
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 13 October 2022
Sec. Plant Bioinformatics
Volume 13 - 2022 | https://doi.org/10.3389/fpls.2022.991114
This article is part of the Research Topic Multi-omics and Computational Biology in Horticultural Plants: From Genotype to Phenotype View all 32 articles
Dioecious plant species have a high genetic variation that is important for coping with or adapting to environmental stress through natural selection. Intensive studies have reported dimorphism morphism in morphology, physiology, as well as biotic and abiotic stress responses in dioecious plants. Here, we demonstrated the dimorphism of metabolic profile and the preference of some microorganisms in the roots and rhizosphere soils of male and female papaya. The metabolic composition of roots were significantly different between the males and females. Some sex hormones occurred in the differential metabolites in roots and rhizosphere soils. For example, testosterone was up-regulated in male papaya roots and rhizosphere soils, whereas norgestrel was up-regulated in the female papaya roots, indicating a possible balance in papaya roots to control the sexual differentiation. Plant hormones such as BRs, JAs, SA and GAs were also detected among the differential metabolites in the roots and rhizosphere soils of dioecious papaya. In addition, some metabolites that have medicinal values, such as ecliptasaponin A, crocin, berberine and sapindoside A were also expressed differentially between the two sexes. Numerous differential metabolites from the papaya roots were secreted in the soil, resulting in the differences in microbial community structure in the roots and rhizosphere soils. Some nitrogen-fixing bacteria such as Allorhizobium-Neorhizobium-Pararhizobium-Rhizobium, Brevundimonas and Microvirga were enriched in the male papaya roots or rhizosphere soils. While Candidatus Solibacter and Tumebacillus, which utilize organic matters, were enriched in the roots or rhizosphere soils of the female papaya. Some differences in the fungi abundance were also observed in both male and female papaya roots. These findings uncovered the effect of sex types on the metabolic and microbiota differences in roots and rhizosphere soils in papaya and will lead to investigations of underlining genomic and molecular mechanisms.
Most angiosperm species are hermaphroditic, with individuals having both male and female sexual organs within the same flower. However, approximately 6% of angiosperm species are dioecious (Ricklefs, 1995; Renner, 2014), which are to have evolved from ancestral hermaphroditism (Endress and Doyle, 2015; Yakimowski and Barrett, 2016). Dioecious plants play an important role in protecting species diversity and maintaining ecosystem stability. Research has shown that males and females typically have dimorphism morphism in morphology, physiology, defence and patterns of allocation to life history (Liu et al., 2021b). Compared with female plants, male plants tend to be more tolerant to environmental stresses, such as drought, salinity, heavy metals, and nutrient deficiency (Melnikova et al., 2017; Xia et al., 2020).
Papaya (Carica papaya.L) is an important fruit crop, known for its nutritional benefits and medicinal applications. It is cultivated in tropical and sub-tropical regions, belonging to the genus Carica of the family Caricaceae. All species in Carica are dioecious, expect one monoecious species (Decraene and Smets, 1999). Papaya is one of the two species in Caricaceae that has three sex types, including female, male and bisexual. Since pear-shaped papayas produced by hermaphrodites are more popular in the market, while the cultivated type is mostly gynodioecious. The wild papayas are dioecious with a strong dimorphism and is an ideal model to study its different responses to sexual identity. However, a number of studies have demonstrated that there are no apparent differences in the morphology between the male and female plants during the vegetative phase. Only after flowering, the males and females show many floral morphological differences. For instance, male inflorescences bear more flowers than female ones, and flowers of female papaya are larger than males in size (Decraene and Smets, 1999).
The sex in wild papaya is controlled by XY chromosomes, XX for females, and XY for males (Ming et al., 2008). Previous studies have suggested that sex differentiation in papaya may be regulated by transcription, epigenetic DNA methylation and phytohormone (Zhou et al., 2020; Liu et al., 2021a; Zhou et al., 2022). However, a vast majority of these studies have focused on the aboveground parts of dioecious plants such as flower buds, and shoot meristems, while few reports focused on the belowground parts. For example, the response of metabolic profiles and microbial communities in roots and rhizosphere soils of dioecious papaya to sex identity is still unclear.
Research on metabolites has been a hotspot in biological research because biochemical phenotype largely reflects the development stages of plants and their interactions with the environment (Carreno-Quintero et al., 2013). It is estimated that there are approximately 200, 000 structurally distinct compounds (Dixon and Strack, 2003). The phytochemical components help us effectively understand the biological processes of plants and their mechanisms. Moreover, the microbiome is strongly associated with host plant by forming a dynamically balanced ecosystem in the process of co-evolution (Compant et al., 2019; Canto et al., 2020). The microorganisms rely on the suitable habitat and nutrients provided by plants. On the other hand, microorganisms promote plant growth, nutrient uptake, photosynthesis, and stress resilience (Yu et al., 2019; Xia et al., 2021). In this study, we demonstrate the sexual differences in metabolomes and microbial communities, which can help us better understand their sex specificity of papaya.
Papaya variety Zhonghuang seeds were planted into the soil at a greenhouse under 30°C, with 50% relative humidity at 16:8 (light: dark) photoperiod. After 3 months, the plants were planted in field at the experimental base of Fujian Agriculture and Forestry University (25°13′15″N, 117°41′55″E), Wufeng Town, Yongchun County, Quanzhou City, Fujian Province, China. About 2 months later, the papaya began to flower, the roots and rhizosphere soils of the male and female plants were collected and stored Whirl-Pak® bags. Each sample consisted of three replicates. The samples were stored in an ice box and transported immediately to the laboratory. The roots were washed using sterilized ddH2O, and then surface-sterilized with 100% ethanol for 1 min, 2.5% fresh bleach for 30 min and 100% ethanol for 1 min before storing at -80°C. The rhizosphere soils was detached from the roots of papaya. The soil of each sample was mixed, and visible roots and stones were removed before storing at -80°C (Pang et al., 2022a).
LC-MS/MS analyses were performed to identify exometabolites across samples using a UHPLC system (1290, Agilent Technologies) with a UPLC BEH Amide column (1.7μm 2.1*100mm, Waters) coupled to TripleTOF 5600 (Q-TOF, AB Sciex). Then 100μg of each sample was extracted with 300μL of methanol, 20μL internal standard substances with vortex was added for 30s, and was subsequently treated with ultrasound for 10min (incubated in ice water) and incubation for 1h at -20C to precipitate proteins. After centrifugation at 13,000rpm for 15 minutes at 4°C, the supernatant was transferred into a fresh 2mL LC/MS glass vial. 20μL supernatant from each sample was pooled and 200μL mixed supernatant was taken for the UHPLC-QTOF-MS analysis.
The mobile phase consisted of 25mM NH4OAc and 25mM NH4OH in water(pH=9.75)(A) and acetonitrile (B) was conducted using elution gradient as follows: 0 min, 95% B; 7min, 65% B; 9 min, 40% B; 9.1 min, 95% B; 12 min, 95% B, which was delivered at 0.5mL min-1. The injection volume was 3μL. The Triple TOF mass spectrometer was used for its ability to acquire MS/MS spectra on an information-dependent basis (IDA) during an LC/MS experiment. In this mode, the acquisition software (Analyst TF 1.7, AB Sciex) continuously evaluates the full scan survey MS data. In each cycle, 12 precursor ions whose intensity greater than 100 were chosen for fragmentation at collision energy (CE) of 30 V. ESI source conditions were set as follows: Ion source gas 1 as 60 Psi, Ion source gas 2 as 60 Psi, Curtain gas as 35 Psi, source temperature 650C, Ion Spray Voltage Floating (ISVF) 5000 V or -4000 V in positive or negative modes, respectively.
The raw data files were converted to the mzXML format using ProteoWizard, and processed by R package XCMS (version 3.2). The preprocessing results generated a data matrix that consisted of the retention time (RT), massto-charge ratio (m/z) values, and peak intensity. R package CAMERA was used for peak annotation after XCMS data processing. In-house MS2 database was applied in metabolites identification.
Total genomic DNA was extracted using the Fast DNATM Spin Kit (MP Biomedicals, LLC, Santa Ana, CA, United States). The DNA purity and quantity were determined by the NanoDrop 2000 spectrophotometer (Thermo Fisher) and agarose gel. Using the genomic DNA as a template, the hypervariable V3-V4 regions of the 16S rRNA gene were amplified by PCR with the primers 341F (5′-CCTACGGGNBGCASCAG-3′) and 785R (5′-CCTACGGGNBGCASCAG-3′) (Fadeev et al., 2021). The fungal ITS1 regions of ITS were amplified by PCR with the primers ITS5-1737F (5′- GGAAGTAAAAGTCGTAACAAGG-3′) and ITS2-2043R (5′- GCTGCGTTCTTCATCGATGC-3′) (Huang et al., 2016). The PCR amplification products were collected from a 2% agarose gel and purified by Vazyme VAHTSTM DNA Clean Beads. The sequencing libraries were established using TruSeq Nano DNA LT Library Prep Kit (Illumina, SD, USA) and then sequenced on an Illumina MiSeq platform (Biomarker Technologies Corporation, Beijing, China).
The Illumina paired-end raw data were quality filtered using Trimmomatic (Bolger et al., 2014), and then the primer sequences were identified and removed using Cutadapt (Martin, 2011), followed by paired-end pairing using USEARCH (Edgar, 2013). Afterwards, the chimera readings were detected and removed by using the UCHIME (Edgar et al., 2011) to obtain high-quality sequences for subsequent analysis (Edgar et al., 2011). Sequences with 97% similarity were clustered at the sequence level using USEARCH with a default threshold of 0.005% of all sequences to filter operational taxonomic units (OTUs) (Edgar, 2013). To annotate the taxonomic information for the sequences, Ribosomal Database Project (RDP) classifier was used to annotate the species of all representative reads with a confidence threshold of 70% according to the Silva database (version 138) (Wang et al., 2007).
Shannon index and richness index (ACE) estimator were used to analyze the alpha diversity by using phyloseq package (McMurdie and Holmes, 2013). For beta diversity analysis, Principal Component Analysis (PCoA) was performed using Bray-Curtis algorithm of Quantitative Insights into Microbial Ecology (QIIME) and R software (version 4.1.3) to compare the similarity of species diversity in the different samples (Suchodolski et al., 2012). We later conducted Permutational Multivariate Analysis of Variance (PERMANOVA) and paired PERMANOVA using vegan package at 999 permutations and α = 0.05 to test metabolites dissimilarities between sexes. The depleted or enriched metabolites and microbial genera in each sex were determined on the criteria having an P value < 0.05 and fold change value >2. Manhattan plot and volcano plot were employed using the R language to illustrate the differential metabolites and differential microbes of the roots and rhizosphere soils between the two sexes. The correlations between differential metabolites and differential microbes were visualized using a correlation matrix by computing all potential pairwise Spearman’s rank using Cytoscape version 3.6.1 (Shannon et al., 2003). We used the peak area intensity of metabolites and the relative abundance of microorganisms Pearson correlation analysis. The co-occurrence network analysis was based on Pearson correlation (p) was >0.9 and the P-value was < 0.05.
We investigated the morphological dimorphism of inflorescences in dioecious papaya. Compared with the females, male papaya showed longer peduncles,beraing numerous flowers at different stages (Figures 1A, B). This morphology ensures male papaya take full advantage of space and produce pollen grains continually. While female papaya showed short but thicker peduncle (Figures 1C, D).
LC-MS/MS non-targeted analyses were conducted to identify metabolites in roots and rhizosphere soils of dioecious papaya based on The Human Metabolome Database (HMDB). A total of 2602 metabolites were detected in the entire sample. The metabolites were classified into 80 taxonomic categories, including fatty acyls, coumarins and derivatives, followed by indoles and derivatives and phenols.
The PCoA analysis showed that there were significant difference in root and rhizosphere soil metabolites between females and males (P < 0.01 in Permanova analysis). The first (PC1) and second (PC2) principal component explained 77% and 8% of the variance in the metabolic composition, respectively. The metabolite compositions of roots and rhizosphere soils were distinctly separated along the first major axis, indicating that the differences in root and rhizosphere soil of the males and females were more significant (Figure 1E). The inter-group difference analysis of PC2 showed that the root metabolic composition of male papaya was significantly different to female papaya.
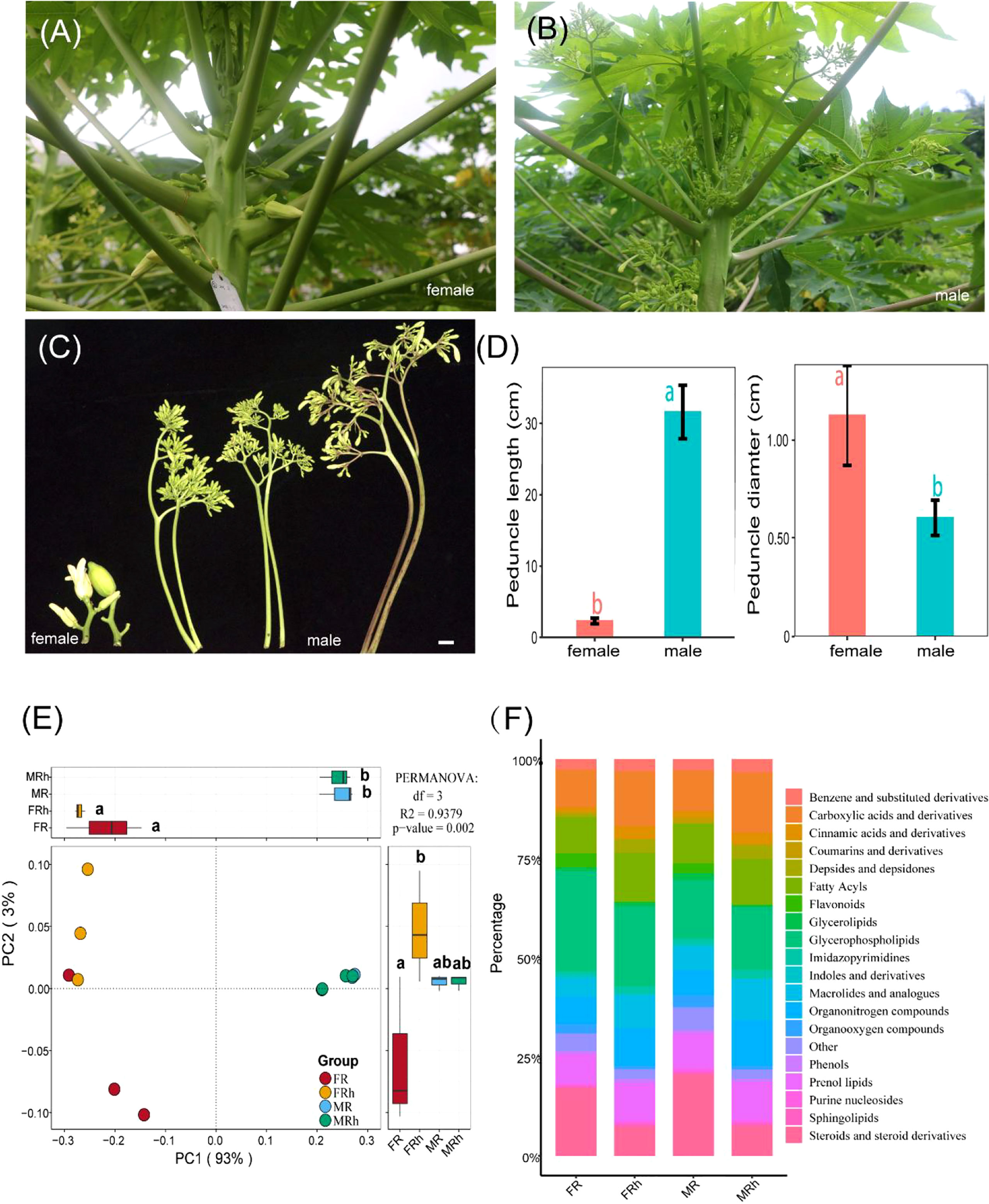
Figure 1 (A-D) Male papaya bears a longer but less thick peduncle with numerous flowers. Bar = 2cm. (E) Principal coordinates analysis (PCoA) plot based on unweighted Unifrac distance (β-diversity) on Bray-Curtis distance showing the separation of the samples according to metabolite profile of the roots and rhizosphere soils of the female and male papaya. PC1 and PC2 together explained 96% of the information in metabolic composition. The differential analysis was performed on PC1 and PC2 to reveal the differences in metabolic composition. Different letters represented significant differences (P < 0.05). Additionally, there were significant differences between every two groups in Permanova analysis (P < 0.01). Each point corresponded to a sample. (F) Relative abundance of Top 19 metabolic taxonomy in the entire sample. FR, female papaya roots; FRh, female papaya rhizosphere soils; MR, male papaya roots; MRh, male papaya rhizosphere soils.
In addition, the differences between the roots and rhizosphere soils of the male papaya were smaller than those observed in the female papaya. Moreover, we also identified the top twenty relative abundance of metabolic taxonomy, including glycerophospholipids (14.8%-25.3%), steroids and steroid derivatives (7.4%-20.5%), carboxylic acids and derivatives (9.6%-15.3%), fatty acyls (9.1%-12.5%), prenol lipids (7.6%-9.9%), organonitrogen compounds (6.3%-11.6%), macrolides and analogues (4.6%-10.3%), flavonoids (0.0%-3.5%), and phenols (0.8%-1.1%) (Figure 1F). We performed multiple difference comparison analysis to test the significant difference of these taxa in different compartments under both groups. Compared with the roots and rhizosphere soils of the males plant, the relative abundance of steroids and steroid derivatives, organooxygen compounds, flavonoids and indoles and derivatives of the female roots and rhizosphere soils were significantly higher (P < 0.05), while the abundance of Cinnamic acids and derivatives showed an opposite trend (Table S1).
To reveal the differences in metabolites of both sexes in the rhizosphere soils and roots, we adopted DESeq2 analysis and later visualized using Manhattan plot. The results showed that there were numerous differential metabolites in papaya roots and rhizosphere soils of both female and male papaya, with the female accounting for 400 metabolites and 173 metabolites for the male. Among them, the abundance of gibberellin A24, gibberellin A36, indole-3-ethanol, squalene, ecliptasaponin A, 3-O-Methylisoetharine and norgestrel were found significantly higher in the female papaya roots than in male papaya. However, the abundance of 26-Hydroxybrassinolide, (-)-Jasmonic acid, glycocholate, gibberellin A14, picroside III, sapindoside A, 6-Keto-prostaglandin E1 and 3-O-Methylisoetharine showed the opposite trend (Figure 2A; Table S2A). The abundance of metabolites such as eriodictyol and fumarate in the female papaya rhizosphere soils was significantly increased, while testosterone, aconitic acid, 5-Hydroxyindoleacetylglycine, betaine, 19-Oxotestosterone and others were significantly increased in the male papaya rhizosphere soils. Some metabolites such as testosterone, ADP, 28-Homobrassinolide and brassinolide were significantly increased in both roots and rhizosphere soils of male papaya (Figure 2B; Table S2B).
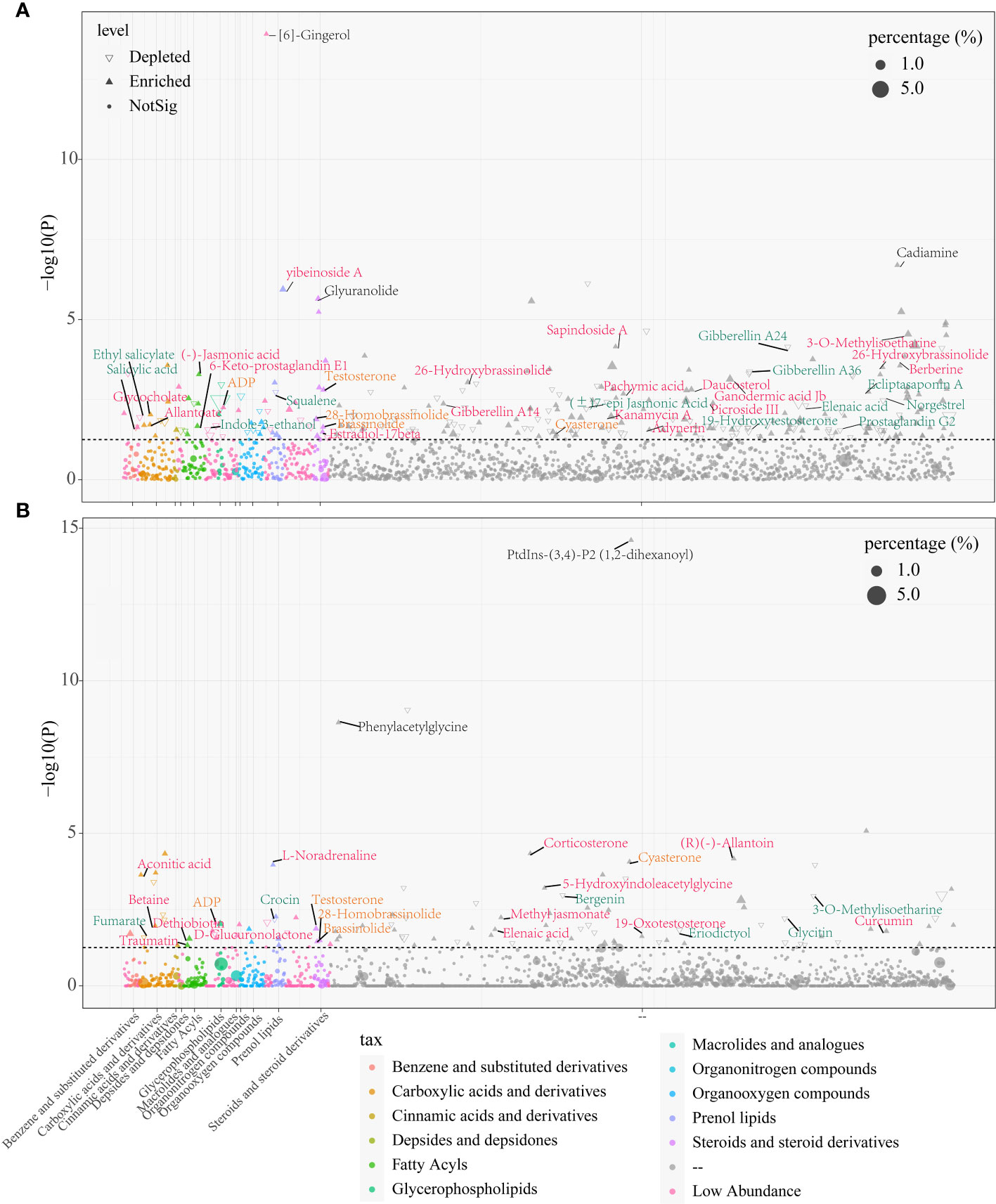
Figure 2 Manhattan plot showing differences in metabolites between the male and female papaya in the roots and rhizosphere soils, (A) papaya roots; (B) papaya rhizosphere soils. Each point represented a metabolite, and its size depicted the relative abundance of the metabolite, and the colors of the point denoted the metabolite categories. The P threshold of the dotted line was about 1.30 [-log10 (0.05)], and metabolites above the dashed line represents the metabolites with significant differences between males and females. The upward triangles showed that the metabolites of the males were significantly higher than those of the females, while the downward triangles of the males represented significantly lower metabolites than those of the females. Each dot signified no difference in substance between the males and females. Metabolites in black font were the ones with the highest change fold. Red font indicated metabolites that were significantly enriched in the males. Orange font indicated metabolites that were significantly increased in both roots and rhizosphere soils of the male, while green font indicated metabolites that were significantly lower in the males. “–” in taxonomy indicated metabolites which was not currently classified in the HMDB database, accounting for more than 75% of the total amount of substances detected.
The sex hormones occurred in the differential metabolites in roots and rhizosphere soils. For example, testosterone was up-regulated in male papaya roots and rhizosphere soils, while norgestrel was up-regulated in the female papaya roots (Figure 3). Plant hormones such as BRs, JAs, SA and GAs were also differentially expressed in the roots and rhizosphere soils of dioecious papaya (Figure 3).
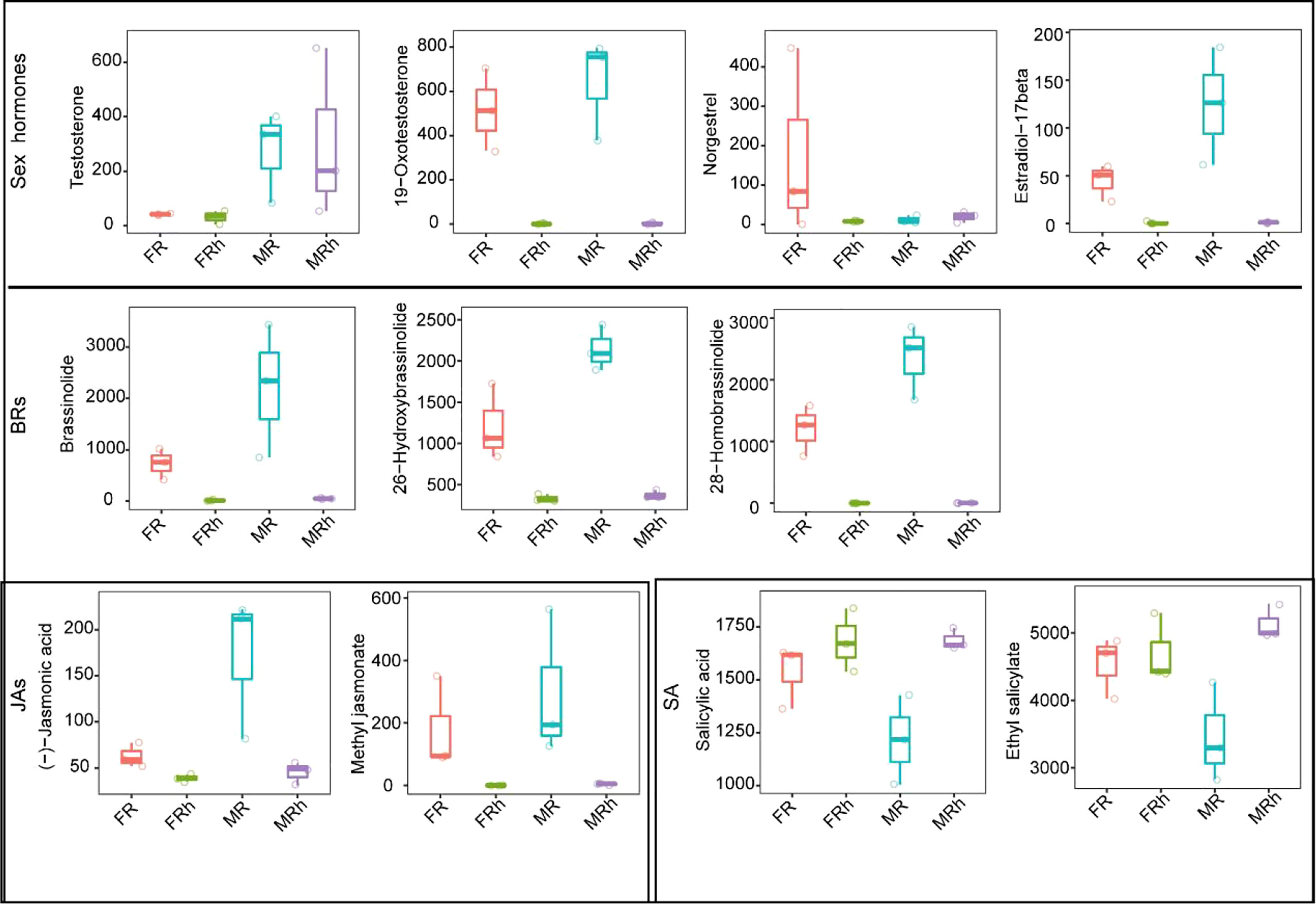
Figure 3 The compare analysis of hormone relative abundance in the roots and rhizosphere soils of papaya.
For 16S rRNA sequencing data, a total of 959,962 pairs of reads were obtained from 12 samples, and 956,415 clean reads were generated after paired-end reads quality control and splicing. Each sample produced at least 76,196 clean reads, with an average of 79,701 clean reads. The average GC content of 16S bacterial rRNA was 55.48%, and the bases with a base quality value greater than or equal to 30 accounted for 96.67% of the total bases. We obtained 1,806 OTUs at 97% identity from the 12 samples, with the number ranging from 824 to 1,767 per sample (Table S3A). The coverage for the observed OTUs was 99.81 ± 0.01% (mean ± sem) and the rarefaction curves showed clear asymptotes (Figure S1A), which demonstrated a near-complete sampling of the community.
To identify fungal species, the ITS1 region of the ITS was amplified and sequenced using the DNA from the roots and rhizosphere soils of papaya. A total of 511,385 pairs of reads were obtained from the 12 samples, and a total of 508,494 clean reads were filtered for splicing. Each sample generated at least 26,121 clean reads, with an average of 42,374 clean reads.
The average GC content of ITS1 was 45.76%, with bases containing a quality value greater than or equal to 30, accounting for 99.14% of the total number of bases. We obtained 1,021 OTUs at 97% identity from the entire sample, with the number of OTUs ranging from 268 to 638 per sample (Table S3B). The coverage for the observed OTUs was 99.84 ± 0.04% (mean ± sem) and the rarefaction curves showed clear asymptotes (Figure S1B), which demonstrated a near-complete sampling of the community.
Numerous bacteria and fungi were detected in papaya roots and rhizosphere soils by16S rRNA/ITS sequencing. Proteobacteria (10.8%-55.1%), Acidobacteriota (4.4%-39.3%) and Actinobacteriota (14.7%-28.2%) were dominant bacteria identified, followed by Firmicutes (1.2%-17.6%) and Gemmatimonadota (1.8%-6.9%) (Figure 4A1). Notably, the relative abundances of Proteobacteria and Myxococcota in papaya roots were significantly increased (p < 0.05) compared with rhizosphere soils, while the opposite results were found for Acidobacteriota (Table S4). On the other hand, Ascomycota (59.5%-66.7%) and Basidiomycota (15.2%-33.3%) were the dominant fungi in papaya root and rhizosphere soils, followed by Chytridiomycota (0.2%-3.5%) (Figure 4A2). The relative abundance of Chytridiomycota in papaya roots was significantly (p < 0.05) lower than in papaya rhizosphere soils (Table S4).
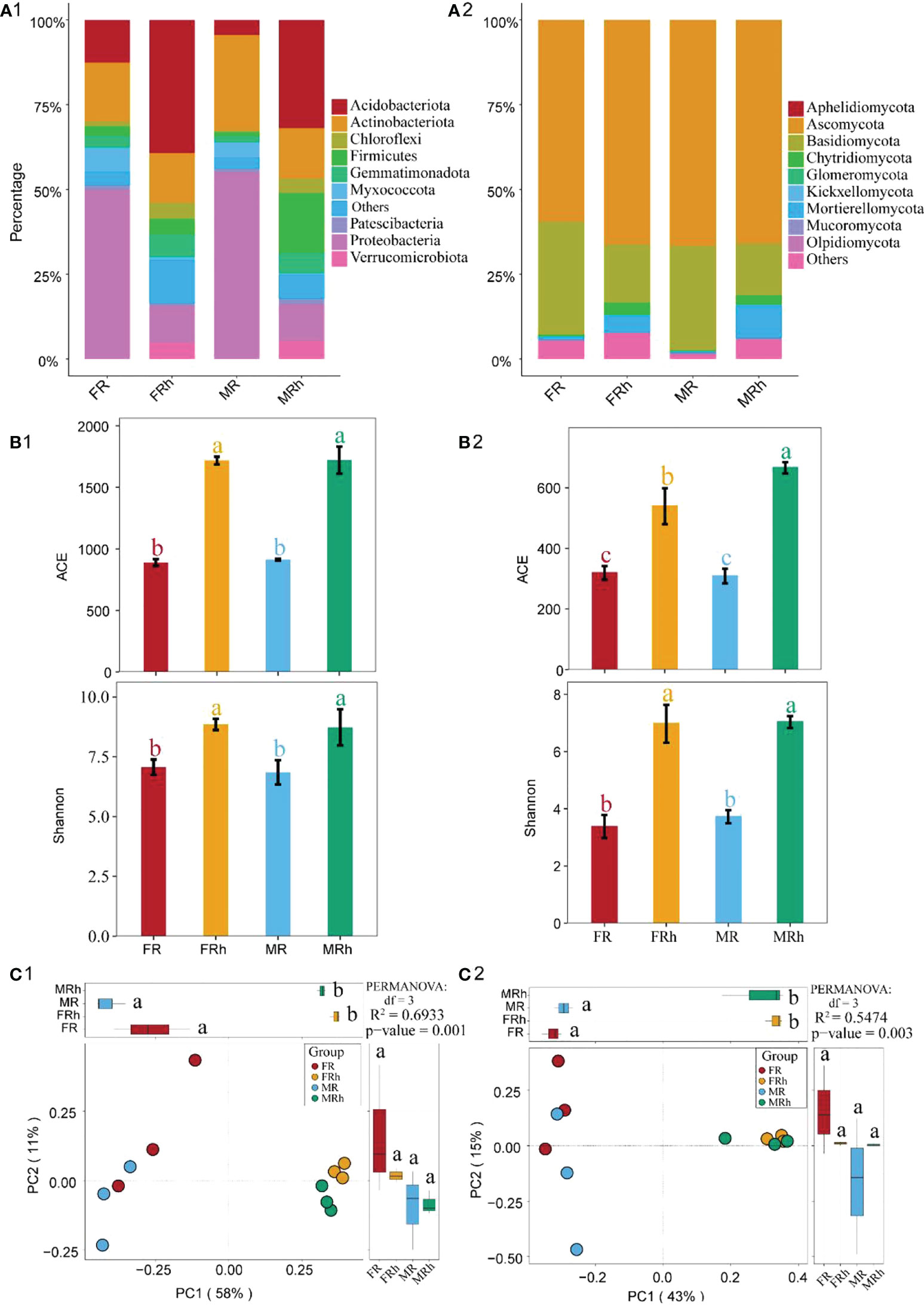
Figure 4 Taxonomic composition of the bacteria (A1) and fungi (A2) in the roots and rhizosphere soils of the female and male papaya. Alpha diversity analysis showed the richness index (ACE index) and diversity index (Shannon index) of bacteria (B1) and fungi (B2) in the roots and rhizosphere soils between the male and female papaya. The different letters meant a significant difference (p< 0.05). PCoA analysis demonstrating bacteria (C1) and fungal (C2) diversity in the roots and rhizosphere soils between the female and male papaya. The differential analysis was performed on PC1 and PC2 to reveal the differences in microbial composition. Different letters represented significant differences (P < 0.05). Permanova analysis revealed significant differences between every two sample groups (P < 0.01).
We further investigated the bacterial and fungal richness (ACE) and diversity (Shannon) indices in roots and rhizosphere soils of the female and male papaya. The results showed that compared with the rhizosphere soils, the ACE and Shannon index of bacteria and fungi in the roots of both female and male papaya were significantly decreased (p< 0.05), while the difference between the two sexes was not significant (Figures 4B1, B2).
The OTUs of 16S rRNA/ITS sequencing were used for PCoA analysis which illustrated the overall similarity of the bacterial and fungal community composition between samples. PCoA plot based on unweighted Unifrac distance (β-diversity) on Bray-Curtis distance showing the separation of the samples according to the composition of bacteria and fungi in roots and rhizosphere soils between the female and male papaya. PC1 and PC2 explained 58% and 11% of the total variance in bacterial community composition, respectively (Figure 4C1). PC1 and PC2 also explained 43% and 15% of the total variance in fungal community composition, respectively (Figure 4C2). The results showed that the microbial compositions in the roots and rhizosphere soils were significantly separated along the first axis, suggesting that the microbial differences in soil and roots were more pronounced (Figures 4C1, C2).
To identify the differential abundance of bacteria and fungi of the two sex types in roots and rhizosphere soils, DESeq2 was employed and visualized using volcano plot. The results showed that the sex-differentiated bacteria in roots and rhizosphere soils were 40 and 119 genera (Figures 5A1, A2, Tables S5A, B), respectively, while the differential fungi were 28 and 9 genera (Figures 5B1, B2, Tables S5C, D), respectively. In the roots, the abundance of bacteria such as Candidatus-Solibacter, Parabacteroides, Dactylosporangium, Escherichia_Shigella, Rodentibacter, unclassified-Burkholderiales and Polycyclovorans were significantly higher than the female papaya’s, whereas the abundance of Allorhizobium-Neorhizobium-Pararhizobium-Rhizobium, Brevundimonas, Microvirga, Streptomyces, Agromyces, Mitsuaria, Uliginosibacterium, Mycobacterium, Sandaracinus, Dyadobacter, and Methylophilus were significantly lower than males (Figure 5A1). The female papaya rhizosphere soils was significantly enriched with Tumebacillus, Roseiarcus, Haliangium, Anaeromyxobacter, Nitrospira, uncultured-Desulfovirga sp., and Rhodoplanes. While male papaya rhizosphere soils was enriched the bacterial genera including Brevundimonas, Bifidobacterium, Flavitalea, Flavisolibacter, Lactobacillus, Microvirga, Cupriavidus, Faecalibacterium, Lysobacte, Variovorax, Bacteroides and Bacillus than male (Figure 5A2). For fungi, the abundance of Candida, Solicoccozyma, Rhodotorula, etc. were significantly higher in the female papaya roots than in the males. Conversely, the abundance of, Paraconiothyrium, Gibellulopsis and Curvularia were significantly lower in the male papaya roots compared with the male papaya roots (Figure 5B1). Moreover, the abundance of Exserohilum, Acrocalymma, Sampaiozyma and Pseudaleuria in the rhizosphere soils of the female papaya was significantly lower than that of the males (Figure 5B2).
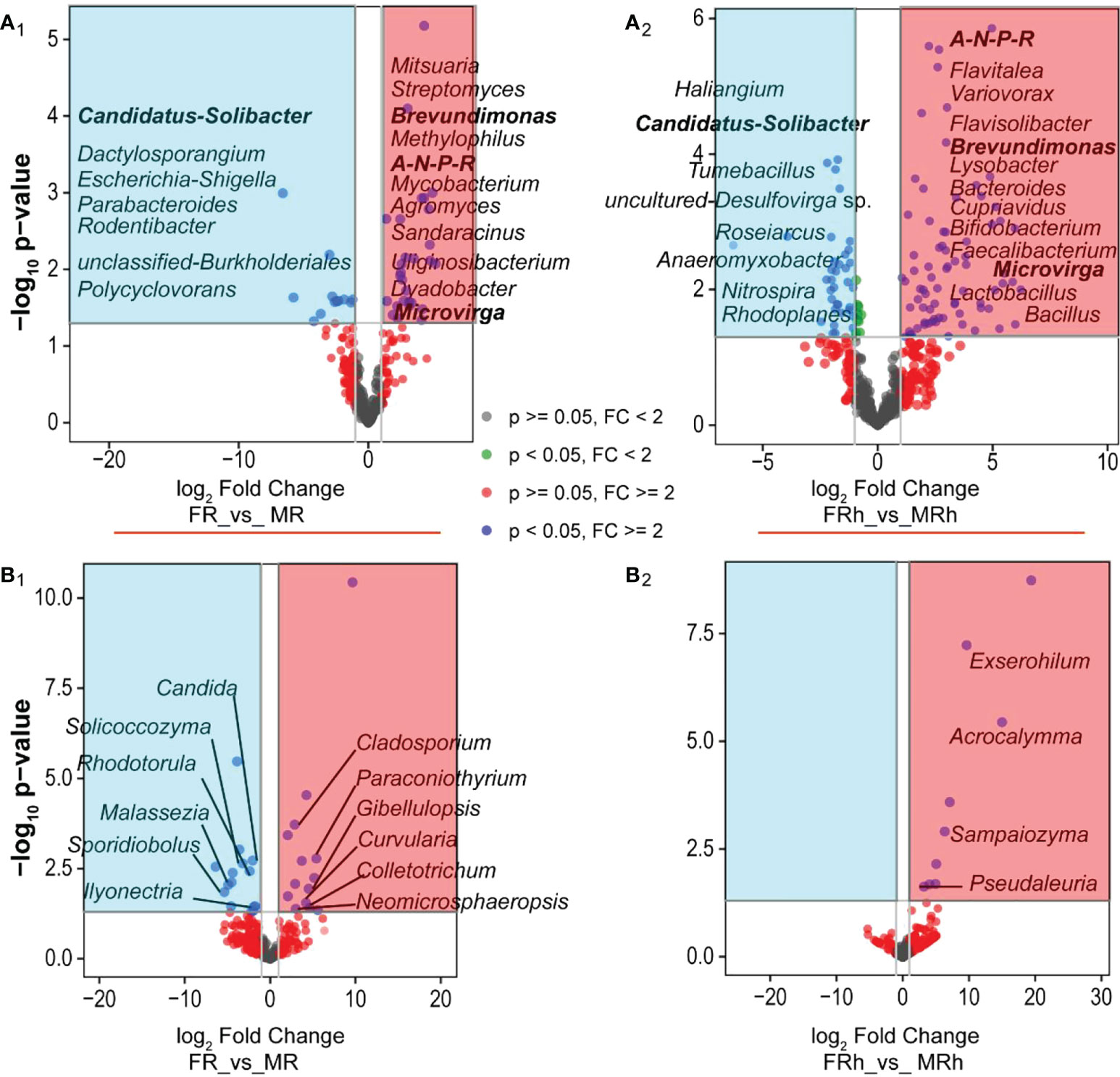
Figure 5 Volcano plots depicting the differential abundance of microbial communities (genus level) in the roots and rhizosphere soils of the female and male. (A), bacteria; (B), fungi; 1, roots; 2, rhizosphere soils. A-N-P-R is the abbreviation of Allorhizobium-Neorhizobium-Pararhizobium-Rhizobium. The spots in the blue background represented significantly up-regulated in the females, while the spots in the red background were significantly up-regulated in the males. The bold black font indicated the significantly changed microorganisms in both male and female papaya roots and rhizosphere soils.
In order to comprehensively explore the possible relationships between differential microorganisms and differential metabolites, co-occurrence network analysis was performed based on Pearson correlation (P< 0.05). The analysis results demonstrated that most differential metabolites were positively correlated with the differential microbes in abundance (Figure 6). In the roots of papaya, adynerin was positively correlated with Paraconiothyrium, Colletotrichum, Xenomyrothecium, whereas allantoate exhibited a positive correlation with Gibellulopsis, while daucosterol showed a positive relationship with Vishniacozyma. Furthermore, norgestrel tended to have a positive association with Malassezia, Rhodotorula, Sporidiobolus and Filobasidium. Both gibberellin A14 and gibberellin A24 had a positive correlation with Neomicrosphaeropsis and Solicoccozyma, respectively. It was also observed that indole-3-ethanol was positively associated with Conocybe, while brassinolide and testosterone had a significant positive correlation with Neomicrosphaeropsis and Mitsuaria.
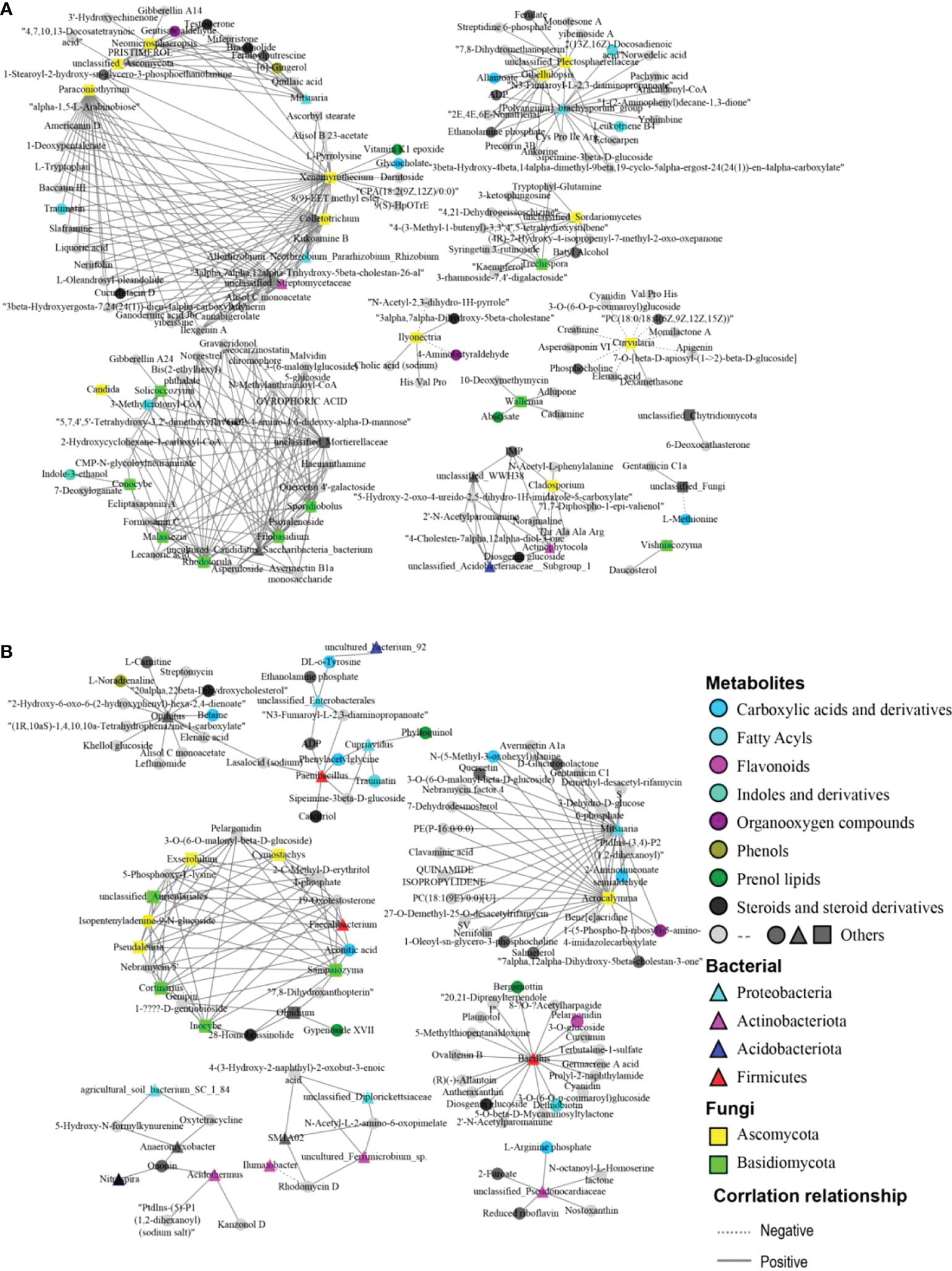
Figure 6 Co-occurrence network analysis among the different microbiota and metabolites from the roots (A) and the rhizosphere soils (B) of the female and male papaya. The triangles, squares and circles represented bacteria, fungi and metabolites, respectively. Different colors represented the classification of bacteria and fungi at the phylum level and metabolite HMDB classification. The straight line represented a significant positive correlation, while the dotted line represented a negative correlation.
In the rhizosphere soils, the network revealing the association between metabolites and microorganisms was distinct compared with the roots of the papaya. For instance, 28-Homobrassinolide had a positive correlation with Olpidium, Sampaiozyma and Inocybe, whereas 19-Oxotestosterone revealed a positive association with Faecalibacterium, Cymostachys, Exserohilum, Cortinarius, Pseudaleuria, and Sampaiozyma, while D-Glucuronolactone was positively correlated with Mitsuaria, Acrocalymma. We also noticed that aconitic acid exhibited apositive relationship with Faecalibacterium, Exserohilum, while ADP and betaine were significantly positively correlated with Paenibacillus and Opitutus in abundance, respectively.
From an evolutionary perspective, hermaphroditic plants have both male and female reproductive organs which results in reduction of genetic variation during the largely self-pollination. However, dioecious plants have high genetic variation because their genetic materials of sexual reproduction were from two or more different individuals. They have the advantage of maintaining genetic diversity of species, which is beneficial to their adaption to different environments by natural selection or artificial selection during domestication of crops (Barrett and Hough, 2013; Lei et al., 2017). Although sexual dimorphism in morphology (Decraene and Smets, 1999; Barrett and Hough, 2013), physiology (Laporte and Delph, 1996), timing of development (Gouker et al., 2021) and resistance to stress (Melnikova et al., 2017; Tonnabel et al., 2017; Liu et al., 2021b) has been overwhelmingly reported, the relationship between metabolic profiles and microbial communities in roots and rhizosphere soils of dioecious plants has rarely been investigated. In the present study, we found that sex-based differences exhibited in metabolites and microbiota in dioecious papaya.
For the metabolic composition, the variations in root rhizosphere soils of the males and females were mainly influenced by niches, and the sexual dimorphism exhibited in the metabolic profiles of male and female papaya. Studies have revealed that sex hormones, including testosterone, progesterone, and the other estrogens were associated with sexual differentiation and reproduction (Bouman et al., 2005; Burger, 2006; Rastrelli et al., 2018). Here, differential metabolites in the roots of papaya and some sex hormones were also detected. For example, male hormone such as testosterone was found significantly up-regulated in the male papaya while the female hormone norgestrel showed a significant up-regulation in the female papaya. These results suggested that sex hormones in papaya may also affect sexual differentiation in a dose-dependent manner.
Besides the sex hormones, the phytohormones such as brassinolide, 28-homobrassinolide, jasmonic acid (JA) and GAs were also identified in the roots and rhizosphere soils. Brassinosteroids (BRs) are a group of steroidal plant hormones that interact with auxin and GAs to regulate many developmental processes including stem, petioles and hypocotyls elongation (Yin et al., 2002; Kim et al., 2009). Hence, males seemed to require a singnificant amount of BRs to promote their strong root system, which resulted in the accumulation of brassinolide and 28-homobrassinolide in the roots and rhizosphere soils. In addition, BRs and JAs were reported to mediate the balance between plant growth and defense responses (Liao et al., 2020). The relatively higher abundance of brassinolide, 28-homobrassinolide and JA in both the roots and rhizosphere soils of the male papaya suggested male papaya had higher resistance to stresses.
Plants have mutualistic relationships with their inhabiting microbiome which is referred to as the host’s second or extended genome (Beckers et al., 2017). They interact and evolve with each other over time for nutrient acquisition, productivity and resistance (Hacquard et al., 2015; Beckers et al., 2017; Xiong et al., 2021). Additionally, the male and female papaya have evolved to have preference for some microbes. For instance, male papaya roots or rhizosphere soils enriched some nitrogen-fixing bacteria such as Allorhizobium-Neorhizobium-Pararhizobium-Rhizobium, Brevundimonas and Microvirga (Mousavi et al., 2014; Radl et al., 2014; Naqqash et al., 2020; Pang et al., 2021; Pang et al., 2022b). The enriched testosterone identified in the roots and rhizosphere soils of the male papaya has also been reported to promote nitrogen retention in human e (Bhasin et al., 2003). These results are consistent with the demands of high nitrogen nutrients for numerous pollens in some species (Lei et al., 2017; Tonnabel et al., 2017; Liu et al., 2021b). The roots or rhizoshphere soil of female papaya were enriched with Candidatus, Solibacter and Tumebacillus that utilize organic carbon sources (Baek et al., 2011; Challacombe et al., 2011; Naqqash et al., 2020), which provided evidence that females consume more carbon nutrient for their reproduction leading to fruit production, a major sink organ (Harris and Pannell, 2008; Lei et al., 2017).
Our finding also revealed that the abundance of some fungi genera exhibited distinct distribution patterns in both the male and female papaya roots. For example, Candida, Solicoccozyma and Rhodotorula which play important role in the degradation of a variety of organic materials (Shailubhai et al., 1985; Luo et al., 2021) showed higher abundance in the female papaya roots, suggesting that they could collaborate with endophytic bacteria to provide carbon nutrient for female papaya reproduction. However, in the roots of male papaya, some fungal species such as Gibellulopsis (Giraldo and Crous, 2019), Curvularia (Cui et al., 2020), Colletotrichum (Gautam, 2014) and Vishniacozyma (Pem et al., 2020) which have been described as plant pathogens were enriched in male papaya. It has also been reported that some phytopathogens in the environment are of endophyte origins and that these innocuous fungal endophytes are beneficial to their host plants against the biotic and abiotic stresses through producing secondary metabolites, which is tigger by a long process of co-evolution (Tan and Zou, 2001). Most differential metabolites demonstrated a positive correlation with the differential microbes, suggesting that these differential metabolites detected in the papaya roots had a positive effect on the density and structure of the microbes in roots and rhizosphere soils.
The original contributions presented in the study are publicly available. This data can be found here: NCBI, PRJNA857495.
RM, YZ and ZP designed the research. YZ and ZP performed the experiment and data analysis, and draft the manuscript. RM, ZY and NF supervised the experiment, revised and improved the manuscript. ZP and HJ collected plant materials and extracted DNA. All authors contributed to the article and approved the submitted version.
This research was funded by National Science Foundation (NSF) Plant Genome Research Program Award DBI-1546890 to RM.
We thank BMKCloud for providing a platform for data analysis.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.991114/full#supplementary-material
Baek, S. H., Cui, Y., Kim, S. C., Cui, C. H., Yin, C., Lee, S. T., et al. (2011). Tumebacillus ginsengisoli sp. nov., isolated from soil of a ginseng field. Int. J. Of Systematic And Evolutionary Microbiol. 61, 1715–1719. doi: 10.1099/ijs.0.023358-0
Barrett, S. C., Hough, J. (2013). Sexual dimorphism in flowering plants. J. Exp. Bot. 64, 67–82. doi: 10.1093/jxb/ers308
Beckers, B., Op De Beeck, M., Weyens, N., Boerjan, W., Vangronsveld, J. (2017). Structural variability and niche differentiation in the rhizosphere and endosphere bacterial microbiome of field-grown poplar trees. Microbiome 5, 25. doi: 10.1186/s40168-017-0241-2
Bhasin, S., Taylor, W. E., Singh, R., Artaza, J., Sinha-Hikim, I., Jasuja, R., et al. (2003). The mechanisms of androgen effects on body composition: Mesenchymal pluripotent cell as the target of androgen action. J Gerontol Ser. a-Biological Sci. Med. Sci. 58, 1103–1110. doi: 10.1093/gerona/58.12.m1103
Bolger, A. M., Lohse, M., Usadel, B. (2014). Trimmomatic: A flexible trimmer for illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Bouman, A., Heineman, M. J., Faas, M. M. (2005). Sex hormones and the immune response in humans. Hum. Reprod. Update 11, 411–423. doi: 10.1093/humupd/dmi008
Burger, H. (2006). How effective is testosterone replacement therapy in premenopausal women with severe androgen deficiency? Nat. Clin. Pract. Endocrinol. Metab. 2, 432–433. doi: 10.1038/ncpendmet0241
Canto, C. D., Simonin, M., King, E., Moulin, L., Bennett, M. J., Castrillo, G., et al. (2020). An extended root phenotype: The rhizosphere, its formation and impacts on plant fitness. Plant J. 103, 951–964. doi: 10.1111/tpj.14781
Carreno-Quintero, N., Bouwmeester, H. J., Keurentjes, J. J. (2013). Genetic analysis of metabolome-phenotype interactions: from model to crop species. Trends Genet. 29, 41–50. doi: 10.1016/j.tig.2012.09.006
Challacombe, J. F., Eichorst, S. A., Hauser, L., Land, M., Xie, G., Kuske, C. R. (2011). Biological consequences of ancient gene acquisition and duplication in the large genome of candidatus solibacter usitatus Ellin6076. PloS One 6, e24882. doi: 10.1371/journal.pone.0024882
Compant, S., Samad, A., Faist, H., Sessitsch, A. (2019). A review on the plant microbiome: Ecology, functions, and emerging trends in microbial application. J. Advanced Res. 19, 29–37. doi: 10.1016/j.jare.2019.03.004
Cui, W. L., Lu, X. Q., Bian, J. Y., Qi, X. L., Li, D. W., Huang, L. (2020). Curvularia spicifera and curvularia muehlenbeckiae causing leaf blight on cunninghamia lanceolata. Plant Pathol. 69, 1139–1147. doi: 10.1111/ppa.13198
Decraene, L. P. R., Smets, E. F. (1999). The floral development and anatomy of carica papaya (Caricaceae). Can. J. Botany-Revue Can. Botanique 77, 582–598. doi: 10.1139/b99-026
Dixon, R. A., Strack, D. (2003). Phytochemistry meets genome analysis, and beyond. Phytochemistry 62, 815–816. doi: 10.1016/s0031-9422(02)00712-4
Edgar, R. C. (2013). UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996–998. doi: 10.1038/nmeth.2604
Edgar, R. C., Haas, B. J., Clemente, J. C., Quince, C., Knight, R. (2011). UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27, 2194–2200. doi: 10.1093/bioinformatics/btr381
Endress, P. K., Doyle, J. A. (2015). Ancestral traits and specializations in the flowers of the basal grade of living angiosperms. Taxon 64, 1093–1116. doi: 10.12705/646.1
Fadeev, E., Cardozo-Mino, M. G., Rapp, J. Z., Bienhold, C., Salter, I., Salman-Carvalho, V., et al. (2021). Comparison of two 16S rRNA primers (V3-V4 and V4-V5) for studies of Arctic microbial communities. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.637526
Gautam, A. (2014). Colletotrichum gloeosporioides: Biology, pathogenicity and management in India. J. Plant Physiol. Pathol. 2, 1–11. doi: 10.4172/2329-955X.1000125
Giraldo, A., Crous, P. W. (2019). Inside plectosphaerellaceae. Stud. Mycology 92, 227–286. doi: 10.1016/j.simyco.2018.10.005
Gouker, F. E., Carlson, C. H., Zou, J., Evans, L., Crowell, C. R., Smart, C. D., et al. (2021). Sexual dimorphism in the dioecious willow salix purpurea. Am. J. Bot. 108, 1374–1387. doi: 10.1002/ajb2.1704
Hacquard, S., Garrido-Oter, R., Gonzalez, A., Spaepen, S., Ackermann, G., Lebeis, S., et al. (2015). Microbiota and host nutrition across plant and animal kingdoms. Cell Host Microbe 17, 603–616. doi: 10.1016/j.chom.2015.04.009
Harris, M. S., Pannell, J. R. (2008). Roots, shoots and reproduction: sexual dimorphism in size and costs of reproductive allocation in an annual herb. Proc. R. Soc. B-Biological Sci. 275, 2595–2602. doi: 10.1098/rspb.2008.0585
Huang, Y. L., Kuang, Z. Y., Wang, W. F., Cao, L. X. (2016). Exploring potential bacterial and fungal biocontrol agents transmitted from seeds to sprouts of wheat. Biol. Control 98, 27–33. doi: 10.1016/j.biocontrol.2016.02.013
Kim, T. W., Guan, S. H., Sun, Y., Deng, Z. P., Tang, W. Q., Shang, J. X., et al. (2009). Brassinosteroid signal transduction from cell-surface receptor kinases to nuclear transcription factors. Nat. Cell Biol. 11, 1254–U1233. doi: 10.1038/ncb1970
Laporte, M. M., Delph, L. F. (1996). Sex-specific physiology and source-sink relations in the dioecious plant silene latifolia. Oecologia 106, 63–72. doi: 10.1007/BF00334408
Lei, Y., Jiang, Y., Chen, K., Duan, B., Zhang, S., Korpelainen, H., et al. (2017). Reproductive investments driven by sex and altitude in sympatric populus and salix trees. Tree Physiol. 37, 1503–1514. doi: 10.1093/treephys/tpx075
Liao, K., Peng, Y. J., Yuan, L. B., Dai, Y. S., Chen, Q. F., Yu, L. J., et al. (2020). Brassinosteroids antagonize jasmonate-activated plant defense responses through BRI1-EMS-SUPPRESSOR1 (BES1). Plant Physiol. 182, 1066–1082. doi: 10.1104/pp.19.01220
Liu, J. A., Chen, L. Y., Zhou, P., Liao, Z. Y., Lin, H., Yu, Q. Y., et al. (2021a). Sex biased expression of hormone related genes at early stage of sex differentiation in papaya flowers. Horticulture Res. 8, 147. doi: 10.1038/s41438-021-00581-4
Liu, M., Korpelainen, H., Li, C. Y. (2021b). Sexual differences and sex ratios of dioecious plants under stressful environments. J. Plant Ecol. 14, 920–933. doi: 10.1093/jpe/rtab038
Luo, Y., Li, J., Zhou, H., Yu, B., He, J., Wu, A., et al. (2021). The nutritional significance of intestinal fungi: Alteration of dietary carbohydrate composition triggers colonic fungal community shifts in a pig model. Appl. Environ. Microbiol. 87, e00038-21. doi: 10.1128/AEM.00038-21
Martin, M. (2011). CUTADAPT removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17, 1138–43. doi: 10.14806/ej.17.1.200
Mcmurdie, P. J., Holmes, S. (2013). Phyloseq: An r package for reproducible interactive analysis and graphics of microbiome census data. PloS One 8, e61217. doi: 10.1371/journal.pone.0061217
Melnikova, N. V., Borkhert, E. V., Snezhkina, A. V., Kudryavtseva, A. V., Dmitriev, A. A. (20171827). Sex-specific response to stress in populus. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.01827
Ming, R., Hou, S., Feng, Y., Yu, Q., Dionne-Laporte, A., Saw, J. H., et al. (2008). The draft genome of the transgenic tropical fruit tree papaya (Carica papaya Linnaeus). Nature 452, 991–996. doi: 10.1038/nature06856
Mousavi, S. A., Osterman, J., Wahlberg, N., Nesme, X., Lavire, C., Vial, L., et al. (2014). Phylogeny of the rhizobium-Allorhizobium-Agrobacterium clade supports the delineation of neorhizobium gen. nov. Systematic Appl. Microbiol. 37, 208–215. doi: 10.1016/j.syapm.2013.12.007
Naqqash, T., Imran, A., Hameed, S., Shahid, M., Majeed, A., Iqbal, J., et al. (2020). First report of diazotrophic brevundimonas spp. as growth enhancer and root colonizer of potato. Sci. Rep. 10, 12893. doi: 10.1038/s41598-020-69782-6
Pang, Z., Dong, F., Liu, Q., Lin, W., Hu, C., Yuan, Z. (2021). Soil metagenomics reveals effects of continuous sugarcane cropping on the structure and functional pathway of rhizospheric microbial community. Front. Microbiol. 12, 627569. doi: 10.3389/fmicb.2021.627569
Pang, Z. Q., Fallah, N., Weng, P. Y., Zhou, Y. M., Tang, X. M., Tayyab, M., et al. (2022a). Sugarcane-peanut intercropping system enhances bacteria abundance, diversity, and sugarcane parameters in rhizospheric and bulk soils. Front. Microbiol. 12, 815129. doi: 10.3389/fmicb.2021.815129
Pang, Z. Q., Huang, J. W., Fallah, N., Lin, W. X., Yuan, Z. N., Hu, C. H. (2022b). Combining n fertilization with biochar affects root-shoot growth, rhizosphere soil properties and bacterial communities under sugarcane monocropping. Ind. Crops Products 182, 114899. doi: 10.1016/j.indcrop.2022.114899
Pem, D., Jeewon, R., Selcuk, F., Ulukapi, M., Bhat, J., Doilom, M., et al. (2020). Ribosomal and protein gene phylogeny reveals novel saprobic fungal species From Juglans regia and Urtica dioica. Front. Microbiol. 11, 1303. doi: 10.3389/fmicb.2020.01303
Radl, V., Simoes-Araujo, J. L., Leite, J., Passos, S. R., Martins, L. M. V., Xavier, G. R., et al. (2014). Microvirga vignae sp nov., a root nodule symbiotic bacterium isolated from cowpea grown in semi-arid Brazil. Int. J. Systematic Evolutionary Microbiol. 64, 725–730. doi: 10.1099/ijs.0.053082-0
Rastrelli, G., Corona, G., Maggi, M. (2018). Testosterone and sexual function in men. Maturitas 112, 46–52. doi: 10.1016/j.maturitas.2018.04.004
Renner, S. S. (2014). The relative and absolute frequencies of angiosperm sexual systems: dioecy, monoecy, gynodioecy, and an updated online database. Am. J. Bot. 101, 1588–1596. doi: 10.3732/ajb.1400196
Ricklefs, R. (1995). Dioecy and its correlates in the flowering plants. Am. J. Bot. 82, 596–606. doi: 10.1002/j.1537-2197.1995.tb11504.x
Shailubhai, K., Rao, N. N., Modi, V. V. (1985). Degradation of petroleum industry oil sludge by rhodotorula rubra and pseudomonas aeruginosa. Oil Petrochemical pollut. 2, 133–136. doi: 10.1016/S0143-7127(85)90478-8
Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., et al. (2003). Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504. doi: 10.1101/gr.1239303
Suchodolski, J. S., Dowd, S. E., Wilke, V., Steiner, J. M., Jergens, A. E. (2012). 16S rRNA gene pyrosequencing reveals bacterial dysbiosis in the duodenum of dogs with idiopathic inflammatory bowel disease. PloS One 7, e39333. doi: 10.1371/journal.pone.0039333
Tan, R. X., Zou, W. X. (2001). Endophytes: A rich source of functional metabolites. Nat. Prod Rep. 18, 448–459. doi: 10.1039/b100918o
Tonnabel, J., David, P., Pannell, J. R. (2017). Sex-specific strategies of resource allocation in response to competition for light in a dioecious plant. Oecologia 185, 675–686. doi: 10.1007/s00442-017-3966-5
Wang, Q., Garrity, G. M., Tiedje, J. M., Cole, J. R. (2007). Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267. doi: 10.1128/AEM.00062-07
Xia, Z. C., He, Y., Yu, L., Li, Z. J., Korpelainen, H., Li, C. Y. (2021). Revealing interactions between root phenolic metabolomes and rhizosphere bacterial communities in populus euphratica plantations. Biol. Fertility Soils 57, 421–434. doi: 10.1007/s00374-020-01527-z
Xia, Z. C., He, Y., Zhou, B., Korpelainen, H., Li, C. Y. (2020). Sex-related responses in rhizosphere processes of dioecious populus cathayana exposed to drought and low phosphorus stress. Environ. Exp. Bot. 175, 104049. doi: 10.1016/j.envexpbot.2020.104049
Xiong, C., Zhu, Y. G., Wang, J. T., Singh, B., Han, L. L., Shen, J. P., et al. (2021). Host selection shapes crop microbiome assembly and network complexity. N Phytol. 229, 1091–1104. doi: 10.1111/nph.16890
Yakimowski, S. B., Barrett, S. C. (2016). The role of hybridization in the evolution of sexual system diversity in a clonal, aquatic plant. Evolution 70, 1200–1211. doi: 10.1111/evo.12941
Yin, Y. H., Wang, Z. Y., Mora-Garcia, S., Li, J. M., Yoshida, S., Asami, T., et al. (2002). BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell 109, 181–191. doi: 10.1016/S0092-8674(02)00721-3
Yu, K., Pieterse, C. M. J., Bakker, P., Berendsen, R. L. (2019). Beneficial microbes going underground of root immunity. Plant Cell Environ. 42, 2860–2870. doi: 10.1111/pce.13632
Zhou, P., Zhang, X., Fatima, M., Ma, X., Fang, H., Yan, H., et al. (2020). DNA Methylome and transcriptome landscapes revealed differential characteristics of dioecious flowers in papaya. Horticulture Res. 7, 81. doi: 10.1038/s41438-020-0298-0
Keywords: root, rhizosphere soil, metabolome, microbiome, dimorphism, papaya
Citation: Zhou Y, Pang Z, Yuan Z, Fallah N, Jia H and Ming R (2022) Sex-based metabolic and microbiota differences in roots and rhizosphere soils of dioecious papaya (Carica papaya L.). Front. Plant Sci. 13:991114. doi: 10.3389/fpls.2022.991114
Received: 11 July 2022; Accepted: 30 September 2022;
Published: 13 October 2022.
Edited by:
Suvendu Mondal, Bhabha Atomic Research Centre (BARC), IndiaReviewed by:
Qiuming Yao, University of Nebraska-Lincoln, United StatesCopyright © 2022 Zhou, Pang, Yuan, Fallah, Jia and Ming. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ray Ming, cm1pbmdAbGlmZS5pbGxpbm9pcy5lZHU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.