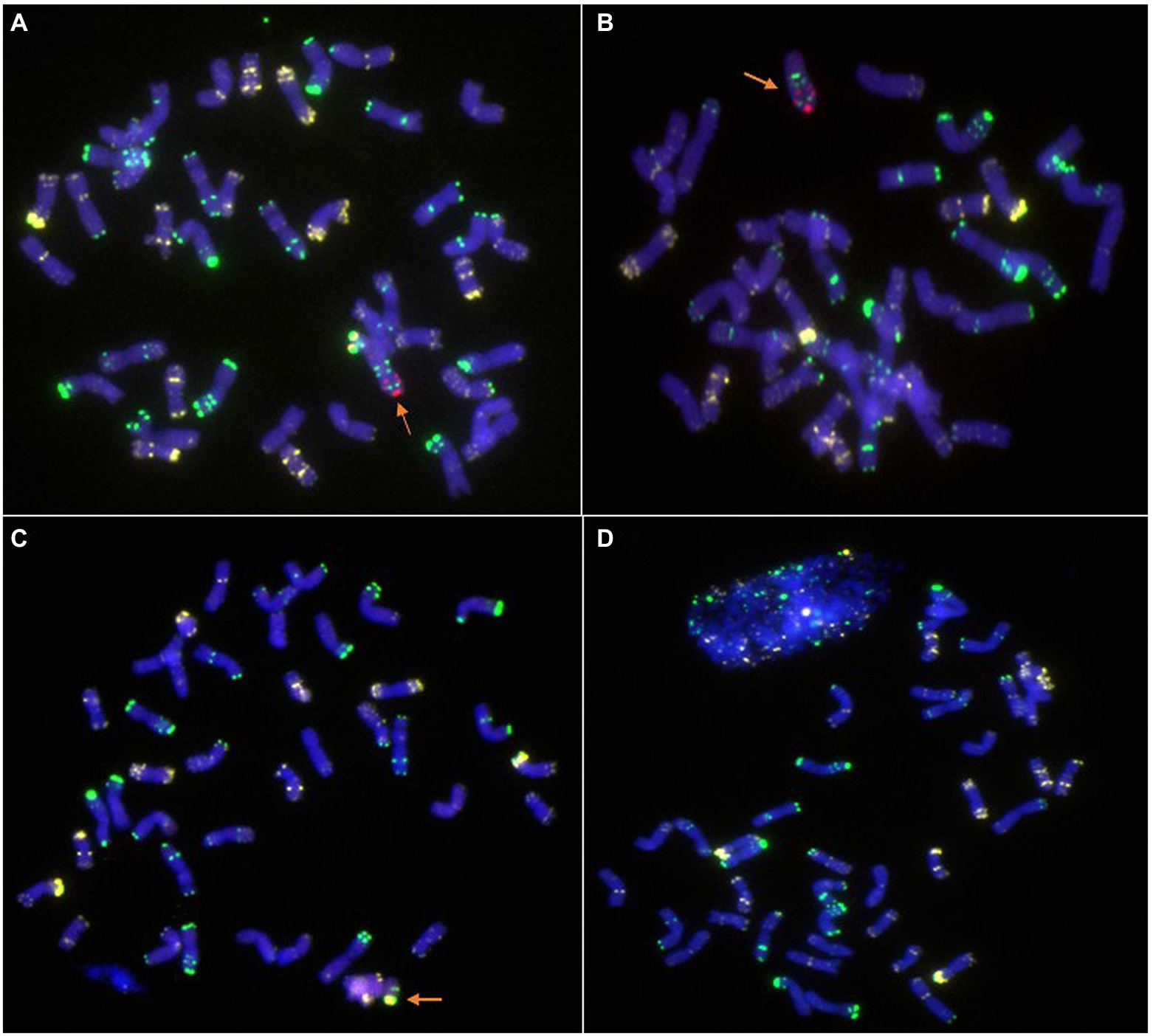
Figure 5. Lines with other types identified by ND-FISH. (A) 6R monosomic addition. (B) 6RL monotelosomic addition. (C) 6RS monotelosomic addition. (D) Lines that missed 6R. The arrows show the 6R, 6RL, or 6RS chromosomes. Oligo-pSc119.2-1: green; Oligo-pTa535-1: white; Oligo-Ku, Oligo-pSc200, and Oligo-pSc250: red.
Discussion
6R chromosome of Qinling rye provides resistance to Pst and Bgt
Related species of wheat are very important for the improvement of wheat genetics (Friebe et al., 1996), and rye is the most important species. According to Weining rye genome sequencing data, 1,909 disease resistance-associated (DRA) genes were identified in the rye genome (Li et al., 2021). Several useful resistance genes were introduced into the wheat genome through wheat-rye distance crosses. For example, Yr9, Pm8, Pm17, Sr31, and Lr26 were derived from the 1RS chromosome arm through T1RS·1BL or T1RS·1AL chromosome translocation (Mago et al., 2005). Sr59 and H21 were derived from the 2RL chromosome arm through T2RL.2BS or T2RL.2DS chromosome translocation (Friebe et al., 1990; Rahmatov et al., 2016). Sr27 was derived from the 3RS chromosome arm through the T3RS.3BL chromosome translocation (Marais and Marais, 1994). Many other rye chromosomes with resistance have been introduced into the wheat genome. For example, Ren et al. reported (Ren et al., 2020) a T7RL.7BS translocation line with resistance to Pst, Bgt, and Fusarium head blight. An et al. reported (An et al., 2019) a 4R disomic addition line with resistance to Pst, Bgt, and sharp eyespot. The 6R chromosome of rye also contains many excellent resistance genes and plays an important role in wheat genetic improvement. According to Weining rye genome sequencing data, 287 disease resistance-associated genes mapped to the 6R chromosome (Li et al., 2021). To date, stripe rust resistance gene Yr83 has been mapped on the 6RL chromosome arm (Li et al., 2020), and powdery mildew stripe rust gene Pm56 has been mapped on the 6RS chromosome arm (Hao et al., 2018). Several other 6R chromosomes have been introduced into the wheat genome. For example, Han et al. (2022) reported a 6R addition line originating from a cross between octoploid triticale and common wheat that exhibited resistance to powdery mildew. Duan et al. (2022) reported a T6RL.6BS translocation line with resistance to stripe rust and originating from rye AR106BONE. Du et al. (2018) reported a 6RL/6D small segment translocation line with powdery mildew resistance that originated from rye Kustro. Schneider et al. (2016) reported a 6R disomic addition line with resistance to stripe rust originating from Secale cereanum. An et al. (2015) reported a 6R disomic addition line with resistance to powdery mildew originating from the rye cultivar., German White. However, most of these new lines were resistant to only one disease. A few new lines exhibit resistance to both stripe rust and powdery mildew. In this study, three 6RS ditelosomic addition lines, three 6RL ditelosomic addition lines, and two 6R disomic addition lines were selected from the cross between wheat CN25 and rye QL2 (Figures 1–3). All eight new lines exhibited high resistance to both stripe rust and powdery mildew (Table 3). Once 6RS or 6RL chromosome arms were added to the wheat genome, resistance to stripe rust and powdery mildew was demonstrated. If 6R chromosomes were missed in the offspring, the resistance was also lost (Figure 5; Table 4). The results indicated that both the 6RS and 6RL chromosome arms of QL2 contained gene(s) associated with resistance to stripe rust and powdery mildew.
Genetic diversity of rye resistance genes
Rye has great potential as a source for favorable genes, such as genes supporting higher yield, stress resistance, disease resistance, and other agronomic traits desirable for wheat genetic improvement (Ren et al., 2011). Rye is a wind-pollinated plant, and high levels of genetic diversity are found not only between cultivars originating from different regions but also within the same cultivar (Persson and von Bothmer, 2002; Ren et al., 2011). A large number of disease resistance genes may also be present in the rye genome (Ren et al., 2022). The results of genome sequencing of Weining rye also revealed that there were many DRA genes that existed in the rye genome and were distributed to seven chromosomes with NBS (nucleotide-binding site), RLK (receptor-like kinase), RLP (receptor-like protein), and CC-TM (coiled-coil plus transmembrane receptor) types (Li et al., 2021). The numbers of these DRA genes along the seven assembled chromosomes of Weining rye (1R to 7R) were 242, 296, 301, 301, 255, 287, and 227, respectively (Li et al., 2021). Considering the crucial importance of DRA genes in plant responses to biotic adversities (Kourelis and van der Hoorn, 2018; Li et al., 2021), they may facilitate efficient genetic studies and molecular improvement of disease resistance in wheat. For example, Ren et al. (2022) reported 166 different T1RS·1BL translocation lines, which originated from three rye cultivars. These T1RS·1BL translocation lines exhibited different resistance patterns when they were tested by different Pst races. The stripe rust resistance gene Yr9 originated from Petkus rye (Mago et al., 2005). Shi et al. (2001) found an allele of Yr9 that exhibited different resistance patterns to the tested Pst pathotypes from a T1RS·1BL line Weique, which also originated from Petkus rye. Ren et al. (2009) reported the Pst resistance gene YrCn17 from the T1RS·1BL cultivar CN17, which originated from an inbred line of Petkus rye, L155. The powdery mildew resistance gene Pm8 was derived from Petkus rye (Mater et al., 2004), and another Bgt resistance gene Pm17 was also mapped on the 1RS chromosome of Amigo, which originated from rye Insave (Mater et al., 2004). Ren et al. (2009) also reported a Bgt resistance gene, PmCn17, from cultivar CN17. In recent years, more 1RS chromosomes have carried more disease-resistance genes into the wheat genome and played a positive role in wheat disease-resistance breeding. Similar to 1RS chromosomes, the other six chromosomes of rye (2R–6R) also contain a large number of DRA genes (Li et al., 2021), which has great potential in wheat disease-resistance breeding in the future. To date, the stripe rust resistance gene Yr83 has been mapped to 6RL, and the powdery mildew resistance gene Pm56 has been mapped to 6RS (Hao et al., 2018; Li et al., 2020). Obviously, there is also abundant genetic diversity on the 6R chromosome, which may contain more disease-resistance genes for wheat genetic improvement. In this study, six monotelosomic addition lines and two disomic addition lines were identified from the distance cross between rye line QL2 and wheat cultivar CN25. The resistance analysis showed that all new lines exhibited high resistance to Pst and Bgt. This finding indicated that similar to 1RS, high genetic diversity of resistance genes existed on the 6RS or the 6RL chromosome arms. The genetic diversity of resistance genes on alien chromosomes could be transferred into the wheat genome to increase the genetic diversity of resistance genes in wheat (Ren et al., 2022). Further utilization of the genetic diversity of resistance genes is an important method to further utilize rye resources to improve wheat genetics. These new lines could be used as a promising bridging parent and valuable genetic resource for wheat disease resistance improvement.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding authors.
Author contributions
TR and ZL: conceived and designed the study. TR and ZR: created the materials. TR, ZR, and ZL: analyzed the data. TR: wrote the manuscript. ZS, YH, FT, and PL: performed the experiments and analyzed the data. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (31801357) and the Foundation of Sichuan Province Science and Technology Support Program (2019YJ0510, 2021YJ0509, and 2021JDRC0127).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
An, D., Ma, P., Zheng, Q., Fu, S., Li, L., Han, F., et al. (2019). Development and molecular cytogenetic identification of a new wheat-rye 4R chromosome disomic addition line with resistances to powdery mildew, stripe rust and sharp eyespot. Theor. Appl. Genet. 132, 257–272. doi: 10.1007/s00122-018-3214-3
An, D. G., Zheng, Q., Luo, Q. L., Ma, P. T., Zhang, H. X., Li, L. H., et al. (2015). Molecular cytogenetic identification of a new wheat-rye 6R chromosome disomic addition line with powdery mildew resistance. PLoS One 10:e0134534. doi: 10.1371/journal.pone.0134534
Chaves, M. S., Martinelli, J. A., Wesp-Guterres, C., Graichen, F. A., Brammer, S. P., Scagliusi, S. M., et al. (2013). The importance for food security of maintaining rust resistance in wheat. Food Secur. 5, 157–176. doi: 10.1007/s12571-013-0248-x
Chen, X. M. (2005). Epidemiology and control of stripe rust (Puccinia striiformis f. sp. tritici) on wheat. Can. J. Plant Pathol. 27, 314–337. doi: 10.1080/07060660509507230
Chen, X. M. (2014). Integration of cultivar resistance and fungicide application for control of wheat stripe rust. Can. J. Plant Pathol. 36, 311–326. doi: 10.1080/07060661.2014.924560
Chen, P. D., Qi, L. L., Zhou, B., Zhang, S. Z., and Liu, D. J. (1995). Development and molecular cytogenetic analysis of wheat-Haynaldia villosa 6VS/6AL translocation lines specifying resistance to powdery mildew. Theor. Appl. Genet. 91, 1125–1128. doi: 10.1007/BF00223930
Chen, W. Q., Wellings, C., Chen, X. M., Kang, Z. S., and Liu, T. G. (2014). Wheat stripe (yellow) rust caused by Puccinia striiformis f. sp. tritici. Mol. Plant Pathol. 15, 433–446. doi: 10.1111/mpp.12116
Cuadrado, Á., and Jouve, N. (2010). Chromosomal detection of simple sequence repeats (SSRs) using nondenaturing FISH (ND-FISH). Chromosoma 119, 495–503. doi: 10.1007/s00412-010-0273-x
Doyle, J. J., and Doyle, J. L. (1987). A rapid DNA isolation procedure from small quantities of fresh leaf tissues. Phytochem. Bull. 19, 11–15.
Du, H., Tang, Z., Duan, Q., Tang, S., and Fu, S. (2018). Using the 6RLKu minichromosome of rye (Secale cereale L.) to create wheat-rye 6D/6RLKu small segment translocation lines with powdery mildew resistance. Int. J. Mol. Sci. 19, 3933. doi: 10.3390/ijms19123933
Duan, Y., Luo, J., Yang, Z., Li, G., Tang, Z., and Fu, S. (2022). The physical location of stripe rust resistance genes on chromosome 6 of Rye (Secale cereale L.) AR106BONE. Front. Plant Sci. 13:928014. doi: 10.3389/fpls.2022.928014
Duveiller, E., Singh, R. P., and Nicol, J. M. (2007). The challenges of maintaining wheat productivity: pests, diseases, and potential epidemics. Euphytica 157, 417–430. doi: 10.1007/s10681-007-9380-z
Friebe, B., Hatchett, J. H., Sears, R. G., and Gill, B. S. (1990). Transfer of hessian fly resistance from ‘Chaupon’ rye to hexaploid wheat via a 2BS/2RL wheat-rye chromosome translocation. Theor. Appl. Genet. 79, 385–389. doi: 10.1007/BF01186083
Friebe, B., Jiang, J., Raupp, W. J., McIntosh, R. A., and Gill, B. S. (1996). Characterization of wheat-alien translocations conferring resistance to diseases and pests: current status. Euphytica 91, 59–87. doi: 10.1007/BF00035277
Han, G. H., Liu, S. Y., Wang, J., Jin, Y. L., Zhou, Y. L., Luo, Q. L., et al. (2020). Identification of an elite wheat-rye T1RS.1BL translocation line conferring high resistance to powdery mildew and stripe rust. Plant Dis. 104, 2940–2948. doi: 10.1094/PDIS-02-20-0323-RE
Han, G., Yan, H., Wang, J., Cao, L., Liu, S., Li, X., et al. (2022). Molecular cytogenetic identification of a new wheat-rye 6R addition line and physical localization of its powdery mildew resistance gene. Front. Plant Sci. 13:889494. doi: 10.3389/fpls.2022.889494
Hao, M., Liu, M., Luo, J., Fan, C., Yi, Y., Zhang, L., et al. (2018). Introgression of powdery mildew resistance gene Pm56 on rye chromosome arm 6RS into wheat. Front. Plant Sci. 9:1040. doi: 10.3389/fpls.2018.01040
Hou, Z. G., Liu, W. C., Shao, Z. R., and Jiang, R. Z. (2000). On developing long-term meteorological prediction research of crops pests and diseases prevailing in China. J. Nat. Disasters 9, 117–121.
Huo, Z., Ye, C., Qian, S., Chen, L., and Liu, W. (2002). Relationship between climatic anomaly and prevailing of the wheat powdery mildew in China. J. Nat. Disasters 11, 85–90.
Johansson, E., Henriksson, T., Prieto-Linde, M. L., Andersson, S., Ashraf, R., and Rahmatov, M. (2020). Diverse wheat-alien introgression lines as a basis for durable resistance and quality characteristics in bread wheat. Front. Plant Sci. 11:1067. doi: 10.3389/fpls.2020.01067
Kourelis, J., and van der Hoorn, R. A. L. (2018). Defended to the nines: 25 years of resistance gene cloning identifies nine mechanisms for R protein function. Plant Cell 30, 285–299. doi: 10.1105/tpc.17.00579
Li, J. B., Dundas, I., Dong, C. M., Li, G. R., Trethowan, R., Yang, Z. J., et al. (2020). Identification and characterization of a new stripe rust resistance gene Yr83 on rye chromosome 6R in wheat. Theor. Appl. Genet. 133, 1095–1107. doi: 10.1007/s00122-020-03534-y
Li, G. R., Gao, D., La, S. X., Wang, H. J., Li, J. B., He, W. L., et al. (2016). Characterization of wheat-Secale africanum chromosome 5R(a) derivatives carrying Secale specific genes for grain hardness. Planta 243, 1203–1212. doi: 10.1007/s00425-016-2472-z
Li, G., Wang, L., Yang, J., He, H., Jin, H., Li, X., et al. (2021). A high-quality genome assembly highlights rye genomic characteristics and agronomically important genes. Nat. Genet. 53, 574–584. doi: 10.1038/s41588-021-00808-z
Luo, P. G., Luo, H. Y., Chang, Z. J., Zhang, H. Y., Zhang, M., and Ren, Z. L. (2009). Characterization and chromosomal location of Pm40, in common wheat: A new gene for resistance to powdery mildew derived from elytrigia intermedium. Theor. Appl. Genet. 118, 1059–1064. doi: 10.1007/s00122-009-0962-0
Mago, R., Miah, H., Lawrence, G. J., Wellings, C. R., Spielmeyer, W., Bariana, H. S., et al. (2005). High-resolution mapping and mutation analysis separate the rust resistance genes Sr31, Lr26 and Yr9 on the short arm of rye chromosome 1. Theor. Appl. Genet. 112, 41–50. doi: 10.1007/s00122-005-0098-9
Marais, G. F., and Marais, A. S. (1994). The derivation of compensating translocations involving homoeologous group 3 chromosomes of wheat and rye. Euphytica 79, 75–80. doi: 10.1007/BF00023578
Mater, Y., Baenziger, S., Gill, K., Graybosch, R., Whitcher, L., Baker, C., et al. (2004). Linkage mapping of powdery mildew and greenbug resistance genes on recombinant 1RS from ‘amigo’ and ‘Kavkaz’ wheat-rye translocations of chromosome 1RS.1AL. Genome 47, 292–298. doi: 10.1139/g03-101
Persson, K., and von Bothmer, R. (2002). Genetic diversity amongst landraces of rye (Secale cereale L.) from northern Europe. Hereditas 136, 29–38. doi: 10.1034/j.1601-5223.2002.1360105.x
Qiu, L., Tang, Z., Li, M., and Fu, S. (2016). Development of new PCR-based markers specific for chromosome arms of rye (Secale cereale L.). Genome 59, 159–165. doi: 10.1139/gen-2015-0154
Rahmatov, M., Rouse, M. N., Nirmala, J., Danilova, T., Friebe, B., Steffenson, B. J., et al. (2016). A new 2DS·2RL Robertsonian translocation transfers stem rust resistance gene Sr59 into wheat. Theor. Appl. Genet. 129, 1383–1392. doi: 10.1007/s00122-016-2710-6
Ren, T. H., Chen, F., Zou, Y. T., Jia, Y. H., Zhang, H. Q., Yan, B. J., et al. (2011). Evolutionary trends of microsatellites during the speciation process and phylogenetic relationships within the genus Secale. Genome 54, 316–326. doi: 10.1139/g10-121
Ren, T., He, M., Sun, Z., Tan, F., Luo, P., Tang, Z., et al. (2019). The polymorphisms of oligonucleotide probes in wheat cultivars determined by ND-FISH. Molecules 24, 1126. doi: 10.3390/molecules24061126
Ren, T. H., Jiang, Q., Sun, Z., Zhao, L., Peng, W., Ren, Z., et al. (2022). Development and molecular cytogenetic characterization of novel primary wheat-rye 1RS.1BL translocation lines from multiple rye sources with resistance to stripe rust. Plant Dis. 106, 2191–2200. doi: 10.1094/PDIS-11-21-2605-RE
Ren, T., Li, Z., Yan, B., Tan, F., Tang, Z., Fu, S., et al. (2017b). Targeted segment transfer from Rye chromosome 2R to wheat chromosomes 2A, 2B, and 7B. Cytogenet. Genome Res. 151, 50–59. doi: 10.1159/000458743
Ren, T., Ren, Z., Yang, M., Yan, B., Tan, F., Fu, S., et al. (2018). Novel source of 1RS from Baili rye conferred high resistance to diseases and enhanced yield traits to common wheat. Mol. Breed. 38, 101. doi: 10.1007/s11032-018-0856-4
Ren, T., Sun, Z., Ren, Z., Tan, F., Luo, P., Tang, Z., et al. (2020). Molecular and cytogenetic characterization of a wheat-rye 7BS.7RL translocation line with resistance to stripe rust, powdery mildew and Fusarium head blight. Phytopathology 110, 1713–1720. doi: 10.1094/PHYTO-02-20-0061-R
Ren, T., Tang, Z., Fu, S., Yan, B., Tan, F., Ren, Z., et al. (2017a). Molecular cytogenetic characterization of novel wheat-rye T1RS.1BL translocation lines with high resistance to diseases and great agronomic traits. Front. Plant Sci. 8:799. doi: 10.3389/fpls.2017.00799
Ren, T. H., Yang, Z. J., Yan, B. J., Zhang, H. Q., Fu, S. L., and Ren, Z. L. (2009). Development and characterization of a new 1BL.1RS translocation line with resistance to stripe rust and powdery mildew of wheat. Euphytica 169, 207–213. doi: 10.1007/s10681-009-9924-5
Schlegel, R., and Korzun, V. (1997). About the origin of 1RS.1BL wheat-rye chromosome translocations from Germany. Plant Breed. 116, 537–540. doi: 10.1111/j.1439-0523.1997.tb02186.x
Schneider, A., Rakszegi, M., Molnár-Láng, M., and Szakács, É. (2016). Production and cytomolecular identification of new wheat-perennial rye (Secale cereanum) disomic addition lines with yellow rust resistance (6R) and increased arabinoxylan and protein content (1R, 4R, 6R). Theor. Appl. Genet. 129, 1045–1059. doi: 10.1007/s00122-016-2682-6
Shi, Z. X., Chen, X. M., Line, R. F., Leung, H., and Wellings, C. R. (2001). Development of resistance gene analog polymorphism markers for the Yr9 gene resistance to wheat stripe rust. Genome 44, 509–516. doi: 10.1139/g01-028
Tang, Z., Yang, Z., and Fu, S. (2014). Oligonucleotides replacing the roles of repetitive sequences pAs1, pSc119.2, pTa-535, pTa71, CCS1, and pAWRC.1 for FISH analysis. J. Appl. Genet. 55, 313–318. doi: 10.1007/s13353-014-0215-z
Wan, A. M., Chen, X. M., and He, Z. H. (2007). Wheat stripe rust in China. Aus. J. Agri. Res. 58, 605–619. doi: 10.1071/AR06142
Wan, A., Zhao, Z., Chen, X., He, Z., Jin, S., Jia, Q., et al. (2004). Wheat stripe rust epidemic and virulence of Puccinia striiformis f. sp. tritici in China in 2002. Plant Dis. 88, 896–904. doi: 10.1094/PDIS.2004.88.8.896
Wang, X., Han, B., Sun, Y., Kang, X., Zhang, M., Han, H., et al. (2022). Introgression of chromosome 1P from Agropyron cristatum reduces leaf size and plant height to improve the plant architecture of common wheat. Theor. Appl. Genet. 135, 1951–1963. doi: 10.1007/s00122-022-04086-z
Xie, C. J., Sun, Q. X., Ni, T., Nevo, E., and Fahima, F. (2004). Identification of resistance gene analogue markers closely linked to wheat powdery mildew resistance gene Pm31. Plant Breed. 123, 198–200. doi: 10.1046/j.1439-0523.2003.00940.x
Yang, H., Zhong, S., Chen, C., Yang, H., Chen, W., Tan, F., et al. (2021). Identification and cloning of a CC-NBS-NBS-LRR gene as a candidate of Pm40 by integrated analysis of both the available transcriptional data and published linkage mapping. Int. J. Mol. Sci. 22:10239. doi: 10.3390/ijms221910239
Keywords: Triticum aestivum, Secale cereale, chromosome addition, resistance, genetic resource
Citation: Ren T, Sun Z, Hu Y, Ren Z, Tan F, Luo P and Li Z (2022) Molecular cytogenetic identification of new wheat-rye 6R, 6RS, and 6RL addition lines with resistance to stripe rust and powdery mildew. Front. Plant Sci. 13:992016. doi: 10.3389/fpls.2022.992016
Edited by:
Cheng Liu, Shandong Academy of Agricultural Sciences, ChinaReviewed by:
Pengtao Ma, Yantai University, ChinaDiaoguo An, Institute of Genetics and Developmental Biology (CAS), China
Copyright © 2022 Ren, Sun, Hu, Ren, Tan, Luo and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tianheng Ren, cmVudGhAc2ljYXUuZWR1LmNu; Zhi Li, bGl6aGlAc2ljYXUuZWR1LmNu
†These authors have contributed equally to this work
 Tianheng Ren
Tianheng Ren Zixin Sun1,2†
Zixin Sun1,2† Peigao Luo
Peigao Luo Zhi Li
Zhi Li