- 1School of Life Sciences, Jiangsu University, Zhenjiang, China
- 2School of Food and Biological Engineering, Jiangsu University, Zhenjiang, China
- 3School of Life Sciences, Henan University, Kaifeng, China
- 4School of Environment, Jiangsu University, Zhenjiang, China
Wheat powdery mildew is a devastating disease leading to severe yield loss. The powdery mildew resistance gene Pm21, encoding a nucleotide-binding leucine-rich repeat receptor (NLR) protein, confers broad-spectrum resistance to powdery mildew and has great potential for controlling this disease. In this study, a large-scale mutagenesis was conducted on wheat cultivar (cv.) Yangmai 18 carrying Pm21. As a result, a total of 113 independent mutant lines susceptible to powdery mildew were obtained, among which, only one lost the whole Pm21 locus and the other 112 harbored one- (107) or two-base (5) mutations in the encoding region of Pm21. From the 107 susceptible mutants containing one-base change, we found that 25 resulted in premature stop codons leading to truncated proteins and 82 led to amino acid changes involving in 59 functional sites. We determined the mutations per one hundred amino acids (MPHA) indexes of different domains, motifs, and non-domain and non-motif regions of PM21 protein and found that the loss-of-function mutations occurred in a tendentious means. We also observed a new mutation hotspot that was closely linked to RNBS-D motif of the NB-ARC domain and relatively conserved in different NLRs of wheat crops. In addition, we crossed all the susceptible mutants with Yangmai 18 carrying wild-type Pm21, subsequently phenotyped their F1 plants and revealed that the variant E44K in the coiled-coil (CC) domain could lead to dominant-negative effect. This study revealed key functional sites of PM21 and their distribution characteristics, which would contribute to understanding the relationship of resistance and structure of Pm21-encoded NLR.
Introduction
Plants have developed a fine two-layered innate immune system to defense pathogens during long-time evolution. The first layer is pattern-triggered immunity (PTI) that is promoted via an activation of plasma membrane-resident pattern recognition receptors (PRRs) by perception of pathogen-associated molecular patterns (PAMPs). The second layer is effector-triggered immunity (ETI) that is elicited by pathogen effectors directly or indirectly recognized by intracellular nucleotide-binding leucine-rich repeat receptors (NLRs), commonly accompanied with hypersensitivity reaction (HR) leading to host cell death at the local site invaded by pathogens. It is considered that PTI contributes to basal defense and ETI provides race-specific resistance to pathogens (Dangl et al., 2013; Sun et al., 2020).
A typical plant NLR protein contains an N-terminal domain, a central nucleotide-binding (NB) domain, and a C-terminal leucine-rich repeat (LRR) domain. The variable N-terminus of NLR usually includes a Toll-interleukin 1 receptor (TIR) domain or a coiled-coil (CC) domain. Correspondingly, NLRs are classified into two types, TIR-type NLRs (TIR-NLRs) and CC-type NLRs (CC-NLRs) (Cui et al., 2015; Cesari, 2018). The central NB domain is highly conserved that works as a molecular switch to alter “off” and “on” state of NLR via binding adenosine diphosphate (ADP) and adenosine triphosphate (ATP) (Bernoux et al., 2016). The LRR domain participates in direct or indirect perception of effectors delivered into host cell by pathogens. The CC or TIR domains of many NLRs perform signal transduction and trigger cell death via homodimerization (Bernoux et al., 2011; Bai et al., 2012). Recently, it was found that TIR domain is a NADase that can cleave NAD+ into nicotinamide and ADP-ribose and transduce signaling into cell death response (Horsefield et al., 2019; Wan et al., 2019). In a plant NLR protein, the above three different domains may interact intramolecularly each other, which can maintain NLR under the status of autoinhibition (Moffett et al., 2002; Wang et al., 2019a,b).
Wheat powdery mildew, caused by the biotrophic fungus Blumeria graminis f. sp. tritici (DC.) Speer (Bgt), is a devastating disease leading to severe yield losses. Up to date, over 80 formally designated wheat powdery mildew resistance genes/alleles have been characterized from wheat and its cultivated and wild relatives. Among them, several genes have been cloned (He et al., 2021a,b). Pm21, originated from wheat wild relative Dasypyrum villosum (L.) Candargy, confers broad-spectrum resistance to Bgt pathogen and displays a great value for controlling wheat powdery mildew. Recently, using a map-based cloning strategy, Pm21 has been cloned that encodes a typical NLR protein (GenBank accession number: MF370199) (He et al., 2018). Transient expression assay suggests that PM21 protein is autoinhibited via intramolecular interactions of its different domains (Gao et al., 2020). Evolutionary analysis, based on a series of Pm21 alleles isolated from different accessions of D. villosum, reveals that the LRR domain has higher genetic diversity than other domains and the solvent-exposed LRR residues have been undergone diversifying selection (dN/dS = 3.20), which is similar to the race-specific powdery mildew resistance genes Pm3 from wheat and Mla from barley (He et al., 2020). In general, we know less about the characteristics of Pm21 so far.
Mutational analysis is a powerful approach for uncovering structure and function of genes of interest. In the past years, several large-scale mutagenesis researches have been conducted on plant NLR genes, such as Arabidopsis RPS2 (Tao et al., 2000) and RPM1 (Tornero et al., 2002), both of which confer resistance against Pseudomonas syringae, and tobacco mosaic virus resistance gene N (Dinesh-Kumar et al., 2000). However, there is no systemic mutagenesis research focused on an NLR gene-derived wheat crops. Here, we report over a hundred Bgt-susceptible mutants of common wheat cv. Yangmai 18 carrying Pm21 via ethyl methanesulfonate (EMS) mutagenesis and identify dozens of mutated sites that lead to lose Pm21 resistance. These mutations will contribute to understanding the molecular basis of PM21 protein.
Materials and methods
Plant materials and growth conditions
Wheat cv. Yangmai 18 is a common wheat-D. villosum translocation line that carries Pm21 and confers effective resistance to powdery mildew. Yangmai 23, a susceptible wheat cv., was used as a negative control and a host for reproducing asexual spores of Bgt. Wheat plants were grown in the field or in a greenhouse.
Ethyl methanesulfonate treatment
About 8,000 seeds of Yangmai 18 carrying Pm21 were soaked with 0.05 M phosphate buffer solution (PBS, pH 7.0) for 4 h. The seeds were then soaked with 0.8% EMS diluted in 0.05 M PBS (pH 7.0) and shook at 20°C for 12 h. The seeds treated with EMS were washed with running water for 4 h and then placed at 4°C for 3 days. After germination, the seeds were sowed in the field for generation. All seeds of each single plant were harvested as an M2 family.
Preliminary screening of mutants susceptible to powdery mildew
About 100 seeds of each M2 family were sowed in a pot. The plants at one-leaf stage were inoculated with Bgt isolate BgtYZ01 in a greenhouse with LED light under long-day condition (16-h light/8–h dark) at 24 ± 2°C. The powdery mildew responses were assessed at 7 days after inoculation. The susceptible individuals were kept and planted in the field. The responses of the harvested susceptible mutants were re-assessed with the same Bgt isolate at the seedling stage. Infection types (IT) were scored according to a 0–4 scale (He et al., 2021b).
Allelism test of candidate susceptible mutants
Each of the obtained candidate susceptible mutants was crossed with the mutant line Y18-S7, who carries a mutation site at position 834 (G to A) of Pm21 leading to a premature stop codon and be completely susceptible to isolate BgtYZ01 at whole growth stages (He et al., 2018). The powdery mildew responses of the derived F1 plants were detected with BgtYZ01 again to understand whether the mutation leading to susceptibility occurs in Pm21.
Detection of mutation sites
Total RNAs were extracted from leaves of each susceptible mutant using the TRIzol reagent (Life Technologies, Carlsbad, CA, United States), and 2 μg of total RNA was used to synthesize the first-strand cDNA using a PrimeScript™ II 1st Strand cDNA Synthesis Kit (TaKaRa, Shiga, Japan). The gene Pm21 was PCR-amplified from the cDNA using the high fidelity PrimeSTAR Max Premix (TaKaRa, Shiga, Japan) and the primer pair (forward primer: 5′-TTACCCGGGCTCACCCGTTGGACTTGGACT-3′; reverse primer: 5′-CCCACTAGTCTCTCTTCGTTACATAATGTAGTG CCT-3′). PCR products were digested with SmaI and SpeI, inserted into the vector pAHC25-MCS1 (He et al., 2018) and sequenced. Each mutation site was further verified by sequencing the independent PCR products.
Annotation of domains and motifs of PM21 protein and statistical analysis of mutation frequency
In PM21 protein, the CC, NB-ARC, and LRR domains and LRR motifs were annotated based on our previous work (Gao et al., 2020; He et al., 2020). The nT motif in the CC domain was characterized as described by Bai et al. (2002). The motifs in the NB-ARC domain were identified as described by Meyers et al. (1999) and Dilbirligi and Gill (2003). Mutation frequency in full-length protein, different domains, and motifs was calculated by the index of mutations per one hundred amino acids, simply named MPHA index. Sequence logos were created by the WebLogo tool (Crooks et al., 2004) using different NLR proteins in wheat crops.
Screening and verification of susceptible mutants with dominant-negative phenotype
Yangmai 18 carrying wild-type Pm21 was used as the female parent to cross, respectively, with different susceptible mutants. The powdery mildew responses of the generated F1 plants were assessed by inoculation with BgtYZ01. The phenotypes of the F2 plants derived from the susceptible F1 were further tested with the same isolate. Chi-squared (χ2) test was used to determine the goodness-of-fit of the observed segregation ratio to theoretical Mendelian ratio.
Results
A collection of loss-of-function of Pm21 consists of 113 independent mutants
Ethyl methanesulfonate -induced mutagenesis was conducted on Yangmai 18 carrying Pm21. From derived 4,462 M2 families, a total of 116 candidate mutants susceptible to Bgt isolate BgtYZ01 were identified. We also investigated the phenotypes of M3 plants of the candidate mutants at the seedling stage and confirmed that all of them lost resistance completely. To exclude loss-of-function mutants caused by non-target mutations, all candidate mutants were crossed with Y18-S7 that had a premature stop codon (G834A) in Pm21 leading to encode a 280-aa truncated protein and completely susceptible to BgtYZ01 at whole growth stages (He et al., 2018). The generated F1 plants derived from 113 crosses were all susceptible whereas those from 3 crosses were resistant, suggesting that the susceptibility of the corresponding 113 susceptible mutants was truly caused by mutated Pm21. Of the 113 susceptible mutants, 58 were reported in our previous work (He et al., 2017, 2018) and 55 were newly obtained in this study (Figure 1 and Supplementary Table 1). Except Y18-S6 that had a deletion of the Pm21 locus, each of the other 112 contained one- (107) or two-base (5) changes in the encoding sequence of Pm21. Due to that it was unclear which change(s) led to loss the function in the 5 mutants harbored two mutation sites in Pm21, only the 107 susceptible mutants contained one mutation site were used for further statistical analysis.
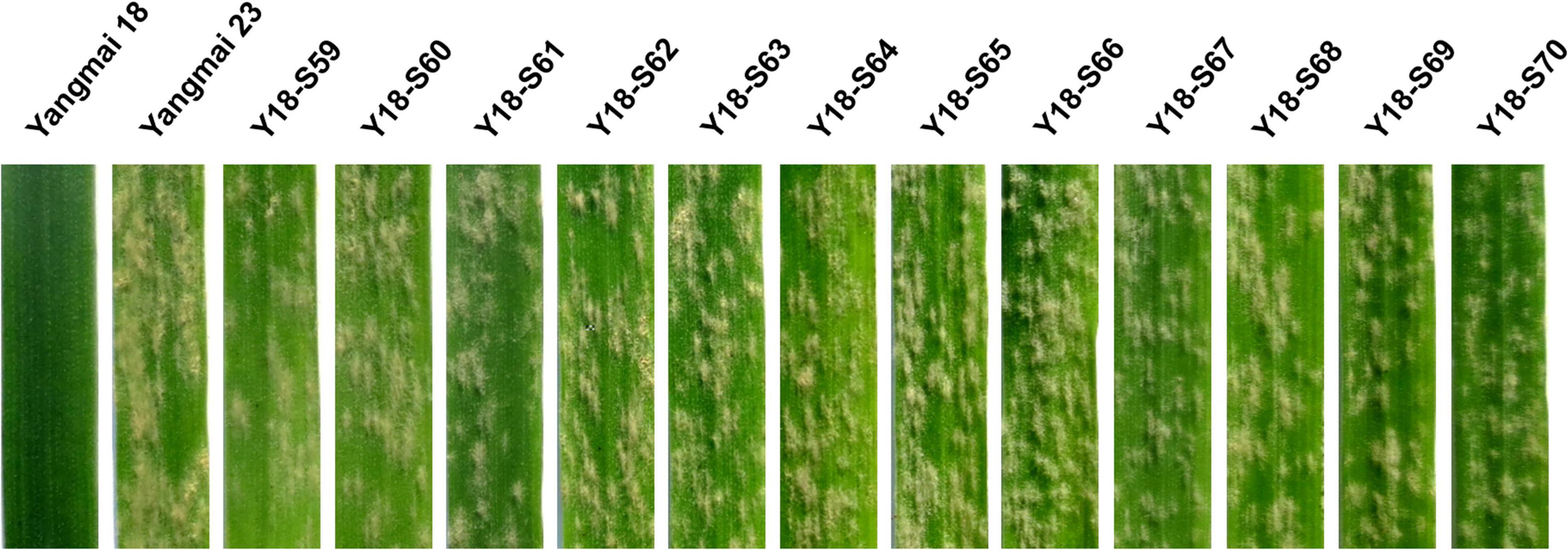
Figure 1. Powdery mildew responses of 12 representative susceptible mutants of Yangmai 18. Responses of the mutant lines Y18-S59∼Y18-S70 to Bgt isolate BgtYZ01 were assessed at one-leaf stage. Wheat cv. Yangmai 18 carrying wild-type Pm21 and cv. Yangmai 23 were used as the resistant and susceptible controls, respectively.
In general, of the 107 mutation sites, 59 and 47 were involved in the transition of G to A (55.2%) and C to T (43.9%), respectively, and only one (Y18-S113) was involved in the transversion of A to T (0.9%). Of the 107 mutations of Pm21, 25 resulted in premature stop codons leading to truncated proteins ranged from 178 to 843 aa. Compared with full-length PM21 protein that contains 16 LRR motifs as described in our previous work (He et al., 2020), the largest truncated protein (843 aa in Y18-S47) retained 13 LRR motifs, suggesting the importance of the last three LRR motifs (LRR14–LRR16) for resistance. Besides, 82 mutations of Pm21 each led to a single amino acid change, of which, 59 mutations were unique and the other 23 were repetitive (Figure 2 and Supplementary Table 1).
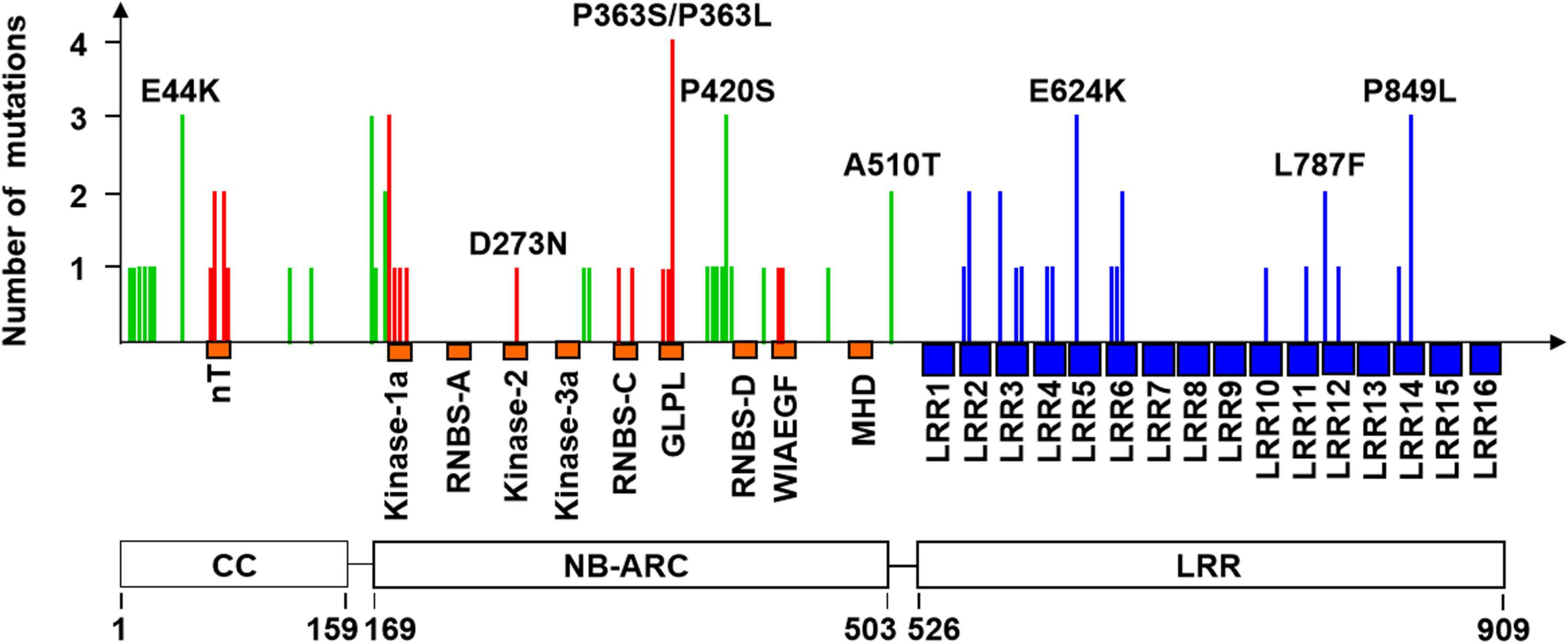
Figure 2. Frequency and distribution of 59 unique mutations leading to amino acid changes. The locations of the coiled-coil (CC), NB-ARC, and leucine-rich repeat (LRR) domains of PM21 are shown at bottom. The known conserved motifs (orange) of the CC and NB-ARC domains and 16 leucine-rich repeat motifs (blue) in the LRR domain are shown below the horizontal axis. The mutations in the motifs and non-motif regions of the CC and NB-ARC domains are shown as vertical red and green lines, respectively. The mutations in the LRR domain are shown as blue lines. A total of eight representative mutations are marked in the figure. More details of all mutation sites are listed in Supplementary Table 1.
Loss-of-function mutations of Pm21 occur in a tendentious means
In further study, the distribution characteristics of the above 59 distinctive mutations that led to single amino acid changes were analyzed using the index mutations per one hundred amino acids (MPHA). The data showed that the MPHA indexes of the CC (10.1) and NB-ARC (7.2) domains were higher than that of the full-length PM21 (6.5), whereas the MPHA index of the LRR domain (4.4) was lower than that of the full-length PM21. In addition, the MPHA indexes of the Linker 1 (the linker between the CC and NB-ARC domains) and the Linker 2 (the linker between the NB-ARC and LRR domains) were 11.1 and 4.5, which were similar to those of the CC and LRR domains, respectively (Table 1). It was indicated that the two linker regions also play the critical roles in powdery mildew resistance.
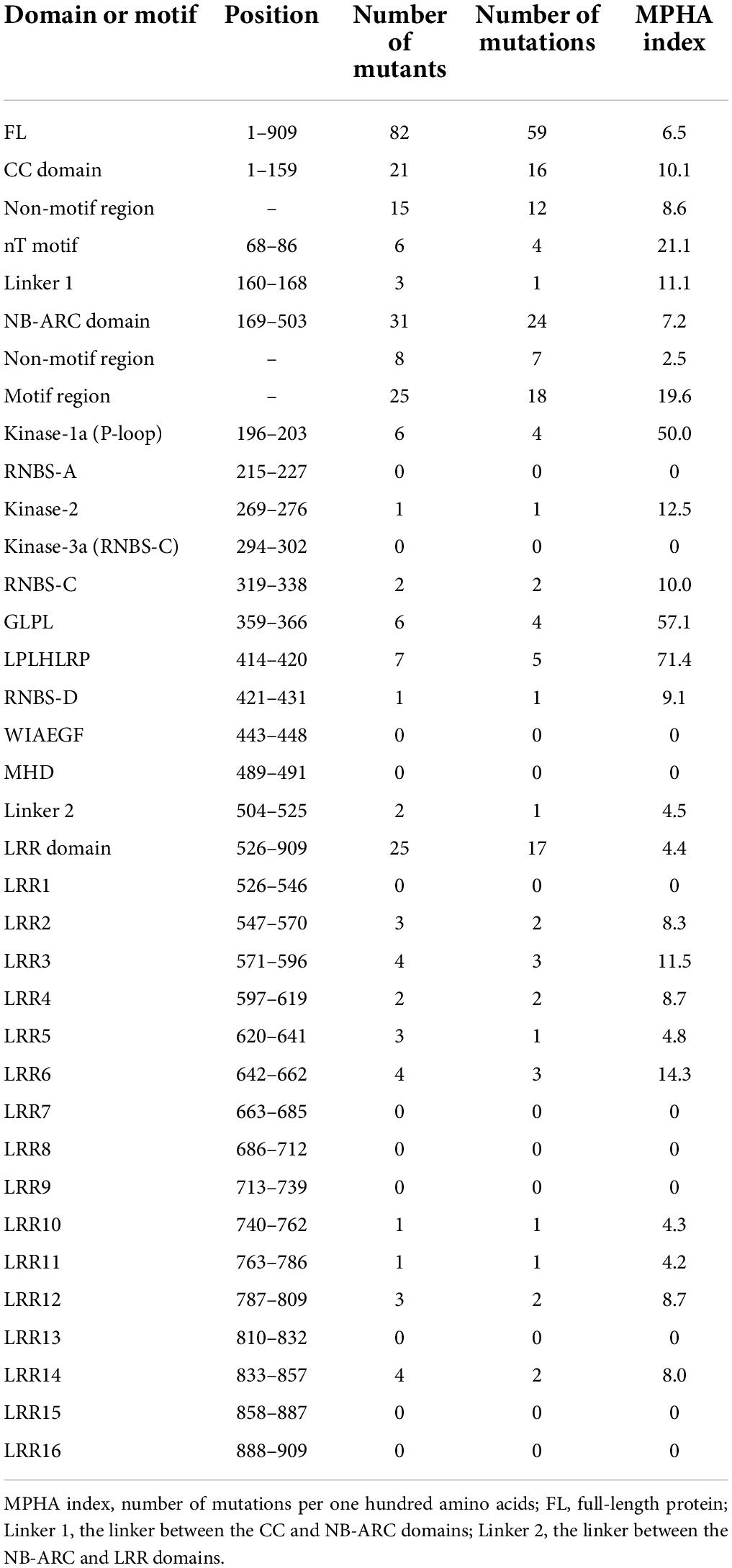
Table 1. Distribution and frequency of loss-of-function mutations altering amino acids in different domains and motifs of PM21.
In the CC domain, the MPHA index of the nT motif (21.1) was obviously higher than that of non-motif region (8.6). In the NB-ARC domain, the MPHA index of motif region (19.6) was significantly higher than that of non-motif region (2.5). Strikingly, the MPHA indexes of Kinase-1a (also called P-loop or Walker A) and GLPL motifs reached 50.0 and 57.1, respectively, which were significantly higher than those of other motifs reported previously. Additionally, no mutation was observed in the motifs RNBS-A, Kinase-3a (also named RNBS-B), WIAEGF, and MHD. All the mutation sites in conserved motifs in the CC and NB-ARC domains are listed in Table 2. In the LRR domain, the MPHA indexes of the motifs LRR2, LRR3, LRR4, LRR6, LRR12, and LRR14 were obviously higher than that of the whole LRR domain (4.4). The MPHA indexes of the motifs LRR5, LRR10, and LRR11 were lower than the average value of the LRR domain, whereas there was no mutation observed in the remaining 7 LRR motifs (Figure 3 and Table 1).
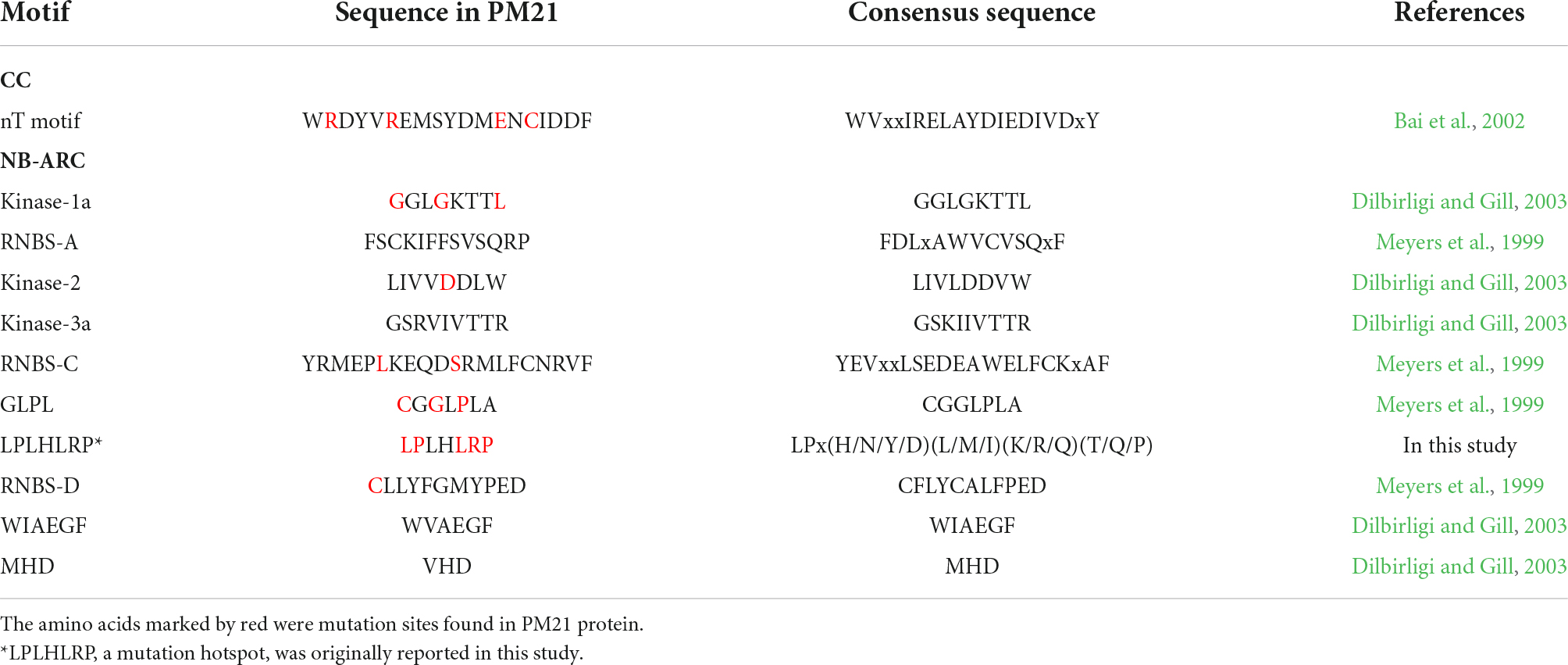
Table 2. Mutation sites in conserved motifs in the coiled-coil (CC) and NB-ARC domains of PM21 protein.

Figure 3. Mutation sites in the leucine-rich repeat (LRR) domain. The LRR domain of PM21 can be divided into 16 LRR motifs and their consensus sequence LxxLxLxx is given at the bottom of this figure. Substituted amino acids (green) and their positions are shown in the below and the above of the mutated ones (red), respectively.
Taken together, these results indicated that the distribution of mutations that led to amino acid changes and loss-of-function of Pm21 resistance is tendentious in some degrees.
LPLHLRP immediately close to RNBS-D motif is a new mutation hotspot
LPLHLRP at position 414–420 in PM21 protein was closely near to the N-terminus of RNBS-D motif in the NB-ARC domain. We observed that 7 susceptible mutants involved in 5 amino acid changes in this region, including L414F (Y18-S12), P415L (Y18-S59), L418F (Y18-S112), R419H (Y18-S58), and P420S (Y18-S19, Y18-S79, and Y18-S82). It meant that the MPHA index of LPLHLRP reached 71.4, significantly higher than those of any other motifs (Table 1). Obviously, LPLHLRP was a mutation hotspot that has not been reported previously. We compared 26 wheat and barley NLR proteins against different diseases and found that the seven amino acids were relatively conserved, with the consensus sequence LPx(H/N/Y/D)(L/M/I)(K/R/Q)(T/Q/P) (Figure 4). It was strongly suggested that this conserved sequence might play an important role(s) in NLR proteins.
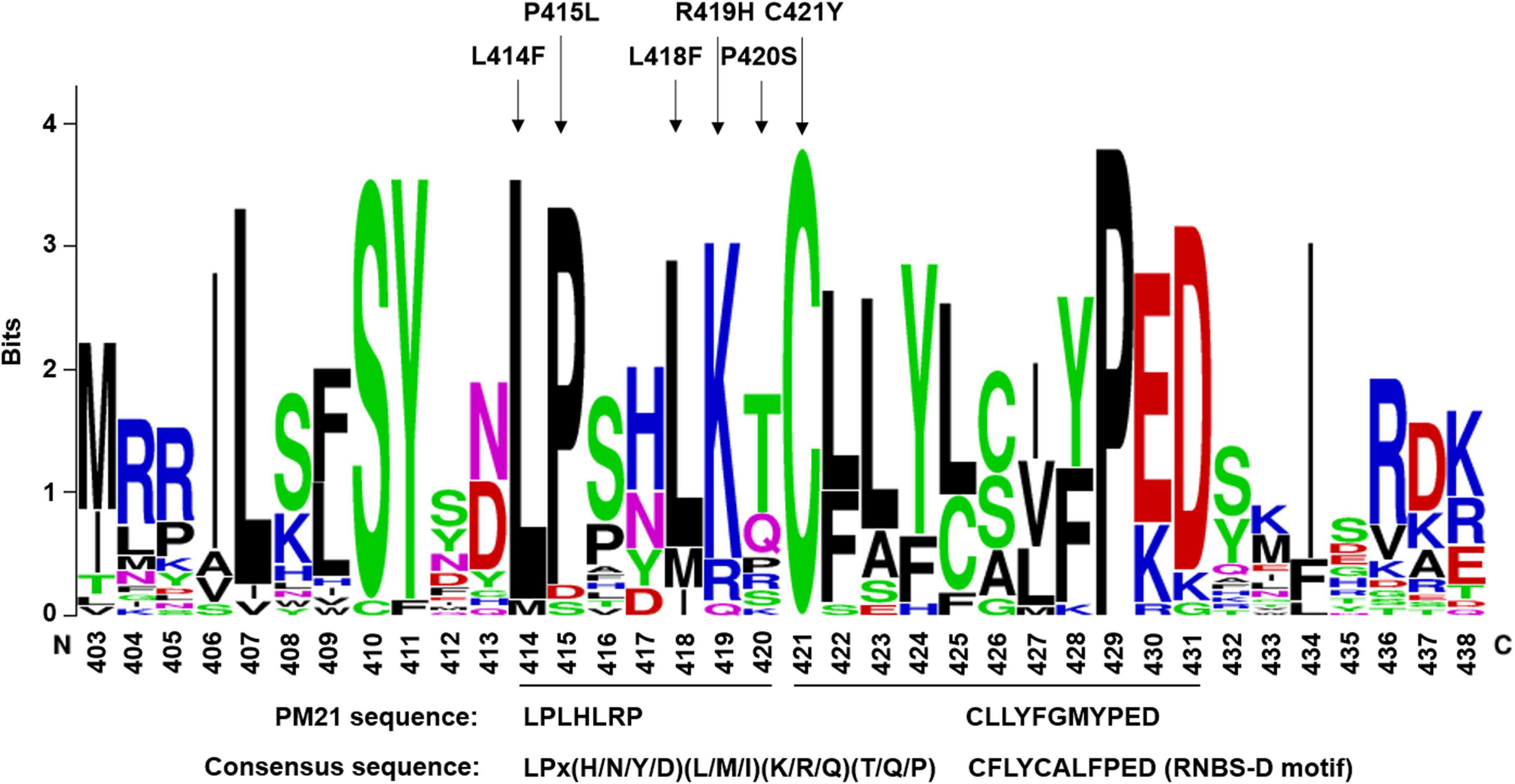
Figure 4. Sequence logos of the mutation hotspot LPLHLRP and its closely linked RNBS-D motif (CFLYCALFPED). Sequence logos were generated from 26 NLR proteins of wheat crops, including PM3, PM8, PM21, LR1, LR21, SR33, SR35, YR10, MLA1, MLA6, and so on. Arrows indicate amino acid changes leading to loss-of-function of PM21. The consensus sequences and the corresponding PM21 sequence are shown at the bottom.
The variant E44K in the coiled-coil domain leads to dominant-negative phenotype
All susceptible mutants were crossed with Yangmai 18 carrying wild-type Pm21, and the powdery mildew responses of the generated F1 plants were then assessed. The results showed that all the F1 plants derived from three crosses, Yangmai 18/Y18-S13, Yangmai 18/Y18-S23, and Yangmai 18/Y18-S100, were susceptible to BgtYZ01 (Figure 5), whereas all F1 plants derived from other crosses were resistant. The susceptible and resistant individuals in the F2 plants derived from the above three crosses were 116 and 35 (χ2 = 0.267, p = 0.605), 145 and 46 (χ2 = 0.086, p = 0.770), and 138 and 43 (χ2 = 0.149, p = 0.699), respectively, which were all fit for the theoretical Mendelian segregation ratio 3:1 (Table 3). Interestingly, all the three independent susceptible mutants (Y18-S13, Y18-S23, and Y18-S100) contained the same mutation G to A at position 130, which led to the amino acid change E to K at position 44 in the CC domain. Therefore, it was strongly suggested that the mutation E44K could lead to dominant-negative phenotype.

Figure 5. Powdery mildew responses of mutant lines and F1 plants involving dominant-negative effect. Responses of the F1 plants of the crosses Yangmai 18/Y18-S13, Yangmai 18/Y18-S23, and Yangmai 18/Y18-S100 were tested at one-leaf stage by inoculation with Bgt isolate BgtYZ01. Wheat cultivar (cv.) Yangmai 18, Yangmai 23, and the mutant lines Y18-S13, Y18-S23, and Y18-S100 were used as controls.
Discussion
Pm21, originated from D. villosum, a wild relative of wheat, is one of the most valuable powdery mildew resistance genes that provides effective resistance to all tested isolates of Bgt (He et al., 2018; Zhang et al., 2019). Our recent research shows that Pm21 encodes a CC-type NLR protein. Usually, disease resistance conferred by such kind of genes is subjected to be overcome by fast-evolved pathogen races. So, uncovering the molecular characteristic of PM21 protein will be very important for sustainable utilization of Pm21 in wheat production. In this study, we performed large-scale mutagenesis on Pm21 to find its key functional site. Nucleotide changes induced by EMS treatment widely occur in a genome, and some changes out of Pm21, such as changes in resistance signal components, may also impede Pm21 resistance in some degrees. To exclude such non-target mutation leading to loss-of-function as possible, all the candidate susceptible mutants were crossed with Y18-S7, which carries a mutated Pm21 only encoding very small fragment of PM21 protein and shows complete susceptibility at whole growth stages, and perform allelism tests. Finally, 113 stable susceptible mutants, whose variants leading to loss-of-function were considered to be occurred in the target gene Pm21, were identified. Among them, 82 independent mutants were involved in 59 different single amino acid substitutions, on which, we focused in this study.
In the CC domain of NLR, a core motif, EDVID motif (consensus sequence: VRELAYDAEDVID), was identified from the nT motif (consensus sequence: WVxxIRELAYDIEDIVDxY) originally predicted in rice (Bai et al., 2002; Rairdan et al., 2008). The EDVID motif, about 110–130 residues N-terminal to the P-loop motif of the NB-ARC domain, is conserved in most NLRs and required for the intramolecular interaction with the NB-ARC domain (Rairdan et al., 2008). Some mutations occurring in the residues in the EDVID motif can lead to lose cell death-inducing activity and disease resistance of potato Rx and barley MLA10 (Rairdan et al., 2008; Maekawa et al., 2011; Bai et al., 2012). In PM21 protein, a sequence, WRDYVREMSYDMENCIDDF, similar to the EDVID motif, was characterized. In total, three variants, R73Q, E80K, and C82Y, could lead to loss-of-function of Pm21. Interestingly, our recent work showed that the CC domain containing any of the variants R73Q and E80K keeps the cell-death inducing activity but inhabits the autoactivation of PM21 (D491V) that harbors a mutation in the MHD motif (Gao et al., 2020). In addition, the variant R69K in the nT motif but not in the EDVID motif could also lead to loss-of-function of Pm21.
A recent research reported another motif, MADA motif, in the CC domain of NLR. The consensus sequence of MADA motif has been characterized as MADAxVSFxVxKLxxLLxxEx that lies in the N-terminus of the CC domain of ∼20% plant NLRs (Adachi et al., 2019). The MADA motif corresponds to the α1 helix of Arabidopsis ZAR1, which is a conformational switch during resistosome activation (Adachi et al., 2019; Wang et al., 2019a,b). Here, no similar sequence of the MADA motif could be found in PM21 protein. However, a mutation hotspot, ATMGAMNPLIGKL (from position 8–20), was observed in the N-terminus of the CC domain. In this hotspot, 7 variants involved in 6 sites could lead to lose Pm21 resistance. So, it was speculated that this region might play the roles in PM21 resistosome activation.
The variant E44K in PM21 protein could lead to lose the cell death-inducing activity of the CC domain (Gao et al., 2020). Here, we found three independent mutants susceptible to powdery mildew that shows dominant-negative effect when crossed them with Yangmai 18 carrying wild-type Pm21 gene. Surprisingly, each of the mutants contained the same mutation E44K in PM21 protein. It strongly indicated that E44K is responsible for this dominant-negative effect. In the previous research, two variants involved in the CC domain of Arabidopsis RPS2, a CC-NLR, can lead to dominant-negative effect. The first variant R2M1 has a truncated CC domain lacking of position 7–25. The second variant R2M2 has two amino acid substitutions, L38R and T40L (Tao et al., 2000).
Dominant-negative effect of NLR is thought to be caused by defective protein complex which consists of wild-type and mutated NLR (Dinesh-Kumar et al., 2000). Recently, plant NLR resistosome is confirmed to be a functional complex for disease resistance (Wang et al., 2019a,b). Therefore, for dominant-negative effect, one reasonable explanation is that mutated NLR could be integrated into resistosome with wild-type NLR and interfere the normal function of resistosome. In this study, a number of PM21 variants were identified; however, only E44K could cause dominant-negative effect, whereas other variants could not. This phenomenon suggested that most PM21 variants could not disturb the function of resistosome. It was also suggested that, in most situations, resistosome might possess strong fault-tolerance and maintain its resistance function when an abnormal peptide is incorporated into it.
The NB-ARC domain is a molecular switch for regulation of the activity of NLR (Bernoux et al., 2016). It consists of at least nine conserved motifs as reported previously (Meyers et al., 1999; Dilbirligi and Gill, 2003). Among them, the motifs P-loop, Kinase-2, and MHD are well known. P-loop, also called Kinase-1a or Walker A motif, has the consensus sequence GGLGKTTL and makes that NLR has the ability to bind ATP (Meyers et al., 1999; Tameling et al., 2002). Kinase-2, also called Walker B, shares the consensus sequence LIVLDDVW, in which, each aspartate (D) is almost conserved in all plant NLRs. The first D takes part in coordination of the Mg2+ ion in the catalytic site, and the second D acts as the catalytic site for ATP hydrolysis and activation of NLR (Meyers et al., 1999; Tameling et al., 2006). MHD lies in the C-terminus of the NB-ARC domain, which is known because the change aspartate (D) to valine (V) can enhance the binding between NLR and ATP and result in the autoactivation of many NLRs (Bendahmane et al., 2002; Tameling et al., 2006; Gao et al., 2020). According to the result of sequence comparison, PM21 protein contains all the nine motifs. Our mutational analysis showed that 10 variants, leading to loss-of-function of PM21, were involved in amino acid substitutions in the core sites of 5 reported motifs of the NB-ARC domain (Table 2).
The sequence of PM21 P-loop motif is GGLGKTTL (position: 196–203), which well matches the consensus sequence of this motif. Mutational analysis showed that each of the six mutations such as G196R, G196E, G197D, G199D, L203F (occurring in P-loop), and G194E (near the N-terminus of P-loop) could lead to loss-of-function of PM21. Mutational research on tobacco N reveals that specific mutations, including G216A/E/V/R, G218R, G219D, K222E/N, and T223A/N, near or in the P-loop not only lead to lose N function but also result in dominant-negative effects (Dinesh-Kumar et al., 2000). However, in another research on Arabidopsis RPM1, no dominant-negative effect was detected in the mutant line rpm1-94 that carries the variant G200E corresponding to G216A/E/V/R of N (Tornero et al., 2002). Here, the variants G194E and G196R/E of PM21 corresponded to G216A/E/V/R and G218R of N, respectively. As our observation, all the F1 plants derived from the crosses of the corresponding susceptible mutants with Yangmai 18 (harboring wild-type Pm21) showed complete resistance to powdery mildew, indicating that these variants could not interfere with PM21 function.
Previously, we found a susceptible natural accession of D. villosum that carries a non-functional Pm21 allele with a transition A821G resulting in the amino acid change D274G in the Kinase-2 (Walker B) motif (He et al., 2020). Here, through EMS treatment, Pm21 in the susceptible mutants Y18-S28 was identified to be containing a transition G817A resulting in the substitution D273N. The results suggested that the two D residues are important for Kinase-2 function and PM21 resistance, which is in accordance with the previous reports on Arabidopsis RPS2 (Tao et al., 2000), tobacco N (Dinesh-Kumar et al., 2000), and tomato I-2 (Tameling et al., 2006).
In the NB-ARC domain, the functions of another six motifs such as RNBS-A, RNBS-B (Kinase-3a), RNBS-C, GLPL, RNBS-D, and WIAEGF are not well known. It is believed that these motifs may play the roles in binding ATP for the regulation of NLR activity (Takken et al., 2006). Mutational analysis of Arabidopsis RPM1 shows that three variants, E340K, A341V, and L344F, respectively located in the 10th, 11th, and 14th residues in the RNBS-C motif, could result in RPM1 function defect (Tornero et al., 2002). Here, we demonstrated that two variants, L324F and S329L, occurred in the first and 11th residues in the RNBS-C motif could lead to lose PM21 resistance. S329L of PM21 and A341V of RPM1 situate in the same position of the RNBS-C motif, whereas L324F of PM21 is a new found functional site.
The GLPL motif has a conserved sequence CGGLPLA. Mutational analyses of flax rust resistance protein P2 (variant: G395E) (Dodds et al., 2001), maize Rp1-D21 (variant: P398L) (Wang et al., 2005), and RPM1 (variant: A379V) (Tornero et al., 2002) indicate the importance of the 3rd residue glycine (G), the 5th residue proline (P) and the last residue alanine (A). In this study, we obtained six independent susceptible mutants involved in four variants of three sites (C359Y, G361R, P363S, and P363L) in the GLPL motif. It was suggested that not only the 3rd residue G and the 5th residue P but also the first residues cysteine (C) are crucial for PM21 resistance.
The RNBS-D motif has a consensus sequence CFLYCALFPED. RPM1 variants (S439F and P442L) at the 6th and 9th residues in this motif result in loss-of-function (Tornero et al., 2002). Here, we revealed that replacement of the first residue cysteine (C) with tyrosine (Y) (C421Y) could lead to hampering PM21 resistance, indicating the importance of the cysteine residues in NLR. Interestingly, near the N-terminus of the RNBS-D motif, seven independent mutations, involved in five sites (L414F, P415L, L418F, R419H, and P420S), were observed in this study, suggesting that the sequence LPLHLRP of PM21 seems to be a mutation hotspot leading to loss-of-function. Comparative analysis showed that both the LPLHLRP-like sequences and the RNBS-D sequences are conserved in different NLRs. Due to that LPLHLRP is immediate before the reported motif RNBS-D, we propose that it should be considered as an extended functional region of the RNBS-D motif rather than as a new independent motif.
The LRR domain is more variant than other domains in NLR, which is usually considered as a sensor for specific recognition of pathogen effector by a direct or indirect means (Dangl et al., 2013). This is supported by several researches. For instance, a single amino acid difference can make that rice blast resistance gene Pi-ta differs from its susceptible allele (Bryan et al., 2000). A total of six amino acid changes can alter flax rust resistance gene P2 to P specificities (Dodds et al., 2001). Natural variations in the LRR domain result in different powdery mildew responses of Pm3 alleles (Srichumpa et al., 2005). A recent research showed that site-directed mutagenesis on the LRR domain of PM3A can obviously control response strength and resistance spectrum (Lindner et al., 2020). In this research, a total of 25 susceptible mutants involved in 17 amino acid changes were obtained. In our recent work, the LRR domain of PM21 has been divided into 16 LRR motif with the consensus sequence LxxLxLxx (where L represents a conserved leucine or other aliphatic residue and x represents any amino acid) (He et al., 2020). These variants were distributed in 9 LRR motifs (LRR2–LRR6, LRR10–LRR12, and LRR14), some of which have been confirmed to be under diversifying selection (He et al., 2020). Of 17 variants, 2 (L571F and L787F), 4 (R551H, E624K, P654S, and E839K), and other 11 occurred in the position of L, x, and the non-core region of LRR motif, respectively. It was suggested the importance of these sites for complete PM21 resistance. Whether the amino acid changes in the LRR domain alter recognition specificity for Bgt isolates needs further investigation.
In conclusion, we performed large-scale mutagenesis on wheat powdery mildew resistance gene Pm21, which revealed the distribution characteristics of variant amino acid residues and discussed their potential impacts on PM21 resistance. To our knowledge, some mutation sites have not been reported in plant NLRs previously. Furthermore, two mutation hotspots, one near the N-terminus of the CC domain and the other immediately close to the N-terminus of RNBS-D motif of the NB-ARC domain and a variant resulting in dominant-negative effect, were identified. This study will contribute to understanding the relationship of resistance and structure of Pm21-encoded NLR, which would be useful for the development of durable resistance for efficiently controlling wheat powdery mildew.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
HH and SZ conceived and designed the experiments and wrote and revised the manuscript. HH, RG, ZC, RL, and XK screened the mutants and identified the mutation sites. HH performed allelism test. HH, AG, and TL analyzed the data. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the grants from National Natural Science Foundation of China (32171990 and U1604116), Key Research and Development Program of Zhenjiang (NY2021001), State Key Laboratory of Plant Cell and Chromosome Engineering (PCCE-KF-2021-05 and PCCE-KF-2022-07), and State Key Laboratory of Crop Biology in Shandong Agricultural University (2021KF01).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.988641/full#supplementary-material
References
Adachi, H., Contreras, M. P., Harant, A., Wu, C., Derevnina, L., Sakai, T., et al. (2019). An N-terminal motif in NLR immune receptors is functionally conserved across distantly related plant species. Elife 8:e49956. doi: 10.7554/eLife.49956
Bai, J., Pennill, L. A., Ning, J., Lee, S. W., Ramalingam, J., Webb, C. A., et al. (2002). Diversity in nucleotide binding site-leucine-rich repeat genes in cereals. Genome Res. 12, 1871–1884. doi: 10.1101/gr.454902
Bai, S., Liu, J., Chang, C., Zhang, L., Maekawa, T., Wang, Q., et al. (2012). Structure-function analysis of barley NLR immune receptor MLA10 reveals its cell compartment specific activity in cell death and disease resistance. PLoS Pathog. 8:e1002752. doi: 10.1371/journal.ppat.1002752
Bendahmane, A., Farnham, G., Moffett, P., and Baulcombe, D. C. (2002). Constitutive gain-of-function mutants in a nucleotide binding site-leucine rich repeat protein encoded at the Rx locus of potato. Plant J. 32, 195–204. doi: 10.1046/j.1365-313x.2002.01413.x
Bernoux, M., Burdett, H., Williams, S. J., Zhang, X., Chen, C., Newell, K., et al. (2016). Comparative analysis of the flax immune receptors L6 and L7 suggests an equilibrium-based switch activation model. Plant Cell 28, 146–159. doi: 10.1105/tpc.15.00303
Bernoux, M., Ve, T., Williams, S., Warren, C., Hatters, D., Valkov, E., et al. (2011). Structural and functional analysis of a plant resistance protein TIR domain reveals interfaces for self-association, signaling, and autoregulation. Cell Host Microbe 9, 200–211. doi: 10.1016/j.chom.2011.02.009
Bryan, G. T., Wu, K., Farrall, L., Jia, Y., Hershey, H. P., McAdams, S. A., et al. (2000). A single amino acid difference distinguishes resistant and susceptible alleles of the rice blast resistance gene Pi-ta. Plant Cell 12, 2033–2045. doi: 10.1105/tpc.12.11.2033
Cesari, S. (2018). Multiple strategies for pathogen perception by plant immune receptors. New Phytol. 219, 17–24. doi: 10.1111/nph.14877
Crooks, G. E., Hon, G., Chandonia, J. M., and Brenner, S. E. (2004). WebLogo: A sequence logo generator. Genome Res. 14, 1188–1190. doi: 10.1101/gr.849004
Cui, H., Tsuda, K., and Parker, J. E. (2015). Effector-triggered immunity: From pathogen perception to robust defense. Annu. Rev. Plant Biol. 66, 487–511. doi: 10.1146/annurev-arplant-050213-040012
Dangl, J. L., Horvath, D. M., and Staskawicz, B. J. (2013). Pivoting the plant immune system from dissection to deployment. Science 341, 746–751. doi: 10.1126/science.1236011
Dilbirligi, M., and Gill, K. S. (2003). Identification and analysis of expressed resistance gene sequences in wheat. Plant Mol. Biol. 53, 771–787. doi: 10.1023/B:PLAN.0000023663.55701.5f
Dinesh-Kumar, S. P., Tham, W. H., and Baker, B. J. (2000). Structure-function analysis of the tobacco mosaic virus resistance gene N. Proc. Natl. Acad. Sci. U.S.A. 97, 14789–14794. doi: 10.1073/pnas.97.26.14789
Dodds, P. N., Lawrence, G. J., and Ellis, J. G. (2001). Six amino acid changes confined to the leucine-rich repeat β-strand/β-turn motif determine the difference between the P and P2 rust resistance specificities in flax. Plant Cell 13, 163–178. doi: 10.1105/tpc.13.1.163
Gao, A., Hu, M., Gong, Y., Dong, R., Jiang, Y., Zhu, S., et al. (2020). Pm21 CC domain activity modulated by intramolecular interactions is implicated in cell death and disease resistance. Mol. Plant Pathol. 21, 975–984. doi: 10.1111/mpp.12943
He, H., Du, H., Liu, R., Liu, T., Yang, L., Gong, S., et al. (2021a). Characterization of a new gene for resistance to wheat powdery mildew on chromosome 1RL of wild rye Secale sylvestre. Theor. Appl. Genet. 134, 887–896. doi: 10.1007/s00122-020-03739-1
He, H., Liu, R., Ma, P., Du, H., Zhang, H., Wu, Q., et al. (2021b). Characterization of Pm68, a new broad-spectrum powdery mildew resistance gene on chromosome 2BS of Greek durum wheat TRI 1796. Theor. Appl. Genet. 134, 53–62. doi: 10.1007/s00122-020-03681-2
He, H., Ji, J., Li, H., Tong, J., Feng, Y., Wang, X., et al. (2020). Genetic diversity and evolutionary analyses reveal the powdery mildew resistance gene Pm21 undergoing diversifying selection. Front. Genet. 11:489. doi: 10.3389/fgene.2020.00489
He, H., Ji, Y., Zhu, S., Li, B., Zhao, R., Jiang, Z., et al. (2017). Genetic, physical and comparative mapping of the powdery mildew resistance gene Pm21 originating from Dasypyrum villosum. Front. Plant Sci. 8:1914. doi: 10.3389/fpls.2017.01914
He, H., Zhu, S., Zhao, R., Jiang, Z., Ji, Y., Ji, J., et al. (2018). Pm21, encoding a typical CC-NBS-LRR protein, confers broad-spectrum resistance to wheat powdery mildew disease. Mol. Plant 11, 879–882. doi: 10.1016/j.molp.2018.03.004
Horsefield, S., Burdentt, H., Zhang, X., Manik, M. K., Shi, Y., Chen, J., et al. (2019). NAD+ cleavage activity by animal and plant TIR domains in cell death pathways. Science 365, 793–799. doi: 10.1126/science.aax1911
Lindner, S., Keller, B., Singh, S. P., Hasenkamp, Z., Jung, E., Müller, M. C., et al. (2020). Single residues in the LRR domain of the wheat PM3A immune receptor can control the strength and the spectrum of the immune response. Plant J. 104, 200–214. doi: 10.1111/tpj.14917
Maekawa, T., Cheng, W., Spiridon, L. N., Töller, A., Lukasik, E., Saijo, Y., et al. (2011). Coiled-coil domain-dependent homodimerization of intracellular barley immune receptors defines a minimal functional module for triggering cell death. Cell Host Microbe 9, 187–199. doi: 10.1016/j.chom.2011.02.008
Meyers, B. C., Dickerman, A. W., MIchelmore, R. W., Sivaramakrishnan, S., Sobral, B. W., and Young, N. D. (1999). Plant disease resistance genes encode members of an ancient and diverse protein family within the nucleotide-binding superfamily. Plant J. 20, 317–332. doi: 10.1046/j.1365-313x.1999.t01-1-00606.x
Moffett, P., Farnham, G., Peart, J., and Baulcombe, D. C. (2002). Interaction between domains of a plant NBS-LRR protein in disease resistance-related cell death. EMBO J. 21, 4511–4519. doi: 10.1093/emboj/cdf453
Rairdan, G. J., Collier, S. M., Sacco, M. A., Baldwin, T. T., Boettrich, T., and Moffett, P. (2008). The coiled-coil and nucleotide binding domains of the potato Rx disease resistance protein function in pathogen recognition and signaling. Plant Cell 20, 739–751. doi: 10.1105/tpc.107.056036
Srichumpa, P., Brunner, S., Keller, B., and Yahiaoui, N. (2005). Allelic series of four powdery mildew resistance genes at the Pm3 locus in hexaploid bread wheat. Plant Physiol. 139, 885–895. doi: 10.1104/pp.105.062406
Sun, Y., Zhu, Y., Balint-kurti, P. J., and Wang, G. (2020). Fine-tuning immunity: Players and regulators for plant NLRs. Trends Plant Sci. 25, 695–713. doi: 10.1016/j.tplants.2020.02.008
Takken, F. L. W., Albrecht, M., and Tameling, W. I. L. (2006). Resistance proteins: Molecular switches of plant defence. Curr. Opin. Plant Biol. 9, 383–390. doi: 10.1016/j.pbi.2006.05.009
Tameling, W. I. L., Elzinga, S. D., Darmin, P. S., Vossen, J. H., Takken, F. L. W., Haring, M. A., et al. (2002). The tomato R gene products I-2 and Mi-1 are functional ATP binding proteins with ATPase activity. Plant Cell 14, 2929–2939. doi: 10.1105/tpc.005793
Tameling, W. I. L., Vossen, J. H., Albrecht, M., Lengauer, T., Berden, J. A., Haring, M. A., et al. (2006). Mutations in the NB-ARC domain of I-2 that impair ATP hydrolysis cause autoactivation. Plant Physiol. 140, 1233–1245. doi: 10.1104/pp.105.073510
Tao, Y., Yuan, F., Leister, R. T., Ausubel, F. M., and Katagiri, F. (2000). Mutational analysis of the Arabidopsis nucleotide binding site-leucine-rich repeat resistance gene RPS2. Plant Cell 12, 2541–2554. doi: 10.1105/tpc.12.12.2541
Tornero, P., Chao, R. A., Luthin, W. N., Goff, S. A., and Dangl, J. L. (2002). Large-scale structure-function analysis of the Arabidopsis RPM1 disease resistance protein. Plant Cell 14, 435–450. doi: 10.1105/tpc.010393
Wan, L., Essuman, K., Anderson, R. G., Sasaki, Y., Monteiro, F., Chung, E., et al. (2019). TIR domains of plant immune receptors are NAD+-cleaving enzymes that promote cell death. Science 365, 799–803. doi: 10.1126/science.aax1771
Wang, G., Ji, J., Ei-Kasmi, F., Dangl, J. L., Johal, G., and Balint-Kurti, P. J. (2005). Molecular and functional analyses of a maize autoactive NB-LRR protein identify precise structural requirements for activity. PLoS Pathog. 11:e1004674. doi: 10.1371/journal.ppat.1004674
Wang, J., Wang, J., Hu, M., Wu, S., Qi, J., Wang, G., et al. (2019a). Ligand-triggered allosteric ADP release primes a plant NLR complex. Science 364:eaav5868. doi: 10.1126/science.aav5868
Wang, J., Hu, M., Wang, J., Qi, J., Han, Z., Wang, G., et al. (2019b). Reconstitution and structure of a plant NLR ressitosome conferring immunity. Science 364:eaav5870. doi: 10.1126/science.aav5870
Keywords: wheat powdery mildew, Pm21, nucleotide-binding leucine-rich repeat receptor (NLR), mutational analysis, mutation hotspot, dominant-negative effect
Citation: He H, Guo R, Gao A, Chen Z, Liu R, Liu T, Kang X and Zhu S (2022) Large-scale mutational analysis of wheat powdery mildew resistance gene Pm21. Front. Plant Sci. 13:988641. doi: 10.3389/fpls.2022.988641
Received: 07 July 2022; Accepted: 19 July 2022;
Published: 09 August 2022.
Edited by:
Handong Su, Huazhong Agricultural University, ChinaReviewed by:
Guohao Han, Institute of Genetics and Developmental Biology (CAS), ChinaSung Un Huh, Kunsan National University, South Korea
Shulan Fu, Sichuan Agricultural University, China
Copyright © 2022 He, Guo, Gao, Chen, Liu, Liu, Kang and Zhu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Huagang He, aGdoZUB1anMuZWR1LmNu; Shanying Zhu, emh1c2hhbnlpbmdAdWpzLmVkdS5jbg==
 Huagang He
Huagang He Rui Guo2
Rui Guo2 Anli Gao
Anli Gao